Abstract
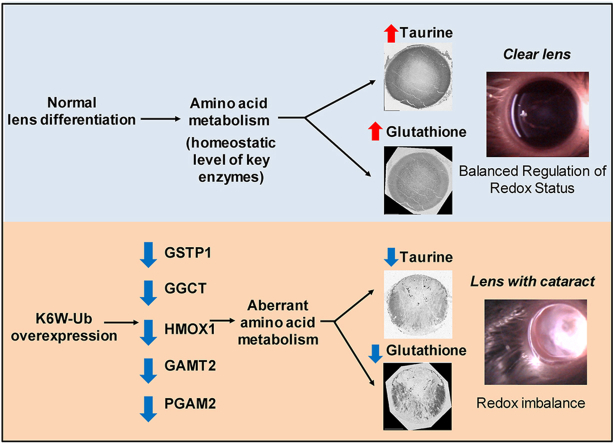
The lens proteome undergoes dramatic composition changes during development and maturation. A defective developmental process leads to congenital cataracts that account for about 30% of cases of childhood blindness. Gene mutations are associated with approximately 50% of early-onset forms of lens opacity, with the remainder being of unknown etiology. To gain a better understanding of cataractogenesis, we utilized a transgenic mouse model expressing a mutant ubiquitin protein in the lens (K6W-Ub) that recapitulates most of the early pathological changes seen in human congenital cataracts. We performed mass spectrometry-based tandem-mass-tag quantitative proteomics in E15, P1, and P30 control or K6W-Ub lenses. Our analysis identified targets that are required for early normal differentiation steps and altered in cataractous lenses, particularly metabolic pathways involving glutathione and amino acids. Computational molecular phenotyping revealed that glutathione and taurine were spatially altered in the K6W-Ub cataractous lens. High-performance liquid chromatography revealed that both taurine and the ratio of reduced glutathione to oxidized glutathione, two indicators of redox status, were differentially compromised in lens biology. In sum, our research documents that dynamic proteome changes in a mouse model of congenital cataracts impact redox biology in lens. Our findings shed light on the molecular mechanisms associated with congenital cataracts and point out that unbalanced redox status due to reduced levels of taurine and glutathione, metabolites already linked to age-related cataract, could be a major underlying mechanism behind lens opacities that appear early in life.
Keywords: Cataracts, Ubiquitin, Redox status, Glutathione, Taurine, Proteomics
Graphical abstract
Highlights
-
•
Unbalanced redox underlies lens opacity in the K6W-Ub congenital cataract model.
-
•
Dynamic proteome changes are seen in K6W-Ub cataracts and during development.
-
•
Proteomic targets are related to glutathione and amino acid metabolic pathways.
-
•
Computational molecular phenotyping shows altered spatial patterns of amino acids.
-
•
Antioxidants taurine and glutathione are reduced and mislocalized in cataracts.
1. Introduction
The lens is a transparent and avascular structure inside the eye that focuses light onto the retina for visual processing. Defective lens development and maturation results in lens opacification or cataracts. Lens clouding is a leading cause of blindness worldwide and cataracts that appear early in life account for one-third of the cases of blindness in children [1]. Although some genetic variants and gene mutations are associated with congenital cataracts and progress has been made with regard to disease etiology, a complete understanding of molecular pathogenic events behind congenital cataracts remains aspirational. Congenital cataracts need to be removed surgically early in life and, unfortunately, there is a higher risk of post-operative complications in infants and children than in adults [2]. A comprehensive understanding of the molecular mechanisms behind lens opacity is crucial to better designed strategies to diminish the prevalence of early-onset forms of cataracts.
Lens differentiation is a complex biological developmental process in which lens fiber cells undergo massive changes including organelle removal and high expression of lens-specific proteins (e.g. crystallins). Terminal lens fiber cell differentiation begins around embryonic day fifteen (E15) in mice, and fiber cell differentiation continues throughout the life of the organism. All these changes require proteome remodeling and a fine-tuned expression of proteins in different developmental stages for proper completion of the lens maturation. Although failures in the lens fiber cell differentiation are associated with many forms of congenital cataracts, to date a systematic comparative proteomics analysis in congenital cataracts has not been reported.
Oxidative stress occurs during the course of lens differentiation and oxidative metabolism is vital to preserve lens transparency. Multiple evidence supports a major role for a balanced redox status network in lens homeostasis. In age-related cataracts, oxidative stress is believed to be an initiating event and a reduction of antioxidant capacity leads to age-related lens opacification [[3], [4], [5], [6]]. γ-Glutamyl-cysteinyl-glycine (glutathione) is a key antioxidant in our body and high concentrations of reduced glutathione (GSH) are found in transparent lenses, although lower amounts of the oxidized form (GSSG) can be also detected [4,5,7]. The GSH/GSSG ratio is an indicator of redox status and a decrease in the ratio is linked to age-related cataracts [4,5,8,9]. In addition to glutathione, a significant antioxidant capacity is bestowed on free amino acids that are found more concentrated in the lens than in other tissues [[10], [11], [12]] and deficit in the enzymes involved in amino acid metabolism are linked to infantile cataract [13,14]. Interestingly, the most abundant amino acid in the lens is taurine, a sulfur β-amino acid with high antioxidant properties whose dietary intake protects against glutathione depletion-derived opacity [15]. Taurine levels are highly reduced in the aging lens, where its deficiency has been connected to age-related cataracts [16]. More recently, taurine deficiency has been broadly linked to age-related disease and shortened lifespan [17].
Ubiquitination is an essential process in proteostasis that determines the protein stability at different levels. Ubiquitination is able to modulate protein-protein interactions, activate/inactivate protein function, and direct proteins substrate to degradative compartments [18]. Ubiquitin biology is also essential for lens maturation and mutations in the ubiquitin system have been associated with human cataracts [[19], [20], [21]]. The lens-specific overexpression of a dominant negative mutant of ubiquitin (K6W, lysine is replaced to tryptophan at position 6) results in defective lens development and congenital cataract formation [22]. The K6W-Ub cataractogenic mouse recapitulates multiple pathogenic characteristics found in human cataracts including accumulation of intracellular aggregates and retained organelles [[23], [24], [25]]. The K6W-Ub mouse thus provides a genetic model to elucidate molecular processes involved in the onset of lens opacification.
Here, we performed a multi-disciplinary approach in order to reveal early pathogenic cataractogenic events using the K6W-Ub mouse as congenital cataract model. MS-based tandem-mass-tag quantitative proteomics (TMT) was carried out in lenses collected in different developmental stages in control and K6W-Ub overexpressing mice. Computational molecular phenotyping (CMP) and high-performance liquid chromatography (HPLC) showed that altered amino acid and glutathione metabolism lead to unbalanced redox status in cataractous lenses. Reduced levels of the antioxidant taurine and glutathione were the most prominent metabolites contributing to the pathological phenotype. In sum, our research documents the dynamic proteome changes in a mouse model of congenital cataracts pointing to the unbalanced redox status network as an early pathological event.
2. Material and methods
2.1. Animal husbandry
All animal experiments were approved by the Institutional Animal Care and Use Committee at Tufts University and were performed according to NIH guidelines for using experimental animals. The transgenic mouse was generated as previously reported [22]. The expression of MRGS(His)6-K6W-Ub is under the control of the αA-crystallin promoter with DCR1 enhancer and expression of the transgene was determined by Western blotting using an antibody specific to the transgene product (MRGS-His4) from Qiagen. All experiments were performed using mice that were homozygous for the K6W-Ub transgene in a C57BL/6J genetic background. Wild-type (WT) mice were C57BL/6J mice purchased from Jackson laboratories.
2.2. Mass spectrometry-based tandem-mass-tag (TMT) quantitative proteomics
Thirty-six pooled lenses from E15 and twelve pooled lenses from newborn (P1) mice and P30 of WT and K6W-Ub mice were used to generate three biological replicates for proteomic analysis. Samples were sonicated in hypotonic buffer and 100 μg from each pooled sample and dried by vacuum centrifugation. The experimental proteomics design utilized 7-plex TMT (Thermo Scientific) [26] to directly compare samples of mouse lens tissue: three-time points (E15, P1, P30) of two mouse lines (WT and K6W-Ub) and a common control sample (mixture of both types). The latter was used in all experiments to normalize between assays of all time points and genotypes. Prior to the analytical run, 20 μg of isotopically labeled peptides from each sample were checked for labeling efficiency, combined, freed of remaining TMT, and analyzed by a multidimensional LC-MS/MS, MudPIT [27], using Dionex pumps coupled to a nanoESI Q Exactive Plus Orbitrap mass spectrometer. HCD fragmentation yields ions that represent the relative amount of peptide present in each sample. Protein identification, assembly, and reporter ion quantitation were performed using Proteome Discoverer (Thermo Scientific). HCD fragmentation spectra was searched against a mouse subset of the Uniprot KB protein database (uniprot.org). For rigor, the TMT experiment was run in triplicate with each 7-plex analysis representing a separate cohort of animals. For statistical analysis of TMT protein ratios, log2 protein ratios were fit to a normal distribution using non-linear (least squares) regression. The mean and standard deviation values from the Gaussian fit were used to calculate p-values, using Z-score statistics. A given log2 TMT protein ratio, with the calculated mean and standard deviation of the fitted data, was transformed to a standard normal variable (z = (x-μ)/σ). Calculated p-values were corrected for multiple comparisons using the Benjamini-Hochberg method [28]. A p-value of 0.01 and FDR of 0.1 was used as the cutoff for identifying differentially expressed proteins in this assay [25]. One-way ANOVA analysis was performed for each protein's abundance. Analyses of differential protein expression between WT and K6W-Ub lenses at each time point was performed using R and the edgeR package [29]. Annotation and function, gene ontology and pathway enrichment of proteins levels of which are statistically different between K6W-Ub and WT lenses, were determined using DAVID and Enrichr [30,31]. The full proteomic data analysis is shown in Supplemental Data 1. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD045040.
2.3. Computational molecular phenotyping (CMP)
Lenses used for CMP analysis were fixed in a 2.5% glutaraldehyde, 1% formaldehyde, 0.1 M cacodylate (pH 7.4), 1 mM MgSO4, and 3% sucrose fixation solution. Following fixation, lenses were dehydrated through increasing concentrations of methanol to acetone and embedded in an epon resin. Embedded lenses were serially sectioned at 100 nm and placed on 12-well plates. CMP used IgG probes to small molecules in serial sections, visualized with a secondary antibody conjugated to a 1.4 nm gold granule, followed by silver intensification to generate spatially preserved, quantitative, metabolic maps of the lens [[32], [33], [34], [35], [36]]. Antibodies against amino acids alanine, cysteine, aspartate, lysine, glutamate, glutamine, glycine, arginine, glutathione, valine, taurine, and serine were used in this study as previously reported [[37], [38], [39]]. K-means clustering was used to identify the metabolic signatures present across the WT or K6W lenses. To perform clustering, immunolabeled, silver-intensified, serial sections were imaged using an Infinity 3 (Teledyne Luminera) camera under constant gain and offset. Captured grayscale images were then computationally aligned using IR-Tweak (SCI-University of Utah) [35]. The resulting image stacks were then clustered using k-means to identify metabolic classes (PCI-Geomatica). Cell classes are then represented as a theme map, in which each color represents an individual metabolic class. Total levels of each analyte were measured as average intensities using ImageJ 1.52 and are reported as arbitrary units (a.u.), that is, the sum of the gray values of all the pixels in the region of interest divided by the number of pixels. Partial least-square regression analysis was performed using MetaboAnalyst [40].
2.4. Immunohistochemistry and western blotting
Lenses dissected from WT and transgenic mice were fixed with 4% paraformaldehyde for 2 h, embedded in optimal cutting temperature compound (Sakura Finetek, Torrance, CA, USA) frozen, and subsequently sectioned, as reported previously [22,23]. Lens cryosections (12 μm) were permeabilized with 0.05% Triton X-100 for 5 min at room temperature, blocked with 5% normal donkey or goat serum (Jackson ImmunoResearch) in PBS for 1 h at room temperature. Primary antibodies against GGCT (Proteintech, Rosemont IL) and GSTP1 (GeneTex, Irvine CA) were incubated at room temperature for 1 h and secondary antibodies (Jackson Immunoresearch. West Grove PA) for 45 min at room temperature. Nuclear DNA was visualized with Prolong Gold Antifade with DAPI (Thermo Fisher, Waltham MA). Fluorescent images were obtained with an inverted fluorescence microscope (Axiovert 200 equipped with AxioVision V 4.5 software; Zeiss, Jena, Germany).
For Western blot analysis, lenses were homogenized directly in loading buffer. Equal quantities of samples were separated by SDS-PAGE. Proteins were transferred by electroblotting onto nitrocellulose membranes (0.2 μm; Bio-Rad, Hercules, CA, USA), after which the membranes were probed with primary antibodies against GGCT, GAMT, and PGAM2 (Proteintech) and HMOX1, GSTP1 (GeneTex) overnight at 4 °C and HRP-conjugated secondary antibodies for 45 min at room temperature. Signals were detected by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientific).
2.5. High performance liquid chromatography (HPLC)
Lens samples were homogenized in prechilled medium containing phosphate buffer (Fluka, Buchs, Switzerland) (pH 7.0) and perchloric acid (PCA) (Pancreac, Barcelona, Spain). Suspensions were centrifuged at 14,000×g and the resulting supernatants were collected, stored at −80 °C and used for both HPLC determinations. GSH and GSSG content of lens homogenates were quantified by the method of Reed et al., as previously reported in lens samples [41,42]. The samples were mixed with a solution of iodoacetic acid (Sigma, St. Louis, MO) and Sanger reagent (1-fluor-2,4-dinitrobencene) (Sigma, St. Louis, MO). These products are quickly separated by HPLC (Gilson, detector UV/VIS 156), which allows the quantification of nanomolar levels.
Free tissue levels of amino acids were assessed as previously [43,44]. Supernatants were derivatized with ortho-phthal-dialdehyde (OPA). The reagent was a mixture of 32 mg OPA in borate buffer 0.4 M pH 9.5 (7140 μl) containing 60 μl of 3-mercaptopropionic acid. The fluorescent derivatized amino acids were separated by a ‘‘Ultrasphere ODS Beckman’’ (150 × 4.6 mm, particle size 5 μm) column using gradient elution. Gradients were performed with two degassed mixture solvents. Solvent A was 0.05 M sodium acetate pH 5.88: methanol (90:10), and solvent B was methanol: H2O (70:30). (Gradient profile: time = 0 min % B 2, time = 0.1 min % B 15, time = 1% B 47, time = 6% B 100, time = 9% B 2); at time = 13 the column is ready for a new sample injection. The solvent flow rate was adjusted to 1 ml/min and the injection volume was 10 μl. Fluorescence detection was accomplished with Jasco detector (model FP-2020) at 240 and 450 nm for excitation and emission wavelengths, respectively. Amino acids were identified by their retention times, and their concentrations were calculated by comparison to calibrated amino acid external standard solutions (1.5 μM).
3. Results
3.1. Quantitative proteomics revealed changes in amino acid and glutathione metabolism in the cataractogenic model
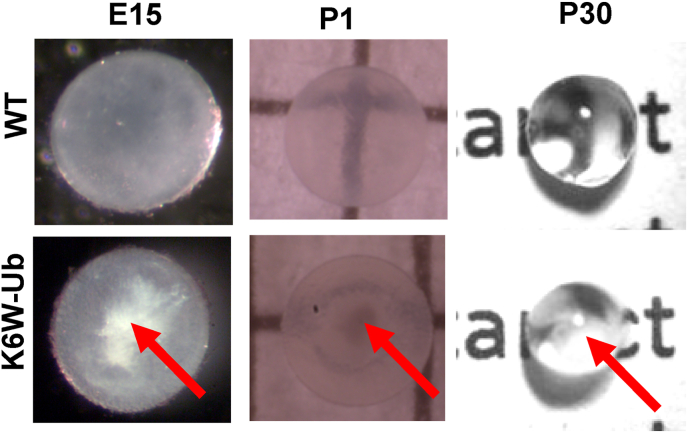
In order to gain molecular insights into the early pathogenic events underlying lens opacity in congenital cataracts, we analyzed the lenses dissected from the cataractogenic animal model overexpressing a dominant negative mutant of ubiquitin (K6W-Ub). Lysine 6 is in the surface of the ubiquitin molecule and, when it is replaced with tryptophan, the formation of K6-kinked ubiquitin chains in protein substrates is blocked [45]. The overexpression of this mutant ubiquitin protein results in defective lens development and nuclear cataract formation [22,24,25] (Fig. 1).
Fig. 1.
Cataract formation in K6W-Ub lenses. Photographs of dissected WT and K6W-Ub lenses at E15, P1, and P30 show nuclear cataracts (arrows) in K6W-Ub lenses.
In this study, we compared the lens proteome at different hallmark stages of lens development: embryonic day fifteen (E15, when the lens is formed but organelle removal has not yet occurred), postnatal day one (P1, when organelle removal and remodeling of fibers are in progress) and postnatal day thirty (P30, the developmental process is advanced, and the maturation of the lens fibers is completed). As observed in dissected WT and K6W-Ub lenses from these timepoints, nuclear cataracts are evident in the K6W-Ub lenses (Fig. 1).
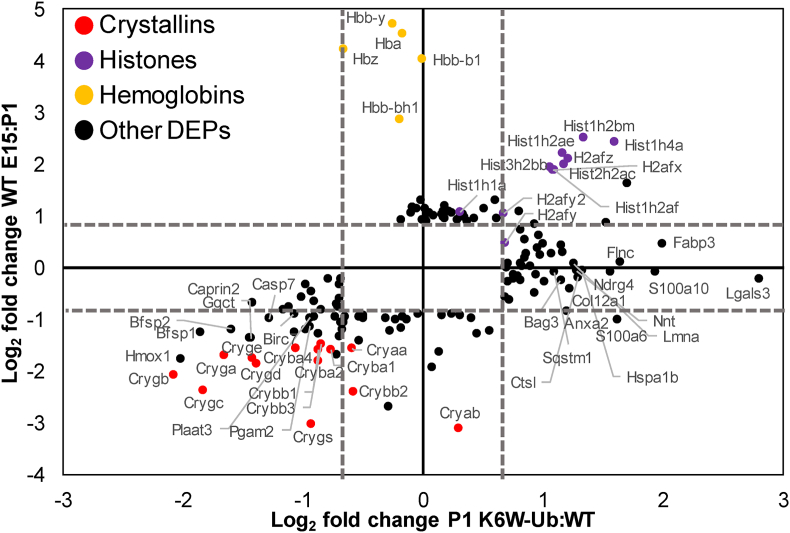
TMT-quantitative proteomics detected a total of 3789 proteins. A total of 91 (53 down- and 38 up-regulated), 97 (43 down- and 54 up-regulated), and 163 (49 down- and 114 upregulated) were differentially expressed proteins (DEPs) in K6W-Ub lenses at the different stages, that is, 2.4%, 2.5% and 4.3% of total protein detected at E15, P1 and P30, respectively (Supplemental Data 1). We also identified DEPs that were upregulated or downregulated developmentally in WT mice between E15-P1 (56 down- and 46 up-regulated) and P1–P30 (21 down- and 62 up-regulated). Comparisons of DEPs between E15-P1 in WT lenses and that were altered in K6W-Ub P1 lenses revealed that many proteins that are developmentally upregulated, particularly lens crystallins and other lens structural proteins were downregulated in K6W-Ub lenses (Fig. 2, Supplemental Data 2). We identified 62 proteins that were only altered developmentally, 57 proteins that were only altered by K6W-Ub expression, and 40 proteins that were altered both developmentally and by K6W-Ub expression (Supplemental Data 2). A small subset of proteins, almost entirely consisting of histones were downregulated developmentally and upregulated in K6W-Ub lenses. These may be connected to the normal process of lens fiber cell denucleation, which is impaired in K6W-Ub lenses [22]. Hemoglobin proteins were present exclusively at E15, likely deriving from both residual hyaloid vasculature and the embryonic lens, which produces hemoglobin, but not heme [46].
Fig. 2.
Developmental proteomic changes in the lens overlap with K6W-Ub proteomic alterations. Comparison of DEPs between WT and K6W-Ub at P1 (X-axis) versus developmental changes in WT lenses between E15 and P1 (Y-axis). Dashed axis lines mark boundaries of statistical significance. Colored dots represent different functional groups of proteins.
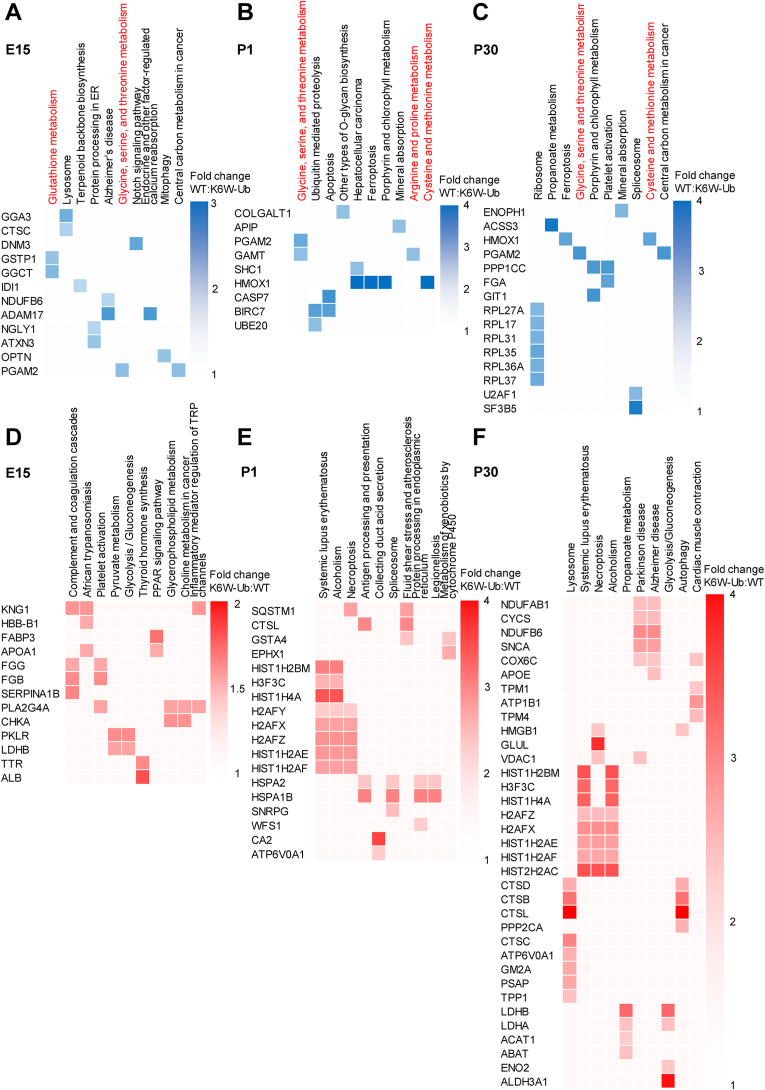
Next, we performed gene ontology analysis and functional annotation clustering to extract biological information. We found that the “glycine, serine, and threonine metabolism” pathway was consistently enriched in the downregulated DEPs in the K6W-Ub background in all the developmental stages (Fig. 3A–C). In addition, we observed that downregulated DEPs in cataractous lenses are enriched in different amino acid-related pathways: glutathione metabolism at E15, arginine and proline metabolism at P1, and cysteine and methionine metabolism at P1 and P30 (Fig. 3A–C). Interestingly, proteins that change during the development of normal lenses are also enriched in these pathways, including glutathione metabolism, glycine, serine and threonine metabolism, cysteine and methionine metabolism, arginine biosynthesis, phenylalanine metabolism and histidine metabolism (Suppl. Fig. 1). Of note, downregulated DEPs in K6W-Ub background at all developmental stages are associated with different abnormalities in lens development including abnormal lens fiber morphology, cataracts, vacuolated lens, small lens (Suppl. Fig. 2) and were involved in different gene ontology processes related to vision at E15 and P1 that include visual perception and sensory perception of light stimulus (Suppl. Fig. 3). By contrast, we did not find biological processes linked to vision-related processes in the upregulated DEPs in K6W-Ub background and the targets were not associated to lens abnormalities or gene ontology processes related to vision (Fi.g 3 D-F, Suppl. Figs. 1–3).
Fig. 3.
Quantitative proteomics reveals changes in amino acid and glutathione metabolism in congenital cataracts. (A–C) Grid visualization of downregulated DEPs identified in K6W-Ub lenses at (A) E15, (B) P1, and (C) P30 and. (D–F) Grid visualization of upregulated DEPs identified in K6W-Ub lenses at (D) E15, (E) P1, and (F) P30. Shown in columns are the top 10 associated KEGG pathways, which are ordered by p-values. KEGG pathways involved in glutathione and amino acid metabolism (highlighted in red). Rows indicate DEPs, based on comparisons of K6W-Ub relative to WT. In A-C, blue shades indicate the degree of downregulation in K6W-Ub lenses, with darker shades corresponding to more downregulation, according to the key next to each grid. In D-F, red shades indicate the degree of upregulation in K6W-Ub lenses, with darker shades corresponding to more upregulation, according to the key next to each grid. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
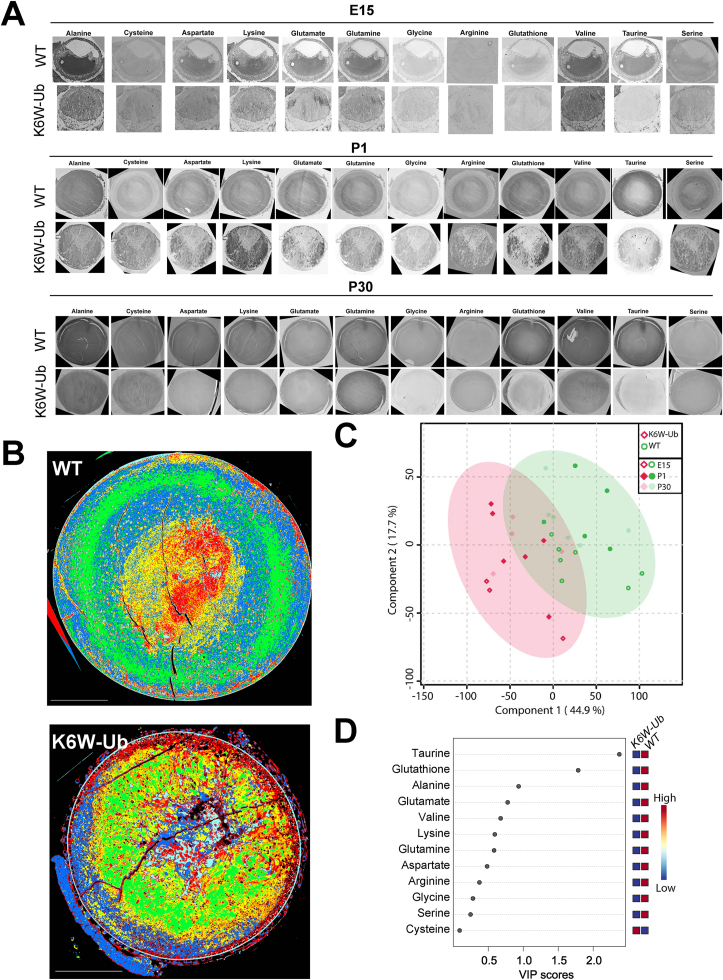
3.2. Computational molecular phenotyping revealed spatiotemporal expression patterns for free amino acids in lenses and altered distribution in congenital cataracts
Given that the overexpression of K6W-Ub altered targets that modulate amino acid metabolism, we performed computational molecular phenotyping (CMP), a technique that allow us to visualize free amino acid fingerprinting at cellular resolution and to preserve the morphological context. Lenses from WT and transgenic animals were harvested at E15, P1, and P30, and the levels of the following metabolites were evaluated: alanine, cysteine, aspartate, lysine, glutamate, glutamine, glycine, arginine, glutathione, valine, taurine, and serine (Fig. 4A). CMP revealed that amino acid distributions were highly altered in K6W-Ub lenses at all developmental times, with altered spatial patterns and different abundances. In order to understand the heterogeneities spatially and by abundance, we turned to multiparametric methods. We first utilized a K-means clustering approach to map spatial patterns of amino acid signatures in the P1 WT or K6W-Ub lens (Fig. 4B). In P1 WT lenses, we identified clusters of amino acids that defined cohesive cortical patterns (blue and green pseudocolors) or nuclear patterns (red and yellow pseudocolors) (Fig. 4B, top). In K6W-Ub lenses, these patterns degraded and hypervariability in metabolic state was observed (Fig. 4B, bottom). We next applied a chemometric method, partial least-square regression discrimination analysis, to identify differences in amino acid composition between WT and K6W-Ub lenses. This analysis revealed that the amino acid metabolomes of WT and K6W-Ub lenses were significantly different at all timepoints (Fig. 4C). We found that taurine and glutathione contributed the most variance based on significant variable-in-project (VIP) scores greater than 1 (Fig. 4D).
Fig. 4.
CMP reveals altered spatiotemporal expression patterns for free amino acids in congenital cataracts. (A) Representative CMP pictures of WT and K6W-Ub lenses collected at E15 (top), P1 (middle), and P30 (bottom) and stained against alanine, cysteine, aspartate, lysine, glutamate, glutamine, glycine, arginine, glutathione, valine, taurine, and serine. (B) Metabolic signatures of WT and K6W-Ub lenses collected at P1 and colored according to metabolic classes derived from K-means clustering. (C) Partial least squares regression analysis discriminated metabolomes of WT (green) and K6W-Ub (red) lenses at all ages. (D) Variable importance in projection (VIP) scores for different amino acids responsible for the separation in the regression plot. All amino acids were higher in WT than K6W-Ub except for cysteine. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
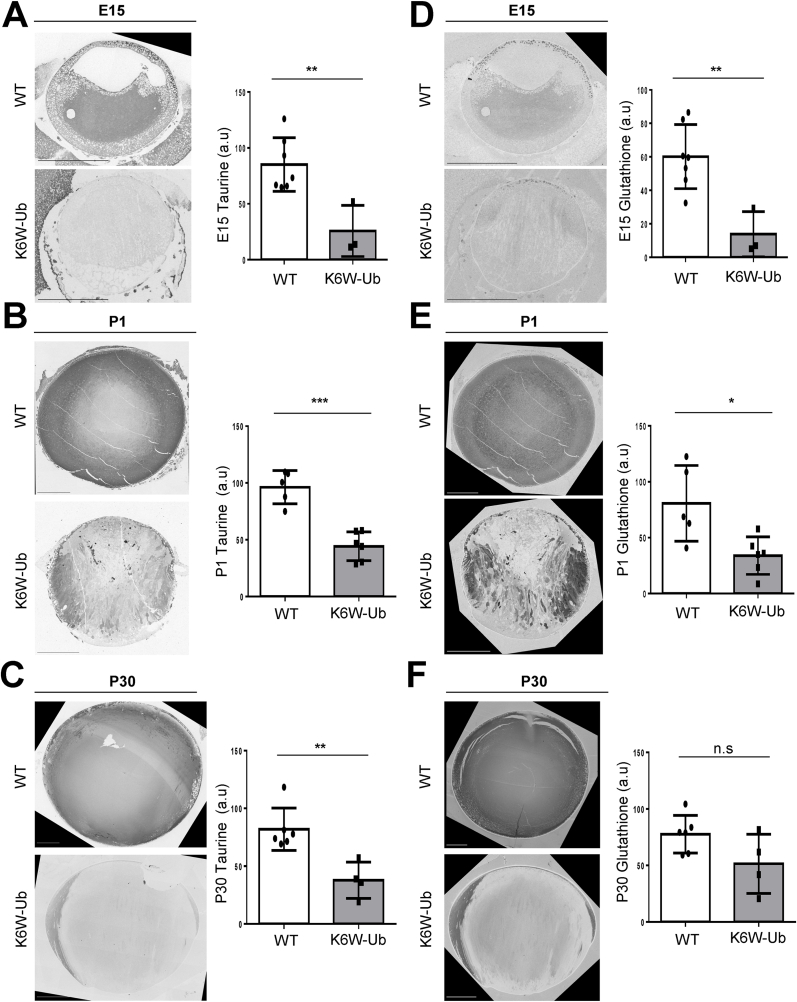
Levels of taurine in K6W-Ub cataractous lenses decreased by 69.8%, 54.0%, and 53.9% at E15, P1 and P30, respectively (Fig. 5A–C), while levels of glutathione decreased by 77.0%, 58.0%, and 33.6% at E15, P1 and P30, respectively (Fig. 5D–F). Taurine and glutathione spatial distributions in K6W-Ub lenses were also highly altered. Glutathione levels were mislocalized equatorially at E15.5 and P30 and were mosaic at P1, with both abnormally high- and low-containing cells in K6W-Ub lenses (Fig. 5D–F). The anterio-central nuclear cataracts contained very low amounts of glutathione and taurine (Fig. 5B and E). No significant changes in total amino acid levels were found in early developmental stages of lens maturation for any other metabolites (Suppl. Fig. 4).
Fig. 5.
Changes in abundance and spatial distributions of taurine and glutathione in the WT and K6W-Ub lenses. (A–C) Representative CMP pictures for taurine staining in WT and K6W-Ub lenses collected at (A) E15, (B) P1, and (C) P30. (D–F) Representative CMP pictures for glutathione staining in WT and K6W-Ub lenses collected at (D) E15, (E) P1, and (F) P30. Quantification of average intensities is shown as arbitrary units (a.u.). All values are mean ± SEM. and differences with WT were significant for (*) p < 0.05; (**) p < 0.01; and (***) p < 0.001.
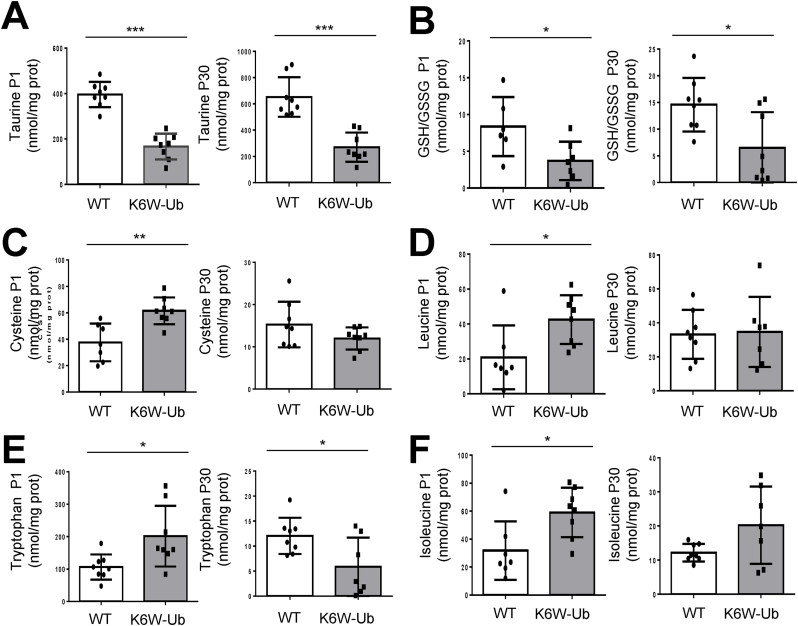
3.3. High-performance liquid chromatography reveals differential changes in taurine and glutathione levels and unbalanced redox in cataractous lenses
In order to corroborate and expand on our CMP findings, we used high-performance liquid chromatography (HPLC) to quantify the total levels of free amino acids in WT and K6W-Ub cataractous lenses, as well as total levels of reduced and oxidized glutathione (GSH, GSSG). Firstly, we found that overexpression of K6W-Ub did not impact all amino acids but only a few sets of amino acids were compromised in K6W-Ub lenses (Fig. 6 and Suppl. Fig.5). Taurine significantly decreased 57.8% and 58.5% at P1 and P30, respectively in K6W-Ub lenses (Fig. 6A). The GSH/GSSG ratio significantly decreased 55.7% and 55.4% in K6W-Ub lenses at P1 and P30, respectively (Fig. 4B), which was driven by decreases in GSH and increases in GSSG, even though not all changes were statistically significant (Suppl. Figs. 5A–B). Cysteine, a precursor of GSH, was augmented 63.8% along with leucine, isoleucine and tryptophan (Fig. 6C–F) in K6W-Ub lenses at P1. The latter amino acids did not show statistically significant changes in P30 lenses, except tryptophan which decreased 51.3% in K6W-Ub lenses. Together with our CMP data, these data support a model by which proteomic changes caused by overexpression of K6W-Ub lead to the imbalance of only a specific set of amino acids involved in the maintenance of redox status could contribute to lens opacity in congenital cataracts.
Fig. 6.
HPLC revealed altered levels of amino acids in K6W-Ub lenses. (A–F) Lens content of (A) taurine, (B) GSH/GSSG, (C) cysteine, (D) leucine, (E) tryptophan and (F) isoleucine in wild types and K6W-Ub lenses collected at P1 (left) and P30 (right) are shown. All values are mean ± SEM. and differences with WT were significant for (*) p < 0.05; (**) p < 0.01; and (***) p < 0.001. Sample size is n = 8.
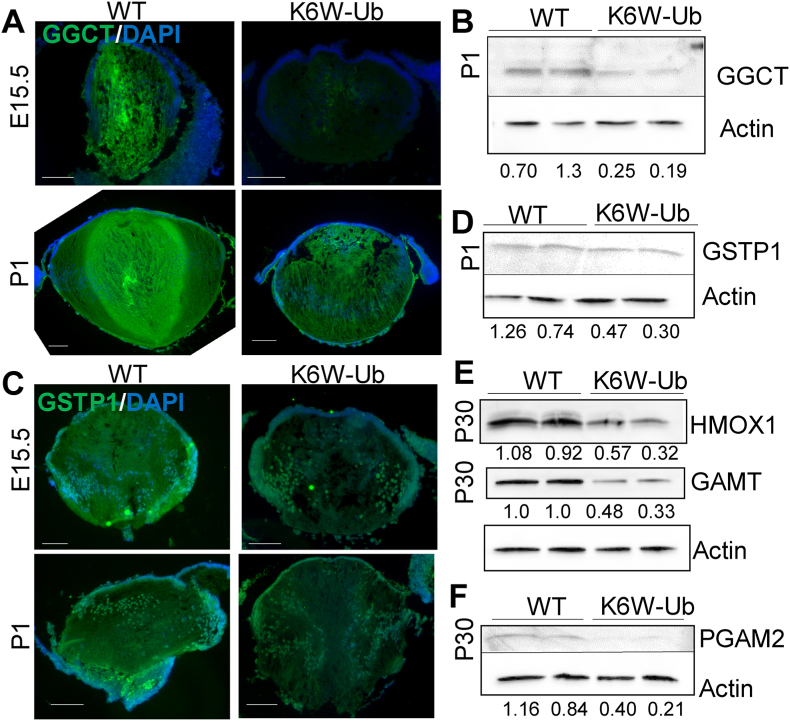
3.4. Reduced levels of proteins related to glutathione and amino acid metabolism in K6W-Ub lenses
In order to mechanistically connect proteomic changes with the altered amino acid distribution we observed using CMP and HPLC, we utilized Western blotting and immunohistochemistry to query select DEPs identified above. We found that γ-glutamylcyclotransferase (GGCT), related to glutathione metabolism, was decreased in E15 and P1 K6W-Ub lenses (Fig. 7A and B). Also in the glutathione pathway, the glutathione s-transferase pi 1 (GSTP1), an enzyme with antioxidant capacity, was decreased in E15 and P1 K6W-Ub lenses (Fig. 7C and D). Proteins related to the glycine, serine and threonine metabolism pathway and cysteine and methionine metabolism pathway, such as phosphoglycerate mutase 2 (PGAM2), heme oxygenase 1 (HMOX1), and guanidinoacetate N-methyltransferase (GAMT) were reduced in P30 K6W-Ub lenses (Fig. 7E and F).
Fig. 7.
Downregulated proteins in K6W-Ub lenses associated with glutathione and amino acids metabolism. (A–B) Representative images of immunofluorescence (A) in E15 and P1 WT and K6W-Ub lenses and (B) Western blotting of lysates of P1 WT and K6W-Ub lenses using antibodies against γ-glutamylcyclotransferase (GGCT). (C–D) Representative images of immunofluorescence (C) in E15 and P1 WT and K6W-Ub lenses and (D) Western blotting of lysates of P1 lenses using antibodies against glutathione s-transferase pi 1 (GSTP1). (E–F) Representative immunoblot against heme oxygenase 1 (HMOX1) (E), guanidinoacetate N-methyltransferase (GAMT) (E), and phosphoglycerate mutase 2 (PGAM2) (F) in P30 WT and K6W-Ub lenses. Actin-normalized densitometric quantitation is shown for each sample. For all Western blots, two biological replicates are shown.
Altogether, micromolecular profiling technology revealed that the overexpression of K6W-Ub disrupts the spatial organization of free amino acids in the lens, with the highest impact in taurine and glutathione, and western blotting and immunohistochemistry revealed that proteins that modulate these free amino acid levels were highly compromised in our model of congenital cataracts.
4. Discussion
Our research analyzed dynamic changes of the lens proteome during different steps of development and maturation in the K6W-Ub cataract mouse model that recapitulates most of the abnormalities seen in human congenital cataracts. Our multi-pronged approach (quantitative proteomics, morphological techniques, and biochemical analysis) identified a limited number of proteins related to taurine and glutathione biology and revealed unbalanced redox status network as early pathological event in congenital cataract.
Molecular genetic studies of congenital cataract have revealed roles for diverse lens fiber cell proteins like crystallins, connexins, HSF4, aquaporins, transcription factors, and cytoskeletal proteins in at least 50% of congenital cataracts [1]. Protein aggregation is a common finding, but it is not clear in cases outside of crystallin mutants, whether it is a cause or effect of other tissue dysfunction. Our research provides evidence pointing out the imbalance in taurine and glutathione levels as a pathological contributor to a metabolic deficit that, ultimately, leads to aberrant redox status regulation and precipitation of lenticular proteins.
In the past, the proteome in different tissues has been explored utilizing techniques that fail to identify less abundant proteins. This is especially relevant in the mature lens where crystallins make up over 90% of the total wet weight proteins and non-crystallins proteins are challenging to characterize. However, new mass spectrometry–based proteome methodologies have recently made possible the analysis of a more complete lens proteome [47,48]. Here, we used TMT quantitative proteomics to identify proteomic changes in the lens of a congenital cataract mouse. In agreement with previous reports in our lab that relied on less sensitive methods [24,25], the number of DEPs in the K6W-Ub background was extremely low. The lens-specific overexpression of the dominant mutant of ubiquitin did not cause a pleiotropic disturbance in the whole lens proteome but has a highly specific impact on molecular pathways. In this study we focused on proteins significantly down-regulated in K6W-Ub lenses because mutations of those proteins are associated to lens abnormalities and these DEPs and associated pathways are also enriched in the course of normal lens development. We have previously described developmental delay in the K6W-Ub lens, which is a characteristic of several other congenital cataract models and may be a critical component of cataract formation in K6W-Ub lenses [22,24]. No literature was found linking the targets significantly upregulated in cataractous lenses and further analysis will be required to evaluate the biological ramifications of these upregulated DEPs in the context of opacity. It is intriguing that some upregulated DEPs have been linked to proteostatic pathways, such as chaperones HSPA2 and HSPA1B (HSP70), and the autophagy receptor p62/SQSTM1.
Only a few down-regulated proteins and biological processes associated with these targets were altered at all stages of development in the cataractogenic models. Those were prominently linked to glutathione and amino acids metabolism. This smaller collection of DEPs allowed us to carefully query specific proteins in the context of K6W-Ub cataracts and their potential role in a broader number of congenital and age-related cataracts. GGCT has been linked to glutathione homeostasis in the context of cancer cells, but not in cataracts [49]. Reduced expression of GSTP1, an antioxidant protein, has been observed in age-related cataracts, possibly connected to promoter hypermethylation [50] or more recently to proteolytic degradation via the Parkin E3 ligase [51]. HMOX1 is a well-characterized downstream target of the NRF2 antioxidant transcription factor and has been extensively studied in lens epithelial cells, particularly because of its roles in regulation of ferroptosis [52,53]. GAMT and PGAM2 have not been directly connected to cataracts, but PGAM2 has been connected through its developmental upregulation in the lens as a key lens signature gene [54].
Surprisingly, we did not identify DEPs linked to taurine biosynthesis in our proteomic analyses, although we did identify DEPs connected to cysteine metabolism, an amino acid precursor needed for de novo biosynthesis of taurine. Since lens taurine is both of dietary source and from biosynthesis in the liver, future studies will be required to carefully measure taurine synthesis, transport, and catabolism in WT and cataractous lenses. Taurine deficiency created by knockout of the Slc6a6 taurine transporter led to age-related diseases, including retinal degeneration, and shortened lifespan [17,55]. Cataracts or other lens abnormalities were not reported in these studies. Although we did not detect Slc6a6 protein in the lens using TMT proteomics, gene expression data in the iSyTE database (integrated Systems Tool for Eye gene discovery) indicate that Slc6a6 levels in the lens increase throughout development, mirroring increases in lens taurine that we observed via CMP analysis [54].
Imbalance of amino acids has been proposed as factor in lens opacification and an early event in the etiology of senile cataracts [[56], [57], [58], [59]]. We found that taurine and glutathione are major determinants in the imbalance of free amino acids along with leucine, isoleucine, tryptophan and cysteine, the latter a precursor for the de novo synthesis of glutathione. Our results indicate that major free amino acids changes in the lens are associated with cataract [10,60], and are also in agreement with other studies in which deficiency of specific amino acids was sufficient to cause lens opacity [56,61,62]. Dietary amino acid supplementation has been shown to prevent cataract development [[63], [64], [65]] and the dietary intake of taurine was protective in a glutathione depletion-derived opacity model [15]. This opens up the possibility that dietary supplementation of taurine could be used as strategy to prevent human congenital cataract in a similar way that folic acid is used to prevent brain and spine birth defects. Further experiments in different congenital cataracts animal models will be required to establish an optimal taurine intake during pregnancy with capacity to prevent opacity.
An important aspect of our study is the spatial distribution of amino acids along the optical and equatorial axes in normal lenses during different developmental stages. Using CMP as a tool that provides a single-cell molecule fingerprint, we did not observe a homogenous distribution for the majority of free amino acids but a differential spatiotemporal pattern. In agreement with other reports, we found a concentric patterns for taurine and glutathione and a nuclear enrichment for other amino acids [66,67]. This could reflect the combination of biological processes modulating amino acid levels, including biosynthesis and influx/efflux transport processes [14]. We found significant changes in the pattern between normal and cataractous lenses in K6W-Ub lenses. Experiments conducted using imaging mass spectrometry methods like MALDI have considered the nuclear depletion of glutathione in aging as a potential initiating factor in age-related cataract [68]. Our findings extend the importance of maintenance of sufficient glutathione levels even in a congenital cataract model, a finding also mirrored in analysis of the Mip-mutant congenital cataract [69].
In addition, our research sheds light on the role of protein ubiquitination in lens biology, and more specifically on the function of unconventional K6-derived ubiquitination. The cellular function of K6-derived ubiquitination is not well understood, and recent papers highlight emerging roles of the lysine on position 6 of the ubiquitin molecule [70]. An increased of K6 linkages was not found upon proteasomal inhibition and K6 has been associated to DNA repair [[71], [72], [73], [74]]. Mitophagy is compromised in a K6R mutant ubiquitin [75] and deubiquitinases that selectively remove K6-linked ubiquitin chains were associated to mitochondrial quality control [76,77]. Our findings here support a critical role for K6-derived ubiquitination in the modulation of specific enzymes involved in amino acid metabolism, although the role of K6 linkages in this context remains to be elucidated. Further experiments are required to reveal which targets of K6-derived ubiquitination are responsible for the differential expression of key enzymes of taurine and glutathione metabolism.
Previously we observed that cells expressing K6W-Ub were highly susceptible to oxidative stress [78]. Our analysis here indicates that redox regulation is impaired, however, the mechanisms for this are unclear. Several of the proteins we have identified as decreased in the K6W-Ub lenses are known to be regulated by the ubiquitin proteasome pathway, including HMOX1 and GSTP1 [51,79]. Additionally, the rate limiting enzyme for taurine synthesis, cysteine dioxygenase (CDO1) is also regulated by the ubiquitin proteasome system [80]. The expression of K6W-Ub could alter not only the turnover of these substrates but also their function. Impaired mitochondrial function in K6W-Ub lenses may also be connected to increased redox stress. As discussed above, K6-linked ubiquitination is critical for mitophagy. K6W-Ub mitochondria are abnormally retained and may be damaged [22]. Upregulated DEPs in the glycolysis and gluconeogenesis pathways might also indicate abnormal mitochondrial function (Fig. 3D,F).
There are several potential limitations to our study. TMT proteomics is a sensitive and quantitative method to perform proteomics but is not able to detect and quantitate all proteins present in the lens. Therefore, it is likely that important proteins that are impacted by K6W-Ub expression or the cataractous state are missing from our dataset. Although we present data that indicate impaired redox status in K6W-Ub lenses, we have not directly measured increased oxidative damage in these lenses. Future studies will be required to carefully measure protein and lipid oxidation and oxidation-related post-translational modifications. K6W-Ub lenses are significantly smaller than WT lenses and contain a reduced amount of total proteins. Therefore, we cannot rule out that alterations in free amino acid levels may be connected to a reduced rate of protein synthesis, which we do not measure in this study. Taurine, however, is not incorporated into proteins – thus its decrease is not a reflection of decreased protein synthesis.
Altogether, the findings of this study reveal that unbalanced redox status derived from an imbalance in taurine and glutathione could be a major determinant for lens opacity that appear early in life and our research identify regulatory protein of amino acids metabolism as potential therapeutical targets in congenital cataracts.
Authors contributions
E.B, E.A.W, S.R, A.T. designed approach, generated primary manuscript, analyzed data and generated figures; R.L.P and B.W.J. carried out CMP approach, analyzed images, generated figures and revised the manuscript; K.L.R and K.L.S. carried out TMT approach, analyzed proteomics data and revised the manuscript; A.C. and I.A. provided HPLC data for GSH, GSSG and cysteine; A.P., M.J.A. and J.A.R. provided HPLC data for most of the free amino acids and revised the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Donald Smith, Min-Lee Chang, and Sarah Francisco for assistance with animal husbandry. This work was supported by RYC2018-024434-I, MINECO PID2020-119466RB-I00 (to E.B.); NIH RO1EY021212, RO1EY028559, and RO1EY026979 (to A.T.); R01-EY015128 R01-EY028927, P30 EY014800 Vision Core Grant (to B.W.J); USDA NIFA 2016–08885 (to A.T. and S.R.); USDA 8050-51000-089-01S and 8050-51000-101-01S (to A.T.); Thome Memorial Foundation (to A.T.); BrightFocus Foundation (to S.R.); an unrestricted grant from Research to Prevent Blindness, New York, NY to the Moran Eye Center; NSF 1835904 (to B.W.J). This material is based upon work supported by the US Department of Agriculture – Agricultural Research Service (ARS), under Agreement Nos. 58-1950-4-003 and 58-8050-9-004.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102869.
Contributor Information
Allen Taylor, Email: allen.taylor@tufts.edu.
Sheldon Rowan, Email: sheldon.rowan@tufts.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request. Proteomic data available at ProteomeXchange with the dataset identifier PXD045040.
References
- 1.Shiels A., Hejtmancik J.F. Mutations and mechanisms in congenital and age-related cataracts. Exp. Eye Res. 2017;156:95–102. doi: 10.1016/j.exer.2016.06.011. Epub 2016/06/24. PubMed PMID: 27334249; PMCID: Pmc5538314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.J., Wong E.S., Yam J.C. In: Pediatric Cataract : for Every Ophthalmologist. Agrawal S., editor. Springer Singapore; Singapore: 2021. Pediatric cataract surgery: post-operative complications and their management; pp. 131–154. [Google Scholar]
- 3.Truscott R.J. Age-related nuclear cataract-oxidation is the key. Exp. Eye Res. 2005;80(5):709–725. doi: 10.1016/j.exer.2004.12.007. Epub 2005/05/03. PubMed PMID: 15862178. [DOI] [PubMed] [Google Scholar]
- 4.Lou M.F. Glutathione and glutaredoxin in redox regulation and cell signaling of the lens. Antioxidants (Basel, Switzerland) 2022;11(10) doi: 10.3390/antiox11101973. Epub 2022/10/28. PubMed PMID: 36290696; PMCID: PMC9598519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giblin F.J. Glutathione: a vital lens antioxidant. J. Ocul. Pharmacol. Therapeut. 2000;16(2):121–135. doi: 10.1089/jop.2000.16.121. Epub 2000/05/10. PubMed PMID: 10803423. [DOI] [PubMed] [Google Scholar]
- 6.Spector A. Oxidative stress-induced cataract: mechanism of action. Faseb. J. : Off. Publ. Feder. Am. Soc. Exp. Biol. 1995;9(12):1173–1182. Epub 1995/09/01. PubMed PMID: 7672510. [PubMed] [Google Scholar]
- 7.Vinson J.A. Oxidative stress in cataracts. Pathophysiology : Off. J. Int. Soc. Pathophysiol. 2006;13(3):151–162. doi: 10.1016/j.pathophys.2006.05.006. Epub 2006/06/13. PubMed PMID: 16765571. [DOI] [PubMed] [Google Scholar]
- 8.Wei M., Xing K.Y., Fan Y.C., Libondi T., Lou M.F. Loss of thiol repair systems in human cataractous lenses. Invest. Ophthalmol. Vis. Sci. 2014;56(1):598–605. doi: 10.1167/iovs.14-15452. Epub 2014/12/30. PubMed PMID: 25537203; PMCID: Pmc4306347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X., Monnier V.M., Whitson J. Lens glutathione homeostasis: discrepancies and gaps in knowledge standing in the way of novel therapeutic approaches. Exp. Eye Res. 2017;156:103–111. doi: 10.1016/j.exer.2016.06.018. Epub 2016/07/05. PubMed PMID: 27373973; PMCID: Pmc5199622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinämäki A.A., Muhonen A.S., Piha R.S. Taurine and other free amino acids in the retina, vitreous, lens, iris-ciliary body, and cornea of the rat eye. Neurochem. Res. 1986;11(4):535–542. doi: 10.1007/bf00965323. Epub 1986/04/01. PubMed PMID: 3724960. [DOI] [PubMed] [Google Scholar]
- 11.Surai P.F., Earle-Payne K., Kidd M.T. Taurine as a natural antioxidant: from direct antioxidant effects to protective action in various toxicological models. Antioxidants (Basel, Switzerland) 2021;10(12) doi: 10.3390/antiox10121876. Epub 2021/12/25. PubMed PMID: 34942978; PMCID: Pmc8698923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu N., Chen G., Liu H. Antioxidative categorization of twenty amino acids based on experimental evaluation. Molecules (Basel, Switzerland) 2017;22(12) doi: 10.3390/molecules22122066. Epub 2017/12/01. PubMed PMID: 29186888; PMCID: PMC6149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rumping L., Tessadori F., Pouwels P.J.W., Vringer E., Wijnen J.P., Bhogal A.A., Savelberg S.M.C., Duran K.J., Bakkers M.J.G., Ramos R.J.J., Schellekens P.A.W., Kroes H.Y., Klomp D.W.J., Black G.C.M., Taylor R.L., Bakkers J.P.W., Prinsen H.C.M.T., van der Knaap M.S., Dansen T.B., Rehmann H., Zwartkruis F.J.T., Houwen R.H.J., van Haaften G., Verhoeven-Duif N.M., Jans J.J.M., van Hasselt P.M. GLS hyperactivity causes glutamate excess, infantile cataract and profound developmental delay. Hum. Mol. Genet. 2018;28(1):96–104. doi: 10.1093/hmg/ddy330. [DOI] [PubMed] [Google Scholar]
- 14.Knöpfel E.B., Vilches C., Camargo S.M.R., Errasti-Murugarren E., Stäubli A., Mayayo C., Munier F.L., Miroshnikova N., Poncet N., Junza A., Bhattacharya S.S., Prat E., Berry V., Berger W., Heon E., Moore A.T., Yanes Ó., Nunes V., Palacín M., Verrey F., Kloeckener-Gruissem B. Dysfunctional LAT2 amino acid transporter is associated with cataract in mouse and humans. Front. Physiol. 2019;10:688. doi: 10.3389/fphys.2019.00688. Epub 2019/06/25. PubMed PMID: 31231240; PMCID: Pmc6558864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sevin G., Kerry Z., Sozer N., Ozsarlak-Sozer G. Taurine supplementation protects lens against glutathione depletion. Eur. Rev. Med. Pharmacol. Sci. 2021;25(13):4520–4526. doi: 10.26355/eurrev_202107_26244. Epub 2021/07/22. PubMed PMID: 34286494. [DOI] [PubMed] [Google Scholar]
- 16.Gupta K., Mathur R.L. Distribution of taurine in the crystalline lens of vertebrate species and in cataractogenesis. Exp. Eye Res. 1983;37(4):379–384. doi: 10.1016/0014-4835(83)90174-4. Epub 1983/10/01. PubMed PMID: 6641821. [DOI] [PubMed] [Google Scholar]
- 17.Singh P., Gollapalli K., Mangiola S., Schranner D., Yusuf M.A., Chamoli M., Shi S.L., Lopes Bastos B., Nair T., Riermeier A., Vayndorf E.M., Wu J.Z., Nilakhe A., Nguyen C.Q., Muir M., Kiflezghi M.G., Foulger A., Junker A., Devine J., Sharan K., Chinta S.J., Rajput S., Rane A., Baumert P., Schonfelder M., Iavarone F., di Lorenzo G., Kumari S., Gupta A., Sarkar R., Khyriem C., Chawla A.S., Sharma A., Sarper N., Chattopadhyay N., Biswal B.K., Settembre C., Nagarajan P., Targoff K.L., Picard M., Gupta S., Velagapudi V., Papenfuss A.T., Kaya A., Ferreira M.G., Kennedy B.K., Andersen J.K., Lithgow G.J., Ali A.M., Mukhopadhyay A., Palotie A., Kastenmuller G., Kaeberlein M., Wackerhage H., Pal B., Yadav V.K. Taurine deficiency as a driver of aging. Science. 2023;380(6649) doi: 10.1126/science.abn9257. Epub 2023/06/08. PubMed PMID: 37289866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tracz M., Bialek W. Beyond K48 and K63: non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021;26(1):1. doi: 10.1186/s11658-020-00245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiuso P., Libondi T., Facchiano A.M., Colicchio P., Ferranti P., Lilla S., Colonna G. Alteration in the ubiquitin structure and function in the human lens: a possible mechanism of senile cataractogenesis. FEBS Lett. 2002;531(2):162–167. doi: 10.1016/s0014-5793(02)03494-4. Epub 2002/11/06. PubMed PMID: 12417305. [DOI] [PubMed] [Google Scholar]
- 20.Tsurusaki Y., Ohashi I., Enomoto Y., Naruto T., Mitsui J., Aida N., Kurosawa K. A novel UBE2A mutation causes X-linked intellectual disability type Nascimento. Hum. Genome Variat. 2017;4 doi: 10.1038/hgv.2017.19. Epub 2017/06/15. PubMed PMID: 28611923; PMCID: Pmc5462939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph T., Sjölander A., Palmér M.S., Minthon L., Wallin A., Andreasen N., Tasa G., Juronen E., Blennow K., Zetterberg H., Zetterberg M. Ubiquitin carboxyl-terminal esterase L1 (UCHL1) S18Y polymorphism in patients with cataracts. Ophthalmic Genet. 2011;32(2):75–79. doi: 10.3109/13816810.2010.544360. Epub 2011/01/28. PubMed PMID: 21268678; PMCID: Pmc3116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caceres A., Shang F., Wawrousek E., Liu Q., Avidan O., Cvekl A., Yang Y., Haririnia A., Storaska A., Fushman D., Kuszak J., Dudek E., Smith D., Taylor A. Perturbing the ubiquitin pathway reveals how mitosis is hijacked to denucleate and regulate cell proliferation and differentiation in vivo. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013331. Epub 2010/10/27. PubMed PMID: 20975996; PMCID: Pmc2958118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu L., Whitcomb E.A., Jiang S., Chang M.L., Gu Y., Duncan M.K., Cvekl A., Wang W.L., Limi S., Reneker L.W., Shang F., Du L., Taylor A. Unfolded-protein response-associated stabilization of p27(Cdkn1b) interferes with lens fiber cell denucleation, leading to cataract. Faseb. J. : Off. Publ. Feder. Am. Soc. Exp. Biol. 2016;30(3):1087–1095. doi: 10.1096/fj.15-278036. Epub 2015/11/22. PubMed PMID: 26590164; PMCID: Pmc4750420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K., Lyu L., Chin D., Gao J., Sun X., Shang F., Caceres A., Chang M.L., Rowan S., Peng J., Mathias R., Kasahara H., Jiang S., Taylor A. Altered ubiquitin causes perturbed calcium homeostasis, hyperactivation of calpain, dysregulated differentiation, and cataract. Proc. Natl. Acad. Sci. U.S.A. 2015;112(4):1071–1076. doi: 10.1073/pnas.1404059112. Epub 2015/01/15. PubMed PMID: 25583491; PMCID: Pmc4313858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang F., Wilmarth P.A., Chang M.L., Liu K., David L.L., Caceres M.A., Wawrousek E., Taylor A. Newborn mouse lens proteome and its alteration by lysine 6 mutant ubiquitin. J. Proteome Res. 2014;13(3):1177–1189. doi: 10.1021/pr400801v. Epub 2014/01/24. PubMed PMID: 24450463; PMCID: Pmc3993935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A.K., Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. Epub 2003/04/26. PubMed PMID: 12713048. [DOI] [PubMed] [Google Scholar]
- 27.Florens L., Washburn M.P. Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-x:159. Epub 2006/06/21. PubMed PMID: 16785648. [DOI] [PubMed] [Google Scholar]
- 28.Thissen D., Steinberg L., Kuang D. Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J. Educ. Behav. Stat. 2002;27(1):77–83. doi: 10.3102/10769986027001077. [DOI] [Google Scholar]
- 29.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. Epub 2009/11/17. PubMed PMID: 19910308; PMCID: Pmc2796818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. Epub 2013/04/17. PubMed PMID: 23586463; PMCID: Pmc3637064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marc R.E., Murry R.F., Basinger S.F. Pattern recognition of amino acid signatures in retinal neurons. J. Neurosci. : Off. J. Soc. Neurosci. 1995;15(7 Pt 2):5106–5129. doi: 10.1523/jneurosci.15-07-05106. Epub 1995/07/01. 1995. PubMed PMID: 7623139; PMCID: Pmc6577898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marc R.E., Cameron D. A molecular phenotype atlas of the zebrafish retina. J. Neurocytol. 2001;30(7):593–654. doi: 10.1023/a:1016516818393. Epub 2002/07/16. PubMed PMID: 12118163. [DOI] [PubMed] [Google Scholar]
- 34.Marc R.E., Jones B.W. Molecular phenotyping of retinal ganglion cells. J. Neurosci. : Off. J. Soc. Neurosci. 2002;22(2):413–427. doi: 10.1523/jneurosci.22-02-00413.2002. Epub 2002/01/11. PubMed PMID: 11784786; PMCID: Pmc6758675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson J.R., Jones B.W., Yang J.H., Shaw M.V., Watt C.B., Koshevoy P., Spaltenstein J., Jurrus E., U V.K., Whitaker R.T., Mastronarde D., Tasdizen T., Marc R.E. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7(3) doi: 10.1371/journal.pbio.1000074. Epub 2009/10/27. PubMed PMID: 19855814; PMCID: PMC2661966 other authors declare no other competing interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeiffer R.L., Marc R.E., Jones B.W. Müller cell metabolic signatures: evolutionary conservation and disruption in disease. Trends Endocrinol. Metabol.: TEM. 2020;31(4):320–329. doi: 10.1016/j.tem.2020.01.005. Epub 2020/03/19. PubMed PMID: 32187524; PMCID: Pmc7188339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeiffer R.L., Marc R.E., Kondo M., Terasaki H., Jones B.W. Müller cell metabolic chaos during retinal degeneration. Exp. Eye Res. 2016;150:62–70. doi: 10.1016/j.exer.2016.04.022. Epub 2016/05/05. PubMed PMID: 27142256; PMCID: Pmc5031519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones B.W., Pfeiffer R.L., Ferrell W.D., Watt C.B., Tucker J., Marc R.E. Retinal remodeling and metabolic alterations in human AMD. Front. Cell. Neurosci. 2016;10:103. doi: 10.3389/fncel.2016.00103. Epub 2016/05/21. PubMed PMID: 27199657; PMCID: Pmc4848316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffer R.L., Anderson J.R., Emrich D.P., Dahal J., Sigulinsky C.L., Morrison H.A.B., Yang J.H., Watt C.B., Rapp K.D., Kondo M., Terasaki H., Garcia J.C., Marc R.E., Jones B.W. Pathoconnectome analysis of Müller cells in early retinal remodeling. Adv. Exp. Med. Biol. 2019;1185:365–370. doi: 10.1007/978-3-030-27378-1_60. Epub 2019/12/31. PubMed PMID: 31884639; PMCID: Pmc7046339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia J., Wishart D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011;6(6):743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 41.Arnal E., Miranda M., Almansa I., Muriach M., Barcia J.M., Romero F.J., Diaz-Llopis M., Bosch-Morell F. Lutein prevents cataract development and progression in diabetic rats. Graefe's Arch. Clin. Exp. Ophthalmol. = Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie. 2009;247(1):115–120. doi: 10.1007/s00417-008-0935-z. Epub 2008/09/04. PubMed PMID: 18766362. [DOI] [PubMed] [Google Scholar]
- 42.Reed D.J., Babson J.R., Beatty P.W., Brodie A.E., Ellis W.W., Potter D.W. High-performance liquid chromatography analysis of nanomole levels of glutathione, glutathione disulfide, and related thiols and disulfides. Anal. Biochem. 1980;106(1):55–62. doi: 10.1016/0003-2697(80)90118-9. Epub 1980/07/15. PubMed PMID: 7416469. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-Navarro J.A., Gonzalo-Gobernado R., Herranz A.S., Gonźlez-Vigueras J.M., Solís J.M. High potassium induces taurine release by osmosensitive and osmoresistant mechanisms in the rat hippocampus in vivo. J. Neurosci. Res. 2009;87(1):208–217. doi: 10.1002/jnr.21818. [DOI] [PubMed] [Google Scholar]
- 44.Solís O., García-Sanz P., Herranz A.S., Asensio M.J., Moratalla R. L-DOPA reverses the increased free amino acids tissue levels induced by dopamine depletion and rises GABA and tyrosine in the Striatum. Neurotox. Res. 2016;30(1):67–75. doi: 10.1007/s12640-016-9612-x. Epub 2016/03/12. PubMed PMID: 26966009. [DOI] [PubMed] [Google Scholar]
- 45.Akutsu M., Dikic I., Bremm A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016;129(5):875–880. doi: 10.1242/jcs.183954. Epub 2016/02/26. PubMed PMID: 26906419. [DOI] [PubMed] [Google Scholar]
- 46.Mansergh F.C., Hunter S.M., Geatrell J.C., Jarrin M., Powell K., Evans M.J., Wride M.A. Developmentally regulated expression of hemoglobin subunits in avascular tissues. Int. J. Dev. Biol. 2008;52(7):873–886. doi: 10.1387/ijdb.082597fm. Epub 2008/10/29. PubMed PMID: 18956317. [DOI] [PubMed] [Google Scholar]
- 47.Whitson J.A., Wilmarth P.A., Klimek J., Monnier V.M., David L., Fan X. Proteomic analysis of the glutathione-deficient LEGSKO mouse lens reveals activation of EMT signaling, loss of lens specific markers, and changes in stress response proteins. Free Radic. Biol. Med. 2017;113:84–96. doi: 10.1016/j.freeradbiomed.2017.09.019. Epub 2017/09/28. PubMed PMID: 28951044; PMCID: Pmc5699945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan S.Y., Ali M., Kabir F., Renuse S., Na C.H., Talbot C.C., Jr., Hackett S.F., Riazuddin S.A. Proteome profiling of developing murine lens through mass spectrometry. Invest. Ophthalmol. Vis. Sci. 2018;59(1):100–107. doi: 10.1167/iovs.17-21601. Epub 2018/01/15. PubMed PMID: 29332127; PMCID: Pmc5769801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kageyama S., Hanada E., Ii H., Tomita K., Yoshiki T., Kawauchi A. Gamma-glutamylcyclotransferase: a novel target molecule for cancer diagnosis and treatment. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/345219. Epub 2015/09/05. PubMed PMID: 26339607; PMCID: Pmc4538363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J., Zhou J., Wu J., Zhang G., Kang L., Ben J., Wang Y., Qin B., Guan H. Aberrant epigenetic alterations of glutathione-S-transferase P1 in age-related nuclear cataract. Curr. Eye Res. 2017;42(3):402–410. doi: 10.1080/02713683.2016.1185129. Epub 2016/06/28. PubMed PMID: 27348130. [DOI] [PubMed] [Google Scholar]
- 51.Wu A., Zhang W., Zhang G., Ding X., Kang L., Zhou T., Ji M., Guan H. Age-related cataract: GSTP1 ubiquitination and degradation by Parkin inhibits its anti-apoptosis in lens epithelial cells. Biochim. Biophys. Acta Mol. Cell Res. 2023;1870(4) doi: 10.1016/j.bbamcr.2023.119450. Epub 2023/03/06. PubMed PMID: 36871745. [DOI] [PubMed] [Google Scholar]
- 52.Ma T., Chen T., Li P., Ye Z., Zhai W., Jia L., Chen W., Sun A., Huang Y., Wei S., Li Z. Heme oxygenase-1 (HO-1) protects human lens epithelial cells (SRA01/04) against hydrogen peroxide (H2O2)-induced oxidative stress and apoptosis. Exp. Eye Res. 2016;146:318–329. doi: 10.1016/j.exer.2016.02.013. Epub 2016/03/20. PubMed PMID: 26992777. [DOI] [PubMed] [Google Scholar]
- 53.Liao S., Huang M., Liao Y., Yuan C. vol. 48. 2023. pp. 25–33. (HMOX1 Promotes Ferroptosis Induced by Erastin in Lens Epithelial Cell through Modulates Fe(2+) Production). 1. PubMed PMID: 36300537. [DOI] [PubMed] [Google Scholar]
- 54.Kakrana A., Yang A., Anand D., Djordjevic D., Ramachandruni D., Singh A., Huang H., Ho J.W.K., Lachke S.A. iSyTE 2.0: a database for expression-based gene discovery in the eye. Nucleic Acids Res. 2018;46(D1):D875–D885. doi: 10.1093/nar/gkx837. Epub 2017/10/17. PubMed PMID: 29036527; PMCID: PMC5753381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heller-Stilb B., van Roeyen C., Rascher K., Hartwig H.G., Huth A., Seeliger M.W., Warskulat U., Haussinger D. Disruption of the taurine transporter gene (taut) leads to retinal degeneration in mice. Faseb. J. : Off. Publ. Feder. Am. Soc. Exp. Biol. 2002;16(2):231–233. doi: 10.1096/fj.01-0691fje. Epub 2002/01/05. PubMed PMID: 11772953. [DOI] [PubMed] [Google Scholar]
- 56.Wegener A., Golubnitschaja O., Breipohl W., Schild H.H., Vrensen G.F. Effects of dietary deficiency of selective amino acids on the function of the cornea and lens in rats. Amino Acids. 2002;23(1–3):337–342. doi: 10.1007/s00726-001-0147-x. Epub 2002/10/10. PubMed PMID: 12373556. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Grenell A., Zhong F., Yam M., Hauer A., Gregor E., Zhu S., Lohner D., Zhu J., Du J. Metabolic signature of the aging eye in mice. Neurobiol. Aging. 2018;71:223–233. doi: 10.1016/j.neurobiolaging.2018.07.024. Epub 2018/09/02. PubMed PMID: 30172221; PMCID: Pmc6162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanshole V.V., Snytnikova O.A., Kiryutin A.S., Yanshole L.V., Sagdeev R.Z., Tsentalovich Y.P. Metabolomics of the rat lens: a combined LC-MS and NMR study. Exp. Eye Res. 2014;125:71–78. doi: 10.1016/j.exer.2014.05.016. Epub 2014/06/10. PubMed PMID: 24910091. [DOI] [PubMed] [Google Scholar]
- 59.Remø S.C., Hevrøy E.M., Breck O., Olsvik P.A., Waagbø R. Lens metabolomic profiling as a tool to understand cataractogenesis in Atlantic salmon and rainbow trout reared at optimum and high temperature. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175491. Epub 2017/04/19. PubMed PMID: 28419112; PMCID: PMC5395160 our adherence to PLOS ONE policies on sharing data and materials. We have made all our data available for publishing and there are no restrictions on sharing of data or competing interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chauhan B.S., Desai N.C., Bhatnagar R., Garg S.P. Study of the relationship between free amino acids and cataract in human lenses. Exp. Eye Res. 1984;38(2):177–179. doi: 10.1016/0014-4835(84)90101-5. Epub 1984/02/01. PubMed PMID: 6143681. [DOI] [PubMed] [Google Scholar]
- 61.Ohrloff C., Stoffel C., Koch H.R., Wefers U., Bours J., Hockwin O. Experimental cataracts in rats due to tryptophan-free diet. Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie Albrecht von Graefe's Arch. Clin. Exp. Ophthalmol. 1978;205(2):73–79. doi: 10.1007/bf00410102. Epub 1978/01/31. PubMed PMID: 305215. [DOI] [PubMed] [Google Scholar]
- 62.Hall W.K., Bowles L.L., et al. Cataracts due to deficiencies of phenylalanine and histidine in the rat; a comparison with other types of cataracts. J. Nutr. 1948;36(2):277–295. doi: 10.1093/jn/36.2.277. Epub 1948/08/01. PubMed PMID: 18876490. [DOI] [PubMed] [Google Scholar]
- 63.Waagbø R., Trösse C., Koppe W., Fontanillas R., Breck O. Dietary histidine supplementation prevents cataract development in adult Atlantic salmon, Salmo salar L., in seawater. Br. J. Nutr. 2010;104(10):1460–1470. doi: 10.1017/s0007114510002485. Epub 2010/08/10. PubMed PMID: 20691125. [DOI] [PubMed] [Google Scholar]
- 64.Sulochana K.N., Punitham R., Ramakrishnan S. Beneficial effect of lysine and amino acids on cataractogenesis in experimental diabetes through possible antiglycation of lens proteins. Exp. Eye Res. 1998;67(5):597–601. doi: 10.1006/exer.1998.0547. Epub 1999/01/08. PubMed PMID: 9878222. [DOI] [PubMed] [Google Scholar]
- 65.Bunce G.E., Caasi P., Hall B., Chaves N. Prevention of cataract in the progeny of rats fed a maternal diet based on vegetable proteins. PSEBM (Proc. Soc. Exp. Biol. Med.) 1972;140(3):1103–1107. doi: 10.3181/00379727-140-36620. [DOI] [PubMed] [Google Scholar]
- 66.Nye-Wood M.G., Spraggins J.M., Caprioli R.M., Schey K.L., Donaldson P.J., Grey A.C. Spatial distributions of glutathione and its endogenous conjugates in normal bovine lens and a model of lens aging. Exp. Eye Res. 2017;154:70–78. doi: 10.1016/j.exer.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamara S.O., Yanshole L.V., Yanshole V.V., Fursova A.Z., Stepakov D.A., Novoselov V.P., Tsentalovich Y.P. Spatial distribution of metabolites in the human lens. Exp. Eye Res. 2016;143:68–74. doi: 10.1016/j.exer.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 68.Lim J.C., Grey A.C., Zahraei A., Donaldson P.J. Age-dependent changes in glutathione metabolism pathways in the lens: new insights into therapeutic strategies to prevent cataract formation—a review. Clin. Exp. Ophthalmol. 2020;48(8):1031–1042. doi: 10.1111/ceo.13801. [DOI] [PubMed] [Google Scholar]
- 69.Zhou Y., Bennett T.M., Shiels A. Lens ER-stress response during cataract development in Mip-mutant mice. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2016;1862(8):1433–1442. doi: 10.1016/j.bbadis.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oltion K., Carelli J.D., Yang T., See S.K., Wang H.Y., Kampmann M., Taunton J. An E3 ligase network engages GCN1 to promote the degradation of translation factors on stalled ribosomes. Cell. 2023;186(2):346–362. doi: 10.1016/j.cell.2022.12.025. e17. Epub 2023/01/14. PubMed PMID: 36638793; PMCID: PMC9994462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morris J.R., Solomon E. BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 2004;13(8):807–817. doi: 10.1093/hmg/ddh095. Epub 2004/02/21. PubMed PMID: 14976165. [DOI] [PubMed] [Google Scholar]
- 72.Nishikawa H., Ooka S., Sato K., Arima K., Okamoto J., Klevit R.E., Fukuda M., Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 2004;279(6):3916–3924. doi: 10.1074/jbc.M308540200. Epub 2003/11/26. PubMed PMID: 14638690. [DOI] [PubMed] [Google Scholar]
- 73.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., Harper J.W., Gygi S.P. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. Epub 2011/09/13. PubMed PMID: 21906983; PMCID: Pmc3200427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M., Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics : MCP. 2011;10(10) doi: 10.1074/mcp.M111.013284. M111.013284. Epub 2011/09/06. PubMed PMID: 21890473; PMCID: Pmc3205876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ordureau A., Heo J.M., Duda D.M., Paulo J.A., Olszewski J.L., Yanishevski D., Rinehart J., Schulman B.A., Harper J.W. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl. Acad. Sci. U.S.A. 2015;112(21):6637–6642. doi: 10.1073/pnas.1506593112. Epub 2015/05/15. PubMed PMID: 25969509; PMCID: Pmc4450373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Durcan T.M., Tang M.Y., Pérusse J.R., Dashti E.A., Aguileta M.A., McLelland G.L., Gros P., Shaler T.A., Faubert D., Coulombe B., Fon E.A. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. The EMBO journal. 2014;33(21):2473–2491. doi: 10.15252/embj.201489729. Epub 2014/09/14. PubMed PMID: 25216678; PMCID: Pmc4283406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cunningham C.N., Baughman J.M., Phu L., Tea J.S., Yu C., Coons M., Kirkpatrick D.S., Bingol B., Corn J.E. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 2015;17(2):160–169. doi: 10.1038/ncb3097. Epub 2015/01/27. PubMed PMID: 25621951. [DOI] [PubMed] [Google Scholar]
- 78.Shang F., Deng G., Liu Q., Guo W., Haas A.L., Crosas B., Finley D., Taylor A. Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. J. Biol. Chem. 2005;280(21):20365–20374. doi: 10.1074/jbc.M414356200. Epub 2005/03/26. PubMed PMID: 15790562; PMCID: PMC1382285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang C., Zhu Y., Zhu X., Chen R., Zhang X., Lian N. USP7 regulates HMOX-1 via deubiquitination to suppress ferroptosis and ameliorate spinal cord injury in rats. Neurochem. Int. 2023;168 doi: 10.1016/j.neuint.2023.105554. Epub 2023/06/01. PubMed PMID: 37257587. [DOI] [PubMed] [Google Scholar]
- 80.Dominy J.E., Jr., Hirschberger L.L., Coloso R.M., Stipanuk M.H. Regulation of cysteine dioxygenase degradation is mediated by intracellular cysteine levels and the ubiquitin-26 S proteasome system in the living rat. Biochem. J. 2006;394(Pt 1):267–273. doi: 10.1042/BJ20051510. Epub 2005/11/03. PubMed PMID: 16262602; PMCID: PMC1386025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request. Proteomic data available at ProteomeXchange with the dataset identifier PXD045040.