To the editor:
Innovation in mass spectrometry (MS) and the rapidly increasing throughput and sensitivity of MS instrumentation require adaptations and innovations in data processing tools. Here, we introduce MZmine 3, a scalable MS data analysis platform that supports hybrid datasets from various instrumental setups, including gas and liquid chromatography (GC and LC)-MS, ion mobility spectrometry (IMS)-MS, and MS imaging. In particular, the integration of IMS-MS imaging and LC-IMS-MS datasets provides opportunities for spatial metabolomics analyses with increased annotation confidence.
Over the past decade, the MZmine project has evolved into a community-driven, collaborative effort. As an open-source ecosystem for MS data processing, MZmine is a cross-platform software (Supplementary Note 1) that can be tuned for robust, scalable, and reproducible data analysis on personal computers as well as high-performance super computers. The project has seen continuous development since its inception in 2004.1,2 Community additions (Fig. 1a) introduced various functions, such as performant feature detection workflows,3,4 modules for lipid annotation,5 and strong ties to other community projects (Fig. 1b). Here, data exchange formats and direct interfaces (listed in Tool integration in the documentation) enable downstream analysis in external tools, such as compound annotation in SIRIUS,6 statistical analysis in MetaboAnalyst,7 and directly bind MZmine results into the molecular networking ecosystem of the Global Natural Products Social Molecular Networking (GNPS) web-platform (Supplementary Note 2).8–10
Fig. 1 |. MZmine, an open-source community project for integrative LC-IMS-MS and IMS-MS data processing.
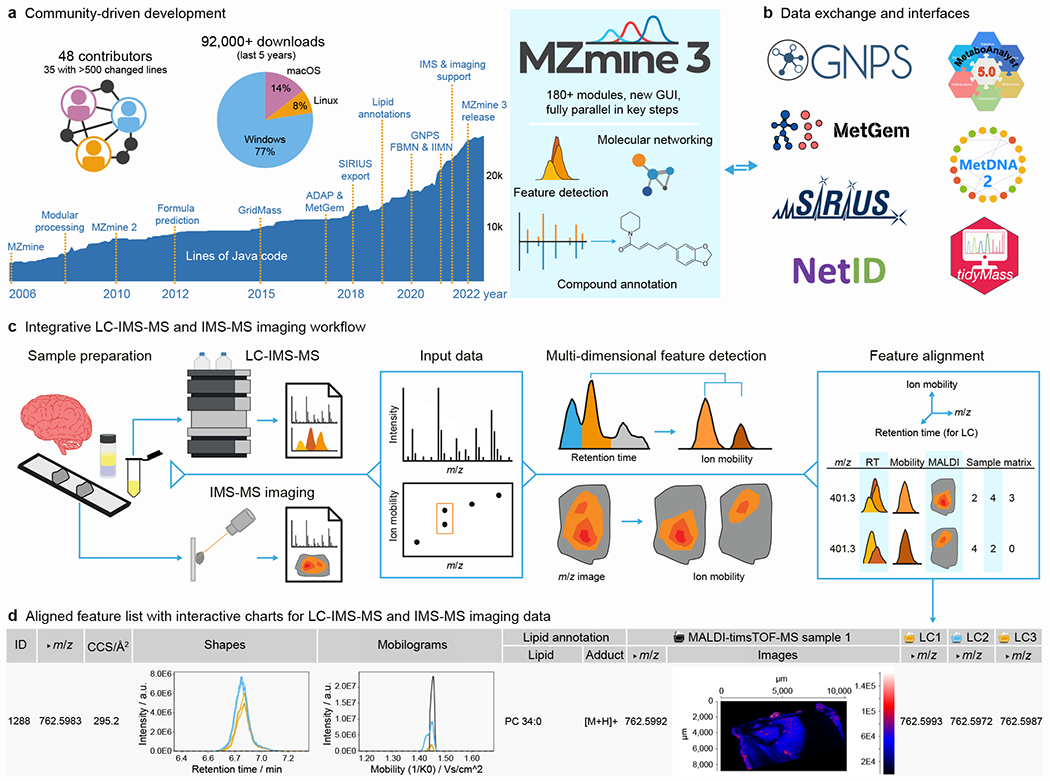
a, Overview of active developments and key additions to MZmine since the first publication, which led to over 180 modules that now drive interactive, reproducible, and efficient data processing and visualization in MZmine 3. b, Data exchange formats and direct interfaces enable downstream analysis with strong ties to projects like GNPS, SIRIUS, and MetaboAnalyst. c, The integrative LC- and IMS-MS imaging workflow applies feature detection in RT, ion mobility, and m/z dimension to MS data stored in open or vendor formats. Comprehensive processing and annotation results are merged into d, an aligned feature list with one ion feature detected in LC-IMS-MS samples and aligned to one MALDI-IMS-MS ion feature image. Annotation results (Lipid annotation column) and interactive charts include the table columns Shapes (extracted ion chromatograms), Mobilograms (extracted ion mobilograms), and Images (extracted ion images).
Recent advances in MS instrumentation push sensitivity, resolving power, and data acquisition speed, resulting in increased data volume and complexity. Notably, IMS gains traction in the field by including an additional separation dimension to LC-MS or imaging-based techniques like matrix-assisted laser desorption/ionization (MALDI)-MS. These advances introduce new acquisition modes (e.g., parallel accumulation-serial fragmentation - PASEF)11, or enable hyphenation of IMS and imaging, which was shown to improve annotation quality in MS imaging.12 Furthermore, the number of large-scale cohort and multifactorial studies in clinical, environmental, and other fields is growing, as registered in the three major metabolomics data repositories, MassIVE/GNPS,8 MetaboLights, and Metabolomics Workbench.13 The need for scalable, reproducible, and flexible data analysis workflows that can combine mass spectrometry data from various sources, remains unaddressed by existing tools. For example, to combine LC- and imaging-(IMS)-MS results from the same sample, users are forced to master multiple software tools12 that divide the workflow and are specialized in either chromatography-MS (e.g., MS-DIAL, XCMS, OpenMS)14–16 or MS imaging (e.g., METASPACE, rMSI, Cardinal MSI, SpectralAnalysis).17
The integrative spatial metabolomics workflow in MZmine 3 (Fig. 1c) imports LC-IMS-MS and IMS-MS imaging datasets stored in either open or vendor-specific formats and processes them by non-targeted feature detection. This entails resolving peak shapes for ion features in both the retention time (RT) and ion mobility dimension in LC-IMS-MS and extracting mobility-resolved ion image features with spatial distributions in IMS-MS imaging (Supplementary Fig. 1). Individual features from both methodologies are subsequently represented and aligned by their RT (LC only), m/z, and ion mobility values. The resulting aligned feature list combines the strengths of the individual analytical methods by integrating the compound annotation capabilities of modern chromatography-based MS with spatial metabolite distributions that can be mapped to histological data, addressing the issue of missing MS2 data in most imaging studies. For data evaluation, MZmine organizes annotations in a feature table with interactive charts, exemplified in Fig. 1d for one ion feature detected in LC-IMS-MS samples and aligned to an ion image from one MALDI-IMS-MS imaging dataset. An exemplary spatial metabolomics workflow leading to LC-IMS-MS resolved molecular networks, enriched with spatial ion feature information is described in Supplementary Note 2 (Supplementary Fig. 4). Additional visualization modules (Supplementary Fig. 5) connect all available data dimensions; a fast memory-mapped data backend enables interactive exploration.
In MZmine 3, special attention was directed towards scalability due to the ever-increasing study sizes that lead to large raw data volumes, particularly in the case of LC-IMS-MS datasets. Efficient memory management and parallelization removed bottlenecks, resulting in an 89% reduction in processing time for 250 dissolved organic matter (DOM) samples when compared to MZmine 2. A stress test demonstrated in high sample throughput, where the mean processing times elapsed to 0.1% to 0.3% of the total data acquisition time for six different LC-MS datasets (Supplementary Note 3; Supplementary Fig. 6). Further, MZmine 3 was benchmarked using 8273 fecal LC-MS2 samples, requiring just 47 min of processing time (see hardware specifications in Supplementary Note 3).
The improved performance of MZmine 3 over previous MZmine versions now allows processing of large datasets, including large-volume LC-IMS-MS data. For new users, the MZmine website contains detailed manuals and video tutorials, and the new processing wizard in MZmine provides starting points for various standard workflows and mass spectrometer types. In addition, a development tutorial is available for potential new contributors, and the modular design of MZmine enables testing and implementing new ideas within the MZmine framework.
Supplementary Material
Acknowledgments
We thank Christopher Jensen and Gauthier Boaglio for their contributions to the MZmine codebase. We thank Jianbo Zhang and Zachary Russ for their donations to MZmine development. The MZmine 3 logo was designed by the Bioinformatics & Research Computing group at the Whitehead Institute for Biomedical Research. T.P. is supported by the Czech Science Foundation (GA CR) grant 21-11563M and by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement 891397. P.C.D. support was from NIH U19 AG063744, P50HD106463, 1U24DK133658 and BBSRC-NSF award 2152526. T.S. acknowledges funding by Deutsche Forschungsgemeinschaft (441958208). Mi.W. acknowledges the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337; a DOE Office of Science User Facility) and is supported by the Office of Science of the U.S. Department of Energy operated under Subcontract NO. 7601660. E.R. and H.H. thank Wen Jiang (HILICON AB) for providing the iHILIC Fusion(+) column for HILIC measurements. M.F., K.D. and S.B. are supported by Deutsche Forschungsgemeinschaft (BO 1910/20). L.-F.N. is supported by the Swiss National Science Foundation (Project 189921). D.P. was supported through the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through the CMFI Cluster of Excellence (EXC-2124 — 390838134 project-ID 1-03.006_0) and the Collaborative Research Center CellMap (TRR 261 - 398967434). J.K.W. acknowledges the U.S. National Science Foundation (MCB-1818132), U.S. Department of Agriculture, and the Chan Zuckerberg Initiative. MZmine developers have received support from the European COST Action CA19105 — Pan-European Network in Lipidomics and EpiLipidomics (EpiLipidNET). We acknowledge the support of the Google Summer of Code (GSoC) program, which has funded the development of several MZmine modules through student projects. We thank Adam Tenderholt for introducing MZmine to the GSoC program.
Footnotes
Code availability
The latest release of MZmine can be downloaded from www.mzmine.org. The complete source codes are available at https://github.com/mzmine/mzmine3/ under the MIT license.18 The MZmine documentation is hosted on GitHub and available at https://mzmine.github.io/mzmine_documentation/.
Competing interests
A.K. is employed at Bruker Daltonics GmbH & Co. KG. S.B., K.D. and M.F. are co-founders of Bright Giant. P.C.D. is a scientific advisor for Cybele and is a scientific advisor and a co-founder of Enveda, Arome and Ometa with prior approval by UC-San Diego. Mi.W. is a co-founder of Ometa Labs LLC. J.K.W. is a member of the Scientific Advisory Board and a shareholder of DoubleRainbow Biosciences, Galixir and Inari Agriculture, which develop biotechnologies related to natural products, drug discovery and agriculture.
Data availability
Datasets are available on MassIVE8 with their accession IDs:
MSV000088054, human cohort study, LC-MS, neg
MSV000087728, diverse plant extracts, LC-MS2, top-3 DDA, pos
MSV000090079, DOM, LC-MS2, top-5 DDA, pos
MSV000090328, sheep brain, LC-tims-MS, PASEF, pos
MSV000090327, piper plant extracts, LC-tims-MS, PASEF, pos
IMS resolved ion identity molecular networking results are available through GNPS: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=7a06fa3dfadd4158bcb4ee300b574747
References
- 1.Katajamaa M, Miettinen J & Oresic M Bioinformatics 22, 634–636 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Pluskal T, Castillo S, Villar-Briones A & Oresic M BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smirnov A. et al. Anal. Chem 91, 9069–9077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du X, Smirnov A, Pluskal T, Jia W & Sumner S Methods Mol. Biol 2104, 25–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korf A, Jeck V, Schmid R, Helmer PO & Hayen H Anal. Chem 91, 5098–5105 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Dührkop K. et al. Nat. Methods 16, 299–302 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Pang Z. et al. Nucleic Acids Res. 49, W388–W396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M et al. Nat. Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nothias L-F et al. Nat. Methods 17, 905–908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmid R et al. Nat. Commun 12, 3832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meier F et al. J. Proteome Res 14, 5378–5387 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Helmer PO et al. Anal. Chem 93, 2135–2143 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Aksenov AA, da Silva R, Knight R, Lopes NP & Dorrestein PC Nature Reviews Chemistry 1, 1–20 (2017). [Google Scholar]
- 14.Smith CA, Want EJ, O’Maille G, Abagyan R & Siuzdak G Anal. Chem 78, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Tsugawa H et al. Nat. Biotechnol 38, 1159–1163 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Röst HL et al. Nat. Methods 13, 741–748 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Weiskirchen R, Weiskirchen S, Kim P & Winkler RJ Cheminform. 11, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. https://github.com/mzmine/mzmine3/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets are available on MassIVE8 with their accession IDs:
MSV000088054, human cohort study, LC-MS, neg
MSV000087728, diverse plant extracts, LC-MS2, top-3 DDA, pos
MSV000090079, DOM, LC-MS2, top-5 DDA, pos
MSV000090328, sheep brain, LC-tims-MS, PASEF, pos
MSV000090327, piper plant extracts, LC-tims-MS, PASEF, pos
IMS resolved ion identity molecular networking results are available through GNPS: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=7a06fa3dfadd4158bcb4ee300b574747


