Abstract
Introduction
The purpose of this feasibility study was to select targeted therapies according to “ESMO Scale for Clinical Actionability of molecular Targets (ESCAT)”. Data interpretation was further supported by a browser-based Treatment Decision Support platform (MH Guide, Molecular Health, Heidelberg, Germany).
Patients
We applied next generation sequencing based whole exome sequencing of tumor tissue and peripheral blood of patients with metastatic breast cancer (n = 44) to detect somatic as well as germline mutations.
Results
In 32 metastatic breast cancer patients, data interpretation was feasible. We identified 25 genomic alterations with ESCAT Level of Evidence I or II in 18/32 metastatic breast cancer patients, which were available for evaluation: three copy number gains in HER2 , two g BRCA1 , two g BRCA2 , six PIK3CA, one ESR1 , three PTEN , one AKT1 and two HER2 mutations. In addition, five samples displayed Microsatellite instability high-H.
Conclusions
Resulting treatment options were discussed in a tumor board and could be recommended in a small but relevant proportion of patients with metastatic breast cancer (7/18). Thus, this study is a valuable preliminary work for the establishment of a molecular tumor board within the German initiative “Center for Personalized Medicine” which aims to shorten time for analyses and optimize selection of targeted therapies.
Keywords: breast cancer, targeted therapy, whole exome sequencing, decision making platform
Zusammenfassung
Einleitung
Ziel dieser Machbarkeitsstudie war es, zielgerichtete Therapien entsprechend der ESCAT-Skala (ESMO Scale for Clinical Actionability of molecular Targets) zu bestimmen. Für die Interpretation der Daten wurde eine browserbasierte Plattform zur Entscheidungsfindung (MH Guide, Molecular Health, Heidelberg, Germany) eingesetzt.
Patientinnen
Es wurde eine Exomsequenzierung von Tumorgewebe und peripherem Blut von Patientinnen mit metastasiertem Mammakarzinom (n = 44) durchgeführt, um somatische sowie Keimbahnmutationen zu identifizieren.
Ergebnisse
Bei 32 Patientinnen mit metastasiertem Mammakarzinom konnte eine Dateninterpretation durchgeführt werden. Es wurden 25 genomische Veränderungen (ESCAT-Evidenzstufe I oder II) bei 18/32 Patientinnen mit metastasiertem Mammakarzinom identifiziert und abschließend ausgewertet: darunter fanden sich 3 Fälle mit höheren Vervielfältigungszahlen bei HER2 , 2 g BRCA1- , 2 g BRCA2- , 6 PIK3CA-, 1 ESR1- , 3 PTEN- , 1 AKT1- und 2 HER2 -Mutationen. Dazu kamen noch 5 Proben, die hochgradige Mikrosatelliteninstabilität aufwiesen.
Schlussfolgerung
Die daraus abzuleitenden Behandlungsoptionen wurden in einer Tumorkonferenz diskutiert und dann einer kleinen, aber relevanten Anzahl von Patientinnen mit metastasiertem Mammakarzinom (7/18) empfohlen. Die hier vorgestellte Arbeit stellt eine wertvolle Vorstudie dar, die dazu beitragen kann, molekulare Tumorboards innerhalb des Deutschen Netzwerks für Personalisierte Medizin zu etablieren. Ziel ist, die für Analysen benötigte Zeit zu verkürzen und die Wahl zielgerichteter Therapien zu optimieren.
Schlüsselwörter: Mammakarzinom, zielgerichtete Therapie, Ganz-Exom-Sequenzierung, Entscheidungsplattform
Abbreviations
- BRCA
BReast CAncer-Gene
- CNG
Copy number gain
- CNL
Copy number loss
- CNV
Copy number variation
- DPD
Dihydropyrimidine dehydrogenase
- ESCAT
ESMO Scale for Clinical Actionability of molecular Targets
- ER
Estrogen receptor
- ESMO
European Society for Medical Oncology
- GA
Genomic alteration
- g
Germline
- HER2
Human epidermal growth factor receptor 2
- HR
Hormone receptor
- LoE
Level of evidence
- MBC
Metastatic breast cancer
- MSI
Microsatellite instability
- PI3K
Phosphoinositide-3-kinase
- PARP
Poly-ADP-Ribose-Polymerase
- PT
Primary tumor
- Rb1
Retinoblastoma 1
- TNBC
Triple negative breast cancer
- WES
Whole-exome sequencing
Introduction
Metastatic breast cancer (MBC) is still an incurable disease 1 . Routine therapy options are limited or accompanied by relevant side effects. Therefore, targeted therapy to optimize treatment adherence and outcome has emerged as the preferred approach in past years.
Alongside the decoding of the human genome “precision medicine” moved into the focus and became a reachable goal. The molecular genetic profile of a tumor, a metastatic lesion or even germline status of a patient can give a valuable information in this context 2 . Whole exome sequencing (WES) by identifying genomic alterations (GAs) can give crucial insights into the cellular features which cause and drive cancer development and progression, respectively, and may inform about potential therapeutic targets 3 . GAs detected by next generation sequencing (NGS) include mutations (single nucleotide variants such as missense, nonsense, splice-site mutations) as well as copy number variations (CNV). Identifying cancer-causing and driving genes activated due to somatic CNV partly also led to specific therapeutic approaches 3 .
The first targeted therapy for BReast CAncer -Gene ( BRCA )-mutated ovarian cancer was approved in 2014 being the Poly-ADP-Ribose-Polymerase (PARP)-inhibitor olaparib 4 . Meanwhile, further PARP-inhibitors were developed to treat ovarian cancer (niraparib 5 and rucaparib 6 ) or approval was gained for g BRCA -mutated breast cancer (olaparib and talazoparib) 7 8 , as well as for BRCA -mutated metastatic castration-resistant prostate 9 and pancreatic 10 cancer (olaparib). Therefore, in individual cases targeted therapies approved in other tumor types may also be an option for off-label use. Other GAs provide hints for ineffectiveness or toxicity of certain therapies. There is for example good evidence for mutations in retinoblastoma 1 (Rb1) causing resistance of CDK4/6 inhibitors in MBC patients 11 12 . Moreover since 2020 it is recommended to test patients for lack of dihydropyrimidine dehydrogenase (DPD) before starting a therapy with fluorouracil or related medicines such as capecitabine 13 . The enzyme DPD is needed to break down fluorouracil, whose enrichment in the blood can lead to severe and life-threatening side effects 13 .
However, obtaining information of molecular genetic properties to choose a targeted therapy is still technically challenging and clinical variant interpretation is not trivial. Further the actual challenge of personalized medicine is to combine the clinical information of a specific patient and the molecular properties of his/her tumor with the existing biomedical knowledge to offer a personalized treatment.
To get a comprehensive picture of a patient’s GAs we present a feasibility study that explores both the somatic (tumor/metastatic tissues) and germline (white blood cells) mutational status. Therefore, we performed WES of patients with MBC and recommended individualized targeted therapy accordingly.
Patients, Materials and Methods
Patients
We identified patients with MBC in our weekly tumor conference. From 2017 to 2020 44 patients with MBC signed written informed consent to participate in our feasibility study “Fighting therapy resistance in patients with solid tumors” (approved by the Ethics Committee of the Medical Faculty of the Heinrich Heine University Düsseldorf; Ref-No: 5673). Inclusion criteria were: metastatic patient with solid tumor; no standard therapy option; Karnowsky-Index > 70%; expected life span > 6 months. Accordingly, our patient collective was heavily pretreated with several lines of chemo- and/ or endocrine therapies.
Hormone receptor (HR) status and Human epidermal growth factor receptor 2 (HER2) amplification was determined according to German clinical routine (immunohistochemistry and/or in situ hybridization analysis). 26/44 (59%) had a HR+ (HER2 non amplified), 7/44 (16%) a HER2 positive (HER2+; HR+ or HR-) and 10/44 (23%) had a triple negative breast cancer (TNBC). They experienced bone and visceral (n = 16), visceral only (n = 9), cerebral (n = 9), bone only (n = 1) or other metastatic lesions. Clinical patient data are shown in Table S1 .
Due to insufficient DNA quality and quantity or failed sequencing procedure only 32 out of the recruited 44 patients were available for WES and data interpretation ( Fig. S1 ).
Blood and tumor tissue
Peripheral blood was obtained by routine vain puncture (two 10 ml EDTA container). Suitable tumor tissue for analyses was identified according to patients’ history (preferable most recent metastatic lesion). The tissue was obtained in clinical routine diagnostics. From archived FFPE tissues 5 µm thick sections were prepared and slides were analyzed by experienced pathologists to determine tumor content (at least 20%) by staining with hematoxylin/ eosin and immunohistochemistry. We analyzed tumor tissue from breast, liver, lymph node, brain, skin, lung, or bone.
DNA extraction and whole-exome sequencing
DNA was purified from FFPE tissue with the GeneRead DNA FFPE Kit (Qiagen, Venlo, the Netherlands). DNA of matched blood and tumor tissue samples were sequenced using the Agilent Sure Select XT V7 or the Illumina IDT Exome Analysis Kit. The libraries were sequenced on a HiSeq 3000 system (Illumina).
Data Analysis
Anonymized NGS data were transferred via secure VPN channels to a server hosted by Molecular Health GmbH, Heidelberg, Germany. Data were analyzed with the CE-marked in vitro diagnostic software (IVD) MH GUIDE (MH Guide). The MH Guide Variant detection pipeline (MH Guide VDP) uses input sequencing data in (raw) FASTQ format, MH Guide requires for the analysis of paired exomes (WES) of somatic samples a minimum average real coverage of > 200× (the average real coverage is defined as on-target coverage after removal of duplicate read pairs), in which at least 80% of the target region has > 100× average real coverage in the tumor sample. SNVs and Indels that passed the quality filters fulfilled the following quality parameters: PHRED score > 28.5, coverage > 20×, allele frequency > 5%, population frequency < 1%. The raw sequencing data (reads) are aligned through the MH Guide VDP using standard (GRCh37, HG19) or proprietary population specific human reference genomes (MH PHREGs) based on data from the 1000 genomes project for alignment, using LoFreq (PMID: 23066108) for variant calling for SNVs and atlas and freebayes ( https://arxiv.org/abs/1207.3907 ) for Indel calling. The MH Guide VDP provides all detected variants in VCF format for transcript and protein mapping by MH Guide. In paired analyses, all variants detected in the control sample are considered to be germline variants.
For MSI detection MH Guide uses the tool MANTIS (Microsatellite Analysis for Normal-Tumor Instability) to detect MSI biomarkers from FASTQ input [MANTIS tool available at: https://github.com/OSU-SRLab/MANTIS ] 14 . If the stepwise difference is ≥ 0.3, an MSI-H biomarker is automatically added to the variants list. This threshold of 0.3 is based on validation of 40 TCGA (Cancer Genome Atlas) cases from three cancer entities.
Data interpretation
MH Guide system screens all GAs identified against the reference information in the proprietary knowledge platform, Dataome. The core of this platform is a manually curated database with evidence-based biomarker information on peer-reviewed published evidence – the so-called clinical variant interpretations (CVIs).
Information captured during the curation process of the CVIs include I) the variant – i.e. the type of genomic aberration (e.g. SNV, Insertion, Deletion etc.); II) the drug or treatment used in the underlying published peer-reviewed evidence (preclinical studies and clinical trials) for which the data source is mainly PubMed; III) the effect of the variant on treatment – i.e. response, resistance or safety; IV) the quantity of effect – e.g. strong, medium, weak; V) the observation context (i.e. the disease/disease stage or model system); VI) a link to the underlying evidence and a grading of its reliability. Based on the information provided by the MH Guide platform a GA was classified as “effective” (potential target with therapeutic options), “ineffective” (GA with evidence of less effectiveness or ineffectiveness when certain drugs are applied) and “safety” (GA raising concerns about potential toxicity when administering some medication).
Evidence was further underpinned using the European Society for Medical Oncology (ESMO) classification “ ESMO Scale for Clinical Actionability of molecular Targets (ESCAT)” 15 . Clinical decision on therapy options were based on these actual ESCAT evidence tier I and II 16 . In short:
ESCAT evidence tier
I: “alteration-drug match is associated with improved outcome in clinical trials”
II: “alteration-drug match is associated with antitumor activity, but magnitude of benefit is unknown”
III: “alteration-drug match suspected to improve outcome based on clinical trial data in other tumor type(s) or with similar molecular alteration”
IV: “pre-clinical evidence of actionability”
X: “lack of evidence for actionability”
Results
GAs detected by WES in patients with MBC
Out of the total number of 44 screened MBC patients 32 were available for data evaluation: 21 of these harbored hormone receptor positive (HR+) disease, five presented with HER2+ MBC and six with TNBC. In total we detected 481 GAs in 77 different genes. This comprises 253 mutations, 223 CNVs and five samples showed high microsatellite instability (MSI-H). Most alterations were found in the XPC gene (n = 22), followed by the genes FCGR3A (n = 19) and MTHFR (n = 17) as well as CYP2C19 (n = 17), which were exclusively mutations. We detected a minimum of three and a maximum of 32 GAs per patient (median 16.5, mean 15.7).
ESCAT Level of evidence (LoE) I or II in MBC patients
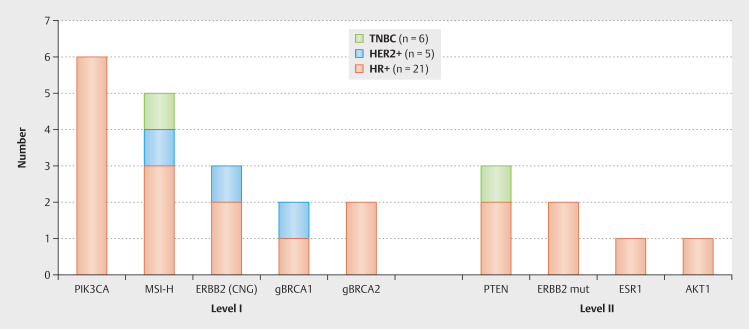
According to ESMO recommendations 25 GAs from 18 patients belong to ESCAT LoE I or II 16 (see Fig. 1 ). Most of the patients (12/18; 67%) had one actionable GA, five had two therapeutic options and one patient had three GAs with two possible therapy recommendations. In Table 1 the detected mutations and the clinical consequences which were taken are listed in detail. The detected alterations of Level IA were: three copy number gains (CNG) in HER2, two g BRCA1 , two g BRCA2 and six PIK3CA mutations. MSI-H as a molecular target of level 1C was identified in five samples. One ESR1 mutation, three PTEN mutations, one AKT1 mutation and two HER2 mutations were grouped in Level II A+B. Fig. 1 shows the detected GAs ESCAT LoE I and II according to tumor subtype.
Fig. 1.
Twenty-five genomic alterations with potential therapeutical consequences (Level of evidence I-II according to ESMO/ESCAT) detected in 18 metastatic breast cancer patients by next generation sequencing based whole exome sequencing. Five patients had two and one patient three different alterations (CNG = copy number gain, mut = mutation).
Table 1 GAs of our MBC patients with LoE I or II according to ESMO/ESCAT and clinical consequences drawn. Table adapted from 14 .
| Alterations | LoE | GA in MBC patient | Variant | g/s | VAF | MBC Subtype | Same patient | Targeted Therapy | Consequences |
| Consequences: 1 = none, known before, 2 = Therapy already taken, 3 = not suitable regarding medical history, 4 = high toxicity expected, 5 = the general health at the time the genetic test result was available did not allow the implementation, # recommended; ♠ ♦ ♥ ♣ • ◊ $ & ~ Ω Θ indicating the same patient (see also Table 2 ). LoE = Level of evidence, CN = copy number, MIS = microsatellite instability scores above MANTIS cutoff threshold 0.3 17 , g = germline, s = somatic, VAF = variant allele frequency. | |||||||||
| ERBB2 amplification | IA | ERBB2 (CNG) | CN: 4 | s | / | HR+ | ♣ | HER2-targeted | 3 |
| ERBB2 (CNG) | CN: 4 | s | / | HR+ | ♥ | 1 and 5 | |||
| ERBB2 (CNG) | CN: 7 | s | / | HER2+ | 1, 2 and 3 | ||||
| Germline BRCA1/2 mutation | IA | BRCA1 p.R1645fs | c.4932_4933dup | g | 47.5 | HR+ | ♥ | Olaparib | 1 and 5 |
| BRCA1 p.Q1756fs | c.5266dup | g | 52.0 | HER2+ | ♦ | 1 | |||
| BRCA2 p.S2835* | c.8504C>G | g | 45.3 | HR+ | ♠ | 1 and 2 | |||
| BRCA2 p.S1271* | c.3812C>G | g | 47.3 | HR+ | 1 and 2 | ||||
| PIK3CA mutation | IA | PIK3CA p.H1047R | c.3140A>G | s | 22.1 | HR+ | $ | Alpelisib | 5 |
| PIK3CA p.H1047R | c.3140A>G | s | 57.5 | HR+ | ♣ | 3 | |||
| PIK3CA p.H1047R | c.3140A>G | s | 39.8 | HR+ | ° | 5 | |||
| PIK3CA p.H1047R | c.3140A>G | s | 47.1 | HR+ | # | ||||
| PIK3CA p.E545K | c.1633G>A | s | 15.1 | HR+ | ∞ | 3 | |||
| PIK3CA p.E545K | c.1633G>A | s | 20.3 | HR+ | Ω | 3 | |||
| Micro-satellite instability (MSI) | IC | MSI-H | MIS: 0,37 | s | / | HR+ | ◊ | Pembro-lizumab | # |
| MSI-H | MIS: 0,35 | s | / | HR+ | • | # | |||
| MSI-H | MIS: 0,48 | s | / | HR+ | 4 | ||||
| MSI-H | MIS: 0,31 | s | / | HER2+ | ♦ | # | |||
| MSI-H | MIS: 0,30 | s | / | TNBC | & | # | |||
| ESR1 mutation | IIA | ESR1 p.Y537C | c.1610A>G | s | 25.5 | HR+ | 5 | ||
| PTEN mutation | IIA | PTEN p.Y225fs | c.673dupT | s | 33.8 | HR+ | ~ | capivasertib plus fulvestrant | 3 |
| PTEN p.T319fs | c.955dupA | s | 43.6 | HR+ | ♠ | 5 | |||
| PTEN p.R130* | c.388C>G | s | 29.5 | TNBC | Θ | # | |||
| AKT1 mutation | IIB | AKT1 p.E17K | c.49G>A | s | 9.0 | HR+ | • | # | |
| ERBB2 mutation | IIB | ERBB2 p.V777L | c.2329G>T | s | 50.0 | HR+ | ◊ | Neratinib | # |
| ERBB2 p.V842I | c.2524G>A | s | 52.0 | HR+ | ◊ | # | |||
Only one out of five patients with HER2+ MBC (HER2 status according to clinical routine assessment) harbored a CNG in HER2, based on NGS CNV analysis. Contrarily, CNG in HER2 , based on NGS CNV analysis, were reported in two of the patients with clinically HR+/HER2− tumor (see Table 1 ). Further, in one patient diagnosed with HR+/HER2− tumors we detected two somatic HER2 mutations ( ERBB2 p.V777L and ERBB2 p.V842I ).
In the HR+/HER2− subgroup one patient was carrier of a gBRCA1 mutation ( BRCA1 p.R1645fs ) and two patients were identified with a gBRCA2 mutation ( BRCA2 p.S2835*, BRCA2 p.S1271* ), respectively. Interestingly the patient with gBRCA1 mutation additionally had a somatic BRCA2 mutation ( BRCA2 p.S3250* , ESCAT LoE IIIA) and one patient with a gBRCA2 mutation ( BRCA2 p.S2835* ) also had an PTEN mutation ( PTEN p.T319fs ) (please refer to Table 1 and Table 2 ). Further, one gBRCA1 mutation ( BRCA1 p.Q1756fs ) was found in a patient with HER2+ MBC. No gBRCA mutations were seen in the subgroup of TNBC patients. All detected gBRCA mutations were also confirmed by routine genetic analysis due to young age or family history according the criteria for genetic counselling in Germany 19 .
Table 2 Genomic alterations of our metastatic breast cancer patients with LoE III and IV according to ESMO/ESCAT. ♥ ∞ * ◊ & ~ Ω Θ indicating the same patient (see also Table 1 ). LoE = level of evidence, g = germline, s = somatic, VAF = variant allele frequency.
| Alterations | LoE | GA in MBC patient | Variant | s/g | VAF | Subtyp | Same patient |
| Somatic BRCA 1/2 mutation | IIIA | BRCA2 p.S3250* | c.9749C>G | s | 30.5 | HR+ | ♥ |
| BRCA2 p.S368* | c.1103C>A | s | 13.8 | HR+ | ∞ | ||
| BRCA2 p.E1879fs | c.5636_5676del | s | 14.6 | HER2+ | * | ||
| ARID1A/B | IVA | ARID1A p.Q1330* | c.3988C>T | s | 30.2 | TNBC | |
| ATM mutation | IVA | ATM p.E2837* | c.8509G>T | s | 6.9 | HR+ | ◊ |
| CDH1 mutation | IVA | CDH1 p.Q23* | c.67C>T | s | 28.4 | HR+ | |
| CDH1 p.G877* | c.2629G>T | s | 5.2 | HR+ | |||
| CDH1 p.S851* | c.2552C>A | s | 30.1 | HR+ | ∞ | ||
| CDH1 p.L466fs | c.1397_1398del | s | 22.3 | HR+ | ∞ | ||
| CDH1 p.P159fs | c.476del | s | 30.7 | HR+ | $ | ||
| MYC | IVA | MYC (CNG) | Copy number 6 | s | / | HR+ | ♣ |
| NF1 mutation | IVA | NF1 p.D1644fs | c.4925_4926ins11 | s | 19.2 | TNBC | & |
| NF1 p.S2687fs | c.8059_8060del | s | 32.8 | HR+ | ~ | ||
| TP53 mutation | IVA | TP53 p.R213* | c.637C>T | s | 39.3 | HR+ | ♥ |
| TP53 p.R248W | c.742C>T | s | 66.2 | HR+ | |||
| TP53 p.E294fs | c.881_885del | s | 57.5 | HR+ | ° | ||
| TP53 p.E285K | c.853G>A | s | 21.6 | HR+ | Ω | ||
| TP53 p.E204* | c.610G>T | s | 30.4 | HER2+ | * | ||
| TP53 p.I232fs | c.694_695ins22 | s | 16.7 | TNBC | & | ||
| TP53 p.Y220C | c.659A>G | s | 37.3 | TNBC | Θ |
All spotted PIK3CA mutations (four times PIK3CA p.H1047R and twice PIK3CA p.E545K ) were seen in the subgroup of HR+ patients (29% of HR+ patients) as listed in Table 1 .
MSI-H tumors were detected in all subgroups (three times HR+, once HER2+ and TNBC respectively). The ESR1 p.Y537C and the AKT1 p.E17K mutation and two of the three PTEN mutations ( p.Y225fs and p.T319fs ) were found in HR+ MBC patients. One PTEN nonsense mutation ( p.R130* ) was detected in a TNBC patient ( Fig. 1 and Table 1 ).
ESCAT LoE III or IV in MBC
We detected 20 GAs grouped in LoE III or IV according to ESCAT. In Level IIIA we detected two somatic BRCA2 mutations ( p.S3250*; p.S368* ) in the subgroup of HR+/HER2− patients and one somatic BRCA2 mutation ( p.E1879fs ) in a HER2+ tumor. In LoE IV we found one ARID1A , one ATM , five CDH1 , two NF1 and seven TP53 mutations, as well as one MYC CNG (for detail see Table 2 ).
CNV in known onco-targets
The following known onco-targets 20 with copy number loss were detected in our MBC patients: RB1 (n = 5) , PTEN (n = 4) , CDKN2B (n = 2) , NF1 (n = 5) , SMAD4 (n = 1) , BRCA1 (n = 9), BRCA2 (n = 4). CNG were identified for HER2 (n = 3, as already described) , EGFR (n = 1) , MYC (n = 1) , PIK3CA (n = 7) , FGFR1 (n = 5), FGFR2 (n = 3) , KRAS (n = 5) , CCND1 (n = 4) , MET (n = 3) , and CDK6 (n = 4).
GAs indicating ineffectiveness or safety concerns according to MH Guide
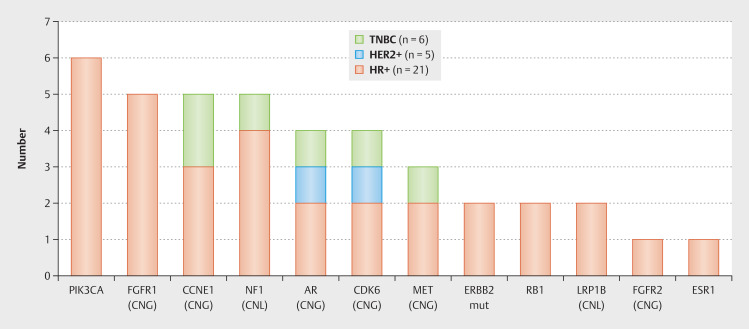
In addition to the actionable targets, we have also identified 92 GAs in 25 different genes that indicate ineffectiveness of a particular drug in patients with MBC (mutations n = 22, MSI-H n = 5, CNV n = 65). From a clinical point of view 11 21 22 the most relevant GAs were: ESR1 (mutation, n = 1; as already described), RB1 (mutation, n = 2), CCNE1 (CNG, n = 5), FGFR1 (CNG, n = 5) and PIK3CA (mutation, n = 6; as stated before). In three patients with a PIK3CA mutation, we saw evidence of endocrine resistance (whereby various endocrine substances were used in multiple lines of therapy). In the other three cases, endocrine therapy was combined with either a CDK4/6 inhibitor or fulvestrant or HER2-targeted therapy when the subtype changed during the course of the disease, and therefore probably showed a good effect of the respective therapy. Other GAs of clinical interest due to possible ineffectiveness 11 23 24 25 26 detected in our collective are: NF1 (CNL, n = 5), AR (CNG, n = 4), CDK6 (CNG, n = 4), MET (CNG, n = 3), HER2 (mutation, n = 2; as specified above), LRP1B (CNL, n = 2) and FGFR2 (CNG, n = 1) (see Fig. 2 ).
Fig. 2.
Genomic alterations with potential ineffectiveness concerning certain therapies according to MH Guide detected by next generation sequencing based whole exome sequencing in our metastatic breast cancer patients (multiple genomic alterations per patient possible; CNG = copy number gain, CNL = copy number loss, mut = mutation).
The RB1 mutations ( RB1p.L199fs and RB1p.E184 ) and three of the CCNE1 CNG were seen in the HR+/HER2− subgroup which may cause failure of CDK4/6 treatment. Further ESR1 mutation and FGFR1 CNG can be associated with endocrine resistance and was also altered exclusively in this subgroup. However, PIK3CA mutations (as mentioned above), which may indicate trastuzumab resistance, were also detected exclusively in the HR+/HER2− subgroup, therefore without clinical consequences in our patient collective ( Fig. 2 shows the GAs with potential ineffectiveness according to MBC subtype).
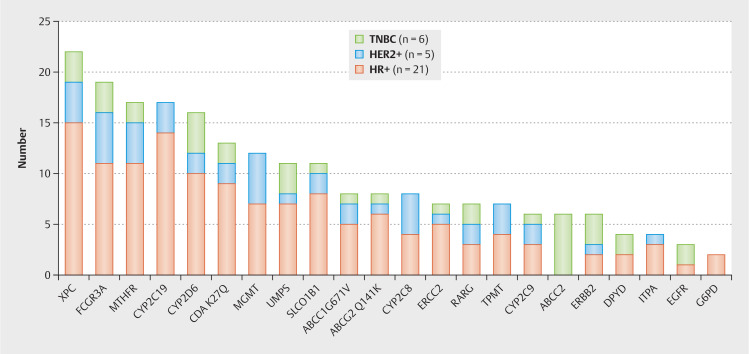
Toxicity concerns were raised for 214 GAs in 22 different genes, with mutations in XPC (n = 22), FCGR3A (n = 19), MTHFR (n = 17), CYP2 C19 (n = 17) and CYP2 D6 (n = 16) being the most altered ones (see Fig. 3 ). A DPD mutation was identified in four patients (two HR+ and two TNBC), which is highly important from a clinical perspective as it increases the likelihood of relevant adverse events during therapy with fluorouracil. Further we detected three EGFR mutations, which may be a driver of tumorigenesis in breast cancer and/or may indicate therapy resistance since these GAs have been found to develop under drug pressure 27 ( Fig. 3 shows the GAs with potential toxicity according to MBC subtype).
Fig. 3.
Genomic alterations with potential toxicity concerning certain therapies according to MH Guide detected by next generation sequencing based whole exome sequencing in our metastatic breast cancer patients (multiple genomic alterations per patient possible).
Discussion
Here we present a feasibility study performing NGS-based WES in patients with MBC. We have established a working procedure for patient recruitment, sample collection, collaboration between different institutions and discussion of the results in a tumor board. Due to the availability of new targeted therapies whose indications are based on GAs, there is an increasing need for genetic analyses at an early timepoint of metastatic/advanced breast cancer. Because of tumor heterogeneity and changes during carcinogenesis or the metastatic process, several analyses during the course of disease may even be useful and necessary (e.g. regarding a somatic PIK3CA mutation) 28 29 . In 2017 the FDA approved two comprehensive mid-size panels for genetic testing in cancer (Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) and FoundationOne CDx) which addressed the unmet need for precision oncology 30 . Nevertheless, it is a question of time, costs, and technical equipment to be able to perform genetic tests and, above all, to make them available for clinical use. For instance the relatively high dropout rate (25%) of our patient population due to sample errors (too little tumor tissue or insufficient DNA quality) is in line with data published by another research group 31 . For clinical decision-making it is further necessary to know the patient’s medical history in detail, which is essential and must be included in the discussion of treatment decisions in a molecular tumor board.
We identified 481 GAs in patients with MBC. These include CNV, mutations and MSI-H. According to the 2019 ESMO criteria for MBC, 25 GAs were classified as ESCAT LoE I–II. In 7/18 patients with MBC (39%), we were able to recommend targeted therapy accordingly 32 with one patient having two options and one patient having two GAs leading to the same therapy recommendation (see Table 1 ). Precisely we suggested four times pembrolizumab if MSI-H was detected, once alpelisib for a PIK3CA mutation, once capivasertib plus fulvestrant if there was a PTEN loss-of-function mutation, once capivasertib plus fulvestrant for an AKT mutation and after reevaluation of the “likely pathogenic” HER2 mutations we could recommend neratinib in one patient diagnosed as HER2- MBC. In six patients, the possible targeted therapy was not suitable due to the individual medical history. This means that concomitant diseases did not allow the administration of the matching drug, or the concomitant medication did not fulfil the approval. Once, the high probability of side effects and the patient’s advanced age spoke against the optional targeted therapy. Unfortunately, the general health of six patients at the time the genetic test result was available did not allow the implementation of potential targeted therapy. In three patients no additional therapeutic recommendations were made after WES, mainly because appropriate targeted therapies were already in use based on findings of routine diagnostic (HER2-targeted therapies or the PARP-inhibitor olaparib). However, this also means that we recognize these findings in clinical routine and do not miss these GAs (please refer to Table 1 for details).
The differences in HER2 status detected by NGS ( HER2 CNG) compared to immunohistochemistry or FISH could be explained by tumor heterogeneity 33 34 or by changes in HER2 status during the course of the disease 35 , which are common phenomena in MBC. In addition, intermetastatic heterogeneity is a described phenomenon that arises under therapy pressure, as different cell clones may respond differently or escape certain therapies. Personalized therapy aims to treat a tumor according to its genetic characteristics, which are homogeneous in the vast majority of cancer cases at diagnosis, rather than applying a standard therapy to all patients with a specific cancer type. 36 . HER2 mutations occur in approximately 2% of patients with breast cancer and could expand treatment options to targeted HER2 therapy in patients with HER2− PT. In our collective, we detected two likely pathogenic HER2 mutations (6%), which is significantly more compared to the current literature 37 . This may be explained mainly due to our small number of patients. Hempel et al. detected an HER2 amplification by NGS in nine out of 41 patients with advanced breast cancer (out of which seven had a HER2+ tumor by immunohistochemistry and/or in situ hybridization) and HER2 mutation in two patients. They conclude that a threshold must be defined on basis of CNV to allow a better interpretation of NGS based amplification analysis. Further, performing NGS analyses they could recommend promising treatment options according ESCAT Level I in 58.5% of their patients ( HER2 mutation n = 9, PIK3CA mutation n = 14, MSI n = 1) 38 .
Consequently, according to our results and those of others, targeted therapy based on molecular findings is appropriate for a significant proportion of patients 39 . In this context, the selection of suitable patients and definition of targets are crucial 31 38 40 41 .
Due to the late stage of disease or irrelevance for the current treatment option none of the inefficacy or safety concerns based on GAs changed our clinical therapy decision. However, in earlier stage of disease the detected GAs may give valuable hints concerning possible ineffectiveness or relevant toxicity of certain therapies. In this context, Reinhardt et al. screened 1270 patients with early breast cancer for the presence of PIK3CA mutations and found a significant influence on the effect of adjuvant aromatase inhibitors but no influence on the efficacy of adjuvant tamoxifen 18 . Similar to ESCAT a validated classification of these GAs is highly needed for clinical routine. According to the experts, due to its high clinical relevance, the ESR1 mutation, for example, although indicating endocrine resistance, was included in ESCAT.
Focus of our feasibility study was to identify possibly druggable molecular targets. Besides, using WES we further detected several CNV which until now have no suitable therapeutic agent or at least there is no sufficient clinical data. Their relevance stems from the fact that the expression level of a gene strongly correlates with its copy number 17 20 . Somatic CNV typically arise during carcinogenesis and, if resulting in the deletion of tumor suppressor genes or the amplification of oncogenes, they are usually pathogenic 20 .
Although we were able to implement the NGS based diagnostic workflow for patients with MBC in our center, several limitations must be considered. Different tumor cell clones harboring different somatic mutations can divide a cancer into several subgroups (heterogeneity). In this context, the prognosis and response to treatment can be very unique for each clone 20 . In addition, the genomic profile of biopsy tissues provides a picture that is limited to only a single point in space and time. This may lead to an under-representation of intratumor heterogeneity 42 , limiting the predictive value of a single tissue biopsy as in our study. Other limitations of our work are, first, the heterogeneous patient population recruited in routine clinical practice without randomization, follow-up, or survival data. Secondly, missing or very late tissue samples of varying quality and highly variable time points in the disease course and thirdly, the long processing time as well as the software for data evaluation, which could overlook GAs.
In line with ESMO recommendations, our data show that large gene panels and routine use of NGS result in few clinically significant responders. Nevertheless, the patient and physician may decide together to perform a large gene panel if the patient is informed that the likelihood of benefit is rather low. According to ESMO, the use of off-label drugs matched to GAs is only recommended when an access program and decision-making process are available 39 .
Consequently, the German initiative “Center for Personalized Medicine” established structured access to a molecular tumor board and interdisciplinary case discussion.
Conclusion
In this feasibility study we demonstrate that WES using NGS for patients with MBC is technically possible and feasible. However, in oncology practice there are no recommendations from scientific societies about its use in daily routine 39 . Due to the low detection rate of truly actionable GAs leading to therapeutic consequences genetic testing is recommended only for a selected patient collective. In this context the molecular tumor board is an essential instrument to choose appropriate patients and discuss the results in an interdisciplinary setting. The identification of drugable molecular alterations among many possible targets and especially among the various alterations of the same target, is crucial. ESCAT provides a useful continuously updated tool for classifying GAs according to their clinical relevance which assists clinicians in delivering accurate and individualized indications for the patients 32 . The browser-based Treatment Decision Support platform MH Guide can further help interpret the abundance of genetic data.
Supplement
Fig. S1 : Study cohort and flowchart (MBC = metastatic breast cancer, WES = whole exome sequencing).
Table S1 : Patient collective.
Acknowledgement
We thank our technicians and the study office for their continuous assistance and dedicated work. Computational support of the “Zentrum für Informations- und Medientechnologie”, especially the HPC team (High Performance Computing) at the Heinrich-Heine University is acknowledged. As this work was funded by the “Förderung Krebsforschung Nordrhein-Westfalen e.V.” ( www.krebsforschung-nrw.de ), we thank them for their financial support. Finally, we would like to express our sincere gratitude to the patients who participated in the study.
Footnotes
Conflict of Interest The authors declare that they have no conflict of interest.
Supporting information
References
- 1.Cardoso F, Senkus E, Costa A et al. 4th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 4) Ann Oncol. 2018;29:1634–1657. doi: 10.1016/j.annonc.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Priestley P, Baber J, Lolkema MP et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beroukhim R, Mermel CH, Porter D et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledermann J, Harter P, Gourley C et al. Olaparib Maintenance Therapy in Patients With Platinum-Sensitive Relapsed Serous Ovarian Cancer. Obstet Gynecol Surv. 2014;69:594–596. [Google Scholar]
- 5.González-Martín A, Pothuri B, Vergote I et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 6.Holloway RW, Gancedo MA, Fong PC et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2018;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson M, Im SA, Senkus E et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 8.Litton JK, Rugo HS, Ettl J et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bono J, Mateo J, Fizazi K et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 10.Golan T, Hammel P, Reni M et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheidemann ER, Shajahan-Haq AN. Resistance to CDK4/6 inhibitors in estrogen receptor-positive breast cancer. Int J Mol Sci. 2021;22:12292. doi: 10.3390/ijms222212292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono M, Oba T, Shibata T et al. The mechanisms involved in the resistance of estrogen receptor-positive breast cancer cells to palbociclib are multiple and change over time. J Cancer Res Clin Oncol. 2021;147:3211–3224. doi: 10.1007/s00432-021-03722-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Medicines Agency . EMA recommendations on DPD testing prior to treatment with fluorouracil, capecitabine, tegafur and flucytosine. Eur Med Agency. 2020;31:3. [Google Scholar]
- 14.Kautto EA, Bonneville R, Miya J et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2017;8:7452–7463. doi: 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateo J, Chakravarty D, Dienstmann R et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2018;29:1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condorelli R, Mosele F, Verret B et al. Genomic alterations in breast cancer: Level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2019;30:365–373. doi: 10.1093/annonc/mdz036. [DOI] [PubMed] [Google Scholar]
- 17.Chang Z, Liu X, Zhao W et al. Identification and Characterization of the Copy Number Dosage-Sensitive Genes in Colorectal Cancer. Mol Ther Methods Clin Dev. 2020;18:501–510. doi: 10.1016/j.omtm.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhardt K, Stückrath K, Hartung C et al. PIK3CA-mutations in breast cancer. Breast Cancer Res Treat. 2022;196:483–493. doi: 10.1007/s10549-022-06637-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kast K, Rhiem K, Wappenschmidt B et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J Med Genet. 2016;53:465–471. doi: 10.1136/jmedgenet-2015-103672. [DOI] [PubMed] [Google Scholar]
- 20.Liu B, Morrison CD, Johnson CS et al. Computational methods for detecting copy number variations in cancer genome using next generation sequencing: Principles and challenges. Oncotarget. 2013;4:1868–1881. doi: 10.18632/oncotarget.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner N, Pearson A, Sharpe R et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majewski IJ, Nuciforo P, Mittempergher L et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015;33:1334–1339. doi: 10.1200/JCO.2014.55.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minuti G, Cappuzzo F, Duchnowska R et al. Increased MET and HGF gene copy numbers are associated with trastuzumab failure in HER2-positive metastatic breast cancer. Br J Cancer. 2012;107:793–799. doi: 10.1038/bjc.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandarlapaty S, Sakr RA, Giri D et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18:6784–6791. doi: 10.1158/1078-0432.CCR-12-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannelli P, Di Donato M, Galasso G et al. The androgen receptor in breast cancer. Front Endocrinol (Lausanne) 2018;9:492. doi: 10.3389/fendo.2018.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serio PAMP, de Lima Pereira GF, Katayama MLH et al. Somatic mutational profile of high-grade serous ovarian carcinoma and triple-negative breast carcinoma in young and elderly patients: Similarities and divergences. Cells. 2021;10:3586. doi: 10.3390/cells10123586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12:3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen JD, Laenkholm A, Knoop A. PIK3CA Mutations May Be Discordant between Primary and Corresponding Metastatic Disease in Breast Cancer. Clin Cancer Res. 2011;17:667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 29.Fusco N, Malapelle U, Fassan M et al. PIK3CA Mutations as a Molecular Target for Hormone Receptor-Positive, HER2-Negative Metastatic Breast Cancer. Front Oncol. 2021;11:644737. doi: 10.3389/fonc.2021.644737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allegretti M, Fabi A, Buglioni S et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J Exp Clin Cancer Res. 2018;37:47. doi: 10.1186/s13046-018-0702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Geelen CT, Savas P, Teo ZL et al. Clinical implications of prospective genomic profiling of metastatic breast cancer patients. Breast Cancer Res. 2020;22:91. doi: 10.1186/s13058-020-01328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crimini E, Repetto M, Aftimos P et al. Precision medicine in breast cancer: From clinical trials to clinical practice. Cancer Treat Rev. 2021;98:102223. doi: 10.1016/j.ctrv.2021.102223. [DOI] [PubMed] [Google Scholar]
- 33.Ohlschlegel C, Zahel K, Kradolfer D et al. HER2 genetic heterogeneity in breast carcinoma. J Clin Pathol. 2011;64:1112–1116. doi: 10.1136/jclinpath-2011-200265. [DOI] [PubMed] [Google Scholar]
- 34.Van Bockstal MR, Agahozo MC, van Marion R et al. Somatic mutations and copy number variations in breast cancers with heterogeneous HER2 amplification. Mol Oncol. 2020;14:671–685. doi: 10.1002/1878-0261.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niikura N, Liu J, Hayashi N et al. Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumors. J Clin Oncol. 2012;30:593–599. doi: 10.1200/JCO.2010.33.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiter JG, Baretti M, Gerold JM et al. An analysis of genetic heterogeneity in untreated cancers. Nat Rev Cancer. 2019;19:639–650. doi: 10.1038/s41568-019-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross JS, Gay LM, Wang K et al. Non-Amplification ERBB2 Genomic Alterations in 5,605 Cases of Relapsed and Metastatic Breast Cancer: an Emerging Opportunity for anti-HER2 Targeted Therapies. Cancer. 2016;122:2654–2662. doi: 10.1002/cncr.30102. [DOI] [PubMed] [Google Scholar]
- 38.Hempel D, Ebner F, Garg A et al. Real world data analysis of next generation sequencing and protein expression in metastatic breast cancer patients. Sci Rep. 2020;10:10459. doi: 10.1038/s41598-020-67393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosele F, Remon J, Mateo J et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Sultova E, Westphalen CB, Jung A et al. NGS-guided precision oncology in metastatic breast and gynecological cancer: first experiences at the CCC Munich LMU. Arch Gynecol Obstet. 2021;303:1331–1345. doi: 10.1007/s00404-020-05881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sivapiragasam A, Ashok Kumar P, Sokol ES et al. Predictive Biomarkers for Immune Checkpoint Inhibitors in Metastatic Breast Cancer. Cancer Med. 2021;10:53–61. doi: 10.1002/cam4.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhardt F, Franken A, Fehm T et al. Navigation through inter- and intratumoral heterogeneity of endocrine resistance mechanisms in breast cancer: A potential role for Liquid Biopsies? Tumor Biol. 2017;39:1.010428317731511E15. doi: 10.1177/1010428317731511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.