Abstract
Kidney fibrosis is associated with tubular injury, oxidative stress and activation of interstitial fibroblasts. However, whether these events are somehow connected is poorly understood. In this study, we show that glutathione peroxidase-3 (GPX3) depletion in renal tubular epithelium after kidney injury plays a central role in orchestrating an oxidatively stressed extracellular microenvironment, which drives interstitial fibroblast activation and proliferation. Through transcriptional profiling by RNA-sequencing, we found that the expression of GPX3 was down-regulated in various models of chronic kidney disease (CKD), which was correlated with induction of nicotinamide adenine dinucleotide phosphate (NAPDH) oxidase-4 (NOX4). By using decellularized extracellular matrix (ECM) scaffold, we demonstrated that GPX3-depleted extracellular microenvironment spontaneously induced NOX4 expression and reactive oxygen species (ROS) production in renal fibroblasts and triggered their activation and proliferation. Activation of NOX4 by advanced oxidation protein products (AOPPs) mimicked the loss of GPX3, increased the production of ROS, stimulated fibroblast activation and proliferation, and activated protein kinase C-α (PKCα)/mitogen-activated protein kinase (MAPK)/signal transducer and activator of transcription 3 (STAT3) signaling. Silencing NOX4 or inhibition of MAPK with small molecule inhibitors hampered fibroblast activation and proliferation. In mouse model of CKD, knockdown of NOX4 repressed renal fibroblast activation and proliferation and alleviated kidney fibrosis. These results indicate that loss of GPX3 orchestrates an oxidatively stressed extracellular microenvironment, which promotes fibroblast activation and proliferation through a cascade of signal transduction. Our studies underscore the crucial role of extracellular microenvironment in driving fibroblast activation and kidney fibrosis.
Keywords: Oxidative stress, Extracellular microenvironment, GPX3, Fibroblast activation, Kidney fibrosis
Graphical abstract
Highlights
-
•
Loss of GPX3 is associated with upregulation of NOX4 in CKD.
-
•
GPX3-depleted extracellular microenvironment triggers NOX4 induction and fibroblast activation.
-
•
AOPPs imitates GPX3 depletion and induces NOX4 expression, promotes ROS production and fibroblast proliferation.
-
•
NOX4 promotes further NOX4 induction by forming amplification loop via ROS/PKCα/MAPK/STAT3 cascade, and promotes fibroblast activation.
-
•
Knockdown of NOX4 inhibits fibroblast activation and ameliorates kidney fibrosis in vivo.
1. Introduction
Kidney fibrosis, characterized by excessive accumulation of extracellular matrix (ECM), is the common outcome of a wide variety of chronic kidney disease (CKD) [1,2]. Studies show that fibrotic lesions are often unevenly distributed throughout renal parenchyma, suggesting that local tissue environment plays a decisive role in the initiation and development of kidney fibrosis. We recently proposed the concept of the fibrogenic niche, which portrays the special tissue microenvironment that promotes fibroblast activation in organ fibrosis [3,4]. Although a series of cellular events may take place in the fibrogenic niche [1,5,6], fibroblast activation is the principal one with particular importance, as it renders quiescent fibroblasts to proliferate, acquire a myofibroblast phenotype by expressing α-smooth muscle actin (α-SMA) and become the principal matrix-producing cells [7,8]. Kidney fibrosis is also associated with oxidative stress, a condition caused by an imbalance between the production of reactive oxygen species (ROS) and antioxidant defenses [9]. However, how oxidative stress is connected to fibroblast activation in the fibrotic kidney remains incompletely understood.
Oxidative stress results from an increased production of ROS and/or a decrease in the effectiveness of antioxidant defenses [10,11]. The best studied cellular antioxidants are the glutathione peroxidases (GPXs), a family of enzymes with peroxidase activity whose main biological role is to protect cells and tissues from oxidative damage [12,13]. There are 8 isozymes in the GPX family, which are encoded by different genes and vary in cellular and subcellular locations and substrate specificity. Together, they catalyze the reduction of hydrogen peroxide or organic hydroperoxides using glutathione. Of these 8 GPXs, GPX3, also known as extracellular or plasma GPX, is quite unique in that it is secreted into extracellular fluid and plasma [14,15]. As a major ROS scavenger, GPX3 plays an imperative role in safeguarding tissue extracellular environment from oxidative damage by eliminating ROS [16]. GPX3 in blood circulation is mainly originated from the proximal tubular epithelium of the kidney [17], and it is secreted to extracellular space and binds to the tubular basement membrane and other matrix proteins [18]. As such, GPX3 acts as an integral component of the extracellular microenvironment in normal kidneys. It is conceivable to speculate that loss of GPX3 may lead to excessive accumulation of ROS in the extracellular compartment, thereby affecting the surrounding cells and changing their phenotypes and trajectories.
How cells sense the oxidative stress in extracellular environment and then make an appropriate response is an intriguing question. As nicotinamide adenine dinucleotide phosphate (NAPDH) oxidase (NOX) is a family of the membrane-bound enzyme complex that faces the extracellular space [[19], [20], [21]], we wondered whether NOX enzymes play a role in connecting oxidative stress in the extracellular environment to intracellular signaling and subsequent cellular actions. In line with this speculation, earlier studies have shown that NOX4 can act as an oxygen sensor and its activity can be regulated by extracellular oxygen levels to control intracellular ROS production [22]. Furthermore, the NOX-derived ROS is related to alterations in cell signaling [23]. However, exactly how NOX4-derived ROS signaling causes fibroblast activation and fibrotic lesions in the kidney deserves further investigation.
In this study, we show that GPX3 was downregulated in the kidney in mouse models of CKD, which was associated with NOX4 induction. Using decellularized kidney tissue scaffolds (KTS), we demonstrate that GPX3-depleted ECM scaffold spontaneously promoted NOX4 expression and ROS production, and induced renal fibroblast proliferation. Furthermore, knockdown of NOX4 inhibited the proliferation and activation of fibroblasts both in vitro and in vivo. Our results suggest that oxidatively stressed extracellular microenvironment plays a vital role in driving fibroblast activation and kidney fibrosis.
2. Materials and methods
2.1. RNA-seq analysis
Male BALB/c mice were divided to two groups: (i) sham group, (ii) unilateral ureteral obstruction (UUO) group. Mice were euthanized at 7 days after UUO, and then the kidneys removed [24]. Total RNA of the kidneys was extracted with Trizol (Invitrogen) and qualified using Agilent 2100 bioanalyzer (Thermo Fisher Scientific). Aliquot of total RNA (200 ng) was used to construct the RNA sequencing (RNA-seq) libraries. RNA-seq was carried out on MGISEQ2000 platforms (BGI Genomics), and approximately 20 M clean reads were generated. Each RNA-seq read was mapped to the mouse genome using HISAT2 v2.0.5 with default values for the parameters [25]. The mappable reads were assembled by cufflinks v2.2.1 with default parameters [26]. FPKM (fragments per kilobase per million mapped reads) values were called using Cufflinks. Differential gene expression was performed by using the cuffdiff. Adjusted P < 0.05 was considered as a significantly differential expression.
2.2. Bioinformatics analyses
The pathways of proteins were elucidated based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/kegg/) using the blast software against the database. Enrichment analyses were applied based on the Fisher’ exact test, considering the whole quantified proteins as background dataset. Benjamini-Hochberg correction for multiple testing was further applied to adjust derived p-values. Only functional categories and pathways with p-values under a threshold of 0.05 were considered as significant. Gene set enrichment analysis (GSEA) was performed by GSEApy software with default parameters against KEGG gene sets.
The prediction of transcription factors binding sites of mouse NOX4 gene was performed by using the JASPAR Predicted Transcription Factor Targets program (http://jaspar.genereg.net/). By analyzing mouse NOX4 gene in the dataset, we found that the signal transducer and activator of transcription 3 (STAT3) could bind to multiple sites in the promoter of mouse NOX4 gene.
2.3. Animal models
Male BALB/c mice were obtained from the Southern Medical University Animal Center (Guangzhou, China). Mice were subjected to UUO surgery and euthanized at 7 days after UUO, as previous reported [27]. In separate experiments, mice were subjected to unilateral ischemia/reperfusion injury (UIRI) [24]. At day 10 after UIRI, the contralateral intact kidney was removed by nephrectomy. Mice were euthanized at 11 days after UIRI. In addition, male CD1 mice were subjected to 5/6 nephrectomy (5/6NX), as described previously [24]. Briefly, renal ablation of the left kidney was produced by amputation of both renal poles. One week later, the mice were subjected to a right nephrectomy via a dorsal lumbotomy incision. After 8 weeks, the mice were euthanized and kidney tissues collected. All animal studies were approved by the Experimental Animal Committee at the Nanfang Hospital, Southern Medical University.
2.4. Preparation of HK-2-derived ECM scaffold
Human kidney proximal tubular epithelial cell line (HK-2) was obtained from the American Type Culture Collection (ATCC; Manassas, VA). HK-2 cells were transfected with control or GPX3-specific siRNA for 24 h. The cells were then treated with EGTA [ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid] (0.5 mM; pH 7.4) in calcium-free phosphate-buffered saline (PBS), followed by shaking for 2 h at 4 °C. The treatment was repeated 3–4 times until all cells were lifted from their underlying matrix. The HK-2-derived extracellular matrix (ECM) scaffold was washed with PBS. Normal rat kidney interstitial fibroblast cells (NRK-49F; ATCC) were then seeded on the ECM scaffold and cultured for 24 h. Some ECM scaffold was collected for subsequent Western blot analysis.
2.5. Preparation of kidney tissue scaffold
Decellularized kidney tissue scaffold (KTS) was prepared according to an established protocol [4]. Briefly, kidneys were cut into 3-4 slices of same thickness along the sagittal plane. The kidney slices were then subjected to multiple steps of decellularization procedures as described [4].
2.6. AOPPs-BSA preparation
The advanced oxidation protein products-bovine serum albumin (AOPPs-BSA) was prepared by incubation of BSA (Sigma-Aldrich, St. Louis, MO) with hypochlorous acid (Fluke, Buchs, Switzerland) in the absence of free amino acid/carbohydrates/lipids to exclude the formation of advanced glycation end products (AGEs)-like structures [28]. Prepared samples were dialyzed against sterile PBS to remove free hypochlorous acid (HOCL) and passed through a Detoxi-Gel column (Pierce, Rockford, IL) to remove contaminated endotoxin. The endotoxin levels in the preparations were found to be < 0.025 EU/ml by the Amebocyte Lysate Kit (Sigma Chemical). The components of AGEs, including N(6)-carboxylmethyllysine, pentosidine, pyridine, or glyoxal-, glycolaldehyde-, and glyceraldehyde-modified proteins were undetectable in the prepared samples as described previously [28].
2.7. Cell culture and treatment
The NRK-49F cells were maintained in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 10% fetal bovine serum (FBS). NRK-49F cells were seeded on six-well culture plates with 70∼80% confluence in the complete medium containing 10% FBS for about 12 h, and then changed to serum-free medium after washing twice with PBS. Cells were then treated with AOPPs at different concentrations (0, 25, 50, and 100 μg/ml) in the serum-free medium for various periods of time (0, 12, 24, 48 h) or transfected with either control siRNA or NOX4-specific siRNA. In some experiments, cells were pretreated for 1 h with various chemical inhibitors at the specified concentrations and then treated with AOPPs (50 μg/ml). Furthermore, cells were also transfected with NOX4 overexpression plasmid (pCMV-NOX4), STAT3 overexpression plasmid (pCMV-STAT3) or STAT3-siRNA for 24 h, respectively, followed by treating without or with AOPPs (50 μg/ml). Control cells were treated with an equal amount of vehicle. Cells were then collected and subjected to various analyses. In independent experiments, HK-2 cells were transfected with control or NOX4 overexpression plasmid (pCMV-NOX4) for 24 h. Moreover, HK-2 cells were transfected with NOX4-siRNA or GPX3-siRNA, followed by treating with AOPPs (50 μg/ml).
2.8. Determination of ROS production
Production of intracellular ROS was analyzed using flow cytometry. After incubation with AOPPs for various periods of time or transfected with either control siRNA or NOX4-specific siRNA, cells were incubated with dichloro-dihydro-fluorescein diacetate (DCFH-DA) (10 μM) for 30 min. Cells were then washed, trypsinized, and resuspended in PBS. The mean fluorescence intensity at 488 nm was measured in the BD Flow Cytometry FACS Canto II equipment (BD Biosciences, Nova Jersey, EUA). In addition, total ROS including superoxide radical anion (O2•−), hydroxyl radical (•OH) and singlet oxygen (1O2) was also assayed by using an enzyme-linked immunosorbent assay (ELISA) Kit (Yuanju Biotechnology Center, Shanghai, China). Cell supernatants from different treatment groups were collected. The kidney tissues of each group were collected for homogenization, and the supernatant collected. The content of ROS in different groups was determined by using ROS ELISA kit according to the manufacturer’s instructions. In addition, to analytically assess the different types of ROS, dihydrorhodamine 123 (DHR123), dihydroethidium (DHE) and singlet oxygen sensor green (SOSG) were utilized as fluorescent probes to measure the superoxide radical anion (O2•−), hydroxyl radical (•OH) and singlet oxygen (1O2), respectively. Following the manufacturer's instructions, 5 μM fluorescent probes were added to the cell supernatant of each group, and the fluorescence intensity was measured using a fluorescence spectrophotometer.
2.9. Cell proliferation assay
Cell mass was assessed by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide] assay [24]. MTT was added at the final concentration of 0.5 mg/ml to individual cultures for 4 h before the end of the experiments. Tetrazolium was released by dimethyl sulfoxide, and the optical density was determined with an ELx800 Absorbance Reader (BioTek Instruments, Winooski, VT) at 570 nm.
2.10. EdU incorporation assay
EdU (5-ethynyl-2’-deoxyuridine) incorporation assay was performed according to an established protocol [29]. In short, cells were inoculated on the six-well plates and treated as needed, and then incubated with EdU (10 μM) for 1 day. At the end of incubation, cells were fixed, followed by incubating with the EdU reaction mixture at room temperature for 30 min. Hoechst reaction solution was then added to each well. The staining samples were observed under the Eclipse E600 epifluorescence microscope equipped with a digital camera (Nikon, Tokyo, Japan).
2.11. Knockdown of NOX4 in vivo
Knockdown of NOX4 expression in vivo was performed using short hairpin RNA (shRNA)-mediated approach as reported [30]. The efficiency of transgene transfection after hydrodynamics-based gene transfer was confirmed by injecting plasmid vector encoding HA-tagged Wnt1 fusion protein, as recently reported [31]. Mice were divided into four groups (n = 5 in each group): (i) sham mice injected with vehicle, (ii) UIRI mice injected with pcDNA3.1 plasmid, (iii) UIRI mice injected with AOPPs and control shRNA, and (iv) UIRI mice injected with AOPPs and NOX4-shRNA. Mice were euthanized at 11 days after UIRI, and serum and kidney tissues were analyzed.
2.12. Isolation and culture of mouse primary renal fibroblasts
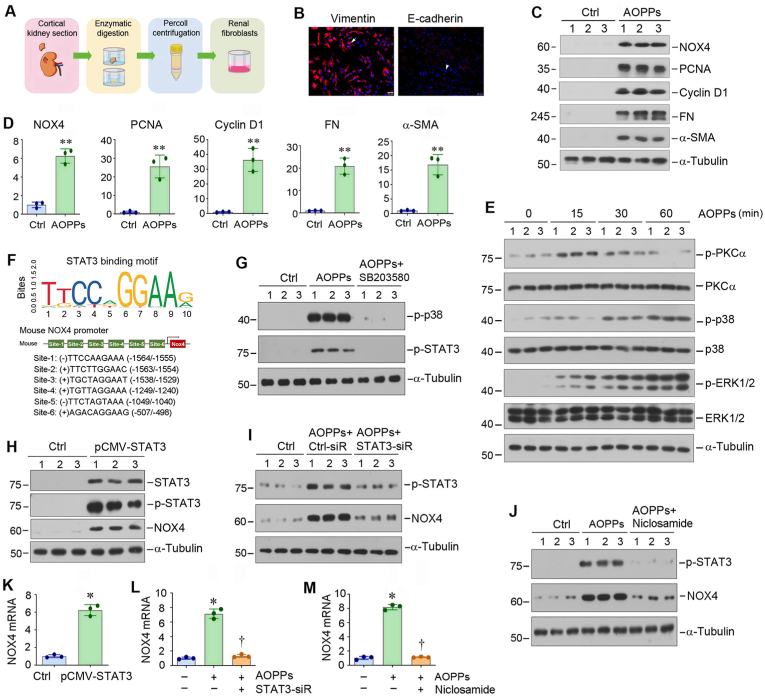
Renal cortex was obtained from six control or UIRI kidneys, then minced after removal of kidney capsule. The cortex was digested in a 1 mg/ml solution of collagenase type IV (Sigma Chemical Co., Poole, Dorset, UK). A small, sterile magnetic stirrer was added and placed on a stirring block in a water bath at 37 °C for 30 min [32]. The digest was passed through a 100-mm mesh, then a 70-mm mesh, to remove contaminating glomeruli and tubules, and then separated on a 50% Percoll density gradient (containing 0.84 g of mannitol and 100 ml of 1 M HCl) in 30 ml by centrifugation at 30,000 g. The cell suspension was plated onto tissue culture dishes or flasks in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 Nutrient Mix (Gibco BRL, Life Technologies Ltd., Paisley, UK) with 10% fetal calf serum (FCS), 50 U/ml penicillin, 50 mg/ml streptomycin, allowed to grow to confluence [33]. The cells were characterized by immunofluorescence staining and stained positively for vimentin and negatively for E-cadherin. The primary fibroblasts were then collected and subjected to various treatments and analyses.
2.13. Western blot analyses
Protein expression was analyzed by Western blot analysis as described previously [24]. The antibodies used are listed in Supplementary Table S1.
2.14. Quantitative real-time RT-PCR
Total RNA was isolated using the TRIzol RNA purification system (Thermo Fisher Scientific, Waltham, MA). The qRT-PCR was performed on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) as described previously [24]. The mRNA levels of different genes were calculated after normalization with β-actin. The sequences of primer pairs are presented as Supplementary Table S2.
2.15. Histology, immunohistochemical and immunofluorescence staining
Paraffin-embedded mouse kidney sections were prepared by a routine procedure. The sections were stained with Masson’s trichrome staining (MTS) reagents. Immunohistochemical staining was performed as described previously [24]. Antibodies used are listed in Supplementary Table S1.
2.16. Statistical analysis
All data examined were expressed as mean ± SEM. Statistical analysis of the data was carried out using IBM SPSS Statistics (SPSS Inc., Chicago, IL). Comparisons between groups were made by t-test or using one-way analysis of variance (ANOVA), followed by Fisher’s least significant difference test or Dunnett T3 test. P < 0.05 was considered significant.
3. Results
3.1. Loss of GPX3 is associated with upregulation of NOX4 in CKD
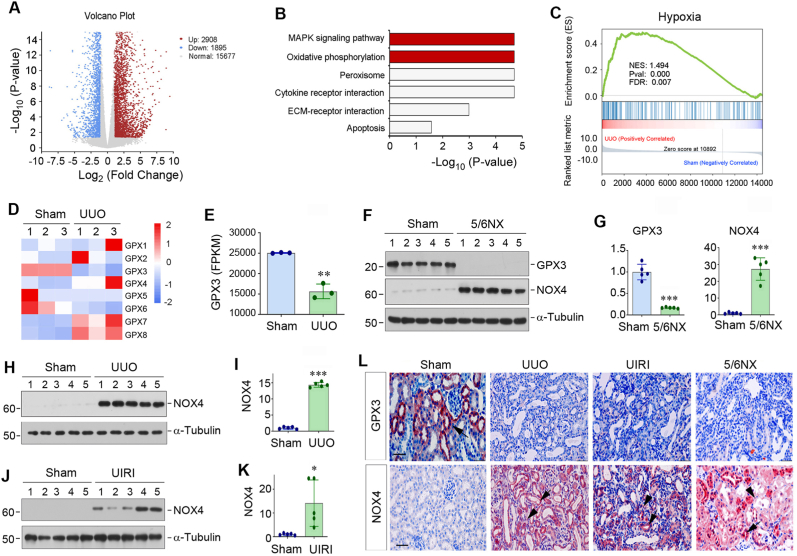
To investigate the mechanism of kidney fibrosis, we sought to identify the differentially expressed genes in fibrotic kidneys by using an unbiased RNA-seq approach. As shown in Fig. 1A, transcriptomic analysis of gene expression profiles demonstrated that there were 2908 mRNAs up-regulated in UUO kidney, whereas 1895 were down-regulated, when compared to sham controls. Kyoto Encyclopedia of Gene and Genomes (KEGG) enrichment analysis of the differentially expressed genes revealed that several signaling pathways were enriched, including mitogen-activated protein kinase (MAPK) signaling, oxidative phosphorylation, peroxisome, cytokine receptor interaction, ECM-receptor interaction and apoptosis (Fig. 1B). Furthermore, we performed gene set enrichment analysis (GSEA) for the differentially expressed genes and found that hypoxia pathway was enriched in the UUO kidney (Fig. 1C). These results suggest that oxidative stress could play a role in kidney fibrogenesis.
Fig. 1.
Gene expression profiling by RNA sequencing identifies loss of GPX3 as a pathologic feature of chronic kidney disease (CKD). (A) The volcano diagram of RNA-seq data displays the differentially expressed genes of sham and UUO kidneys. The red dots represent the genes up-regulated in UUO group, while blue dots show genes down-regulated. The grey dots denote mRNAs with no change in abundance between the two groups. (B) KEGG enrichment analysis reveals that several signaling pathways as indicated were enriched. (C) GSEA enrichment analysis shows that hypoxia was enriched in UUO group. NES, normalized enrichment score; FDR, false discovery rate. (D) The heat map shows differential expression of GPX family (GPX1-GPX8) genes in sham and UUO kidneys. (E) RNA-seq analysis shows the expression level (FPKM) of GPX3 in normal and UUO kidneys. FPKM, fragments per kilobase per million mapped reads. (F, G) Western blot analyses show the levels of GPX3 and NOX4 in different groups as indicated. Representative Western blot (F) and quantitative data (G) are shown. (H, I) Western blot analyses show the levels of NOX4 in sham and UUO mice. Representative Western blot (H) and quantitative data (I) are shown. (J, K) Changes of NOX4 expression in the kidney after UIRI. Representative Western blot (J) and quantitative data (K) are shown. *P < 0.05, **P < 0.01 and ***P < 0.001 versus sham (n = 5). (L) Immunohistochemical staining shows renal expression and localization of GPX3 (upper panel) and NOX4 (lower panel) proteins in different groups as indicated. Arrows indicate positive staining in renal tubular epithelial cells, whereas arrowheads indicate positive staining in renal interstitium. Scale bar, 50 μm.
We found that the mRNA expression of several members of the GPX family of antioxidant enzymes were changed (Fig. 1D). Among GPX1-8, the mRNA levels of GPX7 and GPX8 were increased in UUO kidney, whereas GPX3 mRNA level was down-regulated, compared to sham group (Fig. 1D and E). As GPX3 is the only extracellular GPX and can be secreted into extracellular fluid and plasma [14], and its downregulation in UUO kidney was consistent and coherent in different animals (Fig. 1D and E), we decided to investigate its role in kidney fibrogenesis in great detail in the subsequent studies.
We examined GPX3 expression in 3 well-characterized models of renal fibrosis induced by 5/6NX, UUO and UIRI. As shown in Fig. 1F and G, GPX3 protein was abundantly expressed in normal control kidney, whereas its expression was markedly downregulated in the fibrotic kidneys induced by 5/6NX. Intriguingly, loss of GPX3 was associated with the induction of NOX4, a member of the NOX family of complex enzymes that generate ROS, in the fibrotic kidneys (Fig. 1F and G). Similarly, GPX3 was also reduced in the fibrotic kidneys induced by UUO and UIRI as previously reported [34], whereas NOX4 was markedly upregulated in the kidneys of both models (Fig. 1H–K). We further examined the expression and localization of GPX3 and NOX4 by immunohistochemical staining. As shown in Fig. 1L, GPX3 protein was predominantly localized in kidney tubular epithelium in control mice, while NOX4 protein was localized in both tubular epithelium and interstitial cells of diseased kidneys. These results indicate that loss of GPX3 is a common feature of CKD, which is closely associated with NOX4 induction.
3.2. GPX3-depleted microenvironment triggers NOX4 expression and fibroblast activation
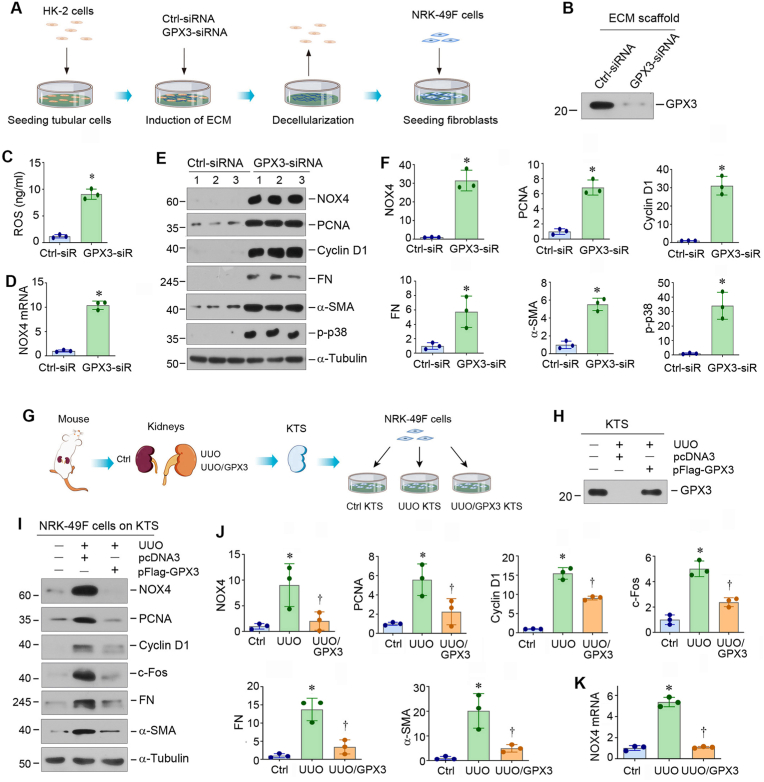
Because GPX3 is the only member of the GPX family enzymes localizing in the extracellular compartment, we sought to delineate how extracellular GPX3 affects cell behaviors. To address this issue, we investigated the effect of GPX3-rich or GPX3-poor ECM scaffolds on renal fibroblasts. As GPX3 is mainly produced by tubular epithelial cells, we prepared GPX3-depleted ECM scaffolds from human kidney proximal tubular cells (HK-2) by transfecting GPX3-specific small interfering RNA (siRNA), followed by decellularization as previously reported [4]. Fig. 2A shows the experimental protocol. The depletion of GPX3 in the HK-2-derived ECM scaffolds was confirmed by Western blot analysis (Fig. 2B). When normal rat kidney interstitial fibroblast (NRK-49F) cells were inoculated on the GPX3-depleted ECM scaffold, ROS level in the supernatant was elevated (Fig. 2C). NOX4 mRNA and protein was markedly induced (Fig. 2D–F). Interestingly, GPX3-depleted ECM scaffold promoted the expression of proliferating cell nuclear antigen (PCNA), cyclin D1, fibronectin (FN), α-smooth muscle actin (α-SMA) and phosphorylated p38 mitogen-activated protein kinase (p-p38) in NRK-49F cells (Fig. 2E and F). These results indicate GPX3-depleted microenvironment can lead to ROS accumulation and NOX4 upregulation and spontaneously induce fibroblast proliferation and activation.
Fig. 2.
GPX3-deprived extracellular microenvironment induces NOX4 expression, cell proliferation and activation in renal interstitial fibroblast. (A) Diagram shows the experimental protocol by which NRK-49F cells were cultured on GPX3-depleted ECM scaffolds. HK-2 cells were transfected with either control siRNA or GPX3 siRNA for 24 h to knockdown GPX3 expression and secretion. The ECM scaffold was then prepared after decellularization. NRK-49F cells were inoculated on ECM scaffold and cultured for 24 h. (B) Knockdown of GPX3 resulted in its depletion in the ECM scaffolds. Western blot analyses show GPX3 protein levels in the ECM scaffolds prepared from HK2 cells after different treatments as indicated. (C) GPX3-depleted ECM scaffold induces ROS production. ROS levels in the supernatant of the NRK-49F cells cultured in different ECM scaffolds were measured by specific ELISA. Quantitative data are shown. (D) GPX3-depleted ECM scaffold induces NOX4 mRNA expression in NRK-49F cells. The qRT-PCR analyses show mRNA levels of NOX4 in different groups as indicated. (E, F) GPX3-depleted ECM scaffold induces NOX4 expression, fibroblast proliferation and activation. Western blots analyses show the expression of NOX4, PCNA, Cyclin D1, FN, α-SMA and phosphorylated p38 (p-p38) in different groups as indicated. Representative Western blot (E) and quantitative data (F) are shown. *P < 0.05 versus controls (n = 3). (G) Diagram shows the experimental protocol by which NRK-49F cells were cultured on GPX3-rich or -depleted KTS. The KTS was prepared from sham and UUO kidneys in mice injected with empty vector (pcDNA3) or Flag-tagged GPX3 expression vector (pFlag-GPX3). NRK-49F cells were inoculated on different KTS for 24 h. (H) Overexpression of GPX3 in vivo restored its presence and abundance in the KTS. Western blot analyses show GPX3 protein levels in the KTS prepared from different groups as indicated. (I, J) GPX3-depleted KTS induces NOX4 expression, fibroblast proliferation and activation. Western blots analyses show the expression of NOX4, PCNA, Cyclin D1, c-Fos, FN and α-SMA in different groups as indicated. Representative Western blot (I) and quantitative data (J) are shown. *P < 0.05 versus controls, †P < 0.05 versus UUO + pcDNA3 (n = 3). (K) GPX3-depleted KTS induces NOX4 mRNA expression. qRT-PCR analyses show mRNA levels of NOX4 in different groups as indicated. *P < 0.05 versus controls, †P < 0.05 versus UUO + pcDNA3 (n = 3).
To further confirm the effect of GPX3-depleted extracellular microenvironment on fibroblast activation, we carried out ex vivo studies using decellularized KTS, which were prepared from the control and UUO kidneys of mice injected with empty vector (pcDNA3) or Flag-tagged GPX3 expression vector (pFlag-GPX3) (Fig. 2G). We then inoculated NRK-49F cells onto these 3 different kinds of KTS (Fig. 2G). Western blotting confirmed the loss of GPX3 protein in the KTS prepared from UUO kidney, compared with the controls. However, it was largely restored in the KTS prepared from UUO mice injected with pFlag-GPX3 (Fig. 2H). As shown in Fig. 2I and J, when NRK-49F cells were cultivated in the GPX3-depleted KTS from UUO mice, NOX4, PCNA, cyclin D1, c-Fos, FN and α-SMA were dramatically induced. However, restoration of GPX-3 in UUO-KTS after injection of pFlag-GPX3 abolished these inductions (Fig. 2I and J). Similar results were obtained when NOX4 mRNA was assessed by qRT-PCR (Fig. 2K). Taken together, these results suggest that loss of GPX3 in extracellular microenvironment triggers NOX4 mRNA and protein expression, oxidative stress and fibroblasts proliferation and activation.
3.3. Induction of NOX4 by AOPPs promotes ROS production and fibroblast proliferation
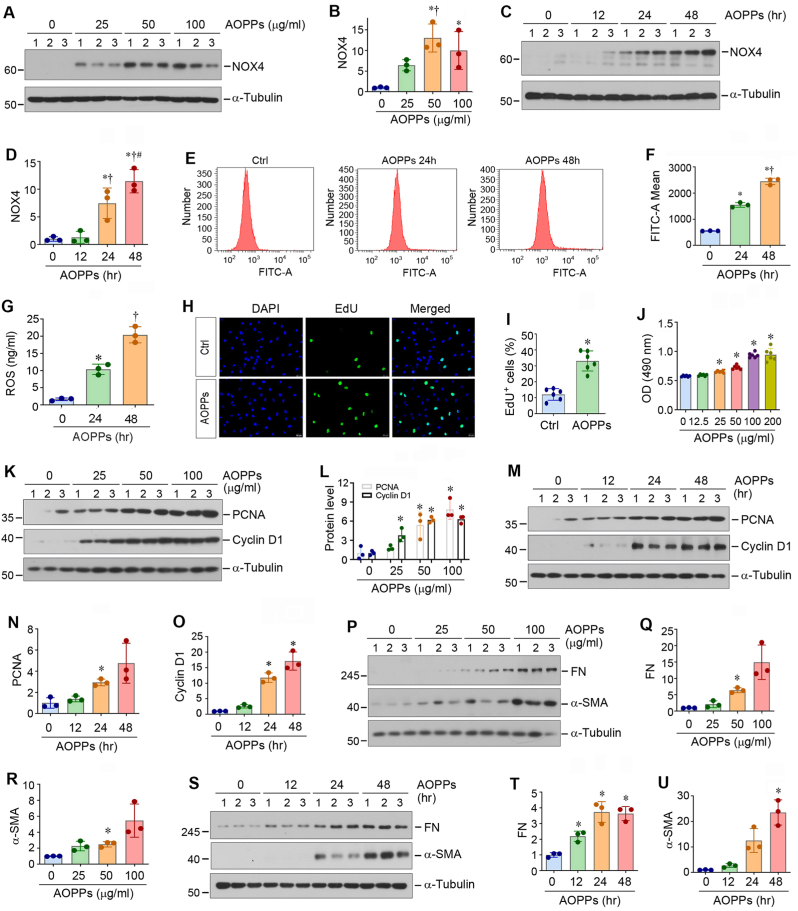
To elucidate the mechanism by which NOX4 upregulation due to the loss of GPX3 in the extracellular microenvironment leads to fibroblast activation, we imitated the condition by incubating the cells with AOPPs, a marker and trigger of oxidative stress [35]. To this end, we first incubated NRK-49F cells with AOPPs at different doses for varying periods of time. As shown in Fig. 3A–D, Western blotting showed that AOPPs upregulated NOX4 expression in a dose- and time-dependent manner. Consistently, qRT-PCR analysis also revealed that AOPPs induced the mRNA expression of various NOX subunits, such as NOX1, NOX2, NOX4 and dual oxidase 1 (DUOX1), but not DUOX2, in NRK-49F cells (Figs. S1A–E). Among them, NOX4 expression was most highly upregulated (Fig. S1C).
Fig. 3.
Activation of NOX4 by AOPPs increases reactive oxidative species (ROS) and induces fibroblast activation and proliferation. (A, B) Western blot analyses show that AOPPs induced NOX4 protein expression in NRK-49F cells in a dose-dependent manner. NRK-49F cells were treated with different concentrations of AOPPs for 24 h. Representative Western blot (A) and quantitative data (B) are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPPs (25 μg/ml) (n = 3). (C, D) Western blot analyses show that AOPPs upregulated NOX4 expression in a time-dependent manner. NRK-49F cells were treated with AOPPs (50 μg/ml) for various periods of time as indicated. Representative Western blot (C) and quantitative data (D) are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPP 12 h, #P < 0.05 versus AOPP 24 h (n = 3). (E, F) AOPPs increased ROS production in NRK-49F cells. Representative flow cytometry histograms (E) and quantitative data (F) are shown. (G) ROS levels in cell supernatant were measured by specific ELISA. Quantitative data are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPP 12 h, #P < 0.05 versus AOPP 24 h (n = 3). (H, I) EdU incorporation assay shows that AOPPs (50 μg/ml) promoted DNA synthesis by increasing EdU incorporation in NRK-49F cells. Representative EdU incorporation assay (H) and quantitative data (I) are shown. *P < 0.05 versus control (n = 3). (J) Quantitative colorimetric MTT assay shows that AOPPs promoted NRK-49F cell proliferation. NRK-49F cells were treated with different concentrations of AOPPs as indicated for 24 h. *P < 0.05 versus controls. OD, optical density. (K-O) Western blot analyses show that AOPPs induced the expression of numerous proliferation-related proteins in NRK-49F cells in a dose- (K, L) and time- (M–O) dependent manner. Representative Western blot (K, M) and quantitative data (L, N and O) are shown. *P < 0.05 versus controls (n = 3). (P–U) Western blot analyses show that AOPPs induced renal fibroblast activation in a dose- and time-dependent manner. NRK-49F cells were treated with different concentrations of AOPPs as indicated for 24 h, or 50 μg/ml of AOPPs for various periods of time as indicated. Cell lysates were subjected to Western blot analyses for FN, α-SMA and α-tubulin. Representative Western blot (P, S) and quantitative data (Q, R, T and U) are shown. *P < 0.05 versus controls (n = 3).
As NOX enzymes are one of the major sources of cellular ROS, we then assessed ROS production in the setting of NOX4 induction by AOPPs in NRK-49F cells, which mimicked the loss of GPX3. As shown in Fig. 3E–G, flow cytometry analysis and ROS ELISA showed that AOPPs induced ROS production in renal fibroblasts in a time-dependent manner. We further analytically measured the different types of ROS using specific fluorescent probes. As shown in Figs. S2A–C, superoxide radical anion (O2•−), hydroxyl radical (•OH) and singlet oxygen (1O2) were all increased after AOPPs treatment. As shown in Fig. 3H and I, AOPPs induced EdU incorporation in NRK-49F cells, indicating its ability to promote DNA synthesis and cell proliferation. Similar results were obtained by using a quantitative colorimetric MTT assay, as AOPPs increased the number of NRK-49F cells in a dose- and time-dependent manner (Fig. 3J and Fig. S2D). Moreover, AOPPs increased the expression of PCNA and cyclin D1 in NRK-49F cells (Fig. 3K–O). Furthermore, AOPPs also increased the expression of FN and α-SMA in NRK-49F cells in a dose- and time-dependent manner (Fig. 3P–U), indicating that AOPPs can promote renal fibroblast transformation into myofibroblasts. Taken together, these results suggest that activation of NOX4 by AOPPs mimics the loss of GPX3 and promotes renal fibroblast proliferation and activation, which leads to excessive ECM accumulation and deposition.
3.4. Upregulation of NOX4 by AOPPs amplifies its action through PKCα/MAPK/STAT3 signaling
To better model the response of renal fibroblasts to NOX4 induction by AOPPs, we used primary fibroblasts isolated from mouse kidney, as depicted in Fig. 4A. These cells were positive for vimentin but negative for E-cadherin by immunofluorescence staining (Fig. 4B). Treatment of mouse primary fibroblasts with AOPPs dramatically induced the expression of NOX4, PCNA, cyclin D1, FN and α-SMA (Fig. 4C and D). These data provide strong evidence that NOX4 induction by AOPPs promotes renal fibroblasts to transform into matrix-producing myofibroblasts.
Fig. 4.
Activation of NOX4 by AOPPs induces renal fibroblast activation and activates PKCα/MAPK/STAT3 signal cascade in vitro. (A) Diagram shows the experimental protocol for isolation and primary culture of renal interstitial fibroblasts. (B) Representative micrographs show the expression of vimentin and E-cadherin by immunofluorescence staining. The primary fibroblasts are stained positively for vimentin (arrow) and negatively for E-cadherin (arrowhead). Scale bar, 50 μm. (C, D) Western blot analyses show that AOPPs induced primary fibroblast proliferation and activation. Primary fibroblasts from mouse kidneys were treated with AOPPs (50 μg/ml) for 24 h. Cell lysates were subjected to Western blot analyses for NOX4, PCNA, cyclin D1, FN, α-SMA and α-tubulin. Representative Western blot (C) and quantitative data (D) are shown. *P < 0.05, **P < 0.01 versus controls (n = 3). (E) Western blot analyses show that AOPPs activated PKCα/MAPK signal cascade. NRK-49F cells were treated with AOPPs (50 μg/ml) for various periods of time as indicated. Cell lysates were subjected to Western blot analyses for p-PKCα, PKCα, p-p38, p38, p-ERK1/2, ERK1/2 and α-tubulin. (F) Bioinformatic analysis shows the presence of putative STAT3 binding sites in the promoter of mouse NOX4 gene. According to the prediction obtained by using the JASPAR database, STAT3 could bind the promoter of mouse NOX4 at six putative sites. (G) Blockade of p38 activation inhibited STAT3 phosphorylation induced by AOPPs in NRK-49F cells. NRK-49F cells were pretreated with p38 inhibitor SB203580 (10 μM) for 1 h, then treated with AOPPs (50 μg/ml) for 30 min. (H) Western blot analyses show that overexpression of STAT3 induced the expression of NOX4 in NRK-49F cells. NRK-49F cells were transfected with pcDNA3 or pCMV-STAT3 plasmids for 24 h. (I) Western blot analyses show that knockdown of STAT3 inhibited AOPPs-induced NOX4 expression. NRK-49F cells were transfected with either control siRNA or STAT3-siRNA for 24 h, and then treated with AOPPs (50 μg/ml) for additional 24 h. (J) Blockade of STAT3 activation inhibited AOPP-induced NOX4 expression. NRK-49F cells were pretreated with STAT3 inhibitor niclosamide (5 μM), and then treated with AOPPs (50 μg/ml) for 24 h. (K–M) qRT-PCR analyses show overexpression of STAT3 (K) induced NOX4 mRNA expression, whereas knockdown (L) or inhibition (M) of STAT3 reduced NOX4 mRNA expression in NRK-49F cells. *P < 0.05 versus controls, †P < 0.05 versus AOPPs (50 μg/ml) (n = 3).
We further investigated the mechanism of AOPPs promoting oxidative stress and activation of fibroblasts. As shown in Fig. 4E and Fig. S3A, AOPPs induced a rapid phosphorylation and activation of protein kinase C-α (PKCα), a unique member of the serine- and threonine-specific protein kinase family, which peaked at 15 min and then returned to the baseline at 60 min. We also found that AOPPs induced the phosphorylation of p38 MAPK and extracellular signal-regulated kinase-1/2 (ERK1/2) in fibroblasts (Fig. 4E and Figs. S3B–C). Collectively, these results suggest that AOPPs activates the PKCα/MAPK signal cascade in renal fibroblasts.
Given the fact that NOX4 upregulation by AOPPs occurred at the mRNA level (Fig. S1C), we further explored the molecular detail by which oxidative stress triggers NOX4 expression. As shown in Fig. 4F, bioinformatics analysis revealed that there were 6 putative STAT3 binding sites in the promoter region of mouse NOX4 gene, suggesting the potential role of STAT3 in mediating NOX4 gene expression. We found that AOPPs could induce STAT3 phosphorylation and activation in NRK-49F cells, which was mediated by p38 MAPK, as SB203580 blocked p38 MAPK and STAT3 activation (Fig. 4G, and Figs. S3D–E). Furthermore, overexpression of STAT3 in NRK-49F cells induced NOX4 expression (Fig. 4H, and Figs. S3F–H). Conversely, knockdown of STAT3 via siRNA approach abolished AOPPs-triggered STAT3 activation and NOX4 induction (Fig. 4I, and Figs. S3I–J). Moreover, inhibition of STAT3 by Niclosamide, a small molecule STAT3 inhibitor, also abolished STAT3 activation and NOX4 upregulation induced by AOPPs (Fig. 4J, and Fig. S3K-L). Similar results were obtained when NOX4 mRNA was assessed by qRT-PCR (Fig. 4K–M). These results suggest that oxidative stress induces NOX4 expression through ROS/PKCα/p38 MAPK/STAT3 signal cascade.
3.5. Knockdown of NOX4 inhibits fibroblast activation via repressing PKCα/MAPK signaling
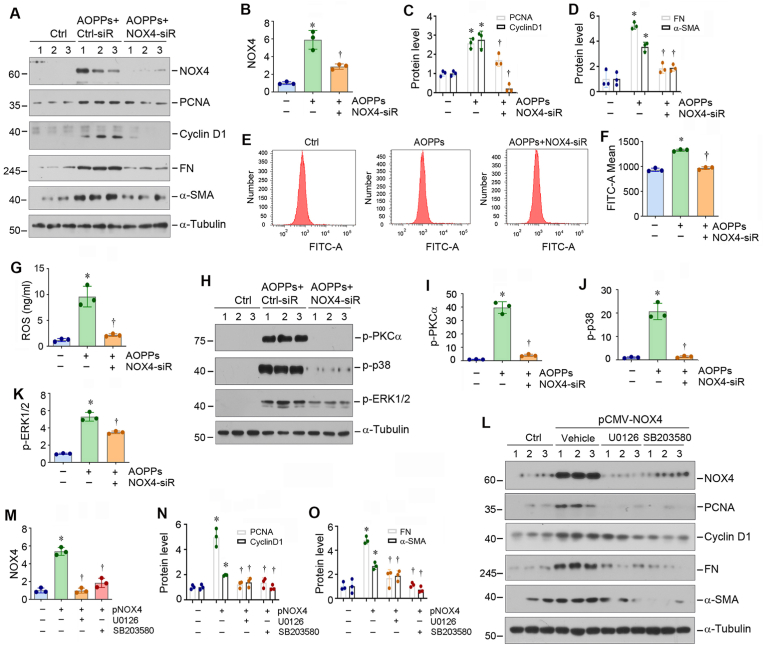
To confirm the role of NOX4 in mediating AOPPs-triggered fibroblast activation, we knocked down its expression by using siRNA-mediated inhibition. As shown in Fig. 5A–D, knockdown of NOX4 abolished AOPPs-induced PCNA, cyclinD1, FN and α-SMA expression in NRK-49F cells. Similarly, silencing NOX4 also blocked total ROS production (Fig. 5E–G), as well as superoxide radical anion (O2•−), hydroxyl radical (•OH) and singlet oxygen (1O2) (Figs. S4A–C), induced by AOPPs. Furthermore, knockdown of NOX4 also suppressed AOPPs-induced PKCα, p38 and ERK1/2 phosphorylation in NRK-49F cells (Fig. 5H–K). Consistently, overexpression of NOX4 by transfection of NRK-9F cells with expression vector (pCMV-NOX4) induced PCNA, cyclin D1, FN and α-SMA expression (Fig. 5L–O), whereas blocking p38 and ERK1/2 activation by SB203580 and U0126, respectively, inhibited NOX4-induced fibroblasts proliferation and activation (Fig. 5L–O). Taken together, these results illustrate a pivotal role for NOX4/PKCα/MAPK signaling in mediating fibroblasts proliferation and activation.
Fig. 5.
Blockade of NOX4/MAPK signaling inhibits renal fibroblast proliferation and activation in vitro. (A–D) Western blot analyses show that knockdown of NOX4 suppressed AOPPs-induced PCNA, cyclin D1, FN and α-SMA expression in NRK-49F cells. NRK-49F cells were transfected with either control siRNA or NOX4-siRNA for 24 h, and then cells were treated with AOPPs (50 μg/ml) for additional 24 h. Representative Western blot (A) and quantitative data (B–D) are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPPs (50 μg/ml) (n = 3). (E, F) Knockdown of NOX4 reduces ROS production in renal fibroblasts. Representative flow cytometry histograms (E) and quantitative data (F) are shown. (G) ROS concentrations in cell supernatant were measured by specific ELISA. Quantitative data are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPPs (50 μg/ml) (n = 3). (H–K) Western blot analyses show that knockdown of NOX4 suppressed AOPPs-induced p-PKCα, p-p38 and p-ERK1/2 activation in NRK-49F cells. NRK-49F cells were transfected with either control siRNA or NOX4 siRNA for 24 h, and then treated with AOPPs (50 μg/ml) for 30 min. Representative Western blot (H) and quantitative data (I–K) are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPPs (50 μg/ml) (n = 3). (L–O) Blockade of MAPK signaling abolished NOX4-induced fibroblasts proliferation and activation. NRK-49F cells were pretreated with MAPK kinase (MEK1/2) inhibitor U0126 or p38 inhibitor SB203580 for 1 h, followed by transfection with pcDNA3 or pCMV-NOX4 plasmids for 24 h, and then cell lysates were subjected to Western blot analyses for NOX4, PCNA, cyclin D1, FN, α-SMA and α-tubulin. Representative Western blot (L) and quantitative data (M–O) are shown. *P < 0.05 versus controls, †P < 0.05 versus pCMV-NOX4 (n = 3).
3.6. Knockdown of NOX4 ameliorates kidney injury and fibrosis and restores GPX3 in vivo
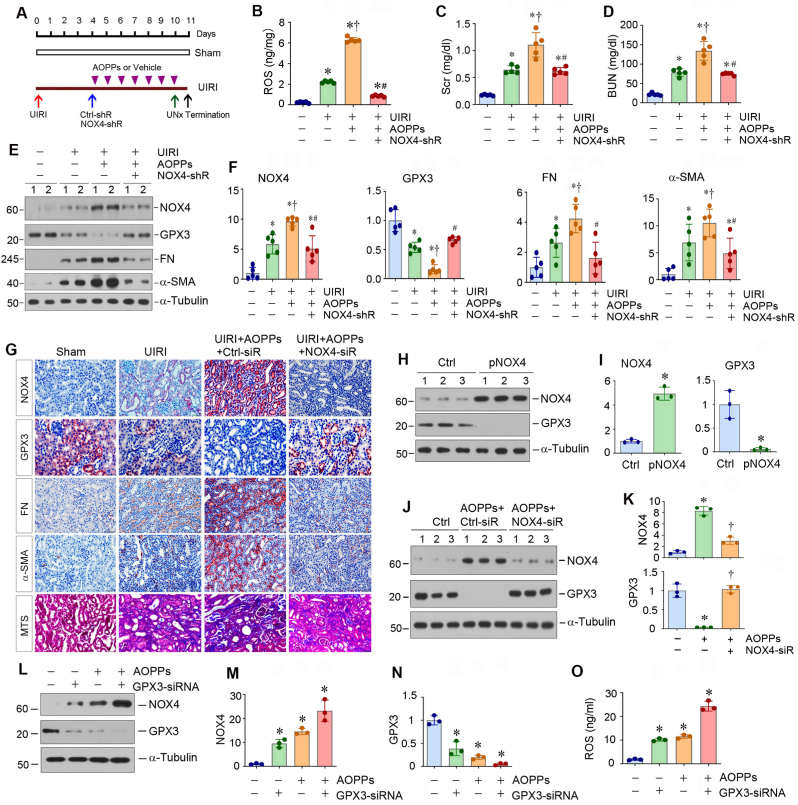
To validate the role of NOX4 in kidney injury and fibrosis in vivo, we carried out animal studies by knocking down NOX4 using short hairpin RNA (shRNA)-mediated inhibition [24]. As shown in Fig. 6A, mice underwent UIRI, followed by intravenously injecting with control-shRNA (Ctrl-shR) or NOX4-specific shRNA (NOX4-shR) vectors at 4 days after surgery. At the same time, mice were daily injected with either AOPPs or vehicle for one week. One day prior to termination of experiment, contralateral intact kidney was removed by unilateral nephrectomy (UNx) (Fig. 6A). We found that knockdown of NOX4 reduced ROS accumulation in the kidneys (Fig. 6B), mitigated kidney dysfunction in AOPPs-treated UIRI mice, as serum creatinine (Scr) and blood urea nitrogen (BUN) levels were reduced after knockdown of NOX4 (Fig. 6C and D). Western blot analysis of whole kidney lysates confirmed the efficient downregulation of NOX4 in the kidneys after injection of NOX4-shRNA vector (Fig. 6E and F). Furthermore, knockdown of NOX4 inhibited renal expression of FN and α-SMA in AOPPs-treated UIRI mice but largely restored renal expression of GPX3 (Fig. 6E and F). Consistently, qRT-PCR analysis showed that knockdown of NOX4 decreased renal mRNA levels of NOX4, FN and α-SMA in AOPPs-treated UIRI mice but restored mRNA level of GPX3 (Figs. S5A–D).
Fig. 6.
Knockdown of NOX4 ameliorates kidney injury and fibrosis in vivo. (A) Diagram shows the experimental protocol. Knockdown of NOX4 was carried out by short hairpin RNA (shRNA)-mediated approach. Mice were injected intravenously with either control-shRNA or NOX4-shRNA plasmids at 4 days after UIRI. At the same time, mice were injected intravenously with AOPPs or vehicle for 7 days. Red arrow indicates the timing of UIRI. Blue arrow indicates the timing of injecting control-shRNA or NOX4-shRNA plasmids. The arrowheads refer to the timing of injecting AOPPs or vehicle. Green arrow indicates the timing of uninephrectomy (UNx). Black arrow depicts the timing of sacrifice. (B) Knockdown of NOX4 ameliorated oxidative stress in the kidney after UIRI. Renal ROS levels were measured by specific ELISA. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 5). (C, D) Graphic presentations show serum creatinine (Scr) (C) and blood urea nitrogen (BUN) (D) levels in different groups as indicated. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 5). (E, F) Western blot analyses show renal expression of NOX4, GPX3, FN and α-SMA proteins in different groups as indicated. Representative Western blot (E) and quantitative data (F) are shown. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 5). (G) Representative micrographs show renal expression and localization of NOX4, GPX3, FN and α-SMA by immunohistochemical staining, and collagens deposition by Masson’s trichrome staining in different groups as indicated. (H, I) Western blot analyses show that overexpression of NOX4 inhibited the expression of GPX3 in tubular epithelial cells. HK-2 cells were transfected with pcDNA3 or pCMV-NOX4 plasmids for 24 h. Representative Western blot (H) and quantitative data (I) are shown. *P < 0.05 versus controls. (J, K) Western blot analyses show that knockdown of NOX4 restored AOPPs-repressed GPX3 expression in HK-2 cells. HK-2 cells were transfected with either control siRNA or NOX4-siRNA for 24 h, and then treated with AOPPs (50 μg/ml) for additional 24 h. Representative Western blot (J) and quantitative data (K) are shown. *P < 0.05 versus controls, †P < 0.05 versus AOPPs (50 μg/ml) (n = 3). (L–N) Western blot analyses show that knockdown of GPX3 aggravated AOPPs-induced NOX4 expression in HK-2 cells. HK-2 cells were transfected with either control siRNA or GPX3 siRNA for 24 h to knockdown GPX3 expression, and then treated with AOPPs (50 μg/ml) for additional 24 h. Representative Western blot (L) and quantitative data (M, N) are shown. *P < 0.05 versus controls (n = 3). (O) Knockdown of GPX3 aggravated AOPPs-induced ROS production in HK-2 cells. ROS levels in cell supernatant were measured by specific ELISA. Quantitative data are shown. *P < 0.05 versus controls (n = 3).
Immunohistochemical staining also showed that AOPPs aggravated renal expression of NOX4 in UIRI mice, which was associated with further downregulation of GPX3 (Fig. 6G, and Figs. S5E–F). However, knockdown of NOX4 largely restored the expression of GPX3 in the kidney of UIRI mice (Fig. 6G and Figs. S5E–F). Knockdown of NOX4 also inhibited renal expression of FN and α-SMA and ameliorated renal collagen accumulation and fibrotic lesions, as shown by Masson’s trichrome staining (MTS) (Fig. 6G, and Figs. S5G–I). These data indicate that upregulated NOX4 due to the loss of GPX3 acts as a pathogenic mediator promoting renal fibrosis.
As knockdown of NOX4 restored renal GPX3 expression in vivo (Fig. 6E), this prompted us to speculate whether loss of GPX3 was a consequence of NOX4 induction. To test this hypothesis, we examined GPX3 regulation by NOX4 in kidney tubular epithelial cells (HK-2), as GPX3 was mainly produced by tubular epithelium in vivo (Fig. 1L). As shown in Fig. 6H and I, overexpression of NOX4 by transfection of pCMV-NOX4 vector suppressed GPX3 expression in HK-2 cells. Conversely, knockdown of NOX4 restored GPX3 expression after AOPPs treatment (Fig. 6J and K). Similar results were obtained when NOX4 and GPX3 mRNA were assessed by qRT-PCR (Figs. S6A–D). Furthermore, NOX4 was negatively correlated with GPX3, and positively correlated with ROS, in HK-2 cells after various treatments as indicated (Fig. 6L–O). Therefore, GPX3 depletion and NOX4 induction could form a vicious cycle after kidney injury.
3.7. Knockdown of NOX4 inhibits fibroblast activation and proliferation in vivo
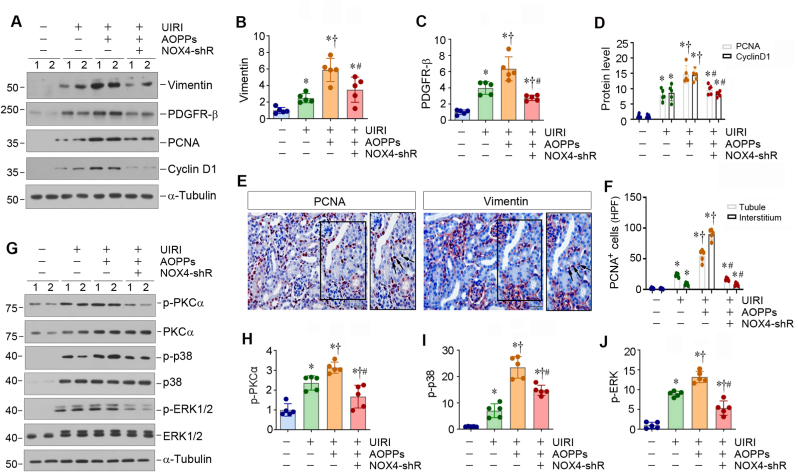
We further investigated the effect of knockdown of NOX4 on fibroblast activation and proliferation in vivo. Western blot analysis of whole kidney lysates revealed that AOPPs aggravated renal expression of vimentin, platelet-derived growth factor receptor-β (PDGFR-β), PCNA and cyclin D1 in UIRI mice, whereas knockdown of NOX4 inhibited the expression of these fibroblast activation- and proliferation-related proteins (Fig. 7A–D). Immunohistochemical staining for PCNA and vimentin on serial sections showed the co-localization of PCNA and vimentin in UIRI mice after AOPPs injections (Fig. 7E, arrows), indicating fibroblast proliferation in the fibrotic kidneys. However, knockdown of NOX4 reduced the number of PCNA-positive cells in both renal interstitial and tubular compartments in AOPPs-treated UIRI mice (Fig. 7F).
Fig. 7.
Knockdown of NOX4 represses fibroblast proliferation and MAPK signaling in vivo. (A–D) Western blot analyses show renal expression of vimentin, PDGFR-β, PCNA and cyclin D1 proteins in different groups as indicated. Representative Western blot (A) and quantitative data (B–D) are shown. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 5). (E) Representative micrographs show renal expression and localization of PCNA and vimentin on serial sections. Arrows indicate positive staining. Scale bar, 50 μm. (F) Graphic presentation shows the number of PCNA-positive cells in renal interstitial and tubular compartment in different groups. At least 10 randomly selected fields were assessed, and results averaged for each kidney. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 5). (G) Western blot analyses show renal expression of p-PKCα, PKCα, p-p38, p38, p-ERK1/2 and ERK1/2 proteins in different groups as indicated. Representative Western blot (G) and quantitative data (H–J) are shown. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 5).
To further investigate fibroblast activation and proliferation in vivo, we isolated primary fibroblast cells from sham and UIRI kidneys. As shown in Figs. S7A–F, the expression of fibrosis-related and oxidative stress-related genes, such as FN, α-SMA, PCNA, cyclin D1, NOX2 and NOX4, were increased in the primary fibroblasts from UIRI kidneys, compared with that from sham group. Consistently, double immunofluorescence staining confirmed the co-localization of NOX4 and α-SMA in these primary fibroblasts from UIRI mice (Fig. S7G).
We also investigated the signal pathway involved in mediating NOX4 action. As shown in Fig. 7G–J, the phosphorylation and activation of PKCα, p38 MAPK and ERK1/2 were evident in the kidney of the AOPPs-treated UIRI mice, which was largely abolished by NOX4 depletion. These results suggest that NOX4/PKCα/MAPK signal cascade plays a role in mediating fibroblast activation and proliferation in fibrotic kidney.
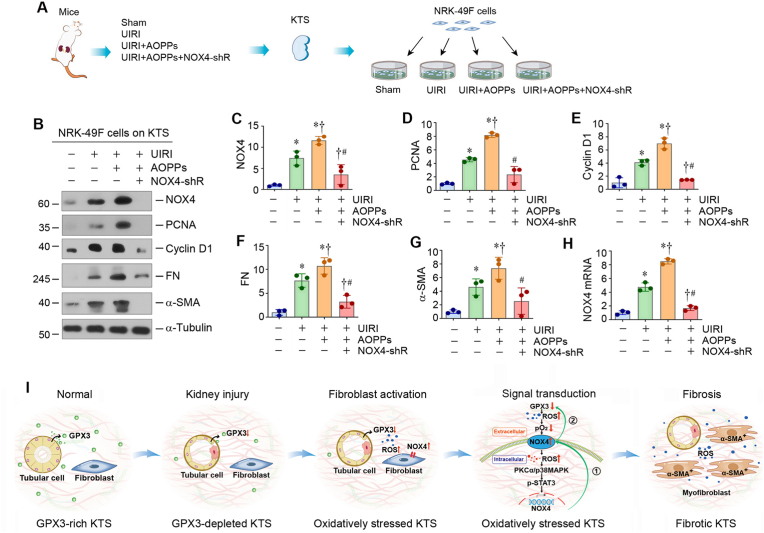
3.8. Knockdown of NOX4 inhibits fibroblast activation by improving extracellular microenvironment
As NOX4 is a catalytic component of the membrane-bound enzyme complex that faces the extracellular space, we speculated that it may function as an oxygen sensor in response to excessive ROS in the extracellular compartment due to the loss of GPX3. To test this hypothesis, we prepared the decellularized KTS from various groups of mice and inoculated NRK-49F cells on them (Fig. 8A). When NRK-49F cells were cultivated on the KTS derived from AOPPs-treated UIRI mice, they expressed high level of NOX4, PCNA, cyclin D1, FN and α-SMA, compared with either sham controls or UIRI alone mice (Fig. 8B–G). However, when NRK-49F cells were seeded on the KTS derived from the NOX4-depleted and AOPPs-treated UIRI mice, they remained quiescent with little expression of NOX4, PCNA, cyclin D1, FN and α-SMA (Fig. 8B–G). Of note, the NOX4 mRNA abundance was consistent with its protein levels in these groups (Fig. 8H). These results suggest that NOX4 depletion improves the oxidatively stressed extracellular microenvironment in the fibrotic kidney, resulting in an inhibition of fibroblast activation and proliferation.
Fig. 8.
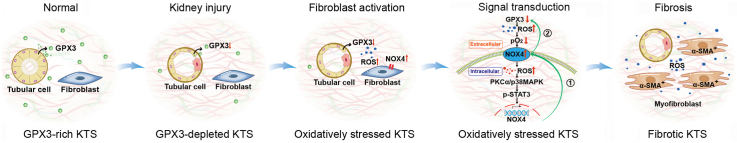
Oxidatively stressed extracellular microenvironment dictates fibroblast proliferation and activation. (A) Diagram shows the experimental protocol in which NRK-49F cells are cultured on different KTS as indicated. (B) Western blots analyses show the expression of NOX4, PCNA, cyclin D1, FN and α-SMA in different groups as indicated. Representative Western blot (B) and quantitative data (C–G) are shown. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 3). (H) qRT-PCR analyses show the mRNA level of NOX4 in different groups as indicated. *P < 0.05 versus sham; †P < 0.05 versus UIRI; #P < 0.05 versus UIRI + AOPPs + Ctrl-shRNA (n = 3). (I) Schematic diagram shows that loss of GPX3 contributes to the formation of an oxidatively stressed extracellular microenvironment, which facilitates fibroblast activation. Under normal conditions, GPX3 in the extracellular microenvironment is abundant. Kidney injury causes GPX3 deficiency, leading to forming an oxidatively stressed microenvironment. This oxidatively stressed microenvironment leads to low tissue oxygen level (pO2), which triggers plasma membrane NOX4, an oxygen sensor, and induces intracellular ROS production and upregulates NOX4 expression via PKCα/p38 MAPK/STAT3 cascade in fibroblasts. As such, this series of events cause the formation of the NOX4 amplification loop (①). In addition, the increased NOX4 also further suppresses GPX3 expression in renal tubules, thereby creating a vicious cycle between GPX3 loss and NOX4 induction (②).
4. Discussion
Kidney fibrosis initiates at certain focal sites where the fibrogenic niche is formed, which provides a specialized tissue microenvironment promoting fibroblast activation. In this study, we demonstrate that oxidative stress due to antioxidant GPX3 depletion is an integral element of the fibrogenic niche. As depicted in Fig. 8I, kidney injury inhibits GPX3 expression and secretion by tubular cells, causing its depletion in the extracellular microenvironment. This triggers NOX4, an oxygen sensor, activation in neighboring interstitial fibroblasts, leading to further NOX4 induction through ROS/PKCα/MAPK/STAT3 cascade. As such, GPX3 loss in renal tubules leads to the formation of NOX4 amplification loop in renal interstitial fibroblasts (Fig. 8I①) and promotes their proliferation and myofibroblastic transformation. In supporting this view, renal fibroblasts cultured on GPX3-depleted ECM scaffold are spontaneously stimulated to proliferate and produce a large amount of matrix proteins. These findings illustrate the fundamental role of extracellular microenvironment in dictating fibroblasts activation and kidney fibrosis.
Oxidative stress is defined as a disturbance in the balance between the production of ROS and antioxidant defenses. Through RNA-seq transcriptome analysis, we found that hypoxia pathway is enriched in the fibrotic kidney (Fig. 1), suggesting that oxidative stress could play a critical role in kidney fibrogenesis. The GPXs are antioxidant enzyme family consisting of eight isozymes (GPX1–8) that catalyze the reduction of hydrogen peroxide, hydroperoxides, and lipid hydroperoxides by reduced glutathione, thereby controlling the cellular redox environment and protecting from oxidative damage [36]. Of all 8 GPXs, GPX3 is quite unique in that it is secreted into the extracellular fluid and plasma [15]. Under normal conditions, plasma GPX3 is mainly originated from the kidney, which accounts for 70% GPX3 in human body [17]. Studies have shown that GPX3 has a protective effect on diabetic nephropathy [37,38]. Of interest, only GPX3 mRNA level is consistently downregulated in UUO, whereas GPX7 and GPX8 are actually upregulated in the fibrotic kidney (Fig. 1). Loss of GPX3 is further confirmed in 3 well-characterized mouse models of renal fibrosis (Fig. 1) [34]. Notably, GPX3 is produced and secreted by normal kidney tubular epithelial cells and stored in the peritubular interstitial compartment of the kidney by binding to tubular basement membrane [18]. In this context, the loss of tubule-derived GPX3 will disrupt the antioxidant defenses machinery, leading to forming an oxidatively stressed interstitial microenvironment that drives fibroblasts activation. In essence, loss of GPX3 in renal tubules mediates fibroblast activation via building an oxidatively stressed extracellular microenvironment in CKD.
The present study further delineates the mechanism in which GPX3-depletion causes fibroblast activation. We found that GPX3-depleted ECM microenvironment induces the expression of NOX4, a subunit of the membrane-bound NOX enzyme complex that faces the extracellular space and functions as an oxygen sensor, in renal fibroblasts. Consistently, loss of GPX3 is associated with NOX4 induction in various models of CKD in vivo (Fig. 1). Notably, when fibroblasts are stimulated with AOPPs, NOX4 is also upregulated, suggesting that loss of GPX3 can be imitated by AOPPs treatment. There are 7 isoforms in the NOX enzyme family including NOX1–5, DUOX1 and DUOX2 [[39], [40], [41]]. Of these NOX isoforms, NOX4 is expressed at its highest levels in the kidney and is an important source of renal ROS [9,20]. We found that although AOPPs induces several NOX enzymes such as NOX1, NOX2, NOX4 and DUOX1, the induction of NOX4 is the highest (Fig. S1), suggesting that AOPPs-induced ROS production is mainly derived from NOX4 in renal fibroblasts.
The ROS produced by NOX is regulated by a variety of factors, depending on the cell types and the variants of NOX homologues [21,42,43]. The most striking difference between NOX4 and other NOX/DUOX enzymes is that NOX4 is constitutively active in the absence of regulatory subunits or calcium-elevating stimuli. Therefore, NOX4-dependent ROS generation is primarily regulated by its expression level. Apart from expression levels, studies also show that the activity of NOX4 can be acutely regulated by tissue oxygen levels (pO2), allowing it to function as an oxygen sensor [22]. Therefore, it is conceivable to speculate that loss of extracellular GPX3 leads to accumulation of ROS formed in normal oxygen metabolism in the peritubular tissue compartment. This decreases tissue oxygen levels, triggers upregulation and activation of the plasma membrane-bound NOX4, and promotes intracellular ROS production, eventually leading to further NOX4 induction and fibroblast activation and proliferation via PKCα/MAPK/STAT3 signaling (Fig. 8I). Furthermore, NOX4 upregulation further represses GPX3 expression in renal tubules, thereby creating a vicious cycle between GPX3 depletion and NOX4 induction (Fig. 6, Fig. 8②).
This study also explores the intracellular signaling by which NOX4-derived ROS induces NOX4 expression and fibroblast activation. We show that NOX4 induction by AOPPs triggers ROS production and activates PKCα/MAPK/STAT3 signaling in renal fibroblasts, resulting in further NOX4 induction and fibroblast proliferation and activation (Fig. 3, Fig. 4). These results are in harmony with previous report that NOX4-derived ROS can activate specific PKC isoforms including PKCα in the kidney, thereby promoting renal injury in experimental diabetes [44]. Furthermore, specific knockout of NOX4 in podocytes is associated with inhibition of PKCα [45], and PKCα activation mediates podocytes apoptosis through activation of p38 MAPK [46]. Furthermore, knockdown of NOX4 abolishes PKCα, p38 and ERK1/2 activation and hinders fibroblasts activation (Fig. 5). Similarly, blockade of p38 and ERK1/2 activation by small molecule MAPK kinase inhibitor U0126 or p38 inhibitor SB203580 represses fibroblast proliferation and activation (Fig. 5), consistent with earlier reports on the role of MAPK in the pathogenesis of CKD [47]. These results suggest that NOX4 promotes fibroblast activation primarily by stimulating PKCα/MAPK signaling cascade, although we cannot exclude the possibility that other signaling pathways may also be involved.
The present study has some limitations and leaves several questions unanswered. For example, while the importance of fibroblast activation by NOX4 is emphasized, the shRNA-mediated knockdown approach is not cell type-specific and also affects tubular epithelial cells expressing NOX4 (Fig. 6). A more precise strategy to manipulate NOX4 specifically in fibroblasts in vivo, such as creating a conditional fibroblast-specific NOX4 knockout mouse model, is needed in the future to address this issue. Of note, inhibition of NOX4 in tubular cells restores GPX3 expression and its secretion to the extracellular space (Fig. 6), thereby indirectly reducing NOX4 induction in fibroblast cells as well. Another unresolved issue is whether the GPX3-NOX4 axis is involved in regulating metabolic reprogram from mitochondrial oxidative phosphorylation to aerobic glycolysis, as emerging evidence suggests that fibroblast activation in kidney fibrosis is associated with the metabolic transition from oxidative phosphorylation to aerobic glycolysis [[48], [49], [50], [51]]. This area of investigation deserves to be pursued in the future.
In summary, by using transcriptomic profiling and decellularized KTS approach, we have presented clear evidence that GPX3-depleted extracellular microenvironment is an integral element of the fibrogenic niche that drives fibroblast proliferation and activation. We show that loss of GPX3 creates an oxidatively stressed peritubular microenvironment that promotes interstitial fibroblast activation, proliferation and further NOX4 induction through the NOX4/ROS/PKCα/MAPK/STAT3 signaling. Although oxidative stress is known to play a role in the evolution of CKD [52], previous studies largely focused on its intracellular signaling and implications. In this regard, the present study is the first to demonstrate how an oxidatively stressed peritubular environment due to GPX3 deficiency drives fibroblast activation and kidney fibrosis. Therefore, it is reasonable to speculate that strategies to target such an oxidatively stressed microenvironment may be a novel approach for prevention and therapy of fibrotic CKD.
Funding
This work was supported by the National Key Research and Development Program of China grant 2022YFC2502504, National Natural Science Foundation of China (NSFC) grants 81920108007 and 82230020, Key Technologies R&D Program of Guangdong Province (2023B1111030004) and the funds from Guangdong Provincial Key Laboratory of Renal Failure and Guangdong Provincial Clinical Research Center for Kidney Disease. L.L. was supported by NSFC grants 82100785 and 82370692, China Postdoctoral Science Foundation grants 2021M691471 and 2023T160294.
Author contributions
Y.L. and L.Z. conceived the project. L.L., L.Z. and Y.L. designed the studies. L.L., M.L., Y.P., J.H., X.T., J.L., J.C., M.H. and X.H. performed the experiments. L.L., M.L., H.F, R.L., F.F.H., L.Z. and Y.L. analyzed and interpreted the data. L.L. and Y.L. wrote and revised the manuscript. All authors read and approved the final paper.
Data availability
Transcriptomic data produced in this study are available at NCBI with accession number PRJNA846588. Data will be made available on request.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102868.
Contributor Information
Li Li, Email: guilinlily3@i.smu.edu.cn.
Lili Zhou, Email: jinli730@smu.edu.cn.
Youhua Liu, Email: liuyh@smu.edu.cn.
Appendix. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 2011;7:684–696. doi: 10.1038/nrneph.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphreys B.D. Mechanisms of renal fibrosis. Annu. Rev. Physiol. 2018;80:309–326. doi: 10.1146/annurev-physiol-022516-034227. [DOI] [PubMed] [Google Scholar]
- 3.Li L., Fu H., Liu Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat. Rev. Nephrol. 2022;18:545–557. doi: 10.1038/s41581-022-00590-z. [DOI] [PubMed] [Google Scholar]
- 4.Fu H., Tian Y., Zhou L., Zhou D., Tan R.J., Stolz D.B., Liu Y. Tenascin-C is a major component of the fibrogenic niche in kidney fibrosis. J. Am. Soc. Nephrol. 2017;28:785–801. doi: 10.1681/ASN.2016020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovisa S., Zeisberg M., Kalluri R. Partial epithelial-to-mesenchymal transition and other new mechanisms of kidney fibrosis. Trends Endocrinol. Metabol. 2016;27:681–695. doi: 10.1016/j.tem.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhou D., Liu Y. Renal fibrosis in 2015: understanding the mechanisms of kidney fibrosis. Nat. Rev. Nephrol. 2016;12:68–70. doi: 10.1038/nrneph.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeBleu V.S., Taduri G., O'Connell J., Teng Y., Cooke V.G., Woda C., Sugimoto H., Kalluri R. Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 2013;19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan Q., Tan R.J., Liu Y. Myofibroblast in kidney fibrosis: origin, activation, and regulation. Adv. Exp. Med. Biol. 2019;1165:253–283. doi: 10.1007/978-981-13-8871-2_12. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Uribe A.P., Gomez-Sierra T., Aparicio-Trejo O.E., Orozco-Ibarra M., Pedraza-Chaverri J. Backstage players of fibrosis: NOX4, mTOR, HDAC, and S1P; companions of TGF-beta. Cell. Signal. 2021;87 doi: 10.1016/j.cellsig.2021.110123. [DOI] [PubMed] [Google Scholar]
- 10.Forman H.J., Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021;20:689–709. doi: 10.1038/s41573-021-00233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costantini D. Understanding diversity in oxidative status and oxidative stress: the opportunities and challenges ahead. J. Exp. Biol. 2019;222:jeb194688. doi: 10.1242/jeb.194688. [DOI] [PubMed] [Google Scholar]
- 12.Thannickal V.J., Jandeleit-Dahm K., Szyndralewiez C., Torok N.J. Pre-clinical evidence of a dual NADPH oxidase 1/4 inhibitor (setanaxib) in liver, kidney and lung fibrosis. J. Cell Mol. Med. 2023;27:471–481. doi: 10.1111/jcmm.17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brigelius-Flohe R., Maiorino M. Glutathione peroxidases. Biochim. Biophys. Acta. 2013;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Chang C., Worley B.L., Phaeton R., Hempel N. Extracellular glutathione peroxidase GPx3 and its role in cancer. Cancers. 2020;12:2197. doi: 10.3390/cancers12082197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauffe R., Stein V., Chudoba C., Flore T., Rath M., Ritter K., Schell M., Wardelmann K., Deubel S., Kopp J.F., Schwarz M., Kappert K., Bluher M., Schwerdtle T., Kipp A.P., Kleinridders A. GPx3 dysregulation impacts adipose tissue insulin receptor expression and sensitivity. JCI Insight. 2020;5 doi: 10.1172/jci.insight.136283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nirgude S., Choudhary B. Insights into the role of GPX3, a highly efficient plasma antioxidant, in cancer. Biochem. Pharmacol. 2021;184 doi: 10.1016/j.bcp.2020.114365. [DOI] [PubMed] [Google Scholar]
- 17.Whitin J.C., Bhamre S., Tham D.M., Cohen H.J. Extracellular glutathione peroxidase is secreted basolaterally by human renal proximal tubule cells. Am. J. Physiol. Ren. Physiol. 2002;283:F20–F28. doi: 10.1152/ajprenal.00014.2001. [DOI] [PubMed] [Google Scholar]
- 18.Olson G.E., Whitin J.C., Hill K.E., Winfrey V.P., Motley A.K., Austin L.M., Deal J., Cohen H.J., Burk R.F. Extracellular glutathione peroxidase (Gpx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am. J. Physiol. Ren. Physiol. 2010;298:F1244–F1253. doi: 10.1152/ajprenal.00662.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 20.Munoz M., Lopez-Oliva M.E., Rodriguez C., Martinez M.P., Saenz-Medina J., Sanchez A., Climent B., Benedito S., Garcia-Sacristan A., Rivera L., Hernandez M., Prieto D. Differential contribution of Nox1, Nox2 and Nox4 to kidney vascular oxidative stress and endothelial dysfunction in obesity. Redox Biol. 2020;28 doi: 10.1016/j.redox.2019.101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babelova A., Avaniadi D., Jung O., Fork C., Beckmann J., Kosowski J., Weissmann N., Anilkumar N., Shah A.M., Schaefer L., Schroder K., Brandes R.P. Role of Nox4 in murine models of kidney disease. Free Radic. Biol. Med. 2012;53:842–853. doi: 10.1016/j.freeradbiomed.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 22.Nisimoto Y., Diebold B.A., Cosentino-Gomes D., Lambeth J.D. Nox4: a hydrogen peroxide-generating oxygen sensor. Biochemistry. 2014;53:5111–5120. doi: 10.1021/bi500331y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vermot A., Petit-Hartlein I., Smith S.M.E., Fieschi F. NADPH oxidases (NOX): an overview from discovery. Molecular Mech. Physiol. Pathol., Antioxidants (Basel) 2021;10:890. doi: 10.3390/antiox10060890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Liao J., Yuan Q., Hong X., Li J., Peng Y., He M., Zhu H., Zhu M., Hou F.F., Fu H., Liu Y. Fibrillin-1-enriched microenvironment drives endothelial injury and vascular rarefaction in chronic kidney disease. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abc7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song D., Shang J., Long Y., Zhong M., Li L., Chen J., Xiang Y., Tan H., Zhu H., Hong X., Hou F.F., Fu H., Liu Y. Insulin-like growth factor 2 mRNA-binding protein 3 promotes kidney injury by regulating beta-catenin signaling. JCI Insight. 2022;8 doi: 10.1172/jci.insight.162060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou L., Chen X., Lu M., Wu Q., Yuan Q., Hu C., Miao J., Zhang Y., Li H., Hou F.F., Nie J., Liu Y. Wnt/beta-catenin links oxidative stress to podocyte injury and proteinuria. Kidney Int. 2019;95:830–845. doi: 10.1016/j.kint.2018.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limsirichaikul S., Niimi A., Fawcett H., Lehmann A., Yamashita S., Ogi T. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU) Nucleic Acids Res. 2009;37:e31. doi: 10.1093/nar/gkp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H., Liao J., Zhou X., Hong X., Song D., Hou F.F., Liu Y., Fu H. Tenascin-C promotes acute kidney injury to chronic kidney disease progression by impairing tubular integrity via alphavbeta6 integrin signaling. Kidney Int. 2020;97:1017–1031. doi: 10.1016/j.kint.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian X.J., Zhou D., Fu H., Zhang R., Wang X., Huang S., Liu Y., Xing J. Sequential Wnt agonist then antagonist treatment accelerates tissue repair and minimizes fibrosis. iScience. 2020;23 doi: 10.1016/j.isci.2020.101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeisberg M., Tampe B., LeBleu V., Tampe D., Zeisberg E.M., Kalluri R. Thrombospondin-1 deficiency causes a shift from fibroproliferative to inflammatory kidney disease and delays onset of renal failure. Am. J. Pathol. 2014;184:2687–2698. doi: 10.1016/j.ajpath.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W., Liu C., Farrar C.A., Ma L., Dong X., Sacks S.H., Li K., Zhou W. Collectin-11 promotes the development of renal tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 2018;29:168–181. doi: 10.1681/ASN.2017050544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L., He M., Tang X., Huang J., Li J., Hong X., Fu H., Liu Y. Proteomic landscape of the extracellular matrix in the fibrotic kidney. Kidney Int. 2023;103:1063–1076. doi: 10.1016/j.kint.2023.01.021. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L.L., Hou F.F., Wang G.B., Yang F., Xie D., Wang Y.P., Tian J.W. Accumulation of advanced oxidation protein products induces podocyte apoptosis and deletion through NADPH-dependent mechanisms. Kidney Int. 2009;76:1148–1160. doi: 10.1038/ki.2009.322. [DOI] [PubMed] [Google Scholar]
- 36.Brigelius-Flohe R., Flohe L. Regulatory phenomena in the glutathione peroxidase superfamily. Antioxidants Redox Signal. 2020;33:498–516. doi: 10.1089/ars.2019.7905. [DOI] [PubMed] [Google Scholar]
- 37.Roumeliotis A., Roumeliotis S., Tsetsos F., Georgitsi M., Georgianos P.I., Stamou A., Vasilakou A., Kotsa K., Tsekmekidou X., Paschou P., Panagoutsos S., Liakopoulos V. Oxidative stress genes in diabetes mellitus type 2: association with diabetic kidney disease. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/2531062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zitouni K., Steyn M., Lyka E., Kelly F.J., Cook P., Ster I.C., Earle K.A. Derepression of glomerular filtration, renal blood flow and antioxidant defence in patients with type 2 diabetes at high-risk of cardiorenal disease. Free Radic. Biol. Med. 2020;161:283–289. doi: 10.1016/j.freeradbiomed.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Sedeek M., Nasrallah R., Touyz R.M., Hebert R.L. NADPH oxidases, reactive oxygen species, and the kidney: friend and foe. J. Am. Soc. Nephrol. 2013;24:1512–1518. doi: 10.1681/ASN.2012111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griendling K.K. Novel NAD(P)H oxidases in the cardiovascular system. Heart. 2004;90:491–493. doi: 10.1136/hrt.2003.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambeth J.D., Kawahara T., Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free Radic. Biol. Med. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asaba K., Tojo A., Onozato M.L., Goto A., Quinn M.T., Fujita T., Wilcox C.S. Effects of NADPH oxidase inhibitor in diabetic nephropathy. Kidney Int. 2005;67:1890–1898. doi: 10.1111/j.1523-1755.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 43.Spanier G., Xu H., Xia N., Tobias S., Deng S., Wojnowski L., Forstermann U., Li H. Resveratrol reduces endothelial oxidative stress by modulating the gene expression of superoxide dismutase 1 (SOD1), glutathione peroxidase 1 (GPx1) and NADPH oxidase subunit (Nox4) J. Physiol. Pharmacol. 2009;60(Suppl 4):111–116. [PubMed] [Google Scholar]
- 44.Thallas-Bonke V., Jha J.C., Gray S.P., Barit D., Haller H., Schmidt H.H., Coughlan M.T., Cooper M.E., Forbes J.M., Jandeleit-Dahm K.A. Nox-4 deletion reduces oxidative stress and injury by PKC-alpha-associated mechanisms in diabetic nephropathy. Phys. Rep. 2014;2 doi: 10.14814/phy2.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jha J.C., Thallas-Bonke V., Banal C., Gray S.P., Chow B.S., Ramm G., Quaggin S.E., Cooper M.E., Schmidt H.H., Jandeleit-Dahm K.A. Podocyte-specific Nox4 deletion affords renoprotection in a mouse model of diabetic nephropathy. Diabetologia. 2016;59:379–389. doi: 10.1007/s00125-015-3796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goncalves G.L., Costa-Pessoa J.M., Thieme K., Lins B.B., Oliveira-Souza M. Intracellular albumin overload elicits endoplasmic reticulum stress and PKC-delta/p38 MAPK pathway activation to induce podocyte apoptosis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-36933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grande M.T., López-Novoa J.M. Therapeutical relevance of MAP-kinase inhibitors in renal diseases: current knowledge and future clinical perspectives. Curr. Med. Chem. 2008;15:2054–2070. doi: 10.2174/092986708785132889. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X., Jiang L., Long M., Wei X., Hou Y., Du Y. Metabolic reprogramming and renal fibrosis. Front. Med. 2021;8 doi: 10.3389/fmed.2021.746920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Q., Su J., Dong G., Zhang M., Huo Y., Dong Z. Glycolysis inhibitors suppress renal interstitial fibrosis via divergent effects on fibroblasts and tubular cells. Am. J. Physiol. Ren. Physiol. 2019;316:F1162–f1172. doi: 10.1152/ajprenal.00422.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding H., Jiang L., Xu J., Bai F., Zhou Y., Yuan Q., Luo J., Zen K., Yang J. Inhibiting aerobic glycolysis suppresses renal interstitial fibroblast activation and renal fibrosis. Am. J. Physiol. Ren. Physiol. 2017;313:F561–f575. doi: 10.1152/ajprenal.00036.2017. [DOI] [PubMed] [Google Scholar]
- 51.Pagano G., Tiano L., Pallardó F.V., Lyakhovich A., Mukhopadhyay S.S., Di Bartolomeo P., Zatterale A., Trifuoggi M. Re-definition and supporting evidence toward Fanconi Anemia as a mitochondrial disease: prospects for new design in clinical management. Redox Biol. 2021;40 doi: 10.1016/j.redox.2021.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thallas-Bonke V., Jandeleit-Dahm K.A., Cooper M.E. Nox-4 and progressive kidney disease. Curr. Opin. Nephrol. Hypertens. 2015;24:74–80. doi: 10.1097/MNH.0000000000000082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic data produced in this study are available at NCBI with accession number PRJNA846588. Data will be made available on request.