Abstract
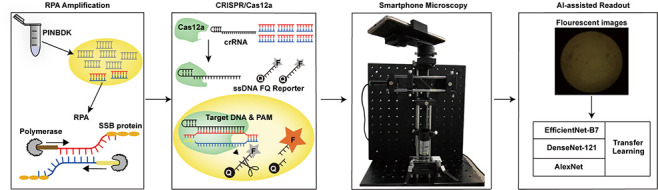
A fast, easy-to-implement, highly sensitive, and point-of-care (POC) detection system for frog virus 3 (FV3) is proposed. Combining recombinase polymerase amplification (RPA) and CRISPR/Cas12a, a limit of detection (LoD) of 100 aM (60.2 copies/μL) is achieved by optimizing RPA primers and CRISPR RNAs (crRNAs). For POC detection, smartphone microscopy is implemented, and an LoD of 10 aM is achieved in 40 min. The proposed system detects four positive animal-derived samples with a quantitation cycle (Cq) value of quantitative PCR (qPCR) in the range of 13 to 32. In addition, deep learning models are deployed for binary classification (positive or negative samples) and multiclass classification (different concentrations of FV3 and negative samples), achieving 100 and 98.75% accuracy, respectively. Without temperature regulation and expensive equipment, the proposed RPA-CRISPR/Cas12a combined with smartphone readouts and artificial-intelligence-assisted classification showcases the great potential for FV3 detection, specifically POC detection of DNA virus.
1. Introduction
Ranaviruses, including the type species frog virus 3 (FV3), are double-stranded DNA (dsDNA) viruses within the family Iridoviridae.1,2 Diseases caused by ranaviruses are a major concern for biodiversity and aquaculture.3−7 FV3 infections induce hematopoietic necrosis in the bone marrow, lesions in the skin and hematopoietic tissue, and necrosis of the lymphoid tissue and mucosal epithelium, resulting in the death of hosts within a few days to several weeks.8,9 FV3 causes 90–100% mortality in a wide range of amphibian species, which significantly contributes to the global decline in the amphibian population.10−12 However, no effective preventative vaccines exist in the market.13 Therefore, a rapid and accurate detection system is urgently required to prevent the spread of FV3. No single gold standard test has been established for FV3 detection.12 Traditional detection methods include histopathology examination, virus isolation, enzyme-linked immunosorbent assay, polymerase chain reaction (PCR), and quantitative real-time PCR (qPCR).14 The major capsid (MCP) gene is highly conserved and generally selected as the target for detecting ranaviruses using PCR. The limits of detection (LoDs) by PCR and qPCR for FV3 have not been reported. As a reference, when targeting the MCP gene, the LoDs for largemouth bass virus (LMBV) by PCR and qPCR are 1000 and 5.8 copies/μL, respectively, in over 60 min.15 Most animal samples are currently inspected and quarantined in the laboratory, which require bulky and expensive instruments.12−15 These common methods are complicated, costly, and time consuming for point-of-care (POC) detection.
Recently, clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) protein have attracted substantial attention for nucleic acid detection and have shown promising results.16−24 The Cas12a protein guided by CRISPR RNA (crRNA) binds and cleaves target DNA, unleashing nonspecific single-stranded DNA (ssDNA) trans-cleavage activity,25−27 which makes it suitable for nucleic acid detection.9,28−30 We previously developed a CRISPR/Cas12a-based automated, integrated, and inexpensive detection system for African swine fever virus (ASFV), achieving a detection limit of 1 pM in 2 h without amplification.28 To improve the sensitivity for trace viral DNA detection, the CRISPR/Cas12a system can be combined with recombinase polymerase amplification (RPA) or loop-mediated isothermal amplification (LAMP), which has been used for detecting multiple types of pathogens such as Plasmodium falciparum, Plasmodium vivax, scale drop disease virus, ASFV, and SARS-CoV-2.30,31,32,33,34 However, LAMP has limited uses because the design of primers is complicated.35 Therefore, this study combines RPA and CRISPR/Cas12a for FV3 detection, which remains unexplored, to the best of our knowledge.
For POC detection, a handheld fluorimeter, lateral flow strips, and smartphone microscopy (SPM) have been developed.32,36,37 SPM uses a smartphone camera to capture images, which can then be uploaded to mobile applications for rapid data analysis as well as the creation of a portable, cheap, and miniaturized signal acquisition system. SPM has shown advantages in the detection of pathogens, such as H5N1, Zika virus, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), because of its portability and high sensitivity.38,39 Therefore, a portable SPM is built to detect fluorescence triggered by RPA-CRISPR/Cas12a-virus recognition.
To detect the presence and concentrations of target pathogens, experts using professional software must analyze the results, which is a time-consuming process.40 The proposed method can accurately and quickly obtain the results from fluorescence images captured by SPM. Recently, machine learning and deep learning have been used to quantify the concentrations of virus DNA from fluorescence images, which integrates the detection system and saves time.41 Conventional neural networks (CNNs) are widely used to learn features from raw pixelated images for a given classification task in an end-to-end manner.42−45 The most popular CNN-based deep learning models, such as AlexNet, DenseNet-121, and EfficientNet-B7, have been widely applied in medical image classification.46−48 Additionally, in most cases, large data sets for specific domains are unavailable, thereby necessitating the use of transfer learning.49,50 Transfer learning pretrains a deep learning model with a large data set, and the pretrained model is then used as the starting point for a new task with a small data set, and this alleviates the demand for large data sets, thereby overcoming overfitting and reducing training time.51 Herein, deep learning models with transfer learning are used to classify the fluorescence images.
In this study, a plate reader is used, and an LoD of 100 aM is achieved for purified MCP fragments with optimized RPA primers and crRNAs in 60 min. For POC detection, a portable SPM is built, and an LoD of 10 aM is achieved in 40 min. SPM outperforms commercial plate readers by systematically optimizing the optical path and components. Additionally, four animal-derived samples are used to confirm the reliability in practical application. To obtain results quickly and conveniently, three deep learning models, namely, AlexNet, DenseNet-121, and EfficientNet-B7, are used with transfer learning to achieve accurate classification based on fluorescence images captured by SPM.
2. Materials and Methods
2.1. Chemicals and Reagents
231 bp DNA fragments of the MCP gene from FV3 and the 240 bp MCP gene from the spleen and kidney necrosis virus (ISKNV, Iridoviridae) were selected as the target and control, respectively. Synthetic DNA fragments (FV3MCP and ISKNV MCP) cloned into the pUC57 vector, crRNAs, and ssDNA–fluorophore quencher (FQ) reporter were purchased from Sangon Biotech. The sequences of the DNA fragments used in this study are listed in Table S1. Pathogen inactive, nucleic acid extraction, direct-to-PCR buffer with proteinase K (PINDBK) was purchased from Ebio Biotechnology Co., Shenzhen, China. Lachnospiraceae bacterium Cas12a (LbCas12a) protein (M0653T) was purchased from New England Biolabs (Beijing), Ltd. A black microplate with 96 wells was purchased from Corning Incorporated, New York, USA. UltraSYBR Mixture (CW0957M) was purchased from CWBIO, Taizhou, China.
2.2. Protein Production and Purification
UvsX, UvsY, GP32, and Bsu plasmids were transformed in Escherichia coli BL21 (DE3) for protein expression. The expression and purification procedures followed the published protocols.51 It was noted that 0.1 mM IPTG was used to induce UvsX, UvsY, and GP32 expression.52 Moreover, 0.5 mM IPTG was used for Bsu expression. The incubation was performed at 16 °C with vigorous agitation (200 rpm) overnight for 20 h. Cells were subsequently collected by centrifugation at 5600g for 15 min at 4 °C. Pelleted cells were lysed via sonication (125 W, 15 min, pulse on for 3 s, and pause for 8 s) in a lysis buffer (25 mM HEPES pH 7.5, 150 mM KCl, 10 mM imidazole, 1 mM TCEP, 0.01% Triton X-100, and 5% glycerol) supplemented with PMSF (1 mM), lysozyme, and DNase I. The lysate was clarified by centrifugation at 10,000g for 25 min at 4 °C. One milliliter of Ni Sepharose High Performance (Cytiva) was added into the column and equilibrated with 5 to 10 column volumes of the wash buffer (500 mM NaCl; 20 mM Tris-HCL; 1 mM DTT; 500 mM Imidazole for UvsX, UvsY, and GP32; 15 mM Imidazole for Bsu; and 5% glycerol). The pretreated samples were then loaded and washed until the absorbance at 280 nm wavelength reached the baseline (about 5 column volumes). Next, the proteins were eluted with 20 to 30 mL elution buffer. To obtain a suitable sample volume for the next stage purification or enzymatic reaction, the eluted protein was pooled and concentrated to ∼40 mg/mL using a 50 kDa cutoff ultrafiltration unit (Millipore). Finally, the purity of proteins was confirmed using SDS-PAGE (Figure S1C).
2.3. Target Amplification
A PCR amplification kit (Takara Bio, Japan) was used to obtain the purified dsDNA target and control with primers listed in Table S1. The PCR was run on a series multi-Block thermal Cycler PCR instrument (LongGene, China) with the following procedures: 95 °C for 5 min (1 cycle); 95 °C for 30 s, 60 °C for 45 s, and 72 °C for 30 s (35 cycles); 72 °C for 5 min (1 cycle); and 4 °C for storage. Agarose gel electrophoresis was used to confirm the DNA size, and dsDNA fragments were extracted and purified from the gel (Omega Bio-tek, D2500-02). The purified DNA fragments were quantified using a NanoDrop 2000 spectrophotometer.
2.4. RPA Reaction
RPA primers were designed as described earlier.44 The sequences of the primers are listed in Table S1. The 50 μL RPA reaction contains 1 μL of the genome sample (with various concentrations), 0.2 μM forward and reverse primer, 16.2 μM UvsX, 5.5 μM UvsY, 7.2 μM Gp32, 4.5 μM Bsu, and reaction buffer (TwistDx, Ltd., UK). Next, 100 mM MgCl2 was used to initiate the reaction. The assay was performed at 37 °C for 30 min.53
2.5. CRISPR/Cas12a Detection
crRNAs were designed as described in our previous work.28 LbCas12a–crRNA complexes were preassembled by incubating 1 μM LbCas12a and 1.25 μM crRNA at 37 °C for 5 min. RPA reaction product (1 μL) was dissolved in 1× binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, and 0.1 mg/mL BSA) mixed with LbCas12a–crRNA complexes and 500 nM ssDNA reporter probe in a reaction volume of 100 μL (Figure 1). Reactions were performed at 37 °C for 30 min. Fluorescence signals were continuously examined using a microplate reader (SPARK, TECAN, Switzerland) at an excitation wavelength of 535 nm, an emission wavelength of 595 nm, and a gain of 60. For POC detection, reactions were performed at 37 °C for 10 min, and fluorescence signals were measured by SPM. Various purified targets were used to obtain a standard curve with SPM, including 10 aM, 100 aM, 1 fM, 10 fM, 1 pM, and 100 pM. Moreover, 100 pM ISKNV MCP was used as the control (before RPA). All data were normalized by negative control (divide the control value by the positive sample’s measurement) and then integrated for the two-sample t-test analysis.
Figure 1.
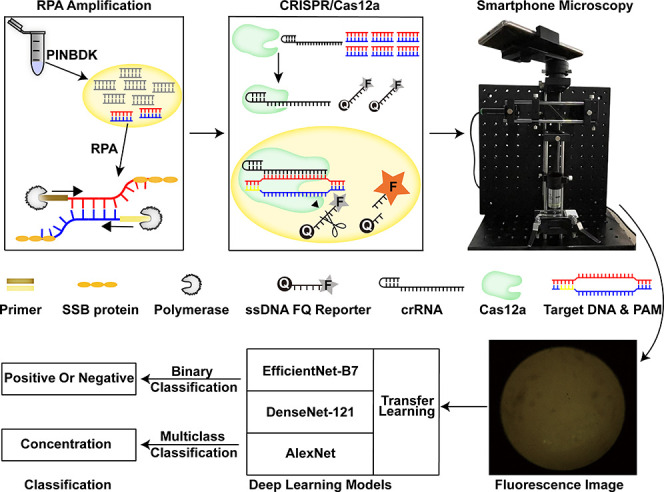
Schematic of the RPA-CRISPR/Cas12-SPM-AI detection system. The nucleic acids of animal-derived samples are released by PINDBK. Target DNA of the virus is amplified and recognized specifically by the RPA-CRISPR/Cas12a system. The Cas12a–crRNA complex binds to the target DNA, which triggers the collateral cleavage of Cas12a on the reporter (FQ-labeled ssDNA probe). The fluorophore on the reporter is then released, and the fluorescence is detected by the developed SPM. Three different deep learning models, including AlexNet, DenseNet-121, and EfficientNet-B7, with transfer learning, are used to classify the captured fluorescence images.
2.6. SPM Setup
The SPM was built using a 532 nm green laser with 0.9 mW output power as the light source (Thorlabs, PL201), aspherical lens (Lubang), transmitted neutral density filters (Thorlabs, ND40A), dichroic mirror (SEMROCK, FF555-Di03-25x36) with a cutoff wavelength of 535 nm (chrome, AT535), objective with 20× amplification (OLYMPUS OPLN20X), triplet achromatic lenses as the external lens (Thorlabs, TRH127-020-A), band-pass filter (SEMROCK, FF01-542/27-25), and a smartphone (Huawei Mate 10). Translation stages, filter holders, and dichroic holders were purchased from Ruicage Company. The instrument consisted of two parts: excitation and emission paths. A laser beam passed through neutral density (ND) filters to modulate the laser intensity. An aspherical lens (L1) was set away from the ND filters to generate a collimated beam. The collimated beam was reflected by a dichroic mirror (DM). The objective directed the beam onto the glass slide to illuminate and excite the sample, forming the excitation path. The stage under the sample allowed for fine adjustment of the focal plane, which directed the beam to the objective’s back focal plane.54 Additionally, the emission beam was collected by the objective. On the other side of the objective, an external lens (L2) was positioned to form an intermediate image.55,56 The objective simultaneously illuminates the sample and collects the emission signal. Particularly, the filter rejected the excited light, allowing only the emitted light from the sample to reach the camera. The smartphone at the end of the emission path, set away from the external lens, recorded the fluorescence signal. The band-pass filter in front of the camera was used to optimize the detection of fluorescence signals. The glass slide and polydimethylsiloxane (PDMS) were treated as previously described.28 The setup was immobilized on a breadboard for portable deployment.
2.7. Animal-Derived Samples
The animal-derived FV3 samples collected from wild or rearing animals were provided by the Animal and Plant Inspection and Quarantine Technical Center, Shenzhen, which include tiger frog virus (TFV), Bohle iridovirus (BIV), soft-shelled turtle iridovirus (STIV), Rana grylio iridovirus (RGV), and a negative homologous sample. The information concerning the pathogens used in this study is summarized in Table S2.57 The viruses were stored with preserving fluid (Biocomma, Shenzhen, China) and inactivated at 56 °C for 30 min. To release the viral DNA, equal volumes of PINDBK were added to the samples and then incubated at 95 °C for 5 min. The number of fluorescence images captured by SPM for the animal-derived samples was 14, 15, 24, 25, and 21 for TFV, BIV, STIV, RGV, and the negative sample, respectively.
2.8. Quantitative PCR Reaction
The standard samples (purified target) and animal-derived samples were detected with the following program: predenaturation at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 10 s, annealing at 56 °C for 15 s, and extension at 72 °C for 30 s. Various concentrations (the highest concentration was 1 nM, and the subsequent samples were diluted ten times each) of the purified target were used to obtain the standard curve (Figure S3B). Moreover, PINDBK was used to obtain the viral nucleic acids from animal-derived samples and to conduct the qPCR reaction with the same procedures.
2.9. Statistical Analysis
Data were displayed as mean ± standard deviation (s.d.). Each experiment was repeated at least three times. The Student’s two-sample unpaired t-test was used for statistical analysis. Statistical significance of the data is categorized as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and N.D. means no difference. GraphPad Prism 9 was used for statistical analysis.
2.10. Data Set and Data Augmentation
The data sets with fluorescence images collected in the detection assay for animal-derived samples and purified fragments are Figure 4A and Figure S3A. ImageJ was used to measure the mean gray value of each image. For data cleaning, an intensity range [median – standard deviation, median + standard deviation] is set, and if the images’ intensities are out of the range, they were considered outliers and excluded. Subsequently, the number of images for the purified target at different concentrations was as follows: 8 for 100 pM; 9 for 1 pM; 10 for 10 fM, 1 fM, and 100 aM; 7 for 10 aM; and 20 for the control. Increasing concentration of the purified target was labeled with 0–6 in ascending order. The number of images after data cleaning for the animal-derived samples was 12, 11, 23, 22, and 20 for TFV, BIV, STIV, RGV, and the negative sample, respectively, which were labeled according to the concentrations determined by qPCR. Next, a data set was obtained comprising 162 fluorescence images. Image augmentation, including horizontal flipping, vertical flipping, and random noise, was implemented before transfer-learning-based deep learning to prevent overfitting and improve system robustness.58 The number of fluorescence images was expanded from 162 to 337 for binary classification and 398 for multiclass classification.
Figure 4.
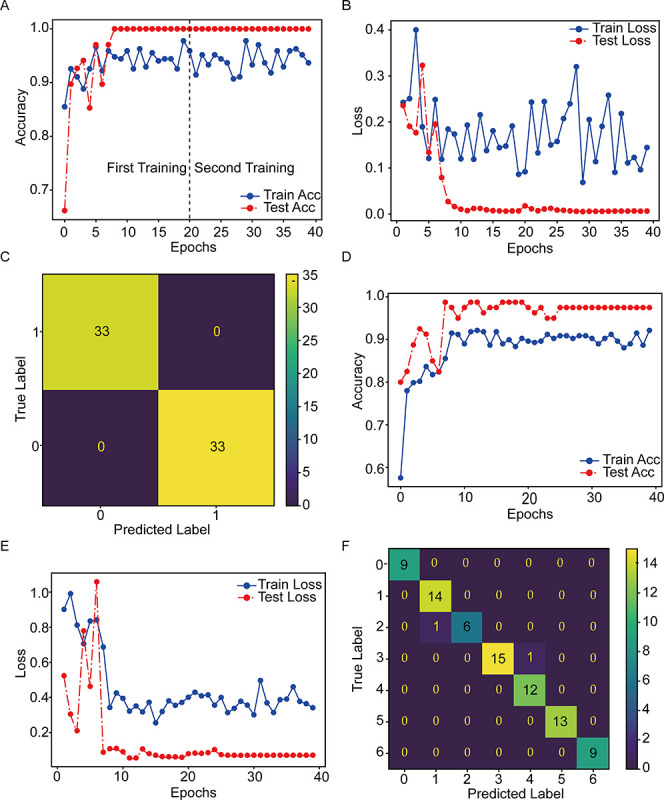
Evaluation of deep learning models with transfer learning for the classification of fluorescence images. (A,B) Training traces of EfficientNet-B7 show the accuracy and loss changes versus epoch number for binary classification. (C) Confusion matrix analysis of EfficientNet-B7 for binary classification. The values on the diagonal of the confusion matrix indicate the number of correct predictions, and the off-diagonal values are the numbers of samples with incorrect predictions. (D,E) Training traces of AlexNet show the accuracy and loss changes versus epoch number for multiclass classification. (F) Confusion matrix analysis of AlexNet for multiclass classification.
2.11. Transfer Learning
Three deep learning models were adopted as the backbone network, namely, AlexNet, DenseNet-121, and EfficientNet-B7, for binary classification and multiclass classification tasks. The input images were reshaped to 224 × 224 × 3 to satisfy the constraint of the pretrained model, which was a common preprocessing step for heterogeneous data in transfer learning. The training process was two steps. For the first stage of training, the weights of intermediate hidden layers were leveraged, and a pretrained backbone network was employed for feature extraction using the ImageNet data set. Next, the last fully connected layer having 1000 neurons for the ImageNet task was replaced with a fully connected layer comprising two or seven neurons for the fluorescence classification task. For the second training, the full-scale backbone network was fine-tuned with the fluorescence data set, and the learning rate of the model was dynamically adjusted to accelerate convergence via a function in PyTorch.59 The performance of the experiments was evaluated by using a series of metrics, including confusion matrix, accuracy, precision, recall, and F1-score.60
3. Results
Detection of FV3MCP with RPA-CRISPR/Cas12a
The purified target and control DNA fragments for crRNA screening and LoD quantitation were extracted (Figure S1A). Amplification and collateral cleavage efficiency contribute to the sensitivity of detection. Thus, the RPA primers and crRNAs were designed and tested for the target DNA.61−64 According to the preliminary results, the fourth pair of RPA primers and crRNA-3 were the most efficient ones for amplification and collateral cleavage, respectively (Figure S1B and Figure 2C). The CRISPR/Cas12a reaction was performed, and LoDs of 10 pM and 100 aM were achieved by crRNA-3 with and without RPA, respectively. These values were higher than those achieved by crRNA-1 and crRNA-2 (Figure 2A,B). The integrated fluorescence signal of CRISPR/Cas12 and RPA-CRISPR/Cas12a by crRNA-3 is presented in Figure 2D. Using crRNA-3 and RPA's fourth primer, the LoD was decreased by approximately 106 times from 100 pM to 100 aM. Therefore, further experiments were performed using crRNA-3 and RPA fourth primer.
Figure 2.
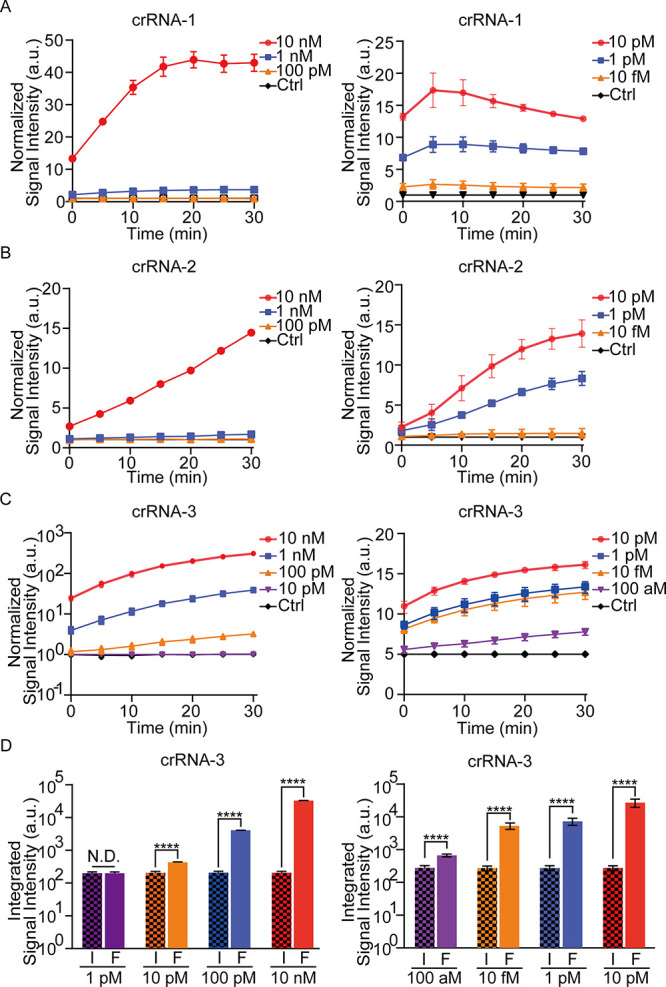
Detection of the purified fragments with three different crRNAs by the RPA-CRISPR/Cas12a system on a plate reader. (A) Normalized signal intensity of CRISPR/Cas12a (left) and RPA- CRISPR/Cas12a (right) by crRNA-1 for 10 nM to 100 pM target DNA versus 10 nM control DNA (left) and for 10 pM to 10 fM target DNA versus 10 pM control DNA (right; incubation time: 30 min). (B,C) Similar experiments are performed by crRNA-2 and crRNA-3. (D) Integrated signal intensity of CRISPR/Cas12a (left) and RPA-CRISPR/Cas12a (right) by crRNA-3 for various concentrations of target DNA versus 10 nM control DNA (left) and 10 pM control DNA (right; incubation time: 30 min): F, FV3; I, ISKNV. Data represent the mean ± s.d. from at least three independent experiments. The Student’s two-sample t-test is used for statistical analysis. *p < 0.05, **p < 0.01, and ****p < 0.001. No error bar appears for certain points because it is shorter than the symbol size.
POC Detection System with SPM
For POC detection, an SPM is built to detect the fluorescence signal, generalized by the RPA-CRISPR/Cas12a reaction. The schematic and physical images of SPM are shown in Figure 3A. Standard samples were used to confirm the practicability of SPM.65 As shown in Figure S2A,B, beads with diameters of ∼1 and ∼4 μm are observed, indicating that the limit of resolution is approximately 1 μm. The images of the potato’s underground stem and stem tuber are shown in Figure S2C,D. A significant change in the SPM signal intensity could be observed in 40 min, including 30 min of RPA and 10 min of CRISPR/Cas12a reaction, as shown in Figure 3B. Therefore, the reaction time of CRISPR/Cas12a was selected to be maintained at 10 min for later experiments. SPM was used to calibrate the fluorescence signal triggered by the RPA-CRISPR/Cas12a reaction, which demonstrated that the fluorescence signal increased with the increasing concentration of target DNA. A significant change in signal intensity could be observed between the 10 aM purified target and the control, which indicated that an LoD of 10 aM was achieved (Figure 3C). Additionally, as shown in Figure S3A, the fluorescence signal linearly increased with an elevated concentration of the purified target DNA (Pearson’s R2 = 0.9404). Next, five animal-derived samples were detected, including four positive samples and one negative sample. The captured fluorescence images and subsequent analysis of the results proved that the proposed system was able to detect the animal-derived samples (Figure 3D,E). To assess the agreement between the proposed system and qPCR, the concentrations of FV3 DNA from four positive samples were compared using these two systems. The calculated concentrations of the TFV, BIV, STIV, and RGV samples when using the proposed system were 4 pM, 59 fM, 1 fM, and 10 aM, respectively. Furthermore, the respective concentrations are 12 pM, 36 fM, 15 fM, and 45 aM with qPCR (Figure 3F and Figure S3B). The Pearson’s correlation coefficient was calculated to evaluate the correlation between qPCR and the proposed system (R2 = 0.9364, p < 0.0001, Figure S3C). The results therefore showed good consistency between the proposed methodology and the standard technique, demonstrating the reliability of the former.
Figure 3.
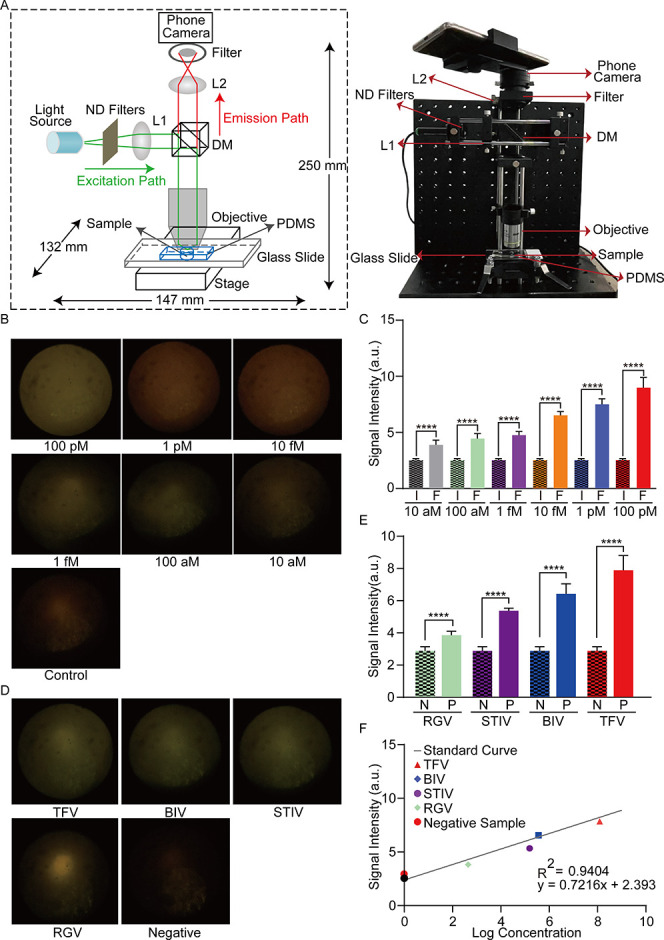
Detection of FV3 with RPA-CRISPR/Cas12a-SPM. (A) Schematic of SPM for fluorescence detection (left). The physical appearance of the assembled device used for fluorescence image collection after the RPA-CRISPR/Cas12 reaction (right). (B) Fluorescence images of RPA-CRISPR reactions with the purified target DNA or control DNA. (C) The statistics of the signal intensity for fluorescence images collected in RPA-CRISPR reactions with various concentrations of purified fragments: F, FV3; I, ISKNV. (D) The SPM images of the positive and negative animal-derived samples in the RPA-CRISPR/Cas12a detection assay. (E) The statistics of the signal intensity of fluorescence images from animal-derived samples: P, positive; N, negative. (F) The fluorescence signal intensity of animal-derived samples and their absolute concentration detected by qPCR. The Student’s two-sample t-test is used for statistical analysis. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and N.D. indicates no difference.
Classification of Fluorescence Images with Deep Learning
The difference in fluorescence signals between positive and negative samples could not be observed clearly with the naked eye (Figure 3B,D). Thus, to further improve the practicability and efficiency of the proposed detection system, the three most popular deep learning models were used along with transfer learning to achieve an accurate classification of the fluorescence images.66,67 As summarized in Table S3, Densenet-121, AlexNet, and EfficientNet-B7 achieved decent performances with 100.00% F1-score, 100.00% recall, 100.00% precision, and 100.00% accuracy for binary classification in the positive or negative group. Both the accuracy and loss curve reached a plateau, indicating an absence of overfitting (Figure 4A,B). The confusion matrix showed that all test samples were correctly classified, proving the model’s reliability (Figure 4C). Next, the ability of the models to classify fluorescence images into multiple classes was evaluated based on the concentration variation of the target DNA. AlexNet performed the best with 98.75% accuracy, 98.85% precision, 98.75% recall, 98.75% F1-score, and 15 ms inference time (Table S4). The confusion matrix showed that only 2 out of 80 test data were wrongly classified, which proves that an approximate concentration could be obtained by using transfer-learning-based deep learning models without overfitting (Figure 4D–F).
4. Discussion
In this study, FV3 is successfully detected using the proposed RPA-CRISPR/Cas12a-SPM-AI system for the first time. Multiple perspectives are investigated to improve the specificity, sensitivity, speed, and integrity of the proposed system. The collateral cleavage efficiency of CRISPR/Cas12a significantly improves the sensitivity of detection.68 A general issue for the application of Cas12a nucleases is the unpredictable success of the crRNA design.69,70 Herein, three crRNAs were designed, and crRNA-3 demonstrably triggered a stronger fluorescence signal intensity (Figure 2). Additionally, RPA-CRISPR/Cas12a decreased the LoDs of crRNA-1, crRNA-2, and crRNA-3 by approximately 104, 104, and 106 times, respectively. RPA generates the amplicons from the end to the middle of the chain with ATP, dNTP, Mg2+, and enzymes.71 Exhaustion of raw materials may lead to the production of amplicons that are not full length. Incomplete amplicons in RPA produce more termini that can amplify terminal regions, and this helps crRNA recognize these regions more efficiently. The crRNA-3 recognizes the more accessible terminal of the target DNA, while crRNA-1 or crRNA-2 binds with the middle region of the target DNA. Both similar and opposite conclusions have been reported, which must be clarified with further experiments.72,73
Another approach involves designing specific RPA primers and screening out the most efficient ones. Different primers influence the amplification efficiency of the RPA reaction.74 However, no guide exists for designing efficient and specific primers, which requires experimental validation.75 In this study, nine pairs of primers were designed, and the fourth pair demonstrated slightly stronger amplification efficiency among all candidate primers (Figure S1B). The specificity of the primers was confirmed by blasting against the standard database of nucleotide sequencing. Recombination between FV3 and the common midwife toad virus (CMTV, Ranavirus, Iridoviridae) is reported,76 which may result in cross detection, but it is very rare in nature. The minor flaw caused by evolution is hypothesized to not limit the practicability of the proposed system. Notably, 10 aM purified targets and five animal-derived samples were successfully detected (Figure 3).
For POC detection, a portable SPM was built to detect the fluorescence signal, because traditional microscopy was bulky and expensive for rapid POC diagnosis. Fluorescence microscopy had been introduced for viral particle imaging and virus detection.77,78 The SPM setup was fixed on a breadboard for portable deployment. Additionally, it cost (approximately $2000) much less than a commercialized plate reader or real-time PCR machine.
A deep-learning-assisted classification algorithm was developed for rapid SPM image classification and virus detection (15 ms for multiclass classification of one image). Transfer learning was adapted, and a maximum of 98.75% accuracy was achieved in multiclass classification (Table S4). To the best of our knowledge, this was the first time that fluorescence images from RPA-CRISPR/Cas12a, captured by SPM, were used to calculate the concentration of pathogenic DNA with deep learning.68,79,80 Three deep learning models based on transfer learning performed well on this task, and classification using fluorescence images did not require tiny features. The results indicated that a larger kernel of AlexNet (7 × 7) performs more efficiently on this task, which was deployed on smartphones for convenient and efficient application.81,82
An LoD of 10 aM was achieved with the optimized RPA primers, crRNA, and SPM, which was comparable to the detection sensitivity of qPCR. FV3 DNA can be detected by PCR in the liver at 4 days postinfection (dpi) and in most organs at 14 dpi.8 The detection sensitivity of the proposed system was much higher than that of traditional PCR. Therefore, this optimized detection system could be deployed for early diagnosis with appropriate modifications. Additionally, the collateral activity of the LbCas12a protein was constantly activated for 24 h.28 The LoD can be further potentially decreased by extending the incubation time.
The development of detection methods for FV3 has been a stagnant issue for many years. However, FV3 is becoming a global threat to biodiversity and aquaculture.83−85 In this study, an RPA-CRISPR/Cas12-AI system is established, optimized, and evaluated, providing a novel POC detection system for FV3. Furthermore, because crRNA is reprogrammable, the integrated system can be used for detecting any viral DNA target. In terms of detection sensitivity, specificity, time, and reliability, the RPA-CRISPR/Cas12a-AI system is expected to play an important role in DNA pathogen detection. In this study, we have developed the POC detection system and demonstrated its feasibility on FV3. Given its low cost, speed, specificity, and absolute quantification capability, we believe it holds promising potential for diagnosing diseases that are highly correlated to the DNA pathogen infections.
5. Conclusions
In this study, a novel, integrated, highly sensitive, and easy-to-implement POC detection system is developed for FV3. With the combination of RPA, CRISPR/Cas12a, SPM, and deep-learning-assisted classification, an LoD of 10 aM is achieved in 40 min. Without temperature regulation, this integrated POC system shows great potential for FV3 or DNA pathogen detection.
Acknowledgments
We thank the support from the National Natural Science Foundation of China 31970752; Science, Technology, Innovation Commission of Shenzhen Municipality JCYJ20220530143014032, JCYJ20190809180003689, JSGG20200225150707332, ZDSYS20200820165400003, WDZC20200820173710001, WDZC20200821150704001, and JSGG20191129110812708; Shenzhen Bay Laboratory Open Funding, SZBL2020090501004; Department of Chemical Engineering-iBHE special cooperation joint fund project, DCE-iBHE-2022-3; Tsinghua Shenzhen International Graduate School Cross-disciplinary Research and Innovation Fund Research Plan, JC2022009; and Bureau of Planning, Land and Resources of Shenzhen Municipality (2022) 207.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c02929.
Additional experimental details, materials, and results; oligonucleotides used in this study; information on animal-derived samples; deep learning models performance for binary and multiclass classification; agarose gel results of purified target and RPA primer amplification efficiency; SDS-PAGE result of purified proteins; images of standard samples with SPM; standard curve of the proposed system and qPCR; and evaluation of several deep learning models for classification of fluorescence images (PDF)
Author Contributions
& Z.L., L.L., and L.Z. contributed equally to this paper. P.Q. and D.Y. supervised the study. Z.L. performed the experiment, collected data, analyzed data, wrote the first draft of the manuscript, and wrote the response letter. L.L. designed and built up the portable smartphone microscope. L.Z. analyzed the fluorescence images using deep learning. S.Z., J.W., H.L., Y.L., Z.D., X.Y., I.G., H.Z., Z.Q., S.Z., P.J., K.D., and L.D. helped revised the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Jancovich J. K.; Bremont M.; Touchman J. W.; Jacobs B. L. Evidence for multiple recent host species shifts among the Ranaviruses (family Iridoviridae). J. Virol. 2010, 84 (6), 2636–2647. 10.1128/JVI.01991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J.; Gregory C. V. ″Ranaviruses: an emerging threat to ectothermic vertebrates″ report of the First International Symposium on Ranaviruses, Minneapolis MN July 8, 2011. Dev. Comp. Immunol. 2012, 36 (2), 259–261. 10.1016/j.dci.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Chen G.; Robert J. Antiviral immunity in amphibians. Viruses 2011, 3 (11), 2065–2086. 10.3390/v3112065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price S. J.; Garner T. W.; Nichols R. A.; Balloux F.; Ayres C.; Mora-Cabello D. A. A.; Bosch J. Collapse of amphibian communities due to an introduced Ranavirus. Curr. Biol. 2014, 24 (21), 2586–2591. [DOI] [PubMed] [Google Scholar]
- Whittington R. J.; Becker J. A.; Dennis M. M. Iridovirus infections in finfish-critical review with emphasis on ranaviruses. J. Fish Dis. 2010, 33 (2), 95–122. 10.1111/j.1365-2761.2009.01110.x. [DOI] [PubMed] [Google Scholar]
- Youker-Smith T. E.; Boersch-Supan P. H.; Whipps C. M.; Ryan S. J. Environmental Drivers of Ranavirus in Free-Living Amphibians in Constructed Ponds. ECOHEALTH 2018, 15 (3), 608–618. 10.1007/s10393-018-1350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Zhang W.; Gu C.; Chen J.; Zhao M.; Fu L.; Han J.; He M.; Xiao Q.; Xiao W.; He L.; Zhang Z. Genomic analysis of Ranavirus and exploring alternative genes for phylogenetics. Trasnbound Emerg. Dis. 2021, 68 (4), 2161–2170. 10.1111/tbed.13864. [DOI] [PubMed] [Google Scholar]
- Forzan M. J.; Jones K. M.; Ariel E.; Whittington R. J.; Wood J.; Markham R.; Daoust P. Y. Pathogenesis of Frog Virus 3 (Ranavirus, Iridoviridae) Infection in Wood Frogs (Rana sylvatica). Vet. Pathol. 2017, 54 (3), 531–548. 10.1177/0300985816684929. [DOI] [PubMed] [Google Scholar]
- Morrison E. A.; Garner S.; Echaubard P.; Lesbarreres D.; Kyle C. J.; Brunetti C. R. Complete genome analysis of a frog virus 3 (FV3) isolate and sequence comparison with isolates of differing levels of virulence. Virology 2014, 11, 46. 10.1186/1743-422X-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossainey M. R. H.; Yaparla A.; Hauser K. A.; Moore T. E.; Grayfer L. The Roles of Amphibian (Xenopus laevis) Macrophages during Chronic Frog Virus 3 Infections. VIRUSES 2021, 13 (11), 2299. 10.3390/v13112299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoverman J. T.; Gray M. J.; Haislip N. A.; Miller D. L. Phylogeny, life history, and ecology contribute to differences in amphibian susceptibility to ranaviruses. ECOHEALTH 2011, 8 (3), 301–319. 10.1007/s10393-011-0717-7. [DOI] [PubMed] [Google Scholar]
- Miller D. L.; Rajeev S.; Gray M. J.; Baldwin C. A. Frog virus 3 infection, cultured American bullfrogs. Emerg. Infect. Dis. 2007, 13 (2), 342–343. 10.3201/eid1302.061073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchar V. G.; Waltzek T. B.; Subramaniam K. Ranaviruses and other members of the family Iridoviridae: Their place in the virosphere. VIROLOGY 2017, 511, 259–271. 10.1016/j.virol.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Fong T. T.; Lipp E. K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol. Biol. Rev. 2005, 69 (2), 357–371. 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Wang Y.; Fan Z.; Zhao X.; Bergmann S. M.; Dong H.; Jin Y.; Sun D.; Mai Q.; Liu W.; Zeng W.. Establishment and evaluation of qPCR and real-time recombinase-aided amplification assays for detection of largemouth bass ranavirus. J. Fish Dis. 2022, 7, 1033, 10.1111/jfd.13627. [DOI] [PubMed] [Google Scholar]
- Bao M.; Chen Q.; Xu Z.; Jensen E. C.; Liu C.; Waitkus J. T.; Yuan X.; He Q.; Qin P.; Du K. Challenges and Opportunities for Clustered Regularly Interspaced Short Palindromic Repeats Based Molecular Biosensing. ACS Sens. 2021, 6 (7), 2497–2522. 10.1021/acssensors.1c00530. [DOI] [PubMed] [Google Scholar]
- Broughton J. P.; Deng X.; Yu G.; Fasching C. L.; Servellita V.; Singh J.; Miao X.; Streithorst J. A.; Granados A.; Sotomayor-Gonzalez A.; Zorn K.; Gopez A.; Hsu E.; Gu W.; Miller S.; Pan C. Y.; Guevara H.; Wadford D. A.; Chen J. S.; Chiu C. Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38 (7), 870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S.; Ma E.; Harrington L. B.; Da C. M.; Tian X.; Palefsky J. M.; Doudna J. A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360 (6387), 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S.; Abudayyeh O. O.; Lee J. W.; Essletzbichler P.; Dy A. J.; Joung J.; Verdine V.; Donghia N.; Daringer N. M.; Freije C. A.; Myhrvold C.; Bhattacharyya R. P.; Livny J.; Regev A.; Koonin E. V.; Hung D. T.; Sabeti P. C.; Collins J. J.; Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356 (6336), 438–442. 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass K. N.; Bao M.; He Q.; Liu L.; He J.; Park M.; Qin P.; Du K. Integrated micropillar polydimethylsiloxane accurate CRISPR detection system for viral DNA sensing. ACS Omega 2020, 5 (42), 27433–27441. 10.1021/acsomega.0c03917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul I.; Liu C.; Yuan X.; Du Z.; Zhai S.; Lei Z.; Chen Q.; Raheem M. A.; He Q.; Hu Q.; Xiao C.; Haihui Z.; Wang R.; Han S.; Du K.; Yu D.; Zhang C. Y.; Qin P. Current and perspective sensing methods for monkeypox virus. Bioengineering 2022, 9 (10), 571. 10.3390/bioengineering9100571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.; Yang C.; He Q.; Chen J.; Yu D.; Li J.; Zhai S.; Qin Z.; Du K.; Chu Z.; Qin P. Current and perspective diagnostic techniques for COVID-19. ACS Infect. Dis. 2020, 6 (8), 1998–2016. 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- Yin L.; Man S.; Ye S.; Liu G.; Ma L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541 10.1016/j.bios.2021.113541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.; Gul I.; Liu C.; Lei Z.; Li X.; Raheem M. A.; He Q.; Haihui Z.; Leeansyah E.; Zhang C. Y.; Pandey V.; Du K.; Qin P. CRISPR-Cas12-based field-deployable system for rapid detection of synthetic DNA sequence of the monkeypox virus genome. J. Med. Virol. 2023, 95 (1), e28385 10.1002/jmv.28385. [DOI] [PubMed] [Google Scholar]
- Liang M.; Li Z.; Wang W.; Liu J.; Liu L.; Zhu G.; Karthik L.; Wang M.; Wang K. F.; Wang Z.; Yu J.; Shuai Y.; Yu J.; Zhang L.; Yang Z.; Li C.; Zhang Q.; Shi T.; Zhou L.; Xie F.; Dai H.; Liu X.; Zhang J.; Liu G.; Zhuo Y.; Zhang B.; Liu C.; Li S.; Xia X.; Tong Y.; Liu Y.; Alterovitz G.; Tan G. Y.; Zhang L. X. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 2019, 10 (1), 3672. 10.1186/1743-422X-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunder E.; Rydzewski K.; Grunow R.; Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int. J. Med. Microbiol. 2013, 303 (2), 51–60. 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Zetsche B.; Gootenberg J. S.; Abudayyeh O. O.; Slaymaker I. M.; Makarova K. S.; Essletzbichler P.; Volz S. E.; Joung J.; van der Oost J.; Regev A.; Koonin E. V.; Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015, 163 (3), 759–771. 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Yu D.; Bao M.; Korensky G.; Chen J.; Shin M.; Kim J.; Park M.; Qin P.; Du K. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens. Bioelectron 2020, 154, 112068 10.1016/j.bios.2020.112068. [DOI] [PubMed] [Google Scholar]
- Li S. Y.; Cheng Q. X.; Liu J. K.; Nie X. Q.; Zhao G. P.; Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28 (4), 491–493. 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P.; Park M.; Alfson K. J.; Tamhankar M.; Carrion R.; Patterson J. L.; Griffiths A.; He Q.; Yildiz A.; Mathies R.; Du K. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4 (4), 1048–1054. 10.1021/acssensors.9b00239. [DOI] [PubMed] [Google Scholar]
- Bai J.; Lin H.; Li H.; Zhou Y.; Liu J.; Zhong G.; Wu L.; Jiang W.; Du H.; Yang J.; Xie Q.; Huang L. Cas12a-Based On-Site and Rapid Nucleic Acid Detection of African Swine Fever. Front. Microbiol. 2019, 10, 2830. 10.3389/fmicb.2019.02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. A.; Puig H.; Nguyen P. Q.; Angenent-Mari N. M.; Donghia N. M.; McGee J. P.; Dvorin J. D.; Klapperich C. M.; Pollock N. R.; Collins J. J. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. U.S.A. 2020, 117 (41), 25722–25731. 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayuramart O.; Nimsamer P.; Rattanaburi S.; Chantaravisoot N.; Khongnomnan K.; Chansaenroj J.; Puenpa J.; Suntronwong N.; Vichaiwattana P.; Poovorawan Y.; Payungporn S. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2021, 246 (4), 400–405. 10.1177/1535370220963793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukonta T.; Senapin S.; Meemetta W.; Chaijarasphong T. CRISPR-based platform for rapid, sensitive and field-deployable detection of scale drop disease virus in Asian sea bass (Lates calcarifer). J. Fish Dis. 2022, 45 (1), 107–120. 10.1111/jfd.13541. [DOI] [PubMed] [Google Scholar]
- Hema M.; Konakalla N. C. Recent developments in detection and diagnosis of plant viruses. Recent Dev. Appl. Microbiol. Biochem. 2021, 163–180. 10.1016/B978-0-12-821406-0.00016-3. [DOI] [Google Scholar]
- Fozouni P.; Son S.; Díaz de León Derby M.; Knott G. J.; Gray C. N.; D’Ambrosio M. V.; Zhao C.; Switz N. A.; Kumar G. R.; Stephens S. I.; Boehm D.; Tsou C. L.; Shu J.; Bhuiya A.; Armstrong M.; Harris A. R.; Chen P. Y.; Osterloh J. M.; Meyer-Franke A.; Joehnk B.; Walcott K.; Sil A.; Langelier C.; Pollard K. S.; Crawford E. D.; Puschnik A. S.; Phelps M.; Kistler A.; DeRisi J. L.; Doudna J. A.; Fletcher D. A.; Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184 (2), 323–333. 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.; Gulati S.; Ansari A. H.; Phutela R.; Acharya S.; Azhar M.; Murthy J.; Kathpalia P.; Kanakan A.; Maurya R.; Vasudevan J. S.; S A.; Pandey R.; Maiti S.; Chakraborty D. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. eLife 2021, 10, e67130 10.7554/eLife.67130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli A.; Ornob A.; Yu H.; Damhorst G. L.; Chen W.; Sun F.; Bhuiya A.; Cunningham B. T.; Bashir R. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed. Microdevices 2017, 19 (4), 73. 10.1007/s10544-017-0209-9. [DOI] [PubMed] [Google Scholar]
- Yeo S. J.; Choi K.; Cuc B. T.; Hong N. N.; Bao D. T.; Ngoc N. M.; Le M. Q.; Hang N. K.; Thach N. C.; Mallik S. K.; Kim H. S.; Chong C. K.; Choi H. S.; Sung H. W.; Yu K.; Park H. Smartphone-Based Fluorescent Diagnostic System for Highly Pathogenic H5N1 Viruses. Theranostics 2016, 6 (2), 231–242. 10.7150/thno.14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Chamier L.; Laine R. F.; Jukkala J.; Spahn C.; Krentzel D.; Nehme E.; Lerche M.; Hernández-Pérez S.; Mattila P. K.; Karinou E.; Holden S.; Solak A. C.; Krull A.; Buchholz T. O.; Jones M. L.; Royer L. A.; Leterrier C.; Shechtman Y.; Jug F.; Heilemann M.; Jacquemet G.; Henriques R. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat Commun 2021, 12 (1), 2276. 10.1038/s41467-021-22518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiaelis N.; Tometzki A.; Peto L.; Mcmahon A.; Hepp C.; Bickerton E.; Favard C.; Muriaux D.; Andersson M.; Oakley S.. Virus detection and identification in minutes using single-particle imaging and deep learning. Cold Spring Harbor Laboratory Press; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J.; Doshi A.; Walker B.; Bloom K. AI-Assisted Forward Modeling of Biological Structures. Front. Cell Dev. Biol. 2019, 7, 279. 10.3389/fcell.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Yan Q.; Huang J.; Chen J.; Guo Z.; Liu Z.; Cai L.; Li R.; Wang Y.; Yang G.; Lan Q. Influence of design probe and sequence mismatches on the efficiency of fluorescent RPA. World J. Microbiol. Biotechnol. 2019, 35 (6), 95. 10.1007/s11274-019-2620-2. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Hu Y.; Zhang X.; Wang S.. Two-Stage Selective Ensemble of CNN via Deep Tree Training for Medical Image Classification. IEEE Trans. Cybern.2021. [DOI] [PubMed]
- Zhang R.; Han X.; Lei Z.; Jiang C.; Gul I.; Hu Q.; Zhai S.; Liu H.; Lian L.; Liu Y.; Zhang Y.; Dong Y.; Zhang C. Y.; Lam T. K.; Han Y.; Yu D.; Zhou J.; Qin P. RCMNet: A deep learning model assists CAR-T therapy for leukemia. Comput. Biol. Med. 2022, 106084 10.1016/j.compbiomed.2022.106084. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhu H.; Wang S.; Zhang Y. D. A Review of Deep Learning on Medical Image Analysis. Mob. Netw. Appl. 2021, 26 (1), 351–380. 10.1007/s11036-020-01672-7. [DOI] [Google Scholar]
- Xie Y.; Yang H.; Yuan X.; He Q.; Zhang R.; Zhu Q.; Chu Z.; Yang C.; Qin P.; Yan C. Stroke prediction from electrocardiograms by deep neural network. Multimed. Tools Appl. 2020, 17291–7. 10.1007/s11042-020-10043-z. [DOI] [Google Scholar]
- Zhang L.; Lei Z.; Xiao C.; Du Z.; Jiang C.; Yuan X.; Hu Q.; Zhai S.; Xu L.; Liu C.; Zhong X.; Guan H.; Hassan M.; Gul I.; Pandey V.; Xing X.; Zhang C. Y.; He Q.; Qin P. AI-boosted CRISPR-Cas13a and total internal reflection fluorescence microscopy system for SARS-CoV-2 detection. Front. Sens. 2022, 35. 10.3389/fsens.2022.1015223. [DOI] [Google Scholar]
- Artoni P.; Piffer A.; Vinci V.; LeBlanc J.; Nelson C. A.; Hensch T. K.; Fagiolini M. Deep learning of spontaneous arousal fluctuations detects early cholinergic defects across neurodevelopmental mouse models and patients. Proc. Natl. Acad. Sci.USA 2020, 117 (38), 23298–23303. 10.1073/pnas.1820847116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Cai W.; Wang X.; Zhou Y.; Feng D. D.; Chen M.. Medical image classification with convolutional neural network. 13th international conference on control automation robotics & vision. ICARCV; 2014; pp 844–848.
- Yosinski J.; Clune J.; Bengio Y.; Lipson H.. How transferable are features in deep neural networks?; MIT Press. Adv Neural Inf Process Syst.2014. [Google Scholar]
- Yonesaki T.; Minagawa T. Synergistic action of three recombination gene products of bacteriophage T4, uvsX, uvsY, and gene 32 proteins. J. Biol. Chem. 1989, 264 (14), 7814–7820. 10.1016/S0021-9258(18)83114-3. [DOI] [PubMed] [Google Scholar]
- Huang Q. J.; Chen Y.; Liu H.; St-Hilaire S.; Gao S.; MacKinnon B.; Zhu S. Q.; Wen Z. Q.; Jia P.; Zheng X. C. Establishment of a real-time Recombinase Polymerase Amplification (RPA) for the detection of decapod iridescent virus 1 (DIV1). J. Virol. Methods. 2022, 300, 114377 10.1016/j.jviromet.2021.114377. [DOI] [PubMed] [Google Scholar]
- Vietz C.; Schutte M. L.; Wei Q.; Richter L.; Lalkens B.; Ozcan A.; Tinnefeld P.; Acuna G. P. Benchmarking Smartphone Fluorescence-Based Microscopy with DNA Origami Nanobeads: Reducing the Gap toward Single-Molecule Sensitivity. ACS Omega. 2019, 4 (1), 637–642. 10.1021/acsomega.8b03136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B.; Yu T.; Zhang S.; Huang Z.; Tian D.; Lin Z.; Niu A.; Golden N.; Hensley K.; Threeton B.; Lyon C. J.; Yin X. M.; Roy C. J.; Saba N. S.; Rappaport J.; Wei Q.; Hu T. Y. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2021, 7 (2), eabe3703 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samacoits A.; Nimsamer P.; Mayuramart O.; Chantaravisoot N.; Sitthi-Amorn P.; Nakhakes C.; Luangkamchorn L.; Tongcham P.; Zahm U.; Suphanpayak S.; Padungwattanachoke N.; Leelarthaphin N.; Huayhongthong H.; Pisitkun T.; Payungporn S.; Hannanta-Anan P. Machine Learning-Driven and Smartphone-Based Fluorescence Detection for CRISPR Diagnostic of SARS-CoV-2. ACS Omega. 2021, 6 (4), 2727–2733. 10.1021/acsomega.0c04929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Teng Y.; Liu H.; Lin X.; Wang K.; Jiang Y.; Chen H. Characterization of a late gene encoding for MCP in soft-shelled turtle iridovirus (STIV). Virus Res. 2007, 129 (2), 135–144. 10.1016/j.virusres.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Chlap P.; Min H.; Vandenberg N.; Dowling J.; Holloway L.; Haworth A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imag. Rad. Oncol. 2021, 65 (5), 545–563. 10.1111/1754-9485.13261. [DOI] [PubMed] [Google Scholar]
- Paszke A.; Gross S.; Chintala S.; Chanan G.; Yang E.; Devito Z.; Lin Z.; Desmaison A.; Antiga L.; Lerer A.. Automatic differentiation in PyTorch. Computer Science 2017. [Google Scholar]
- Lawton S.; Viriri S.; Versaci M. Detection of COVID-19 from CT Lung Scans Using Transfer Learning. Comput. Intel. Neurosci. 2021, 5527923. 10.1155/2021/5527923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R.; Long J.; Yuan M.; Zheng X.; Shen Y.; Jin Y.; Yang H.; Li H.; Chen S.; Duan G.. CRISPR/Cas12-Based Ultra-Sensitive and Specific Point-of-Care Detection of HBV. Int. J. Mol. Sci. 2021, 22 ( (9), ) 4842. 10.3390/ijms22094842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Duan J. J.; Wei X. Y.; Hu H.; Wang Y. B.; Jia P. P.; Pei D. S. Generation and application of a novel high-throughput detection based on RPA-CRISPR technique to sensitively monitor pathogenic microorganisms in the environment. Sci. Total Environ. 2022, 838 (Pt 2), 156048 10.1016/j.scitotenv.2022.156048. [DOI] [PubMed] [Google Scholar]
- Lu S.; Li F.; Chen Q.; Wu J.; Duan J.; Lei X.; Zhang Y.; Zhao D.; Bu Z.; Yin H. Rapid detection of African swine fever virus using Cas12a-based portable paper diagnostics. Cell Discovery 2020, 6, 18. 10.1038/s41421-020-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan Nair V.; El Salhat H.; Taha R. Z.; John A.; Ali B. R.; Elkord E. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clinical Epigenetics 2018, 10, 78. 10.1186/s13148-018-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.; Breshears L. E.; Perea S.; Morrison C. M.; Betancourt W. Q.; Reynolds K. A.; Yoon J. Y. Smartphone-Based Paper Microfluidic Particulometry of Norovirus from Environmental Water Samples at the Single Copy Level. ACS Omega. 2019, 4 (6), 11180–11188. 10.1021/acsomega.9b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosny K. M.; Kassem M. A.; Fouad M. M. Classification of Skin Lesions into Seven Classes Using Transfer Learning with AlexNet. J. Digit. Imaging. 2020, 33 (5), 1325–1334. 10.1007/s10278-020-00371-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talo M. Automated classification of histopathology images using transfer learning. Artif. Intell. Med. 2019, 101, 101743 10.1016/j.artmed.2019.101743. [DOI] [PubMed] [Google Scholar]
- Habimana J. D.; Huang R.; Muhoza B.; Kalisa Y. N.; Han X.; Deng W.; Li Z. Mechanistic insights of CRISPR/Cas nucleases for programmable targeting and early-stage diagnosis: A review. Biosens. Bioelectron. 2022, 203, 114033 10.1016/j.bios.2022.114033. [DOI] [PubMed] [Google Scholar]
- Creutzburg S.; Wu W. Y.; Mohanraju P.; Swartjes T.; Alkan F.; Gorodkin J.; Staals R.; van der Oost J. Good guide, bad guide: spacer sequence-dependent cleavage efficiency of Cas12a. Nucleic Acids Res. 2020, 48 (6), 3228–3243. 10.1093/nar/gkz1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T.; Smith B. M.; Jain P. K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020, 11 (1), 4906. 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato I. M.; O’Sullivan C. K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt. Chem. 2018, 98, 19–35. 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Huang Y.; Xiao B.; Deng H.; Gong K.; Li K.; Li L.; Hao W. Development of a RPA-CRISPR-Cas12a Assay for Rapid, Simple, and Sensitive Detection of Mycoplasma hominis. Front. Microbiol. 2022, 13, 842415 10.3389/fmicb.2022.842415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Ma J.; Li Y.; Kang L.; Yuan B.; Li S.; Chao J.; Wang L.; Wang J.; Su S.; Yuan Y. A general RPA-CRISPR/Cas12a sensing platform for Brucella spp. detection in blood and milk samples. Sens. Actuators B: Chem. 2022, 364, 131864 10.1016/j.snb.2022.131864. [DOI] [Google Scholar]
- Chen Z.; Huang J.; Zhang F.; Zhou Y.; Huang H. Detection of shrimp hemocyte iridescent virus by recombinase polymerase amplification assay. Mol. Cell 2020, 49, 101475 10.1016/j.mcp.2019.101475. [DOI] [PubMed] [Google Scholar]
- Fu X.; Sun J.; Ye Y.; Zhang Y.; Sun X. A rapid and ultrasensitive dual detection platform based on Cas12a for simultaneous detection of virulence and resistance genes of drug-resistant Salmonella. Biosens. Bioelectron. 2022, 195, 113682 10.1016/j.bios.2021.113682. [DOI] [PubMed] [Google Scholar]
- Vilaça S. T.; Bienentreu J. F.; Brunetti C. R.; Lesbarrères D.; Murray D. L.; Kyle C. J.; Shisler J. L. Frog Virus 3 Genomes Reveal Prevalent Recombination between Ranavirus Lineages and Their Origins in Canada. J. Virol. 2019, 93 (20), e00765–19. 10.1128/JVI.00765-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaraman D.; Biswas P.; Cella L. N.; Yates M. V.; Chen W. Detecting RNA viruses in living mammalian cells by fluorescence microscopy. Trends Biotechnol. 2011, 29 (7), 307–313. 10.1016/j.tibtech.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Wang I. H.; Burckhardt C.; Yakimovich A.; Greber U. Imaging, Tracking and Computational Analyses of Virus Entry and Egress with the Cytoskeleton. Viruses 2018, 10 (4), 166. 10.3390/v10040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T.; Qiu Z.; Jiang Y.; Zhu D.; Zhou X. Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. 2022, 196, 113701 10.1016/j.bios.2021.113701. [DOI] [PubMed] [Google Scholar]
- Zavvar T. S.; Khoshbin Z.; Ramezani M.; Alibolandi M.; Abnous K.; Taghdisi S. M. CRISPR/Cas-engineered technology: Innovative approach for biosensor development. Biosens. Bioelectron. 2022, 214, 114501 10.1016/j.bios.2022.114501. [DOI] [PubMed] [Google Scholar]
- Alkhulaifi A.; Alsahli F.; Ahmad I. Knowledge distillation in deep learning and its applications. PeerJ. Comput. Sci. 2021, 7, e474 10.7717/peerj-cs.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujit S. J.; Bonfante E.; Aein A.; Coronado I.; Riascos-Castaneda R.; Giancardo L. Deep learning enabled brain shunt valve identification using mobile phones. Comput. Methods Programs Biomed. 2021, 210, 106356 10.1016/j.cmpb.2021.106356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozad R. A.; Norton T. M.; Aresco M. J.; Allender M. C.; Hernandez S. M. Pathogen Surveillance and Detection of Ranavirus (Frog virus 3) in Translocated Gopher Tortoises (Gopherus polyphemus). J. Wildl. Dis. 2020, 56 (3), 679–683. 10.7589/2019-02-053. [DOI] [PubMed] [Google Scholar]
- Ferreira C. M.; Subramaniam K.; de Sousa R.; Tavares L. S.; Correa T. C.; Waltzek T. B. Genomic sequencing of a frog virus 3 strain from cultured American bullfrogs (Lithobates catesbeianus) in Brazil. Arch. Virol. 2021, 166 (7), 1961–1964. 10.1007/s00705-021-05094-y. [DOI] [PubMed] [Google Scholar]
- Oliveira C. R.; Alfaia S. R.; Ikari F. L.; Tavares L. S.; Ferreira C. M. Detection and molecular characterization of Frog virus 3 in bullfrogs from frog farms in Brazil. Aquaculture. 2019, 516, 734575 10.1016/j.aquaculture.2019.734575. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


