Abstract
Despite the widespread adoption of wastewater surveillance, more research is needed to understand the fate and transport of viral genetic markers in wastewater. This information is essential for optimizing monitoring strategies and interpreting wastewater surveillance data. In this study, we examined the solid–liquid partitioning behavior of four viruses in wastewater: SARS-CoV-2, respiratory syncytial virus (RSV), rhinovirus (RV), and F+ coliphage/MS2. We used two approaches: (1) laboratory partitioning experiments using lab-grown viruses and (2) distribution experiments using endogenous viruses in raw wastewater. Partition experiments were conducted at 4 and 22 °C. Wastewater samples were spiked with varying concentrations of each virus, solids and liquids were separated via centrifugation, and viral RNA concentrations were quantified using reverse-transcription-digital droplet PCR (RT-ddPCR). For the distribution experiments, wastewater samples were collected from six wastewater treatment plants and processed without spiking exogenous viruses; viral RNA concentrations were measured in wastewater solids and liquids. In both experiments, RNA concentrations were higher in the solid fraction than the liquid fraction by approximately 3–4 orders of magnitude. Partition coefficients (KF) ranged from 2000–270,000 mL·g–1 across viruses and temperature conditions. Distribution coefficients (Kd) were consistent with results from partitioning experiments. Further research is needed to understand how virus and wastewater characteristics might influence the partitioning of viral genetic markers in wastewater.
Keywords: virus, partitioning, wastewater, RSV, rhinovirus, SARS-CoV-2, MS2, F+ coliphage
Short abstract
We examined the solid−liquid partitioning behavior of SARS-CoV-2, RSV, RV, and F+ coliphage/MS2 RNA in wastewater influent. Overall, partition/distribution coefficients were similar across viruses and temperature conditions.
Introduction
Multiple countries are currently monitoring the spread of COVID-19 by measuring the genetic markers of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants in wastewater and primary settled solids (hereafter referred to as wastewater matrices). A few wastewater surveillance programs also monitor the genetic markers of common respiratory diseases like influenza, respiratory syncytial virus (RSV), rhinovirus (RV), and human metapneumovirus (HMPV).1−5 This information can be used by public health officials to monitor infection trends, complement clinical surveillance data, and strengthen public health responses.6 Despite the widespread adoption of wastewater surveillance, research is still needed to understand the fate and transport of viral genetic markers in wastewater matrices. This information is essential for optimizing monitoring strategies and interpreting wastewater surveillance data. Also, efforts to estimate the number of infected individuals in a sewershed have been increasingly proposed;7−9 however, the feasibility and efficacy of these models remain uncertain, particularly given the paucity of data on virus fate in wastewater.
Viral adsorption can be influenced by the physical, chemical, and biological characteristics of wastewater (e.g., temperature, pH, organic matter) and the characteristics of viruses (e.g., virus structure and size).10−13 Previous studies suggest that viruses and their genetic markers tend to partition more favorably into the solid fraction of wastewater matrices than the liquid fraction.2,14−17 For example, Mercier et al.2 studied the distribution of endogenous influenza A virus (IAV) in wastewater influent and primary sludge and found that the majority of IAV RNA was in settled solids compared to suspended solids (larger than 0.45 μm) and the liquid fraction of these matrices. Note that in this context, we use the term “endogenous” to refer to viruses that are naturally present in wastewater samples (i.e., samples that have not been spiked with an external source of viruses). Li et al.16 also examined the distribution of endogenous SARS-CoV-2 RNA (N1, N2, and E gene targets) in wastewater influent and found that the majority of viral genetic markers were in the solid fraction of wastewater. A few studies have also reported higher concentrations of viral genetic markers in primary sludge compared to paired wastewater influent samples. For instance, Wolfe et al.18 found that IAV RNA concentrations were 1000 times higher in paired primary sludge samples than wastewater influent samples, on mass equivalent basis. Similar results have been reported for SARS-CoV-2, MPOX virus (formerly known as monkeypox), and pepper mild mottle virus (PMMoV) RNA, where viral RNA concentrations were enriched by 3–4 orders of magnitude in primary sludge compared to wastewater influent.14−16,19 Yin et al.20 summarized the solid–liquid distribution of different strains of enteroviruses, hepatitis A, adenovirus, rotavirus, and bacteriophages in wastewater and activated sludge and found that viral adsorption can vary greatly between viruses and wastewater matrices. Still, in all cases, viruses tended to partition into wastewater solids.
A few lab studies have also examined the equilibrium and kinetic adsorption of viruses and their genetic markers in wastewater matrices. For example, Ye et al.12 studied the sorption kinetics of four infectious lab-grown virus surrogates (MHV, Phi6, MS2, and T3) in wastewater influent and found that enveloped viruses partitioned more to the solid fraction of wastewater compared to non-enveloped viruses. A similar study was conducted by Yang et al.11 using quantitative PCR to measure the concentrations of four lab-grown surrogates (Phi6, MS2, T4, and Phix174) in activated sludge. Partition coefficients (converted from log KF; also known as the Freundlich coefficient) were 4.1 × 106, 5.4 × 105, 1.2 × 105, and 8.5 × 103 mL·g–1 for Phi6, MS2, T4, and Phix174 in sludge, respectively. Yin et al.20 also evaluated the sorption of human Adenovirus 40 (HAV40) in primary and secondary sludge and found that the majority of HAV40 DNA was adsorbed onto the solid fraction of these matrices. Partition coefficients (reported as Kp in the paper) were 3.7 × 104 and 4.0 × 104 mL·g–1 in primary and secondary sludge, respectively. Researchers have also examined the equilibrium and kinetic adsorption of SARS-CoV-2 RNA onto passive samplers designed for wastewater surveillance.21
In our review of the previous work described above, we noted the lack of laboratory partitioning and/or distribution data on key respiratory viruses in wastewater, including SARS-CoV-2, RV, and RSV, which have been shown to be present in wastewater matrices and correlate to community disease occurrence.22,23 In addition, we noted that the experiments typically did not include temperature as an experimental factor and that laboratory and field experiments were rarely coupled. In the present study, we fill these knowledge gaps by examining the partitioning behavior of four viruses in wastewater: SARS-CoV-2, RSV, RV, and MS2/F+ coliphage. We achieve this through laboratory partitioning experiments and examination of the distribution of these viruses in raw wastewater influent samples. SARS-CoV-2, RSV, and RV were chosen for the study because their equilibrium partitioning and distribution in wastewater have not been previously studied. MS2 was chosen because it is widely used as a surrogate for pathogenic respiratory viruses in fate and persistence lab experiments. These viruses represent both enveloped (SARS-CoV-2 and RSV) and non-enveloped (RV and MS2) viruses. Additionally, the human pathogenic viruses chosen in this study are targets for wastewater-based epidemiology monitoring efforts. Besides establishing the partitioning coefficients for these viruses, we also tested the hypotheses that temperature and virus type affected the partitioning coefficient, and we tested whether partitioning (derived from lab experiments with spiked viruses) and distribution coefficients (derived from actual samples) were different. Understanding the partitioning behavior of viral genetic markers could inform wastewater sampling strategies and help optimize methods for processing wastewater and primary sludge samples. Partition and distribution coefficients can also help inform complex mathematical models that aim to estimate or predict the number of positive cases in communities.24
Materials and Methods
Overview
We conducted two sets of experiments to examine the partition and distribution of SARS-CoV-2, RSV, RV, and F+ coliphage in wastewater influent. The characteristics of these viruses are shown in Table 1. The partitioning experiment was conducted using lab-grown SARS-CoV-2, RSV-A, RV-B, and MS2; wastewater influent samples were spiked with varying concentrations of each virus and incubated at two different temperatures (4 and 22 °C) to allow the system to equilibrate. After incubation, influent samples were centrifuged and decanted to obtain an aliquot from the liquid and solid fractions. RNA was extracted from the aliquots and quantified using reverse-transcription-digital droplet PCR (RT-ddPCR). The distribution experiment examined the distribution of endogenous SARS-CoV-2, RSV, RV, and F+ coliphage in raw wastewater influent samples. Influent samples were collected from six wastewater plants and processed using the same pre-analytical procedures (i.e., centrifugation to separate solid and liquid fractions) but without spiking with exogenous (lab-grown) viruses. The following sections provide a detailed description of the experiments. Reporting of methods follows EMMI guidelines25 (Figure S1 provides EMMI checklist and details).
Table 1. Characteristics of SARS-CoV-2, RSV, RV, and MS239.
| virus | family/genus | genome type | structure | shape | genome size (kb) | virion size (nm) |
|---|---|---|---|---|---|---|
| SARS-CoV-2 | coronaviridae | + ssRNA | enveloped | spherical | 30 | 50–140 |
| RSV | pneumoviridae | – ssRNA | enveloped | spherical | 15 | 150–250 |
| rhinovirus | picornavirus | + ssRNA | nonenveloped | icosahedral | 7 | 15–30 |
| MS2 | leviviridae | + ssRNA | nonenveloped | icosahedral | 3.6 | 23–28 |
Wastewater Sample Collection
The partitioning experiment was conducted using two raw wastewater influent samples from the Palo Alto Regional Water Quality Control Plant (PA). The plant serves approximately 215,000 people and treats an annual average daily flow of 19.8 million gallons per day (MGD). Total suspended solids (TSS) and pH levels range from 220–360 mg/L and 7.5–7.8, respectively. We collected approximately two liters of a 24 h raw influent composite sample on September 20, 2022, for the 4 °C experiment and on October 28, 2022, for the 22 °C experiment. Samples were collected in 10% HCl acid-washed plastic containers, acclimated to the respective experimental temperatures, and spiked with a mixture of lab-grown SAR-CoV-2, RSV-A, RV-B, and MS2 within 24 h of sample collection (see detailed methods below) for experiments.
Virus Purification and Spike Cocktails
Heat-inactivated SARS-CoV-2 (Isolate: USA-WA1/2020; catalog no. 0810587CFHI), RSV-A (catalog no. 0810040ACF), and RV-B (catalog no. 0810284CF) were purchased from ZeptoMetrix (Buffalo, New York). The manufacturer inactivates SARS-CoV-2 by heating the virus at 60 °C for 1 h. RSV-A and RV-B are viable viruses suspended in cell culture fluids. Escherichia coli phage MS2 (DMS no. 13767) was purchased from the DSMZ German Collection of Microorganisms and Cell Cultures. Viruses were purified to remove viral culture fluid using Amicon Ultra-0.5 mL centrifugal filters (100 kDa MWCO; Millipore UFC5100) following the manufacturer’s instructions. Briefly, 0.5 mL of virus stock, as received from the vendor, was added to individual centrifugal filters and centrifuged at 14,000 × g for 5 min. The filters were immediately flipped and centrifuged at 1000 × g for 2 min to recover the retentate. A series of dilutions were prepared using the filter retentate and autoclaved phosphate-buffered saline (PBS; Fisher BioReagents, Pittsburgh, Pennsylvania) to achieve a viral gene concentration of approximately 1 × 103, 1 × 104, 1 × 105, 1 × 106, and 1 × 107 cp/μL PBS. A total of five spike cocktails were prepared by mixing equal volumes of purified, heat-inactivated SARS-CoV-2, RSV-A, RV-B, and MS2 stock. The final concentrations of the five stock cocktails ranged from approximately 1 × 103–1 × 107 cp/μL PBS (103, 104, 105, 106, 107) for each virus.
Preanalytical Processing
Wastewater samples were consistently mixed by inverting 3–4 times and aliquoted into eighteen 50 mL centrifuge tubes (hereafter referred to as subsamples). Five sets of three subsamples were then spiked with one of the five different concentrations of spiked cocktails to achieve a final concentration of approximately 103, 104, 105, 106, or 107 cp/mL wastewater for each virus. Three additional subsamples were reserved to measure the background concentration of endogenous SARS-CoV-2, RSV, RV, and F+ coliphage RNA. After spiking, subsamples were stored at 4 or 22 °C, depending on the isotherm experiment, and gently mixed (∼20 rpm) using a tube roller (Globe Scientific, GSCI-GTR-AVS) for approximately three hours to allow the system to equilibrate. The time needed to reach equilibrium (<3 h) was determined based on a preliminary experiment (see Figures S2 and S3), the results of which are consistent with other virus adsorption studies conducted in wastewater.11,12 After three hours, subsamples were centrifuged at 24,500 × g for 20 min at the temperatures of the experiments. This process removes solid particles with hydrodynamic radii greater than 0.3 μm in diameter.26 200 μL of the supernatant was transferred to a 2 mL collection tube. These aliquots represent the liquid fraction of the wastewater sample. Note that the liquid fraction was not further processed (concentrated) given the relatively high concentration of spiked viruses and to prevent potential losses of RNA signal typically associated with additional pre-analytical processing. Liquid aliquots were then spiked with 5 μL of bovine coronavirus vaccine (BCoV; Zoetis, CALF-GUARD; Parsippany-Troy Hills, NJ) as an extraction recovery control. The BCoV vaccine contains attenuated strains of the live virus and comes in a lyophilized form. We reconstituted the vaccine by adding 3 mL of molecular-grade water.
The remaining supernatant was decanted, and the pellet represents dewatered solids. A portion of the dewatered solids was reserved to calculate the dry weight. Dewatered solids were weighed before and after drying at 105 °C for 24 h. To collect solids for viral analysis, approximately 0.1 g of dewatered solids was collected from the bottom of the centrifuge tube and aliquoted into 2 mL microcentrifuge tubes using a disposable spatula (Fisher Scientific, catalog no. 50-476-569). Dewatered solids were resuspended in a BCoV-spiked DNA/RNA shield (Zymo Research, catalog no. R1100-250) at a concentration of approximately 75 mg/mL; this concentration of solids in the buffer has been shown to alleviate downstream RT-ddPCR inhibition.27 Spiked DNA/RNA shield was pre-prepared using 1.5 μL of BCoV per ml of DNA/RNA shield. Three to five grinding balls (OPS DIAGNOSTICS, GBSS 156-5000-01) were added to a 2 mL microcentrifuge tube, and the mixture was homogenized at 4 m/s for 1 min using the MP Bio Fastprep-24TM (MP Biomedicals, Santa Ana, CA). The homogenized aliquots were then centrifuged for 5 min at 5250 × g, and 200 μL of the supernatant was transferred to a 2 mL collection tube. This aliquot contains viral targets from the solid fraction of the wastewater sample. Liquid and solid aliquots were stored at 4 °C overnight, and the nucleic acids were extracted from them the next day.
RNA Extraction
RNA was extracted from the solid and liquid aliquots using the Qiagen AllPrep PowerViral DNA/RNA kit and further purified using the Zymo OneStep PCR inhibitor removal columns (Zymo Research, Irvine, CA) following the manufacturer’s instructions. RNA extracts were aliquoted into 1.5 mL DNA LoBind tubes, stored at −80 °C for less than two weeks, and thawed once (1 freeze–thaw cycle) before quantification. Nuclease-free water and BCoV-spiked DNA/RNA shields were used as negative and positive extraction controls, respectively. These controls were carried throughout the extraction process, with one set of positive and negative controls per extraction batch (∼15 solid or liquid aliquots/batch of extraction).
RNA Quantification
SARS-CoV-2 and RSV were quantified using a duplex assay described in Hughes et al.22 and RV, MS2, and BCoV were quantified using singleplex assays from previous studies.1,15,28 Primers and probes were purchased from Integrated DNA Technologies (IDT, San Diego, CA) and are provided in Table S1. Using NCBI Blast, we determined that the MS2 assay will detect, in addition to MS2, the following sequenced and deposited strains of genotype group 1 (GI) F+ RNA coliphages: JP501, M12, DL16, DL52, and DL54, which could potentially be present in wastewater. There could be additional unsequenced/undeposited endogenous wastewater F+ RNA coliphages that are also detectable using this assay. Therefore, detections with MS2 in wastewater will be referred to as F+ coliphage, hereafter.
All viral targets were quantified using the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad 1863021). For the RT-ddPCR, 20 μL of a 22 μL reaction mix was prepared for each well. The reaction consisted of 5.5 μL of template RNA, 5.5 μL of Supermix, 2.2 μL of 200 U/μL Reverse Transcriptase (RT), 1.1 μL of 300 mM dithiothreitol (DDT), 4.4 μL of nuclease-free water, and 3.3 μL of primer and probe mixture with a final concentration of 900 and 250 nM, respectively. RNA extracts were processed undiluted and in duplicate (two technical replicates). Nuclease-free water and viral RNA extracts for each target were used as negative and positive PCR controls, respectively, and each run in three wells per plate. Positive controls were extracted from the SARS-CoV-2, RSV, RV, and MS2 purified stocks, which are the same virus stocks used to prepare the spike cocktails. Droplets were generated using the AutoDG Automated Droplet Generator (Bio-Rad, Hercules, CA) and amplified using the C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA). The thermal cycling conditions for each RT-ddPCR assay are shown in Table S2. After amplification, droplets were analyzed using the QX200 droplet reader (Bio-Rad, Hercules, CA) and the Quantasoft Analysis Pro Software. Wells with less than 10,000 droplets were excluded, and technical PCR replicates (wells) were merged before performing dimensional analysis. Merged wells needed to have at least three positive droplets to be considered positive. The estimated lower measurement limits for solids and liquid aliquots were 3500 cp/g and 0.7 cp/mL, respectively.
Distribution of Endogenous Viruses in Wastewater
The second experiment examined the distribution of endogenous SAR-CoV-2, RSV, RV, and F+ coliphage between liquid and solid fractions in wastewater samples. Raw influent samples (∼1 L of a 24 h composite sample) were collected from six wastewater treatment plants on November 18, 2022. Samples were stored at 4 °C and processed within 24 h. These plants are part of an ongoing wastewater surveillance program and include the following: Oceanside Water Pollution Control Plant (OS), Southeast Water Pollution Control Plant (SE), Silicon Valley Clean Water Wastewater Treatment Plant (SV), Sunnyvale Water Pollution Control Plant (SU), San Jose-Santa Clara Regional Wastewater Facility (SJ), and South County Regional Wastewater Treatment Plant (GI). The location, estimated population served, annual daily average flow, pH, and TSS for each plant can be found in Table S3. Figure S4 provides a map of the sewershed (area served) for each wastewater treatment plant.
Wastewater samples were consistently mixed, poured into 50 mL centrifuge tubes, and processed. Three subsamples were prepared for each wastewater treatment plant, for a total of 18 subsamples. Solid and liquid aliquots were obtained from the wastewater samples, and nucleic acids were extracted. Viral targets were quantified from the aliquots using the methods described above for the partitioning experiments.
Dimensional Analysis, Adsorption Models, and Statistical Analysis
SARS-CoV-2, RSV, RV, and MS2/F+ coliphage RNA concentrations were expressed in units of copies per gram dry weight solids (cp/g) or per ml liquid (cp/mL) for the solid and liquid fractions of wastewater, respectively, using dimensional analysis. Note that the volume used in the dimensional analysis only refers to the liquid fraction (i.e., the supernatant without solids). BCoV recovery, used as an extraction and inhibition control, was calculated for the solid and liquid fractions as follows:
| 1 |
where Ncopies is the number of gene copies in the solid or liquid fraction and Ncopies,spiked is the number of gene copies in the spiked DNA/RNA solid or liquid fraction.
For the partition experiment using lab-grown viruses, 36 RNA concentrations (solids: N = 18 and liquids: N = 18) were obtained for each virus and isotherm experiment (4 and 22 °C). These concentrations represent triplicate measurements for six different initial conditions: one unspiked and five different spiked concentrations of viruses. For each virus and temperature condition, average viral RNA concentrations (solids: N = 6 and liquids: N = 6) and standard deviations were calculated across triplicate subsamples. To calculate the recovery of spiked viruses, we first subtracted the background concentrations of viral genetic markers from liquid and solid fractions of spiked subsamples and estimated the total RNA recovery in spiked subsamples as follows:
| 2 |
where CRNA,s and CRNA,l are the concentrations of RNA in the solid and liquid fractions, respectively, ms is the mass of solids, Vl is the volume of the liquid sample, and Ncopies,spiked,tot is the total number of gene copies spiked into the sample.
Partition Experiment
Viral RNA concentrations in the solid and liquid fractions of wastewater were fit to the Linear, Langmuir, and Freundlich isotherm models. These models have been used in previous virus partitioning studies11,14,20,21 and describe a multi-layer (Freundlich) or monolayer (Langmuir) adsorption process. A linear model is generally used when the coverage ratio of adsorption sites is minimal.29 The Linear, Freundlich, and Langmuir isotherm parameters were determined for each virus and temperature condition. The average relative error (ARE) was calculated for each model and compared to identify the model with the best fit. The Freundlich model produced the smallest ARE overall and therefore is discussed in the main paper (see Table S4 for results of other models). The nonlinear and linear forms of the Freundlich model are described as follows:
| 3 |
| 4 |
where qe is the equilibrium concentration of viral gene copies in solids (cp/g), Ce is the equilibrium concentration of viral gene copies in the liquid fraction of wastewater, KF is the Freundlich adsorption capacity parameter, and n is the adsorption intensity. A higher n indicates a stronger interaction between the adsorbent (i.e., wastewater solids) and the adsorbate (i.e., spiked viruses). We determined the Freundlich isotherm parameters, log KF (y-intercept) and n (slope), using a linear regression model (lm function) in R. Standard errors (SE) were also obtained from the linear regression model in R (summary function). Finally, we compared the KF obtained for the viruses under different temperature conditions by examining whether their values and standard deviations overlapped.
For the distribution experiment (examining endogenous viruses in wastewater samples), six RNA concentrations (solids: N = 3 and liquids: N = 3) were obtained for each virus and wastewater treatment plant. Only a few liquid fractions resulted in non-detects (ND). NDs were substituted with half of the lower measurement limit (0.35 cp/mL for concentrations measured in liquid fractions). The average concentration and standard deviation for each virus were calculated using data from the three replicate subsamples. The distribution coefficient was calculated as the ratio of average viral gene concentrations detected in the solid and liquid fractions of wastewater influent; Kd = Cs/Cw; errors were determined by propagating errors on the numerator and denominator. We tested the null hypothesis that Kd were the same for endogenous viruses using a Kruskal–Wallis test; p < 0.05 was used to assess statistical significance. Statistical analysis was performed in R (version 4.1.2). Finally, we compared the partition coefficients (KF) for spiked viruses at 4 and 22 °C to the distribution coefficients (Kd) for endogenous viruses in wastewater for each virus to determine if KF and Kd values and errors overlapped. We also summarized the results from previous experiments measuring the concentration of viral genetic markers in wastewater matrices to see if our results were consistent with previously reported KF and Kd values (see Table 2).
Table 2. Results from Previous Experiments Measuring the Concentration of Viral Genetic Markers in the Solid and Liquid Fractions of Wastewater Matrices.
| study | virus | wastewater matrix | sample processing | KF or Kd (mL·g–1) |
|---|---|---|---|---|
| Graham et al.15 | SARS-CoV-2 | paired wastewater influent and primary sludge samples | solid and liquid fractions were separated by centrifugation; the supernatant from influent samples was further processed using PEG precipitation method; viral concentrations (N1 and N2) were measured using RT-ddPCR | Kd: 350–3100 |
| calculated/reported as the solid-influent concentration ratio | ||||
| Li et al.16 | SARS-CoV-2 | wastewater influent | solid and liquid fractions were separated by centrifugation; liquids were further processed using PEG precipitation method; viral concentrations (N1, N2, and E gene) were measured using qPCR | Kd: 4000–20,000 |
| calculated/reported as the solid–liquid concentration ratio | ||||
| Kim et al.40 | SARS-CoV-2 | paired wastewater influent and primary sludge samples | solid and liquid fractions were separated using different processing techniques; viral concentrations (N1 and N2) were measured using RT-ddPCR | KF: 1000–100,000 |
| calculated using the Freundlich model; solid to influent ratio | ||||
| Kim et al.31 | SARS-CoV-2 | wastewater collected from sewer network | solid and liquid fractions were separated by centrifugation; liquids were further processed using a 0.45 μm pore size filter; viral concentrations (N and S gene) were measured using RT-ddPCR | Kd: 8600, 16,000 |
| calculated/reported as the solid–liquid concentration ratio | ||||
| Wolfe et al.41 | influenza A | paired wastewater influent and primary sludge samples | solid and liquid fractions were separated by centrifugation; the supernatant from influent samples was further processed using PEG precipitation method; viral concentrations (N1 and N2) were measured using RT-ddPCR | Kd: 1000 |
| calculated/reported as the solid-influent concentration ratio | ||||
| Mercier et al.2 | influenza A | wastewater and primary sludge samples | solid and liquid fractions were separated by centrifugation and filtration using a 0.45 μm pore size filter; viral concentrations were measured in settled solids, suspended solids, and liquid fractions (supernatant) using RT-qPCR | authors only reported the percent of viral RNA adsorbed onto wastewater solids, Kd was not reported |
| Wolfe et al.19 | Mpox | paired wastewater influent and primary sludge samples | influent samples were processed using an affinity-based capture method with magnetic hydrogel Nanotrap particles with enhancement reagent 1; primary sludge samples were centrifuged to obtain solids; viral concentrations were measured using RT-ddPCR | Kd: 1000 |
| calculated/reported as the solid-influent concentration ratio | ||||
| Yin et al.20 | adenovirus | primary and secondary sludge | solid and liquid fractions were separated by centrifugation; viral concentrations were measured in the liquid fraction (supernatant) using qPCR; viral concentrations in solids were estimated using a mass balance equation | KF: 37,000, 40,000 |
| calculated using the Freundlich model | ||||
| Yang et al.11 | MS2, Phi6, Phix174, T4 | primary sludge | solid and liquid fractions were separated by centrifugation and filtration using a 0.22 μm pore size filter; viral concentrations were measured using qPCR | KF: 4.1 × 106, 5.4 × 105, 1.2 × 105, and 8.5 × 103 for Phi6, MS2, T4, and Phix174, respectively |
Results
Extraction and PCR Controls
Positive and negative extraction and PCR controls were positive and negative, respectively. BCoV was used as a process recovery and gross inhibition control. It is important to note that measuring virus recovery in wastewater presents certain limitations and challenges, which are comprehensively discussed in Kantor et al.30 In our study, BCoV recoveries were similar between the solid and liquid fractions of wastewater. Solid and liquid aliquots had a median BCoV recovery of 0.40 and 0.35, respectively. Viral gene concentrations were not adjusted by recovery given the complexities associated with estimating recovery using surrogate viruses30 and given that the recoveries were similar. The total recovery of spiked SARS-CoV-2, RSV-A, RV-B, and MS2 RNA was approximately 35, 45, 22, and 33%, respectively. These recoveries are similar to those of BCoV. Although inhibition can vary between assays, our previous work has determined that there is minimal inhibition with the pre-analytical and analytical workflow,27 and the similar and high detection of BCoV and spiked viruses indicates that inhibition is minimal.
Partitioning of Lab-Grown Viruses in Wastewater Influent
Endogenous SARS-CoV-2, RSV, RV, and MS2/F+ coliphage RNA were detected in the liquid and solid fractions of wastewater influent samples from PA (Table S5). Concentrations of endogenous virus make up 22–95% of viral RNA in subsamples spiked with the lowest concentration of virus cocktail (i.e., 103 cp/mL), but otherwise represent a negligible percentage of the RNA in subsamples spiked with a higher concentration of virus cocktail.
At equilibrium, the results from the partitioning experiments showed that RNA concentrations of spiked viruses were higher in solids than the liquid fraction of wastewater influent on a mass equivalent basis, by approximately 3–4 orders of magnitude. In the 4 and 22 °C isotherm experiments, qe ranged from 1.3 × 104–5.6 × 107 cp/g for SARS-CoV-2, 4.2 × 103–1.9 × 108 cp/g for RSV, 2.3 × 104–1.3 × 107 cp/g for RV, and 4.1 × 103–3.2 × 107 cp/g for MS2. In the liquid fraction, Ce concentrations ranged from 0.1–1.3 × 103 cp/mL for SARS-CoV-2, 0.2–4.5 × 102 cp/mL for RSV, 1.6–4.8 × 104 cp/mL for RV, and 0.8–4.3 × 104 cp/mL for MS2. The reported ranges represent measurements across spiking conditions and the two experimental temperatures.
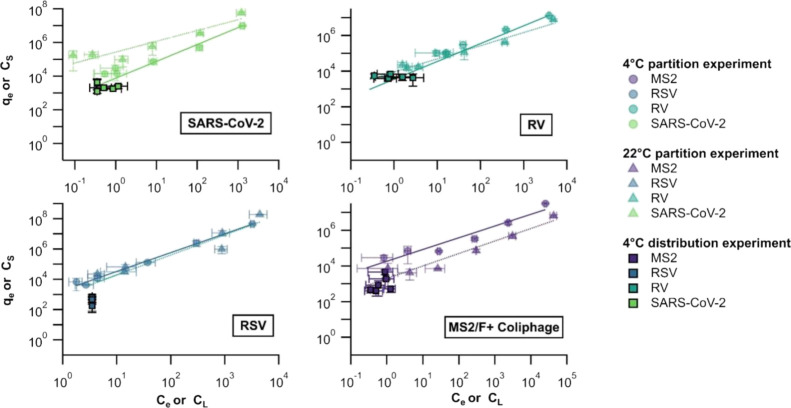
Viral RNA concentrations (qe and Ce, see Figure 1) were fit to a Linear, Freundlich, and Langmuir model. The Freundlich isotherm models produced the lowest ARE compared to the other models based on the calculated partition coefficient (see Table S4 for results of the Linear and Langmuir models). Table 3 shows the Freundlich isotherm parameters (KF and n) for each virus and temperature condition. In the 4 °C experiment, KF and n ranged from 1.8 × 103–3.2 × 103 mL·g–1 and 0.66–1.24, respectively. Similar results were obtained in the 22 °C experiments except for the partition coefficient of SARS-CoV-2. In the 22 °C experiment, KF and n ranged from 2.0 × 103–2.7 × 105 mL·g–1 and 0.64–1,32 respectively. The partition coefficient of SARS-CoV-2 in the 22 °C experiment was significantly higher (approximately one order of magnitude) compared to other viruses and temperature conditions. However, partition coefficients were not different across other viruses and temperatures (see Figure S5).
Figure 1.
qe and Ce from partition experiments at 4 °C (circles) and 22 °C (triangles) and CS and CL from distribution experiments at 4 °C (squares, with black edges). Lines represent the Freundlich isotherm model and error bars represent the standard deviation across triplicate subsamples. Note that the values for CL for RSV represent an upper bound since the concentration of RSV RNA was below the lower detection limit; a value of 1/2 the lower detection limit was used for the plot. If a smaller value had been used, it is possible that the points would fall in the same line as those from the RSV partition experiment.
Table 3. Isotherm Parameters (KF and n) and Average Relative Error (ARE) of Freundlich Models for the Adsorption of SARS-CoV-2, RSV-A, RV-B, and MS2 in Wastewater at 4 and 22 °Ca.
| lab-grown viruses | virus structure | temperature (°C) | KF (LE-UE) (mL·g–1) | n ± SE | ARE |
|---|---|---|---|---|---|
| SARS-CoV-2 | enveloped | 4 | 18,000 (4100–41,000) | 0.81 ± 0.07 | 0.40 |
| 22 | 270,000 (74,000–630,000) | 0.64 ± 0.09 | 0.91 | ||
| RSV-A | enveloped | 4 | 32,000 (2000–67,000) | 1.24 ± 0.02 | 0.25 |
| 22 | 19,200 (10,000–60,000) | 1.32 ± 0.21 | 1.18 | ||
| RV-B | nonenveloped | 4 | 13,000 (1500–28,000) | 0.84 ± 0.03 | 0.15 |
| 22 | 8900 (2400–21,000) | 0.74 ± 0.07 | 0.39 | ||
| MS2 | nonenveloped | 4 | 18,000 (7400–49,000) | 0.66 ± 0.09 | 0.64 |
| 22 | 2000 (760–5200) | 0.70 ± 0.08 | 0.78 |
SE, LE, and UE are the standard error, the lower SE bound, and the upper SE bound as reported by lm function in R, respectively. n and ARE are dimensionless.
Distribution of Endogenous Viruses in Wastewater Influent
Across samples collected from the six wastewater treatment plants, we observed results similar to those obtained in the laboratory partitioning experiment. Viral RNA concentrations were higher in the solid fraction than the liquid fraction of wastewater by approximately 3–4 orders of magnitude (see Figure 1). Across six wastewater treatment plants, Cs ranged from 1.2 × 103–4.2 × 103 cp/g (median = 2.2 × 103 cp/g) for SARS-CoV-2, 9.1 × 102–3.7 × 103 cp/g (median = 1.7 × 103 cp/g) for RSV, 2.4 × 102–6.3 × 102 cp/g (median = 3.7 × 102 cp/g) for RV, and 4.9 × 102–6.1 × 103 cp/g (median = 1.1 × 103 cp/g) for F+ coliphage. CL ranged from 0.4–1.1 cp/mL (median = 0.4 cp/mL) for SARS-CoV-2, 0.4–2.7 cp/mL (median = 0.8 cp/mL) for RV, and 0.4–1.3 cp/mL (median = 0.7 cp/mL) for F+ coliphage. For RSV, CL were ND across wastewater treatment plants; NDs were replaced with half of the lower measurable limit (0.35 cp/mL for viral concentrations in liquid fractions) to calculate the distribution coefficient (Kd).
Kd was calculated as the ratio of Cs/CL. Across wastewater treatment plants, Kd ranged from 3.5 × 103–1.2 × 104 mL·g–1 (median = 5.0 × 103 mL·g–1) for SARS-CoV-2, 2.1 × 103–1.6 × 104 mL·g–1 (median = 7.6 × 103 mL·g–1) for RV, and 4.9 × 102–7.4 × 103 mL·g–1 (median = 1.2 × 103 mL·g–1) for F+ coliphage (see Table 4). Since RSV concentrations were ND for the liquid phase, we could only estimate lower bounds for Kd: 5.0 × 102–2.0 × 103 mL·g–1 (median = 1.3 × 103 mL·g–1. RV had the highest solid–liquid distribution, followed by SARS-CoV-2 and F+ coliphage. RSV cannot be compared as our estimates are lower bounds; Kd for RSV could be higher. RV Kd was statistically higher than F+ coliphage Kd (Kruskal–Wallis and Dunn’s post hoc test, both p < 0.05). Note that RSV was excluded from the statistical analysis because we were only able to estimate a lower bound and a direct comparison with other Kd values might result in misleading conclusions.
Table 4. Distribution Coefficient (Kd = Cs/Cw) of Endogenous SARS-CoV-2, RSV, RV, and F+ Coliphage RNA in Wastewater Influenta.
| wastewater treatment plant | Kd ± SD (mL·g–1) | |||
|---|---|---|---|---|
| SARS-CoV-2 | RSV | RV | F+ coliphage | |
| Gilroy | 3500 ± 820 | 1300 ± 650 | 7200 ± 1400 | 490 ± 380 |
| San Jose | 11,000 ± 6200 | 2000 ± 760 | 16,000 ± 4100 | 2800 ± 1800 |
| Sunnyvale | 12,000 ± 7600 | 1800 ± 460 | 8000 ± 4800 | 950 ± 710 |
| SVCW | 5600 ± 4300 | 1300 ± 450 | 11,000 ± 7200 | 7400 ± 6300 |
| SEP | 4400 ± 1400 | 500 ± 320 | 2100 ± 1400 | 1200 ± 760 |
| OSP | 3000 ± 2500 | 700 ± 470 | 2800 ± 1000 | 1300 ± 470 |
SD is the standard deviation across triplicate subsamples. Endogenous refers to viruses present in wastewater (not seeded). Note that the values for RSV represent lower bound estimates since the concentrations in the liquid phase were below the lower limit of detection.
Discussion
This is the first batch experiment examining the solid–liquid partitioning of SARS-CoV-2, RSV, and RV in wastewater and the first experiment to examine the distribution of endogenous RSV and RV in this matrix. Higher concentrations of viral RNA were observed in solids compared to the liquid fraction of wastewater, for all viruses and temperature conditions; viral RNA concentrations were higher in solids by 3–4 orders of magnitude on a mass equivalent basis. Partition and distribution coefficients were similar across viruses and temperature conditions, with KF and Kd ranging from 490–270,000 mL·g–1. Our results are consistent with previously reported partition/distribution coefficients for viral genetic markers in wastewater. For example, Li and co-workers31 evaluated the distribution of endogenous SARS-CoV-2 RNA (N1, N2, and E genes) in wastewater samples and found higher concentrations of SARS-CoV-2 RNA in solids compared to the liquid fraction; Kd (reported in their paper as the solid–liquid concentration ratio) ranged from 4000–20,000 mL·g–1. Kim et al.31 also studied the distribution of endogenous SARS-CoV-2 RNA (S and N genes) in wastewater samples from two K-12 schools and observed similar results; viral RNA concentrations were higher in solids by three orders of magnitude, and Kd (reported in their paper as the concentration ratio in solid to liquid samples) were 8600 and 16,000 mL·g–1 for SARS-CoV-2N and S genes, respectively. We observed similar partitioning behavior for SARS-CoV-2, RSV, RV, and MS2 RNA in our study; except for SARS-CoV-2 RNA at 22 °C, which had a higher partition (by approximately one order of magnitude) compared to other viruses and temperature conditions. A study by Breadner et al.32 found that a large proportion of virus RNA may remain in the liquid fraction of wastewater samples after centrifugation. However, it is important to note that in their study, samples were spiked and incubated for 10 min before processing, which might not have allowed enough time for the system to equilibrate.33
There are limited studies in the literature on the effects of temperature on virus adsorption to particles. One study examined how temperature may influence the isotherm and kinetic adsorption processes of viral genetic markers in wastewater solids. Yang et al.11 examined the kinetic adsorption of Phi6, MS2, T4, and Phix174 RNA in primary sludge at two temperature conditions (4 and 25 °C). They found that the rate of virus adsorption increased with increasing temperature (i.e., the time needed to reach equilibrium was reduced). Other studies have examined how temperature affects viral adsorption to clays. Syngouna et al. examined the isotherm and kinetic adsorption of infectious MS2 and ΦX174 in clay particles at 4 and 25 °C and found results similar to Yang et al.34 However, Bellou et al.35 evaluated the adsorption of MS2, ΦX174, and human adenovirus (hAdV) nucleic-acids on clay particles at 4 and 25 °C and found that the adsorption of hAdV was inversely proportional to temperature, whereas adsorption of MS2 and ΦX174 was directly proportional to temperature. In our isotherm (equilibrium) experiments, wastewater temperature did not seem to have an impact on the adsorption of viral genetic markers, except for the case of SARS-CoV-2. For SARS-CoV-2, a higher partition coefficient was observed at 22 than at 4 °C. The equivocal results described here suggest that additional work is needed to better understand if and how temperature affects viral adsorption.
Limited previous research suggests that the presence of a lipid envelope outside the viral protein capsid of a virus may impact the solid–liquid partitioning of viruses and viral genetic markers in wastewater. For example, Ye et al.12 measured the adsorption of infectious MHV, ϕ6, MS2, and T3 in wastewater samples and found that enveloped viruses (MHV and ϕ6) were more strongly associated with solids than non-enveloped viruses (MS2 and T3). Similar results were reported in the study by Yang et al.,11 where they examined the solid–liquid partitioning behavior of Phi6, MS2, T4, and Phix174 nucleic-acids in primary sludge samples; researchers found that the majority of viral genetic markers were in the solid fraction of primary sludge. KF ranged from 8500–4,100,000 mL·g–1 for Phi6, MS2, T4, and Phix174 RNA. In our study, the partition and distribution behavior across enveloped (SARS-CoV-2 and RSV) and non-enveloped (RV and F+ coliphage/MS2) viruses appeared to be similar. Our results were also similar across the partition and distribution experiments, which suggest that lab-grown viruses and endogenous viruses for SARS-CoV-2, RSV, RV, and F+ coliphage/MS2 RNA may exhibit similar solid–liquid partitioning behavior in wastewater.
Efforts to estimate the number of infected individuals in a sewershed have been increasingly proposed. For example, Soller et al.36 developed a mechanistic model that estimates the fraction of a sewershed population actively infected with SARS-CoV-2. Wolfe et al.37 also developed a mass balance model that links SARS-CoV-2 RNA concentrations in solids to the number of individuals shedding SAR-CoV-2 RNA in stool within the sewershed. These models can potentially be applied to other viruses of interest; however, a key parameter of these mechanistic models is the solid–liquid partitioning coefficient of viral genetic markers in wastewater solids. The availability of viral-specific partitioning data is limited, which hinders the application of these models and the interpretation of wastewater surveillance data. Our results presented here fill a knowledge gap by providing information on virus partitioning that can be used in these modeling applications. It should be noted that other model input variables, such as the longitudinal shedding of viruses by infected individuals, are also needed.38
Understanding the fate and transport of viral genetic markers could inform wastewater sampling strategies and help optimize methods for processing wastewater and primary sludge samples. For example, a study by Kim et al.14 showed that methods for processing influent and settled solids have comparable sensitivity. However, targeting the solid fraction of wastewater matrices might be a more advantageous medium in sewersheds that have a low level of active infections because it requires less sample volume compared to influent methods and may be more scalable and automatable than processing liquids due to reduced pre-analytical steps.14,37 Targeting wastewater solids could be a valuable strategy for wastewater-based epidemiology (WBE) efforts aiming to achieve early detection and timely warning of infectious diseases. Understanding the solid–liquid partitioning of viruses may also provide valuable insights into the mechanisms through which viruses may be removed from wastewater. For example, primary treatment units may contribute to the removal of solid-associated viruses by settling out wastewater solids. Before WBE efforts begin at a particular plant, it may be advantageous to conduct experiments to investigate the distribution of the infectious disease target between the liquid and solid phases of wastewater. Further research should be conducted to examine the solid–liquid partitioning of other viruses of interest for WBE efforts and evaluate how their characteristics (e.g., virus size and structure) might influence their partitioning behavior in wastewater. Additional research is also needed to evaluate how solid characteristics (e.g., particle size and biological/chemical composition) and extracellular polymeric substances (EPS) might influence virus adsorption. Enhanced understanding of the properties (e.g., virus and wastewater matrix) and mechanisms underpinning partitioning behavior may improve and inform sampling and monitoring strategies.
Environmental Implications
This study fills an important knowledge gap on the partitioning of respiratory viruses in wastewater and indicates that they partition preferentially to the wastewater solids. This information is useful in WBE applications, including both sampling and modeling. These findings add to a growing body of evidence suggesting that the solids in wastewater (defined as material generally larger than 0.3 μm in hydrodynamic diameter) are enriched with infectious disease targets like viral RNA relative to the liquid phase on a mass-equivalent basis (note that intact bacteria and fungi, given their sizes, automatically fall into this size class). There has been some confusion among researchers and practitioners in interpreting this statement. A given volume of raw wastewater typically contains a small mass of solids (typically on the order of 102 mg/L), so it can be true that most of the infectious disease target (mass or number) in that volume is in the liquid phase, and that the concentration of the infectious disease target is enriched by orders of magnitude in the solid phase relative to the liquid phase. The fact that infectious disease targets are enriched in the solid phase indicates that sample efforts that enrich solids and choose the solids as a measurement matrix will improve the sensitivity of measurement approaches.
Acknowledgments
This work is supported by gifts from the CDC Foundation and the Sergey Brin Family Foundation. This research was performed on the ancestral and unceded lands of the Muwekma Ohlone people. We pay our respects to them and their elders, past and present, and are grateful for the opportunity to live and work here. Graphical abstract was created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.3c03376.
Additional details about wastewater treatment plants, isotherm models, and ddPCR assays in this experiment, primers and probes for RT-ddPCR assays; thermal cycling conditions for SARS-CoV-2, RSV, RV, MS2, and BCoV; served population and annual average daily flow of wastewater treatment plants; Langmuir and Linear isotherm parameters for SARS-CoV-2, RSV-A, RV-B, and MS2 into wastewater solids and their respective ARE; background concentration of endogenous of SARS-CoV-2, RSV, RV, and F+ coliphage/MS2 in wastewater influent samples; EMMI checklist for reporting; MS2 RNA concentration in liquid fraction; MS2 RNA concentration in solid fraction; outline of wastewater treatment plant service area (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Boehm A. B.; Hughes B.; Doung D.; Chan-Herur V.; Buchman A.; Wolfe M. K.; White B. J. Wastewater Surveillance of Human Influenza, Metapneumovirus, Parainfluenza, Respiratory Syncytial Virus (RSV), Rhinovirus, and Seasonal Coronaviruses during the COVID-19 Pandemic; preprint; Infectious Diseases (except HIV/AIDS). Lancet Microbe 2022, 4, e340–e348. 10.1016/S2666-5247(22)00386-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier E.; D’Aoust P. M.; Thakali O.; Hegazy N.; Jia J.-J.; Zhang Z.; Eid W.; Plaza-Diaz J.; Kabir M. P.; Fang W.; Cowan A.; Stephenson S. E.; Pisharody L.; MacKenzie A. E.; Graber T. E.; Wan S.; Delatolla R. Municipal and Neighbourhood Level Wastewater Surveillance and Subtyping of an Influenza Virus Outbreak. Sci. Rep. 2022, 12, 15777. 10.1038/s41598-022-20076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Bivins A.; Stephens M.; Metcalfe S.; Smith W. J. M.; Sirikanchana K.; Kitajima M.; Simpson S. L. Occurrence of Multiple Respiratory Viruses in Wastewater in Queensland, Australia: Potential for Community Disease Surveillance. Sci. Total Environ. 2023, 864, 161023 10.1016/j.scitotenv.2022.161023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A.; Bloemen M.; Thijssen M.; Pussig B.; Beuselinck K.; Van Ranst M.; Wollants E. Epidemiological Surveillance of Respiratory Pathogens in Wastewater in Belgium; preprint. Epidemiology 2022, 10.1101/2022.10.24.22281437. [DOI] [Google Scholar]
- Dumke R.; Geissler M.; Skupin A.; Helm B.; Mayer R.; Schubert S.; Oertel R.; Renner B.; Dalpke A. H. Simultaneous Detection of SARS-CoV-2 and Influenza Virus in Wastewater of Two Cities in Southeastern Germany, January to May 2022. IJERPH 2022, 19, 13374. 10.3390/ijerph192013374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A. E.; Walters M. S.; Jennings W. C.; Fugitt R.; LaCross N.; Mattioli M.; Marsh Z. A.; Roberts V. A.; Mercante J. W.; Yoder J.; Hill V. R. Using Wastewater Surveillance Data to Support the COVID-19 Response — United States, 2020–2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1242–1244. 10.15585/mmwr.mm7036a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I. Can We Predict Viral Outbreaks Using Wastewater Surveillance?. J. Environ. Eng. 2020, 146, 01820003 10.1061/(ASCE)EE.1943-7870.0001831. [DOI] [Google Scholar]
- Xagoraraki I.; O’Brien E.. Wastewater-Based Epidemiology for Early Detection of Viral Outbreaks. In Women in Water Quality; O’Bannon D. J., Ed.; Women in Engineering and Science; Springer International Publishing: Cham, 2020; pp 75–97. [Google Scholar]
- Kitajima M.; Ahmed W.; Bibby K.; Carducci A.; Gerba C. P.; Hamilton K. A.; Haramoto E.; Rose J. B. SARS-CoV-2 in Wastewater: State of the Knowledge and Research Needs. Sci. Total Environ. 2020, 739, 139076 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.; Flury M.. Fate and Transport of Viruses in Porous Media. In Advances in Agronomy; Elsevier, 2002; vol 77, pp 39–102. [Google Scholar]
- Yang W.; Cai C.; Dai X. Interactions between Virus Surrogates and Sewage Sludge Vary by Viral Analyte: Recovery, Persistence, and Sorption. Water Res. 2022, 210, 117995 10.1016/j.watres.2021.117995. [DOI] [PubMed] [Google Scholar]
- Ye Y.; Ellenberg R. M.; Graham K. E.; Wigginton K. R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Armanious A.; Aeppli M.; Jacak R.; Refardt D.; Sigstam T.; Kohn T.; Sander M. Viruses at Solid–Water Interfaces: A Systematic Assessment of Interactions Driving Adsorption. Environ. Sci. Technol. 2016, 50, 732–743. 10.1021/acs.est.5b04644. [DOI] [PubMed] [Google Scholar]
- Kim S.; Kennedy L.; Wolfe M.; Criddle C.; Duong D.; Topol A.; White B. J.; Kantor R.; Nelson K.; Steele J.; Langlois K.; Griffith J.; Zimmer-Faust A.; McLellan S.; Schussman M.; Armmerman M.; Wigginton K.; Bakker K.; Boehm A. SARS-CoV-2 RNA Is Enriched by Orders of Magnitude in Solid Relative to Liquid Wastewater at Publicly Owned Treatment Works; preprint; Infectious Diseases (except HIV/AIDS). Environ. Sci.: Water Res. Technol. 2022, 8, 757–770. 10.1039/D1EW00826A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K. E.; Loeb S. K.; Wolfe M. K.; Catoe D.; Sinnott-Armstrong N.; Kim S.; Yamahara K. M.; Sassoubre L. M.; Mendoza Grijalva L. M.; Roldan-Hernandez L.; Langenfeld K.; Wigginton K. R.; Boehm A. B. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2021, 55, 488–498. 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Li B.; Di D. Y. W.; Saingam P.; Jeon M. K.; Yan T. Fine-Scale Temporal Dynamics of SARS-CoV-2 RNA Abundance in Wastewater during A COVID-19 Lockdown. Water Res. 2021, 197, 117093 10.1016/j.watres.2021.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K.; Sadamasu K.; Muramatsu M.; Yoshida H. Efficient Detection of SARS-CoV-2 RNA in the Solid Fraction of Wastewater. Sci. Total Environ. 2021, 763, 144587 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. K.; Duong D.; Bakker K. M.; Ammerman M.; Mortenson L.; Hughes B.; Martin E. T.; White B. J.; Boehm A. B.; Wigginton K. R. Wastewater-Based Detection of an Influenza Outbreak; preprint; Public and Global Health. Environ. Sci. Technol. Lett. 2022, 9, 687–692. 10.1021/acs.estlett.2c00350. [DOI] [Google Scholar]
- Wolfe M. K.; Yu A. T.; Duong D.; Rane M. S.; Hughes B.; Chan-Herur V.; Donnelly M.; Chai S.; White B. J.; Vugia D. J.; Boehm A. B. Use of Wastewater for Mpox Outbreak Surveillance in California. N. Engl. J. Med. 2023, 388, 570–572. 10.1056/NEJMc2213882. [DOI] [PubMed] [Google Scholar]
- Yin Z.; Voice T. C.; Tarabara V. V.; Xagoraraki I. Sorption of Human Adenovirus to Wastewater Solids. J. Environ. Eng. 2018, 144, 06018008 10.1061/(ASCE)EE.1943-7870.0001463. [DOI] [Google Scholar]
- Hayes E. K.; Sweeney C. L.; Fuller M.; Erjavec G. B.; Stoddart A. K.; Gagnon G. A. Operational Constraints of Detecting SARS-CoV-2 on Passive Samplers Using Electronegative Filters: A Kinetic and Equilibrium Analysis. ACS EST Water 2022, 11, 1910–1920. 10.1021/acsestwater.1c00441. [DOI] [PubMed] [Google Scholar]
- Hughes B.; Duong D.; White B. J.; Wigginton K. R.; Chan E. M. G.; Wolfe M. K.; Boehm A. B. Respiratory Syncytial Virus (RSV) RNA in Wastewater Settled Solids Reflects RSV Clinical Positivity Rates; preprint; Infectious Diseases (except HIV/AIDS). Environ. Sci. Technol. Lett. 2021, 9, 173–178. 10.1021/acs.estlett.1c00963. [DOI] [Google Scholar]
- Keshaviah A.; Diamond M. B.; Wade M. J.; Scarpino S. V.; Ahmed W.; Amman F.; Aruna O.; Badilla-Aguilar A.; Bar-Or I.; Bergthaler A.; Bines J. E.; Bivins A. W.; Boehm A. B.; Brault J.-M.; Burnet J.-B.; Chapman J. R.; Chaudhuri A.; De Roda Husman A. M.; Delatolla R.; Dennehy J. J.; Diamond M. B.; Donato C.; Duizer E.; Egwuenu A.; Erster O.; Fatta-Kassinos D.; Gaggero A.; Gilpin D. F.; Gilpin B. J.; Graber T. E.; Green C. A.; Handley A.; Hewitt J.; Holm R. H.; Insam H.; Johnson M. C.; Johnson R.; Jones D. L.; Julian T. R.; Jyothi A.; Keshaviah A.; Kohn T.; Kuhn K. G.; La Rosa G.; Lesenfants M.; Manuel D. G.; D’Aoust P. M.; Markt R.; McGrath J. W.; Medema G.; Moe C. L.; Murni I. K.; Naser H.; Naughton C. C.; Ogorzaly L.; Oktaria V.; Ort C.; Karaolia P.; Patel E. H.; Paterson S.; Rahman M.; Rivera-Navarro P.; Robinson A.; Santa-Maria M. C.; Scarpino S. V.; Schmitt H.; Smith T.; Stadler L. B.; Stassijns J.; Stenico A.; Street R. A.; Suffredini E.; Susswein Z.; Trujillo M.; Wade M. J.; Wolfe M. K.; Yakubu H.; Zanoli Sato M. I. Wastewater Monitoring Can Anchor Global Disease Surveillance Systems. Lancet Glob. Health 2023, 11, e976–e981. 10.1016/S2214-109X(23)00170-5. [DOI] [PubMed] [Google Scholar]
- Shah S.; Gwee S. X. W.; Ng J. Q. X.; Lau N.; Koh J.; Pang J. Wastewater Surveillance to Infer COVID-19 Transmission: A Systematic Review. Sci. Total Environ. 2022, 804, 150060 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M. A.; Boehm A. B.; Salit M.; Spencer S. K.; Wigginton K. R.; Noble R. T. The Environmental Microbiology Minimum Information (EMMI) Guidelines: QPCR and DPCR Quality and Reporting for Environmental Microbiology. Environ. Sci. Technol. 2021, 55, 10210–10223. 10.1021/acs.est.1c01767. [DOI] [PubMed] [Google Scholar]
- Hejkal T. W.; Wellings F. M.; Lewis A. L.; LaRock P. A. Distribution of Viruses Associated with Particles in Waste Water. Appl. Environ. Microbiol. 1981, 41, 628–634. 10.1128/aem.41.3.628-634.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman J. S.; Scire J.; Caduff L.; Fernandez-Cassi X.; Ganesanandamoorthy P.; Kull A.; Scheidegger A.; Stachler E.; Boehm A. B.; Hughes B.; Knudson A.; Topol A.; Wigginton K. R.; Wolfe M. K.; Kohn T.; Ort C.; Stadler T.; Julian T. R. Wastewater-Based Estimation of the Effective Reproductive Number of SARS-CoV-2. Environ. Health Perspect. 2022, 130, 057011 10.1289/EHP10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standard Operating Procedures for Interlaboratory and Methods Assessment of the SARS-CoV-2 Genetic Signal in Wastewater.
- Kalam S.; Abu-Khamsin S. A.; Kamal M. S.; Patil S. Surfactant Adsorption Isotherms: A Review. ACS Omega 2021, 6, 32342–32348. 10.1021/acsomega.1c04661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R. S.; Nelson K. L.; Greenwald H. D.; Kennedy L. C. Challenges in Measuring the Recovery of SARS-CoV-2 from Wastewater. Environ. Sci. Technol. 2021, 55, 3514–3519. 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kim S.; Boehm A. B. Wastewater Monitoring of SARS-CoV-2 RNA at K-12 Schools: Comparison to Pooled Clinical Testing Data. PeerJ 2023, 11, e15079 10.7717/peerj.15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breadner P. R.; Dhiyebi H. A.; Fattahi A.; Srikanthan N.; Hayat S.; Aucoin M. G.; Boegel S. J.; Bragg L. M.; Craig P. M.; Xie Y.; Giesy J. P.; Servos M. R. A Comparative Analysis of the Partitioning Behaviour of SARS-CoV-2 RNA in Liquid and Solid Fractions of Wastewater. Sci. Total Environ. 2023, 895, 165095 10.1016/j.scitotenv.2023.165095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik A. H. S.; Glier M. B.; Servos M.; Mangat C. S.; Pang X.-L.; Qiu Y.; D’Aoust P. M.; Burnet J.-B.; Delatolla R.; Dorner S.; Geng Q.; Giesy J. P.; McKay R. M.; Mulvey M. R.; Prystajecky N.; Srikanthan N.; Xie Y.; Conant B.; Hrudey S. E. Comparison of Approaches to Quantify SARS-CoV-2 in Wastewater Using RT-QPCR: Results and Implications from a Collaborative Inter-Laboratory Study in Canada. J. Environ. Sci. 2021, 107, 218–229. 10.1016/j.jes.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syngouna V. I.; Chrysikopoulos C. V. Interaction between Viruses and Clays in Static and Dynamic Batch Systems. Environ. Sci. Technol. 2010, 44, 4539–4544. 10.1021/es100107a. [DOI] [PubMed] [Google Scholar]
- Bellou M. I.; Syngouna V. I.; Tselepi M. A.; Kokkinos P. A.; Paparrodopoulos S. C.; Vantarakis A.; Chrysikopoulos C. V. Interaction of Human Adenoviruses and Coliphages with Kaolinite and Bentonite. Sci. Total Environ. 2015, 517, 86–95. 10.1016/j.scitotenv.2015.02.036. [DOI] [PubMed] [Google Scholar]
- Soller J.; Jennings W.; Schoen M.; Boehm A.; Wigginton K.; Gonzalez R.; Graham K. E.; McBride G.; Kirby A.; Mattioli M. Modeling Infection from SARS-CoV-2 Wastewater Concentrations: Promise, Limitations, and Future Directions. J. Water Health 2022, 20, 1197–1211. 10.2166/wh.2022.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. K.; Archana A.; Catoe D.; Coffman M. M.; Dorevich S.; Graham K. E.; Kim S.; Grijalva L. M.; Roldan-Hernandez L.; Silverman A. I.; Sinnott-Armstrong N.; Vugia D. J.; Yu A. T.; Zambrana W.; Wigginton K. R.; Boehm A. B. Scaling of SARS-CoV-2 RNA in Settled Solids from Multiple Wastewater Treatment Plants to Compare Incidence Rates of Laboratory-Confirmed COVID-19 in Their Sewersheds. Environ. Sci. Technol. Lett. 2021, 8, 398–404. 10.1021/acs.estlett.1c00184. [DOI] [PubMed] [Google Scholar]
- Lowry S. A.; Wolfe M. K.; Boehm A. B. Respiratory Virus Concentrations in Human Excretions That Contribute to Wastewater: A Systematic Review and Meta-Analysis. J. Water Health 2023, 21, 831–848. 10.2166/wh.2023.057. [DOI] [PubMed] [Google Scholar]
- Montiel-Garcia D.; Santoyo-Rivera N.; Ho P.; Carrillo-Tripp M.; Brooks C. L. III; Johnson J. E.; Reddy V. S. VIPERdb v3.0: A Structure-Based Data Analytics Platform for Viral Capsids. Nucleic Acids Res. 2021, 49, D809–D816. 10.1093/nar/gkaa1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Kennedy L. C.; Wolfe M. K.; Criddle C. S.; Duong D. H.; Topol A.; White B. J.; Kantor R. S.; Nelson K. L.; Steele J. A.; Langlois K.; Griffith J. F.; Zimmer-Faust A. G.; McLellan S. L.; Schussman M. K.; Ammerman M.; Wigginton K. R.; Bakker K. M.; Boehm A. B. SARS-CoV-2 RNA Is Enriched by Orders of Magnitude in Primary Settled Solids Relative to Liquid Wastewater at Publicly Owned Treatment Works. Environ. Sci.: Water Res. Technol. 2022, 8, 757–770. 10.1039/D1EW00826A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. K.; Duong D.; Bakker K. M.; Ammerman M.; Mortenson L.; Hughes B.; Arts P.; Lauring A. S.; Fitzsimmons W. J.; Bendall E.; Hwang C. E.; Martin E. T.; White B. J.; Boehm A. B.; Wigginton K. R. Wastewater-Based Detection of Two Influenza Outbreaks. Environ. Sci. Technol. Lett. 2022, 9, 687–692. 10.1021/acs.estlett.2c00350. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.