Abstract
The lifecycle of Zymoseptoria tritici requires a carefully regulated asymptomatic phase within the wheat leaf following penetration of the mesophyll via stomata. Here we compare the roles in this process of two key fungal signalling pathways, mutants of which were identified through forward genetics due to their avirulence on wheat. Whole‐genome resequencing of avirulent Z. tritici T‐DNA transformants identified disruptive mutations in ZtBCK1 from the kinase cascade of the cell wall integrity (CWI) pathway, and the adenylate cyclase gene ZtCYR1. Targeted deletion of these genes abolished the pathogenicity of the fungus and led to similar in vitro phenotypes to those associated with disruption of putative downstream kinases, both supporting previous studies and confirming the importance of these pathways in virulence. RNA sequencing was used to investigate the effect of ZtBCK1 and ZtCYR1 deletion on gene expression in both the pathogen and host during infection. ZtBCK1 was found to be required for the adaptation to the host environment, controlling expression of infection‐associated secreted proteins, including known virulence factors. Meanwhile, ZtCYR1 is implicated in controlling the switch to necrotrophy, regulating expression of effectors associated with this transition. This represents the first study to compare the influence of CWI and cAMP signalling on in planta transcription of a fungal plant pathogen, providing insights into their differential regulation of candidate effectors during invasive growth.
Keywords: cAMP signalling, cell wall integrity, effectors, plant pathogen, virulence, wheat, Zymoseptoria tritici
The importance of Zymoseptoria tritici cell wall integrity and cAMP signalling on virulence and in planta gene expression was compared, demonstrating their differential regulation of candidate effectors during invasive growth.
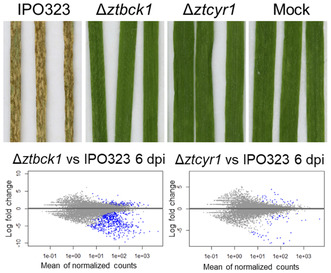
1. INTRODUCTION
Signalling cascades are integral for the ability of cells to adapt to their environment, enabling them to couple the recognition of external cues at the surface with appropriate intracellular processes, such as regulation of gene expression and protein activity. For fungal pathogens of plants, this involves regulating crucial changes in morphology and expression of virulence factors in response to the host environment. This is essential for controlling developmental transitions during infection, involving host surface recognition and penetration, invasive growth, and differentiation of reproductive structures. It is also important for responding to the stresses imposed by the host environment, such as nutrient deprivation and the plant immune response.
The cell wall integrity (CWI) pathway is a conserved fungal signalling cascade that regulates remodelling of the cell wall (Levin, 2011). This pathway transduces cell wall perturbation signals recognized by mechanosensors at the cell surface to the conserved mitogen‐activated protein kinases (MAPK) Slt2. Modulation of cell wall composition is required for fungal growth and morphological transitions (Cabib & Arroyo, 2013; Riquelme et al., 2018), as well as responses to various stresses, including those encountered in the host environment (Geoghegan et al., 2017; Hopke et al., 2018). Furthermore, fungal pathogens adapt their cell wall composition to avoid host detection, as both chitin and β‐1,3‐glucan are recognized as pathogen‐associated molecular patterns (PAMPs) by host immune systems (Ballou et al., 2016; El Gueddari et al., 2002; Fujikawa et al., 2012).
The CWI pathway has been found to play a crucial role in the virulence of plant fungal pathogens (Turrà et al., 2014), with functions in host penetration (Kojima et al., 2002; Rui & Hahn, 2007; Xu et al., 1998) and invasive growth (Joubert et al., 2011; Mehrabi et al., 2006; Mey et al., 2002; Rui & Hahn, 2007). The latter function has been attributed to the role of the CWI pathway in withstanding host defences, as Slt2 deletion was shown to increase sensitivity to hydrolytic enzymes (Mey et al., 2002; Rui & Hahn, 2007; Xu et al., 1998), as well as plant‐derived antimicrobial compounds (Joubert et al., 2011; Ramamoorthy et al., 2007).
Another signalling cascade that has emerged as crucial for the virulence of fungal plant pathogens is the cyclic adenosine monophosphate (cAMP)‐dependent protein kinase A (PKA) pathway (Turrà et al., 2014). In Saccharomyces cerevisiae, cAMP generated by the adenylate cyclase enzyme binds to the regulatory subunits (encoded by BCY1) of the PKA complex, allowing the catalytic subunits (encoded by TPK1‐3) to dissociate and phosphorylate downstream targets. This pathway functions in nutrient sensing in yeast to regulate carbohydrate metabolism, the cell cycle, growth, and development (Zaman et al., 2008). However, the cAMP‐PKA pathway is known to regulate an extremely diverse range of fungal processes, including morphological transitions (D'Souza & Heitman, 2001), sexual reproduction (Hu et al., 2014), secondary metabolism (Studt et al., 2013), and responses to diverse nutritional signals (Caza & Kronstad, 2019). cAMP‐PKA signalling is required for development of host penetration structures in many plant‐pathogenic fungi, including Magnaporthe oryzae (Choi & Dean, 1997; Xu et al., 1997) and Fusarium graminearum (Bormann et al., 2014). Furthermore, there is also evidence for the role of this pathway in post‐penetration invasive growth, with strains lacking cAMP‐PKA components unable to cause disease following inoculation into experimental wounded leaf tissue (Li et al., 2017; Yamauchi et al., 2004).
The ascomycete pathogen Zymoseptoria tritici causes the most economically important disease of wheat in Europe (Jørgensen et al., 2014). Colonization of wheat by Z. tritici involves complex regulation of morphological development and the molecular interaction with the host, requiring dramatic shifts in gene expression across different infection stages (Rudd et al., 2015). Initial colonization involves hyphal germination on the leaf surface and invasion of the host stomata (Kema et al., 1996). Z. tritici then grows asymptomatically in the apoplast whilst suppressing the host immune response through expression of secreted effectors, including essential repression of chitin recognition by the LysM effectors (Marshall et al., 2011; Rudd et al., 2015). Finally, Z. tritici switches to necrotrophic growth, instigating host programmed cell death through up‐regulation of a distinct set of effectors and acquiring nutrients from dying host tissue through expression of secreted enzymes and membrane transporters (Rudd et al., 2015).
The CWI pathway has been proposed to control Z. tritici resistance to apoplastic wheat defence compounds during asymptomatic invasion, as Δmgslt2 mutants were unable to colonize the mesophyll tissue following stomatal penetration and showed increased sensitivity to cell wall‐degrading enzymes (Mehrabi et al., 2006). Furthermore, deletion of the PKA catalytic subunit MgTpk2 led to delayed symptom development and abolished pycnidia formation, despite successful host penetration and invasive growth, suggesting its involvement in triggering the switch to necrotrophy and asexual development (Mehrabi & Kema, 2006). However, despite these investigations, links between these regulatory pathways and the transcription of genes involved in host manipulation by Z. tritici remain to be elucidated.
In this study, forward genetic investigation of avirulent T‐DNA insertion transformants identified disruptions to Z. tritici genes encoding adenylate cyclase and the CWI MAPK cascade enzyme BCK1. Targeted deletion of these genes was used to confirm that loss of their functions led to abolishment of pathogenicity. Despite showing defects during in vitro vegetative growth, deletion strains were able to penetrate host stomata, supporting previous evidence for the role of the CWI and cAMP‐PKA pathways during infection post‐penetration. Finally, RNA sequencing was used to characterize transcription in both Z. tritici and wheat during infection by these strains to investigate the enigmatic in planta functions of these pathways. This revealed different suites of candidate effectors involved in invasive growth regulated by these two pathways and provided insight into how this differential regulation impacts the host defence response.
2. RESULTS
2.1. Nonsynonymous SNP identified in ZtCYR1 gene of avirulent T‐DNA insertion strains
Two transformants with severely reduced pathogenicity, designated L2 and C5, were identified during in planta phenotyping of targeted deletion mutants that aimed to inactivate the genes ZtritIPO323_04g10737 (Mycgr3G43288) and ZtritIPO323_04g13543 (Mycgr3G18212), encoding putative secreted lipase (ZtL2) and cutinase (ZtCUT5) proteins, respectively. L2 and C5 did not cause necrosis or develop pycnidia, although patches of mild chlorosis were observed at 21 days postinoculation (dpi) (Figure S1a). This phenotype was contrary to two PCR‐validated independent deletion mutants in these target genes, which displayed wild‐type virulence (Figure S1a). Whole‐genome resequencing of C5 and L2 found no evidence for ectopic insertion of transformed T‐DNA (Figure S2a,b), suggesting that avirulence in these strains was caused by random mutation(s). Variant calling found these strains to share 15 polymorphisms compared to the isogenic IPO323 strain (Table S2), suggesting that they originate from the same genotype in the background population. These included two within the coding regions of genes; a 3 bp deletion was identified in the first exon of ZtAGO1 (ZtritIPO323_04g08085/Mycgr3G38035), causing a P76del deletion, and a nonsynonymous single‐nucleotide polymorphism (SNP) in the second exon of the Z. tritici homologue of adenylate cyclase enzyme MgCYR1 (ZtritIPO323_04g11209/Mycgr3G86659), hereafter ZtCYR1 (Table S2). The latter substitution causes a missense mutation of E1663K at a predicted nucleotidyl binding site in the cyclase domain (IPR001054) of ZtCYR1 (Figure S2c). Previous studies have found ZtAGO1 to have a mild or no contribution to virulence (Habig et al., 2021; Kettles et al., 2019), while other components of the cAMP‐PKA pathway are known to contribute to the virulence of Z. tritici (Mehrabi et al., 2009; Mehrabi & Kema, 2006). Considering this, disruption of ZtCYR1 was hypothesized to cause the loss of virulence in the strains C5 and L2, prompting further functional characterization of this gene.
2.2. Ectopic T‐DNA insertion causes disruption of ZtBCK1 gene and promotor region
A third avirulent T‐DNA insertion mutant, referred to as T21, was identified during screening of transformants generated for targeted deletion of ZtritIPO323_04g07737 (Mycgr3G108617), encoding a triacylglycerol lipase (ZtTGL1). T21 was identified as having a defect in melanization during subculturing. Unlike three independent Δzttgl1 mutants, which displayed wild‐type virulence, T21 was avirulent (Figure S1b). Whole‐genome resequencing confirmed the absence of T‐DNA insertion at the ZtTGL1 locus but identified insertion of the full T‐DNA sequence at a locus on chromosome 11 (Figure S3). The ectopic insertion site lies 133 bp upstream of the gene ZtritIPO323_04g03043 (Figure S3), hereafter ZtBCK1, encoding the Z. tritici homologue yeast mitogen‐activated protein kinase kinase kinase regulating Slt2 in the CWI pathway (Lee et al., 1992). Furthermore, variant calling identified a 12 bp deletion (chr11:240,721‐240,732) in the coding sequence of ZtBCK1, causing a four amino acid deletion G6_R9del (Table S2 and Figure S3) outside any predicted conserved domains. Variant calling also identified a nonsynonymous SNP in the second exon of ZtritIPO323_04t11145 (Mycgr3G60318), encoding a putative major facilitator superfamily (MFS) transporter (Table S2). However, considering the previous evidence for the virulence‐related function of the CWI pathway in Z. tritici (Mehrabi et al., 2006), the disruption of ZtBCK1 was chosen for further investigation into the cause of avirulence in T21.
2.3. Targeted deletion of ZtBCK1 and ZtCYR1 abolishes virulence in Z. tritici
To investigate the function of ZtBCK1 and ZtCYR1 in Z. tritici development and virulence, three independently constructed null mutants were generated for both genes (Figure S4a,b), and these were shown to display comparable phenotypes both in vitro and in planta. The resulting Δztcyr1 and Δztbck1 strains showed equivalent attenuation of virulence to that displayed by strains L2/C5 and T21, with all deletion strains unable to induce necrosis or develop pycnidia at 21 dpi (Figure 1a). Although the mild chlorosis caused by L2 and C5 was not observed on Δztcyr1‐infected leaves at 21 dpi, continuation of infection to 35 dpi led to the appearance of chlorosis on these leaves (Figure 1a). Chlorosis was not observed on mock‐inoculated or Δztbck1‐infected leaves, confirming that this was not the result of environmental conditions or senescence (Figure 1a). These findings provide supporting evidence that the avirulence of L2/C5 and T21 is probably caused by disruption of ZtCYR1 and ZtBCK1, respectively.
FIGURE 1.
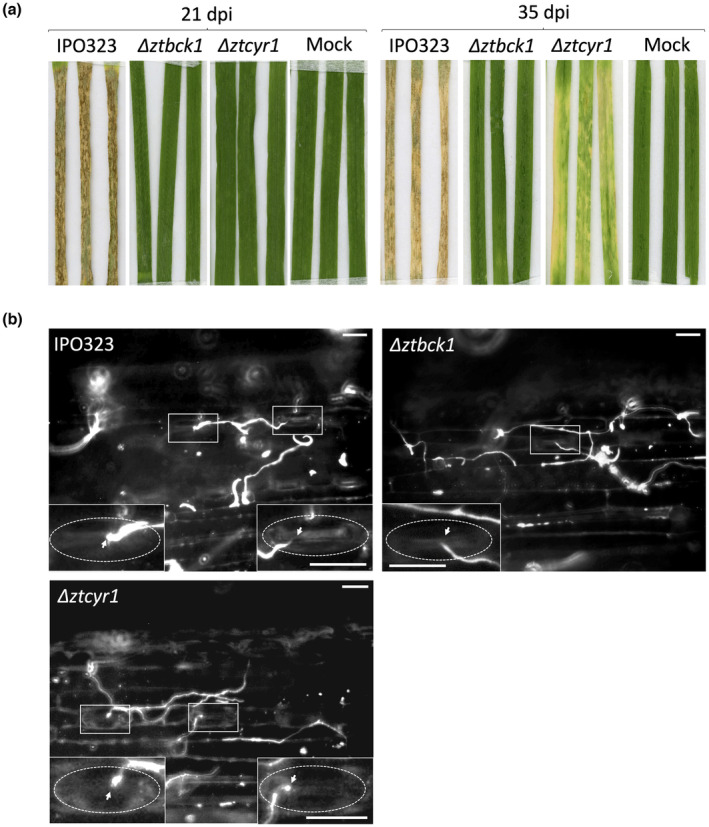
Targeted deletion of ZtBCK1 and ZtCYR1 leads to abolition of Zymoseptoria tritici virulence. (a) Disease symptoms on wheat leaves after 21 and 35 days postinoculation (dpi) by Δztcyr1 and Δztbck1 strains compared to wild‐type IPO323. (b) Epiphytic Z. tritici hyphae stained with calcofluor white on the leaf surface 3 dpi. Magnified images detailing hypha–stomata interactions (arrows), with guard cells indicated by dotted oval. Scale bars = 40 μm.
Microscopic observations revealed no difference in stomatal interactions between both Δztcyr1 and Δztbck1 strains and wild‐type IPO323 (Figure 1b). Spores of both mutants germinated on the leaf surface to produce epiphytic hyphae, and numerous stomatal interactions were observed for all strains at 3 dpi (Figure 1b). These findings establish that avirulence in these strains is not caused by disruption of the morphological switch to hyphal growth, sensory perception of the host surface or recognition of stomatal apertures. Instead, this suggests that signalling via ZtCYR1 and ZtBCK1 is required during infection stages after host penetration.
2.4. Deletion of ZtBCK1 impacts vegetative growth and the cell wall stress response
Considering previous reports of the CWI pathway influencing both yeast‐like and hyphal growth of Z. tritici (Mehrabi et al., 2006), the vegetative growth characteristics of Δztbck1 strains were investigated. While the growth rate by blastosporulation on yeast peptone dextrose (YPD) agar was consistent between Δztbck1 strains and IPO323, a defect in melanization was seen in Δztbck1 mutants after a prolonged growth period (Figure 2a). Furthermore, swollen cells were observed regularly in the blastospores of Δztbck1 strains after 6 days of growth on YPD, which were absent in IPO323 cells under the same conditions (Figure 2b). Although germination efficiency and early colony formation on water agar (WA) was unchanged in Δztbck1 strains, their growth rate was reduced over time as radial growth continued, resulting in smaller colonies than the wild type after 14 days (Figure 2c). In addition, while the wild type developed abundant aerial hyphae at 28°C on potato dextrose agar (PDA), Δztbck1 strains displayed equivalent growth by blastosporulation at 19 and 28°C (Figure 2c). These findings suggest that the Z. tritici CWI pathway is involved in regulating the morphological switch to hyphal growth in response to heat stress, as well as in prolonged hyphal growth under starvation. Finally, the in vitro phenotypes of Δztbck1 were entirely consistent with those of T21, providing further evidence for ZtBCK1 disruption being the sole genetic cause of the phenotypes of this strain.
FIGURE 2.
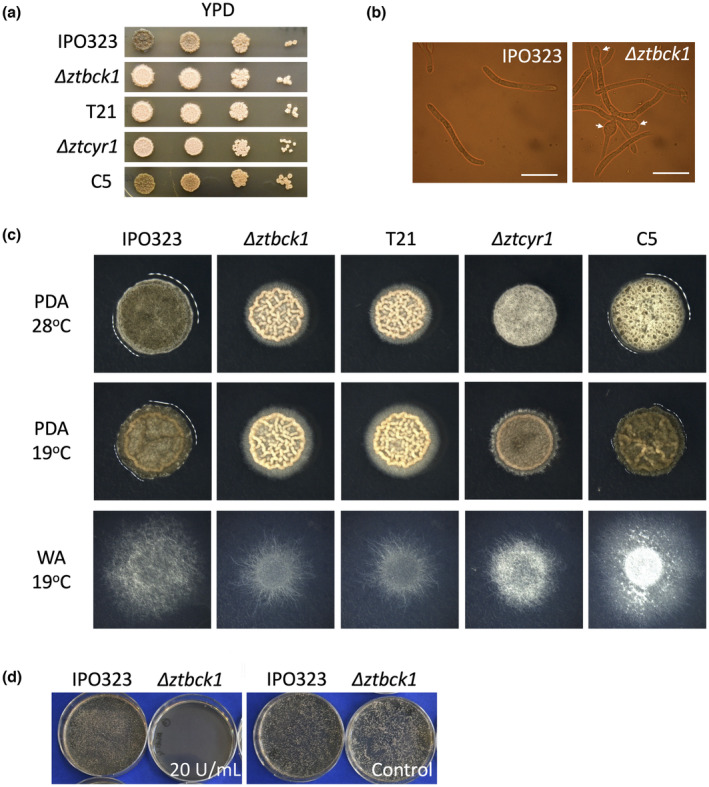
Vegetative growth phenotypes of ZtBCK1 and ZtCYR1 deletion strains. (a) Zymoseptoria tritici grown for 10 days on yeast peptone dextrose (YPD) at 19°C, 5‐μL droplets of a 10‐fold serial dilution starting at a concentration of 5 × 106 spores/mL. (b) Blastospores grown on YPD for 6 days, with appearance of swollen cells (arrows) in Δztbck1. Scale bar = 20 μm. (c) Growth on potato dextrose agar (PDA) at 28 and 19°C for 7 days and radial hyphal growth on water agar (WA) at 19°C for 14 days. (d) Z. tritici cell suspensions exposed 20 U/mL of lyticase enzyme from Arthrobacter luteus for 2 h at 25°C before being cultured on YPD agar for 5 days at 19°C, showing loss of viability of Δztbck1 strains at lower enzyme concentrations than the wild type IPO323.
The sensitivity of wild‐type and Δztbck1 strains to cell wall stress was assessed by exposure to enzyme mixtures possessing β‐1,3‐glucanase activity. The wild type was able to maintain viability after exposure to a higher concentration of lyticase enzymes than Δztbck1, providing evidence that deletion of ZtBCK1 leads to enhanced sensitivity to cell wall stress (Figure 2d). To investigate whether this effect was specific to disruption of β‐1,3‐glucan, the sensitivity of Δztbck1 to calcofluor white, which disrupts chitin synthesis, and caspofungin, which inhibits β‐1,3‐glucan synthesis, was assessed. While Δztbck1 strains displayed no change in sensitivity to calcofluor white, they were found to have elevated sensitivity to caspofungin (Figure S5a). Together, these results suggest that the ZtBCK1 is specifically involved in regulation of the Z. tritici response to perturbation of β‐1,3‐glucan integrity. Furthermore, Δztcyr1 strains, but not strain C5, also displayed increased sensitivity to caspofungin (Figure S5a), suggesting that cAMP signalling may also play a role in this response.
2.5. Deletion of ZtCYR1 impairs melanization and increases osmosensitivity
Vegetative growth phenotypes of Δztcyr1 strains were also investigated for comparison with previously characterized other components of the PKA signalling pathway (Mehrabi et al., 2009; Mehrabi & Kema, 2006). The growth rate by blastosporulation on YPD was indistinguishable from the wild type (Figure 2a). However, melanization was impaired in Δztcyr1 when grown on YPD and PDA at 19°C in the dark (Figure 2a,c). Similarly, melanization of aerial hyphae was not observed in Δztcyr1 strains when grown on PDA at 28°C. Furthermore, while the germination efficiency of Δztcyr1 strains on WA was unchanged from the wild type, subsequent hyphal growth proceeded at a much slower rate in these mutants (Figure 2c). Interestingly, the disruption of melanization and hyphal growth rate was not observed in the C5 strain. Finally, sensitivity to high osmolarity of the growth media was increased in the C5 and Δztcyr1 strains, which displayed a reduced growth rate compared to the wild type on PDA amended with 1.5 M sorbitol (Figure S5b). These findings suggest that ZtCYR1 is involved in the regulation of melanization, hyphal growth, and the osmotic stress responses, but that the phenotype potentially caused by the point mutation in ZtCYR1 in the C5 strain is distinct from that resulting from deletion of the whole coding sequence.
2.6. Deletion of ZtBCK1 and ZtCYR1 impacts in planta growth and gene transcription
Considering the observation of successful epiphytic growth and stomatal interactions during infection by Δztcyr1 and Δztbck1 strains, we hypothesized that avirulence in these strains was caused by defective regulation of genes and processes required for intercellular colonization of the leaf tissue. To investigate this, transcriptome profiling of both Z. tritici and wheat during infection by the wild‐type IPO323, Δztcyr1, and Δztbck1 was carried out by RNA sequencing. Infected leaf samples were taken at 6 dpi to assess differences in gene expression during the late symptomless phase of wild‐type infection, when stomatal penetration by most epiphytic hyphae has occurred and invasive growth is being established in the mesophyll. Samples were also taken at 9 dpi at the start of the transition to necrotrophic growth by the wild‐type IPO323, which has been shown previously to coincide with dramatic transcriptional changes in both the pathogen and host (Rudd et al., 2015). It must be noted that leaf samples infected by all Z. tritici strains remained asymptomatic at 9 dpi before the wild type began to induce chlorosis between 10 and 12 dpi.
The progress of leaf colonization by each strain between the sampled time points was assessed quantitatively through the relative abundance of Z. tritici reads as a measure of fungal biomass. No significant difference in the proportion of Z. tritici transcripts was identified between the strains at 6 dpi, suggesting that fungal growth rate up to this point of infection was largely equivalent (Figure 3a). IPO323 displayed significant increases in the proportion of reads mapped to the Z. tritici genome at 9 dpi (Figure 3a). This is consistent with previous reports of measurable increases in fungal biomass as Z. tritici transitions to necrotrophy, changes which are difficult to detect during early symptomless growth (Keon et al., 2007; Palma‐Guerrero et al., 2016; Rudd et al., 2015). This provides evidence that the infection time points where samples were taken effectively encompass this transition in the infection stage.
FIGURE 3.
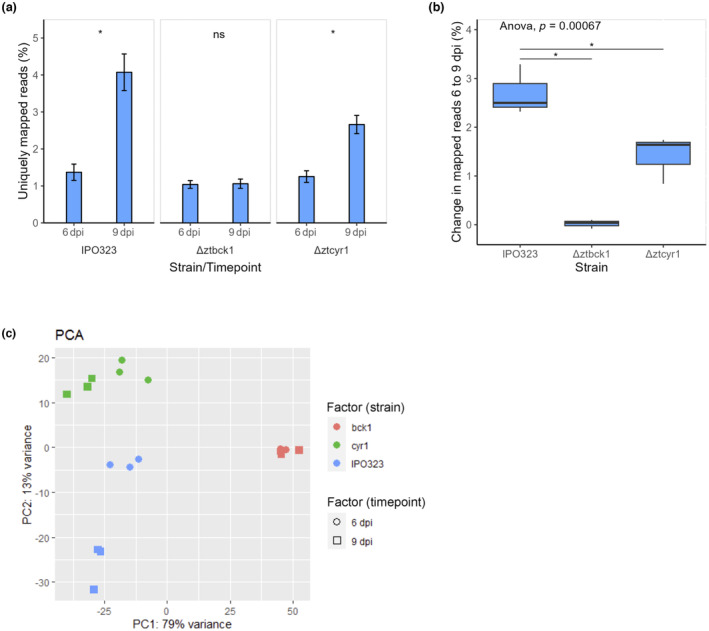
In planta growth and global gene transcription are impacted by deletion of ZtBCK1 and ZtCYR1. (a) Percentage of RNA sequencing reads uniquely mapped to the Zymoseptoria tritici genome across infection time points in each strain, as an indication of in planta fungal biomass. Mean values across each repeated sample are plotted along with the standard error. Statistically significant differences between percentages across time points in each strain from paired t test are indicated (*p < 0.05). (b) Change in the percentage of uniquely mapped reads between 6 and 9 days postinoculation (dpi) in each experimental repeat grouped by strain, as an indication of growth between these time points. p value from one‐way analysis of variance is given and statistically significant differences from pairwise t test comparison of mutants to IPO323 are indicated (*p < 0.05). (c) Principal component analysis (PCA) of global normalized gene expression data.
In contrast to the wild type, no significant increase in the percentage of reads mapped to the Z. tritici genome was identified between 6 and 9 dpi for the Δztbck1 samples, indicating that there was no significant increase in the relative biomass of this strain in infected leaf tissue (Figure 3a). This suggests that colonization of the wheat leaf by Δztbck1 is inhibited during the symptomless phase of infection. Furthermore, while Δztcyr1 samples did display an increase in Z. tritici mapped reads between 6 and 9 dpi (Figure 3a), the magnitude of this increase was significantly greater in IPO323 than in Δztcyr1‐infected leaves (Figure 3b). This suggests that the increase in fungal biomass between these time points was less pronounced for the Δztcyr1 strain.
Principal component analysis (PCA) using global normalized gene expression data revealed that the Z. tritici transcriptome of each sample could be most easily distinguished by the strain used for infection (Figure 3c). The first principal component (PC1) encompasses most of the variation in gene expression between Δztbck1 and both Δztcyr1 and IPO323, explaining 76% of the variation in the dataset (Figure 3c). The second principal component (PC2), which explains 16% of the variation in gene expression, appears to comprise the majority of the variation between Δztcyr1 and IPO323, which is accentuated by the progression of infection to 9 dpi (Figure 3c). Although global gene expression in Δztcyr1 does appear to change between the time points, with clear grouping of Δztcyr1 6 and 9 dpi samples, this change is not as dramatic as the wild type (Figure 3c). These results suggest that divergence in global gene expression between IPO323 and Δztcyr1 increases as the wild type transitions to necrotrophic growth. This led to the hypothesis that disruption of cAMP signalling impairs the ability of Z. tritici to undergo this necrotrophic switch. Furthermore, Δztbck1 shows no variation between the sampled time points (Figure 3c), which supports the hypothesis that colonization by Δztbck1 does not progress beyond the initial invasion of the wheat leaf.
2.7. ZtBCK1 and ZtCYR1 regulate distinct sets of putative secreted proteins and candidate effectors in planta
Differential expression analysis led to the identification of 682 differentially expressed genes (DEGs) in Δztbck1 at 6 dpi, which increased to 1652 DEGs at 9 dpi compared to the expression profile in IPO323 (Figure 4a and Table S3). Furthermore, only three genes were found to be differentially expressed between the sampling time points in Δztbck1, compared to 198 DEGs identified between 6 and 9 dpi in IPO323 (Figure 4b and Table S3). These results suggest that not only is gene expression in the Δztbck1 mutant significantly different to the wild type during early infection, but that it also displays none of the transcriptional changes associated with the transition to necrotrophy.
FIGURE 4.
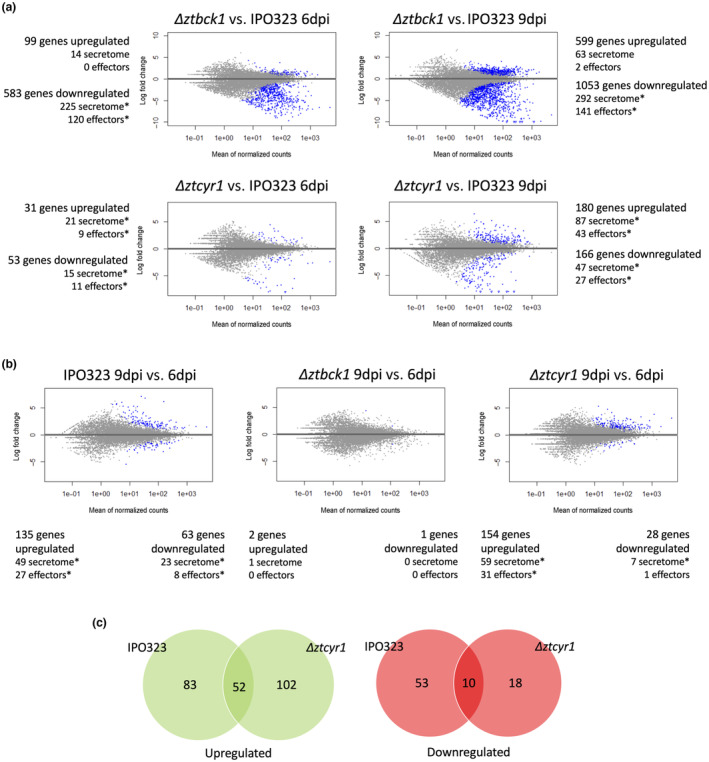
Δztbck1 shows widespread down‐regulation of secreted effectors and Δztcyr1 gene expression diverges from IPO323 as infection progresses. MA plots, of the log fold change in expression versus expression level (mean of normalized counts) are shown, displaying differentially expressed genes (blue points) between (a) mutant strains (Δztbck1 and Δztcyr1) and IPO323 at 6 and 9 days postinoculation (dpi), and (b) between 9 and 6 dpi in IPO323 (left), Δztbck1 (middle), and Δztcyr1 (right). The number of differentially expressed genes in each comparison are detailed, as well as the number of secretome and predicted effector genes in those differentially expressed sets. Asterisks indicate significant enrichment of these gene categories amongst differentially expressed gene sets determined using Fisher's exact tests (p < 0.01). (c) Comparison of genes found to be differentially expressed between 6 and 9 dpi in IPO323 and Δztcyr1.
Contrastingly, differential expression analysis found there to be 84 DEGs between Δztcyr1 and IPO323 at 6 dpi, which increased to 346 DEGs at 9 dpi (Figure 4a and Table S3). Furthermore, a set of 182 genes was found to be differentially expressed between 6 and 9 dpi in Δztcyr1 (Figure 4b and Table S3). Although this set showed some similarity to the genes found to be differentially expressed between the time points in IPO323, the majority (66%) were distinct (Figure 4c). These results support those from PCA in suggesting that while global transcription is similar between Δztcyr1 and IPO323 at 6 dpi, the progression of infection to 9 dpi leads to divergence in gene expression between these strains.
Given the importance of secreted proteins in the molecular interaction between Z. tritici and wheat, the differential expression of genes within the recently updated Z. tritici secretome was investigated (King et al., 2017). The secretome was defined as those genes within the Rothamsted genome annotation that contain a predicted signal peptide and do not contain a predicted transmembrane domain. Δztbck1 displayed widespread down‐regulation of genes within the secretome compared to IPO323 at both time points during infection (Figures 4a and 5a, and Table S3). Secretome genes were significantly enriched in the down‐regulated genes identified in Δztbck1 at both 6 and 9 dpi, making up 38.6% and 27.7% in these sets, respectively (Figure 4a). Functional annotation based on InterPro domains revealed that these down‐regulated secreted proteins include many with enzymatic activities that have been shown to be induced during wild‐type Z. tritici infection (Palma‐Guerrero et al., 2016; Rudd et al., 2015). This includes a host of secreted proteases, plant cell wall‐degrading enzymes, peroxidases, and superoxide dismutases (Figure 5b–d, and Table S3). Overall, these results suggest that Δztbck1 is unable to induce the transcription of a broad range of infection‐related secreted proteins in response to colonization of the host environment.
FIGURE 5.
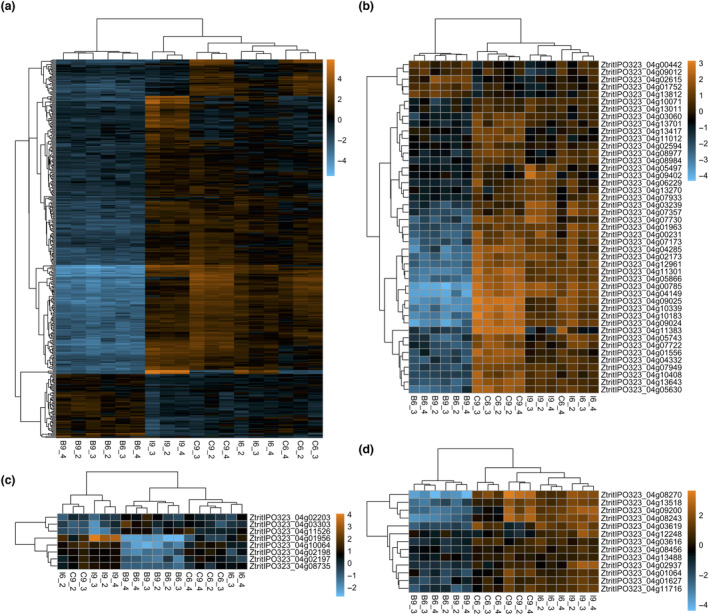
Differential expression of secreted proteins in Δztbck1 and Δztcyr1. Heat maps displaying log‐fold change in normalized expression values relative to the mean normalized expression for differentially expressed (a) secretome genes, (b) proteases, (c) peroxidase/superoxide dismutases, and (d) plant cell wall‐degrading enzymes. Column labels (e.g., B6_2) indicate the sample strain (B, Δztbck1; C, Δztcyr1; I, IPO323), time point (6 and 9 days postinocuation [dpi]), and replicate (2, 3, and 4).
Furthermore, Δztbck1 displayed widespread down‐regulation of genes encoding effector proteins (Figures 4a and 6a), predicted from the secretome using EffectorP v. 2.0 (Sperschneider et al., 2018). This includes the necrosis‐ and ethylene‐inducing protein 1 (Nep1)‐like protein MgNLP (Figure 6b), which is known to be highly up‐regulated during the late symptomless phase but is dispensable for virulence (Motteram et al., 2009). Crucially, all three LysM‐domain containing effectors, which are known to be Z. tritici virulence factors (Tian et al., 2021), are strongly down‐regulated in Δztbck1 at both 6 and 9 dpi (Figure 6b). This finding suggests that host colonization by Δztbck1 is inhibited by the inability of this strain to suppress the host defence response.
FIGURE 6.
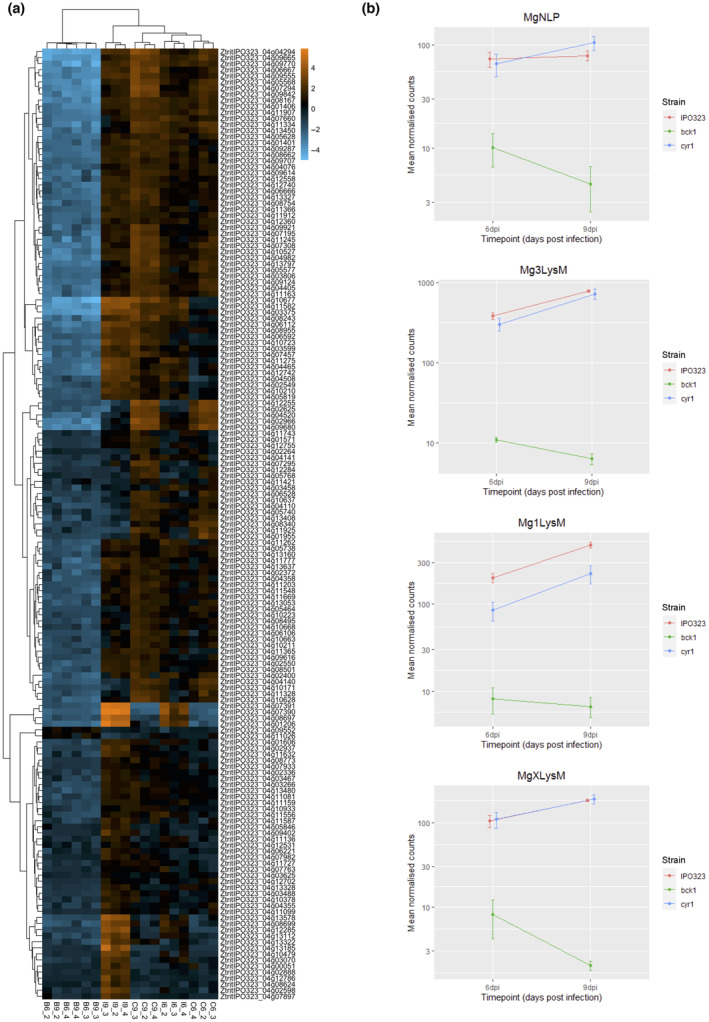
Putative secreted effector genes show differential regulation in Δztbck1 and Δztcyr1. (a) Heat map displaying log‐fold change in normalized expression values relative to the mean normalized expression for differentially expressed effectors in each sample. Column labels (e.g., B6_2) indicate the sample strain (B, Δztbck1; C, Δztcyr1; I, IPO323), time point (6 and 9 days postinoculation [dpi]), and replicate (2, 3, and 4). (b) Mean of the normalized count values for MgNLP (ZtritIPO323_04g04358), Mg3LysM (ZtritIPO323_04g03143), Mg1LysM (ZtritIPO323_04g12742), and MgXLysM (ZtritIPO323_04g12740) in each strain at 6 and 9 dpi. Error bars represent standard error.
The sets of up‐ and down‐regulated genes in Δztcyr1 compared to IPO323 from both time points were also significantly enriched with secretome genes (Figure 4a), the majority of which were functionally uncharacterized. All sets of Δztcyr1 DEGs were significantly enriched for predicted effectors, the number of which increased between 6 and 9 dpi (Figure 4a). However, compared to their widespread down‐regulation in Δztbck1 strains, the number of candidate effector DEGs was relatively few, indicating regulation of a more specific infection‐related gene set by the cAMP‐PKA pathway. Nineteen of the 27 effectors down‐regulated in Δztcyr1 at 9 dpi were up‐regulated between 6 and 9 dpi in IPO323 (Figures 6a and S6a, and Table S3), suggesting that they function in the onset of necrotrophy. To support this, it was found that all of the effectors in this set of 27 displayed peak expression at the transition to, or during, the necrotrophic phase in a previous study (Rudd et al., 2015). As well as protein effectors, two polyketide synthase (PKS) genes and the hybrid PKS ribosomal peptide synthase HSP1 (Rudd et al., 2015), which could function in biosynthesis of secondary metabolite effectors, were down‐regulated in Δztcyr1 at 9 dpi (Figure S6b). These results indicate that the expression of effectors is diverging between Δztcyr1 and the wild type as infection moves towards the necrotrophic phase.
Another striking difference in expression between Δztcyr1 and IPO323 during infection was the up‐regulation of secreted proteases in the mutant (Figure 5b), which increased from 5 to 16 between 6 and 9 dpi (Table S3). All of these secreted proteases were found previously to show peak expression at the transition to necrotrophy during IPO323 infection, which was proposed to represent a switch to the use of host proteins as a carbon source (Rudd et al., 2015). Also heavily represented in the DEGs in Δztcyr1 were the major facilitator superfamily (MFS) transporters, 18 and six of which were down‐ and up‐regulated, respectively, at 9 dpi in this strain (Table S2). Of these, 13 were annotated as sugar transporters (Figure S6c). Also down‐regulated at 9 dpi were three predicted amino acid transporters (Table S3). Together, these results suggest that genes potentially involved in acquisition of nutrients from the host are differentially regulated in Δztcyr1 as infection progresses.
As well as showing differential expression of infection‐related secreted proteins, genes involved in cell wall biosynthesis were also found to be differentially regulated in Δztbck1 during infection (Figure S7). A Z. tritici homologue of the Aspergillus fumigatus β‐1‐3‐glucanosyltransferase ZtGel2, involved in the formation of β‐1,3‐glucan branches, and an α‐1,3‐glucan synthase were both down‐regulated in Δztbck1 compared to IPO323 at both time points (Figure S7a). Along with the sensitivity of Δztbck1 to caspofungin and glucanase enzymes, this suggests that the Z. tritici CWI pathway is responsible for responding to β‐1,3‐glucan perturbation. Conversely, four chitin synthase genes were found to show significantly higher expression in Δztbck1 during infection (Figure S7b). Enhanced expression of chitin synthases in Δztbck1 could occur in response to cell wall damage caused by the unsuppressed host defence response, regulated by signalling mechanisms other than the CWI pathway.
2.8. Transcriptome profiling of wheat reveals different defence response to infection by Δztcyr1 and Δztbck1
Analysis of RNA sequencing reads aligned to the wheat genome was used to investigate whether the host response to Δztcyr1 and Δztbck1 infection was altered compared to colonization by the wild type. Clustering of samples based on global wheat gene expression data was investigated by multidimensional scaling (MDS) analysis (Figure 7a). This revealed that 40% of the variance between the samples can be summarized by the first dimension, which broadly segregates the samples by time point during infection at which they were taken (Figure 7a). When comparing wheat infected with each strain, a total of 3432 genes were differentially regulated between 6 and 9 dpi in the same direction (Figure 7b). TaMPK3, which is known to be involved in responses to various abiotic stresses in wheat (Goyal et al., 2018; Zhan et al., 2017), was significantly down‐regulated between 6 and 9 dpi in leaves infected with all the strains (Figure S8a). Furthermore, many RLK genes showed significant down‐regulation between 6 and 9 dpi in all infection experiments (Figure S8b and Table S4), expression of which is also known to respond to diverse abiotic stresses (Lehti‐Shiu et al., 2009; Shumayla, Kumar, et al., 2016; Shumayla, Pandey, et al., 2016). These findings suggest that all plants were under environmental stress at 6 dpi, which could have caused the large proportion of shared variation in global wheat gene expression observed between the time points in all experiments.
FIGURE 7.
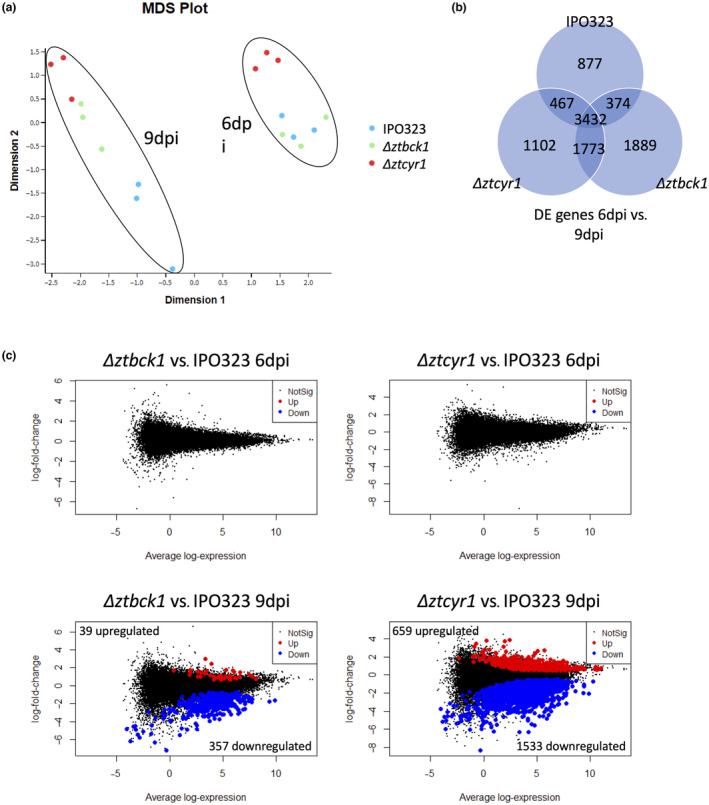
Global wheat gene expression analysis during infection. (a) Multidimensional scaling (MDS) plot on global wheat gene expression data. (b) Comparison of genes differentially expressed between 6 and 9 days postinoculation (dpi) in leaves infected with IPO323, Δztbck1, and Δztcyr1. (c) MA plots displaying differentially expressed genes between leaves infected with Δztbck1 at 6 dpi (top left) and 9 dpi (bottom left), and Δztcyr1 at 6 dpi (top right) and 9 dpi (bottom right), using leaves infected with IPO323 each time point as a reference.
Despite this, samples did cluster depending on the inoculated Z. tritici strain; leaf samples infected with IPO323 and Δztbck1 clustered separately from Δztcyr1 at 6 dpi, before samples infected by IPO323 diverged further from Δztcyr1 and Δztbck1 at 9 dpi (Figure 7b). Despite the apparent difference in global transcription between samples infected by Δztcyr1 and IPO323 at 6 dpi from MDS analysis, no statistically significant DEGs were identified between any strains at this time point (Figure 7c). However, a set of 396 DEGs were identified between Δztbck1‐ and IPO323‐infected samples at 9 dpi (Figure 7c and Table S4). Furthermore, 2192 DEGs were identified between Δztcyr1‐ and IPO323‐infected samples at 9 dpi (Figure 7c and Table S4).
Further analysis of these DEG sets focused on wheat genes with predicted functions in defence responses against pathogens. Four genes characterized as responding to Z. tritici infection in a previous study (Ray et al., 2003) were significantly down‐regulated in Δztcyr1 at 9 dpi (Figure 8a). This included the pathogenesis‐related (PR) proteins the PR1 protein PR‐1‐4 (TraesCS7D02G161200), the β‐1,3‐glucanase PR2 (TraesCS7D02G551400), and the thaumatin‐like protein PR5 (TraesCS7D02G551400), as well as the protein disulphide isomerase PDI2 (TraesCS4B02G101800). In addition to these genes, two other PR1 proteins, 12 other β‐1,3‐glucanases (PR2), 10 chitinases (PR3), and five other thaumatin‐like proteins (PR5) were found to be down‐regulated in Δztcyr1‐infected samples at 9 dpi (Figure 8b). A subset of β‐1,3‐glucanases and chitinases was also down‐regulated in Δztbck1‐infected samples at 9 dpi (Figure 8b).
FIGURE 8.
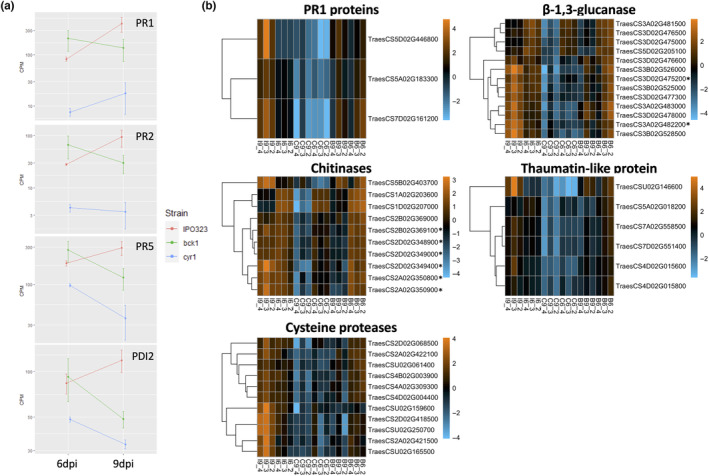
Defence‐related wheat genes show differential regulation in Δztbck1‐ and Δztcyr1‐infected leaves. (a) The mean counts per million (CPM) of each gene across samples infected with each strain at 6 and 9 days postinoculation (dpi) is plotted on a log scale, with error bars representing the standard error. (b) Heat maps displaying log‐CPM values relative to the mean of each row, scaled so that the standard deviation is 1 (z‐score). Plots display genes of defence‐related families that were differentially expressed in Δztcyr1‐infected samples at 9 dpi. Asterisks indicate differential expression in Δztbck1‐infected samples at 9 dpi. Column labels (e.g., B6_2) indicate the sample strain (B, Δztbck1; C, Δztcyr1; I, IPO323) and time point (6 and 9 dpi).
Other protein annotations that were highly represented in the down‐regulated genes in Δztcyr1 were proteases, including Cys‐ (Figure 8b), metallo‐ and subtilisin‐like families, ubiquitination proteins, and drug resistance ABC transporters (Table S3). Also highly represented in the DEGs within Δztcyr1‐infected leaves at 9 dpi were receptor‐like kinases (RLKs), including many from the wall‐associated, leucine‐rich repeat, cysteine‐rich, and lectin kinase families (Table S4). These protease and RLK families were previously characterized as transcriptionally associated with the transition to necrotrophy (Rudd et al., 2015). Furthermore, three LysM domain GPI‐anchored proteins of the chitin elicitor‐binding protein (CEBiP), which have been characterized in chitin‐induced defence responses to Z. tritici (Lee et al., 2014), were also down‐regulated at 9 dpi in Δztcyr1 (Table S4). TaMPK3, a plant stress‐responsive mitogen activated protein kinase, which is induced by Z. tritici at the switch to necrotrophy (Rudd et al., 2008), was also significantly down‐regulated in Δztcyr1 compared to IPO323‐infected leaves at 9 dpi. These findings strongly suggest that Δztcyr1 is unable to induce the wheat defence response seen during wild‐type infection, despite continuing to grow in the host environment.
Regarding the host transcriptional response to Δztbck1, many of the aforementioned defence‐related genes were found amongst those down‐regulated between 6 and 9 dpi specifically in Δztbck1 samples, and not in IPO323‐ or Δztcyr1‐infected leaves (Table S4). This suggests that the host defence response was induced in Δztbck1‐infected leaves at 6 dpi and subsequently declined at 9 dpi. This is supported by the fact that many of these defence‐related genes then become significantly down‐regulated in Δztbck1‐infected leaves compared to IPO323‐infected samples at 9 dpi (Figure 8b and Table S4), as the wild type progresses towards the necrotrophic phase and induction of the hypersensitive host response. Along with the finding that Δztbck1 fungal biomass does not increase between the time points, this suggests that Δztbck1 colonization is inhibited early on by the host defence response, which starts to subside as less fungal penetration events occur.
3. DISCUSSION
The hyphal growth rate of Δztbck1 was found to be reduced during prolonged growth on WA, similar to the previously characterized Δmgslt2 mutant (Mehrabi et al., 2006). This indicates that disruption of the CWI pathway may have a negative impact on Z. tritici polarized growth through deregulation of proteins involved in cell wall biosynthesis at the hyphal apex. However, despite causing defects in vegetative growth in vitro, deletion of ZtBCK1 did not influence the ability of Z. tritici spores to germinate on the leaf surface and grow to the stomatal aperture. Furthermore, no evidence was found for the growth of Δztbck1 mutants beyond the symptomless phase, with no increase in fungal biomass detected after 6 dpi. These findings are consistent with the phenotypes identified in Δmgslt2 mutants, which were unable to colonize the mesophyll after entering the substomatal cavities (Mehrabi et al., 2006), and point towards the role of the CWI pathway in regulating adaptation to the host environment.
Transcriptome profiling revealed that Δztbck1 was severely inhibited in the expression of predicted secreted proteins during infection. This included the three Z. tritici LysM effectors, which are known to have partially redundant functions in the evasion and tolerance of the chitin‐induced wheat defence response, and together are essential for full Z. tritici virulence (Marshall et al., 2011; Tian et al., 2021). Furthermore, wheat expression analysis revealed the high expression of numerous defence‐related genes in response to infection by Δztbck1, including many bona fide PR proteins. Together, these findings suggest that the CWI pathway is involved in regulation of virulence‐associated secreted proteins in response to the host apoplast, and that this is required for suppression of the host immune response to enable mesophyll colonization.
In S. cerevisiae, the CWI pathway is required for controlling cell wall homeostasis in response to stress caused by hydrolytic enzymes and cell wall‐perturbing agents, as well as heat, osmotic, and pH stress (Garcia et al., 2009; Kamada et al., 1995; Reinoso‐Martín et al., 2003). Here, Δztbck1 mutants were found to have increased sensitivity to β‐1,3‐glucanase enzymes and the β‐1,3‐glucan synthesis‐inhibiting echinocandin fungicide caspofungin, but not to chitin disruption by calcofluor white. This corroborates previous findings that deletion of MgSLT2 increases sensitivity to glucanase but not chitinase enzymes (Mehrabi et al., 2006). Furthermore, genes involved in biosynthesis of β‐1,3‐glucan and α‐1,3‐glucan were found to be down‐regulated in Δztbck1 during infection, while chitin synthases were up‐regulated in this strain. Increased chitin synthase expression in Δztbck1 could occur in response to the cell wall perturbation caused by the enhanced host defence response to this strain, without the protective influence of LysM effectors against host chitinases (Marshall et al., 2011). These findings suggest that the Z. tritici CWI signalling specifically regulates responses to glucan perturbation, while responses to chitin disruption are regulated by a separate pathway, such as the HOG1 MAPK and calcineurin pathways (Bruder Nascimento et al., 2016; Fortwendel et al., 2010; Munro et al., 2007).
Considering this, we hypothesize that the CWI pathway in Z. tritici has adapted to co‐regulate secreted proteins involved in cell wall remodelling and virulence‐related functions, explaining the down‐regulation of these genes in Δztbck1 during infection. These findings raise the possibility that ZtBCK1 regulates secreted protein expression during infection following cell wall perturbation by host β‐1,3‐glucanases in the apoplast. Alternatively, this could occur on recognition of other environmental stimuli in the apoplast, such as the acidic pH, response to which is known to be controlled by the CWI pathway in yeast (Claret et al., 2005).
Unlike Δztbck1 mutants, Δztcyr1 strains continued to grow within the host late into the symptomless phase. This is consistent with the phenotypes of deletion mutants in the catalytic and regulatory PKA subunit genes, which were able to extensively colonize the mesophyll tissue (Mehrabi & Kema, 2006). This suggests that the function of ZtCYR1 in Z. tritici infection is distinct from the role of cAMP‐PKA signalling in host penetration by appressorium‐forming M. oryzae and Colletotrichum sp. (Choi & Dean, 1997; Yamauchi et al., 2004), and even fungal phytopathogens that do not develop true appressoria such as F. graminearum (Bormann et al., 2014).
While the Δztcyr1 mutants have completely abolished virulence, previously characterized deletion strains in the Z. tritici PKA subunit MgTPK2 are still able to cause delayed necrosis, but do not develop pycnidia (Mehrabi & Kema, 2006). Furthermore, the sensitivity to high osmolarity and defective melanization of Δztcyr1 is more similar to those previously observed in Z. tritici mutants lacking the PKA regulatory subunit (MgBCY1) than the catalytic subunit MgTPK2 (Mehrabi & Kema, 2006). This suggests that MgTPK2 has redundant function with the second PKA catalytic subunit gene in Z. tritici MgTPK1 (ZtritIPO323_04g08063), as in other ascomycete pathogens (Fuller et al., 2011; Hu et al., 2014; Li et al., 2017). Similar phenotypes of regulatory PKA subunit and adenylate cyclase mutants may be surprising, such as those observed here and in M. oryzae (Selvaraj et al., 2017), as BCY1 inhibits the activity of PKA catalytic subunits, while CYR1 activates this pathway. However, BCY1 is known to control the nuclear localization of PKA in S. cerevisiae and Candida albicans (Cassola et al., 2004; Griffioen et al., 2001). Combined, these results suggest that MgBCY1 has additional roles in regulating the PKA pathway beyond its inhibitory effect, which are required for correct PKA function.
The Δztcyr1 strains were also found to display increased sensitivity to caspofungin and osmotic stress. Although PKA signalling is inhibited during cell wall stress in S. cerevisiae (García et al., 2017, 2019), positive regulation of the cell wall stress response through crosstalk between the CWI and PKA pathways was identified in Cryptococcus neoformans (Donlin et al., 2014). Furthermore, PKA has been shown to facilitate cell wall remodelling in response to cell wall and osmotic stress in A. fumigatus through its involvement in carbohydrate mobilization in conjunction with the high osmolarity glycerol (HOG) pathway (de Assis et al., 2018; Shwab et al., 2017). This provides a potential mechanism by which PKA signalling may contribute to the response to cell wall and osmotic stress in Z. tritici.
Along with the previously characterized role of PKA in Z. tritici asexual development (Mehrabi & Kema, 2006), the results presented here suggest that cAMP signalling is required for the induction of necrosis during infection. A similar function of the cAMP‐PKA pathway in controlling invasive growth has been identified in M. oryzae, in which ΔcpkaΔcpk1 mutants were unable to cause necrosis when injected into the rice leaf (Li et al., 2017). Transcriptome analysis found that gene expression in Δztcyr1 was similar to IPO323 during the symptomless phase at 6 dpi but diverged as infection moved towards the transition to necrotrophy. This divergence in expression was particularly noticeable in predicted effector genes, including many which display peak expression around the necrotrophic switch (Rudd et al., 2015). Furthermore, analysis of wheat gene expression revealed that Δztcyr1‐infected leaves show widespread down‐regulation of defence‐related genes compared to wild‐type‐infected leaves at 9 dpi. This strongly suggests that Δztcyr1 is unable to induce the immune response required for the onset of necrosis. Those effector genes that are strongly down‐regulated in Δztcyr1 at 9 dpi may therefore be involved in inducing the host hypersensitive response, warranting further functional characterization.
In conclusion, this study provides evidence that the CWI pathway regulates the transcriptional response to the host environment in Z. tritici, which goes beyond genes involved in cell wall remodelling to activate expression of effector proteins. This includes virulence factors known to be required for suppression of the chitin‐triggered host immune response. This points towards the possible co‐regulation of the cell wall stress response with virulence‐related genes in response to the host mesophyll environmental. Furthermore, this study implicates the PKA pathway in controlling the switch to necrotrophic growth in addition to its previously characterized function in asexual development. The absence of ZtCYR1 influences the expression of putative effectors and genes involved in accessing nutrients around the transition to necrotrophy, which may disrupt the induction of host necrosis. These findings further our understanding of how CWI and cAMP signalling contribute to Z. tritici pathogenicity. Further study is required of the signals that activate both pathways during infection, as well as the regulatory components that detect these signals.
4. EXPERIMENTAL PROCEDURES
4.1. Strains and growth conditions
The wild type Z. tritici strain IPO323 was used in this study. Yeast‐like Z. tritici cells were stored long‐term in suspensions of 50% glycerol at −80°C and cultured on yeast extract peptone dextrose (YPD; 1% yeast extract, 2% peptone, 2% glucose) agar at 19°C under darkness for 5 days before use in transformations, infection experiments, and in vitro phenotypic assays.
4.2. Whole‐genome resequencing
To extract DNA, 100 mg of Z. tritici yeast‐like cells was collected from YPD agar cultures, snap frozen in liquid nitrogen, and ground in a pestle and mortar. DNA was extracted using the illustra Nucleon Phytopure Genomic DNA Extraction Kit (GE Healthcare) following the manufacturer's protocol, including the RNAse A digestion step. Extracted DNA was then further purified using the DNeasy Plant Mini Kit (Qiagen). Whole‐genome resequencing was carried out by BGI Tech Solutions Co., Ltd (Hong Kong) using their BGISEQ‐500 platform for paired‐end 150 bp reads. Bioinformatic analysis is described in detail in File S1. In brief, reads were trimmed using fastp (Chen et al., 2018) and aligned with the IPO323 genome with Bowtie 2 (Langmead & Salzberg, 2012), before variant calling using FreeBayes (Garrison & Marth, 2012).
4.3. Construction of plasmid vectors
The plasmids pC‐HYG‐CYR1KO and pC‐HYG‐BCK1KO were constructed by homologous recombination in yeast using the plasmid pC‐HYG‐YR following the previously described method (Sidhu et al., 2015). PCR amplification of left flank (LF) and right flank (RF) regions from either side of the ZtBCK1 and ZtCYR1 coding sequences was carried out using the primer pairs ZtCYR1‐LF‐F/R, ZtCYR1‐RF‐F/R, ZtBCK1‐LF‐F/R and ZtBCK1‐RF‐F/R (Table S1). LF and RF amplicons were transformed into S. cerevisiae alongside the pC‐HYG‐YR vector, which had been linearized by restriction digestion with the enzymes EcoRI and HindIII (New England Biolabs). The sequences of flanking regions were confirmed to be correct in the completed vectors by Sanger sequencing. These vectors were transformed into Z. tritici via Agrobacterium tumefaciens‐mediated transformation following the protocol deposited on protocols.io (Child & Helmstetter, 2022), which describes modifications to the original method (Zwiers & de Waard, 2001).
4.4. Enzymatic spore lysis assays
The ability of Z. tritici strains to withstand disruption of cell wall integrity was assessed using lyticase from Arthrobacter luteus (Sigma‐Aldrich). Cells were harvested from YPD agar cultures and suspended at a concentration of 5 × 106 spores/mL in sterile water containing a range of concentrations of lyticase enzyme, following a two‐fold dilution from 40 to 0.625 U/mL. Suspensions were incubated at 25°C for 2 h on an orbital shaker (120 rpm) before cell suspensions were diluted 40 fold and 100 μL plated onto YPD agar plates. The viability of Z. tritici after lyticase treatment was assessed by observing growth after 5 days at 19°C. The experiment was repeated three times.
4.5. Wheat infection assays
Wheat leaves (cultivar Riband) were infected with Z. tritici strains IPO323, Δztcyr1.4 and Δztbck1.4 at a concentration of 5 × 106 spores/mL following the previously described method (Motteram et al., 2009). Leaf samples for RNA extractions were harvested at 6 and 9 dpi, with each technical replicate being generated from six independent seedling (biological replicate) leaf sections (c.6 cm each) infected with the same Z. tritici strain, and frozen immediately in liquid nitrogen before storage at −80°C. Three technical replicates of each experiment were carried out.
4.6. RNA sequencing
RNA was extracted from Z. tritici‐infected wheat leaf tissue using TRIzol reagent (Fisher Scientific). Samples were ground in liquid nitrogen using a pestle and mortar and RNA was extracted with TRIzol reagent following the manufacturer's protocol apart from phase separation with 1‐bromo‐3‐chloropropane. RNA was purified using the RNeasy Plant Mini Kit (Qiagen), following the RNA Cleanup protocol including an on‐column DNA digestion step using the RNase‐free DNase set (Qiagen).
Libraries were prepared using the TruSeq HT stranded mRNA preparation kit following the manufacturer's protocol and sequenced as paired‐ends using an Illumina Novaseq 6000 on an S4 flow cell at 2 × 150 bp.
4.7. Alignments and differential expression analysis
Details of RNA sequencing analysis are given in File S1. In brief, reads were adapter‐ and quality‐trimmed with fastp before alignment to the Z. tritici genome (indexed with gene annotations from Rothamsted Research; King et al., 2017) and calculation of gene counts was carried out with STAR (Dobin et al., 2013). Normalization of gene counts and differential expression analysis was carried out using DESeq2 (Love et al., 2014), identifying DEGs between each mutant strain and the wild type at each time point, and between 6 and 9 dpi for each strain. Wald test p values were adjusted for multiple testing using the Benjamin–Hochberg (BH) correction method, and genes were deemed as differentially expressed when p adj <0.01.
RSEM (with alignment using STAR) was chosen for wheat transcript quantification as it factors in the high number of multimapped reads, which result from the polyploidy of the wheat genome, using an expectation maximization (EM) algorithm (Deschamps‐Francoeur et al., 2020; Li & Dewey, 2011). Expected gene‐level counts were filtered and normalized using the trimmed mean of M‐values (TMM) method in EdgeR (Robinson et al., 2010; Robinson & Oshlack, 2010), before the limma package was used for differential expression analysis (Ritchie et al., 2015). Genes were identified as differentially expressed between treatments using the moderated t test within the topTable function, testing whether the log2‐fold‐change values for a particular contrast differ from 0 at a BH‐corrected p adj threshold of 0.01.
Supporting information
FIGURE S1 Identification of avirulent Zymoseptoria tritici T‐DNA insertion strains
FIGURE S2 Strains C5 and L2 contain T‐DNA insertions at targeted loci and a nonsynonymous point mutation in the ZtCYR1 gene
FIGURE S3 Identification of T‐DNA insertion upstream of, and deletion within, ZtBCK1 in the avirulent Zymoseptoria tritici strain T21
FIGURE S4 Diagrams showing strategy for deletion of ZtCYR1 and ZtBCK1
FIGURE S5 In vitro phenotypes of Δztbck1 and Δztcyr1 under cell wall and osmotic stress
FIGURE S6 Differentially expressed effector, polyketide synthase, and major facilitator superfamily genes in Δztcyr1
FIGURE S7 Cell wall biosynthesis enzymes are differentially expressed in Δztbck1
FIGURE S8 Receptor‐like kinases and TaMPK3 expression is down‐regulated in Δztbck1 and Δztcyr1 at 9 days postinoculation
TABLE S1 Primers used in this study
TABLE S2 Polymorphisms identified in avirulent T‐DNA insertion strains
TABLE S3 Differentially expressed Zymoseptoria tritici genes during infection by Δztbck1 and Δztcyr1 compared to IPO323 at 6 and 9 days postinoculation (dpi), and between the time points for each strain
TABLE S4 Differentially expressed wheat genes during infection by Δztbck1 and Δztcyr1 compared to IPO323 at 9 days postinoculation (dpi), and between 6 and 9 dpi for each strain
FILE S1 Supplementary methods. Details of bioinformatics methods used in this study
ACKNOWLEDGEMENTS
This work was supported by the BBSRC‐funded South West Biosciences Doctoral Training Partnership (BB/M009122/1) and the Designing Future Wheat Institute Strategic Programme (BB/P016855/1) awarded to Rothamsted Research. J.J.R. is additionally supported by the BBSRC ISP grants Delivering Sustainable Wheat (DSW) BB/X011003/1 and Growing Health (GH) BB/X010953. We thank the University of Exeter Sequencing Facility for RNA library preparation and sequencing. This project used equipment funded by the Wellcome Trust (Multi‐User Equipment Grant award number 218247/Z/19/Z).
Child, H.T. , Deeks, M.J. , Rudd, J.J. & Bates, S. (2023) Comparison of the impact of two key fungal signalling pathways on Zymoseptoria tritici infection reveals divergent contribution to invasive growth through distinct regulation of infection‐associated genes. Molecular Plant Pathology, 24, 1220–1237. Available from: 10.1111/mpp.13365
DATA AVAILABILITY STATEMENT
RNA sequencing reads have been deposited on the European Nucleotide Archive (ENA) at www.ebi.ac.uk/ena under the accession number PRJEB58154.
REFERENCES
- Ballou, E.R. , Avelar, G.M. , Childers, D.S. , Mackie, J. , Bain, J.M. , Wagener, J. et al. (2016) Lactate signalling regulates fungal β‐glucan masking and immune evasion. Nature Microbiology, 2, 16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann, J. , Boenisch, M.J. , Brückner, E. , Firat, D. & Schäfer, W. (2014) The adenylyl cyclase plays a regulatory role in the morphogenetic switch from vegetative to pathogenic lifestyle of Fusarium graminearum on wheat. PLoS One, 9, e0091135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder Nascimento, A.C. , Dos Reis, T.F. , de Castro, P.A. , Hori, J.I. , Bom, V.L.P. , de Assis, L.J. et al. (2016) Mitogen activated protein kinases SakA HOG1 and MpkC collaborate for Aspergillus fumigatus virulence. Molecular Microbiology, 100, 841–859. [DOI] [PubMed] [Google Scholar]
- Cabib, E. & Arroyo, J. (2013) How carbohydrates sculpt cells: chemical control of morphogenesis in the yeast cell wall. Nature Reviews Microbiology, 11, 648–655. [DOI] [PubMed] [Google Scholar]
- Cassola, A. , Parrot, M. , Silberstein, S. , Magee, B.B. , Passeron, S. , Giasson, L. et al. (2004) Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryotic Cell, 3, 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza, M. & Kronstad, J.W. (2019) The cAMP/protein kinase a pathway regulates virulence and adaptation to host conditions in Cryptococcus neoformans . Frontiers in Cellular and Infection Microbiology, 9, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Zhou, Y. , Chen, Y. & Gu, J. (2018) Fastp: An ultra‐fast all‐in‐one FASTQ preprocessor. Bioinformatics, 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child, H.T. & Helmstetter, N. (2022) Agrobacterium‐mediated transformation of Zymoseptoria tritici. Protocols.io . Available from: 10.17504/protocols.io.b5ukq6uw. [DOI]
- Choi, W. & Dean, R.A. (1997) The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. The Plant Cell, 9, 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret, S. , Gatti, X. , Thoraval, D. & Crouzet, M. (2005) The Rgd1p rho GTPase‐activating protein and the Mid2p cell wall sensor are required at low pH for protein kinase C pathway activation and cell survival in Saccharomyces cerevisiae . Eukaryotic Cell, 4, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis, L.J. , Manfiolli, A. , Mattos, E. , Fabri, J.H.T.M. , Malavazi, I. , Jacobsen, I.D. et al. (2018) Protein kinase a and high‐osmolarity glycerol response pathways cooperatively control cell wall carbohydrate mobilization in Aspergillus fumigatus . mB Mbio, 9, e01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps‐Francoeur, G. , Simoneau, J. & Scott, M.S. (2020) Handling multi‐mapped reads in RNA‐seq. Computational and Structural Biotechnology Journal, 18, 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C.A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. et al. (2013) STAR: Ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin, M.J. , Upadhya, R. , Gerik, K.J. , Lam, W. , Vanarendonk, L.G. , Specht, C.A. et al. (2014) Cross talk between the cell wall integrity and cyclic AMP/protein kinase a pathways in Cryptococcus neoformans . mBio, 5, e01573‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, C.A. & Heitman, J. (2001) Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiology Reviews, 25, 349–364. [DOI] [PubMed] [Google Scholar]
- El Gueddari, N.E. , Rauchhaus, U. , Moerschbacher, B.M. & Deising, H.B. (2002) Developmentally regulated conversion of surface‐exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytologist, 156, 103–112. [Google Scholar]
- Fortwendel, J.R. , Juvvadi, P.R. , Perfect, B.Z. , Rogg, L.E. , Perfect, J.R. & Steinbach, W.J. (2010) Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrobial Agents and Chemotherapy, 54, 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa, T. , Sakaguchi, A. , Nishizawa, Y. , Kouzai, Y. , Minami, E. , Yano, S. et al. (2012) Surface α‐1,3‐glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathogens, 8, e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, K.K. , Richie, D.L. , Feng, X. , Krishnan, K. , Stephens, T.J. , Wikenheiser‐Brokamp, K.A. et al. (2011) Divergent protein kinase a isoforms co‐ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus . Molecular Microbiology, 79, 1045–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, R. , Bravo, E. , Diez‐Muñiz, S. , Nombela, C. , Rodríguez‐Peña, J.M. & Arroyo, J. (2017) A novel connection between the cell wall integrity and the PKA pathways regulates cell wall stress response in yeast. Scientific Reports, 7, 5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, R. , Pulido, V. , Orellana‐Muñoz, S. , Nombela, C. , Vázquez de Aldana, C.R. , Rodríguez‐Peña, J.M. et al. (2019) Signalling through the yeast MAPK cell wall integrity pathway controls P‐body assembly upon cell wall stress. Scientific Reports, 9, 3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, R. , Rodríguez‐Pena, J.M. , Bermejo, C. & Arroyo, J. (2009) The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase‐induced cell wall stress in Saccharomyces cerevisiae . Journal of Biological Chemistry, 284, 10901–10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E. & Marth, G. (2012) Haplotype‐based variant detection from short‐read sequencing. ArXiv, 1207.3907 [preprint]. Available from: 10.48550/arXiv.1207.3907 [DOI]
- Geoghegan, I. , Steinberg, G. & Gurr, S. (2017) The role of the fungal cell wall in the infection of plants. Trends in Microbiology, 25, 957–967. [DOI] [PubMed] [Google Scholar]
- Goyal, R.K. , Tulpan, D. , Chomistek, N. , Fundora, D.G. , West, C. , Ellis, B.E. et al. (2018) Analysis of MAPK and MAPKK gene families in wheat and related Triticeae species. BMC Genomics, 19, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen, G. , Branduardi, P. , Ballarini, A. , Anghileri, P. , Norbeck, J. , Baroni, M.D. et al. (2001) Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1‐dependent phosphorylation of its targeting domain. Molecular and Cellular Biology, 21, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig, M. , Schotanus, K. , Hufnagel, K. , Happel, P. & Stukenbrock, E.H. (2021) Ago1 affects the virulence of the fungal plant pathogen Zymoseptoria tritici . Genes, 12, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopke, A. , Brown, A.J.P. , Hall, R.A. & Wheeler, R.T. (2018) Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends in Microbiology, 26, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S. , Zhou, X. , Gu, X. , Cao, S. , Wang, C. & Xu, J.R. (2014) The cAMP‐PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum . Molecular Plant‐Microbe Interactions, 27, 557–566. [DOI] [PubMed] [Google Scholar]
- Jørgensen, L.N. , Hovmøller, M.S. , Hansen, J.G. , Lassen, P. , Clark, B. , Bayles, R. et al. (2014) IPM strategies and their dilemmas including an introduction to www.eurowheat.org. Journal of Integrative Agriculture, 13, 265–281. [Google Scholar]
- Joubert, A. , Bataille‐Simoneau, N. , Campion, C. , Guillemette, T. , Hudhomme, P. , Iacomi‐Vasilescu, B. et al. (2011) Cell wall integrity and high osmolarity glycerol pathways are required for adaptation of Alternaria brassicicola to cell wall stress caused by brassicaceous indolic phytoalexins. Cellular Microbiology, 13, 62–80. [DOI] [PubMed] [Google Scholar]
- Kamada, Y. , Jung, U.S. , Piotrowski, J. & Levin, D.E. (1995) The protein kinase C‐activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes and Development, 9, 1559–1571. [DOI] [PubMed] [Google Scholar]
- Kema, G.H.J. , Yu, D.Z. , Rijkenberg, F.H.J. , Shaw, M.W. & Baayen, R.P. (1996) Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 86, 777–786. [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. et al. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Molecular Plant‐Microbe Interactions, 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Kettles, G.J. , Hofinger, B.J. , Hu, P. , Bayon, C. , Rudd, J.J. , Balmer, D. et al. (2019) sRNA profiling combined with gene function analysis reveals a lack of evidence for cross‐kingdom RNAi in the wheat–Zymoseptoria tritici pathosystem. Frontiers in Plant Science, 10, e00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R. , Urban, M. , Lauder, R.P. , Hawkins, N. , Evans, M. , Plummer, A. et al. (2017) A conserved fungal glycosyltransferase facilitates pathogenesis of plants by enabling hyphal growth on solid surfaces. PLoS Pathogens, 13, e1006672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, K. , Kikuchi, T. , Takano, Y. , Oshiro, E. & Okuno, T. (2002) The mitogen‐activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium . Molecular Plant‐Microbe Interactions, 15, 1268–1276. [DOI] [PubMed] [Google Scholar]
- Langmead, B. & Salzberg, S.L. (2012) Fast gapped‐read alignment with bowtie 2. Nature Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.S. , Levin, D.E. & Eg, M. (1992) Dominant mutations in a gene encoding a putative protein kinase (BCKI) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Molecular and Cellular Biology, 12, 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.S. , Rudd, J.J. , Hammond‐Kosack, K.E. & Kanyuka, K. (2014) Mycosphaerella graminicola LysM effector‐mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Molecular Plant‐Microbe Interactions, 27, 236–243. [DOI] [PubMed] [Google Scholar]
- Lehti‐Shiu, M.D. , Zou, C. , Hanada, K. & Shiu, S.‐H. (2009) Evolutionary history and stress regulation of plant receptor‐like kinase/Pelle genes. Plant Physiology, 150, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D.E. (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics, 189, 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. & Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhang, X. , Hu, S. , Liu, H. & Xu, J.R. (2017) PKA activity is essential for relieving the suppression of hyphal growth and appressorium formation by MoSfl1 in Magnaporthe oryzae . PLoS Genetics, 13, e1006954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. et al. (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiology, 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. & Kema, G.H.J. (2006) Protein kinase a subunits of the ascomycete pathogen Mycosphaerella graminicola regulate asexual fructification, filamentation, melanization and osmosensing. Molecular Plant Pathology, 7, 565–577. [DOI] [PubMed] [Google Scholar]
- Mehrabi, R. , M'Barek, S.B. , van der Lee, T.A.J. , Waalwijk, C. , de Wit, P.J.G.M. & Kema, G.H.J. (2009) Gα and Gβ proteins regulate the cyclic AMP pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola . Eukaryotic Cell, 8, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. , van der Lee, T. , Waalwijk, C. & Kema, G.H.J. (2006) MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Molecular Plant‐Microbe Interactions, 19, 389–398. [DOI] [PubMed] [Google Scholar]
- Mey, G. , Held, K. , Scheffer, J. , Tenberge, K.B. & Tudzynski, P. (2002) CPMK2, an SLT2‐homologous mitogen‐activated protein (MAP) kinase, is essential for pathogenesis of Claviceps purpurea on rye: evidence for a second conserved pathogenesis‐related MAP kinase cascade in phytopathogenic fungi. Molecular Microbiology, 46, 305–318. [DOI] [PubMed] [Google Scholar]
- Motteram, J. , Küfner, I. , Deller, S. , Brunner, F. , Hammond‐Kosack, K.E. , Nürnberger, T. et al. (2009) Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain‐containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola . Molecular Plant‐Microbe Interactions, 22, 790–799. [DOI] [PubMed] [Google Scholar]
- Munro, C.A. , Selvaggini, S. , de Bruijn, I. , Walker, L. , Lenardon, M.D. , Gerssen, B. et al. (2007) The PKC, HOG and Ca2+ signalling pathways co‐ordinately regulate chitin synthesis in Candida albicans . Molecular Microbiology, 63, 1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma‐Guerrero, J. , Torriani, S.F.F. , Zala, M. , Carter, D. , Courbot, M. , Rudd, J.J. et al. (2016) Comparative transcriptomic analyses of Zymoseptoria tritici strains show complex lifestyle transitions and intraspecific variability in transcription profiles. Molecular Plant Pathology, 17, 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy, V. , Zhao, X. , Snyder, A.K. , Xu, J.R. & Shah, D.M. (2007) Two mitogen‐activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum . Cellular Microbiology, 9, 1491–1506. [DOI] [PubMed] [Google Scholar]
- Ray, S. , Anderson, J.M. , Urmeev, F.I. & Goodwin, S.B. (2003) Rapid induction of a protein disulfide isomerase and defense‐related genes in wheat in response to the hemibiotrophic fungal pathogen Mycosphaerella graminicola . Plant Molecular Biology, 53, 701–714. [DOI] [PubMed] [Google Scholar]
- Reinoso‐Martín, C. , Schu, C. , Schuetzer‐muehlbauer, M. & Kuchler, K. (2003) The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen‐activated protein kinase signaling. Eukaryotic Cell, 2, 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme, M. , Aguirre, J. , Bartnicki‐García, S. , Braus, G.H. , Feldbrügge, M. , Fleig, U. et al. (2018) Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiology and Molecular Biology Reviews, 82, e00068‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E. , Phipson, B. , Wu, D. , Hu, Y. , Law, C.W. , Shi, W. et al. (2015) Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Research, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. & Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. & Oshlack, A. (2010) A scaling normalization method for differential expression analysis of RNA‐seq data. Genome Biology, 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Kanyuka, K. , Hassani‐Pak, K. , Derbyshire, M. , Andongabo, A. , Devonshire, J. et al. (2015) Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle. Plant Physiology, 167, 1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Keon, J. & Hammond‐Kosack, K.E. (2008) The wheat mitogen‐activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola . Plant Physiology, 147, 802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui, O. & Hahn, M. (2007) The Slt2‐type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Molecular Plant Pathology, 8, 173–184. [DOI] [PubMed] [Google Scholar]
- Selvaraj, P. , Tham, H.F. , Ramanujam, R. & Naqvi, N.I. (2017) Subcellular compartmentation, interdependency and dynamics of the cyclic AMP‐dependent PKA subunits during pathogenic differentiation in rice blast. Molecular Microbiology, 105, 484–504. [DOI] [PubMed] [Google Scholar]
- Shumayla, S.S. , Kumar, R. , Mendu, V. & Singh, K. (2016) Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Frontiers in Plant Science, 7, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumayla, Sharma, S. , Pandey, A.K. & Singh, K. (2016) Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PLoS One, 11, e0153925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab, E.K. , Juvvadi, P.R. , Waitt, G. , Soderblom, E.J. , Moseley, M.A. , Nicely, N.I. et al. (2017) Phosphorylation of Aspergillus fumigatus PkaR impacts growth and cell wall integrity through novel mechanisms. FEBS Letters, 591, 3730–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu, Y.S. , Cairns, T.C. , Chaudhari, Y.K. , Usher, J. , Talbot, N.J. , Studholme, D.J. et al. (2015) Exploitation of sulfonylurea resistance marker and non‐homologous end joining mutants for functional analysis in Zymoseptoria tritici . Fungal Genetics and Biology, 79, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Dodds, P.N. , Gardiner, D.M. , Singh, K.B. & Taylor, J.M. (2018) Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Molecular Plant Pathology, 19, 2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt, L. , Humpf, H.U. & Tudzynski, B. (2013) Signaling governed by G proteins and cAMP is crucial for growth, secondary metabolism and sexual development in Fusarium fujikuroi . PLoS One, 8, e0058185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H. , MacKenzie, C.I. , Rodriguez‐Moreno, L. , van den Berg, G.C.M. , Chen, H. , Rudd, J.J. et al. (2021) Three LysM effectors of Zymoseptoria tritici collectively disarm chitin‐triggered plant immunity. Molecular Plant Pathology, 22, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrà, D. , Segorbe, D. & di Pietro, A. (2014) Protein kinases in plant‐pathogenic fungi: conserved regulators of infection. Annual Review of Phytopathology, 52, 267–288. [DOI] [PubMed] [Google Scholar]
- Xu, J.R. , Staiger, C.J. & Hamer, J.E. (1998) Inactivation of the mitogen‐activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proceedings of the National Academy of Sciences of the United States of America, 95, 12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.R. , Urban, M. , Sweigard, J.A. & Hamer, J.E. (1997) The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Molecular Plant‐Microbe Interactions, 10, 187–194. [Google Scholar]
- Yamauchi, J. , Takayanagi, N. , Komeda, K. , Takano, Y. & Okuno, T. (2004) cAMP‐PKA signaling regulates multiple steps of fungal infection cooperatively with Cmk1 MAP kinase in Colletotrichum lagenarium . Molecular Plant‐Microbe Interactions, 17, 1355–1365. [DOI] [PubMed] [Google Scholar]
- Zaman, S. , Lippman, S.I. , Zhao, X. & Broach, J.R. (2008) How Saccharomyces responds to nutrients. Annual Review of Genetics, 42, 27–81. [DOI] [PubMed] [Google Scholar]
- Zhan, H. , Yue, H. , Zhao, X. , Wang, M. , Song, W. & Nie, X. (2017) Genome‐wide identification and analysis of MAPK and MAPKK gene families in bread wheat (Triticum aestivum L.). Genes, 8, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers, L.H. & de Waard, M.A. (2001) Efficient Agrobacterium tumefaciens‐mediated gene disruption in the phytopathogen Mycosphaerella graminicola . Current Genetics, 39, 388–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Identification of avirulent Zymoseptoria tritici T‐DNA insertion strains
FIGURE S2 Strains C5 and L2 contain T‐DNA insertions at targeted loci and a nonsynonymous point mutation in the ZtCYR1 gene
FIGURE S3 Identification of T‐DNA insertion upstream of, and deletion within, ZtBCK1 in the avirulent Zymoseptoria tritici strain T21
FIGURE S4 Diagrams showing strategy for deletion of ZtCYR1 and ZtBCK1
FIGURE S5 In vitro phenotypes of Δztbck1 and Δztcyr1 under cell wall and osmotic stress
FIGURE S6 Differentially expressed effector, polyketide synthase, and major facilitator superfamily genes in Δztcyr1
FIGURE S7 Cell wall biosynthesis enzymes are differentially expressed in Δztbck1
FIGURE S8 Receptor‐like kinases and TaMPK3 expression is down‐regulated in Δztbck1 and Δztcyr1 at 9 days postinoculation
TABLE S1 Primers used in this study
TABLE S2 Polymorphisms identified in avirulent T‐DNA insertion strains
TABLE S3 Differentially expressed Zymoseptoria tritici genes during infection by Δztbck1 and Δztcyr1 compared to IPO323 at 6 and 9 days postinoculation (dpi), and between the time points for each strain
TABLE S4 Differentially expressed wheat genes during infection by Δztbck1 and Δztcyr1 compared to IPO323 at 9 days postinoculation (dpi), and between 6 and 9 dpi for each strain
FILE S1 Supplementary methods. Details of bioinformatics methods used in this study
Data Availability Statement
RNA sequencing reads have been deposited on the European Nucleotide Archive (ENA) at www.ebi.ac.uk/ena under the accession number PRJEB58154.


