Abstract
The ESKAPE bacteria are the six highly virulent and antibiotic-resistant pathogens that require the most urgent attention for the development of novel antibiotics. Detailed knowledge of target proteins specific to bacteria is essential to develop novel treatment options. The methylerythritol-phosphate (MEP) pathway, which is absent in humans, represents a potentially valuable target for the development of novel antibiotics. Within the MEP pathway, the enzyme 1-deoxy-D-xylulose-5-phosphate synthase (DXPS) catalyzes a crucial, rate-limiting first step and a branch point in the biosynthesis of the vitamins B1 and B6. We report the high-resolution crystal structures of DXPS from the important ESKAPE pathogens Pseudomonas aeruginosa and Klebsiella pneumoniae in both the co-factor-bound and the apo forms. We demonstrate that the absence of the cofactor thiamine diphosphate results in conformational changes that lead to disordered loops close to the active site that might be important for the design of potent DXPS inhibitors. Collectively, our results provide important structural details that aid in the assessment of DXPS as a potential target in the ongoing efforts to combat antibiotic resistance.
Keywords: 1-deoxy-D-xylulose 5-phosphate synthase, Pseudomonas aeruginosa, X-ray crystallography, DXPS, conformational changes, Klebsiella pneumonia
The widespread use of antibiotics over the past five decades has caused bacteria to develop resistance mechanisms to evade the detrimental effect of these agents (1, 2). This effect is especially critical for infections caused by ESKAPE pathogens, which are difficult to treat and result in increasing fatality rates. Pseudomonas aeruginosa and Klebsiella pneumoniae are both Gram-negative pathogens with the ability to integrate exogenous DNA to obtain antibiotic resistance (horizontal gene transfer) (3). Both organisms can adapt to the environment of human airways (4) and are major causes of opportunistic infections, predominantly, pneumonia and sepsis in hospitalized patients and patients with cystic fibrosis (5, 6). Measures such as the antibiotic stewardship program to improve the way antibiotics are prescribed, and the synthesis of novel chemical entities with new modes of action need to be implemented to tackle the antimicrobial-resistance crisis.
The methylerythritol-phosphate (MEP) pathway has attracted attention as it represents a rich source of potentially attractive anti-infective drug targets. The MEP pathway, also called the mevalonate-independent pathway, is considered to be the main pathway for the synthesis of isoprenoid building blocks in most bacteria, plants, and protozoa (7, 8, 9). Its absence in humans makes the enzymes of this pathway, particularly interesting targets for anti-infective drug discovery. The complete pathway consists of seven enzymes, each of which could be targeted individually. 1-Deoxy-D-xylulose-5-phosphate synthase (DXPS) catalyzes the first rate-limiting step of the MEP pathway. The enzyme is responsible for the formation of 1-deoxy-D-xylulose 5-phosphate (DXP) by condensation of pyruvate and glyceraldehyde 3-phosphate (D-GAP) in the presence of thiamine diphosphate (ThDP), Figure 1. DXP, in and of itself, is an important metabolite in not only the biosynthesis of isoprenoid precursors but also in vitamin B1 (thiamine) and vitamin B6 (pyridoxine) biosynthesis (10). DXPS is a promising anti-infective drug target (11, 12, 13).
Figure 1.
DXPS reaction mechanism with pyruvate and fluoropyruvate. Suggested formation of the fluoropyruvate adduct (2-acetyl ThDP) catalyzed by paDXPS in combination with ThDP in comparison to the reported reaction with the natural substrate pyruvate. The point of difference between fluoropyruvate and pyruvate is highlighted in gray.
The first DXPS structures of Deinococcus radiodurans and Escherichia coli were solved via X-ray crystallography and published in 2007 (14). Subsequent studies on these structures have shown that DXPS undergoes a number of conformational changes during catalysis which takes place near the substrate-entry site (15, 16, 17). The ThDP-bound enzyme, in the presence of pyruvate, forms a pre-decarboxylation intermediate, lactyl thiamine diphosphate (LThDP) (15, 18). The enzyme adopts a closed conformation when bound to LThDP and exposure to D-GAP or DXP triggers a conformational change to an open conformation, which seemingly initiates LThDP decarboxylation (17, 19). Chen et al. coined the name spoon-fork motif to describe the shape of the region that shows conformational changes during catalysis. They have shown that binding of LThDP results in a closed conformation of the spoon motif that opens when the subsequent intermediate enamine is formed (17). These results highlight the important role of conformational changes in DXPS catalysis. Further investigation is needed to expand our knowledge about the dynamic catalysis and structural differences of this unique enzyme.
Given that the binding site of the cofactor ThDP is the most promising site for drugs targeting ThDP-dependent enzymes including DXPS (16, 20, 21), it is also vital to understand the precise location of ThDP and the general three-dimensional shape of the pocket. It is also considered the main druggable pocket in DXPS enzymes (22). Thus far, few experimentally determined structures of DXPS have been reported (23), but due to the generally conserved active sites, the available crystal structures of DXPS are used as models for structure-based drug design (16, 20, 21). However, this has proven to be very limiting, as three-dimensional active sites can typically not be deduced from sequence homology alone. To improve our understanding of DXPS active-site architecture and to provide a better framework for drug-discovery efforts addressing this promising target, we determined the crystal structures of both the apo and the cofactor-bound forms from the ESKAPE pathogens P. aeruginosa and K. pneumoniae. We demonstrate that the binding of ThDP leads to conformational changes involving a loop in domain I, which alters the ThDP biding site and contributes to the binding of the ThDP diphosphate. Together with the structure of P. aeruginosa DXPS bound to the less reactive 2-acetylThDP, we provide a firm basis for future drug design and -discovery efforts targeting this important enzyme from the MEP pathway (2).
Results and discussion
Expression, purification, and kinetic characterization of DXPS
Our group recently reported a truncated D. radiodurans DXPS (drDXPS) construct in which a flexible loop has been modified to produce a protein that crystallizes readily and yields high-quality crystals that diffract to a resolution better than 2.0 Å (24). This loop typically has very low evolutionary conservation and neither shapes the active site nor is it involved in the formation of a secondary structure (24). Our construct served as a blueprint for the alteration in DXPS enzymes from different organisms (23). After multiple sequence alignments of published sequences, we identified the loop to be corresponding to residues 206 to 245 in paDXPS and residues 197 to 240 in kpDXPS (Fig. S1) and replaced them with a six glycine unit. We then heterologously expressed the enzymes in E. coli, purified and analyzed them as detailed in the Experimental procedures section. Next, we analyzed the activity of new homolog constructs, evaluated, and compared the enzyme kinetics for both versions of the enzyme i.e. truncated and full-length, of the enzyme. The results summarized in Table 1 demonstrate comparable activities of the native and truncated paDXPS, both versions of the enzyme show similar affinities for the substrate and co-factor. Although native kpDXPS was not tested, the truncated version is showing comparable kinetics for both substrates. No kinetics values for ThDP were reported for other homologs, which makes our data impossible to compare to literature values. However, kpDXPS shows a slightly higher affinity for the substrates and elevated catalytic activity in comparison to paDXPS, which may be due to the different shape of the binding site. The kinetic parameters remain within a similar range for paDXPSs, and the preserved enzymatic activity of kpDXPS suggests that the truncation does not affect any catalytically significant residues. These results allowed us to use these optimized and more stable constructs for biological testing as well as structure determination. After confirming the identity and activity of our purified enzymes, we opted to elucidate the structures of these novel homologs from ESKAPE pathogens.
Table 1.
The steady-state kinetics parameters of ThDP, pyruvate, and D-GAP for the wild-type and mutated paDXPS and mutated kpDXPS
| Enzyme | Native paDXPS 0.15 μmol/L |
Truncated paDXPS 0.15 μmol/L |
Truncated kpDXPS 0.20 μmol/L |
|---|---|---|---|
| Pyruvate |
Km: 184 ± 25 μM Kcat: 1.6 ± 0.01 × 103 min−1 |
Km: 122 ± 8.2 μM Kcat: 1.2 ± 0.02 × 103 min−1 |
Km: 113 ± 34 μM Kcat: 2.1 ± 0.03 × 103 min−1 |
| D-GAP |
Km: 357 ± 77 μM Kcat: 1.8 ± 0.02 ×103 min−1 |
Km: 336 ± 98.0 Kcat: 1.5 ± 0.03 × 103 min−1 |
Km: 170 ± 43 μM Kcat: 2.4 ± 0.01 × 103 min−1 |
| ThDP |
Km: 70 ± 9.0 nM Kcat: 1.6 ± 0.01 × 103 min−1 |
Km: 96.5 ± 14 nM Kcat: 0.94 ± 0.01 × 103 min−1 |
Km: 110 ± 17.0 nM Kcat: 2.6 ± 0.03 × 103 min−1 |
When measuring Km, the other substrate and/or cofactor concentrations were kept in excess to ensure the enzyme reaches maximum velocity.
Structure determination of DXPS from K. pneumoniae and P. aeruginosa
The optimized constructs of DXPS from K. pneumoniae and P. aeruginosa were used for crystallization trials. The structures were determined using molecular replacement with the published DXPS structure of E. coli (PDB ID: 2o1s) as a search model. The two proteins in the asymmetric unit form the expected biological dimer known from other DXPS enzymes, which was supported by native MS measurements for paDXPS (Fig. S2). The low solubility of kpDXPS precluded native MS data acquisition, but an analysis of the dimer observed in the crystal structure using the PISA server (25) and analytical gel filtration (Fig. S3) confirmed a stable dimer interface, which is highly similar between all DXPS structures. Although we determined a comparable affinity for ThDP, we can only observe electron density for ThDP in one of the two protomers of the dimer of kpDXPS with low occupancy. This may be the result of the longer crystal formation time of 20 days and the consequent degradation of the cofactor. Intriguingly, the N-terminus of this protomer extends into the cofactor binding site of the dimer partner up to Ser186. Given that this would lead to an inactive enzyme and the absence of this arrangement from the apo structures (see below), we consider this arrangement a crystallographic artifact representing a low-energy state of this particular crystal form. The addition of ThDP was essential to obtain this crystal form, and we thus screened new conditions to obtain an apo structure. Crystals of apo kpDXPS were obtained at a considerably higher concentration than the cofactor-bound version and crystals grew more quickly. They belonged to space group C 2221 and a complete 2.1 Å dataset was collected at ESRF beamline ID23 to 1. The structure was solved using molecular replacement with a refined ThDP-bound kpDXPS structure as the search model.
Similar to kpDXPS, we chose an optimized construct to obtain a crystal structure of DXPS from P. aeruginosa (paDXPS). The structure was determined to 2.0 Å with molecular replacement using the published structure of drDXPS (PDB ID: 6OUV) as a search model. Subsequently, a refined paDXPS structure was used as the search model for the molecular replacement of structures of ThDP-bound paDXPS (2.3 Å), a structure with 2-acetylThDP (2.0 Å), and a co-crystal structure with a selected ThDP analog (2.2 Å). In general, paDXPS crystallized in orthorhombic crystal form, space group P212121. The asymmetric unit of all paDXPS structures consisted of six protomers, four of which formed dimers with proteins within the asymmetric unit, while the remaining two protomers formed dimers with symmetry mates. In most obtained paDXPS structures, approximately the first 30 N-terminal residues are disordered. Native MS data on paDXPS confirmed that the enzyme exists as a dimer in solution (Fig. S2), which is consistent with the crystallographic results and suggests that the dimer is the biological unit in vivo. All data collection and refinement statistics can be found in Table S2, and the structures were deposited in the PDB.
Structural comparison of kpDXPS and paDXPS
kpDXPS and paDXPS share high sequence homology with previously identified homologs from E. coli (ecDXPS) and D. radiodurans (drDXPS) and Mycobacterium tuberculosis (mtDXPS) (14, 23). kpDXPS and paDXPS share 62.5% sequence identity and 84.3% sequence similarity. Consequently, the structures are also highly similar with a Cα-RMSD of only 0.716 when comparing both ThDP-bound crystal structures. Each DXPS monomer in all structures we determined consists of three domains. The domain organization is shown in Figure 2A and is highly similar to the other homologs. The loop connecting domains I and II (residues 307–315 in paDXPS) is highly flexible and appears to be disordered in most structures. Similar to other reported homologs, the binding site of ThDP is located at the interface between domains I and II with the pyrimidine ring facing domain II and the diphosphate moiety pointing toward domain I. ThDP adopts a chair-like conformation; the pyrimidine ring is perpendicular to the thiazole ring, which is, in turn, perpendicular to the diphosphate. In the paDXPS structure, the pyrimidine ring of ThDP is π-π stacked against Phe395, while N3 of the pyrimidine is forming a backbone hydrogen bond (2.9 Å) with Ser158. Residues His115, Asp187, Ala189, Asn214, Asn216, and Lys286 are coordinating the diphosphate/Mg2+ part of ThDP (Fig. 2D). The active site of kpDXPS shows an almost identical binding mode of ThDP, and similar residues are involved in binding. The diphosphate is coordinated in an asparagine-rich region, with the contribution of Ser52, which forms an H bond with the diphosphate. This residue is an alanine in the paDXPS structure. Among other structurally characterized DXPS homologs, paDXPS, together with mtDXPS, seem to be the only homologs in which this hydrogen bond cannot be formed. In mtDXPS, this residue is changed to proline (Fig. S4). Further comparison of the two DXPS structures, reveals a key difference in the active site. His115, in paDXPS, is positioned to form a hydrogen bond with the diphosphate of ThDP (Fig. 2B). This interaction cannot be formed in kpDXPS as the histidine side chain is now 7 Å away from the diphosphate (Fig. 2B). This may be caused by Ser52 preventing His80 from adopting the conformation seen in paDXPS (steric hindrance). The H bonding interaction with His115 specific to paDXPS seems to be compensating for the loss of the interaction with diphosphate caused by the change from serine to alanine in position 87 (Fig. 2D). This residue might be an important distinction for drug design, as the hydroxyl group of serine adds a targetable nucleophilic moiety to the active site. Fig. S4 shows the active site differences between the structures of the published DXPS homologs from E. coli, D. radiodurans, and M. tuberculosis and our structures. Of these, the most noteworthy is the change of Ala189 placed to make direct backbone interaction with the N2 of the pyrimidine in the homolog from the non-pathogenic bacterium D. radiodurans to serine (Ser158 in paDXPS and Ser123 in kpDXPS, Fig. 2, C and D) as well as the homologs from the pathogenic E. coli, where the hydroxyl group of the serine side chain can again be targeted by hydrogen bonding with a ligand.
Figure 2.
Overview of the overall structures of paDXPS and kpDXPS.A, domain organization; domain I (cyan; residues 1–306) contains a five-stranded parallel β-sheet, domain II (violet; residues 317–493) contains a six-stranded parallel β-sheet, domain III (green; residues 496–620) contains a five-stranded β-sheet, in paDXPS, aligned with kpDXPS structure in gray. B, comparison of ThDP-bound kpDXPS and paDXPS structures highlighting differences in the active site. paDXPS is shown in cyan, overlaid with kpDXPS in gray, different residues from kpDXPS are labeled in blue. C and D, important interactions with ThDP in paDXPS and kpDXPS, respectively. Interactions are shown as black, dashed lines (below 5 Å). All figures of this type were created in PyMol.
Structural analysis of the conformational difference in the binding site between ThDP-bound and apo DXPS
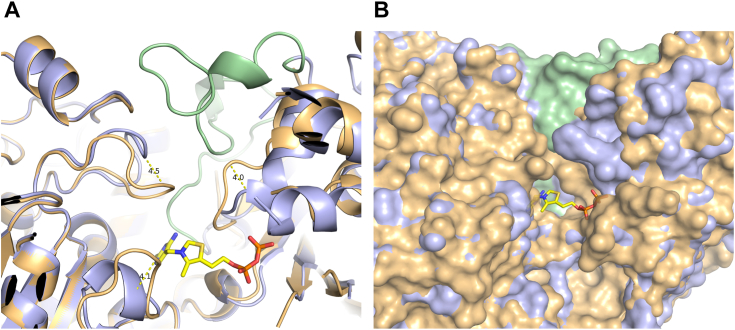
The influence of cofactor binding on the shape of the active site was evaluated by obtaining structural information on the apo paDXPS and kpDXPS and comparing it to the cofactor-bound proteins. The loop spanning from Asn216 to Trp250, contributing to the ThDP binding site, exhibits a conformational change between the holo and the apo structure of paDXPS (Fig. 3A). In the cofactor-bound version, residues As217, Met218, Ser219, and Ile220 fold around the diphosphate moiety (Fig. 3). The coordination of the co-factor bound Mg2+ by the side chain of Asn216 and the main chain of Met218 appear to stabilize the loop. The ThDP-bound structure also shows the participation of the ser219 and Ile220 side chains, with the latter showing the potential to form an arene-H interaction with the thiazole ring (Fig. 3D). Most other DXPS structures exhibit disorder in this particular region. Furthermore, our study presents the first report of an apo DXPS structure, making it challenging to directly compare our results with those of other homologs. In the kpDXPS structure regions near the active site seem to undergo various conformational changes. We observe loop rearrangement around the active site: Three loops are partially occluding the active site in the apo structure, and appear to get displaced by the bound co-factor, resulting in larger movements averaging around 4 Å (Fig. 4). The loop belonging to the domain I of DXPS (amino acids 91–124) is completely disordered in the apo structure of kpDXPS, leading to a more open conformation towards the other side of the substrate entrance channel (pale green region in (Fig. 4, A and B). Unfortunately, this part of the protein could not be resolved sufficiently to allow us to compare it to the structure of paDXPS. Furthermore, while the structure obtained for the apo kpDXPS has a Rfree value of 20%, indicating a reasonable fit to the experimental data, it is important to state that the Wilson B factor is relatively high at 35 Å2. This could be linked to the disordered regions observed in the absence of the cofactor, showcasing the role of ThDP in stabilizing the overall structure of DXPS. The distinct behavior of the loops surrounding the active sites of the two homologs may have important implications for inhibitor design, as a different three-dimensional fold needs to be targeted.
Figure 3.
Diphosphate loop movement in in the active site of paDXPS.A, structure of ThDP-bound paDXPS in yellow is superimposed with the structure of apo paDXPS in cyan, showing the conformational change of the active site in the bound and unbound state. B, loop in the ThDP bound structure extending from Asn216 to Trp250, in orange exhibiting a closed conformation; C, the open conformation of the same loop. D, highlighting residues, Asn216, Asp217, and Met218, involved in the coordination with the diphosphate. Interactions are shown as black, dashed lines (below 5 Å). All figures of this type were created in PyMol.
Figure 4.
Loops rearrangement close to the active site in kpDXPS.A, the two versions of kpDXPS are visualized as a cartoon representation in light blue (ThDP-bound) and light orange (apo). An N-terminal disordered loop in apo kpDXPS (amino acids 91–124) is shown in pale green. B, comparison of the surface representation of the kpDXPS active site, with the same color-coding as in (A). Loop movement leads to a much narrower pocket in apo kpDXPS, however, the missing N-terminal loop opens up a tunnel on the other side of the cofactor-binding site.
The structure of paDXPS with thiamine analog provides evidence for the contribution of the diphosphate moiety in stabilizing a loop close to the active site
Thiamine analogs were previously reported as potent inhibitors of ThDP-dependent enzymes (26, 27). Several thiamine analogs had been synthesized in our group as inhibitors of DXPS (27, 28). We used one of these compounds to co-crystalize with our homologs as a diphosphate-free version of ThDP. The compounds were tested against paDXPS and kpDXPS. The co-crystalized thiamine analog compound (2-{3-[(4-Amino-2-Methylpyrimidin-5-Yl)methyl]phenyl}ethanol) showed a half-maximal inhibitory concentration (IC50) of 197 μM for paDXPS and 94 μM for kpDXPS. To further study the conformational changes in the active site loop, we set out to obtain an X-ray structure of either paDXPS or kpDXPS with the thiamine analog. We were unsuccessful to generate cocrystals for kpDXPS with the inhibitor. We were, however, able to obtain a 2.2 Å co-crystal structure of paDXPS (PDB ID: 8A4D).
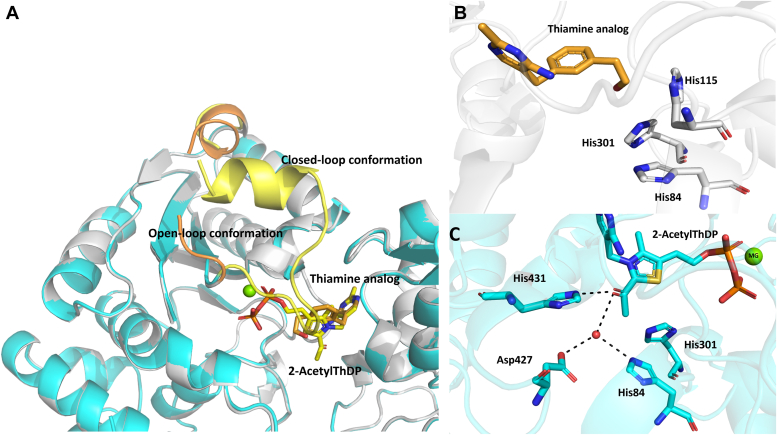
As anticipated, the binding of this analog is similar to the binding of the thiamine moiety of ThDP (pyrimidine and thiazole) with the pyrimidine ring stacking against Phe395. However, the phenyl ring is showing a slight deviation from the plane of thiazole binding seen with ThDP. The hydroxyl group of the ligand is pointing toward the entry site of the substrates while the flexibility of the hydroxyethyl group resulted in poor density for this part of the inhibitor. Nonetheless, it is surrounded by a histidine-rich region and shows the potential of H bonding to these residues (Fig. 5B). Compounds with shorter hydroxyl linkages show less activity (results not shown). Interestingly, the binding of this inhibitor excludes the part of Domain I that is involved in coordinating the diphosphate/Mg+2; the diphosphate loop shows the same open conformation as seen in the apo enzyme (Fig. 5A). This adds to the notion that this loop is stabilized by the interaction with this specific part of ThDP. This is an especially critical observation for drug discovery, as a different three-dimensional shape of the active site could enable the exploration of different regions of the DXPS in search for inhibitors.
Figure 5.
Binding of the thiamine analog and 2-acetyl-ThDP with paDXPS.A, thiamine analog bound structure in gray (loop in orange) and 2-acetyl ThDP structure in cyan (loop in yellow). B, zoomed in view of the Interactions between thiamine analog (orange) and binding residues of paDXPS, the structure shows the hydroxyethyl part of the ligand pointing towards a histidine-rich region in the substrate binding site. The diphosphate loop shows an open conformation in the thiamine-bound structure, in orange. C, interactions between 2-acetyl-ThDP (cyan) and binding residues of paDXPS, the structure shows a closed diphosphate loop conformation, in yellow. Interactions are shown as black, dashed lines (below 5 Å).
Structure of paDXPS in the presence of the substrate analog fluoropyruvate shows the binding of acetylated ThDP and competitive inhibition
Fluoropyruvate is an analog of the natural substrate pyruvate; it was also described as the first inhibitor of DXPS from P. aeruginosa (9). DXPS decarboxylates pyruvate in an interaction mediated by ThDP in a stepwise manner: pyruvate first interacts with ThDP to form the intermediate lactyl ThDP (LThDP), this intermediate was shown to be stabilized by a histidine network near the C2 of ThDP. This reaction promotes a closed conformation of a loop close to the active site and the addition of D-GAP leads to a more open conformation, which eventually encourages decarboxylation (15, 17).
We performed an inhibition assay and a mode-of-inhibition (MOI) study on paDXPS using fluoropyruvate. In the inhibition assay, fluoropyruvate inhibited the enzyme with an (IC50) value of around 27 μM. The MOI study indicated competitive inhibition with the natural substrate pyruvate (Table S3). Fluoropyruvate has also been reported to inactivate the E1 subunit of the pyruvate dehydrogenase complex, a mammalian ThDP-dependent enzyme. It was originally proposed that the fluoropyruvate interaction with the ThDP-bound enzyme results in a modification of a sulfhydryl group in the active site (8). Illustrated in Figure 1 is the interaction of fluoropyruvate with ThDP-bound paDXPS as compared to pyruvate. To determine whether the inhibition of the enzyme is the result of the acetylation of a reactive residue in the active site after the addition of fluoropyruvate, we performed intact MS measurements to detect the acetylation of the enzyme by mass shift, intact mass results show similar masses for fluoropyruvate treated and untreated proteins (Fig. S5). These results, together with the MOI assay, suggest that no covalent modification is involved in inhibition by fluoropyruvate.
We then opted to get structural information on the fluoropyruvate-treated paDXPS. The enzyme was incubated and crystallized with excess fluoropyruvate in the presence of ThDP and MgCl2. The structure was solved using molecular replacement and refined to 2.0 Å (PDB ID: 8A45). We found no evidence of an acetylated residue within the active site. On the other hand, we observed convincing electron density for the reaction intermediate 2-acetyl-ThDP in the active site (Fig. 5), consistent with the proposed reaction mechanism (Fig. 1). These findings indicate that inhibition with fluoropyruvate results rather from the acetylated ThDP, which is expected to be less reactive than ThDP and competes with the reactive enamine formed with pyruvate.
The structure of paDXPS in the presence of fluoropyruvate provides a high-resolution view of acetylThDP-bound enzyme, a reaction product related to the enamine intermediate. The pyrimidine and diphosphate moiety of 2-acetyl-ThDP is bound in a highly similar way to ThDP (Fig. 5A), with the diphosphate loop also stabilized by coordinating residues Asp187, Asn216, and Met218 to the diphosphate/Mg part. Hydrogen bonds are formed between His-431 and the carbonyl of acetyl-ThDP. This histidine residue is conservatively preserved in other ThDP-dependent enzymes and has been shown to contribute to a relaxed/packed state, that ultimately drives decarboxylation (29). The conformation of this region is different in the structure of mtDXPS. M. tuberculosis replaces this interaction with a network of water-coordinated histidine, serine, and tyrosine (23). Interestingly, the carbonyl is also involved in coordinating water molecules with Asp427 and His84, while preserving the his431 conformation (Fig. 5C). The presence of these interactions results in a tetrahedral geometry around C2 of 2-acetylThDP, causing it to deviate from the plane of the thiazole, enabled by the single bond in the acetyl-ThDP compared to the double bond in the enamine. Our structure also shows the presence of His301, adding a positive charge to the pocket, which is missing in the structure with enamine in drDXPS. This histidine is replaced by tyrosine in ecDXPS (14). His301 is also part of the spoon-fork motif, the flexibility of this site facilitates binding to the substrates and stabilizes intermediates (17). This region is significantly different from the DXPS structures. Figure 6 shows only some differences within the aligned fork motifs of the 2-acetyl ThDP paDXPS structure and drDXPS structure. In kpDXPS residues 246 to 284 are disordered and are also part of the spoon-fork motif. These differences might result in even more different conformational changes in this region during catalysis.
Figure 6.
Spoon-fork motif of paDXPS.A, spoon-fork motif in paDXPS in the presence of 2-acetyl THDP colored yellow, superimposed with drDXPS in the presence of PLThDP, colored in orange showing the open conformation of the spoon motif in both structures. B, zoomed-in view of the fork motif highlighting changes in residues involved in stabilizing conformational changes in this region. Residues from drDXPS are labeled in orange, from paDXPS in black.
Conclusion
This study introduces the first apo structures of DXPS from P. aeruginosa and K. pneumoniae, along with their cofactor ThDP-bound structures. These findings contribute to expanding our understanding of this important enzyme class and provide crucial structural insights for the advancement of innovative drug development targeting ESKAPE pathogens.
Upon ThDP binding, DXPS undergoes conformational changes that lead to an open and closed loop conformation near the active site. In the paDXPS structure, the closed conformation is stabilized by interactions of the ThDP diphosphate/Mg2+ moiety with an asparagine-rich region of near the active site. In contrast, an open conformation is seen when this interaction is lost in the apo structure. This hypothesis was also confirmed by analyzing the crystal structure of paDXPS with a thiamine analog lacking the diphosphate moiety, in which the same loop adopts the open conformation seen with the apo enzyme. The DXPS structure from K. pneumonia also shows loop movement deep in the active site, which leads to a much narrower pocket in the absence of ThDP.
From a drug-design perspective, these results open the door to different modes of DXPS inhibition. Our structures reveal the important role of the diphosphate moiety of ThDP in stabilizing part of the active site. Targeting specific residues in this loop may prevent the enzyme from binding to ThDP by not providing enough space for ThDP to bind, especially with the rearrangement of the loops in kpDXPS. In paDXPS, one strategy could be to prevent the diphosphate-binding loop from closing, thereby preventing the enzyme from entering its active conformation. This could perhaps be achieved by using the pyrimidyl ring present in ThDP as a warhead and optimizing the rest of the compound to bind to the loop in its “open” conformation. Additionally, the binding mode of the thiamine analog indicates that it is also possible to explore compounds that target the substrate-binding channel in addition to the ThDP binding pocket. Following this approach, a three-headed inhibitor could perhaps be developed, which would likely show increased selectivity for DXPS over other ThDP-dependent enzymes due to the exploitation of DXPS’s unique binding-site geometry. Furthermore, we demonstrated that a competitive inhibition could be achieved by blocking further interactions of ThDP-bound enzyme with molecules that can form direct, less reactive intermediates with ThDP, therefore trapping the enzyme in this rate-determining kinetic step and preventing subsequent interactions with the natural substrates. These additional DXPS X-ray crystal structures from important ESKAPE pathogens lay the foundation for future structure-based drug design of potent and selective small-molecule inhibitors with a novel mode of action to treat bacterial infections.
Experimental procedures
Protein expression and purification
paDXPS
The construct genes were synthesized commercially, cloned into pET28a, and transformed into BL21(DE3) for overexpression. The transformed cells were grown in a selective LB medium supplemented with Kanamycin (50 mg/ml) up to an OD600 of 0.6, then induced with 1 mM isopropyl-L-D-thiogalactoside (IPTG) and were subsequently grown at 18 °C for 16 h. The harvested cells were then disrupted in a microfluidizer after resuspension in a washing buffer consisting of 50 mM HEPES, 100 mM NaCl, 20 mM imidazole, and 2 mM β-mercaptoethanol (100 ml buffer per 25 g of wet cell pellet). After centrifugation, the supernatant was loaded onto a 5 ml HisTrap HP column equilibrated in washing buffer. After an extensive washing step with 30 column volumes of washing buffer, the bound fraction was then eluted with 50 mM HEPES, 100 mM NaCl, 300 mM Imidazole, and 2 mM β-mercaptoethanol. His-tag purification was followed by size-exclusion chromatography in 20 mM HEPES, 200 mM NaCl, and 2 mM β-mercaptoethanol. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and protein mass spectrometry were used to confirm the purity of protein purifications.
kpDXPS
The construct gene was synthesized commercially, cloned into pET28b+, and transformed into Lemo21(DE3) for overexpression. The transformed cells were grown in a selective LB medium at 37 °C supplemented with kanamycin (50 mg/ml) and chloramphenicol (34 mg/ml) up to an OD600 of 0.9, then induced with 0.1 mM isopropyl-L-D-thiogalactoside (IPTG), and were subsequently grown at 18 °C for 16 h. The harvested cells were then disrupted in a microfluidizer after resuspension in a washing buffer consisting of 20 mM Tris pH 8.0, 200 mM NaCl, 20 mM Imidazole pH 8.0, and 5% glycerol (100 ml buffer per 25 g of wet cell pellet). After centrifugation, the supernatant was loaded onto a 5 ml HisTrap HP column equilibrated in washing buffer. After an extensive washing step with 30 column volumes of washing buffer, the bound fraction was then eluted with an elution buffer consisting of 20 mM Tris pH 8.0, 200 mM NaCl, 250 mM imidazole pH 8.0, and 5% glycerol. Eluted protein was passed over a Hiprep 26/10 desalting column equilibrated in washing buffer to remove excess imidazole, after which TEV digestion was carried out by adding TEV protease in a 1:10 protein: TEV ratio and incubating the solution at 4 °C for 16 h. The N-terminal 6x His-Tag was removed by passing the protein over a 5 ml His-Trap HP column and collection of the flow-through. Purification was finalized by size-exclusion chromatography in 20 mM Tris, 200 mM NaCl, final pH 8.0 for apo kpDXPS and 10 mM HEPES, 200 mM NaCl, 5% glycerol, final pH 7.4 for ThDP-bound kpDXPS. SDS-PAGE and protein mass spectrometry were used to confirm the purity of protein purifications.
Crystallization
paDXPS
Crystallization trials were carried out with a protein solution containing 15 mg/ml paDXPS in a storage buffer using commercially available screens from NeXtalbiotech. Ligand-bound structures were obtained after incubating paDXPS with an excess amount of the ligand for 2 h at RT. In every case, crystals were grown in hanging drops with a reservoir solution consisting of 100 mM HEPES, 12% PEG-8000, and 200 mM calcium acetate. Plates were observed every 3 days and optimization of the identified conditions was carried out. 32% Glycerol was added as a cryo-protectant before the crystals were mounted in a cryo-loop and stored in liquid nitrogen until data collection.
kpDXPS
Crystallization trials were carried out with a protein solution containing 12.5 mg/ml kpDXPS (ligand-bound) and 47.5 mg/ml kpDXPS (apo) in the respective storage buffer using commercially available screens from NeXtalbiotech. Ligand-bound structures of kpDXPS were obtained by the addition of 1 mM ThDP/MgCl2 and incubating the solution for 4 h on ice before setting up the crystallization plates. For the protein complex, crystals were grown in sitting-drop optimization plates at 18 °C with a reservoir solution consisting of 100 mM HEPES pH 7.0 to 7.5 and 18 to 28% PEG-3000. For the apo version, crystals were grown in hanging-drop optimization plates at 18 °C with a reservoir solution consisting of 200 to 400 mM MgCl2, 100 mM HEPES pH 7.5, and 14 to 18% PEG-8000. In both cases, 15% 2R,3R-butanediol was added as a cryoprotectant before the crystals were mounted in a cryo-loop and stored in liquid nitrogen until data collection.
Data collection and refinement
X-ray diffraction data for DXPS structures were collected at the Swiss Light Source located at the Paul Scherrer Institute in Switzerland. Specific information about each beamline can be found in the specific sections, all data collection and refinement statistics can be found in Table S2. Data reduction and scaling were done using AIMLESS (30) in CCP4i (31). Structures were solved using molecular replacement using the program Phaser.MR (32) in Phenix software (33). Repetitive cycles of refinement were done in COOT (34) and Phenix refine (35) to obtain final structures (Tables S1 and S2). Structures were validated using the Molprobity server and all figures were rendered using PyMoL.
Kinetics and inhibition assays
To measure DXPS activity and kinetic parameters, a coupled spectrophotometric enzyme assay was adapted from the assay protocol described in the literature (36). A continuous kinetic photometric assay was used to measure DXPS activity. NADPH consumption by the auxiliary enzyme 1-deoxy-D-xylulose 5-phosphate reductoisomerase (IspC) was measured in a microplate reader (PHERAstar), monitoring the absorbance at 340 nm. In all cases (IspC) concentration was kept in excess (1 μM) to ensure DXPS is the only rate-limiting enzyme in the reaction mix. In the inhibition assays, in the case of the thiamine analog inhibitor, ThDP concentration was kept around 4X Km (400 nM) to ensure the enzyme reaches maximum velocity while allowing competitive inhibition. In the case of fluoropyruvate, pyruvate concentration was also kept at 4X Km (500 μM) for the same reason. For inhibition studies, DXPS activity was analyzed at RT for 30 min as follows; reaction mixture A (100 mM HEPES pH 7.5, 100 mM NaCl, 1 mM MgCl2, 0.5 μM NAPDH, 1 μM IspC, 400 nM ThDP, and 0.15 μM and 0.2 μM paDXPS and kpDXPS respectively) was pre-incubated with different concentrations of inhibitor for 5 min at 37 °C in 10% DMSO. The reaction was then started by adding mixture B (100 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DL-glyceraldehyde-3-phosphate and 2 mM pyruvate (thiamine analog inhibitor) and 500 μM pyruvate (fluoropyruvate). For kinetic assays, the cofactor ThDP and substrates pyruvate and D-GAP were used in varying concentrations. When a substrate or cofactor was kept constant, a concentration of 400 nM was used for ThDP and 2 mM for pyruvate and D-GAP. Blank correction and linear fitting of the absorption data were performed using the program Origin 2019 (OriginLab). The initial velocities obtained were plotted against the substrate concentrations, and the Km values were determined by nonlinear curve fitting using the Michaelis–Menten model of the enzyme kinetics, Figs. S6 and S7.
Data availability
Data supporting this study are included within the article and/or supporting materials.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Dr Nicolas Frank and Dr Timo Risch for native MS and intact MS measurements. We acknowledge the Paul Scherrer Institute, Villigen, Switzerland for the provision of synchrotron radiation beamtime at beamline X06DA-PXIII and X06SA-PXI of the SLS and the European Synchrotron Radiation Facility (ESRF), Grenoble, France for provision of beamline ID23 to 1.
Author contributions
A. K. H. H., R. H., and S. A. conceptualization; R. H. and S. A. writing–original draft; A. L. and L. M. synthesis. A. K. H. H., R. H., S. A., A. L., L. M., and J. K., writing–review and editing; A. K. H. H. supervision.
Funding and additional information
The Helmholtz Association’s Initiative and Networking Fund (to A. K. H. H.) and the Schlumberger Foundation faculty for the future (FFTF) (to R. H.) funded this work.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
Supporting information
References
- 1.Lin J., Nishino K., Roberts M.C., Tolmasky M., Aminov R.I., Zhang L. Mechanisms of antibiotic resistance. Front. Microbiol. 2015;6:34. doi: 10.3389/fmicb.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miethke M., Pieroni M., Weber T., Brönstrup M., Hammann P., Halby L., et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021;5:726–749. doi: 10.1038/s41570-021-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nolan L.M., Turnbull L., Katrib M., Osvath S.R., Losa D., Lazenby J.J., et al. Pseudomonas aeruginosa is capable of natural transformation in biofilms. Microbiology (Reading) 2020;166:995–1003. doi: 10.1099/mic.0.000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camus L., Briaud P., Vandenesch F., Moreau K. How bacterial adaptation to cystic fibrosis environment shapes interactions between Pseudomonas aeruginosa and Staphylococcus aureus. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.617784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riquelme S.A., Ahn D., Prince A. Pseudomonas aeruginosa and Klebsiella pneumoniae adaptation to innate immune clearance mechanisms in the lung. J. Innate Immun. 2018;10:442–454. doi: 10.1159/000487515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Concepcion M. The MEP pathway: a new target for the development of herbicides, antibiotics and antimalarial drugs. CPD. 2004;10:2391–2400. doi: 10.2174/1381612043384006. [DOI] [PubMed] [Google Scholar]
- 8.Flournoy D.S., Frey P.A. Inactivation of the pyruvate dehydrogenase complex of escherichia coli by fluoropyruvate. Biochemistry. 1989;28:9594–9602. doi: 10.1021/bi00451a007. [DOI] [PubMed] [Google Scholar]
- 9.Altincicek B., Hintz M., Sanderbrand S., Wiesner J., Beck E., Jomaa H. Tools for discovery of inhibitors of the 1-deoxy-D-xylulose 5-phosphate (DXP) synthase and DXP reductoisomerase: an approach with enzymes from the pathogenic bacterium Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2000;190:329–333. doi: 10.1111/j.1574-6968.2000.tb09307.x. [DOI] [PubMed] [Google Scholar]
- 10.Gräwert T., Groll M., Rohdich F., Bacher A., Eisenreich W. Biochemistry of the non-mevalonate isoprenoid pathway. Cell. Mol. Life Sci. 2011;68:3797–3814. doi: 10.1007/s00018-011-0753-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Dowd C.S. The methylerythritol phosphate pathway: promising drug targets in the Fight against tuberculosis. ACS Infect. Dis. 2018;4:278–290. doi: 10.1021/acsinfecdis.7b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J.M., Vierling R.J., Meyers C.F. Selective inhibition of E. coli1-deoxy-D-xylulose-5-phosphate synthase by acetylphosphonates. Med. Chem. Commun. 2012;3:65–67. doi: 10.1039/C1MD00233C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J.M., Warrington N.V., Vierling R.J., Kuhn M.L., Anderson W.F., Koppisch A.T., et al. Targeting DXP synthase in human pathogens: enzyme inhibition and antimicrobial activity of butylacetylphosphonate. J. Antibiot. (Tokyo) 2014;67:77–83. doi: 10.1038/ja.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang S., Usunow G., Lange G., Busch M., Tong L. Crystal structure of 1-Deoxy-d-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 2007;282:2676–2682. doi: 10.1074/jbc.M610235200. [DOI] [PubMed] [Google Scholar]
- 15.Patel H., Nemeria N.S., Brammer L.A., Freel Meyers C.L., Jordan F. Observation of thiamin-bound intermediates and microscopic rate constants for their Interconversion on 1-deoxy- d -xylulose 5-phosphate synthase: 600-fold rate Acceleration of pyruvate decarboxylation by d -Glyceraldehyde-3-phosphate. J. Am. Chem. Soc. 2012;134:18374–18379. doi: 10.1021/ja307315u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartee D., Freel Meyers C.L. Toward understanding the chemistry and biology of 1-deoxy- d -xylulose 5-phosphate (DXP) synthase: a unique antimicrobial target at the Heart of bacterial metabolism. Acc. Chem. Res. 2018;51:2546–2555. doi: 10.1021/acs.accounts.8b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P.Y.-T., DeColli A.A., Freel Meyers C.L., Drennan C.L. X-ray crystallography–based structural elucidation of enzyme-bound intermediates along the 1-deoxy-d-xylulose 5-phosphate synthase reaction coordinate. J. Biol. Chem. 2019;294:12405–12414. doi: 10.1074/jbc.RA119.009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brammer Basta L.A., Patel H., Kakalis L., Jordan F., Freel Meyers C.L. Defining critical residues for substrate binding to 1-deoxy-d-xylulose 5-phosphate synthase – active site substitutions stabilize the predecarboxylation intermediate C2α-lactylthiamin diphosphate. FEBS J. 2014;281:2820–2837. doi: 10.1111/febs.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J., Yang L., DeColli A., Freel Meyers C., Nemeria N.S., Jordan F. Conformational dynamics of 1-deoxy-D-xylulose 5-phosphate synthase on ligand binding revealed by H/D exchange MS. Proc. Natl. Acad. Sci. U. S. A. 2017;114:9355–9360. doi: 10.1073/pnas.1619981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartee D., Freel Meyers C.L. Targeting the unique mechanism of bacterial 1-deoxy-D-xylulose 5-phosphate (DXP) synthase. Biochemistry. 2018;57:4349–4356. doi: 10.1021/acs.biochem.8b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gierse R.M., Redeem E., Diamanti E., Wrenger C., Groves M.R., Hirsch A.K. DXS as a target for structure-based drug design. Future Med. Chem. 2017;9:1277–1294. doi: 10.4155/fmc-2016-0239. [DOI] [PubMed] [Google Scholar]
- 22.Masini T., Kroezen B.S., Hirsch A.K.H. Druggability of the enzymes of the non-mevalonate-pathway. Drug Discov. Today. 2013;18:1256–1262. doi: 10.1016/j.drudis.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Gierse R.M., Oerlemans R., Reddem E.R., Gawriljuk V.O., Alhayek A., Baitinger D., et al. First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization. Sci. Rep. 2022;12:7221. doi: 10.1038/s41598-022-11205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gierse R.M., Reddem E.R., Alhayek A., Baitinger D., Hamid Z., Jakobi H., et al. Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties. Biochem. Biophys. Res. Commun. 2021;539:42–47. doi: 10.1016/j.bbrc.2020.12.069. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Versées W., Spaepen S., Wood M.D.H., Leeper F.J., Vanderleyden J., Steyaert J. Molecular mechanism of allosteric substrate activation in a thiamine diphosphate-dependent Decarboxylase. J. Biol. Chem. 2007;282:35269–35278. doi: 10.1074/jbc.M706048200. [DOI] [PubMed] [Google Scholar]
- 27.Mann S., Perez Melero C., Hawksley D., Leeper F.J. Inhibition of thiamin diphosphate dependent enzymes by 3-deazathiamin diphosphate. Org. Biomol. Chem. 2004;2:1732. doi: 10.1039/b403619k. [DOI] [PubMed] [Google Scholar]
- 28.Masini T., Lacy B., Monjas L., Hawksley D., de Voogd A.R., Illarionov B., et al. Validation of a homology model of Mycobacterium tuberculosis DXS: rationalization of observed activities of thiamine derivatives as potent inhibitors of two orthologues of DXS. Org. Biomol. Chem. 2015;13:11263–11277. doi: 10.1039/c5ob01666e. [DOI] [PubMed] [Google Scholar]
- 29.Arjunan P., Sax M., Brunskill A., Chandrasekhar K., Nemeria N., Zhang S., et al. A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J. Biol. Chem. 2006;281:15296–15303. doi: 10.1074/jbc.M600656200. [DOI] [PubMed] [Google Scholar]
- 30.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi S., Kuzuyama T., Watanabe H., Seto H. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting this study are included within the article and/or supporting materials.