Abstract
Coronavirus disease 2019 (COVID-19) vaccines are highly efficacious at preventing symptomatic infection, severe disease, and death. Most of the evidence that COVID-19 vaccines also reduce transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is based on retrospective, observational studies. Specifically, an increasing number of studies are evaluating vaccine effectiveness against the secondary attack rate of SARS-CoV-2 using data available in existing health-care databases or contact-tracing databases. Since these types of databases were designed for clinical diagnosis or management of COVID-19, they are limited in their ability to provide accurate information on infection, infection timing, and transmission events. We highlight challenges with using existing databases to identify transmission units and confirm potential SARS-CoV-2 transmission events. We discuss the impact of common diagnostic testing strategies, including event-prompted and infrequent testing, and illustrate their potential biases in estimating vaccine effectiveness against the secondary attack rate of SARS-CoV-2. We articulate the need for prospective observational studies of vaccine effectiveness against the SARS-CoV-2 secondary attack rate, and we provide design and reporting considerations for studies using retrospective databases.
Keywords: COVID-19, retrospective studies, SARS-CoV-2
Abbreviations
- COVID-19
coronavirus disease 2019
- PCR
polymerase chain reaction
- SAR
secondary attack rate
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- VE
vaccine effectiveness
Coronavirus disease 2019 (COVID-19) vaccines were developed primarily to prevent symptomatic infection and, most importantly, severe disease and death. They have shown high efficacy against these endpoints in experimental and observational studies (1–13). Evidence suggests that these vaccines also prevent infection (5, 14–18) and potentially reduce transmission (19–23), albeit with smaller effects against the highly transmissible Omicron variant compared with wildtype severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and earlier variants (24–26).
While randomized controlled trials play a key role in understanding efficacy against COVID-19 disease, they are inefficient for capturing asymptomatic SARS-CoV-2 infection or transmission, and so most evidence for COVID-19 vaccine effects against these endpoints comes from observational studies. Recent literature has evaluated study designs and analytical approaches for estimating vaccine efficacy against infection (27–30). Here, we focus on another key component of a vaccine’s effect on the overall burden of the epidemic: reducing secondary transmission (30–33).
The secondary attack rate (SAR)—the rate of transmission to susceptible contacts within a transmission unit over a well-defined time period (34)—is a common measure of transmission risk. Vaccine efficacy against the SAR is 1 minus the relative SAR for infected vaccinated individuals versus the SAR for infected unvaccinated individuals (31, 34, 35). Evaluating vaccine efficacy against the SAR of SARS-CoV-2 is challenging in the experimental setting, particularly in the context of an ongoing pandemic (28, 36). It requires frequent surveillance of trial participants to detect all incident infections, even those that are asymptomatic, and enrollment and surveillance of all contacts of infected participants to detect secondary transmission. Phase III COVID-19 vaccine efficacy trials generally captured asymptomatic infections only through periodic SARS-CoV-2 serology, and infectiousness was assessed through the proxy of SARS-CoV-2 viral load in symptomatic individuals; transmission events from infected trial participants were not captured (2, 3, 5–8, 33, 37–40). Phase III efficacy trials also provide limited evidence on variants of concern because the blinded follow-up was largely complete before their emergence.
Given these challenges, most evidence on COVID-19 vaccine effects against the SARS-CoV-2 SAR come from retrospective observational, postlicensure studies, where systematic diagnostic testing is often absent (33). In particular, studies are often done using linked national registries of vaccination and infection data based on household addresses (21–23), and of contact-tracing databases (19, 20). Observational studies can be used to evaluate vaccine effectiveness (VE) against the SARS-CoV-2 SAR. VE against the SAR is influenced not only by biological effects on transmission but also by potential behavioral effects such as behavioral disinhibition and by effects of “real world” vaccine delivery such as variation in timing of dose administration (41, 42). We illustrate the limitations of retrospective, observational study designs for studying VE against the SARS-CoV-2 SAR. First, we highlight the main classes of study designs. Next, we discuss challenges in the ascertainment of infections and transmission events using these designs. We derive an analytical formula for the bias in VE against the SAR introduced by common diagnostic testing strategies in these designs. Finally, we articulate the need for well-designed prospective observational studies of VE against the SARS-CoV-2 SAR, provide study design and reporting considerations for retrospective designs, and highlight areas for methodological innovation that may enable more accurate estimation of VE of the SARS-CoV-2 SAR when retrospective databases are the only option.
RETROSPECTIVE STUDY DESIGNS FOR EVALUATING VE AGAINST THE SARS-COV-2 SAR
Existing studies use 2 primary retrospective designs to evaluate VE against the SARS-CoV-2 SAR: record-linked database studies and contact-tracing database studies. We discuss the features of these designs with a focus on identifying transmission units and transmission events (see Table 1 for examples).
Table 1.
Statistical Designs of Example Studies of COVID-19 Vaccine Efficacy Against SARS-CoV-2 Transmissiona
| First Author, Year (Reference No.) | Design | Testing Strategy | Transmission Unit and Susceptible Contacts | Strategy to Exclude Community Transmission | Strategy to Restrict to Transmissions from Primary Subject Only |
|---|---|---|---|---|---|
| Eyre, 2022 (19) | Analysis of contact-tracing database. | Event-prompted testing for primary subjects. Only close contacts that received a test were included. | Relative risk of infection in close contacts that received a PCR test within 1–10 days of the index case, comparing index cases who were unvaccinated, partially vaccinated, and fully vaccinated, controlling for vaccination status of contact. | N/A: No predefined transmission unit. | Included only test results from contacts tested within 1–10 days of index case |
| de Gier, 2021 (20) | Analysis of contact-tracing database. Person can be both a contact and an index. | Event-prompted testing for primary subjects. Encouraged contacts of index case to get tested as soon as possible, and on the fifth day after last exposure. | Relative risk of infection in close contacts of unvaccinated adults compared with adults with confirmed infection who have received partial or full vaccination in the Netherlands. Both pooled and stratified analyses by vaccination status of the contact. | N/A: No predefined transmission unit. | Excluded indexes where their most likely source of infection was in the home. Included only test results from contacts tested within 1–14 days of index case. Required quarantining of contacts after exposure. |
| Harris, 2022 (21) | Analysis of health-care databases with linked records for households. | Event-prompted testing for primary subject and contacts. | Relative risk of infection in unvaccinated household contacts of vaccinated individuals compared with unvaccinated individuals who also test positive. | Only include contact infections that occurred within 2–14 days of index. | Exclude households with more than 1 person testing positive within 2 days. |
| Prunas, 2021 (22) | Analysis of health-care databases with linked records for households. Does not assign an index case; models acquisition and transmission events within households over time by imputing infection times. | Event-prompted testing for primary subject and contacts. | Relative risk of infection in household contacts of vaccinated subjects compared with unvaccinated subjects, controlling for vaccination status of contact. | Allow for baseline risk of community-acquired infection in modeling. | Imputation of infection times allows for within-household sources of infections that are not the primary subject in modeling. |
Table 1.
Continued
| First Author, Year (Reference No.) | Design | Testing Strategy | Transmission Unit and Susceptible Contacts | Strategy to Exclude Community Transmission | Strategy to Restrict to Transmissions from Primary Subject Only |
|---|---|---|---|---|---|
| Lyngse, 2022 (23) | Analysis of health-care databases with linked records for households. | Event-prompted testing for primary subjects. Substantial effort put into contact tracing. | Relative risk of infection in household contacts (controlling for vaccination status) of fully vaccinated and boosted individuals who test positive compared with partially or unvaccinated individuals who also test positive. Paper specifically focuses on comparing this relative risk between households with primary Omicron infection versus primary Delta infection. | Only include contact infections that occurred within 1–7 days of index. | Exclude households with more than 1 person testing positive on same day. |
Abbreviations: COVID-19, coronavirus disease 2019; N/A, not applicable; PCR, polymerase chain reaction; SAR, secondary attack rate; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
a Studies were selected as examples to illustrate key elements of the retrospective designs discussed. This is not an exhaustive list of retrospective studies of vaccine efficacy against the SARS-CoV-2 SAR.
Identifying transmission units in retrospective designs
Key to studying SARS-CoV-2 transmission is identifying susceptible contacts of infected individuals. The group of individuals that includes an infectious person and their susceptible contacts is commonly referred to as a transmission unit (34). We use the term index case for the first detected case and primary case for the first infected person in the transmission unit (34, 43). Transmission units may be defined based on a characteristic at the group level, such as a shared residence, or in reference to an individual (e.g., close contacts identified through contact tracing). Identifying susceptible contacts requires defining “susceptible.” Given data suggesting that breakthrough SARS-CoV-2 infections (44–46) and reinfections (47–50) are not uncommon, there are arguments for considering all contacts to be susceptible.
Health-care record linkage to infer transmission units
An important class of retrospective designs defines a household as the transmission unit and identifies SARS-COV-2 transmission events within households by leveraging databases of diagnostic test results; vaccination status, type, and timing; and household addresses. Prunas et al. (22) used data from household members in a database from a health maintenance organization covering 2.5 million individuals in Israel that includes demographic characteristics, health-care utilization data, vaccination data, and SARS-CoV-2 test results. Lyngse et al. (23) used unique identification numbers for residents of Denmark to link reverse-transcription polymerase chain reaction (PCR) and antigen test results from the Danish Microbiology Database with vaccination records in the Danish Vaccination Register and identified transmission units based on residential address. Similarly, Harris et al. (21) linked the national immunization database in England to a data set with all laboratory-confirmed SARS-CoV-2 infections and identified persons sharing the same address. Transmission units captured via record linkage may incorporate some degree of measurement error due to changes in households over the course of the study or nontraditional household structures (51, 52).
Contact-tracing databases to infer transmission units
An alternative retrospective design links existing SARS-CoV-2 contact-tracing databases that include diagnostic test results of contacts of confirmed cases with databases of vaccination records. One example is de Gier et al. (20), where close contacts of confirmed COVID-19 cases were sought out for PCR testing by the Municipal Health Services contact monitoring in the Netherlands, and the test results were combined with the national infectious disease notification registry that contains vaccination records. Similarly, Eyre et al. (19) used data from England’s National Health Service Test and Trace Service, which performs a similar contact monitoring service for individuals who test positive in England. Vaccination status was obtained from the National Immunization Management Service. Transmission units inferred from contact-tracing databases are specific to the definitions, strategies, and function of the tracing service and may be incompletely captured if tracing services are overly conservative in defining contacts of infected individuals, if infected individuals are reticent to disclose their contacts, or if contacts cannot be contacted or tested. Incomplete capture of contacts is a pervasive challenge for contact-tracing systems (53–55). Mutually exclusive transmission units, which are ideal for direct estimation of the SAR, may be difficult to define using contact-tracing databases, where “strings” of infections and contacts are identified.
IMPACT OF TESTING STRATEGY IN RETROSPECTIVE DESIGNS
Retrospective designs, by definition, rely on testing designed for clinical diagnosis or management as opposed to research. One common challenge is “event-prompted” SARS-CoV-2 diagnostic testing, where testing is triggered by occurrence of an event, such as onset of symptoms, potential SARS-CoV-2 exposure, or mandate by social institutions or circumstances, such as travel, employment, or medical procedures (33). Another challenge is infrequent testing (29). In this section, we describe the pitfalls of event-prompted and infrequent diagnostic testing and illustrate their potential biases. In what follows, we consider tests with no potential for false positives for people in the transmission unit. We revisit this issue in the discussion.
Limitations of event-prompted testing
Symptom-prompted testing is common and guaranteed to miss many SARS-CoV-2 infections, given the large burden of asymptomatic infection (56, 57). The proportion of infections that are asymptomatic varies with age, comorbidities, and preexisting immunity (56, 58); method for ascertaining symptoms (59); and infecting variant (60). The extent to which existing databases capture primarily symptomatic testing may vary over time and geography as routine asymptomatic testing of individuals becomes more common or is mandated by social institutions. Importantly, many studies have no information on what prompted the tests captured in the database, which prevents exploration of the impact of the missed infections on VE estimates.
In addition to missed asymptomatic infections, symptom-prompted testing poses challenges in inferring transmission chains because infections are not detected until (and if) symptoms occur, typically several days after acquisition of infection. The incubation period for SARS-CoV-2—the time between onset of infection and symptoms—has been estimated as 6 days on average for ancestral strains, shorter with new variants, and is variable across individuals (61–64).
Limitations of infrequent testing
Even with routine testing, infections may be missed if testing occurs infrequently. The duration of SARS-CoV-2 positivity by diagnostic testing can be as short as 1 day (65–67), especially when vaccinated or previously infected. And yet, even individuals who shed for a short period may still have transmission potential if shedding large amounts of virus (68). Daily PCR testing is likely necessary to capture all SARS-CoV-2 infections, and twice-weekly PCR testing likely captures a supermajority of infections (69–72).
SARS-CoV-2 serology is another tool for capturing current or past infections based on periodic blood collection. While commercial assays detecting antibodies against the nucleocapsid protein (anti-N antibodies) have demonstrated high sensitivity and specificity (73), and anti-N antibody responses are reported to be durable (74) and not elicited by COVID-19 vaccines that target the spike protein, serology has limited value for studies of transmission because it does not inform on timing of infection. Furthermore, serological assays may be insensitive to past infection in vaccinated individuals (75).
Potential impact of event-prompted or infrequent testing on estimation of VE
To illustrate the potential impact of event-prompted or infrequent testing on estimation of VE against transmission, we compare the “target” estimand—the expectation of the statistical estimate of VE against the SARS-CoV-2 SAR—with the “actual” estimands under imperfect testing. We assume a range of scenarios around SARS-CoV-2 infection and transmission (see Web Appendix 1, available at https://doi.org/10.1093/aje/kwad046) that illustrate the issues of symptom-prompted testing (see Web Appendix 2) and infrequent testing (see Web Appendix 3) in isolation; in practice there may be several issues at play.
As shown in Figure 1, under the scenarios we consider, for both symptom-prompted testing and infrequent testing, the actual estimand is no larger than the target estimand. The target and actual estimands agree when either vaccinated individuals have no transmission potential (VE = 1) or when asymptomatic and symptomatic individuals have the same transmission potential. As the transmission potential of breakthrough infections increases, the difference between the estimands increases. This relationship is more pronounced for larger reductions in transmission for asymptomatic infections (Figure 1A). The implication is that, since asymptomatic individuals likely transmit less frequently than symptomatic individuals (76), testing only symptomatic infections may tend to underestimate VE against the SARS-CoV-2 SAR.
Figure 1.
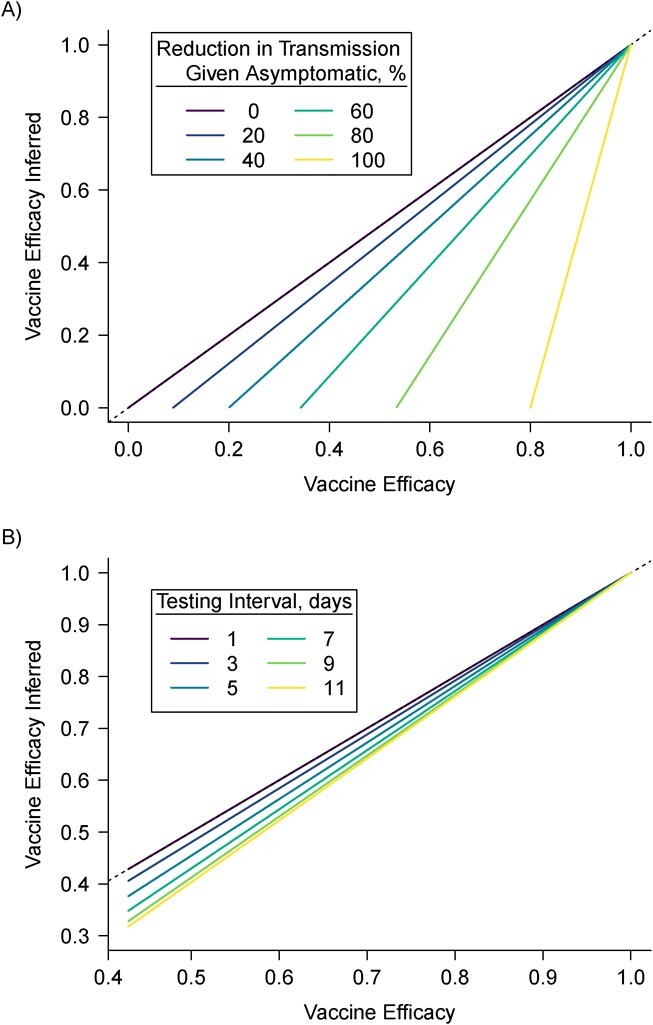
Analytical comparison of actual (y-axis) and target (x-axis) vaccine efficacy against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission estimands. The dashed line indicates equality. A) Difference between target and actual estimands under symptom-prompted testing under varying reduction in transmission potential for asymptomatic versus symptomatic infections. B) Difference between target and actual estimands under infrequent testing under varied frequency of testing. Note the different scales of the 2 x- and y-axes. In (A), the color indicates percent reduction in the secondary attack rate comparing asymptomatic to symptomatic infected people: A value of 100% indicates that asymptomatic infected people cannot transmit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and a value of 0% indicates that the transmission potential for asymptomatic and symptomatic people is the same. In (B), the color indicates interval between subsequent tests (e.g., a value of 7 means that individuals were tested once every 7 days). “Actual” estimand refers to the estimand that is available given the sampling design, and is derived analytically (see Web Material).
Under infrequent testing, the difference between estimands is a function of the testing interval: When the testing interval is longer than the minimum duration of infection (1 day), the actual estimand is smaller than the target estimand (Figure 1B). In Figure 1B, we show only testing intervals that are shorter than the maximum duration of infection in the vaccinated group (15 days). Generally, the relationship between the testing interval and the difference between the actual and target estimands is not monotonic (see Web Figure 1), and when the interval is longer than the longest duration of infection, the difference between the estimands does not depend on the testing interval. The implication is that infrequent testing would tend to yield an underestimate of VE against the SARS-CoV-2 SAR.
In Figure 1, we assumed correct classification of who infected whom. Figure 2 illustrates how irregular and infrequent testing for SARS-CoV-2 may also misclassify transmission events. In the hypothetical transmission units shown (Figure 2A), both symptom-prompted testing (Figure 2B) and infrequent testing (Figure 2C) underestimate the number of transmission events and result in misclassifying the primary case in at least 1 of the transmission units. Under symptom-prompted testing (Figure 2B), the misclassification is due to a longer presymptomatic period for the primary case vs. the secondary infection. The impact of misclassification of transmission events will depend on the extent and nature of the misclassification; the actual estimand may be higher or lower than the target estimand.
Figure 2.
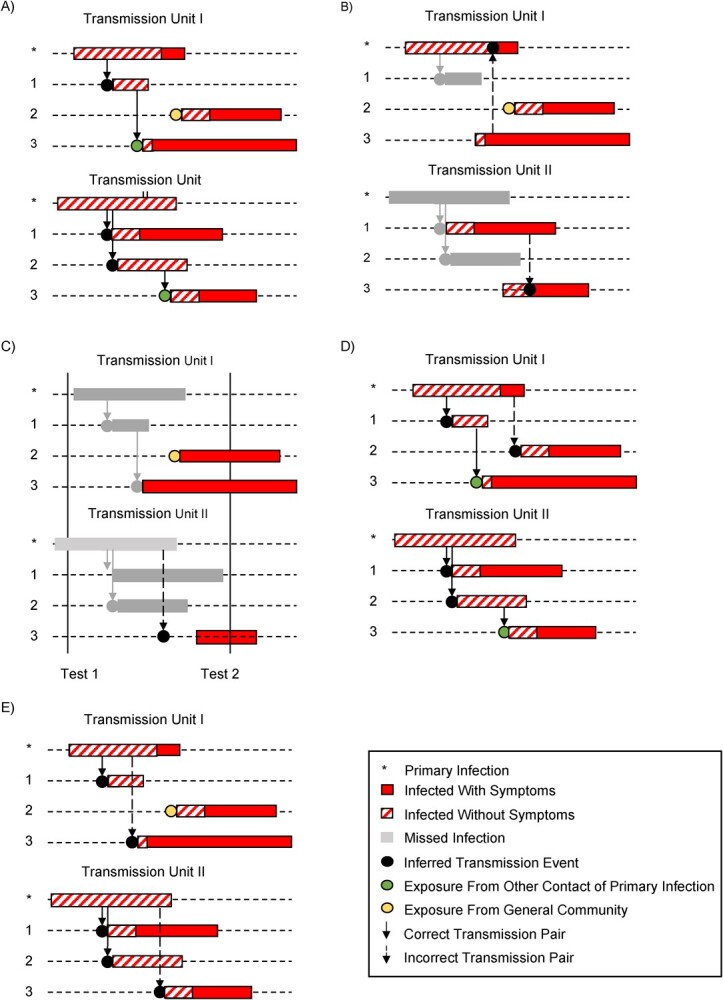
True and inferred transmission chains for 2 hypothetical transmission units, each with 1 primary case and 3 susceptible contacts. Black dots represent true or inferred transmission events. A) True infection and transmission histories. B) Inferred transmission events based on symptom-prompted testing. C) Inferred transmission events based on infrequent testing. D) Inferred transmission events if community acquisition events are incorrectly included as transmission events. E) Inferred transmission events if contact-to-contact transmissions are incorrectly included as transmission events.
Impact of incomplete testing of transmission unit
Some individuals in a transmission unit may “opt out” of testing entirely or during certain time periods, or there may be limited testing availability. Such incomplete testing is likely nearly universal and is not always reported. Two out of 3 of the household studies listed in Table 1 did not report testing rates; Lyngse et al. (23) reported that about 10% of household members in their study were not tested within 1 week after the index case tested positive. For records-based studies, the most common approach is to assume that individuals not tested are SARS-CoV-2 negative. However, the outcome for untested individuals is missing, resulting in incomplete capture of both primary case and secondary transmission events. For contract-tracing studies, the number of susceptible contacts—the denominator of the SAR—may also be underestimated due to incomplete testing. Without being able to identify whether missingness is “completely at random” (77) or depends on factors such as vaccination or demographic characteristics, it is not possible to determine the direction of the induced bias. We revisit this issue when discussing analytical strategies.
Strategies for confirming potential transmission events
Correct estimation of the SAR requires knowledge of transmission from a primary case to susceptible contacts in the transmission unit. To confirm an infection in a contact as a transmission event, acquisition of infection from the community and contact-to-contact transmission within a transmission unit need to be ruled out. Including community acquisition or contact-to-contact transmission events generally biases the calculation of the SAR upwards (78). The direction and magnitude of the bias in the VE against the SAR will depend on the extent to which contacts of vaccinated vs. unvaccinated primary cases acquire infection from the community and from other transmission unit members.
Community-acquired infections in susceptible contacts.
Figure 2D illustrates the issue with community-acquired infections: We may classify infections diagnosed after the index case as transmission events when in fact the infections are community-acquired. As an example, 2 people may be infected from the same source outside of a household but test positive at different times, so we incorrectly infer that the second infection was a within-household transmission event. One way to account for community acquisition is through modeling, with assumptions around the latent and infectious periods for both vaccinated and unvaccinated individuals (78–81), as applied to SARS-CoV-2 by Prunas et al. (22). Households without any infections other than the index case contribute information about community transmission under this estimation strategy. An alternative approach is to leverage additional data to distinguish between community-acquired and transmission events. One source of information is the timing of diagnosis; for example, de Gier et al. (20) and Harris et al. (21) include only infected contacts who developed symptoms within 2–14 days of the index case. Viral genetic sequence data may also assist in ruling out potential transmission events (82, 83). High-quality SARS-CoV-2 genome sequencing data and epidemiologic data have been used during outbreak investigations (84, 85). Studies of infection clustering have used viral genetic testing to construct and infer phylogenetic trees, including studies of SARS-CoV-2 transmission among university students (86) and US Marine recruits (87). Finally, epidemiologic data can assist in restricting attention to likely transmission events based on close contact. For example, Sikkens et al. (88) used test results and behavioral data to rule out cases of health-care worker–to-patient transmission. Note that these data on viral sequences and epidemiologic data are rarely available in records-linkage studies.
The choices of transmission unit and study duration can also assist in distinguishing between community acquisition events and transmission events. For household studies, the within-household SAR for SARS-CoV-2 during Omicron has been estimated at 50%–70% (24). In contrast, the likelihood of community-acquired infection over a short time period, even in the context of a local outbreak, is generally much smaller. Even during the peak of the Omicron epidemic in New York City, with nearly 700,000 recorded incident infections in January 2022, about 8% of residents were diagnosed with infection (89). In general, the greater the ability of the study to restrict attention to populations with tightly interacting transmission units over the duration of infectiousness for the primary case, the better the chances are that community acquisition events can be excluded. However, this may limit generalizability of the estimates of VE.
Contact-to-contact transmission within the same transmission unit.
A susceptible contact may acquire a SARS-CoV-2 infection from a different contact within the transmission unit, other than the primary case (Figure 2E). Given that a transmission unit is selected as a group of individuals who have frequent, close physical contact, it is difficult to rule out the possibility of contact-to-contact transmission even with timing of diagnosis, viral genetic data, and behavioral data. Failing to account for contact-to-contact transmission may introduce bias in the estimation of the SARS-CoV-2 SAR, and thus bias in the estimation of VE against the SAR (78). One strategy is a model-based approach such as that explored by Prunas et al. (22), where infection times and infection durations are simulated to infer pairwise transmission risk (79). These strategies may help limit misclassification of transmission events.
The SARS-CoV-2 viral load in the index case and infected contact, which is a likely surrogate for transmission potential (90–93), may assist in excluding contact-to-contact transmission events. Prospectively designed studies have the important merit that they can capture full viral load curves, thus greatly enhancing accurate timing of acquisition in the contact and of infectiousness of the index. However, retrospective studies seldom capture viral load in indexes or contacts, as this is not a routine measurement in clinical practice. Some qualitative PCR assays report cycle threshold (Ct) as a surrogate of viral load; however, there is considerable variability across assays, platforms, and specimen types in terms of the reference range and reliability of this measure (91, 92). Even when measured, retrospective studies generally only have the cycle threshold measurement at a single time point—at infection diagnosis—which greatly limits the utility of the measure for classifying transmission events.
DISCUSSION
More than 2 years into the pandemic, SARS-CoV-2 remains a major global public health challenge. As more transmissible variants have arisen, in the context of high rates of vaccine hesitancy in some populations and scarce access to vaccine in other populations, definitive evidence on the overall ability of COVID-19 vaccines to reduce onward transmission of infection is needed.
While there remain arguments for randomized trial designs in settings without access to COVID-19 vaccines (94, 95) and for randomized rollout of vaccines in settings where vaccines are starting to be distributed (33, 36, 96, 97), future studies with randomized designs and clinical endpoints are likely to be few. Furthermore, because capturing transmission events through prospective and frequent testing is resource-intensive, future studies of COVID-19 vaccine effects on transmission will likely be observational.
Observational designs that follow potential transmission units prospectively provide the most rigorous answers. This may be accomplished by prospective enrollment and testing of entire transmission units, prior to any infection diagnosis, and follow-up of all members over a long period spanning an outbreak (33, 98, 99). Cohen et al. initiated such a design in the context of seasonal influenza (100) and pivoted to study SARS-CoV-2 at the onset of the pandemic (101), where individuals within household transmission units performed twice-weekly nasal swabs for 13 months to permit PCR testing regardless of symptoms. Alternatively, a “case-ascertained” approach enrolls transmission units with an incident infection, all of whose members are subsequently tested for a short period (98, 102). Clifford et al. (103) implemented a case-ascertained approach in which household contacts of index cases in the United Kingdom were tested for SARS-CoV-2 on days 1, 3, and 7 after enrollment. Prospective testing of contacts will provide the most accurate capture and classification of transmission events but is resource-intensive.
While health-care record–linkage and contact-tracing designs often leverage large databases that are rich sources of data, they incorporate limitations due to their retrospective nature and the fact that the associated data are collected for nonresearch purposes. Infrequent and irregular testing is likely to miss infections and to selectively capture longer-duration and symptomatic infections (29). Misclassification of the primary cases and secondary transmission events is also possible. There is likely to be incomplete testing of both indexes and contacts, as well as a limited understanding of drivers of testing utilization. Even with correct identification of primary cases, there may be bias due to the inclusion of community-acquired infections and contact-to-contact transmission. We did not consider the potential for false positives, which could occur for a variety of reasons (104). The false-positive rate of PCR (where a false positive is defined as positive for only a single gene at a cycle threshold of >35) has been estimated to be around 0.5% (105). The impact of false positives on estimation of SAR will depend on the underlying incidence of SARS-CoV-2 circulating and the rate of testing.
Finally, factors that are associated with both risk of acquisition or transmission of SARS-CoV-2 and with uptake and timing of COVID-19 vaccination, or with both receipt of SARS-CoV-2 tests and vaccination, are potential cofounders. Behavioral factors that influence pathogen exposure and transmission, uptake of vaccination, and testing strategies are likely not captured in retrospective databases and may lead to unmeasured confounding.
Considerations for statistical analysis
Improved statistical analyses may help in addressing some of the challenges encountered in retrospective studies of VE against the SAR for SARS-CoV-2 and other respiratory pathogens. The fundamental challenge encountered for both record-linked and contact-tracing databases is informatively missing data: missed primary cases and transmission events and misclassified transmission events. Given additional information or knowledge on the mechanism of missingness, it is possible to employ statistical corrections. If drivers of missingness are known or can be inferred from the data, statistical models may be leveraged to model the probability of missing data as a function of measured variables, and inverse-probability weighted (IPW)—or more efficient “augmented” IPW—analyses may be performed to attempt to account for the informative missingness (106, 107). Latent variable analyses that allow for the modeling of unobserved variables may also be helpful (108). Sensitivity analyses are critical tools for evaluating the extent to which study conclusions depend on potential errors introduced by missing or mismeasured primary cases and transmission events (109, 110).
Recommendations for study design
We recommend leveraging studies in populations that have controlled diagnostic testing prompts over the duration of study, such as health-care workers or university students, as this can mitigate the effects of infrequent and event-driven testing. Performing studies in homogeneous study populations, with tightly interacting transmission units, will assist in evaluating the relative likelihood of community acquisition vs. within-unit transmission. Authors should collect contextual information on factors influencing testing uptake and timing, such as relevant public health guidelines and institutional policies and practices, as well as factors influencing transmission, such as guidelines on masking, social distancing, isolation, and quarantine. Contextual data are helpful for interpreting study reliability and generalizability, even if only available at the population level. Finally, using epidemiologic and sequence data in addition to data on the timing of infection diagnosis, will improve the ability to classify transmission events.
Recommendations for reporting results
STROBE guidelines for observational studies (111, 112), and RECORD guidelines for observational routinely collected health data (113) should be followed. Authors should describe the testing program in place during the study, including drivers of testing utilization and timing and the extent of testing completion (e.g., percent of household members receiving a test within 1 week of the index case, see Lyngse et al. Figure 1A (23)), and summarize how these drivers vary by vaccination status. This may include performing sensitivity analyses around factors related to the testing program to describe directions of bias in vaccine efficacy estimates, as in our Figure 1. Authors should provide details about how transmission events were ascertained, the expected level of community transmission that occurred during the study, and how the authors accounted for potential contact-to-contact transmission. In addition to describing which variables were used to control for confounding, we recommend including a discussion of which unmeasured variables may contribute to residual confounding given the study population, time, and local epidemic dynamics. We advise that authors provide careful interpretation of the VE estimand that was estimated given the testing program. Employing these strategies will assist readers in interpreting the VE parameters in context and in gauging the reliability and generalizability of the results.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Biostatistics, School of Public Health, University of Washington, Seattle, Washington, United States (Marlena S. Bannick, Fei Gao, Elizabeth R. Brown, Holly E. Janes); Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Center (Fei Gao, Elizabeth R. Brown, Holly E. Janes); and Public Health Sciences Division, Fred Hutchinson Cancer Center (Fei Gao, Elizabeth R. Brown, Holly E. Janes).
Supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (grants R56 AAI143418 and UM1 AI068635 to Fred Hutchinson Cancer Center). Supported by the Infectious Diseases Clinical Research Consortium through the National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health, under award number UM1AI148684. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The code to produce figures is available at https://github.com/mbannick/ve-transmission-retrospective.
A preprint of this article has been published online. Bannick MS, Gao F, Brown ER, Janes HE. Retrospective, observational studies for estimating vaccine effects on the secondary attack rate of SARS-CoV-2. arXiv. 2022. (https://doi.org/10.48550/arXiv.2206.07495).
E.R.B. declares support within the last 36 months from the Bill and Melinda Gates Foundation (paid to her institution) and has received payments in the last 36 months for consulting from the Adolescent Trials Network at UNC Chapel Hill and for participation on a data safety and monitoring board from Merck. The other authors report no conflicts.
REFERENCES
- 1. Emary KRW, Golubchik T, Aley PK, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397(10282):1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shinde V, Bhikha S, Hoosain Z, et al. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(20):1899–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(20):1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frenck Jr RW, Klein NP, Kitchin N, et al. Safety, immunogenicity, and efficacy of the BNT162b2 Covid-19 vaccine in adolescents. N Engl J Med. 2021;385(3):239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voysey M, Costa Clemens SA, Madhi SA, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397(10277):881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384(23):2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baden LR, el Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al Kaabi N, Zhang Y, Xia S, et al. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA. 2021;326(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vasileiou E, Simpson CR, Shi T, et al. Interim findings from first-dose mass COVID-19 vaccination roll-out and COVID-19 hospital admissions in Scotland: a national prospective cohort study. Lancet. 2021;397(10285):1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: test negative case-control study. BMJ. 2021;373:n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jara A et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N Engl J Med. 2021;385(10):875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrejko KL, Pry J, Myers JF, et al. Prevention of coronavirus disease 2019 (COVID-19) by mRNA-based vaccines within the general population of California. Clin Infect Dis. 2022;74(8):1382–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swift MD, Breeher LE, Tande AJ, et al. Effectiveness of mRNA COVID-19 vaccines against SARS-CoV-2 infection in a cohort of healthcare personnel. Clin Infect Dis. 2021;73(6):e1376–e1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thompson MG. Interim estimates of vaccine effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines in preventing SARS-CoV-2 infection among health care personnel, first responders, and other essential and frontline workers—eight U.S. locations, December 2020–March 2021. MMWR Morb Mortal Wkly Rep. 2021;70:13, 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall VJ, Foulkes S, Saei A, et al. COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. Lancet. 2021;397(10286):1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones NK, Rivett L, Seaman S, et al. Single-dose BNT162b2 vaccine protects against asymptomatic SARS-CoV-2 infection. Elife. 2021;10:e68808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang L, Hijano DR, Gaur AH, et al. Asymptomatic and symptomatic SARS-CoV-2 infections after BNT162b2 vaccination in a routinely screened workforce. JAMA. 2021;325(24):2500–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eyre DW, Taylor D, Purver M, et al. Effect of Covid-19 vaccination on transmission of Alpha and Delta variants. N Engl J Med. 2022;386(8):744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Gier B, Andeweg S, Joosten R, et al. Vaccine effectiveness against SARS-CoV-2 transmission and infections among household and other close contacts of confirmed cases, the Netherlands, February to May 2021. Euro Surveill. 2021;26(31):2100640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris RJ, Hall JA, Zaidi A, et al. Effect of vaccination on household transmission of SARS-CoV-2 in England. N Engl J Med. 2021;385(8):759–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prunas O, Warren JL, Crawford FW, et al. Vaccination with BNT162b2 reduces transmission of SARS-CoV-2 to household contacts in Israel. Science. 2022;375(6585):1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lyngse FP, Mortensen LH, Denwood MJ, et al. Household transmission of the SARS-CoV-2 omicron variant in Denmark. Nat Commun. 2022;13(1):5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baker JM, Nakayama JY, O’Hegarty M, et al. SARS-CoV-2 B.1.1.529 (Omicron) variant transmission within households—four U.S. jurisdictions, November 2021–February 2022. MMWR Morb Mortal Wkly Rep. 2022;71(9):341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jørgensen SB, Nygård K, Kacelnik O, et al. Secondary attack rates for Omicron and Delta variants of SARS-CoV-2 in Norwegian households. JAMA. 2022;327(16):1610–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Allen H, Tessier E, Turner C, et al. Comparative transmission of SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants and the impact of vaccination: national cohort study, England [preprint]. medRxiv. 2022. 10.1101/2022.02.15.22271001. Accessed April 6, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Accorsi EK, Qiu X, Rumpler E, et al. How to detect and reduce potential sources of biases in studies of SARS-CoV-2 and COVID-19. Eur J Epidemiol. 2021;36(2):179–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Madewell ZJ, Dean NE, Berlin JA, et al. Challenges of evaluating and modelling vaccination in emerging infectious diseases. Epidemics. 2021;37:100506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Follmann DA, Fay MP. Vaccine efficacy at a point in time [published online ahead of print on March 17, 2022]. Biostatistics. 2022. 10.1093/biostatistics/kxac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lipsitch M, Krammer F, Regev-Yochay G, et al. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat Rev Immunol. 2022;22(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Halloran ME, Longini IM, Struchiner CJ. Overview of vaccine effects and study designs. In: Halloran ME, Longini IM, Struchiner CJ, eds. Design and Analysis of Vaccine Studies. New York, NY: Springer; 2010:19–45. [Google Scholar]
- 32. Lipsitch M, Kahn R. Interpreting vaccine efficacy trial results for infection and transmission. Vaccine. 2021;39(30):4082–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lipsitch M, Dean NE. Understanding COVID-19 vaccine efficacy. Science. 2020;370(6518):763–765. [DOI] [PubMed] [Google Scholar]
- 34. Halloran ME, Préziosi MP, Chu H. Estimating vaccine efficacy from secondary attack rates. J Am Stat Assoc. 2003;98(461):38–46. [Google Scholar]
- 35. Ainslie KEC, Haber MJ, Malosh RE, et al. Maximum likelihood estimation of influenza vaccine effectiveness against transmission from the household and from the community. Stat Med. 2018;37(6):970–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dean NE, Gsell PS, Brookmeyer R, et al. Design of vaccine efficacy trials during public health emergencies. Sci Transl Med. 2019;11(499):eaat0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dunkle LM, Kotloff KL, Gay CL, et al. Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N Engl J Med. 2022;386(6):531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Falsey AR, Sobieszczyk ME, Hirsch I, et al. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N Engl J Med. 2021;385(25):2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heath PT, Galiza EP, Baxter DN, et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med. 2021;385(13):1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Logunov DY, Dolzhikova IV, Shcheblyakov DV, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schaper C, Fleming TR, Self SG, et al. Statistical issues in the design of HIV vaccine trials. Annu Rev Public Health. 1995;16(1):1–22. [DOI] [PubMed] [Google Scholar]
- 42. Mohammed I, Nauman A, Paul P, et al. The efficacy and effectiveness of the COVID-19 vaccines in reducing infection, severity, hospitalization, and mortality: a systematic review. Hum Vaccin Immunother. 2022;18(1):2027160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Giesecke J. Primary and index cases. Lancet. 2014;384(9959):2024. [DOI] [PubMed] [Google Scholar]
- 44. Mizrahi B, Lotan R, Kalkstein N, et al. Correlation of SARS-CoV-2-breakthrough infections to time-from-vaccine. Nat Commun. 2021;12(1):6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gharpure R, Sami S, Vostok J, et al. Multistate outbreak of SARS-CoV-2 infections, including vaccine breakthrough infections, associated with large public gatherings, United States. Emerg Infect Dis. 2022;28(1):35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goldberg Y, Mandel M, Bar-On YM, et al. Waning immunity after the BNT162b2 vaccine in Israel. N Engl J Med. 2021;385(24):e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abu-Raddad LJ, Chemaitelly H, Bertollini R, et al. Severity of SARS-CoV-2 reinfections as compared with primary infections. N Engl J Med. 2021;385(26):2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vitale J, Mumoli N, Clerici P, et al. Assessment of SARS-CoV-2 reinfection 1 year after primary infection in a population in Lombardy. JAMA Intern Med. 2021;181(10):1407–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hansen CH, Michlmayr D, Gubbels SM, et al. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397(10280):1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malhotra S, Mani K, Lodha R, et al. SARS-CoV-2 reinfection rate and estimated effectiveness of the inactivated whole virion vaccine BBV152 against reinfection among health care workers in new Delhi. JAMA Netw Open. 2022;5(1):e2142210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doidge JC, Harron KL. Reflections on modern methods: linkage error bias. Int J Epidemiol. 2019;48(6):2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Casey JA, Schwartz BS, Stewart WF, et al. Using electronic health records for population health research: a review of methods and applications. Annu Rev Public Health. 2016;37(1):61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lewis D. Why many countries failed at COVID contact-tracing—but some got it right. Nature. 2020;588(7838):384–387. [DOI] [PubMed] [Google Scholar]
- 54. Shelby T, Schenck C, Weeks B, et al. Lessons learned from COVID-19 contact tracing during a public health emergency: a prospective implementation study. Front Public Health. 2021;9:721952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lash RR, Moonan PK, Byers BL, et al. COVID-19 case investigation and contact tracing in the US, 2020. JAMA Netw Open. 2021;4(6):e2115850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sah P, Fitzpatrick MC, Zimmer CF, et al. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc Natl Acad Sci USA. 2021;118(34):e2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Johansson MA, Quandelacy TM, Kada S, et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4(1):e2035057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Syangtan G, Bista S, Dawadi P, et al. Asymptomatic SARS-CoV-2 carriers: a systematic review and meta-analysis. Front Public Health. 2021;8:587374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jeong TH, Pak C, Ock M, et al. Real asymptomatic SARS-CoV-2 infection might be rare: importance of careful interviews and follow-up. J Korean Med Sci. 2020;35(37):e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garrett N, Tapley A, Andriesen J, et al. High asymptomatic carriage with the Omicron variant in South Africa. Clin Infect Dis. 2022;75(1):e289–e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jansen L, Tegomoh B, Lange K, et al. Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) variant cluster—Nebraska, November–December 2021. MMWR Morb Mortal Wkly Rep. 2021;70(5152):1782–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Elias C, Sekri A, Leblanc P, et al. The incubation period of COVID-19: a meta-analysis. Int J Infect Dis. 2021;104:708–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Salzberger B, Buder F, Lampl B, et al. Epidemiology of SARS-CoV-2. Infection. 2021;49(2):233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wei Y, Wei L, Liu Y, et al. Comprehensive estimation for the length and dispersion of COVID-19 incubation period: a systematic review and meta-analysis. Infection. 2022;50(4):803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Johnston C, Brown ER, Stewart J, et al. Hydroxychloroquine with or without azithromycin for treatment of early SARS-CoV-2 infection among high-risk outpatient adults: a randomized clinical trial. EClinicalMedicine. 2021;33:100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stankiewicz Karita HC, Dong TQ, Johnston C, et al. Trajectory of viral RNA load among persons with incident SARS-CoV-2 G614 infection (Wuhan strain) in association with COVID-19 symptom onset and severity. JAMA Netw Open. 2022;5(1):e2142796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mallett S, Allen AJ, Graziadio S, et al. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Med. 2020;18(1):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Singanayagam A, Hakki S, Dunning J, et al. Community transmission and viral load kinetics of the SARS-CoV-2 Delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2021;22(2):183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chin ET, Huynh BQ, LAC C, et al. Frequency of routine testing for coronavirus disease 2019 (COVID-19) in high-risk healthcare environments to reduce outbreaks. Clin Infect Dis. 2021;73:e3127–e3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lopman B, Liu CY, le Guillou A, et al. A modeling study to inform screening and testing interventions for the control of SARS-CoV-2 on university campuses. Sci Rep. 2021;11(1):5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hellewell J, Russell TW, the SAFER Investigators and Field Study Team, the Crick COVID-19 Consortium, et al. Estimating the effectiveness of routine asymptomatic PCR testing at different frequencies for the detection of SARS-CoV-2 infections. BMC Med. 2021;19(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Larremore DB, Wilder B, Lester E, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2021;7(1):eabd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tan SS, Saw S, Chew KL, et al. Comparative clinical evaluation of the Roche Elecsys and Abbott severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serology assays for coronavirus disease 2019 (COVID-19). Arch Pathol Lab Med. 2021;145(1):32–38. [DOI] [PubMed] [Google Scholar]
- 74. Gallais F, Gantner P, Bruel T, et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. 2021;71:103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Follmann D, Janes HE, Buhule OD, et al. Antinucleocapsid antibodies after SARS-CoV-2 infection in the blinded phase of the randomized, placebo-controlled mRNA-1273 COVID-19 vaccine efficacy clinical trial. Ann Intern Med. 2022;175(9):1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Madewell ZJ, Yang Y, Longini IM, et al. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw Open. 2020;3(12):e2031756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Little RJA. A test of missing completely at random for multivariate data with missing values. J Am Stat Assoc. 1988;83(404):1198–1202. [Google Scholar]
- 78. Sharker Y, Kenah E. Estimating and interpreting secondary attack risk: binomial considered biased. PLoS Comput Biol. 2021;17(1):e1008601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Longini IM, Koopman JS. Household and community transmission parameters from final distributions of infections in households. Biometrics. 1982;38(1):115–126. [PubMed] [Google Scholar]
- 80. Haber M, Longini IM, Cotsonis GA. Models for the statistical analysis of infectious disease data. Biometrics. 1988;44(1):163–173. [PubMed] [Google Scholar]
- 81. Petrie JG, Eisenberg MC, Ng S, et al. Application of an individual-based transmission Hazard model for estimation of influenza vaccine effectiveness in a household cohort. Am J Epidemiol. 2017;186(12):1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lythgoe KA, Hall M, Ferretti L, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372(6539):eabg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Perera D, Perks B, Potemkin M, et al. Reconstructing SARS-CoV-2 infection dynamics through the phylogenetic inference of unsampled sources of infection. PLoS One. 2021;16(12):e0261422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang X, Wu C, Li X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7(6):1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Siddle KJ, Krasilnikova LA, Moreno GK, et al. Transmission from vaccinated individuals in a large SARS-CoV-2 Delta variant outbreak. Cell. 2022;185(3):485–492.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Weil AA et al. SARS-CoV-2 epidemiology on a public university campus in Washington state. Open forum. Infect Dis. 2021;8:ofab464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Letizia AG, Ramos I, Obla A, et al. SARS-CoV-2 transmission among marine recruits during quarantine. N Engl J Med. 2020;383(25):2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sikkens JJ, Buis DTP, Peters EJG, et al. Serologic surveillance and phylogenetic analysis of SARS-CoV-2 infection among hospital health care workers. JAMA Netw Open. 2021;4(7):e2118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. The New York Times . Coronavirus (Covid-19) data in the United States. https://github.com/nytimes/covid-19-data. 2022. Accessed March 30, 2022.
- 90. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. [DOI] [PubMed] [Google Scholar]
- 91. Lee MJ. Quantifying SARS-CoV-2 viral load: current status and future prospects. Expert Rev Mol Diagn. 2021;21(10):1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Buchta C, Görzer I, Chiba P, et al. Variability of cycle threshold values in an external quality assessment scheme for detection of the SARS-CoV-2 virus genome by RT-PCR. Clin Chem Lab Med. 2021;59(5):987–994. [DOI] [PubMed] [Google Scholar]
- 93. Walker AS, Pritchard E, House T, et al. Ct threshold values, a proxy for viral load in community SARS-CoV-2 cases, demonstrate wide variation across populations and over time. Elife. 2021;10:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fleming TR, Nason M, Krause PR, et al. COVID-19 vaccine trials: the potential for ‘hybrid’ analyses. Clin Trials. 2021;18(4):391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. WHO Ad Hoc Expert Group on the Next Steps for Covid-19 Vaccine Evaluation . Placebo-controlled trials of Covid-19 vaccines—why we still need them. N Engl J Med. 2021;384(2):e2. [DOI] [PubMed] [Google Scholar]
- 96. Herzog LM, Norheim OF, Emanuel EJ, et al. Covax must go beyond proportional allocation of covid vaccines to ensure fair and equitable access. BMJ. 2021;372:m4853. [DOI] [PubMed] [Google Scholar]
- 97. Hemkens LG, Goodman SN. Randomized COVID-19 vaccination rollout can offer direct real-world evidence. J Clin Epidemiol. 2021;138:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang Y, Longini IM, Halloran ME. Design and evaluation of prophylactic interventions using infectious disease incidence data from close contact groups. J R Stat Soc Ser C Appl Stat. 2006;55(3):317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Klick B, Nishiura H, Leung GM, et al. Optimal design of studies of influenza transmission in households II: comparison between cohort and case-ascertained studies. Epidemiol Infect. 2014;142(4):744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cohen C, Kleynhans J, Moyes J, et al. Asymptomatic transmission and high community burden of seasonal influenza in an urban and a rural community in South Africa, 2017–18 (PHIRST): a population cohort study. Lancet Glob Health. 2021;9(6):e863–e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cohen C, Kleynhans J, von Gottberg A, et al. SARS-CoV-2 incidence, transmission, and reinfection in a rural and an urban setting: results of the PHIRST-C cohort study, South Africa, 2020–21. Lancet Infect Dis. 2022;22(6):821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Klick B, Leung GM, Cowling BJ. Optimal design of studies of influenza transmission in households. I: case-ascertained studies. Epidemiol Infect. 2012;140(1):106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Clifford S et al. Effectiveness of BNT162b2 and ChAdOx1 against SARS-CoV-2 household transmission: a prospective cohort study in England [preprint]. medRxiv. 2021. ( 10.1101/2021.11.24.21266401). Accessed March 26, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Braunstein GD, Schwartz L, Hymel P, et al. False positive results with SARS-CoV-2 RT-PCR tests and how to evaluate a RT-PCR-positive test for the possibility of a false positive result. J Occup Environ Med. 2021;63(3):e159–e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Healy B, Khan A, Metezai H, et al. The impact of false positive COVID-19 results in an area of low prevalence. Clin Med (Lond). 2021;21(1):e54–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Robins JM, Rotnitzky A. Semiparametric efficiency in multivariate regression models with missing data. J Am Stat Assoc. 1995;90(429):122–129. [Google Scholar]
- 107. Rotnitzky A, Lei Q, Sued M, et al. Improved double-robust estimation in missing data and causal inference models. Biometrika. 2012;99(2):439–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rabe-Hesketh S, Skrondal A. Classical latent variable models for medical research. Stat Methods Med Res. 2008;17(1):5–32. [DOI] [PubMed] [Google Scholar]
- 109. Austin PC, Stuart EA. Moving towards best practice when using inverse probability of treatment weighting (IPTW) using the propensity score to estimate causal treatment effects in observational studies. Stat Med. 2015;34(28):3661–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res. 2013;22(3):278–295. [DOI] [PubMed] [Google Scholar]
- 111. Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. PLoS Med. 2007;4(10):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. von Elm E, Altman DG, Egger M, et al. The Strengthening The reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med. 2007;4(10):e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Benchimol EI, Smeeth L, Guttmann A, et al. The Reporting of Studies Conducted Using Observational Routinely-Collected Health Data (RECORD) statement. PLoS Med. 2015;12(10):e1001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


