Abstract
Numerous in vitro biofilm model systems are available to study oral biofilms. Over the past several decades, increased understanding of oral biology and advances in technology have facilitated more accurate simulation of intraoral conditions and have allowed for the increased generalizability of in vitro oral biofilm studies. The integration of contemporary systems with confocal microscopy and 16S rRNA community profiling has enhanced the capabilities of in vitro biofilm model systems to quantify biofilm architecture and analyse microbial community composition. In this review, we describe several model systems relevant to modern in vitro oral biofilm studies: the constant depth film fermenter, Sorbarod perfusion system, drip–flow reactor, modified Robbins device, flowcells and microfluidic systems. We highlight how combining these systems with confocal microscopy and community composition analysis tools aids exploration of oral biofilm development under different conditions and in response to antimicrobial/anti-biofilm agents. The review closes with a discussion of future directions for the field of in vitro oral biofilm imaging and analysis.
Keywords: biofilms, bioinformatics, disease processes, microbial physiology, microbial structure
INTRODUCTION: IMPORTANCE OF IN VITRO MODEL SYSTEMS TO THE STUDY OF ORAL BIOFILMS
Micro-organisms form dynamic multi-species biofilm communities on numerous surfaces in the human oral cavity (Marsh, 2009). Over time, oral biofilms change in composition and architecture as component microbes interact with each other, the environment and the host (Lamont et al., 2018). Oral biofilm communities can be extremely resilient; redeveloping rapidly after physical perturbations (e.g. brushing or flossing) and chemical treatments (e.g. application of mouthwash) (Marsh, 2010). Furthermore, certain ecological and environmental conditions can alter the microbial composition and behaviour of oral biofilm communities resulting in dental caries and periodontal disease (Aas et al., 2008; Marsh, 2018; Peterson et al., 2013). Dental caries and periodontal disease are among the most prevalent of human diseases (Petersen et al., 2005) ranking 1 and 11 in a 2016 ranking of global health burden of 328 diseases (Vos et al., 2017). In 2016, an estimated 2.44 billion people had active dental caries while about 750 million suffered from periodontal disease worldwide (Vos et al., 2017).
While clinical studies are the gold standard for evaluating approaches to control oral biofilms, implementing such studies can be costly and logistically demanding (Martin-Kerry et al., 2015). By contrast, in vitro biofilm systems offer a relatively less challenging platform for exploratory, fundamental and applied studies to close knowledge gaps in human oral biofilms prior to clinical studies. For example, in vitro biofilm model systems have been used to demonstrate how biofilm formation, succession and/or architecture respond to environmental challenges (Hojo et al., 2009; Kolenbrander et al., 2006), and to evaluate candidate antimicrobials (Corbin et al., 2011). Many of the available in vitro biofilm systems can be adapted to simulate multiple in vivo conditions representative of the human oral cavity (Coenye & Nelis, 2010; Yu et al., 2017). The closer the in vivo mimicry, the more generalizable the results gathered from in vitro model systems are likely to be.
An additional advantage of in vitro model systems is the ability to alter one parameter at a time, thus providing a powerful strategy for studying how biofilms develop (Fernandez et al., 2017). These experiments can provide clues into how component species interact with each other within the oral cavity and enable the characterization of potential keystone pathogenic species in biofilm development (Hajishengallis et al., 2012). For example, when considering investigations into understanding how oral species interact with one another, using a two-stage chemostat system and a defined 10-species biofilm community, Bradshaw and colleagues showed the absence of the promiscuous coaggregating organism Fusobacterium nucleatum resulted in significant changes in biofilm community representation (Bradshaw et al., 1998). Other examples of how in vitro model systems have been used in fundamental and applied oral biofilm research are detailed in Table 1.
TABLE 1.
Examples of fundamental and applied research of in vitro oral biofilms. Studies that improve the understanding of the biology of oral biofilms are considered fundamental. Applied studies, on the other hand, are studies that focus on interventions to control oral biofilms.
| Outcomes | Fundamental study (Reference) | Applied study (Reference) | Model system(s) used |
|---|---|---|---|
| Cariogenesisa | d-Glucose and sucrose induce caries (Pigman et al., 1962) | Fluoride slurry inhibits enamel softening (Pigman & Newbrun, 1962) | Artificial mouth |
| Single-species biofilm | S. mutans biofilms fed sucrose induces caries (Deng and Cate, 2004) | Chlorhexidine in dentin bonding systems may inhibit S. mutans biofilm formation (Brambilla et al., 2017) | Constant Depth Film Fermenter, Drip-Flow Reactor |
| Defined-species biofilm | S. oralis and A. oris biofilm growth was enhanced when co-cultured compared to when alone (Palmer et al., 2001) | C. albicans, L. casei, S. mutans mixed-species biofilm growth inhibited 10-fold on MRD coupons containing fluoride compared to coupons containing no fluoride (Yassin et al., 2016) | Flowcells, modified Robbins device |
| Microcosm biofilm | Community composition of in vitro biofilms can reflect that of microcosm donor (McBain et al., 2005) | Nisin retarded multi-species biofilm development without cytotoxicity to human cells (Shin et al., 2015) | Sorbarod Perfusion, Bioflux™ |
Not an oral biofilm outcome but listed to provide historical context and highlight the shift of focus to oral biofilm outcomes
In this review, we describe the relevance of in vitro biofilm models to oral health and disease research and provide a distillation of previously established models used to develop defined single-species, defined multi-species and complex multi-species (i.e. microcosm) oral biofilms. We also focus on select biofilm models that can be integrated with confocal microscopy and 16S rRNA community profiling. This integration enables the study of biofilm growth under conditions representative of the oral cavity. A particular focus of discussion will be on biofilm models that are open (constant delivery of fresh media), multiple-throughput (allowing for concurrent side-by-side testing) and that use small volumes to conduct experiments. Furthermore, we discuss the impact and potential clinical relevance of in vitro oral biofilm model systems, their limitations and future directions for in vitro oral biofilm model research.
PAST AND PRESENT: ORAL IN VITRO BIOFILM MODELS
From early oral biofilm models developed in the mid-1900s (Dietz, 1943; Pigman et al., 1952), that followed from relatively primitive models in the late 19th century (Tang et al., 2003), and throughout the ensuing decades, newer conceptual designs improved upon their predecessors. From a historical perspective, in vitro oral biofilm studies using model systems can be characterized by transitions in foci from fundamental to applied studies within three main arenas: (1) understanding the development of single-species biofilms, (2) exploring environmental and cell–cell interactions in defined multi-species biofilms and finally (3) studies of complex multi-species biofilms. In each arena, fundamental studies of biofilm development provide the framework for applied studies, such as the effects of antimicrobial or anti-biofilm interventions, resulting in insights into potential approaches to improve oral healthcare. It should be noted that there are fewer fundamental and applied in vitro periodontal disease models compared to cariogenic models, partly because of the increased complexity of simulating subgingival plaque (Velsko & Shaddox, 2018; Walker & Sedlacek, 2007). Research in multi-species (microcosm) biofilms has recently gained traction due to technological advancements and methodologies that enable investigators to measure biofilm outcomes such as community membership with 16S rRNA profiling and measuring biofilm architecture captured by a confocal microscope (Fernandez et al., 2017; Rudney et al., 2012).
Among the earliest examples of in vitro oral biofilm model systems was an ‘artificial mouth’ developed by Pigman and colleagues to study early carious lesions using extracted teeth (Pigman et al., 1952). This model was particularly notable because it was arranged vertically, and sterile media was drip-fed over an extracted human tooth inoculated with pooled human saliva and housed in an acrylic box. The media reservoir was positioned above the extracted tooth and media delivered with a hypodermic needle. This experimental setup focused on identifying conditions that favour cariogenesis; Pigman’s model is arguably an ancestor to contemporary drip-fed systems (discussed later in this review). From the 1950s to the 1960s, many in vitro oral studies improved Pigman’s artificial mouth system by including an incubator cabinet and sterilization with ethylene oxide (Pigman et al., 1955, 1962; Pigman & Newbrun, 1962). From a fundamental perspective, these studies linked common dietary sugars, for example, glucose and sucrose, to cariogenicity. From an applied standpoint, anti-cariogenic effects of compounds and dentifrice slurries could be evaluated by treating tooth enamel with anti-caries agents concomitantly with conditions that would favour cariogenesis.
Artificial mouth model variants have been used extensively over the years since the mid-1980s, most frequently by Sissons’ group (Sissons et al., 1985, 1991, 2000). Their artificial mouth system, called the ‘Multiple-plaque Artificial Mouth’ (MAM), was developed from designs by Russell and Coulter (1975) and Dibdin et al. (1976). The MAM is experimentally flexible and reproducible, and is compatible with computer-controlled systems (Sissons et al., 1991, 2000; Wong et al., 1994). Contributions and advancements by Sisson’s group and other research groups to the development of artificial mouth systems and oral biofilm research (and in particular, dental caries research) are described in further detail in an informative review by Tang and colleagues (Tang et al., 2003).
From the 1960s onwards, investigators identified and characterized many key microbial species associated with oral diseases (Gibbons & Fitzgerald, 1969; Keyes, 1968; Listgarten, 1965; Tanner et al., 1979). Consequently, biofilm model studies from the 1970s to present often focused on single-species surface-attachment/biofilm development or dual-species interaction studies using key microbial species (Bos et al., 1996; Noorda et al., 1986; Russell & Coulter, 1977; Wright et al., 1997). For example, biofilm model systems have improved understanding of coaggregation. Notably, using an in vitro flowcell biofilm model that used 25% pooled human saliva as the sole nutrient source, Palmer and colleagues evaluated biofilm development by three species known to coaggregate with one another: Streptococcus gordonii, Streptococcus oralis and Actinomyces oris. Independently, A. oris and S. oralis were shown to poorly form biofilms within the model system; however, dual-species cultures of A. oris and S. oralis formed more abundant biofilms (Palmer et al., 2001). The role of coaggregation in biofilm development has since been further explored, using in vitro biofilm models (Foster & Kolenbrander, 2004; Nagaoka et al., 2008; Periasamy & Kolenbrander, 2009).
In part due to limitations with the ability to identify micro-organisms in complex microcosm communities, as well as the interest in the behaviour of specific oral pathogens/species, many studies in the 1990s and 2000s were restricted to the development of oral biofilms containing one or a few species. While single or small consortium biofilm model systems can play an important role in uncovering the behaviour of individual or small groups of species (as mentioned above), studies of such communities provide limited understanding of how natural oral multi-species microbial communities function in their native environment (Rudney et al., 2012). Natural oral biofilms exist as a dynamic ecosystem with estimates of the total number of indigenous species ranging in the hundreds (Avila et al., 2009). In complex multi-species communities, the behaviour of a single species can be modified by other species in a community to behave in a way distinct from its behaviour when alone. Emphasizing this point, Sissons (1997) remarked in his review of oral biofilm model systems: ‘an attempt to explain plaque behavior based on the properties of monocultures can be regarded somewhat as heroic’. However, through broad technological advancements in the last decade, most notably advances in microscopy and 16S community profiling, investigators have acquired tools and methods to better characterize multi-species or microcosm biofilms (Tan et al., 2017). In recent years, many fundamental validation and protocol studies emerged to gauge reproducibility and provide preliminary microbiological results from in vitro oral microcosm biofilm (Edlund et al., 2013; Klug et al., 2016; Samarian et al., 2014). Specifically, studies using in vitro oral microcosm biofilm models have enabled the measurement of different biofilm outcomes, such as biofilm architecture, microbial community profiles and taxonomic spatial distribution (Luo et al., 2019; Roder et al., 2020).
To provide historical context, this review describes in vitro model systems that have been developed and adapted over the last 50 years. Particular attention is given to selected drip-fed and flow-fed model systems which have been used in oral biofilm studies by various research groups (Figure 1). Static microplate-based systems, which generally expose developing biofilms to minimal fluid flow, are not discussed as these types of biofilm systems were recently reviewed by Azeredo et al. (2017). Drip-fed systems deliver nutrient semi-continuously, whereas flow-fed systems deliver a constant flow of nutrients. The drip-fed systems discussed are the constant depth film fermenter (CDFF), the Sorbarod perfusion system and the drip–flow biofilm reactor. The flow-fed systems that are discussed are the modified Robbins device (MRD), flowcells and microfluidic systems, of which we describe the Bioflux™ in detail. Many of these systems possess attributes that make them appealing candidates as model systems for modern oral biofilm studies. All the model systems discussed in this review are compatible to varying degrees with confocal microscopy and have or can conceivably be manipulated to harvest biofilm cells for microbial community profiling using culture-dependent techniques and/or modern culture-independent (next-generation sequencing [NGS]) methods (Figure 1). Finally, all systems can be set up for multiple-throughput studies, and some require only relatively small volumes for experiments. A summary of the discussed model systems is presented in Table 2.
FIGURE 1.
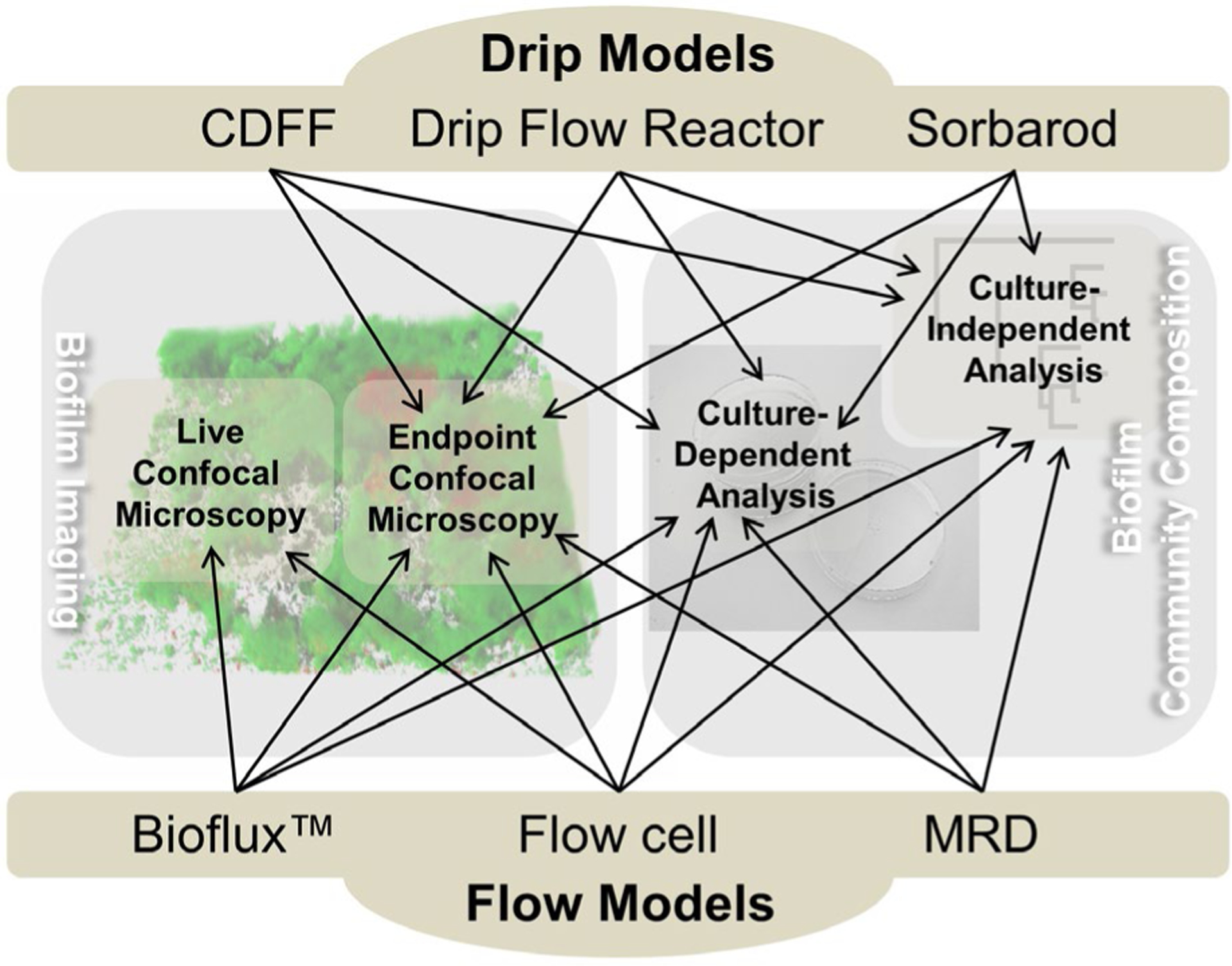
Diagram highlighting the integrative potential of different types of drip models and flow models described in this review. These models have been, or could conceptually be, integrated with a confocal microscope to image biofilms in 3D at the end of an experiment (‘Endpoint Confocal Microscopy’) and/or image repeatedly in 3D over time during an experiment for spatiotemporal analyses (‘Live Confocal Microscopy’). Both confocal microscopy approaches have the potential for the spatial analysis of single or multiple species. Some of these systems have or could also conceivably be combined with culture-dependent approaches and/or culture-independent techniques to study the community composition of oral biofilms.
TABLE 2.
Examples of biofilm model systems that have been used for the study of oral biofilms. General properties of each system are described, along with each system’s nutrient delivery classification, number of biofilms that can be grown per model and volumetric scale.
| Biofilm model | Classification | General properties | Number of biofilms grown per modela | Volumetric range |
|---|---|---|---|---|
| Constant depth film fermenter (Peters & Wimpenny, 1988) | Drip-fed |
|
25 (Peters & Wimpenny, 1988), 75 (Deng et al., 2005) | Litres |
| Sorbarod perfusion system (Hodgson et al., 1995) | Drip-fed |
|
1 (Hodgson et al., 1995), 5 (McBain et al., 2005) | Litres to millilitres |
| Drip-flow biofilm reactor (Xu et al., 1998) | Drip-fed |
|
4–6b | Litres to millilitres |
| Modified Robbins device (McCoy et al., 1981) | Flow-fed |
|
12, 25b,c | Litres |
| Flowcells (Palmer, 1999) | Flow-fed |
|
1–4b,d | Litres to millilitres |
| Bioflux™ (Benoit et al., 2010) | Flow-fed |
|
3,8,24e | Millilitres to microliters |
Refers to the number of biofilms that can be grown for sampling per device and these can be in the same channel/vessel (constant depth film fermenter, modified Robbins device or sorbarod perfusion system) or spread across multiple channels/vessels in one device (drip–flow biofilm reactor, flowcells and Bioflux™ system).
Commercially available through Biosurfaces Technologies Corporation.
Commercially available through Tyler Research Corporation
Commercially available through Stovall Life Science, Inc.
Commercially available through Fluxion Biosciences.
ADVANCEMENTS IN IN VITRO MODEL SYSTEMS FOR ORAL BIOFILM RESEARCH
Over the years, in vitro biofilm models, including drip-fed and flow-fed model systems, have been modified to better reflect the characteristics of the oral environment. One particularly important modification replaced traditional bacteriologic culture medium with either artificial saliva such as ‘McBain medium’, variations of ‘SHI medium’ (Lamont et al., 2021; McBain et al., 2005; Tian et al., 2010), other artificial saliva types such as those highlighted by Pratten et al. (1998) or human saliva (Palmer et al., 2001; Yaari & Bibby, 1976). Biofilms grown in artificial saliva or pooled human saliva will likely better represent in vivo plaque as the bacterial composition is influenced by selective pressure of the physical–chemical properties and nutrients of human saliva, rather than artificial media. Indeed, over 10 years of research published by the Kolenbrander group using in vitro oral biofilm models has highlighted the utility of using pooled 25% human saliva as a growth medium to study complex interactions between oral bacteria in biofilms (Kolenbrander, 2011).
In addition to the relevance of growth medium composition, growth of biofilms under different shear is important for simulating salivary or gingival crevicular flow (Blanc et al., 2014; Fernandez et al., 2017). The composition of exhaled breath can also be mimicked by delivering a gas mixture consisting of 95% atmospheric air and 5% carbon dioxide (Dibdin et al., 1976). Lastly, the choice of a substratum that represents human enamel or dentin should be considered. Hydroxyapatite and glass are two surfaces commonly used to represent oral hard surfaces. While glass may seem to be less relevant than hydroxyapatite for oral biofilm studies, a study comparing the differences of S. sanguinis biofilm growth on both surfaces, on which an acquired pellicle (i.e. conditioning film) had also formed, found no difference in resultant biofilm development (Elliott et al., 2005). The authors concluded that the generation of a conditioning film reduced the influence of differences in substratum surface properties. Indeed, many papers have described the use of saliva (artificial or pooled human saliva) to ‘condition’ glass surfaces to generate an acquired pellicle to enhance bacterial adhesion for subsequent biofilm studies (Foster & Kolenbrander, 2004; Tsutsumi et al., 2016). With the development of in vitro biofilms that are increasingly representative of biofilms in the oral cavity, investigators will gain a better platform to observe the role oral biofilm plays in disease.
Once an in vitro model system has been validated and optimized for a dental biofilm study, the cost to maintain the system and serially perform multiple runs decreases significantly. Compared to in vivo based research (Martin-Kerry et al., 2015), proof of concept and testing for efficacy of new anti-biofilm agents through in vitro model systems will likely be time- and cost-effective. Another advantage of using in vitro oral biofilm models is that oral biofilm communities can be relatively easily developed. In vitro systems can be extremely versatile: nutrient availability, flow, the introduction of defined species and time can be strategically controlled to help answer specific research questions regarding biofilm architecture, cellular organization and mechanisms associated with biofilm growth (Roder et al., 2020).
DRIP-FED BIOFILM MODELS
Constant depth film fermenter
The CDFF was first described by Peters and Wimpenny (1988) as a means to develop freshwater biofilms at a defined thickness. The reason for maintaining biofilms at a constant depth is to achieve a steady-state biofilm within a reactor where measurable properties do not change significantly over time (Kinniment et al., 1996). Mechanically, the CDFF is a chamber housing a rotating turntable on the bottom (for a graphical representation, see McBain, 2009). The rotating turntable holds customizable sampling pans where each pan contains plugs made of a material on which biofilms develop. To distribute media to each plug, media is drip-fed from above via inlets as the disc rotates. Spent media is collected in a waste outlet located below the disc. The CDFF keeps biofilms at a constant depth using a scraper blade that removes excess biofilm biomass and spent media as the disc rotates. The initial model described by Peters and Wimpenny held 25 plugs to support biofilm development (Peters & Wimpenny, 1988), while later models had the capacity of up to 75 plugs (Deng et al., 2005).
While initially used to study freshwater biofilms (Peters & Wimpenny, 1988), the CDFF has been applied successfully to the development of in vitro oral biofilms (Hope et al., 2012; McBain, 2009). The CDFF has been used extensively for single species (Metcalf et al., 2006; Zanin et al., 2005), defined consortia (Fan et al., 2012) and oral microcosm studies (Abdulkareem et al., 2015; Hope et al., 2002; McBain et al., 2003). CDFFs are particularly well-equipped to conduct studies of antimicrobial challenges on mature oral biofilms and for monitoring the growth of biofilms. Biofilm can be grown on the plugs in the same chamber and assigned to treatment or control groups during or post-growth. Specifically, plugs can be removed from the device and then treated (Hope et al., 2002) or treatment(s) can occur while the plugs are within the device (Deng et al., 2005). For example, Deng and colleagues grew S. mutans on dentin plugs in a split CDFF chamber that was simultaneously treated with sodium fluoride or sodium fluoride/ chlorhexidine formulations after the biofilm had matured (Deng et al., 2005). Sodium fluoride/chlorhexidine formulations conferred the greatest kill, lactic acid reduction and remineralization of dentin compared to sodium fluoride alone. In another study, Feldman and coworkers monitored dual-species C. albicans and S. mutans biofilm development on pre-treated hydroxyapatite discs (Feldman et al., 2017). The discs were coated with a membrane designed to slowly release thiazolidinedione-8, a quorum sensing quencher. Biofilm development was hindered on discs containing the quorum sensing quencher. When considering these and other papers using the CDFF, it has been, and still is, a valued in vitro model system to study oral biofilms.
Sorbarod perfusion system
In the mid-1990s, Hodgson and colleagues developed a perfused in vitro model system that was called the Sorbarod perfusion system (also referred to as a Sorbarod biofilm fermenter system) (Hodgson et al., 1995). There are multiple structural variations of this system that have been published, but all use Sorbarod filters as the material on which biofilms develop. Sorbarod filters are cylinders that contain a roll of cellulose fibres and the cylinders are approximately 10mm in diameter and 20 mm in length (Budhani & Struthers, 1997; McBain, 2009). Sorbarods can be loaded into supports such as tubing (Hodgson et al., 1995), syringes (Rickard et al., 2008) or an engineered device that can support multiple Sorbarods (McBain et al., 2005), and exposed to media. Harvested Sorbarods can be used to perform viable counts and biofilms on the Sorbarod fibres can be imaged. Another benefit of this model system is the high surface area to volume ratio, which maximizes the amount of biofilm that can form. During an experiment, gas or fluid can be collected to track cell numbers, volatile sulphur compounds and cell-signalling molecules (Hodgson et al., 1995; Rickard et al., 2008; Spencer et al., 2007).
A Sorbarod perfusion system can be used for anaerobic and microcosm biofilm studies which require extended run times to achieve dynamic steady states (McBain, 2009). In a study by McBain et al. (2005), multiple Sorbarod devices were inoculated with saliva from human volunteers and supplied with artificial saliva nutrient. Dynamic stability was achieved after 2–3days, with high bacterial diversity and presence of anaerobic species. McBain et al. (2005) concluded that the Sorbarod system was effective at maintaining a stable and reproducible oral biofilm community over multiple days. In an oral malodor study by Spencer et al. (2007), a microcosm derived from dorsal tongue scraping was used as inoculum to grow representative communities that produce volatile sulphur compounds. Biofilm development was studied over 96 h and quasi-steady states were achieved by 48 h. The community composition of developed biofilms resembled that of the original dorsal tongue scrapings. Overall, Spencer and colleagues demonstrated the viability of the Sorbarod system for maintaining a stable tongue microcosm community.
Drip–flow biofilm reactor
The drip–flow biofilm reactor was first described by Xu et al. (1998) in the late 1990s as a means to develop P. aeruginosa biofilms. Unlike the CDFF and Sorbarod systems, the drip–flow biofilm reactor is unique in that it is positioned at an angle and media is dripped from above at the apex of the reactor. During use, the media flows downward, coating a glass microscope slide or a detachable coupon. The coupon can be made from various materials, allowing investigators the flexibility of choosing a substratum on which a biofilm can develop (Gomes et al., 2018). The gravity-assisted flow of media creates a low shear environment that can be adjusted by elevating or depressing the angle of the system. At the bottom of the reactor is an outlet where effluent media traverses into a waste receptacle. An excellent review with informative diagrams and detailed descriptions of the use of drip–flow biofilm reactors is presented by Goeres et al. (2009). When considering analysis of biofilms developed in the system, care must be applied in sampling biofilms over a large surface area whether it be imaging or harvesting biomass for further testing. As demonstrated by Xu et al. (1998), oxygen availability can influence heterogeneity of P. aeruginosa biofilms and if media flow across the slide is not uniform, then the development of a heterogeneous biofilm is possible.
Several studies have used the drip–flow reactor to model single-species and multi-species oral biofilms. For example, two single-species studies used the drip–flow reactor to test the efficacy of antimicrobial agents on S. mutans biofilm development (Brambilla et al., 2017; Williams et al., 2017). Williams and colleagues used silver loaded into polymethyl methacrylate (PMMA) sheets which were cut into rectangular coupons; Brambilla and colleagues used chlorhexidine loaded into dentin bonding systems. Williams and colleagues demonstrated that silver PMMA coupons were able to resist S. mutans biofilm formation in short-term washouts but not long-term washouts. As described by Brambilla and colleagues, chlorhexidine-loaded dentin adhesion bonding agents demonstrated variable results, leading authors to suspect the variable chemical composition of the dentin binding systems masked the effects of chlorhexidine. Drip–flow reactors have also been used for dentifrice studies on mature oral multi-species microcosm biofilms (Ledder & McBain, 2012; Ledder et al., 2010). In those studies, oral microcosm biofilms were grown over 24 or 48 h, followed by treatment regimens delivering dentifrice slurries every 6 h for 6 days. The dentifrice treatments reduced culture counts and affected oral biofilm community alpha diversity.
FLOW-FED BIOFILM MODELS
Modified Robbins device
Based on an earlier design called the Robbins device, the MRD (McCoy et al., 1981) facilitates the study of biofilms under flow. The MRD uses individual coupons affixed to plugs that then can be inserted into ports that run along the length of a device. The coupons can be made of different materials such as those used in dental prostheses or hydroxyapatite (Blanc et al., 2014). A peristaltic pump provides unidirectional media flow across all ports after coupons are inoculated. Biofilm development occurs on the surfaces of the coupons as the system runs. Plugs containing coupons can be removed aseptically over time and replaced with plugs containing fresh coupons. The number of sampling ports of the MRD varies by design. For example, commercially available low pressure and small volume MRDs are available that range from 12 to 25 ports. Thus, longitudinal studies of biofilms can be performed, although, as with the CDFF, Sorbarod system, and the drip–flow biofilm reactor, it is not possible to perform repeated in situ biofilm visualizations of the same biofilm sample over time and only endpoint imaging can be performed (Figure 1). Coupons with the supporting plug and associated biofilm must be removed to be visualized microscopically.
The MRD has been used extensively to study oral biofilms, with many studies demonstrating its reproducibility at developing oral biofilms (Blanc et al., 2014; Coenye et al., 2008; Honraet & Nelis, 2006; Noiri et al., 2008; Sliepen et al., 2010; Yassin et al., 2016). The system and its detachable coupons proved to be particularly useful in evaluating the efficacy of antimicrobials and materials primed with antimicrobial. For example, in the study by Yassin et al. (2016), MRD coupons were prepared from a mixture of PMMA and sodium fluoride to create a copolymer that can be used for dentures while also releasing fluoride ions passively while worn. The investigators observed that three-species (C. albicans, L. casei and S. mutans) biofilm growth was inhibited by 10-fold on coupons containing the fluoride compared to biofilm growth on coupons that did not. Conversely, biofilm can be treated after biofilm development to evaluate effectiveness of an antimicrobial (Coenye et al., 2008). Coenye et al. (2008) grew mono-species biofilms of C. albicans, S. mutans, S. aureus and P. aeruginosa in a stainless steel MRD. After growth, the biofilms were treated with NitrAdine™, sonicated to remove biofilm from the coupons and plated to determine efficacy of treatment in preventing regrowth. Similarly, Blanc et al. developed multi-species biofilms on hydroxyapatite coupons to test antimicrobial efficacy of chlorhexidine, cetylpyridinium chloride (CPC) and sodium fluoride mouthwash rinses (Blanc et al., 2014).
Flowcells
Of the six model systems described in this review, flowcells are among the smallest in physical size (Table 2). Due to the compactness of the system, flowcells use small volumes of inocula and media for biofilm experiments. Oral biofilms can be studied at the end of an experiment using a confocal microscope (endpoint studies, for example by Foster et al., 2004) or at different times, for example during treatment with antimicrobials (Corbin et al., 2011) (Figure 1). An example of a flowcell system built in-house for oral biofilm studies was described by Palmer and Caldwell (1995) in the mid-1990s. The main advantage of using flowcells to study oral biofilms is the capability of studying changes to biofilm community composition and architecture over time (Figure 1). For imaging, this can be accomplished because the substratum of the flowcell is often glass. Using confocal or even epifluorescence microscopy (for less-detailed studies), the accumulation of biofilm biomass can be monitored at different times following inoculation.
The flowcell has played a prominent role in oral biofilm research. For example, in 2004, Foster and colleagues used flowcells to test the efficacy of antimicrobials on oral biofilms. The authors grew single species S. gordonii biofilms in saliva-conditioned flowcells and treated them with commercially available mouthwashes (Foster et al., 2004). The study indicated that different active ingredients within mouthwashes differed in antimicrobial efficacy. Later, Foster and Kolenbrander (2004) used the same type of saliva-conditioned flowcells for consortia biofilms containing four oral species and showed that biofilm formation can depend on whether the micro-organisms form coaggregates with each other in the planktonic phase. The flowcell has also been used in studies to test pellicle formation on glass compared to hydroxyapatite. Elliott et al. (2005) showed that the two surfaces were similar and had no effect on biofilm attachment. Another study used flowcells to image in real-time biofilm development of the oral pathogen Candida albicans (McCall & Edgerton, 2017). McCall and Edgerton compared wild-type and hyperfilamentous Δhog1 C. albicans strains in their ability to attach to the flowcells and develop biomass during the 18-h growth. The gene hog1 is activated by oxidative stress, osmotic stress and heavy metal stress resulting in hyphal filamentation (Su et al., 2013). McCall and Edgerton demonstrated that the wild-type C. albicans had twice the attachment rate of the Δhog1 mutant, but formed biofilms of lesser biomass, suggesting that cellular detachment is integral for biomass accumulation.
Microfluidic model systems
Microfluidics involves the engineered delivery of fluids on the sub-millilitre levels through microchannels (Sackmann et al., 2014). A significant advantage of in vitro microfluidics systems over other in vitro model biofilm systems is the much smaller amounts of inoculum that are needed (Samarian et al., 2014). This is especially advantageous if sample volume is limited or reagents are expensive. Additionally, the systems are compact and require low energy costs to run. Microfluidic biofilm model systems have become increasingly popular in oral biofilm studies as they can be used to perform culturing, bioinformatics and microscopy (Gashti et al., 2016; Mira, 2018).
One commercially available microfluidic system is the Bioflux™ system, manufactured by Fluxion Biosciences. The Bioflux™ is a continuous flow microfluidic system that has been used by investigators to model oral biofilms (Ding et al., 2014; Tao et al., 2011; Volgenant et al., 2016). The system consists of three main parts: consumable microfluidic plates, a controller and a software control interface (Samarian et al., 2014). The software control interface regulates the flow rate, the total runtime and determines which pumps are active. A pressure top that is fixed to the top of the consumable plates creates an airtight environment within the Bioflux™ plate, allowing pressure to be applied only from the controller. This forces fluid from inlet well to output well at a fixed rate. A viewing port exists between the inlet and outlet wells, where biofilms develop under the prescribed flow rate.
Of all the systems described in this review, the Bioflux™ requires the least amount of media and inocula. Oral biofilms have been developed overnight at 0.2 dynes/cm2, requiring 380 μl of media per sample and as little as 50 μl of inoculum. Volumes required were calculated from the Bioflux™ software interface. The low volumes required are especially advantageous for studies using donations of bodily fluid for media and/or inoculum. Another advantage of the Bioflux™ system is its throughput. With evenly distributed flow supplied by a computerized pneumatic pump and a heating plate that covers the base of the plate, multiple biofilms can be produced in parallel under the same environmental parameters. Additionally, the atmospheric composition of the airtight environment within the Bioflux™ can be controlled by fitting a Bioflux™ controller with a pressurized gas cylinder containing a defined gaseous mixture. Different plate formats contain 3, 8 or 24 channels which enable replicates of oral biofilms to be developed in parallel. Given the dimensions of the Bioflux™ plates, which are compatible with microplate holders, both endpoint and live imaging of oral biofilm development are possible (Figure 1).
First described in 2010, Benoit et al. (2010) used the throughput advantage of the Bioflux™ system to screen the effectiveness of several antimicrobials on P. aeruginosa PAO1 biofilms. Over the last decade, the Bioflux™ system has been adapted for oral biofilm architecture and community studies (Ding et al., 2010; Dong et al., 2012; Fernandez et al., 2017; Samarian et al., 2014). Nance et al. (2013) developed overnight microcosm biofilms seeded from salivary inoculum and tested the antimicrobial effectiveness of CPC. Using LIVE/DEAD™ staining, a dose–response viability gradient was observed between 0.001% and 0.5% w/v CPC. Also, in the study, Nance and coworkers established that the Bioflux™ system was capable of developing an oral biofilm that was compositionally similar to early supragingival plaque. A standardized protocol for developing oral multi-species biofilms using the Bioflux™ system was described by Samarian et al. (2014). The Bioflux™ system also has been used to study the effects of different antimicrobial compounds on oral biofilms. For example, Luo et al. (2019) evaluated the effect of stannous fluoride on oral multi-species biofilm architecture. Lastly, the Bioflux™ system has been used in single-species studies. Ding and coworkers grew single-species S. mutans biofilms with flowing media and tested the antimicrobial peptide bactenecin (Ding et al., 2014). The authors observed a significant decrease in viability. In another study using the Bioflux™, Dong et al. (2012) showed that development of S. mutans biofilms in subminimum inhibitory concentrations of chlorhexidine or sodium fluoride altered the biofilm architecture, and development in subminimum inhibitory concentrations of tea polyphenols reduced biofilm biomass.
INTEGRATION OF IN VITRO ORAL MODEL SYSTEMS WITH MICROSCOPY AND BIOINFORMATICS
Since the first biofilm model systems were described in the mid-1900s, innovations in methodologies have enhanced the generalizability of oral biofilms grown in vitro. Today, investigators can cultivate an in vitro oral biofilm that is compositionally similar to the microbial community of plaque (Nance et al., 2013; Rudney et al., 2012). The ability to generate representative communities is critical if the desired outcome is to generalize results to human subjects. Two disciplines where technological advancements have significantly augmented the value of laboratory model systems are microscopy and bioinformatics, particularly in the domain of 16S rRNA bacterial community profiling. Microscopy is essential for the study of biofilm architecture, whereas bioinformatics techniques are becoming increasingly popular for characterizing the taxonomic diversity and function of microbial biofilm communities as a whole.
Confocal microscopy
Several different microscope technologies are available to study in vitro oral biofilms each with advantages and disadvantages. While not the focus of this review, a useful review of microscopy and image analysis has been published by McNamara et al. (2017). Here, we will focus on the use of the confocal microscope, which was first used to describe biofilms in 1991 (Lawrence et al., 1991). Using a confocal microscope, investigators can capture oral biofilm architecture and simultaneously gain insight into cell viability or species location (Cuadra-Saenz et al., 2012; Ruangcharoen et al., 2017; Zaura-Arite et al., 2001). Instead of destructively removing oral biofilm for downstream quantification, confocal microscopy enables in situ quantification by taking optical sections of a biofilm and subsequently generating 3D renderings using the optical sections. This can be performed for single-species biofilms, a defined multi-species consortium, or complex microcosm biofilms. For example, instead of culturing and harvesting biofilm to determine colony forming units (CFU), a confocal microscope can take digital snapshots of a biofilm stained with viability stains (e.g. a mixture of SYTO-9 stain and propidium iodide stain, which are part of the commercially available LIVE/DEAD™ staining system). In this scenario, the amount of viable (membrane intact) and inactive/dead (membrane compromised) cells or biofilm biomass can be quantified while the biofilm remains attached to the substratum. This approach has advantages because determining CFUs may underestimate true viability due to the destructive nature of the biofilm harvesting process and/or inadequate cell removal from the surface. However, it should be noted that the use of viability stains is not without potential problems, which include possible issues with differential staining (Netuschil et al., 2014).
A key advantage of confocal microscopy over other forms of microscopy is the ability to discern complex biofilm architecture, the properties of the contained cells and spatial arrangement of biofilm species. In non-targeted (i.e. non species-specific) fluorescence studies, confocal microscopy has been used to identify distribution of viable and non-viable cells in multi-species oral microcosm biofilms developed within a CDFF (Hope et al., 2002). Using LIVE/DEAD staining, Hope and colleagues demonstrated that the basal layer of an untreated oral multi-species biofilm contained more non-viable cells compared to the surface. In targeted (i.e. species-specific) fluorescence studies, the spatial position of a specific species within a multi-species biofilm can be determined (Palmer et al., 2001; Thurnheer et al., 2019). For example, Palmer and coworkers used fluorescently labelled antibodies to discern the spatial arrangement of oral Streptococcus gordonii, Streptococcus oralis and Actinomyces oris in single-species and dual-species biofilms developed in pooled human saliva (Palmer et al., 2001). These biofilms were grown in flowcells where the only potential perturbation to the biofilms was from labelling with antibodies after growth.Another notable study using an in vitro model system and confocal microscopy was performed by Thurnheer et al. (2019), who grew biofilms containing six species on hydroxyapatite disks in 24-well polystyrene cell culture plates, and used fluorescent in situ hybridization (FISH) to discern their spatial arrangement. This work showed that FISH, in combination with the optical sectioning capabilities of a confocal microscope, enabled the analysis of spatial arrangement of numerous species and had the potential to investigate alterations in biofilm species arrangement in response to environmental challenges. Understanding these biofilm structures and cellular arrangements could be important to biofilm control. Thus, considerable effort has been dedicated to identify a disease-associated motif seen in biofilm architecture and its possible role in pathogenesis. With this in mind, a recent paper by Kim and colleagues identified corona-like biofilm architectures formed when S. mutans developed biofilms with other oral species, and these architectures could enhance the pathogenic potential of S. mutans in biofilm communities (Kim et al., 2020).
With modification, certain in vitro model systems can be adapted to monitor changes in biofilm architecture over time (Figure 1). To image a developing biofilm over time, the model system must be capable of growing an oral biofilm on a surface that can be simultaneously imaged with microscopy techniques as the system is running. Indeed, a recent study by Paula and coworkers explored the dynamics of S. mutans biofilm formation from micro-colonies to biofilm superstructures (Paula et al., 2020). Using a modified flowcell that can house hydroxyapatite discs containing attached S. mutans, biofilm development was monitored with a confocal microscope taking images every 20 min.
To maximize information derived from imaged in vitro biofilms, the application of appropriate downstream computational analytics is required to describe the arrangement of fluorescently labelled biofilm species. Many analytical software packages are publicly available and offer a multitude of outcome measures. Alternatively, customized in-house analysis can be performed. A computing environment such as MATLAB is necessary for the latter alternative and its successful implementation is described in more detail by Beyenal et al. (2004). Furthermore, the commonly used biofilm image analysis program COMSTAT, which was originally coded in MATLAB (Heydorn et al., 2000), provides users a graphical user interface to analyse confocal data. A more recent analytical tool built using the MATLAB environment is the Biofilm Architecture Inference Tool (BAIT), developed by Luo and colleagues (Luo et al., 2019). BAIT can import confocal image stack data and perform various image thresholding algorithms prior to image analysis. One method, named the biovolume elasticity method, identifies thresholds that more accurately define biofilm edges (Luo et al., 2018). Post-processed image stacks can then be quantified for various architectural descriptors including: biovolume, surface area, fluffiness, total number of objects, connectivity and convex hull porosity. Viability can also be evaluated if the confocal stack possesses two channels. For combining optical sections collected by confocal microscopy and the subsequent image rendering of biofilms, commercially available software such as Imaris (Zurich, Switzerland) and Volocity (Puslinch, Ontario) can be used to give further insight into architectural features of oral biofilms. Open-source software imaging program software such as ICY (de Chaumont et al., 2012) and BioimageXD (Kankaanpaa et al., 2012) are also available to render biofilms from confocal image stacks.
16S rRNA community profiling
Since its inception in the 1970s, 16S rRNA gene sequencing technology has become extremely useful in studying bacterial phylogeny and taxonomy (Konstantinidis & Tiedje, 2007; Weisburg et al., 1991; Woese & Fox, 1977). Given that all bacteria possess and require the 16S rRNA gene, it is an excellent target for identifying and analysing community membership (Aas et al., 2005; Clarridge, 2004; Petti et al., 2005). Furthermore, 16S rRNA sequences from bacterial species are readily available on public and curated repositories such as GenBank, Greengenes, RDP and SILVA for comparative sequence analyses (Balvociute & Huson, 2017; Benson et al., 2018). Depending on the length of the 16S rRNA gene sequence that is analysed and the variable regions covered, for which there are nine ‘hypervariable regions’ (labelled V1–V9) in the 16S rRNA gene, identities can be assigned to a taxonomic rank often to the genus or species level (Chakravorty et al., 2007; Janda & Abbott, 2007). With more hypervariable regions sequenced within a read, a higher-resolution taxonomic assignment can be achieved. Prior to the advent of NGS, investigators relied upon culture-dependent techniques, such as culturing on agar to isolate bacteria for identification, or older culture-independent (molecular) technologies (e.g. Sanger sequencing of cloned 16S rRNA gene sequences or denaturing gradient gel electrophoresis) that produced relatively low read counts of 16S rRNA sequences and/or limited species resolution for in vitro oral microcosm biofilm studies (Figure 1). With NGS, massively parallel and deep sequencing capabilities have emerged, enabling the oral microbiome to be quickly characterized (Behjati & Tarpey, 2013).
The development of NGS and recent endeavours to study complex in vitro oral biofilm communities has coincided with a shift in the focus on the pathogenicity of natural oral biofilms; from individual species associated with disease to understanding the disease-causing ability of microbial communities (Li et al., 2016; Lamont et al., 2018). Substantial evidence indicates that multiple species, and their interactions with the host and one another, are involved in pathways for soft and hard tissue destruction seen in periodontal disease and caries (Negrini et al., 2019; Wade, 2013). For instance, Whitmore and Lamont reviewed the role mitis group streptococci play in the recruitment of successional pathogenic species such as Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans (Whitmore & Lamont, 2011). Another review by Banas and Drake (2018) discussed the perspective shift away from S. mutans being the lone causative agent to caries, but rather a relative contributor within a complex oral microbiome. Thus, the present challenge is to identify microbial community profiles most associated with disease.
Pertinent to this review, the incorporation of NGS approaches with biofilm model systems is relatively new and there are a variety of factors and challenges that must be considered when considering NGS studies of in vitro biofilm model systems. Critically, there have been numerous NGS platforms used for the 16S rRNA profiling of biofilm communities. Choice of sequencing platform depends on the investigator’s research questions and involves trade-offs between read length, read depth, sequencing depth and accuracy. Sequencing platforms relevant to oral biofilm studies are listed in Table 3, although this is not an exhaustive list since NGS technologies that offer insufficient or unnecessary read length (e.g. 20 Kb read lengths offered by PacBio) for 16S rRNA gene sequencing are excluded. The choice of sequencing platform heavily influences which hypervariable regions can be included in one contiguous read. The longer the read length, the more hypervariable regions can be included. Some platforms offer paired-end reads (Table 3), which can be joined to create a longer fragment, but a trade-off between read length and sequence overlap for accuracy must be considered. Hypervariable region selection can also influence interpretation of results and taxonomic resolution (Barb et al., 2016; Bukin et al., 2019; Teng et al., 2018). This consideration is accentuated for oral streptococci where species are difficult to differentiate due to the limited amount of variation in the hypervariable regions of the 16S gene (Mukherjee et al., 2018).
TABLE 3.
Sequencing platforms for 16S rRNA community profiling. Compatible next-generation sequencers that have been used to characterize an oral microcosm biofilm grown in vitro are listed. The sequencing chemistry, expected read length, sequencing depth and consensus accuracy of each platform are also described.
| Sequencing platform (Reference) | Sequencing chemistry | Read length | Sequencing depth | Consensus accuracy |
|---|---|---|---|---|
| 454 GS FLX+a (Kistler et al., 2015; Koopman et al., 2015; Nance et al., 2013) | Pyrosequencing | Up to 1000 bp | 700 Mb | 99.997 |
| Illumina MiSeq (Agnello et al., 2017; Koopman et al., 2016) | Sequencing by synthesis | 2 × 150 2 × 250 2 × 300 |
4.5–5.1 Gb 7.5–8.5 Gb 13.2–15 Gb |
80% bases >99.9 75% bases >99.9 70% bases >99.9 |
| Illumina HiSeq (Edlund et al., 2013) | Sequencing by synthesis | 2 × 125 | 450–500 Gb | 80% bases >99.9 |
| IonTorrent PGM (Fernandez et al., 2017) | Ion semiconductor | Up to 400 bp | Up to 2 Gb | >99.0 |
Technology is no longer supported by manufacture.
When considering the collection of biofilm material to analyse the community composition of an oral biofilm grown in vitro, investigators must first harvest and prepare biofilm cells from their model system to be analysed with NGS technologies. This process will vary by model system and may involve using physical treatments to harvest biofilm cells. For example, in the Bioflux™ system, this involves removing biofilm material from substratum with high shear (Samarian et al., 2014). With the MRD, sonication could be used to remove biofilm cells from coupons (Coenye et al., 2008). Unlike cell culturing techniques, the destructive nature of removing biofilm is less of a concern for 16S rRNA community profiling. Ultimately, the objective is to retrieve a cross-sectional snapshot of the oral biofilm community composition at the time of harvesting.
Several oral biofilm studies have utilized NGS technologies to characterize the microbial community within biofilms that were developed using in vitro model systems. Velsco and Shaddox described a static system where they collected plaque samples from healthy and periodontitis-affected individuals (Velsko & Shaddox, 2018). Plaque samples were used to inoculate hydroxyapatite discs and grown statically over eight days. The resultant communities were sequenced with Illumina MiSeq and characterized with the software QIIME (Caporaso et al., 2010; Velsko & Shaddox, 2018). They concluded that periodontitis-derived plaque resulted in communities that differed from communities derived from healthy individuals’ plaque samples, as determined by weighted UniFrac measures. In another study, Klug et al. (2016) used 454 pyrosequencing to determine community diversity and survivorship after enamel–dentin slabs worn by volunteers were removed and placed in biofilm reactors. They discovered general survivorship of the biofilm community and diversity was maintained from after removal to 48 h after growth in the biofilm reactor. Fernandez and colleagues studied the effect of shear force on oral communities derived from saliva, tongue and plaque-based inoculum (Fernandez et al., 2017). After harvesting biofilm communities grown in a microfluidics in vitro model system, the samples were sequenced with Ion Torrent sequencing platform. The group discovered that, after overnight growth, bacterial communities shifted to a community with less alpha diversity compared to its starting inoculum. Taken together, these studies highlight the application of different NGS technologies and demonstrate its relevance in various in vitro oral biofilm model system studies.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
The miniaturization of in vitro platforms operating on the microscale, combined with integration with imaging and ‘omic’ technologies, and a greater understanding of the biology of oral biofilms have reinvigorated the appeal of laboratory biofilm model systems. A PubMed search using the search terms ‘in vivo model system oral biofilm’ and ‘in vitro model system oral biofilm’ indicates that laboratory-based models are more commonly used in the realm of oral biofilm research than animal-based models. This observation has held steady in the last 25 years. This popularization of laboratory-based systems is likely owed to technologies that can be tethered to model systems, such as confocal microscopes (Valm et al., 2012) and 16S community profiling approaches (Azevedo et al., 2009). Combined with decreasing costs, in vitro biofilm model systems have become an appealing option for multi-species oral biofilm studies.
The future directions of in vitro model systems could involve a shift from developing representative dental plaque within the system to transplanting already-developed in vivo plaque into the system. For example, Fernandez et al. (2016) described a cariogenic model using ex situ methods that involve human participants wearing non-invasive oral prostheses housing enamel specimens. In vitro model systems could also incorporate a biological substratum for biofilm development, such as that developed using tissue culture techniques.
There are multiple surfaces in the intraoral cavity including hard and soft palate, tongue, subgingival, buccal and teeth. Glass and hydroxyapatite are representative of the hard surfaces of teeth but are a poor model for attachment and development of subgingival plaque (Cieplik et al., 2019). There is a disparity in volume of research involving epithelial substratum in oral diseases; thus, periodontal biofilm models are lacking (Walker & Sedlacek, 2007). This is due to the relative difficulty of cell culture techniques over use of glass or hydroxyapatite. Epithelial cells are the preferred substratum for periodontal models, as they more adequately represent the substratum of subgingival plaque (Guggenheim et al., 2009). As demonstrated by Guggenheim and colleagues, an epithelial substratum can actively model the interaction between host immune cells and oral microbial biofilm cells. This is important to consider in periodontal models where in vivo microbial cells at the periodontal tissue interface trigger host immune response and then mount evasion or defence mechanisms.
In conclusion, the development and validation of new in vitro biofilm model systems for oral biofilm research is a continual effort, especially with changing paradigms, perspectives and capabilities in microbiological research techniques. The biggest challenge thus far in translating in vitro model system findings into clinical practice has been the difficulty to form in vivo-like biofilms in a laboratory setting. Enhancing older ‘classic’ model systems or creating newer model systems and combining such models with new or improved technologies is allowing investigators to move closer to mimicking natural oral biofilm states and providing tools to measure oral biofilm outcomes more accurately.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
REFERENCES
- Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE et al. (2008) Bacteria of dental caries in primary and permanent teeth in children and young adults. Journal of Clinical Microbiology, 46, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas JA, Paster BJ, Stokes LN, Olsen I & Dewhirst FE (2005) Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology, 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulkareem EH, Memarzadeh K, Allaker RP, Huang J, Pratten J & Spratt D (2015) Anti-biofilm activity of zinc oxide and hydroxyapatite nanoparticles as dental implant coating materials. Journal of Dentistry, 43, 1462–1469. [DOI] [PubMed] [Google Scholar]
- Agnello M, Cen L, Tran NC, Shi W, McLean JS & He X (2017) Arginine improves pH homeostasis via metabolism and microbiome modulation. Journal of Dental Research, 96, 924–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila M, Ojcius DM & Yilmaz O (2009) The oral microbiota: living with a permanent guest. DNA and Cell Biology, 28, 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeredo J, Azevedo NF, Briandet R, Cerca N, Coenye T, Costa AR et al. (2017) Critical review on biofilm methods. Critical Reviews in Microbiology, 43, 313–351. [DOI] [PubMed] [Google Scholar]
- Azevedo NF, Lopes SP, Keevil CW, Pereira MO & Vieira MJ (2009) Time to “go large” on biofilm research: advantages of an omics approach. Biotechnology Letters, 31, 477–485. [DOI] [PubMed] [Google Scholar]
- Balvociute M & Huson DH (2017) SILVA, RDP, Greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genomics, 18, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA & Drake DR (2018) Are the mutans streptococci still considered relevant to understanding the microbial etiology of dental caries? BMC Oral Health, 18, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb JJ, Oler AJ, Kim HS, Chalmers N, Wallen GR, Cashion A et al. (2016) Development of an analysis pipeline characterizing multiple hypervariable regions of 16S rRNA using mock samples. PLoS One, 11, e0148047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S & Tarpey PS (2013) What is next generation sequencing? Archives of Disease in Childhood - Education & Practice Edition, 98, 236–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit MR, Conant CG, Ionescu-Zanetti C, Schwartz M & Matin A (2010) New device for high-throughput viability screening of flow biofilms. Applied and Environment Microbiology, 76, 4136–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Ostell J, Pruitt KD et al. (2018) GenBank. Nucleic Acids Research, 46, D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyenal H, Donovan C, Lewandowski Z & Harkin G (2004) Three-dimensional biofilm structure quantification. Journal of Microbiol Methods, 59, 395–413. [DOI] [PubMed] [Google Scholar]
- Blanc V, Isabal S, Sanchez MC, Llama-Palacios A, Herrera D, Sanz M et al. (2014) Characterization and application of a flow system for in vitro multispecies oral biofilm formation. Journal of Periodontal Research, 49, 323–332. [DOI] [PubMed] [Google Scholar]
- Bos R, van der Mei HC & Busscher HJ (1996) Co-adhesion of oral microbial pairs under flow in the presence of saliva and lactose. Journal of Dental Research, 75, 809–815. [DOI] [PubMed] [Google Scholar]
- Bradshaw DJ, Marsh PD, Watson GK & Allison C (1998) Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infection and Immunity, 66, 4729–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla E, Ionescu AC, Cazzaniga G, Ottobelli M, Mazzonni A, Cadenaro M et al. (2017) In vitro Streptococcus mutans biofilm formation on surfaces of chlorhexidine-containing dentin bonding systems. International Journal of Adhesion and Adhesives, 75, 23–30. [Google Scholar]
- Budhani RK & Struthers JK (1997) The use of Sorbarod biofilms to study the antimicrobial susceptibility of a strain of Streptococcus pneumoniae. Journal of Antimicrobial Chemotherapy, 40, 601–602. [DOI] [PubMed] [Google Scholar]
- Bukin YS, Galachyants YP, Morozov IV, Bukin SV, Zakharenko AS & Zemskaya TI (2019) The effect of 16S rRNA region choice on bacterial community metabarcoding results. Scientific Data, 6, 190007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S, Helb D, Burday M, Connell N & Alland D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of Microbiol Methods, 69, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplik F, Zaura E, Brandt BW, Buijs MJ, Buchalla W, Crielaard W et al. (2019) Microcosm biofilms cultured from different oral niches in periodontitis patients. Journal of Oral Microbiology, 11, 1551596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarridge JE 3rd (2004) Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clinical Microbiology Reviews, 17(4), 840–862. 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, De Prijck K, De Wever B & Nelis HJ (2008) Use of the modified Robbins device to study the in vitro biofilm removal efficacy of NitrAdine, a novel disinfecting formula for the maintenance of oral medical devices. Journal of Applied Microbiology, 105, 733–740. [DOI] [PubMed] [Google Scholar]
- Coenye T & Nelis HJ (2010) In vitro and in vivo model systems to study microbial biofilm formation. Journal of Microbiol Methods, 83, 89–105. [DOI] [PubMed] [Google Scholar]
- Corbin A, Pitts B, Parker A & Stewart PS (2011) Antimicrobial penetration and efficacy in an in vitro oral biofilm model. Antimicrobial Agents and Chemotherapy, 55, 3338–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadra-Saenz G, Rao DL, Underwood AJ, Belapure SA, Campagna SR, Sun Z et al. (2012) Autoinducer-2 influences interactions amongst pioneer colonizing streptococci in oral biofilms. Microbiology, 158, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T et al. (2012) Icy: an open bioimage informatics platform for extended reproducible research. Nature Methods, 9, 690–696. [DOI] [PubMed] [Google Scholar]
- Deng DM & ten Cate JM (2004) Demineralization of dentin by Streptococcus mutans biofilms grown in the constant depth film fermentor. Caries Research, 38, 54–61. [DOI] [PubMed] [Google Scholar]
- Deng DM, van Loveren C & ten Cate JM (2005) Caries-preventive agents induce remineralization of dentin in a biofilm model. Caries Research, 39, 216–223. [DOI] [PubMed] [Google Scholar]
- Dibdin GH, Shellis RP & Wilson CM (1976) An apparatus for the continuous culture of micro-organisms on solid surfaces with special reference to dental plaque. Journal of Applied Bacteriology, 40, 261–268. [DOI] [PubMed] [Google Scholar]
- Dietz VH (1943) In vitro production of plaques and caries. Journal of Dental Research, 22, 423–440. [Google Scholar]
- Ding AM, Palmer RJ Jr, Cisar JO & Kolenbrander PE (2010) Shear-enhanced oral microbial adhesion. Applied and Environment Microbiology, 76, 1294–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang W, Fan M, Tong Z, Kuang R, Jiang W et al. (2014) Antimicrobial and anti-biofilm effect of Bac8c on major bacteria associated with dental caries and Streptococcus mutans biofilms. Peptides, 52, 61–67. [DOI] [PubMed] [Google Scholar]
- Dong L, Tong Z, Linghu D, Lin Y, Tao R, Liu J et al. (2012) Effects of sub-minimum inhibitory concentrations of antimicrobial agents on Streptococcus mutans biofilm formation. International Journal of Antimicrobial Agents, 39, 390–395. [DOI] [PubMed] [Google Scholar]
- Edlund A, Yang Y, Hall AP, Guo L, Lux R, He X et al. (2013) An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome, 1, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D, Pratten J, Edwards M, Crowther J, Petrie A & Wilson M (2005) Bacterial biofilm development on hydroxyapatite-coated glass. Current Microbiology, 51, 41–45. [DOI] [PubMed] [Google Scholar]
- Fan Y, Wen ZT, Liao S, Lallier T, Hagan JL, Twomley JT et al. (2012) Novel amelogenin-releasing hydrogel for remineralization of enamel artificial caries. Journal of Bioactive and Compatable Polymers, 27, 585–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Shenderovich J, Lavy E, Friedman M & Steinberg D (2017) A sustained-release membrane of thiazolidinedione-8: effect on formation of a candida/bacteria mixed biofilm on hydroxyapatite in a continuous flow model. BioMed Research International, 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CE, Aspiras MB, Dodds MW, Gonzalez-Cabezas C & Rickard AH (2017) The effect of inoculum source and fluid shear force on the development of in vitro oral multispecies biofilms. Journal of Applied Microbiology, 122, 796–808. [DOI] [PubMed] [Google Scholar]
- Fernandez CE, Tenuta LM & Cury JA (2016) Validation of a cariogenic biofilm model to evaluate the effect of fluoride on enamel and root dentine demineralization. PLoS One, 11, e0146478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JS & Kolenbrander PE (2004) Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Applied and Environment Microbiology, 70, 4340–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JS, Pan PC & Kolenbrander PE (2004) Effects of antimicrobial agents on oral biofilms in a saliva-conditioned flowcell. Biofilms, 1, 5–12. [Google Scholar]
- Gashti MP, Asselin J, Barbeau J, Boudreau D & Greener J (2016) A microfluidic platform with pH imaging for chemical and hydrodynamic stimulation of intact oral biofilms. Lab on a Chip, 16, 1412–1419. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ & Fitzgerald RJ (1969) Dextran-induced agglutination of Streptococcus mutans, and its potential role in the formation of microbial dental plaques. Journal of Bacteriology, 98, 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres DM, Hamilton MA, Beck NA, Buckingham-Meyer K, Hilyard JD, Loetterle LR et al. (2009) A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nature Protocols, 4, 783–788. [DOI] [PubMed] [Google Scholar]
- Gomes IB, Meireles A, Goncalves AL, Goeres DM, Sjollema J, Simoes LC et al. (2018) Standardized reactors for the study of medical biofilms: a review of the principles and latest modifications. Critical Reviews in Biotechnology, 38, 657–670. [DOI] [PubMed] [Google Scholar]
- Guggenheim B, Gmur R, Galicia JC, Stathopoulou PG, Benakanakere MR, Meier A et al. (2009) In vitro modeling of host-parasite interactions: the ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiology, 9, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP & Curtis MA (2012) The keystone-pathogen hypothesis. Nature Reviews Microbiology, 10, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK et al. (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology, 146(Pt 10), 2395–2407. [DOI] [PubMed] [Google Scholar]
- Hodgson AE, Nelson SM, Brown MR & Gilbert P (1995) A simple in vitro model for growth control of bacterial biofilms. Journal of Applied Bacteriology, 79, 87–93. [DOI] [PubMed] [Google Scholar]
- Hojo K, Nagaoka S, Ohshima T & Maeda N (2009) Bacterial interactions in dental biofilm development. Journal of Dental Research, 88, 982–990. [DOI] [PubMed] [Google Scholar]
- Honraet K & Nelis HJ (2006) Use of the modified Robbins device and fluorescent staining to screen plant extracts for the inhibition of S. mutans biofilm formation. Journal of Microbiol Methods, 64, 217–224. [DOI] [PubMed] [Google Scholar]
- Hope C.k., Bakht K, Burnside G, Martin GC, Burnett G, Josselin de Jong E et al. (2012) Reducing the variability between constant-depth film fermenter experiments when modelling oral biofilm. Journal of Applied Microbiology, 113, 601–608. [DOI] [PubMed] [Google Scholar]
- Hope CK, Clements D & Wilson M (2002) Determining the spatial distribution of viable and nonviable bacteria in hydrated microcosm dental plaques by viability profiling. Journal of Applied Microbiology, 93, 448–455. [DOI] [PubMed] [Google Scholar]
- Janda JM & Abbott SL (2007) 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. Journal of Clinical Microbiology, 45, 2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankaanpaa P, Paavolainen L, Tiitta S, Karjalainen M, Paivarinne J, Nieminen J et al. (2012) BioImageXD: an open, general-purpose and high-throughput image-processing platform. Nature Methods, 9, 683–689. [DOI] [PubMed] [Google Scholar]
- Keyes PH (1968) Research in dental caries. Journal of the American Dental Association, 76, 1357–1373. [DOI] [PubMed] [Google Scholar]
- Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y et al. (2020) Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proceedings of the National Academy of Sciences, 117, 12375–12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinniment SL, Wimpenny JW, Adams D & Marsh PD (1996) Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiology, 142(Pt 3), 631–638. [DOI] [PubMed] [Google Scholar]
- Kistler JO, Pesaro M & Wade WG (2015) Development and pyrosequencing analysis of an in-vitro oral biofilm model. BMC Microbiology, 15, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug B, Santigli E, Westendorf C, Tangl S, Wimmer G & Grube M (2016) From mouth to model: combining in vivo and in vitro oral biofilm growth. Frontiers in Microbiology, 7, 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE (2011) Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. International Journal of Oral Science, 3, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI & Diaz PI (2000) (2006) Bacterial interactions and successions during plaque development. Periodontology 2000, 42, 47–79. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT & Tiedje JM (2007) Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Current Opinion in Microbiology, 10, 504–509. [DOI] [PubMed] [Google Scholar]
- Koopman JE, Buijs MJ, Brandt BW, Keijser BJ, Crielaard W & Zaura E (2016) Nitrate and the origin of saliva influence composition and short chain fatty acid production of oral microcosms. Microbial Ecology, 72, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JE, Roling WF, Buijs MJ, Sissons CH, ten Cate JM, Keijser BJ et al. (2015) Stability and resilience of oral microcosms toward acidification and Candida outgrowth by arginine supplementation. Microbial Ecology, 69, 422–433. [DOI] [PubMed] [Google Scholar]
- Lamont EI, Gadkari A, Kerns KA, To TT, Daubert D, Kotsakis G et al. (2021) Modified SHI medium supports growth of a disease-state subgingival polymicrobial community in vitro. Molecular Oral Microbiology, 36, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Koo H & Hajishengallis G (2018) The oral microbiota: dynamic communities and host interactions. Nature Reviews Microbiology, 16, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JR, Korber DR, Hoyle BD, Costerton JW & Caldwell DE (1991) Optical sectioning of microbial biofilms. Journal of Bacteriology, 173, 6558–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledder RG & McBain AJ (2012) An in vitro comparison of dentifrice formulations in three distinct oral microbiotas. Archives of Oral Biology, 57, 139–147. [DOI] [PubMed] [Google Scholar]
- Ledder RG, Sreenivasan PK, DeVizio W & McBain AJ (2010) Evaluation of the specificity and effectiveness of selected oral hygiene actives in salivary biofilm microcosms. Journal of Medical Microbiology, 59, 1462–1468. [DOI] [PubMed] [Google Scholar]
- Li Y, Zou CG, Fu Y, Li Y, Zhou Q, Liu B et al. (2016) Oral microbial community typing of caries and pigment in primary dentition. BMC Genomics, 17, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listgarten MA (1965) Electron microscopic observations on the bacterial flora of acute necrotizing ulcerative gingivitis. Journal of Periodontology, 36, 328–339. [DOI] [PubMed] [Google Scholar]
- Luo TL, Eisenberg MC, Hayashi MAL, Gonzalez-Cabezas C, Foxman B, Marrs CF et al. (2018) A sensitive thresholding method for confocal laser scanning microscope image stacks of microbial biofilms. Scientific Reports, 8, 13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo TL, Hayashi M, Zsiska M, Circello B, Eisenberg M, Gonzalez-Cabezas C et al. (2019) Introducing BAIT (Biofilm Architecture Inference Tool): a software program to evaluate the architecture of oral multi-species biofilms. Microbiology, 165, 527–537. [DOI] [PubMed] [Google Scholar]
- Marsh PD (2009) Role of the oral microflora in health. Microbial Ecology in Health and Disease, 12, 130–137. [Google Scholar]
- Marsh PD (2010) Controlling the oral biofilm with antimicrobials. Journal of Dentistry, 38(Suppl 1), S11–S15. [DOI] [PubMed] [Google Scholar]
- Marsh PD (2018) In sickness and in health-what does the oral microbiome mean to us? An ecological perspective. Advances in Dental Research, 29, 60–65. [DOI] [PubMed] [Google Scholar]
- Martin-Kerry JM, Lamont TJ, Keightley A, Calache H, Martin R, Floate R et al. (2015) Practical considerations for conducting dental clinical trials in primary care. British Dental Journal, 218, 629–634. [DOI] [PubMed] [Google Scholar]
- McBain AJ (2009) Chapter 4: In vitro biofilm models: an overview. Advances in Applied Microbiology, 69, 99–132. [DOI] [PubMed] [Google Scholar]
- McBain AJ, Bartolo RG, Catrenich CE, Charbonneau D, Ledder RG & Gilbert P (2003) Growth and molecular characterization of dental plaque microcosms. Journal of Applied Microbiology, 94, 655–664. [DOI] [PubMed] [Google Scholar]
- McBain AJ, Sissons C, Ledder RG, Sreenivasan PK, De Vizio W & Gilbert P (2005) Development and characterization of a simple perfused oral microcosm. Journal of Applied Microbiology, 98, 624–634. [DOI] [PubMed] [Google Scholar]
- McCall A & Edgerton M (2017) Real-time approach to flow cell imaging of Candida albicans biofilm development. Journal of Fungi, 3(1), 13– 10.3390/jof3010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy WF, Bryers JD, Robbins J & Costerton JW (1981) Observations of fouling biofilm formation. Canadian Journal of Microbiology, 27, 910–917. [DOI] [PubMed] [Google Scholar]
- McNamara G, Difilippantonio M, Ried T & Bieber FR (2017) Microscopy and Image Analysis. Current Protocols in Human Genetics, 94(1), 1–4. 10.1002/cphg.42. [DOI] [PubMed] [Google Scholar]
- Metcalf D,Robinson C, evine D & Wood S (2006) Enhancement of erythrosine-mediated photodynamic therapy of Streptococcus mutans biofilms by light fractionation. Journal of Antimicrobial Chemotherapy, 58, 190–192. [DOI] [PubMed] [Google Scholar]
- Mira A (2018) Oral microbiome studies: potential diagnostic and therapeutic implications. Advances in Dental Research, 29, 71–77. [DOI] [PubMed] [Google Scholar]
- Mukherjee C, Beall CJ, Griffen AL & Leys EJ (2018) High-resolution ISR amplicon sequencing reveals personalized oral microbiome. Microbiome, 6, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S, Hojo K, Murata S, Mori T, Ohshima T & Maeda N (2008) Interactions between salivary Bifidobacterium adolescentis and other oral bacteria: in vitro coaggregation and coadhesion assays. FEMS Microbiology Letters, 281, 183–189. [DOI] [PubMed] [Google Scholar]
- Nance WC, Dowd SE, Samarian D, Chludzinski J, Delli J, Battista J et al. (2013) A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials. Journal of Antimicrobial Chemotherapy, 68, 2550–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini TC, Koo H & Arthur RA (2019) Candida-bacterial biofilms and host-microbe interactions in oral diseases. Advances in Experimental Medicine and Biology, 1197, 119–141. [DOI] [PubMed] [Google Scholar]
- Netuschil L, Auschill TM, Sculean A & Arweiler NB (2014) Confusion over live/dead stainings for the detection of vital microorganisms in oral biofilms–which stain is suitable? BMC Oral Health, 14, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiri Y, Katsumoto T, Azakami H & Ebisu S (2008) Effects of Er:YAG laser irradiation on biofilm-forming bacteria associated with endodontic pathogens in vitro. Journal of Endodontics, 34, 826–829. [DOI] [PubMed] [Google Scholar]
- Noorda WD, van Montfort AM, Purdell-Lewis DJ & Weerkamp AH (1986) Developmental and metabolic aspects of a monobacterial plaque of Streptococcus mutans C 67–1 grown on human enamel slabs in an artificial mouth model. I. Plaque data. Caries Research, 20, 300–307. [DOI] [PubMed] [Google Scholar]
- Palmer RJ Jr (1999) Microscopy flowcells: perfusion chambers for real-time study of biofilms. Methods in Enzymology, 310, 160–166. [DOI] [PubMed] [Google Scholar]
- Palmer RJ & Caldwell DE (1995) A flowcell for the study of plaque removal and regrowth. Journal of Microbiol Methods, 24, 171–182. [Google Scholar]
- Palmer RJ Jr, Kazmerzak K, Hansen MC & Kolenbrander PE (2001) Mutualism versus independence: strategies of mixedspecies oral biofilms in vitro using saliva as the sole nutrient source. Infection and Immunity, 69, 5794–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paula AJ, Hwang G & Koo H (2020) Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nature Communications, 11, 1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S & Kolenbrander PE (2009) Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infection and Immunity, 77, 3542–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AC & Wimpenny JW (1988) A constant-depth laboratory model film fermentor. Biotechnology and Bioengineering, 32, 263–270. [DOI] [PubMed] [Google Scholar]
- Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S & Ndiaye C (2005) The global burden of oral diseases and risks to oral health. Bulletin of the World Health Organization, 83, 661–669. [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ et al. (2013) The dental plaque microbiome in health and disease. PLoS One, 8, e58487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti CA, Polage CR & Schreckenberger P (2005) The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. Journal of Clinical Microbiology, 43, 6123–6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigman W, Brasher J & Koulourides T (1962) Cariogenicity of common sugars as evaluated in the artificial mouth. Nature, 195, 190–191. [DOI] [PubMed] [Google Scholar]
- Pigman W, Elliott HC Jr & Laffre RO (1952) An artificial mouth for caries research. Journal of Dental Research, 31, 627–633. [DOI] [PubMed] [Google Scholar]
- Pigman W, Hawkins WL, Watson J, Powell R & Gaston C (1955) The effect of the concentration of D-glucose on the attack of tooth substances in the artificial mouth. Journal of Dental Research, 34, 537–545. [DOI] [PubMed] [Google Scholar]
- Pigman W & Newbrun E (1962) Evaluation of anticaries agents by the use of the artificial mouth. Journal of Dental Research, 41, 1304–1311. [DOI] [PubMed] [Google Scholar]
- Pratten J, Wills K, Barnett P & Wilson M (1998) In vitro studies of the effect of antiseptic-containing mouthwashes on the formation and viability of Streptococcus sanguis biofilms. Journal of Applied Microbiology, 84, 1149–1155. [DOI] [PubMed] [Google Scholar]
- Rickard AH, Campagna SR & Kolenbrander PE (2008) Autoinducer-2 is produced in saliva-fed flow conditions relevant to natural oral biofilms. Journal of Applied Microbiology, 105, 2096–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roder HL, Olsen NMC, Whiteley M & Burmolle M (2020) Unravelling interspecies interactions across heterogeneities in complex biofilm communities. Environmental Microbiology, 22, 5–16. [DOI] [PubMed] [Google Scholar]
- Ruangcharoen S, Suwannarong W, Lachica M, Bolscher JGM, Nazmi K, Khunkitti W et al. (2017) Killing activity of LFchimera on periodontopathic bacteria and multispecies oral biofilm formation in vitro. World Journal of Microbiology & Biotechnology, 33, 167. [DOI] [PubMed] [Google Scholar]
- Rudney JD, Chen R, Lenton P, Li J, Li Y, Jones RS et al. (2012) A reproducible oral microcosm biofilm model for testing dental materials. Journal of Applied Microbiology, 113, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C & Coulter WA (1975) Continuous monitoring of pH and Eh in bacterial plaque grown on a tooth in an artificial mouth. Applied Microbiology, 29, 141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C & Coulter WA (1977) Plaque formation by streptococci in an artificial mouth and factors influencing colonization. Journal of Applied Bacteriology, 42, 337–344. [DOI] [PubMed] [Google Scholar]
- Sackmann EK, Fulton AL & Beebe DJ (2014) The present and future role of microfluidics in biomedical research. Nature, 507, 181–189. [DOI] [PubMed] [Google Scholar]
- Samarian DS, Jakubovics NS, Luo TL & Rickard AH (2014) Use of a high-throughput in vitro microfluidic system to develop oral multi-species biofilms. Journal of Visualized Experiments: JoVE, 10.3791/52467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JM, Ateia I, Paulus JR, Liu H, Fenno JC, Rickard AH et al. (2015) Antimicrobial nisin acts against saliva derived multi-species biofilms without cytotoxicity to human oral cells. Frontiers in Microbiology, 6, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissons CH (1997) Artificial dental plaque biofilm model systems. Advances in Dental Research, 11, 110–126. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Cutress TW, Hoffman MP & Wakefield JS (1991) A multi-station dental plaque microcosm (artificial mouth) for the study of plaque growth, metabolism, pH, and mineralization. Journal of Dental Research, 70, 1409–1416. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Cutress TW & Pearce EI (1985) Kinetics and product stoichiometry of ureolysis by human salivary bacteria and artificial mouth plaques. Archives of Oral Biology, 30, 781–790. [DOI] [PubMed] [Google Scholar]
- Sissons CH, Wong L & An YH (2000) Laboratory culture and analysis of microbial biofilms. In: An YH & Friedman RJ (Eds.) Handbook of bacterial adhesion: principles, methods, and applications. Totowa, NJ: Humana Press, pp. 133–169. [Google Scholar]
- Sliepen I, Van Essche M, Quirynen M & Teughels W (2010) Effect of mouthrinses on Aggregatibacter actinomycetemcomitans biofilms in a hydrodynamic model. Clinical Oral Investigations, 14, 241–250. [DOI] [PubMed] [Google Scholar]
- Spencer P, Greenman J, McKenzie C, Gafan G, Spratt D & Flanagan A (2007) In vitro biofilm model for studying tongue flora and malodour. Journal of Applied Microbiology, 103, 985–992. [DOI] [PubMed] [Google Scholar]
- Su C, Lu Y & Liu H (2013) Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Molecular Biology of the Cell, 24, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CH, Lee KW, Burmolle M, Kjelleberg S & Rice SA (2017) All together now: experimental multispecies biofilm model systems. Environmental Microbiology, 19, 42–53. [DOI] [PubMed] [Google Scholar]
- Tang G, Yip HK, Cutress TW & Samaranayake LP (2003) Artificial mouth model systems and their contribution to caries research: a review. Journal of Dentistry, 31, 161–171. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Haffer C, Bratthall GT, Visconti RA & Socransky SS (1979) A study of the bacteria associated with advancing periodontitis in man. Journal of Clinical Periodontology, 6, 278–307. [DOI] [PubMed] [Google Scholar]
- Tao R, Tong Z, Lin Y, Xue Y, Wang W, Kuang R et al. (2011) Antimicrobial and antibiofilm activity of pleurocidin against cariogenic microorganisms. Peptides, 32, 1748–1754. [DOI] [PubMed] [Google Scholar]
- Teng F, Darveekaran Nair SS, Zhu P, Li S, Huang S, Li X et al. (2018) Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Scientific Reports, 8, 16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurnheer T, Karygianni L, Flury M & Belibasakis GN (2019) Fusobacterium species and subspecies differentially affect the composition and architecture of supra- and subgingival biofilms models. Frontiers in Microbiology, 10, 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, He X, Torralba M, Yooseph S, Nelson KE, Lux R et al. (2010) Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Molecular Oral Microbiology, 25, 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi C, Takakuda K & Wakabayashi N (2016) Reduction of Candida biofilm adhesion by incorporation of prereacted glass ionomer filler in denture base resin. Journal of Dentistry, 44, 37–43. [DOI] [PubMed] [Google Scholar]
- Valm AM, Mark Welch JL & Borisy GG (2012) CLASI-FISH: principles of combinatorial labeling and spectral imaging. Systematic and Applied Microbiology, 35, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velsko IM & Shaddox LM (2018) Consistent and reproducible long-term in vitro growth of health and disease-associated oral subgingival biofilms. BMC Microbiology, 18, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volgenant CM, Hoogenkamp MA, Krom BP, Janus MM, Ten Cate JM, de Soet JJ et al. (2016) Red and green fluorescence from oral biofilms. PLoS One, 11, e0168428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, AbdAllah F et al. (2017) Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet, 390(10100), 1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG (2013) The oral microbiome in health and disease. Pharmacological Research, 69 (1), 137–143. 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Walker C & Sedlacek MJ (2007) An in vitro biofilm model of subgingival plaque. Oral Microbiology and Immunology, 22, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA & Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173, 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore SE & Lamont RJ (2011) The pathogenic persona of community-associated oral streptococci. Molecular Microbiology, 81, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DL, Epperson RT, DeGrauw JP, Nielsen MB, Taylor NB & Jolley RD (2017) Effect of silver-loaded PMMA on Streptococcus mutans in a drip flow reactor. Journal of Biomedical Materials Research Part A, 105, 2632–2639. [DOI] [PubMed] [Google Scholar]
- Woese CR & Fox GE (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences USA, 74, 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, Sissons CH & Cutress TW (1994) Control of multiple dental plaque culture system and long-term, continuous, plaque pH measurement using LabVIEW. Binary, 6, 173–180. [Google Scholar]
- Wright TL, Ellen RP, Lacroix JM, Sinnadurai S & Mittelman MW (1997) Effects of metronidazole on Porphyromonas gingivalis biofilms. Journal of Periodontal Research, 32, 473–477. [DOI] [PubMed] [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT & McFeters GA (1998) Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Applied and Environment Microbiology, 64, 4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari A & Bibby BG (1976) Production of plaques and initiation of caries in vitro. Journal of Dental Research, 55, 30–36. [DOI] [PubMed] [Google Scholar]
- Yassin SA, German MJ, Rolland SL, Rickard AH & Jakubovics NS (2016) Inhibition of multispecies biofilms by a fluoride-releasing dental prosthesis copolymer.Journal of Dentistry, 48, 62–70. [DOI] [PubMed] [Google Scholar]
- Yu OY, Zhao IS, Mei ML, Lo EC & Chu CH (2017) Dental biofilm and laboratory microbial culture models for cariology research. Dentistry Journal (Basel), 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin IC, Goncalves RB, Junior AB, Hope CK & Pratten J (2005) Susceptibility of Streptococcus mutans biofilms to photodynamic therapy: an in vitro study. Journal of Antimicrobial Chemotherapy, 56, 324–330. [DOI] [PubMed] [Google Scholar]
- Zaura-Arite E, van Marle J & ten Cate JM (2001) Confocal microscopy study of undisturbed and chlorhexidine-treated dental biofilm. Journal of Dental Research, 80, 1436–1440. [DOI] [PubMed] [Google Scholar]


