Abstract
Plant terrestrialization brought forth the land plants (embryophytes). Embryophytes account for most of the biomass on land and evolved from streptophyte algae in a singular event. Recent advances have unravelled the first full genomes of the closest algal relatives of land plants; among the first such species was Mesotaenium endlicherianum. Here we used fine-combed RNA sequencing in tandem with a photophysiological assessment on Mesotaenium exposed to a continuous range of temperature and light cues. Our data establish a grid of 42 different conditions, resulting in 128 transcriptomes and ~1.5 Tbp (~9.9 billion reads) of data to study the combinatory effects of stress response using clustering along gradients. Mesotaenium shares with land plants major hubs in genetic networks underpinning stress response and acclimation. Our data suggest that lipid droplet formation and plastid and cell wall-derived signals have denominated molecular programmes since more than 600 million years of streptophyte evolution—before plants made their first steps on land.
Subject terms: Plant evolution, Evolution, Transcriptomics
Dadras et al. exposed Mesotaenium, an alga of the sister lineage to land plants, to a bifactorial gradient of environmental cues and use comparative analyses to pinpoint conserved circuits in plant genetic response networks.
Main
Plant terrestrialization changed the face of our planet. It gave rise to land plants (Embryophyta), the major constituents of Earth’s biomass1 and founders of the current levels of atmospheric oxygen2. Land plants belong to the Streptophyta, a monophyletic group that includes the paraphyletic freshwater and terrestrial streptophyte algae and the monophyletic land plants. Meticulous phylogenomic efforts have established the relationships of land plants to their algal relatives3–6. These data brought a surprise: the filamentous and unicellular Zygnematophyceae—and not other morphologically more elaborate algae—are the closest algal relatives of land plants. Now, the first genomes of major orders of Zygnematophyceae (see ref. 7) are at hand: Mesotaenium endlicherianum8, Spirogloea muscicola8, Zygnema circumcarinatum9, Closterium peracerosum–strigosum–littorale10 and Penium margaritaceum11. Using these, we are beginning to redefine the molecular chassis shared by land plants and their closest algal relatives. Included in this shared chassis will be those genes that facilitated plant terrestrialization. In this Article, we focus on one critical aspect: the molecular toolkit for the response to environmental challenges. For this, we used the unicellular freshwater/subaerial alga Mesotaenium endlicherianum.
Land plants use a multi-layered system for the adequate response to environmental cues. This involves sensing, signalling and response, mainly by the production of, for example, protective compounds. Some of the most versatile patterns in land plant genome evolution concern genes for environmental adaptation12–14. That said, there is a shared core of key regulatory and response factors that are at the heart of plant physiology. These include phytohormones such as abscisic acid (ABA) found in non-vascular and vascular plants15,16, protective compounds resting on specialized metabolic routes such as phenylpropanoid-derived compounds and proteins such as LATE EMBRYOGENESIS ABUNDANT (LEA)17,18. Many of the genes integrated into these stress-relevant metabolic routes have homologues in streptophyte algae19. Taking angiosperms as reference, such stress-relevant pathways are often patchy. Whether these are also used under the relevant conditions is currently unknown. For example, while Zygnematophyceae have a homologue to the ABA-receptor PYL8,20, this homologue works in a different, ABA-independent fashion21. Thus, it is important to put the genetic chassis that could act under environmental shifts to the test.
Here we used a fine grid of a bifactorial gradient for two key terrestrial stressors, variation in irradiance and temperature, to probe the genetic network that the closest algal relatives of land plants possess for the responsiveness to abiotic cues. Correlating environmental parameters, physiology and global differential gene expression patterns from 128 transcriptomes (9,892,511,114 reads, 1.5 Tbp of data) across 126 distinct samples covering a temperature range of >20 °C and light range of >500 µmol photons m−2 s−1, we pinpoint hubs in the circuits that have been shared along more than 600 million years of streptophyte evolution.
Results
A physiological grid: co-dependency of eurythermy and euryphoty
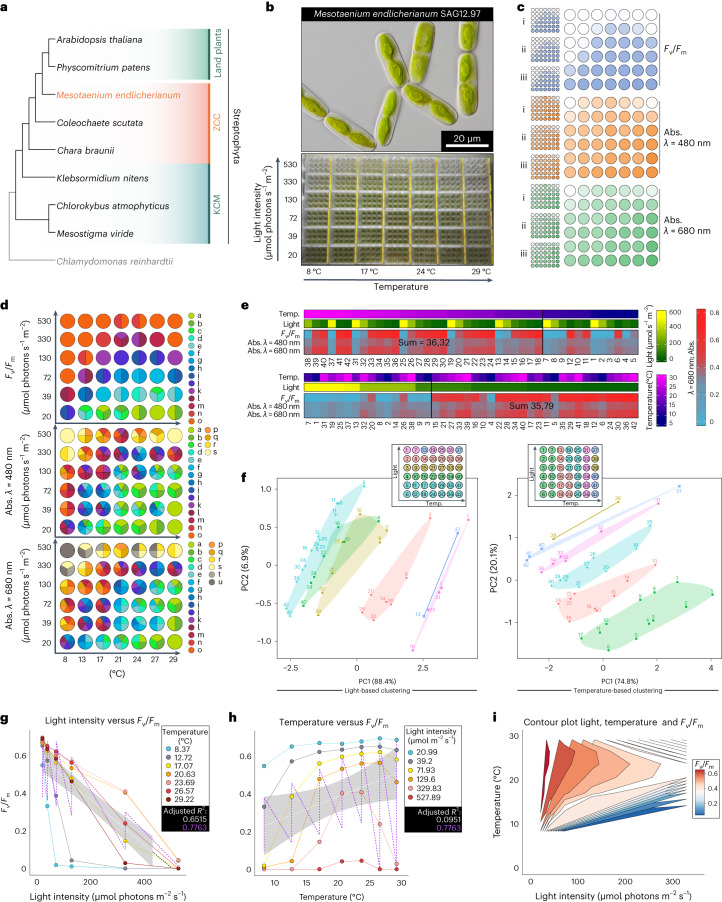
We studied the genome-sequenced strain SAG 12.97 of the freshwater alga Mesotaenium endlicherianum, a member of the Zygnematophyceae, the closest algal relatives of land plants8 (Fig. 1a,b). Natural habitats for Mesotaenium, belonging to the order Serritaeniales, are diverse—ranging from plankton to aeroterrestrial7,8. We cultivated Mesotaenium in a large-scale setup in 1.5 l of C-medium up to a cell density of 0.33 AU at 680 nm and distributed the culture across 504 wells (42 12-well plates, 2.5 ml of culture per well). Well plates were placed on a table with a temperature gradient from 8.6 ± 0.5 °C to 29.2 ± 0.5 °C on the x axis; from above, light-emitting diode (LED) lamps created an irradiance gradient from 21.0 ± 2.0 to 527.9 ± 14.0 µmol photons m−2 s−1 across the y axis, thus creating a two-dimensional gradient table (Fig. 1b, Supplementary Table 1 and for light quality, see Extended Data Fig. 4); the conditions were chosen to strike a balance between cell viability and environmental challenge, as determined in a set of pre-experiments (Extended Data Figs. 4–6 and Methods). The 504 cultures were exposed to this gradient setup for 65 h. The physiological status of the algae was assessed by determining the maximum quantum yield of photosystem II (Fv/Fm) using pulse amplitude modulation fluorometry (IMAGING-PAM, Walz) and a microplate reader with absorption at 480, 680 and 750 nm (Fig. 1c, Extended Data Fig. 5 and Supplementary Fig. 1a); the entire procedure was repeated in three successive biological replicates (that is, three runs of the table, 504 Fv/Fm and 4,536 absorption measurements per replicate).
Fig. 1. A fine-combed setup for assessing environmental responses in Mesotaenium.
a, Cladogram of Streptophyta, highlighting that Mesotaenium endlicherianum SAG 12.97 is a representative of the closest algal relatives of land plants. KCM, the grade of Klebsormidiophyceae, Chlorokybophyceae and Mesostigmatophyceae; ZCC, the grade of Zygnematophyceae, Coleochaetophyceae and Charophyceae. b, M. endlicherianum grown in C-medium in 42 12-well plates on a gradient table that produces a temperature range of 8.6 ± 0.5 °C to 29.2 ± 0.5 °C on the x axis and an irradiance gradient of 21.0 ± 2.0 to 527.9 ± 14.0 µmol photons m−2 s−1 on the y axis; for phenotyping per well, at least ten micrographs were taken, all showing similar phenotypes of the cells. c, Overview of the measured maximum quantum yield Fv/Fm as a proxy for gross physiology (blue) and absorption (abs.) at 480 (orange) and 680 nm (green); individual replicates of the biological triplicates are shown on the left and the average values are shown on the right. d, Statistical analysis of the physiological values (Fv/Fm, abs. 480 nm, abs. 680 nm). Numbers correspond to environmental conditions on the table. Biological triplicates were grouped into significant groups (a–o, a–s and a–u) with R (version 4.1.3) using a Kruskal–Wallis test coupled with Fisher’s least significance; P values were Bonferroni corrected. Significant differences at P ≤ 0.001 are shown as letters. e, Heat maps displaying averaged physiological values of the 42 conditions sorted either by temperature (temp.) or light. A cut-off was set (black vertical line) on the basis of the distribution of the highest values, which were then summed to determine a positive correlation with temperature or light conditions. f, Two PCAs showing the correlation of light conditions (left) or temperature conditions (right) to physiological values (Fv/Fm, abs. 480, 680 nm). Clusters are shown in different colours, which are also visualized in an overview scheme of the gradient table at the top of the plots. g,h, Unifactorial regression analysis of light intensity (g) and temperature (h) versus Fv/Fm; note the unifactorial linear regression curves (white) versus the bifactorial (violet). i, Contour plot of the bifactorial impact of light and temperature on Fv/Fm (gradient colour).
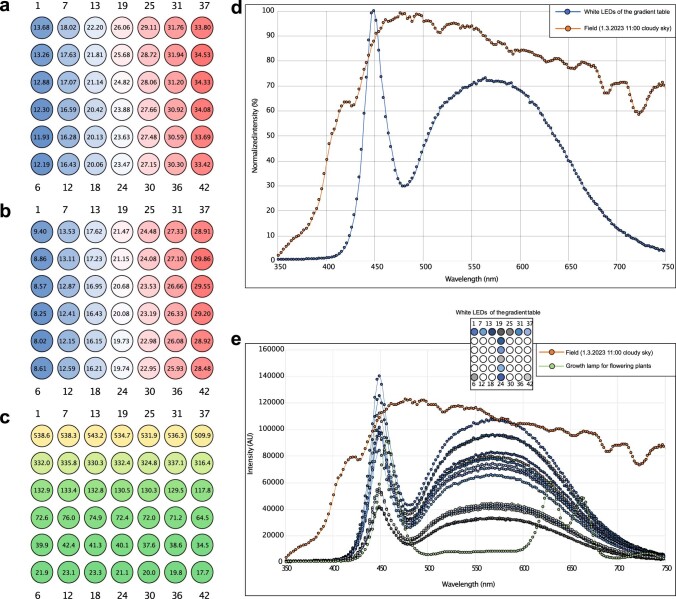
Extended Data Fig. 4. Pre-experimental setups: temperature conditions comparison, light intensities, and light spectra.
(a) Temperature conditions for the first experimental setup (n1(I)) depicted by a blue to red color gradient (see also supplementary table ST 1.4). (b) Temperature conditions for the second and the final experimental setup (n1(II), n2(II), n1(III), n2(III), and n3(III)) depicted by a blue to red color gradient (see also supplementary table ST 1.2). (c) Light intensity/irradiance values depicted by a green to yellow color gradient (see also supplementary table ST 1.1). (d) Average light spectra of the gradient table (blue) assessed using SpectraPen (PSI, Brno, CZ) compared to a spectra assessed of natural sunlight (orange). (e) Light spectra of various plates of the gradient table (see overview) in various shades of blue assessed using SpectraPen (PSI, Brno, CZ) compared to a light spectrum assessed of natural sunlight (orange) and a light spectrum from a growth lamp used for flowering plants.
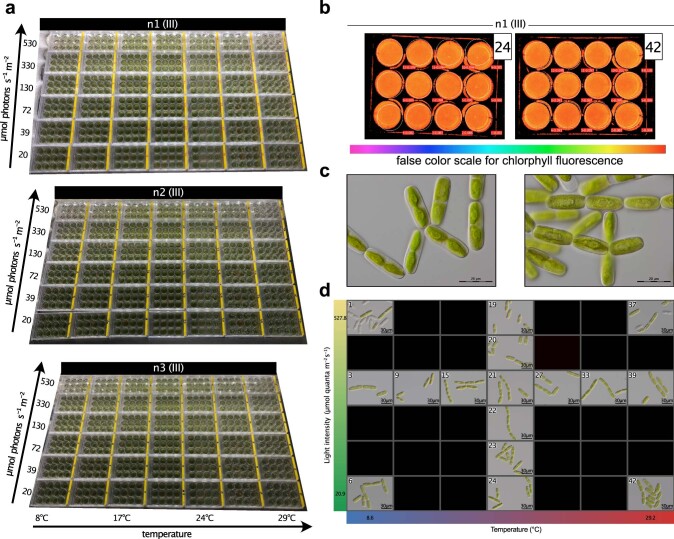
Extended Data Fig. 6. Main-experimental setup (n1,2,3 (III)): Morphology and growth.
(a) Photographs of the main experimental setups n1, n2, and n3 (III) with temperature conditions ranging from 8.6 to 29.0 °C after incubation on the table for 65 h. (b) Fm measurements (maximal fluorescence) using IMAGING-PAM in various table conditions, legend on the right is a false color gradient indicating fluorescence intensity. (c) Differential interference contrast (DIC) micrographs of SAG12.97 cells grown on C-Medium (growth conditions see methods: growth conditions prior to exposure to environmental conditions); at least 10 micrographs were taken, all showing similar phenotypes of the cells. (d) Differential interference contrast (DIC) micrographs of SAG12.97 under most extreme environmental conditions (four corners: samples 1, 6, 37, and 42) as well as along an irradiance gradient at 21 °C (samples 19–24) and a temperature gradient at 130 µmol photons m-2 s-1 (samples 3, 9, 15, 21, 27, 33, and 39); for each well, at least 10 micrographs were taken, all showing similar phenotypes of the cells.
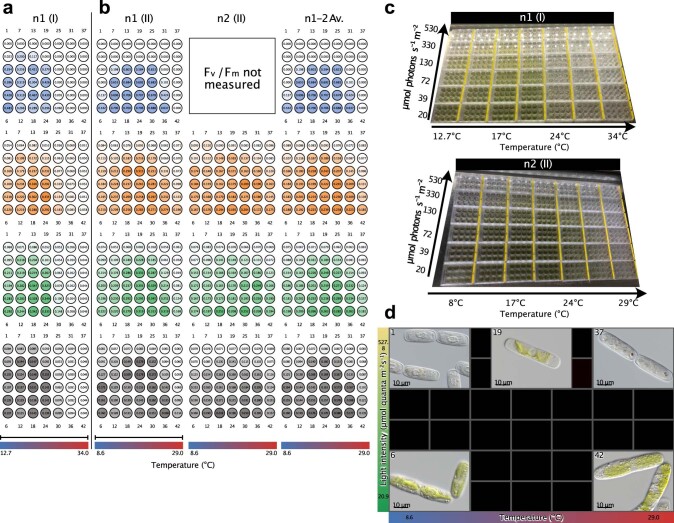
Extended Data Fig. 5. Pre-experimental setup II: Fv/Fm, absorption, and morphology.
(a) Fv/Fm values (blue gradient) and absorption values (orange, green, grey color gradient, Colors indicate measured wavelength: orange gradients = Absorption measured at λ 480 nm, green gradients = Absorption measured at λ 680 nm, grey gradients = Absorption measured at λ 750 nm) of the first pre-experimental setup (n1(I)) with temperature settings ranging from 12.7–34 °C. (b) Fv/Fm and absorption values of second pre- experimental setup (n1(II) and n2(II)) and averaged values (n1-2 Av.) with new temperature settings ranging from 8.6 -29.0 °C. (c) Photographs of the pre-experimental setups n1(I) with temperature conditions ranging from 12.7–34 °C and n2(II) with temperature conditions ranging from 8.6 to 29.0 °C after incubation on the table for 216 h (n1(I)) or 191 h (n2(II)) respectively. The photograph of pre-experiment n2(I) is not shown. (d) Differential interference contrast (DIC) micrographs of SAG 12.97 cells (pre-experimental setup n1(II)) under most extreme environmental conditions (four corners: samples 1, 6, 37, and 42) as well as under high irradiance 527.8 µmol photons m-2 s-1 at 20.5 °C; for each well, at least 10 micrographs were taken, all showing similar phenotypes of the cells.
The algae showed significant differences (P ≤ 0.001) in Fv/Fm values as well as absorption values, both decreasing with rising intensities of irradiance (for Fv/Fm values at 20.5 ± 1.0 °C: from 0.66 ± 0.02 at a light intensity of 21.14 µmol photons m−2 s−1 to 0.042 ± 0.04 at a light intensity of 534.7 µmol photons m−2 s−1) (Fig. 1d, Supplementary Fig. 1 and Supplementary Table 2); despite ample growth at 29.2 ± 0.5 °C and low irradiance, higher temperatures (that is, above 29 °C) were out of the tolerable scope of Mesotaenium (Extended Data Fig. 5). We recorded the lowest Fv/Fm values (down to zero) at conditions of highest irradiance and lowest temperature. Under the ranges tested here, the low temperature had a stronger negative impact on physiology than light. For example, Fv/Fm values at 8.6 ± 0.5 °C and 133 ± 27 µmol photons m−2 s−1 are in a different significance group (P ≤ 0.001) (group o in Fig. 1d) than Fv/Fm values at 29.2 ± 0.5 °C at 118 ± 25 µmol photons m−2 s−1 (purple, group k in Fig. 1d). Values on physiology clustered by light were less broadly distributed than if clustered by temperature (Fig. 1e,f). Even the highest light intensity (527.9 ± 14.0 µmol photons m−2 s−1) was stressful but tolerable for the physiology of Mesotaenium at temperatures between 20.5 ± 0.1 °C (Fv/Fm = 0.042 ± 0.04) and 25.3 ± 0.1 °C (Fv/Fm = 0.045 ± 0.04); more extreme temperatures resulted in undetectable Fv/Fm values. On the basis of the environmental parameters tested herein, eurythermy (broad viable tolerance of temperature) might establish the foundation for euryphoty (broad viable tolerance of light intensities) in M. endlicherianum. Thus, we used regression analysis to understand the effect and importance of the independent values of light and temperature on the dependent physiological values (Fig. 1g–i, Supplementary Fig. 1b and Supplementary Table 2). We find that physiology was always better explained by a combination of temperature and light than a single parameter alone (for example, for Fv/Fm, R2 of 0.776 versus 0.652 and 0.095; Fig. 1g–i).
Fine-combed global gene expression profiles and gene models
To shed light on the molecular mechanisms that underpin the switch from tolerable conditions to adverse environmental cues in Mesotaenium, we applied global gene expression analyses using RNA sequencing (RNA-seq). We pooled all 12 wells per plate and extracted RNA from a total of 126 samples (42 plates, three biological replicates). A total of 114 samples yielded usable RNA that was used to build 128 libraries for sequencing on the Illumina NovaSeq 6000 platform (a minimum of three biological replicates and additional technical replicates; see cartoon of grid in Fig. 2). We generated a total of 1.5 Tbp of 150 bp paired read data at an average depth of 37.7 million reads per sample (~9.9 billion reads in total). Building on this wealth of data, we updated the Mesotaenium gene models to V2. V2 has an increased number of protein-coding messenger RNAs, from 11,080 in the original annotation8 (V1) to 40,326 protein-coding mRNAs (26,009 high confidence, 14,317 low confidence; including splice variants) in 19,233 genes; we labelled an additional 4,408 mRNAs (in 4,312 genes) as ‘predicted gene’ (Supplementary Table 3). With V2, we bring the number of genes in Mesotaenium closer to other Zygnematophyceae with similar genome sizes; V2 has 43 more complete and single copy Benchmarking Universal Single-Copy Orthologs (BUSCO) genes than V1 (+10%; 21 less fragmented, 22 less missing; viridiplantae_odb10; Supplementary Fig. 2). To assess the congruence between biological evidence and V1 and V2, we calculated annotation edit distance metrics (AED; 0 to 1, with 0 being the best). In the cumulative fraction of annotation against AED score, V2 has more mRNAs with AED <0.5. For example, 70% of mRNAs in V1 (7,756 mRNAs) have an AED score <0.5 compared with 60% in V2 (26,840 mRNAs). This is sensible since V2 was built based on the same set of evidence used to calculate AED and it shows higher congruence with them (Supplementary Fig. 3a). Thus, we pseudo-aligned our data onto the new Mesotaenium transcriptome V2 (average alignment rate was 87.31%; Supplementary Table 4a).
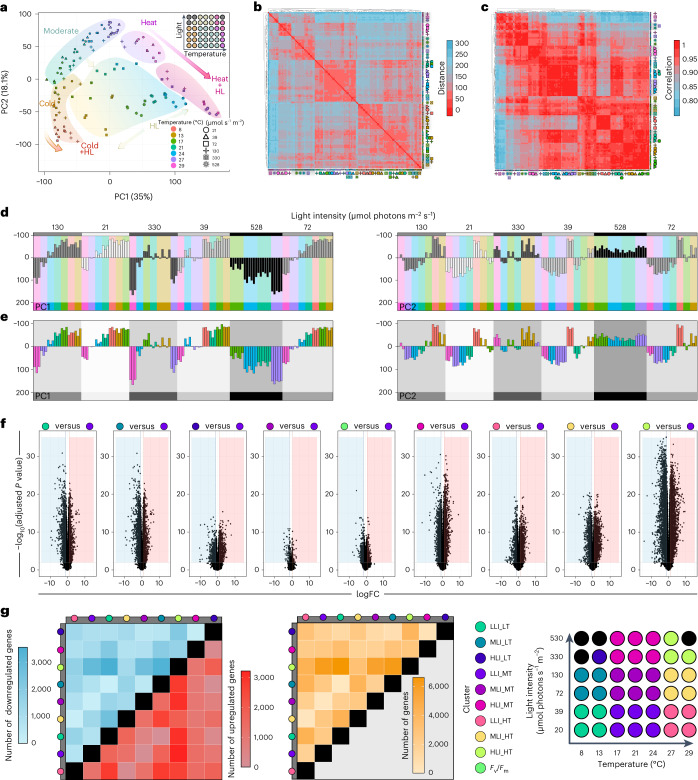
Fig. 2. Global profiles of environment-governed gene expression response.
a, PCA visualizing PC1 and PC2. Backgrounds were drawn to highlight our interpretation of the observed trends; samples are coded by colour (temperature) and symbols (irradiance in µmol photons m−2 s−1). Samples that did not yield usable RNA are indicated as grey dots in the top-right overview of the experimental setup. b, Visualization of Euclidean distances between samples via heat map, from red, zero distance, to blue, furthest distance (a distance of 300). c, Heat map of Spearman correlation between samples, from red, maximum correlation (1.0), to blue, least correlation (<0.8). The clusters were calculated via the Euclidean distance. d,e, PC1 and PC2 scrutinized using a small multiples method of light intensity (d) and temperature (e). In d, shades of grey correspond to different light intensities. In e, different colours represent different temperatures and were mapped with the same colours as a. To perform differential gene expression analysis, we divided the table into nine sectors (see scheme of the table); additionally, a tenth group was raised based on Fv/Fm < 0.5. Linear models were fitted for each gene and empirical Bayes statistics computed for DEGs by the limma package. In total, 37 comparisons were made. DEGs were defined as genes with an absolute fold change (FC) ≥2 and Benjamini–Hochberg-adjusted P value less than 0.01. f, Volcano plots of DEGs for nine selected comparisons based on the sectors and the Fv/Fm < 0.5 criterion. g, Heat maps of numbers of DEGs for all sector-based comparisons (blue, downregulation; red, upregulation; yellow, sum of up- and downregulated genes); grey bars label the first component (treatment) for calculating the contrasts (treatment versus control).
Cheng et al.8 reported that 33.2% of the genome was impacted by transposable elements. We surveyed V2 for protein domains related to transposon biology, retrieving 6,186 entries in 1,748 unique genes (Supplementary Table 4b). Among the 96 that passed the expression threshold, high temperature (29 °C) appeared to have had the strongest effect on transposable element mobilization (Supplementary Fig. 3b).
To understand the gross profile of the gene expression data, we performed a principal component analysis (PCA; Fig. 2a). Independent biological replicates from the same condition clustered in close proximity. High temperature followed by irradiance brought forth clear separation of the data, with PC1 describing 35% and PC2 describing 18.1% of the variance. We evaluated the distance (Fig. 2b) and Spearman correlation (Fig. 2c) using all genes to look for trends among different environmental conditions. The data can be grouped into at least three categories: (1) samples with high light and/or high temperature, (2) a collection of low-temperature (8, 13 and 17 °C) samples, and (3) samples at moderate conditions. Large clusters included low to medium light + medium temperature (‘moderate’ conditions), high light + high temperature, and high light (Fig. 2a). Most distinct was the cluster formed by samples from the high temperature + high light (small multiples; Fig. 2d,e).
Plastid-related genes stand out in differential gene expression profiles
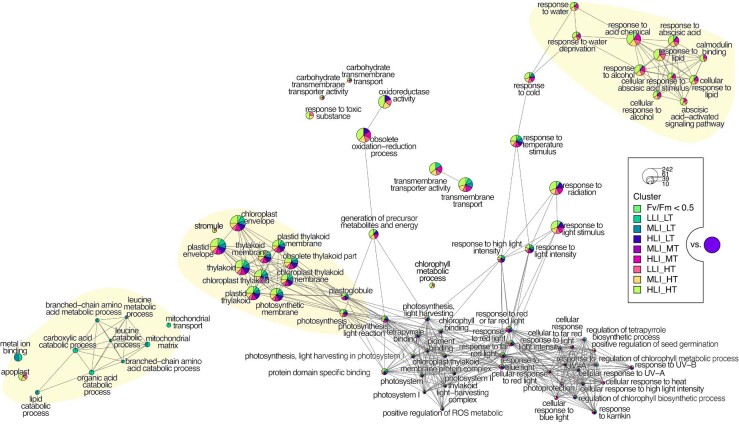
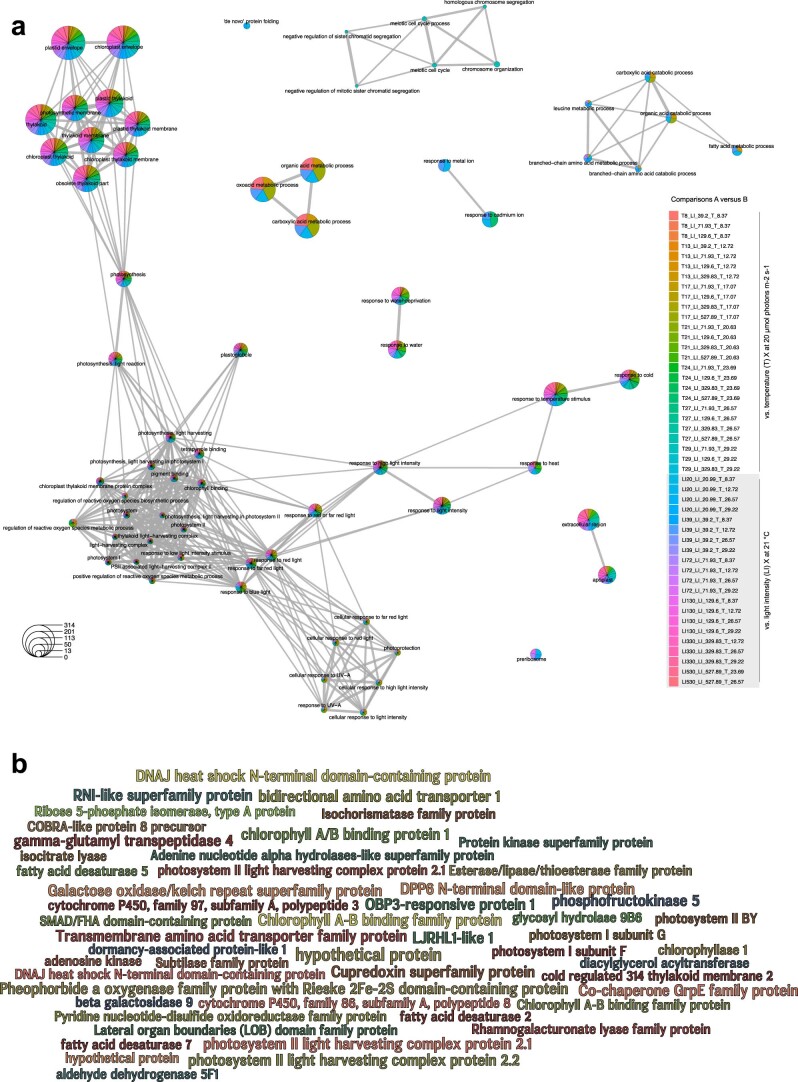
For dissecting the differential gene expression responses, we divided the table into nine sectors and, additionally, a cohort of stressed algae based on Fv/Fm < 0.5 (Fig. 2f,g). We performed 36 comparisons, among which we focused on nine, which additionally included the Fv/Fm-based comparison. Genes were considered to be differentially expressed between groups at an absolute fold change ≥2 and a Benjamini–Hochberg-corrected P ≤ 0.01 (Fig. 2f,g). The intensity of environmental cues governed gross gene expression profiles as increasing disparity between conditions yielded more differentially expressed genes (DEGs), generally following the pattern of the PCA (compare Fig. 2a,g). The most differentially regulated genes (6,578) were pinpointed by comparing low light and low temperature (LLI_LT) versus high light and high temperature (HLI_HT). Enriched Gene Ontology (GO) terms among regulated genes most frequently included plastid biology-associated genes (Extended Data Fig. 1); similar patterns were recovered in 63 unifactorial comparisons where we kept one environmental parameter constant (Extended Data Fig. 7a). To scrutinize our data for specific genes that show a robust and universal response to alterations in the environment, we intersected all 8,157 significantly regulated genes pinpointed by the different comparisons: 3, 30 and 124 genes overlapped among all 9, 8 and 7 comparisons, respectively. These concertedly pinpointed genes were mostly light harvesting genes, corroborating the importance of plastids in the overall cell biology of Mesotaenium (Extended Data Fig. 7b). Indeed, the 30 genes found in all comparisons included, for example, reactive oxygen species (ROS)-relevant genes such as EARLY LIGHT-INDUCIBLE PROTEIN (ELIP) and fatty acid metabolic genes.
Extended Data Fig. 1. Biological theme comparison summarizing all GO term enrichment analysis with adjusted p value ≤ 0.01 of DEGs against all genes that were expressed and passed the filtering in our analyses as background.
The size of each circle is proportional to the count of each GO-term. Only the top 30 enriched terms are shown.
Extended Data Fig. 7. Differential gene expression comparisons highlight plastid-related responses.
(a) Biological theme comparison of GO terms enriched in differential gene expression analyses in which one factor was always kept constant; the top 10 different connected graphs are shown. (b) Wordle of the 124 genes that showed significant regulation across multiple comparisons shown in main Fig. 2f,g and Extended Data Fig. 1; word size correspond to the number of comparisons in which a gene appeared. Color serves to increase the contrast between words.
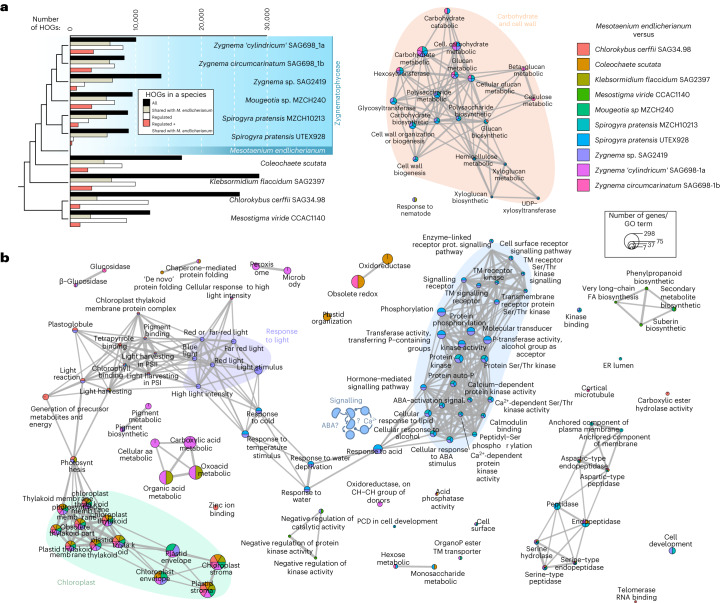
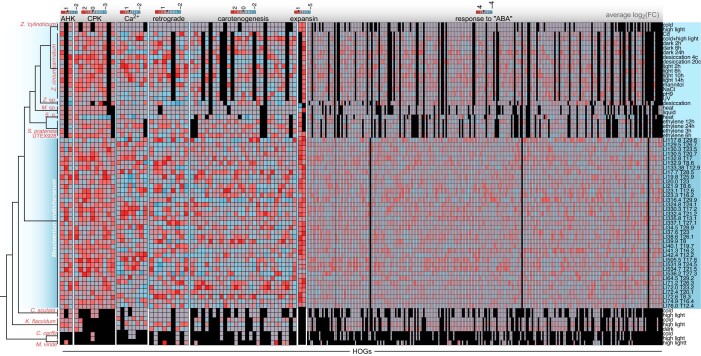
How do these responses compare across land plants’ close relatives? To answer this, we downloaded major stress transcriptome data from streptophyte algae9,20,22–25, inferred significant differential gene expression between stress treatment and control per species, and asked whether regulated genes belong to the same phylogenetic hierarchical orthogroups (HOGs, inferred with Orthofinder26). Depending on the phylogenetic distance of the species, we found between 3,107 and 6,458 HOGs shared with Mesotaenium, with 46.6–73.0% shared within and 15.8–30.4% outside of the clade Zygnematophyceae (Fig. 3a). Of these shared HOGs, between 4.6% and 59.8% show shared regulation with Mesotaenium. The degree of similarity depends on treatment not phylogenetic position. The most common responses across species were related to chloroplasts and photosynthesis (Fig. 3b and Extended Data Fig. 2). However, within Zygnematophyceae, additional signalling processes such as kinase activities and calcium-dependent signalling stood out (Fig. 3b and Extended Data Fig. 2), corroborating (1) their noted importance in Zygnematophyceae23,24 and (2) the concept that important steps in the evolution of streptophyte calcium signalling system (Extended Data Fig. 2) occurred before plant terrestrialization27.
Fig. 3. Comparative analyses of global differential gene expression profiles across stress-treated streptophyte algae.
Publicly available data on stress transcriptomes from ten different streptophyte algae were downloaded and significant differential gene expression between stress treatment and control per species were calculated. Phylogenetic HOGs were inferred with Orthofinder. a, Bar graph of the number of all HOGs detected (black), HOGs shared with Mesotaenium endlicherianum (tan), all regulated HOGs in a given species (white) and, of those regulated, which are in the same HOG as significantly regulated genes in Mesotaenium (red); the relationship between the streptophyte algae is shown by the cladogram on the left. b, GO term-based biological theme comparison of these shared significantly regulated genes in HOGs. Note the recurrent pattern of chloroplast-associated differential gene expression (green), light quality (purple) and the putative integration of calcium signalling with pathways known from phytohormone signalling, including ABA, in land plants (blue, also note the little sketch of a hypothetical model). PCD, programmed cell death.
Extended Data Fig. 2. Heat maps of average differential gene expression in log2(fold change) per HOG.
From the strongest upregulation in red to the strongest downregulation in blue; black means that no HOG was found. The heat maps were sorted by phylogeny (see the cladogram on the left) and treatment (written on the right); light teal highlights the data on Zygnematophyceae, dark teal on Mesotaenium endlicherianum.
To understand whether these genes integrate into the context of molecular programmes, we next analysed gene co-expression.
Gene expression clusters recover ancient genetic programmes
The environmental gradients triggered changes in the expression of gene cohorts. We wanted to understand their concerted action independent of any prioritization guided by homology to any land plant genes—solely from the molecular programmes that operated in the algae. To do so, we applied a weighted gene co-expression network analysis28 (WGCNA) for unsupervised clustering (Fig. 4, Supplementary Figs. 4–7 and 10–13 and Extended Data Fig. 8). To then understand the driving forces behind these changes, we turned to the highly connected genes (nodes) in the network (hubs) (Fig. 5).
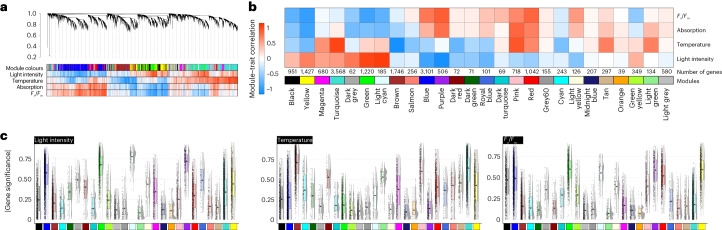
Fig. 4. Unsupervised gene expression clusters recover genetic programmes separated by environmental cues.
Gene expression clustering into 26 coloured modules was performed using WGCNA; grey is the module of unclustered genes. a, Hierarchical cluster tree of 17,095 genes. The heat map below the dendrogram and module colour assignment shows the gene significance measure (from red, positive correlation, to white, no correlation, to blue, negative correlation) for the four different conditions or physiological parameters. b, Heat map of the module–trait correlation based on eigengenes (from red, positive correlation, to white, no correlation, to blue, negative correlation); see Supplementary Fig. 7. c, Box plots of the mean gene significance across modules (given in the corresponding module colour) towards the parameters light intensity, temperature and Fv/Fm. The box plots display the interquartile range (IQR) of the data, compactly displaying the distribution of a continuous variable. They visualize five summary statistics (the median, two hinges and two whiskers). The upper whiskers extends from the hinges to the largest/smallest value no further than 1.5× IQR from the hinge. Each data point (n) is a gene, and the total n of genes is the same as shown in b. We calculated the gene significance for each gene using the WGCNA package and Pearson method.
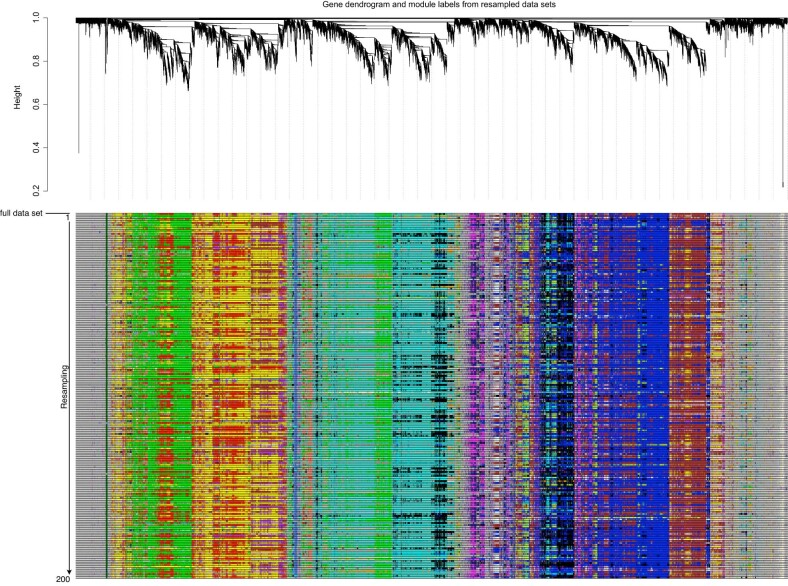
Extended Data Fig. 8. Module stability analysis.
We performed 200 simulations by sampling from our input expression profiles under different conditions using the WGCNA package. Each row in the heat map represents a simulation and stable modules form a ‘column’ of similar color on the heat map.
Fig. 5. Molecular programmes for environmental responses around recurrent plant hubs.
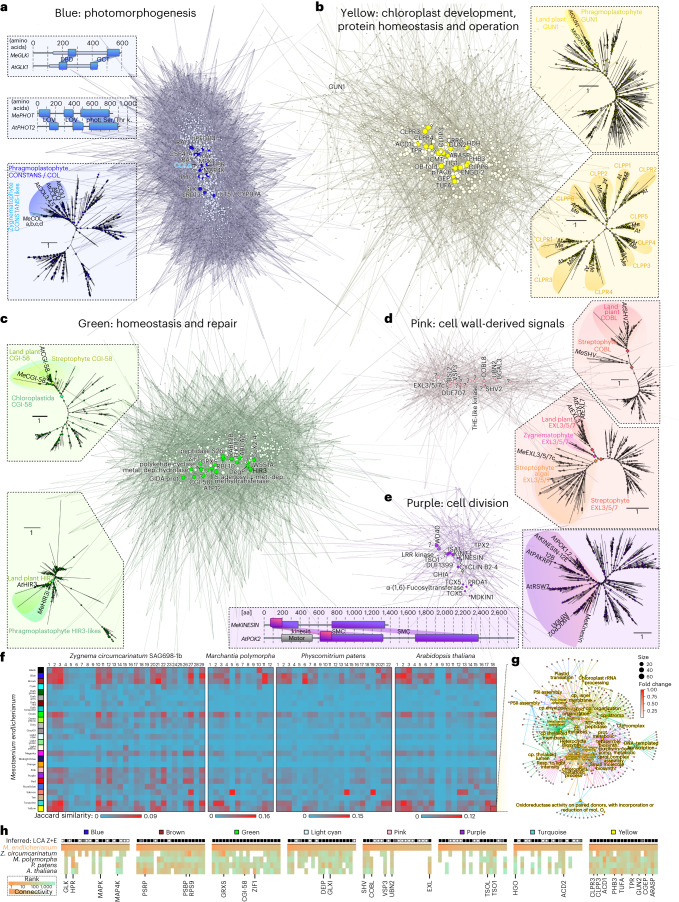
a–e, Visualization of the co-expression networks clustered by WGCNA into the modules blue (3,101) (a), yellow (1,427) (b), green (1,220) (c), pink (718) (d) and purple (506 genes) (e). Nodes (circles) represent genes connected by edges whose weight (light to dark colour) is based on a weighted TOM. Brightly coloured nodes represent the 20 most connected genes (hubs) and are annotated based on homology; all other nodes are depicted in the corresponding paler colour. Around the clusters, different protein-coding hub genes are highlighted, giving information such as predicted domain structures or phylogenetic relationships; for fully labelled phylogenies, see Supplementary Fig. 26b. Circles in phylogenies give a scale of the ultrafast bootstrap support values; diamonds indicate high (>90%) support for branches separating highlighted clades. An alignment of GLK homologues can be found in Supplementary Fig. 8. f, Using WGCNA, co-expression networks were computed from 212 publicly available RNA-seq datasets from Z. circumcarinatum, M. polymorpha, P. patens and A. thaliana exposed to diverse abiotic challenges, yielding between 12 and 29 modules (labelled above the heat map), and orthogroups for all genes in the modules of these different species were determined. The heat map shows the similarity, based on Jaccard indices, between the modules of Mesotaenium (same colours as throughout the paper, see Fig. 4b) and the co-expression modules in the three land plants as well as Zygnema; red to blue colour gradients indicate high to low Jaccard similarity. g, Cnet plot of the enriched GO terms in the module ‘Arabidopsis 18’, which has high Jaccard similarity to the M. endlicherianum module yellow—note the recurrent terms of plastid operation and, especially, the Clp complex. h, Heat map of the connectivity ranks across all five species for homologues of hub genes of Mesotaenium, from orange (high) to green (low connectivity). Black boxes (top row) indicate if our phylogenies (see data on Zenodo) suggest that the hub genes fall into families that were present in the last common ancestor of Zygnematophyceae and land plants, and hence emerged before plant terrestrialization; white boxes signify the absence of such indication and grey boxes highlight ambiguous relationships.
We clustered the 17,905 genes expressed in our samples (passing the minimum expression threshold) into 26 modules, which we refer to with colours (Fig. 4a). Orange is the smallest module (39 genes), and the largest modules are turquoise, blue and brown with 3,568, 3,101 and 1,746 genes, respectively (Fig. 4b). The samples reflect a range of distinct physiological conditions and resulting data are a combined expression of the different environmental cues and the modulation of the algal physiology. To investigate the biological role of each module, we used their eigengenes as representatives for the modules’ gene expression profiles and correlated their behaviour with the two environmental cues (light intensity and temperature), as well as the physiological parameters absorption and Fv/Fm (Fig. 4b,c). One of the foremost general patterns in cellular response to stress are ROS. ROS act as signals as well as culprits that, if not quenched, damage biomolecules; GO terms capture ROS biology (Extended Data Fig. 3), especially in module green that positively correlates with light intensity (r = 0.88, P = 6 × 10−43) and negatively with Fv/Fm (r = −0.79, P = 6 × 10−29) (Extended Data Fig. 3, Supplementary Figs. 4–7 and Supplementary Tables 5 and 6).
Extended Data Fig. 3.
Enriched GO-terms for eight of the 26 modules; each inset shows the gene expression profiles of all genes in a given module. (a–i) Arabidopsis homologs for key processes were mined based on keywords; they were retrieved from a look-up table of BLASTp hits in a search of Mesotaenium V2 against A. thaliana representative protein sequences. Bar charts show the percentage of detected Mesotaenium homologs across the modules relative to the number of all Arabidopsis IDs assigned to the terms. No BLAST hit was not depicted. Abbreviations: proc. = process; reg. = regulation; biogen. = biogenesis; develop. = development; pos. = positive; neg. = negative; init. = initiation; GEP = Gene expression profile; med. = mediated; dep. = dependent; modif. = modification; conjug. = conjugation; anneal. = annealing; compl. = complex; synth. = synthesis; resp. = response; transf. = transferring.
The clusters also recover the genetic signatures of thriving algae. Module purple negatively correlates with increasing light (r = −0.94, P = 3 × 10−60) and positively with absorption and Fv/Fm (r = 0.71, P = 3 × 10−21 and r = 0.79, P = 4 × 10−28). These dense and physiologically healthy cell populations (experiencing no light stress) likely ramped up cell division (Extended Data Fig. 3 and Supplementary Table 6), signified by homologues of cyclin and TPX2 appearing as hub genes (Fig. 5e). The ninth most connected hub is a kinesin homologous to genes coding for proteins such as PHRAGMOPLAST ORIENTING KINESIN 2, and homologues of the important growth regulators29 Tesmin and TSO1 ranked at positions 7, 15 and 17 of the most connected hubs in module purple (Fig. 5e and Supplementary Table 7).
To understand the evolutionary conservation of the genetic programmes in these modules, we processed 212 publicly available RNA-seq datasets from Zygnema circumcarinatum9, M. polymorpha, P. patens and A. thaliana exposed to diverse abiotic challenges using the same WGCNA pipeline, which yielded between 12 and 29 modules. We determined orthogroups between the modules of these different species and compared the similarity in modules by calculating Jaccard indices (Fig. 5f) and GO-term enrichment in these modules (Fig. 5g and Supplementary Fig. 14b–e). Also here, blue, brown, turquoise and yellow stand out as important and likely conserved environmental response modules (compare Figs. 4b and 5f and Extended Data Fig. 3). We further analysed shared connectivity of hub orthogroups. For the mentioned Tesmin and TSO1 orthogroups (Fig. 5h), this reveals that they are likely connected regulators of cell division since about 600 million years of streptophyte evolution. To scrutinize this aspect, we inferred the evolutionary history of the 160 hubs using maximum likelihood phylogenetic analyses (Fig. 5h; data on Zenodo). We retrieved 135 phylogenies, 107 of which indicate that the hubs are in gene families that were present in (or before) the last common ancestor of Zygnematophyceae and land plants. Thus, they pre-date plant terrestrialization.
Conserved hubs: integration of plastid and cell physiology
Chloroplasts act as environmental sensors in land plant cells30. In concert with this, many of the modules we identified were associated with plastid biology and/or physiology (Extended Data Fig. 3, Supplementary Figs. 4–7 and Supplementary Table 6). Module brown is enriched in GO terms related to plastids, general transcription and translation, and negatively correlates with temperature (r = −0.95, P = 7 × 10−65; Extended Data Fig. 3 and Supplementary Fig. 5). Among the top 20 hub genes in module brown, 12 are associated with translation and ribosomes (Supplementary Table 7). As expected, this cluster shows conservation in enriched functions of its related modules in the other four streptophytes, including shared high connectivity of hubs (Fig. 5f,h). The module light cyan positively correlates with increasing light (r = 0.93, P = 1 × 10−56; Supplementary Fig. 6) and negatively with Fv/Fm (r = −0.67, P = 5 × 10−18; Supplementary Fig. 4). It features not only hubs related to ROS homeostasis from the thioredoxin superfamily and other light-induced proteins, but also pigment and apocarotenoid metabolism (Extended Data Fig. 3); these are the source of important signals from the chloroplast that likely have deep evolutionary roots19 and are also formed by light-dependent oxidative reactions31. The blue module negatively correlates with increasing light (r = −0.76, P = 10−25) and positively with Fv/Fm (r = 0.67, P = 2 × 10−18). Concomitantly, the blue module has a high number of enriched GO terms, many of which are plastid-related terms, cellular signalling and terms that tie the two together—that is, signalling processes emanating from the plastid (Extended Data Fig. 3 and Supplementary Table 6). Such responses align with similar clusters in other species (Fig. 5f), where the related Arabidopsis modules 2 and 10 show terms for light intensity and quality (Supplementary Fig. 14b).
The hubs of many modules, including those in blue, light cyan and yellow mentioned before, reflect an association with plastid-related processes. To highlight a few, the second most connected gene in module blue is a homologue of GOLDEN2-LIKE 1 (GLK1) (Supplementary Fig. 8). GLK1 is a transcription factor (TF) that regulates chloroplast development and the activity of nuclear genes involved in photosynthetic light reaction and chlorophyll biosynthesis32–34; indeed, genes in the GLK orthogroup are highly connected throughout the modules of land plants, and in the zygnematophyte, Zygnema a GLK homologue is the eighth most connected gene in its module (Fig. 5h). Blue also features hydroxypyruvate reductase-coding gene, important in photorespiration35, as the fourth most connected hub, which appeares in the top-five most connected genes in the bryophytes (Fig. 5h). A CYP450 gene homologous to LUTEIN DEFICIENT 5 (LUT5), is the seventh most connected gene, suggesting the involvement of pigment-related signalling. Module 21 in P. patens is dominated by ABA signalling (Supplementary Fig. 14d) and it is similar to Mesotaenium modules turquoise and blue (Fig. 5f), enriched in homologues of ABA-activated signalling (Extended Data Fig. 3), featuring a highly connected homologue of ABA-RESPONSIVE ELEMENT-BINDING FACTOR 2 (ABF2). Thus, parts of the ABA signalling module consist of ancient wires whose relevance in environmental response pre-date plant terrestrialization, and ABA dependency20,21,36.
Next to GLK1—the most connected TF-coding gene—other highly connected transcriptional regulators appear in module blue. These include homologues of photomorphogenesis-regulating genes such as CONSTANS-like 3 (COL3, the fourth most connected TF-coding gene) and CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1); CO/COL and GLKs are both degradation targets of COP1 (refs. 37–39). Further, the circadian regulator40 BROTHER OF LUX ARRHYTHMO is the second most connected TF-coding gene in module blue. All of this aligns with the similarity to the Arabidopsis module 2 and the P. patens module 6, featuring, next to light quality, also photoperiodism (Fig. 5f and Supplementary Fig. 14e,f). Further, homologues of ETHYLENE-INSENSITIVE3-like 1 (the sixth most connected TF-coding gene) and several ETHYLENE RESPONSE FACTORs (ERFs) are among the most connected TF-coding genes. Previous investigations of the Zygnematophyceae Spirogyra pratensis have shown that SpEIN3 can rescue Arabidopsis ein3-1 mutant plants41; exogenous application of ethylene on Spirogyra triggers stress-, plastid- and photosynthesis-associated gene expression responses similar to land plants22, which we recover, as outlined, across co-expression modules (Fig. 5f, Extended Data Fig. 3 and Supplementary Fig. 14a–f) and shared differential patterns (Fig. 3b and Extended Data Fig. 2). This speaks for a conserved regulatory framework that involves the plastid, photosynthesis, ethylene-associated factors, and maybe ethylene itself, in environmental signalling cascades in the common ancestor of land plants and their closest algal relatives.
Module yellow correlates positively with light intensity (r = 0.62, P = 10−14) and negatively with absorption and Fv/Fm (r = −0.79, P = 10−28 and r = −0.81, P = 3 × 10−31; Fig. 4b); GO terms are associated with plastids and proteolytic enzymes42,43 (FtsH and ClpP), recapitulating well-known ties of protein homeostasis and plastid maintenance (Extended Data Fig. 3). Indeed, yellow features five hubs that are homologous to genes coding for CLP proteases, critical for chloroplast protein homeostasis44,45, and hubs homologous to genes that orchestrate the coordination of transcriptional activity between chloroplasts and the nucleus (Fig. 5b); the latter includes homologues of (1) pTAC6, which is essential for plastid gene expression and thus chloroplast development in Arabidopsis46, and (2) a homologue of GENOMES UNCOUPLED 2, one of the foremost genes in the classical plastid–nucleus communication pathway47. Among the TF-coding genes in module yellow is a homologue of the bZIP light signalling master regulator ELONGATED HYPOCOTYL 5 (ref. 48) (HY5). Module yellow is among those with the most consistency in similar modules across the analysed streptophyte co-expression networks and hubs (Fig. 5f,h), as exemplified by the GO term similarities between yellow and Arabidopsis module 18 (compare Fig. 5g and Extended Data Fig. 3) and the consistency of the plastid operational genes as hubs (Fig. 5h). Hence, hallmark genes for plastid operation and its integration into molecular cell physiology probably acted in concert since before the dawn of embryophytes.
Of ancient signalling cascades and cell wall perturbance
Mitogen-activated protein kinases (MAPK) constitute environmental response pathways in all eukaryotes49. In land plants, several abiotic and biotic cues have been described to trigger MAPK-mediated signalling50–53. Genes coding for MAPK and phototropin kinases appear as hubs in module blue. Moreover, plant MAPK-based signalling is interwoven with wound response and brassinosteroid signalling50; the MAPK orthologue in Zygnema is also highly connected (Fig. 5h) and blue is similar to the kinase-rich module 17 of Arabidopsis (Fig. 5f). Stress often coincides with a perturbance of plant cell wall homeostasis. Module pink includes hubs for such wounding and cell wall-derived signals. This pairs with the GO term brassinosteroid signalling, which balances growth, cell wall homeostasis and stress in Arabidopsis54,55. Among the hubs in pink are homologues for (1) diverse receptor kinases known from Arabidopsis to sense alterations in cell wall integrity56 and (2) EXORDIUM-like (EXL; Mesotaenium has 12 EXL homologues), which integrates growth with environmental signalling57 (Fig. 5d). This pairs with genes coding for the COBRA family proteins being the most and third most connected hubs in the module. COBRA proteins are known to be involved in cell expansion and balancing pathogen response with growth58–60. It appears that Mesotaenium bears parts of a loop that senses physico-chemical perturbance of cell wall homeostasis; in land plants, these loops include brassinosteroid signalling61 and wiring of the core genes mentioned here are ancient, evident by the recurrent high connectivity of EXL and COBL homologues (Fig. 5h) throughout 600 million years of streptophyte evolution.
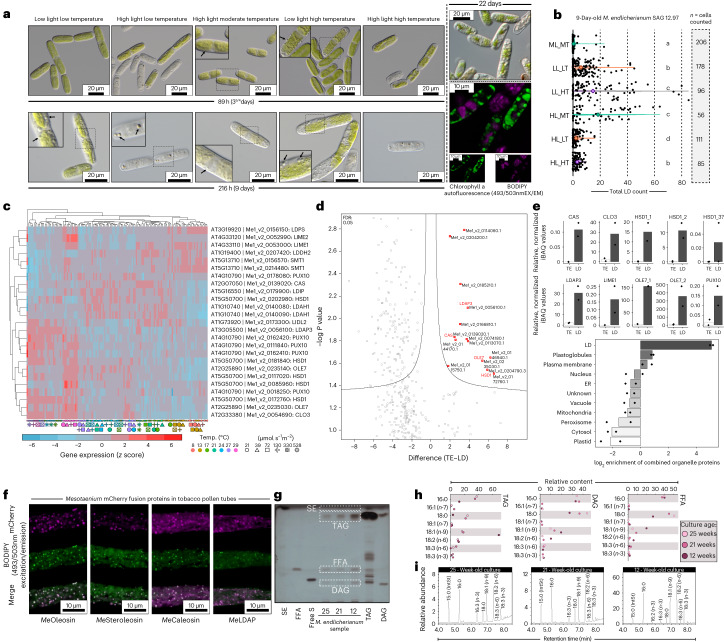
LDs: a response pre-dating plant terrestrialization
In land plants, lipid droplet (LD) formation and triacylglycerol (TAG) accumulation are common to many stress responses, including heat, cold and drought62–66. We observed that cells of Mesotaenium accumulated inclusions resembling LDs upon prolonged exposure to stress (Fig. 6a). Consistently, these globular structures were stained by BODIPY 493/503 (EM/EX), a common dye for lipid- and oil-rich compartments67,68. Under different temperature and light conditions, counts of LDs per cell showed significant differences (Fig. 6b and Supplementary Table 8). A CGI-58 homologue is the tenth most connected hub in module green (Fig. 5c). CGI-58 is key to lipid homeostasis, causing, if perturbed, Chanarin–Dorfman syndrome in humans and LD overaccumulation in Arabidopsis69,70 (Fig. 5c); CGI-58 is the 22nd most connected gene in Arabidopsis module 5 (Fig. 5h). Further, differential gene expression profiles pinpointed elevation of transcripts for characteristic LD protein homologues such as steroleosin (HSD1) and oleosin (OLE7) under high temperature and moderate light conditions (29 °C, 21–130 µmol photons m−2 s−1) and LD-associated protein (LDAP) and PUX10 under high temperature and light conditions (21–29 °C, 130–528 µmol photons m−2 s−1; Fig. 6c).
Fig. 6. LDs accumulate in Mesotaenium upon changing environments.
a, DIC and confocal micrographs of Mesotaenium endlicherianum SAG 12.97 cells accumulating LDs (arrows) upon exposure to different temperature/light conditions (abbreviations) of the gradient table for 89 h or 216 h. For confocal microscopy, algae were cultured independent of table conditions at 75 µmol photons m−2 s−1 and 22 °C for 22 days. LDs are visible as distinct globular structures and were stained with BODIPY (false-coloured green; 493 nm excitation, 503 nm emission); chlorophyll autofluorescence in false-coloured purple; for each condition, at least ten micrographs were taken, all showing similar phenotypes of the cells. b, Violin plots of LD quantification after 9 days of exposure to different environmental conditions; significance grouping (Mann–Whitney U) is based on P < 0.05; see also Supplementary Fig. 27. c, Heat map of row-scaled z scores of the expression of homologues for LD biogenesis and function (see also Supplementary Fig. 28). Conditions are displayed at the bottom as symbols in different colours; best Arabidopsis hits (via BLASTp) are shown on the right. d,e, Proteomic investigation into lipid-enriched phases extracted from Mesotaenium; note the enrichment in hallmark proteins of LDs. Volcano plot showing significantly (false discovery rate (FDR) <0.05) enriched Mesotaenium proteins in the lipid-enriched (LD) versus the TE (d). Bar plots show the relative, normalized iBAQ values for ten LD signature proteins detected in Mesotaenium (e). Bottom bar plot shows the log2 enrichment of proteins characteristic for subcellular compartments. LL, ML and HL, low, moderate and high light; LT, MT and HT, low, moderate and high temperature, respectively; ER, endoplasmic reticulum. f, LD proteins of M. endlicherianum localize to LDs in tobacco pollen tubes: cLSM images of transiently expressed proteins appended to mCherry in transiently transformed N. tabacum pollen tubes. LDs were stained with BODIPY 493/503; for each construct, the images are representative of at least nine micrographs of transformed pollen tubes per fusion construct. Scale bars, 10 µm. g,h, Lipid composition in M. endlicherianum LDs of 12- to 25-week-old cultures and standards for sterol esters (SE), FFA, free sterols (free S), TAG and DAG via analytical TLC (g) and preparative TLC followed by GC for profiling (h). i, Full lipid profiles assessed via GC.
To scrutinize whether these structures are comparable to LDs of land plants, we performed subcellular fractionizations, obtained lipid-rich phases and subjected them to proteomics using liquid chromatography–mass spectrometry (LC–MS). We identified 739 proteins in the putative LD fraction and 1,574 proteins in the total extract (TE) (Supplementary Table 9). Of these, 14 were significantly enriched in the putative LD fraction (Fig. 6d) including hallmark LD proteins71 such as OLE, caleosin (CLO), HSD and LDAP (Fig. 6e). We confirmed the localization to LDs for these four proteins by transiently expressing mCherry-tagged variants in tobacco pollen tubes; mCherry clearly overlapped with BODIPY 493/503 fluorescence (Fig. 6f). Resembling LDs of seeds71, we found predominantly TAG in the lipid content of the LDs (Fig. 6g and Supplementary Fig. 27b); the lipid profiles of Mesotaenium LDs varied with age of the cultures (Fig. 6h,i). Overall, Mesotaenium responds to stress conditions by formation of LDs containing signature proteins typical of embryophytic LDs.
Discussion
Owing to their plain morphology, Zygnematophyceae emerged as the unexpected closest algal relatives of land plants4–7. That said, the molecular programmes of Zygnematophyceae speak of their close relationship to land plants. These point to a conserved chassis that probably operated in the last common ancestor of land plants and algae, featuring the proposed action of various hallmark proteins (for example, PYL homologues20, GRAS family TFs8 and more) that were once considered land plant innovations. During plant terrestrialization, challenges did not come in isolation. The aim of this work was to define stress responses to temperature and irradiance combinations in a close algal relative of land plants. In the approach we have chosen, we made sure to capture both tolerable ranges of cues and those that go beyond tipping points, to allow for robust definition of where stress starts and to pinpointing molecular programmes whose expression dynamics follow the kinetics of their environmental trigger (for example, light intensity in the case of programmes for high light response); this included capturing the well-known double assault of low temperature and high light on the photosynthesis machinery. Building on the genomic resources for Mesotaenium, we have here delved into the molecular physiology and genetic programmes of this alga, revealing which programmes bear out when challenged with environmental cues.
Recent studies have proposed homology for the chassis of plastid–nucleus communication upon adverse environmental conditions between land plants and phragmoplastophytic streptophyte algae20,72,73. The GUN pathway probably has a conserved role in chloroplast transcription and streptophyte algal GUN1 homologues can rescue chloroplast retrograde signalling of Arabidopsis Atgun1 mutants74; the degree of evolutionary conservation in the retrograde signalling pathway across streptophytes remains obscure74. Signals from damaged chloroplasts inhibit GLK1 expression in Arabidopsis75. The negative correlation of module blue (featuring MeGLK) with high light (leading to damaged chloroplasts) supports a role of MeGLK in operational retrograde signalling. Indeed, our comparative analyses revealed a consistency in plastidial integration on the basis of similar networks in land plants and Zygnema (Fig. 5f) and with regard to highly connected hub genes associated with ROS and plastid protein homeostasis (Fig. 5g,h). Altogether, these insights point to operational processes of the plastid of the closest relatives of land plants, governed by nuclear gene expression for dealing with light regimes and adjustment of photosynthetic performance. On balance, our data underscore that the wires between components in plastid–nucleus communication are probably shared across more than 600 million years of streptophyte evolution.
In land plants, the formation of LDs is known to occur under a variety of adverse environmental conditions63,64,76. Stress-dependent formation of LDs probably evolved before land plants came to be24,77,78, but their molecular underpinnings outside of land plants remain unclear. Here, we confirmed the identity of these Mesotaenium LDs using confocal microscopy, LD-specific staining and proteomics. Our comprehensive transcriptomic data illuminate co-expressed modules that might constitute a homologous programme for stress-dependent LDs that acted before plants conquered land.
Methods
Algal culturing and gradient table setup
We used the axenic and genome-sequenced Mesotaenium endlicherianum SAG 12.97 (ref. 79) from the Algal Culture Collection, Göttingen (SAG80). Mesotaenium was cultivated in C-medium81 for an average of 12 days in an aerated culture glass flask (SCHOTT) at 80 µmol photons m−2 s−1 (12h/12h light/dark cycle (light from 6 am to 6 pm, Central European winter time) at 18 °C). Before the experiment, cell density was analysed using a LUNA Automated Cell Counter (Logos Biosystems) and set to 2.03 × 107 cells ml−1 (diluting with C-medium if needed; settings for cell counting: cell roundness, 60%; minimum size, 3 µm; maximum size, 60 µm), corresponding to Abs680nm = 0.33 (Epoch Microplate reader, BioTek Instruments). For the gradient table setup, the algal suspension was distributed across 504 wells (42 12-well plates (tissue culture testplates 12 no. 92412, TPP); 2.5 ml of culture per well). Plates were sealed with surgical Micropore tape (3M) to minimize evaporation. The 42 12-well plates were then placed on a table that generates a cross-gradient of temperature (8.6 ± 0.5 °C to 29.2 ± 0.5 °C on the x axis) and irradiance (21.0 ± 2.0 to 527.9 ± 14.0 µmol photons m−2 s−1 on the y axis) (Supplementary Table 1). The temperature gradient was generated using a custom-made table (Labio) equipped with true-daylight LEDs (sTube 2 W 120 ver 11:11, Snaggi) set to a 16 h/8 h light/dark cycle (light from 6 am to 10 pm, Central European winter time). Mesotaenium samples exposed to the 504 different conditions for 65 h (for sampling for RNA-seq and physiological measurements) and 89 h (for detailed light microscopy) on the gradient table. Condensed water at the top of the 12-well plates lids was removed three times in the 65 h by lightly tapping the lids twice.
Algal culturing and gradient table setup (pre-experiments n1(I), n 1,2(II))
To assess optimal, stress and lethal culture conditions for Mesotaenium endlicherianum SAG 12.97 three pre-experiments were performed (n1(I), performed once, and n1,2(II), performed twice). We assessed Mesotaenium performance in Woods Hole Medium (WHM)82,83 and C-medium81 for an average of 23.6 days in aerated culture glass flasks (SCHOTT) at 80 µmol photons m−2 s−1 (12h/12h light/dark cycle (light from 6 am to 6 pm, Central European winter time) at 18 °C). Before the experiment, different cell densities were analysed using a microplate reader and adjusting the culture to Abs680nm 0.06 (n1(I) or 0.12 (n1,2(II)) (Epoch Microplate reader, BioTek Instruments). For the gradient table setup, the algal suspension was distributed across 504 wells (42 12-well plates (tissue culture testplates 12 no. 92412, TPP); 2.5 ml of culture per well). Plates were sealed with surgical Micropore tape (3M) to minimize evaporation. The 42 12-well plates were then placed on a table that generates a cross-gradient of temperature (for n1(I): 12.7 ± 1.0 °C to 34.0 ± 0.8 °C on the x axis; for n1,2(II): 8.6 ± 0.5 °C to 29.2 ± 0.5 °C on the x axis) and irradiance (21.0 ± 2.0 to 527.9 ± 14.0 µmol photons m−2 s−1 on the y axis) (Supplementary Table 1 and Supplementary Fig. 1a). The temperature gradient was generated using a custom-made table (Labio) equipped with true-daylight LEDs (sTube 2 W 120 ver 11:11, Snaggi) set to a 16 h/8 h light/dark cycle (light from 6 am to 10 pm, Central European winter time). Mesotaenium samples were exposed to the 504 different conditions either for 191 h (n1,2(II)) or 216 h (n1(I)) (for performing physiological measurements) and 216 h (n1,2(II)) (for absorption spectra measurements). Condensed water at the top of the 12-well plates lids was removed by lightly tapping the lids twice.
Plate reader
In vivo absorbance at 480, 680 and 750 nm of all 42 plates was measured after 65 h exposition (4–6 h after light on) with an absorbance microplate reader Epoch (BioTek Instruments). Nine data points per well were analysed and averaged using Gen5 2.0 software (Biotek), resulting in 108 measurements per 12-well plate per wavelength. For downstream analyses, these values were averaged resulting in one value per 12-well plate per wavelength (Supplementary Fig. 1). After 89 h exposition, 16 plates were chosen from the prominent gradients (the four most extreme conditions in the corners and a cross of vibrant growth along the two gradients) for analysing a full absorption spectrum (300–900 nm) using the same setup (Supplementary Fig. 9 and Supplementary Table 10).
Photophysiological measurements
For maximum quantum yield measurements (Fv/Fm) the maxi version of the IMAGING-PAM (ImagMAX/L, M-series, Walz) with an IMAG-K5 CCD camera, controlled with the ImagingWinGigE (V2.32) software, was used. The Mesotaenium cultures in the 12-well plates were dark adapted for 10–30 min before measuring. Before measurements, the lid was removed. For the Fv/Fm measurement, a short saturation pulse (intensity 3) was applied. The measurement settings on the IMAGING-PAM were as follows: measuring light 1, gain 3, damping 2 and mean over area of interest was turned off. No special saturation pulse routine was applied to modify the signal-to-noise ratio of the chlorophyll fluorescence measurement.
Statistical analysis of absorption and Fv/Fm values and temperature/light cluster analysis
Statistical analysis of the absorption and the Fv/Fm values was done using a Kruskal–Wallis test with a post hoc Fisher’s least significant difference test84 using R (version 4.1.3). P values were Bonferroni corrected and grouped into significant groups using R packages ‘agricolae’ version 1.3–5 and ‘dplyr’ version 1.0.9. For heat map generation of physiological values plotted against temperature or light, the R package ‘pheatmap’ version 1.0.12 was used. For cluster analysis, the R package ‘factoextra’ version 1.0.7 was used. Clusters were generated using the eclust function with clustering function ‘kmeans’ with the number of clusters set to six and for hierarchical clustering; ‘euclidean’ was used as the distance measure. Clusters were visualized with PCA in R.
RNA extraction and sequencing
After absorption measurements, the 12-well plates were put back on the table to let cells adjust to the table conditions again for a minimum of 5 min before collecting them. For RNA extraction 0.4 ml was taken from every well of the 42 12-well plates on the table after pipetting the cells up and down twice to homogenize them. In total, 4.8 ml liquid culture was taken per condition on the table (that is, pooling 0.4 ml of each 12 wells per each of the 42 conditions). The samples were then centrifuged for 5 min at 20 °C and 4,200g. The supernatant was removed and the pellet was frozen at −80 °C. To extract RNA, the Spectrum Plant Total RNA Kit (STRN250-1KT, Sigma-Aldrich) was used according to the manufacturer’s instructions. For cell disruption, samples in lysis buffer were ultrasonicated for 1 min and vortexed. RNA samples were treated with DNAse I (EN0521; Thermo Fisher) and shipped on dry ice to Novogene where they were quality checked with a Bioanalyzer (Agilent Technologies). Libraries were built on the basis of total RNA using poly-T oligo-attached magnetic beads. Following fragmentation, synthesis of the first-strand complementary DNA was carried out using random hexamer primers and second-strand cDNA using dUTP, instead of dTTP. A directional size-selected library was built that included PCR-based amplification. Sequencing adaptors were 5′ adaptor: 5′-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT-3′ and 3′ adaptor: 5′-GATCGGAAGAGCACACGTCTGAACTCCAGTCACGGATGACTATCTCGTATGCCGTCTTCTGCTTG-3′. The library was sequenced on an Illumina NovaSeq 6000 platform and data were downloaded using wget (GNU Wget 1.14).
Quality control of reads
We checked the quality of our raw reads via FastQC84 (v0.11.9) and summarized the results via MultiQC85 (v1.11). On the basis of these and the used adaptor sequence, we filtered and trimmed reads via Trimmomatic86 (v 0.36) with these parameters: (‘ILLUMINACLIP: novogene_adapter_sequences_Trimmomatic.fa:2:30:10:2:True LEADING:26 TRAILING:26 SLIDINGWINDOW:4:20 MINLEN:36′). We checked the quality of the trimmed reads with FastQC and MultiQC again.
Genome annotation
The original annotation of M. endlicherianum8 had a lower number of genes compared with other Zygnematophyceae algae. We took advantage of our newly generated RNA-seq dataset to improve genome annotation. Trimmed reads were mapped via HISAT2 (ref. 87) and assembled via StringTie87. The StringTie results showed many novel isoforms as well as novel transcripts. We also used BUSCO V5 (ref. 88) to measure the completeness of the gene models in annotation V1 independent of StringTie. Although the gene prediction method used by BUSCO at the genome level is very efficient, it is not unexpected if it misses some proteins that were annotated in a genome via experimental data, based on bioinformatic methods and next-generation sequencing data, or ab initio-based gene prediction methods. Therefore, we expect that the BUSCO score based on the proteins of a gene model should be equal to or greater than the BUSCO score of the genome. When we compared the BUSCO score between the genome and protein sequences for M. endlicherianum with ‘viridiplantae.odb.10-2020-09-10’, we noticed that they show similar numbers (Supplementary Fig. 2). Therefore, we decided to re-annotate the genome of M. endlicherianum with our comprehensive RNA-seq datasets as well as public protein and genome sequences published for its close relatives.
We annotated the M. endlicherianum genome using REAT (v0.6.1). Various gene models were predicted based on different types of evidence and methods. The final gene models and annotation V2 were based on agreement with the experimental evidence. At the end, we tried to quantify ‘completeness’ and quality of the new annotation V2 and the old V1.
First, we used RNA-seq evidence with REAT’s ‘Transcriptome Workflow’ with HISAT2 (v2.2.1), Scallop89 (v0.10.5) and StringTie (v2.1.5). We also used Portcullis90 (v1.2.4) to identify genuine junctions based on short reads alignments. This workflow uses Mikado91 (v2.3.4) to identify the ‘best’ set of transcripts from multiple transcript assemblies.
Then, we used gene homology information from representative streptophytes in REAT’s ‘Homology Workflow’. SPALN92,93 (v2.4.7) was used to align representative protein sequences onto the M. endlicherianum genome. The representative dataset consisted on genome, gene models and protein sequences of Anthoceros agrestis94 (Oxford strain), Arabidopsis thaliana95, Azolla filiculoides96, Chara braunii72, Chlorokybus melkonianii97 (for naming, see ref. 98), Chlamydomonas reinhardtii99 (v5.6), Klebsormidium nitens100, Mesostigma viride101, Marchantia polymorpha102 (v6.1r1), Penium margaritaceum11, Physcomitrium patens103 (v3.3), Selaginella moellendorffii104 and Spirogloea muscicola8. We also used the junction file produced by Portcullis. Since there were no close relatives of M. endlicherianum on the SPALN species-specific parameter set, we used three different closest possibilities (Angiosp, Chlospec and MossWorts) and built three models. These alignments are filtered using a predefined set of criteria (compare code on GitHub) including exon length, intron length and internal stop codon, among others. The final gene models of V2 were prepared by Mikado.
Afterwards, we used REAT’s ‘Prediction Workflow’ to predict gene models ab initio and based on RNA-seq and homology evidence. This uses Augustus105–107 (v 3.4.0), SNAP108 (version 2006-07-28), Glimmer109 (v0.3.2) and CodingQuarry110 (v2.0), which generate different gene models as the raw material for EVidenceModeler111 (v1.1.1) that chooses the best set of exons and combine them in a gene model using weights (see GitHub) that could be adjusted for each sort of prediction and evidence. To include untranslated regions where possible, the EVidenceModeler output is then processed by Mikado using untranslated region-containing gene models from the transcriptome and homology workflows as inputs, as well as gene models classified by REAT as gold, silver and bronze based on their agreement with the set of protein sequences from other streptophyte genomes (streptophyte algae and land plants), transcriptome alignment, homology alignment and junctions. To train ab initio predictors, a user-defined number of models are randomly chosen in a user-defined ratio between mono-exonic (10%) and multi-exonic (90%). These models were chosen from best-classified models (gold and silver). For Augustus, we performed meta parameter optimization and train a model with kfold of 8. Beside ab initio predictions, we used Augustus to predict gene models with three different weights for each evidence type as suggested by REAT authors (compare code on GitHub).
At last, we used Minos112 (v1.8.0), which is gene model consolidation pipeline and produces external metrics based on DIAMOND113 (v0.9.34) ‘BLASTp/BLASTx’, Kallisto114 (v0.46.2) expression quantification, coding potential calculator115 (CPC2 v0.1) and BUSCO assessments. These metrics pass through Mikado in combination with various gene models produced with different methods (as mentioned above); Minos determines the best gene model for each region based on user-defined criteria (for details, see GitHub) and external metrics. Minos also puts a tag on each gene model to categorize them based on a user-defined threshold (we used default values) for sequence similarity coverage of homologues, BUSCO score, coding potential calculator score, transcript per million expression and transcript score into ‘high confidence’, ‘low confidence’ and ‘predicted genes’.
Genome annotation assessment
We used two methods to compare the quality of the new gene model with the published one. We compared the BUSCO scores of the annotated protein sequences as well as genome sequence using the reference ‘viridiplantae.odb.10-2020-09-10’ dataset. We also used maker116 (v3.01.04) to calculate the AED117 to evaluate the agreement of the gene models with external evidences. Maker-P was used to build the M. endlicherianum gene model V1.
Further, we used the maker package to perform functional annotation via InterProScan and BLAST using the agat118 package (v0.9.2). Additionally, we performed a BLAST (v2.11.0+) search against A. thaliana protein sequences (Araport11) and reported the best hit for each sequence (Supplementary Table 11) and used eggNOG-mapper119,120 (v2.1.8) to perform functional annotation. We used DIAMOND113 (v2.0.15) with ultra-sensitive mode, with e value cut-off of 1e−7 and in an iterative manner. We used the protein sequences as our inputs and Viridiplantae (33090) as our taxonomy scope.
RNA-seq analysis: pseudoalignment
To quantify gene expression, we used a Snakemake-managed pipeline (7.7.0) that hinged on Kallisto114 (v0.45.0). We indexed the transcriptome file with —kmer-size=31 parameter, and used —bootstrap-samples 100 and —rf-stranded to quantify gene expression based on pseudo-aligned reads. We used MultiQC to obtain an overview of alignment for each condition.
Filtering, normalization, modelling mean–variance relationship and data exploration
Kallisto quantification files were imported into R (v4.2.0; tidyverse v1.3.1) with tximport121 (v1.24.0) to calculate the counts from abundance via ‘lengthScaledTPM’ based on our study design file (Supplementary Table 12). We used edgeR122 (v3.38.1) for filtering and trimmed mean of M-values normalization123 of the reads (genes with >1 count per million at log2 scale in at least three samples—the number of replicates—were kept). Then, we used the voom function from limma124–127 (v3.52.2) to model the mean–variance relationship. The normalized expression table on the log2 scale is available in Supplementary Table 13. We performed PCA based on the expression table output of voom and visualized the result with ggplot2 (ref. 128) (v3.3.6). We visualized the heat map of distance and Spearman correlation between all samples considering all genes via pheatmap (v1.0.12), and calculated clusters via the Euclidian method.
RNA-seq analysis: WGCNA
We used the WGCNA28,129 package (v1.71) with the expression table produced by limma. We checked for and filtered out outliers as suggested by WGCNA authors (Supplementary Fig. 10). Then, we visualized the scale-free topology model fit (R2) against the soft thresholds (β) to pick a β for our network construction (Supplementary Fig. 11). We used signed network type and ‘bicor’ as our correlation function for WGCNA. On the basis of these results, we picked 16 as our soft threshold ‘β’. We experimentally chose a merging threshold of 0.25 after exploring different values from 0.2 to 0.4 and investigating the relationship between eigengenes and temperature, light intensity, Fv/Fm and absorption (Supplementary Fig. 12). We built the gene co-expression network using a merging threshold of 0.25 for modules, maximum portion of outliers as 0.05 and minimum module size of 30. Then, we visualized the correlation between each module’s eigengene and temperature, light intensity, Fv/Fm and absorption to identify which modules are more related to each treatment (Fig. 4c). We provided a table for all genes, their module assignment, inter- and intramodular connectivity, gene significance for temperature and light intensity, correlation with temperature and light intensity, and their module membership (that is, signed eigengene-based connectivity) (Supplementary Table 5). We also visualized the graphical representation of the topological overlap matrix (TOM) of our samples (Supplementary Fig. 13). To have a visual representation of gene expression in each module, we drew heat maps for each module via pheatmap (using the Euclidean method for calculating the distance and complete method clustering) (Supplementary Fig. 14). GO enrichment analysis was performed via the clusterProfiler package130,131 (v4.4.4) using the output of eggNOG-mapper and adjusted P value cut-off of 0.05 and q value cut-off of 0.05, considering only genes that are present in our GO term-to-gene table, which was expressed and passed filtering as our background gene universe (Supplementary Table 6). Determining the proper background gene list has major importance in enrichment analysis132.
To see how A. thaliana’s well-known genes in stress-response mechanisms (downloaded from the TAIR database via keyword search) were distributed across different modules, we performed BLASTp searches against the new M. endlicherianum annotated proteins. We visualized the distribution of these IDs for different stress-related keywords (Supplementary Fig. 15) and the expression of these genes across different samples via pheatmap (Supplementary Fig. 16). We defined as module hubs the top 20 genes (nodes) with the highest connectivity within each module (Supplementary Tables 5 and 14).
Differential gene expression analysis
We performed differential gene expression analysis using the limma package. We divided samples into multiple groups as follows: low light intensity (21 and 39 µmol photons m−2 s−1), medium light intensity (72 and 129 µmol photons m−2 s−1), high light intensity (329 and 527 µmol photons m−2 s−1), low temperature (8 and 12 °C), medium temperature (17, 20 and 23 °C) and high temperature (26 and 29 °C; see grid/coloured table layout in Fig. 2). We performed all-against-all comparisons and an additional comparison of those samples from an Fv/Fm < 0.5 versus low light intensity + medium temperature. We used duplicateCorrelation as suggested by Smyth et al.133 to consider technical replicates. We used cluster Profiler for GO enrichment analysis131 with an adjusted P value and q value cut-off of 0.01 and only genes that were expressed and passed filtering as our background universe. The heat map of gene expression profiles, dot plot and cnetplot of enriched GO terms for each comparison is available in Supplementary Table 14 and Supplementary Figs. 17–25).
Phylogenetic analyses
We assembled a protein database based on the protein releases from the genomes of: Anthoceros agrestis BONN94, Anthoceros punctatus94, Amborella trichopoda134, Arabidopsis thaliana135, Azolla filiculoides96, Bathycoccus prasinos136, Brassica oleracea137, Brassica rapa138, Brachypodium distachyon139, Capsella grandiflora140, Chara braunii72, Chlorokybus melkonianii97 (for naming, see ref. 98), Chlamydomonas reinhardtii99, Coccomyxa subellipsoidea141, Gnetum montanum142, Klebsormidium nitens100, Marchantia polymorpha143, Mesostigma viride97, Micromonas pusilla144, Micromonas sp.144, Oryza sativa145, Picea abies146, Physcomitrium patens103, Salvinia cucullata96, Selaginella moellendorffii104, Solanum lycopersicum147, Theobroma cacao148, Mesotaenium endlicherianum8, Ostreococcus lucimarinus149, Penium margaritaceum11, Spirogloea muscicola8, Ulva mutabilis150, Volvox carteri151, Isoetes taiwanensis152 and Ceratopteris richardii153.
Homologues for proteins were detected using BLASTp with Arabidopsis and Mesotaenium proteins as query against the aforementioned proteins as database. Alignments were computed using MAFFT v7.490 (ref. 154). All phylogenies were computed with IQ-TREE155 multicore version 1.5.5; their respective best-fit model for protein evolution was determined using ModelFinder156 (integrated in IQ-TREE multicore version 1.5.5 for Linux 64-bit built 2 June 2017) according to Bayesian Information Criterion; and 1,000 ultrafast bootstrap157 pseudoreplicates were carried out and 100 non-parametric bootstrap158 pseudoreplicates for the LDAP phylogeny. We coloured phylogeny trees via ggtree (v3.9.0).
DIC and confocal laser scanning microscopy
Differential interference contrast (DIC) imaging was done for all replicates from the table with an Olympus BX-60 microscope (Olympus, Japan) with a ProgRes C14plus camera and the ProgRes CapturePro Software (version 2.9.01) (JENOPTIK AG). The morphology of chosen conditions (Fig. 1, Extended Data Figs. 4–6 and Supplementary Fig. 1) of Mesotaenium cells that were 89 h on the table was analysed.
For algae that were used for quantifying the abundance of LD per cell, a ZEISS Axioscope 7 microscope (Carl Zeiss) was used including the Zen software (Carl Zeiss). The LD count was carried out in Fiji159. For statistical analysis of the LD count data, we first used a Shapiro–Wilk test160 to assess normality and used Mann–Whitney U tests161 with R (version 3.6.1) accordingly.
Confocal laser scanning microscope was done on a Zeiss LSM780 (Carl Zeiss) set as in Müller et al.162. For the staining of the LD structures, we used the neutral lipid specific stain BODIPY 493/503 (EM/EX) (Merck). Mesotaenium cells were grown for 22 days on WHM medium at 70–80 µmol photons m−2 s−1 and 22 °C. These cells were ultrasonicated for 1 min with 1:500 BODIPY and incubated on a shaker for 5 min before visualization.
LD isolation and proteomics
For LD isolation 23-day-old Mesotaenium cells grown on WHM medium at 70–80 µmol photons m−2 s−1 and 22 °C were homogenized using a Tenbroeck or potter homogenizer in LD isolation buffer (10 mM sodium phosphate buffer pH 7.5, 200 µM phenylmethylsulfonyl fluoride, 0.5 mM dithiobis(succinimidyl propionate) and 10 mM N-ethylmaleimide). The resulting centrifuged supernatant of a 100g spin for 1 min was considered as TE. After two further high-speed centrifugations (SW40 Ti for 1 h, 4 °C at 100,000g, TLA120 for 1 h at 100,000g and 4 °C) the floating fat pad was precipitated at −20 °C using 100% ethanol overnight. The precipitated pellet was washed with 80% ethanol twice, dried and then suspended in 6 M urea. Protein concentration was determined using the bicinchoninic acid assay. An in-gel sodium dodecyl sulphate gel digestion was done with trypsin adapted from Shevchenko et al.163. C18 Stage tip purification was done according to Rappsilber et al.164,165. Protein samples were analysed using LC–MS. For this, peptide samples were reconstituted in 20 µl LC–MS sample buffer (2% acetonitrile and 0.1% formic acid). Then, 2 µl of each sample were subjected to reverse-phase liquid chromatography for peptide separation using an RSLCnano UltiMate 3000 system (Thermo Fisher Scientific). Peptides were loaded on an Acclaim PepMap 100 pre-column (100 µm × 2 cm, C18, 5 µm, 100 Å; Thermo Fisher Scientific) with 0.07% trifluoroacetic acid at a flow rate of 20 µl min−1 for 3 min. Analytical separation of peptides was done on an Acclaim PepMap RSLC column (75 µm × 50 cm, C18, 2 µm, 100 Å; Thermo Fisher Scientific) at a flow rate of 300 nl min−1. The solvent composition was gradually changed within 94 min from 96% solvent A (0.1% formic acid) and 4% solvent B (80% acetonitrile and 0.1% formic acid) to 10% solvent B within 2 min, to 30% solvent B within the next 58 min, to 45% solvent B within the following 22 min and to 90% solvent B within the last 12 min of the gradient. All solvents and acids had Optima grade for LC–MS (Thermo Fisher Scientific). Eluting peptides were on-line ionized by nano-electrospray using the Nanospray Flex Ion Source (Thermo Fisher Scientific) at 1.5 kV (liquid junction) and transferred into a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Full scans in a mass range of 300–1,650 m/z were recorded at a resolution of 30,000 followed by data-dependent top ten higher energy collisional dissociation fragmentation at a resolution of 15,000 (dynamic exclusion enabled). LC–MS method programming and data acquisition was performed with the XCalibur 4.0 software (Thermo Fisher Scientific). Afterwards, the raw proteome data were analysed using Max Quant software166 version 1.6.2.10. The database for this analysis was our new V2 gene model data. The data were then further processed by the Perseus (1.6.2.2) software166,167.
Lipid analysis of LDs
LDs (200–300 µl) were extracted with 10 ml of methanol/chloroform/formic acid (20:10:1, vol/vol/vol), 5 ml of 0.2 M phosphoric acid and 1 M potassium chloride168. After vortexing and centrifugation at 50g for 2 min, the lower chloroform phases were dried under streaming nitrogen and redissolved in chloroform/methanol (2:1, vol/vol). For analytical analysis, one-fifth of the lipid extracts were spotted on a thin layer chromatography (TLC) silica plate (TLC Silica gel 60, 20 × 20 cm, Merck KGaG) and separated with petroleum ether/diethyl ether/acetic acid (70:30:0.5, vol/vol/vol)169. The lipid composition was identified after incubation in copper sulphate solution (0.4 M CuSO4 in 6.8 % (vol/vol) phosphoric acid) and heating at 180 °C. For preparative analysis, half of the lipid extracts were additionally separated by TLC. After development, the lipid spots were visualized after spraying with 0.05% (wt/vol) primuline in 80% (vol/vol) acetone. The silica gel spots containing TAG, diacylglycerol (DAG) and free fatty acids (FFA) were used for preparation of fatty acid methyl esters as already described170 with some modifications. For acidic hydrolysis, 1 ml of methanol/toluene (2:1, vol/vol) containing 2.75% (vol/vol) sulphuric acid (95–97%) and 2% (vol/vol) dimethoxypropane was added to the scraped silica gel. For quantification, 1 µg of tripentadecanoate was added and the samples were incubated for 1 h at 80 °C. To extract the resulting fatty acid methyl esters, 1.5 ml of saturated aqueous sodium chloride solution and 1.2 ml of hexane were added and centrifuged at 450g for 10 min. The hexane phase was dried under streaming nitrogen and redissolved in 10 µl acetonitrile. Gas chromatography (GC) analysis was performed with an Agilent (Waldbronn, Germany) 6890 gas chromatograph fitted with a capillary DB-23 column (30 m × 0.25 mm; 0.25 µm coating thickness; J&W Scientific, Agilent) modified from Hornung et al.171. Helium was used as carrier gas at a flow rate of 1 ml min−1. The temperature gradient was 150 °C for 1 min, 150–200 °C at 4 K min−1, 200–250 °C at 5 K min−1 and 250 °C for 6 min. The peak area was collected with the ChemStation software (Agilent). From the absolute fatty acid contents, relative fatty acid profiles for TAG, DAG and FFA were calculated.
Pollen tube transformation and microscopy
Coding sequences for Mesotaenium homologues salient to LD biology were MeCaleosinf 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCATGTCGAAGCTCAGTCTTGCC-3′, MeCaleosinr 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGACTGCTTCTTCCTCTGCTT-3′, MeLDAPf 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCATGGCCGAAAGTCAGGGCCC-3′, MeLDAPr 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCGACTTCTTGAGGGCGTCGGC-3′, MeSteroleosinf 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCATGGGGTTACTTAATGCCCTTGC-3′, MeSteroleosinr 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGCCATTGGACTTGACGAGGG-3′, MeOleosinf 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCATGCCTCAGGATCAGCAGCAAG-3′, and MeOleosinr 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTTCCTCTCCTTCTCAACCTTGT-3′. Constructs for expression in pollen tubes were cloned into the pLatMCC-GW vector using the fast Gateway method as described previously162. Pollen transformation, pollen tube growth and fixation were also performed according to this protocol. LDs were stained with BODIPY 493/503 at a final concentration of 1.3 µg ml−1. Microscopy images of transformed tobacco pollen tubes were acquired using an LSM 980 confocal laser scanning microscope using the objective C-Apochromat 40×/1.20 W Korr (both Carl Zeiss). mCherry was excited at 561 nm and detected at 600–640 nm. BODIPY 493/503 was excited at 488 nm and detected at 490–535 nm. In both cases, the major beam splitter MBS 488/561 was used. Both channels were recorded independently using the line mode.
Comparative evolutionary analyses
To perform comparative evolutionary analyses among M. endlicherianum, other streptophyte algae and embryophytes, we used two separate workflows based on one criterion: the availability of at least 15 raw RNA-seq samples for a given species challenged with abiotic stresses and control conditions. This is the minimum requirement to build a co-expression network using the WGCNA package. If a species passed this criterion, we used them in two approaches; all results from the comparative analyses can be found in Supplementary Table 15.
Approach 1: to compare co-expression networks computed based on control and abiotic stress samples, we first used Orthofinder and protein sequences of A. agrestis94, A. filiculoides96, A. thaliana135, B. distachyon139, C. braunii72, Closterium sp. NIES 67 (ref. 10), C. melkonianii97, C. reinhardtii99, K. nitens100, M. endlicherianum8, M. polymorpha102, M. viride97, O. sativa145, P. margaritaceum11, P. patens103, S. lycopersicum147, S. moellendorffii104, S. muscicola8, Z. mays172 and Zygnema circumcarinatum9 as well as a species cladogram to find phylogenetic HOGs using these parameters: -S mmseqs -M msa -A mafft -s species_tree.txt -y. For A. thaliana, we downloaded a gene-GO table from arabidopsis.org. For P. patens, M. polymorpha and Zygnema 1b, we used eggNOG-mapper and their protein sequences to create a gene-GO table using these parameters: -m diamond -dmnd_iterate yes -evalue 1e-10 -sensmode ultra-sensitive -tax_scope 33090. We downloaded raw RNA-seq reads for A. thaliana173–177, P. patens178 and accessions PRJNA277025 and PRJNA192876, M. polymorpha179–181 and Zygnema 1b (ref. 9) from the National Center for Biotechnology Information (NCBI). We followed the same quantification as Mesotaenium for each species here. In short, we used FastQC, MultiQC and Trimmomatic to check the quality of each read and filter and trim the raw reads. Then, we used Kallisto to pseudoalign the reads to the transcriptome of that species. Then, we imported gene counts for each species into R and performed similar exploratory analyses to Mesotaenium for each species. An additional layer of analysis here was to check for batch effect when we looked at all samples from different sources for a species. We used hierarchical clustering and PCA to pick the best expression profile from (i) uncorrected, (ii) batch-corrected as a covariate using limma, and (iii) batch-corrected using ComBat-seq182 to adjust for batch effects (if there were any). There is a debate in the community about which method is the best practice; therefore, we did all for every species and picked the best (less confounding effect between batches and maximum similarity between similar conditions) for each species. Then, we used the expression profile and built a signed co-expression network using the WGCNA package for each species. We followed the same procedure as Mesotaenium. We performed a GO enrichment analysis for each module in the co-expression networks. Then, we used the Orthofinder-based orthogroups to find genes that have a counterpart in Mesotaenium for each species and then we calculated the Jaccard similarity and dissimilarity between each Mesotaenium modules and each module of A. thaliana, P. patens, M. polymorpha and Zygnema circumcarinatum SAG698-1b. For each module in these co-expression networks, we looked for the connectivity of genes that share a HOG with Mesotaenium hubs.
Approach 2: to determine the shared DEGs under abiotic stresses across streptophyte algae, we first downloaded raw RNA-seq reads from NCBI as follows: (1) Mougeotia24,25 sp. MZCH240 and S. pratensis MZCH10213 (ref. 24), (2) M. viride, C. cerffii, K. flaccidum, C. globularis, C. scutata, Zygnema ‘cylindricum’20 SAG698-1a, (3) Zygnema sp.23 SAG2419, (4) S. pratensis22 UTEX92 and (5) Z. circumcarinatum9. If it was possible, we also obtained the transcriptome or genome file for each species. Then, we used Orthofinder and protein sequences of Mesotaenium and the protein sequences of these species as well as a species cladogram to find phylogenetic hierarchical orthogroups using these parameters: -S mmseqs -M msa -A mafft -s species_tree.txt -y. For species for which only the transcriptome was available, we used TransDecoder (v5.7.0) using TransDecoder.LongOrfs and TransDecoder.Predict scripts to get a protein-coding sequence for our Orthofinder run. For those that we did not have a transcriptome, we built one using Trinity (v2.15.1) (ref. 183) and the settings -seqType fq –trimmomatic. We followed the same quantification steps as for Mesotaenium and workflow A to pseudoalign reads to the transcriptome. Then, we followed similar steps to Mesotaenium to calculate DEGs for each species. Finally, we compared these DEGs with DEGs in Mesotaenium using HOGs from Orthofinder run in this workflow. We used BioNERO package184 to aggregate log2(fold change) values for each gene in each species to the corresponding HOGs and then used cluster Profiler to perform GO enrichment analyses and visualized the heat maps. In all comparisons, we considered adjusted P values <0.05 as significant enrichment.
TEs
We used InterProScan185 (v5.59-91.0) on all predicted proteins in Mesotaenium endlicherianum V2 and filtered the results for transposon-related domains. This resulted in 6,186 entries in 1,748 unique gene IDs, among which only 96 were expressed in our RNA-seq data (that is, passing an expression cut-off of at least 1 count per million in at least three samples); all results are presented in Supplementary Table 4b).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Overview of supplementary contents and Supplementary Figs. 1–28.
Supplementary Tables 1–15.
Acknowledgements
We thank R. Heise for excellent technical support. J.d.V. thanks the European Research Council for funding under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 852725; ERC-StG ‘TerreStriAL’). J.d.V., U.H., I.F. and H.B. are grateful for support through the German Research Foundation (DFG), on the grant SHOAL (514060973; VR132/11-1) and within the framework of the Priority Programme ‘MAdLand – Molecular Adaptation to Land: Plant Evolution to Change’ (SPP 2237; 440231723 VR 132/4-1; BU 2301/6-1; HO 2793/5-1; FE 446/14-1), in which T.P.R. and M.H. are PhD students and A.D., J.M.R.F.-J and I.I. partake as associate members. A.D. is grateful for being supported through the International Max Planck Research School (IMPRS) for Genome Science. J.M.R.F.-J. and T.P.R. gratefully acknowledge support by the PhD programme ‘Microbiology and Biochemistry’ within the framework of the ‘Göttingen Graduate Center for Neurosciences, Biophysics, and Molecular Biosciences’ (GGNB) at the University of Goettingen. P.S. was supported by the GGNB in frame of the PRoTECT programme at the University of Goettingen. T.I. acknowledges funding from DFG (GRK 2172-PRoTECT). M.M. is supported by Singaporean Ministry of Education grant T2EP30122-0001. P.S. is grateful for support from the Studienstiftung des Deutschen Volkes. This work was further supported by the DFG through the infrastructure grant INST 211/903-1 FUGG for the used confocal microscope as operated by the Imaging Network of the University of Münster (RI_00497). We thank C. Gatz and G. Kriete for giving us access to the ImagMAX/L PAM in the Department of Plant Molecular Biology and Physiology. We are grateful to T. Friedl for supporting us with access to the facilities of the Department of Experimental Phycology and SAG Culture Collection of Algae, including cultivation facilities, the Olympus BX-60 microscope and the absorbance microplate reader Epoch (BioTek Instruments).
Extended data
Source data
Whole scan of the TLC, shown in Fig. 6g.
Author contributions
J.d.V. and M.L. conceived the project. J.d.V. coordinated the project with M.M. M.L. provided the plant material. J.M.R.F.-J., T.D., S.S., M.L., and T.P.R. performed the experimental work. A.D. carried out the computational analysis. O.V., J.M.R.F.-J., P.S., T.I., D.K. and G.H.B. performed the proteomics. D.K., C.H. and I.F. performed the lipid profiling. H.B. investigated the cell division patterns. M.H. and U.H. investigated the photomorphogenesis patterns. A.D. and R.S. built the web resources. J.d.V., A.D. and J.M.R.F.-J. contributed to writing the manuscript. J.d.V. organized the manuscript. All authors commented, discussed and provided input on the final manuscript.
Peer review
Peer review information
Nature Plants thanks Xin Liu, Mingbing Zhou and Haim Treves for their contribution to the peer review of this work.
Data availability
All RNA-seq reads have been uploaded to the NCBI Sequence Read Archive and can be accessed under BioProject PRJNA832564 and Sequence Read Archive accessions SRR18936040 to SRR18936170. Furthermore, data can be interactively explored at https://mesotaenium.uni-goettingen.de and proteomic data have been uploaded to EMBL-EBI PRIDE (accession PXD037847). On Zenodo, we have deposited (1) raw light and confocal micrographs generated, for example, for LD assessment in Mesotaenium and pollen tubes 10.5281/zenodo.7921367 and (2) raw and visualized phylogenetic data 10.5281/zenodo.7950653. The additional previously published RNA-seq datasets that were used for comparisons are: (1) A. thaliana: SRR2302908 to SRR2302919, ERR754084, ERR754066, ERR754077, ERR754069, ERR754087, ERR754064, ERR754059, SRR7659142, SRR7659143, SRR7659144, SRR7659145 to SRR7659150, SRR5197904, to SRR5197909; (2) M. polymorpha: SRR12076853, SRR12076855, SRR12076857, SRR12076859, SRR12076861, SRR12076863, SRR12076865, SRR12076867, SRR12076869, SRR12076871, SRR12076873, SRR12076875, SRR12076877, SRR12076879, SRR12076917 to SRR12076925, SRR15186078 to SRR15186125, DRR093991 to DRR093996; (3) P. patens: SRR1824306 to SRR1824320, SRR10235460 to SRR10235483, SRR787291, SRR787292, SRR787293, SRR787294, SRR787295; (4) Z. circumcarinatum SAG698-1b: SRR24939299, SRR24940177, SRR24909175, SRR24757807, SRR24757829, SRR24757830, SRR24757831, SRR24205691 to SRR24205702, SRR24286545 to SRR24286562, SRR24576622, SRR24576623, SRR24385702, SRR24450996, SRR24450997, SRR24451196, SRR24480449, SRR24707416, SRR24707417, SRR24952091, SRR21891679 to SRR21891705; (5) C. cerffii (at the time, C. atmophyticus, see ref. 98): SRR5949009, SRR5949013 to SRR5949016, SRR5949027 to SRR5949030; (6) C. scutata: SRR5948993, SRR5948995 to SRR5948998, SRR5949001, SRR5949004, SRR5949005, SRR5949007; (7) K. flaccidum: SRR5949010, SRR5949011, SRR5949012, SRR5990072 to SRR5990080; (8) M. viride: SRR5949021 to SRR5949026; (9) Mougeotia sp. MZCH240: SRR9083681, SRR9083682, SRR9083688, SRR9083692 to SRR9083701; (10) S. pratensis MZCH10213: SRR9083685, SRR9083686, SRR9083687, SRR9083689, SRR9083690, SRR9083696; (11) S. pratensis UTEX928: SRR4018077 to SRR4018100; (12) Z. circumcarinatum SAG698-1a: SRR5948999, SRR5949000, SRR5949002, SRR5949003, SRR5949006, SRR5949008, SRR5949017, SRR5949018; and (13) Z. circumcarinatum SAG2419: SRR6047298, SRR6047299, SRR6047302 to SRR6047305. Source data are provided with this paper.
Code availability
Codes and data used for genome re-annotation, WGCNA and differential gene expression analysis are available on our GitHub page https://github.com/deVries-lab/Response_to_a_gradient_of_environmental_cues_in_mesotaenium_endlicherianum.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Armin Dadras, Janine M. R. Fürst-Jansen.
Extended data
is available for this paper at 10.1038/s41477-023-01491-0.
Supplementary information
The online version contains supplementary material available at 10.1038/s41477-023-01491-0.
References
- 1.Bar-On YM, Phillips R, Milo R. The biomass distribution on Earth. Proc. Natl Acad. Sci. USA. 2018;115:6506–6511. doi: 10.1073/pnas.1711842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenton TM, et al. Earliest land plants created modern levels of atmospheric oxygen. Proc. Natl Acad. Sci. USA. 2016;113:9704–9709. doi: 10.1073/pnas.1604787113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wodniok S, et al. Origin of land plants: do conjugating green algae hold the key? BMC Evol. Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. USA. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puttick MN, et al. The interrelationships of land plants and the nature of the ancestral embryophyte. Curr. Biol. 2018;28:733–745. doi: 10.1016/j.cub.2018.01.063. [DOI] [PubMed] [Google Scholar]
- 6.One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature574, 679–685 (2019). [DOI] [PMC free article] [PubMed]
- 7.Hess S, et al. A phylogenomically informed five-order system for the closest relatives of land plants. Curr. Biol. 2022;32:4473–4482. doi: 10.1016/j.cub.2022.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng S, et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell. 2019;179:1057–1067.e14. doi: 10.1016/j.cell.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Feng, X. et al. Chromosome-level genomes of multicellular algal sisters to land plants illuminate signaling network evolution. Preprint at bioRxiv10.1101/2023.01.31.526407 (2023). [DOI] [PMC free article] [PubMed]
- 10.Sekimoto H, et al. A divergent RWP‐RK transcription factor determines mating type in heterothallic Closterium. N. Phytol. 2023 doi: 10.1111/nph.18662. [DOI] [PubMed] [Google Scholar]
- 11.Jiao C, et al. The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell. 2020;181:1097–1111.e12. doi: 10.1016/j.cell.2020.04.019. [DOI] [PubMed] [Google Scholar]
- 12.Golicz AA, Batley J, Edwards D. Towards plant pangenomics. Plant Biotechnol. J. 2016;14:1099–1105. doi: 10.1111/pbi.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon SP, et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017;8:2184. doi: 10.1038/s41467-017-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D. Plant pan-genomes are the new reference. Nat. Plants. 2020;6:914–920. doi: 10.1038/s41477-020-0733-0. [DOI] [PubMed] [Google Scholar]
- 15.Umezawa T, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman JL, Briginshaw LN, Fisher TJ, Flores-Sandoval E. Something ancient and something neofunctionalized—evolution of land plant hormone signaling pathways. Curr. Opin. Plant Biol. 2019;47:64–72. doi: 10.1016/j.pbi.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118–122. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carella P, et al. Conserved biochemical defenses underpin host responses to oomycete infection in an early-divergent land plant lineage. Curr. Biol. 2019;29:2282–2294.e5. doi: 10.1016/j.cub.2019.05.078. [DOI] [PubMed] [Google Scholar]
- 19.Rieseberg TP, et al. Crossroads in the evolution of plant specialized metabolism. Sem. Cell Dev. Biol. 2023;134:37–58. doi: 10.1016/j.semcdb.2022.03.004. [DOI] [PubMed] [Google Scholar]
- 20.de Vries J, Curtis BA, Gould SB, Archibald JM. Embryophyte stress signaling evolved in the algal progenitors of land plants. Proc. Natl Acad. Sci. USA. 2018;115:E3471–E3480. doi: 10.1073/pnas.1719230115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, et al. A ligand-independent origin of abscisic acid perception. Proc. Natl Acad. Sci. USA. 2019;116:24892–24899. doi: 10.1073/pnas.1914480116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van de Poel B, Cooper ED, Van Der Straeten D, Chang C, Delwiche CF. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016;172:533–545. doi: 10.1104/pp.16.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rippin M, Becker B, Holzinger A. Enhanced desiccation tolerance in mature cultures of the streptophytic green alga Zygnema circumcarinatum revealed by transcriptomics. Plant Cell Physiol. 2017;58:2067–2084. doi: 10.1093/pcp/pcx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Vries J, et al. Heat stress response in the closest algal relatives of land plants reveals conserved stress signaling circuits. Plant J. 2020;103:1025–1048. doi: 10.1111/tpj.14782. [DOI] [PubMed] [Google Scholar]
- 25.Fürst-Jansen JMR, et al. Submergence of the filamentous Zygnematophyceae Mougeotia induces differential gene expression patterns associated with core metabolism and photosynthesis. Protoplasma. 2022;259:1157–1174. doi: 10.1007/s00709-021-01730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edel KH, Marchadier E, Brownlee C, Kudla J, Hetherington AM. The evolution of calcium-based signalling in plants. Curr. Biol. 2017;27:R667–R679. doi: 10.1016/j.cub.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song J-Y, Leung T, Ehler LK, Wang C, Liu Z. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development. 2000;127:2207–2217. doi: 10.1242/dev.127.10.2207. [DOI] [PubMed] [Google Scholar]
- 30.Kleine T, et al. Acclimation in plants—the Green Hub consortium. Plant J. 2021;106:23–40. doi: 10.1111/tpj.15144. [DOI] [PubMed] [Google Scholar]
- 31.Moreno JC, Mi J, Alagoz Y, Al‐Babili S. Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant J. 2021;105:351–375. doi: 10.1111/tpj.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossini L, Cribb L, Martin DJ, Langdale JA. The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell. 2001;13:1231–1244. doi: 10.1105/tpc.13.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasumura Y, Moylan EC, Langdale JA. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell. 2005;17:1894–1907. doi: 10.1105/tpc.105.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters MT, et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timm S, et al. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell. 2008;20:2848–2859. doi: 10.1105/tpc.108.062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fürst-Jansen JMR, de Vries S, de Vries J. Evo-physio: on stress responses and the earliest land plants. J. Exp. Bot. 2020;71:3254–3269. doi: 10.1093/jxb/eraa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L-J, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarid-Krebs L, et al. Phosphorylation of CONSTANS and its COP 1‐dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 2015;84:451–463. doi: 10.1111/tpj.13022. [DOI] [PubMed] [Google Scholar]
- 39.Ordoñez-Herrera N, et al. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol. 2018;176:1327–1340. doi: 10.1104/pp.17.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai S, et al. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell. 2011;23:961–972. doi: 10.1105/tpc.111.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju C, et al. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants. 2015;1:14004. doi: 10.1038/nplants.2014.4. [DOI] [PubMed] [Google Scholar]
- 42.Kato Y, Sakamoto W. Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 2009;146:463–469. doi: 10.1093/jb/mvp073. [DOI] [PubMed] [Google Scholar]
- 43.Kato Y, Sun X, Zhang L, Sakamoto W. Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 2012;159:1428–1439. doi: 10.1104/pp.112.199042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjögren LLE, Stanne TM, Zheng B, Sutinen S, Clarke AK. Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell. 2006;18:2635–2649. doi: 10.1105/tpc.106.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura K, Kato Y, Sakamoto W. Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiol. 2016;171:2280–2293. doi: 10.1104/pp.16.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfalz J, Liere K, Kandlbinder A, Dietz K-J, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 48.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 49.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez MCS, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 52.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013;51:245–266. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- 53.Chen X, et al. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021;63:53–78. doi: 10.1111/jipb.13061. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Planas-Riverola A, et al. Brassinosteroid signaling in plant development and adaptation to stress. Development. 2019;146:dev151894. doi: 10.1242/dev.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hématy K, et al. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 2007;17:922–931. doi: 10.1016/j.cub.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 57.Schröder F, Lisso J, Lange P, Müssig C. The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 2009;9:20. doi: 10.1186/1471-2229-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelman G, et al. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roudier F, Schindelman G, DeSalle R, Benfey PN. The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiol. 2002;130:538–548. doi: 10.1104/pp.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko J-H, Kim JH, Jayanty SS, Howe GA, Han K-H. Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defence responses in Arabidopsis thaliana. J. Exp. Bot. 2006;57:2923–2936. doi: 10.1093/jxb/erl052. [DOI] [PubMed] [Google Scholar]
- 61.Wolf S, et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl Acad. Sci. USA. 2014;111:15261–15266. doi: 10.1073/pnas.1322979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higashi Y, Okazaki Y, Myouga F, Shinozaki K, Saito K. Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 2015;5:10533. doi: 10.1038/srep10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mueller SP, Krause DM, Mueller MJ, Fekete A. Accumulation of extra-chloroplastic triacylglycerols in Arabidopsis seedlings during heat acclimation. J. Exp. Bot. 2015;66:4517–4526. doi: 10.1093/jxb/erv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gidda SK, et al. Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 2016;170:2052–2071. doi: 10.1104/pp.15.01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doner NM, et al. Arabidopsis thaliana EARLY RESPONSIVE TO DEHYDRATION 7 localizes to lipid droplets via its senescence domain. Front. Plant Sci. 2021;12:658961. doi: 10.3389/fpls.2021.658961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krawczyk HE, et al. Heat stress leads to rapid lipid remodeling and transcriptional adaptations in Nicotiana tabacum pollen tubes. Plant Physiol. 2022;189:490–515. doi: 10.1093/plphys/kiac127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Listenberger LL, Brown DA. Fluorescent detection of lipid droplets and associated proteins. Curr. Protoc. Cell Biol. 2007;35:24.2.1–24.2.11. doi: 10.1002/0471143030.cb2402s35. [DOI] [PubMed] [Google Scholar]
- 68.Kretzschmar FK, et al. Identification of low-abundance lipid droplet proteins in seeds and seedlings. Plant Physiol. 2020;182:1326–1345. doi: 10.1104/pp.19.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lass A, et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin–Dorfman syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 70.James CN, et al. Disruption of the Arabidopsis CGI-58 homologue produces Chanarin–Dorfman-like lipid droplet accumulation in plants. Proc. Natl Acad. Sci. USA. 2010;107:17833–17838. doi: 10.1073/pnas.0911359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guzha A, Whitehead P, Ischebeck T, Chapman KD. Lipid droplets: packing hydrophobic molecules within the aqueous cytoplasm. Annu. Rev. Plant Biol. 2023;74:195–223. doi: 10.1146/annurev-arplant-070122-021752. [DOI] [PubMed] [Google Scholar]
- 72.Nishiyama T, et al. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell. 2018;174:448–464.e24. doi: 10.1016/j.cell.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 73.Zhao C, et al. Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proc. Natl Acad. Sci. USA. 2019;116:5015–5020. doi: 10.1073/pnas.1812092116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Honkanen S, Small I. The GENOMES UNCOUPLED1 protein has an ancient, highly conserved role but not in retrograde signalling. New Phytol. 2022;236:99–113. doi: 10.1111/nph.18318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martín G, et al. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat. Commun. 2016;7:11431. doi: 10.1038/ncomms11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gasulla F, et al. The response of Asterochloris erici (Ahmadjian) Skaloud et Peksa to desiccation: a proteomic approach. Plant Cell Environ. 2013;36:1363–1378. doi: 10.1111/pce.12065. [DOI] [PubMed] [Google Scholar]
- 77.Li-Beisson Y, Thelen JJ, Fedosejevs E, Harwood JL. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019;74:31–68. doi: 10.1016/j.plipres.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 78.de Vries J, Ischebeck T. Ties between stress and lipid droplets pre-date seeds. Trends Plant Sci. 2020;25:1203–1214. doi: 10.1016/j.tplants.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 79.The Culture Collection of Algae at the University of Göttingen, Germany (SAG) (The University of Göttingen); https://sagdb.uni-goettingen.de/detailedList.php?str_number=12.97
- 80.Friedl T, Lorenz M. The Culture Collection of Algae at Göttingen University (SAG): a biological resource for biotechnological and biodiversity research. Procedia Environ. Sci. 2012;15:110–117. doi: 10.1016/j.proenv.2012.05.015. [DOI] [Google Scholar]
- 81.Ichimura, T. Sexual cell division and conjugation-papilla formation in sexual reproduction of Closterium strigosum. In Proc. 7th International Seaweed Symposium 208–214 (Univ. of Tokyo Press, 1971).
- 82.Nichols, H. W. in Handbook of Phycological Methods (ed. Stein J. R.) p. 16–17 (Cambridge Univ. Press, 1973).
- 83.Conover, W. J. Practical Nonparametric Statistics 3rd edn (John Wiley & Sons, 1999).
- 84.Simon, A. FastQC: a quality control tool for high throughput sequence data (Babraham Bioinformatics, Babraham Institute, 2010); https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 85.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021;38:4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shao M, Kingsford C. Accurate assembly of transcripts through phase-preserving graph decomposition. Nat. Biotechnol. 2017;35:1167–1169. doi: 10.1038/nbt.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mapleson D, Venturini L, Kaithakottil G, Swarbreck D. Efficient and accurate detection of splice junctions from RNA-seq with Portcullis. Gigascience. 2018;7:giy131. doi: 10.1093/gigascience/giy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Venturini L, Caim S, Kaithakottil GG, Mapleson DL, Swarbreck D. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. GigaScience. 2018;7:giy093. doi: 10.1093/gigascience/giy093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gotoh O. Direct mapping and alignment of protein sequences onto genomic sequence. Bioinformatics. 2008;24:2438–2444. doi: 10.1093/bioinformatics/btn460. [DOI] [PubMed] [Google Scholar]
- 93.Gotoh O. A space-efficient and accurate method for mapping and aligning cDNA sequences onto genomic sequence. Nucleic Acids Res. 2008;36:2630–2638. doi: 10.1093/nar/gkn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li F-W, et al. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants. 2020;6:259–272. doi: 10.1038/s41477-020-0618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng C-Y, et al. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- 96.Li F-W, et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants. 2018;4:460–472. doi: 10.1038/s41477-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S, et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants. 2020;6:95–106. doi: 10.1038/s41477-019-0560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Irisarri I, et al. Unexpected cryptic species among streptophyte algae most distant to land plants. Proc. Biol. Sci. 2021;288:20212168. doi: 10.1098/rspb.2021.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hori K, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liang Z, et al. Mesostigma viride genome and transcriptome provide insights into the origin and evolution of Streptophyta. Adv. Sci. 2019;7:1901850. doi: 10.1002/advs.201901850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montgomery SA, et al. Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Curr. Biol. 2020;30:573–588.e7. doi: 10.1016/j.cub.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lang D, et al. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2018;93:515–533. doi: 10.1111/tpj.13801. [DOI] [PubMed] [Google Scholar]
- 104.Banks JA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stanke M, Tzvetkova A, Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006;7:S11. doi: 10.1186/gb-2006-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hoff KJ, Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr. Protoc. Bioinformatics. 2019;65:e57. doi: 10.1002/cpbi.57. [DOI] [PubMed] [Google Scholar]
- 108.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kelley DR, Liu B, Delcher AL, Pop M, Salzberg SL. Gene prediction with glimmer for metagenomic sequences augmented by classification and clustering. Nucleic Acids Res. 2012;40:e9. doi: 10.1093/nar/gkr1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Testa AC, Hane JK, Ellwood SR, Oliver RP. CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics. 2015;16:170. doi: 10.1186/s12864-015-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Minos—a gene model consolidation pipeline for genome annotation projects. GitHubhttps://github.com/EI-CoreBioinformatics/minos (2019).
- 113.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 114.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 115.Kang Y-J, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Campbell MS, Holt C, Moore B, Yandell M. Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics. 2014;48:4.11.1–4.11.39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eilbeck K, Moore B, Holt C, Yandell M. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics. 2009;10:67. doi: 10.1186/1471-2105-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dainat, J. et al. AGAT: another gff analysis toolkit to handle annotations in any gtf/gff format. (Version v0.9.2). Zenodo 10.5281/zenodo.6621429 (2022). [DOI]
- 119.Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huerta-Cepas J, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research10.12688/f1000research.7563.2 (2016). [DOI] [PMC free article] [PubMed]
- 122.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu R, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43:e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2022).
- 129.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012;46:1–17. doi: 10.18637/jss.v046.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu G, Wang L-G, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu T, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wijesooriya K, Jadaan SA, Perera KL, Kaur T, Ziemann M. Urgent need for consistent standards in functional enrichment analysis. PLoS Comput. Biol. 2022;18:e1009935. doi: 10.1371/journal.pcbi.1009935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 134.Amborella Genome Project. et al. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 135.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moreau H, et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012;13:R74. doi: 10.1186/gb-2012-13-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu S, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 139.The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 140.Slotte T, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 2013;45:831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 141.Blanc G, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wan T, et al. A genome for gnetophytes and early evolution of seed plants. Nat. Plants. 2018;4:82–89. doi: 10.1038/s41477-017-0097-2. [DOI] [PubMed] [Google Scholar]
- 143.Bowman JL, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304.e15. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 144.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes. Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 145.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nystedt B, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- 147.The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Argout X, et al. The genome of Theobroma cacao. Nat. Genet. 2011;43:101–108. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- 149.Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl Acad. Sci. USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Clerck O, et al. Insights into the evolution of multicellularity from the sea lettuce genome. Curr. Biol. 2018;28:2921–2933.e5. doi: 10.1016/j.cub.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 151.Prochnik SE, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wickell D, et al. Underwater CAM photosynthesis elucidated by Isoetes genome. Nat. Commun. 2021;12:6348. doi: 10.1038/s41467-021-26644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Marchant DB, et al. Dynamic genome evolution in a model fern. Nat. Plants. 2022;8:1038–1051. doi: 10.1038/s41477-022-01226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 159.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 161.Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947;1:50–60. doi: 10.1214/aoms/1177730491. [DOI] [Google Scholar]
- 162.Müller AO, Blersch KF, Gippert AL, Ischebeck T. Tobacco pollen tubes—a fast and easy tool for studying lipid droplet association of plant proteins. Plant J. 2017;89:1055–1064. doi: 10.1111/tpj.13441. [DOI] [PubMed] [Google Scholar]
- 163.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 164.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 165.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 166.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 167.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 168.Wang, Z. & Benning, C. Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp.10.3791/2518 (2011). [DOI] [PMC free article] [PubMed]
- 169.Reich M, et al. Fatty acid metabolism in the ectomycorrhizal fungus Laccaria bicolor. New Phytol. 2009;182:950–964. doi: 10.1111/j.1469-8137.2009.02819.x. [DOI] [PubMed] [Google Scholar]
- 170.Miquel M, Browse J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem. 1992;267:1502–1509. doi: 10.1016/S0021-9258(18)45974-1. [DOI] [PubMed] [Google Scholar]
- 171.Hornung E, et al. Production of (10E,12Z)-conjugated linoleic acid in yeast and tobacco seeds. Biochim. Biophys. Acta. 2005;1738:105–114. doi: 10.1016/j.bbalip.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 172.Jiao Y, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Clauw P, et al. Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 2015;167:800–816. doi: 10.1104/pp.114.254284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu Z, et al. Genome-wide DNA mutations in Arabidopsis plants after multigenerational exposure to high temperatures. Genome Biol. 2021;22:160. doi: 10.1186/s13059-021-02381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Suzuki N, et al. ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE. 2016;11:e0147625. doi: 10.1371/journal.pone.0147625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang L, et al. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020;20:86. doi: 10.1186/s12870-020-2292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang S-S, et al. Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell. 2017;29:1007–1023. doi: 10.1105/tpc.16.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Elzanati O, Mouzeyar S, Roche J. Dynamics of the transcriptome response to heat in the moss, Physcomitrella patens. IJMS. 2020;21:1512. doi: 10.3390/ijms21041512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jahan A, et al. Archetypal roles of an abscisic acid receptor in drought and sugar responses in liverworts. Plant Physiol. 2019;179:317–328. doi: 10.1104/pp.18.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lagercrantz U, et al. DE‐ETIOLATED1 has a role in the circadian clock of the liverwort Marchantia polymorpha. New Phytol. 2021;232:595–609. doi: 10.1111/nph.17653. [DOI] [PubMed] [Google Scholar]
- 181.Wu T-Y, et al. Evolutionarily conserved hierarchical gene regulatory networks for plant salt stress response. Nat. Plants. 2021;7:787–799. doi: 10.1038/s41477-021-00929-7. [DOI] [PubMed] [Google Scholar]
- 182.Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020;2:lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Almeida-Silva F, Venancio TM. cageminer: an R/Bioconductor package to prioritize candidate genes by integrating genome-wide association studies and gene coexpression networks. In Silico Plants. 2022;4:diac018. doi: 10.1093/insilicoplants/diac018. [DOI] [Google Scholar]
- 185.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of supplementary contents and Supplementary Figs. 1–28.
Supplementary Tables 1–15.
Data Availability Statement
All RNA-seq reads have been uploaded to the NCBI Sequence Read Archive and can be accessed under BioProject PRJNA832564 and Sequence Read Archive accessions SRR18936040 to SRR18936170. Furthermore, data can be interactively explored at https://mesotaenium.uni-goettingen.de and proteomic data have been uploaded to EMBL-EBI PRIDE (accession PXD037847). On Zenodo, we have deposited (1) raw light and confocal micrographs generated, for example, for LD assessment in Mesotaenium and pollen tubes 10.5281/zenodo.7921367 and (2) raw and visualized phylogenetic data 10.5281/zenodo.7950653. The additional previously published RNA-seq datasets that were used for comparisons are: (1) A. thaliana: SRR2302908 to SRR2302919, ERR754084, ERR754066, ERR754077, ERR754069, ERR754087, ERR754064, ERR754059, SRR7659142, SRR7659143, SRR7659144, SRR7659145 to SRR7659150, SRR5197904, to SRR5197909; (2) M. polymorpha: SRR12076853, SRR12076855, SRR12076857, SRR12076859, SRR12076861, SRR12076863, SRR12076865, SRR12076867, SRR12076869, SRR12076871, SRR12076873, SRR12076875, SRR12076877, SRR12076879, SRR12076917 to SRR12076925, SRR15186078 to SRR15186125, DRR093991 to DRR093996; (3) P. patens: SRR1824306 to SRR1824320, SRR10235460 to SRR10235483, SRR787291, SRR787292, SRR787293, SRR787294, SRR787295; (4) Z. circumcarinatum SAG698-1b: SRR24939299, SRR24940177, SRR24909175, SRR24757807, SRR24757829, SRR24757830, SRR24757831, SRR24205691 to SRR24205702, SRR24286545 to SRR24286562, SRR24576622, SRR24576623, SRR24385702, SRR24450996, SRR24450997, SRR24451196, SRR24480449, SRR24707416, SRR24707417, SRR24952091, SRR21891679 to SRR21891705; (5) C. cerffii (at the time, C. atmophyticus, see ref. 98): SRR5949009, SRR5949013 to SRR5949016, SRR5949027 to SRR5949030; (6) C. scutata: SRR5948993, SRR5948995 to SRR5948998, SRR5949001, SRR5949004, SRR5949005, SRR5949007; (7) K. flaccidum: SRR5949010, SRR5949011, SRR5949012, SRR5990072 to SRR5990080; (8) M. viride: SRR5949021 to SRR5949026; (9) Mougeotia sp. MZCH240: SRR9083681, SRR9083682, SRR9083688, SRR9083692 to SRR9083701; (10) S. pratensis MZCH10213: SRR9083685, SRR9083686, SRR9083687, SRR9083689, SRR9083690, SRR9083696; (11) S. pratensis UTEX928: SRR4018077 to SRR4018100; (12) Z. circumcarinatum SAG698-1a: SRR5948999, SRR5949000, SRR5949002, SRR5949003, SRR5949006, SRR5949008, SRR5949017, SRR5949018; and (13) Z. circumcarinatum SAG2419: SRR6047298, SRR6047299, SRR6047302 to SRR6047305. Source data are provided with this paper.
Codes and data used for genome re-annotation, WGCNA and differential gene expression analysis are available on our GitHub page https://github.com/deVries-lab/Response_to_a_gradient_of_environmental_cues_in_mesotaenium_endlicherianum.