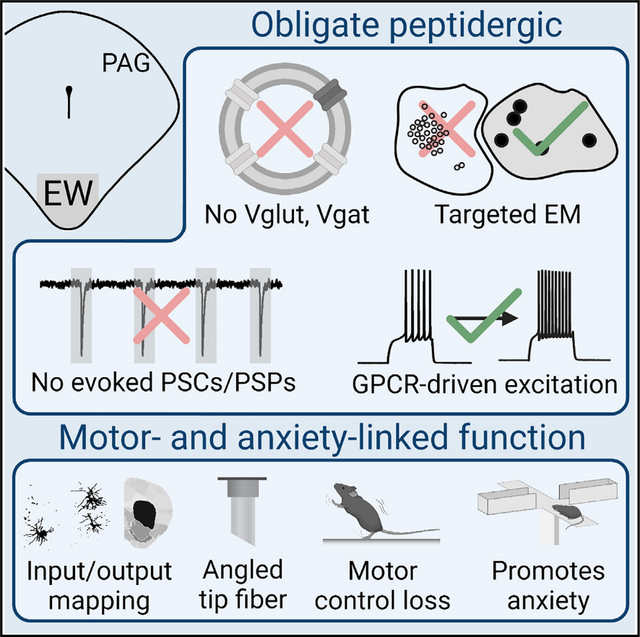
SUMMARY
Many neuronal populations that release fast-acting excitatory and inhibitory neurotransmitters in the brain also contain slower-acting neuropeptides. These facultative peptidergic cell types are common, but it remains uncertain whether neurons that solely release peptides exist. Our fluorescence in situ hybridization, genetically targeted electron microscopy, and electrophysiological characterization suggest that most neurons of the non-cholinergic, centrally projecting Edinger-Westphal nucleus in mice are obligately peptidergic. We further show, using anterograde projection mapping, monosynaptic retrograde tracing, angled-tip fiber photometry, and chemogenetic modulation and genetically targeted ablation in conjunction with canonical assays for anxiety, that this peptidergic population activates in response to loss of motor control and promotes anxiety responses. Together, these findings elucidate an integrative, ethologically relevant role for the Edinger-Westphal nucleus and functionally align the nucleus with the periaqueductal gray, where it resides. This work advances our understanding of peptidergic modulation of anxiety and provides a framework for future investigations of peptidergic systems.
In brief
Most neuronal signaling is rapid. Here, Priest et al. provide evidence for neurons that signal only through slow-acting neuropeptides. These neurons reside in the Edinger-Westphal nucleus, projecting broadly through the brain and spinal cord, receiving diverse motor-related inputs, activating in response to loss of motor control, and promoting anxiety.
Graphical Abstract
INTRODUCTION
Numerous glutamatergic and GABAergic neurons in the mammalian brain have been shown to be facultatively peptidergic.1–6 That is, they can package and release one or more neuropeptides, but the neurons also release fast-acting neurotransmitters independently of peptide release. Similar phenomena are observed in other classes of neurons that release fast neurotransmitters along with small-molecule neuromodulators including biogenic amines or acetylcholine.7–13 Other neurons release biogenic amines as well as neuropeptides.1–3,5 Moreover, different chemical signaling molecules released from the same neuronal population can produce distinct behavioral responses.14 Furthermore, distinct cellular machineries exist for the packaging and release of neuropeptides versus neurotransmitters, e.g., large dense core vesicles for peptides and small clear vesicles for single amino acid-derived transmitters.1,3–5
As opposed to these well-described neuronal types, the existence of obligate peptidergic neurons, which release no fast-acting canonical neurotransmitters, is a matter of debate. Indeed, recent reviews expressed skepticism toward the existence of obligate peptidergic neurons3,5 or have suggested that this class may be limited to the neurosecretory cells of the paraventricular nucleus of the hypothalamus (PVN).1 Recent single-cell RNA sequencing (scRNA-seq) of the mouse brain suggests there may be obligate peptidergic neuronal populations within and outside the PVN.15–17 Due to technical limitations associated with sequencing depth in scRNA-seq, the absence of a given transcript from a cell type does not preclude its existence. Nevertheless, scRNA-seq data highlight several neuronal populations that represent obligate peptidergic candidates (Figure 1A). One potential candidate is the cocaine- and amphetamine-regulated transcript (CART)-positive neuronal population of the Edinger-Westphal (EW) nucleus.
Figure 1. The CART+ EW lacks molecules required for fast neurotransmission.
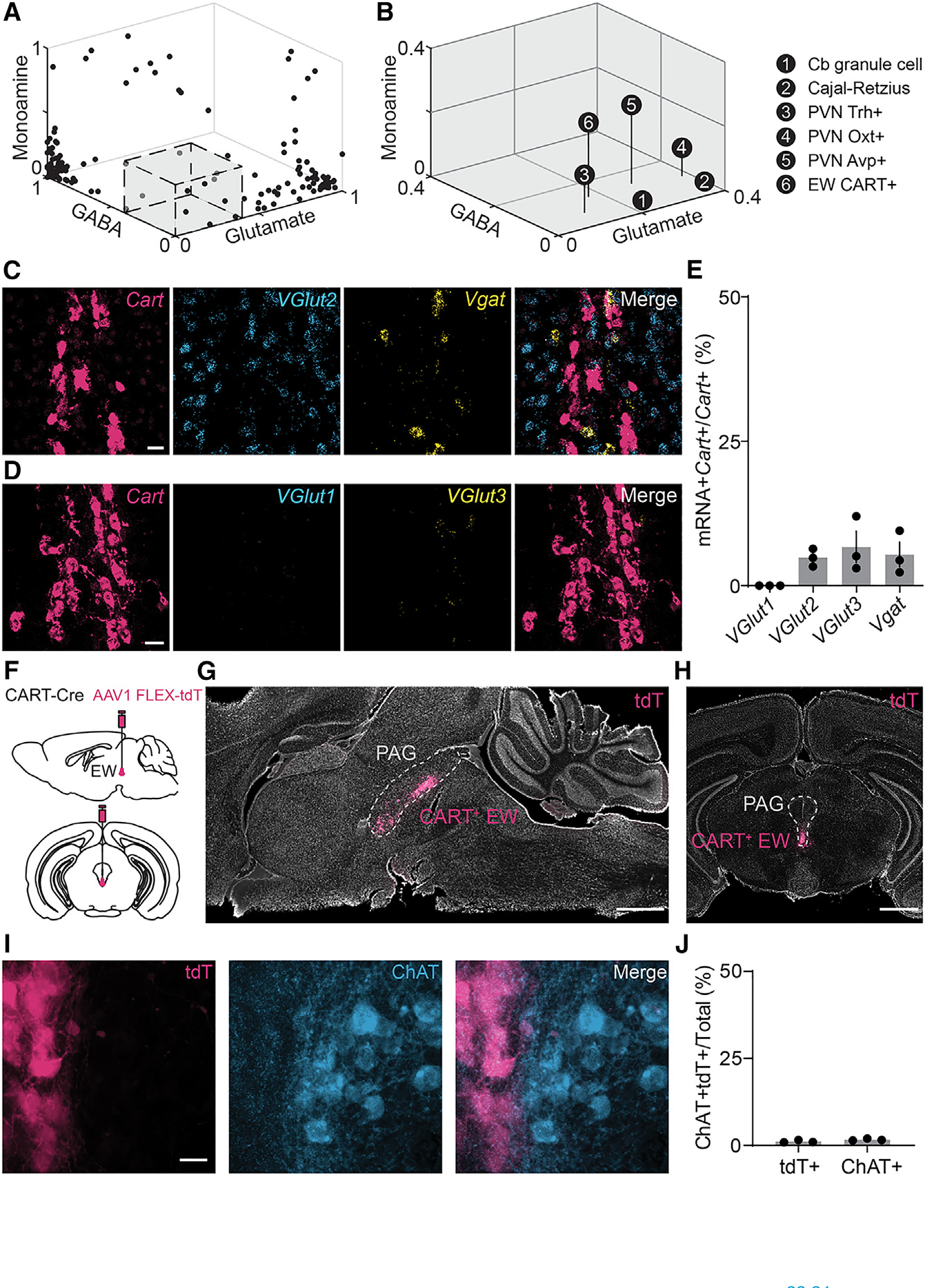
(A) Neuronal types in the mouse brain, by the proportion of cells in each group that contain markers for GABA, glutamate, and monoamines inclusive of acetylcholine. Many neuronal types have high proportions of cells that contain markers of inhibitory or excitatory neurons. The gray box, enlarged in (B), delineates neuronal types with very few cells identified as glutamatergic, GABAergic, or monoaminergic. Data are reanalyzed from publicly available scRNA datasets.15
(B) As in (A), but for neuronal types with <0.4 of any neurotransmitter or neuromodulator marker. The six neuronal types are cerebellar (Cb) granule cells, Cajal-Retzius cells, Trh+, Oxt+, and Avp+ neurons of the hypothalamic paraventricular nucleus (PVN), and CART+ neurons of the Edinger-Westphal (EW) nucleus.
(C) Example FISH against mRNA encoding Cartpt (red, Cart), Slc17a6 (blue, VGlut2), and Slc32a1 (yellow, Vgat) in the EW. Scale bar, 20 μm.
(D) As in (C), but against VGlut1 (blue, Slc17a7 or VGlut1) and VGlut3 (yellow, Slc17a8 or VGlut3). Scale bar, 20 μm.
(E) Colocalization of vesicular transporters with Cart in the EW (VGlut1, 0.00% ± 0.00%; VGlut2, 4.83% ± 0.90%; VGlut3, 6.70% ± 2.72%; Vgat, 5.40% ± 2.14%; n = 3 mice for all; 297 Cart+ cells for VGlut2 and Vgat; 404 Cart+ cells for VGlut1 and VGlut3).
(F) Schematic of AAV1 FLEX-tdTomato (tdT) injection into the mouse CART-Cre EW.
(G) Selective tdT (red) expression in the CART+ EW, sagittal slice. The PAG is marked by the dashed gray line. Hoechst nuclear stain, white. Scale bar, 1 mm.
(H) As in (G), but for a midbrain coronal slice. Scale bar, 1 mm.
(I) tdT+ neurons of the EW (red) compared to immunofluorescence against ChAT (blue). Scale bar, 20 μm.
(J) CART+ EW neurons do not colocalize with ChAT+ neurons (ChAT+CART+/CART+, 1.1% ± 0.2%, n = 3 mice, 784 cells;ChAT+CART+/ChAT+,1.7%± 0.2%, n = 3 mice, 879 cells).
Error bars represent SEM. See also Figure S1.
The CART+ EW resides in the rostral ventromedial periaqueductal gray (PAG), aligning with the anatomical regions labeled as EW and pre-EW nucleus in rodent brain atlases.18,19 The CART+ EW contains multiple neuropeptides including CART, urocortin (Ucn), and cholecystokinin (CCK), and it projects to the spinal cord.20 These properties distinguish the CART+ EW, also known as the centrally projecting EW, from both the cholinergic EW21 that projects to the ciliary ganglion and controls lens accommodation22 and the glutamatergic CCK+ peri-EW neuronal population that promotes non-REM (rapid eye movement) sleep and lacks the descending projections that are characteristic of CART+ EW neurons.23
Whether the CART+ EW contains neurons that are capable of rapid neurotransmission has been unclear. The CART+ EW was recently classified as peptidergic in an scRNA-seq study of the mouse dorsal raphe.16 In contrast, another scRNA-seq study of cells from across the entire mouse brain classified the CART+ EW as cell type MEGLU14: glutamatergic projection neurons in the dorsal midbrain that are enriched in Cart and Ucn.15 In addition, prior optogenetic experiments showed light-evoked glutamate-mediated excitatory postsynaptic currents (EPSCs) in the medial prefrontal cortex following Cre-dependent viral vector-driven channelrhodopsin (ChR2) expression in CCK-Cre+ neurons of the EW, which largely colocalize with CART.24 To resolve this discrepancy, we combined fluorescence in situ hybridization (FISH), genetically targeted electron microscopy, whole central nervous system anterograde mapping, and electrophysiological characterization to assess whether the CART+ EW is an obligate peptidergic nucleus.
At the behavioral level, the CART+ EW has been shown to respond to stressors,25–28 although this response has not been consistently demonstrated in mice.29 Furthermore, it remains unknown whether CART+ EW activity modulates stress-associated behaviors such as anxiety-like responses. Indeed, prior reports focusing on consumptive and maternal preparatory nesting behavior found that activity on an elevated maze was unchanged following lesions or genetically targeted ablation of this nucleus.30,31 In an attempt to move toward a unified framework for understanding the behavioral roles of the CART+ EW, we used monosynaptic retrograde labeling and in vivo calcium fiber photometry to interrogate the circuits and stimuli that regulate the CART+ EW. Further, we performed chemogenetic modulation and genetically targeted ablation to test whether the CART+ EW mediates anxiety responses. Together, our data define the CART+ EW as a predominantly peptidergic nucleus that responds to loss of motor control and promotes anxiety responses.
RESULTS
The CART+ EW lacks molecules required for fast neurotransmission
We defined candidate obligate peptidergic nuclei using an unbiased scRNA-seq study of the mouse nervous system that identified 181 neuronal clusters, or cell types, in the central nervous system.15 We excluded neuroblast-like cell types and spinal cord cells, focusing on the proportion of neurons in each of 148 neuronal brain cell types that contained canonical markers of the excitatory neurotransmitter glutamate, the inhibitory neurotransmitters GABA and glycine, and monoamines or acetylcholine (Figure 1A). For the remainder of this paper, we group acetylcholine, which is a monoammonium, with the canonical monoamines serotonin, dopamine, epinephrine, and norepinephrine. Additionally, although glutamate, GABA, and glycine are chemically monoamines, we categorize glutamate and GABA/glycine separately. Inhibitory cell types that contain markers for either GABA or glycine are referred to for simplicity as “GABAergic.” Glutamatergic markers were the vesicular glutamate transporters Slc17a6, Slc17a7, and Slc17a8. GABAergic markers were the vesicular inhibitory amino acid transporter, Slc32a1, the glycine transporters Slc6a5 and Slc6a9, and the GABA-synthesizing glutamate decarboxylase enzymes GAD1 and GAD2. Monoaminergic markers were the transporters Slc5a7, Slc6a3, Slc6a4, Slc18a1, Slc18a2, and Slc18a3, biosynthetic enzymes Chat, Dbh, Ddc, Hdc, Pnmt, Th, Tph1, and Tph2, and the serotonergic cell marker Fev. For each cell, the presence of any amount of transcript was used to identify a cell as glutamatergic, GABAergic, and/or monoaminergic, and any cell could be categorized as positive or negative across all three categories. As expected, many neuronal types previously defined by clustering15 had large proportions of cells that contained markers for glutamate, GABA, or a monoamine (Figure 1A).
To identify candidates for obligate peptidergic neuronal types, we focused on neuronal classes with a minority (<40%) of their cells positive for any of the 23 markers (Figure 1B). We chose 40% as a threshold, as it captured the core PVN populations broadly acknowledged as peptidergic. Only six neuronal types fit these criteria: the Cajal-Retzius cell of the hippocampus, the granule cells of the cerebellum, the vasopressin-positive (Avp+), oxytocin-positive (Oxt+), and thyrotropin-releasing hormone-positive (Trh+)/urocortin 3-positive neurons in or near the PVN, and the CART+ neurons of the EW nucleus. It is unclear why granule cells of the cerebellum appear here, as they are known to be glutamatergic, but their extremely small size32 likely contributes to technical difficulties in sequencing depth15; their inclusion underscores the importance of validating scRNA-seq results of interest with lower-throughput approaches. Cajal-Retzius cells are crucial for proper cortical development in early life33 and may serve a predominantly developmental role rather than one dependent on neurotransmission in established circuits.34 The neurohypophysiotropic populations of the PVN have been previously suggested as candidate obligate neuropeptidergic populations.1
The remaining candidate is the CART+ population of the EW, which expressed glutamatergic markers in 17.0% of its 47 cells, GABAergic markers in 12.8% of cells, and monoaminergic or cholinergic markers in 23.4% of cells. Only 4.3% of the CART+ EW contained both a biosynthetic enzyme and a related transporter, e.g., tyrosine hydroxylase with a vesicular monoamine transporter. Another scRNA-seq study also found that the CART+ EW lacked monoaminergic or cholinergic markers,16 and the CART+ EW is not considered a monoaminergic or cholinergic population.20 However, despite the low frequency in EW single-cell sequencing data of mRNA-encoding transporters required for vesicular packaging, one recent report has characterized the CART+ EW as glutamatergic.24
One possibility is that scRNA-seq failed to pick up sparse transcripts for vesicular glutamate or GABA transporters. We tested the presence of these transporters in the CART+ EW using quantitative FISH against mRNAs for CART (Cartpt or Cart) and vesicular transporters for glutamate (Slc17a6/VGlut2, Slc17a7/VGlut1, Slc17a8/VGlut3) and GABA (Slc32a1/Vgat) (Figures 1C and 1D). Male and female mice were included in all experiments. The vast majority of CART+ EW neurons lack canonical transporters for glutamate (VGlut1, 0.00% ± 0.00%; VGlut2, 4.83% ± 0.90%; VGlut3, 6.70% ± 2.72%, n = 3 mice, 404 Cart+ cells for VGlut1 and VGlut3, and 297 Cart+ cells for VGlut2) or GABA (Vgat, 5.40% ± 2.14%, n = 3 mice and 297 Cart+ cells) (Figures 1C–1E and S1A–S1D). This FISH quantification shows that approximately 83% of CART+ EW neurons in mice lack any canonical vesicular transporter for glutamate, GABA, or glycine release, suggesting that much of the CART+ EW may function as an obligate peptidergic neuronal population. As multiple canonical vesicular glutamate or GABA transporters may colocalize, this percentage reflects a lower bound on the proportion of obligate peptidergic neurons in the CART+ EW. The absence of VGlut1+ cells in subcortical areas was contrasted by hippocampal neurons in the same tissue, which expressed VGlut1 robustly (Figure S1E). Immediately adjacent to and within the EW nucleus, we observed numerous Cart-negative VGlut2+ (n = 771 cells, 3 mice) and Vgat+ (n = 256 cells, 3 mice) neurons, as well as fewer VGlut3+ neurons (n = 11 cells, 3 mice), underscoring the necessity of distinguishing the genetically defined CART+ EW population from the anatomically defined EW.
Therefore, to genetically target the CART+ EW, we used the Cart-IRES2-Cre mouse line (CART-Cre) for experiments throughout the study.35 Following injection of adeno-associated viral vector (AAV) packaged with a Cre-dependent tdTomato (tdT) gene into the EW nucleus (Figure 1F), we observed robust tdT expression that was limited to cells in this region (Figures 1G and 1H). Quantification of tdT+ cells revealed that the CART+ EW is composed of approximately 1,200 CART-Cre neurons (1,185 ± 90.6 cells, n = 3 mice). As expected, genetically defined CART+ EW shows virtually no overlap (1.15% ± 0.21%, n = 3 mice, 1,185 cells) with preganglionic, cholinergic neurons of the EW or other nearby oculomotor nuclei that are labeled with choline acetyltransferase (ChAT) (Figures 1I and 1J).21,36 This confirms that the genetically targetable CART+ EW lacks the required biosynthetic enzyme for producing the fast-acting neurotransmitter acetylcholine. We also confirmed that the AAV-targeted CART+ EW colocalizes heavily with immunolabeled CART (85.12% ± 1.52%, n = 3 mice, 784 cells)37 and urocortin (80.01% ± 1.44%, n = 3 mice, 631 cells) (Figures S2A–S2D). The CART and urocortin populations of the EW appear to be largely identical, consistent with prior work.38,39
The CART+ EW contains numerous neuropeptides and large vesicles
In addition to CART and urocortin, the EW has been suggested to release other neuropeptides. To refine our understanding of peptidergic motifs in the EW, we examined the colocalization of EW Cart with peptides previously suggested to localize to the EW: substance P (Tac1),40,41 cholecystokinin (Cck),15,16,24,42 pituitary adenylate cyclase-activating peptide (Adcyap1, or Pacap), and neuromedin B (Nmb)43 (Figures 2A, S2E, and S2F). We found co-expression of Cart with Cck (72.69% ± 6.08%, n = 3 mice, 261 cells), Nmb (67.17% ± 6.28%, n = 3 mice, 636 cells), and Pacap (54.12% ± 6.16%, n = 3 mice, 233 cells), but not with Tac1 (3.34% ± 2.63%, n = 2 mice, 208 cells) or canonical endogenous opioids (Pomc, 1.40% ± 0.65%, n = 3 mice, 636 cells; Penk, 4.44% ± 1.93%, n = 3 mice, 385 cells; Pdyn, 3.22% ± 1.55%, n = 3 mice, 385 cells) (Figures 2A, 2B, and S2E–S2K). Thus, the CART+ EW contains numerous neuropeptides, including CART, urocortin, CCK, neuromedin B, and PACAP, but lacks molecules required for fast neurotransmission, including glutamate and GABA vesicular transporters as well as ChAT.
Figure 2. The CART+ EW contains numerous neuropeptides and large vesicles.
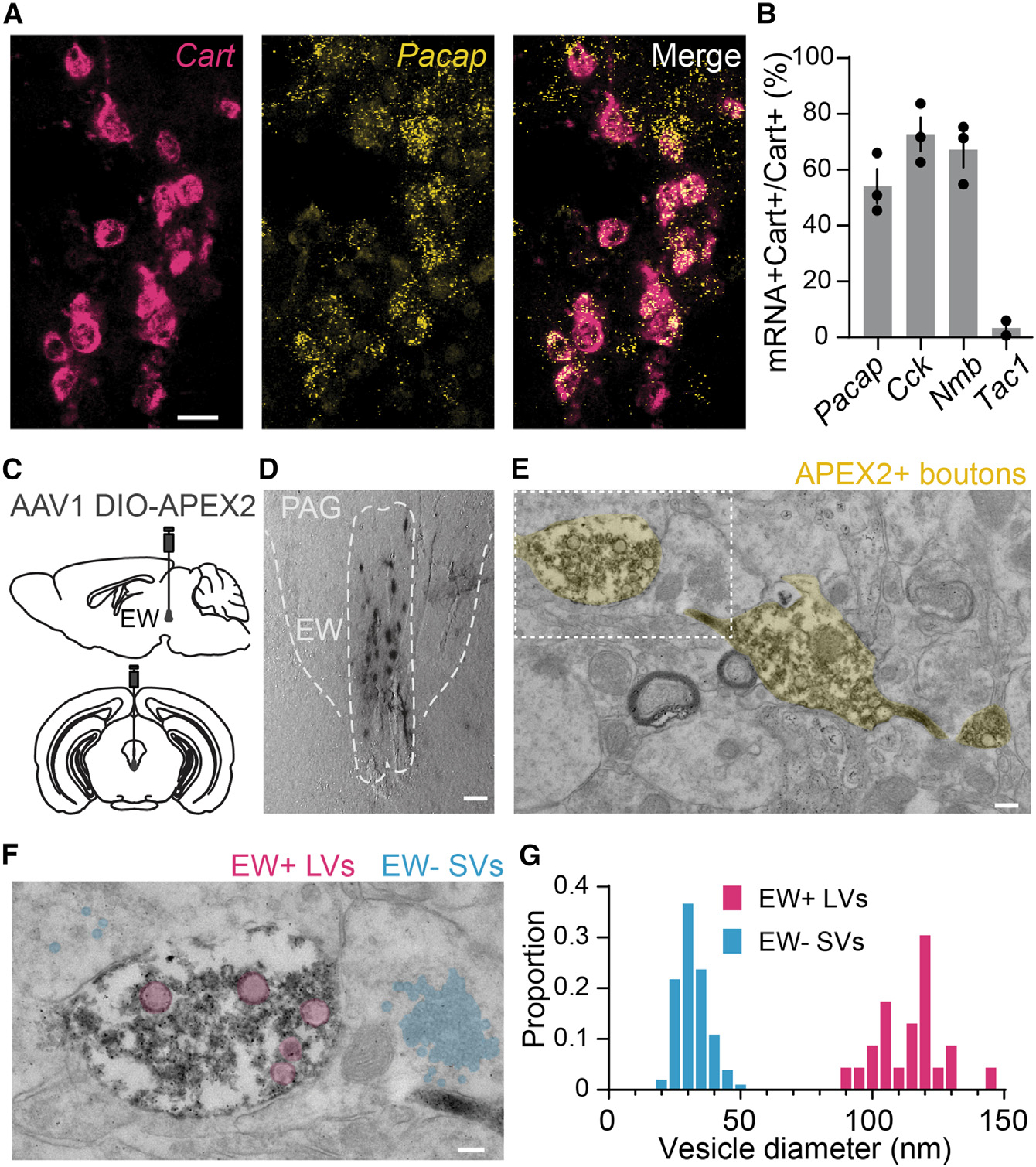
(A) FISH against Cart (red) and Adcyap1/Pacap (yellow). Scale bar, 20 μm.
(B) Quantification of Cart colocalization with Pacap, Cck, Nmb, and Tac1. Pacap, 54.12% ± 6.16%; Cck, 72.69% ± 6.08%; Nmb, 67.17% ± 6.28%; Tac1, 3.34% ± 2.63%; n = 2 mice for Tac1, 3 mice for all others; >200 cells counted for each mouse.
(C) Schematic of AAV1 DIO-APEX2 injection into the mouse CART-Cre EW.
(D) Brightfield image of genetically targeted APEX DAB staining of CART+ EW cells. Scale bar, 50 μm.
(E) Transmission electron micrograph of APEX2+ synaptic boutons in the midbrain. Scale bar, 200 nm.
(F) Higher-magnification image of the white box in (E). Multiple small clear vesicles (blue) are observed in synaptic processes around the APEX2+ bouton, which contains multiple large vesicles (red). Scale bar, 100 nm.
(G) Diameters of dense core vesicles (red) observed in APEX+ boutons and small clear vesicles (blue) found in regions adjacent to the APEX2+ boutons; n = 23 large vesicles from 16 APEX2+ boutons, diameter = 114.1 nm ± 2.6 nm; n = 101 small vesicles near 9 APEX2+ boutons, diameter = 31.8 nm ± 0.6 nm. Histogram bin size is 5 nm. Error bars represent SEM. See also Figure S2.
Next, we targeted the CART+ EW with Cre-dependent LCK-APEX2 (Figure 2C), an engineered peroxidase (APEX) capable of performing proximity labeling and deposition of electrondense diaminobenzidine.44 APEX is fused to a membrane anchor sequence (LCK) for membranal localization with a separately transcribed GFP for identifying expression.44 Following APEX expression in the CART+ EW (Figure 2D), fixed and stained sections were imaged under a transmission electron microscope. In addition to labeling structures in the soma, APEX+ boutons were abundant in the imaged peri-EW midbrain regions (Figure 2E). These boutons contained numerous large vesicles (n = 23 vesicles from 16 boutons, diameter = 114.1 ± 2.6 nm) matching the size of large dense core vesicles5 (Figures 2F and 2G). No small clear vesicles were observed in these boutons, although nearby structures (<2 μm from an APEX+ bouton) frequently contained vesicles the size of small clear vesicles (n = 101 vesicles near nine APEX+ boutons, diameter = 31.8 nm ± 0.6 nm) (Figures 2F and 2G). We cannot preclude the possibility that our sample preparation may have obscured the presence of small clear vesicles in the APEX+ boutons. However, the marked presence of CART+ EW boutons containing large vesicles necessary for peptidergic release and the apparent absence of the small clear vesicles used for fast neurotransmission1,3–5 support the hypothesis that the CART+ EW is predominantly obligately peptidergic.
The CART+ EW projects to the spinal cord and multiple subcortical regions
To functionally assess whether a population is obligately or facultatively peptidergic, it is important to know its efferent targets. However, the targets of EW projections within the brain24,45 and spinal cord45–47 remain disputed. To resolve conflicting reports and facilitate functional characterization of CART+ EW signaling, we mapped the CART+ EW anterograde projections using genetically targeted AAV tdT injections into the EW of CART-Cre mice, with subsequent immunofluorescence against tdT (Figure 3A). While it is broadly acknowledged that the EW projects to the spinal cord, this canonical descending projection has been localized to different laminae by different anterograde tracers or axonal markers and has not been characterized using genetically targeted approaches.45–47 For example, prominent EW-spinal projections have been described as going to laminae I and V,47 laminae VII and X,46 and laminae I through IV and VII through X.45 We found robust projections to laminae III, IV, V, VII, VIII, and X of the spinal cord (Figure 3B, n = 3 mice) using isolectin B4 labeling to demarcate lamina II (Figure S3A). We registered brain sections (Figure 3C) to the Allen Brain Atlas for automated quantification of CART+ EW projections in each brain region by density (the abundance of projections in a given brain region divided by the size of the brain region) and abundance (the quantification of projections in a given brain region) (Figures 3D–3F, S3B, and S3C; Table S1; n = 3 mice). Projections were found across numerous subcortical brain regions. This finding partially aligns with non-genetically defined anterograde tracing of the rat anatomical EW45 but contradicts genetically defined trans-synaptic viral expression from a CCK+ EW population, which targeted neurons primarily in the medial prefrontal and cingulate cortices.24
Figure 3. The CART+ EW projects to the spinal cord and multiple subcortical regions.
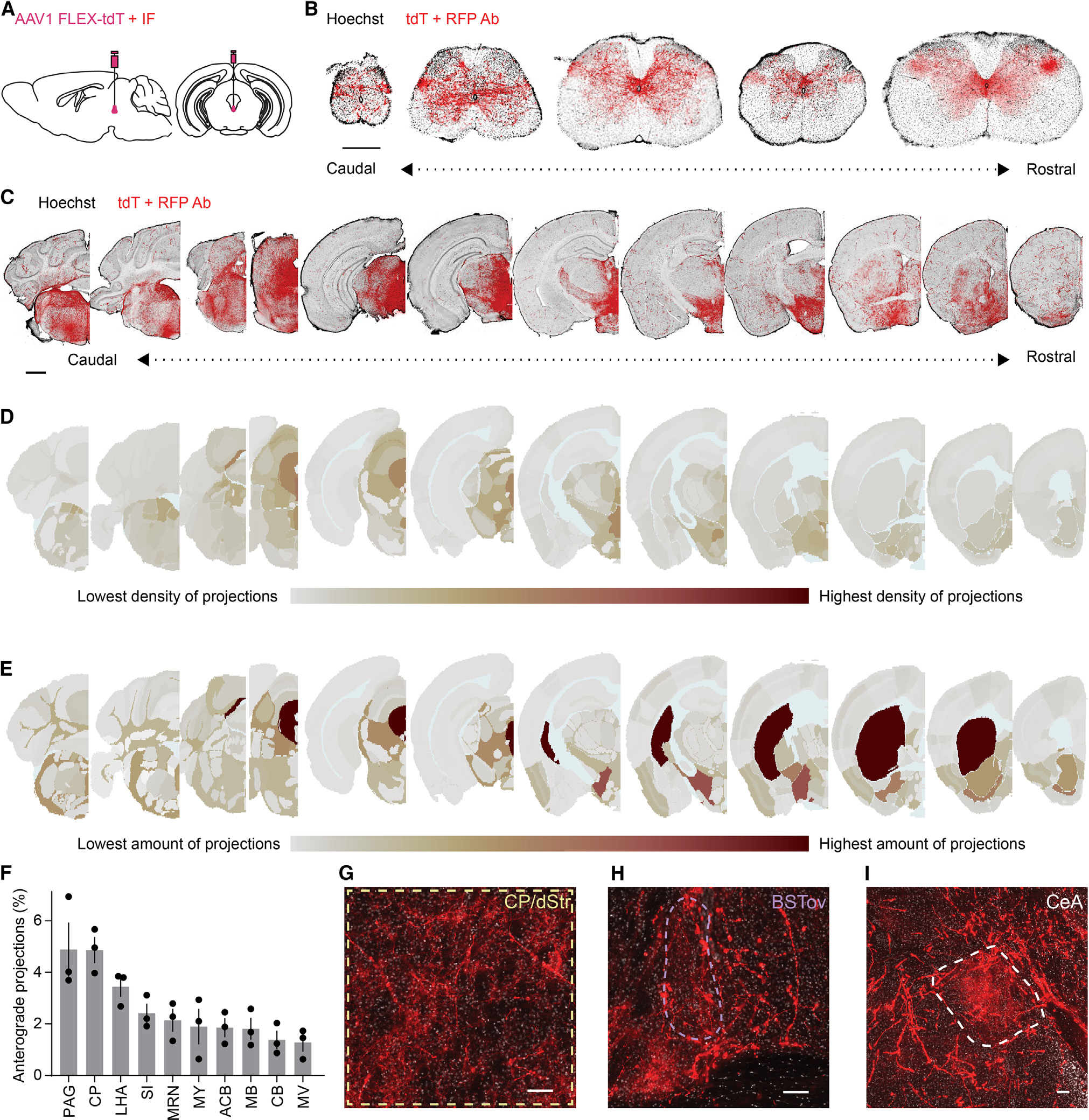
(A) Schematic of AAV1 FLEX-tdT EW injection with immunoenhancement for tdT.
(B) Coronal sections of CART+ EW projections to the spinal cord (coccygeal through cervical; Hoechst, black; immunoenhanced tdT, red; scale bar, 0.5 mm).
(C) As in (B), but brain sections. Scale bar, 1 mm.
(D) Projection density heatmap across brain regions aligned to Allen Brain Atlas. Red, high expression; gray, low expression; light blue, undefined areas, white matter, and ventricles.
(E) As in (D), but projection abundance instead of density.
(F) Brain regions with the most CART+ EW projections (n = 3 mice), displayed as percentage of total quantified projections in the brain.
(G–I) Immunoenhanced CART+ EW tdT in the (G) dorsal striatum/caudoputamen (dStr/CP), (H) oval nucleus of the bed nucleus of the stria terminalis (BSTov), and (I) the central nucleus of the amygdala (CeA). Scale bar, 100 μm.
Error bars represent SEM. See also Figure S3.
The two brain regions with the greatest proportion of observed CART+ EW neuronal processes were the PAG and the caudoputamen (CP), also called the dorsal striatum (dStr) in mice (PAG: 4.89% ± 1.03%; CP/dStr: 4.87% ± 0.49%; n = 3 mice, Figures 3D–3F and Table S1). Given that the EW is located within the PAG, one possibility is that these local projections are fibers of passage. In the dStr we observed wispy terminal fields in addition to rarer varicosity-studded axonal fibers (Figure 3G). We found similar terminal fields in the oval bed nucleus of the stria terminalis (BSTov) (Figure 3H) and in the central nucleus of the amygdala (CeA) (Figure 3I). While the CeA has many sizable neuronal populations,48,49 the BSTov has roughly three predominant populations of projection neurons,50 and the dStr contains two large classes of striatal spiny projection neurons. Consequently, we focused on the dStr and BSTov to determine whether the CART+ EW alters the activity of neurons in these regions and whether the modulation is consistent with glutamatergic signaling or neuropeptide-mediated effects.
The CART+ EW is functionally peptidergic
To evaluate neurotransmitter release from CART+ EW neurons, we performed whole-cell current-clamp recordings of putative postsynaptic neurons in the dStr/CP and the BSTov. Using established high-frequency optogenetic stimulation protocols for evoking peptide release from oxytocinergic neurons,51–55 we stimulated channelrhodopsin (ChR2) in CART+ EW terminals with a single bout of blue-light pulses (10 ms pulses delivered at 30 Hz for 20 s, 10 mW) (Figure 4A). ChR2-driven stimulation of CART+ EW fibers more than doubled the firing rate in a subset of neurons in both the dStr/CP (8 of 17 SPNs) and the BSTov (3 of 11 cells) (Figures 4B, 4C, and S4A). The time constant of the increase in firing rate (Figure S4B) was approximately 3 min in BSTov cells (170.1 ± 82.1 s, n = 3) and approximately 7 min in SPNs (427.4 ± 86.5 s, n = 8). This slow time constant of firing-rate increase is more consistent with peptidergic release from CART+ EW terminals rather than glutamatergic or GABAergic effects. Using retroAAVs packaged with GFP, we anatomically validated the projections from the CART+ EW to the striatum and tested whether nearby glutamatergic and GABAergic populations of the EW projected to the dStr (Figure S4C); CART-Cre mice showed robust GFP expression in the EW while VGlut2-IRES-Cre and Vgat-IRES-Cre mice did not (Figures S4D–S4G). In agreement with our FISH data, there was zero colocalization between CART and retrogradely labeled VGlut2-IRES-Cre or Vgat-IRES-Cre neurons in or near the EW.
Figure 4. The CART+ EW is functionally peptidergic.
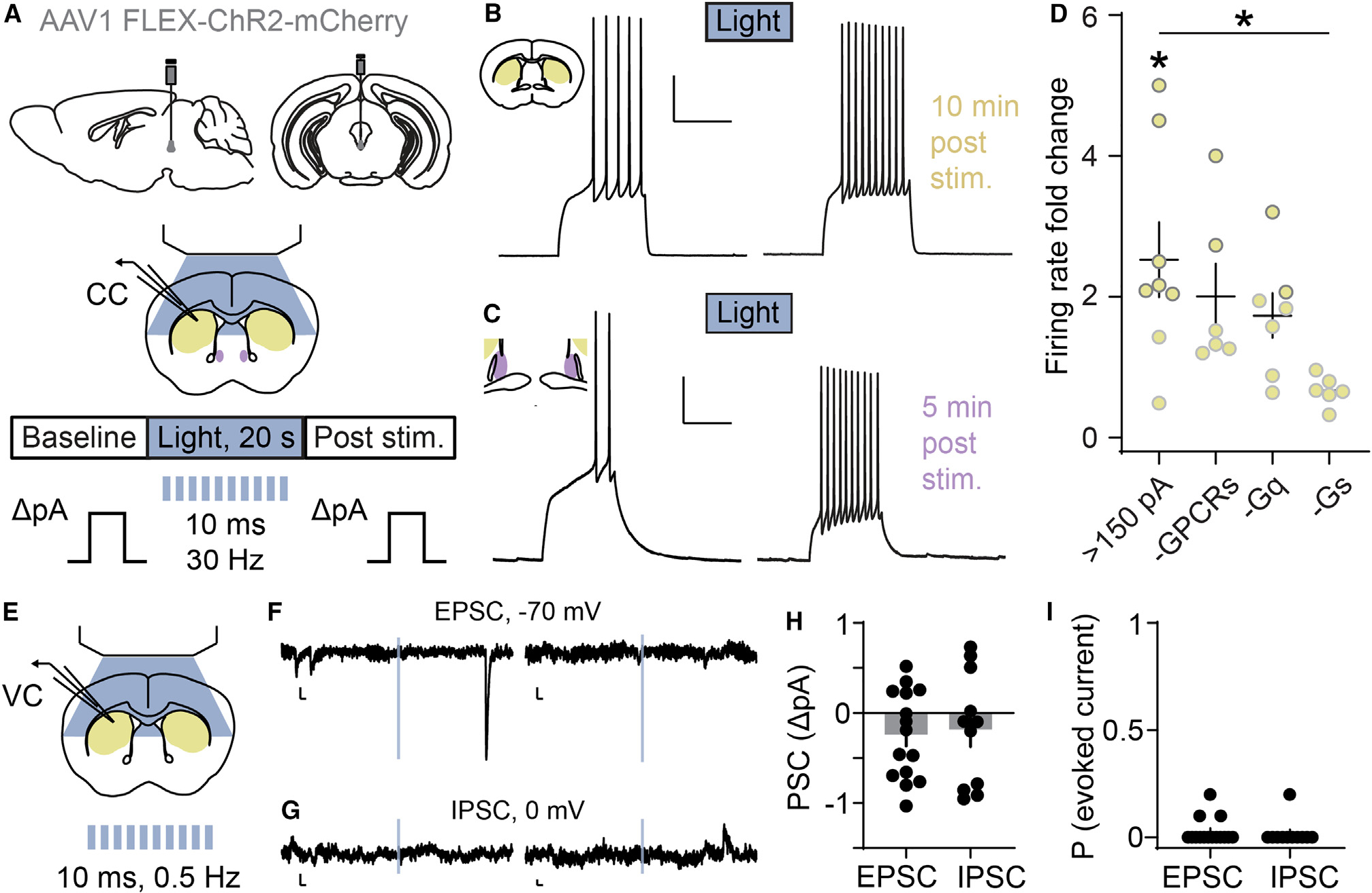
(A) Schematic of AAV1 FLEX-ChR2-mCherry EW injection and whole-cell current-clamp recordings of CART+ EW target regions (CP/dStr, yellow; BSTov, purple). An optical stimulation bout (20 s, 30 Hz) evokes vesicular release from ChR2-expressing CART+ EW terminals.
(B) Following ChR2 stimulation, a striatal SPN fires more in response to a depolarizing current injection. Scale bars, 25 mV and 500 ms.
(C) As in (B) but recorded from the BSTov. Scale bars, 20 mV and 500 ms.
(D) SPNs with rheobase >150 pA generally increased their firing rate compared to baseline. *p < 0.05, paired t test, t(7) = 2.860, p = 0.0243, n = 8 neurons from 8 mice. Neither application of a cocktail of GPCR antagonists (-GPCRs, n = 6 neurons from 4 mice) nor bath-applied PLC blocker (-Gq, n = 7 neurons from 4 mice) altered firing-rate increases, but intracellular PKA block (-Gs, n = 6 neurons from 5 mice) significantly decreased the light-evoked firing-rate increase. *p < 0.05, one-way ANOVA with post hoc Dunnett’s test (F(3,23) = 3.483; p = 0.0322; >150 pA vs. >150 pA-GPCRs, p = 0.7172; >150 pA vs. >150 pA-Gq, p = 0.3814; >150 pA vs. >150 pA-Gs, p = 0.0114).
(E) Schematic of whole-cell voltage-clamp recordings of CP/dStr with 0.5 Hz blue light applied in 10 ms pulses.
(F) Example current traces from two SPNs as a 10 ms light pulse (blue) was applied while holding at −70 mV. Evoked EPSCs were not observed. Spontaneous EPSCs were observed. Scale bars, 5 pA and 20 ms.
(G) As in (F), from SPNs held at 0 mV. Evoked IPSCs were not observed. Scale bars, 5 pA and 20 ms.
(H) Summary of evoked current data, shown as the amplitude of postsynaptic current taken from the onset of the light pulse to 15 ms after the offset, compared tobaseline (n = 15 neurons for EPSCs, t(14) = 1.876, p = 0.0817; 11 neurons for IPSCs, t(10) = 0.9562, p = 0.3615, from 6 mice).
(I) Summary of evoked current data, shown as proportion of ten applied light pulses generating evoked EPSCs (n = 15 neurons) or IPSCs (n = 11 neurons). Error bars represent SEM. See also Figure S4.
In current-clamp recordings, spiny projection neurons (SPNs) are generally silent and require injections of positive current to elicit action potential firing56; different classes of SPNs likely differ in the minimum amount of current that must be injected to produce an action potential. We found that dStr SPNs with a rheobase over 150 pA significantly increased excitability following CART+ EW stimulation (Figure 4D), while SPNs with a rheobase less than 150 pA did not increase excitability (Figure S4H). Therefore, we further pharmacologically interrogated the multipeptidergic release of CART+ EW neurons in the dStr by selectively recording from SPNs with rheobase over 150 pA.
As shown previously (Figure 2B), CART+ EW neurons contain numerous excitatory peptides including urocortin, CCK, and PACAP, and each peptide binds to multiple G protein-coupled receptors (GPCRs). Using a cocktail of five pharmacological GPCR antagonists, we investigated whether blockade of a subset of molecular targets of CART+ EW neurons (CRFR1, CRFR2, CCKAR, CCKBR, PAC1R, VPAC1R, and VPAC2R) prevents SPN excitation following optogenetic stimulation of CART+ EW fibers. The cocktail of antagonists was insufficient to abolish SPN excitation (Figure 4D). Notably, although a receptor for CART has recently been proposed,57 no cell-based activity assays have been performed to conclusively demonstrate its functionality, nor have pharmacological antagonists to the receptor been reported. Instead of receptor blockade, we tested whether CART is sufficient to alter excitability of dStr SPNs. Application of CART peptide to SPNs with a rheobase greater than 150 pA had no effect on excitability (Figure S4I). CART+ EW neurons have also been shown to contain nesfatin,39,58 which, like CART, lacks a druggable candidate receptor. Application of nesfatin increased the firing rate in a subset of SPNs with a rheobase above 150 pA (Figure S4I).
Since CART+ EW neurons contain peptides that bind to unknown or pharmacologically intractable receptors, we tested whether its effects on excitability of SPNs depended on GPCRs by blocking intracellular signaling pathways downstream of GPCR activation. We observed that Gαq-signaling block using bath-applied phospholipase C inhibitor (U 73122) did not abolish SPN excitability induced by optogenetic stimulation of CART+ EW fibers (Figure 4D). However, increases in firing rate in SPNs were abolished by the intracellular block of Gαs signaling with protein kinase A inhibitor (PKI) (Figure 4D). Increases in firing rate were also not found in the absence of ChR2 in CART+ EW neurons (Figure S4J). Our findings do not preclude a role of Gαq- or urocortin-, CCK-, or PACAP-coupled signaling in the excitability of SPNs by the CART+ EW.
To test whether the CART+ EW releases glutamate onto its striatal targets, we pooled all neurons with a 2-fold or greater firing-rate increase following light-evoked vesicle release and examined membrane potential time-locked to optogenetic stimulation. In agreement with an absence of glutamatergic release, we observed no evidence of time-locked excitatory postsynaptic potentials (Figures S4K and S4L). Finally, time-locked excitatory and inhibitory postsynaptic currents (EPSCs and IPSCs) were not observed in whole-cell voltage-clamp recordings of SPNs in response to light stimulation (10 ms, 10 mW, 0.5 Hz, 5 pulses given in the first 10 s of a 24-s sweep, repeated twice) to evoke fast neurotransmitter release (Figures 4E–4I). The absence of time-locked IPSCs and EPSCs is notably different from a similar study examining projections from dopaminergic neurons of the dorsal raphe to the BST, which elicited EPSCs in roughly 50% of downstream neurons.59
Taken together, our results suggest that the CART+ EW increases the excitability of subpopulations of neurons in the BSTov and dStr and that the CART+ EW modulation of striatal projection neurons is dependent on intracellular Gαs signaling cascades. The importance of Gαs intracellular cascades for CART+ EW signaling aligns with the presence of numerous Gαs-linked peptides in the CART+ EW, including urocortin, PACAP, and CCK.60–62 We see no evidence that the CART+ EW is functionally glutamatergic, as all anatomical and electrophysiological data support the hypothesis that the CART+ EW is a predominantly obligate peptidergic nucleus.
The CART+ EW receives inputs from brain regions related to motor control and threat responses
Next, we characterized inputs to the CART+ EW from the brain and spinal cord using monosynaptic retrograde mapping (Figure 5A). GFP-expressing helper virus colocalized with tdT-expressing pseudotyped rabies virus strain CVS-N2c63 in the EW (Figure S5A). Previously, the ventral hippocampus, medial septum, dorsal raphe, locus coeruleus,24 and the ventral tegmental area64 had been proposed to project to the genetically defined neuropeptidergic EW. We did not find consistent evidence (≥2 neurons in each mouse) of projections from any of these areas despite observing sparse labeling in nearby areas, e.g., the dorsal subiculum and lateral septum (Figures S5B–S5E, n = 3 mice, 190–233 neurons per mouse). Notably, despite a prior report that glutamatergic neurons in the ventral hippocampus provide monosynaptic excitatory input to the EW,24 we found no retrogradely labeled neurons in this area (Figures S5B and S5E). We did, however, observe robust and consistent evidence of CART+ EW inputs in numerous cortical and subcortical regions of the brain (Figure 5B).
Figure 5. The CART+ EW receives inputs from brain regions related to motor control and threat responses.
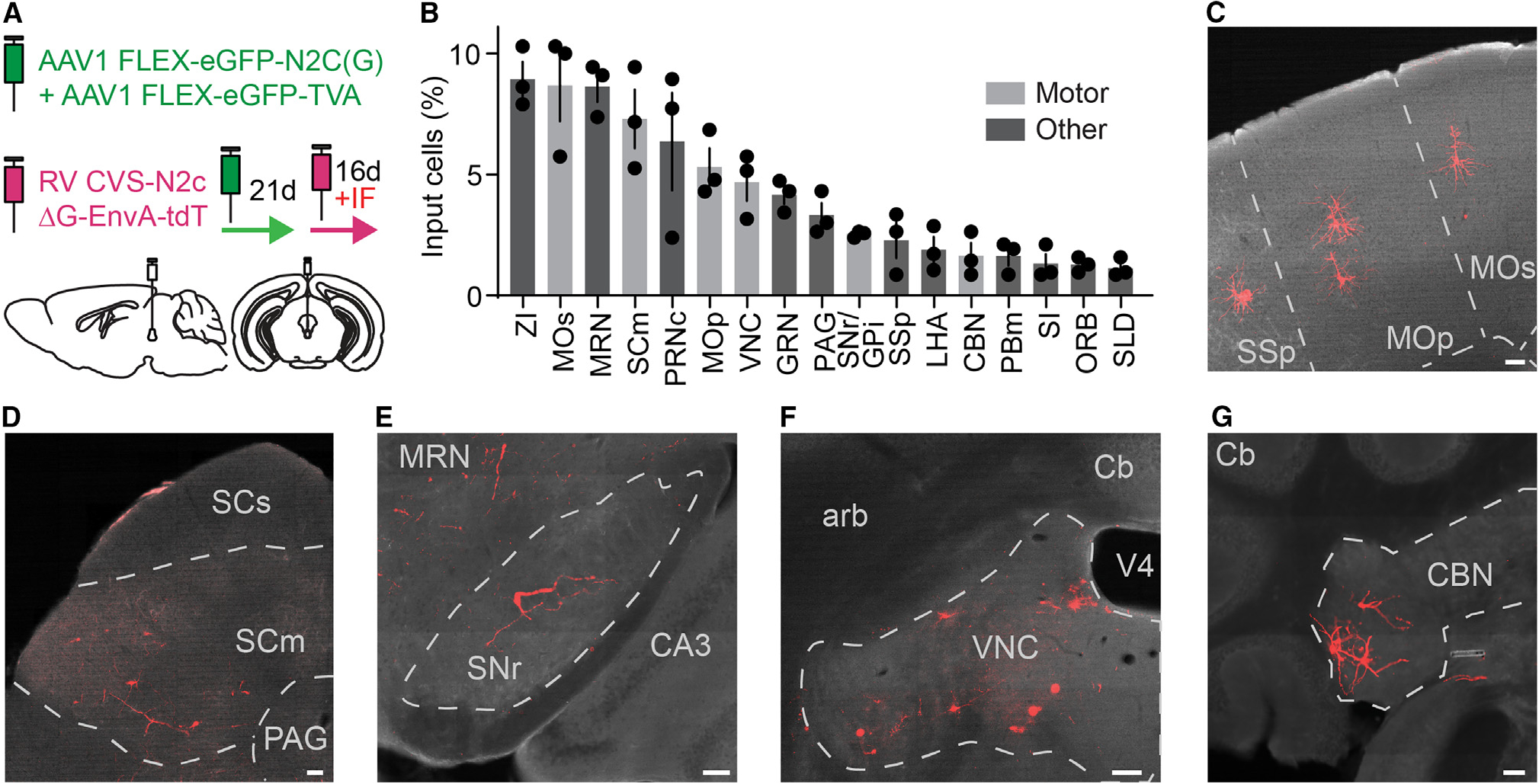
(A) Schematic of monosynaptic retrograde tracing from CART+ EW. tdT fluorescence is immunoenhanced.
(B) Retrogradely labeled tdT+ neurons by brain region, as percentage of all retrogradely labeled tdT+ neurons (n = 3 mice, 190–233 neurons per mouse). Motor control brain regions are shown in gray. ZI, zona incerta; MOs, secondary motor cortex; MRN, midbrain reticular nucleus; SCm, motor-related areas of superior colliculus; PRNc, caudal pontine reticular nucleus; MOp, primary motor cortex; VNC, vestibular nuclei; GRN, gigantocellular reticular nucleus; PAG, periaqueductal gray; SNr/GPi, substantia nigra pars reticulata/globus pallidus internal; SSp, primary somatosensory cortex; LHA, lateral hypothalamus; CBN, cerebellar nuclei; PBm, medial parabrachial nucleus; SI, substantia innominata; ORB, orbital cortex; SLD, sublaterodorsal nucleus.
(C) Image of retrogradely labeled neurons (red) in primary and secondary motor cortex and primary somatosensory cortex. Scale bar, 100 μm.
(D–G) as in (C), but retrogradely labeled neurons in (D) motor-related areas of superior colliculus, (E) substantia nigra pars reticulata, (F) vestibular nuclei, and (G) cerebellar nuclei. Scale bars, 100 μm.
Error bars represent SEM. See also Figure S5.
Genetically targeted retrograde tracing does not provide information on synaptic input strength.65 Therefore, we examined whether any of the 17 brain regions consistently found to provide input to the CART+ EW shared a common function. We found monosynaptic CART+ EW inputs from six brain regions directly related to motor control (Figure 5B): pyramidal neurons in layer V of the primary and secondary motor cortices (Figure 5C), motor-related superior colliculus (Figure 5D), substantia nigra pars reticulata (Figure 5E), vestibular nuclei (Figure 5F), and cerebellar nuclei (Figure 5G). This enrichment of input nuclei involved in motor control is similar to that observed for the input nuclei of the substantia nigra pars compacta (SNc), a neuromodulatory nucleus considered vital for motor control. Six of the 17 brain regions with the greatest number of labeled cells found upstream of the SNc using monosynaptic retrograde labeling are closely tied to motor control.66
In addition to receiving direct inputs from cortical and subcortical regions that transmit information about motor commands or outcomes, we found that the CART+ EW also received projections from multifunctional regions involved in responses to threat or pain, including the somatosensory cortex,67 lateral hypothalamus,68,69 zona incerta,70,71 and PAG72,73 (Figure 5B).
CART+ EW neurons respond to loss of motor control
To determine the relationship between CART+ EW neuronal activity and movement, we expressed GCaMP6s and measured in vivo CART+ EW activity with angled mirror tipped photometry fibers to avoid the cerebral aqueduct (Figures 6A and S6A). Conventional flat-tipped photometry fibers occlude the cerebral aqueduct, which leads to high surgical mortality rates. Under blue light (470 nm) excitation of GCaMP6s, we observed that gentle tail restraint elicited calcium transients that were time-locked with the restraint and were reproducible across tail restraint trials and different mice (Figure 6B). To control for the possibility of motion artifact-induced transients, we performed separate trials using light (405 nm) that excites near the isosbestic point. No transients were observed under isosbestic illumination (Figures S6B–S6F), suggesting that the transients we observed under blue-light excitation are due to changes in intracellular calcium concentration in the CART+ EW. Similar fluorescence signals were observed in response to tail suspension (Figures 6C and S6C).
Figure 6. CART+ EW neurons respond to loss of motor control.
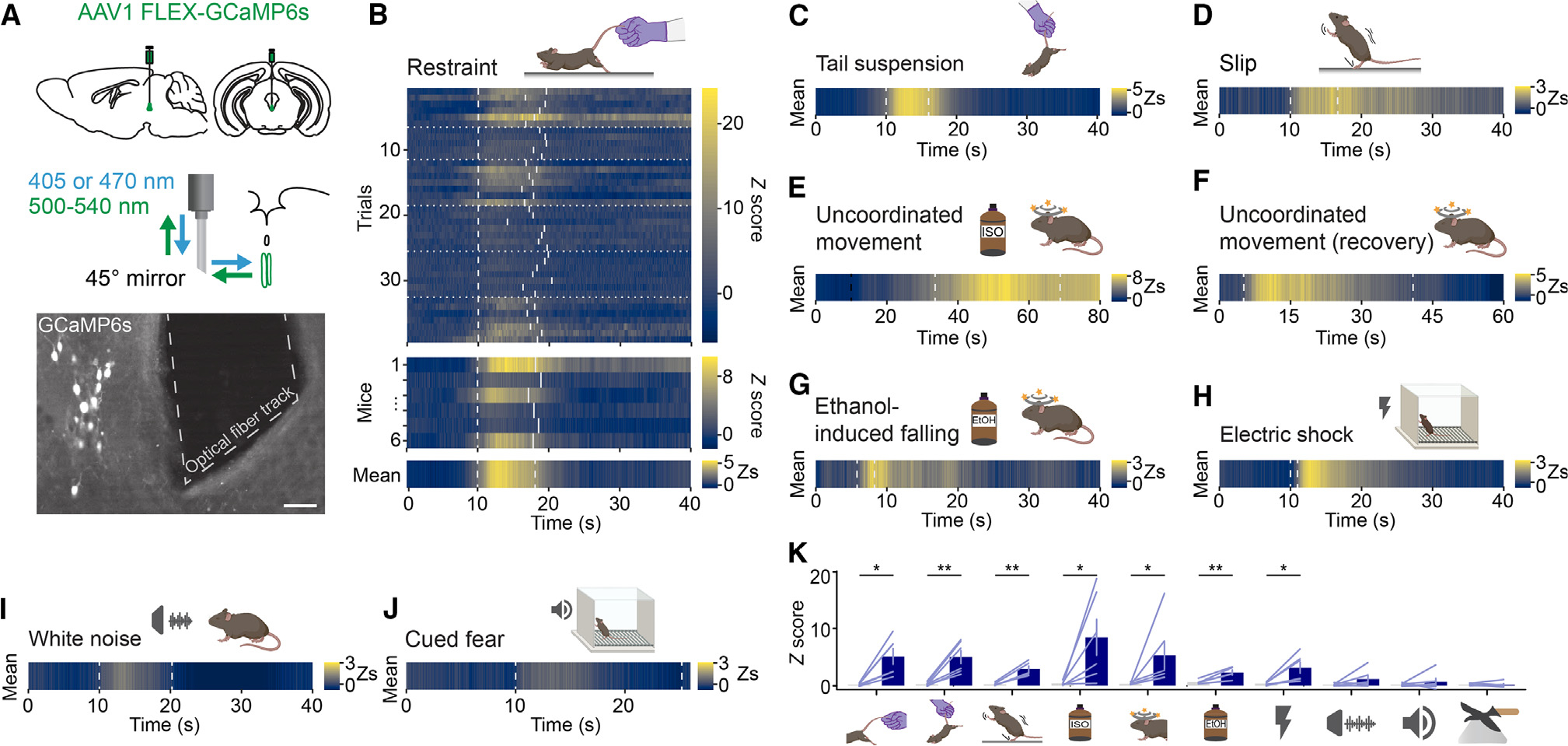
(A) (Top) Schematic of CART+ EW fiber photometry for in vivo calcium recordings. (Middle) A 45° angled mirror tip fiber is used to avoid occluding the cerebral aqueduct dorsal to the CART+ EW. 405- and 470-nm light measures motion artifact-induced fluorescence and calcium concentration-dependent fluorescence, respectively. (Bottom) GCaMP6s expression and 45° angled mirror tip fiber placement. Scale bar, 50 μm.
(B) Time-locked fluorescence signals recorded from the CART+ EW in response to mild tail restraint. Heatmap of Z scores is from blue (little to no calcium signal) to yellow (high calcium signal). (Upper) Cartoon of tail restraint. (Lower) Each trial (n = 5–7) from each mouse (n = 6). The start and end of each manual tail restraint are shown by dashed gray lines. Data from different mice are separated by dotted gray lines. Beneath are fluorescence signals for each mouse, averaged across all trials. The stimulus time points are shown by dashed gray lines marking the average start and stop point of all trials for a given animal. Bottom: the fluorescence signal Z score (Zs) averaged across all mice, with the stimulus time points shown as a dashed gray line at the average start and stop point across all animals.
(C) As in the bottom panel of (B), but in response to tail suspension (n = 6 mice, 6–9 trials each).
(D) As in the bottom panel of (B), but in response to slipping following placement in an arena with a thin layer of corn oil (n = 5 mice, 4 slipping bouts each). The dashed gray lines mark the average start and stop point of slipping bouts.
(E) As in the bottom panel of (B), but in response to anesthetic isoflurane exposure (n = 6 mice, 3–4 trials each). Exposure to isoflurane (dashed black line) does not elicit time-locked fluorescence signals. Uncoordinated movement following isoflurane exposure (dashed gray line) coincides with GCaMP6s fluorescence transients.
(F) As in the bottom panel of (B), but in response to uncoordinated movement during recovery from anesthesia (n = 6 mice, 3–4 trials). Average start and stop times of uncoordinated movement are marked by dashed gray lines.
(G) As in the bottom panel of (B), but in response to stumbles or falls elicited following intraperitoneal administration of ethanol (2.5 g/kg) (n = 5 mice, 2–5 trials each).
(H) As in the bottom panel of (B), but in response to an electric shock (n = 6 mice, 7 trials per mouse).
(I) As in the bottom panel of (B), but in response to a stressful, loud white noise (n = 6 mice, 6–8 trials each).
(J) As in the bottom panel of (B), but in response to a conditioned fear tone cue (n = 6 mice, 6 trials per mouse). For (I) and (J) the Zs heatmap is set to match the heatmap used in the slipping data (~−1 Z to 3 Z), as this was the stimulus that reproducibly activated the CART+ EW with the smallest fluorescence changes. (K) Peak Z score of stimulus-driven fluorescence changes versus baseline for, from left to right, restraint (*p < 0.05, t(5) = 3.80, p = 0.0126), tail suspension (**p < 0.01, t(5) = 4.469, p = 0.0066), slip (**p < 0.01, t(4) = 5.83, p = 0.0043), isoflurane-induced uncoordinated movement (*p < 0.05, t(5) = 2.64, p = 0.0459), uncoordinated movement during recovery from anesthesia (*p < 0.05, p = 0.0313), falling and stumbling following ethanol (**p < 0.01, t(4) = 4.887, p = 0.0081), shock (*p < 0.05, t(5) = 3.229, p = 0.0232), white noise (t(5) = 1.526, p = 0.1876), conditioned fear cue (p = 0.56), and looming stimulus (t(5) = 0.2596, p = 0.8055). Paired t test was used for all comparisons except uncoordinated movement during recovery from anesthesia and cued fear (Wilcoxon matched-pairs signed rank test), based on a Shapiro-Wilk test of normality.
Error bars represent SEM. See also Figure S6.
We also observed that CART+ EW calcium transients were sporadic while the mouse moved freely in its home cage or an open field (0.017 ± 0.008 Hz, n = 6 mice, Z score ≥3). We noticed that transients did not coincide with locomotor initiation or cessation, but they did coincide with brief losses of balance or slipping by the animal (29.2% ± 14.3% of observed transients, n = 5 mice, 17 of 50 observed transients). We confirmed this by placing mice in an arena with a thin film of corn oil; we observed time-locked fluorescence signals when mice slipped (Figure 6D).
Induction of CART+ EW activity has previously been observed through increases in c-fos following anesthetic exposure.25 We found that CART+ EW neurons activate during the transient motor dysfunction that occurs following anesthetic exposure (Figure 6E). Furthermore, CART+ EW fluorescence signal also increased as the mice moved in an uncoordinated fashion while recovering from anesthesia (Figure 6F). There is a rich literature linking CART+ EW activity and increases in neuropeptide expression to alcohol administration and consumption.74–76 CART+ EW neurons were activated during bouts of motor incoordination following intraperitoneal injection of ethanol at a dosage of 2.5 g/kg (Figure 6G). We also saw CART+ EW GCaMP6s transients following electric footshocks, which produce rapid defensive movements (Figure 6H). Thus, painful stimuli, or defensive motions that they elicit, may also activate the CART+ EW.
In summary, we observed increases in CART+ EW activity following multiple stimuli that induce a loss of motor control, whether passively (slipping on oil), actively (restraint or suspension), or chemically (anesthetic, alcohol) (Figure 6K). Importantly, other stressful stimuli that did not alter the motor control of the animal, such as a loud white noise (Figure 6I), a conditioned fear cue (Figure 6J), or a looming stimulus (Figure S6G), did not reproducibly elicit time-locked fluorescence responses (Figure 6K).
The CART+ EW promotes anxiety responses
Finally, we evaluated how the CART+ EW influences behavior. Numerous roles for the CART+ EW have been suggested, including alcohol consumption,75 maternal preparatory nesting behavior,31 and alertness.24,37 The CART+ EW has also been implicated in stress adaptation27 on the basis of increased c-fos and neuropeptide production following stressful stimuli. However, whether CART+ EW function is linked to anxiety responses remains uncertain. Therefore, we tested whether CART+ EW activation or ablation alters anxiety-related behaviors. In separate experiments we conditionally expressed the Gαs-coupled, clozapine N-oxide (CNO)-activated rM3Ds for enhancing neuronal activity, diphtheria toxin for ablation, and the Gαi-coupled hM4Di for inhibition of CART+ EW neurons (Figures 7A–7D). Cell-attached voltage-clamp recordings of rM3Ds+ CART+ EW neurons showed significant increases in spontaneous firing rate following CNO application (Figures 7E and S7A). Diphtheria toxin expression routinely ablated >90% of EW neurons (Figure S7B). Current-clamp recordings of hM4Di+ CART+ EW neurons showed significant decreases in firing rate following CNO application (Figures 7F and S7C). For controls, littermate mice of both sexes were transduced with a virus encoding an inert control fluorophore, such as enhanced yellow fluorescent protein (EYFP) or tdT. Anxiety-related behavior was measured ≥3 weeks following AAV infection on an elevated maze or in an open field. No changes in general locomotion in the open field were observed (Figures S7D and S7E).
Figure 7. The CART+ EW promotes anxiety responses.
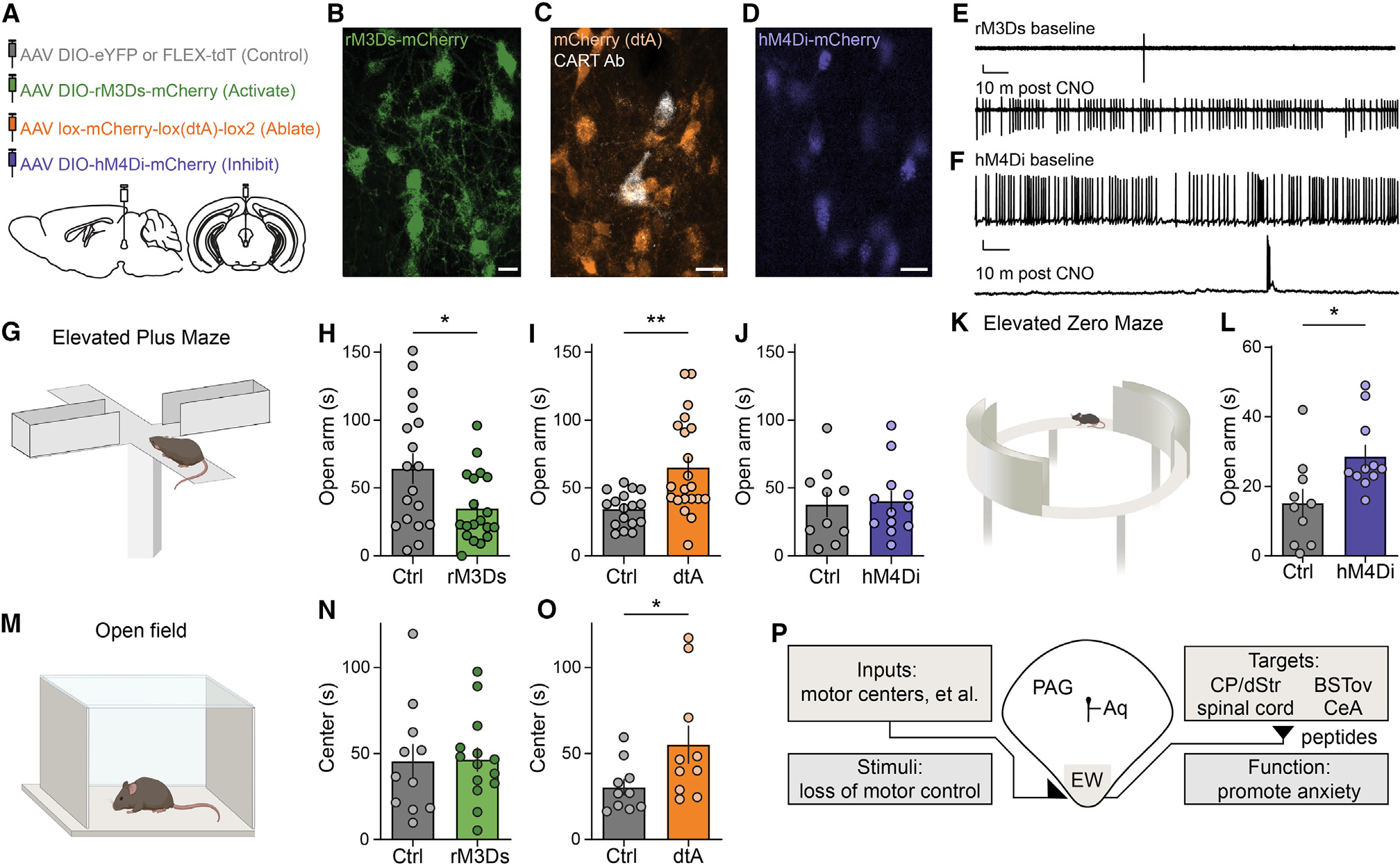
(A) Schematic of injection of AAVs expressing Cre-dependent control fluorophore (gray), rM3Ds (green), diphtheria toxin (dtA, orange), or hM4Di (blue).
(B) rM3Ds-mCherry (green) in CART+ EW. Scale bar, 20 μm.
(C) Cre-off mCherry (orange) in the EW; CART immunofluorescence (white) confirms dtA ablates most CART+ EW neurons. Scale bar, 20 μm.
(D) hM4Di-mCherry (blue) in CART+ EW. Scale bar, 20 μm.
(E) Cell-attached voltage-clamp recording of an rM3Ds-expressing EW neuron at baseline (top) and 10 min after addition of CNO (bottom). Scale bars, 50 pA and 5 s.
(F) Current-clamp recording of an hM4Di-expressing EW neuron at baseline (top) and 10 min after addition of CNO (bottom). Scale bars, 10 mV and 5 s.
(G) Schematic of elevated plus maze.
(H) CNO-injected rM3Ds-expressing mice spend less time than control mice on maze open arm. *p < 0.05, unpaired t test, t(35) = 2.417, p = 0.0210, n = 18 Ctrl mice, n = 19 rM3Ds mice.
(I) dtA mice spend more time than control mice on maze open arm. **p < 0.01, unpaired t test, t(36) = 3.391, p = 0.0017, n = 17 Ctrl mice, n = 21 dtA mice.
(J) Acutely CNO-injected hM4Di-expressing mice and control mice spend similar times on elevated plus maze open arms. Unpaired t test, t(20) = 0.2076, p = 0.8377, n = 10 Ctrl mice, n = 12 hM4Di mice.
(K) Schematic of elevated zero maze.
(L) Chronically CNO-injected hM4Di-expressing mice spend more time than control mice on elevated zero maze open arms. *p < 0.05, unpaired t test, t(19) = 2.668, p = 0.0152, n = 10 Ctrl mice, n = 11 hM4Di mice.
(M) Schematic of open field test.
(N) Time spent in center of the open field is similar for rM3Ds and control mice. Unpaired t test, t(23) = 0.08149, p = 0.9358, n = 11 Ctrl mice, n = 14 rM3Ds mice.
(O) dtA mice spend more time than control mice in the center of the open field. *p < 0.05, unpaired t test, t(18) = 2.113, p = 0.0488, n = 10 Ctrl mice, n = 10 dtA mice.
(P) An anatomical-, circuit-, and function-based model for the EW as a ventromedial column of the PAG.
Error bars represent SEM. See also Figure S7.
On an elevated plus maze, a common behavioral assay for anxiety responses (Figure 7G), mice with increased CART+ EW activity spent significantly less time on the open arms (Figure 7H), suggesting that the CART+ EW may be anxiogenic. Mice with genetically targeted ablation of CART+ EW neurons spent significantly more time on the open arms as compared with littermate control AAV-injected mice (Figure 7I). We next examined whether acute inhibition of CART+ EW neurons using CNO with the Gαi-coupled hM4Di alters elevated maze behavior. Despite the acute reduction by CNO of CART+ EW action potential firing rate in cells expressing hM4Di, a single administration of CNO failed to elicit changes in behavior on an elevated plus maze (Figure 7J). We then chronically administered CNO (twice daily, ~12 h apart, for 10–14 days) to this same cohort of mice. Because changes in anxiety behavior are commonly observed during repeated measures on an elevated plusmaze,77–79 we used an elevated zero maze to measure anxiety following chronic CNO administration (Figure 7K). Chronically CNO-injected mice spent more time on the open arms of an elevated zero maze in comparison with littermate control-injected mice (Figure 7L). The observation that acute administration of CNO produced robust inhibition of CART+ EW electrical activity while chronic administration of CNO was needed to alter behavioral responses is inconsistent with rapid glutamatergic or GABAergic signaling.
Mice were also tested in an open field assay for anxiety behavior (Figure 7M). Activation of CART+ EW neurons did not alter the amount of time mice spent in the center of the open field (Figure 7N), but genetic ablation of CART+ EW neurons increased the amount of time mice spent in the center (Figure 7O). Taken together, these findings are consistent with the possibility that the CART+ EW behaves as a peptidergic anxiogenic locus. Summarizing our data, we propose that the CART+ EW responds to stimuli that induce a loss of motor control. It then signals in a predominantly obligate peptidergic fashion to numerous downstream targets throughout the central nervous system, enhancing anxiety responses (Figure 7P).
DISCUSSION
We hypothesize that neuronal populations in the brain including the CART+ EW could be obligately peptidergic, i.e., incapable of fast neurotransmission. Proving the absence of something is intrinsically difficult, especially given the existence of non-canonical purinergic and gaseous transmitters, and further research will be needed to formally define any neuronal population as obligately peptidergic. For example, prior studies of Trh+, Oxt+, and Avp+ neurons of the PVN have potentially ruled out these populations as being obligately peptidergic. Trh+/urocortin 3+ neurons of the PVN/perifornical area colocalize with VGlut2 and produce EPSCs in multiple downstream areas.80 Robust monosynaptic EPSCs have been recorded in agouti-related peptide (AgRP) neurons of the arcuate hypothalamus upon optogenetic stimulation of Trh+ PVN terminals, but glutamatergic transmission from Oxt+ or Avp+ neurons was not observed.81 Previously, we did not observe glutamatergic transmission from Oxt+ terminals in the ventral tegmental area.82 Following ChR2 expression in the PVN, fibers that projected to the dorsal motor nucleus of the vagus included Oxt+ fibers and released glutamate.83 Similarly, expression of ChR2 in a mixed population of Oxt+ and oxytocin receptor+ neurons of the PVN could elicit light-evoked action potentials and EPSCs in a subpopulation of oxytocin receptor+ neurons of the parabrachial nucleus.84 Others have found that the Oxt+ PVN is glutamatergic, based on VGLUT2 immunolabeling in Oxt+ PVN fibers and glutamate-dependent modulation of disynaptic IPSCs following light-evoked excitation of these fibers.51,85,86 Finally, the Avp+ PVN has recently been found to colocalize with VGlut2 based on FISH.87 However, all our FISH, immunofluorescence, electron microscopy, and electrophysiology data suggest that the bulk of the CART+ EW may be an obligate peptidergic population.
One piece of evidence against our hypothesis is a prior report of evoked EPSCs on parvalbumin interneurons of the medial prefrontal cortex (mPFC) after optogenetic stimulation of ChR2+ fibers following delivery of ChR2 to the EW of a CCK-Cre transgenic mouse line.24 We cannot preclude the possibility that the small proportion of CART+ neurons we found containing mRNA for a vesicular glutamate transporter (~11.5%) project to the mPFC, although we never observed robust projections to this brain region. Another possible explanation is that the use of AAVs with a CCK-Cre mouse line may not selectively target the CART+ EW, even though CART and CCK colocalize in these neurons. For example, AAV injection in the midbrain of a CCK-Cre mouse line drove expression of ChR2 in sleep-promoting glutamatergic CCK+ neurons in the nearby perioculomotor region.23 Additionally, characterization of a CCK-Cre mouse line88 shows expression in the mediodorsal nucleus of the thalamus. Mediodorsal thalamic neurons supply glutamatergic inputs to parvalbumin interneurons in medial prefrontal cortex89 and could underlie the putatively EW-derived EPSCs previously recorded from the mPFC.24 Use of the transgenic CART-Cre line to target the EW, therefore, is likely to produce different results from using the CCK-Cre line to target the EW, even though both peptides colocalize in the CART+ EW.
The CART+ EW has been shown to respond to numerous stressors,25–28 and stress responses are highly interconnected with anxiety.90 The CART+ EW does not appear to behave as a multimodal stress- or threat-alarm system, contrary to previously described neurons of the parabrachial nucleus.91,92 However, it is clearly activated by numerous stressors and stimuli that induce disruptions in motor control (Figure 6). We also found evidence for CART+ EW terminal projections to the BSTov and CeA (Figure 3), which each mediate anxiety and/or fear responses. Additionally, many neuropeptides contained in the CART+ EW have been implicated in anxiety. CART itself has been shown to be anxiogenic,93 as have PACAP,94 CCK,95 and nesfatin.96 PACAP receptors have also been localized to the BSTov where they mediate anxiety responses.97,98 Other candidate obligate peptidergic neurons in the PVN have been shown to modulate fear or anxiety,51,85,99 so peptidergic modulation of anxiety may be widespread.
The CART+ EW has a history of being mischaracterized. The anatomical area now shown to contain the CART+ EW was incorrectly defined in the 19th and much of the 20th century as the site of the cholinergic EW oculomotor nucleus.20 As a result, despite residing within the PAG, the CART+ EW has typically been considered distinct.100 Both the CART+ EW and the PAG are phylogenetically ancient, with strong conservation through mammals20,101 and proposed homologous structures found in teleost fish.37,101 The PAG is known to mediate defensive behaviors in response to threats, producing anxiety and fear responses,72,73,100–102 and we show here that the CART+ EW responds to loss of motor control and promotes anxiety responses. The PAG canonically comprises four columns—dorsomedial, dorsolateral, lateral, and ventrolateral72,101—with different columns potentially responding preferentially to specific classes of threat.100,102 The phylogenetic and anatomical similarities between the CART+ EW and the PAG, as well as our in vivo fiber photometry and behavioral assay results, yield a model in which the CART+ EW may form a ventromedial column of the PAG. Additionally, the results of this study clarify opposing characterizations of CART+ EW function, advance our understanding of neuromodulatory mechanisms underlying anxiety behavior, and support the classification of the CART+ EW as an obligate peptidergic nucleus.
Numerous questions surrounding peptidergic signaling remain, including details of the genesis and maturation of large vesicles,103,104 to what extent peptides are colocalized or segregated within individual vesicles or release sites,105 the mechanisms underpinning peptide vesicle release,106–110 and the modulatory capabilities of neuronal processing in the absence of faster-acting neurotransmitters.111 Our characterization of a putatively obligate peptidergic population in the mammalian midbrain, together with associated ethologically relevant behaviors, provides an effective system for investigation of neuronal peptide transmission.
Limitations of the study
Our in vivo fiber photometry recordings were acquired in freely moving animals. This allowed us to examine behavior in a more ethologically relevant fashion and to reveal that a loss of motor control underlies increases in CART+ EW calcium activity in response to disparate stimuli (e.g., restraint, anesthesia, alcohol). However, this approach does preclude a rigorous quantification of the loss of motor control. Additionally, animals can make unplanned movements while retaining motor control, and further experiments could help determine whether these unplanned movements also activate the CART+ EW.
Neuropeptidergic signaling can range from seconds- or minutes-long neurophysiological changes to days-long changes in gene expression or structural plasticity,3 and the exact time course of the neuropeptidergic signaling we observe remains unclear. Our electrophysiological data showed that downstream cellular activity is increased on timescales on the order of minutes. Our behavioral data suggested that acute CNO-induced inhibition was insufficient to alter behavior, while CNO-induced inhibition or ablation for longer than 10 days did alter behavior. It remains unknown whether shorter bouts of inhibition (e.g., two administrations of CNO spaced 2 h or 12 h apart) would suffice to alter behavior. These experiments, along with investigations into CART+ EW-induced neuroplasticity, could further reveal the full time course and mechanisms of obligate neuropeptidergic signaling.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yevgenia Kozorovitskiy (yevgenia.kozorovitskiy@northwestern.edu).
Materials availability
This study did not generate new unique reagents. Previously published lab-generated plasmids (e.g., CBA-FLEX-rM3Ds-mCherry) will be deposited on Addgene.
Data and code availability
Data in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://github.com/KozorovitskiyLaboratory and is publicly available as of the date of publication. DOIs are available in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-CART | Phoenix Pharmaceuticals | H-003–62; RRID:AB_2313614 |
| Anti-Urocortin | Sigma | U4757; RRID:AB_261834 |
| Anti-RFP (e.g., tdTomato, mCherry) | Rockland Immunochemicals | 600–401-379; RRID:AB_2209751 |
| Anti-ChAT | Millipore | AB144P; RRID:AB_2079751 |
| Anti-GFP (e.g., EGFP) | Abcam | ab13970; RRID:AB_300798 |
| Goat Anti-Rabbit Alexa Fluor 488 | Life Technologies | A11008; RRID:AB_143165 |
| Goat Anti-Rabbit Alexa Fluor 647 | Life Technologies | A21244; RRID:AB_2535812 |
| Donkey Anti-Goat Alexa Fluor 647 | Life Technologies | A21447; RRID:AB_2535864 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| AAV1-CAG-FLEX-tdTomato-WPRE | University of Pennsylvania Vector Core; Addgene; Oh et al.112 | Addgene 51503 |
| AAV1-EF1 α-DIO-LCK-APEX2-P2A-EGFP | University of North Carolina Vector Core; Dumrongprechachan et al.44 | Addgene 182826 |
| AAV1-CBA-FLEX-ChR2-mCherry | University of Pennsylvania Vector Core; Vigene; Atasoy et al.113 | Addgene 18916 |
| AAVrg-CAG-FLEX-EGFP | Addgene; Oh et al.112 | Addgene 51502-AAVrg |
| AAV1-CBA-DIO-rM3Ds-mCherry-WPRE | Vigene; Wu et al.114 | N/A |
| AAV1-EF1 α-Lox-mCherry-lox(dtA)-lox2 | University of North Carolina Vector Core; Wu et al.115 | Addgene 58536 |
| AAV1-CBA-DIO-hM4Di-mCherry-WPRE | Vigene; Hou et al.116 | Addgene 81008 |
| AAV9-EF1 α-DIO-eYFP-WPRE | University of Pennsylvania Vector Core; Addgene | Addgene 27056 |
| AAV1-CAG-Flex-H2B-eGFP-N2c(G) | Zuckerman Institute Virology Core; Reardon et al.63 | N/A |
| AAV1-EF1 α-FLEX-GT | Salk Institute Viral Vector Core | Addgene 26198 |
| Rabies virus CVS-N2cΔG tdTomato EnvA | Zuckerman Institute Virology Core; Reardon et al.63 | Addgene 73462 |
| AAV1-CAG-FLEX-GCaMP6s-WPRE | University of Pennsylvania Vector Core; Chen et al.117 | Addgene 100842 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Streptavidin AF488 | Invitrogen | S32354 |
| Biotinylated IB4 | Sigma | L2140; RRID:AB_2313663 |
| Hoechst 33342 | ThermoFisher Scientific | H1399 |
| Prolong Gold Antifade Mountant with DAPI | ThermoFisher Scientific | P36931 |
| Low-melting point agarose | Sigma | A9414 |
| 3,3′-Diaminobenzidine (DAB) with metal enhancer | Sigma | D0426 |
| Bovine serum albumin | Sigma | A3059 |
| Triton X-100 | Sigma | T8787 |
| Paraformaldehyde | Electron Microscopy Sciences | 15714 |
| Glutaraldehyde | Fisher Scientific | 5026223 |
| [D-p-Cl-Phe6,Leu17]-VIP | Tocris | 3054 |
| PACAP 6–38 | Tocris | 3236 |
| Proglumide sodium salt | Tocris | 1478 |
| Astressin 2B | Tocris | 2391 |
| NBI 35965 hydrochloride | Tocris | 3100 |
| CART (55–102) | Phoenix Pharmaceuticals | 003–62 |
| Nesfatin-1 (1–82) | Phoenix Pharmaceuticals | 003–22A |
| U 73122 | Tocris | 1268 |
| PKI (5–24) | Tocris | 6221 |
| Clozapine N-oxide | Enzo Life Sciences | BML-NS105 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAScope Fluorescence Multiplex Reagent Kit | ACDBio/BioTechne | 320850 |
| Cartpt-C1 | ACDBio/BioTechne | 432001 |
| Adcyap1-C1 | ACDBio/BioTechne | 405911 |
| Penk-C1 | ACDBio/BioTechne | 318761 |
| Nmb-C1 | ACDBio/BioTechne | 459931 |
| Slc17a6-C2 | ACDBio/BioTechne | 319171-C2 |
| Slc17a7-C2 | ACDBio/BioTechne | 416631-C2 |
| Cck-C2 | ACDBio/BioTechne | 402271-C2 |
| Pdyn-C2 | ACDBio/BioTechne | 318771-C2 |
| Pomc-C2 | ACDBio/BioTechne | 314081-C2 |
| Cartpt-C3 | ACDBio/BioTechne | 432001-C3 |
| Slc17a8-C3 | ACDBio/BioTechne | 431261-C3 |
| Tac1-C3 | ACDBio/BioTechne | 410351-C3 |
| Slc32a1-C3 | ACDBio/BioTechne | 319191-C3 |
|
| ||
| Deposited data | ||
|
| ||
| scRNAseq data | Linnarsson group, Zeisel et al.15 | mousebrain.org/adolescent; Sequence Read Archive: SRP135960 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6 | Charles River | 000664; RRID: IMSR_JAX:000664 |
| Mouse: B6; 129S-Cartpttm1.1(cre)Hze/J | Jackson Laboratory | 028533; RRID:IMSR_JAX:028533 |
| Mouse: Slc17a6tm2(cre)Lowl/J | Jackson Laboratory | 016963; RRID:IMSR_JAX:016963 |
| Mouse: Slc32a1tm2(cre)Lowl/J | Jackson Laboratory | 016962; RRID:IMSR_JAX:016962 |
|
| ||
| Software and algorithms | ||
|
| ||
| MATLAB | Mathworks | mathworks.com |
| FIJI | Schindelin et al.118 | https://imagej.net/software/fiji/downloads |
| AtlasQuantifier | This paper | https://zenodo.org/record/8172932 |
| LoomLab | This paper | https://zenodo.org/record/8172935 |
| StCamSWare | Sentech | N/A |
| Yamb | Kurtnoise | http://yamb.unite-video.com/download.html |
| ToxTrac | Rodriguez et al.119 | https://sourceforge.net/projects/toxtrac/ |
| BioRender | BioRender | biorender.com |
| Prism 8.4 | GraphPad | graphpad.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice used in all experiments except fluorescence in situ hybridization were heterozygous, formed by crossing B6; 129S-Cartpttm1.1(cre)Hze/J+/− (CART-Cre, #028533, The Jackson Laboratory)35 mice to each other, or, more commonly, crossing CART-Cre+/+ mice to wild-type C57BL/6J mice (Charles River, Wilmington, MA). Wild-type C57BL/6J mice were used for fluorescence in situ hybridization. For retrograde tracing experiments, heterozygous Slc17a6tm2(cre)Lowl/J (VGlut2-Cre, #028863 or #016963, The Jackson Laboratory) or Slc32a1tm2(cre)Lowl/J (Vgat-Cre, #016962, The Jackson Laboratory) were backcrossed to wild-type C57BL/6J mice. After weaning, experimental mice were put in single-sex housing with ad libitum food and water. Mouse were generally maintained on a 12h:12h light-dark cycle, with a subset of mice maintained on a 12h:12h reverse light-dark cycle following surgery prior to behavioral experiments. Mice were maintained on the reverse light-dark cycle for >3 weeks prior to behavioral experiments. Male and female mice were used for all experiments, and all experiments were performed on adult (>P40) mice. Littermates of the same sex were randomly assigned to experimental groups, when applicable. All mouse handling, surgeries, and behavioral experiments were performed according to protocols approved by the Northwestern University Animal Care and Use Committee.
METHOD DETAILS
Single-cell RNAseq database analysis
The dataset ‘l6_r2_cns_neurons.loom’ was downloaded from mousebrain.org.15 Neuronal types were taken from the clusters defined within the scRNAseq database. A custom MATLAB (MathWorks) script tabulated the existence of user-defined transcripts in each cell, where any evidence of a transcript in a cell was sufficient to positively-identify a cell as containing that transcript. If a cell was positively identified as containing a transcript that was a marker for any population (i.e., glutamatergic, GABAergic, or monoaminergic), it was considered positively identified within that population. The proportion of cells in each neuronal type that were positively identified for a given neurotransmitter-defined population were calculated, and neuronal types were then plotted in three dimensions based on these proportions.
Fluorescence in situ hybridization
Wild-type C57BL/6J mice were deeply anesthetized prior to decapitation and brain extraction. Brains were rapidly frozen in Tissue-Tek O.C.T. Compound (VWR) using a slurry of dry ice and ethanol and then transferred to −80°C overnight. 20 μm thick brain slices for fluorescence in situ hybridization were cut from fresh frozen brains at −15°C to −25°C using a Leica CM1850 cryostat (Leica Biosystems). Slices were mounted on Superfrost Plus microscope slides (Fisher Scientific) and processed and labeled with fluorescence in situ hybridization probes according to the manufacturer (ACDBio) instructions. Both male and female mice were included in each dataset. Probes used included Cartpt-C1, Adcyap1-C1, Penk-C1, Nmb-C1, Slc17a6-C2, Slc17a7-C2, Cck-C2, Pdyn-C2, Pomc-C2, Cartpt-C3, Tac1-C3, and Slc32a1-C3. Labeled slices were covered with Prolong Gold Antifade Mountant with DAPI (ThermoFisher Scientific) and coverslipped.
FISH data were collected and analyzed similarly to previous descriptions.55,82 Stacks were taken at a 0.5 mm interval on a Zeiss 880 or Leica SP8 confocal microscope at 40x, with imaging for DAPI, Alexa Fluor 488, Atto 550, and Atto 647. Five consecutive z-plane images were merged for analysis, and cells were found based on DAPI signal and a watershed algorithm on fluorescent signal. Cartpt transcript was frequently so abundant that individual puncta could not be observed at this or higher magnification levels; a conservative checkerboard counting system was used,82 assuming that the fluorescence signals from puncta were approximately 2 pixels (1.248 μm) in diameter.
Intracranial injections and implants
Viral vectors were stored at −80°C prior to use and were backfilled into Wiretrol II pipettes (Drummond Scientific Company) pulled on a P-1000 Flaming/Brown micropipette puller (Sutter Instruments). Mice were anesthetized with vaporized isoflurane and positioned on a stereotactic apparatus (Kopf Instruments) so the skull was lying flat with the vertical positions of lambda and bregma within 0.1 mm of each other. For injections to the EW, the pipette was placed at the midline (±0.0 mm M/L), 0.7 mm rostral to lambda (+0.7 mm A/P), and, generally, both 3.5 and 3.1 mm ventral to the pial surface (−3.5 and −3.1 D/V). Injection at two depths was performed to ensure sufficient transduction of the EW, which varies by > 1 mm across its rostrocaudal axis. For injections to the dorsal striatum, the pipette was placed at −2.6 mm M/L, 0.6 mm rostral to bregma (+0.6 mm A/P), and 2.6 mm ventral to the pial surface (−2.6 D/V). Virus was injected using a microsyringe pump controller (World Precision Instruments); >2 min elapsed before the pipette was moved from the ventral injection site to the more dorsal injection site, and >5 min elapsed before the pipette was slowly retracted fully from the brain following the second injection. Approved postoperative analgesia protocols were performed.
For anterograde fluorescent projection mapping, 50–100 nL of AAV1-CAG-FLEX-tdTomato-WPRE112 (1.9 × 1013 gc/mL, University of Pennsylvania Vector Core) was injected at −3.5 and −3.1 D/V at a rate of 50 nL/min. For APEX-mediated electron microscopy, 200 nL of AAV1-EF1α-DIO-LCK-APEX2-P2A-EGFP (University of North Carolina Vector Core, 5.0 × 1012 gc/mL) was injected at −3.3 D/V at a rate of 100 nL/min. For light-induced excitation, 200–250 nL of AAV1-CBA-FLEX-ChR2-mCherry113 (5–6 × 1012 gc/mL, University of Pennsylvania Vector Core or Vigene) was injected at −3.5 and −3.1 D/V at a rate of 100 nL/min. For retrograde validation of EW projections to striatum, 800 nL of AAVrg-CAG-FLEX-EGFP112 was injected into the dorsal striatum at a rate of 150 nL/min120,121 (Addgene, 5.4 × 1012 gc/mL). For pharmacogenetics experiments, 225–300 nL of AAV1-CBA-DIO-rM3Ds-mCherry-WPRE114 (Vigene, 1.08 to 4.3 × 1013 gc/mL) or 225 nL of AAV1-CBA-DIO-hM4Di-mCherry-WPRE116 (Vigene, 1.28 × 1013 gc/mL), was injected at −3.5 and −3.1 D/V at a rate of 100 nL/min.
For retrograde rabies tracing, AAV1-CAG-Flex-H2B-eGFP-N2c(G) (1.43 × 1012 gc/mL, Zuckerman Institute Virology Core) and AAV1-EF1α-FLEX-GT (6.14 × 1011 gc/mL, Salk Institute Viral Vector Core) were injected into adult mice in a single 150–200 nL injection at −3.4 D/V at a rate of 100 nL/min. After three weeks, rabies virus CVS-N2cΔG tdTomato EnvA63 (Zuckerman Institute Virology Core) was injected at −3.6, −3.3, and −3.0 D/V with 400 nL released at each site at a rate of 100 nL/min.
For fiber photometry, 200–250 nL of AAV1-CAG-FLEX-GCaMP6s-WPRE117 (University of Pennsylvania Vector Core, 4 × 1012 to 1 × 1013 gc/mL) was injected at −3.5 and −3.1 D/V at a rate of 100 nL/min. For genetically targeted ablation, 300–400 nL of AAV1-EF1α-Lox-mCherry-lox(dtA)-lox2115 (University of North Carolina Vector Core, Canadian Neurophotonics Platform Viral Vector Core Facility, 5.2 × 1012 gc/mL) was injected at −3.5 and −3.1 D/V at a rate of 100 nL/min. Control behavioral animals were injected with AAV1-CAG-FLEX-tdTomato-WPRE or AAV9-EF1α-DIO-eYFP-WPRE in a titer and volume matched manner (Addgene or University of Pennsylvania Vector Core).
AAV1-CAG-FLEX-tdTomato for anterograde tracing was packaged from AAV pCAG-FLEX-tdTomato-WPRE, which was a gift from Hongkui Zeng (Addgene plasmid # 51503); additional AAV1-CAG-FLEX-tdTomato was packaged from pAAV-FLEX-tdTomato, which was a gift from Edward Boyden (Addgene plasmid # 28306). AAV1-EF1α-DIO-LCK-APEX2-P2A-EGFP was packaged from AAV-EF1a-DIO-LckAPEX-P2A-EGFP plasmid, which we have deposited at Addgene (plasmid # 182826). AAV1-CBA-FLEX-ChR2-mCherry was packaged from AAV-FLEX-rev-ChR2(H134R)-mCherry plasmid, which was a gift from Scott Sternson (Addgene plasmid #18916). AAVrg-CAG-FLEX-EGFP was packaged from AAV pCAG-FLEX-EGFP-WPRE, which was a gift from Hongkui Zeng (Addgene viral prep # 51502-AAVrg). AAV1-EF1α-FLEX-GT was packaged from pAAV-EF1a-FLEX-GT, which was a gift from Edward Callaway (Addgene plasmid # 26198). Rabies virus CVS-N2cΔG tdTomato EnvA was packaged from RabV CVS-N2c(deltaG)-tdTomato, which was a gift from Thomas Jessell (Addgene plasmid # 73462). AAV1-CAG-FLEX-GCaMP6s-WPRE was packaged from plasmid pAAV.CAG.Flex.GCaMP6s.WPRE.SV40, which was a gift from Douglas Kim & GENIE Project (Addgene plasmid #100842). AAV1-CBA-DIO-rM3Ds-mCherry-WPRE was modified from plasmid pAAV-hSyn-DIO-rM3D(Gs)-mCherry, which was a gift from Bryan Roth (Addgene plasmid #50458). AAV1-CBA-DIO-hM4Di-mCherry-WPRE was packaged from pAAV-CBA-DIO-hM4Di-mCherry, which was a gift from Bernardo Sabatini (Addgene plasmid #81008). AAV1-EF1α-Lox-mCherry-lox(dtA)-lox2 was packaged from pAAV-mCherry-flex-dtA, which was a gift from Naoshige Uchida (Addgene plasmid # 58536). AAV9-EF1α -DIO-eYFP-WPRE was packaged from pAAV-Ef1a-DIO EYFP, which was a gift from Karl Deisseroth (Addgene plasmid # 27056).
For retrograde and anterograde tracing, adult mice > P60 were injected. Mice between P24 and P45 were injected for electrophysiological recordings. Mice between P60 and P65 were injected for behavioral experiments.
GCaMP6s-injected mice were implanted with a 400 μm diameter, 0.48 NA photometry fiber with a mirrored tip angled at 45° (MFC_400/430–0.48_4.5mm_MF2.5_MA45, Doric Lenses). The angled tip allowed chronic placement of the fiber in a position that could image the EW without blocking the cerebral aqueduct found immediately dorsal. Fiber placement coordinates were +0.5 to 0.6 mm A/P of lambda, ±0.3 to 0.4 mm M/L, and −3.4 to −3.6 mm D/V. Following behavioral experiments, post hoc confirmation of appropriate fiber placement was performed.
Fixed tissue preparation
Mice were anesthetized with isoflurane and perfused transcardially with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) in phosphate buffered saline (PBS). Brains and when applicable, spinal columns, were extracted and stored in 4% PFA overnight. Brain slices were made at 80 μm thickness for fluorescent projection mapping and 60 μm for all other analyses. Some brains and all spinal cords were embedded in a gel of 4% low-melting point agarose (Sigma) for slicing. All slices were made using a Leica VT1000 S vibratome (Leica Biosystems).
Immunofluorescence
Slices were stored in PBS and sampled at between 1 in 6 and 1 in 3 for immunofluorescence and expression validation and sampled at 1 in 2 for projection mapping. Immunofluorescence staining protocols varied based on primary antibody. For CART, urocortin, red fluorescent protein (RFP, e.g., tdTomato), green fluorescent protein (GFP, e.g., EGFP), and isolectin B4 staining, slices were permeabilized in 0.2% Triton X-100 for 1–2 h, blocked with 10% bovine serum albumin (BSA) with 0.05% Triton X-100 for 2 h, washed, stained with 1:1000 primary antibody (anti-CART Ab, Phoenix Pharmaceuticals, H-003–62; anti-urocortin Ab, Sigma, U4757; anti-RFP Ab, Rockland Immunochemicals, 600–401-379), 1:2000 primary antibody (anti-GFP Ab, Abcam, ab13970), or 1:1500 biotinylated IB4 (Sigma, L2140) while shaking at 4°C overnight in solution with 0.2% Triton X-100, washed, stained with 1:500 secondary antibody (Goat Anti-Rabbit 488 or 647 or Goat Anti-Chicken 488, Life Technologies) or 1:500 streptavidin AF488 (Invitrogen S32354), and washed a final time. For choline acetyltransferase (ChAT) staining, the protocol used was from a prior report.122 Briefly, slices were rinsed in Tris-HCl buffer and then blocked and permeabilized in Tris-HCl buffer with 5% donkey serum and 0.2% Triton X-100 for 60 min, stained with goat anti-ChAT antibody (AB144P, Millipore) overnight with shaking at 4°C, rinsed in Tris-HCl buffer with 0.2% Triton X-100, and incubated for 2 h in 1:500 secondary antibody (Donkey Anti-Goat 647, Life Technologies). Slices were then mounted on Superfrost Plus microscope slides (Fisher Scientific), dried, and coverslipped with Hoechst 33342 nuclear stain (ThermoFisher Scientific).
Anatomical imaging
Virally fluorescently expressing brain slices and immunofluorescently labeled brain sections were imaged on an Olympus VS.110 imaging system at 10x. Regions of interest were subsequently imaged on a Leica TCS SPE confocal microscope (Leica Microsystems) at 40x or a Zeiss LSM800 confocal microscope at 63x. Colocalization counts of immunofluorescence and genetically encoded fluorescent protein signal were performed manually. Quantitative two-dimensional projection mapping was performed using the CCFv3 of the Allen Mouse Brain Atlas,123 downloaded in November 2017. Atlas images were manually registered in Adobe Illustrator against raw data images that were obtained by extracting slices imaged on the Olympus VS110 imaging system using the Bioimaging and Optics Platform VSI Reader ActionBar plugin within FIJI.118 Custom-written MATLAB scripts quantified the number of pixels found with a Sobel edge detector in anatomical regions defined in the atlas.
Transmission electron microscopy (TEM)
For preparation of TEM tissue, mice were transcardially perfused with ice-cold PBS, followed by ice-cold PBS containing 2% glutaraldehyde and 2% PFA. Following overnight post-fixation at 4°C in the same fixative, coronal brain slices were made at 100 μm on a Leica VT1000 vibratome. Slices were incubated with 3,3′-Diaminobenzidine (DAB) with metal enhancer (D0426, Sigma). DAB solution (0.25 mg/mL DAB, 0.1 mg/mL CoCl2, 0.15 mg/mL H2O2) was prepared by dissolving DAB and hydrogen peroxide tablets separately in 5 mL of PBS for each. Solutions were mixed immediately before use. Brain slices were incubated in DAB solution for ~3 min to selectively label APEX-containing cellular structures. After DAB precipitation, slices were washed several times with 0.05 M sodium phosphate buffer (PB), and then processed for TEM with 2 exchanges of fixative that consisted of 2.5% glutaraldehyde, 2% PFA in 0.1 M PB.
Slices were washed 3x with buffer followed by a secondary fixation in 1.5% osmium tetroxide (aqueous). Samples were washed 3x with DI water before beginning an acetone dehydration series. Osmium staining, washes, and acetone dehydration series were carried out in a Pelco Biowave Microwave with Cold Spot and vacuum. EMBed 812 embedding media by EMS was gradually infiltrated with acetone for flat embedding. Selected ROIs were cut out and mounted on a blank stub for sectioning. 90 nm thin sections were collected on copper grids using a Leica Ultracut S ultramicrotome and DiATOME 45° diamond knife. Images were acquired at 100 kV on a 1230 JEOL TEM and Gatan Orius camera with Digital Micrograph software. Magnification for quantified images was 8000x. All quantified images came from multiple sample sections taken from a single mouse. This work made use of the BioCryo facility of Northwestern University’s NUANCE Center, which has received support from the SHyNE Resource (NSF ECCS-2025633), the IIN, and Northwestern’s MRSEC program (NSF DMR-1720139).
Fresh tissue preparation
Coronal brain slices were prepared from P40 to P65 mice that had been deeply anesthetized with isoflurane and then perfused transcardially with cold, oxygenated ACSF containing, in mM, 127 NaCl, 2.5 KCl, 25 NaHCO3, 1.25 NaH2PO4, 2 CaCl2, 1 MgCl2, and 25 glucose with a final osmolarity of around 310 mOsm/L. Extracted brains were sliced in cold ACSF with the support of a small piece of 4% agar. Slices were made at 250 μm thickness for striatum and 300 μm for all others using a Leica VT1000s vibratome and then transferred into a holding chamber with ACSF equilibrated with 95% O2/5% CO2, where they were incubated at 34°C for 20–30 min prior to recording. Slices for recording optogenetic stimulation of EW terminals in the bed nucleus of the stria terminalis were prepared similarly, but with a cold sucrose cutting solution in place of ACSF for the perfusion and slicing steps; slices were then incubated in ACSF at 34°C for 60 min prior to recording. Sucrose cutting solution contained, in mM, 194 sucrose, 20 NaCl, 4.4 KCl, 26 NaHCO3, 1.2 NaH2PO4, 2 CaCl2, 1 MgCl2, and 10 glucose.124
Electrophysiology
The recording chamber was perfused with oxygenated ACSF. Neurons were visualized with QIClick CCD camera (QImaging) under the control of MicroManager,125 using infrared DODT contrast under a 40x or 60× water-immersion objective (LUMPlan FL, Olympus) with a PE300 CoolLED illumination system (CoolLED Ltd.) providing illumination for fluorescence visualization and optogenetic stimulation, as required. For current clamp experiments, optogenetic stimulation was performed with a 460 nm LED at a power of 10 mW, at 30 Hz with a 10 ms pulse duration.51 For SPNs, the amplitude of minimal current injection necessary for evoking action potentials was determined in intervals of 25 pA. Action potentials were also evoked at 50 pA below and above this value. In the BST, minimal current injection was determined in intervals of 5 pA. For all current clamp optogenetic experiments, current injections were performed with 10 s between current injections. The order of the three amplitudes of injected current was varied randomly between cells. For voltage clamp experiments to measure fast neurotransmitter release, optogenetic stimulation was performed at a power of 10 mW at 0.5 Hz with a 10 ms pulse duration. Excitatory postsynaptic currents were measured at a holding potential of −70 mV and inhibitory postsynaptic currents were measured at a holding potential of 0 mV. Recordings with a leak current >150 pA for EPSCs and >300 pA for IPSCs were excluded from analysis.
Electrophysiological recordings were obtained using an Axon 700B amplifier (Axon Instruments), sampled at 10–20 kHz, and filtered at 3–5 kHz with ScanImage, an adapted MATLAB-based acquisition package.126 BNC-2110 data acquisition boards (National Instruments) were used for data acquisition and amplifier and LED pulse control. For whole cell current clamp and cell-attached recordings, the internal solution contained, in mM, 135 K-gluconate, 4 KCl, 10 HEPES, 10 Na2-phosphocreatine, 4 MgATP, 0.4 Na2GTP, and 0.5 to 1 EGTA (pH 7.2, ~295 mOsm/L). For some recordings, compounds were added to the internal solution to visualize cell morphology or confirm cell identity and location: Alexa Fluor 488 (10–20 μM, Thermo Fisher Scientific), Neurobiotin (0.1%, Neurobiotin 488 tracer, Vector Laboratories), Neurobiotin-Plus (0.5%, Vector Laboratories). To block G protein-coupled receptors, we added a cocktail of pharmacological antagonists to the bath. The cocktail included: 1 μM [D-p-Cl-Phe6,Leu17]-VIP to block VPACRs,127 200 nM PACAP 6–38 to block PAC1R,128 2 mM proglumide to block CCKRs,129 300 nM astressin 2B to block CRFR2,130 and 1 μM NBI 35965 hydrochloride to block CRFR1130 (all from Tocris). For flow-in peptide experiments we added 40 nM CART (55–102) or 10 nM nesfatin-1 (1–82) (both from Phoenix Pharmaceuticals). To block Gαq signaling pathways, 10 μM U 73122 (Tocris) was added to the bath solution. To block Gαs signaling pathways, 20 μM PKI (5–24) (Tocris) was added to the internal solution. For rM3Ds validation, spontaneous currents were recorded from mCherry+ EW neurons in cell-attached mode. For hM4Di validation, membrane potentials were recorded from mCherry+ EW neurons under current-clamp. Some neurons fired spontaneously at baseline; others were silent at baseline and were injected with a small amount of current (<50 pA) to elicit action potentials. 10–20 μM clozapine N-oxide (Enzo Life Sciences) dissolved in either ACSF or saline was added to the bath solution. For whole cell voltage-clamp recordings, the bath solution contained 5 μM CPP and the internal solution contained, in mM, 120 CsMeSO3, 15 CsCl, 10 HEPES, 2 QX-314 Cl, 2 MgATP, 0.3 Na2GTP, and 1 EGTA (pH ~7.2, ~295 mOsm/L).
Electrophysiological data were analyzed in MATLAB. For measurements of current-evoked action potentials, 0 mV threshold was imposed for identification. Action potentials were averaged across five trials for optogenetic experiments and across three trials for ligand application experiments. To calculate the time-course of optogenetic stimulation-evoked excitation of SPNs and BSTov neurons, action potential rates were plotted over time in seconds post-light application. This data was then fit with the formula , with the reciprocal of b equal to the time constant of the increase in firing.131 For cell-attached mode, action potential current rates were averaged across 60 s. For optically-evoked EPSCs and IPSCs, currents were compared to the 50 ms prior to the light stimulation and the duration of light stimulation combined with the 5 ms after the cessation of the light. The existence of a peak >5 pA above or below these baselines was classified as an EPSC or IPSC. Under this analysis, spontaneous EPSCs and IPSCs were observed throughout the traces. Positive EPSC/IPSC identification was then manually validated based on the latency between the onset of light stimulus and the onset of the PSC. Excitatory currents with onset latencies between 0 and 8 ms were considered ‘evoked’; inhibitory currents with onset latencies between 8 and 15 ms were considered ‘evoked’. For quantification of PSC amplitude, the mean of the 15 ms following the onset of the light was compared to the mean of the 15 ms preceding light onset.
Photometry recording
Recording of GCaMP6s was performed >6 days after fiber implantation and 3–6 weeks post-viral injection. Excitation was performed with a 470 nm LED or a 405 nm LED at a power of 15–30 μW at the fiber tip (ThorLabs, M470F3, M405FP1) coupled through a fiber optic patch cable (ThorLabs, 200 μm, 0.39 NA) to a 6 port fluorescence Mini-cube (Doric Lenses). Emission was collected through a fiber optic patch cable (Doric Lenses, 400 μm, 0.48 NA) coupled to a rotary joint commutator (Doric Lenses). A fiber optic patch cable (ThorLabs, 600 μm, 0.48 NA) transmitted emission light to the detector.132 Emission was detected with a Newport visible femtowatt photoreceiver (Doric Lenses) and collected at 250 Hz through a BNC-2110 data acquisition board using electrophysiology acquisition scripts. Acquisition of fluorescence triggered a simultaneous 25 fps video recording with a Raspberry Pi camera module v2. Behavioral recordings were taken from a square open field box, the mouse home cage, an isoflurane induction chamber, or from one side of an Active/Passive Avoidance Shuttle Box (MazeEngineers).
Uncoordinated movements were quantified by duration (i.e., time of start and end); movements that were scored as uncoordinated included a) the mouse falling or slipping following rearing, b) a hindpaw slipping out from under the animal (generally while grooming), or c) gross gait dysfunction or stumbling while walking. For corn oil and ethanol, a single exposure to corn oil or ethanol was generally sufficient to induce multiple bouts of slipping or uncoordinated movement. For isoflurane administration and recovery, one bout of uncoordinated movement was recorded from each trial. Spontaneous slipping was taken from 4 to 6 slipping events for each mouse at 470 nm excitation. A thin film of corn oil was applied to a plastic open field. Tail restraint was performed for 8–10 s, 5 to 7 times for each mouse at 470 nm excitation and once at 405 nm excitation. Tail suspension was performed for ~6 s, 6 to 9 times for each mouse at 470 nm excitation and 3 times at 405 nm excitation. Anesthetic induction and recovery were performed 3 to 4 times for each mouse at 470 nm excitation and once at 405 nm excitation. Alcohol was administered intraperitoneally at a dosage of 2.5 g/kg, using ethanol dissolved in sterile saline to produce a 20% ethanol solution by volume. This results in an expected blood alcohol level of around 300–400 mg/dL.133 Thermal support was provided. Looming stimuli were presented by a dark paper shape repeatedly moved downwards toward the mouse for ~10 s with an interstimulus interval of ~1 s. Each looming stimulus trial was given 6 to 7 times to each mouse at 470 nm excitation and once at 405 nm excitation. Stressful white noise stimulus was given at ~90 dB for 10 s.134 White noise was presented 6 to 8 times at 470 nm excitation and once at 405 nm excitation.
For shock and fear responses, an acoustic cue (15 s, 7.5 kHz tone at 70 dB) was given 3 to 6 times. This acoustic cue was then paired with a 1 s electric shock (0.6 mA) immediately following the cessation of the tone. Eight cue-shock pairs were given, with 7 presentations at 470 nm excitation and 1 at 405 nm excitation. The acoustic cue was then given in the absence of the shock an additional three times at 470 nm excitation. Analysis of fear photometry was averaged from the final three cue-shock pair trials and the three cue-only trials following fear conditioning. For all others, fluorescence responses were averaged across trials for each mouse. Fluorescence traces were generally aligned to the time-locking stimulus and baselined using the time period 20 s–10 s prior to the stimulus. For motor dysfunction following recovery from anesthesia and motor dysfunction following ethanol administration, traces were baselined using the time period 5 s–0 s prior to the stimulus. Peak Z scores were calculated from the average of 250 values surrounding the maximum found during the baseline and during the stimulus. For baseline behavior recordings, in the absence of extraneous stimuli, 8 min of behavior were acquired. Z scores were calculated across the recordings and thresholded at a Z score of 3 for at least 100 ms. Analysis was performed in MATLAB.
Behavior
Male and female mice were used for all experiments. Experimental animals were injected with AAV1-CBA-DIO-rM3Ds-mCherry-WPRE, AAV1-EF1α-Lox-mCherry-lox(dtA)-lox2, or AAV1-CBA-DIO-hM4Di-mCherry-WPRE. Control animals were injected with volume-matched AAV1-CAG-FLEX-tdTomato-WPRE or AAV9-EF1α-DIO-eYFP-WPRE. In a single cage of littermates, experimental and control animals were counterbalanced. Post hoc validation of viral expression was performed on all experimental animals, and animals lacking signs of expression were excluded. Researchers were blinded to condition during all animal handling and analysis of behavior. Following the surgery, mice were housed for three weeks prior to any behavioral tests or clozapine N-oxide (CNO) administration to allow for sufficient viral expression. Prior to being run in any behavioral tests, the mice were acclimated to handling and the researcher running the behavioral tests. Acclimation consisted of letting the mouse sit in the palm of the researcher’s hand for two to 3 min and took across two days. Mice that would later be injected with CNO were scruffed briefly (~10 s) immediately following acclimation, but no injection was given.
Video recordings were made for all behavioral tests. Cameras were an STC-MC33USB (Sentech) with a YV2.8×2.8LA-2 lens (Fujinion, Fujifilm) with StCamSWare software (Sentech) for the dtA elevated plus maze experiments, and a Raspberry Pi camera module v2 for all other experiments, acquiring at 25 or 30 frames per second. Recordings were converted from.h264 to.mp4 files with Yamb. For open field locomotion, mouse position was tracked using ToxTrac v2.96.119 Care was taken to not perform background subtraction, as this produced incorrect outputs of the behavioral tracking. For elevated maze tests, behavior was scored manually by an investigator blinded to the condition of the animal. To minimize circadian influences, behavioral tests were performed at least 1 h from zeitgeber of the lights turning off (i.e., the lights were off for at least 1 h before the animals are run on any behavioral assay), and all animals were evaluated within a 3 h time window for any given behavioral test. Between individual animals the behavioral setup was cleaned with 70% ethanol, with residual ethanol allowed to dissipate for about 5 min.
For experiments comparing rM3Ds or hM4Di to control injected mice, all mice were given a 3 mg/kg intraperitoneal (i.p.) injection of CNO 40–45 min prior to behavioral testing. CNO (Enzo Life Sciences) was diluted in sterile saline to 3 mg/kg for 0.1 mL injected per 20 g. For chronic CNO administration to hM4Di mice, i.p. injections of 1 mg/kg CNO were given twice daily at 10–14 h intervals, with CNO administered for 10–14 days. The final dose of CNO was given such that it came 40–45 min prior to behavioral testing, in order to minimize experimental variability between acute and chronic injection conditions. Control mice for rM3Ds and hM4Di experimental groups were given injections during handling, while control mice for dtA experimental groups are not.
Elevated maze
Mice were acclimated near the behavioral room for at least 15 min. Mice were placed on a 58 cm × 58 cm elevated plus maze raised 61 cm off the ground. Chronically CNO-injected hM4Di mice and their controls were placed on a 56.5 cm diameter zero maze at the same height. Exploratory behavior was recorded for 5 min. An entrance to an open arm was marked when all four paws were on the open arm. Light intensity was ~100–150 lux. Experiments were performed during the animals’ dark cycle.
Open field locomotion
Mice were acclimated near the behavioral room for at least 15 min. Mice were placed in the corner of a square open field 54 cm on a side with walls 30 cm high. Mice were allowed to explore the box freely for 5 min. The center was defined as the center half of the arena, i.e., the four middle squares of the arena when divided into a 4×4 grid. Light intensity was ~250 lux. Experiments were performed during the animals’ dark cycle.
Data display
All behavioral schematics depicted in Figures 6, 7, and S6 were created with BioRender.com. Whole brain RNAseq data15 was downloaded from mousebrain.org and analyzed using custom MATLAB scripts.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed in Prism 8.4 (GraphPad Software). All statistical tests were two-sided. Statistical details of experiments can be found in the figure legends. Data are reported as mean ± SEM.
Supplementary Material
Highlights.
Most CART+ EW neurons signal only through neuropeptides
Quantitative whole CNS mapping of the CART+ EW clarifies inputs and outputs
Angled-fiber tip photometry records activity of cells below the cerebral aqueduct
The CART+ EW responds to loss of motor control and promotes anxiety-like behaviors
ACKNOWLEDGMENTS
We thank the Northwestern University Biological Imaging Facility as well as Dr. Tiffany Schmidt and Dr. Reza Vafabakhsh for confocal microscope access, Dr. Martha Vitaterna for loaning behavioral apparatuses, Dr. Gregory Scherrer for a gift of biotinylated IB4, Lindsey Butler for mouse colony management, and Jordan Nasenbeny and Sara Boyle for technical assistance in early stages of the study. For transmission electron microscopy sample preparation and microscope use, this work made use of the BioCryo facility of Northwestern University’s NUANCE Center, which has received support from the SHyNE Resource (NSF ECCS-2025633), the IIN, and Northwestern’s MRSEC program (NSF DMR-1720139). This work was supported by NIH R01MH117111, NIH R01NS107539, NSF CAREER award 1846234, and the following awards: Beckman Young Investigator, Rita Allen Foundation Scholar, Searle Scholar, William and Bernice E. Bumpus Young Innovator, NARSAD Young Investigator/P&S Fund, and One Mind Nick LeDeit Rising Star award (to Y.K.). M.F.P. was supported by the Arnold O. Beckman Postdoctoral Fellowship and NIH T32AG20506. S.N.F. was supported by NIH 2T32MH067564. V.D. was supported by an American Heart Association predoctoral fellowship (19PRE34380056) and by NIH T32GM15538.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112992.
REFERENCES
- 1.Merighi A, Salio C, Ferrini F, and Lossi L (2011). Neuromodulatory function of neuropeptides in the normal CNS. J. Chem. Neuroanat. 42, 276–287. 10.1016/j.jchemneu.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Nusbaum MP, Blitz DM, and Marder E (2017). Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci. 18, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Den Pol AN (2012). Neuropeptide transmission in brain circuits. Neuron 76, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schöne C, and Burdakov D (2012). Glutamate and GABA as rapid effectors of hypothalamic “peptidergic” neurons. Front. Behav. Neurosci. 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hökfelt T, Barde S, Xu Z-QD, Kuteeva E, Rüegg J, Le Maitre E, Risling M, Kehr J, Ihnatko R, Theodorsson E, et al. (2018). Neuropeptide and small transmitter coexistence: fundamental studies and relevance to mental illness. Front. Neural Circuits 12, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dicken MS, Tooker RE, and Hentges ST (2012). Regulation of GABA and glutamate release from proopiomelanocortin neuron terminals in intact hypothalamic networks. J. Neurosci. 32, 4042–4048. 10.1523/JNEUROSCI.6032-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granger AJ, Wang W, Robertson K, El-Rifai M, Zanello AF, Bistrong K, Saunders A, Chow BW, Nuñez V, Turrero García M, et al. (2020). Cortical ChAT+ neurons co-transmit acetylcholine and GABA in a target- and brain-region-specific manner. Elife 9, e57749. 10.7554/eLife.57749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnasko TS, Chuhma N, Zhang H, Goh GY, Sulzer D, Palmiter RD, Rayport S, and Edwards RH (2010). Vesicular glutamate transport promotes dopamine storage and glutamate corelease in vivo. Neuron 65, 643–656. 10.1016/j.neuron.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuber GD, Hnasko TS, Britt JP, Edwards RH, and Bonci A (2010). Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J. Neurosci. 30, 8229–8233. 10.1523/JNEUROSCI.1754-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tritsch NX, Ding JB, and Sabatini BL (2012). Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature 490, 262–266. 10.1038/nature11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H-L, Zhang S, Qi J, Wang H, Cachope R, Mejias-Aponte CA, Gomez JA, Mateo-Semidey GE, Beaudoin GMJ, Paladini CA, et al. (2019). Dorsal raphe dual serotonin-glutamate neurons drive reward by establishing excitatory synapses on VTA mesoaccumbens dopamine neurons. Cell Rep. 26, 1128–1142.e7. 10.1016/j.celrep.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Qi J, Li X, Wang H-L, Britt JP, Hoffman AF, Bonci A, Lupica CR, and Morales M (2015). Dopaminergic and glutamatergic microdomains in a subset of rodent mesoaccumbens axons. Nat. Neurosci. 18, 386–392. 10.1038/nn.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaaga CE, Borisovska M, and Westbrook GL (2014). Dual-transmitter neurons: functional implications of co-release and co-transmission. Curr. Opin. Neurobiol. 29, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomrenze MB, Giovanetti SM, Maiya R, Gordon AG, Kreeger LJ, and Messing RO (2019). Dissecting the roles of GABA and neuropeptides from rat central amygdala CRF neurons in anxiety and fear learning. Cell Rep. 29, 13–21.e4. 10.1016/j.celrep.2019.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22. 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang KW, Ochandarena NE, Philson AC, Hyun M, Birnbaum JE, Cicconet M, and Sabatini BL (2019). Molecular and anatomical organization of the dorsal raphe nucleus. Elife 8, e46464. 10.1101/573923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R,Alpár A, Mulder J, Clotman F, Keimpema E, et al. (2017). Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176–188. 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paxinos G, and Franklin KBJ (2019). Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates (Academic press; ). [Google Scholar]
- 19.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 20.Kozicz T, Bittencourt JC, May PJ, Reiner A, Gamlin PDR, Palkovits M, Horn AKE, Toledo CAB, and Ryabinin AE (2011). The Edinger-Westphal nucleus: a historical, structural, and functional perspective on a dichotomous terminology. J. Comp. Neurol. 519, 1413–1434. 10.1002/cne.22580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasconcelos LAP, Donaldson C, Sita LV, Casatti CA, Lotfi CFP, Wang L, Cadinouche MZA, Frigo L, Elias CF, Lovejoy DA, et al. (2003). Urocortin in the central nervous system of a primate (Cebus apella): Sequencing, immunohistochemical, and hybridization histochemical characterization. J. Comp. Neurol. 463, 157–175. 10.1002/cne.10742. [DOI] [PubMed] [Google Scholar]
- 22.Toyoshima K, Kawana E, and Sakai H (1980). On the neuronal origin of the afferents to the ciliary ganglion in cat. Brain Res. 185, 67–76. 10.1016/0006-8993(80)90671-X. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Zhong P, Hu F, Barger Z, Ren Y, Ding X, Li S, Weber F, Chung S, Palmiter RD, et al. (2019). An excitatory circuit in the perioculomotor midbrain for Non-REM sleep control. Cell 177, 1293–1307.e16. 10.1016/j.cell.2019.03.041. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Chen W, Pan K, Li H, Pang P, Guo Y, Shu S, Cai Y, Pei L, Liu D, et al. (2018). Serotonin receptor 2c-expressing cells in the ventral CA1 control attention via innervation of the Edinger–Westphal nucleus. Nat. Neurosci. 21, 1239–1250. 10.1038/s41593-018-0207-0. [DOI] [PubMed] [Google Scholar]
- 25.Gaszner B, Csernus V, and Kozicz T (2004). Urocortinergic neurons respond in a differentiated manner to various acute stressors in the Edinger-Westphal nucleus in the rat. J. Comp. Neurol. 480, 170–179. 10.1002/cne.20343. [DOI] [PubMed] [Google Scholar]
- 26.Korosi A, Schotanus S, Olivier B, Roubos EW, and Kozicz T (2005). Chronic ether stress-induced response of urocortin 1 neurons in the Edinger–Westphal nucleus in the mouse. Brain Res. 1046, 172–179. 10.1016/J.BRAINRES.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Okere B, Xu L, Roubos EW, Sonetti D, and Kozicz T (2010). Restraint stress alters the secretory activity of neurons co-expressing urocortin-1, cocaine- and amphetamine-regulated transcript peptide and nesfatin-1 in the mouse Edinger–Westphal nucleus. Brain Res. 1317, 92–99. 10.1016/J.BRAINRES.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 28.Kozicz T, Li M, and Arimura A (2001). The activation of urocortin immunoreactive neurons in the Edinger-Westphal nucleus following acute pain stress in rats. Stress 4, 85–90. 10.3109/10253890109115724. [DOI] [PubMed] [Google Scholar]
- 29.Spangler E, Cote DM, Anacker AMJ, Mark GP, and Ryabinin AE (2009). Differential sensitivity of the perioculomotor urocortin-containing neurons to ethanol, psychostimulants and stress in mice and rats. Neuroscience 160, 115–125. 10.1016/j.neuroscience.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weitemier AZ, and Ryabinin AE (2005). Lesions of the Edinger-Westphal nucleus alter food and water consumption. Behav. Neurosci. 119, 1235–1243. 10.1037/0735-7044.119.5.1235. [DOI] [PubMed] [Google Scholar]
- 31.Topilko T, Diaz SL, Pacheco CM, Verny F, Rousseau CV, Kirst C, Deleuze C, Gaspar P, and Renier N (2022). Edinger-Westphal peptidergic neurons enable maternal preparatory nesting. Neuron 110, 1385–1399.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lackey EP, Heck DH, and Sillitoe RV (2018). Recent advances in understanding the mechanisms of cerebellar granule cell development and function and their contribution to behavior. F1000Res. 7. 10.12688/f1000research.15021.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, and Mikoshiba K (1995). The reeler gene-associated antigen on cajal-retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron 14, 899–912. 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 34.Causeret F, Moreau MX, Pierani A, and Blanquie O (2021). The multiple facets of Cajal-Retzius neurons. Development 148, dev199409. 10.1242/dev.199409. [DOI] [PubMed] [Google Scholar]
- 35.Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen R, Takeno MM, Huang L, Gu H, Larsen RS, et al. (2018). A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitemier AZ, Tsivkovskaia NO, and Ryabinin AE (2005). Urocortin 1 distribution in mouse brain is strain-dependent. Neuroscience 132, 729–740. 10.1016/J.NEUROSCIENCE.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 37.Lovett-Barron M, Andalman AS, Allen WE, Vesuna S, Kauvar I, Burns VM, and Deisseroth K (2017). Ancestral circuits for the coordinated modulation of brain state. Cell 171, 1411–1423.e17. 10.1016/J.CELL.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozicz T (2003). Neurons colocalizing urocortin and cocaine and amphetamine-regulated transcript immunoreactivities are induced by acute lipopolysaccharide stress in the Edinger-Westphal nucleus in the rat. Neuroscience 116, 315–320. 10.1016/S0306-4522(02)00772-8. [DOI] [PubMed] [Google Scholar]
- 39.Xu L, Bloem B, Gaszner B, Roubos EW, and Kozicz T (2009). Sex-specific effects of fasting on urocortin 1, cocaine- and amphetamine-regulated transcript peptide and nesfatin-1 expression in the rat Edinger–Westphal nucleus. Neuroscience 162, 1141–1149. [DOI] [PubMed] [Google Scholar]
- 40.Phipps BS, Maciewicz R, Sandrew BB, Poletti CE, and Foote WE (1983). Edinger-Westphal neurons that project to spinal cord contain substance P. Neurosci. Lett. 36, 125–131. 10.1016/0304-3940(83)90253-7. [DOI] [PubMed] [Google Scholar]
- 41.Skirboll L, Hökfelt T, Rehfeld J, Cuello AC, and Dockray G (1982). Coexistence of substance P- and cholecystokinin-like immunoreactivity in neurons of the mesencephalic periaqueductal central gray. Neurosci. Lett. 28, 35–39. 10.1016/0304-3940(82)90204-X. [DOI] [PubMed] [Google Scholar]
- 42.Maciewicz R, Phipps BS, Grenier J, and Poletti CE (1984). Edinger-Westphal nucleus: cholecystokinin immunocytochemistry and projections to spinal cord and trigeminal nucleus in the cat. Brain Res. 299, 139–145. 10.1016/0006-8993(84)90796-0. [DOI] [PubMed] [Google Scholar]
- 43.Burnell J, Ng L, and Guillozet-Bongaarts A (2008). Edinger-Westphal nucleus. Nat. Prec. 10.1038/npre.2008.2198.1. [DOI] [Google Scholar]
- 44.Dumrongprechachan V, Salisbury RB, Soto G, Kumar M, MacDonald ML, and Kozorovitskiy Y (2021). Cell-type and subcellular compartment-specific APEX2 proximity labeling reveals activity-dependent nuclear proteome dynamics in the striatum. Nat. Commun. 12, 4855. 10.1038/s41467-021-25144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dos Santos Júnior ED, Da Silva AV, Da Silva KRT, Haemmerle CAS, Batagello DS, Da Silva JM, Lima LB, Da Silva RJ, Diniz GB, Sita LV, et al. (2015). The centrally projecting Edinger–Westphal nucleus—I: efferents in the rat brain. J. Chem. Neuroanat. 68, 22–38. 10.1016/j.jchemneu.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Bittencourt JC, Vaughan J, Arias C, Rissman RA, Vale WW, and Sawchenko PE (1999). Urocortin expression in rat brain: evidence against a pervasive relationship of urocortin-containing projections with targets bearing type 2 CRF receptors. J. Comp. Neurol. 415, 285–312. [PubMed] [Google Scholar]
- 47.Loewy AD, and Saper CB (1978). Edinger-Westphal nucleus: Projections to the brain stem and spinal cord in the cat. Brain Res. 150, 1–27. 10.1016/0006-8993(78)90650-9. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Zhang X, Muralidhar S, LeBlanc SA, and Tonegawa S(2017). Basolateral to central amygdala neural circuits for appetitive behaviors. Neuron 93, 1464–1479.e5. 10.1016/j.neuron.2017.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCullough KM, Morrison FG, Hartmann J, Carlezon WA Jr., and Ressler KJ (2018). Quantified coexpression analysis of central amygdala subpopulations. eNeuro 5, ENEURO.0010–18.2018. 10.1523/ENEURO.0010-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniel SE, and Rainnie DG (2016). Stress modulation of opposing circuits in the bed nucleus of the Stria Terminalis. Neuropsychopharmacology 41, 103–125. 10.1038/npp.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knobloch HS, Charlet A, Hoffmann LC, Eliava M, Khrulev S, Cetin AH, Osten P, Schwarz MK, Seeburg PH, Stoop R, et al. (2012). Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73, 553–566. 10.1016/J.NEURON.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 52.Choe HK, Reed MD, Benavidez N, Montgomery D, Soares N, Yim YS, and Choi GB (2015). Oxytocin mediates entrainment of sensory stimuli to social cues of opposing valence. Neuron 87, 152–163. 10.1016/j.neuron.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eliava M, Melchior M, Knobloch-Bollmann HS, Wahis J, da Silva Gouveia M, Tang Y, Ciobanu AC, Triana del Rio R, Roth LC, Althammer F, et al. (2016). A new population of parvocellular oxytocin neurons controlling magnocellular neuron activity and inflammatory pain processing. Neuron 89, 1291–1304. 10.1016/j.neuron.2016.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marlin BJ, Mitre M, D’amour JA, Chao MV, and Froemke RC (2015). Oxytocin enables maternal behaviour by balancing cortical inhibition. Nature 520, 499–504. 10.1038/nature14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao L, Priest MF, Nasenbeny J, Lu T, and Kozorovitskiy Y (2017). Biased oxytocinergic modulation of midbrain dopamine systems. Neuron 95, 368–384.e5. 10.1016/J.NEURON.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugimori M, Preston RJ, and Kitai ST (1978). Response properties and electrical constants of caudate nucleus neurons in the cat. J. Neurophysiol. 41, 1662–1675. 10.1152/jn.1978.41.6.1662. [DOI] [PubMed] [Google Scholar]
- 57.Yosten GL, Harada CM, Haddock C, Giancotti LA, Kolar GR, Patel R, Guo C, Chen Z, Zhang J, Doyle TM, et al. (2020). GPR160 de-orphanization reveals critical roles in neuropathic pain in rodents. J. Clin. Invest. 130, 2587–2592. 10.1172/JCI133270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foo KS, Brismar H, and Broberger C (2008). Distribution and neuropeptide coexistence of nucleobindin-2 mRNA/nesfatin-like immunoreactivity in the rat CNS. Neuroscience 156, 563–579. 10.1016/j.neuroscience.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 59.Yu W, Pati D, Pina MM, Schmidt KT, Boyt KM, Hunker AC, Zweifel LS, McElligott ZA, and Kash TL (2021). Periaqueductal gray/dorsal raphe dopamine neurons contribute to sex differences in pain-related behaviors. Neuron 109, 1365–1380.e5. 10.1016/j.neuron.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dautzenberg FM, Grigoriadis DE, Hauger RL, Risbrough VB, Steckler T, Vale WW, and Valentino RJ (2019). Corticotropin-releasing factor receptors (version 2019.4) in the IUPHAR/BPS guide to pharmacology database. In IUPHAR/BPS Guide to Pharmacology CITE 2019. 10.2218/gtopdb/F19/2019.4. [DOI] [Google Scholar]
- 61.Fahrenkrug J, Goetzl EJ, Gozes I, Harmar A, Laburthe M, May V, Pisegna JR, Said SI, Vaudry D, Vaudry H, et al. (2019). VIP and PACAP receptors (version 2019.4) in the IUPHAR/BPS guide to pharmacology database. In IUPHAR/BPS Guide to Pharmacology CITE 2019. 10.2218/gtopdb/F67/2019.4. [DOI] [Google Scholar]
- 62.Beinfeld M, Chen Q, Gao F, Liddle RA, Miller LJ, and Rehfeld J (2019). Cholecystokinin receptors (version 2019.4) in the IUPHAR/BPS guide to pharmacology database. In IUPHAR/BPS Guide to Pharmacology CITE 2019. 10.2218/gtopdb/F15/2019.4. [DOI] [Google Scholar]
- 63.Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, and Losonczy A (2016). Rabies virus CVS-N2cΔG strain enhances retrograde synaptic transfer and neuronal viability. Neuron 89, 711–724. 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryabinin AE, Cocking DL, and Kaur S (2013). Inhibition of VTA neurons activates the centrally projecting Edinger–Westphal nucleus: Evidence of a stress–reward link? J. Chem. Neuroanat. 54, 57–61. 10.1016/J.JCHEMNEU.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ugolini G (2011). Chapter 10 - Rabies Virus as a Transneuronal Tracer of Neuronal Connections. In Research Advances in Rabies, Jackson AC, ed. (Academic Press; ), pp. 165–202. 10.1016/B978-0-12-387040-7.00010-X. [DOI] [PubMed] [Google Scholar]
- 66.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, and Uchida N (2012). Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74, 858–873. 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 67.Iwamoto S, Tamura M, Sasaki A, and Nawano M (2021). Dynamics of neuronal oscillations underlying nociceptive response in the mouse primary somatosensory cortex. Sci. Rep. 11, 1667. 10.1038/s41598-021-81067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L, Cai P, Wang R-F, Lu Y-P, Chen H-Y, Guo Y-R, Huang S-N, Hu L-H, Chen J, Zheng Z-H, et al. (2020). Glutamatergic lateral hypothalamus promotes defensive behaviors. Neuropharmacology 178, 108239. 10.1016/j.neuropharm.2020.108239. [DOI] [PubMed] [Google Scholar]
- 69.Siemian JN, Arenivar MA, Sarsfield S, Borja CB, Erbaugh LJ, Eagle AL, Robison AJ, Leinninger G, and Aponte Y (2021). An excitatory lateral hypothalamic circuit orchestrating pain behaviors in mice. Elife 10, e66446. 10.7554/eLife.66446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou M, Liu Z, Melin MD, Ng YH, Xu W, and Südhof TC (2018). A central amygdala to zona incerta projection is required for acquisition and remote recall of conditioned fear memory. Nat. Neurosci. 21, 1515–1519. 10.1038/s41593-018-0248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chou XL, Wang X, Zhang ZG, Shen L, Zingg B, Huang J, Zhong W, Mesik L, Zhang LI, and Tao HW (2018). Inhibitory gain modulation of defense behaviors by zona incerta. Nat. Commun. 9, 1151. 10.1038/s41467-018-03581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.George DT, Ameli R, and Koob GF (2019). Periaqueductal gray sheds light on dark areas of psychopathology. Trends Neurosci. 42, 349–360. 10.1016/j.tins.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 73.Lefler Y, Campagner D, and Branco T (2020). The role of the periaqueductal gray in escape behavior. Curr. Opin. Neurobiol. 60, 115–121. 10.1016/j.conb.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 74.Bachtell RK, Tsivkovskaia NO, and Ryabinin AE (2002). Strain differences in urocortin expression in the Edinger–Westphal nucleus and its relation to alcohol-induced hypothermia. Neuroscience 113, 421–434. 10.1016/S0306-4522(02)00174-4. [DOI] [PubMed] [Google Scholar]
- 75.Giardino WJ, Rodriguez ED, Smith ML, Ford MM, Galili D, Mitchell SH, Chen A, and Ryabinin AE (2017). Control of chronic excessive alcohol drinking by genetic manipulation of the Edinger–Westphal nucleus urocortin-1 neuropeptide system. Transl. Psychiatry 7, e1021. 10.1038/tp.2016.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryabinin AE, Galvan-Rosas A, Bachtell RK, and Risinger FO(2003). High alcohol/sucrose consumption during dark circadian phase in C57BL/6J mice: involvement of hippocampus, lateral septum and urocortin-positive cells of the Edinger-Westphal nucleus. Psychopharmacology (Berl) 165, 296–305. 10.1007/s00213-002-1284-y. [DOI] [PubMed] [Google Scholar]
- 77.Treit D, Menard J, and Royan C (1993). Anxiogenic stimuli in the elevated plus-maze. Pharmacol. Biochem. Behav. 44, 463–469. 10.1016/0091-3057(93)90492-C. [DOI] [PubMed] [Google Scholar]
- 78.Espejo EF (1997). Effects of weekly or daily exposure to the elevated plus-maze in male mice. Behav. Brain Res. 87, 233–238. 10.1016/S0166-4328(97)02286-9. [DOI] [PubMed] [Google Scholar]
- 79.Tucker LB, and McCabe JT (2017). Behavior of male and female C57BL/6J mice is more consistent with repeated trials in the elevated zero maze than in the elevated plus maze. Front. Behav. Neurosci. 11, 13. 10.3389/fnbeh.2017.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Autry AE, Wu Z, Kapoor V, Kohl J, Bambah-Mukku D, Rubinstein ND, Marin-Rodriguez B, Carta I, Sedwick V, Tang M, et al. (2021). Urocortin-3 neurons in the mouse perifornical area promote infant-directed neglect and aggression. eLife 10, e64680. 10.7554/eLife.64680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, et al. (2014). An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242. 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao L, Priest MF, and Kozorovitskiy Y (2018). Oxytocin functions as a spatiotemporal filter for excitatory synaptic inputs to VTA dopamine neurons. Elife 7, e33892. 10.7554/eLife.33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piñ ol RA, Jameson H, Popratiloff A, Lee NH, and Mendelowitz D (2014). Visualization of oxytocin release that mediates paired pulse facilitation in hypothalamic pathways to brainstem autonomic neurons. PLoS One 9, e112138. 10.1371/journal.pone.0112138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ryan PJ, Ross SI, Campos CA, Derkach VA, and Palmiter RD (2017). Oxytocin-receptor-expressing neurons in the parabrachial nucleus regulate fluid intake. Nat. Neurosci. 20, 1722–1733. 10.1038/s41593-017-0014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hasan MT, Althammer F, Silva da Gouveia M, Goyon S, Eliava M, Lefevre A, Kerspern D, Schimmer J, Raftogianni A, Wahis J, et al. (2019). A fear memory engram and its plasticity in the hypothalamic oxytocin system. Neuron 103, 133–146.e8. 10.1016/j.neuron.2019.04.029. [DOI] [PubMed] [Google Scholar]
- 86.Grinevich V, and Ludwig M (2021). The multiple faces of the oxytocin and vasopressin systems in the brain. J. Neuroendocrinol. 33, e13004. 10.1111/jne.13004. [DOI] [PubMed] [Google Scholar]
- 87.Zhang L, Hernández VS, Zetter MA, and Eiden LE (2020). VGLUT-VGAT expression delineates functionally specialised populations of vasopressin-containing neurones including a glutamatergic perforant path-projecting cell group to the hippocampus in rat and mouse brain. J. Neuroendocrinol. 32, e12831. 10.1111/jne.12831. [DOI] [PubMed] [Google Scholar]
- 88.Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delevich K, Tucciarone J, Huang ZJ, and Li B (2015). The mediodorsal thalamus drives feedforward inhibition in the anterior cingulate cortex via parvalbumin interneurons. J. Neurosci. 35, 5743–5753. 10.1523/JNEUROSCI.4565-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daviu N, Bruchas MR, Moghaddam B, Sandi C, and Beyeler A (2019). Neurobiological links between stress and anxiety. Neurobiol. Stress 11, 100191. 10.1016/j.ynstr.2019.100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kang SJ, Liu S, Ye M, Kim D-I, Pao GM, Copits BA, Roberts BZ, Lee K-F, Bruchas MR, and Han S (2022). A central alarm system that gates multi-sensory innate threat cues to the amygdala. Cell Rep. 40, 111222. 10.1016/j.celrep.2022.111222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim J, Lee S, Fang Y-Y, Shin A, Park S, Hashikawa K, Bhat S, Kim D, Sohn J-W, Lin D, et al. (2019). Rapid, biphasic CRF neuronal responses encode positive and negative valence. Nat. Neurosci. 22, 576–585. 10.1038/s41593-019-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stanek LM (2006). Cocaine- and amphetamine related transcript (CART) and anxiety. Peptides 27, 2005–2011. 10.1016/j.peptides.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 94.Hammack SE, and May V (2015). Pituitary adenylate cyclase activating polypeptide in stress-related disorders: data convergence from animal and human studies. Biol. Psychiatr. 78, 167–177. 10.1016/j.biopsych.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bowers ME, Choi DC, and Ressler KJ (2012). Neuropeptide regulation of fear and anxiety: implications of cholecystokinin, endogenous opioids, and neuropeptide Y. Physiol. Behav. 107, 699–710. 10.1016/j.physbeh.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Merali Z, Cayer C, Kent P, and Anisman H (2008). Nesfatin-1 increases anxiety- and fear-related behaviors in the rat. Psychopharmacology (Berl) 201, 115–123. 10.1007/s00213-008-1252-2. [DOI] [PubMed] [Google Scholar]
- 97.Kash TL, Pleil KE, Marcinkiewcz CA, Lowery-Gionta EG, Crowley N, Mazzone C, Sugam J, Hardaway JA, and McElligott ZA (2015). Neuropeptide regulation of signaling and behavior in the BNST. Mol. Cells 38, 1–13. 10.14348/molcells.2015.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roman CW, Lezak KR, Hartsock MJ, Falls WA, Braas KM, Howard AB, Hammack SE, and May V (2014). PAC1 receptor antagonism in the bed nucleus of the stria terminalis (BNST) attenuates the endocrine and behavioral consequences of chronic stress. Psychoneuroendocrinology 47, 151–165. 10.1016/j.psyneuen.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rigney N, Whylings J, de Vries GJ, and Petrulis A (2021). Sex differences in the control of social investigation and anxiety by vasopressin cells of the paraventricular nucleus of the hypothalamus. Neuroendocrinology 111, 521–535. 10.1159/000509421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keay KA, and Bandler R (2015). Periaqueductal Gray. The Rat Nervous System, Fourth Edition, pp. 207–221. 10.1016/B978-0-12-374245-2.00010-3. [DOI] [Google Scholar]
- 101.Silva C, and McNaughton N (2019). Are periaqueductal gray and dorsal raphe the foundation of appetitive and aversive control? A comprehensive review. Prog. Neurobiol. 177, 33–72. 10.1016/j.pneurobio.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 102.Gross CT, and Canteras NS (2012). The many paths to fear. Nat. Rev. Neurosci. 13, 651–658. 10.1038/nrn3301. [DOI] [PubMed] [Google Scholar]
- 103.Kim T, Gondré-Lewis MC, Arnaoutova I, and Loh YP (2006). Dense-core secretory granule biogenesis. Physiology 21, 124–133. 10.1152/physiol.00043.2005. [DOI] [PubMed] [Google Scholar]
- 104.Ailion M, Hannemann M, Dalton S, Pappas A, Watanabe S, Hegermann J, Liu Q, Han H-F, Gu M, Goulding MQ, et al. (2014). Two Rab2 interactors regulate dense-core vesicle maturation. Neuron 82, 167–180. 10.1016/j.neuron.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merighi A (2018). Costorage of high molecular weight neurotransmitters in large dense core vesicles of mammalian neurons. Front. Cell. Neurosci. 12, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chang C-W, Hsiao Y-T, and Jackson MB (2021). Synaptophysin regulates fusion pores and exocytosis mode in chromaffin cells. J. Neurosci. 41, 3563–3578. 10.1523/JNEUROSCI.2833-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moro A, van Nifterick A, Toonen RF, and Verhage M (2021). Dynamin controls neuropeptide secretion by organizing dense-core vesicle fusion sites. Sci. Adv. 7, eabf0659. 10.1126/sciadv.abf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Puntman DC, Arora S, Farina M, Toonen RF, and Verhage M (2021). Munc18–1 is essential for neuropeptide secretion in neurons. J. Neurosci. 41, 5980–5993. 10.1523/JNEUROSCI.3150-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Persoon CM, Hoogstraaten RI, Nassal JP, van Weering JRT, Kaeser PS, Toonen RF, and Verhage M (2019). The RAB3-RIM pathway is essential for the release of neuromodulators. Neuron 104, 1065–1080.e12. 10.1016/j.neuron.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ding K, Han Y, Seid TW, Buser C, Karigo T, Zhang S, Dickman DK, and Anderson DJ (2019). Imaging neuropeptide release at synapses with a genetically engineered reporter. Elife 8, e46421. 10.7554/eLife.46421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nusbaum MP, and Blitz DM (2012). Neuropeptide modulation of microcircuits. Curr. Opin. Neurobiol. 22, 592–601. 10.1016/j.conb.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Atasoy D, Aponte Y, Su HH, and Sternson SM (2008). A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 28, 7025–7030. 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu M, Minkowicz S, Dumrongprechachan V, Hamilton P, and Kozorovitskiy Y (2021). Ketamine rapidly enhances glutamate-evoked dendritic spinogenesis in medial prefrontal cortex through dopaminergic mechanisms. Biol. Psychiatr. 89, 1096–1105. 10.1016/j.biopsych.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu Z, Autry AE, Bergan JF, Watabe-Uchida M, and Dulac CG (2014). Galanin neurons in the medial preoptic area govern parental behaviour. Nature 509, 325–330. 10.1038/nature13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hou XH, Hyun M, Taranda J, Huang KW, Todd E, Feng D, Atwater E, Croney D, Zeidel ML, Osten P, et al. (2016). Central control circuit for context-dependent micturition. Cell 167, 73–86.e12. 10.1016/J.CELL.2016.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodriguez A, Zhang H, Klaminder J, Brodin T, Andersson PL, and Andersson M (2018). ToxTrac: a fast and robust software for tracking organisms. Methods Ecol. Evol. 9, 460–464. 10.1111/2041-210X.12874. [DOI] [Google Scholar]
- 120.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A Designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382. 10.1016/J.NEURON.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gritton HJ, Howe WM, Mallory CS, Hetrick VL, Berke JD, and Sarter M (2016). Cortical cholinergic signaling controls the detection of cues. Proc. Natl. Acad. Sci. USA 113, E1089–E1097. 10.1073/pnas.1516134113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Q, Ding S-L, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, et al. (2020). The allen mouse brain common coordinate framework: a 3D reference atlas. Cell 181, 936–953.e20. 10.1016/j.cell.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kash TL, and Winder DG (2006). Neuropeptide Y and corticotropin-releasing factor bi-directionally modulate inhibitory synaptic transmission in the bed nucleus of the stria terminalis. Neuropharmacology 51, 1013–1022. 10.1016/j.neuropharm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 125.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, and Stuurman N (2014). Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pologruto TA, Sabatini BL, and Svoboda K (2003). ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jones JR, Tackenberg MC, and McMahon DG (2015). Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat. Neurosci. 18, 373–375. 10.1038/nn.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Le N, Hernandez J, Gastelum C, Perez L, Vahrson I, Sayers S, and Wagner EJ (2021). Pituitary adenylate cyclase activating polypeptide inhibits A10 dopamine neurons and suppresses the binge-like consumption of palatable food. Neuroscience 478, 49–64. 10.1016/j.neuroscience.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hahne WF, Jensen RT, Lemp GF, and Gardner JD (1981). Proglumide and benzotript: members of a different class of cholecystokinin receptor antagonists. Proc. Natl. Acad. Sci. USA 78, 6304–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lemos JC, Shin JH, and Alvarez VA (2019). Striatal Cholinergic interneurons are a novel target of corticotropin releasing factor. J. Neurosci. 39, 5647–5661. 10.1523/JNEUROSCI.0479-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Priest MF, Lee EE, and Bezanilla F (2021). Tracking the movement of discrete gating charges in a voltage-gated potassium channel. Elife 10, e58148. 10.7554/eLife.58148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu M, Minkowicz S, Dumrongprechachan V, Hamilton P, Xiao L, and Kozorovitskiy Y (2021). Attenuated dopamine signaling after aversive learning is restored by ketamine to rescue escape actions. Elife 10, e64041. 10.7554/eLife.64041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smolen TN, and Smolen A (1989). Blood and brain ethanol concentrations during absorption and distribution in long-sleep and short-sleep mice. Alcohol 6, 33–38. [DOI] [PubMed] [Google Scholar]
- 134.Kim JS, Han SY, and Iremonger KJ (2019). Stress experience and hormone feedback tune distinct components of hypothalamic CRH neuron activity. Nat. Commun. 10, 5696. 10.1038/s41467-019-13639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://github.com/KozorovitskiyLaboratory and is publicly available as of the date of publication. DOIs are available in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-CART | Phoenix Pharmaceuticals | H-003–62; RRID:AB_2313614 |
| Anti-Urocortin | Sigma | U4757; RRID:AB_261834 |
| Anti-RFP (e.g., tdTomato, mCherry) | Rockland Immunochemicals | 600–401-379; RRID:AB_2209751 |
| Anti-ChAT | Millipore | AB144P; RRID:AB_2079751 |
| Anti-GFP (e.g., EGFP) | Abcam | ab13970; RRID:AB_300798 |
| Goat Anti-Rabbit Alexa Fluor 488 | Life Technologies | A11008; RRID:AB_143165 |
| Goat Anti-Rabbit Alexa Fluor 647 | Life Technologies | A21244; RRID:AB_2535812 |
| Donkey Anti-Goat Alexa Fluor 647 | Life Technologies | A21447; RRID:AB_2535864 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| AAV1-CAG-FLEX-tdTomato-WPRE | University of Pennsylvania Vector Core; Addgene; Oh et al.112 | Addgene 51503 |
| AAV1-EF1 α-DIO-LCK-APEX2-P2A-EGFP | University of North Carolina Vector Core; Dumrongprechachan et al.44 | Addgene 182826 |
| AAV1-CBA-FLEX-ChR2-mCherry | University of Pennsylvania Vector Core; Vigene; Atasoy et al.113 | Addgene 18916 |
| AAVrg-CAG-FLEX-EGFP | Addgene; Oh et al.112 | Addgene 51502-AAVrg |
| AAV1-CBA-DIO-rM3Ds-mCherry-WPRE | Vigene; Wu et al.114 | N/A |
| AAV1-EF1 α-Lox-mCherry-lox(dtA)-lox2 | University of North Carolina Vector Core; Wu et al.115 | Addgene 58536 |
| AAV1-CBA-DIO-hM4Di-mCherry-WPRE | Vigene; Hou et al.116 | Addgene 81008 |
| AAV9-EF1 α-DIO-eYFP-WPRE | University of Pennsylvania Vector Core; Addgene | Addgene 27056 |
| AAV1-CAG-Flex-H2B-eGFP-N2c(G) | Zuckerman Institute Virology Core; Reardon et al.63 | N/A |
| AAV1-EF1 α-FLEX-GT | Salk Institute Viral Vector Core | Addgene 26198 |
| Rabies virus CVS-N2cΔG tdTomato EnvA | Zuckerman Institute Virology Core; Reardon et al.63 | Addgene 73462 |
| AAV1-CAG-FLEX-GCaMP6s-WPRE | University of Pennsylvania Vector Core; Chen et al.117 | Addgene 100842 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Streptavidin AF488 | Invitrogen | S32354 |
| Biotinylated IB4 | Sigma | L2140; RRID:AB_2313663 |
| Hoechst 33342 | ThermoFisher Scientific | H1399 |
| Prolong Gold Antifade Mountant with DAPI | ThermoFisher Scientific | P36931 |
| Low-melting point agarose | Sigma | A9414 |
| 3,3′-Diaminobenzidine (DAB) with metal enhancer | Sigma | D0426 |
| Bovine serum albumin | Sigma | A3059 |
| Triton X-100 | Sigma | T8787 |
| Paraformaldehyde | Electron Microscopy Sciences | 15714 |
| Glutaraldehyde | Fisher Scientific | 5026223 |
| [D-p-Cl-Phe6,Leu17]-VIP | Tocris | 3054 |
| PACAP 6–38 | Tocris | 3236 |
| Proglumide sodium salt | Tocris | 1478 |
| Astressin 2B | Tocris | 2391 |
| NBI 35965 hydrochloride | Tocris | 3100 |
| CART (55–102) | Phoenix Pharmaceuticals | 003–62 |
| Nesfatin-1 (1–82) | Phoenix Pharmaceuticals | 003–22A |
| U 73122 | Tocris | 1268 |
| PKI (5–24) | Tocris | 6221 |
| Clozapine N-oxide | Enzo Life Sciences | BML-NS105 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAScope Fluorescence Multiplex Reagent Kit | ACDBio/BioTechne | 320850 |
| Cartpt-C1 | ACDBio/BioTechne | 432001 |
| Adcyap1-C1 | ACDBio/BioTechne | 405911 |
| Penk-C1 | ACDBio/BioTechne | 318761 |
| Nmb-C1 | ACDBio/BioTechne | 459931 |
| Slc17a6-C2 | ACDBio/BioTechne | 319171-C2 |
| Slc17a7-C2 | ACDBio/BioTechne | 416631-C2 |
| Cck-C2 | ACDBio/BioTechne | 402271-C2 |
| Pdyn-C2 | ACDBio/BioTechne | 318771-C2 |
| Pomc-C2 | ACDBio/BioTechne | 314081-C2 |
| Cartpt-C3 | ACDBio/BioTechne | 432001-C3 |
| Slc17a8-C3 | ACDBio/BioTechne | 431261-C3 |
| Tac1-C3 | ACDBio/BioTechne | 410351-C3 |
| Slc32a1-C3 | ACDBio/BioTechne | 319191-C3 |
|
| ||
| Deposited data | ||
|
| ||
| scRNAseq data | Linnarsson group, Zeisel et al.15 | mousebrain.org/adolescent; Sequence Read Archive: SRP135960 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6 | Charles River | 000664; RRID: IMSR_JAX:000664 |
| Mouse: B6; 129S-Cartpttm1.1(cre)Hze/J | Jackson Laboratory | 028533; RRID:IMSR_JAX:028533 |
| Mouse: Slc17a6tm2(cre)Lowl/J | Jackson Laboratory | 016963; RRID:IMSR_JAX:016963 |
| Mouse: Slc32a1tm2(cre)Lowl/J | Jackson Laboratory | 016962; RRID:IMSR_JAX:016962 |
|
| ||
| Software and algorithms | ||
|
| ||
| MATLAB | Mathworks | mathworks.com |
| FIJI | Schindelin et al.118 | https://imagej.net/software/fiji/downloads |
| AtlasQuantifier | This paper | https://zenodo.org/record/8172932 |
| LoomLab | This paper | https://zenodo.org/record/8172935 |
| StCamSWare | Sentech | N/A |
| Yamb | Kurtnoise | http://yamb.unite-video.com/download.html |
| ToxTrac | Rodriguez et al.119 | https://sourceforge.net/projects/toxtrac/ |
| BioRender | BioRender | biorender.com |
| Prism 8.4 | GraphPad | graphpad.com |


