Abstract
BrainForge is a cloud-enabled, web-based analysis platform for neuroimaging research. This website allows users to archive data from a study and effortlessly process data on a high-performance computing cluster. After analyses are completed, results can be quickly shared with colleagues. BrainForge solves multiple problems for researchers who want to analyze neuroimaging data, including issues related to software, reproducibility, computational resources, and data sharing. BrainForge can currently process structural, functional, diffusion, and arterial spin labeling MRI modalities, including preprocessing and group level analyses. Additional pipelines are currently being added, and the pipelines can accept the BIDS format. Analyses are conducted completely inside of Singularity containers and utilize popular software packages including Nipype, Statistical Parametric Mapping, the Group ICA of fMRI Toolbox, and FreeSurfer. BrainForge also features several interfaces for group analysis, including a fully automated adaptive ICA approach.
Keywords: neuroimaging, cloud computing, containerization, high performance computing
I. Introduction
Over the past few decades, the neuroscience research community has witnessed a meteoric rise in the use of neuroimaging to understand neurodevelopment and aging, study the structure and function of the brain, and answer questions about mental disorders and brain pathologies1, 2. The advent of quality fast magnetic resonance imaging (MRI) to probe function and connectivity has resulted in an immense amount of high-resolution data collected across multiple modalities, including structural MRI (sMRI), functional MRI (fMRI), and diffusion MRI (dMRI). This imaging data is frequently used in conjunction with data from electroencephalography (EEG), magnetoencephalography (MEG), genomics, neuropsychological and behavioral testing, and others. This substantial increase in information has led researchers toward novel discoveries that contribute to our knowledge of the brain. However, this advanced technology also requires immense computational resources, and neuroscientists must master complex software to effectively process their data. BrainForge3 offers a solution to these problems with a cloud-enabled, web-based analysis platform for neuroimaging research. BrainForge is a new and powerful platform that is already being used by many researchers. As of this writing, BrainForge has 47 users, who have run a total of 3865 analyses in 17 studies. These users are distributed over 16 research groups across 10 sites.
II. Challenges and Motivation
There are several key challenges that researchers encounter when performing neuroimaging research, including issues with computing, software, reproducibility, and data sharing. These challenges motivated us to create BrainForge and are described below. In the next section, we cover how BrainForge addresses these challenges.
A. Computational Resource Issues
First, a researcher must gain access to computational resources that enable the timely processing of neuroimaging data for a study. Without sufficient computational resources such as parallel computing and GPUs, neuroimaging pipelines sometimes require up to a day or more to finish for a single subject. Additionally, the amount of data collected across all modalities can sum to more than 1 GB of raw data for a single subject, and derived data may be one hundred-fold greater. With hundreds or thousands of subjects needing to be processed, it is clear that a single workstation or even small computing cluster may not be sufficient even for a single study, let alone a research group.
B. Software Issues
Modern neuroimaging packages are ever-advancing, with multiple software dependencies that vary between platforms. Installing and using neuroimaging software can be difficult, as a moderate degree of computer literacy is required for complete and proper installation. Furthermore, to process subjects at scale, a researcher must be proficient in basic operating system programming and scripting to programmatically call the neuroimaging packages. In large neuroimaging analyses that utilize hundreds or thousands of subjects, the ability to analyze many sessions with minimal manual intervention is necessary. Finally, computer resources are often strictly controlled by IT in a university setting, which can add delays or make installation of some software impossible.
C. Reproducibility Issues
In addition to challenges related to software and computational resources, reproducibility in research has attracted significant attention in recent years4–6. It is important that researchers publish results that are transparent, correct, and replicable. A lack of disclosure about details of parameters, software versions, and operating systems can prevent results from being replicated and validated. Thus, it is important to track and organize the pipelines and analyses that have been run and be able to inspect them later.
D. Data Sharing Issues
Because of the vast amounts of data involved, sharing data and results can be onerous. Typical file sharing systems, such as Google Drive, One Drive, and Dropbox, do not handle many large files well, as there are often problems with compression and transfer at the terabyte scale. Additionally, transferring large amounts of data both to and from one of these services requires considerable time.
III. Comparison
Given that there are several other major platforms built for neuroimaging data analysis, one might ask why build a new platform? When surveying the landscape of neuroimaging platforms, we did not find any platform that fulfilled all of our requirements. We wanted a platform that (1) integrated well with the Collaborative Informatics and Neuroimaging Suite (COINS)7 or some other study management and data acquisition platform, (2) performed a variety of group analyses, especially in the fMRI modality, and (3) was interoperable with our on-premise hardware using the Slurm Workload Manager8. Two other neuroimaging platforms, namely, Brainlife9 and Flywheel10, were examined. Both platforms can be utilized with on-premise or cloud resources and have general-purpose interfaces for numerous preprocessing pipelines in multiple MRI modalities. However, we found that they did not feature many group analyses. Each platform has a generic interface that is dynamically constructed based on the specifications of an analysis container, which allows their platform to scale well with increasing numbers of pipelines. However, this interface provides a constrained UI, which is noticeable for complicated pipelines and analyses. Modern web design frameworks offer many UI options to create intuitive and interactive interfaces, which offer flexibility, reduced errors, and time savings for neuroimaging researchers.
Because we could not find an option that satisfied our requirements at the Center for Translational Research in Neuroimaging and Data Science (TReNDS), we decided to build a new platform. Instead of a generic, dynamically generated interface that is specified by an analysis container, BrainForge has a dedicated and customizable interface for each pipeline. This allows for more design freedom on the front end, enhancing the user experience. Additionally, BrainForge features group analyses including SPM statistical models and GIFT analyses. As mentioned above, interoperability with COINS was a design requirement, so BrainForge procures metadata from COINS via a REST API and then downloads data from the COINS repository on the S3 service on Amazon Web Services (AWS). Finally, integration with our on-premise hardware was a requirement, so BrainForge controls our high-performance computing (HPC) cluster via Slurm.
IV. Solutions
To address these challenges, we have created a platform called BrainForge that connects researchers to large-scale computing resources behind a user-friendly web interface, offering complete standard and customized processing pipelines.
A. Computing
BrainForge runs on the TReNDS HPC cluster at Georgia State University. As of this writing, this cluster includes 49 nodes with a total of 2448 cores, 32.5 TB of memory, and 60 GPUs. A detailed breakdown is shown in Table 1. The computing resources at TReNDS enable simultaneous processing of thousands of subjects. BrainForge interfaces with Slurm to schedule jobs on the cluster. Furthermore, BrainForge will provide the option of running jobs on AWS if there is not sufficient capacity on the cluster to run all jobs by their deadlines. BrainForge can be adapted to other on-premise and cloud environments as well.
Table 1:
Computing resources currently available on the TReNDS cluster available to BrainForge. The total number of cores, amount of memory, and number of GPUs on the entire cluster are shown on the bottom row. Cluster resources are expanding every year.
| Manufacturer | Number | Cores Per Node | Memory Per Node | GPUs |
|---|---|---|---|---|
| Intel | 20 | 32 | 768 GB | |
| Intel | 3 | 96 | 1.5 TB | |
| AMD | 20 | 64 | 512 GB | 1xNvidia 2080 |
| Nvidia DGX-1 | 4 | 40 | 512 GB | 8xNvidia V100 |
| Dell | 2 | 40 | 192 GB | 4xNvidia V100 |
| Total | 49 | 2448 | 32.5 TB | 60 |
B. Software
BrainForge executes predesigned and customizable neuroimaging pre- and postprocessing pipelines, containerized using Singularity11. Containerization encapsulates the operating system, software libraries, and scripts, obviating the need to replicate or build specific software environments required for analysis. Singularity is similar to Docker12 but is designed to adhere to university HPC IT policies. BrainForge does not require programming expertise, allowing users to focus on their research. A web graphical user interface (GUI) provides all of the parameters as inputs to the pipelines, including those running industry-standard packages, such as Statistical Parametric Mapping (SPM)13, the FMRIB Software Library (FSL)14, Analysis of Functional NeuroImages (AFNI)15, 16, and the Group ICA for fMRI Toolbox (GIFT)17. BrainForge is also planning to provide a generic, dynamically generated interface that can easily utilize new, user-created containers.
C. Reproducibility
Because BrainForge analyses are encapsulated within Singularity containers, they produce the same results on every platform, including HPC clusters. Of course, run time will depend on the underlying hardware and can be affected more strongly if the software uses MPI or hardware optimizations. However, even a worklflow that runs more slowly on some platforms is still reproduceable. Furthermore, these containers can be introspected and are located on DockerHub18, with their code located on GitHub19, in the corresponding “trendscenter” organizations. The repositories are currently private but can be made available to BrainForge users on request. Many of the workflows are programmed using the Neuroimaging in Python Pipelines and Interfaces (Nipype)20 software libraries, adding another layer of reproducibility. Provenance, complete lists of runtime parameters, input data, start and end times, computational resources, and user information are also retained.
D. Data Sharing
To solve the challenges of data sharing, BrainForge allows primary investigators (PIs) to share data and results with others. PIs can also share data and invite others to contribute data and run analyses. This eliminates the need for large data transfers to collaborators for the purpose examining results. A sensible and effective permissions scheme is key for data sharing. Permissions are centered around sites and studies, and include the super admin, site admin, study PI, and user roles. PIs have full control over their studies, and site admins have control over all studies within their sites. Data are organized using the Brain Imaging Data Structure (BIDS) standard21, which eases sharing and reuse. In addition, both raw data and results are stored on the TReNDS cluster and can be downloaded for viewing and organizing offline using several different methods, including a web interface, a command-line interface (CLI), and Globus22, 23. Storage is included in the cost of analyzing data on BrainForge.
BrainForge can be operated standalone or optionally can interface with COINS, a neuroinformatics data management platform. COINS is a full study management system, able to pull DICOM files directly from a scanner, manage subjects, and store assessments, among other features. BrainForge procures data and associated metadata from COINS via a REST application programming interface (API), providing seamless integration of data acquisition, curation, and analysis.
V. Features
Brainforge offers many features that help a neuroimaging researcher analyze their data, including preprocessing and analysis pipelines and advanced analysis management features.
A. Analyses
Preprocessing pipelines and group-level analyses available in BrainForge operate in a wide range of modalities, including sMRI, fMRI, dMRI, and arterial spin labeling (ASL) modalities. These pipelines, designed and regularly utilized by TReNDS lab members and collaborators, have been validated through numerous publications24–26. Examples include:
Analyses for sMRI include voxel-based morphometry (VBM)27 using SPM12 and FreeSurfer (v6 and v7)28.
dMRI workflows for generation of diffusion derivative images (FA, MD, RD, AD) using AFNI and diffusion kurtosis imaging (DKI) using Python. All diffusion data is corrected prior to analysis for artifacts such as susceptibility distortion, head movement, outlier detection, and slice-to-volume movement artifacts using FSL-6.014, 29–31. Many quality assurance (QA) measures provided by the FSL program eddy are plotted for quick review within BrainForge (Fig. 1). A comprehensive QA report using FSL’s eddy_quad is generated for each subject and is available for download in PDF form. The DTI container utilizes GPUs via the Nvidia Container Toolkit, which is required to run the FSL eddy_quad program.
Task and resting-state fMRI data can be processed using two different workflows—the first using FSL-6.0 to correct echoplanar imaging (EPI) distortion followed by AFNI for realignment and registration of EPI to a standard space utilizing a T1 image. The second pipeline offers processing using SPM12, with optional features enabling registration to single band reference (SBRef) images and distortion correction using FSL-6.0. Plots of motion over time and frame displacement (FD) metrics are provided as QA measures.
fMRI first-level analysis using SPM12, including event-related and block-design modeling of task fMRI data. A user-friendly GUI has been created to run this analysis.
fMRI second (group)-level analyses using SPM12 for results from any of the BrainForge pipelines or from other precompiled group data results, featuring the complete set of SPM12 statistical models. Input to this analysis is controlled by a GUI that includes group selection based on subject metadata, such as age and gender.
A containerized version of GIFT, a MATLAB toolbox that implements multiple algorithms for independent component analysis (ICA) and blind source separation of group and single-subject fMRI data. This includes twenty group ICA (GICA)24 algorithms, dynamic functional network connectivity (dFNC)26, 32, and ICAstats25. There is also a fully automated group ICA pipeline, which simplifies the application of ICA to large studies and facilitates comparison across analyses33. An example of GICA results is shown in Fig. 2.
White matter hyperintensity (WMH) lesion quantification using the BIANCA tool in FSL-6.034. A training dataset can be uploaded and incorporated into the pipeline.
Arterial spin labeling (ASL) perfusion analysis using the BASIL tool in FSL-6.035. Output includes normalization to standard space.
Magnetic resonance spectroscopy (MRS) is a noninvasive technique used to assess neurometabolites in vivo. Owing to their abundant concentrations in the CNS, the most commonly measured spectral peaks in the brain are N-acetyl aspartate, creatine, choline, glutamate, glutamine, and myoinositol. Brainforge uses the LCModel36 and Gannet37 software packages and offers the option to process MEGA-PRESS (edited spectroscopy) data to assess GABA and glutathione.
Regression analyses for FreeSurfer results, written in Python.
Ordinary least squares regression, written in Python.
Polyssifier38, 39, a software tool to quickly benchmark multiple classification approaches on a single generic data set.
Figure 1:
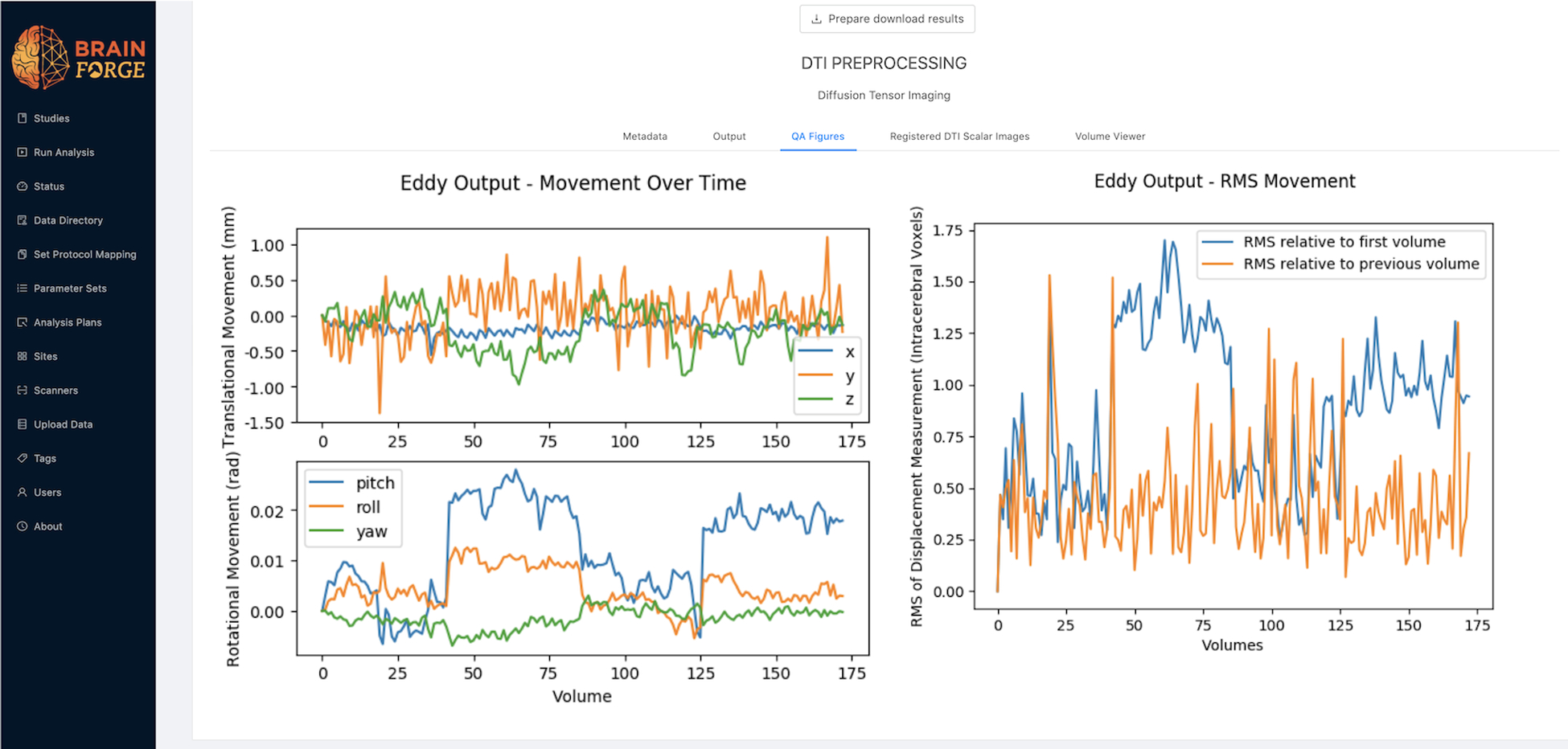
QA measures produced by DTI pipeline related to movement as provided by FSL eddy program.
Figure 2:
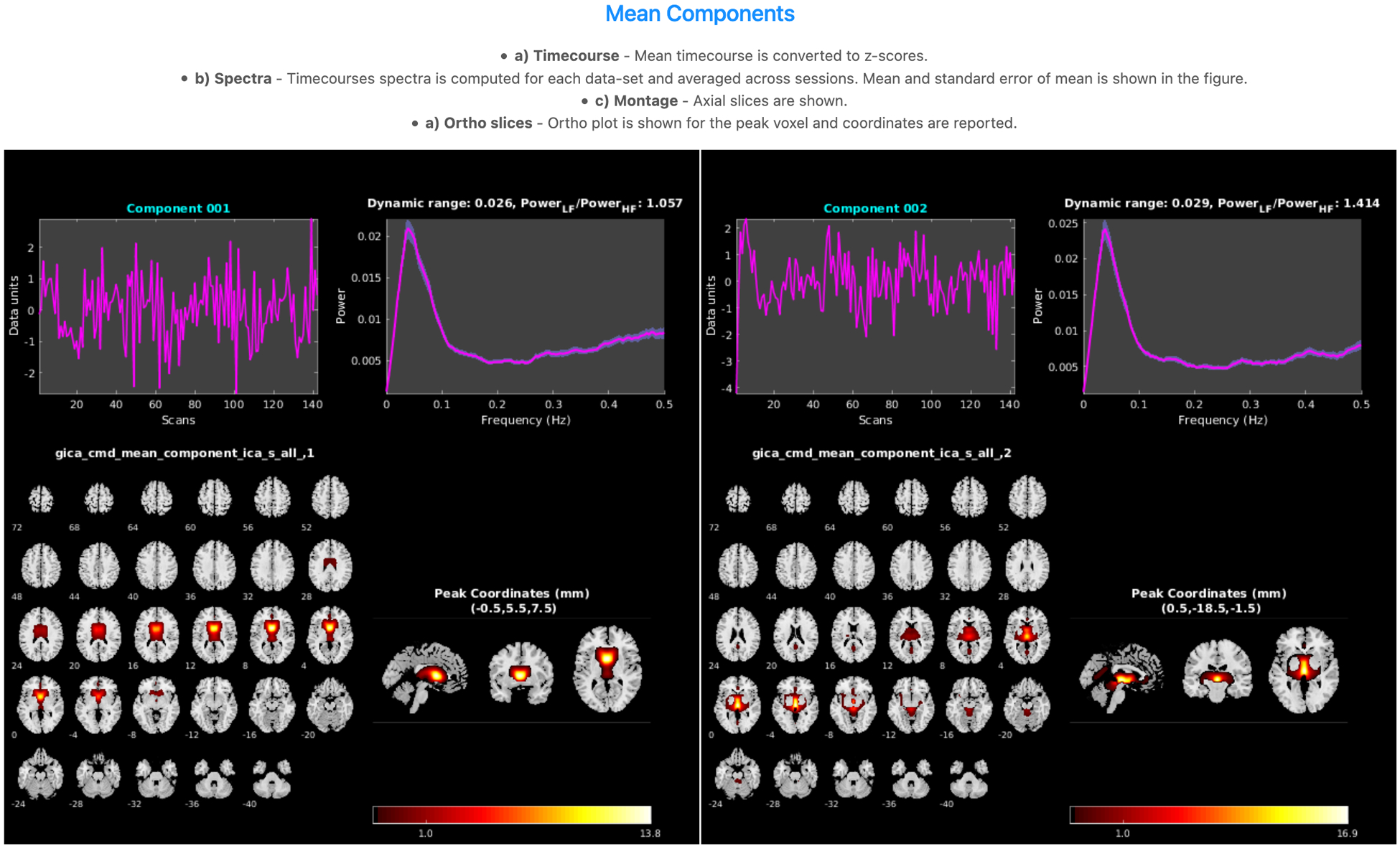
Sample components in group ICA results, including timecourses, spectra, montage, and ortho slices.
BrainForge is utilized by researchers from many institutions with different scanner vendors and software versions. To ensure accurate multistudy analyses, scanner signal quality is critical. BrainForge provides a QA pipeline that runs on phantom data and includes signal intensity, image signal-to-noise ratio (SNR), peak-to-peak signal percentage, and ghost levels, using methods similar to the TIM Trio EPI stability QA routine by Siemens40. These measures are derived from a common protocol across sites using an fMRI phantom and can be viewed interactively as longitudinal plots (Fig. 3).
Figure 3:
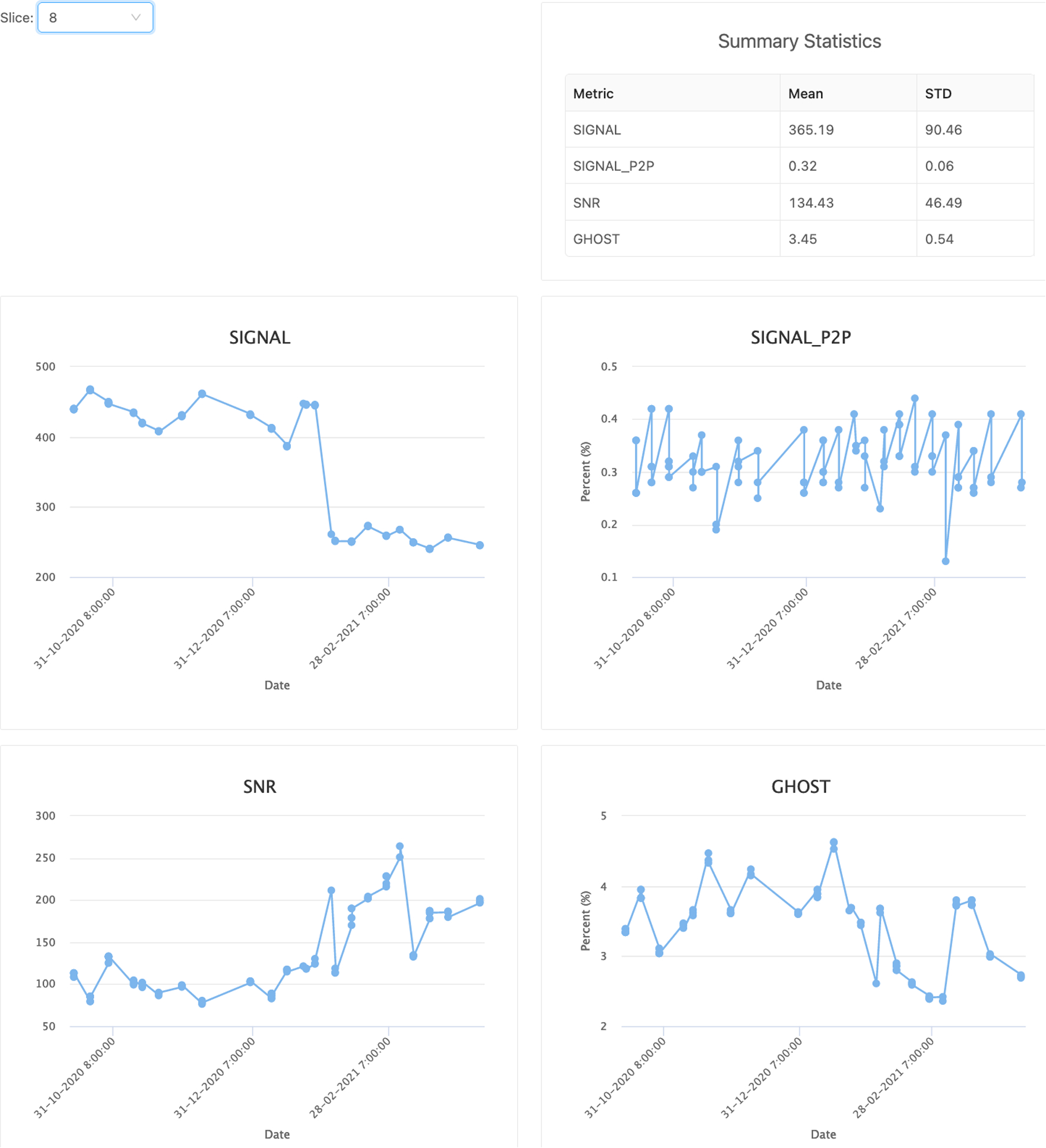
Longitudinal plots of scanner fMRI quality metrics derived from phantom scans.
BrainForge pipelines are streamlined for compatibility with NeuroMark33, an ICA-based framework designed to elucidate functional connectivity features and link brain changes among different datasets, studies, and disorders. NeuroMark leverages the benefits of a data-driven approach and provides comparability across multiple sites and analyses, making BrainForge among the first cloud-based data management platforms for integrative neuroimaging acquisition, analysis, and clinical phenotypes.
B. Analysis Management
BrainForge provides multiple features to aid in large-scale processing of neuroimaging data, including parameter set reuse, analysis plans, preprocessing summaries, and group selection. For each pipeline or analysis, BrainForge provides a GUI exposing a variety of parameters to customize the process to suit the data and task at hand. These parameter sets can be saved, versioned, reused, and shared with others, contributing to reproducibility. An analysis plan associates the data collected via a protocol within a study with an analysis using a specific container and parameter set. This analysis plan can either be executed manually, queued in batch mode, or made automatic for ongoing analysis as acquisitions are added to a study, The automatic plans immediately execute the analysis on all acquisitions within the study with a matching protocol, as well. The status of preprocessing across all sessions in a study is displayed on a preprocessing summary page, providing an overview of the status of analysis for each subject. An example of the preprocessing summary page in shown in Fig. 4. Each row is a session, and each column is a combination of an analysis, parameter set, and acquisition protocol. Cells in the table are color-coded according to the analysis status (set up = white, complete = green, error = red, and no data = gray) and contain a link leading to the analysis result. Near the top, there is also a row showing the number of analyses successfully completed against the total number of sessions present in the study. Results can further be grouped according to demographic data or imaging protocol. Additional features visually flag outliers, annotate data, remove or archive errors, and set rules for how data can be shared or used.
Figure 4:
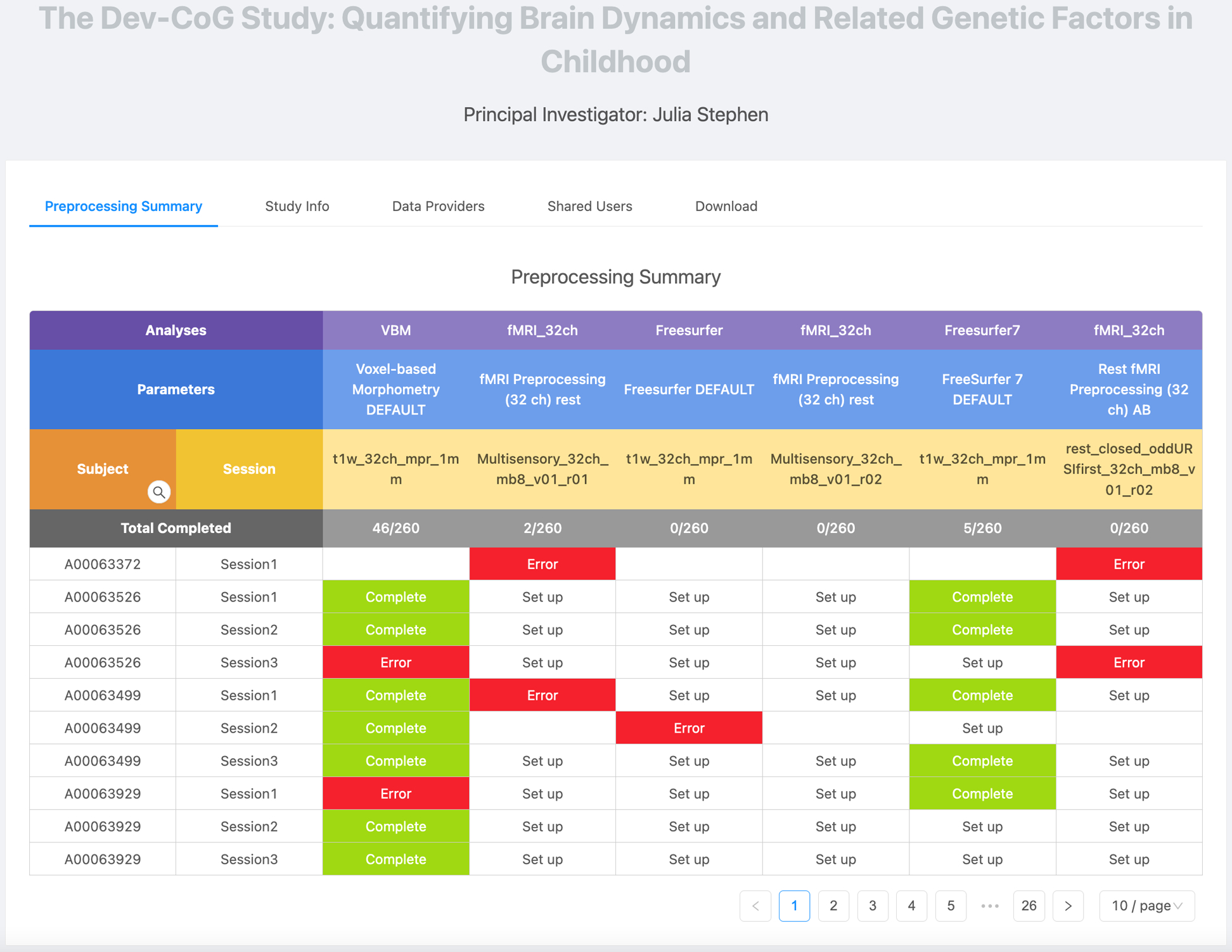
Preprocessing summary page for Dev-Cog study. Rows represent each session, and columns represent each combination of analysis, parameter set, and protocol.
VI. Implementation
A. Architecture
The BrainForge architecture (Fig. 5) is divided into a back end written in Python Django and a front end written in React.js using the Ant Design framework. The back end uses the Django Rest Framework to expose an API that is accessed by the front end. Django was chosen because it is easy to work with, feature rich, and scalable. Many high-profile, high-traffic websites have used Django, including Instagram, Pinterest, and Bitbucket41. Additionally, nginx is used as a reverse proxy server, and Redis is used for web socket communication and as a queue. Jobs are executed via a service account on an HPC architecture using the Slurm Workload Manager, a cluster management and job scheduling system for Linux, or in the cloud on AWS. Slurm is controlled via PySlurm42, a low-level Python wrapper around the Slurm C-API. A Postgres database is used to store metadata, analysis provenance, and user information. A relational database structure is primarily employed, utilizing JSON fields to store parameters and analysis information, as this type of information varies too widely to be practically encoded in database columns. Tables are normalized with some exceptions to improve performance, and database backups are saved daily.
Figure 5:
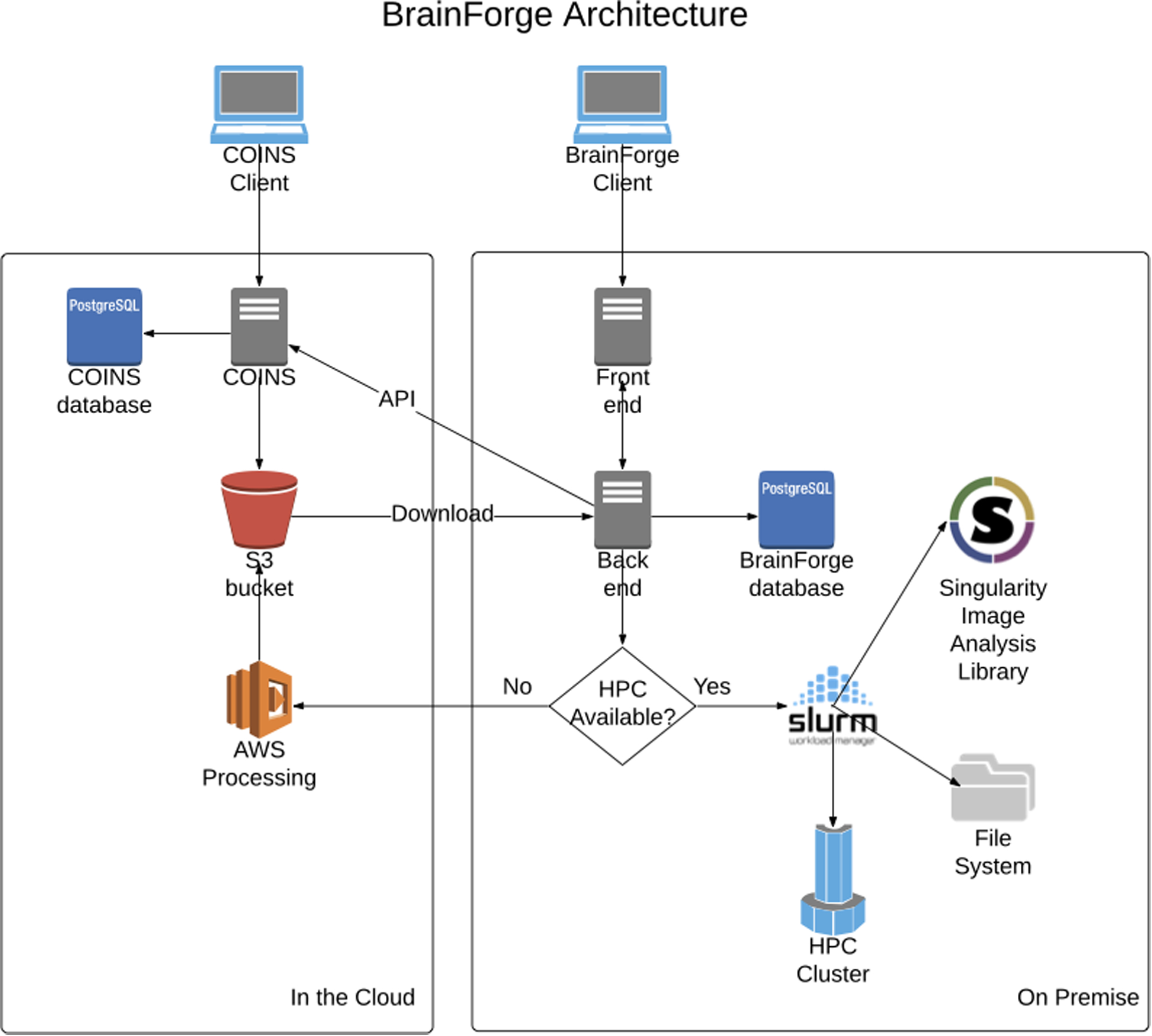
BrainForge architecture diagram showing on-premise components and COINS integration.
BrainForge runs on a CentOS 7 virtual machine (VM) in the TReNDS cluster. The VM has 4 virtual CPU cores and 16 GB of memory. The code for the BrainForge back end and the Singularity containers reside on a 200-GB volume on a network file system. Postgres, nginx, and Redis are deployed using Docker Compose. The back end resides on the host system so that it can run as a non-root user when interacting with Slurm, which is a security requirement at TReNDS. The code for the JavaScript front end is bundled using GitHub Actions and served from an AWS S3 bucket. The promotional website is at brainforge.trendscenter.org, and the platform can be accessed at https://brainforge.rs.gsu.edu. BrainForge is currently proprietary software.
B. Security
BrainForge uses best practices in its security. This includes protection against cross site scripting (XSS), cross site request forgery (CSRF), SQL injection, and clickjacking. Additionally, all interactions with the website are encrypted via HTTPS. User passwords are also hashed with the password-based key derivation function 2 (PBKDF2) algorithm43 with an SHA256 hash, as recommended by the National Institute of Standards and Technology (NIST)44. Furthermore, BrainForge uses JSON web tokens (JWTs) for authenticated sessions.
VII. Future Work
There are many future endeavors in line for BrainForge. We plan to allow users to compare their scans against the NeuroMark database using relevant imaging biomarkers derived by our algorithms. This feature can be used to help select subjects for clinical trials or to test future clinical decision support tools. Additional neuroimaging pipelines for genetics, electroencephalography (EEG), and magnetoencephalography (MEG) will be incorporated. Furthermore, we will add approaches for additional advanced analysis including data fusion (joint ICA and multivariate canonical correlation analysis with joint ICA45), and deep learning. We will also create a public collection of containers, such as MRIQC46 and fMRIPrep47, that are compatible with BrainForge, with built-in interfaces to accept them on the platform. The platform will also allow users to use and share their own pipelines on BrainForge. We also plan to allow the option of running analyses using the CLI. This will be important for users who want to programmatically run analyses and integrate them with other scripts or who simply feel more comfortable using a CLI rather than a GUI. To further serve users who are strong programmers, we will implement the ability to spawn Jupyter notebooks48 on our servers with access to study data. In the near term, we will create a feature to dynamically generate a text file describing analysis methods based on workflow parameters. This will be useful for researchers who want to publish analyses performed on BrainForge in scientific journals.
Recently, the neuroimaging research community has been moving towards adoption of the FAIR Data Principles, namely, findability, accessibility, interoperability, and reusability49. We plan to move BrainForge in this direction as well. To improve findability and accessibility to study results, we plan to create public pages with digital object identifiers (DOIs) for analyses and datasets and annotation with metadata according to Schema.org50. Additionally, we are working to link BrainForge with the Collaborative Informatics and Neuroimaging Suite Toolkit for Anonymous Computation (COINSTAC)51, a platform for federated analysis of neuroimaging data to promote reusability for data that cannot be shared.
VIII. Conclusion
BrainForge is a cloud-enabled web-based platform designed for neuroimaging research that addresses several urgent needs in the scientific community. Standard and customizable processing pipelines, implemented with industry-standard neuroimaging software, provide trusted outcomes. These pipelines are containerized, ensuring reproducibility and transparency, and are executed on an HPC grid, providing timely results. Flexible data management allows independent researchers to upload data, share their data selectively with other PIs, or to merge multistudy data for postprocessing analyses. Data analyses can be small in scale or batched to process thousands of subjects. BrainForge is built with modern web development tools, resulting in an intuitive and user-friendly neuroinformatics suite that enables prospective management, sharing, and analysis of studies, assessments, and neuroimaging data.
IX. Acknowledgements
BrainForge was funded by NIH R41MH122201, NIH R01MH118695, NSF #2112455, and internal funds from Georgia State University.
References
- 1.Morita T, Asada M, Naito E. Contribution of Neuroimaging Studies to Understanding Development of Human Cognitive Brain Functions. Review. Frontiers in Human Neuroscience. 2016-September-15 2016;10(464)doi: 10.3389/fnhum.2016.00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandettini PA. What’s new in neuroimaging methods? Ann N Y Acad Sci. 2009/March// 2009;1156:260–293. doi: 10.1111/j.1749-6632.2009.04420.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verner E, Baker BT, Bockholt J, Fries J, Petropoulos H, Raja R, Kalyanam R, Calhoun V Accelerating Neuroimaging Research with BrainForge. presented at: Gateways 2020; October 21, 2020. 2020; Session Improving Data Use for Biomedical Research. [Google Scholar]
- 4.Poldrack RA, Baker CI, Durnez J, et al. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nature Reviews Neuroscience. 2017/February/01 2017;18(2):115–126. doi: 10.1038/nrn.2016.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorgolewski K, Poldrack R. A Practical Guide for Improving Transparency and Reproducibility in Neuroimaging Research. PLOS Biology. July/07 2016;14:e1002506. doi: 10.1371/journal.pbio.1002506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M 1,500 scientists lift the lid on reproducibility. Nature. 2016/May/01 2016;533(7604):452–454. doi: 10.1038/533452a [DOI] [PubMed] [Google Scholar]
- 7.Scott A, Courtney W, Wood D, et al. COINS: An Innovative Informatics and Neuroimaging Tool Suite Built for Large Heterogeneous Datasets. Original Research. Frontiers in Neuroinformatics. 2011-December-23 2011;5(33)doi: 10.3389/fninf.2011.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo AB JMA, Grondona M SLURM: Simple Linux Utility for Resource Management. In: Feitelson DRL, Schwiegelshohn U, ed. Job Scheduling Strategies for Parallel Processing JSSPP 2003 Lecture Notes in Computer Science. Springer; 2003. [Google Scholar]
- 9.Avesani P, McPherson B, Hayashi S, et al. The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services. Scientific Data. 2019/May/23 2019;6(1):69. doi: 10.1038/s41597-019-0073-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flywheel. Powering the Digital Transformation in Healthcare and Research. Accessed May 14, 2021, flywheel.io
- 11.Kurtzer GM, Sochat V, Bauer MW. Singularity: Scientific containers for mobility of compute. PLOS ONE. 2017;12(5):e0177459. doi: 10.1371/journal.pone.0177459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merkel D Docker: lightweight linux containers for consistent development and deployment. Linux Journal. 2014;239:2. [Google Scholar]
- 13.Ashburner JB G; Chen C-C; Daunizeau J; Flandin G; Friston K; Kiebel S; Kilner J; Litvak V; Moran R; et al. SPM12 Manual. Vol. vol. 2464. 2014. [Google Scholar]
- 14.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23 Suppl 1:S208–19. doi: 10.1016/j.neuroimage.2004.07.051 [DOI] [PubMed] [Google Scholar]
- 15.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014 [DOI] [PubMed] [Google Scholar]
- 16.Cox RW, Hyde JS. Software tools for analysis and visualization of fMRI data. NMR in Biomedicine. 1997;10(4‐5):171–178. doi: [DOI] [PubMed] [Google Scholar]
- 17.Group ICA of fMRI Toolbox (v4.0c). Center for Translational Research in Neuroimaging and Data Science. Accessed May 6, 2021, https://trendscenter.org/software/gift/
- 18.TReNDS Center. Docker, Inc. https://hub.docker.com/orgs/trendscenter/repositories
- 19.Center for Translational Research in Neuroimaging and Data Science. GitHub, Inc. https://github.com/trendscenter/
- 20.Gorgolewski K, Burns CD, Madison C, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform. 2011;5:13. doi: 10.3389/fninf.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorgolewski KJ, Auer T, Calhoun VD, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data. 2016/June/21 2016;3(1):160044. doi: 10.1038/sdata.2016.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster I Globus Online: Accelerating and Democratizing Science through Cloud-Based Services. IEEE Internet Computing. 2011;15(3):70–73. doi: 10.1109/MIC.2011.64 [DOI] [Google Scholar]
- 23.Allen B, Bresnahan J, Childers L, et al. Software as a service for data scientists. Commun ACM. 2012;55(2):81–88. doi: 10.1145/2076450.2076468 [DOI] [Google Scholar]
- 24.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapping. 2001;14(3):140–151. doi: 10.1002/hbm.1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen E, Erhardt E, Damaraju E, et al. A Baseline for the Multivariate Comparison of Resting-State Networks. Original Research. Frontiers in Systems Neuroscience. 2011-February-04 2011;5(2)doi: 10.3389/fnsys.2011.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. Mar 2014;24(3):663–76. doi: 10.1093/cercor/bhs352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashburner J, Friston KJ. Voxel-Based Morphometry—The Methods. NeuroImage. 2000/June/01/ 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582 [DOI] [PubMed] [Google Scholar]
- 28.Fischl B FreeSurfer. NeuroImage. 2012/August/15/ 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage. 2016/January/15/ 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. Oct 2003;20(2):870–88. doi: 10.1016/s1053-8119(03)00336-7 [DOI] [PubMed] [Google Scholar]
- 31.Andersson JLR, Graham MS, Drobnjak I, Zhang H, Filippini N, Bastiani M. Towards a comprehensive framework for movement and distortion correction of diffusion MR images: Within volume movement. Neuroimage. May 15 2017;152:450–466. doi: 10.1016/j.neuroimage.2017.02.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calhoun Vince D, Miller R, Pearlson G, Adalı T. The Chronnectome: Time-Varying Connectivity Networks as the Next Frontier in fMRI Data Discovery. Neuron. 2014/October/22/ 2014;84(2):262–274. doi: 10.1016/j.neuron.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Y, Fu Z, Sui J, et al. NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. NeuroImage: Clinical. 2020/January/01/ 2020;28:102375. doi: 10.1016/j.nicl.2020.102375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffanti L, Zamboni G, Khan A, et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. NeuroImage. 2016/November/01/ 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chappell MA, Groves AR, Whitcher B, Woolrich MW. Variational Bayesian Inference for a Nonlinear Forward Model. IEEE Transactions on Signal Processing. 2009;57(1):223–236. doi: 10.1109/TSP.2008.2005752 [DOI] [Google Scholar]
- 36.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993/December// 1993;30(6):672–679. doi: 10.1002/mrm.1910300604 [DOI] [PubMed] [Google Scholar]
- 37.Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid–edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40(6):1445–1452. doi: 10.1002/jmri.24478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulloa A, Verner E, Plis S Application of a Universal Multi-model Classification Tool to Structural Magnetic Resonance Imaging. 2016:
- 39.Polyssifier. Center for Translational Research in Neuroimaging and Data Science. Accessed May 14, 2021, https://trendscenter.org/software/polyssifier/
- 40.TIM Trio EPI Stability Quality Assurance. Center for Brain Science. Accessed May 14, 2021, http://cbs.unix.fas.harvard.edu/science/core-facilities/neuroimaging/facilities/stability
- 41.Korsun J 10 Popular Websites Built With Django. Django Stars Blog. Updated September 21, 2021. Accessed November 1, 2021, https://djangostars.com/blog/10-popular-sites-made-on-django/
- 42.Roberts M, Torres G. PySlurm: Slurm Interface to python. Updated Feb 3, 2019. Accessed May 14, 2021, https://pyslurm.github.io/
- 43.Force IET. PKCS #5: Password-Based Cryptography Specification Version 2.1. 2017.
- 44.Recommendation for Password-Based Key Derivation, Part 1: Storage Applications (Department of Commerce) (2010).
- 45.Sui J, He H, Pearlson GD, et al. Three-way (N-way) fusion of brain imaging data based on mCCA+jICA and its application to discriminating schizophrenia. Article. NeuroImage. February/1 2013;66:119–132. doi: 10.1016/j.neuroimage.2012.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esteban O, Birman D, Schaer M, Koyejo OO, Poldrack RA, Gorgolewski KJ. MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLoS One. 2017;12(9):e0184661. doi: 10.1371/journal.pone.0184661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esteban O, Markiewicz CJ, Blair RW, et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature Methods. 2019/January/01 2019;16(1):111–116. doi: 10.1038/s41592-018-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kluyver T, Ragan-Kelley B, Pérez F, et al. Jupyter Notebooks – a publishing format for reproducible computational workflows. presented at: 20th International Conference on Electronic Publishing (01/01/16); 2016; https://eprints.soton.ac.uk/403913/ [Google Scholar]
- 49.Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific data. 2016;3:160018–160018. doi: 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welcome to Schema.org. Accessed May 14, 2021, https://schema.org/
- 51.Plis SM, Sarwate AD, Wood D, et al. COINSTAC: A Privacy Enabled Model and Prototype for Leveraging and Processing Decentralized Brain Imaging Data. Methods. Frontiers in Neuroscience. 2016-August-19 2016;10(365)doi: 10.3389/fnins.2016.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]


