Abstract
Zinc oxide nanoparticles (ZnO-NPs) synthesized through biogenic methods have gained significant attention due to their unique properties and potential applications in various biological fields. Unlike chemical and physical approaches that may lead to environmental pollution, biogenic synthesis offers a greener alternative, minimizing hazardous environmental impacts. During biogenic synthesis, metabolites present in the biotic sources (like plants and microbes) serve as bio-reductants and bio-stabilizers. Among the biotic sources, microbes have emerged as a promising option for ZnO-NPs synthesis due to their numerous advantages, such as being environmentally friendly, non-toxic, biodegradable, and biocompatible. Various microbes like bacteria, actinomycetes, fungi, and yeast can be employed to synthesize ZnO-NPs. The synthesis can occur either intracellularly, within the microbial cells, or extracellularly, using proteins, enzymes, and other biomolecules secreted by the microbes. The main key advantage of biogenic synthesis is manipulating the reaction conditions to optimize the preferred shape and size of the ZnO-NPs. This control over the synthesis process allows tailoring the NPs for specific applications in various fields, including medicine, agriculture, environmental remediation, and more. Some potential applications include drug delivery systems, antibacterial agents, bioimaging, biosensors, and nano-fertilizers for improved crop growth. While the green synthesis of ZnO-NPs through microbes offers numerous benefits, it is essential to assess their toxicological effects, a critical aspect that requires thorough investigation to ensure their safe use in various applications. Overall, the presented review highlights the mechanism of biogenic synthesis of ZnO-NPs using microbes and their exploration of potential applications while emphasizing the importance of studying their toxicological effects to ensure a viable and environmentally friendly green strategy.
Keywords: biological applications, green synthesis, microbial synthesis, nanoparticles, toxicological effects, zinc oxide
1. Introduction
Recently, the ZnO-NPs have gained recognition for their application in several industrial areas, including pharmaceuticals, food, photocatalyst, cosmetics, and agriculture, due to their distinctive properties (Jain et al., 2020; Murali et al., 2021a; Akhtar et al., 2022). As mentioned earlier, the large surface area-to-volume ratio of ZnO-NPs makes them more efficient than their counterparts, i.e., bulk ZnO, for their applications. Physical and chemical techniques have traditionally been employed for synthesizing ZnO-NPs, which provide high production rates with control sizes. However, the physical approach uses much energy, pressure, and heat, whereas the chemical method uses toxic and hazardous chemicals that contribute to environmental pollution and adversely affect human and animal health (Basnet et al., 2018). Moreover, the chemically synthesized ZnO-NPs have limited clinical and biological applications because they are toxic, less biocompatible, and may also bind or reside in the final NP products that interfere with the biological applications (Anjum et al., 2021). Hence, it is necessary to develop non-toxic, bio-safe, cost-effective, more eco-friendly, biocompatible ZnO-NPs as a substitute.
The green synthesis of ZnO-NPs, which uses diverse bioactive chemicals from plants and microbes as reductants and stabilizers, has recently come into the spotlight as a replacement for chemically and physically manufactured ZnO-NPs without altering their properties. Apart from plants, both unicellular and multicellular organisms (such as bacteria, actinomycetes, fungi, and yeast) are involved in the biological synthesis of ZnO-NPs (Kundu et al., 2014; Moghaddam et al., 2017; Shamsuzzaman et al., 2017; Kalaba et al., 2021). The green synthesis process is devoid of toxic chemicals and materials, energy-efficient, eco-friendly, and makes their applications in living organisms completely safe (Weldegebrieal, 2020). The plant-mediated synthesis of ZnO-NPs has been extensively reviewed to understand their unique properties and biological applications (Ahmed et al., 2017; Murali et al., 2021a); in contrast, only a few studies have noted the importance of microbial synthesis of ZnO-NPs and their applications. Hence the present study was focused on providing a holistic view of the microbe-mediated synthesis of ZnO-NPs and their potential applications, with specific objectives that include synthesis mechanisms and biological properties apart from their toxicological effects on the environmental ecosystem.
2. Microbes for ZnO-NP synthesis
Since the microbes are easily reproducible compared to plants, it offers an advantage over plant-mediated ZnO-NP synthesis. In the new millennium, using microbes, viz., bacteria, actinomycetes, fungi and yeast, has attracted considerable interest in synthesizing the ZnO-NPs. These organisms serve as miniature nano-factories wherein the enzymes, proteins or biomolecules secreted by them help to selectively reduce metal ions into their corresponding metal or metal oxide NPs. The formation of mono- and poly-dispersed NPs with different sizes and shapes is attributed to the numerous organic compounds released into the growing media or suspension cultures (Yusof et al., 2020a). Not all microbes are involved in the NPs synthesis process due to unique enzyme activity and metabolic processes, which differ from organism to organism. Due to these properties, selecting appropriate microbes independent of their enzyme activity or metabolic and biosynthetic pathways (intracellular or extracellular) is critical for forming ZnO-NPs, which remains unexplored. In addition, these microbes must also be able to tolerate heavy metals to synthesize the ZnO-NPs, as it is noted that the high metal stress is well documented to alter various microbial activities (Jain et al., 2020). Under this stress, the microbes reduce metal ions into appropriate metal NPs, demonstrating their potential to act as natural nano-factories (Lahiri et al., 2021). Microbes generally inhabit metal-rich environments and have strong metal resistance owing to their metal chelation and adsorption by intracellular and extracellular proteins (Yusof et al., 2019). As a result, mimicking the natural biomineralization process for synthesizing ZnO-NPs could be considered a promising strategy. Many Zn metal-tolerating microbes have been isolated from native metal-rich soils and mines to synthesize the ZnO-NPs (Jain et al., 2013).
In addition, metal (Zn) precursors (such as zinc acetate, zinc chloride, zinc nitrate, zinc sulphate, etc.) are needed for the microbe-mediated ZnO-NP synthesis. These salts are typically provided as soluble salts precipitating in the microbial cell suspension or its extract with bioactive components during the synthesis, usually taking minutes or hours (Murali et al., 2021b). The successful transformation is indicated by precipitation/ white deposition or gradual changes in the suspension color. Once the synthesis process is complete, the mixture contains microbial cells and ZnO-NPs, separated using various techniques, including centrifugation, filtration, or calcination. In addition, to obtain white crystalline powder of ZnO-NPs, the obtained particles are dried in a hot air oven for an extended time at 60°C after being completely washed with distilled water, followed by ethanol to remove any contaminants that were present (Kundu et al., 2014; Kalpana et al., 2018; Yusof et al., 2019). Different physicochemical methods characterize the synthesized NPs to ascertain their distinctive characteristics, such as shape, particle size, purity, composition, surface charge, and active functional groups. These methods include Ultraviolet–Visible (UV–Vis) spectroscopy, Fourier Transform Infrared (FTIR) spectroscopy, X-Ray Diffraction (XRD), Dynamic Light Scattering (DLS), Scanning Electron and Transmission Electron Microscopy (Faridvand et al., 2021; Nayeri et al., 2021; Thakur et al., 2022). Furthermore, several factors, like temperature, pH, reaction period and precursor concentration, are essential for determining the production rate, yield, and morphologies of NPs. The microbe-assisted synthesis of ZnO-NPs and their potential biological applications are listed in Table 1.
Table 1.
Microbe-mediated ZnO-NPs and their potential biological applications.
| Microbes | Precursor | Size (nm) | Shape | Purification methods | Application | Reference |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Aeromonas hydrophila | Zinc oxide | 57 | Spherical/oval | Centrifugation | Antimicrobial | Jayaseelan et al. (2012) |
| Lactobacillus sporogens | Zinc sulphate | 145.7 | Hexagonal | Centrifugation, washing and drying in oven at 40°C for 8 h | Antibacterial | Mishra et al. (2013) |
| Serratia ureilytica | Zinc acetate | 170–600 | Spherical | – | Dhandapani et al. (2014) | |
| Rhodococcus pyridinivorans | ZnSO4.H2O | 100–120 | Roughly spherical | Centrifugation, filtration, washing and drying at 70°C in hot air oven for 4 h | Antibacterial, Anticancer, Photocatalytic and Drug delivery | Kundu et al. (2014) |
| Pseudomonas aeruginosa | Zinc nitrite | 35–80 | Spherical | Centrifugation, washing and drying at 70°C under vacuum oven | Antioxidant | Singh et al. (2014) |
| Bacillus licheniformis | Zinc acetate dihydrate | 320–400 | Rod-like | Centrifugation | Photocatalytic | Tripathi et al. (2014) |
| Acinetobacter schindleri | Zinc nitrate | 20–100 | Polydispersed and spherical | Centrifugation, washing and drying in hot air oven at 60°C for 6 h | Antibacterial | Busi et al. (2016) |
| Streptomyces sp. | Zinc chloride | 20–50 | Spherical | – | Antibacterial and anticancer | Balraj et al. (2017) |
| Sphingobacterium thalpophilum | Zinc nitrate | 112 | Triangle chips-like | Drying at 120°C and annealing at 700°C for 5 h | Antibacterial | Rajabairavi et al. (2017) |
| Staphylococcus aureus | Zinc acetate | 10–50 | Acicular | Centrifugation, washing, lyophilization and storage at 0°C | Rauf et al. (2017) | |
| P. aeruginosa | Zinc nitrate | 50–100 | Pseudospherical | – | Antimicrobial | Barsainya and Singh (2018) |
| L. paracasei | Zinc nitrate | 1,179 | Spherical | – | Antibacterial | Król et al. (2018) |
| B. megaterium | Zinc nitrate | 45–95 | Rod and cubic | Centrifugation and repeated washing | Saravanan et al. (2018) | |
| S. enissocaesilis | Zinc sulfate | 5–20 | Spherical | Centrifugation, washing and drying | Shaaban and El-Mahdy (2018) | |
| Halomonas elongata | Zinc chloride | 18 | Spherical | – | Taran et al. (2018) | |
| P. putida | Zinc nitrate | 44.5 | Spherical | Centrifugation and drying in hot air furnace 400°C for 2 h | Jayabalan et al. (2019) | |
| B. haynesii | Zinc sulphate | 50 | Spherical | Drying at 80°C and annealing at 700°C for 5 h | Rehman et al. (2019) | |
| B. cereus | Zinc sulphate hepta-hydrate | 58–63 | Irregular | – | Antibacterial | Iqtedar et al. (2020) |
| S. nematodiphila | Zinc sulfate | 15–30 | Roughly spherical | Washing through multiple centrifugations and drying at 120°C | Antimicrobial and Photocatalytic | Jain et al. (2020) |
| L. plantarum | Zinc nitrate | 191–291 | Flower-like and irregular | Cell biomass: Ultrasonic disruption at 30°C for 30 min, centrifugation and drying at 100°C. Cell-free supernatant: Centrifugation, washing and drying at 100°C |
Antibacterial | Yusof et al. (2020b) |
| B. cereus | Zinc sulfate heptahydrate | 21–35 | Spherical | Centrifugation, washing and freeze-drying | Ahmed et al. (2021) | |
| Lactobacillus spp. | Zinc acetate | 32 | Spherical | Interruption by ultrasound cycles in 100 W for 5 min, centrifugation, washing and air-drying at 60°C | Antimicrobial and anticancer | Suba et al. (2021) |
| Actinomycetes | ||||||
| Streptomyces sp. | Zinc acetate dihydrate | 16–25 | Spherical | Centrifugation, drying at 120°C overnight and heating at 450°C for 5 h | Antibacterial, and anticancer | Shanmugasundaram and Balagurunathan (2017) |
| S. plicatus | Zinc sulfate | 21.72 | Spherical | Centrifugation, washing and drying in an oven at 90°C | Antimicrobial | Kalaba et al. (2021) |
| Pseudochrobactrum sp. | Zinc acetate | 90–110 | Granular | Oven drying at 85°C, calcination for 7 h in muffle furnace at 700°C and grinding | Photocatalytic | Siddique et al. (2021) |
| S. enisocaesilis | Zinc sulphate | 12–326 | Needle shape | Washing, centrifugation and drying in muffle furnace at 400°C for 8 h | Antibacterial | Rajivgandhi et al. (2022) |
| Fungi | ||||||
| Aspergillus terreus | Zinc sulfate | 54.8–82.6 | Spherical | Centrifugation and lyophilization | Antifungal | Baskar et al. (2013) |
| A. aeneus | Zinc acetate | 100–140 | Spherical | – | – | Jain et al. (2013) |
| A. fumigatus | Zinc sulfate | 60–80 | Spherical | Centrifugation and lyophilization | Antibacterial | Rajan et al. (2016) |
| A. niger | Zinc chloride | 41–75 | Spherical | Centrifugation and drying in oven at 60°C | Ibrahem et al. (2017) | |
| Candida albicans | Zinc oxide | ∼20 | Quasi-spherical | Centrifugation | – | Shamsuzzaman et al. (2017) |
| A. terreus | Zinc acetate | 10–45 | Spherical | Separation and drying at 150°C for 48 h | Antibacterial and anticancer | Fouda et al. (2018) |
| A. niger | Zinc nitrate | 53–69 | Spherical | Centrifugation | Antibacterial and photocatalytic | Kalpana et al. (2018) |
| A. niger | Zinc nitrate | 30–70 | Spherical | – | Antioxidant and anticancer | Es-haghi et al. (2019) |
| A. niger | Zinc acetate | 80–130 | Rod and cluster | Collection and drying at 150°C for 48 h | Antibacterial, antioxidant, and anticancer | Gao et al. (2019) |
| Cordyceps militaris | Zinc nitrate hexahydrate | 10.15 | Flower | Centrifugation, washing and drying at 60°C | Photocatalytic | Li et al. (2019) |
| Fusarium keratoplasticum | Zinc acetate | 10–42 | Hexagonal | Collection and drying at 150°C for 48 h | Antibacterial and anticancer | Mohamed et al. (2019) |
| A. niger | Zinc acetate | 8–38 | Nano-rod | Collection and drying at 150°C for 48 h | Antibacterial and anticancer | |
| Alternaria tenuissima | Zinc sulphate | 15.45 | Spherical | Ultra-centrifugation, washing and drying at 50°C in hot air | Antimicrobial, antioxidant, anticancer and photocatalytic | Abdelhakim et al. (2020) |
| Penicillium corylophilum | Zinc acetate dihydrate | 9–51 | Spherical | Centrifugation, washing and oven-drying at 80°C for 48 h | Photocatalytic | Fouda et al. (2020) |
| Periconium sp. | Zinc nitrate | 16–78 | Quasi spherical | Drying at 125°C for 12 h and calcination at 700°C for 4 h in muffle furnace | Antimicrobial and antioxidant | Ganesan et al. (2020) |
| Agaricus bisporus | Zinc acetate dehydrate | <40 nm | Hexagonal | Filtration, washing and drying at 80°C in hot air oven | Antibacterial and antioxidant | Singh and Dutta (2020) |
| Trichoderma harzianum, and T. reesei | Zinc nitrate hexahydrate | 60–70 | Crystal planes | Calcination at 700°C for 2 h | Antibacterial | Shobha et al. (2020) |
| Xylaria acuta | Zinc nitrate hexahydrate | 34–55 | Hexagonal | Calcination at 700°C for 2 h | Antimicrobial and anticancer | Sumanth et al. (2020) |
| Acremonium potronii | Zinc acetate hexahydrate | 13–15 | Spherical | Washing, centrifugation and drying at 150°C for 30 min | Photocatalytic | Ameen et al. (2021) |
| A. niger | Zinc acetate dihydrate | 35 | Spherical | Centrifugation, washing and drying | Antibacterial and anticancer | Es-haghi et al. (2021) |
| A. niger | Zinc acetate | 20 | Hexagonal | Washing and centrifugation | Antimicrobial and anticancer | Mekky et al. (2021) |
| P. chrysogenum | Zinc acetate dihydrate | 9–35 | Hexagonal | Separation and drying at 80°C for 48 h | Antimicrobial | Mohamed et al. (2021) |
| A. terreus | Zinc acetate | 30.45 | Almost spherical with irregular margins | Ultra-centrifugation, washing and drying at 50°C | Antimicrobial and Antioxidant | Mousa et al. (2021) |
| A. niger | Zinc nitrate | 82–176 | Irregular, individual, and quaternary | – | Antibacterial and Wound healing | Rasha et al. (2021) |
| Yeast | ||||||
| Pichia fermentans | Zinc oxide | – | Smooth, and elongated | – | Antimicrobial | Chauhan et al. (2015) |
| P. kudriavzevii | Zinc acetate dihydrate | ~10–61 | Hexagonal | Centrifugation and drying at 150°C for 6 h | Antibacterial, Antioxidant, and Anticancer | Moghaddam et al. (2017) |
| Marine yeast | Zinc oxide | 86.27 | Almost round | – | Antioxidant | Aswathy et al. (2017) |
3. Proposed mechanisms of microbe-mediated ZnO-NPs synthesis
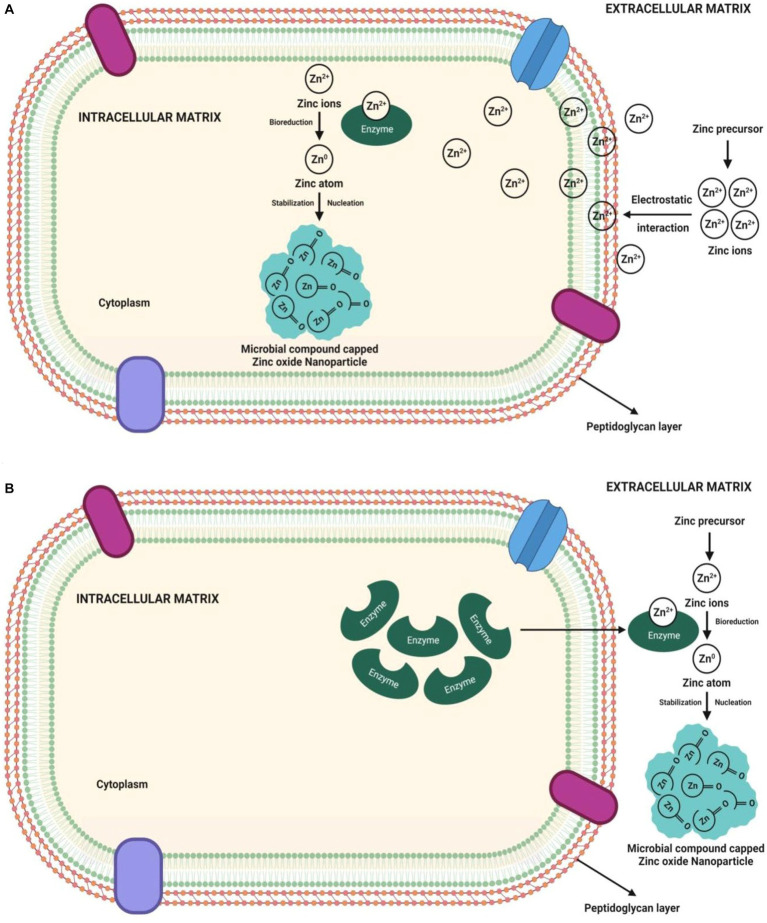
Usually, the microbes have the inherent ability to synthesize the ZnO-NPs directed via intra or extracellular pathways. Among the two, synthesis of the nanoparticles through extracellular mode is more favorable and has been extensively employed than intracellular synthesis since it can be utilized to synthesize large extent and involves a considerably simple downstream processing protocol that avoids several synthesis steps as well as easy cell separation and industrialization (Kundu et al., 2014; Busi et al., 2016; Sumanth et al., 2020). To obtain NPs with purity, the recovery of intracellular synthesized NPs requires extra purification steps, such as collecting cell biomass and ultrasonication for cell lysis (Yusof et al., 2020b). It has been well documented that the pH affects the synthesis of NPs through surface or intra or extracellular. Although microbial components, such as proteins, enzymes, and other substances, are essential to the synthesis process, only a few chemical elements are involved in NP production (Figure 1).
Figure 1.
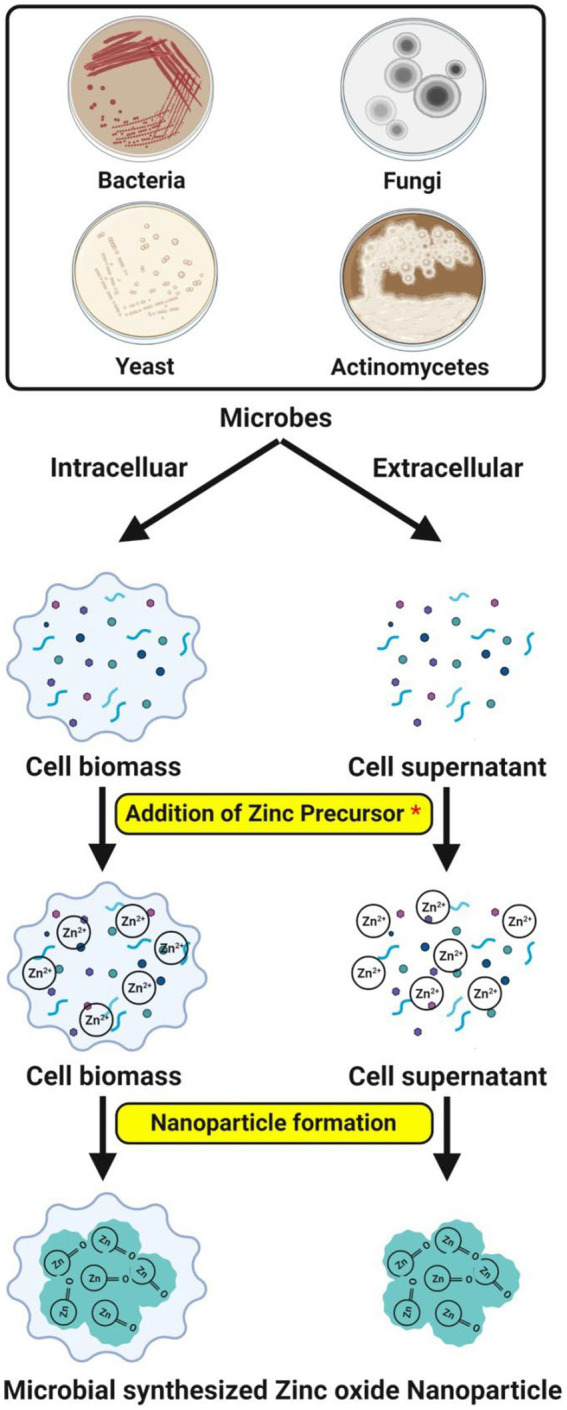
Possible mechanisms of microbe-mediated ZnO-NPs synthesis.
By giving Pi (π) electrons from carbonyl groups, bioactive chemicals, such as amino acids and polyphenolic compounds, serve as effective reducing agents during ZnO-NPs synthesis. Following the reduction of zinc ions (Zn2+) to zero-valent zinc atoms (Zn0), the zinc complex is broken down into ZnO-NPs during annealing. The ZnO nano-rods formed are stabilized by certain extracellular proteins secreted by the bacteria to detoxify the stress condition (Tripathi et al., 2014). The investigations showed that the protein’s amino acids interact with Zn2+ ions to create NPs, showing that the protein’s natural state is not necessary to create ZnO-NPs (Jain et al., 2013). The interaction between hydrogen bonds may explain the disruption of non-polar hydrophobic molecules during heating. As a result, zinc ions were exposed to amino acids, leading to the formation of ZnO-NPs. Microbial-secreted amino acids have hydroxyl groups that allow the complexation of zinc ions, hydrolysis, and synthesis of ZnO-NPs via thermal decomposition. The amino acids assist in the stability of ZnO-NPs by aggregation inhibition and control of crystal growth (Moghaddam et al., 2017).
With the aid of microbes and water molecules in an acidic milieu, zinc’s complexation commenced the ZnO-NP production process. The development of the zinc aqua-hydroxo complex, which later changed into ZnO-NPs, was caused by the zinc aqua complex accepting an electron from the deprotonated carboxyl group of bioactive molecules (such as enzymes and peptidoglycan) released by the bacteria (Król et al., 2018). In some cases, the specific organic functional groups present on the surface of the cell wall serve as the building blocks for a non-enzymatic synthesis that aids in reducing the zinc ions. The organic molecules (as reducing agents) are released from the microbial cell due to cell membrane rupture and cell lysis during the heat-killed process. The zinc ions made more organic molecules available, leading to a higher reduction (Yusof et al., 2019). The size, chemical composition, shape, solubility, dispersion factor and surface area of NPs have all significantly impacted their biological responses. The NPs synthesis depends on physicochemical factors such as temperature, pH, precursor concentration, age of microbes, reaction time, stirring, and irradiation, which affect the particle size, monodispersity, shape and yield (Jain et al., 2020).
3.1. Mechanism of microbe-mediated intracellular ZnO-NP synthesis
The microbial cell wall structure and ionic charges play a crucial role in the intracellular synthesis pathway of NPs. This technique sends zinc ions into the microbial cell to combine with other molecules, including enzymes and coenzymes, to produce ZnO-NPs. The microbial cell wall comprises various polysaccharides and proteins that activate metal ion binding sites. Heavy metal ions have been demonstrated to pose a significant threat to microbes as they react to the metal stress by penetrating or absorbing the metal ions onto the cell wall structure via electrostatic interactions (Selvarajan and Mohanasrinivasan, 2013). It is because specific enzymes, polypeptides, and cysteine on the microbial cell membrane contain negatively charged carboxylate groups that draw metal ions. Likewise, the trapped ions are reduced into elemental atoms by NADH-dependent reductase, wherein an electron carrier enmeshed in the plasma membrane transfers an electron from NADH to the latter (Kundu et al., 2014). Finally, the nuclei transform into NPs that amass in the cytoplasm or periplasmic region bordered by the cell wall. The greatest contributors to NP stability in cells are the proteins, peptides, and amino acids cysteine, tryptophan, and tyrosine (Balraj et al., 2017).
The intracellular synthesis mechanism involves the electrostatic transfer of zinc ions into the microbial cell, where the zinc ions (Zn2+) are reduced to zinc atoms (Zn0) by cell wall enzymes, which subsequently expand the nuclei to form the ZnO-NPswithin the periplasmic space of the cell wall or cytoplasm (Tripathi et al., 2014; Król et al., 2018). The NPs formed are intracellularly penetrated out of the cell. Ultrasonication is required to obtain the pure NPs from intracellular synthesis. The mechanism of microbe-mediated intracellular ZnO-NP synthesis is depicted in Figure 2A. The pH dependent membrane-bound oxidoreductases of probiotic bacteria Lactobacillus sporogens employed to synthesize the ZnO-NPs were active at low pH, suggesting a low pH environment required for ZnO NPs synthesis. Similarly, Selvarajan and Mohanasrinivasan (2013) reported that the biosynthesized ZnO-NPs might have resulted from carbon source-dependent rH2 and pH-sensitive membrane-bound oxidoreductases in L. plantarum culture solution.
Figure 2.
Pictorial representation of mechanisms of microbe-mediated (A) intracellular ZnO-NP synthesis and (B) extracellular ZnO-NPs synthesis.
3.2. Mechanism of microbe-mediated extracellular ZnO-NP synthesis
Contrary to intracellular synthesis, the extracellular synthesis mechanism involves either enzymatic synthesis on the microbial cell membrane or the release of microbial enzymes (such as cofactor NADH and NADH-dependent enzymes) as extracellular enzymes into the growth medium. The electron transfer from NADH through NADH-dependent reductase (like nitrate reductase enzyme) started the bioreduction of Zn2+, which then reduced to Zn0, resulting in the ZnO-NP formation (Kundu et al., 2014; Saravanan et al., 2018; Shaaban and El-Mahdy, 2018). Microbes release extracellular proteins that act as capping agents for NPs stability (Balraj et al., 2017). The synthesis of NPs is demonstrated by the appearance of white precipitation in the media. The participation of bacterial extracellular enzymes/ proteins not only allows for the ZnO-NP synthesis but these enzymes/ proteins also control the size and stability of NPs (Kundu et al., 2014; Chauhan et al., 2015; Shaaban and El-Mahdy, 2018). Figure 2B shows a pictorial representation of the microbe-mediated extracellular ZnO-NP synthesis mechanism. The extracellular ZnO-NP synthesis from the culture filtrate of Aspergillus niger would be important to identify the responsible reducing agents (Ibrahem et al., 2017; Kalpana et al., 2018). Bacterial secreted metabolites outside the cell carry out the extracellular ZnO-NP synthesis. The siderophore pyoverdine (a secondary metabolite) released extracellularly by Pseudomonas aeruginosa comprises amino and hydroxamate groups that reduce Zn2+ to form ZnO-NPs (Barsainya and Singh, 2018).
4. Bacterial-mediated ZnO-NP synthesis
Several microbes have been used for ZnO-NP synthesis, but bacteria are most preferred due to their genetic manipulation capability and ease of handling over other eukaryotic microbes (Jayabalan et al., 2019; Iqtedar et al., 2020; Ahmed et al., 2021). The reproducible bacteria (like lactic acid producing bacteria) have gained more interest in bacterial-mediated NP synthesis because of their high production of diverse enzymes and non-pathogenic nature. The lactic acid bacteria (Gram-positive) has a thick cell wall layer comprising proteins, polysaccharides, lipoteichoic and teichoic acid, etc., which helps in the bioreduction and biosorption of metal ions (Yusof et al., 2020b; Suba et al., 2021). They can also produce exopolysaccharides that defend cells from metal stress and serve as an additional site for metal ion biosorption (Yusof et al., 2020a). Moreover, lactic acid bacteria are recognized as health-beneficial bacteria for their pessimistic electrokinetic potential (Yusof et al., 2020b), which permits them to attract metal ions for the synthesis of NPs under oxidative as well as reductive conditions (Yusof et al., 2020a,b). Selvarajan and Mohanasrinivasan (2013) have reported that moderately stable ZnO-NPs were produced in which the lactic acid bacteria secreted biomolecules acted as capping agents in intracellular synthesis.
5. Actinomycetes-mediated ZnO-NP synthesis
Actinomycetes are regarded as superior among commercially important microbial species because of their saprophytic behavior, allowing them to produce various extracellular enzymes and bioactive components. Streptomyces sp. is a member of the actinomycetes, recognized for their soil degrading properties and as the potent source of secondary metabolites, especially antibiotics (Rajivgandhi et al., 2022). Many reports have suggested that actinomycetes can synthesize ZnO-NPs via extracellular and intracellular methods (Shanmugasundaram and Balagurunathan, 2017; Kalaba et al., 2021; Rajivgandhi et al., 2022) and needs to be exploited further to utilize them as an alternative to plants. The advantage of using actinomycetes for NP synthesis lies in their ability to produce specific enzymes and metabolites that can control the size, shape, and stability of the NPs formed. It offers potential advantages in tailoring the properties of ZnO-NPs for specific biological applications.
6. Fungal-mediated ZnO-NP synthesis
The mechanical method for producing ZnO-NPs from the fungal culture supernatant or biomass is similar to bacterial-mediated synthesis. But, the fungal-mediated ZnO-NP synthesis is a potential strategy owing to their greater metal tolerance, excellent metal binding capacity, metal bioaccumulation ability and higher productivity over bacteria (Baskar et al., 2013; Gao et al., 2019; Sumanth et al., 2020). In addition, the fungi efficiently secrete an excess of extracellular redox enzymes and proteins as bioactive phytochemicals to the culture media than bacteria, which helps reduce the zinc ions (Zn2+) into ZnO-NPs in greater quantities (Fouda et al., 2018; Ganesan et al., 2020). The fungal extracellular enzymes and proteins secreted in the media, which act as reducing and capping agents, are bound and encapsulated on the surface of ZnO-NPs, ensuring their stability without agglomeration (Raliya and Tarafdar, 2013; Sarkar et al., 2014).
The fungal strains are typically grown in sterilized media and incubated at the appropriate temperature to prepare the fungal extracts. The fungal metabolite-enriched cell-free supernatant is then harvested by filtration and centrifugation for further ZnO-NP synthesis (Ibrahem et al., 2017; Abdelhakim et al., 2020; Ganesan et al., 2020). The ability of fungal cell-free extracts to potentially cap and reduce the size of ZnO-NPs formed in a definite size and shape was demonstrated. Thus, the shape of fungal-mediated ZnO-NPs synthesized depends on the species type (Mohamed et al., 2019). The plausible mechanism of fungal-mediated ZnO-NPs synthesis has been postulated that the synthesis process involves the initial transformation of zinc acetate (as a precursor) into ZnO-NPs, followed by capping fungal extracellular proteins on the surface of NPs (Kadam et al., 2019).
7. Yeast-mediated ZnO-NP synthesis
Like fungi, yeast has been shown to synthesize the ZnO-NPs because of their better zinc stress tolerance. Specific yeast strains are chosen for their ability to interact with zinc ions and facilitate the reduction of zinc ions to form ZnO-NPs. The yeast cells act as a biological reducing agent, converting the zinc ions in the solution into ZnO-NPs through a reduction reaction. The process is usually carried out under controlled conditions to regulate the size and shape of the NPs. The yeast is preferred due to its availability, ease of handling, and biocompatibility. Moghaddam et al. (2017) have reported that ZnO-NPs (~10–61 nm) formed from Pichia kudriavzevii yeast strain were found to depend on the reaction time, which plays a main role in the size, distribution and shape of NPs. In addition, studies have revealed that ZnO-NP synthesized from P. fermentans extracellularly possessed potent inhibitory effects against various pathogenic bacteria and fungi, thereby contributing to the beneficial effect of the pharmaceutical industry. Due to the toxic metals absorbing and accumulating ability, the diverse yeast species could be used as carriers for synthesizing the ZnO-NPs (Chauhan et al., 2015; Aswathy et al., 2017).
8. Applications of microbial-mediated ZnO-NPs
The microbial-mediated ZnO-NPs possess prospective biomedical applications, especially antimicrobial, antioxidant, anticancer, wound healing, drug delivery system, photocatalytic activity, etc., which are described below.
8.1. Antibacterial activity
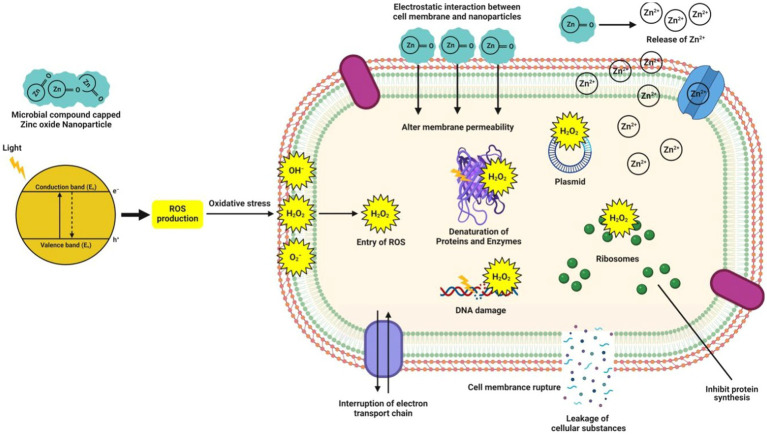
The microbial-mediated ZnO-NPs could be emerged as potential antimicrobial agents mostly because of their unique physiochemical properties in combating a wide range of bacterial pathogens (Moghaddam et al., 2017; Saravanan et al., 2018). Some bacteria can synthesize various antimicrobial chemicals known as bacteriocin (Perez et al., 2014). The microbial-derived bacteriocin might act as a reducing agent in the ZnO-NP synthesis and a capping agent for binding to the NPs surface, thereby increasing their antibacterial properties (Sidhu and Nehra, 2019). The microbial-mediated ZnO-NPs possess more than one mechanism responsible for their antibacterial activity. The possible bactericidal mechanism proposed that the smaller NPs have a higher surface reactivity to penetrate the cells and release free Zn2+ from ZnO-NPs easily. The release of Zn2+ ions on the biomolecules (DNA/ proteins/ enzymes) found in bacterial cells is one of the major antibacterial mechanisms. It is well known that the formed ZnO-NPs disrupt numerous bacterial cell functions such as metabolism, active transport, and enzyme activity and subsequently induce bacterial cell death (Rauf et al., 2017; Taran et al., 2018; Iqtedar et al., 2020).
While the ZnO-NPs frequently exploit the production of reactive oxygen species (ROS) [such as superoxide anion (O2), hydroxyl ion (OH), and hydrogen peroxide (H2O2)], when they are exposed to UV light to kill bacteria by inducing oxidative stress and cell death (Rehman et al., 2019). These ROS are generated when hydroxyl groups on the NPs surface react with water (H2O) to produce hydroxyl radicals (OH−) and hydrogen ions (H+), which then produce superoxide anion (O2−) in the presence of oxygen (O2) (Kalpana et al., 2018). Eventually, the bacterial cell membrane barrier is breached by the H2O2, damaging cellular elements (such as DNA, proteins, and lipids), leading to cell injury and death (Król et al., 2018; Abdelhakim et al., 2020). However, due to their negative charge, OH− and O2− are only found on the surface of bacteria and cannot enter the cell membrane except for H2O2 (Sumanth et al., 2020). The electrostatic adherence of the ZnO-NPs to the bacterial cell membrane is another mechanism underlying the antibacterial potential of these particles. Because ZnO-NPs include amine surface groups, their positive zeta potential facilitates their electrostatic adherence to the negatively charged bacterial cell membrane, which allows NPs to enter the bacterial cells. This direct contact can potentially compromise the integrity of the bacterial cell and rupture the plasma membrane structure, leading to intracellular content leakage and cell death (Jayaseelan et al., 2012; Taran et al., 2018). Figure 3 illustrates the potential antibacterial processes of microbe-mediated ZnO-NPs.
Figure 3.
Mechanisms predicted during the expression of antibacterial potential of microbe-mediated ZnO-NPs.
Due to the structure of their cell walls, Gram-negative bacteria are more susceptible to the microbe-mediated ZnO-NPs, while in contrast, Gram-positive bacteria have an extra outer cell membrane that contains lipopolysaccharides that can enhance the outer membrane barrier characteristics, exert a potent aversion to the NPs, and increase their resistance to ZnO-NPs (Gao et al., 2019; Yusof et al., 2020b). The size, composition and shape of NPs influenced their biological activity (i.e., antibacterial activity) because different particle surfaces have varied thicknesses of surface atoms and electronic structures, resulting in various physical and chemical properties (Mohamed et al., 2019). The findings of Mousa et al. (2021) suggested that the ZnO-NPs synthesized using the endophytic A. terreus confirmed the broad spectrum of antibacterial activity against four different human pathogenic bacteria (such as Escherichia coli, Klebsiella pneumoniae, P. aeruginosa and Staphylococcus aureus) with the recorded MIC value of 100 μg/mL. Suba et al. (2021) have reported that the ZnO-NPs biosynthesized from Lactobacillus spp. extract was found to show a strong microbicidal effect with the maximum zone of inhibition ranging from 20 to 24 mm against Clostridium difficile, C. perfringens, E. coli, and Salmonella typhi. Recently, Rajivgandhi et al. (2022) suggested that the ZnO-NPs biosynthesized from an actinomycete, Streptomyces enisocaesilis possessed antibacterial properties against multi-drug resistant K. pneumoniae at increasing concentrations. Additionally, Table 2 lists the antibacterial effectiveness of microbe-mediated ZnO-NPs against bacterial pathogens.
Table 2.
Microbe-mediated ZnO-NPs exhibiting potential Antibacterial properties.
| Microbes | Pathogen | MIC | Reference |
|---|---|---|---|
| Bacteria | |||
| A. hydrophila | Aeromonas hydrophila; E. coli; S. aureus; Enterococcus faecalis; P. aeruginosa and Streptococcus pyogenes | 1.2–25 μg mL−1 | Jayaseelan et al. (2012) |
| L. sporogens | S. aureus | 20 mM | Mishra et al. (2013) |
| S. ureilytica | E. coli and S. aureus | – | Dhandapani et al. (2014) |
| R. pyridinivorans | Staphylococcus epidermidis | – | Kundu et al. (2014) |
| A. schindleri | E. coli and Salmonella enterica | 100 μg mL−1 | Busi et al. (2016) |
| Streptomyces sp. | E. coli and Bacillus subtilis | 100 μg mL−1 | Balraj et al. (2017) |
| S. thalpophilum | P. aeruginosa and Enterobacter aerogens | 0.3 g disc−1 | Rajabairavi et al. (2017) |
| S. aureus | S. aureus; MRSA 1; MRSA 2; E. faecalis; P. aeruginosa; S. epidermidis; Listeria monocytogenes; Klebsiella pneumonia; E. coli; MREC 1 and MREC 2 | 32–256 μg mL−1 | Rauf et al. (2017) |
| P. aeruginosa | S. aureus; Bacillus sp. and E. coli | 100 μg well−1 | Barsainya and Singh (2018) |
| L. paracasei | S. aureus and Acinetobacter baumannii | – | Król et al. (2018) |
| B. megaterium | Helicobacter pylori strains | 16–17 μg mL−1 | Saravanan et al. (2018) |
| H. elongata | E. coli and S. aureus | 0.01–0.1 M | Taran et al. (2018) |
| P. putida | A. baumannii; P. otitidis; P. oleovorans; E. faecalis and B. cereus | 30–120 μg μL−1 | Jayabalan et al. (2019) |
| B. haynesii | E. coli and S. aureus | 4–16 mg mL−1 | Rehman et al. (2019) |
| B. cereus | E. coli; S. aureus and Salmonella typhi | 0.6 μg mL−1 | Iqtedar et al. (2020) |
| S. nematodiphila | Xanthomonas oryzae | 100 μg mL−1 | Jain et al. (2020) |
| L. plantarum | E. coli; Salmonella sp.; S. aureus and S. epidermidis | 625–2,500 μg mL−1 | Yusof et al. (2020b) |
| B. cereus | Burkholderia glumae and B. gladioli | 50 μg mL−1 | Ahmed et al. (2021) |
| Actinomycetes | |||
| Streptomyces sp. | B. subtilis; S. aureus; K. pneumoniae; E. coli; E. aerogenes; S. typhi and Proteus vulgaris | 50 μg mL−1 | Shanmugasundaram and Balagurunathan (2017) |
| S. plicatus | Erwinia amylovora | 15.6 μg mL−1 | Kalaba et al. (2021) |
| S. enisocaesilis | K. pneumoniae | 50–250 μg mL−1 | Rajivgandhi et al. (2022) |
| Fungi | |||
| A. fumigatus | K. pneumoniae; P. aeruginosa; E. coli; S. aureus and B. subtilis | 250–1,000 μg mL−1 | Rajan et al. (2016) |
| A. niger | S. aureus and E. coli | 1–1.5 mg mL−1 | Ibrahem et al. (2017) |
| A. terreus | P. aeruginosa; S. aureus; B. subtilis and E. coli | 250–500 μg mL−1 | Fouda et al. (2018) |
| A. niger | E. aerogenes; E. coli; K. pneumoniae and P. aeruginosa | 500 μg mL−1 | Gao et al. (2019) |
| F. keratoplasticum | E. coli; S. aureus; B. subtilis and P. aeruginosa | 250–500 ppm | Mohamed et al. (2019) |
| A. niger | |||
| A. tenuissima | P. aeruginosa, K. pneumoniae, E. coli and S. aureus | 100–200 μg mL−1 | Abdelhakim et al. (2020) |
| Periconium sp. | S. aureus and E. coli | 50 μg/mL | Ganesan et al. (2020) |
| A. bisporus | B. subtilis and E. coli | 10–100 μg mL−1 | Singh and Dutta (2020) |
| T. harzianum and T. reesei | Xanthomonas oryzae | 25–50 μg mL−1 | Shobha et al. (2020) |
| X. acuta | E. coli; B. cereus; S. aureus and P. aeruginosa | 15.6–31.3 μg mL−1 | Sumanth et al. (2020) |
| A. niger | E. coli | – | Es-haghi et al. (2021) |
| A. niger | A. baumannii; K. pneumoniae; P. aeruginosa; E. coli; S. aureus and S. haemolyticus | 12.5–50 μg mL−1 | Mekky et al. (2021) |
| P. chrysogenum | B. subtilis; S. aureus; S. typhimurium; E. coli and P. aeruginosa | 2–3 mg mL−1 | Mohamed et al. (2021) |
| A. terreus | S. aureus; P. aeruginosa; E. coli and K. pneumoniae | 100 μg mL−1 | Mousa et al. (2021) |
| A. niger | K. pneumoniae | 0.7 mg mL−1 | Rasha et al. (2021) |
| Yeast | |||
| P. fermentans | P. aeruginosa | – | Chauhan et al. (2015) |
| P. kudriavzevii | B. subtilis; S. aureus; S. epidermidis; S. marcescens and E. coli | 40–100 μg mL−1 | Moghaddam et al. (2017) |
8.2. Antifungal activity
The mechanism of antifungal activity of microbe-mediated ZnO-NPs was similar to the antibacterial mechanism. ROS production could promote the antifungal activity of ZnO-NPs as the hydroxyl radicals are produced in the water suspension of NPs, which trigger the breakdown of fungal cell membranes and inhibit cell growth (Jain et al., 2020). The release of Zn2+ ions by ZnO-NPs, when they come into close contact with the fungal cell membrane could be another mechanism of action. The positive charge of Zn2+ ions is easily attracted to the negative charge of the cell membrane, which reacts further with the sulfhydryl groups of protein inside the cell membrane and damages the synthetase activity. As a result, the fungi lose their cell division and growth, which finally leads to cause their cell death (Figure 4; Jayaseelan et al., 2012; Chauhan et al., 2015; Jain et al., 2020).
Figure 4.
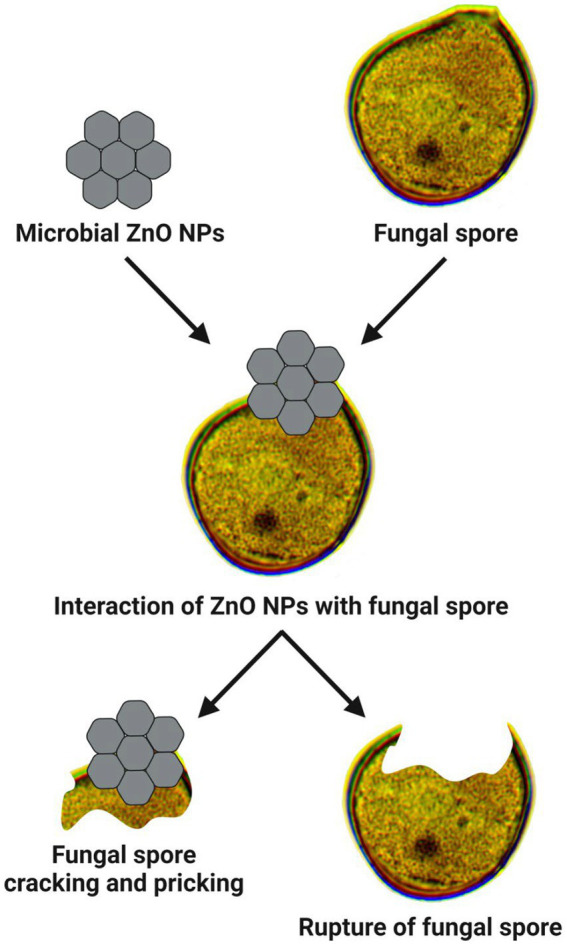
Mode of action involved in antifungal properties of microbe-mediated ZnO-NPs.
Jain et al. (2020) have demonstrated that the synthesized ZnO-NPs using Serratia nematodiphila significantly offered antifungal activity against the phytopathogenic Alternaria sp. in a dose-dependent manner. Sumanth et al. (2020) have revealed that the extracellular ZnO-NPs synthesized from Xylaria acuta extract exhibited antifungal activity against A. flavus, Cladosporium cladosporioides, Fusarium oxysporum, and Phomopsis sp., with the higher inhibition percentage of fungal mycelia in a dose-dependent manner. Mohamed et al. (2021) reported that the ZnO-NPs synthesized using Penicillium chrysogenum showed antifungal potential against phytopathogenic fungi (such as A. terreus, F. oxysporum, F. solani, and Sclerotium sclerotia) at 10 mg/mL. Recently, Mousa et al. (2021) revealed that the synthesized ZnO-NPs from the endophytic A. terreus showed promising antifungal potential against two human pathogenic fungi (such as A. brasiliensis and Candida albicans) and two plant pathogenic fungi (such as Alternaria alternata and F. oxysporum) with the recorded MIC value of 100 μg/mL. Besides, Suba et al. (2021) have reported that the ZnO NPs biosynthesized from Lactobacillus spp. extract also exhibited the antifungal effect with the maximum inhibition zone (i.e., 18–21 mm) against A. flavus and C. albicans. The antifungal activity of microbe-mediated ZnO-NPs against various fungal pathogens is listed in Table 3.
Table 3.
Microbe-mediated ZnO-NPs exhibiting potential antifungal properties.
| Microbes | Pathogen | MIC | Reference |
|---|---|---|---|
| Bacteria | |||
| A. hydrophila | A. flavus; A.niger and C. albicans | 0.9–25 μg mL−1 | Jayaseelan et al. (2012) |
| P. aeruginosa | Rhizoctonia solani; Fusarium sp., and Penicillium sp. | 100 μg well−1 | Barsainya and Singh (2018) |
| S. nematodiphila | Alternaria sp. | 250 μg mL−1 | Jain et al. (2020) |
| Lactobacillus spp. | C. albicans and A. flavus | – | Suba et al. (2021) |
| Actinomycetes | |||
| S. plicatus | A. flavus; A. niger, F. oxysporum; F. moniliform and A. alternata | 62.5–500 μg mL−1 | Kalaba et al. (2021) |
| Fungi | |||
| A. tenuissima | C. albicans; A. solani; A. niger and F. oxysporum | 50–200 μg mL−1 | Abdelhakim et al. (2020) |
| Periconium sp. | C. albicans | 40 μg mL−1 | Ganesan et al. (2020) |
| X. acuta | A. flavus; Phomopsis sp., F. oxysporum and Cladosporium cladosporioides | 100–400 μg mL−1 | Sumanth et al. (2020) |
| A. niger | A. niger; P. marneffei; C. glabrata and C. parapsilosis | 12.5–25 μg mL−1 | Mekky et al. (2021) |
| P. chrysogenum | F. solani; F. oxysporum; Sclerotium sclerotia and A. terreus | 10 mg mL−1 | Mohamed et al. (2021) |
| A. terreus | C. albicans; A. brasiliensis; A. alternata and F. oxysporum | 100 μg mL−1 | Mousa et al. (2021) |
| Yeast | |||
| P. fermentans | Fusarium sp.; Ganoderma sp. and A. terreus | – | Chauhan et al. (2015) |
8.3. Antioxidant activity
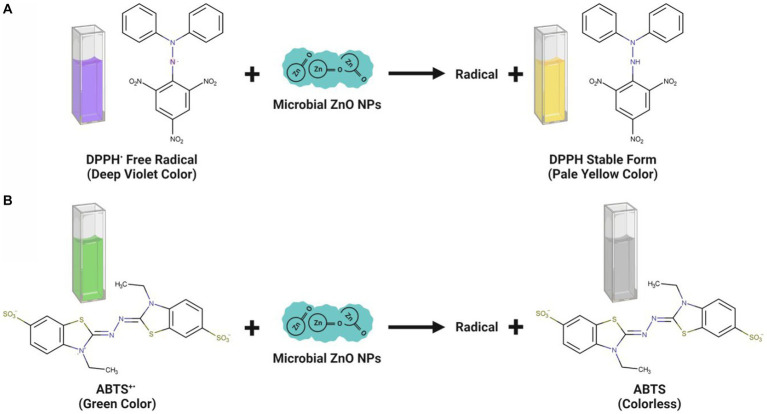
The microbe-mediated ZnO-NPs showed promising antioxidant properties, paving the way for their use as a new antioxidant source. The antioxidant potential is mainly due to the inhibition and neutralization of DPPH and ABTS free radical formation (Moghaddam et al., 2017; Abdelhakim et al., 2020). The DPPH is a stable nitrogen-centered, lipophilic free radical extensively employed to assess antioxidant activity. It can easily accept an electron or hydrogen radical from the corresponding donor to become a stable diamagnetic molecule. The odd electron located at the nitrogen atom in DPPH is reduced to hydrazine by receiving a hydrogen atom from the antioxidants. The DPPH solution color gradually changes from its characteristic deep purple/ violet color to pale yellow in the presence of ZnO-NPs. The DPPH radical scavenging activity was calculated by measuring the DPPH solution color change from deep purple to pale yellow spectrophotometrically at 517 nm, showing a strong absorption band (Figure 5A; Gao et al., 2019).
Figure 5.
Antioxidant mechanisms observed upon interaction of microbe-mediated ZnO-NPs during DPPH (A) and ABTS (B) assay.
The findings of the DPPH experiment revealed that the ZnO-NPs samples effectively inhibited free radicals. The many phytochemicals attached to the NP surface may play a significant role in the antioxidant activity of microbe-mediated ZnO-NPs samples (Ganesan et al., 2020). The DPPH radical scavenging activity also depends on the solubility of ZnO-NPs. The scavenging activity of •OH radicals was also evaluated to determine whether the synthesized ZnO-NPs could protect deoxyribose sugar from damage by scavenging •OH radicals (Singh et al., 2014). The functional groups and bioactive components in the fungal biomass may be responsible for the significant antioxidant property and free radical quenching capacity of microbe-mediated ZnO-NPs (Gao et al., 2019). The bioactive components are thought to be involved in hydrogen atom donation to prevent the free radical process.
Conversely, the ABTS assay has also proved that the microbe-mediated ZnO-NPs have a strong potential for scavenging activity of the harmful free radicals (Figure 5B). Abdelhakim et al. (2020) have suggested that ZnO-NPs synthesized using the culture filtrate of endophytic fungus (A. tenuissima) exhibited promising antioxidant activity with 50% inhibitory concentration (IC50) of 102.13 μg/mL, which was mainly due to the inhibition and neutralization of DPPH free radical formation. Ganesan et al. (2020) have reported that the endophytic fungal (Periconium sp.) extract mediated ZnO-NPs exhibited good antioxidant properties at 100 μg mL−1 concentration with 85.52% of DPPH free radical quenching. Mousa et al. (2021) have indicated that the ZnO-NPs synthesized from the endophytic A. terreus had a promising antioxidant activity with the recorded DPPH radical scavenging activity (IC50 value) of 108.67 μg mL−1. Besides, the antioxidant activity of microbe-mediated ZnO-NPs is represented in Table 4.
Table 4.
Microbe-mediated ZnO-NPs exhibiting potential antioxidant properties.
| Microbes | Half-maximal inhibitory concentration (IC50) | Maximum activity | Reference |
|---|---|---|---|
| Bacteria | |||
| P. aeruginosa | 200 μg mL−1 | 40–70% | Singh et al. (2014) |
| Actinomycetes | |||
| Streptomyces sp. | 500 μg mL−1 | 75–80% | Shanmugasundaram and Balagurunathan (2017) |
| Fungi | |||
| A. niger | 1,000 μg mL−1 | 50% | Es-haghi et al. (2019) |
| A. niger | 100 μg mL−1 | 57.74–73.58% | Gao et al. (2019) |
| A. tenuissima | 102.13 μg mL−1 | 50% | Abdelhakim et al. (2020) |
| Periconium sp. | 100 μg mL−1 | 85.52% | Ganesan et al. (2020) |
| A. bisporus | 100 μg mL−1 | 67–79% | Singh and Dutta (2020) |
| A. terreus | 108.67 μg mL−1 | 50% | Mousa et al. (2021) |
| Yeast | |||
| P. kudriavzevii | 5.26–25.46 μg mL−1 | 50% | Moghaddam et al. (2017) |
| Marine yeast | 80–100 μg mL−1 | 48.95–54.1% | Aswathy et al. (2017) |
8.4. Anticancer activity
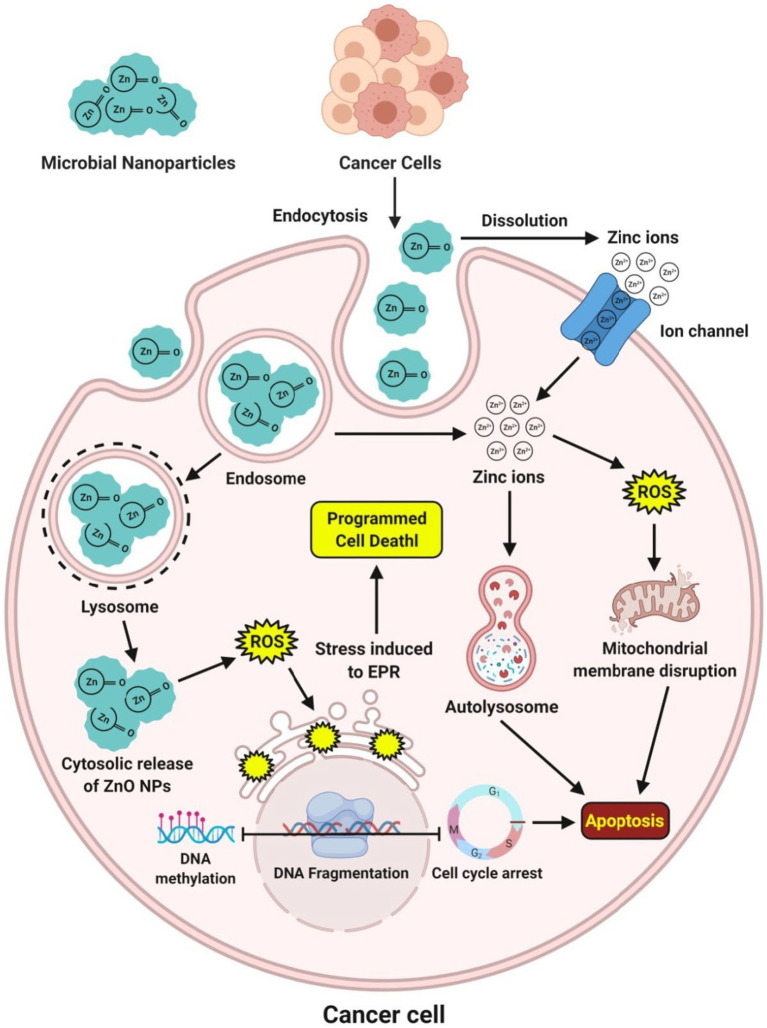
Microbe-mediated ZnO-NPs are gaining much more interest in exploring as an excellent anticancer therapeutic candidate with low toxicity than chemically synthesized ZnO-NPs (Gao et al., 2019). The ZnO-NPs positive surface charge interacts with the cancerous cells’ negative charge cell membranes, resulting in the membrane’s rupture, oxidative stress, and cell death. The interaction between the positive surface charge of ZnO-NPs and the negatively charged cancer cell membrane caused the cell membrane to rupture, allowing ROS to enter the cancer cell and causing oxidative stress and cell death. An extensive study has reported that microbe-mediated ZnO-NPs can be selectively targeted and display remarkable apoptotic features in the cytoplasm and nucleus. They regulated oxidative stress, cell cycle progression, DNA replication, DNA repair, and finally induced apoptosis in a dose-dependent manner in many cancer cell lines (Figure 6). Cell dynamic behavior is expected to be based on cell sorting and varies in response to different shapes of NPs. However, interestingly the cytotoxicity of microbe-mediated ZnO-NPs is shape-dependent and dose-dependent (Mohamed et al., 2019).
Figure 6.
Mechanism of action observed during the expression of anticancer potential of microbe-mediated ZnO-NPs.
Abdelhakim et al. (2020) have reported that the ZnO-NPs synthesized from the culture filtrate of endophytic fungus, A. tenuissima, were found to be more active against both malignant MCF-7 and HepG-2 cancer cell lines as well as the non-malignant Hbf-4 cell line with the IC50 concentrations from 16.87–55.76 μg mL−1. Sumanth et al. (2020) have suggested that the extracellular ZnO-NPs synthesized from X. acuta extract showed anticancer activity at 1 μg mL−1 concentration, which internalized and distributed without disturbing the morphology of cancer cells. Moreover, Suba et al. (2021) have reported that the MTT test using biosynthesized ZnO-NPs from Lactobacillus spp. extract confirmed the excellent biocompatibility activity and exhibited a potential toxic effect of 54.16 μg mL−1 in the human colon cancer (HT-29) cell line. Additionally, the anticancer activity of microbe-mediated ZnO-NPs is summarized in Table 5.
Table 5.
Anticancer activity of microbe-mediated ZnO-NPs.
| Microbes | Toxicity against cell lines | Activity (IC50 Value) | Reference |
|---|---|---|---|
| Bacteria | |||
| R. pyridinivorans | Colon Cancer (HT-29) | – | Kundu et al. (2014) |
| Streptomyces sp. | Lung Cancer (A549) | 15.6 μg mL−1 | Balraj et al. (2017) |
| Lactobacillus spp. | Colon Cancer (HT-29) | 54.16 μg mL−1 | Suba et al. (2021) |
| Actinomycetes | |||
| Streptomyces sp. | Human Osteosarcoma (MG63), Vero cells | 200 μg mL−1 | Shanmugasundaram and Balagurunathan (2017) |
| Fungi | |||
| A. alternata | Human Lymphocyte Cells | ≥500 μg mL−1 | Sarkar et al. (2014) |
| A. terreus | Human Colorectal Adenocarcinoma (Caco-2), Normal Vero (kidney of African green monkey), Normal Rat Liver Epithelial (Colne-9) | 26.85–79.57 μgmL−1 | Fouda et al. (2018) |
| A. niger | Human Breast Carcinoma (MCF-7) | 38–50 μg mL−1 | Es-haghi et al. (2019) |
| A. niger | Liver Cancer (HepG2), Human Embryonic Kidney (HEK-293) (non-cancerous) | 19.16–26.75 μgmL−1 | Gao et al. (2019) |
| F. keratoplasticum | Human Colorectal Adenocarcinoma (Caco-2), Normal Vero (kidney of African green monkey), Normal Rat Liver Epithelial (Colne-9) | 20.1–104.3 ppm | Mohamed et al. (2019) |
| A. niger | 57.6–131 ppm | ||
| A. tenuissima | Normal Human Melanocytes (HFB-4), Hepatocelluar Carcinoma (HepG-2), Human Breast Carcinoma (MCF-7) | 16.87–55.76 μg mL−1 | Abdelhakim et al. (2020) |
| X. acuta | Human Mammary Gland Carcinoma (MDA-MB 134) | 1 μgmL−1 | Sumanth et al. (2020) |
| A. niger | Human Breast Carcinoma (MCF-7), Skin Fibroblast Cells | – | Es-haghi et al. (2021) |
| A. niger | Wi-38 Cells | ~800.42 μg mL−1 | Mekky et al. (2021) |
| Yeast | |||
| P. kudriavzevii | Vero Cells | 190–238 μg mL−1 | Moghaddam et al. (2017) |
The biocompatibility of microbe-mediated ZnO-NPs is essential, especially when exploring their potential anticancer activity. Biocompatibility refers to the ability of the NPs to interact with biological systems without causing harm or adverse effects. In cancer therapy, it is crucial to ensure that the NPs selectively target cancer cells while sparing normal cells to minimize side effects; thus, researchers must assess their effects on both cancerous and normal cells (Sumanth et al., 2021). The ideal scenario is to find a therapeutic concentration of ZnO-NPs that effectively kills cancer cells while having minimal toxicity toward healthy cells. Achieving this selective targeting can be challenging, as many anticancer agents can also affect normal cells. It is important to note that while microbe-mediated ZnO-NPs may offer promising anticancer properties, thorough biocompatibility studies are necessary before considering their use at clinical levels (Rauf et al., 2017; Murali et al., 2021b; Faisal et al., 2022).
8.5. Drug delivery system
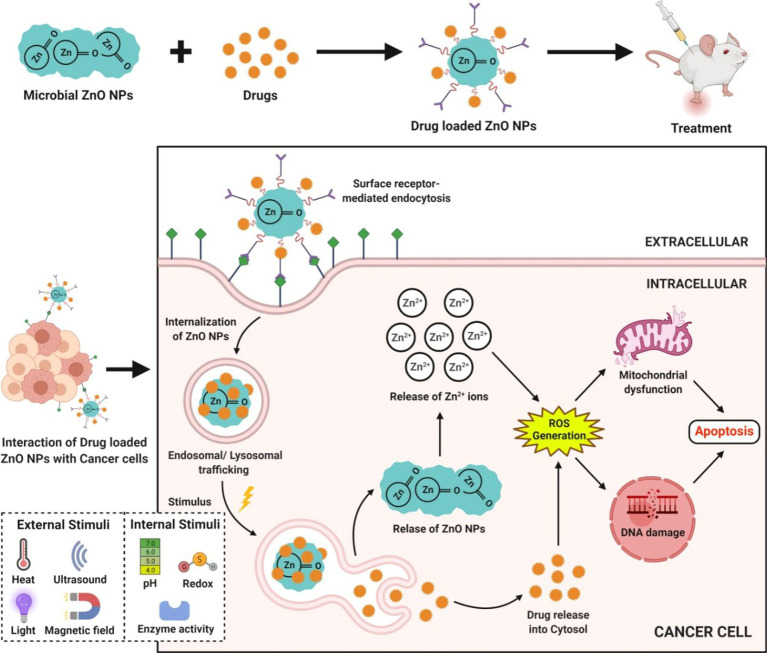
In addition to their anticancer properties, the microbe-mediated ZnO-NPs are being used as promising drug delivery vehicles because of their biocompatibility and extremely low toxicity to normal cells. The mechanism behind the cytotoxicity of microbe-mediated ZnO-NPs against cancer cells is the targeted drug delivery to the cancer cells with decreased toxicity to non-target organs. The microbe-mediated ZnO-NPs are utilized to deliver targeted drugs with the anticancer agent released once it reaches the diseased zone of the body. ZnO-NPs are also employed in targeted drug delivery to extend the drug retention time. The anthraquinone-loaded ZnO-NPs synthesized from Rhodococcus pyridinivorans exhibited a preferential ability to kill HT-29 cancer cells compared with the normal peripheral blood mononuclear cells and anthraquinone loaded ZnO-NPs could be used as anticancer drug delivery vehicles (Figure 7; Kundu et al., 2014). These findings highlight the exciting potential of microbe-mediated ZnO-NPs as a viable tool for delivering bioactive anticancer drugs over time. Thus, they can be used as safe biological and exact drug delivery vehicles for cancer treatment in the near future.
Figure 7.
Action of microbe-mediated ZnO-NPs during drug delivery.
8.6. Wound healing
Additionally, the biologically synthesized ZnO-NPs using A. niger improve the carbapenem-resistant K. pneumonia infected wound healing property in experimental rats (Rasha et al., 2021). This recent advance shows that this microbial nanotherapeutics is promising prospects for treating burn wounds.
8.7. Photocatalytic activity
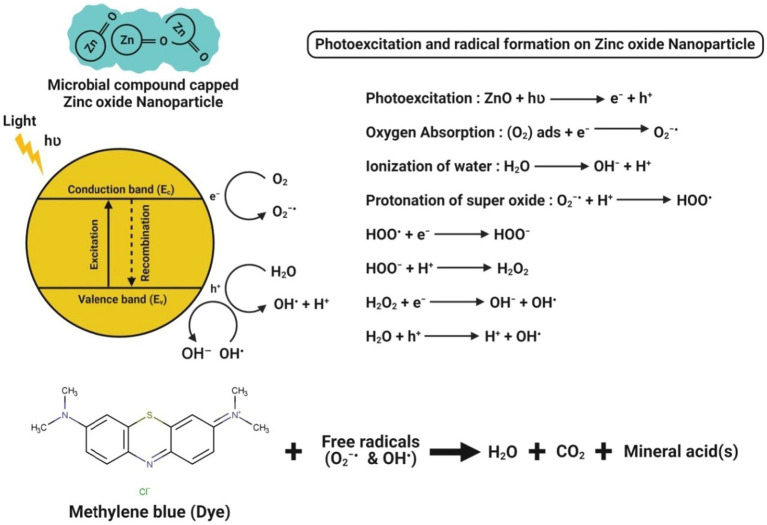
The microbe-mediated ZnO-NPs could also be applied as an excellent photocatalyst in degrading many organic dyes under UV irradiation (Tripathi et al., 2014). The proposed mechanism of photocatalytic activity of microbe-mediated ZnO-NPs is illustrated in Figure 8. When light with energy greater than or equal to the bandgap illuminates ZnO-NPs, an electron (e−) can be excited from the valence band (VB) to the conduction band (CB). At the same time, a hole (h+) is simultaneously generated in the VB, thereby forming electron–hole (e−h+) pairs. The hole oxidizes the water molecules adsorbed on the surface of NPs into hydroxyl radicals (•OH) and hydrogen ions (H+), while the photoexcited electron reacts with dissolved oxygen (O2) to form superoxide radicals (O2•–). The reaction between H+ and O2•– further produces hydroperoxyl radicals (•OOH), which can produce hydrogen peroxide (H2O2). These radicals have extremely high oxidative properties that easily degrade the toxic organic compounds into carbon dioxide and water molecules (Fouda et al., 2020).
Figure 8.
Chemical interactions observed during the photocatalytic activity of microbe-mediated ZnO-NPs.
Li et al. (2019) have reported that Cordyceps militaris fungus-mediated ZnO-NPs had the degradation ability of hazardous dye (like methylene blue) about 97% within 180 min irradiation of UV light. Abdelhakim et al. (2020) have suggested that ZnO-NPs synthesized from the culture filtrate of endophytic A. tenuissima further efficiently showed their photocatalytic behavior by the complete (100%) degradation of methylene blue dye in a concentration-dependent manner after 20 min exposure to sunlight. Jain et al. (2020) demonstrated that the ZnO-NPs synthesized from S. nematodiphila showed ~90% of methyl orange degradation after 80 min of UV light irradiation, which was evident by the visual color observation and absorbance peak intensity at 463 nm. Recently, Siddique et al. (2021) efficiently bioprospecting Pseudochrobactrum sp. for the green synthesis of ZnO-NPs and reported that the biosynthesized ZnO-NPs significantly showed a better photocatalytic degradation potential of various dyes (viz., methylene blue, 4-nitriophenol, brilliant blue R, brilliant yellow, reactive black 5, and reactive red 120) after 10 h exposure to sunlight as compared to the chemically synthesized ZnO-NPs. This higher photocatalytic degradation might be due to the relatively smaller size, more stability and higher surface area of the biosynthesized NPs. Moreover, the photocatalytic activity of microbe-mediated ZnO-NPs is represented in Table 6.
Table 6.
Photocatalytic activity of microbe-mediated ZnO-NPs.
| Microbes | Dye degraded | Irradiation type and Intensity range | Irradiation time | pH range | Degradation efficiency (%) | Reference |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| R. pyridinivorans | Malachite green | UV irradiation at 300–800 nm | 2 h | – | 95% | Kundu et al. (2014) |
| B. licheniformis | Methylene blue | UV irradiation at 365 nm | 60 min | – | 83% | Tripathi et al. (2014) |
| S. nematodiphila | Methyl orange | UV light | 80 min | 8.06 | ~90% | Jain et al. (2020) |
| Fungi | ||||||
| A. niger | Bismarck brown | UV light at 365 nm | – | – | 90% | Kalpana et al. (2018) |
| C. militaris | Methylene blue | UV light | 180 min | – | 97% | Li et al. (2019) |
| A. tenuissima | Methylene blue | Sunlight | 20 min | – | 100% | Abdelhakim et al. (2020) |
| P. corylophilum | Methylene blue | Visible light | 85 min | – | 98% | Fouda et al. (2020) |
| A. potronii | Methylene blue | UV–vis light | 30 min | Nearly 6.5 | 93% | Ameen et al. (2021) |
| Pseudochrobactrum sp. | Methylene blue, 4-nitriophenol, brilliant blue R, brilliant yellow, reactive black 5, reactive red 120 | Sunlight | 280 min – 10 h | – | 44.9–95.2% | Siddique et al. (2021) |
9. Toxicological effects
The toxicological effects of microbe-mediated ZnO-NPs are an important area of research and concern. When the NPs are synthesized using microbes such as bacteria, actinomycetes, fungi, or yeast, their potential toxicity can differ from conventionally produced NPs due to size, shape, surface charge, and chemical composition variations. It is important to note that the toxicological effects of NPs can be highly dependent on factors such as specific microbial synthesis methods, distinctive characteristics of NPs, dose, exposure route, and the vulnerability of target organisms. Research on the toxicity of microbial-mediated ZnO-NPs is ongoing, and it is crucial to conduct comprehensive studies to understand and assess their potential risks fully. It was reported that the larger NPs also tend to accumulate in the kidneys for a longer period due to the slower glomerular filtration excretion mechanisms, and this prolonged retention might result in organ damage (Rauf et al., 2017; Sumanth et al., 2021; Faisal et al., 2022). Furthermore, the different morphologies (such as nanorods, nanoflowers, nanosheets and nanoplates) of NPs also contribute to their toxicity due to their larger surface area. The fungal-mediated ZnO-NPs were significantly found to show in vitro cytotoxicity and genotoxicity in human lymphocyte cells, wherein they demonstrated a concentration-dependent decrease in mitochondrial activity at ≥0.5 mg mL−1 and induced DNA fragmentation at 1 mg mL−1 (Sarkar et al., 2014). Many NPs toxicity studies have focused on the plants due to their critical functions in ecosystems as primary producers of organic compounds from atmospheric or aqueous carbon dioxide. Güllüce et al. (2020) have reported that the ZnO-NPs synthesized by Rhodococcus erythropolis using the precursors (such as zinc sulfate heptahydrate, zinc nitrate hexahydrate, zinc chloride and zinc acetate) were found to show their toxicological potentials on Triticum aestivum, where they significantly affected the seed germination and seedling growth along with their genotoxic potentials. If released into the environment, the microbe-mediated ZnO-NPs may interact with living organisms and ecosystems, potentially causing ecological toxicity and disrupting natural processes. Therefore, it is further necessary to investigate any potential long-term toxicological effects of microbe-mediated ZnO-NPs on plants, animals and human health due to these multiple uses in numerous industries. To ensure more effective and safe use of ZnO-NPs, it is also essential to adopt responsible manufacturing practices, perform rigorous toxicity assessments, and follow safety assessments and regulatory guidelines. In addition, Table 7 provides the advantages and disadvantages of microbe-mediated synthesis of ZnO-NPs.
Table 7.
Advantages and disadvantages of microbial-mediated ZnO-NPs over plant-mediated synthesis.
| Advantages | Disadvantages |
|---|---|
|
|
10. Future prospects
The major drawback of microbe-mediated synthesis of ZnO-NPs is that the microbes may lose their capacity to synthesize NPs because of mutations over time. In addition, not all microbes can produce ZnO-NPs; thus, those potential microbes need to be explored through rigorous screening programs. It is still unknown how exactly microbes synthesize NPs through extracellular and intracellular processes. There is a lack of knowledge regarding the reducing and capping agents involved in bioreduction and stabilization, respectively, during NPs synthesis. Uncertainty exists regarding the identity of reducing and capping agents and their function in influencing both the size and shape of NPs. Particularly because of the requirement of a complete aseptic environment and specific maintenance, the microbe-mediated NPs are not suitable for large-scale production. In the future, extensive optimisation studies are required before scaling up the industrial production of NPs to understand the impact of every aspect completely. Further, cooperation between major stakeholders in basic sciences, chemical engineering, and industry is required to exploit the microbe-mediated ZnO-NPs economically. Future research studies must address the concentration and duration of exposure required for causing the potential toxicologic effects of ZnO-NPs to understand the therapeutic effects better and prevent unintended cytotoxic effects.
11. Conclusion
The present review highlights the need for green synthesis approaches in producing ZnO-NPs due to the environmental pollution caused by conventional chemical synthesis methods. Using biological sources like plants and microbes makes the process more environmentally friendly and safer. The advantages of microbial synthesis include using bioactive metabolites released by microbes as reducing and stabilizing agents, which enhance the biological properties of the ZnO-NPs. The extracellular biosynthesis method is simpler since the enzymes and proteins outside the cells directly reduce or chelate Zn2+ ions. In contrast, the intracellular method requires an additional cell lysis step to release the NPs inside the microbe, making it more expensive and time-consuming. Among the microbial synthesis methods, fungal-mediated synthesis shows promise due to its ability to produce more bioactive compounds, while bacteria have an advantage in cell growth activity.
The review also suggests that microbial synthesis has excellent biotechnological potential and could pave the way for large-scale industrial production of ZnO-NPs. Since the biological components are natural, biodegradable, and safer, this approach is expected to be a promising alternative for plant-mediated ZnO-NPs for future industrial and commercial production. However, further research is still needed to understand the biochemical and molecular mechanisms of NP synthesis among different microbes. Investigating these mechanisms could unlock even more potential applications of microbe-mediated ZnO-NPs in various fields, including medicine, industry, environment, and agriculture. Overall, this review emphasizes the importance of green synthesis methods and highlights the promising role of microbe-mediated synthesis in producing ZnO-NPs with exciting applications in diverse areas. It encourages further research in this area to exploit the full potential of these eco-friendly nanomaterials and extend the next level of their applications.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the University of Mysore for providing facilities to conduct the research work. CS and RA are thankful to the leadership of JSS AHER, Mysuru, for infrastructure and high-end computational facilities. SK thanks the Director, Amrita Vishwa Vidyapeetham, Mysuru Campus, for infrastructure support. The authors thank the Deanship of Scientific Research at King Khalid University for financial support under the project grant number R.G.P. 2/213/44. Part of the work was done in accordance with the memorandum of understanding between the JSS AHER and I.M. Sechenov First Moscow State Medical University of the Ministry of Health of the Russian Federation (Moscow, Russia).
References
- Abdelhakim H. K., El-Sayed E. R., Rashidi F. B. (2020). Biosynthesis of zinc oxide nanoparticles with antimicrobial, anticancer, antioxidant and photocatalytic activities by the endophytic Alternaria tenuissima. J. Appl. Microbiol. 128, 1634–1646. doi: 10.1111/jam.14581, PMID: [DOI] [PubMed] [Google Scholar]
- Ahmed S., Annu, Chaudhry S. A., Ikram S. (2017). A review on biogenic synthesis of ZnO nanoparticles using plant extracts and microbes: a prospect towards green chemistry. J. Photochem. 166, 272–284. doi: 10.1016/j.jphotobiol.2016.12.011, PMID: [DOI] [PubMed] [Google Scholar]
- Ahmed T., Wu Z., Jiang H., Luo J., Noman M., Shahid M., et al. (2021). Bioinspired green synthesis of zinc oxide nanoparticles from a native Bacillus cereus strain RNT6: characterization and antibacterial activity against Rice panicle blight pathogens Burkholderia glumae and B. gladioli. Nano 11:884. doi: 10.3390/nano11040884, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar N., Ilyas N., Meraj T. A., Pour-Aboughadareh A., Sayyed R. Z., Mashwani Z., et al. (2022). Improvement of plant responses by Nanobiofertilizer: a step towards sustainable agriculture. Nano 12:965. doi: 10.3390/nano12060965, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameen F., Dawoud T., AlNadhari S. (2021). Ecofriendly and low-cost synthesis of ZnO nanoparticles from Acremonium potronii for the photocatalytic degradation of azo dyes. Environ. Res. 202:111700. doi: 10.1016/j.envres.2021.111700 [DOI] [PubMed] [Google Scholar]
- Anjum S., Hashim M., Malik S. A., Khan M., Lorenzo J. M., Abbasi B. H., et al. (2021). Recent advances in zinc oxide nanoparticles (ZnO NPs) for cancer diagnosis, target drug delivery, and treatment. Cancers 13:4570. doi: 10.3390/cancers13184570, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswathy R., Gabylis B., Anwesha S., Bhaskara Rao K. (2017). Green synthesis and characterization of marine yeast-mediated silver and zinc oxide nanoparticles and assessment of their antioxidant activity. Asian J. Pharm. Clin. Res. 10, 235–240. doi: 10.22159/ajpcr.2017.v10i10.19979 [DOI] [Google Scholar]
- Balraj B., Senthilkumar N., Siva C., Krithikadevi R., Julie A., Potheher I. V., et al. (2017). Synthesis and characterization of zinc oxide nanoparticles using marine Streptomyces sp. with its investigations on anticancer and antibacterial activity. Res. Chem. Intermed. 43, 2367–2376. doi: 10.1007/s11164-016-2766-6 [DOI] [Google Scholar]
- Barsainya M., Singh D. P. (2018). Green synthesis of zinc oxide nanoparticles by Pseudomonas aeruginosa and their broad-spectrum antimicrobial effects. J. Pure Appl. Microbiol. 12, 2123–2134. doi: 10.22207/JPAM.12.4.50 [DOI] [Google Scholar]
- Baskar G., Chandhuru J., Sheraz Fahad K., Praveen A. S. (2013). Mycological synthesis, characterization and antifungal activity of zinc oxide nanoparticles. Asian J. Pharm. Tech. 3, 142–146. doi: 10.3390/cryst13020171 [DOI] [Google Scholar]
- Basnet P., Chanu T. I., Samanta D., Chatterjee S. (2018). A review on bio-synthesized zinc oxide nanoparticles using plant extracts as reductants and stabilizing agents. J. Photochem. Photobiol. B Biol. 183, 201–221. doi: 10.1016/j.jphotobiol.2018.04.036, PMID: [DOI] [PubMed] [Google Scholar]
- Busi S., Rajkumari J., Pattnaik S., Parasuraman P., Hnamte S. (2016). Extracellular synthesis of zinc oxide nanoparticles using Acinetobacter schindleri SIZ7 and its antimicrobial property against foodborne pathogens. J. Microbiol. Biotech. Food Sci. 5, 407–411. doi: 10.15414/jmbfs.2016.5.5.407-411 [DOI] [Google Scholar]
- Chauhan R., Reddy A., Abraham J. (2015). Biosynthesis of silver and zinc oxide nanoparticles using Pichia fermentans JA2 and their antimicrobial property. Appl. Nanosci. 5, 63–71. doi: 10.1007/s13204-014-0292-7 [DOI] [Google Scholar]
- Dhandapani P., Siddarth A. S., Kamalasekaran S., Maruthamuthu S., Rajagopal G. (2014). Bio-approach: Ureolytic bacteria mediated synthesis of ZnO nanocrystals on cotton fabric and evaluation of their antibacterial properties. Carbohydr. Polym. 103, 448–455. doi: 10.1016/j.carbpol.2013.12.074, PMID: [DOI] [PubMed] [Google Scholar]
- Es-haghi A., Soltani M., Karimi E., Namvar F., Homayouni-Tabrizi M. (2019). Evaluation of antioxidant and anticancer properties of zinc oxide nanoparticles synthesized using aspergillus Niger extract. Mater. Res. Express 6:125415. doi: 10.1088/2053-1591/ab5f72 [DOI] [Google Scholar]
- Es-haghi A., Taghavizadeh Yazdi M. E., Sharifalhoseini M., Baghani M., Yousefi E., Rahdar A., et al. (2021). Application of response surface methodology for optimizing the therapeutic activity of ZnO nanoparticles biosynthesized from aspergillus Niger. Biomimetics 6:34. doi: 10.3390/biomimetics6020034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal S., Rizwan M., Ullah R., Alotaibi A., Khattak A., Bibi N., et al. (2022). Paraclostridium benzoelyticum bacterium-mediated zinc oxide nanoparticles and their in vivo multiple biological applications. Oxid. Med. Cell. Longev. 2022:5994033. doi: 10.1155/2022/5994033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridvand S., Amirnia R., Tajbakhsh M., El Enshasy H. A., Sayyed R. Z. (2021). The effect of foliar application of magnetic water and nano-fertilizers on phytochemical and yield characteristics of fennel. Horticulturae 7:475. doi: 10.3390/horticulturae7110475 [DOI] [Google Scholar]
- Fouda A., Hassan S. E. D., Salem S. S., Shaheen T. I. (2018). In-vitro cytotoxicity, antibacterial, and UV protection properties of the biosynthesized zinc oxide nanoparticles for medical textile applications. Microb. Pathog. 125, 252–261. doi: 10.1016/j.micpath.2018.09.030, PMID: [DOI] [PubMed] [Google Scholar]
- Fouda A., Salem S. S., Wassel A. R., Hamza M. F., Shaheen T. I. (2020). Optimization of green biosynthesized visible light active CuO/ZnO nano-photocatalysts for the degradation of organic methylene blue dye. Heliyon 6:e04896. doi: 10.1016/j.heliyon.2020.e04896, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan V., Hariram M., Vivekanandhan S., Muthuramkumar S. (2020). Periconium sp. (endophytic fungi) extract mediated sol-gel synthesis of ZnO nanoparticles for antimicrobial and antioxidant applications. Mater. Sci. Semicond. Process. 105:104739. doi: 10.1016/j.mssp.2019.104739 [DOI] [Google Scholar]
- Gao Y., Anand M. A. V., Ramachandran V., Karthikkumar V., Shalini V., Vijayalakshmi S., et al. (2019). Biofabrication of zinc oxide nanoparticles from aspergillus Niger, their antioxidant, antimicrobial and anticancer activity. J. Clust. Sci. 30, 937–946. doi: 10.1007/s10876-019-01551-6 [DOI] [Google Scholar]
- Güllüce M., Karadayı M., Demir A. Y., Işık C., Alaylar B., İspirli N. H. (2020). Genotoxic potentials of biosynthesized zinc oxide nanoparticles. Pol. J. Environ. Stud. 29, 111–119. doi: 10.15244/pjoes/99239 [DOI] [Google Scholar]
- Ibrahem E. J., Thalij K. M., Saleh M. K., Badawy A. S. (2017). Biosynthesis of zinc oxide nanoparticles and assay of antibacterial activity. Am. J. Biochem. Biotechnol. 13, 63–69. doi: 10.3844/ajbbsp.2017.63.69 [DOI] [Google Scholar]
- Iqtedar M., Riaz H., Kaleem A., Abdullah R., Aihetasham A., Naz S., et al. (2020). Biosynthesis, optimization and characterization of ZnO nanoparticles using Bacillus cereus MN181367 and their antimicrobial activity against multidrug resistant bacteria. Rev. Mex. Ing. Quim. 19, 253–266. doi: 10.24275/rmiq/Bio1605 [DOI] [Google Scholar]
- Jain N., Bhargava A., Tarafdar J. C., Singh S. K., Panwar J. (2013). A biomimetic approach towards synthesis of zinc oxide nanoparticles. Appl. Microbiol. Biotechnol. 97, 859–869. doi: 10.1007/s00253-012-3934-2 [DOI] [PubMed] [Google Scholar]
- Jain D., Shivani, Bhojiya A. A., Singh H., Daima H. K., Singh M., et al. (2020). Microbial fabrication of zinc oxide nanoparticles and evaluation of their antimicrobial and photocatalytic properties. Front. Chem. 8:778. doi: 10.3389/fchem.2020.00778, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan J., Mani G., Krishnan N., Pernabas J., Devadoss J. M., Jang H. T. (2019). Green biogenic synthesis of zinc oxide nanoparticles using Pseudomonas putida culture and its in vitro antibacterial and anti-biofilm activity. Biocatal. Agric. Biotechnol. 21:101327. doi: 10.1016/j.bcab.2019.101327 [DOI] [Google Scholar]
- Jayaseelan C., Rahuman A. A., Kirthi A. V., Marimuthu S., Santhoshkumar T., Bagavan A., et al. (2012). Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta A Mol. Biomol. Spectrosc. 90, 78–84. doi: 10.1016/j.saa.2012.01.006, PMID: [DOI] [PubMed] [Google Scholar]
- Kadam V. V., Ettiyappan J. P., Balakrishnan R. M. (2019). Mechanistic insight into the endophytic fungus mediated synthesis of protein capped ZnO nanoparticles. Mater. Sci. Eng. B 243, 214–221. doi: 10.1016/j.mseb.2019.04.017 [DOI] [Google Scholar]
- Kalaba M. H., Moghannem S. A., El-Hawary A. S., Radwan A. A., Sharaf M. H., Shaban A. S. (2021). Green synthesized ZnO nanoparticles mediated by Streptomyces plicatus: characterizations, antimicrobial and nematicidal activities and cytogenetic effects. Plan. Theory 10:1760. doi: 10.3390/plants10091760, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana V. N., Kataru B. A. S., Sravani N., Vigneshwari T., Panneerselvam A., Rajeswari V. D. (2018). Biosynthesis of zinc oxide nanoparticles using culture filtrates of aspergillus Niger: antimicrobial textiles and dye degradation studies. Open Nano. 3, 48–55. doi: 10.1016/j.onano.2018.06.001 [DOI] [Google Scholar]
- Król A., Railean-Plugaru V., Pomastowski P., Złoch M., Buszewski B. (2018). Mechanism study of intracellular zinc oxide nanocomposites formation. Colloids Surf. A Physicochem. Eng. Asp. 553, 349–358. doi: 10.1016/j.colsurfa.2018.05.069 [DOI] [Google Scholar]
- Kundu D., Hazra C., Chatterjee A., Chaudhari A., Mishra S. (2014). Extracellular biosynthesis of zinc oxide nanoparticles using Rhodococcus pyridinivorans NT2: multifunctional textile finishing, biosafety evaluation and in vitro drug delivery in colon carcinoma. J. Photochem. Photobiol. B 140, 194–204. doi: 10.1016/j.jphotobiol.2014.08.001, PMID: [DOI] [PubMed] [Google Scholar]
- Lahiri D., Nag M., Sheikh H. I., Sarkar T., Edinur H. A., Pati S., et al. (2021). Microbiologically-synthesized nanoparticles and their role in silencing the biofilm signaling cascade. Front. Microbiol. 12:636588. doi: 10.3389/fmicb.2021.636588, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. F., Rupa E. J., Hurh J., Huo Y., Chen L., Han Y., et al. (2019). Cordyceps militaris fungus mediated zinc oxide nanoparticles for the photocatalytic degradation of methylene blue dye. Optik 183, 691–697. doi: 10.1016/j.ijleo.2019.02.081 [DOI] [Google Scholar]
- Mekky A. E., Farrag A. A., Hmed A. A., Sofy A. R. (2021). Preparation of zinc oxide nanoparticles using aspergillus Niger as antimicrobial and anticancer agents. J. Pure Appl. Microbiol. 15, 1547–1566. doi: 10.22207/JPAM.15.3.49 [DOI] [Google Scholar]
- Mishra M., Paliwal J. S., Singh S. K., Selvarajan E., Subathradevi C., Mohanasrinivasan V. (2013). Studies on the inhibitory activity of biologically synthesized and characterized zinc oxide nanoparticles using Lactobacillus sporogens against Staphylococcus aureus. J. Pure Appl. Microbiol. 7, 1263–1268. [Google Scholar]
- Moghaddam A. B., Moniri M., Azizi S., Rahim R. A., Ariff A. B., Saad W. Z., et al. (2017). Biosynthesis of ZnO nanoparticles by a new Pichia kudriavzevii yeast strain and evaluation of their antimicrobial and antioxidant activities. Molecules 22:872. doi: 10.3390/molecules22060872, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A. A., Abu-Elghait M., Ahmed N. E., Salem S. S. (2021). Eco-friendly mycogenic synthesis of ZnO and CuO nanoparticles for in vitro antibacterial, antibiofilm, and antifungal applications. Biol. Trace Elem. Res. 199, 2788–2799. doi: 10.1007/s12011-020-02369-4, PMID: [DOI] [PubMed] [Google Scholar]
- Mohamed A. A., Fouda A., Abdel-Rahman M. A., Hassan S. E. D., El-Gamal M. S., Salem S. S., et al. (2019). Fungal strain impacts the shape, bioactivity and multifunctional properties of green synthesized zinc oxide nanoparticles. Biocatal. Agric. Biotechnol. 19:101103. doi: 10.1016/j.bcab.2019.101103 [DOI] [Google Scholar]
- Mousa S. A., El-Sayed E. S. R., Mohamed S. S., El-Seoud M. A. A., Elmehlawy A. A., Abdou D. A. M. (2021). Novel mycosynthesis of Co3O4, CuO, Fe3O4, NiO, and ZnO nanoparticles by the endophytic aspergillus terreus and evaluation of their antioxidant and antimicrobial activities. Appl. Microbiol. Biotechnol. 105, 741–753. doi: 10.1007/s00253-020-11046-4, PMID: [DOI] [PubMed] [Google Scholar]
- Murali M., Kalegowda N., Gowtham H. G., Ansari M. A., Alomary M. N., Alghamdi S., et al. (2021a). Plant-mediated zinc oxide nanoparticles: advances in the new millennium towards understanding their therapeutic role in biomedical applications. Pharmaceutics 13:1662. doi: 10.3390/pharmaceutics13101662, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali M., Naziya B., Brijesh Singh S., Chandrashekar S., Udayshankar A. C., Amruthesh K. N. (2021b). “Management of Plant Fungal disease by microbial nanotechnology” in Microbial Nanotechnology: Green Synthesis and Applications. eds. Ansari M. A., Rehman S. (Singapore: Springer Nature; ). [Google Scholar]
- Nayeri F., Mafakheri S., Mirhosseini M., Sayyed R. Z. (2021). Phyto-mediated silver nanoparticles via Melissa officinalis aqueous and methanolic extracts: synthesis, characterization and biological properties against infectious bacterial strains. Int. J. Adv. Biol. Biomed. Res. 9, 270–285. doi: 10.22034/ijabbr.2021.525079.1349 [DOI] [Google Scholar]
- Perez R. H., Zendo T., Sonomoto K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Factories 13:S3. doi: 10.1186/1475-2859-13-S1-S3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabairavi N., Raju C. S., Karthikeyan C., Varutharaju K., Nethaji S., Hameed A. S. H., et al. (2017). “Biosynthesis of novel zinc oxide nanoparticles (ZnO NPs) using endophytic bacteria Sphingobacterium thalpophilum” in Recent Trends in Materials Science and Applications. ed. Ebenezar J. (Cham: Springer; ) [Google Scholar]
- Rajan A., Cherian E., Baskar G. (2016). Biosynthesis of zinc oxide nanoparticles using Aspergillus fumigatus JCF and its antibacterial activity. Int. J. Mod. Sci. Technol. 1, 52–57. [Google Scholar]
- Rajivgandhi G., Gnanamangai B. M., Prabha T. H., Poornima S., Maruthupandy M., Alharbi N. S., et al. (2022). Biosynthesized zinc oxide nanoparticles (ZnO NPs) using actinomycetes enhance the antibacterial efficacy against K. pneumoniae. J. King Saud Univ. Sci. 34:101731. doi: 10.1016/j.jksus.2021.101731 [DOI] [Google Scholar]
- Raliya R., Tarafdar J. C. (2013). ZnO nanoparticle biosynthesis and its effect on phosphorous-mobilizing enzyme secretion and gum contents in Clusterbean (Cyamopsis tetragonoloba L.). Agric. Res. 2, 48–57. doi: 10.1007/s40003-012-0049-z [DOI] [Google Scholar]
- Rasha E., Alkhulaifi M. M., AlOthman M., Khalid I., Doaa E., Alaa K., et al. (2021). Effects of zinc oxide nanoparticles synthesized using aspergillus Niger on carbapenem-resistant Klebsiella pneumonia in vitro and in vivo. Front. Cell. Infect. Microbiol. 11:748739. doi: 10.3389/fcimb.2021.748739, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf M. A., Owais M., Rajpoot R., Ahmad F., Khan N., Zubair S. (2017). Biomimetically synthesized ZnO nanoparticles attain potent antibacterial activity against less susceptible S. aureus skin infection in experimental animals. RSC Adv. 7, 36361–36373. doi: 10.1039/C7RA05040B [DOI] [Google Scholar]
- Rehman S., Jermy B. R., Akhtar S., Borgio J. F., Azeez S. A., Ravinayagam V., et al. (2019). Isolation and characterization of a novel thermophile; Bacillus haynesii, applied for the green synthesis of ZnO nanoparticles. Artif. Cells Nanomed. Biotechnol. 47, 2072–2082. doi: 10.1080/21691401.2019.1620254 [DOI] [PubMed] [Google Scholar]
- Saravanan M., Gopinath V., Chaurasia M. K., Syed A., Ameen F., Purushothaman N. (2018). Green synthesis of anisotropic zinc oxide nanoparticles with antibacterial and cytofriendly properties. Microb. Pathog. 115, 57–63. doi: 10.1016/j.micpath.2017.12.039, PMID: [DOI] [PubMed] [Google Scholar]
- Sarkar J., Ghosh M., Mukherjee A., Chattopadhyay D., Acharya K. (2014). Biosynthesis and safety evaluation of ZnO nanoparticles. Bioprocess Biosyst. Eng. 37, 165–171. doi: 10.1007/s00449-013-0982-7 [DOI] [PubMed] [Google Scholar]
- Selvarajan E., Mohanasrinivasan V. (2013). Biosynthesis and characterization of ZnO nanoparticles using Lactobacillus plantarum VITES07. Mater. Lett. 112, 180–182. doi: 10.1016/j.matlet.2013.09.020 [DOI] [Google Scholar]
- Shaaban M., El-Mahdy A. M. (2018). Biosynthesis of ag, se, and ZnO nanoparticles with antimicrobial activities against resistant pathogens using waste isolate Streptomyces enissocaesilis. IET Nanobiotechnol. 12, 741–747. doi: 10.1049/iet-nbt.2017.0213, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsuzzaman, Mashrai A., Khanam H., Aljawfi R. N. (2017). Biological synthesis of ZnO nanoparticles using C. albicans and studying their catalytic performance in the synthesis of steroidal pyrazolines. Arab. J. Chem. 10:S1530-S 1536. doi: 10.1016/j.arabjc.2013.05.004 [DOI] [Google Scholar]
- Shanmugasundaram T., Balagurunathan R. (2017). Bio-medically active zinc oxide nanoparticles synthesized by using extremophilic actinobacterium, Streptomyces sp. (MA30) and its characterization. Artif. Cells Nanomed. Biotechnol. 45, 1521–1529. doi: 10.1080/21691401.2016.1260577 [DOI] [PubMed] [Google Scholar]
- Shobha B., Lakshmeesha T. R., Ansari M. A., Almatroudi A., Alzohairy M. A., Basavaraju S., et al. (2020). Mycosynthesis of ZnO nanoparticles using Trichoderma spp. isolated from rhizosphere soils and its synergistic antibacterial effect against Xanthomonas oryzae pv. Oryzae. J. Fungi. 6:181. doi: 10.3390/jof6030181, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique K., Shahid M., Shahzad T., Mahmood F., Nadeem H., Rehman M. S. U., et al. (2021). Comparative efficacy of biogenic zinc oxide nanoparticles synthesized by Pseudochrobactrum sp. C5 and chemically synthesized zinc oxide nanoparticles for catalytic degradation of dyes and wastewater treatment. Environ. Sci. Pollut. Res. 28, 28307–28318. doi: 10.1007/s11356-021-12575-9, PMID: [DOI] [PubMed] [Google Scholar]
- Sidhu P. K., Nehra K. (2019). Bacteriocin-nanoconjugates as emerging compounds for enhancing antimicrobial activity of bacteriocins. J. King Saud Univ. Sci. 31, 758–767. doi: 10.1016/j.jksus.2017.12.007 [DOI] [Google Scholar]
- Singh A., Dutta P. K. (2020). Green synthesis, characterization and biological evaluation of chitin glucan based zinc oxide nanoparticles and its curcumin conjugation. Int. J. Biol. Macromol. 156, 514–521. doi: 10.1016/j.ijbiomac.2020.04.081, PMID: [DOI] [PubMed] [Google Scholar]
- Singh B. N., Rawat A. K. S., Khan W., Naqvi A. H., Singh B. R. (2014). Biosynthesis of stable antioxidant ZnO nanoparticles by Pseudomonas aeruginosa rhamnolipids. PLoS One 9:e106937. doi: 10.1371/journal.pone.0106937, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suba S., Vijayakumar S., Vidhya E., Punitha V. N., Nilavukkarasi M. (2021). Microbial mediated synthesis of ZnO nanoparticles derived from Lactobacillus spp: characterizations, antimicrobial and biocompatibility efficiencies. Sens. Int. 2:100104. doi: 10.1016/j.sintl.2021.100104 [DOI] [Google Scholar]
- Sumanth B., Balagangadharaswamy S., Chowdappa S., Ansari M. A., Salim S. H., Murali M., et al. (2021). “Fungal biogenesis of NPs and their limitations” in Microbial Nanotechnology: Green Synthesis and Applications. eds. Ansari M. A., Rehman S. (Singapore: Springer Nature; ). [Google Scholar]
- Sumanth B., Lakshmeesha T. R., Ansari M. A., Alzohairy M. A., Udayashankar A. C., Shobha B., et al. (2020). Mycogenic synthesis of extracellular zinc oxide nanoparticles from Xylaria acuta and its nanoantibiotic potential. Int. J. Nanomedicine 15, 8519–8536. doi: 10.2147/IJN.S271743, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taran M., Rad M., Alavi M. (2018). Biosynthesis of TiO2 and ZnO nanoparticles by Halomonas elongata IBRC-M 10214 in different conditions of medium. Bioimpacts 8, 81–89. doi: 10.15171/bi.2018.10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur P., Thakur S., Kumari P., Shandilya M., Sharma S., Poczai P., et al. (2022). Nano-insecticide: synthesis, characterization, and evaluation of insecticidal activity of ZnO NPs against Spodoptera litura and Macrosiphum euphorbiae. Appl. Nanosci. 12, 3835–3850. doi: 10.1007/s13204-022-02530-6 [DOI] [Google Scholar]
- Tripathi R. M., Bhadwal A. S., Gupta R. K., Singh P., Shrivastav A., Shrivastav B. R. (2014). ZnO nanoflowers: novel biogenic synthesis and enhanced photocatalytic activity. J. Photochem. Photobiol. B, Biol. 141, 288–295. doi: 10.1016/j.jphotobiol.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Weldegebrieal G. K. (2020). Synthesis method, antibacterial and photocatalytic activity of ZnO nanoparticles for azo dyes in wastewater treatment: a review. Inorg. Chem. Commun. 120:108140. doi: 10.1016/j.inoche.2020.108140 [DOI] [Google Scholar]
- Yusof H. M., Abdul Rahman N., Mohamad R., Zaidan U. H., Samsudin A. A. (2020b). Biosynthesis of zinc oxide nanoparticles by cell-biomass and supernatant of Lactobacillus plantarum TA4 and its antibacterial and biocompatibility properties. Sci. Rep. 10:19996. doi: 10.1038/s41598-020-76402-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof H. M., Mohamad R., Zaidan U. H., Rahman N. A. A. (2019). Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: a review. J. Anim. Sci. Biotechnol. 10:57. doi: 10.1186/s40104-019-0368-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof H. M., Mohamad R., Zaidan U. H., Rahman N. A. A. (2020a). Sustainable microbial cell nanofactory for zinc oxide nanoparticles production by zinc-tolerant probiotic Lactobacillus plantarum strain TA4. Microb. Cell Factories 19:10. doi: 10.1186/s12934-020-1279-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]