Abstract
Oral squamous cell carcinoma (OSCC) develops on the mucosal epithelium of the oral cavity. It accounts for approximately 90% of oral malignancies and impairs appearance, pronunciation, swallowing, and flavor perception. In 2020, 377,713 OSCC cases were reported globally. According to the Global Cancer Observatory (GCO), the incidence of OSCC will rise by approximately 40% by 2040, accompanied by a growth in mortality. Persistent exposure to various risk factors, including tobacco, alcohol, betel quid (BQ), and human papillomavirus (HPV), will lead to the development of oral potentially malignant disorders (OPMDs), which are oral mucosal lesions with an increased risk of developing into OSCC. Complex and multifactorial, the oncogenesis process involves genetic alteration, epigenetic modification, and a dysregulated tumor microenvironment. Although various therapeutic interventions, such as chemotherapy, radiation, immunotherapy, and nanomedicine, have been proposed to prevent or treat OSCC and OPMDs, understanding the mechanism of malignancies will facilitate the identification of therapeutic and prognostic factors, thereby improving the efficacy of treatment for OSCC patients. This review summarizes the mechanisms involved in OSCC. Moreover, the current therapeutic interventions and prognostic methods for OSCC and OPMDs are discussed to facilitate comprehension and provide several prospective outlooks for the fields.
Subject terms: Cancer, Oral cancer
Introduction
Oral squamous cell carcinoma (OSCC), which develops in the oral mucosa, is a common type of head and neck malignancy.1–3 According to data collected by the Global Cancer Observatory (GCO), there were 377,713 cases of OSCC worldwide in 2020, with the majority occurring in Asia.4 OSCC affects more males than females, with middle-aged to elderly men being the most susceptible.5 OSCC results in disfiguration and functional impairments, including swallowing, speech, and taste, which have a substantial impact on the life quality of patients.6,7
Clinically, OSCC is characterized by a red and white or red lesion with a slightly uneven surface and distinct borders.8,9 Early-stage lesions are typically painless,10 but they may cause discomfort and exhibit features such as ulceration, nodularity, and tissue attachment as they progress.11 Ulceration is a typical symptom of OSCC, which appears with an irregular floor and margins and is hard upon palpation.11,12 The posterior lateral border of the tongue has the highest incidence of OSCC, accounting for an estimated 50% of all OSCC cases,13 followed by the mouth floor, the soft palate, the gingiva, the buccal mucosa, and the hard palate.14 OSCC spreads predominantly to ipsilateral lymph nodes of the neck via lymphatic outflow, but can also invade contralateral or bilateral lymph nodes. Lungs, bones, and the liver are typical locations for OSCC metastases.15
Patients with oral potentially malignant disorders (OPMDs) are more likely than those with healthy mucosa to develop invasive oral carcinomas.16–20 At the time of diagnosis, the majority of patients with OPMDs are asymptomatic;21 however, some patients may exhibit symptoms of suspected malignancy, such as erythema, pain, tingling sensations, or ulceration.16 Consequently, the diagnosis of OPMDs is a crucial method for clinicians to evaluate the risk of OSCC and guide appropriate treatments (Table 1). OPMDs include oral leukoplakia (OL), oral erythroplakia (OE), oral submucosal fibrosis (OSMF), and oral lichen planus (OLP).22,23
Table 1.
The risks factors, characteristics, epidemiology, and diagnosis of OPMDs
| OPMDs | Risk factors | Histopathology | Epidemiology | Diagnosis |
|---|---|---|---|---|
| Homogeneous OL |
▪ Tobacco ▪ Alcohol ▪ BQ ▪ HPV |
▪ Superficial surface ▪ White surface ▪ Flat surface ▪ Sharp boundaries |
▪ Men aged over 40 ▪ Women non-smoking |
▪ Biopsy ▪ Toluidine blue ▪ Salivary diagnostics ▪ Brush biopsy |
| Non-homogeneous OL |
▪ Speckled red lesions ▪ Irregular white lesions ▪ Corrugated epidermis ▪ Wrinkled epidermis ▪ Verrucous surface ▪ Exophytic growth ▪ Nodular outgrowths ▪ Polypoid outgrowths |
▪ Men aged 50-70 |
▪ Biopsy ▪ Toluidine blue ▪ Salivary diagnostics ▪ Brush biopsy |
|
| PVL |
▪ Verrucous surface ▪ Keratotic surface ▪ Multifocal |
▪ Women aged over 60 | ▪ Biopsy | |
| OE |
▪ Tobacco ▪ Alcohol ▪ HPV |
▪ Fiery red patch ▪ Smooth surface ▪ Velvety surface ▪ Atrophic epithelium ▪ Thin epithelium ▪ Tough texture (Speckled) ▪ Granular texture (Speckled) |
▪ People susceptible to HPV | ▪ By exclusion |
| OSMF |
▪ BQ ▪ Areca nuts |
▪ Vesicles ▪ Blanching mucosa ▪ Fibrosis ▪ Diffuse boundaries |
▪ People aged 20–50 |
▪ Solid biopsy ▪ Liquid biopsy |
| Reticular OLP |
▪ Psychological stress ▪ Medications ▪ Dental materials ▪ EBV |
▪ Wickham striae ▪ Hyperkeratotic plaques |
▪ Women aged over 40 | ▪ Direct immunofluorescence |
| Erosive OLP |
▪ Atrophic ulcers ▪ White striae ▪ Keratinization |
|||
| Erythematous OLP | ▪ Atrophic mucosa | |||
| Plaque-like OLP |
▪ White lesions ▪ Hyperkeratotic surface |
|||
| Bullous OLP |
▪ Bullae ▪ Ulcerative surface |
The World Health Organization (WHO) Collaborating Center defines OL as a permanent, white, and non-scrapable lesion that appears to be “a predominantly white plaque of questionable risk having excluded (other) known diseases or disorders that carry no increased risk for cancer”.24–27 OL patients have a prospective risk of malignancy ranging between 1% and 30%.28 OL could present as homogeneous or non-homogeneous depending on the color and the texture of the surface.29 In non-homogeneous OLs, malignant transformation is more prevalent. Proliferative verrucous leukoplakia (PVL) is a rare form of multifocal OL. PVL plaque has a verrucous and keratotic surface and is asymptomatic and non-homogeneous. PVL exhibits invasive behavior and recurrence following excision,30 of which 60% to 100% develop into oral carcinomas.31 In addition to PVL, OE displays an elevated potential for malignant transformation, with approximately 50% of patients at risk of progressing to dysplasia, cancer in situ, or aggressive cancer.32
OE is an isolated condition described as a “predominantly fiery red patch that cannot be characterized clinically or pathologically as any definable disease”.33–35 85%–90% of early OSCC manifests initially as OE.36
OSMF is characterized by burning sensations or intolerance to spicy food, as well as the presence of vesicles on the palate. Histopathological features of OSMF include alterations in epithelial cell morphology and changes in the composition and structure of the connective tissue.37–39 OSMF has a 7.6% rate of malignant transformation over 17 years and may be accompanied by OL and other potentially malignant lesions and carcinomas.40
OLP is an inflammatory mucocutaneous disorder affecting between 1% and 2% of the general population, of which 0.07% to 5.8% undergo malignant transformation.41,42 There are three clinical subtypes of OLP: erythematous or atrophic, reticular, and erosive. Erosive OLP is the most prevalent clinical subtype associated with malignant transformation.43–45
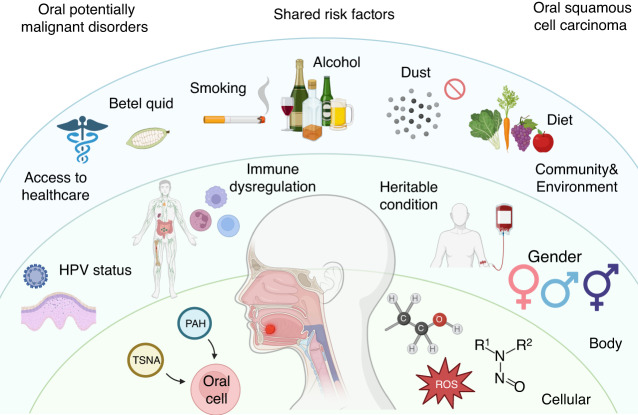
OPMD and OSCC have complex etiologies, including smoking,46,47 alcohol abuse,48–50 betel quid (BQ) chewing,51,52 human papillomavirus (HPV) infection,53,54 nutritional insufficiency,55 immune deficiency,56 and hereditary conditions (Fig. 1).57 The carcinogenicity of polycyclic aromatic hydrocarbons (PAH) and tobacco-specific nitrosamines (TSNA) in tobacco,58 ethanol in alcohol,59 and nitrosamines in BQ has been demonstrated.52,60 Additionally, exposure to dust and heavy metals can cause chronic inflammation or serve as carriers for other oncogenic compounds, thereby increasing the incidence of oral cancer.61,62 HPV can cause precancerous squamous intraepithelial neoplasia, which has the potential to become malignant,63 and has been hypothesized to assist in OSCC progression.64 Nutritional insufficiency, particularly in plant foods and vitamin D, is also related to an elevated potential of oral carcinomas.65 Individuals with suppressed immune systems and rare hereditary diseases, such as Fanconi anemia (FA) and dyskeratosis congenital (DC), are more susceptible to OSCC than those with normal physiological function.66
Fig. 1.
Risk factors of OPMDs and OSCC. The initiation and development of OPMDs and OSCC share similar risk factors, including smoking, alcohol abuse, betel quid (BQ) chewing, human papillomavirus (HPV) infection, nutritional insufficiency, immune deficiency, and hereditary conditions. OPMDs oral potentially malignant disorders, OSCC oral squamous cell carcinoma, PAH polycyclic aromatic hydrocarbons, ROS reactive oxygen species, TSNA tobacco-specific nitrosamines
Persistent exposure to these risk factors results in genetic alterations, epigenetic modifications, and a dysregulated tumor microenvironment, all of which contribute to the occurrence and transformation of OPMDs to OSCC. The genetic alterations result in the aberrant activation of oncogenic pathways, such as EGFR,67 Wnt/β-catenin,68 JAK/STAT,69 NOTCH,70 PI3K/AKT/mTOR,71 MET,72 and RAS/RAF/MAPK, as well as disruptions of suppressor pathways, such as TP53/RB,73 p16/Cyclin D1/Rb,74 which significantly contribute to the progression of OSCC. Epigenetic modifications, such as DNA methylation,75 histone covalent modification,76 chromatin remodeling,77 and gene regulation by non-coding RNAs (ncRNAs),78 participate in OSCC formation and development. In addition, immune suppression, stromal alteration, hypoxia, and an imbalanced oral microbiome can contribute to the dysregulated tumor microenvironment, thus facilitating OSCC progression.79–81
As mentioned above, OSCC may be induced by various risk factors. Chronic exposure to these stimuli promotes carcinogenesis and cancer metastasis by causing genetic mutations, altered epigenetic modification, and a dysregulated tumor microenvironment. Here, we briefly review the mechanisms involved in the occurrence of OSCC. We also discuss therapeutic interventions and the clinical prognosis of OSCC and OPMDs, followed by perspectives for future advancements in the field.
Genetic alterations drive the occurrence of OSCC
There exist a multitude of risk factors that have been identified as being capable of inducing genomic alterations, which are commonly observed in both OSCC and OPMDs.82,83 Genetic mutations contribute to aberrant activation of oncogenic signaling and inactivation of suppressor signaling, promoting the transformation and uncontrolled proliferation of OSCC cells (Fig. 2).70,84–86
Fig. 2.
Genetic alterations in OSCC. Genetic alteration in the TP53/RB, p16/Cyclin D1/Rb, EGFR, Wnt/β-catenin, JAK/STAT, NOTCH, PI3K/AKT/mTOR, MET, RAS/RAF/MASK signaling pathways contribute to OSCC progression. EGFR epidermal growth factor receptors, JAK Janus-activated kinase, MAPK mitogen-activated protein kinase, OSCC oral squamous cell carcinoma, RB retinoblastoma, Rb retinoblastoma tumor suppressor protein, STAT signal transducer and activator of the transcription, TP53 tumor protein p53
Aberrant activation of oncogenic signaling
Oncogenic signaling pathways, including the EGFR pathway, PI3K/AKT/mTOR pathway, JAK/STAT pathway, MET pathway, Wnt/β-catenin pathway, and RAS/RAF/MAPK pathway, are aberrantly activated and upregulated to promote the progression of OSCC.87
EGFR pathway
80%-90% of head and neck squamous cell carcinoma (HNSCC) is found to overexpress epidermal growth factor receptors (EGFR), a member of the HER/ErbB family of receptor tyrosine kinases (RTKs).88–90 It has been reported that OSCC shows increased EGFR (42% to 58%),91 which is associated with poor treatment outcomes and prognosis.92
The EGFR pathway prompts OSCC cell proliferation, metastasis, invasion, and apoptosis resistance.93 Radiation triggers the translocation of EGFR into the nucleus, where it functions as a transcription factor and leads to radiotherapy resistance in oral cancer.94 The EGFR also interacts with other receptors, such as Axl, increasing its carcinogenic potential on the mucosal surface of the oral cavity.95
Meanwhile, various factors are implicated in the EGFR pathway to drive the malignancy of OSCC. For instance, downregulated hsa_circ_0005379 facilitates the proliferation and metastasis of OSCC cells by regulating the EGFR pathway.96 The overexpression of distal-less homeobox 6 (DLX6) enhances proliferation and inhibits apoptosis in OSCC cells through the EGFR-CCND1 axis.97 Upregulated bone marrow stromal cell antigen 2 (BST2) promotes tumor growth and confers gefitinib resistance in OSCC patients via activating the EGFR pathway.98
PI3K/AKT/mTOR pathway
Thirty-seven percent of HNSCC cases, more specifically 34% of HPV- and 56% of HPV+ patients, exhibit overexpression or mutation of PIK3CA, as reported by the Cancer Genome Atlas (TCGA) study.99,100 Furthermore, patients with OSCC are more likely to exhibit somatic copy number alterations in genes encoding components of the PI3K/AKT/mTOR network.71,101–103
The PI3K/AKT/mTOR pathway leads to the metastasis and proliferation of OSCC cells.104,105 The PI3K-AKT pathway is frequently activated in OSCC malignancies due to the evaluated phosphorylation levels of AKT and associated mTOR. Then, by stimulating AKT, PDK1, and mTOR, a cascade of downstream biological processes, such as cell metabolism, cell proliferation, cell death, protein synthesis, and transcription, are increased to drive OSCC.106 In the meantime, extracellular ATP stimulates the PI3K-AKT pathway through the P2Y2-Src-EGFR axis to prompt OSCC cell metastasis.107 Also, the circEPSTI1/miR-942-5p/LTBP2 axis phosphorylates the components of the PI3K-AKT-mTOR pathway and facilitates epithelial–mesenchyme transition (EMT) to accelerate the metastasis and proliferation of OSCC cells.108 Moreover, ITGB2 high cancer-associated fibroblasts (CAFs) stimulate the PI3K-AKT-mTOR pathway to promote the progression of OSCC malignancy via NADH oxidation.109 Additionally, a variety of factors, including ZNF703,110 PDGF-D,111 CCL18,112 and Muc1,113 activate the PI3K/AKT/mTOR pathway, resulting in OSCC cell survival, invasion, and drug resistance.105,114 Collectively, targeting the PI3K/AKT/mTOR pathway could be a potent method to prevent OSCC.
JAK/STAT pathway
Both HPV+ and HPV- HNSCC display abnormal activation of the signal transducer and activator of the transcription (STAT) pathway.69,115 Upregulated STAT3 is associated with HNSCC malignancies and resistance to chemotherapy, radiotherapy, and EGFR-targeted therapy.116,117 The STAT3 signaling pathway causes immune suppression and protects OSCC cells from being recognized and destroyed by cytotoxic T cells by stimulating the release of cytokines, such as transforming growth factor (TGF)-β1, vascular endothelial growth factor (VEGF), interleukin (IL)-6, and IL-10.118 Moreover, in response to upstream signals from the IL-6 receptor family and RTKs including EGFR, VEGFRs, Jenus-activated kinases (JAKs), and Src family kinases (SFKs), STAT3 is activated and translocated to the nucleus,119 thereby inducing the expression of cyclin D1, Bcl-xL, and other pro-survival factors.118 In addition, various factors, such as miR-548d-3p and long non-coding RNA (lncRNA) P4713, participate in the JAK/STAT pathway. Specifically, miR-548d-3p binds to the 3’UTR of SOCS5 and SOCS6 to downregulate their expression, regulating the JAK/STAT pathway and serving as an oncogene in OSCC.120 lncRNA P4713 activates the JAK/STAT pathway and drives the metastasis and proliferation of OSCC cells.121
MET pathway
Mutations and gene amplifications in hepatocyte growth factor (HGF) receptor (MET or c-Met) and its ligand HGF are uncommon, occurring in 6% and 2%-13% of HNSCC, respectively.122–125 Immunohistochemistry analysis reveals MET and/or HGF are upregulated in approximately 80% of HNSCC.126 Lymph node metastases with elevated MET levels can also be present.127 Overexpression of MET is recognized as a cause of EGFR inhibitor resistance, as it compensates for PI3K and MAPK inhibition in EGFR signaling.123,128 Anoikis resistance is enhanced in HNSCC as a consequence of HGF amplification, which is essential for developing nodal metastasis.127 KRT16 overexpression is associated with metastasis, increased mortality rate, unfavorable pathological differentiation, and advanced stages in OSCC patients. c-Met was also discovered to correlate with KRT16 through β5-integrin.129
Wnt/β-catenin pathway
Various components of the Wnt/β-catenin signaling pathway, including Wnt ligands, Wnt inhibitors, membrane receptors, and intracellular mediators, are regularly impaired by genetic alterations in malignant tumors, such as OSCC.130–133 The Wnt/β-catenin signaling pathway determines cell fate and proliferation in OSCC, whereas aberrant Wnt/β-catenin signaling promotes oncogenesis, typically via various mechanisms related to abnormal β-catenin stimulation.134 For example, KMT2D, one of the most frequently mutated genes in OSCC cells, collaborates with MEF2A to boost the transcription activity of β-catenin.135 In addition, the SNHG17/miR-384/ELF1 axis stimulates the Wnt/β-catenin pathway by upregulating CTNNB1 expression to drive the proliferation and metastasis of OSCC cells.136 DEP domain containing 1 (DEPDC1) is also essential for OSCC progression, which drives OSCC metastasis and aerobic glycolysis through the WNT/β-catenin pathway.137 Mutations that inactivate NOTCH1 and FAT1 diminish their capability to suppress the expression of β-catenin.138 In addition to mutations in Wnt/β-catenin signaling components, OSCC exhibits an overexpression of Wnt ligands.139 A high level of Wnt-7b in OSCC activates Wnt/β-catenin and facilitates cancer cell invasion and proliferation.140 Moreover, Wnt7a enhances the expression of MMP-9 to facilitate OSCC progression.141 Overall, Wnt/β-catenin signaling has a vital role to play in the formation of oral malignancies.
RAS/RAF/MAPK pathway
Only 4% of HNSCC cases exhibit mutations of mitogen-activated protein kinase (MAPK) signaling, which modulates cell proliferation, death, differentiation, angiogenesis, and dissemination.142–144 It comprises four sub-pathways, namely extracellular signal-regulated kinase (ERK1/2), c-Jun N-terminal kinase (JNK), p38, and ERK5 sub-pathways.144 In oral cancer, the ERK1/2 pathway has generated significant interest due to the fact that ERK1/2 is activated mechanistically by binding the growth factor EGF. Erl1/2 separates from RAS-RAF-MEK-ERK1/2 and induces the phosphorylation of OSCC-causing transcription factors, such as c-Myc, ETS-1, AP-1, NF-κB, and others.144 Meanwhile, OSCC cell growth is influenced by various proteins serving as targets, including SH3 domain-containing kinase binding protein 1 (SH3KBP1),145 annexin A10,146 fibroblast activation protein,147 EGFR,148 parathyroid hormone-related protein,149 angiopoietin-like 3 (ANGPTL3),150 quaking 5,151 and 70-kDa ribosomal S6 kinase.152
Aberrant inactivation of suppressor signaling
TP53/RB, p16/Cyclin D1/Rb, and NOTCH are examples of suppressor signaling pathways. During the malignant transformation of OPMDs to OSCC, they become abnormally inactivated and downregulated.
TP53/RB pathway
Approximately 80% of HPV- HNSCC have muted tumor protein p53 (TP53), resulting in gene dysfunction.138,153,154 Exon 4 or intron 6 is the location of TP53 mutations that occur early in the progression of HNSCC, especially OSCC.155 Consistently, p53 mutation is commonly observed in HPV- OSCC, as the HPV E6 oncoprotein degrades p53.156 In both subtypes, mutations in p53 are correlated with a lower overall survival rate, treatment resistance, and an increased risk of relapse.138 In early OSCC, TP53 expression is also associated with tumor stage and grade, as well as surgical margin dysplasia. It is not clear, however, whether TP53 expression and lymph node metastasis are related.157,158 p53 modulates cell death, apoptosis, and differentiation in OSCC cells by interacting with a complicated network of proteins.138,159 APR-246 targets GSTP1 to reactivate p53 and induce cell dealth.160 Co-expression of platelet-derived growth factor receptor A (PDGFRα) and p53 stimulates cell growth in poorly differentiated OSCC.161 Accordingly, TP53 and p53 collaborate to enhance OSCC invasion.162
Similar to mutations in the TP53 pathway, retinoblastoma (RB) pathway mutations are early manifestations of HNSCC. Both p53 and RB pathway mutations contribute to the unrestricted replication of HNSCC cells.100 In HPV+ neoplasms, the degradation of pRb by E7 contributes to the secretion of E2F and unregulated HNSCC cell proliferation.163 In persistent HPV infection, E2-regulated expression of E6 and E7 is responsible for p53 degradation and Rb functional suppression.150 When pRb is dysregulated, oral epithelial dysplasia has an increased likelihood of transforming into malignant carcinomas.164
p16/Cyclin D1/Rb pathway
In most HNSCC, the tumor suppressor p16 is inactivated, resulting in aberrant cell cycle control and cell proliferation, a deficiency in cell senescence, and ultimately dysplasia.165 Similar to HNSCC, OSCC frequently has a low level of p16.166,167 OSCC patients with inactive p16 tend to have a lower survival rate than those with normal or augmented p16 levels.168
Cyclin D1 (CCND1) amplification occurs in 25–43% of OSCC cases.169–171 In the early stages of OSCC, CCND1 is upregulated and contributes to the proliferation of OSCC cells.172 Particularly, CCND1 is more likely cytoplasmically expressed in advanced OSCC with deleterious differentiation, increased mitosis, and invasive cell morphology.173 Elevated expression of CCND1 is also related to reduced overall survival and poor prognosis among patients with OSCC.174,175 CCND1 deficiency inhibits the cyclin-dependent kinases CDK4 and CDK6, which are responsible for the cell cycle progression by dephosphorylating and inactivating pRb and then hindering G1 to S transition.176
NOTCH pathway
Notch signaling shows diverse effects on different cell types.177–181 Therefore, the suppressive or oncogenic functions of Notch in tumorigenesis have a contextual basis. According to a 2015 TCGA analysis, NOTCH1-3 is inactivated in 17% HPV+ and 26% HPV- HNSCC.138 The majority of these aberrations are found in NOTCH1, including nonsense mutations causing truncated proteins, missense mutations within functional regions, and frameshift deletions and insertions. On the basis of mutational features, such as the absence of mutational hotspots and the presence of nonsense mutations, it is hypothesized that NOTCH1 acts as a tumor suppressor in HNSCC.182 Notch signaling has been demonstrated as a tumor suppressor in epithelial SCC malignancies (lung, bladder, and esophageal tumors) and several in vivo models.159
Nevertheless, in vitro assays with HNSCC cell lines have shown a requirement for an increase in Notch signaling activity to maintain malignant behavior.183 Recent research has shown that 43% of OSCC cases from a Chinese population are associated with activating mutations in NOTCH1, including novel mutations in heterodimerization and abrupted domains likely to obtain function.184 Furthermore, the mutation of NOTCH1 can result in a poor prognosis and lymph node metastasis in OSCC.185 NOTCH1 is responsible for sustaining the characteristics of cancer stem cells (CSCs), which are essential for cancer relapse and migration, via Wnt signaling; and 32% of HNSCC showed overexpression of downstream Notch effectors (measured by methylation, DNA copy number, and expression of 47 genes involved in Notch signaling).186
Overall, it remains unclear whether NOTCH mutations in HNSCC are typically activating or inactivating.184,187 Various mutations may be present in distinct subtypes of HNSCC.177 Since in vitro assays may not accurately reflect the disease progression in patients, it is imperative to investigate the functional role of Notch signaling in OSCC using robust in vivo models. Clinical trials of inhibitors or stimulators of the Notch pathway must be carefully considered.188,189
Epigenetic modifications promote the development of OSCC
Epigenetic regulation refers to heritable and stable alterations in gene expression that do not modify the DNA sequence and are responsible for the formation and progression of OSCC neoplasms by regulating gene expression.190–194 Epigenetic modifications comprise DNA methylation, histone covalent modification, chromatin remodeling, and the impact of ncRNAs on gene expression (Table 2).195
Table 2.
Epigenetic modifications in OSCC
| Epigenetic modifications | Targets | Exhibitions | Outcomes |
|---|---|---|---|
| DNA methylation |
▪ p16 ▪ MGMT ▪ MLH1 ▪ p15INK4B ▪ E-cadherin ▪ PTEN ▪ APC ▪ P14ARF ▪ P16INK4A ▪ miR-137 ▪ miR193a |
▪ Hypermethylation | ▪ Oncogenic |
|
▪ AIM2 ▪ CEACAM1 ▪ LINE-1 ▪ PI3 ▪ PTHLH |
▪ Hypomethylation | ▪ Oncogenic | |
| Histone acetylation |
▪ H3K9ac ▪ H3K4ac ▪ miR-154-5p |
▪ Downregulation | ▪ Oncogenic |
|
▪ H3K27me3 ▪ HDAC6 ▪ HDAC8 ▪ HDAC1 ▪ HDAC2 |
▪ Upregulation | ▪ Oncogenic | |
| Chromatin modification |
▪ SATB1 ▪ ZSCAN4 ▪ CSC factors ▪ RSF-1 |
▪ Upregulation | ▪ Oncogenic |
| ncRNAs |
▪ miR-1246 ▪ miR-31 ▪ miR-214 ▪ miR-23a ▪ miR-372 |
▪ Upregulation | ▪ Oncogenic |
|
▪ miR-181a ▪ miR-17-92 cluster ▪ miR-329 ▪ miR-410 ▪ miR-211 ▪ TCF12 ▪ miR-214 ▪ miR-23a ▪ miR-372 |
▪ Upregulation | ▪ Tumor suppression | |
|
▪ lncRNA HOXA11-AS ▪ ZBTB7A ▪ miR-98-5p ▪ miR-214-3p ▪ circITCH |
▪ Downregulation | ▪ Oncogenic |
DNA methylation
The development and prognosis of OSCC are affected by DNA methylation abnormalities.196 Both hypomethylation and hypermethylation increase the prevalence of oral malignancies. In addition to the correlation between smoking and global hypomethylation,197 alcohol consumption is linked to evaluated levels of CpG hypermethylation in genes associated with oral cancers.198 Several OSCC samples have been found to manifest both hypermethylation and hypomethylation, leading to abnormal expression of genes, the majority of which are implicated in the tumorigenic process of OSCC by stimulating the Wnt and MAPK pathways.199,200
Particularly, DNA methylation silences suppressor genes in OSCC.201,202 CpG island hypermethylation inhibits over 40 tumor suppressor genes, which regulate the cell cycle, programmed cell death, Wnt pathway, cell-to-cell adhesion, and DNA repair in OSCC.203 The specific pattern of gene methylation leads to hypermethylated promoters in 22%-76% of OSCC patients.193,204,205 In contrast, the p16 promoter region was found to be methylated in only 5.4% of normal mucosa samples, indicating the epigenetic silencing of p16 in the development of OSCC.193 Moreover, DNA is methylated throughout the entire genome in 28-58% of premalignant oral tissues of tobacco users, with methylation levels increasing as the cancer progressed.193 Continued smoking increases DNA methyltransferase activity, enhancing methylation of the p16 promoter.206,207
OSCC also demonstrates hypermethylation in the promoter regions of many other genes, including O-6-methylguanine-DNA methyltransferase (MGMT), mutL homolog 1 (MLH1), and p15INK4B. MGMT functions in DNA repair by eliminating guanine DNA adducts, and maintaining the integrity of the genome.191 Therefore, increased MGMT levels render normal cells more resistant to carcinogens and spontaneous mutations.208 Silencing MGMT is associated with a poor prognosis in the early stages of OSCC development.198,200 Furthermore, the hypermethylation of MLH1 is essential for DNA mismatch repair and prevents the accumulation of DNA mutations, which are linked to the initiation of OSCC.209 Due to its significance, methylation patterns of MLH1 have been extensively investigated.200
In addition, p15INK4B significantly contributes to tumor suppression. It inhibits cell proliferation and, consequently, cell cycle progression at the G1 stage, which is induced by stimulation of extracellular TGF-β and IFN-α.200 Hypermethylation of p15INK4B may render cells less sensitive to these external stimuli, thereby influencing the progression of OSCC. Normal tissues lack methylation of p15INK4B; therefore, its abnormal methylation can serve as an indicator for OSCC.210 Additionally, hypermethylated E-cadherin,211 phosphatase and tensin homolog (PTEN),212 adenomatous polyposis coli (APC),213 p14ARF,214 p16INK4A,215 miR-137,216 and miR-193a217 prevent oral cells from promoting OSCC.218
Hypermethylation can also lead to the suppression of genes involved in the progression and metastasis of OSCC. For example, DNA methyltransferase (DNMT) levels have been linked to OSCC progression, growth, poor prognosis, and a higher risk of metastasis.219 DNMT3a immunoreactivity increased significantly in OSCC tissues compared to normal tissues.220 Despite some reports of normal DNMT1 expression in OSCC, the preponderance of studies have demonstrated that OSCC development is associated with DNMT overexpression.221 In general, DNMT1 regulates the prognosis of OSCC patients in a manner that decreases their overall survival.222
In alcoholic beverages, ethanol and acetaldehyde cause DNA hypomethylation.223 Global hypomethylation may facilitate tumorigenesis by diminishing the methylation of CpG dinucleotides across the entire genome.194 Moreover, global hypomethylation may promote the progression of cancer by demethylating previously methylated promoter regions of numerous oncogenes, thereby altering their expression.200 The presence of these characteristics is linked to the progression of malignant tumors, as they can increase the instability of the genome.191 AIM2,224 CEACAM1,225 LINE-1,226 PI3,227 and PTHLH228 are found as hypomethylated genes that contribute to OSCC.218
Histone and chromatin modification
There are two distinct forms of chromatin: heterochromatin, which is highly compressed and transcriptionally silent, and euchromatin, which is less dense and transcriptionally active. The organization of chromatin and the expression of genes depend heavily on histone modifications, particularly histone acetylation. It is therefore not surprising that abnormalities in histone acetylation correlate with the progression of oral malignancies.229 Recent studies have identified reduced histone H3K9ac as an indicator of chemoresistance related to the NFκB pathway and CSCs recruitment230–232; it is also associated with enhanced cell proliferation and disruption of EMT in oral tumorigenesis.233 In light of these findings, H3K9ac assists in HNSCC development. Furthermore, low H3K4ac and high H3K27me3 levels are related to disease-free survival (DFS) and cancer-specific survival (CSS) and influence the progression of OSCC.234
In contrast to other histone modifications (such as methylation and phosphorylation), which have been extensively investigated in various cancer types, studies on the role of acetylation in the formation of OSCC are relatively limited. The acetylation of histones is governed by the equilibrium between histone acetyltransferases (HATs) and histone deacetylases (HDACs). Multiple HDACs are correlated with differentiation, cell cycle-associated genes, programmed cell death, angiogenesis, and dissemination of cancer cells, which have been observed to be altered in OSCC.235,236 OSCC exhibits overexpression of HDAC1,237 which contributes to OSCC growth and progression by regulating miR-154-5p/PCNA signaling.238 HDAC2 mRNA and protein levels are higher in OSCC and premalignant lesions than in controls. Meanwhile, the protein level of HDAC2 is related to histological differentiation and the stage of tumor-node-metastasis (TNM) in OSCC patients, emphasizing its significance in the progression of premalignant to malignant carcinomas.239
Chromatin remodelers are crucial molecules in charge of modulating nucleosome positioning and chromatin accessibility in response to DNA-driven stimulation of biological processes in OSCC.240 For instance, special AT-rich sequence binding protein 1 (SATB1) is a genome-organizing protein that modifies chromatin structure and directs chromatin remodeling enzymes to specific chromatin regions to regulate gene expression. High SATB1 expression is related to HNSCC metastasis, poor prognosis, and decreased survival rate.241 The zinc finger and SCAN domain containing 4 (ZSCAN4) is another example of a protein that can alter the epigenetic profile and chromatin state in tumors.242 ZSCAN4 induces the functional hyperacetylation of histone 3 at the OCT3/4 and NANOG promoters, thereby upregulating CSC factors.77 In contrast, ZSCAN4 exhaustion contributes to the downregulation of CSC markers, the diminished capacity to develop tumorspheres, and the restriction of tumor growth.243 Therefore, ZSCAN4 is essential for maintaining the CSC phenotype and tumor progression in HNSCC.77 Furthermore, the chromatin remodeler RSF-1, a member of the ISWI family,244 is upregulated in OSCC and linked to enhanced invasion, lymph node metastasis, and advanced stages of carcinomas. RSF-1 exhibits the ability to increase the resistance of OSCC cells to both radiotherapy and chemotherapy.245
Non-coding RNAs
Similar to proteins, microRNA (miRNA) dysfunction can arise from abnormalities in miRNA expression caused by genetic mutations, epigenetic modifications, or processing deficiencies. MicroRNAs are a class of ncRNAs that have been intensively investigated in HNSCC. Due to genetic and epigenetic alterations, a number of miRNAs have been linked to OSCC.246 miR-26,247 -137,248 and -203249 are repressed by CpG hypermethylation in OSCC. Notably, miR-26a binds to the DNMT3B enzyme, which stimulates cell proliferation, indicating an intimate relationship between epigenetic modifications and the progression of oral malignancies.250
miRNAs are capable of functioning as either tumor oncogenes or suppressors to modulate cell growth.251 For instance, miR-1246 functions as an oncogene and is abundant in exosomes derived from the oral carcinoma cell line HOC313-LM, which is highly metastatic.252 After the transfer of miR-1246 via exosomes, inadequately metastatic cells displayed enhanced cell motility and invasion capability. miR-1246 facilitates cell mobility by directly binding to DENN/MADD Domain Containing 2D (DENND2D) in OSCC.253 In addition, patients with oral malignancies had increased levels of miR-31 in their saliva throughout the entire process of the disease.254 Following oral tumor excision, the level of salivary miR-31 decreased significantly, indicating that the majority of the raised level of salivary miR-31 originated from tumor tissues.255
OSCC is frequently associated with the downregulation of numerous tumor-suppressing miRNAs. For example, ectopic miR-181a expression inhibits the proliferation and anchorage-independent growth of OSCC.256 miR-181a significantly inhibits OSCC formation in three-dimensional organotypic raft cultures.257 Mechanistically, miR-181a reduced K-ras protein levels and luciferase activity in receptor vectors containing the 3’-untranslated region of the K-ras gene. By inhibiting the oncogene K-ras, miR-181a could suppress OSCC.257 Similar results are observed when the miR-17-92 cluster, comprising miR-17, miR-19b, miR-20a, and miR-92a, is induced to overexpress in OSCC cell lines.258 miR-17/20a within the miR-17-92 cluster modulates OSCC migration predominantly and is inversely associated with TNM stage and lymphatic metastasis in clinical investigations; it is known to suppress tumor migration in OSCC.258 Other examples include miR-329, miR-410, and miR-211. Increased levels of miR-329 and miR-410 inhibit the proliferation and dissemination of OSCC cells. Specifically, miR-329 and miR-410 could bind to Wnt-7b and then attenuate the Wnt-β-catenin pathway in OSCC.140 miR-211 is upregulated in OSCC cells by arecoline and 4-nitroquinoline 1-oxide (4NQO), and it directly inhibits OSCC cell growth by targeting transcription factor 12 (TCF12). However, miR-211 levels are drastically reduced in tumor tissues, resulting in an enhanced oncogenicity for OSCC.259
The progression of OSCC cells toward a drug-resistant state appears to be mediated by multifunctional ncRNAs, including miRNA, lncRNA, and circular RNA (circRNA).260 Over 10% of all identified miRNAs correlate with chemotherapy resistance in cancer cells, particularly those implicated in OSCC cell chemoresistance.261–263 In the presence of cisplatin (CDDP) resistance, miR-214 and miR-23a levels were promoted, as determined by microarray analysis.264–266 miR-372 is overexpressed in OSCC and inhibits zinc finger and BTB domain-containing 7A protein (ZBTB7A), promoting tumorigenesis and CDDP resistance in OSCC cells.267,268 OSCC cells and tissues overexpress the lncRNA HOXA11-AS relative to adjacent normal tissues and oral keratinocytes.269 lncRNA HOXA11-AS consumes miR-98-5p, which is capable of inhibiting OSCC cell proliferation,270 and suppresses miR-214-3p expression, which may lead to the establishment of drug resistance in OSCC.271 In addition, when circITCH is upregulated, chemotherapy agents are more effective against drug-resistant myeloma cells. However, OSCC tissues and cells express less tumor-suppressing circITCH than adjacent normal tissues and human oral keratinocytes.272
In addition to the aforementioned characteristics, OSCC exhibits phenotypic plasticity. EMT is an epigenetically regulated process that induces plasticity in OSCC and leads to the transition of cancerous cells into distinct phenotypic forms with increased motility and survival. Notably, oral CSC plasticity interacts with EMT to enhance treatment resistance in OSCC.273–276
Dysregulated tumor microenvironment in OSCC
OSCC occurrence and progression are influenced by a dysregulated tumor microenvironment. Specifically, a suppressed immune system, stromal alteration, hypoxia, and an unbalanced oral microbiome all contribute to the development and metastasis of OSCC, so their underlying mechanisms present therapeutic opportunities.
Immune suppression
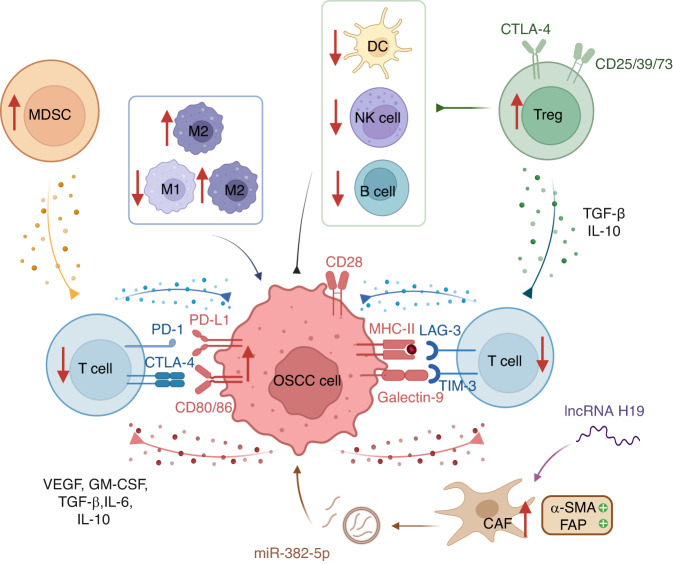
Patients with suppressed immune states, such as HIV+ patients and organ transplant recipients, are more likely to develop oral malignancies,277–279 suggesting that immune response plays a crucial role in OSCC development (Fig. 3).92,280–283
Fig. 3.
Immunosuppressive TME in OSCC. The tumor microenvironment (TME) contains various immunomodulatory cells, including cancer-associated fibroblasts (CAFs), regulatory T cells (Tregs), tumor-associated macrophages (TAMs), and myeloid-derived suppressor cells (MDSCs). The interaction between programmed death 1 (PD-1) and programmed death ligand-1 (PD-L1) leads to T-cell suppression and adaptive immunity tolerance, whereas cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) competes with CD28 by interacting with CD80/86 on OSCC cells. Other immune checkpoints also play a role, including LAG-3 and TIM-3. OSCC cells produce immune suppressive factors such as vascular endothelial growth factor (VEGF), granulocyte-macrophage colony-stimulating factor (GM-CSF), transforming growth factor (TGF)-β, interleukin (IL)-6, and IL-10, which block effector cells. Tregs generate TGF-β and IL-10 to diminish the functions of T cells. CAFs express α-smooth muscle actin (α-SMA) and fibroblast activation protein (FAP) and promote tumor growth by overexpressing miR-385-5p in their exosomes. In the TME, there is a greater proportion of M2 macrophages to M1 macrophages, with M2 TAMs possessing carcinogenic properties. OSCC oral squamous cell carcinoma
OSCC evades the immune surveillance of their hosts by employing a variety of molecular-level strategies. First, a high mutational burden in OSCC caused by smoking and alcohol abuse-induced DNA damage facilitates immune evasion.86 Particularly, OSCC is linked to mutations in human leukocyte antigen (HLA) and antigen processing machinery (APM) that are critical for evading immune surveillance.284,285 Second, immune checkpoints contribute to immune evasion of OSCC. An estimated 83% of OSCC expresses programmed death ligand-1 (PD-L1), which interacts with programmed death 1 (PD-1) on T cells and induces T-cell suppression and tolerance to adaptive immunity.286 The expression of cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) is elevated in OSCC for evading immune surveillance. CTLA-4 interacts with CD80/86 on antigen-presenting cells (APCs) to compete against its stimulatory counterpart CD28 to block the differentiation of naïve T cells.287 Additionally, significant immune checkpoints such as lymphocyte activation gene-3 (LAG-3),288 T-cell immunoglobulin and mucin-containing protein-3 (TIM-3),289 and B7 Homolog 3 (B7-H3)290 are overexpressed in OSCC. Third, cancer cells can secrete cytokines that suppress adaptive immunity and promote tumor growth. Particularly, OSCC cells generate immunosuppressive inflammatory cytokines, such as VEGF, granulocyte-macrophage colony-stimulating factor (GM-CSF), TGF-β, IL-6, and IL-10, which affect T cells.291 Moreover, a significant reduction of immune-activating cytokines, like IL-2, inhibits the stimulation of the innate and adaptive immune response against OSCC.292
Microenvironment regulators such as hypoxia, abnormal vasculature and lymphatics, and high interstitial pressure, may also manipulate the immune response to OSCC by influencing the secretion of cytokines, the trafficking of immune cells, and the function of the immune system. In addition, patients with uncommon inherited diseases such as FA and DC exhibit immune deficiencies that promote OSCC progression.293,294
FA is an autosomal recessive genetic disorder. It manifests as aplastic anemia, progressive pancytopenia, congenital anomalies, and an elevated incidence of developing malignancies. In a study conducted in Brazil, 121 cases of oral malignancies in FA patients were identified.293 Hematopoietic stem cell transplant (HSCT), the only current treatment option for the hematological complications of FA patients, is linked to a 500-fold increased risk of head and neck malignancies and a risk factor for a more rapid progression of oral malignancy in comparison to non-transplanted patients.295 DC is a rare genetic disorder characterized by premature telomere shortening that leads to bone marrow failure. DC-associated mucocutaneous illness symptoms include reticulated pigmentation of the skin, nail dysplasia, and oral leukoplakia. Multiple malignancies may develop in patients with DC, such as a transition from leukoplakia to HNSCC.294,296
Taken together, the above-mentioned immunosuppressive tumor microenvironment enables OSCC to evade immune recognition and elimination, thereby presenting therapeutic opportunities.
Stromal alteration
Due to stromal alteration, HPV- and HPV+ OSCC can escape the cytotoxic mechanisms despite overexpressing CD8+ cytotoxic T cells and activated NK cells. This is influenced by various stromal cells, including CAFs, tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), and regulatory T cells (Tregs) (Fig. 3).297 These stromal cells can also secrete cytokines and immune checkpoint inhibitors, which act together to constitute an immunosuppressive microenvironment and allow the growth of neoplasms.291
Active CAFs express α-smooth muscle actin (α-SMA) and fibroblast activation protein (FAP), which promote OSCC metastasis.298 CAFs boost OSCC development via miR-382-5p overexpression in their exosomes and lncRNA-regulated RUNX2/GDF10 signaling. Meanwhile, lncRNA H19 enhances glycolysis of CAFs in the oral cavity through the miR-382-5p/PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6 biphosphatase 3) axis.299 In addition, senescent CAFs facilitate the rapid progression of oral malignancies in genetically unstable OSCC by promoting keratinocyte migration, inhibiting epithelial adhesions, and releasing active matrix metalloproteinases-2 (MMP-2).300–303 CXCL1304 and peroxiredoxin1 (PRDX1)305 drive the development of OSCC through the induction of cell senescence.
TAMs consist of M1 TAMs with antitumor properties and M2 TAMs with tumorigenic properties.306–308 A higher ratio of M2 to M1 TAMs is commonly observed in the peritumoral microenvironment surrounding OSCC, promoting carcinogenesis.309,310 MDSCs are immature myeloid-derived cells which can suppress T cells.311–315
Patients with OSCC have an elevated quantity of Tregs in their peripheral circulation, lymph nodes, and neoplasms.277–279,316 Tregs are able to suppress various stromal cells, including CD4+ and CD8+ T cells, B cells, dendritic cells (DCs), and natural killer cells (NKs),317,318 due to the expression of CTLA-4, CD73, CD39, and CD25, and the production of TGF-β, IL-10, and perforin/granzyme B.319–322 It has been demonstrated that mice with OSCC have elevated levels of immunosuppressive CD11b+Gr-1+ cells in the peripheral circulation, spleen, and tumors.323 Tumor CD11b+Gr-1+ cells express more PD-L1 than cells from other tissues, disrupt T-cell proliferation in vitro, and ultimately suppress immunity.323 On the other hand, researchers hypothesize that elevated T-helper 2 cells in the immune system may shield individuals with allergies or asthma from tumor development;324 however, additional investigation is required to verify if this is true.
Hypoxia
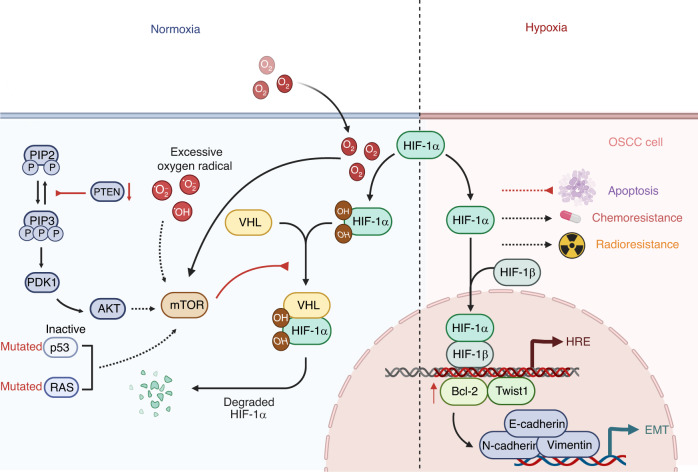
OSCC is a locally aggressive tumor with an elevated hypoxia level, resulting in dissemination, relapse, and poor therapeutic response (Fig. 4).285,325–328 Of note, hypoxia is induced by hypoxia-inducible factors (HIF) in OSCC.329,330 HIFs 1-3 are the principal hypoxia response mediators. Under normoxic conditions, the E3 ubiquitin ligase Von Hippel-Lindau (VHL) protein degrades the HIFα subunits.331,332 In the presence of hypoxia, HIFα becomes stable and binds to HIFβ in the nucleus, adhering to hypoxia response elements (HREs) to facilitate tumor adaptation.333 HREs are present in genes involved in metabolism, extracellular matrix remodeling, angiogenesis, immune modulation, and inflammation.334–337 In addition, hypoxia promotes Bcl-2/Twist1 interaction by enhancing Bcl-2 attachment to Twist1, which is related to the poor prognosis of OSCC patients.325,338
Fig. 4.
Hypoxia in OSCC. Under normoxic conditions, VHL degrades the HIFα subunits. In the condition of hypoxia, HIFα becomes stable and binds to HIFβ within the nucleus, adhering to hypoxia HREs to enable tumor adaptation. Hypoxia also promotes Bcl-2/Twist1 interaction by enhancing Bcl-2 attachment to Twist. Particularly, the mTOR pathway has been demonstrated to increase the level of HIFα in tumor regions that do not experience significant hypoxia. Several oncogenic mechanisms, such as inactive p53 mutations, RAS mutations, excessive oxygen radical accumulation, suppression of PTEN, and the infective HIF-1α degradation by VHL mutation, have been identified as contributing to this development. In addition, HIF-1α stimulates Twist1 to transactivate EMT-related genes, including Vimentin, N-cadherin, and E-cadherin, in order to drive EMT. Moreover, HIF-1α blocks apoptosis and confers higher resistance to chemotherapy and radiotherapy on OSCC. EMT epithelial–mesenchyme transition, HERs hypoxia response elements, HIF hypoxia-inducible factors, OSCC oral squamous cell carcinoma, PTEN phosphatase and tensin homolog, VHL Von Hippel-Lindau
Several examples demonstrate that HIFs can stabilize under normoxic conditions, suggesting that hypoxia is not clearly defined. In particular, the mTOR pathway raises the level of HIF-1α in tumor regions that do not experience significant hypoxia. A variety of oncogenic mechanisms, such as inactive p53 mutations, RAS mutations, excessive oxygen radical accumulation, suppression of PTEN, and infective HIF-1α degradation due to VHL mutations, have been identified as contributing to this development. HIF signaling is synergized with activating mutations in p53.335,339,340
Hypoxia and EMT have a correlation with OSCC metastasis and invasion.325 Hypoxia-induced decreases in E-cadherin mRNA levels boost the migration capability of OSCC cells.341 HIF-1α drives EMT by stimulating Twist1 to transactivate EMT-related genes, such as Vimentin, N-cadherin, and E-cadherin.342 Moreover, HIF-1α blocks apoptosis and imparts increased chemoresistance and radio-resistance in OSCC, thereby contributing to the aggressiveness of the disease.343
BQ chewers exhibit endogenous nitrosation, which generates potentially carcinogenic nitrosamines, including 3-methylnitrosopropionitrile.344 As a result of the auto-oxidation of polyphenols found in areca nuts, reactive oxygen species (ROS) are present in the mouths of patients who consume BQ, which is exacerbated by the alkaline pH of slaked lime345 and can stimulate hypoxic adaptation in OSCC cells.346
Taken together, as HPV- HNSCC are susceptible to p53 mutations and are hypoxic, the synergy between p53 mutants and HIF-1 signaling may provide a valuable avenue for future research. Hypoxia tumors are a subtype of OSCC characterized by a poor prognosis and resistance to treatment.287
Oral microbiome
Independent of other risk factors, oral microbiome alterations may facilitate the progression of oral malignancies in 7–15% of cases.347–351 The microbiome is comprised of a complicated community of bacteria, fungi, protists, archaea, and viruses, all of which contribute to the maintenance of microbial diversity.352–357 Statistical evidence establishes a correlation between dysbiosis and the incidence of various cancers, thereby elevating the clinical significance of the oral microbiome (Fig. 5).358 In the diverse oral cavity environment, including mucosal surfaces and deep tissue crevices, there are distinct microbial species present in both healthy and malignant.359
Fig. 5.
Dysbiosis in OSCC. a A correlation exists between dysbiosis and the occurrence of various cancers. OSCC could be caused by Lactobacillus, Porphyomonas gingivalis, Fusobacterium nucleatum, and Prevotella intermedia. OSCC patients have elevated levels of Prevotella melanogenic, Streptococcus mitis, and Capnocytophaga gingivalis in their saliva, while Rothia, Leptotrichia, Haemophilus, Aggregatibacter, and Neisseria are diminished. b P. gingivalis stimulates the expression of B7-H1 receptors on OSCC cells. B7-H1 activates the development of Tregs, thereby inhibiting effector T cells. S. mitis has been demonstrated to trigger mutations and epithelial hyperplasia by generating acetaldehyde. OSCC oral squamous cell carcinoma, Tregs regulatory T cells
Certain bacterial species have been linked to the development of oral carcinoma. Some periodontal bacteria, such as Porphyomonas gingivalis, Fusobacterium nucleatum, and Prevotella intermedia, may be responsible for OSCC.360 The presence of elevated levels of Lactobacillus,361 Prevotella melanogenic, Streptococcus mitis, and Capnocytophaga gingivalis362 in saliva can also be used to diagnose OSCC. OSCC patients have been found to lack Rothia,363 Leptotrichia,364 Haemophilus,363 Aggregatibacter,365 and Neisseria.361
P. gingivalis can stimulate OSCC cells to express the B7-H1 and B7-DC receptors.366 Expression of B7-H1 activates Tregs development, thereby inhibiting effector T cells. Consequently, the expression of B7-H1 by oral carcinomas may aid their ability to evade immunity.366 Infection with P. gingivalis induces ERK1/2- Ets1, p38/HSP27, and PAR2/NF-KB signaling to promote expression of promatrix metalloproteinase (proMMP-9).360 In this process, P. gingivalis produces gingipains, which are cysteine proteinases with a dual function, and then binds to the PAR2 receptor, resulting in the maturation of the proenzyme MMP-9 into its active form. The destruction of the basement membrane facilitates the invasion and metastasis of OSCC cells through blood vessels and lymphatic systems. Collectively, P. gingivalis contributes to the spread of OSCC.367
The oral microbiome is affected by OSCC risk factors such as smoking, alcohol abuse, and HPV infection.368 There is a correlation between exposure to these risk factors and a shift in diverse bacterial genera of bacteria. Also, oral microbes have been demonstrated to trigger mutations and epithelial hyperplasia by generating acetaldehyde, a cancer-causing derivative of ethanol.369 The ability of different bacterial strains to produce acetaldehyde varies considerably. For instance, S. mitis produces a substantial quantity of acetaldehyde and is an active alcohol dehydrogenase. OSCC has been found to contain elevated levels of S. mitis.370
In addition, OSCC has been observed in immunocompromised patients with chronic mucocutaneous candidiasis and rarely in patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy.371 Candida albicans is more prevalent in the oral cavity of patients with OSCC or leukoplakia compared to those without oral pathology.372,373
Therapeutic interventions and prognostic factors for OPMDs and OSCC
The goal of multidisciplinary therapeutic strategies for OPMDs is to impede the development of OSCC and reduce mortality and morbidity. However, due to the complexity and diversity of OPMDs, there has yet to be a consensus regarding the optimal treatment approach. Meanwhile, OSCC management is crucial for improving survival rates, especially considering the possibility of malignant transformation of OPMDs.
Therapeutic interventions for OPMDs
OPMDs are treated differently depending on their classification. Although there are numerous methods for preventing and managing OL, there is no standard approach.26 According to the severity of the dysplasia, OL is typically treated surgically by excision of the lesions. Nevertheless, a study on leukoplakia surgery reveals that surgery decreases but does not completely eliminate the risk of developing leukoplakia. Also, randomized controlled trials have yet to be conducted to determine whether removing the OL reduces the likelihood of OSCC. According to a previous study, 20% of OL patients may recover if risk factors are eliminated and antifungal therapy is administered. Based on a retrospective study of 94 patients whose OL was surgically removed and 175 patients who did not undergo surgery, OSCC was found to occur in 12% and 4% of patients, respectively.374 Overall, surgery did not safeguard the inhibition of transformation.
As an alternative to OPMD surgery, chemoprevention has the potential to diminish the risk of developing cancer. Chemoprevention is effective when vitamin A, bleomycin, β-carotene, or retinoids are utilized.375,376 Despite their ability to induce therapeutic responses, frequent relapses were observed. Additionally, laser ablation, cryosurgery, and photodynamic therapy are effective treatments for OL. Recent studies have demonstrated statistically significant improvements in the clinical outcomes of OL following erbium: yttrium aluminum garnet laser compared to cold scalpel excision.376
In terms of OE, early treatment is recommended due to its high incidence of malignant transformation.377 Lesions exhibiting severe epithelial dysplasia on excisional or incisional biopsies should be completely removed under microscopic examination.32 Laser surgery is also a recommended treatment option for OE.378 OE can be effectively ablated with CO2 lasers which contributes to a low morbidity rate.32 Using natural agents as chemopreventative agents is an additional method of treating OE. Several natural agents, including curcumin, green tea extract, and Bowman-Birk inhibitor concentrate, are beneficial in the treatment of OE.379,380 Continuous monitoring is advised for lesions exhibiting minimal to moderate dysplasia. In addition, postoperative recurrences are more likely to occur when the initial lesion size is larger, with lesions more than 80 mm2 regarded as significant predictors of recurrence.32
Due to the complicated pathogenesis of OSF, it is difficult to ascertain the most effective treatment. OSF cannot be completely cured by a singular treatment modality. Therefore, the primary objective of OSF treatment is to improve the condition of the oral mucosa, alleviate the burning sensation, and facilitate mouth opening. There are a variety of therapies available, including medical, physical, and surgical treatments. In the initial stages of OSF, cessation of BQ chewing and therapeutic interventions may reduce or eliminate symptoms. In the intermediate or advanced phases of OSF, surgical treatments are necessary to alleviate pain in the mucosal epithelium of the mouth and to facilitate mouth opening.40,381,382
In general, OLP is treated with palliative care as opposed to curative care. Asymptomatic reticular lesions are unnecessary to treat, but continued observation is advised. The main objective of treatment is to diminish inflammation and relieve the symptoms.383 In most cases, topical steroids are required to treat symptomatic OLP. Corticosteroids are used intralesionally to combat erosive OLP. In cases that are more severe or resistant, systemic steroids are prescribed.384,385 Topical calcineurin inhibitors can be administered to patients who do not respond to corticosteroids.384 Furthermore, topical cyclosporine and systemic immunosuppressants have been used to treat OLP.386
Therapeutic interventions for OSCC
OSCC treatments include surgical intervention followed, if necessary, by postoperative radiation or chemotherapy. In most cases, surgery is the first-line treatment for oral carcinomas. In advanced cases, postoperative radiation, chemoradiation, oncogene-targeted therapy, and immunotherapy may be administered (Fig. 6).
Fig. 6.
Treatments of OSCC. a Oncogene-targeted drugs facilitate OSCC chemotherapy. Cetuximab is an antibody that suppresses the EGFR pathway. NVP-BEZ235 is a PI3K/AKT/mTOR pathway inhibitor. Flavopereirine silences the JAK/STAT pathway and upregulates LASP1 expression. FLI-06 inactivates the Notch pathway. In the Wnt/β-catenin pathway, OMP-18R5 inhibits Fzd receptors; PRI-724 interrupts the interaction between β-catenin and CBP; and LGK974 targets PORCN. b Immunotherapy is an alternative treatment. Monoclonal antibodies pembrolizumab, nivolumab, and lgG4 have been authorized to target PD-1. LAG-3 and TIM-3 are blocked by their antibodies. IRX-2, comprising IL-2, IL-1β, IFN-γ, and TNF-α, is proven effective against inflammatory immune suppression cytokines in OSCC. Gemtuzmab ozogamicin promotes the differentiation of MDSCs into mature phenotypes, thereby reducing their immunosuppressive properties. EGFR epidermal growth factor receptors, PD-1 programmed death 1, OSCC oral squamous cell carcinoma
Surgical procedures, such as open procedures, endoscopic procedures, and robotic surgery, are used to treat the majority of oral carcinomas. The purpose of surgical resection is to eliminate sufficient tumor tissue. Inadequate removal of tumor cells increases the likelihood of local and regional recurrences, thereby reducing long-term survival rates.387,388 In oral carcinoma surgery, a 1 cm margin of three-dimensional dissection is deemed appropriate.389 During primary tumor dissection, iodine solution staining is recommended to identify and delineate the dysplastic epithelium. However, a greater resection margin may enhance the risk of esthetic and functional complications in OSCC.390,391
Following the excision of the primary tumor, reconstructive surgery is typically required to restore oral cavity function and head and neck aspect. Postoperative oral disabilities can be reduced through routine surgical reconstruction. The choice of an appropriate method of reconstruction is affected by a variety of factors, such as the features of the primary defects, the medical history and general health of the patient, the skills of the surgeon, and the prognosis. In general, reconstructive procedures adhere to a “reconstructive ladder” consisting of a skin graft followed by a microvascular free flap. In the field of oral reconstruction, unrestricted tissue transfers are regarded as one of the most reliable and widely used techniques. There are various options for free flaps; however, no singular flap is capable of resolving the entire range of oral defects at present.14,392
Radiotherapy is typically administered postoperatively, as irradiated tissue cannot be removed surgically. Tissue fibrosis diminishes the effectiveness of regeneration. Radiotherapy at the primary site is determined by variables such as the primary tumor size, positive surgical margins, and the presence of perineural, lymphatic, and vascular invasion. Nevertheless, it is also common to treat the neck to prevent the possibility of metastasis and recurrence, particularly in lymph nodes with extracapsular dissemination. It is recommended to begin radiotherapy within six weeks after surgery. There are variations in the radiation doses, but an approximate cumulative dose of 60 Gy is typically provided.393 In addition, chemotherapy has recently become a popular adjunct treatment for locally advanced OSCC. Even though chemotherapy is not considered a curative treatment for oral carcinomas, it can be administered prior to surgery or in conjunction with irradiation before or after surgery. Adjuvant chemotherapy and radiotherapy are becoming standard remedies for advanced oral cancers. Other variants, such as daily low-dose and weekly intermediate-dose of CDDP, are also effective in improving survival rates.394,395 Typically, EGRF inhibitors are utilized to treat metastatic HNSCC.14
Furthermore, oncogene-targeted drugs can improve chemotherapy to OSCC.396 The antibody cetuximab has been shown to suppress the EGFR pathway and is approved for HNSCC treatment.397 NVP-BEZ235 inhibits the PI3K/AKT/mTOR pathway to sensitize OSCC cells to infrared radiation and diminish their resistance to radiotherapy.398 Flavopereirine silences JAK/STAT signaling and upregulates LASP1 to block the development of oral carcinomas.399 FLI-06 inactivates the Notch signaling pathway to disrupt the proliferation and self-renewal of oral malignancy cells.400 In the Wnt/β-catenin pathway, OMP-18R5 inhibits Fzd receptors,401 whereas PRI-724 interrupts the interaction between β-catenin and CBP.402 LGK974 targets PORCN, an acyltransferase vital for producing Wnt proteins in various carcinomas.403 Targeted drugs hold great potential for OSCC treatments in the clinic.
Immunotherapy is an alternative treatment. OSCC is considered to be an immunosuppressive disease. It has been suggested that a malfunctioning immune system participates in the development or recurrence of OSCC. Immunotherapy has shown promise in the handling of OSCC. For instance, anti-PD-1/PD-L1 agents promote an immune response against tumors by blocking the suppression signals of the immune checkpoints.404 Pembrolizumab, nivolumab, and lgG4 monoclonal antibodies that target PD-1 have been authorized for therapeutic interventions of metastatic HNSCC based on proven efficacy in clinical trials.405,406 Cytotoxic CD8+ T cells are recruited to suppress the growth of tumors in vivo by blocking LAG-3 with its antibodies.407 Also, anti-TIM-3 therapy appears to inhibit neoplasm growth in in vivo models.408 IRX-2, a multi-cytokine biologic preparation derived from homologous cells and comprised of IL-2, IL-1β, IFN-γ, and TNF-α, is under investigation and has been shown to be effective against inflammatory immune suppressive cytokines. To boost both adaptive and innate immune defenses against tumor cells present in the host, IRX-2 blocks tumor-induced apoptosis of T cells and facilitates their effector function in regional lymph nodes.409 In addition, anti-Treg receptor monoclonal antibodies are being developed to decrease the quantity of Tregs in the tumor microenvironment.410 Gemtuzumab ozogamicin facilitates MDSCs maturation, thereby reducing their immunosuppressive properties.411 An alternative strategy is to inhibit the recruitment of TAMs and MDSCs into the TME by blocking their chemotactic receptors.412
Prognosis of OSCC
As mentioned above, multidisciplinary team care is essential to the effective treatment of OSCC. However, malignant tumors have a poor prognosis, with limited improvements in survival over many decades.413–415 After malignant transformation, it is crucial to evaluate the progression and development of OSCC in terms of angiogenesis, tumor budding, perineural invasion, staging, HPV status, and the presence of specific biomarkers. These prognostic factors help to assess the mortality rate of OSCC patients and guide treatment decisions (Table 3).416–418
Table 3.
Prognostic factors for OSCC
| Prognostic factors | Indicators | Exhibitions | Outcomes |
|---|---|---|---|
| Staging |
Clinical stages: ▪ Stage I ▪ Stage II ▪ Stage III ▪ Stage IV |
5-year survival rate: ▪ 79.8% ▪ 70.0% ▪ 57.6% ▪ 53.9% |
▪ Poor prognosis |
| Biomarkers |
▪ G3BP1 ▪ B7-H6 ▪ FAM3C |
▪ Upregulation | ▪ Poor prognosis |
| Angiogenesis |
▪ MVD ▪ HMGA2 ▪ Id-1 |
▪ Upregulation | ▪ Poor prognosis |
| HPV status |
▪ HPV+ ▪ p16+ |
▪ Upregulation | ▪ Favorable prognosis |
| Tumor budding |
▪ Snail ▪ Twist |
▪ Upregulation | ▪ Poor prognosis |
| Perineural invasion |
▪ miR-21/phosphatase ▪ Tensin homologs ▪ MMP-2 ▪ NGF ▪ Tyrosine kinase A |
▪ Upregulation | ▪ Poor prognosis |
Clinicopathological factors
Several immunohistochemical staining protocols can be used on patient tissue samples to identify clinicopathological alterations such as angiogenesis, tumor budding, perineural invasion, and staging. Oral malignancies have the capability of inducing blood vessel formation, which is crucial to the dissemination of tumors.
Angiogenesis in tumors is generally evaluated by quantifying the blood vessels (microvessel density, MVD) present in regions of the tissue.419 In addition to immunohistochemical staining techniques for identifying vessels, angiogenesis can be investigated by other methods, such as the Chalkley method and flow cytometry.420 The presence of marked angiogenesis was related to an elevated possibility of nodal metastases and may indicate the requirement for intensified adjunctive treatment following surgery. Moreover, angiogenesis in OSCC is associated with the parameters of size (T) and lymph node involvement (N), a reliable indicator of tumor relapse.11 HMGA2 regulates OSCC angiogenesis-related genes and correlates with both distant and lymph node metastasis.421 Patients with high HMGA2 expression have a worse 5-year survival rate. HMGA2-high samples exhibit more CD34-stained blood vessels and higher expression of VEGF-A, VEGF-C, and fibroblast growth factor (FGF)-2, which are associated with new blood vessel formation in vitro.421 In addition, Id-1 expression is associated with intratumoral MVD, and there is a positive correlation between Id-1 overexpression and angiogenesis as well as poor clinical outcomes in OSCC.422
There is a significant correlation between tumor budding and shorter overall survival.423,424 Also, tumor budding correlates positively with lymph node metastasis.425,426 A study revealed that 33.9% of OSCC specimens displayed tumor budding, 58.9% of 56 OSCC patients have died, and the 5-year survival rate is 44.6%.423 However, no other clinicopathological factors are associated with tumor budding. Moreover, tumor budding is correlated with a rise in Snail expression and a tendency toward higher Twist expression. 46.4% of 56 OSCC specimens exhibit a positive expression of Snail, and 32.1% display a positive expression of Twist.423 Specifically, Snail is primarily located in cytoplasm and nuclei, whereas Twist is only present in a small number of nuclei. Expression of Snail and Twist are associated with lymph node metastasis in OSCC.423 However, well-differentiated OSCC expresses significantly less Twist, and there is no correlation between Snail or Twist expression and other clinicopathological factors. The overall survival rate of patients expressing Snail or Twist decreases dramatically.423 Taken together, tumor budding is strongly related to an unfavorable prognosis in patients with OSCC and correlates with the process of EMT.
Perineural invasion (PNI) may influence the progression of malignant cells and lead to poor prognosis. PNI has been detected in 17.4% of OSCC samples.427 In OSCC patients, miR-21/phosphatase and tensin homologs are abundant, and their dysregulation correlates with PNI and a poor prognosis.428 MMP-2 expressed by fibroblasts in the microenvironment of PNI is associated with a poorer prognosis in the treatment of OSCC and may be a contributing factor in OSCC PNI.429 Moreover, tumors with PNI have substantially higher levels of nerve growth factor (NGF) and tyrosine kinase than tumors without PNI (84% and 92%, respectively). PNI is associated with advanced carcinomas and worse DSS. Therefore, PNI in OSCC can be predicted by a high expression of NGF and tyrosine kinase A. The overexpression of PNI and NGF can also lead to pain in OSCC patients.427 Taken together, the expression of PNI and NGF is capable of determining the aggressiveness and prognosis of oral cancers in patients.427
Typically, prognosis has been correlated with the stage of the tumor. The 5-year survival rate for oral cancer patients is 64.4% overall, and 79.8%, 70.0%, 57.6%, and 53.9% for stages I–IV, respectively, with clinical stages II-IV having a reduced survival.430 In a research of 274 patients with oral malignancies, the survival rate among them after 12, 24, 36, 48, and 60 months is approximately 80%, 60%, 46%, 40%, and 39%, respectively.431 However, over 60% of oral carcinomas are detected in the advanced phases.432 In conclusion, the low survival rate obtained can be attributed primarily to the high proportion of OSCC cases diagnosed at an advanced stage.
Biological factors
Several biological indicators can be used to assess the progression of oral malignancies in patients. Accumulating evidence suggests a causal relationship between HPV and OSCC. Independent of cigarette smoking and alcohol abuse, HPV is linked to an elevated possibility of suffering from oral malignancies.433 This association applies to high-risk HPV samples, including subtypes 16, 18, 33, and 35.434 Over 80% of HPV+ OSCC may be due to HPV-16.435
HPV infection categorizes tumors into two distinct groups with varying prognoses and therapeutic implications.436 Generally, HPV+OSCC patients have a better treatment response, a higher two-year overall survival rate, a reduced disease progression risk and an improved prognosis, and lower death and recurrence rate than HPV- patients.437 p16 is one of the most investigated prognostic biomarkers of OSCC. Of note, HPV+ and p16+ patients have a higher overall survival rate than HPV- or HPV+ but p16- patients.438 When p53 interacts with E6 encoded by carcinogenic types of HPV (such as HPV-16 and HPV-18), it is proteolyzed by ubiquitin-dependent proteases.434 There is a significant difference between the level of wild-type p53 in HPV+ neoplasms and the elevated possibility of p53 mutations in HPV- tumors, which is related to a favorable prognosis for HPV+ OSSC patients.53
Additional OSCC prognostic biomarkers include G3BP1, B7-H6, and FAM3C. Patients with overexpressed G3BP1 mRNA exhibit a lower overall survival rate. In OSCC, mRNA and protein levels of G3BP1 are significantly higher than in normal tissues.439 G3BP1 has a direct relationship with Ki67 and an inverse relationship with Cleaved-caspase 3. The correlation between CD4+ T-cell infiltration and G3BP1 mRNA levels is positive. Enrichment analysis reveals that G3BP1 participates in helicase/catalytic/ATPase activity functions as well as spliceosome/RNA transport/cell cycle pathways and can be used as a biomarker to predict the prognosis of OSCC.440 Moreover, B7-H6 is identified as a distinct prognostic factor in OSCC involving DFS and CSS. OSCC tissues express significantly more B7-H6 protein than normal oral mucosa. B7-H6 expression correlates with differentiation; OSCC patients with less B7-H6 expression or more differentiated tumor tissue may have a better prognosis.441 Family with sequence similarity 3 member C (FAM3C) is an additional prognostic indicator that is essential for EMT. Immunohistochemical staining of OSCC samples with FAM3C, EMT markers, CSC markers, and co-inhibitory immune checkpoints is utilized to evaluate FAM3C levels and pathological features of OSCC. Compared to healthy mucosa and epithelial dysplasia, the level of FAM3C in OSCC specimens increases, and patients with a higher FAM3C expression are more likely to have a poor prognosis.442 In addition, the expression of FAM3C correlates positively with immune checkpoints such as PD-L1, VISTA, and B7-H4, the EMT marker Slug, and the CSC markers SOX2 and ALDH1.442
Conclusion and perspectives
OSCC is typically associated with oral mucosa and a variety of risk factors. To mitigate risks, it is now understood that electronic cigarettes must be regulated similarly to traditional cigarettes.443,444 It is also necessary to limit the consumption of alcohol445 and BQ.446 Given that HPV is an influential risk factor for OSCC,447 HPV vaccinations should be promoted globally.448 However, there is insufficient molecular evidence to support the hypothesis that HPV+ OSCC is driven by HPV, as HPV is not inherently an indicator of a biologically active virus.449 Besides, OSCC risk factors include Epstein–Barr virus (EBV), which makes early diagnosis of OSCC patients essential.450
Due to these risk factors, OPMDs may develop prior to the onset of OSCC. To better stratify patients and follow their risk of malignancy, pathology assessments of OPMDs must go beyond subjective evaluation and be standardized.451 Furthermore, additional data from epidemiologic studies are required to elucidate the population of patients at current and future risk for OPMDs and subsequently the progression of OSCC.452 Future research on the incidence of OPMDs and OSCC will necessitate advances in molecular biology and genetics to uncover more distinct indicators, as interventional strategies based solely on histopathology are insufficient.453 In addition, more exploration into the aberrant metabolism of OPMDs and OSCC may shed new light on their pathogenesis.109,454
Through early diagnosis, oral cancer survival rates could reach up to 80%-90%.455–460 It will be feasible to make more sensitive and specific diagnoses of premalignancy and cancer through the development and rigorous testing of new diagnostic tools.461–463 Specifically, accelerated advancements in artificial intelligence (AI) hold promise for mass oral cancer screening. Currently, research is being conducted to develop AI-based technologies for the identification of oral malignancies with improved sensitivity and specificity, and in the future, the use of AI-based mobile applications will be advantageous for both frontline healthcare workers and the general public.464–468 These technological advances may allow for the early detection and management of suspicious lesions.469,470 Further explorations into the mechanisms of oncogenesis will assist and promote the accuracy of early diagnosis. Through the availability of a vast amount of information from transcriptomics, genomics, proteomics, epigenomics, and metabolomics, high-throughput sequencing technology will enable the development of novel therapeutic approaches for the treatment of OSCC. Furthermore, nanomedicine will provide efficient OSCC therapies by generating multiple synergetic therapeutics.471–479 Already, oral cancer treatment has undergone a substantial transformation, leading to improved patient outcomes.480 A multidisciplinary team will be required to manage these tumors, consisting not only of surgeons and oncologists, but also of specialists evaluating the nutritional, mental, social, and oral status of the individuals before, during, and after treatment.481
Acknowledgements
This work was supported by the National Key Research and Development Project of China (2020YFA0509400), Guangdong Basic and Applied Basic Research Foundation(2019B030302012), National Natural Science Foundation of China (81821002, 82130082), 1·3·5 project for disciplines of excellence (ZYGD22007, ZYGC21004). Figures were created with BioRender (www.biorender.com).
Author contributions
C.H. and J.T. conceived and organized the manuscript. Y.T. and Z.W. wrote the manuscript. Y.T. drew the figures. M.X. organized the tables. C.H., J.T., B.L., E.C.N., Z.H., and S.Q. reviewed and edited the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yunhan Tan, Zhihan Wang
Contributor Information
Jing Tang, Email: tangjing@wchscu.cn.
Canhua Huang, Email: hcanhua@hotmail.com.
References
- 1.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac. Surg. Clin. 2014;26:123–141. doi: 10.1016/j.coms.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng JH, Iyer NG, Tan M-H, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39:297–304. doi: 10.1002/hed.24589. [DOI] [PubMed] [Google Scholar]
- 3.Mody MD, Rocco JW, Yom SS, Haddad RI, Saba NF. Head and neck cancer. Lancet. 2021;398:2289–2299. doi: 10.1016/S0140-6736(21)01550-6. [DOI] [PubMed] [Google Scholar]
- 4.Romano A, et al. Noninvasive imaging methods to improve the diagnosis of oral carcinoma and its precursors: state of the art and proposal of a three-step diagnostic process. Cancers. 2021;13:2864. doi: 10.3390/cancers13122864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safi A-F, et al. Clinicopathological parameters affecting nodal yields in patients with oral squamous cell carcinoma receiving selective neck dissection. J. Cranio-Maxillofac. Surg. 2017;45:2092–2096. doi: 10.1016/j.jcms.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Linsen SS, Gellrich N-C, Krüskemper G. Age-and localization-dependent functional and psychosocial impairments and health related quality of life six months after OSCC therapy. Oral Oncol. 2018;81:61–68. doi: 10.1016/j.oraloncology.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Meier JK, et al. Health-related quality of life: a retrospective study on local vs. microvascular reconstruction in patients with oral cancer. BMC Oral Health. 2019;19:1–8. doi: 10.1186/s12903-019-0760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maymone MBC, et al. Premalignant and malignant oral mucosal lesions: Clinical and pathological findings. J. Am. Acad. Dermatol. 2019;81:59–71. doi: 10.1016/j.jaad.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 9.Odell E, Kujan O, Warnakulasuriya S, Sloan P. Oral epithelial dysplasia: recognition, grading and clinical significance. Oral Dis. 2021;27:1947–1976. doi: 10.1111/odi.13993. [DOI] [PubMed] [Google Scholar]
- 10.Brandizzi, D., Gandolfo, M., Velazco, M. L., Cabrini, R. L. & Lanfranchi, H. Clinical features and evolution of oral cancer: a study of 274 cases in Buenos Aires, Argentina. Med. Oral Patol. Oral Cir. Bucal. 13, E544-8 (2008). [PubMed]
- 11.Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncol. 2010;46:414–417. doi: 10.1016/j.oraloncology.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Scully C, Bagan JV. Oral squamous cell carcinoma: overview of current understanding of aetiopathogenesis and clinical implications. Oral Dis. 2009;15:388–399. doi: 10.1111/j.1601-0825.2009.01563.x. [DOI] [PubMed] [Google Scholar]
- 13.Harada H, et al. Characteristics of oral squamous cell carcinoma focusing on cases unaffected by smoking and drinking: a multicenter retrospective study. Head Neck. 2023;45:1812–1822. doi: 10.1002/hed.27398. [DOI] [PubMed] [Google Scholar]
- 14.Omura K. Current status of oral cancer treatment strategies: surgical treatments for oral squamous cell carcinoma. Int. J. Clin. Oncol. 2014;19:423–430. doi: 10.1007/s10147-014-0689-z. [DOI] [PubMed] [Google Scholar]
- 15.Jerjes W, et al. Clinicopathological parameters, recurrence, locoregional and distant metastasis in 115 T1-T2 oral squamous cell carcinoma patients. Head Neck Oncol. 2010;2:1–21. doi: 10.1186/1758-3284-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warnakulasuriya S. Oral potentially malignant disorders: A comprehensive review on clinical aspects and management. Oral Oncol. 2020;102:104550. doi: 10.1016/j.oraloncology.2019.104550. [DOI] [PubMed] [Google Scholar]
- 17.Yap T, et al. Non-invasive screening of a microRNA-based dysregulation signature in oral cancer and oral potentially malignant disorders. Oral Oncol. 2019;96:113–120. doi: 10.1016/j.oraloncology.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Chuang S-L, et al. Malignant transformation to oral cancer by subtype of oral potentially malignant disorder: a prospective cohort study of Taiwanese nationwide oral cancer screening program. Oral Oncol. 2018;87:58–63. doi: 10.1016/j.oraloncology.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Warnakulasuriya S. Clinical features and presentation of oral potentially malignant disorders. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018;125:582–590. doi: 10.1016/j.oooo.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Dionne KR, Warnakulasuriya S, Binti Zain R, Cheong SC. Potentially malignant disorders of the oral cavity: Current practice and future directions in the clinic and laboratory. Int. J. Cancer. 2015;136:503–515. doi: 10.1002/ijc.28754. [DOI] [PubMed] [Google Scholar]
- 21.Tarakji B. Dentists’ perception of oral potentially malignant disorders. Int. Dent. J. 2022;72:414–419. doi: 10.1016/j.identj.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerr AR, Lodi G. Management of oral potentially malignant disorders. Oral Dis. 2021;27:2008–2025. doi: 10.1111/odi.13980. [DOI] [PubMed] [Google Scholar]
- 23.Mello FW, et al. Prevalence of oral potentially malignant disorders: a systematic review and meta‐analysis. J. Oral Pathol. Med. 2018;47:633–640. doi: 10.1111/jop.12726. [DOI] [PubMed] [Google Scholar]
- 24.Warnakulasuriya S, Johnson NW, van der Waal I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J. Oral Pathol. Med. 2007;36:575–580. doi: 10.1111/j.1600-0714.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- 25.Villa A, Sonis S. Oral leukoplakia remains a challenging condition. Oral Dis. 2018;24:179–183. doi: 10.1111/odi.12781. [DOI] [PubMed] [Google Scholar]
- 26.Holmstrup P, Dabelsteen E. Oral leukoplakia—to treat or not to treat. Oral Dis. 2016;22:494–497. doi: 10.1111/odi.12443. [DOI] [PubMed] [Google Scholar]
- 27.Carrard VC, van der Waal I. A clinical diagnosis of oral leukoplakia; A guide for dentists. Med. Oral Patol. Oral Cir. Bucal. 2018;23:e59. doi: 10.4317/medoral.22292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warnakulasuriya S, et al. Oral potentially malignant disorders: a consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2021;27:1862–1880. doi: 10.1111/odi.13704. [DOI] [PubMed] [Google Scholar]
- 29.Speight PM, Khurram SA, Kujan O. Oral potentially malignant disorders: risk of progression to malignancy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018;125:612–627. doi: 10.1016/j.oooo.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Pentenero M, Meleti M, Vescovi P, Gandolfo S. Oral proliferative verrucous leucoplakia: are there particular features for such an ambiguous entity? A systematic review. Br. J. Dermatol. 2014;170:1039–1047. doi: 10.1111/bjd.12853. [DOI] [PubMed] [Google Scholar]
- 31.Cabay RJ, Morton TH, Jr, Epstein JB. Proliferative verrucous leukoplakia and its progression to oral carcinoma: a review of the literature. J. Oral Pathol. Med. 2007;36:255–261. doi: 10.1111/j.1600-0714.2007.00506.x. [DOI] [PubMed] [Google Scholar]
- 32.Yang S-W, Lee Y-S, Chang L-C, Hsieh T-Y, Chen T-A. Outcome of excision of oral erythroplakia. Br. J. Oral Maxillofac. Surg. 2015;53:142–147. doi: 10.1016/j.bjoms.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Boy, S. C. Leukoplakia and erythroplakia of the oral mucosa—a brief overview. SADJ67, 558–560 (2012). [PubMed]
- 34.Holmstrup P. Oral erythroplakia—what is it? Oral Dis. 2018;24:138–143. doi: 10.1111/odi.12709. [DOI] [PubMed] [Google Scholar]
- 35.Yang S-W, et al. Clinical characteristics of narrow-band imaging of oral erythroplakia and its correlation with pathology. BMC Cancer. 2015;15:1–8. doi: 10.1186/s12885-015-1422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boy SC. Leukoplakia and erythroplakia of the oral mucosa-a brief overview: clinical review. SADJ. 2012;67:558–560. [PubMed] [Google Scholar]
- 37.Tilakaratne WM, Ekanayaka RP, Warnakulasuriya S. Oral submucous fibrosis: a historical perspective and a review on etiology and pathogenesis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016;122:178–191. doi: 10.1016/j.oooo.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Qin X, Ning Y, Zhou L, Zhu Y. Oral submucous fibrosis: etiological mechanism, malignant transformation, therapeutic approaches and targets. Int. J. Mol. Sci. 2023;24:4992. doi: 10.3390/ijms24054992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y-W, Shih Y-H, Fuh L-J, Shieh T-M. Oral submucous fibrosis: a review on biomarkers, pathogenic mechanisms, and treatments. Int. J. Mol. Sci. 2020;21:7231. doi: 10.3390/ijms21197231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Q, Li H, Chen J, Wang Y, Tang Z. Oral submucous fibrosis in Asian countries. J. Oral Pathol. Med. 2020;49:294–304. doi: 10.1111/jop.12924. [DOI] [PubMed] [Google Scholar]
- 41.Al‐Hassiny A, et al. Upregulation of angiogenesis in oral lichen planus. J. Oral Pathol. Med. 2018;47:173–178. doi: 10.1111/jop.12665. [DOI] [PubMed] [Google Scholar]
- 42.De Rossi SS, Ciarrocca K. Oral lichen planus and lichenoid mucositis. Dent. Clin. 2014;58:299–313. doi: 10.1016/j.cden.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Aghbari SMH, et al. Malignant transformation of oral lichen planus and oral lichenoid lesions: a meta-analysis of 20095 patient data. Oral Oncol. 2017;68:92–102. doi: 10.1016/j.oraloncology.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 44.González‐Moles MÁ, et al. Worldwide prevalence of oral lichen planus: a systematic review and meta‐analysis. Oral Dis. 2021;27:813–828. doi: 10.1111/odi.13323. [DOI] [PubMed] [Google Scholar]
- 45.Nogueira PA, Carneiro S, Ramos‐e‐Silva M. Oral lichen planus: an update on its pathogenesis. Int. J. Dermatol. 2015;54:1005–1010. doi: 10.1111/ijd.12918. [DOI] [PubMed] [Google Scholar]
- 46.Ahmadi N, et al. Association of PD-L1 expression in oral squamous cell carcinoma with smoking, sex, and p53 expression. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019;128:631–638. doi: 10.1016/j.oooo.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Xu J, Wang L, Liu C, Wang H. The role of cigarette smoking and alcohol consumption in the differentiation of oral squamous cell carcinoma for the males in China. J. Cancer Res. Therapeut. 2015;11:141–145. doi: 10.4103/0973-1482.137981. [DOI] [PubMed] [Google Scholar]
- 48.Dong J, Thrift AP. Alcohol, smoking and risk of oesophago-gastric cancer. Best Pract. Res. Clin. Gastroenterol. 2017;31:509–517. doi: 10.1016/j.bpg.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Matejcic M, Gunter MJ, Ferrari P. Alcohol metabolism and oesophageal cancer: a systematic review of the evidence. Carcinogenesis. 2017;38:859–872. doi: 10.1093/carcin/bgx067. [DOI] [PubMed] [Google Scholar]
- 50.Ghantous Y, Schussel JL, Brait M. Tobacco and alcohol induced epigenetic changes in oral carcinoma. Curr. Opin. Oncol. 2018;30:152. doi: 10.1097/CCO.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P, et al. Molecular mechanisms of malignant transformation of oral submucous fibrosis by different betel quid constituents—does fibroblast senescence play a role? Int. J. Mol. Sci. 2022;23:1637. doi: 10.3390/ijms23031637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ko AM-S, Lee C-H, Ko Y-C. Betel quid–associated cancer: Prevention strategies and targeted treatment. Cancer Lett. 2020;477:60–69. doi: 10.1016/j.canlet.2020.02.030. [DOI] [PubMed] [Google Scholar]
- 53.Singh V, et al. p16 and p53 in HPV‐positive versus HPV‐negative oral squamous cell carcinoma: do pathways differ? J. Oral Pathol. Med. 2017;46:744–751. doi: 10.1111/jop.12562. [DOI] [PubMed] [Google Scholar]
- 54.Purwanto DJ, et al. The prevalence of oral high‐risk HPV infection in Indonesian oral squamous cell carcinoma patients. Oral Dis. 2020;26:72–80. doi: 10.1111/odi.13221. [DOI] [PubMed] [Google Scholar]
- 55.Wolfer S, Foos T, Kunzler A, Ernst C, Schultze-Mosgau S. Association of the preoperative body mass index with postoperative complications after treatment of oral squamous cell carcinoma. J. Oral Maxillofac. Surg. 2018;76:1800–1815. doi: 10.1016/j.joms.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 56.Eckert AW, et al. Clinical relevance of the tumor microenvironment and immune escape of oral squamous cell carcinoma. J. Transl. Med. 2016;14:1–13. doi: 10.1186/s12967-016-0828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ram H, et al. Oral cancer: risk factors and molecular pathogenesis. J. Maxillofac. Oral Surg. 2011;10:132–137. doi: 10.1007/s12663-011-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng T. Chemical evaluation of electronic cigarettes. Tob. Control. 2014;23:ii11–ii17. doi: 10.1136/tobaccocontrol-2013-051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7:149–156. doi: 10.1016/S1470-2045(06)70577-0. [DOI] [PubMed] [Google Scholar]
- 60.Jeng J-H, Chang MC, Hahn LJ. Role of areca nut in betel quid-associated chemical carcinogenesis: current awareness and future perspectives. Oral Oncol. 2001;37:477–492. doi: 10.1016/s1368-8375(01)00003-3. [DOI] [PubMed] [Google Scholar]
- 61.Lagoa, R. et al. (eds.). Molecular Mechanisms Linking Environmental Toxicants to Cancer Development: Significance for Protective Interventions with Polyphenols (Elsevier, 2020). [DOI] [PubMed]
- 62.Rehman K, Fatima F, Waheed I, Akash MSH. Prevalence of exposure of heavy metals and their impact on health consequences. J. Cell. Biochem. 2018;119:157–184. doi: 10.1002/jcb.26234. [DOI] [PubMed] [Google Scholar]
- 63.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 64.Kouketsu A, et al. Detection of human papillomavirus infection in oral squamous cell carcinoma: a cohort study of Japanese patients. J. Oral Pathol. Med. 2016;45:565–572. doi: 10.1111/jop.12416. [DOI] [PubMed] [Google Scholar]
- 65.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer. 2014;14:342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 66.Stechschulte SA, Kirsner RS, Federman DG. Vitamin D: bone and beyond, rationale and recommendations for supplementation. Am. J. Med. 2009;122:793–802. doi: 10.1016/j.amjmed.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 67.Ribeiro FAP, Noguti J, Oshima CTF, Ribeiro DA. Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: a promising approach. Anticancer Res. 2014;34:1547–1552. [PubMed] [Google Scholar]
- 68.Liu F, Millar S. Wnt/β-catenin signaling in oral tissue development and disease. J. Dent. Res. 2010;89:318–330. doi: 10.1177/0022034510363373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J-S, et al. Honokiol inhibits sphere formation and xenograft growth of oral cancer side population cells accompanied with JAK/STAT signaling pathway suppression and apoptosis induction. BMC Cancer. 2016;16:1–13. doi: 10.1186/s12885-016-2265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali J, et al. Genetic etiology of oral cancer. Oral Oncol. 2017;70:23–28. doi: 10.1016/j.oraloncology.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 71.Simpson DR, Mell LK, Cohen EEW. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2015;51:291–298. doi: 10.1016/j.oraloncology.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 72.Speight PM, et al. Screening for oral cancer—a perspective from the Global Oral Cancer Forum. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017;123:680–687. doi: 10.1016/j.oooo.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, et al. Attenuated TRAF3 fosters activation of alternative NF-κB and reduced expression of antiviral interferon, TP53, and RB to promote HPV-positive head and neck cancers decreased TRAF3 promotes HPV+ HNSCC. Cancer Res. 2018;78:4613–4626. doi: 10.1158/0008-5472.CAN-17-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vijayalakshmi N, Selvaluxmi G, Majhi U, Rajkumar T. Alterations found in p16/RB/Cyclin D1 pathway in the dysplastic and malignant cervical epithelium. Oncol. Res. 2007;16:527–533. doi: 10.3727/096504007783438367. [DOI] [PubMed] [Google Scholar]
- 75.Shridhar K, et al. DNA methylation markers for oral pre-cancer progression: A critical review. Oral Oncol. 2016;53:1–9. doi: 10.1016/j.oraloncology.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, et al. The histone demethylase LSD1 is a novel oncogene and therapeutic target in oral cancer. Cancer Lett. 2016;374:12–21. doi: 10.1016/j.canlet.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 77.Portney BA, et al. ZSCAN4 facilitates chromatin remodeling and promotes the cancer stem cell phenotype. Oncogene. 2020;39:4970–4982. doi: 10.1038/s41388-020-1333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irimie AI, et al. A looking-glass of non-coding RNAs in oral cancer. Int. J. Mol. Sci. 2017;18:2620. doi: 10.3390/ijms18122620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Elmusrati A, Wang J, Wang C-Y. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int. J. Oral Sci. 2021;13:1–11. doi: 10.1038/s41368-021-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haas L, et al. Acquired resistance to anti-MAPK targeted therapy confers an immune-evasive tumor microenvironment and cross-resistance to immunotherapy in melanoma. Nat. Cancer. 2021;2:693–708. doi: 10.1038/s43018-021-00221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teles FR, Alawi F, Castilho RM, Wang Y. Association or causation? Exploring the oral microbiome and cancer links. J. Dent. Res. 2020;99:1411–1424. doi: 10.1177/0022034520945242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salahshourifar I, Vincent-Chong VK, Kallarakkal TG, Zain RB. Genomic DNA copy number alterations from precursor oral lesions to oral squamous cell carcinoma. Oral Oncol. 2014;50:404–412. doi: 10.1016/j.oraloncology.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 83.Chai, A. W. Y., Lim, K. P. & Cheong, S. C. in Seminars in Cancer Biology. 71–83 (Elsevier, 2020). 10.1016/j.semcancer.2019.09.011. [DOI] [PubMed]
- 84.Ishida K, et al. Current mouse models of oral squamous cell carcinoma: genetic and chemically induced models. Oral Oncol. 2017;73:16–20. doi: 10.1016/j.oraloncology.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 85.Fadlullah MZH, et al. Genetically-defined novel oral squamous cell carcinoma cell lines for the development of molecular therapies. Oncotarget. 2016;7:27802. doi: 10.18632/oncotarget.8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gillison ML, et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 2019;29:1–17. doi: 10.1101/gr.241141.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Q, Tie Y, Alu A, Ma X, Shi H. Targeted therapy for head and neck cancer: Signaling pathways and clinical studies. Signal. Transduct. Target. Ther. 2023;8:31. doi: 10.1038/s41392-022-01297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chung CH, et al. Genomic alterations in human epidermal growth factor receptor 2 (HER2/ERBB2) in head and neck squamous cell carcinoma. Head Neck. 2017;39:E15–E19. doi: 10.1002/hed.24587. [DOI] [PubMed] [Google Scholar]
- 89.Concha-Benavente F, et al. Identification of the cell-intrinsic and-extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res. 2016;76:1031–1043. doi: 10.1158/0008-5472.CAN-15-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alsahafi E, et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10:540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laimer K, et al. High EGFR expression predicts poor prognosis in patients with squamous cell carcinoma of the oral cavity and oropharynx: a TMA-based immunohistochemical analysis. Oral Oncol. 2007;43:193–198. doi: 10.1016/j.oraloncology.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 92.Solomon, B., Young, R. J. & Rischin, D. in Seminars in Cancer Biology. 228–240 (Elsevier, 2018). 10.1016/j.semcancer.2018.01.008. [DOI] [PubMed]
- 93.Szturz P, Vermorken JB. Management of recurrent and metastatic oral cavity cancer: Raising the bar a step higher. Oral Oncol. 2020;101:104492. doi: 10.1016/j.oraloncology.2019.104492. [DOI] [PubMed] [Google Scholar]
- 94.Brand TM, et al. Nuclear EGFR as a molecular target in cancer. Radiother. Oncol. 2013;108:370–377. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 95.Vouri M, et al. Axl-EGFR receptor tyrosine kinase hetero-interaction provides EGFR with access to pro-invasive signalling in cancer cells. Oncogenesis. 2016;5:e266–e266. doi: 10.1038/oncsis.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Su W, et al. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer. 2019;19:1–13. doi: 10.1186/s12885-019-5593-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Liang J, Liu J, Deng Z, Liu Z, Liang L. DLX6 promotes cell proliferation and survival in oral squamous cell carcinoma. Oral Dis. 2022;28:87–96. doi: 10.1111/odi.13728. [DOI] [PubMed] [Google Scholar]
- 98.Jin H, Zhang L, Wang S, Qian L. BST2 promotes growth and induces gefitinib resistance in oral squamous cell carcinoma via regulating the EGFR pathway. Arch. Med. Sci. 2021;17:1772. doi: 10.5114/aoms.2019.86183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cochicho D, et al. PIK3CA gene mutations in HNSCC: systematic review and correlations with HPV status and patient survival. Cancers. 2022;14:1286. doi: 10.3390/cancers14051286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat. Rev. Clin. Oncol. 2015;12:11–26. doi: 10.1038/nrclinonc.2014.192. [DOI] [PubMed] [Google Scholar]
- 101.Peng C-H, et al. Somatic copy number alterations detected by ultra-deep targeted sequencing predict prognosis in oral cavity squamous cell carcinoma. Oncotarget. 2015;6:19891. doi: 10.18632/oncotarget.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marquard FE, Jücker M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem. Pharmacol. 2020;172:113729. doi: 10.1016/j.bcp.2019.113729. [DOI] [PubMed] [Google Scholar]
- 103.Ghafouri-Fard S, et al. Role of PI3K/AKT pathway in squamous cell carcinoma with an especial focus on head and neck cancers. Cancer Cell Int. 2022;22:1–27. doi: 10.1186/s12935-022-02676-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 105.Ito K, et al. Inhibition of Nox1 induces apoptosis by attenuating the AKT signaling pathway in oral squamous cell carcinoma cell lines. Oncol. Rep. 2016;36:2991–2998. doi: 10.3892/or.2016.5068. [DOI] [PubMed] [Google Scholar]
- 106.Kupferman ME, Myers JN. Molecular biology of oral cavity squamous cell carcinoma. Otolaryngol. Clin. North Am. 2006;39:229–247. doi: 10.1016/j.otc.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 107.Zhou Q, et al. ATP promotes oral squamous cell carcinoma cell invasion and migration by activating the PI3K/AKT pathway via the P2Y2-Src-EGFR axis. ACS Omega. 2022;7:39760–39771. doi: 10.1021/acsomega.2c03727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis. 2020;11:682. doi: 10.1038/s41419-020-02851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang X, et al. ITGB2-mediated metabolic switch in CAFs promotes OSCC proliferation by oxidation of NADH in mitochondrial oxidative phosphorylation system. Theranostics. 2020;10:12044. doi: 10.7150/thno.47901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang H, et al. Elevated expression of zinc finger protein 703 promotes cell proliferation and metastasis through PI3K/AKT/GSK-3β signalling in oral squamous cell carcinoma. Cell. Physiol. Biochem. 2017;44:920–934. doi: 10.1159/000485360. [DOI] [PubMed] [Google Scholar]
- 111.Zhang H, Sun J-D, Yan L, Zhao X-P. PDGF-D/PDGFRβ promotes tongue squamous carcinoma cell (TSCC) progression via activating p38/AKT/ERK/EMT signal pathway. Biochem. Biophys. Res. Commun. 2016;478:845–851. doi: 10.1016/j.bbrc.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 112.Jiang X, et al. Elevated autocrine chemokine ligand 18 expression promotes oral cancer cell growth and invasion via Akt activation. Oncotarget. 2016;7:16262. doi: 10.18632/oncotarget.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li P, Xiao LY, Tan H. Muc-1 promotes migration and invasion of oral squamous cell carcinoma cells via PI3K-Akt signaling. Int. J. Clin. Exp. Pathol. 2015;8:10365. [PMC free article] [PubMed] [Google Scholar]
- 114.Yang H, et al. FoxM1 promotes epithelial–mesenchymal transition, invasion, and migration of tongue squamous cell carcinoma cells through a c-Met/AKT-dependent positive feedback loop. Anti-Cancer Drugs. 2018;29:216. doi: 10.1097/CAD.0000000000000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal. Transduct. Target. Ther. 2021;6:402. doi: 10.1038/s41392-021-00791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geiger JL, Grandis JR, Bauman JE. The STAT3 pathway as a therapeutic target in head and neck cancer: Barriers and innovations. Oral Oncol. 2016;56:84–92. doi: 10.1016/j.oraloncology.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Avalle L, et al. STAT3 localizes to the ER, acting as a gatekeeper for ER-mitochondrion Ca2+ fluxes and apoptotic responses. Cell Death Differ. 2019;26:932–942. doi: 10.1038/s41418-018-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bu LL, et al. STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. J. Dent. Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 120.Tan J, Xiang L, Xu G. LncRNA MEG3 suppresses migration and promotes apoptosis by sponging miR‐548d‐3p to modulate JAK–STAT pathway in oral squamous cell carcinoma. IUBMB Life. 2019;71:882–890. doi: 10.1002/iub.2012. [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, et al. Long non-coding RNA P4713 contributes to the malignant phenotypes of oral squamous cell carcinoma by activating the JAK/STAT3 pathway. Int. J. Clin. Exp. Pathol. 2017;10:10947. [PMC free article] [PubMed] [Google Scholar]
- 122.Cho YA, et al. Alteration status and prognostic value of MET in head and neck squamous cell carcinoma. J. Cancer. 2016;7:2197. doi: 10.7150/jca.16686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seiwert TY, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009;69:3021–3031. doi: 10.1158/0008-5472.CAN-08-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hartmann S, Bhola NE, Grandis JR. HGF/Met signaling in head and neck cancer: impact on the tumor microenvironment. Clin. Cancer Res. 2016;22:4005–4013. doi: 10.1158/1078-0432.CCR-16-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moosavi F, Giovannetti E, Saso L, Firuzi O. HGF/MET pathway aberrations as diagnostic, prognostic, and predictive biomarkers in human cancers. Crit. Rev. Clin. Lab. Sci. 2019;56:533–566. doi: 10.1080/10408363.2019.1653821. [DOI] [PubMed] [Google Scholar]
- 126.Knowles LM, et al. HGF and c-Met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin. Cancer Res. 2009;15:3740–3750. doi: 10.1158/1078-0432.CCR-08-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szturz P, et al. Understanding c-MET signalling in squamous cell carcinoma of the head & neck. Crit. Rev. Oncol./Hematol. 2017;111:39–51. doi: 10.1016/j.critrevonc.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 128.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 129.Huang W-C, et al. A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing β5-integrin/c-met signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:1–17. doi: 10.1186/s13046-019-1091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alamoud KA, Kukuruzinska MA. Emerging insights into Wnt/β-catenin signaling in head and neck cancer. J. Dent. Res. 2018;97:665–673. doi: 10.1177/0022034518771923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moon JH, Lee SH, Lim YC. Wnt/β-catenin/Slug pathway contributes to tumor invasion and lymph node metastasis in head and neck squamous cell carcinoma. Clin. Exp. Metastasis. 2021;38:163–174. doi: 10.1007/s10585-021-10081-3. [DOI] [PubMed] [Google Scholar]
- 132.Xie J, et al. Cancer‐associated fibroblasts secrete hypoxia‐induced serglycin to promote head and neck squamous cell carcinoma tumor cell growth in vitro and in vivo by activating the Wnt/β-catenin pathway. Cell. Oncol. 2021;44:661–671. doi: 10.1007/s13402-021-00592-2. [DOI] [PubMed] [Google Scholar]
- 133.Hiremath IS, et al. The multidimensional role of the Wnt/β‐catenin signaling pathway in human malignancies. J. Cell. Physiol. 2022;237:199–238. doi: 10.1002/jcp.30561. [DOI] [PubMed] [Google Scholar]
- 134.Chamoli A, et al. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol. 2021;121:105451. doi: 10.1016/j.oraloncology.2021.105451. [DOI] [PubMed] [Google Scholar]
- 135.Wang X, et al. Histone methyltransferase KMT2D cooperates with MEF2A to promote the stem-like properties of oral squamous cell carcinoma. Cell Biosci. 2022;12:1–13. doi: 10.1186/s13578-022-00785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qiao C, Qiao T, Yang S, Liu L, Zheng M. SNHG17/miR-384/ELF1 axis promotes cell growth by transcriptional regulation of CTNNB1 to activate Wnt/β-catenin pathway in oral squamous cell carcinoma. Cancer Gene Ther. 2022;29:122–132. doi: 10.1038/s41417-021-00294-9. [DOI] [PubMed] [Google Scholar]
- 137.Huang G, et al. Glycolysis-Related Gene Analyses Indicate That DEPDC1 Promotes the Malignant Progression of Oral Squamous Cell Carcinoma via the WNT/β-Catenin Signaling Pathway. Int. J. Mol. Sci. 2023;24:1992. doi: 10.3390/ijms24031992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsuchiya R, et al. Expression of adenomatous polyposis coli (APC) in tumorigenesis of human oral squamous cell carcinoma. Oral Oncol. 2004;40:932–940. doi: 10.1016/j.oraloncology.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 140.Shiah S-G, et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7bmiR329 and miR410 regulate Wnt–β-Catenin signaling pathway. Cancer Res. 2014;74:7560–7572. doi: 10.1158/0008-5472.CAN-14-0978. [DOI] [PubMed] [Google Scholar]
- 141.Xie H, et al. WNT7A promotes EGF-induced migration of oral squamous cell carcinoma cells by activating β-catenin/MMP9-mediated signaling. Front. Pharmacol. 2020;11:98. doi: 10.3389/fphar.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chiba T, et al. MALT1 inhibition of oral carcinoma cell invasion and ERK/MAPK activation. J. Dent. Res. 2016;95:446–452. doi: 10.1177/0022034515621740. [DOI] [PubMed] [Google Scholar]
- 143.Rezatabar S, et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J. Cell. Physiol. 2019;234:14951–14965. doi: 10.1002/jcp.28334. [DOI] [PubMed] [Google Scholar]
- 144.Peng Q, et al. Mitogen-activated protein kinase signaling pathway in oral cancer. Oncol. Lett. 2018;15:1379–1388. doi: 10.3892/ol.2017.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tao Y, et al. SH3-domain binding protein 1 in the tumor microenvironment promotes hepatocellular carcinoma metastasis through WAVE2 pathway. Oncotarget. 2016;7:18356. doi: 10.18632/oncotarget.7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shimizu T, et al. Annexin A10 in human oral cancer: biomarker for tumoral growth via G1/S transition by targeting MAPK signaling pathways. PLoS ONE. 2012;7:e45510. doi: 10.1371/journal.pone.0045510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 148.Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018;12:3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu W, et al. Extracellular vesicles-released parathyroid hormone-related protein from Lewis lung carcinoma induces lipolysis and adipose tissue browning in cancer cachexia. Cell Death Dis. 2021;12:1–14. doi: 10.1038/s41419-020-03382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Carbone C, et al. Angiopoietin-like proteins in angiogenesis, inflammation and cancer. Int. J. Mol. Sci. 2018;19:431. doi: 10.3390/ijms19020431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang S, et al. Quaking 5 suppresses TGF‐β‐induced EMT and cell invasion in lung adenocarcinoma. EMBO Rep. 2021;22:e52079. doi: 10.15252/embr.202052079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Artemenko M, Zhong SSW, To SKY, Wong AST. p70 S6 kinase as a therapeutic target in cancers: More than just an mTOR effector. Cancer Lett. 2022;535:215593. doi: 10.1016/j.canlet.2022.215593. [DOI] [PubMed] [Google Scholar]
- 153.Kim MP, Lozano G. Mutant p53 partners in crime. Cell Death Differ. 2018;25:161–168. doi: 10.1038/cdd.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lindemann A, Takahashi H, Patel AA, Osman AA, Myers JN. Targeting the DNA damage response in OSCC with TP 53 mutations. J. Dent. Res. 2018;97:635–644. doi: 10.1177/0022034518759068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ara N, Atique M, Ahmed S, Ali Bukhari SG. Frequency of p53 gene mutation and protein expression in oral squamous cell carcinoma. J. Coll. Phys. Surg. Pak. 2014;24:749–753. doi: 10.2014/JCPSP.749753. [DOI] [PubMed] [Google Scholar]
- 156.Castellsagué X, et al. HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J. Natl Cancer Inst. 2016;108:djv403. doi: 10.1093/jnci/djv403. [DOI] [PubMed] [Google Scholar]
- 157.Yang L, et al. The expression and correlation of iNOS and p53 in oral squamous cell carcinoma. BioMed. Res. Int. 2015;2015:637853. doi: 10.1155/2015/637853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang X-H, et al. p53-positive expression in dysplastic surgical margins is a predictor of tumor recurrence in patients with early oral squamous cell carcinoma. Cancer Manag. Res. 2019;11:1465. doi: 10.2147/CMAR.S192500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hang W, et al. Piperlongumine and p53-reactivator APR-246 selectively induce cell death in HNSCC by targeting GSTP1. Oncogene. 2018;37:3384–3398. doi: 10.1038/s41388-017-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jun HJ, et al. A PDGFRα-driven mouse model of glioblastoma reveals a stathmin1-mediated mechanism of sensitivity to vinblastine. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-05036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cierpikowski P, Lis-Nawara A, Gajdzis P, Bar J. PDGFRα/HER2 and PDGFRα/p53 co-expression in oral squamous cell carcinoma. Anticancer Res. 2018;38:795–802. doi: 10.21873/anticanres.12286. [DOI] [PubMed] [Google Scholar]
- 163.Gipson BJ, Robbins HA, Fakhry C, D’Souza G. Sensitivity and specificity of oral HPV detection for HPV-positive head and neck cancer. Oral Oncol. 2018;77:52–56. doi: 10.1016/j.oraloncology.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Soni S, et al. Alterations of rb pathway components are frequent events in patients with oral epithelial dysplasia and predict clinical outcome in patients with squamous cell carcinoma. Oncology. 2005;68:314–325. doi: 10.1159/000086970. [DOI] [PubMed] [Google Scholar]
- 165.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int. J. Cancer. 2012;130:1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nemes JA, Deli L, Nemes Z, Márton IJ. Expression of p16INK4A, p53, and Rb proteins are independent from the presence of human papillomavirus genes in oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2006;102:344–352. doi: 10.1016/j.tripleo.2005.10.069. [DOI] [PubMed] [Google Scholar]
- 167.Salehinejad J, et al. Immunohistochemical expression of p16 protein in oral squamous cell carcinoma and lichen planus. Ann. Diagn. Pathol. 2014;18:210–213. doi: 10.1016/j.anndiagpath.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 168.Suzuki H, Sugimura H, Hashimoto K. p16INK4A in oral squamous cell carcinomas—a correlation with biological behaviors: immunohistochemical and FISH analysis. J. Oral Maxillofac. Surg. 2006;64:1617–1623. doi: 10.1016/j.joms.2005.11.097. [DOI] [PubMed] [Google Scholar]
- 169.Hanken H, et al. CCND1 amplification and cyclin D1 immunohistochemical expression in head and neck squamous cell carcinomas. Clin. Oral Investig. 2014;18:269–276. doi: 10.1007/s00784-013-0967-6. [DOI] [PubMed] [Google Scholar]
- 170.Monteiro LS, et al. Prognostic significance of cyclins A2, B1, D1, and E1 and CCND1 numerical aberrations in oral squamous cell carcinomas. Anal. Cell. Pathol. 2018;2018:7253510. doi: 10.1155/2018/7253510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ramos-García P, et al. Clinicopathological significance of tumor cyclin D1 expression in oral cancer. Arch. Oral Biol. 2019;99:177–182. doi: 10.1016/j.archoralbio.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 172.Ramos-García P, et al. Asymmetrical proliferative pattern loss linked to cyclin D1 overexpression in adjacent non-tumour epithelium in oral squamous cell carcinoma. Arch. Oral Biol. 2019;97:12–17. doi: 10.1016/j.archoralbio.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 173.Ramos‐García P, Bravo M, González‐Ruiz L, González‐Moles MÁ. Significance of cytoplasmic cyclin D1 expression in oral oncogenesis. Oral Dis. 2018;24:98–102. doi: 10.1111/odi.12752. [DOI] [PubMed] [Google Scholar]
- 174.Jayasurya R, et al. Phenotypic alterations in Rb pathway have more prognostic influence than p53 pathway proteins in oral carcinoma. Mod. Pathol. 2005;18:1056–1066. doi: 10.1038/modpathol.3800387. [DOI] [PubMed] [Google Scholar]
- 175.Ramos-Garcia P, et al. Prognostic and clinicopathological significance of cyclin D1 expression in oral squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2018;83:96–106. doi: 10.1016/j.oraloncology.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 176.Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 177.Sun W, et al. Activation of the NOTCH Pathway in Head and Neck CancerNOTCH in Head and Neck Cancer. Cancer Res. 2014;74:1091–1104. doi: 10.1158/0008-5472.CAN-13-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yap LF, et al. The opposing roles of NOTCH signalling in head and neck cancer: a mini review. Oral Dis. 2015;21:850–857. doi: 10.1111/odi.12309. [DOI] [PubMed] [Google Scholar]
- 179.Kałafut J, et al. Shooting at moving and hidden targets—tumour cell plasticity and the notch signalling pathway in head and neck squamous cell carcinomas. Cancers. 2021;13:6219. doi: 10.3390/cancers13246219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grilli G, et al. Impact of notch signaling on the prognosis of patients with head and neck squamous cell carcinoma. Oral Oncol. 2020;110:105003. doi: 10.1016/j.oraloncology.2020.105003. [DOI] [PubMed] [Google Scholar]
- 181.D’assoro AB, Leon-Ferre R, Braune E-B, Lendahl U. Roles of notch signaling in the tumor microenvironment. Int. J. Mol. Sci. 2022;23:6241. doi: 10.3390/ijms23116241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat. Rev. Cancer. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 183.Ishida T, Hijioka H, Kume K, Miyawaki A, Nakamura N. Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol. Lett. 2013;6:1201–1206. doi: 10.3892/ol.2013.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Song X, et al. Common and complex Notch1 mutations in chinese oral squamous cell carcinoma complexity of notch1 mutations in Chinese OSCC. Clin. Cancer Res. 2014;20:701–710. doi: 10.1158/1078-0432.CCR-13-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kwon C, et al. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat. Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee SH, et al. Notch1 signaling contributes to stemness in head and neck squamous cell carcinoma. Lab. Investig. 2016;96:508–516. doi: 10.1038/labinvest.2015.163. [DOI] [PubMed] [Google Scholar]
- 187.Nyman PE, Buehler D, Lambert PF. Loss of function of canonical notch signaling drives head and neck carcinogenesis effects of loss of function of Notch in HNSCC. Clin. Cancer Res. 2018;24:6308–6318. doi: 10.1158/1078-0432.CCR-17-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kozakiewicz P, Grzybowska‑Szatkowska L. Application of molecular targeted therapies in the treatment of head and neck squamous cell carcinoma. Oncol. Lett. 2018;15:7497–7505. doi: 10.3892/ol.2018.8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhao Y-Y, Yu G-T, Xiao T, Hu J. The Notch signaling pathway in head and neck squamous cell carcinoma: a meta-analysis. Adv. Clin. Exp. Med. 2017;26:881–887. doi: 10.17219/acem/64000. [DOI] [PubMed] [Google Scholar]
- 190.Castilho RM, Squarize CH, Almeida LO. Epigenetic modifications and head and neck cancer: implications for tumor progression and resistance to therapy. Int. J. Mol. Sci. 2017;18:1506. doi: 10.3390/ijms18071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hema KN, Smitha T, Sheethal HS, Mirnalini SA. Epigenetics in oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 2017;21:252. doi: 10.4103/jomfp.JOMFP_150_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bais MV. Impact of epigenetic regulation on head and neck squamous cell carcinoma. J. Dent. Res. 2019;98:268–276. doi: 10.1177/0022034518816947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.D’Souza W, Saranath D. Clinical implications of epigenetic regulation in oral cancer. Oral Oncol. 2015;51:1061–1068. doi: 10.1016/j.oraloncology.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 194.Irimie AI, et al. Current insights into oral cancer epigenetics. Int. J. Mol. Sci. 2018;19:670. doi: 10.3390/ijms19030670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 196.Basu B, et al. Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India. Clin. Epigenet. 2017;9:1–15. doi: 10.1186/s13148-017-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Guerrero-Preston R, et al. Global DNA methylation: a common early event in oral cancer cases with exposure to environmental carcinogens or viral agents. P R Health Sci. J. 2009;28:24–29. [PubMed] [Google Scholar]
- 198.Supic G, Kozomara R, Jovic N, Zeljic K, Magic Z. Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol. 2011;47:702–708. doi: 10.1016/j.oraloncology.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 199.Towle R, et al. Global analysis of DNA methylation changes during progression of oral cancer. Oral Oncol. 2013;49:1033–1042. doi: 10.1016/j.oraloncology.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 200.Gasche JA, Goel A. Epigenetic mechanisms in oral carcinogenesis. Fut. Oncol. 2012;8:1407–1425. doi: 10.2217/fon.12.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 202.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 203.Mascolo M, et al. Epigenetic disregulation in oral cancer. Int. J. Mol. Sci. 2012;13:2331–2353. doi: 10.3390/ijms13022331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schussel J, et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions EDNRB and DCC methylation distinguish oral malignant lesion. Clin. Cancer Res. 2013;19:3268–3275. doi: 10.1158/1078-0432.CCR-12-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shaw RJ, et al. Promoter methylation of P16, RARβ, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br. J. Cancer. 2006;94:561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lin R-K, et al. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J. Clin. Investig. 2010;120:521–532. doi: 10.1172/JCI40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jayaprakash C, Radhakrishnan R, Ray S, Satyamoorthy K. Promoter methylation of MGMT in oral carcinoma: A population-based study and meta-analysis. Arch. Oral Biol. 2017;80:197–208. doi: 10.1016/j.archoralbio.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 209.Guerrero-Preston R, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva differential methylation and oncogenic pathways in OSCC. Cancer Prev. Res. 2011;4:1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yeh K-T, et al. Epigenetic changes of tumor suppressor genes, P15, P16, VHL and P53 in oral cancer. Oncol. Rep. 2003;10:659–663. [PubMed] [Google Scholar]
- 211.Mendonsa AM, Na T-Y, Gumbiner BM. E-cadherin in contact inhibition and cancer. Oncogene. 2018;37:4769–4780. doi: 10.1038/s41388-018-0304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Álvarez-Garcia, V., Tawil, Y., Wise, H. M. & Leslie, N. R. in Seminars in Cancer Biology. 66–79 (Elsevier, 2019). 10.1016/j.semcancer.2019.02.001. [DOI] [PubMed]
- 213.Flanagan DJ, et al. NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature. 2021;594:430–435. doi: 10.1038/s41586-021-03525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Repenning A, et al. PRMT1 promotes the tumor suppressor function of p14ARF and is indicative for pancreatic cancer prognosis. EMBO J. 2021;40:e106777. doi: 10.15252/embj.2020106777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Olesen TB, et al. Prevalence of human papillomavirus DNA and p16INK4a in penile cancer and penile intraepithelial neoplasia: a systematic review and meta-analysis. Lancet Oncol. 2019;20:145–158. doi: 10.1016/S1470-2045(18)30682-X. [DOI] [PubMed] [Google Scholar]
- 216.Du F, et al. miR-137 alleviates doxorubicin resistance in breast cancer through inhibition of epithelial-mesenchymal transition by targeting DUSP4. Cell Death Dis. 2019;10:1–10. doi: 10.1038/s41419-019-2164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Khordadmehr M, Shahbazi R, Sadreddini S, Baradaran B. miR‐193: a new weapon against cancer. J. Cell. Physiol. 2019;234:16861–16872. doi: 10.1002/jcp.28368. [DOI] [PubMed] [Google Scholar]
- 218.Flausino CS, Daniel FI, Modolo F. DNA methylation in oral squamous cell carcinoma: from its role in carcinogenesis to potential inhibitor drugs. Crit. Rev. Oncol./Hematol. 2021;164:103399. doi: 10.1016/j.critrevonc.2021.103399. [DOI] [PubMed] [Google Scholar]
- 219.Nikitakis NG, et al. Molecular markers associated with development and progression of potentially premalignant oral epithelial lesions: Current knowledge and future implications. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018;125:650–669. doi: 10.1016/j.oooo.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 220.Daniel FI, Rivero ERC, Modolo F, Lopes TG, Salum FG. Immunohistochemical expression of DNA methyltransferases 1, 3a and 3b in oral leukoplakias and squamous cell carcinomas. Arch. Oral Biol. 2010;55:1024–1030. doi: 10.1016/j.archoralbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 221.Baba S, et al. Global DNA hypomethylation suppresses squamous carcinogenesis in the tongue and esophagus. Cancer Sci. 2009;100:1186–1191. doi: 10.1111/j.1349-7006.2009.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Supic G, Kozomara R, Zeljic K, Jovic N, Magic Z. Prognostic value of the DNMTs mRNA expression and genetic polymorphisms on the clinical outcome in oral cancer patients. Clin. Oral Investig. 2017;21:173–182. doi: 10.1007/s00784-016-1772-9. [DOI] [PubMed] [Google Scholar]
- 223.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 224.Wu C-S, et al. ASC contributes to metastasis of oral cavity squamous cell carcinoma. Oncotarget. 2016;7:50074. doi: 10.18632/oncotarget.10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ling Y, et al. Roles of CEACAM1 in cell communication and signaling of lung cancer and other diseases. Cancer Metastasis Rev. 2015;34:347–357. doi: 10.1007/s10555-015-9569-x. [DOI] [PubMed] [Google Scholar]
- 226.Wu B, Xiong X, Jia J, Zhang W-F. MicroRNAs: new actors in the oral cancer scene. Oral Oncol. 2011;47:314–319. doi: 10.1016/j.oraloncology.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 227.Facompre ND, Harmeyer KH, Basu D. Regulation of oncogenic PI3-kinase signaling by JARID1B. Oncotarget. 2017;8:7218. doi: 10.18632/oncotarget.14790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Urosevic J, et al. Colon cancer cells colonize the lung from established liver metastases through p38 MAPK signalling and PTHLH. Nat. Cell Biol. 2014;16:685–694. doi: 10.1038/ncb2977. [DOI] [PubMed] [Google Scholar]
- 229.Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 2016;6:a026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Almeida LO, et al. NFκB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC) FEBS Open Bio. 2014;4:96–104. doi: 10.1016/j.fob.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Almeida LO, et al. Unlocking the chromatin of adenoid cystic carcinomas using HDAC inhibitors sensitize cancer stem cells to cisplatin and induces tumor senescence. Stem Cell Res. 2017;21:94–105. doi: 10.1016/j.scr.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Guimarães DM, et al. Sensitizing mucoepidermoid carcinomas to chemotherapy by targeted disruption of cancer stem cells. Oncotarget. 2016;7:42447. doi: 10.18632/oncotarget.9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Webber LP, et al. Hypoacetylation of acetyl‐histone H3 (H3K9ac) as marker of poor prognosis in oral cancer. Histopathology. 2017;71:278–286. doi: 10.1111/his.13218. [DOI] [PubMed] [Google Scholar]
- 234.Chen Y-W, Kao S-Y, Wang H-J, Yang M-H. Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer. 2013;119:4259–4267. doi: 10.1002/cncr.28356. [DOI] [PubMed] [Google Scholar]
- 235.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 236.Rastogi B, et al. Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol. Cell. Biochem. 2016;415:183–196. doi: 10.1007/s11010-016-2690-5. [DOI] [PubMed] [Google Scholar]
- 237.Kumar B, Yadav A, Lang JC, Teknos TN, Kumar P. Suberoylanilide hydroxamic acid (SAHA) reverses chemoresistance in head and neck cancer cells by targeting cancer stem cells via the downregulation of nanog. Genes Cancer. 2015;6:169. doi: 10.18632/genesandcancer.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lv Y, et al. Histone deacetylase 1 regulates the malignancy of oral cancer cells via miR-154-5p/PCNA axis. Biol. Chem. 2020;401:1273–1281. doi: 10.1515/hsz-2020-0189. [DOI] [PubMed] [Google Scholar]
- 239.Chang H-H, et al. Histone deacetylase 2 expression predicts poorer prognosis in oral cancer patients. Oral Oncol. 2009;45:610–614. doi: 10.1016/j.oraloncology.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 240.Kumar R, Li D-Q, Müller S, Knapp S. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene. 2016;35:4423–4436. doi: 10.1038/onc.2015.513. [DOI] [PubMed] [Google Scholar]
- 241.Panchal O, et al. SATB1 as oncogenic driver and potential therapeutic target in head & neck squamous cell carcinoma (HNSCC) Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-65077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vitale I, Manic G, Maria R, de, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol. Cell. 2017;66:306–319. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 243.Hung MH, et al. Tumor methionine metabolism drives T-cell exhaustion in hepatocellular carcinoma. Nat. Commun. 2021;12:1–15. doi: 10.1038/s41467-021-21804-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Aydin ÖZ, Vermeulen W, Lans H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle. 2014;13:3016–3025. doi: 10.4161/15384101.2014.956551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Fang F-M, et al. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. Am. J. Pathol. 2011;178:2407–2415. doi: 10.1016/j.ajpath.2011.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.D’ANGELO B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5575. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 247.Deng M, et al. TET-mediated sequestration of miR-26 drives EZH2 expression and gastric carcinogenesis. Cancer Res. 2017;77:6069–6082. doi: 10.1158/0008-5472.CAN-16-2964. [DOI] [PubMed] [Google Scholar]
- 248.Zhang W, et al. miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation. Lab. Investig. 2018;98:1397–1407. doi: 10.1038/s41374-018-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Viticchiè G, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- 250.Jia L-F, et al. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN–AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50:1062–1071. doi: 10.1016/j.oraloncology.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 251.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 252.Kanlikilicer P, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine. 2018;38:100–112. doi: 10.1016/j.ebiom.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep38750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Liu C-J, et al. Increase of microRNA miR‐31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16:360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 255.Liu C-J, Lin S-C, Yang C-C, Cheng H-W, Chang K-W. Exploiting salivary miR‐31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck. 2012;34:219–224. doi: 10.1002/hed.21713. [DOI] [PubMed] [Google Scholar]
- 256.Rezaei T, et al. microRNA-181 serves as a dual-role regulator in the development of human cancers. Free Radic. Biol. Med. 2020;152:432–454. doi: 10.1016/j.freeradbiomed.2019.12.043. [DOI] [PubMed] [Google Scholar]
- 257.Shin K-H, et al. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem. Biophys. Res. Commun. 2011;404:896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 258.Chang C-C, et al. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncol. 2013;49:923–931. doi: 10.1016/j.oraloncology.2013.03.430. [DOI] [PubMed] [Google Scholar]
- 259.Chen Y-F, et al. MicroRNA-211 enhances the oncogenicity of carcinogen-induced oral carcinoma by repressing TCF12 and increasing antioxidant activity miR-211-TCF12-FAM213A activation in OSCC. Cancer Res. 2016;76:4872–4886. doi: 10.1158/0008-5472.CAN-15-1664. [DOI] [PubMed] [Google Scholar]
- 260.Chen, S., Thorne, R. F., Zhang, X. D., Wu, M. & Liu, L. in Seminars in Cancer Biology. 72–83 (Elsevier, 2021). 10.1016/j.semcancer.2020.09.002. [DOI] [PubMed]
- 261.Ayers D, Vandesompele J. Influence of microRNAs and long non-coding RNAs in cancer chemoresistance. Genes. 2017;8:95. doi: 10.3390/genes8030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer. 2007;6:1–11. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sun L, et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31:432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]
- 264.Chen Q, Qin R, Fang Y, Li H. Berberine sensitizes human ovarian cancer cells to cisplatin through miR-93/PTEN/Akt signaling pathway. Cell. Physiol. Biochem. 2015;36:956–965. doi: 10.1159/000430270. [DOI] [PubMed] [Google Scholar]
- 265.Komatsu S, et al. Plasma microRNA profiles: identification of miR-23a as a novel biomarker for chemoresistance in esophageal squamous cell carcinoma. Oncotarget. 2016;7:62034. doi: 10.18632/oncotarget.11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Meng X, et al. The role of non‐coding RNAs in drug resistance of oral squamous cell carcinoma and therapeutic potential. Cancer Commun. 2021;41:981–1006. doi: 10.1002/cac2.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Liu J, et al. ZBTB7A, a miR-144-3p targeted gene, accelerates bladder cancer progression via downregulating HIC1 expression. Cancer Cell Int. 2022;22:1–14. doi: 10.1186/s12935-022-02596-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Yeh L-Y, Chou C-H, Liu C-J, Lin S-C, Chang K-W. miR-372 enhances tumorigenesis and drug resistance in oral carcinoma by targeting ZBTB7A transcription factor. Cancer Res. 2018;78:479. [Google Scholar]
- 269.Feller L, Altini M, Lemmer J. Inflammation in the context of oral cancer. Oral Oncol. 2013;49:887–892. doi: 10.1016/j.oraloncology.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 270.Niu X, Yang B, Liu F, Fang Q. LncRNA HOXA11-AS promotes OSCC progression by sponging miR-98-5p to upregulate YBX2 expression. Biomed. Pharmacother. 2020;121:109623. doi: 10.1016/j.biopha.2019.109623. [DOI] [PubMed] [Google Scholar]
- 271.Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed. Pharmacother. 2017;95:1504–1513. doi: 10.1016/j.biopha.2017.08.097. [DOI] [PubMed] [Google Scholar]
- 272.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 2018;11:1–20. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Chatterjee R, et al. Pathophysiological relationship between hypoxia associated oxidative stress, Epithelial-mesenchymal transition, stemness acquisition and alteration of Shh/Gli-1 axis during oral sub-mucous fibrosis and oral squamous cell carcinoma. Eur. J. Cell Biol. 2021;100:151146. doi: 10.1016/j.ejcb.2020.151146. [DOI] [PubMed] [Google Scholar]
- 274.Li Z, et al. The Hippo transducer TAZ promotes epithelial to mesenchymal transition and cancer stem cell maintenance in oral cancer. Mol. Oncol. 2015;9:1091–1105. doi: 10.1016/j.molonc.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Biddle A, Gammon L, Liang X, Costea DE, Mackenzie IC. Phenotypic plasticity determines cancer stem cell therapeutic resistance in oral squamous cell carcinoma. EBioMedicine. 2016;4:138–145. doi: 10.1016/j.ebiom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Vig N, Mackenzie IC, Biddle A. Phenotypic plasticity and epithelial‐to‐mesenchymal transition in the behaviour and therapeutic response of oral squamous cell carcinoma. J. Oral Pathol. Med. 2015;44:649–655. doi: 10.1111/jop.12306. [DOI] [PubMed] [Google Scholar]
- 277.Liu S, Liu D, Li J, Zhang D, Chen Q. Regulatory T cells in oral squamous cell carcinoma. J. Oral Pathol. Med. 2016;45:635–639. doi: 10.1111/jop.12445. [DOI] [PubMed] [Google Scholar]
- 278.Gaur P, Singh AK, Shukla NK, Das SN. Inter-relation of Th1, Th2, Th17 and Treg cytokines in oral cancer patients and their clinical significance. Hum. Immunol. 2014;75:330–337. doi: 10.1016/j.humimm.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 279.Schwarz S, Butz M, Morsczeck C, Reichert TE, Driemel O. Increased number of CD25+ FoxP3+ regulatory T cells in oral squamous cell carcinomas detected by chromogenic immunohistochemical double staining. J. Oral Pathol. Med. 2008;37:485–489. doi: 10.1111/j.1600-0714.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 280.Duray A, Demoulin S, Hubert P, Delvenne P, Saussez S. Immune suppression in head and neck cancers: a review. Clin. Dev. Immunol. 2010;2010:701657. doi: 10.1155/2010/701657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Whiteside, T. L. in Seminars in Cancer Biology. 3–15 (Elsevier, 2021). 10.1016/j.semcancer.2005.07.008.
- 282.Sathiyasekar AC, Chandrasekar P, Pakash A, Kumar KG, Jaishlal MS. Overview of immunology of oral squamous cell carcinoma. J. Pharm. Bioallied Sci. 2016;8:S8. doi: 10.4103/0975-7406.191974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Caponio VCA, Zhurakivska K, Lo Muzio L, Troiano G, Cirillo N. The immune cells in the development of oral squamous cell carcinoma. Cancers. 2023;15:3779. doi: 10.3390/cancers15153779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ling, D. C., Bakkenist, C. J., Ferris, R. L. & Clump, D. A. in Seminars in Radiation Oncology. 12–16 (Elsevier, 2018). 10.1016/j.semradonc.2017.08.009. [DOI] [PubMed]
- 285.de Ruiter EJ, Ooft ML, Devriese LA, Willems SM. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: a systematic review and meta-analysis. Oncoimmunology. 2017;6:e1356148. doi: 10.1080/2162402X.2017.1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zitvogel, L. & Kroemer, G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology1, 1223–1225 (2012). [DOI] [PMC free article] [PubMed]
- 287.Alsahafi E, et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10:1–17. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lythgoe MP, Liu DSK, Annels NE, Krell J, Frampton AE. Gene of the month: Lymphocyte-activation gene 3 (LAG-3) J. Clin. Pathol. 2021;74:543–547. doi: 10.1136/jclinpath-2021-207517. [DOI] [PubMed] [Google Scholar]
- 289.Zhou K, et al. Immunosuppression of human adipose-derived stem cells on T cell subsets via the reduction of NF-kappaB activation mediated by PD-L1/PD-1 and Gal-9/TIM-3 pathways. Stem Cells Dev. 2018;27:1191–1202. doi: 10.1089/scd.2018.0033. [DOI] [PubMed] [Google Scholar]
- 290.Zhou X, et al. The novel non‐immunological role and underlying mechanisms of B7‐H3 in tumorigenesis. J. Cell. Physiol. 2019;234:21785–21795. doi: 10.1002/jcp.28936. [DOI] [PubMed] [Google Scholar]
- 291.Davis RJ, van Waes C, Allen CT. Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and Tregs as mediators of the immunosuppressive microenvironment in head and neck cancer. Oral Oncol. 2016;58:59–70. doi: 10.1016/j.oraloncology.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wolf GT, et al. Novel neoadjuvant immunotherapy regimen safety and survival in head and neck squamous cell cancer. Head Neck. 2011;33:1666–1674. doi: 10.1002/hed.21660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Furquim CP, Pivovar A, Amenábar JM, Bonfim C, Torres-Pereira CC. Oral cancer in Fanconi anemia: Review of 121 cases. Crit. Rev. Oncol./Hematol. 2018;125:35–40. doi: 10.1016/j.critrevonc.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 294.Alter BP, et al. Squamous cell carcinomas in patients with Fanconi anemia and dyskeratosis congenita: a search for human papillomavirus. Int. J. Cancer. 2013;133:1513–1515. doi: 10.1002/ijc.28157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Amenábar JM, Torres‐Pereira CC, Tang KD, Punyadeera C. Two enemies, one fight: an update of oral cancer in patients with Fanconi anemia. Cancer. 2019;125:3936–3946. doi: 10.1002/cncr.32435. [DOI] [PubMed] [Google Scholar]
- 296.Bongiorno M, Rivard S, Hammer D, Kentosh J. Malignant transformation of oral leukoplakia in a patient with dyskeratosis congenita. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017;124:e239–e242. doi: 10.1016/j.oooo.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 297.Horton JD, Knochelmann HM, Day TA, Paulos CM, Neskey DM. Immune evasion by head and neck cancer: foundations for combination therapy. Trends Cancer. 2019;5:208–232. doi: 10.1016/j.trecan.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Wang L, et al. Cancer-associated fibroblasts enhance metastatic potential of lung cancer cells through IL-6/STAT3 signaling pathway. Oncotarget. 2017;8:76116. doi: 10.18632/oncotarget.18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Zhang D, et al. Cancer‐associated fibroblasts promote tumor progression by lncRNA‐mediated RUNX2/GDF10 signaling in oral squamous cell carcinoma. Mol. Oncol. 2022;16:780–794. doi: 10.1002/1878-0261.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Prime SS, et al. Fibroblast activation and senescence in oral cancer. J. Oral Pathol. Med. 2017;46:82–88. doi: 10.1111/jop.12456. [DOI] [PubMed] [Google Scholar]
- 301.Tan ML, Parkinson EK, Yap LF, Paterson IC. Autophagy is deregulated in cancer-associated fibroblasts from oral cancer and is stimulated during the induction of fibroblast senescence by TGF-β1. Sci. Rep. 2021;11:584. doi: 10.1038/s41598-020-79789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hassona Y, Cirillo N, Heesom K, Parkinson EK, Prime SS. Senescent cancer-associated fibroblasts secrete active MMP-2 that promotes keratinocyte dis-cohesion and invasion. Br. J. Cancer. 2014;111:1230–1237. doi: 10.1038/bjc.2014.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Bijai LK, Muthukrishnan A. Potential role of fibroblast senescence in malignant transformation of oral submucous fibrosis. Oral Oncol. 2022;127:105810. doi: 10.1016/j.oraloncology.2022.105810. [DOI] [PubMed] [Google Scholar]
- 304.Kim EK, Moon S, Kim DK, Zhang X, Kim J. CXCL1 induces senescence of cancer-associated fibroblasts via autocrine loops in oral squamous cell carcinoma. PLoS ONE. 2018;13:e0188847. doi: 10.1371/journal.pone.0188847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Lu Y, et al. Peroxiredoxin1 knockdown inhibits oral carcinogenesis via inducing cell senescence dependent on mitophagy. OncoTargets Ther. 2021;14:239–251. doi: 10.2147/OTT.S284182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Chen Y, et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J. Biomed. Sci. 2019;26:1–13. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yang Q, et al. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharmaceut. Sin. B. 2020;10:2156–2170. doi: 10.1016/j.apsb.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Goswami KK, et al. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell. Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 309.Jiang C, Yuan F, Wang J, Wu L. Oral squamous cell carcinoma suppressed antitumor immunity through induction of PD-L1 expression on tumor-associated macrophages. Immunobiology. 2017;222:651–657. doi: 10.1016/j.imbio.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 310.Zhu Y, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models CSF1R blockade improves checkpoint immunotherapy. Cancer Res. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Chen J, et al. Suppression of T cells by myeloid-derived suppressor cells in cancer. Hum. Immunol. 2017;78:113–119. doi: 10.1016/j.humimm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 312.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Groth C, et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br. J. Cancer. 2019;120:16–25. doi: 10.1038/s41416-018-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Qu P, Wang L, Lin PC. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. 2016;380:253–256. doi: 10.1016/j.canlet.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Pyzer AR, Cole L, Rosenblatt J, Avigan DE. Myeloid‐derived suppressor cells as effectors of immune suppression in cancer. Int. J. Cancer. 2016;139:1915–1926. doi: 10.1002/ijc.30232. [DOI] [PubMed] [Google Scholar]
- 316.Aggarwal S, Sharma SC, N Das S. Dynamics of regulatory T cells (Tregs) in patients with oral squamous cell carcinoma. J. Surg. Oncol. 2017;116:1103–1113. doi: 10.1002/jso.24782. [DOI] [PubMed] [Google Scholar]
- 317.Munn DH, Bronte V. Immune suppressive mechanisms in the tumor microenvironment. Curr. Opin. Immunol. 2016;39:1–6. doi: 10.1016/j.coi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ni X, et al. YAP is essential for treg-mediated suppression of antitumor immunity. Cancer Discov. 2018;8:1026–1043. doi: 10.1158/2159-8290.CD-17-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Meng X, et al. Regulatory T cells in cardiovascular diseases. Nat. Rev. Cardiol. 2016;13:167–179. doi: 10.1038/nrcardio.2015.169. [DOI] [PubMed] [Google Scholar]
- 320.Miyara M, Sakaguchi S. Natural regulatory T cells: mechanisms of suppression. Trends Mol. Med. 2007;13:108–116. doi: 10.1016/j.molmed.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 321.Ohue Y, Nishikawa H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019;110:2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer. 2020;19:1–23. doi: 10.1186/s12943-020-01234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Fuse H, et al. Enhanced expression of PD-L1 in oral squamous cell carcinoma-derived CD11b+ Gr-1+ cells and its contribution to immunosuppressive activity. Oral Oncol. 2016;59:20–29. doi: 10.1016/j.oraloncology.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 324.Wynn TA. Type 2 cytokines: mechanisms and therapeutic strategies. Nat. Rev. Immunol. 2015;15:271–282. doi: 10.1038/nri3831. [DOI] [PubMed] [Google Scholar]
- 325.Joseph JP, Harishankar MK, Pillai AA, Devi A. Hypoxia induced EMT: a review on the mechanism of tumor progression and metastasis in OSCC. Oral Oncol. 2018;80:23–32. doi: 10.1016/j.oraloncology.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 326.Kumar A, Deep G. Hypoxia in tumor microenvironment regulates exosome biogenesis: Molecular mechanisms and translational opportunities. Cancer Lett. 2020;479:23–30. doi: 10.1016/j.canlet.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 327.Muz B, de La Puente P, Azab F, Kareem Azab A. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia. 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bhandari V, et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 2019;51:308–318. doi: 10.1038/s41588-018-0318-2. [DOI] [PubMed] [Google Scholar]
- 329.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N. Engl. J. Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Pérez-Sayáns M, et al. Hypoxia-inducible factors in OSCC. Cancer Lett. 2011;313:1–8. doi: 10.1016/j.canlet.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 331.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Corn PG, McDonald ER, Herman JG, El-Deiry WS. Tat-binding protein-1, a component of the 26S proteasome, contributes to the E3 ubiquitin ligase function of the von Hippel–Lindau protein. Nat. Genet. 2003;35:229–237. doi: 10.1038/ng1254. [DOI] [PubMed] [Google Scholar]
- 333.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia‐inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1: interaction between H-ras and hypoxia. J. Biol. Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 335.Gammon L, Mackenzie IC. Roles of hypoxia, stem cells and epithelial–mesenchymal transition in the spread and treatment resistance of head and neck cancer. J. Oral Pathol. Med. 2016;45:77–82. doi: 10.1111/jop.12327. [DOI] [PubMed] [Google Scholar]
- 336.Koukourakis MI, et al. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin. Cancer Res. 2001;7:3399–3403. [PubMed] [Google Scholar]
- 337.Tsai Y-P, Wu K-J. Hypoxia-regulated target genes implicated in tumor metastasis. J. Biomed. Sci. 2012;19:1–7. doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Duan Y, et al. Hypoxia induced Bcl-2/Twist1 complex promotes tumor cell invasion in oral squamous cell carcinoma. Oncotarget. 2017;8:7729. doi: 10.18632/oncotarget.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Amelio I, et al. p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc. Natl Acad. Sci. USA. 2018;115:E10869–E10878. doi: 10.1073/pnas.1808314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem. Sci. 2015;40:425–434. doi: 10.1016/j.tibs.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 341.Domingos PLB, et al. Hypoxia reduces the E-cadherin expression and increases OSCC cell migration regardless of the E-cadherin methylation profile. Pathol. Res. Pract. 2017;213:496–501. doi: 10.1016/j.prp.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 342.Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Zheng Y, Ni Y, Huang X, Wang Z, Han WE. Overexpression of HIF-1α indicates a poor prognosis in tongue carcinoma and may be associated with tumour metastasis. Oncol. Lett. 2013;5:1285–1289. doi: 10.3892/ol.2013.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Xie L, et al. Association between dietary nitrate and nitrite intake and site-specific cancer risk: evidence from observational studies. Oncotarget. 2016;7:56915. doi: 10.18632/oncotarget.10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Nair U, Bartsch H, Nair J. Alert for an epidemic of oral cancer due to use of the betel quid substitutes gutkha and pan masala: a review of agents and causative mechanisms. Mutagenesis. 2004;19:251–262. doi: 10.1093/mutage/geh036. [DOI] [PubMed] [Google Scholar]
- 346.Jing X, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer. 2019;18:1–15. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Banerjee S, et al. Microbial signatures associated with oropharyngeal and oral squamous cell carcinomas. Sci. Rep. 2017;7:1–20. doi: 10.1038/s41598-017-03466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Perera M, Al-Hebshi NN, Speicher DJ, Perera I, Johnson NW. Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio-pathogenic bacteria. J. Oral Microbiol. 2016;8:32762. doi: 10.3402/jom.v8.32762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Irfan M, Delgado RZR, Frias-Lopez J. The oral microbiome and cancer. Front. Immunol. 2020;11:591088. doi: 10.3389/fimmu.2020.591088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Yost S, et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. Int. J. Oral Sci. 2018;10:32. doi: 10.1038/s41368-018-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Eun Y-G, et al. Oral microbiome associated with lymph node metastasis in oral squamous cell carcinoma. Sci. Rep. 2021;11:23176. doi: 10.1038/s41598-021-02638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Rizzetto L, de Filippo C, Cavalieri D. Richness and diversity of mammalian fungal communities shape innate and adaptive immunity in health and disease. Eur. J. Immunol. 2014;44:3166–3181. doi: 10.1002/eji.201344403. [DOI] [PubMed] [Google Scholar]
- 353.Deo PN, Deshmukh R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019;23:122. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Mosaddad SA, et al. Oral microbial biofilms: an update. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:2005–2019. doi: 10.1007/s10096-019-03641-9. [DOI] [PubMed] [Google Scholar]
- 355.Willis JR, Gabaldón T. The human oral microbiome in health and disease: from sequences to ecosystems. Microorganisms. 2020;8:308. doi: 10.3390/microorganisms8020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Idris A, Hasnain SZ, Huat LZ, Koh D. Human diseases, immunity and the oral microbiota—Insights gained from metagenomic studies. Oral Sci. Int. 2017;14:27–32. [Google Scholar]
- 357.Li Q, et al. Role of oral bacteria in the development of oral squamous cell carcinoma. Cancers. 2020;12:2797. doi: 10.3390/cancers12102797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Mao W-M, Zheng W-H, Ling Z-Q. Epidemiologic risk factors for esophageal cancer development. Asian Pac. J. Cancer Prev. 2011;12:2461–2466. [PubMed] [Google Scholar]
- 359.Wang H, et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017;9:1–10. doi: 10.1186/s13073-017-0405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Gholizadeh P, et al. Role of oral microbiome on oral cancers, a review. Biomed. Pharmacother. 2016;84:552–558. doi: 10.1016/j.biopha.2016.09.082. [DOI] [PubMed] [Google Scholar]
- 361.Frank DN, et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene. 2022;41:1269–1280. doi: 10.1038/s41388-021-02137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Mager DL, et al. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005;3:1–8. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Yang K, et al. Oral microbiota analysis of tissue pairs and saliva samples from patients with oral squamous cell carcinoma–a pilot study. Front. Microbiol. 2021;12:719601. doi: 10.3389/fmicb.2021.719601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Delaney C, et al. Limitations of using 16S rRNA microbiome sequencing to predict oral squamous cell carcinoma. APMIS. 2023;131:262–276. doi: 10.1111/apm.13315. [DOI] [PubMed] [Google Scholar]
- 365.Hashimoto K, et al. Feasibility of oral microbiome profiles associated with oral squamous cell carcinoma. J. Oral Microbiol. 2022;14:2105574. doi: 10.1080/20002297.2022.2105574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology. 2011;216:1302–1310. doi: 10.1016/j.imbio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 367.Lafuente Ibáñez de Mendoza I, Maritxalar Mendia X, Garcia de la Fuente AM, Quindos Andres G, Aguirre Urizar JM. Role of Porphyromonas gingivalis in oral squamous cell carcinoma development: a systematic review. J. Periodontal. Res. 2020;55:13–22. doi: 10.1111/jre.12691. [DOI] [PubMed] [Google Scholar]
- 368.Börnigen D, et al. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-17795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Marttila E, et al. Acetaldehyde production and microbial colonization in oral squamous cell carcinoma and oral lichenoid disease. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013;116:61–68. doi: 10.1016/j.oooo.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 370.Kurkivuori J, et al. Acetaldehyde production from ethanol by oral streptococci. Oral Oncol. 2007;43:181–186. doi: 10.1016/j.oraloncology.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 371.Rautemaa R, Hietanen J, Niissalo S, Pirinen S, Perheentupa J. Oral and oesophageal squamous cell carcinoma–a complication or component of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED, APS-I) Oral Oncol. 2007;43:607–613. doi: 10.1016/j.oraloncology.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 372.Solis NV, Swidergall M, Bruno VM, Gaffen SL, Filler SG. The aryl hydrocarbon receptor governs epithelial cell invasion during oropharyngeal candidiasis. MBio. 2017;8:e00025–17. doi: 10.1128/mBio.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Zhu W, et al. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc. Natl Acad. Sci. USA. 2012;109:14194–14199. doi: 10.1073/pnas.1117676109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Warnakulasuriya S, Ariyawardana A. Malignant transformation of oral leukoplakia: a systematic review of observational studies. J. Oral Pathol. Med. 2016;45:155–166. doi: 10.1111/jop.12339. [DOI] [PubMed] [Google Scholar]
- 375.Nadeau C, Kerr AR. Evaluation and management of oral potentially malignant disorders. Dent. Clin. 2018;62:1–27. doi: 10.1016/j.cden.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 376.Monteiro L, et al. Type of surgical treatment and recurrence of oral leukoplakia: a retrospective clinical study. Med. Oral Patol. Oral Cir. Bucal. 2017;22:e520. doi: 10.4317/medoral.21645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Brignardello-Petersen R. Proliferative verrucous leukoplakia and erythroplakia are probably the disorders with the highest rate of malignant transformation. J. Am. Dent. Assoc. 2020;151:e62. doi: 10.1016/j.adaj.2020.01.035. [DOI] [PubMed] [Google Scholar]
- 378.van der Waal I. Potentially malignant disorders of the oral and oropharyngeal mucosa; terminology, classification and present concepts of management. Oral Oncol. 2009;45:317–323. doi: 10.1016/j.oraloncology.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 379.Armstrong WB, et al. Single-dose administration of Bowman-Birk inhibitor concentrate in patients with oral leukoplakia. Cancer Epidemiol. Biomark. Prev. 2000;9:43–47. [PubMed] [Google Scholar]
- 380.Armstrong WB, et al. Clinical modulation of oral leukoplakia and protease activity by Bowman-Birk inhibitor concentrate in a phase IIa chemoprevention trial. Clin. Cancer Res. 2000;6:4684–4691. [PubMed] [Google Scholar]
- 381.Chaudhry Z, Gupta SR, Oberoi SS. The efficacy of ErCr: YSGG laser fibrotomy in management of moderate oral submucous fibrosis: a preliminary study. J. Maxillofac. Oral Surg. 2014;13:286–294. doi: 10.1007/s12663-013-0511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Shah PH, Venkatesh R, More CB, Vassandacoumara V. Comparison of therapeutic efficacy of placental extract with dexamethasone and hyaluronic acid with dexamethasone for oral submucous fibrosis-a retrospective analysis. J. Clin. Diagn. Res. 2016;10:ZC63. doi: 10.7860/JCDR/2016/20369.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Cheng Y-SL, Gould A, Kurago Z, Fantasia J, Muller S. Diagnosis of oral lichen planus: a position paper of the American Academy of Oral and Maxillofacial Pathology. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016;122:332–354. doi: 10.1016/j.oooo.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 384.Liu C, et al. Efficacy of intralesional betamethasone for erosive oral lichen planus and evaluation of recurrence: a randomized, controlled trial. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013;116:584–590. doi: 10.1016/j.oooo.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 385.Olson MA, Rogers RS, III, Bruce AJ. Oral lichen planus. Clin. Dermatol. 2016;34:495–504. doi: 10.1016/j.clindermatol.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 386.Halonen P, et al. Cancer risk of Lichen planus: a cohort study of 13,100 women in Finland. Int. J. Cancer. 2018;142:18–22. doi: 10.1002/ijc.31025. [DOI] [PubMed] [Google Scholar]
- 387.Sutton DN, Brown JS, Rogers SN, Vaughan ED, Woolgar JA. The prognostic implications of the surgical margin in oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2003;32:30–34. doi: 10.1054/ijom.2002.0313. [DOI] [PubMed] [Google Scholar]
- 388.Brands MT, Brennan PA, Verbeek AL, Merkx MA, Geurts SM. Follow-up after curative treatment for oral squamous cell carcinoma. A critical appraisal of the guidelines and a review of the literature. Eur. J. Surg. Oncol. 2018;44:559–565. doi: 10.1016/j.ejso.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 389.Mistry RC, Qureshi SS, Kumaran C. Post‐resection mucosal margin shrinkage in oral cancer: quantification and significance. J. Surg. Oncol. 2005;91:131–133. doi: 10.1002/jso.20285. [DOI] [PubMed] [Google Scholar]
- 390.Petruzzi M, Lucchese A, Baldoni E, Grassi FR, Serpico R. Use of Lugol’s iodine in oral cancer diagnosis: an overview. Oral Oncol. 2010;46:811–813. doi: 10.1016/j.oraloncology.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 391.Xiao T, Kurita H, Shimane T, Nakanishi Y, Koike T. Vital staining with iodine solution in oral cancer: iodine infiltration, cell proliferation, and glucose transporter 1. Int. J. Clin. Oncol. 2013;18:792–800. doi: 10.1007/s10147-012-0450-4. [DOI] [PubMed] [Google Scholar]
- 392.Sakuraba M, et al. Recent advances in reconstructive surgery: head and neck reconstruction. Int. J. Clin. Oncol. 2013;18:561–565. doi: 10.1007/s10147-012-0513-6. [DOI] [PubMed] [Google Scholar]
- 393.Tsao AS, et al. Phase II randomized, placebo-controlled trial of green tea extract in patients with high-risk oral premalignant lesions. Cancer Prev. Res. 2009;2:931–941. doi: 10.1158/1940-6207.CAPR-09-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Furusaka T, Asakawa T, Tanaka A, Matsuda H, Ikeda M. Efficacy of multidrug superselective intra-arterial chemotherapy (docetaxel, cisplatin, and 5-fluorouracil) using the Seldinger technique for tongue cancer. Acta Otolaryngol. 2012;132:1108–1114. doi: 10.3109/00016489.2012.684702. [DOI] [PubMed] [Google Scholar]
- 395.Mitsudo K, et al. Organ preservation with daily concurrent chemoradiotherapy using superselective intra-arterial infusion via a superficial temporal artery for T3 and T4 head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 2011;79:1428–1435. doi: 10.1016/j.ijrobp.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 396.Colli LM, et al. Landscape of combination immunotherapy and targeted therapy to improve cancer management prioritizing immunotherapy and targeted drug combinations. Cancer Res. 2017;77:3666–3671. doi: 10.1158/0008-5472.CAN-16-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Vermorken JB, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 398.Yu C-C, et al. Targeting the PI3K/AKT/mTOR signaling pathway as an effectively radiosensitizing strategy for treating human oral squamous cell carcinoma in vitro and in vivo. Oncotarget. 2017;8:68641. doi: 10.18632/oncotarget.19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Xu J, Wu Z, Huang J. Flavopereirine suppresses the progression of human oral cancer by inhibiting the JAK-STAT signaling pathway via targeting LASP1. Drug Des. Dev. Ther. 2021;15:1705. doi: 10.2147/DDDT.S284213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Gan R, et al. FLI-06 intercepts notch signaling and suppresses the proliferation and self-renewal of tongue cancer cells. OncoTargets Ther. 2019;12:7663. doi: 10.2147/OTT.S221231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Smith DC, et al. First-in-human evaluation of the human monoclonal antibody vantictumab (OMP-18R5; anti-Frizzled) targeting the WNT pathway in a phase I study for patients with advanced solid tumors. J. Clin. Oncol. 2013;31:2540–2540. [Google Scholar]
- 402.El-Khoueiry, A. B. et al. A Phase I First-in-Human Study of PRI-724 in Patients (pts) with Advanced Solid Tumors (American Society of Clinical Oncology, 2013).
- 403.Liu J, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl Acad. Sci. USA. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Brahmer JR, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Burtness B, et al. KEYNOTE-048: Phase III study of first-line pembrolizumab (P) for recurrent/metastatic head and neck squamous cell carcinoma (R/M HNSCC) Ann. Oncol. 2018;29:viii729. [Google Scholar]
- 406.Cohen EEW, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet. 2019;393:156–167. doi: 10.1016/S0140-6736(18)31999-8. [DOI] [PubMed] [Google Scholar]
- 407.Burova E, et al. Preclinical development of the anti-LAG-3 antibody REGN3767: Characterization and activity in combination with the anti-PD-1 antibody cemiplimab in human PD-1xLAG-3–knockin mice. Mol. Cancer Therapeut. 2019;18:2051–2062. doi: 10.1158/1535-7163.MCT-18-1376. [DOI] [PubMed] [Google Scholar]
- 408.Huang J, Barbera L, Brouwers M, Browman G, Mackillop WJ. Does delay in starting treatment affect the outcomes of radiotherapy? A systematic review. J. Clin. Oncol. 2003;21:555–563. doi: 10.1200/JCO.2003.04.171. [DOI] [PubMed] [Google Scholar]
- 409.Lee W-HS, et al. Effective killing of acute myeloid leukemia by TIM-3 targeted chimeric antigen receptor T CellsmRNA encoding anti–TIM-3 CAR T cells to treat AML. Mol. Cancer Therapeut. 2021;20:1702–1712. doi: 10.1158/1535-7163.MCT-20-0155. [DOI] [PubMed] [Google Scholar]
- 410.Maeda S, Murakami K, Inoue A, Yonezawa T, Matsuki N. CCR4 blockade depletes regulatory T cells and prolongs survival in a canine model of bladder cancer anti-treg immunotherapy for canine bladder cancer. Cancer Immunol. Res. 2019;7:1175–1187. doi: 10.1158/2326-6066.CIR-18-0751. [DOI] [PubMed] [Google Scholar]
- 411.Fultang L, et al. MDSC targeting with Gemtuzumab ozogamicin restores T cell immunity and immunotherapy against cancers. EBioMedicine. 2019;47:235–246. doi: 10.1016/j.ebiom.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Wolf GT, et al. Tumor infiltrating lymphocytes after neoadjuvant IRX-2 immunotherapy in oral squamous cell carcinoma: Interim findings from the INSPIRE trial. Oral Oncol. 2020;111:104928. doi: 10.1016/j.oraloncology.2020.104928. [DOI] [PubMed] [Google Scholar]
- 413.Yete S, Saranath D. MicroRNAs in oral cancer: biomarkers with clinical potential. Oral Oncol. 2020;110:105002. doi: 10.1016/j.oraloncology.2020.105002. [DOI] [PubMed] [Google Scholar]
- 414.Zanoni DK, et al. Survival outcomes after treatment of cancer of the oral cavity (1985–2015) Oral Oncol. 2019;90:115–121. doi: 10.1016/j.oraloncology.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Almangush A, et al. Staging and grading of oral squamous cell carcinoma: an update. Oral Oncol. 2020;107:104799. doi: 10.1016/j.oraloncology.2020.104799. [DOI] [PubMed] [Google Scholar]
- 416.Rivera C, Oliveira AK, Costa RAP, Rossi Tde, Leme AFP. Prognostic biomarkers in oral squamous cell carcinoma: a systematic review. Oral Oncol. 2017;72:38–47. doi: 10.1016/j.oraloncology.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 417.Sasahira T, Kirita T. Hallmarks of cancer-related newly prognostic factors of oral squamous cell carcinoma. Int. J. Mol. Sci. 2018;19:2413. doi: 10.3390/ijms19082413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Suresh GM, Koppad R, Prakash BV, Sabitha KS, Dhara PS. Prognostic indicators of oral squamous cell carcinoma. Ann. Maxillofac. Surg. 2019;9:364. doi: 10.4103/ams.ams_253_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Kather JN, et al. Continuous representation of tumor microvessel density and detection of angiogenic hotspots in histological whole-slide images. Oncotarget. 2015;6:19163. doi: 10.18632/oncotarget.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Mărgăritescu C, et al. VEGF expression and angiogenesis in oral squamous cell carcinoma: an immunohistochemical and morphometric study. Clin. Exp. Med. 2010;10:209–214. doi: 10.1007/s10238-010-0095-4. [DOI] [PubMed] [Google Scholar]
- 421.Sakata J, et al. HMGA2 contributes to distant metastasis and poor prognosis by promoting angiogenesis in oral squamous cell carcinoma. Int. J. Mol. Sci. 2019;20:2473. doi: 10.3390/ijms20102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Dong Z, et al. Overexpression of Id-1 is associated with tumor angiogenesis and poor clinical outcome in oral squamous cell carcinoma. Oral Oncol. 2010;46:154–157. doi: 10.1016/j.oraloncology.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 423.Hong K-O, et al. Tumor budding is associated with poor prognosis of oral squamous cell carcinoma and histologically represents an epithelial-mesenchymal transition process. Hum. Pathol. 2018;80:123–129. doi: 10.1016/j.humpath.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 424.Almangush A, et al. Tumour budding in oral squamous cell carcinoma: a meta-analysis. Br. J. Cancer. 2018;118:577–586. doi: 10.1038/bjc.2017.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Staton CA, Reed MWR, Brown NJ. A critical analysis of current in vitro and in vivo angiogenesis assays. Int. J. Exp. Pathol. 2009;90:195–221. doi: 10.1111/j.1365-2613.2008.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Seki M, Sano T, Yokoo S, Oyama T. Tumour budding evaluated in biopsy specimens is a useful predictor of prognosis in patients with cN0 early stage oral squamous cell carcinoma. Histopathology. 2017;70:869–879. doi: 10.1111/his.13144. [DOI] [PubMed] [Google Scholar]
- 427.Alkhadar H, Macluskey M, White S, Ellis I. Perineural invasion in oral squamous cell carcinoma: Incidence, prognostic impact and molecular insight. J. Oral Pathol. Med. 2020;49:994–1003. doi: 10.1111/jop.13069. [DOI] [PubMed] [Google Scholar]
- 428.Yu E-H, Tu H-F, Wu C-H, Yang C-C, Chang K-W. MicroRNA-21 promotes perineural invasion and impacts survival in patients with oral carcinoma. J. Chin. Med. Assoc. 2017;80:383–388. doi: 10.1016/j.jcma.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 429.Tassone P, et al. The role of matrixmetalloproteinase-2 expression by fibroblasts in perineural invasion by oral cavity squamous cell carcinoma. Oral Oncol. 2022;132:106002. doi: 10.1016/j.oraloncology.2022.106002. [DOI] [PubMed] [Google Scholar]
- 430.Zollo M, et al. Targeting monocyte chemotactic protein-1 synthesis with bindarit induces tumor regression in prostate and breast cancer animal models. Clin. Exp. Metastasis. 2012;29:585–601. doi: 10.1007/s10585-012-9473-5. [DOI] [PubMed] [Google Scholar]
- 431.Rylands J, Lowe D, Rogers SN. Outcomes by area of residence deprivation in a cohort of oral cancer patients: survival, health-related quality of life, and place of death. Oral Oncol. 2016;52:30–36. doi: 10.1016/j.oraloncology.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 432.Liu F, et al. Prospective study on factors affecting the prognosis of oral cancer in a Chinese population. Oncotarget. 2017;8:4352. doi: 10.18632/oncotarget.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Lambert R, Sauvaget C, Camargo Cancela Mde, Sankaranarayanan R. Epidemiology of cancer from the oral cavity and oropharynx. Eur. J. Gastroenterol. Hepatol. 2011;23:633–641. doi: 10.1097/MEG.0b013e3283484795. [DOI] [PubMed] [Google Scholar]
- 434.Jayaprakash V, et al. Human papillomavirus types 16 and 18 in epithelial dysplasia of oral cavity and oropharynx: a meta-analysis, 1985–2010. Oral Oncol. 2011;47:1048–1054. doi: 10.1016/j.oraloncology.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Ramqvist T, et al. Studies on human papillomavirus (HPV) 16 E2, E5 and E7 mRNA in HPV-positive tonsillar and base of tongue cancer in relation to clinical outcome and immunological parameters. Oral Oncol. 2015;51:1126–1131. doi: 10.1016/j.oraloncology.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 436.Sousa LGde, et al. Human papillomavirus status and prognosis of oropharyngeal high-grade neuroendocrine carcinoma. Oral Oncol. 2023;138:106311. doi: 10.1016/j.oraloncology.2023.106311. [DOI] [PubMed] [Google Scholar]
- 437.Fakhry C, D’Souza G. Discussing the diagnosis of HPV-OSCC: Common questions and answers. Oral Oncol. 2013;49:863–871. doi: 10.1016/j.oraloncology.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Wang F, et al. A systematic investigation of the association between HPV and the clinicopathological parameters and prognosis of oral and oropharyngeal squamous cell carcinomas. Cancer Med. 2017;6:910–917. doi: 10.1002/cam4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Xiao W, et al. Circular RNAs in cell cycle regulation: mechanisms to clinical significance. Cell Prolif. 2021;54:e13143. doi: 10.1111/cpr.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Hu X, Xia K, Xiong H, Su T. G3BP1 may serve as a potential biomarker of proliferation, apoptosis, and prognosis in oral squamous cell carcinoma. J. Oral Pathol. Med. 2021;50:995–1004. doi: 10.1111/jop.13199. [DOI] [PubMed] [Google Scholar]
- 441.Wang J, et al. The prognostic value of B7‐H6 protein expression in human oral squamous cell carcinoma. J. Oral Pathol. Med. 2017;46:766–772. doi: 10.1111/jop.12586. [DOI] [PubMed] [Google Scholar]
- 442.Wu C-C, et al. Overexpression of FAM3C is associated with poor prognosis in oral squamous cell carcinoma. Pathol. Res. Pract. 2019;215:772–778. doi: 10.1016/j.prp.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 443.Yu V, et al. Electronic cigarettes induce DNA strand breaks and cell death independently of nicotine in cell lines. Oral Oncol. 2016;52:58–65. doi: 10.1016/j.oraloncology.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Hawk ET, Colbert Maresso K. E-Cigarettes: unstandardized, under-regulated, understudied, and unknown health and cancer risks e-cigarettes: unknown health and cancer risks. Cancer Res. 2019;79:6079–6083. doi: 10.1158/0008-5472.CAN-19-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Reidy J, McHugh E, Stassen LF. A review of the relationship between alcohol and oral cancer. Surgeon. 2011;9:278–283. doi: 10.1016/j.surge.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 446.Madathil SA, et al. Nonlinear association between betel quid chewing and oral cancer: Implications for prevention. Oral Oncol. 2016;60:25–31. doi: 10.1016/j.oraloncology.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 447.Chaturvedi AK, et al. Effect of prophylactic human papillomavirus (HPV) vaccination on oral HPV infections among young adults in the United States. J. Clin. Oncol. 2018;36:262. doi: 10.1200/JCO.2017.75.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Sathish N, Wang X, Yuan Y. Human papillomavirus (HPV)-associated oral cancers and treatment strategies. J. Dent. Res. 2014;93:29S–36S. doi: 10.1177/0022034514527969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Hübbers CU, Akgül B. HPV and cancer of the oral cavity. Virulence. 2015;6:244–248. doi: 10.1080/21505594.2014.999570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Gondivkar SM, et al. Involvement of viral factors with head and neck cancers. Oral Oncol. 2012;48:195–199. doi: 10.1016/j.oraloncology.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 451.de La Cour CD, Sperling CD, Belmonte F, Syrjänen S, Kjaer SK. Human papillomavirus prevalence in oral potentially malignant disorders: systematic review and meta‐analysis. Oral Dis. 2021;27:431–438. doi: 10.1111/odi.13322. [DOI] [PubMed] [Google Scholar]
- 452.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br. J. Cancer. 2016;114:237–242. doi: 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Farah, C. S. et al. Oral cancer and oral potentially malignant disorders. Int. J. Dent.2014, 853479 (2014). [DOI] [PMC free article] [PubMed]
- 454.Hu Q, et al. Obesity and genes related to lipid metabolism predict poor survival in oral squamous cell carcinoma. Oral Oncol. 2019;89:14–22. doi: 10.1016/j.oraloncology.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 455.Sinevici N, O’sullivan J. Oral cancer: deregulated molecular events and their use as biomarkers. Oral Oncol. 2016;61:12–18. doi: 10.1016/j.oraloncology.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 456.Dumache R. Early diagnosis of oral squamous cell carcinoma by salivary microRNAs. Clin. Lab. 2017;63:1771–1776. doi: 10.7754/Clin.Lab.2017.170607. [DOI] [PubMed] [Google Scholar]
- 457.Dikova V, Jantus-Lewintre E, Bagan J. Potential non-invasive biomarkers for early diagnosis of oral squamous cell carcinoma. J. Clin. Med. 2021;10:1658. doi: 10.3390/jcm10081658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Bolandparva F, et al. Early diagnosis of oral squamous cell carcinoma (OSCC) by miR-138 and miR-424-5p expression as a cancer marker. Asian Pac. J. Cancer Prev. 2021;22:2185. doi: 10.31557/APJCP.2021.22.7.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Sujir N, Ahmed J, Pai K, Denny C, Shenoy N. Challenges in early diagnosis of oral cancer: Cases series. Acta Stomatol. Croat. 2019;53:174. doi: 10.15644/asc53/2/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Roi A, et al. The challenges of OSCC diagnosis: salivary cytokines as potential biomarkers. J. Clin. Med. 2020;9:2866. doi: 10.3390/jcm9092866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Lingen MW, Kalmar JR, Karrison T, Speight PM. Critical evaluation of diagnostic aids for the detection of oral cancer. Oral Oncol. 2008;44:10–22. doi: 10.1016/j.oraloncology.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Carreras-Torras C, Gay-Escoda C. Techniques for early diagnosis of oral squamous cell carcinoma: Systematic review. Med. Oral Patol. Oral Cir. Bucal. 2015;20:e305. doi: 10.4317/medoral.20347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Javaid MA, Ahmed AS, Durand R, Tran SD. Saliva as a diagnostic tool for oral and systemic diseases. J. Oral Biol. Craniofacial. Res. 2016;6:67–76. doi: 10.1016/j.jobcr.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.McKinney SM, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577:89–94. doi: 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- 465.Kar A, et al. Improvement of oral cancer screening quality and reach: The promise of artificial intelligence. J. Oral Pathol. Med. 2020;49:727–730. doi: 10.1111/jop.13013. [DOI] [PubMed] [Google Scholar]
- 466.Alabi RO, et al. Machine learning in oral squamous cell carcinoma: current status, clinical concerns and prospects for future—et systematic review. Artif. Intell. Med. 2021;115:102060. doi: 10.1016/j.artmed.2021.102060. [DOI] [PubMed] [Google Scholar]
- 467.Wang X, et al. A personalized computational model predicts cancer risk level of oral potentially malignant disorders and its web application for promotion of non‐invasive screening. J. Oral Pathol. Med. 2020;49:417–426. doi: 10.1111/jop.12983. [DOI] [PubMed] [Google Scholar]
- 468.de Souza LL, et al. Machine learning for detection and classification of oral potentially malignant disorders: a conceptual review. J. Oral Pathol. Med. 2023;52:197–205. doi: 10.1111/jop.13414. [DOI] [PubMed] [Google Scholar]
- 469.Elmakaty I, Elmarasi M, Amarah A, Abdo R, Malki MI. Accuracy of artificial intelligence-assisted detection of oral squamous cell carcinoma: a systematic review and meta-analysis. Crit. Rev. Oncol./Hematol. 2022;178:103777. doi: 10.1016/j.critrevonc.2022.103777. [DOI] [PubMed] [Google Scholar]
- 470.Musulin J, et al. An enhanced histopathology analysis: an ai-based system for multiclass grading of oral squamous cell carcinoma and segmenting of epithelial and stromal tissue. Cancers. 2021;13:1784. doi: 10.3390/cancers13081784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Tian H, Zhang M, Jin G, Jiang Y, Luan Y. Cu-MOF chemodynamic nanoplatform via modulating glutathione and H2O2 in tumor microenvironment for amplified cancer therapy. J. Colloid Interface Sci. 2021;587:358–366. doi: 10.1016/j.jcis.2020.12.028. [DOI] [PubMed] [Google Scholar]
- 472.Tian H, et al. Enhancing the therapeutic efficacy of nanoparticles for cancer treatment using versatile targeted strategies. J. Hematol. Oncol. 2022;15:1–40. doi: 10.1186/s13045-022-01320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Tian H, et al. A cascaded copper-based nanocatalyst by modulating glutathione and cyclooxygenase-2 for hepatocellular carcinoma therapy. J. Colloid Interface Sci. 2022;607:1516–1526. doi: 10.1016/j.jcis.2021.09.049. [DOI] [PubMed] [Google Scholar]
- 474.Tian H, et al. A targeted nanomodulator capable of manipulating tumor microenvironment against metastasis. J. Control. Release. 2022;348:590–600. doi: 10.1016/j.jconrel.2022.06.022. [DOI] [PubMed] [Google Scholar]
- 475.Zhu Y, et al. Recent advances of nano-drug delivery system in oral squamous cell carcinoma treatment. Eur. Rev. Med. Pharmacol. Sci. 2019;23:9445–9453. doi: 10.26355/eurrev_201911_19438. [DOI] [PubMed] [Google Scholar]
- 476.Gharat SA, Momin MM, Bhavsar C. Oral squamous cell carcinoma: current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit. Rev. Therapeut. Drug Carr. Syst. 2016;33:363–400. doi: 10.1615/CritRevTherDrugCarrierSyst.2016016272. [DOI] [PubMed] [Google Scholar]
- 477.Fan H, et al. Light stimulus responsive nanomedicine in the treatment of oral squamous cell carcinoma. Eur. J. Med. Chem. 2020;199:112394. doi: 10.1016/j.ejmech.2020.112394. [DOI] [PubMed] [Google Scholar]
- 478.Zheng D-W, et al. Biomaterial-mediated modulation of oral microbiota synergizes with PD-1 blockade in mice with oral squamous cell carcinoma. Nat. Biomed. Eng. 2022;6:32–43. doi: 10.1038/s41551-021-00807-9. [DOI] [PubMed] [Google Scholar]
- 479.Mapanao AK, Santi M, Voliani V. Combined chemo-photothermal treatment of three-dimensional head and neck squamous cell carcinomas by gold nano-architectures. J. Colloid. Interface Sci. 2021;582:1003–1011. doi: 10.1016/j.jcis.2020.08.059. [DOI] [PubMed] [Google Scholar]
- 480.Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol. Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Pillay B, et al. The impact of multidisciplinary team meetings on patient assessment, management and outcomes in oncology settings: a systematic review of the literature. Cancer Treat. Rev. 2016;42:56–72. doi: 10.1016/j.ctrv.2015.11.007. [DOI] [PubMed] [Google Scholar]