Abstract
There are five fibroblast growth factor receptors (FGFRs), namely, FGFR1–FGFR5. When FGFR binds to its ligand, namely, fibroblast growth factor (FGF), it dimerizes and autophosphorylates, thereby activating several key downstream pathways that play an important role in normal physiology, such as the Ras/Raf/mitogen‐activated protein kinase kinase/extracellular signal‐regulated kinase, phosphoinositide 3‐kinase (PI3K)/AKT, phospholipase C gamma/diacylglycerol/protein kinase c, and signal transducer and activator of transcription pathways. Furthermore, as an oncogene, FGFR genetic alterations were found in 7.1% of tumors, and these alterations include gene amplification, gene mutations, gene fusions or rearrangements. Therefore, FGFR amplification, mutations, rearrangements, or fusions are considered as potential biomarkers of FGFR therapeutic response for tyrosine kinase inhibitors (TKIs). However, it is worth noting that with increased use, resistance to TKIs inevitably develops, such as the well‐known gatekeeper mutations. Thus, overcoming the development of drug resistance becomes a serious problem. This review mainly outlines the FGFR family functions, related pathways, and therapeutic agents in tumors with the aim of obtaining better outcomes for cancer patients with FGFR changes. The information provided in this review may provide additional therapeutic ideas for tumor patients with FGFR abnormalities.
Keywords: fibroblast growth factor, fibroblast growth factor receptor, signaling pathway, tumor, tyrosine kinase inhibitors (TKIs)
FGFR amplification, mutations, fusions, or rearrangements can activate many pathways that lead to tumor development, and FGFR inhibitory drugs, especially TKIs, exert antitumor effects by blocking these pathways, but at the same time, secondary drug resistance inevitably arises in some patients, as overcoming drug resistance becomes a challenge that we must overcome.
1. INTRODUCTION
A tumor is formed by the abnormal proliferation of cells due to the loss of normal regulation of local tissue cells at the gene level under the action of various tumorigenic factors, such as viruses, pollution, tobacco, alcohol, and chemical carcinogens, 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 which is attributed to the action of these factors in stimulating or inactivating key signaling pathways and genes, such as the phosphoinositide 3‐kinase (PI3K)/AKT, Wnt/β‐catenin, Ras/extracellular signal‐regulated kinase (ERK), nuclear factor kappa B (NF‐κB), phospholipase C gamma (PLCγ), sting, signal transducer and activator of transcription (STAT), fibroblast growth factor (FGF)/fibroblast growth factor receptor (FGFR), c‐Myc, p53, and so on. 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 Among them, the FGF/FGFR pathway plays a significant role in maintaining normal physiological balance of the human body.
FGF is the key ligand of FGFR. Normally, when FGFR is triggered by FGF, a lot of pathways involved in many physiological activities, such as cell growth, cell migration, cell survival, angiogenesis, embryonic organ development, tissue repair, wound healing, and metabolism are activated. The event is caused by the auto‐phosphorylation of FGFR. 43 , 44 , 45 , 46 , 47 The FGFR family includes five genes, namely, FGFR1, FGFR2, FGFR3, FGFR4, and FGFR5 (FGFRL1). FGFR1–4 auto‐phosphorylated tyrosine residues bind and phosphorylate adaptor proteins, such as FGFR substrate 2 (FRS2). FRS2 consists of a phosphotyrosine‐binding domain and a C‐terminal tail with multiple binding sites for the SH2 domains of growth factor receptor‐bound 2 (GRB2) and Src homology phosphatase 2 (Shp2). 48 , 49 Phosphorylated FRS2 recruits GRB2 and son of sevenless (SOS) to activate Ras and its downstream mitogen‐activated protein kinase (MAPK) pathway. 50 GRB2‐associated binding protein 1 (GAB1) binds to GRB2 and activates the AKT pathway. In addition to FRS2, PLCγ binds to the Y766 phosphorylated tyrosine residue of FGFR to activate the PLCγ/diacylglycerol/protein kinase C (PKC) pathway. 51 These pathways are involved in many physiological processes. For example, FGF2/FGFR regulates cumulus expansion and oocyte meiosis in mouse, which may be achieved due to the activation of the c‐Mos/MAPK pathway. 52 Under apoptotic stress, cells release FGF2, which promotes transcriptional upregulation of B‐cell lymphoma‐2 (BCL‐2) protein in a non‐cell‐autonomous manner through activation of the mitogen‐activated protein kinase kinase (MEK)/ERK axis, which protects neighboring cells from apoptotic damage. 53 The FGFR1/Janus kinase 2 (JAK2)/STAT3 signaling pathway, which is enhanced by human b‐defensin‐3, promotes wound healing, angiogenesis, and fibroblast activation. 43 FGF4 activates AMP‐activated protein kinase/Caspase 6 signal axis through FGFR4 to regulate liver stress response, reduce stem cell apoptosis and mitigate liver injury. 54
Notably, in addition to the typical FGF/FGFR pathways mentioned above, there are also some atypical examples. For instance, in Xenopus embryos, FGFR is activated by family with sequence similarity 3 member B to activate downstream ERK signaling and promote posterior development, forming an anterior–posterior axial pattern. 55 Furthermore, activation of ERK induces remodeling of cytoskeletal proteins, including F‐actin, embryonic calmodulin C‐cadherin, and the tight junction protein zonula occluden‐1, which is required for enhanced cellular junctions and tissue sclerosis during early embryogenesis. It is worth noting that, here, the FGFR1/ERK pathway is not FGF‐dependent due to mechanical stress. 56 FGF/FGFR2 was shown to enhance the SHH–BMP4 signaling axis in the mouse, thereby promoting uroepithelial and mesenchymal development in the early ureter. 57 In the liver, FGF15 acts on FGFR4, recruits and phosphorylates NF2, deregulates the inhibition of Hippo kinases Mst1/2 by Raf, which activates the Hippo pathway, downregulates the expression of bile acid synthase Cyp7a1, and limits bile acid synthesis. 58
At the same time, abnormal regulation of these pathways is also involved in the development and progression of many diseases, especially tumors. For example, FGFR in oligodendrocytes downregulates brain‐derived neurotrophic factor/tropomyosin receptor kinase B signaling via the ERK/AKT pathway, which may be associated with the development of multiple sclerosis. 59 Overexpression of high molecular weight FGF2 heterodimer (HMWFGF2), which activates Wnt signaling by targeting FGFR, is involved in the development of osteoarthrosis. 60 In tumor cells, FGFR inhibits the JAK/STAT signaling pathway activated by T cell‐produced interferon gamma and then reduces the expression of its target genes, namely, beta‐2 microglobulin (B2M), C‐X‐C motif chemokine ligand 10 (CXCL10), and programmed death‐ligand 1 (PD‐L1), thereby mediating immune escape. 60 In non‐small cell lung cancer (NSCLC), FGFR activates the MAPK/c‐Fos pathway to promote tumor cell proliferation, migration, and resistance to erlotinib. 61 Similarly, FGFR promotes cholangiocarcinoma cell progression and resistance to gemcitabine through upregulation of AKT/mammalian target of rapamycin (mTOR) and STAT3 signaling. 62 The AKT/SRY‐box 2 (SOX2) axis, mediated by FGFR2, also controls the stemness of pancreatic cancer. 63
In this review, we will summarize the role of the FGFR family involved in human tumors, related regulatory signaling, and possible targeted interventions. The information offered in this review will help to highlight the role of FGFR proteins in tumor pathogenesis as well as the possibility of FGFR proteins as targets for tumor therapy.
2. OVERVIEW OF THE FGFR FAMILY
2.1. Major ligands of FGFR
FGFs are the main ligands of FGFR. The FGF family has 18 canonical isoforms in human, namely, FGF1 to FGF10 and FGF16 to FGF23, and their molecular weights range from 15 to 38 kDa. The FGF family can be divided into the following six subfamilies: FGF1 (FGF1 and FGF2); FGF4 (FGF4, FGF5, and FGF6); FGF7 (FGF3, FGF7, FGF10, and FGF22); FGF8 (FGF8, FGF17, and FGF18); FGF9 (FGF9, FGF16, and FGF20); and FGF19 (FGF19, FGF21, and FGF23). Of these, five subfamilies are paracrine subfamilies, and one subfamily (FGF19 subfamily) is an endocrine subfamily. 64 , 65 , 66 , 67 These FGFs act by binding, dimerizing, and activating the tyrosine kinase of the FGFR and its downstream pathways. 68 , 69 It is worth noting that paracrine subfamilies also bind to heparin or heparan sulfate (HS) proteoglycan, which does not transmit signals but acts as a cofactor to shorten the diffusion distance between FGF and its secretory cells as well as regulates the binding and signaling of FGF to its receptor (FGFR). 70 , 71 , 72 , 73 In addition, the binding of FGF to HS stabilizes growth factors, limits growth factor diffusion, and provides cells with growth factor reserves. 74 , 75 , 76 The HS binding site on FGF consists of the β1‐β2 loop and a region composed of the β10 strand, β10‐β11 loop, β11 strand, and β11‐β12 loop. 69
2.2. The structure of FGFR
The conventional FGFR has four different members, namely, FGFR1 to FGFR4, which consist of an extracellular domain consisting of three immunoglobulin‐like domains (D1, D2, and D3), a single transmembrane domain, and an intracellular domain containing a tyrosine kinase domain. 77 , 78 In particular, D1 and D2 are linked by a sequence of seven to eight amino acids rich in glutamate, aspartate, and serine, which is known as the acid box (AB). D1 and AB play a key role in the auto‐inhibition of FGFR. 79 , 80 D2 has an HS‐binding site, which consists of a conserved positively charged region. 81 The D2‐D3 regions are critical for ligand binding and specificity. 82 FGFR1 to FGFR3 are selectively spliced to produce b and c isoforms, 83 , 84 , 85 and this splicing event modulates the specificity of FGFR ligand binding by altering the primary sequence of the critical ligand binding region in the C‐terminal half of D3. 86 , 87
There is a unique member of the FGFR family, known as FGFR like 1 (FGFRL1) or FGFR5. The extracellular domain of FGFRL1 is similar to the conventional extracellular domain of FGFR1–4, but the intracellular domain lacks the functional tyrosine kinase domain and contains only a short tail with a unique histidine‐rich motif. 88 , 89 , 90 FGFRL1 was initially thought to act as a decoy receptor to negatively regulate the canonical FGFR signal pathway. 91 , 92 However, later evidence has demonstrated that FGFRL1 also participates in the canonical FGFR pathway and exerts a number of functions, such as promoting cell differentiation. The expression levels of FGFRL1 are elevated together with FGFR2 and FGFR1 during the differentiation of mesenchymal cells into osteoblasts and adipocytes, respectively. 93 In pancreatic β cells, FGFRL1 interacts with SHP‐1 to enhance the ERK1/2 signaling pathway. 94 Moreover, FGFRL1 is induced by inflammation and acts as a coreceptor for FGFR1, promoting FGFR1‐induced survival. 95 (Figure 1)
FIGURE 1.
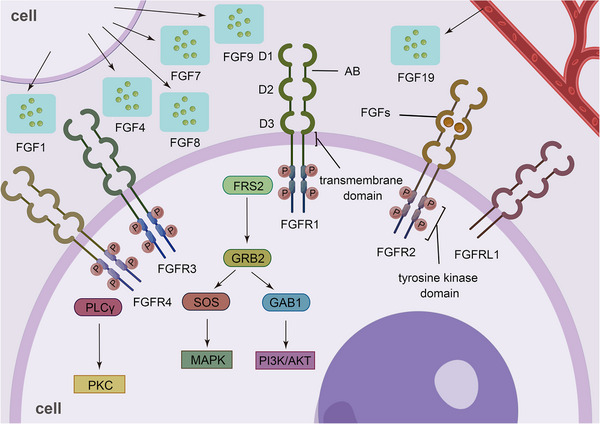
Basic structure of the FGFR family. There are five members of the FGFR family, FGFR1‐FGFR5, which all have an extracellular domain consisting of three immunoglobulin‐like domains (D1, D2, and D3), and a single transmembrane domain, differing in that FGFR5 lacks the intracellular tyrosine kinase domain.
3. ROLE OF FGFR IN NORMAL PHYSIOLOGY
During embryonic development, FGFR promotes cell growth, cell proliferation, cell survival, cell migration, angiogenesis, and organ development. In adult cells, FGFR contributes to tissue repair, wound healing, and metabolism. 96 , 97 , 98 , 99 , 100 , 101
3.1. Effect of FGFR on embryonic development
The FGFR pathway plays an active role in the development of almost all tissues and organs. When FGFR in mouse embryonic stellate or myofibroblastic cells is activated by FGF10, β‐catenin is activated, thus promoting the survival of hepatoblast cells. 102 Furthermore, FGFR is involved in the regulation of the anterior–posterior pattern of zeugopod in chicken limb development, and it plays a role in the early critical period of eye field formation. 103 , 104 FGFR2‐mediated reciprocal regulatory loops between FGF8 and FGF10 are essential for limb induction. 84 FGFR2 is the major FGFR expressed in Wolffian duct epithelial cells and is essential for the maintenance of the Wolffian duct, which is a primordium of the male reproductive tract, ureter, and kidney collecting duct system. 105 Development of the stomach is dependent on FGFR2b. 106 FGFR1 contributes to proliferation of mouse hippocampal progenitor cells. 107 The FGFR1 signaling pathway promotes mitosis, leading to proliferation of myoblast cells and delaying their differentiation. 108 , 109 It is worth noting that the role of FGFR in cell differentiation remains unclear, but it may involve various FGFR members in different tissues. It has been shown that FGFRs, especially the most expressed FGFR1 and FGFR2, and their downstream ERK signaling pathway have a negative effect on odontogenic differentiation. 110 FGFR3 has a negative effect on cartilage differentiation, which may be due to the interference of FGFR3 with Indian hedgehog/parathyroid hormone‐related protein signaling. At the same time, the Wnt/Lrp6 pathway is also activated. 111 , 112 , 113 In contrast, FGFR2 expression is increased during the differentiation of mesenchymal stromal cells to osteoblasts, while FGFR1 expression in the process of adipocyte differentiation is increased; both processes are accompanied by an increase in FGFRL1 levels. 93 During mouse embryonic development, the FGFRIIIc isoform drives the differentiation of embryonic stem cells into the endoderm. 114
N‐cadherin targets the FGFR to inhibit its ubiquitination and lysosomal degradation, leading to ERK1/2 phosphorylation, which is essential for the migration of neocortical projection neurons. 115 FGFR also regulates the proliferation and migration of lymphatic vessel endothelial cells by promoting glycolysis. 116 In Xenopus laevis, it has been found that FGFR2–4 promote the expression of Sema3a in the forebrain, while the expression of Slit1 is dependent on FGFR1; both are mediated by the PIK3/AKT pathway, which synergistically directs retinal ganglion cell axons from the mid‐mesencephalon to the parietal membrane, thus forming a functional neural circuit. 117 , 118 , 119 FGFR1 is the dominant FGFR expressed in endothelial cells, which promotes angiogenesis through downstream ERK1/2, c‐Jun N‐terminal kinase, p38 MAPK, and focal adhesion kinase. 120 , 121 , 122 Because endothelial cells also have the ability to proliferate in adults, the above processes of lymphangiogenesis or angiogenesis may also occur in adult cells and participate in processes, such as inflammation or wound healing.
3.2. Effect of FGFR on adult cells
In adult cells, the promotion of FGFR1 for neovascularization, tissue repair, and wound healing is achieved by activating the JAK2/STAT3 pathway. 43 In addition, FGFR1 may also be involved in angiogenesis and plaque instability within inflammatory plaques in rabbit atherosclerosis. 123 Bile acids are important mediators of liver regeneration, while FGFR, activated by FGF15, maintains bile acid homeostasis and is an important mediator of bile acids for liver regeneration. 124
4. FGFR IS INVOLVED IN HUMAN TUMORS
It has been shown that FGFR genetic alterations are present in 7.1% of tumors, of which 66% are amplification, 26% are mutations, and 8% are rearrangements. Alterations in FGFR1 are the most common (3.5%) followed by FGFR3 (2.0%), FGFR2 (1.5%), and FGFR4 (0.5%). 125 In addition, some epigenetic alterations may also affect the expression levels or function of FGFR, thus activating significant signaling pathways to promote tumor cell proliferation, invasion, migration, epithelial–mesenchymal transition (EMT), angiogenesis, metabolic changes, chemoradiotherapy resistance, and tumor cell stemness maintenance 126 , 127 , 128 , 129 (Table 1).
TABLE 1.
FGFR genetic alterations are associated with tumor development.
| FGFRs | Tumors | Major genetic alterations | Diagnostic or prognostic makers | References |
|---|---|---|---|---|
| FGFR1 | Breast cancer | Amplification | + | 133, 134 |
| Lung cancer | + | 152, 153 | ||
| Ovarian cancer | ‐ | 163 | ||
| Bladder cancer | + | 59 | ||
| Renal cell carcinoma | ‐ | 164 | ||
| Prostate cancer | ‐ | 165 | ||
| Esophageal carcinoma | ‐ | 167 | ||
| Gastric cancer | + | 168 | ||
| Colorectal cancer | ‐ | 169 | ||
| Pancreatic cancer | ‐ | 63, 170 | ||
| Head and neck squamous cell carcinoma | ‐ | 171 | ||
| Osteosarcoma | ‐ | 172 | ||
| Acute myeloid leukemia | Fusions or rearrangements | ‐ | 188 | |
| 8p11 myeloproliferative syndrome | ‐ | 189, 190 | ||
| Stem cell leukemia/lymphoma syndrome | ‐ | 191 | ||
| Gliomas | Mutations | + | 193 | |
| FGFR2 | Gastric cancer | Amplification | + | 208 |
| Colorectal cancer | ‐ | 210 | ||
| Intrahepatic cholangiocarcinoma | Fusions or rearrangements | + | 196, 197 | |
| Gliomas | ‐ | 205, 206 | ||
| Endometrial cancer | Mutations | + | 212 | |
| Melanomas | ‐ | 213 | ||
| Breast cancer | Gene polymorphisms | + | 229, 230 | |
| Prostate cancer | Altered expression of splice isoform | + | 237 | |
| Renal cell carcinoma | ‐ | 242 | ||
| Bladder cancer | ‐ | 244 | ||
| FGFR3 | Gliomas | Fusions or rearrangements | ‐ | 266, 267 |
| Lung cancer | ‐ | 268, 269 | ||
| Bladder cancer | Mutations | + | 251, 252 | |
| Hepatocellular carcinoma | ‐ | 261 | ||
| Renal cell carcinoma | ‐ | 262 | ||
| Colorectal cancer | Overexpression | + | 274 | |
| FGFR4 | Hepatocellular carcinoma | Overexpression | + | 276, 277 |
| Gastric cancer | + | 285, 286 | ||
| Colorectal cancer | ‐ | 287, 288 | ||
| Breast cancer | ‐ | 289, 290 | ||
| Thyroid cancer | ‐ | 291 | ||
| Nasopharyngeal carcinoma | ‐ | 292 | ||
| FGFRL1 | Small cell lung cancer | Overexpression | ‐ | 298 |
| Oral squamous cell carcinoma | ‐ | 299 | ||
| Ovarian carcinoma | + | 300 | ||
| Prostate cancer | + | 301 |
Gene amplification, mutations, fusions, or rearrangements of FGFRs and unknown upregulation of FGFRs expression are associated with the development and progression of many tumors. The table shows the main genetic alterations of FGFR1–4 in various tumors, where “+” indicates that the genetic alteration can be used as a diagnostic or prognostic marker for the tumor, and “‐” indicates that it cannot or is not clear.
4.1. Regulatory network of FGFR1
FGFR1 is located at 8p11.23 and has a total length of 68,620 bases. FGFR1 is expressed in all tissues and organs throughout the body, especially in plasma, heart, and cerebrospinal fluid. FGFR1 is the most widely reported and studied gene among the FGFR family members, and it has been widely reported as an oncogene to promote tumor initiation and development, 130 indicating its importance in tumorigenesis. The abnormal activation of FGFR1 is mainly associated with its genetic amplification, mutations, fusions, and rearrangements.
The most frequent abnormal activation of FGFR1 is gene amplification. The amplification rate of FGFR1 is 10−20% in estrogen receptor positive (ER+) breast cancer 131 , 132 , 133 and 5% in triple negative breast cancer (TNBC). 134 In TNBC, FGF2, as a ligand for FGFR1, activates the ERK/AKT pathway and promotes nuclear translocation of c‐Rel through FGFR1, which promotes the expression and secretion of S100A4. Paracrine activation of the S100A4/receptor for advanced glycation end products (RAGE) pathway promotes angiogenic effects in vascular endothelial cells and migration of tumor‐associated fibroblasts. 135 Activation of the ERK/AKT pathway also phosphorylates or reprograms the ER, leading to chemoresistance to fulvestrant and the cyclin‐dependent kinase (CDK)4/6 inhibitor, palbociclib, in ER+ breast cancer. 136 , 137 Moreover, resistance to metformin of ER+ breast cancer cells is caused by FGFR1‐activated insulin receptor substrate 1/ERK signaling. 138 Activation of the ERK2/c‐Fos/forkhead box Q1 (FOXQ1) pathway plays a key role in the growth of breast cancer cells. 139 FGFR1 has been reported to enhance the expression of vascular endothelial growth factor A through tumor necrosis factor alpha‐induced protein 3, thus contributing to breast cancer angiogenesis. 140 By gene set enrichment analysis, Riaz et al. 141 found that FGFR1 is strongly correlated with GLI1 family zinc finger 1 (GLI1), a member of the Sonic hedgehog (SHH) pathway, and that the FGFR/GLI1 axis significantly promotes the metastasis of tumor cells. Furthermore, nuclear FGFR1, interacting with RNA‐Polymerase II (Pol II) and forkhead box protein A1 (FOXA1), regulates the transcription of target genes, which is a process independent of tyrosine kinase activity and mediates resistance of ER+ breast cancer cells to estrogen inhibitors and fulvestrant. 142 Some noncoding RNAs regulate FGFR1 expression. For example, microRNA (miR)−338‐3p directly targets the 3′‐untranslated region of FGFR1 to reduce its levels, while circular RNA (circRNA)‐TFF1 acts as a sponge for miRNA‐338‐3p, indirectly restoring the expression of FGFR1. 143 MiR‐133b inhibits breast cancer cell growth and resistance to cisplatin by targeting FGFR1 to inactivate the Wnt/β‐catenin pathway. 144 MiR‐136 and miR‐326 directly bind to FGFR1, and circ‐0000518 competitively inhibits miR‐326. 145 , 146 Using bioinformatics, Boothby‐Shoemaker et al. 147 found that leptin mRNA is positively correlated with FGFR1 mRNA in breast cancer. Subsequently, Pang et al. 148 demonstrated that leptin‐induced pre‐B‐cell leukemia transcription factor 3 binds to the promoter of FGFR1, thereby activating the FGFR1 pathway and leading to letrozole resistance in tumor cells. The negative regulator of FGFR1, namely, circFGFR1‐encoded circFGFR1p, inhibits tumor cell development due to the absence of the tyrosine kinase structural domain. 149
FGFR1 amplification is in 10−20% of patients with NSCLC 150 , 151 , 152 and 5−7% of patients with small cell lung cancer. 153 , 154 , 155 In KRAS mutant NSCLC, FGFR1 activates the MAPK/mTOR pathway, leading to increased expression of D‐cyclins and CDK6, resulting in resistance to palbociclib. In this case, the activation of mTOR is independent of AKT and is due to ERK phosphorylation of its key regulator, namely, tuberous sclerosis complex 2. 156 Activation of the MAPK pathway has also been reported to lead to resistance to EGFR inhibitors and EMT of lung cancer cells, which was induced by hypoxia. 157 Moreover, this drug resistance is also induced by activation of the AKT pathway. 158 ERK downregulates beclin‐1 to inhibit autophagy of tumor cells. 159 CircFGFR1 acts as a sponge for miR‐381‐3p to upregulate C‐X‐C motif chemokine receptor 4, thereby promoting the progression of NSCLC and resistance to anti‐PD‐1‐based therapy. 160 In addition, miR‐22 in vascular endothelial cells in NSCLC tissues directly targets sirtuin 1 and FGFR1, inactivating their common AKT/mTOR pathway to inhibit angiogenesis and tumor cell growth. 161 In small cell lung cancer, the promotion of tumor development by FGFR1 is mediated by the phospholipase C gamma 1 pathway, and retinoblastoma‐like protein 2 is negatively correlated with the expression of FGFR1. 162
FGFR1 amplification also plays a role in ovarian cancer, 163 bladder cancer, 59 renal cell carcinoma, 164 prostate cancer, 165 , 166 esophageal carcinoma, 167 gastric cancer, 168 colorectal cancer, 169 pancreatic cancer, 63 , 170 head and neck squamous cell carcinoma, 62 , 171 and osteosarcoma. 172 L1 cell adhesion molecule has been demonstrated to facilitate ovarian cancer cell spheroid formation, tumor initiation, and chemoresistance by triggering FGFR1 and its downstream SRC/STAT3 pathway. 173 Glycosyltransferase 8 domain containing 2 induces resistance to Cis‐dichlorodiammine‐platinum treatment in ovarian cancer cells through activation of the FGFR/PI3K/AKT signaling pathway. 174 In bladder cancer, tweety family member 3 inhibits phosphorylation of FGFR1 and downregulates the H‐Ras/A‐Raf/MEK/ERK signaling pathway (downstream targets include c‐Jun and c‐Fos) to inhibit the invasion and migration of bladder cancer cells. 175 In prostate cancer, yes‐associated protein (YAP)/T‐box transcription factor 5 activates FGFR1 to mediate resistance to MET inhibitors. 176 FGFR1 has also been shown to be associated with downregulation of choline kinase α and dysregulated choline metabolism, which promotes the progression of prostate cancer. 177 HOTAIR promotes migration and invasion of prostate cancer cells by competitively inhibiting miR‐520b, which indirectly increases the expression of FGFR1. 178 MiR‐1205 directly targets FGFR1 to inhibit colony formation, metastasis, and resistance to cisplatin in gastric cancer cells, while circARVCF reverses this effect. 179 MiR‐198 and miR‐497 have also been found to target FGFR1. 180 , 181 Furthermore, AGAP2‐AS1 is highly expressed in colon cancer tissues, and it promotes colon cancer cell proliferation, migration, invasion, and resistance to gemcitabine via sponging miR‐497 to regulate FGFR1. 182 In pancreatic cancer, SNHG1 also competitively inhibits miR‐497. 183 The FGFR1/SRC/NF‐κB signaling axis plays a critical role in maintaining the stemness of pancreatic cancer cells. 184 Interestingly, the nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells favors pancreatic cancer cell invasion. 185 MiR‐573 also directly interacts with FGFR1 and reduces its expression levels, thereby inhibiting tumor cell migration, invasion, and EMT. 186
Although occurring infrequently, the gene mutations, fusions, or rearrangements of FGFR1 also lead to the development of some tumors. For example, FGFR gene fusions or rearrangements have been reported in squamous NSCLC, 187 acute myeloid leukemia, 188 8p11 myeloproliferative syndrome, 189 , 190 and stem cell leukemia/lymphoma syndrome. 191 The common fusion partners of NSCLC, acute myeloid leukemia, and 8p11 myeloproliferative syndrome are transforming acidic coiled‐coil‐containing protein 1 (TACC1), FGFR1 oncogene partner 2, and breakpoint cluster region, respectively. The mutation rate of FGFR1 p.N546K or p.K656E in low‐grade gliomas and mixed neuronal‐glial tumors is 6%, which is the fifth most common gene alteration in these tumors. 192 In glioblastoma multiforme, Notch2 enhances the activity of FGFR1, thereby enhancing the AKT/glycogen synthase kinase 3 (GSK3) pathway to inhibit apoptosis. 193 Notably, miR‐3116 inactivates the PI3K/AKT pathway by downregulating FGFR1, thus sensitizing glioma cells to temozolomide 194 (Figure 2).
FIGURE 2.
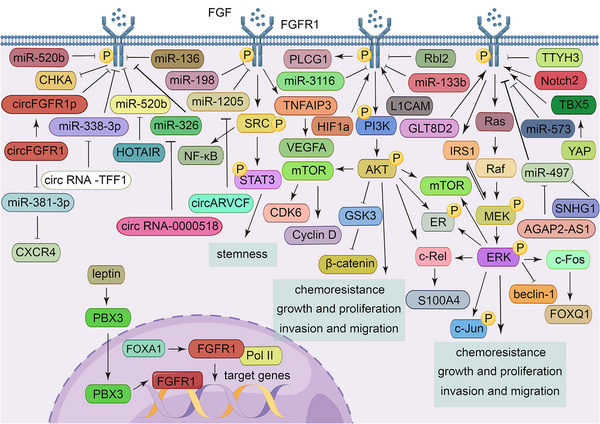
FGFR1 regulatory networks. In the cytoplasm, FGFR1 exerts its cancer‐promoting effects, mainly by activating the PI3K/AKT, Ras/ERK, and STAT pathways, to promote tumor cell proliferation, invasion, and metastasis as well as maintain tumor cell stemness and improve resistance to chemotherapy. In the nucleus, FGFR exerts its cancer‐promoting effects by targeting genes in a process independent of tyrosine kinase activity.
4.2. Role and signal network of FGFR2 in tumors
FGFR2 is located at 10q26.13 and has a length of 120,129 bases, and it is mainly expressed in the brain and spinal cord. Gene fusions or rearrangements of FGFR2 are the most common FGFR gene alteration events, and these are found almost exclusively in intrahepatic cholangiocarcinoma to facilitate its progress. 195
In patients with intrahepatic cholangiocarcinoma, the incidence of FGFR2 rearrangements or fusions is 9−16%, and this is often accompanied by mutational inactivation of tumor protein P53, cyclin‐dependent kinase inhibitor 2A (CDKN2A), or BRCA1 associated protein‐1. 196 , 197 , 198 , 199 FGFR2 fusions usually involve exons 1−17, and the common fusion partners are BicC family RNA‐binding protein 1 (BICC1), KIAA1217, TACC2, coiled‐coil domain‐containing protein 6 (CCDC6), and adenosylhomocysteinase like 1. The FGFR2‐N/A fusion is when intron 17 or exon 18 of FGFR2 is fused to an intergenic region. 200 The domain of FGFR2 is replaced by various fusion partners, and fusion partners then dimerize adjacent to FGFR2 to activate the tyrosine kinase domain along with a series of downstream signaling pathways. 201 It has been reported that 32.9% of patients have a unique fusion partner, while 15.7% of patients have a fusion partner shared with only one other patient. 200 This fusion diversity has been suggested to result in different biological effects. 50 In addition, FGFR2 extracellular domain in‐frame deletions and the truncation of exon 18 also activate the downstream pathways of FGFR2. 201 , 202 SOX9, which is stimulated by the Wnt/β‐catenin pathway, enhances the transcription and expression of FGF7 and FGFR2, thereby stimulating the downstream pathway of FGFR2 to promote the proliferation of cholangiocarcinoma cells and resistance to pemigatinib. 203 Through activation of the AKT/mTOR pathway, FGFR2 reduces the sensitivity of cholangiocarcinoma to gemcitabine. 204 The correct folding of FGFR2–TACC3, FGFR2‐meningioma‐expressed antigen 5, and FGFR2–BICC1 requires the help of heat shock protein 90 (HSP90). 205
FGFR2 fusions and rearrangements have also been found in gliomas. 205 , 206 In gliomas, FGFR2 phosphorylates phosphatase and tensin homolog on tyrosine 240, which interacts with Ki‐67 when exposed to ionizing radiation, resulting in a rapid increase and binding to chromosomes, thereby promoting the recruitment of RAD51 to facilitate DNA repair, ultimately leading to radiation resistance. 207
FGFR2 is also subjected to gene amplification or mutations in gastric cancer, 208 , 209 colorectal cancer, 210 , 211 endometrial cancer, 171 , 212 and melanomas. 213 FGFR2 promotes gastric cancer progression by downregulating thrombospondin‐4 (TSP4) through the PI3K/AKT/mTOR signaling axis. 214 Similarly, FGFR2 activates the PI3K/AKT/mTOR axis and its downstream TSP1, which regulates migration and invasion of gastric cancer cells. 215 FGF18, a ligand for FGFR2, interacts with FGFR2, enhances F‐actin, and then promotes nuclear aggregation of YAP1; FGFR2 also activates the MAPK pathway and its downstream molecule, c‐Jun, which upregulates YAP1 transcription to promote the progression of gastric cancer. 216 Furthermore, some molecules act by regulating the levels of FGFR2 in gastric cancer. For example, there is a mutual positive regulation between CD44 and FGFR2. 217 The posttranscriptional process of FGFR2 is regulated by the human DEAD/H‐box RNA helicase, DDX6, a protein encoded by a fusion gene. 218 MiR‐381‐3p, miR‐494, miR‐5701, and miR‐519e‐5p also directly target FGFR2 to inhibit its expression and gastric cancer progression, while methyl‐CpG‐binding domain protein 1 and histone deacetylase 3 bind to form a complex that inhibits miR‐5701 expression, thereby restoring FGFR2 levels. 219 , 220 , 221 Long noncoding RNA (lncRNA) ASNR also competitively inhibits miR‐519e‐5p. 222 FGFR2 has been reported to mediate immune tolerance in colorectal cancer cells by inducing PD‐L1 expression through the JAK/STAT3 pathway. 223 In endometrial cancer, FGFR2 has been reported to be activated in an autocrine manner, which activates AKT signaling and its downstream hairy and enhancer of split‐1 (HES1), thereby promoting tumor cell proliferation. 224 Moreover, activating mutations in FGFR2 also induce Golgi fragmentation, loss of polarity, and directional migration. 225
In addition, gene polymorphisms of FGFR2, including rs2981582, 226 , 227 , 228 , 229 rs1219648, 230 , 231 rs35054928, and rs45631563, 232 have been found to be associated with susceptibility to breast cancer. In breast cancer, FGFR2 activates multiple pathways. Activation of the FGFR2/STAT3 pathway promotes proliferation, invasion, migration, and EMT of tumor cells. 233 Ribosomal s6 kinase 2 interacts with FGFR2 to form an indirect complex that promotes FGF2‐triggered internalization of FGFR2. 234 FGFR2 positively regulates PD‐L1 and helps tumor cells undergo immune escape. 235 CD151 downregulates the posttranscriptional levels of FGFR2 via PKC, a process that requires HuR, a multifunctional RNA‐binding protein, and the assembly of processing bodies (P‐bodies). 236
In some prostate cancers, the FGFR2 splice isoform, FGFR2 IIIc, has been found to be overexpressed, while FGFR2 IIIb expression is reduced. 237 , 238 FGFR2IIIb reverses the EMT of tumor cells and enhances sensitivity to docetaxel. 239 MiR‐628 directly targets FGFR2 to inhibit the proliferation and invasion of prostate cancer cells. 240 In contrast, Lee et al. 241 suggested that nuclear FGFR2 plays an anticancer role by interacting with the transactivation domain of hypoxia‐inducible factor 1 (HIF1α) under hypoxic conditions, blocking the recruitment of coactivator p300 and thus leading to transcriptional repression of HIF target genes, which may be related to different subcellular localizations of FGFR. Conversion of FGFR2IIIb to FGFR2IIIc has also been identified in renal cell carcinoma and bladder cancer. 242 , 243 , 244 MiR‐148b‐3p inhibits the growth and angiogenesis of renal cell carcinoma upon binding to FGFR2. 245 Similarly, miR‐223 directly targets FGFR2 to promote bladder cancer growth and metastasis, an event that is counteracted by circUVRAG 246 (Figure 3).
FIGURE 3.
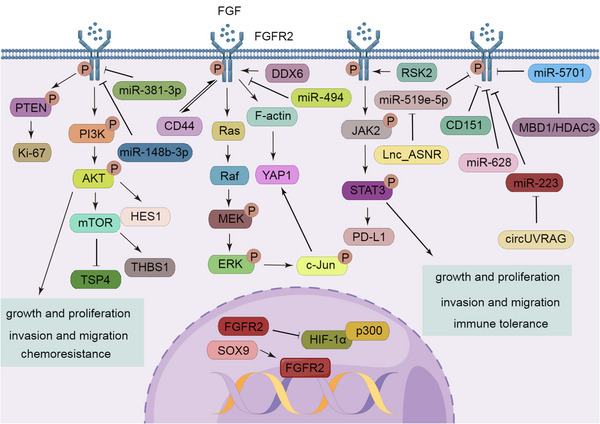
FGFR2 regulatory networks. In the cytoplasm, FGFR2 promotes tumor cell growth, tumor cell migration, and immune resistance through activation of the PI3K/AKT, Ras/ERK, and STAT pathways. In the nucleus, FGFR2 exerts tumor suppressive effects by inhibiting the recruitment of P300.
4.3. FGFR3 functions as a promoter in tumors
FGFR3 is located at 4p16.3 and contains 15,580 bases. FGFR3 is expressed in the brain, kidney, and testis, but it has low or no expression in the spleen, heart, and muscle. Abnormal activation of FGFR3 has been recognized to play a cancer‐promoting role in patients with urothelial cancer, especially bladder cancer, in which FGFR3 mutations, rearrangements, and fusions are the most common. 247 , 248 In addition, the tyrosine kinase inhibitor (TKI) has been shown to have a tangible effect, and its use has been approved in urothelial patients with FGFR abnormalities. 249 , 250
Bladder cancer is the most frequently occurring tumor with FGFR3 mutations, with an incidence of 9−11%. 251 , 252 , 253 In bladder cancer, activation of the SRC pathway by FGFR3 leads to resistance of tumor cells to FGFR inhibitors, such as infigratinib or erdafitinib. 254 FGFR3 upregulates ETS translocation variant 5 (ETV5) via the MAPK/ERK pathway, leading to elevated transcriptional coactivator with PDZ‐binding motif (TAZ), a cotranscriptional regulator of the Hippo signaling pathway involved in cell contact inhibition, which promotes the proliferation of bladder cancer cells. 255 Moreover, activation of the FGFR3/RAS/MAPK pathway has also been found to promote the invasion of upper tract urothelial cancer. 256 FGFR3 phosphorylates and activates the E3 ubiquitin ligase, neuronal precursor cell expressed developmentally downregulated 4, thereby inducing ubiquitinated degradation of PD‐L1 to regulate CD8+ T cell‐mediated immune detection. 257 Acrolein from cigarette smoke induces cisplatin resistance in muscle invasive bladder cancer via the FGFR pathway. 258 MiR‐181a‐5p directly targets FGFR3 to inhibit its expression and downstream STAT3 pathway, while circ_0068871 reverses this condition. 259 MiR‐99a‐5p also directly binds to FGFR3 to inhibit mTOR signaling. 260
Mutations in FGFR3 are also present in hepatocellular carcinoma (HCC) 261 and renal cell carcinoma. 262 FGFR3 regulates angiogenesis and metastasis in HCC by increasing the level of monocyte chemotactic protein 1. 263 In addition, the FGFR3 splice isoform, FGFR3 IIIb/IIIc, and its ligand, FGF9, have been found to be upregulated in HCC, promoting tumor cell growth and aggressiveness. 264 Similarly, a newly identified splicing mutant, FGFR3Δ7–9, which directly connects exon 6 and exon 10, directly phosphorylates the ten‐eleven translocation‐2 DNA demethylase and causes its ubiquitinated degradation, activating the AKT pathway to promote the proliferation of HCC. 125 , 265
FGFR3 may also undergo rearrangements or amplifications in gliomas 266 , 267 and lung cancer. 268 , 269 In glioma, the most common fusion partner of FGFR3 is TACC3, and the fusion process of FGFR3–TACC3, which requires HSP90, requires the involvement of cell division cycle 37 (CDC37). 270 Furthermore, lncRNA CCAT1 promotes the proliferation, migration, and EMT of glioma cells through sponging miR‐181b. 271 MiR‐99b targets FGFR3 to inhibit lung cancer progression. 272 Interestingly, circFGFR3 has been reported to enhance galectin‐1 expression by competitively binding to miR‐22‐3p, thereby activating the AKT and ERK pathways to promote the proliferation and invasion of NSCLC cells. 273
Notably, FGFR3 is overexpressed in colorectal cancer, but this overexpression is not associated with genetic alterations but may be due to epigenetic modifications. 274 In addition, protein arginine methyltransferase 5 methylates histone 4 to promote the transcription of FGFR3 275 (Figure 4).
FIGURE 4.
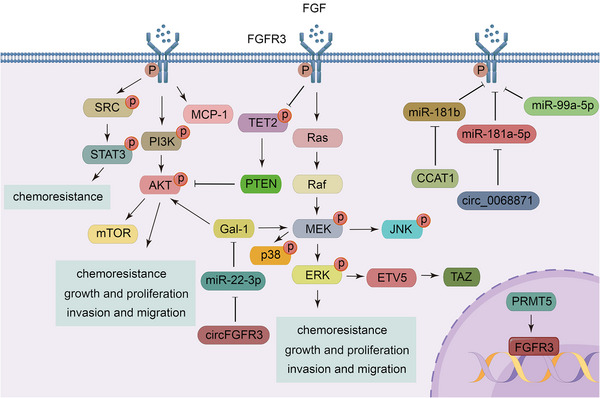
FGFR3 regulatory networks. FGFR3 promotes tumor cell growth, tumor cell migration, and chemoresistance through activation of the PI3K/AKT, Ras/ERK, and STAT pathways, and FGFR3 is regulated by various molecules, including numerous miRNAs.
4.4. Oncogenic role and signal modulation of FGFR4
FGFR4 is located at 5q35.2 and has 11,230 bases, and it is mainly expressed in the lung and liver. As low probability events, the genetic alterations of FGFR4 have not been thoroughly investigated, but FGFR4 and its ligand, FGF19, have been found to be overexpressed and induce tumor development in many tumors, especially in HCC. 276 , 277 , 278
Shin et al. 279 found that FGFR4 activates the SRC/STAT3 pathway, and FGFR4 forms an endosomal complex with SRC and STAT3, which enters the nucleus in HCC, representing a potential mechanism of action for the FGFR4 pathway complex. Moreover, endoplasmic reticulum stress induces FGF19, which in turn activates GSK3β/Nrf2 signaling via FGFR4 to resist this event. 280 Activation of GSK3β/β‐catenin signaling is also associated with EMT. 281 FGFR4 activates store‐operated Ca2+ entry and its downstream nuclear factor of activated T cells‐c2 via PLCγ or the ERK pathway to promote self‐renewal of HCC stem cells. 282 FGFR4 activates the PI3K/AKT/HIF1α pathway to promote transcription of homeobox B5, which in turn upregulates FGFR4, thereby promoting tumor cell metastasis. 283 Furthermore, miR‐486‐3p directly targets FGFR4 to mediate growth inhibition and overcome sorafenib resistance in HCC. 284
Overexpression of FGFR4 also occurs in gastric cancer, 285 , 286 colorectal cancer, 287 , 288 breast cancer, 289 , 290 thyroid cancer, 291 and nasopharyngeal carcinoma. 292 Helicobacter pylori infection increases the expression of FGFR4 in gastric cancer cells through activation of the STAT3 pathway by FGF19, and STAT3 binds directly to the FGFR4 promoter, forming a feed‐forward response loop. 293 In addition, miR‐491‐5p indirectly downregulates FGFR4 levels. 294 In colon cancer tissues, tumor‐associated fibroblasts generate chemokine ligand 2, which acts on its receptor on the tumor surface to enhance the transcription of FGFR4 in an Ets‐1‐dependent manner, thereby activating the β‐catenin pathway to lead to EMT. 295 The expression of FGFR4 is also regulated by forkhead box C1 (FOXC1), which directly targets FGFR4, promotes its transcription, and drives metastasis of colorectal cancer cells. 287 In human epidermal growth factor receptor 2 (HER2)+ breast cancer, epigenetic alteration of m6A hypomethylation causes FGFR4 to phosphorylate GSK3β and then activate the β‐catenin/transcription factor 4 signaling pathway, leading to anti‐HER2 therapeutics in tumor cells. 296 Activation of the MAPK/ERK pathway leads to elevated glucose metabolism and resistance to adriamycin in breast cancer 297 (Figure 5).
FIGURE 5.
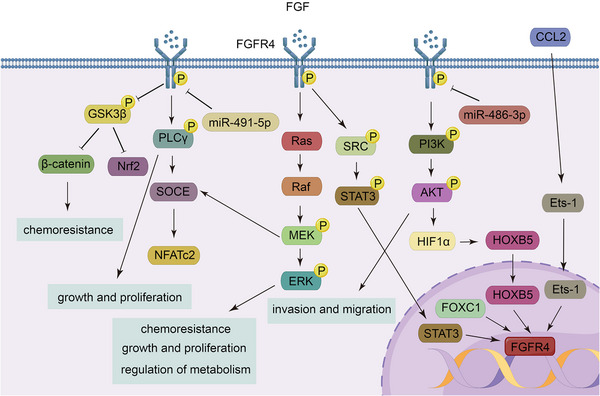
FGFR4 regulatory networks. Through activation of the PI3K/AKT, Ras/ERK, STAT, PLCγ, and Wnt/β‐catenin pathways, FGFR4 promotes the progression of a variety of tumors, while its expression levels are regulated by many molecules via transcriptional or posttranscriptional processes.
4.5. Action and molecular basis of FGFRL1 in tumors
FGFRL1 is located at 4p16.3 with a length of 16,963 bases; it is preferentially expressed in cartilage tissue and pancreas, and weakly expressed in lung, small intestine and spleen. FGFRL1 has been reported to have elevated expression in small cell lung cancer, 298 oral squamous cell carcinoma, 299 ovarian carcinoma, 300 and prostate cancer. 301 Despite lacking the intracellular tyrosine kinase domain, FGFRL1 interacts with a number of proteins to indirectly activate several signaling pathways, thereby promoting tumor development. For instance, in small cell lung cancer, enhanced FGFRL1 interacts with alpha‐enolase to activate its downstream PI3K/AKT pathway, leading to chemotherapy resistance. 298 Under hypoxic conditions, HIF1α binds to the promoter of FGFRL1 to promote it expression levels. Subsequently, FGFRL1 upregulates Gil1 and activates the Hedgehog (Hh) pathway, thereby promoting the growth, proliferation, and migration of ovarian cancer cells. 302 Moreover, the expression of FGFRL1 is regulated by a number of molecules. In esophageal squamous cell carcinoma, miR‐210 directly targets FGFRL1 to induce cell death and cell cycle arrest in G1/G0 and G2/M, thereby inhibiting cancer cell survival and proliferation. 303 Similarly, miR‐210 plays the same role in HCC. 304 In NSCLC, lncRNA FGD5‐AS1 indirectly upregulates FGFRL1 levels and promotes cancer cell proliferation through sponging hsa‐miR‐107 305 (Figure 6).
FIGURE 6.
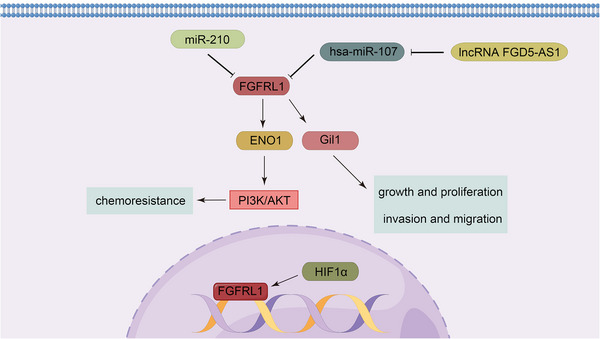
FGFRL1 regulatory networks. Despite the lack of an intracellular tyrosine kinase structural domain, FGFRL1 promotes tumor progression by interacting with various molecules and indirectly activating a number of pathways.
5. FGFR ACTS AS A THERAPEUTIC TARGET
FGFR amplification, mutations, rearrangements, or fusions are considered as potential biomarkers of FGFR therapeutic response. However, it is important to note that the amplification of FGFR is not equivalent to overexpression. There have been many findings indicating that the association between FGFR amplification and overexpression is not strong. For example, Gonzalez‐Ericsson et al. 131 found that only 50% of FGFR‐amplified breast cancers exhibit protein overexpression. And, there is evidence that it is the high expression levels of FGFR1–4 rather than copy‐number alteration that is strongly associated with the efficacy of FGFR inhibitors. 306 Similarly, Bogatyrova et al. 307 suggested that the overexpression of FGFR1 in NSCLC is better explained by promoter demethylation or downregulation of specific miRNAs, which may be related to the poor effect of FGFR TKIs in some patients with FGFR amplification. The difference in FGFR amplification fragments may be the cause of the above events. Therefore, it may be better to use FGFR amplification and overexpression as a combined treatment indicator. In addition, a series of genetic alterations in FGFR2 have been found to result in transcription of exon 18‐truncated FGFR2, which should perhaps also be considered as an indicator of the effectiveness of FGFR‐targeted therapy. 308
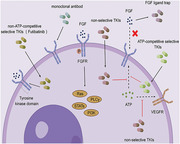
TKIs can be divided into nonselective and selective inhibitors. Nonselective inhibitors include ponatinib (AP24534), lenvatinib, ninetedanib (BIBF 1120), olverembatinib (GZD824, HQP1351), dovitinib (TKI258, CHIR‐258), lucitanib (AL3810), and derazantinib (ARQ 087). Selective inhibitors include futibatinib (TAS120), erdafitinib (JNJ‐42756493), LY2874455, pemigatinib (INCB054828, FIGHT‐101), infigratinib (BGJ398), AZD4547, rogaratinib (BAY1163877), E7090 (tasurgratinib), and debio 1347 (CH5183284) (Table 2).
TABLE 2.
Common TKIs and the cancers they treat and their adverse effects.
| Category | Drugs | Target points | Drug status | Main types of cancer treated | Side effects | clinical trial number | References |
|---|---|---|---|---|---|---|---|
| Non‐selective TKIs | Ponatinib | FGFR 1–4 | Marketed | Philadelphia chromosome‐positive acute lymphoblastic leukemia, chronic myeloid leukemia | Pancreatitis, hypertension, hyperlipidemia, liver dysfunction, and AOE | ‐ | 309, 310 |
| Lenvatinib | Hepatocellular carcinoma, thyroid cancer, renal cell carcinoma | Fatigue, loss of appetite, diarrhea, weight loss, hypertension, and some liver‐related side effects | ‐ | 311, 312 | |||
| Nintedanib | FGFR 1–3 | Non‐small cell lung cancer | Gastrointestinal reactions, decreased platelets and hypertension | ‐ | 313, 314 | ||
| Olverembatinib | Chronic myeloid leukemia | Skin pigmentation, thrombocytopenia, hypocalcemia, proteinuria, and hyperuricemia | ‐ | 315, 316 | |||
| Dovitinib | Phase 3 clinical trial | Breast cancer, hepatocellular carcinoma, renal cell carcinoma prostate cancer | Nausea, vomiting, fatigue, anorexia, and diarrhea | NCT02116803, NCT01223027 | 317, 318 | ||
| Lucitanib | FGFR1 and FGFR2 | Small cell lung cancer, breast cancer, nasopharyngeal carcinoma | Hypertension, hypothyroidism, nausea, and proteinuria | NCT04254471 | 319, 320 | ||
| Derazantinib | FGFR 1–3 | Phase 2 clinical trial | Intrahepatic cholangiocarcinoma | Fatigue, ocular toxicity, and hyperphosphatemia | NCT03230318, NCT05174650 | 321, 322 | |
| Selective TKIs | Futibatinib | FGFR 1–4 | Marketed | Intrahepatic cholangiocarcinoma | Hyperphosphatemia, diarrhea, and nausea | ‐ | 341, 342 |
| Erdafitinib | Uroepithelial carcinoma | Hyponatremia, stomatitis, hyperphosphatemia, and weakness | ‐ | 343 | |||
| Pemigatinib | FGFR 1–3 | Cholangiocarcinoma | Hyperphosphatemia, hypophosphatemia, arthralgia, stomatitis, hyponatremia, abdominal pain, fatigue, and fever | ‐ | 195, 346 | ||
| Infigratinib | Cholangiocarcinoma | Hyperphosphatemia, fatigue, stomatitis, and alopecia | ‐ | 347, 348 | |||
| AZD4547 | Phase 3 clinical trial | Breast cancer, squamous cell lung cancer | Hyperphosphatemia, dry mouth, hair loss, taste disturbance, constipation, nausea, and retinal pigment epithelial detachment | NCT02965378 | 349, 350 | ||
| Rogaratinib | Uroepithelial carcinoma | Diarrhea, decreased appetite, fatigue, and asymptomatic lipase elevation | NCT03410693 | 351, 352 | |||
| E7090 | Phase 2 clinical trial | Cholangiocarcinoma | NCT04238715, NCT04962867 | 353, 354 | |||
| Debio 1347 | Breast cancer, cholangiocarcinoma | Hyperphosphatemia, diarrhea, constipation, fatigue, loss of appetite, and dry mouth | NCT03834220, NCT03344536 | 355, 356 | |||
| LY2874455 | FGFR 1–4 | Phase 1 clinical trial | Gastric cancer, head and neck squamous cell carcinoma | Hyperphosphatemia, diarrhea, and stomatitis | NCT01212107 | 344, 345 |
TKIs are usually classified as selective or nonselective, with different mechanisms of action, targets, and side effects.
5.1. TKIs
5.1.1. Nonselective TKIs
Non‐selective inhibitors include ponatinib, 309 , 310 lenvatinib, 311 , 312 ninetedanib, 313 , 314 olverembatinib, 315 , 316 dovitinib, 317 , 318 lucitanib, 319 , 320 and derazantinib. 321 , 322 Nonselective inhibitors are characterized by their ability to inhibit FGFR as well as some other kinases, such as vascular endothelial growth factor receptor (VEGFR), platelet‐derived growth factor receptor (PDGFR), and fetal liver tyrosine kinase receptor (FLT).
Ponatinib has been shown to be active in a variety of tumors, such as philadelphia chromosome‐positive acute lymphoblastic leukaemia, chronic myeloid leukemia, endometrial cancer, bladder cancer, gastric cancer, breast cancer, lung cancer, and colon cancer, and it acts on more than 40 kinases, including FGFR, SFKs, VEGFR, and PDGFR. 323 , 324 Ponatinib has significant adverse effects, mainly including pancreatitis, hypertension, hyperlipidemia, liver dysfunction, and arterial occlusive events (AOEs). 325 Lenvatinib is a first‐line systemic chemotherapeutic agent used in patients with unresectable HCC that inhibits FGFR1–4, thereby reducing tumor stem cells in HCC. 326 It can also be used for thyroid cancer and renal cell carcinoma. Lenvatinib also targets VEGFR1–3, PDGFRα, RET, and KIT. The most common adverse effects of lenvatinib treatment include fatigue, loss of appetite, diarrhea, weight loss, hypertension, and some liver‐related side effects, such as ascites and hepatic encephalopathy. 327 Nintedanib is approved in combination with docetaxel for the treatment of locally advanced, metastatic or locally recurrent non‐small cell lung cancer after chemotherapy due to its ability to block FGFR1–3, EGFR1–3, and PDGFFRα and β. Common adverse reactions include gastrointestinal reactions, decreased platelets and hypertension. 328 , 329 Olverembatinib binds to FGFR1–3, FLT3, and PDGFRα, and it overcomes the drug resistance activity of some mutations, such as FGFR1‐V561F/M, indicating that it is a new drug to target FGFR. 330 In addition, as an approved drug, it can treat T315I‐mutated chronic myeloid leukemia. 315 , 331 Common adverse events with olverembatinib include skin pigmentation, thrombocytopenia, hypocalcemia, proteinuria, and hyperuricemia. 332 The primary target of dovitinib is VEGFR1–3, but it also targets FGFR1–3, PDGFRβ, FMS‐like tyrosine kinase 3 (FLT3), KIT, RET, TrkA, and colony stimulating factor 1 (Csf‐1). Dovitinib has shown therapeutic effects in breast cancer, HCC, prostate cancer, renal cell carcinoma, melanoma, multiple myeloma, and gastrointestinal mesenchymal tumors and its major drug‐related toxicities include nausea, vomiting, fatigue, anorexia, and diarrhea. 333 , 334 , 335 Lucitanib strongly inhibits FGFR1, FGFR2, and VEGFR1–3, and it has antiangiogenic and broad‐spectrum antitumor activity against several cancers, including small cell lung cancer, breast cancer, nasopharyngeal carcinoma, colorectal cancer, ovarian cancer, and kidney cancer. 336 , 337 Common adverse events of lucitanib include hypertension, hypothyroidism, nausea, and proteinuria. 338 Derazantinib inhibits FGFR1–3, CSF1R ahd VEGFR2. It has shown antitumor activity in advanced, unresectable intrahepatic cholangiocarcinoma that has progressed after chemotherapy, and the treatment‐related adverse events of derazantinib include fatigue, ocular toxicity, and hyperphosphatemia. 339 , 340
As nonselective inhibitors, drug toxicity is a major problem that the above mentioned drugs have to face, but it is undeniable that some drugs have shown resistance to mutations, such as the above mentioned third generation TKIs olverembatinib.
5.1.2. Selective TKIs
Selective inhibitors include futibatinib, 341 , 342 erdafitinib, 343 LY2874455, 344 , 345 pemigatinib, 195 , 346 infigratinib, 347 , 348 AZD4547, 349 , 350 rogaratinib, 351 , 352 E7090, 353 , 354 and debio 1347. 355 , 356 Selective inhibitors target FGFR to inhibit downstream signaling pathways, thereby acting as cancer suppressors. Futibatinib, 357 , 358 erdafitinib, 359 , 360 and LY2874455 are pan‐FGFR inhibitors. Futibatinib, a non‐ATP‐competitive FGFR1–4 inhibitor that binds covalently and irreversibly to the conserved cysteine in the P‐loop of the FGFR kinase domain, has resulted in remission in various cancers, including cholangiocarcinoma, gastric cancer, uroepithelial cancer, central nervous system tumors, head and neck cancer, and breast cancer, with the greatest activity in intrahepatic cholangiocarcinoma with FGFR2 rearrangements or fusions. And, it has been approved in September 2022 for the treatment of adult, previously treated, unresectable, locally advanced or metastatic FGFR2 fusions or rearrangements intrahepatic cholangiocarcinoma patients. In addition, as a third‐generation irreversible TKI, futibatinib is able to overcome resistance caused by some mutations in the FGFR kinase domain, including FGFR2‐K660M, N550K, and L618V. 361 The most common adverse events of futibatinib are hyperphosphatemia, diarrhea, and nausea. 362 Erdafitinib has been approved for patients with uroepithelial carcinoma with abnormal FGFR; its common adverse effects include hyponatremia, stomatitis, hyperphosphatemia, and weakness. 363 LY2874455 has been found to be useful in gastric and head and neck squamous cell carcinoma, and, notably, as a novel drug, it is resistant to most FGFR mutations resulting in drug resistance, including FGFR4 V550L and FGFR1–561 M, which partially compensates for the lack of futibatinib action, but its specific action needs to be further clarified. Its main side effects include hyperphosphatemia, diarrhea, and stomatitis. 364 , 365 The inhibitors of FGFR1–3 are pemigatinib, 366 infigratinib, AZD4547, 367 rogaratinib, E7090 and debio 1347. Pemigatinib and infigratinib have been approved for cholangiocarcinoma with FGFR fusions or rearrangements. The most common adverse effect of pemigatinib is hyperphosphatemia, and other adverse effects of pemigatinib include hypophosphatemia, arthralgia, stomatitis, hyponatremia, abdominal pain, fatigue, and fever. 195 Adverse events associated with infigratinib include hyperphosphatemia, fatigue, stomatitis, and alopecia. 368 AZD4547 has significant efficacy in breast cancer and squamous cell lung cancer patients, and the common adverse events of AZD4547 include hyperphosphatemia, dry mouth, hair loss, taste disturbance, constipation, nausea, and retinal pigment epithelial detachment. 369 Rogaratinib has been shown to be effective in patients with uroepithelial and head and neck squamous cell carcinoma with overexpressed levels of FGFR. Common adverse effects include diarrhea, decreased appetite, fatigue, and asymptomatic lipase elevation. 370 , 371 E7090 selectively inhibits FGFR1–3 and has been found to be effective in patients with cholangiocarcinoma with FGFR2 gene fusions and in patients with gastric cancer with FGFR2 gene amplification or increased expression, but more information is needed for a larger sample size. 372 Debio 1347 is an ATP‐competitive, highly selective FGFR1–3 inhibitor, and it is mainly used to treat breast cancer and cholangiocarcinoma. The common adverse effects of debio 1347 include hyperphosphatemia, diarrhea, constipation, fatigue, loss of appetite, and dry mouth. 373
5.2. Non‐TKIs FGFR‐targeted drugs
In addition to the TKIs mentioned above, here are some other drugs that target FGFR. For example, bemarituzumab (FPA144), a monoclonal antibody against FGFR2IIb, is still in clinical trials and is used to treat patients with FGFR2IIb overexpression, including gastroesophageal cancer. It reduces the risk of hyperphosphatemia compared with the TKIs mentioned above. Of course, it still has some side effects, such as neutropenia, corneal disease, and stomatitis. 374 , 375 , 376 And there is the FGF ligand trap, FP‐1039 (GSK3052230), which is the extracellular ligand binding domain of FGFR1 fused to the Fc segment of human immunoglobulin G1, thereby inhibiting the activation of FGFR by FGF. Its common adverse effects include neutropenia, hair loss, nausea, joint pain, weakness and diarrhea 377 , 378 , 379 (Figure 7).
FIGURE 7.
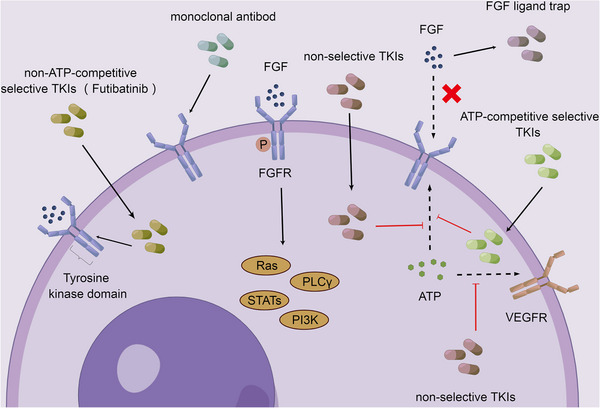
Major FGFR‐targeted drugs in tumors. The target drugs of FGFR in tumors are mainly TKIs, among which, futibatinib as a non‐ATP‐competitive inhibitor can directly bind to the P‐loop of kinase domain, while other drugs are ATP kinase inhibitors, in addition, there are also monoclonal antibody drugs and FGF ligand trap.
6. SUMMARY AND OUTLOOK
In tumor tissues, some genetic or epigenetic changes in FGFR, such as gene amplification, mutations, fusions, or rearrangements, as well as histone methylation or noncoding RNAs mediate the elevation of FGFR expression, lead to aberrant activation of FGFR, which upregulates a number of unique pathways, such as the PI3K/AKT, Ras/ERK, PLCγ, STAT, and Wnt/β‐catenin signaling pathways, to promote tumorigenesis and progression, including tumor cell proliferation, tumor cell invasion, tumor cell migration, EMT, angiogenesis, metabolic changes, chemoradiotherapy resistance, and tumor cell stemness. Because auto‐phosphorylation of FGFR plays a key role in the activation of these pathways, TKIs have become an important strategy for inhibiting the action of FGFR.
Several drugs are now approved for use in cancer treatment. For example, erdafitinib is approved for patients with uroepithelial carcinoma with abnormal FGFR, and pemigatinib and infigratinib are approved for patients with cholangiocarcinoma with FGFR fusions or rearrangements. 362 However, TKIs may have several problems throughout the treatment process. Among some patients with alterations in FGFR, especially FGFR amplification, TKIs are less effective. First, as mentioned above, FGFR amplification is not strongly associated with elevated FGFR expression in some cancers. 307 Therefore, it remains unclear whether FGFR amplification and increased expression should be used as a combined TKIs treatment indicator. Second, the regulatory mechanism of nuclear FGFR on its target genes is independent of tyrosine kinase activity. 142 On this point, the role of other non‐TKIs FGFR‐targeting agents needs to be further confirmed. Therefore, the exploration of new targets for FGFR is necessary. Third, because FGFR mutations are accompanied by changes in some other tumor promoter, tumor suppressor, or pathways, such as P53, EGFR, or MAPK signaling, combination therapy may be a new breakthrough point. There have been some studies. 380 , 381 , 382 For example, the nonselective TKI lenvatinib is thought to inhibit FGFR with feedback activation of the EGFR–PAK2–ERK5 signaling axis, while the combination of lenvatinib and the EGFR inhibitor gefitinib has been found to be clinically meaningful in patients with advanced HCC. 311 In KRAS‐mutated lung adenocarcinoma, the use of the MEK inhibitor trametinib leads to abnormal activation of FGFR1, which leads to the development of drug resistance. However, in combination with FGFR inhibitors, this resistance was suppressed. 383 In addition, in a mouse model of lung cancer with p53mut, erdafitinib was found to promote T cell cloning and expansion and promote anti‐tumor immunity when combined with PD‐1 blockers. 384 But the specific efficacy and toxic effects need to be studied in depth. In addition, some patients have developed resistance to TKIs. There are many mechanisms for the development of drug resistance, the most common of which are FGFR kinase and especially gatekeeper mutations. Gatekeeper residues are located in the hinge region of the ATP‐binding pocket of the kinase, and when they are mutated, they cause spatial inhibition of the ATP pocket, thus blocking entry of TKIs and causing drug resistance. 385 Common gatekeeper mutations include FGFR1–V561F/M, FGFR2–V564F/I, V565I, and FGFR3–V555M. 362 , 386 In addition, the development of FGFR resistance is associated with the activation of some related pathways, that is, bypassing the inhibitory effect of TKIs on the FGF/FGFR signaling pathway, which may result from other mutations in nongatekeepers. For instance, FGFR2 N550H mutation was found to upregulate the PI3K/AKT/mTOR signaling pathway. 387 Some new, mutation‐targeting drugs have emerged, such as futibatinib and GZD824, but they still have some limitations. 330 In summary, there is a demand for the development of new FGFR inhibitory drugs as well as the identification of combinations of drugs (related pathway molecular inhibitors) and other covariant gene inhibitors.
AUTHOR CONTRIBUTION
Q. L., J. Y. H., and W.W.Y. collected the related reports. Q. L. drafted the manuscript and discussed the concepts of the manuscript. J. Y. H. and W.W.Y. provided valuable discussion. Z. L., S. L., and W. Y. F., provided valuable discussion and revised the manuscript. W. Y. F. participated in designing the review. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Not applicable.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant number 81872198) and the Nature Science Foundation of Guangdong Province (grant number 2022A1515012624). We use Figdraw (https://www.figdraw.com/static/index.html) to create our figures.
Liu Q, Huang J, Yan W, Liu Z, Liu S, Fang W. FGFR families: biological functions and therapeutic interventions in tumors. MedComm. 2023;4:e367. 10.1002/mco2.367
Contributor Information
Zhen Liu, Email: narcissus_jane@163.com.
Shu Liu, Email: lius6963@sina.com.
Weiyi Fang, Email: fangweiyi1975@163.com.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Parida S, Drewes JL. Unwanted passengers: microbes hitchiking in breast cancer metastases. Cell Host Microbe. 2022;30(6):875‐877. [DOI] [PubMed] [Google Scholar]
- 2. Wang Z, Zhai Z, Chen C, et al. Air pollution particles hijack peroxidasin to disrupt immunosurveillance and promote lung cancer. Elife. 2022;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu N, Xiong W. Early life exposure to tobacco smoke and lung cancer in adulthood. Am J Respir Crit Care Med. 2022;207(3):370–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouras E, Tsilidis KK, Triggi M, Siargkas A, Chourdakis M, Haidich AB. Diet and risk of gastric cancer: an umbrella review. Nutrients. 2022;14(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mahajan V, Gujral P, Jain L, Ponnampalam AP. Differential expression of steroid hormone receptors and ten eleven translocation proteins in endometrial cancer cells. Front Oncol. 2022;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gapstur SM, Bandera EV, Jernigan DH, et al. Alcohol and Cancer: existing knowledge and evidence gaps across the cancer continuum. Cancer Epidemiol Biomarkers Prev. 2022;31(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia C, Su J, Liu C, et al. Human microbiomes in cancer development and therapy. MedComm. 2023;4(2):e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin X, Li A‐M, Li Y‐H, et al. Silencing MYH9 blocks HBx‐induced GSK3β ubiquitination and degradation to inhibit tumor stemness in hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meng Q, Sun H, Wu S, et al. Epstein‐Barr virus‐encoded microRNA‐BART18‐3p promotes colorectal cancer progression by targeting de novo lipogenesis. Adv Sci (Weinh). 2022;9(35):e2202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu Q‐Y, Zhao G‐X, Li Y, et al. Advances in pathogenesis and precision medicine for nasopharyngeal carcinoma. MedComm. 2021;2(2):175‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ho SR, Lee YC, Ittmann MM, Lin FT, Chan KS, Lin WC. RNF144A deficiency promotes PD‐L1 protein stabilization and carcinogen‐induced bladder tumorigenesis. Cancer Lett. 2021;520:344‐360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang JY, Du Y, Gong LP, et al. ebv‐circRPMS1 promotes the progression of EBV‐associated gastric carcinoma via Sam68‐dependent activation of METTL3 (vol 535, 215646, 2022). Cancer Lett. 2022;545:4. [DOI] [PubMed] [Google Scholar]
- 13. Citro S, Miccolo C, Medda A, et al. HPV‐mediated regulation of SMAD4 modulates the DNA damage response in head and neck cancer. J Exp Clin Cancer Res. 2022;41(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen QQ, Wang YL, Yang L, et al. PM2.5 promotes NSCLC carcinogenesis through translationally and transcriptionally activating DLAT‐mediated glycolysis reprograming. J Exp Clin Cancer Res. 2022;41(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan Y, Cai XS, Shen FR, Ma F. HPV post‐infection microenvironment and cervical cancer. Cancer Lett. 2021;497:243‐254. [DOI] [PubMed] [Google Scholar]
- 16. Rao S, Hossain T, Mahmoudi T. 3D human liver organoids: An in vitro platform to investigate HBV infection, replication and liver tumorigenesis. Cancer Lett. 2021;506:35‐44. [DOI] [PubMed] [Google Scholar]
- 17. Paolini F, Amici C, Carosi M, et al. Intrabodies targeting human papillomavirus 16 E6 and E7 oncoproteins for therapy of established HPV‐associated tumors. J Exp Clin Cancer Res. 2021;40(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boffetta P, Hecht S, Gray N, Gupta P, Straif K. Smokeless tobacco and cancer. Lancet Oncol. 2008;9(7):667‐675. [DOI] [PubMed] [Google Scholar]
- 19. Hecht SS, Hatsukami DK. Smokeless tobacco and cigarette smoking: chemical mechanisms and cancer prevention. Nat Rev Cancer. 2022;22(3):143‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stasiewicz M, Karpiński TM. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 2022;86(Pt 3):633‐642. [DOI] [PubMed] [Google Scholar]
- 21. Krump NA, You J. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol. 2018;16(11):684‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marongiu L, Allgayer H. Viruses in colorectal cancer. Mol Oncol. 2022;16(7):1423‐1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaglia MM, Munger K. Editorial overview: Viruses and cancer. Curr Opin Virol. 2019;39:iii‐iiv. [DOI] [PubMed] [Google Scholar]
- 24. Chen S, Cao XF, Zhang JY, Wu WY, Zhang B, Zhao FQ. circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c‐Myc translation. Adv Sci. 2022;9(8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang R, Liu J, Li K, et al. An SETD1A/Wnt/β‐catenin feedback loop promotes NSCLC development. J Exp Clin Cancer Res. 2021;40(1):318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cui YB, Zhang CY, Ma SS, et al. RNA m6A demethylase FTO‐mediated epigenetic up‐regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ouyang SM, Li HX, Lou LL, et al. Inhibition of STAT3‐ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol. 2022;52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi XH, Yang JX, Liu MY, et al. Circular RNA ANAPC7 inhibits tumor growth and muscle wasting via PHLPP2‐AKT‐TGF‐I3 signaling axis in pancreatic cancer. Gastroenterology. 2022;162(7):2004‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiong W, Zhang B, Yu HX, Zhu L, Yi L, Jin X. RRM2 regulates sensitivity to sunitinib and PD‐1 blockade in renal cancer by stabilizing ANXA1 and activating the AKT pathway. Adv Sci. 2021;8(18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Jiang Q, Liu X, et al. Cinobufotalin powerfully reversed EBV‐miR‐BART22‐induced cisplatin resistance via stimulating MAP2K4 to antagonize non‐muscle myosin heavy chain IIA/glycogen synthase 3β/β‐catenin signaling pathway. EBioMedicine. 2019;48:386‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kornepati AVR, Vadlamudi RK, Curiel TJ. Programmed death ligand 1 signals in cancer cells. Nat Rev Cancer. 2022;22(3):190‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hou RT, Liu X, Yang HL, et al. Chemically synthesized cinobufagin suppresses nasopharyngeal carcinoma metastasis ENKUR to stabilize. Cancer Lett. 2022;531:57‐70. [DOI] [PubMed] [Google Scholar]
- 33. Lou JC, Hao YC, Lin KF, et al. Circular RNACDR1asdisrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol Cancer. 2020;19(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hou R, Li Y, Luo X, et al. ENKUR expression induced by chemically synthesized cinobufotalin suppresses malignant activities of hepatocellular carcinoma by modulating β‐catenin/c‐Jun/MYH9/USP7/c‐Myc axis. Int J Biol Sci. 2022;18(6):2553‐2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu J‐H, Yang H‐L, Deng S‐T, et al. The small molecule chemical compound cinobufotalin attenuates resistance to DDP by inducing ENKUR expression to suppress MYH9‐mediated c‐Myc deubiquitination in lung adenocarcinoma. Acta Pharmacol Sin. 2022;43(10):2687‐2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang PCT, Chung JYF, Xue VWW, et al. Smad3 promotes cancer‐associated fibroblasts generation via macrophage‐myofibroblast transition. Adv Sci. 2022;9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mirzaei S, Paskeh MDA, Okina E, et al. Molecular landscape of LncRNAs in prostate cancer: a focus on pathways and therapeutic targets for intervention. J Exp Clin Cancer Res. 2022;41(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu Y, Yu QF, Wang P, et al. A selective small‐molecule c‐Myc degrader potently regresses lethal c‐Myc overexpressing tumors. Adv Sci. 2022;9(8):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen Y‐L, Wang Y‐M, Zhang Y‐X, et al. Targeting cyclin‐dependent kinase 9 in cancer therapy. Acta Pharmacol Sin. 2022;43(7):1633‐1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng XJ, Chen WL, Yi J, et al. Apolipoprotein C1 promotes glioblastoma tumorigenesis by reducing KEAP1/NRF2 and CBS‐regulated ferroptosis. Acta Pharmacol Sin. 2022;43(11):3011‐3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dhanasekaran R, Deutzmann A, Mahauad‐Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene ‐ the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2022;19(1):23‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi M, Umehara Y, Yue H, et al. The antimicrobial peptide human β‐defensin‐3 accelerates wound healing by promoting angiogenesis, cell migration, and proliferation through the FGFR/JAK2/STAT3 signaling pathway. Front Immunol. 2021;12:712781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patel VN, Pineda DL, Berenstein E, et al. Loss of Hs3st3a1 or Hs3st3b1 enzymes alters heparan sulfate to reduce epithelial morphogenesis and adult salivary gland function. Matrix Biol. 2021;103:37‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watson J, Ferguson HR, Brady RM, et al. Spatially resolved phosphoproteomics reveals fibroblast growth factor receptor recycling‐driven regulation of autophagy and survival. Nat Commun. 2022;13(1):6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu XL, Qiu C, Wang YR, et al. FGFR1 SUMOylation coordinates endothelial angiogenic signaling in angiogenesis. Proc Natl Acad Sci USA. 2022;119(26):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao Y, Wang Q, Xie C, et al. Peptide ligands targeting FGF receptors promote recovery from dorsal root crush injury via AKT/mTOR signaling. Theranostics. 2021;11(20):10125‐10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ong SH, Guy GR, Hadari YR, et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Research Support, Non‐U.S. Gov't. Mol Cell Biol. 2000;20(3):979‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dhalluin C, Yan KS, Plotnikova O, et al. Structural basis of SNT PTB domain interactions with distinct neurotrophic receptors. Research Support, U.S. Gov't, P.H.S. Mol Cell. 2000;6(4):921‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cristinziano G, Porru M, Lamberti D, et al. FGFR2 fusion proteins drive oncogenic transformation of mouse liver organoids towards cholangiocarcinoma. J Hepatol. 2021;75(2):351‐362. [DOI] [PubMed] [Google Scholar]
- 51. Mohammadi M, Dionne CA, Li W, et al. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Research Support, Non‐U.S. Gov't. Nature. 1992;358(6388):681‐684. [DOI] [PubMed] [Google Scholar]
- 52. Du C, Davis JS, Chen C, et al. FGF2/FGFR signaling promotes cumulus‐oocyte complex maturation in vitro. Reproduction. 2021;161(2):205‐214. [DOI] [PubMed] [Google Scholar]
- 53. Bock FJ, Sedov E, Koren E, et al. Apoptotic stress‐induced FGF signalling promotes non‐cell autonomous resistance to cell death. Nat Commun. 2021;12(1):6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song L, Wang L, Hou Y, et al. FGF4 protects the liver from nonalcoholic fatty liver disease by activating the AMP‐activated protein kinase‐Caspase 6 signal axis. Hepatology (Baltimore, Md). 2022;76(4):1105‐1120. [DOI] [PubMed] [Google Scholar]
- 55. Zhang FF, Zhu XC, Wang P, et al. The cytokine FAM3B/PANDER is an FGFR ligand that promotes posterior development in Xenopus. Proc Natl Acad Aci USA. 2021;118(20):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brown WS, Tan L, Smith A, Gray NS, Wendt MK. Covalent targeting of fibroblast growth factor receptor inhibits metastatic breast cancer. Mol Cancer Ther. 2016;15(9):2096‐2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meuser M, Deuper L, Rudat C, et al. FGFR2 signaling enhances the SHH‐BMP4 signaling axis in early ureter development. Development. 2022;149(1) [DOI] [PubMed] [Google Scholar]
- 58. Ji S, Liu Q, Zhang S, et al. FGF15 activates Hippo signaling to suppress bile acid metabolism and liver tumorigenesis. Dev Cell. 2019;48(4) [DOI] [PubMed] [Google Scholar]
- 59. Ross JS, Wang K, Al‐Rohil RN, et al. Advanced urothelial carcinoma: next‐generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2014;27(2):271‐280. [DOI] [PubMed] [Google Scholar]
- 60. Xiao LP, Williams D, Hurley MM. Inhibition of FGFR signaling partially rescues osteoarthritis in mice overexpressing high molecular weight FGF2 isoforms. Endocrinology. 2020;161(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu C, Zhou A, Hu X, et al. LMNA reduced acquired resistance to erlotinib in NSCLC by reversing the epithelial‐mesenchymal transition via the FGFR/MAPK/c‐fos signaling pathway. Int J Mol Sci. 2022;23(21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clauditz TS, Bottcher A, Hanken H, et al. Prevalence of fibroblast growth factor receptor 1 (FGFR1) amplification in squamous cell carcinomas of the head and neck. J Cancer Res Clin Oncol. 2018;144(1):53‐61. [DOI] [PubMed] [Google Scholar]
- 63. Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie YL, Su N, Yang J, et al. FGF/FGFR signaling in health and disease. Signal Transduct Target Ther. 2020;5(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xie YL, Zinkle A, Chen L, Mohammadi M. Fibroblast growth factor signalling in osteoarthritis and cartilage repair. Nat Rev Rheumatol. 2020;16(10):547‐564. [DOI] [PubMed] [Google Scholar]
- 66. Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8(3):235‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149(2):121‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139‐149. [DOI] [PubMed] [Google Scholar]
- 69. Goetz R, Beenken A, Ibrahimi OA, et al. Molecular insights into the klotho‐dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol. 2007;27(9):3417‐3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ornitz DM, Yayon A, Flanagan JG, Svahn CM, Levi E, Leder P. Heparin is required for cell‐free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Research Support, Non‐U.S. Gov't. Mol Cell Biol. 1992;12(1):240‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin X, Buff EM, Perrimon N, Michelson AM. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126(17):3715‐3723. [DOI] [PubMed] [Google Scholar]
- 72. Spivak‐Kroizman T, Lemmon MA, Dikic I, et al. Heparin‐induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, Non‐P.H.S.; Research Support, U.S. Gov't, P.H.S. Cell. 1994;79(6):1015‐1024. [DOI] [PubMed] [Google Scholar]
- 73. Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin‐like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. Cell. 1991;64(4):841‐848. [DOI] [PubMed] [Google Scholar]
- 74. Saksela O, Moscatelli D, Sommer A, Rifkin DB. Endothelial cell‐derived heparan sulfate binds basic fibroblast growth factor and protects it from proteolytic degradation. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. J Cell Biol. 1988;107(2):743‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Flaumenhaft R, Moscatelli D, Saksela O, Rifkin DB. Role of extracellular matrix in the action of basic fibroblast growth factor: matrix as a source of growth factor for long‐term stimulation of plasminogen activator production and DNA synthesis. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. J Cell Physiol. 1989;140(1):75‐81. [DOI] [PubMed] [Google Scholar]
- 76. Moscatelli D. Metabolism of receptor‐bound and matrix‐bound basic fibroblast growth factor by bovine capillary endothelial cells. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. J Cell Biol. 1988;107(2):753‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Roghani M, Moscatelli D. Prostate cells express two isoforms of fibroblast growth factor receptor 1 with different affinities for fibroblast growth factor‐2. Prostate. 2007;67(2):115‐124. [DOI] [PubMed] [Google Scholar]
- 78. Servetto A, Formisano L, Arteaga CL. FGFR signaling and endocrine resistance in breast cancer: Challenges for the clinical development of FGFR inhibitors. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kalinina J, Dutta K, Ilghari D, et al. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure. 2012;20(1):77‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schlessinger J, Plotnikov AN, Ibrahimi OA, et al. Crystal structure of a ternary FGF‐FGFR‐heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. Mol Cell. 2000;6(3):743‐750. [DOI] [PubMed] [Google Scholar]
- 82. Farooq M, Khan AW, Kim MS, Choi S. The role of fibroblast growth factor (FGF) signaling in tissue repair and regeneration. Cells. 2021;10(11):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jin CL, Wang F, Wu XC, Yu CD, Luo YD, McKeehan WL. Directionally specific paracrine communication mediated by epithelial FGF9 to stromal FGFR3 in two‐compartment premalignant prostate tumors. Cancer Res. 2004;64(13):4555‐4562. [DOI] [PubMed] [Google Scholar]
- 84. Xu X, Weinstein M, Li C, et al. Fibroblast growth factor receptor 2 (FGFR2)‐mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. Development. 1998;125(4):753‐765. [DOI] [PubMed] [Google Scholar]
- 85. Orr‐Urtreger A, Bedford MT, Burakova T, et al. Developmental localization of the splicing alternatives of fibroblast growth factor receptor‐2 (FGFR2). Research Support, Non‐U.S. Gov't. Dev Biol. 1993;158(2):475‐486. [DOI] [PubMed] [Google Scholar]
- 86. Olsen SK, Li JYH, Bromleigh C, et al. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20(2):185‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yeh BK, Igarashi M, Eliseenkova AV, et al. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc Natl Acad Sci USA. 2003;100(5):2266‐2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68(6):951‐964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kim I, Moon S, Yu K, Kim U, Koh GY. A novel fibroblast growth factor receptor‐5 preferentially expressed in the pancreas(1). Biochim Biophys Acta. 2001;1518(1‐2):152‐156. [DOI] [PubMed] [Google Scholar]
- 90. Sleeman M, Fraser J, McDonald M, et al. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271(2):171‐182. [DOI] [PubMed] [Google Scholar]
- 91. Steinberg F, Zhuang L, Beyeler M, Kalin RE, Mullis PE, Brandli AW. The FGFRL1 receptor is shed from cell membranes, binds fibroblast growth factors (FGFs), and antagonizes FGF signaling in xenopus embryos. J Biol Chem. 2010;285(3):2193‐2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Trueb B, Amann R, Gerber SD. Role of FGFRL1 and other FGF signaling proteins in early kidney development. Cell Mol Life Sci. 2013;70(14):2505‐2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kahkonen TE, Ivaska KK, Jian M, Buki KG, Vaananen HK, Harkonen PL. Role of fibroblast growth factor receptors (FGFR) and FGFR like‐1 (FGFRL1) in mesenchymal stromal cell differentiation to osteoblasts and adipocytes. Mol Cell Endocrinol. 2018;461(C):194‐204. [DOI] [PubMed] [Google Scholar]
- 94. Silva PN, Altamentova SM, Kilkenny DM, Rocheleau JV. Fibroblast growth factor receptor like‐1 (FGFRL1) interacts with SHP‐1 phosphatase at insulin secretory granules and induces beta‐cell ERK1/2 protein activation. J Biol Chem. 2013;288(24):17859‐17870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Regeenes R, Silva PN, Chang HH, et al. Fibroblast growth factor receptor 5 (FGFR5) is a co‐receptor for FGFR1 that is up‐regulated in beta‐cells by cytokine‐induced inflammation. J Biol Chem. 2018;293(44):17218‐17228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lin KR, Bieri G, Gontier G, et al. MHC class I H2‐K‐b negatively regulates neural progenitor cell proliferation by inhibiting FGFR signaling. PLoS Biol. 2021;19(6):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Iseki S, Wilkie AO, Morriss‐Kay GM. Fgfr1 and Fgfr2 have distinct differentiation‐ and proliferation‐related roles in the developing mouse skull vault. Research Support, Non‐U.S. Gov't. Development. 1999;126(24):5611‐5620. [DOI] [PubMed] [Google Scholar]
- 98. Boilly B, Vercoutter‐Edouart AS, Hondermarck H, Nurcombe V, Le Bourhis X. FGF signals for cell proliferation and migration through different pathways. Cytokine Growth Factor Rev. 2000;11(4):295‐302. [DOI] [PubMed] [Google Scholar]
- 99. Prudovsky I. Cellular mechanisms of FGF‐stimulated tissue repair. Cells. 2021;10(7):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Padrissa‐Altes S, Bachofner M, Bogorad RL, et al. Control of hepatocyte proliferation and survival by Fgf receptors is essential for liver regeneration in mice. Gut. 2015;64(9):1444‐1453. [DOI] [PubMed] [Google Scholar]
- 101. Zhu X, Qiu C, Wang Y, et al. FGFR1 SUMOylation coordinates endothelial angiogenic signaling in angiogenesis. Proc Natl Acad Sci USA. 2022;119(26):e2202631119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Berg T, Rountree CB, Lee L, et al. Fibroblast growth factor 10 is critical for liver growth during embryogenesis and controls hepatoblast survival via beta‐catenin activation. Hepatology (Baltimore, Md). 2007;46(4):1187‐1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Horakova D, Cela P, Krejci P, et al. Effect of FGFR inhibitors on chicken limb development. Dev Growth Diff. 2014;56(8):555‐572. [DOI] [PubMed] [Google Scholar]
- 104. Atkinson‐Leadbeater K, Hehr CL, McFarlane S. Fgfr signaling is required as the early eye field forms to promote later patterning and morphogenesis of the eye. Dev Dyn. 2014;243(5):663‐675. [DOI] [PubMed] [Google Scholar]
- 105. Okazawa M, Murashima A, Harada M, et al. Region‐specific regulation of cell proliferation by FGF receptor signaling during the Wolffian duct development. Dev Biol. 2015;400(1):139‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Spencer‐Dene B, Sala FG, Bellusci S, Gschmeissner S, Stamp G, Dickson C. Stomach development is dependent on fibroblast growth factor 10/fibroblast growth factor receptor 2b‐mediated signaling. Gastroenterology. 2006;130(4):1233‐1244. [DOI] [PubMed] [Google Scholar]
- 107. Ohkubo Y, Uchida AO, Shin D, Partanen J, Vaccarino FM. Fibroblast growth factor receptor 1 is required for the proliferation of hippocampal progenitor cells and for hippocampal growth in mouse. J Neurosci. 2004;24(27):6057‐6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gospodarowicz D, Weseman J, Moran J. Presence in brain of a mitogenic agent promoting proliferation of myoblasts in low density culture. Research Support, U.S. Gov't, P.H.S. Nature. 1975;256(5514):216‐219. [DOI] [PubMed] [Google Scholar]
- 109. Kardami E, Spector D, Strohman RC. Selected muscle and nerve extracts contain an activity which stimulates myoblast proliferation and which is distinct from transferrin. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. Dev Biol. 1985;112(2):353‐358. [DOI] [PubMed] [Google Scholar]
- 110. Lu CM, Huguley S, Cui C, et al. Effects of FGFR signaling on cell proliferation and differentiation of apert dental cells. Cells Tissues Organs. 2015;201(1):26‐37. [DOI] [PubMed] [Google Scholar]
- 111. Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125(24):4977‐4988. [DOI] [PubMed] [Google Scholar]
- 112. Chen L, Adar R, Yang X, et al. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104(11):1517‐1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Andrade AC, Nilsson O, Barnes KM, Baron J. Wnt gene expression in the post‐natal growth plate: Regulation with chondrocyte differentiation. Bone. 2007;40(5):1361‐1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Peterslund JML, Serup P. Activation of FGFR(IIIc) isoforms promotes activin‐induced mesendoderm development in mouse embryonic stem cells and reduces Sox17 coexpression in EpCAM(+) cells. Stem Cell Res. 2011;6(3):262‐275. [DOI] [PubMed] [Google Scholar]
- 115. Kon E, Calvo‐Jimenez E, Cossard A, Na Y, Cooper JA, Jossin Y. N‐cadherin‐regulated FGFR ubiquitination and degradation control mammalian neocortical projection neuron migration. Elife. 2019;8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Song HY, Zhu J, Li P, Han F, Fang LH, Yu PC. Metabolic flexibility maintains proliferation and migration of FGFR signaling‐deficient lymphatic endothelial cells. J Biol Chem. 2021;297(4):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yang JLJ, Bertolesi GE, Dueck S, Hehr CL, McFarlane S. The expression of key guidance genes at a forebrain axon turning point is maintained by distinct Fgfr isoforms but a common downstream signal transduction mechanism. eNeuro. 2019;6(2):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yang JLJ, Bertolesi GE, Hehr CL, Johnston J, McFarlane S. Fibroblast growth factor receptor 1 signaling transcriptionally regulates the axon guidance cue slit1. Cell Mol Life Sci. 2018;75(19):3649‐3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Atkinson‐Leadbeater K, Bertolesi GE, Hehr CL, Webber CA, Cechmanek PB, McFarlane S. Dynamic expression of axon guidance cues required for optic tract development is controlled by fibroblast growth factor signaling. J Neurosci. 2010;30(2):685‐693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Esser JS, Rahner S, Deckler M, Bode C, Patterson C, Moser M. Fibroblast growth factor signaling pathway in endothelial cells is activated by BMPER to promote angiogenesis. Arterioscler Thromb Vasc Biol. 2015;35(2):358‐367. [DOI] [PubMed] [Google Scholar]
- 121. Ardi VC, Van den Steen PE, Opdenakker G, Schweighofer B, Deryugina EI, Quigley JP. Neutrophil MMP‐9 proenzyme, unencumbered by TIMP‐1, undergoes efficient activation in vivo and catalytically induces angiogenesis via a basic fibroblast growth factor (FGF‐2)/FGFR‐2 pathway. J Biol Chem. 2009;284(38):25854‐25866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Glyceollins Lee SH., a novel class of soy phytoalexins, inhibit angiogenesis by blocking the VEGF and bFGF signaling pathways (vol 57, pg 225, 2013). Mol Nutr Food Res. 2013;57(3):556‐556. [DOI] [PubMed] [Google Scholar]
- 123. Mao Y, Liu XQ, Song Y, Zhai CG, Zhang L. VEGF‐A/VEGFR‐2 and FGF‐2/FGFR‐1 but not PDGF‐BB/PDGFR‐beta play important roles in promoting immature and inflammatory intraplaque angiogenesis. PLoS One. 2018;13(8):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Schaap FG, Leclercq IA, Jansen PLM, Damink SWO. Prometheus' little helper, a novel role for fibroblast growth factor 15 in compensatory liver growth. J Hepatol. 2013;59(5):1121‐1123. [DOI] [PubMed] [Google Scholar]
- 125. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next‐generation sequencing. Clin Cancer Res. 2016;22(1):259‐267. [DOI] [PubMed] [Google Scholar]
- 126. Bao Y, Gabrielpillai J, Dietrich J, et al. Fibroblast growth factor (FGF), FGF receptor (FGFR), and cyclin D1 (CCND1) DNA methylation in head and neck squamous cell carcinomas is associated with transcriptional activity, gene amplification, human papillomavirus (HPV) status, and sensitivity to tyrosine kinase inhibitors. Clin Epigen. 2021;13(1):228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Orlando KA, Wade PA. Epigenetic remodelling upon FGFR inhibition. Nat Cell Biol. 2021;23(11):1115‐1116. [DOI] [PubMed] [Google Scholar]
- 128. Wang X, Sun M, Gao Z, et al. N‐nitrosamines‐mediated downregulation of LncRNA‐UCA1 induces carcinogenesis of esophageal squamous by regulating the alternative splicing of FGFR2. Sci Total Environ. 2023;855:158918. [DOI] [PubMed] [Google Scholar]
- 129. Li Y, Qiu X, Wang X, et al. FGFR‐inhibitor‐mediated dismissal of SWI/SNF complexes from YAP‐dependent enhancers induces adaptive therapeutic resistance. Nat Cell Biol. 2021;23(11):1187‐1198. [DOI] [PubMed] [Google Scholar]
- 130. Bourrier C, Pierga JY, Xuereb L, et al. Shallow whole‐genome sequencing from plasma identifies FGFR1 amplified breast cancers and predicts overall survival. Cancers. 2020;12(6):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gonzalez‐Ericsson PI, Servetto A, Formisano L, et al. FGFR1 antibody validation and characterization of FGFR1 protein expression in ER plus breast cancer. Appl Immunohistochem. 2022;30(9):600‐608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Chen Z, Tong L‐J, Tang B‐Y, et al. C11, a novel fibroblast growth factor receptor 1 (FGFR1) inhibitor, suppresses breast cancer metastasis and angiogenesis. Acta Pharmacol Sin. 2019;40(6):823‐832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Santolla MF, Talia M, Maggiolini M. S100A4 is involved in stimulatory effects elicited by the FGF2/FGFR1 signaling pathway in triple‐negative breast cancer (TNBC) cells. Int J Mol Sci. 2021;22(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Cheng Q, Ma ZK, Shi YJ, Parris AB, Kong LF, Yang XH. FGFR1 overexpression induces cancer cell stemness and enhanced Akt/Erk‐ER signaling to promote palbociclib resistance in luminal a breast cancer cells. Cells. 2021;10(11):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mao P, Cohen O, Kowalski KJ, et al. Acquired FGFR and FGF alterations confer resistance to estrogen receptor (ER) targeted therapy in ER+ metastatic breast cancer. Clin Cancer Res. 2020;26(22):5974‐5989. [DOI] [PubMed] [Google Scholar]
- 138. Shi YJ, Ma ZK, Cheng Q, et al. FGFR1 overexpression renders breast cancer cells resistant to metformin through activation of IRS1/ERK signaling. Biochim Biophys Acta Mol Cell Res. 2021;1868(1):13. [DOI] [PubMed] [Google Scholar]
- 139. Lin Y, Lin FK, Zhang ZR, et al. The FGFR1 signaling pathway upregulates the oncogenic transcription factor FOXQ1 to promote breast cancer cell growth. Int J Biol Sci. 2023;19(3):744‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gao MD, Li X, Yang M, Feng WR, Lin Y, He T. TNFAIP3 mediates FGFR1 activation‐induced breast cancer angiogenesis by promoting VEGFA expression and secretion. Clin Transl Oncol. 2022;24(12):2453‐2465. [DOI] [PubMed] [Google Scholar]
- 141. Riaz SK, Khan W, Wang F, et al. Targeted inhibition of fibroblast growth factor receptor 1‐GLI through AZD4547 and GANT61 modulates breast cancer progression. Front Cell Dev Biol. 2021;9:758400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Servetto A, Kollipara R, Formisano L, et al. Nuclear FGFR1 regulates gene transcription and promotes antiestrogen resistance in ER+ breast cancer. Clin Cancer Res. 2021;27(15):4379‐4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wan L, Han Q, Zhu BS, Kong ZH, Feng ER. Circ‐TFF1 facilitates breast cancer development via regulation of miR‐338‐3p/FGFR1 axis. Biochem Genet. 2022;60(1):315‐335. [DOI] [PubMed] [Google Scholar]
- 144. Lin Y, Lin F, Anuchapreeda S, et al. Effect of miR‐133b on progression and cisplatin resistance of triple‐negative breast cancer through FGFR1‐Wnt‐β‐catenin axis. Am J Transl Res. 2021;13(6):5969‐5984. [PMC free article] [PubMed] [Google Scholar]
- 145. Zhou LJ, Yang J, Ma SY, Gao H. Effect of miRNA‐136‐targeted regulation o FGFR1 on proliferation and apoptosis of triple‐negative breast cancer cells. Am J Transl Res. 2021;13(7):7723‐7729. [PMC free article] [PubMed] [Google Scholar]
- 146. Jiang J, Lin H, Shi S, Hong Y, Bai X, Cao X. Hsa_circRNA_0000518 facilitates breast cancer development via regulation of the miR‐326/FGFR1 axis (vol 11, pg 3181, 2020). Thorac Cancer. 2021;12(15):2229‐2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Boothby‐Shoemaker W, Benham V, Paithankar S, Shankar R, Chen B, Bernard JJ. The relationship between leptin, the leptin receptor and FGFR1 in primary human breast tumors. Cells. 2020;9(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Pang ZY, Wei YT, Shang MY, et al. Leptin‐elicited PBX3 confers letrozole resistance in breast cancer. Endocr Relat Cancer. 2021;28(3):173‐189. [DOI] [PubMed] [Google Scholar]
- 149. Chen CK, Cheng R, Demeter J, et al. Structured elements drive extensive circular RNA translation. Mol Cell. 2021;81(20):4300‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Cho BC. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in resected squamous cell lung cancer patients. J Thorac Oncol. 2012;7(11):S455‐S455. [DOI] [PubMed] [Google Scholar]
- 151. Heist RS, Mino‐Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7(12):1775‐1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Peifer M, Fernandez‐Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small‐cell lung cancer. Nat Genet. 2012;44(10):1104‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Schultheis AM, Bos M, Schmitz K, et al. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small‐cell lung cancer. Mod Pathol. 2014;27(2):214‐221. [DOI] [PubMed] [Google Scholar]
- 156. Haines E, Chen T, Kommajosyula N, et al. Palbociclib resistance confers dependence on an FGFR‐MAP kinase‐mTOR‐driven pathway in KRAS‐mutant non‐small cell lung cancer. Oncotarget. 2018;9(60):31572‐31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Lu YH, Liu YF, Oeck S, Zhang GJ, Schramm A, Glazer PM. Hypoxia induces resistance to EGFR inhibitors in lung cancer cells via upregulation of FGFR1 and the MAPK pathway. Cancer Res. 2020;80(21):4655‐4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Terp MG, Jacobsen K, Molina MA, et al. Combined FGFR and Akt pathway inhibition abrogates growth of FGFR1 overexpressing EGFR‐TKI‐resistant NSCLC cells. npj Precis Oncol. 2021;5(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Yuan H, Li ZM, Shao JX, Ji WX, Xia WL, Lu S. FGF2/FGFR1 regulates autophagy in FGFR1‐amplified non‐small cell lung cancer cells. J Exp Clin Cancer Res. 2017;36:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti‐PD‐1 resistance by sponging miR‐381‐3p in non‐small cell lung cancer cells (vol 18, 179, 2019). Mol Cancer. 2020;19(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gu Y, Pais G, Becker V, et al. Suppression of endothelial miR‐22 mediates non‐small cell lung cancer cell‐induced angiogenesis. Mol Ther‐Nucl Acids. 2021;26:849‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kim KB, Kim Y, Rivard CJ, Kim DW, Park KS. FGFR1 is critical for RBL2 loss‐driven tumor development and requires PLCG1 activation for continued growth of small cell lung cancer. Cancer Res. 2020;80(22):5051‐5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Gorringe KL, Jacobs S, Thompson ER, et al. High‐resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13(16):4731‐4739. [DOI] [PubMed] [Google Scholar]
- 164. Tsimafeyeu I, Demidov L, Stepanova E, Wynn N, Ta H. Overexpression of fibroblast growth factor receptors FGFR1 and FGFR2 in renal cell carcinoma. Scand J Urol Nephrol. 2011;45(3):190‐195. [DOI] [PubMed] [Google Scholar]
- 165. Edwards J, Krishna NS, Witton CJ, Bartlett JMS. Gene amplifications associated with the development of hormone‐resistant prostate cancer. Clin Cancer Res. 2003;9(14):5271‐5281. [PubMed] [Google Scholar]
- 166. Yang F, Zhang YY, Ressler SJ, et al. FGFR1 is essential for prostate cancer progression and metastasis. Cancer Res. 2013;73(12):3716‐3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Kim HS, Lee SE, Bae YS, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget. 2015;6(4):2562‐2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Schäfer MH, Lingohr P, Sträßer A, et al. Fibroblast growth factor receptor 1 gene amplification in gastric adenocarcinoma. Hum Pathol. 2015;46(10):1488‐1495. [DOI] [PubMed] [Google Scholar]
- 169. Kawamata F, Patch AM, Nones K, et al. Copy number profiles of paired primary and metastatic colorectal cancers. Oncotarget. 2018;9(3):3394‐3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Guan ZH, Lan HR, Sun D, Wang XW, Jin KT. A potential novel therapy for FGFR1‐amplified pancreatic cancer with bone metastasis, screened by next‐generation sequencing and a patient‐derived xenograft model. Oncol Lett. 2019;17(2):2303‐2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Jeske YW, Ali S, Byron SA, et al. FGFR2 mutations are associated with poor outcomes in endometrioid endometrial cancer: an NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol. 2017;145(2):366‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Amary MF, Ye HT, Berisha F, et al. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo‐adjuvant chemotherapy. Cancer Med. 2014;3(4):980‐987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Giordano M, Decio A, Battistini C, et al. L1CAM promotes ovarian cancer stemness and tumor initiation via FGFR1/SRC/STAT3 signaling. J Exp Clin Cancer Res. 2021;40(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Huang ST, Liang SY, Chen GD, et al. Overexpression of glycosyltransferase 8 domain containing 2 confers ovarian cancer to CDDP resistance by activating FGFR/PI3K signalling axis. Oncogenesis. 2021;10(7):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Biswas PK, Kwak Y, Kim A, et al. TTYH3 modulates bladder cancer proliferation and metastasis via FGFR1/H‐Ras/A‐Raf/MEK/ERK pathway. Int J Mol Sci. 2022;23(18):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Koinis F, Corn P, Parikh N, et al. Resistance to MET/VEGFR2 inhibition by cabozantinib is mediated by YAP/TBX5‐dependent induction of FGFR1 in castration‐resistant prostate cancer. Cancers. 2020;12(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Fan ZC, Ma JS, Pan XB, et al. Crosstalk of FGFR1 signaling and choline metabolism promotes cell proliferation and survival in prostate cancer cells. Int J Cancer. 2022;150(9):1525‐1536. [DOI] [PubMed] [Google Scholar]
- 178. Zhang JJ, Zhou XH, Zhou Y, et al. Bufalin suppresses the migration and invasion of prostate cancer cells through HOTAIR, the sponge of miR‐520b. Acta Pharmacol Sin. 2019;40(9):1228‐1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhang RR, Zhao HY, Yuan HM, et al. CircARVCF contributes to cisplatin resistance in gastric cancer by altering miR‐1205 and FGFR1. Front Genet. 2021;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Gu JX, Li XZ, Li H, Jin Z, Jin JJ. MicroRNA‐198 inhibits proliferation and induces apoptosis by directly suppressing FGFR1 in gastric cancer. Biosci Rep. 2019;39:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Xie G, Ke Q, Ji YZ, Wang AQ, Jing M, Zou LL. FGFR1 is an independent prognostic factor and can be regulated by miR‐497 in gastric cancer progression. Braz J Med Biol Res. 2019;52(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hong S, Yan ZK, Song YM, Bi MM, Li SQ. LncRNA AGAP2‐AS1 augments cell viability and mobility, and confers gemcitabine resistance by inhibiting miR‐497 in colorectal cancer. Aging‐Us. 2020;12(6):5183‐5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Chen S, Guo W, Meng M, et al. LncRNA SNHG1 promotes the progression of pancreatic cancer by regulating FGFR1 expression via competitively binding to miR‐497. Front Oncol. 2022;12:813850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Lai S‐W, Bamodu OA, Tsai W‐C, et al. The therapeutic targeting of the FGFR1/Src/NF‐κB signaling axis inhibits pancreatic ductal adenocarcinoma stemness and oncogenicity. Clin Exp Metastasis. 2018;35(7):663‐677. [DOI] [PubMed] [Google Scholar]
- 185. Coleman SJ, Chioni AM, Ghallab M, et al. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med. 2014;6(4):467‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wang L, Song GH, Tan WW, et al. miR‐573 inhibits prostate cancer metastasis by regulating epithelial‐mesenchymal transition. Oncotarget. 2015;6(34):35978‐35990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Moes‐Sosnowska J, Skupinska M, Lechowicz U, et al. FGFR1‐4 RNA‐based gene alteration and expression analysis in squamous non‐small cell lung cancer. Int J Mol Sci. 2022;23(18):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Marchwicka A, Corcoran A, Berkowska K, Marcinkowska E. Restored expression of vitamin D receptor and sensitivity to 1,25‐dihydroxyvitamin D‐3 in response to disrupted fusion FOP2‐FGFR1 gene in acute myeloid leukemia cells. Cell Biosci. 2016;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Huang C, Zou LL, Wang DW, Zhang XY, Li JY, Tu CQ. 8p11 myeloproliferative syndrome with t(8;22) (p11; q11): a case report. Zhonghua xue ye xue za zhi. 2019;40(11):923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Wehrli M, Leibundgut EO, Gattiker HH, Manz MG, Muller AMS, Goede JS. Response to tyrosine kinase inhibitors in myeloproliferative neoplasia with 8p11 translocation and CEP110‐FGFR1 rearrangement. Oncologist. 2017;22(4):480‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Cai B, Liu Y, Chong Y, et al. IRAK1‐regulated IFN‐γ signaling induces MDSC to facilitate immune evasion in FGFR1‐driven hematological malignancies. Mol Cancer. 2021;20(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Engelhardt S, Behling F, Beschorner R, et al. Frequent FGFR1 hotspot alterations in driver‐unknown low‐grade glioma and mixed neuronal‐glial tumors. J Cancer Res Clin Oncol. 2022;148(4):857‐866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Tome M, Tchorz J, Gassmann M, Bettler B. Constitutive activation of Notch2 signalling confers chemoresistance to neural stem cells via transactivation of fibroblast growth factor receptor‐1. Stem Cell Res. 2019;35:9. [DOI] [PubMed] [Google Scholar]
- 194. Kong SQ, Cao YX, Li X, Li ZZ, Xin YL, Meng Y. MiR‐3116 sensitizes glioma cells to temozolomide by targeting FGFR1 and regulating the FGFR1/PI3K/AKT pathway. J Cell Mol Med. 2020;24(8):4677‐4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Abou‐Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open‐label, phase 2 study. Lancet Oncol. 2020;21(5):671‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next‐generation sequencing. Oncologist. 2014;19(3):235‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Farshidfar F, Zheng SY, Gingras MC, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH‐mutant molecular profiles (vol 18, pg 2780, 2017). Cell Rep. 2017;19(13):2878‐2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Graham RP, Barr Fritcher EG, Pestova E, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45(8):1630‐1638. [DOI] [PubMed] [Google Scholar]
- 199. Silverman IM, Li MJ, Murugesan K, et al. Validation and characterization of FGFR2 rearrangements in cholangiocarcinoma with comprehensive genomic profiling. J Mol Diagn. 2022;24(4):351‐364. [DOI] [PubMed] [Google Scholar]
- 200. Silverman IM, Hollebecque A, Friboulet L, et al. Clinicogenomic analysis of FGFR2‐rearranged cholangiocarcinoma identifies correlates of response and mechanisms of resistance to pemigatinib. Cancer Discov. 2021;11(2):326‐339. [DOI] [PubMed] [Google Scholar]
- 201. Cleary JM, Raghavan S, Wu Q, et al. FGFR2 extracellular domain in‐frame deletions are therapeutically targetable genomic alterations that function as oncogenic drivers in cholangiocarcinoma. Cancer Discov. 2021;11(10):2488‐2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Zingg D, Bhin J, Yemelyanenko J, et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers (vol 608, pg 609, 2022). Nature. 2022;609(7929):E13‐E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Liu ZL, Liu JL, Chen TL, et al. Wnt‐TCF7‐SOX9 axis promotes cholangiocarcinoma proliferation and pemigatinib resistance in a FGF7‐FGFR2 autocrine pathway. Oncogene. 2022;41(20):2885‐2896. [DOI] [PubMed] [Google Scholar]
- 204. Jaidee R, Kukongviriyapan V, Senggunprai L, et al. Inhibition of FGFR2 enhances chemosensitivity to gemcitabine in cholangiocarcinoma through the AKT/mTOR and EMT signaling pathways. Life Sci. 2022;296:11. [DOI] [PubMed] [Google Scholar]
- 205. Lamberti D, Cristinziano G, Porru M, et al. HSP90 inhibition drives degradation of FGFR2 fusion proteins: implications for treatment of cholangiocarcinoma (vol 69, pg 131, 2019). Hepatology. 2019;69(2):925‐925. [DOI] [PubMed] [Google Scholar]
- 206. Zanazzi G, Liechty BL, Pendrick D, et al. Diffuse midline glioma with novel, potentially targetable, FGFR2‐VPS35 fusion. Case Reports; ; Research Support, Non‐U.S. Gov't. Cold Spring Harb Mol Case Stud. 2020;6(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Ma JH, Benitez JA, Li J, et al. Inhibition of nuclear PTEN tyrosine phosphorylation enhances glioma radiation sensitivity through attenuated DNA repair (vol 35, pg 504, 2019). Cancer Cell. 2019;36(6):690‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Kim H, Park S, Kang SY, Ahn S, Kim KM. Peritoneal seeding is more common in gastric cancer patients with FGFR2 amplification or high tumor mutation burden. Diagnostics. 2022;12(10):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Jogo T, Nakamura Y, Shitara K, et al. Circulating tumor DNA analysis detects FGFR2 amplification and concurrent genomic alterations associated with FGFR inhibitor efficacy in advanced gastric cancer. Clin Cancer Res. 2021;27(20):5619‐5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Cha Y, Kim HP, Lim Y, Han SW, Song SH, Kim TY. FGFR2 amplification is predictive of sensitivity to regorafenib in gastric and colorectal cancers in vitro. Mol Oncol. 2018;12(7):993‐1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Mathur A, Ware C, Davis L, Gazdar A, Pan B‐S, Lutterbach B. FGFR2 is amplified in the NCI‐H716 colorectal cancer cell line and is required for growth and survival. PLoS One. 2014;9(6):e98515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Lin YC, Er TK, Yeh KT, Hung CH, Chang JG. Rapid identification of FGFR2 gene mutations in Taiwanese patients with endometrial cancer using high‐resolution melting analysis. Appl Immunohistochem. 2015;23(7):532‐537. [DOI] [PubMed] [Google Scholar]
- 213. Gaudy‐Marqueste C, Macagno N, Loundou A, et al. Molecular characterization of fast‐growing melanomas. J Am Acad Dermatol. 2022;86(2):312‐321. [DOI] [PubMed] [Google Scholar]
- 214. Huang TT, Liu D, Wang YH, et al. FGFR2 promotes gastric cancer progression by inhibiting the expression of thrombospondin4 via PI3K‐Akt‐Mtor pathway. Cell Physiol Biochem. 2018;50(4):1332‐1345. [DOI] [PubMed] [Google Scholar]
- 215. Huang TT, Wang L, Liu D, et al. FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin‐1. Int J Oncol. 2017;50(5):1501‐1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Zhang JL, Wong CC, Leung KT, et al. FGF18‐FGFR2 signaling triggers the activation of c‐Jun‐YAP1 axis to promote carcinogenesis in a subgroup of gastric cancer patients and indicates translational potential. Oncogene. 2020;39(43):6647‐6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Park J, Kim SY, Kim HJ, Kim KM, Choi EY, Kang MS. A reciprocal regulatory circuit between CD44 and FGFR2 via c‐myc controls gastric cancer cell growth. Oncotarget. 2016;7(19):28670‐28683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Tajirika T, Tokumaru Y, Taniguchi K, et al. DEAD‐box protein RNA‐helicase DDX6 regulates the expression of HER2 and FGFR2 at the post‐transcriptional step in gastric cancer cells. Int J Mol Sci. 2018;19(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Zhao CA, Miao JY, Sun RF, et al. MBD1/HDAC3‐miR‐5701‐FGFR2 axis promotes the development of gastric cancer. Aging‐Us. 2022;14(14):5878‐5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Yu YX, Yu XJ, Liu H, Song QX, Yang YM. miR‐494 inhibits cancer‐initiating cell phenotypes and reverses resistance to lapatinib by downregulating FGFR2 in HER2‐positive gastric cancer. Int J Mol Med. 2018;42(2):998‐1007. [DOI] [PubMed] [Google Scholar]
- 221. Gao X, Liu H, Wu Q, et al. miRNA‐381‐3p functions as a tumor suppressor to inhibit gastric cancer by targeting fibroblast growth factor receptor‐2. Cancer Biother Radiopharm. 2022; [DOI] [PubMed] [Google Scholar]
- 222. Chen Z, Li Y, Tan B, et al. Long non‐coding RNA ASNR targeting miR‐519e‐5p promotes gastric cancer development by regulating FGFR2. Front Cell Dev Biol. 2021;9:679176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Li P, Huang TT, Zou Q, et al. FGFR2 promotes expression of PD‐L1 in colorectal cancer via the JAK/STAT3 signaling pathway. J Immunol. 2019;202(10):3065‐3075. [DOI] [PubMed] [Google Scholar]
- 224. Mori M, Mori T, Yamamoto A, Takagi S, Ueda M. Proliferation of poorly differentiated endometrial cancer cells through autocrine activation of FGF receptor and HES1 expression. Hum Cell. 2019;32(3):367‐378. [DOI] [PubMed] [Google Scholar]
- 225. Stehbens SJ, Ju RJ, Adams MN, et al. FGFR2‐activating mutations disrupt cell polarity to potentiate migration and invasion in endometrial cancer cell models. J Cell Sci. 2018;131(15):16. [DOI] [PubMed] [Google Scholar]
- 226. AlRaddadi RIR, Alamri RJN, Shebli WTY, et al. Fibroblast growth factor receptor 2 gene (FGFR2) rs2981582T/C polymorphism and susceptibility to breast cancer in Saudi women. Saudi J Biol Sci. 2021;28(11):6112‐6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Purnomosari D, Raharjo C, Kalim AS, et al. P21 Ser31Arg and FGFR2 rs2981582 polymorphisms as risk factors for early onset of breast cancer in Yogyakarta, Indonesia. Asian Pacific J Cancer Prevent. 2019;20(11):3305‐3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Shu J, Hui XL, Zheng X, et al. Correlation of FGFR2 rs2981582 polymorphisms with susceptibility to breast cancer: a case‐control study in a Chinese population. J Int Med Res. 2019;47(10):4753‐4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Zanna I, Silvestri V, Palli D, et al. Smoking and FGFR2 rs2981582 variant independently modulate male breast cancer survival: A population‐based study in Tuscany, Italy. Breast. 2018;40:85‐91. [DOI] [PubMed] [Google Scholar]
- 230. Wang Y, Zhang HY, Lin MZ, Wang YS. Association of FGFR2 and PI3KCA genetic variants with the risk of breast cancer in a Chinese population. Cancer Manag Res. 2018;10:1305‐1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Mazhar A, Jamil F, Bashir Q, et al. Genetic variants in FGFR2 and TNRC9 genes are associated with breast cancer risk in Pakistani women. Mol Med Rep. 2016;14(4):3443‐3451. [DOI] [PubMed] [Google Scholar]
- 232. Campbell TM, Castro MAA, de Santiago I, et al. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis. 2016;37(8):741‐750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Du JX, Zhao Q, Liu K, et al. FGFR2/STAT3 signaling pathway involves in the development of MMTV‐related spontaneous breast cancer in TA2 mice. Front Oncol. 2020;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Czaplinska D, Mieczkowski K, Supernat A, et al. Interactions between FGFR2 and RSK2‐implications for breast cancer prognosis. Tumor Biol. 2016;37(10):13721‐13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Lei JH, Lee MH, Miao K, et al. Activation of FGFR2 signaling suppresses BRCA1 and drives triple‐negative mammary tumorigenesis that is sensitive to immunotherapy. Adv Sci. 2021;8(21):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Sadej R, Lu XH, Turczyk L, et al. CD151 regulates expression of FGFR2 in breast cancer cells via PKC‐dependent pathways. J Cell Sci. 2018;131(21):10. [DOI] [PubMed] [Google Scholar]
- 237. Kwabi‐Addo B, Ropiquet F, Giri D, Ittmann M. Alternative splicing of fibroblast growth factor receptors in human prostate cancer. Research Support, U.S. Gov't, Non‐P.H.S. Prostate. 2001;46(2):163‐172. [DOI] [PubMed] [Google Scholar]
- 238. Carstens RP, Eaton JV, Krigman HR, Walther PJ, Garcia‐Blanco MA. Alternative splicing of fibroblast growth factor receptor 2 (FGF‐R2) in human prostate cancer. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. Oncogene. 1997;15(25):3059‐3065. [DOI] [PubMed] [Google Scholar]
- 239. Shoji K, Teishima J, Ohara S, Matsubara A. Restoration of fibroblast growth factor receptor 2IIIB enhances the chemosensitivity of human prostate cancer cells. J Urol. 2014;191(4):E266‐E266. [DOI] [PubMed] [Google Scholar]
- 240. Chen J, Hao P, Zheng T, Zhang Y. miR‐628 reduces prostate cancer proliferation and invasion via the FGFR2 signaling pathway. Exp Ther Med. 2019;18(2):1005‐1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Lee JE, Shin SH, Shin HW, Chun YS, Park JW. Nuclear FGFR2 negatively regulates hypoxia‐induced cell invasion in prostate cancer by interacting with HIF‐1 and HIF‐2. Sci Rep. 2019;9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Zhao Q, Caballero OL, Davis ID, et al. Tumor‐specific isoform switch of the fibroblast growth factor receptor 2 underlies the mesenchymal and malignant phenotypes of clear cell renal cell carcinomas. Clin Cancer Res. 2013;19(9):2460‐2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Chaffer CL, Brennan JP, Slavin JL, Blick T, Thompson EW, Williams ED. Mesenchymal‐to‐epithelial transition facilitates bladder cancer metastasis: Role of fibroblast growth factor receptor‐2. Cancer Res. 2006;66(23):11271‐11278. [DOI] [PubMed] [Google Scholar]
- 244. Ricol D, Cappellen D, El Marjou A, et al. Tumour suppressive properties of fibroblast growth factor receptor 2‐IIIb in human bladder cancer. Research Support, Non‐U.S. Gov't; Research Support, U.S. Gov't, P.H.S. Oncogene. 1999;18(51):7234‐7243. [DOI] [PubMed] [Google Scholar]
- 245. Zhang H, Ye Q, Du ZF, Huang M, Zhang M, Tan HF. MiR‐148b‐3p inhibits renal carcinoma cell growth and pro‐angiogenic phenotype of endothelial cell potentially by modulating FGF2. Biomed Pharmacother. 2018;107:359‐367. [DOI] [PubMed] [Google Scholar]
- 246. Yang C, Wu SQ, Wu XB, Zhou XJ, Jin SM, Jiang HW. Silencing circular RNA UVRAG inhibits bladder cancer growth and metastasis by targeting the microRNA‐223/fibroblast growth factor receptor 2 axis. Cancer Sci. 2019;110(1):99‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Flippot R, Loriot Y. The FGFR3 story in bladder cancer: another piece of the puzzle? Eur Urol. 2020;78(5):688‐689. [DOI] [PubMed] [Google Scholar]
- 248. Matuszczak M, Salagierski M. Diagnostic and prognostic potential of biomarkers CYFRA 21.1, ERCC1, p53, FGFR3 and TATI in bladder cancers. Int J Mol Sci. 2020;21(9):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Cathomas R, Lorch A, Bruins HM, et al. The 2021 Updated European Association of Urology Guidelines on metastatic urothelial carcinoma. Eur Urol. 2022;81(1). [DOI] [PubMed] [Google Scholar]
- 250. Tripathi A, MacDougall K, Sonpavde GP. Therapeutic landscape beyond immunotherapy in advanced urothelial carcinoma: moving past the checkpoint. Drugs. 2022;82(17):1649‐1662. [DOI] [PubMed] [Google Scholar]
- 251. Monteiro CRDA, Korkes F, Krutman‐Zveibil D, Glina S. Fibroblast growth factor receptor 3 gene (FGFR3) mutations in high‐grade muscle‐invasive urothelial bladder cancer in a Brazilian population: evaluation and prevalence. Einstein (Sao Paulo, Brazil). 2022;20:eAO6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Mertens LS, Claps F, Mayr R, et al. Prognostic markers in invasive bladder cancer: FGFR3 mutation status versus P53 and KI‐67 expression: a multi‐center, multi‐laboratory analysis in 1058 radical cystectomy patients. Urol Oncol‐Semin Orig Investig. 2022;40(3):9. [DOI] [PubMed] [Google Scholar]
- 253. van Rhijn BWG, Mertens LS, Mayr R, et al. FGFR3 mutation status and FGFR3 expression in a large bladder cancer cohort treated by radical cystectomy: implications for anti‐FGFR3 treatment?†. Eur Urol. 2020;78(5):682‐687. [DOI] [PubMed] [Google Scholar]
- 254. Lima NC, Atkinson E, Bunney TD, Katan M, Huang PH. Targeting the Src pathway enhances the efficacy of selective FGFR inhibitors in urothelial cancers with FGFR3 alterations. Int J Mol Sci. 2020;21(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. di Martino E, Alder O, Hurst CD, Knowles MA. ETV5 links the FGFR3 and Hippo signalling pathways in bladder cancer. Sci Rep. 2019;9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Huang G‐K, Huang C‐C, Kang C‐H, et al. Genetic interference of FGFR3 impedes invasion of upper tract urothelial carcinoma cells by alleviating RAS/MAPK signal activity. Int J Mol Sci. 2023;24(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Jing WQ, Wang GY, Cui ZW, et al. FGFR3 destabilizes PD‐L1 via NEDD4 to control T‐cell‐ mediated bladder cancer immune surveillance. Cancer Res. 2022;82(1):114‐129. [DOI] [PubMed] [Google Scholar]
- 258. Hong JH, Tong ZJ, Wei TE, et al. Cigarette smoke containing acrolein contributes to cisplatin resistance in human bladder cancers through the regulation of HER2 pathway or FGFR3 pathway. Mol Cancer Ther. 2022;21(6):1010‐1019. [DOI] [PubMed] [Google Scholar]
- 259. Mao WP, Huang X, Wang LS, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR‐181a‐5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Lin JF, Tsai TF, Lin YC, Chen HE, Chou KY, Hwang TIS. Benzyl isothiocyanate suppresses IGF1R, FGFR3 and mTOR expression by upregulation of miR‐99a‐5p in human bladder cancer cells. Int J Oncol. 2019;54(6):2106‐2116. [DOI] [PubMed] [Google Scholar]
- 261. Yan XP, Shao C, Chen C, et al. Mutation detection of fibroblast growth factor receptor 3 for infiltrative hepatocellular carcinoma by whole‐exome sequencing. Dig Dis Sci. 2017;62(2):407‐417. [DOI] [PubMed] [Google Scholar]
- 262. Stoehr CG, Stoehr R, Hartmann A, et al. Mutational activation of FGFR3: no involvement in the development of renal cell carcinoma. J Cancer Res Clin Oncol. 2012;138(2):359‐361. [DOI] [PubMed] [Google Scholar]
- 263. Liu XY, Jing XQ, Cheng X, et al. FGFR3 promotes angiogenesis‐dependent metastasis of hepatocellular carcinoma via facilitating MCP‐1‐mediated vascular formation. Med Oncol. 2016;33(5):11. [DOI] [PubMed] [Google Scholar]
- 264. Paur J, Valler M, Sienel R, et al. Interaction of FGF9 with FGFR3‐IIIb/IIIc, a putative driver of growth and aggressive behaviour of hepatocellular carcinoma. Liver Int. 2020;40(9):2279‐2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Jin ZJ, Feng HR, Liang JY, et al. FGFR3(Delta 7–9) promotes tumor progression via the phosphorylation and destabilization of ten‐eleven translocation‐2 in human hepatocellular carcinoma. Cell Death Dis. 2020;11(10):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Di Stefano AL, Picca A, Saragoussi E, et al. Clinical, molecular, and radiomic profile of gliomas with FGFR3‐TACC3 fusions. Neuro Oncol. 2020;22(11):1614‐1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Métais A, Tauziède‐Espariat A, Garcia J, et al. Clinico‐pathological and epigenetic heterogeneity of diffuse gliomas with FGFR3::TACC3 fusion. Acta Neuropathol Commun. 2023;11(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Li B, Qu H, Zhang J, et al. Genomic characterization and outcome evaluation of kinome fusions in lung cancer revealed novel druggable fusions. npj Precis Oncol. 2021;5(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Pros E, Saigi M, Alameda D, et al. Genome‐wide profiling of non‐smoking‐related lung cancer cells reveals common RB1 rearrangements associated with histopathologic transformation in EGFR‐mutant tumors. Ann Oncol. 2020;31(2):274‐282. [DOI] [PubMed] [Google Scholar]
- 270. Li T, Mehraein‐Ghomi F, Forbes ME, et al. HSP90‐CDC37 functions as a chaperone for the oncogenic FGFR3‐TACC3 fusion. Mol Ther. 2022;30(4):1610‐1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Cui BZ, Li BS, Liu Q, Cui YQ. lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR‐181b. J Cell Biochem. 2017;118(12):4548‐4557. [DOI] [PubMed] [Google Scholar]
- 272. Du XM, Qi F, Lu SY, Li YC, Han W. Nicotine upregulates FGFR3 and RB1 expression and promotes non‐small cell lung cancer cell proliferation and epithelial‐to‐mesenchymal transition via downregulation of miR‐99b and miR‐192. Biomed Pharmacother. 2018;101:656‐662. [DOI] [PubMed] [Google Scholar]
- 273. Qiu BQ, Zhang PF, Xiong D, et al. CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non‐small cell lung cancer by regulating Galectin‐1‐AKT/ERK1/2 signaling. J Cell Physiol. 2019;234(7):11256‐11264. [DOI] [PubMed] [Google Scholar]
- 274. Fromme JE, Schmitz K, Wachter A, et al. FGFR3 mRNA overexpression defines a subset of oligometastatic colorectal cancers with worse prognosis. Oncotarget. 2018;9(63):32204‐32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Zhang BL, Dong SH, Zhu RM, et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget. 2015;6(26):22799‐22811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Lin ZZ, Hsu C, Jeng YM, et al. Klotho‐beta and fibroblast growth factor 19 expression correlates with early recurrence of resectable hepatocellular carcinoma (vol 39, pg 1682, 2019). Liver Int. 2020;40(9):2309‐2309. [DOI] [PubMed] [Google Scholar]
- 277. Li Y, Zhang WZ, Doughtie A, et al. Up‐regulation of fibroblast growth factor 19 and its receptor associates with progression from fatty liver to hepatocellular carcinoma. Oncotarget. 2016;7(32):52329‐52339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Motylewska E, Stępień T, Borkowska M, et al. Alteration in the serum concentrations of FGF19, FGFR4 and βKlotho in patients with thyroid cancer. Cytokine. 2018;105:32‐36. [DOI] [PubMed] [Google Scholar]
- 279. Shin JY, Ahn SM. Src is essential for the endosomal delivery of the FGFR4 signaling complex in hepatocellular carcinoma. J Transl Med. 2021;19(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Teng Y, Zhao H, Gao L, Zhang W, Shull AY, Shay C. FGF19 protects hepatocellular carcinoma cells against endoplasmic reticulum stress via activation of FGFR4‐GSK3β‐Nrf2 signaling. Cancer Res. 2017;77(22):6215‐6225. [DOI] [PubMed] [Google Scholar]
- 281. Zhao H, Lv F, Liang G, et al. FGF19 promotes epithelial‐mesenchymal transition in hepatocellular carcinoma cells by modulating the GSK3β/β‐ catenin signaling cascade via FGFR4 activation. Oncotarget. 2016;7(12):13575‐13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Wang JC, Zhao HK, Zheng L, et al. FGF19/SOCE/NFATc2 signaling circuit facilitates the self‐renewal of liver cancer stem cells. Theranostics. 2021;11(10):5045‐5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. He Q, Huang WJ, Liu DF, et al. Homeobox B5 promotes metastasis and poor prognosis in hepatocellular carcinoma, via FGFR4 and CXCL1 upregulation. Theranostics. 2021;11(12):5759‐5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Ji L, Lin ZJ, Wan Z, et al. miR‐486‐3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. 2020;11(4):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Li JJ, Ye YW, Wang M, et al. The over‐expression of FGFR4 could influence the features of gastric cancer cells and inhibit the efficacy of PD173074 and 5‐fluorouracil towards gastric cancer. Tumor Biol. 2016;37(5):6881‐6891. [DOI] [PubMed] [Google Scholar]
- 286. Ye YW, Zhang XF, Zhou Y, et al. The correlations between the expression of FGFR4 protein and clinicopathological parameters as well as prognosis of gastric cancer patients. J Surg Oncol. 2012;106(7):872‐879. [DOI] [PubMed] [Google Scholar]
- 287. Liu J, Zhang Z, Li XW, et al. Forkhead box Cl promotes colorectal cancer metastasis through transactivating ITGA7 and FGFR4 expression. Oncogene. 2018;37(41):5477‐5491. [DOI] [PubMed] [Google Scholar]
- 288. Pelaez‐Garcia A, Barderas R, Torres S, et al. FGFR4 role in epithelial‐mesenchymal transition and its therapeutic value in colorectal cancer. PLoS One. 2013;8(5):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Dallol A, Buhmeida A, Merdad A, et al. Frequent methylation of the KLOTHO gene and overexpression of the FGFR4 receptor in invasive ductal carcinoma of the breast. Tumor Biol. 2015;36(12):9677‐9683. [DOI] [PubMed] [Google Scholar]
- 290. Levine KM, Priedigkeit N, Basudan A, et al. FGFR4 overexpression and hotspot mutations in metastatic ER+ breast cancer are enriched in the lobular subtype. npj Breast Cancer. 2019;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Ezzat S, Huang P, Dackiw A, Asa SL. Dual inhibition of RET and FGFR4 restrains medullary thyroid cancer cell growth. Clin Cancer Res. 2005;11(3):1336‐1341. [PubMed] [Google Scholar]
- 292. Shi S, Li X, You B, Shan Y, Cao X, You Y. High expression of FGFR4 enhances tumor growth and metastasis in nasopharyngeal carcinoma. J Cancer. 2015;6(12):1245‐1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Zhang X, Soutto M, Chen Z, et al. Induction of fibroblast growth factor receptor 4 by Helicobacter pylori via signal transducer and activator of transcription 3 with a feedforward activation loop involving steroid receptor coactivator signaling in gastric cancer. Gastroenterology. 2022;163(3):620‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Yu T, Wang LN, Li W, et al. Downregulation of miR‐491‐5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109(5):1393‐1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Liu R, Li JY, Xie K, et al. FGFR4 promotes stroma‐induced epithelial‐to‐mesenchymal transition in colorectal cancer. Cancer Res. 2013;73(19):5926‐5935. [DOI] [PubMed] [Google Scholar]
- 296. Zou YT, Zheng SQ, Xie XH, et al. N6‐methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2‐positive breast cancer. Nat Commun. 2022;13(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Xu M, Chen SZ, Yang WB, et al. FGFR4 links glucose metabolism and chemotherapy resistance in breast cancer. Cell Physiol Biochem. 2018;47(1):151‐160. [DOI] [PubMed] [Google Scholar]
- 298. Chen R, Li DY, Zheng M, et al. FGFRL1 affects chemoresistance of small‐cell lung cancer by modulating the PI3K/Akt pathway via ENO1. J Cell Mol Med. 2020;24(3):2123‐2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Zheng C, Shi C‐J, Du L‐J, Jiang Y‐H, Su J‐M. Expression of fibroblast growth factor receptor like 1 protein in oral squamous cell carcinoma and its influence on tumor cell proliferation and migration. Hua xi kou qiang yi xue za zhi. 2020;38(5):558‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Schild C, Trueb B. Aberrant expression of FGFRL1, a novel FGF receptor, in ovarian tumors. Int J Mol Med. 2005;16(6):1169‐1173. [PubMed] [Google Scholar]
- 301. Yu L, Toriseva M, Afshan S, et al. Increased expression and altered cellular localization of fibroblast growth factor receptor‐like 1 (FGFRL1) are associated with prostate cancer progression. Cancers. 2022;14(2):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Tai HY, Wu ZY, Sun SA, Zhang ZG, Xu CJ. FGFRL1 promotes ovarian cancer progression by crosstalk with hedgehog signaling. J Immunol Res. 2018;2018:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Tsuchiya S, Fujiwara T, Sato F, et al. MicroRNA‐210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor‐like 1 (FGFRL1). J Biol Chem. 2011;286(1):420‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Yang Y, Zhang J, Xia T, et al. MicroRNA‐210 promotes cancer angiogenesis by targeting fibroblast growth factor receptor‐like 1 in hepatocellular carcinoma. Oncol Rep. 2016;36(5):2553‐2562. [DOI] [PubMed] [Google Scholar]
- 305. Fan YF, Li HX, Yu ZP, et al. Long non‐coding RNA FGD5‐AS1 promotes non‐small cell lung cancer cell proliferation through sponging hsa‐miR‐107 to up‐regulate FGFRL1. Biosci Rep. 2020;40:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Sánchez‐Guixé M, Hierro C, Jiménez J, et al. High FGFR1‐4 mRNA expression levels correlate with response to selective FGFR inhibitors in breast cancer. Clin Cancer Res. 2022;28(1):137‐149. [DOI] [PubMed] [Google Scholar]
- 307. Bogatyrova O, Mattsson JSM, Ross EM, et al. FGFR1 overexpression in non‐small cell lung cancer is mediated by genetic and epigenetic mechanisms and is a determinant of FGFR1 inhibitor response. Eur J Cancer. 2021;151:136‐149. [DOI] [PubMed] [Google Scholar]
- 308. Zingg D, Bhin J, Yemelyanenko J, et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers. Nature. 2022;608(7923):609‐617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Cortes J, Apperley J, Lomaia E, et al. Ponatinib dose‐ranging study in chronic‐phase chronic myeloid leukemia: a randomized, open‐label phase 2 clinical trial. Blood. 2021;138(21):2042‐2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Jabbour E, Haddad FG, Short NJ, Kantarjian H. Treatment of adults with philadelphia chromosome‐positive acute lymphoblastic leukemia‐from intensive chemotherapy combinations to chemotherapy‐free regimens: a review. JAMA Oncol. 2022;8(9):1340‐1348. [DOI] [PubMed] [Google Scholar]
- 311. Jin H, Shi Y, Lv Y, et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature. 2021;595(7869):730‐734. [DOI] [PubMed] [Google Scholar]
- 312. Yi C, Chen L, Lin Z, et al. Lenvatinib targets FGF receptor 4 to enhance antitumor immune response of anti‐programmed cell death‐1 in HCC. Hepatology (Baltimore, Md). 2021;74(5):2544‐2560. [DOI] [PubMed] [Google Scholar]
- 313. Wells AU, Flaherty KR, Brown KK, et al. Nintedanib in patients with progressive fibrosing interstitial lung diseases‐subgroup analyses by interstitial lung disease diagnosis in the INBUILD trial: a randomised, double‐blind, placebo‐controlled, parallel‐group trial. Lancet Respir Med. 2020;8(5):453‐460. [DOI] [PubMed] [Google Scholar]
- 314. Baldini C, Danlos F‐X, Varga A, et al. Safety, recommended dose, efficacy and immune correlates for nintedanib in combination with pembrolizumab in patients with advanced cancers. J Exp Clin Cancer Res. 2022;41(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Dhillon S. Olverembatinib: first approval. Drugs. 2022;82(4):469‐475. [DOI] [PubMed] [Google Scholar]
- 316. Jiang K, Tang X, Guo J, et al. GZD824 overcomes FGFR1‐V561F/M mutant resistance in vitro and in vivo. Cancer Med. 2021;10(14):4874‐4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Crawford K, Bontrager E, Schwarz MA, et al. Targeted FGFR/VEGFR/PDGFR inhibition with dovitinib enhances the effects of nab‐paclitaxel in preclinical gastric cancer models. Cancer Biol Ther. 2021;22(10‐12):619‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Motzer RJ, Porta C, Vogelzang NJ, et al. Dovitinib versus sorafenib for third‐line targeted treatment of patients with metastatic renal cell carcinoma: an open‐label, randomised phase 3 trial. Lancet Oncol. 2014;15(3):286‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Formisano L, Lu Y, Servetto A, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun. 2019;10(1):1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Roskoski R. The role of fibroblast growth factor receptor (FGFR) protein‐tyrosine kinase inhibitors in the treatment of cancers including those of the urinary bladder. Pharmacol Res. 2020;151:104567. [DOI] [PubMed] [Google Scholar]
- 321. Gourd E. Derazantinib for intrahepatic cholangiocarcinoma. Lancet Oncol. 2019;20(1):e11. [DOI] [PubMed] [Google Scholar]
- 322. Mazzaferro V, El‐Rayes BF, Droz Dit Busset M, et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion‐positive intrahepatic cholangiocarcinoma. Br J Cancer. 2019;120(2):165‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24534), a multitargeted pan‐FGFR inhibitor with activity in multiple FGFR‐amplified or mutated cancer models. Mol Cancer Ther. 2012;11(3):690‐699. [DOI] [PubMed] [Google Scholar]
- 324. Guo M, Duan YK, Dai SY, et al. Structural study of ponatinib in inhibiting SRC kinase. Biochem Biophys Res Commun. 2022;598:15‐19. [DOI] [PubMed] [Google Scholar]
- 325. Pulte ED, Chen H, Price LSL, et al. FDA approval summary: revised indication and dosing regimen for ponatinib based on the results of the OPTIC trial. Oncologist. 2022;27(2):149‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Shigesawa T, Maehara O, Suda G, et al. Lenvatinib suppresses cancer stem‐like cells in HCC by inhibiting FGFR1‐3 signaling, but not FGFR4 signaling. Carcinogenesis. 2021;42(1):58‐69. [DOI] [PubMed] [Google Scholar]
- 327. Welland S, Leyh C, Finkelmeier F, et al. Real‐world data for lenvatinib in hepatocellular carcinoma (ELEVATOR): a retrospective multicenter study. Liver Cancer. 2022;11(3):219‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Iyer RV, Konda B, Fountzilas C, et al. Multicenter phase 2 trial of nintedanib in advanced nonpancreatic neuroendocrine tumors. Cancer. 2020;126(16):3689‐3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Addition of nintedanib to neoadjuvant chemotherapy is safe in bladder cancer. Cancer Discov. 2022;12(7):OF9. [DOI] [PubMed] [Google Scholar]
- 330. Jiang KL, Tang X, Guo J, et al. GZD824 overcomes FGFR1‐V561F/M mutant resistance in vitro and in vivo. Cancer Med. 2021;10(14):4874‐4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Jiang Q, Li Z, Qin Y, et al. Olverembatinib (HQP1351), a well‐tolerated and effective tyrosine kinase inhibitor for patients with T315I‐mutated chronic myeloid leukemia: results of an open‐label, multicenter phase 1/2 trial. J Hematol Oncol. 2022;15(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Qian H, Gang D, He X, Jiang S. A review of the therapeutic role of the new third‐generation TKI olverembatinib in chronic myeloid leukemia. Front Oncol. 2022;12:1036437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Trudel S, Li ZH, Wei E, et al. CHIR‐258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood. 2005;105(7):2941‐2948. [DOI] [PubMed] [Google Scholar]
- 334. Woei‐A‐Jin FJSH, Weijl NI, Burgmans MC, et al. Neoadjuvant treatment with angiogenesis‐inhibitor dovitinib prior to local therapy in hepatocellular carcinoma: a phase II study. Oncologist. 2021;26(10):854‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Choi YJ, Kim HS, Park SH, et al. Phase II study of dovitinib in patients with castration‐resistant prostate cancer (KCSG‐GU11‐05). Cancer Res Treat. 2018;50(4):1252‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Bello E, Colella G, Scarlato V, et al. E‐3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011;71(4):1396‐1405. [DOI] [PubMed] [Google Scholar]
- 337. Soria JC, DeBraud F, Bahleda R, et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol. 2014;25(11):2244‐2251. [DOI] [PubMed] [Google Scholar]
- 338. Hui R, Pearson A, Cortes J, et al. Lucitanib for the Treatment of HR+/HER2‐ Metastatic Breast Cancer: Results from the Multicohort Phase II FINESSE Study. Clin Cancer Res. 2020;26(2):354‐363. [DOI] [PubMed] [Google Scholar]
- 339. Papadopoulos KP, El‐Rayes BF, Tolcher AW, et al. A Phase 1 study of ARQ 087, an oral pan‐FGFR inhibitor in patients with advanced solid tumours. Br J Cancer. 2017;117(11):1592‐1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Braun S, McSheehy P, Litherland K, et al. Derazantinib: an investigational drug for the treatment of cholangiocarcinoma. Expert Opin Investig Drugs. 2021;30(11):1071‐1080. [DOI] [PubMed] [Google Scholar]
- 341. Meric‐Bernstam F, Bahleda R, Hierro C, et al. Futibatinib, an irreversible FGFR1‐4 inhibitor, in patients with advanced solid tumors harboring FGF/FGFR aberrations: a phase i dose‐expansion study. Cancer Discov. 2022;12(2):402‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Goyal L, Meric‐Bernstam F, Hollebecque A, et al. Futibatinib for FGFR2‐rearranged intrahepatic cholangiocarcinoma. N Engl J Med. 2023;388(3):228‐239. [DOI] [PubMed] [Google Scholar]
- 343. Siefker‐Radtke AO, Necchi A, Park SH, et al. Efficacy and safety of erdafitinib in patients with locally advanced or metastatic urothelial carcinoma: long‐term follow‐up of a phase 2 study. Lancet Oncol. 2022;23(2):248‐258. [DOI] [PubMed] [Google Scholar]
- 344. Hanes R, Munthe E, Grad I, et al. Preclinical evaluation of the pan‐FGFR inhibitor LY2874455 in FRS2‐amplified liposarcoma. Cells. 2019;8(2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Michael M, Bang Y‐J, Park YS, et al. A phase 1 study of LY2874455, an oral selective pan‐FGFR inhibitor, in patients with advanced cancer. Target Oncol. 2017;12(4):463‐474. [DOI] [PubMed] [Google Scholar]
- 346. Subbiah V, Iannotti NO, Gutierrez M, et al. FIGHT‐101, a first‐in‐human study of potent and selective FGFR 1–3 inhibitor pemigatinib in pan‐cancer patients with FGF/FGFR alterations and advanced malignancies. Ann Oncol. 2022;33(5):522‐533. [DOI] [PubMed] [Google Scholar]
- 347. Javle M, Roychowdhury S, Kelley RK, et al. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: mature results from a multicentre, open‐label, single‐arm, phase 2 study. Lancet Gastroenterol Hepatol. 2021;6(10):803‐815. [DOI] [PubMed] [Google Scholar]
- 348. Kang C. Infigratinib: first approval. Drugs. 2021;81(11):1355‐1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Chae YK, Hong F, Vaklavas C, et al. Phase II study of AZD4547 in patients with tumors harboring aberrations in the FGFR pathway: results from the NCI‐MATCH trial (EAY131) subprotocol W. J Clin Oncol. 2020;38(21):2407‐2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Aytatli A, Barlak N, Sanli F, et al. AZD4547 targets the FGFR/Akt/SOX2 axis to overcome paclitaxel resistance in head and neck cancer. Cell Oncol (Dordr). 2022;45(1):41‐56. [DOI] [PubMed] [Google Scholar]
- 351. Addeo A, Rothschild SI, Holer L, et al. Fibroblast growth factor receptor (FGFR) inhibitor rogaratinib in patients with advanced pretreated squamous‐cell non‐small cell lung cancer over‐expressing FGFR mRNA: the SAKK 19/18 phase II study. Lung Cancer. 2022;172:154‐159. [DOI] [PubMed] [Google Scholar]
- 352. Grünewald S, Politz O, Bender S, et al. Rogaratinib: A potent and selective pan‐FGFR inhibitor with broad antitumor activity in FGFR‐overexpressing preclinical cancer models. Int J Cancer. 2019;145(5):1346‐1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Koyama T, Shimizu T, Iwasa S, et al. First‐in‐human phase I study of E7090, a novel selective fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. Cancer Sci. 2020;111(2):571‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Watanabe Miyano S, Yamamoto Y, Kodama K, et al. E7090, a novel selective inhibitor of fibroblast growth factor receptors, displays potent antitumor activity and prolongs survival in preclinical models. Mol Cancer Ther. 2016;15(11):2630‐2639. [DOI] [PubMed] [Google Scholar]
- 355. Goyal L, Shi L, Liu LY, et al. TAS‐120 overcomes resistance to ATP‐competitive FGFR inhibitors in patients with FGFR2 fusion‐positive intrahepatic cholangiocarcinoma. Cancer Discov. 2019;9(8):1064‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Nakanishi Y, Akiyama N, Tsukaguchi T, et al. The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Mol Cancer Ther. 2014;13(11):2547‐2558. [DOI] [PubMed] [Google Scholar]
- 357. Kalyukina M, Yosaatmadja Y, Middleditch MJ, Patterson AV, Smaill JB, Squire CJ. TAS‐120 cancer target binding: defining reactivity and revealing the first fibroblast growth factor receptor 1 (FGFR1) irreversible structure. ChemMedChem. 2019;14(4):494‐500. [DOI] [PubMed] [Google Scholar]
- 358. Sootome H, Fujita H, Ito K, et al. Futibatinib is a novel irreversible FGFR 1–4 inhibitor that shows selective antitumor activity against FGFR‐deregulated tumors. Cancer Res. 2020;80(22):4986‐4997. [DOI] [PubMed] [Google Scholar]
- 359. Perera TPS, Jovcheva E, Mevellec L, et al. Discovery and pharmacological characterization of JNJ‐42756493 (Erdafitinib), a functionally selective small‐molecule FGFR family inhibitor. Mol Cancer Ther. 2017;16(6):1010‐1020. [DOI] [PubMed] [Google Scholar]
- 360. Soria JC, Italiano A, Cervantes A, et al. Safety and activity of the pan‐fibroblast growth factor receptor (FGFR) inhibitor erdafitinib in phase 1 study patients with advanced urothelial carcinoma. Meeting Abstract. Ann Oncol. 2016;27:1.32645814 [Google Scholar]
- 361. Syed YY. Futibatinib: first approval. Drugs. 2022;82(18):1737‐1743. [DOI] [PubMed] [Google Scholar]
- 362. Meric‐Bernstam F, Bahleda R, Hierro C, et al. Futibatinib, an irreversible FGFR1‐4 inhibitor, in patients with advanced solid tumors harboring FGF/FGFR aberrations: a phase I dose‐expansion study. Cancer Discov. 2022;12(2):402‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Loriot Y, Necchi A, Park SH, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381(4):338‐348. [DOI] [PubMed] [Google Scholar]
- 364. Darwis NDM, Horigome E, Li S, et al. Radiosensitization by the selective pan‐FGFR inhibitor LY2874455. Cells. 2022;11(11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Zhao G, Li W‐Y, Chen D, et al. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther. 2011;10(11):2200‐2210. [DOI] [PubMed] [Google Scholar]
- 366. Liu PCC, Koblish H, Wu LX, et al. INCB054828 (pemigatinib), a potent and selective inhibitor of fibroblast growth factor receptors 1, 2, and 3, displays activity against genetically defined tumor models. PLoS One. 2020;15(4):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Gavine PR, Mooney L, Kilgour E, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72(8):2045‐2056. [DOI] [PubMed] [Google Scholar]
- 368. Javle M, Lowery M, Shroff RT, et al. Phase II study of BGJ398 in patients with FGFR‐altered advanced cholangiocarcinoma. J Clin Oncol. 2018;36(3):276‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Coombes RC, Badman PD, Lozano‐Kuehne JP, et al. Results of the phase IIa RADICAL trial of the FGFR inhibitor AZD4547 in endocrine resistant breast cancer. Nat Commun. 2022;13(1):3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Schuler M, Cho BC, Sayehli CM, et al. Rogaratinib in patients with advanced cancers selected by FGFR mRNA expression: a phase 1 dose‐escalation and dose‐expansion study. Lancet Oncol. 2019;20(10):1454‐1466. [DOI] [PubMed] [Google Scholar]
- 371. Sternberg CN, Petrylak DP, Bellmunt J, et al. FORT‐1: phase II/III study of rogaratinib versus chemotherapy in patients with locally advanced or metastatic urothelial carcinoma selected based on FGFR1/3 mRNA expression. J Clin Oncol. 2023;41(3):629‐639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Chiba Y, Sudo K, Kojima Y, et al. A multicenter investigator‐initiated Phase 2 trial of E7090 in patients with advanced or recurrent solid tumor with fibroblast growth factor receptor (FGFR) gene alteration: FORTUNE trial. BMC Cancer. 2022;22(1):869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Voss MH, Hierro C, Heist RS, et al. A phase I, open‐label, multicenter, dose‐escalation study of the oral selective FGFR inhibitor Debio 1347 in patients with advanced solid tumors harboring FGFR gene alterations. Clin Cancer Res. 2019;25(9):2699‐2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Wainberg ZA, Enzinger PC, Kang Y‐K, et al. Bemarituzumab in patients with FGFR2b‐selected gastric or gastro‐oesophageal junction adenocarcinoma (FIGHT): a randomised, double‐blind, placebo‐controlled, phase 2 study. Lancet Oncol. 2022;23(11):1430‐1440. [DOI] [PubMed] [Google Scholar]
- 375. Catenacci DVT, Rasco D, Lee J, et al. Phase I escalation and expansion study of bemarituzumab (FPA144) in patients with advanced solid tumors and FGFR2b‐selected gastroesophageal adenocarcinoma. J Clin Oncol. 2020;38(21):2418‐2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Bemarituzumab is active in FGFR2b‐high gastroesophageal adenocarcinoma. Cancer Discov. 2020;10(5):638. [DOI] [PubMed] [Google Scholar]
- 377. Deal watch: HGS and FivePrime in FGF ‘ligand trap’ deal. Nat Rev Drug Discov. 2011;10(5):328. [DOI] [PubMed] [Google Scholar]
- 378. Tolcher AW, Papadopoulos KP, Patnaik A, et al. A phase I, first in human study of FP‐1039 (GSK3052230), a novel FGF ligand trap, in patients with advanced solid tumors. Ann Oncol. 2016;27(3):526‐532. [DOI] [PubMed] [Google Scholar]
- 379. Morgensztern D, Karaseva N, Felip E, et al. An open‐label phase IB study to evaluate GSK3052230 in combination with paclitaxel and carboplatin, or docetaxel, in FGFR1‐amplified non‐small cell lung cancer. Lung Cancer. 2019;136:74‐79. [DOI] [PubMed] [Google Scholar]
- 380. Yang Z, Liang SQ, Saliakoura M, et al. Synergistic effects of FGFR1 and PLK1 inhibitors target a metabolic liability in KRAS‐mutant cancer. EMBO Mol Med. 2021;13(9):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Yang Z, Liang SQ, Yang HT, et al. CRISPR‐mediated kinome editing prioritizes a synergistic combination therapy for FGFR1‐amplified lung cancer. Cancer Res. 2021;81(11):3121‐3133. [DOI] [PubMed] [Google Scholar]
- 382. Wu QB, Zhen YL, Shi L, et al. EGFR inhibition potentiates FGFR inhibitor therapy and overcomes resistance in FGFR2 fusion‐positive cholangiocarcinoma. Cancer Discov. 2022;12(5):1378‐1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Bernat‐Peguera A, Navarro‐Ventura J, Lorenzo‐Sanz L, et al. FGFR inhibition overcomes resistance to EGFR‐targeted therapy in epithelial‐like cutaneous carcinoma. Clin Cancer Res. 2021;27(5):1491‐1504. [DOI] [PubMed] [Google Scholar]
- 384. Palakurthi S, Kuraguchi M, Zacharek SJ, et al. The combined effect of FGFR inhibition and PD‐1 blockade promotes tumor‐intrinsic induction of antitumor immunity. Cancer Immunol Res. 2019;7(9):1457‐1471. [DOI] [PubMed] [Google Scholar]
- 385. Yue S, Li Y, Chen X, et al. FGFR‐TKI resistance in cancer: current status and perspectives. J Hematol Oncol. 2021;14(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Sohl CD, Ryan MR, Luo B, Frey KM, Anderson KS. Illuminating the molecular mechanisms of tyrosine kinase inhibitor resistance for the FGFR1 gatekeeper mutation: the Achilles' heel of targeted therapy. ACS Chem Biol. 2015;10(5):1319‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Krook MA, Lenyo A, Wilberding M, et al. Efficacy of FGFR inhibitors and combination therapies for acquired resistance in FGFR2‐fusion cholangiocarcinoma. Mol Cancer Ther. 2020;19(3):847‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


