Abstract
Most variants associated with complex traits and diseases identified by genome-wide association studies (GWAS) map to noncoding regions of the genome with unknown effects. Using ancestrally diverse biobank-scale GWAS data, massively parallel CRISPR screens, and single cell transcriptomic and proteomic sequencing, we discovered 124 cis-target genes of 91 noncoding blood trait GWAS loci. Using precise variant insertion via base editing, we connected specific variants with gene expression changes. We also identified trans-effect networks of noncoding loci when cis target genes encoded transcription factors or microRNAs. Networks were themselves enriched for GWAS variants and demonstrated polygenic contributions to complex traits. This platform enables massively-parallel characterization of the target genes and mechanisms of human noncoding variants in both cis and trans.
One-Sentence Summary
High-throughput single cell CRISPR screens to understand noncoding human genetic variants for blood cell traits.
A major goal for the study of common diseases is to identify causal genes, which can clarify biological mechanisms and inform drug targets for these diseases. To this end, genome-wide association studies (GWAS) have identified thousands of genetic variants associated with disease outcomes and disease-relevant phenotypes. However, since these associations are nearly always found in noncoding regions, their target genes and functions often remain elusive. This is commonly referred to as the variant-to-function (V2F) problem (1, 2).
Recent studies have used statistical fine-mapping to identify plausibly causal GWAS variants and functional genomics to find candidate cis-regulatory elements (cCREs) and their putative target genes (3-6). Other studies have performed CRISPR-based silencing or mutagenesis screens of noncoding regulatory elements to identify target genes (7-9). Here, we combine these approaches in a modular workflow, Systematic Targeted Inhibition of Noncoding GWAS loci coupled with single-cell sequencing (STING-seq), to identify target genes at noncoding GWAS loci using single-cell pooled CRISPR screens. We first prioritize cCREs by functional annotation and overlap with fine-mapped GWAS variants. We then test for gene regulatory function using pooled CRISPR inhibition (CRISPRi) and single-cell RNA-sequencing and cell surface protein measurements (Fig. 1A). For a subset of validated CREs, we also inserted specific GWAS variants using base editing STING-seq (BeeSTING-seq), which couples base editing with single-cell multiomics. We demonstrate the utility of these approaches in blood cell traits by targeted perturbation of ~500 cCREs at noncoding GWAS loci, identifying target genes in cis and trans for 134 of these CREs, and further explore the effects of 46 fine-mapped noncoding C-to-T variants using precise variant insertion.
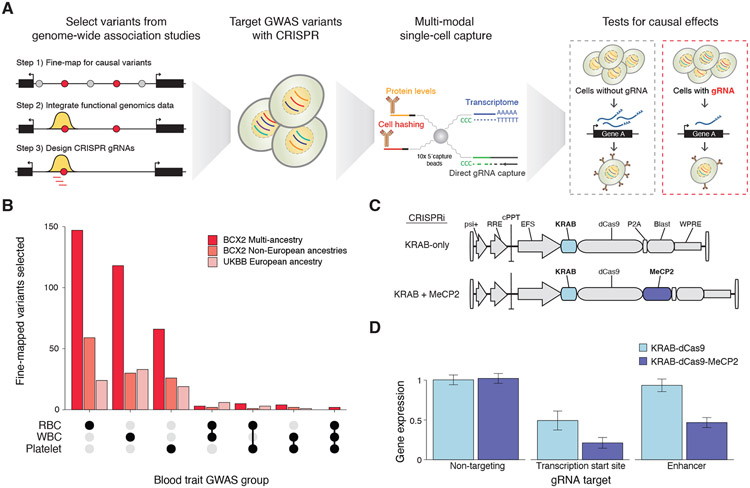
Figure 1. Overview of Systematic Targeting and Inhibition of Noncoding GWAS loci with single-cell sequencing (STING-seq).
(A) STING-seq pipeline for perturbation and single-cell analysis of human genetic variants from genome-wide association studies (GWAS). First, plausibly causal variants are identified via statistical fine-mapping of GWAS. After further refinement of candidate cis-regulatory elements (cCREs) using key molecular hallmarks of regulatory elements, CRISPR guide RNAs (gRNAs) are designed to target cCREs and lentivirally transduced at a high multiplicity-of-infection into human cells. Using multimodal single-cell sequencing, target genes for GWAS variants are identified using differential transcript or protein expression. (B) The number of targeted GWAS variants mapping to cCREs across 29 blood traits in UK Biobank (n = 361,194 participants) and 15 blood traits in the Blood Cell Consortium (n = 746,667 participants). (C) Lentiviral CRISPR inhibition (CRISPRi) vector with a single effector domain (KRAB-dCas9) or dual effector domains (KRAB-dCas9-MeCP2). (D) Mean digital PCR gene expression in human erythrocyte cells (K562) by targeting the transcription start sites (TSS) and known enhancers of three genes (MRPS23, SLC25A27 and FSCN1) with either single-effector KRAB-dCas9 or dual-effector KRAB-dCas9-MeCP2 CRISPRi. Error bars indicate s.e.m.
Results
Fine-mapping multi-ancestry blood trait GWAS to identify candidate CREs
We elected to study blood cell traits due to their high polygenicity, links to multiple common diseases, and the large number of genotyped individuals available in ancestrally diverse biobank-scale data repositories with measured blood traits (10-12). We examined 29 blood trait GWASs in the UK Biobank (UKBB) and 15 traits from the Blood Cell Consortium (BCX) (11), including traits from platelets, red blood cells (RBCs), and white blood cells (WBCs) (Table S1A). The UKBB GWASs include 361,194 participants with European ancestries. The BCX multi-ancestry GWASs include 746,667 participants (76% European, 20% Asian, 2% African, 1% Hispanic/Latino and 1% South Asian ancestries) with both multi-ancestry and individual population analyses. We performed statistical fine-mapping for the 29 UKBB blood trait GWASs, identifying a median of 469 conditionally independent signals and 3,328 fine-mapped variants per trait (13, 14). Multi-ancestry BCX meta-analyses identified a median of 384 conditionally independent signals and 3,586 fine-mapped variants per trait. Across all BCX population-specific GWASs, excluding European ancestries, there were 42 conditionally independent signals and 418 fine-mapped variants per trait (Table S1A-B). In all cases, we found that greater than 90% of fine-mapped variants were in noncoding regions of the genome.
For our study, we targeted candidate cis-regulatory elements (cCREs) from different GWASs — 543 variants in 254 loci — by intersecting fine-mapped noncoding variants with biochemical hallmarks of enhancer activity, such as chromatin accessibility (ATAC-seq and DNase I hypersensitivity) and canonical histone modifications (H3K27ac ChIP-seq) from a human erythroid progenitor cell line (K562). K562 cells are an established and well-characterized model for blood traits: In these cells, reporter assays have identified genetic variants with erythroid-specific effects (15), transcription factor (TF) occupancy is strongly conserved with human proerythroblasts (16), gene expression and open chromatin profiles are similar to human erythrocyte progenitors (17), and promoter-interacting regions defined from Hi-C data are enriched for blood trait GWAS variants (18). The integration of functional genomic data yielded a large set of targetable variants from UKBB and BCX GWASs (Table S1C-D). The variants we selected were often the highest probability variant in a fine-mapped GWAS locus (294 variants) or among the 10 most probable variants (249 variants). We also prioritized variants from non-European ancestries: In total, we selected variants from BCX multi-ancestry analyses (339 variants), BCX non-European ancestries (118 variants), and UKBB European ancestries (86 variants) (Fig. 1B, Table S1C-E).
Optimized dual-repressor CRISPRi system
To perturb the selected cCREs, we designed (Table S1F) a dual-repressor KRAB-dCas9-MeCP2 system (19) that yielded 50 – 60% greater gene repression when targeting transcription start sites (TSSs) or previously described enhancer loci (7) than a single-repressor (KRAB-dCas9) system (Fig. 1C-D, Fig. S1, Table S2). We further characterized the dual-repressor CRISPRi using a pooled library of ~2,000 CRISPR guide RNAs (gRNAs) that target sites at different distances from the TSSs of ~250 essential genes. We found that dual-repressor CRISPRi had a focused activity window with minimal repression beyond 1 kb and that a majority of active gRNAs were located between −400 to +850 nt from the TSS (Fig. S2) (20).
A massively parallel assay to perturb CREs and find their target genes
We designed STING-seq gRNA libraries to target each blood trait cCRE with up to three gRNAs using the dual-repressor CRISPRi (KRAB-dCas9-MeCP2). These gRNAs were optimized for minimal off-target activity (21, 22). We also embedded in the STING-seq library several control gRNAs: negative (non-targeting) controls (23), positive controls (targeting highly-expressed genes at TSSs), and, to estimate the average number of perturbations per cell via flow cytometry, multiple gRNAs targeting a gene encoding a ubiquitously-expressed cell surface protein (CD55) (Table S3A).
We transduced K562 cells with pooled library virus at a high multiplicity of infection (MOI), which we verified via flow cytometry for CD55 (Fig. S3). We then simultaneously captured four different modalities from single cells: CRISPR gRNAs, transcriptomes, cell-surface proteomes via oligo-tagged antibodies, and cell hashing (Table S3B) (24, 25). We recovered 46,583 single cells with a median of 13 gRNAs per cell and with each cCRE targeted in a median of 978 cells (Fig. S4A-B, Table S3C). To perform differential expression testing, we recently developed a conditional resampling approach (SCEPTRE) that yields state-of-the-art calibration on CRISPR single-cell datasets to connect perturbations with changes in gene and protein expression (26). Using SCEPTRE, we grouped together gRNAs targeting each cCRE, performing 4,627 pairwise tests with a median of 7 genes tested per cCRE within 500 kb for cis-effects (27). We observed good calibration for positive and negative controls: Non-targeting gRNAs had no effect, and control genes had decreased expression or protein levels at a 5% false discovery rate (FDR) (Fig. 2A, Fig. S5, Table S3C-E). In most cases, target genes in cis for GWAS variants were more likely to be identified when both H3K27ac and open chromatin peaks were present (Fig. 2B).
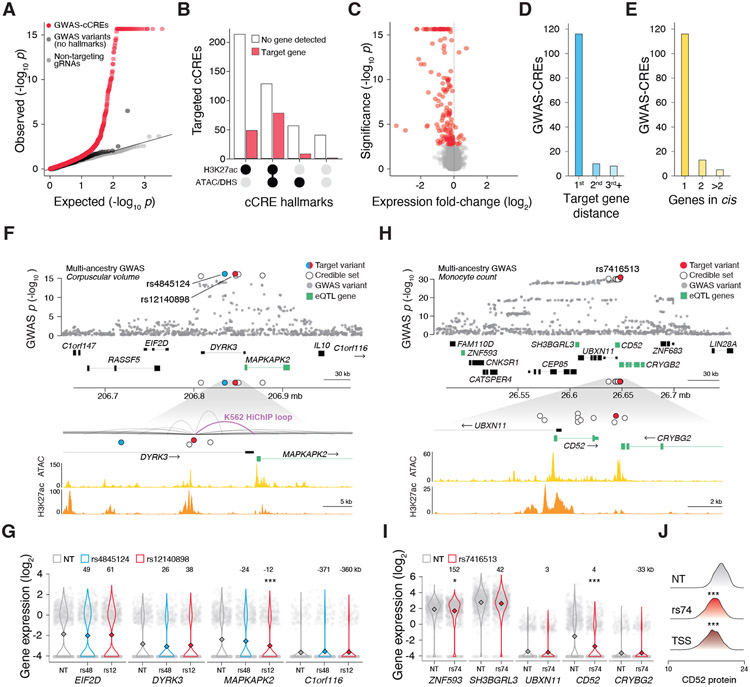
Figure 2. Mapping cis-regulatory target genes for blood trait GWAS variants.
(A) Quantile-quantile plot of cis-effects (within 500 kb) of 531 candidate CREs (cCREs, defined as regions with regulatory hallmarks: ATAC/DHS or H3K27ac) overlapping 535 GWAS variants (GWAS-cCREs), 41 GWAS variants without CRE hallmarks, and 32 non-targeting (NT) gRNAs. Genes for NT tests were randomly sampled from the set of genes in cis for targeting gRNAs. We identified 154 pairs of target genes and CREs for GWAS variants with CRE hallmarks, 1 target gene-CRE pair for GWAS variants without CRE hallmarks and no target genes with NT gRNAs significant at a 5% FDR (Benjamini-Hochberg adjusted SCEPTRE p-value). (B) Targeted GWAS-cCREs with and without target genes detected, and their functional hallmarks of enhancer activity (ATAC/DHS or H3K27ac) in K562 cells. (C) Volcano plot of cis-regulatory effects. Significant pairs of genes and GWAS-CREs are indicated in red. (D) Distance to gene rank for GWAS-CREs and target genes, where genes were ranked according to closest TSS to a given GWAS-CRE. (E) Number of target genes in cis per GWAS-CRE. (F) (top) For a multi-ancestry corpuscular volume locus, two fine-mapped variants were targeted, the lead variant, rs4845124 (blue), and rs12140898 (red). MAPKAPK2 (green) was nominated as the target gene by fine-mapped blood cell eQTLs for both variants. (bottom) rs12140898 mapped to a K562 HiChIP loop, connecting its GWAS-CRE to the MAPKAPK2 promoter. (G) Single-cell gene expression for cells with gRNAs targeting GWAS-cCREs (rs4845124 or rs12140898) or NT. Only rs12140898 had a target gene within 500 kb, MAPKAPK2. (H) For a multi-ancestry monocyte count locus, one fine-mapped variant was targeted, the lead variant, rs741613 (red). ZNF593, SH3BGRL3, CD52, and CRYBG2 (green) were nominated as target genes by fine-mapped blood cell eQTLs. (I) Single-cell gene expression for cells with gRNAs targeting the GWAS-cCRE rs741613 or NT. CD52 and ZNF593 were both identified as target genes. (J) Single-cell protein expression for cells with gRNAs targeting the GWAS-cCRE rs741613, the CD52 transcription start site (TSS) or NT. Asterisks denote significant q-values, Benjamini-Hochberg adjusted SCEPTRE p-values (* q < 0.05, ** q < 0.01, *** q < 0.001).
Out of 539 targeted cCREs (from 254 loci), we found 134 CREs (from 91 loci) had a target gene within 500 kb (Fig. 2C, Table S3F). When examining gRNAs that target the same CRE, the number of cells was most directly responsible for statistical power, and not distance between gRNAs or predicted off-target effects (Fig. S6). We found minimal differences in target gene identification when looking at potential cis-effects within a smaller (100 kb) or larger (1 Mb) window surrounding the targeted cCRE (Table S3F) (28-30).
Most cis-target genes were also the closest gene to the variant; however, there were 10 cis-target genes that were the second closest, and eight that were further away (Fig. 2D). We identified a single cis-target gene for 116 CREs and identified 18 CREs with two or more cis-target genes (Fig. 2E). We also targeted 41 variants that were the most plausibly causal variants at their respective loci but did not overlap biochemical hallmarks of enhancers. From the 41 variants we targeted that did not overlap called peaks for biochemical hallmarks of enhancers, there was one variant (rs106585 for WBC counts) with a significant target gene, LTBR (log2 fold-change [FC] = −0.38, SCEPTRE p = 3.1x10−7) (Fig. 2A, Fig. S7, Table S3G). Upon further inspection, we found a weak enhancer-associated histone modification (H3K27ac) at this locus despite the lack of a called peak, suggesting that biochemical hallmarks of enhancer activity are required and that spurious signals from inactive chromatin are rare (Fig. S8).
We next sought to characterize concordance between cis-target genes identified via STING-seq and other methods, such as physical contact mapping and allele-specific expression. To identify gene promoters anchored in 3-dimensional space to H3K27ac-bound chromatin, we generated H3K27ac HiChIP libraries in K562 cells. Of the 134 STING-seq CREs and their 124 target genes, we observed 32 CREs where the same gene was identified with H3K27ac HiChIP contacts, 27 CREs where the same gene was identified through expression quantitative trait loci (eQTL) mapping of the same fine-mapped variant (31), and 73 CREs where the same gene was identified through a transcriptome-wide association study (TWAS) of a blood trait (32). Although the sensitivity of TWAS for target gene identification is reasonably high (54%), we and others have found that specificity can be low using this approach (33). Additionally, 54 CREs with fine-mapped GWAS variants had allele-specific effects on enhancer activity or transcription factor binding (34, 35), suggesting these variants are causal at their respective CREs (Table S3F).
Identification of causal variants and their impact on gene and protein expression
In the STING-seq dataset, we identified examples where multiple lines of orthogonal evidence converged to explain how a CRE regulates a cis-target gene. For example, the lead variant (rs4845124) at a locus associated with mean corpuscular volume in multi-ancestry meta-analyses (GWAS p = 6.9x10−17) was fine-mapped as plausibly causal (in the 95% credible set with posterior probability ≥ 1%); however, upon CRISPR inhibition of the cCRE, there was no target gene (Fig. 2F-G). Fine-mapping of this locus nominated a second plausibly causal variant mapping to a cCRE (rs12140898) whereupon inhibition identified MAPKAPK2 as the target gene (log2 FC = −0.64, SCEPTRE p = 2.2x10−16). Notably, both variants were fine-mapped eQTLs for MAPKAPK2 in neutrophils. However, only rs12140898 had predicted allele-specific effects, on SPI1 binding, and mapped to a HiChIP contact domain for the MAPKAPK2 promoter. Therefore, while eQTL studies nominated the correct target gene for this locus, it was through experimental CRE-gene mapping that we pinpointed the most likely causal GWAS variant. Importantly, the majority of targeted GWAS variants did not have supporting evidence from eQTL data but were within proximity (500 kb) of a TWAS gene, demonstrating that we can uncover genes that may be underpowered by eQTL mapping and refine TWAS results that may have high false-positive rates (Table S3F) (33).
To disentangle loci with multiple target genes in cis, we can combine targeted CRE inhibition and gene inhibition. For example, the lead variant (rs7416513) at a locus associated with monocyte count in multi-ancestry meta-analyses (GWAS p = 3.8x10−32) was fine-mapped as plausibly causal (Fig. 2H). This variant maps to an intergenic region, between the gene bodies of CRYBG2 and CD52, and the gene with the closest TSS is UBXN11. Given this, it is unclear which of these genes — if any — might be the target gene. The variant is also a fine-mapped blood cell eQTL for multiple genes in the locus (CD52, CRYBG2, SH3BGRL3, and ZNF593), further obscuring the target gene. Upon inhibiting the rs7416513-CRE, we detected CD52 as the most significantly altered gene (log2 FC = −1.6, SCEPTRE p = 2.2x10−16) (Fig. 2I), and ZNF593 also had a weak change in expression (log2 FC = −0.04, SCEPTRE p = 1.3x10−3) with no effect on SH3BGRL3 or CRYBG2. Directly targeting CD52 does not influence ZNF593 (SCEPTRE p = 0.65) expression, suggesting the rs7416513-CRE has a pleiotropic regulatory effect on multiple genes.
Using single-cell proteomics, we also detected a significant decrease in cell surface CD52 protein expression upon rs7416513-CRE inhibition (log2 FC = −0.1, SCEPTRE p = 1.2x10−15) (Fig. 2J), demonstrating that CREs with GWAS variants modulate not only cis-target gene expression but also protein expression. CD52 protein can be targeted with alemtuzumab to improve clinical outcomes in patients with myelodysplastic syndrome, suggesting that this may be the causal gene for the monocyte count GWAS association (36). The rs7416513 derived C allele is associated with increased monocyte count in multi-ancestry meta-analyses (GWAS effect = 0.025, p = 3.8x10−32) (11) and also with increased CD52 expression in monocytes (eQTL estimate = 0.71, p = 4.5x10−31) (37), highlighting the power of STING-seq to connect variants to druggable genes and identify those variants that may impact response to drugs like alemtuzumab.
Target gene discovery in STING-seq using non-European and multi-ancestry GWAS
Historically, the majority of GWAS loci have been identified using individuals of European ancestry (38). Recent efforts to use non-European ancestries and to combine multiple ancestries for GWAS have yielded numerous new associations (11, 39, 40). By leveraging ancestry-specific and multi-ancestry GWAS, we increased the discovery space of CREs and target genes for STING-seq: We identified 16 CREs with cis-target genes from GWAS variants in non-European ancestries. For example, we identified ATP1A1 as the target gene for a locus associated with neutrophil counts exclusively in African ancestries (Fig. S9A-B). The lead variant (rs6674304) was fine-mapped as plausibly causal in individuals with African ancestries (GWAS p = 3.4x10−44) but not in individuals with European ancestries (GWAS p = 0.58). Although rs6674304 did not map to any cCREs, statistical fine-mapping nominated three additional variants that did map to cCREs (rs6660743, rs12087680, and rs7544679) (Fig. S9A). We targeted all three variants using STING-seq and found that targeting the rs12087680-CRE revealed the cis-target gene ATP1A1 (log2 FC = −0.35, SCEPTRE p = 2.0x10−10) (Fig. S9B). ATP1A1 maintains electrochemical gradients of sodium and potassium ions, and prior work has linked both ATP1A1 and neutrophil counts with hypertension (41-43). As the ATP1A1 CRE demonstrates, STING-seq using non-European and multi-ancestry GWAS can identify new trait genes.
A pleiotropic CRE in the APOE and APOC1 locus
In a minority of STING-seq CREs, we identified multiple cis-target genes, which may occur through direct regulation of multiple genes or indirect effects on other nearby genes driven by a single cis-target gene. These outcomes can be difficult to distinguish without additional perturbations or a known gene regulatory network.
For example, we found that rs1065853 was the lead variant and fine-mapped as plausibly causal for an immature red blood cell trait (high light scatter reticulocyte percentage) at its locus (GWAS p = 5.8x10−48) (Fig. S9C). This variant mapped to an intergenic region, between the gene bodies of APOE and APOC1, with APOE being the closest gene and also associated with high and low density lipoprotein levels (44). Upon inhibiting the rs1065853-CRE, we observed significant decreases in expression for both APOE (log2 FC = −0.63, SCEPTRE p = 2.8x10−6) and APOC1 (log2 FC = −0.27, SCEPTRE p = 3.5x10−6) (Fig. S9D). Previous studies have shown that APOE and APOC1, which encode apolipoproteins E and C1, influence blood lipids and diverse ailments including cardiovascular disease and Alzheimer’s disease (45, 46). To help distinguish direct and indirect regulation, we used a prior genome-wide Perturb-seq (GWPS) study in the same cell line (K562) to infer whether APOE or APOC1 regulate one another (47): APOC1 expression was unchanged upon APOE inhibition (GWPS z = 0.02) but APOE expression was decreased upon APOC1 inhibition (GWPS z = −1.4). APOE and APOC1 direct inhibition suggests that rs1065853-CRE may target either APOC1 alone — even though APOE is the closest gene — or both APOC1 and APOE. Since these genes work in a coordinated fashion to regulate lipid metabolism (48), the co-regulation of these genes is a notable observation of regulatory pleiotropy that may contribute to trait associations.
Targeting multiple CREs in the PTPRC locus reveals non-functional LD proxies
We also examined loci with several fine-mapped variants near a single gene. At the PTPRC locus, we targeted nine variants that were fine-mapped variants for 10 traits (Fig. S10A, Table S1E) and mostly not in strong linkage disequilibrium (LD) as quantified by pairwise R2 from 1000 Genomes (49) (Fig. S10B). The nine variants mapped to distinct cCREs: One was 5 kb before the PTPRC TSS and the remaining eight were in the first intron, from 2 kb to 42 kb after the TSS (Fig. S10C). We observed modulation in PTPRC when targeting six of the cCREs (Fig. S10D). For the cCREs with no effect, we found that two variants were in high LD (R2 ≥ 0.95) with variants mapping to PTPRC CREs, suggesting that these may be non-functional variants in LD with functional variants (i.e., non-functional LD proxies). For all CREs, PTPRC was the only significant target gene and thus very likely the causal GWAS gene (Table S1E).
The high allelic heterogeneity — driven by multiple independent regulatory variants in distinct CREs modulating PTPRC expression — and the 10 blood trait associations suggest that the CREs may have cell-type specific activity. That is, different CREs may regulate PTPRC in different contexts, given that the 10 trait associations include RBCs, WBCs, and platelet traits (Fig. S10A).
We found that experimental evidence (e.g., STING-seq) is required to link these CREs to PTPRC expression: None of the targeted variants are fine-mapped blood eQTLs and only a single targeted variant, rs1326279, showed evidence of allele-specific effects on transcription factor binding (31, 35). Thus, in silico methods that use eQTL data are insufficient to measure the impact of the CREs on PTPRC expression.
Direct GWAS variant insertion with beeSTING-seq
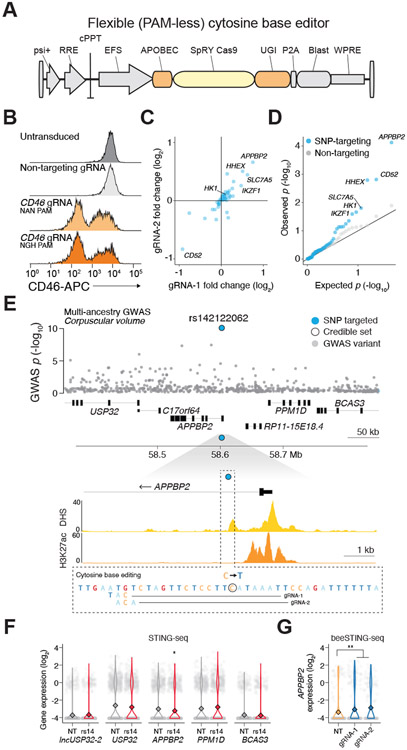
Next, we sought to expand the STING-seq approach to precise insertion of fine-mapped GWAS variants with base editing. We fused a cytosine base editor (FNLS-BE3) to a PAM-flexible Cas9 variant (SpRY) (Table S1F) and validated activity using gRNAs designed to disrupt splice junctions in CD46, which encodes a ubiquitously-expressed cell surface protein, in an arrayed fashion (Fig. 3A-B, Table S3H) (50, 51). We observed up to ~70% knockdown of CD46 when targeting splice sites with diverse PAM sequences, and an average knockdown of 27% (n = 12 target sites), similar to prior pooled screens using base editing (52, 53), (Fig. S11, Table S3H). We then performed a single-cell pooled base editing screen (beeSTING-seq) targeting 46 C>T fine-mapped GWAS variants mapping to 42 STING-seq-identified CREs with three gRNAs each (Table S3I). We tested for direct effects on known target genes and found that 32 out of 46 had at least two gRNAs with concordant effects, and that all three gRNAs had concordant effects for 17 variants (Fig. 3C, Table S3K). We identified three sets of beeSTING-seq gRNAs with cis-regulatory effects on the same target genes identified using STING-seq (5% FDR) with no enrichment of non-targeting (negative control) gRNAs (Fig. 3D, Table S3L-M).
Figure 3. Precise GWAS variant editing with base editing STING-seq (BeeSTING-seq).
(A) Lentiviral CRISPR base editor (FNLS-BE3) with a relaxed protospacer-adjacent motif (PAM) SpCas9 variant, SpRY, for base editing STING-seq (beeSTING-seq). (B) Flow cytometry of CD46 cell surface protein after base editing at CD46 splice donor sites. CD46 knockdown was compared to untransduced and non-targeting (NT) controls. (C) Target gene fold-change for the two gRNAs with the most concordant effects for each variant. (D) Quantile-quantile plot of NT gRNAs and gRNAs targeting 46 fine-mapped GWAS variants mapping to STING-seq GWAS-CREs with cis-effect genes. Genes for NT tests were randomly sampled from the set of genes in cis for targeting gRNAs. (E) (top) For a multi-ancestry corpuscular volume locus, one fine-mapped variant was targeted, the lead variant, rs142122062 (blue). (bottom) Base editing by gRNA-1 and gRNA-2 changes the rs142122062 allele from reference to alternative; for both gRNAs, this is the only cytosine in the base editing window. (F) Single-cell gene expression for cells with gRNAs targeting the GWAS-cCRE rs142122062 or NT. APPBP2 was identified as a cis-target gene. (G) BeeSTING-seq of rs142122062 increases APPBP2 expression with two independent gRNAs whose positions are shown in panel E. Asterisks denote significant q-values, Benjamini-Hochberg adjusted SCEPTRE p-values (* q < 0.05, ** q < 0.01, *** q < 0.001).
In one case, beeSTING-seq gRNAs target the lead variant (rs142122062) at a locus associated with RBC volume in multi-ancestry meta-analyses (GWAS p = 8.2x10−11) (Fig. 3E, Table S3M). Targeted inhibition of the rs142122062-CRE decreased APPBP2 expression (log2 FC = −0.46, SCEPTRE p = 2.5x10−4) and identified it as the target gene for this locus (Fig. 3F). For beeSTING-seq, we were able to design multiple gRNAs capable of inserting the same single-nucleotide edit by capitalizing on the targeting flexibility of SpRY Cas9 (51). With direct insertion of the rs142122062-T allele with two independent gRNAs, we observed a significant increase in APPBP2 expression (combined log2 FC = 0.74, SCEPTRE p = 7.6x10−5) (Fig. 3G), demonstrating the ability of beeSTING-seq to identify GWAS variants that act to increase expression. Both gRNAs exclusively edit the GWAS variant, as it is the only C nucleotide within the editing window (50). Using TWAS, we found that amyloid precursor protein, which APPBP2 binds, has the strongest association with RBC counts (54), suggesting a possible mechanism of how altered APPBP2 expression impacts RBC traits. In this manner, beeSTING-seq can more precisely interrogate the impact of GWAS variants, moving beyond CRE inhibition to reveal the impact of specific alleles on target gene expression.
CRE-driven, dosage-dependent transcriptome-wide changes in gene expression
To understand the impact of GWAS-CREs on gene expression across the genome, we performed transcriptome-wide differential expression tests. We applied a strict (1%) FDR to identify target genes in trans and again found good calibration with non-targeting gRNAs (Fig. 4A, Table S3C). We observed trans-effects for CREs that targeted in cis the transcription factors (TFs) GFI1B, NFE2, IKZF1, HHEX, and RUNX1 and the host genes of microRNAs (miRNAs) miR-142 and miR-144/451 (Fig. 4A, Table S3F, Table S4A). These TFs and miRNAs are known to play key roles in hematopoietic stem cell differentiation (55-61).
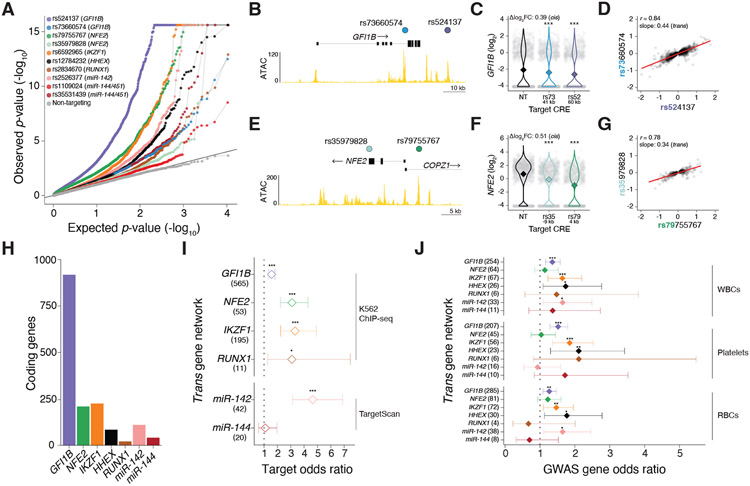
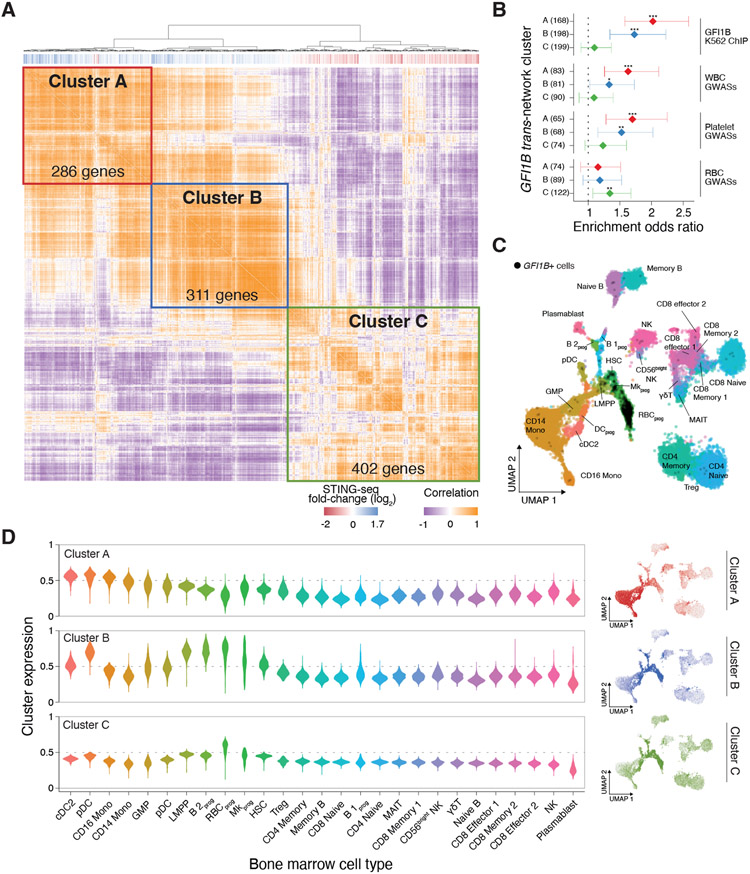
Figure 4. Trans-regulatory network discovery of genes that impact diverse blood cell traits.
(A) Quantile-quantile plots of trans-effects (whole transcriptome) of GWAS-CREs and non-targeting (NT) gRNAs. We identified significant genes at a 1% FDR (Benjamini-Hochberg adjusted SCEPTRE p-value) for GWAS-CREs with transcription factors (GFI1B, NFE2, IKZF1, HHEX, and RUNX1) and microRNAs (miR-142 and miR-144/451) as cis-regulatory target genes. No trans-effects were found for NT gRNAs. (B) Two GWAS-CREs targeted at the GFI1B locus, rs524137 and rs79755767. (C) Single-cell gene expression for cells with gRNAs targeting GWAS-cCREs at the GFI1B locus (rs524137 or rs79755767) or NT. (D) Expression of rs524137-CRE significant trans-target genes in cells with perturbation of either GWAS-CRE at the GFI1B locus (rs524137 or rs79755767) (n = 1,161 genes at a 1% FDR). (E) Two GWAS-CREs targeted at the NFE2 locus, rs79755767 and rs35979828. (F) Single-cell gene expression for cells with gRNAs targeting GWAS-cCREs at the NFE2 locus (rs79755767 or rs35979828) or NT. (G) Expression of rs79755767-CRE significant trans-target genes in cells with perturbation of either GWAS-CRE at the GFI1B locus (rs79755767 or rs35979828) (n = 343 genes at a 1% FDR). (H) Protein-coding genes with changes in expression for trans-regulatory networks. (I) Gene set enrichment odds ratios (diamonds) and 95% confidence intervals (lines) for transcription factor and microRNA targets within each trans-regulatory network. Targets for transcription factors (TFs) were given as the closest gene to each TF-specific ENCODE K562 ChIP-seq peak and for microRNAs as TargetScan-predicted targets based on sequence. (J) For each trans-regulatory network, gene set enrichment odds ratios (diamonds) and 95% confidence intervals (lines) of closest genes to fine-mapped variants from WBC, platelet, and RBC GWASs from 29 UK Biobank blood traits and 15 Blood Cell Consortium blood traits. Asterisks in panels C and F denote significant Benjamini-Hochberg adjusted SCEPTRE p-values and in panels I and J denote logistic regression p-values (* p < 0.05, ** p < 0.01, *** p < 0.001).
For GFI1B, we identified two independent CREs with trans-effects. One variant (rs524137), associated with monocyte percentage and basophil counts, maps to an intergenic CRE 11.5 kb downstream of GFI1B (Fig. 4B). The other variant (rs73660574), associated with several RBC traits (mean sphered corpuscular volume, immature reticulocyte fraction, mean reticulocyte volume, and mean corpuscular hemoglobin), maps to a CRE in an intron of GFI1B (Fig. 4B). These CREs exhibited independent dosage effects on GFI1B expression, with the rs524137-CRE having a ~70% stronger effect than the rs73660574-CRE. Thus, perturbing either rs73660574- or rs524137-CREs led to changes in the expression of GFI1B (Fig. 4C) and its target genes. To better understand the trans-effects of these two GFI1B CREs, we examined gene-expression changes in all 1,161 differentially expressed genes identified from the rs524137-CRE (Fig. 4D). For these genes, we observed a high correlation between perturbations targeting each CRE (r = 0.84), even though many of the gene expression changes were more modest when perturbing the rs73660574-CRE. We found a linear dosage relationship between the trans regulatory effects for the CREs that agreed with the difference in their effect on cis (GFI1B) expression (~1.3-fold) (Fig. 4C-D). Using single-cell proteomics in the same cells, we observed changes in protein levels for nine of the genes in the GFI1B network; for these, changes in transcript expression and protein levels were highly correlated (r = 0.9) (Fig. S12). This example demonstrates how GWAS variants mapping to CREs perturb regulatory networks and that these changes at the RNA level also alter protein expression.
In addition to GFI1B, we also observed CRE dosage effects on target gene expression and regulatory networks for NFE2 (rs79755767, associated with hematocrit and red cell distribution width, and rs35979828, associated with eosinophil count, mean corpuscular hemoglobin, and monocyte count) (Fig. 4E). When targeting these variants, we observed dosage effects on NFE2 expression (rs79755767-CRE log2 FC = −1.1, SCEPTRE p = 2.2x10−16; rs35979828-CRE log2 FC = −0.6, SCEPTRE p = 2.2x10−16) (Fig. 4F) and on a 343 gene regulatory network (r = 0.78) (Fig. 4G). These results reinforce our findings that fine-mapped GWAS variants at independent CREs have independent effects not only on target gene expression, but on entire regulatory networks in trans.
A limitation of many GWAS functional interpretation approaches is that they focus on nearby protein-coding genes and overlook relevant noncoding RNAs. With STING-seq, we also identified regulatory networks for microRNAs, which can have a broad impact on gene regulation. For example, STING-seq at the CRE for rs2526377, the most plausibly causal variant for a locus associated with platelet count locus, revealed no protein coding cis-target genes (Fig. S13A). However, when examining noncoding transcripts, we found a differentially expressed noncoding transcript AC004687.1, which is also known as the miR-142 host gene (log2 FC = −1.8, SCEPTRE p = 2.2x10−16) (Fig. S13B). This finding is further supported by prior work in the context of Alzheimer’s disease showing that the risk allele decreases miR-142 host gene promoter activity (62, 63).
For STING-seq perturbation of rs2526377, we detected a 119 gene trans-regulatory network (Fig. S13C). The top upregulated genes within the rs2526377 trans-regulatory network (WASL and CFL2) were also the top upregulated genes in miR-142 knockout mice (60). This lends further support that the trans-regulatory effects of rs2526377 perturbation are due to cis effects on miR-142, as found in STING-seq. This cis-target microRNA and its regulatory network can be easily missed when considering only protein-coding genes for target gene annotation.
We also analyzed trans effects with direct variant insertion using beeSTING-seq. We could detect changes in regulatory network expression in the expected direction upon inserting the rs12784232-A allele (associated with lymphocyte percentage) and rs6592965-A allele (corpuscular hemoglobin), which mapped to the HHEX and IKZF1 GWAS-CREs, respectively. In contrast to GWAS-CRE inhibition which decreased expression of HHEX and IKZF1 (Fig. 4A), direct variant insertion resulted in increased expression of the cis-target genes and, accordingly, trans-effects for genes tended to switch directions in differential expression, as compared to STING-seq. Specifically, we observed that 60 - 70% of HHEX and IKZF1 network genes had reversed directions of effect, demonstrating that GWAS variants which act to increase expression can impact networks in discordant directions from CRE silencing.
Enrichment of cis-target binding sites and GWAS genes in trans-regulatory networks
To better characterize how CREs with target genes in trans alter blood cell phenotypes, we examined genome-wide binding for GFI1B, NFE2, IKZF1, and RUNX1 (ChIP-seq) (64, 65) and sequence-based predicted targets of miR-142 and miR-144/451 (TargetScan) (66, 67). We asked whether the closest genes to each ChIP-seq peak or predicted microRNA target genes were enriched in STING-seq trans-regulatory networks (Fig. 4H, Table S4B). We observed enrichments of predicted target genes for GFI1B, NFE2, IKZF1, RUNX1, and miR-142 (OR = 2.4 ± 1.9, mean ± sem) (Fig. 4I, Table S4C). Thus, perturbing CREs can reveal second-order interactions for regulatory networks driven by TFs or microRNAs.
A related and pertinent question is whether the genes in the trans-regulatory networks identified by STING-seq may also play a role in blood traits and whether they also harbor cis-regulatory genetic variants. To answer this question, we constructed a set of putatively causal genes for each of the 29 UKBB and 15 BCX GWASs by selecting the closest protein-coding genes to fine-mapped variants of GWAS loci. We then grouped them by cell type, generating gene sets for platelets, RBCs, and WBCs that were mostly distinct (Fig. S14, Table S4B). For nearly all trans-regulatory networks, we found enrichments for blood cell GWAS genes (Fig. 4J, Table S4C). These blood cell trait GWAS loci enrichments indicate that the known roles of these genes in hematopoiesis and cell differentiation are mediated by their effects on regulatory networks. Furthermore, identification of the trans genes with STING-seq pinpointed regulatory networks whose polygenic perturbation by distinct variants across the genome appears to contribute to the GWAS signal. This suggests a mechanistic importance for networks themselves, where we do not need to functionally dissect V2F per locus if we know the pathway through which they are likely to act, similar to recent work that focuses on perturbation of target genes (68).
Trans-regulated genes reveal biological mechanisms and cell types of trait associations
Given these relationships between trans-regulated genes and GWAS loci, we analyzed the structure of these regulatory networks to better understand the mechanistic roles of specific genes in blood traits. Using single-cell gene co-expression and clustering, we identified co-expressed gene clusters for each of the loci (Fig. 5A, Fig. S15). For the trans-acting gene GFI1B, we identified two clusters (A and B) of genes with increased expression upon GFI1B CRE repression with STING-seq. These clusters were the most strongly enriched for GFI1B binding sites (Fig. 5B, Table S4B-C). A third cluster (C) consisted primarily of genes with decreased expression, which were not enriched for GFI1B binding sites. Interestingly, clusters A and B were enriched for genes from platelet and WBC GWASs whereas cluster C was only enriched for genes from RBC GWASs.
Figure 5. Subnetworks of GFI1B target genes are expressed in specific hematopoietic progenitors and differentiated cells.
(A) Co-expression matrix of rs524137-CRE GFI1B network genes in K562 with hierarchical clustering. Three clusters (A, B, and C) are indicated. The vertical bars below the dendrogram indicate if genes had increased (blue) or decreased (red) expression upon inhibiting the GFI1B CRE. (B) For each trans-regulatory GFI1B subnetwork (cluster), gene set enrichment odds ratios (diamonds) and 95% confidence intervals (lines) of closest genes to (top) GFI1B K562 ChIP-seq peaks and (bottom) fine-mapped variants from WBC, platelet, and RBC GWASs from 29 UK Biobank blood traits and 15 Blood Cell Consortium blood traits. Asterisks denote logistic regression p-values (* p < 0.05, ** p < 0.01, *** p < 0.001). (C) Uniform Manifold Approximation and Projection (UMAP) of human bone marrow cell gene expression from 35 Human Cell Atlas donors. Labels and colors indicate cell types (B 2prog: progenitor B-2 cells; RBCprog: red blood cell progenitors; DCprog: dendritic cell progenitors; full list in Table S4D). The black dots denote cells expressing GFI1B. GFI1B is most highly expressed in RBC progenitors, megakaryocyte progenitors, and hematopoietic stem cells. (D) Expression of genes from Clusters A, B, and C in each human bone marrow cell type (left) and in each cell in the UMAP space from panel C (right).
To further refine and validate the individual cell types involved with different clusters of co-regulated genes, we integrated the GFI1B co-expression network with primary cells from the Human Cell Atlas, which includes progenitors and/or differentiated cell types for platelets, WBCs, and RBCs. Specifically, we used single-cell RNA-sequencing from 35 bone marrow donors (69, 70), as bone marrow includes a rich sample of multipotent progenitor cells crucial for hematopoiesis. We first confirmed that GFI1B was expressed in hematopoietic stem cells and progenitor cells for RBCs and megakaryocytes — in line with GFI1B’s well-established role as a transcriptional repressor in early and lineage-specific progenitors (Fig. 5C) (55, 71-73). As expected, GFI1B is not expressed in granulocytes and lymphocytes (73, 74). Genes from Cluster A were highly enriched for GFI1B binding sites and had increased expression upon inhibiting GFI1B, suggesting that these genes are actively being repressed in cells where GFI1B is expressed (Fig. 5B). We next observed that genes from Cluster A were highly expressed in granulocyte-monocyte progenitors (GMP) and differentiated WBC types, including monocytes and dendritic cells (Fig. 5D, Table S4D). For example, CD33 is a well-known marker for myeloid cells that is commonly used to diagnose acute myeloid leukemia, and its expression increases upon inhibiting the GFI1B CRE (Fig. S12) (75, 76). GFI1B directly binds the promoter of CD33 (Fig. S16A) and, upon inhibiting GFI1B, we found that CD33 transcript and protein expression were both increased (Fig. S16B-C). CD33 is expressed in myeloid progenitors and differentiated cells such as dendritic cells or monocytes (Fig. S16D). Overall, Cluster A is comprised of genes that GFI1B directly represses, and their downstream targets, to prevent differentiation of hematopoietic stem cells into WBCs.
Like Cluster A, genes in Cluster B were also enriched for GFI1B binding sites and had increased expression upon inhibiting GFI1B (Fig. 5A-B). However, genes in Cluster B were not expressed in differentiated WBCs, but rather in a broad set of progenitor cell types (Fig. 5D), suggesting that these may be genes that are repressed in hematopoietic stem cells to maintain a multipotent cell state. Cluster C differed from Clusters A and B in that it was not enriched for GFI1B binding sites and had decreased expression upon inhibiting GFI1B. Genes in Cluster C were expressed most highly in RBC progenitors, suggesting that these genes are secondary targets of GFI1B that act in a lineage-specific manner to differentiate hematopoietic stem cells into erythrocytes. These findings are supported by this cluster being enriched for RBC GWAS genes (Fig. 5B), and pathway analysis identifying these genes as part of the heme biosynthesis pathway (Table S4E). The identification of these trans-regulatory networks in a homogeneous blood progenitor-like cell type (K562) demonstrates the utility of STING-seq in studying diverse effects of CREs on target genes.
Trade-offs between CRE effect sizes, number of cells and sequencing depth in STING-seq
Given the large number of GWASs performed over the past 15 years, with numbers of trait-associated loci per GWAS ranging from tens to thousands (44), we wanted to understand the scale of cells needed to perform STING-seq under various settings. By performing statistical down-sampling experiments on the cis-regulatory effects identified with STING-seq, we computed the number of cells required for nominal significance (SCEPTRE p < 10−3) for target genes with different expression levels, different CRE perturbation effect sizes, and different per-cell sequencing depths (Fig. S17). For CREs with large effects, STING-seq requires as few as 100 cells and 5,000 reads per cell, comparable to methods like Perturb-seq and ECCITE-seq which target genes directly (47, 68, 77, 78). For CREs with moderate effects, STING-seq requires about 400 cells per gRNA or, if cell number is fixed at 100 cells, 15,000 reads per cell. This downsampling analysis provides a useful set of guidelines for estimating the resources required for applying STING-seq to other GWASs beyond blood traits.
Discussion
In summary, we have developed an approach for characterization of functional effects of GWAS loci that takes noncoding human genetic variants and integrates fine-mapping, pooled CRISPR screens, and single-cell RNA- and protein-sequencing to identify target genes in cis and trans. We demonstrated the utility of STING-seq to identify target genes of CREs overlapping GWAS variants and described complex regulatory architectures of CREs. We found that 77% of blood trait GWAS loci have at least one fine-mapped variant overlapping an enhancer region and can be targeted with STING-seq. Notably, we identified target genes for 25% of tested cCREs, and 36% of tested loci, a high yield over previous studies that studied regulatory effects of noncoding genomic loci (7, 8). We also found that CRE activity is needed for CRISPRi-based target detection and that spurious signals from inactive chromatin are rare. Additionally, we identified CREs with GWAS variants for TFs and miRNAs, and, through their perturbation, identified trans-regulatory network clusters with distinct biological functions. The enrichment of genes in independent blood cell trait GWAS loci in these networks implies a polygenic contribution to the cellular functions that underlie diverse blood cell traits. We also identified target genes for non-European associations where functional genomics data are typically sparse. For example, we nominated ATP1A1 as a causal gene for neutrophil counts through targeting a locus identified exclusively in African ancestries. Importantly, targeting loci identified from ancestry-specific GWAS in cell models is ancestry-agnostic, provided the GWAS variant maps to a candidate regulatory element, and can lead to target gene identification.
We also performed direct variant insertion with beeSTING-seq, identifying noncoding GWAS variants with causal effects on target gene expression. Given incomplete editing efficiencies (many studies reporting ~30% (79)), that the biological effect of individual GWAS variants are expected to be small, and that single-cell transcriptome data are sparse, it was not unexpected that we were only able to identify few loci and future work is needed to further optimize base editors for studying the effects of GWAS variants. Targeted enrichment panels will have utility in improving the sparsity of single-cell sequencing, however further innovation will be necessary to improve base editing efficiency, through directed evolution of existing base editors and the discovery of additional ones. However, the trade-off between higher yield from blunt perturbations, such as CRISPRi, versus highly precise base editing with smaller functional effects is likely to persist, and the ideal approach depends on the goals and design of each study.
A key feature of recent CRISPRi screens of cCREs (7, 8), including STING-seq, is the introduction of multiple perturbations per cell. This substantially increases the number of loci that can be feasibly analyzed. While this is feasible for immortalized cell lines, expanding multiple perturbations (via either high MOI transduction or innovative vector designs) to other cell lines and primary cells will be instrumental for the next stage of target gene identification and characterization for diverse GWAS traits. However, caution is warranted in study designs where a large proportion of gRNAs are likely to have trans effects, as their potential interactions may complicate interpretation of the data. In these cases, reducing the number of perturbations per cell may be necessary.
Our results demonstrate the power of single-cell sequencing for sensitive and scalable readout of regulatory effects of GWAS loci in cis and trans. While we have a high yield in cis target gene discovery, we note that identification of a cis gene alone with STING-seq does not prove its mechanistic causal role driving the GWAS association, nor exclude other potential causal variants, CREs, and genes, including in other cell types. Indeed, our observation of multiple CREs with highly correlated cis and trans effects but GWAS associations for different blood traits suggests that they might have distinct additional effects in other cellular contexts. In loci where cis-effects are coupled with trans-network effects, STING-seq can be highly informative of potential cellular mechanisms, which also provides strong support for the causal role of the cis-target gene. Given these network enrichments, we suggest that GWAS loci that putatively target TFs or miRNAs should be high priority targets for STING-seq given the wealth of information we can gain. Furthermore, integration of STING-seq with cellular phenotype screens (80-82) will be an invaluable next step to connect genetic variants with cellular mechanisms driving GWAS associations.
Altogether, the STING-seq workflow provides a roadmap to address V2F challenges and identify target genes for GWAS loci in a high-throughput fashion, enabling deeper understanding of human noncoding genome function and translation of these insights into new therapies.
Materials and Methods
UK Biobank genome-wide association studies of blood cell traits
UK Biobank data were used upon ethical approval from the Northwest Multi-Centre Research Ethics Committee and informed consent was obtained from all participants prior to participation. We used genome-wide association study (GWAS) summary statistics for 29 blood cell traits from 361,194 white British UK Biobank participants: WBC (leukocyte) count, RBC (erythrocyte) count, hemoglobin concentration, hematocrit percentage, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, RBC (erythrocyte) distribution width, platelet count, platelet crit, mean platelet (thrombocyte) volume, platelet distribution width, lymphocyte count, monocyte count, neutrophil count, eosinophil count, basophil count, lymphocyte percentage, monocyte percentage, neutrophil percentage, eosinophil percentage, basophil percentage, reticulocyte percentage, reticulocyte count, mean reticulocyte volume, mean sphered cell volume, immature reticulocyte fraction, high light scatter reticulocyte percentage, and high light scatter reticulocyte count (Table S1A). Each GWAS was performed by fitting the following covariates to inverse normal transformed traits with linear regression models: Principal components 1 through 20, sex, age, age2, sex and age interaction, and sex and age2 interaction. The summary statistics were generated by the Neale Lab (www.nealelab.is/uk-biobank).
Statistical fine-mapping of UK Biobank blood cell traits
The 29 UK Biobank GWASs of blood cell traits were uniformly processed with a statistical fine-mapping pipeline. First, each GWAS was analyzed with GCTA-COJO v.1.93.1 (13, 14) to identify conditionally independent lead variants (pj < 6.6x10−9) and define 1 Mb regions for statistical fine-mapping. All variants within 500 kb of a lead variant were analyzed with FINEMAP v.1.3.1 (83), a Bayesian fine-mapping method that assigns each variant a Bayes factor for being plausibly causal. Both GCTA-COJO and FINEMAP require population-matched covariance matrices, therefore we generated these with PLINK v.2.0 (84), QCTOOL v.2.0.2, BGENIX v.1.1.5 (85), and LDstore v.1.1 (86), using a subset of 50,000 UK Biobank white British participants (UK Biobank accession code 47976). FINEMAP allows for a maximum number of causal configurations to test for each input set of variants, therefore we set the maximum to 10 causal configuration variants per fine-mapped region and excluded cases where FINEMAP failed to converge. We then retained noncoding variants with a high Bayes factor (log10 BF ≥ 2) and that were at least 1% likely to be causal for a set of causal variants. Fine-mapped variants that had more than one Bayes factor, due to being within 500 kb of multiple lead variants, had their highest value retained. Across all 29 GWASs, we identified 827 loci, separated by at least 500 kb, and 57,531 fine-mapped variants. The Variant Effect Predictor (VEP) tool (87) was used to identify 53,874 noncoding variants.
Fine-mapped Blood Cell Consortium blood cell trait GWAS
The Blood Cell Consortium (BCX) generated GWAS summary statistics and fine-mapped 95% credible sets for 15 blood traits from 746,667 participants from five global populations (European ancestries, South Asian ancestries, Hispanic ancestries, East Asian ancestries, and African ancestries): RBC count, hemoglobin, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, MCH concentration, RBC distribution width, WBC count, neutrophils, monocytes, lymphocytes, basophils, eosinophils, platelet count, and mean platelet volume (11). Each GWAS was performed within each global population by fitting linear mixed models, adjusting for cohort-specific covariates, to generate population-specific GWAS summary statistics. Population-specific GWAS were fine-mapped using an approximate Bayesian approach (88) to construct 95% credible sets from all variants within 250 kb of a lead variant. 95% credible sets are generated by ordering marginal variant posterior probabilities from highest to lowest and retaining variants until the probabilities sum 95%. Population-specific GWAS for each trait were then meta-analyzed using a multi-ancestry meta-analysis method (89) that also generates marginal variant posterior probabilities, from which multi-ancestry 95% credible sets were generated. We additionally required that variants were at least 1% likely to be causal. Across all 15 multi-ancestry meta-analyzed GWASs, we identified 1,91 loci, separated by at least 500 kb, and 62,494 fine-mapped variants. VEP (87) was used to identify 58,573 noncoding variants.
Functional annotation of causal noncoding SNPs
We integrated multiple functional genomics datasets for K562 cells. Specifically, we used DNase I hypersensitive sites (DHS) from ENCODE (65), H3K27ac ChIP-seq peak calls from ENCODE, and ATAC-seq peak calls that we generated previously (81) to identify candidate cis-regulatory elements (cCREs). We used bedtools v.2.25.0 (90) and bedops v.2.4.3 (91) to identify variants mapping directly to cCREs. We also required variants to be further than 1 kb from any gene TSS. We analyzed the UK Biobank and BCX GWAS variants separately. For UK Biobank GWASs, we identified 10,628 distinct variants mapping cCREs in 629 loci. We then selected 88 variants from 56 loci for targeting based on whether a variant was targetable and more plausibly causal than others for a given GWAS and locus by ranking FINEMAP log10 Bayes factors and manual inspection of loci. For the 88 selected variants, 32 were the most probable variant for at least one GWAS locus, and 52 were in the top-10 most probable variants. For the 56 loci, there was a median of 10.5 (± 8.6) targetable SNPs. Elements of manual inspection included selecting variants that mapped to intergenic regions between gene TSSs or selecting multiple variants that map proximal to the same gene. For BCX GWASs, we identified 10,446 variants mapping to 886 loci. We selected 507 variants mapping to 265 loci for targeting, including 41 variants mapping to closed chromatin. Of the cCRE-mapping variants, we targeted 137 that were the sole variant within the 95% credible set and 239 variants that comprised all targetable 95% credible set variants for 112 loci. The remaining 131 variants were selected because they were identified by GWASs from non-European ancestries and either fine-mapped in a population-specific GWAS or in the multi-ancestry meta-analysis. K562 DHS peaks and H3K27ac, RUNX1, IKZF1, and NFE2 ChIP-seq peaks are available from the ENCODE Project (www.encodeproject.org). K562 ATAC-seq peaks are available from GEO accession number GSE161002. K562 GFI1B ChIP-seq peaks are available from GEO accession number GSE117944.
Plasmid cloning for lentiviral CRISPRi, cytosine base editor, and modified gRNA scaffold vectors
To generate the KRAB-dCas9 (lentiCRISPRi(v1)-Blast) and KRAB-dCas9-MeCP2 (lentiCRISPRi(v2)-Blast) plasmids, KRAB and dCas9 were PCR amplified from pCC_09 (Addgene 139094) (92) and the MeCP2 effector domain was synthesized as a gBlock (IDT). KRAB and MeCP2 were linked to dCas9 with flexible glycine-serine linkers and cloned into lentiCas9-Blast (Addgene 52962) (23). To generate the FNLS-BE3-SpRY (lentiBE3-SpRY-Blast) plasmid, we used Gibson cloning to replace the puromycin resistance gene in pLenti-FNLS-P2A-Puro (Addgene 110841) with blasticidin resistance from lentiCRISPRi(v2)-Blast. We then used Gibson cloning to replaced SpCas9(D10A) with the SpRY nickase from pCAG-CBE4max-SpRY-P2A-EGFP (Addgene 139999) (51). To generate the gRNA vector (lentiGuideFE-Puro), we digested pCC_09 with NheI and KpnI to isolate the U6 promoter and Cas9 guide RNA scaffold with the F+E scaffold modification (93). After gel extraction (Qiagen 28706), we ligated this piece into NheI and KpnI-digested pLentiRNAGuide_001 (Addgene 138150) vector using T4 ligase (NEB M0202M) (94). Primer sequences for Gibson cloning reactions are available in Table S1F.
Cell culture and monoclonal cell line generation
HEK293FT cells were acquired from Thermo Fisher (R70007). HEK293FT (human) cells were maintained at 37°C with 5% CO2 in D10 medium: DMEM with high glucose and stabilized L-glutamine (Caisson DML23) supplemented with 10% fetal bovine serum (Thermo Fisher 16000044). K562 cells were acquired from ATCC (CCL-243) and were maintained at 37°C with 5% CO2 in R10 medium: RPMI with stabilized L-glutamine (Thermo Fisher 11875119) supplemented with 10% fetal bovine serum (Thermo Fisher 16000044). Cells were regularly passaged and tested for presence of mycoplasma contamination with MycoAlert Plus Mycoplasma Detection Kit (Lonza).
Lentivirus was produced by polyethylenimine linear MW 25000 (Polysciences 23966) transfection of HEK293FT cells with the transfer plasmid containing a Cas9 effector, or gRNA library, packaging plasmid psPAX2 (Addgene 12260) and envelope plasmid pMD2.G (Addgene 12259). After 72 hours post-transfection, cell medium containing lentiviral particles was harvested and filtered through 0.45 mm filter Steriflip-HV (Millipore SE1M003M00). K562 cells were transduced with lentiCRISPRi(v1)-Blast, lentiCRISPRi(v2)-Blast, or lentiBE3-SpRY-Blast at a low multiplicity-of-infection (MOI < 1). Transduced K562 cells were selected with 10 μg/μL blasticidin (Thermo A1113903) for 10 days to enrich for expression of the Cas9 effector proteins. To isolate individual clones, K562 polyclonal lines were serially diluted to 50 cells per 10 mL medium. We then plated 100 μL of this cell-media mix in 96-well round bottom plates (~0.5 cells/well).
Digital PCR for CRISPRi gene repression
We compared the single-repressor CRISPRi (KRAB-dCas9) and dual-repressor CRISPRi (KRAB-dCas9-MeCP2) systems by targeting the transcription start sites and known enhancers of three genes (MRPS23, SLC25A37 and FSCN1) with two gRNAs per targeted region. We synthesized gRNAs as top and bottom strand oligos (IDT) and cloned them into BsmBI-digested lentiGuideFE-Puro. We transduced the cells in biological triplicate with gRNA lentiviruses at a low MOI and after 24 hours selected for cells with gRNAs using puromycin (1 μg/μL, Thermo Fisher A1113803). We harvested the cells 10 days after transduction and extracted RNA using TRIzol (ThermoFisher 15596026). We quantified RNA concentration by spectrophotometry (NanoDrop). To measure gene expression, we performed digital PCR (Formulatrix Consellation) with Cy5/Iowa Black RQ target gene probes (IDT), FAM/ZEN/Iowa Black FQ for the actin normalizer (IDT), and Luna Universal One-Step RT qPCR Master Mix kit (NEB E3005L) and Tween-20 (Sigma-Aldrich P1379). We first normalized the target gene expression by actin expression per sample and then normalized this ratio to the ratio from cells transduced with non-targeting control gRNAs.
KRAB-dCas9-MeCP2 CRISPRi pooled screen for essential gene gRNA depletion
We performed CRISPRi pooled screens to quantify the KRAB-dCas9-MeCP2 inhibitory effect window in HCT116 and MCF7 cell lines. Both lines were acquired from ATCC (CCL-247 and and HTB-22, respectively) maintained in the appropriate media (McCoy’s 5A Medium and Dulbecco′s Modified Eagle′s Medium, respectively) supplemented with 10% serum and 1% penicillin/streptomycin. These cell lines were cultured at 37 °C, 5% CO2, and ambient oxygen levels. Monoclonal HCT116 KRAB-dCas9-MeCP2 and MCF7 KRAB-dCas9-MeCP2 cell lines were generated as previously described for K562 cells. Expression was confirmed via western blot.
For screening, HEK293 cells were plated in Dulbecco’s Modified Eagle Medium (DMEM) + 10% FBS (D10) in a 15 cm dish so that the following day, cells were 90% confluent. Half of the media was removed from the flask, and cells in each flask were transfected with 13.8 μg of a cCRE/TSS-targeting library specific to HCT116 and MCF7, 6.6 μg pMD2.G (envelope plasmid), and 9.6 μg psPAX2 (packaging plasmid) using 1.2 mL Opti-MEM and 112.5 μL polyethylenimine linear 25K (Polysciences 23966). The following morning, the media was removed and fresh D10 + 1%BSA was added. Then, 48 hours later, we collected the viral supernatant and put it immediately on ice. We concentrated the supernatant by centrifugation at 100,000 g (Thermo Sorvall LYNX) for 2 hours at 4 °C. The resulting pellet was resuspended in cold DMEM and stored at −80 °C until use.
We determined the appropriate titer of virus before the experimental transduction. We transduced 3M cells with a standard spinfection protocol with different dilutions of virus in a 12-well plate as well as a no virus control well. After adding virus, we spun the cells at 2000 rpm for 1 hour at 37 °C (Beckman Coulter Allegra X-14R) and incubated overnight. The next day, we plated half of the cells in each well into two new wells of a 6-well plate. In one set of wells, we added the appropriate puromycin concentration (1.5 μg/mL for HCT116 and 3 μg/mL for MCF7). After all the cells in the no virus well had died, cells in the corresponding wells (with puromycin) were counted to determine the viral volume that results in 20 to 40% cell survival, corresponding to a MOI of 0.2 to 0.5.
We cultured each cell line in the appropriate media and transduced 2x108 of them with the CRISPR lentiviral library via spinfection with the viral volume determined from the previous spinfection. As before, after adding virus, we spun cells at 2000 rpm for 1 hour at 37 °C and incubated them overnight. The following day, cells were plated at 30% confluence and selected in the appropriate puromycin concentration for 3 days. After selection, we passaged cells in 15 cm dishes for 21 days and split at ~80% confluence. We isolated genomic DNA from cells using a modified salting-out precipitation. The gRNA readout was performed using two rounds of PCR. For PCR1, we used 10 μg of gDNA in each 100ul reaction. We pooled the PCR1 products and used the mixture for a second PCR reaction. This second PCR adds on Illumina sequencing adaptors and barcodes. We performed PCR1 reactions using TaqB polymerase (Enzymatics P7250L) and PCR2 reactions with Q5 (NEB M0491). We pooled and purified PCR2 reactions with Illumina Purification Beads. We quantified the concentration of the gel-extracted PCR products using Qubit dsDNA HS Assay Kit (Thermo Fisher Q32851), then diluted and sequenced it on NextSeq 500 high-output (Illumina). We demultiplexed the samples using bcl2fastq v2.20.0.422 (Illumina), trimmed off adapters and aligned to our guides with bowtie v.1.1.2 (95). We library normalized the resulting reads (each read divided by the total number of reads). We then used the Robust Rank Aggregation algorithm (96) and estimataed log2 fold changes as log2(Day 21 / Day 1). We targeted +/− 5 kb of the transcription start site (TSS) essential genes (DepMap Chronos scores < −1) (97-100). In total we screened 1,992 gRNAs targeting 263 essential human genes. As negative controls, we embedded 1,000 non-targeting gRNAs into this library.
Flow sorting for near PAM-less base editing
We verified cytosine base editing by designing 12 gRNAs targeting CD46 splice sites using SpliceR v1.2.0 (101). SpliceR designed gRNAs that were predicted to disrupt CD46 splice sites through C>T nucleotide changes. These included gRNAs that would recognize a diverse set of non-canonical PAMs, such as NGN, NAN, NCN, and NTN (Table S3H). We also used four non-targeting gRNAs from the GeCKOv2 library (23) as negative controls. We synthesized gRNAs as top and bottom strand oligos (IDT) and cloned them into BsmBI-digested lentiGuideFE-Puro. We transduced the cells with gRNA lentiviruses at a low MOI in an arrayed fashion and after 24 hours selected for cells with gRNAs using puromycin (1 μg/μL, Thermo Fisher A1113803). After six days of selection, we proceeded to flow cytometry to measure CD46 protein. For flow cytometry, 1x106 cells per condition were harvested and washed with PBS after selection. The cells were stained for 5 minutes at room temperature with LIVE/DEAD Fixable Violet Dead Stain Kit (ThermoFisher L34864). Subsequently, the cells were stained with antibodies for 20 minutes on ice with 1 μL CD46-APC (clone TRA-2-10) (BioLegend 352405). Cells were washed with PBS to remove unbound antibodies prior to sorting. Cell acquisition and sorting was performed using a Sony SH800S cell sorter. Sequential gating was performed as follows: 1) exclusion of debris based on forward and side scatter cell parameters, 2) dead cell exclusion. The sorting gates were set such that 90% of live K562 cells would be considered CD46 positive.
CRISPR inhibition and base editing library design and cloning
Two individual CRISPR inhibition libraries were designed and cloned, termed STING-seq v1 and STING-seq v2, and one base editing library was designed and cloned, termed beeSTING-seq. For STING-seq v1, we designed 20 nt gRNAs to target within 200 bp of the 88 selected plausibly causal noncoding variants from UK Biobank GWASs of blood traits. We used FlashFry v1.10.0 (22) to retain gRNAs with the lowest predicted off-target activity, as estimated by the Hsu-Scott score (21). Each variant was targeted by two different gRNAs. In addition, we also included in our library 12 non-targeting gRNAs from the GeCKOv2 library (23) as negative controls and 12 gRNAs targeting the TSSs of six non-essential genes as positive controls. The six non-essential genes (CD46, CD52, HSPA8, NMU, PPIA and RPL22) were identified by a CRISPR knock-out screen in K562 cells using the PICKLES database (102). We additionally included 10 gRNAs targeting the CD55 TSS for our FACS-based MOI estimator, bringing the total number of gRNAs to 210. For STING-seq v2, we designed 20 nt gRNAs to target within 200 bp of the 507 selected plausibly causal variants from the Blood Cell Consortium multi-ancestry and ancestry-specific blood trait GWASs. We again retained gRNAs with the lowest predicted off-target activity and each variant was targeted by three different gRNAs. In addition, we included 30 non-targeting gRNAs from the GeCKOv2 library and 32 groups of three TSS-targeting gRNAs for positive controls. We additionally included 45 CD55 TSS-targeting gRNAs for FACS-based MOI estimation. For beeSTING-seq, we designed three sets of gRNAs for each of 46 C>T select GWAS variants mapping to CREs with cis-target genes. We followed recommended gRNA design instructions, and positioned the target nucleotide within a 5 nt window (103). We also included 28 non-targeting gRNAs from the GeCKOv2 library.
To clone the STING-seq v1 gRNA library, top and bottom strand oligos (IDT) were resuspended in water at 100 μM and then mixed at 1:1 ratio for each gRNA. Then, 1 μL of the oligo mix was added to a master mix containing 1x T4 ligase buffer (NEB M0202M), 0.5 μL T4 PNK (NEB M0201L) and water to a final concentration of 10 μL per reaction. For oligo annealing, we incubated the oligo mix at 37°C for 30 minutes, then 95°C for 5 minutes with a temperature change of 1 °C every 5 seconds until reaching 4 °C. To create the oligo pool, we pooled together 3 μL of each annealed oligo. The oligo pool was diluted 1:10 with water and then cloned in the lentiGuideFE-Puro, which was linearized with BsmBI (Thermo ER0451) and dephosphorylated. The ligation was performed in 11 reactions with each reaction consisting of 5 μL Rapid Ligation Buffer (Enzymatics B101), 0.5 μL T7 ligase (Enzymatics L602L), digested plasmid at 25 ng per reaction, 1 μL diluted oligo mix and ddH2O to final volume of 10 μL. The ligation was performed at room temperature for 15 minutes.
Next, 100 μL of the combined ligation reactions were mixed with 100 μL isopropanol, 1 μL GlycoBlue (Thermo Fisher AM9515) and 2 μL of 5 M NaCl (50 mM final concentration), incubated for 15 minutes at room temperature, and spun at 12,000 g for 15 minutes. The pellet was washed twice with prechilled 70% ethanol, air dried for 15 minutes or until dried completely, resuspended in 5 μL 1x TE buffer (Sigma). Next, 2 μL of library ligation was added to 50 μL Endura cells (Lucigen) then electroporated, recovered and plated. The following day bacterial colonies were scraped, plasmids were isolated using a maxi prep (Qiagen 12965) and library representation was determined by MiSeq (Illumina).
The STING-seq v2 and beeSTING-seq pooled gRNA libraries were synthesized as single-stranded oligonucleotide pools (Twist Biosciences) and diluted to 0.5 ng/μL in molecular-grade water. Then, 2 μL of the diluted pooled oligos were added to a master mix containing forward and reverse primer mixes (10 μM) and NEBNext High-Fidelity 2X PCR Master Mix (M0541S). We then PCR purified the product and Gibson cloned in pLentiGuideFE-Puro, which was linearized as described above. We used 500 ng of the digested vector, maintained a 1:10 molar ratio of library and incubated at 50 °C for 1 hour. We concentrated DNA using isopropanol precipitation, washed and resuspended the DNA, then transformed 1 μL of library in 25 μL of Endura cells (Endura #60242-2) according to protocol specifications. We then plated the transformed cells on LB-Ampicillin plates to get at least 100 to 500 colonies per gRNA.
The quality of all pooled libraries was verified by sequencing with a MiSeq (Illumina) to estimate the 90:10 quantile ratio. To generate and concentrate all pooled libraries, lentivirus was generated as described above. Briefly, we seeded 10 x 225 cm2 flasks with HEK293FT cells and, at 70% confluency, we co-transfected the pooled gRNA library, psPAX2 and pMD2.G. Lentivirus was collected 72 hours post-transfection and filtered using a 0.45 μm filter. The supernatant was then ultracentrifuged for 2 hours at 100,000 g (Sorvall Lynx 6000), and the pellet was resuspended overnight at 4 °C in phosphate-buffered saline with 1% bovine serum albumin.
Multiplicity-of-infection estimation via flow cytometry
When transducing cells at a high MOI, it is not possible to estimate the MOI by traditional methods (e.g., survival after drug selection) or without the time and cost of single-cell sequencing. By including multiple gRNAs that target the CD55 TSS (10 gRNAs for STING-seq v1, 45 gRNAs for STING-seq v2), we were able to estimate the number of gRNAs per cell (MOI) using flow cytometry for CD55 cell surface protein knockdown over a range of viral transduction volumes. We performed two transductions for STING-seq v1 with concentrated lentivirus (4 μL and 6 μL) and, after 48 hours, we selected with puromycin for 10 days. We performed five transductions for STING-seq v2 with concentrated lentivirus (1, 5, 10, 20, 30 μL) and, after 48 hours, we selected with puromycin for 10 days. We included three positive control transductions with different CD55 TSS-targeting gRNAs and three negative control transductions with three different non-targeting gRNAs for both experiments. For beeSTING-seq, we performed five transductions with concentrated lentivirus (1, 5, 10, 25, 50 μL), and, after 48 hours, we selected with puromycin for 10 days. We used the most viable cell culture for beeSTING-seq for sequencing (10 μL) with MACS dead cell removal kit (Miltenyi Biotec #130-090-101), as we observed high cell death at higher lentivirus concentrations.
For flow cytometry, 1x106 cells per condition were harvested and washed with PBS after selection. The cells were stained for 5 minutes at room temperature with LIVE/DEAD Fixable Violet Dead Stain Kit (ThermoFisher L34864). Subsequently, the cells were stained with antibodies for 20 minutes on ice with 1 μL CD55-FITC (clone JS11) (BioLegend 311306). Cells were washed with PBS to remove unbound antibodies prior to sorting. Cell acquisition and sorting was performed using a Sony SH800S cell sorter. Sequential gating was performed as follows: 1) exclusion of debris based on forward and side scatter cell parameters, 2) dead cell exclusion. The sorting gates were set such that 90% of live K562 cells would be considered CD55 positive. From this estimation, we can estimate MOI using X = 1 - NY, where X is the proportion of cells with CD55 targeting gRNAs, N is the inverse of the number of CD55 targeting gRNAs divided by the total library size, leaving X as the predicted MOI. For the STING-seq v1 library, N = 1 - (10/210), and for the STING-seq v2 library, N = 1 - (45/1695). We estimated that 6 μL STING-seq v1 viral volume yielded an MOI of ~13.5 and 30 μL STING-seq v2 viral volume yielded MOI of ~30 and elected to use these conditions for our STING-seq assay (Fig. S2).
Expanded CRISPR-compatible Cellular Indexing of Transcriptomes and Epitopes (ECCITE-seq)
For ECCITE-seq and the STING-seq v1 experiment, we ran one lane of a 10x Genomics 5’ kit (Chromium Single Cell Immune Profiling Solution v1.0, 1000014, 1000020, 1000151) with superloading and recovered 15,285 total cells (including multiple cells per droplet counts, or “multiplets”). Cell hashing was performed as described in a previously published protocol using four hashtag-derived oligonucleotides (HTOs) using hyperconjugation (24). Gene expression (cDNA), hashtags (HTOs) and guide RNA (Guide-derived oligos, GDOs) libraries were constructed by following 10x Genomics and ECCITE-seq protocols. We sequenced the cDNA, HTO and GDO libraries with two NextSeq 500 high-output runs (Illumina). For ECCITE-seq and the STING-seq v2 experiment, we ran four lanes of a 10x Genomics 5’ v2 kit (Chromium Next GEM Single Cell 5' Kit v2 1000265) with superloading. We recovered 82,339 total cells (including multiplets). Cell hashing was performed using eight HTOs followed by staining with a 188 antibody-tagged oligonucleotides (ADTs) panel (Biolegend) (Table S3B). cDNA, HTO, ADT, and GDO libraries were constructed by following 10x Genomics and ECCITE-seq protocols. We sequenced the cDNA, HTO, ADT and GDO libraries with one NovaSeq 6000 S1 run and two NovaSeq 6000 S2 runs (Illumina). For ECCITE-seq and the beeSTING-seq experiment, we ran three lanes of a 10x Genomics 5’ v2 kit with superloading and recovered 39,049 total cells, including multiplets. Cell hashing was performed using nine HTOs. cDNA, HTO, and GDO libraries were constructed by following 10x Genomics and ECCITE-seq protocols. We sequenced the cDNA, HTO, and GDO libraries with one NextSeq 500 mid-output run, one NovaSeq 6000 SP run, and one NovaSeq 6000 S1 run (Illumina).
Single cell data processing
UMI count matrices were generated for all single-cell sequencing libraries with 10x Cell Ranger v.6.0.0 (104). We generated outputs using the Gene Expression Output, Antibody Capture Output, and CRISPR Guide Capture Output functions. We then analyzed the UMI count matrices in R v.4.0.2 with Seurat v.4.0.0 (105) and tested for differential gene expression and protein levels within the SCEPTRE framework (26). The distributions of cDNA, GDO, HTO, and ADT UMIs were inspected manually for each lane of 10x sequenced. Custom thresholds were set to remove outliers for total cDNA count, unique genes detected, mitochondrial percentage, total gRNA count, unique gRNAs detected, total HTO count, unique HTOs detected, total ADT count, and unique ADTs detected. Lanes were merged for STING-seq v2 and beeSTING-seq only after quality control was completed. For STING-seq v1, we processed cDNA UMI count matrices and retained cells between the 15th to 99th percentiles for unique gene count, between the 20th and 99th percentiles for total cDNA UMI count, and between the 5th and 90th percentile for mitochondrial percentage. Next, we center-log-ratio (CLR) transformed the HTO UMI counts and demultiplexed cells by their transformed HTO counts to identify singlets. We used the HTODemux function implemented in Seurat v.4.0.0 to maximize the number of singlets detected. We used then processed the GDO UMI count matrix, keeping cells between the 1st and 99th percentiles for total GDO count and used 10x Cell Ranger predicted GDO thresholds per cell, but required at least 3 UMIs per GDO to assign a GDO to a given cell. This resulted a high confidence set of 7,667 single cells for the STING-seq v1 experiment. For STING-seq v2, we uniformly processed all four cDNA UMI count matrices and retained cells between the 1st and 99th percentile for unique gene count, between the 10th and 99th percentile for total cDNA UMI count, and between the 1st and 90th percentile for mitochondrial percentage. Next, we CLR transformed the HTO UMI counts and maximized singlet count using the HTODemux function. We then processed the GDO UMI count matrices, keeping cells between the 1st and 99th percentiles for total GDO count and again used the 10x Cell Ranger predicted GDO thresholds per cell, but required at least 3 UMIs per GDO. This resulted in a high confidence set of 38,916 cells for differential expression testing. We further applied quality control filters for ADTs, retaining cells with between the 1st and 99th percentiles for total ADT count. This resulted in 38,133 cells for differential protein testing. For beeSTING-seq, we uniformly processed all three cDNA UMI count matrices and retained cells between the 10th and 90th percentiles for unique gene count, between the 10th and 90th percentiles for total cDNA count, and between the 10th and 90th percentiles for mitochondrial percentage. We then CLR transformed the HTO counts and used the HTODemux function to maximize singlets and retained cells between the 1st and 99th percentiles for total GDO counts. 10x Cell Ranger set the majority of UMI thresholds to 1, therefore we generated a series of GDO UMI count matrices with thresholds from 1 to 5 to iteratively test optimal GDO thresholds for each gRNA. This resulted in a series of UMI count matrices for each GDO threshold. We had sets of 12,068 cells (GDO threshold = 1), 11,235 cells (GDO threshold = 2), 9,739 (GDO threshold = 3), 7,869 (GDO threshold = 4), and 5,896 (GDO threshold = 5) for differential expression testing.
Differential gene expression and protein level testing with SCEPTRE
We utilized the processed UMI count matrices for gene expression or protein levels and gRNA expression, along with accompanying single cell meta-data to use as covariates in model fitting (Table S3B). For STING-seq analyses, we defined for each cCRE targeted by 2 to 3 gRNAs a list of genes within 500 kb to be tested for differential expression in cis. For each gene per set of gRNAs, we extracted that gene’s UMI counts and labeled the cells with the given gRNAs. We then tested for differential outcomes within the SCEPTRE framework (26), adjusting for the following single cell covariates for expression tests: total gene expression UMIs, unique genes, total gRNA expression UMIs, unique gRNAs, percentage of mitochondrial genes, and 10x lane (for STING-seq v2 and beeSTING-seq). For protein tests, we adjusted for: total ADT count, total HTO count, total gRNA expression UMIs, unique gRNAs, and ADTs for four mouse-specific antibody controls to represent non-specific binding. We developed SCEPTRE as a statistical framework to analyze high MOI CRISPR screens in single cells with state-of-the-art calibration. First, SCEPTRE fits a negative binomial distribution to measure the effect of a single gRNA on a given gene via Z-score. Then, the distribution of gRNAs to cells is randomly sampled to build a gRNA-specific null distribution, recomputing a negative binomial Z-score. A skew-t distribution is fit to compare the test Z-score and the null distribution, and a two-sided p-value is derived, allowing for significance tests of increased or decreased gene expression or protein levels (26). To test for differential expression in trans, we defined for each set of gRNAs a list of all genes detected in at least 5% of cells and repeated the test above. Non-targeting gRNAs were tested against all genes used in the cis and trans settings discussed previously and randomly sampled to match the number of cis and trans tests displayed on QQ-plots. For each set of gRNAs with a cis-effect target gene we then performed marginal gRNA-gene pair testing, observing that the number of cells bearing gRNAs is the main driver behind statistical power (Fig. S6). To determine significance for multiple hypotheses (genes) tested in cis, SCEPTRE p-values were adjusted with the Benjamini-Hochberg procedure. For beeSTING-seq differential expression tests, we tested each gRNA against its known target gene from STING-seq analyses. We used for each gRNA the lowest GDO UMI threshold that resulted in at least 100 cells per gRNA and repeated this strategy for all non-targeting gRNAs against the same set of known cis-effect target genes. We then repeated differential expression testing grouping together GWAS-CRE targeting gRNAs if they shared concordant effects and UMI thresholds, evaluating their combined effects on target gene expression.
To report significant results for STING-seq analyses, we identified cis-target genes if they were significant at a 5% FDR (Benjamini-Hochberg adjusted SCEPTRE p < 0.05). We defined trans-target genes of each GWAS-CRE as those significant at a stricter 1% FDR. For beeSTING-seq analyses, we identified cis-target genes if they were significant at a 5% FDR. We examined all STING-seq genes significant at a 10% FDR and beeSTING-seq genes with SCEPTRE p < 0.05 to compare the trans-regulatory network effects from perturbing HHEX and IKZF1 GWAS-CREs with direct variant insertion.
Fine-mapped eQTL credible set integration
We examined 31 fine-mapped eQTL studies from the eQTL Catalogue (31) specific to blood traits. Specifically, we used eQTLs identified from human macrophages (106, 107), monocytes (37, 108, 109), neutrophils (37), lymphoblastoid cell lines (110-113), whole blood (110, 114, 115), induced pluripotent stem cells (116-118) , T-cells (37, 109, 112), B-cells (109), and natural killer cells (109). We then retained eQTL variants that were at least 1% plausibly causal and asked if our fine-mapped GWAS variants were in these data. eQTL summary statistics are available from the eQTL Catalog (www.ebi.ac.uk/eqtl).
K562 HiChIP for H3K27ac-interacting promoters
AQuA-HiChIP cell libraries were prepared as described previously (119). Briefly, NIH3T3 cells (mouse) and K562 cells were grown in the appropriate media. Cells were fixed in 1% formaldehyde for 10 minutes and quenched to a final concentration of 125 nM glycine. 2 million fixed mouse cells were mixed with 10 millions of fixed K562 cells. The cells were lysed in 0.5% SDS, quenched with 10% Triton X-100, and digested with MboI (NEB R0147M). The DNA overhangs were blunted, biotinylated (ThermoFisher 19524016), and ligated. Nuclei were spun down, resuspended in nuclear lysis buffer, and sonicated using a Covaris LE220 with the following conditions: Fill level 10, PIP 450, Duty factor 30, CPB 200. The sheared DNA was incubated with Dynabeads Protein A (ThermoFisher 10001D) for 2 hours at 4 °C. The tubes were placed on a magnet and the supernatant was kept. Immunoprecipitation was performed with a cross-species reactive H3K27ac antibody (Active Motif 39133). The samples were incubated with the antibody overnight at 4 °C. The samples were then washed, eluted, and treated with Proteinase K. The samples were purified using Zymo DNA Clean & Concentrator. Biotin capture was performed with Dynabeads M-280 Streptavidin (ThermoFisher 11205D), followed by library preparation. The amplified libraries were purified with Illumina Sample Purification Beads. The libraries were sequenced using paired-end reads with a NovaSeq 6000 S2 (Illumina) to generate 100 to 200 million read pairs per sample.
HiChIP paired end reads were mapped to the hg19 genome using HiC-Pro v.2.10.1 (120). Default settings were using to remove duplicate reads, identify valid interactions, and generate contact maps. Statistically significant contacts were identified using FitHiChIP v.9.1 (121) at a 5% FDR. H3K27ac ChIP-seq data (65) were used as a reference set of peaks in the FitHiChIP pipeline.
Trans-regulated network gene set enrichments
We used chromatin immunoprecipitation sequencing (ChIP-seq) datasets in K562 cells to identify GFI1B (64), NFE2, IKZF1, and RUNX1 (65) transcription factor binding sites. There we no publicly available HHEX K562 ChIP-seq datasets. We assigned the closest protein-coding gene to each ChIP-seq peak with bedtools v2.25.0 (90). For predicted miRNA targets we used the TargetScan database (66, 67). To test for enrichment of ChIP-seq peak or TargetScan genes in trans-regulatory gene sets, we fit logistic regression models adjusting for K562 expression (gene expression counts from scRNA-seq data) and computed odds ratios with 95% confidence intervals. To construct GWAS-identified sets of genes, we used all fine-mapped SNPs from the 29 UKBB GWASs and 15 BCX GWASs previously described (categorized by cell type) with a high Bayes factor for being plausibly causal (log10 BF ≥ 2) and that were at least 1% plausibly causal. GWAS gene enrichment was performed in a similar fashion as for ChIP-seq peaks.
Gene co-expression analyses and bone marrow single cell gene expression
To compute co-expression matrices for each trans-regulatory network, we used cDNA UMI count matrices with missing genes per cell imputed with the MAGIC algorithm (122). As a measure of co-expression, the biweight midcorrelation, a weighted correlation analysis, was calculated for each pair of genes (123). Genes were then clustered based on their co-expression patterns by hierarchical clustering. Transcription factor binding site, direct miRNA target, and GWAS gene enrichment was performed as described above. We used Human Cell Atlas single cell RNA-sequencing from 35 bone marrow donors (69) and identified 27 cell types as described previously (70). Single cell data were processed with Seurat v.4.0.0 to generate UMAP plots and heatmaps. To visualize entire trans-regulatory network clusters on a UMAP plot, we plotted the mean expression of all cluster genes within each cell.
STING-seq power estimations
We down-sampled 136 cis-effects of gRNAs targeting CREs on their target genes across two key conditions for experimental design: sequencing read depth per cell and the number of cells per gRNA. We sequenced all STING-seq libraries to a depth of approximately 55,000 to 65,000 reads per cell, therefore we repeated the entire STING-seq quality control and differential expression testing pipeline with 5,000, 15,000, 25,000, 35,000, 45,000 and 55,000. Sequencing reads were down-sampled to generate cDNA UMI count matrices with DropletUtils v.1.18.0 (124, 125) and repeated 10-times with different seed numbers. For each set of 10 randomly down-sampled UMI count matrices at each read depth, we repeated differential expression testing with SCEPTRE. We required at least 500 cells bearing each set of gRNAs, then at each set of 10 randomly down-sampled UMI count matrices at each read depth, we randomly down-sampled the number of cells bearing each set of gRNA from at least 500 cells to 50, and repeated this process 10-times at each stage. We averaged the SCEPTRE skew fit t-test p-values within replicates at each to compute precise measurements for each stage in the down-sampling procedures. We then divided all genes by their expression level and cis-effects by their log2 fold-changes into tertiles to examine at what number of cells and read depth could nominal significance (skew fit t-test p < 0.0001) be attained.
Supplementary Material
Acknowledgments
We thank the entire Sanjana and Lappalainen laboratories for support and advice. We also thank the New York Genome Center for flow cytometry resources, E. Hoelzli for support with digital PCR, T. Stuart and H. Wessels for advice on single-cell sequencing quality control, and S. Kasela for help with library design. This research has been conducted using the UK Biobank Resource under Application Number 47976.
Funding
J.A.M is supported by a Canadian Institutes of Health Research Banting Postdoctoral Fellowship and NIH/NHGRI (K99HG012792). Z.D is supported by an American Heart Association Postdoctoral Fellowship (20POST35220040). J.D is supported by a European Molecular Biology Organization Postdoctoral Fellowship (ALTF 345-2021). K.R. is supported by NIH/NIMH (R01MH123184). E.K. is supported by NSF (DMS-2113072) and the Wharton Data Science and Business Analytics Fund. T.L. is supported by NIH/NIMH (R01MH106842), NIH/NHGRI (UM1HG008901) and NIH/NIGMS (R01GM122924). N.E.S. is supported by NIH/NHGRI (DP2HG010099, including a supplemental award for STING-seq), NIH/NCI (R01CA279135, R01CA218668), NIH/NIAID (R01AI176601), the Simons Foundation for Autism Research, the MacMillan Center for the Study of the Non-Coding Cancer Genome, New York University and New York Genome Center funds.
Footnotes
Competing interests
N.E.S. is an advisor to Qiagen and is a co-founder and advisor of OverT Bio. T.L. is an advisor to Goldfinch Bio and GSK and, with equity, Variant Bio. All other authors declare that they have no competing interests.
Data and materials availability
Plasmids (lentiCRISPRi(v1)-Blast, lentiCRISPRi(v2)-Blast, lentiGuideFE-Puro, and lentiBE3-SpRY-Blast) have been deposited with Addgene (plasmid nos. 170067, 170068, 170069, 199303). Raw and processed single cell sequencing files are available from Gene Expression Omnibus (GEO) accession number GSE171452.
Code availability
SCEPTRE and related tutorials are available on Github (www.github.com/Katsevich-Lab/sceptre).
References and notes
- 1.Wright JB, Sanjana NE, CRISPR Screens to Discover Functional Noncoding Elements. Trends Genet. TIG 32, 526–529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lappalainen T, MacArthur DG, From variant to function in human disease genetics. Science (2021), doi: 10.1126/science.abi8207. [DOI] [PubMed] [Google Scholar]
- 3.Ulirsch JC, Lareau CA, Bao EL, Ludwig LS, Guo MH, Benner C, Satpathy AT, Kartha VK, Salem RM, Hirschhorn JN, Finucane HK, Aryee MJ, Buenrostro JD, Sankaran VG, Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat. Genet 51, 683–693 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, Vulpescu NA, Forgetta V, Kleinman A, Mohanty ST, Sergio CM, Quinn J, Nguyen-Yamamoto L, Luco A-L, Vijay J, Simon M-M, Pramatarova A, Medina-Gomez C, Trajanoska K, Ghirardello EJ, Butterfield NC, Curry KF, Leitch VD, Sparkes PC, Adoum A-T, Mannan NS, Komla-Ebri DSK, Pollard AS, Dewhurst HF, Hassall TAD, Beltejar M-JG, Adams DJ, Vaillancourt SM, Kaptoge S, Baldock P, Cooper C, Reeve J, Ntzani EE, Evangelou E, Ohlsson C, Karasik D, Rivadeneira F, Kiel DP, Tobias JH, Gregson CL, Harvey NC, Grundberg E, Goltzman D, Adams DJ, Lelliott CJ, Hinds DA, Ackert-Bicknell CL, Hsu Y-H, Maurano MT, Croucher PI, Williams GR, Bassett JHD, Evans DM, Richards JB, An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet 51, 258–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasser J, Bergman DT, Fulco CP, Guckelberger P, Doughty BR, Patwardhan TA, Jones TR, Nguyen TH, Ulirsch JC, Lekschas F, Mualim K, Natri HM, Weeks EM, Munson G, Kane M, Kang HY, Cui A, Ray JP, Eisenhaure TM, Collins RL, Dey K, Pfister H, Price AL, Epstein CB, Kundaje A, Xavier RJ, Daly MJ, Huang H, Finucane HK, Hacohen N, Lander ES, Engreitz JM, Genome-wide enhancer maps link risk variants to disease genes. Nature. 593, 238–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Q, Crowley CA, Huang L, Wen J, Chen J, Bao EL, Auer PL, Lettre G, Reiner AP, Sankaran VG, Raffield LM, Li Y, From GWAS variant to function: A study of ~148,000 variants for blood cell traits. Hum. Genet. Genomics Adv 3, 100063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, Trapnell C, Ahituv N, Shendure J, A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell. 176, 377–390.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie S, Armendariz D, Zhou P, Duan J, Hon GC, Global Analysis of Enhancer Targets Reveals Convergent Enhancer-Driven Regulatory Modules. Cell Rep. 29, 2570–2578.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wünnemann F, Tadjo TF, Beaudoin M, Lalonde S, Lo KS, Kleinstiver BP, Lettre G, Multimodal CRISPR perturbations of GWAS loci associated with coronary artery disease in vascular endothelial cells. PLOS Genet. 19, e1010680 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, Freson K, Garner SF, Grassi L, Guerrero J, Haimel M, Janssen-Megens EM, Kaan A, Kamat M, Kim B, Mandoli A, Marchini J, Martens JHA, Meacham S, Megy K, O’Connell J, Petersen R, Sharifi N, Sheard SM, Staley JR, Tuna S, van der Ent M, Walter K, Wang S-Y, Wheeler E, Wilder SP, Iotchkova V, Moore C, Sambrook J, Stunnenberg HG, Di Angelantonio E, Kaptoge S, Kuijpers TW, Carrillo-de-Santa-Pau E, Juan D, Rico D, Valencia A, Chen L, Ge B, Vasquez L, Kwan T, Garrido-Martín D, Watt S, Yang Y, Guigo R, Beck S, Paul DS, Pastinen T, Bujold D, Bourque G, Frontini M, Danesh J, Roberts DJ, Ouwehand WH, Butterworth AS, Soranzo N, The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 167, 1415–1429.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M-H, Raffield LM, Mousas A, Sakaue S, Huffman JE, Moscati A, Trivedi B, Jiang T, Akbari P, Vuckovic D, Bao EL, Zhong X, Manansala R, Laplante V, Chen M, Lo KS, Qian H, Lareau CA, Beaudoin M, Hunt KA, Akiyama M, Bartz TM, Ben-Shlomo Y, Beswick A, Bork-Jensen J, Bottinger EP, Brody JA, van Rooij FJA, Chitrala K, Cho K, Choquet H, Correa A, Danesh J, Di Angelantonio E, Dimou N, Ding J, Elliott P, Esko T, Evans MK, Floyd JS, Broer L, Grarup N, Guo MH, Greinacher A, Haessler J, Hansen T, Howson JMM, Huang QQ, Huang W, Jorgenson E, Kacprowski T, Kähönen M, Kamatani Y, Kanai M, Karthikeyan S, Koskeridis F, Lange LA, Lehtimäki T, Lerch MM, Linneberg A, Liu Y, Lyytikäinen L-P, Manichaikul A, Martin HC, Matsuda K, Mohlke KL, Mononen N, Murakami Y, Nadkarni GN, Nauck M, Nikus K, Ouwehand WH, Pankratz N, Pedersen O, Preuss M, Psaty BM, Raitakari OT, Roberts DJ, Rich SS, Rodriguez BAT, Rosen JD, Rotter JI, Schubert P, Spracklen CN, Surendran P, Tang H, Tardif J-C, Trembath RC, Ghanbari M, Völker U, Völzke H, Watkins NA, Zonderman AB, Wilson PWF, Li Y, Butterworth AS, Gauchat J-F, Chiang CWK, Li B, Loos RJF, Astle WJ, Evangelou E, van Heel DA, Sankaran VG, Okada Y, Soranzo N, Johnson AD, Reiner AP, Auer PL, Lettre G, Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell. 182, 1198–1213.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuckovic D, Bao EL, Akbari P, Lareau CA, Mousas A, Jiang T, Chen M-H, Raffield LM, Tardaguila M, Huffman JE, Ritchie SC, Megy K, Ponstingl H, Penkett CJ, Albers PK, Wigdor EM, Sakaue S, Moscati A, Manansala R, Lo KS, Qian H, Akiyama M, Bartz TM, Ben-Shlomo Y, Beswick A, Bork-Jensen J, Bottinger EP, Brody JA, van Rooij FJA, Chitrala KN, Wilson PWF, Choquet H, Danesh J, Di Angelantonio E, Dimou N, Ding J, Elliott P, Esko T, Evans MK, Felix SB, Floyd JS, Broer L, Grarup N, Guo MH, Guo Q, Greinacher A, Haessler J, Hansen T, Howson JMM, Huang W, Jorgenson E, Kacprowski T, Kähönen M, Kamatani Y, Kanai M, Karthikeyan S, Koskeridis F, Lange LA, Lehtimaki T, Linneberg A, Liu Y, Lyytikäinen L-P, Manichaikul A, Matsuda K, Mohlke KL, Mononen N, Murakami Y, Nadkarni GN, Nikus K, Pankratz N, Pedersen O, Preuss M, Psaty BM, Raitakari OT, Rich SS, Rodriguez BAT, Rosen JD, Rotter JI, Schubert P, Spracklen CN, Surendran P, Tang H, Tardif J-C, Ghanbari M, Völker U, Völzke H, Watkins NA, Weiss S, Cai N, Kundu K, Watt SB, Walter K, Zonderman AB, Cho K, Li Y, Loos RJF, Knight JC, Georges M, Stegle O, Evangelou E, Okada Y, Roberts DJ, Inouye M, Johnson AD, Auer PL, Astle WJ, Reiner AP, Butterworth AS, Ouwehand WH, Lettre G, Sankaran VG, Soranzo N, The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell. 182, 1214–1231.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Lee SH, Goddard ME, Visscher PM, GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM, Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sankaran VG, Ludwig LS, Sicinska E, Xu J, Bauer DE, Eng JC, Patterson HC, Metcalf RA, Natkunam Y, Orkin SH, Sicinski P, Lander ES, Lodish HF, Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 26, 2075–2087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulirsch JC, Lacy JN, An X, Mohandas N, Mikkelsen TS, Sankaran VG, Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation. PLOS Genet. 10, e1004890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, Melnikov A, McDonel P, Do R, Mikkelsen TS, Sankaran VG, Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell. 165, 1530–1545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen J, Lagler TM, Sun Q, Yang Y, Chen J, Harigaya Y, Sankaran VG, Hu M, Reiner AP, Raffield LM, Li Y, Super interactive promoters provide insight into cell type-specific regulatory networks in blood lineage cell types. PLOS Genet. 18, e1009984 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo NC, Chavez A, Lance-Byrne A, Chan Y, Menn D, Milanova D, Kuo C-C, Guo X, Sharma S, Tung A, Cecchi RJ, Tuttle M, Pradhan S, Lim ET, Davidsohn N, Ebrahimkhani MR, Collins JJ, Lewis NE, Kiani S, Church GM, An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 15, 611–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS, Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F, DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Shendure J, FlashFry: a fast and flexible tool for large-scale CRISPR target design. BMC Biol. 16, 74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P, Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mimitou EP, Cheng A, Montalbano A, Hao S, Stoeckius M, Legut M, Roush T, Herrera A, Papalexi E, Ouyang Z, Satija R, Sanjana NE, Koralov SB, Smibert P, Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barry T, Wang X, Morris JA, Roeder K, Katsevich E, SCEPTRE improves calibration and sensitivity in single-cell CRISPR screen analysis. Genome Biol. 22, 344 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A, An atlas of active enhancers across human cell types and tissues. Nature. 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fauman EB, Hyde C, An optimal variant to gene distance window derived from an empirical definition of cis and trans protein QTLs. BMC Bioinformatics. 23, 1–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao D, Tycko J, Oh JW, Bounds LR, Gosai SJ, Lataniotis L, Mackay-Smith A, Doughty BR, Gabdank I, Schmidt H, Youngworth I, Andreeva K, Ren X, Barrera A, Luo Y, Siklenka K, Yardımcı GG, Consortium TE, Tewhey R, Kundaje A, Greenleaf WJ, Sabeti PC, Leslie C, Pritykin Y, Moore JD, Beer MA, Gersbach CA, Reddy TE, Shen Y, Engreitz JM, Bassik MC, Reilly SK, Multi-center integrated analysis of non-coding CRISPR screens (2022), p. 2022.12.21.520137, , doi: 10.1101/2022.12.21.520137. [DOI] [Google Scholar]
- 30.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, Mohammadi P, Park Y, Parsana P, Segrè AV, Strober BJ, Zappala Z, Cummings BB, Gelfand ET, Hadley K, Huang KH, Lek M, Li X, Nedzel JL, Nguyen DY, Noble MS, Sullivan TJ, Tukiainen T, MacArthur DG, Getz G, Addington A, Guan P, Koester S, Little AR, Lockhart NC, Moore HM, Rao A, Struewing JP, Volpi S, Brigham LE, Hasz R, Hunter M, Johns C, Johnson M, Kopen G, Leinweber WF, Lonsdale JT, McDonald A, Mestichelli B, Myer K, Roe B, Salvatore M, Shad S, Thomas JA, Walters G, Washington M, Wheeler J, Bridge J, Foster BA, Gillard BM, Karasik E, Kumar R, Miklos M, Moser MT, Jewell SD, Montroy RG, Rohrer DC, Valley D, Mash DC, Davis DA, Sobin L, Barcus ME, Branton PA, Abell NS, Balliu B, Delaneau O, Frésard L, Gamazon ER, Garrido-Martín D, Gewirtz ADH, Gliner G, Gloudemans MJ, Han B, He AZ, Hormozdiari F, Li X, Liu B, Kang EY, McDowell IC, Ongen H, Palowitch JJ, Peterson CB, Quon G, Ripke S, Saha A, Shabalin AA, Shimko TC, Sul JH, Teran NA, Tsang EK, Zhang H, Zhou Y-H, Bustamante CD, Cox NJ, Guigó R, Kellis M, McCarthy MI, Conrad DF, Eskin E, Li G, Nobel AB, Sabatti C, Stranger BE, Wen X, Wright FA, Ardlie KG, Dermitzakis ET, Lappalainen T, Aguet F, Ardlie KG, Cummings BB, Gelfand ET, Getz G, Hadley K, Handsaker RE, Huang KH, Kashin S, Karczewski KJ, Lek M, Li X, MacArthur DG, Nedzel JL, Nguyen DT, Noble MS, Segrè AV, Trowbridge CA, Tukiainen T, Abell NS, Balliu B, Barshir R, Basha O, Battle A, Bogu GK, Brown A, Brown CD, Castel SE, Chen LS, Chiang C, Conrad DF, Cox NJ, Damani FN, Davis JR, Delaneau O, Dermitzakis ET, Engelhardt BE, Eskin E, Ferreira PG, Frésard L, Gamazon ER, Garrido-Martín D, Gewirtz ADH, Gliner G, Gloudemans MJ, Guigo R, Hall IM, Han B, He Y, Hormozdiari F, Howald C, Kyung Im H, Jo B, Yong Kang E, Kim Y, Kim-Hellmuth S, Lappalainen T, Li G, Li X, Liu B, Mangul S, McCarthy MI, McDowell IC, Mohammadi P, Monlong J, Montgomery SB, Muñoz-Aguirre M, Ndungu AW, Nicolae DL, Nobel AB, Oliva M, Ongen H, Palowitch JJ, Panousis N, Papasaikas P, Park Y, Parsana P, Payne AJ, Peterson CB, Quan J, Reverter F, Sabatti C, Saha A, Sammeth M, Scott AJ, Shabalin AA, Sodaei R, Stephens M, Stranger BE, Strober BJ, Sul JH, Tsang EK, Urbut S, van de Bunt M, Wang G, Wen X, Wright FA, Xi HS, Yeger-Lotem E, Zappala Z, Zaugg JB, Zhou Y-H, Akey JM, Bates D, Chan J, Chen LS, Claussnitzer M, Demanelis K, Diegel M, Doherty JA, Feinberg AP, Fernando MS, Halow J, Hansen KD, Haugen E, Hickey PF, Hou L, Jasmine F, Jian R, Jiang L, Johnson A, Kaul R, Kellis M, Kibriya MG, Lee K, Billy Li J, Li Q, Li X, Lin J, Lin S, Linder S, Linke C, Liu Y, Maurano MT, Molinie B, Montgomery SB, Nelson J, Neri FJ, Oliva M, Park Y, Pierce BL, Rinaldi NJ, Rizzardi LF, Sandstrom R, Skol A, Smith KS, Snyder MP, Stamatoyannopoulos J, Stranger BE, Tang H, Tsang EK, Wang L, Wang M, Van Wittenberghe N, Wu F, Zhang R, Nierras CR, Branton PA, Carithers LJ, Guan P, Moore HM, Rao A, Vaught JB, Gould SE, Lockart NC, Martin C, Struewing JP, Volpi S, Addington AM, Koester SE, Little AR, GTEx Consortium, Lead analysts:, D. A. & C. C. (LDACC): Laboratory, NIH program management:, Biospecimen collection:, Pathology:, eQTL manuscript working group:, D. A. & C. C. (LDACC)—Analysis W. G. Laboratory, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI, Genetic effects on gene expression across human tissues. Nature. 550, 204–213 (2017).29022597 [Google Scholar]
- 31.Kerimov N, Hayhurst JD, Peikova K, Manning JR, Walter P, Kolberg L, Samoviča M, Sakthivel MP, Kuzmin I, Trevanion SJ, Burdett T, Jupp S, Parkinson H, Papatheodorou I, Yates AD, Zerbino DR, Alasoo K, A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet 53, 1290–1299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowland B, Venkatesh S, Tardaguila M, Wen J, Rosen JD, Tapia AL, Sun Q, Graff M, Vuckovic D, Lettre G, Sankaran VG, Voloudakis G, Roussos P, Huffman JE, Reiner AP, Soranzo N, Raffield LM, Li Y, Transcriptome-wide association study in UK Biobank Europeans identifies associations with blood cell traits. Hum. Mol. Genet, ddac011 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Veturi Y, Verma A, Bradford Y, Daar ES, Gulick RM, Riddler SA, Robbins GK, Lennox JL, Haas DW, Ritchie MD, Tissue specificity-aware TWAS (TSA-TWAS) framework identifies novel associations with metabolic, immunologic, and virologic traits in HIV-positive adults. PLOS Genet. 17, e1009464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurano MT, Haugen E, Sandstrom R, Vierstra J, Shafer A, Kaul R, Stamatoyannopoulos JA, Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet 47, 1393–1401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abramov S, Boytsov A, Bykova D, Penzar DD, Yevshin I, Kolmykov SK, Fridman MV, Favorov AV, Vorontsov IE, Baulin E, Kolpakov F, Makeev VJ, Kulakovskiy IV, Landscape of allele-specific transcription factor binding in the human genome. Nat. Commun 12, 2751 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blatt K, Herrmann H, Hoermann G, Willmann M, Cerny-Reiterer S, Sadovnik I, Herndlhofer S, Streubel B, Rabitsch W, Sperr WR, Mayerhofer M, Rülicke T, Valent P, Identification of campath-1 (CD52) as novel drug target in neoplastic stem cells in 5q-patients with MDS and AML. Clin. Cancer Res. Off J. Am. Assoc. Cancer Res 20, 3589–3602 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Ge B, Casale FP, Vasquez L, Kwan T, Garrido-Martín D, Watt S, Yan Y, Kundu K, Ecker S, Datta A, Richardson D, Burden F, Mead D, Mann AL, Fernandez JM, Rowlston S, Wilder SP, Farrow S, Shao X, Lambourne JJ, Redensek A, Albers CA, Amstislavskiy V, Ashford S, Berentsen K, Bomba L, Bourque G, Bujold D, Busche S, Caron M, Chen S-H, Cheung W, Delaneau O, Dermitzakis ET, Elding H, Colgiu I, Bagger FO, Flicek P, Habibi E, Iotchkova V, Janssen-Megens E, Kim B, Lehrach H, Lowy E, Mandoli A, Matarese F, Maurano MT, Morris JA, Pancaldi V, Pourfarzad F, Rehnstrom K, Rendon A, Risch T, Sharifi N, Simon M-M, Sultan M, Valencia A, Walter K, Wang S-Y, Frontini M, Antonarakis SE, Clarke L, Yaspo M-L, Beck S, Guigo R, Rico D, Martens JHA, Ouwehand WH, Kuijpers TW, Paul DS, Stunnenberg HG, Stegle O, Downes K, Pastinen T, Soranzo N, Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ, Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C-Y, Chen T-T, Feng Y-CA, Longchamps RJ, Lin S-C, Wang S-H, Hsu Y-H, Yang H-I, Kuo P-H, Daly MJ, Chen WJ, Huang H, Ge T, Lin Y-F, Analysis across Taiwan Biobank, Biobank Japan and UK Biobank identifies hundreds of novel loci for 36 quantitative traits (2021), p. 2021.04.12.21255236, , doi: 10.1101/2021.04.12.21255236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W, Kanai M, Wu K-HH, Rasheed H, Tsuo K, Hirbo JB, Wang Y, Bhattacharya A, Zhao H, Namba S, Surakka I, Wolford BN, Lo Faro V, Lopera-Maya EA, Läll K, Favé M-J, Partanen JJ, Chapman SB, Karjalainen J, Kurki M, Maasha M, Brumpton BM, Chavan S, Chen T-T, Daya M, Ding Y, Feng Y-CA, Guare LA, Gignoux CR, Graham SE, Hornsby WE, Ingold N, Ismail SI, Johnson R, Laisk T, Lin K, Lv J, Millwood IY, Moreno-Grau S, Nam K, Palta P, Pandit A, Preuss MH, Saad C, Setia-Verma S, Thorsteinsdottir U, Uzunovic J, Verma A, Zawistowski M, Zhong X, Afifi N, Al-Dabhani KM, Al Thani A, Bradford Y, Campbell A, Crooks K, de Bock GH, Damrauer SM, Douville NJ, Finer S, Fritsche LG, Fthenou E, Gonzalez-Arroyo G, Griffiths CJ, Guo Y, Hunt KA, Ioannidis A, Jansonius NM, Konuma T, Lee MTM, Lopez-Pineda A, Matsuda Y, Marioni RE, Moatamed B, Nava-Aguilar MA, Numakura K, Patil S, Rafaels N, Richmond A, Rojas-Muñoz A, Shortt JA, Straub P, Tao R, Vanderwerff B, Vernekar M, Veturi Y, Barnes KC, Boezen M, Chen Z, Chen C-Y, Cho J, Smith GD, Finucane HK, Franke L, Gamazon ER, Ganna A, Gaunt TR, Ge T, Huang H, Huffman J, Katsanis N, Koskela JT, Lajonchere C, Law MH, Li L, Lindgren CM, Loos RJF, MacGregor S, Matsuda K, Olsen CM, Porteous DJ, Shavit JA, Snieder H, Takano T, Trembath RC, Vonk JM, Whiteman DC, Wicks SJ, Wijmenga C, Wright J, Zheng J, Zhou X, Awadalla P, Boehnke M, Bustamante CD, Cox NJ, Fatumo S, Geschwind DH, Hayward C, Hveem K, Kenny EE, Lee S, Lin Y-F, Mbarek H, Mägi R, Martín HC, Medland SE, Okada Y, Palotie AV, Pasaniuc B, Rader DJ, Ritchie MD, Sanna S, Smoller JW, Stefansson K, van Heel DA, Walters RG, Zöllner S, Martin AR, Willer CJ, Daly MJ, Neale BM, Global Biobank Meta-analysis Initiative: Powering genetic discovery across human disease. Cell Genomics. 2, 100192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shull MM, Lingrel JB, Multiple genes encode the human Na+,K+-ATPase catalytic subunit. Proc. Natl. Acad. Sci. U. S. A 84, 4039–4043 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glorioso N, Herrera VLM, Bagamasbad P, Filigheddu F, Troffa C, Argiolas G, Bulla E, Decano JL, Ruiz-Opazo N, Association of ATP1A1 and Dear Single-Nucleotide Polymorphism Haplotypes With Essential Hypertension. Circ. Res 100, 1522–1529 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Araos P, Figueroa S, Amador CA, The Role of Neutrophils in Hypertension. Int. J. Mol. Sci 21, 8536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H, The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leduc V, Jasmin-Bélanger S, Poirier J, APOE and cholesterol homeostasis in Alzheimer’s disease. Trends Mol. Med 16, 469–477 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Suchindran S, Rivedal D, Guyton JR, Milledge T, Gao X, Benjamin A, Rowell J, Ginsburg GS, McCarthy JJ, Genome-Wide Association Study of Lp-PLA2 Activity and Mass in the Framingham Heart Study. PLOS Genet. 6, e1000928 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Replogle JM, Saunders RA, Pogson AN, Hussmann JA, Lenail A, Guna A, Mascibroda L, Wagner EJ, Adelman K, Lithwick-Yanai G, Iremadze N, Oberstrass F, Lipson D, Bonnar JL, Jost M, Norman TM, Weissman JS, Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell (2022), doi: 10.1016/j.cell.2022.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuior EV, Gafencu AV, Apolipoprotein C1: Its Pleiotropic Effects in Lipid Metabolism and Beyond. Int. J. Mol. Sci 20 (2019), doi: 10.3390/ijms20235939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, Flicek P, Gabriel SB, Gibbs RA, Green ED, Hurles ME, Knoppers BM, Korbel JO, Lander ES, Lee C, Lehrach H, Mardis ER, Marth GT, McVean GA, Nickerson DA, Schmidt JP, Sherry ST, Wang J, Wilson RK, Gibbs RA, Boerwinkle E, Doddapaneni H, Han Y, Korchina V, Kovar C, Lee S, Muzny D, Reid JG, Zhu Y, Wang J, Chang Y, Feng Q, Fang X, Guo X, Jian M, Jiang H, Jin X, Lan T, Li G, Li J, Li Y, Liu S, Liu X, Lu Y, Ma X, Tang M, Wang B, Wang G, Wu H, Wu R, Xu X, Yin Y, Zhang D, Zhang W, Zhao J, Zhao M, Zheng X, Lander ES, Altshuler DM, Gabriel SB, Gupta N, Gharani N, Toji LH, Gerry NP, Resch AM, Flicek P, Barker J, Clarke L, Gil L, Hunt SE, Kelman G, Kulesha E, Leinonen R, McLaren WM, Radhakrishnan R, Roa A, Smirnov D, Smith RE, Streeter I, Thormann A, Toneva I, Vaughan B, Zheng-Bradley X, Bentley DR, Grocock R, Humphray S, James T, Kingsbury Z, Lehrach H, Sudbrak R, Albrecht MW, Amstislavskiy VS, Borodina TA, Lienhard M, Mertes F, Sultan M, Timmermann B, Yaspo M-L, Mardis ER, Wilson RK, Fulton L, Fulton R, Sherry ST, Ananiev V, Belaia Z, Beloslyudtsev D, Bouk N, Chen C, Church D, Cohen R, Cook C, Garner J, Hefferon T, Kimelman M, Liu C, Lopez J, Meric P, O’Sullivan C, Ostapchuk Y, Phan L, Ponomarov S, Schneider V, Shekhtman E, Sirotkin K, Slotta D, Zhang H, McVean GA, Durbin RM, Balasubramaniam S, Burton J, Danecek P, Keane TM, Kolb-Kokocinski A, McCarthy S, Stalker J, Quail M, Schmidt JP, Davies CJ, Gollub J, Webster T, Wong B, Zhan Y, Auton A, Campbell CL, Kong Y, Marcketta A, Gibbs RA, Yu F, Antunes L, Bainbridge M, Muzny D, Sabo A, Huang Z, Wang J, Coin LJM, Fang L, Guo X, Jin X, Li G, Li Q, Li Y, Li Z, Lin H, Liu B, Luo R, Shao H, Xie Y, Ye C, Yu C, Zhang F, Zheng H, Zhu H, Alkan C, Dal E, Kahveci F, Marth GT, Garrison EP, Kural D, Lee W-P, Fung Leong W, Stromberg M, Ward AN, Wu J, Zhang M, Daly MJ, DePristo MA, Handsaker RE, Altshuler DM, Banks E, Bhatia G, del Angel G, Gabriel SB, Genovese G, Gupta N, Li H, Kashin S, Lander ES, McCarroll SA, Nemesh JC, Poplin RE, Yoon SC, Lihm J, Makarov V, Clark AG, Gottipati S, Keinan A, Rodriguez-Flores JL, Korbel JO, Rausch T, Fritz MH, Stütz AM, Flicek P, Beal K, Clarke L, Datta A, Herrero J, McLaren WM, Ritchie GRS, Smith RE, Zerbino D, Zheng-Bradley X, Sabeti PC, Shlyakhter I, Schaffner SF, Vitti J, Cooper DN, Ball EV, Stenson PD, Bentley DR, Barnes B, Bauer M, Keira Cheetham R, Cox A, Eberle M, Humphray S, Kahn S, Murray L, Peden J, Shaw R, Kenny EE, Batzer MA, Konkel MK, Walker JA, MacArthur DG, Lek M, Sudbrak R, Amstislavskiy VS, Herwig R, Mardis ER, Ding L, Koboldt DC, Larson D, Ye K, Gravel S, The 1000 Genomes Project Consortium, Corresponding authors, Steering committee, Production group, Baylor College of Medicine, BGI-Shenzhen, Broad Institute of MIT and Harvard, Coriell Institute for Medical Research, E. B. I. European Molecular Biology Laboratory, Illumina, Max Planck Institute for Molecular Genetics, McDonnell Genome Institute at Washington University, US National Institutes of Health, University of Oxford, Wellcome Trust Sanger Institute, Analysis group, Affymetrix, Albert Einstein College of Medicine, Bilkent University, Boston College, Cold Spring Harbor Laboratory, Cornell University, European Molecular Biology Laboratory, Harvard University, Human Gene Mutation Database, Icahn School of Medicine at Mount Sinai, Louisiana State University, Massachusetts General Hospital, McGill University, N. National Eye Institute, A global reference for human genetic variation. Nature. 526, 68–74 (2015).26432245 [Google Scholar]
- 50.Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, Simon A, Han T, Goswami S, Montgomery E, Thibado J, Kastenhuber ER, Sánchez-Rivera FJ, Shi J, Vakoc CR, Lowe SW, Tschaharganeh DF, Dow LE, Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol 36, 888–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP, Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science (2020), doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanna RE, Hegde M, Fagre CR, DeWeirdt PC, Sangree AK, Szegletes Z, Griffith A, Feeley MN, Sanson KR, Baidi Y, Koblan LW, Liu DR, Neal JT, Doench JG, Massively parallel assessment of human variants with base editor screens. Cell. 184, 1064–1080.e20 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Cuella-Martin R, Hayward SB, Fan X, Chen X, Huang J-W, Taglialatela A, Leuzzi G, Zhao J, Rabadan R, Lu C, Shen Y, Ciccia A, Functional interrogation of DNA damage response variants with base editing screens. Cell. 184, 1081–1097.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B, Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet 100, 473–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orkin SH, Zon LI, Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell. 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasiorek JJ, Blank V, Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells. Cell. Mol. Life Sci 72, 2323–2335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis KL, Ikaros: master of hematopoiesis, agent of leukemia. Ther. Adv. Hematol 2, 359–368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan RJ, Hromas R, Yoder MC, The role of Hex in hemangioblast and hematopoietic development. Methods Mol. Biol. Clifton NJ 330, 123–133 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Ichikawa M, Yoshimi A, Nakagawa M, Nishimoto N, Watanabe-Okochi N, Kurokawa M, A role for RUNX1 in hematopoiesis and myeloid leukemia. Int. J. Hematol 97, 726–734 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Chapnik E, Rivkin N, Mildner A, Beck G, Pasvolsky R, Metzl-Raz E, Birger Y, Amir G, Tirosh I, Porat Z, Israel LL, Lellouche E, Michaeli S, Lellouche J-PM, Izraeli S, Jung S, Hornstein E, miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. eLife. 3, e01964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang T, Wu F, Yu D, miR-144/451 in hematopoiesis and beyond. ExRNA. 1, 16 (2019). [Google Scholar]
- 62.Ghanbari M, Munshi ST, Ma B, Lendemeijer B, Bansal S, Adams HH, Wang W, Goth K, Slump DE, van den Hout MCGN, van IJcken WFJ, Bellusci S, Pan Q, Erkeland SJ, de Vrij FMS, Kushner SA, Ikram MA, A functional variant in the miR-142 promoter modulating its expression and conferring risk of Alzheimer disease. Hum. Mutat 40, 2131–2145 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, Naj AC, Campos-Martin R, Grenier-Boley B, Andrade V, Holmans PA, Boland A, Damotte V, van der Lee SJ, Costa MR, Kuulasmaa T, Yang Q, de Rojas I, Bis JC, Yaqub A, Prokic I, Chapuis J, Ahmad S, Giedraitis V, Aarsland D, Garcia-Gonzalez P, Abdelnour C, Alarcón-Martín E, Alcolea D, Alegret M, Alvarez I, Álvarez V, Armstrong NJ, Tsolaki A, Antúnez C, Appollonio I, Arcaro M, Archetti S, Pastor AA, Arosio B, Athanasiu L, Bailly H, Banaj N, Baquero M, Barral S, Beiser A, Pastor AB, Below JE, Benchek P, Benussi L, Berr C, Besse C, Bessi V, Binetti G, Bizarro A, Blesa R, Boada M, Boerwinkle E, Borroni B, Boschi S, Bossù P, Bråthen G, Bressler J, Bresner C, Brodaty H, Brookes KJ,Brusco LI, Buiza-Rueda D, Bûrger K, Burholt V, Bush WS, Calero M, Cantwell LB, Chene G, Chung J, Cuccaro ML, Carracedo Á, Cecchetti R, Cervera-Carles L, Charbonnier C, Chen H-H, Chillotti C, Ciccone S, Claassen JAHR, Clark C, Conti E, Corma-Gómez A, Costantini E, Custodero C, Daian D, Dalmasso MC, Daniele A, Dardiotis E, Dartigues J-F, de Deyn PP, de Paiva Lopes K, de Witte LD, Debette S, Deckert J, Del Ser T, Denning N, DeStefano A, Dichgans M, Diehl-Schmid J, Diez-Fairen M, Rossi PD, Djurovic S, Duron E, Düzel E, Dufouil C, Eiriksdottir G, Engelborghs S, Escott-Price V, Espinosa A, Ewers M, Faber KM, Fabrizio T, Nielsen SF, Fardo DWL, Marotti C Fenoglio, Fernández-Fuertes M, Ferrari R, Ferreira CB, Ferri E, Fin B, Fischer P, Fladby T, Fliesßach K, Fongang B, Fornage M, Fortea J, Foroud TM, Fostinelli S, Fox NC, Franco-Macías E, Bullido MJ, Frank-García A, Froelich L, Fulton-Howard B, Galimberti D, García-Alberca JM, García-González P, Garcia-Madrona S, Garcia-Ribas G, Ghidoni R, Giegling I, Giorgio G, Goate AM, Goldhardt O, Gomez-Fonseca D, González-Pérez A, Graff C, Grande G, Green E, Grimmer T, Grünblatt E, Grunin M, Gudnason V, Guetta-Baranes T, Haapasalo A, Hadjigeorgiou G, Haines JL, Hamilton-Nelson KL, Hampel H, Hanon O, Hardy J, Hartmann AM, Hausner L, Harwood J, Heilmann-Heimbach S, Helisalmi S, Heneka MT, Hernández I, Herrmann MJ, Hoffmann P, Holmes C, Holstege H, Vilas RH, Hulsman M, Humphrey J, Biessels GJ, Jian X, Johansson C, Jun GR, Kastumata Y, Kauwe J, Kehoe PG, Kilander L, Ståhlbom AK, Kivipelto M, Koivisto A, Kornhuber J, Kosmidis MH, Kukull WA, Kuksa PP, Kunkle BW, Kuzma AB, Lage C, Laukka EJ, Launer L, Lauria A, Lee C-Y, Lehtisalo J, Lerch O, Lleó A, Longstreth W, Lopez O, de Munain AL, Love S, Löwemark M, Luckcuck L, Lunetta KL, Ma Y, Macías J, MacLeod CA, Maier W, Mangialasche F, Spallazzi M, Marquié M, Marshall R, Martin ER, Montes AM, Rodríguez CM, Masullo C, Mayeux R, Mead S, Mecocci P, Medina M, Meggy A, Mehrabian S, Mendoza S, Menéndez-González M, Mir P, Moebus S, Mol M, Molina-Porcel L, Montrreal L, Morelli L, Moreno F, Morgan K, Mosley T, Nöthen MM, Muchnik C, Mukherjee S, Nacmias B, Ngandu T, Nicolas G, Nordestgaard BG, Olaso R, Orellana A, Orsini M, Ortega G, Padovani A, Paolo C, Papenberg G, Parnetti L, Pasquier F, Pastor P, Peloso G, Pérez-Cordón A, Pérez-Tur J, Pericard P, Peters O, Pijnenburg YAL, Pineda JA, Piñol-Ripoll G, Pisanu C, Polak T, Popp J, Posthuma D, Priller J, Puerta R, Quenez O, Quintela I, Thomassen JQ, Rábano A, Rainero I, Rajabli F, Ramakers I, Real LM, Reinders MJT, Reitz C, Reyes-Dumeyer D, Ridge P, Riedel-Heller S, Riederer P, Roberto N, Rodriguez-Rodriguez E, Rongve A, Allende IR, Rosende-Roca M, Royo JL, Rubino E, Rujescu D, Sáez ME, Sakka P, Saltvedt I, Sanabria Á, Sánchez-Arjona MB, Sanchez-Garcia F, Juan PS, Sánchez-Valle R, Sando SB, Sarnowski C, Satizabal CL, Scamosci M, Scarmeas N, Scarpini E, Scheltens P, Scherbaum N, Scherer M, Schmid M, Schneider A, Schott JM, Selbæk G, Seripa D, Serrano M, Sha J, Shadrin AA, Skrobot O, Slifer S, Snijders GJL, Soininen H, Solfrizzi V, Solomon A, Song Y, Sorbi S, Sotolongo-Grau O, Spalletta G, Spottke A, Squassina A, Stordal E, Tartan JP, Tárraga L, Tesí N, Thalamuthu A, Thomas T, Tosto G, Traykov L, Tremolizzo L, Tybjærg-Hansen A, Uitterlinden A, Ullgren A, Ulstein I, Valero S, Valladares O, Broeckhoven CV, Vance J, Vardarajan BN, van der Lugt A, Dongen JV, van Rooij J, van Swieten J, Vandenberghe R, Verhey F, Vidal J-S, Vogelgsang J, Vyhnalek M, Wagner M, Wallon D, Wang L-S, Wang R, Weinhold L, Wiltfang J, Windle G, Woods B, Yannakoulia M, Zare H, Zhao Y, Zhang X, Zhu C, Zulaica M, EADB, GR@ACE, DEGESCO, EADI, GERAD, Demgene, FinnGen, ADGC, CHARGE, Farrer LA, Psaty BM, Ghanbari M, Raj T, Sachdev P, Mather K, Jessen F, Ikram MA, de Mendonça A, Hort J, Tsolaki M, Pericak-Vance MA, Amouyel P, Williams J, Frikke-Schmidt R, Clarimon J, Deleuze J-F, Rossi G, Seshadri S, Andreassen OA, Ingelsson M, Hiltunen M, Sleegers K, Schellenberg GD, van Duijn CM, Sims R, van der Flier WM, Ruiz A, Ramirez A, Lambert J-C, New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet 54, 412–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shooshtarizadeh P, Helness A, Vadnais C, Brouwer N, Beauchemin H, Chen R, Bagci H, Staal FJT, Coté JF, Möröy T, Gfi1b regulates the level of Wnt/β-catenin signaling in hematopoietic stem cells and megakaryocytes. Nat. Commun 10, 1270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee B-K, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kellis M, Khatun J, Kheradpour P, Kundaje A, Lassmann T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SCJ, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M M. Pazin J, Lowdon RF, Dillon LAL, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elnitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigó R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Stamatoyannopoulos JA, Tenenbaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandhu KS, Schaeffer L, See L-H, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee B-K, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Ki Kim S, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge E, Trout D, Varley KE, Gasper C, The ENCODE Project Consortium, Overall coordination (data analysis coordination), Data production leads (data production), Lead analysts (data analysis), Writing group, NHGRI project management (scientific management), Principal investigators (steering committee), Boise State University and University of North Carolina at Chapel Hill Proteomics groups (data production and analysis), Broad Institute Group (data production and analysis), U. of G. Cold Spring Harbor Center for Genomic Regulation, Barcelona, RIKEN, Sanger Institute, University of Lausanne, Genome Institute of Singapore group (data production and analysis), Data coordination center at UC Santa Cruz (production data coordination), E. Duke University University of Texas, Austin, University of North Carolina-Chapel Hill group (data production and analysis), Genome Institute of Singapore group (data production and analysis), C. HudsonAlpha Institute UC Irvine, Stanford group (data production and analysis), An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74 (2012).22955616 [Google Scholar]
- 66.Agarwal V, Bell GW, Nam J-W, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs. eLife. 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP, The biochemical basis of microRNA targeting efficacy. Science. 366, eaav1741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schnitzler GR, Kang H, Lee-Kim VS, Ma RX, Zeng T, Angom RS, Fang S, Vellarikkal SK, Zhou R, Guo K, Sias-Garcia O, Bloemendal A, Munson G, Guckelberger P, Nguyen TH, Bergman DT, Cheng N, Cleary B, Aragam K, Mukhopadhyay D, Lander ES, Finucane HK, Gupta RM, Engreitz JM, Mapping the convergence of genes for coronary artery disease onto endothelial cell programs (2022), p. 2022.11.01.514606, , doi: 10.1101/2022.11.01.514606. [DOI] [Google Scholar]
- 69.Science Forum: The Human Cell Atlas ∣ eLife, (available at https://elifesciences.org/articles/27041).
- 70.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moray T, Vassen L, Wilkes B, Khandanpour C, From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood. 126, 2561–2569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anguita E, Candel FJ, Chaparro A, Roldán-Etcheverry JJ, Transcription Factor GFI1B in Health and Disease. Front. Oncol 7 (2017), doi: 10.3389/fonc.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beauchemin H, Möröy T, Multifaceted Actions of GFI1 and GFI1B in Hematopoietic Stem Cell Self-Renewal and Lineage Commitment. Front. Genet 11 (2020), doi: 10.3389/fgene.2020.591099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osawa M, Yamaguchi T, Nakamura Y, Kaneko S, Onodera M, Sawada K, Jegalian A, Wu H, Nakauchi H, Iwama A, Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 100, 2769–2777 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Garnache-Ottou F, Chaperot L, Biichle S, Ferrand C, Remy-Martin J-P, Deconinck E, de Tailly PD, Bulabois B, Poulet J, Kuhlein E, Jacob M-C, Salaun V, Arock M, Drenou B, Schillinger F, Seilles E, Tiberghien P, Bensa J-C, Plumas J, Saas P, Expression of the myeloid-associated marker CD33 is not an exclusive factor for leukemic plasmacytoid dendritic cells. Blood. 105, 1256–1264 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Propris MSD, Raponi S, Diverio D, Milani ML, Meloni G, Falini B, Foà R, Guarini A, High CD33 expression levels in acute myeloid leukemia cells carrying the nucleophosmin (NPM1) mutation. Haematologica. 96, 1548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A, Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS, A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu S-Y, Birkenshaw A, Thomson T, Carlaw T, Zhang L-H, Ross CJD, Increasing the Targeting Scope of CRISPR Base Editing System Beyond NGG. CRISPR J. 5, 187–202 (2022). [DOI] [PubMed] [Google Scholar]
- 80.Daniloski Z, Jordan TX, Wessels H-H, Hoagland DA, Kasela S, Legut M, Maniatis S, Mimitou EP, Lu L, Geller E, Danziger O, Rosenberg BR, Phatnani H, Smibert P, Lappalainen T, tenOever BR, Sanjana NE, Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 184, 92–105.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liscovitch-Brauer N, Montalbano A, Deng J, Méndez-Mancilla A, Wessels H-H, Moss NG, Kung C-Y, Sookdeo A, Guo X, Geller E, Jaini S, Smibert P, Sanjana NE, Profiling the genetic determinants of chromatin accessibility with scalable single-cell CRISPR screens. Nat. Biotechnol 39, 1270–1277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Legut M, Gajic Z, Guarino M, Daniloski Z, Rahman JA, Xue X, Lu C, Lu L, Mimitou EP, Hao S, Davoli T, Diefenbach C, Smibert P, Sanjana NE, A genome-scale screen for synthetic drivers of T cell proliferation. Nature. 603, 728–735 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benner C, Spencer CCA, Havulinna AS, Salomaa V, Ripatti S, Pirinen M, FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC, PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Band G, Marchini J, BGEN: a binary file format for imputed genotype and haplotype data. bioRxiv, 308296 (2018). [Google Scholar]
- 86.Benner C, Havulinna AS, Järvelin M-R, Salomaa V, Ripatti S, Pirinen M, Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. Am. J. Hum. Genet 101, 539–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F, The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K, Su Z, Howson JMM, Auton A, Myers S, Morris A, Pirinen M, Brown MA, Burton PR, Caulfield MJ, Compston A, Farrall M, Hall AS, Hattersley AT, Hill AVS, Mathew CG, Pembrey M, Satsangi J, Stratton MR, Worthington J, Craddock N, Hurles M, Ouwehand W, Parkes M, Rahman N, Duncanson A, Todd JA, Kwiatkowski DP, Samani NJ, Gough SCL, McCarthy MI, Deloukas P, Donnelly P, Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet 44, 1294–1301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mägi R, Horikoshi M, Sofer T, Mahajan A, Kitajima H, Franceschini N, McCarthy MI, COGENT-Kidney Consortium, T2D-GENES Consortium, Morris AP, Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet 26, 3639–3650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, Sandstrom R, Humbert R, Stamatoyannopoulos JA, BEDOPS: high-performance genomic feature operations. Bioinformatics. 28, 1919–1920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Legut M, Daniloski Z, Xue X, McKenzie D, Guo X, Wessels H-H, Sanjana NE, High-Throughput Screens of PAM-Flexible Cas9 Variants for Gene Knockout and Transcriptional Modulation. Cell Rep. 30, 2859–2868.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B, Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wessels H-H, Méndez-Mancilla A, Guo X, Legut M, Daniloski Z, Sanjana NE, Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol 38, 722–727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kolde R, Laur S, Adler P, Vilo J, Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 28, 573–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A, Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, Tsherniak A, Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines (2019), p. 720243. [Google Scholar]
- 99.Dempster JM, Boyle I, Vazquez F, Root D, Boehm JS, Hahn WC, Tsherniak A, McFarland JM, Chronos: a CRISPR cell population dynamics model (2021), p. 2021.02.25.432728, , doi: 10.1101/2021.02.25.432728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pacini C, Dempster JM, Boyle I, Gonçalves E, Najgebauer H, Karakoc E, van der Meer D, Barthorpe A, Lightfoot H, Jaaks P, McFarland JM, Garnett MJ, Tsherniak A, Iorio F, Integrated cross-study datasets of genetic dependencies in cancer. Nat. Commun 12, 1661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kluesner MG, Lahr WS, Lonetree C-L, Smeester BA, Qiu X, Slipek NJ, Claudio Vazquez PN, Pitzen SP, Pomeroy EJ, Vignes MJ, Lee SC, Bingea SP, Andrew AA, Webber BR, Moriarity BS, CRISPR-Cas9 cytidine and adenosine base editing of splice-sites mediates highly-efficient disruption of proteins in primary and immortalized cells. Nat. Commun 12, 2437 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lenoir WF, Lim TL, Hart T, PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 46, D776–D780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell. 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alasoo K, Rodrigues J, Mukhopadhyay S, Knights AJ, Mann AL, Kundu K, Hale C, Dougan G, Gaffney DJ, Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet 50, 424–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nédélec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, Dumaine A, Grenier J-C, Freiman A, Sams AJ, Hebert S, Sabourin AP, Luca F, Blekhman R, Hernandez RD, Pique-Regi R, Tung J, Yotova V, Barreiro LB, Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 167, 657–669.e21 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Quach H, Rotival M, Pothlichet J, Loh Y-HE, Dannemann M, Zidane N, Laval G, Patin E, Harmant C, Lopez M, Deschamps M, Naffakh N, Duffy D, Coen A, Leroux-Roels G, Clément F, Boland A, Deleuze J-F, Kelso J, Albert ML, Quintana-Murci L, Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 167, 643–656.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, Seumois G, Rao A, Kronenberg M, Peters B, Vijayanand P, Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 175, 1701–1715.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buil A, Brown AA, Lappalainen T, Viñuela A, Davies MN, Zheng H-F, Richards JB, Glass D, Small KS, Durbin R, Spector TD, Dermitzakis ET, Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet 47, 88–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lappalainen T, Sammeth M, Friedländer MR, ‘t Hoen PAC, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlöf J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HPJ, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo Á, Antonarakis SE, Häsler R, Syvänen A-C, van Ommen G-J, Brazma A, Meitinger T, Rosenstiel P, Guigó R, Gut IG, Estivill X, Dermitzakis ET, Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 501, 506–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, Romano L, Planchon A, Falconnet E, Bielser D, Gagnebin M, Padioleau I, Borel C, Letourneau A, Makrythanasis P, Guipponi M, Gehrig C, Antonarakis SE, Dermitzakis ET, Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife. 2, e00523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Theusch E, Chen Y-DI, Rotter JI, Krauss RM, Medina MW, Genetic variants modulate gene expression statin response in human lymphoblastoid cell lines. BMC Genomics. 21, 555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.T. Gte. Consortium, The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lepik K, Annilo T, Kukuškina V, eQTLGen Consortium, Kisand K, Kutalik Z, Peterson P, Peterson H, C-reactive protein upregulates the whole blood expression of CD59 - an integrative analysis. PLOS Comput. Biol 13, e1005766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ, Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 546, 370–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Panopoulos AD, D’Antonio M, Benaglio P, Williams R, Hashem SI, Schuldt BM, DeBoever C, Arias AD, Garcia M, Nelson BC, Harismendy O, Jakubosky DA, Donovan MKR, Greenwald WW, Farnam K, Cook M, Borja V, Miller CA, Grinstein JD, Drees F, Okubo J, Diffenderfer KE, Hishida Y, Modesto V, Dargitz CT, Feiring R, Zhao C, Aguirre A, McGarry TJ, Matsui H, Li H, Reyna J, Rao F, O’Connor DT, Yeo GW, Evans SM, Chi NC, Jepsen K, Nariai N, Müller F-J, Goldstein LSB, Belmonte JCI, Adler E, Loring JF, Berggren WT, D’Antonio-Chronowska A, Smith EN, Frazer KA, iPSCORE: A Resource of 222 iPSC Lines Enabling Functional Characterization of Genetic Variation across a Variety of Cell Types. Stem Cell Rep. 8, 1086–1100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD, Musunuru K, Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell. 20, 558–570.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gryder BE, Khan J, Stanton BZ, Measurement of differential chromatin interactions with absolute quantification of architecture (AQuA-HiChIP). Nat. Protoc 15, 1209–1236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P, Heard E, Dekker J, Barillot E, HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhattacharyya S, Chandra V, Vijayanand P, Ay F, Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun 10, 4221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, Bierie B, Mazutis L, Wolf G, Krishnaswamy S, Pe’er D, Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell. 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Griffiths JA, Richard AC, Bach K, Lun ATL, Marioni JC, Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun 9, 2667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, Marioni JC, participants in the 1st Human Cell Atlas Jamboree, EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.