Summary
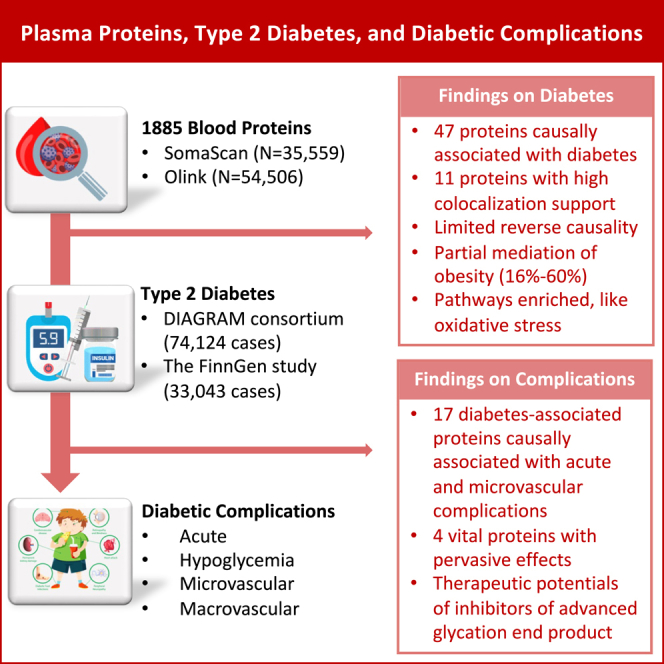
We conduct proteome-wide Mendelian randomization and colocalization analyses to decipher the associations of blood proteins with the risk of type 2 diabetes and diabetic complications. Genetic data on plasma proteome are obtained from 54,306 UK Biobank participants and 35,559 Icelanders. Summary-level data on type 2 diabetes are obtained from the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis consortium) consortium (74,124 cases) and FinnGen study (33,043 cases). Data on 10 diabetic complications are obtained from FinnGen and corresponding studies. Among 1,886 proteins, genetically predicted levels of 47 plasma proteins are associated with type 2 diabetes. Eleven of these proteins have strong support of colocalization. Seventeen proteins are associated with at least one diabetic complication, although a few have colocalization support. HLA-DRA, AGER, HSPA1A, and HSPA1B are associated with most microvascular complications. This study reveals causal proteins for the onset of type 2 diabetes and diabetic complications, which enhances the understanding of molecular etiology and development of therapeutics.
Keywords: colocalization, diabetic complication, Mendelian randomization, plasma protein, type 2 diabetes
Graphical abstract
Highlights
-
•
A proteome-wide analysis has been conducted for type 2 diabetes and complications
-
•
Forty-seven plasma proteins are causally associated with type 2 diabetes
-
•
Seventeen proteins are associated causally with at least one diabetic complication
-
•
HLA-DRA, AGER, HSPA1A, and HSPA1B are associated with microvascular complications
To determine the causal associations of blood proteins with type 2 diabetes and its complications, Yuan et al. perform a proteome-wide MR investigation and identify many circulating proteins in relation to type 2 diabetes and four proteins with universal effects on microvascular complications.
Introduction
Type 2 diabetes is an emerging pandemic affecting 1 in 10 adults globally.1 This number is projected to continue to increase in coming decades, especially in low-income and middle-income countries,1 which will cause a heavy burden to the medical system and the whole of society.
Studies have been conducted from different perspectives to understand type 2 diabetes and thus to better prevent and manage the disease. Population-based and clinical studies have indicated that obesity, physical inactivity, a sedentary lifestyle, smoking, and unhealthy diets are the major modifiable risk factors for the diabetes epidemic.2,3,4 To deepen the genetic understanding, several large-scale genome-wide association studies (GWASs) were conducted to decipher the genetic architecture of type 2 diabetes5,6 and identified more than 300 loci, which increased the genetic prediction usefulness.6 Because of the development of high-throughput detection and quantification of serum proteins, an increasing number of studies were also conducted to explore the associations of proteins with the risk of type 2 diabetes aiming at revealing molecular pathological basis.7,8 A population-based study found 24 plasma proteins consistently associated with prevalent type 2 diabetes in discovery and replication studies; however, only 3 proteins were found to be associated with incident type 2 diabetes.7 Another study with a larger sample size identified 47 proteins associated with incident type 2 diabetes after 19 years of follow-up.8 Still, merely three associations were confirmed by Mendelian randomization (MR) analysis.8 These findings may convey that observational studies on proteomic research in type 2 diabetes are prone to be influenced by reverse causation, as well as confounding given that the factors influencing proteomic profiles remain unestablished.
Diabetic complications lead to a heavy disease burden, such as, blindness, kidney dysfunction, an impaired quality of life, and premature mortality among those with diabetes. Several drugs, like metformin, sodium-glucose co-transporter-2 (SGLT2) inhibitors, and glucagon-like peptide-1 receptor agonists, have been used to lower the risk of developing complications or delay their onset among diabetic patients. An appraisal of shared proteomic basis between diabetes and diabetic complications is also of importance to better understand underlying pathophysiology and possibly to manage disease progression. It was found in a cohort of 528 individuals that 15 plasma proteins shared by type 2 diabetes and coronary artery disease (a major macrovascular complication of diabetes).9 However, the shared proteomic etiology between diabetes and diabetic microvascular complications remains to be fully explored.
Proteomic research can not only deepen molecular understanding, but also help to reveal potential therapeutic targets, since many circulating proteins always act as the principal regulators of molecular pathways.10 For example, some classes of drugs used for type 2 diabetes target proteins, like alpha-glucosidase inhibitors that inhibit alpha-amylase and alpha-glucosidase, the key enzymes involved in the digestion of carbohydrates, and SGLT2 inhibitors that inhibit sodium-glucose transport proteins in the nephron.11 MR design is an epidemiological approach that can strength causal inference by using genetic variants as an instrumental variable for the exposure (e.g., circulating levels of a protein). There are three key assumptions of MR: (1) the genetic variants used as the instrumental variable should be robustly associated with the exposure of interest; (2) the genetic instruments should not be associated with any confounders; and (3) the genetic instruments should impact the outcome only via the exposure of interest and not via alternative pathways. Using MR analysis to explore the associations between plasma proteins and health outcomes has been widely used and found to be assumption satisfied when using the cis-variant in the protein-encoding gene as the genetic instrument for the protein.12 Some previous MR studies have been conducted to estimate the association between blood proteins and the risk of type 2 diabetes.13,14 Based on a larger number of blood proteins with data from two large-scale studies, we conducted a protein-wide MR study supplemented by colocalization analysis to explore nearly 2,000 plasma proteins in relation to the onset of type 2 diabetes and diabetic complications.
Results
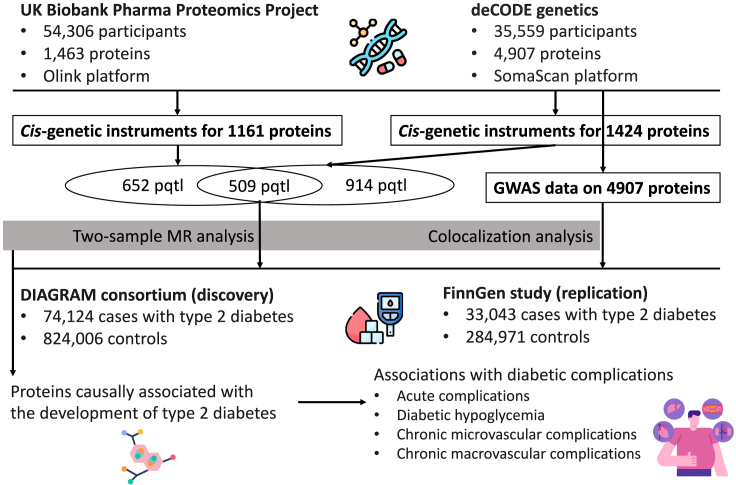
An overview of the study design is shown in Figure 1. All analyses were based on summary-level data listed in Table 1. We replaced 132 missing SNPs with proxy SNPs in FinnGen. After removing plasma proteins without genetic instruments or SNP proxies in type 2 diabetes data, MR analysis included 1,797 proteins in the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis consortium) consortium, 1,835 proteins in the FinnGen R8 study, and 1,885 proteins in the combined analysis. The minimal F statistic of used genetic instruments was more than 300 in all outcome data. The statistical power in the analyses of diabetic complications was generally low because of small sample sizes (Figure S1).
Figure 1.
Study design overview
Table 1.
Data sources for studied phenotypes
| Study | Phenotype | Cases | Controls | PMID | Adjustment |
|---|---|---|---|---|---|
| UKB-PPP | Plasma protein | 54,306 | – | Preprint | age, gender, batch, UKB center, UKB genetic array, time between blood sampling and measurement and the first 20 genetic principal components |
| deCODE | Plasma protein | 35,559 | – | 34857953 | age, gender, and sample age |
| DIAGRAM | Type 2 diabetes | 74,124 | 824,006 | 30297969 | principal components, relatedness, and study-specific covariates |
| FinnGen R8 | Type 2 diabetes | 33,043 | 284,971 | NA | age, gender, and ≤20 genetic principal components |
| GIANT | BMI | 681,275 | – | 30124842 | age, gender, recruitment center, genotyping batches, and 10 PCs |
| Neale Lab | BMI | 336,107 | – | NA | age, gender, 10 genetic principal components, and genotyping batch |
| FinnGen R8 | Diabetic ketoacidosis | 7,201 | 249,480 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Diabetic retinopathy | 8,942 | 283,545 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Diabetic nephropathy | 3,676 | 283,456 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Diabetic neuropathy | 2,444 | 249,480 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Diabetic maculopathy | 3,115 | 283,472 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Diabetic hypoglycemia | 6,500 | 249,480 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Diabetes peripheral circulatory complications | 1,975 | 283,224 | NA | age, gender, and ≤20 genetic principal components |
| FinnGen R8 | Cardiomyopathy | 5,344 | 237,263 | NA | age, gender, and ≤20 genetic principal components |
| CARDIoGRAMplusC4D | Coronary artery disease | 60,801 | 123,504 | 26343387 | NA |
| Malik et al. GWAS15 | Ischemic stroke | 34,217 | 406,111 | 29531354 | age and gender |
CARDIoGRAMplusC4D, Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) Genetics; GIANT, The Genetic Investigation of ANthropometric Traits consortium; NA, not available; UKB-PPP, The UK Biobank Pharma Proteomics Project.
Associations between plasma proteins and type 2 diabetes
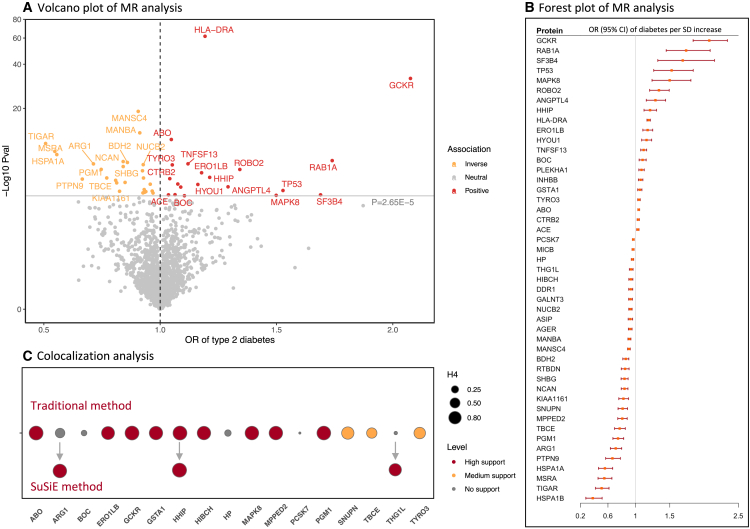
Figure 2 displays the result summary of the analyses of type 2 diabetes. In the combined analysis of two outcomes data, genetically predicted levels of 47 proteins were significantly associated with the risk of type 2 diabetes after Bonferroni correction for multiple testing (p < 2.65 × 10−5) (Figure 2A). Per SD increase in genetically predicted levels of protein, the odds ratio of type 2 diabetes ranged from 0.38 (95% confidence interval [CI], 0.28–0.51) for heat shock 70 kDa protein 1B (HSPA1B) to 2.08 (95% CI, 1.84–2.34) for glucokinase regulatory protein (GCKR) (Figure 2B and Table S1). The identified associations were directionally consistent between the discovery and replication studies for all proteins except heat shock 70 kDa protein 1A (HSPA1A) (Tables S2 and S3). Among identified associations, eight proteins were measured in both outcome datasets and generally showed consistent associations with type 2 diabetes, despite the proteins being measured on two different profiling platforms (Table S4). Genetically predicted levels of the other studied proteins were not associated with the risk of type 2 diabetes (Table S1).
Figure 2.
Result summary of MR and colocalization analysis on the associations between plasma proteins and the risk of type 2 diabetes
(A–C) OR, odds ratio. Full name of proteins: ABO, histo-blood group ABO system transferase; ANGPTL4, angiopoietin-related protein 4; ARG1, arginase-1; ASIP, Agouti-signaling protein; BDH2, 3-hydroxybutyrate dehydrogenase type 2; BOC, brother of CDO; CTRB2, chymotrypsinogen B2; DDR1, epithelial discoidin domain-containing receptor 1; GALNT3, polypeptide N-acetylgalactosaminyltransferase 3; GCKR, glucokinase regulatory protein chain; HP, haptoglobin; HSPA1A, heat shock 70 kDa protein 1A; HSPA1B, heat shock 70 kDa protein 1B; HYOU1, hypoxia up-regulated protein 1; INHBB, inhibin beta B chain; KIAA1161, uncharacterized family 31 glucosidase KIAA1161; MPPED2, metallophosphoesterase MPPED2; MSRA, mitochondrial peptide methionine sulfoxide reductase; NCAN, neurocan core protein; NUCB2, nucleobindin-2; PCSK7, proprotein convertase subtilisin/kexin type 7; PLEKHA1, pleckstrin homology domain-containing family A member 1; PTPN9, tyrosine-protein phosphatase non-receptor type 9; RAB1A, Ras-related protein Rab-1A; ROBO2, roundabout homolog 2; RTBDN, retbindin; SF3B4, splicing factor 3B subunit 4; SNUPN, Snurportin-1; TBCE, tubulin-specific chaperone E; TIGAR, fructose-2,6-bisphosphatase TIGAR; TNFSF13, tumor necrosis factor ligand superfamily member 13; TP53, cellular tumor antigen p53; TYRO3, tyrosine-protein kinase receptor TYRO3.
Among 47 MR-identified proteins in relation to type 2 diabetes, 9 proteins had high support of colocalization analysis (PH4 ≥0.8) (Figure 2C and Table S5), which were GCKR, histo-blood group ABO system transferase (ABO), hedgehog-interacting protein (HHIP), phosphoglucomutase-1 (PGM1), mitogen-activated protein kinase 8 (MAPK8), metallophosphoesterase domain-containing protein 2 (MPPED2), 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial (HIBCH), glutathione S-transferase A1 (GSTA1), and ERO1-like protein beta. Three proteins had medium support of colocalization analysis (0.8 ≥ PH4 ≥ 0.5) (Figure 2C). Sum of single effects (SuSiE) additionally identified strong colocalization support for the associations for arginase 1 (ARG1) and probable tRNA(His) guanylyltransferase (THG1L) (Figure 2C).
Function and network prediction of diabetes-associated proteins
Identified diabetes-associated proteins had networks, in particular co-expression and physical interactions (Figure S2). Based on cis-genes for these proteins, many pathways were enriched, including protein-folding and catabolism, response to temperature stimulus/heat, intrinsic apoptotic signaling pathway, and response to oxidative stress (Table S6).
Reverse MR analysis of type 2 diabetes liability with levels of identified 47 proteins
Genetic liability to type 2 diabetes was associated with levels of six blood proteins (THG1L, sex hormone-binding globulin [SHBG], inhibin beta B chain, chymotrypsinogen B2, major histocompatibility complex class I polypeptide-related sequence B [MICB], and HIBCH) after Bonferroni multiple testing correction. The associations were consistent in a series of analyses (Table S7).
Associations between diabetes-associated proteins and body mass index
Among 47 diabetes-associated proteins, genetically predicted levels of 18 proteins were associated with body mass index (BMI) in the GIANT (The Genetic Investigation of ANthropometric Traits consortium) consortium or the UK Biobank study (Figure S3). The associations were overall consistent between the two sources. Per SD increase, the changes of BMI ranged from −0.29 (95% CI, –0.37 to −0.20) for mitochondrial peptide methionine sulfoxide reductase to 0.12 (95% CI, 0.08–0.16) for GCKR. The two-stage network MR analysis identified pathways mediated by BMI from protein to type 2 diabetes. The mediation by BMI ranged from 15.8% for the GCKR-diabetes association to 60.6% for the hypoxia up-regulated protein 1-diabetes association (Table S8). Mainly because of the high co-expression of these proteins (Figure S4), the sum of mediation proportion was greater than 100%.
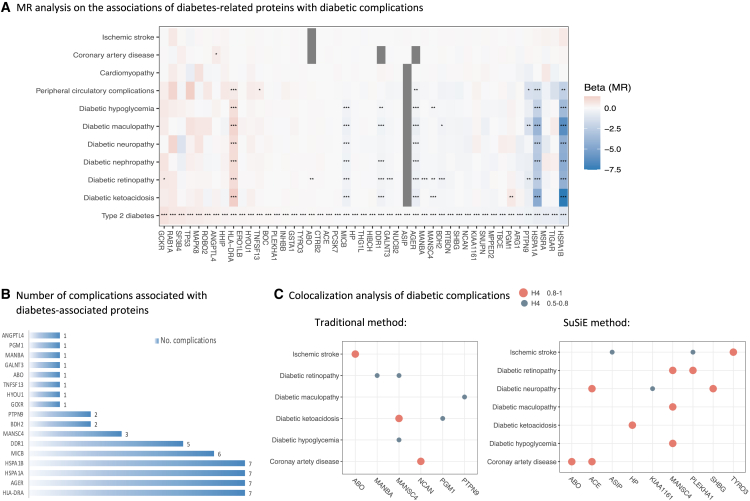
Associations between diabetes-associated proteins and diabetic complications
Figure 3 displays the result summary of the analyses of 47 diabetes-associated plasma proteins in relation to major diabetic complications. After adjusting for multiple testing, genetically predicted levels of 17 proteins were associated with at least one diabetic complication in MR analysis (Figures 3A and 3B). The directions of the protein-diabetes associations were overall consistent with that of the associations between proteins and diabetic complications (Figure 3A), except for genetically predicted levels of PGM1 with an inverse association with type 2 diabetes, but a positive association with diabetic ketoacidosis (Figure 3A). Diabetes-associated proteins were more likely to be associated with acute and chronic microvascular complications instead of macrovascular complications (Figure 3A). Four of these proteins that included human leukocyte antigen class II histocompatibility antigen, DR alpha chain (HLA-DRA), advanced glycosylation end product-specific receptor (AGER), HSPA1A, and HSPA1B were directionally consistently associated with most studied diabetic complications (Figure 3B). Genetically predicted levels of the other proteins were not associated with any of studied diabetic complications (Table S9).
Figure 3.
Result summary of MR and colocalization analysis on the associations between diabetes-related proteins and the risk of diabetic complications
(A–C) The gray squares in (A) indicate missing data. Full name of proteins: ABO, histo-blood group ABO system transferase; ANGPTL4, angiopoietin-related protein 4; ARG1, arginase-1; ASIP, Agouti-signaling protein; BDH2, 3-hydroxybutyrate dehydrogenase type 2; BOC, brother of CDO; CTRB2, chymotrypsinogen B2; DDR1, epithelial discoidin domain-containing receptor 1; GALNT3, polypeptide N-acetylgalactosaminyltransferase 3; GCKR, glucokinase regulatory protein; HP, haptoglobin; HSPA1A, heat shock 70 kDa protein 1A; HSPA1B, heat shock 70 kDa protein 1B; HYOU1, hypoxia up-regulated protein 1; INHBB, inhibin beta B chain; KIAA1161, uncharacterized family 31 glucosidase KIAA1161; MPPED2, metallophosphoesterase MPPED2; MSRA, mitochondrial peptide methionine sulfoxide reductase; NCAN, neurocan core protein; NUCB2, nucleobindin-2; PCSK7, proprotein convertase subtilisin/kexin type 7; PLEKHA1, pleckstrin homology domain-containing family A member 1; PTPN9, tyrosine-protein phosphatase non-receptor type 9; RAB1A, Ras-related protein Rab-1A; ROBO2, roundabout homolog 2; RTBDN, retbindin; SF3B4, splicing factor 3B subunit 4; SNUPN, snurportin-1; TBCE, tubulin-specific chaperone E; TIGAR, fructose-2,6-bisphosphatase TIGAR; TNFSF13, tumor necrosis factor ligand superfamily member 13; TP53, cellular tumor antigen p53; TYRO3, tyrosine-protein kinase receptor TYRO3.
Some of above MR associations were supported by traditional and SuSiE colocalization analyses with PH4 >0.5 (Figure 3C and Table S10). MANSC domain-containing protein 4 (MANSC4) had moderate to strong support of colocalization with diabetic ketoacidosis (PH4 = 0.92), diabetic hypoglycemia (PH4 = 0.67 in traditional and 0.96 in SuSiE), and diabetic retinopathy (PH4 = 0.63 in traditional and 0.85 in SuSiE). Beta-mannosidase (MANBA) had moderate support of colocalization with diabetic retinopathy (PH4 = 0.65 in traditional). PGM1 had moderate support of colocalization with diabetic ketoacidosis (PH4 = 0.59 in traditional). PTPN9 had moderate support of colocalization with diabetic maculopathy (PH4 = 0.63 in traditional).
Discussion
This MR study examined the associations of 1,885 plasma proteins with the risk of type 2 diabetes and was supplemented by a colocalization analysis. We found 47 plasma proteins with a potential causal association with type 2 diabetes, of which 11 proteins had a strong support of colocalization. Function prediction based on these proteins enriched many pathways, including protein folding and catabolism, response to temperature stimulus/heat, intrinsic apoptotic signaling pathway, and response to oxidative stress. Several of these proteins were associated with BMI in a consistent direction with that for diabetes and the two-stage network MR indicated partial mediation of BMI in these protein-diabetes associations. The subsequent analysis revealed the roles of 17 diabetes-associated proteins in diabetic complications, although only some proteins were identified in colocalization analysis. These diabetes-associated proteins were more likely to be associated with microvascular complications compared with macrovascular complications. This analysis further found a pervasive effect of four proteins (i.e., HLA-DRA, AGER, HSPA1A, and HSPA1B) on acute and chronic microvascular complications.
Our study corroborated some previously identified protein-diabetes associations, such as the associations of diabetes with TYRO3 (tyrosine-protein kinase receptor TYRO3), ARG1, SHBG, MANSC4, HP, MANBA, ABO, angiotensin-converting enzyme (ACE), ERO1-like protein beta (ERO1LB), MICB, and GSTA1.8,14 The associations of some well-studied proteins, like GCKR,5 SHBG,16 ABO,17 and ACE18 with the risk of developing type 2 diabetes were also identified, which indicated a good validity of data sources used in the current analysis. However, except for ABO, ARG1, ERO1LB, GSTA1, and TYRO3, we observed weak colocalization support of the associations for other above-mentioned proteins, which implies that these associations may be influenced by linkage disequilibrium. However, a lack of strong colocalization support may be caused by an inadequate power, even though the current colocalization analysis was based on a large-scale meta-analysis GWAS including 107,167 type 2 diabetes cases. In addition, this study did not pinpoint certain proteins associated with diabetes described in previous studies, like growth-differentiation factor-15 and matrix metallopeptidase 12,13 which did not indicate that our findings were against these associations. Instead, these associations were also observed in our analysis, albeit non-significantly, which might be caused by a heavy multiple testing burden. However, this multiple-testing correction strategy suits one of the study’s aims, which is to discover proteins strongly associated with type 2 diabetes.
Except for the GCKR protein, a well-known protein affecting diabetes risk,5 our study revealed many other plasma proteins that might be causally associated with the risk of type 2 diabetes based on a larger pool of plasma proteins. For example, HHIP has been found to be involved in the development of cancers.19 Our study in line with an observational study20 established a positive association between HHIP protein levels and the risk of type 2 diabetes, which indicates that the Hedgehog signaling pathway may play a role in glycemic homeostasis. HIBCH plays a role in the catabolism of the branched-chain amino acid valine that has been positively associated with the risk of type 2 diabetes.21 In addition, this protein also determines the metabolism of 3-hydroxyisobutyrate, which was associated with incident type 2 diabetes.22 Altogether, support our finding on a causal inverse association between HIBCH protein and type 2 diabetes. Likewise, MAPK8-related signaling was found to be related to the development of type 2 diabetes in animal studies.23 The causality of this association in human has now been strengthened by our study. Another interesting protein associated with diabetes was MPPED2, which was found to impact the development of cancers24 with functions of regulating cell proliferation, differentiation, and apoptosis. However, whether the MPPED2-diabetes association was caused by pleiotropy needs assessment. PGM1 protein has been proven to act in glucose metabolism and its encoding gene has been identified as a shared loci between type 1 and type 2 diabetes.25 Thus, the association between PGM1 and type 2 diabetes can be regarded as a positive control. A double catch of this association in our MR and colocalization analyses partly reflects a good validity of the overall analysis. In addition to the above-mentioned proteins, we identified more proteins associated with type 2 diabetes but with weak evidence of colocalization. More studies are needed to verify our findings.
In reverse MR analysis, we observed associations of genetic liability to type 2 diabetes with six proteins, including some well-established diabetes-associated proteins (i.e., SHBG, MICB, and HIBCH), which raises the concern that these proteins may be associated with type 2 diabetes due to reverse causation. However, we noticed that the directions of protein-diabetes and diabetes-protein associations were opposite, except that for chymotrypsinogen B2, which minimizes the possibility of reverse causation and imply potential bidirectional associations caused by a feedback mechanism.26
Our analysis of diabetes-associated proteins in relation to diabetic complications added insights on diabetic progression from the molecular perspective. We found that diabetes-associated proteins seemed to be more associated with microvascular complications compared to macrovascular complications, which supported the findings that the associations of diabetes with macrovascular outcomes were less pronounced, and significant than those with microvascular.27 However, given that data used on three major macrovascular complications were obtained from general populations instead of diabetic patients, our result might be influenced by this.
We found that four proteins (i.e., HLA-DRA, AGER, HSPA1A, and HSPA1B) were associated with most studied diabetic complications. However, it is important to note that these protein associations with diabetic complications lacked strong colocalization support, possibly due to limited statistical power. HLA-DR consisted of HLA-DRA and HLA-DRB1 (HLA class II histocompatibility antigen, DRB1 beta chain) has been associated with the risk of type 1 diabetes.28 A few studies linked this protein to the development of type 2 diabetes as well as its progression. A study in the Han ethnicity of China showed that alleles of HLA-DRA and HLA-DRB1 genes were associated with the susceptibility of type 2 diabetes and diabetic nephropathy.29 These associations were replicated in a dataset-based multi-omics study.30 Another study found that HLA-DR-related inflammation might be associated with beta-cell destruction in patients with type 2 diabetes, which may explain the associations with diabetic complications.31 Still, the immune response to increased levels of HLA-DRA is complex. Our study can only convey that HLA-DRA-related immune reactions may play a role in diabetic progression and more studies are needed to explore in detail the underlying mechanistic pathways. AGER is a cell-surface receptor of AGEs (advanced glycation end products) by glycation. It will activate pro-inflammatory pathways when AGEs bind to AGER.32 Thus, low levels of AGER may decrease glycation-related inflammation and thus decrease the risk of diabetes and its complications, which is supported by our findings. Many small molecule AGER inhibitors or antagonists have been proposed to treat diabetic complications and cancer progression with a good tolerance in humans.33 Heat shock proteins were associated with hypoglycemia34 and diabetic foot ulcer.35 However, the mechanisms underlying the inverse associations of HSPA1A and HSPA1B with diabetes and its complications are unclear, but possibly related to attenuation of oxidative stress.36
Except for the above four proteins, we also observed GCKR and ANGPTL4 associated with some complications, which was consistent with recent trial evidence that GCKR and ANGPTL4 are currently in therapeutic development for metabolic disorders. Our data demonstrated potential involvement of these two proteins in the development of type 2 diabetes and related complications; thus, these drugs under development hold also promise for these indications.
This study has several strengths, including a large pool of plasma proteins, a large number of type 2 diabetes cases, a mutual validation in two independent outcome data, and a supportive analysis of colocalization. Limitations deserve to be discussed when interpreting our results. First, this analysis was confined to Europeans, which limited the generalizability of our findings to other populations. Second, we did colocalization analysis only for proteins included in the deCODE Health study37 due to data availability. Thus, whether the MR associations for proteins with instruments from the UKB-PPP study are supported by colocalization analysis remain unknown. Third, even though we meta-analyzed GWAS data on type 2 diabetes from two independent sources to increase the power, we may have overlooked some weak associations, especially in colocalization analyses. Fourth, the power may have been inadequate in the analysis of diabetic complications due to a small number of cases. Fifth, data on coronary artery disease, ischemic stroke, and cardiomyopathy were not obtained from a diabetic population, which might influence the comparison between the associations for macrovascular and microvascular complications with diabetes-associated proteins. Sixth, even though we were not able to differentiate the diabetic complications of type 1 or type 2 diabetes, given that the analysis was based on the summary-level data, our results should reflect more on the complications of type 2 diabetes due to a small number of complication cases of type 1 in the FinnGen study. For example, there were 1,897 cases of type 1 diabetes with ketoacidosis among 7,201 individuals with ketoacidosis. Seventh, even though cis-MR analysis can minimize the influence of horizontal pleiotropy, the observed associations might be affected by this phenomenon, especially for a certain protein with an encoding gene exerting a broad range of effects. Eighth, our study could not assess the risk of non-specific binding for proteins or the variability of protein measurement since this study was based on summary-level data. However, including proteins with cis-SNPs might decrease the risk of measurement error due to non-specific binding and results were overall consistent for overlapping proteins with data measured by SomaScan and Olink. Last, but not least, we used proteomic data from Icelanders and outcome data from Finnish, which might introduce bias by violating the second assumption of MR because the genetic backgrounds of these two populations may differ from other European populations. However, this potential bias is likely to be minimal as we observed directionally consistent protein-type 2 diabetes associations with outcome data from DIAGRAM and FinnGen studies. Furthermore, we observed that more than 85% of the proteins-type 2 diabetes associations with a p < 0.05 were directionally consistent among proteins with genetic instruments from both deCODE (Icelanders) and UKB-PPP (British). The around 15% inconsistency might be attributed to different genetic architectures but more likely to different proteomic profiling platforms (SomaScan and Olink) used in the two cohorts. This hypothesis is supported by studies that observed that at least 14.7% of proteins had poor correlations when measured in SomaScan and Olink platforms.38,39,40
In summary, this proteome-wide MR and colocalization analysis identified many plasma proteins highly causally associated with type 2 diabetes. Among these proteins, almost one-third were associated with diabetic complications, in particular acute and microvascular complications. Universal effects of four diabetes-associated proteins (HLA-DRA, AGER, HSPA1A, and HSPA1B) on most studied diabetic complications may suggest the roles of inflammation and oxidative stress in diabetic progression. Our findings also suggest that AGER inhibitors or antagonists may be a promising therapeutic target for diabetic complications.
Limitations of the study
Our findings, in relation to the analysis itself, may be subject to the following potential limitations: (1) bias due to population structure; (2) macrovascular complications in non-diabetic populations; (3) the presence of complications from both type 1 and type 2 diabetes; (4) horizontal pleiotropy; (5) heterogeneity introduced by the usage of two protein profiling platforms; and (6) limited generalizability. Each of these factors has been thoroughly addressed in the text. Furthermore, as these associations were established through in silico analyses, they require further validation through animal studies and population-based research. Despite the inclusion of a vast number of blood proteins in this study, we may have inadvertently overlooked significant proteins that lack genetic instruments. The risk prediction ability of these identified proteins also warrants further studies.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Shuai Yuan (shuai.yuan@ki.se).
Materials availability
This study did not generate new unique reagents.
Experimental models and subject details
The analysis was based on summary-level GWAS data in European populations. We first examined the associations of genetically predicted plasma proteins with risk of type 2 diabetes in two independent large-scale studies. To explore whether the diabetes-associated proteins play a role in the development of diabetic complications, we estimated the associations of diabetes-associated proteins with ten diabetic complications. We also tested the associations of the diabetes-associated proteins with BMI and conducted a two-stage network MR analysis to reveal whether these protein-diabetes associations were mediated via obesity. The used studies had been approved by corresponding ethical review committees and participants had signed the consent form. This study was approved by the Swedish Ethical Review Authority.
Method details
Data sources for plasma proteins
We selected index cis-SNPs associated with the levels of plasma proteins at the genome-wide significance level (p < 5 × 10−8) as instrumental variables from two large-scale GWASs in the UK Biobank Pharma Proteomics Project (UKB-PPP)41 and the deCODE Health study.37 Cis-SNPs were defined as SNPs within 1Mb from the gene encoding the protein, and linkage disequilibrium was estimated based on 1000 Genomes European panel. UKB-PPP conducted proteomic profiling on blood plasma samples from 54,306 participants using the Olink platform and collected data on 1,463 proteins.41 For the two-sample MR analysis, we selected index cis-SNPs as instrumental variables for 1161 proteins.41 Likewise, index cis-SNPs for 1423 plasma proteins were obtained from the deCODE Health study where 4,907 aptamers were measured among 35,559 Icelanders using the SomaScan platform.37 There were 509 proteins with index cis-SNPs that overlapped between the two studies. Given that colocalization analysis could only be performed for proteins from the deCODE Health study due to the availability of the summary-level data, we used genetic instrumental variables selected from the deCODE Health study for these overlapped proteins in the main results. We have presented results for the overlapping proteins with genetic instruments from two studies to examine the consistency of results based on different proteomic profiling platforms.
Data sources for type 2 diabetes
Data on the associations of protein-associated SNPs with type 2 diabetes were obtained from the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) consortium5 and the FinnGen study.42 The DIAGRAM consortium included 32 studies with a total of 74,124 cases and 824,006 controls of European descent. We used the latest release data on type 2 diabetes from the FinnGen study R8 in this analysis, which comprised 33,043 cases 284,971 controls. There were no sample overlaps between two outcome datasets. In MR analysis, we treated the DIGRAM consortium as the discovery study and the FinnGen R8 study as the replication. To increase the power, we meta-analyzed the two GWASs and performed colocalization analysis based on the GWAS meta-analysis data (107,167 cases and 1,108,976 controls). The GWAS meta-analysis was performed by the METAL software. Genetic variants associated with type 2 diabetes at p < 5 × 10−8 in this GWAS meta-analysis and with low linkage disequilibrium (R2 < 0.001) were selected as the instrument variable for type 2 diabetes in the reverse MR analysis.
Data sources for BMI
Obesity is an important causal risk factor for type 2 diabetes. We estimated the associations of diabetes-associated proteins with BMI to reveal whether these associations may be mediated via obesity. Summary-level data on BMI were available from a GWAS in the GIANT (Genetic Investigation of ANthropometric Traits) consortium that included 681,275 individuals.43 Given a comparatively small number of SNPs analyzed in this genome-wide association analysis (causing missing in MR analysis), we also used GWAS data on BMI (N = 336,107) from the Neale Lab. The Neale Lab GWAS on BMI was based on 336,107 participants from the UK Biobank study (http://www.nealelab.is/uk-biobank/). For proteins associated with BMI and type 2 diabetes in a consistent direction, we performed a two-stage network MR analysis using the Neale Lab data and GWAS meta-analysis of type 2 diabetes to estimate the mediation of BMI in the protein-diabetes associations. Genetic variants associated with BMI at p < 5 × 10−8 in Neale Lab data and with low linkage disequilibrium (R2 < 0.001) were selected as the instrument variable for BMI in this network MR analysis. Detailed methods of the two-stage network MR analysis and mediation calculation can be found somewhere else.44
Data sources for diabetic complications
The analysis included ten diabetic complications, which can be classified into four categories that are acute complications (diabetic ketoacidosis), diabetic hypoglycemia, chronic microvascular complications (diabetic retinopathy, diabetic nephropathy, diabetic neuropathy, and diabetic maculopathy), and chronic macrovascular complications (diabetes peripheral circulatory complications, cardiomyopathy, coronary artery disease, and ischemic stroke). Given no available data on coronary artery disease and ischemic stroke among diabetic patients, we used data on coronary artery disease (60,801 cases and 123,504 controls) and ischemic stroke (34,217 cases and 406,111 controls) from corresponding genome-wide association studies among general populations, respectively.15,45 Likewise, data on general cardiomyopathy were obtained from the FinnGen R8 study (5344 cases). Summary-level data on other diabetic complications were obtained from the FinnGen R8 study with the number of cases ranging from 1975 for peripheral circulatory complications to 8942 for diabetic retinopathy.
Quantification and statistical analysis
MR analysis
Protein-associated SNPs unavailable in outcome data were replaced by SNP proxies with a high linkage disequilibrium (R2 ≥ 0.8) based on 1000 Genomes European reference panel. Missing SNPs without suitable SNP proxies were removed from the analysis (N = 190). The F statistic was calculated to assess the strength of the instrumental variables. We estimated statistical power for the analyses assuming a disease prevalence of 2% in general.46 The ORs and corresponding CIs of the associations between plasma proteins and studied outcomes were estimated by the Wald ratio and the delta method (a general method for deriving the variance of a function of asymptotically normal random variables with known variance and commonly used to estimate standard error of MR ratio estimate),47 respectively. The associations for one protein from two outcome data were meta-analyzed using the fixed-effect model. The MR associations were scaled to one SD increase in genetically predicated levels of circulating proteins. Bonferroni method was used to correct for multiple testing in the analysis for type 2 diabetes. The combined association with a p value <2.65 × 10−5 (0.05/1885 proteins) was deemed significant. For type 2 diabetes, we performed a reverse MR analysis to estimate the associations of type 2 diabetes liability with the levels of identified proteins to explore potential reverse causation. For the MR analysis of diabetic complications, we used the false discovery rate approach to correct for multiple testing given a comparative small sample size. MR analyses were performed using TwoSampleMR and MendelianRandomization packages in R software (4.4.1).
Colocalization analysis
We performed the colocalization analysis to test whether identified associations of proteins with type 2 diabetes and diabetic complications were driven by linkage disequilibrium. The analysis was based on a Bayesian model that assesses the support for five exclusive hypotheses: 1) no association with either trait; 2) association with trait 1 only; 3) association with trait 2 only; 4) both traits are associated, but distinct causal variants were for two traits; and 5) both traits are associated, and the same shares causal variant for both traits.48 A posterior probability is provided for each hypothesis testing (H0, H1, H2, H3, and H4). In this analysis, we used the setting that is the prior probabilities of the SNP being associated with trait 1 only (p1) at 1 × 10−4; the probability of the SNP being associated with trait 2 only (p2) at 1 × 10−4; and the probability of the SNP being associated with both traits (p12) at 1 × 10−5. Two signals were considered to have a strong support of colocalization if the posterior probability for shared causal variants (PH4) was ≥0.8. Medium colocalization indication was defined as 0.5< PH4 <0.8. The analysis was performed using the coloc package in R software (4.4.1). Given that the traditional colocalization approach cannot detect the scenario where the exposure and outcome traits share more than one causal hit, we, therefore, utilized SuSiE (Sum of Single Effects)49 colocalization method by integrating proteomics GWAS summary statistics and genetic correlation matrix reference panels based on individuals of European ancestry from the 1000 Genomes Project Phase 3 to identify multiple causal variants.
Function and network prediction
We used GeneMANIA (http://www.genemania.org) to predict the functions and networks of cis-genes for the diabetes-associated proteins and both diabetes- and BMI-associated proteins. Detailed information on included datasets in GeneMANIA is described somewhere else.50 Enriched function pathways were considered to be significant with false discovery rate <0.05.
Acknowledgments
The current study received no funding. We acknowledge investigators and participants in the UK Biobank study, the deCODE project, the FinnGen study, and cited genome-wide association studies for sharing data.
Author contributions
S.Y., F.X., C.S.M., and S.C.L. conceived and designed the study. S.Y. and F.X. undertook the statistical analyses. S.Y. wrote the first draft of the manuscript. S.Y., F.X., X.L., J.C., J.Z., C.S.M., and S.C.L. interpreted the data, provided important comments to the manuscript, and approved the final version of the manuscript.
Declaration of interests
All authors declare no conflict of interest.
Published: August 30, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101174.
Contributor Information
Shuai Yuan, Email: shuai.yuan@ki.se.
Susanna C. Larsson, Email: susanna.larsson@ki.se.
Supplemental information
Data and code availability
This paper analyzes existing, publicly available data. All GWAS are referenced. This paper does not report the original code. Any additional information required to reanalyze the data reported in this report is available from the lead contact upon request.
References
- 1.Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B.B., Stein C., Basit A., Chan J.C.N., Mbanya J.C., et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183:109119. doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng Y., Ley S.H., Hu F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 3.Yuan S., Merino J., Larsson S.C. Causal factors underlying diabetes risk informed by Mendelian randomisation analysis: evidence, opportunities and challenges. Diabetologia. 2023;66:800–812. doi: 10.1007/s00125-023-05879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knowler W.C., Barrett-Connor E., Fowler S.E., Hamman R.F., Lachin J.M., Walker E.A., Nathan D.M., Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vujkovic M., Keaton J.M., Lynch J.A., Miller D.R., Zhou J., Tcheandjieu C., Huffman J.E., Assimes T.L., Lorenz K., Zhu X., et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhadad M.A., Jonasson C., Huth C., Wilson R., Gieger C., Matias P., Grallert H., Graumann J., Gailus-Durner V., Rathmann W., et al. Deciphering the plasma proteome of type 2 diabetes. Diabetes. 2020;69:2766–2778. doi: 10.2337/db20-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rooney M.R., Chen J., Echouffo-Tcheugui J.B., Walker K.A., Schlosser P., Surapaneni A., Tang O., Chen J., Ballantyne C.M., Boerwinkle E., et al. Proteomic predictors of incident diabetes: results from the atherosclerosis risk in communities (ARIC) study. Diabetes Care. 2023;46:733–741. doi: 10.2337/dc22-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrannini G., Manca M.L., Magnoni M., Andreotti F., Andreini D., Latini R., Maseri A., Maggioni A.P., Ostroff R.M., Williams S.A., Ferrannini E. Coronary artery disease and type 2 diabetes: a proteomic study. Diabetes Care. 2020;43:843–851. doi: 10.2337/dc19-1902. [DOI] [PubMed] [Google Scholar]
- 10.Finan C., Gaulton A., Kruger F.A., Lumbers R.T., Shah T., Engmann J., Galver L., Kelley R., Karlsson A., Santos R., et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017;9:eaag1166. doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padhi S., Nayak A.K., Behera A. Type II diabetes mellitus: a review on recent drug based therapeutics. Biomed. Pharmacother. 2020;131:110708. doi: 10.1016/j.biopha.2020.110708. [DOI] [PubMed] [Google Scholar]
- 12.Zheng J., Haberland V., Baird D., Walker V., Haycock P.C., Hurle M.R., Gutteridge A., Erola P., Liu Y., Luo S., et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020;52:1122–1131. doi: 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudmundsdottir V., Zaghlool S.B., Emilsson V., Aspelund T., Ilkov M., Gudmundsson E.F., Jonsson S.M., Zilhão N.R., Lamb J.R., Suhre K., et al. Circulating protein signatures and causal candidates for type 2 diabetes. Diabetes. 2020;69:1843–1853. doi: 10.2337/db19-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghanbari F., Yazdanpanah N., Yazdanpanah M., Richards J.B., Manousaki D. Connecting genomics and proteomics to identify protein biomarkers for adult and youth-onset type 2 diabetes: a two-sample mendelian randomization study. Diabetes. 2022;71:1324–1337. doi: 10.2337/db21-1046. [DOI] [PubMed] [Google Scholar]
- 15.Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., Rutten-Jacobs L., Giese A.K., van der Laan S.W., Gretarsdottir S., et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan S., Wang L., Sun J., Yu L., Zhou X., Yang J., Zhu Y., Gill D., Burgess S., Denny J.C., et al. Genetically predicted sex hormone levels and health outcomes: phenome-wide Mendelian randomization investigation. Int. J. Epidemiol. 2022;51:1931–1942. doi: 10.1093/ije/dyac036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höglund J., Karlsson T., Johansson T., Ek W.E., Johansson Å. Characterization of the human ABO genotypes and their association to common inflammatory and cardiovascular diseases in the UK Biobank. Am. J. Hematol. 2021;96:1350–1362. doi: 10.1002/ajh.26307. [DOI] [PubMed] [Google Scholar]
- 18.Pigeyre M., Sjaarda J., Chong M., Hess S., Bosch J., Yusuf S., Gerstein H., Paré G. ACE and type 2 diabetes risk: a mendelian randomization study. Diabetes Care. 2020;43:835–842. doi: 10.2337/dc19-1973. [DOI] [PubMed] [Google Scholar]
- 19.Bartl J., Zanini M., Bernardi F., Forget A., Blümel L., Talbot J., Picard D., Qin N., Cancila G., Gao Q., et al. The HHIP-AS1 lncRNA promotes tumorigenicity through stabilization of dynein complex 1 in human SHH-driven tumors. Nat. Commun. 2022;13:4061. doi: 10.1038/s41467-022-31574-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A.C., Hung H.C., Chen Y.W., Cheng K.P., Li C.H., Lin C.H., Chang C.J., Wu H.T., Ou H.Y. Elevated Hedgehog-interacting protein levels in subjects with prediabetes and type 2 diabetes. J. Clin. Med. 2019;8:1635. doi: 10.3390/jcm8101635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan S., Larsson S.C. An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia. 2020;63:2359–2371. doi: 10.1007/s00125-020-05253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardinoglu A., Gogg S., Lotta L.A., Stančáková A., Nerstedt A., Boren J., Blüher M., Ferrannini E., Langenberg C., Wareham N.J., et al. Elevated plasma levels of 3-hydroxyisobutyric acid are associated with incident type 2 diabetes. EBioMedicine. 2018;27:151–155. doi: 10.1016/j.ebiom.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim A.K.H., Nikolic-Paterson D.J., Ma F.Y., Ozols E., Thomas M.C., Flavell R.A., Davis R.J., Tesch G.H. Role of MKK3-p38 MAPK signalling in the development of type 2 diabetes and renal injury in obese db/db mice. Diabetologia. 2009;52:347–358. doi: 10.1007/s00125-008-1215-5. [DOI] [PubMed] [Google Scholar]
- 24.Gu S., Lin S., Ye D., Qian S., Jiang D., Zhang X., Li Q., Yang J., Ying X., Li Z., et al. Genome-wide methylation profiling identified novel differentially hypermethylated biomarker MPPED2 in colorectal cancer. Clin. Epigenet. 2019;11:41. doi: 10.1186/s13148-019-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inshaw J.R.J., Sidore C., Cucca F., Stefana M.I., Crouch D.J.M., McCarthy M.I., Mahajan A., Todd J.A. Analysis of overlapping genetic association in type 1 and type 2 diabetes. Diabetologia. 2021;64:1342–1347. doi: 10.1007/s00125-021-05428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S., Swanson S.A., Labrecque J.A. Are Mendelian randomization investigations immune from bias due to reverse causation? Eur. J. Epidemiol. 2021;36:253–257. doi: 10.1007/s10654-021-00726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoungas S., Woodward M., Li Q., Cooper M.E., Hamet P., Harrap S., Heller S., Marre M., Patel A., Poulter N., et al. Impact of age, age at diagnosis and duration of diabetes on the risk of macrovascular and microvascular complications and death in type 2 diabetes. Diabetologia. 2014;57:2465–2474. doi: 10.1007/s00125-014-3369-7. [DOI] [PubMed] [Google Scholar]
- 28.Alviggi L., Johnston C., Hoskins P.J., Tee D.E., Pyke D.A., Leslie R.D., Vergani D. Pathogenesis of insulin-dependent diabetes: a role for activated T lymphocytes. Lancet. 1984;2:4–6. doi: 10.1016/s0140-6736(84)91994-9. [DOI] [PubMed] [Google Scholar]
- 29.Ma Z.J., Sun P., Guo G., Zhang R., Chen L.M. Association of the HLA-DQA1 and HLA-DQB1 alleles in type 2 diabetes mellitus and diabetic nephropathy in the Han ethnicity of China. J. Diabetes Res. 2013;2013:452537. doi: 10.1155/2013/452537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Liu S., Yu Z., Qiu X., Jiang R., Li W. Uncovering the gene regulatory network of type 2 diabetes through multi-omic data integration. J. Transl. Med. 2022;20:604. doi: 10.1186/s12967-022-03826-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groop L., Miettinen A., Groop P.H., Meri S., Koskimies S., Bottazzo G.F. Organ-specific autoimmunity and HLA-DR antigens as markers for beta-cell destruction in patients with type II diabetes. Diabetes. 1988;37:99–103. doi: 10.2337/diab.37.1.99. [DOI] [PubMed] [Google Scholar]
- 32.Sims G.P., Rowe D.C., Rietdijk S.T., Herbst R., Coyle A.J. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 33.Hudson B.I., Lippman M.E. Targeting RAGE signaling in inflammatory disease. Annu. Rev. Med. 2018;69:349–364. doi: 10.1146/annurev-med-041316-085215. [DOI] [PubMed] [Google Scholar]
- 34.Moin A.S.M., Nandakumar M., Kahal H., Sathyapalan T., Atkin S.L., Butler A.E. Heat shock-related protein responses and inflammatory protein changes are associated with mild prolonged hypoglycemia. Cells. 2021;10:3109. doi: 10.3390/cells10113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mir K.A., Pugazhendhi S., Paul M.J., Nair A., Ramakrishna B.S. Heat-shock protein 70 gene polymorphism is associated with the severity of diabetic foot ulcer and the outcome of surgical treatment. Br. J. Surg. 2009;96:1205–1209. doi: 10.1002/bjs.6689. [DOI] [PubMed] [Google Scholar]
- 36.Levada K., Guldiken N., Zhang X., Vella G., Mo F.R., James L.P., Haybaeck J., Kessler S.M., Kiemer A.K., Ott T., et al. Hsp72 protects against liver injury via attenuation of hepatocellular death, oxidative stress, and JNK signaling. J. Hepatol. 2018;68:996–1005. doi: 10.1016/j.jhep.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferkingstad E., Sulem P., Atlason B.A., Sveinbjornsson G., Magnusson M.I., Styrmisdottir E.L., Gunnarsdottir K., Helgason A., Oddsson A., Halldorsson B.V., et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021;53:1712–1721. doi: 10.1038/s41588-021-00978-w. [DOI] [PubMed] [Google Scholar]
- 38.Haslam D.E., Li J., Dillon S.T., Gu X., Cao Y., Zeleznik O.A., Sasamoto N., Zhang X., Eliassen A.H., Liang L., et al. Stability and reproducibility of proteomic profiles in epidemiological studies: comparing the Olink and SOMAscan platforms. Proteomics. 2022;22:e2100170. doi: 10.1002/pmic.202100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietzner M., Wheeler E., Carrasco-Zanini J., Kerrison N.D., Oerton E., Koprulu M., Luan J., Hingorani A.D., Williams S.A., Wareham N.J., Langenberg C. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat. Commun. 2021;12:6822. doi: 10.1038/s41467-021-27164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney M.R., Chen J., Ballantyne C.M., Hoogeveen R.C., Tang O., Grams M.E., Tin A., Ndumele C.E., Zannad F., Couper D.J., et al. Comparison of proteomic measurements across platforms in the atherosclerosis risk in communities (ARIC) study. Clin. Chem. 2023;69:68–79. doi: 10.1093/clinchem/hvac186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun B.B., Chiou J., Traylor M., Benner C., Hsu Y.-H., Richardson T.G., Surendran P., Mahajan A., Robins C., Vasquez-Grinnell S.G., et al. Genetic regulation of the human plasma proteome in 54,306 UK Biobank participants. bioRxiv. 2022 doi: 10.1101/2022.06.17.496443. Preprint at. [DOI] [Google Scholar]
- 42.Kurki M.I., Karjalainen J., Palta P., Sipilä T.P., Kristiansson K., Donner K., Reeve M.P., Laivuori H., Aavikko M., Kaunisto M.A., et al. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv. 2022 doi: 10.1101/2022.03.03.22271360. Preprint at. [DOI] [Google Scholar]
- 43.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M., GIANT Consortium Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larsson S.C., Woolf B., Gill D. Appraisal of the causal effect of plasma caffeine on adiposity, type 2 diabetes, and cardiovascular disease: two sample mendelian randomisation study. BMJ Med. 2023;2:e000335. doi: 10.1136/bmjmed-2022-000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brion M.J.A., Shakhbazov K., Visscher P.M. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce B.L., Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 2013;178:1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foley C.N., Staley J.R., Breen P.G., Sun B.B., Kirk P.D.W., Burgess S., Howson J.M.M. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun. 2021;12:764. doi: 10.1038/s41467-020-20885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 2021;17:e1009440. doi: 10.1371/journal.pgen.1009440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. All GWAS are referenced. This paper does not report the original code. Any additional information required to reanalyze the data reported in this report is available from the lead contact upon request.