Abstract
In 2021, breast cancer accounted for a substantial proportion of cancer cases and represented the second leading cause of cancer deaths among women worldwide. Although tumor cells originate from normal cells in the human body, they possess distinct biological characteristics resulting from changes in gene structure and function of cancer cells in contrast with normal cells. These distinguishing features, known as hallmarks of cancer cells, differ from those of normal cells. The hallmarks primarily include high metabolic activity, mitochondrial dysfunction, and resistance to cell death. Current evidence suggests that the fundamental hallmarks of tumor cells affect the tissue structure, function, and metabolism of tumor cells and their internal and external environment. Therefore, these fundamental hallmarks of tumor cells enable tumor cells to proliferate, invade and avoid apoptosis. Modifying these hallmarks of tumor cells represents a new and potentially promising approach to tumor treatment. The key to breast cancer treatment lies in identifying the optimal therapeutic agent with minimal toxicity to normal cells, considering the specific types of tumor cells in patients. Some herbal medicines contain active ingredients which can precisely achieve this purpose. In this review, we introduce Ginsenoside's mechanism and research significance in achieving the therapeutic effect of breast cancer by changing the functional hallmarks of tumor cells, providing a new perspective for the potential application of Ginsenoside as a therapeutic drug for breast cancer.
Keywords: Ginsenoside, Breast cancer, Hallmarks of cancer, Apoptosis, Epithelial-mesenchymal transition, Cell growth
Introduction
In 2021, breast cancer became the most common malignancy in women worldwide, accounting for more than 30% of the total number of newly diagnosed cancers. According to the American Cancer Society, about 281,500 women were diagnosed with breast cancer in 2021 in the United States, resulting in 43,600 deaths. Predictive modeling suggests that around 287,850 new cases of breast cancer and approximately 43,250 deaths are projected in the United States in 2022 [1]. The high incidence and number of deaths caused by breast cancer impose a severe economic burden on countries worldwide, placing immense importance on ensuring public safety and well-being.
It has been found that breast cancer can be classified into three main groups according to whether or not the patient's breast cancer cells express estrogen receptor/progesterone receptor, human epidermal growth factor 2 receptor (HER2) [2]: ER- or PR- positive (also known as luminal type breast cancer), HER2 positive cancer and triple-negative breast cancer (TNBC) (ER-/PR-/HER2-). Among them, luminal B breast cancer can be divided into two main categories, HER2 positive and HER2 negative, based on the presence or absence of positive expression of HER2. It is generally believed that triple-negative breast cancer has large heterogeneity with large differences in cell molecular types. However, Lehmann et al. [3] performed a cluster analysis of 21 groups of gene expression profiling data in 587 cases of TNBC. They found that TNBC could be further classified into six subtypes, including basal-like type 1 (BL1), basal-like type 2 (BL2), immune modulatory type (IM), luminal androgen receptor type (LAR), mesenchymal type (M), and mesenchymal stem-like cell type (MSL). These six subtypes represent distinct molecular features of TNBC and different therapeutic strategies (Fig. 1, Table 1).
Fig. 1.
Molecular typing and treatment of breast cancer. The current molecular typing of breast cancer is mainly based on the surface receptors of the breast cancer cell membrane, which is mainly divided into luminal type (A), HER2 type (B), and Triple-negative type (C). At present, due to the expression of hormone receptors on the membrane surface, anti-hormone therapy (AI, TAM, CDK4/6i) is indicated for luminal breast cancer. The primary treatment of HER2 breast cancer is targeted anti-HER2 monoclonal antibody therapy (HER2-TKI, HER2-ADC, HER2 inhibitor). Triple-negative breast cancer can also be divided into six categories (BL1, BL2, IM, LAR, M, MSL, MSL) according to the different activation characteristics of intracellular signaling pathways. Chemotherapy is the main treatment choice for triple-negative breast cancer. However, with the deepening of research, some new anti-tumor drugs (PI3K inhibitors, PD-1 inhibitors, Trop2-ADC, anti-angiogenic drugs) also show the dawn in the clinical treatment of triple-negative breast cancer
Table 1.
Molecular typing of breast cancer
| Molecular typing (Molecular subtype) | Molecular characteristics | Possible treatment strategies | Cell line names represented | ||||
|---|---|---|---|---|---|---|---|
| HER2 | ER | PgR | Ki67 | Other molecular features | |||
| HER2 positive | Positive | – | – | Whatever | / | Anti-Her2 treatment | SK-BR-3, AU-565, MDA-MB-453 |
| Luminal A | – | Positive | Positive and high expression of | Low expression | / | Endocrine therapy | MCF-7, T47D, SUM185 |
| Luminal B (HER2 negative) | – | Positive | Low expression or - | High expression | / | Endocrine therapy combined with chemotherapy | / |
| Luminal B (HER2 positive) | Positive | Positive | Whatever | Whatever | / | Anti-Her2 treatment with endocrine therapy combined | BT-474, ZR-75-1 |
| Triple-negative | – | – | – | Whatever | / | Chemotherapy based comprehensive treatment | MDA-MB-468, SUM190, BT549, MDA-MB-231, Hs578T, SUM1315, BT483,MCF-12A, HBL101, HS598T (Appropriate cell lines were selected based on different populations and genetic circumstances) |
| Triple-negative (BL1) | / | / | / | / | Highly expressed cell cycle and DNA damage response genes | Platinum based chemotherapeutics, PARP inhibitors | |
| Triple-negative (BL2) | / | / | / | / | Increased activity of growth factor pathways and PI3K pathway | mTOR inhibitors, growth factor inhibitors | |
| Triple-negative (IM) | / | / | / | / | Highly expressed immune-related genes | Immune checkpoint inhibitors | |
| Triple-negative (LAR) | / | / | / | / | AR signaling pathway activation; enrichment of PIK3CA mutations | Antiandrogens, PI3K inhibitors | |
| Triple-negative (M) | / | / | / | / | EMT characteristics; growth factor pathway activation | mTOR inhibitors, growth factor inhibitors | |
| Triple-negative (MSL) | / | / | / | / | EMT characteristics; growth factors and PI3K pathway activation | mTOR inhibitors, PI3K inhibitors, growth factor inhibitors | |
In clinical practice, breast cancer cells in patients are classified and treated based on various factors, including tumor growth type (in situ or invasive) and specific classifications. Treatment interventions typically involve a combination of surgical resection, radiation therapy, immunotherapy, targeted therapy, and chemotherapy [4, 5]. However, determining the optimal treatment for patients is often quite challenging because of the difficulty in determining the appropriate dose of therapeutic drugs for different patients and the optimal surgical resection margin. Moreover, different therapeutic agents can adversely affect normal tissue cells, leading to reduced bone mineral density, immune-related inflammation, and diarrhea, all of which negatively impact the patient’s quality of life. And the effects that different therapeutic agents bring to normal tissue cells, such as reduced bone density, immune related inflammation and diarrhea, all give a reduced quality of life of patient [6]. Additionally, patients may experience ineffective treatment if the therapeutic drug dosage needs to be reduced due to intolerance to side effects, resulting in suboptimal therapeutic levels [7]. As research in breast cancer-related molecular biology advances, an increasing number of molecular mechanisms that contribute to the proliferation and invasion of breast cancer are being discovered. This research has led to the development of more small-molecule drugs that specifically induce the death of breast cancer cells while sparing normal cells, offering targeted treatment options for breast cancer [8]. Additionally, experiments have demonstrated the promising potential of various active ingredients found in natural products to treat breast cancer [9]. Further exploration of these natural compounds has revealed their ability to partially replace chemotherapy drugs in killing tumor cells. Moreover, they can help reduce the side effects associated with chemotherapy, enhance the efficacy of tumor cell eradication, and potentially lower the required doses of chemotherapeutic drugs used in clinical practice [10].
The fundamental characteristics of tumor cells encompass their distinct biological capabilities acquired during the progressive transformation from normal cells to tumor cells [11]. These characteristics serve as a framework for understanding the intricacies of neoplastic diseases and account for the significant heterogeneity observed among tumor cells. The specific biological abilities of tumor cells consist of fourteen fundamental traits, including sustaining proliferative signals, evading growth inhibitory factors, resisting cell death, and achieving replicative immortality. These hallmarks are underpinned by genomic instability, which further contributes to the growing genetic diversity observed among tumor cells. Consequently, tumor cells exhibit increasingly disparate hallmarks compared to normal cells [12]. The hallmarks of tumor cells play a vital role in maintaining tumor survival and development conditions and promoting their growth and development. Currently, research on tumor treatment extends beyond focusing solely on tumor cells and encompasses the various components within the tumor microenvironment. The aim is to induce tumor cell apoptosis by targeting and inhibiting the fundamental characteristics of tumors. It has been established that the proliferation, invasion, metastasis, immune evasion, and angiogenesis of tumor cells are outcomes of the intricate interplay between tumor cells and the tumor microenvironment. These processes are considered vital hallmarks of tumor cells [13]. However, targeting one or more of the basic hallmarks of tumors can effectively inhibit tumor angiogenesis, inhibit tumor release of pro-inflammatory mediators, and so on, to induce tumor apoptosis [14]. At present, the most commonly used anti-angiogenic drugs in the clinic are drugs targeted at inhibiting the angiogenesis of tumor tissues and inducing tumor cell apoptosis. For instance, anti-angiogenic drugs can block neoendothelial angiogenesis necessary for tumor cell proliferation by inhibiting various angiogenic factors such as VEGF and PDGF and cause tumor cells to lack the nutrients required for growth to induce apoptosis [15]. There is a rich literature substantiating the ability of Radix Ginseng and its active components to effectively modify the fundamental characteristics of tumor cells, thereby achieving therapeutic effects in tumor treatment [16].
Over the years, significant progress has been achieved in identifying and understanding the active components present in Radix Ginseng. These bioactive components encompass a variety of compounds, including saponins, polysaccharides, flavonoids, volatile oils, and fatty acids [17]. Ginsenosides, which are the most abundant and diverse active compounds in Radix Ginseng, have received considerable attention for their potential in the treatment of breast cancer. They have been extensively studied and reported for their anti-tumor properties, immune-enhancing effects, and stress-relieving capabilities [18]. This manuscript provides a comprehensive review of the mechanism and research significance of ginsenosides in treating breast cancer by modulating the tumor microenvironment. Our findings offer a novel perspective on the potential therapeutic application of ginsenosides as a valuable drug in breast cancer treatment.
The basic hallmarks of tumor cells are necessary for the survival of breast cancer cells
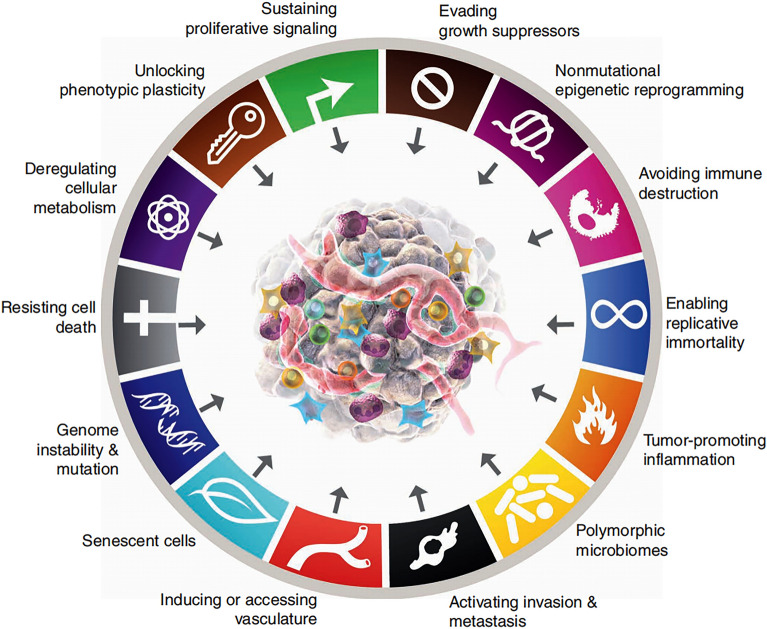
The fundamental hallmark features of cancer encompass a series of functional capabilities that human cells acquire during their transition from a normal state to tumor growth. These functional abilities are crucial for tumor cells to develop into malignant tumors. Breast cancer cells possess these functional capabilities acquired during the growth phase of malignant tissues. Three versions of Hanahan’s classic reviews on cancer features totaling 14 basic cancer features have been published, and these reviews have become wind vanes in basic cancer research and clinical research (Fig. 2) [12, 19, 20]. The basic hallmarks of cancer summarized so far include: Sustaining proliferative signaling, Evolving growth suppressors, Deregulating cellular metabolism, Avoiding immune destruction, Resisting cell death, Enabling replicative immortality, Genome instability & mutation, Tumor-promoting inflammation, Inducing or accessing vascular, Activating invasion & metastasis, Unlocking phenotypic plasticity, Nonmutational epigenetic reprogramming, Senescent cells, Polymorphic microbiomes. The fundamental hallmarks of breast cancer are a response to the complexity of the pathogenesis of breast cancer, but these characteristics still cannot accurately explain the complex pathogenesis of breast cancer. The precise molecular and cellular mechanisms underlying the development and acquisition of these aberrant phenotypic capabilities during the progression of breast cancer have been found to originate from pre-existing breast cancer progenitor cells. These mechanisms have evolved and acquired over time, leading to the manifestation of these capabilities in breast cancer cells [21]. Like other tumor cells, breast cancer cells do not have these basic hallmarks during the early stages of tumor development. Breast cancer cells also acquire these malignant features gradually during malignant progression [22]. In advanced breast cancer, cancerous cells possess comprehensive abilities to regulate abnormal phenotypes, posing a greater challenge for effective eradication [23]. However, it has been found that various ginsenosides can play a single or comprehensive targeted therapeutic role according to different characteristics of breast cancer [24]. Therefore, ginsenoside and its metabolic components hold significant potential in relevant fields focused on targeting the fundamental characteristics of breast cancer cells to treat breast cancer [25].
Fig. 2.
Schematic of Hallmarks of cancer. Hallmarks of Cancer is a process of continuous discovery. Currently, 14 Hallmarks of Cancer have been found: sustaining proliferative signaling, Evolving growth suppressors, Deregulating cellular metabolism, Avoiding immune destruction, Resisting cell death, Enabling replicative immortality, Genome instability & mutation, Tumor-promoting inflammation, Inducing or accessing vascular, Activating invasion & metastasis, Unlocking phenotypic plasticity, Nonmutational epigenetic reprogramming, Senescent cells, Polymorphic microbiomes. The first version of Hallmarks of Cancer was proposed in 2000; six basic Hallmarks of cancer were proposed at that time (Sustaining proliferative signaling, Evolving growth suppressors, Resisting cell death, Enabling replicative immortality, Inducing or accessing vascular, Activating invasion & metastasis). In 2011, four basic Hallmarks were added to the Hallmarks of Cancer (Avoiding immune destruction, Genome instability & mutation, Tumor-promoting inflammation, and Deregulating cellular metabolism). In 2022, four basic Hallmarks were added again (Senescent cells, Polymorphic microbiomes, Unlocking phenotypic plasticity, Nonmutational epigenetic reprogramming). In the future, more Hallmarks of Cancer will be found and added, which will help cancer researchers to continuously discuss and experimentally elaborate relevant treatments for cancer
Ginsenoside is the main active ingredient of Radix Ginseng, exhibiting anti-breast cancer effects
Radix Ginseng is the dried root or rhizome of the Araliaceae plant ginseng, officially known as “Panax ginseng C. A. Meyer” [26]. In East Asia, Radix Ginseng has long been used to treat and prevent various diseases mainly manifested as body energy deficiency. Approximately 2500 years ago, the first book in China was written, documenting the usage of ginseng. The records indicate that the primary function of Radix Ginseng is its tonic effect, which can nourish the body and provide energy [27]. These beneficial effects can be attributed to its active components, with ginsenoside and its metabolites being the predominant substances found in Radix Ginseng. These components are considered the primary active constituents responsible for the pharmacological effects of Radix Ginseng. Ginsenoside, a water-soluble sterol compound, is commonly referred to as “saponin” because it produces a long-lasting soap-like foam when shaken, which persists even after heating [28].
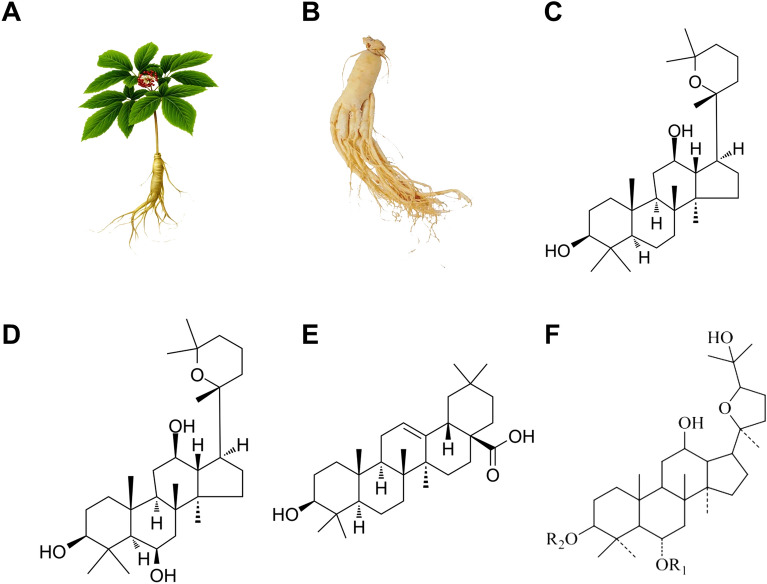
It is now understood that ginsenoside has a dammarane triterpenoid structure. Currently, ginsenoside can be divided into four categories according to the number of steroid backbone and attached glycosyl or hydroxyl groups (Fig. 3, Table 2) [29]. The protopanaxadiol-type ginsenoside comprises ginsenoside Ra1, Ra2, and Ra3, while protopanaxatriol-type ginsenoside includes ginsenoside Re, Rf, and Rg1. Oleanolic Acid-type ginsenoside consists of ginsenoside R0, Rh3, and R1, while Ocotillol-type ginsenoside includes ginsenoside F11. At present, research on Radix Ginseng has revealed that most ginsenosides contained in Radix Ginseng belong to protopanaxadiol type saponin and protopanaxatriol type saponin, while oleanolic acid type ginsenoside accounts for only a smaller proportion [18]. Current evidence suggests that most ginsenoside protopanaxadiol-type and protopanaxatriol-type saponins have definite anti-breast cancer activity [25]. Besides, laboratory studies have confirmed that besides protopanaxadiol and protopanaxatriol, the secondary metabolic derivatives of ginsenoside yield a significant anti-breast cancer effect. Furthermore, laboratory studies have confirmed that secondary metabolic derivatives of ginsenosides, apart from the protopanaxadiol-type and protopanaxatriol-type, also considerably impact anti-breast cancer properties [30]. Although different ginsenosides exhibit varying pharmacological actions against breast cancer, the molecular signals and mechanisms of action differ among the different ginsenosides, rendering the molecular mechanism complex. Consequently, developing ginsenoside preparations with anti-breast cancer effects has become an important aspect of ginsenoside commercialization.
Fig. 3.
Formula of four chemical structures of ginsenoside, Panax ginseng C. A. Meyer plant morphology, and Panax ginseng C. A. Meyer medicinal part. A Panax ginseng C. A. Meyer plant morphology; B the main medicinal part of Panax ginseng C. A. Meyer—Morphological diagram of Panax ginseng C. A. Meyer; C a chemical structure diagram of Protopanaxadiol-type ginsenoside; D a chemical structure diagram of Protopanaxatriol-type ginsenoside; E a chemical structure diagram of Oleanolic Acid-type ginsenoside; F a chemical structure diagram of Ocotillol-type ginsenoside
Table 2.
Structural classification of ginsenosides
| Structure type | Ginsenoside specific name | References |
|---|---|---|
| Panaxadiol | Compound K | [130, 159] |
| Rh2 | [66, 89, 103, 115, 116, 118, 119, 132, 150, 157, 169] | |
| Rg3 | [58, 95, 101, 102, 113, 114, 117, 131, 147, 148, 156] | |
| Rb1 | [78, 79] | |
| Rg5 | [60, 61] | |
| Rk1 | [62, 63] | |
| Rd | [94, 158] | |
| 20(S)-PPD | [160] | |
| Rg6 | [91] | |
| 20(S)-Rh3 | [59] | |
| 1f | [69] | |
| Rb3 | [149] | |
| Panaxatriol | Ginsenoside panaxatriol | [49] |
| Rh1 | [70–72] | |
| Rg2 | [75, 76, 167, 168] | |
| Rh4 | [68] | |
| Rg1 | [64, 65, 90, 120, 170] | |
| 20(S)-PPT | [77] |
Ginsenosides can be divided into four categories according to different chemical structures. But there is no indication that the different structures of ginsenosides will affect their treatment in different molecular types of breast cancer. Different types of ginsenosides have different steroid skeletons and the number of attached sugar groups or hydroxyl groups, but the main body of ginsenosides is still dammarane triterpenoid structure. Therefore, the next research direction should focus on the related research of the biological impact of different chemical structures of ginsenosides on different molecular types of breast cancer.
Metabolic shift and pharmacokinetic changes of ginsenosides in vivo
Given that ginsenosides represent major active components in traditional Chinese Radix Ginseng, oral administration remains the predominant route for their administration until other routes are proven effective and safe [31]. Indeed, when conducting in vivo experiments with ginsenosides, it is crucial to consider their pharmacokinetics. Although studies on the pharmacokinetics of ginsenosides have primarily focused on in vitro cellular models, animal models, and healthy human volunteers [32], the most active research areas are in vitro cellular models and animal models. However, it remains uncertain whether the metabolism of different ginsenosides differs significantly. The general understanding of ginsenoside metabolism is that, after oral administration, ginsenosides undergo partial or complete hydrolysis under acidic conditions by gastric and intestinal microflora, producing active ingredients [33]. The most well-studied ginsenoside, Rg3 and other protopanaxadiol ginsenosides undergo deglycosylation upon metabolism by anaerobic intestinal bacteria. On the other hand, compound K, the end metabolite of some ginsenosides, can be detected in human plasma as early as seven hours after ingestion [34]. In vitro studies have shown that Rg3 in ginsenosides interacts with cytochrome P450 isoenzymes, indicating that some ginsenosides may affect the metabolism of normal anti-tumor drugs. Ginsenoside Rg3 from Radix Ginseng was found to have a weak inhibitory effect on CYP3A4, a moderate inhibitory effect on CYP2C19, CYP1A2, and a potent inhibitory effect on CYP2D4 [35]. Therefore the possible effects of ginsenosides on their production need to be considered when using drugs metabolized via these isozymes. Gut flora is also very important for the metabolism of ginsenosides. Current evidence suggests that some ginsenosides are transformed into other types of ginsenosides after decomposition by the gut flora. For example, ginsenoside Rg3 can be transformed into ginsenoside Rh2 by incubation with intestinal flora in vivo [36]. Although in vitro studies have shown that deglycosylation is one of the major pathways for ginsenoside Rg3 metabolism, studies have not found ginsenosides with complete deglycosylation in animals [37]. The effects of different modes of administration on the metabolic half-life of ginsenosides are highly heterogeneous. When ginsenoside Rg3 was administered intravenously in rats, its half-life was 14 min [38] and 18.5 min [39] in separate studies. The variations in the metabolic time of ginsenoside Rg3 in rats can be attributed to the dissimilar solubility of ginsenoside Rg3 and heterogeneity in its absorption and utilization within rats. These studies overlap in their assertion that ginsenoside Rg3 has high metabolic clearance and low bioavailability when administered intravenously in rats. However, it is important to consider that species differences between rats and humans may influence these findings. In healthy humans, the plasma half-life of ginsenoside Rg3 can reach up to 8 h [40] and 216 h [41] following oral and intramuscular administration, respectively. These results suggest that ginsenoside Rg3 exhibits a longer half-life and potentially higher bioavailability in humans compared to rats. Nevertheless, oral administration remains the most predominant mode of administration of ginsenosides in clinical studies and basic research at present.
The tissue distribution concentration of a drug is a critical factor influencing the treatment of breast cancer with ginsenosides after oral administration [42]. Following the oral administration of ginsenoside 20(S)-Rg3 to rats at a dosage of 68 mg/kg, the concentration of 20(S)-Rg3 in gastrointestinal tissues was the highest, suggesting its significant impact on the digestive tract. However, 20(S)-Rg3 concentrations in other tissues such as muscle, spleen, lung, and adipose were similar to or lower than plasma concentrations. Additionally, 20(S)-Rg3 trace amounts were detected in the brain, heart, and kidney [42]. This illustrates that through oral administration, ginsenoside 20(S)-Rg3 may exert effects on tissues throughout the body. Interestingly, when 20(R)-Rg3 was administered using various routes of administration, its accumulation was predominantly observed in the liver and gastrointestinal tract, without its presence detected in plasma, suggesting that oral administration should remain the primary route for ginsenoside administration in future studies, with the gastrointestinal tract being the most directly affected organ by ginsenosides [42]. However, there is currently no available research on the metabolism of ginsenosides in tumor patients. Therefore, future research should investigate whether there are any differences in the metabolism of ginsenosides between tumor patients and healthy individual (Tables 3, 4).
Table 3.
Classification of anti-breast cancer effects of ginsenosides based on the basic hallmarks of tumors (2015–2023)
| Ginsenoside type | Anti-breast cancer activity | Cell line | In Vivo/In Vitro | Type of tumor hallmarks | Target spot/signaling pathway | Refs. |
|---|---|---|---|---|---|---|
| Rg3 | Activation of exogenous death pathway | MDA-MB-231, MDA-MB-453, BT-549 | In Vivo/In Vitro | Resisting cell death | NF-κB p65, Bcl-2, Bax, caspase-3 | [58] |
| 20(S)-Rg3 | Increase cell radiosensitivity and induce cell apoptosis | MDA-MB-231 | In Vitro | Resisting cell death | Unknown | [59] |
| Rg5 | Inducing apoptosis and autophagy death | MCF-7 | In Vivo | Resisting cell death | PI3K/Akt pathway | [60] |
| Rg5 | Caspase-dependent apoptosis can be induced by activating external death receptors and internal mitochondrial signaling pathways, and autophagy can be promoted | MCF-7 | In Vitro | Resisting cell death | PI3K/Akt signaling pathway | [61] |
| Rk1 | Activation of mitochondrial-mediated endogenous death pathway | MCF-7 | In Vitro | Resisting cell death | PTEN/PI3K/Akt/mTOR signaling pathway | [62] |
| Rk1 | Activation of mitochondrial-mediated endogenous death pathway | MDA-MB-231 | In Vitro | Resisting cell death | ROS/PI3K/Akt pathway | [63] |
| Rg1 | Increase cell DNA damage and induce cell apoptosis | MDA-MB-231 | In Vitro | Resisting cell death | Akt, ERK, MAPK,ATM, H2AX, Rad51, TP53, BCl2, CDK2, NF-κB, STAT-3, MAPK, iNOS, MMP2, MMP9, TGFB1, VEGFA, EGFR, SOD, Catalase, GPx, GSH XRCC1, p21, TP53, apaf1, Bax, CASP3, CASP9, ROS, mitochondrial membrane potential | [64] |
| Rg1 | Promote cell DNA damage, prevent the elevation of oxidative damage markers, restore antioxidant enzymes to near normal levels, inhibit the expression of cell proliferation and survival-related markers, regulate apoptosis markers, and downregulate invasion and angiogenesis markers | MB-MD-231 | In Vitro | Resisting cell death, Tissue Invasion And Metastasis, Sustained Angiogenesis, Deregulating Cellular Energetics | TP53, Bcl-2, CDK2, NF-κB, STAT-3, MAPK, iNOS, MMP2, MMP9, TGFB1, VEGFA, EGFR, SOD, Catalase, GPx, GSH | [65] |
| Rh2 | Upregulation of tumor suppressor gene expression, activation of extrinsic death pathway, and induction of apoptosis | MCF-7 | In Vitro | Resisting cell death | caspase-9/p38 MAPK, p53, Bax, Bcl-2、PRAP | [66] |
| Rh2 | Activation of the mitochondrial-mediated endogenous death pathway | MCF-7 | In Vitro | Resisting cell death | Mitochondrial pathway | [67] |
| Rh4 | Activation of extrinsic death pathways | MCF-7 | In Vivo/In Vitro | Resisting cell death | Bcl-2, Bax, caspase-8, caspase-3, PARP | [68] |
| 1f | Activation of the mitochondrial-mediated endogenous death pathway | MCF-7 | In Vitro | Resisting cell death | Unknown | [69] |
| Rh1 | Induction of apoptosis and autophagic death | MCF-7, HCC1428 | In Vivo/In Vitro | Resisting cell death | ROS/PI3K/Akt pathway | [70] |
| Rh1 | Induce mitochondrial dysfunction, activate mitochondria-mediated endogenous and exogenous death pathways | MDA-MB-231, BT549 | In Vivo/In Vitro | Resisting cell death | PERK/eIF2α/ATF4/CHOP pathway, mtROS, caspase-3 | [71] |
| Rh1 | Activation of the mitochondrial-mediated endogenous death pathway | MD-MB-231 | In Vitro | Resisting cell death, tissue invasion, and metastasis | STAT3/NF-κB pathway, MMP2, MMP9, ROS, VEGF-A | [72] |
| Rg2 | Induction of autophagic cell death | MCF-7 | In Vitro | Resisting cell death | p-p53, p-AMPK, p-ACC, Atg-7, LC3-II, p62 | [75] |
| Rg2 | Activation of the mitochondrial-mediated endogenous death pathway | MCF-7 | In Vitro | Resisting cell death | AMPKpathway, mTOR | [76] |
| 20(S)-PPT | Induction of apoptosis and nonprotective autophagy | MDA-MB-231, SUM-15-PT | In Vivo/In Vitro | Resisting cell death | Akt/mTOR signaling pathway | [77] |
| Rb1 | Increase irreversible cell death under short infrared low light | MCF-7, 4T1 | In Vitro | Resisting cell death | Unknown | [78] |
| Rb1 | Induction of apoptosis | MCF-7 | In Vitro | Resisting cell death | Unknown | [79] |
| Rh2 | Inhibit cell proliferation, promote apoptosis, and cycle arrest | MCF-7, MD-MB-231 | In Vivo/In Vitro | Self-sufficiency in growth signals | ERβ-TNFα pathway | [89] |
| Rg1 | Exert estrogen regulation antagonism and inhibit cell proliferation | MCF-7 | In Vivo/In Vitro | Self-sufficiency in growth signals | ER signalosome, EGFR, c-SrcER, cavolin-1 | [90] |
| Rg6 | It can change the paclitaxel resistance of cells by changing the chromosomal instability induced by stress hormones or steroid hormones, reduce the mitotic speed of cells, and inhibit cell proliferation | MCF-7, MDA-MB-468 | In Vitro | Self-sufficiency in growth signals, Non-mutational epigenetic reprogramming | γ-tubulin, MTOC, GR, ER-α pathway | [91] |
| Rd | Eliminated VEGF-induced sprouting of blood vessels and inhibited the formation of blood vessels, and inhibited cell proliferation | MDA-MB-231 | In Vivo/In Vitro | Sustained angiogenesis | Akt/mTOR/P70S6 pathway | [94] |
| Rg3 | Inhibit proliferation, invasion, and angiogenesis, and enhance autophagy | MCF-7 | In Vivo | Sustained angiogenesis, resisting cell death, tissue invasion and metastasis | EGFA, VEGFB, VEGFC, MMP2, MMP9, p62, mTOR, PI3K, Akt, JNK, Beclin-1, LC3-II/LC3-3 I | [95] |
| Rg3 | Reduces cell stem-like properties and inhibits proliferation | MDA-MB-231, MCF-7 | In Vitro | Unlocking phenotypic plasticity, tumor promotion inflammation, resisting cell death, tissue invasion, and metastasis | Akt-mediated self-renewal signaling | [101] |
| Rg3 | Reduce the level of stemness markers of tumor cells, inhibit proliferation and metastasis, and promote apoptosis | MDA-MB-231, HCC1143 | In Vivo/In Vitro | Unlocking phenotypic plasticity, tissue invasion and metastasis, resisting cell death | Akt/mTOR pathway, CD44, ALDH | [102] |
| Rh2 | Inhibit the senescence phenotype and secretion phenotype of normal breast epithelial cells caused by doxorubicin treatment, | MCF-7, MCF-10A | In Vitro | Unlocking phenotypic plasticity, tumor promotion inflammation | NF-κB pathway, ROSj, SIRT 3, SIRT 5, SOD1, SOD2 | [103] |
| Panaxatriol | It can change the phenotype of paclitaxel-resistant cells, inhibit the expression of inflammatory factors and stem cell-related genes, inhibit proliferation and invasion, and induce apoptosis | MDA-MB-231 PTX, SUM159-PR | In Vivo/In Vitro | Unlocking phenotypic plasticity, tumor promotion inflammation, resisting cell death, tissue invasion, and metastasis | IRAK1/NF-κB and ERK pathways | [49] |
| Rg3 | Regulating promoter methylation of noncoding RNA inhibits cell proliferation and induces apoptosis | MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, resisting cell death, deregulating cellular energetics, tissue invasion and metastasis | lncRNA STXBP5-AS1, lncRNA RFX3-AS1, STXBP5, GRM1, RFX3, SLC1A1 | [113] |
| Rg3 | Regulating promoter methylation of noncoding RNA inhibits cell proliferation and induces apoptosis | MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, resisting cell death, deregulating cellular energetics | lnc RNA ATXN8OS, miR-424-5p, EYA1, DACH1, CHRM3 | [114] |
| Rh2 | Regulating promoter methylation of noncoding RNA inhibits cell proliferation and induces apoptosis | MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, resisting cell death, deregulating cellular energetics | ACOX2, FAM107A, lncRNA C3orf67-AS1 | [115] |
| Rh2 | Regulation of cell noncoding RNA levels inhibits cell proliferation | MCF-7, MDA-MB-231, T47D | In Vitro | Non-mutational epigenetic reprogramming, deregulating cellular energetics | lnc RNA CFAP20DC-AS1, BBX, TNFAIP3 | [116] |
| Rg3 | It inhibits tumor cell proliferation and promotes apoptosis by affecting gene methylation levels to affect the expression levels of tumor-associated proteins | MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, resisting cell death, deregulating cellular energetics | cell morphology-related pathway, TRMT1L, PSMC6, NOX4, ST3GAL4, RNLS, KDM5A | [117] |
| Rh2 | It inhibits tumor cell proliferation and promotes apoptosis by affecting gene methylation levels to affect the expression levels of tumor-associated proteins | MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, resisting cell death, deregulating cellular energetics, avoiding immune destruction | CASP1, INSL5, OR52A1, CLINT1, ST3GAL4, C1orf198 | [118] |
| Rh2 | Regulating m6A methylation levels regulates the transcriptional activity and subcellular localization of oncogenic proteins | MDA-MB-157 , MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, deregulating cellular energetics | KIF26B, m6A RNA, ZC3H13/CBLL1 | [119] |
| Rg1 | Promoting mitotic defects in cells leads to delayed mitotic progression to inhibit cell proliferation | MDA-MB-231, MCF-7 | In Vitro | Non-mutational epigenetic reprogramming, deregulating cellular energetics | Haspin, H3T3ph, Aurora B | [120] |
| CK | Reduce cellular glutamine utilization levels to inhibit proliferation and trigger apoptosis | MCF-7, BT474, MDA-MB-231, SUM159, HCC 1806 | In Vivo/In Vitro | Deregulating cellular energetics, resisting cell death, deregulating cellular energetics | ATP, GLS1, GSH, ROS | [130] |
| Rg3 | Inhibit glucose uptake by binding glucose transporters, reverse the level of immunosuppression in the tumor microenvironment, and reduce the levels of CAFs and collagen in the tumor microenvironment | 4T1 | In Vivo | Deregulating cellular energetics, avoiding immune destruction | Glut1, TGF-β/Smad pathway, CAFs, Collagen level, CD4, CD8, CD86, CD206, CD11b+/Gr-1+, CD4+FoxP3+, CD45 | [131] |
| Rh2 | Regulates the mitochondrial apoptotic pathway, inhibits glycolysis, and inhibits mitochondrial respiration | MCF-7 | In Vitro | Deregulating cellular energetics, resisting cell death, deregulating cellular energetics | ROS, ATP, caspase-9, Bax, HK II | [132] |
| Rg3 | Reverse drug resistance, rebuild TME, change macrophage phenotype, inhibit the expression of MDSC, AFs, and collagen fibers, improve tumor-associated inflammation, and promote tumor cell apoptosis | MCF-7 | In Vivo/In Vitro | Avoiding immune destruction, Tumor promotion inflammation, resisting cell death | IL-6/STAT3/p-STAT3 pathway, MDSC, macrophages, TAFs, collagen fibers | [147] |
| Rg3 | Reverses the level of immunosuppression in the tumor microenvironment and kills tumor cells by binding glucose-specific transporters | 4T1 | In Vivo | Avoiding immune destruction, tissue invasion, and metastasis | STAT3, CCL2, CD4, CD8, CD86, CD206, CD11b+/Gr-1+, CD4+FoxP3+, CD45, NF-κB, Bcl-2, Bax | [148] |
| Rg3 | Reduce PD-1 expression of activated T cells and increase cytokine levels to promote T cell recognition and killing of breast cancer cells | MDA-MB-231 , BT-549 | In Vivo/In Vitro | Avoiding immune destruction, resisting cell death | IFN-γ, IL-2, IL-9, IL-10, IL-22, IL-23, PD-1, PD-L1 | [149] |
| Rh2 | Improve tumor-associated inflammation, remodel the structure of TME, and reverse the immunosuppressive environment | 4T1 | In Vivo/In Vitro | Avoiding immune destruction, resisting cell death, Tumor promotion inflammation | α-SMA, TAFs, CD31, CD11b+/F4/80+/CD86+, M1 macrophage, M2 macrophage, IL-6, CD4, CD8 | [150] |
| Rg3 | Inhibit angiogenesis and cell invasion, enhance autophagy, and inhibit cell proliferation | MCF-7 | In Vitro | Tissue invasion and metastasis, sustained angiogenesis, resisting cell death, deregulating cellular energetics | VEGFA, VEGFB, VEGFC, MMP2, MMP9, p62, mTOR, PI3K, Akt, JNK, Beclin-1, LC3-II/LC3-I | [156] |
| Rh2 | Reduce cell invasion-related proteins to inhibit cell invasion | MDA-MB-231, MCF-7 | In Vitro | Tissue invasion and metastasis | Anxa2-K301A, NF-κB, E-cadherin, N-cadherin, Snail1, Twist, Slug, SIP1, MMP-2, MMP-9, Myc | [157] |
| Rd | Changing the expression level of non-coding RNA inhibits cell invasion | 4T1 | In Vivo/In Vitro | Tissue invasion and metastasis | microRNA-18a, Smad2 | [158] |
| CK | Inhibit cell proliferation and invasion, induce apoptosis | MCF-7 | In Vitro | Tissue invasion and metastasis, resisting cell death | N-cadherin, vimentin, FN, E-cadherin, PI3K/Akt pathway | [159] |
| 20(S)-PPD | Reducing the expression level of epidermal growth factor inhibits cell proliferation, metastasis, invasion, and metastasis | MDA-MB-231 , SUM159 | In Vivo/In Vitro | Tissue invasion and metastasis, resisting cell death, deregulating cellular energetics | ERK1/2, p38, JNK pathway, EGFR-mediated MAPK pathway | [160] |
Table 4.
Related effects of ginsenoside in attenuating adverse reactions induced by chemical antitumor drugs
| Ginsenoside type | Anti-breast cancer activity | Cell line | In vivo/in vitro | Drugs that cause adverse reactions | Signaling pathway/target spot | Refs. |
|---|---|---|---|---|---|---|
| Rg2 | Reducing cardiomyocyte apoptosis induced by trastuzumab therapy | / | In Vivo | Trastuzumab | caspase-3, caspase-9, Bax | [167] |
| Rg2 | Promote the protective autophagy of myocardial cells and avoid the apoptosis of myocardial cells caused by trastuzumab therapy | Human primary HCMs | In Vitro | Trastuzumab | p-Akt, p-mTOR, beclin 1, LC3, ATG5 | [168] |
| Rh2 | Cardiotoxicity is reduced by inhibiting cardiac histopathological changes, apoptosis and necrosis, and consequent inflammation. Pathological remodeling is attenuated by reducing fibroblast to myofibroblast transformation (FMT) and endothelial-mesenchymal transformation (EndMT). It can promote the senescence of myofibroblasts and reverse the differentiation of myofibroblasts established in EndMT to alleviate fibrosis | MDA-MB-231, HUVEC | In Vivo | Doxorubicin | caspase-3, caspase-7, caspase-9, TNF-α, IL-6, IL-1β, CD31, CD206, fα-SMA, Vimentin, Smad2, Smad3 | [169] |
| Rg1 | Promote that specific combination of doxorubicin and tumor cells and avoid the apoptosis of myocardial cells induced by doxorubicin | / | In Vivo | Doxorubicin | ROS, p53, caspase-3 | [170] |
| Panaxatriol | Reversing paclitaxel resistance | MDA-MB-231, SUM159 | In Vivo | Paclitaxel | RAK1/NF-κB and ERK signaling pathways, S100A7/9, inflammatory factors (IL6, IL8, CXCL1, CCL2), cancer stem cell-related (OCT4, SOX2, NANOG, ALDH1, CD44) | [49] |
At present, there is no research on the metabolism of ginsenoside in tumor patients. Therefore, it is speculated that ginsenoside is also metabolized in human body through liver-kidney pathway. In the clinical study with only a few ginsenoside, the relationship between the dosage of ginsenoside and adverse reactions was reported [43–45]. In vivo studies of healthy volunteers found that ginsenoside at therapeutic dose (1–2g/day) would not cause serious adverse reactions. After taking ginsenoside orally to healthy volunteers, the reported side effects are mainly mild, mainly dizziness, insomnia, nervousness and uterine bleeding. However, the study found that the adverse reactions, such as dizziness, insomnia, nervousness and uterine bleeding, would disappear after one month of treatment. The most common adverse reactions after ginsenoside administration for one month are constipation, dyspepsia, insomnia and hot flashes, and these adverse reactions are mild. Therefore, it can be concluded that ginsenoside will not cause serious treatment-related adverse reactions when it is treated in vivo at therapeutic dose. This also means that ginsenoside is safe and tolerable in the treatment of breast cancer patients.
Mechanism of action of ginsenosides altering breast cancer hallmarks
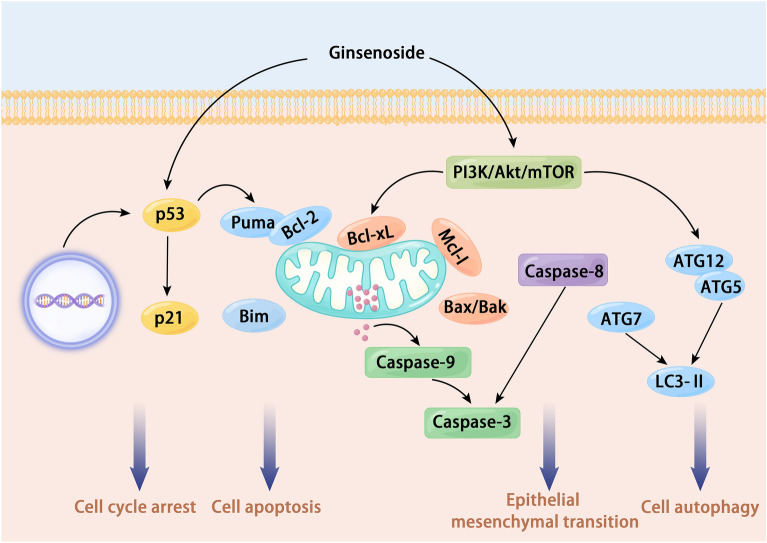
Regardless of the stereotype, ginsenosides have been studied in several in vivo and in vitro models of breast cancer, and various mechanisms of the therapeutic effect of ginsenosides have been revealed. The mechanisms of action of ginsenosides are achieved by altering the characteristics of breast cancer cells (Fig. 4). These mechanisms include induction of apoptosis, inhibition of cell proliferation, induction of autophagy through upregulation of autophagy-related molecules, inhibition of metastasis and angiogenesis, cell cycle arrest, immunomodulatory effects, induction of cellular phenotypic changes, prompting cellular epigenetic reprogramming, remodeling of cellular abnormal energy metabolism, and suppression of tumor cell-induced inflammation.
Fig. 4.
The main molecular mechanism of ginsenosides anti-breast cancer. Ginsenosides can treat breast cancer through various targets and multiple signaling pathways. Cell cycle arrest, cell apoptosis, EMT, and cell autophagy are the most common phenotypic changes after ginsenoside intervention in breast cancer cells, and these phenotypic changes can achieve the purpose of treating breast cancer
Although most currently discovered ginsenosides have been identified as naturally occurring compounds or compounds that can be metabolically produced, some can be chemically synthesized [46]. The current generation of ginsenosides with definite anti-breast cancer activity has become a major research hotspot, especially Rg3 (12 studies), Rh2 (11 studies), Rg1 (5 studies), and Rg2 (4 studies). Compound K is the in vivo metabolite of natural ginsenosides, and many ginsenosides undergo metabolism in the body to eventually form compound K. In Asia, boiling is one of the most common methods for processing natural Radix Ginseng. It has been found that Radix Ginseng, processed by steaming, decreases the major ginsenosides Rb1, Rb2, Rb3, Rc, Rd, Re, and Rg1 and increases the specific ginsenosides Rg2, Rg3, Rg5, Rh1, Rh2, and Rh4, leading to an increase in the ginsenoside components in Radix Ginseng that have anti-breast cancer activity [47]. The less abundant ginsenosides, such as 20(S)- and 20(R)-ginsenosides, are scarce in natural Radix Ginseng but are equally cytotoxic to breast cancer cells. The structural differences between 20(S)-Rg3 and 20(R)-Rg3 are attributed to the position of the C-20 hydroxyl group. Studies have indicated that 20(S)-Rg3 exhibits superior inhibitory activity against breast cancer compared to 20(R)-Rg3 [48]. Most anti-breast cancer studies involving ginsenosides have focused on in vitro assays using breast cancer cell lines such as MCF-7 (luminal type A) and MDA-MB-231 (triple-negative breast cancer). However, there are limited in vivo studies available. Some studies have explored the potential of ginsenosides in reversing drug resistance in breast cancer cell lines specifically resistant to certain chemotherapeutic agents [49]. Clinical studies have shown that Ginsenoside Rg3 treatment could enhance the anti-tumor efficacy of chemotherapy agents (such as Capecitabine, Docetaxel, and Cisplatin) in patients with advanced breast cancer [50–52]. Additionally, ginsenoside therapy has been found to reduce the adverse effects caused by chemotherapy, improve immune function, and enhance patient quality of life [53]. However, it is important to note that current studies on ginsenoside therapy for breast cancer are predominantly conducted in vitro, with only a limited number of in vivo models being explored. The following mechanisms associated with ginsenosides in breast cancer are discussed below.
Ginsenosides can induce apoptosis in breast cancer cells
Resisting cell death is one of the fundamental characteristics of tumor cells. Cells that experience irreversible DNA damage undergo programmed cell death, also known as apoptosis [54]. However, tumor cells have developed mechanisms to evade this program, making them apoptosis-resistant. Apoptosis is a fundamental type of programmed cell death and plays a crucial role in the mechanism of action of various anti-tumor drugs. Apoptosis occurs mainly through the extrinsic death receptor pathway and the intrinsic mitochondrial pathway [55]. These pathways lead to the activation of cysteine proteases, which can cleave to generate different substrates and then cause cell death [56]. Promoting breast cancer cell pyroptosis by extrinsic death receptor pathway to induce cellular cysteine proteases activation is one of the main and most directly effective mechanisms of ginsenosides anti-breast cancer. Several studies have investigated the effects of different ginsenosides on breast cancer cells, and they have identified several ginsenosides, such as Rh2, Rh4, Rk1, Rg3, and Rg5, that are capable of inducing apoptosis in various subtypes of breast cancer cells, including MCF-7 and MDA-MB-231 cells.
Ginsenoside Rg3 is currently recognized as one of the most promising agents for tumor treatment among the various ginsenosides. Numerous basic and clinical studies have demonstrated its therapeutic efficacy against various tumors [57]. Moreover, several studies have confirmed that ginsenoside Rg3 exerts therapeutic effects against breast cancer through multiple targets and mechanisms. In laboratory studies, both in vivo and in vitro experiments have consistently shown the ability of ginsenoside Rg3 to induce apoptosis in breast cancer cells, highlighting its potential as an effective treatment option for breast cancer. After ginsenoside Rg3 intervention in breast cancer cells [58], cell proliferation viability was inhibited, and apoptotic cells with DNA fragmentation cells increased. For ginsenoside-induced apoptosis in triple-negative breast cancer cells, the mechanism is mainly through inhibiting NF-κB activation, decreasing NF-κB p65 and Bcl-2 protein expression, and increasing the ratio of Bax and caspase-3 protein expression to Bax/Bcl-2 to achieve therapeutic efficacy. The elucidation of the mechanism of ginsenoside Rg3 in breast cancer treatment has contributed to its success in clinical trials as a potential breast cancer drug. Interestingly, the intervention of 20(S)-Rg3 has been shown to increase the sensitivity of MDA-MB-231 cells to radiotherapy. However, the precise mechanism behind this effect remains unclear [59]. Ginsenoside Rg5 is also an active component that can induce breast cancer cells apoptosis in various ways. Ginsenoside Rg5 [60, 61] and ginsenoside Rk1 [62, 63] have shown promising anti-tumor effects on MCF-7 cells by binding to active domains of PI3K proteins and increasing the levels of reactive oxygen species expression to inhibit PI3K/Akt signaling and via the extrinsic apoptotic pathway, intrinsic apoptotic pathway, and autophagic pathway. Other ginsenosides can also induce apoptosis in breast cancer cells through multiple pathways. For example, Rg1 [64, 65] can induce apoptosis by enhancing the expression levels of apoptotic genes and altering mitochondrial membrane potential, Rh2 [66, 67] composites induce apoptosis by activating the caspase-9/p38 MAPK signaling pathway and mitochondrial pathway, and Rh4 [68] induces apoptosis by blocking the cell cycle and upregulating the expression levels of apoptotic proteins. The above findings demonstrate that despite their heterogeneous chemical structures, different ginsenosides can induce apoptosis in breast cancer cells, offering potential treatment options. Additional ginsenosides, such as 1f [69] and Rh1 [70–72], have been found to induce apoptosis in breast cancer cells by affecting multiple signaling pathways associated with both the intrinsic and extrinsic pathways.
Cell autophagy is a cellular process where damaged proteins or organelles are enclosed within double-membrane autophagic vesicles, which are then delivered to lysosomes for degradation and recycling [73]. Traditionally, autophagy and apoptosis have been regarded as mutually inhibitory processes. However, research has shown that certain pharmacological interventions can simultaneously activate both autophagy and apoptosis, synergistically promoting cell death [74]. It has been observed that some ginsenosides can induce irreversible autophagy and apoptosis in cells, leading to therapeutic effects against breast cancer. Ginsenoside Rg5 [60, 61] enhanced the expression and activation of autophagy-related and apoptotic proteins to promote the apoptosis of MCF-7 cell xenografts by suppressing the activation level of the PI3K/Akt signaling pathway in in vivo experiments. Treatment of MCF-7 cells with ginsenoside Rg2 [75] at the in vitro level can lead to upregulation of p-p53, p-AMPK, p-ACC, Atg-7, and LC3-II levels and reduction of p62 levels in a concentration-dependent manner to promote autophagy and induce apoptosis. Likewise, intervention with Rg2 [76] was found to sensitize breast cancer cells to autophagic death and apoptosis. The application of (20S)-PPT [77] in vivo and in vitro can enhance the expression levels of autophagy-related proteins, autophagosomes, and apoptotic protein families (Bcl-2, Bax) by inhibiting the expression level of Akt/mTOR pathway, and achieve the dual effects of autophagy induction and apoptosis on the premise of ensuring the safety of the drugs. The nanoparticle carriers of Rb1 [78] and Rb1 [79] all showed cytotoxicity to breast cancer cells in vitro, although it remains unclear how they induce apoptosis.
Ginsenosides alter self-sufficient growth signaling in breast cancer cells
It is well-established that normal mammary epithelial cells require the activation of mitogenic growth signals to progress from a quiescent to an active proliferative state [80]. However, breast cancer cells do not require mitogenic growth signals to continuously maintain an active proliferative state [81]. These signals are transmitted into the cell through transmembrane receptors, which combine different types of signaling molecules: diffusible growth factors, extracellular matrix components, and intercellular adhesion/interaction molecules [82]. Without this stimulatory signal, normal mammary epithelial cells of any type fail to proliferate. In contrast, many oncogenes in breast cancer cells mimic normal growth signals in various ways to stimulate the proliferation of tumor cells [83].
Although most soluble mitogenic growth factors are synthesized and secreted by one cell type to stimulate the proliferation of another, tumor cells can produce mitogenic growth factors to eliminate the reliance on mitogenic growth factors for other cells within the tissue [84]. In approximately 25% of patients with tumors, Ras proteins exist in structurally altered forms that allow them to release numerous mitogenic signals into the cell without sustained stimulation by their normal upstream regulators [85]. An example of the interplay between signaling pathways in promoting cell proliferation involves the direct interaction of Ras proteins with PI3K, leading to the simultaneous activation of growth signals and survival signals within the cell [86]. This phenomenon is driven by the persistent activation of oncogenes and the inactivation of tumor suppressor genes. In estrogen receptor-positive breast cancer cells, the primary reliance for proliferation is on estrogen receptor signaling, which activates growth factor signaling [87]. When estrogen expression and signaling are inhibited, the intracellular growth signal is blocked, ultimately inducing apoptosis [88]. Several recent studies have shown that ginsenoside intervention can inhibit the sustained proliferation of breast cancer cells by inhibiting estrogen receptors and their signaling.
Ginsenoside Rh2 has been studied in MCF-7 cells and shown to reduce the protein level of estrogen receptor alpha (ERα) while increasing the mRNA and protein levels of ER-beta (ER-β) and tumor necrosis factor-alpha (TNF-α). Through the β-TNF-α signaling pathway, ginsenoside Rh2 induces apoptosis and G1/S arrest, leading to anti-tumor effects in xenograft mice [89]. On the other hand, ginsenoside Rg1 has been found to induce the translocation of ER to the plasma membrane through caveolin-1 in MCF-7 cells, leading to the formation of a signaling complex and phosphorylation of extracellular signal-regulated protein kinase (ERK) and mitogen-activated protein kinase (MAPK). Additionally, it activates epidermal growth factor receptor and cellular nonreceptor tyrosine kinase independently of ER, exerting estrogenic effects by rapidly activating membrane-associated ER and G protein-coupled estrogen receptors. This, in turn, inhibits the activation of proliferative signals in breast cancer cells [90]. Another study found [91] that intervention of Rg6 could block cortisol/cortisone-induced deregulation of microtubule organizing centers as well as ER signaling in breast cancer cells, alter stress hormone or steroid hormone-induced chromosomal instability, and inhibit aberrant activation of tumor cell proliferative signaling. The stress hormone cortisol and cortisone have been shown to elevate the expression of γ-tubulin, increasing the number of microtubule organizing centers and promoting resistance to paclitaxel in breast cancer cells. However, the intervention of ginsenoside Rg6 has been found to block the dysregulation of MTOCs induced by cortisol/cortisone and inhibit ER-alpha signaling.
Ginsenosides block breast cancer angiogenesis
Targeting angiogenesis inhibition has emerged as a promising therapeutic approach for treating breast cancer, yielding favorable results across different subtypes. Cancer growth and metastasis rate can be controlled by inhibiting the formation of new blood vessels in breast tumors. Additionally, blocking the supply of oxygen and nutrients necessary for breast cancer cell proliferation can induce apoptosis or autophagy in these cells [92]. Although commonly used anti-angiogenic drugs in clinical practice (such as Bevacizumab, Apatinib, Anlotinib, etc.) can effectively inhibit neoangiogenesis in breast cancer, they can also lead to adverse effects such as hypertension, thrombosis, and hemorrhage, thereby impacting patients' quality of life [93]. However, certain ginsenosides have shown potential as inhibitors of breast tumor angiogenesis, offering a more targeted and potentially safer approach to breast cancer treatment.
Ginsenoside Rg1 [65] exhibited significant anti-breast cancer activity in in vitro experiments by promoting tumor cell DNA damage and undergoing apoptosis through alteration of mitochondrial membrane potential and ROS levels, and in vivo experiments by downregulating angiogenesis, invasion, and EMT markers mediated by anti-oxidant enzymes. Ginsenoside Rd [94] could dependently inhibit vascular endothelial growth factor-induced migration, tube formation, and proliferation of primary cultured human umbilical vascular endothelial cells, and the activation of Akt/mammalian target of the rapamycin signaling cascade in VEGF-induced human umbilical vascular endothelial cells was inhibited under normoxic or hypoxic conditions to abolish aortic ring vessel sprouting and vessel formation. In an intraperitoneal xenograft mouse model constructed with MDA-MB-231 cells, Rd significantly reduced the volume and weight of breast cancer tumor tissues, decreased tumor angiogenesis, and inhibited breast cancer cell proliferation by suppressing Akt/mTOR/p70S6 kinase signaling in a dose-dependent manner. Rg3 [95] yielded a strong inhibitory effect on tumor growth in MDA-MB-231 cells tumor-bearing mice when combined with recombinant human endostatin, as well as inhibiting angiogenesis and cell invasion and enhancing cell autophagy.
Ginsenosides can transform breast cancer cell phenotypes
In normal tissues, the transformation of progenitor cells into mature tissue cells allows them to assume specialized functions and achieve homeostasis by ceasing their growth. This process is facilitated through development and terminal differentiation, typically resulting in an anti-proliferative state. However, in the context of breast carcinogenesis, the capacity for phenotypic plasticity becomes deregulated, allowing breast cancer cells to evade or escape from the terminally differentiated state. This abnormal phenotypic plasticity is a key factor in the development of breast cancer [96]. Breast cancer cells, which originate from normal cells, may undergo a reversal of their differentiation state, dedifferentiating back to a progenitor-like state as a potential therapeutic strategy [97]. Conversely, breast cancer cells arising from progenitor cells programmed to follow a pathway of end-stage differentiation may disrupt this process and maintain themselves in a partially differentiated, progenitor-like state. Some breast cancer cells may differentiate into fully formed tumor cells, referred to as breast cancer stem-like cells, while others retain characteristics of progenitor cells [98]. Breast cancer stem-like cells are more proliferative, invasive, and metastatic than normal breast cancer cells [99]. Therefore, inducing breast cancer cells to differentiate into a progenitor-like state or inducing breast cancer stem-like cells into normal breast cancer cells by drug or gene therapy may be an unusual approach to treating breast cancer.
Inducing differentiation in tumor cells can modulate the expression levels of selective regulators involved in cellular plasticity and signal transduction, such as Wnt signaling, Hippo signaling, Notch signaling, and Hedgehog signaling. This modulation ultimately affects the cellular phenotype [98]. Altering phenotypic plasticity in tumor cells can be achieved through three main subclasses: dedifferentiation of mature cells to progenitor states, blocking differentiation to maintain cells in a progenitor/stem cell state, and transdifferentiation to alternative cell lineages [100]. Inducing differentiation has significant implications for breast cancer formation, malignant progression, and response to therapy and appears effective across different breast cancer types [100]. While gene therapy is currently limited in clinical applications due to ethical considerations, drug-induced differentiation is an attractive direction in the research of induced differentiation of tumor stem-like cells. Some ginsenosides have shown the ability to modulate various regulators of cell plasticity and signal transduction, leading to the induction of differentiation in breast cancer stem-like cells. These ginsenosides can transform breast cancer stem cells into normal breast cancer cells, exerting cytotoxic effects and contributing to breast cancer treatment.
Ginsenoside Rg3 [101] has been shown to reduce the clonogenic capacity of breast cancer stem-like cells by decreasing the expression levels of CD44, a surface marker of breast cancer stem-like cells, to induce their differentiation through Akt-mediated self-renewal signaling to decrease the expression and localization of SOX2, BMI-1, and hypoxia-inducible factor-1α. The combination of C3 [102], a distinct isoform of Rg3, with Rg3, can convert CD44+ MD-MB-231 cells into normal CD44− MD-MB-231 cells and significantly inhibit cell migration ability. C3 alone also achieves therapeutic effects on breast cancer by binding to IGF-1R, Akt, and mTOR, reducing the primary focus and metastatic load in tumor-bearing mice. Rh2 [103] decreased the protein expression levels of TRAF6, p62, and phosphorylated IKK and IKB and inactivated NF-κB activity by inhibiting IL-8 secretion mediated by regulation of ROS levels and mitochondrial autophagy. Ginsenoside panaxatriol [49] induces the transition of breast cancer stem cells into normal breast cancer cells by repressing paclitaxel-resistant breast cancer stem cell-associated genes (OCT4, SOX2, NANOG, CD44, ALDH1) and by inhibiting IRAK1/NF-κB and ERK pathway reduced inflammatory cytokines (IL-6, IL-8, CXCL1, CCL2) expression, which contributed to resensitization of paclitaxel-resistant cells to paclitaxel.
Ginsenosides can reverse nonmutational epigenetic reprogramming in breast cancer cells
Genome instability and mutation are fundamental in breast cancer pathogenesis [104]. There is an increasing consensus that cancer cells can regulate gene expression in various ways, such as regulating noncoding RNAs, altering chromatin states, and affecting epigenetic modifications [105]. It is well-established that the genomes (DNA of normal individuals and breast cancer patients are identical. In normal adults, the formation of long-term memory relies on various mechanisms, including modifications to genes and histones, changes in chromatin structure, and the activation or repression of specific genes through intricate feedback loops. However, in breast cancer patients, these regulatory switches are dysregulated, leading to the activation of numerous oncoproteins and the suppression of tumor suppressor proteins, ultimately contributing to the initiation and progression of breast cancer. This model involves alterations in gene expression that are controlled purely through epigenetic mechanisms, a phenomenon referred to as “nonmutational epigenetic reprogramming” [106]. Nonmutational epigenetic reprogramming mainly includes chromatin remodeling complexes, histone modifications, non-coding RNAs, and other epigenetic mechanisms. The nonmutational epigenetic reprogramming of breast cancer cells is not restricted to breast cancer cells themselves but comprises three important aspects: epigenetic regulation of stromal cell types in the breast cancer cell microenvironment, epigenetic regulation of breast cancer cell heterogeneity, and epigenetic regulation of the breast cancer cell microenvironment [107].
Advanced research techniques have enabled a better understanding of breast cancer epigenomic heterogeneity. These techniques include profiling genomewide DNA methylation, histone modification, chromatin accessibility, posttranscriptional modification, and translation of RNA [108]. These technologies provide us with a better understanding of how nonmutational epigenetic reprogramming plays a role in the development of breast cancer. For example, non-coding RNAs can overexpress or degrade target proteins; chromatin remodeling leads to changes in the structural position of nucleosomes affecting transcriptional regulation of genes, and modifications of histones through acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation of histones [109]. Nonmutational epigenetic reprogramming ultimately suppresses tumor suppressor protein expression and (or) the activation of oncoprotein expression [110]. Interventions targeting nonmutational epigenetic inheritance have shown promise for breast cancer treatment, with two main approaches being gene therapy and drug therapy [111]. While gene therapy faces limitations due to ethical considerations in clinical applications, pharmacotherapy aimed at regulating breast cancer through nonmutational epigenetic reprogramming has attracted significant interest. Importantly, modulating the expression of oncoproteins and/or tumor suppressor proteins through chromatin remodeling complexes, histone modifications, non-coding RNAs, and other mechanisms can suppress tumor growth, invasion, and metastasis [112]. Breast cancer epigenetic drugs currently under investigation include DNA methyltransferase inhibitors, histone methyltransferase inhibitors, histone demethylase inhibitors, histone deacetylase inhibitors, bromodomain, and extra-terminal, isocitrate dehydrogenase inhibitors. However, studies on ginsenoside treatment for breast cancer have revealed its potential to intervene in breast cancer cells through various nonmutational epigenetic mechanisms, including chromatin remodeling complexes, histone modifications, and non-coding RNAs.
Ginsenosides Rg3 [113, 114] and Rh2 [115, 116] have been found to regulate the expression levels of breast cancer-related genes and proteins through the involvement of various non-coding RNAs. For example, they can modulate the expression of genes such as BBX, TNFAIP3, and SLC1A1 through non-coding RNAs like lnc STXBP5-AS1, lnc RFX3-AS1, miR-3614-3p, and others, thereby exerting anti-tumor activity. Additionally, Rg3 [117] and Rh2 [118, 119] have been shown to regulate the methylation levels and demethylation levels of breast cancer-related genes and proteins, including TRMT1l, KDM5A, and CAS1, through mechanisms such as N6-adenylate methylation, thus altering the breast cancer-associated microenvironment, enhancing immunogenicity, and inhibiting cancer cell growth. Furthermore, Rg1 [120] has been demonstrated to inhibit the phosphorylation of histone H3Thr3 mediated by Haspin kinase, leading to an increase in the width of the metaphase plate and spindle instability during cancer cell division, resulting in a delay in the progression of mitotic differentiation and inhibition of proliferation in breast cancer cells. Similarly, intervention with Rg6 [91] can induce chromosomal instability, which resensitizes paclitaxel-resistant breast cancer cells to paclitaxel treatment.
Ginsenosides drive remodeling of energy metabolism reprogramming in breast cancer cells
In recent years, many reports have indicated a close relationship between breast cancer-related cancer signaling pathways and energy metabolism activities. Breast tumor cells undergo metabolic reprogramming during tumorigenesis, including enhanced glycolysis, the tricarboxylic acid cycle, glutaminolysis, and fatty acid biosynthetic processes [121]. It is important to note that the specific metabolic changes can vary between different subtypes of breast cancer cells. However, this metabolic reprogramming is considered a crucial characteristic of breast cancer occurrence and progression. The continuous proliferation and invasive behavior of breast cancer cells necessitate a substantial energy supply to sustain their activities. In normal breast cancer cells with sufficient oxygen, glucose undergoes oxidative phosphorylation in the cytoplasm to produce carbon dioxide and energy via glycolysis to produce pyruvate, followed by oxidative phosphorylation in the mitochondria. However, when oxygen is insufficient, glucose generates lactate and energy in mitochondria undergoing anaerobic glycolysis [122]. Otto Warburg first observed the abnormal energy metabolism of cancer cells, noting that tumor cells can take up glucose in large quantities and undergo glycolysis even under sufficient oxygen conditions [123]. Recent studies have found that tumor cells increase the uptake of glucose and energy substrates, such as amino acids and fat, in large amounts to meet the energy demand of rapid tumor cell proliferation [124]. Oncogenes associated with breast cancer promote increased uptake of energy substances mainly by promoting breast cancer cell proliferation, inducing cell transfer, and reducing apoptosis. Furthermore, breast cancer cells can take up certain metabolites, such as lactate, and serve as signaling molecules that promote proliferation and invasion [125].
The reprogramming of energy metabolism is largely controlled by key proteins involved in energy metabolism that participate in programming cancer's fundamental hallmarks [126]. Targeted inhibition of these key proteins could achieve anti-tumor effects by regulating the mode of energy metabolism reprogramming. Over the past decade, targeting tumor energy metabolism has emerged as a new hotspot in developing novel anti-tumor drugs. Tumor-associated alterations in bioenergetic metabolism include aerobic glycolysis and tricarboxylic acid cycle, mitochondrial respiration, glutamine metabolism, and so on. These important metabolic processes can serve as potential anti-tumor drug targets [127]. However, there is significant heterogeneity in metabolic reprogramming among different tumor cells, emphasizing the need to develop drugs according to the metabolic reprogramming characteristics of different tumor cells. For instance, the anti-diabetic drug metformin is currently being evaluated for its potential anti-tumor effects in breast cancer patients. Metformin can interfere with mitochondrial complex I action, thus enabling AMPK activation. In parallel, the inhibitory effect of metformin on mitochondrial respiration has the potential to contribute to its anti-tumor effects [128]. Of note, the mechanism by which metformin reduces cancer incidence and mortality does not appear to be related to its pharmacological effects on glycemia. It is widely thought that its anti-tumor effects are mainly mediated through inhibiting AMPK-dependent and independent hepatic gluconeogenesis [129]. Ginsenosides can also contribute to remodeling energy metabolism reprogramming in breast cancer cells by regulating key energy metabolism proteins.
It has been shown that ginsenoside CK [130], when administered to TNBC cells, inhibits glutamine consumption and glutamate production by downregulating the expression of glutaminase 1. This leads to reduced cellular ATP production and the utilization of amino acids involved in glutamine metabolism. Consequently, TNBC cells experience glutathione depletion and accumulation of reactive oxygen species, leading to the regulation of apoptosis-related protein expression levels and induction of TNBC cell apoptosis. Rg3 and DTX synthesized DTX-loaded Rg3 liposomes (Rg3-Lp/DTX) [131] exhibit enhanced accumulation at the tumor site due to the interaction between the glycosyl moiety of Rg3 exposed on the liposome surface and the overexpressed glucose transporter 1 on breast cancer cells. This allows Rg3-Lp/DTX to accumulate at the tumor site and block glucose transport by glucose transporter 1. Additionally, it reverses activated cancer-associated fibroblasts (CAFs) to a resting phase and attenuates the dense stromal barrier by inhibiting the secretion of TGF in tumor cells and modulating TGF/Smad signaling. As a result, it achieves a dual effect of modulating the tumor microenvironment and enhancing the anti-tumor effect of DTX. 2-Deoxy-Rh2 [132] exhibits dual anti-tumor activities by inhibiting glycolysis and mitochondrial respiration. It achieves this by binding to the active site of hexokinase II, thereby reducing the activation of anaerobic glycolysis compared to mitochondrial respiration. Furthermore, it regulates the mitochondrial apoptotic pathway.
Ginsenosides prevent breast cancer cells from evading immunogenic cell death
The immune system constantly monitors normal cells and tissues and is responsible for the recognition and elimination of the vast majority of potential cancer cells and early tumors [133]. However, when immune surveillance functions are impaired, cancer cells can evade detection, and the immune system may mistakenly perceive tumor tissue as normal, allowing the uncontrolled proliferation of tumor cells [134]. Studies using carcinogens to induce tumor development in mice have shown that immunodeficient mice develop tumors more frequently and at a faster rate than mice with intact immune activity [135]. Similar results have been observed in experiments involving transplanting human tumor cells into animals. Clinical research also supports the presence of anti-tumor immune responses in human cancers. For instance, immune cell infiltration, including natural killer cells, can be observed in the tumor tissues of colon and ovarian cancer patients [136]. Patients with a higher degree of immune cell infiltration, known as “hot” tumors, tend to have a better prognosis compared to tumors with limited immune cell infiltration, referred to as “cold” tumors [137]. Various immune cells are present in the tumor microenvironment, including tumor-infiltrating lymphocytes, macrophages, and neutrophils [138]. Among these, CD8+ T cells play a critical role in targeted killing of tumor cells. Upon activation, CD8+ T cells release intracellular granule toxins that induce tumor cell death [139]. The progression of tumors is viewed immunologically as consisting of three phases: tumor elimination, tumor equilibrium, and tumor escape [140]. During the tumor escape phase, tumor cells employ mechanisms to evade recognition and killing by secreting immunosuppressive factors and converting T cells into immunosuppressive T cells, such as exhausted T cells and anergic T cells that do not have tumor recognition or killing functions [141]. Additionally, tumor cells may recruit immunosuppressive inflammatory cells, such as regulatory T cells and myeloid-derived suppressor cells, which inhibit the action of cytotoxic lymphocytes targeting tumor cells [142].
Approaches to treating tumors by targeting tumor immune escape-related mechanisms have been demonstrated in laboratory studies and clinical studies. The methods of tumor immunotherapy can mainly be divided into two main categories, immune cell therapy and cytokine therapy [143]. Immune cell therapy involves increasing the number of immune cells or modifying the antigen presentation of immune cells to enhance their recognition and induction of immunogenic cell death in tumor cells [144]. However, the complexity of the preparation process, high cost, and potential side effects limit the widespread use of immune cell therapy in clinical settings. Conversely, cytokine therapy involves using cytokines to block the secretion of immunosuppressive factors by cancer cells, allowing immune cells to exert their normal immunogenic cell death effects on tumor cells [145]. Cytokine therapy is currently the most commonly used immunotherapy in the clinic. The expression of these immunosuppressive factors by cancer cells can be blocked by monoclonal antibodies against proteins such as PD-1, PD-L1, and CTAL-4, which prompt normal recognition by immune cells to kill cancer cells [146]. Breast cancer cells often possess robust and complex immune escape mechanisms that hinder the normal recognition and elimination of cancer cells by immune cells. However, certain natural products, including ginsenosides, have been shown to enhance immunogenic cell death of breast cancer cells by inhibiting immune factors secreted by cancer cells, modifying antigen presentation by immune cells, and increasing immune cell infiltration.
Modifying ginsenoside Rg3 into Rg3 liposomes [147] can regulate the expression levels of immunosuppressive factors such as TGF-β and IL-6, as well as related signaling pathways. This leads to reshaping the expression levels of immune cells and fibroblasts in the immune microenvironment, improving the immunosuppression observed in breast cancer tissues, and enhancing immune cells' recognition and killing function against breast cancer cells. Rg3 liposomes, combined with chemotherapeutic agents [148], can inhibit breast cancer proliferation and invasion by binding to specific sites on breast cancer cells and altering the components of the tumor microenvironment. Rb3 incorporated into carbon nanotubes [149] can impact the PD-1/PD-L1 axis in a co-culture system of T cells and triple-negative breast cancer cells. This reduces the expression of T cell-related inhibitors, promotes the recognition and killing of T cells targeting breast cancer cells, and achieves an anti-cancer effect. Rh2 [118] can increase immunogenic cell death and inhibit the growth of breast cancer cells by modifying the epigenetic methylation of genes involved in immune response and tumorigenesis. Multifunctional liposomes containing Rh2 [150] can easily interact with tumor cells through glucose transporters (GLUT), effectively inhibiting the growth of breast cancer by reshaping the cellular composition of the tumor microenvironment and reversing the immunosuppressive environment.
Ginsenosides block breast cancer cell invasion and metastasis
In general, most cells in the body have a fixed spatial localization within the tissues they belong to and cannot detach and survive elsewhere in the body. However, during the early stages of tumor development, tumor cells proliferate uncontrollably in the primary tumor site and disrupt the organ's normal functions. The majority of cancer-related deaths, more than 90%, are attributed to multi-organ failure caused by the development of metastatic lesions in other organs [151]. Current research suggests that cancer cells when they detach from the primary tumor site and undergo invasion and metastasis, undergo a process called “epithelial-mesenchymal transition” (EMT) [152]. This program allows cancer cells to acquire invasive properties, resist apoptosis, and disseminate to other tissues.
EMT is a physiological and pathological phenomenon that can plays a crucial role in the loss and gain of epithelial characteristics of cells, essential in breast cancer cell invasion and metastasis [153]. In the absence of EMT, breast cancer cells are columnar or cuboidal in shape, more polar, form tight cell–cell junctions, and remain stationary. However, when stimulated by external factors such as growth factors and cytokines, breast cancer cells can undergo EMT, transitioning into cells with a mesenchymal phenotype lacking polarity and gaining motility. During this EMT process [153], breast cancer cells gradually acquire a spindle-like fibrous shape, lose cell polarity, exhibit weak cell adhesion, become more loosely packed, and become prone to metastasis. EMT in breast cancer is a complex and highly regulated process [154]. Transcription factors, including zinc finger transcription factors, cell transformation regulators, E-box binding zinc finger proteins, and signaling pathways such as TGF-β, MAPK, and NF-κB, play a role in inducing and regulating EMT [155]. Activation of these transcription factors and signaling pathways can induce alterations in the biological characteristics of breast cancer cells, mainly including loss of adherens junctions and transition from polygonal/epithelial to elongated/fibroblastic morphology, expression of matrix-degrading enzymes, increased cell motility, and enhanced resistance to cell death. These biologically tailored alterations all contribute to breast cancer cells' invasive and metastatic processes. Activation of these transcription factors and signaling pathways can directly repress the expression of EMT suppressors, including E-cadherin, leading to the loss of this key suppressor and promoting invasiveness. However, some active ingredients in ginsenosides, including Rg3, Rg1, and CK, could effectively prevent the occurrence and development of breast cancer by targeting the related genes and proteins of EMT.
The intervention of ginsenoside Rh1 [72] on TNBC cells can affect the migration and invasion mediated by STAT3 and NF-κB signaling through the regulation of mtROS and downregulated the expression of metastasis factors (e.g., MMP2, MMP9, and VEGF-A), thus exerting a powerful anti-cancer effect. In MDA-MB-231 cells, the intervention of Rg1 [65] downregulated invasion and angiogenesis markers and modulated EMT markers, controlling breast cancer progression. As the most intensively studied active ingredient in the field of ginsenosides against breast cancer, Rg3 [95, 113, 156] reportedly inhibits breast cancer cell invasion by modulating immune activity, altering breast cancer stem cell phenotype, and affecting non-coding RNA expression after intervening in breast cancer cells. Some other rare ginsenoside components, such as 0(S)-Rh2 [157], Rd [158], CK [159], and 20(S)-PPD [160], can inhibit EMT in breast cancer cells through multiple targets and mechanisms. These studies suggest that EMT inhibition is also an important mechanism by which ginsenosides exert therapeutic effects anti-breast cancer.
Ginsenosides reduce adverse effects from other medical treatments for breast cancer
The current standard treatment for breast cancer which is based on chemotherapy, radiotherapy, and targeted therapy, can cause damage to normal tissue organs while killing breast cancer cells. One of the main adverse reactions caused by these standard treatments is myocardial cell damage [161]. Anthracyclines are commonly used chemotherapy drugs for breast cancer, with significant cardiotoxicity, including cardiac dysfunction and heart failure [162]. These adverse effects may contribute to the inability of breast cancer patients to use adequate drug doses during treatment, rendering breast cancer a higher recurrence risk [163]. On the other hand, treatment with chemotherapeutic and targeted agents has the potential to cause drug resistance in breast cancer cells, leading to the failure of subsequent anti-breast cancer treatment.
At present, although the specific mechanism of action of anthracyclines leading to cardiotoxicity is not well defined, it has been established that the mechanism of injury mainly involves chelation of iron ions by anthracyclines, generation of reactive oxygen species that are mainly hydroxyl radicals and induction of lipid peroxidation in cardiomyocytes and mitochondrial damage in the heart machine [164]. On the other hand, the cardiotoxicity associated with anti-HER2 therapy is primarily attributed to the high expression of HER2 in cardiomyocytes. HER2 plays a crucial role in maintaining cardiomyocyte function and repairing cardiomyocyte damage. When anti-HER2 therapy blocks HER2 signaling, intracellular reactive oxygen species accumulate in cardiomyocytes, leading to injury and apoptosis [165]. Therefore, cardiotoxicity from medical treatment of breast cancer is mainly due to reactive oxygen species accumulation in cardiomyocytes due to treatment [166]. Ginsenosides, when combined with standard treatments, have shown the potential to inhibit adverse effects from standard treatments and prevent them.
Treatment with trastuzumab, an anti-HER2 therapy, can induce apoptosis in cardiomyocytes by upregulating the expression of apoptotic genes such as caspase-3, caspase-9, and Bax. However, ginsenoside Rg2 has been shown to reduce the expression of these apoptotic genes and attenuate trastuzumab-induced apoptosis in cardiomyocytes Rg2 [167]. Interestingly, Rg2 [168] induced protective autophagy in cardiomyocytes to avoid the apoptotic damage caused by trastuzumab by upregulating p-Akt, p-mTOR, Beclin 1, LC3, and ATG5 expression levels. As previously mentioned, doxorubicin can lead to cardiotoxic effects when treating breast cancer. Since doxorubicin causes histopathological changes, apoptosis and necrosis, and subsequent inflammation in the heart when treating breast cancer, Rh2 can reverse these changes [169]. Rh2, when administered with doxorubicin, alters the cell cycle and microtubule attachment, promotes TNF, chemokine, and interferon production, responses to cytokines and chemokines, and genes involved in T-cell activation, enhancing the efficacy of doxorubicin. In contrast, Rg1-conjugated [170] nanoparticles with doxorubicin can specifically bind to breast cancer cells to induce apoptosis and activate ROS and caspase-3 expression of tumor cells without damage to cardiomyocytes. Paclitaxel resistance is a common mechanism to reduce the benefit of paclitaxel treatment in breast cancer treatment. However, intervention with Panaxatriol can target paclitaxel-resistant breast cancer cells by blocking the activation of IRAK1/NF-κB and ERK signaling pathways, downregulating the expression of S100A7/9, inflammatory factors (IL-6, IL-8, CXCL1, CCL2), and cancer stem cell-related markers (OCT4, SOX2, NANOG, ALDH1, CD44), and restoring the sensitivity of breast cancer cells to paclitaxel [49].
Ginsenoside has good anti-cancer activity in clinical application
At present, only four clinical studies on ginsenoside in cancer patients have been reported, including two studies on patients with non-small cell lung cancer [50, 171], one study on hepatocellular carcinoma [51] and one study on breast cancer [52]. In a randomized controlled study of ginsenoside Rg3 combined with capecitabine in the treatment of triple negative breast cancer [52], 30 patients with stage IV triple negative breast cancer received capecitabine alone (15 cases) or Rg3 combined with capecitabine (15 cases). Rg3 was administered twice a day (40 mg/day) for at least 2 weeks. The results showed that compared with chemotherapy with capecitabine alone, Rg3 + Capecitabine could achieve a higher short-term effective rate (P < 0.05). Patients treated with Rg3 can also get lower incidence of adverse reactions and higher quality of life. This proves that ginsenoside combined with chemotherapy is a very potential treatment mode for patients with triple negative breast cancer.
In the study of Rg3 combined with chemotherapy in the treatment of stage II-III non-small cell lung cancer [50], 133 patients received Rg3(43 cases), Rg3 + chemotherapy (46 cases) or chemotherapy alone (44 cases). Rg3 was administered twice a day (40–50 mg/day) for at least 6 months. The results showed that compared with Rg3 or chemotherapy alone, Rg3 + chemotherapy improved the 3-year survival rate (54.3% versus 46.5% or 47.7%, respectively; P > 0.05). In the course of treatment, patients receiving Rg3 therapy have lower incidence of adverse reactions and better immune system function (increased activity of NK cells and CD4+ T cells and normal CD4+/CD8+ T cell ratio), which indicates that the combination of Rg3 therapy and chemotherapy is a very potential treatment strategy. In another study of Rg3 combined with TKI in the treatment of patients with unresectable EGFR mutation NSCLC in stage III–IV [171], 124 patients were divided into TKI + Rg3 (20 mg/day) or TKI monotherapy, and the administration lasted for at least 6 months. The results showed that Rg3 increased the median progression-free survival time of patients by 2.5 months, delayed acquired TKI drug resistance, and reduced TKI drug-related adverse reactions. Among the treatment-related adverse reactions, rash is the worst side effect in the two groups, while nausea, diarrhea and anorexia are the most common side effects in the two groups. However, the use of Rg3 reduces these adverse reactions caused by TKI drugs.
In another study of Rg3 in the treatment of patients with advanced hepatocellular carcinoma [51], 228 patients were randomly divided into two groups and received TACE alone or combined with Rg3 (40 mg/day). The results showed that the median overall survival time of patients receiving TACE + Rg3 was longer than that of patients receiving TACE alone (13.2 months compared with 10 months; P = 0.002). During the treatment, Rg3 was well tolerated, and the reported adverse effects were grade 1 or grade 2 constipation, nosebleed and hypertension. More importantly, the treatment of Rg3 is helpful to alleviate the adverse effects caused by TACE treatment and improve the quality of life of patients.
Although there are not many clinical studies on the treatment of cancer patients at present, it can be seen from the published studies that ginsenoside has a very bright prospect in the treatment of malignant patients. It has been found that ginsenoside can treat breast cancer through multi-targets, multi-channels and multi-mechanisms in vivo and in vitro, and the next stage of research should focus on transforming these research results into clinical use.
Discussion
The fundamental hallmarks of a tumor are the defining characteristics that differentiate normal mammary epithelium from breast cancer cells. They are the underlying causes and driving forces behind the development and progression of breast cancer. These hallmarks of breast cancer cells, when combined, promote continuous proliferation, invasion, and metastasis. They also aid in the survival of breast cancer cells during conventional treatment methods and the acquisition of drug resistance. As a result, targeting breast cancer therapeutics modifies the basic hallmarks of breast cancer cells, causing them to lose the crucial features on which they depend for survival.
Therapeutic approaches and drugs targeting the general characteristics of tumor cells have shown success in breast cancer treatment. Ongoing basic research focused on tumors has led to the discovery of new fundamental hallmarks of tumors over the years. Therefore, therapeutic drugs and approaches that target these novel hallmarks hold significant potential in breast cancer treatment. In clinical breast cancer treatment, the commonly used therapeutic modalities aim to induce apoptosis (programmed cell death), block self-sufficient growth signals in cells, alter the ability of cells to replicate indefinitely, and enhance immune cells' recognition and killing of tumor cells, among other strategies [172]. However, both traditional and novel antitumor drugs may have side effects that can reduce patients’ quality of life and increase the risk of treatment resistance [161]. Natural compounds, which are naturally occurring antitumor agents, offer an alternative approach. By identifying potential antitumor drugs from informative data containing natural compounds, these compounds have the potential to be a class of antitumor therapeutics with fewer side effects, significant therapeutic benefits, and relative affordability [9].
Radix Ginseng, a widely utilized natural medicine, has been recognized for its therapeutic effects in modern pharmacological studies. It has demonstrated positive outcomes in anti-inflammation, antioxidant activity, antitumor properties, and energy supplementation [173]. Among the active components of Radix Ginseng, ginsenosides have been identified as the most predominant and abundant compounds, exhibiting significant potential in antitumor therapy [17]. Over 180 ginsenosides have been discovered so far, and most of these active components have demonstrated varying degrees of inhibition against breast cancer cells [174].
Ginsenosides, the primary active components with anticancer properties found in Radix Ginseng, are typically administered orally. However, some ginsenosides require metabolic transformation by gut microbes after oral intake to generate ginsenosides with enhanced anticancer activity [175]. This metabolic conversion primarily involves the conversion of protopanaxadiol-type ginsenosides into compound K and ginsenoside Rh2, as well as the conversion of protopanaxatriol-type ginsenosides into ginsenosides Rh1 and protopanaxatriol. Interestingly, ginsenosides can also influence the characteristics of breast cancer cells by modulating the species and abundance of intestinal flora and the levels of various metabolites in breast cancer patients. This modulation can have both attenuating and enhancing effects on breast cancer treatment [176].
Ginsenoside has been proved to have great potential in the treatment of breast cancer, but at present, the mainstream research direction is still ginsenoside combined with anti-tumor drugs (chemotherapy drugs, targeted therapy drugs, immunotherapy drugs, antibody-coupled conjugate), which proves that ginsenoside can enhance the efficacy of anti-tumor drugs. At present, many studies have confirmed that ginsenoside can delay the acquired drug resistance of tumor cells, but there is no relevant report on the drug resistance of tumor cells to ginsenoside. As a natural product, ginsenoside may acquire drug resistance in tumor cells for a longer time than other synthetic chemicals. At present, there is no research on the contraindications of ginsenoside in clinical use in the reports. Therefore, in the next research, the contraindications of ginsenoside in the treatment of malignant tumor patients should be clarified to facilitate the clinical use of ginsenoside.
Many studies have established that ginsenosides yield heterogeneous effects in breast cancer treatment, altering the key hallmarks of breast cancer cells, including apoptosis, tumor angiogenesis, EMT, and epigenetic changes [174]. While basic research has demonstrated the anti-breast cancer effects of ginsenosides, clinical research in this area is still ongoing [177]. Clinical studies have mainly focused on reducing toxic side effects and improving the efficacy of ginsenosides combined with anti-tumor drugs. Novel studies have also been conducted to improve the solubility and stability of ginsenosides in vivo by changing the carriers and modes of administration of ginsenosides [178]. Therefore, the application of ginsenosides in the treatment of breast cancer represents a promising strategy that has the potential to make significant breakthroughs in the future.
Ginsenoside has been proved to have great potential in the treatment of breast cancer, but at present, the mainstream research direction is still ginsenoside combined with anti-tumor drugs (chemotherapy drugs, targeted therapy drugs, immunotherapy drugs, antibody-coupled conjugate), which proves that ginsenoside can enhance the efficacy of anti-tumor drugs. At present, many studies have confirmed that ginsenoside can delay the acquired drug resistance of tumor cells, but there is no relevant report on the drug resistance of tumor cells to ginsenoside. As a natural product, ginsenoside may acquire drug resistance in tumor cells for a longer time than other synthetic chemicals. At present, there is no research on the contraindications of ginsenoside in clinical use in the reports. Therefore, in the next research, the contraindications of ginsenoside in the treatment of malignant tumor patients should be clarified.
Acknowledgements
Not applicable.
Abbreviations
- Akt
Protein kinase B
- AMPK
Adenosine 5′-monophosphate (AMP)-activated protein kinase
- CAF
Cancer-associated fibroblast
- DTX
Docetaxel
- EMT
Epithelial–mesenchymal transition
- ER
Estrogen receptor
- HER2
Human epidermal growth factor receptor 2
- IL
Interleukin
- lncRNA
Long non-coding RNA
- NF-κB
PINuclear factor kappa-B
- MAPK
Mitogen-activated protein kinase
- mTOR
Mammalian target of rapamycin
- miRNA
Micro RNA
- MMP
Matrix metallo proteinase
- MDSC
Myeloid-derived suppressor cells
- PR
Progesterone receptor
- PI3K
Phosphatidylinositol-3 kinase
- PD-1
Programmed cell death protein 1
- PDGF
Platelet derived growth factor
- ROS
Reactive oxygen species
- STAT
Signal transducer and activator of transcription
- TNBC
Triple negative breast cancer
- TNF-α
Tumor necrosis factor-α
- TME
Tumor microenvironment
- VEGF
Vascular endothelial growth factor
Author contributions
R-YJ: conceptualized the study, wrote original draf. H-PZ: developed methodology. Z-RF: developed software. J-YZ: curated data. K-YC: validated the study. WW: performed formal analysis. XJ: reviewed and edited the article. XJW: supervised the study, administered the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Key Projects of Traditional Chinese Medicine in Zhejiang Province (No.2015zz004), Major Science and Technology Projects of Zhejiang Province (2012C13019-1), Beijing Xisike Clinical Oncology Research Foundation(Y-pierrefabre202101-0126).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to the publication of this work in Chinese Medicine.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rui-yuan Jiang, Zi-ru Fang and Huan-ping Zhang are co-first authors.
Contributor Information
Xiao Jiang, Email: jiangx1985@gxtcmu.edu.cn.
Xiao-jia Wang, Email: wxiaojia0803@163.com.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Wang GS, Zhu H, Bi SJ. Pathological features and prognosis of different molecular subtypes of breast cancer. Mol Med Rep. 2012;6(4):779–782. doi: 10.3892/mmr.2012.981. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E, Farahmand L. Breast cancer: biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535. doi: 10.1016/j.intimp.2020.106535. [DOI] [PubMed] [Google Scholar]
- 5.Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, Zackrisson S, Senkus E, ESMO Guidelines Committee Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann Oncol. 2019;30(8):1194–1220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 6.Greenlee H, DuPont-Reyes MJ, Balneaves LG, Carlson LE, Cohen MR, Deng G, Johnson JA, Mumber M, Seely D, Zick SM, Boyce LM, Tripathy D. Clinical practice guidelines on the evidence-based use of integrative therapies during and after breast cancer treatment. CA Cancer J Clin. 2017;67(3):194–232. doi: 10.3322/caac.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zanuso V, Fregoni V, Gervaso L. Side effects of adjuvant chemotherapy and their impact on outcome in elderly breast cancer patients: a cohort study. Future Sci OA. 2020;6(9):FSO617. doi: 10.2144/fsoa-2020-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau KH, Tan AM, Shi Y. New and emerging targeted therapies for advanced breast cancer. Int J Mol Sci. 2022;23(4):2288. doi: 10.3390/ijms23042288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan L, Cai Y, Zhang L, Liu S, Li P, Li X. Promoting apoptosis, a promising way to treat breast cancer with natural products: a comprehensive review. Front Pharmacol. 2021;12:801662. doi: 10.3389/fphar.2021.801662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Li H, Zhang J, Zhao C, Lu S, Qiao J, Han M. The combinatory effects of natural products and chemotherapy drugs and their mechanisms in breast cancer treatment. Phytochem Rev. 2019;19(5):1179–1197. [Google Scholar]
- 11.Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017;7(5):1016–1036. [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 13.Ravi S, Alencar AM, Jr, Arakelyan J, Xu W, Stauber R, Wang CI, Papyan R, Ghazaryan N, Pereira RM. An update to hallmarks of cancer. Cureus. 2022;14(5):e24803. doi: 10.7759/cureus.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molnar-Stanciu D, Guimas V, Bensalem A, Thiery-Vuillemin A. Targeted therapy and breast cancer: state of the art. Pathol Biol. 2012;60(4):254–263. doi: 10.1016/j.patbio.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Ruan L, Zhang S, Chen X, Liang W, Xie Q. Role of anti-angiogenic factors in the pathogenesis of breast cancer: a review of therapeutic potential. Pathol Res Pract. 2022;236:153956. doi: 10.1016/j.prp.2022.153956. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Wang X, Wang Y, Bao S, Chang Q, Liu L, Zhang S, Sun L. Strategies for remodeling the tumor microenvironment using active ingredients of Ginseng—a promising approach for cancer therapy. Front Pharmacol. 2021;12:797634. doi: 10.3389/fphar.2021.797634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun TK, Lee YS, Lee YH, Kim SI, Yun HY. Anticarcinogenic effect of Panax ginseng C.A. Meyer and identification of active compounds. J Korean Med Sci. 2001;16(Suppl):S6–18. doi: 10.3346/jkms.2001.16.S.S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo YH, Kuruganti R, Gao Y. Recent advances in ginsenosides as potential therapeutics against breast cancer. Curr Top Med Chem. 2019;19(25):2334–2347. doi: 10.2174/1568026619666191018100848. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Dontu G, El-Ashry D, Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15(5):193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Ebrahimi M, Ebrahimie E, Shamabadi N, Ebrahimi M. Are there any differences between features of proteins expressed in malignant and benign breast cancers? J Res Med Sci. 2010;15(6):299–309. [PMC free article] [PubMed] [Google Scholar]
- 23.Linn SC, Pinedo HM, van Ark-Otte J, van der Valk P, Hoekman K, Honkoop AH, Vermorken JB, Giaccone G. Expression of drug resistance proteins in breast cancer, in relation to chemotherapy. Int J Cancer. 1997;71(5):787–795. doi: 10.1002/(sici)1097-0215(19970529)71:5<787::aid-ijc16>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Hashemi F, Zarrabi A, Zabolian A, Saleki H, Farahani MV, Sharifzadeh SO, Ghahremaniyeh Z, Bejandi AK, Hushmandi K, Ashrafizadeh M, Khan H. Novel strategy in breast cancer therapy: revealing the bright side of ginsenosides. Curr Mol Pharmacol. 2021;14(6):1093–1111. doi: 10.2174/1874467214666210120153348. [DOI] [PubMed] [Google Scholar]
- 25.Younas M, Hano C, Giglioli-Guivarc'h N, Abbasi BH. Mechanistic evaluation of phytochemicals in breast cancer remedy: current understanding and future perspectives. RSC Adv. 2018;8(52):29714–29744. doi: 10.1039/c8ra04879g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhan Y, Xie HZ, Yan N, Li Q, Zhang T. Ginseng: history, cultivation, industry and future prospects. Allelopathy J. 2021;54(1):81–94. [Google Scholar]
- 27.Xiang YZ, Shang HC, Gao XM, Zhang BL. A comparison of the ancient use of ginseng in traditional Chinese medicine with modern pharmacological experiments and clinical trials. Phytother Res. 2008;22(7):851–858. doi: 10.1002/ptr.2384. [DOI] [PubMed] [Google Scholar]
- 28.Kim IW, Sun WS, Yun BS, Kim NR, Min D, Kim SK. Characterizing a full spectrum of physico-chemical properties of (20S)- and (20R)-ginsenoside Rg3 to be proposed as standard reference materials. J Ginseng Res. 2013;37(1):124–134. doi: 10.5142/jgr.2013.37.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tung NH, Song GY, Park YJ, Kim YH. two new dammarane-type saponins from the leaves of Panax ginseng. Chem Pharm Bull. 2009;57(12):1412–1414. doi: 10.1248/cpb.57.1412. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Kim N, Jeong J, Lee S, Kim W, Ko S-G, Kim B. Anti-cancer effect of panax ginseng and its metabolites: from traditional medicine to modern drug discovery. Processes. 2021;9(8):1344. [Google Scholar]
- 31.Wang ZL, Liu R, Chen L, Wang HH, Zhou M, Wang YS, Qin YP. Pharmacokinetics of Ginsenoside Rh2, the major anticancer ingredient of ginsenoside H dripping pills, in healthy subjects. Clin Pharmacol Drug Dev. 2021;10(6):669–674. doi: 10.1002/cpdd.877. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P, Fu P, Xiang L, Wang SP, Liu XJ, Yang L, Tao JF, Chen ZL, Zhan CS, Huang XM, Liu RH, Zhang WD. The effectiveness of borneol on pharmacokinetics changes of four ginsenosides in Shexiang Baoxin Pill in vivo. Biomed Chromatogr. 2014;28(3):419–427. doi: 10.1002/bmc.3037. [DOI] [PubMed] [Google Scholar]
- 33.Kang A, Zhang SJ, Shan JJ, Di LQ. Gut microbiota-mediated deglycosylation of ginsenoside Rb-1 in rats: in vitro and in vivo insights from quantitative ultra-performance liquid chromatography-mass spectrometry analysis. Anal Methods. 2015;7(15):6173–6181. [Google Scholar]
- 34.Lee J, Lee E, Kim D, Lee J, Yoo J, Koh B. Studies on absorption, distribution and metabolism of ginseng in humans after oral administration. J Ethnopharmacol. 2009;122(1):143–148. doi: 10.1016/j.jep.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Hao M, Zhao YQ, Chen PZ, Huang H, Liu H, Jiang HL, Zhang RW, Wang H. Structure-activity relationship and substrate-dependent phenomena in effects of ginsenosides on activities of drug-metabolizing P450 enzymes. PLoS ONE. 2008;3(7):e2697. doi: 10.1371/journal.pone.0002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian T, Cai Z. Biotransformation of ginsenosides Rb1, Rg3 and Rh2 in rat gastrointestinal tracts. Chin Med. 2010;5:19–19. doi: 10.1186/1749-8546-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K, Chen X, Xu J, Li X, Zhong D. Liquid chromatography/tandem mass spectrometry for pharmacokinetic studies of 20(R)-ginsenoside Rg3 in dog. Rapid Commun Mass Spectrom: RCM. 2005;19(6):813–817. doi: 10.1002/rcm.1862. [DOI] [PubMed] [Google Scholar]
- 38.Yang D, Li X, Fu YH, Tao XY, Zheng F, Yu JB, Yue H, Dai YL. Metabolic study of ginsenoside Rg3 and glimepiride in type 2 diabetic rats by liquid chromatography coupled with quadrupole-Orbitrap mass spectrometry. Rapid Commun Mass Spectrom. 2021;35(11):e9083. doi: 10.1002/rcm.9083. [DOI] [PubMed] [Google Scholar]
- 39.Qian T, Cai Z, Wong RNS, Mak NK, Jiang Z-H. In vivo rat metabolism and pharmacokinetic studies of ginsenoside Rg3, Journal of chromatography. B Anal Technol Biomed Life Sci. 2005;816(1–2):223–232. doi: 10.1016/j.jchromb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 40.Pang H, Wang HL, Fu L, Su CY. Pharmacokinetic studies of 20(R)-ginsenoside RG3 in human volunteers. Yao xue xue bao = Acta Pharm Sin. 2001;36(3):170–3. [PubMed] [Google Scholar]
- 41.Zhao Q, Zheng X, Jiang J, Zhou H, Hu P. Determination of ginsenoside Rg3 in human plasma and urine by high performance liquid chromatography-tandem mass spectrometry. J Chromatogr B-Anal Technol Biomed Life Sci. 2010;878(24):2266–2273. doi: 10.1016/j.jchromb.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 42.Bae SH, Park JB, Zheng YF, Jang MJ, Kim SO, Kim JY, Yoo YH, Yoon KD, Oh E, Bae SK. Pharmacokinetics and tissue distribution of ginsenoside Rh2 and Rg3 epimers after oral administration of BST204, a purified ginseng dry extract, in rats. Xenobiotica. 2014;44(12):1099–1107. doi: 10.3109/00498254.2014.929192. [DOI] [PubMed] [Google Scholar]
- 43.Kim H-G, Yoo S-R, Park H-J, Lee N-H, Shin J-W, Sathyanath R, Cho J-H, Son C-G. Antioxidant effects of Panax ginseng C.A. Meyer in healthy subjects: a randomized, placebo-controlled clinical trial. Food Chem Toxicol. 2011;49(9):2229–2235. doi: 10.1016/j.fct.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Seo SK, Hong Y, Yun BH, Chon SJ, Jung YS, Park JH, Cho S, Choi YS, Lee BS. Antioxidative effects of Korean red ginseng in postmenopausal women: a double-blind randomized controlled trial. J Ethnopharmacol. 2014;154(3):753–757. doi: 10.1016/j.jep.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 45.Lee N-H, Yoo S-R, Kim H-G, Cho J-H, Son CG. Safety and tolerability of panax ginseng root extract: a randomized, placebo-controlled, clinical trial in healthy korean volunteers. J Altern Complement Med. 2012;18(11):1061–1069. doi: 10.1089/acm.2011.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren S, Sun Q, Zhang L, Sun W, Li Y, Feng X, Li C. Sustainable production of rare oleanane-type ginsenoside Ro with an artificial glycosylation pathway in Saccharomyces cerevisiae. Green Chem. 2022;24(21):8302–8313. [Google Scholar]
- 47.Lee SA, Jo HK, Im BO, Kim S, Whang WK, Ko SK. Changes in the contents of prosapogenin in the red ginseng (Panax ginseng) depending on steaming batches. J Ginseng Res. 2012;36(1):102–106. doi: 10.5142/jgr.2012.36.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X-J, Zhang X-J, Shui Y-M, Wan J-B, Gao J-L. Anticancer activities of protopanaxadiol- and protopanaxatriol-type ginsenosides and their metabolites. Evid-Based Compl Altern Med. 2016;2016:5738694. doi: 10.1155/2016/5738694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, Song D, Wan D, Li L, Mei W, Li X, Han L, Zhu X, Yang L, Cai Y, Zhang R. Ginsenoside panaxatriol reverses TNBC paclitaxel resistance by inhibiting the IRAK1/NF-kappaB and ERK pathways. PeerJ. 2020;8:e9281. doi: 10.7717/peerj.9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu P, Su W, Miao ZH, Niu HR, Liu J, Hua QL. Effect and mechanism of ginsenoside Rg3 on postoperative life span of patients with non-small cell lung cancer. Chin J Integr Med. 2008;14(1):33–36. doi: 10.1007/s11655-007-9002-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhou B, Yan Z, Liu R, Shi P, Qian S, Qu X, Zhu L, Zhang W, Wang J. Prospective study of transcatheter arterial chemoembolization (TACE) with Ginsenoside Rg3 versus TACE alone for the treatment of patients with advanced hepatocellular carcinoma. Radiology. 2016;280(2):630–639. doi: 10.1148/radiol.2016150719. [DOI] [PubMed] [Google Scholar]
- 52.Li X. Clinical efficacy of Ginsenoside Rg3 combined with capecitabine in the treatment of advanced triple negative breast cancer. Hebei Med J. 2015;37(16):2445–2447. [Google Scholar]
- 53.Choi J-S, Chun K-S, Kundu J, Kundu JK. Biochemical basis of cancer chemoprevention and/or chemotherapy with ginsenosides. Int J Mol Med. 2013;32(6):1227–1238. doi: 10.3892/ijmm.2013.1519. [DOI] [PubMed] [Google Scholar]
- 54.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16(1):20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 55.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17(7):395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green DR, Llambi F. Cell death signaling. Cold Spring Harbor Perspect Biol. 2015;7(12):a006080. doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh H-M, Cho C-K, Son C-G. Experimental evidence for the anti-metastatic action of ginsenoside Rg3: a systematic review. Int J Mol Sci. 2022;23(16):9077. doi: 10.3390/ijms23169077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan Z, Jiang H, Zhu X, Liu X, Li J. Ginsenoside Rg3 promotes cytotoxicity of paclitaxel through inhibiting NF-kappaB signaling and regulating Bax/Bcl-2 expression on triple-negative breast cancer. Biomed Pharmacother. 2017;89:227–232. doi: 10.1016/j.biopha.2017.02.038. [DOI] [PubMed] [Google Scholar]
- 59.Changizi V, Gharekhani V, Motavaseli E. Co-treatment with ginsenoside 20(S)-Rg3 and curcumin increases radiosensitivity of MDA-MB-231 cancer cell line. Iran J Med Sci. 2021;46(4):291–297. doi: 10.30476/ijms.2020.83977.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, Fan D. Ginsenoside Rg5 induces apoptosis and autophagy via the inhibition of the PI3K/Akt pathway against breast cancer in a mouse model. Food Funct. 2018;9(11):5513–5527. doi: 10.1039/c8fo01122b. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y, Fan D. The preparation of ginsenoside Rg5, its antitumor activity against breast cancer cells and its targeting of PI3K. Nutrients. 2020;12(1):246. doi: 10.3390/nu12010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong Y, Fan D. Ginsenoside Rk1 induces cell death through ROS-mediated PTEN/PI3K/Akt/mTOR signaling pathway in MCF-7 cells. J Funct Foods. 2019;57:255–265. [Google Scholar]
- 63.Hong Y, Fan D. Ginsenoside Rk1 induces cell cycle arrest and apoptosis in MDA-MB-231 triple negative breast cancer cells. Toxicology. 2019;418:22–31. doi: 10.1016/j.tox.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 64.Liu S, Huang J, Gao F, Yin Z, Zhang R. Ginsenoside RG1 augments doxorubicin-induced apoptotic cell death in MDA-MB-231 breast cancer cell lines. J Biochem Mol Toxicol. 2022;36(1):e22945. doi: 10.1002/jbt.22945. [DOI] [PubMed] [Google Scholar]
- 65.Chu Y, Zhang W, Kanimozhi G, Brindha GR, Tian D. Ginsenoside Rg1 induces apoptotic cell death in triple-negative breast cancer cell lines and prevents carcinogen-induced breast tumorigenesis in sprague dawley rats. Evid Based Compl Alternat Med. 2020;2020:8886955. doi: 10.1155/2020/8886955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim YJ, Perumalsamy H, Castro-Aceituno V, Kim D, Markus J, Lee S, Kim S, Liu Y, Yang DC. Photoluminescent and self-assembled hyaluronic acid-zinc oxide-ginsenoside Rh2 nanoparticles and their potential caspase-9 apoptotic mechanism towards cancer cell lines. Int J Nanomed. 2019;14:8195–8208. doi: 10.2147/IJN.S221328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng Y, Mao J, Wang X, Yin B, Shen Z, Di C, Gu W, Wu M. Mechanism for ginsenoside Rh2-induced apoptosis of triple-negative breast cancer MDA-MB-231 cells. Clin Exp Obstet Gynecol. 2020;47(1):99–104. [Google Scholar]
- 68.Duan Z, Wei B, Deng J, Mi Y, Dong Y, Zhu C, Fu R, Qu L, Fan D. The anti-tumor effect of ginsenoside Rh4 in MCF-7 breast cancer cells in vitro and in vivo. Biochem Biophys Res Commun. 2018;499(3):482–487. doi: 10.1016/j.bbrc.2018.03.174. [DOI] [PubMed] [Google Scholar]
- 69.Ma L, Wang X, Li W, Li T, Xiao S, Lu J, Xu J, Zhao Y. Rational design, synthesis and biological evaluation of triphenylphosphonium-ginsenoside conjugates as mitochondria-targeting anti-cancer agents. Bioorg Chem. 2020;103:104150. doi: 10.1016/j.bioorg.2020.104150. [DOI] [PubMed] [Google Scholar]
- 70.Huynh DTN, Jin Y, Myung CS, Heo KS. Ginsenoside Rh1 induces MCF-7 cell apoptosis and autophagic cell death through ROS-mediated Akt signaling. Cancers. 2021;13(8):1892. doi: 10.3390/cancers13081892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin Y, Huynh DTN, Heo K-S. Ginsenoside Rh1 inhibits tumor growth in MDA-MB-231 breast cancer cells via mitochondrial ROS and ER stress-mediated signaling pathway. Arch Pharm Res. 2022;45(3):174–184. doi: 10.1007/s12272-022-01377-3. [DOI] [PubMed] [Google Scholar]
- 72.Jin Y, Huynh DTN, Myung CS, Heo KS. Ginsenoside Rh1 prevents migration and invasion through mitochondrial ROS-mediated inhibition of STAT3/NF-kappaB signaling in MDA-MB-231 cells. Int J Mol Sci. 2021;22(19):10458. doi: 10.3390/ijms221910458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol. 2018;20(10):1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yun CW, Lee SH. The roles of autophagy in cancer. Int J Mol Sci. 2018;19(11):3466. doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chung Y, Jeong S, Choi HS, Ro S, Lee JS, Park JK. Upregulation of autophagy by Ginsenoside Rg2 in MCF-7 cells. Anim Cells Syst. 2018;22(6):382–389. doi: 10.1080/19768354.2018.1545696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeon H, Jin Y, Myung CS, Heo KS. Ginsenoside-Rg2 exerts anti-cancer effects through ROS-mediated AMPK activation associated mitochondrial damage and oxidation in MCF-7 cells. Arch Pharm Res. 2021;44(7):702–712. doi: 10.1007/s12272-021-01345-3. [DOI] [PubMed] [Google Scholar]
- 77.Li Y, Wang P, Zou Z, Pan Q, Li X, Liang Z, Li L, Lin Y, Peng X, Zhang R, Tian H, Han L. Ginsenoside (20S)-protopanaxatriol induces non-protective autophagy and apoptosis by inhibiting Akt/mTOR signaling pathway in triple-negative breast cancer cells. Biochem Biophys Res Commun. 2021;583:184–191. doi: 10.1016/j.bbrc.2021.10.067. [DOI] [PubMed] [Google Scholar]
- 78.Luo Z, An J, Shi W, Li C, Gao H. One step assembly of ginsenoside Rb1-based nanovehicles with fast cellular transport in photothermal-chemical combined cancer therapy. Nanotechnology. 2021;32(19):195103. doi: 10.1088/1361-6528/abe1f0. [DOI] [PubMed] [Google Scholar]
- 79.My PLT, My HTK, Phuong NTX, Dat TD, Thanh VH, Nam HM, Thanh Phong M, Hieu NH. Optimization of enzyme-assisted extraction of ginsenoside Rb1 from Vietnamese Panax notoginseng (BURK.) F.H. Chen roots and anticancer activity examination of the extract. Sep Sci Technol. 2020;56(10):1687–1698. [Google Scholar]
- 80.Mayer D, Shukla A, Enzmann H. Proliferative effects of insulin analogues on mammary epithelial cells. Arch Physiol Biochem. 2008;114(1):38–44. doi: 10.1080/13813450801900645. [DOI] [PubMed] [Google Scholar]
- 81.Menon SS, Guruvayoorappan C, Sakthivel KM, Rasmi RR. Ki-67 protein as a tumour proliferation marker. Clin Chim Acta. 2019;491:39–45. doi: 10.1016/j.cca.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 82.Feitelson MA, Arzumanyan A, Kulathinal RJ, Blain SW, Holcombe RF, Mahajna J, Marino M, Martinez-Chantar ML, Nawroth R, Sanchez-Garcia I, Sharma D, Saxena NK, Singh N, Vlachostergios PJ, Guo S, Honoki K, Fujii H, Georgakilas AG, Bilsland A, Amedei A, Niccolai E, Amin A, Ashraf SS, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Mohammed SI, Azmi AS, Bhakta D, Halicka D, Keith WN, Nowsheen S. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin Cancer Biol. 2015;35:S25–S54. doi: 10.1016/j.semcancer.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136(2):331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pickup M, Novitskiy S, Moses HL. The roles of TGF beta in the tumour microenvironment. Nat Rev Cancer. 2013;13(11):788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aksamitiene E, Kiyatkin A, Kholodenko BN. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans. 2012;40:139–146. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- 86.York RD, Molliver DC, Grewal SS, Stenberg PE, McCleskey EW, Stork PJ. Role of phosphoinositide 3-kinase and endocytosis in nerve growth factor-induced extracellular signal-regulated kinase activation via Ras and Rap1. Mol Cell Biol. 2000;20(21):8069–8083. doi: 10.1128/mcb.20.21.8069-8083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohammed H, Russell IA, Stark R, Rueda OM, Hickey TE, Tarulli GA, Serandour AAA, Birrell SN, Bruna A, Saadi A, Menon S, Hadfield J, Pugh M, Raj GV, Brown GD, D'Santos C, Robinson JLL, Silva G, Launchbury R, Perou CM, Stingl J, Caldas C, Tilley WD, Carroll JS. Progesterone receptor modulates ER alpha action in breast cancer. Nature. 2015;523(7560):313. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spring LM, Wander SA, Andre F, Moy B, Turner NC, Bardia A. Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: past, present, and future. Lancet. 2020;395(10226):817–827. doi: 10.1016/S0140-6736(20)30165-3. [DOI] [PubMed] [Google Scholar]
- 89.Peng K, Luo T, Li J, Huang J, Dong Z, Liu J, Pi C, Zou Z, Gu Q, Liu O, Zhang J-T, Luo Z-Y. Ginsenoside Rh2 inhibits breast cancer cell growth via ER beta-TNF alpha pathway. Acta Biochim Biophys Sin. 2022;54(5):647–656. doi: 10.3724/abbs.2022039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao Q-G, Zhou L-P, Lee VH-Y, Chan H-Y, Man CW-Y, Wong M-S. Ginsenoside Rg1 activates ligand-independent estrogenic effects via rapid estrogen receptor signaling pathway. J Ginseng Res. 2019;43(4):527–538. doi: 10.1016/j.jgr.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cho JH, Chun HY, Lee JS, Lee JH, Cheong KJ, Jung YS, Woo TG, Yoon MH, Oh AY, Kang SM, Lee C, Sun H, Hwang J, Song GY, Park BJ. Prevention effect of rare ginsenosides against stress-hormone induced MTOC amplification. Oncotarget. 2016;7(23):35144–35158. doi: 10.18632/oncotarget.9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:1–4. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teleanu RI, Chircov C, Grumezescu AM, Teleanu DM. Tumor angiogenesis and anti-angiogenic strategies for cancer treatment. J Clin Med. 2020;9(1):84. doi: 10.3390/jcm9010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang E, Shi H, Yang L, Wu X, Wang Z. Ginsenoside Rd regulates the Akt/mTOR/p70S6K signaling cascade and suppresses angiogenesis and breast tumor growth. Oncol Rep. 2017;38(1):359–367. doi: 10.3892/or.2017.5652. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y, Liu QZ, Xing SP, Zhang JL. Inhibiting effect of Endostar combined with ginsenoside Rg3 on breast cancer tumor growth in tumor-bearing mice. Asian Pac J Trop Med. 2016;9(2):180–183. doi: 10.1016/j.apjtm.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 96.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang C-Y, Yaswen P, Goga A, Werb Z. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, Ji X, Liu W, Huang B, Luo W, Liu B, Lei Y, Du S, Vuppalapati A, Luu HH, Haydon RC, He T-C, Ren G. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5(2):77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12(8):445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mohammadalipour A, Burdick MM, Tees DFJ. Viscoelasticity measurements reveal rheological differences between stem-like and non-stem-like breast cancer cells. Cell Mol Bioeng. 2017;10(3):235–248. doi: 10.1007/s12195-017-0485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beug H. Breast cancer stem cells: eradication by differentiation therapy? Cell. 2009;138(4):623–625. doi: 10.1016/j.cell.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 101.Oh J, Yoon HJ, Jang JH, Kim DH, Surh YJ. The standardized Korean Red Ginseng extract and its ingredient ginsenoside Rg3 inhibit manifestation of breast cancer stem cell-like properties through modulation of self-renewal signaling. J Ginseng Res. 2019;43(3):421–430. doi: 10.1016/j.jgr.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakhjavani M, Smith E, Palethorpe HM, Tomita Y, Yeo K, Price TJ, Townsend AR, Hardingham JE. Anti-cancer effects of an optimised combination of ginsenoside Rg3 epimers on triple negative breast cancer models. Pharmaceuticals. 2021;14(7):633. doi: 10.3390/ph14070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hou J, Yun Y, Xue J, Jeon B, Kim S. Doxorubicin-induced normal breast epithelial cellular aging and its related breast cancer growth through mitochondrial autophagy and oxidative stress mitigated by ginsenoside Rh2. Phytother Res. 2020;34(7):1659–1669. doi: 10.1002/ptr.6636. [DOI] [PubMed] [Google Scholar]
- 104.Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168(4):644. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomas ML, Marcato P. epigenetic modifications as biomarkers of tumor development, therapy response, and recurrence across the cancer care continuum. Cancers. 2018;10(4):101. doi: 10.3390/cancers10040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kan RL, Chen J, Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022;38(2):182–193. doi: 10.1016/j.tig.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sarvari P, Sarvari P, Ramirez-Diaz I, Mahjoubi F, Rubio K. Advances of epigenetic biomarkers and epigenome editing for early diagnosis in breast cancer. Int J Mol Sci. 2022;23(17):9521. doi: 10.3390/ijms23179521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jankowska AM, Millward CL, Caldwell CW. The potential of DNA modifications as biomarkers and therapeutic targets in oncology. Expert Rev Mol Diagn. 2015;15(10):1325–1337. doi: 10.1586/14737159.2015.1084229. [DOI] [PubMed] [Google Scholar]
- 109.Nakasuka F, Yamada Y. iPS cell technology for dissecting mechanisms of cancer development, gan to kagaku ryoho. Cancer Chemother. 2020;47(10):1407–1410. [PubMed] [Google Scholar]
- 110.Richly H, Lange M, Simboeck E, Di Croce L. Setting and resetting of epigenetic marks in malignant transformation and development. BioEssays. 2010;32(8):669–679. doi: 10.1002/bies.201000016. [DOI] [PubMed] [Google Scholar]
- 111.Brown LJ, Achinger-Kawecka J, Portman N, Clark S, Stirzaker C, Lim E. Epigenetic therapies and biomarkers in breast cancer. Cancers. 2022;14(3):474. doi: 10.3390/cancers14030474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sher G, Salman NA, Khan AQ, Prabhu KS, Raza A, Kulinski M, Dermime S, Haris M, Junejo K, Uddin S. Epigenetic and breast cancer therapy: promising diagnostic and therapeutic applications. Semin Cancer Biol. 2022;83:152–165. doi: 10.1016/j.semcancer.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 113.Ham J, Jeong D, Park S, Kim HW, Kim H, Kim SJ. Ginsenoside Rg3 and korean red ginseng extract epigenetically regulate the tumor-related long noncoding RNAs RFX3-AS1 and STXBP5-AS1. J Ginseng Res. 2019;43(4):625–634. doi: 10.1016/j.jgr.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim H, Ji HW, Kim HW, Yun SH, Park JE, Kim SJ. Ginsenoside Rg3 prevents oncogenic long noncoding RNA ATXN8OS from inhibiting tumor-suppressive microRNA-424–5p in breast cancer cells. Biomolecules. 2021;11(1):118. doi: 10.3390/biom11010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jeong D, Ham J, Park S, Kim HW, Kim H, Ji HW, Kim SJ. Ginsenoside Rh2 suppresses breast cancer cell proliferation by epigenetically regulating the long noncoding RNA C3orf67-AS1. Am J Chin Med. 2019;47(7):1643–1658. doi: 10.1142/S0192415X19500848. [DOI] [PubMed] [Google Scholar]
- 116.Park JE, Ji HW, Kim HW, Baek M, Jung S, Kim SJ. Ginsenoside Rh2 regulates the CFAP20DC-AS1/MicroRNA-3614-3p/BBX and TNFAIP3 axis to induce apoptosis in breast cancer cells. Am J Chin Med. 2022;50(6):1703–1717. doi: 10.1142/S0192415X22500720. [DOI] [PubMed] [Google Scholar]
- 117.Ham J, Lee S, Lee H, Jeong D, Park S, Kim SJ. Genome-wide methylation analysis identifies NOX4 and KDM5A as key regulators in inhibiting breast cancer cell proliferation by ginsenoside Rg3. Am J Chin Med. 2018;46(06):1333–1355. doi: 10.1142/S0192415X18500702. [DOI] [PubMed] [Google Scholar]
- 118.Lee H, Lee S, Jeong D, Kim SJ. Ginsenoside Rh2 epigenetically regulates cell-mediated immune pathway to inhibit proliferation of MCF-7 breast cancer cells. J Ginseng Res. 2018;42(4):455–462. doi: 10.1016/j.jgr.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu C, Yang L, Wang Y, Zhou S, Luo J, Gu Y. Ginsenoside Rh2 reduces m6A RNA methylation in cancer via the KIF26B-SRF positive feedback loop. J Ginseng Res. 2021;45(6):734–743. doi: 10.1016/j.jgr.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hong J, Gwon D, Jang CY. Ginsenoside Rg1 suppresses cancer cell proliferation through perturbing mitotic progression. J Ginseng Res. 2022;46(3):481–488. doi: 10.1016/j.jgr.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang D, Xu X, Ye Q. Metabolism and immunity in breast cancer. Front Med. 2021;15(2):178–207. doi: 10.1007/s11684-020-0793-6. [DOI] [PubMed] [Google Scholar]
- 122.Wu Z, Wu J, Zhao Q, Fu S, Jin J. Emerging roles of aerobic glycolysis in breast cancer. Clin Transl Oncol. 2020;22(5):631–646. doi: 10.1007/s12094-019-02187-8. [DOI] [PubMed] [Google Scholar]
- 123.Liberti MV, Locasale JW. The warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Seyfried TN, Flores RE, Poff AM, D'Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35(3):515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Elia I, Haigis MC. Metabolites and the tumour microenvironment: from cellular mechanisms to systemic metabolism. Nat Metab. 2021;3(1):21–32. doi: 10.1038/s42255-020-00317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab. 2022;34(3):355–377. doi: 10.1016/j.cmet.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim S-Y. Targeting cancer energy metabolism: a potential systemic cure for cancer. Arch Pharmacal Res. 2019;42(2):140–149. doi: 10.1007/s12272-019-01115-2. [DOI] [PubMed] [Google Scholar]
- 128.Skuli SJ, Alomari S, Gaitsch H, Bakayoko A, Skuli N, Tyler BM. Metformin and cancer, an ambiguanidous relationship. Pharmaceuticals. 2022;15(5):626. doi: 10.3390/ph15050626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Micic D, Cvijovic G, Trajkovic V, Duntas LH, Polovina S. Metformin: Its emerging role in oncology. Horm-Int J Endocrinol Metab. 2011;10(1):5–15. doi: 10.14310/horm.2002.1288. [DOI] [PubMed] [Google Scholar]
- 130.Zhang B, Fu R, Duan Z, Shen S, Zhu C, Fan D. Ginsenoside CK induces apoptosis in triple-negative breast cancer cells by targeting glutamine metabolism. Biochem Pharmacol. 2022;202:115101. doi: 10.1016/j.bcp.2022.115101. [DOI] [PubMed] [Google Scholar]
- 131.Xia J, Zhang S, Zhang R, Wang A, Zhu Y, Dong M, Ma S, Hong C, Liu S, Wang D, Wang J. Targeting therapy and tumor microenvironment remodeling of triple-negative breast cancer by ginsenoside Rg3 based liposomes. J Nanobiotechnology. 2022;20(1):414. doi: 10.1186/s12951-022-01623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao H, Liang D, Li C, Xu G, Jiang M, Li H, Yin J, Song Y. 2-Deoxy-Rh2: a novel ginsenoside derivative, as dual-targeting anti-cancer agent via regulating apoptosis and glycolysis. Biomed Pharmacother. 2020;124:109891. doi: 10.1016/j.biopha.2020.109891. [DOI] [PubMed] [Google Scholar]
- 133.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14(3):155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 134.Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 2015;5(9):915–919. doi: 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Teng MWL, Swann JB, Koebel CM, Schreiber RD, Smyth MJ. Immune-mediated dormancy: an equilibrium with cancer. J Leukoc Biol. 2008;84(4):988–993. doi: 10.1189/jlb.1107774. [DOI] [PubMed] [Google Scholar]
- 136.Del Prete A, Wu Q. Tissue-resident immune cells in tumor immunity and immunotherapy. Front Cell Dev Biol. 2022;10:1068720. doi: 10.3389/fcell.2022.1068720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang J, Huang D, Saw PE, Song E. Turning cold tumors hot: from molecular mechanisms to clinical applications. Trends Immunol. 2022;43(7):523–545. doi: 10.1016/j.it.2022.04.010. [DOI] [PubMed] [Google Scholar]
- 138.Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The tumor microenvironment: a milieu hindering and obstructing antitumor immune responses. Front Immunol. 2020;11:940. doi: 10.3389/fimmu.2020.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maimela NR, Liu S, Zhang Y. Fates of CD8+ T cells in tumor microenvironment, computational and structural. Biotechnol J. 2019;17:1–13. doi: 10.1016/j.csbj.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baxevanis CN, Fortis SP, Perez SA. The balance between breast cancer and the immune system: challenges for prognosis and clinical benefit from immunotherapies. Semin Cancer Biol. 2021;72:76–89. doi: 10.1016/j.semcancer.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 141.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang D, DuBois RN. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis. 2015;36(10):1085–1093. doi: 10.1093/carcin/bgv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Adams S, Gatti-Mays ME, Kalinsky K, Korde LA, Sharon E, Amiri-Kordestani L, Bear H, McArthur HL, Frank E, Perlmutter J, Page DB, Vincent B, Hayes JF, Gulley JL, Litton JK, Hortobagyi GN, Chia S, Krop I, White J, Sparano J, Disis ML, Mittendorf EA. Current landscape of immunotherapy in breast cancer: a review. JAMA Oncol. 2019;5(8):1205–1214. doi: 10.1001/jamaoncol.2018.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12(4):269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol. 2022;19(4):237–253. doi: 10.1038/s41571-021-00588-9. [DOI] [PubMed] [Google Scholar]
- 146.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33(17):1974–U161. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhu Y, Wang A, Zhang S, Kim J, Xia J, Zhang F, Wang D, Wang Q, Wang J. Paclitaxel-loaded ginsenoside Rg3 liposomes for drug-resistant cancer therapy by dual targeting of the tumor microenvironment and cancer cells. J Adv Res. 2022 doi: 10.1016/j.jare.2022.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xia J, Ma S, Zhu X, Chen C, Zhang R, Cao Z, Chen X, Zhang L, Zhu Y, Zhang S, Li S, Gu G, Wei X, Yu K, Wang J. Versatile ginsenoside Rg3 liposomes inhibit tumor metastasis by capturing circulating tumor cells and destroying metastatic niches. Sci Adv. 2022;8(6):eabj1262. doi: 10.1126/sciadv.abj1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Luo X, Wang H, Ji D. Carbon nanotubes (CNT)-loaded ginsenosides Rb3 suppresses the PD-1/PD-L1 pathway in triple-negative breast cancer. Aging. 2021;13(13):17177–17189. doi: 10.18632/aging.203131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hong C, Liang J, Xia J, Zhu Y, Guo Y, Wang A, Lu C, Ren H, Chen C, Li S, Wang D, Zhan H, Wang J. One stone four birds: a novel liposomal delivery system multi-functionalized with ginsenoside Rh2 for tumor targeting therapy. Nanomicro Lett. 2020;12(1):129. doi: 10.1007/s40820-020-00472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dillekas H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8(12):5574–5576. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 153.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ribatti D, Tamma R, Annese T. Epithelial-mesenchymal transition in cancer: a historical overview. Transl Oncol. 2020;13(6):100773. doi: 10.1016/j.tranon.2020.100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochim Biophys Acta-Rev Cancer. 2017;1868(2):584–591. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 156.Song JH, Eum DY, Park SY, Jin YH, Shim JW, Park SJ, Kim MY, Park SJ, Heo K, Choi YJ. Inhibitory effect of ginsenoside Rg3 on cancer stemness and mesenchymal transition in breast cancer via regulation of myeloid-derived suppressor cells. PLoS ONE. 2020;15(10):e0240533. doi: 10.1371/journal.pone.0240533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang YS, Li H, Li Y, Zhang S, Jin YH. (20S)G-Rh2 inhibits NF-kappaB regulated epithelial-mesenchymal transition by targeting annexin A2. Biomolecules. 2020;10(4):528. doi: 10.3390/biom10040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang P, Du X, Xiong M, Cui J, Yang Q, Wang W, Chen Y, Zhang T. Ginsenoside Rd attenuates breast cancer metastasis implicating derepressing microRNA-18a-regulated Smad2 expression. Sci Rep. 2016;6:33709. doi: 10.1038/srep33709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang K, Li Y. Effects of ginsenoside compound K combined with cisplatin on the proliferation, apoptosis and epithelial mesenchymal transition in MCF-7 cells of human breast cancer. Pharm Biol. 2016;54(4):561–8. doi: 10.3109/13880209.2015.1101142. [DOI] [PubMed] [Google Scholar]
- 160.Peng B, He R, Xu Q, Yang Y, Hu Q, Hou H, Liu X, Li J. Ginsenoside 20(S)-protopanaxadiol inhibits triple-negative breast cancer metastasis in vivo by targeting EGFR-mediated MAPK pathway. Pharmacol Res. 2019;142:1–13. doi: 10.1016/j.phrs.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 161.Odle TG. Adverse effects of breast cancer treatment. Radiol Technol. 2014;85(3):297M–319M. [PubMed] [Google Scholar]
- 162.Fradley MG. Heart failure in patients with cancer treated with anthracyclines-revisiting the foundation of cardio-oncology. JAMA Netw Open. 2023;6(2):e2254677–e2254677. doi: 10.1001/jamanetworkopen.2022.54677. [DOI] [PubMed] [Google Scholar]
- 163.Cardinale D, Iacopo F, Cipolla CM. Cardiotoxicity of anthracyclines. Front Cardiovasc Med. 2020;7:26. doi: 10.3389/fcvm.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Henriksen PA. Anthracycline cardiotoxicity: an update on mechanisms, monitoring and prevention. Heart. 2018;104(12):971–977. doi: 10.1136/heartjnl-2017-312103. [DOI] [PubMed] [Google Scholar]
- 165.Tarantini L, Feola M, Albini A, Gori S, Foglietta J, Cicoira MA, Pulignano G. Heart failure in women treated with adjuvant trastuzumab for breast cancer. G Ital Cardiol. 2006;13(5 Suppl 1):54S–62S. [PubMed] [Google Scholar]
- 166.Miao J-X, Gao S, Fan L, Cao F. Progress in prevention and treatment of myocardial injury induced by cancer therapy. Chin Med J. 2019;132(22):2724–2728. doi: 10.1097/CM9.0000000000000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu G, Zhang J, Sun F, Ma J, Qi X, Carbone F. Ginsenoside Rg2 attenuated trastuzumab-induced cardiotoxicity in rats. Biomed Res Int. 2022;2022:1–6. doi: 10.1155/2022/8866660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu G, Qi X, Li X, Sun F. Ginsenoside Rg2 protects cardiomyocytes against trastuzumab-induced toxicity by inducing autophagy. Exp Ther Med. 2021;21(5):1–7. doi: 10.3892/etm.2021.9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hou J, Yun Y, Cui C, Kim S. Ginsenoside Rh2 mitigates doxorubicin-induced cardiotoxicity by inhibiting apoptotic and inflammatory damage and weakening pathological remodelling in breast cancer-bearing mice. Cell Prolif. 2022;55(6):e13246. doi: 10.1111/cpr.13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li C, Gou X, Gao H. Doxorubicin nanomedicine based on ginsenoside Rg1 with alleviated cardiotoxicity and enhanced antitumor activity. Nanomedicine. 2021;16(29):2587–2604. doi: 10.2217/nnm-2021-0329. [DOI] [PubMed] [Google Scholar]
- 171.Li Y, Wang Y, Niu K, Chen X, Xia L, Lu D, Kong R, Chen Z, Duan Y, Sun J. Clinical benefit from EGFR-TKI plus ginsenoside Rg3 in patients with advanced non-small cell lung cancer harboring EGFR active mutation. Oncotarget. 2016;7(43):70535–70545. doi: 10.18632/oncotarget.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Queiroga F, Cogliati B. Translational and comparative research on innovative anti-cancer therapies. Cancers. 2023;15(4):1335. doi: 10.3390/cancers15041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lei F, Zhang A, Zhang Q, Zhang L. Advances in research on allelopathy of ginseng and American ginseng. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi =China J Chin Mater Med. 2010;35(17):2221–6. [PubMed] [Google Scholar]
- 174.Jin Y, Huynh DTN, Nguyen TLL, Jeon H, Heo K-S. Therapeutic effects of ginsenosides on breast cancer growth and metastasis. Arch Pharmacal Res. 2020;43(8):773–787. doi: 10.1007/s12272-020-01265-8. [DOI] [PubMed] [Google Scholar]
- 175.Kim D-H. Gut microbiota-mediated pharmacokinetics of ginseng saponins. J Ginseng Res. 2018;42(3):255–263. doi: 10.1016/j.jgr.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen Z, Zhang Z, Liu J, Qi H, Li J, Chen J, Huang Q, Liu Q, Mi J, Li X. Gut microbiota: therapeutic targets of ginseng against multiple disorders and ginsenoside transformation. Front Cell Infect Microbiol. 2022;12:853981. doi: 10.3389/fcimb.2022.853981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nakhjavani M, Hardingham JE, Palethorpe HM, Tomita Y, Smith E, Price TJ, Townsend AR. Ginsenoside Rg3: potential molecular targets and therapeutic indication in metastatic breast cancer. Medicines. 2019;6(1):17. doi: 10.3390/medicines6010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dai L, Liu K, Si C, Wang L, Liu J, He J, Lei J. Ginsenoside nanoparticle: a new green drug delivery system. J Mater Chem B. 2016;4(3):529–538. doi: 10.1039/c5tb02305j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.