Abstract
The mitochondrion serves a critical role as a platform for energy transduction, signaling, and cell death pathways relevant to common diseases of the myocardium such as heart failure. This review focuses on the molecular regulatory events and downstream effector pathways involved in mitochondrial energy metabolic derangements known to occur during the development of heart failure.
Introduction
All cellular processes are driven by ATP-dependent pathways. The heart has perpetually high energy demands related to the maintenance of specialized cellular processes, including ion transport, sarcomeric function, and intracellular Ca2+ homeostasis. Myocardial workload (energy demand) and energy substrate availability (supply) are in continual flux, yet the heart has a limited capacity for substrate storage. Thus, ATP-generating pathways must respond proportionately to dynamic fluctuations in physiological demands and fuel delivery. The time frame of such metabolic regulatory responses ranges from seconds to minutes (acute) or hours to days (chronic) and involves regulation at multiple levels, including allosteric control of enzyme activity via metabolic intermediates, signal transduction events, and the regulation of genes encoding rate-limiting enzymes and proteins.
Metabolic regulation is inextricably linked with cardiac function. This metabolism-function relationship is relevant to diseases that lead to cardiac hypertrophy and heart failure. The progression to heart failure of any cause is associated with a gradual but progressive decline in the activity of mitochondrial respiratory pathways leading to diminished capacity for ATP production. Reduced capacity for energy transduction leads to secondary dysregulation of cellular processes critical for cardiac pump function, including Ca2+ handling and contractile function, which results in a downward spiral of increased energy demand and diminished function. Evidence has emerged that energy deficiency can be a cause and an effect of heart failure. Cardiac metabolic regulatory events may also be adaptive in certain disease states. For example, in the ischemic heart, a reduction in mitochondrial oxidative capacity serves to reduce oxygen consumption in the context of limited O2 availability. Many of the metabolic regulatory events that dictate fuel selection and capacity for ATP production in the normal and failing heart occur at the level of gene expression. The consequence of specific metabolic gene regulatory events as adaptive versus maladaptive in the context of myocardial disease is an area of intense investigation.
The purposes of this review are to provide a brief overview of cardiac energy metabolic pathways with emphasis on the mitochondrion, to describe the gene regulatory circuitry involved in the regulation of cardiac mitochondrial energy metabolism in the normal heart, to summarize the current knowledge about how this metabolic regulatory network is altered during the development of heart failure, and to review the evidence that links altered energy metabolism to the development of heart failure. Emphasis will be given to gene regulatory mechanisms and upstream signaling pathways.
Cardiac energy metabolic pathways
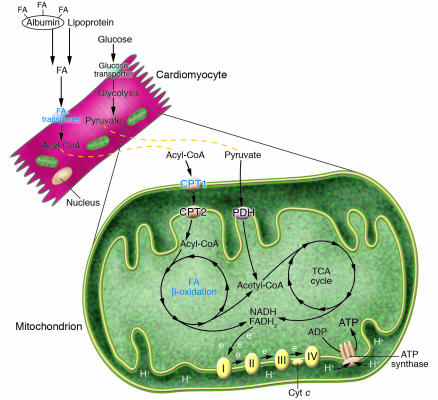
Oxidation of fatty acids (FAs) and glucose in mitochondria accounts for the vast majority of ATP generation in the healthy adult heart (1, 2). FAs are the preferred substrate in the adult myocardium, supplying about 70% of total ATP (3–5). FAs derived from circulating triglyceride-rich lipoproteins and albumin-bound nonesterified FAs are oxidized in the mitochondrial matrix by the process of FA β-oxidation (FAO), whereas pyruvate derived from glucose and lactate is oxidized by the pyruvate-dehydrogenase (PDH) complex, localized within the inner mitochondrial membrane (Figure 1). Acetyl-CoA, derived from both pathways, enters the tricarboxylic acid (TCA) cycle. Reduced flavin adenine dinucleotide (FADH2) and NADH are generated via substrate flux through the β-oxidation spiral and the TCA cycle, respectively. The reducing equivalents enter the electron transport chain, producing an electrochemical gradient across the mitochondrial membrane that drives ATP synthesis in the presence of molecular oxygen (oxidative phosphorylation).
Figure 1.
Pathways involved in cardiac energy metabolism. FA and glucose oxidation are the main ATP-generating pathways in the adult mammalian heart. Acetyl-CoA derived from FA and glucose oxidation is further oxidized in the TCA cycle to generate NADH and FADH2, which enter the electron transport/oxidative phosphorylation pathway and drive ATP synthesis. Genes encoding enzymes involved at multiple steps of these metabolic pathways (i.e., uptake, esterification, mitochondrial transport,and oxidation) are transcriptionally regulated by PGC-1α with its nuclear receptor partners, including PPARs and ERRs (blue text). Glucose uptake/oxidation and electron transport/oxidative phosphorylation pathways are also regulated by PGC-1α via other transcription factors, such as MEF-2 and NRF-1. Cyt c, cytochrome c.
Mitochondrial enzymes are encoded by both nuclear and mitochondrial genes (reviewed in ref. 6). All of the enzymes of β-oxidation and the TCA cycle, and most of the subunits of electron transport/oxidative phosphorylation, are encoded by nuclear genes. The mitochondrial genome is comprised of 1 circular double-stranded chromosome that encodes 13 electron transport chain subunits within complexes I, III, and IV (7). Since mitochondrial number and function require both nuclear and mitochondrial-encoded genes, coordinated mechanisms exist to regulate the 2 genomes and determine overall cardiac oxidative capacity. In addition, distinct pathways exist to coordinately regulate nuclear genes encoding component mitochondrial pathways.
The transcriptional network regulating cardiac mitochondrial biogenesis and respiratory function
PGC-1 α: an inducible integrator of transcriptional circuits regulating mitochondrial biogenesis and function.
The PPARγ coactivator-1 (PGC-1) family of transcriptional coactivators is involved in regulating mitochondrial metabolism and biogenesis. PGC-1α was the first member discovered through its functional interaction with the nuclear receptor PPARγ in brown adipose tissue (BAT), a mitochondrial-rich tissue involved in thermogenesis (8). Two PGC-1α–related coactivators, PGC-1β (also called PERC) and PGC-1–related coactivator (PRC), have since been identified (9–11). PRC is ubi-quitously expressed and coactivates transcription factors involved in mitochondrial biogenesis; however, to date there is no direct empirical evidence that PRC drives or is necessary for mitochondrial biogenesis (9, 12). PGC-1α and PGC-1β share some regulatory overlap. Both are preferentially expressed in tissues with high oxidative capacity, such as heart, slow-twitch skeletal muscle, and BAT, where they serve critical roles in the regulation of mitochondrial functional capacity (8, 10, 13–15). PGC-1α regulates additional metabolic pathways, including hepatic gluconeogenesis and skeletal muscle glucose uptake (16–18). Based on its tissue expression pattern, PGC-1β probably plays a role in regulating energy metabolism in the heart, although its precise role in the normal heart and in mediating alterations in energy metabolism observed in heart failure has not been investigated.
PGC-1α is distinct from other PGC-1 family members, indeed from most coactivators, in its broad responsiveness to developmental alterations in energy metabolism and physiological and pathological cues at the level of expression and transactivation. In the heart, PGC-1α expression increases at birth coincident with an increase in cardiac oxidative capacity and a perinatal shift from reliance on glucose metabolism to the oxidation of fats for energy (19). PGC-1α is induced by physiological stimuli that increase ATP demand and stimulate mitochondrial oxidation, including cold exposure, fasting, and exercise (8, 19–23). Activation of the PGC-1α regulatory cascade increases cardiac mitochondrial oxidative capacity in the heart. In cardiac myocytes in culture, PGC-1α increases mitochondrial number, upregulates expression of mitochondrial enzymes, and increases rates of FA oxidation and coupled respiration (19, 24). A mouse cardiac-specific conditional transgenic expression system was used to demonstrate that PGC-1α triggers mitochondrial biogenesis in vivo in a developmental stage–dependent manner: permissive during the neonatal period but less so in the adult heart (25). In summary, PGC-1α is an inducible coactivator that coordinately regulates cardiac fuel selection and mitochondrial ATP-producing capacity.
How does PGC-1α exert its pleiotropic effects on mitochondrial biogenesis and respiratory function? Recent work by several laboratories has provided insight into the downstream transcriptional regulatory circuits through which PGC-1α mediates its effects (Figure 2) (6). PGC-1α activates expression of nuclear respiratory factor-1 (NRF-1) and NRF-2 and directly coactivates NRF-1 on its target gene promoters (13). NRF-1 and NRF-2 regulate expression of mitochondrial transcription factor A (Tfam), a nuclear-encoded transcription factor that binds regulatory sites on mitochondrial DNA and is essential for replication, maintenance, and transcription of the mitochondrial genome (26–28). Furthermore, NRF-1 and NRF-2 regulate the expression of nuclear genes encoding respiratory chain subunits and other proteins required for mitochondrial function (29, 30).
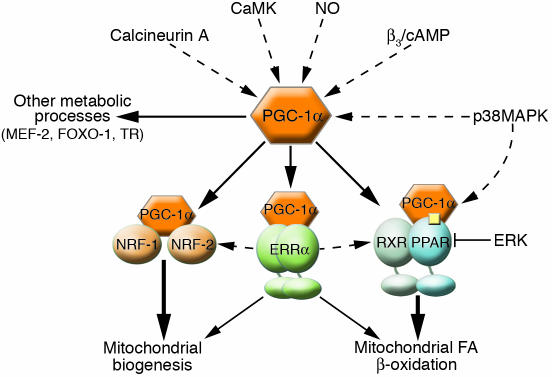
Figure 2.
PGC-1α is an integrator of the transcriptional network regulating mitochondrial biogenesis and function. Numerous signaling pathways, including Ca2+-dependent, NO, MAPK, and β-adrenergic pathways (β3/cAMP), activate the PGC-1α directly by increasing either PGC-1α expression or activity. Additionally, the p38MAPK pathway selectively activates PPARα, which may bring about synergistic activation in the presence of PGC-1α, whereas ERK-MAPK has the opposite effect. These signaling pathways transduce physiological stimuli, such as stress, fasting, and exercise, to the PGC-1α pathway. PGC-1α, in turn, coactivates transcriptional partners, including NRF-1 and -2, ERRα, and PPARα, which regulate mitochondrial biogenesis and FA-oxidation pathways. Dashed lines indicate activation mediated by signal transduction pathways in contrast to the coactivation by PGC-1α, which is denoted by solid lines. The arrows from ERRα to the NRFs and the PPAR complex indicate that ERRα activates these pathways at the level of expression.
PGC-1 α coactivates the PPAR and ERR nuclear receptors, critical regulators of myocardial FA utilization.
PGC-1α regulates genes involved in the cellular uptake and mitochondrial oxidation of FAs through direct coactivation of PPARs and estrogen-related receptors (ERRs) (Figure 2) (31–34). PPARs are FA-activated members of the nuclear receptor superfamily of transcription factors that serve as central regulators of cardiac FA metabolism. There are 3 PPAR isoforms (α, β/δ, and γ); PPARα and PPARβ are the primary regulators of FA metabolism in the heart. PPARs function by binding as obligate heterodimers with the retinoid X receptor α (RXRα) and recruiting coactivators, including PGC-1α, in response to direct binding and activation by FAs and their derivatives, which serve as ligands. PPARα regulates genes involved in virtually every step of cardiac FA utilization (reviewed in ref. 35). PPARα-null mice have reduced cardiac expression of genes involved in the cellular uptake, mitochondrial transport, and mitochondrial (and peroxisomal) oxidation of FAs (36–38). Myocardial FA uptake and oxidation rates are decreased in these mice, while glucose oxidation rates are increased (39). PPARα-null mice have also provided evidence that PPARα serves an important homeostatic function in the context of physiological and dietary stressors. When subjected to fasting or pharmacological inhibition of FAO, PPARα–/– mice develop massive cardiac myocyte lipid accumulation and hepatic steatosis (40, 41). These results have defined PPARα as an important regulator of myocardial energy substrate preference and FAO capacity within the PGC-1α transcriptional regulatory cascade.
In contrast to the biological function of PPARα, that of PPARβ (also known as PPARδ) in the heart has not been extensively studied. However, recent evidence indicates that cardiac PPARβ target genes significantly overlap with those of PPARα. PPARβ selective ligands induce mitochondrial FAO enzyme genes and increase palmitate oxidation rates in cardiac myocytes from both wild-type and PPARα–/– mice (42, 43). Consistent with findings from activation studies, cardiac-specific deletion of the PPARβ gene results in reduced expression of FAO enzyme genes and diminished palmitate oxidation rates, which is similar to what occurs in PPARα-null mice (44). In contrast to PPARα–/– mice, however, cardiac-specific PPARβ-null mice do not exhibit a fasting-induced phenotype. Rather, PPARβ-deficient mice develop a cardiomyopathy under basal conditions. Collectively, these results suggest that, although PPARα and PPARβ drive similar gene targets, PPARβ probably serves to regulate basal metabolism whereas PPARα is perhaps more important in the response to physiological conditions that increase FA delivery.
ERRs are a subfamily of orphan nuclear receptors for which a role in cardiac energy metabolism has recently been described. There are 3 members of the ERR family: ERRα, ERRβ, and ERRγ (45–47). Cardiac ERRα expression increases dramatically following birth, coincident with the switch to FAs as an energy substrate and the upregulation of PPARα and PGC-1α and of enzymes involved in FA uptake and mitochondrial oxidation (33, 48). Both ERRα and ERRγ are directly coactivated by PGC-1α while ERRα is further regulated by PGC-1α at the gene expression level (33, 34, 49, 50). Recent evidence suggests that ERRα cross-regulates other transcription factors within the PGC-1α network. In cardiac myocytes, ERRα activates expression of many known PPARα and NRF target genes involved in cellular FA utilization and mitochondrial electron transport/oxidative phosphorylation, respectively (24). This apparently overlapping function was recently shown to occur, in part, via direct transactivation of the PPARα and Gabpa (NRF-2 subunit) gene promoters. (Figure 2) (24, 51). In addition, ERRα cooperates with NRF-1 and NRF-2 to regulate mitochondrial biogenesis and expression of oxidative phosphorylation enzyme genes (51, 52). Collectively, these results support a role for ERRα as an amplifier of cardiac oxidative energy metabolism downstream of PGC-1α (Figure 2).
Signaling pathways trigger changes in mitochondrial function through the PGC-1 α regulatory cascade.
How do PGC-1α and its transcription factor partners receive input from physiological and pathological stimuli? Insights into the physiological responsiveness of the PGC-1α pathway come from identification of signal transduction pathways that modulate the activity of PGC-1α and its downstream partners (Figure 2). PGC-1α is upregulated in response to β-adrenergic signaling, consistent with the involvement of this pathway in thermogenesis (8, 53). The stress-activated MAPK p38 activates PGC-1α by both increasing PGC-1α protein stability and promoting dissociation of a repressor (21, 54, 55). p38 also increases mitochondrial FAO through selective activation of the PGC-1α partner, PPARα (56). Conversely, the ERK-MAPK pathway inactivates the PPARα/RXRα complex via direct phosphorylation (57). Therefore, distinct limbs of the MAPK pathway exert opposing regulatory influences on the PGC-1α cascade. Recently, NO has emerged as a novel signaling molecule proposed to integrate pathways involved in regulating mitochondrial biogenesis. NO induces mitochondrial proliferation and increases coupled respiration and ATP content in numerous cell types via a cGMP-dependent pathway (58, 59). The effects of NO/cGMP appear to be mediated, at least in part, by transcriptional activation of PGC-1α expression levels (58).
Recent evidence has also implicated Ca2+ signaling in the control of the PGC-1α/PPARα regulatory pathway. Transgenic mice overexpressing constitutively active calcineurin (Cn) or Ca2+/calmodulin-dependent kinase (CaMK) in skeletal muscle displayed increased PGC-1α expression and expansion of slow-twitch oxidative fibers and mitochondrial biogenesis (22, 60, 61). Activation of PGC-1α by Cn and CaMK is mediated transcriptionally through myocyte enhancer factor-2 (MEF-2) and cAMP response element–binding protein (CREB), respectively (62). In the heart, exercise training activates the Cn and CaMK pathways coincident with maintained or enhanced mitochondrial function; however, paradoxically, overexpression of constitutively activated forms of Cn or CaMK results in reduced mitochondrial oxidation and heart failure (63–65). Recent studies have shown that Cn and CaMK both activate PGC-1α expression in cardiac myocytes but have distinct effects on downstream targets of the PGC-1α pathway (66). CaMK activates genes involved in glucose oxidation and mitochondrial electron transport while Cn selectively activates mitochondrial FAO enzyme genes. The selective effects of Cn are thought to be mediated, in part, through direct transcriptional activation of the PPARα gene. Consistent with the gain-of-function results, reduction in Cn catalytic activity through cardiac-specific deletion of the CnB regulatory subunit results in reduced PGC-1α and PPARα expression and downregulation of FAO enzymes (66).
Perturbations in PGC-1/PPAR signaling in the hypertrophied and failing heart
Numerous studies have demonstrated altered cardiac substrate preference in the hypertrophied and failing heart. However, the direction of the “fuel shifts” varies with the etiology and severity of ventricular dysfunction. Studies using animal models of ventricular hypertrophy due to hypertension or imposed pressure overload have consistently demonstrated a myocardial shift from FAO toward glucose oxidation (67–72). Changes in gene expression in the hypertrophied heart, including downregulation of mitochondrial FAO enzymes, are consistent with the observed metabolic alterations (73, 74).
The results of studies of energy substrate shifts in the failing heart, particularly in humans, have not led to a clear conclusion. Some investigators have shown that expression of FAO genes and corresponding enzymatic activities are reduced in the failing rodent and human heart (70, 73, 75). Consistent with these findings, recent cardiac positron emission tomography (PET) studies in humans have shown a shift away from FA utilization with hypertensive cardiac hypertrophy and idiopathic cardiomyopathy (76, 77). However, others have demonstrated the opposite metabolic profile or no change in substrate utilization in humans and animal models of heart failure of mild-to-moderate severity (78–80). Animal models of failure induced by pacing or myocardial infarction show that earlier stages are not associated with a switch from FAs to glucose as the primary energy substrate. Dogs with moderate severity coronary microembolism–induced heart failure exhibited FA and glucose uptake patterns that were indistinguishable from those of controls (81). These apparent discrepancies have been attributed to both the severity of failure and temporal differences during the progressive remodeling that characterizes the transition to heart failure. The specific etiology of the myocardial disease may also play an important role.
Significant progress has been made in delineating the gene regulatory events driving the reduction of myocardial FA utilization in the hypertrophied, failing, and hypoxic heart. Given the importance of PPARα for the transcriptional control of cardiac lipid metabolism, this nuclear receptor has served as an important focus for such studies. In rodent models of pressure overload hypertrophy, expression of PPARα and PGC-1α is reduced in the hypertrophied and failing heart and in hypertrophied cardiac myocytes in culture (82–87). Levels of the PPAR partner, RXRα, are also reduced in a canine pacing-induced model of heart failure and by hypoxia in cardiac myocytes (88, 89). Interestingly, a reciprocal induction of transcription factors, including chicken ovalbumin upstream promoter-transcription factor (COUP-TF) and Sp1 and Sp3, which repress promoter activity of key FAO enzyme genes, is observed in hypertrophy and heart failure models and is thought to contribute to the reduction in myocardial FAO (82, 90). Moreover, PPARα activity is inhibited posttranslationally in the hypertrophied cardiac myocyte in culture through ERK-MAPK–dependent phosphorylation (83). The results of these studies suggest that one key mechanism involved in the energy substrate switches in the hypertrophied and failing heart involves deactivation of the PGC-1α/PPARα complex at both transcriptional and posttranscriptional levels (Figure 3).
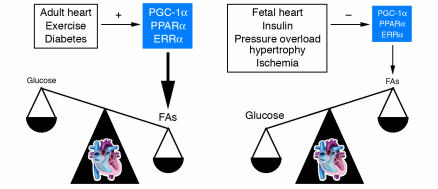
Figure 3.
Cardiac energy substrate selection is a dynamic balance influenced by developmental, physiological, and pathological cues. In the fetal heart, glucose oxidation is favored, whereas FA oxidation serves as the major ATP-generating pathway in the adult myocardium. Significant shifts in substrate preference occur in response to dietary (insulin) and physiological (exercise) stimuli. Certain pathophysiological contexts, such as hypertrophy and ischemia, drive metabolism toward glucose utilization, whereas in uncontrolled diabetes, the heart utilizes FAs almost exclusively. In some cases, as in early response to pressure overload–induced hypertrophy, these metabolic shifts are thought to be protective. Alterations in activity or expression of nuclear receptors (PPARs and ERRs) and PGC-1α mediate these shifts in energy substrate utilization.
In contrast to heart failure related to pressure overload or ischemia, the cardiomyopathy that develops in the context of insulin resistance and frank diabetes is associated with increased cardiac reliance on FAs as the primary energy substrate (Figure 3) (91–94). This fuel switch is linked to the combined effects of myocyte insulin resistance and high-circulating free FAs. We have found that, consistent with this metabolic profile, the expression and activity of PPARα, PGC-1α, and the enzymes of mitochondrial FAO are induced in both insulin-deficient and insulin-resistant forms of diabetes in mouse models (95, 96). The mechanism involved in the activation of PPARα signaling in the diabetic heart is unknown but likely involves increased cellular import of FAs, which serve as ligands for this nuclear receptor. In contrast, other studies have demonstrated a reduction in PPARα transcript levels in diabetic myocardium (97, 98). This apparent discrepancy could reflect temporal-dependent regulatory events during progression of diabetic myocardial disease. Future studies aimed at defining the precise time-course of PPARα-driven regulatory events during the development of diabetic cardiomyopathy are needed.
Do derangements in mitochondrial energy metabolism cause heart failure?
Myocardial fuel shifts as a cause of heart failure.
Do deactivation of PGC-1α/PPARα and the switch away from FAO in the hypertrophied heart contribute to the pathological remodeling that leads to heart failure? Similarly, does chronic activation of PPARα and myocardial FAO in the diabetic heart lead to cardiac dysfunction? Alternatively, do these myocardial substrate shifts serve adaptive functions in the diseased heart? The answers to these questions have not been resolved, but evidence provided largely by observations in rare genetic human diseases and genetically modified mice offers some insight. However, perhaps the strongest evidence supporting deleterious effects of reduced capacity for cardiac FA utilization comes from the observations of the cardiac phenotype of human genetic defects in FAO pathway enzymes. Children with deficiencies in enzymes involved in mitochondrial long-chain FAO often develop a stress-induced cardiomyopathy associated with myocardial lipid accumulation (99). In addition, mouse models in which the FAO enzymes, very-long-chain acyl-CoA dehydrogenase (VLCAD) or long-chain acyl-CoA dehydrogenase (LCAD), have been disrupted exhibit cardiomyopathies similar to that observed in humans (100, 101). Furthermore, the cardiomyopathic phenotype of mice with cardiac-specific deletion of the PPARβ gene also supports the conclusion that a reduction in capacity to oxidize FAs in the heart has deleterious effects (44). The PPARβ–/– hearts accumulate lipids in association with the development of cardiac hypertrophy, which ultimately leads to dilated cardiomyopathy, myocyte apoptosis, and death.
One proposed mechanism for cardiac dysfunction in these models is excess intracellular lipid accumulation resulting in myocyte dysfunction or death, termed “lipotoxicity” (reviewed in ref. 102). The heart and other high-energy flux organs are adapted to closely match energy substrate import and utilization, not storage. A mismatch can result from increased lipid delivery, such as occurs in obesity or diabetes, or impaired FA oxidation, as in the aforementioned models. The resulting derangements in cellular lipid homeostasis can lead to accumulation of lipid intermediates such as acyl-CoA thioesters, acylcarnitines, ceramides, and triglycerides, molecules that could confer cellular toxicity. Transgenic models in which proteins involved in FA uptake/delivery are overexpressed also exhibit lipid accumulation and systolic dysfunction (103, 104). These hearts display evidence of myocyte dropout due to activation of apoptotic pathways. Indeed, direct treatment with saturated long-chain FAs has been shown to trigger apoptosis in cardiac myocytes as well as in other cell types in culture via mechanisms that may involve generation of reactive oxygen species (105–107). Collectively, observations in mice and humans indicate that severe reduction in mitochondrial FAO capacity sets the stage for cardiac lipotoxic effects related to lipid intermediates that accumulate in the context of impaired catabolism.
Despite compelling evidence for the deleterious effects of reduced mitochondrial FAO in genetic models and human deficiency states, there is some data to suggest that a shift from FAO to glucose utilization in the hypertrophied heart may be adaptive, at least in the short term. Taegtmeyer and coworkers have shown that reactivation of FAO in a rat ventricular-pressure overload model causes ventricular dysfunction (108). In addition, despite reduced cardiac FAO rates, PPARα-null mice do not exhibit overt ventricular dysfunction (39). It is likely that the degree and duration of the pathophysiological stimulus as well as the systemic metabolic state (e.g., levels of circulating lipids) ultimately determine whether alterations in FAO capacity contribute to the pathogenesis of heart failure.
As described above, the insulin resistant and diabetic heart is characterized by increased FAO rates due, perhaps in part, to chronic activation of the PPARα gene regulatory pathway. Studies of mice genetically modified to mimic the metabolic derangements of the diabetic heart have provided evidence that chronically increased reliance on FAs for energy leads to pathological signatures of the diabetic heart. Mice with cardiac-specific overexpression of PPARα (MHC-PPARα mice) exhibit increased expression of PPARα target genes involved in cellular FA import and peroxisomal and mitochondrial FAO, coincident with lipid accumulation and increased rates of FAO (96, 109). Interestingly, myocardial glucose uptake and oxidation rates are reciprocally decreased in the MHC-PPARα mouse, a metabolic phenotype that mimics the diabetic heart. These results demonstrate that a primary drive on the PPARα gene regulatory pathway triggers cross-talk suppression of glucose utilization pathways. Importantly, the metabolic derangements of MHC-PPARα mice are associated with ventricular diastolic/systolic dysfunction at baseline, which becomes more severe in the context of increased delivery of FAs to the heart such as occurs with high-fat feeding or insulinopenia (95). Thus, restricting the heart to reliance on FAs to the virtual exclusion of glucose oxidation leads to development of cardiomyopathy (109).
The metabolic dysregulation in the MHC-PPARα heart is associated with neutral lipid accumulation and increased production of reactive species consistent with the importance of oxidative stress and mitochondrial dysfunction in diabetic cardiomyopathy (95). Direct characterization of mitochondria from hearts in type 1 diabetic models has revealed evidence of damaged mitochondria and impaired mitochondrial respiration, presumably due to oxidative stress (110). In the same type 1 model, overexpression of antioxidant proteins such as metallothionein and catalase reduced reactive oxygen species and rescued cardiac contractility (111–113). A similar effect was observed with catalase overexpression in a mouse model of type 2 diabetes (112). The observations from elevated FA flux and FAO-deficient states, which indicate that either a chronic increase (diabetes) or decrease (pressure overload hypertrophy) in myocardial FAO can lead to heart failure, emphasize the importance of substrate flexibility for normal cardiac function (Figure 3).
Derangements in mitochondrial ATP generation.
Evidence for a link between mitochondrial respiratory dysfunction and heart failure is compelling. The cardiomyopathic phenotype of humans with mitochondrial genome defects underscores the importance of high-capacity mitochondrial ATP production for normal striated-muscle function. Mutations in both nuclear- and mitochondrial-encoded genes account for heritable respiratory chain defects (for an extensive review of this topic, see refs. 114–117). Respiratory chain defects typically present as multisystem dysfunction disproportionately affecting organs with high ATP demand, such as the heart, skeletal muscle, and the central nervous system. Cardiomyopathy may develop during childhood or at later ages. Mouse models of mitochondrial dysfunction have also provided important information about the role of mitochondrial proteins in regulating mitochondrial number and function as well as the way in which altered mitochondrial energetics contribute to the development of heart failure. Tfam is a nuclear-encoded mitochondrial transcription factor necessary for mitochondrial biogenesis and gene expression. Cardiac-specific Tfam–/– mice exhibit reduced respiratory capacity and mitochondrial DNA in the heart before birth coincident with a high neonatal mortality (118). Surviving animals develop hypertrophy, progressing to dilated cardiomyopathy and conduction abnormalities, and die by 4 months of age. Interestingly, cardiac dysfunction is accompanied by a metabolic shift from FAO to glucose oxidation (119).
Adenine nucleotide translocators (Ants) are mitochondrial-membrane proteins involved in the transport of cytoplasmic ADP in exchange for mitochondrial ATP, so mitochondria deficient in Ant have reduced capacity for substrate level phosphorylation. Mice targeted for the Ant1 gene, the cardiac/skeletal muscle expressed isoform, exhibit reduced ADP-dependent respiration rates in heart and skeletal muscle in spite of increased mitochondrial number (120, 121). The mitochondrial proliferation is likely a compensatory response to cellular energy deficiency but is thought to contribute to progressive cardiac hypertrophy that develops in these mice (121, 122). ATP deficiency is thought to be a primary cause for the observed pathophysiology, but these models also support a role for mitochondrial-derived ROS in mediating cellular damage (123). Although the precise cellular insults are still unknown, these genetic models demonstrate that mitochondrial dysfunction is sufficient for the development of heart failure.
Several recently developed mouse models have identified exciting potential links between PGC-1α–mediated control of mitochondrial function and the development of heart failure. Sano et al. recently found that overexpression of cyclin T/Cdk9, an RNA polymerase kinase, triggers cardiac hypertrophy (124). Further studies revealed that in the context of ventricular pressure overload, mice overexpressing Cdk9 in the heart develop a fulminant apoptotic cardiomyopathy (125). Gene expression–profiling studies demonstrated that Cdk9 suppresses expression of PGC-1α and its downstream targets involved in mitochondrial respiratory function. Rescue of PGC-1α expression in cardiac myocytes in culture prevented Cdk9-triggered apoptosis. In a separate mouse model, chronic activation of PGC-1α in the heart, as occurs in diabetes, led to ventricular dysfunction. In a study using an inducible, cardiac-specific transgenic system, chronic overexpression of PGC-1α protein in the adult mouse heart caused mitochondrial ultrastructural abnormalities and reduced myofibrillar density, which led to cardiomyopathy and diastolic dysfunction (25). Interestingly, the mitochondrial proliferative response is reversible and the cardiomyopathy rescued upon cessation of transgene expression. The basis for this reversible cardiomyopathy is unknown but could involve the accumulation of reactive intermediates or abnormalities in ATP generation. Taken together, these recent findings suggest that, in the context of a mechanical stress such as pressure overload, reduced levels of PGC-1α predispose the heart to pathological remodeling related to mitochondrial dysfunction and apoptosis. Conversely, chronic activation of PGC-1α and its downstream targets, such as PPARα, that mimic the diabetic state also leads to ventricular dysfunction through mechanisms that are reversible.
Despite strong evidence for a link between mitochondrial dysfunction and heart failure in genetic models, the role of altered mitochondrial ATP generation in the pathogenesis of acquired forms of heart failure is less clear. Phosphocreatine (pCR) serves as the main energy store in myocardium (for review, see ref. S1; see Supplemental References; supplemental material available online with this article; doi:10.1172/JCI200524405DS1). Early studies revealed that myocardial pCR/ATP ratios are reduced in the hypertrophied and failing heart while absolute ATP levels are only detectably reduced in heart failure (S2–S4). However, the concentration of ATP measured in the failing myocardium is still above the Km for most cellular ATPases. Consistent with a gradual fall in intracellular high-energy phosphates, the “energy sensor kinase,” AMP-activated protein kinase (AMPK), is upregulated and activated during hypertrophy (S5). These results suggest that myocardial energy reserves are disproportionately reduced during hypertrophy and heart failure progression compared to absolute ATP levels. In dog models of pacing-induced heart failure, ATP concentrations fall gradually coincident with a reduction in mitochondrial respiration rates during the progression to heart failure (S6, S7). These functional changes are associated with evidence of structural abnormalities in mitochondria from these hearts (S8). Reduced mitochondrial oxidative capacity has also been observed in rodent heart failure models (87). Further studies will be necessary to accurately delineate the temporal pattern of alterations in bioenergetics during the development of heart failure. Murine genetic loss-of-function and pharmacological rescue strategies in larger animal models should serve as useful experimental strategies to determine cause-and-effect relationships.
Metabolic modulators as a new treatment strategy for heart failure? A question of balance
In summary, evidence is emerging to support the concept that alterations in myocardial fuel selection and energetics are linked to the development and progression of heart failure. Accordingly, metabolic pathways involved in cardiac FA and glucose utilization or ATP generation are attractive targets of novel therapeutic strategies aimed at the prevention or early treatment of heart failure. Indeed, specific activators for each of the PPARs have been developed and are currently used for treatment of hyperlipidemia (e.g., fibrates) and diabetes (thiazolidinediones). Activation of the PPAR pathway in heart or extracardiac tissues, such as adipose or liver, could theoretically reduce cardiac lipotoxicity by reducing lipid delivery or increasing mitochondrial oxidation. However, a strong word of caution is necessary. As described above, it is now clear that the degree of metabolic modulation is an important determinant of whether shifts in energy substrate utilization serve adaptive or maladaptive functions in the context of disease states that predispose patients to heart failure. Conversely, chronic activation of PPARα could lead to deleterious effects, particularly in the context of diabetes, hyperlipidemic states, or the ischemic heart, given the potential for increased mitochondrial oxidative flux, which could generate ROS and increase oxygen consumption. It is likely that the response will be disease-specific and dependent on the systemic metabolic phenotype. Studies in large animals and humans will be particularly informative. Such studies will require rigorous metabolic phenotyping approaches with imaging modalities such as PET and spectroscopy to define baseline myocardial substrate utilization and energetic profiles, respectively. Similarly, both metabolic and functional endpoints will be necessary to assess response to therapy. If metabolic modulator therapy proves useful, the standard diagnostic approach to the heart failure patient will likely include metabolic imaging or surrogate metabolic biomarkers to guide therapeutic decisions and assess the patient’s response. In this regard, an interdisciplinary approach involving cardiologists, endocrinologists, and radiologists might be envisioned.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01-DK45416, R01-HL58493, and P01-HL57278 and Digestive Diseases Core Center grant P30-DK52574 (to D.P. Kelly). J.M. Huss is supported by NIH grant K01-DK063051 and Washington University School of Medicine Diabetes Research Training Center grant P60-DK20579. The authors thank Adam Wende for valuable contributions to the figures and Mary Wingate for expert assistance in preparing this manuscript.
Footnotes
Nonstandard abbreviations used: Ant, adenine nucleotide translocator; BAT, brown adipose tissue; ERR, estrogen-related receptor; FA, fatty acid; FADH2, reduced flavin adenine dinucleotide; FAO, FA β-oxidation; NRF, nuclear respiratory factor; PDH, pyruvate dehydrogenase; PET, positron emission tomography; PGC-1, PPARγ coactivator-1; PRC, PGC-1–related coactivator; RXRα, retinoid X receptor α; TCA, tricarboxylic acid; Tfam, mitochondrial transcription factor A.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Stanley WC, Chandler MP. Energy metabolism in the normal and failing heart: potential for therapeutic interventions. Heart Fail. Rev. 2002;7:115–130. doi: 10.1023/a:1015320423577. [DOI] [PubMed] [Google Scholar]
- 2.Taegtmeyer H. Energy metabolism of the heart: from basic concepts to clinical applications. Curr. Probl. Cardiol. 1994;19:59–113. doi: 10.1016/0146-2806(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 3.Bing RJ, Siegel A, Ungar I, Gilbert M. Metabolism of the human heart. II. Studies on fat, ketone and amino acid metabolism. Am. J. Med. 1954;16:504–515. doi: 10.1016/0002-9343(54)90365-4. [DOI] [PubMed] [Google Scholar]
- 4.Shipp JC, Opie LH, Challoner D. Fatty acid and glucose metabolism in the perfused heart. Nature. 1961;189:1018–1019. [Google Scholar]
- 5.Wisnecki JA, Gertz EQ, Neese RA, Mayr M. Myocardial metabolism of free fatty acids: studies with 14C labelled substrates in humans. J. Clin. Invest. 1987;79:359–366. doi: 10.1172/JCI112820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 7.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 8.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 9.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor-1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 11.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 12.Savagner F, et al. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem. Biophys. Res. Commun. 2003;310:779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 14.Kamei Y, et al. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St-Pierre J, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 16.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 17.Rhee J, et al. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1α): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael LF, et al. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehman JJ, et al. PPARγ coactivator-1 (PGC-1) promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baar K, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1α. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 21.Puigserver P, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, et al. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 23.Goto M, et al. cDNA cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem. Biophys. Res. Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 24.Huss JM, Pinéda Torra I, Staels B, Giguère V, Kelly DP. ERRα directs PPAR α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell LK, et al. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-actived receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 26.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 27.Garesse R, Vallejo CG. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263:1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- 28.Larsson N-G, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 29.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002; 1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 30.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 31.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dressel U, et al. The peroxisome proliferator-activated receptor β/δ agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Mol. Endocrinol. 2003;17:2477–2493. doi: 10.1210/me.2003-0151. [DOI] [PubMed] [Google Scholar]
- 33.Huss JM, Kopp RP, Kelly DP. PGC-1α coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα) J. Biol. Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 35.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 36.Lee SST, et al. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe K, et al. Constitutive regulation of cardiac fatty acid metabolism through peroxisome proliferator-activated receptor α associated with age-dependent cardiac toxicity. J. Biol. Chem. 2000;275:22293–22299. doi: 10.1074/jbc.M000248200. [DOI] [PubMed] [Google Scholar]
- 38.Djouadi F, et al. The role of the peroxisome proliferator-activated receptor α (PPARα) in the control of cardiac lipid metabolism. Prostaglandins Leukot. Essent. Fatty Acids. 1999;60:339–343. doi: 10.1016/s0952-3278(99)80009-x. [DOI] [PubMed] [Google Scholar]
- 39.Campbell FM, et al. A role for PPARα in the control of cardiac malonyl-CoA levels: reduced fatty acid oxidation rates and increased glucose oxidation rates in the hearts of mice lacking PPARα are associated with higher concentrations of malonyl-CoA and reduced expression of malonyl-CoA decarboxylase. J. Biol. Chem. 2002;277:4098–4103. doi: 10.1074/jbc.M106054200. [DOI] [PubMed] [Google Scholar]
- 40.Kersten S, et al. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J. Clin. Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilde AJ, et al. PPARα and PPARβ/δ, but not PPARγ, modulate the expression of genes involved in cardiac lipid metabolism. Circ. Res. 2003;92:518–524. doi: 10.1161/01.RES.0000060700.55247.7C. [DOI] [PubMed] [Google Scholar]
- 43.Cheng L, et al. Peroxisome proliferator-activated receptor δ activates fatty acid oxidation in cultured neonatal and adult cardiomyocytes. Biochem. Biophys. Res. Commun. 2004;313:277–286. doi: 10.1016/j.bbrc.2003.11.127. [DOI] [PubMed] [Google Scholar]
- 44.Cheng L, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-δ deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat. Med. 2004;10:1245–1250. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 45.Giguère V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 46.Heard DJ, Norbu PL, Holloway J, Vissing H. Human ERRγ, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: tissue-specific isoforms are expressed during development and in the adult. Mol. Endocrinol. 2000;14:383–392. doi: 10.1210/mend.14.3.0431. [DOI] [PubMed] [Google Scholar]
- 47.Hong H, Yang L, Stallcup MR. Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J. Biol. Chem. 1999;274:22618–22626. doi: 10.1074/jbc.274.32.22618. [DOI] [PubMed] [Google Scholar]
- 48.Sladek R, Bader J-A, Giguère V. The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme a dehydrogenase gene. Mol. Cell. Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hentschke M, Susens U, Borgmeyer U. PGC-1 and PERC, coactivators of the estrogen receptor-related receptor gamma. Biochem. Biophys. Res. Commun. 2002;299:872–879. doi: 10.1016/s0006-291x(02)02753-5. [DOI] [PubMed] [Google Scholar]
- 50.Laganiere J, et al. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor α (ERRα) promoter dictates peroxisome proliferator-activated receptor γ coactivator-1α control of ERRα expression. J. Biol. Chem. 2004;279:18504–18510. doi: 10.1074/jbc.M313543200. [DOI] [PubMed] [Google Scholar]
- 51.Mootha VK, et al. ERRα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreiber SN, et al. The estrogen-related receptor alpha (ERRα) functions in PPARγ coactivator 1α (PGC-1α) - induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boss O, et al. Role of the β3-adrenergic receptor and/or putative β4-adrenergic receptor on the expression of uncoupling proteins and peroxisome proliferator-activated receptor-γ coactivator-1. Biochem. Biophys. Res. Commun. 1999;261:870–876. doi: 10.1006/bbrc.1999.1145. [DOI] [PubMed] [Google Scholar]
- 54.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a corepressor. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan M, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barger PM, Browning AC, Garner AN, Kelly DP. p38 MAP kinase activates PPARα: a potential role in the cardiac metabolic stress response. J. Biol. Chem. 2001;276:44495–44501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- 57.Ballal K, Sekiguchi K, Nanda S, Barger P. ERK MAPK regulation of cardiac retinoid X receptor is involved in impaired fatty acid β-oxidation during hypertrophic growth [abstract] Circulation. 2004;110(Suppl.):946. [Google Scholar]
- 58.Nisoli E, et al. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 59.Nisoli E, Clementi E, Moncada S, Carruba MO. Mitochondrial biogenesis as a cellular signaling framework. Biochem. Pharmacol. 2004;67:1–15. doi: 10.1016/j.bcp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 60.Naya FJ, et al. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 61.Ryder JW, Bassel-Duby R, Olson EN, Zierath JR. Skeletal muscle reprogramming by activation of calcineurin improves insulin action on metabolic pathways. J. Biol. Chem. 2003;278:44298–44304. doi: 10.1074/jbc.M304510200. [DOI] [PubMed] [Google Scholar]
- 62.Handschin C, Rhee J, Lin J, Tam PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eto Y, et al. Calcineurin is activated in rat hearts with physiological left ventricular hypertrophy induced by voluntary exercise training. Circulation. 2000;101:2134–2137. doi: 10.1161/01.cir.101.18.2134. [DOI] [PubMed] [Google Scholar]
- 64.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Passier R, et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaeffer PJ, et al. Calcineurin and calcium/calmodulin-dependent protein kinase activate distinct metabolic gene regulatory programs in cardiac muscle. J. Biol. Chem. 2004;279:39593–39603. doi: 10.1074/jbc.M403649200. [DOI] [PubMed] [Google Scholar]
- 67.Bishop SP, Altschuld RA. Increased glycolytic metabolism in cardiac hypertrophy and congestive failure. Am. J. Physiol. 1970;218:153–159. doi: 10.1152/ajplegacy.1970.218.1.153. [DOI] [PubMed] [Google Scholar]
- 68.Allard MF, Schonekess BO, Henning SL, English DR, Lopaschuk GD. Contribution of oxidative metabolism and glycolysis to ATP production in hypertrophied hearts. Am. J. Physiol. 1994;267:H742–H750. doi: 10.1152/ajpheart.1994.267.2.H742. [DOI] [PubMed] [Google Scholar]
- 69.Christe MD, Rodgers RL. Altered glucose and fatty acid oxidation in hearts of the spontaneously hypertensive rat. J. Mol. Cell. Cardiol. 1994;26:1371–1375. doi: 10.1006/jmcc.1994.1155. [DOI] [PubMed] [Google Scholar]
- 70.Taegtmeyer H, Overturf ML. Effects of moderate hypertension on cardiac function and metabolism in the rabbit. Hypertension. 1988;11:416–426. doi: 10.1161/01.hyp.11.5.416. [DOI] [PubMed] [Google Scholar]
- 71.Massie BM, et al. Myocardial high-energy phosphate and substrate metabolism in swine with moderate left ventricular hypertrophy. Circulation. 1995;91:1814–1823. doi: 10.1161/01.cir.91.6.1814. [DOI] [PubMed] [Google Scholar]
- 72.Sambandam N, Lopaschuk GD, Brownsey RW, Allard MF. Energy metabolism in the hypertrophied heart. Heart Fail. Rev. 2002;7:161–173. doi: 10.1023/a:1015380609464. [DOI] [PubMed] [Google Scholar]
- 73.Sack MN, et al. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 74.Razeghi P, et al. Metabolic gene expressin in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 75.Razeghi P, Young ME, Abbasi S, Taegtmeyer H. Hypoxia in vivo decreases peroxisome proliferator-activated receptor alpha-regulated gene expression in rat heart. Biochem. Biophys. Res. Commun. 2001;287:5–10. doi: 10.1006/bbrc.2001.5541. [DOI] [PubMed] [Google Scholar]
- 76.Davila-Roman VG, et al. Altered myocardial fatty acid and glucose metabolism in idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol. 2002;40:271–277. doi: 10.1016/s0735-1097(02)01967-8. [DOI] [PubMed] [Google Scholar]
- 77.de las Fuentes L, et al. Myocardial fatty acid metabolism: independent predictor of left ventricular mass in hypertension and in left ventricular dysfunction. Hypertension. 2003;41:83–87. doi: 10.1161/01.hyp.0000047668.48494.39. [DOI] [PubMed] [Google Scholar]
- 78.Recchia FA, et al. Reduced nitric oxide production and altered myocardial metabolism during the decompensation of pacing-induced heart failure in the conscious dog. Circ. Res. 1998;83:969–979. doi: 10.1161/01.res.83.10.969. [DOI] [PubMed] [Google Scholar]
- 79.Paolisso G, et al. Total-body and myocardial substrate oxidation in congestive heart failure. Metabolism. 1994;43:174–179. doi: 10.1016/0026-0495(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 80.Wallhaus TR, et al. Myocardial free fatty acid and glucose use after carvedilol treatment in patients with congestive heart failure. Circulation. 2001;103:2441–2446. doi: 10.1161/01.cir.103.20.2441. [DOI] [PubMed] [Google Scholar]
- 81.Chandler MP, et al. Moderate severity heart failure does not involve a downregulation of myocardial fatty acid oxidation. Am. J. Physiol. Heart Circ. Physiol. 2004;287:H1538–H1543. doi: 10.1152/ajpheart.00281.2004. [DOI] [PubMed] [Google Scholar]
- 82.Sack MN, Disch DL, Rockman HA, Kelly DP. A role for Sp and nuclear receptor transcription factors in a cardiac hypertrophic growth program. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6438–6443. doi: 10.1073/pnas.94.12.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-α during cardiac hypertrophic growth. J. Clin. Invest. 2000;105:1723–1730. doi: 10.1172/JCI9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin. Exp. Pharmacol. Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 85.Barger PM, Kelly DP. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc. Med. 2000;10:238–245. doi: 10.1016/s1050-1738(00)00077-3. [DOI] [PubMed] [Google Scholar]
- 86.Kanda H, Nohara R, Hasegawa K, Kishimoto C, Sasayama S. A nuclear complex containing PPARα/RXRα is markedly downregulated in the hypertrophied rat left ventricular myocardium with normal systolic function. Heart Vessels. 2000;15:191–196. doi: 10.1007/s003800070022. [DOI] [PubMed] [Google Scholar]
- 87.Garnier A, et al. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J. Physiol. 2003;551:491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huss JM, Levy FH, Kelly DP. Hypoxia inhibits the PPARα/RXR gene regulatory pathway in cardiac myocytes. J. Biol. Chem. 2001;276:27605–27612. doi: 10.1074/jbc.M100277200. [DOI] [PubMed] [Google Scholar]
- 89.Osorio JC, et al. Impaired myocardial fatty acid oxidation and reduced protein expression of retinoid X receptor-alpha in pacing-induced heart failure. Circulation. 2002;106:606–612. doi: 10.1161/01.cir.0000023531.22727.c1. [DOI] [PubMed] [Google Scholar]
- 90.Tian R. Transcriptional regulation of energy substrate metabolism in normal and hypertrophied heart. Curr. Hypertens. Rep. 2003;5:454–458. doi: 10.1007/s11906-003-0052-7. [DOI] [PubMed] [Google Scholar]
- 91.Wall SR, Lopaschuk GD. Glucose oxidation rates in fatty acid perfused isolated working hearts from diabetic rat. Biochim. Biophys. Acta. 1989; 1006:97–103. doi: 10.1016/0005-2760(89)90328-7. [DOI] [PubMed] [Google Scholar]
- 92.Saddik M, Lopaschuk GD. Triacylglycerol turnover in isolated working hearts of acutely diabetic rats. Can. J. Physiol. Pharmocol. 1994;72:1110–1119. doi: 10.1139/y94-157. [DOI] [PubMed] [Google Scholar]
- 93.Belke DD, Larsen TS, Gibbs EM, Severson DL. Altered metabolism causes cardiac dysfunction in perfused hearts from diabetic (db/db) mice. Am. J. Physiol. 2000;279:E1104–E1113. doi: 10.1152/ajpendo.2000.279.5.E1104. [DOI] [PubMed] [Google Scholar]
- 94.Neitzel AS, Carley AN, Severson DL. Chylomicron and palmitate metabolism by perfused hearts from diabetic mice. Am. J. Physiol. Endocrinol. Metab. 2003;284:E357–E365. doi: 10.1152/ajpendo.00380.2002. [DOI] [PubMed] [Google Scholar]
- 95.Finck B, et al. A critical role for PPARα-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation of phenotype by dietary fat content. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Finck B, et al. The cardiac phenotype induced by PPARα overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 2002;109:121–130. doi:10.1172/JCI200214080. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Depre C, et al. Streptozotocin-induced changes in cardiac gene expression in the absence of severe contractile dysfunction. J. Mol. Cell. Cardiol. 2000;32:985–996. doi: 10.1006/jmcc.2000.1139. [DOI] [PubMed] [Google Scholar]
- 98.Young ME, et al. Uncoupling protein 3 transcription is regulated by peroxisome proliferator-activated receptor α in the adult rodent heart. FASEB J. 2001;15:833–845. doi: 10.1096/fj.00-0351com. [DOI] [PubMed] [Google Scholar]
- 99.Kelly DP, Strauss AW. Inherited cardiomyopathies. N. Engl. J. Med. 1994;330:913–919. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- 100.Exil VJ, et al. Very-long-chain Acyl-coenzyme A dehydrogenase deficiency in mice. Circ. Res. 2003;93:448–455. doi: 10.1161/01.RES.0000088786.19197.E4. [DOI] [PubMed] [Google Scholar]
- 101.Kurtz DM, et al. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schaffer JE. Lipotoxicity: when tissues overeat. Curr. Opin. Lipidol. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 103.Chiu H-C, et al. A novel mouse model of lipotoxic cardiomyopathy. J. Clin. Invest. 2001;107:813–822. doi: 10.1172/JCI10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yagyu H, et al. Lipoprotein lipase (LpL) on the surface of cardiomyocytes increases lipid uptake and produces a cardiomyopathy. J. Clin. Invest. 2003;111:419–426. doi:10.1172/JCI200316751. doi: 10.1172/JCI16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Listenberger LL, Ory DS, Schaffer JE. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J. Biol. Chem. 2001;276:14890–14895. doi: 10.1074/jbc.M010286200. [DOI] [PubMed] [Google Scholar]
- 106.deVries JE, et al. Saturated but not mono-unsaturated fatty acids induce apoptotic cell death in neonatal rat ventricular myocytes. J. Lipid Res. 1997;38:1384–1394. [PubMed] [Google Scholar]
- 107.Kong JY, Rabkin SW. Palmitate-induced cardiac apoptosis is mediated through CPT-1 but not influenced by glucose and insulin. Am. J. Physiol. Heart Circ. Physiol. 2002;282:717–725. doi: 10.1152/ajpheart.00257.2001. [DOI] [PubMed] [Google Scholar]
- 108.Young ME, Laws FA, Goodwin GW, Taegtmeyer H. Reactivation of peroxisome proliferator-activated receptor alpha is associated with contractile dysfunction in hypertrophied rat heart. J. Biol. Chem. 2001;276:44390–44395. doi: 10.1074/jbc.M103826200. [DOI] [PubMed] [Google Scholar]
- 109.Hopkins TA, et al. Control of cardiac pyruvate dehydrogenase activity in peroxisome proliferator-activated receptor-α transgenic mice. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H270–H276. doi: 10.1152/ajpheart.00852.2002. [DOI] [PubMed] [Google Scholar]
- 110.Shen X, et al. Cardiac mitochondrial damage and biogenesis in a chronic model of type I diabetes. Am. J. Physiol. Endocrinol. Metab. 2004;287:896–905. doi: 10.1152/ajpendo.00047.2004. [DOI] [PubMed] [Google Scholar]
- 111.Ye G, Metreveli NS, Ren J, Epstein PN. Metallothionein prevents diabetes-induced deficits in cardiomyocytes by inhibiting reactive oxygen species production. Diabetes. 2003;52:777–783. doi: 10.2337/diabetes.52.3.777. [DOI] [PubMed] [Google Scholar]
- 112.Ye G, et al. Catalase protect cardiomyocyte function in models of type 1 and type 2 diabetes. Diabetes. 2004;53:1336–1343. doi: 10.2337/diabetes.53.5.1336. [DOI] [PubMed] [Google Scholar]
- 113.Liang Q, et al. Overexpression of metallothionein reduces diabetic cardiomyopathy. Diabetes. 2002;51:174–181. doi: 10.2337/diabetes.51.1.174. [DOI] [PubMed] [Google Scholar]
- 114.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 115.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am. J. Med. Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 116.Larsson NG, Oldfors A. Mitochondrial myopathies. Acta Physiol. Scand. 2001;171:385–393. doi: 10.1046/j.1365-201x.2001.00842.x. [DOI] [PubMed] [Google Scholar]
- 117.Kirby DM, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial comlex I deficiency. J. Clin. Invest. 2004;114:837–845. doi:10.1172/JCI200420683. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H, et al. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3467–3472. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hansson A, et al. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient hearts. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stepien G, Torroni A, Chung AB, Hodge JA, Wallace DC. Differential expression of adenine nucleotide translocator isoforms in mammalian tissues and during muscle cell differentiation. J. Biol. Chem. 1992;267:14592–14597. [PubMed] [Google Scholar]
- 121.Graham BH, et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 122.Murdock DG, Boone BE, Esposito LA, Wallace DC. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart/muscle isoform of the adenine nucleotide translocator. J. Biol. Chem. 1999;274:14429–14433. doi: 10.1074/jbc.274.20.14429. [DOI] [PubMed] [Google Scholar]
- 123.Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sano M, et al. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 125.Sano M, et al. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 2004;23:3559–3569. doi: 10.1038/sj.emboj.7600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.