ABSTRACT
Short-chain fatty acids (SCFAs) are products of bacterial fermentation that help maintain important gut functions such as maintenance of the intestinal barrier, cell signaling, and immune homeostasis. The main SCFAs acetate, propionate, and butyrate have demonstrated beneficial effects for the host, including its importance in alleviating infections caused by pathogens such as Clostridioides difficile. Despite the potential role of SCFAs in mitigating C. difficile infection, their direct effect on C. difficile remains unclear. Through a set of in vitro experiments, we investigated how SCFAs influence C. difficile growth, sporulation, and toxin production. Similar to previous studies, we observed that butyrate decreased growth of C. difficile strain 630 in a dose-dependent manner. The presence of butyrate also increased C. difficile sporulation, with minimal increases in toxin production. RNA-Seq analysis validated our experimental results, demonstrating increased expression of sporulation-related genes in conjunction with changes in metabolic and regulatory genes, such as a putative carbon starvation protein, CstA. Collectively, these data suggest that butyrate may induce alternative C. difficile survival pathways, modifying its growth ability and virulence to persist in the gut environment.
IMPORTANCE
Several studies suggest that butyrate may modulate gut infections, such as reducing inflammation caused by the healthcare-associated Clostridioides difficile. While studies in both animal models and human studies correlate high levels of butyrate with reduced C. difficile burden, the direct impact of butyrate on C. difficile remains unclear. Our study demonstrates that butyrate directly influences C. difficile by increasing its sporulation and modifying its metabolism, potentially using butyrate as a biomarker to shift survival strategies in a changing gut environment. These data point to additional therapeutic approaches to combat C. difficile in a butyrate-directed manner.
KEYWORDS: Clostridioides difficile, butyrate, growth assay, metabolism, sporulation
INTRODUCTION
Clostridioides (Clostridium) difficile is an anaerobic, gram-positive bacterium of serious concern, causing nearly half a million infections and 30,000 deaths each year in the United States (1). C. difficile infection (CDI) causes inflammation and colitis in the gut, with symptoms ranging from diarrhea to pseudomembranous colitis and megacolon in extreme cases (2). Recurrence occurs in up to 30% of individuals, resulting in higher patient mortality and increased healthcare costs, thus making CDI an important public health threat (3). Risk factors for CDI include advanced age, pre-existing gastrointestinal issues, immunocompromised status, and antibiotic exposure, making CDI highly prevalent in healthcare-associated environments (4).
The gut microbiota, the microbial community residing in the gastrointestinal tract, provides colonization resistance against C. difficile within the colon (5). In most healthy individuals, contact with metabolically inert C. difficile spores from the environment does not result in disease. However, antibiotics and other environmental perturbations have been demonstrated to disrupt the microbiota (6, 7), allowing for C. difficile spores to colonize the gut environment and produce toxins after germination and outgrowth (8). During initial colonization, removal of microbes that transform cholic acid and conjugated derivatives (primary bile acids), known to induce C. difficile germination (9), into deoxycholic acid (secondary bile acids), known to reduce C. difficile growth (10, 11), has been correlated with the development of CDI (6). Once colonized, C. difficile can use a variety of metabolic approaches to persist in the gut. For instance, the ability of C. difficile to use a multitude of carbohydrate sources for growth (12), as well as amino acids via Stickland fermentation (13, 14), likely supports its ability to colonize multiple nutritional niches following microbial perturbation in the gut.
Metabolic flexibility of C. difficile is also connected to virulence mechanisms. The nutritional regulators CodY (15, 16) and CcpA (12) are known to decrease toxin production and sporulation by sensing nutrient deprivation or carbohydrate availability, respectively. Other regulators include PrdR, a proline regulator important for C. difficile growth in vivo (17), and Rex, a global redox-sensing regulator, both of which influence toxin and spore production (14, 18). The main toxins produced by C. difficile include TcdA and TcdB, expressed by the tcdA and tcdB genes located on the PaLoc, or pathogenicity locus (19). Production of TcdA/B is regulated by the additional PaLoc genes tcdR and tcdC, which are further regulated by global transcriptional networks that respond to environmental cues (20). Part of successful colonization also includes sporulation, which is controlled by the master transcriptional regulator, Spo0A, leading to further persistence in the gut and potential transmission of C. difficile to new hosts (21).
The high recurrence rate observed for CDI following standard antibiotic treatment has led to interest in developing microbial-mediated treatments that aim to recover colonization resistance against C. difficile . In addition to bile acid transformation, other microbiota-mediated mechanisms hypothesized to control C. difficile infection include short-chain fatty acids (SCFAs), which are fermentation end products produced by select microbes that are generally regarded as beneficial to the host (22, 23). The SCFA, butyrate, has been correlated with recovery from CDI following treatment with fecal microbiota transplantation (FMT) (24, 25), which aims to restore microbial functions that provide colonization resistance against C. difficile. Butyrate has also been demonstrated to decrease C. difficile growth in vitro (26), as well as alleviate toxin-based inflammation in an animal model of CDI without directly reducing C. difficile burden (27, 28). Yet, the mechanism by which butyrate might control C. difficile pathogenesis is relatively undefined.
This study sought to identify how SCFAs might directly influence C. difficile pathogenesis. Using an in vitro platform, we observed that in addition to attenuating growth, butyrate and propionate increased sporulation of C. difficile strain 630. Butyrate’s effects were dependent on the nutritional environment, suggesting its effects might be metabolically regulated. RNA-Seq validated the observed experimental effects of butyrate and further identified involvement of the major regulators CcpA and Spo0A, as well as a putative carbon starvation gene, CstA, in butyrate-dependent control of C. difficile. Collectively, these results point to additional considerations in targeting butyrate as a therapeutic strategy to prevent or treat C. difficile.
MATERIALS AND METHODS
In vitro growth of C. difficile
In an anaerobic chamber (Coy), a 10-2 dilution of the noted spore stock [C. difficile strain 630 (ATCC BAA-1382), VPI10463 (ATCC 43255-FZ), and R20291 (29)], prepared as described previously (30), was plated on pre-reduced taurocholate-cefoxitin-cycloserine-fructose agar (TCCFA) plate (31) and incubated overnight at 37°C. A single colony was inoculated into 5 mL of 1× of brain heart infusion broth supplemented with 5 g/L yeast extract and 0.1% l-cysteine (BHI) (32), in biological triplicates per treatment group, and incubated at 37°C. After 18 h of growth, tubes were centrifuged at 1500 × g for 10 min. After discarding the supernatant, each pellet was resuspended in 1 mL of 2× BHI. For each technical triplicate (three per biological replicate), 250 µL of the resuspended pellet was added into 5 mL of 2× BHI. In a 96-well plate, 100 µL of the prepared inoculum was added into each of the wells, making the final volume 200 µL. For negative control wells (no C. difficile), 2× BHI was added instead.
To test the effect of acetate, propionate, and butyrate on the growth of the prepared inoculum above, previously prepared 500 mM SCFA stocks of each (frozen until use) were diluted to a final working concentration of 50 mM in the anaerobic chamber a day prior. A 96-well plate was set up to test 5 mM and 25 mM acetate, propionate, and butyrate from 50 mM concentration. The plate was then placed into a Sunrise plate reader (Tecan) for 24 h at 37°C, where optical density (OD600) was measured every 15 min. After 24 h, colony-forming units (CFUs) were assessed, as described below. The plate was covered in parafilm and placed in the −80°C freezer for storage for toxin assays.
For assessment of C. difficile growth at multiple timepoints, two replicate 96-well plates were prepared simultaneously, one placed in the plate reader and one in the incubator at 37°C to acquire matched OD600 and CFU measurements. At 6, 12, 18, and 24 h, three wells per treatment were sampled and diluted for calculating CFUs. CFUs were quantified for each corresponding treatment or timepoint. For three biological triplicates per treatment, bacterial growth was serially diluted from 10−1 to 10−5. CFUs per milliliter was determined by the log10 of colonization, determined by number of colonies × dilution of plate counted × dilution factor. For assessment of spores, 20 µL of the sample was added to PCR tubes and the tubes were heated for 20 min at 65°C to kill off vegetative cells. After heating, the samples were plated on TCCFA plates using the same dilutions as above.
For assessing butyrate dose response on C. difficile growth, a working stock of butyrate from a previously frozen 500 mM stock was prepared initially. A 96-well plate was set up as described above, comprising of C. difficile inoculum in BHI supplemented with final concentrations of 5 mM, 10 mM, 25 mM, and 50 mM butyrate in the wells.
To investigate the effect of pH, BHI was prepared with a final pH of 6.2, 7.2, and 8.0. Using a pH probe (Mettler Toledo FiveEasy Plus), pH values were confirmed before autoclaving, after autoclaving, and after preparing growth conditions (including SCFA addition).
For assessing the effect of butyrate on C. difficile growth under single carbohydrate sources, 96-well plates were prepared as described above except using 2× C. difficile minimal media (CDMM) (33, 34) supplemented with 1% of the indicated sugar. A stock concentration of 2% weight by volume of the carbohydrates [glucose (35), fructose (36), lactose (37), maltose (38), trehalose (39), cellobiose (40), sucrose (20), mannitol (41), mannose (35), and raffinose (6)] was prepared and 100 µL of each sugar stock was added to 2× CDMM. Growth of C. difficile was assessed using OD600 as described above.
C. difficile toxin assay
This protocol was adapted from Theriot et al. (42). Briefly, filtered media consisting of Dulbecco’s Modified Eagle Medium (DMEM) (Gibco DMEM #11965-092), with 5% fetal bovine serum (Fisher, Gibco Fetal Bovine Serum, qualified, heat inactivated, US Origin #16-140-071), and PenStrep (Life Technologies, Gibco Penicillin Streptomycin 5,000 U/mL (penicillin 5,000 U/mL; streptomycin 5,000 µg/mL; #15070063) was used to propagate Green African monkey kidney epithelial (Vero) cells to confluence within a 96-well plate, at a density of 103 cells per well based on the number of viable cells observed in a 1:1 mixture of cell suspension in trypan blue. The seeded plate was incubated for an hour at room temperature before being placed at 37°C overnight. Prior to addition of samples for toxin assessment, old media was replaced with fresh media. For the toxin assay, cell growth samples (i.e., spent media from C. difficile growth assays in BHI supplemented with the indicated SCFAs) were filtered with a 20 µm filter. A dilution plate was prepared serial with dilutions of cell culture filtrate and diluted down to 10−6. For a positive control, 0.01 µg/µL Toxin A (Invitrogen #10977-015) was added to PBS. The seeded plate was incubated for 40 min at room temperature to allow for anti-toxin activity, then placed at 37°C overnight. Toxin activity was determined by the presence of confluence (>75% confluent) under a microscope for the last dilution of each sample. The amount of toxin was quantified as log10(anti-toxin dilution factor × Vero cell dilution factor × last dilution with cell rounding × initial PBS dilution). For visualization, log10 of the calculated toxin above was divided by log10 of average of colonization per condition to normalize the toxin activity per CFU.
Assessment of spore production using phase contrast
Spore stock was streaked anaerobically onto TCCFA and incubated at 37°C for 24 h. An isolated colony was inoculated into BHI for 18 h at 37°C (in triplicate). Tubes were centrifuged at 1500 × g for 10 min. After discarding the supernatant, the pellet was resuspended with 1 mL of 2 × 70:30 sporulation media (43). For each replicate, resuspended pellet was added into 2 × 70:30 media at 1:200. For conditions supplemented with 25 mM butyrate, a working stock of 50 mM was prepared as described above. A 96-well plate was set up to test sporulation efficiency of 70:30 media with or without butyrate, with each well for a final volume of 200 µL. The 96-well plate was then placed into the Tecan plate reader for 24 h at 37°C to assess growth as described above.
After 24 h, the C. difficile grown in 70:30 and butyrate were added together in separate tubes and centrifuged for 30 s at 13,000 rpm at room temperature. The cells were then resuspended in 25 µL of BHI. A microscope slide was then prepared for both conditions by adding 5 µL of the resuspended culture to a microscope slide and adding a coverslip. Phase contrast images were captured at 100× on Ph3 on a phase contrast microscope (Leica DM750 fitted with Leica ICC50W camera). Sporulation efficiency was calculated through the following equation: (spores)/(vegetative cells + spores) × 100 (43). At least 1,000 cells (from >5 frames per condition) were counted to get an accurate efficiency per experiment (n = 3). Original cell density was also calculated using a spectrophotometer.
RNA extraction
C. difficile strain 630 was grown with or without butyrate (25 mM) as described above. C. difficile culture was collected and immediately frozen for RNA extraction at points representing early log (~0.2 OD600) and late log (~0.5 OD600). For growth in BHI without butyrate, culture was collected at 7 h (early log) and 10 h (late log); for growth in BHI supplemented with butyrate, culture media was collected at 10 h (early log) and 13 h (late log). Before RNA extractions, samples were thawed and centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was then discarded, and the pellet was resuspended in 1 mL of 1:100 β-mercaptoethanol/water dilution. The samples were then centrifuged at 14,000 rpm for 1 min at 4°C. The supernatant was discarded, and the cell pellet was resuspended in 1 mL of Trizol. The samples were incubated at room temperature for 15 min. The samples were then centrifuged at 5,000 rpm for 15 min at 4°C. All further extraction steps required the aqueous phase (44). The Zymo Direct-zol RNA Miniprep Plus extraction kit (Zymo Research #R2071) was used to extract the RNA. Qubit (ThermoFisher Scientific #Q33230) was then performed to confirm the concentration of RNA before sequencing.
RNA-Seq and data analysis
RNA collected from growth experiments above was sent to the Microbial Genome Sequencing Center [MiGS, Pittsburgh (www.migscenter.com)] for Illumina sequencing (NextSeq 2000). Raw reads were quality checked and adapter-trimmed using Trim-galore (45). Metaphlan was used to identify relative species abundance of sequence reads (46). Sequences were aligned using RNA-Seq by Expectation Maximization (RSEM) (47) to the C. difficile strain 630 reference GCA_000009205.2 under the accession number AM180355 (PRJNA78) (48, 49). FeatureCounts from subread was utilized to quantify reads (50). The DESeq2 package (51) was used then to analyze the differential expression, identifying genes that were significant. The RNA-Seq data in R using ggplot2 (52) was used to visualize results. Gene set enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of the top ranked genes were analyzed using clusterprofiler with Wald’s statistic (53).
RT-qPCR
Real-time quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) was used to assess the expression of select genes. Following RNA extractions, cDNA of the samples was made following the NEB M-MuLV Reverse Transcriptase protocol (NEB #M0253). Qubit was used to assess the concentration of cDNA in the samples. Primers used included rpoC (housekeeping gene) (FP: CCAGTCTCTCCTGGATCAACTA, RP: CTAGCTGCTCCTATGTCTCACATC) (54), tcdR (FP: TTATTAAATCTGTTTCTCCCTCTTCA, RP: AGCAAGAAATAACTCAGTAGATGATT) (55), tcdC (FP: GAGCACAAAGGGTATTGCTCTA, RP: AAATGACCTCCTCATGGTCTTC) (56), codY (FP: CTCATCTTCTATAACTGAACTGTCTTGAAC, RP: TTTGATTTACTGGCCGGAGCATTG) (16), ccpA (FP: TCTTGTTCAACTATCCATGAAATCATAAC, RP: AAATGGGATAGAAGAGGTTGCTAAA) (57), and rex (FP: TGGTGGATTTGGACAACAAGGA, RP: TGCTCCTACAAGAACTGCGT) (generated for this study). Reactions were made using iQ SYBR Green Supermix (BioRad #1708880) using manufacturer’s directions with a final cDNA concentration of 5 ng. Samples were run in triplicate with biological replicates using manufacturer’s directions. Expression levels were quantified/normalized using the housekeeping gene rpoC (44). The 2-ΔΔCT method was used to calculate relative expression fold change between the control (C. difficile + BHI) and treatment groups (C. difficile + BHI + butyrate 25 mM) in the genes of interest (tcdR, tcdC, codY, ccpA, and rex) compared to the housekeeping gene (rpoC) (58).
Statistical analysis
Significance was determined using one-way analysis of variance (ANOVA) for area under the curve (AUC; calculated using growthcurver in R) followed by post-hoc Dunnett’s test (using DescTools in R) for the growth curves. The significance on plate counts and toxin activity at different time points was tested using one-way ANOVA followed by Dunnett’s test. Significance on the spore efficiency was tested using Bonferroni pairwise t-test. Significance on the transcriptomics was calculated by DESeq2 using Wald’s test.
RESULTS
Butyrate decreases C. difficile growth
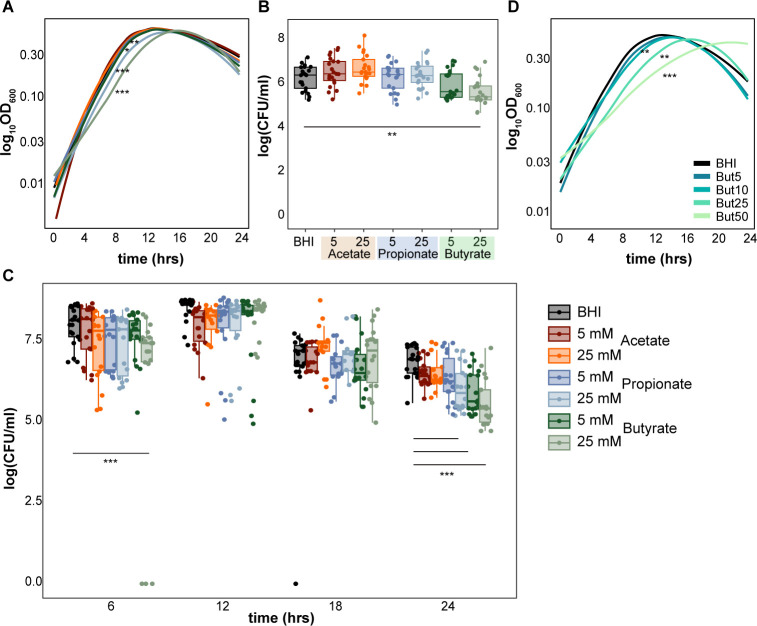
To assess the effect of the predominant SCFAs on C. difficile growth in vitro, we grew different strains of C. difficile (630, VPI10463, R20291) in the presence of low (5 mM) and high (25 mM) concentrations of acetate, propionate, and butyrate supplemented in the rich medium, BHI (Fig. 1; Fig. S1). We observed significantly decreased growth of C. difficile strain 630 in the presence of both butyrate and propionate (5 and 25 mM) concentrations (Dunnett’s test on AUC, P < 0.001 and P < 0.05) using OD600 measurements (Fig. 1A). At 24 h, we observed significantly decreased growth (Dunnett’s test, P < 0.01) of C. difficile strain 630 using CFU enumeration in 25 mM butyrate-supplemented BHI (Fig. 1B). We also observed decreased growth of C. difficile strain 630 using CFU enumeration at 6 h and 24 h in butyrate- and propionate-supplemented BHI (Dunnett’s test, P < 0.001) (Fig. 1C). While a previous study has demonstrated butyrate-induced growth defects across multiple C. difficile strains (26), we did not observe significant differences in growth with any SCFA for two additional strains, C. difficile VPI 10463 and R20291 (Fig. S1).
Fig 1.
Butyrate inhibits the growth of C. difficile strain 630. (A) Growth curves [log10(OD600)] over 24 h (n = 7 per condition). (B) CFUs after 24 h (n = 21 per condition) and (C) at 6, 12, 18, and 24 h of growth in BHI supplemented with 5 and 25 mM acetate, propionate, or butyrate (n > 15 per condition). (D) Growth curves [log10(OD600)] over 24 h in BHI supplemented with increasing concentrations of butyrate (0, 5, 10, 25, 50 mM; n = 3 per condition). Statistical significance calculated using Dunnett’s test: *P-value <0.05; **P-value <0.01; ***P-value <0.001.
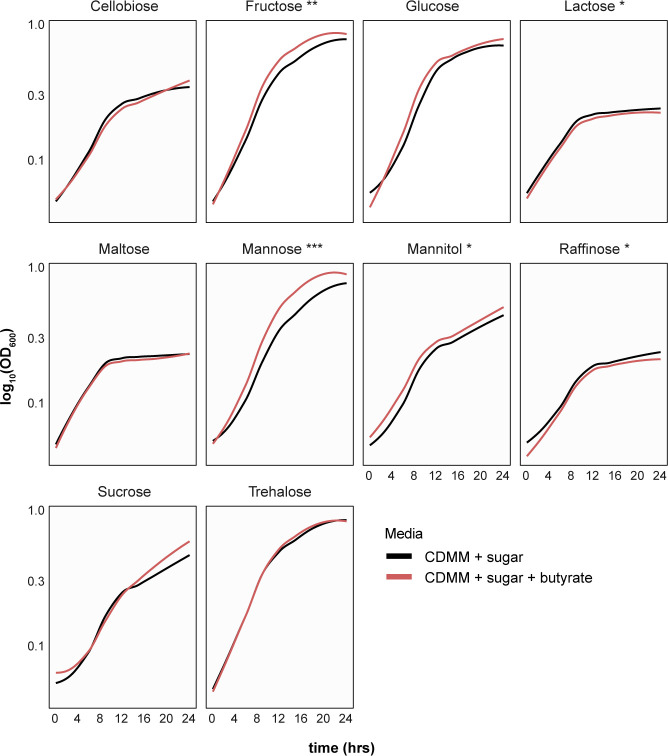
Given these strain-specific results, we mainly focused on the effect of butyrate on C. difficile strain 630 for the remaining experiments. Decreased growth was also dose-dependent, as increased concentrations of butyrate up to 50 mM concentrations increasingly impaired C. difficile strain 630 growth (Fig. 1D). To preclude the possibility that butyrate’s impact on C. difficile growth was pH-dependent, media with and without butyrate was adjusted to 7.2, as well as tested at pH 6.2 and 8. Significant growth decrease in the presence of 25 mM butyrate was still observed at pH 6.2 and 7.2 levels, with no significance at a more basic pH of 8.0 (Fig. S2). To test whether butyrate’s ability to modulate C. difficile growth is dependent on its metabolic environment, we grew C. difficile with or without butyrate in minimal media (CDMM) supplemented with different single carbohydrate sources known to support C. difficile growth (Fig. 2) (59, 60). We observed decreased growth of C. difficile with butyrate in CDMM supplemented with 1% lactose and raffinose only (Dunnett’s test, P < 0.05) (Fig. 2). The addition of butyrate did not significantly influence growth of C. difficile in CDMM supplemented with cellobiose, maltose, or trehalose. Surprisingly, butyrate increased the growth of C. difficile in CDMM supplemented with fructose, mannose, and mannitol (Dunnett’s test, P < 0.01), and trended toward increase in growth in the presence of glucose and sucrose (not significant). These results suggest metabolism-dependent impacts of butyrate on C. difficile growth.
Fig 2.
Butyrate-induced inhibition of C. difficile growth is dependent on the metabolic environment. Growth curves of C. difficile strain 630 [log10(OD600)] over 24 h in minimal media (CDMM) in the presence of a single sugar supplemented with (red) and without (black) 25 mM butyrate (n = 3 per condition). Statistical significance calculated using Dunnett’s test: *P-value <0.05; **P-value <0.01; ***P-value <0.001.
Butyrate increases toxin and spore production in C. difficile
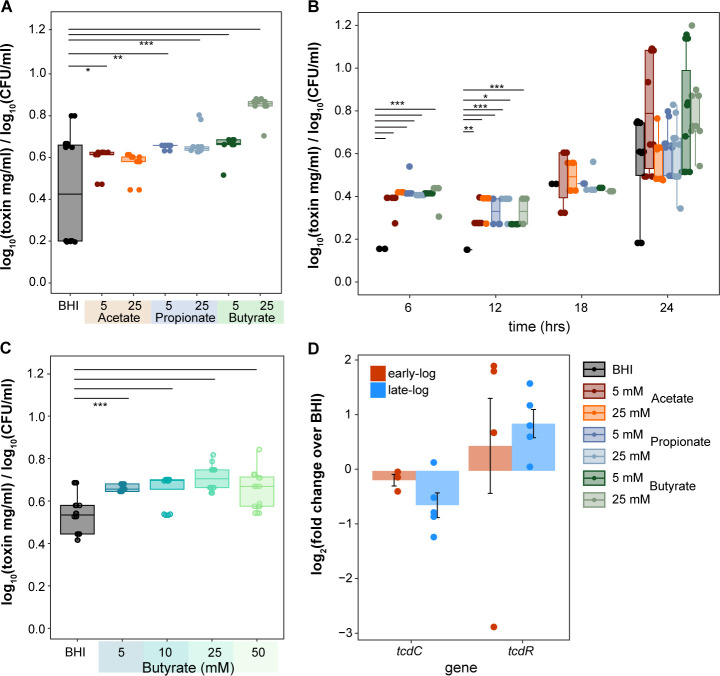
We next assessed the impact of SCFAs on C. difficile toxin and spore production, which typically occur during the stationary growth phase (61). We assessed toxin production in the presence of SCFAs using an in vitro cell rounding assay (62). After 24 h of growth, we observed increased toxin by C. difficile strain 630 in the presence of all SCFAs, compared to BHI alone (Fig. 3A, Dunnett’s test, P < 0.001). All SCFAs significantly increased toxin activity as early as 6 to 12 h in strain 630 (Fig. S3C, Dunnett’s test, P < 0.05, P < 0.01, P < 0.001). Despite minimal effects on growth, SCFAs increased toxin activity in C. difficile strains VPI10463 and R20291 (Fig. S3A and S3B, Dunnett’s test P < 0.05), which typically produce more toxin than C. difficile strain 630 (63). Enhanced and significant toxin activity was also observed at increasing butyrate concentrations starting at 12 h as low as 5 mM, and at all concentrations except at 10 mM at 24 h (Fig. 3B, Dunnett’s test, P < 0.01). To further validate these observations, we used RT-qPCR to assess the expression of tcdR and tcdC, the positive and negative regulators of C. difficile toxins located on the PaLoc (64), during early log (~0.2 OD600) and late log (~0.5 OD600) growth of C. difficile with or without butyrate (Fig. 3C). In the presence of butyrate, tcdC expression was decreased during both early and late log growth, whereas tcdR expression was increased during both late and early log growth, albeit not significantly.
Fig 3.
Short-chain fatty acids increase C. difficile toxin production. Toxin activity (log10 of toxin mg/mL) of C. difficile strain 630 normalized to the average C. difficile load [log10(CFU/mL)] per condition, as measured by an in vitro cell assay (A) after 24 h of growth in BHI supplemented with 5 and 25 mM acetate, propionate, and butyrate (n = 21 per condition), and (B) at 6, 12, 18, and 24 h in BHI supplemented with 5 and 25 mM acetate, propionate, or butyrate; n > 6 per condition. (C) Toxin activity (log10 of toxin mg/mL) of C. difficile strain 630 after 24 h in BHI with increasing concentrations of butyrate (0, 5, 10, 25, 50 mM; n > 3 per condition). (D) Log2 fold change of tcdC and tcdR expressions in C. difficile strain 630 growing in BHI with 25 mM butyrate over without butyrate (measured at early and late log growth using RT-qPCR, n = 5 per condition). Statistical significance calculated using Dunnett’s test, *P-value <0.05; **P-value <0.01; ***P-value <0.001.
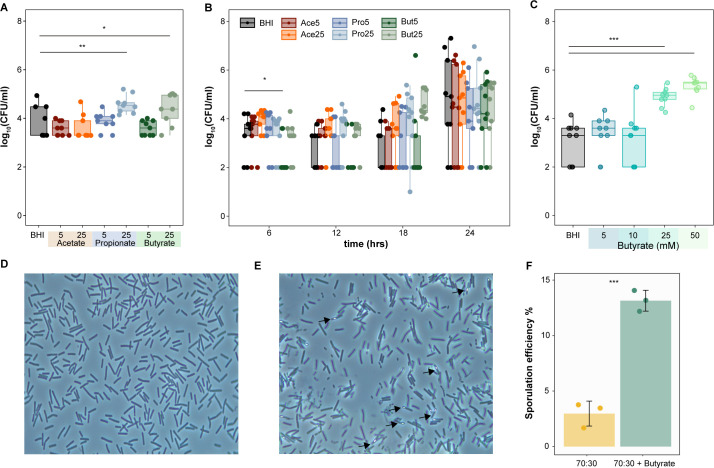
To assess spore CFUs, C. difficile 630 cultures were heated at 65°C for 20 min to kill the vegetative cells prior to plating anaerobically on TCCFA. Compared to BHI alone, we observed significantly higher spore counts in the presence of 25 mM propionate and butyrate after 24 h (Dunnett’s test, P < 0.05, P < 0.01) (Fig. 4A). Significantly increased spores were also observed as early as 6 h and later at 18 h specifically in the presence of butyrate, even as low as 5 mM (Fig. 4B, Dunnett’s test, P < 0.05). Additionally, more spores were observed at increasing butyrate concentrations compared to BHI alone (Fig. 4C, Dunnett’s Test, P < 0.001). The ability of butyrate to increase spore production was also observed using phase contrast microscopy. Using a modified sporulation assay and phase contrast microscopy (43), we calculated the sporulation efficiency with and without butyrate after 24 h of growth (Fig. 4D through F). We observed higher sporulation efficiency in the presence of butyrate (13.26%) compared to BHI alone (3%). (Fig. 4F, Welch’s two-sample test, P < 0.001).
Fig 4.
Butyrate increases C. difficile spore production. (A) Growth (log10 of colony-forming units) of C. difficile strain 630 spores (A) after 24 h of growth (n = 9 per condition) and (B) throughout growth at 6, 12, 18, and 24 h in BHI supplemented with 5 and 25 mM acetate, propionate, and butyrate (n = 9 per condition per timepoint), or (C) after 24 h of growth in BHI supplemented with increasing concentrations of butyrate (5, 10, 25, and 50 mM; n = 9 per condition). Cultures were collected at indicated timepoints and heated at 65°C for 20 min to kill off vegetative cells, reflecting spore CFUs. Representative phase contrast images (100×) of C. difficile strain 630 cells grown for 24 h in (D) 70:30 media alone (E) supplemented with 25 mM butyrate. (F) Sporulation efficiency calculated over 1,000 cells in 70:30 media with or without 25 mM butyrate (n = 3 experiments; >5 frames per experiment per condition). Welch’s two-sample test, *P-value <0.05; **P-value <0.01; ***P-value <0.001.
Butyrate induces the expression of genes related to spore production and alternate metabolic pathways
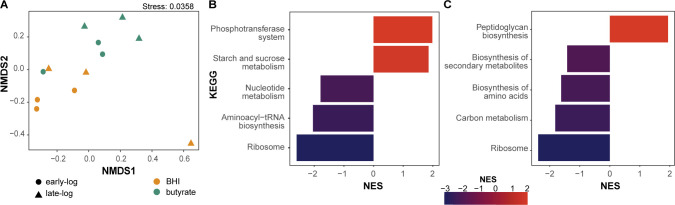
To determine global expression changes induced by butyrate, we conducted RNA-Seq of C. difficile strain 630 grown in BHI with and without 25 mM butyrate at both early (~0.2 OD600) and late log (~0.5 OD600) growth, and mapping the sequencing reads to C. difficile strain 630 (GCA_000009205.2; BioProject: PRJNA78). Non-metric multidimensional scaling (NMDS) of all identified genes demonstrated clustering of samples based on the presence of butyrate (Fig. 5A). Initial gene set enrichment analysis on genes expressed during early or late log growth identified differential pathways with or without butyrate (cutoff > log2 fold change, with adjusted P < 0.05 calculated with Wald’s test). During early log growth, genes related to the phosphotransferase system (PTS) and sucrose and starch metabolism were over-represented in the presence of butyrate, whereas genes related to nucleotide metabolism, aminoacyl-tRNA biosynthesis, and ribosomal functions were under-represented when the genes are in decreasing order of log2 fold change (Fig. 5B). During late log growth, genes related to peptidoglycan biosynthesis were over-represented in the presence of butyrate, whereas genes related to secondary metabolites, amino acid, and carbon metabolism were under-represented (Fig. 5C).
Fig 5.
Butyrate modulates C. difficile gene expression. (A) NMDS of C. difficile strain 630 transcriptomic sequences (n = 3 per condition; total n = 12) using Bray-Curtis dissimilarity and normalized enrichment scores (NES) for KEGG assignments significantly upregulated and downregulated genes at (B) early log (~0.2 OD600), and (C) late log (~0.5 OD600) for C. difficile strain 630 grown in BHI with or without 25 mM butyrate. NES was calculated using clusterprofiler (gseKEGG) in R with P-values adjusted post hoc using false discovery rate.
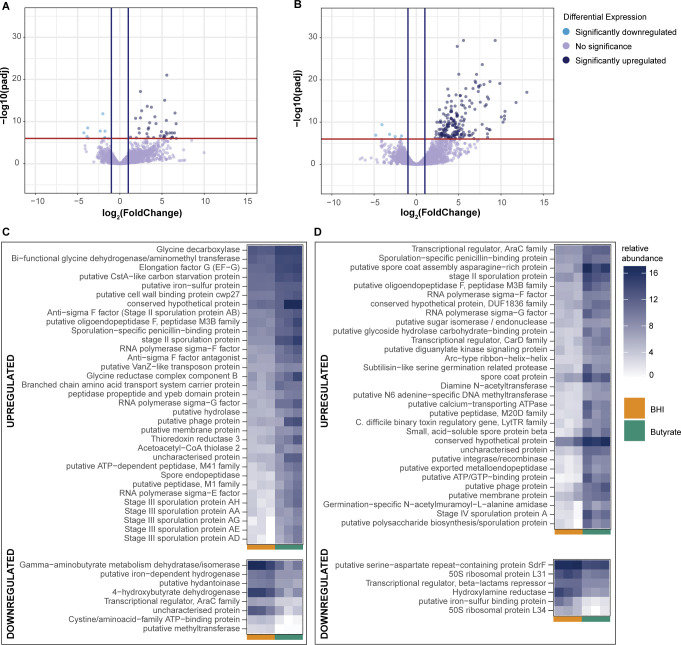
At the level of individual genes, 38 and 8 genes were significantly over- or under-expressed during early log growth in the presence of butyrate (Fig. 6A and C, Wald’s test, log2 fold change >1 and adjusted P < 10−6). Many of these included genes related to sporulation, such as stages II, III, or even V sporulation proteins, as well as spore endopeptidases, which are required for spore germination and produced during sporulation (Fig. 6C). The toxin gene TcdA was significantly over-expressed (Wald’s test, adjusted P < 0.05) during early log growth but not late log, validating earlier production of toxin in the presence of butyrate (Fig. S4 and S5). Several sigma factors (sigma E, F, and G) that are involved in transcription regulation of sporulation typically expressed during later log growth (65 - 67) were also over-expressed during early log growth in the presence of butyrate (Fig. 6C). Several genes expressed significantly differentially during early log growth were also related to metabolism. Genes related to glycine metabolism, such as the bi-functional glycine dehydrogenase/aminomethyl transferase protein (gcvTPA), and glycine decarboxylase (gcvPB) were upregulated in the presence of butyrate (Fig. 6C). Other upregulated genes included phosphotransferase (PTS) genes related to lactose (PTS, lactose/cellobiose family IIBC), fructose (PTS, fructose/mannitol family IIB), mannose (PTS, mannose specific IIBC), and mannitol (PTS, fructose/mannitol family IIB), which collectively aid in non-glucose-related carbohydrate metabolism (Fig. S5) (68). Genes related to butyrate metabolism were also downregulated in the presence of butyrate, such as gamma-aminobutyrate dehydratase gamma-aminobutyrate-dehydratase (abfD) and 4-hydroxybutyrate dehydrogenase (4hbD) (Fig. 6C), both involved in the succinate to butanoate fermentation pathway (69, 70).
Fig 6.
Butyrate upregulates genes related to spore formation and metabolism. Volcano plots of significantly upregulated and downregulated genes (Wald’s test, adjusted P < 10−6, log2 fold change > 1) at (A) early log and (B) late log growth of C. difficile strain 630 grown in BHI with or without 25 mM butyrate. Heatmap depicting normalized transcript counts of the top 50 significantly upregulated (top panels) and downregulated (bottom panels) genes in the presence of butyrate at (C) early log and (D) late log growth.
At late log growth, 100 and 6 genes were over- and under-expressed, respectively (Fig. 6B and D). Many sporulation-related genes remained over-represented during later log growth, including sporulation stage IV proteins, stage II sporulation proteins, subtilisin-like proteases, oligoendopeptidases, and others, very likely due to the late log also being the stationary phase where sporulation is observed en masse. Butyrate metabolism-associated genes remained downregulated in the presence of butyrate, gamma-aminobutyrate-dehydrogenase, and 4-hydroxy dehydrogenase. Most notably, genes associated with sporulation were upregulated in the presence of butyrate, many of which are involved in later steps of sporulation, such as spore coat and maturation proteins (spore coat proteins sipL, cotB; spore maturation proteins spmA, spmB; stage V sporulation proteins spoIIIAB, spoIIIAC, spoIIIAF). The binary toxin regulatory gene from the LytR family of proteins, cdtR, was also upregulated. Other significantly downregulated genes included the ribosomal proteins L31 and L34, as well as a putative iron-sulphur binding protein, and genes related to antibiotic stress, including a transcriptional repressor for the beta-lactams, and hydroxylamine reductase, which is upregulated in response to metronidazole and fidaxomicin stress (71, 72).
We also investigated the involvement of known C. difficile global regulators in butyrate-induced growth changes, such as CcpA, Rex, PrdR, and CodY, which are known modulators of C. difficile virulence in response to its environment (12). None of these global regulators were significantly differentially expressed (Fig. S4 and S5). Additionally, we did identify over-representation of transcripts encoding for a putative carbon starvation protein, CstA, in early log (Fig. 6C; Fig. S4 and S5), and for a histidine-kinase of Spo0A (CD630_15790) in late log (Fig. S4) that was recently identified to encode for an inhibitor of Spo0A (73).
DISCUSSION
Butyrate has shown major promise in alleviating prominent intestinal diseases, such as graft-versus-host disease (74) or inflammatory bowel disease (75). In the context of CDI, higher butyrate levels are correlated with successful FMT in human studies (24) and inversely correlated with C. difficile burden in mice (76). While recent studies, including the data presented here, have demonstrated an inhibitory effect on in vitro growth of C. difficile (26, 76), the mechanism by which butyrate could inhibit C. difficile remains unknown. Our current results suggest a more complex role for butyrate in directly influencing C. difficile. Indeed, exogenous butyrate supplementation, while capable of attenuating disease via host effects, has not demonstrably reduced C. difficile burden in infected mice (27, 28). Furthermore, recent studies in mice also suggest that the presence of butyrate-producing bacteria alone is not sufficient to inhibit C. difficile colonization (77).
Our results support previous observations that butyrate can inhibit growth of C. difficile. These results were observed at different pH conditions, which have been previously demonstrated to influence C. difficile pathogenesis in vitro (78). However, the ability of butyrate to inhibit C. difficile was not observed for all strains tested in our study, nor was it universally observed across different media. In contrast to a recent study observing butyrate-induced growth inhibition of various clinical strains (albeit in reinforced clostridial media rather than BHI) (26), we observed limited inhibition by butyrate against two commonly used lab strains, C. difficile strains VPI10463 and R20291. For strain 630, growth inhibition by butyrate was also context-dependent; when grown in vitro with CDMM and a single carbohydrate source, C. difficile growth was significantly inhibited by butyrate only in the presence of raffinose and lactose. Other tested sugars, including mannitol, fructose, and mannose, demonstrated increased growth of C. difficile with butyrate compared to without. These observations make sense in an in vivo context, where a diverse milieu of metabolites and energy sources could possibly negate the inhibitory effects of butyrate. For instance, we also observed increased growth of C. difficile in vitro in the presence of acetate, another prominent SCFA in the gut, although these differences were not significant.
Our results also demonstrate the ability of butyrate to modulate C. difficile pathogenesis via spore and toxin production. To our knowledge, increased spore production by butyrate has not been previously demonstrated, although higher spore and toxin production has been predicted in response to increasing SCFAs, which were shown to decrease biofilm production (79). A previous study reported enhanced toxin production by C. difficile in the presence of butyrate, similar to our current study (80). This study also observed a correlation between toxin and butyrate production by C. difficile itself, whereby the addition of different amino acids downregulated the production of both. Furthermore, toxin production has been correlated with increased expression of butyrate metabolism in C. difficile in subsequent studies (81). This contrasts with our results, where we observed higher toxin production in the presence of butyrate, yet downregulation of 3-hydroxybutyryl-CoA dehydrogenase, an enzyme known to be involved in the production of butyrate and butanol in Clostridium acetobutylicum (82). Although these results might initially seem contradictory, we take these observations as further evidence for coordination of metabolism and toxin production by C. difficile, whereby the presence of butyrate might initially increase toxin production but later downregulate its production, either within the same cell or in different populations.
Perhaps more importantly, both our phenotypic and RNA-Seq data demonstrated a significant increase in C. difficile spore production in the presence of butyrate, which is connected to toxin production and metabolism (48). Indeed, a recent study demonstrated higher spore counts and increased disease severity in mice mono-colonized with a butyrate-producing bacterium, Clostridium sardiniense, prior to C. difficile infection (77). This is in contrast to impeded growth and attenuated disease in mice mono-colonized with Paraclostridium bifermentans, which can compete for amino acids via Stickland fermentation. Interestingly, our results mimic in vivo C. difficile RNA-Seq profiles of mice infected with pathogenic strains compared to strains deficient in toxin (33, 83), whereby PTS transport of alternative carbohydrate metabolic pathways, such as mannose, lactose or fructose, is preferred instead of glucose-focused pathways or other alternate carbon sources. Our RNA-Seq data in rich media also match what we observed phenotypically in the presence of carbohydrate supplementation of CDMM, in which butyrate only had a positive growth impact in the presence of certain carbohydrates, such as mannose, lactose, or fructose, reflected by the increased expression of PTS transporters of these carbohydrates in our RNA-Seq data. Though our data also show an increase in expression of PTS transporters of mannitol, cellobiose, and xylose, we did not test these carbohydrates due to an expectation that these will also show similar results to mannose, lactose, and fructose in vitro.
In terms of how butyrate may impact regulation of C. difficile virulence (70), we might expect decreased codY, ccpA, and rex expression in the presence of butyrate, given that we observed increased toxin in the presence of butyrate. CodY and CcpA typically decrease toxin and butyrate production (84, 85), whereas Rex is an important global regulator that responds to NAD+/NADH ratios in the cell, particularly when glucose or other rapidly metabolized sugars are not around (12, 86, 87). While we observed increased ccpA expression during early and late log growth from our RT-qPCR data (~0.8 mean log fold change) but significantly decreased in our RNA-Seq (−1.23 log fold change) only in early log, our results from both RNA-Seq and RT-qPCR for the canonical virulence regulators (codY, rex, prdR) were not significant in the presence of butyrate. The involvement of these genes cannot be resolved from our data alone, and it is possible that regulatory responses to butyrate may be independent of these regulators, even though our data suggest a metabolic connection. Interestingly, we observed significant upregulation of a putative, non-canonical, carbon starvation gene, cstA, during early log growth. The canonical gene cstA has been demonstrated to be involved in peptide utilization, agglutination, and motility in another gut pathogen, Campylobacter jejuni (88), which was not upregulated in our data. Given the effect of butyrate on toxin and spore production, as well as the types of genes that were upregulated by it, it is possible that butyrate induces a collective stress response via alternate regulatory mechanisms leading to premature induction of sporulation in vegetative cells.
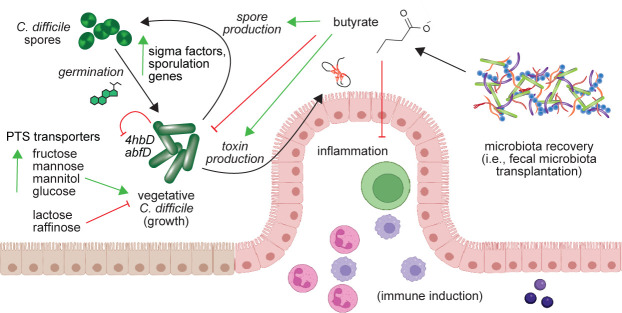
Independent of the potential regulatory mechanisms, the effect of butyrate on C. difficile has important clinical implications (Fig. 7). While butyrate and C. difficile levels are consistently negatively regulated following successful FMT and have been demonstrated to attenuate inflammation via the host, our results suggest a complicated outcome for butyrate-focused treatments. For instance, treating patients with CDI with butyrate alone (either with butyrate-producing bacteria or exogenous application) may have a detrimental effect on the patient, as has been observed in vivo in mice (77). Yet, combining this approach with additional microbiota members that can compete with C. difficile for nutrients may appropriately supplement the anti-inflammatory, and potentially inhibitory, effect by butyrate in the gut environment. Identification of the regulatory elements that dictate the effects of butyrate may expedite these findings.
Fig 7.
Model for potential mechanism of butyrate effectiveness against C. difficile strain 630. Increasing butyrate may alleviate host inflammation during recovery of the microbiota (such as via fecal microbiota transplantation) but also signals C. difficile to change metabolic strategies to increase survival. This may involve increased expression of PTS transporters for mannose, fructose, and mannitol and decreasing expression of butyrate-producing genes (4hbD, abfD), inducing alternate metabolic pathways for carbohydrate utilization in metabolically active cells. While growth of vegetative cells may be inhibited, sporulation and toxin genes are upregulated to optimize colonization. Figure illustrated in part with Biorender.
ACKNOWLEDGMENTS
We would like to acknowledge the Clemson University for generous allotment of compute time on Palmetto cluster. This publication was made possible, in part, with support from the Clemson University Genomics and Bioinformatics Facility, which receives support from an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM109094.
A.M.S. was supported by grant number K01-DK111794 from the National Institute of Diabetes and Digestive and Kidney Diseases. We would like to thank the Clemson University Department of Biological Sciences Microbiology prep labs for the use of their phase contrast microscope. A.M.S. has received consultation fees from Finch Therapeutics and Rebiotix/Ferring Pharmaceuticals.
M.B.: Conceptualization, Formal analysis, Data curation, Investigation, Methodology, Validation, Writing—original draft; J.C.: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing—original draft; D.B.: Formal analysis, Methodology, Software, Writing—original draft, and Writing—review and editing; S.N.: Methodology, Investigation; A.M.: Methodology, Investigation; A.M.S.: Supervision, Project Administration, Funding Acquisition, Conceptualization, Writing – original draft, Writing – review and editing.
Contributor Information
Anna M. Seekatz, Email: aseekat@clemson.edu.
Tina M. Henkin, Ohio State University, Columbus, Ohio, USA
DATA AVAILABILITY
All code used to analyze data and preliminary statistics generated during RNA-Seq analysis are available at Github. The raw reads from RNA-Seq are available under BioProject number: PRJNA955248 (BioSamples SAMN34190971 – SAMN34190982).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00138-23.
All supplemental figures with their respective legends.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, Beldavs ZG, Kainer MA, Karlsson M, Gerding DN, McDonald LC, Emerging Infections Program Clostridioides difficile Infection Working Group . 2020. Trends in U.S. burden of Clostridioides difficile infection and outcomes. N Engl J Med 382:1320–1330. doi: 10.1056/NEJMoa1910215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham C-A, Dunne WM. 2011. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol 49:2887–2893. doi: 10.1128/JCM.00891-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodrigues R, Barber GE, Ananthakrishnan AN. 2017. A comprehensive study of costs associated with recurrent Clostridium difficile infection. Infect Control Hosp Epidemiol 38:196–202. doi: 10.1017/ice.2016.246 [DOI] [PubMed] [Google Scholar]
- 4. Allegretti JR, Marcus J, Storm M, Sitko J, Kennedy K, Gerber GK, Bry L. 2020. Clinical predictors of recurrence after primary Clostridioides difficile infection: a prospective cohort study. Dig Dis Sci 65:1761–1766. doi: 10.1007/s10620-019-05900-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. 1977. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in Hamsters. J Infect Dis 136:701–705. doi: 10.1093/infdis/136.5.701 [DOI] [PubMed] [Google Scholar]
- 6. Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu D, Sorg JA, Sun X. 2018. Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol 8. doi: 10.3389/fcimb.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Usui Y, Ayibieke A, Kamiichi Y, Okugawa S, Moriya K, Tohda S, Saito R. 2020. Impact of deoxycholate on Clostridioides difficile growth, toxin production, and sporulation. Heliyon 6:e03717. doi: 10.1016/j.heliyon.2020.e03717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouillaut L, Dubois T, Sonenshein AL, Dupuy B. 2015. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol 166:375–383. doi: 10.1016/j.resmic.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jenior ML, Leslie JL, Young VB, Schloss PD. 2018. Clostridium difficile alters the structure and metabolism of distinct cecal microbiomes during initial infection to promote sustained colonization. mSphere 3:e00261-18. doi: 10.1128/mSphere.00261-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bouillaut L, Dubois T, Francis MB, Daou N, Monot M, Sorg JA, Sonenshein AL, Dupuy B. 2019. Role of the global regulator Rex in control of NAD+-regeneration in Clostridioides (Clostridium) difficile. Mol Microbiol 111:1671–1688. doi: 10.1111/mmi.14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x [DOI] [PubMed] [Google Scholar]
- 16. Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM, Henkin TM. 2016. CodY-dependent regulation of sporulation in Clostridium difficile. J Bacteriol 198:2113–2130. doi: 10.1128/JB.00220-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janoir C, Denève C, Bouttier S, Barbut F, Hoys S, Caleechum L, Chapetón-Montes D, Pereira FC, Henriques AO, Collignon A, Monot M, Dupuy B. 2013. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 81:3757–3769. doi: 10.1128/IAI.00515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sickmier EA, Brekasis D, Paranawithana S, Bonanno JB, Paget MSB, Burley SK, Kielkopf CL. 2005. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure 13:43–54. doi: 10.1016/j.str.2004.10.012 [DOI] [PubMed] [Google Scholar]
- 19. Chandrasekaran R, Lacy DB. 2017. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41:723–750. doi: 10.1093/femsre/fux048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x [DOI] [PubMed] [Google Scholar]
- 21. Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yadegar A, Pakpoor S, Ibrahim FF, Nabavi-Rad A, Cook L, Walter J, Seekatz AM, Wong K, Monaghan TM, Kao D. 2023. Beneficial effects of fecal Microbiota transplantation in recurrent Clostridioides difficile infection. Cell Host & Microbe 31:695–711. doi: 10.1016/j.chom.2023.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gregory AL, Pensinger DA, Hryckowian AJ. 2021. A short chain fatty acid-centric view of Clostridioides difficile pathogenesis. PLoS Pathog 17:e1009959. doi: 10.1371/journal.ppat.1009959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seekatz AM, Theriot CM, Rao K, Chang Y-M, Freeman AE, Kao JY, Young VB. 2018. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 53:64–73. doi: 10.1016/j.anaerobe.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, Allegretti JR, Smith MB, Xavier RJ, Alm EJ. 2018. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23:229–240. doi: 10.1016/j.chom.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pensinger DA, Fisher AT, Dobrila HA, Van Treuren W, Gardner JO, Higginbottom SK, Carter MM, Schumann B, Bertozzi CR, Anikst V, Martin C, Robilotti EV, Chow JM, Buck RH, Tompkins LS, Sonnenburg JL, Hryckowian AJ. 2023. Butyrate differentiates permissiveness to Clostridioides difficile infection and influences growth of diverse C. difficile isolates. Infect Immun 91:e0057022. doi: 10.1128/iai.00570-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang S, Deng W, Li F, Xiang L, Lv P, Chen Y. 2023. Treatment with butyrate alleviates dextran sulfate sodium and Clostridium difficile-induced colitis by preventing activity of Th17 cells via regulation of SIRT1/mTOR in mice. J Nutr Biochem 111:109155. doi: 10.1016/j.jnutbio.2022.109155 [DOI] [PubMed] [Google Scholar]
- 28. Fachi JL, Felipe J de S, Pral LP, da Silva BK, Corrêa RO, de Andrade MCP, Basso PJ, da Fonseca DM, Câmara NOS, de Sales e SÉ, Guima SES, Thomas AM, Setubal JC, Magalhães YT, Forti FL, Candreva T, Rodrigues HG, de Jesus MB, Consonni SR, Farias A dos S, Varga-Weisz P, Vinolo MAR. 2019. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep 27:750–761. doi: 10.1126/sciimmunol.aay7501 [DOI] [PubMed] [Google Scholar]
- 29. Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102. doi: 10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theriot CM, Schumacher CA, Bassis CM, Seekatz AM, Young VB. 2015. Effects of tigecycline and vancomycin administration on established Clostridium difficile infection. Antimicrob Agents Chemother 59:1596–1604. doi: 10.1128/AAC.04296-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhattacharjee D, Francis MB, Ding X, McAllister KN, Shrestha R, Sorg JA, Schneewind O. 2016. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J Bacteriol 198:777–786. doi: 10.1128/JB.00908-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit9A.1. doi: 10.1002/9780471729259.mc09a01s12 [DOI] [PubMed] [Google Scholar]
- 33. Fletcher JR, Pike CM, Parsons RJ, Rivera AJ, Foley MH, McLaren MR, Montgomery SA, Theriot CM. 2021. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun 12:462. doi: 10.1038/s41467-020-20746-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karasawa T, Ikoma S, Yamakawa K, Nakamura S. 1995. A defined growth medium for Clostridium difficile. Microbiology 141 ( Pt 2):371–375. doi: 10.1099/13500872-141-2-371 [DOI] [PubMed] [Google Scholar]
- 35. Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. 2012. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res 40:10701–10718. doi: 10.1093/nar/gks864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stewart ML, Timm DA, Slavin JL. 2008. Fructooligosaccharides exhibit more rapid fermentation than long-chain inulin in an in vitro fermentation system. Nutr Res 28:329–334. doi: 10.1016/j.nutres.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 37. Keohane PP, Attrill H, Jones BJ, Brown I, Frost P, Silk DB. 1983. The roles of lactose and Clostridium difficile in the pathogenesis of enteral feeding associated diarrhoea. Clin Nutr 1:259–264. doi: 10.1016/0261-5614(83)90003-1 [DOI] [PubMed] [Google Scholar]
- 38. Lozniewski A, Rabaud C, Dotto E, Weber M, Mory F. 2001. Laboratory diagnosis of Clostridium difficile-associated diarrhea and colitis: usefulness of Premier Cytoclone A+B enzyme immunoassay for combined detection of stool toxins and toxigenic C. difficile strains. J Clin Microbiol 39:1996–1998. doi: 10.1128/JCM.39.5.1996-1998.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins J, Danhof H, Britton RA. 2019. The role of trehalose in the global spread of epidemic Clostridium difficile. Gut Microbes 10:204–209. doi: 10.1080/19490976.2018.1491266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Theriot CM, Young VB. 2014. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes 5:86–95. doi: 10.4161/gmic.27131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kazamias MT, Sperry JF. 1995. Enhanced fermentation of mannitol and release of cytotoxin by Clostridium difficile in alkaline culture media. Appl Environ Microbiol 61:2425–2427. doi: 10.1128/aem.61.6.2425-2427.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Theriot CM, Koumpouras CC, Carlson PE, Bergin II, Aronoff DM, Young VB. 2011. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2:326–334. doi: 10.4161/gmic.19142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edwards AN, McBride SM. 2016. Isolating and purifying Clostridium difficile spores, p 117–128. In Methods in molecular biology. Humana Press Inc. doi: 10.1007/978-1-4939-6361-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reed AD, Nethery MA, Stewart A, Barrangou R, Theriot CM. 2020. Strain-dependent inhibition of Clostridioides difficile by Commensal Clostridia carrying the bile acid-inducible (bai) operon. J Bacteriol 202:39. doi: 10.1128/jb.00039-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 46. Blanco-Míguez A, Beghini F, Cumbo F, McIver LJ, Thompson KN, Zolfo M, Manghi P, Dubois L, Huang KD, Thomas AM, Nickols WA, Piccinno G, Piperni E, Punčochář M, Valles-Colomer M, Tett A, Giordano F, Davies R, Wolf J, Berry SE, Spector TD, Franzosa EA, Pasolli E, Asnicar F, Huttenhower C, Segata N. 2023. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat Biotechnol. doi: 10.1038/s41587-023-01688-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, Dougan G, Choudhary JS, Lawley TD. 2014. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics 15:160. doi: 10.1186/1471-2164-15-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Monot M, Boursaux-Eude C, Thibonnier M, Vallenet D, Moszer I, Medigue C, Martin-Verstraete I, Dupuy B. 2011. Reannotation of the genome sequence of Clostridium difficile strain 630. J Med Microbiol 60:1193–1199. doi: 10.1099/jmm.0.030452-0 [DOI] [PubMed] [Google Scholar]
- 50. Liao Y, Smyth GK, Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. doi: 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wickham H. 2016. Ggplot2: elegant graphics for data analysis. Springer-Verlag New York. https://ggplot2.tidyverse.org [Google Scholar]
- 53. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. 2021. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2:100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McBride SM, Sonenshein AL. 2011. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79:167–176. doi: 10.1128/IAI.00731-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. 2013. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol 195:5174–5185. doi: 10.1128/JB.00501-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen K-Y, Rathod J, Chiu Y-C, Chen J-W, Tsai P-J, Huang I-H. 2019. The transcriptional regulator Lrp contributes to toxin expression, sporulation, and swimming motility in Clostridium difficile. Front Cell Infect Microbiol 9:356. doi: 10.3389/fcimb.2019.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Edwards AN, Tamayo R, McBride SM. 2016. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol 100:954–971. doi: 10.1111/mmi.13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rao X, Huang X, Zhou Z, Lin X. 2013. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 59. Simonida G, David AG, William WM. 2020. Diverse energy-conserving pathways in Clostridium difficile: growth in the absence of amino acid Stickland Acceptors and the role of the wood-Ljungdahl pathway. J Bacteriol 202:233. doi: 10.1128/jb.00233-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakamura S, Nakashio S, Yamakawa K, Tanabe N, Nishida S. 1982. Carbohydrate fermentation by Clostridium difficile. Microbiol Immunol 26:107–111. doi: 10.1111/j.1348-0421.1982.tb00159.x [DOI] [PubMed] [Google Scholar]
- 61. Hofmann JD, Otto A, Berges M, Biedendieck R, Michel AM, Becher D, Jahn D, Neumann-Schaal M. 2018. Metabolic reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front Microbiol 9:1970. doi: 10.3389/fmicb.2018.01970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hamo Z, Azrad M, Fichtman B, Peretz A. 2021. The cytopathic effect of different toxin concentrations from different Clostridioides difficile sequence types strains in vero cells. Front Microbiol 12:763129. doi: 10.3389/fmicb.2021.763129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. López-Ureña D, Orozco-Aguilar J, Chaves-Madrigal Y, Ramírez-Mata A, Villalobos-Jimenez A, Ost S, Quesada-Gómez C, Rodríguez C, Papatheodorou P, Chaves-Olarte E. 2019. Toxin B variants from Clostridium difficile strains VPI 10463 and Nap1/027 share similar substrate profile and cellular intoxication Kinetics but use different host cell entry factors. Toxins (Basel) 11:348. doi: 10.3390/toxins11060348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Martin-Verstraete I, Peltier J, Dupuy B. 2016. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins (Basel) 8:153. doi: 10.3390/toxins8050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li Z, Di Donato F, Piggot PJ. 2004. Compartmentalization of gene expression during sporulation of Bacillus subtilis is compromised in mutants blocked at stage III of sporulation. J Bacteriol 186:2221–2223. doi: 10.1128/JB.186.7.2221-2223.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nicholson WL, Sun DX, Setlow B, Setlow P. 1989. Promoter specificity of sigma G-containing RNA polymerase from sporulating cells of Bacillus subtilis: identification of a group of forespore-specific promoters. J Bacteriol 171:2708–2718. doi: 10.1128/jb.171.5.2708-2718.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A, Viollier PH. 2013. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9:e1003660. doi: 10.1371/journal.pgen.1003660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Marshall A, McGrath JW, Graham R, McMullan G. 2023. Food for thought-the link between Clostridioides difficile metabolism and pathogenesis. PLoS Pathog 19:e1011034. doi: 10.1371/journal.ppat.1011034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W. 2000. Fermentation of 4-aminobutyrate by Clostridium aminobutyricum: cloning of two genes involved in the formation and dehydration of 4-hydroxybutyryl-CoA. Arch Microbiol 174:189–199. doi: 10.1007/s002030000195 [DOI] [PubMed] [Google Scholar]
- 70. Ternan NG, Moore ND, Smyth D, McDougall GJ, Allwood JW, Verrall S, Gill CIR, Dooley JSG, McMullan G. 2018. Increased sporulation underpins adaptation of Clostridium difficile strain 630 to a biologically–relevant faecal environment, with implications for pathogenicity. Sci Rep 8:16691. doi: 10.1038/s41598-018-35050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maaß S, Otto A, Albrecht D, Riedel K, Trautwein-Schult A, Becher D. 2018. Proteomic signatures of Clostridium difficile stressed with metronidazole, vancomycin, or fidaxomicin. Cells 7:213. doi: 10.3390/cells7110213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kumar M, Adhikari S, Hurdle JG. 2014. Action of nitroheterocyclic drugs against Clostridium difficile. Int J Antimicrob Agents 44:314–319. doi: 10.1016/j.ijantimicag.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Edwards AN, Wetzel D, DiCandia MA, McBride SM. 2022. Three orphan histidine kinases inhibit Clostridioides difficile sporulation. J Bacteriol 204:e0010622. doi: 10.1128/jb.00106-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, Oravecz-Wilson K, Wu S-R, Sun Y, Rossi C, Fujiwara H, Byun J, Shono Y, Lindemans C, Calafiore M, Schmidt TM, Honda K, Young VB, Pennathur S, van den Brink M, Reddy P. 2016. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 17:505–513. doi: 10.1038/ni1016-1235b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. 2021. Short chain fatty acids and its producing organisms: an overlooked therapy for IBD? EBioMedicine 66:103293. doi: 10.1016/j.ebiom.2021.103293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. 2018. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 3:662–669. doi: 10.1038/s41564-018-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Girinathan BP, DiBenedetto N, Worley JN, Peltier J, Arrieta-Ortiz ML, Immanuel SRC, Lavin R, Delaney ML, Cummins CK, Hoffman M, Luo Y, Gonzalez-Escalona N, Allard M, Onderdonk AB, Gerber GK, Sonenshein AL, Baliga NS, Dupuy B, Bry L. 2021. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29:1693–1708. doi: 10.1016/j.chom.2021.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wetzel D, McBride SM, Ercolini D. 2020. The impact of pH on Clostridioides difficile sporulation and Physiology. Appl Environ Microbiol 86:e02706-19. doi: 10.1128/AEM.02706-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tremblay YDN, Durand BAR, Hamiot A, Martin-Verstraete I, Oberkampf M, Monot M, Dupuy B. 2021. Metabolic adaption to extracellular pyruvate triggers biofilm formation in Clostridioides difficile. ISME J 15:3623–3635. doi: 10.1038/s41396-021-01042-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun 68:5881–5888. doi: 10.1128/IAI.68.10.5881-5888.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Karlsson S, Burman LG, Åkerlund T. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology (N Y) 154:3430–3436. doi: 10.1099/mic.0.2008/019778-0 [DOI] [PubMed] [Google Scholar]
- 82. Dürre P, Fischer RJ, Kuhn A, Lorenz K, Schreiber W, Stürzenhofecker B, Ullmann S, Winzer K, Sauer U. 1995. Solventogenic enzymes of Clostridium acetobutylicum : catalytic properties, genetic organization, and transcriptional regulation. FEMS Microbiol Rev 17:251–262. doi: 10.1111/j.1574-6976.1995.tb00209.x [DOI] [PubMed] [Google Scholar]
- 83. Pruss KM, Sonnenburg JL. 2021. C. difficile exploits a host metabolite produced during toxin-mediated disease. Nature 593:261–265. doi: 10.1038/s41586-021-03502-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem 73:245–259. doi: 10.1271/bbb.80479 [DOI] [PubMed] [Google Scholar]
- 85. Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr Opin Microbiol 8:203–207. doi: 10.1016/j.mib.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 86. Müh U, Ellermeier CD, Weiss DS. 2022. The WalRK two-component system is essential for proper cell envelope biogenesis in Clostridioides difficile. J Bacteriol 204:e0012122. doi: 10.1128/jb.00121-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Franza T, Rogstam A, Thiyagarajan S, Sullivan MJ, Derré-Bobillot A, Bauer MC, Goh KGK, Da Cunha V, Glaser P, Logan DT, Ulett GC, von Wachenfeldt C, Gaudu P. 2021. NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence. PLoS Pathog 17:e1009791. doi: 10.1371/journal.ppat.1009791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rasmussen JJ, Vegge CS, Frøkiær H, Howlett RM, Krogfelt KA, Kelly DJ, Ingmer H. 2013. Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J Med Microbiol 62:1135–1143. doi: 10.1099/jmm.0.059345-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All supplemental figures with their respective legends.
Data Availability Statement
All code used to analyze data and preliminary statistics generated during RNA-Seq analysis are available at Github. The raw reads from RNA-Seq are available under BioProject number: PRJNA955248 (BioSamples SAMN34190971 – SAMN34190982).