Abstract
Psyllids (Hemiptera: Sternorrhyncha: Psylloidea) are plant sap-sucking insects that are closely associated with various microbes. To obtain a more detailed understanding of the ecological and evolutionary behaviors of microbes in Psylloidea, the bacterial populations of six psyllid species, belonging to the family Carsidaridae, were analyzed using high-throughput amplicon sequencing of the 16S rRNA gene. The majority of the secondary symbionts identified in the present study were gammaproteobacteria, particularly those of the order Enterobacterales, including Arsenophonus and Sodalis, which are lineages found in a wide variety of insect hosts. Additionally, Symbiopectobacterium, another Enterobacterales lineage, which has recently been recognized and increasingly shown to be vertically transmitted and mutualistic in various invertebrates, was identified for the first time in Psylloidea. This lineage is closely related to Pectobacterium spp., which are plant pathogens, but forms a distinct clade exhibiting no pathogenicity to plants. Non-Enterobacterales gammaproteobacteria found in the present study were Acinetobacter, Pseudomonas (both Pseudomonadales), Delftia, Comamonas (both Burkholderiales), and Xanthomonas (Xanthomonadales), a putative plant pathogen. Regarding alphaproteobacteria, three Wolbachia (Rickettsiales) lineages belonging to supergroup B, the major group in insect lineages, were detected in four psyllid species. In addition, a Wolbachia lineage of supergroup O, a minor group recently found for the first time in Psylloidea, was detected in one psyllid species. These results suggest the pervasive transfer of bacterial symbionts among animals and plants, providing deeper insights into the evolution of the interactions among these organisms.
Keywords: Symbiopectobacterium, Wolbachia supergroup O, Arsenophonus, Sodalis, Xanthomonas
Psyllids (Psylloidea) are plant sap-sucking hemipteran insects that encompass ~4,000 described species worldwide, constituting the suborder Sternorrhyncha along with whiteflies (Aleyrodoidea), aphids (Aphidoidea), phylloxera (Phylloxeroidea), and scale insects (Coccoidea) (Burckhardt et al., 2021). Similar to other sternorrhynchan insects (International Aphid Genomics Consortium, 2010; Nakabachi and Miyagishima, 2010; Tamborindeguy et al., 2010), psyllids feed exclusively on phloem sap using their needle-like mouthpart called the stylet (Hodkinson, 1974; Burckhardt et al., 2021). Due to this feeding habit, some psyllid species transmit plant pathogens, including “Candidatus Liberibacter spp.” (Alphaproteobacteria: Rhizobiales) and “Ca. Phytoplasma spp.” (Bacilli: Acholeplasmatales), making them important agricultural pests (Jarausch and Jarausch, 2010; Grafton-Cardwell et al., 2013; Mora et al., 2021). Moreover, phloem sap is deficient in nutrients, including essential amino acids and some vitamins (Ziegler et al., 1975; Sandström and Moran, 1999). Therefore, these deficiencies are compensated by transovarially transmitted bacterial mutualists harbored in a specialized organ called the bacteriome (Nakabachi et al., 2010b; Sloan et al., 2014). Although in situ hybridization has revealed the symbiont localization of only a few psyllid species (Fukatsu and Nikoh, 1998, 2000; Subandiyah et al., 2000; Nakabachi et al., 2013b; Dan et al., 2017), the psyllid bacteriome is considered to typically harbor two distinct intracellular symbionts, based on microscopic observations with classical staining methods (Profft, 1937; Buchner, 1965), followed by bacterial gene amplicon analyses with cloning (Spaulding and von Dohlen, 1998, 2001; Thao et al., 2000a; Hansen et al., 2007) and next-generation sequencing methods (Arp et al., 2014; Hall et al., 2016; Fromont et al., 2017; Morrow et al., 2017, 2020; Nakabachi et al., 2020a, 2022a, 2022b), along with metagenomic analyses (Sloan and Moran, 2012; Nakabachi et al., 2013b, 2020b). The primary symbiont, the nutritional mutualist conserved among host species and, thus, considered to be essential in distinct insect clades, is “Ca. Carsonella ruddii” (Gammaproteobacteria: Oceanospirillales, hereafter Carsonella) in Psylloidea (Thao et al., 2000b), which provides the host with essential amino acids (Nakabachi et al., 2006; Sloan and Moran, 2012; Nakabachi et al., 2013b, 2020b; Nakabachi and Moran, 2022). Molecular phylogenetic analyses have repeatedly demonstrated cospeciation between host psyllids and Carsonella, resulting from the single acquisition of a Carsonella ancestor by a psyllid common ancestor and its subsequent stable vertical transmission (Thao et al., 2000b; Spaulding and von Dohlen, 2001; Hall et al., 2016; Nakabachi et al., 2020a, 2022a, 2022b). Another bacterial lineage harbored in the bacteriome is categorized as a ‘secondary symbiont’, meaning an additional symbiont. Although secondary symbionts in diverse insect taxa form various host-symbiont relationships across the mutualism-parasitism continuum (Gherna et al., 1991; Dale and Maudlin, 1999; Nakabachi et al., 2003; Russell et al., 2003; Thao and Baumann, 2004; Moran et al., 2008; Werren et al., 2008; Oliver et al., 2010; Johnson, 2015), those in the psyllid bacteriome consistently appear to have obligate mutualistic relationships with the host psyllid (Sloan and Moran, 2012; Nakabachi et al., 2013b, 2020b). Nevertheless, these secondary symbionts are phylogenetically diverse among host lineages, suggesting their repeated infection and replacement during the evolution of Psylloidea (Thao et al., 2000a; Spaulding and von Dohlen, 2001; Sloan and Moran, 2012; Hall et al., 2016; Morrow et al., 2017; Nakabachi et al., 2020a, 2022a, 2022b). They are mostly considered to have a nutritional basis (Spaulding and von Dohlen, 2001; Sloan and Moran, 2012; Morrow et al., 2017), with the unique exception of “Ca. Profftella armatura” (Gammaproteobacteria: Burkholderiales) (Nakabachi et al., 2013b, 2020a, 2020b; Dan et al., 2017), the main role of which appears to protect the holobiont (host psyllid plus bacteriome-associated mutualists) from natural enemies, including predators, parasitoids, and pathogens (Nakabachi et al., 2013b, 2020b; Nakabachi and Fujikami, 2019; Nakabachi and Okamura, 2019; Yamada et al., 2019; Tanabe et al., 2022). Besides these bacteriome-associated obligate mutualists, psyllids may harbor various secondary symbionts of a facultative nature, including Wolbachia (Alphaproteobacteria: Rickettsiales) and Rickettsia (Alphaproteobacteria: Rickettsiales), which cause systemic infection in host insects (Spaulding and von Dohlen, 2001; Sloan and Moran, 2012; Arp et al., 2014; Chu et al., 2016, 2019; Jain et al., 2017; Kruse et al., 2017; Morrow et al., 2017; Nakabachi et al., 2020a, 2022a, 2022b). Similar to other hemipteran insects (Nakabachi and Ishikawa, 1997, 1999, 2000, 2001; Nakabachi et al., 2003, 2005, 2010a, 2014; Moran et al., 2008; Kikuchi, 2009; Nikoh and Nakabachi, 2009; Gerardo et al., 2010; Nikoh et al., 2010; Ramsey et al., 2010; Shigenobu et al., 2010; Tamborindeguy et al., 2010; Nakabachi, 2015; Uchi et al., 2019), evidence is accumulating to show that interactions among symbiotic microbes, including those associated with the bacteriome, facultative symbionts, and plant pathogens, along with interactions between host psyllids and these bacterial populations (Nakabachi et al., 2006, 2010b, 2013b; Sloan et al., 2014; Dan et al., 2017; Nakabachi and Fujikami, 2019), are important for psyllid biology and host plant pathology (Nakabachi et al., 2013a, 2020a, 2022b; Chu et al., 2016, 2019; Dan et al., 2017; Jain et al., 2017; Kruse et al., 2017; Killiny, 2022; Tanabe et al., 2022). Therefore, the identification of microbiomes in various psyllid lineages, which mirror the ecological and evolutionary behaviors of bacterial populations in Psylloidea, will help establish a basis for the better control of pest species.
According to a definition revised by Burckhardt et al., psyllids are classified into seven extant families: Aphalaridae, Calophyidae, Carsidaridae, Liviidae, Mastigimatidae, Psyllidae, and Triozidae (Burckhardt et al., 2021). Carsidaridae is a relatively small family consisting of three subfamilies, Carsidarinae, Homotominae, and Pachypsyllinae, which comprise 148 species of 23 genera (Ouvrard, 2023). This family encompasses agricultural pests, including Mesohomotoma tessmanni (Carsidarinae), Allocarsidara malayensis (Carsidarinae), and Homotoma ficus (Homotominae), which feed on cacao (Yana et al., 2015), durian (Ketsa et al., 2020), and fig (Tasi et al., 2021) trees, respectively. A recent study performed a relatively comprehensive analysis of 16S rRNA gene amplicons using 44 psyllid species of five families, including four Carsidaridae species: H. ficus, Macrohomotoma gladiata (Homotominae), Mesohomotoma hibisci (Carsidarinae), and Protyora sterculiae (Carsidarinae) (Kwak et al., 2021). The analysis revealed various bacteria, including two lineages of “Ca. Liberibacter” that are potential plant pathogens. One of them was a close relative of “Ca. Liberibacter asiaticus”, which is a notorious pathogen of the most devastating citrus disease, Huanglongbing or greening disease, and the other was “Ca. Liberibacter capsica”, a novel species that may potentially cause diseases in solanaceous crops. Although this study provided important insights into the evolution of microbiomes in Psylloidea, the analysis was performed using (1) only one individual or a pool of two individuals for each psyllid species, making the analysis less reliable; (2) primers unsuitable for detecting bacteria with AT-rich 16S rRNA genes, including Carsonella; and (3) the analytical method that clusters sequence reads with a similarity threshold of 97%, resulting in a lower resolution of the analysis. Moreover, (4) none of the representative sequences were deposited in public databases, making phylogenetic analyses difficult to reproduce.
In the present study, amplicon analyses were performed based on Illumina sequencing of the V3 and V4 regions of 16S rRNA genes to assess the microbiomes of six Carsidaridae species collected in Japan (Table 1) using (1) ten (five adult males and five adult females) pooled individuals for each species, (2) primers optimized to detect 16S rRNA genes with a wider variety of G+C contents, and (3) the method to remove sequencing errors and resolve sequence variants (SVs) down to the level of single-nucleotide differences. Furthermore, (4) all of the main SVs (>1% of the total reads) have been deposited in public databases, providing the research community with a basis for the further investigation and reexamination of the analytical results.
Table 1.
Psyllid species used in the present study
| Species | Subfamily | Sampling site | Collection date | Host plant |
|---|---|---|---|---|
| Carsidara limbata | Carsidarinae | Hakozaki, Fukuoka city, Fukuoka Pref., Kyushu, Japan (33.6262 N 130.4248 E) | November 15, 2015 | Firmiana simplex (Malvaceae) |
| Mesohomotoma camphorae | Carsidarinae | Gusuku, Sumiyô, Amami-oshima Is., Kagoshima Pref., Ryukyus, Japan (28.2956 N 129.4572 E) | May 23, 2009 | Hibiscus hamabo (Malvaceae) |
| Tyora ornata | Carsidarinae | Komi, Iriomote Is., Okinawa Pref., Ryukyus, Japan (24.3129 N 123.9053 E) | May 5, 2000 | Heritiera littoralis (Malvaceae) |
| Homotoma radiata | Homotominae | Oniike, Itsuwa-machi, Amakusa City, Kumamoto Pref., Amakusa-shimoshima Is., Kyushu, Japan (32.5470 N 130.1865 E) | April 9, 2015 | Ficus subpisocarpa (Moraceae) |
| Homotoma unifasciata | Homotominae | Fukuregi, Amakusa city, Kumamoto Pref., Amakusa-shimoshima Is., Kyushu, Japan (32.4029 N 130.0800 E) | May 22, 2013 | Ficus erecta (Moraceae) |
| Celtisaspis japonica | Pachypsyllinae | Edosaki, Inasiki city, Ibaraki Pref., Honshu, Japan (35.9518 N 140.3216 E) | May 27, 2016 | Celtis sinensis (Cannabaceae) |
Materials and Methods
Insect sampling and DNA extraction
Adults of six psyllid species belonging to the family Carsidaridae were collected from their host plants at various locations in Japan (Table 1). Insect samples were stored in acetone (Carsidara limbata, Homotoma radiata, and Celtisaspis japonica) or 99.5% ethanol (the other species) at –20°C until DNA extraction. DNA was extracted from the whole bodies of pooled individuals of five adult males and five adult females of each psyllid species using the DNeasy Blood & Tissue Kit (Qiagen). The quality of extracted DNA was assessed using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). Its quantity was assessed using a Qubit 2.0 fluorometer with the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific).
Construction and sequencing of amplicon libraries
Bacterial populations in psyllids were analyzed in accordance with the instructions provided by Illumina (Illumina, 2013) with some modifications (Nakabachi et al., 2020a, 2022a, 2022b). An amplicon polymerase chain reaction (PCR) was performed using DNA extracted from psyllids, KAPA HiFi HotStart ReadyMix (KAPA Biosystems), and the primer set 16S_341Fmod (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGYYTAMGGRNGGCWGCAG-3′) and 16S_805R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′), targeting the V3 and V4 regions of the 16S rRNA gene. Underlined areas indicate regions corresponding to target 16S rRNA gene sequences. Other nucleotides were the overhangs required for preparing and sequencing Illumina libraries. Although both primers were based on the instructions by Illumina (Illumina, 2013), 16S_341F was modified (bold italics), where the original CC, C, and G were replaced with the mixed bases YY (C or T), M (A or C), and R (A or G), respectively. This modification was based on an alignment of 16S rRNA genes encoded in divergent bacterial genomes with various G+C contents. Referenced sequences included those of 44 diverse lineages of Carsonella derived from 38 psyllid species from 23 genera, belonging to five families, which were available in the nucleotide database of the National Center for Biotechnology Information (NCBI) as of August 2014. The modification achieved a 100% match of the primers to most Carsonella sequences, retaining a single mismatch each in four lineages (Supplementary Fig. S1). In our reanalysis in May 2023, we identified 11 more Carsonella sequences deposited in the NCBI nucleotide database, nine of which showed no mismatch to the primers. One sequence had a single mismatch and another showed two mismatches (single each for 16S_341Fmod and 16S_805R) (Supplementary Fig. S1). This modification increased sensitivity to detect symbionts with AT-rich genomes, including Carsonella, without reducing sensitivity to those with GC-rich genomes (Nakabachi et al., 2020a, 2022a, 2022b). Dual indices and Illumina sequencing adapters were attached to the amplicons with index PCR using Nextera XT Index Kit v2 (Illumina). The libraries were combined with PhiX Control v3 (Illumina), and 250 bp of both ends were sequenced on the MiSeq platform (Illumina) with MiSeq Reagent Kit v2 (500 cycles; Illumina).
Computational analysis of bacterial populations
Output sequences were processed using the QIIME2 platform (version 2022.8) (Bolyen et al., 2019). Primer sequences were removed from the demultiplexed sequence reads using the cutadapt plugin (Martin, 2011). The denoising and joining of paired-end reads and removal of low-quality or chimeric reads were subsequently performed using the DADA2 plugin (Callahan et al., 2016). In this process, parameters were set to --p-trunc-len-f 230 and --p-trunc-len-r 225 to remove 3′-end nucleotides with quality scores below the Phred score 30, referring to the quality histograms drawn with fastQC (version 0.11.9) (Andrews, 2023) and multiQC (version1.12) (Ewels et al., 2016). Dereplicated amplicon reads were classified, and taxonomic information was assigned using the q2-feature-classifier (Bokulich et al., 2018), which was trained with the V3 and V4 regions of the 16S rRNA gene (Silva 138 SSURef NR99) (Glöckner et al., 2017). The SVs obtained were manually checked by BLASTN searches against the NCBI non-redundant database (Camacho et al., 2009). After SVs were aligned to related sequences using SINA (version 1.2.11) (Pruesse et al., 2012), phylogenetic trees were inferred by the maximum likelihood (ML) method using RAxML (version 8.2.12) (Stamatakis, 2014). The GTR+Γ model was used with no partitioning of the data matrix, with 1,000 bootstrap iterations (options -f a -m GTRGAMMA -# 1000).
Data availability
Nucleotide sequence data are available in the DDBJ/EMBL/GenBank databases under the accession numbers DRR420941–DRR420946 (raw fastq files) and TAAE01000001–TAAE01000035 (dereplicated SVs).
Results
Overview
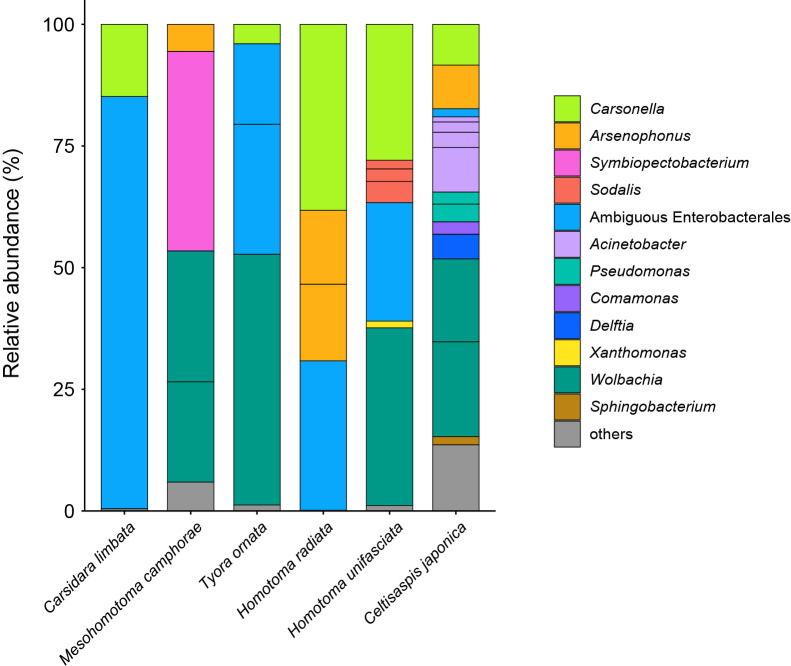
Raw libraries after demultiplexing yielded 48,874–83,223 pairs of forward and reverse reads for the six psyllid species. The denoising and joining of paired-end reads and removal of low-quality or chimeric reads resulted in 42,709–64,067 non-chimeric high-quality reads (Supplementary Table S1). The dereplication of these reads resulted in 342 independent SVs, 33 of which accounted for >1% of the total reads (Supplementary Table S2 and Fig. S2). The present study focused on the 33 main SVs unless otherwise noted because (1) filtering with a threshold of 1% was previously demonstrated to be among the most accurate and effective methods to eliminate potential contaminants (Karstens et al., 2019) and (2) the targets of the present study were relatively abundant symbionts with close associations with the host psyllids. SVs with a relative abundance of <1% were collectively categorized as ‘others’ (Fig. 1) and accounted for 0.1 (H. radiata) - 13.6% (C. japonica) of the total reads in each psyllid species (Supplementary Table S2).
Fig. 1.
Composition of bacterial populations in psyllids of the family Carsidaridae. The relative abundance of Illumina reads belonging to assigned bacterial taxa are shown.
Identification of Carsonella
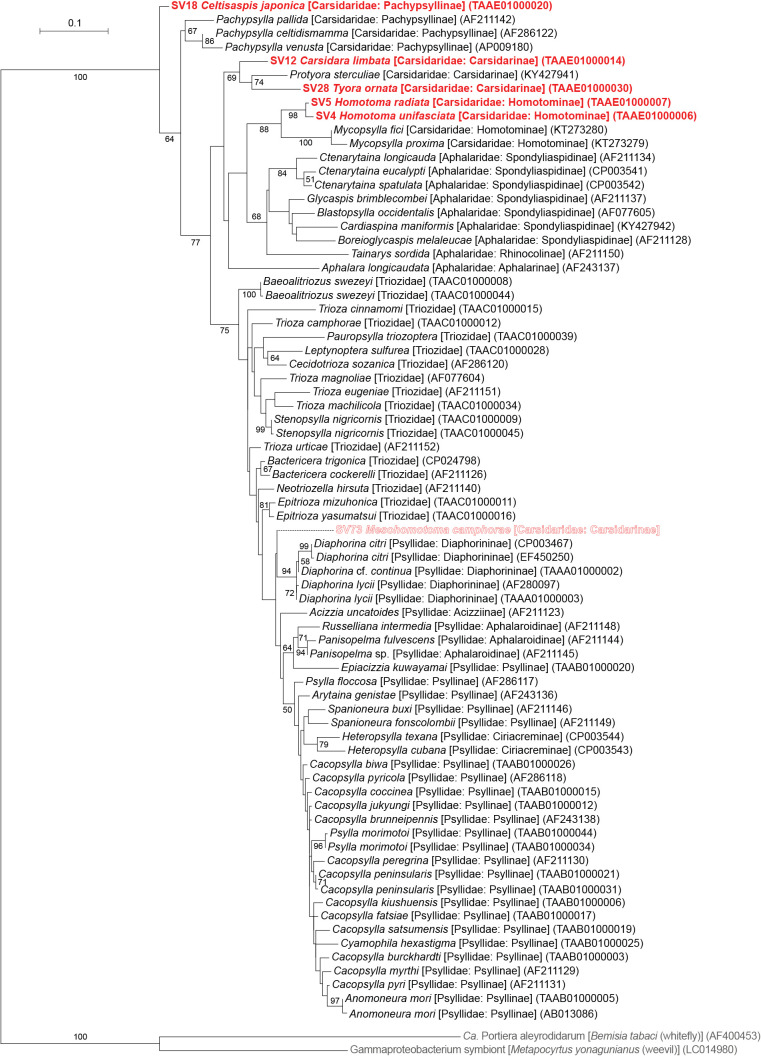
The taxonomic classification by QIIME2 (Supplementary Table S2) followed by independent BLAST searches and phylogenetic analyses showed that all psyllid species, except for Mesohomotoma camphorae, possess distinct lineages of Carsonella (Fig. 1). The ML tree placed these Carsonella sequences at positions that are largely consistent with the host psyllid phylogeny inferred by mitochondrial and nuclear gene analyses with the aid of morphological analyses (Percy et al., 2018; Cho et al., 2019; Burckhardt et al., 2021) (Fig. 2). SV4 and SV5, derived from the congeneric species Homotoma unifasciata (27.9% of the reads) and H. radiata (38.2% of the reads; both Homotominae), respectively, formed a robustly supported clade (bootstrap: 98%). This clade further formed a well-supported clade (bootstrap: 88%) with other psyllid species belonging to the same subfamily Homotominae (Fig. 2). SV12 and SV28, derived from C. limbata (14.8% of the reads) and Tyora ornata (4.1% of the reads; both Carsidarinae), respectively, also formed a moderately supported clade (bootstrap: 69%) with a sequence from P. sterculiae (Carsidaridae: Carsidarinae, KY427941). SV18, which accounted for 8.4% of C. japonica (Pachypsyllinae) reads, was placed at the basal position in the Carsonella tree (Fig. 2). Although SV18 did not form a clade with sequences derived from other Pachypsyllinae species of the New World endemic genus Pachypsylla, the phylogenetic position was proximal to the moderately supported clade consisting of these sequences (bootstrap: 67%) (Fig. 2). These results were consistent with previous findings showing that host psyllids and Carsonella cospeciated due to the stable vertical transmission of Carsonella (Thao et al., 2000b; Spaulding and von Dohlen, 2001; Hall et al., 2016; Nakabachi et al., 2020a, 2022a, 2022b).
Fig. 2.
Maximum likelihood phylogram of Carsonella. A total of 427 unambiguously aligned nucleotide sites of 16S rRNA genes were subjected to the analysis. On each branch, bootstrap support values of >50% are shown. Designations other than those for outgroups refer to psyllid hosts. Families and subfamilies (if applicable) of host psyllids are shown in brackets. Sequences from this study are shown in bold red. SV73 derived from Mesohomotoma camphorae is outlined. DDBJ/EMBL/GenBank accession numbers for sequences are provided in parentheses. The bar represents nucleotide substitutions per position. The outgroups were Ca. Portiera aleyrodidarum; the primary symbiont of the whitefly Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea), and a gammaproteobacterium symbiont of the weevil Metapocyrtus yonagunianus (Coleoptera: Curculionidae).
In contrast, no Carsonella-like SVs with a relative abundance of ≥1% were detected in M. camphorae (Supplementary Table S2). This was unexpected because Carsonella was considered to be essential in Psylloidea, and the primer set (Supplementary Fig. S1) used in the present study was shown to be suitable for detecting Carsonella (Nakabachi et al., 2020a, 2022a, 2022b). Therefore, the search was extended to include SVs with a relative abundance of <1% in M. camphorae. SV73, the most abundant SV in M. camphorae showing similarity to Carsonella sequences, accounted for only 0.16% of M. camphorae reads (Supplementary Table S2). The sequence was only 93.3% identical to the most similar sequences in the database, which were derived from Carsonella of Diaphorina cf. continua (AP023214, TAAA01000009) and Diaphorina lycii (TAAA01000012). These psyllid species belong to the family Psyllidae and are distantly related to M. camphorae. Moreover, the ML tree placed this SV at a position apart from Carsonella sequences found in other Carsidaridae species (Fig. 2). No sequence reads analyzed in the same MiSeq flow cell showed higher similarity to SV73 than the sequences found in Diaphorina spp. These results imply that SV73 was derived from a chimeric PCR artifact that QIIME2 failed to remove or a minor contaminant of an unknown source, not a true symbiont in M. camphorae. Similarly, all other M. camphorae SVs showing similarity to Carsonella, namely, SV125 (0.06% of the reads), SV213 (0.02%), SV219 (0.02%), and SV293 (0.01%), appeared to be derived from artifacts or contaminants. These types of minor SVs that showed similarity to Carsonella sequences, but appeared to correspond to PCR artifacts or contaminants of unknown sources were also detected in C. limbata, T. ornata, and C. japonica (Supplementary Table S2).
Enterobacterales symbionts in Carsidaridae
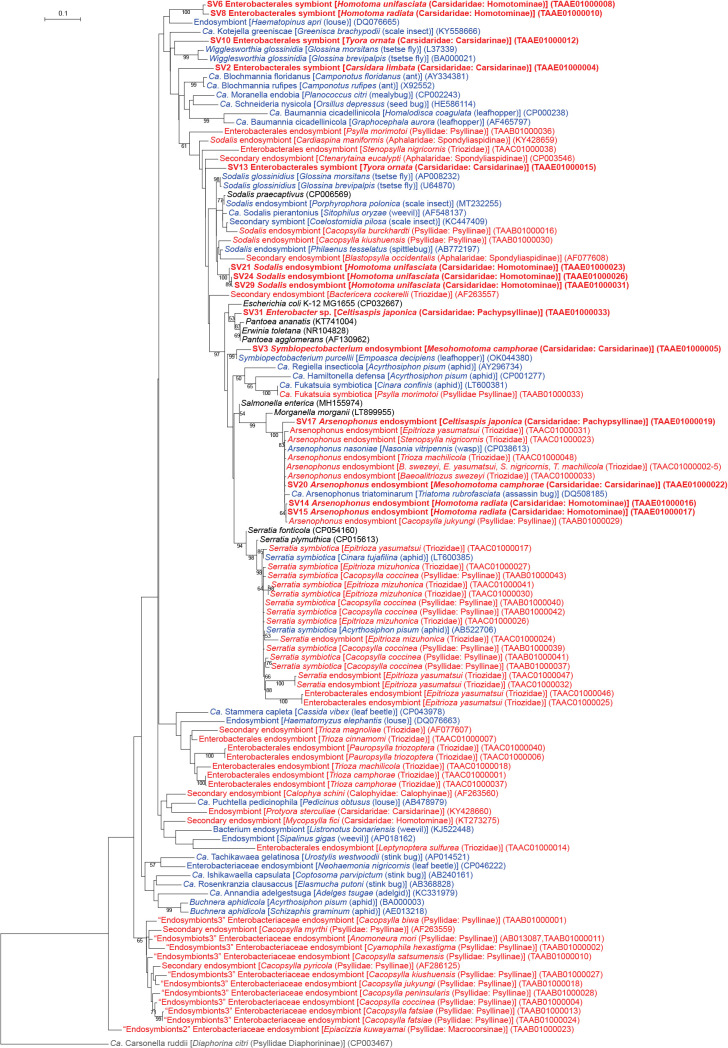
Of the 33 main SVs obtained in the present study, 28 corresponded to gammaproteobacteria, 14 of which belonged to the order Enterobacterales (Supplementary Table S2). Enterobacterales is a bacterial taxon that comprises numerous insect symbionts, including those associated with the bacteriome (Moran et al., 2008; McCutcheon et al., 2019). Enterobacterales bacteria identified in the present study included Arsenophonus, Sodalis, Symbiopectobacterium, and several lineages with ambiguous phylogenetic placements (Fig. 1 and 3, Supplementary Table S2).
Fig. 3.
Maximum likelihood phylogram of Enterobacterales. A total of 427 unambiguously aligned nucleotide sites of 16S rRNA genes were subjected to the analysis. On each branch, bootstrap support values of >50% are shown. The scale bar indicates substitutions per site. Regarding symbiotic bacteria, host organisms are shown in brackets. Symbionts of animals other than psyllids are shown in blue, while symbionts of psyllids are shown in red. Sequences from the present study are shown in bold. DDBJ/EMBL/GenBank accession numbers are provided in parentheses. Carsonella was used as an outgroup.
Arsenophonus symbionts
Four distinct SVs corresponding to Arsenophonus lineages were detected in three of the six Carsidaridae species: H. radiata, C. japonica, and M. camphorae (Fig. 1 and 3, Supplementary Table S2). SV14 (15.8% of H. radiata reads), SV15 (15.2% of H. radiata reads), SV17 (9.0% of C. japonica reads), and SV20 (5.5% of M. camphorae reads) were 97.2–100% identical to the sequences of Arsenophonus nasoniae (CP038613), the type species of Arsenophonus found in the parasitoid wasp Nasonia vitripennis (Hymenoptera: Pteromalidae) (Gherna et al., 1991), and Arsenophonus symbionts detected in various insect lineages. Host insects included aphids (Russell et al., 2003), whiteflies (Thao and Baumann, 2004), louse flies (Diptera: Hippoboscoidea) (Nováková et al., 2009), and the psyllid species Baeoalitriozus swezeyi, Epitrioza yasumatsui, Stenopsylla nigricornis, Trioza machilicola (all Triozidae; TAAC01000002–5), Cacopsylla jukyungi (Psyllidae; TAAB01000029), Cardiaspina tenuitela (Aphalaridae; KY428657), and Glycaspis brimblecombei (Aphalaridae; EU043378) (Hansen et al., 2007; Morrow et al., 2017; Nakabachi et al., 2022a, 2022b). These sequences formed a robustly supported clade (bootstrap: 100%) in the ML tree (Fig. 3). SV14 and SV15, both of which were derived from H. radiata, were 99.8% identical to each other. The similarities observed in both nucleotide sequences and read frequencies (see above) implied that these SVs corresponded to multiple copies of the 16S rRNA gene encoded in a single Arsenophonus genome. Although we cannot exclude the possibility that the nucleotide difference was caused by PCR/sequencing errors, the latter is less likely because the DADA2 plugin corrects sequencing errors during the denoising process (Callahan et al., 2016; Prodan et al., 2020).
Sodalis symbionts
SV21, SV24, and SV29, which accounted for 4.3, 2.6, and 1.8%, respectively, of H. unifasciata reads (Supplementary Table S2), formed a clade with the Sodalis endosymbionts found in various insects, including the other psyllid species Blastopsylla occidentalis (Aphalaridae: Spondyliaspidinae; AF077608) (Spaulding and von Dohlen, 1998), Cacopsylla burckhardti (Psyllidae: Psyllinae; TAAB01000016), and Cacopsylla kiushuensis (Psyllidae: Psyllinae; TAAB01000030) (Fig. 3) (Nakabachi et al., 2022b). These SVs were 96.5 (SV29)–97.0% (SV21) identical to the sequence of the type species Sodalis glossinidius (AP008232), a secondary symbiont of the tsetse fly Glossina morsitans (Diptera: Hippoboscoidea) (Dale and Maudlin, 1999). Although the clade was only poorly supported (bootstrap: <50%) (Fig. 3), these SVs were tentatively named “Sodalis endosymbionts” (Fig. 1 and 3) because their similarities to the type species were above the generally used arbitrary genus threshold of 94.5–95% (Yarza et al., 2014; Barco et al., 2020). SV21, SV24, and SV29 were 99.1–99.8% identical to one another.
First report on a Symbiopectobacterium symbiont in Psylloidea
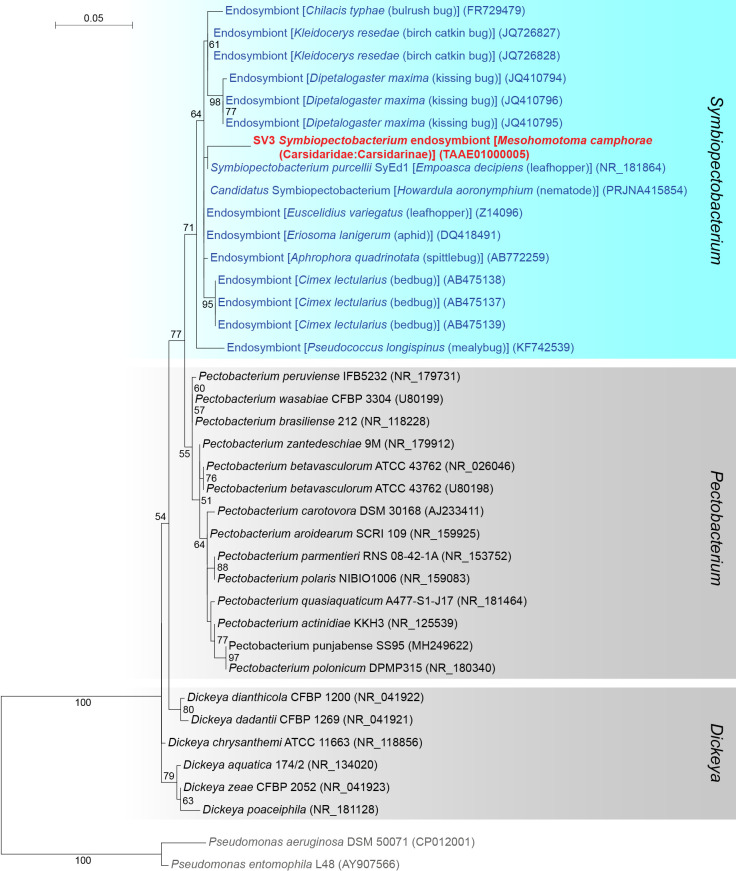
SV3, which accounted for as much as 41.0% of M. camphorae reads (Supplementary Table S2), was 97.4% identical to the sequence of Symbiopectobacterium purcellii, an endosymbiont found in the leafhopper Empoasca decipiens (CP081864) (Nadal-Jimenez et al., 2022b). These sequences formed a robustly supported clade (bootstrap: 99%) in the ML tree of the order Enterobacterales (Fig. 3). Since the genus Symbiopectobacterium is closely related to the genus Pectobacterium, which includes important plant pathogens (Rossmann et al., 2018; Oulghazi et al., 2021), we performed a more comprehensive analysis that focused on Symbiopectobacterium and related bacterial genera. The results obtained placed SV3 in a well-supported clade (bootstrap: 71%) of vertically transmitted Symbiopectobacterium symbionts recently recognized and identified in various invertebrate lineages (Fig. 4) (Hosokawa et al., 2010; Kuechler et al., 2011, 2012; da Mota et al., 2012; Koga et al., 2013; Lopez-Madrigal et al., 2014; Husnik and McCutcheon, 2016; Martinson et al., 2020; Nadal-Jimenez et al., 2022b). This clade is distinct from that of Pectobacterium spp., which are plant pathogens that cause soft rot disease in various economically important crops (Rossmann et al., 2018; Oulghazi et al., 2021) (Fig. 4). Bacteria belonging to this newly emerging Symbiopectobacterium clade have mainly been discovered in hemipteran insects, including plant-sap feeders and vertebrate-blood feeders. However, to the best of our knowledge, this is the first study to report Symbiopectobacterium in Psylloidea.
Fig. 4.
Maximum likelihood phylogram of Symbiopectobacterium and related genera. A total of 427 unambiguously aligned nucleotide sites of 16S rRNA genes were subjected to the analysis. On each branch, bootstrap support values of >50% are shown. The scale bar indicates substitutions per site. Regarding symbiotic bacteria, host organisms are shown in brackets. Symbionts of animals other than psyllids are shown in blue, while symbionts of psyllids are shown in red. The sequence from the present study is shown in bold. DDBJ/EMBL/GenBank accession numbers are provided in parentheses. Pseudomonas aeruginosa and Pseudomonas entomophila were used as an outgroup.
Other Enterobacterales symbionts
SV2 (84.7% of C. limbata reads), SV6 (24.4% of H. unifasciata reads), SV8 (30.7% of H. radiata reads), SV10 (26.2% of T. ornata reads), and SV13 (16.6% of T. ornata reads) were placed at ambiguous positions in the ML tree (Fig. 1 and 3, Supplementary Table S2). Since their branching patterns were mostly poorly supported (bootstrap: <50%) and their sequence identities with those of bacteria with a genus name were low (<94.5%), they were collectively referred to as “Enterobacterales symbionts” (Fig. 1 and 3). Among them, SV6 and SV8, which were derived from congeneric psyllid species, formed a robustly supported clade (bootstrap: 100%) in the ML tree (Fig. 3), suggesting that the corresponding symbionts are sister lineages that share a common ancestor in the common ancestral host. This phylogenetic relationship implies the important and conserved roles of these symbionts in host psyllids. SV31, which accounted for 1.7% of C. japonica reads, was 100% identical to the sequence of Enterobacter spp. (Fig. 1 and 3, Supplementary Table S2). This low relative abundance and 100% identity with free-living bacteria implied that the corresponding bacterium was a transient associate, not a stable symbiont.
Bacteria of Pseudomonadales, Burkholderiales, and Xanthomonadales
Six SVs detected in C. japonica were placed in the clade of the order Pseudomonadales (Fig. 1 and 5, Supplementary Table S2). SV16, SV23, SV27, and SV33, which accounted for 9.2, 3.2, 2.1, and 1.1%, respectively, of C. japonica reads, were resolved as distinct lineages in the robustly supported clade (bootstrap: 99%) of the genus Acinetobacter (Fig. 5). SV22 and SV26, which accounted for 3.7 and 2.5%, respectively, of C. japonica reads, were shown as distinct lineages in the robustly supported clade (bootstrap: 100%) of the genus Pseudomonas (Fig. 5).
Fig. 5.
Maximum likelihood phylogram of Pseudomonadales. A total of 427 unambiguously aligned nucleotide sites of 16S rRNA genes were subjected to the analysis. On each branch, bootstrap support values of >50% are shown. The scale bar indicates substitutions per site. Regarding symbiotic bacteria, host organisms are shown in brackets. Symbionts of animals other than psyllids are shown in blue, while symbionts of psyllids are shown in red. The sequence from the present study is shown in bold. DDBJ/EMBL/GenBank accession numbers are provided in parentheses. Escherichia coli was used as an outgroup.
SV19 and SV25, which accounted for 5.1 and 2.5%, respectively, of C. japonica reads, were inferred as distinct lineages in the robustly supported clade (bootstrap: 100%) within the order Burkholderiales (Fig. 1 and 6, Supplementary Table S2). Although Profftella, a unique organelle-like defensive symbiont (Nakabachi et al., 2013b), also belongs to this order, the ML tree showed that these SVs were distantly related to Profftella. The SVs placed in Pseudomonadales or Burkholderiales were 100% identical to the sequences of free-living species, and their abundance was relatively low. Therefore, these SVs may be derived from transient associates, not stable symbionts of C. japonica.
Fig. 6.
Maximum likelihood phylogram of Burkholderiales. A total of 427 unambiguously aligned nucleotide sites of 16S rRNA genes were subjected to the analysis. On each branch, bootstrap support values of >50% are shown. The scale bar indicates substitutions per site. Regarding symbiotic bacteria, host organisms are shown in brackets. Symbionts of animals other than psyllids are shown in blue, while symbionts of psyllids are shown in red. The sequence from the present study is shown in bold. DDBJ/EMBL/GenBank accession numbers are provided in parentheses. Escherichia coli was used as an outgroup.
SV32, which accounted for 1.4% of H. unifasciata reads, was 100% identical to the sequence of Xanthomonas spp. (Xanthomonadales), including X. campestris (type species), X. oryzae, X. citri, X. arboricola, X. hortorum, and X. vasicola, which are pathogens of important agricultural crops (Timilsina et al., 2020) (Fig. 1 and Supplementary Table S2).
Wolbachia of supergroups B and O
The analysis identified four SVs corresponding to distinct lineages of Wolbachia (Alphaproteobacteria: Rickettsiales) (Fig. 1 and 7, Supplementary Table S2), which are rickettsial bacteria found in various arthropods and nematodes (Werren et al., 2008). Wolbachia lineages are currently classified into supergroups A–Q (Lindsey et al., 2016). Supergroups A and B are the most widespread supergroups in arthropods, and most Wolbachia strains previously found in psyllids belonged to supergroup B (Spaulding and von Dohlen, 2001; Sloan and Moran, 2012; Arp et al., 2014; Jain et al., 2017; Morrow et al., 2017; Chu et al., 2019; Nakabachi et al., 2020a, 2022a, 2022b). In contrast, SV7, which accounted for 26.9% of M. camphorae reads (Supplementary Table S2), was 100% identical to the sequence of Wolbachia belonging to supergroup O. This sequence was recently detected in another psyllid species Trioza cinnamomi (Triozidae), which was the first report of this supergroup in Psylloidea (Nakabachi et al., 2022a). Moreover, the sequence was identical to that of Wolbachia detected in two aphid species, Kaburagia rhusicola (MT554837) and Schlechtendalia chinensis (MT554838) (Ren et al., 2020). The ML analysis placed the sequence within a robustly supported clade (bootstrap: 91%) of Wolbachia supergroup O (Fig. 7). All other Wolbachia strains found in the present study (SV1, SV9, and SV11) belonged to supergroup B (Fig. 7).
Fig. 7.
Maximum likelihood phylogram of Wolbachia. A total of 402 unambiguously aligned nucleotide sites of 16S rRNA genes were subjected to the analysis. On each branch, bootstrap support values of >50% are shown. Host organisms are shown in brackets. Symbionts of animals other than psyllids are shown in blue, while symbionts of psyllids are shown in red. The sequence from this study is shown in bold. DDBJ/EMBL/GenBank accession numbers for sequences are provided in parentheses. Supergroups of Wolbachia are shown in angle brackets. The scale bar represents nucleotide substitutions per position. Liberibacter was used as an outgroup.
Discussion
The present study identified various bacterial populations in six psyllid species of the family Carsidaridae collected in Japan. Although distinct lineages of Carsonella were identified in five psyllid species, essentially no sequence for Carsonella was detected in M. camphorae (Fig. 1 and 2, Supplementary Table S2). The universal primers suggested by Illumina (Illumina, 2013) tend to fail to detect AT-rich 16S rRNA genes of Carsonella (Fromont et al., 2017; Morrow et al., 2017, 2020; Kwak et al., 2021). However, the primer set used in the present study was modified to detect sequences with a wider variety of G+C contents (Supplementary Fig. S1) and successfully identified Carsonella in diverse psyllid lineages (Nakabachi et al., 2020a, 2022a, 2022b). Therefore, the present results may imply the absence of Carsonella at least in the M. camphorae individuals analyzed in the present study. However, the failure of PCR detection may also be attributed to other reasons, including exceptionally diverged nucleotide sequences at primer annealing sites. Further studies, including comprehensive histological analyses of a large set of specimens from several sampling points, are required to establish whether M. camphorae truly lacks Carsonella.
As shown in other psyllid lineages (Thao et al., 2000a; Spaulding and von Dohlen, 2001; Sloan and Moran, 2012; Hall et al., 2016; Morrow et al., 2017; Nakabachi et al., 2020a, 2022a, 2022b; Kwak et al., 2021), the majority of secondary symbionts identified in the present study belong to Gammaproteobacteria, particularly the order Enterobacterales (Fig. 1 and 3, Supplementary Table S2). These include Arsenophonus, Sodalis, and Symbiopectobacterium, as well as several lineages with ambiguous phylogenetic placements. Although Arsenophonus and Sodalis are among the symbionts most frequently observed in arthropod hosts including psyllid lineages (Thao et al., 2000a; Spaulding and von Dohlen, 2001; Hansen et al., 2007; Moran et al., 2008; Sloan and Moran, 2012; Arp et al., 2014; Hall et al., 2016; Morrow et al., 2017; Nakabachi et al., 2022a, 2022b), Symbiopectobacterium is the lineage identified for the first time in Psylloidea. Arsenophonus have been found in diverse insect groups, including wasps (Gherna et al., 1991; Nadal-Jimenez et al., 2023), bees (Hymenoptera: Apoidea) (Nadal-Jimenez et al., 2022a), aphids (Russell et al., 2003; Wulff and White, 2015; Tian et al., 2019; Zhang et al., 2021; Yorimoto et al., 2022), psyllids (Spaulding and von Dohlen, 1998, 2001; Subandiyah et al., 2000; Thao et al., 2000a; Hansen et al., 2007; Hall et al., 2016; Morrow et al., 2017; Nakabachi et al., 2022a, 2022b), whiteflies (Thao and Baumann, 2004; Chiel et al., 2007; El Hamss et al., 2021), triatomine bugs (Hemiptera: Heteroptera) (Hypsa and Dale, 1997), lice (Psocodea: Anoplura) (Sasaki-Fukatsu et al., 2006; Allen et al., 2007; Kirkness et al., 2010), louse flies (Dale et al., 2006; Nováková et al., 2009, 2016), and ticks (Arachnida: Ixodida) (Grindle et al., 2003). The types of symbiotic relationships with hosts exhibit a wide diversity from parasitism (e.g. killing male progeny to drive the spread of maternally inherited symbionts in the host population [Gherna et al., 1991]) through facultative mutualism (e.g. increasing the host population growth rate possibly through nutrient supplementation [Wulff and White, 2015; Tian et al., 2019]) to bacteriome-associated obligate mutualism in order to provide essential nutrients to the host (e.g. essential amino acids and B vitamins for sap feeders and blood feeders, respectively) (Dale et al., 2006; Sasaki-Fukatsu et al., 2006; Allen et al., 2007; Nováková et al., 2009, 2016; Kirkness et al., 2010; Yorimoto et al., 2022). Phylogenetic analyses by Nováková et al. revealed two contrasting evolutionary patterns in Arsenophonus lineages: random associations with distantly related hosts in diverse insect taxa and host-symbiont co-cladogenesis in the lineages of lice and louse flies (Nováková et al., 2009). These findings imply at least two transitions from facultative symbionts capable of infecting new host lineages to obligate mutualists that have lost this ability. Sodalis symbionts have also been found in various insect taxa, including tsetse flies (Dale and Maudlin, 1999), weevils (Coleoptera: Curculionoidea) (Toju et al., 2013; Oakeson et al., 2014), psyllids (Arp et al., 2014; Hall et al., 2016; Morrow et al., 2017; Nakabachi et al., 2022b), scale insects (Dhami et al., 2013; Husnik and McCutcheon, 2016), spittlebugs (Hemiptera: Auchenorrhyncha: Cercopoidea) (Koga et al., 2013; Koga and Moran, 2014), stinkbugs (Hemiptera: Heteroptera) (Kaiwa et al., 2010), lice (Fukatsu et al., 2007), and louse flies (Nováková and Hypša, 2007). Although the symbiotic types of Sodalis vary, their main role appears to be the provision of nutrients in many insect hosts (McCutcheon et al., 2019). Sodalis symbionts were hypothesized to have replaced more ancient antecedent symbionts in spittlebugs (Koga et al., 2013; Koga and Moran, 2014) and weevils (Toju et al., 2013; Oakeson et al., 2014). Physiological and genomic analyses have suggested they provide the hosts with nutrients formerly supplied by the antecedents (Koga and Moran, 2014; Oakeson et al., 2014). Since similar symbiont replacements by Sodalis have been suggested in psyllid lineages (Nakabachi et al., 2022b), their functional roles in Psylloidea are of interest. The clade of Symbiopectobacterium was relatively recently recognized in comparison to Arsenophonus and Sodalis (Martinson et al., 2020). Bacteria belonging to this clade have mainly been found in hemipteran insects. These encompass not only plant-feeding taxa, including aphids, mealybugs (Sternorrhyncha: Coccoidea) (Lopez-Madrigal et al., 2014; Husnik and McCutcheon, 2016), leafhoppers (Auchenorrhyncha: Membracoidea) (Nadal-Jimenez et al., 2022b), spittlebugs (Koga et al., 2013), and seed bugs (Heteroptera) (Kuechler et al., 2011, 2012), but also blood-feeding taxa, including kissing bugs (da Mota et al., 2012) and bedbugs (both Heteroptera) (Hosokawa et al., 2010). A recent analysis discovered Symbiopectobacterium in the nematode Howardula aoronymphium (Secernentea: Tylenchida) that parasitizes Drosophila flies (Diptera: Drosophilidae) (Martinson et al., 2020). Symbiopectobacterium symbionts are vertically transmitted through host generations, and lineages in mealybugs, seed bugs, and nematodes were demonstrated to be obligate mutualists (Kuechler et al., 2011, 2012; Husnik and McCutcheon, 2016; Martinson et al., 2020). All of the clades of Arsenophonus (Zreik et al., 1998; Bressan, 2014), Sodalis (Chari et al., 2015; Tláskal et al., 2021), and Symbiopectobacterium (Leite et al., 2023) include lineages that have close associations with plants, implying their evolutionary history of horizontal transfer among insects, at least partly through plants. Since the physiological and ecological roles of Arsenophonus, Sodalis, and Symbiopectobacterium symbionts identified in Psylloidea remain unknown, future studies need to focus on clarifying their localization within the insect body (e.g. whether they are harbored in the bacteriome) and their genomic structures to obtain an understanding of their metabolic potential.
Regarding Wolbachia (Rickettsiales), the present study identified three lineages belonging to supergroup B, the major group in insect lineages, in four psyllid species. Moreover, supergroup O, another Wolbachia lineage, which is a relatively minor taxon and was recently found for the first time in Psylloidea (Nakabachi et al., 2022a), was identified in M. camphorae. This implies that supergroup O is widespread in Psylloidea, although it has only recently been recognized. Wolbachia is among the most widely distributed symbionts worldwide, estimated to infect 40% of terrestrial arthropods (Werren et al., 2008; Brinker et al., 2019). Although limited lineages are recognized to be obligate mutualists (Hosokawa et al., 2010), they are mostly parasites in insects, in which they manipulate host reproduction to drive their dissemination (Werren et al., 2008; Brinker et al., 2019). They induce resistance to various microbes, including viruses and protozoans, in some insect taxa (Brinker et al., 2019; Wang et al., 2021). Moreover, a genome editing technique using the CRISPR-Cas9 system to transform Wolbachia has recently been developed (Pelz-Stelinski, 13 May 2021, United States Patent and Trademark Office). Therefore, they are regarded as promising agents in the control of pest insects and the microbes that they transmit (Brinker et al., 2019; Pelz-Stelinski, 13 May 2021, United States Patent and Trademark Office; Wang et al., 2021). A high incidence of Wolbachia in pest psyllids worldwide (Spaulding and von Dohlen, 2001; Sloan and Moran, 2012; Arp et al., 2014; Chu et al., 2016, 2019; Morrow et al., 2017; Nakabachi et al., 2020a, 2022b) and the interactions observed between Wolbachia and other symbionts (Chu et al., 2016, 2019; Jain et al., 2017; Kruse et al., 2017; Killiny, 2022) have led researchers to anticipate using Wolbachia to manipulate pest psyllids (e.g. providing them with resistance to plant pathogens) and/or plant pathogens (e.g. preventing them from infecting plants) (Chu et al., 2016, 2019; Kruse et al., 2017; Pelz-Stelinski, 13 May 2021, United States Patent and Trademark Office). The present study suggested the pervasive horizontal transmission of various Wolbachia strains among various insects, including psyllids (Fig. 7). This implies that Wolbachia may be artificially infected into psyllids, forming the basis for the use of this bacterial group for application purposes.
Furthermore, the present study detected potential plant pathogens, including Xanthomonas sp., in H. unifasciata. Since H. unifasciata feeds on Ficus spp. (Moraceae), further studies are warranted to establish whether the host plants are infected with Xanthomonas and if the infection causes disease symptoms.
Conclusions
The present study identified various bacterial symbionts in six psyllid species of the family Carsidaridae. The majority of the secondary symbionts were gammaproteobacteria, particularly those of the order Enterobacterales, including Arsenophonus and Sodalis. In addition, Symbiopectobacterium, another lineage belonging to Enterobacterales, was detected for the first time in Psylloidea. Regarding non-Enterobacterales gammaproteobacteria, Acinetobacter, Pseudomonas (both Pseudomonadales), Delftia, Comamonas (both Burkholderiales), and Xanthomonas (Xanthomonadales), a putative plant pathogen, were identified. Regarding alphaproteobacteria, three Wolbachia (Rickettsiales) lineages belonging to supergroup B, the major group in insect lineages, were detected in four psyllid species. Moreover, a Wolbachia lineage of supergroup O, a minor group that was recently found for the first time in Psylloidea, was detected in M. camphorae, suggesting that this supergroup is widespread in Psylloidea. These results provide deeper insights into the interactions among insects, bacteria, and plants, which will help establish a basis for the better control of pest species.
Citation
Maruyama, J., Inoue, H., Hirose, Y., and Nakabachi, A. (2023) 16S rRNA Gene Sequencing of Six Psyllid Species of the Family Carsidaridae Identified Various Bacteria Including Symbiopectobacterium. Microbes Environ 38: ME23045.
https://doi.org/10.1264/jsme2.ME23045
Supplementary Material
Acknowledgements
We thank Dr. Vincent G. Martinson (University of New Mexico) for his support in the phylogenetic analysis of Symbiopectobacterium. This work was supported by the Japan Society for the Promotion of Science (https://www.jsps.go.jp) KAKENHI (grant numbers 26292174 and 20H02998 to AN, and 25850035 to HI), and research grants from Tatematsu Foundation and Nagase Science and Technology Foundation to AN. The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation. The DNA sequencing facility was supported by the Department of Applied Chemistry and Life Science and the University-Community Partnership Promotion Center at the Toyohashi University of Technology.
References
- Allen, J.M., Reed, D.L., Perotti, M.A., Braig, H.R., Al, A.E.T., and Icrobiol, A.P.P.L.E.N.M. (2007) Evolutionary relationships of “Candidatus Riesia spp.,” endosymbiotic enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol 73: 1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2023) FastQC: A quality control tool for high throughput sequence data. URL https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Arp, A., Munyaneza, J.E., Crosslin, J.M., Trumble, J., and Bextine, B. (2014) A global comparison of Bactericera cockerelli (Hemiptera: Triozidae) microbial communities. Environ Entomol 43: 344–352. [DOI] [PubMed] [Google Scholar]
- Barco, R.A., Garrity, G.M., Scott, J.J., Amend, J.P., Nealson, K.H., and Emerson, D. (2020) A genus definition for Bacteria and Archaea based on a standard genome relatedness index. mBio 11: e02475-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N.A., Kaehler, B.D., Rideout, J., Dillon, M., Bolyen, E., Knight, R., et al. (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E., Rideout, J.R., Dillon, M.R., Bokulich, N., Abnet, C., Al-Ghalith, G.A., et al. (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, A. (2014) Emergence and evolution of Arsenophonus bacteria as insect-vectored plant pathogens. Infect Genet Evol 22: 81–90. [DOI] [PubMed] [Google Scholar]
- Brinker, P., Fontaine, M.C., Beukeboom, L.W., and Salles, J.F. (2019) Host, symbionts, and the microbiome: the missing tripartite interaction. Trends Microbiol 27: 480–488. [DOI] [PubMed] [Google Scholar]
- Buchner, P. (1965) Endosymbiosis of Animals with Plant Microorganisms. New York, NY: Interscience. [Google Scholar]
- Burckhardt, D., Ouvrard, D., and Percy, D.M. (2021) An updated classification of the jumping plant-lice (Hemiptera: Psylloidea) integrating molecular and morphological evidence. Eur J Taxon 736: 137–182. [Google Scholar]
- Callahan, B.J., McMurdie, P.J., Rosen, M.J., Han, A.W., Johnson, A.J.A., and Holmes, S.P. (2016) DADA2 : High-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., and Madden, T.L. (2009) BLAST+: architecture and applications. BMC Bioinf 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari, A., Oakeson, K.F., Enomoto, S., Grant Jackson, D., Fisher, M.A., and Dale, C. (2015) Phenotypic characterization of Sodalis praecaptivus sp. nov., a close non-insect-associated member of the Sodalis-allied lineage of insect endosymbionts. Int J Syst Evol Microbiol 65: 1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiel, E., Gottlieb, Y., Zchori-Fein, E., Mozes-Daube, N., Katzir, N., Inbar, M., and Ghanim, M. (2007) Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull Entomol Res 97: 407–413. [DOI] [PubMed] [Google Scholar]
- Cho, G., Malenovský, Igor, and Lee, S. (2019) Higher-level molecular phylogeny of jumping plant lice (Hemiptera: Sternorrhyncha: Psylloidea). Syst Entomol 44: 638–651. [Google Scholar]
- Chu, C., Hoffmann, M., Braswell, W.E., and Pelz-Stelinski, K.S. (2019) Genetic variation and potential coinfection of Wolbachia among widespread Asian citrus psyllid (Diaphorina citri Kuwayama) populations. Insect Sci 26: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, C.C., Gill, T.A., Hoffmann, M., and Pelz-Stelinski, K.S. (2016) Inter-population variability of endosymbiont densities in the Asian citrus psyllid (Diaphorina citri Kuwayama). Microb Ecol 71: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Mota, F.F., Marinho, L.P., de Moreira, C.J.C., Lima, M.M., Mello, C.B., Garcia, E.S., et al. (2012) Cultivation-independent methods reveal differences among bacterial gut microbiota in triatomine vectors of Chagas disease. PLoS Neglected Trop Dis 6: e1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, C., and Maudlin, I. (1999) Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol 49: 267–275. [DOI] [PubMed] [Google Scholar]
- Dale, C., Beeton, M., Harbison, C., Jones, T., and Pontes, M. (2006) Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl Environ Microbiol 72: 2997–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan, H., Ikeda, N., Fujikami, M., and Nakabachi, A. (2017) Behavior of bacteriome symbionts during transovarial transmission and development of the Asian citrus psyllid. PLoS One 12: e0189779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhami, M.K., Buckley, T.R., Beggs, J.R., and Taylor, M.W. (2013) Primary symbiont of the ancient scale insect family Coelostomidiidae exhibits strict cophylogenetic patterns. Symbiosis 61: 77–91. [Google Scholar]
- El Hamss, H., Ghosh, S., Maruthi, M.N., Delatte, H., and Colvin, J. (2021) Microbiome diversity and reproductive incompatibility induced by the prevalent endosymbiont Arsenophonus in two species of African cassava Bemisia tabaci whiteflies. Ecol Evol 11: 18032–18041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016) MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont, C., Riegler, M., and Cook, J.M. (2017) Relative abundance and strain diversity in the bacterial endosymbiont community of a sap-feeding insect across its native and introduced geographic range. Microb Ecol 74: 722–734. [DOI] [PubMed] [Google Scholar]
- Fukatsu, T., and Nikoh, N. (1998) Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (insecta Homoptera). Appl Environ Microbiol 64: 3599–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu, T., and Nikoh, N. (2000) Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera). Appl Environ Microbiol 66: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu, T., Koga, R., Smith, W.A., Tanaka, K., Nikoh, N., Sasaki-Fukatsu, K., et al. (2007) Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl Environ Microbiol 73: 6660–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo, N.M., Altincicek, B., Anselme, C., Atamian, H., Barribeau, S.M., de Vos, M., et al. (2010) Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol 11: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherna, R.L., Werren, J.H., Weisburg, W., Cote, R., Woese, C.R., Mandelco, L., and Brenner, D.J. (1991) Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int J Syst Bacteriol 41: 563–565. [Google Scholar]
- Glöckner, F.O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., et al. (2017) 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol 261: 169–176. [DOI] [PubMed] [Google Scholar]
- Grafton-Cardwell, E.E., Stelinski, L.L., and Stansly, P.A. (2013) Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu Rev Entomol 58: 413–432. [DOI] [PubMed] [Google Scholar]
- Grindle, N., Tyner, J.J., Clay, K., and Fuqua, C. (2003) Identification of Arsenophonus-type bacteria from the dog tick Dermacentor variabilis. J Invertebr Pathol 83: 264–266. [DOI] [PubMed] [Google Scholar]
- Hall, A.A.G., Morrow, J.L., Fromont, C., Steinbauer, M.J., Taylor, G.S., Johnson, S.N., et al. (2016) Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts. Environ Microbiol 18: 2591–2603. [DOI] [PubMed] [Google Scholar]
- Hansen, A.K., Jeong, G., Paine, T.D., and Stouthamer, R. (2007) Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl Environ Microbiol 73: 7531–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodkinson, I.D. (1974) The biology of the Psylloidea (Homoptera): a review. Bull Entomol Res 64: 325–338. [Google Scholar]
- Hosokawa, T., Koga, R., Kikuchi, Y., Meng, X.-Y., and Fukatsu, T. (2010) Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A 107: 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik, F., and McCutcheon, J.P. (2016) Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A 113: E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hypsa, V., and Dale, C. (1997) In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum,” an intracellular bacterium from the triatomine bug, Triatoma infestans. Int J Syst Bacteriol 47: 1140–1144. [DOI] [PubMed] [Google Scholar]
- Illumina (2013) 16S metagenomic sequencing library preparation Part#15044223 Rev.B. URL http://jp.support.illumina.com/content/dam/illumin
- International Aphid Genomics Consortium (2010) Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8: e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, M., Fleites, L.A., and Gabriel, D.W. (2017) A small Wolbachia protein directly represses phage lytic cycle genes in “Candidatus Liberibacter asiaticus” within psyllids. mSphere 2: e00171-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarausch, B., and Jarausch, W. (2010) Psyllid vectors and their control. In Phytoplasmas: Genomes, Plant Hosts and Vectors. Weintraub, P.G., and Jones, P. (eds). Wallingford, Oxfordshire: CAB International, pp. 250–271. [Google Scholar]
- Johnson, K.N. (2015) The impact of Wolbachia on virus infection in mosquitoes. Viruses 7: 5705–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiwa, N., Hosokawa, T., Kikuchi, Y., Nikoh, N., Meng, X.Y., Kimura, N., et al. (2010) Primary gut symbiont and secondary, Sodalis-allied symbiont of the Scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol 76: 3486–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karstens, L., Asquith, M., Davin, S., Fair, D., Gregory, W.T., Wolfe, A.J., et al. (2019) Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems 4: e00290-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketsa, S., Wisutiamonkul, A., Palapol, Y., and Paull, R.E. (2020) The durian: botany, horticulture, and utilization. In Horticultural Reviews. Hoboken, NJ: John Wiley & Sons, pp. 125–211. [Google Scholar]
- Kikuchi, Y. (2009) Endosymbiotic bacteria in insects: Their diversity and culturability. Microbes Environ 24: 195–204. [DOI] [PubMed] [Google Scholar]
- Killiny, N. (2022) Made for each other: Vector-pathogen interfaces in the Huanglongbing pathosystem. Phytopathology 112: 26–43. [DOI] [PubMed] [Google Scholar]
- Kirkness, E.F., Haas, B.J., Sun, W., Braig, H.R., Perotti, M.A., Clark, J.M., et al. (2010) Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A 107: 12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, R., Bennett, G.M., Cryan, J.R., and Moran, N.A. (2013) Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15: 2073–2081. [DOI] [PubMed] [Google Scholar]
- Koga, R., and Moran, N.A. (2014) Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 8: 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse, A., Fattah-Hosseini, S., Saha, S., Johnson, R., Warwick, E., Sturgeon, K., et al. (2017) Combining ’omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS One 12: e0179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler, S.M., Dettner, K., and Kehl, S. (2011) Characterization of an obligate intracellular bacterium in the midgut epithelium of the bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae). Appl Environ Microbiol 77: 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler, S.M., Renz, P., Dettner, K., and Kehl, S. (2012) Diversity of symbiotic organs and bacterial endosymbionts of lygaeoid bugs of the families Blissidae and Lygaeidae (Hemiptera: Heteroptera: Lygaeoidea). Appl Environ Microbiol 78: 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, Y., Sun, P., Meduri, V.R.S., Percy, D.M., Mauck, K.E., and Hansen, A.K. (2021) Uncovering symbionts across the psyllid tree of life and the discovery of a new Liberibacter species, “Candidatus” Liberibacter capsica. Front Microbiol 12: 739763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, L.N., Visnovsky, S.B., Wright, P.J., and Pitman, A.R. (2023) Draft genome sequences of three ‘Candidatus Symbiopectobacterium’ isolates collected from potato tubers grown in New Zealand. Microbiol Resour Announc 12: e0114822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey, A.R.I., Bordenstein, S.R., Newton, I.L.G., and Rasgon, J.L. (2016) Wolbachia pipientis should not be split into multiple species: A response to Ramírez-Puebla et al., Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola.’ Syst Appl Microbiol 39: 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Madrigal, S., Beltra, A., Resurreccion, S., Soto, A., Latorre, A., Moya, A., et al. (2014) Molecular evidence for ongoing complementarity and horizontal gene transfer in endosymbiotic systems of mealybugs. Front Microbiol 5: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. [Google Scholar]
- Martinson, V.G., Gawryluk, R.M.R., Gowen, B.E., Curtis, C.I., Jaenike, J., and Perlman, S.J. (2020) Multiple origins of obligate nematode and insect symbionts by a clade of bacteria closely related to plant pathogens. Proc Natl Acad Sci U S A 117: 31979–31986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon, J.P., Boyd, B.M., and Dale, C. (2019) The life of an insect endosymbiont from the cradle to the grave. Curr Biol 29: R485–R495. [DOI] [PubMed] [Google Scholar]
- Mora, V., Ramasamy, M., Damaj, M.B., Irigoyen, S., Ancona, V., Ibanez, F., et al. (2021) Potato zebra chip: An overview of the disease, control strategies, and prospects. Front Microbiol 12: 700663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N.A., McCutcheon, J.P., and Nakabachi, A. (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42: 165–190. [DOI] [PubMed] [Google Scholar]
- Morrow, J.L., Hall, A.A.G., and Riegler, M. (2017) Symbionts in waiting : the dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 5: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, J.L., Om, N., Beattie, G.A.C., Chambers, G.A., Donovan, N.J., Liefting, L.W., et al. (2020) Characterization of the bacterial communities of psyllids associated with Rutaceae in Bhutan by high throughput sequencing. BMC Microbiol 20: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Jimenez, P., Siozios, S., Frost, C.L., Court, R., Chrostek, E., Drew, G.C., et al. (2022a) Arsenophonus apicola sp. nov., isolated from the honeybee Apis mellifera. Int J Syst Evol Microbiol 72: 005469. [DOI] [PubMed] [Google Scholar]
- Nadal-Jimenez, P., Siozios, S., Halliday, N., Cámara, M., and Hurst, G.D.D. (2022b) Symbiopectobacterium purcellii, gen. nov., sp. nov., isolated from the leafhopper Empoasca decipiens. Int J Syst Evol Microbiol 72: 005440. [DOI] [PubMed] [Google Scholar]
- Nadal-Jimenez, P., Parratt, S.R., Siozios, S., and Hurst, G.D.D. (2023) Isolation, culture and characterization of Arsenophonus symbionts from two insect species reveal loss of infectious transmission and extended host range. Front Microbiol 14: 1189143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., and Ishikawa, H. (1997) Differential display of mRNAs related to amino acid metabolism in the endosymbiotic system of aphids. Insect Biochem Mol Biol 27: 1057–1062. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., and Ishikawa, H. (1999) Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol 45: 1–6. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., and Ishikawa, H. (2000) Polyamine composition and expression of genes related to polyamine biosynthesis in an aphid endosymbiont, Buchnera. Appl Environ Microbiol 66: 3305–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., and Ishikawa, H. (2001) Expression of host S-adenosylmethionine decarboxylase gene and polyamine composition in aphid bacteriocytes. Insect Biochem Mol Biol 31: 491–496. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Ishikawa, H., and Kudo, T. (2003) Extraordinary proliferation of microorganisms in aposymbiotic pea aphids, Acyrthosiphon pisum. J Invertebr Pathol 82: 152–161. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Shigenobu, S., Sakazume, N., Shiraki, T., Hayashizaki, Y., Carninci, P., et al. (2005) Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci U S A 102: 5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., Yamashita, A., Toh, H., Ishikawa, H., Dunbar, H.E., Moran, N.A., and Hattori, M. (2006) The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314: 267. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., and Miyagishima, S. (2010) Expansion of genes encoding a novel type of dynamin in the genome of the pea aphid, Acyrthosiphon pisum. Insect Mol Biol 19: 165–173. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Shigenobu, S., and Miyagishima, S. (2010a) Chitinase-like proteins encoded in the genome of the pea aphid, Acyrthosiphon pisum. Insect Mol Biol 19: 175–185. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Koshikawa, S., Miura, T., and Miyagishima, S. (2010b) Genome size of Pachypsylla venusta (Hemiptera: Psyllidae) and the ploidy of its bacteriocyte, the symbiotic host cell that harbors intracellular mutualistic bacteria with the smallest cellular genome. Bull Entomol Res 100: 27–33. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Nikoh, N., Oshima, K., Inoue, H., Ohkuma, M., Hongoh, Y., et al. (2013a) Horizontal gene acquisition of Liberibacter plant pathogens from a bacteriome-confined endosymbiont of their psyllid vector. PLoS One 8: e82612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., Ueoka, R., Oshima, K., Teta, R., Mangoni, A., Gurgui, M., et al. (2013b) Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23: 1478–1484. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Ishida, K., Hongoh, Y., Ohkuma, M., and Miyagishima, S. (2014) Aphid gene of bacterial origin encodes protein transported to obligate endosymbiont. Curr Biol 24: R640-641. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A. (2015) Horizontal gene transfers in insects. Curr Opin Insect Sci 7: 24–29. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., and Fujikami, M. (2019) Concentration and distribution of diaphorin, and expression of diaphorin synthesis genes during Asian citrus psyllid development. J Insect Physiol 118: 103931. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., and Okamura, K. (2019) Diaphorin, a polyketide produced by a bacterial symbiont of the Asian citrus psyllid, kills various human cancer cells. PLoS One 14: e0218190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., Malenovský, I., Gjonov, I., and Hirose, Y. (2020a) 16S rRNA sequencing detected Profftella, Liberibacter, Wolbachia, and Diplorickettsia from relatives of the Asian citrus psyllid. Microb Ecol 80: 410–422. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A., Piel, J., Malenovský, I., and Hirose, Y. (2020b) Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: Providing toxins, vitamins, and carotenoids. Genome Biol Evol 12: 1975–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., and Moran, N.A. (2022) Extreme polyploidy of Carsonella, an organelle-like bacterium with a drastically reduced genome. Microbiol Spectrum 10: e0035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., Inoue, H., and Hirose, Y. (2022a) High-resolution microbiome analyses of nine psyllid species of the family Triozidae identified previously unrecognized but major bacterial populations, including Liberibacter and Wolbachia of supergroup O. Microbes Environ 37: ME22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A., Inoue, H., and Hirose, Y. (2022b) Microbiome analyses of 12 psyllid species of the family Psyllidae identified various bacteria including Fukatsuia and Serratia symbiotica, known as secondary symbionts of aphids. BMC Microbiol 22: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh, N., and Nakabachi, A. (2009) Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh, N., McCutcheon, J.P., Kudo, T., Miyagishima, S., Moran, N.A., and Nakabachi, A. (2010) Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet 6: e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková, E., and Hypša, V. (2007) A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett 269: 131–135. [DOI] [PubMed] [Google Scholar]
- Nováková, E., Hypša, V., and Moran, N.A. (2009) Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol 9: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková, E., Hypša, V., Nguyen, P., Husník, F., and Darby, A.C. (2016) Genome sequence of Candidatus Arsenophonus lipopteni, the exclusive symbiont of a blood sucking fly Lipoptena cervi (Diptera: Hippoboscidae). Stand Genomic Sci 11: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeson, K.F., Gil, R., Clayton, A.L., Dunn, D.M., von Niederhausern, A.C., Hamil, C., et al. (2014) Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol 6: 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M., Degnan, P.H., Burke, G.R., and Moran, N.A. (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55: 247–266. [DOI] [PubMed] [Google Scholar]
- Oulghazi, S., Sarfraz, S., Zaczek-Moczydłowska, M.A., Khayi, S., Ed-Dra, A., Lekbach, Y., et al. (2021) Pectobacterium brasiliense: Genomics, host range and disease management. Microorganisms 9: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouvrard, D. (2023) Psyl’list – The World Psylloidea Database. URL https://data.nhm.ac.uk/dataset/psyl-list/resource/8746ceec-4846-4899-b607-9ba603002033
- Percy, D.M., Crampton-Platt, A., Sveinsson, S., Lemmon, A.R., Lemmon, E.M., Ouvrard, D., and Burckhardt, D. (2018) Resolving the psyllid tree of life: phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Syst Entomol 43: 762–776. [Google Scholar]
- Prodan, A., Tremaroli, V., Brolin, H., Zwinderman, A.H., Nieuwdorp, M., and Levin, E. (2020) Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS One 15: e0227434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profft, J. (1937) Beiträge zur Symbiose der Aphiden und Psylliden. Z Morphol Okol Tiere 32: 289–326. [Google Scholar]
- Pruesse, E., Peplies, J., and Glöckner, F.O. (2012) SINA : Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, J.S., MacDonald, S.J., Jander, G., Nakabachi, A., Thomas, G.H., and Douglas, A.E. (2010) Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol 19: 241–248. [DOI] [PubMed] [Google Scholar]
- Ren, W., Wei, H., Yang, Y., Shao, S., Wu, H., Chen, X., and Yang, Z. (2020) Molecular detection and phylogenetic analyses of Wolbachia in natural populations of nine galling aphid species. Sci Rep 10: 12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann, S., Dees, M.W., Perminow, J., Meadow, R., and Brurberg, M.B. (2018) Soft rot Enterobacteriaceae are carried by a large range of insect species in potato fields. Appl Environ Microbiol 84: e00281-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J.A., Latorre, A., Sabater-Muñoz, B., Moya, A., Moran, N.A., Sabater-Munoz, B., et al. (2003) Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Sandström, J., and Moran, N. (1999) How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl 91: 203–210. [Google Scholar]
- Sasaki-Fukatsu, K., Koga, R., Nikoh, N., Yoshizawa, K., Kasai, S., Mihara, M., et al. (2006) Symbiotic bacteria associated with stomach discs of human lice. Appl Environ Microbiol 72: 7349–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu, S., Richards, S., Cree, A.G., Morioka, M., Fukatsu, T., Kudo, T., et al. (2010) A full-length cDNA resource for the pea aphid, Acyrthosiphon pisum. Insect Mol Biol 19: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, D.B., and Moran, N.A. (2012) Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol 29: 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, D.B., Nakabachi, A., Richards, S., Qu, J., Murali, S.C., Gibbs, R.A., and Moran, N.A. (2014) Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol 31: 857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding, A.W., and von Dohlen, C.D. (1998) Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha). Mol Biol Evol 15: 1506–1513. [DOI] [PubMed] [Google Scholar]
- Spaulding, A.W., and von Dohlen, C.D. (2001) Psyllid endosymbionts exhibit patterns of co-speciation with hosts and destabilizing substitutions in ribosomal RNA. Insect Mol Biol 10: 57–67. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subandiyah, S., Nikoh, N., Tsuyumu, S., Somowiyarjo, S., and Fukatsu, T. (2000) Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera : Psylloidea). Zool Sci 17: 983–989. [Google Scholar]
- Tamborindeguy, C., Monsion, B., Brault, V., Hunnicutt, L., Ju, H.J., Nakabachi, A., and Van Fleet, E. (2010) A genomic analysis of transcytosis in the pea aphid, Acyrthosiphon pisum, a mechanism involved in virus transmission. Insect Mol Biol 19: 259–272. [DOI] [PubMed] [Google Scholar]
- Tanabe, N., Takasu, R., Hirose, Y., Kamei, Y., Kondo, M., and Nakabachi, A. (2022) Diaphorin, a polyketide produced by a bacterial symbiont of the Asian citrus psyllid, inhibits the growth and cell division of Bacillus subtilis but promotes the growth and metabolic activity of Escherichia coli. Microbiol Spectrum 10: e0175722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasi, A., Halbert, S., and Morawo, T. (2021) Fig psyllid Homotoma ficus (L.) (Insecta: Hemiptera: Homotomidae); curtain fig psyllid Macrohomotoma gladiata Kuwayama (Insecta: Hemiptera: Homotomidae); and ficus leaf-rolling psyllid Trioza brevigenae Mathur (Insecta: Hemiptera: Triozidae). IFAS Ext doi: https://doi.org/10.32473/edis-in1329-2021
- Thao, M.L., Clark, M.A., Baumann, L., Brennan, E.B., Moran, N.A., and Baumann, P. (2000a) Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol 41: 300–304. [DOI] [PubMed] [Google Scholar]
- Thao, M.L., Moran, N.A., Abbot, P., Brennan, E.B., Burckhardt, D.H., and Baumann, P. (2000. b) Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol 66: 2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao, M.L.L., and Baumann, P. (2004) Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae). Curr Microbiol 48: 140–144. [DOI] [PubMed] [Google Scholar]
- Tian, P.-P., Chang, C.-Y., Miao, N.-H., Li, M.-Y., and Liu, X.-D. (2019) Infections with Arsenophonus facultative endosymbionts alter performance of aphids (Aphis gossypii) on an amino-acid- deficient diet. Appl Environ Microbiol 85: e01407-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timilsina, S., Potnis, N., Newberry, E.A., Liyanapathiranage, P., Iruegas-Bocardo, F., White, F.F., et al. (2020) Xanthomonas diversity, virulence and plant-pathogen interactions. Nat Rev Microbiol 18: 415–427. [DOI] [PubMed] [Google Scholar]
- Tláskal, V., Pylro, V.S., Žifčáková, L., and Baldrian, P. (2021) Ecological divergence within the enterobacterial genus Sodalis: From insect symbionts to inhabitants of decomposing deadwood. Front Microbiol 12: 668644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju, H., Tanabe, A.S., Notsu, Y., Sota, T., and Fukatsu, T. (2013) Diversification of endosymbiosis: replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J 7: 1378–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchi, N., Fukudome, M., Nozaki, N., Suzuki, M., Osuki, K.I., Shigenobu, S., and Uchiumi, T. (2019) Antimicrobial activities of cysteine-rich peptides specific to bacteriocytes of the pea aphid Acyrthosiphon pisum. Microbes Environ 34: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.H., Gamez, S., Raban, R.R., Marshall, J.M., Alphey, L., Li, M., et al. (2021) Combating mosquito-borne diseases using genetic control technologies. Nat Commun 12: 4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J.H., Baldo, L., and Clark, M.E. (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741–751. [DOI] [PubMed] [Google Scholar]
- Wulff, J.A., and White, J.A. (2015) The endosymbiont Arsenophonus provides a general benefit to soybean aphid (Hemiptera: Aphididae) regardless of host plant resistance (Rag). Environ Entomol 44: 574–581. [DOI] [PubMed] [Google Scholar]
- Yamada, T., Hamada, M., Floreancig, P., and Nakabachi, A. (2019) Diaphorin, a polyketide synthesized by an intracellular symbiont of the Asian citrus psyllid, is potentially harmful for biological control agents. PLoS One 14: e0216319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yana, W., Ndankeu Mveyo, Y.P., Dzokou, V.J., and Tamesse, J.L. (2015) Jumping plant lice of the family Carsidaridae (Hemiptera : Psylloidea) from Cameroon: taxonomic, faunisitic, phenology and host plants. J Biodivers Environ Sci 6: 1–20. [Google Scholar]
- Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.-H., et al. (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12: 635–645. [DOI] [PubMed] [Google Scholar]
- Yorimoto, S., Hattori, M., Kondo, M., and Shigenobu, S. (2022) Complex host/symbiont integration of a multi-partner symbiotic system in the eusocial aphid Ceratovacuna japonica. iScience 25: 105478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Su, H., Jiang, W., Hu, D., Ali, I., Jin, T., et al. (2021) Symbiotic microbial studies in diverse populations of Aphis gossypii, existing on altered host plants in different localities during different times. Ecol Evol 11: 13948–13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, H., Pirson, A., and Zimmermann, M.H. (1975) Nature of transported substances. In Transport in Plants I. Zimmermann, M.H., and Milburn, J.A. (eds). New York, NY: Springer-Verlag, pp. 59–100. [Google Scholar]
- Zreik, L., Bové, J.M., and Garnier, M. (1998) Phylogenetic characterization of the bacterium-like organism associated with marginal chlorosis of strawberry and proposition of a Candidatus taxon for the organism, “Candidatus Phlomobacter fragariae.” Int J Syst Bacteriol 48: 257–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequence data are available in the DDBJ/EMBL/GenBank databases under the accession numbers DRR420941–DRR420946 (raw fastq files) and TAAE01000001–TAAE01000035 (dereplicated SVs).