Abstract
Prostate cancer treatment decisions are heavily reliant upon subjective visual interpretation (assigning Gleason patterns, or International Society of Urological Pathology (ISUP) grade groups) of limited numbers of 2D histology sections. Under this paradigm, inter-observer variance is high, with ISUP grades not correlating well with outcome for individual patients, and which contributes to the over- and under-treatment of patients. Recent studies have demonstrated improved prognostication of prostate cancer outcomes based on computational analyses of glands and nuclei within 2D whole slide images. Our group has also shown that the computational analysis of 3D glandular features, extracted from 3D pathology datasets of whole intact biopsies, can allow for improved recurrence prediction compared to corresponding 2D features. Here we seek to expand on these prior studies by exploring the prognostic value of 3D shape-based nuclear features in prostate cancer (e.g., nuclear size, sphericity). 3D pathology datasets were generated using open-top light-sheet (OTLS) microscopy of 102 cancer-containing biopsies extracted ex vivo from the prostatectomy specimens of 46 patients. A deep learning-based workflow was developed for 3D nuclear segmentation within the glandular epithelium versus stromal regions of the biopsies. 3D shape-based nuclear features were extracted, and a nested cross-validation scheme was used to train a supervised machine classifier based on 5-year biochemical recurrence (BCR) outcomes. Nuclear features of the glandular epithelium were found to be more prognostic than stromal cell nuclear features (AUC = 0.72 versus 0.63). 3D shape-based nuclear features of the glandular epithelium were also more strongly associated with risk of BCR than analogous 2D features (AUC = 0.72 versus 0.62). The results of this preliminary investigation suggest that 3D shape-based nuclear features are associated with prostate cancer aggressiveness and could be of value for the development of decision-support tools.
Keywords: Light-sheet microscopy, Non-destructive 3D pathology, Digital pathology, Prostate cancer, 3D segmentation, Prognosis
Introduction
Prostate cancer (PCa) accounts for 1 in 8 of the newly detected cancer cases in the United States, affecting nearly 250,000 patients annually [1]. Currently, grading of PCa for prognosis and treatment planning relies on 2D histology, where a set of core-needle biopsies is formalin-fixed and paraffin-embedded (FFPE), sectioned, and mounted on glass slides and stained for microscopic analysis. To assess the aggressiveness of the cancer, pathologists use the International Society of Urological Pathology (ISUP) grade group system [2–4] which involves visual examination and interpretation of complex glandular morphologies in a limited number of 2D histological tissue sections (4-μm thick) cut from each biopsy. In this workflow, typically ~1% of the volumetric extent of each biopsy is analyzed. In part due to this limited “sampling” of the biopsies in 2D, grading of prostate cancer suffers from high levels of inter- and intra-observer variability [5–8]. A related issue is that ISUP grade groups do not precisely correlate with individual patient outcomes, especially for moderate-risk cases (ISUP Grade Group 2) that account for approximately 50% of the newly detected PCa cases in the USA. These problems contribute to the overtreatment of indolent cases (with surgery or radiation), which are often accompanied by life-changing side effects (incontinence or impotence) and potentially avoidable healthcare costs. Conversely, they also contribute to the undertreatment of aggressive cases, which can lead to potentially avoidable metastasis and death [9].
By combining recent advances in tissue-clearing techniques [10–12] and high-throughput three-dimensional microscopy [13–19], there is now the ability to generate non-destructive 3D pathology datasets of large clinical specimens (surgical excisions or whole biopsies). In particular, it is possible to rapidly label thick tissues with a small-molecule fluorescent analog of H&E, and to false color the datasets so that they closely mimic the appearance of standard slide-based H&E histology (e.g. supplementary material, Figure S1) [14,18,20]. Unlike conventional histology, non-destructive 3D pathology achieves orders-of-magnitude greater microscopic sampling of tissues than conventional slide-based histology and enables volumetric quantification of diagnostically significant microarchitectures. The non-destructive imaging process also preserves tissue specimens for downstream molecular assays [21]. However, since a single 3D pathology dataset of a biopsy can be tens to hundreds of gigabytes in size, there are significant challenges for the human interpretation of such datasets. Manual examination of these datasets can be tedious, which motivates the development of computational methods for analysis. Here, we employ interpretable “hand-crafted” features, computationally derived from segmentation masks of tissue structures (e.g., glands and nuclei), to facilitate the clinical acceptance of 3D pathology as a new diagnostic approach.
A recent study by our group demonstrated the benefits of the computational analysis of 3D glandular features for PCa risk stratification [19]. In that study, ex vivo biopsies were extracted from archived radical prostatectomy specimens obtained from patients with known 5-year biochemical recurrence (BCR) outcomes. These patients were originally diagnosed with low- to moderate-risk PCa (Grade Groups 1 – 3), with 50% of cases exhibiting BCR within 5 years of prostatectomy [22]. Biopsies were stained with small-molecule fluorescent analogs of hematoxylin and eosin (H&E), optically cleared with a dehydration and solvent-immersion protocol, and then non-destructively imaged with open-top light-sheet microscopy (OTLS) to generate whole-biopsy 3D pathology datasets (Figure 1A). For 3D segmentation of prostate glands (lumen, epithelium, and stromal compartments), our group developed a computational pipeline called “image-translation-assisted segmentation in 3D” (ITAS3D). 3D glandular features (e.g., gland curvature, eccentricity, volume ratios) were quantified from the cancer-containing regions of the biopsies. We showed that these quantitative features are more strongly associated with BCR outcomes than analogous 2D glandular features.
Figure 1. Workflow to evaluate the prognostic value of 3D nuclear features for prostate cancer risk assessment.
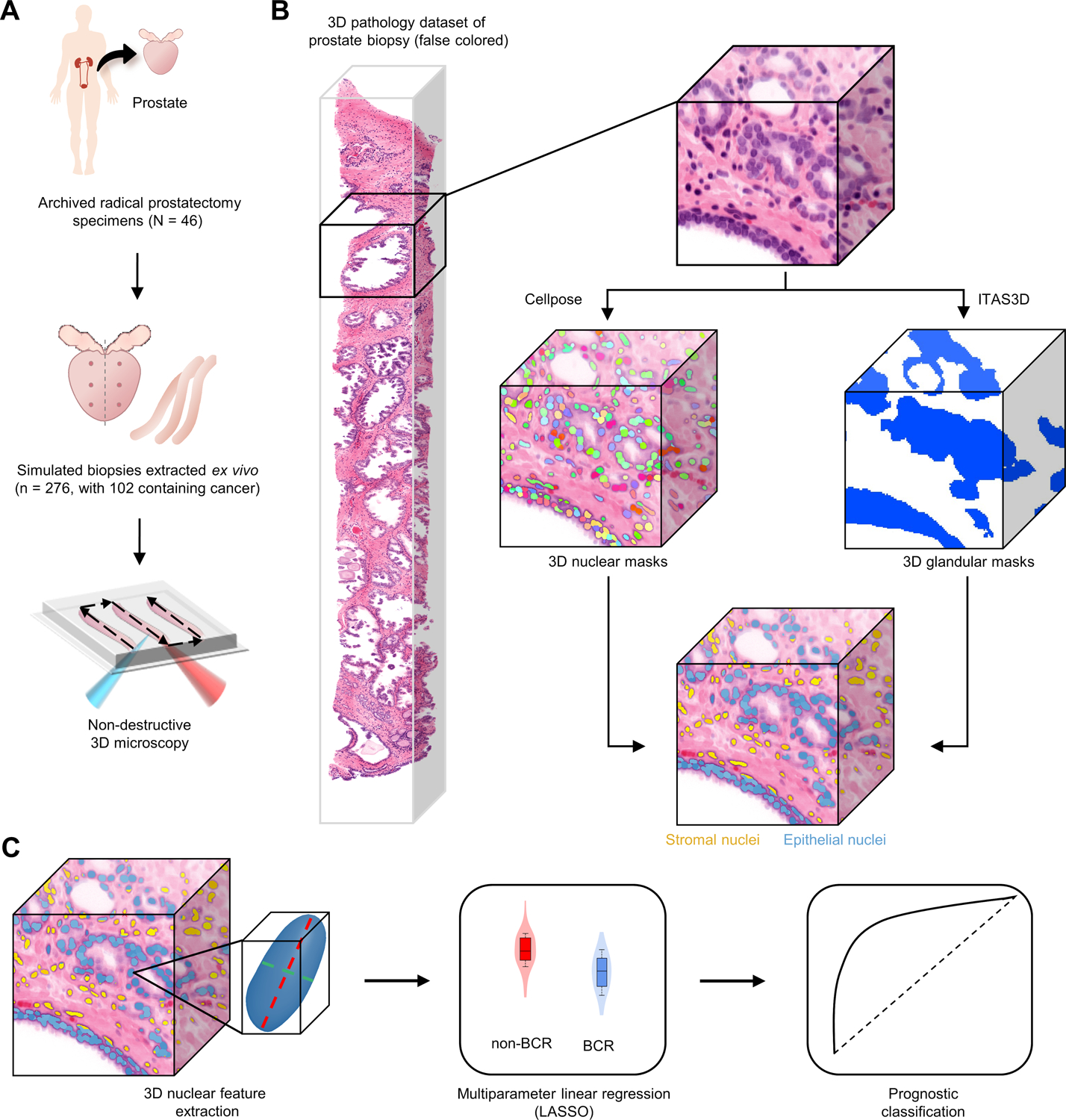
(A) Archived (FFPE) radical prostatectomy specimens were obtained from a 46-patient cohort with known 5-year biochemical recurrence (BCR) outcomes, from which simulated (ex vivo) biopsies were extracted (6 biopsies per case, per sextant biopsy protocol). The biopsies were labeled with a fluorescent analog of H&E, optically cleared to render them transparent to light, and comprehensively imaged in 3D via OTLS microscopy. (B) 3D nuclear segmentation masks of cancer-containing regions were generated using the deep-learning based cellpose model and 3D glandular segmentations were generated by the previously published ITAS3D pipeline. This enabled us to distinguish between nuclei in the glandular epithelium (blue) versus the surrounding stroma (yellow). (C) 3D shape-based features of epithelial and stromal nuclei were extracted and passed into a multiparameter classifier (LASSO) to stratify patients based on 5-year BCR outcomes.
In addition to glandular morphologies, which are currently the sole basis for Gleason grading (nuclear morphologies are not looked at for standard-of-care Gleason grading), studies using 2D whole-slide images (conventional 2D pathology) have shown that there is prognostic value in computationally extracted nuclear features within PCa [22–33]. Note that prior to Gleason grading becoming the widely accepted standard for grading PCa, 2D cytology, including nuclear morphologies, were also used by pathologists in various grading schemes [30–35]. However, the prognostic value of 3D (volumetric) nuclear features has not been previously investigated. Therefore, we sought to quantify a preliminary set of 3D shape-based nuclear features and their 2D counterparts to directly compare their ability to predict BCR outcomes. Our underlying hypothesis is that 2D cross sections of a nucleus (as seen with conventional histology) cannot fully elucidate certain complex changes in 3D nuclear morphology and therefore may be limited for patient risk stratification. For example, supplementary material, Figure S2 provides a simple illustration of how an irregularly shaped 3D nucleus may appear quite regular and circular when viewed as 2D cross sections.
For 3D nuclear and glandular segmentation, we used cellpose [36] and our previously reported ITAS3D pipeline [19], respectively, which allowed us to distinguish between nuclei within the gland epithelium versus surrounding stromal compartments (Figure 1B). 3D shape-based nuclear features and 2D analogs were extracted from each biopsy, such that the prognostic value of 3D versus 2D nuclear features could be compared. These 3D and 2D features were used to train a multi-parameter machine classifier to stratify patients based on their BCR outcomes, which serves as a proxy for aggressive versus indolent PCa (Figure 1C).
Materials and methods
Tissue preparation
This study was approved by the institutional review board (IRB) of the University of Washington (Seattle, WA, USA; Study 00004980), where research specimens were previously obtained from patients with informed consent. Archived FFPE prostatectomy specimens from a prior case-cohort research study (Canary TMA) [22] were collected from 46 patients with PCa, all of which were identified as ISUP Grade Group 1–3 (Gleason Grade 3+3, 3+4, or 4+3). These patients were followed for at least 5 years after radical prostatectomy as part of the prior study [22], in which the primary study endpoints were 5-year BCR outcomes and time to BCR, which were also used as endpoints for this study. FFPE specimens were identified for each case corresponding to the six regions of the prostate targeted in standard biopsy procedures (Figure 1A). Specimens were deparaffinized by heating them at 75 °C for 1 h until the paraffin wax melted. The specimens were then placed in 65 °C xylene for 48 h. Next, a simulated core-needle biopsy (~ 1 mm in width) was taken from each of the six deparaffinized blocks per case, resulting in 300 total biopsy cores (Figure 1A).
We used a previously developed “T&E” staining protocol (nuclear dye TO-PRO3 and Eosin) and imaged each biopsy core using OTLS microscopy [15,19]. Simulated biopsies underwent two 1-h washes in 100% ethanol to remove excess xylene and were partially rehydrated in 70% ethanol for 1 h. Individual biopsies were stained for 48 h in 70% ethanol at pH 4 with a 1:200 dilution of Eosin-Y (catalog no. 3801615, Leica Biosystems) and a 1:500 dilution of TO-PRO3 (catalog no. T3605, Thermo Fisher Scientific) at room temperature with gentle agitation. After staining was complete, the biopsies were dehydrated in 100% ethanol for 2 hours. Samples were optically cleared by individually placing them in ethyl cinnamate (catalog no. 112372, Sigma-Aldrich) (refractive index of n = 1.56) for 8 h before imaging using OTLS microscopy.
For imaging, ethyl cinnamate was used as the immersion medium and samples were mounted within a custom HIVEX (n = 1.55) biopsy holder, as described previously [15]. A four-channel digitally controlled laser module (Skyra, Cobolt Lasers, Hübner Photonics, Kassel, Germany) was used for illumination. TO-PRO3 fluorescence was excited at 638 nm and Eosin fluorescence was excited at 561 nm. Fluorescence bandpass filters at 721 nm ± 65 nm and 618 nm ± 50 nm were used for collection of TO-PRO3 fluorescence and Eosin fluorescence, respectively. The lateral and axial resolution of the OTLS microscope used for this study was ~0.95 μm and ~3.5 μm, respectively (full width at half maximum of the point spread function). Tissues were imaged at an isotropic sampling pitch of ~0.44 μm/pixel. The volumetric imaging time was approximately 0.5 minute per cubic mm of tissue for each illumination wavelength.
Data preparation
Two-channel OTLS microscopy datasets were stored on disk in the HDF5 format with microscopy metadata in an XML file. A custom compression filter (B3D) was used to provide 10x lossless compression. Raw OTLS images were reviewed in BigDataViewer [37] by board-certified pathologists to identify cancer-containing biopsies (Figure 1A). Of the initial 276 biopsy cores extracted, 102 contained cancer. Visual examination of the 3D datasets enabled us to identify regions of the biopsies in which most of the glands were cancer. Continuous (stitched and fused) 3D volumes were created of these cancer regions using the BigStitcher plugin for ImageJ [38]. H&E-like false coloring was performed on our OTLS datasets using FalseColor-python, a previously published method that mimics Beer–Lambert absorption of light as a function of staining concentration [20].
Nuclear segmentation and feature extraction
Given the memory requirements necessary for 3D segmentation, fused 3D images were broken into discrete blocks for processing. Each block was 2048 × 1432 × 500 voxels in size, or approximately 1024 by 716 by 200 μm in the axial direction (along the length of the needle core) (Figure 2A,B). Prior to segmentation, data blocks underwent median filtering with a circular structuring element (r = 2 voxels) from scikit-image [39] followed by contrast-limited adaptive histogram equalization (CLAHE) (Figure 2C, supplementary material, Figures S3,S4). After preprocessing, data blocks were passed into the cellpose [36] 3D nuclear segmentation model using the following parameters: an average diameter of 17 voxels, net averaging, and a batch size of 6 (Figure 2D, Supplementary materials and methods). All processing was done on a desktop computer equipped with 512 GB of RAM and a Nvidia Quadro 5000 RTX GPU.
Figure 2. 3D nuclear segmentation pipeline for biopsies imaged with OTLS microscopy.
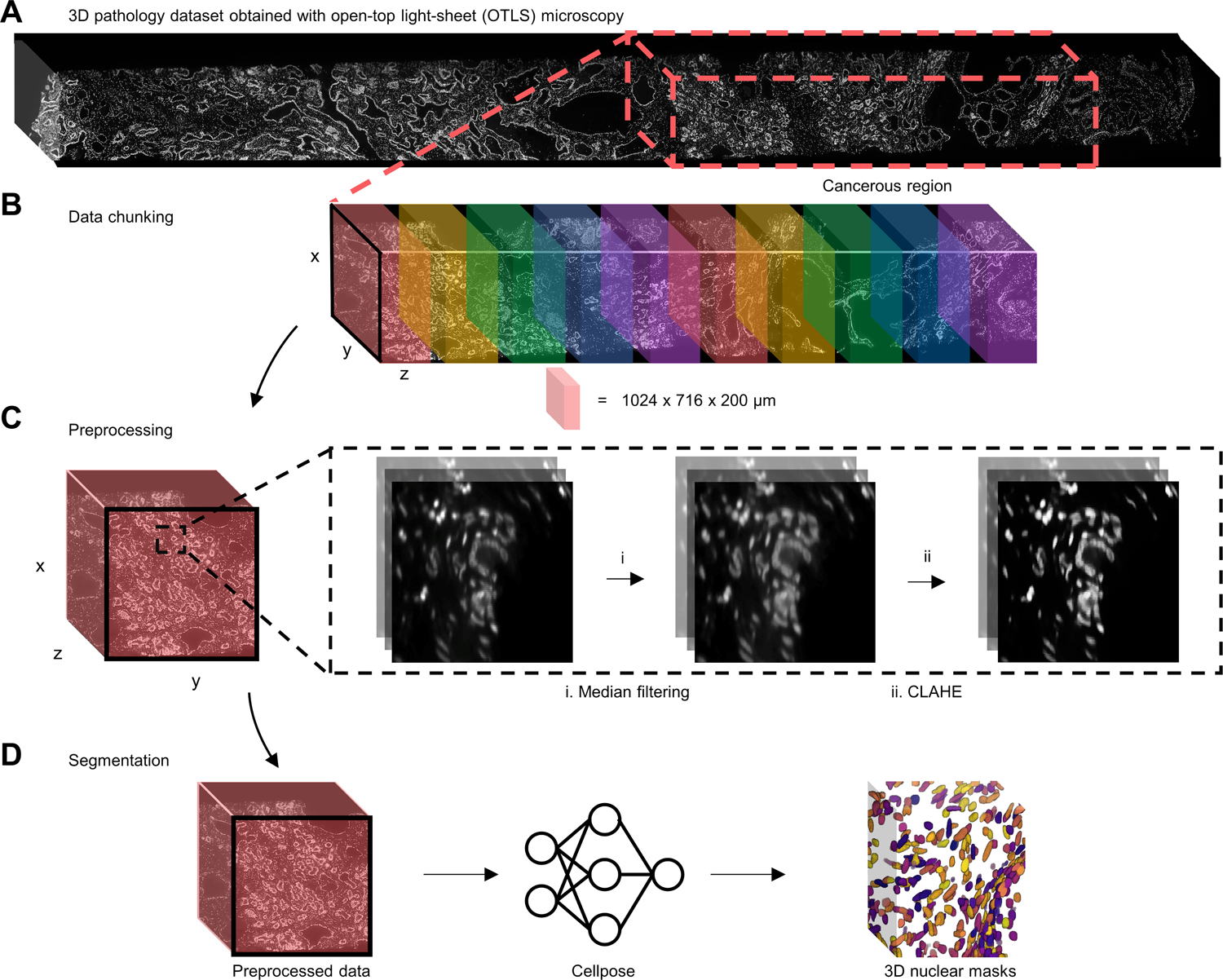
(A) Nuclear channel (To-Pro-3) of a PCa biopsy imaged by OTLS microscopy with the cancerous region outlined with a dashed red box. (B) The cancerous region is broken up into discrete data blocks before processing. (C) Each data block is passed into a two-step preprocessing procedure before segmentation (see text for details). (D) Preprocessed data blocks are passed into cellpose to generate 3D nuclear segmentation masks, where each segmented nucleus is assigned a unique label.
To avoid quantifying the properties of nuclei that were artificially fragmented/cropped at the edges of each data block, segmented nuclei touching the boundaries of each data block were removed using the clear_border method from the skimage.segmentation python library [39]. Before feature extraction, small objects within each segmentation mask measuring less than the 1st volume percentile were removed as noise (Supplementary materials and methods, supplementary material, Figure S5). Quantitative features (e.g. volume, sphericity, major/minor axis length) were extracted from the segmentations using the regionprops method from the skimage.measure python library. A full set of 3D nuclear features (and 2D counterparts) used is listed in supplementary material, Figure S6. Glandular masks of each cancerous region, generated via our ITAS3D pipeline [19], were used to classify segmented nuclei as belonging to epithelial or stromal compartments based on the centroid location of each segmented nucleus within the glandular mask (Supplementary materials and methods). Biopsy-level feature sets were collected by averaging the extracted features from all data blocks within a biopsy.
To compare the prognostic value of 2D and 3D nuclear features, 2D nuclear features were extracted from the 3D dataset at three cross-sectional levels separated by 20 μm, and then averaged (supplementary material, Figure S7). This replicates the standard-of-care pathology practice at many institutions in which histology sections are cut (and viewed) at three such levels [40].
Statistical analysis
Patients who experienced BCR within 5 years after radical prostatectomy were labeled as belonging to the ‘BCR’ group, and all other patients as ‘non-BCR.’ Biochemical recurrence (BCR) is defined as a rise in prostate-specific antigen (PSA) levels of >0.2 ng/ml after treatment (>8 weeks after radical prostatectomy). To assess the ability of different nuclear features to distinguish between BCR and non-BCR groups, we applied ROC analysis from which the area-under-the-curve (AUC) could be extracted.
Using the binary 5-year BCR category as the endpoint, a multiparameter classifier was developed to stratify patient risk. The least absolute shrinkage and selection operator (LASSO) linear regression model was used [41]. LASSO minimizes overfitting by making use of an L1 regularization term and identifies a subset of the most predictive input features. The LASSO tuning parameter was optimized using an internal 4-fold cross validation (CV) scheme, where the dataset was randomly separated into four equally sized groups: three training datasets, and one to validate the model’s performance. Due to the lack of an external validation cohort, a nested CV scheme was used to develop multiparameter classifiers based on 3D and 2D features, and to determine the performance of the multiparameter models without bias or data leakage between parameter estimation and validation. Hyperparameter tuning was performed during each iteration of the outer CV, and LASSO regression was applied on the training set of the outer CV once an optimal was identified in the inner CV. AUC values were calculated from the validation dataset of the outer CV. This nested CV was performed 200 times to determine an AUC (average and SD) [42].
Kaplan–Meier (KM) analysis was carried out to compare BCR-free survival rates for high- versus low-risk groups of patients. This analysis used a subset of 45 cases for which time-to-recurrence data was available. The performance of the models based on 2D or 3D features for epithelial nuclei was quantified with p-values (via log-rank test), hazard ratios (HR), and concordance index (C-index) metrics. For the multiparameter classification model used for KM analysis, the outer 4-fold CV in our nested CV schema was replaced by a leave-one-out approach, where one case was left out each iteration (45 total iterations, i.e. one iteration for each of the cases) to calculate the probability of 5-year BCR for that patient [43]. The samples were labeled as low- or high-risk via a posterior class probability threshold of 0.5. MATLAB was used for KM analysis and all other statistical analyses was performed in Python using the scipy and scikit-learn libraries [44].
Results
3D nuclear segmentation
Generating ground-truth annotations to train a 3D nuclear-segmentation algorithm can be tedious. Therefore, we chose cellpose as our segmentation framework for two reasons: first, as a “generalist” segmentation algorithm, cellpose requires minimal retraining or annotations to operate on unseen data, and secondly, cellpose is compatible with 3D datasets and can efficiently segment densely packed nuclei within large tissue volumes. To assess segmentation quality, we generated 3D ground truth annotations of nuclei within small sub-volumes (n = 6) of PCa biopsies imaged with OTLS microscopy and found that cellpose successfully generated 3D nuclear segmentations with an average dice coefficient of 0.81 (supplementary material, Figure S8). When evaluating cellpose segmentation quality with our moderate-resolution OTLS datasets (~0.9-micron lateral resolution), the best segmentation results were achieved with the native-resolution datasets without any down sampling (supplementary material, Video S1).
At full resolution, each individual (single biopsy) 3D pathology dataset was too large to segment at once. Thus, cancerous biopsy regions were divided into smaller blocks for 3D segmentation (Figure 2A,B). Each data block underwent a two-step preprocessing routine (Figure 2C) (Supplementary materials and methods, supplementary material, Figures S3,S4) that improved segmentation quality. This process decreased the rate of over-segmentation and improved the detection of nuclei in low-signal regions of the tissue. Images were then passed into the cellpose nuclear-segmentation model to generate a final 3D segmentation mask (Figure 2D, supplementary material, Videos S2 and S3). An image atlas of example 3D nuclear-segmentation results is shown in Figure 3, including the stratification of nuclei into epithelial and stromal compartments by using ITAS3D-generated gland-segmentation masks.
Figure 3. Image atlas of 3D nuclear segmentation results.
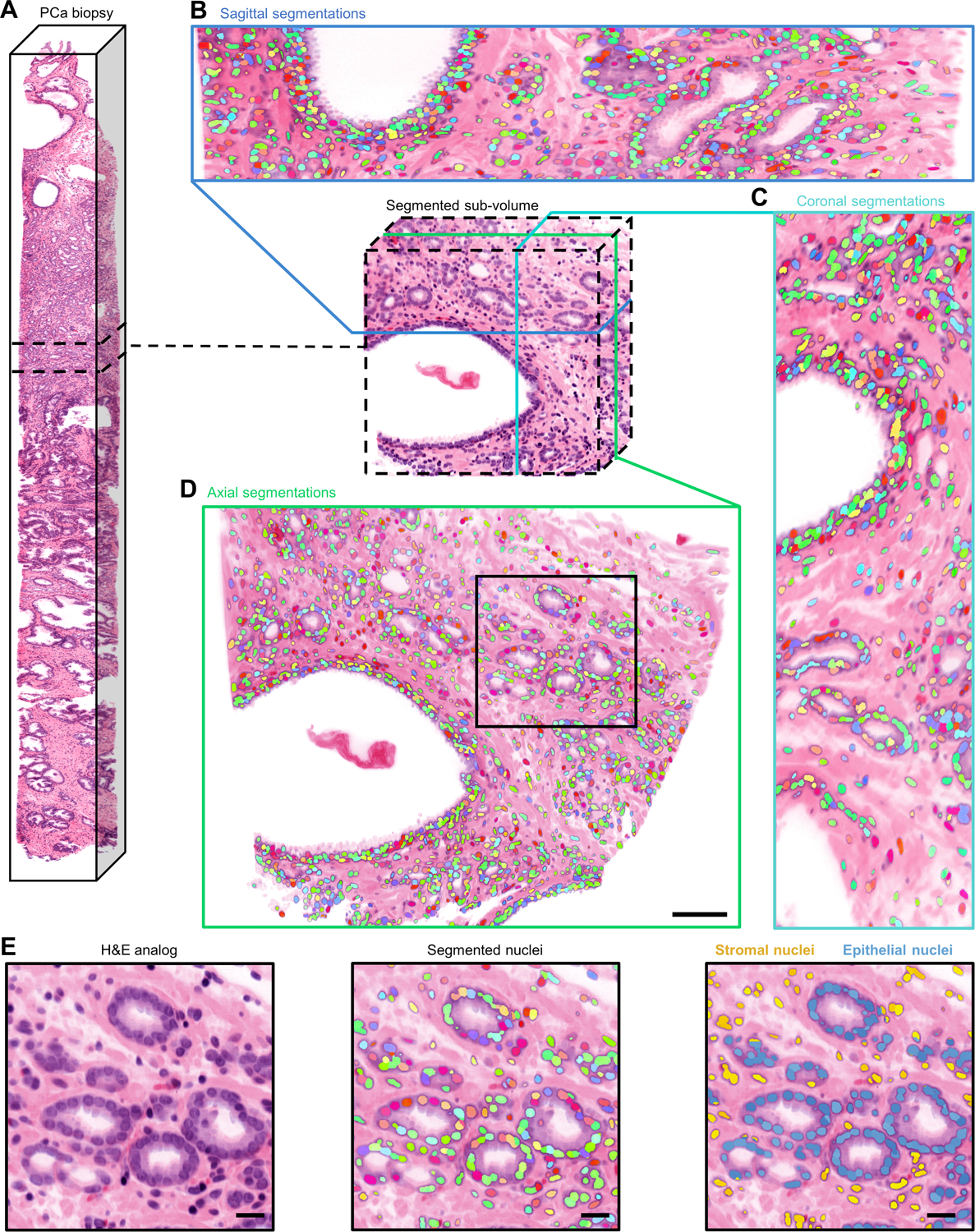
(A) Non-destructive 3D pathology dataset of a PCa biopsy and segmented sub-volume. Sagittal (B), coronal (C), and axial (D) views of 3D nuclear segmentations of the segmented sub-volume. During segmentation, each identified nucleus is given a unique integer label, which is represented by a color for visualization purposes (scale bar, 100 μm). (E) Larger field of view showing cancerous glands and surrounding stroma (left). colorized segmentation masks overlaid onto H&E false-colored OTLS images (middle). Segmented nuclei colored by their location within the prostate microarchitecture (right) (scale bar, 50 μm).
Preliminary clinical study
To evaluate the prognostic value of 3D versus 2D nuclear features (see materials and methods), our study consisted of 46 PCa cases in which the patients were followed for a minimum of 5 years after radical prostatectomy as part of the Canary TMA case-cohort study, which recruited primarily low- to intermediate-risk patients [22]. The primary endpoints of the Canary TMA study were 5-year biochemical recurrence (BCR) outcomes and time to BCR, which were also used as endpoints in our study. Roughly half of the cases in the Canary TMA study had BCR within 5 years.
Six simulated core needle biopsies were extracted from the 46 cases and imaged with OTLS (see materials and methods). Review of the 3D pathology datasets by genitourinary pathologists (L.D. True and N.P. Reder) identified the cancer-enriched regions within each of the 276 biopsies (i.e. regions in which most glands were cancerous), in which only 102 of the biopsies contained appreciable amounts of cancer. The nuclear-segmentation and ITAS3D (gland-segmentation) pipelines were applied to the cancer-enriched regions of these 102 biopsies. By combining the 3D nuclear segmentations (instance segmentations) with the glandular segmentation masks (semantic segmentation), nuclei were identified as contained in either the glandular epithelium or stromal tissue compartments (see Materials and Methods).
2D and 3D shape-based nuclear features were quantified and analyzed in terms of their association with BCR outcomes (supplementary material, Figure S6). For 2D analysis, average values from a total of 3 cross-sectional levels were calculated, in which the three levels were separated by 20 μm (mimicking clinical practice at many institutions, supplementary material, Figure S7). A total of 18 shape-based nuclear features were used as the inputs into a multiparameter risk classifier based on 5-year BCR outcomes. Separate risk-classification models were trained for the nuclei in the glandular epithelium versus stromal compartments. The average area-under-the-ROC-curve (AUC) was used as a measure of the ability of the combined nuclear features to predict 5-year BCR outcomes as a proxy for indolent versus aggressive disease.
3D shape-based nuclear features consistently outperformed their analogous 2D features, both for epithelial nuclei and stromal nuclei (Figure 4A,B). From comparison of Figure 4A and Figure 4B, it was apparent that there was better performance with models trained on shape-based nuclear features from the glandular epithelium than models trained with nuclei from the stromal regions. 3D shape-based nuclear features in the stroma were found to be weakly prognostic with an average AUC = 0.63 ± 0.06, while 2D shape-based nuclear features in the stroma were not prognostic with an average AUC = 0.50 ± 0.05.
Figure 4. Clinical analysis comparing the prognostic value of 3D & 2D nuclear features.
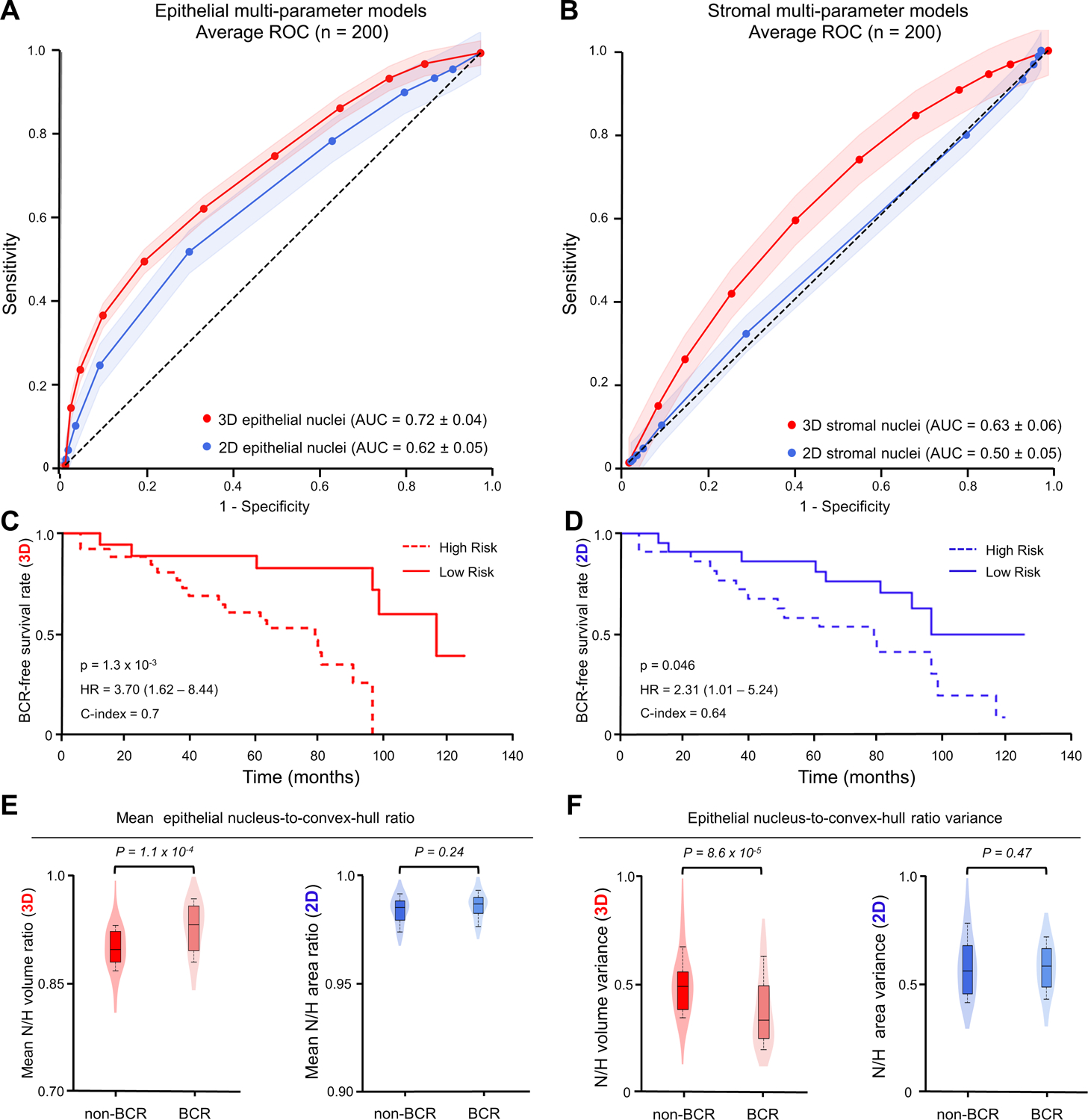
(A) ROC curves are shown for multiparameter models trained on 3D (red) and 2D (blue) nuclear features in epithelial regions. (B) ROC curves of multiparameter models trained on 3D (red) and 2D (blue) nuclear features in stromal regions. (C,D) Kaplan–Meier curves are shown for BCR-free survival, showing that the model trained on epithelial 3D nuclear features (C) can better stratify patients into low- and high-risk categories than the model trained on epithelial 2D nuclear features (D). (E,F) Violin and box plots are shown for two examples of epithelial 3D nuclear features, along with their analogous 2D features, for cases in which BCR occurred within 5 years of radical prostatectomy (“BCR”) and for cases in which there was no BCR within 5 years of radical prostatectomy (“non-BCR”). For both example features, “Mean epithelial nucleus-to-convex-hull ratio” in (E), and “epithelial nucleus-to-convex-hull variance” in (F), the 3D feature shows improved patient risk stratification compared to its 2D counterpart.
Kaplan–Meier (KM) curves of BCR-free survival were constructed for a subset of cases in which time-to-recurrence (BCR) data was available (Figure 4C,D). Compared with models based on 2D shape-based nuclear features, models based on 3D shape-based nuclear features are associated with a higher HR and C-index, along with a significant P value (P < 0.05). This suggests a stronger association with outcome for 3D shape-based nuclear features compared to corresponding 2D features.
For this early-stage study, 14 out of 18 of the shape-based nuclear features were independently associated with BCR outcomes (p < 0.05). Of these 14 features, 5 were prognostically significant in 3D but not in 2D. As one example, Figure 4E,F shows the mean and variance of the nucleus-to-convex hull ratio of epithelial nuclei (3D versus 2D). The convex hull is the minimum polyhedron (or polygon in 2D) that encloses a nucleus. Figure 4E shows that the nuclei in BCR cases (i.e., aggressive PCa) typically had a higher mean nucleus-to-convex-hull ratio (P = 1.1 × 10−4). Physically, this means that the nuclei were more spherical or ovaloid rather than irregular and/or curved in shape. However, this relationship is not apparent in the 2D case (P = 0.24). Similarly, Figure 4F demonstrates that the variance of the nucleus-to-convex-hull ratio was greater for recurrent versus non-recurrent cases when examined in 3D with P = 8.6 × 10−6, while this difference was less in 2D (P = 0.47).
Overall, our analysis of a variety of nuclear shape features consistently reveals less heterogeneity (variance) in aggressive (BCR) cancers compared to non-aggressive cases (supplementary material, Figure S9). This parallels the results of a previous study in which higher-grade PCa cases had smaller and more homogeneous glandular structures (less spatial variance) [19].
Discussion
Improvements in resolution, scale, and multiplexing capacity for non-destructive 3D imaging technologies are leading to disease insights that can inform treatment decisions [21]. However, given the growing size of datasets generated by modern “spatial biology” techniques, computational tools must also be developed to enable pathologists and oncologists to efficiently comprehend such large datasets. An attractive initial approach is to rely on intuitive features familiar to pathologists (e.g., glandular and cytologic features), which will improve interpretability and facilitate clinical acceptance. As in our previous study examining 3D glandular features [19], our goal in this study has been to demonstrate the value of 3D pathology by providing a direct comparison of intuitive 3D versus 2D nuclear features analyzed computationally. Our work leverages the fact that computational 2D pathology has already been demonstrated to improve disease prognostication [29,45,46], and explores the additional value that computational 3D pathology can offer for certain applications such as risk stratification of PCa. We have avoided comparing our computational results with risk classifiers or nomograms that rely on human interpretation of histology images [43,47,48]. Our motivation for this is that by directly comparing 3D versus 2D computational pathology, we remove the subjectivity introduced by human interpretation. Such human-observer studies, to be performed in the future, will require a significantly larger cohort of patients and a large panel of pathologists to mitigate interobserver discordance.
Previous studies have used 3D imaging of in vitro cancer models to examine tumorigenesis, drug response, and cancer-associated alterations in cellular development [49–51]. However, to our knowledge, this is the first report to analyze the prognostic significance of nuclei within their native 3D context in human cancer specimens. Given the relatively small number of cases in this preliminary analysis, we limited the number of 3D nuclear features to those that we deemed would most likely have prognostic significance based on previous studies [23–29,52–58]. Shape-based nuclear features are an attractive choice for several reasons: they are the most frequently used features for prognostication based on 2D whole slide images [18–23, 42, 43], they are intuitive for clinical and biological interpretation [59], they have been considered in the past for PCa grading [30–35], and there are relatively straightforward analogs between 2D and 3D shape-based features. Rather than exhaustively mining all possible nuclear features to maximize classifier performance, we have deliberately based our classifiers on a limited set of intuitive features to show that 3D nuclear features have obvious prognostic value. In other words, with some effort, we believe that better 3D features and models can be developed in the future.
Our analysis shows that for PCa, epithelial nuclei hold the most prognostic significance (versus stromal nuclei) for stratifying patients based on known clinical outcomes (Figure 4A). Given that PCa is typically a disease of epithelial cells that form glands, this result is not surprising. However, we find that 3D shape-based features of stromal nuclei are also somewhat prognostic (Figure 4B), which is supported by previous studies using 2D histology [4,28,29,60–62]. Since these stromal nuclei are from a mixture of cell types, it is not surprising that they are less prognostic than the epithelial nuclei in the cancer glands. Most importantly, for both epithelial and stromal models, our results demonstrated that 3D shape-based nuclear features were more strongly associated with BCR compared to their 2D counterparts for risk stratification (Figure 4A–D). Even with the limited number of samples and extracted features in this preliminary analysis, certain differences between nuclei in indolent and aggressive cases were only statistically significant when examined in 3D (Figure 4E,F). These findings support the value of 3D digital pathology methods for clinical management of PCa. Overall, the prognostic value of the 3D nuclear features analyzed in this study is less than the prognostic value of the 3D glandular features we previously explored19. This is expected and consistent with the fact that PCa glandular structures are the current basis for clinical Gleason grading. Therefore, the ideal clinical classifier should holistically combine both glandular and cytologic/nuclear features in the future.
In addition to combining cytologic and architectural features (e.g., nuclei and glands), there are clear opportunities to extend our work. For example, adding 3D features from additional tissue structures, such as vasculature, could reveal novel signatures of aggressiveness. Having a comprehensive spatial and molecular view of tumors in 3D would also be of obvious clinical value63,64. Finally, obtaining higher-resolution datasets with the latest generations of OTLS microscopy systems16,17 should allow for more accurate quantification of 3D nuclear features as well as the extraction of sub-nuclear features (e.g. chromatin texture, nucleoli). This preliminary study motivates future exploratory directions using computational 3D pathology, as well as larger-scale clinical studies to guide treatment decisions, such as deciding which PCa patients should be placed on active surveillance versus treated with surgery/radiation, or which patients should receive adjuvant therapies after surgery/radiation. Ultimately, we aim to demonstrate that computational 3D pathology can improve the long-term outcomes and quality of life for patients with PCa and many other diseases.
Supplementary Material
Figure S1. Example 2D images of benign (top) and cancerous (bottom) prostate tissues imaged with 2nd-generation OTLS microscopy versus slide-based H&E histology
Figure S2. Morphological complexities of nuclei cannot be fully captured in 3D
Figure S3. Effect of median filter kernel size on segmentation results
Figure S4. Effect of preprocessing procedure on cellpose segmentation results in low signal regions
Figure S5. Sensitivity of nuclear properties and features with respect to volume threshold
Figure S6. Set of shape-based nuclear features used for prognostic analysis
Figure S7. 2D segmentation pipeline
Figure S8. Comparison of nuclear segmentation methods on manually annotated OTLS volumes
Figure S9. Comparison of the epithelial nuclear features (variance) between BCR groups
Video S1. Effect of down sampling on segmentation results
Video S2. 3D segmentation results
Video S3. Colorized segmentation results for a PCa biopsy
Acknowledgements
The authors acknowledge funding support from the NIH National Center for Advancing Translational Sciences through TL1 TR002318 (Serafin), the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) through W81WH-18-10358 (Liu and True), W81XWH-19-1-0589 (Reder), and W81XWH-20-1-0851 (Madabhushi and Liu). Support was also provided by the National Cancer Institute (NCI) through R01CA268207 (Liu and Madabhushi), K99 CA240681 (to A.K. Glaser), U24CA199374 (Madabhushi), R01CA249992 (Madabhushi), R01CA202752 (Madabhushi), R01CA208236 (Madabhushi), R01CA216579 (Madabhushi), R01CA220581 (Madabhushi), R01CA257612 (Madabhushi), U01CA239055 (Janowczyk and Madabhushi), U01CA248226 (Madabhushi and Janowczyk), U54CA254566 (Madabhushi), and the Pacific Northwest Prostate Cancer SPORE - P50CA097186 (True). Additional support was provided by the National Heart, Lung and Blood Institute (NHLBI) through R01HL151277 (Madabhushi), the National Institute of Biomedical Imaging and Bioengineering (NIBIB) through R01EB031002 (Liu) and R43EB028736 (Madabhushi), the National Institute of Digestive Diseases and Kidney (NIDDK) through U01DK133090 (Janowczyk) and R01DK118431 (Janowczyk), the National Library of Medicine (NLM) through R01LM013864 (Janowczyk), the VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs (Madabhushi), the Prostate Cancer Foundation Young Investigator Award (Reder), the Prostate Cancer United Kingdom (PCUK) charity (Liu), the National Science Foundation (NSF) through 1934292 HDR: I-DIRSE-FW (Liu and Serafin), and the Canary Foundation.
A.K. Glaser is a cofounder of Alpenglow Biosciences, Inc. N.P. Reder is a cofounder and CEO of Alpenglow Biosciences, Inc. A. Janowczyk reports personal fees from Roche, Merck, and personal fees from Lunaphore outside the submitted work. L.D. True is a cofounder of Alpenglow Biosciences, Inc. A. Madabhushi is an equity holder in Picture Health, Elucid Bioimaging, and Inspirata Inc. Currently he serves on the advisory board of Picture Health, Aiforia Inc, and SimBioSys. He also currently consults for Biohme, SimBioSys and Castle Biosciences. He also has sponsored research agreements with AstraZeneca, Boehringer-Ingelheim, Eli-Lilly and Bristol Myers-Squibb. His technology has been licensed to Picture Health and Elucid Bioimaging. He is also involved in 3 different R01 grants with Inspirata Inc. J.T.C. Liu is a cofounder and board member of Alpenglow Biosciences, Inc., which has licensed the OTLS microscopy portfolio developed in his lab at the University of Washington. No other conflicts of interests were declared.
Footnotes
Disclaimer
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NIH, NSF, Prostate Cancer Foundation, Prostate Cancer United Kingdom (PCUK) charity, Department of Defense, the Department of Veterans Affairs, or the United States Government.
References 65–67 are cited only in the supplementary material.
Data availability statement
The original 3D pathology datasets used in this study may be found at: https://doi.org/10.7937/44MA-GX21 [68]
The base code for the cellpose segmentation framework may be found at: https://github.com/mouseland/cellpose
The code base for the segmentation, feature extraction, and analysis pipelines may be found at: https://github.com/serrob23/OTLS-cellpose
References
- 1.Siegel RL, Miller KD, Fuchs HE & Jemal A Cancer Statistics, 2021. CA. Cancer J. Clin 71, 7–33 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Egevad L, Delahunt B, Srigley JR & Samaratunga H International Society of Urological Pathology (ISUP) grading of prostate cancer - An ISUP consensus on contemporary grading. APMIS 124, 433–435 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Srigley JR et al. One is the new six: The International Society of Urological Pathology (ISUP) patient-focused approach to Gleason grading. Can. Urol. Assoc. J 10, 339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Leenders GJLH et al. The 2019 International Society of Urological Pathology (ISUP) Consensus Conference on Grading of Prostatic Carcinoma. Am. J. Surg. Pathol 44, e87–e99 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah RB et al. Diagnosis of Gleason Pattern 5 Prostate Adenocarcinoma on Core Needle Biopsy: An Interobserver Reproducibility Study Among Urologic Pathologists. Am. J. Surg. Pathol 39, 1242–1249 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Kweldam CF et al. Gleason grade 4 prostate adenocarcinoma patterns: an interobserver agreement study among genitourinary pathologists. Histopathology 69, 441–449 (2016). [DOI] [PubMed] [Google Scholar]
- 7.McKenney JK et al. The Potential Impact of Reproducibility of Gleason Grading in Men With Early Stage Prostate Cancer Managed by Active Surveillance: A Multi-Institutional Study. J. Urol 186, 465–469 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Zhou M et al. Diagnosis of “Poorly Formed Glands” Gleason Pattern 4 Prostatic Adenocarcinoma on Needle Biopsy: An Interobserver Reproducibility Study Among Urologic Pathologists With Recommendations. Am. J. Surg. Pathol 39, 1331–1339 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Etzioni R et al. Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J. Natl. Cancer Inst 94, 981–990 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Renier N et al. iDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Susaki EA et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Azaripour A et al. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog. Histochem. Cytochem 51, 9–23 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Power RM & Huisken J A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 14, 360–373 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Glaser AK et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng 1, 0084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser AK et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat. Commun 10, 2781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barner LA, Glaser AK, Huang H, True LD & Liu JTC Multi-resolution open-top light-sheet microscopy to enable efficient 3D pathology workflows. Biomed. Opt. Express 11, 6605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser AK et al. A hybrid open-top light-sheet microscope for versatile multi-scale imaging of cleared tissues. Nat. Methods 19, 613–619 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reder NP et al. Open-Top Light-Sheet Microscopy Image Atlas of Prostate Core Needle Biopsies. Arch. Pathol. Lab. Med 143, 1069–1075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie W et al. Prostate Cancer Risk Stratification via Nondestructive 3D Pathology with Deep Learning–Assisted Gland Analysis. Cancer Res. 82, 334–345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serafin R, Xie W, Glaser AK & Liu JTC FalseColor-Python: A rapid intensity-leveling and digital-staining package for fluorescence-based slide-free digital pathology. PLOS ONE 15, e0233198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu JTC et al. Harnessing non-destructive 3D pathology. Nat. Biomed. Eng 5, 203–218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawley S et al. A Model for the Design and Construction of a Resource for the Validation of Prognostic Prostate Cancer Biomarkers: The Canary Prostate Cancer Tissue Microarray. Adv. Anat. Pathol 20, 39–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Poel HG, Oosterhof GON, Schaafsma HE, Debruyne FMJ & Schalken JA Intratumoral nuclear morphologic heterogeneity in prostate cancer. Urology 49, 652–657 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Leo P et al. Stable and discriminating features are predictive of cancer presence and Gleason grade in radical prostatectomy specimens: a multi-site study. Sci. Rep 8, 14918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji M-Y et al. Nuclear shape, architecture and orientation features from H&E images are able to predict recurrence in node-negative gastric adenocarcinoma. J. Transl. Med 17, 92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee G et al. Nuclear Shape and Architecture in Benign Fields Predict Biochemical Recurrence in Prostate Cancer Patients Following Radical Prostatectomy: Preliminary Findings. Eur. Urol. Focus 3, 457–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carleton NM, Lee G, Madabhushi A & Veltri RW Advances in the computational and molecular understanding of the prostate cancer cell nucleus. J. Cell. Biochem 119, 7127–7142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandramouli S et al. Computer Extracted Features from Initial H&E Tissue Biopsies Predict Disease Progression for Prostate Cancer Patients on Active Surveillance. Cancers 12, 2708 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhargava HK et al. Computationally Derived Image Signature of Stromal Morphology Is Prognostic of Prostate Cancer Recurrence Following Prostatectomy in African American Patients. Clin. Cancer Res 26, 1915–1923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mostofi FK Grading of prostatic carcinoma. Cancer Chemother. Rep 59, 111–117 (1975). [PubMed] [Google Scholar]
- 31.Böcking A & Sinagowitz E Histologic Grading of Prostatic Carcinoma. Pathol. - Res. Pract 168, 115–125 (1980). [DOI] [PubMed] [Google Scholar]
- 32.Böcking A, Kiehn J & Heinzel-Wach M Combined histologic grading of prostatic carcinoma. Cancer 50, 288–294 (1982). [DOI] [PubMed] [Google Scholar]
- 33.Fujikawa K, Sasaki M, Aoyama T & Itoh T Prognostic criteria in patients with stage D2 prostate cancer. Correlation with mean nuclear volume. Cancer 76, 91–95 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Diamond DA, Berry SJ, Jewett HJ, Eggleston JC & Coffey DS A New Method to Assess Metastatic Potential of Human Prostate Cancer: Relative Nuclear Roundness. J. Urol 128, 729–734 (1982). [DOI] [PubMed] [Google Scholar]
- 35.Epstein JI, Berry SJ & Eggleston JC Nuclear roundness factor. A predictor of progression in untreated stage A2 prostate cancer. Cancer 54, 1666–1671 (1984). [DOI] [PubMed] [Google Scholar]
- 36.Stringer C, Wang T, Michaelos M & Pachitariu M Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Pietzsch T, Saalfeld S, Preibisch S & Tomancak P BigDataViewer: visualization and processing for large image data sets. Nat. Methods 12, 481–483 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Hörl D et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874 (2019). [DOI] [PubMed] [Google Scholar]
- 39.van der Walt S et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King CR & Long JP Prostate biopsy grading errors:A sampling problem? Int. J. Cancer 90, 326–330 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Tibshirani R Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B Methodol 58, 267–288 (1996). [Google Scholar]
- 42.Kuhn M & Johnson K Applied predictive modeling. (Springer, 2013). [Google Scholar]
- 43.Stephenson AJ et al. Postoperative Nomogram Predicting the 10-Year Probability of Prostate Cancer Recurrence After Radical Prostatectomy. J. Clin. Oncol 23, 7005–7012 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virtanen P et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leo P et al. Computer extracted gland features from H&E predicts prostate cancer recurrence comparably to a genomic companion diagnostic test: a large multi-site study. Npj Precis. Oncol 5, 35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leo P et al. Computationally Derived Cribriform Area Index from Prostate Cancer Hematoxylin and Eosin Images Is Associated with Biochemical Recurrence Following Radical Prostatectomy and Is Most Prognostic in Gleason Grade Group 2. Eur. Urol. Focus 7, 722–732 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooperberg MR et al. Multiinstitutional validation of the UCSF cancer of the prostate risk assessment for prediction of recurrence after radical prostatectomy. Cancer 107, 2384–2391 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Newcomb LF et al. Outcomes of Active Surveillance for Clinically Localized Prostate Cancer in the Prospective, Multi-Institutional Canary PASS Cohort. J. Urol 195, 313–320 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberge CL et al. Non-Destructive Tumor Aggregate Morphology and Viability Quantification at Cellular Resolution, During Development and in Response to Drug. Acta Biomater. 117, 322–334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kingsley DM et al. Laser-based 3D bioprinting for spatial and size control of tumor spheroids and embryoid bodies. Acta Biomater. 95, 357–370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen Y et al. Quantitative analysis and comparison of 3D morphology between viable and apoptotic MCF-7 breast cancer cells and characterization of nuclear fragmentation. PLOS ONE 12, e0184726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nickerson JA Nuclear dreams: The malignant alteration of nuclear architecture. J. Cell. Biochem 70, 172–180 (1998). [PubMed] [Google Scholar]
- 53.Zink D, Fischer AH & Nickerson JA Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Uhler C & Shivashankar GV Nuclear Mechanopathology and Cancer Diagnosis. Trends Cancer 4, 320–331 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Bhargava R & Madabhushi A Emerging Themes in Image Informatics and Molecular Analysis for Digital Pathology. Annu. Rev. Biomed. Eng 18, 387–412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hveem TS et al. Chromatin changes predict recurrence after radical prostatectomy. Br. J. Cancer 114, 1243–1250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee G et al. Cell Orientation Entropy (COrE): Predicting Biochemical Recurrence from Prostate Cancer Tissue Microarrays. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013 (eds. Mori K, Sakuma I, Sato Y, Barillot C & Navab N) vol. 8151 396–403 (Springer; Berlin Heidelberg, 2013). [DOI] [PubMed] [Google Scholar]
- 58.Ali S, Veltri R, Epstein JA, Christudass C & Madabhushi A Cell cluster graph for prediction of biochemical recurrence in prostate cancer patients from tissue microarrays. in (eds. Gurcan MN & Madabhushi A) 86760H (2013). doi: 10.1117/12.2008695. [DOI] [Google Scholar]
- 59.Fischer EG Nuclear Morphology and the Biology of Cancer Cells. Acta Cytol. 64, 511–519 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Tuxhorn JA et al. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 8, 2912–2923 (2002). [PubMed] [Google Scholar]
- 61.Ayala GE et al. Determining prostate cancer-specific death through quantification of stromogenic carcinoma area in prostatectomy specimens. Am. J. Pathol 178, 79–87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Epstein JI et al. The 2019 Genitourinary Pathology Society (GUPS) White Paper on Contemporary Grading of Prostate Cancer. Arch. Pathol. Lab. Med 145, 461–493 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Lewis SM et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Berger MF & Mardis ER The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol 15, 353–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasse TM, Hollandi R & Horvath P OpSeF: Open Source Python Framework for Collaborative Instance Segmentation of Bioimages. Front. Bioeng. Biotechnol 8, 558880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mandracchia B et al. Fast and accurate sCMOS noise correction for fluorescence microscopy. Nat. Commun 11, 94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang TC et al. “Video-to-Video Synthesis.” arXiv, December 3, 2018. http://arxiv.org/abs/1808.06601. [Google Scholar]
- 68.Xie W, Reder NP, Koyuncu CF, et al. 3D pathology of prostate biopsies with biochemical recurrence outcomes: raw H&E-analog datasets and image translation-assisted segmentation in 3D (ITAS3D) datasets (PCa_Bx_3Dpathology). 2023
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Example 2D images of benign (top) and cancerous (bottom) prostate tissues imaged with 2nd-generation OTLS microscopy versus slide-based H&E histology
Figure S2. Morphological complexities of nuclei cannot be fully captured in 3D
Figure S3. Effect of median filter kernel size on segmentation results
Figure S4. Effect of preprocessing procedure on cellpose segmentation results in low signal regions
Figure S5. Sensitivity of nuclear properties and features with respect to volume threshold
Figure S6. Set of shape-based nuclear features used for prognostic analysis
Figure S7. 2D segmentation pipeline
Figure S8. Comparison of nuclear segmentation methods on manually annotated OTLS volumes
Figure S9. Comparison of the epithelial nuclear features (variance) between BCR groups
Video S1. Effect of down sampling on segmentation results
Video S2. 3D segmentation results
Video S3. Colorized segmentation results for a PCa biopsy
Data Availability Statement
The original 3D pathology datasets used in this study may be found at: https://doi.org/10.7937/44MA-GX21 [68]
The base code for the cellpose segmentation framework may be found at: https://github.com/mouseland/cellpose
The code base for the segmentation, feature extraction, and analysis pipelines may be found at: https://github.com/serrob23/OTLS-cellpose


