SUMMARY
Allogeneic hematopoietic stem cell transplantation (alloHSCT) from donors lacking C-C chemokine receptor 5 (CCR5Δ32/Δ32) can cure HIV, yet mechanisms remain speculative. To define how alloHSCT mediates HIV cure, we performed MHC-matched alloHSCT in SIV+, anti-retroviral therapy (ART)-suppressed Mauritian cynomolgus macaques (MCMs) and demonstrated that allogeneic immunity was the major driver of reservoir clearance, occurring first in peripheral blood, then peripheral lymph nodes, and finally in mesenteric lymph nodes draining the gastrointestinal tract. While allogeneic immunity could extirpate the latent viral reservoir, and did so in two alloHSCT-recipient MCMs that remained aviremic >2.5 years after stopping ART, in other cases it was insufficient without protection of engrafting cells afforded by CCR5-deficiency, as CCR5-tropic virus spread to donor CD4+ T cells despite full ART suppression. These data demonstrate the individual contributions of allogeneic immunity and CCR5-deficiency to HIV cure, and support defining targets of alloimmunity for curative strategies independent of HSCT.
Keywords: HSCT, HIV, GVHD
Graphical Abstract
eTOC
The only cases of HIV cure were achieved through CCR5-deficient allogeneic stem cell transplantation, but the precise mechanisms mediating cure remain unknown. Wu et al. use a nonhuman primate model to demonstrate that allogeneic immunity is the major driver of virus clearance following alloHSCT, but can be thwarted by viral spread to engrafting CCR5+ donor cells despite suppressive anti-retroviral therapy.
INTRODUCTION
The major barrier to cure of human and simian immunodeficiency virus (HIV/SIV) infections is the reservoir of proviruses stably integrated into the genomes of host CD4+ T cells that persist despite combination antiretroviral therapy (ART) and inevitably cause viral rebound upon ART cessation in the vast majority of individuals.1-10 However, there are four published cases of HIV cure, referred to as the Berlin, London, Düsseldorf, and New York City patients, which were achieved through allogeneic hematopoietic stem cell transplantation (alloHSCT) from CCR5Δ32/Δ32 donors, individuals homozygous for a 32 base-pair deletion within CCR5 that ablates surface expression.11-16 While alloHSCT is not a treatment suitable for widespread application, defining how this cure is achieved has profound implications for the future of the millions of people living with HIV (PLWH).
The following three potential mechanisms are hypothesized for how alloHSCT-mediated HIV cure in the four documented cases: (1) pre-transplant immune conditioning ablated the reservoir, (2) donor cell-mediated allogeneic immunity eliminated latently infected cells, and (3) the reconstituted CCR5Δ32/Δ32 immune system was resistant to HIV rebound events. Notably, the London and Düsseldorf patients received a reduced intensity conditioning (RIC) regimen prior to alloHSCT, suggesting myeloablative conditioning like that administered to the Berlin patient is not required for transplant-mediated HIV cure. Additional studies of autologous HSCT in PLWH and SIV/SHIV-infected monkeys further suggest that myeloablative conditioning without accompanying allogeneic immunity has limited impact on the viral reservoir.17-25 Therefore, while allogeneic immunity and CCR5 deficiency are likely the major determinants of HIV cure following alloHSCT, their individual mechanistic contributions remain undefined. This is due to the inherent challenges of studying HIV cure in clinical settings including low case numbers, differences in infection history and reservoir size, diverse ART and transplant regimens, limited blood and tissue availability, and the necessity of long-term ART interruption to demonstrate cure.13,14,26-31 Thus, a physiologically-relevant, pre-clinical model of alloHSCT would provide unparalleled insight into the immunological processes that clear latent HIV and result in cure, thereby opening new avenues for non-alloHSCT therapeutic curative approaches.
In order to investigate mechanisms of HSCT-mediated HIV cure in an experimentally controlled setting, we developed a clinically-relevant nonhuman primate model of fully MHC-matched alloHSCT using Mauritian cynomolgus macaques (MCM)32-34, a Macaca fascicularis population with limited MHC diversity that accurately models HIV infection and immunity.35-46 Here, we sought to define mechanisms by which alloHSCT can cure HIV by performing reduced-intensity, CCR5wt/wt alloHSCT on MCM infected with pathogenic SIVmac239 and maintained on ART. Our results define the individual contributions of allogeneic immunity and CCR5 deficiency to alloHSCT-mediated HIV cure, and support the development of HIV curative modalities designed to mimic allogeneic immune responses without the need for the complex and dangerous process of alloHSCT.
RESULTS
AlloHSCT of SIVmac239-infected Mauritian cynomolgus macaques on suppressive combination antiretroviral therapy yields durable donor engraftment
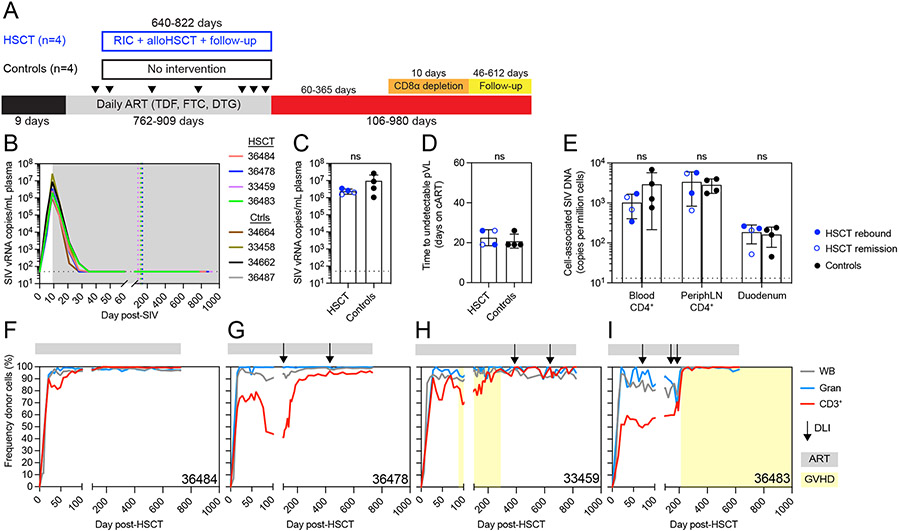
To investigate the impact of alloHSCT and its sequelae on HIV persistence in an experimentally controlled setting, we performed alloHSCT in SIV-infected, ART-suppressed MCM using reduced intensity conditioning (RIC) and peripheral blood stem cell grafts from CCR5wt/wt donors. We infected eight MCM intravenously with 100 TCID50 SIVmac239 and initiated daily ART (TDF, FTC, DTG) at day nine post-infection, resulting in peak viremia of 106-107 copies/mL at day nine post-infection followed by complete suppression of plasma viremia (<50 copies/mL) by day 35 post-infection (Figure 1A and B). After 9-17 weeks of undetectable SIV plasma viremia, four MCM underwent RIC followed by fully MHC-matched alloHSCT, a regimen previously described to achieve long-term donor engraftment 32,33 (Figure S1A and S2). The remaining four MCM received only ART and served as no-transplant controls, with each ART-only control time matched to one of the four HSCT recipients in length of ART treatment and timepoints of SIV reservoir measurements. We observed no significant differences in peak SIV plasma viral load or time to full viral suppression between HSCT recipient and no-transplant control MCM (Figure 1C and D). In addition, cell-associated SIV DNA copies in blood and peripheral lymph node CD4+ T cells and in bulk duodenum cells did not differ significantly between recipient and control MCM prior to HSCT (Figure 1E). Full suppression of SIV plasma viremia was maintained for the remainder of the ART treatment period in all eight MCM until analytical treatment interruption (ATI, day 771-918 post-SIV, day 640-822 post-HSCT for recipient macaques).
Figure 1. Reduced intensity alloHSCT in ART-suppressed SIVmac239-infected MCMs.
(A) Study outline for a cohort of alloHSCT (n=4) and time-matched no-transplant control (n=4) MCM. RIC = reduced intensity conditioning, ATI = analytic treatment interruption. Black arrows indicate biopsy timepoints. (B) Longitudinal SIVmac239 plasma viral loads from infection until ATI. Gray box denotes ART treatment. Colored dotted vertical lines indicate the day of HSCT for each recipient. Black dotted horizontal lines in B-D indicate the limit of quantification (LOQ = 50 copies/mL). Undetectable values are graphed at the LOQ. (C) Peak SIVmac239 plasma viral loads (day 9 post-infection, day of ART initiation). ns = not significantly different by Mann-Whitney test (C and D). Bars show mean ±SD (C-E). (D) Time to suppression of plasma viremia (days on ART). (E) Cell-associated SIV DNA copies in blood and tissues from HSCT recipients (blue, n=4) prior to alloHSCT and controls (black, n=4). HSCT recipients in SIV remission post-ATI are shown in open symbols. Black dotted horizontal lines indicate the LOQ for SIV DNA (13 copies/million cells). Undetectable values are graphed at 1. ns = not significantly different by repeated measures ANOVA. (F-I) Longitudinal donor chimerism in whole blood, blood granulocytes (Gran), and blood T cells (CD3+) in the alloHSCT recipients, from HSCT until ATI. Donor lymphocyte infusions (DLIs) are shown in the black arrows (see Figure S1C for doses). Gray bars above each graph denote ART treatment. Yellow boxes denote clinical GVHD. See also figures S1, S2, S3, S4.
Successful long-term donor engraftment was achieved in all four HSCT recipients as evidenced by neutrophil, lymphocyte, and platelet reconstitution (Figure S1B) and stable donor chimerism in whole blood and peripheral blood granulocytes through 600 days post-transplant (Figure 1F-I). To measure donor-mediated replacement of recipient T cells, we monitored donor chimerism in CD3+ T cells isolated from longitudinal blood samples. Blood T cells were completely donor-derived (100%) in one recipient (36484) by day 177 post-HSCT and remained >99% donor-derived throughout the remainder of the ART treatment period without the appearance of GVHD (Figure 1F). In contrast, the other three recipients exhibited mixed donor chimerism in peripheral blood T cells early post-transplant and thus we administered donor lymphocyte infusions (DLIs) to increase T cell donor chimerism (Figure S1C). One recipient, 36478, exhibited incomplete blood T cell donor chimerism despite infusion of all remaining donor cells, peaking at 96.5% donor on day 715 post-HSCT (Figure 1G). The final two recipients, 33459 and 36483, achieved complete blood T cell donor chimerism post-DLI. Recipient 33459 experienced clinical GVHD prior to DLI, but following DLI, slowly achieved full donor T cell chimerism without recurrence of GVHD (Figure 1H). In contrast, donor T cell chimerism rapidly rose from 50% to 100% in recipient 36483 following the final DLI, and was accompanied by GVHD. (Figure 1I).
We monitored for GVHD by physical examination and bloodwork as previously described.32 All four recipients were successfully tapered off tacrolimus immunosuppression between days 57 and 132 post-transplant. Recipients 36484 and 36478 did not show any signs of GVHD throughout the monitoring period, which continued past ART interruption and until euthanasia, for over 900 days post-transplant. In contrast, recipients 33459 and 36483 developed clinical GVHD. Recipient 33459 developed a skin rash at day 88 post-HSCT, 31 days after the final tacrolimus dose and prior to receiving DLI (Figure 1H). Histological assessment of a skin biopsy read by a pathologist blinded to the animal’s treatment history confirmed cutaneous GVHD, which was controlled with oral prednisone. During subsequent prednisone taper, recipient 33459 began wheezing (day 213 post-HSCT) and lung radiographs showed a diffuse bronchial pattern with tubular opacities in the caudodorsal thorax and bronchial plugs, observations consistent with bronchiolitis obliterans, a lung alloreaction associated with chronic GVHD.47 While lung GVHD was not confirmed by biopsy, bronchoalveolar lavage was negative for infectious agents and wheezing resolved after increasing the prednisone dose. Prednisone was successfully tapered for recipient 33459 in the second attempt and discontinued on day 289 post-HSCT without cutaneous or pulmonary GVHD recurrence, including after administration of DLIs on days 388 and 640 post-HSCT. Recipient 36483 developed long-term chronic GVHD, which first presented as a skin rash at day 214 post-HSCT, 28 days after receiving the third, and highest dose DLI (Figure 1H and Figure S1C). Cutaneous GVHD was confirmed by histology of a skin biopsy read by a pathologist blinded to the animal’s treatment history. Similar to 33459, cutaneous GVHD in 36483 was controlled with oral prednisone. However, multiple attempts to taper prednisone resulted in skin rash and/or pruritis recurrence at days 274, 479, and 628 post-HSCT. A final attempt to taper prednisone at day 1,175 post-HSCT resulted in hepatic GVHD characterized by elevated liver enzymes, and confirmed via histology of a liver biopsy taken on day 1289 post-HSCT (Figure S3A-B). Hepatic GVHD resolved following resumption of higher dose prednisone and re-initiation of tacrolimus, and this MCM remains on prednisone.
Latent SIV is cleared as recipient CD4+ T cells are replaced by donor cells
In order to assess engraftment across tissues and cell subsets, we measured donor chimerism in extensive sorted immune cell subsets from blood and biopsies of peripheral (axillary/inguinal) lymph nodes (LN), mesenteric LN, spleen, bone marrow, and colon taken after blood T cell donor chimerism stabilized and prior to ART interruption. We observed high donor chimerism across all tissues in NK cells, B cells, monocytes, and CD8+ T cell memory subsets, ranging from 75 to 100%, with the vast majority of immune cell populations composed of >90% donor-derived cells (Figure S4A). Bulk CD45+ immune cells and CD34+ stem cells present in bone marrow biopsies were fully donor-derived, with the exception of 36484 where ~10% of residual stem cells remained of recipient origin (Figure S4B). Examination of donor chimerism in CD4+ T cell subsets across the same tissues described above revealed that very few recipient-derived CD4+ T cells persisted in 33459, 36483, and 36484, with 95-100% of CD4+ T cells identified as donor-derived (Figure S4C). Even in 36478, the recipient that failed to reach 100% T cell donor chimerism in blood, we observed ~90% replacement of recipient CD4+ T cells by donor cells.
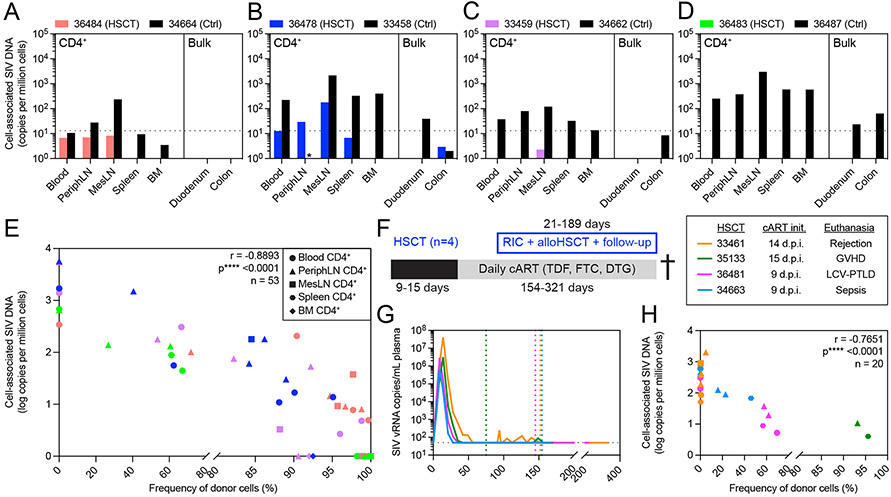
Given the high frequencies of donor-derived CD4+ T cells observed in multiple tissue sites known to harbor the latent viral reservoir, we next investigated what impact replacement of recipient CD4+ T cells had on the size of the cell-associated SIV reservoir in the same tissues described above prior to ATI. In recipient 36484 we found detectable cell-associated SIV DNA in CD4+ T cells from blood, peripheral LN, and mesenteric LN that were below the assay limit of quantification (Figure 2A). CD4+ T cell-associated SIV DNA was found at higher copy numbers and across more tissues in 36478, the recipient that never achieved full donor T cell chimerism, with the highest copy number found in CD4+ T cells isolated from mesenteric LN (Figure 2B). 36478 was also the only alloHSCT recipient with SIV DNA detectable in bulk colon biopsies. Lower SIV DNA copies were found in the tissues taken from the two alloHSCT recipients that experienced GVHD. In 33459, CD4+ T cell-associated SIV DNA was present in only one tissue with two copies measured in mesenteric LN, and no SIV DNA found in any other tissues sampled (Figure 2C). In 36483, the recipient that experienced GVHD simultaneously with achieving 100% donor chimerism following DLI and who had among the highest frequencies of donor CD4+ T cells across tissues, SIV DNA was completely undetectable across all tissues, raising the possibility of eradication of the latent viral reservoir by allogeneic immunity (Figure 2D). In contrast to the MCM that received alloHSCT, SIV DNA was readily detected across all tissues sampled in the no-transplant control MCM (Figure 2A-D), underscoring the significant reduction in the cell-associated viral reservoir size achieved by alloHSCT, particularly in recipients 33459 and 36483 with GVHD.
Figure 2. Elimination of recipient-derived CD4+ T cells after alloHSCT results in profound decreases in cell-associated SIV DNA across blood and tissue reservoirs.
(A-D) Cell-associated SIV DNA in blood and tissues from alloHSCT recipients (colored bars) and time-matched controls (black bars) from Figure 1. Black dotted horizontal lines indicate the LOQ for SIV DNA (13 copies/million cells). Bars not shown represent undetectable SIV DNA copies. Asterisks (*) indicate measurements not performed due to technical issues. (E) Correlation of CD4+ cell-associated SIV DNA copy number and CD4+ donor chimerism in blood and tissues sampled from the HSCT recipients shown in A. Symbol colors and shapes correspond to a particular recipient (see A-D) and tissue, respectively. Undetectable SIV DNA were considered 0 for this analysis. r and p-values calculated by Spearman test: *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. PeriphLN = peripheral (axillary/inguinal) lymph nodes, MesLN = mesenteric lymph nodes, BM = bone marrow. (F) Study outline for alloHSCT macaques (n=4) euthanized post-HSCT during ART treatment. RIC = reduced intensity conditioning. (G) Longitudinal SIVmac239 plasma viral loads from infection until euthanasia. Colored dotted vertical lines indicate the day of HSCT for each HSCT recipient. Black dotted horizontal line indicates the LOQ (50 copies/mL). Undetectable values are graphed at the LOQ. Macaques were maintained on daily ART from the indicated initiation timepoint until euthanasia. (H) Correlation of CD4+ T cell-associated SIV DNA copy number and CD4+ T cell donor chimerism in blood and tissues sampled from the recipients shown in F and G. Symbol colors and shapes correspond to a particular recipient macaque and tissue (see part E legend), respectively. r and p-values calculated by Spearman test using log-transformed SIV DNA copies: *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Undetectable SIV DNA were considered 1 for this analysis. See also figures S1, S2, S4.
These results suggested that the major mechanism of viral reservoir clearance after HSCT was allogeneic immunity, defined as the elimination of recipient-derived CD4+ T cells and replacement with donor cells. Thus, we next investigated the relationship between donor CD4+ T cell engraftment and latent reservoir size by simultaneously measuring both donor engraftment and SIV DNA copies in purified CD4+ T cells from 53 distinct longitudinal samples isolated from the four alloHSCT recipients. We observed a statistically significant inverse correlation between donor chimerism and cell-associated SIV DNA in CD4+ T cells from blood, peripheral LN, mesenteric LN, spleen, and bone marrow sampled longitudinally in the four HSCT recipients prior to ATI (Figure 2E). To validate this observation, we performed the same analysis using 20 longitudinal biopsies from a separate cohort of four SIVmac239-infected, ART-suppressed alloHSCT MCM recipients that were euthanized pre-ATI due to HSCT complications (Figure 2F-G). In this second cohort, we again found the same statistically significant inverse correlation between donor chimerism and cell-associated SIV DNA in CD4+ T cells isolated from various tissues (Figure 2H), further linking allogenic immunity to reservoir clearance.
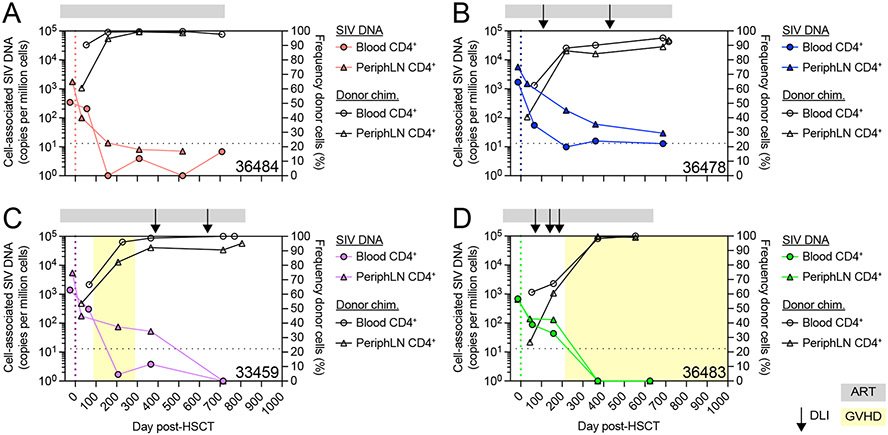
To more precisely understand the kinetics of allogeneic immunity-mediated replacement of recipient CD4+ T cells and loss of the SIV cell-associated DNA reservoir in these cells, we followed these two parameters longitudinally in the blood and peripheral LN of the four alloHSCT recipients. In every recipient, CD4+ T cell-associated SIV DNA in blood and peripheral lymph nodes decreased by ~102 SIV copies per million cells in the approximately 100 days between pre-transplant and early post-transplant measurements, which included the period of RIC and early engraftment before complete blood T cell donor chimerism (Figure 3). As expected, these early post-alloHSCT results indicated that while immune conditioning can shrink the latent viral reservoir, it is not curative. Indeed, despite this initial decrease early after transplant, a substantial viral reservoir of ~101-103 SIV DNA copies per million CD4+ T cells persisted in both blood and LN in all four MCM. Further longitudinal measurements revealed a consistent trend across the four HSCT recipient MCM, whereby increases in CD4+ T cell donor chimerism over time post-transplant were accompanied by profound decreases in CD4+ T cell-associated SIV DNA in each compartment. In 36484, the recipient that achieved full blood T cell donor chimerism but did not experience GVHD, as CD4+ T cells became fully donor-derived, CD4+ T cell-associated SIV DNA fell below the limit of quantitation, but remained present at low copy numbers (Figure 3A). In 36478, the recipient with incomplete blood T cell donor chimerism that also never experienced GVHD, quantifiable SIV DNA persisted in blood and peripheral lymph node CD4+ T cells throughout the ART treatment period (Figure 3B). In 33459 and 36483, the two recipients that achieved full blood T cell donor chimerism and experienced GVHD, post-DLI increases in CD4+ donor chimerism resulted in completely undetectable cell-associated SIV DNA in blood and peripheral LN CD4+ cells by the end of the ART treatment period (Figure 3C-D). In particular, in 36483 following the third and final DLI, cell-associated SIV DNA in both blood and lymph node CD4+ T cells dropped below the limit of detection concomitant with full donor CD4+ T cell engraftment in those compartments, both events temporally occurring with the onset of clinical GVHD as described above (Figure 3D). These results suggested that while RIC could diminish the latent viral reservoir, it was insufficient to clear it. In contrast, allogeneic immunity, as measured by active replacement of recipient CD4+ T cells by donor CD4+ T cells, was temporally associated with reductions in the reservoir, and in some cases, as seen with 33459 and 36483, led to complete or near complete loss of latently infected cells across lymphoid tissues.
Figure 3. SIV DNA persists early post-HSCT but decreases as CD4+ T cell donor chimerism increases.
(A-D) Longitudinal CD4+ T cell-associated SIV DNA copy number (colored symbols, left axis) and CD4+ T cell donor chimerism (open black symbols, right axis) in the alloHSCT recipients from Figure 1. DLIs are shown as black arrows. Colored dotted vertical lines indicate the day of HSCT. Gray bars above each graph denote ART treatment. Yellow boxes denote clinical GVHD. Black dotted horizontal lines indicate the LOQ for SIV DNA (13 copies/million cells). Undetectable SIV DNA values are graphed at 1.
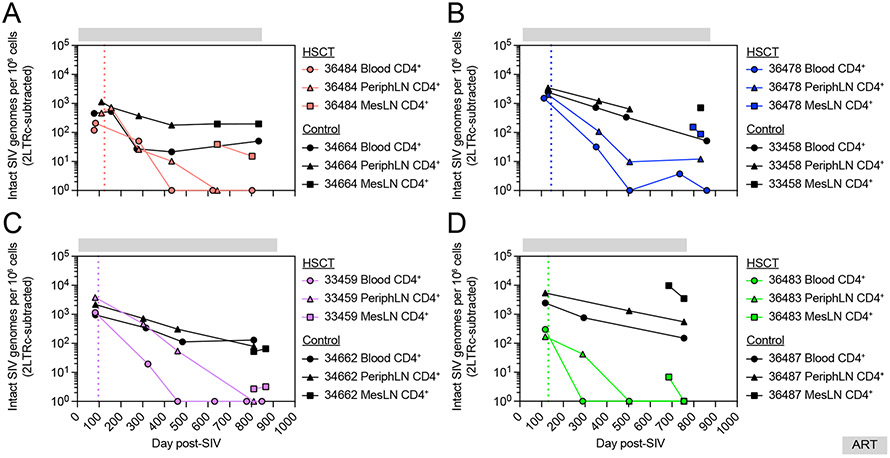
A large percentage of HIV/SIV proviruses that persist during ART harbor large deletions and hypermutations that render them incapable of producing infectious virus.48,49 In order to specifically quantify viruses capable of causing viral rebound in alloHSCT recipient and no-transplant control MCM, we performed intact proviral DNA assays (IPDA) on CD4+ T cells isolated from longitudinal blood and peripheral and mesenteric LN samples. In all four no-transplanted control MCM, intact SIV remained detectable throughout the approximately two-year ART treatment period, with substantial reservoirs of intact SIV uniformly detected across blood and peripheral and mesenteric LN prior to ART discontinuation (Figure 4). In contrast to the no-transplant control MCM, and in line with the total cell-associated SIV DNA measurements, intact SIV proviruses persisted early post-transplant in HSCT recipients and then decreased over time as CD4+ T cell donor chimerism increased. We observed an additional tissue-specific trend post-transplant, whereby intact SIV was cleared first from CD4+ T cells in peripheral blood, then from peripheral LN, and lastly from mesenteric LN. In 36484, the recipient that achieved full donor T cell engraftment without clinical GVHD, intact provirus was first lost in the peripheral blood as no intact SIV was found in peripheral blood CD4+ T cells following day 433 post-SIV, a timepoint where intact SIV persisted in peripheral LN CD4+ T cells (Figure 4A). Intact SIV in peripheral LN CD4+ T cells then disappeared, but 15 copies of intact SIV remained in mesenteric LN CD4+ T cells prior to ART release. In 36478, the recipient that never achieved full donor T cell chimerism, intact SIV was absent from peripheral blood CD4+ T cells beginning at day 506 post-SIV, yet detected in CD4+ T cells from peripheral (12 copies) and mesenteric (87 copies) LN prior to ART release (Figure 4B). In 33459, intact SIV was first lost in peripheral blood CD4+ T cells beginning at day 461 post-SIV, a timepoint when replication competent virus remained detectable in peripheral LN CD4+ T cells (Figure 4C). At the end of the ART treatment period for 33459, intact SIV was undetectable in CD4+ T cells isolated from peripheral blood and peripheral LN, and detected only at near background (2 copies) in mesenteric LN CD4+ T cells, indicating a substantial reduction in the latent reservoir by engrafting donor cells. Finally, in 36483 we also observed stepwise loss of intact SIV, occurring first in peripheral blood at day 289 post-SIV despite intact SIV remaining present in peripheral LN CD4+ T cells at that timepoint (Figure 4D). Intact SIV then became undetectable in peripheral LN CD4+ T cells, yet remained detectable in CD4+ T cells from mesenteric LNs. However, prior to ART release, no intact SIV was detectable in CD4+ T cells from any tissue sampled, including mesenteric LN, indicating that allogeneic immunity had eradicated the viral reservoir in this recipient during the engraftment period. These results further linked allogeneic immunity to clearance of latent virus and suggested that the two recipients experiencing GVHD may have been functionally (33459) or fully (36483) cured of SIV.
Figure 4. AlloHSCT mediates profound decreases in intact SIV proviruses.
Longitudinal intact SIV provirus copies in CD4+ T cells from blood and LN of alloHSCT recipients (colored symbols) and time-matched no-transplant controls (black symbols) from Figure 1. Measurements shown are corrected for double positive 2-LTR circles. Undetectable values are graphed at 0. Colored dotted vertical lines indicate the day of HSCT for each recipient macaque. Gray bars above each graph denote ART treatment. See also figure S4.
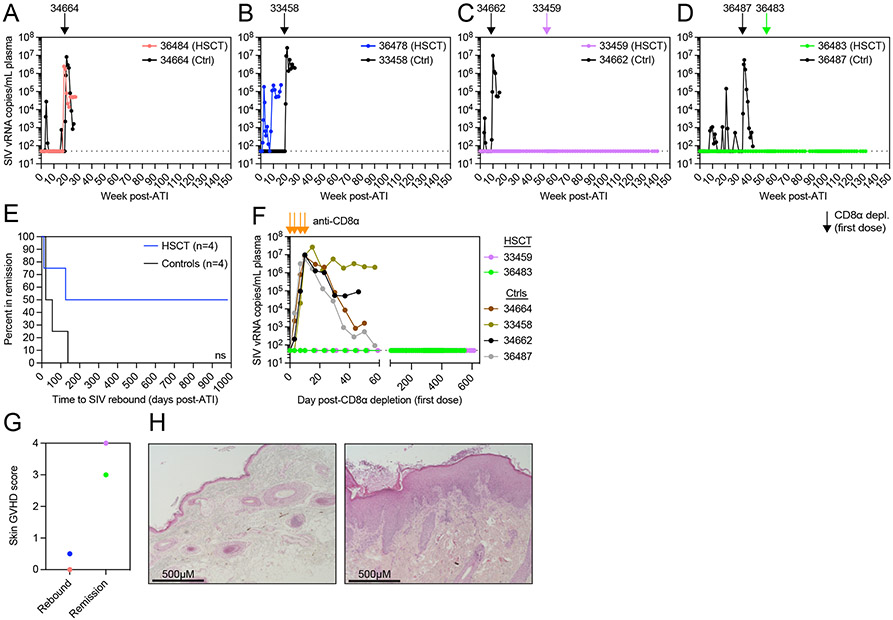
Two HSCT recipient macaques with GVHD manifest long-term ART-free SIV remission
To determine if the alloHSCT-induced reductions in the replication-competent SIV reservoir described above were sufficient to mediate long-term ART-free remission, we performed an ATI and discontinued antiretroviral treatment for all alloHSCT recipient and no-transplant control MCM between day 771 and 918 post-SIV (762-909 days on ART, 640-822 days post-HSCT for recipients). We detected SIV viremia within 8 weeks post-ATI for three of the four no-transplant control MCM (34664, 34662, 36487) that was subsequently spontaneously controlled (Figure 5A,C-D). Although we did not initially detect rebound SIV viremia in no-transplant control MCM 33458 (Figure 5B), this MCM harbored the largest cell-associated latent reservoir pre-ATI (Figure 2 and 4) and, accordingly, antibody-mediated CD8α+ cell depletion, a treatment shown to induce increases in plasma viremia through depletion of CD8α+ cell-mediated immunity and activation of CD4+ T cells50-52, resulted in plasma viremia that peaked above 107 copies/mL (Figure 5B and S5). Indeed, after antibody-mediated CD8α+ cell depletion, we observed SIV plasma viremia that peaked above 106 copies/mL in all four no-transplant control MCM, confirming the presence of replication-competent SIV (Figure 5A-D and S5). We observed distinctly different outcomes in the alloHSCT recipients. 36484, the HSCT recipient macaque that attained complete blood T cell donor chimerism without signs of GVHD, remained in ART-free SIV remission for 17 weeks, but eventually experienced SIV plasma viral rebound at week 18 post-ATI (Figure 5A). 36478, the alloHSCT recipient MCM with the lowest T cell donor chimerism and largest pre-ATI SIV reservoir, experienced SIV rebounded within 11 days of ART discontinuation (Figure 5B). In contrast, 33459 and 36483, the two recipient macaques that achieved complete blood T cell donor chimerism accompanied by GVHD, remained in aviremic ART-free SIV remission for one year after ART discontinuation (Figure 5C-D). To more stringently test for the presence of any residual replication-competent SIV in recipients 33459 and 36483, we performed antibody-mediated CD8α+ cell depletion at one year post-ATI. Despite effective depletion of CD8α+ T cells and NK cells similar to that achieved in the no-transplant control MCM, both 33459 and 36483 remained aviremic following CD8α+ cell depletion (Figure 5C-F and S5). 33459 and 36483 remain in ART-free SIV remission at the time of this report, with both surpassing 2.5 years of ART-free remission from viremia (Figure 5C-F). Finally, to examine how allogenic immunity might relate to this long-term ART-free remission observed, skin biopsies from all four alloHSCT recipients were scored by an expert in GVHD histopathology who was blinded to the treatment of the MCM.53 Comparing ATI outcomes among the four alloHSCT recipient macaques, SIV remission was associated with higher skin GVHD histopathology scores (Figure 5G-H), linking allogeneic immunity to long-term ART-free remission.
Figure 5. Long-term ART-free SIV remission despite CD8α+ cell depletion in two alloHSCT recipients.
(A-D) SIVmac239 plasma viral loads after ATI. Arrows indicate the first dose of anti-CD8α depleting antibody for alloHSCT recipients (colored) and time-matched controls (black) from Figure 1. Black dotted horizontal lines in A-D,F indicate the LOQ (50 copies/mL). Undetectable values are graphed at the LOQ. (E) Kaplan-Meier curve of time in remission. ns = not significantly different by Log-rank (Mantel-Cox) test. (F) SIVmac239 plasma viral loads during period of CD8α+ cell depletion. Orange arrows indicate doses of anti-CD8α depleting antibody. (G) Skin biopsy GVHD histopathology scores for HSCT recipients that rebounded after ATI or remain in SIV remission. Symbol colors correspond to recipients shown in A. (H) Representative images of hematoxylin and eosin stained tissue sections from HSCT recipient skin biopsies scored in Figure 5G. 36478 (left) shows minimal findings, GVHD score = 0.5. 33459 (right) shows hyperkeratosis and epidermal hyperplasia with mononuclear cell infiltrate, GVHD score = 4. Images taken with 10X objective lens (total 100X magnification). See also figure S5.
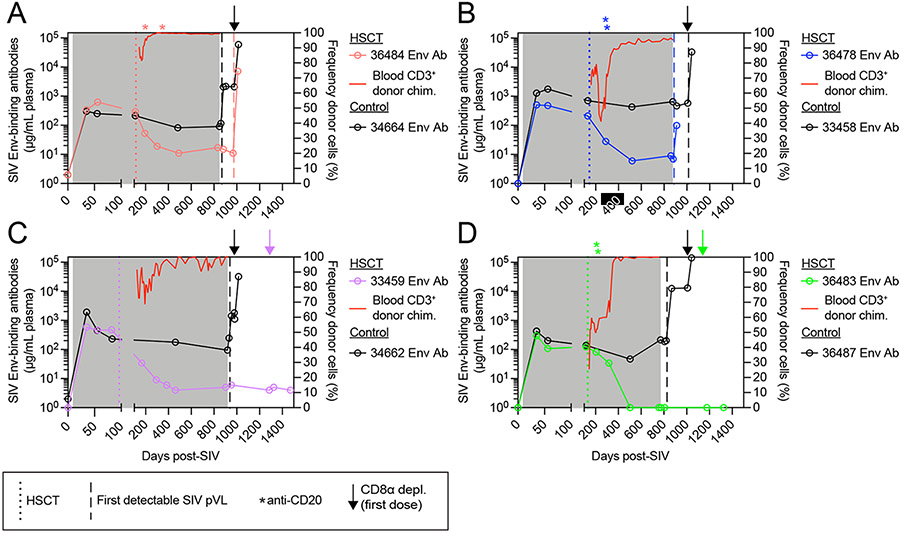
Next, we examined the impact of alloHSCT and ATI on anti-SIV immunity. In documented cases of HSCT-mediated HIV cure, anti-HIV immunity decreased post-transplant, and, importantly, did not increase after ATI, suggesting a complete lack of HIV antigenic stimulation.11,13,14 To monitor changes in anti-SIV immunity, we measured SIV Env-binding antibody titers in longitudinal plasma samples from both alloHSCT recipient and no-transplant control MCM. We observed no difference in the plasma concentration of SIV Env-binding antibodies between the four alloHSCT recipients and the no-transplant controls prior to HSCT (Figure S6A). However, SIV Env-binding antibody titers were significantly higher in no-transplant control MCM prior to ATI (Figure S6B). Thus, we longitudinally measured plasma Env-specific antibody titers in order to link their decrease in alloHSCT recipients temporally to the events pre-ATI. In all four no-transplant control MCM, plasma Env-specific antibody concentrations remained stable throughout the ART treatment period, increased post-ATI after SIV viral rebound, and then increased further concomitant with increases in SIV viremia post-CD8α depletion (Figure 6A-D). Analysis of the pre-ATI plasma SIV Env-specific antibody titers in the four no-transplant controls revealed that time to rebound directly and significantly correlated to the antibody titer (Figure S6C), indicating that the late rebound observed in control MCM 33458 might be due to its robust anti-SIV humoral immunity. In the alloHSCT recipient MCM, Env-specific antibody titers decreased post-transplant and then stabilized at low or undetectable concentrations as blood donor chimerism stabilized. Of note, recipients 36484 (Figure 6A), 36478 (Figure 6B), and 36483 (Figure 6D) received anti-CD20 depletion as treatment for post-transplant complications as described previously.33 While B cell depletion may have contributed to decreased antibody titers in these three MCM, Env-specific antibody titers were already waning prior to anti-CD20 depletion in all three recipients, and they continued to decrease after blood CD20+ B cells returned in 36483 as donor chimerism continued to increase (Figure 6D and Figure S1B). In 36483, who possessed detectable Env-specific antibodies immediately prior to administration of the third DLI, Env-specific antibodies became undetectable concomitant with achieving complete blood T cell donor chimerism and manifestation of clinical GVHD (Figure 6D). During the ATI period, Env-specific antibody titers increased in recipient macaques 36484 and 36478 after rebound of SIV plasma viremia (Figure 6A-B), but did not change in recipients 33459 and 36483 (Figure 6C-D), even after CD8α depletion, suggesting that no SIV antigen was present after ART interruption. The loss of anti-SIV humoral immunity in these two alloHSCT recipient MCM is consistent with the lack of cell-associated SIV and long-term ART-free remission despite CD8α+ cell depletion described above, which, cumulatively, support the notion that SIV was successfully extirpated by allogeneic immunity.
Figure 6. SIV Env-binding antibody titers wane in alloHSCT recipient macaques as T cell donor chimerism increases and remain low after ATI only in aviremic HSCT recipients.
(A-D) Longitudinal plasma titers of SIV Env-binding antibodies in alloHSCT recipients (left axis, colored symbols) and time-matched controls (left axis, black symbols) from Figure 1, alongside blood CD3+ T cell donor chimerism in HSCT recipients (right axis, red traces). Colored dotted vertical lines indicate the day of HSCT for each recipient macaque. Asterisks indicate anti-CD20 depleting antibody administration in alloHSCT recipients. Gray boxes denote ART treatment. Vertical dashed lines indicate the timepoint of first detectable SIV plasma viral load for alloHSCT recipients (colored) and controls (black). Arrows indicate the first dose of anti-CD8α depleting antibody for alloHSCT recipients (colored) and controls (black). See also figure S6.
Virus spreads to donor-derived cells early post-transplant despite ART
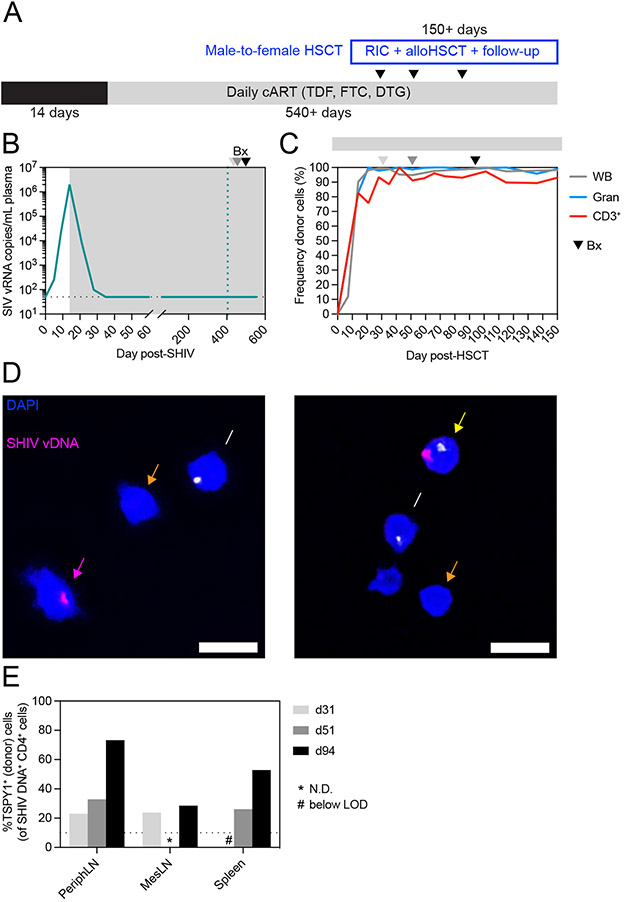
The data described above suggested that allogeneic immunity, the elimination of recipient-derived CD4+ T cells by donor cells, was the major mechanism of SIV reservoir clearance after allogeneic HSCT and could result in SIV cure even without a CCR5-deficient graft. However, cure did not occur in every case, and particularly curious was the case of recipient 36484. While recipient 36484 did not experience clinical GVHD, enough allogeneic immunity existed to eliminate the vast majority of recipient-derived CD4+ T cells, comparable to that observed in 33459 and 36483. Despite this, 36484 harbored a small reservoir of intact SIV in mesenteric lymph nodes and eventually rebounded with SIV viremia at 18 weeks post-ATI. One potential explanation is that the very small frequencies of residual recipient-derived CD4+ T cells in 36484 harbored intact SIV, which eventually reactivated and led to viral rebound. However, we also questioned whether ART had failed to confine SIV proviruses to recipient-derived cells and instead the virus had spread to donor cells post-transplant, and thus even complete elimination of every recipient-derived cell would not be curative. This would suggest that CCR5 deficiency is critically important in the peri-transplant period as lack of CCR5 on engrafting donor cells would shield them from infection while mediating allogeneic immunity via cell-to-cell contact. In order to address this question, we next sought to determine if latent virus could be found in donor-derived CD4+ T cells post-transplant. Unfortunately, no biopsy material remained from recipient 36484 for study, but because this recipient received a sex-mismatched alloHSCT, it raised the possibility that we could distinguish between donor and recipient based on the presence of the Y chromosome. Thus, we developed a microscopy assay using a DNAscope probe specific for Testis Specific Protein Y-Linked 1 (TSPY1) DNA, a sequence on the Y chromosome, and validated the ability to reliably distinguish between male and female cells using cell mixtures with defined ratios of male-to-female CD4+ T cells (Figure S7). Next, we performed an additional reduced-intensity CCR5wt/wt allogeneic HSCT of a SHIV-AD8EOM-infected female macaque using a male donor (Figure 7A). We selected SHIV-AD8EOM as this virus contains a CCR5-tropic HIV Env.54 Female macaque 38142 was infected intravenously with 10,000 TCID50 SHIV-AD8EOM and initiated the same daily ART regimen (TDF, FTC, DTG) at day 14 post-infection, which resulted in viremia closely mirroring the SIVmac239 cohort, with peak viremia of 2 x 106 copies/mL at day 14 post-infection and complete suppression of plasma viremia (<50 copies/mL) by day 35 post-infection (Figure 7B). The transplant of macaque 38142 was performed identically to the transplants of the SIVmac239 cohort, but additional biopsies were performed early post-transplant, prior to complete blood T cell donor chimerism (Figure 7C). Dual SHIV/TSPY1 DNAscope of isolated CD4+ T cells from biopsies revealed the presence of male (donor) cells harboring SHIV DNA (Figure 7D), suggesting virus had spread to donor cells despite daily ART and full suppression of plasma viremia. Indeed, the proportion of residual SHIV DNA harbored in donor-derived CD4+ T cells versus recipient CD4+ T cells increased within the peripheral LN, mesenteric LN, and spleen as donor CD4+ T cells engrafted early after alloHSCT (Figure 7E). Thus, while allogeneic immunity is the major mechanism of viral reservoir clearance post-transplant, it can be thwarted by the spread of virus to engrafting donor cells. These results highlight the importance of CCR5 deficiency to protect engrafting donor cells early following alloHSCT.
Figure 7. Virus spreads to donor CD4+ T cells despite ART.
(A) Study outline for a female HSCT recipient macaque (38142) transplanted with cells from a male donor. RIC = reduced intensity conditioning. (B) Longitudinal SHIV-AD8-EOM plasma viral loads from HSCT recipient 38142. Gray box denotes ART treatment. Colored dotted vertical line indicates the day of alloHSCT. Arrowheads in B-C denote biopsy timepoints. Black dotted horizontal line indicates LOQ (50 copies/mL). Undetectable values are graphed at the LOQ. (C) Longitudinal donor chimerism in whole blood, blood granulocytes (Gran), and blood T cells (CD3+) in HSCT recipient 38142. Gray bar above the graph denotes ART treatment. (D) Representative images from DNAscope cytospin assay of peripheral LN CD4+ cells from HSCT recipient 38142 at day 51 post-HSCT. Yellow arrow indicates an infected (male) donor cell (TSPY1+SHIV+), pink arrow indicates an infected (female) recipient cell (TSPY1−SHIV+), white arrow indicates an uninfected (male) donor cell (TSPY1+SHIV−), and orange arrow indicates an uninfected (female) recipient cell (TSPY1−SHIV−). (E) Frequency of SHIV DNA+ cells that are TSPY1+ (male). Asterisks (*) indicate measurements not determined due to insufficient cell numbers. Hashtags (#) indicate measurements below the limit of detection. See also figure S1, S2, S7.
DISCUSSION
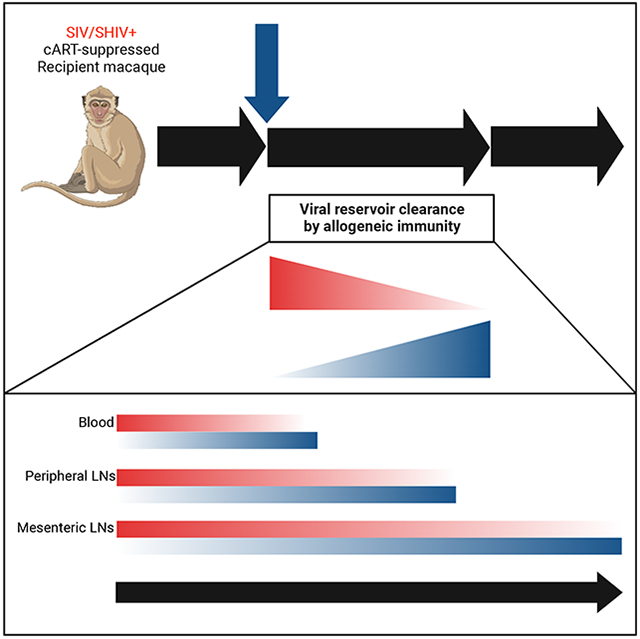
Novel strategies for HIV cure are the subject of intense research, yet CCR5Δ32/Δ32 alloHSCT remains the only approach proven capable of purging the latent viral reservoir. Defining the mechanisms by which alloHSCT achieves HIV cure has been precluded by inherent limitations of studying HIV+ individuals undergoing alloHSCT. Thus, to delineate how alloHSCT clears the latent viral reservoir, we used our physiologically-relevant pre-clinical macaque model of alloHSCT in the setting of SIV infection and long-term ART suppression. We monitored the four alloHSCT recipient MCM intensively, at least every two weeks, for four years, including longitudinal biopsies of anatomical locations like spleen and mesenteric LN that are not readily accessible in humans, thereby opening a window into the dynamic processes that impact the viral reservoir following alloHSCT. Our results link allogeneic immunity-mediated elimination of recipient-derived CD4+ T cells by donor T cells to clearance of viral reservoirs. This was most prominent in 36483, where, following the final DLI, donor T cell chimerism rapidly rose in blood from ~50% to 100%, which was accompanied temporally by full replacement of recipient CD4+ T cells in blood and peripheral LN simultaneously with clearance of cell-associated SIV in these tissues and loss of Env-specific humoral immunity. SIV remains undetectable in all tissues sampled in 36483, who is virologically and immunologically indistinguishable from an SIV-naïve MCM, suggesting this alloHSCT recipient has been cured. This result, along with the approximate thousand-fold reduction of the SIV reservoir in all four alloHSCT recipients in peripheral blood and lymphoid tissues evidences the power of graft-versus-host immunity to eliminate immunodeficiency virus reservoirs.
Our results substantiate allogeneic immunity as the major mechanism responsible for clearance of the latent viral reservoir following alloHSCT, mediating graft-versus-reservoir (GVR) effects in HIV infection similar to the beneficial effect of graft-versus-leukemia (GVL) in cancer. However, allogeneic immunity also causes the pathogenic off-target destruction of healthy cells that manifests as GVHD, and it is difficult to disentangle the two phenomena.11-14,29-31 Encouragingly, the New York City patient who received CCR5Δ32/Δ32 alloHSCT achieved HIV cure without clinical GVHD15, suggesting that GVR effects can indeed be disentangled from GVHD. Harnessing the beneficial GVL effects of allogeneic immunity while avoiding GVHD is a major goal for cancer treatment.55,56 We propose similar efforts should be focused on safely enhancing GVR in PLWH undergoing alloHSCT as well as harnessing allogeneic immunity outside of transplantation for novel approaches to HIV cure that can be applied more broadly across PLWH. Detailed characterization of alloreactive T cells from transplanted individuals, such as analysis of T cell receptors, antigen specificity, and target recognition, would be highly informative and could form the basis for new T cell therapies to HIV cure58,59. If harnessed safely, allogeneic mechanisms of HIV reservoir clearance have a distinct advantage over “shock-and-kill” cure strategies that rely on expression of viral antigens for immune recognition, which has proven difficult to universally induce across latent proviral reservoirs.57,58
Our results also demonstrate that following alloHSCT, latent virus is cleared in a stepwise fashion across anatomical compartments and potentially explains the outcome of the Boston and Minnesota patients who experienced HIV rebound following ATI, despite undetectable cell-associated HIV in peripheral blood CD4+ T cells.29-31 In particular, the rebounding virus in the Minnesota patient was phylogenetically distinct from viral sequences present in PBMC pre-transplant. Furthermore, a follow-up study of the London patient identified cell-associated HIV DNA in the axillary LN, although no intact provirus was found.14 Post-mortem analysis of HIV+ recipients who died following CCR5Δ32/Δ32 alloHSCT found no HIV DNA in PBMC, while HIV DNA was readily detected in LN.59 These outcomes are in line with our results, which suggest that future HIV cure-focused studies should monitor for viral eradication in the following phased fashion: first in peripheral blood, then in peripheral LN, and finally in mesenteric LN.
The precise role of the CCR5Δ32/Δ32 donor immune system in alloHSCT-mediated HIV cure remains undefined, but our results suggest a protective mechanism for CCR5 deficiency early during engraftment. Specifically, CCR5 deficiency protects donor CD4+ T cells from infection via cell-to-cell viral spread while the engrafting donor immune system mediates allogenic immunity and clears infected recipient CD4+ T cells. It should be noted that while the Berlin, London, Düsseldorf, and New York City patients received CCR5Δ32/Δ32 grafts, our alloHSCT recipient MCM received CCR5wt/wt grafts, similar to the Boston and Minnesota patients who underwent RIC and received CCR5wt/wt grafts.29-31 These three HIV+ alloHSCT recipients achieved full donor T cell chimerism in blood, developed GVHD, and experienced complete disappearance of cell-associated HIV in blood and a brief period of ART-free remission before HIV viremia rebounded following ATI. These three cases are remarkably similar to the outcome observed in 36484, the MCM that achieved full blood donor T cell chimerism with no intact SIV provirus detectable in peripheral blood. In particular, the timing of events following 36484’s alloHSCT closely resembled that of Boston patient A. Recipient 36484 achieved 100% blood T cell donor chimerism without DLI by day 177 post-transplant compared to day 216 post-transplant in Boston patient A, and viral remission was maintained until rebound weeks 18 and 12 post-ATI, respectively. It is unclear why the Boston and Minnesota patients were not cured despite full donor T cell chimerism and lack of detectable cell-associated HIV in peripheral blood, but the case of MCM 36484, where intact SIV provirus persisted in mesenteric LN despite CD4+ T cells being 99% donor-derived, implies the possibility of virus transfer to donor cells and maintenance of the reservoir in tissues such as the mesenteric LN that are not easily sampled in humans. Indeed, in 38142, the female alloHSCT recipient that received a sex-mismatched stem cell graft from a CCR5wt/wt donor, we documented progressive transfer of CCR5-tropic SHIV DNA to donor-derived male CD4+ T cells during the peri-transplant period in multiple tissues. Thus, we hypothesize that CCR5 deficiency is most critical early after alloHSCT, protecting donor CD4+ T cells during engraftment mediated by allogeneic immunity. Although we cannot discount a role for CCR5-deficient CD4+ T cells resisting localized HIV rebound events that subsequently extinguish themselves once the donor immune system is fully engrafted, our results suggest that lack of CCR5 is most critical during the period that allogeneic immunity is most active. A corollary of this hypothesis is that mimicking CCR5 deficiency with therapeutics that block HIV entry through CCR5 during the peri-transplant period in alloHSCT of HIV+ individuals with CCR5wt/wt donors may be sufficient to recapitulate the cure achieved in the four documented clinical cases. We recently demonstrated that the CCR5-blocking antibody Leronlimab is able to pharmacologically mimic the protective CCR5Δ32/Δ32 phenotype and sterilely protect macaques from intra-rectal challenge with CCR5-tropic SHIV, control ongoing SIV infection, and maintain multi-year suppression of HIV.60-62 Given the rarity of CCR5Δ32/Δ32, MHC-matched donors, most HIV+ individuals requiring alloHSCT must utilize a CCR5wt/wt donor. If, however, the addition of Leronlimab during the peri-transplant period successfully mimics a CCR5Δ32/Δ32 donor, then HIV cure could be attempted in any HIV+ patient undergoing allogeneic HSCT with an appropriately MHC-matched donor regardless of their CCR5 genotype. Clinical studies are currently planned to test this hypothesis63, which if successful would generate invaluable insights into the requirements of reservoir eradication and expand the pool of HIV+ individuals eligible to attempt a cure. Furthermore, given that CCR5 blockade can lower the risk of acute GVHD while preserving protective GVL effects64, this approach may help further disentangle GVHD from the reservoir clearing effects of allogeneic immunity.
Limitations of the study
There are a few important limitations to this study. First, the RIC regimens used in this study are distinct from clinical regimens due to inherent differences in drug efficacy between macaques and humans. However, every individual element of the conditioning regimen utilized here is used or is mirrored in clinical approaches, and we previously established that this model mirrors outcomes observed in clinical reduced-intensity alloHSCT.32 For example, anti-thymocyte globulin (ATG) is not effective macaques32, and thus we employed CD3-immunotoxin as an alternative method of T cell depletion. While the regimen employed here may differ slightly from those used in the clinic, clinical conditioning regimens vary among medical centers and no conditioning regimen alone, including the one described here, is sufficient to purge the HIV/SIV reservoir. Second, despite intensive sampling, these are small cohorts of alloHSCT recipient MCM. In particular, the observation of virus spread to donor cells post-transplant despite ART warrants further investigation in additional macaques infected with other viral strains. As with alloHSCT in humans, macaque alloHSCT is accompanied by clinical complications, including opportunistic infections and GVHD32,33, and poor clinical prognosis necessitates euthanasia in ~50% of transplanted macaques. Despite small case numbers, we would argue that due to the ability to rigorously sample and interrogate viral reservoir dynamics across multiple tissues and timepoints, even a single macaque alloHSCT recipient can offer insight into the biological processes involved in HSCT-mediated cure. Another caveat is the initiation of ART beginning at 9-15 days post-infection, which is earlier than most PLWH start ART. However, this timing is sufficient to fully establish a stable reservoir that consistently leads to plasma viral rebound upon ART cessation after years of treatment.65-67 Finally, the no-transplant control MCM in this study exhibited enhanced control of SIV viremia post-ATI, with one control remaining aviremic until CD8α depletion. This may be due to more effective SIV-specific immunity.68 Indeed, among control MCM, Env-specific antibody titers prior to ATI were strongly correlated with time to viral rebound. Further, SIV Env-specific antibody titers prior to ATI were higher in no-transplant control macaques compared to transplanted macaques 36484 and 36478, who manifested higher and more sustained SIV plasma viremia after viral rebound. These findings are in line with previous autologous HSCT and alloHSCT studies in rhesus and pigtailed macaques, where transplant had deleterious effects on SIV- and SHIV-specific immune responses and was associated with higher viral load during rebound compared to no-transplant control macaques.22,23,69 Despite this caveat, the ability of CD8α depletion to induce high SIV viremia in all four no-transplant control MCM, but not in alloHSCT recipients 33459 and 36483, suggests the SIV reservoir was well-established and that alloHSCT facilitated functional SIV cure in these two MCM.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jonah Sacha (sacha@ohsu.edu)
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Research animals
A total of 6 male and 13 female Mauritian-origin cynomolgus macaques (Macaca fascicularis) between 3-9 years of age at study initiation were housed at the ONPRC and utilized for studies under the approval of the Oregon Health and Science University (OHSU) West Campus Institutional Animal Care and Use Committee (IACUC). All macaques in this study were managed according to the ONPRC animal care program, which is fully accredited by AAALAC International and is based on the laws, regulations, and guidelines set forth by the United States Department of Agriculture (e.g., the Animal Welfare Act and Animal Welfare Regulations, the Guide for the Care and Use of Laboratory Animals, 8th edition (Institute for Laboratory Animal Research), and the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The nutritional plan utilized by the ONPRC is based on National Research Council recommendations and supplemented with a variety of fruits, vegetables, and other edible objects as part of the environmental enrichment program established by the Behavioral Services Unit. Animals were socially housed, when possible. All efforts were made to minimize suffering through the use of minimally invasive procedures, anesthetics, analgesics, and environmental enrichment. During the peri-transplant period, recipient macaques had indwelling subclavian catheters and were placed on a tether catheter protection system, which allowed for full range of motion as well as blood collection and intravenous drug delivery without sedation. Recipient macaques were acclimated to the tether catheter protection system prior to catheterization and subsequent transplantation. Where indicated, animals were painlessly euthanized with sodium pentobarbital followed by exsanguination and bilateral pneumothorax, consistent with the recommendations of the American Veterinary Medical Guidelines on Euthanasia (2020).
Method details
MHC typing, ABO typing, and blood and tissue processing
All macaques were MHC-typed by Illumina Miseq as previously described.32 MHC haplotypes can be found in Figure S1A. HSCT recipient and donor macaques were blood typed by PCR (GP100 assay, Zoologix). In rare cases, PCR yielded inconclusive results, and thus these macaques were typed by agglutination assays instead as previously described.34 Macaque blood and tissues were collected and processed as previously described.60,70-72 For serum chemistry, non-anticoagulated blood was centrifuged at 1860rcf for 10 minutes to separate serum from clotted blood, and then run on an ABX Pentro 400 Chemistry Analyzer. For complete blood counts, EDTA blood was run on an ABX Pentro 60C+ Hematology Analyzer. For plasma isolation, EDTA blood was centrifuged at 1860rcf for 10 minutes, and plasma was removed and clarified at 830rcf for 4 minutes. For PBMC isolation, EDTA blood was layered onto Ficoll-paque for density gradient centrifugation at 1860rcf for 30 minutes, and buffy coats were removed and washed. For bulk leukocyte isolation for sorting blood granulocytes, EDTA blood was ACK-treated and washed. Lymph node and spleen tissues were diced and mashed through 70-micron cell strainers to collect single cell suspensions. For bone marrow mononuclear cell preparations, bone marrow was pelleted, resuspended in PBS with 2mM EDTA with vigorous shaking, pelleted, resuspended in 70% isotonic Percoll, underlayed below 37% isotonic Percoll, and centrifuged at 500rcf for 20 minutes. Bone marrow buffy coats containing mononuclear cells were collected and washed. For colon leukocyte preparations, colon biopsies were incubated in R3 (RPMI160 with 3% fetal bovine serum) with 0.25 mg/mL collagenase and 0.25 mg/mL DNase for 1 hour at 37C, shaking at 250 rpm. EDTA was added for a final concentration of 6.25 mM to inactivate digestion enzymes, and contents were filtered through a metal tea strainer to collect cells in the flow thru. In cases of significant contamination by red blood cells, an additional ACK treatment step was performed for peripheral blood and bone marrow mononuclear cell and spleen cell preparations.
Virus stocks, antiretroviral therapy, and viral detection assays
The SIVmac239 stock (stock 20082, 9 x104 TCID50/mL, 2.15 x 10^8 copies/mL, 318 ng/mL p27, harvested February 5, 2011) was generated by transfection of 293T cells and subsequent infection of rhesus macaque PBMC with 293T supernatant. The SHIV-AD8-EOM stock (SP2105A_2019, 2 x105 TCID50/mL in TZMbl cells) was provided by Dr. Brandon Keele. Combination antiretroviral therapy (ART) was administered in a once-daily subcutaneous injection of tenofovir disoproxil fumarate (TDF; 5.1 mg/kg), emtricitabine (FTC; 40 mg/kg), and dolutegravir (DTG; 2.5 mg/kg) initially provided by Gilead and subsequently purchased from APIChem and formulated as described.73 Plasma and cell-associated SIV/SHIV viral loads were performed as previously described.74 Briefly, nucleic acid was extracted from plasma, tissue homogenates (bulk), or cell pellets (isolated CD4+ cells). Viral DNA/RNA copies were measured by qPCR and RT-qPCR, respectively, targeting a conserved sequence of gag. The limit of quantification was 50 copies viral RNA per mL for plasma viral loads and 13 copies viral DNA per million cells for cell-associated viral loads. Intact proviral DNA assays were performed as previously described.49 Briefly, DNA from isolated CD4+ T cells was subject to digital droplet PCR to quantify intact SIV genomes (targets in pol and env), host gene RPP30 (for input cell number quantitation and DNA shearing correction), and 2-LTR circles (for background subtraction of unintegrated viral DNA). For CD4+ cell-associated viral loads and IPDA assays, CD4+ cells were isolated from samples prior to assays using positive selection (nonhuman primate CD4 microbeads, Miltenyi Biotec).
Reduced intensity conditioning, immunosuppression, and peri-transplant care
HSCT recipient Mauritian cynomolgus macaques were transplanted with leukapheresis product from mobilized, MHC-matched unrelated donors collected and infused as previously described.32-34 Figure S1A details transplant doses of CD34+ hematopoietic stem cell progenitors and CD3+ T cells. Detailed transplant regimens for each HSCT recipient macaque studied are shown in Figure S2. Briefly, all HSCT recipient macaques except 33461 underwent reduced intensity conditioning with a previously described regimen consisting of CD3-immunotoxin (acquired through the NHP Reagent Resource), busulfan, and low-dose total body irradiation (TBI, delivered at 10-25 cGy/min using an Elektra Synergy system). Recipient macaque 33461 received CD3-immunotoxin and busulfan, but did not receive TBI. One recipient macaque, 33459, received a slightly altered regimen of CD3-immunotoxin and a single dose of anti-CD8α depleting monoclonal antibody M-T807R1 (acquired through NHP Reagent Resource) prior to HSCT. All recipient macaques received anti-GVHD prophylaxis consisting of cyclophosphamide (single dose, administered with bladder protectant MESNA), belatacept (7 doses over 31 days), and daily tacrolimus with dose adjusted to maintain a tacrolimus trough concentration of 5-15 ng/ml in whole blood (measured by immunoassay, Abbott Architect i2000). Tacrolimus was tapered as previously described.33 Enrofloxacin was administered (10 mg/kg IV, SID) when absolute neutrophil count was less than 1,000/ml of whole blood and continued until absolute neutrophil count exceeded 1,000/ml of whole blood for two consecutive complete blood counts. Some macaques received ondansetron (up to 2 mg/kg), dronabinol (up to 5 mg), and maropitant citrate (up to 2 mg/kg) to treat nausea and anorexia. Serum chemistry and complete blood counts were performed 2-3 times weekly early post-transplant, then once every 1-4 weeks or as clinically indicated. Leukoreduced whole blood or packed red blood cells were transfused when hemoglobin was less than 6 g/dL or hematocrit was less than 20%. Platelet-rich plasma was transfused when platelet counts were less than 20,000/μL blood or if signs of coagulopathy were observed on physical examination. All blood product transfusions were matched according to the ABO blood group antigen system. Animals received appropriate supportive veterinary care under the direction of an ONPRC veterinarian. Any animal suspected of being in discomfort received analgesics.
Post-transplant complications
Recipient macaques experienced reactivations of opportunistic viruses and were treated with antivirals as previously described.33 Four recipient macaques (33459, 36483, 36481, 35133) were treated with prednisone (up to 4 mg/kg/day) and/or methylprednisolone sodium succinate (up to 5 mg/kg/day) to treat suspected or confirmed GVHD. Graft rejection was suspected in one macaque, 36478, due to rapidly decreasing T cell donor chimerism (Figure 1G), and anti-CD8α depleting monoclonal antibody M-T807R1 (acquired through the NHP Reagent Resource) was administered prior to DLI #1 in an attempt to deplete alloreactive recipient-derived cells. Four recipient macaques received anti-CD20 depleting monoclonal antibody 2B8R1F8-afucosylated (acquired through the NHP Reagent Resource) due to suspicion of or confirmed lymphocryptovirus-associated post-transplant lymphoproliferative disease (LCV-PTLD, 36483, 36478, 36481) or post-transplant anemia suspected to be mediated by recipient B cells specific for “B” blood antigen on donor-derived red blood cells (36484, donor blood type AB, recipient blood type A). Four macaque recipients were euthanized prior to ART discontinuation due to graft rejection (33461), GVHD (35133), LCV-PTLD (36481), and sepsis (34663) (Figure 2F).
Donor chimerism measurements
Donor chimerism was measured by Illumina Miseq as previously described.32 Briefly, donor and recipient macaques were typed for a previously defined panel of single nucleotide polymorphisms (SNPs). To measure donor chimerism in post-transplant samples, SNPs of interest (i.e. different between donor and recipient) were amplified from extracted genomic DNA, PCR-purified, prepared for deep sequencing with NEXTflex Rapid DNA Sequencing Bundle kit, run on an Illumina Miseq with a 2x250 kit, and analyzed with Geneious. Sequencing of each sample resulted in 10,000-100,000 reads covering the SNP of interest. Donor chimerism was calculated as previously described.32 Where indicated, specific cell populations were isolated by either magnetic isolation (CD4+, CD4 NHP microbeads, Miltenyi) or flow cytometric sorting (see below) prior to gDNA extraction.
Flow cytometry assays
Blood lymphocyte frequencies were monitored by whole blood staining as previously described.32 Briefly, EDTA-treated whole blood (50–100 μL) was washed twice with PBS and stained for surface markers and viability (Live/dead Yellow Fixable, Invitrogen) at room temperature for 30 min. After staining, whole blood was resuspended in 1 ml 1X FACS Lysing Solution (BD) to lyse red blood cells and fix remaining cells, incubated at room temperature for 8 min, washed three times with 1X PBS with 10% bovine growth serum, and collected on a BD LSRII instrument. All flow cytometric analysis was performed using Flow Jo (BD). The following staining panels were used: Panel 1 (T/B cell, Figure S1B), CD45, CD3, CD20, CD4, CD8α, Live/dead fixable yellow (Life Technologies); Panel 2 (CD8α depletion period, Figure S5), CD45, CD3, CD20, CD14, CD16, CD4, CD8α (clone SK1), CD8α (clone DK25), CD8β, Live/dead fixable yellow. Unlike clone SK1, anti-CD8α clone DK25 can resolve CD8α+ cells in the presence of depleting antibody M-T807R1.75,76 Cell subsets within the live CD45+ gate were defined as follows: CD4+ T cells (CD3+, CD20−, CD4+), CD8+ T cells (CD3+, CD20−, CD4−, CD8α+), NK cells (CD3−, CD20−, CD8α+), CD8αα+ T cells (CD3+, CD20−, clone DK25 CD8α+, CD8β−), CD8αα+ T cells (CD3+, CD20−, clone DK25 CD8α+, CD8β+), CD8α+ CD16+ NK cells (CD3−, CD20−, CD45-mid/lo, clone DK25 CD8α+, CD16+), and CD8α+ CD16− NK cells (CD3−, CD20−, CD45-mid/lo, clone DK25 CD8α+, CD16−). Absolute lymphocyte counts (Figure S1B and S5) were calculated by multiplying whole blood staining frequencies within the live CD45+ gate by the white blood cell count (WBC), as determined by complete blood count (CBC) performed on an additional aliquot of EDTA-treated whole blood.
To assess donor chimerism in immune cell subsets, specific cell populations were sorted from blood and tissue cell preparations using a BD Aria or Fusion instrument as previously described.32 Briefly, cells were stained for surface markers and viability for 30 min at 4 °C and subsequently washed with cold PBS prior to sorting. The following staining panels were used: Panel 1 (longitudinal blood T cells/granulocytes, Figure 1F-I and 7C), CD45, CD3, CD20, Live/dead fixable near-IR (Life Technologies); Panel 2 (T follicular helper cells, Figure S4C), CD45, CD3, CD4, CD95, CD28, CXCR5, PD-1, Live/dead fixable near-IR; Panel 3 (NK cells, T cells, B cells, monocytes, macrophages, Figure S4A), CD45, CD3, CD14, CD20, CD8α, Live/dead fixable near-IR; Panel 4 (T cell subsets, Figure S4A and S4C), CD45, CD3, CD20, CD4, CD8α, CD28, CD95, Live/dead fixable near-IR; Panel 5 (bone marrow HSCs/T cells, Figure S4B), CD45, CD3, CD20, CD4, CD8α, CD34, Live/dead fixable near-IR; Panel 6 (colon T cells, Figure S4B), CD45, CD3, CD20, CD4, CD8α, Live/dead fixable near-IR. Immune subsets within the live singlets population were defined and sorted as follows: bulk T cells (CD45-hi, SSC-lo, CD3+, CD20−), granulocytes (CD45-lo/mid, SSC-hi, CD3−CD20−), T follicular helper cells (CD45-hi, SSC-lo, CD3+, CD4+, CD95+, PD1-hi, CXCR5-hi), NK cells (CD45-hi, SSC-lo, CD14−, CD20−, CD3−, CD8α+), B cells (CD45-hi, SSC-lo, CD14−, CD20+, CD3−), monocytes/macrophages (CD45-mid, SSC-mid/high, CD3−, CD20−, CD14+, CD8α−), bulk CD4+ T cells (CD45-hi, SSC-lo, CD3+, CD20−, CD4+, CD8α−), and bulk CD8+ T cells (CD45-hi, SSC-lo, CD3+, CD20−, CD4−, CD8α+). Memory T cell subsets within bulk CD4+ or CD8+ T cell populations were defined and sorted as follows: naive (CD95−, CD28+), central memory (CD95+, CD28+), and effector memory (CD95+, CD28−). For sorted populations, genomic DNA was extracted from 25,000-150,000 sorted cells (>95% pure) with the exception of low frequency cell populations (CD4+ T follicular helper cells, NK cells, macrophages, effector memory T cells in lymph nodes), for which gDNA was extracted from at least 2,000 cells.
GVHD scores and histology
GVHD was monitored clinically using a previously described macaque GVHD clinical staging scale.32,53 Briefly, this evaluation consisted of semi-quantitative individual scores assessing GvHD-mediated abnormalities of skin (presence and extent of skin rash), liver (extent of alterations in serum bilirubin concentration), gastrointestinal tract (presence and extent of diarrhea), and activity (animal well-being and behavior). GVHD was also monitored by histopathology using paraffin-embedded biopsy tissues. Extent of GvHD was scored on hematoxylin/eosin-stained slides by a GVHD histopathology expert blinded to the clinical history of the animals.
Post-ATI CD8α+ cell depletion
For post-ATI depletion of CD8α+ cells, macaques received four doses of anti-CD8α monoclonal antibody M-T807R1 (acquired through NHP Reagent Resource). Macaques received 10 mg/kg subcutaneously on day 0 followed by 5 mg/kg intravenously on days 3, 7, and 10. Frequencies and absolute counts of CD8α+ cell subsets were monitored as described above.
SIV Env-binding antibody endpoint ELISA
SIVmac239 Env-binding antibody concentrations were measured in longitudinal plasma samples after heat-inactivation at 56°C for 30 minutes. Plates were coated with 1.3 mg/mL of SIVmac239 gp140 trimer, washed with 1X PBS with 0.1% Triton X-100, blocked with 1X PBS with 1% normal goat serum and 5% dry milk, and dilutions of heat-inactivated plasma and SIVIG standard were added. After a one-hour incubation, plates were washed three times and a 1:5000 dilution of goat anti-human IgG HRP (Southern Biotech) was added. After another one-hour incubation, plates were washed and developed by adding TMB substrate, incubating exactly 10 minutes, and then adding 1N H2SO4. Absorbance (450nm with 650nm background subtraction) was measured on a Synergy HTX Multi-Mode Microplate Reader (BioTek). The limit of detection for this assay is 1 μg/mL.
Immunofluorescent microscopy and quantitative image analysis
CD4+ cells were isolated from samples prior to assays using positive selection (nonhuman primate CD4 microbeads, Miltenyi Biotec). Purified CD4+ T cells were fixed in freshly prepared neutral-buffered 4% paraformaldehyde for 2 hours, washed twice in PBS, resuspended in Cytospin collection fluid (Shandon; 6768315), and spun onto ColorFrost Plus slides (Fisher Scientific; 12-550-17). Slides were dried at room temperature and stored at 4°C until all samples were collected. Using a Leica Bond Rx, the carbowax was removed with 100% ethanol and heat induced epitope retrieval (HIER) was performed with ER2 (Leica; AR9640) at 97°C for 25 min. SIVmac239-sense DNAscope (Advanced Cell Diagnostics (ACD); 314078), targeting integrated SIV/SHIV vDNA, was performed per the standard Leica Bond RNAscope protocol using an RNAscope 2.5 HD Detection kit-BROWN (ACD; 322100) and developed using tyramide Alexa Fluor 568 (Invitrogen; B40956). To remove the in situ amplification tree and/or inactivate HRP, slides were gently boiled in citrate pH 6 retrieval buffer for 5 minutes. Slides were incubated with probe TSPY1 (ACD; 839311), targeting the TSPY1 gene on the Y chromosome, for ten hours at 40°C and amplification was performed with RNAscope 2.5 HD Detection kit-BROWN (ACD; 322310) using 0.5X wash buffer (ACD; 310091). Slides were developed with Alexa-fluor 647 conjugated tyramide (Invitrogen; B40958), counterstained with DAPI (Invitrogen; D1306) at a dilution of 0.5ug/mL in ddH2O for 10 minutes, cover slipped using Prolong Gold antifade mounting media (Invitrogen; P36930), and scanned as Z-stacks at 20x using a Zeiss AxioScan Z1. The multiplex DNAscope images were analyzed for the presence of SIV vDNA and TSPY1 using the FISH module (v3.0.4) within Halo software (v3.2.1851.393; Indica Labs). The entire viewable cytospin, containing at least 2 x 104 cells, was quantified for SIV vDNA and/or TSPY1 and the results were manually reviewed to ensure accurate binning. The TSPY1 false positive rate was 0.48%, and the SIV vDNA false positive rate was 0.12%, as determined across 5 slides containing >2.5 x 105 cells.
QUANTIFICATION AND STATISTICAL ANALYSIS
Group differences in peak SIV plasma viral loads, time to full suppression of plasma viremia on daily ART, and SIV Env-binding antibody titers were evaluated by non-parametric Mann-Whitney test. Group differences in cell-associated SIV DNA copy numbers across tissues were evaluated by repeated measures ANOVA of log-transformed data. Correlations between CD4+ donor chimerism and cell-associated SIV DNA copy numbers were evaluated by Spearman test using log-transformed cell-associated SIV DNA copy numbers. For log transformation, a constant value of 1 was added to each value prior to log transformation in order to include values of zero in the analysis. Correlation between SIV-Env binding antibody titers and time to rebound of SIV plasma viremia post-ATI was evaluated by Pearson test. All statistical analyses were performed using Prism software (version 9.4.0) with the exception of repeated measures ANOVA, which was performed using SAS (version 9.4, PROC MIXED).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC, anti-NHP CD45, clone D058-1283 | BD Biosciences | Cat #561290; RRID: AB_10613814 |
| PE-Cy7, mouse anti-NHP CD45, clone D058-1283 | BD Biosciences | Cat #563861; RRID: AB_10612014 |
| FITC, mouse anti-human CD20, clone 2H7 | BD Biosciences | Cat #555622; RRID: AB_395988 |
| Pacific Blue, mouse anti-human CD3, clone SP34-2 | BD Biosciences | Cat #558124, RRID: AB_397044 |
| ECD, mouse anti-human CD14, clone RMO52 | Beckman Coulter | Cat #IM2707U |
| PerCP-Cy5.5, mouse anti-human CD4, clone L200 | BD Biosciences | Cat #552838, RRID: AB_394488 |
| APC-H7, mouse anti-human CD8alpha, clone SK1 | BD Biosciences | Cat #561423, RRID: AB_10682894 |
| FITC, mouse anti-human CD16, clone 3G8 | BD Biosciences | Cat #555406 |
| Pacific Blue, mouse anti-human CD8, clone DK25 | Agilent | Cat #PB98401-1 |
| Alexa Fluor 700, mouse anti-human CD3, clone SP34-2 | BD Biosciences | Cat #557917, RRID: AB_396938 |
| PerCP-Cy5.5, mouse anti-human CD8alpha, clone SK1 | BD Biosciences | Cat #341051, RRID: AB_400209 |
| PE, mouse anti-human CD8beta, clone 2ST8.5H7 | Beckman Coulter | Cat #IM2217U |
| APC-H7, mouse anti-human CD20, clone 2H7 | BD Biosciences | Cat #560853, RRID: AB_10561681 |
| PE-Cy7, mouse anti-human CD4, clone OKT4 | Biolegend | Cat #317414, RRID: AB_571959 |
| PE, mouse anti-human CD28, clone 28.2 | BD Biosciences | Cat #556622, RRID: AB_396494 |
| PE-Cy7, mouse anti-human CD95, clone DX2 | BD Biosciences | Cat #561636, RRID: AB_10896323 |
| Pacific Blue, mouse anti-human CD8alpha, clone RPA-T8 | BD Biosciences | Cat #558207, RRID: AB_397058 |
| FITC, mouse anti-human CD95, clone DX2 | BD Biosciences | Cat #556640, RRID: AB_396506 |
| PerCP-Cy5.5, mouse anti-human CD279/PD-1, clone EH12.2H7 | Biolegend | Cat #329914, RRID: AB_1595461 |
| Alexa Fluor 700, mouse anti-human CD4, clone L200 | BD Biosciences | Cat #560836, RRID: AB_10563410 |
| PE-Cy7, mouse anti-human CD185/CXCR5, clone MU5UBEE | eBioscience | Cat #25-9185-42, RRID: AB_2573540 |
| PE, mouse anti-human CD34, clone 561 | Biolegend | Cat #343606, RRID: AB_1732008 |
| Bacterial and Virus Strains | ||
| SIVmac239 | Lab-generated | Stock 20082 |
| SHIV-AD8-EOM | Laboratory of Dr. Brandon Keele | Stock SP2105A_2019 |
| Experimental Models: Organisms/Strains | ||
| Mauritian-origin Macaca fascicularis | Accredited vendors: Worldwide Primates, Inc., PrimeGen, LLC, or Charles River Primates, Inc. | N/A |
| Oligonucleotides | ||
| SIV/SHIV qPCR detection primers and probe: SGAG21 forward primer 5'-GTCTGCGTCATPTGGTGCATTC-3', SGAG22 reverse primer 5'-CACTAGKTGTCTCTGCACTATPTGTTTTG-3', pSGAG23 probe 5′-6-carboxyfluorescein [FAM]-CTTCPTCAGTKTGTTTCACTTTCTCTTCTGCG-black hole quencher [BHQ1]-3′ | Integrated DNA Technologies | N/A |
| MHC typing primers: MHC I 5′-GCTACGTGGACGACACG and 5′-TCGCTCTGGTTGTAGTAGC, MHC II 5′-CGCTTCGACAGCGAC and 5′-ACTCGCCGCTGCA | Integrated DNA Technologies, see reference 30 | N/A |
| Donor chimerism SNP-specific primers, see reference 30 Supplementary Table 1 for sequences | Integrated DNA Technologies | N/A |
| Software and Algorithms | ||
| FlowJo v10.8.1 | BD Biosciences | N/A |
| Prism v9.4.0 | GraphPad Software | N/A |
| Halo v3.2.1851.393 (FISH module v3.0.4) | Indica Labs | N/A |
| SAS v9.4 | SAS Institute | N/A |
| Geneious Prime 2022.1.1 | Biomatters Ltd | N/A |
Highlights.
Allogeneic immunity clears SIV reservoirs after allogeneic stem cell transplant
Post-transplant SIV reservoir clearance occurs stepwise, first in blood, then tissues
CCR5+ allogeneic HSCT can functionally cure macaques of SIV
SIV spreads to engrafting CCR5+ donor cells despite fully suppressive therapy
ACKNOWLEDGMENTS
We thank Amgen, Inc. for providing Neupogen, Bristol Myers Squibb for providing Nulojix, Gilead Sciences, Inc. for providing TDF and FTC, ViiV Healthcare for providing DTG, Brandon Keele for providing the SHIV-AD8-EOM virus, William Sutton and Nancy Haigwood for providing SIVmac239 gp140 trimer, and the ONPRC research support and animal care staff for their work with the study animals. We also wish to acknowledge the animals that contributed to this study and express our respect and gratitude for their invaluable role in this research. This work was supported by the National Institutes of Health grants AI112433 and AI129703 awarded to J.B.S. and P51 OD011092 awarded to the Oregon National Primate Research Center (ONPRC). The ONPRC Molecular Virology Support Core and Integrated Pathology Core provided support services for this research. This work was also supported by The Foundation for AIDS Research (amfAR) grant 108832 awarded to J.B.S. with support from Foundation for AIDS Immune Research (FAIR). Graphical abstract created with BioRender.com.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
Dr. Sacha has a significant financial interest in and serves on the scientific advisory board of CytoDyn, a company that may have a financial interest in the results of this research and technology. This potential individual conflict of interest has been reviewed and managed by OHSU.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. (1997). Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science 278, 1295–1300. 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Chun T-W, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, and Fauci AS (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc National Acad Sci 94, 13193–13197. 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. (1997). Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188. 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 4.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. (1999). Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5, 512–517. 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 5.Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. (1999). HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. P Natl Acad Sci Usa 96, 15109–15114. 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, and Swanstrom R (2016). HIV reservoirs: what, where and how to target them. Nat Rev Microbiol 14, 55–60. 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 7.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel M-R, Ghattas G, Brenchley JM, et al. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15, 893–900. 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, et al. (2015). Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. P Natl Acad Sci Usa 112, E1126–34. 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, Zink MC, and Clements JE (2009). A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J Virol 83, 9247–9257. 10.1128/jvi.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, Li M, Clements JE, and Siliciano RF (2003). Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol 77, 4938–4949. 10.1128/jvi.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hütter G, Nowak D, Mossner M, Ganepola S, Müßig A, Allers K, Schneider T, Hofmann J, Kücherer C, Blau O, et al. (2009). Long-Term Control of HIV by CCR5 Delta32/Delta32 Stem-Cell Transplantation. New Engl J Medicine 360, 692–698. 10.1056/nejmoa0802905. [DOI] [PubMed] [Google Scholar]
- 12.Allers K, Hütter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, and Schneider T (2011). Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117, 2791–2799. 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 13.Gupta RK, Abdul-Jawad S, McCoy LE, Mok HP, Peppa D, Salgado M, Martinez-Picado J, Nijhuis M, Wensing AMJ, Lee H, et al. (2019). HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 568, 244–248. 10.1038/S41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RK, Peppa D, Hill AL, Gálvez C, Salgado M, Pace M, McCoy LE, Griffith SA, Thornhill J, Alrubayyi A, et al. (2020). Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet Hiv 7, e340–e347. 10.1016/s2352-3018(20)30069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu J, Besien KV, Glesby MJ, Pahwa S, Coletti A, Warshaw MG, Petz L, Moore TB, Chen YH, Pallikkuth S, et al. (2023). HIV-1 remission and possible cure in a woman after haplo-cord blood transplant. Cell 186, 1115–1126.e8. 10.1016/j.cell.2023.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen B-EO, Knops E, Cords L, Lübke N, Salgado M, Busman-Sahay K, Estes JD, Huyveneers LEP, Perdomo-Celis F, Wittner M, et al. (2023). In-depth virological and immunological characterization of HIV-1 cure after CCR5Δ32/Δ32 allogeneic hematopoietic stem cell transplantation. Nat Med 29, 583–587. 10.1038/s41591-023-02213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cillo AR, Krishnan A, Mitsuyasu RT, McMahon DK, Li S, Rossi JJ, Zaia JA, and Mellors JW (2013). Plasma Viremia and Cellular HIV-1 DNA Persist Despite Autologous Hematopoietic Stem Cell Transplantation for HIV-Related Lymphoma. Jaids J Acquir Immune Defic Syndromes 63, 438–441. 10.1097/qai.0b013e31828e6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Resino S, Prez A, Seoane E, Serrano D, Berenguer J, Balsalobre P, Gomz-Chacon GF, Dez-Martin JL, and Muoz-Fernndez M. ngeles (2007). Short Communication Immune Reconstitution after Autologous Peripheral Blood Stem Cell Transplantation in HIV-Infected Patients Might Be Better Than Expected. Aids Res Hum Retrov 23, 543–548. 10.1089/aid.2006.0071. [DOI] [PubMed] [Google Scholar]
- 19.Simonelli C, Zanussi S, Pratesi C, Rupolo M, Talamini R, Caffau C, Bortolin MT, Tedeschi R, Basaglia G, Mazzucato M, et al. (2010). Immune Recovery after Autologous Stem Cell Transplantation Is Not Different for HIV-Infected versus HIV-Uninfected Patients with Relapsed or Refractory Lymphoma. Clin Infect Dis 50, 1672–1679. 10.1086/652866. [DOI] [PubMed] [Google Scholar]
- 20.Cillo AR, Krishnan S, McMahon DK, Mitsuyasu RT, Para MF, and Mellors JW (2014). Impact of Chemotherapy for HIV-1 Related Lymphoma on Residual Viremia and Cellular HIV-1 DNA in Patients on Suppressive Antiretroviral Therapy. Plos One 9, e92118. 10.1371/journal.pone.0092118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavigner M, Watkins B, Lawson B, Lee ST, Chahroudi A, Kean L, and Silvestri G (2014). Persistence of Virus Reservoirs in ART-Treated SHIV-Infected Rhesus Macaques after Autologous Hematopoietic Stem Cell Transplant. Plos Pathog 10, e1004406. 10.1371/journal.ppat.1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson CW, Benne C, Polacino P, Kaur J, McAllister CE, Filali-Mouhim A, Obenza W, Pecor TA, Huang M-L, Baldessari A, et al. (2017). Loss of immune homeostasis dictates SHIV rebound after stem-cell transplantation. Jci Insight 2, e91230. 10.1172/jci.insight.91230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves DB, Peterson CW, Kiem H-P, and Schiffer JT (2017). Autologous Stem Cell Transplantation Disrupts Adaptive Immune Responses during Rebound Simian/Human Immunodeficiency Virus Viremia. J Virol 91. 10.1128/jvi.00095-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhen A, Peterson CW, Carrillo MA, Reddy SS, Youn CS, Lam BB, Chang NY, Martin HA, Rick JW, Kim J, et al. (2017). Long-term persistence and function of hematopoietic stem cell-derived chimeric antigen receptor T cells in a nonhuman primate model of HIV/AIDS. Plos Pathog 13, e1006753. 10.1371/journal.ppat.1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabarre J, Marcelin A-G, Azar N, Choquet S, Lévy V, Lévy Y, Tubiana R, Charlotte F, Norol F, Calvez V, et al. (2004). High-dose therapy plus autologous hematopoietic stem cell transplantation for human immunodeficiency virus (HIV)-related lymphoma: results and impact on HIV disease. Haematologica 89, 1100–1108. [PubMed] [Google Scholar]
- 26.Salgado M, Kwon M, Gálvez C, Badiola J, Nijhuis M, Bandera A, Balsalobre P, Miralles P, Buño I, Martinez-Laperche C, et al. (2018). Mechanisms That Contribute to a Profound Reduction of the HIV-1 Reservoir After Allogeneic Stem Cell Transplant. Ann Intern Med 169, 674. 10.7326/m18-0759. [DOI] [PubMed] [Google Scholar]
- 27.Eberhard JM, Angin M, Passaes C, Salgado M, Monceaux V, Knops E, Kobbe G, Jensen B, Christopeit M, Kröger N, et al. (2020). Vulnerability to reservoir reseeding due to high immune activation after allogeneic hematopoietic stem cell transplantation in individuals with HIV-1. Sci Transl Med 12. 10.1126/scitranslmed.aay9355. [DOI] [PubMed] [Google Scholar]
- 28.Koelsch KK, Rasmussen TA, Hey-Nguyen WJ, Pearson C, Xu Y, Bailey M, Marks KH, Sasson SC, Taylor MS, Tantau R, et al. (2017). Impact of Allogeneic Hematopoietic Stem Cell Transplantation on the HIV Reservoir and Immune Response in 3 HIV-Infected Individuals. Jaids J Acquir Immune Defic Syndromes 75, 328–337. 10.1097/qai.0000000000001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cummins NW, Rizza S, Litzow MR, Hua S, Lee GQ, Einkauf K, Chun T-W, Rhame F, Baker JV, Busch MP, et al. (2017). Extensive virologic and immunologic characterization in an HIV-infected individual following allogeneic stem cell transplant and analytic cessation of antiretroviral therapy: A case study. Plos Med 14, e1002461. 10.1371/journal.pmed.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee T-H, Robles YP, Davis BT, Li JZ, Heisey A, et al. (2014). Antiretroviral-Free HIV-1 Remission and Viral Rebound After Allogeneic Stem Cell Transplantation: Report of 2 Cases. Ann Intern Med 161, 319. 10.7326/m14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, Gallien S, Lin NH, Giguel FF, Lavoie L, et al. (2013). Long-Term Reduction in Peripheral Blood HIV Type 1 Reservoirs Following Reduced-Intensity Conditioning Allogeneic Stem Cell Transplantation. J Infect Dis 207, 1694–1702. 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burwitz BJ, Wu HL, Abdulhaqq S, Shriver-Munsch C, Swanson T, Legasse AW, Hammond KB, Junell SL, Reed JS, Bimber BN, et al. (2017). Allogeneic stem cell transplantation in fully MHC-matched Mauritian cynomolgus macaques recapitulates diverse human clinical outcomes. Nat Commun 8, 1418. 10.1038/s41467-017-01631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu HL, Weber WC, Shriver-Munsch C, Swanson T, Northrup M, Price H, Armantrout K, Robertson-LeVay M, Reed JS, Bateman KB, et al. (2020). Viral opportunistic infections in Mauritian cynomolgus macaques undergoing allogeneic stem cell transplantation mirror human transplant infectious disease complications. Xenotransplantation 27, e12578. 10.1111/xen.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu HL, Greene JM, Swanson T, Shriver-Munsch C, Armantrout K, Weber WC, Bateman KB, Maier NM, Northrup M, Legasse AW, et al. (2021). Terumo spectra optia leukapheresis of cynomolgus macaques for hematopoietic stem cell and T cell collection. J Clin Apheresis 36, 67–77. 10.1002/jca.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O’Connor SL, and O’Connor DH (2006). Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol 81, 349–361. 10.1128/jvi.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budde ML, Greene JM, Chin EN, Ericsen AJ, Scarlotta M, Cain BT, Pham NH, Becker EA, Harris M, Weinfurter JT, et al. (2012). Specific CD8+ T Cell Responses Correlate with Control of Simian Immunodeficiency Virus Replication in Mauritian Cynomolgus Macaques. J Virol 86, 7596–7604. 10.1128/jvi.00716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohns MS, Greene JM, Cain BT, Pham NH, Gostick E, Price DA, and O’Connor DH (2015). Expansion of Simian Immunodeficiency Virus (SIV)-Specific CD8 T Cell Lines from SIV-Naive Mauritian Cynomolgus Macaques for Adoptive Transfer. J Virol 89, 9748–9757. 10.1128/jvi.00993-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greene JM, Lhost JJ, Hines PJ, Scarlotta M, Harris M, Burwitz BJ, Budde ML, Dudley DM, Pham N, Cain B, et al. (2013). Adoptive transfer of lymphocytes isolated from simian immunodeficiency virus SIVmac239Δnef-vaccinated macaques does not affect acute-phase viral loads but may reduce chronic-phase viral loads in major histocompatibility complex-matched recipients. J Virol 87, 7382–7392. 10.1128/jvi.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene JM, Lhost JJ, Burwitz BJ, Budde ML, Macnair CE, Weiker MK, Gostick E, Friedrich TC, Broman KW, Price DA, et al. (2010). Extralymphoid CD8 + T Cells Resident in Tissue from Simian Immunodeficiency Virus SIVmac239Δnef-Vaccinated Macaques Suppress SIVmac239 Replication Ex Vivo. J Virol 84, 3362–3372. 10.1128/jvi.02028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burwitz BJ, Pendley CJ, Greene JM, Detmer AM, Lhost JJ, Karl JA, Piaskowski SM, Rudersdorf RA, Wallace LT, Bimber BN, et al. (2009). Mauritian Cynomolgus Macaques Share Two Exceptionally Common Major Histocompatibility Complex Class I Alleles That Restrict Simian Immunodeficiency Virus-Specific CD8 + T Cells. J Virol 83, 6011–6019. 10.1128/jvi.00199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Omange RW, Czarnecki C, Correia-Pinto JF, Crecente-Campo J, Richmond M, Li L, Schultz-Darken N, Alonso MJ, Whitney JB, et al. (2017). Mauritian cynomolgus macaques with M3M4 MHC genotype control SIVmac251 infection. J Med Primatol 46, 137–143. 10.1111/jmp.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antony JM, and MacDonald KS (2015). A critical analysis of the cynomolgus macaque, Macaca fascicularis, as a model to test HIV-1/SIV vaccine efficacy. Vaccine 33, 3073–3083. 10.1016/j.vaccine.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Lawler SH, Sussman RW, and Taylor LL (1995). Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): An example of the founder effect. Am J Phys Anthropol 96, 133–141. 10.1002/ajpa.1330960203. [DOI] [PubMed] [Google Scholar]
- 44.Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, and O’Connor DH (2010). Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62, 773–780. 10.1007/S00251-010-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, and O’Connor DH (2007). Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 59, 449–462. 10.1007/S00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malouli D, Gilbride RM, Wu HL, Hwang JM, Maier N, Hughes CM, Newhouse D, Morrow D, Ventura AB, Law L, et al. Cytomegalovirus Vaccine-induced Unconventional T cell Priming and Control of SIV Replication is Conserved Between Primate Species. Cell Host & Microbe (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chien JW, Duncan S, Williams KM, and Pavletic SZ (2010). Bronchiolitis Obliterans Syndrome After Allogeneic Hematopoietic Stem Cell Transplantation—An Increasingly Recognized Manifestation of Chronic Graft-versus-Host Disease. Biol Blood Marrow Tr 16, S106–S114. 10.1016/j.bbmt.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, et al. (2019). A novel quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125. 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bender AM, Simonetti FR, Kumar MR, Fray EJ, Bruner KM, Timmons AE, Tai KY, Jenike KM, Antar AAR, Liu P-T, et al. (2019). The Landscape of Persistent Viral Genomes in ART-Treated SIV, SHIV, and HIV-2 Infections. Cell Host Microbe 26, 73–85.e4. 10.1016/j.chom.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okoye A, Park H, Rohankhedkar M, Coyne-Johnson L, Lum R, Walker JM, Planer SL, Legasse AW, Sylwester AW, Piatak M, et al. (2009). Profound CD4+/CCR5+ T cell expansion is induced by CD8+ lymphocyte depletion but does not account for accelerated SIV pathogenesis. J Exp Medicine 206, 1575–1588. 10.1084/jem.20090356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, Kaizu M, Soma T, León EJ, MacNair C, et al. (2008). Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Medicine 205, 2537–2550. 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, León EJ, et al. (2007). Subdominant CD8 + T-Cell Responses Are Involved in Durable Control of AIDS Virus Replication. J Virol 81, 3465–3476. 10.1128/jvi.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller WP, Srinivasan S, Panoskaltsis-Mortari A, Singh K, Sen S, Hamby K, Deane T, Stempora L, Beus J, Turner A, et al. (2010). GVHD after haploidentical transplantation: a novel, MHC-defined rhesus macaque model identifies CD28− CD8+ T cells as a reservoir of breakthrough T-cell proliferation during costimulation blockade and sirolimus-based immunosuppression. Blood 116, 5403–5418. 10.1182/blood-2010-06-289272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khanal S, Fennessey CM, O’Brien SP, Thorpe A, Reid C, Immonen TT, Smith R, Bess JW, Swanstrom AE, Prete GQD, et al. (2019). In Vivo Validation of the Viral Barcoding of Simian Immunodeficiency Virus SIVmac239 and the Development of New Barcoded SIV and Subtype B and C Simian-Human Immunodeficiency Viruses. J Virol 94. 10.1128/jvi.01420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radujkovic A, Guglielmi C, Bergantini S, Iacobelli S, van Biezen A, Milojkovic D, Gratwohl A, Schattenberg AVMB, Verdonck LF, Niederwieser DW, et al. (2015). Donor Lymphocyte Infusions for Chronic Myeloid Leukemia Relapsing after Allogeneic Stem Cell Transplantation: May We Predict Graft-versus-Leukemia Without Graft-versus-Host Disease? Biol Blood Marrow Tr 21, 1230–1236. 10.1016/j.bbmt.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Lulla PD, Naik S, Vasileiou S, Tzannou I, Watanabe A, Kuvalekar M, Lulla S, Carrum G, Ramos CA, Kamble R, et al. (2021). Clinical effects of administering leukemia-specific donor T cells to patients with AML/MDS after allogeneic transplant. Blood 137, 2585–2597. 10.1182/blood.2020009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, and Siliciano RF (2013). Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell 155, 540–551. 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DIS, Keele BF, Ho Y-C, Siliciano JD, and Siliciano RF (2017). Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med 214, 959–972. 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huyveneers LEP, Bruns A, Stam A, Ellerbroek P, Jong D. de, Nagy NA, Gumbs SBH, Tesselaar K, Bosman K, Salgado M, et al. (2022). Autopsy Study Defines Composition and Dynamics of the HIV-1 Reservoir after Allogeneic Hematopoietic Stem Cell Transplantation with CCR5Δ32/Δ32 Donor Cells. Viruses 14, 2069. 10.3390/v14092069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang XL, Webb GM, Wu HL, Greene JM, Abdulhaqq S, Bateman KB, Reed JS, Pessoa C, Weber WC, Maier N, et al. (2021). Antibody-based CCR5 blockade protects Macaques from mucosal SHIV transmission. Nat Commun 12, 3343. 10.1038/s41467-021-23697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang XL, Reed JS, Webb GM, Wu HL, Le J, Bateman KB, Greene JM, Pessoa C, Waytashek C, Weber WC, et al. (2022). Suppression of human and simian immunodeficiency virus replication with the CCR5-specific antibody Leronlimab in two species. Plos Pathog 18, e1010396. 10.1371/journal.ppat.1010396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang XL, Wu HL, Webb GM, Tiwary M, Hughes C, Reed JS, Hwang J, Waytashek C, Boyle C, Pessoa C, et al. (2021). CCR5 Receptor Occupancy Analysis Reveals Increased Peripheral Blood CCR5+CD4+ T Cells Following Treatment With the Anti-CCR5 Antibody Leronlimab. Front Immunol 12, 794638. 10.3389/fimmu.2021.794638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.amfAR amfAR Partners with CytoDyn to Expand HIV Cure Strategy (press release). https://www.amfar.org/press-releases/amfar-partners-with-cytodyn-to-expand-hiv-cure-strategy/.
- 64.Reshef R, Luger SM, Hexner EO, Loren AW, Frey NV, Nasta SD, Goldstein SC, Stadtmauer EA, Smith J, Bailey S, et al. (2012). Blockade of Lymphocyte Chemotaxis in Visceral Graft-versus-Host Disease. New Engl J Medicine 367, 135–145. 10.1056/nejmoa1201248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okoye AA, Hansen SG, Vaidya M, Fukazawa Y, Park H, Duell DM, Lum R, Hughes CM, Ventura AB, Ainslie E, et al. (2018). Early antiretroviral therapy limits SIV reservoir establishment to delay or prevent post-treatment viral rebound. Nat Med 24, 1430–1440. 10.1038/s41591-018-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, et al. (2014). Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77. 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strain MC, Little SJ, Daar ES, Havlir DV, Günthard HF, Lam RY, Daly OA, Nguyen J, Ignacio CC, Spina CA, et al. (2005). Effect of Treatment, during Primary Infection, on Establishment and Clearance of Cellular Reservoirs of HIV-1. J Infect Dis 191, 1410–1418. 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- 68.Reimann KA, Parker RA, Seaman MS, Beaudry K, Beddall M, Peterson L, Williams KC, Veazey RS, Montefiori DC, Mascola JR, et al. (2005). Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J Virol 79, 8878–8885. 10.1128/jvi.79.14.8878-8885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colonna L, Peterson CW, Schell JB, Carlson JM, Tkachev V, Brown M, Yu A, Reddy S, Obenza WM, Nelson V, et al. (2018). Evidence for persistence of the SHIV reservoir early after MHC haploidentical hematopoietic stem cell transplantation. Nat Commun 9, 4438. 10.1038/S41467-018-06736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moats C, Cook K, Armantrout K, Crank H, Uttke S, Maher K, Bochart RM, Lawrence G, Axthelm MK, and Smedley JV (2022). Antimicrobial prophylaxis does not improve post-surgical outcomes in SIV/SHIV-uninfected or SIV/SHIV-infected macaques (Macaca mulatta and Macaca fascicularis) based on a retrospective analysis. Plos One 17, e0266616. 10.1371/journal.pone.0266616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smedley J, Macalister R, Wangari S, Gathuka M, Ahrens J, Iwayama N, May D, Bratt D, O’Connor M, Munson P, et al. (2016). Laparoscopic Technique for Serial Collection of Para-Colonic, Left Colic, and Inferior Mesenteric Lymph Nodes in Macaques. Plos One 11, e0157535. 10.1371/journal.pone.0157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zevin AS, Moats C, May D, Wangari S, Miller C, Ahrens J, Iwayama N, Brown M, Bratt D, Klatt NR, et al. (2017). Laparoscopic Technique for Serial Collection of Liver and Mesenteric Lymph Nodes in Macaques. J Vis Exp Jove, 55617. 10.3791/55617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitney JB, Lim S-Y, Osuna CE, Kublin JL, Chen E, Yoon G, Liu P-T, Abbink P, Borducci EN, Hill A, et al. (2018). Prevention of SIVmac251 reservoir seeding in rhesus monkeys by early antiretroviral therapy. Nat Commun 9, 5429. 10.1038/s41467-018-07881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Webb GM, Molden J, Busman-Sahay K, Abdulhaqq S, Wu HL, Weber WC, Bateman KB, Reed JS, Northrup M, Maier N, et al. (2020). The human IL-15 superagonist N-803 promotes migration of virus-specific CD8+ T and NK cells to B cell follicles but does not reverse latency in ART-suppressed, SHIV-infected macaques. Plos Pathog 16, e1008339. 10.1371/journal.ppat.1008339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitz JE, Simon MA, Kuroda MJ, Lifton MA, Ollert MW, Vogel C-W, Racz P, Tenner-Racz K, Scallon BJ, Dalesandro M, et al. (1999). A Nonhuman Primate Model for the Selective Elimination of CD8+ Lymphocytes Using a Mouse-Human Chimeric Monoclonal Antibody. Am J Pathology 154, 1923–1932. 10.1016/s0002-9440(10)65450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martins MA, Tully DC, Shin YC, Gonzalez-Nieto L, Weisgrau KL, Bean DJ, Gadgil R, Gutman MJ, Domingues A, Maxwell HS, et al. (2017). Rare Control of SIVmac239 Infection in a Vaccinated Rhesus Macaque. Aids Res Hum Retrov 33, 843–858. 10.1089/aid.2017.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.