Abstract
Telomeres and other single-stranded regions of the genome require specialized management to maintain stability and for proper progression of DNA metabolism pathways. Human Replication Protein A and CTC1-STN1-TEN1 are structurally similar heterotrimeric protein complexes that have essential ssDNA-binding roles in DNA replication, repair, and telomeres. Yeast and ciliates have related ssDNA-binding proteins with strikingly conserved structural features to these human heterotrimeric protein complexes. Recent breakthrough structures have extended our understanding of these commonalities by illuminating a common mechanism used by these proteins to act as processivity factors for their partner polymerases through their ability to manage ssDNA.
Introduction
Telomere maintenance and replication are highly coordinated processes regulated by a suite of proteins to ensure proper telomere length homeostasis, which, if dysfunctional can lead to cancers and premature aging phenotypes. The critical job of managing single-stranded DNA (ssDNA) during telomere replication is largely performed by conserved heterotrimeric protein complexes. The nucleic acid sequence, length, and structure of telomeres across organisms varies, and as a result the protein players are exquisitely tuned to recognize and manage ssDNA. Recent breakthroughs have shown that these ssDNA-management proteins are extremely structurally similar across humans, yeast, and ciliates. Moreover, these heterotrimeric protein complexes appear to act as processivity factors by a common mechanism made possible by their shared architecture. Here we explore the structural similarities and differences that define these heterotrimeric protein complexes as a specific class of proteins. Furthermore, we propose that these proteins have evolved as specialized activators and processivity factors for the enzymes responsible for replicating telomeres and the rest of the genome.
RPA-like protein complexes are essential managers of ssDNA
The protein complexes analyzed in this review are human CST (hCST) and RPA (hRPA), S. cerevisiae CST (ScCST) and RPA (ScRPA), and Tetrahymena CST (TtCST) and Teb (Figure 1A; See figure for full names). These protein complexes are often described as RPA-like because of their similar domain structures and organization leading to shared functions.
Figure 1.
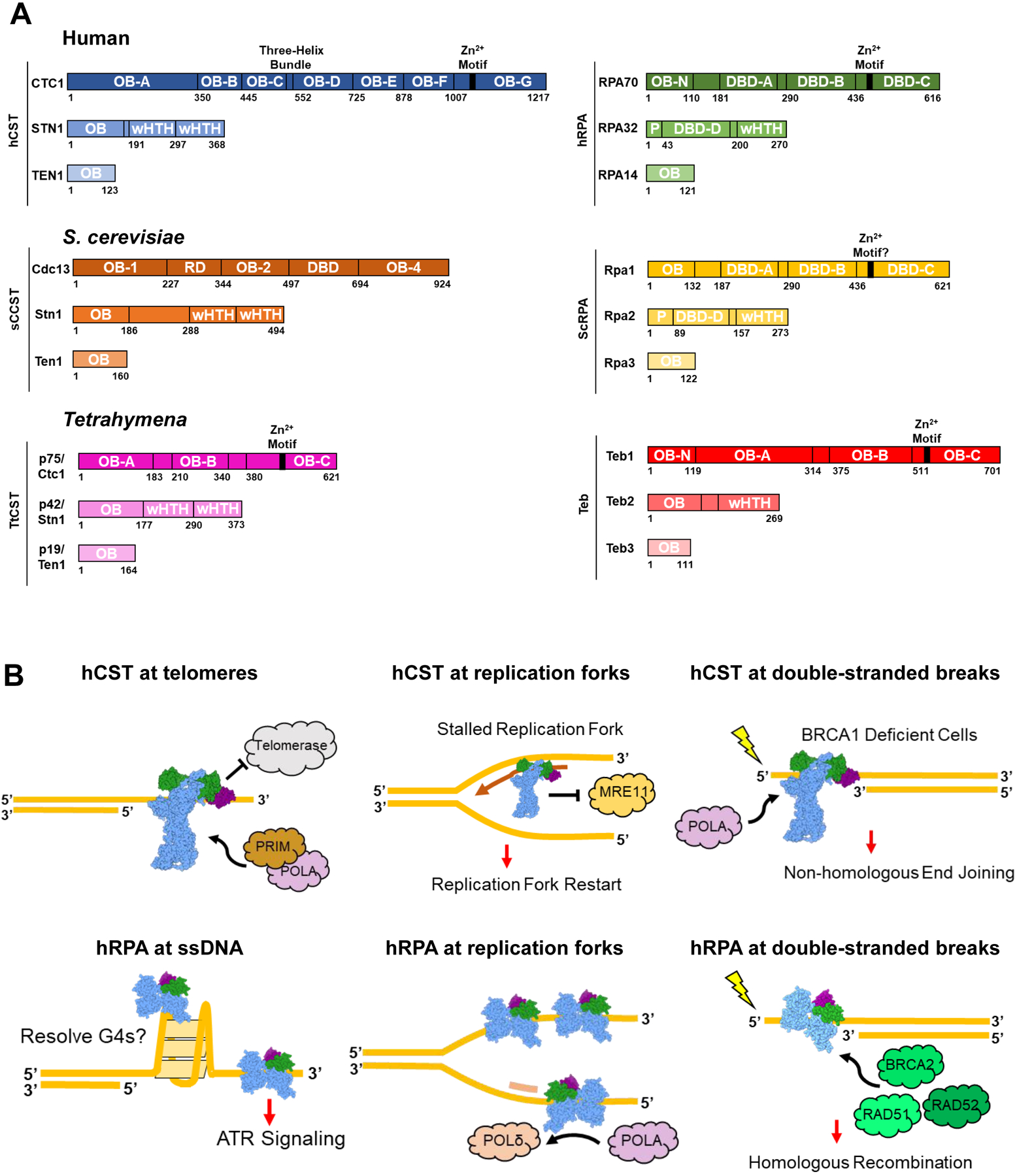
CST, RPA, and similar protein complexes across organisms. (A) Domain maps of the protein complexes explored in this review. hCST [37] in blue and hRPA [38,39] in green. ScCST [15,40,41] in orange and ScRPA [42] in yellow. TtCST [16] in pink and Teb [34,36] in red. OB = oligonucleotide/oligosaccharide binding fold; wHTH = winged helix-turn-helix; DBD = DNA-binding domain; P = phosphorylation domain; RD = recruitment domain. (B) Summary of hCST and hRPA functions at sites of ssDNA. Top left: hCST binds to the 3’ overhang of telomeres and terminates telomerase activity by blocking the binding site. hCST then facilitates the switch to C-strand fill-in by polymerase-α-primase (POLA, PRIM). Top middle: hCST aids in restarting stalled replication forks by recruiting polymerase-α-primase and blocking MRE11 from degrading nascent DNA. Top right: In BRCA1 deficient cells, hCST binds to single-stranded overhangs at double-stranded breaks which prevents end resection and facilitates fill-in by polymerase-α-primase. Ultimately this promotes non-homologous end joining as the DNA repair pathway. Bottom left: hRPA can bind and unfold G-quadruplexes as well as induce the ATR-repair pathway at long stretches of ssDNA. Bottom middle: hRPA stabilizes ssDNA at replication forks and facilitates the switch from polymerase-α-primase to polymerase-δ on the lagging strand. Bottom right: At sites of DNA damage, hRPA binds to the ssDNA and recruits repair proteins such as BRCA2, RAD51 and RAD52 which lead to the homologous recombination repair pathway.
A major activity of the CST protein complexes is to coordinate the switch between G-strand elongation by telomerase and C-strand fill-in. hCST terminates telomerase activity by binding ssDNA which occludes the binding site for telomerase to continue elongation [1,2]. Then, CST acts as a processivity factor for C-strand fill-in by polymerase-α-primase [3]. In addition to the human telomeric sequence, hCST binds other G-rich sequences across the genome [4]. Here, hCST binding to ssDNA aids in replication fork stalling events by blocking MRE11 from degrading the nascent strand, thus, promoting replication fork restart [5,6]. Loss of hCST is also implicated in overcoming Poly-(ADP)-ribose-polymerase (PARP) inhibitor resistance in BRCA1-deficient cells [7]. hCST binds the single-stranded portion of a DSB, blocking access to nucleases, thereby preventing end-resection and promoting repair via non-homologous end joining. Loss of CST restores end resection and homologous recombination as the predominant DNA repair pathway (Figure 1B) [7]. Comparatively, the ScCST complex (also known as tRPA) only is known to act at telomeres and is essential for cell viability [8–12]. ScCST recruits telomerase to the telomere, positively regulating its activity [13,14]. Like the human complex, it then facilitates the switch to C-strand fill-in by recruiting polymerase-α-primase [14,15]. It is not known if ScCST acts solely as a recruiter or by additional mechanisms to mediate telomerase and polymerase-α-primase activity. TtCst, also known as p75-p45-p19 or 7–4-1, binds telomeric ssDNA and activates polymerase-α-primase for C-strand fill-in in Tetrahymena [16,17].
RPA is highly abundant and the primary non-specific ssDNA-binding protein complex across eukaryotes. [18–20]. hRPA binding to telomeres during G2-phase has long been described as adversarial because induction of DNA-damage signaling may cause chromosome end-to-end fusions, leading to genome instability [21]. However, recently, hRPA has been shown to localize to telomeres during replication [22]. One possible beneficial function of hRPA at telomeres is to unfold G-quadruplexes which are a barrier for telomere replication [23,24]. hRPA binding also unfolds G-quadruplexes which aids in the activity of helicases that also disrupt these structures [23,24]. At sites of DNA replication across the genome, hRPA is important for recruiting replication machinery and facilitating the switch between polymerases [25–27]. Additionally, hRPA promotes homologous recombination as the DNA repair pathway by recruiting repair proteins, such as BRCA2 which mediates the switch between RPA and RAD51 occupying the ssDNA[18,26,28–31] (Figure 1B). Like hRPA, ScRPA acts as the main ssDNA-binding protein in the cell. ScRPA stabilizes and protects exposed ssDNA and recruits important proteins to sites of DNA replication, repair and recombination pathways [32,33] In Tetrahymena, Teb is a telomere-specific RPA-like protein complex that recruits telomerase to telomeric ssDNA and, in partnership with the cofactor p50, acts as a processivity factor for telomerase elongation of telomeres [34,35]. Notably, the Tetrahymena Rpa (not included in Figure 1A) and Teb share their medium and small subunits for their full complexes, so while Teb1 is telomere-specific, Teb2 and Teb3 are not [36]. To our knowledge, this exchange is unique to Teb/ttRPA and not seen in any other CST or RPA-like complexes across these three organisms.
hCST and hRPA share a strikingly similar structural architecture
These protein complexes share extremely similar structural architecture composed of the three subunits. (Figure 2A). They are characterized by a large subunit that is composed of several oligonucleotide/oligosaccharide-binding folds (OB folds), only some of which engage with ssDNA. OB folds are common protein domains comprised of five or six beta-sheets that form a compressed beta-barrel capped by an α-helix and are canonical ssDNA-binders [43]. The large subunit of hCTC1 uniquely contains additional N-terminal OB folds that are currently functionally uncharacterized. The most C-terminal OB fold contains a zinc ribbon motif comprised of four cysteine residues in all the large subunits except Cdc13. The medium subunit contains an N-terminal OB fold, sometimes preceded by a phosphorylation domain, and either one or two C-terminal winged helix-turned-helix domains. The structures of hCST and TtCST show that the medium subunit undergoes a conformational change upon ssDNA binding where a flexible linker swings the wHTH domain(s) to contact the N-terminal region of the large subunit [16,37]. This conformational change could be important for the wHTH to be in place for important protein-protein interactions. The smallest subunit is comprised of a single OB fold. Complex formation is generally implemented through a triple-helix bundle assembled from the Cterminal helices of each of the three subunits (Figure 2B) [16,39,42]. The exception is hCST, in which STN1 mediates complex formation by C-terminal helix-helix interactions with CTC1 and TEN1, with no direct interaction between CTC1 and TEN1 [37,44]. The first structure of hCST revealed various oligomerization states with up to ten monomers forming a ring structure, termed the decamer [37]. The functional relevance of this state is still to be determined.
Figure 2.
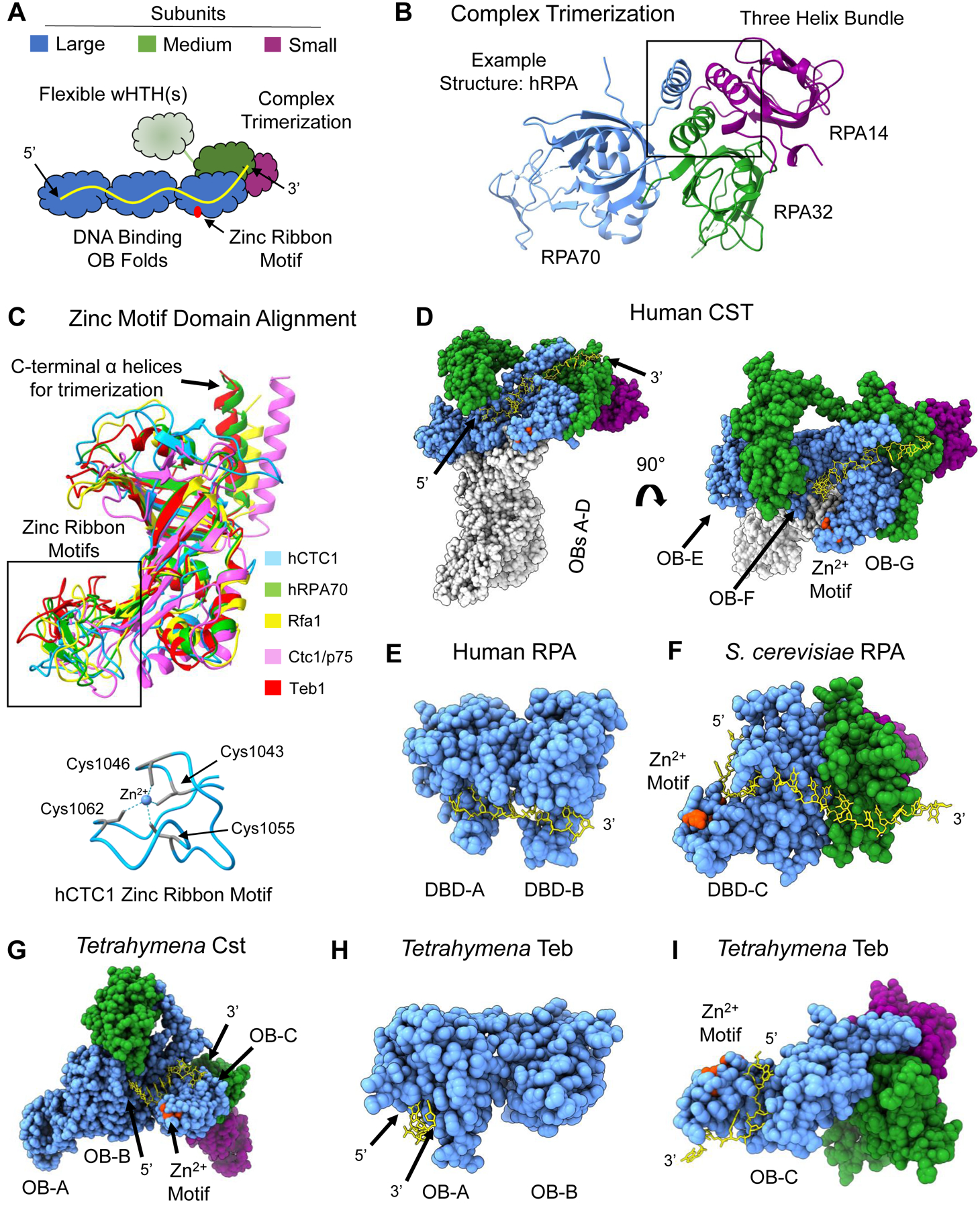
Shared structural features of RPA-like protein complexes. (A) Schematic of general architecture of RPA and RPA-like complexes. Three potential ssDNA-binding OB-folds are part of the largest subunit (blue) with a zinc ribbon motif (red) in the most C-terminal OB fold. The medium subunit (green) contains an OB fold that participates in complex trimerization with the large subunit and the smallest subunit (purple). The medium subunit contains either one or two wHTH domains that may undergo a conformational change via a flexible linker upon ssDNA (yellow) binding. (B) Three-helix bundle formation mediates trimerization between the three subunits. Example structure is from hRPA (PDB 1L1O). (C) Structure alignment of the zinc-ribbon motif containing OB folds of the five protein complexes in this figure. hCST in blue, hRPA in green, ScRPA in yellow, TtCst in pink and Teb in red. Below is the structure of the zinc ribbon motif of hCTC1 with the coordinating cysteine residues highlighted in teal with dashed lines representing interaction with the zinc ion in the center. An additional depiction of a zinc ribbon motif can be seen in RPA70 in panel B.(D) Structure of hCST bound to ssDNA (PDB 8D0B) from a face view (left) and a top view (right). The ssDNA spans OB-F and OB-G of CTC1 and the OB fold of STN1. It should be noted that this structure of hCST was solved as a co-complex with polymerase-a-primase, which is not shown here, and hCST may engage with ssDNA in a different manner without this protein-protein interaction. (E) Structure of hRPA with bound ssDNA resolved (PDB 1JMC). The ssDNA spans DBD-A and DBD-B of RPA70. The structure does not include DBD-C or either of the other subunits. (F) ScRPA bound to ssDNA (PDB 6I52). The ssDNA is bound to DBD-C of Rpa1 and the OB fold of Rpa2. (G) TtCST bound to ssDNA (PDB 7UY7). The ssDNA is bound to OB-B and OB-C. In this structure, the ssDNA does not contact Stn1 as seen in hCST. It should be noted that this structure of ttCST was solved in complex with polymerase-a-primase, which is not shown here. (H) Teb bound to ssDNA (PDB 3U58). The structure is only of OB-A and OB-B of Teb1 and depicts ssDNA in contact with OB-A alone. (I) Teb complex bound to ssDNA (PDB 6D6V). The structure only includes OB-C of Teb1 where the ssDNA is bound and the OB domain of Teb2 and Teb3. The zinc-ribbon motif is highlighted in red in D-I structures. Structure images and alignments generated in UCSF ChimeraX. Dashed lines in a structure refer to unresolved amino acids.
The zinc ribbon motif appears to be a relatively conserved structural feature of these RPA-like complexes, but the function is not fully understood (Figure 2C). Notably, the structure of S. cerevisiae Rpa1 does not show a zinc ion and a zinc ribbon motif is not described in the literature [42], however, we predict that this zinc ribbon motif does exist in Rpa1 based on structural and sequence homology. Whether the zinc ribbon motif is important for ssDNA binding is unclear; some of the ssDNA paths come near the zinc ribbon motif while others do not (Figure 2D–I). While mutagenesis data showed that deletion of the zinc ribbon motif had no effect on Teb binding ssDNA, strikingly, it was shown that the deletion drastically reduced its ability to act as a processivity factor for the telomerase enzyme [45]. It will be exciting to explore why this conserved zinc motif is important for processivity and if this is a conserved function across these proteins.
A defining function for these proteins is their ability to tightly bind ssDNA. They do so with a variety of interfaces that share some unifying features. The ssDNA always engages with the OB fold(s) of the large subunit, with the 5’ end of the DNA localized to the N-terminal region, but the number of OB folds making DNA contacts differs (Figure 2D–I) [16,42,44–47]. Some, but not all, structures show ssDNA extending to the OB fold of the medium subunit. hCST binds an 18 nucleotide (nt) telomeric ligand with a reported KD ranging between 6–22nM with mostly hydrogen bonding to both the phosphate backbone and bases [4,37,44]. TtCST binds to d(GTTGGG)5 with a KD of 180nM [16]. The side chain interactions were not resolved in this Cryo-EM structure, but mutagenesis to three charged and aromatic residues reduced the binding affinity by 5-fold [16]. hRPA and ScRPA utilize aromatic residues to form base-stacking interactions with ssDNA with single-digit nM affinity [19,42,48,49]. hRPA has been shown to be a dynamic binder, with DBDs A and B being flexible while DBDs C, D and E are more static [50]. Teb OB-A binds to dGGGT with aromatic residues via base-stacking interactions and the full-length Teb binds tightly to dGGGTTGGGGTTG with a KD of 1.8nM [35,45]. The ssDNA-interaction surfaces of these proteins have allowed them to bind telomeres and other regions of ssDNA with high affinity and finely tuned specificity.
yCST is an elaboration on this theme
Interestingly, ScCdc13 is the most divergent of the large subunits of these CST and RPA-like complexes, while the yeast Stn1 and Ten1 subunits maintain extremely similar structure to the other Stn1 and Ten1 proteins [51]. Cdc13 contains several OB folds like the other members of the family, however it binds to the heterogenous yeast telomere sequence using just a single OB fold. Remarkably, it still binds an 11-nt ssDNA nearly 1000-fold more tightly than hCST with a KD of 2 pM [41]. Cdc13 also does not contain a zinc-ribbon motif and has a unique telomerase recruitment domain downstream of the N-terminal OB fold. Lastly, yCST is the only known member of this family that acts as a dimer at telomeres [15,52].
Structural information for ScCST is available only at the domain level, and we anxiously await a full complex structure (Figure 3A). OB-1 is solved as a dimer and with a small peptide derived from the putative binding region of polymerase-α-primase [15]. OB-2 and the DBD, bound to ssDNA, have both been solved individually [53,54]. In the absence of a structure of the heterotrimer, genetic and biochemical studies have provided insights into complex trimerization and other protein-protein interactions, suggesting commonalities with other members of the family particularly with the presence of the three-helix bundle [14,55].
Figure 3.
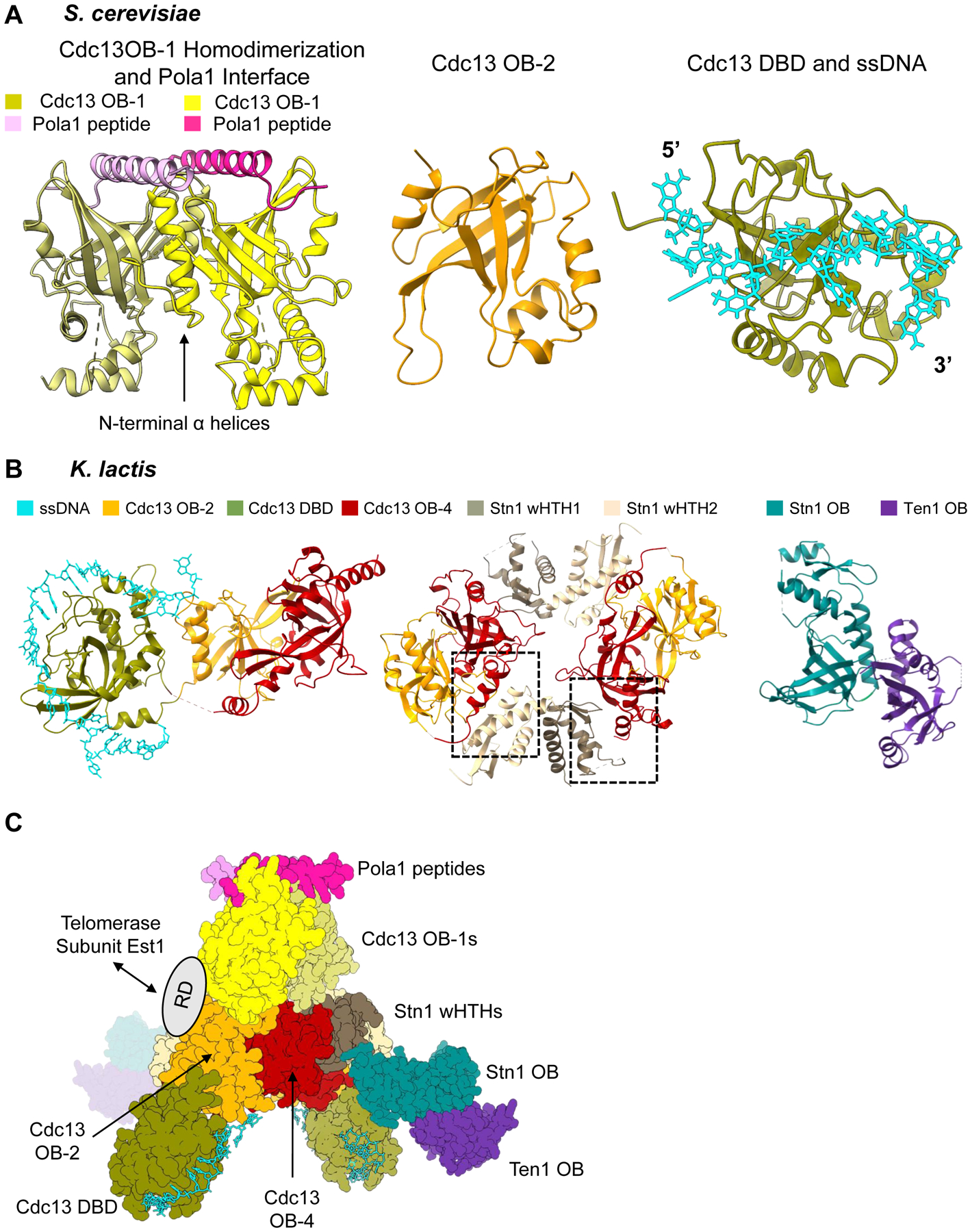
Piecing together the overall architecture of S. cerevisiae Cdc13-Stn1-Ten1. (A) Available structural information on S. cerevisiae Cdc13. Left: Dimerization of Cdc13 OB-1 and polymerase-α interaction site (PDB 3OIQ). N-terminal α-helices of OB-1 mediate the dimerization interface. The α-helix of a small polymerase-α peptide interacts with the Beta-barrel of the OB fold. Middle: OB-2 of Cdc13 (PDB 4HCE). Right: Cdc13 DBD bound to 11-nt ssDNA ligand 5’- GTGTGGGTGTG −3’ (PDB 1S40). (B) Structures of K. lactis Cdc13-Stn1-Ten1. Left: Cdc13 OB-2, DBD bound to 25-nt ssDNA ligand, and OB-4 (PDB 6LBR). The three domains form a horseshoe shape, allowing Cdc13 OB-2 and Cdc13 OB-4 to interact with each other. Middle: OB-2 and OB-4 of Cdc13 and the two winged-helix-turn-helix domains of Stn1 (PDB 6LBT). There are two unique hydrophobic interfaces (boxed) between Cdc13 OB-4 and each of the wHTHs that mediate a dimerization interaction. Right: Heterodimerization of OB fold domain of Stn1 and Ten1 (PDB 6LBU). (C) Overall architecture of Cdc13-Stn1-Ten1 proposed by Ge et al. 2020 using the solved structures of K. lactis and S. cerevisiae together. The ring dimerization interface of K. lactis Cdc13 OB-2 and OB-4 and Stn1 wHTHs is the center of the protein complex. The K. lactis DBD bound to ssDNA is then docked in based on its relationship with OB-2 and OB-4. The K. lactis Stn1 OB fold interacts with Stn1 wHTH-2 in the center ring, leaving Ten1 protruding from the center of the protein complex. The authors then used the S. cerevisiae dimerized OB-1 structure to show the possible interaction with the OB-2 domains in the ring. The polymerase-α interaction site with the OB-1 dimer is then accessible at one end of the protein complex. The flexible recruitment domain of Cdc13 would be accessible for interaction with Est1 on the outside of the dimerized protein complex. Structure images generated in UCSF ChimeraX. Dashed lines in a structure refer to unresolved amino acids.
A crystal structure of yeast CST from the distantly related Kluyveromyces lactis unexpectedly reveals a vastly different protein architecture than any other member of the CST and RPA family (Figure 3B) [56]. Using the structures of K. lactis and S. cerevisiae together, Ge et al. 2020 proposed an overall architecture for yeast CST (Figure 3C). This structure shows several interfaces that mediate dimerization, with a core ring of OB-2, OB-4 and the wHTHs of Stn1 in the center. The DBDs from each monomer protrude outward from the ring in the same direction. The interaction site between OB-1 and polymerase-α is accessible at a single pole of the protein complex. Novel interactions include the OB fold of Stn1 interacting directly with wHTH-2 in the ring and Ten1 association with the protein complex is solely mediated by interaction with Stn1 OB with no interaction with Cdc13. The recruitment domain is not resolved in any of these structures but is likely accessible for interaction with the Est1, which is a subunit of yeast telomerase, or other protein-protein interactions. A supplemental CryoEM structure would confirm this unique architecture is not due to crystal packing.
RPA-like proteins thread ssDNA into the active site of associated polymerase enzymes
Recent structures have revealed a common mechanism among RPA-like protein complexes that allow them to function as activators of polymerase enzymes by precise positioning of the 3’ end of ssDNA. Cryo-EM structures of hCST bound to polymerase-α-primase shows that hCST recruits and acts as a scaffold for polymerase-α-primase binding [44,57]. hCST first recruits polymerase-α-primase in an inactive interaction mode (Figure 4A) [57]. A transformation into a productive state is accompanied by a significant conformational change of polymerase-α-primase and the two enzymatic active sites are separated and bound to distinct sites on hCST which position them ideally for their respective activities (Figure 4B). The 5’ end of ssDNA is bound to the C-terminal OB folds of CTC1 and the OB fold of STN1. The interaction between hCST and the POLA1 subunit of polymerase-α-primase form a tunnel for the ssDNA 3’ end to thread perfectly into active site of primase (Figure 4C) [44]. The ssDNA is in position for primase to use as a template to synthesize and RNA primer before switching to DNA polymerase activity. Notably, TEN1 stabilizes the primase association with hCST which could be a conserved role for the smallest subunit of these protein complexes to participate in increasing processivity. While the first Cryo-EM structure of hCST showed it can form a complex of ten monomers in a ring structure, this oligomerization does not appear to be compatible with either hCST-polymerase-α-primase complex [37,44].
Figure 4.
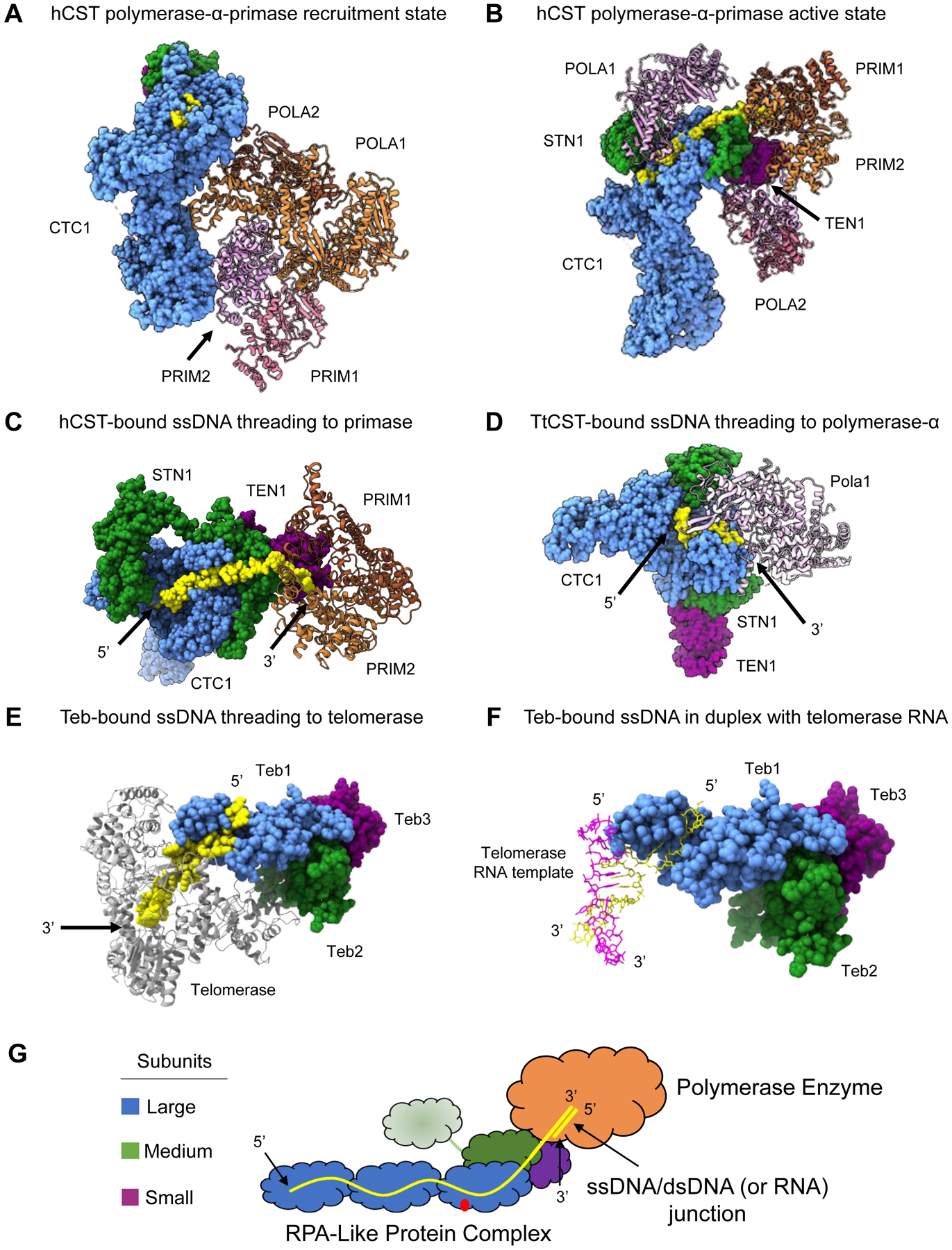
RPA-like protein complexes as processivity factors for polymerase enzymes. (A) hCST bound to polymerase-α-primase in the recruitment state. POLA1, POLA2 and PRIM2 make contacts with the OB folds of CTC1 (PDB 7U5C). (B) Polymerase-α-primase undergoes a sweeping conformational change into the active state in complex with hCST. PRIM1 and PRIM2 are stabilized by STN1 and TEN1 of hCST. POLA1 is bound to hCST in two distinct sites and POLA2 is in complex via interaction with POLA1 (PDB 8D0K). (C) hCST bound to polymerase-α-primase with telomeric ssDNA oriented to show the threading of ssDNA into the active site of primase (PDB 8D0K). This is the same complex as shown in Panel B, the POLA1 and POLA2 subunits are not shown to facilitate viewing the ssDNA (D) ttCST bound to the 5’ end telomeric ssDNA while the 3’ end is interacting with the active site of polymerase-α (PDB 7UY7). (E) ttTeb bound to telomeric ssDNA that is threaded into the active site of telomerase (PDB 6D6V). (F) ttTeb bound to telomeric ssDNA that forms a duplex with the telomerase RNA (PDB 6D6V). (G) Proposed model for how these RPA-like complexes act as a processivity factors to polymerase enzymes. The ssDNA is bound to the large subunit OB folds and may also bind the medium subunit, where the 3’ end is then threaded into the active site of the polymerase where duplex DNA may be generated by the enzyme. Structure images generated in UCSF ChimeraX. Dashed lines in a structure refer to unresolved amino acids.
The structure of TtCst bound to (GTTGGG)10 ssDNA and in complex with polymerase-α shows a similar threading mechanism. In this complex the 5’ end of ssDNA is bound across the large subunit and rather than extending to Stn1, the 3’ end is threaded into the active site of polymerase-α (Figure 4D) [16]. TtCST in complex with primase was not captured, so presumably, this structure represents a snapshot after primase had synthesized an RNA primer and polymerase-α is poised for DNA synthesis. Additionally, it was observed that TtCst is tethered to the telomerase enzyme via p50, where p50 bridges telomerase to Ctc1 [16,58]. Interestingly, in a second complex structure, Teb binds nascent telomeric ssDNA as it exits the telomerase enzyme (Figure 4E–F) [47]. Telomerase uses an RNA template to synthesize six-nucleotides of G-strand telomeric ssDNA at a time. Teb binding the exiting ssDNA could help position the new 3’ end for telomerase to continue elongation. Thus, it appears TtCst and Teb work together in Tetrahymena telomere replication, with Teb stabilizing the nascent G-strand ssDNA and TtCst then delivering the ssDNA to polymerase-α-primase for C-strand synthesis. Taken together, these structures suggest this class of protein complexes perform their role as processivity factors by threading the 3’ end of ssDNA into the active site of an enzyme (polymerase-α, primase, telomerase, etc) and stabilizing it for the enzyme to generate duplex nucleic acid (Figure 4G). If this is the common mechanism across this class of protein complexes, we would expect hRPA to act in a similar way with its interacting polymerase partners.
Conclusion
RPA-like protein complexes, including the human and yeast CSTs, have evolved as specialized polymerase cofactors at telomeres and other sites of ssDNA in the genome. Recent structures reveal that they not only share a similar structural architecture but that this architecture allows them to optimally position ssDNA into the active site of a polymerase enzyme (Figure 4G). Future work will have to determine if this threading mechanism represents how hRPA activates polymerases at sites of DNA replication, repair, and recombination. Another outstanding question is how malleable ssDNA is across the protein interaction interface and if the ssDNA can be repositioned upon polymerase binding. These structural studies reveal how a common protein architecture has created essential managers of ssDNA. All together, these contributions have provided a better understanding of how the cell maintains, replicates, and repairs its extensive genome.
Highlights.
CST and RPA share similar protein structure architecture
Human, Saccharomyces cerevisiae and Tetrahymena all have RPA-like protein complexes
RPA-like proteins use an ssDNA threading mechanism to act as processivity factors
Acknowledgements
We thank Conner Olson and Halley Steiner for careful reading and insightful comments on this manuscript. We apologize that many important contributions to this field couldn’t be addressed due to space limitations.
Funding
National Institutes of Health [R01 GM139274 to D.S.W.] and the National Science Foundation [MCB 1716425 to D.S.W.]. A.T.B. is supported by a fellowship provided by the NIH-University of Colorado Boulder (T32GM008759).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Alexandra Barbour, Declaration of Interest: none
Dr. Deborah Wuttke, Declaration of Interest: none
References
- 1.Chen L-Y, Redon S, Lingner J: The human CST complex is a terminator of telomerase activity. Nature 2012, 488:540–544. [DOI] [PubMed] [Google Scholar]
- 2.*.Zaug AJ, Lim CJ, Olson CL, Carilli MT, Goodrich KJ, Wuttke DS, Cech TR: CST does not evict elongating telomerase but prevents initiation by ssDNA binding. Nucleic Acids Res 2021, 49: 11653–11665. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study combines quantitative experimental measurement of telomerase inhibition by CST with rigorous computational modeling to unambiguously show that inhibition of telomerase by CST is mediated by competition for the ssDNA primer. In contrast, CST is incapable of inhibition ongoing telomerase action, suggesting a mechanism for the switch from telomerase extension to C-strand fill in.
- 3.*.Zaug AJ, Goodrich KJ, Song JJ, Sullivan AE, Cech TR: Reconstitution of a telomeric replicon organized by CST. Nature 2022, 608:819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides a benchmark in the reconstitution of the full replication of the telomere. Combination of telomerase-mediated extension of a telomeric primer with C-strand fill in by CST and polaprimase on telomeric templates results in an overhang structure similar to that found in cells.
- 4.Hom RA, Wuttke DS: Human CST Prefers G-Rich but Not Necessarily Telomeric Sequences. Biochemistry 2017, 56:4210–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyu X, Lei K-H, Biak Sang P, Shiva O, Chastain M, Chi P, Chai W: Human CST complex protects stalled replication forks by directly blocking MRE11 degradation of nascent-strand DNA. EMBO J 2021, 40:e103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD, Wright WE, Price CM: Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 2012, 31:3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barazas M, Annunziato S, Pettitt SJ, de Krijger I, Ghezraoui H, Roobol SJ, Lutz C, Frankum J, Song FF, Brough R, et al. : The CST Complex Mediates End Protection at Double-Strand Breaks and Promotes PARP Inhibitor Sensitivity in BRCA1-Deficient Cells. Cell Rep 2018, 23:2107–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YC, Shih JW, Hsu CL, Lin JJ: Binding and partial denaturing of G-quartet DNA by Cdc13p of Saccharomyces cerevisiae. J Biol Chem 2001, 276:47671–47674. [DOI] [PubMed] [Google Scholar]
- 9.Lin JJ, Zakian VA: The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci U S A 1996, 93:13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YC, Hsu CL, Shih JW, Lin JJ: Specific binding of single-stranded telomeric DNA by Cdc13p of Saccharomyces cerevisiae. J Biol Chem 2001, 276:24588–24593. [DOI] [PubMed] [Google Scholar]
- 11.Lin Y-C, Wu Lee Y-H, Lin J-J: Genetic analysis reveals essential and non-essential amino acids within the telomeric DNA-binding interface of Cdc13p. Biochem J 2007, 403:289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nugent CI, Hughes TR, Lue NF, Lundblad V: Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 1996, 274:249–252. [DOI] [PubMed] [Google Scholar]
- 13.Evans SK, Lundblad V: Est1 and Cdc13 as comediators of telomerase access. Science 1999, 286:117–120. [DOI] [PubMed] [Google Scholar]
- 14.Chandra A, Hughes TR, Nugent CI, Lundblad V: Cdc13 both positively and negatively regulates telomere replication. Genes Dev 2001, 15:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Yang Y, Wan K, Mao N, Yu T-Y, Lin Y-C, DeZwaan DC, Freeman BC, Lin J-J, Lue NF, et al. : Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res 2011, 21:258–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.**.He Y, Song H, Chan H, Liu B, Wang Y, Sušac L, Zhou ZH, Feigon J: Structure of Tetrahymena telomerase-bound CST with polymerase α-primase. Nature 2022, 608:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]; A major contribution of this study is the first full structure of ttCtc1. Additionally, they show the architecture of ttCst in complexes with telomerase and polymerase-α, the latter complex with a global resolution of 4.2Å. Polymerase-α-primase undergoes major conformational changes upon ttCst binding and ttCst threads ssDNA into active site of polymerase-α.
- 17.Min B, Collins K: An RPA-Related Sequence-Specific DNA-Binding Subunit of Telomerase Holoenzyme Is Required for Elongation Processivity and Telomere Maintenance. Mol Cell 2009, 36:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R, Wold MS: Replication Protein A: Single-stranded DNA’s first responder Dynamic DNA-interactions allow Replication Protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays News Rev Mol Cell Dev Biol 2014, 36:1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.*.Wieser TA, Wuttke DS: Replication Protein A Utilizes Differential Engagement of Its DNA-Binding Domains to Bind Biologically Relevant ssDNAs in Diverse Binding Modes. Biochemistry 2022, doi: 10.1021/acs.biochem.2c00504. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive biochemical study that highlights how RPA recognizes and tightly binds a set of ssDNAs presented in a range of biologically relevant structural contexts. Specific inhibition of individual DNA-binding modules enforces alternate modes of recognition that uncovers multiple dynamic modes of recognition.
- 20.Kim C, Snyder RO, Wold MS: Binding properties of replication protein A from human and yeast cells. Mol Cell Biol 1992, 12:3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn RL, Chang S, Zou L: RPA and POT1: friends or foes at telomeres? Cell Cycle Georget Tex 2012, 11:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.*.Lin C-YG, Näger AC, Lunardi T, Vančevska A, Lossaint G, Lingner J: The human telomeric proteome during telomere replication. Nucleic Acids Res 2021, 49:12119–12135. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a strategy to isolate proteins associated with nascent DNA, this study identifies the suite of proteins specifically associated with replicating telomeres. In addition to the core replisome, replicating telomeres show enrichment and depletion of specific factors whose subsequent analysis is poised to reveal the novel features of telomere replication.
- 23.Ray S, Qureshi MH, Malcolm DW, Budhathoki JB, Celik U, Balci H: RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys J 2013, 104:2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lancrey A, Safa L, Chatain J, Delagoutte E, Riou J-F, Alberti P, Saintomé C: The binding efficiency of RPA to telomeric G-strands folded into contiguous G-quadruplexes is independent of the number of G4 units. Biochimie 2018, 146:68–72. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Nasmyth K: Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J 1998, 17:5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wold MS: Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 1997, 66:61–92. [DOI] [PubMed] [Google Scholar]
- 27.Oakley GG, Patrick SM: Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci Landmark Ed 2010, 15:883–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krasikova YS, Rechkunova NI, Lavrik OI: Replication protein A as a major eukaryotic single-stranded DNA-binding protein and its role in DNA repair. Mol Biol 2016, 50:649–662. [DOI] [PubMed] [Google Scholar]
- 29.Maréchal A, Zou L: RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res 2015, 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Doty T, Gibson B, Heyer W-D: Human BRCA2 protein promotes RAD51 filament formation on RPA-covered ssDNA. Nat Struct Mol Biol 2010, 17:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filippo JS, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P: Recombination Mediator and Rad51 Targeting Activities of a Human BRCA2 Polypeptide. J Biol Chem 2006, 281:11649–11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heyer WD, Rao MR, Erdile LF, Kelly TJ, Kolodner RD: An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J 1990, 9:2321–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.U K, S N, C C, H Je, K Rd: Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 1998, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upton HE, Hong K, Collins K: Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol Cell Biol 2014, 34:4200–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min B, Collins K: Multiple Mechanisms for Elongation Processivity within the Reconstituted Tetrahymena Telomerase Holoenzyme. J Biol Chem 2010, 285:16434–16443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upton HE, Chan H, Feigon J, Collins K: Shared Subunits of Tetrahymena Telomerase Holoenzyme and Replication Protein A Have Different Functions in Different Cellular Complexes. J Biol Chem 2017, 292:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.**.Lim CJ, Barbour AT, Zaug AJ, Goodrich KJ, McKay AE, Wuttke DS, Cech TR: The structure of human CST reveals a decameric assembly bound to telomeric DNA. Science 2020, 368:1081–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reports the first full structure of human CTC1 revealing seven OB fold domains and the complex architecture with STN1 and TEN1, which closely resembles human RPA. This CryoEM structure (global resolution of 3.0Å) identifies the anchor telomeric ssDNA-binding site on CTC1. A striking decameric oligomerization of CST was seen upon ssDNA binding that could be biologically relevant.
- 38.Bochkareva E, Belegu V, Korolev S, Bochkarev A: Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J 2001, 20:612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A: Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 2002, 21: 1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg-Neifach O, Lue NF: Telomere DNA recognition in Saccharomycotina yeast: potential lessons for the co-evolution of ssDNA and dsDNA-binding proteins and their target sites. Front Genet 2015, 6:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis KA, Pfaff DA, Earley JN, Altschuler SE, Wuttke DS: The tenacious recognition of yeast telomere sequence by Cdc13 is fully exerted by a single OB-fold domain. Nucleic Acids Res 2014, 42:475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates LA, Aramayo RJ, Pokhrel N, Caldwell CC, Kaplan JA, Perera RL, Spies M, Antony E, Zhang X: A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Nat Commun 2018, 9:5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flynn RL, Zou L: Oligonucleotide/Oligosaccharide-Binding (OB) Fold Proteins: A Growing Family of Genome Guardians. Crit Rev Biochem Mol Biol 2010, 45:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.**.He Q, Lin X, Chavez BL, Agrawal S, Lusk BL, Lim CJ: Structures of the human CST-Polα-primase complex bound to telomere templates. Nature 2022, 608:826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]; CryoEM structures of hCST in an active conformation with polymerase-α-primase with a global resolution of 3.4Å and 4.3Å. This is breakthrough insight into the mechanism of how CST acts as processivity factor for C-strand fill-in at telomeres. Highlights include CST threading ssDNA into the active site of primase and TEN1 acting to stabilize primase after the conformational change into the active state.
- 45.Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M: Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci 2011, 108:20357–20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L: Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 1997, 385:176–181. [DOI] [PubMed] [Google Scholar]
- 47.*.Jiang J, Wang Y, Sušac L, Chan H, Basu R, Zhou ZH, Feigon J: Structure of telomerase with telomeric DNA. Cell 2018, 173:1179–1190.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]; CryoEM structure at 4.8 Å of the Tetrahymena telomerase bound to telomeric DNA reveals an intricate arrangement of protein/RNA/DNA that informs the roles of RNA and accessory proteins in mediating catalytic activitiy.
- 48.Kim C, Paulus BF, Wold MS: Interactions of human replication protein A with oligonucleotides. Biochemistry 1994, 33:14197–14206. [DOI] [PubMed] [Google Scholar]
- 49.Alani E, Thresher R, Griffith JD, Kolodner RD: Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol 1992, 227:54–71. [DOI] [PubMed] [Google Scholar]
- 50.Ahmad F, Patterson A, Deveryshetty J, Mattice JR, Pokhrel N, Bothner B, Antony E: Hydrogen–deuterium exchange reveals a dynamic DNA-binding map of replication protein A. Nucleic Acids Res 2021, 49:1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M: Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev 2009, 23:2900–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell MT, Smith JS, Mason M, Harper S, Speicher DW, Johnson FB, Skordalakes E: Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol Cell Biol 2010, 30:5325–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mason M, Wanat JJ, Harper S, Schultz DC, Speicher DW, Johnson FB, Skordalakes E: Cdc13 OB2 dimerization required for productive Stn1 binding and efficient telomere maintenance. Struct Lond Engl 1993 2013, 21:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS: Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol 2004, 338:241–255. [DOI] [PubMed] [Google Scholar]
- 55.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V: RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 2007, 14:208–214. [DOI] [PubMed] [Google Scholar]
- 56.**.Ge Y, Wu Z, Chen H, Zhong Q, Shi S, Li G, Wu J, Lei M: Structural insights into telomere protection and homeostasis regulation by yeast CST complex. Nat Struct Mol Biol 2020, 27:752–762. [DOI] [PubMed] [Google Scholar]; Crystal structure that describes the first overall architecture of yeast Cdc13-Stn1-Ten1. The structure is a dimer that forms an unusual ring structure composed of OB folds of Cdc13 and the wHTHs of Stn1. Cdc13 is poised for DNA binding and interaction with polymerase-α-primase.
- 57.**.Cai SW, Zinder JC, Svetlov V, Bush MW, Nudler E, Walz T, de Lange T: Cryo-EM structure of the human CST–Polα/primase complex in a recruitment state. Nat Struct Mol Biol 2022, 29:813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]; This CryoEM structure shows a recruitment state between hCST and polymerase-α-primase with a global resolution of 4.6Å. This is the first structural evidence describing a role for OB-D of CTC1 as interaction partners with polymerase-α-primase.
- 58.Ma Y, Huang C, Tang T, Wu B, Xue H, Cao Y, Wu J, Wan B, Lei M: Structural and functional insights into CST tethering in Tetrahymena thermophila telomerase. Structure 2022, 30:1565–1572.e4. [DOI] [PubMed] [Google Scholar]


