Abstract
Objective:
Psoriasis and multiple sclerosis (MS) are complex immune diseases that are mediated by T-cells and share multiple comorbidities. Previous studies have suggested psoriatic patients are at higher risk of MS; however, causal relationships between the two conditions remain unclear. Through epidemiology and genetics, we provide a comprehensive understanding of the relationship and shared molecular factors between psoriasis and MS.
Methods:
We used logistic regression, trans-disease meta-analysis (TDMA) and Mendelian randomization (MR). Medical claims data was included from 30 million patients, including 141,544 with MS and 742,919 with psoriasis. We used GWAS summary statistics from 11,024 psoriatic, 14,802 MS cases and 43,039 controls for TDMA, with additional summary statistics from 5 million individuals for MR.
Results:
Psoriatic patients have significantly higher risk of MS (4,637 patients with both diseases; OR=1.07, p=1.2×10−5), after controlling for potential confounders. Using inverse variance and equally weighted TDMA, we revealed more than 20 shared and opposing (direction of effect) genetic loci outside the MHC which show significant genetic colocalization (in COLOC and COLOC-SuSiE v5.1.0). Co-expression analysis of genes from these loci further identified distinct clusters which were enriched among pathways for IL-17/TNFα (p<1.6×10−3, OR>39) and JAK-STAT (p=1.1×10−5, OR=35), including genes such as TNFAIP3, TYK2 and TNFRSF1A. MR found psoriasis as an exposure has a significant causal effect on MS (OR=1.04, p=5.8×10−3), independent of T1D (p=4.3×10−7, OR=1.05), T2D (p=2.3×10−3, OR=1.08), IBD (p=1.6×10−11, OR=1.11) and vitamin D level (p=9.4×10−3, OR=0.75).
Interpretation:
By investigating the shared genetics of psoriasis and MS, along with their modifiable risk factors, our findings will advance innovations in treatment for patients suffering from comorbidities.
INTRODUCTION
Multiple sclerosis (MS), a chronic central nervous system disease that damages central nervous system myelin or white matter through immune dysregulation1, is associated with multiple comorbidities and risk factors that can increase susceptibility and/or accelerate neurodegeneration2,3, such as smoking, cardiovascular disease and low vitamin D. While receiving less attention, psoriasis, a complex chronic skin disease4 associated with systemic inflammation5, has been found to impose a higher (1.29 odds ratio6) risk of MS. Notably, the risk of MS increases with psoriasis severity7. Certain cytokines have been implicated in the pathogenesis of both psoriasis and MS, including IL-178,9, IFNγ10,11 and TNF12,13. Clinical responses were observed in both diseases for IL-17 regulating drugs dimethyl fumarate14,15 and secukinumab16,17, as well as α4-integrin antagonist, natalizumab18; however, even though TNF inhibitors work well for psoriasis, they have been found to exacerbate MS12. Similarly, IFNβ is a common treatment for MS but can trigger psoriasis19. Until now, there has been limited study to discern the effects of shared genetic and modifiable risk factors on the psoriasis/MS comorbidity and how they are associated with the aforementioned cytokine signaling pathways. However, this is essential for understanding the mechanisms and identifying effective treatments for many patients suffering from both conditions.
Psoriasis and MS each have a substantial genetic component (~70% heritability for psoriasis20 and 50–64% for MS21,22), with genetic signals from both diseases enriched among regulatory regions for CD4+ and CD8+ T-cells1,4. Infiltration of activated CD4+/CD8+ T-cells in the skin increases proliferation of keratinocytes to produce psoriatic plaques23, while in MS, T-cells are involved in an inflammatory process that damages myelin nerve insulation24. Myelin-specific CD4+ T-cells are over four times more abundant in MS patients than controls25, and there is a 10-fold increase of dermal T-cells in psoriasis lesions compared to healthy skin26. Psoriasis and MS are both characterized by TH1 and TH17 cells23,27, with TH17 demonstrating greater ability to cross the choroid plexus (in MS) than other CD4+ subsets28, whereas in psoriasis, neutrophil extracellular traps (NETs) help enhance TH17 induction29. Both diseases are associated with class I HLA alleles (e.g. B*44 is protective for MS30–32 and C*06:02 increases the risk for psoriasis33), although the primary association for MS in the MHC is with class II HLA alleles (including DRB1*15:01)34. However, outside the major histocompatibility complex (MHC), little is known about the genetic components they share.
Psoriasis and MS have also been associated with overlapping modifiable risk factors. For instance, both are more prevalent in northern latitudes35,36 and are connected with vitamin D deficiency37,38. There is evidence that vitamin D may be involved in modulating immune responses, including the activation of CD4+/CD8+ T-cells39,40 and it has also been found to suppress IL-17 induction41. However, much remains to be known regarding its precise role in inflammatory diseases, such as psoriasis and MS42. In psoriasis, vitamin D analogs are regularly used as topical treatments38, while there have been multiple inconclusive trials for its use as an oral supplement in MS43,44. Ultraviolet radiation is an effective treatment for psoriasis45, and early trials suggest it may be beneficial for MS46. Obesity is another key risk factor for immune-mediated diseases in general47, and previous Mendelian randomization studies have suggested it can causally affect both psoriasis48 and MS49. Metabolic dysfunction resulting from obesity impacts the immune system50, for example through the effect of adipokines on TNF51. Further modifiable risk factors reported by the literature include smoking52,53, triggering events54,55, infections56,57 and the microbiome58,59. These risk factors should be taken into consideration when assessing the causal relationship between psoriasis and MS.
Understanding the pathophysiology of comorbidities is essential for precision medicine and optimal disease management, as it can provide clues to their underlying molecular mechanisms and common etiology. In this study, we conduct epidemiological analysis on a large medical claims dataset to reveal risk factors common to both diseases, and then we apply trans-disease meta-analysis to identify more than 20 shared genetic loci. Finally, we apply Mendelian randomization using genetic variants as instruments to establish a causal relationship between psoriasis and MS independent of their comorbidities and modifiable risk factors.
METHODS
The genetic cohorts involved in both the psoriasis and MS GWAS were IRB approved (details in previous publications). The Optum Clinformatics® data was exempt from IRB approval.
Epidemiology
We investigated the association between psoriasis and MS, in the context of potential confounders, through an epidemiological analysis of 30,445,892 patients from Optum’s deidentified Clinformatics® Data Mart60. All included individuals had a recorded year of birth, self-reported sex and race; the majority (75%) were White and just over half (55%) were female, with a mean age of 45 (at the most recent encounter) and mean enrollment (follow-up) of 5 years. The status of the following traits among the patients were evaluated by presence/absence of ICD-9/10 codes, as indicated in Supplementary Table 1: type 1 diabetes (T1D), type 2 diabetes (T2D), coronary artery disease (CAD), rheumatoid arthritis (RA), inflammatory bowel disease (IBD), asthma, vitamin D deficiency, obesity / morbid obesity, smoking and alcohol use disorder (AUD). In total, our analysis included 141,544 patients with MS, 742,919 patients with psoriasis and 4,637 patients with both diseases.
Logistic regression was applied to medical claims data extracted for the years 2001 to 2018 from 30,445,892 patients in Optum’s deidentified Clinformatics® Data Mart60 (version 7.2). Patients were included that had a recorded year of birth (from which we calculated the age at most recent encounter), sex and race, then we added up the total enrollment periods, to give the total enrollment for each patient. Traits were ascertained by ICD-9/10 codes, as indicated in Supplementary Table 1, including obesity, smoking and alcohol use disorder, as no quantitative data on BMI, smoking or alcohol consumption was readily available.
Trans-disease meta-analysis (TDMA)
To compare genetic signals between psoriasis and MS, we performed trans-disease meta-analysis (TDMA) on GWAS summary statistics from psoriasis (11,024 cases, 16,336 controls)61 and MS (14,802 cases, 26,703 controls)1 cohorts. Meta-analyses were prepared as per the data collection and processing steps described in their respective GWAS1,61. We applied the standard fixed effects inverse variance weighted (IVW) approach62 to meta-analysis summary statistics, and also implemented TDMA using an equally weighted combination63 of effect sizes () and variances (). Loci were considered independent if they are separated by >500kb or revealed through approximate conditional analysis (GCTA-COJO64), with the largest psoriasis cohort (11,675 cases and controls) being used as the reference dataset for LD computation. We also applied HESS65 to identify locally correlated regions, using the European 1000 Genomes data provided with the software as reference, and the recommended partitions66. Colocalization analysis was performed using COLOC67 and its SuSiE extension68 on markers within ±100kb of each lead TDMA marker, with the same reference cohort as COJO.
Mendelian randomization (MR)
We applied MR to investigate potential causal relationships between psoriasis, MS and their comorbidities. We employed six different MR techniques (MR-PRESSO69, MR-Egger70, MR-Robust71, MR-RAPS72, MR-Median73 and MR-Mode74) to provide confidence in the results, by minimizing the risk of spurious findings due to the weaknesses of any one approach. First, MR-PRESSO69 was used to correct for horizontal pleiotropy by removing outliers, which addressed distortion for asthma (p=0.012), T1D (p=0.002) and RA (p<3.3×10−5) on MS. Then, the other techniques were used to test assumptions in different ways: MR-Egger70 includes the intercept in its model; MR-Robust71 uses Tukey’s loss function for robust regression; MR-RAPS72 accounts for variance in effect sizes and uses a random effects model; MR-Median73 and MR-Mode74 control for heterogeneity by calculating weighted median- or mode-based estimates, respectively. We extracted genetic data from full GWAS summary statistics for 10 traits (Supplementary Table 6), in addition to psoriasis and MS. Genetic instruments were selected from the intersection of markers across traits, through LD clumping (p≤1×10−4, LD≥0.001, window size=10Mbp), using the European 1000 Genomes data as reference. The six different MR techniques were applied in univariable analysis, to estimate the causal effect of each trait on psoriasis and MS; and then we conducted multivariable analysis using GRAPPLE75, by pooling the genetic markers from each trait.
RESULTS
Epidemiology
Unlike psoriasis, MS was more strongly associated with female sex (Table 1; OR=2.36, p=1.2×10−4,323), and both MS and psoriasis had reduced prevalence in Asian (OR=0.36, p=2.7×10−513) and Hispanic (OR=0.62, p=1.7×10−488) patients, while psoriasis also had lower prevalence among Black patients (OR=0.67, p=7.2×10−1,693). Vitamin D deficiency (OR=3.02, p=1.9×10−368) had the strongest association for MS and was also associated with psoriasis (OR=1.77, p=9.2×10−8,302). The strongest association for psoriasis was RA (OR=3.82, p=1.2×10−21,815), which was also associated with MS (OR=2.64, p=9.1×10−1,792). Both MS and psoriasis were associated with AUD, smoking, asthma, T1D, T2D, CAD, IBD and morbid obesity. When including all covariates in a conditional model, the effect of psoriasis on MS was OR=1.07 (p=1.2×10−5), while the effect of MS on psoriasis was OR=1.10 (p=9.4×10−10).
Table 1: Epidemiological Analysis.
The top portion of the table shows the effects of different demographic variables on MS/psoriasis; the bottom portion of the table illustrates the effects of other traits/diseases on MS/psoriasis, after adjusting for the demographic variables
| Covariate | ||||||||
|---|---|---|---|---|---|---|---|---|
| All | MS | Psor | Both | OR (95% CI) | P-value | OR (95% CI) | P-value | |
| Age in Years Mean (SD) |
45 (23) | 54 (15) | 53 (18) | 56 (14) | 1.47 (1.46–1.48) | 1.2×10−4,442 | 1.49 (1.48–1.49) | 1.2×10−24,027 |
| – Female Sample Size (%) |
16,800,019 (55.18) | 105,242 (0.63) | 387,577 (2.31) | 3,343 (0.02) | 2.36 (2.34–2.39) | 1.2×10−4,323 | 0.88 (0.88–0.89) | 8.8×10−608 |
| – Hispanic Sample Size (%) |
3,391,756 (11.14) | 10,461 (0.31) | 70,748 (2.09) | 317 (0.01) | 0.62 (0.60–0.63) | 1.7×10−488 | 0.80 (0.79–0.81) | 4.5×10−676 |
| Trait | ||||||||
|---|---|---|---|---|---|---|---|---|
| All | MS | Psor | Both | OR (95% CI) | P-value | OR (95% CI) | P-value | |
| Psoriasis | 742,919 (2.44) | - | - | 4,637 (0.02) | 1.22 (1.20–1.24) | 5.3×10−41 | - | - |
| MS | 141,544 (0.46) | - | - | 4,637 (0.02) | - | - | 1.24 (1.22–1.26) | 8.0×10−47 |
| Type 1 Diabetes | 923,543 (3.03) | 7,158 (0.78) | 33,256 (3.60) | 331 (0.04) | 1.34 (1.33–1.36) | 3.9×10−126 | 1.14 (1.13–1.15) | 4.7×10−115 |
| Type 2 Diabetes | 4,767,871 (15.66) | 32,595 (0.68) | 172,729 (3.62) | 1,405 (0.03) | 1.24 (1.23–1.25) | 1.5×10−211 | 1.19 (1.19–1.20) | 4.8×10−747 |
| Coronary Artery Disease | 3,170,317 (10.41) | 21,996 (0.69) | 119,635 (3.77) | 991 (0.03) | 1.13 (1.12–1.14) | 1.1×10−53 | 1.03 (1.02–1.03) | 8.5×10−15 |
| Rheumatoid Arthritis | 813,738 (2.67) | 9,495 (1.17) | 66,613 (8.19) | 813 (0.10) | 1.85 (1.83–1.87) | 1.0×10−699 | 3.11 (3.09–3.12) | 6.8×10−14,924 |
| Inflammatory Bowel Disease | 403,112 (1.32) | 3,935 (0.98) | 18,686 (4.64) | 206 (0.05) | 1.76 (1.73–1.79) | 6.5×10−265 | 1.65 (1.64–1.67) | 2.8×10−948 |
| Asthma | 4,026,381 (13.22) | 25,643 (0.64) | 116,560 (2.89) | 1,128 (0.03) | 1.36 (1.35–1.37) | 3.9×10−433 | 1.24 (1.24–1.24) | 1.0×10−949 |
| Vitamin D Deficiency | 3,937,160 (12.93) | 43,555 (1.11) | 152,644 (3.88) | 1,846 (0.05) | 2.30 (2.29–2.32) | 1.5×10−3,974 | 1.42 (1.42–1.43) | 1.1×10−2,867 |
| – Morbidly Obese | 1,686,290 (5.54) | 13,077 (0.78) | 67,983 (4.03) | 646 (0.04) | 1.47 (1.46–1.48) | 1.1×10−370 | 1.68 (1.67–1.69) | 4.5×10−3,350 |
| Smoking | 4,605,396 (15.13) | 38,163 (0.83) | 174,563 (3.79) | 1,587 (0.03) | 1.78 (1.77–1.79) | 3.5×10−1,857 | 1.38 (1.38–1.39) | 2.6×10−2,738 |
| Alcohol Use Disorder | 836,527 (2.75) | 5,491 (0.66) | 31,853 (3.81) | 238 (0.03) | 1.53 (1.51–1.55) | 1.8×10−204 | 1.43 (1.42–1.44) | 4.7×10−803 |
Shared and opposing genetic loci
Table 2 shows the results of TDMA using fixed effects inverse variance weighted (IVW) meta-analysis, while Supplementary Tables 2–3 shows the results using the equally weighted approach. TDMA signals with shared or opposing direction of effect are presented as circular Manhattan plot tracks (Figure 1a and 1b for IVW, Supplementary Figure 1a and 1b for equally weighted). In total, 22 and 20 genetic loci were identified for IVW and equally weighted approaches respectively (counting the major histocompatibility complex, MHC, as a single locus due to its complex linkage disequilibrium, LD) in which the TDMA lead marker was: 1) genome-wide significant (p<5×10−8) in TDMA; 2) suggestively significant (p<1×10−4) for each trait; and 3) more significant in TDMA than in both traits. IVW and equally weighted approaches were highly concordant, with only two loci identified by each approach not by the other (two loci appear both shared and opposing in IVW). Indicated in red on Figure 1a and 1b are 11 shared and 11 opposing loci, revealed using IVW TDMA, of which Figure 2 illustrates two shared and two opposing loci using regional association plots. We then performed conditional analysis separately on psoriasis and MS outside the MHC using conditional and joint analysis (GCTA-COJO64) and applied the TDMA criteria to each independent signal identified, discovering an additional two shared and one opposing IVW TDMA locus (indicated in blue in Figure 1), of which one of the shared loci was identified by the equally weighted approach, leading to 27 independent TDMA signals across the three approaches (Supplementary Figures 2–24); we also confirmed seven shared and six opposing loci. Table 2 and Supplementary Tables 4–5 present the loci identified by TDMA and COJO, respectively.
Table 2: Loci Identified by Trans-Disease Meta-Analysis (TDMA).
Susie # indicates the number of pairs of fine-mapped signals with evidence of colocalization (PP>0.7)
| Shared (same direction of effect) loci: | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyt. Band | rsID | Position (hg19) | RA/NR | MS | Psoriasis | TDMA | Heterogeneity | Colocalization | Nearby Genes | |||||
| OR | P | OR | P | OR | P | Q | P | COLOC PP | SuSiE # | |||||
| 5q33.3 | rs2546890 | 158759900 | A/G | 1.12 | 1.0×10−12 | 1.33 | 6.4×10−51 | 1.21 | 7.1×10−58 | 1.4×10−2 | 0.91 | 4.4×10−7 | 0 | IL12B |
| 6p21.1 | rs59024520 | 42238973 | C/T | 1.16 | 6.3×10−5 | 1.19 | 8.2×10−5 | 1.17 | 2.3×10−8 | 4.4×10−4 | 0.98 | 0.83 | 0 | USP49 |
| 6q23.3 | rs9321623 | 137958265 | C/T | 1.08 | 4.5×10−6 | 1.10 | 4.5×10−7 | 1.09 | 9.5×10−12 | 1.7×10−4 | 0.99 | 1.5×10−3 | 1 | TNFAIP3 |
| 7p14.1 | rs11767350 | 37385365 | A/G | 1.07 | 6.5×10−6 | 1.09 | 7.4×10−6 | 1.08 | 2.2×10−10 | 6.4×10−5 | 0.99 | 0.80 | 3 | ELMO1 * |
| 10q22.2 | rs2459446 | 75601596 | C/T | 1.07 | 7.9×10−5 | 1.13 | 1.5×10−10 | 1.09 | 8.9×10−14 | 1.8×10−3 | 0.97 | 0.87 | 3 | CAMK2G * |
| 11q13.1 | rs479777 | 64107477 | T/C | 1.08 | 3.5×10−5 | 1.13 | 2.1×10−9 | 1.10 | 6.0×10−13 | 9.1×10−4 | 0.98 | 0.87 | 0 | PRDX5 * , RPS6KA4 * |
| 12p13.31 | rs4149576 | 6449115 | T/C | 1.11 | 3.5×10−9 | 0.07 | 8.8×10−5 | 1.09 | 6.8×10−12 | 3.4×10−4 | 0.99 | 0.07 | 6 | CD27, TNFRSF1A * |
| 13q14.2 | rs9591325 | 50811220 | T/C | 1.24 | 4.2×10−10 | 1.24 | 6.6×10−9 | 1.24 | 2.0×10−17 | 1.7×10−5 | 1.00 | 0.99 | 2 | DLEU1 * |
| 16p13.13 | rs243324 | 11354970 | A/G | 1.12 | 5.9×10−12 | 1.08 | 6.6×10−5 | 1.10 | 5.5×10−15 | 6.6×10−4 | 0.98 | 3.1×10−4 | 0 | SOCS1, RMI2 * |
| 17q21.2 | rs957970 | 40519890 | A/G | 1.14 | 1.1×10−13 | 1.11 | 7.3×10−8 | 1.13 | 6.4×10−20 | 2.2×10−4 | 0.99 | 0.97 | 7 | STAT3 * , STAT5A * /B |
| 19p13.2 | rs55677033 | 11166293 | T/C | 1.09 | 2.6×10−6 | 1.08 | 9.0×10−5 | 1.09 | 9.5×10−10 | 1.0×10−5 | 1.00 | 0.17 | 2 | ILF3, CARM1 |
| Opposing (opposite direction of effect) loci: | ||||||||||||||
| Cyt. Band | rsID | Position (hg19) | RA/NR | MS | Psoriasis | TDMA | Heterogeneity | Colocalization Prob. | Nearby Genes | |||||
| OR | P | OR | P | OR | P | Q | P | COLOC PP | SuSiE # | |||||
| 1p36.11 | rs6672420 | 25291010 | A/T | 1.07 | 2.2×10−5 | 0.86 | 8.8×10−15 | 1.11 | 6.3×10−18 | 2.9×10−3 | 0.96 | 0.93 | 3 | RUNX3 |
| 2p16.1 | rs1177213 | 61079090 | A/G | 0.93 | 4.6×10−6 | 1.16 | 2.1×10−15 | 1.11 | 5.2×10−18 | 3.0×10−3 | 0.96 | 0.73 | 6 | REL * , PUS10 * |
| 5q31.1 | rs3843503 | 131466629 | T/A | 1.08 | 1.9×10−5 | 0.92 | 2.1×10−5 | 1.08 | 1.6×10−9 | 1.5×10−6 | 1.00 | 0.68 | 3 | CSF2, P4HA2 * |
| 6p22.1 | rs1611653 | 29841702 | G/C | 1.31 | 3.9×10−50 | 0.73 | 1.5×10−61 | 1.34 | 6.4×10−109 | 1.5×10−3 | 0.97 | - | - | HLA-B * /C, TNF |
| 6q23.3 | rs7746779 | 138154501 | A/G | 0.90 | 8.8×10−7 | 1.15 | 1.1×10−9 | 1.13 | 1.1×10−14 | 7.8×10−4 | 0.98 | 3.2×10−3 | 0 | TNFAIP3, WAKMAR2 |
| 6q25.3 | rs2451279 | 159515077 | G/A | 1.10 | 6.4×10−8 | 0.91 | 3.6×10−6 | 1.10 | 1.8×10−12 | 4.3×10−6 | 1.00 | 2.6×10−3 | 2 | TAGAP * |
| 7q36.1 | rs10243355 | 150356318 | G/A | 1.09 | 3.4×10−5 | 0.89 | 1.8×10−5 | 1.10 | 3.4×10−9 | 6.2×10−4 | 0.98 | 0.81 | 1 | GIMAP2 * /6 * |
| 10q22.3 | rs1250565 | 81047015 | A/G | 1.12 | 1.0×10−10 | 0.89 | 3.3×10−9 | 1.12 | 2.1×10−18 | 2.4×10−5 | 1.00 | 0.83 | 2 | ZMIZ1 * |
| 11p11.2 | rs12574410 | 47169228 | C/G | 1.11 | 3.9×10−6 | 0.90 | 9.3×10−5 | 1.11 | 1.5×10−9 | 1.3×10−5 | 1.00 | 0.88 | 0 | MYBPC3 * , AGBL2 * |
| 16p13.13 | rs3862471 | 11113463 | G/T | 1.17 | 3.2×10−23 | 0.92 | 6.4×10−6 | 1.14 | 1.3×10−25 | 3.0×10−3 | 0.96 | 0.85 | 0 | CLEC16A * |
| 22q12.3 | rs5756405 | 37310954 | A/G | 1.07 | 2.4×10−5 | 0.93 | 4.3×10−5 | 1.07 | 4.3×10−9 | 4.7×10−5 | 0.99 | 0.76 | 3 | CSF2RB * , NCF4 |
eQTL evidence in eQTLGen or GTEx v8
Figure 1: IVW trans-disease meta-analysis (TDMA).
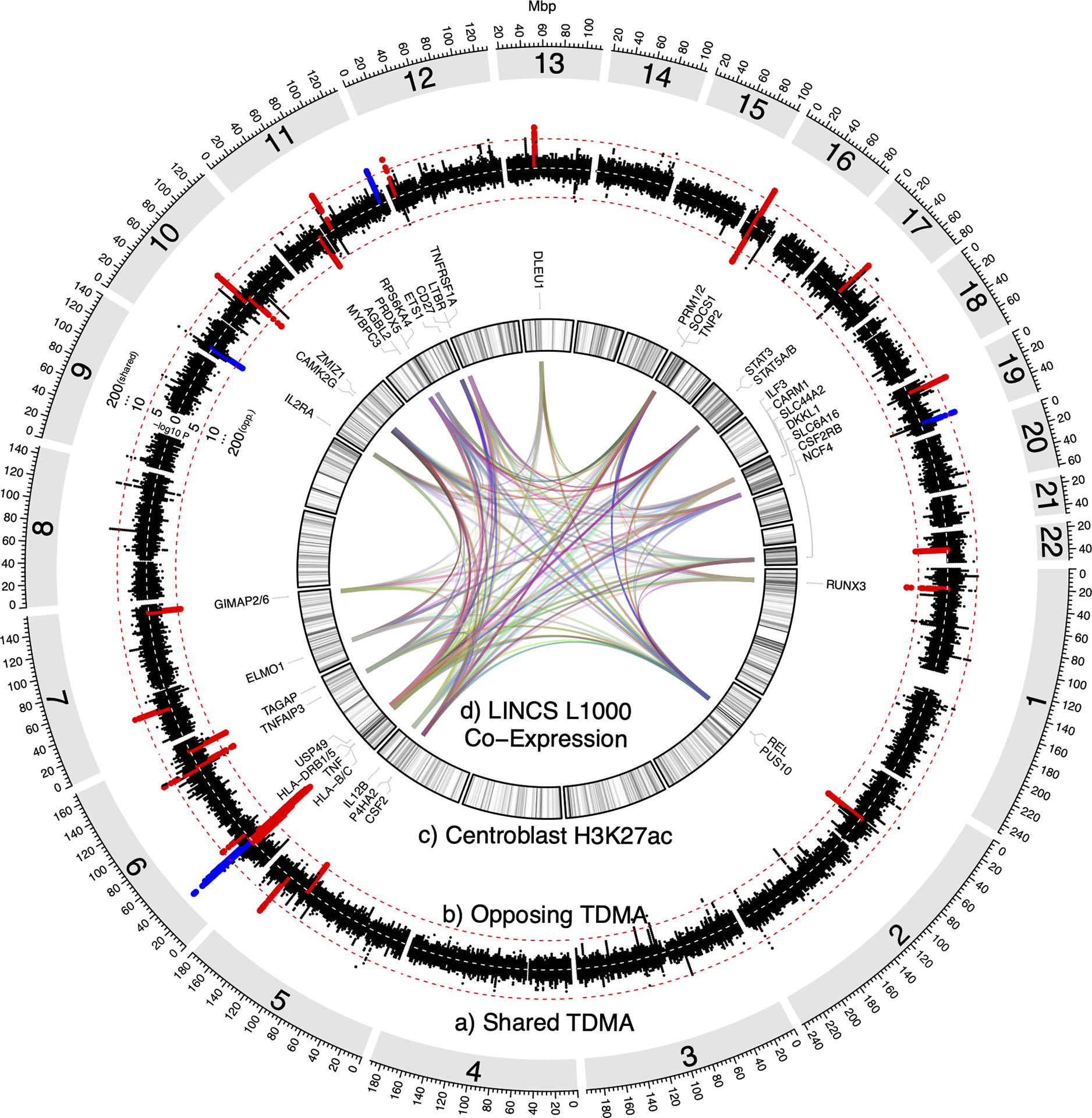
Circular diagram including the following: a) Manhattan plot of shared (same direction of effect) psoriasis/MS TDMA signals, showing markers more significant in TDMA than for either trait. b) Manhattan plot of opposing (opposite direction of effect) psoriasis/MS trans-disease meta-analysis (TDMA) signals, showing markers more significant in TDMA than for either trait. Red dashed lines indicate the genome-wide significance (p<5×10−8) threshold for shared and opposing signals, respectively. Loci which meet this threshold and are suggestively significant (p<1×10−4) for both traits are highlighted in red (if identified through our original TDMA approach) or blue (for additional loci identified using GCTA-COJO). c) Density of H3K27ac active enhancer marks for B-cell centroblasts (the most enriched cell type among the TDMA loci, compared to other established loci for psoriasis and MS). The darker the color, the higher the proportion of regulatory marks overlapping each 2Mbp region. Genes reported by previous psoriasis and MS GWAS1,4,96–99 are labeled for each locus. d) Links between genes, according to co-expression in L1000 assay perturbation experiments from NIH’s Library of Integrated Network-Based Cellular Signatures (LINCS). Each link has a random color, with transparency (alpha) values set proportional to the log-scaled number of experiments in which at least one gene from a locus is co-expressed with at least one gene from another locus, such that more opaque links represent pairs of loci with genes co-expressed in more experiments.
Figure 2: Regional association plots for four loci identified by IVW TDMA.
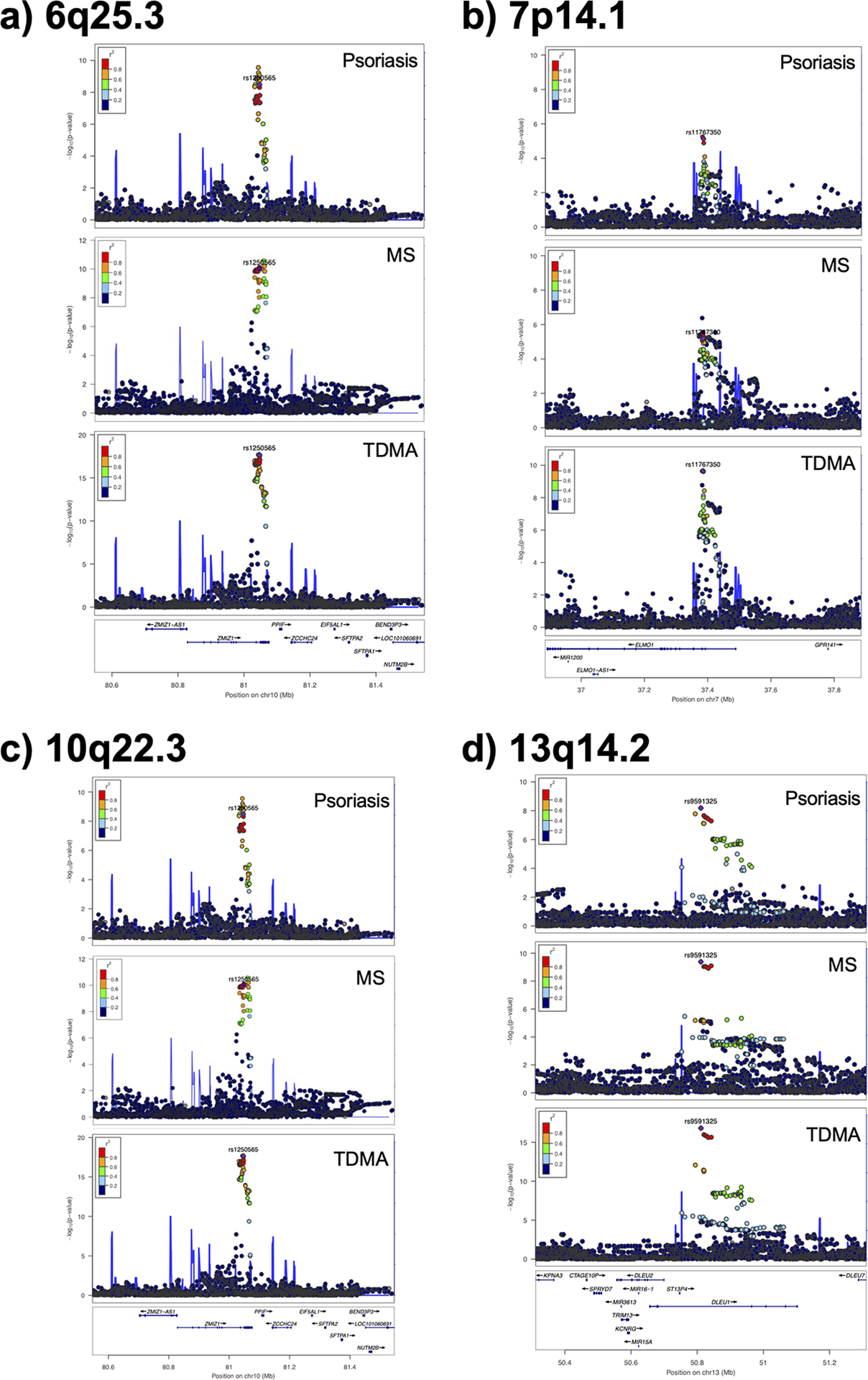
For each of the following loci, the lead TDMA marker is indicated in purple and the other markers are colored according to their LD with the lead marker: a) 6q25.3 opposing locus, with rs2451279 lead marker; b) 7p14.1 shared locus, with rs17259252 lead marker; c) 10q22.3 opposing locus, with rs1108618 lead marker; d) 13q14.2 shared locus, with rs9591325 lead marker.
We additionally evaluated broader genetic correlations between psoriasis and MS using Heritability Estimation from Summary Statistics (HESS)65. Three LD-independent regions66 had significant correlation between psoriasis and MS (FDR≤5%): a chromosome 5 (157–159Mbp; p=4.3×10−17) and chromosome 10 (81–82Mbp; p=8.6×10−5) region, which encompass loci identified by our previous two approaches, and the adjacent chromosome 5 region (159–160Mbp; p=1.3×10−6), which does not. Marker rs72804018 from this region (Table 2) is in low LD (r2=0.03, D’=0.32) with the shared locus 154kb upstream and meets our three criteria (MS p=4.6×10−7; psoriasis p=1.8×10−7; IVW TDMA p=5.2×10−13; equally weighted TDMA p=4.6×10−13), suggesting it may be a secondary signal.
Colocalization analysis
We investigated whether the shared and opposing loci colocalize to the same causal signals in psoriasis and MS, using COLOC67 (Supplementary Table 6). 16 of the 25 non-MHC loci (identified by the three approaches) showed strong evidence of colocalization, with posterior probabilities ranging from 0.68 (for the opposing locus, centered on rs3843503 in chromosome 5) to 0.99 (for the rs9591325 shared locus in chromosome 13). For the remaining loci, we used the Sum of Single Effects (SuSiE) COLOC extension68, which applies fine-mapping to provide more accurate inference by assuming the potential presence of multiple causal variants per locus. Significantly, all of the loci identified by the equally weighted TDMA approach had evidence (PP=1.0) of colocalization with SuSiE (Supplementary Table 7), and interestingly it was the secondary signal (i.e., rs72804018) identified by HESS that colocalized for the chromosome 5 locus. However, none of the additional loci from the GCTA-COJO analysis had evidence of colocalization, suggesting there may be different causal variants in psoriasis and MS.
To evaluate which TDMA loci might be explained by comorbidities, we retrieved full summary statistics from the largest available GWAS of European-ancestry individuals for each comorbidity. Supplementary Table 8 presents the shared and opposing TDMA loci that had at least suggestively significant association with each trait. Traits with the most associated loci (e.g., RA, with 7 loci, and IBD, with 5 loci) are primarily mediated by immunology rather than modifiable risk factors. T1D (an autoimmune disorder) has 7 associated loci, whereas T2D (a metabolic condition) has none. The risk allele locus associated with the most traits, rs413024, is positively associated with IBD, T1D and RA. All but one of the risk alleles for the shared loci exhibit increased risk of the comorbidities, while for the opposing loci, psoriasis and MS impart increasing/decreasing risk on the same traits. These results suggest no one comorbidity dominates the genetic relationship between psoriasis and MS, with the loci instead pertaining to complex imbalances in systemic inflammation.
Functional analysis
Using H3K27ac marks for active enhancers in 33 different cell types77, we conducted binomial enrichment tests to identify how the genetic signals can play regulatory roles in the specific cells involved in psoriasis/MS. Supplementary Table 9 compares enriched cell types for the 23 equally weighted TDMA plus GCTA-COJO loci outside the MHC, against the 62 MS and 20 psoriasis genome-wide significant loci, identified from their respective GWAS outside these regions. Immune cells were the highest enriched among TDMA, psoriasis and MS loci, with stimulated Th17 cells ranking among the most enriched in each (TDMA p=2.1×10−8, MS p=2.0×10−12, psoriasis p=0.011). Other CD4+/CD8+ T-cell subsets were highly enriched in TDMA, including Th0 (p=1.5×10−8), Th1 (p=1.4×10−7) and CD8+ memory T-cells (p=6.3×10−5). However, centroblasts were the most enriched cell type for TDMA (p=7.7×10−12), while they were less enriched in MS (p=4.6×10−7) and psoriasis (p=0.038), respectively. Figure 1c presents the H3K27ac active enhancer marks for B-cell centroblasts as a density plot, showing that regions of higher density (darker color on the plot) often co-occur with TDMA loci. We compared enrichments in TDMA against the other psoriasis and MS loci using binomial tests and found centroblasts to be the most significant cell type compared to psoriasis (p=3.4×10−5) and MS (p=9.4×10−4). As a sensitivity check, we repeated the enrichment analysis excluding the four TDMA loci identified using the conditional analysis approach (that did not colocalize) and found once again that centroblasts were more enriched in TDMA than the other psoriasis (p=1.4×10−4) and MS (p=2.3×10−3) loci. Interestingly, brain cell types were only significantly enriched (adjusting for FDR) among the TDMA loci, and not the MS- or psoriasis-only loci. Of these, mid-frontal lobe (p=1.9×10−4), hippocampus middle (p=1.0×10−3) and inferior temporal lobe (p=1.1×10−3) had the strongest enrichment.
Gene Co-Expression
Previous studies highlight the value of integrating gene co-expression networks with GWAS results to infer biological functions. Therefore, we utilized the L1000 assay perturbation experiment from NIH’s Library of Integrated Network-Based Cellular Signatures (LINCS)78 to understand the molecular network regulated by the TDMA loci. Links between these loci shown in Figure 1d indicate at least one gene from the first locus is co-expressed with a gene from the other. The opacity of each link is proportional to the log-scaled number of experiments in which the genes are co-expressed, such that pairs of loci with stronger evidence of connection are more clearly visible. As might be expected, links between the MHC and other loci are among the strongest; however, there are also connections between many of the non-MHC loci. For example, the 17q21.2 (STAT3, STAT5A/B) shared locus is highly connected with the 7p14.1 (ELMO1) shared locus (co-expressing in 642 experiments), while the 5q31.1 (CSF2, P4HA2) opposing locus is highly connected with the 2p16.1 (REL, PUS10) opposing locus (in 505 experiments).
We inferred distinct groups of loci/genes from the log-weighted LINCS data by applying four different community detection algorithms (leading eigenvector, Louvain, optimal integer programming and spin-glass) in the iGraph software package79. Three co-expressing clusters (Supplementary Table 10) were consistently identified by all four algorithms and were unaffected by including or excluding the MHC. We applied pathway enrichment analysis (excluding the MHC) using Enrichr80, which aggregates annotations from multiple sources. In the Kyoto Encyclopedia of Genes and Genomes (KEGG), the most significant pathway for cluster 1 (including CSF2 and TNFAIP3) was IL-17 signaling (p=1.6×10−3, OR=39.32), for cluster 2 (including IL12B and TYK2) it was JAK-STAT signaling (p=1.1×10−5, OR=35.85) and for cluster 3 (including CAMK2G and TNFRSF1A), it was necroptosis (p=3.7×10−9, OR=161.01).
Cluster 1 was also significantly enriched for TNFα signaling pathways in the National Center for Advancing Translational Sciences (NCATS) BioPlanet (p=1.4×10−6, OR=67.37) and the Molecular Signatures Database (MSigDB) (p=2.7×10−4, OR=30.14), while cluster 2 and 3 were both significant for IL-6/JAK/STAT3 signaling in MSigDB (p=6.2×10−5, OR=47.38 and p=6.6×10−6, OR=118.49, respectively). Cluster 2 was also significantly enriched for IL-12 signaling in BioPlanet (p=3.0×10−7, OR=93.31). Overall, there appear to be two main mechanisms in which the TDMA loci are involved: IL-17/TNFα signaling (cluster 1) and JAK-STAT signaling (clusters 2 and 3). We annotated Supplementary Table 10 to indicate which loci have genes involved in each pathway.
Mendelian randomization (MR)
Figure 3 provides estimates of causal effects on MS for each MR technique we applied, while Supplementary Figure 25 presents the same for psoriasis. Psoriasis was estimated to have a significant (FDR<0.05) effect on MS by four of the six techniques, while it was nominally significant for the remaining two (MR-Egger and MR-Robust). By contrast, none of the techniques indicated a significant effect for MS on psoriasis, and only one (MR-RAPS) was nominally significant. Consistent with the (covariate adjusted) epidemiological analysis, estimates of the effect of psoriasis on MS ranged from p=7.1×10−3, OR=1.05 for MR-PRESSO to p=7.9×10−3, OR=1.07 for MR-Mode. We confirmed the causal effect of psoriasis on MS with a significant Steiger test result (p=6.6×10−298), indicating higher correlation between the genetic instruments with psoriasis (r2=0.144) than MS (r2=0.011).
Figure 3: Mendelian randomization (MR) results for the effects of psoriasis and other comorbidities on multiple sclerosis.
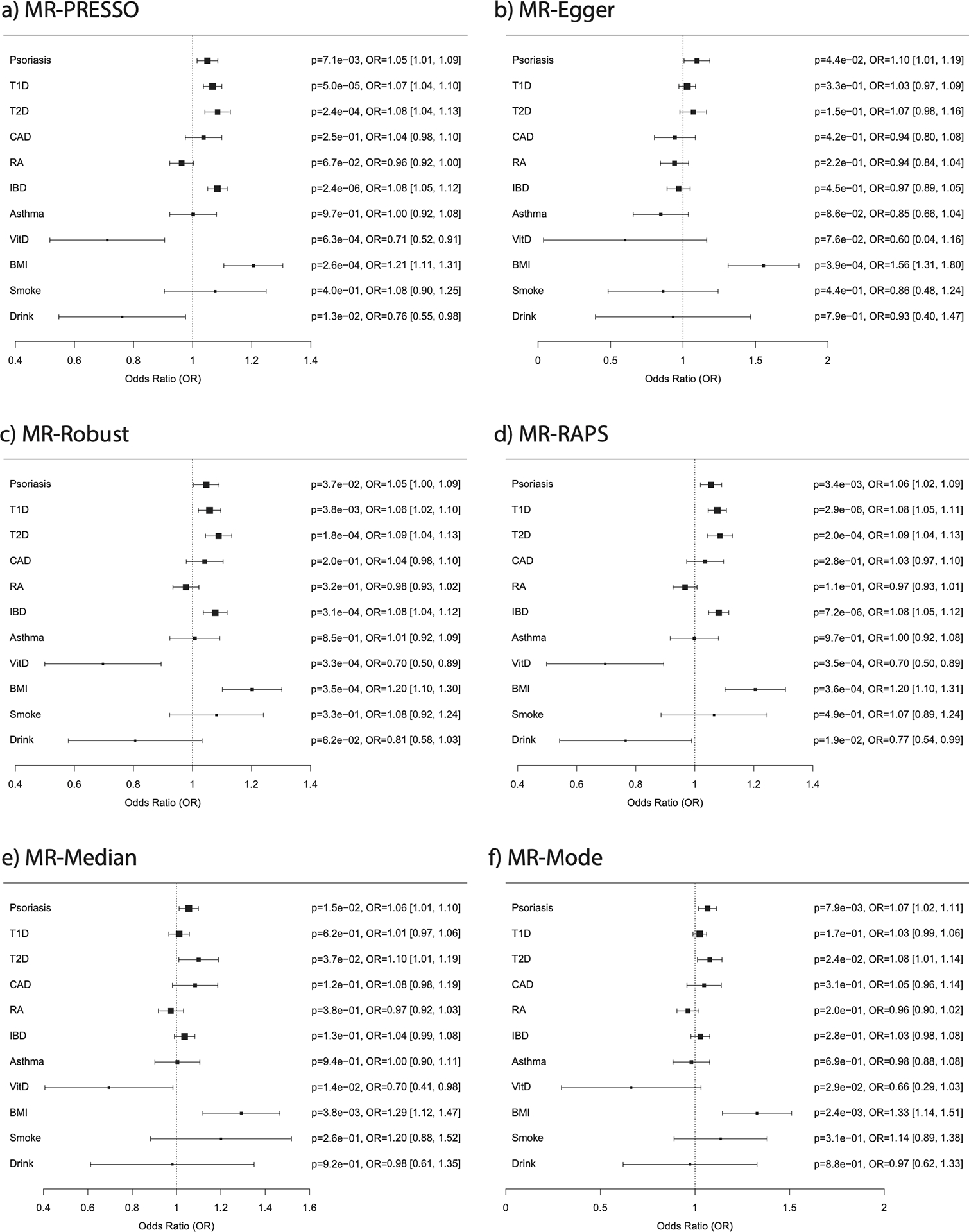
Forest plots generated from the results of six MR techniques (a-f). Abbreviations are as follows: T1D, type 1 diabetes; T2D, type 2 diabetes; CAD, coronary artery disease; RA, rheumatoid arthritis; IBD, inflammatory bowel disease; VitD, vitamin D (25OHD); BMI, body mass index; Smoke, cigarettes per day; Drink, drinks per week; p, p-value; OR, odds ratio.
Selecting the six comorbidities/traits (T1D, T2D, IBD, vitamin D, BMI and drinks/week) estimated to have a significant causal effect by at least one technique, we conducted a multivariable analysis using MR-GRAPPLE75. The causal effect of psoriasis on MS remained significant (p=5.8×10−3, OR=1.04), after conditioning on effects of T1D (p=4.3×10−7, OR=1.05), T2D (p=2.3×10−3, OR=1.08), IBD (p=1.6×10−11, OR=1.11) and vitamin D levels (p=9.4×10−3, OR=0.75); however, BMI and drinks/per week were no longer even nominally significant. While by univariable analysis BMI was the only trait (apart from psoriasis) estimated to have at least a nominally significant effect on MS by all six techniques, it is known to have a causal effect on other traits, so it is possible that it affects MS indirectly.
DISCUSSION
By combining large-scale epidemiological analysis with genetics, we confirmed a significant and causal association between psoriasis and MS that is independent of different confounding factors. A fully adjusted odds ratio of 1.07 (p=1.2×10−5) was estimated using medical claims data from ~900 thousand patients with psoriasis and/or MS and ~30 million controls. Mendelian randomization (MR) techniques gave comparable effect sizes (OR=1.05–1.07), with OR=1.04 (p=5.8×10−3) when conditioning on T1D, T2D, IBD, vitamin D, BMI and drinks/per week (traits significant in univariable analysis), while no significant causal effect was observed for MS on psoriasis. In total, more than 20 non-MHC genome-wide significant shared or opposing genetic loci were identified between psoriasis and MS that were at least suggestively significant (p<1×10−4) for each trait and more significant in TDMA than both traits. In an independent replication study for MS, two of the suggestively significant loci (rs6672420 and rs5756405) were genotyped, and both were confirmed to be genome-wide significant (p=1.5×10−9, OR=1.06; p=5.4×10−11, OR=1.07)1. No significant heterogeneity was identified for these markers between the main MS and replication cohorts, nor between the individual psoriasis cohorts. When combining the MS and psoriasis cohorts together (rather than applying TDMA), rs6672420 has nominally significant (p=0.01) heterogeneity, while for rs5756405 it is not significant (p=0.98). The mixture of shared and opposing loci is interesting, as it suggests a complex genetic relationship between psoriasis and MS. This could help explain why certain treatments (e.g., TNFα inhibitors and IFNβ) are beneficial in only one of the two diseases. Our analysis of the co-expression of genes at these loci suggests IL-17/TNFα and JAK-STAT signaling to be particularly important mechanisms for the psoriasis/MS comorbidity.
We evaluated multiple different approaches for TDMA: inverse weighted (IVW) meta-analysis62, typically used for single trait GWAS, can favor loci that are dominated by one or the other trait, while equally weighted TDMA avoids this bias, however it does not fully take advantage of the greater accuracy provided by larger studies; we also applied approximate conditional analysis (GCTA-COJO). It is reassuring that most of the loci were found by all three approaches; however, the loci from our equally weighted TDMA all had evidence of colocalization (through COLOC and SuSiE), whereas not all of the additional loci from the other approaches did. This does not rule out these loci from affecting the same genes and pathways in psoriasis and MS, but it suggests they may have different causal variants that could affect these pathways in different ways (i.e., pleiotropy). We addressed potential pleiotropy in MR by removing any outliers detected by MR-PRESSO and using five other techniques that test the assumptions of MR. Furthermore, by applying multivariable analysis in addition to the traditional univariable approach, we controlled for the effect of confounding factors.
The TDMA loci we identified are enriched in H3K27ac active enhancer marks for B-cell centroblasts, both compared to the rest of the genome and to the psoriasis/MS-specific loci. While the role of B-cells has been elucidated in MS1, they have been less well studied in psoriasis, potentially because they are detected in smaller numbers than T-cells in lesional skin81. Nevertheless, regulatory B-cell involvement in responses to the phosphodiesterase 4 inhibitor apremilast has recently been reported82, and they are able to suppress IL-23-mediated inflammation83. Previous research showed tonsils from psoriasis patients had lower germinal center to marginal zone area ratio84, and germinal center affinity maturation plays an important role in MS85. B-cell activation is believed to be enhanced by neutrophil extracellular traps (NETs) in MS86 as well as in lupus87. While NETs have been found to promote psoriatic inflammation, particularly through Th1729,88, it has yet to be investigated whether they assist B-cell maturation in a similar way to other diseases. Biological effects of genetic signals are challenging to identify, and require mechanistic study, for example through multiomic analysis. In Table 2, we indicate which genes have eQTL support in two large datasets (eQTLGen89 and GTEx90), finding the lead marker of 80% of the loci to be an eQTL, however further work is required to pinpoint and validate specific gene targets.
The use of medical claims data can have limitations, as they are primarily collected for billing purposes rather than research. We also did not have access to quantitative data on obesity, smoking and alcohol use and so used ICD-9/10 codes for these covariates instead. It is conceivable that this information would only be recorded if the physician considers these details to be relevant to the patient’s health – for example, ICD codes reflect alcohol use disorder, rather than the number of drinks consumed, and only 16% of patients were indicated as obese, whereas other studies suggest the proportion may be almost twice as high in the USA91. The overall consistency with genetic and MR results was reassuring. However, psoriatic arthritis (PsA) occurs in up to 30% of psoriasis patients92 and can sometimes be misdiagnosed as rheumatoid arthritis (RA). This could explain the strong effect sizes observed for RA in epidemiology, and lack of significance in MR, in which patients were assessed by rheumatologists. A recent survey93 found the ICD-9/10 codes for MS have up to 92.4% sensitivity and 92.6% specificity, while for psoriasis, 81% of patients who have an ICD-10 code had a confirmed diagnosis94, with 88% sensitivity for ICD-9 codes95. However, ICD codes can still be inaccurate, especially for diseases such as MS that have variable symptoms, and future work will focus on developing and applying more rigorous case definitions, for example based on prescriptions for disease-modifying therapies, or multiple visits to relevant specialists (as is recorded in Optum’s deidentified Clinformatics® Data Mart60).
These limitations notwithstanding, our study provides genetic and epidemiological evidence for similarity and causal relationship between MS and psoriasis immunomes, while identifying important differences between these two complex diseases that should help guide future research.
Supplementary Material
Summary for Social Media If Published.
What is the current knowledge on the topic?
Both psoriasis and multiple sclerosis (MS) are complex genetic diseases, and previous studies have illustrated that psoriatic patients are at higher risk of MS.
What question did this study address?
By using a multi-omic approach, we aimed to reveal shared genetic and genomic components for psoriasis and MS.
What does this study add to our knowledge?
Our trans-disease meta-analysis (TDMA) revealed multiple genetic loci shared between psoriasis and MS, highlighting the involvement of IL17 and JAK-STAT signaling.
How might this potentially impact on the practice of neurology?
A better understanding of the shared mechanisms between psoriasis and MS can advance our management for patients that suffer from one or both conditions.
Acknowledgements
This work was supported by the Dermatology Foundation (MTP), National Psoriasis Foundation (LCT, MTP, and JEG), and awards from the National Institutes of Health (K01AR072129 to LCT; P30AR075043 to LCT, MTP, and JEG; UC2 AR081033 to LCT and JEG; R01AR042742, R01AR050511, R01AR054966, R01AR063611, and R01AR065183 to JTE).
Footnotes
Potential Conflicts of Interests
LCT has received support from Galderma, Janssen, and Novartis. J. E. Gudjonsson has served as a consultant to Almirall, BMS, Sanofi, AbbVie, Novartis, Eli Lilly, Pfizer, and Galderma; and has received research support from Almirall, Janssen, Novartis, Pfizer, BMS/Celgene, Timberpharma, and Galderma. S. Ständer has served as a consultant to Almirall, Bayer, Beiersdorf, Bellus, Bionorice, Cara Therapeutics, Celgene, Clexio, DS Biopharma, Galderma, Menlo Therapeutics, Novartis, Perrigo, Trevi Therapeutics, Dermasence, Galderma, Kiniksa, Sanofi, and Vanda Therapeutics.
Data Availability
The medical claims data is available by applying for access to Optum’s deidentified Clinformatics® Data Mart. Access to the GWAS summary statistics is detailed in the manuscript for each study. Pathway and LINCS L1000 gene sets are available through the Enrichr website (https://maayanlab.cloud/Enrichr/).
REFERENCES
- 1.Patsopoulos A Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsson T, Barcellos LF & Alfredsson L Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol 13, 25–36 (2017). [DOI] [PubMed] [Google Scholar]
- 3.McGinley MP, Goldschmidt CH & Rae-Grant AD Diagnosis and Treatment of Multiple Sclerosis: A Review. Jama 325, 765–779 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Tsoi LC et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Commun 8, 15382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich K The concept of psoriasis as a systemic inflammation: implications for disease management. J Eur Acad Dermatol Vener 26, 3–11 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Liu CY et al. Association of Multiple Sclerosis with Psoriasis: A Systematic Review and Meta-Analysis of Observational Studies. Am J Clin Dermatol 20, 201–208 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Egeberg A, Mallbris L, Gislason GH, Skov L & Hansen PR Risk of Multiple Sclerosis in Patients with Psoriasis: A Danish Nationwide Cohort Study. J Invest Dermatol 136, 93–8 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Luchtman DW, Ellwardt E, Larochelle C & Zipp F IL-17 and related cytokines involved in the pathology and immunotherapy of multiple sclerosis: Current and future developments. Cyt Growth Factor Rev 25, 403–13 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Blauvelt A & Chiricozzi A The Immunologic Role of IL-17 in Psoriasis and Psoriatic Arthritis Pathogenesis. Clin Rev Allergy Immunol 55, 379–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lees JR & Cross AH A little stress is good: IFN-gamma, demyelination, and multiple sclerosis. J Clin Invest 117, 297–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson-Huang LM et al. A single intradermal injection of IFN-γ induces an inflammatory state in both non-lesional psoriatic and healthy skin. J Invest Dermatol 132, 1177–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lozeron P, Denier C, Lacroix C & Adams D Long-term course of demyelinating neuropathies occurring during tumor necrosis factor-alpha-blocker therapy. Arch Neurol 66, 490–7 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Mylonas A & Conrad C Psoriasis: Classical vs. Paradoxical. The Yin-Yang of TNF and Type I Interferon. Front Immunol 9, 2746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold R et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N Engl J Med 367, 1098–107 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Altmeyer PJ et al. Antipsoriatic effect of fumaric acid derivatives. Results of a multicenter double-blind study in 100 patients. J Am Acad Dermatol 30, 977–81 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Langley RG et al. Secukinumab in plaque psoriasis--results of two phase 3 trials. N Engl J Med 371, 326–38 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Havrdova E et al. Activity of secukinumab, an anti-IL-17A antibody, on brain lesions in RRMS: results from a randomized, proof-of-concept study. J Neurol 263, 1287–95 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Berkovich R, Yakupova A, Eskenazi J, Carlson NG & Steinman L Improvement of Comorbid Psoriasis in Patients With MS Treated With Natalizumab. Neurol Neuroimmunol Neuroinflamm 8(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toussirot E, Bereau M, Bossert M, Malkoun I & Lohse A Occurrence of Psoriatic Arthritis during Interferon Beta 1a Treatment for Multiple Sclerosis. Case Rep Rheumatol 2014, 949317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonnberg AS et al. Heritability of psoriasis in a large twin sample. Br J Dermatol 169, 412–6 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Fagnani C et al. Twin studies in multiple sclerosis: A meta-estimation of heritability and environmentality. Mult Scler 21, 1404–13 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Westerlind H et al. Modest familial risks for multiple sclerosis: a registry-based study of the population of Sweden. Brain 137, 770–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Y, Fleming C & Yan J New insights of T cells in the pathogenesis of psoriasis. Cell Mol Immunol 9, 302–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinman L Multiple sclerosis: a coordinated immunological attack against myelin in the central nervous system. Cell 85, 299–302 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Bielekova B et al. Expansion and functional relevance of high-avidity myelin-specific CD4+ T cells in multiple sclerosis. J Immunol 172, 3893–904 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Cheuk S et al. Epidermal Th22 and Tc17 cells form a localized disease memory in clinically healed psoriasis. J Immunol 192, 3111–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dendrou CA, Fugger L & Friese MA Immunopathology of multiple sclerosis. Nat Rev Immunol 15, 545–58 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Nishihara H et al. Human CD4(+) T cell subsets differ in their abilities to cross endothelial and epithelial brain barriers in vitro. Fluids Barriers CNS 17, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert S et al. Neutrophil Extracellular Traps Induce Human Th17 Cells: Effect of Psoriasis-Associated TRAF3IP2 Genotype. J Invest Dermatol 139, 1245–1253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cree BA et al. A major histocompatibility Class I locus contributes to multiple sclerosis susceptibility independently from HLA-DRB1*15:01. PLoS One 5, e11296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack SJ et al. Correction: High resolution HLA analysis reveals independent class I haplotypes and amino-acid motifs protective for multiple sclerosis. Genes Immun 20, 340 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Osoegawa K et al. High Resolution Haplotype Analyses of Classical HLA Genes in Families With Multiple Sclerosis Highlights the Role of HLA-DP Alleles in Disease Susceptibility. Front Immunol 12, 644838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart PE et al. Transethnic analysis of psoriasis susceptibility in South Asians and Europeans enhances fine mapping in the MHC and genome wide. Human Genetics and Genomics Advances 3, 100069 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollenbach JA & Oksenberg JR The immunogenetics of multiple sclerosis: A comprehensive review. J Autoimmun 64, 13–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisi R, Symmons DP, Griffiths CE & Ashcroft DM Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol 133, 377–85 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Simpson S Jr. et al. Latitude continues to be significantly associated with the prevalence of multiple sclerosis: an updated meta-analysis. J Neurol Neurosurg Psych 90, 1193–1200 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Sintzel MB, Rametta M & Reder AT Vitamin D and Multiple Sclerosis: A Comprehensive Review. Neurol Ther 7, 59–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrea L et al. Vitamin D and its role in psoriasis: An overview of the dermatologist and nutritionist. Rev Endocr Metab Disord 18, 195–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams JS & Hewison M Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab 4, 80–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Essen MR et al. Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat Immunol 11, 344–9 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Tang J et al. Calcitriol suppresses antiretinal autoimmunity through inhibitory effects on the Th17 effector response. J Immunol 182, 4624–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giustina A et al. Controversies in Vitamin D: Summary Statement From an International Conference. J Clin Endocrinol Metab 104, 234–240 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Darwish H et al. Effect of Vitamin D Replacement on Cognition in Multiple Sclerosis Patients. Sci Rep 7, 45926 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burton JM et al. A phase I/II dose-escalation trial of vitamin D3 and calcium in multiple sclerosis. Neurology 74, 1852–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boswell K et al. Narrowband ultraviolet B treatment for psoriasis is highly economical and causes significant savings in cost for topical treatments. Br J Dermatol 179, 1148–1156 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Hart PH et al. A randomised, controlled clinical trial of narrowband UVB phototherapy for clinically isolated syndrome: The PhoCIS study. Mult Scler J Exp Transl Clin 4, 2055217318773112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Versini M, Jeandel PY, Rosenthal E & Shoenfeld Y Obesity in autoimmune diseases: not a passive bystander. Autoimmun Rev 13, 981–1000 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Budu-Aggrey A et al. Evidence of a causal relationship between body mass index and psoriasis: A mendelian randomization study. PLoS Med 16, e1002739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mokry LE et al. Obesity and Multiple Sclerosis: A Mendelian Randomization Study. PLoS Med 13, e1002053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zmora N, Bashiardes S, Levy M & Elinav E The Role of the Immune System in Metabolic Health and Disease. Cell Metab 25, 506–521 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Zhao XY et al. The obesity-induced adipokine sST2 exacerbates adipose T(reg) and ILC2 depletion and promotes insulin resistance. Sci Adv 6, eaay6191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hedstrom AK, Olsson T & Alfredsson L Smoking is a major preventable risk factor for multiple sclerosis. Mult Scler 22, 1021–6 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Armstrong AW, Harskamp CT, Dhillon JS & Armstrong EJ Psoriasis and smoking: a systematic review and meta-analysis. Br J Dermatol 170, 304–14 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Saul A et al. Stressful life events and the risk of initial central nervous system demyelination. Mult Scler 23, 1000–1007 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Stewart TJ, Tong W & Whitfeld MJ The associations between psychological stress and psoriasis: a systematic review. Int J Dermatol 57, 1275–1282 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Ascherio A & Munger KL Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann Neurol 61, 288–99 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Fry L & Baker BS Triggering psoriasis: the role of infections and medications. Clin Dermatol 25, 606–15 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Jangi S et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun 7, 12015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang HW et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 6, 154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gunaseelan V, Kenney B, Lee JS & Hu HM Databases for surgical health services research: Clinformatics Data Mart. Surgery 165, 669–671 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Patrick MT et al. Genetic signature to provide robust risk assessment of psoriatic arthritis development in psoriasis patients. Nat Commun 9, 4178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patrick MT et al. Causal Relationship and Shared Genetic Loci between Psoriasis and Type 2 Diabetes through Trans-Disease Meta-Analysis. J Invest Dermatol 141, 1493–1502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 44, 369–75, s1–3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi H, Mancuso N, Spendlove S & Pasaniuc B Local Genetic Correlation Gives Insights into the Shared Genetic Architecture of Complex Traits. Am J Hum Genet 101, 737–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berisa T & Pickrell JK Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics 32, 283–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giambartolomei C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wallace C A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet 17, e1009440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verbanck M, Chen CY, Neale B & Do R Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bowden J, Davey Smith G & Burgess S Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44, 512–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burgess SB,J; Dudbridge F; Thompson SG Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization . arXiv (2016). [Google Scholar]
- 72.Zhao Q, Wang J, Hemani G, Bowden J & Small DS Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. arXiv (2018). [Google Scholar]
- 73.Bowden J, Davey Smith G, Haycock PC & Burgess S Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol 40, 304–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hartwig FP, Davey Smith G & Bowden J Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46, 1985–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J et al. Causal inference for heritable phenotypic risk factors using heterogeneous genetic instruments. PLoS Genet 17, e1009575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patrick MT et al. Associations between COVID-19 and skin conditions identified through epidemiology and genomic studies. J Allergy Clin Immunol 147, 857–869.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farh KK et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian A et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Csardi G & Nepusz T The Igraph Software Package for Complex Network Research. InterJournal Complex Systems, 1695 (2005). [Google Scholar]
- 80.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grän F, Kerstan A, Serfling E, Goebeler M & Muhammad K Current Developments in the Immunology of Psoriasis. Yale J Biol Med 93, 97–110 (2020). [PMC free article] [PubMed] [Google Scholar]
- 82.Mavropoulos A et al. Apremilast increases IL-10-producing regulatory B cells and decreases proinflammatory T cells and innate cells in psoriatic arthritis and psoriasis. Rheumatology (Oxford) 58, 2240–2250 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Mizumaki K, Horii M, Kano M, Komuro A & Matsushita T Suppression of IL-23-mediated psoriasis-like inflammation by regulatory B cells. Sci Rep 11, 2106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sigurdardottir SL, Thorleifsdottir RH, Valdimarsson H & Johnston A The association of sore throat and psoriasis might be explained by histologically distinctive tonsils and increased expression of skin-homing molecules by tonsil T cells. Clin Exp Immunol 174, 139–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corcione A et al. B-cell differentiation in the CNS of patients with multiple sclerosis. Autoimmun Rev 4, 549–54 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Parker Harp CR et al. Neutrophils promote VLA-4-dependent B cell antigen presentation and accumulation within the meninges during neuroinflammation. Proc Natl Acad Sci U S A 116, 24221–24230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gestermann N et al. Netting Neutrophils Activate Autoreactive B Cells in Lupus. J Immunol 200, 3364–3371 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Herster F et al. Neutrophil extracellular trap-associated RNA and LL37 enable self-amplifying inflammation in psoriasis. Nat Commun 11, 105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Võsa U et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet 53, 1300–1310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aguet F The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inoue Y, Qin B, Poti J, Sokol R & Gordon-Larsen P Epidemiology of Obesity in Adults: Latest Trends. Curr Obes Rep 7, 276–288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mease PJ et al. Prevalence of rheumatologist-diagnosed psoriatic arthritis in patients with psoriasis in European/North American dermatology clinics. J Am Acad Dermatol 69, 729–735 (2013). [DOI] [PubMed] [Google Scholar]
- 93.St Germaine-Smith C et al. Recommendations for optimal ICD codes to study neurologic conditions: a systematic review. Neurology 79, 1049–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Löfvendahl S et al. Validity of diagnostic codes and prevalence of physician-diagnosed psoriasis and psoriatic arthritis in southern Sweden--a population-based register study. PLoS One 9, e98024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Asgari MM et al. Validity of diagnostic codes and prevalence of psoriasis and psoriatic arthritis in a managed care population, 1996–2009. Pharmacoepidemiol Drug Saf 22, 842–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsoi LC et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 44, 1341–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beecham AH et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45, 1353–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bowes J et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat Commun 6, 6046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheng Y et al. Sequencing-based approach identified three new susceptibility loci for psoriasis. Nat Commun 5, 4331 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The medical claims data is available by applying for access to Optum’s deidentified Clinformatics® Data Mart. Access to the GWAS summary statistics is detailed in the manuscript for each study. Pathway and LINCS L1000 gene sets are available through the Enrichr website (https://maayanlab.cloud/Enrichr/).


