Abstract
The genetic diversity of porB genes from meningococcal isolates characterized as serotype 22 was investigated by gene sequencing. This procedure identified seven distinct porB sequences, demonstrating variation in the PorB protein recognized by the serotype 22 monoclonal antibody. This is consistent with the genetic heterogeneity of serotype 22 meningococci reported previously.
Neisseria meningitidis causes bacterial meningitis and septicemia worldwide (3). For routine epidemiological surveillance, meningococci are classified by immunological reagents into serogroups (by type of capsular polysaccharide), serotypes (PorB, class 2 or 3 outer membrane protein [OMP]), and serosubtypes (PorA, class 1 OMP) (7). Many meningococcal isolates are nonserotypeable (NT), or nonserosubtypeable (NST), as a result of antigenic variation in the PorA and PorB proteins (5, 11, 14) and the assay-dependent reactivity of some of the monoclonal antibodies (MAbs) used (17, 20, 23).
A MAb identifying a new serotype, 22, was produced in the Czech Republic in 1994 to combat the large proportion (50 to 80%) of NT meningococci isolated there between 1973 and 1994 (9). Use of this MAb in the National Reference Laboratory for Meningococcal Infections, Prague, Czech Republic, showed that 44% of the meningococci previously characterized as B:NT during 1995, and 37% of such isolates obtained between 1973 and 1994, were serotype 22 (10). Testing of meningococcal isolates in other European countries gave the following rates of serotype 22 for isolates previously classified as NT: Austria, 5.4%; Germany, 11.3%; and Greece, 9.5% (19). Addition of this reagent to the serotyping panel used at the Meningococcal Reference Unit for England and Wales in 1995 identified serotype 22 organisms among invasive and noninvasive serogroup B and C isolates of diverse serosubtype. A study of 22 Czech serogroup B, serotype 22 (B:22), meningococci by PCR-restriction endonuclease pattern analysis of the pilA gene and multilocus enzyme electrophoresis concluded that these organisms were highly heterogeneous, with 17 clonal complexes and 14 pilA alleles identified among serotype 22 isolates (16).
In the present work, the antigenic heterogeneity of the PorB proteins recognized by the serotype 22 MAb was examined by nucleotide sequence determination of the porB genes of serotype 22 meningococci. The study included 10 Czech B:22 isolates (isolates 312204 to 312213) and 3 United Kingdom (U.K.) isolates, 1 B:22 (isolate 312664) and 2 C:22 (isolates 312472 and 312597) (Table 1). The meningococcal template DNA preparation was as described previously (20). Amplification of porB genes by PCR (in 100-μl reaction mixtures) was carried out with reaction buffer (Gibco BRL); 200 μM (each) dATP, dCTP, dGTP, and dTTP; 1 μM concentrations of PCR primers PB1 (5′-TAAATGCAAAGCTAAGCGGCTTG-3′) and PB2 (5′-TTTGTTGATACCAATCTTTTCAG); 0.5 U of Taq polymerase (Gibco BRL); and 1 μl of template DNA (approximately 50 ng μl−1). Reaction conditions were 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min, followed by incubation at 72°C for a further 2 min. Purification and direct nucleotide sequence determination of amplified porB genes were done as described previously (20).
TABLE 1.
Meningococcal serotype 22 strains examined and GenBank accession numbers of their porB gene sequences
| Isolate no. | Country of origin | Serological data | porB sequence accession no. |
|---|---|---|---|
| 312204 | Czech Republic | B:22:NST | AF065125 |
| 312205 | Czech Republic | B:22:NST | AF065126 |
| 312206 | Czech Republic | B:22:P1.9,10 | AF065125 |
| 312207 | Czech Republic | B:22:NST | AF065126 |
| 312208 | Czech Republic | B:22:P1.4 | AF065126 |
| 312209 | Czech Republic | B:22:NST | AF065125 |
| 312210 | Czech Republic | B:22:NST | AF065126 |
| 312211 | Czech Republic | B:22:P1.14 | AF065126 |
| 312212 | Czech Republic | B:22:P1.14 | AF065126 |
| 312213 | Czech Republic | B:22:P1.1,7 | AF065126 |
| 312472 | U.K. | C:22:P1.15,10 | AF065126 |
| 312597 | U.K. | C:22:NST | AF065126 |
| 312664 | U.K. | B:22:NST | AF065126 |
| 315/85 | Germany | B:22:P1.7,14 | AF065127 |
| EG 011 | Germany | B:22:P1.3 | AF065128 |
| NG H38 | Norway | NG:22:P1.3,6 | AF065129 |
| 528 | USSR | B:22:P1.14 | AF065130 |
| 503/93 | Czech Republic | B:22:NST | U92906a |
Published previously (17).
Two class 2 OMP-encoding porB allele sequences were identified among the 13 isolates (GenBank accession no. AF065125 and AF065126). The peptide sequences deduced from these alleles were consistent with the porin model of PorB structure (22), with eight surface exposed loop regions (loops I to VIII) in which the serotype-specific peptide sequences reside (6). The new sequences were aligned with 92 meningococcal PorB protein sequences covering all known serotypes, including a distinct sequence from an additional serotype 22 isolate (GenBank accession no. U92906 [17]) and many PorB sequences from isolates described as NT, most of which had not been tested with the serotype 22 MAb (2, 6, 17, 21).
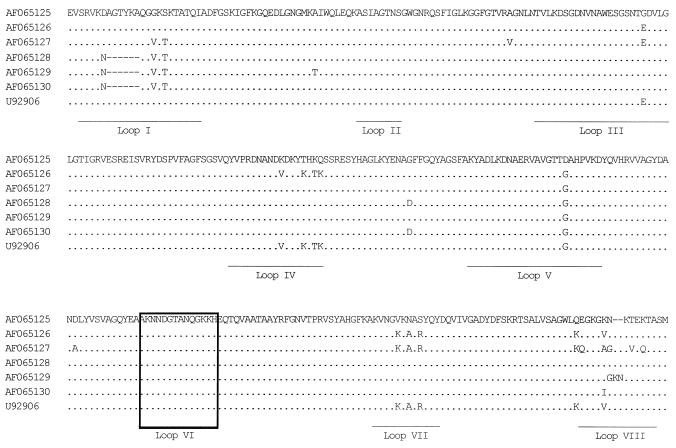
Comparison of the alignment showed that the only peptide sequence unique to the serotype 22 PorB sequences was AKNNDGTANQGKKH, located in putative loop VI, all other loop sequences being diverse among serotype 22 isolates or shared with PorB sequences from isolates with different serotypes (Fig. 1 and data not shown). This loop VI sequence was also encoded by the porB genes from isolates 315/85, EG 011, NG H38, and 528 (GenBank no. AF065127, AF065128, AF065129, and AF065130, respectively) (Fig. 1). These isolates had not been typed with a reagent panel including the serotype 22 MAb and were classified as NT. To test the hypothesis that the loop VI peptide sequence was required for serotype 22 MAb recognition, the isolates were reserotyped, in a blinded fashion, with a MAb panel including the serotype 22 MAb. All four meningococci were characterized as serotype 22 (Table 1), strengthening the evidence that loop VI is the critical loop for serotype 22 recognition.
FIG. 1.
Alignment of seven meningococcal PorB protein sequences obtained by translation of the nucleotide sequences of each of the porB alleles identified among serotype 22 meningococci. The locations of the putative surface loops (I to VIII) of the porin are indicated. The boxed area defines the putative surface loop VI that is likely to be important for recognition of the PorB proteins by the serotype 22 MAb. Sequences AF065126 and U92906 were different at the nucleotide sequence level but possessed identical peptide sequences.
The relationships among the seven porB allele sequences identified in serotype 22 meningococci were represented graphically by the split-decomposition method (1) (Fig. 2). The split graph obtained illustrates a network of possible pathways linking the porB allele sequences obtained from serotype 22 meningococci, suggesting that genetic recombination, which occurs within and between Neisseria species (12, 15, 18), has resulted in the circulation of a sequence encoding the serotype 22 epitope among the porB alleles present in populations of N. meningitidis.
FIG. 2.
Split-decomposition analysis of the seven porB sequences obtained from serotype 22 meningococcal isolates. Split graphs were drawn from Hamming (uncorrected) distance matrices of aligned porB allele sequences by using SplitsTree version 2.4 (8). Branch lengths are drawn to scale.
In conclusion, these data demonstrate that while meningococcal strains that react with the serotype 22 MAb possess an identical peptide sequence in variable surface loop VI of the PorB protein, the PorB proteins may be encoded by mosaic gene structures that are highly diverse in one or more of the other variable surface loops. Furthermore, the serotype 22 epitope is encoded by only a small part of the meningococcal genome, the remainder of which has been shown previously to be highly heterogeneous among serotype 22 meningococci (16). Positive reactions with the serotype 22 MAb therefore do not provide a robust indication of the genetic relatedness of meningococcal isolates unless they are supported by additional epidemiological information, obtained, for example, from multilocus sequence typing (13) or multilocus enzyme electrophoresis analyses (4).
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under accession no. AF065125 to AF065130.
Acknowledgments
M.C.J.M. is a Wellcome Senior Research Fellow in Biodiversity, and R.U. and M.C.J.M. are grateful to the Wellcome Trust for financial support. P.K. and M.M. acknowledge the support of research grant 310/96/K102 from the Grant Agency of the Czech Republic and research grant 3982-3 from the Internal Grant Agency of the Ministry of Health of the Czech Republic.
We thank Steve Gray at the Meningococcal Reference Unit for England and Wales for serological characterization of meningococcal isolates.
REFERENCES
- 1.Bandelt H J, Dress A W. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 2.Bash M C, Lesiak K B, Banks S D, Frasch C E. Analysis of Neisseria meningitidis class 3 outer membrane protein gene variable regions and type identification using genetic techniques. Infect Immun. 1995;63:1484–1490. doi: 10.1128/iai.63.4.1484-1490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartwright K A V. Meningococcal disease. Chichester, United Kingdom: Wiley; 1995. [Google Scholar]
- 4.Caugant D A, Mocca L F, Frasch C E, Frøholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feavers I M, Fox A J, Gray S, Jones D M, Maiden M C J. Antigenic diversity of meningococcal outer membrane protein PorA has implications for epidemiological analysis and vaccine design. Clin Diagn Lab Immunol. 1996;3:444–450. doi: 10.1128/cdli.3.4.444-450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feavers I M, Suker J, McKenna A J, Heath A B, Maiden M C J. Molecular analysis of the serotyping antigens of Neisseria meningitidis. Infect Immun. 1992;60:3620–3629. doi: 10.1128/iai.60.9.3620-3629.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 8.Huson D H. Splits Tree: a program for analysing and visualising evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 9.Krizova P, Musilek M. Changing epidemiology of meningococcal invasive disease in the Czech Republic caused by new clone Neisseria meningitidis C:2a:P1.2(P1.5), ET-15/37. Central Eur J Public Health. 1995;3:189–194. [PubMed] [Google Scholar]
- 10.Krizova P, Musilek M, Danielova V, Holubova J. New serotype candidate of Neisseria meningitidis. Central Eur J Public Health. 1996;4:169–172. [PubMed] [Google Scholar]
- 11.Maiden M C J. The impact of molecular techniques on the study of meningococcal disease. In: Woodford N, Johnson A P, editors. Molecular bacteriology: protocols and clinical applications. Totowa, N.J: Humana Press; 1998. pp. 265–291. [DOI] [PubMed] [Google Scholar]
- 12.Maiden M C J. Population genetics of a transformable bacterium: the influence of horizontal genetical exchange on the biology of Neisseria meningitidis. FEMS Microbiol Lett. 1993;112:243–250. doi: 10.1111/j.1574-6968.1993.tb06457.x. [DOI] [PubMed] [Google Scholar]
- 13.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden M C J, Feavers I M. Meningococcal typing. J Med Microbiol. 1994;40:157–158. doi: 10.1099/00222615-40-3-157. [DOI] [PubMed] [Google Scholar]
- 15.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musilek M, Giorgini D, Hamadouche N, Kriz P, Taha M-K. Genetic heterogeneity of strains of Neisseria meningitidis belonging to serotype 22 isolated in the Czech Republic. J Clin Microbiol. 1998;36:563–565. doi: 10.1128/jcm.36.2.563-565.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacchi C T, Lemos A P S, Whitney A M, Solari C A, Brandt M E, Melles C E A, Frasch C E, Mayer L W. Correlation between serological and sequencing analyses of the PorB outer membrane protein in the Neisseria meningitidis serotyping system. Clin Diagn Lab Immunol. 1998;5:348–354. doi: 10.1128/cdli.5.3.348-354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spratt B G, Smith N H, Zhou J, O’Rourke M, Feil E. The population genetics of the pathogenic Neisseria. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 143–160. [Google Scholar]
- 19.Tzanakaki G, Kriz P, Kremastinou J, Musilek M, Smart L E, Blackwell C C. Reactivity of the new monoclonal antibody ‘22’ with meningococcal strains isolated from patients and carriers in Greece. FEMS Immunol Med Microbiol. 1997;19:1–5. doi: 10.1111/j.1574-695X.1997.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 20.Urwin R, Feavers I M, Jones D M, Maiden M C J, Fox A J. Molecular analysis of meningococcal serotype 4 antigen genes. Epidemiol Infect. 1998;1998:95–101. doi: 10.1017/s0950268898008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urwin, R., and M. C. J. Maiden. Unpublished observations.
- 22.van der Ley P, Heckels J E, Virji M, Hoogerhout P, Poolman J T. Topology of outer membrane porins in pathogenic Neisseria spp. Infect Immun. 1991;59:2963–2971. doi: 10.1128/iai.59.9.2963-2971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wedege E, Caugant D A, Frøholm L O, Zollinger W D. Characterization of serogroup A and B strains of Neisseria meningitidis with serotype 4 and 21 monoclonal antibodies and by multilocus enzyme electrophoresis. J Clin Microbiol. 1991;29:1486–1492. doi: 10.1128/jcm.29.7.1486-1492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]