Abstract
BACKGROUND & AIMS:
The amino acid hypusine, synthesized from the polyamine spermidine by the enzyme deoxyhypusine synthase (DHPS), is essential for the activity of eukaryotic translation initiation factor 5A (EIF5A). The role of hypusinated EIF5A (EIF5AHyp) remains unknown in intestinal homeostasis. Our aim was to investigate EIF5AHyp in the gut epithelium in inflammation and carcinogenesis.
METHODS:
We utilized human colon tissue mRNA samples and publicly-available transcriptomic datasets, tissue microarrays, and patient-derived colon organoids. Mice with intestinal epithelial-specific deletion of Dhps were investigated at baseline and in models of colitis and colon carcinogenesis.
RESULTS:
We found that patients with ulcerative colitis and Crohn’s disease exhibit reduced colon levels of DHPS mRNA and DHPS protein, and reduced levels of EIF5AHyp. Similarly, colonic organoids from colitis patients also show downregulated DHPS expression. Mice with intestinal epithelial-specific deletion of Dhps develop spontaneous colon hyperplasia, epithelial proliferation, crypt distortion, and inflammation. Furthermore, these mice are highly susceptible to experimental colitis and show exacerbated colon tumorigenesis when treated with a carcinogen. Transcriptomic and proteomic analysis on colonic epithelial cells demonstrated that loss of hypusination induces multiple pathways related to cancer and immune response. Moreover, we found that hypusination enhances translation of numerous enzymes involved in aldehyde detoxification, including glutathione S-transferases and aldehyde dehydrogenases. Accordingly, hypusination-deficient mice exhibit increased levels of aldehyde adducts in the colon, and their treatment with a scavenger of electrophiles reduces colitis.
CONCLUSIONS:
Hypusination in intestinal epithelial cells has a key role in prevention of colitis and colorectal cancer, and enhancement of this pathway via supplementation of spermidine could have therapeutic impact.
Keywords: Hypusine, Intestinal epithelial cells, Inflammation, Colon cancer
Graphical Abstract
Introduction
The prevalence of inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is increasing considerably worldwide, especially in countries with the largest populations, such as USA, India, and China.1,2 Newer biologic therapies for IBD have been developed and extensively used, but they induce remission in only about half of patients, are expensive, and have various side effects. Moreover, colonic inflammation is also a risk factor for colitis-associated carcinoma (CAC) that accounts for approximately 15% of all deaths in IBD patients.3 Thus, a better understanding of the molecular events contributing to inflammation and neoplastic transformation would benefit IBD patients.
Polyamines are organic cations and the first one, putrescine, is generated by the enzymatic decarboxylation of arginine and ornithine by the arginine decarboxylase/agmatinase pathway and the enzyme ornithine decarboxylase, respectively. Putrescine is converted into spermidine (Spd), which is then transformed into spermine. The enzyme spermine oxidase (SMOX) catalyzes the back-conversion of spermine to Spd.4 We have shown that patients with colitis have reduced SMOX expression and that mice with Smox deletion exhibit decreased colonic Spd and an exacerbation of colitis5,6 and CAC.6 Treatment of animals with Spd ameliorates both diseases and protects mice with epithelial deletion of Apc from tumorigenesis.6 The enzyme deoxyhypusine synthase (DHPS) transfers the N-terminal moiety of Spd to the lysine-50 residue of the protein eukaryotic translation initiation factor 5A (EIF5A) to form the intermediate deoxyhypusine residue;7 then deoxyhypusine hydroxylase (DOHH) catalyzes the hydroxylation of this intermediate8 to complete the synthesis of the nonproteinogenic amino acid hypusine. Thus, EIF5A is the only known protein that contains hypusine. Hypusinated EIF5A (EIF5AHyp) enters the nucleus via passive diffusion and binds specific mRNAs exhibiting a 5′-AAAUGU-3′ consensus sequence.9,10 EIF5AHyp/mRNA complexes are then translocated to the cytoplasm by the nuclear transporter exportin-411 and reach the ribosomes for translation.12 EIF5AHyp also alleviates ribosome pausing during the translation of peptides enriched in diprolyl and diglycyl motifs.13,14 Hypusination is crucial for embryogenesis and deletion of Eif5a1 or Dhps is lethal.15 Because hypusination supports cell differentiation and proliferation,16,17 it has been proposed that this pathway could represent a potential target to limit tumor progression.18 Chemical inhibition of DHPS using the competitive Spd analog N1-guanyl-1,7-diamineheptane (GC7) or knockdown of the genes encoding for EIF5A or DHPS reduces the growth of established cancer cell lines in vitro and in xenograft models.18,19 However, the effect of hypusination on inflammation and carcinogenesis in non-transformed epithelial cells, notably in the gastrointestinal tract, remains unknown.
We found that mice with specific deletion of Dhps in intestinal epithelial cells (IECs) exhibit spontaneous intestinal inflammation, are highly susceptible to dextran sulfate sodium (DSS)-induced colitis and to the azoxymethane (AOM)-DSS protocol and develop more tumors in a model of colorectal cancer driven by AOM alone. We demonstrate that epithelial EIF5AHyp regulates the translation of enzymes involved in reactive aldehyde detoxification, leading to the development of a proinflammatory and procarcinogenic profile in colonic epithelial cells (CECs) lacking DHPS. Human translational relevance is supported by our findings that the levels of DHPS mRNA are decreased in IBD patient colon tissues, levels of DHPS and EIF5AHyp in CECs are reduced in these tissues, and colon organoids from patients with colitis exhibit decreased DHPS expression compared to controls. Moreover, administration of a scavenger of reactive aldehydes ameliorated DSS-induced colitis in Dhps epithelial-specific knockout mice. Thus, enhancement of the hypusination pathway by supplementation with the DHPS substrate, Spd, and/or detoxification of reactive aldehydes are potential treatments for IBD and chemopreventive methods for prevention of CAC and CRC.
Methods
Ethics Statement
The ethics statements regarding human tissues and animal experiments are provided in the Supplementary Methods.
Database Analysis
Series matrix files were analyzed by GEO2R from the GEO database (http://www.ncbi.nlm.nih.gov/geo/).
Mice and Models of Colitis and Colon Cancer
Animals were house bred in our animal facility and were fed with 5L0D chow (LabDiet). To achieve specific deletion of Dhps in IECs, we crossed C57BL/6J Dhpsfl/fl mice, in which exons 2 to 7 of the Dhps gene were flanked by Cre recombinase recognition sequences,20 to transgenic mice overexpressing the epithelial driver Vil1cre (Strain 021504; The Jackson Laboratory). The resulting Dhps+/fl;Vil1cre mice were then crossed to Dhpsfl/fl mice to create Dhpsfl/fl;Vil1cre mice (46% of offspring), termed DhpsΔepi, and the littermate Dhpsfl/fl mice (54% of offspring), which were used as controls. Littermate co-housed Dhpsfl/fl and DhpsΔepi female and male mice (8–12 wk) were used for all the experiments.
For the colitis model, age-matched (8–12 wk) female and male mice were treated with 2%–4% DSS (mol. wt. 36,000–50,000; TdB Labs) in the drinking water for 5 days; DSS was then removed, and mice were maintained on regular water until euthanasia. For the CAC model, animals received one intraperitoneal (i.p.) injection of 12.5 mg/kg AOM (Millipore) followed by 3 cycles of 1.7%–3% DSS for 4 days beginning at day 5, 26, and 476,21 and mice were euthanized on day 56 or sooner if moribund. In an additional model, mice were treated every wk with an i.p. injection of 12.5 mg/kg AOM for 3 or 6 consecutive wks and were euthanized 127 days after the first injection.22 Animals were also treated with 1.5 mg/ml 5-ethyl-2-hydroxybenzylamine (EtHOBA), a potent scavenger of dicarbonyl electrophiles,21,23,24 in the drinking water one wk before DSS and were also maintained on EtHOBA during DSS treatment and the recovery period.29
Mice were monitored daily and weighed daily for the DSS model and weekly for the CAC model. Animals showing extreme distress, that became moribund, or lost more than 20% of initial body weight were euthanized. After sacrifice, colons were removed, measured, dissected, and Swiss-rolled for histology. In the cancer models, tumors were also counted and measured in two dimensions in the distal colon with electronic calipers under a dissecting microscope.6,21,25 Tumor burden was determined by the sum of areas of all tumors.
Histology
Swiss-rolled colons were fixed in formalin and embedded in paraffin, and 5 μm sections were stained with hematoxylin and eosin (H&E) and examined in a blinded manner by a gastrointestinal pathologist (M.K.W.). Histological injury score (0–40) and severity of dysplasia in the AOM model were determined as described.6,21,25
Immunostaining
The details regarding immunohistochemistry (IHC) and immunofluorescence (IF) are provided in the Supplementary Methods.
Isolation of CECs
The details are provided in the Supplementary Methods.
Colon Organoids
The methods used to generate and stimulate colon organoids are provided in the Supplementary Methods.
Analysis of mRNA Expression
RNA sequencing (RNA-Seq) and RT-PCR are described in the Supplementary Methods.
Protein Expression Analysis
Proteomic analysis and Western blotting are described in the Supplementary Methods.
Measurement of Glutathione-S-transferase (GST) Activity
Colonoids were treated or not with 25 μM GC7 for 24 h. Proteins were then extracted and quantified as reported.26 The activity of the total GST activity in protein lysates (10 μg) was measured using the GST Assay Kit (LSBio).
Quantification of Dicarbonyl Electrophile Adducts
The measurement of the crosslinks of malondialdehyde (MDA) to lysine (dilysyl-MDA) was performed by mass spectrometry, as described in the Supplementary Methods.
Fecal Microbiota Analysis
Littermate co-housed Dhpsfl/fl and DhpsΔepi mice (9 wks) were sacrificed, and feces present in the colon were harvested. Fecal DNA isolation, sequencing of 16S rRNA genes, and analysis were performed as described.6
Statistics
Figures and statistics were performed using GraphPad Prism 9.4.1 software. Significance level was set as P < .05 and all statistical tests were two-sided. All the data represent the mean ± SEM. Outliers were identified using the ROUT test (Q = 5%) and removed from the analysis. Data that were not normally distributed according to the D’Agostino & Pearson normality test were log or square root transformed, and distribution was re-assessed. Student’s t test or one-way/two-way ANOVA with the Tukey or Dunnett tests were used to determine significant differences between two or multiple groups, respectively. Contingency and correlation were analyzed by the Chi-square or the Fisher’s exact test and the Pearson test, respectively. Survival was analyzed by the Log-rank (Mantel-Cox) test.
For the RNA-Seq, the DEGs were identified using a Benjamini-Hochberg test adjusted for false discovery rate.
The relative abundance of the genera of the intestinal microbiota was analyzed by the Wilcoxon rank sum test after multiple comparison adjustment by the Benjamini & Hochberg method.
Data Availability
RNA-Seq data can be accessed on the GEO repository using the accession number GSE201556 (token: yxybcsecfxundih). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD033228 (Reviewer account details: Username: reviewer_pxd033228@ebi.ac.uk; Password: c5NVRzhi). The 16S rRNA sequencing of the fecal microbiota has been deposited on the Sequence Read Archive website with the BioProject accession number PRJNA814820 (Temporary Submission ID: SUB11173798).
Results
DHPS Level Is Reduced in the Colon During Colitis
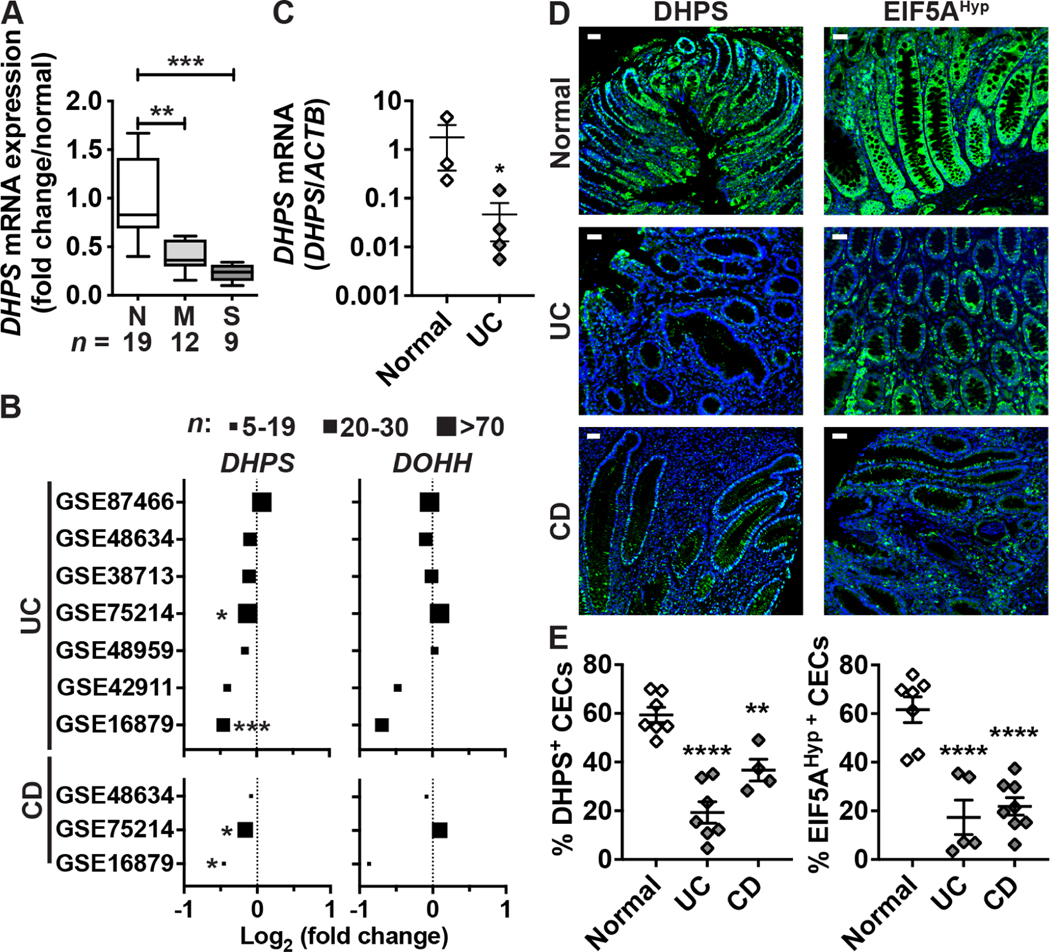
Utilizing RNA samples from a previous study,27,28 we found that DHPS expression was reduced in colon tissues of patients with moderate and severe UC compared to normal controls (Figure 1A). This result was confirmed by previously published RNA-Seq publicly available data on the GEO website; among 7 studies with UC patients, 6 exhibited a reduced expression of DHPS in the colon, including 3 with significant differences (Figure 1B). In contrast, the expression of DOHH was not affected in UC patients (Figure 1B). Also, DHPS expression was reduced in each of 3 studies in the colon of CD patients, with 2 showing significant differences, while DOHH did not show this pattern (Figure 1B). Accordingly, we found that DHPS level in colonoids from patients with UC was lower compared to those from normal tissues (Figure 1C). Further, when we immunostained IBD tissue microarrays (TMAs) we observed strong DHPS and EIF5AHyp expression in CECs from normal individuals (Figure 1D), while the levels of DHPS-positive and EIF5AHyp-positive CECs were significantly reduced in patients with UC or CD (Figures 1D–E).
Figure 1.
Expression of DHPS in patients with colitis. (A) Total RNA was extracted from colon biopsies from normal (N; n = 19) or UC patients with moderate (M; n = 12) or severe (S; n = 9) colitis. DHPS mRNA levels were determined by RT-real-time PCR. **P < .01, ***P < .001 by ANOVA and Tukey test. (B) RNA-Seq data was interrogated from published studies using the GEO data repository to determine the expression of the genes DHPS and DOHH in patients with UC or CD compared to normal individuals. n indicates the number of patients with colitis in each study. *P < .05, ***P < .001 by the Benjamini & Hochberg test adjusted for false discovery rate. (C) DHPS mRNA expression was assessed by quantitative RT-PCR in colon organoids from normal or UC tissues. Each dot represents the mean of 2–3 experiments performed with colonoids from each patient; *P < .05 using the unpaired t test. (D-E) Tissues sections of two TMAs were immunostained for DHPS or EIF5AHyp and representative images of normal tissues (n = 7) or tissues from patients with UC (n = 7) or CD (n = 4) are shown in (D) with scale bar, 50 μm; (E) Quantification using QuPath of DHPS+ and EIF5AHyp+ epithelial cells among total CECs on the TMAs. **P < .01, ****P < .0001 by ANOVA and Dunnett’s test.
We then verified the level of hypusination in the colon of mice with CAC. There was a reduction of EIF5AHyp in the non-tumor tissues from AOM-DSS-treated C57BL/6 mice, and hypusination level was significantly augmented in these mice that were given the DHPS substrate, Spd (Supplementary Figure 1).
Mice with Specific Deletion of Dhps in IECs Develop Colonic Inflammation
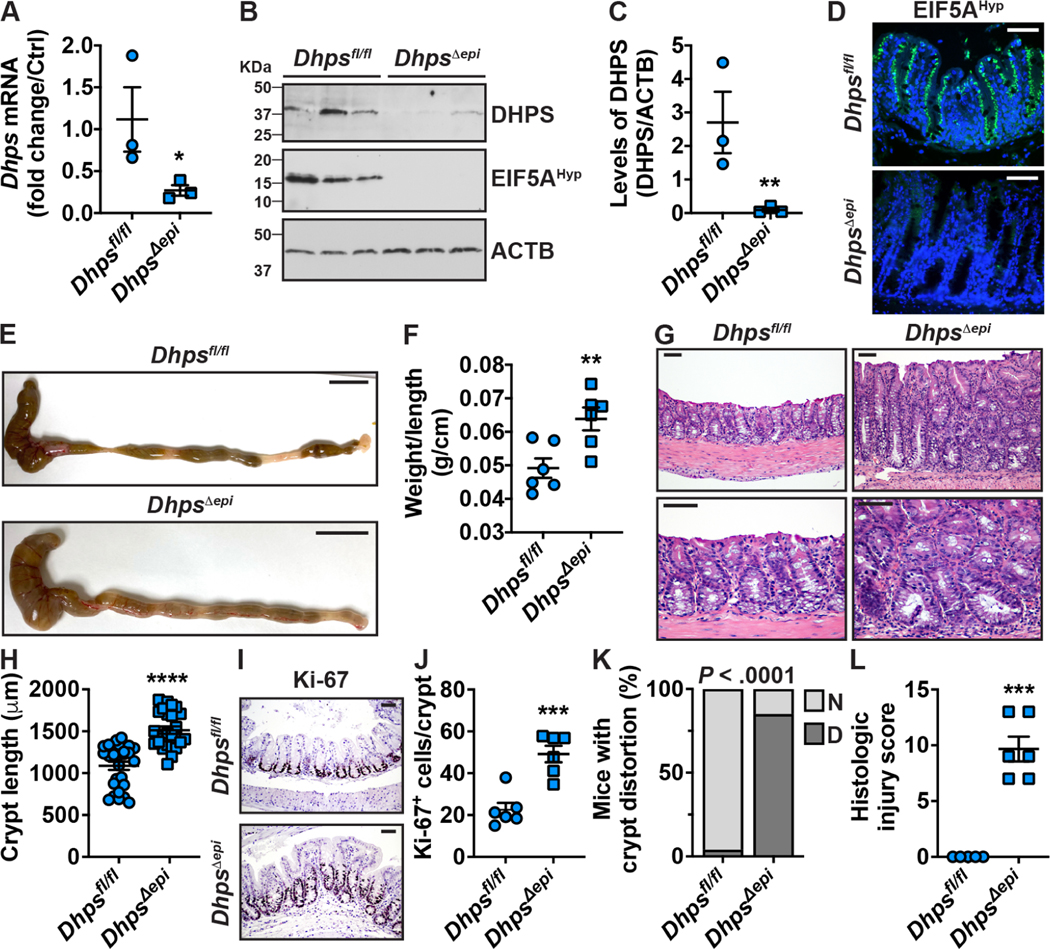
Since patients with IBD exhibit reduced hypusination in their colon, we sought to assess the effect of epithelial-specific deletion of Dhps on intestinal homeostasis. Isolated CECs from DhpsΔepi mice exhibited a significant reduction of Dhps mRNA (Figure 2A) and DHPS protein (Figures 2B–C) compared to Dhpsfl/fl mice. EIF5AHyp levels were also reduced in DhpsΔepi CECs (Figure 2B). Using IF, we observed that EIF5AHyp was localized all along the crypts and that the staining was markedly reduced in the epithelium of DhpsΔepi mice (Figure 2D).
Figure 2.
Effect on the colon of the selective depletion of Dhps in IECs. CECs from 7-wk-old Dhpsfl/fl and DhpsΔepi mice were isolated and the expression of Dhps mRNA was assessed by RT-real time PCR (A). The level of DHPS and EIF5AHyp was analyzed by Western blot (B) followed by densitometry (C). (D) IF for EIF5AHyp in the colon; representative image of 3 mice/genotype. The colon of these animals (3 males and 3 females per genotype) was removed (E) and length and weight were measured (F). Longitudinal sections were stained by H&E (G). Crypt length was determined using ImageJ 1.53a (n = 25–27 crypts counted from 6 different mice per genotype) (H). Proliferation was determined by IHC for Ki-67 (I) and quantification of Ki-67-positive nuclei (J); a total of 100 crypts were counted per mouse. The number of animals (n = 26/genotype) exhibiting normal crypt (N) or crypt distortion (D) (K) and the histologic injury scores (L) were evaluated by our pathologist in a blinded manner. In all the panels with dot plots, *P < .05, **P < .01, ***P < .001, ****P < .0001 by unpaired Student’s t test; in K, P was calculated using the Fisher’s exact test. Scale bars represent 1 cm (E) or 50 μm (D, G, H).
Strikingly, the colon and the cecum of DhpsΔepi mice exhibited a marked thickening (Figure 2E) that was confirmed by the significant increase of the colon weight/length ratio (Figure 2F). Histologically, we observed a marked expansion of the mucosal compartment (Figure 2G), confirmed by increased crypt length (Figure 2H) and by increased proliferation assessed by immunodetection (Figure 2I) and quantification (Figure 2J) of cells expressing Ki-67. The mucosal architecture of the colon of DhpsΔepi mice was abnormal, showing variation in crypt size and shape (Figure 2G). Crypts showed basal dilatation and bifurcation, and the luminal surface was undulating and villiform (Figure 2G); thus, crypt distortion was found in 84.6% of DhpsΔepi mice, whereas only 4% of Dhpsfl/fl exhibited this phenotype (Figure 2K). Patchy accumulation of neutrophils and chronic inflammatory cell infiltrates were present in the lamina propria of DhpsΔepi mice (Figure 2G), indicating mild colitis. Accordingly, the histologic injury score was significantly higher in DhpsΔepi mice (Figure 2L). This was further evidenced by the significant increase of i) myeloperoxidase (MPO)-positive cells in the mucosa of DhpsΔepi mice (Supplementary Figures 2A–B) and ii) the expression of the genes encoding for the chemokine CXCL2, the innate effectors NOS2, TNF-α and IL-1β, and the Th1 and Th17 prototype cytokines, IFN-γ and IL-17, respectively, in the colon of DhpsΔepi mice (Supplementary Figure 2C). The expression of the tight junction protein ZO1 was similar in the colon of Dhpsfl/fl and DhpsΔepi mice (Supplementary Figure 2D), suggesting that the inflammation observed in DhpsΔepi mice is not associated with a loss of barrier function.
In addition, we found that epithelial distortion and inflammation was also observed along the small intestine of DhpsΔepi mice, progressively from the duodenum and jejunum to the ileum (Supplementary Figure 3).
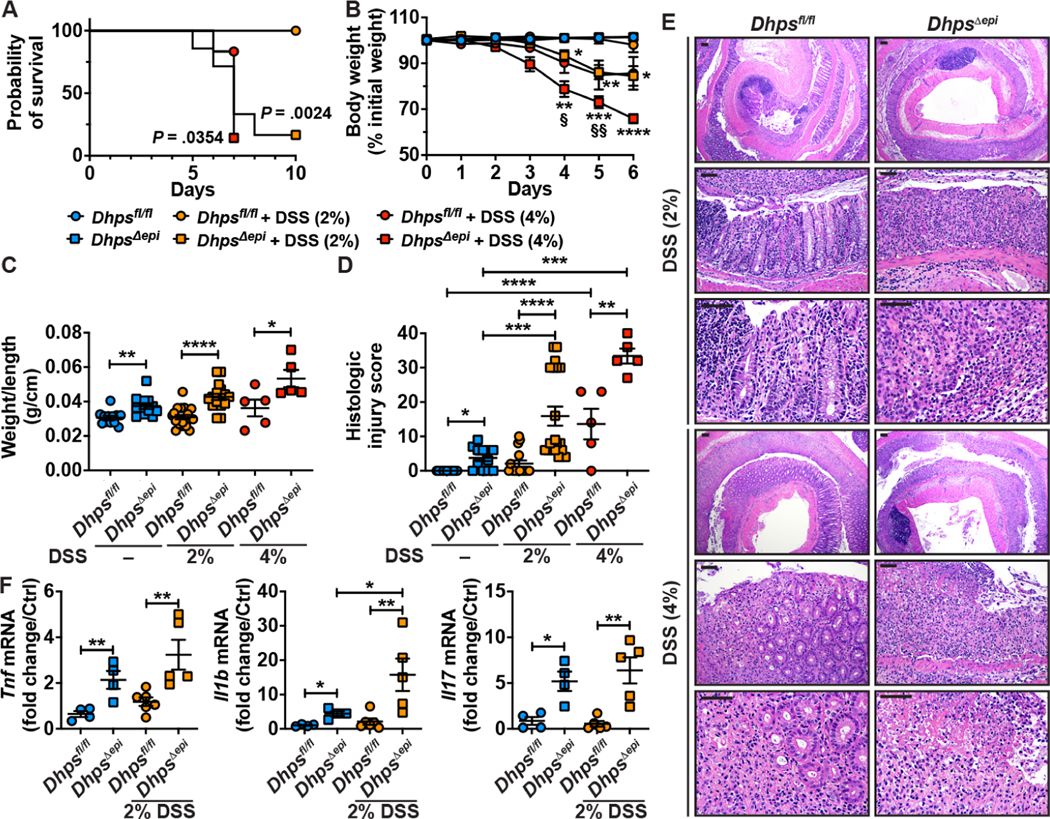
Deletion of Dhps in IECs Worsens DSS Colitis
Dhpsfl/fl and DhpsΔepi mice were then treated with DSS to induce epithelial injury and colitis. Mortality of DhpsΔepi mice treated with 2% DSS was significantly increased beginning day 6 post-treatment, whereas Dhpsfl/fl mice were not affected (Figure 3A). DhpsΔepi mice began losing weight on day 4 after starting 2% DSS, whereas Dhpsfl/fl mice did not lose weight (Figure 3B). When 4% DSS was used, DhpsΔepi mice exhibited a rapid worsening of their condition, characterized by high mortality level (Figure 3A) and a marked body weight loss (Figure 3B). In this context, mice treated with 2% and 4% DSS were sacrificed at day 9 and day 6, respectively. The weight/length ratio of the colon (Figure 3C) and colonic inflammation and epithelial damage (Figures 3D–E) were significantly increased in DhpsΔepi mice vs. Dhpsfl/fl animals. We also found significant increases in the expression of the proinflammatory genes Tnf, Il1b, and Il17 in DhpsΔepi mice that were given 2% DSS (Figure 3F).
Figure 3.
DSS-induced colitis in Dhpsfl/fl and DhpsΔepi mice. Animals were treated or not with 2% or 4% DSS. (A) Kaplan-Meier curves of DSS-treated mice; the P value was obtained using the Log-rank test (n = 7–20 mice/group). (B) Body weights were measured daily and are depicted as percentage of initial body weight; *P < .05, **P < .01, ***P < .001, ****P < .0001 compared to untreated DhpsΔepi mice, and §P < .05, §§P < .01 versus Dhpsfl/fl + 4% DSS, by two-way ANOVA and Tukey test. (C) Colons were harvested, washed, weighed, and measured. (D) Histological injury was determined from H&E staining (E); the scale bars on the low-power and high-power photomicrographs correspond to 100 μm and 50 μm, respectively. (F) mRNA expression from the colonic mucosa; Dhpsfl/fl mice were used as control for the semi-quantitative analysis. In all panels with dot plots, *P < .05, **P < .01, ***P < .001 by two-way ANOVA and Tukey test.
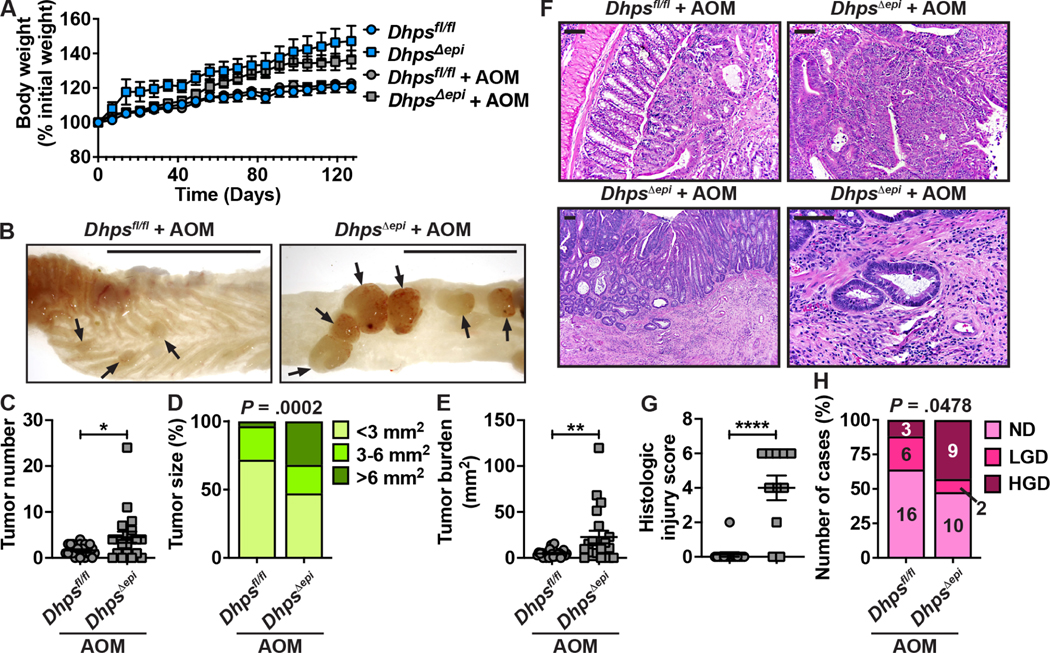
Increased Tumorigenesis in DhpsΔepi Mice
Since DhpsΔepi mice exhibit spontaneous colitis and are highly susceptible to DSS, we hypothesized that these animals would develop more tumors in the AOM-DSS model of CAC. Since we observed a high mortality rate with 2% and 4% DSS, we used 1.7% for DhpsΔepi mice. In fact, we observed a 100% mortality rate in DhpsΔepi mice after the first cycle of 1.7% DSS, whereas 90% of Dhpsfl/fl animals survived the 3 cycles of 3% DSS (Supplementary Figure 4). Therefore, it was not possible to assess tumorigenesis in the DhpsΔepi mice in this model since the disease was so severe. We thus reasoned that the spontaneous colonic inflammation of DhpsΔepi mice would be sufficient to support tumor development in animals treated only with the carcinogen AOM. When DhpsΔepi mice received 3 injections of AOM, we observed no effect on body weight (Supplementary Figure 5A). While only 1 out of 10 Dhpsfl/fl mice treated with AOM developed a tumor in the colon, 50% of the DhpsΔepi mice exhibited tumors (Supplementary Figure 5B) and, therefore, tumor number (Supplementary Figure 5B) and burden (Supplementary Figure 5C) were significantly increased in DhpsΔepi animals. After 6 injections of AOM, we did not observe a significant effect on body weight (Figure 4A). However, there was a marked increase in the number (Figures 4B and 4C) and the size (Figures 4B and 4D) of the tumors in DhpsΔepi mice, yielding a significant increase in the tumor burden (Figure 4E). Histologically, the injury score of the non-tumor area of the colon was significantly higher in DhpsΔepi mice (Figures 4F and 4G). Importantly, high-grade dysplasia (HGD) was observed in 43% of the DhpsΔepi mice, while only 12% of the Dhpsfl/fl mice exhibited HGD (Figures 4F and 4H). Notably, we also observed one case of invasive carcinoma in the AOM-treated DhpsΔepi group (Figure 4F), which was included in the HGD group in Figure 4H.
Figure 4.
Effect of epithelial Dhps deletion on colon carcinogenesis. Dhpsfl/fl (n = 25) and DhpsΔepi mice (n = 22) were given 6 i.p. injections of 12.5 mg/kg AOM. Note that two DhpsΔepi mice died between day 119 and 127 (P = 0.132), and that none of the control animals that did not receive AOM developed tumors. (A) Body weights are depicted as percentage of initial body weight. (B-E) Colons were harvested, opened longitudinally (B) and tumors (arrow) were counted (C) and measured (D); in D, the percentages were determined from a total of 57 and 91 tumors in Dhpsfl/fl and DhpsΔepi mice, respectively. Tumor burden (E) corresponds to the sum of the surface area of all the tumors in each mouse. (F-H) H&E staining (F) was used to assess histologic injury (G) and the grade of dysplasia (H). In F, the photomicrographs in the upper panels for Dhpsfl/fl and DhpsΔepi mice show LGD and HGD, respectively; the lower panels depict an invasive carcinoma; scale bars, 50 μm. *P < .05, **P < .01, ****P < .0001 by Student’s t test; in D and H, the P values were calculated using the Chi-square test.
Dhps Deletion in IECs Does not Modify the Gut Microbiome
We assessed the fecal microbiome of littermate Dhpsfl/fl and DhpsΔepi mice by sequencing the V4 region of the 16S rRNA gene. Bacterial community diversity, measured by Shannon and inverse Simpson indexes (Supplementary Figure 6A), and total richness, assessed by the Chao1 index (Supplementary Figure 6B), were similar between both genotypes. As observed in C57BL/6 mice,6 the fecal microbiota of Dhpsfl/fl mice was dominated by the Bacteroidetes phylum (Supplementary Figure 6C), including the genera Prevotella, Porphyromonadaceae, and Bacteroides (Supplementary Figure 6D). Proteobacteria, including Helicobacter and Sutterellaceae, and Firmicutes represented by the Oscillibacter, Lachnospiraceae, and Akkermansia genera were also found in the fecal microbiome (Supplementary Figures 6C and 6D). The relative abundance of these genera was not significantly different between Dhpsfl/fl and DhpsΔepi mice (Supplementary Figure 6E).
Hypusination in CECs Regulates the Translation of Enzymes Involved in Reactive Aldehyde Detoxification
To understand the mechanism by which the deletion of Dhps in IECs supports colitis and carcinogenesis, we analyzed the proteome of isolated CECs from 3 different mice each from the Dhpsfl/fl and DhpsΔepi genotypes, using the isobaric tag for relative and absolute quantification (iTRAQ)-based proteomics approach. A total of 4729 proteins were identified. Among them, 99 and 102 were significantly upregulated and downregulated, respectively, in DhpsΔepi versus Dhpsfl/fl mice (Supplementary Table 1). The proteins induced in DhpsΔepi CECs included i) classical markers of the immune response, such as S100A8/9, NGP, and PERM, but also ii) signaling molecules involved in oncogenic transformation (PTN11, MMP9, RAC2), iii) the integrins ITB2, ITB6, and ITAM, and iv) Histone H2AX, a marker of DNA double strand breaks (Supplementary Table 1). The proteins downregulated with Dhps deletion, and thus potentially direct targets of EIF5AHyp, included i) MPTX and S26A2/3 that have been shown to be downregulated in colonic tumors29 and ii) the keratins K2C6A, K1C16, and K2C79, which maintain normal function of the gastrointestinal epithelium (Figure 5A, Supplementary Table 1). Remarkably, we also found two types of enzymes involved in aldehyde detoxification: the aldehyde dehydrogenases (ALDHs) AL1A7, AL1B1, and ALDH2, which oxidize aldehydes to carboxylates,30 and 6 GSTs, which catalyze the conjugation of aldehydes to glutathione,31 including GSTA4, GSTM3, GSTM2, GSTP1, GSTO1, and GSTM1 (Figure 5A, Supplementary Table 1). Note that the expression of 3 other ALDHs (AL3B2, AL3A2, AL9A1) and 5 other GSTs (GSTK1, GSTT2, LANCL1, GSTM5, GSTT3) was reduced in DhpsΔepi CECs, but did not reach statistical significance (Supplementary Table 1). Importantly, all these proteins exhibited the molecular features of EIF5AHyp-regulated proteins (Supplementary Table 2), i.e., an AAAUGU consensus sequence in their mRNA9,10,32 and/or diprolyl/diglycyl motifs in their protein.13,14 Lastly, thioredoxin (THIO) and the peroxiredoxins 1 and 6 (PRDX1/6), which are involved in peroxide detoxification, were also significantly less expressed in DhpsΔepi CECs.
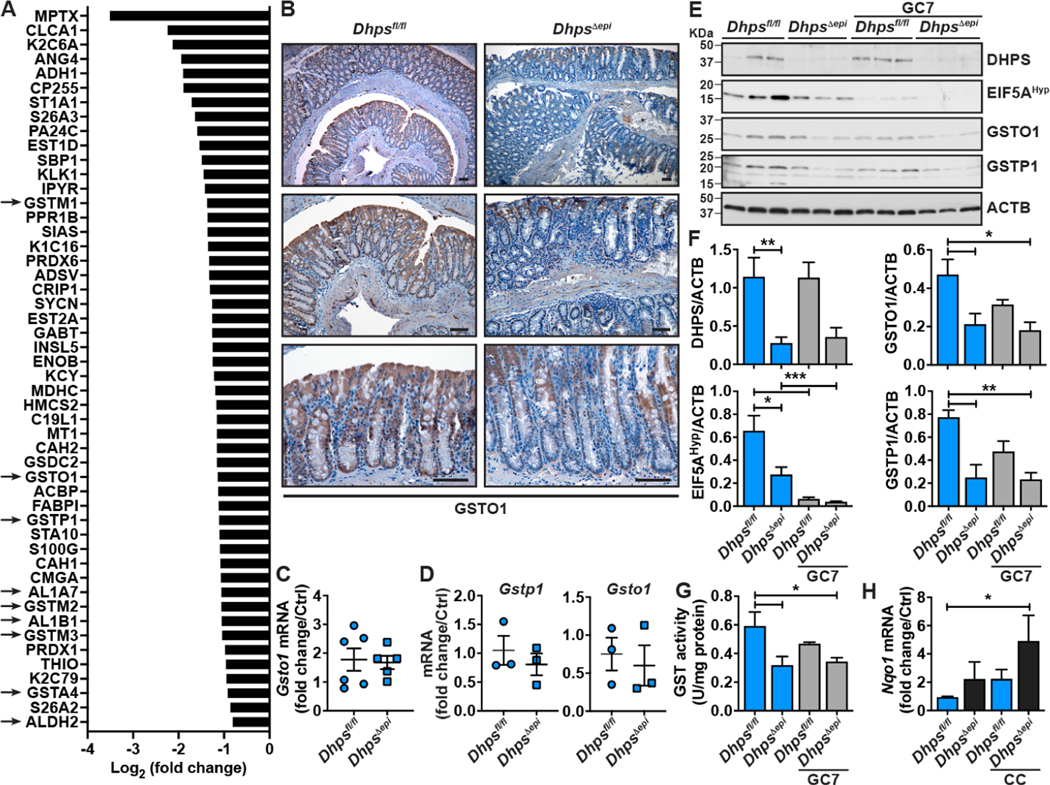
Figure 5.
Effect of hypusination on the proteome of CECs. (A) iTRAQ was used to determine the differential expression of the proteins from isolated Dhpsfl/fl and DhpsΔepi CECs (n = 3 mice per group). This figure depicts a list of 50 proteins significantly downregulated in DhpsΔepi CECs compared to Dhpsfl/fl control CECs; the arrows highlight the proteins involved in aldehyde detoxification. (B) IHC for GSTO1 in the colon of Dhpsfl/fl and DhpsΔepi mice. These photomicrographs are representative of 6 animals per genotype; scale bars, 50 μm. The images are representatives of 5 mice/condition. (C-D) Gsto1 and Gstp1 mRNA levels in the colon (C) and in colonoids (D) of Dhpsfl/fl and DhpsΔepi mice. (E-F) The level of DHPS, EIF5AHyp, GSTO1, and GSTP1 was assessed by Western blot in colonoids from Dhpsfl/fl and DhpsΔepi mice, cultured or not for 24 h with 25 μM GC7 (E), followed by densitometry (F). (G) Total GST activity was determined in the same colonoids ± GC7. (H) Expression of the gene Nqo1 in Dhpsfl/fl and DhpsΔepi colonoids stimulated for 24 h with a cytokine cocktail (CC) composed of IFN-γ, IL-1β, and TNF-α. In bar graphs, *P < .05, **P < .01, ***P < .001 by one-way ANOVA and Dunnett’s test.
We found by IHC that the level of the protein GSTO1 was reduced in DhpsΔepi mice compared to Dhpsfl/fl mice (Figure 5B), thus validating the results obtained by proteomic analysis. Importantly, the expression of Gsto1 mRNA in isolated CECs was not affected by Dhps deletion (Figure 5C), further demonstrating that DHPS regulates only GST translation in CECs. Similarly, Gsto1 and Gstp1 mRNA levels were similar in Dhpsfl/fl and DhpsΔepi colon organoids (Figure 5D), whereas GSTO1 and GSTP1 proteins were less expressed in colonoids derived from DhpsΔepi mice and in Dhpsfl/fl colonoids treated with GC7 (Figures 5E and 5F). Consequently, the total GST activity was reduced with Dhps deletion or inhibition (Figure 5G) and the expression of Nqo1, a marker of electrophilic and oxidative stress,33 was increased in naïve or stimulated DhpsΔepi colonoids (Figure 5H).
The differential proteome dataset from Dhpsfl/fl and DhpsΔepi mice was then subjected to Ingenuity Pathway Analysis (IPA). Strikingly, numerous pathways related to cancer and to the immune response were significantly induced by Dhps deletion (Supplementary Figure 7 and Supplementary Table 3). There was also a significant dampening of only two pathways, one of which was “Conjugation of glutathione” (Supplementary Figure 7 and Supplementary Table 3), which is related to the inhibition of GST translation.
The Transcriptome of DhpsΔepi Colon Is Reshaped Toward a Pro-neoplastic State
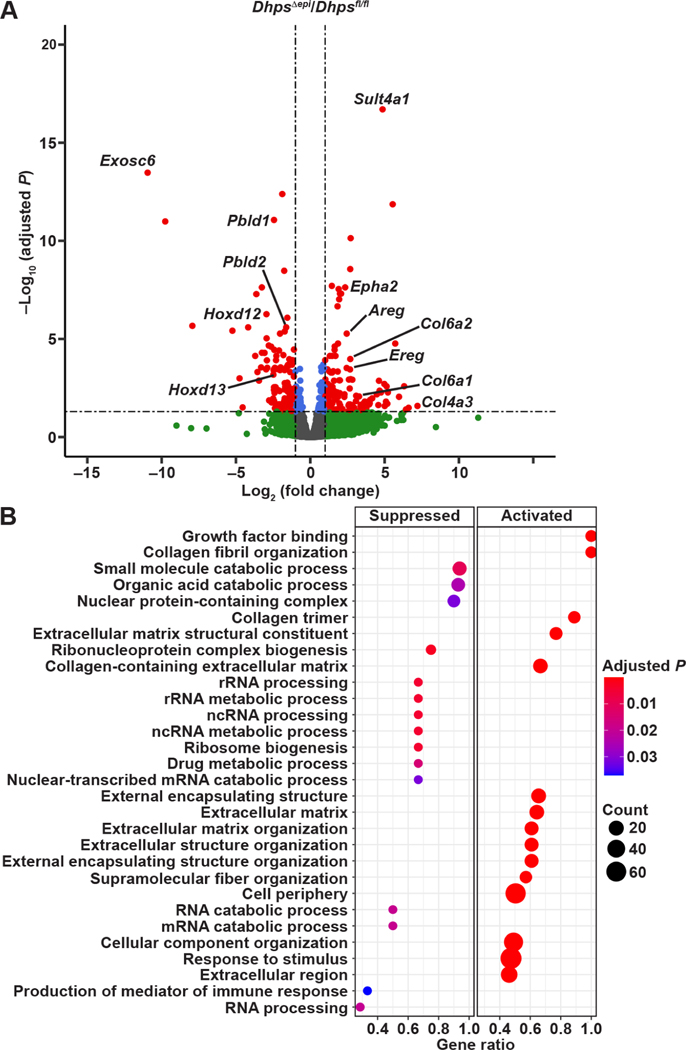
By RNA-Seq, we identified 25,182 sequences comprising 19,498 known mRNAs and 5,684 unknown sequences (Supplementary Table 4). There were 135 and 133 differentially expressed genes (DEGs) upregulated and downregulated (2-fold or more; P < .05) in DhpsΔepi mice, respectively. We observed that several genes encoding for different proteins of the extracellular matrix, including the collagen genes Col6a1, Col6a2, and Col4a3, the cadherin-encoding gene Cdh11, and the laminin gene Lama5 were among the most induced in DhpsΔepi CECs (Figure 6A and Supplementary Table 4). Genes involved in signaling such as i) Epha2, encoding the receptor tyrosine kinase ephrin type-A receptor 2 that plays a role in colon carcinogenesis34 or ii) Areg and Ereg, which encode amphiregulin and epiregulin, respectively, two EGF receptor ligands involved in the maintenance and progression of the tumorigenic state,35 were also significantly induced in mice with Dhps deletion (Figure 6A and Supplementary Table 4). The most downregulated gene was Exosc6, encoding for the exosome complex component MTR3, which mediates mRNA degradation (Figure 6A and Supplementary Table 4). Additionally, different homeobox genes (Hoxd11/12/13), whose expression has been shown to be reduced in colitis,6 and the genes Pbld1/2, encoding for the tumor suppressor phenazine biosynthesis-like domain-containing protein, were less expressed in CECs from DhpsΔepi mice (Figure 6A and Supplementary Table 4). None of the genes encoding for the GSTs and ALDHs were significantly affected by Dhps deletion (Supplementary Table 4), demonstrating that these effectors are regulated at the post-transcriptional level.
Figure 6.
Transcriptomic changes orchestrated by DHPS in CECs. (A) RNA extracted from CECs from Dhpsfl/fl and DhpsΔepi (n = 3 mice per group) was sequenced, and the volcano plot corresponding to the DEGs in DhpsΔepi mice compared to Dhpsfl/fl animals was generated using R; red dots correspond to genes significantly (P < .05) up- or downregulated by 2–fold or more. All the genes are provided in Supplementary Table 4. (B) Pathway analysis of DEGs by GSEA; the complete list of pathways is shown in Supplementary Table 5.
The transcriptional function of Dhps deletion was performed by gene set enrichment analysis (GSEA) of DEGs (Supplementary Table 5). The top signal pathways enriched in mice lacking Dhps in IECs were related to growth factors and to the organization of the extracellular matrix (Figure 6B), which are associated with cancer development and progression.36 As expected, the pathways connected to RNA processing were reduced (Figure 6B).
Increased Formation of Aldehyde Adducts in DhpsΔepi Mice Supports Colonic Inflammation
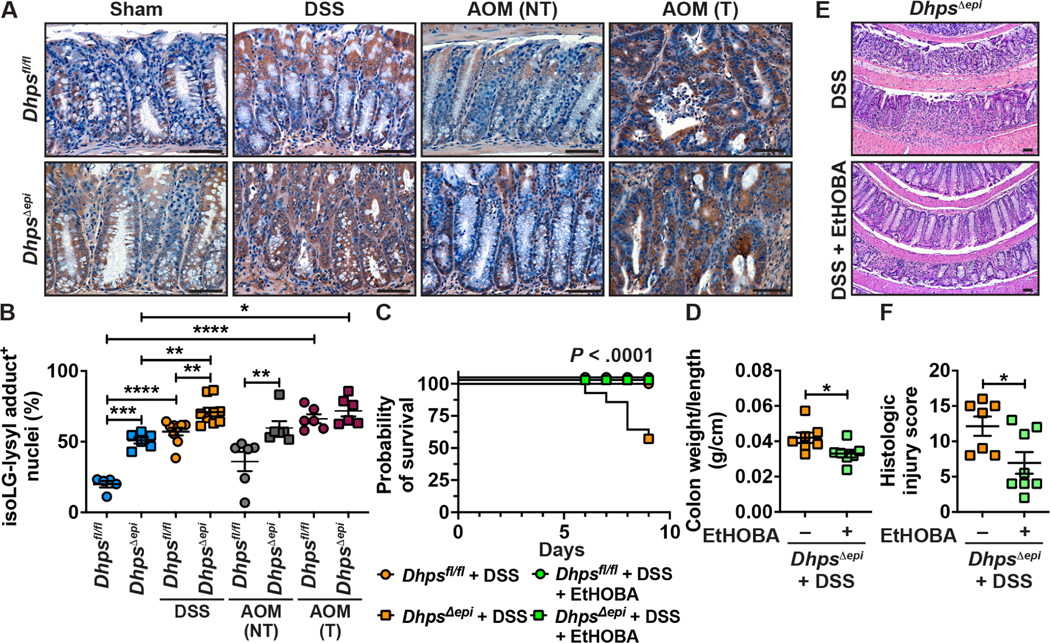
Because enzymes that detoxify reactive electrophiles are less expressed in the colon of DhpsΔepi mice, we reasoned that reduced hypusination in IECs would be associated with oxidative covalent adduction of these aldehydes, such as isolevuglandins (isoLGs) or malondialdehydes (MDA). Using a specific single-chain fragment variable Ab (D11) that specifically recognizes adducts of isoLGs to lysine (isoLG-lysyl),21,37 we observed increased adducts in the nuclei of CECs from naive DhpsΔepi mice compared to Dhpsfl/fl animals (Figures 7A–B). Furthermore, we found by LC-MS that the level of adducts of MDA-dilysyl crosslinks was also significantly increased in the colon of DhpsΔepi mice (Supplementary Figure 8). Moreover, we observed an increase of isoLG-lysyl adducts in Dhpsfl/fl animals treated with DSS, and a further enhancement in DSS-treated DhpsΔepi mice (Figures 7–7B). The number of isoLG-lysyl adduct-positive nuclei in the non-tumor areas was also higher in AOM-treated DhpsΔepi mice, whereas strong nuclear staining was observed in tumors from both genotypes (Figures 7A–B). These data indicate that dicarbonyl electrophiles are more abundant in the colon of DhpsΔepi mice and are more reactive in the nuclei of CECs under inflammatory stress but are present at the same level when tumors are formed.
Figure 7.
Levels and role of reactive aldehydes associated with Dhps deletion. (A-B) The levels of isoLG-lysyl adducts in the colon of Dhpsfl/fl and DhpsΔepi mice (n = 2–3 per group), treated or not with DSS or AOM, was assessed by immunostaining with D11 Ab and the images presented are representative of 2–3 mice/group. (A); the quantification of cells with positive staining for isoLG-lysyl adducts in the nucleus is shown in B. Each dot represents a crypt; *P < .05, **P < .01, ***P < .001, and ****P < .0001 by one-way ANOVA and Tukey test. (C-F) DhpsΔepi mice were given 2% DSS (n = 15) or DSS ± EtHOBA (n = 19). Survival was assessed by Kaplan-Meier curves and the P value was calculated by the Log-rank test (C); only the female mice died in this experiment. The weight and length of the colons of DhpsΔepi mice + DSS ± EtHOBA were measured and expressed as a ratio (D). H&E staining (E) was used to score histologic injury in male mice (F). *P < .05 by Student’s t test. Dhpsfl/fl treated with 2% DSS did not develop significant increase in histologic injury score compared to control animals, and EtHOBA had no significant effect on these mice. In A and E, the scale bars represent 50 μm.
To verify whether reactive aldehydes play a role in the pathogenesis of the colon of DhpsΔepi mice, we used EtHOBA, a potent scavenger of reactive aldehydes.21,23,24 We found that EtHOBA reduced the concentration of MDA-dilysyl crosslinks in the colon of DhpsΔepi mice (Supplementary Figure 8). These animals were then given 2% DSS and were also maintained with or without EtHOBA. Survival (Figure 7C), colon weight/length ratio (Figure 7D), plus inflammation and epithelial damage (Figures 7E and 7F) were significantly improved when DSS-treated DhpsΔepi mice were given EtHOBA.
Discussion
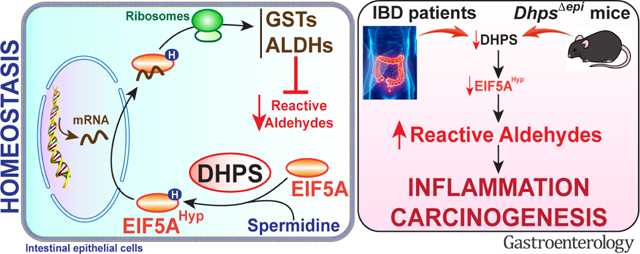
Our study establishes that the hypusination of EIF5A in IECs is essential to limit inflammation and carcinogenesis. Mice with specific deletion of Dhps in IECs exhibit spontaneous colonic inflammation and crypt distortion, a hallmark of IBD histopathology. These animals are highly susceptible to DSS and are prone to develop more tumors in response to carcinogens. Mechanistically, we found that i) hypusination in CECs controls the translation of enzymes involved in reactive aldehyde detoxification, ii) there are high levels of electrophile adducts in the nuclei of DhpsΔepi CECs, and iii) treatment of DhpsΔepi mice with an electrophile scavenger reduces susceptibility to colitis, strongly suggesting that one of the major functions of hypusination in IECs is to prevent aldehyde-mediated oxidative damage and dampen inflammation, epithelial injury, and malignant transformation. Since we also found that UC and CD patients exhibit low levels of colonic DHPS mRNA, DHPS protein, and EIF5AHyp, our data imply that reduced hypusination in CECs in IBD patients may lead to increased inflammation and risk for CAC.
Supporting the concept that the DHPS-EIF5AHyp axis prevents intestinal inflammation, mice with specific deletion of Dhps or Dohh in CD4+ T cells develop signs of colitis and die at 10–20 wk of age.38 However, we have reported that the deletion of Dhps in myeloid cells does not cause spontaneous colitis.26 It has also been reported that putrescine produced by the gut microbiota is utilized by CECs to form Spd and EIF5AHyp, and this is associated with dampening of DSS colitis.39 Furthermore, it has been shown that heterozygous deletion of the genes encoding for EIF5A and adenosylmethionine decarboxylase 1, which is essential for the biosynthesis of Spd, are associated with lymphoma in humans and supports lymphomagenesis in mice.40 However, EIF5A and DHPS expression is increased in tumors18 and hypusination correlates with poor prognosis and metastasis in several cancer types.41,42. In fact, knockdown of the genes encoding for proteins involved in the hypusination pathway and/or inhibition of DHPS reduce the growth of established human cancerous gastrointestinal cell lines19,41,43,44. Further, tumor growth and metastasis in mice are reduced when xenografts of human carcinoma cell lines are subjected to knockdown of DHPS19,45 or when animals are treated with GC7 when the tumors are already formed.19 To explain this phenotype, it has been shown hypusination supports the translation of carcinoma-specific signaling molecules such as the MYC proto-oncogene,19 the transforming protein RHOA,45 the inactive tyrosine-protein kinase PEAK1,44 or the histone-lysine-specific demethylase LSD1,43 which have been all involved in cell proliferation and metastasis. We did not find these targets in our proteomic analysis comparing CECs of Dhpsfl/fl and DhpsΔepi mice, evidencing that the proteins regulated by hypusination in non-transformed and cancerous cells are different and may thus have varying effects. We therefore propose that hypusination in CECs is critical to reduce colon carcinogenesis, but that once tumors are formed, EIF5AHyp in cancerous cells may sustain their growth. Fascinatingly, a similar paradox exists for polyamines: on one hand, we found that Spd dampens colon tumorigenesis,6 but polyamines in general are required for the growth of cancerous cells.46
In this report, we also assessed the molecular mechanism by which Dhps deletion in IECs leads to intestinal inflammation and creates a pro-oncogenic state in the colon. We determined that the translation of numerous enzymes involved in detoxification of peroxide (THIO, PRDXs) and reactive aldehydes (ALDHs, GSTs), are regulated by hypusination and are thus less expressed in the colon of DhpsΔepi mice. Consequently, these animals exhibit increased oxidative stress/damage and inflammation. GSTs are principally localized in CECs.47 The null genotypes of GSTM1 or GSTT1 have been associated with increased risk for IBD;48 and the frequency of the GSTT1 null genotype is significantly increased in colorectal cancer cases, but not in lung, oral, or gastric cancer.49 Moreover, it has been shown that the protein GSTM1 is also absent in 30% and 50% of the patients with UC and colon adenocarcinoma, respectively, that harbor a GSTM1+ genotype,50 demonstrating that this enzyme is also regulated post-transcriptionally. Although numerous redundant GTSs exist, the deletion of Gsto1 in mice is associated with susceptibility to DSS colitis51,52 and to AOM-DSS-induced tumorigenesis.52 Reciprocally, the local delivery of the Gstt1 gene to mice treated with DSS dampens colitis.53 We reported that the aldehyde isoLG is generated in the inflamed stomach and colon, and that the electrophile scavenger EtHOBA prevents dysplasia in the murine AOM-DSS model.21 Herein, we observed increased levels of nuclear isoLG-lysyl adducts in CECs from naïve or DSS- or AOM-treated DhpsΔepi mice as well as a reduction of DSS-induced inflammation in DhpsΔepi mice treated with the electrophile scavenger EtHOBA. We also found that the adducts of the electrophile MDA to lysine were increased in the colon of DhpsΔepi mice and reduced by EtHOBA. All these data and our results emphasize that the defense system against reactive aldehydes in CECs is orchestrated by hypusination and help in the prevention of inflammation/carcinogenesis.
Our data establish that hypusination in CECs is essential to lower the level of spontaneous reactive aldehydes in the colonic mucosa and protects mice from colitis and colon carcinogenesis. We previously reported that Spd dampens colitis and prevents tumorigenesis in mice6 and in the present report we further observed that hypusination is enhanced in the colonic mucosa of mice treated with Spd. Since patients with IBD exhibit reduced levels of DHPS and EIF5AHyp, restoring hypusination by Spd supplementation and/or treatment with an electrophile scavenger can be considered as rational strategies to limit oxidative damage in the colon and the risk for neoplasia.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
Hypusination is a process that controls translation of specific proteins. We determined the effect of hypusination in intestinal epithelial cells in colon homeostasis, inflammation, and cancer.
NEW FINDINGS
Patients with inflammatory bowel disease exhibit reduced hypusination, which is required to support translation of proteins involved in detoxification of reactive aldehydes, thus preventing electrophilic damage, colonic inflammation, and carcinogenesis.
LIMITATIONS
Our animal experiments focus mainly on inflammation-mediated carcinogenesis of the colon, and the role of hypusination in sporadic colorectal cancer is not addressed.
BASIC RESEARCH/CLINICAL RELEVANCE
These findings provide insights into inflammatory bowel disease pathogenesis and implicate a new mechanism by which patients with colitis have increased risk of colon cancer. Our results also suggest that restoration of hypusination or treatment with an electrophile scavenger in patients with diminished hypusination in colonic epithelial cells could represent a clinical strategy to limit colitis and prevent carcinogenesis.
LAY SUMMARY
Hypusine is a specific amino acid that is essential to limit oxidative damage in intestinal epithelial cells, thus preventing inflammation and cancer development.
Acknowledgements
We thank Dr. Venkataraman Amarnath (VUMC) for the synthesis of EtHOBA. Sequencing was performed by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core at VUMC.
Grant Support:
This work was funded by NIH grants R01DK128200 (KTW), P01CA116087 (KTW), P01CA028842 (KTW), R01DK060581 (RGM), and R01DK124906 (RGM); Veterans Affairs Merit Review grant I01CX002171 (KTW); Senior Research Award 703003 from the Crohn’s & Colitis Foundation (APG and KTW); Department of Defense Peer Reviewed Cancer Research Program Impact Award W81XWH-21-1-0617 (KTW); a gift from CURE for IBD (KTW); a gift from the James Rowen Fund (KTW); the Thomas F. Frist Sr. Endowment (KTW); and the Vanderbilt Center for Mucosal Inflammation and Cancer (KTW). LAC was supported by Veterans Affairs Merit Review grant I01BX004366. YLL was supported by T32AI138932, and KMM was supported by T32CA009592. The Mass Spectrometry Core (KLS) and the Tissue Morphology Core (MKW, MBP) of the Vanderbilt Digestive Disease Research Center are supported by P30DK058404 and the Vanderbilt Ingram Cancer Center supported by NIH grant P30CA068485. Proteomics experiments were supported in part by P30DK058404 and P01CA116087. The Cooperative Human Tissue Network is supported by NIH grant UM1CA183727 (MKW).
Abbreviations used in this paper:
- ALDH
aldehyde dehydrogenase
- AOM
azoxymethane
- CAC
colitis-associated carcinoma
- CD
Crohn’s disease
- CECs
colonic epithelial cells
- DEGs
differentially expressed genes
- DHPS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
- DSS
dextran sulfate sodium
- EIF5A
eukaryotic translation initiation factor 5A
- EIF5AHyp
hypusinated EIF5A
- EtHOBA
5-ethyl-2-hydroxybenzylamine
- GC7
N1-guanyl-1,7-diamineheptane
- GSEA
gene set enrichment analysis
- GST
glutathione-S-transferase
- H&E
hematoxylin and eosin
- HGD
high-grade dysplasia
- IBD
inflammatory bowel disease
- IEC
intestinal epithelial cells
- IF
immunofluorescence
- IHC
immunohistochemistry
- i.p.
intraperitoneal
- IPA
Ingenuity Pathway Analysis
- isoLG
isolevuglandins
- LGD
low-grade dysplasia
- RNA-Seq
RNA sequencing
- SMOX
spermine oxidase
- Spd
spermidine
- TMA
tissue microarray
- UC
ulcerative colitis
Footnotes
Disclosures: APG and KTW are named inventors on a patent application for the use of electrophile scavengers. In addition, APG and KTW are named on a licensing agreement between Vanderbilt University and MTI Biotech, Inc. for the future use of electrophile scavengers. All other authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dahlhamer JM, Zammitti EP, Ward BW, et al. Prevalence of Inflammatory Bowel Disease Among Adults Aged >/=18 Years - United States, 2015. MMWR Morb Mortal Wkly Rep 2016; 65:1166–1169. [DOI] [PubMed] [Google Scholar]
- 2.Singh P, Ananthakrishnan A, Ahuja V. Pivot to Asia: inflammatory bowel disease burden. Intestinal research 2017; 15:138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munkholm P. Review article: the incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther 2003; 18 Suppl 2:1–5. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Devereux W, Woster PM, et al. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res 2001; 61:5370–5373. [PubMed] [Google Scholar]
- 5.Gobert AP, Al-Greene NT, Singh K, et al. Distinct immunomodulatory effects of spermine oxidase in colitis induced by epithelial injury or infection. Front Immunol 2018; 9:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gobert AP, Latour YL, Asim M, et al. Protective role of spermidine in colitis and colon carcinogenesis. Gastroenterology 2022; 162:813–827 e818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci U S A 1981; 78:2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JH, Aravind L, Wolff EC, et al. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci U S A 2006; 103:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu A, Chen KY. Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J Biol Chem 2001; 276:2555–2561. [DOI] [PubMed] [Google Scholar]
- 10.Maier B, Ogihara T, Trace AP, et al. The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J Clin Invest 2010; 120:2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipowsky G, Bischoff FR, Schwarzmaier P, et al. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J 2000; 19:4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park MH, Wolff EC. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J Biol Chem 2018; 293:18710–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez E, Shin BS, Woolstenhulme CJ, et al. eIF5A promotes translation of polyproline motifs. Mol Cell 2013; 51:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuller AP, Wu CC, Dever TE, et al. eIF5A functions globally in translation elongation and termination. Mol Cell 2017; 66:194–205 e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura K, Lee SB, Park JH, et al. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 2012; 42:703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park MH, Lee YB, Joe YA. Hypusine is essential for eukaryotic cell proliferation. Biol Signals 1997; 6:115–123. [DOI] [PubMed] [Google Scholar]
- 17.Ganapathi M, Padgett LR, Yamada K, et al. Recessive rare variants in deoxyhypusine synthase, an enzyme involved in the synthesis of hypusine, are associated with a neurodevelopmental disorder. Am J Hum Genet 2019; 104:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakanishi S, Cleveland JL. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 2016; 48:2353–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coni S, Serrao SM, Yurtsever ZN, et al. Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation. Cell Death Dis 2020; 11:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levasseur EM, Yamada K, Pineros AR, et al. Hypusine biosynthesis in beta cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci Signal 2019; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobert AP, Boutaud O, Asim M, et al. Dicarbonyl electrophiles mediate inflammation-induced gastrointestinal carcinogenesis. Gastroenterology 2021; 160:1256–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgueno JF, Fritsch J, Gonzalez EE, et al. Epithelial TLR4 signaling activates DUOX2 to induce microbiota-driven tumorigenesis. Gastroenterology 2021; 160:797–808 e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrier EJ, Zagol-Ikapitte I, Amarnath V, et al. Levuglandin forms adducts with histone H4 in a cyclooxygenase-2-dependent manner, altering its interaction with DNA. Biochemistry 2014; 53:2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagol-Ikapitte I, Amarnath V, Bala M, et al. Characterization of scavengers of gamma-ketoaldehydes that do not inhibit prostaglandin biosynthesis. Chem Res Toxicol 2010; 23:240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polosukhina D, Singh K, Asim M, et al. CCL11 exacerbates colitis and inflammation-associated colon tumorigenesis. Oncogene 2021; 40:6540–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gobert AP, Finley JL, Latour YL, et al. Hypusination orchestrates the antimicrobial response of macrophages. Cell Rep 2020; 33:108510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coburn LA, Horst SN, Chaturvedi R, et al. High-throughput multi-analyte Luminex profiling implicates eotaxin-1 in ulcerative colitis. PloS One 2013; 8:e82300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coburn LA, Horst SN, Allaman MM, et al. L-arginine availability and metabolism is altered in ulcerative colitis. Inflamm Bowel Dis 2016; 22:1847–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Femia AP, Luceri C, Toti S, et al. Gene expression profile and genomic alterations in colonic tumours induced by 1,2-dimethylhydrazine (DMH) in rats. BMC Cancer 2010; 10:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Zavala JS, Calleja LF, Moreno-Sanchez R, et al. Role of aldehyde dehydrogenases in physiopathological processes. Chem Res Toxicol 2019; 32:405–420. [DOI] [PubMed] [Google Scholar]
- 31.Berhane K, Widersten M, Engstrom A, et al. Detoxication of base propenals and other alpha, beta-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci U S A 1994; 91:1480–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem J 2004; 384:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross D, Siegel D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol 2021; 41:101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herath NI, Boyd AW. The role of Eph receptors and ephrin ligands in colorectal cancer. Int J Cancer 2010; 126:2003–2011. [DOI] [PubMed] [Google Scholar]
- 35.Guzman MJ, Shao J, Sheng H. Pro-neoplastic effects of amphiregulin in colorectal carcinogenesis. J Gastrointest Cancer 2013; 44:211–221. [DOI] [PubMed] [Google Scholar]
- 36.Xu S, Xu H, Wang W, et al. The role of collagen in cancer: from bench to bedside. J Transl Med 2019; 17:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies SS, Talati M, Wang X, et al. Localization of isoketal adducts in vivo using a single-chain antibody. Free Radic Biol Med 2004; 36:1163–1174. [DOI] [PubMed] [Google Scholar]
- 38.Puleston DJ, Baixauli F, Sanin DE, et al. Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 2021; 184:4186–4202 e4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura A, Kurihara S, Takahashi D, et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat Commun 2021; 12:2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scuoppo C, Miething C, Lindqvist L, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 2012; 487:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng QB, Kang WM, Yu JC, et al. Overexpression of eukaryotic translation initiation factor 5A2 (EIF5A2) correlates with cell aggressiveness and poor survival in gastric cancer. PLoS One 2015; 10:e0119229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Chen Z, Nie L, et al. Extracellular signal-regulated kinases associate with and phosphorylate DHPS to promote cell proliferation. Oncogenesis 2020; 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Wu BK, Kanchwala M, et al. YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis. Nat Cell Biol 2022; 24:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimura K, Wright T, Strnadel J, et al. A hypusine-eIF5A-PEAK1 switch regulates the pathogenesis of pancreatic cancer. Cancer Res 2014; 74:6671–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muramatsu T, Kozaki KI, Imoto S, et al. The hypusine cascade promotes cancer progression and metastasis through the regulation of RhoA in squamous cell carcinoma. Oncogene 2016; 35:5304–5316. [DOI] [PubMed] [Google Scholar]
- 46.Tutton PJ, Barkla DH. Comparison of the effects of an ornithine decarboxylase inhibitor on the intestinal epithelium and on intestinal tumors. Cancer Res 1986; 46:6091–6094. [PubMed] [Google Scholar]
- 47.Edalat M, Mannervik B, Axelsson LG. Selective expression of detoxifying glutathione transferases in mouse colon: effect of experimental colitis and the presence of bacteria. Histochem Cell Biol 2004; 122:151–159. [DOI] [PubMed] [Google Scholar]
- 48.Ye X, Jiang Y, Wang H, et al. Genetic polymorphisms of glutathione S-transferases are associated with ulcerative colitis in central China. Cell Biochem Biophys 2011; 60:323–328. [DOI] [PubMed] [Google Scholar]
- 49.Deakin M, Elder J, Hendrickse C, et al. Glutathione S-transferase GSTT1 genotypes and susceptibility to cancer: studies of interactions with GSTM1 in lung, oral, gastric and colorectal cancers. Carcinogenesis 1996; 17:881–884. [DOI] [PubMed] [Google Scholar]
- 50.Barker HJ, Alpert LC, Compton CC, et al. Loss of glutathione S-transferase (GST) mu phenotype in colorectal adenocarcinomas from patients with a GSTM1 positive genotype. Cancer Lett 2002; 177:65–74. [DOI] [PubMed] [Google Scholar]
- 51.Menon D, Innes A, Oakley AJ, et al. GSTO1–1 plays a pro-inflammatory role in models of inflammation, colitis and obesity. Sci Rep 2017; 7:17832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tummala P, Rooke M, Dahlstrom JE, et al. Glutathione transferase Omega 1 confers protection against azoxymethane-induced colorectal tumour formation. Carcinogenesis 2021; 42:853–863. [DOI] [PubMed] [Google Scholar]
- 53.Kim JH, Ahn JB, Kim DH, et al. Glutathione S-transferase theta 1 protects against colitis through goblet cell differentiation via interleukin-22. Faseb J 2020; 34:3289–3304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data can be accessed on the GEO repository using the accession number GSE201556 (token: yxybcsecfxundih). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD033228 (Reviewer account details: Username: reviewer_pxd033228@ebi.ac.uk; Password: c5NVRzhi). The 16S rRNA sequencing of the fecal microbiota has been deposited on the Sequence Read Archive website with the BioProject accession number PRJNA814820 (Temporary Submission ID: SUB11173798).