Abstract
Cosmic radiation is the most serious risk that will be encountered during the planned missions to the Moon and Mars. There is a compelling need to understand the effects, safety thresholds, and mechanisms of radiation damage in human tissues, in order to develop measures for radiation protection during extended space travel. As animal models fail to recapitulate the molecular changes in astronauts, engineered human tissues and “organs-on-chips” are valuable tools for studying effects of radiation in vitro. We have developed a bioengineered tissue platform for studying radiation damage in individualized settings. To demonstrate its utility, we determined radiation effects using engineered models of two human tissues known to be affected by radiation: engineered cardiac tissues (eCT, a target of chronic radiation damage) and engineered bone marrow (eBM, a target of acute radiation damage). We report the effects of high-dose neutrons, as a proxy for simulated galactic cosmic rays, on the expression of key genes implicated in tissue responses to ionizing radiation, phenotypic and functional changes in both tissues, and proof-of-principle application of radioprotective agents. We further determined the extent of inflammatory, oxidative stress, and matrix remodeling gene expression changes, and found that these deviations were associated with a hypertrophic phenotype in eCT and myeloid skewing in eBM. We propose that individualized models of human tissues have potential to provide insights into the effects and mechanisms of radiation during deep-space missions and allow testing of radioprotective measures.
One Sentence Summary:
Engineered human tissue models can be used to study and counter the effects of cosmic radiation.
INTRODUCTION
Among the “red risks” identified by NASA for the anticipated deep space mission to Mars, the space radiation is considered most critical for the human body. Notably, animal models that are routinely used to assess the effects of radiation do not recapitulate the effects seen in humans. Galactic cosmic rays (GCR) are comprised primarily of protons, alpha particles, and a small (<1%) fraction of high charge and energy (HZE) particles (1). Secondary radiation fragments are produced when primary components of GCR interact with the spacecraft or human tissue, leading to production of secondary protons and neutrons that form a unique environment of high-linear energy transfer (high-LET) radiations (2). While HZE particles are only a small fraction of GCR, they are a component that is most damaging to human tissues, resulting in clustered damage when passing through the cells (3). We focused on modeling the effects of this most damaging fraction of cosmic radiation using “organs-on-chip” (OoC) human tissue platforms.
Radiation exposure can result in a host of acute and chronic symptoms, including the increased risk of cancer and cardiovascular disease (CVD) (1, 4). The most notorious short-term effect of radiation exposure is acute radiation syndrome (ARS), which can be broken down into hematopoietic, cerebrovascular, and gastrointestinal sub-syndromes (5). In an anticipated 3-year long Mars mission, astronauts are expected to be exposed to a cumulative dose of 0.3 to 0.45 Gy (6), though Mars rover missions have estimated doses may be as high as 1 Gy (7). Such protracted exposure may result in an increase in incidence of CVD, cognitive impairment, reproductive issues, aberrant hematopoiesis, and cancers (1).
Previous ground-level work on HZE particles revealed unique patterns of double- and single-strand breaks within human cell nuclei, increased genome-level mutations and oxidative damage, leading to epigenetic changes contributing to inability to perform DNA repair, as well as to aberrant signaling and development of cancer (8). Retrospective analysis of Hiroshima and Nagasaki survivors indicated an increased risk of acute lymphoblastic or myeloid leukemia diagnoses within 2–10 years post-exposure, with decreasing prognosis with increasing bomb victim age (9). High cumulative doses of radiation during cancer therapy have also resulted in the decline in functionality of cardiovascular tissues, such as atherosclerosis and myocardial fibrosis (4, 10), extending the relevance of the human tissue models to clinical settings.
Studies of the effects of cosmic radiation have been limited by the complex logistics and high costs associated with conducting experiments in space. NASA’s Brookhaven National Laboratory (BNL) has developed a terrestrial galactic cosmic ray simulator (GCRsim) providing seven different ion types with several levels of energy and a series of 33 separate beams (6). Simpler radiation source systems, comprised of one or two high-LET radiation types (i.e., mixed neutron, Fe-ion, etc.), have also been used to simulate space radiation with less experimental constraints. In particular, secondary neutrons from cosmic radiation exposures with spacecrafts and regolith on the surfaces of the Moon and Mars are an area of intense interest, as damaging high-LET neutron exposures are the greatest radiation risk for long space missions (11, 12). In these cases, GCRs are not directly affecting astronauts, rather the secondary production of neutrons may negatively impact astronaut health on the total journey to Mars or once on the surface. Studying the neutron-specific effects is a secondary but critical component in preventing radiation-induced damage on missions to deep space.
Previous work has demonstrated dose-dependent effects of neutron radiation on differentially-expressed genes, as compared to gamma and proton sources. Over a few days post-neutron radiation exposure, many groups have reported an immediate increase in circulating inflammatory signals, by serum lipidomic/metabolomic analysis or circulating blood cell counts (13). Early studies of mice exposed to neutron radiation indicated an increased incidence of myelogenous leukemias as compared to exposure to gamma rays of similar intensity (14–17). However, these studies were conducted in small animal and cell monolayer models. Since human and animal tissues respond differently to radiation damage, especially with respect to the contributions of immune cells to injury and repair, data obtained in animal models have limited translational value.
Over the past decade, engineered human tissue models, including organoids and OoCs, have emerged as new tools for investigating disease progression and therapeutic modalities (18–21). Engineered tissues generated from a combination of primary human cells and induced pluripotent stem cell (iPSC)-derived cells have been used to recapitulate the microenvironmental diversity (i.e., supporting cells, biomaterials to mimic extracellular matrix (ECM) components and architecture) of micro-sized human tissues, designed to mimic functional features of human organs (22). To date, models have emerged for studies of human patho/physiology using the heart, liver, bone marrow, skin, gut, kidney, and brain tissues, individually or in combinations (18, 22, 23). A few groups have recently reported the use of engineered tissue models for studying space health, during short flights to the International Space Station (ISS) (24), studies at NASA’s BNL (25), or with clinical sources of ionizing radiation (26–29). As deep space missions are at the planning stages, there is a pressing need to assess and counter the effects of exposure of human body to deep space radiation.
We describe here the first use of engineered human tissues to model the effects of neutron radiation exposures expected during the Mars mission. Engineered human cardiac tissue (eCT, selected as a site of long-term radiation damage) and human bone marrow (eBM, selected as a site of acute radiation damage) were formed from cells and tissue-specific extracellular matrix. Both tissues were matured to acquire functional characteristics of the respective native organs and exposed to the types and dosages of radiation relevant to cosmic radiation expected during long space missions, based off of recommendations from previous NASA missions and NASA’s Space Radiation Laboratory at BNL (6, 7). We assessed the effects of radiation over a period of 3 weeks post-exposure, using comprehensive molecular, structural and functional assays.
RESULTS
Human tissue platform for modeling and countering the effects of radiation
eCTs were derived by encapsulation of human iPSC-derived cardiomyocytes and primary fibroblasts into fibrin hydrogel stretched between two flexible pillars, and matured over a period of 4 weeks by electromechanical stimulation (Fig. 1A). eBMs were derived by infusing mesenchymal stem/stromal cells (MSCs), endothelial cells, and cord blood-derived hematopoietic stem-progenitor cells (HSPCs) into decellularized bone scaffolds (Fig. 1A).
Figure 1. Experimental design for engineered human tissue models for assessing the effects of galactic cosmic rays.
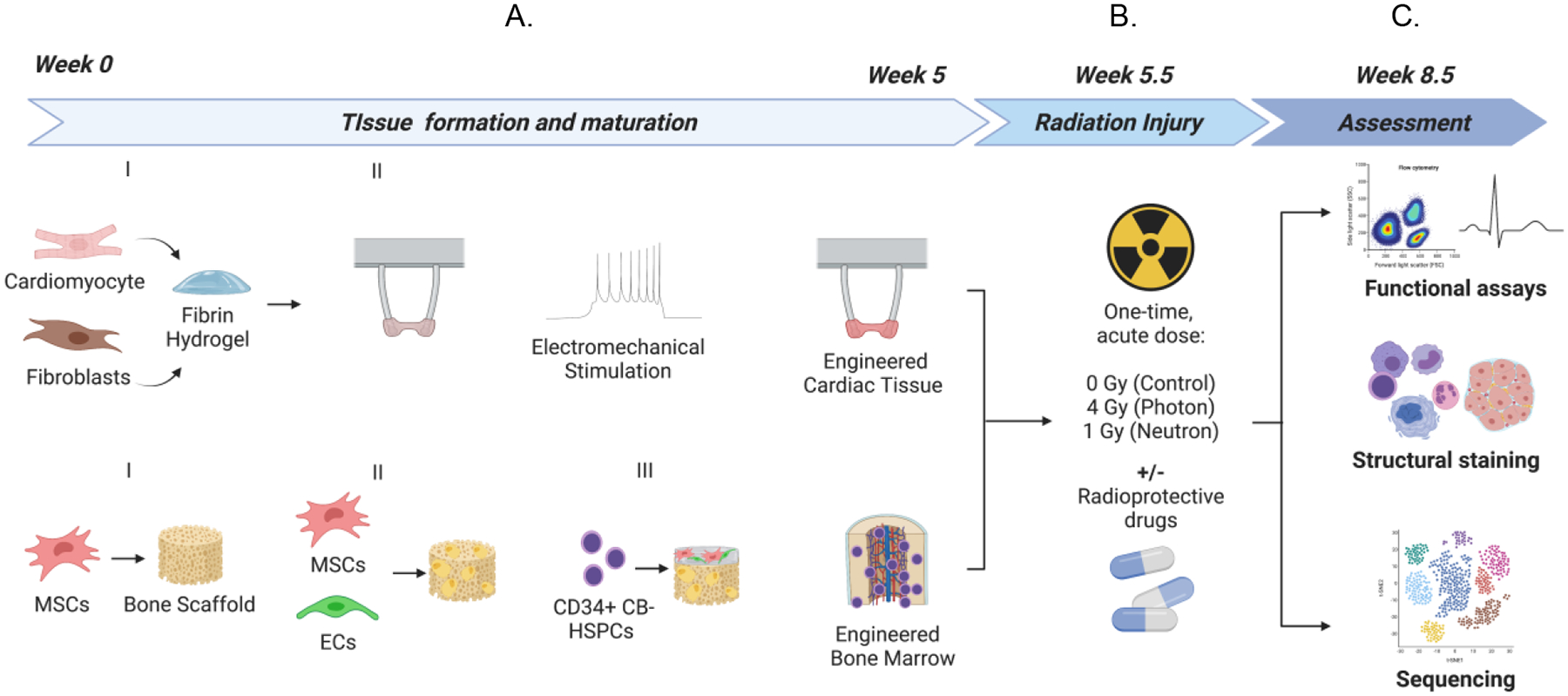
(A) Breakdown of engineered human cardiac muscle tissue and engineered human bone marrow tissue models and timeline for formation/maturation. (B) Exposure of engineered tissues to acute doses of photons or neutron irradiation and endpoint assays to assess tissue health 3-weeks post-irradiation.
After tissue formation and maturation (4 weeks for eCTs and 5 weeks for eBMs), tissues were exposed to an acute dose of radiation. We irradiated eCTs and eBMs with either (i) mixed neutrons (1 Gy acute dose, approximately ~17% concomitant photons) at Columbia University’s Radiological Research Accelerator Facility (RARAF) (30) or (ii) photons (4 Gy acute dose), with and without radiation protective agents (Fig. 1B). We chose a photon dose (4 Gy) comparable to the dose of neutron exposure (1 Gy), based on previous studies that have showed an approximately four times greater dose effect of neutrons than photons for a similar relative biological effectiveness (RBE) in lymphocytes (31, 32).”
After 3 weeks of tissue culture post-radiation exposure, we assessed the structural, functional, and molecular changes associated with radiation damage, relative to non-irradiated controls (Fig. 1C). Using single-cell RNA sequencing, we were able to identify the emergence of unique myeloid populations and myeloid lineage skewing in irradiated eBMs, and phenotypic changes in eCTs. Irradiated eCT and eBM tissues were compared to published data for radiotherapy, astronauts returning to Earth, and animals exposed to high-LET radiation.
We propose that this model lays a foundation for using engineered human tissues to study the acute and chronic effects of GCRs and for testing novel radioprotective measures. By changing the sources and dosages of radiation, the same model can be readily applied to translational studies of clinical and accidental radiation exposures, and for developmental testing of radiation protection measures.
Engineered cardiac muscle and bone marrow tissues mimic human physiological functions
To engineer human cardiac tissues (eCT), we followed a previously published methodology where iPSC-derived cardiomyocytes and primary human cardiac fibroblasts are encapsulated in a fibrin hydrogel stretched between two flexible pillars (Fig. 1A, Fig. 2A–2B) (33, 34). The resulting cell-hydrogel constructs were matured by electromechanical conditioning at 2 Hz, over a total of 4 weeks of culture, towards human tissue phenotype (33, 34). Tissues were made using iPSC-derived cardiomyocytes with a GCaMP reporter, to allow for longitudinal online functional analysis of tissues by bright field and calcium fluorescent imaging. We confirmed the hallmarks of tissue maturation, including cell alignment, striations (Fig. 2C), positive force-frequency relationship (Fig. 2D), and contractile responses to adrenergic stimulation (Fig. 2E).
Figure 2. Baseline properties of engineered human tissues. Engineered cardiac muscle tissues (eCT).
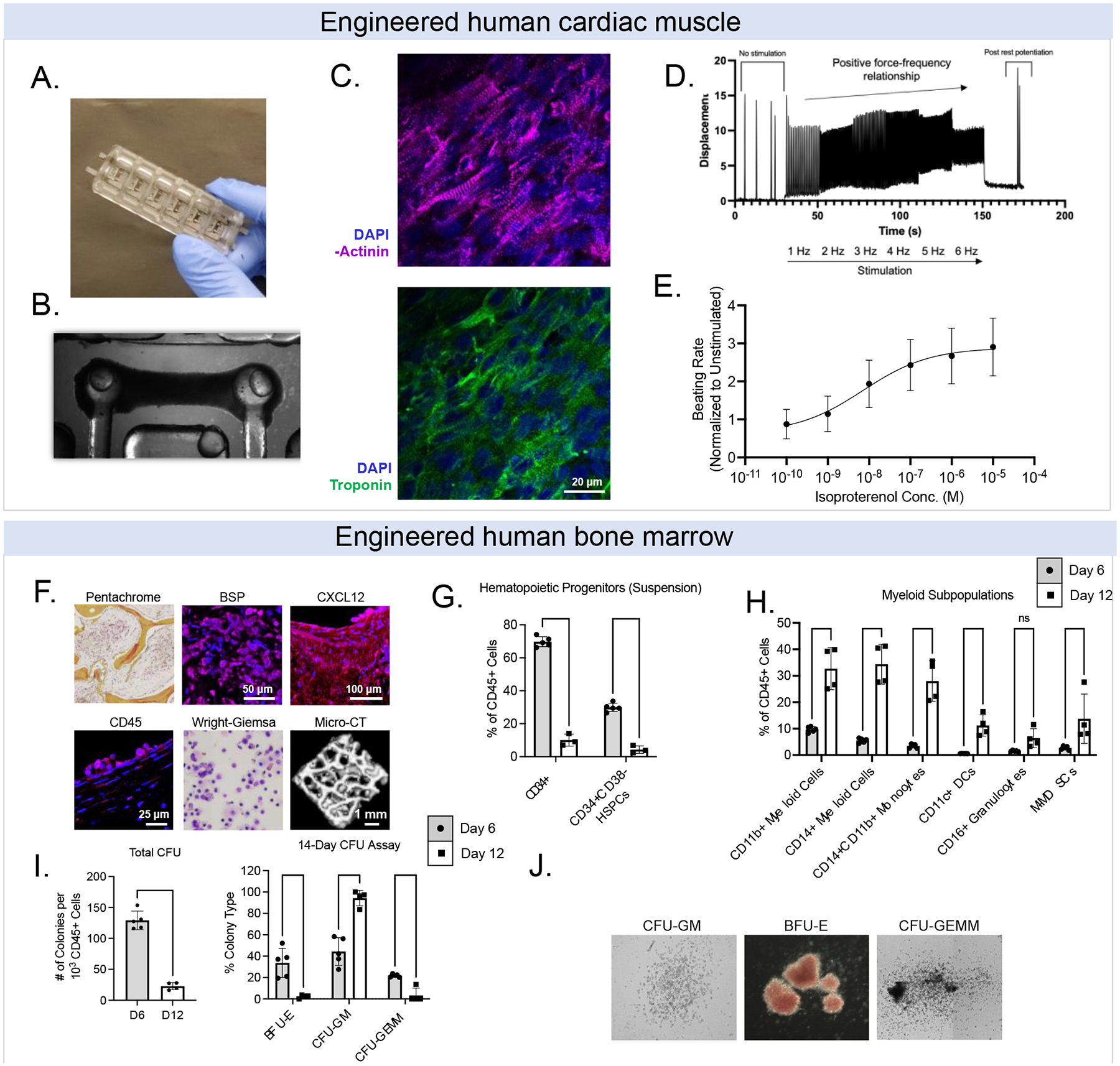
(A) Bioreactor platforms. (B) Example bright field image of eCT on flexible pillars. (C) Example image of aligned cardiomyocytes stained for a-actinin (magenta), cardiac troponin (green), and DAPI (blue); scale = 20 μm. (D) Force-frequency relationship for eCTs. (E) Responses of eCTs to isoproterenol, a beta-adrenergic agonist. Engineered bone marrow tissues (eBM). (F) Histological staining of key hematopoietic and downstream blood/immune markers. (G) Characterization of CB-derived CD34+ cells and CD34+CD38− hematopoietic stem progenitor cells (HSPCs) in eBM tissues over 1–2 weeks. (H) HSPCs in human eBM begin to differentiate into myeloid cells (i.e., monocytes, dendritic cells, granulocytes) over 1–2 weeks. (I, J) Multipotency of HSPCs over 1–2 weeks using colony forming unit assays. Data are shown as mean +/− SD. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
To engineer human bone marrow tissues (eBM), we first engineered bone by infusing iPSC-derived mesenchymal stromal cells (iMSCs) into decellularized bone matrix scaffolds, to induce their differentiation into osteoblasts (35), over a period of 4 weeks (Fig. 1A). Then, we infused iMSCs and human umbilical vein endothelial cells (HUVECs) suspended in fibrin hydrogels into the forming bone, followed by sequential seeding of cord blood derived CD34+ hematopoietic cells (HSPCs). We confirmed the presence of key stromal/hematopoietic markers, including bone sialoprotein (BSP), CXCL12, and CD45 (by immunostaining). We further confirmed key morphological features by pentachrome and Wright-Giemsa staining, as well as micro-computed tomography (μCT) (Fig. 2F).
Over a period of two weeks after introducing HSPCs into the eBM tissue, we validated the maintenance of CD34+ HSPCs (Fig. 2G), as well as cell differentiation into myeloid subpopulations, including monocytes (CD11b+, CD14+), dendritic cells (CD11c+), and granulocytes (CD16+) (Fig. 2H). Consistent with the published studies in the field, we observed inverse relationships between the maintenance and expansion of CD34+ cells and the increase in myeloid progenitors over time (Fig. 2G–2H). Unlike other studies, we cultured the eBMs without adding high concentrations of hematopoietic cytokines (such as stem cell factor, SCF; thrombopoietin, TPO; FMS-like tyrosine kinase 3, FLT3), and instead relied on the endogenous signals produced by the stromal cells. The model supported the natural differentiation trajectory of primary CB-derived HSPCs in culture, as shown by flow cytometry analysis revealing the production of CD11b+ cells by day 12 (Supp. Fig. 1). We confirmed the presence of multipotent progenitors in eBMs after 2 weeks of culture at low cytokine concentrations (5 ng/mL SCF/TPO/FLT-3L), after introduction of CD34+ HSPCs, with approximately 10% of colony forming CD45+ cells at Day 6, and 3% at Day 12 (Fig. 2I–2J).
Effects of acute radiation on cardiac tissues (eCTs)
Functional analysis was performed for all eCT tissues before radiation and after 3 weeks post-irradiation, using a combination of bright field and fluorescent calcium imaging (based on the endogenous GCaMP reporter) to reveal tissue contractility, excitability, and force generation by measuring muscle movement and pillar displacement.
Contractility analysis showed differences in both contraction and relaxation (Fig. 3A). Irradiated tissues had a greater full width half maximum (FWHM) than control tissues (p<0.001), and required longer times for relaxation (p<0.0001, Fig. 3A). There was a significant change in contraction and relaxation lengths between tissues irradiated by photon and neutron sources (p<0.05) (Fig. 3A). Irradiation also caused significant changes in excitability and conduction metrics, including the decreased maximum capture rate and increased excitation thresholds (Fig. 3B). Notably, both the active stress and contraction velocity of neutron-irradiated tissues increased, indicating greater force generation (Fig. 3C). By assessing the deviation of the rest-relaxation interval, we did not observe onset of arrythmias in the eCTs at the 3 weeks post-irradiation timepoint (Supp. Fig. 10D).
Figure 3. Acute radiation causes structural, functional, and molecular changes in engineered cardiac tissues (eCT).
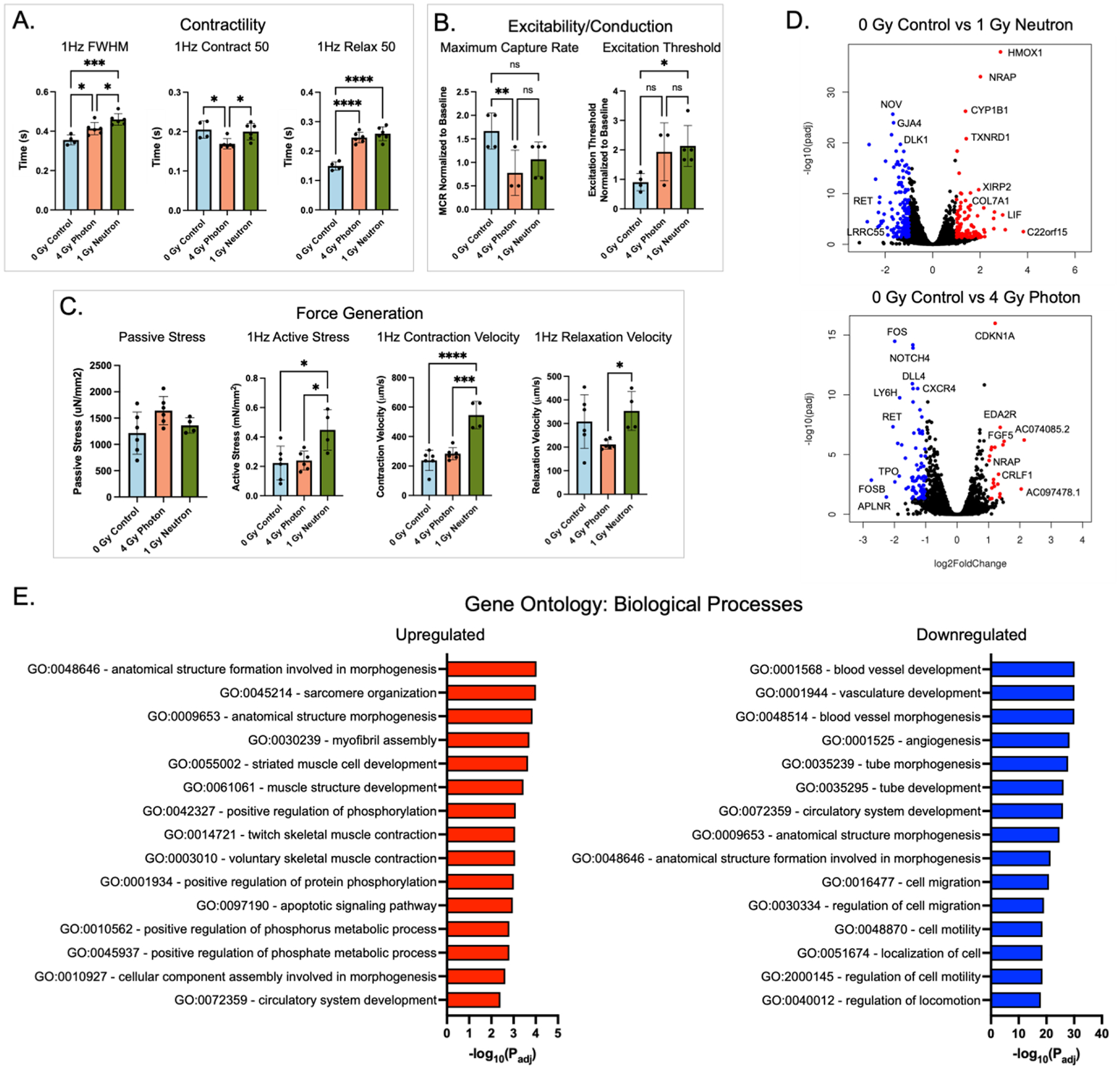
(A) Contractility metrics, including full-width half-max (FWHM), contraction and relaxation lengths at 50% of peak, measured by bright field imaging and computational analysis. (B) Tissue excitability and conduction metrics, including maximum capture rate and excitation threshold, measured by calcium imaging using a GCaMP reporter line and normalized to baseline for each tissue. (C) Force generation metrics: passive and active stress, contraction and relaxation velocities, measured by bright field imaging. (D) Volcano plots showing differentially expressed genes between the irradiated and control eCTs, by RNA sequencing. (E) Biological pathways associated with significant genes in the control and neutron-irradiated groups. Data are shown as mean ± SD. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001
Neutron-irradiated eCTs revealed a number of genes that were differentially expressed relatively to the controls (p<0.05, log2foldchange >1), including the genes implicated in oxidative stress (HMOX1), metabolism (CYP1B1), matrix remodeling (DLK1, NRK, DLL4, COL6A6), cardiac function (TNNT, NRAP, NOV, VASH1) and radiation exposure (CDKN1A). Most notably, HMOX1, a gene implicated in oxidative stress response and mitigation of cytotoxic injury, was significantly increased in neutron-irradiated eCTs in comparison to the non-irradiated control (Fig. 3D).
Gene ontology (GO) analysis revealed that neutron irradiated eCT tissues displayed upregulation of cardiac pathways of sarcomere organization and striated muscle development, as well as increases in apoptosis and metabolic pathways (Fig. 3E). Pathway network analysis revealed significant increases in connections between cardiomyocyte-specific pathways and ECM remodeling, indicating reorganization of the tissue in irradiated groups. Downregulated pathways included those associated with blood vessel development and regulation of cell motility (Fig. 3E), indicating a preferential increase in cardiomyocyte-specific pathways relative to non-myocyte pathways in eCT tissues that contain up to ~15–25% non-myocytes (i.e., fibroblasts, endothelial cells, pericytes).
In analysis of key genes implicated in cardiac muscle function, fibrosis, and inflammation, there was a clear difference in responses to photon and neutron radiation, including the expression of MYH6/MYH7 and IL6/IL1B (Supp. Fig. 2A-2C). Photon and neutron sources appeared to differentially affect DNA damage genes, with increases in CDC25A, RAD17, BRCA1, and BRCA2 in photon irradiated tissues and increases in XRCC3, RAD51, and H2AX in neutron irradiated tissues (Supp. Fig. 2D). Pathway analysis and cytoscape clustering revealed GO terms associated with muscle development, smooth muscle cell proliferation, matrix organization, and positive regulation of apoptotic processes (Supp. Fig. 2E). Only a few of these pathways were found in photon-irradiated tissues (Supp. Fig. 3).
Effects of acute radiation on bone marrow tissues (eBM)
In parallel to eCTs, we expose matured eBM tissues to the same acute doses of radiation (either 4 Gy of photons or 1 Gy of neutrons), and studied the effects on their hematopoietic progeny and microenvironment over 3 weeks post-irradiation. Immediately after exposure, we analyzed double-stranded DNA breaks in CD34+ HSPCs, using an ImageStream analysis pipeline to visualize γH2AX in the nuclei of irradiated HSPCs (36). These data showed early signs of radiation damage, evidenced by the changes in the number and intensity of dsDNA breaks, as well as a dose-dependent effect with increasing dose (Fig. 4A–C, Supp. Fig. 4).
Figure 4. Ionizing radiation causes changes to engineered bone marrow (eBM).
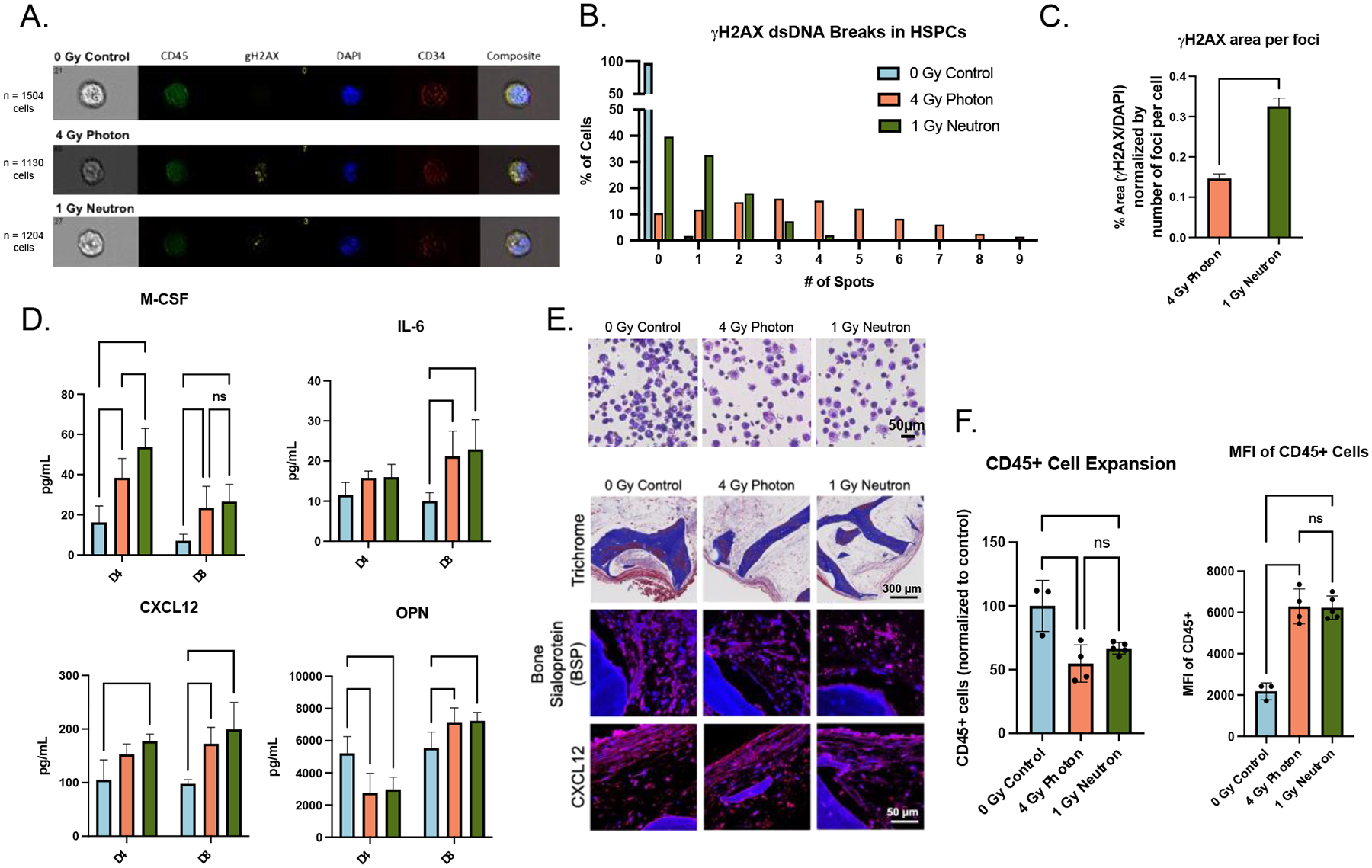
(A) Representative images of DNA-damage marker γH2AX in hematopoietic cells isolated 1-hour post-radiation. (B-C) Quantitative analysis of the numbers and average area of dsDNA breaks (normalized to foci per cell) with unpaired t-test. (D) Acute doses of radiation caused early signs of inflammation (M-CSF, IL-6) and stromal damage (CXCL12, OPN) in supernatant. (E) Histological staining of eBMs 3-weeks post-radiation. (F) CD45+ cell proliferation and median fluorescent intensity 3-weeks post-radiation using flow cytometry. Data are shown as mean ± SD. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Neutron-irradiated tissues had a larger percentage of blood cells concentrated within 1–3 γH2AX dsDNA foci, as compared to photon-irradiated tissues that had a greater distributed spread in the numbers of foci in blood cells (Fig. 4B). Further, there were significant differences in γH2AX foci area between all groups, though these γH2AX foci are representative of a single 1-hour post-irradiation timepoint (Fig. 4C). Right after radiation exposure, eBM tissues exhibited increased secretion of BM stress factors, including inflammatory cytokines M-CSF and IL-6, stromal-support growth factors CXCL12 and osteopontin (OPN) (Fig. 4D). After 3 weeks of culture, histological analysis confirmed morphological changes in BSP+ osteoblasts, and the decrease in their numbers, without evident changes in tissue-level CXCL12 expression (Fig. 4E). Using flow cytometry, we observed a significant reduction in total blood cell numbers, and an increase in CD45+ median fluorescent intensity (MFI), in irradiated groups (Fig. 4F). The increased CD45+ MFI indicates further skewing towards more differentiated progeny, that was confirmed morphologically by Wright-Giemsa staining of blood cells (Fig. 4E).
Effects of acute radiation on blood cells released from eBMs
To understand the downstream and long-term changes to radiation exposure, we performed single-cell RNA sequencing of CD45+ cells isolated from culture medium 3 weeks after acute radiation exposure (12,513 cells in non-radiated controls; 17,515 cells in photon irradiated group; 14,050 cells in neutron irradiated group). UMAP unsupervised clustering helped identify a number of subclusters in isolated samples (Fig. 5A), as well as differential responses, including the new clusters emerging during radiation treatment (Fig. 5B).
Figure 5. Single-cell transcriptomics reveals myeloid skewing in irradiated eBMs.
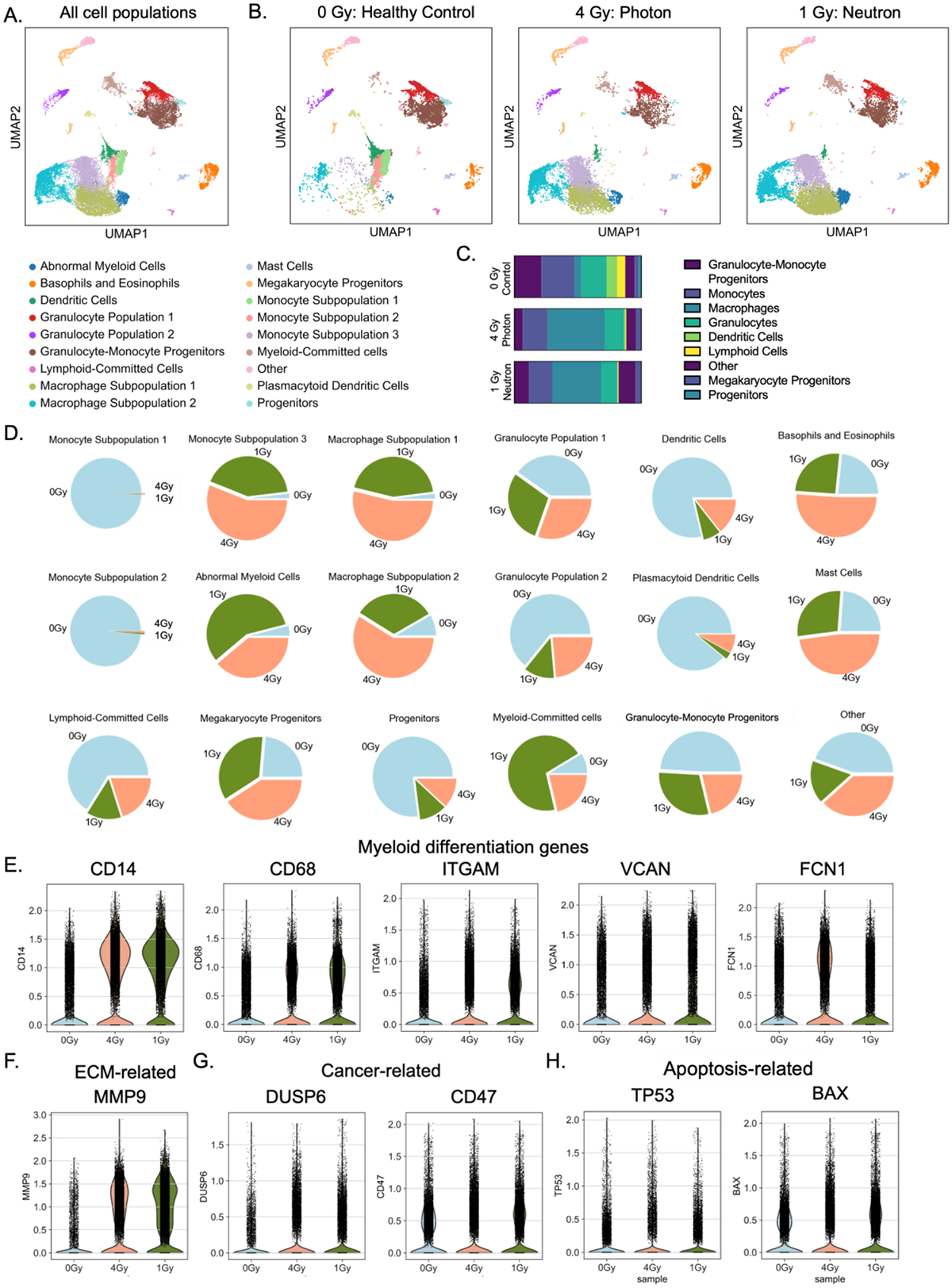
(A) Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction to identify blood and immune cell progeny after 3 weeks of culture. (B) Breakdown of UMAP clusters for each experimental condition. (C) Distribution of the main classes of immune cells, showing increased myeloid population (mainly in macrophages) in irradiated groups. (D) Cell distributions for each experimental condition within the 18 clusters. (E-H) Differential expression analysis of neutron irradiated eBMs reveals increased matrix remodeling. Violin plots showed increased expression of genes related to (E) myeloid differentiation, (F) matrix degradation, (G) cancer, and (H) apoptosis.
We compared the top differentially expressed genes between individual subclusters and the rest of the cell populations, in combination with the expression of cell-type specific markers (37–39) (Fig. 5C–5D; Supp. Fig 5) to identify and name different cell populations (Fig 5A). We first assessed the key markers for each cell type: hematopoietic progenitors (CD34, CD43, PROM1), megakaryocyte progenitors (PPBP, VWF), granulocytes (MPO, ELANE), eosinophils/basophils (CPA3), dendritic cells (CD1C, CD14), lymphoid cells (IL7R), monocytes (CD14, VCAN, ITGAM), and macrophages (CD68, CD163) (Supp Fig. 5). Unique cell populations that emerged from this analysis were labeled as subpopulations within a certain group (e.g., macrophage subpopulations 1 and 2). Using the top genes for each group, we identified cell clusters, such as the myeloid-committed cluster (abnormal myeloid cells) with high expression of matrix metalloproteases (MMPs) and cell cycle proliferative markers (Fig. 5D; Supp. Fig. 6).
When comparing irradiated groups to healthy controls, there was an increase in myeloid cell subtypes (Fig. 5C; Supp. Fig. 7), with a reduction of IL7R+ lymphoid-committed cells and CD1C+ dendritic cells. The majority of cells in the progenitor and lymphoid subgroups were from the non-irradiated control group (Fig. 5D). In the myeloid subtypes, there were large shifts in CD14+ subclusters from monocytes to downstream lineages and in differentiated macrophages: monocyte subpopulation 3 (1.9% in control, 56.1% in photon group, 42% in neutron group), macrophage subpopulation 1 (2.0% in control, 53.8% in photon group, 44.2% in neutron group), macrophage subpopulation 2 (8.4% in control, 58.9% in photon group, 32.8% in Neutron group), and the abnormal myeloid cells (4.0% in control, 38.9% in photon group, 57.1% in neutron group) with clusters mostly present in the irradiated groups (Fig. 5D; Supp Fig. 6).
In analysis of gene expression, we found significant increases in key myeloid differentiation markers following 1 Gy neutron exposure compared to the 0 Gy control, including CD14 (p <10−−306, L2FC = 1.48), CD68 (p <10−214, L2FC = 0.78), ITGAM (p <10−306, L2FC = 1.14), VCAN (p <10−24, L2FC = 0.40), and FN1 (p <10−306, L2FC = 3.44) (Fig. 5E; Supp. Fig. 7). There was also an increase in expression of ECM-associated MMPs in the neutron group, including increases in cancer-related genes, including DUSP6, traditionally associated with myeloid abnormalities (p <10−206, L2FC = 1.67), CD47 (p <0.05, L2FC = 0.06), evasion of tumor cell uptake by immune cells, and TET2 (p<10−40, L2FC = 0.24), associated with numerous human cancers (Fig. 5G).
As expected, there was also an increase in a number of radiation-associated genes in the neutron-irradiated group, including CDKN2A (p<10−5, L2FC = 2.80), TP53 (p<10−48, L2FC = 1.10), and BAX (p<10−4, L2FC = 0.11) (Fig. 5H). In comparing the neutron groups with the control, the highest differentially expressed genes were among those associated with ECM remodeling, including MMP9 and FN1, lipid metabolism, including FBP1, APOE, and G0S2, and macrophage activation/inflammatory-resolve associated genes C1QA and APOC1 (Supp. Fig. 6A). GO molecular functions revealed an increase in MMP activity and growth factor, proteoglycan and chemokine receptor binding, all associated with ECM degradation by activated myeloid cells (Supp. Fig. 5B). This finding matches closely the most upregulated GO processes, associated with ECM remodeling and leukocyte chemotaxis, and the most downregulated pathways, associated with immune response and pro-inflammatory signaling (Supp. Fig. 6C). Notably, all myeloid markers were still increased when compared in single subpopulations (i.e. CD14+ cells or CD68+ cells) between irradiation groups (Supp Fig. 8).
In the emerging cell populations in neutron-irradiated eBM tissues, we found a shift in myeloid differentiation towards macrophage phenotypes implicated in matrix degradation and remodeling (Supp Fig. 6A-6D). Both the macrophage subpopulation 1 (MAS1) and macrophage subpopulation 2 (MAS2) exhibited characteristics associated with an M2 macrophage phenotype, including expression of key pan- and M2-specific macrophage markers (CD68, CD163, MS4A4A, PLXDC2, NEAT1), as well as genes implicated in ECM remodeling and microenvironmental metabolism (MMP9, RAL15, RASAL2, CSTB) (Supp. Fig. 6A).
Notably, pathway analysis of MAS1 revealed the signatures of leukocyte migration and ECM organization, as well as lipid catabolic processes. In MAS2, GO processes indicated pathways related to neural development, morphogenesis, and ECM remodeling, potentially hinting at a pathologic role of these cells in addition to their autophagic phenotype in vitro (Supp. Fig. 6B). Monocyte subpopulation 3 (MOS3) expressed many canonical markers of myeloid- and monocyte-specific cell fate, including FCN1, VCAN, and CD14, but these cells also had high levels of genes associated with myeloid-derived suppressor cell (MDSC) monocytic populations (M-MDSCs), including S100A8, S100A9 and S100A10.
Pathway analysis for the MAS3 population indicated immune responses, including myeloid leukocyte activation and neutrophil degranulation, potentially indicating a role in these cells in regulating other immune progeny (Supp. Fig. 6C). Most interestingly, there was a population of “abnormal myeloid cells” that appeared in irradiated groups, exhibiting canonical myeloid markers of CD14 and CD68, but had significantly higher expression of genes associated with cell proliferation (MKI67, PCLAF, DTYMK), cell cycle regulation, and DNA repair (Supp. Fig. 6D). This group, in addition to MAS1 and MAS2, also exhibited significantly high levels of MMP9, as compared to all other cells (Supp. Fig. 6).
Neutron-irradiated eBMs exhibit increased ECM remodeling
We also used CellChat computational algorithms to predict the cell-cell, cell-ECM, and secreted cell interactions in cell subpopulations, showing decreases in the number and strength of cell-cell interactions and signals following radiation (40). The number of ECM interactions was much higher (709 molecules) in the neutron group than in controls (Fig. 6A). This finding was corroborated with increased remodeling-related GO terms in myeloid subpopulations emerging after radiation exposures (Supp. Fig. 6A-D).
Figure 6. Computationally predicted cell-cell, cell-ECM, and cell-secretome interactions confirm myeloid matrix-remodeling cell phenotype in irradiated eBMs.
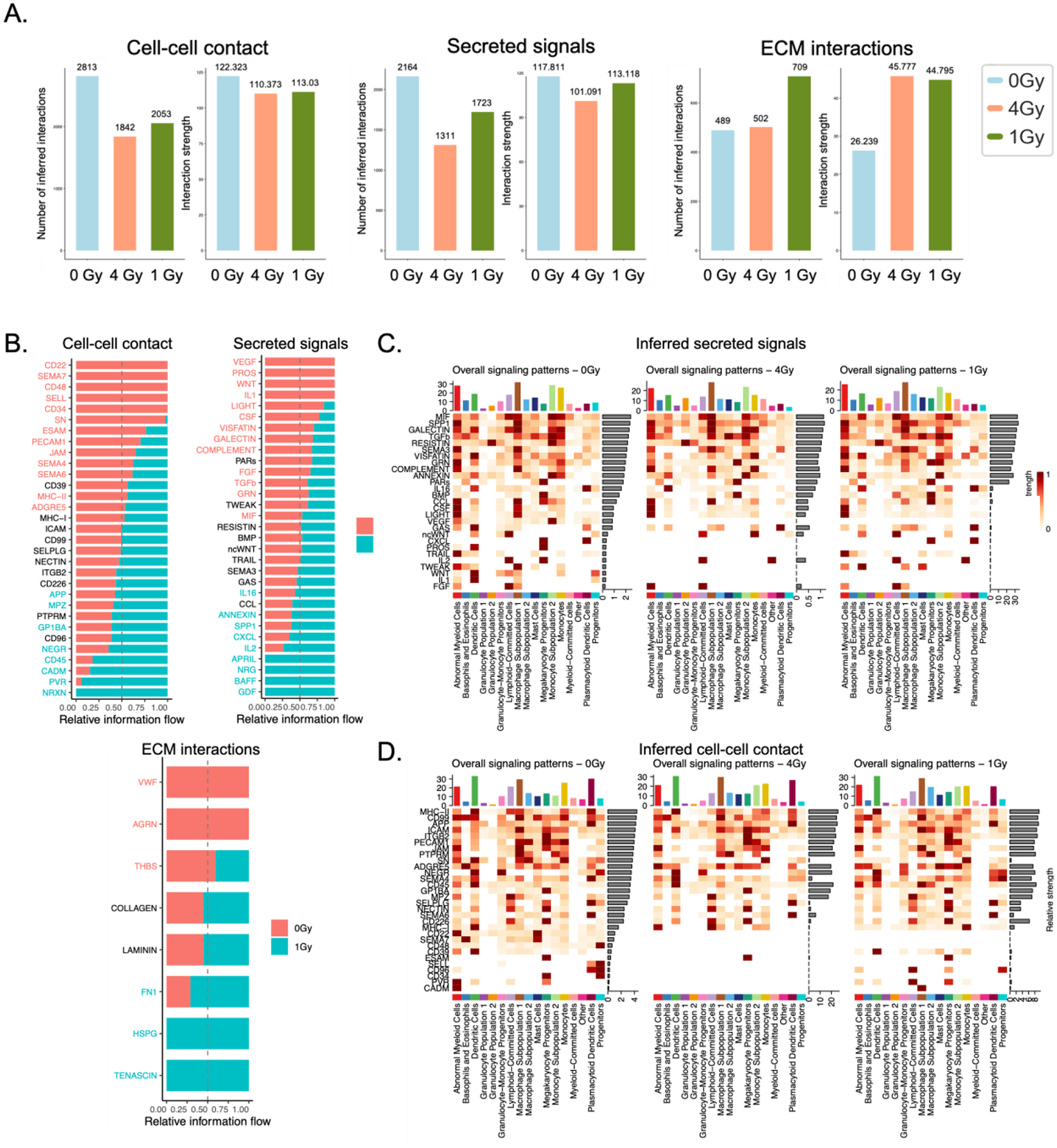
(A) Comparative quantification and (B) protein breakdown of total inferred cell-cell, cell-ECM, and cell-secretome signals via CellChat algorithms. (C-D) Inferred (C) secreted signals and (D) cell-cell interactions within each subpopulation over each experimental group.
When comparing the 1 Gy neutron-irradiated tissues with the non-irradiated controls, 32 distinct cell-cell contact proteins of interest were revealed, with differential expression of a subset of proteins, including APP, MPZ, GP1BA, NEGR, CD45, CADM, PVR, and NRXN, many of which are critical in leukocyte adhesion and motility (Fig. 6B). Among secreted signals from neutron-irradiated tissues, differentially expressed factors include IL16, ANNEXIN, SPP1, CXCL, IL2, APRIL, NRG, BAFF, and GDF (Fig. 6B).
M2 macrophages, present at high concentrations in the MAS3 group, also secrete high levels of SPP1, APRIL, and CXCL. In addition, high levels of secreted ANNEXIN indicated cellular stress in tumor-associated macrophages. The increased M2 phenotype was reflected in higher inferred cell-ECM interactions with matrix proteins fibronectin, heparan sulfate proteoglycan (HSPG), and tenascin – all implicated in MMP9-induced cancer stem cell invasion (Fig. 6B). These predicted interactions can further be broken down into cell subtypes, highlighting similarities between cell types (Fig. 6C–D, Supp. Fig. 9A-9C).
Biomarkers of neutron-induced injury of eCT and eBM tissues
To identify potential biomarkers of neutron-associated radiation injury, we identified differentially expressed (p<0.05, L2FC>1) genes in neutron- and photon-exposed eCT and eBM tissues, trying to elucidate differences between the photon and neutron responses (Fig. 7A–7C; Supp Fig. 10). In eCT models, we observed increases in expression of known radiation-related genes, including PTGS2, LIF, ICAM1, FOS, and OSGIN1 (Fig. 7A). We also found increased expression in genes associated with cardiac hypertrophy (NPPB, MYH7, NEAT1, FLNC, TNNT2, and CSRP3). Pathway analysis revealed increases in smooth muscle cell proliferation, CYP450 metabolism, and cellular responses to hypoxia and inflammatory stimuli (Fig. 7B–C).
Figure 7. Parallel analysis of photon and neutron sources of radiation for engineered cardiac (eCT) and bone marrow (eBM) tissues.
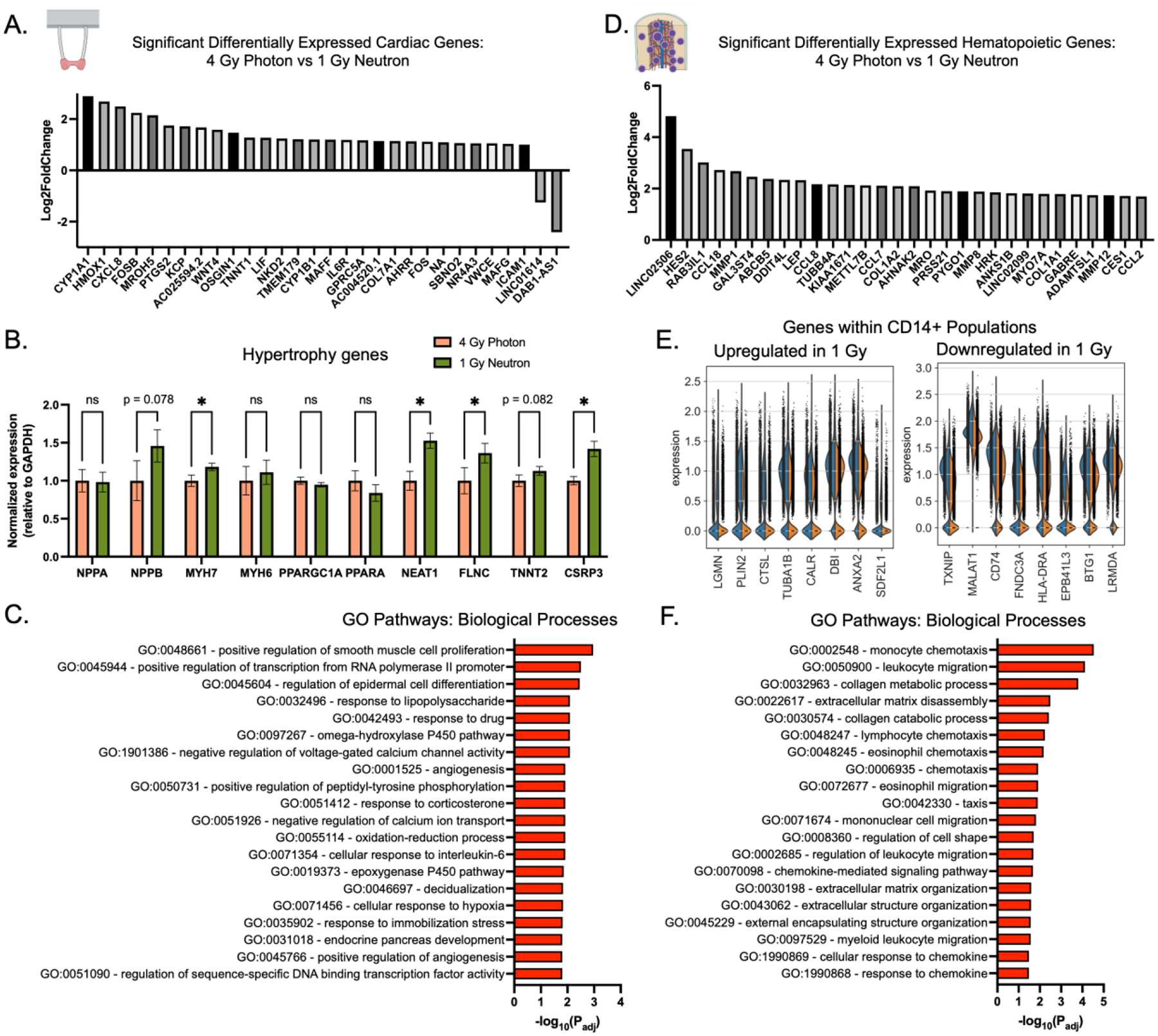
(A) Top differentially expressed genes with highest absolute fold change in eCM tissues. (B) Selected key hypertrophy-related genes are increased in neutron-irradiated eCTs, normalized to GAPDH and photon controls. Data are shown as mean ± SD with unpaired t-tests with *p<0.05. (C) GO biological processes pathways enriched in differentially-expressed genes in eCT 4 Gy Photon versus 1 Gy Neutron comparative analysis. (D) Top differentially expressed genes with highest absolute fold change in eBM tissues. (E) Highest up- and down-regulated genes in CD14+ fraction of eBM-derived cells. (F) GO biological processes pathways enriched in differentially-expressed genes in eBM 4 Gy Photon versus 1 Gy Neutron comparative analysis.
CD45+ cells isolated from eBMs exposed to neutron radiation, relatively to those from photon-irradiated tissues, showed increased expression of genes related to matrix remodeling (MMP1, MMP8, MMP12), chemotaxis (CCL18, CCL8, CCL7, CCL2), collagens (COL1A2, COL1A1), and radiation (HES2, GAL3ST4, DDIT4, HRK), (Fig. 7D). For CD14+ cells, exposure to neutrons enhanced the ECM-remodeling phenotype of the myeloid cells, with downregulation of antigen-presentation genes HLA-DRA and CD74 (Fig. 7E). Using CellChat and secreted signal analysis, TWEAK appeared to be secreted by neutron-irradiated cells, indicating a possible role in the TNF-stimulated apoptosis (Supp. Fig. 8E). Differential gene expression revealed similar changes in ECM remodeling from GO pathway responses to neutron exposure, suggesting neutron-specific effects on macrophage remodeling capacity (Fig. 7F).
We were also interested in understanding whether there were common effects of neutron radiation on hematopoietic cells and cardiac muscle. Comparing the gene expressions in eCT and eBM tissues (Fig. 8A), we observed 25 genes that responded to neutron exposure in both tissues, including genes associated with tumor suppressor p53 (e.g., MIR34AHG, PHLDA3). Expression of COL24A1 suggested the pro-fibrotic and senescence-related radiation injury. Likewise, for tissues exposed to photon radiation, we identified 12 common genes between eCT and eBM models, including a number of genes related to oxidative stress, such as HMOX1, HRK, and MIR22HG (Fig. 8B).
Figure 8. Significant genes of interest between engineered cardiac (eCT) and bone marrow (eBM) tissues in neutron-specific radiation responses.
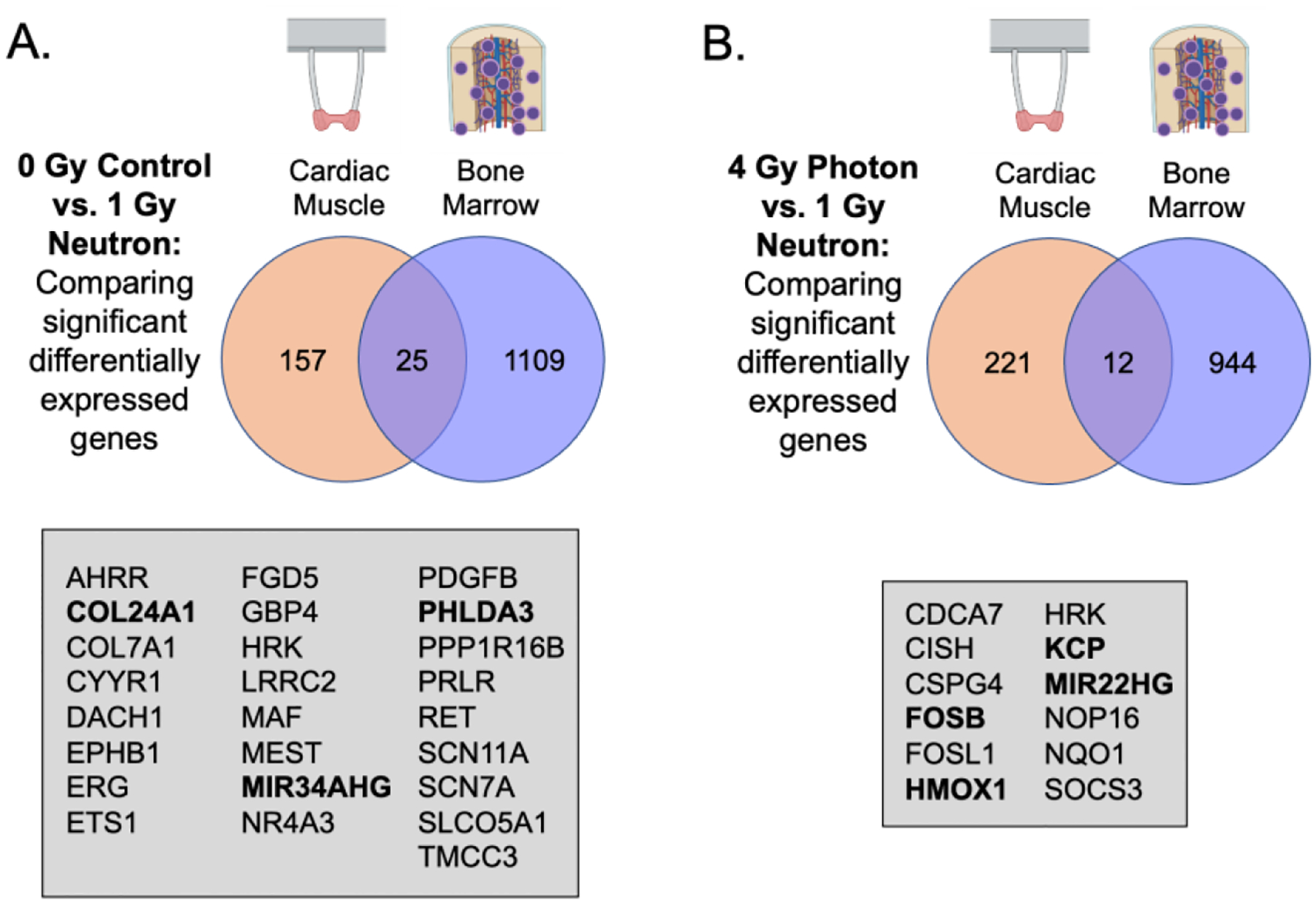
(A) Shared genes between control and neutron-irradiated tissues between eCM and eBM bulk analyses. (B) Shared genes between photon- and neutron-irradiated tissues between eCT and eBM bulk analyses.
Radioprotective drugs mitigate radiation damage in eCT and eBM tissues
To demonstrate utility of our platform for evaluation of radioprotective agents, we studied the functional changes in eCTs following pre-treatment with Amifostine (FDA-approved radioprotective drug, (41), and in eBMs following post-radiation treatment with cytokine granulocyte-colony stimulating factor (G-CSF) (FDA-approved radiomitigator that stimulates marrow recovery post-radiation in vivo (42–44).
eCTs pre-treated with 1 mM Amifostine right before radiation exposures demonstrated remarkable restoration of functional metrics to the levels seen in healthy controls, in contrast to irradiated eCTs not treated with Amifostine (Supp. Fig. 11A). In particular, the Amifostine pre-treatment maintained the excitation threshold and maximum beating frequency at normal levels. The only notable metric that was not restored was the muscle relaxation time, which decreased only slightly in the Amifostine-treated groups but not to the levels of healthy controls (Supp. Fig. 11A).
For eBMs, G-CSF treatment administered at a low dose (5 ng/mL) increased the total CD45+ blood cell production (Supp. Fig. 11B) through increases in neutrophils and subsequent proliferation of multipotent progenitors in vitro (Supp Fig. 11C).
DISCUSSION
We demonstrate that engineered human tissue models can be used to study the effects of high energy radiation designed to mimic that encountered during long-range space travel such as the Mars mission. By using a combination of primary cells and iPSCs, we were able to recapitulate organ level responses of human bone marrow and heart to radiation, including the generation of downstream blood and immune progeny in the eBM and contractile behavior of eCT. These human tissue models captured the functional and molecular changes associated with radiation injury, including the emergence of unique myeloid populations following neutron radiation exposure, and differential expression of key genes in both the eBM and eCT following neutron vs photon radiation exposure (Fig. 7). We also identified a number of shared genes (Fig. 8) associated with neutron-specific radiation exposures between eCTs and eBMs. These genes may be useful for identifying biomarkers of secondary radiation damage in human body during deep space missions (i.e., MIR22HG, HMOX1), as neutrons are considered to be amongst the greatest radiation risks for astronauts on spacecrafts headed to Mars (45, 46). We believe that our study is the first to use engineered human tissue models for studying the effects of high-LET neutrons to mimic space radiation, and establishing a proof-of-concept platform and framework for using engineered human tissue models for mitigating the radiation “red risks.”
As the fields of stem cell biology and OoC models of human physiology continue to develop, we are starting to address the need for human-relevant, and eventually individualized models for studying and countering the radiation damage. Animal models and cell monolayer studies, the current gold-standards for assessing the effects of radiation, are limited in their ability to recapitulate human condition.
Using human tissue models, we found the biological effects of neutrons to be stronger than those of photons, and to vary greatly between endpoints and the two organ types studied. Because of the differences in responses to radiation from one individual to another, developing individualized, organ-specific human tissue models is of great interest to the space-science community. Previous work has shown utility of human OoC models for mimicking the acute radiosensitivity of the gut and marrow in response to low levels of gamma radiation (26–28, 47). Most recently, Verma et al. studied the role of astrocytes in simulated GCR injury for up a week using a blood-brain barrier OoC model (25). However, these studies are limited to short-term (hours to days) responses to radiation, showing immediate loss of viability of the most proliferative cells in response to radiation injury. Studies with tissue model systems that we present here allow uniquely to parse out the individual factors contributing to radiotoxicity and radioprotection for radiation sources and dosages relevant to space travel.
Neutron-irradiated cardiac tissues had impaired conduction and excitability, as evidenced by the decreased maximum capture rate and increased excitation threshold, indicating that more stimulation would be necessary to excite the irradiated eCT tissues. However, these irradiated eCT tissues also demonstrated remarkably higher force generation and contractility than their control counterparts, which may indicate an early hypertrophic phenotype of the neutron-irradiated tissues. The increased force generation was corroborated by the increase in cardiac-specific genes and ontology pathway data, as well as by increases in hypertrophy-related genes MYH7, NPPB, and NEAT1 (48–50). Interestingly, HMOX1 was significantly upregulated in neutron-irradiated muscle as compared to both untreated controls and photon-irradiated eCTs, indicating that the neutron-specific dose was critical in activating the stress-response pathways implicated in oxidative stress and hypoxic injury to the myocardium (51).
Other groups have reported increases in oxidative stress of cardiomyocytes in response to radiation, and increases in cardiac-specific hypertrophic gene expression levels in response to high (>20 Gy) photon doses (52, 53). During neutron-specific radiation injury, there may be compensatory mechanisms transitioning the healthy cardiac muscle to early stages of hypertrophy (54), though this effect must further be investigated at longer culture timepoints. To date, many studies of radiation-induced cardiac dysfunction are focused on microvascular changes in the heart. Although there was no vascular compartment in our model, neutron-irradiated eCTs exhibited downregulated ontology pathways associated with vascular maturation, potentially indicating a repressed role of angiogenic factors in the irradiated eCT tissues (55, 56).
We observed the expected responses of the radiosensitive hematopoietic system to neutron doses, most notably the decreased proliferation of CD45+ cells and increased inflammatory signatures at early time points, similar to cell monolayers and animal studies of similar doses (15, 16, 57). Previous studies have also shown a Poisson distribution for photon exposures, as compared to overdispersed foci numbers in neutron exposures, similar to our findings (58). Notably, the average area of γH2AX foci per cell was larger in neutron irradiated groups, suggesting that the double strand breaks may be overlapping along the neutron beam track and producing larger breaks with less frequency. In addition, the reduction of the hematopoietic lymphoid compartment and skewing towards myeloid lineages is a unique aspect of the models described here, in line with the available data from human studies, including the development of acute myeloid leukemia (AML) as a second cancer in radiotherapy patients, in atomic bomb survivors, and in non-human primate studies (59–61).
In humanized mouse models, radiation with heavy ions (56Fe) promoted the development of a phenotype similar to T-acute lymphoblastic leukemia (T-ALL) in mice by 6–9 months post-irradiation (62). In contrast, human data from recent spaceflight studies indicate skewing of hematopoiesis into myeloid progenitors and increased development of pre-malignant myelogenous clonal hematopoiesis (63). Also, data from ex vivo irradiation of peripheral blood indicates a decrease in the lymphoid compartment in response to neutrons over the course of a few days (64). These previous studies, in tandem with our data, suggest the role of high-LET exposure in promoting a pre-malignant phenotype in HSPCs, skewed towards myeloid-biased outputs and eventual progression into an AML-like manifestation. Unique myeloid populations emerging in our system 3 weeks post-irradiation include M2-macrophage-like and MDSC-like cells, indicating a role in preventing tissue inflammation and promoting matrix remodeling seen in pre-malignant and malignant patient tissues. However, more work is needed to better understand the phenotypes of these pre-malignant cells, and whether they are able to restore normal hematopoiesis post radiation.
Our results show both functional and molecular changes in human OoC models of the bone marrow and heart after exposures to photons or neutron radiation. As we chose two organs known to cause immediate (bone marrow and acute radiation syndrome) and long-term (heart and radiation-induced heart disease) changes in response to radiation, there are many more radiosensitive organs that may be target areas of investigation in the future (i.e., brain, gastrointestinal system). In preparation for a future Mars mission, this model can be used to develop new radioprotective countermeasures that are effective against high-LET radiation. A number of radioprotective agents have been studied over the past 50 years to prevent damage to the body, but none of these drugs was successful in preventing phenotypic changes in all organs. We show in “proof-of-concept” experiments that exposure to radioprotective drugs Amifostine (pre-treatment of the eCTs) and G-CSF (post-treatment of the eBMs) was able to ameliorate some of the neutron-associated changes. Although these drugs may provide some alleviation of injury, long-rage Mars missions with radiation from multiple ion species delivered over a multi-month journey, will require new therapeutics to be developed to provide protection of all sensitive organs, in particular the hematopoietic and cardiovascular systems. OoCs provide an ideal platform to help validate new therapeutic modalities, as those that work in animals may not work in humans.
OoC models may also allow for studies of other NASA “red risks”, including hypoxia and microgravity, both individually and in tandem with radiation. The ability to personalize these model systems may allow for unprecedented understanding of how an astronaut’s health may be impacted by space travel. It is critical, however, to compare data obtained in humanized radiation studies on Earth with those of past nuclear exposures (i.e., Hiroshima, Chernobyl) and clinical data. Recently, new data were collected from short-term studies of human cells in low earth orbit (LEO) experiments on the ISS, and astronauts after returning to Earth (i.e., NASA Twins Study) (65).
Our study also has several limitations. We studied the responses of the cardiac and bone marrow tissues to neutron radiation over 3 weeks post-radiation, a period providing just a snapshot of the time-dependent effects of acute radiation exposure on human tissues. In future studies, these changes should be studied over longer periods of time, and extended to the cellular repair mechanisms. To this end, we are currently extending the lifetime of OoC platforms to as long as 6 months. Also, we studied the effect of radiation on a hematopoietic system without high levels of exogenous stimuli (i.e. cytokines), allowing for hematopoietic differentiation into downstream progeny, rather than the maintenance of the HSPC phenotype. Uniquely, this approach allowed for careful characterization of the downstream populations emerging in response to radiation. However, future work should also address the functional changes to HSPCs in the early (hours/days) and late (weeks/months) periods post radiation. In addition, we focused the single cell transcriptomic analyses on released cells from the eBM tissues, rather than the cells from within the tissue, which is a limitation of the work that may be investigated further in future studies. As we observed a shift in phenotype towards a myeloid-biased hematopoietic system, also seen in clinical exposures of radiotherapy (59) and astronauts returning to Earth (63), we must further characterize the effects of these acute doses on the emergence of myeloid pre/malignancies. Mutational analysis of irradiated HSPCs may help identify genetic and epigenetic-level changes leading to the development of clonal hematopoiesis or acute myeloid leukemias, as shown in the NASA Twins Study (63, 65).
Future studies would greatly benefit from increased throughput and evaluation of individualized responses to radiation. There are a number of biological variables that can influence an individual’s response to radiation, including sex, race, and genetic background. The model systems described here can be personalized, by developing an entire tissue model for the same individual using iPSCs, though are limited in the current study by number of individuals studied.
Finally, linking the engineered human tissues into a multi-organ context may allow for studies of systemic radiation toxicity, showing cross-talk between human organs to mimic the most realistic effects of radiation (19, 66). Extension to a “patient-on-a-chip” model could provide a personalized platform for optimizing protection from the harmful effects of radiation.
MATERIALS AND METHODS
Study design:
Our objective was to engineer multiple multicellular tissue models, capable of recapitulating key features of each target organ (i.e. bone marrow, cardiac muscle), in response to radiation injury. In particular, we were interested to understand the longer-term (weeks) changes acute doses of radiation may have on human tissues, mimicking doses that will have large enough doses to affect tissue function and physiology. In addition, we chose both photon radiation (involved in cancer therapy) and neutron sources (secondary rays from cosmic radiation), to mimic the potential changes associated with human tissues on a mission to deep space. We chose the two tissue models of interest to represent a tissue known to be radiosensitive (bone marrow), with acute changes to hematopoietic cells, and a tissue affected by chronic changes (cardiac muscle), with downstream changes to function. Further, we hoped to identify potential genes altered in radiation exposures, particularly those that are maintained 3 weeks post-radiation in our in vitro culture platforms, as well as those that may be shared amongst the two target organs (bone marrow and cardiac muscle). In addition, we were interested in neutron-specific changes associated with the type of radiation rather than the dose, as neutrons are approximately four times as potent as photon sources.
Radiation dosing:
Photon irradiations were performed at Columbia University’s Center for Radiological Research using a Gammacell 40 137Cs irradiator (Atomic Energy of Canada Ltd) with a dose rate of 0.68 Gy/min for varying low-energy doses (i.e., 1, 2, 4, 6 Gy). Neutron irradiations were performed at the Columbia University Radiological Research Accelerator Facility (RARAF), using an accelerator-based neutron irradiator mimicking the neutron energy spectrum from an Improvised Nuclear Device (67). Briefly, a mixed beam of atomic and molecular ions of hydrogen and deuterium is accelerated to 5 MeV and used to bombard a thick beryllium target. The energy spectrum of neutrons emitted at 60° to the ion beam axis closely mimics the Hiroshima spectrum at 1–1.5 km from the epicenter (68) . During irradiation, samples were placed below and in front of the beryllium target, at an angle of 60° to the particle beam and a distance of 17.5 cm. Irradiations were performed with a total beam current of 20–30 μA, resulting in a dose rate of 2–3 Gy/h of neutrons with an additional 20% of concomitant γ rays, for varying high-energy doses of (i.e., 1, 2, 4, and 6 Gy). Dosimetry is performed at the beginning of each irradiation day, using a custom built Tissue Equivalent Proportional Counter (TEPC) (69), which measures total dose and a compensated Geiger-Mueller dosimeter (70), which has a very low response to neutrons, and thus measures only the photon component.
Cardiomyocyte differentiation from human iPSCs:
hiPSCs (WTC11-GCaMP6f line was obtained through material transfer agreements from B. Conklin, Gladstone Institutes. Cardiomyocytes were differentiated as previously described (33). On Day 10, RPMI-no glucose (Life Technologies, 11879020) supplemented with B27 (Thermo Fisher Scientific, 17504044) and 213 μg/mL ascorbic acid (Sigma-Aldrich, A445), was used to purify the iPSC-CMs population and eliminate any contaminating mesodermal and endodermal populations. Medium was replaced on day 13 with RPMI-B27 supplemented with 213 μg/ml ascorbic acid until day 16. On day 17 cells were pretreated with rock-inhibitor (y-27632 dihydrochloride, 5 μM) for 4 hours before dissociation. Cells were dissociated by enzyme digestion with collagenase type II (95 U/mL; Worthington, LS004176) and pancreatin (0.6 mg/mL; Sigma-Aldrich, P7545) in dissociation buffer (Glucose (5.5 mM), CaCl2·2H20 (1.8 mM), KCl (5.36 mM), MgSO4·7H20 (0.81 mM), NaCl (0.1 M), NaHCO3 (0.44 mM), NaH2PO4 (0.9 mM)) on a shaker in a 37°C incubator. Flow cytometry for cTnT+ (BD BioSciences, 565744) was performed prior to cell use for tissue fabrication to ensure cell purity (>90% cTnT+).
Engineering of cardiac tissues:
We generated eCTs using our previously-published “milliPillar” cardiac microtissue platform (33). Briefly, primary human ventricular cardiac fibroblasts (NHCF-V; Lonza, CC-2904) were cultured according to the manufacturer’s recommendation. Differentiated hiPS-CMs were dissociated and mixed with supporting cardiac fibroblasts in a 75:25 (hiPS-CM:NHCF-V) ratio. The cells were subsequently resuspended in fibrinogen by mixing the cell solution with 33 mg/mL stock human fibrinogen (Sigma-Aldrich, F3879) and RPMI-B27 (RPMI 1640 basal medium, L-ascorbic acid 2-phosphate, and B27 supplement) to a final fibrinogen concentration of 6.25 mg/mL and a cell concentration of 45,833 cells/μL. 3 μL of thrombin solution (2U/mL) were added to each well, followed by 12 μL of the cell/fibrinogen solution. The solutions were mixed and allowed to polymerize at 37 °C for 15 min, so that the tissues readily formed around the pillars. These resulting tissues each contained 550,000 cells in a 5 mg/ml fibrin hydrogel.
400 uL of RPMI-B27 with 10uM Rock inhibitor and 5 mg/mL 6-aminocaproic acid (Sigma-Aldrich, A7824) were added to each well. After 24 hours, the medium was changed to RPMI-B27 with 5 mg/mL 6-aminocaproic acid and replaced every other day. On the 5th day following tissue formation, the medium was changed to RPMI-B27 without 6-aminocaproic acid and replaced every other day. On the 7th day following tissue formation, electrical stimulation was initiated to promote tissue maturation. Tissues were stimulated at 2 Hz with a 5 V/cm electric field provided by 2 ms biphasic pulses throughout culture period, as done in our previous work (33). Tissues administered with Amifostine trihydrate (generously donated by Clinigen and Zambon Chemicals; 112901–68-5) were incubated with 1 mM Amifostine for 30 minutes at 37°C prior to irradiation, with all tissues receiving fresh media prior to irradiation.
Derivation of bone scaffolds:
Bovine calf metacarpals were purchased in bulk and stored at - 40°C (Lampire Biological Laboratories, #19D24003). A band saw is used to obtain a section (~4 cm tall) from the distal end of the metacarpal, as this region contains an enriched concentration of trabecular bone. We then used a CNC Milling machine to acquire smaller rectangular bone cores with a cross section of 4 mm x 8 mm. These pieces were then placed into an IsoMet low speed wafering saw to then cut each individual piece into 1 mm thick bone scaffolds. Each scaffold is approximately 4 mm (width) x 8 mm (height) x 1 mm (depth), and subsequently processed for removal of cell debris.
Decellularization protocols were adapted from our previously established protocols (35), which removes all cellular material but preserves the matrix composition and architecture of the bone. Bone scaffolds were processed in batch with the following step-wise protocol all completed on an orbital shaker: (i) PBS with 0.1% EDTA (w/v) for 1 hour at room temperature; (ii) 10 mM tris, 0.1% EDTA (w/v) in DI water overnight at 4°C; (iii) 10 mM Tris, 0.5% sodium dodecyl sulfate (w/v) in DI water for 24 hours at room temperature; (iv) 100 U/ml DNase, 1 U/ml RNase, 10 mM Tris in DI water for 6 hours at 37°C. After decellularization was complete, bone scaffolds were lyophilized until freeze-dried using a Labconco freezone lyophilizer (7740020), and cut to a final scaffold size of 4 mm x 4 mm x 1 mm. Scaffolds were `weighed to ensure each piece was at the appropriate density for cell seeding (5–7 mg per scaffold), and cut in half to reach a final scaffold. For sterilization, bone scaffolds were subjected to 70% ethanol treatment overnight, and then washed with DMEM basal media overnight.
Engineering of bone niche and bone marrow tissues:
Induced pluripotent stem cells (iPSCs) were expanded on Matrigel-coated plates (Corning) for 2 weeks prior to differentiation. WTC-11 iPSCs were differentiated using the STEMdiff™ Mesenchymal Progenitor Kit according to manufacturer’s instructions into iPSC-derived MSCs (iMSCs) over a period of three weeks (Stem Cell Technologies, 05240). iMSCs were expanded and seeded into the bone scaffolds at a concentration of 2 × 105 cells per scaffold, using 15 μL of medium (4.5 g/L DMEM supplemented with 10% (v/v) HyClone FBS, 1% penicillin/streptomycin, and 1 ng/mL of basic fibroblast growth factor, bFGF), according to established protocols (19, 35, 71). The cells were allowed to attach for 2 hours, and then supplemented with additional MSC medium overnight. After 72 hours, osteogenic differentiation of the seeded cells was initiated with the addition of low glucose (1 g/L) DMEM supplemented with 1 μm dexamethasone (Sigma Aldrich), 10 mm β-glycerophosphate (Sigma Aldrich), and 50 μM L-ascorbic acid-2-phosphate (Sigma Aldrich). Each scaffold was incubated in 1 mL of osteogenic media, with media changes 3 times a week for 4 weeks, allowing for the iMSCs to differentiate into functional, maturing osteoblasts.
Following osteogenic differentiation, human umbilical vein endothelial cells (HUVECs; Lonza, C2519A) and additional iMSCs were added within a 10 uL fibrin hydrogel (11 mg/mL fibrinogen, Sigma Aldrich, F3879; 33 U/mL thrombin, Sigma Aldrich, T6884) to each scaffold. Microtissues were then incubated in endothelial cell growth medium-2 (EGM-2; Lonza, CC-3162) for 1 week with addition of 33 mg/mL of the protease inhibitor aprotinin (Sigma-Aldrich, A3428) and placed on a rocker (Cole Parmer; 51401–00). After 1 week, media was removed from the microtissues and 1.1 × 104 cells/tissue of mixed donor cord blood (CB)-derived, CD34+ human HSPCs (Stem Cell Technologies, 70008) were allowed to attach for 2 hours at 37°C in a humidified incubator at 5% CO2. After seeding, tissues were replenished with StemSpan™ SFEM II medium with 1% P/S (Stem Cell Technologies, 09655) with 10 ng/mL stem cell factor (SCF), thrombopoietin (TPO), and FMS-like tyrosine kinase 3 ligand (FLT-3L) (Peprotech) for the first four days (prior to radiation exposure). After Day 4, tissues underwent half media changes every four days with 5 ng/mL SCF, TPO, FLT-3L in SFEM II medium. Microtissues were kept in culture for 21 days post-irradiation. Tissues administered with recombinant human Granulocyte colony stimulating factor (G-CSF) (Peprotech; 300–23) were treated with 5 ng/mL G-CSF for 8 days and analyzed on Day 12.
Cardiac calcium imaging and analysis:
To visualize calcium handling in real-time, WTC11-GCaMP6f iPSCs that contain a constitutively expressed GCaMP6f calcium-responsive fluorescent protein inserted into a single allele of the AAVS1 safe harbor locus were used (72). Tissues were imaged in a live-cell chamber (STX Temp & CO2 Stage Top Incubator, Tokai Hit, Fujinomiya, Japan) using a sCMOS camera (Zyla 4.2, Andor Technology) connected to an inverted fluorescence microscope with a standard GFP filter set (Olympus IX-81). Tissues were electrically stimulated, and videos were acquired at 20 frames to measure tissue excitability and calcium flux as previously described (33). Calcium signals were analyzed from calcium imaging videos as previously described (33). Briefly, a custom Python script was developed to average the pixel intensities for each frame. This transient was then corrected for fluorescent decay. The SDRR, Tau, FWHM, FW90M, Contract 90, Contract 50, Relax 50, and the Relax 90 were calculated for every transient.
Cardiac brightfield imaging and analysis:
For force generation measurements, videos were acquired at 20 fps using a custom program to stimulate cardiac tissues as previously described and force generation was analyzed from brightfield videos (33). Briefly, a custom Python script was developed to track the motion of the pillar heads and to calculate the force by multiplying the displacement of the pillars with the coefficient determined from the force-displacement calibration curve generated for the pillars.
Cardiac histology:
Tissues were fixed in 4% paraformeldehyde, washed 3 times in 1X PBS, and either kept for whole-mount imaging in PBS or processed for histological sectioning with paraffin embedding (Columbia HICCC Pathology core). Whole mount imaging was performed with primary antibodies for anti-α-actinin antibody (Sigma; A7811), anti-cardiac troponin T (ThermoFisher; MS-295-P1), or anti-COL1A1 (Abcam; ab34710), and mounted in CoverWell™ Imaging Chambers (Grace BioLabs; 631021).
eBM flow cytometry:
The extent of γ-H2AX in irradiated cells from eBM was characterized using the Amnis ImageStream Mk II cytometer to characterize the intensity and number of γ-H2AX foci per cell. 1-hour post-exposure to radiation, cells were fixed with BD Cytofix/Cytoperm according to manufacturer’s instructions and stored at 4°C. When staining, cells were incubated with PE anti-human H2AX antibody (Milteyni), FITC anti-human CD45 (BioLegend), Alexa Fluor 647 anti-human CD34 (BioLegend), and Hoechst 33342 (ThermoFisher) at room temperature for 1 hour and then washed with 1X PBS three times. Images of 1,000–1,500 cells per group were acquired at 60X with extended depth of field (EDF) for clearer visualization and identification of the fluorescent nuclei foci. Compensation coefficients between wavelengths of light were calculated using the compensation wizard in the Image Data Exploration and Analysis Software (IDEAS). After imaging, the spot wizard in IDEAS was used to quantify total foci per cell, total and mean fluorescence intensity each focus, area of foci, and area of nucleus.
eBM flow cytometry: other analyses of cells from the eBM tissues.
Cells were collected in FACS Buffer (2% FBS, 0.5 mM EDTA in PBS), washed, blocked with human Fc blocker solution (Milteyni; 130–059-901), and prepared for flow cytometry with the following antibodies: BV421 anti-human CD45 (Biolegend; 368522), APC anti-human CD34 (BioLegend; 343608), BV605 anti-human CD38 (BioLegend; 356642), BUV395 anti-human CD90 (BD Biosciences), PE anti-human CD45RA (BioLegend; 304108), propidium iodide (Invitrogen/ThermoFisher; P3566), CD14 (BV605), APC anti-human CD15 (BioLegend; 125617), BUV395 CD11c (BD Biosciences; 563787), PE anti-human CD16 (BioLegend; 360704), and BV421 anti-human CD11b (BioLegend; 301324). Flow cytometry was performed on a BioRad ZE5 machine and analyzed with FlowJo (BD Biosciences).
Colony forming unit assay:
Cells were collected from the suspension compartment of eBM, counted, and plated in MethoCult™ SF H4636 (Stem Cell Technologies) in SmartDish™ 6-well, meniscus-free plates (Stem Cell Technologies) and placed within humidified chamber dishes (245 mm x 245 mm Square Dishes; Stem Cell Technologies; 27141). Plates were imaged on Day 14 using a BioTek Cytation 5 automated live-cell fluorescence imaging system with BioSpa automatic, incubated chamber (Columbia HICCC Confocal and Specialized Microscopy Core). Colonies were blindly and manually analyzed from images.
Histology:
eBM tissues were fixed in 4% paraformaldehyde overnight at 4°C, washed three times with 1X PBS, decalcified with Osteosoft (EMD Millipore; 101728) overnight at room temperature (only bone marrow tissues), washed three times with 1X PBS, and paraffin-embedded for histological sectioning. All tissues were processed for hematoxylin and eosin (H&E), trichrome, or pentachrome by the Columbia University HICCC Molecular Pathology Lab. Paraffin-embedded tissue blanks were hydrated, processed for antigen-retrieval using a 10 mM sodium citrate buffer for 20 min in heat, and permeabilized with 0.25% (v/v) Triton-X for 20 minutes. Samples were then blocked for 2 hours with 10% FBS, and individual staining protocols followed for different tissues. Tissues were stained for immunofluorescence with Rabbit anti-human Bone sialoprotein antibody (ThermoFisher; PA5–79424), anti-Human/Mouse CXCL12/SDF-1 antibody (R&D Systems; MAB350), and Rabbit anti-human CD45 antibody (Sigma Aldrich; SAB4502541). After washing with PBS, samples were incubated with fluorophore-conjugated secondary antibodies (Invitrogen) for 2 hours at room temperature. Slides were covered with cover-slips using ProLong™ Diamond Antifade Mountant with 4’,6-diamidino-2-phenylindole (DAPI) (Thermofisher; P36962). Cells in suspension were cytospun onto glass slides using a Shandon Cytospin cytocentrifuge, fixed with methanol, and stained using a Wright-Giemsa Stain Kit (Abcam; ab245888). Histological stains were imaged using an Olympus Upright Microscope / Slide Scanner (Olympus; BX61V5F) and immunofluorescent images were taken with a Nikon Ti Eclipse inverted microscope at the Columbia HICCC Confocal and Specialized Imaging Core.
eCT bulk transcriptomics:
Cardiac microtissues were snap frozen and processed by Genewiz/Azenta Bio for RNA extraction, sequencing, and analysis. Briefly, RNA was processed using RNA depletion library preparation, and sequenced using the Illumina HiSeq 2×150 bp sequencing index. Tissues of poor RIN value (<6) and below usable concentration were not included in analysis (n = 1 control; n = 3 photon; n = 3 neutron). Differentially expressed genes were analyzed using DESeq2 (73), and processed with g:Profiler, Revigo, or Cytoscape with top differentially expressed genes (padj<0.05; log2FC>1) for gene ontology analysis. The full list of differentially expressed genes and ontology pathways is available in Supplementary Tables 1, 2, and 4.
Single cell transcriptomic analysis:
Cell preparation:
Cells were isolated from eBM cultures 21 days post-radiation exposure by collecting the suspension fraction in FACS buffer and counted to ensure viability of >90%. Two technical replicates per experimental group were then mixed at equal amounts at a concentration of 1000 cells per μL. Cells were super-loaded onto the Chromium Next GEM Chip G (PN-1000120) targeting 15,000 cells. Libraries were prepared using the following reagents from the Chromium Single Cell 3’ Reagent Kit (v3.1): Single Cell 3’ Library & Gel Bead Kit v3.1 (PN-1000121), Chromium Next GEM Chip G Single Cell Kit (PN-1000120) and Single Index Kit T Set A (PN-1000213) (10x Genomics). The Chromium Next GM Single Cell 3’ Reagent Kits v3.1 User Guide with Feature Barcoding technology for Cell Surface Protein (CG000206 Rev D) was followed for GEM generation, cDNA amplification and library construction. Libraries were run on either an Illumina HiSeq 4000 as 150-bp paired-end reads at a sequencing depth of at least 20,000 read pairs per cell for the 3’ Gene Expression library and 5,000 read pairs per cell for the Cell Surface Protein library.
Single cell transcriptomic analysis:
Preprocessing and clustering:
CellRanger was applied on the raw sequencing data to generate unique molecular identifier (UMI) matrix. The UMI matrix was then imported into Scanpy. Uniform Manifold Approximation and Projection (UMAP) for dimensionality reduction was performed with Leiden algorithm-based clustering prior to any cell filtering to first identify granulocytes, which require a separate preprocessing method as they have relatively lower RNA content. For all cell types, cells expressing less than 200 genes and expressed in less than 3 cells were excluded from analysis. Mitochondrial and ribosomal DNA was excluded. Cells with over 20% mitochondrial content, indicated by the fraction of mitochondrial transcripts over the total transcript counts, were removed from further analysis. Cells were filtered out for low library size of 500 for granulocytes and 1000 for all other cell types. 15,000 most highly variable genes were identified for further analysis. Principal component analysis (PCA) was performed with 50 components, and nearest neighbors were identified with 12 nearest neighbors. UMAP was generated and Leiden clustering was applied on the filtered dataset to visualize and identify specific cell types. To annotate each cluster, previous studies were referenced to identify marker genes expressed (37, 38). Differential gene expression analyses were done using the Python package Scanpy. Wilcoxon’s t-test was applied to rank differential genes. P-values and log fold changes were exported and used for gene ontology analyses with either g:Profiler, Revigo, or Cytoscape using top differentially expressed genes (p<0.05; log2FC>1), and reported in the Figures and presented in Supplemental Tables 3 and 4.
Single cell transcriptomic analysis:
CellChat:
CellChat was used for inferring cell-cell communications through analyzing the ligand and receptor signaling pairs from the single cell transcriptomics (40). Based on mass action models, social network analysis, pattern recognition methods and manifold learning, CellChat is able to identify the specific signaling roles and determine the intercellular communication among different cell populations and sample groups. After annotating the cell identities for each group in our datasets, we constructed the CellChat objects for 0Gy, 4Gy photon, and 1Gy neutron respectively. And three types of communication including ECM-receptor, secreted signaling, and cell-cell contacts were analyzed separately and compared among different radiation affected groups.
Statistics:
All cardiac imaging functional data, bone marrow flow cytometry, cytokine, and individual cardiac gene expression data were plotted using GraphPad Prism. All statistical analyses for these were performed using GraphPad Prism with One Way or Two Way ANOVA with Multiple Comparisons, as indicated in each Figure. Cardiac Bulk RNA sequencing differential gene expression analysis was analyzed with Wald Chi-Squared tests for significance. Single-cell RNA sequencing analysis from bone marrow cultures were analyzed using Scanpy’s Rank Genes function using t-tests with Bonferroni corrections.
Supplementary Material
Acknowledgements:
We thank the staff at Columbia’s Center for Radiological Research (Mikhail Repin) for assistance in running ionizing radiation equipment. We also thank Keith Yeager and Max Summers for assistance in fabricating the pillar platforms and scaffolds. These studies used the resources of the Herbert Irving Comprehensive Cancer Center Confocal and Specialized Microscopy Shared Resource funded in part through Center Grant P30CA013696. Research reported in this publication using the ImageStreamX MkII imaging cytometer was performed in the Columbia University Stem Cell Initiative Flow Cytometry core facility at Columbia University Irving Medical Center, under the direction of Michael Kissner and Rose Gordon-Schneider, and was supported by the Office of the Director, National Institutes of Health under Award Number S10OD026845.
Funding:
We gratefully acknowledge the funding support of this work by the National Institutes of Health (EB027062, HL076485, HL120046, CA249799 to G.V.-N.; U19-AI067773, 1S10OD025190 to D.J.B.; K99 CA267168 to J.A.B.; NRSA 1F31CA275733 to D.N.T.), the National Science Foundation (1644869 to D.N.T. and ERC16478 to G.V.-N.), the Empire State Development Corporation (AC710 to D.J.B), the Translational Research in Space Health (TRISH/NASA) agency for their support (RAD0104 and NNX16A069A to G.V.-N., D.J.B., K.L.), and the Columbia SEAS-HICCC Pilot Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: The authors do not have any personal or financial relationships that would constitute conflict of interest.
Data availability:
Any additional data will be made available on request.
References:
- 1.Patel ZS et al. , Red risks for a journey to the red planet: The highest priority human health risks for a mission to Mars. NPJ Microgravity 6, 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong TW, Colborn BL, Predictions of secondary neutrons and their importance to radiation effects inside the international space station. Radiation Measurements 33, 229–234 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Furukawa S et al. , Space Radiation Biology for “Living in Space”. BioMed Research International 2020, 4703286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughson RL, Helm A, Durante M, Heart in space: effect of the extraterrestrial environment on the cardiovascular system. Nature Reviews Cardiology 15, 167–180 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Dörr H, Meineke V, Acute radiation syndrome caused by accidental radiation exposure - therapeutic principles. BMC Medicine 9, 126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonsen LC, Slaba TC, Guida P, Rusek A, NASA’s first ground-based Galactic Cosmic Ray Simulator: Enabling a new era in space radiobiology research. PLOS Biology 18, e3000669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassler DM et al. , Mars’ Surface Radiation Environment Measured with the Mars Science Laboratory’s Curiosity Rover. Science 343, 1244797 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Sridharan DM et al. , Understanding cancer development processes after HZE-particle exposure: roles of ROS, DNA damage repair and inflammation. Radiat Res 183, 1–26 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Preston DL et al. , Cancer incidence in atomic bomb survivors. Part III. Leukemia, lymphoma and multiple myeloma, 1950–1987. Radiat Res 137, S68–97 (1994). [PubMed] [Google Scholar]
- 10.Belzile-Dugas E, Eisenberg MJ, Radiation-Induced Cardiovascular Disease: Review of an Underrecognized Pathology . Journal of the American Heart Association 10, e021686 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilbronn L et al. , Production of neutrons from interactions of GCR-like particles. Acta Astronaut 42, 363–373 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Litvak ML et al. , Observations of neutron radiation environment during Odyssey cruise to Mars. Life Sciences in Space Research 29, 53–62 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Laiakis EC et al. , Serum lipidomic analysis from mixed neutron/X-ray radiation fields reveals a hyperlipidemic and pro-inflammatory phenotype. Sci Rep 9, 4539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Upton AC et al. , Late Effects of Fast Neutrons and Gamma-Rays in Mice as Influenced by the Dose Rate of Irradiation: Induction of Neoplasia. Radiation Research 41, 467–491 (1970). [PubMed] [Google Scholar]
- 15.Broustas CG, Harken AD, Garty G, Amundson SA, Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation. BMC Genomics 19, 504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broustas CG et al. , Impact of Neutron Exposure on Global Gene Expression in a Human Peripheral Blood Model. Radiation Research 187, 443–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ossetrova NI et al. , Biomarkers for Radiation Biodosimetry and Injury Assessment after Mixed-field (Neutron and Gamma) Radiation in the Mouse Total-body Irradiation Model. Health Physics 115, 727–742 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Tavakol DN, Fleischer S, Vunjak-Novakovic G, Harnessing organs-on-a-chip to model tissue regeneration. Cell Stem Cell 28, 993–1015 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronaldson-Bouchard JDTK, Yeager K, Tavakol DN, Zhao Y, Chramiec A, Tagore S, Summers M, Stylianos S, Tamargo M, Lee BM, Halligan SP, Abaci EH, Guo Z, Jacków J, Pappalardo A, Shih J, Soni RK, Sonar S, German C, Christiano AM, Califano A, Hirschi KK, Chen CS, Przekwas A, Vunjak-Novakovic G, InterOrgan platform with tissue-specific niches linked by vascular flow. Nature Biomedical Engineering in press, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingber DE, Human organs-on-chips for disease modelling, drug development and personalized medicine. Nature Reviews Genetics 23, 467–491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavakol DN et al. , Injectable, scalable 3D tissue-engineered model of marrow hematopoiesis. Biomaterials 232, 119665 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Sharma A, Sances S, Workman MJ, Svendsen CN, Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 26, 309–329 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vunjak-Novakovic G, Ronaldson-Bouchard K, Radisic M, Organs-on-a-chip models for biological research. Cell 184, 4597–4611 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wnorowski A et al. , Effects of Spaceflight on Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Structure and Function. Stem Cell Reports 13, 960–969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma SD et al. , Astrocytes regulate vascular endothelial responses to simulated deep space radiation in a human organ-on-a-chip model. Frontiers in Immunology 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou DB et al. , On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nature Biomedical Engineering 4, 394–406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jalili-Firoozinezhad S et al. , Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis 9, 223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glaser DE et al. , Organ-on-a-chip model of vascularized human bone marrow niches. Biomaterials 280, 121245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson MR et al. , A multi-niche microvascularized human bone marrow (hBM) on-a-chip elucidates key roles of the endosteal niche in hBM physiology. Biomaterials 270, 120683 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Xu Y et al. , Novel neutron sources at the Radiological Research Accelerator Facility. Journal of Instrumentation 7, C03031–C03031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y et al. , Accelerator-Based Biological Irradiation Facility Simulating Neutron Exposure from an Improvised Nuclear Device. Radiat Res 184, 404–410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q et al. , Cytogenetic Damage of Human Lymphocytes in Humanized Mice Exposed to Neutrons and X Rays 24 h After Exposure. Cytogenet Genome Res 161, 352–361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamargo MA et al. , milliPillar: A Platform for the Generation and Real-Time Assessment of Human Engineered Cardiac Tissues. ACS Biomaterials Science & Engineering 7, 5215–5229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronaldson-Bouchard K et al. , Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villasante A et al. , Tissue-Engineered Model of Human Osteolytic Bone Tumor. Tissue Eng Part C Methods 23, 98–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y, Wang Q, Shuryak I, Brenner DJ, Turner HC, Development of a high-throughput γ-H2AX assay based on imaging flow cytometry. Radiat Oncol 14, 150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N, The Human Cell Atlas bone marrow single-cell interactive web portal. Experimental Hematology 68, 51–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellin D et al. , A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nature Communications 10, 2395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tikhonova AN et al. , The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin S et al. , Inference and analysis of cell-cell communication using CellChat. Nature Communications 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wasserman TH, Brizel DM, The role of amifostine as a radioprotector. Oncology (Williston Park) 15, 1349–1354; discussion 1357–1360 (2001). [PubMed] [Google Scholar]
- 42.Neta R, Oppenheim JJ, Douches SD, Interdependence of the radioprotective effects of human recombinant interleukin 1 alpha, tumor necrosis factor alpha, granulocyte colony-stimulating factor, and murine recombinant granulocyte-macrophage colony-stimulating factor. The Journal of Immunology 140, 108–111 (1988). [PubMed] [Google Scholar]
- 43.Kruse JJ, Strootman EG, Wondergem J, Effects of amifostine on radiation-induced cardiac damage. Acta Oncol 42, 4–9 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Dorr RT, Lagel K, McLean S, Cardioprotection of rat heart myocytes with amifostine (Ethyol) and its free thiol, WR-1065, in vitro. Eur J Cancer 32A Suppl 4, S21–25 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Hedblom A et al. , Heme detoxification by heme oxygenase-1 reinstates proliferative and immune balances upon genotoxic tissue injury. Cell Death & Disease 10, 72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S et al. , First measurements of the radiation dose on the lunar surface. Science Advances 6, eaaz1334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Modeling Hematopoiesis and Responses to Radiation Countermeasures in a Bone Marrow-on-a-Chip. Tissue Engineering Part C: Methods 22, 509–515 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Hinson JT et al. , Integrative Analysis of PRKAG2 Cardiomyopathy iPS and Microtissue Models Identifies AMPK as a Regulator of Metabolism, Survival, and Fibrosis. Cell Rep 17, 3292–3304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun XL et al. , LncRNA NEAT1 promotes cardiac hypertrophy through microRNA-19a-3p/SMYD2 axis. Eur Rev Med Pharmacol Sci 24, 1367–1377 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Vigil-Garcia M et al. , Gene expression profiling of hypertrophic cardiomyocytes identifies new players in pathological remodelling. Cardiovascular Research 117, 1532–1545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayer A, Zarjou A, Agarwal A, Stocker R, Heme Oxygenases in Cardiovascular Health and Disease. Physiol Rev 96, 1449–1508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker BV et al. , Impact of Ionizing Radiation on Electrophysiological Behavior of Human-induced Ipsc-derived Cardiomyocytes on Multielectrode Arrays. Health Physics 115, 21–28 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Sárközy M et al. , Selective Heart Irradiation Induces Cardiac Overexpression of the Pro-hypertrophic miR-212. Front Oncol 9, 598 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimizu I, Minamino T, Physiological and pathological cardiac hypertrophy. Journal of Molecular and Cellular Cardiology 97, 245–262 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Stewart FA, Hoving S, Russell NS, Vascular Damage as an Underlying Mechanism of Cardiac and Cerebral Toxicity in Irradiated Cancer Patients. Radiation Research 174, 865–869 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Azimzadeh O et al. , Integrative Proteomics and Targeted Transcriptomics Analyses in Cardiac Endothelial Cells Unravel Mechanisms of Long-Term Radiation-Induced Vascular Dysfunction. Journal of Proteome Research 14, 1203–1219 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Yamaguchi M, Kashiwakura I, Role of Reactive Oxygen Species in the Radiation Response of Human Hematopoietic Stem/Progenitor Cells. PLOS ONE 8, e70503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandersickel V et al. , Induction and disappearance of γH2AX foci and formation of micronuclei after exposure of human lymphocytes to 60Co γ-rays and p(66)+ Be(40) neutrons. International Journal of Radiation Biology 90, 149–158 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Godley LA, Larson RA, Therapy-related myeloid leukemia. Semin Oncol 35, 418–429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu WL et al. , The incidence of leukemia, lymphoma and multiple myeloma among atomic bomb survivors: 1950–2001. Radiat Res 179, 361–382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zalusky R, Ghidoni JJ, McKinley J, Leffingwell TP, Melville GS, Leukemia in the Rhesus Monkey (Macaca mulatta) Exposed to Whole-Body Neutron Irradiation. Radiation Research 25, 410–416 (1965). [PubMed] [Google Scholar]
- 62.Rodman C et al. , In vitro and in vivo assessment of direct effects of simulated solar and galactic cosmic radiation on human hematopoietic stem/progenitor cells. Leukemia 31, 1398–1407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mencia-Trinchant N et al. , Clonal Hematopoiesis Before, During, and After Human Spaceflight. Cell Rep 33, 108458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vral MA. Cornelissen H. Thierens H. Louagie J. Philippe K. Strijckmans L. Ridder De, Apoptosis induced by fast neutrons versus 60Co gamma -rays in human peripheral blood lymphocytes. International Journal of Radiation Biology 73, 289–295 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Garrett-Bakelman FE et al. , The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 364, eaau8650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lacombe J, Soldevila M, Zenhausern F, in Progress in Molecular Biology and Translational Science, Pandya A, Singh V, Eds. (Academic Press, 2022), vol. 187, pp. 41–91. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y et al. , Accelerator-Based Biological Irradiation Facility Simulating Neutron Exposure from an Improvised Nuclear Device. Radiat Res 184, 404–410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y et al. , Broad Energy Range Neutron Spectroscopy using a Liquid Scintillator and a Proportional Counter: Application to a Neutron Spectrum Similar to that from an Improvised Nuclear Device. Nuclear Instruments & Methods in Physics Research Section A-Accelerators Spectrometers Detectors and Associated Equipment 794, 234–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossi HH, Bateman JL, Bond VP, Goodman LJ, Stickley EE, The dependence of RBE on the energy of fast neutrons: 1. Physical design and measurement of absorbed dose. Radiation Research 13, 503–520 (1960). [PubMed] [Google Scholar]
- 70.Wagner EB, Hurst GS, A Geiger-Mueller γ-ray dosimeter with low neutron sensitivity. Health Phys. 5, 20–26 (1961). [DOI] [PubMed] [Google Scholar]
- 71.Chramiec A et al. , Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip 20, 4357–4372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mandegar Mohammad A. et al. , CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional data will be made available on request.


