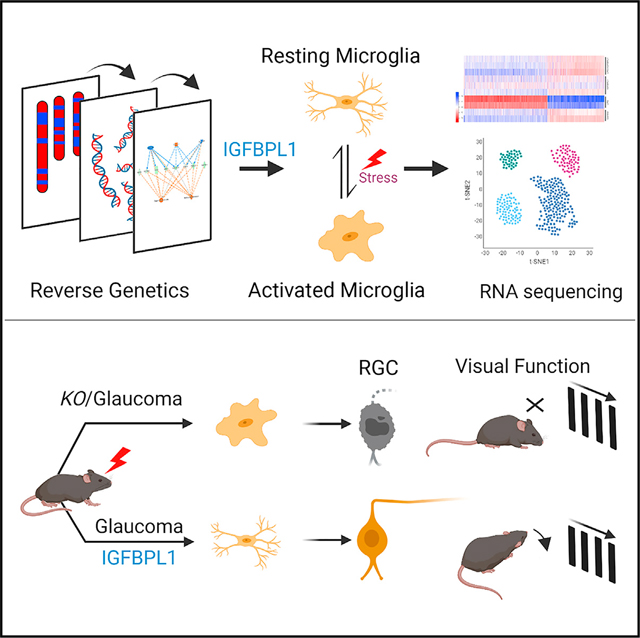
SUMMARY
Microglia shift toward an inflammatory phenotype during aging that is thought to exacerbate age-related neurodegeneration. The molecular and cellular signals that resolve neuroinflammation post-injury are largely undefined. Here, we exploit systems genetics methods based on the extended BXD murine reference family and identify IGFBPL1 as an upstream cis-regulator of microglia-specific genes to switch off inflammation. IGFBPL1 is expressed by mouse and human microglia, and higher levels of its expression resolve lipopolysaccharide-induced neuroinflammation by resetting the transcriptome signature back to a homeostatic state via IGF1R signaling. Conversely, IGFBPL1 deficiency or selective deletion of IGF1R in microglia shifts these cells to an inflammatory landscape and induces early manifestation of brain tauopathy and retinal neurodegeneration. Therapeutic administration of IGFBPL1 drives pro-homeostatic microglia and prevents glaucomatous neurodegeneration and vision loss in mice. These results identify IGFBPL1 as a master driver of the counter-inflammatory microglial modulator that presents an endogenous resolution of neuroinflammation to prevent neurodegeneration in eye and brain.
In brief
In the present study, Li et al. identify an immune regulator, IGFBPL1, that plays a pivotal role as a master switch, turning inflammatory microglia to their homeostatic state to prevent excessive neuroinflammation. IGFBPL1 presents potent and lasting therapeutic effects of neuroprotection, delivering functional benefits in the treatment for neurodegenerative disorders.
Graphical Abstract
INTRODUCTION
The brain and retina undergo inflammatory transition after injures and aging, but the molecular and cellular signals responsible for resolving neuroinflammation are largely undefined. Over the past two decades, microglia have emerged as key contributors to CNS aging and the neurodegenerative disorders, including Alzheimer’s disease (AD) and glaucoma.1,2 Homeostatic microglial activity has housekeeping functions, including synaptic clearance and pathogen surveillance.3–5 Uncontrolled or sustained microglial activation can damage and kill neurons.6,7 Microglial hyperreactivity is regarded as a strong contributor to some facets of age-related neurodegenerative diseases. While triggering receptor expressed on myeloid cells 2 (Trem2) and Cx3cr1 presented the early sensors that mediate the induction phases of microglial activation,8 the work we describe here defines a molecular target that is responsible for terminating pernicious long-term inflammatory hypervigilance.
Glaucoma is the leading cause of irreversible blindness and is the most frequent age-related neurodegenerative disease.9 Despite the prevalence and considerable morbidity, currently, therapeutic options are limited to controlling the intraocular pressure.10 Glaucoma features an immunological component comparable to other neurodegenerative diseases11–13 and an early event of microglial activation14,15 that are shown to initiate a T cell-mediated response and drive neural damage in glaucoma.16–18 However, molecular controls for such microglial initiated events, particularly those that can terminate the process and resolve neuroinflammation, remain unclear.
Systems genetics approaches present powerful tools for the identification of targeted molecules through exploring complex genetic networks. Methods now commonly exploit entire global multiomics datasets that enable efficient screening and causal analysis of candidate genes and networks.19–21 In our work, we leveraged the extended BXD family that consists of about 150 fully sequenced mouse strains descended from crosses between C57BL/6J (B6) and the glaucoma-prone DBA/2J mice (D2).22,23 This single family segregates for over 6 million DNA variants with near-optimal minor allele frequencies of 0.4.20 Importantly, almost all D2 mice and many BXD strains develop glaucoma by 12 months of age.24,25 Since the D alleles that modulate glaucomatous phenotypes segregate among BXD family members, the family as a whole is ideal for modeling genetic variation of age-related glaucomatous neurodegeneration and identifying the potential molecular players.25–29
This systems genetics approach led to our predication and identification of the predominant involvement of insulin-like growth factor-binding protein like protein 1 (IGFBPL1) in neurodegeneration and neuroinflammatory modulation. We discovered that IGFBPL1, as a member of the IGFBP superfamily, is a Trem2-related master regulator of microglial homeostasis. These findings define an endogenous mechanism of neuroinflammation resolution and termination of reactive microglial responses that suggests new therapeutic interventions for glaucoma and age-related neurodegenerative disorders.
RESULTS
Systems genetics approach identifies IGFBPL1 as an immune and microglial regulator
To define the retinal function and phenotypic effects of IGFBPL1—a member of the IGFBP superfamily—we took the approach of systems genetics by correlating Igfbpl1 expression with the genome-wide transcriptome data derived from the retinas of 55 BXD strains (Figure 1A).26,30 The expression of Igfbpl1 varies to a significant degree among the 55 BXD strains (Figure S1A). Gene correlation analysis identified 6,855 transcripts that co-varied with Igfbpl1 expression in the retina (p < 0.05). Mammalian phenotype ontology (MPO) analysis associated these co-varied genes with 263 phenotypes; five of the top seven most significant MPOs were classified to immune-related or inflammatory responses (Figure 1B), strongly pointing toward IGFBPL1’s role as an immune regulator of the adult retina. Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis highlighted cytokine-cytokine and neuroactive ligand-receptor interactions as the top two signaling events associated with Igfbpl1 expression (false discovery rate [FDR] < 0.01) (Figure 1C), supporting its involvement in neuroinflammation and neuron-immune modulation. In line with the previous report,31 expression of Igfbpl1 is strongly associated with calcium signaling and PI3K-Akt pathways (Figure 1C).
Figure 1. Systems genetics approach identifies IGFBPL1 as an immune and microglial regulator in the mouse retina.
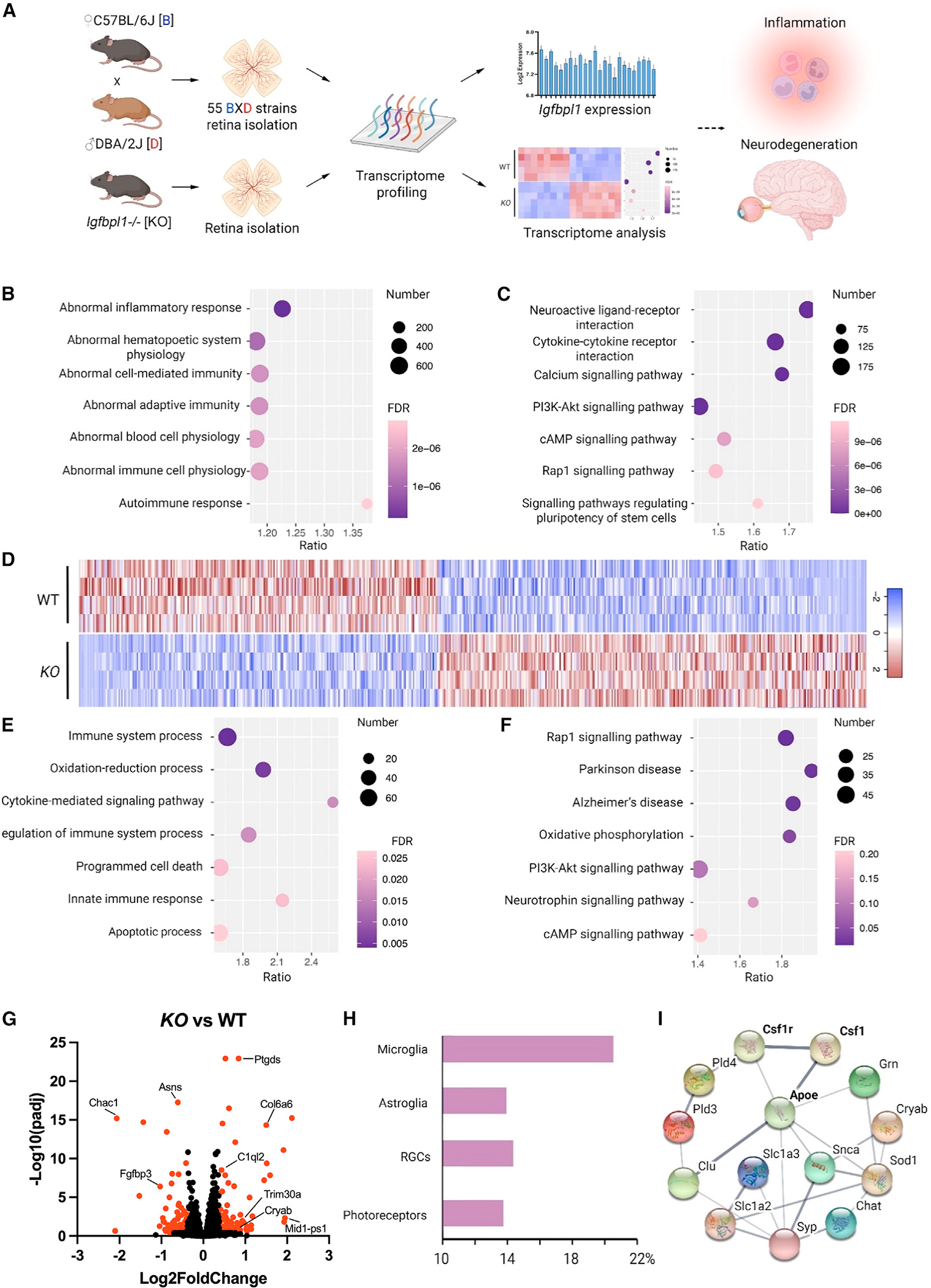
(A) Schematic of the reverse genetics approach that predicts the functions of Igfbpl1 based on genome-wide transcriptome profiles of BXD and KO mouse retinas.
(B and C) Dot plots of top 7 most significant MPO (B) and enriched KEGG (C) terms and projected based on expression covaried with IGFBPL1 in the BXD mouse retinas.
(D) Heatmap of DEGs between KO and WT mouse retinas.
(E and F) Dotplots of top 7 most relevant MPO (E) and enriched KEGG (F) terms projected using DEGs of KO mice compared with WT mice.
(G) Volcano plot showing DEGs of KO mice compared with WT mice. Red dots: FDR <0.05 and fold change (FC) >1.3.
(H) Percentages of DEGs clustered to cell-type-specific genes.
(I) Protein-protein interaction network of top 14 microglial associated DEGs visualized in String.
See also Figure S1.
To validate Igfbpl1-associated gene pathways predicted above, we examined transcriptome profiles of Igfbpl1−/− (knockout [KO]) mouse retinas by bulk RNA sequencing (RNA-seq) and identified 2,305 differentially expressed genes (DEGs) between adult KO and age- and sex-matched wild-type (WT) mice (p < 0.05) (Figure 1D). Consistent with the analysis drawn above, MPO and enriched KEGG analyses revealed strong associations between Igfbpl1 expression and immune system processing, cytokine-mediated pathways, and PI3K-Akt signaling (Figures 1E–1G). We next grouped the DEGs based on their functional relevance to different retinal cell types, including microglia, astroglia (astrocytes and Müller glia), retinal ganglion cells (RGCs), and photoreceptors. In agreement with its association with retinal immune regulation, the analysis unveiled a far greater number of microglia-related genes among DEGs of the KO retinas than of genes associated with any other cell types (Figure 1H). String analysis of protein-protein interaction (PPI) networks connected microglial-specific genes, such as colony-stimulating factor 1 (Csf1), CSF1 receptor (Csf1r), and Apoe, to DEGs in the KO mouse retinas (Figure 1I). Search of a public database of single-cell RNA-seq (scRNA-seq) from mouse and human retinas further supports the expression of Igfbpl1 in microglia, as well as in neurons and other cell types.32 These analyses predicted a regulatory role of IGFBPL1 in microglial function and neuroinflammation in the retina.
To test the association of IGFBPL1 with microglial functionality, GFP+ microglia in the retinas of adult Cx3cr1GFP mice33 were immunolabeled with anti-CD11b antibody and sorted by Miltenyi Tyto sorter (fluorescence-activated cell sorting [FACS]). Results of quantitative real-time PCR showed significantly higher levels of Igfbpl1 mRNAs in microglia that were CD11b-positive (CD11b+) than CD11b-negative retinal cell fractions (Figure S1B). To investigate if the expression of IGFBPL1 changes after injury or in neurodegenerative conditions, we induced glaucoma by injecting microbeads (MBs) into the anterior chamber of the mouse eye (Figure S1C).34 At 4 weeks, but not at 2 weeks, post-MB injection, the levels of Igfbpl1 were upregulated in the microglia of glaucomatous retinas (Figure S1D). We speculated that IGFBPL1 may have a functional role in a later stage of insults or neuroinflammation. We next labeled the retinal microglia using a traditional prototypical marker for microglia/macrophage, ionized calcium-binding adaptor molecule 1 (IBA-1),35,36 and confirmed the co-localization of IGFBPL1 and IBA-1 in both mouse and human retinas (Figures 2A and 2B). Four weeks after MB injection, 49.3% IBA-1+ microglia in the retinas of glaucomatous mice expressed IGFBPL1, while IBA-1+/IGFBPL1+ cells were barely detectable in normal mouse retinas (Figure 2A). Moreover, aged healthy human retina displayed ~33.3% IBA-1+ IGFBPL1+ cells, and this number increased to 94.4% in that of a donor with glaucoma (Figure 2B). We confirmed microglial expression of IGF1R, IGFBPL1’s signaling receptor,31,37 using quantitative real-time PCR and double immunolabeling of IGF1R and CD11b (Figures S1E and S1F). These results support a plausible role of IGFBPL1 in microglial regulation, particularly after the induction of neuroinflammation.
Figure 2. IGFBPL1 resets the transcriptional signature of inflammatory microglia to a homeostatic state.
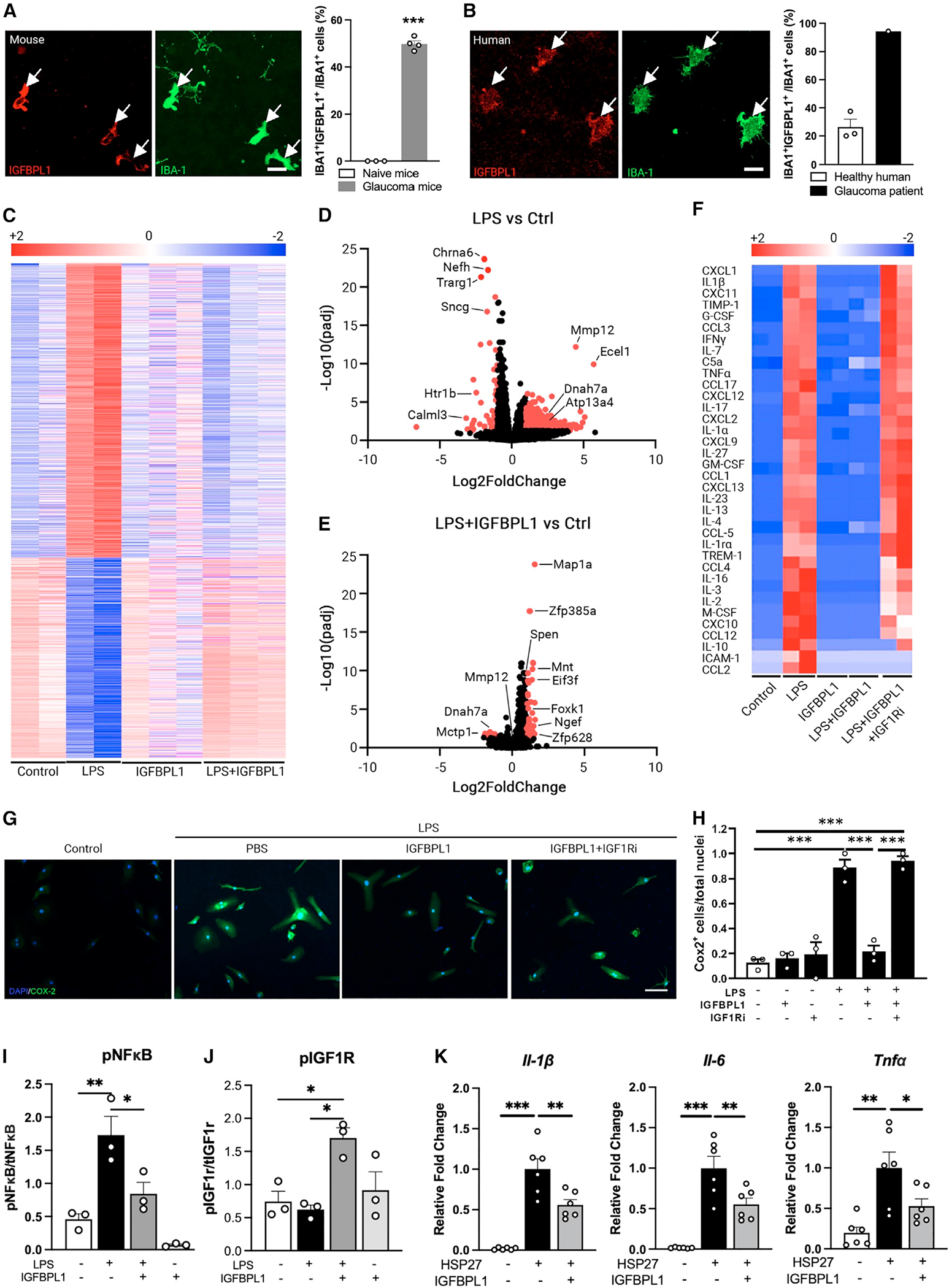
(A and B) Photomicrographs of mouse (A) and human (B) retinal flat mounts double immunolabeled for IGFBPL1 (red) and IBA-1 (green) and percentage of IGFBPL1-expressing microglia. Arrows indicate IGFBPL1+/IBA-1+ cells. Scale bar, 20 μm. ***p < 0.001; Student’s t test. n = 3–4 mice/group, 3 healthy donors and 1 glaucoma donor.
(C) Heatmap of DEGs of cultured retinal microglia treated with control, LPS, IGFBPL1, and LPS+IGFBPL1.
(D and E) Volcano plot showing DEGs of microglia treated with LPS (D) or LPS+IGFBPL1 vs. control (E). Red dots represent DEGs with padj <0.05 and |log2[FC]|>1.
(F) Heatmap of cytokine levels in microglial cultures treated with control (control), LPS, IGFBPL1, LPS+IGFBPL1, and LPS+IGFBPL1+IGF1Ri assessed by cytokine arrays.
(G and H) Images (G) and COX2+ cell counts (H) in microglial cultures treated with control or LPS and PBS, IGFBPL1, or IGFBPL1+IGF1Ri. Cultures were counterstained with DAPI. Scale bar, 20 μm. ***p < 0.001; one-way ANOVA with Dunnett’s multiple comparisons test. n = 3/group. Data are mean ± SEM.
(I and J) Quantifications of pNF-κB (I) and pIGF1R (J) in mouse microglial cultures treated with PBS, LPS, IGFBPL1, or LPS+IGFBPL1. *p < 0.05, **p < 0.01; one-way ANOVA with Dunnett’s multiple comparisons test. n = 3/group.
(K) Relative mRNA levels of pro-inflammatory cytokines in HSP27-primed microglial cultures treated with or without IGFBPL1. *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA with Dunnett’s multiple comparisons test. n = 6 replicates/group. Data are mean ± SEM.
See also Figure S2.
IGFBPL1 resets the transcriptional signature of pro-inflammatory microglia to homeostasis
Microglia exist in resting and activated states depending on the inflammatory milieu. Lipopolysaccharides (LPS) is a known potent trigger of inflammation, causing the resting microglia to lose homeostatic molecular signature and upregulate pro-inflammatory cytokines and cyclooxygenase 2 (COX2).38 To comprehensively investigate the roles of IGFBPL1, we performed bulk RNA-seq in IGFBPL1-treated normal and LPS-primed mouse retinal microglia. Cells from 6–8 retinas of adult Cx3cr1GFP mice were pooled, and microglia were sorted using the Miltenyi Tyto cell sorter under a low microfluidic pressure (150 mPa) that minimizes ex vivo activation of microglia. Purified microglia were incubated with vehicle, LPS, IGFBPL1 alone, or LPS treatment followed by IGFBPL1 6 h later, and cells were cultured for a total of 48 h. RNAs were isolated from at least 2 biological replicates per group; thus, RNAs representing microglia from at least 15 retinas per group were sequenced. Using the cutoff of adjusted p value [padj] <0.05, we detected 2,163 DEGs induced by LPS alone compared to vehicle controls (Figure 2C). These included enriched KEGG pathways related to oxidative stress (e.g., Atp13a4), immune/inflammation signaling (e.g., Mmp12, Mtcp1, and Ncf2), and AD/Parkinson’s disease age-related neurodegeneration (e.g., Dnah7a, Sncg, Ndufb8, and Prkn) (Figures 2C, 2D, and S2A). In the absence of LPS, to our surprise, IGFBPL1 alone induced barely any significant transcriptional changes (except Spen, a Notch signaling protein) in microglia compared to the vehicle control group. As Spen upregulation is suspected to mediate the phagocytotic function, we performed zymogen uptake assay, which detected no significant differences between the PBS- and IGFBPL1-alone-treatment groups (Figures S2B–S2D), suggesting a minimal effect of IGFBPL1 on resting/homeostatic microglia. In contrast, addition of IGFBPL1 6 h after LPS treatment reverted ~97% (2,100) of the LPS-induced DEGs, including Mmp12, Dnah7a, and Mctp1, to a normal level (Figure 2C). This led to only 65 DEGs in the LPS+IGFBPL1-treated group compared to naive cultures treated with PBS controls. These included Spen and genes related to DNA binding (e.g., Mnt and Zfp628) and protein metabolism (e.g., Eif3f and Foxk1) (Figure 2E), with no identifiable KEGG terms (FDR > 0.05). The results unveiled a remarkable effect of IGFBPL1 that selectively resets the molecular signatures of inflammatory microglia to their homeostatic state, suggesting a potent counter-inflammatory effect of IGFBPL1.
The anti-inflammatory effects of IGFBPL1 in microglia were corroborated by assays of cytokine arrays. In agreement with the RNA-seq data, treatment of IGFBPL1 alone did not induce significant changes in any of the 36 pro-inflammatory cytokines/chemokines examined compared to untreated cultures (Figure 2F). As LPS drastically upregulated the levels of the entire 36 cytokines/chemokines examined compared to the vehicle group when examined at 48 h of incubation, administration of IGFBPL1 to LPS-primed microglia 6 h later completely reverted the cytokine profile to the control level (Figure 2F). To determine if IGFBPL1 signals through IGF1R, an IGF1R antagonist derived from an IGF1 analog that competitively binds IGF1R (IGF1Ri) was added simultaneously with IGFBPL1 6 h after LPS administration. IGF1Ri largely abolished the activity of IGFBPL1: in the presence of IGF1Ri, IGFBPL1 failed to revert LPS-induced cytokine productions to the control levels, except for interleukin-10 (IL-10), ICAM-1, and CCL2 (Figure 2F). As LPS induced COX2 upregulation in cultured retinal microglia, the addition of IGFBPL1 6 h later reverted LPS-induced COX2 to the resting level at 24–48 h of incubation (Figures 2G and 2H). This effect of IGFBPL1 was abolished by the presence of IGF1Ri (Figures 2G and 2H). Similar inhibitory effects of IGFBPL1 were also observed in LPS-primed mouse brain microglia and the human HMC3 microglial cell line (Figures S2E and S2F), and IGF1Ri blocked its effect in all cultures (Figure S2E). LPS is reported to induce inflammation through Toll-like receptor (TLR)/nuclear factor κB (NF-κB) pathways, and IGF1R functions upstream in microglia to inhibit TLR4/NF-κB signaling.37 We noted that administration of IGFBPL1 to LPS-treated cultures alleviated LPS-induced NF-κB activation via IGF1R phosphorylation (Figures 2I, 2J, and S2G), whereas IGFBPL1 alone showed a minimal effect on NF-κB or IGF1R phosphorylation. To validate that IGFBPL1 works through TLR signaling, we tested the effects of IGFBPL1 in HSP27-primed mouse microglia39,40 and found that IGFBPL1 effectively alleviated the expression of pro-inflammatory cytokines induced by HSP27 stimulation, including Il-1β, Il-6, and Tnfα (Figure 2K). These data demonstrate a potent effect of IGFBPL1 that resets the molecular signatures of activated/inflammatory microglia to homeostasis via the IGF1R-NF-κB axis without interfering with the function of resting microglia in vitro.
IGFBPL1 drives microglial homeostasis in vivo
To study the effect of IGFBPL1 in vivo, we induced glaucoma in mice by anterior chamber injection of MB to elevate intraocular pressure (IOP) (Figure 3A). Retinal microglia responded to IOP elevation by becoming activated, displaying enlarged cell bodies and fewer ramifications (Figure 3B). We treated glaucomatous mice with IGFBPL1 or PBS (as controls) on days 3 and 10 post-MB injection. By day 14 post-MB administration, as expected, PBS-treated glaucomatous mice exhibited significant increases of CD68+IBA-1+ cells, showing larger cell bodies and shorter processes and expressing an activated microglia marker (CD68) compared with naive mice (Figures 3B–3D). Treatment of IGFBPL1 significantly reduced activated microglia compared with PBS-treated mice (Figures 3B–3D and S2H). Quantitative real-time PCR confirmed the suppression of activated microglial markers (Iba-1, Cox2, Cd163, and Nox2) (Figure 3E) and inflammatory cytokines (IL-1β, IL-1α, Cxcl10, C1q, and Tnfα) by IGFBPL1 compared to the PBS-treated group (Figure 3F). Upregulation of glial fibrillary acidic protein (GFAP) is a hallmark of reactive gliosis and a critical event downstream of microglial activation.41 Elevated IOP induced a significant increase in GFAP expression as shown by immunolabeling and western blots (Figures 3G, 3H, and S2I), whereas mice that received IGFBPL1 treatment maintained a steady level of GFAP similar to that in naive (day 0) mice throughout the 14 day period post-MB injection (Figures 3G and 3H). Thus, IGFBPL1 effectively suppresses microglial and astroglial activation and retinal neuroinflammation in glaucomatous mice in vivo.
Figure 3. IGFBPL1 suppresses retinal microglial activation in mice with glaucoma.
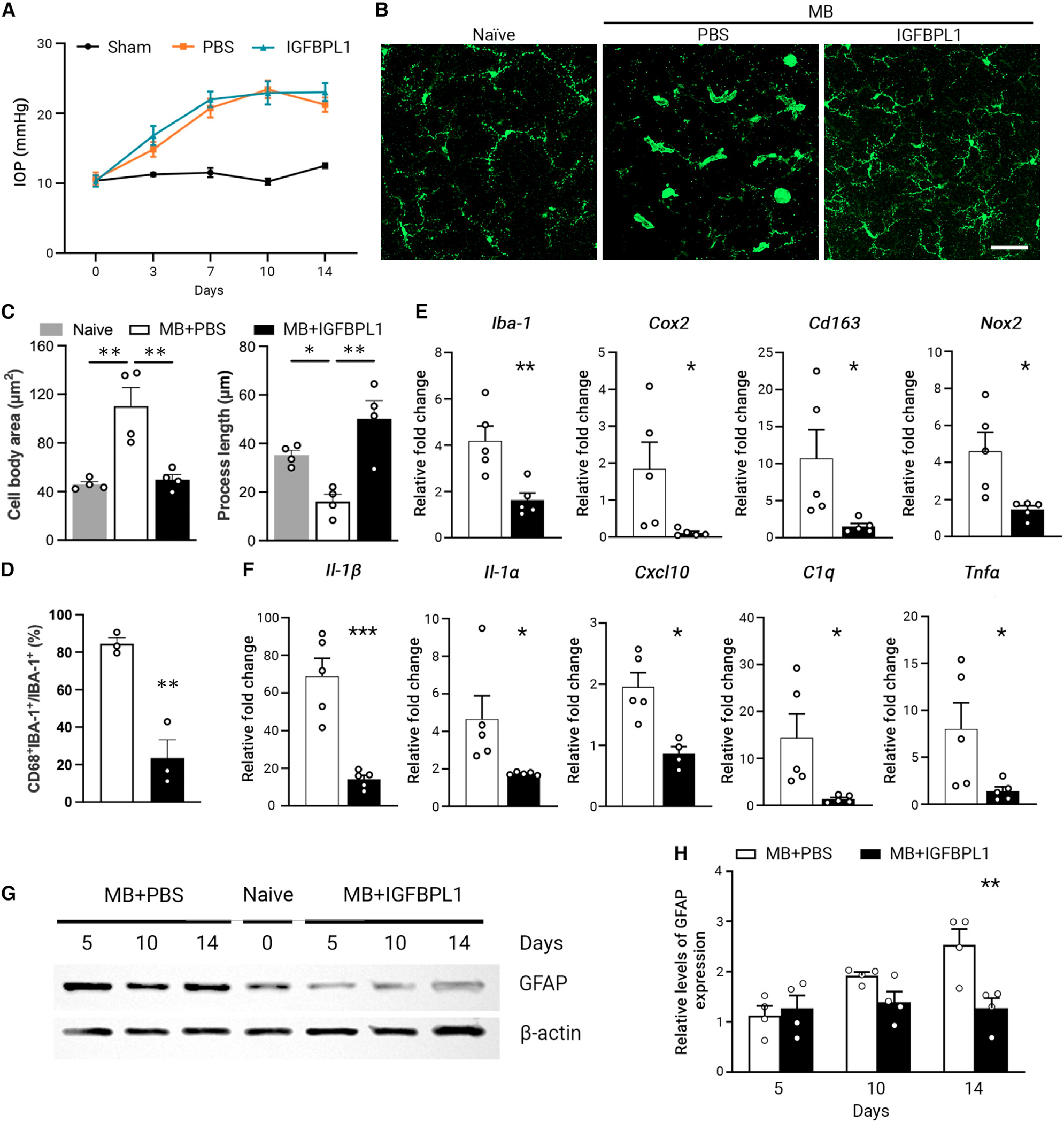
(A) IOP profiles in sham-operated (sham) and MB-injected mice injected with PBS or IGFBPL1. n = 10 mice/group.
(B) Images of IBA-1+ cells in retinal flat mounts of naive or MB-injected mice that received PBS or IGFBPL1 treatment. Scale bar, 50 μm.
(C) Quantification of microglial cell body area and process length in retinal flat mounts of MB-injected mice that received PBS or IGFBPL1 treatment. **p < 0.01; Student’s t test. n = 4 mice/group.
(D) Percentage of CD68+ microglia in retinal flat mounts of MB-injected mice that received PBS or IGFBPL1 treatment. **p < 0.01; Student’s t test. n = 3 mice/group.
(E and F) Relative mRNA levels of activated microglia markers (E) and pro-inflammatory cytokines (F) in PBS- or IGFBPL1-treated retinas normalized to that of naive mice. *p < 0.05, **p < 0.01, ***p < 0.001; Student’s t test. n = 5 mice/group.
(G and H) Western blot image (G) and quantification (H) of GFAP levels in retinal flat mounts of PBS- or IGFBPL1-treated mice after MB injection. GFAP expression was normalized to the corresponding β-actin intensity and presented relative to that of naive mouse retinas. **p < 0.01; two-way ANOVA with Šídák multiple comparisons test. n = 4 mice/group.
Data are mean ± SEM.
See also Figure S2.
We next investigated if IGFBPL1 is required for normal microglial function. KO (Igfbpl1−/−) mice grow normally and are fertile without apparent phenotypes.31 We found that KO mouse retinas showed consistent increases of CD68+IBA-1+ microglia with activated cell morphology (enlarged cell bodies and fewer cellular processes) compared with their WT littermate controls (Figures 4A and S3A). Significant upregulations of the activated microglial markers Cox2, Cd163, and Nox2 (Figure 4B) and pro-inflammatory cytokines, including Il-1β, Il-1α, Cxcl10, and Ccl2 (Figure 4C), were also detected in the retinas of KO mice compared with WT mice. We sorted out the microglia from adult KO and WT mouse retinas as described, showing an efficiency of >99% as demonstrated by IBA-1+ immunolabeling (Figure S3B). Isolated microglia of KO mice exhibited decreased homeostatic microglial marker P2ry12 and increased levels of pro-inflammatory cytokine (Ccl2 and Il-1β) and activated microglial marker Cd163 in culture compared with those of WT mice (Figures S3C–S3F). Importantly, addition of IGFBPL1 to the KO microglia reverted the expression of all markers mentioned above to the control levels. The data support that IGFBPL1 is crucial for maintaining microglial homeostasis in vivo and that its absence shifts microglia toward an inflammatory phenotype.
Figure 4. IGFBPL1 is required for sustaining microglial homeostasis in healthy and glaucomatous retinas.

(A) Quantifications of microglial cell body area and process length and percentages of CD68+ microglia in retinal flat mounts of WT and KO mice. *p < 0.05, **p < 0.01, ***p < 0.001; Student’s t test. n = 4 mice/group.
(B and C) Relative mRNA levels of activated microglia markers (B) and pro-inflammatory cytokines (C) 7-month-old KO retinas normalized to those of WT mice. *p < 0.05, ***p < 0.001; Student’s t test. n = 4 mice/group.
(D) tSNE map of microglial pools of 7-month-old mouse retinas. A total of 4 clusters of microglia were identified. n = 10 mice.
(E) tSNE plots showing distributions of pan-microglial (Cx3cr1, Tmem119), C1 (Apoe, Igf1), C2 (Cxcl10), and C3 (Ki67) markers.
(F) Dot plot of microglial signature genes identified in C0–C3.
(G) Bar plot representing the relative distribution of C0–C3 microglial subsets in WT, KO, and MB-induced glaucomatous WT mice that received PBS or IGFBPL1 treatment. The abundance was normalized to that of the naive WT mice.
(H) RNA velocity ForceAtlas2-based embeddings showing microglial cell-fate directions in all clusters (top panel) or in inflammatory C1 subset (bottom panel) in WT, KO, MB+PBS, and MB+IGFBPL1 retinas. Black arrows indicate the local differentiation direction from individual progenitor to progeny cell. The large red and green arrowheads indicate summed directions of cell-fate transition in C1 subset. Colors of subclusters are in correspondence to those in (D).
Data are mean ± SEM.
See also Figure S3.
Microglia are a heterogeneous population of cells. We sought to investigate if IGFBPL1 deficiency shifts microglia toward specific subset(s) using scRNA-seq. Microglia were isolated from retinas of 10 KO and 10 sex- and age-matched WT mice (at 8–10 months of age) using FACS. As a total of ~3,500 isolated cells were sequenced to an average of 115,000 reads/cell, dimensionality reduction and clustering analysis showed that more than 90% purified cells were microglia, as identified using gene markers of Cx3cr1, Siglech, and Tmem119.42–44 A minimal proportion of non-microglia macrophages and monocytes were noted as determined by their expression of Lyve1, Ccr2, and Ly6c1. The small percentages of non-microglial cells contained ~2.3% natural killer (NK) cells, ~1.6% dendritic cells and neutrophils, and ~2.5% T and B cells. t-Distributed stochastic neighbor embedding (tSNE) analysis of the WT and KO microglia identified four major clusters, consistent with the other reports5,45: the canonical microglial genes Cx3cr1 and Csf1r were expressed by cells of all clusters; cluster 0 (C0) represented the resting microglia that expressed the highest levels of Tmem119, P2ry12, and Trem2 and that were the large majority (86.4%); C1 (7.9%) was classified as inflammatory microglia that expressed Cd68, Tyrobp, Apoe, Cd63, etc.; C2 (3.5%) was interferon-responsive microglia (Stat1/2+ and Ifit1/2/3+); and C3 (2.2%) was identified as proliferating microglia that expressed cell-cycle markers, including Mki67, Cdk1, and Pcna (Figures 4D–4F and S3G). Gene Ontology (GO) enrichment for DEGs between C1 or C2 vs. C0 revealed upregulation of inflammatory and innate immune response gene pathways in C1 and C2 subsets, with C2 trending toward cellular responses to virus- and interferon (IFN)-responsive pathways (Figure S3H). In line with the above finding, IGFBPL1 deficiency resulted in a slight reduction of C0 microglia and shifted microglia toward inflammatory C1 and C2 subsets compared to WT mice (Figure 4G), supporting an essential role of IGFBPL1 in sustaining microglial homeostasis.
We tested further if IGFBPL1 plays a homeostatic function when microglia are hyperactivated in a diseased condition by scRNA-seq using microglia isolated from MB-induced glaucomatous mice that received PBS or IGFBPL1 treatment, respectively. scRNA-seq identified microglial subsets C0–C3. The glaucomatous retinas showed a greater reduction of C0 microglia and even greater increases of C1 and C2 subsets than the KO mice did over the WT controls (Figure 4G), indicating stronger inflammatory responses in glaucomatous retinas than that in KO mice. Notably, IGFBPL1 treatment largely attenuated MB-induced increases of C1 and C2 microglia, which even resulted in a moderate increase of C0 microglia compared to the naive mice (Figure 4G). To investigate the relationships of the microglial subsets, we employed RNA velocity analysis, which predicts the future state of individual cells through calculating the time derivative of gene expression states based on time-dependent abundances of precursor and mature mRNAs.46 Analysis predicted the bidirectional transcription dynamics of microglia in naive WT mice, transitioning back and forth between C0 and inflammatory C1 and C2 subsets (Figure 4H). Intriguingly, while microglia in KO and MB-induced glaucomatous mice exhibited an aggressive phenotype that progressed primarily toward an inflammatory state, IGFBPL1 treatment drove the cells moving from inflammatory C1 and C2 subsets toward the C0 state in glaucomatous mice (Figure 4H). These results demonstrate unequivocally that IGFBPL1 acts a master driver that is essential for sustaining the homeostatic status of microglia in the healthy retina and resolving neuroinflammation after injury or disease.
Microglia-specific deficiency of Igfbpl1/Igf1r signaling leads to progressive neuron loss
Sustained microglial activation and neuroinflammation are suspected to underlie the pathogenesis of age-related neurodegeneration. We therefore asked if microglia shifting toward an inflammatory landscape in KO mice causes neurodegeneration in the retina. RGC counts in BRN3a-immunolabeled retinal flat mounts revealed significant and progressive neuron loss in 1- to 7-month-old KO mice compared with sex- and age-matched WT littermates (Figures 5A and 5B). RGC loss in 7-month-old KO retinas was significantly greater than those at 1 month of age, while the RGC numbers in WT mice remained constant. Double immunolabeling of BRN3a and cleaved caspase-3 in retinal flat mounts of 7-month-old KO mice confirmed RGC apoptosis (Figure S4A) without IOP elevation (Figure S4B), excluding an IOP-dependent mechanism of RGC loss.
Figure 5. IGFBPL1 signaling in microglia is required for neuronal homeostasis in the retina and brain.
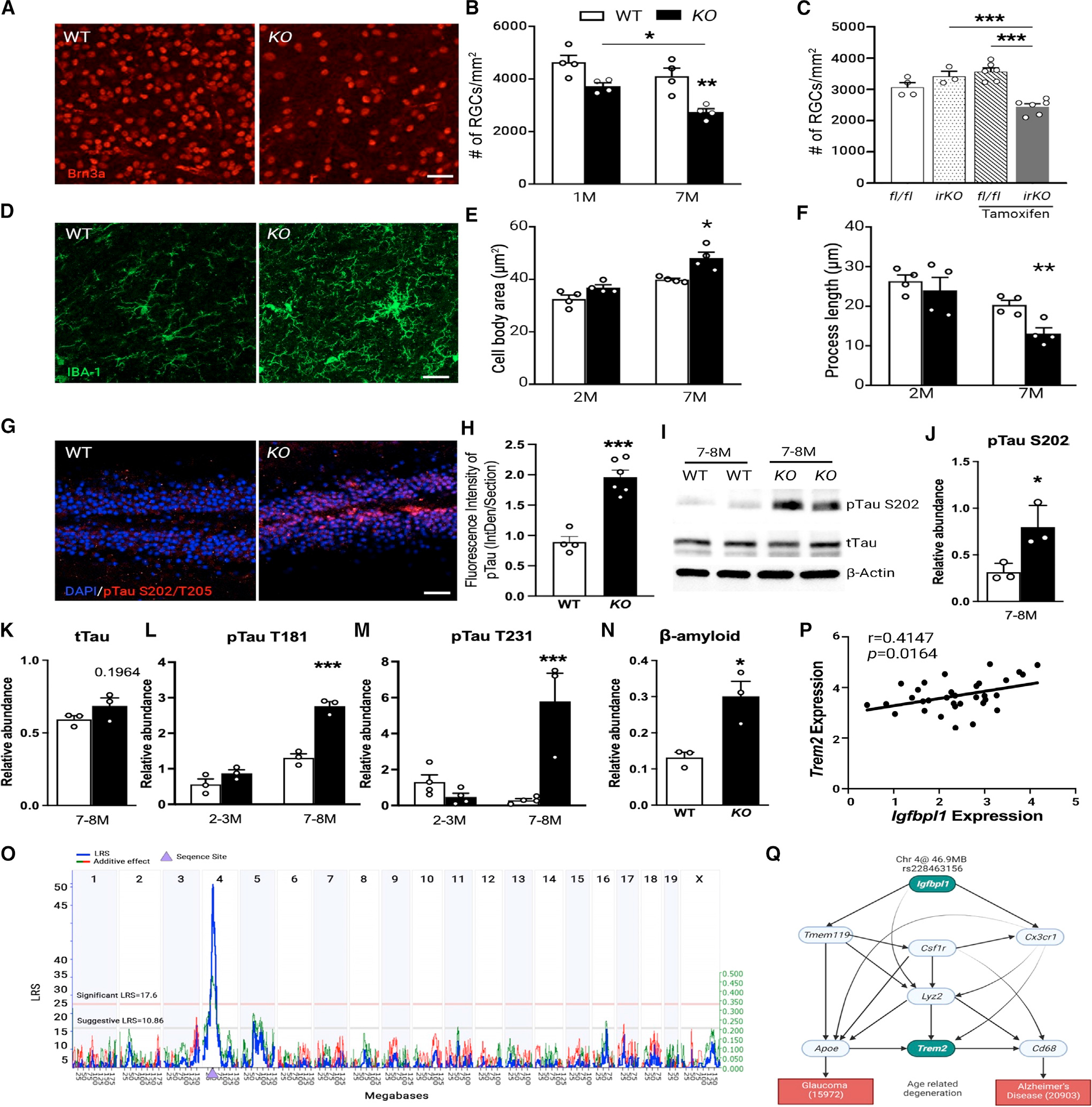
(A and B) Image (A) and counts (B) of Brn3a+ cells in retinal flat mounts of WT and KO mice. Scale bar, 30 μm. *p < 0.05, **p < 0.01, ***p < 0.001; two-way ANOVA with Šídák multiple comparisons test. n = 4 mice/group.
(C) RGC counts in fl/fl and IrKO littermates without or at 3 months after tamoxifen injection. **p < 0.01, ***p < 0.001; one-way ANOVA with Tukey’s multiple comparisons test. n > 4 mice/group.
(D) Images of IBA-1+ cells in the hippocampus of WT and KO mice. Scale bar, 20 μm. n = 4 mice/group.
(E and F) Quantification of microglial cell body area (E) and process length (F) in the hippocampus of WT and KO mice. *p < 0.05, ***p < 0.001; two-way ANOVA with Šídák multiple comparisons test. n = 4 mice/group.
(G and H) Images (G) and fluorescence intensity (H) of pTau S202/T205 immunolabeling in hippocampal dentate gyrus of WT and KO mice. Scale bar, 50 μm.
(I–M) Western blot images (I) and quantification (J–M) of pTau and total tau in the hippocampus of WT and KO mice. The relative abundance of pTau was normalized to total Tau. *p < 0.05, ***p < 0.001; two-way ANOVA with Šídák multiple comparisons test. n = 3–4 mice/group.
(N) Densitometry analysis of β-amyloid levels in the CA1 region and dentate gyrus of WT and KO mice. *p < 0.05, Student’s t test. n = 3 mice/group.
(O) Manhattan plot showing a genome-wide significant eQTL. The x axis denotes the chromosomal position in megabases on the mouse genome, and the y axis indicates the LRS score. The pink and gray horizontal lines indicate significant and suggestive LRSs, respectively. The purple triangle indicates the genomic position of Igfbpl1. LRS is shown by blue line, and additive effects are shown by green line.
(P) Scatterplots of the correlations between Igfbpl1 and Trem2 expression in BXD mouse eyes. Pearson correlation r and p values are indicated. Gene expression values are log2 transformed.
(Q) Bayesian network structure connecting SNP at peak of Igfbpl1 eQTL (rs228463156), Igfbpl1 and Trem2 correlated genes, and glaucoma (15972: IOP) and AD (20903: y-maze performance) phenotypes. Bayesian network structure was recreated using BioRender.com design tools and assets.
Data are mean ± SEM.
To test further that microglial, but not neuronal, deficiency of IGFBPL1/IGF-1R signaling contributes to RGC degeneration in KO mice, we generated inducible microglial-specific deletion of Igf1r by crossing Igf1rfl/fl (fL/fL) mice with Cx3cr1CreERT mice (Cx3cr1CreERT/+:Igf1rfl/fl or IrKO) (Figures S4C and S4D).47 Microglial-specific expression of Cre driven by a Cx3cr1 protomer was reconfirmed recently.48 IrKO mice without any treatment had a similar number of RGCs as fl/fl mice. In contrast, tamoxifen induction of microglia-specific deletion of Igf1r in IrKO mice resulted in significant loss of RGCs at 3 months post-injection compared with tamoxifen-treated littermate fl/fl mice (Figures 5C and S4E). Induction of microglial inflammatory responses in tamoxifen-treated IrKO mice was confirmed by the upregulation of Nox2, Il-1β, and Cxcl10 mRNAs (Figure S4F). A hallmark of reactive gliosis—increased GFAP immunolabeling—was also noted in retinas of IrKO mice compared with fl/fl littermate controls (Figures S4G and S4H). Both tamoxifen-treated IrKO and fl/fl mice displayed similar IOP levels (Figure S4I), supporting that RGC loss in IrKO mice was IOP independent. These results demonstrate a key role for microglial IGF1R signaling in RGC survival in adult mice.
Igfbpl1 is a Trem2-related gene that mediates brain tauopathy
Given the effect of IGFBPL1 on brain microglia, we investigated neuron and microglial changes in the KO mouse brains. Genome-wide studies have associated genes that are common risk factors for AD progression with the microglia.49,50 Chronic activation of microglia in the hippocampus of KO mice was confirmed by their increased IBA-1+ cell counts, enlarged IBA-1+ cell body area, and shortened dendritic processes at both 2 and 7 months of age (Figures 5D–5F and S4J). Since brain tauopathy is considered an essential hallmark of neurodegeneration,51–53 we performed immunolabeling for pTau S202/T205 in the dentate gyrus of the hippocampus of 7-month-old KO mice. Increased expression of pTau S202/T205 was detected in the KO brain compared to age- and sex-matched WT mice (Figures 5G and 5H). Western blot analyses of pTau S202/T205, T181, and T131 demonstrated significant increases in the hippocampus of 7- to 8-month-old, but not 2- to 3-month-old, KO mice compared with age- and sex-matched WT mice without altering the level of total tau (Figures 5I–5M), indicating that IGFBPL1 deficiency resulted in pTau accumulation without affecting tau expression. Upregulation of pTau S202/T205 was also found in the KO retinas (Figures S5A–S5D). Associated with increased pTau, elevated Aβ42 immunolabeling was observed in the dentate gyrus and CA1 regions of the hippocampus of KO mice (Figures 5N, S5E, and S5F). The results show that IGFBPL1 is required for maintaining microglial homeostasis and preventing age-related pTau and Aβ accumulation in the adult mouse brain.
Trem2 is a microglial-specific gene—a lipid-sensing receptor required for microglial responses to Aβ.54 Soluble TREM2 usually correlates with pTau and Aβ accumulation.55 We speculated that Igfbpl1 and Trem2 are functionally connected and hence performed gene network analysis using the bulk RNA-seq data from the aging eyes (12–18 months old) of 91 BXD strains. KEGG and MPO terms revealed co-varied genes of Igfbpl1 similar to those shown above (Figures 1B and 1C), including cytokine-cytokine receptor interaction, chemokine signaling pathway, neuroactive ligand-receptor interaction, PI3K-Akt pathways, etc. (Figures S6A and S6B). Genetic mapping of gene loci conducted with Web quantitative trait loci (WebQTLs) demonstrated a significant expression QTL (eQTL; likelihood ratio statistic [LRS] = 56, genome-wide p < 0.01) for Igfbpl1 at 44–47 Mb of chromosome 4, where the Igfbpl1 gene resides. This suggested that Igfbpl1 is cis-regulated (Figures 5O and S6C), likely acting as an upstream regulator of other genes. The expression of Igfbpl1 and Trem2 genes was found to be highly correlated (p = 0.0164; Figure 5P) across the 91 aged BXD mouse strains, a strong indication of functional coordination. We then employed Bayesian network models that integrated the single-nucleotide polymorphism (SNP) at the peak of Igfbpl1 cis-eQTLs (chromosome 4 [Chr4]: rs228463156) with 7 highly expressed microglial-specific genes, including Trem2, Tmem119, Csf1r, Cx3cr1, Lyz2, Apoe, and CD68, to inform their relations with two behavior traits characteristic of glaucoma (record ID: 15972; IOP) and AD (record ID:20903; y-maze performance for learning and memory) (Figure 5Q). The analysis unveiled significant coordination between Igfbpl1 and Trem2 activity via connections of Tmem119, Csf1r, and Cx3cr1 to affect the behavior trait of AD, while Igfbpl1 verges through Apoe to impact the glaucoma trait. In support of the eQTL analyses, downregulation of TREM2 expression in the KO retinas was confirmed by western blots (Figures S6D and S6E). Moreover, LPS downregulated TREM2 expression, and administration of IGFBPL1 restored the level of TREM2 expression in cultured microglia (Figures S6F and S6G). Collectively, these results suggest that Igfbpl1 functions in association with Trem2 to support microglial function and alleviate age-related neurodegeneration in the eye and brain.
IGFBPL1 exhibits potent neuroprotective and functional benefits in mouse models of glaucoma and age-related neurodegeneration
To determine if administration of IGFBPL1 effectively suppresses microglial activation and prevents neurodegeneration in glaucoma, the recombinant protein was injected intravitreally into MB-induced glaucomatous mice at day 3 after MB injection and then weekly for 3 weeks. Mice that received PBS injections served as controls (Figures 6A and 6B). The positive scotopic threshold response (pSTR) is the most sensitive electroretinogram measurement for detecting functional abnormalities of RGCs in rodents.56,57 As expected, IOP elevation induced significant reductions of pSTR amplitudes (Figure 6C) and loss of visual acuity (VA) and contrast sensitivity (CS), assessed using optomotor reflex assays58 (Figures 6D and 6E) in the PBS-treated group. Therapeutic administrations of IGFBPL1 for 3 weeks significantly improved pSTR amplitudes as well as VA and CS that lasted up to 8 weeks post-MB injection (Figures 6C–6E). RGC quantification at 8 weeks post-MB injection revealed nearly 90% RGC survival (3,625.7 ± 186.4 RGCs/mm2) in the IGFBPL1-treated group compared with ~50% RGC survival (2,126 ± 99.5 RGCs/mm2) in PBS-treated controls (Figures 6F and 6G). We found that a single dose of IGFBPL1 administration on day 3 post-MB induction of IOP elevation sufficed suppression of IL-1β and Cox2 by 3 weeks after MB injection (Figure S7A), indicative of a potent and lasting anti-inflammatory effect. We thus delivered IGFBPL1 monthly in another glaucomatous mouse model, DBA/2J mice, starting at 7 months old—a month after the mice developed spontaneous elevation of IOP (Figure S7B). IGFBPL1 treatment significantly improved pSTR amplitudes (Figures 6H and 6I) and enhanced RGC and axon survival (Figures 6J and S7C–S7E) compared with PBS-treated controls. Quantifications of microglial cell body size and process length in retinas of DBA/2J mice indicated suppressed microglial activation compared with PBS-treated mice (Figures S7F and S7G). Therefore, therapeutic administration of IGFBPL1 presents a potent and lasting neuroprotective effect and functional benefits against glaucomatous neural damage.
Figure 6. IGFBPL1 rescues RGCs and visual function in glaucomatous mice.
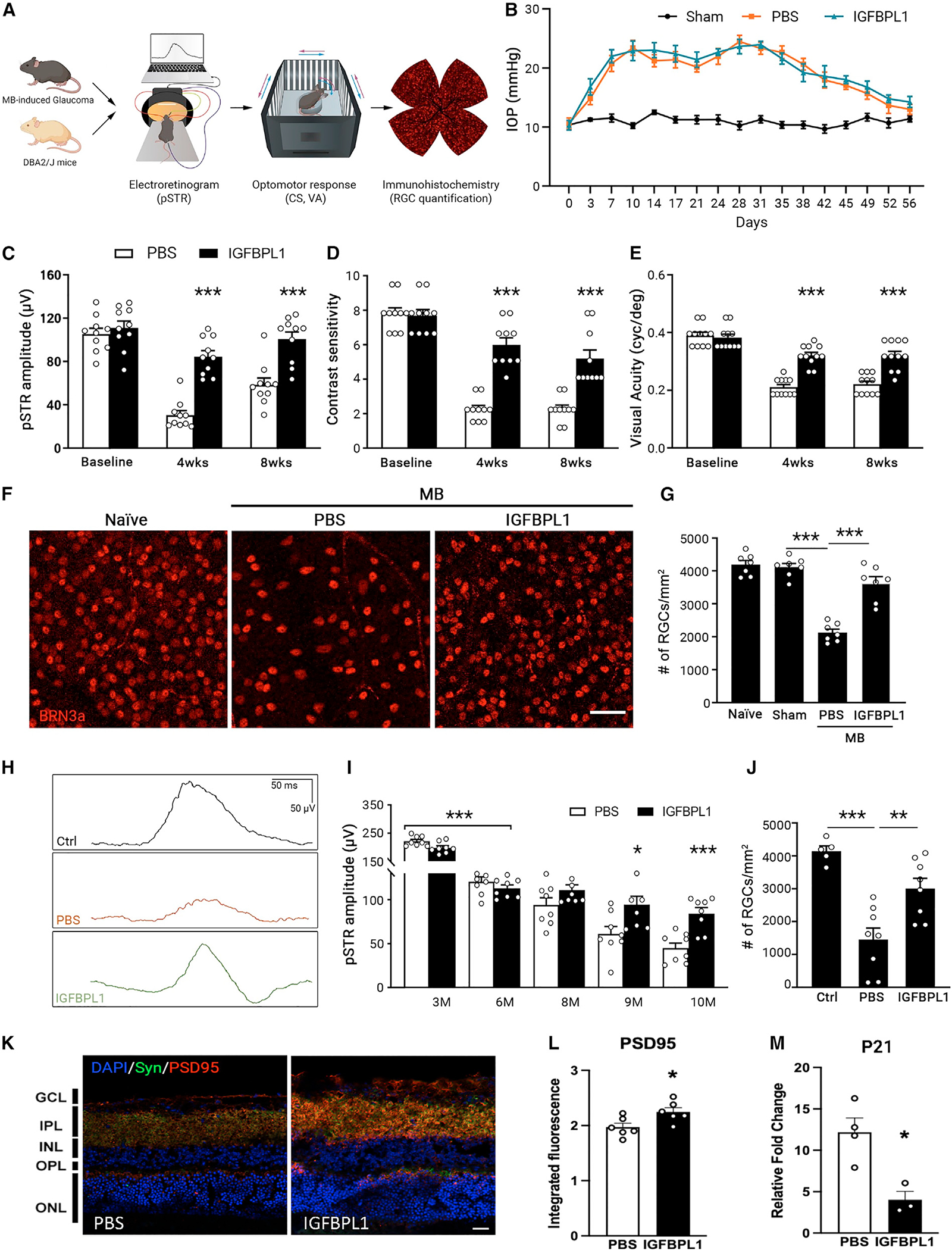
(A) Illustration of electroretinogram (ERG), optomotor response (OMR), and morphological assessments in MB-injected C57BL/6J or DBA/2J mice.
(B) IOP profiles of shamor MB-injected mice that received PBS or IGFBPL1 treatments. n = 10 mice/group.
(C–E) pSTR amplitudes (C) and values of CS (D) and VA (E) in MB-injected mice that received PBS or IGFBPL1 treatment. ***p < 0.001; two-way ANOVA with Šídák multiple comparisons test. n = 10 mice/group.
(F and G) Images (F) and counts (G) of BRN3a+ cells in retinal flat mounts of naive and sham- and MB-induced mice that received PBS or IGFBPL treatment. Scale bar, 50 μm. n = 7 mice/group.
(H) Representative pSTR waveforms in untreated 6- (control [Ctrl]) and 10-month-old DBA/2J mice that received monthly injections of PBS or IGFBPL1.
(I) pSTR amplitudes in DBA/2J mice. Mice that received a monthly injection of PBS or IGFBPL1 at 7–9 months old were sacrificed at 10 months old. *p < 0.05, ***p < 0.001; two-way ANOVA with Šídák multiple comparisons test. n = 8 mice/group.
(J) RGC counts in untreated 6- (Ctrl) and 10-month-old DBA/2J mice that received monthly treatments of PBS or IGFBPL1 at 7–9 months of age. **p < 0.01, ***p < 0.001; one-way ANOVA with Tukey’s multiple comparisons test. n = 8 mice/group.
(K) Image (K) of PSD95 (red) and synaptophysin (Syn; green) immunolabeling in retinas of 15-month-old WT mice that received PBS or IGFBPL1 injections. Scale bar, 15 μm.
(L and M) Quantification of PSD95 immunofluorescence intensity (L) and quantitative real-time PCR of p21 (M) in PBS- and IGFBPL1-treated groups. n = 3–6 mice/ group. *p < 0.05; Student’s t test.
Data are mean ± SEM.
See also Figure S7.
Glaucoma is thought to initiate a neurodegenerative program mimicking the natural aging in the retina that includes microglial activation,59,60 increased senescence cell marker cyclin-dependent kinase inhibitor p21,61 and reduction of synaptic proteins.62 We therefore asked if administration of IGFBPL1 by weekly injection into the vitreous of aged (15-month-old) C57BL/6 mice alleviates the biological changes of the aging eye. By 4 weeks after the first injection, IGFBPL1 significantly enhanced immunofluorescence signal of post-synaptic protein PSD95 and presynaptic protein synaptophysin (SYN) (Figures 6K, 6L, and S7H) as well as decreased the expression of p21 (Figure 6M) compared to PBS-injected mice. These data support the potential of IGFBPL1 in attenuating age-related changes of the mouse retina.
To determine if IGFBPL1 drives microglial function to prevent neurodegeneration, we tested IrKO mice following MB-induced IOP elevation (Figure 7A). Tamoxifen-injected littermate fl/fl mice served as controls. IGFBPL1 or PBS was administered intravitreally on days 3, 10, and 17 post-MB injections. As elevated IOP induced significant reduction of pSTR as well as VA and CS values in PBS-treated mice, administration of IGFBPL1 to control fl/fl mice resulted in significantly improved pSTR amplitudes and VA and CS values (Figures 7B–7D). In contrast, IrKO mice that received tamoxifen injection failed to respond to IGFBPL1 and exhibited reduced pSTR, VA, and CS values comparable to those of PBS-treated fl/fl mice after IOP elevation (Figures 7B–7D). No significant differences in baseline pSTR, VA, and CS values were detected among all groups (Figures S7I–S7K). RGC counts confirmed the protective effects of IGFBPL1 treatment in fl/fl mice but not in IrKO mice (Figure 7E). Together, selective disruption of IGF1R signaling in microglia abolishes the neuroprotective effects of IGFBPL1 against glaucomatous neurodegeneration.
Figure 7. Microglial-specific deletion of Igf1r abolishes the neuroprotective effect of IGFBPL1 in glaucomatous mice.
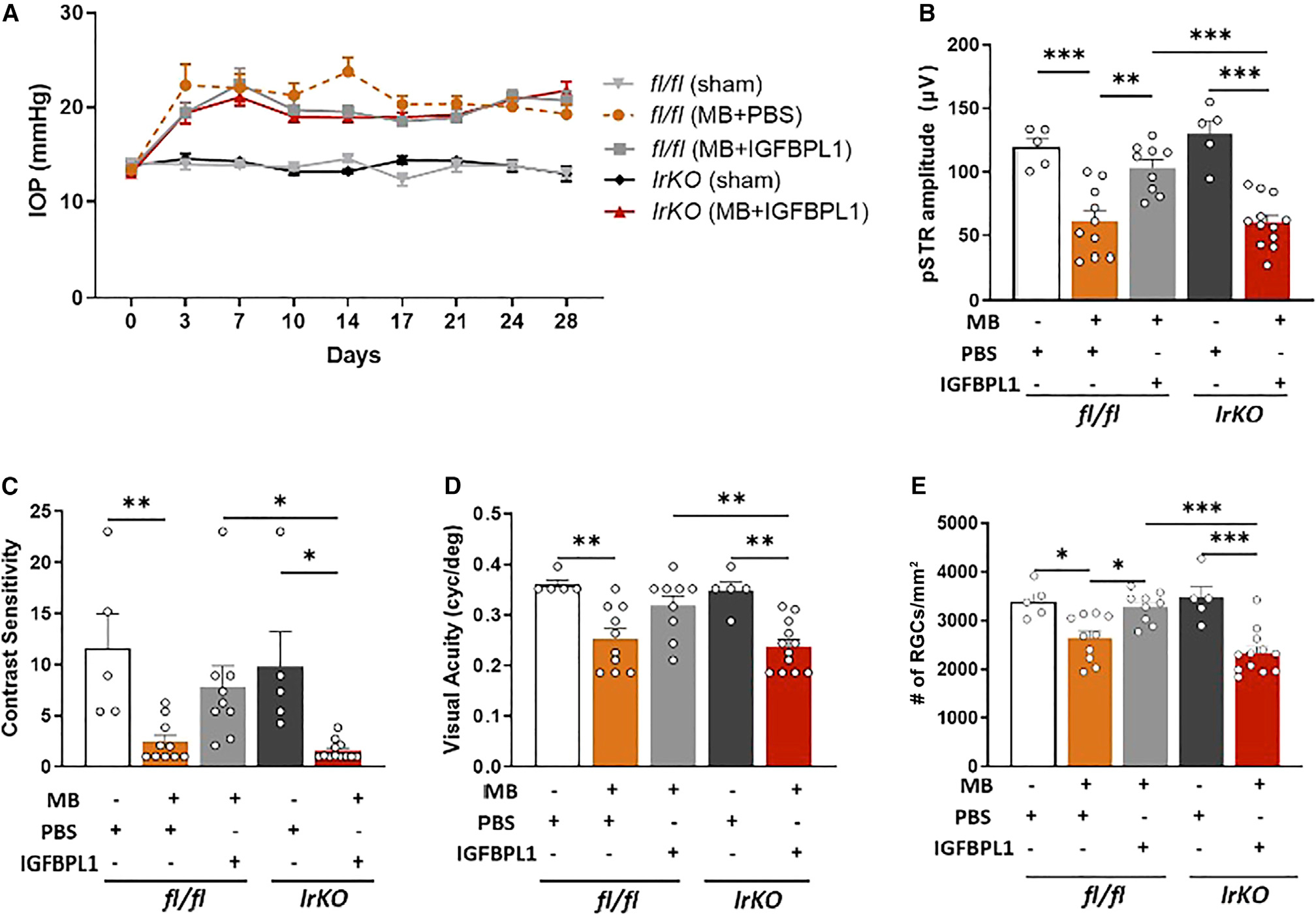
(A) IOP profiles of tamoxifen-treated fl/fl and IrKO mice that received PBS or IGFBPL1 treatments.
(B–E) pSTR amplitudes (B), CS (C), VA (E), and RGC counts in retinal flat mounts of sham- or MB-injected fl/fl and IrKO mice that received PBS or IGFBPL1 treatments. *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA with Tukey’s multiple comparisons test (n > 5 mice/group).
Data are mean ± SEM.
See also Figure S7.
DISCUSSION
Activated microglia are thought to contribute to many neurological and psychiatric disorders. However, our limited understanding of the molecular players that drive the transformation of activated microglia to a homeostatic state or the resolution of neuroinflammation has hindered the development of effective therapies via targeting this process. In the current study, we have identified a secretory protein, IGFBPL1, that functions as a master switch of pro-homeostatic microglia. In the absence of Igfbpl1, mice spontaneously develop microglial activation and chronic neuroinflammation, leading to progressive neurodegeneration and brain tauopathy. In contrast, administration of IGFBPL1 resets the molecular signatures of inflammatory microglia to a homeostatic state in vitro and in vivo via IGF1R signaling. The association mapping and gene network analyses further identified Igfbpl1 as a Trem2-associated gene that acts as an endogenous regulator responsible for “switching off” the inflammatory reactions of microglia to prevent excessive neuroinflammation and neurodegeneration.
It is increasingly evident that activated microglia may not be mere bystanders of CNS pathologies but key players in neurodegenerative diseases.63 Thus, initiation of microglial activation must be counter-balanced by an active process to restore homeostasis. Our previous reports demonstrated the critical involvement of microglia in the pathogenesis of glaucoma and other forms of optic neuropathies.16,17,64 Enriched KEGG analyses of RNA-seq data in the present study connect gene pathways regulating microglial inflammation with neurodegenerative diseases, including AD, Parkinson’s disease, and Huntington’s disease. Accordingly, KO and IrKO mice developed increased microglial activation, leading to progressive retinal neurodegeneration and brain manifestation of tauopathy. While scRNA-seq analysis of KO microglia revealed only a slight shift of inflammatory (C1 and C2) cell fractions, morphometric analyses in retinal flat mounts showed much more pronounced induction of microglial activation. Likely, this is because morphometric quantification of microglial activation focused on the ganglion cell layer (GCL)—the most affected region in glaucoma. Remarkably, administration of IGFBPL1 presents a potent and lasting neuroprotective effect and functional benefits in mouse models of glaucoma in an IGF1R-dependent manner. The fact that the expression of IGF1R in RGCs of adult mice is below detection supports a key role for microglial IGF1R signaling in retinal homeostasis.
Microglia in adult mice exhibit distinctive subpopulations or transition states, which can be defined by unique transcriptional signatures.33 Aging or AD pathology is reported to shift brain microglia toward more inflammatory or immunogenic and IFN-responsive subpopulations,65 termed disease-associated microglia (DAM). DAM exhibit increased expression of Apoe, Lyz2, and Cxcl10 and IFN-response genes.33,66 These markers are consistent with the C1 and C2 microglia identified in the present study. Seemingly, the much-simplified structure of the retina presents fewer subclusters of microglia than that in the brain. IGFBPL1 deficiency results in slight increases of inflammatory and IFN-responsive (C1 and C2) microglia, accompanied by progressive RGC loss and brain manifestation of tauopathy, similar to what was seen in the aging brain. The cell fate and trajectory tracing by analysis of RNA velocity—the time derivative of the gene expression state in scRNA-seq data—further supports that these subclusters are interchangeable functional states of the microglia.66–68 IGFBPL1 serves to drive the inflammatory microglia to their homeostasis; thus, its administration offers a possibility of precisely targeting the inflammatory cells, yielding an effective and safe strategy to limit the negative effects of activated microglia while retaining their beneficial and housekeeping functions.
An intriguing discovery in the present study is the identification of IGFBPL1 as a master driver of microglial homeostasis by imposing a “brake” to neuroinflammation. Previous research reported the involvement of CSF1, IL-34, and the pleiotropic cytokine transforming growth factor β (TGF-β) in microglial maturation and homeostasis in the CNS69–71 and the retina.72 Our studies with the KO mice, using bulk and scRNA-seq, revealed no developmental defects of microglia, suggesting that IGFBPL1 mediates microglial activity only in the adult. We noted that the expression of IGFBPL1 is upregulated with a 4 week temporal delay after IOP elevation, which may serve a role to allow initiation of microglial activation to defend the insult or injury before it is reset to the resting state for mitigating excessive/chronic neuroinflammation. Our studies with genetic mapping and gene networks analyses predict that IGFBPL1 signals upstream as a master switch to reset the expression of Cx3cr1, Trem2, and other microglial genes when transition from inflammatory (C1 and C2) to resting (C0) microglia. This is supported by our data that, as the expression of TREM2 was reduced in the KO retina and in LPS-primed microglia, IGFBPL1 treatment restored the level of TREM2 in culture. Trem2 mutation was shown to be closely associated with AD pathology through mediating microglial function.73,74 Accordingly, increased pTau and Aβ accumulation was detected in the hippocampus and retina of KO mice. Thus, IGFBPL1 deficiency exacerbates pTau and Aβ pathology in the brain and eye, that in turn may accelerate the progression of AD or age-related macular degeneration. As the number of inflammatory microglia increases with age or in the AD brain,75 upregulation of Lyz2, Apoe, and CD68 and concomitant downregulation of microglia homeostatic genes, such as Cx3cr1, are common features of the microglial transition into an inflammatory or DAM state. Likely, inflammatory subtypes of microglia can be induced in the healthy brain in response to neural stress/insult or by misfolded protein aggregations rather than being associated only with the disease etiology. A counter-regulatory mechanism, such as that mediated by IGFBPL1 to keep microglia under tight control, is thus essential for their proper functioning and ensuring risk-free activation.
In summary, our results indicate that IGFBPL1 drives a master switch to reset the process and the molecular identity of inflammatory microglia and actively resolves neuroinflammation to resume tissue homeostasis. IGFBPL1 may thus represent an attractive target for therapeutic interventions of glaucoma and other neurodegenerative disorders. Considering the broader expression of IGF1R in microglia and other immune cells, the counter-inflammatory effect of IGFBPL1 may extend far beyond the eye and the CNS.
Limitations of the study
Currently, there are limited opportunities for direct validation of the effects of IGFBPL1 in human microglial activities. Availability of the human transformed microglial cell line HMC3 and human inducible pluripotent stem cell-derived microglia offers an option for exploration of the human biology in this context. While studies report that microglia express a conserved core program from rodents to humans, particularly for those linked to the homeostatic gene signature,76 it remains to be investigated if IGFBPL1 acts in the same manner in human microglia as what is observed in mice. Moreover, the precise mechanisms through which IGFBPL1 drives the homeostatic function of microglia and interacts with other microglia regulators, such as CX3CR1 and TREM2, are yet to be elucidated. It is important to bear in mind that microglial activation serves as the CNS’s first line of defense and as an adaptive response to infection and/or injury, where controlled neuroinflammation is essential and generally beneficial for neuroprotection. Therefore, constant overexpression of IGFBPL1 may not always be desirable. It is crucial to strike a balance between neuroinflammation and the repair process, which entails determining the timing, context, and duration of microglial activation, to ensure proper CNS function or recovery.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Dong Feng Chen (Dongfeng_chen@meei.harvard.edu).
Material availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table. To provide the possible updates, troubleshooting, and code optimizations, we have GitHub repository available at https://github.com/mcrewcow/Pan_et_al_DFChen_IGFPL1_paper/tree/main.
The scRNA-seq datasets are deposited in the Genome Expression Omnibus under the accession numbers GEO: GSE176204, and the bulk RNA-seq datasets are deposited in the Sequence Read Archive (SRA) with BioProject ID: PRJNA739974.
The code and processed annotated datasets generated in this study for reproducing the bioinformatic analysis can be found on the following GitHub repository: https://github.com/mcrewcow/Pan_et_al_DFChen_IGFPL1_paper. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-BRN3a | Millipore | Cat# MAB1585, RRID: AB_94166 |
| Rabbit anti-IBA-1 | FUJIFILM Wako Shibayagi | Cat# 019–19741, RRID: AB_839504 |
| Goat anti-IGFBPL1 | R&D systems | Cat# AF4130, RRID: AB_2279980 |
| Goat anti-IGF-1R | R&D systems | Cat# AF-305-NA, RRID: AB_354457 |
| Rabbit anti-COX2 | Thermo Fisher Scientific | Cat# MA5–14568, RRID: AB_10984436 |
| anti-pNFκB | Cell Signaling Technology | Cat# 3033; RRID: AB_331284 |
| anti-tNFκB | Cell Signaling Technology | Cat# 9460, RRID:AB_2797704 |
| anti-pIGF1r | Cell Signaling Technology | Cat# 3027, RRID:AB_2122378 |
| anti-IGF1r | Millipore | Cat# ABE332, RRID:AB_10918911 |
| Mouse anti-CD68 | Abcam | Cat# ab201340, RRID: AB_2920880 |
| Mouse anti-phospho-Tau (Ser202, Thr205, AT8) | Thermo Fisher Scientific | Cat# MN1020, RRID: AB_223647 |
| Mouse anti-phospho-Tau (Thr181, AT270) | Thermo Fisher Scientific | Cat# MN1050, RRID: AB_223651 |
| Rabbit anti-phosphor-Tau (Thr231, AT180) | Thermo Fisher Scientific | Cat# 44–746G, RRID: AB_2533742 |
| Mouse anti-Tau | Thermo Fisher Scientific | Cat# MN1000, RRID: AB_2314654 |
| Rabbit anti-β-amyloid1–42 | Thermo Fisher Scientific | Cat# 700254, RRID: AB_2532306 |
| Rabbit anti-cleaved Caspase3 | Cell Signaling Technology | Cat# 9661, RRID: AB_2341188 |
| Mouse anti-GFAP-CY3 | Sigma-Aldrich | Cat# C9205, RRID: AB_476889 |
| Mouse anti-β-actin | Thermo Fisher Scientific | Cat# MA5–15739, RRID: AB_10979409 |
| Rabbit anti-GFAP | Abcam | Cat# ab7260, RRID:AB_305808 |
| Rabbit anti-TREM2 | Thermo Fisher Scientific | Cat# PA5–116068, RRID: AB_2900702 |
| Rat anti-anti-mouse CD11b | Biolegend | Cat# 101224, RRID: AB_755986 |
| Guinea pig anti-RBPMS | PhosphoSolutions | Cat# 1832-RBPMS, RRID: AB_2492226 |
| Rabbit anti-PSD95 | Abcam | Cat# ab18258, RRID: AB_444362 |
| Mouse anti-Syn | Millipore | Cat# MAB5258, RRID: AB_2313839 |
| Goat Anti-Mouse IgG (H + L), Biotinylated | Vector Laboratories | Cat# BA-9200, RRID: AB_2336171 |
| Goat Anti-Rabbit IgG (H + L), Biotinylated | Vector Laboratories | Cat# BA-1000, RRID: AB_2313606 |
| Rabbit Anti-Goat IgG (H + L), Biotinylated | Vector Laboratories | Cat# BA-5000, RRID: AB_2336126 |
| Streptavidin, Alexa Fluor 546 conjugate | Thermo Fisher Scientific | Cat# S-11225, RRID: AB_2532130 |
| Streptavidin, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat# S32354, RRID: AB_2315383 |
| Cy2 Donkey Anti-rabbit | Jackson ImmunoResearch Labs | Cat# 711–225-152, RRID: AB_2340612 |
| Cy3 Donkey Anti-mouse | Jackson ImmunoResearch Labs | Cat# 715–165-150, RRID: AB_2340813 |
| Cy3 Donkey Anti-goat | Jackson ImmunoResearch Labs | Cat# 705–165-147, RRID: AB_2307351 |
|
| ||
| Biological samples | ||
|
| ||
| Human Eye: Retina (Normal) | Florida Lions Eye Bank, Bascom Palmer Eye Institute University of Miami; Miller School of Medicine | Case# 2021–32323; Donor #21–12-050 |
| Human Eye: Retina (Glaucoma) | Lions Gift of Sight University of Minnesota | Patient ID: 22–0250 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat# A7030–100G |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| Tween 20 | Sigma-Aldrich | Cat# P5927 |
| Normal Donkey Serum | Jackson ImmunoResearch Labs | Cat# 017–000-121, RRID: AB_2337258 |
| Normal Goat Serum | Jackson ImmunoResearch Labs | Cat# 005–000-121, RRID: AB_2336990 |
| Collagenase/Dispase | Roche | Cat# 11097113001 |
| rmCSF1 | PeproTech | Cat# 315–02 |
| LPS | Sigma Aldrich | Cat# L6529–1MG |
| rmIGFBPL1 | R&D systems | Cat# 4130-BL |
| IGF1Ri H-1356 | BACHEM | Cat# 4018631 |
| ATP | Thermo Fisher Scientific | Cat# R0441 |
| Anti-CD11b Magnetic Particles | BD Biosciences | Cat# 558013, RRID: AB_398649 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat# 1000269 |
| Proteome Profiler Mouse Cytokine Array Kit | R&D Systems | Cat# ARY006 |
|
| ||
| Deposited data | ||
|
| ||
| Searchable microglia gene expression website | This paper | www.genecards.org |
|
| ||
| Experimented models: Cell lines | ||
|
| ||
| Human microglial cell line HMC3 | ATCC | Cat# CRL-3304, RRID: CVCL_II76 |
| Mouse microglial cell line BV2 | ATCC | Cat# CRL-2469, RRID: CVCL_5745 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | The Jackson Laboratory | Stock No: 000664, RRID: IMSR_JAX:000664 |
| Cx3cr1GFP/GFP | The Jackson Laboratory | B6.129P-Cx3cr1tm1Litt/J, Stock No: 005582, RRID: IMSR_JAX:005582 |
| DBA/2J | The Jackson Laboratory | Stock No: 000671, RRID: IMSR_JAX:000671 |
| Igf1r(flox/flox) | The Jackson Laboratory | B6; 129-Igf1rtm2Arge/J, Stock No: 012251, RRID: IMSR_JAX:012251 |
| Cx3cr1tm2.1(cre/ERT2) | The Jackson Laboratory | B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J, Stock No: 020940, RRID: IMSR_JAX:020940 |
| Igfbpl1−/− (KO) | Velocigene Regeneron Pharmaceuticals | C57BL/6N-Igfbpl1tm1(KOMP)Vlcg/Mmucd, MGI ID: MGI:1933198, Stock No: 048159- UCD, RRID: MMRRC_048159-UCD |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 | N/A | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Cell Ranger | 10x Genomics | Version 3.1.0 |
| R | https://www.r-project.org/ | 4.2.1 |
| RStudio | https://posit.co/ | 2022.07.2 Build 576 |
| biomaRt | https://bioconductor.org/packages/release/bioc/html/biomaRt.html | 2.48.3 |
| cowplot | https://cran.r-project.org/web/packages/cowplot/ | 1.1.1 |
| dittoSeq | https://bioconductor.org/packages/release/bioc/html/dittoSeq.html | 1.4.4 |
| dplyr | https://dplyr.tidyverse.org/ | 1.08 |
| EnhancedVolcano | https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html | 1.10.0 |
| escape | https://github.com/ncborcherding/escape | 1.2.0 |
| ggplot2 | https://ggplot2.tidyverse.org/ | 3.3.5 |
| GSEABase | https://bioconductor.org/packages/release/bioc/html/GSEABase.html | 1.54.0 |
| htmlwidgets | https://www.htmlwidgets.org/ | 1.5.4 |
| Matrix | https://cran.r-project.org/web/packages/Matrix/index.html | 1.3–4 |
| patchwork | https://patchwork.data-imaginist.com/ | 1.1.1 |
| plotly | https://plotly.com/r/ | 4.10.0 |
| reticulate | https://rstudio.github.io/reticulate/ | 1.24 |
| Seurat | https://satijalab.org/seurat/ | 3.0.0/4.1.0 |
| SeuratDisk | https://github.com/mojaveazure/seurat-disk | 0.0.0.9019 |
| SeuratWrappers | https://github.com/satijalab/seurat-wrappers | 0.3.0 |
| Python | https://www.python.org/ | 3.8 |
| Jupyter Notebook | https://jupyter.org/ | 6.4.11 |
| scanpy | https://scanpy.readthedocs.io/en/stable/ | 1.9.1 |
| scFates | https://scfates.readthedocs.io/en/latest/index.html | 0.9.0 |
| anndata | https://anndata.readthedocs.io/en/latest/ | 0.8.0 |
| numpy | https://numpy.org/ | 1.21.6 |
| pandas | https://pandas.pydata.org/ | 1.1.5 |
| palantir | https://www.palantir.com/docs/foundry/transforms-python/use-python-libraries/ | 1.0.1 |
| matplotlib | https://matplotlib.org/ | 3.5.1 |
| seaborn | https://seaborn.pydata.org/ | 0.11.2 |
| scvelo | https://scvelo.readthedocs.io/en/stable/ | 0.2.4 |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/ |
| ImageJ | National Institutes of Health | https://ImageJ.nih.gov/ij/ |
| Gene Ontology | Webgestalt | http://webgestalt.org/option.php |
| Protein-protein interaction | String | https://www.string-db.org/ |
| Zenodo Code Repository | Zenodo (https://zenodo.org/record/8118833) | https://doi.org/10.5281:zenodo/8118833 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Postmortem human specimen
Frozen human eye globe specimens from the postmortem donors were procured from biobank Tissue Solutions LLC (Glasgow, United Kingdom). As reviewed and defined by Mass General Brigham Institutional Review Board policies and Health and Human Services regulations set forth in 45 CFR 46, this research activity was deemed not human subject research as it does not involve human subjects.
Animals
All experimental procedures and use of animals were approved and monitored by the Mass Eye and Ear Institutional Animal Use and Care Committee and conform to the standards set forth by the National Institute of Health and the Association for Research in Vision and Ophthalmology. Adult (2–8 months) male and female C57BL/6J, Cx3cr1GFP,48,77 DBA/2J,78 Igfbpl1−/− (KO),31 Igf-1rfl/fl mice,79 Cx3cr1tm2.1(cre/ERT2), 47 and IrKO mice that were generated from Igf-1rfl/fl and Cx3cr1tm2.1(cre/ERT2) breeding were used in the study at the ages denoted in the manuscript. All mice were on a B6 background, and mice of both genders were used. Selective Igf-1r gene deletion in microglia in IrKO mice was induced by 3 consecutive days injection (i.p.) of 100 mg/kg tamoxifen that was reconstituted in corn oil.
METHOD DETAILS
Generation of gene expression data from BXD mouse retinas
The mice (3–4 mice per strain at 60–100 days old) from 55 BXD strains including their parental strains B6 and D2 mice were killed by rapid cervical dislocation. The retinas were removed and was placed immediately in 1mL of 160 U/ml RiboLock (Thermo Scientific Waltham, MA) for 1 min at room temperature. The retina was then transferred to Hank’s Balanced Salt solution with RiboLock in 50 μL RiboLock (Thermo Scientific, RiboLock RNase #EO0381 40 U/μL 2500U) and stored in −80°C. The RNA was isolated using a QiaCube (Hilden, Germany) and the in-column DNase procedure. All RNA samples were checked for quality before the microarrays were run. The samples were analyzed using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). The RNA integrity values ranged from 7.0 to 10. The gene expression data was generated using Affymetrix Mouse Gene 2.0 ST Array. The detail information about samples, tissue preparation, RNA extraction, microarray, normalization can be found in our previous publication (Molecular Vision 2015; 21:1235–1251 <http://www.molvis.org/molvis/v21/1235>) and at our GeneNetwork (www.genenetwork.org) Website with the accession number of GN709 or through the following link: http://gn1.genenetwork.org/webqtl/main.py?FormID=sharinginfo&GN_AccessionId=709.
Generation of RNA-seq data from the aging BXD mouse eyes
For the aging study, the mouse eyes of 91 aged (1–1.5 years old) BXD strains including their parental strains B6 and D2 were included. The animals (two mice per strain) were sacrificed under saturated isoflurane. Eyeballs from the animals were dissected and stored at −80°C. Total RNA was extracted using Trizol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. 1 μg of RNA was used for cDNA library construction at Novogene using an NEBNext Ultra RNA Library Prep Kit for Illumina (cat# E7420S, New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocol. Briefly, mRNA was enriched using oligo(dT) beads followed by two rounds of purification and fragmented randomly by adding fragmentation buffer. The first strand cDNA was synthesized using random hexamers primer, after which a custom second-strand synthesis buffer (Illumina, San Diego, CA, USA), dNTPs, RNase H and DNA polymerase I were added to generate the second strand (ds cDNA). After a series of terminal repair, poly-adenylation, and sequencing adaptor ligation, the double-stranded cDNA library was completed following size selection and PCR enrichment. The resulting 250–350 bp insert libraries were quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and quantitative PCR. Size distribution was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Qualified libraries were sequenced on an Illumina Novaseq Platform (Illumina, San Diego, CA, USA) using a paired-end 150 run (2 × 150 bases). An average of 40 million raw reads were generated from each library.
Read mapping and normalization
Mus musculus (mouse) reference genome (GRCm38) and gene model annotation files were downloaded from the Ensembl genome browser (https://useast.ensembl.org/). Indices of the reference genome were built using STAR v2.5.0a80 and paired-end reads were aligned to the reference genome. STAR used the method of Maximal Mappable Prefix, which can generate a precise mapping result for junction reads. FeatureCount v0.6.181 was used to count the number of read mapped to each gene. Transcripts Per Million (TPM) was calculated for each gene based on the length of the gene and reads mapped to that gene. In this normalization, the sum of all TPMs (genes-level) are equal to 1,000,000. The TPM was further rescaled to log2(TPM+1).
Expression QTL mapping
Expression QTL (eQTL) mapping was conducted with WebQTL in GeneNetwork (http://www.genenetwork.org/).82 Likelihood ratio statistics (LRS) were used to assess the association or linkage between differences in traits and differences in particular genotype markers. We report QTLs achieving significance (genome wide p value <0.05) based on 2,000 permutation tests.
Genetic correlation and functional enrichment analysis
We performed genetic and literature correlation analysis on GeneNetwork, in which, the genetic correlation was determined by Pearson product correlations value p, while literature correlation determined by R value which described by similar terminology in published papers.83 Genes with p < 0.05 of genetic correlation and r > 0 of literature correlation were selected for further analysis. Genes with significant correlations with Igfbpl1 were used for gene set over representation analysis for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Mammalian Phenotype Ontology (MPO) on Webgestalt website (http://webgestalt.org/option.php).84 Mouse genome was used as reference gene set with a minimum of 7 genes for each category. The p values generated from the test were automatically adjusted to account for multiple comparisons using the Benjamin and Hochberg correction (BH).85
Causative analysis with bayesian network modeling
Bayesian networks are graphical models that describe the conditional dependencies between data variables, where directed edges between network variables imply causal relationships. In order to explore plausible causal models among SNP at Igfbpl1 eQTL location, gene expression, and vision or learning memory related phenotypes, we created Bayesian network models using the Bayesian Network Webserver (BNW: http://compbio.uthsc.edu/BNW/).86,87 The model structures were learned from the data with the following structure learning settings. Each variable had a maximum of 4 direct parents in any potential model, model averaging of the 1,000 highest scoring networks was performed and directed edges with weights greater than 0.5 were included in the final network model.
Microglial isolation and culture
Adult C57BL/6J or Cx3cr1GFP mice were anesthetized by ketamine (120 mg/kg) and xylazine (20 mg/kg) and then transcardiacally perfused with ice-cold PBS. Mouse retinas and brains were dissected. Retinal microglia were sorted from the retinas of adult Cx3cr1GFP mice following the procedure described below: Retinas were digested with papain (20 units), dissociated cell suspension was immunolabeled with pacific blue conjugated anti-CD11b antibody for 30 min at 4°C, and sorted using both pacific blue and GFP signals by Miltenyi Tyto cell sorter under a low microfluidic pressure (150 mPa). Isolated microglia were plated at 30,000 cells/well in 24-well plates in DMEM supplemented with fetal bovine serum (FBS; 10%), Pen/Strep (1%) and recombinant CSF-1 (50 ng/ml). Before experiments, retinal or brain microglial cultures were treated with vehicle control, IGFBPL1 (400 ng/mL), LPS (1 μg/mL), or LPS (1 μg/mL) plus IGFBPL1 (400 ng/mL) which was added with a 6-h delay. The concentration of IGFBPL1 used here was based on previously reported effective doses of IGFBPL1 in neuronal cultures.31 Cultures were maintained for a total of 48 h before cells were washed with PBS and collected for bulk RNA-seq analysis and qPCR or fixed for immunohistochemistry. For primary retina and brain microglia taken from KO and littermate control mice, dissociated cell suspension was immunolabeled with magnetic microbead-conjugated CD11b antibody (Miltenyi Biotec, Cat. No. 130–049-601). CD11b+ cells were isolated according to the manufacture’s protocol and evenly seeded in a 24-well plate. IGF1 (100 ng/mL) or IGFBPL1 (400 ng/mL) was added, respectively, for a total of 48 h before RNAs were collected for qPCR.
Bulk RNA sequencing and analysis
Total RNA was extracted from retinas of ageand sex-matched Igfbpl1−/− and B6 mice or from cultured microglia at 48 h after treatment with vehicle control, IGFBPL1 (400 ng/mL), LPS (1 μg/mL) or LPS (1 μg/mL) plus IGFBPL1 (400 ng/mL) which was administered with a 6-h delay using RNeasy Plus Mini kit or RNeasy Micro kit following manufacturer’s instruction. The raw data was quality controlled and aligned on Mus musculus (mouse) reference genome (GRCm38) with STAR v2.5.0a80; FeatureCount (v0.6.1)81 was used to count the number of read mapped to each gene. Transcripts Per Million (TPM) was calculated for each gene based on the length of the gene and reads mapped to that gene. Differential expression analysis was performed using the DESeq2 R package (v1.22.2).88 Lists of genes with functions specifically related to “Microglia”, “Astroglia”, “neurons”, “retinal ganglion cells”, and “photoreceptors” were downloaded from Genecard (www.genecards.org) with relevance score >5, and further determined the DEGs enriched in each retinal cell type by calculating the number of DEGs as a percentage of the total number of the genes in each retinal cell type (p < 0.05). The percentages of DEGs between KO and B6 mouse retinas under each of the gene list categories were calculated accordingly and used to determine the perturbations to retinal cell types.
Immunohistochemistry, image processing and quantification
Mouse eyeballs and brains were dissected after mice were sacrificed by CO2 inhalation and transcardiacally perfused with PBS. Tissues were fixed in 4% paraformaldehyde in PBS for 3 h at room temperature or overnight at 4°C. Tissue samples were cryoprotected with 30% sucrose in PBS overnight at 4°C and embedded in OCT. Eyeballs were cryo-sectioned at 10μm thickness, and brain at 30μm thickness. Eyeball sections were incubated with blocking buffer (PBS supplemented with 3% donkey serum, 1% BSA, 0.1% Triton, and 0.1%Tween) for 1–3 h at room temperature, followed by incubation with primary antibody for overnight at 4°C, using Goat anti-IGF-1R (1:50), Rabbit anti-COX2 (1:500), Mouse anti-phospho-Tau (S202/T205) (1:100), mouse anti-GFAP-CY3 (1:400), mouse anti-mouse-Brn3a (1:500), Rabbit anti-mouse IBA-1 (1:250), goat anti-IGFBPL1 (1:50), mouse anti-CD68 (1:50), rabbit anti-GFAP (1:200), rabbit anti-cleaved Caspase3 (1:50), Rabbit anti-β-amyloid1–42(1:50), rabbit anti-PSD95 (1:200), mouse anti-Syn (1:50), RBPMS (1:50). After washing three times with a washing buffer (0.1% Triton-100 in PBS), samples were incubated with respective secondary antibodies at room temperature for 2 h. Secondary antibodies used included Streptavidin Alexa Fluor 546 conjugated (1:1000), Streptavidin Alexa Fluor 488 conjugated (1:1000), Cy2 Donkey Anti-rabbit (1:1000), Cy3 Donkey Anti-mouse (1:1000), and Cy3 Donkey Anti-goat (1:1000). For anti-Brn3a, IBA-1, IGFBPL1 immunostaining, samples were incubated with biotin-conjugated antibodies at 1:250 dilution before Streptavidin conjugated Alexa Fluor secondary antibody. Biotin-conjugated antibodies used included goat anti-mouse biotinylated (1:250), goat anti-rabbit biotinylated (1:250), rabbit anti-goat biotinylated (1:250). Brain sections were incubated with a blocking buffer containing 3% donkey serum and 0.5% Triton in PBS, followed by labeling with primary antibody in a blocking buffer on a shaker for overnight at 4°C. After washing three times with a solution containing PBS and 0.5% Triton for 10 min each on a shaker, sections were incubated with biotin-conjugated anti-rabbit antibody (1:250) at room temperature for 1 h. Following 3 washes in PBS on a shaker overnight at 4°C, sections were reacted with streptavidin conjugated Alexa 488 (1:2000) for 2 h at room temperature. After repeated washing, brain sections were incubated with CY3 Donkey anti-mouse (1:1000) for 1 h at room temperature on a shaker. After washing three times in a washing buffer, slides were mounted with Fluoroshield Mounting Medium containing DAPI. At least 3 sections per eyeball or brain were imaged. For RGC quantification, BRN3a-immunolabeled retinal flat-mounts counterstained with DAPI were divided into quadrants: superior, temporal, nasal, and inferior, and four standard regions were selected from each quadrant: one peripheral, two intermediate, and one central. In total, 16 rectangular regions of each retinal flat-mount were photographed at ×40 magnification with a confocal microscope (Leica SP8, Leica, Wetzlar, Germany). The images of IBA-1+ microglia in BRN3a+ RGC layer were taken at a depth of ~6–8 μm. ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to quantify the number of BRN3a+ RGCs and measurement of the longest process length of IBA-1+/DAPI+ microglia and cell body size as previously described.16,89 All quantification procedures were carried out by 2 investigators in a masked fashion, and all cell quantifications were performed according to appropriate counterstains of their markers with DAPI.
Immunolabeling in human retinal flat-mounts
The whole eye globes deceased by natural causes without underlying eye pathology were provided by Florida Lions Eye Bank, Bascom Palmer Eye Institute University of Miami, Miller School of Medicine. The eye globe was fixed in 4% paraformaldehyde in PBS for 48 h 4°C. The retinas were then carefully dissected and cut into fragments ~0.5 cm each. The retinal specimens were incubated with blocking buffer (PBS supplemented with 5% donkey serum, 0.1% Triton) overnight at 4°C. The blocked and permeabilized retinal specimens were incubated with primary antibodies Rabbit anti-mouse IBA-1 (1:200), goat anti-IGFBPL1 (1:50) at 4°C for 24 h. Following 3 washing cycles (0.1% Triton-100 in PBS) for 30 min (1.5 h in total), specimens were incubated with Cy2 Donkey Anti-rabbit (1:1000), Cy3 Donkey Anti-goat (1:1000) overnight at 4°C. Following another 1.5 h of washing, specimens were mounted with Fluoroshield Mounting Medium and imaged.
Zymosan experiment
The evaluation of phagocytosis was performed as reported previously.90 Primary microglia were seeded into 24 well plate treated with PBS or IGFBPL1 (400 ng/mL) and incubated with 50 μg/mL of pHrodo Green Zymosan bioparticles (P35365; ThermoFisher) for 12 h. The resulting cultures were imaged, and ratio of zymosan positive cells to total cells were determined by “masked” observers from the images. The cultures were then incubated with hoechst nuclear staining (62249; ThermoFisher, 2 μM) for 10 min and washed with PBS. The plate was then scanned with florescent plate reader in plate surface scan mode. Averaged signal at 350/420 nm excitation/emission of hoechst nuclear staining was used as the total cell number signal, and 485/520 nm excitation/emission of absorbed zymosan particles was determined. The background florescence was determined against negative cultures that was not treated with zymosan or Hoechst and subtracted from the experimental values. The quantification of the florescence ratios between total and zymosan signals was performed as follows (zymosan particles florescence/Hoechst florescence) to normalize the signal to the cell number.
Cytokine arrays and analysis
Retinal microglia were isolated and cultured as described above. The cultures were divided into 5 treatment groups: vehicle control, IGFBPL1 (400 ng/mL), LPS (1 μg/ml), LPS (1 μg/mL) followed by IGFBPL1 (400 ng/mL) which was administered with a 6-h delay, and LPS (1 μg/mL) followed by IGFBPL1 (400 ng/mL) and IGF1Ri (12.5 μg/mL), both of which were administered with a 6-h delay. After a total of 48 h of incubation, culture media were collected for cytokine array, using Proteome profiler mouse cytokine array Panel A kit and according to the manufacturer’s protocol as previously described.16 The array membranes were imaged with the iBright CL1500 imaging system, and the results were analyzed using ImageJ software.
Quantitative real-time PCR
Total RNA was extracted from the retinas of adult mouse brain and retinas or microglia using RNeasy Plus Mini kit or RNeasy Micro kit following manufacturer’s instruction and reversely transcribed using Takara PrimeScript RT Master Mix. A mixture of master mix containing cDNA, 2x Master Mix from KAPA SYBR Fast qPCR kit, and 10 mM of specific primers was applied to detect specific mRNA expression level using the Mastercycler ep realplex real-time PCR system (Eppendorf). The temperature of initial denaturation was set at 95°C for 2 min followed by 45 cycles of 15 s denaturation (95°C), 15 s annealing (59°C), and 20 s extension (68°C), and lastly holding at 4°C. Relative amount of specific mRNA transcript was presented in fold changes by normalization to the expression level of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (Gapdh). All primers were synthesized by Integrated DNA Technologies. Primers used in this study are listed in Table S1.
Western blots
WB was performed as previously noted.91 Cell or tissue samples were sonicated in a cold RIPA buffer supplemented with a 1:100 Halt protease and phosphatase inhibitor (1861261, Thermo Fisher Scientific). Disruption of the material was performed by a Q55 Sonicator (Qsonica) with four pulses for 22 kHz, 5 s each at 20% power output, and on ice. The lysates were centrifuged at 17,000 for 5 min at 4°C. The total Protein concentration of resulting supernatants was determined using the Qubit 4 Fluorometer (Thermo Fisher Scientific). The 30 μg total protein loaded per lane was separated by SDS-PAGE (4–20% polyacrylamide gel; Biorad) before electrophoretic transfer to 0.45 μm pore nitrocellulose membranes. The membranes were blocked with 2.5% BSA (A7096, Sigma-Aldrich, St Louis, MO, USA) at RT for 1 h and then incubated overnight at 4°C with the primary antibodies against phosphorylated Tau (pTau) (1:1000), GFAP (1:1000), TREM2 (1:500), β-amyloid (1:1000). After washing with PBS-T buffer, the blots were incubated with horseradish peroxidase (HRP)-conjugated 1:2000 secondary antibody in 2.5% BSA in PBS-T (Goat anti-Rabbit IgG; Goat anti-Mouse IgG) for 1 h at room temperature. Signals were developed with ECL using a Super Signal West Pico kit and detected with iBright CL 1500 (Thermo Fisher Scientific). The densitometric analysis of WBs was performed with ImageJ software using 8-bit grayscale chemiluminescent membrane images. All the quantification results were averaged from at least three protein blots and expressed as the mean ratio of the values (target protein/housekeeping protein) ± standard deviation (SD).
Single cell RNA sequencing and computational analysis
Single-cell suspensions of retinal CD45+CD11b+ cells taken from Igfbpl1−/− and age- and sex-matched B6 mice that were transcardially perfused with PBS. Immune cells were stained by PE conjugated anti-mouse CD45 and sorted by flow cytometry (BD FACSAriaII); sorted cells (~20,000 CD45+ cells per sample) were resuspended and washed in 0.05% RNase-free BSA in PBS for single-cell library preparation with 10x Chromium Next GEM Single Cell 3′ Kit (10XGenomics according to the manufacturer’s instructions. The single-cell cDNA libraries were sequenced by NexSeq500 (Illumina). Raw sequences were demultiplexed, aligned, filtered, barcode counting, unique molecular identifier (UMI) counting with Cell Ranger software v3.1 (10XGenomics) to digitalize the expression of each gene for each cell. The analysis was performed using the Seurat 3.0 package.92 We first processed each individual dataset separately prior to combining data from multiple samples. The outlier cells with extremely low number (<500) or high number (>5,000) of gene features as doublets, or low total UMI (<1,000) and high mitochondrial ratio (>10%) from each dataset were removed. Subsequently, samples were combined based on the identified anchors for the following integrated analysis. We ran principal component analysis (PCA) and used the first 15 principal components (PCs) to perform tSNE clustering (resolution = 0.3). We checked well-defined marker genes for each cluster to identify potential cell populations, such as T cells (Cd3d, Cd3e, and Cd3g), B cells (Cd19, Cd20), microglia (Cd68, Cx3cr1). For microglia analysis, Cd68 and Cx3cr1 positive clusters were selected for subsequent analyses. We repeated PCA, tSNE clustering (resolution = 0.08 on the integrated datasets of microglia. Finally, we performed differential expression tests on the integrated datasets to identify the genes significantly upregulated in each cluster compared with all other cells (adjusted p < 0.05) as well as the genes differentially expressed between WT and KO retinal microglial cells. For gene sets representing specific cellular functions or pathways, we performed functional enrichment analysis with the biological process of Gene Ontology by the online tool DAVID (http://david.ncifcrf.gov/).93
For the scRNA-seq analysis, we loaded the previously annotated objects using scanpy read_10x_mtx() function.94 Upon data filtering, normalizing, log-transformation, and highly variable genes search, we used an approach for dimensionality reduction, ForceAtlas2, a force-directed layout used for network spatialization.95 We utilized scFates96 package approach to preprocess the objects by generating PCA projections (scanpy.pp.pca()), palantir package functions to run diffusion maps and determine multiscale space. We then generated the ForceAtlas2 embedding based on neighbor graph in multiscale diffusion space (scanpy.pp.neighbors() with use_rep = ‘X_palantir’) by utilizing scanpy.tl.draw_graph() with initial position based on PCA projections. We used Leiden algorithm97 for further cluster analysis and microglia annotation.
For cell clustering and dataset analyses, we removed photoreceptors (Rho+; Pde6h+), neurons (Pcp2+; Syt1+, Lsamp+; Nrxn3+), retinal ganglion cells (Cplx3+), astrocytes (Car2+), Müller glia (Gpm6b+, Glul+, Vim+), abnormal cells (Runix1+, Mt+, Rho+), T-cells (Trdc+; Rps18+, Inpp4b+, Cd3d+; Lkzf2+; Tmsb10+; Ccr7+), and lymphocytes (Cd74+, Lgkc+; Ly6d+). Upon subsetting and reclustering the dataset, we generated the expression scores (scanpy.tl.score_genes()) for the Homeostatic/Proinflammatory, Proliferating/IFN-response/Inflammatory patterns. The genes used for generating the scores are: Homeostatic – P2ry12, Ccr5, Cd33, Csf1r, Cx3cr1, Glul, Gpr34, Adgrg1, Tgfb1, Tgfbr1, Serinc3, Siglech, Mertk, Bin1, Tmem119, Spi1, Sall1, Mafb, Smad3, Mef2a, Egr1, Jun; Proinflammatory – Apoe, Axl, Clec7a, Csf1, Cst7, Ctsb, Ctsd, Cybb, Fabp5, Fth1, Gnas, Gpnmb, Grn, Il1b, Lgals3, Lilrb4a, Lpl, Lyz2, Msr1, Spp1, Trem2, Tyrobp, Vegfa, H2-Ab1; Proliferating – Mki67, Cdc20; IFN-response – Cxc110, Stat1, Ifit1; inflammatory – Apoe, Igf1, H2-Aa, Cd74.
Embeddings with gene scores shown via scanpy.pl.draw_graph(), bar plot with microglia populations contribution was generated in RStudio using ggplot298 geom_bar() function. Relative contribution amount for the populations was received by utilizing value_counts() function for adata.obs.annotation, and the population contribution was quantified as individual population cells amount divided by total cells amount for every dataset.
RNA velocity analysis of single cell RNA sequencing data
Upon the microglia clusters to states annotation, we utilized Cellranger, samtools, velocyto, and scvelo packages to perform the RNA Velocity analysis.46
First, by using cellranger count function on.fastq files in Ubuntu WSL 20.04 we generated Cellranger output folder. We used samtools sort function in samtools package (-t CB -O BAM) to generate cellsorted_possorted_genome_bam.bam file from possorted_genome_bam.bam file. The generated file was used for creating the.loom file upon utilizing velocyto run10x function of velocyto package. The reference genome used for generating the.loom file is GENCODE (https://www.gencodegenes.org/) annotation v.M27, and the proper repeat annotation for the reference genome was obtained from UCSC genome browser in.gtf format.
Once the.loom file is generated, we used scvelo99 package for the RNA Velocity analysis. We merged.loom file with the respective adata file that was previously generated, processed and annotated. The following file was filtered and normalized, and the first and second order moments (means and uncentered variances) were computed among nearest neighbors in PCA space with scvelo.pp.moments() function. Then we estimated velocity (scvelo.tl.velocity()), built the velocity_graph (scvelo.tl.velocity_graph()) and projected the velocities using scvelo.pl.velocity_embedding_stream() and scvelo.pl.velocity_embedding_grid() on ForceAtlas2 (basis = ‘draw_graph_fa’) embedding.
Induction of intraocular pressure (IOP) elevation in mice
Elevated IOP was induced unilaterally in adult mice as previously described.34 Briefly, the mice were anesthetized by intraperitoneal injection of ketamine (120 mg/kg)/xylazine (20 mg/kg) mixture. The pupils were dilated with topical application of 1% Tropicamide followed by proparacaine HCl (0.5%; Bausch & Lomb Incorporated, Tampa, FL) to numb the eye. A 30-gauge needle generated an entry port on the cornea, and a small air bubble was injected into the anterior chamber via a glass micropipette which was connected with a Hamilton syringe. Two microliters of sterile PBS containing 10 μm microspheres (9.0×107 beads/ml) were injected into the anterior chamber with care to avoid damaging the iris and the lens. Antibiotic ointment was applied topically on the cornea after the injection. Operated mice were allowed to recover on a warm pad and monitored every 15 min until awake and fully sternal.
Measurement of IOP
Mice were anesthetized by isoflurane inhalation (2–4%) delivered in 95% Oxygen with a precision Vaporizer, and IOP was measured by TonoLab tonometer (Colonial Medical Supply) before and post-MB injection. An average of 6 IOP readings will be taken as one reading. At least 6 readings of IOP level were taken per mouse eye, and the means were used as the final readout of the IOP level. IOP measurement was always taken at the same time in the morning to minimize variations due to circadian change.
Intravitreal injection
Adult mice were anesthetized by intraperitoneal injection of ketamine (120 mg/kg)/xylazine (20 mg/kg) mixture using a 25G needle. The pupils were dilated with topical application of 1% Tropicamide followed by proparacaine HCl (0.5%) to numb the eye. A hole on the sclera at about 0.5 mm from the limbus was punctured using a 30G needle. Two microliters of sterile PBS or a solution containing IGFBPL1 (50 ng/μL) was injected into the vitreous via a glass micropipette with care not to damage the lens. Antibiotic ointment was applied on the entry site after injection. Mice were allowed to recover on a warm pad and monitored every 15 min until awake and fully sternal.
Electroretinogram (ERG)
Mice were dark-adapted in a dark adaptation chamber overnight before ERG. Anesthetized by intraperitoneal injection of ketamine (120 mg/kg)/xylazine (20 mg/kg) mixture using a 25G needle, both pupils were dilated by 1% Tropicamide, and a drop of Genteal was applied to keep the corneal moist. The mouse was placed on a warmed platform of the ERG machine (Diagnosys LLC). The reference and ground electrodes were inserted beneath the skin over the forehead and tail, respectively. Two gold-ring recording electrodes were gently placed on the corneas with a drop of artificial tear covered without blocking the pupil. The positive scotopic threshold response (pSTR) were obtained with flash intensities at 6.57E-5[cd.s/m2] and 1.7E-4[cd.s/m2] by averaging 40 responses per intensities. The pSTR was measured from the baseline to the peak of the positive deflection. After ERG recording, antibiotic ointment and artificial tear were applied to the cornea, and mice were allowed to recover on a warming pad with circulating heated water to maintain body temperature until recovery.
Optomotor response (OMR)
The mouse was placed on a small platform in the middle of the optomotor chamber. Each trial was begun with a gray homogeneous stimulus projected to all screens followed by visual stimuli projected such that a virtual cylinder with rotating gratings was produced. Rotating black and white bar visual stimuli were displayed on all computer monitors using a 4-port video splitter (www.Startech.com, model#ST124PROA) as previously described.58 The luminance of individual stripes was measured using a 371 R Optical Power Meter (Graseby Optronics) from the level of the eyes. The contrast at a given spatial frequency was calculated using the formula (Lmax − Lmin)/(Lmax + Lmin) where Lmax was the brightness (cd/m2) of the white stripe, and Lmin is the brightness of the black stripe (cd/m2). Contrast level, stripe width, grating speed, and direction of stripe movement (clockwise vs. counterclockwise) were measured. By changing the stripe width and the contrast, visual acuity and contrast sensitivity of mice were assessed. Quantifications were carried out by 2 investigators in a masked fashion.
Semi-thin optic nerve sectioning and axon quantification
As previously reported,17 the optic nerves were collected after euthanizing mice and placed in ½ Karnovsky’s fixative in a 0.1M sodium cacodylate buffer overnight. The optic nerve was placed evenly between two pieces of a small filter pad to straighten the nerve, semi-thin sectioned and stained for myelin using 2% paraphenylenediamine solution (Fisher Scientific). The sections were imaged and photographed under 100X bright field Leica microscope. Quantification of axon numbers was carried using the ImageJ software by 2 investigators under a masked fashion.
Mouse genotyping
Mice were anesthetized by isoflurane inhalation (2–4%) that was delivered in 95% Oxygen with a precision Vaporizer. Mouse tail was clipped from each mouse, and care was taken between each mouse to prevent gene contamination in each sample. Collected mouse tails were lysed using Proteinase K solution (Invitrogen) at 56°C overnight. After stopping reactions by boiling at 95°C 30 min, a mixture of DNA lysates per sample, 2x Hotstar mix and specific primer were applied for PCR amplification as following: DNA size were examined through electrophoresis and then imaged under iBright CL1500 imaging system (ThermoFisher Scientific). All primers were referenced to the information on the website of JAX laboratory and then synthesized by Integrated DNA Technologies. Igfbpl1−/− (KO) mice were genotyped with assistance from Transnetyx (www.transnetyx.com) using two RT-PCR probes for WT IGFBPL1 sequence and lac operon (LACZ) insert to determine homo and heterozygotes.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis, including mean values, SEM and p values, was performed using GraphPad Prism 9 software (GraphPad Inc., La Jolla, CA, USA). Statistical significance between multiple time point groups was evaluated by One-way ANOVA and Dunnett’s multiple comparisons test, and statistical significance among treatment groups was determined by two-way ANOVA and Tukey’s multiple comparisons test. Probability value of p < 0.05 was considered to be statistically significant, and outlier was excluded after ROUT analysis by Prism 9. Data were expressed as mean ± S.E.M.
Supplementary Material
Highlights.
Systems genetics methods identify IGFBPL1 as a modulator of microglia
IGFBPL1 resets the gene signatures of activated microglia to a homeostatic state
IGFBPL1 deficiency results in microglial activation and progressive neurodegeneration
Therapeutic administration of IGFBPL1 rescues vision in glaucoma mice
ACKNOWLEDGMENTS
We thank Mellissa A. Pottinger (Recovery Technician) at the Lions Eye Bank for providing post-mortem donor eyes, R. Huang for assistance in flow cytometry, P. Seifert for assistance in morphometric analysis, and S. Li for insightful discussions. BioRender.com design tools and graphical assets were used in some of the figure designs. This work was supported by grants from the Massachusetts Eye and Ear Summit Fund (D.F.C.); NIH/NEI grants R01EY025250 (D.F.C.) and R01EY031696 (D.F.C.); a Harvard NeuroDiscovery Center grant (D.F.C.); R01EY021200 (L.L.); NS104315 (J.C.); PolyU grant 1-WZ25 (C.-W.D.); PolyU Postgraduate Studentship (L.P.); BrightFocus Foundation and the Glaucoma Foundation grants (K.-S.C.); and NIH/NEI core Grant for vision Research P30EY03790 (Schepens Eye Research Institute). Patent applications have been filed related to this work.
Footnotes
DECLARATION OF INTERESTS
D.F.C. is a co-founder and SAB member of FireCyte Therapeutics and a consultant to i-Lumen Scientific and Sichuan PriMed. K.-S.C. is a consultant to FireCyte Therapeutics. D.F.C., K.-S.C., and P.L. are inventors of patents on the neuroprotective roles of IGFBPL1, which have been licensed to FireCyte Therapeutics.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112889.
REFERENCES
- 1.Butovsky O, and Weiner HL (2018). Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 19, 622–635. 10.1038/s41583-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond TR, Marsh SE, and Stevens B (2019). Immune Signaling in Neurodegeneration. Immunity 50, 955–974. 10.1016/j.immuni.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang SK, Xue Y, and Cepko CL (2020). Microglia modulation by TGF-beta1 protects cones in mouse models of retinal degeneration. J. Clin. Invest. 130, 4360–4369. 10.1172/JCI136160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman SM, Ma W, Wang X, Zhao L, and Wong WT (2019). C3- and CR3-dependent microglial clearance protects photoreceptors in retinitis pigmentosa. J. Exp. Med. 216, 1925–1943. 10.1084/jem.20190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Koren EG, Yu C, Klingeborn M, Wong AYW, Prigge CL, Mathew R, Kalnitsky J, Msallam RA, Silvin A, Kay JN, et al. (2019). Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity 50, 723–737.e7. 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusch N, Ravichandran KA, Olabiyi BF, Komorowska-Müller JA, Hansen JN, Ulas T, Beyer M, Zimmer A, and Schmöle AC (2022). Cannabinoid receptor 2 is necessary to induce toll-like receptor-mediated microglial activation. Glia 70, 71–88. 10.1002/glia.24089. [DOI] [PubMed] [Google Scholar]
- 7.Block ML, Zecca L, and Hong JS (2007). Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat. Rev. Neurosci. 8, 57–69. 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 8.Hickman S, Izzy S, Sen P, Morsett L, and El Khoury J (2018). Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369. 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tham YC, Li X, Wong TY, Quigley HA, Aung T, and Cheng CY (2014). Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090. 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Weinreb RN, Aung T, and Medeiros FA (2014). The pathophysiology and treatment of glaucoma: a review. JAMA 311, 1901–1911. 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosco A, Steele MR, and Vetter ML (2011). Early microglia activation in a mouse model of chronic glaucoma. J. Comp. Neurol. 519, 599–620. 10.1002/cne.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gramlich OW, Beck S, von Thun Und Hohenstein-Blaul N, Boehm N, Ziegler A, Vetter JM, Pfeiffer N, and Grus FH (2013). Enhanced insight into the autoimmune component of glaucoma: IgG autoantibody accumulation and pro-inflammatory conditions in human glaucomatous retina. PLoS One 8, e57557. 10.1371/journal.pone.0057557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell GR, Soto I, Zhu X, Ryan M, Macalinao DG, Sousa GL, Caddle LB, MacNicoll KH, Barbay JM, Porciatti V, et al. (2012). Radiation treatment inhibits monocyte entry into the optic nerve head and prevents neuronal damage in a mouse model of glaucoma. J. Clin. Invest. 122, 1246–1261. 10.1172/JCI61135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosco A, Romero CO, Breen KT, Chagovetz AA, Steele MR, Ambati BK, and Vetter ML (2015). Neurodegeneration severity can be predicted from early microglia alterations monitored in vivo in a mouse model of chronic glaucoma. Dis. Model. Mech. 8, 443–455. 10.1242/dmm.018788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X, Cho KS, Thee EF, Jager MJ, and Chen DF (2019). Neuroinflammation and microglia in glaucoma: time for a paradigm shift. J. Neurosci. Res. 97, 70–76. 10.1002/jnr.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Xiao Z, Pan L, Zhuang D, Cho KS, Robert K, Chen X, Shu L, Tang G, Wu J, et al. (2020). Therapeutic Targeting of Retinal Immune Microenvironment With CSF-1 Receptor Antibody Promotes Visual Function Recovery After Ischemic Optic Neuropathy. Front. Immunol. 11, 585918. 10.3389/fimmu.2020.585918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Cho KS, Vu THK, Shen CH, Kaur M, Chen G, Mathew R, McHam ML, Fazelat A, Lashkari K, et al. (2018). Commensal microflora-induced T cell responses mediate progressive neurodegeneration in glaucoma. Nat. Commun. 9, 3209. 10.1038/s41467-018-05681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman SM, and Wong WT (2018). Microglia in the Retina: Roles in Development, Maturity, and Disease. Annu. Rev. Vis. Sci. 4, 45–77. 10.1146/annurev-vision-091517-034425. [DOI] [PubMed] [Google Scholar]
- 19.Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, et al. (2016). Systems proteomics of liver mitochondria function. Science 352, aad0189. 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashbrook DG, Arends D, Prins P, Mulligan MK, Roy S, Williams EG, Lutz CM, Valenzuela A, Bohl CJ, Ingels JF, et al. (2021). A platform for experimental precision medicine: The extended BXD mouse family. Cell Syst. 12, 235–247.e9. 10.1016/j.cels.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams EG, Pfister N, Roy S, Statzer C, Haverty J, Ingels J, Bohl C, Hasan M, Čuklina J, Bühlmann P, et al. (2022). Multiomic profiling of the liver across diets and age in a diverse mouse population. Cell Syst. 13, 43–57.e6. 10.1016/j.cels.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson MG, Nair KS, Amonoo LA, Mehalow A, Trantow CM, Masli S, and John SWM (2008). GpnmbR150X allele must be present in bone marrow derived cells to mediate DBA/2J glaucoma. BMC Genet. 9, 30. 10.1186/1471-2156-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, et al. (2016). Joint mouse-human phenome-wide association to test gene function and disease risk. Nat. Commun. 7, 10464. 10.1038/ncomms10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang B, Smith RS, Hawes NL, Anderson MG, Zabaleta A, Savinova O, Roderick TH, Heckenlively JR, Davisson MT, and John SW (1999). Interacting loci cause severe iris atrophy and glaucoma in DBA/2J mice. Nat. Genet. 21, 405–409. [DOI] [PubMed] [Google Scholar]
- 25.Geisert EE, and Williams RW (2020). Using BXD mouse strains in vision research: A systems genetics approach. Mol. Vis. 26, 173–187. [PMC free article] [PubMed] [Google Scholar]
- 26.Geisert EE, Lu L, Freeman-Anderson NE, Templeton JP, Nassr M, Wang X, Gu W, Jiao Y, and Williams RW (2009). Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol. Vis. 15, 1730–1763. [PMC free article] [PubMed] [Google Scholar]
- 27.King R, Lu L, Williams RW, and Geisert EE (2015). Transcriptome networks in the mouse retina: An exon level BXD RI database. Mol. Vis. 21, 1235–1251. [PMC free article] [PubMed] [Google Scholar]
- 28.Chintalapudi SR, and Jablonski MM (2017). Systems Genetics Analysis to Identify the Genetic Modulation of a Glaucoma-Associated Gene. Methods Mol. Biol. 1488, 391–417. 10.1007/978-1-4939-6427-7_18. [DOI] [PubMed] [Google Scholar]
- 29.Chintalapudi SR, Maria D, Di Wang X, Bailey JNC, NEIGHBOR-HOOD consortium; International Glaucoma Genetics consortium; Hysi PG, Wiggs JL, Williams RW, and Jablonski MM (2017). Systems genetics identifies a role for Cacna2d1 regulation in elevated intraocular pressure and glaucoma susceptibility. Nat. Commun. 8, 1755. 10.1038/s41467-017-00837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman NE, Templeton JP, Orr WE, Lu L, Williams RW, and Geisert EE (2011). Genetic networks in the mouse retina: growth associated protein 43 and phosphatase tensin homolog network. Mol. Vis. 17, 1355–1372. [PMC free article] [PubMed] [Google Scholar]
- 31.Guo C, Cho KS, Li Y, Tchedre K, Antolik C, Ma J, Chew J, Utheim TP, Huang XA, Yu H, et al. (2018). IGFBPL1 Regulates Axon Growth through IGF-1-mediated Signaling Cascades. Sci. Rep. 8, 2054. 10.1038/s41598-018-20463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e6. 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Wei X, Cho KS, Chen G, Sappington R, Calkins DJ, and Chen DF (2011). Optic neuropathy due to microbead-induced elevated intraocular pressure in the mouse. Invest. Ophthalmol. Vis. Sci. 52, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prinz M, and Priller J (2014). Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15, 300–312. 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 36.Kambhampati SP, Clunies-Ross AJM, Bhutto I, Mishra MK, Edwards M, McLeod DS, Kannan RM, and Lutty G (2015). Systemic and Intravitreal Delivery of Dendrimers to Activated Microglia/Macrophage in Ischemia/Reperfusion Mouse Retina. Invest. Ophthalmol. Vis. Sci. 56, 4413–4424. 10.1167/iovs.14-16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Z, Wu K, Gu L, Huang L, Zhuge Q, Yang S, and Wang Z (2020). IGF-1R stimulation alters microglial polarization via TLR4/NF-kappaB pathway after cerebral hemorrhage in mice. Brain Res. Bull. 164, 221–234. 10.1016/j.brainresbull.2020.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Weiwer M (2018). Exploring COX-2 as a biomarker of microglia-dependent neuroinflammation. J. Nucl. Med. 59, 1628. [Google Scholar]
- 39.Dukay B, Csoboz B, and Tó th ME. (2019). Heat-Shock Proteins in Neuroinflammation. Front. Pharmacol. 10, 920. 10.3389/fphar.2019.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin C, Cleveland JC, Ao L, Li J, Zeng Q, Fullerton DA, and Meng X (2014). Human myocardium releases heat shock protein 27 (HSP27) after global ischemia: the proinflammatory effect of extracellular HSP27 through toll-like receptor (TLR)-2 and TLR4. Mol. Med. 20, 280–289. 10.2119/molmed.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung W-S, Peterson TC, € et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia 541, pp. 481–487.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 113, E1738–E1746. 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. (2014). Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143. 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128. 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ronning KE, Karlen SJ, Miller EB, and Burns ME (2019). Molecular profiling of resident and infiltrating mononuclear phagocytes during rapid adult retinal degeneration using single-cell RNA sequencing. Sci. Rep. 9, 4858. 10.1038/s41598-019-41141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al.(2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao XF, Alam MM, Liao Y, Huang T, Mathur R, Zhu X, and Huang Y (2019). Targeting Microglia Using Cx3cr1-Cre Lines: Revisiting the Specificity. eNeuro 6, ENEURO.0114, 19.2019. 10.1523/ENEURO.0114-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen DV, Hanson JE, and Sheng M (2018). Microglia in Alzheimer’s disease. J. Cell Biol. 217, 459–472. 10.1083/jcb.201709069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohlen CJ, Friedman BA, Dejanovic B, and Sheng M (2019). Microglia in Brain Development, Homeostasis, and Neurodegeneration. Annu. Rev. Genet. 53, 263–288. 10.1146/annurev-genet-112618-043515. [DOI] [PubMed] [Google Scholar]
- 51.Selkoe DJ (1996). Amyloid beta-protein and the genetics of Alzheimer’s disease. J. Biol. Chem. 271, 18295–18298. 10.1074/jbc.271.31.18295. [DOI] [PubMed] [Google Scholar]
- 52.Yankner BA, and Lu T (2009). Amyloid beta-protein toxicity and the pathogenesis of Alzheimer disease. J. Biol. Chem. 284, 4755–4759. 10.1074/jbc.R800018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pickett EK, Herrmann AG, McQueen J, Abt K, Dando O, Tulloch J, Jain P, Dunnett S, Sohrabi S, Fjeldstad MP, et al. (2019). Amyloid beta and tau cooperate to cause reversible behavioral and transcriptional deficits in a model of Alzheimer’s disease. Cell Rep. 29, 3592–3604.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. (2015). TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071. 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142. 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bui BV, and Fortune B (2004). Ganglion cell contributions to the rat full-field electroretinogram. J. Physiol. 555, 153–173. 10.1113/jphysiol.2003.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fortune B, Bui BV, Morrison JC, Johnson EC, Dong J, Cepurna WO, Jia L, Barber S, and Cioffi GA (2004). Selective ganglion cell functional loss in rats with experimental glaucoma. Invest. Ophthalmol. Vis. Sci. 45, 1854–1862. 10.1167/iovs.03-1411. [DOI] [PubMed] [Google Scholar]
- 58.Shi C, Yuan X, Chang K, Cho KS, Xie XS, Chen DF, and Luo G (2018). Optimization of Optomotor Response-based Visual Function Assessment in Mice. Sci. Rep. 8, 9708. 10.1038/s41598-018-27329-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Damani MR, Zhao L, Fontainhas AM, Amaral J, Fariss RN, and Wong WT (2011). Age-related alterations in the dynamic behavior of microglia. Aging Cell 10, 263–276. 10.1111/j.1474-9726.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mouton PR, Long JM, Lei DL, Howard V, Jucker M, Calhoun ME, and Ingram DK (2002). Age and gender effects on microglia and astrocyte numbers in brains of mice. Brain Res. 956, 30–35. 10.1016/s0006-8993(02)03475-3. [DOI] [PubMed] [Google Scholar]
- 61.Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, and Roninson IB (2000). Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc. Natl. Acad. Sci. USA 97, 4291–4296. 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang LYL, Ardiles AO, Tapia-Rojas C, Araya J, Inestrosa NC, Palacios AG, and Acosta ML (2020). Evidence of Synaptic and Neurochemical Remodeling in the Retina of Aging Degus. Front. Neurosci. 14, 161. 10.3389/fnins.2020.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prinz M, Jung S, and Priller J (2019). Microglia Biology: One Century of Evolving Concepts. Cell 179, 292–311. 10.1016/j.cell.2019.08.053. [DOI] [PubMed] [Google Scholar]
- 64.Khanh Vu TH, Chen H, Pan L, Cho KS, Doesburg D, Thee EF, Wu N, Arlotti E, Jager MJ, and Chen DF (2020). CD4(+) T-Cell Responses Mediate Progressive Neurodegeneration in Experimental Ischemic Retinopathy. Am. J. Pathol. 190, 1723–1734. 10.1016/j.ajpath.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, and Tsai LH (2017). Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell Rep. 21, 366–380. 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 67.Song WM, and Colonna M (2018). The identity and function of microglia in neurodegeneration. Nat. Immunol. 19, 1048–1058. 10.1038/s41590-018-0212-1. [DOI] [PubMed] [Google Scholar]
- 68.Mifflin L, Hu Z, Dufort C, Hession CC, Walker AJ, Niu K, Zhu H, Liu N, Liu JS, Levin JZ, et al. (2021). A RIPK1-regulated inflammatory microglial state in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 118, e2025102118. 10.1073/pnas.2025102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spittau B, Dokalis N, and Prinz M (2020). The Role of TGFbeta Signaling in Microglia Maturation and Activation. Trends Immunol. 41, 836–848. 10.1016/j.it.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kündig TM, Frei K, Ginhoux F, Merad M, and Becher B (2012). Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060. 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma W, Silverman SM, Zhao L, Villasmil R, Campos MM, Amaral J, and Wong WT (2019). Absence of TGFbeta signaling in retinal microglia induces retinal degeneration and exacerbates choroidal neovascularization. Elife 8, e42049. 10.7554/eLife.42049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, et al. (2013). TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368, 117–127. 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ulland TK, and Colonna M (2018). TREM2 - a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 14, 667–675. 10.1038/s41582-018-0072-1. [DOI] [PubMed] [Google Scholar]
- 75.He S, and Sharpless NE (2017). Senescence in Health and Disease. Cell 169, 1000–1011. 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, Balic A, Giladi A, Sheban F, Dutertre CA, et al. (2020). Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 181, 746. 10.1016/j.cell.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 77.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, and Littman DR (2000). Analysis of fractalkine receptor CX(3) CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 20, 4106–4114. 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Howell GR, Libby RT, and John SWM (2008). Mouse genetic models: an ideal system for understanding glaucomatous neurodegeneration and neuroprotection. Prog. Brain Res. 173, 303–321. 10.1016/S0079-6123(08)01122-9. [DOI] [PubMed] [Google Scholar]
- 79.Oherle K, Acker E, Bonfield M, Wang T, Gray J, Lang I, Bridges J, Lewkowich I, Xu Y, Ahlfeld S, et al. (2020). Insulin-like Growth Factor 1 Supports a Pulmonary Niche that Promotes Type 3 Innate Lymphoid Cell Development in Newborn Lungs. Immunity 52, 275–294.e9. 10.1016/j.immuni.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 82.Mulligan MK, Mozhui K, Prins P, and Williams RW (2017). GeneNetwork: A Toolbox for Systems Genetics. Methods Mol. Biol. 1488, 75–120. 10.1007/978-1-4939-6427-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Homayouni R, Heinrich K, Wei L, and Berry MW (2005). Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics 21, 104–115. 10.1093/bioinformatics/bth464. [DOI] [PubMed] [Google Scholar]
- 84.Liao Y, Wang J, Jaehnig EJ, Shi Z, and Zhang B (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205. 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benjamini Y, Drai D, Elmer G, Kafkafi N, and Golani I (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284. 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 86.Ziebarth JD, Bhattacharya A, and Cui Y (2013). Bayesian Network Webserver: a comprehensive tool for biological network modeling. Bioinformatics 29, 2801–2803. [DOI] [PubMed] [Google Scholar]
- 87.Ziebarth JD, and Cui Y (2017). Precise network modeling of systems genetics data using the Bayesian network webserver. In Systems Genetics (Springer; ), pp. 319–335. [DOI] [PubMed] [Google Scholar]
- 88.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Avignone E, Lepleux M, Angibaud J, and Nägerl UV (2015). Altered morphological dynamics of activated microglia after induction of status epilepticus. J. Neuroinflammation 12, 202. 10.1186/s12974-015-0421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lennikov A, Mukwaya A, Saddala MS, and Huang H (2021). Deficiency of C-X-C chemokine receptor type 5 (CXCR5) gene causes dysfunction of retinal pigment epithelium cells. Lab. Invest. 101, 228–244. 10.1038/s41374-020-00491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang H, Lennikov A, Saddala MS, Gozal D, Grab DJ, Khalyfa A, and Fan L (2019). Placental growth factor negatively regulates retinal endothelial cell barrier function through suppression of glucose-6-phosphate dehydrogenase and antioxidant defense systems. Faseb. J. : official publication of the Federation of American Societies for Experimental Biology 33, 13695–13709. 10.1096/fj.201901353R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, and Lempicki RA (2007). The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, R183. 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jacomy M, Venturini T, Heymann S, and Bastian M (2014). ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One 9, e98679. 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Faure L, Soldatov R, Kharchenko PV, and Adameyko I (2023). scFates: a scalable python package for advanced pseudotime and bifurcation analysis from single-cell data. Bioinformatics 39. 10.1093/bioinformatics/btac746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wickham H (2016). Data Analysis. In ggplot2: Elegant Graphics for Data Analysis, Wickham H, ed. (Springer International Publishing; ), pp. 189–201. 10.1007/978-3-319-24277-4_9. [DOI] [Google Scholar]
- 99.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table. To provide the possible updates, troubleshooting, and code optimizations, we have GitHub repository available at https://github.com/mcrewcow/Pan_et_al_DFChen_IGFPL1_paper/tree/main.
The scRNA-seq datasets are deposited in the Genome Expression Omnibus under the accession numbers GEO: GSE176204, and the bulk RNA-seq datasets are deposited in the Sequence Read Archive (SRA) with BioProject ID: PRJNA739974.
The code and processed annotated datasets generated in this study for reproducing the bioinformatic analysis can be found on the following GitHub repository: https://github.com/mcrewcow/Pan_et_al_DFChen_IGFPL1_paper. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-BRN3a | Millipore | Cat# MAB1585, RRID: AB_94166 |
| Rabbit anti-IBA-1 | FUJIFILM Wako Shibayagi | Cat# 019–19741, RRID: AB_839504 |
| Goat anti-IGFBPL1 | R&D systems | Cat# AF4130, RRID: AB_2279980 |
| Goat anti-IGF-1R | R&D systems | Cat# AF-305-NA, RRID: AB_354457 |
| Rabbit anti-COX2 | Thermo Fisher Scientific | Cat# MA5–14568, RRID: AB_10984436 |
| anti-pNFκB | Cell Signaling Technology | Cat# 3033; RRID: AB_331284 |
| anti-tNFκB | Cell Signaling Technology | Cat# 9460, RRID:AB_2797704 |
| anti-pIGF1r | Cell Signaling Technology | Cat# 3027, RRID:AB_2122378 |
| anti-IGF1r | Millipore | Cat# ABE332, RRID:AB_10918911 |
| Mouse anti-CD68 | Abcam | Cat# ab201340, RRID: AB_2920880 |
| Mouse anti-phospho-Tau (Ser202, Thr205, AT8) | Thermo Fisher Scientific | Cat# MN1020, RRID: AB_223647 |
| Mouse anti-phospho-Tau (Thr181, AT270) | Thermo Fisher Scientific | Cat# MN1050, RRID: AB_223651 |
| Rabbit anti-phosphor-Tau (Thr231, AT180) | Thermo Fisher Scientific | Cat# 44–746G, RRID: AB_2533742 |
| Mouse anti-Tau | Thermo Fisher Scientific | Cat# MN1000, RRID: AB_2314654 |
| Rabbit anti-β-amyloid1–42 | Thermo Fisher Scientific | Cat# 700254, RRID: AB_2532306 |
| Rabbit anti-cleaved Caspase3 | Cell Signaling Technology | Cat# 9661, RRID: AB_2341188 |
| Mouse anti-GFAP-CY3 | Sigma-Aldrich | Cat# C9205, RRID: AB_476889 |
| Mouse anti-β-actin | Thermo Fisher Scientific | Cat# MA5–15739, RRID: AB_10979409 |
| Rabbit anti-GFAP | Abcam | Cat# ab7260, RRID:AB_305808 |
| Rabbit anti-TREM2 | Thermo Fisher Scientific | Cat# PA5–116068, RRID: AB_2900702 |
| Rat anti-anti-mouse CD11b | Biolegend | Cat# 101224, RRID: AB_755986 |
| Guinea pig anti-RBPMS | PhosphoSolutions | Cat# 1832-RBPMS, RRID: AB_2492226 |
| Rabbit anti-PSD95 | Abcam | Cat# ab18258, RRID: AB_444362 |
| Mouse anti-Syn | Millipore | Cat# MAB5258, RRID: AB_2313839 |
| Goat Anti-Mouse IgG (H + L), Biotinylated | Vector Laboratories | Cat# BA-9200, RRID: AB_2336171 |
| Goat Anti-Rabbit IgG (H + L), Biotinylated | Vector Laboratories | Cat# BA-1000, RRID: AB_2313606 |
| Rabbit Anti-Goat IgG (H + L), Biotinylated | Vector Laboratories | Cat# BA-5000, RRID: AB_2336126 |
| Streptavidin, Alexa Fluor 546 conjugate | Thermo Fisher Scientific | Cat# S-11225, RRID: AB_2532130 |
| Streptavidin, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat# S32354, RRID: AB_2315383 |
| Cy2 Donkey Anti-rabbit | Jackson ImmunoResearch Labs | Cat# 711–225-152, RRID: AB_2340612 |
| Cy3 Donkey Anti-mouse | Jackson ImmunoResearch Labs | Cat# 715–165-150, RRID: AB_2340813 |
| Cy3 Donkey Anti-goat | Jackson ImmunoResearch Labs | Cat# 705–165-147, RRID: AB_2307351 |
|
| ||
| Biological samples | ||
|
| ||
| Human Eye: Retina (Normal) | Florida Lions Eye Bank, Bascom Palmer Eye Institute University of Miami; Miller School of Medicine | Case# 2021–32323; Donor #21–12-050 |
| Human Eye: Retina (Glaucoma) | Lions Gift of Sight University of Minnesota | Patient ID: 22–0250 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat# A7030–100G |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| Tween 20 | Sigma-Aldrich | Cat# P5927 |
| Normal Donkey Serum | Jackson ImmunoResearch Labs | Cat# 017–000-121, RRID: AB_2337258 |
| Normal Goat Serum | Jackson ImmunoResearch Labs | Cat# 005–000-121, RRID: AB_2336990 |
| Collagenase/Dispase | Roche | Cat# 11097113001 |
| rmCSF1 | PeproTech | Cat# 315–02 |
| LPS | Sigma Aldrich | Cat# L6529–1MG |
| rmIGFBPL1 | R&D systems | Cat# 4130-BL |
| IGF1Ri H-1356 | BACHEM | Cat# 4018631 |
| ATP | Thermo Fisher Scientific | Cat# R0441 |
| Anti-CD11b Magnetic Particles | BD Biosciences | Cat# 558013, RRID: AB_398649 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat# 1000269 |
| Proteome Profiler Mouse Cytokine Array Kit | R&D Systems | Cat# ARY006 |
|
| ||
| Deposited data | ||
|
| ||
| Searchable microglia gene expression website | This paper | www.genecards.org |
|
| ||
| Experimented models: Cell lines | ||
|
| ||
| Human microglial cell line HMC3 | ATCC | Cat# CRL-3304, RRID: CVCL_II76 |
| Mouse microglial cell line BV2 | ATCC | Cat# CRL-2469, RRID: CVCL_5745 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | The Jackson Laboratory | Stock No: 000664, RRID: IMSR_JAX:000664 |
| Cx3cr1GFP/GFP | The Jackson Laboratory | B6.129P-Cx3cr1tm1Litt/J, Stock No: 005582, RRID: IMSR_JAX:005582 |
| DBA/2J | The Jackson Laboratory | Stock No: 000671, RRID: IMSR_JAX:000671 |
| Igf1r(flox/flox) | The Jackson Laboratory | B6; 129-Igf1rtm2Arge/J, Stock No: 012251, RRID: IMSR_JAX:012251 |
| Cx3cr1tm2.1(cre/ERT2) | The Jackson Laboratory | B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J, Stock No: 020940, RRID: IMSR_JAX:020940 |
| Igfbpl1−/− (KO) | Velocigene Regeneron Pharmaceuticals | C57BL/6N-Igfbpl1tm1(KOMP)Vlcg/Mmucd, MGI ID: MGI:1933198, Stock No: 048159- UCD, RRID: MMRRC_048159-UCD |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 | N/A | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Cell Ranger | 10x Genomics | Version 3.1.0 |
| R | https://www.r-project.org/ | 4.2.1 |
| RStudio | https://posit.co/ | 2022.07.2 Build 576 |
| biomaRt | https://bioconductor.org/packages/release/bioc/html/biomaRt.html | 2.48.3 |
| cowplot | https://cran.r-project.org/web/packages/cowplot/ | 1.1.1 |
| dittoSeq | https://bioconductor.org/packages/release/bioc/html/dittoSeq.html | 1.4.4 |
| dplyr | https://dplyr.tidyverse.org/ | 1.08 |
| EnhancedVolcano | https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html | 1.10.0 |
| escape | https://github.com/ncborcherding/escape | 1.2.0 |
| ggplot2 | https://ggplot2.tidyverse.org/ | 3.3.5 |
| GSEABase | https://bioconductor.org/packages/release/bioc/html/GSEABase.html | 1.54.0 |
| htmlwidgets | https://www.htmlwidgets.org/ | 1.5.4 |
| Matrix | https://cran.r-project.org/web/packages/Matrix/index.html | 1.3–4 |
| patchwork | https://patchwork.data-imaginist.com/ | 1.1.1 |
| plotly | https://plotly.com/r/ | 4.10.0 |
| reticulate | https://rstudio.github.io/reticulate/ | 1.24 |
| Seurat | https://satijalab.org/seurat/ | 3.0.0/4.1.0 |
| SeuratDisk | https://github.com/mojaveazure/seurat-disk | 0.0.0.9019 |
| SeuratWrappers | https://github.com/satijalab/seurat-wrappers | 0.3.0 |
| Python | https://www.python.org/ | 3.8 |
| Jupyter Notebook | https://jupyter.org/ | 6.4.11 |
| scanpy | https://scanpy.readthedocs.io/en/stable/ | 1.9.1 |
| scFates | https://scfates.readthedocs.io/en/latest/index.html | 0.9.0 |
| anndata | https://anndata.readthedocs.io/en/latest/ | 0.8.0 |
| numpy | https://numpy.org/ | 1.21.6 |
| pandas | https://pandas.pydata.org/ | 1.1.5 |
| palantir | https://www.palantir.com/docs/foundry/transforms-python/use-python-libraries/ | 1.0.1 |
| matplotlib | https://matplotlib.org/ | 3.5.1 |
| seaborn | https://seaborn.pydata.org/ | 0.11.2 |
| scvelo | https://scvelo.readthedocs.io/en/stable/ | 0.2.4 |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/ |
| ImageJ | National Institutes of Health | https://ImageJ.nih.gov/ij/ |
| Gene Ontology | Webgestalt | http://webgestalt.org/option.php |
| Protein-protein interaction | String | https://www.string-db.org/ |
| Zenodo Code Repository | Zenodo (https://zenodo.org/record/8118833) | https://doi.org/10.5281:zenodo/8118833 |


