Abstract
Induction of cyclin-dependent kinase inhibitor p21Waf1/Cip1/Sdi1 triggers cell growth arrest associated with senescence and damage response. Overexpression of p21 from an inducible promoter in a human cell line induces growth arrest and phenotypic features of senescence. cDNA array hybridization showed that p21 expression selectively inhibits a set of genes involved in mitosis, DNA replication, segregation, and repair. The kinetics of inhibition of these genes on p21 induction parallels the onset of growth arrest, and their reexpression on release from p21 precedes the reentry of cells into cell cycle, indicating that inhibition of cell-cycle progression genes is a mechanism of p21-induced growth arrest. p21 also up-regulates multiple genes that have been associated with senescence or implicated in age-related diseases, including atherosclerosis, Alzheimer's disease, amyloidosis, and arthritis. Most of the tested p21-induced genes were not activated in cells that had been growth arrested by serum starvation, but some genes were induced in both forms of growth arrest. Several p21-induced genes encode secreted proteins with paracrine effects on cell growth and apoptosis. In agreement with the overexpression of such proteins, conditioned media from p21-induced cells were found to have antiapoptotic and mitogenic activity. These results suggest that the effects of p21 induction on gene expression in senescent cells may contribute to the pathogenesis of cancer and age-related diseases.
Induction of the cyclin-dependent kinase (CDK) inhibitor p21Waf1/Cip1/Sdi1 is a common mechanism of growth arrest in different physiological situations. p21 is transiently induced in the course of replicative senescence, reversible and irreversible forms of damage-induced growth arrest, and terminal differentiation of postmitotic cells; its induction is regulated through p53-dependent and -independent mechanisms (1). Ectopic overexpression of p21 leads to cell growth arrest in G1 and G2 (2); this arrest is accompanied by phenotypic markers of senescence in some or all cells (3–5). Although p21 is not a transcription factor, it is conceivable that some of its functions may be mediated by indirect effects of p21 on cellular gene expression. Thus, CDK inhibition by p21 results in dephosphorylation of Rb and the inhibition of E2F transcription factors that regulate many genes involved in DNA replication and cell-cycle progression (6). Accordingly, p21 was shown to be involved in radiation-induced inhibition of several E2F-regulated genes (7). Transient transfection assays showed that p21 can stimulate NFκB-mediated transcription; this effect of p21 has been explained through the interaction of Cdk2 with transcriptional cofactor p300 that augments NFκB and other inducible transcription factors (8). p21 interactions with proteins other than CDK may also have a potential effect on gene expression. For example, p21 was reported to bind c-Jun amino-terminal kinases, apoptosis signal-regulating kinase 1 and Gadd45 (1, 9). Furthermore, the C-terminal portion of p21, which binds the proliferating cell nuclear antigen and is not involved in CDK inhibition, is required for the inhibition of keratinocyte differentiation markers by p21 (10). In the present paper, we report that p21 selectively inhibits or induces sets of genes with distinct biological functions in cell division and aging, suggesting a role for p21 in the pathogenesis of cancer and age-related diseases.
Materials and Methods
Cell Growth and Apoptosis Assays.
All cell lines were propagated in DMEM with 10% FC2 serum (HyClone). Derivation of HT1080 p21–9 cell line that carries p21 in an isopropyl-β-d-thiogalactoside (IPTG)-inducible retroviral vector has been previously described (5). This cell line is p16 deficient and expresses wild-type Rb and p53, as we have shown by PCR sequencing of all of the exons of p53 in the cell line from which p21–9 was derived (5). [3H]Thymidine labeling and mitotic index were measured as previously described (11). Conditioned media were prepared by plating 106 p21–9 cells per 15-cm plate, adding 50 μM IPTG the next day, and replacing the media 3 days later with media containing IPTG and 0.5% serum; the conditioned media were collected 2 days later and stored at 4°C up to 20 days. Control IPTG-free conditioned media containing 0.5% serum were collected from untreated cells grown to the same density as IPTG-treated cells.
HS 15.T cells were from the American Type Culture Collection. For mitogenic activity assays, HS 15.T cells were plated in 12-well plates at 15,000 cells per well and 2 days later given different types of media. After 60 h of growth, [3H]-thymidine (3.13 μCi/ml) was added for 24 h, cells were collected, and [3H]thymidine incorporation determined as described (12). C8 cells were kindly provided by Andrei Gudkov (University of Illinois at Chicago). For apoptosis assays, 3 × 105 C8 cells were plated per 6-cm plate and exposed the next day to fresh media with 0.4% serum or to conditioned media (no fresh serum added). Floating cells recovered from media supernatant and attached cells collected by trypsinization were counted. Cells were analyzed for apoptotic morphology after staining with 5 μg/ml 4′,6-diamidino-2-phenylindole, and FACS analysis of the DNA content was carried out as described (13). DNA ladder formation was analyzed by electrophoresis in 1% agarose gel. The number of attached surviving cells was determined 48 h after media change by cell counting or methylene blue staining.
cDNA Array Hybridization and Gene Expression Assays.
Poly(A)+ RNA was isolated from untreated p21–9 cells and from cells treated for 3 days with 50 μM IPTG. cDNA probe synthesis, hybridization with the Human UniGEM V cDNA microarray, and signal analysis were conducted by Genome Systems (St. Louis), as described at the company's website, http://www.genomesystems.com. More than 2,500 genes and expressed sequence tags (ESTs) showed measurable hybridization signals with both probes. The sequences of clones that showed differential hybridization were analyzed by blast homology search against all available sequence databases.
Reverse transcription–PCR (RT-PCR) analysis was carried out essentially as described (14); sequences of RT-PCR primers and PCR conditions will be provided on request. Northern hybridization was carried out by using inserts of sequence-verified cDNA clones (from Genome Systems) as probes. For immunoblotting, protein concentrations in all samples were equalized after measurement with the Bio-Rad protein assay kit. The following primary antibodies were used: mouse monoclonal antibodies against Cdc2 (Santa Cruz Biotechnology), cyclin A (NeoMarkers, Fremont, CA), Plk1 (polo-like kinase) (Zymed), Rb (PharMingen), and fibronectin (Transduction Laboratories, Lexington, KY); rabbit polyclonal antibodies against Mad2 (BabCo, Richmond, CA), p107 (Santa Cruz), connective tissue growth factor (CTGF) (a gift of L. Lau, University of Illinois at Chicago), Prc1 (a gift of W. Jiang and T. Hunter, The Salk Institute), topoisomerase IIα (Ab-284; a gift of W. T. Beck, University of Illinois at Chicago) and serum amyloid (SAA) (a gift of B. M. Schreiber, Boston University) and sheep polyclonal antibody against SOD2 (Calbiochem). Horseradish peroxidase-conjugated secondary antibodies were goat anti-mouse and goat anti-rabbit IgG (Santa Cruz) and rabbit anti-sheep IgG (Kirkegaard & Perry Laboratories).
Results and Discussion
cDNA Array Analysis of p21 Effects on Cellular Gene Expression.
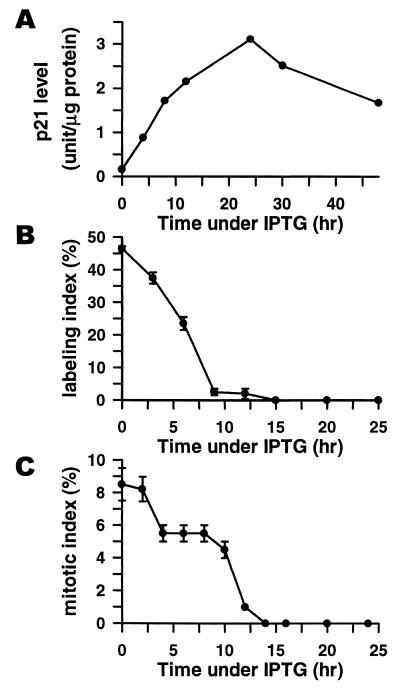
Cell line p21–9 is a derivative of HT1080 human fibrosarcoma cells, where p21 expression can be turned on or off by using a physiologically neutral agent IPTG (5). p21 is rapidly induced in p21–9 cells after the addition of 50 μM IPTG (Fig. 1A), and this induction is accompanied by rapid (within 14 h) cessation of DNA replication (Fig. 1B) and mitosis (Fig. 1C), with approximately equal numbers of cells arresting in G1 and G2 (11). All IPTG-treated p21–9 cells develop morphological and enzymatic markers of senescence, including enlarged and flattened morphology and senescence-associated β-galactosidase activity (5). The senescent phenotype develops subsequently to cell growth arrest, starting at about 48 h after the addition of IPTG. p21 induction is also accompanied by the loss of clonogenicity, a process that correlates with the duration and level of p21 induction and is associated with endoreduplication and abnormal mitosis after release from IPTG (11).
Figure 1.
(A) Time course of p21 induction after the addition of 50 μM IPTG. p21 levels (in arbitrary units) were determined by ELISA by using WAF1 ELISA kit (Oncogene Science). (B) Time course of changes in [3H]thymidine labeling index (determined by autoradiography) after the addition of 50 μM IPTG. (C) Time course of changes in mitotic index (determined microscopically after 4′,6-diamidino-2-phenylindole staining) after the addition of 50 μM IPTG.
To analyze the effects of p21 on gene expression, cDNA probes were generated from the RNA of untreated p21–9 cells and cells treated with IPTG for 3 days, a period that allows for full development of the senescent phenotype. These probes were used for differential hybridization with the Human UniGEM V cDNA microarray (Genome Systems), which contains over 4,000 sequence-verified known human genes and 3,000 ESTs. Genes that were down-regulated with balanced differential expression ≥2.5 or up-regulated with balanced differential expression ≥2.0 are listed in the supplemental data, Tables 1 and 2 (www.pnas.org). Expression of 69 genes was individually tested by RT-PCR or Northern hybridization (see examples in Fig. 2) by using RNA preparations from independent experiments, and the predicted changes were confirmed for 38/39 down-regulated and 27/30 up-regulated genes. The observed signal differences in Northern hybridization or RT-PCR for most of the tested genes appeared to be higher than the values of balanced differential expression determined from the cDNA array, suggesting that cDNA array hybridization tends to underestimate the magnitude of changes in gene expression. Changes in the expression of six down-regulated and six up-regulated genes were also tested at the protein level (see examples in Fig. 2 A and B) and confirmed in all cases.
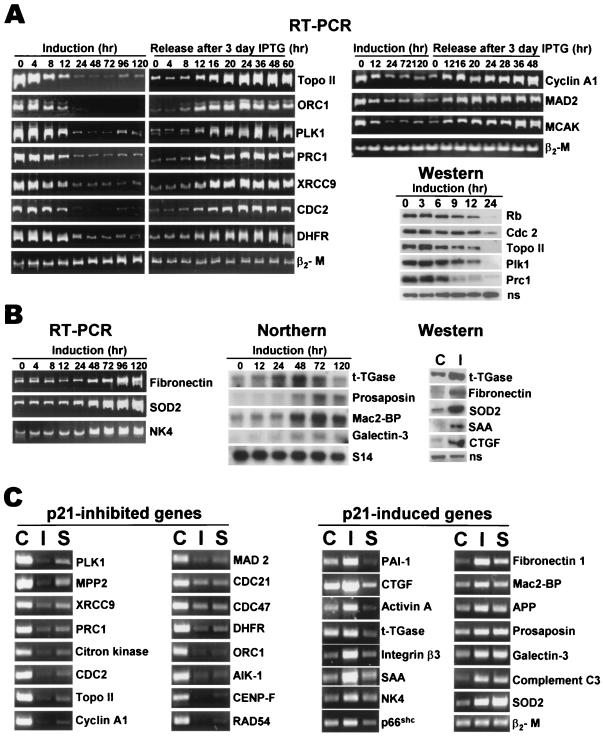
Figure 2.
(A) RT-PCR and immunoblotting analysis of the time course of changes in the expression of p21-inhibited genes on IPTG addition and release. β2-microglobulin (β2-M) was used as a normalization control for RT-PCR. A nonspecific 110-kDa band that crossreacts with the Plk1 antibody (ns) is shown as a normalization control for Western blots. (B) RT-PCR and Northern hybridization analysis of the time course of changes in the expression of p21-induced genes on IPTG addition and immunoblotting assays for IPTG-induced gene products. Control untreated p21–9 cells; I, cells treated for 3 days with 50 μM IPTG. S14 ribosomal protein gene was used as a normalization control for Northern hybridization. (C) RT-PCR analysis of changes in gene expression in IPTG-treated (I) and serum-starved (S) cells relative to exponentially growing control (C) cells. β2-M was used as a normalization control.
p21 Selectively Inhibits Genes Involved in Cell-Cycle Progression and DNA Repair.
Sixty-nine genes and three ESTs were identified by the cDNA array as down-regulated in p21-induced cells, with balanced differential expression of 2.5–12.6; five additional genes were found to be down-regulated by separate RT-PCR assays. As expected, some p21-inhibited genes (e.g., CDC2, ORC1, dihydrofolate reductase) contained E2F sites in their promoters. On the other hand, no E2F sites could be found in the promoters of other p21-inhibited genes (e.g., cyclin B1), and some E2F-dependent genes (e.g., cyclin E) were unaffected by p21, suggesting that the inhibitory effects of p21 are not mediated entirely through E2F. Most of the down-regulated genes identified by the cDNA array (43 of 69) have been associated with cell-cycle progression and DNA repair, indicating a highly selective nature of p21-mediated inhibition of gene expression. To the best of our knowledge, such biological selectivity is unprecedented in large-scale expression profiling studies. A corollary to this observation is that differential cloning of p21-inhibited genes is likely to yield novel genes that play a role in cell-cycle progression. Indeed, six p21-inhibited genes were originally listed in the cDNA array as ESTs or genes with unknown function, but database search has linked three of their products to cell division or DNA repair.
Eighteen p21-inhibited genes are involved in DNA replication, segregation, and chromatin assembly. Some of these genes encode enzymes involved in nucleotide biosynthesis (e.g., thymidine kinase, dihydrofolate reductase, ribonucleotide reductase). Other proteins are involved in DNA replication (e.g., origin recognition complex protein Orc1, DNA polymerase α, DNA ligase I, replication licensing factor components Mcm7 and Mcm4), segregation (topoisomerase IIα), and chromatin formation (e.g., p60 subunit of chromatin assembly factor-I and high-mobility group proteins 1 and 2). Products of 20 other p21-down-regulated genes function in mitosis. Some of these proteins are Cdc2 and cyclin B1 that initiate mitosis, Plk1 involved in several different mitotic stages, Cdc2-interacting protein CKsHs1, centrosome-associated Aik1 kinase, spindle checkpoint-control proteins Mad2, BubR1, and Chl1, mitotic centromere-associated kinesin, and cytokinesis proteins Prc1 and citron kinase.
To determine whether inhibition of cell-cycle progression genes was a cause or consequence of p21-induced cell growth arrest, we have investigated the kinetics of changes in the RNA levels of p21-inhibited genes after the addition and removal of IPTG. We have also used immunoblotting to follow the time course of p21-induced changes in Rb phosphorylation and in the cellular levels of Rb and several p21-inhibited gene products (Fig. 2A). Rb became dephosphorylated (as indicated by increased electrophoretic mobility) as early as 6 h after the addition of IPTG, and Rb protein levels decreased sharply between 12–24 h (Fig. 2A), although no significant changes were detected in RB mRNA (not shown). All of the tested p21-inhibited genes showed a rapid response to p21 induction and release. Five genes showed significant inhibition at both RNA and protein levels between 4 and 8 h after the addition of IPTG, concurrently with the onset of cell growth arrest (Fig. 1 B and C) and Rb dephosphorylation (Fig. 2A). Some other genes showed a slower response, with a major decrease in mRNA levels detectable only 12 h after the addition of IPTG. All p21-inhibited genes, however, resumed their expression 12–16 h after the removal of IPTG (Fig. 2A), which precedes the resumption of DNA replication (20–24 h) and mitosis (30–36 h) (11). Considering the role of p21-inhibited genes in cell-cycle progression, this time course suggests strongly that regulation of such genes by p21 plays a causal role in p21-induced cell cycle arrest and is not a consequence of p21-mediated cell growth arrest and recovery. In addition, we have observed that p21-inhibited mitosis-control proteins were resynthesized asynchronously after release from IPTG, providing the likely cause for mitotic abnormalities in the released cells (11).
Several p21-inhibited genes are associated with DNA repair, including XRCC9, which may play a role in cell-cycle checkpoint control, Rad54 recombination repair protein, exonuclease Hex1/Rad2, a homolog of Rad21 repair protein involved in sister chromatid cohesion and mitotic recombination, and DNA ligase I. Inhibition of such genes agrees with the reports that p21 inhibits DNA repair (15). Inhibition of DNA repair, together with impaired mitosis control, is likely to increase the genetic instability of cells that reenter the cycle after p21 induction, with potential consequences for carcinogenesis and tumor progression.
We have noted some parallels between p21-inhibited genes and changes that occur in replicative senescence. Thus, senescent cells were reported to have reduced expression of cell-cycle control genes (16) and lower levels of the Rb protein (17), as we have also observed on p21 induction. It is also interesting that three p21-inhibited genes, CHL1, CDC21, and RAD54, encode members of the helicase family. A deficiency in another protein of the helicase group has been identified as the cause of Werner syndrome, a clinical condition associated with premature aging and, at the cellular level, accelerated senescence of cells in culture (18). More significant correlations with the senescent phenotype came, however, from the analysis of p21-induced genes (see below).
Induction of Gene Expression by p21: Correlations with Senescence and p21 Specificity of Induction.
Forty-eight known genes and six ESTs or genes with unknown functions were identified as up-regulated in p21-induced cells, with balanced differential expression of 2.0–7.8. The spectrum of p21-induced genes showed numerous correlations with cell senescence and organism aging. A very high fraction (20/48) of p21-induced genes encode extracellular matrix (ECM) components (e.g., fibronectin-1, Mac-2-binding protein), ECM receptors (integrin β3), or other secreted proteins. Overexpression of ECM proteins, including p21-induced gene products fibronectin-1, plasminogen activator inhibitor-1 (PAI-1), tissue-type plasminogen activator (t-PA), and integrin β3, is a hallmark of replicative senescence in normal fibroblasts (19–21). p21 also induced Alzheimer's β-amyloid precursor protein (APP) and cathepsin B, which have been shown to increase their expression in senescent cells (22, 23). p21 further induced several proteins that are known to increase their levels during organism aging, including t-PA, PAI-1, cathepsin B, activin A, prosaposin, APP, SAA, and tissue transglutaminase (t-TGase) (24–29). Senescent cells were also reported to overproduce lysosomal enzymes (30) and mitochondrial proteins (31, 32), and we have found that p21 up-regulates the expression of five lysosomal and three mitochondrial genes. Most interestingly, p21 increased the expression of p66Shc, inactivation of which was recently reported to increase stress resistance and to extend the lifespan in knockout mice (33). These parallels between p21-induced genes and known markers of senescence suggest that different features of the senescent phenotype result at least in part from the induction of gene expression by p21.
The kinetics of induction of p21-up-regulated genes also supports a role for such genes in the senescent phenotype. All of the tested genes (except for t-TGase) showed maximal RNA levels between 24 and 48 h after the addition of IPTG (Fig. 2B), and their expression (also except for t-TGase) remained elevated for at least 3 days after release from IPTG (not shown). This timing lags behind p21-induced inhibition of cell-cycle progression genes (Fig. 2A) and growth arrest (Fig. 1 B and C), but it matches the time course of the development of the senescent phenotype in p21-arrested cells.
To determine whether p21-up-regulated gene expression is a specific effect of p21 induction or a general consequence of cell growth arrest, we have compared p21-induced changes in gene expression with the corresponding changes in p21–9 cells that were growth arrested by incubation in serum-free media. Serum starvation of p21–9 cells induced strong growth arrest, as evidenced by the lack of significant increase in the cell number after 3 or more days of serum starvation and approximately 60-fold decrease in [3H]thymidine incorporation. This growth arrest was associated with only 2-fold increase in cellular p21 levels (not shown), much lower than the extent of p21 induction in senescent fibroblasts (17, 34, 35) or in IPTG-treated p21–9 cells (Fig. 1A). RT-PCR was used to compare changes in the expression of 15 p21-induced and 16 p21-inhibited genes in cells that were growth arrested for the same period by 50 μM IPTG or by serum-free media. As shown in Fig. 2C, all IPTG-inhibited genes were also inhibited in serum-starved cells; about one-half of these genes were inhibited as strongly in serum-starved as in IPTG-treated cells. In contrast, 8 of 15 p21-induced genes, including PAI-1, SAA, t-TGase, integrin β3, CTGF, activin A, natural killer cell protein 4, and p66Shc were not up-regulated at all (and even slightly down-regulated) in serum-starved cells. Three other genes were induced to a lesser extent in serum-starved than in IPTG-treated cells, and only four genes were equally induced in both types of growth arrest (Fig. 2C). These results indicate that the activation of most (but not all) p21-induced genes is not a general consequence of cell growth arrest. NFκB-responsive genes (e.g., t-TGase, SAA, APP, and SOD2) are found among those that are induced by p21 alone or by both p21 and serum starvation, suggesting that NFκB activation by p21 (8) does not determine the p21 specificity of the induction of gene expression.
Paracrine Mitogenic and Antiapoptotic Effects of p21 Induction.
To our surprise, analysis of p21-activated genes suggested that p21-induced growth arrest may be accompanied by a paracrine growth-stimulatory effect, because several of these genes encode secreted proteins with known mitogenic or antiapoptotic activity. In particular, CTGF, activin A, epithelin/granulin, and galectin-3 were reported to act as mitogens (36–39), whereas galectin-3 and prosaposin inhibit apoptosis (40, 41). p21 also induced intracellular proteins SOD2 and R-Ras with reported antiapoptotic activity (42, 43), as well as t-TGase and cathepsin B ascribed a proapoptotic function (27). The observed induction of both antiapoptotic and proapoptotic genes by p21 may explain the contradictory reports on both positive (44) and negative (45, 46) effects of p21 on apoptosis.
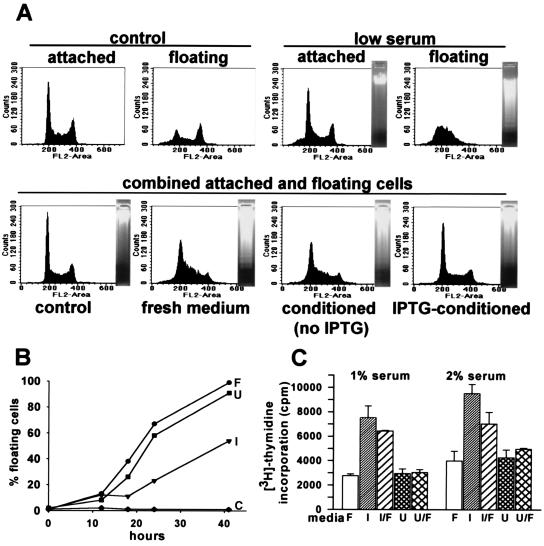
We have investigated whether conditioned media from IPTG-treated p21–9 cells would have an effect on apoptosis or the cell growth. In one set of assays, we tested whether such media would influence apoptosis in C8 cells, a line of mouse embryo fibroblasts transformed by E1A and H-Ras. This cell line is highly susceptible to apoptosis induced by different stimuli, including serum starvation (47–49). The addition of low-serum fresh media rapidly induced apoptosis in C8 cells, as evidenced by cell detachment and apoptotic morphology. The detached (floating) cells showed apoptosis-specific DNA ladder and lack of cells with G2/M DNA content (Fig. 3A), which is characteristic for apoptosis in C8 cells (49). The addition of IPTG to fresh media had no effect on apoptosis (not shown). Conditioned media from IPTG-treated but not from untreated cells strongly decreased the induction of apoptosis, by the criteria of cell detachment (Fig. 3B), ladder formation, and alteration in the FACS profiles of DNA content (Fig. 3A). Furthermore, cells incubated for 48 h in different batches of conditioned media from IPTG-treated cells showed 23–24% survival relative to control cells grown in 10% serum, whereas conditioned media from untreated cells produced only 1–3% survival, and fewer than 1% of the cells survived in low-serum fresh media.
Figure 3.
(A) (Upper) FACS profiles of DNA content and electrophoretic patterns of DNA degradation in the attached or floating C8 cells in 10% serum (control) or 24 h after transfer in 0.4% serum fresh media (low-serum). (Lower) DNA content and electrophoretic pattern of combined attached and floating C8 cells after 24 h incubation in 10% serum (control), in low-serum fresh medium, or in conditioned media from untreated or IPTG-treated p21–9 cells. (B) Effects of control media containing 10% serum (C) and low-serum fresh media (F), conditioned media from IPTG-treated (I) or untreated p21–9 cells (U) on apoptosis of C8 cells, as measured by cell detachment. (C) Effects of fresh media (F), conditioned media from IPTG-treated (I), or untreated (U) p21–9 cells, and 1:1 mixtures of conditioned and fresh media (I/F and U/F), supplemented with 1% or 2% serum, on [3H]thymidine incorporation by HS 15.T cells.
In another set of experiments, we asked whether serum-supplemented conditioned media from IPTG-treated cells would stimulate the growth of two slow-growing human fibrosarcoma cell lines. One of these lines, HS 15.T, showed a small (20–30%) but reproducible increase in the number of cells grown for 5 days in conditioned media from IPTG-treated cells, supplemented with 4–8% serum, relative to conditioned media from untreated cells. We have also used [3H]thymidine incorporation as a more sensitive assay that could be conducted at lower serum concentrations. We have compared [3H]thymidine incorporation by HS 15.T cells in fresh media, in conditioned media from IPTG-treated or untreated cells, and in mixtures of conditioned and fresh media, supplemented with 1% or 2% serum. Conditioned media from IPTG-treated p21–9 cells, but not fresh media or media conditioned by untreated cells, increased [3H]thymidine incorporation by HS 15.T cells up to 3-fold (Fig. 3C), indicating apparent mitogenic activity. The addition of IPTG to fresh media had no effect in this assay (not shown). The above experiments, however, do not allow us to tell whether the effects of the conditioned media are due to the direct stimulation of cell growth or the inhibition of cell death in proliferating HS15.T cells.
As noted by Campisi (19), normal senescent cells overproduce different growth factors, providing a further link between p21 induction and the senescent phenotype. Some paracrine factors that we find to be induced by p21 are also induced by serum starvation (prosaposin, galectin-3), whereas the induction of other genes (CTGF, activin A) is p21-specific (Fig. 2C). The induction of paracrine mitogenic and antiapoptotic factors by p21 raises a possibility that terminal growth arrest of senescent or postmitotic cells may stimulate the proliferation of their neighbors. This paracrine effect may be one of the unselected physiological consequences of senescence or, alternatively, it might play a role in tissue homeostasis. It is interesting to note in this regard that p21 expression in mammalian development has been localized to narrow zones of postmitotic cells, adjacent to the proliferative compartments (50, 51). Together with the potential for genetic destabilization in p21-induced cells (discussed above), the paracrine antiapoptotic and mitogenic effects of p21 induction may also contribute to carcinogenesis and tumor progression.
Involvement of p21-Induced Proteins in Age-Related Diseases.
Strikingly, products of many genes that we found to be induced by p21 play a causal role or have been associated with different age-related diseases. Thus, p21-induced APP gives rise to β-amyloid peptide, the main component of Alzheimer's amyloid plaques. p21 strongly induces the inflammatory protein SAA, deposition of which causes amyloidosis and contributes to atherosclerosis, osteoarthritis, and rheumatoid arthritis (52). p21 also induces t-TGase, which has been described as a pleiotropic mediator of cell differentiation, carcinogenesis, apoptosis, and aging and which plays a role in plaque formation in both Alzheimer's disease and amyloidosis (28, 29). p21-induced CTGF and galectin-3 have been implicated in atherosclerosis (53, 54), whereas p21-up-regulated complement C3 and AMP deaminase were suggested to play a role in Alzheimer's disease (56, 57). In addition, expression of p21-induced proteins cathepsin B, PAI-1, fibronectin, N-acetylgalactosamine-6-sulfate sulfatase, and Mac2-BP has been associated with osteoarthritis and/or rheumatoid arthritis (58–60).
Replicative senescence of normal cells has been associated with changes in protein expression that may contribute to the pathogenesis of cancer and age-related diseases (19). In the present study, we have found that similar changes in gene expression can result from p21 induction, which is a common event in the programs of senescence and damage response. It remains to be determined whether the same effects of p21 that we observed in a fibrosarcoma cell line would also occur in different types of normal cells, and which of these effects would be specific consequences of p21 expression. The elucidation of the mechanisms and the specificity of the effects of p21 on gene expression may suggest new approaches to the prevention of different diseases that are associated with human aging.
Supplementary Material
Acknowledgments
We thank Drs. W. Jiang, T. Hunter (The Salk Institute, La Jolla, CA), B. M. Schreiber (Boston University), L. Lau, W. T. Beck, and H. Kiyokawa and A. V. Gudkov (University of Illinois at Chicago) for gifts of antibodies and cell lines, K. Hagen for assistance with flow sorting, R. Abdryashitov, Y. Chen, and S. Salov for help with some experiments, and A. Ruth and Drs. T. Primiano, A. V. Gudkov, and R. L. Davidson for helpful discussions. This work was supported by National Cancer Institute grants R01CA62099 and R37CA40333 (I.B.R.) and National Institute for Dental and Craniofacial Research grant K08 DE00398 (K.W.).
Abbreviations
- CDK
cyclin-dependent kinase
- IPTG
isopropyl-β-d-thiogalactoside
- SAA
serum amyloid A
- RT-PCR
reverse transcription–PCR
- ESTs
expressed sequence tags
- APP
Alzheimer's β-amyloid precursor protein
- t-TGase
tissue transglutaminase
- CTGF
connective tissue growth factor
References
- 1.Gartel A L, Tyner A L. In: Molecular and Subcellular Biology. Macieir-Coelho A, editor. Vol. 20. Berlin: Springer; 1998. pp. 43–71. [DOI] [PubMed] [Google Scholar]
- 2.Niculescu A B, III, Chen X, Smeets M, Hengst L, Prives C, Reed S I. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogt M, Haggblom C, Yeargin J, Christiansen-Weber T, Haas M. Cell Growth Differ. 1998;9:139–146. [PubMed] [Google Scholar]
- 4.McConnell B B, Starborg M, Brookes S, Peters G. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 5.Chang B D, Xuan Y, Broude E V, Zhu H, Schott B, Fang J, Roninson I B. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 6.Nevins J R. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 7.de Toledo S M, Azzam E I, Keng P, Laffrenier S, Little J B. Cell Growth Differ. 1998;9:887–896. [PubMed] [Google Scholar]
- 8.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 9.Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. EMBO J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cunto F, Topley G, Calautti E, Hsiao J, Ong L, Seth P K, Dotto G P. Science. 1998;280:1069–1072. doi: 10.1126/science.280.5366.1069. [DOI] [PubMed] [Google Scholar]
- 11.Chang, B. D., Broude, E. V., Fang, J., Kalinichenko, T. V., Abdryashitov, R., Poole, J. C. & Roninson, I. B. (2000) Oncogene, in press. [DOI] [PubMed]
- 12.Mosca P J, Dijkwel P A, Hamlin J L. Mol Cell Biol. 1992;12:4375–4383. doi: 10.1128/mcb.12.10.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan M A, Wendell K, Gardiner S, Derry W B, Copp H, Wilson L. Cancer Res. 1996;56:816–825. [PubMed] [Google Scholar]
- 14.Noonan K E, Beck C, Holzmayer T A, Chin J E, Wunder J S, Andrulis I L, Gazdar A F, Willman C L, Griffith B, Von Hoff D D, et al. Proc Natl Acad Sci USA. 1990;87:7160–7164. doi: 10.1073/pnas.87.18.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Z Q, Reardon J T, Li L, Flores-Rozas H, Legerski R, Sancar A, Hurwitz J. J Biol Chem. 1995;270:22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- 16.Shelton D N, Chang E, Whittier P S, Choi D, Funk W D. Curr Biol. 1999;9:939–945. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 17.Stein G H, Drullinger L F, Soulard A, Dulic V. Mol Cell Biol. 1999;19:2109–2117. doi: 10.1128/mcb.19.3.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray M D, Shen J C, Kamath-Loeb A S, Blank A, Sopher B L, Martin G M, Oshima J, Loeb L A. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 19.Campisi J. J Invest Dermatol Symp Proc. 1998;3:1–5. [PubMed] [Google Scholar]
- 20.West M D, Shay J W, Wright W E, Linskens M H. Exp Gerontol. 1996;31:175–193. doi: 10.1016/0531-5565(95)02013-6. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto Y, Kohri K, Akita H, Mitani K, Ikeda K, Nakanishi M. Biochem Biophys Res Commun. 1997;240:88–92. doi: 10.1006/bbrc.1997.7534. [DOI] [PubMed] [Google Scholar]
- 22.Adler M J, Coronel C, Shelton E, Seegmiller J E, Dewji N N. Proc Natl Acad Sci USA. 1991;88:16–20. doi: 10.1073/pnas.88.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiPaolo B R, Pignolo R J, Cristofalo V J. Exp Cell Res. 1992;201:500–505. doi: 10.1016/0014-4827(92)90300-w. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto Y, Kobayashi A, Yamazaki N, Sugawara Y, Takada Y, Takada A. Thromb Res. 1987;46:625–633. doi: 10.1016/0049-3848(87)90264-7. [DOI] [PubMed] [Google Scholar]
- 25.Loria P, Petraglia F, Concari M, Bertolotti M, Martella P, Luisi S, Grisolia C, Foresta C, Volpe A, Genazzani A R, et al. Eur J Endocrinol. 1998;139:487–492. doi: 10.1530/eje.0.1390487. [DOI] [PubMed] [Google Scholar]
- 26.Mathur P P, Mo M Y, Panzironi C, Silvestrini B, Bardin C W, Grima J, Cheng C Y. Biochem Mol Biol Int. 1994;34:1063–1071. [PubMed] [Google Scholar]
- 27.Singhal P C, Reddy K, Franki N, Sanwal V, Kapasi A, Gibbons N, Mattana J, Valderrama E. J Invest Med. 1997;45:567–575. [PubMed] [Google Scholar]
- 28.Dudek S M, Johnson G V. Brain Res. 1994;651:129–133. doi: 10.1016/0006-8993(94)90688-2. [DOI] [PubMed] [Google Scholar]
- 29.Park S C, Yeo E J, Han J A, Hwang Y C, Choi J Y, Park J S, Park Y H, Kim K O, Kim I G, Seong S C, et al. J Gerontol A Biol Sci. 1999;54:B78–B83. doi: 10.1093/gerona/54.2.b78. [DOI] [PubMed] [Google Scholar]
- 30.Christofalo V J, Kabakjian J. Mech Ageing Dev. 1975;4:19–28. doi: 10.1016/0047-6374(75)90004-4. [DOI] [PubMed] [Google Scholar]
- 31.Kodama S, Yamada H, Annab L, Barrett J C. Exp Cell Res. 1995;219:82–86. doi: 10.1006/excr.1995.1207. [DOI] [PubMed] [Google Scholar]
- 32.Kumazaki T, Sakano T, Yoshida T, Hamada K, Sumida H, Teranishi Y, Nishiyama M, Mitsui Y. Mech Ageing Dev. 1998;101:91–99. doi: 10.1016/s0047-6374(97)00159-0. [DOI] [PubMed] [Google Scholar]
- 33.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi P P, Lanfrancone L, Pelicci P G. Nature (London) 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 34.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barrett J C. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robles S J, Adami G R. Oncogene. 1998;16:1113–1123. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- 36.Bradham D M, Igarashi A, Potter R L, Grotendorst G R. J Cell Biol. 1991;114:1285–1294. doi: 10.1083/jcb.114.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakurai T, Abe Y, Kasuya Y, Takuwa N, Shiba R, Yamashita T, Endo T, Goto K. J Biol Chem. 1994;269:14118–14122. [PubMed] [Google Scholar]
- 38.Shoyab M, McDonald V L, Byles C, Todaro G J, Plowman G D. Proc Natl Acad Sci USA. 1990;87:7912–7916. doi: 10.1073/pnas.87.20.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inohara H, Akahani S, Raz A. Exp Cell Res. 1998;245:294–302. doi: 10.1006/excr.1998.4253. [DOI] [PubMed] [Google Scholar]
- 40.Akahani S, Nagia-Makker P, Inohara H, Kim H R, Raz A. Cancer Res. 1997;57:5272–5276. [PubMed] [Google Scholar]
- 41.Hiraiwa M, Taylor E M, Campana W M, Darin S J, O'Brien J S. Proc Natl Acad Sci USA. 1997;94:4778–4781. doi: 10.1073/pnas.94.9.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manna S K, Zhang H J, Yan T, Oberley L W, Aggarwal B B. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki J, Kaziro Y, Koide H. FEBS Lett. 1998;437:112–116. doi: 10.1016/s0014-5793(98)01213-7. [DOI] [PubMed] [Google Scholar]
- 44.Tsao Y P, Huang S J, Chang J L, Hsieh J T, Pong R C, Chen S L. J Virol. 1999;73:4983–4990. doi: 10.1128/jvi.73.6.4983-4990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi M C, Holbrook N J. Oncogene. 1997;14:929–935. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- 46.Lu Y, Yamagishi N, Yagi T, Takebe H. Oncogene. 1998;16:705–712. doi: 10.1038/sj.onc.1201585. [DOI] [PubMed] [Google Scholar]
- 47.Lowe S W, Jacks T, Housman D E, Ruley H E. Proc Natl Acad Sci USA. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe S W, Bodis S, McClatchey A, Remington L, Ruley H E, Fisher D E, Housman D E, Jacks T. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 49.Nikiforov M A, Hagen K, Ossovskaya V S, Connor T M, Lowe S W, Deichman G I, Gudkov A V. Oncogene. 1996;13:1709–1719. [PubMed] [Google Scholar]
- 50.El-Deiry W S, Tokino T, Waldman T, Oliner J D, Velculescu V E, Burrell M, Hill D E, Healy E, Rees J L, Hamilton S R, et al. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 51.Gartel A L, Serfas M S, Gartel M, Goufman E, Wu G S, Tyner A L. Exp Cell Res. 1996;227:171–181. doi: 10.1006/excr.1996.0264. [DOI] [PubMed] [Google Scholar]
- 52.Jensen L E, Whitehead A S. Biochem J. 1998;334:489–503. doi: 10.1042/bj3340489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oemar B S, Werner A, Garnier J M, Do D D, Godoy N, Nauck M, Marz W, Rapp J, Pech M, Luscher T F. Circulation. 1997;95:831–839. doi: 10.1161/01.cir.95.4.831. [DOI] [PubMed] [Google Scholar]
- 54.Nachtigal M, Al-Assaad Z, Mayer E P, Kim K, Monsigny M. Am J Pathol. 1998;152:1199–1208. [PMC free article] [PubMed] [Google Scholar]
- 55.Veerhuis R, van der Valk P, Janssen I, Zhan S S, Van Nostrand W E, Eikelenboom P. Virchows Arch. 1995;426:603–610. doi: 10.1007/BF00192116. [DOI] [PubMed] [Google Scholar]
- 56.Sims B, Powers R E, Sabina R L, Theibert A B. Neurobiol Aging. 1998;19:385–391. doi: 10.1016/s0197-4580(98)00083-9. [DOI] [PubMed] [Google Scholar]
- 57.Howie A J, Burnett D, Crocker J. J Pathol. 1985;145:307–314. doi: 10.1002/path.1711450404. [DOI] [PubMed] [Google Scholar]
- 58.Cerinic M M, Generini S, Partsch G, Pignone A, Dini G, Konttinen Y T, Del Rosso M. Life Sci. 1998;63:441–453. doi: 10.1016/s0024-3205(98)00293-8. [DOI] [PubMed] [Google Scholar]
- 59.Chevalier X. Semin Arthritis Rheum. 1993;22:307–318. doi: 10.1016/s0049-0172(05)80010-1. [DOI] [PubMed] [Google Scholar]
- 60.Seki T, Selby J, Haupl T, Winchester R. Arthritis Rheum. 1998;41:1356–1364. doi: 10.1002/1529-0131(199808)41:8<1356::AID-ART4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.