Abstract
Mayaro virus (MAYV) is an emerging mosquito-borne alphavirus that causes clinical symptoms similar to those caused by Chikungunya virus (CHIKV), Dengue virus (DENV), and Zika virus (ZIKV). To differentiate MAYV from these viruses diagnostically, we have developed a portable device that integrates sample preparation with real-time, reverse-transcription, loop-mediated isothermal amplification (rRT-LAMP). First, we designed a rRT-LAMP assay targeting MAYV’s non-structural protein (NS1) gene and determined the limit of detection of at least 10 viral genome equivalents per reaction. The assay was specific for MAYV, without cross-reactions with CHIKV, DENV, or ZIKV. The rRT-LAMP assay was integrated with a sample preparation device (SPD) wherein virus lysis and RNA enrichment/purification were carried out on the spot, without requiring pipetting, while subsequent real-time amplification device (RAD) enables virus detection at the point of care (POC). The functions of our platform were demonstrated using purified MAYV RNA or blood samples containing viable viruses. We have used the devices for detection of MAYV in as short as 13 min, with limit of detection to as low as 10 GEs/reaction.
Keywords: Mayaro virus, rRT-LAMP, Point of care, Valve, Real-time amplification
Introduction
Mayaro virus (MAYV), genus Alphavirus, is an RNA virus that is transmitted by mosquitoes [1–3]. Human infections with MAYV may result in Mayaro fever, which is an acute febrile illness that is often accompanied by joint and muscle pain. The clinical presentation and course of MAYV is similar to that seen with another alphavirus, Chikungunya virus (CHKV); there are also similarities with the clinical presentation seen with arboviruses, Dengue viruses 1 to 4 (DENV-1 to DENV-4), and Zika virus (ZIKV), making it difficult to differentiate MAYV infection clinically from these other viruses [4–6]. MAYV has been identified as an emerging virus in the Americas, highlighting the need for development of effective detection methods for diagnostic evaluations [1, 7]. This challenge is particularly true in regions where the aforementioned viruses are endemic, resulting in the requirement of specific diagnosis at the point of care (POC) for clinical management and development of appropriate public health interventions.
The standard techniques for detection of MAYV infection include serological tests such as enzyme-linked immunosorbent assay (ELISA) [4, 6, 8–10] and molecular tests such as reverse transcription polymerase chain reactions (RT-PCR) [6, 8, 11–13]. Traditional serological tests are of limited value for detection of acute infections (due to time required to produce antibodies in the blood of patients), and there are potential issues with cross reactivity with other closely related viruses. Acute diagnosis of MAYV infections generally requires RT-PCR, but standard RT-PCR techniques can be time-consuming and require skilled personal and costly equipment. As a result, they are typically carried out in laboratories, making them impractical for diagnostics at the POC, and are not affordable in clinical settings with limited resources, such as those regions where MAYV infections are usually encountered. Antigen-test strips based on lateral flow assays, such as those commonly used for coronavirus disease 2019 (COVID-19), influenza, and other virus’ diagnostic tests, are not yet available for MAYV. They also tend to be less sensitive than molecular tests.
Loop-mediated isothermal amplification (LAMP) has proven to be a rapid and sensitive diagnostic technique for the detection of various pathogens [14–19]. Compared with PCR, LAMP requires simpler thermal management due to the isothermal condition, is faster (~ 30 min. versus 2 h for PCR) and has similar sensitivity. LAMP requires 4–6 primers that target 6–8 distinct sequences, providing high specificity. For RNA viruses such as MAYV, reverse transcription LAMP (RT-LAMP) can be used. RT-LAMP has been demonstrated to detect a wide range of RNA viruses including CHIKV [20–22], DENV-1 to DENV-4 [20–24], Ebola virus [25], human immunodeficiency virus (HIV) [26, 27], influenza virus [28–30], severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [28, 29, 31–38], and ZIKV [20–22, 39–41]. A few commercially available instruments based on LAMP or RT-LAMP also exist, including NINA from PATH, BioRanger from Diagenetix, and Lucira Check It from Lucira Health [42–44].
A few real-time LAMP-based POC platforms have been reported in the literature. Although they are well-designed, sample preparation was not included in their detection systems, either using RNA/DNA purified in laboratory or directly adding a sample to the reaction mixture. It has been shown that LAMP assay without sample preparation reduced the detection sensitivity [31] and increased the number of false negatives [45]. We have aimed to integrate sample preparation with real-time RT-LAMP (rRT-LAMP) for virus detection at POC.
In this work, we report the development of a highly sensitive, specific, and rapid rRT-LAMP assay for the detection of MAYV. Additionally, we have developed POC platforms for carrying out the rRT-LAMP assay.
To achieve this, we designed a sample preparation device that performs virus lysis and RNA enrichment/purification without requiring pipetting. This device follows similar concepts to our previously developed device [40] and is capable of processing blood samples at the POC. Note that the previous device was done with colorimetric detection only, which was carried out at the end of 30-min amplification. The colorimetric detection gives only binary results: the presence or absence of viruses of interest. The real-time POC detection platform, integrated with the sample preparation device, enables the quantitative detection of MAYV (i.e., virus burden) at the POC.
We have assessed the sensitivity and specificity of the rRT-LAMP assay and demonstrated the utility of our platforms using purified MAYV RNA, plasma containing virus, or blood samples containing virus. Our results indicate that the rRT-LAMP assay is specific and highly sensitive, and the POC platform has the potential to be used in the field, including resource-limited regions.
Materials and methods
Cell culture for MAYV
MAYV strain Guyane was obtained from BEI Resources (Manassas, VA; Catalog. no. NR-49911) and propagated in Vero E6 cells, which were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cell culture took place at 37 °C and 5% CO2 using advanced Dulbecco’s essential medium (aDMEM) and 10% fetal bovine serum (FBS, Thermo Fisher Scientific) as previously described [46].
After about 80% of the cells displayed virus-induced cytopathic effects, virus RNA was extracted from spent cell culture medium using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions [47].
rRT-LAMP mix
A solution of 25 μL of mixture was used for each RT-LAMP assay. The 25 μL of mixture contains 2.5 μL of 10X isothermal amplification buffer (ISO), 8 U Bst 2.0 WarmStart® DNA polymerase, 7.5 U WarmStart® RTx reverse transcriptase (RTx), 2.5 μL of 10X concentrated primer mix (PMX), 1.4 mM deoxynucleotide triphosphate (dNTPs), and 6 mM MgSO4. To eliminate possible carryover contamination, 0.5 units of antarctic thermolabile uracil-DNA glycosylase (UDG) and 0.7 mM of deoxyuridine triphosphate (dUTP) were also added to the mixture [48, 49]. Nuclease-free water was then added to fill the 25 μL volume. The nuclease-free water and dNTPs were purchased from Thermo Fisher Scientific (MA, USA). All other reagents were obtained from New England Biolabs (NEB, Ipswich, MA, USA). The final volume of each component for preparation of 25 μL RT-LAMP reaction mixture is tabulated in Electronic Supplementary Material Table S2. When real-time detection was used, an appropriate dye (either SYBR Green I or SYTO9) was added as detailed below.
rRT-LAMP using a commercial cycler
We used a QuantStudio-3 real-time PCR (qPCR) instrument to study the effects of the primer concentrations on the incubation time and to determine the detection limit of our rRT-LAMP assay. rRT-LAMP assays were carried out by adding 1 μL of purified RNA and 0.5 μL of 10X concentrate SYBR Green I nucleic acid gel stain in dimethyl sulfoxide (Thermo Fisher Scientific) to the 25 μL RT-LAMP reaction mixture. A no-template control (NTC) was included as a negative control in each test. The reactions were carried out at 62.5 °C for 60 min. The fluorescence signals obtained by the PCR machine were analyzed through the QuantStudio-3 (Thermo Fisher Scientific). We carried out three replicates for each RNA concentration, and the results presented in this work are the average of the three replicates.
POC sample preparation device
The sample preparation device (SPD) is an improved version of our previously developed device for the detection of ZIKV [40]. As shown in Electronic Supplementary Material Figure S1, SPD consists of buffer, mixing, detection, and drain units. The buffer and mixing units were 3D printed from polylactic acid (PLA). The detection unit is a paper-based amplification device consisting of a polycarbonate well layer and a laminated paper pad. The laminated paper pad is a 4-mm Whatman chromatography paper (Fisher Scientific, Pittsburgh, PA, USA) that is sandwiched between two 75-μm-thick polyester lamination films. Chromatography paper was chosen in the device because it showed a lower limit of detection than FTA card and glass-fiber paper [50]. The SPD is developed to enable RNA purification and enrichment at the POC without the need for laboratory equipment. The RNA enrichment process in the previous device relied on the wicking effects of chromatography paper in the detection unit, which was adequate for processing low viscous samples such as saliva and urine. However, it took long time to process a viscous blood sample. To improve the device, we have developed a drain unit made of polydimethylsiloxane (PDMS), including a 1.9 cm × 1.9 cm chamber for the detection unit and a 6 mm drain well in the center connected to a 1-mm drain channel (Figure S1). This unit allows for the use of external forces such as a syringe to connect with the drain channel to discharge both the lysed sample and buffers quickly when needed. The size of the chamber was optimized to ensure PDMS’s intrinsic sealing capability without the use of any sealant during the integration of the detection unit into the drain unit. The new design significantly reduced the sample preparation time to as low as 7 min (5-min incubation for lysis and 2-min flushing of all reagents in the buffer unit). A comparison between previously reported device (named VLEAD [40]) and SPD is presented in Electronic Supplementary Material Table S1.
Real-time amplification device
As shown in Fig. 1 and Figure S2, our real-time amplification device (RAD) for rRT-LAMP at POC is composed of a portable stand, digital microscope (AM4117MT-G2FBW, Dino-Lite, USA), a temperature controller, a heater, and two 9-V rechargeable batteries. The stand was 3D printed from PLA, with a base containing three chambers for the heater, batteries, and temperature controller. Three caps were also 3D-printed for covering the three chambers. The cap for the heater chamber was designed to contain a 2 × 2 cm2 cavity, which properly fits the detection unit in Figure S1 (which is also described later). In addition, two 5-mm holes were created in the cap, enabling real-time detection of the rRT-LAMP reactions (one hole for the sample and the other for the negative control).
Fig. 1.
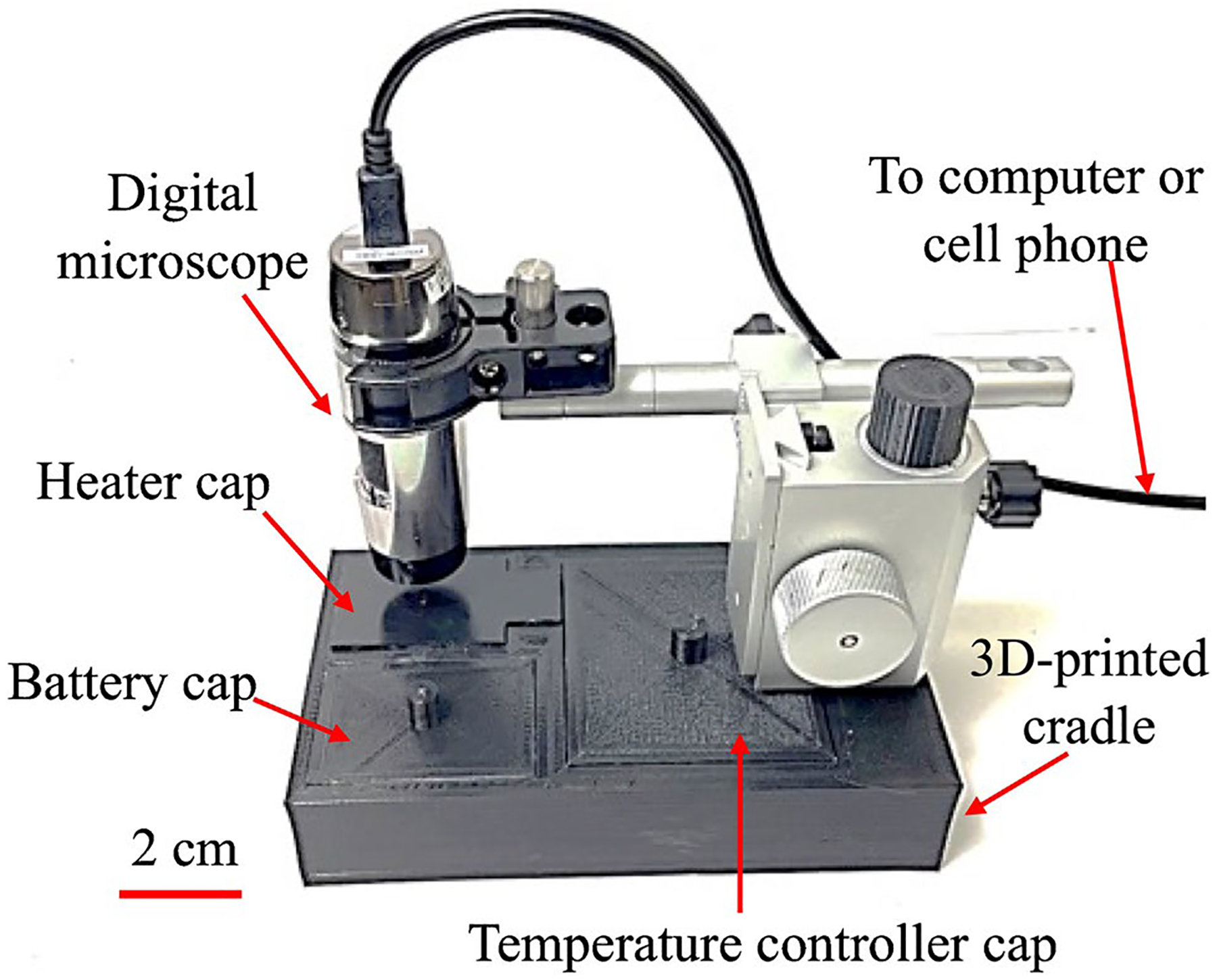
Picture of a real-time amplification device (RAD) consisting of a flashlight-shaped microscope and 3D-printed chambers for a heater, battery, and integrated electrical circuit for temperature control. A scale bar of 2 cm is shown on the bottom left
The digital microscope possesses two bandpass filters which allow only desired excitation and emission wavelength to be transmitted through. The peak excitation wavelength is 465 nm while the emission band is 510–545 nm, as shown in Figure S3b. These wavelengths are compatible with the excitation/emission wavelengths of SYTO9 (Figure S3a), which was used as fluorescent dye in our real-time detection platform. We replaced SYBR Green with SYTO9 in the RAD since SYTO9 has far less inhibitory effects on rRT-LAMP than SYBR Green [51]. We found that 4 μM of SYTO9 is optimal for obtaining highest fluorescence intensity without inhibition on rRT-LAMP.
The heater used for rRT-LAMP is a positive temperature coefficient (PTC) heater (Bolsen Tech, USA), which can be powered by batteries, a laptop, or a smartphone. A commercially available, low-cost temperature controller was used to switch the PTC heater on or off. The controller’s thermostat sensor was attached to the PTC heater, and the sensor temperature was calibrated to set the rRT-LAMP reaction at 62.5 °C. We characterized the heating system by placing a T-type thermocouple in the rRT-LAMP well, and the resultant temperature profile is shown in Figure S4. The results illustrate that the rRT-LAMP temperature was maintained between 62.5 and 63.1 °C, which is within the standard temperature range (60–65 °C) recommended for LAMP. The batteries can last for about 3 h, sufficient for at least 5 experiments. The total weight of RAD is 983 g, which is portable.
Results and discussion
POC testing
Detection can be performed at the end of the amplification reactions by direct visual observation of color change, which is known as endpoint detection. The endpoint detection is suitable for POC applications when the test can be performed for a specific, predetermined time, without the need for an expensive and complex detector. This method provides a binary result (“yes” or “no”), representing the presence or absence of a target virus, without any quantitative information. Alternatively, real-time detection can be employed to continuously monitor and analyze RT-LAMP, enabling dynamic observation of the RT-LAMP results. For positive samples, signals can be detected way before the predetermined time used for the endpoint detection, resulting in a much shorter analysis time. Additionally, this analysis time can be used to infer quantitative information about the amount of virus present in the sample tested because the less the amount of virus, the longer the amplification time required to have detectable signals.
We have employed the SPD for either endpoint detection or real-time detection of MAYV at the POC. To perform real-time detection of MAYV, we have integrated the SPD with RAD for sample preparation and real-time amplification. The SPD was designed for sample preparation and RNA enrichment/purification at POC. After RNA purification on the paper pad of the detection unit, the detection unit of the SPD was separated from mixing unit and then placed in the cavity designed in the cap of the heater chamber in the RAD shown in Fig. 1 and Figure S2. The digital microscope in Fig. 1 was programed to acquire fluorescence images every minute. The light-emitting diode (LED) in the microscope was programed to turn on for 5 s and off for 55 s to avoid possible photobleaching. Two Python-based apps (one compatible with the computer operating system while the other with the smartphone operating system) were developed for measuring fluorescence intensity and displaying the data in real time.
Figure 2 shows the process flow of our portable pathogen detection platform. For the detection of MAYV, a sample (e.g., virus, plasma, or blood) is introduced to the lysis buffer in the first reservoir of the buffer unit (Fig. 2a). By sliding the mixing unit along the buffer unit (detailed in Figure S1), the pin in the mixing unit pushes up the ball at the bottom of the first reservoir when they are aligned (Figure S1b and Figure S1c), discharging the solution in the reservoir from the buffer unit to the mixing unit. This ball-based valve design was inspired by a ballpoint pen, in which ink is dispensed onto paper when the metal ball at the tip is pressed while writing. Further sliding of the mixing unit along the buffer unit discharges the binding buffer in the second reservoir, resulting in retention of viral RNA on the paper pad when the solution in the mixing unit flows through the detection unit. The discharge process is sped up by pulling the plunger of the syringe that is connected to the drain unit (Figure S1). The enriched RNA on the paper pad is then purified after two wash buffers in the third and fourth reservoirs are discharged in sequence by further sliding the mixing unit. After RNA purification, the detection unit is taken apart from SPD, followed by adding 25 μL rRT-LAMP mixture (Fig. 2b). Afterwards, the detection unit is placed in the cavity inside the cap of the heater chamber (while the temperature controller and the PTC heater have been switched on about 5 min earlier). The microscope is then positioned on top of the rRT-LAMP reaction well and takes pictures with 1-min intervals (Fig. 2c). The Python-coded software converts the images from the microscope into a graph. The sample preparation using SPD and rRT-LAMP using RAD are shown in the Electronic Supplementary Material (Video 1).
Fig. 2.
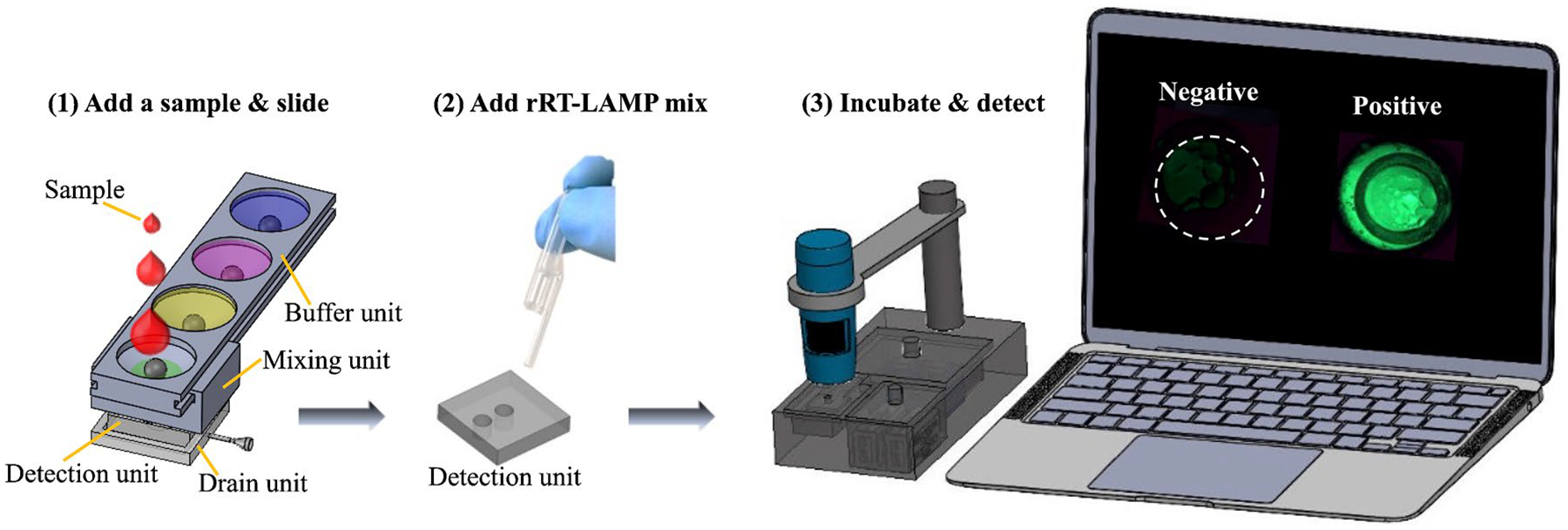
Process flow of using SPD and RAD for virus detection at POC. (1) A sample is loaded into the first reservoir of the buffer unit, followed by virus lysis, RNA binding, and washing steps by sliding the mixing unit along the buffer unit. All buffers pass through the paper in the detection unit due to the vacuum created by pulling out the plunger of a syringe connected to the drain unit. (2) The detection unit is separated from SPD, and the rRT-LAMP mix is added into the reaction wells. (3) The detection unit is placed in the seat of the heater chamber cap (Figure S1), followed by rRT-LAMP
In some situations when binary detection is adequate, we performed isothermal amplification using endpoint detection. In this case, 25 μL RT-LAMP mixture is added into the detection unit, which is placed in a battery-powered coffee mug as illustrated in Figure S5. The mug functions as an isothermal water bath incubator, enabling RT-LAMP at 62.5 °C for 30 min. The colorimetric detection is carried out at the end using SYBR Green I dye; color change is observed by naked eye or recorded by a smartphone [40].
rRT-LAMP assay
To design LAMP primers, we obtained the genome sequence of MAYV strain MAYLC from French Guiana from National Center for Biotechnology Information (NCBI, accession no. DQ001069). This virus strain was chosen as its genomic sequence has high identity with MAYV strains from Haiti and Venezuela that we have recently analyzed [46, 52] and thus serves as a contemporary reference strain for the type of MAYV we have tested in this work. Primer Explorer V5 program (https://primerexplorer.jp) was used to design the specific primers for MAYV under default control parameters. Two sets of rRT-LAMP primers were selected, targeting the conserved NS1 region of MAYV (Figure S6). Each primer set consists of forward inner primer (FIP), backward inner primer (BIP), forward loop primer (FLP), backward loop primer (BLP), forward external primer (F3), and backward external primer (B3), and their sequences are listed in Table 1 [14, 15].
Table 1.
Sequences of the rRT-LAMP primers used for detection of MAYV RNA
| Primer set #1 sequences (5′→3′) | Primer set #2 sequence (5′→3′) | ||
|---|---|---|---|
| F3 | ACTCATCCTGGATATCGGCA | F3 | CGGACATTTTGCCTTCACAC |
| B3 | ACTCATTGTCCGGGGTCG | B3 | GGCCCAGTTGGTTGCATAT |
| FIP | AGACGCTCTGGGTCCTCAGCAATGATGTCTGAGCACACGT | FIP | TCGGTGCGTGAACTGCATAGACATCAGACATGCAGGACTCCA |
| BIP | AGCCAAGGCATCAGGTGAAGTACTGACTGCAGGTCGTCTA | BIP | GGAGTACGCACAGCGTACTGGCCCCGGCCATTGTATCGA |
| FLP | CATTGGGCACACACAATGGT | FLP | TGATAGACTGCCACCTCAGC |
| BLP | CGTTGACAGAAATATTGCAGCAAAG | BLP | ATTGGGTTCGACACTACCCC |
Both primer set#1 and primer set#2 were checked for possible primer dimerization using Multiple Primer Analyzer from Thermo Fisher Scientific, and we found no problematic dimer formation among primers. Possible hairpin structure formation was analyzed using OligoAnalyzer from Integrated DNA Technologies (IDT); no hairpin structures were found to exist at 62.5 °C, the reaction temperature that was used for rRT-LAMP in this work.
First, we employed the qPCR instrument to study the rRT-LAMP conditions such as assay time. We also used the qPCR instrument to compare primer set #1 with primer set #2 in Table 1 in terms of rRT-LAMP assay performance. In addition, we studied the effects of the primer concentrations on the assay performance since they are known to affect LAMP assays [53–55]. We compared the primer concentrations recommended by the LAMP kit manufacturer, New England Biolabs (NEB), with the primer concentrations at half the recommended concentrations, referred to as “1/2NEB,” based on our experience and the literature [28]. The concentrations of all primers used under these two conditions are detailed in Table S3. rRT-LAMP assays were carried out by using 1 μL RNA at concentrations from 5 × 103 to 5 × 106 GEs/μl. Figure S7 shows rRT-LAMP curves using primer set #1 and #2. In either primer sets, inferior assay performance was obtained at NEB than at 1/2NEB. For example, for primer set #1, all four concentrations of viral RNA tested were detectable at 1/2NEB, whereas only two highest concentrations (5 × 106 GEs/μl and 5 × 105 GEs/μl) were detectable at NEB. In addition, although all four concentrations were detected using both primer sets under the 1/2NEB condition, the detection time with primer set #1 (threshold time of 8–14 min.) was shorter than primer set #2 (threshold time of 18–26 min.). Therefore, we chose primer set #1 and 1/2NEB primer concentrations for all subsequent experiments.
Assay sensitivity and specificity
We investigated the limit of detection of the rRT-LAMP assay for MAYV detection. First, we used the qPCR machine to determine the required amplification time for low amount of viral RNA. Figure 3a shows real-time amplification curves of viral RNA ranging from 5 × 106 to 50 GEs/μl. The results showed that all samples containing various concentrations of RNA reached a plateau within 30 min, indicating that 30 min RT-LAMP reaction is sufficient if we use the endpoint detection. The calibration curve between the threshold time (reported by the qPCR machine) and viral RNA concentration (Fig. 3b) indicates the feasibility of semiquantitative MAYV detection.
Fig. 3.
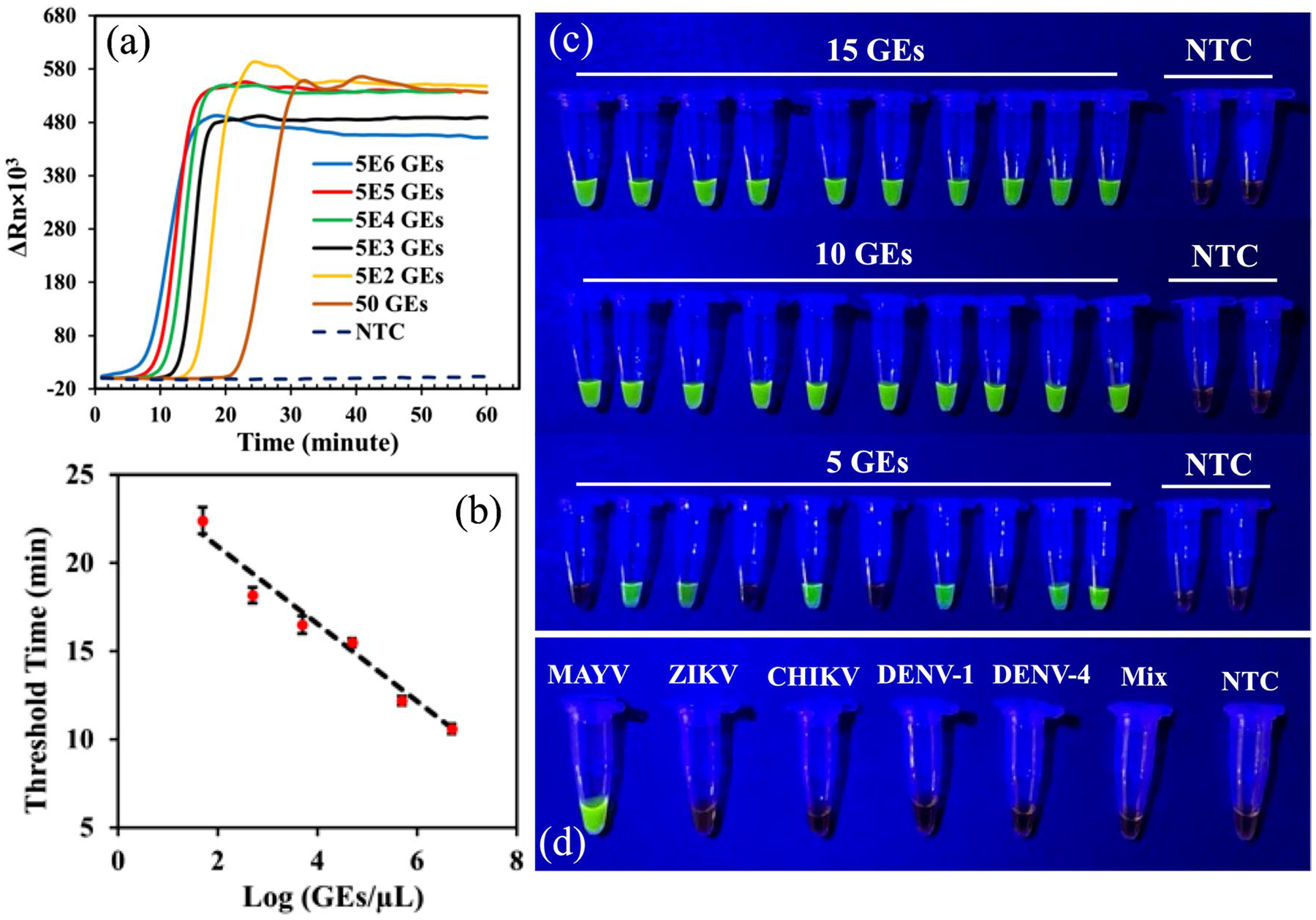
a. rRT-LAMP assay of 5 × 106 to 50 GEs MAYV RNA per reaction. All plots are an average of three replicates. b. Calibration curve between threshold time and MAYV concentration. c. RT-LAMP endpoint detection of 15, 10, and 5 GEs of MAYV RNA. The endpoint detection was carried out after 30-min RT-LAMP reactions. NTC stands for no-template control. d. No cross-reactivity was observed for MAYV-targeted RT-LAMP assay when it was carried out against ZIKV, CHIKV, DENV-1, DENV-4, and a mixture containing these four viruses.
By fixing the amplification time at 30 min and not using the qPCR machine as an optical detector, we explored the colorimetric detection at the end. This method provides “yes (the presence of MAYV)” or “no (the absence of MAYV)” results observed with the naked eye. For this endpoint detection, 0.5 μL of 10000X concentrate SYBR Green I was added to the mixture after the reaction. We first performed the endpoint detection of serial dilutions of the MAYV RNA, ranging from 5 to 5 × 106, to compare with the real-time detection results shown in Fig. 3a and determine the visual limit of detection of the assay. The results shown in Figure S8 confirmed that the visual limit of detection of the 30-min RT-LAMP is between 50 and 5 GEs. To further narrow down the limit of detection of 30 min assay, we performed the endpoint detection in tube using 15, 10, and 5 GEs/μL of MAYV RNA. Ten experiments at each RNA concentration were conducted to ensure reproducibility of the assay and the results shown in Fig. 3c. The results in Fig. 3c show that all samples containing 10 GEs and 15 GEs of MAYV RNA were detected, whereas only six out of 10 samples with 5 GEs were detected, indicating that visual limit of detection of our RT-LAMP assay is at least 10 GEs/reaction of MAYV RNA (i.e., 1 μL of 10 GEs/μL). This limit of detection is at least comparable to, if not better than, the limit of detection of 8.2 copies/μL with 5 μL of MAYV RNA (i.e., 41 copies total) using rRT-PCR [56].
To study the specificity of the RT-LAMP assay for MAYV detection, we tested it against four different mosquito-borne RNA viruses: CHIKV, two subtypes of DENV: DENV-1 and DENV-4, and ZIKV. Before experiments, we employed the basic local alignment search tool (BLAST) from NCBI to evaluate the similarities of the primer sequences designed for MAYV with the genomic sequences of the other four viruses. No similarities between the MAYV primers and sequences of other viruses were found. Isothermal amplifications were subsequently performed using the MAYV primers, and each of the other viruses while the endpoint detection was used. For each test, 1 μL of 106 GEs/μL virus RNA was tested per 25-μL RT-LAMP reaction. As shown in Fig. 3d, the RT-LAMP assay generated positive signals only for MAYV RNA, remaining negative for RNA of the other four viruses, their mixture, and the no-template control (NTC). The results indicate that there is no cross-reactivity between the MAYV RT-LAMP assays for the four mosquito-borne viruses tested.
MAYV endpoint detection using SPD
Our platform, which utilizes molecular amplification for the detection of MAYV, offers improved sensitivity and specificity compared to other LAMP-based POC platforms. This is due to the ability to process larger sample volumes (50 to 200 μL) than typical microfluidic platforms which can only handle a few microliters or less [57]. This leads to a lower detection limit, as more virus RNA can be enriched onto the detection unit’s paper pad. Additionally, compared to those devices that benefit from the use of direct sample load, our detection system enables sample preparation and RNA purification at POC. It has been shown that LAMP assay without sample preparation reduced the detection sensitivity [31] and increased the number of false negatives [45].
A number of reports have documented erroneous outcomes in blood tests employing the LAMP method [58–62]. The LAMP technique has shown to generate false positive outcomes in turbidity assays due to the presence of inhibitors in the anticoagulant-containing blood samples [63] and protein precipitation [63, 64]. Unlike other POC LAMP-based devices developed for blood analysis in which the lysed blood is directly added to either RT-LAMP or LAMP reaction mixture [22, 65], our POC device eliminates all blood components by washing them out using washing buffers, as described above. This approach potentially eliminates false positive results stemming from blood components.
To carry out POC testing, we first employed SPD (Figure S1) for sample preparation and used the MAYV RT-LAMP for the endpoint detection. Prior to the test, for 1 volume (e.g., 50 μL) of the sample, SPD was pre-loaded with 4 volume of lysis buffer (e.g., 200 μL), 4 volume of binding buffer (molecular biology grade ethanol), 4 volume of washing buffer 1 (AW1, QIAGEN), and 4 volume washing buffer 2 (AW2, QIAGEN), followed by sealing reservoirs using a thermoplastic film. The lysis buffer was made by mixing 1 volume of AVL (AVL, QIAGEN) and 2 volume of red blood cell lysis buffer (2.5 mM KHCO3, 37.5 mM NH4Cl, and 0.025 mM EDTA) (1 × RBC lysis buffer, Thermo Fisher Scientific). Note that AVL was used for lysing viruses [28, 50], and 1 × RBC lysis buffer was used for lysing red blood cells [66]. The operation of the SPD started with introducing 50 μL of MAYV sample. After a 5 min of incubation for sample lysis, the binding step was carried out by the sliding mechanism and ball-based valving incorporated in SPD as explained above. Washing steps were then performed in sequence by sliding between the buffer and mixing units. Enriched and purified RNA in the detection unit was subsequently subjected to 25 μL RT-LAMP and placed in a battery-powered coffee mug, for the endpoint detection. For the safety of the operator, virus samples were processed in a BSL-2+ (biosafety level 2 enhanced) laboratory. The device, syringe, and others in contact with the virus sample were disposed of as biohazard waste.
In order to assess the detection limit of our devices for sample-to-result testing at POC, we created whole blood samples by adding viable MAYV virus to healthy whole blood that were obtained from Innovative Research (Novi, MI, USA). The cultured virus used for spiking had a titer of 6 × 104 GEs/μL. We then prepared tenfold serial dilutions of this spiked sample using healthy blood to evaluate the device’s performance. The SPD operation began by introducing 50 μL of the blood samples, followed by the SPD operation described above. As depicted in Fig. 4, we were able to detect MAYV from 6 GEs/μL (300 GEs/reaction), but no signals were observed for the 0.6 GEs/μL (30 GEs/reaction) and the negative blood control (NB, blood samples with no MAYV added). There were no false positives with the five negative blood samples tested. To ensure the robustness of our platform, ten replicates at two lowest MAYV amount were tested. As shown in Figure S9, all samples containing 6 GEs/μL (300 GEs/reaction) were detected, whereas only five samples with 0.6 GEs/μL (30 GEs/reaction) were detected, indicating that our POC system can detect at least 300 GEs/reaction in 37 min (7 min for sample preparation using SPD and 30 min for RT-LAMP reaction) when the endpoint detection is used. The detection limit of the device for blood samples spiked with cultured MAYV (viable viruses) is lower than the in-tube assay using purified MAYV RNA. The possible reasons include (1) some RNA molecules might be lost during the processing; (2) inhibitory substances in blood samples might not be completely removed, thus having adverse effects on RT-LAMP assays; or (3) combination of both.
Fig. 4.
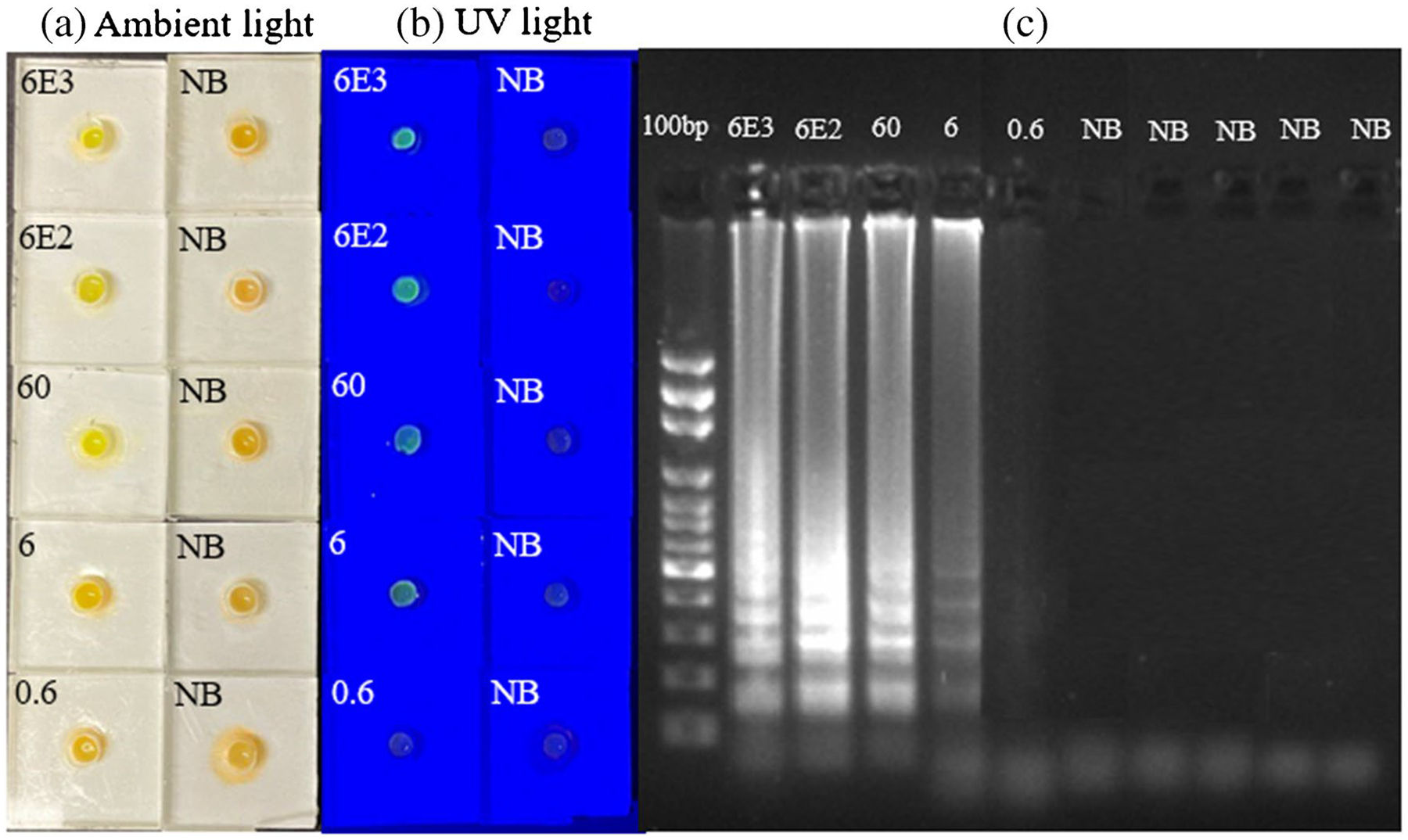
Endpoint detection of MAYV in whole blood samples using SPD. a. Pictures of the detection units under ambient light after RT-LAMP assay of whole blood samples spiked with cultured MAYV (viable virus) and blood samples without MAYV as negative controls (NB). b. Pictures of the same detection units in (a), illuminated by a blue LED flashlight. c. Gel electrophoresis image of the amplicons from each device in (a). The first lane is a DNA ladder while other lanes are for samples in (a)
To evaluate our detection platform for detection of MAYV in clinical samples, we tested three patient plasma samples from either Haiti or Venezuela. These samples were collected according to protocols approved by US and country-specific Institutional Review Boards (IRB), as described previously [67]. One patient from Haiti was infected with MAYV and one from Venezuela had DENV-1 and MAYV infections [67]. The third plasma sample was from a Haitian patient that has tested free of mosquito-borne viruses and served as a negative control sample. The analyst performing the work was initially blinded as to the identity of these samples. RT-LAMP assays were first performed in tubes, with three replicate tests for each sample. Positive MAYV signals resulted for the Haiti and Venezuela samples, but no signals were generated for the negative-control plasma samples and the NTC samples (Figure S10). We then performed the RT-LAMP assay for the detection of MAYV in these clinical samples using SPD by following the procedure explained above. Three replicates were carried out for each sample. Images of the detection unit for each sample are shown in Fig. 5a and b. The RT-LAMP results were also confirmed by gel electrophoresis, shown in Fig. 5c. These results demonstrated that our RT-LAMP assay with SPD could detect MAYV at POC. The test results agree with those obtained by RT-LAMP assay in tubes (Fig. S10) and those obtained by standard RT-PCR [67].
Fig. 5.
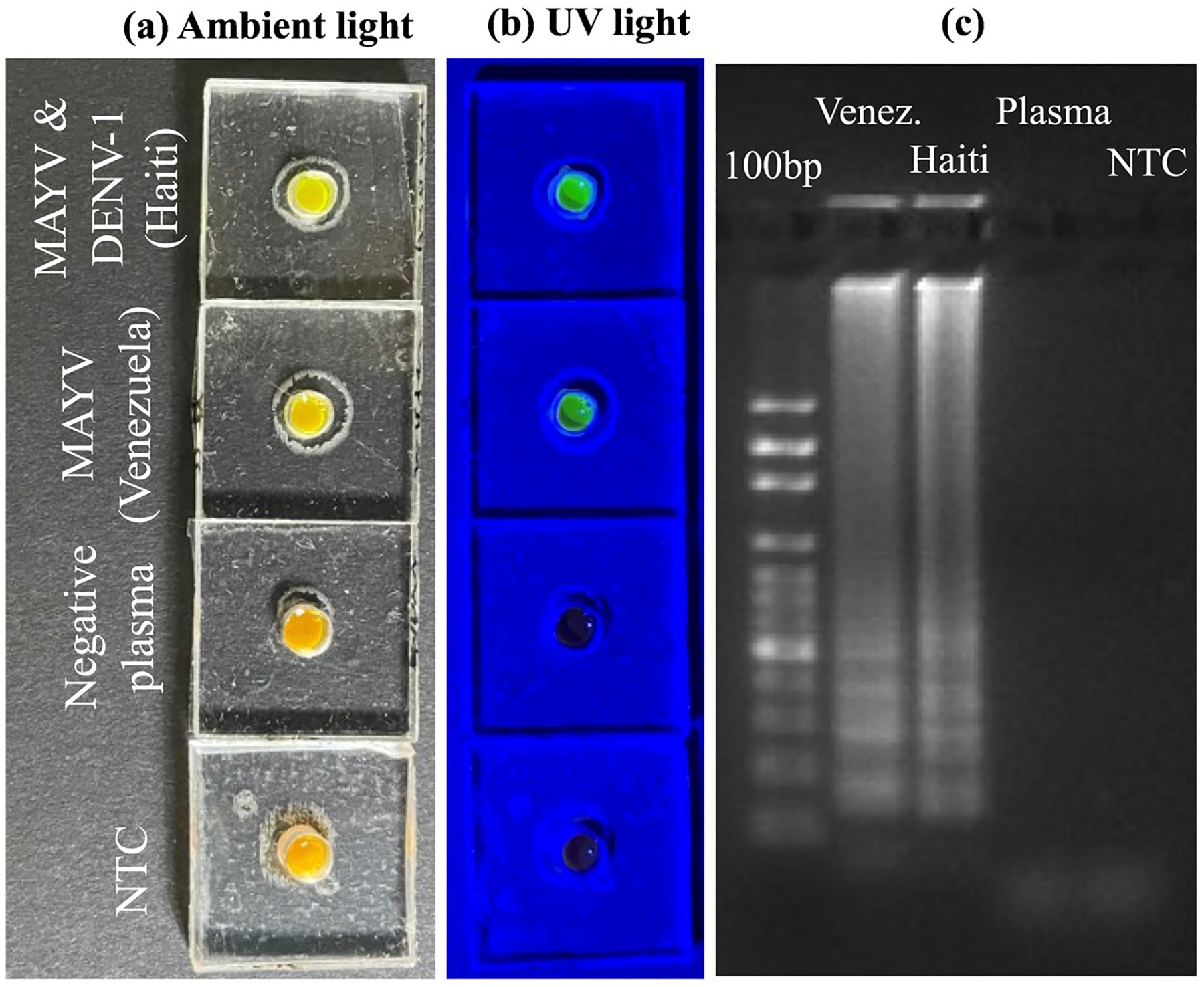
Detection of MAYV in clinical samples using SPD. a. Pictures of the detection units under ambient light after RT-LAMP assay of three clinical samples containing MAYV, MAYV and DENV-1, and free of viruses, respectively, as indicated on the left. The NTC stands for no-template control. b. Pictures of the same detection units in (a), illuminated by a blue LED flashlight. c. Gel electrophoresis image of the amplicons from each device. The first lane is a DNA ladder while other lanes are for clinical samples and NTC
Real-time detection of MAYV
We developed a POC platform for rRT-LAMP, which was designed to assess the MAYV viral load at POC by combining the SPD (Figure S1) with RAD (Fig. 1). We first used four MAYV RNA samples containing 5 × 106, 5 × 104, 5 × 102, and 50 GEs to evaluate the RAD. Each RNA sample was enriched using SPD, and the detection unit was then prepared for rRT-LAMP and placed in the heater chamber in Fig. 1. The amplification curves obtained from the flashlight-shaped digital microscope via the Python-based app are shown in Fig. 6a. The threshold time of each rRT-LAMP assay was calculated by the app, following the same definition as in real-time PCR (the time when the fluorescence signal reached 10 times the standard deviation of the baseline signals). We plotted the threshold time with the MAYV RNA amount, and a linear relationship between them was obtained as shown in Fig. 6b. This result indicates that our RAD system has the potential to provide quantitative information associated with MAYV virus load. We also performed rRT-LAMP assays of the same RNA samples using the commercial real-time PCR machine, and the threshold times were given by the software associated with the PCR machine. The results of amplification curves and the calibration curves obtained from the PCR machine are shown in Figure S11; they are comparable to that in Fig. 6, indicating that our POC platform is capable of providing with quantitative analysis of MAYV. The copy number of a gene of interest can be calculated from the threshold time using a standard curve [68]. This capability is crucial when/where the viral load of the virus in a patient needs to be monitored for clinical management and for taking appropriate measures to prevent virus transmission. Figure 6 also depicts that the assay time was reduced to as low as 6 min if a positive signal was detected using the RAD. In other words, the time from the sample to result can be as short as 13 min (7 min for sample preparation using SPD and 6 min for rRT-LAMP reaction using RAD).
Fig. 6.
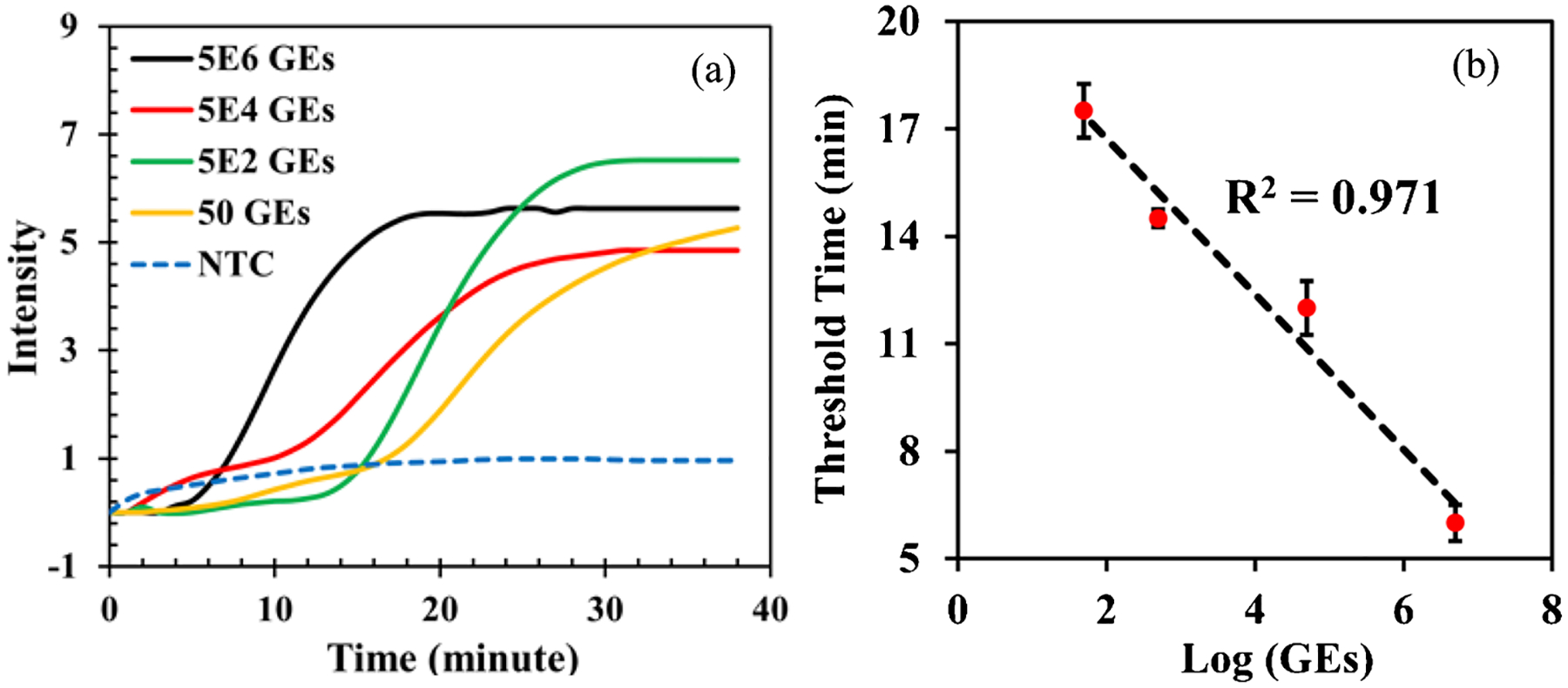
a. Real-time amplification curves obtained from RAD in Fig. 1. MAYV RNA concentrations of 5 × 106, 5 × 104, 5 × 102, and 50 GEs were used in the rRT-LAMP assay. NTC stands for no-template control. b. Calibration curve of the rRT-LAMP assay between the threshold time and RNA amount of MAYV.
To further evaluate our RAD for POC testing, we used whole blood samples spiked with cultured MAYV virus. Two virus loads of 6000 and 600 GEs/μL were tested. Figure 7 shows the amplification curve generated by the app, showing the fluorescence intensity as a function of time for different concentrations of MAYV. These results indicate that our platform can be used to obtain the MAYV viral load in whole blood samples at POC.
Fig. 7.
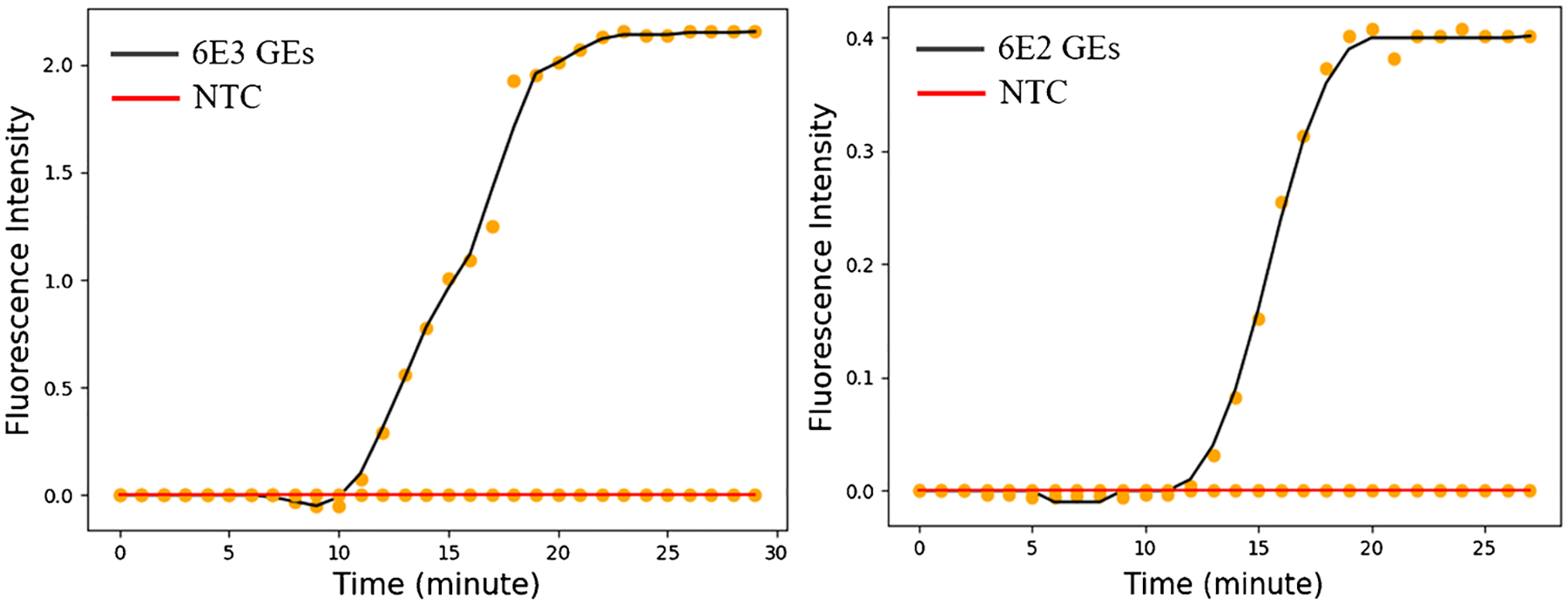
Amplification curves obtained from the RAD in Fig. 1. Two whole blood samples spiked with two different amounts of MAYV were tested. One replicate was carried out for each concentration. NTC stands for no-template control
To evaluate the feasibility of RAD for quantitative detection of other pathogens, we performed the real-time detection of ZIKV, DENV-4, and SARS-CoV-2 using RAD, and the amplification curves were shown in Figure S12. The results demonstrated that our detection system can also detect other pathogens.
In our current real-time detection platform, the detection unit consists of two reaction wells: one for processing a sample and the other for the negative control (NTC). However, it is possible to accommodate multiple samples by adjusting the microscope to have a larger field of view.
The cost of a POC test plays a crucial role in its potential commercialization. Each SPD costs $1.3, while the over-all cost of reagents and the RT-LAMP mixture amounts to $4.45. Therefore, the total cost for a POC test using endpoint detection is $5.75, which is lower than other POC devices in the literature [42, 44, 69].
For real-time detection, however, there is a one-time cost of RAD at $1003. Although the RAD cost is higher than other real-time platforms reported in the literature [36, 43, 65, 70], our system is reusable, capable of extending for other functions. Note that our real-time detection platform is still more than one order of magnitude cheaper than those commercially available real-time PCR machines.
Conclusion
One of the main barriers for preventing and reducing virus-associated disease outbreaks, such as MAYV infection, is the lack of adequate tools for diagnosis, especially those providing fast and reliable detection at POC. Rapid testing is very crucial for clinical management and transmission reduction because the clinical symptoms of MAYV infection overlap with that of other mosquitoes-borne viruses such as CHIKV, DENV, and ZIKV.
LAMP is ideal for POC testing since it is an isothermal amplification technique, enabling it to be carried out using a constant heat source such as a heat block, water bath, and chemical reaction heat [71]. This advantage, as well as its faster reactions (30 min of endpoint detection versus 2 h of PCR) and similar sensitivity, represents the key advantages of LAMP over the PCR-based methods.
Our team developed a rRT-LAMP assay for detecting MAYV and determined that its visual limit of detection was at least 10 GEs/reaction using MAYV RNA. The assay exhibited no cross-reactivity with CHIKV, DENV, or ZIKV. To enable detection of MAYV at POC, we incorporated the assay with SPD capable of detecting at least 300 GEs/reaction of MAYV in a blood sample. Additionally, we developed a portable RAD capable of quantitatively detecting MAYV and demonstrated its capacity for POC quantitative analysis of the virus. The results showed that the integrated devices have the potential for addressing challenges associated with infectious disease outbreaks caused by MAYV.
Supplementary Material
Funding
This work was supported in part by the US National Institutes of Health (R01AI155735 and R01AI158868) and the University of Florida, USA.
Biographies

Morteza Alipanah earned his Ph.D. in mechanical engineering in 2020 from the University of Florida, Gainesville, FL, USA. He has worked on microfluidics technologies, surface sciences, membrane sciences, and microelectronic cooling systems. He is currently working at the University of Florida as a postdoctoral fellow, working on the development of molecular amplification assays and molecular amplification-based point-of-care testing devices for the detection of airborne and mosquito-borne viruses and microfluidic detection devices for the detection of cancer cells in blood.

Carlos Manzanas received his Bachelor of Science degree in mechanical engineering from Northern Illinois University in 2017 and his PhD degree in mechanical engineering from the University of Florida in 2022. He has worked on the development of point-of-care devices for detection of pathogens by combining sample preparation and nucleic acid isothermal amplification assays, and he currently works in industry in Spain.

Xin Hai currently a master’s student in mechanical engineering at Purdue University, served as a lab assistant at the Interdisciplinary Microsystems Group (IMG) during his undergraduate studies at the University of Florida, Gainesville, FL, USA. His research centers on object recognition and the unmapped navigation of robots.

John A. Lednicky is Professor in the Department of Environmental and Global Health and is affiliated with the Emerging Pathogen Institute (EPI) at the University of Florida. Gainesville, FL, USA. He has training and experience in aerovirology, microbiology, and molecular biology and researches various viruses, including mosquito-borne viruses and respiratory viruses such as influenza and coronaviruses.

Alberto Paniz-Mondolfi is Associate Professor at the Icahn School of Medicine at Mount Sinai, New York, NY, USA, and is an internist and pathologist affiliated with multiple hospitals in the greater New York area, including Mount Sinai, Morning-side, West, and Beth Israel Hospitals where he serves as Director of Molecular Microbiology. Additionally, he serves as Academic Director and Founder of the Venezuelan Science Incubator (incubadorave.org), a group dedicated to infectious diseases research and raising awareness, located in Venezuela.

J. Glenn Morris is Director of the Emerging Pathogen Institute (EPI) and Professor of Infectious Diseases in the College of Medicine at the University of Florida, Gainesville, FL, USA. He has worked in public health and pathogen-related fields for more than 40 years and has had a continuing fascination with emerging pathogens.

Z. Hugh Fan is Professor of the Department of Mechanical and Aerospace Engineering, J. Crayton Pruitt Family Department of Biomedical Engineering, and Department of Chemistry at University of Florida (UF), Gainesville, FL, USA. His research focus is to develop microfluidics technologies and apply them to biomedical applications, including cancer diagnosis/prognosis and pathogen detection.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00216-023-04856-8.
Conflict of interest The authors declare no competing interests.
References
- 1.Pezzi L, Rodriguez-Morales AJ, Reusken CB, Ribeiro GS, LaBeaud AD, Lourenco-de-Oliveira R, et al. GloPID-R report on Chikungunya, O’nyong-nyong and Mayaro virus, part 3: Epidemiological distribution of Mayaro virus. Antiviral Res. 2019;172: 104610. [DOI] [PubMed] [Google Scholar]
- 2.Tesh RB, Watts DM, Russell KL, Damodaran C, Calampa C, Cabezas C, et al. Mayaro virus disease: an emerging mosquito-borne zoonosis in tropical South America. Clin Infect Dis. 1999;28(1):67–73. [DOI] [PubMed] [Google Scholar]
- 3.de Thoisy B, Gardon J, Salas RA, Morvan J, Kazanji M. Mayaro virus in wild mammals, French Guiana. Emerg Infect Dis. 2003;9(10):1326–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halsey ES, Siles C, Guevara C, Vilcarromero S, Jhonston EJ, Ramal C, et al. Mayaro virus infection, Amazon Basin region, Peru, 2010–2013. Emerg Infect Dis. 2013;19(11):1839–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich-Jänicke B, Emmerich P, Tappe D, Günther S, Cadar D, Schmidt-Chanasit J. Genome analysis of Mayaro virus imported to Germany from French Guiana. Emerg Infect Dis. 2020;14:1255–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinheiro FP, Freitas RB, Travassos da Rosa JF, Gabbay YB, Mello WA, LeDuc JW. An outbreak of Mayaro virus disease in Belterra, Brazil. I. Clinical and virological findings. Am J Trop Med Hyg. 1981;30(3):674–81. [DOI] [PubMed] [Google Scholar]
- 7.Acosta-Ampudia Y, Monsalve DM, Rodríguez Y, Pacheco Y, Anaya JM, Ramírez-Santana C. Mayaro: an emerging viral threat? Emerg Microbes Infect. 2018;7(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lima WG, Pereira RS, da Cruz Nizer WS, Brito JCM, Godói IP, Cardoso VN, et al. Rate of exposure to Mayaro virus (MAYV) in Brazil between 1955 and 2018: a systematic review and meta-analysis. Arch Virol. 2021;166(2):347–61. [DOI] [PubMed] [Google Scholar]
- 9.Llagonne-Barets M, Icard V, Leparc-Goffart I, Prat C, Perpoint T, André P, et al. A case of Mayaro virus infection imported from French Guiana. J Clin Virol. 2016;77:66–8. [DOI] [PubMed] [Google Scholar]
- 10.Cai Q, Mu J, Lei Y, Ge J, Aryee AA, Zhang X, et al. Simultaneous detection of the spike and nucleocapsid proteins from SARS-CoV-2 based on ultrasensitive single molecule assays. Anal Bioanal Chem. 2021;413(18):4645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long KC, Ziegler SA, Thangamani S, Hausser NL, Kochel TJ, Higgs S, et al. Experimental transmission of Mayaro virus by Aedes aegypti. Am J Trop Med Hyg. 2011;85(4):750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naveca FG, Nascimento VAD, Souza VC, Nunes BTD, Rodrigues DSG, Vasconcelos P. Multiplexed reverse transcription real-time polymerase chain reaction for simultaneous detection of Mayaro, Oropouche, and Oropouche-like viruses. Mem Inst Oswaldo Cruz. 2017;112(7):510–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botes M, de Kwaadsteniet M, Cloete TE. Application of quantitative PCR for the detection of microorganisms in water. Anal Bioanal Chem. 2013;405(1):91–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015;53(1):1–5. [DOI] [PubMed] [Google Scholar]
- 16.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–9. [DOI] [PubMed] [Google Scholar]
- 17.Lin Q, Ye X, Yang B, Fang X, Chen H, Weng W, et al. Real-time fluorescence loop-mediated isothermal amplification assay for rapid and sensitive detection of Streptococcus gallolyticus subsp. gallolyticus associated with colorectal cancer. Anal Bioanal Chem. 2019;411(26):6877–87. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi M, Mashiko T, Wilisiani F, Hartono S, Nishigawa H, Natsuaki T, et al. Development of a RT-LAMP assay for real-time detection of criniviruses infecting tomato. J Virol Methods. 2023;312: 114662. [DOI] [PubMed] [Google Scholar]
- 19.Jiang J, Feindel W, Swisher Grimm K, Harding M, Feindel D, Bajema S, et al. Development of a loop-mediated isothermal amplification (LAMP) method to detect the potato zebra chip pathogen ‘Candidatus Liberibacter solanacearum’ (Lso) and differentiate haplotypes A and B. Plant Dis. 2023;107(6):1697–702. [DOI] [PubMed] [Google Scholar]
- 20.Priye A, Bird SW, Light YK, Ball CS, Negrete OA, Meagher RJ. A smartphone-based diagnostic platform for rapid detection of Zika, Chikungunya, and dengue viruses. Sci Rep. 2017;7:44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaren O, Alto BW, Gangodkar PV, Ranade SR, Patil KN, Bradley KM, et al. Point of sampling detection of Zika virus within a multiplexed kit capable of detecting dengue and Chikungunya. BMC Infect Dis. 2017;17(1):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganguli A, Ornob A, Yu H, Damhorst GL, Chen W, Sun F, et al. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and dengue at point-of-care. Biomed Microdevices. 2017;19(4):73. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Jimena B, Bekaert M, Bakheit M, Frischmann S, Patel P, Simon-Loriere E, et al. Development and validation of four one-step real-time RT-LAMP assays for specific detection of each dengue virus serotype. PLoS Negl Trop Dis. 2018;12(5): e0006381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teoh BT, Sam SS, Tan KK, Johari J, Danlami MB, Hooi PS, et al. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect Dis. 2013;13:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oloniniyi OK, Kurosaki Y, Miyamoto H, Takada A, Yasuda J. Rapid detection of all known ebolavirus species by reverse transcription-loop-mediated isothermal amplification (RT-LAMP). J Virol Methods. 2017;246:8–14. [DOI] [PubMed] [Google Scholar]
- 26.Curtis KA, Morrison D, Rudolph DL, Shankar A, Bloomfield LSP, Switzer WM, et al. A multiplexed RT-LAMP assay for detection of group M HIV-1 in plasma or whole blood. J Virol Methods. 2018;255:91–7. [DOI] [PubMed] [Google Scholar]
- 27.Seok Y, Yin Q, Li R, Mauk MG, Bai H, Bau HH. Manually-operated, slider cassette for multiplexed molecular detection at the point of care. Sens Actuators, B Chem. 2022;369: 132353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manzanas C, Alam MM, Loeb JC, Lednicky JA, Wu CY, Fan ZH. A valve-enabled sample preparation device with isothermal amplification for multiplexed virus detection at the point-of-care. ACS Sens. 2021;6(11):4176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez NM, Linnes JC, Fan A, Ellenson CK, Pollock NR, Klapperich CM. Paper-based RNA extraction, in situ isothermal amplification, and lateral flow detection for low-cost, rapid diagnosis of influenza A (H1N1) from clinical specimens. Anal Chem. 2015;87(15):7872–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang M, Kim S, Song J. Rapid and simple detection of influenza virus via isothermal amplification lateral flow assay. Anal Bioanal Chem. 2022;414(16):4685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Translat Med. 2020;12(556):eabc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garneret P, Coz E, Martin E, Manuguerra JC, Brient-Litzler E, Enouf V, et al. Performing point-of-care molecular testing for SARS-CoV-2 with RNA extraction and isothermal amplification. PLoS ONE. 2021;16(1): e0243712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang B, Xu J, Liu Y, Peng H, Feng W, Cao Y, et al. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology. Anal Chem. 2020;92(24):16204–12. [DOI] [PubMed] [Google Scholar]
- 34.Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci U S A. 2020;117(39):24450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Zheng T, Wang H, Chen W, Huang X, Liang J, et al. Rapid one-pot detection of SARS-CoV-2 based on a lateral flow assay in clinical samples. Anal Chem. 2021;93(7):3325–30. [DOI] [PubMed] [Google Scholar]
- 36.Panpradist N, Kline EC, Atkinson RG, Roller M, Wang Q, Hull IT, et al. Harmony COVID-19: a ready-to-use kit, low-cost detector, and smartphone app for point-of-care SARS-CoV-2 RNA detection. Sci Adv. 2021;7(51):eabj1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganguli A, Mostafa A, Berger J, Aydin MY, Sun F, Ramirez SAS, et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(37):22727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jhou YR, Wang CH, Tsai HP, Shan YS, Lee GB. An integrated microfluidic platform featuring real-time reverse transcription loop-mediated isothermal amplification for detection of COVID-19. Sens Actuators, B Chem. 2022;358: 131447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C. Instrument-free point-of-care molecular detection of Zika virus. Anal Chem. 2016;88(14):7289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Loeb JC, Manzanas C, Lednicky JA, Fan ZH. Valve-enabled sample preparation and RNA amplification in a coffee mug for Zika virus detection. Angew Chem Int Ed Engl. 2018;57(52):17211–4. [DOI] [PubMed] [Google Scholar]
- 41.Calvert AE, Biggerstaff BJ, Tanner NA, Lauterbach M, Lanciotti RS. Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP). PLoS ONE. 2017;12(9): e0185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohon AN, Lee LD, Bayih AG, Folefoc A, Guelig D, Burton RA, et al. NINA-LAMP compared to microscopy, RDT, and nested PCR for the detection of imported malaria. Diagn Microbiol Infect Dis. 2016;85(2):149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz LM, Johnson BE, Jenkins DM. Real-time optical analysis of a colorimetric LAMP assay for SARS-CoV-2 in saliva with a handheld instrument improves accuracy compared with endpoint assessment. J Biomol Tech. 2021;32(3):158–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahavi M, Rohana H, Azrad M, Shinberg B, Peretz A. Rapid SARS-CoV-2 detection using the lucira check it COVID-19 test kit. Diagnostics (Basel). 2022;12(8):1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon YJ, Lee SY, Oh SW. A review of isothermal amplification methods and food-origin inhibitors against detecting food-borne pathogens. Foods. 2022;11(3):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blohm GM, Marquez-Colmenarez MC, Lednicky JA, Bonny TS, Mavian C, Salemi M, et al. Isolation of Mayaro virus from a venezuelan patient with febrile illness, arthralgias, and rash: further evidence of regional strain circulation and possible long-term endemicity. Am J Trop Med Hyg. 2019;101(6):1219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lednicky J, De Rochars VM, Elbadry M, Loeb J, Telisma T, Chavannes S, et al. Mayaro virus in child with acute febrile illness, Haiti, 2015. Emerg Infect Dis. 2016;22(11):2000–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93(1):125–8. [DOI] [PubMed] [Google Scholar]
- 49.Hsieh K, Mage PL, Csordas AT, Eisenstein M, Soh HT. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem Commun (Camb). 2014;50(28):3747–9. [DOI] [PubMed] [Google Scholar]
- 50.Jiang X, Loeb JC, Pan M, Tilly TB, Eiguren-Fernandez A, Lednicky JA, et al. Integration of sample preparation with RNA-amplification in a hand-held device for airborne virus detection. Anal Chim Acta. 2021;1165: 338542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monis PT, Giglio S, Saint CP. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal Biochem. 2005;340(1):24–34. [DOI] [PubMed] [Google Scholar]
- 52.White SK, Mavian C, Elbadry MA, Beau De Rochars VM, Paisie T, Telisma T, et al. Detection and phylogenetic characterization of arbovirus dual-infections among persons during a Chikungunya fever outbreak, Haiti 2014. PLoS Negl Trop Dis. 2018;12(5):e0006505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Xu L, Guo J, Chen R, Grisham MP, Que Y. Development of loop-mediated isothermal amplification for detection of Leifsonia xyli subsp. xyli in sugarcane. Biomed Res Int. 2013;2013:357692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh HY, Shoemaker CA, Klesius PH. Evaluation of a loop-mediated isothermal amplification method for rapid detection of channel catfish Ictalurus punctatus important bacterial pathogen Edwardsiella ictaluri. J Microbiol Methods. 2005;63(1):36–44. [DOI] [PubMed] [Google Scholar]
- 55.Foo PC, NurulNajian AB, Muhamad NA, Ahamad M, Mohamed M, Yean Yean C, et al. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: a comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020;20(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waggoner JJ, Rojas A, Mohamed-Hadley A, de Guillen YA, Pinsky BA. Real-time RT-PCR for Mayaro virus detection in plasma and urine. J Clin Virol. 2018;98:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo HJ, Baek C, Lee MH, Min J. Integrated microsystems for the in situ genetic detection of dengue virus in whole blood using direct sample preparation and isothermal amplification. Analyst. 2020;145(6):2405–11. [DOI] [PubMed] [Google Scholar]
- 58.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, et al. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol. 2003;41(12):5517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Q, Wang F, Prinyawiwatkul W, Ge B. Robustness of Salmonella loop-mediated isothermal amplification assays for food applications. J Appl Microbiol. 2014;116(1):81–8. [DOI] [PubMed] [Google Scholar]
- 60.Yang Q, Chen S, Ge B. Detecting Salmonella serovars in shell eggs by loop-mediated isothermal amplification. J Food Prot. 2013;76(10):1790–6. [DOI] [PubMed] [Google Scholar]
- 61.Suleman E, Mtshali MS, Lane E. Investigation of false positives associated with loop-mediated isothermal amplification assays for detection of Toxoplasma gondii in archived tissue samples of captive felids. J Vet Diagn Invest. 2016;28(5):536–42. [DOI] [PubMed] [Google Scholar]
- 62.Stridiron AK. Development and testing of LAMP assay for diagnosis of Plasmodium falciparum malaria. Ethnic Dis. 2009;19(2 suppl. 3):23–4. [Google Scholar]
- 63.Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, et al. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol. 2011;62(1):41–8. [DOI] [PubMed] [Google Scholar]
- 64.Berth M, Delanghe J. Protein precipitation as a possible important pitfall in the clinical chemistry analysis of blood samples containing monoclonal immunoglobulins: 2 case reports and a review of the literature. Acta Clin Belg. 2004;59(5):263–73. [DOI] [PubMed] [Google Scholar]
- 65.Jankelow AM, Lee H, Wang W, Hoang TH, Bacon A, Sun F, et al. Smartphone clip-on instrument and microfluidic processor for rapid sample-to-answer detection of Zika virus in whole blood using spatial RT-LAMP. Analyst. 2022;147(17):3838–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curtis KA, Rudolph DL, Owen SM. Sequence-specific detection method for reverse transcription, loop-mediated isothermal amplification of HIV-1. J Med Virol. 2009;81(6):966–72. [DOI] [PubMed] [Google Scholar]
- 67.Blohm G, Elbadry MA, Mavian C, Stephenson C, Loeb J, White S, et al. Mayaro as a Caribbean traveler: evidence for multiple introductions and transmission of the virus into Haiti. Int J Infect Dis. 2019;87:151–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol. 2012;78(12):4481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Torezin Mendonça G, Cassaboni Stracke M, de Oliveira Coelho B, Bruna SoligoSanchuki H, Klassen de Oliveira V, KleryntonMarchini F, et al. A new RT-LAMP-on-a-chip instrument for SARS-CoV-2 diagnostics. Microchem J. 2022;180:107600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larrea-Sarmiento A, Dhakal U, Boluk G, Fatdal L, Alvarez A, Strayer-Scherer A, et al. Development of a genome-informed loop-mediated isothermal amplification assay for rapid and specific detection of Xanthomonas euvesicatoria. Sci Rep. 2018;8(1):14298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


