Abstract
Modern toxicology’s throughput has dramatically increased due to alternative models, laboratory automation, and machine learning. This has enabled comparative studies across species and assays to prioritize chemical hazard potential and to understand how different model systems might complement one another. However, such comparative studies of high-throughput data are still in their infancy, with more groundwork needed to firmly establish the approach. Therefore, this study aimed to compare the bioactivity of the NIEHS Division of Translational Toxicology’s (DTT) 87-compound developmental neurotoxicant (DNT) library in zebrafish and an in vitro high-throughput cell culture system. The early life-stage zebrafish provided a whole animal approach to developmental toxicity assessment. Chemical hits for abnormalities in embryonic zebrafish morphology, mortality, and behavior (ZBEscreen™) were compared with chemicals classified as high-risk by the Cell Health Index (CHI™), which is an outcome class probability from a machine learning classifier using 12 parameters from the SYSTEMETRIC® Cell Health Screen (CHS). The CHS was developed to assess human toxicity risk using supervised machine learning to classify acute cell stress phenotypes in a human leukemia cell line (HL60 cells) following a 4-h exposure to a chemical of interest. Due to the design of the screen, the zebrafish assays were more exhaustive, yielding 86 total bioactive hits, whereas the SYSTEMETRIC® CHS focusing on acute toxicity identified 20 chemicals as potentially toxic. The zebrafish embryonic and larval photomotor response assays (EPR and LPR, respectively) detected 40 of the 47 chemicals not found by the zebrafish morphological screen and CHS. Collectively, these results illustrate the advantages of using two alternative models in tandem for rapid hazard assessment and chemical prioritization and the effectiveness of CHI™ in identifying toxicity within a single multiparametric assay.
Keywords: Hazard assessment, alternative models, high-throughput, comparative analyses, developmental neurotoxicity
INTRODUCTION
The U.S. National Research Council’s (NRC) conception of “Toxicity Testing in the 21st Century” urged a paradigm shift to increase the use of in vitro approaches and reduce the cost and time of chemical risk assessments. HTS in vitro studies have yielded expansive amounts of data providing valuable new knowledge (Burgoon et al., 2017; Villeneuve et al., 2019; Zhu et al., 2014). With the continued advancement of automation and data science, HTS assays can generate robust data more efficiently than the NRC anticipated in 2007, particularly for cross-species comparisons. (LaLone et al., 2018). HTS cross-species comparative analyses have gained more attention as an efficient and comprehensive chemical hazard and risk assessment strategy (Darde et al., 2018; George et al., 2011; LaLone et al., 2018). Integrating different model systems and comparative screenings provide greater weight of evidence for chemical prioritization and hazard assessments as response to a chemical exposure shared, or even distinct, across species provides valuable information during the decision-making process (Bell et al., 2018; Hagstrom et al., 2019; Krewski et al., 2020). Many HTS assays are unidimensional and cannot account for whole-system responses to chemical exposure nor address the complexity of chemical bioactivity across species, particularly due to metabolic inactivity within in vitro assays (Bell et al., 2018; Bieberich et al., 2021). A robust and time/resource-efficient approach to chemical screening is needed to keep pace with the production and release of novel chemicals in consumer products, pharmaceuticals, industrial processes, and the environment.
In 2019, the National Institute of Environmental Health Sciences (NIEHS), National Toxicology Program (NTP) now known as the Division of Translation Toxicology (DTT), identified the limitations of traditional toxicity assessments, specifically regarding developmental neurotoxicity (DNT) (Behl et al., 2019). To address this limitation, the NTP curated a chemical library composed of either known or suspected DNT chemicals as a first attempt to establish collaborations between different labs that screen chemicals using alternative models (Behl et al., 2019; Hagstrom et al., 2019). This collaboration among 40 different labs resulted in the production of extensive in vitro and in vivo datasets that permitted cross-system comparative analyses. Comparing developmental zebrafish and a freshwater planarian screen of the DNT library revealed that the zebrafish model was more sensitive. Nonetheless, the endpoint coverage of the multiple systems provided a more exhaustive method for assessing developmental neurotoxicity (Hagstrom et al., 2019). Other screening studies included in the NTP’s DNT library screenings demonstrated that cross-system comparative analyses are powerful in that each system has unique endpoint(s) indicative of developmental neurotoxicity or bioactivity that would be missed if only one system was screened (Crofton et al., 2022). In agreement with this reasoning, this study compares the bioactivity of the NTP’s DNT library in a high-throughput live-cell system and the developmental zebrafish.
AsedaSciences SYSTEMETRIC® Cell Health Screen (CHS) applies a data processing pipeline that automates the detection of predictive features within multiparametric readouts gathered through an established flow cytometry analysis enhanced with proprietary feature extraction techniques based on distances between readout distributions (Edwards et al., 2004,1999; Robinson et al., 2013, 2012; Bernas et al., 2008). In this case, the term “predictive feature” has a mathematical meaning and does not simply designate a biological characteristic. However, the biological characteristics driving these extracted features can briefly be listed as cell morphology, cell membrane integrity, reactive oxygen species (mostly mitochondrial superoxide), glutathione depletion, nuclear membrane integrity, cell cycle, and mitochondrial membrane polarization. The methodology of CHS has been previously described (Bieberich et al., 2021, 2022). The CHS quantifies and utilizes statistical modifications in multivariate distributions created by biological measures of single cells’ characteristics obtained through a multiparametric flow cytometry (FC) system. The resulting biological response vectors (population change trajectories) can be formed by extracting descriptors of cell population changes (inter-distribution divergences) that emerge during exposure to increasing concentrations of test compounds. These vectors of divergences are utilized to train an elastic-net logistic regression model, which leverages a database of carefully curated known compound characteristics. For this study, the classifier used a 300-compound training set comprising on-market drugs, withdrawn drugs, and research compounds. During training, the compounds were assigned to either a “high toxicity risk” or “low toxicity risk” class for human use based upon an extensive review of curated literature. However, when evaluating the risk of unknown compounds, a continuous value indicating the probability of class membership is provided. These values, ranging from 0 to 1, are defined as cell health indices (CHI™), which forecast the human-use risk associated with the tested compounds. The basic premise of the assay is that compounds with similar risk characteristics are likely to induce similar population-wide cell-response trajectories in the context of the associated FC assay. The assay used in the system has been designed to reveal immediate cell toxicity by using the above-listed biological characteristics as proxyies. Thus, the CHI™ primarily assesses the danger to humans posed by compounds that may potentially exhibit these important mechanisms of toxicity (Will and Dykens, 2014). The raw underlying FC parameters utilized with acute cell stress are shown in Table S1.
The CHS is unique in that, rather than reporting only a series of unidimensional assessments associated with each of the FC-measured biomarkers, it also provides the CHI™, which is a single summary predictor based on a multidimensional classifier. Additionally, none of the employed FC data processing pathways require manually-executed tasks such as cytometry gating (Bieberich et al., 2022, 2021). The CHI™, along with other machine learning assigned endpoint scores produced by sparser versions of the model, are collected in the AsedaSciences cloud-based SaaS platform, where further comparisons to other chemicals/structures can be made. The SYSTEMETRIC® CHS is a novel approach for integrating flow cytometry into HTS studies cost-effectively and comprehensively.The zebrafish embryo is an excellent whole-animal model, particularly in developmental and neurotoxicity research (d’Amora and Giordani, 2018). The zebrafish is particularly powerful in that it can be used in HTS studies due to its external fertilization, small size, and rapid development while sharing similar and trackable developmental physiology with humans which includes brain and central nervous system development (Tal et al., 2014; Truong et al., 2014). Embryonic and larval zebrafish assays provide the added dimension of quantitative behaviors that are incorporated into HTS and produce robust endpoints for toxicants below the threshold of morphological effects ( Truong et al., 2013; Tal et al., 2014; Basnet et al., 2019; Shen et al., 2020; Zhang et al., 2021). The Tanguay laboratory’s ZBEscreen™ combines mortality, morphological, and behavioral endpoints to produce multidimensional data outputs for chemical hazard assessments.
Zebrafish exposures began at 6 hours post fertilization (hpf) and animals were assessed for chemical effects at 24 and 120 hpf in order to assess different stages of central nervous system development, as well as exploit the sensitivity of larval behavior assays (Truong et al., 2013). HL60 cell exposures, on the other hand, lasted 4 hours, and CHS was conducted immediately after. In this study, the zebrafish identified 86 of the 87 DNT chemicals as bioactive, while the CHS detected 20 bioactives, according to CHI™. Comparisons made to available in vitro data collected through TOXcast and TOX21 screens revealed severe data gaps but also interesting correlations (and lack thereof) between CHI™, ZBEscreen™, and 42 endpoint readouts across the 1,385 assays curated on the EPA’s CompTox Dashboard. The integration of AsedaSciences SYSTEMETRIC® Cell Health and ZBE™ screens provided a rigorous and comprehensive evaluation of developmental and acute toxicity, with each assay yielding distinct and unique information.
MATERIALS AND METHODS
DNT Library Acquisition
Table S2 lists the chemicals, CASRN, and chemical class within the NTP’s 87-compound DNT library consisting of drugs, industrial chemicals, flame retardants, polycyclic aromatic hydrocarbons (PAHs), and pesticides. NTP’s DNT library was chosen due to its size and scope, covering 6 different chemical classes for a more rigorous interrogation of both model systems’ ability to detect bioactivity. All chemical stocks were 20 mM concentrations in dimethyl sulfoxide (DMSO). Further information about the DNT library can be found in Behl et al. (2019).
AsedaSciences SYSTEMTRIC® Cell Health Screen
HL60 cells were produced in suspension cultures in non-treated 850 cm2 roller bottles with vented caps in RPMI 1640 without glucose, supplemented with 10 mM galactose and 10% dialyzed heat-inactivated FBS, at 1 RPM, 5% CO2, and 37°C. In a 384-well platform, HL60 cells were exposed to a 10-step, 3X dilution series of each chemical (5 nM-100μM) for 4 h at 37°C with 5% CO2. Prior to cell deposition, assay plates containing chemicals were sealed and stored at room temperature in the dark for 2 h in order to allow binding equilibrium between serum components and exposure chemicals. Following the 4 h chemical exposures, the assay plates were removed and cells were gently pelleted at 300 xg for 2 min. A Biomek NXP was then used to aspirate 20 μL of each well volume, after which 20 μL of fluorescent dye mix was deposited, plate sealed, and shaken 2X at 2,200 RPM for 5 s each time. Chemical formatting, cell deposition, and dye application were performed robotically, where final assay conditions comprised 100,000 cells in 40 μL volumes. Flow cytometry data were acquired with a CyAn™ ADP flow cytometer (Beckman Coulter) with automated sampling performed by a HyperCyt® autosampler (Intellicyt). Well-specific flow cytometry data files combined with a map of well contents were then moved to cloud infrastructure where the automated algorithm for quality control and machine learning classification is triggered. A complete description of the CHS can be found in the SI material.
ZBEscreen™
Tropical 5D wildtype adult zebrafish were housed at Oregon State University, Sinnhuber Aquatic Research Laboratory (SARL) in a standard 14h light/10 h dark-light cycle at 28°C. Embryos were collected, cleaned, and staged prior to dechorionation at 4 hours post fertilization (hpf). The chorions were enzymatically removed, and at 6 hpf, 1 embryo was placed in each well of round bottom 96-well plates prefilled will 100 μL of embryo media. The chemicals were digitally dispensed directly from the 20 mM stocks into the test wells using a Hewlett Packard D300e, with all wells normalized to 0.64% DMSO (vol/vol). Up to 1% DMSO has no adverse effects on developing zebrafish (Maes et al., 2012). Each chemical was tested at 0, 1, 2, 4.5, 9, 18, 34, and 67 μM, sealed with parafilm to minimize evaporation, and shaken gently overnight at 235 rpm (Truong et al., 2016). Embryos were not exposed to visible light until 24 hpf assessments, after which they were again placed in the dark until the final 120 hpf assessments. Mortality and behavioral assessments were performed at 24 hpf using the Tanguay Lab’s zebrafish acquisition and analysis program (ZAAP) to record mortality, and embryo photomotor response (EPR) was assessed using the Photo-motor Response Assessment Tool (Reif et al., 2016; Hagstrom et al., 2019). At 120 hpf, morphological assessments included 9 morphological endpoints recorded using ZAAP (following euthanasia using tricaine methanesulfonate), and behavioral assessments were done through larval photomotor response (LPR) using Viewpoint LifeScience Zebraboxes (Truong et al., 2014; Zhang et al., 2021). The EPR test consisted of 30 s darkness; first, 1 s VIS light pulse, 9 s darkness; second, 1 s VIS light pulse; 10 s darkness (Truong et al., 2014). Statistical significance was calculated for each interval using a Kolmogorov-Smirnov test (K-S) with a threshold of p < 0.01. LPR was performed prior to 120 hpf morphological evaluations, which consisted of 4 total light cycles; each cycle was 3 min of alternating light and dark. Malformed or dead animals were excluded from subsequent analyses. Statistical significance was determined using a K-S test (p < 0.01) and a relative ratio of >10% or <10%. All analyses were performed using custom R scripts previously described (R Core Team, 2022). The ZBEscreen™ exposure paradigm is described in more detail in Hagstrom et al. (2019).
Individual System and Comparative Analyses
The ZBEscreen™ provides whole systems approach, while the CHS assesses human safety risk by applying a machine learning classifier to multidimensional cellular stress phenotype vectors, resulting in an overall probability that the chemical will or will not be toxic (CHI™). Hit calls were initially compared for either system, regardless of individual endpoint and/or assay, as it was impossible to compare individual endpoints between the zebrafish and HL60 cells due to their functional and biological differences (Table 1). The lowest effect levels (LELs) for each endpoint screened were identified in the zebrafish when there were 2 consecutive concentration hits for an endpoint. In contrast, individual endpoint scores and CHI™ were collected to assess the bioactivity of the DNT chemical library using CHS (Table S3). CHS analyses examined scores assigned to CHI™ (bioactive) and non-CHI™ (nonbioactive) chemicals to identify patterns and chemical concordance with zebrafish. CHI™ scores of ≥ 0.5 for chemicals were assigned as “bioactive” in line with the classifier training set of chemicals (Table S2). The concentration range chosen in the CHS was based upon previous investigations that utilized flow cytometry-based screening techniques (Bieberich et al., 2022; Robinson, et al., 2012). It’s essential to note that the concentrations used in the assay were set to obtain informative trajectories rather than to replicate the physiologically relevant concentrations observed in vivo. Consequently, it can be inferred that low-risk (low CHI) would mostly pertain to compounds that don’t exhibit cytotoxic characteristics even in high concentrations during human use. Aberrant endpoints (i.e., abnormal zebrafish morphology) identified in the ZBEscreen™ and CHI™ chemicals identified by the CHS were considered “bioactive,” regardless of the concentration in which it occurred. Due to the different ranges of concentrations tested in both systems and the disparities in system characteristics, a direct comparison of concentration in the tested assays was not considered (i.e., whole model vs. in vitro, short vs. prolonged exposure, different identifiable endpoints). Accordingly, ZBEscreen™ LEL data were converted to binary representation: bioactive or nonbioactive (any chemical with LEL value is considered bioactive).
Table 1.
Endpoint classes used in SYSTEMETRIC® Cell Health Screen and the ZBEscreen™.
| Classes of endpoints used in in vitro (SYSTEMETRIC®) and in vivo (zebrafish) systems | ||
|---|---|---|
|
| ||
| Endpoint Class | Zebrafish Endpoints | SYSTEMETRIC® Endpoints |
| Mortality | Mortality (24 hpf, 120 hpf) | |
| Morphology | Craniofacial malformation Curved/bent axis Yolk/heart edema Muscular malformation Lateral trunk malformation Brain malformation or necrosis Hyper/hypo pigmentation Notochord distortion Abnormal touch response |
|
| Behavior | Embryo photomotor response (EPR: 24 hpf) Larval photomotor response (LPR: 120 hpf) |
|
| Cytotoxicity | Cell morphology Cell membrane integrity ROS superoxide Glutathione depletion Nuclear membrane integrity 1 Cell cycle Nuclear membrane integrity 2 Mitochondrial membrane potential CHI probability |
|
System analyses began with chemical classifications (drug, flame retardant, industrial, PAH, pesticide negative) as defined by the NTP to identify chemical classes in which either system/system assay was most sensitive. Available in vitro data from EPA’s CompTOX Dashboard was batch exported for hit calls (binary) for all 87 chemicals and AC50 values. It is important to note that AC50s were averaged per assay endpoint as multiple assays assessed the same endpoint(s), sometimes in different in vitro systems, and correlation tests/calculations could not be run on assay endpoints with multiple values. The data was processed using custom R scripts to identify bioactivity and correlations with CHS across 1385 assays and 46 endpoints (Table S4).
The medians for the provided endpoints were used to impute the missing CompTOX data entries. CompTOX assay values were log-transformed, whereas the ZBEscreen™ assay readouts were transformed using hyperbolic arcsine transformation. The entries on the (0,1) scale of the CHS were transformed using the inverse logit transformation. The transformed data were used to compute pairwise correlation matrix. Subsequently, the resultant matrix was employed as an adjacency matrix for an undirected graph built using the igraph library (Csardi and Nepusz, 2006). The edges associated with correlations with absolute values lower than 0.5 were removed from network for clarity. R-package bootnet and mgm were used to calculate the adjacency matrix representing the mixed graphical model (Epskamp et al., 2018; Haslbeck and Waldorp, 2020). The communities of nodes representing clusters of connected readouts were identified by the walktrap algorithm included in the igraph package. This approach uses random walks to locate communities in large networks. Using these random walks, the distance between nodes is then determined. Nodes are then assigned to groups with smaller intra-community distances and bigger inter-community distances using bottom-up hierarchical clustering (Pons and Latapy, 2005). We conducted such analyses to demonstrate the relationships between executed assays, and to visualize the existence or lack thereof of causal links between simple high-throughput assays, and tests performed with a whole organism, the ZBEscreen™.
The simplest method for analyzing the similarities between the tested screens was to compare their binary prediction results directly. In this regard, as previously discussed, the ZBEscreen™ detected bioactivity in a greater number of compounds than the CHS, which provided a more comprehensive coverage of the library. However, the ground truth is uncertain due to zebrafish sensitivity to various routes of toxicity, not only neurotoxicity (i.e., the bioactivity of the DNT library’s curated subset of negatives found in the ZBEscreen™). Therefore, it is impossible to estimate statistical specificity, negative predictive values, and likelihood ratio for the employed screens (Rajwa, 2017; Shreffler and Huecker, 2022; Behl et al., 2019).
RESULTS AND DISCUSSION
Bioactivity Across the SYSTEMETRIC® Cell Health Screen and ZBEScreen™
Between both model systems, 86 out of 87 chemicals in the DNT chemical library were identified as bioactive. The CHS identified 20 chemicals by binarizing the CHI™ produced by the machine learning classifier using multiple flow cytometry-based readouts characterizing changes in a population of single HL-60 cells. The ZBEscreen™ identified 38 chemicals through mortality/morphological assessments and 86 chemicals through different behavioral assessments (Figure 1). The chemical classes identified by CHI™ spanned industrial, flame retardants, pesticides, and drugs, while ZBEscreen™ bioactivity occurred across all chemical classes, including NTP-categorized negatives. The high sensitivity of zebrafish to other forms of toxicity likely accounts for the detected bioactivity of the four chemicals classified by NTP as negative for DNT. CHI™ classification and morphological assessments within the ZBEscreen™ revealed the greatest pesticide sensitivity, with 7 of 16 and 13 of 16 pesticides identified as bioactive, respectively (Table S5). Behavioral evaluations within the ZBEscreen™ demonstrated that all 16 pesticides were bioactive (in either EPR or LPR assays). The higher number of pesticides identified in both systems suggests that the pesticides likely induced additional forms of toxicity besides DNT, as seen in studies that showed different pesticide classes produced specific forms of toxicity (carcinogenicity, metabolic toxicity etc.) (Gonςalves et al., 2020).
Figure 1. Bioactivity across the SYSTEMETRIC® Cell Health Screen and the ZBEscreen™.
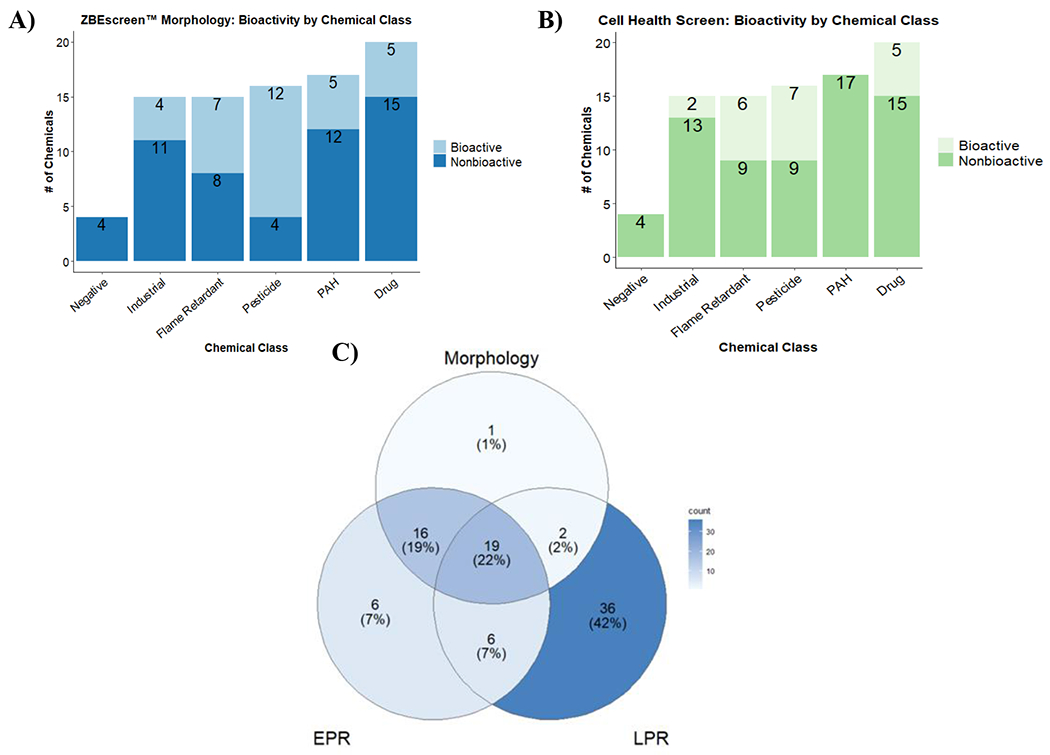
Bar plots of DNT chemical library bioactivity as identified by A) ZBEscreen™ mortality/morphological assessments and B) CHI™ categorized by chemical class C) Venn diagram of bioactivity across the morphological and behavioral (EPR and LPR) assessments of the DNT chemical library in the ZBEscreen™.
ZBEscreen™ behavioral assessments were the most sensitive endpoint(s) among all assays performed between the two systems. LPR alone identified 42% of the total DNT chemical library as bioactive in the zebrafish (Figure 1, C). LPR-related atypical behavior is determined through the quantification of hyperactivity and hypoactivity in both light and dark (infrared) cycles in 120 hpf larvae compared to controls. LPR has become a common endpoint in many pharmacological, toxicological, and developmental studies using zebrafish (Basnet et al., 2019; Maeda et al., 2021; Shen et al., 2020; Zhang et al., 2021). EPR had the most concordance with atypical morphological endpoints, with 35 chemical overlaps with morphology (Figure 1, C). Route of exposure, chemical uptake, and metabolic activity (HL60 cells are not metabolically active) are key differences that could alter the detection of chemical bioactivity between the assay systems. It is important to note that in the current analyses, there was no value judgment regarding specific atypical endpoints; only bioactivity was identified.
Complementarity Between SYSTEMETRIC® Cell Health Screen and the ZBEscreen™
There were 20 bioactive chemicals identified by CHI™ within the CHS and 86 bioactives identified by the ZBEscreen™, which resulted in 20 concordant chemicals between the two systems (Figure 2, A). The 86 chemicals identified as bioactive in the ZBEscreen™ were active in either one or both of the behavioral assays. One chemical, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), was detected only through abnormal zebrafish morphology (Figure 2, B). Additionally, 16 of 20 CHI™ bioactives caused significant atypical morphologies in the ZBEscreen™, but 4 of 20 CHI™ bioactives were identified by at least one ZBEscreen™ behavior assay (Table S6, S7). These results demonstrate the enhanced sensitivity of the behavioral assays within the ZBEscreen™ where EPR and LPR can recognize chemical effects that morphological assessments would miss.
Figure 2. Concordance between the SYSTEMETRIC® Cell Health Screen and ZBEscreen™.
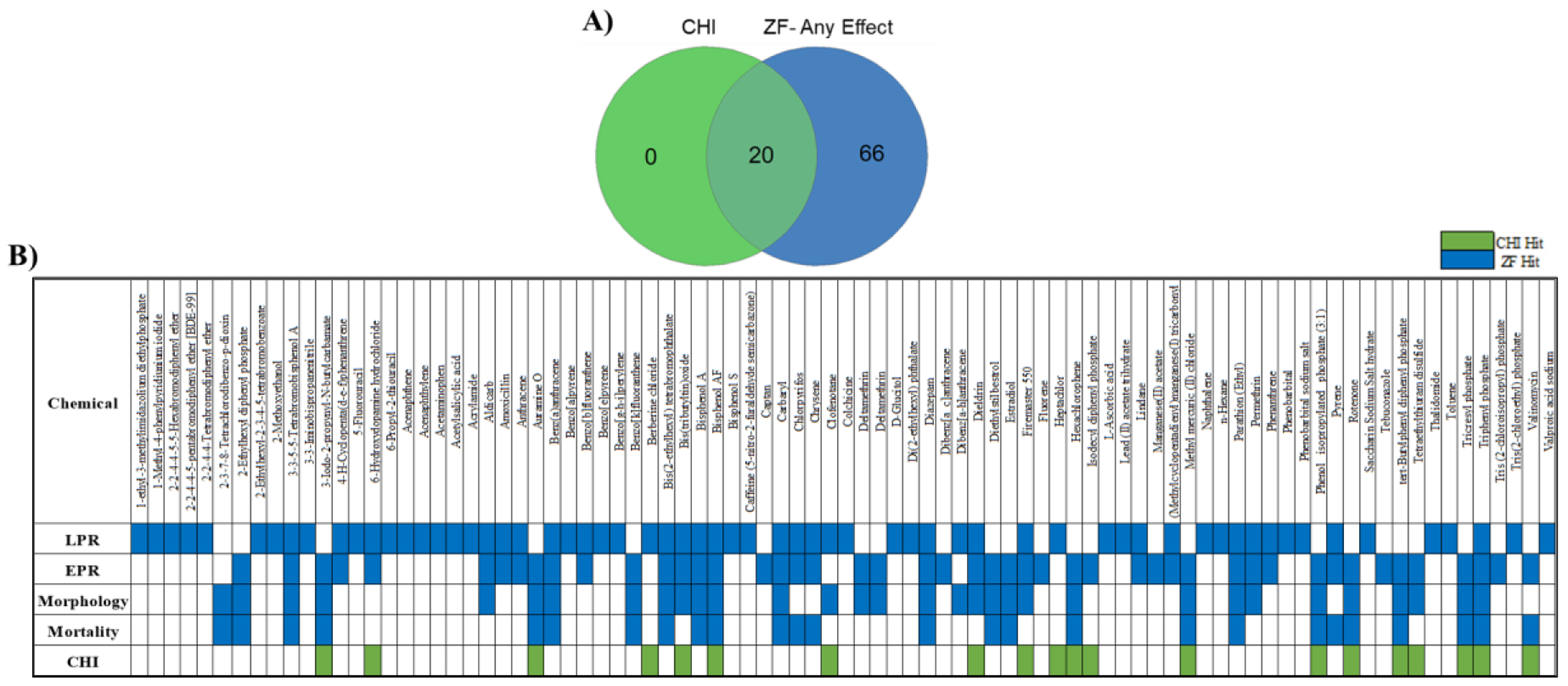
A) Venn diagram showing the number of bioactive chemicals between the ZBEscreen™ and SYSTEMETRIC® CHS B) Distribution of CHI™ bioactives and ZBEscreen™ endpoints across the 87 DNT chemical library where the zebrafish endpoints were separated by mortality, morphology, and the behavioral assessments EPR and LPR (green indicates bioactivity identified by CHI, blue indicates bioactivity identified by any of the 4 zebrafish readouts). Likewise, DNT chemicals not identified through the CHI™ classification were detected by the ZBEscreen™, demonstrating the complementarity between the systems used in tandem.
ZBEscreen™ EPR-related bioactivity correlated more with CHI™ bioactive chemicals (17 of 20) than LPR (9 of 20). Berberine chloride, clofenotane. and heptachlor were the 3 CHI™ chemicals not bioactive in the EPR assay but were identified in the LPR assay. Additionally, 34 DNT chemicals were identified by LPR alone among the CHS, ZBEscreen™ EPR and morphological assessments. The ZBEscreen™ behavioral assessments can identify DNT chemicals that produce gross morphological changes, while the CHI™ classification combines acute cellular stress endpoints to identify cytotoxicity. These observations, collectively, demonstrate the comprehensive power of combining the SYSTEMETRIC® CHS and ZBEscreen™.
SYSTEMETRIC® Cell Health Screen Compared to Available Data from EPA CompTox Chemical Dashboard
Across the 87-chemical library, there were 56,669 total hit calls across 1,385 assays and 48 endpoint readouts curated by CompTOX; 9,043 bioactive hit calls, and 47,626 non-bioactive hit calls. (Figure 3, Table S8). The US EPA CompTOX Dashboard contains data for 906,511 different chemicals, which includes the 87 chemicals present in NTP’s DNT chemical library (https://comptox.epa.gov/dashboard/). There were stark differences in the number and types of assay endpoints found in CompTOX across the DNT chemicals, revealing data gaps. For example, bisphenol A (BPA) is the most well-tested chemical in the library, with 1,205 total conclusive assays (180 untested assays). Valinomycin, Firemaster 550, and 1-methyl-4-phenyl pyridinium iodide were the least tested chemicals, with 36 conclusive assays performed, leaving 1,349 assays untested for each chemical (Table S8). BPA is a well-known endocrine disruptor with considerable downstream effects that result in reproductive, immune, developmental, hepatic and renal damage, and potentially carcinogenic and mutagenic activity (Ma et al., 2019). BPA was not bioactive in 946 of the 1205 assays tested in CompTOX, and the CHI™ classifier did not classify BPA as bioactive. Moreover, 22 of 46 neurodevelopment assay readouts were bioactive (approximately 48%) within CompTOX. In contrast, nuclear receptor assays composed the highest number of bioactive hit calls, with 87 out of the 250 total nuclear receptor assays tested (257 total in CompTOX). In comparison, cell cycle-related assays had the second most hit calls, with 26 bioactives out of the 176 assays tested (185 total cell cycle assays in CompTOX) (Table S9). However, each ZBEscreen™ assay identified bioactivity caused by BPA exposure (mortality, morphology, EPR, and LPR). The least tested chemicals within CompTOX, Firemaster 550 and valinomycin, were bioactive in the CHS and the ZBEscreen™. Although CompTOX provides exceptionally curated, high-quality data, limitations exist in the form of missing information and inconclusive hit calls. For instance, the bioactivity identified in 87 of 250 nuclear receptor assays did not provide enough information to estimate or determine the toxicological risk of BPA. Moreover, this begs the question; which assay endpoint(s) among the 48 within CompTOX are the most accurate indicators of toxicity? The CompTOX Dashboard data, while extensive, are not exhaustive, and thus no single assay of the 1,385 was capable of estimating chemical hazard.
Figure 3. Hit call distribution of 87-DNT chemical library among 1,365 in vitro assays from the EPA CompTox Dashboard.
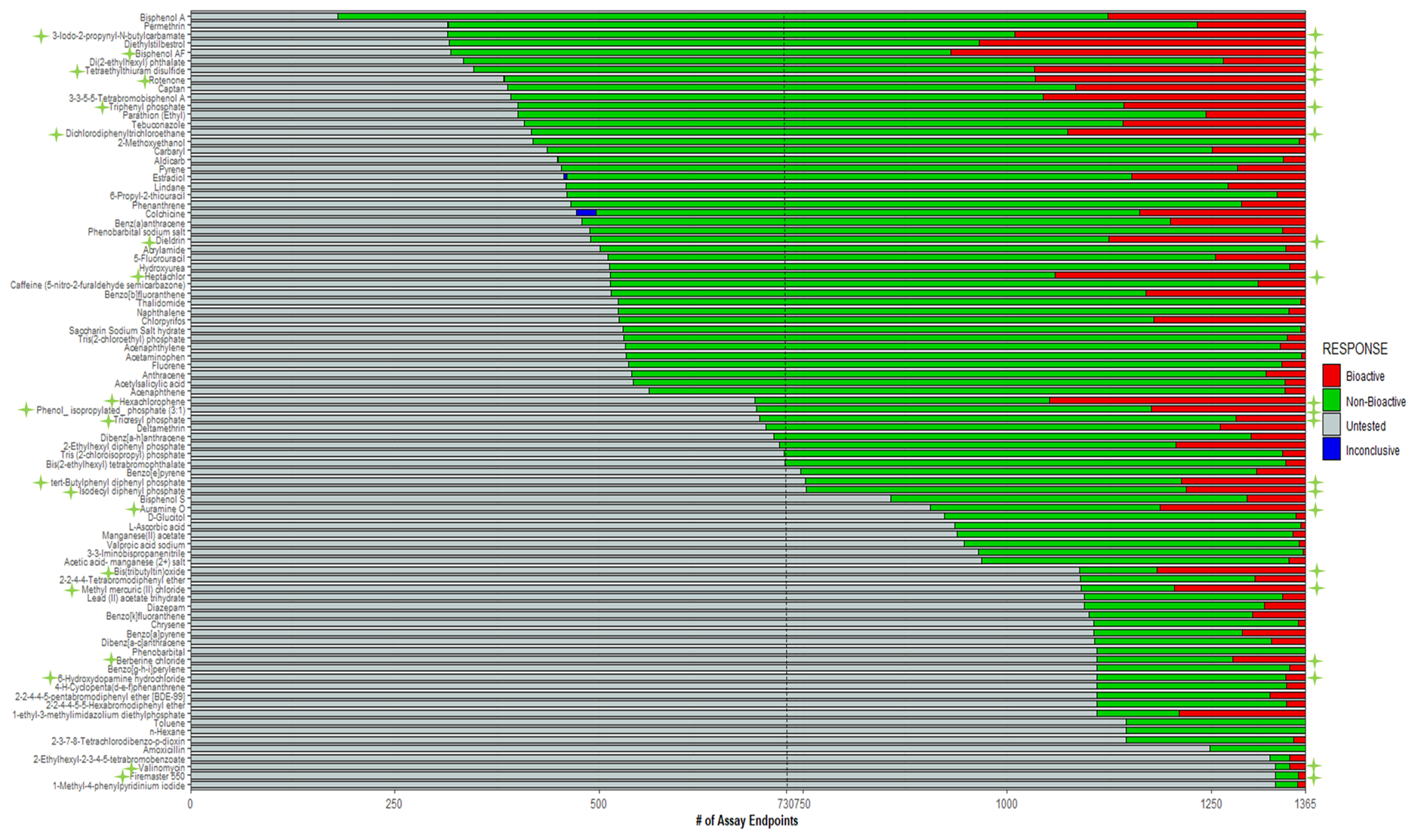
The black dashed line indicates the average number of untested assays across all 87 chemicals, and the green stars indicate CHI™ bioactive chemicals. Among available in vitro data, 9 CHI™ chemicals were undertested (49 of the chemical library), and 11 CHI™ chemicals were thoroughly tested (36 of the chemical library).
The CHS data and the CHI™ classification generated from this assay are based on a different set of assumptions than CompTOX screens. The CHS collects and classifies phenotypic signatures of analyzed compounds instead of utilizing a large number of individual assays based on preconceived mechanistic models covering multiple different aspects of toxicity. Consequently, the accuracy of the CHS is based on the premise that a set of compounds that produces similar patterns of response in tested cell populations (population phenotype) will elicit corresponding responses when used within a more complex system. However, the responses in the model system do not need to resemble the responses in the test system. Additionally, it is essential to note that the phenotypic patterns observed in the model system need not be identical or similar for the compounds to be classified as belonging to the same classification (i.e., bioactive, non-bioactive). The “bioactive” group may be highly multimodal. In other words, the only condition is that two compounds classified as functionally similar in the screened environment are likely to be functionally similar in humans. The classification is performed on the basis of training a statistical model rather than a mechanistic biological model. Therefore, even though elastic net regression is ante hoc explainable (one can examine the model’s coefficients to determine why a particular compound was assigned to a particular class), statistical explainability does not imply mechanistic explainability. The CHS may help formulate the hypothesis regarding the mechanism of action, but only a comprehensive follow-up study would reveal why a specific population phenotype indicates a high probability of toxicity. Given that the CHS is intended to detect relatively rapidly occurring acute responses, it should be viewed as a low-cost, rapid, and reliable pre-selection step, complementing more comprehensive, costly, and time-consuming assays such as the ZBEscreen™.
Corrleation analyses of CHS, ZBEscreen™, and CompTOX assays formed distinct groups that are only distantly related (Figure 4). This demonstrated that the underlying assay readouts are orthogonal and capture complementary data which emphasizes the advantages of pairing an organism-based assay with a much simpler, but rapid pre-screening step utilizing singe cell phenotypes.
Figure 4. Correlation network representation of assay endpoints from CHS, ZBEscreen™, and the EPA’s CompTOX dashboard.
Correlation analyses and clustering of the network vertices showed the formation of distinct groups of assay endpoints. Blue points are assay endpoints curated within CompTOX, red and green points represent endpoint readouts from the ZBEscreen™ and CHS, respectively. The edges indicating correlations with absolute value less than 0.5 are not shown.
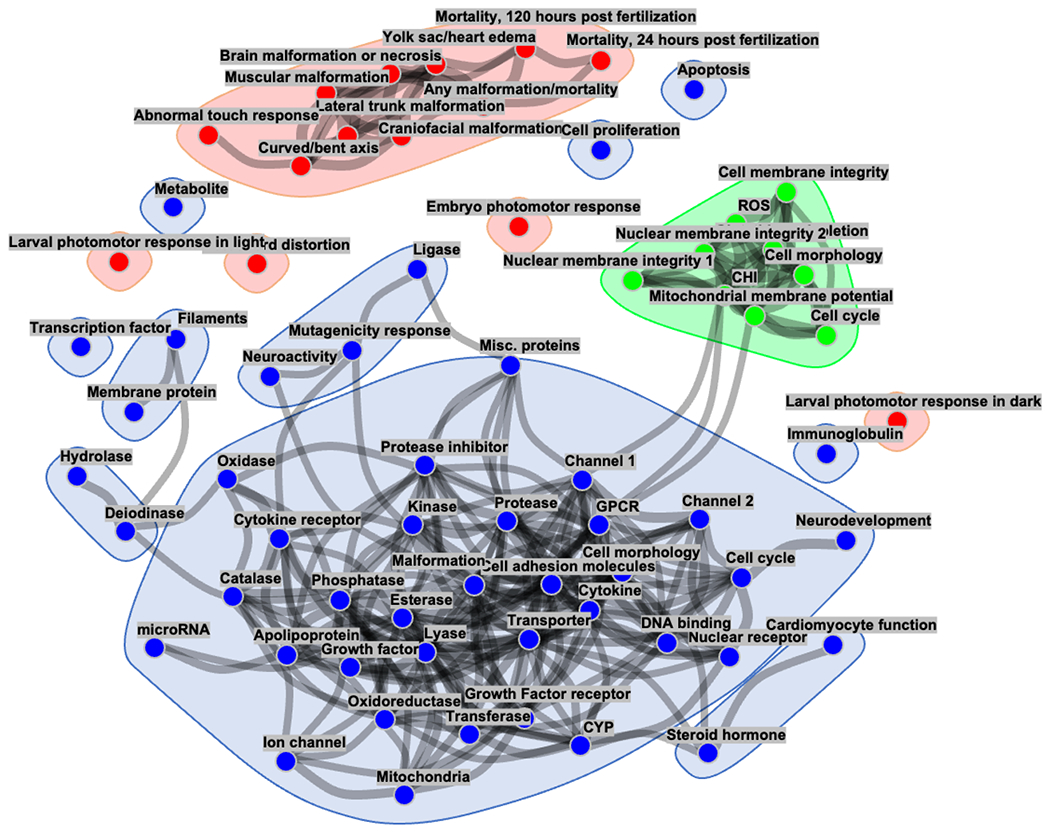
In the next stage we attempted to reconstruct a graphical model network which could be used to suggest hypotheses on the potential representation of causal links between distinct physiological subsystems tested by particular experiments (Figure 5). The employed graphical model allows us to infer dependencies between test endpoints by calculating the conditional dependence structure between readouts (Altenbuchinger et al., 2020). This differs from the correlation analysis mentioned above, which solely analyzes the strength of linear connections between pairs of assay readouts without accounting for the impact of additional readouts or biological characteristics. In order to promote sparsity, the employed method used ℓ1 (LASSO) regularization. The result demonstrates once more that the results of the three comparative assays capture essentially distinct aspects of physiology and are therefore likely to detect distinct toxicity characteristics.
Figure 5. Links between the distinct physiological subsystems tested.
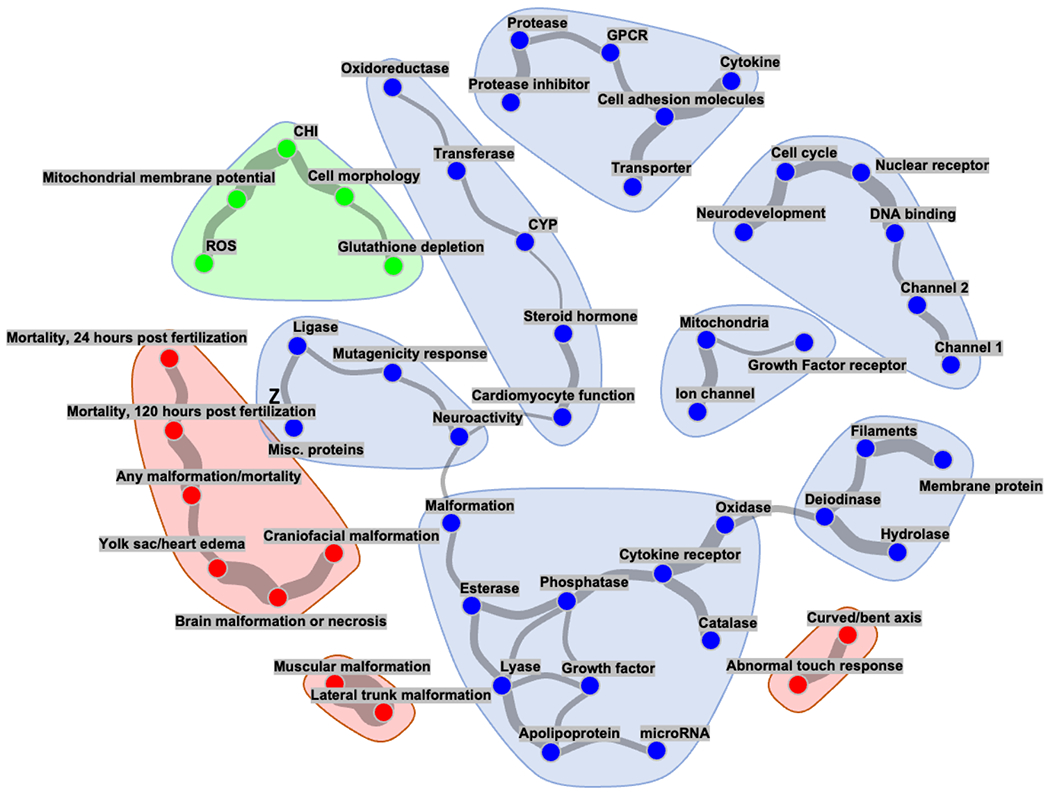
The graphical network model shows each assay readout as a node in a graph, and edges between nodes represent conditional dependencies between assays’ endpoints. The presence and line thickness show the strength of these dependencies (a thicker line indicates stronger association). For the sake of clarity, readouts without corresponding edges are omitted from the network. The blue points indicate CompTOX-curated assay endpoints, whereas the red and green points represent ZBEscreen™ and CHS endpoint readouts, respectively.
Future research evaluating, comparing, and expanding the described screening approaches should include a more thorough cross-validation performed within the context of the employed downstream machine learning tools, which ingest the raw readouts characterizing biological endpoints and link them via a statistical model to an estimate of the risk associated with a tested compound. This may necessitate the creation of a specialized validation library, which will include not only drugs with known toxicities and recognized mechanisms of action, but also a vast selection of inert compounds that are well-tolerated by humans.
CONCLUSIONS
The analysis of the binary classification of the DNT chemical library revealed that the ZBEscreen™ is more sensitive than the AsedaSciences SYSTEMETRIC® CHS in detecting bioactivity potentially associated with neurotoxicity. The ZBEscreen™ included a greater number and variety of endpoints, providing greater weight of evidence regarding the comprehensive chemical hazard evaluation. More specifically, the behavioral assessments within the ZBEscreen™ demonstrated the highest sensitivity to chemical exposure but it is important to add that these behavioral readouts are not restricted to DNT chemicals or responses. In contrast, the CHS encompassed more specific cellular stress endpoints associated with acute cell stress. The CHS, conceived as a quick pre-screening technique, was tailored for high specificity and was not designed to identify specific classes of toxicants that generate cumulative effects or effects depending on fully functional metabolic pathways. Subsequently, the use of both systems, in combination or a tiered approach, would result in more informed decision-making during the drug discovery and development process.
Although none of these assays alone can provide definitive evidence regarding particular mechanisms of action, when combined with modern data analysis (causal inference, explainable artificial intelligence) they could be utilized in the future to elucidate mechanisms by which certain bioactive chemicals act through.
The chemicals comprising the NTP’s DNT chemical library were well-characterized by the EPA’s CompTOX Dashboard’s curated in vitro data; however, the coverage was limited, and huge knowledge gaps still existed across the 1,385 assays. Additionally, the reliance on a high number of endpoints raises the issue of the subsequent classification approach that would be employed with CompTOX. These data could be used to train, test, and possibly validate a number of different classifiers optimized to detect various mechanisms of toxicity. Regardless of the potential outcome, it is evident that CompTOX and our tested assays, approach hazard prediction in radically different ways. CHS is narrow in scope and focuses on acute responses. ZBEscreen™ has a broader target, and it is driven by phenotypic outcomes. In contrast, the single end-point assays in CompTOX attempt to cover the broadest range of possible known responses. However, combining a basic and rapid cell-based model (CHS) with a comprehensive phenotypic organism-based system (ZBEscreen™) may be more efficient and cost-effective, allowing for a higher throughput and a broader coverage of chemicals for hazard assessment.
Although our work shed some light on the sensitivity of the tested assays (defined as the ability to detect subtle changes in phenotype that may be indicative of toxicity or other effects), the limited scope of the employed chemical library and the sparsity of the CompTOX database prevent us from comparing the robustness andreproducibilityof single-readout assays in CompTOX versus ZBEscreen™ and CHS. The executed comparison demonstrates the usefulness of combining several screening techniques, and assays developed to address distinct toxicity features which would ultimately reduce the cost and use of mammal models for hazard assessments in the future.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the screening staff at Sinnhuber Aquatic Research Laboratory for fish husbandry and screening support. The research reported in this manuscript was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Numbers R35 ES031709, T32 ES007060, and P30 ES030287. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Credit Author Statement
Robyn Tanguay and Lisa Truong conceived of the present idea. Lindsey St. Mary and Bartek Rajwa performed the formal analyses of the data curated and collected by Lisa Truong, Andrew Bieberich, and Raymond O. Fatig III. Robyn Tanguay and Lisa Truong supervised the findings of this work. All authors discussed the results and contributed to the final manuscript.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- Altenbuchinger M, Weihs A, Quackenbush J, Grabe HJ, Zacharias HU, 2020. Gaussian and Mixed Graphical Models as (multi-)omics data analysis tools. Biochim. Biophys. Acta BBA - Gene Regul. Mech, Transcriptional Profiles and Regulatory Gene Networks 1863, 194418. 10.1016/j.bbagrm.2019.194418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnet RM, Zizioli D, Taweedet S, Finazzi D, Memo M, 2019. Zebrafish larvae as a behavioral model in neuropharmacology. Biomedicines 7, 23. 10.3390/biomedicines7010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behl M, Ryan K, Hsieh J-H, Parham F, Shapiro AJ, Collins BJ, Sipes NS, Birnbaum LS, Bucher JR, Foster PMD, Walker NJ, Paules RS, Tice RR, 2019. Screening for developmental neurotoxicity at the National Toxicology Program: the future is here. Toxicol. Sci. Off. J. Soc. Toxicol 167, 6–14. 10.1093/toxsci/kfy278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM, Chang X, Wambaugh JF, Allen DG, Bartels M, Brouwer KLR, Casey WM, Choksi N, Ferguson SS, Fraczkiewicz G, Jarabek AM, Ke A, Lumen A, Lynn SG, Paini A, Price PS, Ring C, Simon TW, Sipes NS, Sprankle CS, Strickland J, Troutman J, Wetmore BA, Kleinstreuer NC, 2018. In vitro to in vivo extrapolation for high throughput prioritization and decision making. Toxicol. In Vitro 47, 213–227. 10.1016/j.tiv.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernas T, Asem EK, Robinson JP, Rajwa B, 2008. Quadratic form: A robust metric for quantitative comparison of flow cytometric histograms. Cytometry A 73A, 715–726. 10.1002/cyto.a.20586 [DOI] [PubMed] [Google Scholar]
- Bieberich AA, Laitinen T, Maffuid K, Fatig RO, Torrice CD, Morris DC, Crona DJ, Asquith CRM, 2022. Optimization of the 4-anilinoquin(az)oline scaffold as epidermal growth factor receptor (EGFR) inhibitors for chordoma utilizing a toxicology profiling assay platform. Sci. Rep 12, 12820. 10.1038/s41598-022-15552-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich AA, Rajwa B, Irvine A, Fatig RO, Fekete A, Jin H, Kutlina E, Urban L, 2021. Acute cell stress screen with supervised machine learning predicts cytotoxicity of excipients. J. Pharmacol. Toxicol. Methods 111, 107088. 10.1016/j.vascn.2021.107088 [DOI] [PubMed] [Google Scholar]
- Bieberich AA, Rajwa B, Irvine A, Fatig RO, Fekete A, Jin H, Kutlina E, Urban L, (2021). Acute cell stress screen with supervised machine learning predicts cytotoxicity of excipients. Journal of Pharmacological and Toxicological Methods, Seventeenth Annual Themed Issue on Methods in Safety Pharmacology 111, 107088. 10.1016/j.vascn.2021.107088 [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Druwe IL, Painter K, Yost EE, 2017. Using in vitro high-throughput screening data for predicting benzo[k]fluoranthene human health hazards. Risk Anal. 37, 280–290. 10.1111/risa.12613 [DOI] [PubMed] [Google Scholar]
- Crofton KM, Bassan A, Behl M, Chushak YG, Fritsche E, Gearhart JM, Marty MS, Mumtaz M, Pavan M, Ruiz P, Sachana M, Selvam R, Shafer TJ, Stavitskaya L, Szabo DT, Szabo ST, Tice RR, Wilson D, Woolley D, Myatt GJ, 2022. Current status and future directions for a neurotoxicity hazard assessment framework that integrates in silico approaches. Comput. Toxicol 22, 100223. 10.1016/j.comtox.2022.100223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T, 2006. The igraph software package for complex network research. InterJournal Complex Syst. 1695, 9. [Google Scholar]
- d’Amora M, Giordani S, 2018. The utility of zebrafish as a model for screening developmental neurotoxicity. Front. Neurosci 12. 10.3389/fnins.2018.00976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darde TA, Gaudriault P, Beranger R, Lancien C, Caillarec-Joly A, Sallou O, Bonvallot N, Chevrier C, Mazaud-Guittot S, Jégou B, Collin O, Becker E, Rolland AD, Chalmel F, 2018. TOXsIgN: a cross-species repository for toxicogenomic signatures. Bioinformatics 34, 2116–2122. 10.1093/bioinformatics/bty040 [DOI] [PubMed] [Google Scholar]
- Edwards BS, Kuckuck F, Sklar LA, 1999. Plug flow cytometry: An automated coupling device for rapid sequential flow cytometric sample analysis. Cytometry 37, 156–159. [DOI] [PubMed] [Google Scholar]
- Edwards BS, Oprea T, Prossnitz ER, Sklar LA, 2004. Flow cytometry for high-throughput, high-content screening. Curr. Opin. Chem. Biol 8, 392–398. 10.1016/j.cbpa.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Epskamp S, Borsboom D, Fried EI, 2018. Estimating psychological networks and their accuracy: A tutorial paper. Behav. Res. Methods 50, 195–212. 10.3758/s13428-017-0862-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves ÍFS, Souza TM, Vieira LR, Marchi FC, Nascimento AP, Farias DF, 2020. Toxicity testing of pesticides in zebrafish—a systematic review on chemicals and associated toxicological endpoints. Environ Sci Pollut Res 27, 10185–10204. 10.1007/s11356-020-07902-5 [DOI] [PubMed] [Google Scholar]
- George S, Xia T, Rallo R, Zhao Y, Ji Z, Lin, Sijie, Wang X, Zhang H, France B, Schoenfeld D, Damoiseaux R, Liu R, Lin, Shuo, Bradley KA, Cohen Y, Nel AE, 2011. Use of a high-throughput screening approach coupled with in vivo zebrafish embryo screening to develop hazard ranking for engineered nanomaterials. ACS Nano 5, 1805–1817. 10.1021/nn102734s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom D, Truong L, Zhang S, Tanguay R, Collins E-MS, 2019. Comparative analysis of zebrafish and planarian model systems for developmental neurotoxicity screens using an 87-compound library. Toxicol. Sci 167, 15–25. 10.1093/toxsci/kfy180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck JMB, Waldorp LJ, 2020. mgm: estimating time-varying mixed graphical models in high-dimensional data. J. Stat. Softw 93, 1–46. 10.18637/jss.v093.i08 [DOI] [Google Scholar]
- Krewski D, Andersen ME, Tyshenko MG, Krishnan K, Hartung T, Boekelheide K, Wambaugh JF, Jones D, Whelan M, Thomas R, Yauk C, Barton-Maclaren T, Cote I, 2020. Toxicity testing in the 21st century: progress in the past decade and future perspectives. Arch. Toxicol 94, 1–58. 10.1007/s00204-019-02613-4 [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Doering JA, Blackwell BR, Transue TR, Simmons CW, Swintek J, Degitz SJ, Williams AJ, Ankley GT, 2018. Evidence for cross species extrapolation of mammalian-based high-throughput screening assay results. Environ. Sci. Technol 52, 13960–13971. 10.1021/acs.est.8b04587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Liu H, Wu J, Yuan L, Wang Y, Du X, Wang R, Marwa PW, Petlulu P, Chen X, Zhang H, 2019. The adverse health effects of bisphenol A and related toxicity mechanisms. Environ. Res 176, 108575. 10.1016/j.envres.2019.108575 [DOI] [PubMed] [Google Scholar]
- Maeda H, Fukushima N, Hasumi A, 2021. Standardized method for the assessment of behavioral responses of zebrafish larvae. Biomedicines 9, 884. 10.3390/biomedicines9080884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes J, Verlooy L, Buenafe OE, Witte P.A.M. de, Esguerra CV, Crawford AD, 2012. Evaluation of 14 organic solvents and carriers for screening applications in zebrafish embryos and larvae. PLOS ONE 7, e43850. 10.1371/journal.pone.0043850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons P, Latapy M, 2005. Computing communities in large networks using random walks (long version). 10.48550/arXiv.physics/0512106 [DOI] [Google Scholar]
- R Core Team, 2022. R: A language and environment for statistical computing (manual). Vienna, Austria. [Google Scholar]
- Rajwa B, 2017. Effect-size measures as descriptors of assay quality in high-content screening: a brief review of some available methodologies. ASSAY Drug Dev. Technol 15, 15–29. 10.1089/adt.2016.740 [DOI] [PubMed] [Google Scholar]
- Reif DM, Truong L, Mandrell D, Marvel S, Zhang G, Tanguay RL, 2016. High-throughput characterization of chemical-associated embryonic behavioral changes predicts teratogenic outcomes. Arch. Toxicol 90, 1459–1470. 10.1007/s00204-015-1554-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JP, Patsekin V, Holdman C, Ragheb K, Sturgis J, Fatig R, Avramova LV, Rajwa B, Davisson VJ, Lewis N, Narayanan P, Li N, Qualls CW, 2013. High-throughput secondary screening at the single-cell level. SLAS Technol., Special Issue: Novel Drug Development and Delivery 18, 85–98. 10.1177/2211068212456978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JP, Rajwa B, Patsekin V, Davisson VJ, 2012. Computational analysis of high-throughput flow cytometry data. Expert Opin. Drug Discov 7, 679–693. 10.1517/17460441.2012.693475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Truong L, Simonich MT, Huang C, Tanguay RL, Dong Q, 2020. Rapid well-plate assays for motor and social behaviors in larval zebrafish. Behav. Brain Res 391, 112625. 10.1016/j.bbr.2020.112625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreffler J, Huecker MR, 2022. Diagnostic testing accuracy: sensitivity, specificity, predictive values and likelihood ratios, in: StatPearls. StatPearls Publishing, Treasure Island (FL). [PubMed] [Google Scholar]
- Tal TL, McCollum CW, Harris PS, Olin J, Kleinstreuer N, Wood CE, Hans C, Shah S, Merchant FA, Bondesson M, Knudsen TB, Padilla S, Hemmer MJ, 2014. Immediate and long-term consequences of vascular toxicity during zebrafish development. Reprod. Toxicol., 42nd Annual Conference of the European Teratology Society 48, 51–61. 10.1016/j.reprotox.2014.05.014 [DOI] [PubMed] [Google Scholar]
- Truong L, Reif DM, St Mary L, Geier MC, Truong HD, Tanguay RL, 2014. Multidimensional in vivo hazard assessment using zebrafish. Toxicol. Sci 137, 212–233. 10.1093/toxsci/kft235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong L, Simonich MT, Tanguay RL, 2016. Better, faster, cheaper: getting the most out of high-throughput screening with zebrafish, in: Zhu H, Xia M (Eds.), High-Throughput Screening Assays in Toxicology, Methods in Molecular Biology. Springer, New York, NY, pp. 89–98. 10.1007/978-1-4939-6346-1_10 [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Coady K, Escher BI, Mihaich E, Murphy CA, Schlekat T, Garcia-Reyero N, 2019. High-throughput screening and environmental risk assessment: State of the science and emerging applications. Environ. Toxicol. Chem 38, 12–26. 10.1002/etc.4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will Y, Dykens J, 2014. Mitochondrial toxicity assessment in industry – a decade of technology development and insight. Expert Opin. Drug Metab. Toxicol 10, 1061–1067. 10.1517/17425255.2014.939628 [DOI] [PubMed] [Google Scholar]
- Zhang K, Liang J, Brun NR, Zhao Y, Werdich AA, 2021. Rapid zebrafish behavioral profiling assay accelerates the identification of environmental neurodevelopmental toxicants. Environ. Sci. Technol 55, 1919–1929. 10.1021/acs.est.0c06949 [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhang J, Kim MT, Boison A, Sedykh A, Moran K, 2014. Big data in chemical toxicity research: the use of high-throughput screening assays to identify potential toxicants. Chem. Res. Toxicol 27, 1643–1651. 10.1021/tx500145h [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


