Abstract
Mutations in SHH and several other genes encoding components of the Hedgehog signaling pathway have been associated with holoprosencephaly syndromes, with craniofacial anomalies ranging in severity from cyclopia to facial cleft to midfacial and mandibular hypoplasia. Studies in animal models have revealed that SHH signaling plays crucial roles at multiple stages of craniofacial morphogenesis, from cranial neural crest cell survival to growth and patterning of the facial primordia to organogenesis of the palate, mandible, tongue, tooth, and taste bud formation and homeostasis. This article provides a summary of the major findings in studies of the roles of SHH signaling in craniofacial development, with emphasis on recent advances in the understanding of the molecular and cellular mechanisms regulating the SHH signaling pathway activity and those involving SHH signaling in the formation and patterning of craniofacial structures.
Keywords: Shh, cholesterol, primary cilium, neural crest, palate, mandible, tongue
1. Introduction
Mutations in SHH and several other genes encoding components of the Hedgehog signaling pathway, including PTCH, DISP1, GAS1, CDON, and GLI2, have been associated with holoprosencephaly (HPE) syndromes, with craniofacial anomalies ranging in severity from cyclopia to facial cleft to midfacial and mandibular hypoplasia (Bae et al., 2011; Belloni et al., 1996; Ming et al., 2002; Ribeiro et al., 2010; Roessler et al., 1996; Roessler et al., 2003; Roessler et al., 2009a; Roessler et al., 2009b). HPE is one of the most severe birth defects and has an estimated prevalence of ~4.8–8.8 per 100,000 live births (Croen et al., 1996; Ming and Muenke, 1998; Olsen et al., 1997; Roach et al., 1975; Urioste et al., 1988). Mutations in SHH have been found in about 23% of HPE patients in clinical studies (Abramyan, 2019; Roessler et al., 1996; Roessler and Muenke, 1998), underscoring the essential roles of SHH signaling in craniofacial development.
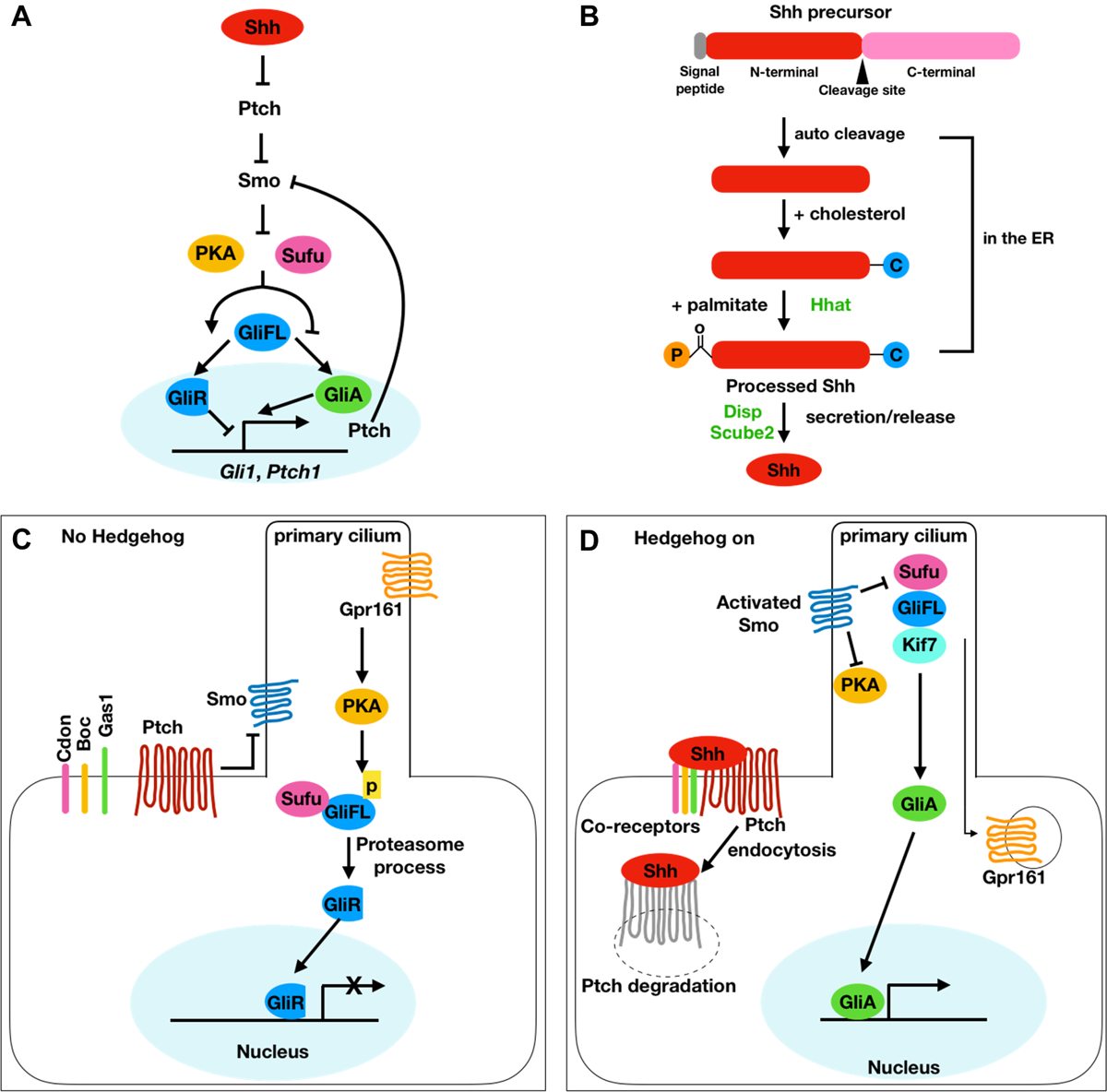
Shh is one of three mammalian homologs of Drosophila Hedgehog, a secreted signaling protein initially discovered through its role in controlling segmental patterning of the Drosophila embryos (Nusslein-Volhard and Wieschaus, 1980; Pereira et al., 2014). The other vertebrate/mammalian HH homologs include desert hedgehog (Dhh) and Indian hedgehog (Ihh) (Ingham, 2022; McMahon et al., 2003). Signaling by all Hedgehog family members shares an evolutionarily conserved but unusual signal transduction cascade composed of a series of inhibitory interactions (Fig. 1A) (Ingham, 2022; Kong et al., 2019). The main cell surface receptor for Hedgehog family ligands is Patched (Ptch), a twelve-pass transmembrane protein, whose function is to inhibit the activity of a transmembrane G-protein coupled receptor (GPCR)-like protein called Smoothened (Smo) in the absence of Hedgehog ligands (Chen and Struhl, 1996; Denef et al., 2000; Goodrich et al., 1996; Jenkins, 2009; Quirk et al., 1997; Stone et al., 1996; Taipale et al., 2002). Binding of Hedgehog ligands to the Ptch receptor relieves its repression of Smo, which then transduces the signaling activity intracellularly to activate the Glioma-associated oncogene (Gli) family of transcription factors (Gli1, Gli2, and Gli3 in vertebrates) by overcoming the negative influences of Suppressor of Fused (Sufu) and Protein Kinase A (PKA) (Huangfu and Anderson, 2006; Tukachinsky et al., 2010). In the absence of Hedgehog signals, Gli2 and Gli3 directly interacts with Sufu and are phosphorylated by PKA, leading to their partial proteolysis into truncated repressor forms (GliR) that function to suppress transcriptional target genes. Activated Smo inhibits PKA and promotes additional modifications of the full-length Gli proteins into the transcriptional activator form (GliA), which translocates into the cell nucleus to drive the expression of downstream target genes (Hui and Angers, 2011) (Fig. 1A). Each step of the Hedgehog signaling pathway, from the biogenesis of the active ligands to ligand-receptor interactions, to Smo and Gli activation, is regulated by multiple biochemical and cellular processes (Ingham, 2022; Kong et al., 2019; Zhang and Beachy, 2023). Thus, in addition to mutations affecting the functions of the core components of the SHH signaling pathway, genetic and/or environmental perturbations of many biochemical or cellular processes can disrupt or alter SHH signaling pathway activity and result in congenital developmental disorders. Before discussing the roles of and mechanisms mediating SHH signaling in craniofacial development, we next briefly review the current understanding of the biochemical and cellular mechanisms regulating SHH signaling pathway activity. More extensive discussions of the biochemical and cellular mechanisms regulating the Hedgehog signaling pathway can be found in several recent review articles (Ingham, 2022; Kong et al., 2019; Zhang and Beachy, 2023) and references therein.
Fig. 1.
Overview of the Shh signaling pathway. (A) A simplified schematic of the core steps of the Shh signaling pathway. Shh ligands bind to its receptor Ptch, resulting in relief of Ptch-mediated inhibition of Smo. Activated Smo transduces the signaling activity intracellularly to activate the Gli family of transcription factors through inhibition of PKA and Sufu. Gli1 and Ptch1 are among the direct transcriptional target genes of the Gli transcription factors. (B) Biogenesis of the Shh ligand. The Shh mRNA encodes a large precursor protein, which undergoes auto-cleavage to release the N-terminal signaling domain with a covalently linked cholesterol moiety at the C-terminus followed by addition of palmitoylation at the N-terminal residue. The dually lipidated mature Shh ligand is released from the signalling cell through the functions of Disp and Scube2. (C, D) Hedgehog signal transduction in vertebrates requires the primary cilium. In the absence of active Hedgehog ligand, Ptch accumulates and inhibits Smo in the primary cilium. PKA and Sufu serve as major negative regulators of Hedgehog signaling and promote Gli proteins to undergo proteolysis into transcriptional repressor (GliR) (C). Binding of Shh ligands to Ptch and co-receptors induces Ptch removal from the ciliary membrane and accumulation of Smo in the ciliary membrane. Activated Smo promotes the transport of the GliSufu complex to the tip of the primary cilium and inhibits PKA. Full-length Gli proteins are post-translationally modified in the primary cilium to the active forms (GliA). GliA then translocates into the nucleus and drives target gene transcription (D).
2. Biochemical and cellular mechanisms regulating SHH signaling activity
2.1. Biogenesis and secretion of the Shh ligand
The Hedgehog family genes encode large precursor proteins containing a signal peptide sequence at the N-terminus, which targets newly synthesized Hedgehog protein to the endoplasmic reticulum where they undergo autocleavage through a cholesterol-mediated nucleophilic attack to release the N-terminal signaling fragment with covalently linked cholesterol at its C-terminus (Fig. 1B) (Hall et al., 1995; Lee et al., 1994; Porter et al., 1996). The HH signaling fragment is further modified by palmitoylation at its N-terminus by the membrane-bound O-acyltransferase HHAT to generate the dually lipid-modified mature ligand (Fig. 1B) (Buglino and Resh, 2008; Jiang et al., 2021; Pepinsky et al., 1998). The cholesterol modification contributes to membrane anchoring and enhances the stability of the Hedgehog ligands, which is critical for both long-distance and local signal transduction (Gallet et al., 2006; Hu and Song, 2019; Ingham, 2000; Kaushal et al., 2022; Li et al., 2006; Peters et al., 2004). Whereas several studies showed that pharmacological inhibition of HHAT blocked SHH signaling in vitro (Petrova et al., 2013; Rodgers et al., 2016), Shh lacking palmitoylation was able to drive expression of a Hedgehog signaling reporter in mammalian cells with lower potency (Chen et al., 2004; Palm et al., 2013; Pepinsky et al., 1998). Mouse embryos lacking Hhat function (Hhatcreface/Creface, description of this and other mutant mouse lines discussed in this article are provided in Supplementary table 1) exhibit severe holoprosencephaly defects including impaired forebrain cleavage and midline facial anomalies (Dennis et al., 2012; Kurosaka et al., 2014). Expression of Patch1, a direct target of SHH signaling, was considerably reduced in the Hhatcreface/Creface embryos, indicating that deficiency in Hhat-mediated palmitoylation diminished the gradient and range of Shh activity in vivo (Dennis et al., 2012).
The lipid modifications render the Hedgehog ligands highly hydrophobic and tethered to the membranes of the producing cells (Kong et al., 2019). Whereas the membrane-associated Hedgehog ligands can signal to neighboring cells through direct cell-cell interactions, long-range signaling requires release of the mature ligands from the cell membrane. The multipass transmembrane protein Dispatched (Disp), which has significant sequence identity to Ptch, functions specifically for Hedgehog ligand release through interacting with the cholesterol moiety (Burke et al., 1999; Caspary et al., 2002; Ma et al., 2002; Tukachinsky et al., 2012). In addition, the Signal Peptide CUB EGF-like domain-containing protein (Scube2), itself a secreted protein, is required for Hedgehog ligand release by interacting with and shielding the lipid moieties to facilitate the dispersal from the secreting cell and enable the mature lipidated ligands to pass through the aqueous extracellular environment (Fig. 1B) (Petrov et al., 2017; Tukachinsky et al., 2012).
2.2. Reception of the SHH signal at the cell surface
Hedgehog family ligands must bind to the Ptch receptor to trigger signaling in target cells (Marigo et al., 1996; Stone et al., 1996). Vertebrates have two Ptch homologs, with Ptch1 acting as the major regulator of SHH signaling and Ptch2 playing a partially redundant role in some developmental processes (Carpenter et al., 1998; Goodrich et al., 1997; Koudijs et al., 2008; Nieuwenhuis et al., 2006). Cryo-electromicroscopy studies of PTCH1 structures indicate that PTCH1 binds to dually lipidated SHH ligand through two interfaces, forming a complex with 1SHH:2PTCH stoichiometry with one PTCH1 binding SHH at its calcium- and zinc-binding surface and the second PTCH1 molecule engaging both the N-terminal palmitoyl and C-terminal cholesterol moieties (Gong et al., 2018; Qi et al., 2018a; Qi et al., 2018b; Qian et al., 2019). Whereas Scube2 is required for SHH release from the secreting cells and shields the lipid moieties of secreted Shh for its transport through the aqueous extracellular environment, the Scube2-SHH complex has significantly reduced affinity for Ptch1 compared to the free lipidated SHH (Wierbowski et al., 2020). Thus, for optimal Ptch1 binding and pathway activation, secreted SHH needs to be dissociated from Scube2 protection. Several co-receptors for the Hedgehog ligands have been identified, including the GPI-linked membrane glycoprotein Gas1 (Growth arrestspecific1) (Martinelli and Fan, 2007), the Ig/fibronectin single-pass membrane-spanning cell adhesion proteins Cdon (cell adhesion associated, oncogene regulated) and Boc (brother of Cdon) (Kang et al., 2002; Okada et al., 2006; Tenzen et al., 2006). Biochemical studies in cultured cells showed that Scube2 binds strongly to Cdon and Boc but not to Ptch1 (Wierbowski et al., 2020). Whereas Gas1 does not bind Scube2, it binds to both the cholesterol and palmitoyl moieties (Wierbowski et al., 2020). A SHH-receptor interaction relay model has been proposed that the Cdon/Boc coreceptors recruit the Shh-Scube2 complex to the cell surface where the SHH ligand is handed off in a dual lipid-dependent manner to Gas1 and then to Ptch1 to initiate signaling (Wierbowski et al., 2020). Studies in cell culture and genetic studies in mice indicate that Cdon, Boc, and Gas1 act partly redundantly (Wierbowski et al., 2020). Cdo mutant (Cdo−/−) mouse embryos display microform holoprosencephaly whereas Boc mutants are adult viable and fertile (Cole and Krauss, 2003; Okada et al., 2006). Cdo−/−;Boc−/− double mutant embryos exhibit severe neural tube patterning defects but many Shh-dependent neural progenitors are still established (Allen et al., 2011). Loss of Gas1 also disrupted neural tube patterning although the defects were less severe than in Shh−/− embryos (Allen et al., 2007). Simultaneous inactivation of all three genes (Gas1−/−;Cdo−/−;Boc−/−) recapitulated mouse phenotypes of complete loss of Hedgehog activity, indicating an obligatory requirement for these co-receptors and differential requirement for each in Hedgehog signaling (Allen et al., 2011).
2.3. Regulation of the functions of Ptch and Smo and crucial roles of cholesterol
Whereas early studies suggested that Ptch might regulate Smo through direct physical interaction (Stone et al., 1996), subsequent studies indicate that Ptch regulates Smo activity in a non-stoichiometric manner (Denef et al., 2000; Ingham, 2000; Taipale et al., 2002). The Ptch proteins have significant sequence homology with the resistance-nodulation-division (RND) family protein pumps that many bacteria use to efflux toxic molecules and antibiotics, and with Niemann-Pick C1 (NPC1) that transports cholesterol out of the lysosomes (Kong et al., 2019). Experiments in cell culture showed that the amino acid residues essential for the transporter function of bacterial RND proteins are conserved in and required for Hedgehog pathway inhibition by Ptch (Taipale et al., 2002). Both Ptch and Disp share sequence homology with the sterol sensing domain of NPC1, which together with the finding that Disp exports the Hedgehog ligands through binding to the cholesterol moiety (Burke et al., 1999; Caspary et al., 2002; Ma et al., 2002; Tukachinsky et al., 2012) suggest that Ptch may regulate Smo activity through transporting cholesterol or other sterols. On the other hand, the discovery that the naturally occurring steroidal alkaloid cyclopamine can bind and inhibit Smo activity (Chen et al., 2002) and subsequent finding that oxysterols, derivatives of cholesterol, can activate the SHH signaling pathway in cell culture (Corcoran and Scott, 2006), suggest that the endogenous Smo regulator is a sterol lipid. Remarkably, crystal structures of the Smo protein revealed a cholesterol molecule bound to the same position that oxysterols were predicted to bind (Byrne et al., 2016). Mutations that prevent Smo binding to cholesterol impair Hedgehog signaling in cultured cells and in mouse embryos (Byrne et al., 2016; Xiao et al., 2017). Additional studies showed that cholesterol is sufficient to activate Smo signaling even in the absence of Hedgehog ligands (Huang et al., 2016; Luchetti et al., 2016). Zhang et al. (2018) showed that cholesterol activity in the inner leaflet of the plasma membrane is reduced by overexpression of PTCH1 but rapidly restored by Hedgehog stimulation (Zhang et al., 2018). More recently, Kinnebrew et al. showed that Ptch1 regulates Smo activity by controlling cholesterol binding to its extracellular cysteine-rich domain (Kinnebrew et al., 2022). Together, these studies provide compelling evidence that Ptch regulates Smo activity by controlling cholesterol availability.
How can Ptch keep cholesterol, the single most abundant lipid in the plasma membrane (Maxfield and van Meer, 2010; Mouritsen and Bagatolli, 2015), away from Smo to prevent inappropriate pathway activation? The solution to this dilemma is based on the key role of the primary cilium in Hedgehog signaling, with the ciliary membrane acting as a subcellular compartment within which the levels of accessible cholesterol can be modulated by Ptch without impacting global cellular cholesterol homeostasis (Ingham, 2022). It has been shown that the ciliary membrane contains lower levels of accessible cholesterol than the plasma membrane but the ciliary cholesterol levels are increased upon Hedgehog stimulation (Kinnebrew et al., 2019). Thus, Ptch would prevent Smo activation within the primary cilium while its inhibition by Shh binding would promote accumulation of active Smo. Whereas Drosophila Hedgehog signaling does not depend on the primary cilia, cholesterol represents a minor fraction of plasma membrane sterols in Drosophila cells and, hence, Hedgehog signaling activity in Drosophila could be regulated through Ptch-mediated regulation of cholesterol availability in the plasma membrane (Ingham, 2022; Kinnebrew et al., 2019).
This model that cholesterol is the endogenous Smo activating ligand regulated by Ptch provides new mechanistic insights into multiple developmental disorders associated with disruption of cholesterol biosynthesis (Kinnebrew et al., 2019; Porter and Herman, 2011). For example, mutations in the gene encoding the cholesterol biosynthetic enzyme 7-dehydrocholesterol reductase (DHCR7) cause Smith–Lemli–Opitz syndrome (SLOS) (OMIM #270400) (Wassif et al., 1998). SLOS patients exhibits craniofacial defects including cleft palate, syndactyly and polydactyly, and HPE (Kelley et al., 1996). Blassberg et al. (2016) showed that SHH signaling activity is reduced in Dhcr7 deficient mouse embryonic fibroblasts (Blassberg et al., 2016). Two SLOS-like syndromes, known as Desmosterolosis (OMIM #602398) and Lathosterolosis (OMIM #607330), respectively, are caused by homozygous mutations in SC5D, encoding sterol-C5-desaturase, and DHCR24, encoding the 24-dehydrocholesterol reductase (Andersson et al., 2002; Brunetti-Pierri et al., 2002; Krakowiak et al., 2003; Waterham et al., 2001). Together with DHCR7, SC5D and DHCR24 catalyze the terminal steps of cholesterol biosynthesis (Porter and Herman, 2011). Cell culture assays showed that loss of function of each of these three genes impaired transcriptional induction of endogenous Gli1, a direct target of Hedgehog signaling (Dai et al., 1999; Kinnebrew et al., 2019). Interestingly, Hedgehog signaling in Dhcr7−/− cells, but not in Dhcr24−/− cells, could be rescued by exogenous cholesterol, suggesting that accumulation of some precursor sterols due to defects in terminal steps of cholesterol biosynthesis may bind to Smo and antagonize Hedgehog signaling (Kinnebrew et al., 2019; Nguyen et al., 2022; Porter and Herman, 2011). Cholesterol biosynthesis involves over 30 biochemical steps and mutations disrupting several other enzymes catalyzing cholesterol biosynthesis have been associated with distinct developmental disorders (Porter and Herman, 2011). Studies in zebrafish showed that disruption of hmgcrb, a homolog of the mammalian Hmgcr gene encoding 3-hydroxy-3-methyl-glutaryl-CoA reductase, the first rate-limiting enzyme in the sterol biosynthesis pathway (Schumacher and DeBose-Boyd, 2021), resulted in orofacial clefts associated with reduction in expression of Gli1 during early facial development (Signore et al., 2016). Interestingly, statins that are commonly used to treat hypercholesterolemia and dyslipidemia are structural analogs of HMG-CoA and competitive inhibitors of HMGCR (Istvan and Deisenhofer, 2001). Thus, better understanding of the effects on cholesterol metabolism and related treatment approaches on Hedgehog and other developmental signaling pathways will directly impact strategies for treatment and/or prevention of developmental disorders.
2.4. Intracellular transduction of SHH signaling and the roles of the primary cilium
Although evolutionarily conserved from Drosophila to human, Hedgehog signaling in vertebrates, but not in Drosophila, depends on the primary cilium, a non-motile microtubule-based flagellar projection on almost all vertebrate cells (Goetz and Anderson, 2010) (Fig. 1C–D). Remarkably, most of the core components of the vertebrate Hedgehog signaling pathway, including the Ptch receptors, Smo, Sufu, and the full-length Gli proteins, are found localized to the primary cilia although the mechanisms localizing these factors to the primary cilia are not well understood yet (reviewed in (Bangs and Anderson, 2017; Ingham, 2022)). Vertebrates have three Gli proteins; Gli2 and Gli3 are expressed in the absence of Hedgehog signaling whereas Gli1 is a direct transcriptional target of Hedgehog signaling and functions mainly as an activator to amplify Hedgehog signaling activity (Bai et al., 2002; Dai et al., 1999; Kong et al., 2019). It is important to note that Gli2 and Gli3 proteins exist in at least three activity states: proteolytically processed transcriptional repressors (Gli2R/Gli3R), transcriptionally inactive full-length Gli2/Gli3 proteins, and full-length transcriptional activators (Gli2A/Gli3A) (Kong et al., 2019). It is also important to know that the Gli proteins travel through the primary cilia whether the cells receive Hedgehog signaling or not (Kong et al., 2019). In the absence of Hedgehog signaling, Sufu binds to Gli2/3 proteins and the Sufu-Gli complex travels through the primary cilia where PKA specifically phosphorylates full-length Gli2/3 to initiate a pathway of proteasomal processing into the truncated GliR forms, which dissociate from Sufu and translocate into the nucleus to repress target gene expression (Ding et al., 1999; Humke et al., 2010; Kogerman et al., 1999; Niewiadomski et al., 2014; Tukachinsky et al., 2010; Tuson et al., 2011). PKA activity is regulated by GPCR signaling (Kong et al., 2019). The orphan rhodopsin GPCR protein Gpr161 is localized to the cilium and contributes to the local activation of PKA (Mukhopadhyay and Rohatgi, 2014; Mukhopadhyay et al., 2013). When Hedgehog signaling is on, activated Smo accumulates in the ciliary membrane and inhibits PKA activity via its C-terminal PKA inhibitor motif (Happ et al., 2022). Activated Smo also promotes the dissociation of full-length Gli2/3 from Sufu, allowing formation of the Gli2/3A transcriptional activators (Kong et al., 2019). Previous studies showed that Sufu-deficient mouse embryos died at ~E9.5 with a ventralized neural tube reminiscent of global activation of SHH signaling but the Sufu−/− embryos exhibited barely detectable Gli2/3 proteins (Chen et al., 2009; Cooper et al., 2005; Svard et al., 2006). Chen et al. (2009) showed that Sufu maintains Gli2 and Gli3 protein levels through antagonizing Spop (speckle-type POZ protein)-mediated ubiquitination (Chen et al., 2009). In addition, Sufu can bind to and modulate Gli-mediated transcriptional activity in the nucleus (Lin et al., 2014). The atypical kinesin Kif7 is strongly enriched at the ciliary tip and plays a crucial role in regulating the processing and stability of Gli proteins (Cheung et al., 2009; Endoh-Yamagami et al., 2009; Liem et al., 2009). The molecular mechanisms converting the full-length Gli proteins to GliA activators and translocating them into the nucleus are still not completely understood.
The assembly of cilia, and trafficking of proteins into and out of the cilia, are mediated by a two-way intraflagellar transport (IFT) system that consists of two IFT complexes, IFT-A and IFT-B (Chinipardaz et al., 2022; Nakayama and Katoh, 2020). The IFT-B complex together with the kinesin II motor proteins regulate anterograde trafficking from the base to the tip of the cilium whereas the IFT-A complex works with the dynein motor proteins to regulate retrograde transport from the tip to the base of cilium (Nakayama and Katoh, 2020). Disruption of the genes encoding IFT-B components, such as Ift88 and Ift172, or disruption of Kif3a, resulted in cilial agenesis and loss of Shh-dependent ventral neural tube cell types (Huangfu and Anderson, 2005; Huangfu et al., 2003). Inactivation of genes encoding IFT-A components exhibited complex effects: mutations in several IFT-A genes, including Ift122 and Ift139, caused bulged cilia and exhibited ligandindependent gain of SHH signaling in neural tube and limb development (Qin et al., 2011; Tran et al., 2008) whereas several other IFT-A mutations resulted in short bloated cilia with a highly disrupted axoneme and were associated with loss of SHH signaling activity in neural tube patterning (Bangs and Anderson, 2017). Production of both Gli2/3R repressors and Gli2/3A activators are affected when ciliogenesis is disrupted (Huangfu and Anderson, 2005; Liu et al., 2005), consistent with the cilium-dependence of PKA mediated phosphorylation of Gli2/3 and Smo-mediated Hedgehog signal transduction. Furthermore, mutations disrupting some ciliary proteins, such as Ift25 and Ift27, apparently did not affect the morphology of the primary cilia but caused mild defects in trafficking of Hedgehog signaling pathway components including Ptch1, Smo, Gli2, through the primary cilia (Eguether et al., 2014; Keady et al., 2012). Hence, the phenotypic effects of mutations in IFT components and other ciliary proteins are dependent on the developmental context of Hedgehog ligand and Gli2/3 expression, mutant effects on ciliary structure and/or function, and may result from the loss of GliR-mediated target gene repression, loss of Hedgehog signaling-dependent GliA-mediated target gene activation, or both.
In summary, SHH signaling pathway is regulated at multiple steps (Fig. 1), from the biogenesis and cellular release of the active ligand, interaction with the receptor and co-receptors, activation of the obligate signaling transducer protein Smo, to multiple mechanisms regulating Gli protein processing and transcription factor activity as well as Gli-independent mediators of downstream target gene regulation (for recent reviews, please see (Ingham, 2022; Kong et al., 2019; Zhang and Beachy, 2023)). Disruption at any of these steps results in dysregulation of target gene expression and may cause developmental defects including craniofacial malformations. It is important to emphasize that Hedgehog signaling is not a binary ON/OFF switch but rather is composed of a series of negative inhibitory interactions of which the outcome depends on the strengths and duration of signaling activity in target cells (Kong et al., 2019). Since the GliA and GliR share the same DNA binding domains, the ratio of GliA and GliR in the target cells is an important determinant for target gene expression. Even modest alterations in SHH signaling strength can lead to developmental defects (Nieuwenhuis and Hui, 2005). In the following sections, we review and summarize advances in the understanding of the roles of and mechanisms involving SHH signaling in major craniofacial developmental processes, including formation and patterning of the facial primordia, palate development, mandibular development and patterning, tongue organogenesis, taste bud formation and homeostasis, and tooth development.
3. SHH function in the formation of facial primordia
At the beginning of facial development in vertebrate embryos, cranial neural crest cells (CNCCs), a population of multipotent progenitor cells that give rise to most of the bones, cartilages, and connective tissues in the head, arise from the lateral edges of the anterior neural plate (Gammill and Bronner-Fraser, 2003; Morales et al., 2005; Steventon et al., 2005). These CNCCs delaminate and migrate ventrally underneath the surface ectoderm where epithelial-mesenchymal interactions between the surface ectoderm and CNCCs result in the formation and outgrowth of the embryonic facial primordia, including the frontonasal prominence (FNP) rostral to the primitive mouth, and the paired maxillary and mandibular arches lateral and caudal, respectively, to the primitive mouth. The FNP grows around the bilateral nasal placodes to form the medial nasal processes (MNP) and lateral nasal processes (LNP). Subsequent growth and convergence of the MNP, LNP, maxillary and mandibular processes result in the formation of the intact embryonic face. During the growth and convergence of the facial primordia, the CNCC-derived facial mesenchyme cells receive multiple signals, including members of the bone morphogenetic protein (BMP) family, fibroblast growth factor (FGF) family, WNT family, and SHH, from the surrounding epithelial cells that regulate their proliferation and differentiation (Helms and Schneider, 2003; Jiang et al., 2006; Santagati and Rijli, 2003). For example, upon arriving in mandibular arches, the CNCCs encounter Fgf8 expressed in the proximal mandibular arch epithelium and Bmp4 expressed by the distal mandibular arch epithelium (Haworth et al., 2004; Shigetani et al., 2000; Tucker et al., 1998; Tucker et al., 1999). Fgf8 and Bmp4 act to pattern the proximal-distal axis of the mandibular arch by activating expression of Barx1 in the proximal domain and expression of Msx1, Msx2, and Alx4, in the distal domain, respectively (Barlow et al., 1999; Ferguson et al., 2000; Parada and Chai, 2015; Tucker et al., 1998). In addition, Fgf8 signalling patterns the rostral-caudal axis of the mandibular arch mesenchyme by restricting the expression of Lhx6 and Lhx8 in the rostral domain (Cobourne and Sharpe, 2003; Grigoriou et al., 1998; Tucker et al., 1999). Several members of the WNT family are also expressed in the early facial ectoderm and activates canonical WNT/β-catenin signaling in both the epithelium and underlying mesenchyme of the MNP, LNP, and maxillary processes (Lan et al., 2006). Mutations in WNT3 and WNT9b have been linked to non-syndromic cleft lip with or without cleft palate (Chiquet et al., 2008; Fontoura et al., 2015; Lu et al., 2015; Mostowska et al., 2012; Nikopensius et al., 2010). Mouse genetic studies showed that Wnt9b from the facial ectoderm signals to the maxillary and nasal mesenchyme to regulate the cell proliferation and ensure lip fusion (Jin et al., 2012; Juriloff et al., 2014; Lan et al., 2006). More information about the roles of BMP, FGF, and WNT signaling pathways in early facial morphogenesis can be found in several recent articles and references therein (Brewer et al., 2016; Graf et al., 2016; Nie et al., 2006; Ray et al., 2020; Reynolds et al., 2019; Reynolds et al., 2020; Stanier and Pauws, 2012; Ueharu and Mishina, 2023). In this review, we focus on discussing the cellular and molecular mechanisms mediating SHH signaling pathway function in regulating craniofacial development.
In both chick and mouse embryos, Shh is expressed in several epithelial regions during early facial morphogenesis, including neuroectoderm of the ventral forebrain, oral ectoderm, and pharyngeal endoderm (Jeong et al., 2004). SHH produced by the forebrain neuroepithelium induces Shh expression in the facial ectoderm (Marcucio et al., 2005). It has been shown that Shh-Gli3 signaling regulates primary mouth opening in both Xenopus and mouse embryos by controlling buccopharyngeal membrane dissolution (Tabler et al., 2014). In the frontonasal region, initial expression of Shh and Fgf8 in the stomodeal ectoderm defines a specific signaling center called frontonasal ectodermal zone (FEZ), with Shh expression in the ectodermal cells comprising the roof of the mouth and with Shh- and Fgf8-expressing cells forming a boundary that spans the mediolateral axis of the FNP (Hu and Marcucio, 2009a, b; Hu et al., 2003). Subsequently, Fgf8 expression becomes restricted to the nasal pits at Hamburger Hamilton stage (HH) 20–21 in chick and at about E9.5 to E10.5 in mice (Hu and Marcucio, 2009a, b, 2012; Hu et al., 2003). BMP signaling is implicated in regulating the onset of Shh expression in the FEZ in chick embryos (Foppiano et al., 2007). Blocking BMP signaling using Noggin at HH~15/16 resulted in decreased Shh expression in the facial ectoderm and a lack of outgrowth of the treated FNP (Hu et al., 2015), but the source of the BMP signal and whether BMP signaling directly activates Shh expression in the facial ectoderm are not known. SHH from the ectoderm in turn regulates expression of several Bmp genes in the adjacent CNCC-derived facial mesenchyme (Hu and Marcucio, 2009a, b; Hu et al., 2015). The BMP-SHH signaling loop is critical for restricting Fgf8 expression to the nasal pits to regulate regional cell proliferation in the FNP (Hu et al., 2015). In chick embryos, inhibiting SHH signaling by using cyclopamine or introduction of dominant-negative Ptch1, in the FNP at HH15 resulted in a decrease in cell proliferation leading to hypoplasia of the lateral edges of the FNP, a failure of the globular process to contact the lateral nasal and maxillary processes, and a cleft of the primary palate (Hu et al., 2015). Together, these findings indicate that interactions of SHH, BMP, and FGF8 signaling pathways form a highly coordinated molecular regulatory network to control early facial patterning and growth although the exact molecular mechanisms regulating the interactions of these signaling pathways remain to be elucidated.
Studies using function-blocking anti-SHH antibody or application of recombinant SHH protein in chick embryos indicated that Shh plays important roles for survival and proliferation of early CNCCs (Ahlgren and Bronner-Fraser, 1999; Hu and Helms, 1999). Using tissue-specific inactivation of Smo in premigratory neural crest cells (Wnt1Cre;Smon/c) in mice, Jeong et al investigated the role of Hedgehog signaling activity in CNCC development and facial primordial outgrowth (Jeong et al., 2004). By E10.5, the Wnt1Cre;Smon/c mouse embryos displayed smaller FNP and mandibular arches, associated with increased cell apoptosis and decreased proliferation of CNCC-derived facial mesenchyme (Jeong et al., 2004). Conversely, the Wnt1-Cre;R26SmoM2 transgenic mouse embryos, which carry a Cre-activated transgene encoding a constitutively active form of Smo (Supplementary Table 1) specifically in the neural crest lineage, exhibited a mild hyperplasia of the facial processes by E10.5 (Jeong et al., 2004). Expression of several genes encoding the Forkhead box (Fox) containing transcription factors, including Foxf1, Foxf2, Foxd1, Foxd2 and Foxc2, were reduced in the developing facial primordia in Wnt1Cre;Smon/c embryos and expanded in Wnt1-Cre;R26SmoM2 embryos (Jeong et al., 2004). Further studies showed that several Fox family transcription factors, including Foxf1 and Foxf2, play crucial roles in cell survival, tissue growth, and patterning of multiple facial developmental processes (Everson et al., 2017; Xu et al., 2019; Xu et al., 2016; Xu et al., 2022). Mice with tissue specific inactivation of both Foxf1 and Foxf2 in cranial neural crest cells (Foxf1c/cFoxf2c/cWnt1Cre) display severe craniofacial defects resembling Wnt1Cre;Smon/c embryos (Jeong et al., 2004; Xu et al., 2019). Furthermore, chromatin immunoprecipitation followed by high throughput sequencing analyses revealed direct binding of Gli3 protein at genomic sites in or around several Fox family genes, including Foxf1 and Foxf2, in multiple tissues including in the developing face (Elliott et al., 2020; Hoffmann et al., 2014). Together, these results indicate that Foxf1 and Foxf2 function as important mediators of SHH signaling during craniofacial development.
Studies of two mutant mouse lines showed that SHH signaling interacts with WNT signaling to regulate upper lip fusion. The Hhatcreface transgenic allele disrupted the Hhat gene, encoding the acyltransferase that adds the palmitoyl moiety to the Hedgehog protein, and resulted in loss of SHH signaling in the facial primordia (Dennis et al., 2012; Kurosaka et al., 2014). The Ptch1wiggable mutant mouse allele generates a truncated Ptch1 protein, leading to enhanced SHH signalling (Kurosaka et al., 2014). The Hhatcreface;Ptch1wiggable double mutant embryos displayed hypoplastic nasal process outgrowth and cleft lip with persistence of epithelial seam between the MNP and LNP (Kurosaka et al., 2014). Interestingly, analysis using the TOPgal transgenic reporter for detecting canonical WNT signaling activity (DasGupta and Fuchs, 1999) showed that WNT signaling activity in the developing facial primordia is reduced in the Ptch1wiggable mutant embryos and increased in Hhatcreface embryos, the Hhatcreface;Ptch1wiggable double mutants exhibited restored TOPgal activity except in the lambdoidal region coinciding with the lack of apoptosis, suggesting that SHH signaling regulates lip fusion at least in part through restricting WNT signaling activity. The balance of SHH-WNT signaling is thought to maintain proper proliferation and apoptosis for removing the epithelial seam at lambdoidal region during lip fusion.
As described above, SHH signaling depends on the primary cilium. Mutations in genes encoding components of the primary cilia cause a class of genetic disorders collectively referred to as ciliopathies, of which many exhibit craniofacial developmental defects (Ferkol and Leigh, 2012). Genetic studies in mice showed that mutations in most of the genes encoding major components of the ciliary IFT machinery or ciliary structural proteins cause disruption of ciliogenesis, resulting in homozygous embryonic lethality at midgestation associated with context-dependent loss or gain of SHH signaling (reviewed in (Bangs and Anderson, 2017)). Interestingly, disruption of the IFT-B genes Ift25 and Ift27 in mice did not overtly disrupt the morphology and structure of the primary cilia but still caused neonatal death with craniofacial defects including hypotelorism, micrognathia, and cleft palate (Eguether et al., 2014; Keady et al., 2012). The developmental defects in Ift25−/− and Ift27−/− embryos suggest decreased SHH signaling activity. Indeed, upon stimulation with Shh or a Smo agonist, the Ift25−/− and Ift27−/− mutant mouse embryonic fibroblast cells exhibited aberrant accumulation of Ptch1 and Gpr61 and reduced Gli2 in the primary cilia, suggesting that these ciliary proteins play highly specialized roles in intraflagellar transport of the Hedgehog pathway components (Eguether et al., 2014; Keady et al., 2012). Tissue specific inactivation of Kif3a, encoding a subunit of the kinesin-II IFT motor for anterograde transport in the cilium, in CNCCs caused gain of SHH signaling activity during midfacial development and resulted in hypertelorism and a midfacial cleft in mice (Brugmann et al., 2010). Chang et al. (2016) showed that the ciliopathic midfacial defects in the Kif3af/f;Wnt1Cre and Ift88f/f;Wnt1Cre mice occur primarily via a de-repression mechanism due to loss of the Gli2R and Gli3R repressors (Chang et al., 2016).
In addition to the IFT machinery, many other proteins play crucial roles in building the primary cilia and their disruption also affect SHH signaling (Bangs and Anderson, 2017). The chick talpid2 mutation disrupts the C2cd3 gene, encoding structural protein required for the centriole to dock on the ciliary vesicle (Ye et al., 2014), and caused facial cleft due to disruption of ciliogenesis (Chang et al., 2014). Post-translational processing of Gli2 and Gli3 was disrupted in the developing facial prominences in talpid2 mutant embryos, resulting in increased accumulation of full-length Gli2 and Gli3 proteins and decreased level of the Gli3R repressor, which correlated with the increased activation/de-repression of the SHH pathway in the developing FNP (Chang et al., 2014). Another ciliopathic mutant mouse, the Fuz−/− mutant mouse, displays high arched palate and a single calvarial bone pair due to expanded CNCCs (Tabler et al., 2013; Tabler et al., 2016). Remarkably, tissue-specific inactivation of Fuz in the CNCCs did not recapitulate the Fuz−/− craniofacial defects, indicating that the effects of Fuz loss of function on CNCCs is cell non-autonomous. Expanded Fgf8 expression was found in the hindbrain and early facial ectoderm associated with reduced Gli3R activity in the Fuz−/− embryos (Tabler et al., 2013; Tabler et al., 2016). Gli3xt-J/xt-J mutants display expanded neural crest cells in the frontal bone mesenchyme and developing larynges similar with the Fuz−/− mutants, suggesting that the expanded Fgf8 expression in the Fuz−/− embryos was due to loss of cilia-mediated production of Gli3R (Tabler et al., 2016; Tabler et al., 2017). Genetically deleting one allele of Fgf8 ameliorated the maxillary and skull phenotypes, indicating that excessive Fgf8 was responsible for the expanded neural crest cells in the Fuz−/− embryos (Tabler et al., 2013; Tabler et al., 2016). Furthermore, mouse embryos deficient in Ofd1, encoding a centriole-associated protein that colocalizes with C2CD3, exhibited strikingly similar maxillary hyperplasia and expanded Fgf8 expression in the facial ectoderm similarly as in Fuz−/− embryos (Tabler et al., 2013). These results identify a crucial role of ciliummediated production of Gli3R and its repression of Fgf8 expression in the embryonic facial ectoderm in the regulation of facial primordial outgrowth (Tabler et al., 2013). Together with other studies that demonstrated crosstalk of SHH, BMP, and FGF8 signaling pathways during facial primordia outgrowth as discussed above, these results indicate that primary cilium mediated GLI processing is a key regulator of the molecular network controlling facial primordial growth.
4. SHH function in palate development and patterning
In mammals, the palate separates the oral and nasal cavities and plays crucial roles in feeding, breathing, and speech. In mouse, the development of secondary palate begins as a pair of outgrowths from the oral side of the paired maxillary at around E11.5 to a pair of palatal shelves that initially grow vertically downward flanking the developing tongue. At around E14.0, the palatal shelves reorient to the horizontal position above the tongue, grow towards and subsequently fuse with each other at the midline. The secondary palate also fuses anteriorly with the primary palate and nasal septum to form the intact roof of the oral cavity (Fig. 2A–F) (reviewed by (Bush and Jiang, 2012; Chai and Maxson, 2006; Dixon et al., 2011; Lan et al., 2015).
Fig. 2.
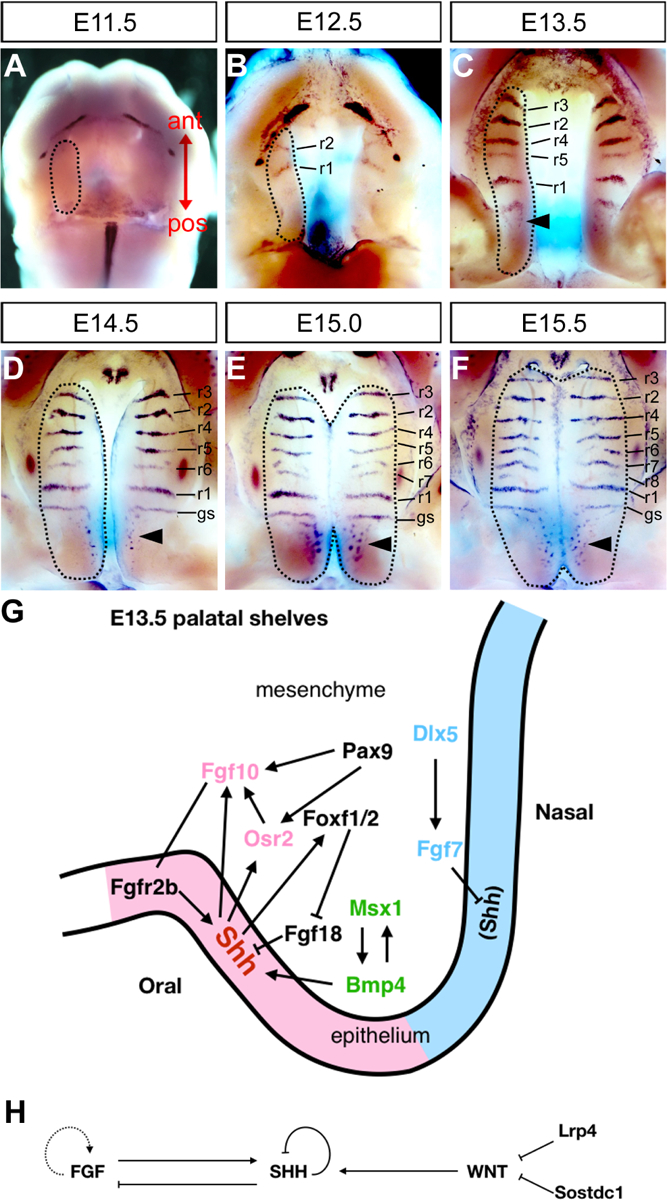
SHH signaling plays a central role in regulating palatal shelf growth and patterning. (A-F) Whole mount in situ hybridization detection of Shh mRNAs showing Shh was initially expressed throughout the oral epithelium at the onset of palatal outgrowth at E11.5 and becomes restricted to the epithelial ridges that develop into the palatal rugae on the oral side of the palatal shelves. Dashed lines in (A-D) outline developing palatal shelves. Dashed lines in (E-F) outline the fused palates. r1–r8 mark the individual rugae in the order of their formation. gs indicates geschmacksstreifen. Arrowheads indicate the sensory papillae. The double headed arrow in (A) shows the anterior-posterior axis of developing palate. (G) Schematic diagram of a coronal section of the developing palatal shelf at E13.5, with the oral side to the left and nasal side to the right, to depict the molecular network involving Shh in the reciprocal epithelial-mesenchymal interactions regulating palatal shelf growth and patterning. The Shh-Osr2-Fgf10-Shh (shown in pink) cascade positively maintains the expression of Shh in oral side of palatal epithelium. The Dlx5-Fgf7 (shown in blue) cascade prevents Shh expression at the nasal side of palatal epithelium. The Msx1-Bmp4 (shown in green) feed-back loop has been shown to positively regulate the expression of Shh in the anterior palatal shelves. The Shh-Foxf-Fgf18-Shh circuit has also been shown to maintain the expression of Shh in the palatal epithelium. (H) Schematic diagram showing the cross regulation of SHH, FGF and WNT signaling during palatal ruga formation. Palatal ruga formation is regulated by a Turing-type activator-inhibitor reaction-diffusion mechanism, with FGF signaling acting as an activator whereas SHH signaling acting as an inhibitor to regulate the periodic ruga formation. WNT signaling is also required to maintain Shh expression and ruga formation.
SHH signalling plays critical roles in regulating growth, patterning, and fusion of palatal shelves. At the onset of palatal outgrowth, Shh expression was detected throughout the early oral epithelium (Rice et al., 2006). During palatal shelf growth, Shh expression becomes restricted to the oral side palatal epithelium, at high levels in the ectodermal ridges forming the palatal rugae in the anterior region and in the sensory papilla in the posterior region of the palatal shelves (Fig. 2A–F) (Pantalacci et al., 2008; Welsh and O’Brien, 2009). Ptch1 and Gli1, known target genes of SHH signalling, are expressed in palatal epithelial and mesenchyme cells, indicating that SHH signalling pathway is active in both the palatal epithelium and mesenchyme (Economou et al., 2012; Lan and Jiang, 2009; Rice et al., 2006). In palatal shelf explants culture, adding exogenous SHH induced, whereas adding anti-SHH antibody inhibited, cell proliferation in the palatal mesenchyme (Han et al., 2009; Rice et al., 2004; Zhang et al., 2002). Tissue-specific inactivation of Smo in the CNCCs (Smon/c;Wnt1Cre) caused complete agenesis of the secondary palate (Jeong et al., 2004). Deletion of Shh in the oral epithelium after E11 (Shhc/n;K14-Cre) caused an incomplete penetrance of cleft palate (Lan and Jiang, 2009; Rice et al., 2004). Loss of function of Gas1 also resulted in an incomplete penetrance of cleft palate phenotype associated with reduced cell proliferation in developing palatal shelves. Deleting one Shh allele or inactivation of Boc increased the penetrance and severity of cleft palate in Gas1 mutant mice (Seppala et al., 2007; Xavier et al., 2016). Tissue-specific inactivation of Smo in the palatal mesenchyme (Smon/c;Osr2-IresCre) resulted in complete penetrance of cleft palate with reduced cell proliferation (Lan and Jiang, 2009). Together, these results indicate that SHH signalling plays a key role in palatal shelf outgrowth.
A series of mouse genetic studies uncovered a positive feedback regulatory loop between Shh expression in the palatal epithelium and Fgf10 expression in the palatal mesenchyme. Mice lacking either Fgf10 or Fgfr2b, an epithelium-specific isoform of the FGF receptor, exhibited cleft palate with failure of maintenance of Shh expression in the palatal epithelium (De Moerlooze et al., 2000; Rice et al., 2004). In palatal explant culture assays, agarose beads containing recombinant FGF10 protein induced Shh expression in wildtype palatal shelves but not in the Fgfr2b−/− palatal shelves (Rice et al., 2004), indicating that FGF10-FGFR2b signaling is required for maintaining Shh expression during palatal shelf outgrowth. Expression of Fgf10 in the palatal mesenchyme was significantly downregulated in the Smon/c;Osr2-IresCre as well as in Osr2−/− embryos (Lan and Jiang, 2009; Zhou et al., 2013). Osr2 expression is also reduced in the Smon/c;Osr2-IresCre palatal mesenchyme and Osr2−/− mice displayed cleft palate with reduced palatal shelf growth, suggesting that Osr2 acts down-stream of SHH signaling and regulates the expression of Fgf10 (Lan and Jiang, 2009; Lan et al., 2004). How SHH signaling regulates, directly or directly, the expression of Osr2 in the developing palatal mesenchyme remains to be determined. Interestingly, mice lacking the Pax9 transcription factor exhibited cleft palate and the Pax9−/− embryos had reduced expression of both Osr2 and Fgf10 in the palatal mesenchyme (Peters et al., 1998; Zhou et al., 2013). Restoring Osr2 expression in the palatal mesenchyme in Pax9 mutants by inserting the Osr2 cDNA into the Pax9 locus partly restored Fgf10 expression and rescued palatal shelf growth and fusion (Zhou et al., 2013). Together, these studies identified a positive regulatory cascade of Shh-Osr2/Pax9-Fgf10-Shh mediating the reciprocal epithelial-mesenchymal interactions driving palatal shelf growth (Fig. 2G).
In contrast to the positive regulation of Shh expression in the palatal epithelium by Fgf10, Fgf7 is also expressed in the palatal mesenchyme, but it inhibited Shh expression in palatal explant culture assays (Han et al., 2009; Rice et al., 2004). Interestingly, Dlx5−/− embryos exhibited reduced Fgf7 expression in the palatal mesenchyme and expansion of Shh expression to the nasal side palatal epithelium. Mice lacking Msx1, a transcription factor with restricted expression in the anterior palatal mesenchyme, exhibited reduced palatal shelf growth accompanied by significant reduction in Shh expression in the anterior palatal epithelium (Zhang et al., 2002). Zhang et al. (2002) showed that Msx1−/− embryos exhibited reduced expression of Bmp4 in the anterior palatal mesenchyme and that ectopic expression of Bmp4 driven by the Msx1 promoter rescued the cleft phenotype of Msx1−/− mutants accompanied by restored palatal cell proliferation and the expression of Shh (Zhang et al., 2002). Remarkably, inactivation of Dlx5 in Msx1−/− mutants also rescued palatal shelf growth and fusion in the Dlx5−/−Msx1−/− embryos compared with cleft palate in the Msx1−/− mutants (Han et al., 2009). These results indicate that multiple molecular pathways converge on the regulation of Shh expression during palate development to regulate palatal shelf growth and patterning (Fig. 2G).
Expression of the Foxf1 and Foxf2 genes in the developing palatal mesenchyme depends on SHH signaling (Jeong et al., 2004; Lan and Jiang, 2009). Foxf2−/− mouse embryos exhibited reduced palatal shelf growth whereas mice with neural crest-specific inactivation of both Foxf1 and Foxf2 (Foxf1c/cFoxf2c/cWnt1Cre) exhibited rudimentary palatal shelves, resembling the severe palatal growth defect observed in Smon/c;Wnt1Cre embryos (Jeong et al., 2004; Xu et al., 2016). Through RNA-seq analysis, we found that Foxf2−/− embryos exhibited aberrantly increased expression of multiple genes, including Fgf18, in the developing palatal shelves (Xu et al., 2016). The domains of ectopic Fgf18 expression in the palatal mesenchyme in Foxf2−/− embryos correlated with where Shh expression was downregulated in the palatal epithelium. Furthermore, Foxf1c/c;Foxf2c/c;Wnt1Cre embryos exhibited ectopic Fgf18 expression throughout the palatal mesenchyme and complete loss of Shh expression in the palatal epithelium (Xu et al., 2016). Addition of recombinant Fgf18 protein to palatal explant cultures inhibited Shh mRNA expression in the palatal epithelium (Xu et al., 2016). Together, these data indicate that the Foxf1/Foxf2 transcription factors act downstream of SHH signaling in the palatal mesenchyme to regulate palatal shelf growth and feedback to maintain Shh expression in the palatal epithelium by preventing ectopic activation of Fgf18 expression (Fig. 2G).
Whereas several FGFs act antagonistically to restrict Shh expression to the oral side of the palatal epithelium during palatal shelf growth (Fig. 2G), studies of gain-of-function mouse models further demonstrated the importance of spatiotemporally regulated SHH signalling activity in palatogenesis. Li et al. analyzed the effects of persistent activation of Smo-mediated Hedgehog signalling throughout the palatal epithelium in K14-Cre;R26SmoM2 embryos and showed that they developed submucous cleft palate with persistence of the medial edge epithelium (MEE) (Li et al., 2018). Cobourne et al. showed that K14-Shh transgenic mouse embryos, expressing Shh throughout the palatal epithelium, also exhibited failure of palatal shelf fusion (Cobourne et al., 2009). These data indicate that Shh is a central node integrating multiple signaling pathways in the palate development regulatory network and that SHH signaling activity is spatiotemporally precisely controlled for proper palatal shelf growth and palatal fusion.
In addition to regulating palatal shelf growth and fusion, the crosstalk between SHH and FGF signaling pathways play a crucial role in the formation and patterning of the palatal rugae, the periodic epithelial ridges on the oral side of the hard palate that are involved in sensing and holding food (Economou et al., 2012). Both Fgf10−/− and Fgfr2b−/− mice lack palatal rugae (Hosokawa et al., 2009; Welsh and O’Brien, 2009). Mice lacking both Spry1 and Spry2, intracellular antagonists of FGF signaling, showed highly disorganized palatal rugae (Economou et al., 2012). Epithelium-specific inactivation of Shh also resulted in highly disorganized rugae. In palatal explant cultures, inhibiting FGF signalling or enhancing SHH signaling inhibited ruga formation, whereas antagonizing SHH signalling by using cyclopamine resulted in dramatic broadening of the Shh-expressing domain at 24 hours of treatment. These results suggest that the formation of palatal rugae is regulated by a Turing-type activator-inhibitor reaction-diffusion mechanism, with FGF signaling acting as an activator whereas SHH signaling acts as an inhibitor to regulate the periodic ruga formation (Economou et al., 2012) (Fig. 2H). Interestingly, deletion of β-Catenin in Shh-expressing palatal epithelial cells resulted in complete loss of palatal rugae formation accompanied by down-regulation of Shh and Ptch1 expression (Lin et al., 2011). On the other hand, mice lacking Sostdc1, a secreted antagonist of BMP and WNT signaling (Ahn et al., 2010; Itasaki et al., 2003; Laurikkala et al., 2003), and mice lacking Lrp4, an antagonist of Lrp6-mediated WNT signaling (Li et al., 2010), displayed disorganized palatal rugae (Kawasaki et al., 2018). Together, these data indicate with both FGF and WNT signaling pathways interact with SHH signaling to regulate palatal rugae formation and patterning.
5. SHH function in mandible development and jaw patterning
5.1. SHH signaling in regulation of mandibular patterning
The mandible is a complex structure, composed of bone, cartilage, teeth, muscle, bone marrow, nerves, vascular and connective tissues. Most of the mandibular tissues arise from the paired first pharyngeal arches in the early embryo. At around E9.5 in mouse, the mandibular processes become morphologically distinguishable from the more dorsally located maxillary processes. During early mandible development, Shh is expressed in the oral epithelium and anterior pharyngeal endoderm whereas Ptch1 is expressed in both oral epithelium and the underlying mandibular mesenchyme, indicating that SHH signals to both cell types (Fig. 3A and B) (Jeong et al., 2004; Xu et al., 2019). Tissue-specific inactivation of Shh in the oropharyngeal epithelium (Shhfx/-;Nkx2.5Cre) or tissue-specific inactivation of Smo in either the pre-migratory CNCCs (Smon/c;Wnt1Cre) or CNCC-derived distal mandibular arch mesenchyme (Smon/c;Hand2Cre) in mouse embryos resulted in increased apoptosis and decreased cell proliferation in the distal mandibular mesenchyme by E10.5, resulting in micrognathia and aglossia at birth (Billmyre and Klingensmith, 2015; Jeong et al., 2004; Xu et al., 2019).
Fig. 3.
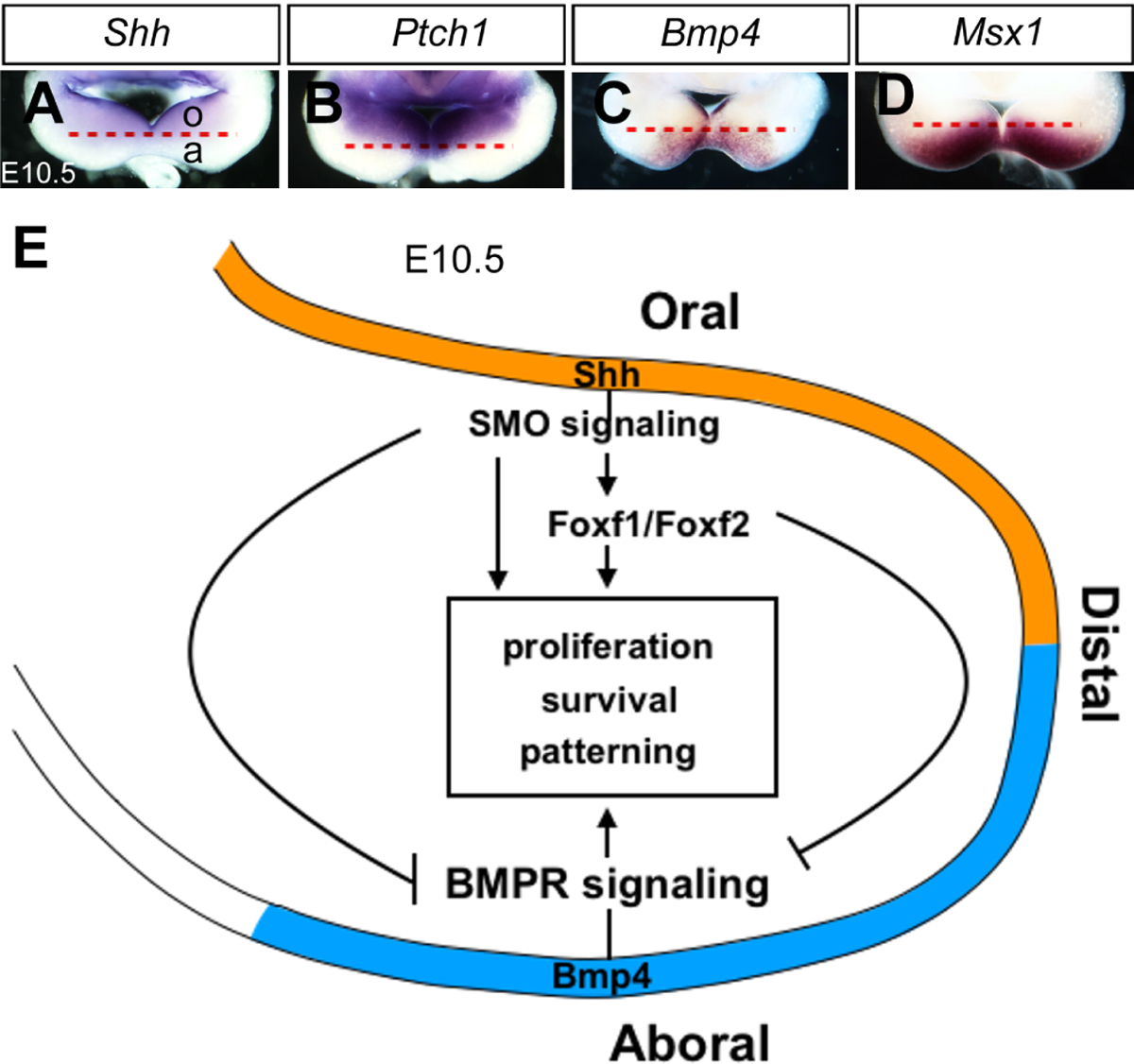
SHH signaling and mandible development. (A-D) Whole mount rostral views of E10.5 embryonic mouse mandibular arches showing the patterns of expression of Shh (A), Ptch1 (B), Bmp4 (C) and Msx1 (D) mRNAs. The mRNA signals are shown in purple/brown color. o, oral, and a, aboral domains of develop mandibular arch. (E) Schematic diagram depicting the crucial roles of SHH signaling in regulating mandibular arch growth and oral-aboral patterning through antagonizing the BMP signaling pathway at E10.5. Shh mRNA is expressed in the oropharyngeal side of the mandibular epithelium at E10.5. Foxf1 and Foxf2, known downstream targets of SHH signaling, are expressed in the oropharyngeal side of the mandibular arch mesenchyme cells. BMP4 signalling activated in an aboral-to-oral gradient in the mandibular arch mesenchyme. SHH signaling antagonize BMP4 signaling and regulate the growth, cell survive and patterning of developing mandibular arch.
Through unbiased single-cell RNA-seq analysis followed by in situ hybridization analysis, we found that SHH and BMP4 signalling pathways are activated in a complementary pattern along the oral and aboral axis in the developing mandibular arch. Shh mRNA is expressed in the oropharyngeal side of the mandibular epithelium at E10.5. Several down-stream target genes of SHH signalling, including Foxf1, Foxf2 and Ptch1, are expressed in the oropharyngeal side of the mandibular arch mesenchyme cells. BMP4 signalling target genes, including Msx1, Msx2 and Alx4, are expressed in an aboral-to-oral gradient in the mandibular arch mesenchyme (Fig. 3A–D). Tissue specific inactivation of Smo in CNCC-derived mesenchyme in the distal mandibular mesenchyme (Smoc/c;Hand2Cre) caused expansion of BMP4 signalling, indicated by phosphorylated Smad1/5/9, to throughout the oral-aboral axis of the distal mandibular mesenchyme. These mutant mice displayed duplication of the dentary bone in the oral side of the mandible at the expense of tongue formation. Furthermore, tissue specific inactivation of Foxf1 and Foxf2 in the CCNC-derived mesenchyme (Foxf1c/c;Foxf2c/c;Wnt1Cre) resulted in similar mandibular defects as the Smoc/c;Hand2Cre mutants, including orally expanded BMP signalling, ectopic bone formation at the oral side of mandible, and oral tongue agenesis, indicating that Foxf1 and Foxf2 are key mediators of SHH signalling in regulating the oral-aboral patterning of mandible (Fig. 3E) (Xu et al., 2019).
5.2. SHH signaling in regulation of cell survival in the developing mandibular arch
We have shown that disruption of SHH signalling in the mandibular mesenchyme (Smoc/c;Hand2-Cre) resulted in increased apoptosis associated with expanded BMP signalling in the distal mandible and that genetically deleting one copy of the Bmp4 gene in the Smoc/c;Hand2-Cre mutants significantly reduced apoptosis in the mandibular mesenchyme (Xu et al., 2019). Billmyre and Klingensmith (2015) showed that inhibition of p53 reduced mandibular mesenchyme apoptosis in the Shhfx/-;Nkx2.5Cre embryos (Billmyre and Klingensmith, 2015). It has also been shown that ectopic activation of BMPR1A signalling in CNCCs caused a significant up-regulation of p53 and p53-mediated apoptosis (Hayano et al., 2015). Together, these results suggest that SHH-SMO signalling regulates mandibular mesenchyme cell survival through inhibition of BMP signalling induced p53-mediated cell apoptosis.
It has been shown that deletion of Islet1 (Isl1), which encodes an LIM homeodomaincontaining transcription factor, in the Shh-expressing oropharyngeal epithelial cells (ShhCre;Isl1fl/fl) resulted in distal mandible truncation (Li et al., 2017), a similar phenotype as in the Smoc/c;Hand2-Cre mice (Xu et al., 2019). The ShhCre;Isl1fl/fl embryos exhibited increased apoptosis and decreased cell proliferation in the distal mandibular arch mesenchyme at E10.5 and E11.5, respectively, with concomitant downregulation of expression of Shh, Ptch1, Gli1 and of a group of Fox genes (Li et al., 2017). Interestingly, expression of several WNT antagonists was upregulated in the distal mandibular arch in the ShhCre;Isl1fl/fl embryos. Cre/loxP-induced reactivation of β-Catenin in the Shh-expressing cells in the Isl1 mutant embryos restored Shh expression, resulting in restored cell survival and partially rescued distal mandible in the ShhCre;Isl1fl/fl;Ctnnbex(3)fl/+ embryos (Li et al., 2017). Together, these data indicate that the Isl1 transcription factor regulates WNT signaling to control expression of Shh in the oropharyngeal epithelium and SHH signaling interacts with BMP4 signaling to regulate survival and patterning of the CNCC-derived mandibular mesenchyme.
5.3. Role of the primary cilia in SHH signaling regulation of mandibular development
Genetic inactivation of several genes encoding components of the primary cilia in CNCCs also caused mandibular defects (Ashe et al., 2012; Brugmann et al., 2010; Eguether et al., 2014; Keady et al., 2012; Kitamura et al., 2020; Millington et al., 2017; Schock et al., 2017). Mice lacking Ift27 and mice with tissue specific inactivation of Kif3a (Kif3afl/fl;Wnt1Cre) exhibited micrognathia and aglossia/microglossia (Brugmann et al., 2010; Eguether et al., 2014; Millington et al., 2017; Schock et al., 2017). Deletion of Ift88 in CNCCs (Ift88fl/fl;Wnt1Cre) resulted in micrognathia, oral tongue agenesis and formation of ectopic mandibular bones, similar as in Smofl/fl;Wnt1Cre mutant mice (Kitamura et al., 2020). Notably, in contrast to the gain of SHH signaling activity due to loss of Gli2R/Gli3R-mediated repression in the developing frontonasal primordia in various ciliopathic mutant mouse lines (Chang et al., 2016), as described earlier, the mandibular defects in these ciliopathic mutant mouse lines primarily reflect a requirement for the primary cilia mediated Smo and Gli activation, further highlighting the context-dependent effects of ciliary function in the regulation of SHH signaling.
5.4. SHH signaling and Meckel’s cartilage formation
SHH signalling has also been implicated to play an important role in Meckel’s cartilage differentiation. During mandible development, a subset of the CNCC-derived mandibular mesenchyme cells condense and then differentiate into chondrocytes to form Meckel’s cartilage, a rod-like structure providing shape to the lower jaw (Yuan and Chai, 2019). Shhfx/-;Nkx2.5Cre mutant mice displayed complete loss of Meckel’s cartilage (Billmyre and Klingensmith, 2015). Cyclopamine treated mouse mandibular explants exhibited a stage-dependent inhibition of Meckel’s cartilage differentiation, and this phenotype could be rescued by exogenous Fgf8 protein treatment (Melnick et al., 2005). In chick embryos, overexpression of Shh in the non-oral ectoderm resulted in the formation of ectopic cartilage associated with expansion of Fgf8 expression (Haworth et al., 2007). Moreover, grafting Shh-expressing cells into the first pharyngeal arch of early chick embryos induced mirror-image duplication of the mandible, including the Meckel’s cartilage (Brito et al., 2008). However, Smoc/c;Wnt1Cre and Smoc/c;Hand2Cre mutant mice developed distally truncated Meckel’s cartilage, likely due to aberrant apoptosis of the distal mandibular mesenchyme prior to Meckel’s cartilage formation (Jeong et al., 2004; Xu et al., 2019). These results suggest that SHH signalling regulates development of Meckel’s cartilage through an indirect mechanism, possibly through regulation of Fgf8 expression in the mandibular arch epithelium.
6. SHH function in tongue organogenesis
The mammalian tongue is composed of mesoderm derived muscles, CNCCs-derived connective tissues, and non-keratinized epithelium. The anterior two-thirds of the tongue arises from the mandibular arch whereas the posterior third of the tongue is derived from the 3rd and 4th pharyngeal arches. At the initiation stage of tongue development, around E10.5 in mouse, the tongue primordium consists of CNCC-derived mesenchymal cells covered by a layer of oral epithelium. By E11.5, myogenic precursor cells from the occipital somites had migrated into the tongue primordium and subsequently give rise to the intrinsic muscular structures as the CNCC-derived mesenchymal cells develop into connective tissues including tendons and ligaments (Noden and Francis-West, 2006; Parada and Chai, 2015; Parada et al., 2012).
Several different mutant mouse models with disruption of SHH signaling during early mandibular development exhibited tongue agenesis (Billmyre and Klingensmith, 2015; Jeong et al., 2004; Xu et al., 2019). In the Smon/c;Wnt1Cre and Smoc/c;Hand2Cre embryos, Myf5-expressing myogenic progenitor cells failed to migrate into the tongue primordium (Jeong et al., 2004; Xu et al., 2019). The ShhCre;Isl1fl/fl mice exhibited tongue agenesis associated with loss of Shh expression in the mandibular arch epithelium and failure of migration of myogenic precursor cells into the tongue primordium (Zhang et al., 2022). Transgenic expression of Ihh in the mandibular arch epithelium was able to partly rescue tongue formation in the ShhCre;Isl1fl/fl mice (Zhang et al., 2022). Tissue-specific inactivation of both Foxf1 and Foxf2 in CNCCs resulted in oral tongue agenesis and severe muscular defects in the remaining rudimentary tongue (Xu et al., 2019). The Myf5-expressing myogenic precursor cells migrated into the mandibular arch by E10.5 but failed to further migrate anteriorly into the tongue primordium in the Foxf1c/cFoxf2c/cWnt1-Cre embryos (Xu et al., 2019; Xu et al., 2022). We recently showed that Hedgehog-Foxf1/Foxf2 signaling in the CNCC derived tongue mesenchyme is required for activation of expression of HGF, a growth factor required for migration of myoblasts into the tongue primordium (Xu et al., 2022). Zhang et al. (2022) found that the ShhCre;Isl1fl/fl embryos exhibited decreased Shh expression and aberrant migration of Cxcl12+ mesenchyme cells in the tongue primordium (Zhang et al., 2022). Further study suggested that SHH signaling acts upstream of WNT5a signaling to attract Cxcl12+ tongue mesenchymal cells to help guide tongue myoblast migration (Zhang et al., 2022). Altogether, these results indicate that SHH signaling regulates the expression of key chemotactic signals in the CNCC-derived tongue mesenchyme to direct myogenic precursor migration and initiation of tongue myogenesis.
Following tongue initiation and myoblast arrival in the tongue primordium, expression of Shh is maintained in the epithelium of the primordial tongue and later becomes restricted to the fungiform papillae of the anterior tongue by E12.5 (Jung et al., 1999). Temporally induced inactivation of Shh after E10.5 in mouse embryos did not affect initial myoblast migration into the tongue primordium but caused disruption and disorganization of the intrinsic muscles, suggesting that SHH signaling continues to regulate tongue myogenesis and tongue muscle organization after initial tongue formation (Okuhara et al., 2019). We recently reported that the Foxf1c/c;Foxf2c/+;Wnt1Cre and Foxf1c/+;Foxf2c/c;Wnt1Cre mutant mouse embryos exhibit disrupted lingual septum tendon formation, leading to an absence or disorganization of specific intrinsic muscles in the tongue (Xu et al., 2022). ChIP-seq analysis identified Hgf, Tgfb2 and Tgfb3 among others, as candidate direct target genes regulated by Foxf1 and Foxf2 in the developing tongue (Xu et al., 2022). Tgfb2 and Tgfb3 expression, and the levels of pSmad2/3, were significantly reduced in the lingual tendon progenitor cells in mouse embryos with temporally induced inactivation of Shh gene at E10.5 as well as in the Foxf1c/c;Foxf2c/+;Wnt1Cre and Foxf1c/+;Foxf2c/c;Wnt1Cre mutant embryos (Okuhara et al., 2019; Xu et al., 2022). Together, these studies identified a SHH/SMO-Foxf1/Foxf2-TGFβ molecular cascade regulating lingual tendon formation and indirectly regulating tongue muscular organization.
7. SHH function in taste bud formation and homeostasis
In addition to its great ability to move in all directions to perform essential coordinated functions in feeding, speech, and vocalization, the tongue is a major sensory organ. The dorsal mucosal surface of the tongue contains numerous papillae and taste buds, which sense and transduce five basic tastes (sweet, bitter, salt, sour, and umami) to signal palatability, nutritional value, and/or danger of substances in the oral cavity (Chaudhari and Roper, 2010; Golden et al., 2021). There are three types of taste papillae in mammals on the tongue surface: the fungiform papillae, the foliate papillae, and the circumvallate papilla. Taste buds are modified lingual epithelial cells that first form during embryogenesis from focal epithelial thickenings, called taste placodes, and are continually renewed postnatally from mitotically active basal keratinocytes (Barlow, 2022; Castillo et al., 2014; Golden et al., 2021). Multiple studies have demonstrated that SHH signaling is a principal and essential regulator of taste bud formation and patterning as well as taste cell differentiation and homeostasis (Barlow, 2022; Golden et al., 2021; Mistretta and Kumari, 2019). Shh is broadly expressed in the embryonic tongue epithelium prior to taste placode formation but its expression becomes restricted to the placode cells and subsequently restricted to the taste-fated cells within growing taste papillae (Bitgood and McMahon, 1995; Hall et al., 1999; Jung et al., 1999). Lineage tracing studies indicate that Shh-expressing embryonic taste placode cells differentiate into the first taste receptor cells around birth but do not contribute to other cells in the taste papillae (Thirumangalathu et al., 2009). Bead implantation studies in embryonic tongue explants showed that exogenous SHH repressed taste placode development, whereas pharmacological inhibition or genetic inactivation of SHH signaling resulted in more and enlarged taste buds (Hall et al., 2003; Iwatsuki et al., 2007; Mistretta et al., 2003). Further investigation revealed that SHH signaling restricts taste placode fate by antagonizing retinoic acid (RA) signaling through maintaining the expression of RA degrading enzymes, Cyp26a1 and Cyp26c1 (El Shahawy et al., 2017). Furthermore, mouse genetic studies showed that taste placode formation requires canonical WNT signaling within the embryonic tongue epithelium (Iwatsuki et al., 2007). In mouse embryonic tongue explants, activation of canonical WNT signaling promoted focal expression of Shh (Iwatsuki et al., 2007). On the other hand, exogenous SHH inhibited WNT signaling activity whereas blocking SHH signaling caused expanded WNT target gene expression in the taste primordia (Iwatsuki et al., 2007). SHH and non-canonical WNT signaling pathways have been involved in the formation of circumvallate papilla (Kim et al., 2009). In an in vitro culturing system, Kim et al., showed that SHH signaling regulates cell proliferation for taste bud and von Ebner’s gland formation, while WNT11/Rock1 signaling modulates cytoskeleton formation for proper structure formation of circumvallate papilla (Kim et al., 2009). Together, these results indicate that the interplay of SHH, RA and WNT signaling pathways control the formation and patterning of lingual taste buds and papillae (Barlow, 2022).
Whereas SHH signaling functions as a repressor of taste bud formation in the embryonic tongue, recent studies show that SHH signaling is required for taste cell renewal and promotes taste receptor cell differentiation in adults (Castillo et al., 2014; Castillo-Azofeifa et al., 2017; Kumari et al., 2015). Genetic lineage tracing studies showed that the embryonic Shh-expressing placode-derived taste receptor cells are lost from taste buds within a few months postnatally and that adult taste progenitors arise from Krt14+ epithelial stem cells after birth (Okubo et al., 2009; Thirumangalathu et al., 2009). Pharmacological inhibition or conditional epithelium-specific inactivation of Smo in adult mice resulted in rapid loss of taste buds due to disruption of taste cell renewal but had no discernable effects on nontaste filiform papillae (Castillo-Azofeifa et al., 2017; Kumari et al., 2015; Kumari et al., 2017), indicating that Smo-mediated Hedgehog signaling in the lingual epithelial cells is selectively required for taste cell differentiation. However, whereas Shh expression is detected in postmitotic taste precursor cells derived from K5+ resident epithelial stem cells in the lingual epithelium (Golden et al., 2021; Miura et al., 2006; Miura et al., 2001; Miura et al., 2014), conditional deletion of Shh in the K5+ lineage resulted in reduction in Shh expression in the taste precursor cells but did not affect taste bud renewal (Castillo-Azofeifa et al., 2017). Castillo-Azofeifa et al. (2017) found that sensory neurons innervating the taste buds express Shh and that SHH supplied by the taste nerves and local taste epithelium act in concert to support continued taste bud differentiation (Castillo-Azofeifa et al., 2017). Whereas recent studies have identified Sox2 and Foxa2 as transcription factors acting downstream of SHH signaling in taste bud renewal (Castillo-Azofeifa et al., 2018; Golden et al., 2021), the exact molecular mechanisms mediating SHH signaling function in taste bud renewal remain to be elucidated.
8. SHH signaling in tooth development
Tooth development begins as a thickening of the oral epithelium, called dental lamina, at around E11 in mice. The dental lamina proliferates and protrudes into the underlying CNCC-derived mesenchyme and induces the mesenchyme to condense around the epithelial bud from E12 to E13. At around E13.5, cells at the tip of the epithelial tooth buds form an epithelial signaling center, termed the primary enamel knot, which secretes a number of growth factors including Shh (Dassule et al., 2000; Seppala et al., 2017). The interactions between tooth bud epithelium and the surrounding mesenchyme drive further tooth morphogenesis through the “cap” and “bell” stages. As development proceeds, the epithelial cells in contact with the dental mesenchyme differentiate into ameloblasts and their adjacent mesenchymal cells differentiate into odontoblasts, and further develop into mature tooth (Cobourne and Sharpe, 2005; Lan et al., 2014; Thesleff and Sharpe, 1997; Tucker and Sharpe, 2004).
During initiation stage of tooth development, the dental epithelium goes through a process of thickening (to form vertically- oriented cells), stratification (to form multiple cells layers), and invagination to become an epithelial bud. Shh is one of the earliest markers of the dental epithelial cells (Cobourne et al., 2001; Keranen et al., 1999). The expression domain of Shh in the dental epithelium is complementary with the expression domain of Wnt7b in the oral epithelium (Sarkar et al., 2000). Transient ectopic expression of Wnt7b in the dental epithelium in mandibular arch explants led to down-regulation of Shh and Ptch1 expression, and arrest of tooth development at the initiation stage (Sarkar et al., 2000). In the Wnt7b-infected mandibular arch explants, treatment with SHH-soaked beads restored tooth development (Sarkar et al., 2000). Furthermore, blocking SHH signaling using 5E1 anti-Shh antibody from E10.5 caused largely reduced dental epithelial thickening and arrested tooth development in mouse (Cobourne et al., 2001). These results led to a hypothesis that Wnt7b interacts with Shh to maintain the boundary of oral-dental epithelium during tooth initiation (Sarkar et al., 2000). In vivo mouse genetic studies further revealed the importance of SHH signaling in tooth development. Enhanced level of Shh in basal epithelium driven by a Keratin-14 promoter (K14-Shh) results in tooth development arrest at the bud stage, secondary to a lack of cell proliferation in this region (Cobourne et al., 2009). The Gli2/Gli3 double homozygous (Gli2−/−;Gli3−/−) mutants did not develop any normal teeth indicating an essential role of SHH signaling pathway during tooth development (Hardcastle et al., 1998). Bead implantation experiments showed that exogenous Shh treatment stimulates, whereas blocking of SHH signaling by cyclopamine inhibits, dental epithelial invagination (Li et al., 2016). Interestingly, inhibition of SHH signaling by cyclopamine starting from E11.5 did not prevent stratification of the dental placode but resulted in shallower and wider tooth buds (Li et al., 2016). In contrast, inhibition of FGF signaling in the same context caused failure of stratification in the dental placodal area. On the other hand, FGF10 treatment at E11.5 induced a prominent thickening of the epithelium. Together, these results suggest that SHH signaling drives cell rearrangement in the prospective dental epithelial tissue for tooth bud formation after FGF signaling induced stratification of the dental placode (Li et al., 2016).
Following tooth bud formation, Shh expression becomes restricted to the tip of the epithelial bud at around E13.5 and in the primary enamel knot at the cap stage (Dassule et al., 2000). At the bell stage, Shh mRNA is expressed in the stellate reticulum, stratum intermedium, pre-ameloblasts, and differentiating ameloblasts. Shh protein, Ptch1 and Gli1 can be observed in a broader domain in the dental epithelium and the dental papilla, indicating that SHH signaling may exert a long-range activity (Gritli-Linde et al., 2002; Gritli-Linde et al., 2001). In mice, tissue specific inactivation of Shh in dental epithelium (Shhc/n;K14-Cre) shortly after ingrowth of the tooth bud resulted in a cap stage tooth rudiment in which the morphology is severely disrupted (Dassule et al., 2000). Although the enamel knot still formed in these mutant mice, the overall size of the tooth is reduced, and the lingual epithelial invagination and the dental cord are absent (Dassule et al., 2000). This study revealed that Shh regulates growth and shape of the tooth. Further study found Ptch1 is more strongly expressed in the lingual side of the cap stage tooth germ (Dassule et al., 2000). Epithelial specific inactivation of Smo in Smoc/n;K14-Cre mutant mice resulted in altered proliferation, differentiation, and polarization of ameloblasts (Gritli-Linde et al., 2002), indicating that SHH signaling regulates the growth and polarization of the dental epithelium. On the other hand, the Smoc/n; Wnt1-Cre mice lack lower incisors and developed only one central upper incisor and malformed small molar tooth germs (Jeong et al., 2004), indicating that SHH signaling plays primary roles in both epithelial and mesenchymal cells to control tooth morphogenesis.
SHH signaling is also plays crucial roles in tooth root formation in postnatal mice. Shh is expressed in Hertwig’s epithelial root sheath (HERS), a bilayered tissue formed from the inner and outer enamel epithelia, during early root morphogenesis (Liu et al., 2015; Nakatomi et al., 2006). Ptch2, a receptor of SHH signaling was also found in HERS (Nakatomi et al., 2006), while Gli1 and Ptch1 were detected in the proliferative dental mesenchyme in addition to the HERS (Liu et al., 2015; Nakatomi et al., 2006). Inhibition of SHH signaling with HH inhibitor or constitutive activation of SHH signaling genetically (Gli1-creERT2;R26SmoM2fl/fl) caused decreased proliferation and shorter roots (Liu et al., 2015). Nfic−/− mice exhibited defect in root formation accompanied by increase SHH signaling (Liu et al., 2015; Steele-Perkins et al., 2003). Treatment with HH inhibitors partially restored root development in Nfic−/− mice (Liu et al., 2015). Interestingly, Shh is also shown to act upstream of Nfic and downstream of BMP/TGFβ-SMAD signaling in HERS controlling tooth root formation (Huang et al., 2010). Moreover, the mesenchymal dysplasia (mes) mice, which express a mutant Ptch1 protein with an abnormal C-terminus, have short tooth roots and reduced proliferation around the HERS (Nakatomi et al., 2006). Together, these studies highlight an important role for SHH signalling in regulating tooth root formation.
In mice, stem cells are maintained in the incisor tooth germs throughout life, resulting in continuous growth of the incisors (Feng et al., 2011; Harada et al., 1999). SHH signaling plays important roles in regulating the maintenance of both epithelial and mesenchymal stem cells in the mouse incisor tooth germs. Previous studies used Gli1, which is a transcription factor and down-stream target of SHH signaling pathway, as a marker of epithelial and mesenchymal stem cells in postnatal mouse incisors (Seidel et al., 2010; Zhao et al., 2014). The epithelial stem niche is located in the cervical loop at the proximal end at the labial side of the incisor (Seidel et al., 2010). Inhibition of SHH signaling pathway disrupts generation of ameloblasts from stem cells, indicating that SHH signaling is essential for generation of ameloblasts (Seidel et al., 2010). Gli1+ cells were also found in the developing molar but decreased significantly after birth (Li et al., 2015). BMP/SMAD4 signaling is implicated in inhibiting the activity of SHH signaling and controlling epithelial stem cell maintenance during molar development (Li et al., 2015). For the incisor mesenchymal stem cell niche, Shh secreted by sensory nerves supports the maintenance of the Gli1-positive stem cells surrounding the arterioles near the cervical loop region and regulates their differentiation into odontoblast and other incisor mesenchyme cells (Zhao et al., 2014).
9. SHH signaling and fetal alcohol exposure
Interactions between environmental stresses and SHH signaling have been shown to contribute to the etiology of craniofacial defects in humans and mice. Alcohol is a common teratogen that could induce a variety of birth defects from exposure during pregnancy (Jones and Smith, 1973). Many of the fetal alcohol syndrome defects, including HPE and other craniofacial defects, have been similarly observed in animal models with dysregulation of SHH signaling. Previous study in chicken showed that embryonic exposure to ethanol during early CNCC migration resulted in increased CNCC apoptosis and decrease in size of the frontonasal mass (Ahlgren et al., 2002). The craniofacial growth defects can be rescued by application of exogenous Shh ligand (Ahlgren et al., 2002). Using zebrafish models, Li et al. showed that alcohol exposure during gastrulation caused inhibition of cholesterol modification of Shh, impaired Hedgehog signal transduction, and a dose-dependent spectrum of defects including HPE (Li et al., 2007). Adding cholesterol rescued the loss of SHH signaling activity and alleviated the developmental defects in the alcohol-treated fish (Li et al., 2007). These studies suggest a role of dysregulation of SHH signaling in mediating the fetal alcohol induced birth defects.
Haploinsufficiency of several SHH pathway components cause structural birth defects in humans. The patients often display incomplete phenotypic penetrance and highly variable expressivity (Solomon et al., 2012), suggesting roles of gene-gene and/or environment-gene interactions. Studies have shown that alcohol exposure synergize with several mutations in SHH pathway components, resulting in significantly enhanced penetrance and severity of birth defects. In mice on the 129S6 background, inactivation of Cdon (Cdon−/−) or in utero ethanol exposure alone resulted in little or no HPE related phenotype, but together they caused inhibition of SHH signaling and a broad spectrum of HPE phenotypes (Hong and Krauss, 2012). Deletion of one Ptch1 allele rescued ethanol induced HPE phenotype in Cdon−/− mice, confirming that the alcohol induced HPE is associated with reduced SHH signaling activity (Hong and Krauss, 2013). Recently, the interaction between Cdon loss-of-function mutation and ethanol exposure was shown to regulate the activity of SHH signaling through a Nodal signaling-associated molecular mechanism (Hong et al., 2020). In wildtype C57BL/6J mice, ethanol exposure in utero alone induced a low penetrance of HPE. However, ethanol exposure significantly enhanced the HPE phenotype in Shh+/− and Gli2+/− mice, respectively, in the C57BL/6J background (Kietzman et al., 2014). Together, these results highlight the role of environmental factors interacting with genetic mutations disrupting SHH signaling in causing the broad spectrums of HPE phenotypes in human patients.
10. Concluding remarks and Perspective
Previous research, especially studies using animal models, revealed essential functions of SHH signaling and interactions between SHH and other signaling pathways during craniofacial development. These studies have provided important insights into the mechanisms underlying SHH related diseases. However, several gaps remain in our knowledge about the molecular mechanisms involving SHH signaling in craniofacial development and patterning. 1) From the onset of facial development, dynamic and tissue specific activation of SHH and other signaling pathways, including members of the FGF, BMP and WNT families, are repeatedly deployed to drive tissue morphogenesis. The molecular mechanisms controlling the specific and dynamic expression patterns of these signaling molecules remains largely unknown. The tissue specific and precise transduction of SHH signaling activity also needs to be further dissected. For example, previous studies have shown that deleting two or more Hedgehog co-receptors caused more severe phenotype than single gene inactivation in certain tissues. More detailed and tissue-specific studies of the co-receptors and modifiers of SHH will advance our knowledge about the precise regulation of SHH signaling activity. 2) During several craniofacial developmental processes, SHH signaling plays key roles in maintaining cell survival and regulating tissue growth and patterning. Previous studies have revealed several important down-stream mediators of SHH signaling at different steps of craniofacial morphogenesis. For example, the Fox family transcription factors are known to be important down-stream mediators. Studies on tissue-specific mutant animal models of Fox genes have provided valuable information between SHH signaling mis-regulation and the related phenotypes. However, much remain to be understood about how disruption of SHH signaling lead to the various phenotypes in mutant animal models and human patients. Future studies involving genome wide screening down-stream targets of SHH signaling at multiple steps of craniofacial development and tissue morphogenesis, including comprehensive single cell genomic/transcriptomic analyses, followed by functional studies of the candidate mediators and effectors of SHH signaling will help fill the gap. 3) Dynamic and highly coordinated interactions between SHH and other signaling pathways have been observed throughout craniofacial development. The positive and negative cross-regulations between SHH and other signaling pathways, involving multiple cell types including CNCCs, orofacial epithelium, and mesoderm derived tissues such as myogenic progenitors, form a complex signaling network driving the developmental processes forming different craniofacial tissues. The mechanisms underlying these cross-regulations are still not well understood. Nowadays cutting-edge technologies such as single cell genomic and proteomic approaches are starting to be widely used in biological and medical studies. Combining these technologies with CRISPR genome edited animal models will help us achieve a better understanding of molecular and cellular mechanisms underlying the precise control of SHH signaling activity, and the gene regulatory networks involving SHH signaling in the multiple stages of craniofacial morphogenesis. Together, these efforts will provide the foundation for new development and improvement of strategies for treatment and prevention of craniofacial birth defects.
Supplementary Material
Acknowledgements
We apologize to colleagues whose relevant research publications were not discussed in this article due to space limitations. The original research on SHH signaling in craniofacial development in our lab was supported by the National Institutes of Health National Institute of Dental and Craniofacial Research (NIH/NIDCR) [grant numbers DE013681, DE027046, and DE029417], and by Shriners Hospitals for Children [grant number 85900].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramyan J, 2019. Hedgehog Signaling and Embryonic Craniofacial Disorders. J Dev Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren SC, Bronner-Fraser M, 1999. Inhibition of sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Curr Biol 9, 1304–1314. [DOI] [PubMed] [Google Scholar]
- Ahlgren SC, Thakur V, Bronner-Fraser M, 2002. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc Natl Acad Sci U S A 99, 10476–10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y, Sanderson BW, Klein OD, Krumlauf R, 2010. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development 137, 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP, 2011. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell 20, 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Tenzen T, McMahon AP, 2007. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev 21, 1244–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson HC, Kratz L, Kelley R, 2002. Desmosterolosis presenting with multiple congenital anomalies and profound developmental delay. Am J Med Genet 113, 315–319. [DOI] [PubMed] [Google Scholar]
- Ashe A, Butterfield NC, Town L, Courtney AD, Cooper AN, Ferguson C, Barry R, Olsson F, Liem KF Jr., Parton RG, Wainwright BJ, Anderson KV, Whitelaw E, Wicking C, 2012. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Hum Mol Genet 21, 1808–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae GU, Domene S, Roessler E, Schachter K, Kang JS, Muenke M, Krauss RS, 2011. Mutations in CDON, encoding a hedgehog receptor, result in holoprosencephaly and defective interactions with other hedgehog receptors. Am J Hum Genet 89, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL, 2002. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761. [DOI] [PubMed] [Google Scholar]
- Bangs F, Anderson KV, 2017. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AJ, Bogardi JP, Ladher R, Francis-West PH, 1999. Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev Dyn 214, 291–302. [DOI] [PubMed] [Google Scholar]
- Barlow LA, 2022. The sense of taste: Development, regeneration, and dysfunction. WIREs Mech Dis 14, e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW, 1996. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet 14, 353–356. [DOI] [PubMed] [Google Scholar]
- Billmyre KK, Klingensmith J, 2015. Sonic hedgehog from pharyngeal arch 1 epithelium is necessary for early mandibular arch cell survival and later cartilage condensation differentiation. Dev Dyn 244, 564–576. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP, 1995. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol 172, 126–138. [DOI] [PubMed] [Google Scholar]
- Blassberg R, Macrae JI, Briscoe J, Jacob J, 2016. Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome. Hum Mol Genet 25, 693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Mazot P, Soriano P, 2016. Genetic insights into the mechanisms of Fgf signaling. Genes Dev 30, 751–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito JM, Teillet MA, Le Douarin NM, 2008. Induction of mirror-image supernumerary jaws in chicken mandibular mesenchyme by Sonic Hedgehog-producing cells. Development 135, 2311–2319. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Allen NC, James AW, Mekonnen Z, Madan E, Helms JA, 2010. A primary cilia-dependent etiology for midline facial disorders. Hum Mol Genet 19, 1577–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Corso G, Rossi M, Ferrari P, Balli F, Rivasi F, Annunziata I, Ballabio A, Russo AD, Andria G, Parenti G, 2002. Lathosterolosis, a novel multiplemalformation/mental retardation syndrome due to deficiency of 3beta-hydroxysteroiddelta5-desaturase. Am J Hum Genet 71, 952–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buglino JA, Resh MD, 2008. Hhat is a palmitoylacyltransferase with specificity for Npalmitoylation of Sonic Hedgehog. J Biol Chem 283, 22076–22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K, 1999. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99, 803–815. [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R, 2012. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, Sansom MSP, Newstead S, Rohatgi R, Siebold C, 2016. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D, Stone DM, Brush J, Ryan A, Armanini M, Frantz G, Rosenthal A, de Sauvage FJ, 1998. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc Natl Acad Sci U S A 95, 13630–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Garcia-Garcia MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, Alcorn HL, Anderson KV, 2002. Mouse Dispatched homolog1 is required for longrange, but not juxtacrine, Hh signaling. Curr Biol 12, 1628–1632. [DOI] [PubMed] [Google Scholar]
- Castillo D, Seidel K, Salcedo E, Ahn C, de Sauvage FJ, Klein OD, Barlow LA, 2014. Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development 141, 2993–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Azofeifa D, Losacco JT, Salcedo E, Golden EJ, Finger TE, Barlow LA, 2017. Sonic hedgehog from both nerves and epithelium is a key trophic factor for taste bud maintenance. Development 144, 3054–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Azofeifa D, Seidel K, Gross L, Golden EJ, Jacquez B, Klein OD, Barlow LA, 2018. SOX2 regulation by hedgehog signaling controls adult lingual epithelium homeostasis. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Maxson RE Jr., 2006. Recent advances in craniofacial morphogenesis. Dev Dyn 235, 2353–2375. [DOI] [PubMed] [Google Scholar]
- Chang CF, Chang YT, Millington G, Brugmann SA, 2016. Craniofacial Ciliopathies Reveal Specific Requirements for GLI Proteins during Development of the Facial Midline. PLoS Genet 12, e1006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CF, Schock EN, O’Hare EA, Dodgson J, Cheng HH, Muir WM, Edelmann RE, Delany ME, Brugmann SA, 2014. The cellular and molecular etiology of the craniofacial defects in the avian ciliopathic mutant talpid2. Development 141, 3003–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N, Roper SD, 2010. The cell biology of taste. J Cell Biol 190, 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA, 2002. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 16, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT, 2004. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev 18, 641–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT, 2009. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev 23, 1910–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G, 1996. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563. [DOI] [PubMed] [Google Scholar]
- Cheung HO, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, Law KK, Briscoe J, Hui CC, 2009. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal 2, ra29. [DOI] [PubMed] [Google Scholar]
- Chinipardaz Z, Liu M, Graves DT, Yang S, 2022. Role of Primary Cilia in Bone and Cartilage. J Dent Res 101, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, Ma D, Stal S, Mulliken JB, Hecht JT, 2008. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet 17, 2212–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobourne MT, Hardcastle Z, Sharpe PT, 2001. Sonic hedgehog regulates epithelial proliferation and cell survival in the developing tooth germ. J Dent Res 80, 1974–1979. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Sharpe PT, 2003. Tooth and jaw: molecular mechanisms of patterning in the first branchial arch. Arch Oral Biol 48, 1–14. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Sharpe PT, 2005. Sonic hedgehog signaling and the developing tooth. Curr Top Dev Biol 65, 255–287. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Xavier GM, Depew M, Hagan L, Sealby J, Webster Z, Sharpe PT, 2009. Sonic hedgehog signalling inhibits palatogenesis and arrests tooth development in a mouse model of the nevoid basal cell carcinoma syndrome. Dev Biol 331, 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Krauss RS, 2003. Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Curr Biol 13, 411–415. [DOI] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE, 2005. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 132, 4407–4417. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Scott MP, 2006. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci U S A 103, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croen LA, Shaw GM, Lammer EJ, 1996. Holoprosencephaly: epidemiologic and clinical characteristics of a California population. Am J Med Genet 64, 465–472. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S, 1999. Sonic Hedgehoginduced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem 274, 8143–8152. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E, 1999. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568. [DOI] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP, 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775–4785. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C, 2000. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483–492. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM, 2000. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102, 521–531. [DOI] [PubMed] [Google Scholar]
- Dennis JF, Kurosaka H, Iulianella A, Pace J, Thomas N, Beckham S, Williams T, Trainor PA, 2012. Mutations in Hedgehog acyltransferase (Hhat) perturb Hedgehog signaling, resulting in severe acrania-holoprosencephaly-agnathia craniofacial defects. PLoS Genet 8, e1002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C, 1999. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol 9, 1119–1122. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC, 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet 12, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou AD, Ohazama A, Porntaveetus T, Sharpe PT, Kondo S, Basson MA, Gritli-Linde A, Cobourne MT, Green JB, 2012. Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat Genet 44, 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, Pazour GJ, 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell 31, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Shahawy M, Reibring CG, Neben CL, Hallberg K, Marangoni P, Harfe BD, Klein OD, Linde A, Gritli-Linde A, 2017. Cell fate specification in the lingual epithelium is controlled by antagonistic activities of Sonic hedgehog and retinoic acid. PLoS Genet 13, e1006914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott KH, Chen X, Salomone J, Chaturvedi P, Schultz PA, Balchand SK, Servetas JD, Zuniga A, Zeller R, Gebelein B, Weirauch MT, Peterson KA, Brugmann SA, 2020. Gli3 utilizes Hand2 to synergistically regulate tissue-specific transcriptional networks. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ, de Sauvage FJ, Peterson AS, 2009. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol 19, 1320–1326. [DOI] [PubMed] [Google Scholar]
- Everson JL, Fink DM, Yoon JW, Leslie EJ, Kietzman HW, Ansen-Wilson LJ, Chung HM, Walterhouse DO, Marazita ML, Lipinski RJ, 2017. Sonic hedgehog regulation of Foxf2 promotes cranial neural crest mesenchyme proliferation and is disrupted in cleft lip morphogenesis. Development 144, 2082–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Mantesso A, De Bari C, Nishiyama A, Sharpe PT, 2011. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci U S A 108, 6503–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Sharpe PT, 2000. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development 127, 403–412. [DOI] [PubMed] [Google Scholar]
- Ferkol TW, Leigh MW, 2012. Ciliopathies: the central role of cilia in a spectrum of pediatric disorders. J Pediatr 160, 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura C, Silva RM, Granjeiro JM, Letra A, 2015. Association of WNT9B Gene Polymorphisms With Nonsyndromic Cleft Lip With or Without Cleft Palate in Brazilian Nuclear Families. Cleft Palate Craniofac J 52, 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foppiano S, Hu D, Marcucio RS, 2007. Signaling by bone morphogenetic proteins directs formation of an ectodermal signaling center that regulates craniofacial development. Dev Biol 312, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet A, Ruel L, Staccini-Lavenant L, Therond PP, 2006. Cholesterol modification is necessary for controlled planar long-range activity of Hedgehog in Drosophila epithelia. Development 133, 407–418. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M, 2003. Neural crest specification: migrating into genomics. Nat Rev Neurosci 4, 795–805. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV, 2010. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden EJ, Larson ED, Shechtman LA, Trahan GD, Gaillard D, Fellin TJ, Scott JK, Jones KL, Barlow LA, 2021. Onset of taste bud cell renewal starts at birth and coincides with a shift in SHH function. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, Yan N, 2018. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 361. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP, 1996. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10, 301–312. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP, 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113. [DOI] [PubMed] [Google Scholar]
- Graf D, Malik Z, Hayano S, Mishina Y, 2016. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev 27, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V, 1998. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 125, 2063–2074. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, Bei M, Maas R, Zhang XM, Linde A, McMahon AP, 2002. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development 129, 5323–5337. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, Lewis P, McMahon AP, Linde A, 2001. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev Biol 236, 364–386. [DOI] [PubMed] [Google Scholar]
- Hall JM, Bell ML, Finger TE, 2003. Disruption of sonic hedgehog signaling alters growth and patterning of lingual taste papillae. Dev Biol 255, 263–277. [DOI] [PubMed] [Google Scholar]
- Hall JM, Hooper JE, Finger TE, 1999. Expression of sonic hedgehog, patched, and Gli1 in developing taste papillae of the mouse. J Comp Neurol 406, 143–155. [DOI] [PubMed] [Google Scholar]
- Hall TM, Porter JA, Beachy PA, Leahy DJ, 1995. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 378, 212–216. [DOI] [PubMed] [Google Scholar]
- Han J, Mayo J, Xu X, Li J, Bringas P Jr., Maas RL, Rubenstein JL, Chai Y, 2009. Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development 136, 4225–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happ JT, Arveseth CD, Bruystens J, Bertinetti D, Nelson IB, Olivieri C, Zhang J, Hedeen DS, Zhu JF, Capener JL, Brockel JW, Vu L, King CC, Ruiz-Perez VL, Ge X, Veglia G, Herberg FW, Taylor SS, Myers BR, 2022. A PKA inhibitor motif within SMOOTHENED controls Hedgehog signal transduction. Nat Struct Mol Biol 29, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I, 1999. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol 147, 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle Z, Mo R, Hui CC, Sharpe PT, 1998. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development 125, 2803–2811. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Healy C, Morgan P, Sharpe PT, 2004. Regionalisation of early head ectoderm is regulated by endoderm and prepatterns the orofacial epithelium. Development 131, 4797–4806. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Wilson JM, Grevellec A, Cobourne MT, Healy C, Helms JA, Sharpe PT, Tucker AS, 2007. Sonic hedgehog in the pharyngeal endoderm controls arch pattern via regulation of Fgf8 in head ectoderm. Dev Biol 303, 244–258. [DOI] [PubMed] [Google Scholar]
- Hayano S, Komatsu Y, Pan H, Mishina Y, 2015. Augmented BMP signaling in the neural crest inhibits nasal cartilage morphogenesis by inducing p53-mediated apoptosis. Development 142, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms JA, Schneider RA, 2003. Cranial skeletal biology. Nature 423, 326–331. [DOI] [PubMed] [Google Scholar]
- Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, Vokes SA, McMahon AP, Kalinichenko VV, Moskowitz IP, 2014. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet 10, e1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Christ A, Christa A, Willnow TE, Krauss RS, 2020. Cdon mutation and fetal alcohol converge on Nodal signaling in a mouse model of holoprosencephaly. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Krauss RS, 2012. Cdon mutation and fetal ethanol exposure synergize to produce midline signaling defects and holoprosencephaly spectrum disorders in mice. PLoS Genet 8, e1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Krauss RS, 2013. Rescue of holoprosencephaly in fetal alcohol-exposed Cdon mutant mice by reduced gene dosage of Ptch1. PLoS One 8, e79269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa R, Deng X, Takamori K, Xu X, Urata M, Bringas P Jr., Chai Y, 2009. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. J Exp Zool B Mol Dev Evol 312B, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu A, Song BL, 2019. The interplay of Patched, Smoothened and cholesterol in Hedgehog signaling. Curr Opin Cell Biol 61, 31–38. [DOI] [PubMed] [Google Scholar]
- Hu D, Helms JA, 1999. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development 126, 4873–4884. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, 2009a. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development 136, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, 2009b. Unique organization of the frontonasal ectodermal zone in birds and mammals. Dev Biol 325, 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, 2012. Neural crest cells pattern the surface cephalic ectoderm during FEZ formation. Dev Dyn 241, 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA, 2003. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development 130, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Hu D, Young NM, Li X, Xu Y, Hallgrimsson B, Marcucio RS, 2015. A dynamic Shh expression pattern, regulated by SHH and BMP signaling, coordinates fusion of primordia in the amniote face. Development 142, 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, Salic A, 2016. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell 166, 1176–1187 e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P Jr., Hung YP, Chai Y, 2010. Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res 25, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV, 2005. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A 102, 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV, 2006. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133, 3–14. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV, 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S, 2011. Gli proteins in development and disease. Annu Rev Cell Dev Biol 27, 513–537. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R, 2010. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev 24, 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, 2000. How cholesterol modulates the signal. Curr Biol 10, R180–183. [DOI] [PubMed] [Google Scholar]
- Ingham PW, 2022. Hedgehog signaling. Curr Top Dev Biol 149, 1–58. [DOI] [PubMed] [Google Scholar]
- Istvan ES, Deisenhofer J, 2001. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Jones CM, Mercurio S, Rowe A, Domingos PM, Smith JC, Krumlauf R, 2003. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development 130, 4295–4305. [DOI] [PubMed] [Google Scholar]
- Iwatsuki K, Liu HX, Gronder A, Singer MA, Lane TF, Grosschedl R, Mistretta CM, Margolskee RF, 2007. Wnt signaling interacts with Shh to regulate taste papilla development. Proc Natl Acad Sci U S A 104, 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D, 2009. Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal 21, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP, 2004. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev 18, 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC, 2006. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn 235, 1152–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Benz TL, Long SB, 2021. Substrate and product complexes reveal mechanisms of Hedgehog acylation by HHAT. Science 372, 1215–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YR, Han XH, Taketo MM, Yoon JK, 2012. Wnt9b-dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development 139, 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL, Smith DW, 1973. Recognition of the fetal alcohol syndrome in early infancy. Lancet 302, 999–1001. [DOI] [PubMed] [Google Scholar]
- Jung HS, Oropeza V, Thesleff I, 1999. Shh, Bmp-2, Bmp-4 and Fgf-8 are associated with initiation and patterning of mouse tongue papillae. Mech Dev 81, 179–182. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Mager DL, Gagnier L, 2014. Epigenetic mechanism causes Wnt9b deficiency and nonsyndromic cleft lip and palate in the A/WySn mouse strain. Birth Defects Res A Clin Mol Teratol 100, 772–788. [DOI] [PubMed] [Google Scholar]
- Kang JS, Mulieri PJ, Hu Y, Taliana L, Krauss RS, 2002. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J 21, 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal JB, Batra SK, Rachagani S, 2022. Hedgehog signaling and its molecular perspective with cholesterol: a comprehensive review. Cell Mol Life Sci 79, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki M, Kawasaki K, Meguro F, Yamada A, Ishikawa R, Porntaveetus T, Blackburn J, Otsuka-Tanaka Y, Saito N, Ota MS, Sharpe PT, Kessler JA, Herz J, Cobourne MT, Maeda T, Ohazama A, 2018. Lrp4/Wise regulates palatal rugae development through Turing-type reaction-diffusion mechanisms. PLoS One 13, e0204126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, Pazour GJ, 2012. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell 22, 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Roessler E, Hennekam RC, Feldman GL, Kosaki K, Jones MC, Palumbos JC, Muenke M, 1996. Holoprosencephaly in RSH/Smith-Lemli-Opitz syndrome: does abnormal cholesterol metabolism affect the function of Sonic Hedgehog? Am J Med Genet 66, 478–484. [DOI] [PubMed] [Google Scholar]
- Keranen SV, Kettunen P, Aberg T, Thesleff I, Jernvall J, 1999. Gene expression patterns associated with suppression of odontogenesis in mouse and vole diastema regions. Dev Genes Evol 209, 495–506. [DOI] [PubMed] [Google Scholar]
- Kietzman HW, Everson JL, Sulik KK, Lipinski RJ, 2014. The teratogenic effects of prenatal ethanol exposure are exacerbated by Sonic Hedgehog or GLI2 haploinsufficiency in the mouse. PLoS One 9, e89448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Lee MJ, Cho KW, Lee JM, Kim YJ, Kim JY, Jung HI, Cho JY, Cho SW, Jung HS, 2009. Shh and ROCK1 modulate the dynamic epithelial morphogenesis in circumvallate papilla development. Dev Biol 325, 273–280. [DOI] [PubMed] [Google Scholar]
- Kinnebrew M, Iverson EJ, Patel BB, Pusapati GV, Kong JH, Johnson KA, Luchetti G, Eckert KM, McDonald JG, Covey DF, Siebold C, Radhakrishnan A, Rohatgi R, 2019. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew M, Woolley RE, Ansell TB, Byrne EFX, Frigui S, Luchetti G, Sircar R, Nachtergaele S, Mydock-McGrane L, Krishnan K, Newstead S, Sansom MSP, Covey DF, Siebold C, Rohatgi R, 2022. Patched 1 regulates Smoothened by controlling sterol binding to its extracellular cysteine-rich domain. Sci Adv 8, eabm5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Kawasaki M, Kawasaki K, Meguro F, Yamada A, Nagai T, Kodama Y, Trakanant S, Sharpe PT, Maeda T, Takagi R, Ohazama A, 2020. Ift88 is involved in mandibular development. J Anat 236, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG, 1999. Mammalian suppressor-of-fused modulates nuclearcytoplasmic shuttling of Gli-1. Nat Cell Biol 1, 312–319. [DOI] [PubMed] [Google Scholar]
- Kong JH, Siebold C, Rohatgi R, 2019. Biochemical mechanisms of vertebrate hedgehog signaling. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudijs MJ, den Broeder MJ, Groot E, van Eeden FJ, 2008. Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev Biol 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak PA, Wassif CA, Kratz L, Cozma D, Kovarova M, Harris G, Grinberg A, Yang Y, Hunter AG, Tsokos M, Kelley RI, Porter FD, 2003. Lathosterolosis: an inborn error of human and murine cholesterol synthesis due to lathosterol 5-desaturase deficiency. Hum Mol Genet 12, 1631–1641. [DOI] [PubMed] [Google Scholar]
- Kumari A, Ermilov AN, Allen BL, Bradley RM, Dlugosz AA, Mistretta CM, 2015. Hedgehog pathway blockade with the cancer drug LDE225 disrupts taste organs and taste sensation. J Neurophysiol 113, 1034–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Ermilov AN, Grachtchouk M, Dlugosz AA, Allen BL, Bradley RM, Mistretta CM, 2017. Recovery of taste organs and sensory function after severe loss from Hedgehog/Smoothened inhibition with cancer drug sonidegib. Proc Natl Acad Sci U S A 114, E10369–E10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka H, Iulianella A, Williams T, Trainor PA, 2014. Disrupting hedgehog and WNT signaling interactions promotes cleft lip pathogenesis. J Clin Invest 124, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Jia S, Jiang R, 2014. Molecular patterning of the mammalian dentition. Semin Cell Dev Biol 25–26, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Jiang R, 2009. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development 136, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R, 2004. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development 131, 3207–3216. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R, 2006. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn 235, 1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Xu J, Jiang R, 2015. Cellular and Molecular Mechanisms of Palatogenesis. Curr Top Dev Biol 115, 59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Kassai Y, Pakkasjarvi L, Thesleff I, Itoh N, 2003. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol 264, 91–105. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA, 1994. Autoproteolysis in hedgehog protein biogenesis. Science 266, 1528–1537. [DOI] [PubMed] [Google Scholar]
- Li F, Fu G, Liu Y, Miao X, Li Y, Yang X, Zhang X, Yu D, Gan L, Qiu M, Chen Y, Zhang Z, Zhang Z, 2017. ISLET1-Dependent beta-Catenin/Hedgehog Signaling Is Required for Outgrowth of the Lower Jaw. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chatzeli L, Panousopoulou E, Tucker AS, Green JB, 2016. Epithelial stratification and placode invagination are separable functions in early morphogenesis of the molar tooth. Development 143, 670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, Park S, Wang S, Chai Y, 2015. BMPSHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev Cell 33, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yuan Y, He J, Feng J, Han X, Jing J, Ho TV, Xu J, Chai Y, 2018. Constitutive activation of hedgehog signaling adversely affects epithelial cell fate during palatal fusion. Dev Biol 441, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pawlik B, Elcioglu N, Aglan M, Kayserili H, Yigit G, Percin F, Goodman F, Nurnberg G, Cenani A, Urquhart J, Chung BD, Ismail S, Amr K, Aslanger AD, Becker C, Netzer C, Scambler P, Eyaid W, Hamamy H, Clayton-Smith J, Hennekam R, Nurnberg P, Herz J, Temtamy SA, Wollnik B, 2010. LRP4 mutations alter Wnt/beta-catenin signaling and cause limb and kidney malformations in Cenani-Lenz syndrome. Am J Hum Genet 86, 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang H, Litingtung Y, Chiang C, 2006. Cholesterol modification restricts the spread of Shh gradient in the limb bud. Proc Natl Acad Sci U S A 103, 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YX, Yang HT, Zdanowicz M, Sicklick JK, Qi Y, Camp TJ, Diehl AM, 2007. Fetal alcohol exposure impairs Hedgehog cholesterol modification and signaling. Lab Invest 87, 231–240. [DOI] [PubMed] [Google Scholar]
- Liem KF Jr., He M, Ocbina PJ, Anderson KV, 2009. Mouse Kif7/Costal2 is a ciliaassociated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci U S A 106, 13377–13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Fisher AV, Yin Y, Maruyama T, Veith GM, Dhandha M, Huang GJ, Hsu W, Ma L, 2011. The inductive role of Wnt-beta-Catenin signaling in the formation of oral apparatus. Dev Biol 356, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yao E, Wang K, Nozawa Y, Shimizu H, Johnson JR, Chen JN, Krogan NJ, Chuang PT, 2014. Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev 28, 2547–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA, 2005. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development 132, 3103–3111. [DOI] [PubMed] [Google Scholar]
- Liu Y, Feng J, Li J, Zhao H, Ho TV, Chai Y, 2015. An Nfic-hedgehog signaling cascade regulates tooth root development. Development 142, 3374–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Han WT, Liu Q, Li JX, Li ZJ, Jiang M, Xu W, 2015. Variations in WNT3 gene are associated with incidence of non-syndromic cleft lip with or without cleft palate in a northeast Chinese population. Genet Mol Res 14, 12646–12653. [DOI] [PubMed] [Google Scholar]
- Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, Covey DF, Siebold C, Rohatgi R, 2016. Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA, 2002. Hedgehogmediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111, 63–75. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA, 2005. Molecular interactions coordinating the development of the forebrain and face. Dev Biol 284, 48–61. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ, 1996. Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176–179. [DOI] [PubMed] [Google Scholar]
- Martinelli DC, Fan CM, 2007. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev 21, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, van Meer G, 2010. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol 22, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ, 2003. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol 53, 1–114. [DOI] [PubMed] [Google Scholar]
- Melnick M, Witcher D, Bringas P Jr., Carlsson P, Jaskoll T, 2005. Meckel’s cartilage differentiation is dependent on hedgehog signaling. Cells Tissues Organs 179, 146–157. [DOI] [PubMed] [Google Scholar]
- Millington G, Elliott KH, Chang YT, Chang CF, Dlugosz A, Brugmann SA, 2017. Cilia-dependent GLI processing in neural crest cells is required for tongue development. Dev Biol 424, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ, Muenke M, 2002. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet 110, 297–301. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M, 1998. Holoprosencephaly: from Homer to Hedgehog. Clin Genet 53, 155–163. [DOI] [PubMed] [Google Scholar]
- Mistretta CM, Kumari A, 2019. Hedgehog Signaling Regulates Taste Organs and Oral Sensation: Distinctive Roles in the Epithelium, Stroma, and Innervation. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistretta CM, Liu HX, Gaffield W, MacCallum DK, 2003. Cyclopamine and jervine in embryonic rat tongue cultures demonstrate a role for Shh signaling in taste papilla development and patterning: fungiform papillae double in number and form in novel locations in dorsal lingual epithelium. Dev Biol 254, 1–18. [DOI] [PubMed] [Google Scholar]
- Miura H, Kusakabe Y, Harada S, 2006. Cell lineage and differentiation in taste buds. Arch Histol Cytol 69, 209–225. [DOI] [PubMed] [Google Scholar]
- Miura H, Kusakabe Y, Sugiyama C, Kawamatsu M, Ninomiya Y, Motoyama J, Hino A, 2001. Shh and Ptc are associated with taste bud maintenance in the adult mouse. Mech Dev 106, 143–145. [DOI] [PubMed] [Google Scholar]
- Miura H, Scott JK, Harada S, Barlow LA, 2014. Sonic hedgehog-expressing basal cells are general post-mitotic precursors of functional taste receptor cells. Dev Dyn 243, 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales AV, Barbas JA, Nieto MA, 2005. How to become neural crest: from segregation to delamination. Semin Cell Dev Biol 16, 655–662. [DOI] [PubMed] [Google Scholar]
- Mostowska A, Hozyasz KK, Biedziak B, Wojcicki P, Lianeri M, Jagodzinski PP, 2012. Genotype and haplotype analysis of WNT genes in non-syndromic cleft lip with or without cleft palate. Eur J Oral Sci 120, 1–8. [DOI] [PubMed] [Google Scholar]
- Mouritsen OG, Bagatolli LA, 2015. Lipid domains in model membranes: a brief historical perspective. Essays Biochem 57, 1–19. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R, 2014. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol 33, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, Jackson PK, 2013. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 152, 210–223. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Morita I, Eto K, Ota MS, 2006. Sonic hedgehog signaling is important in tooth root development. J Dent Res 85, 427–431. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y, 2020. Architecture of the IFT ciliary trafficking machinery and interplay between its components. Crit Rev Biochem Mol Biol 55, 179–196. [DOI] [PubMed] [Google Scholar]
- Nguyen TD, Truong ME, Reiter JF, 2022. The Intimate Connection Between Lipids and Hedgehog Signaling. Front Cell Dev Biol 10, 876815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P, 2006. FGF signalling in craniofacial development and developmental disorders. Oral Dis 12, 102–111. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E, Hui CC, 2005. Hedgehog signaling and congenital malformations. Clin Genet 67, 193–208. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E, Motoyama J, Barnfield PC, Yoshikawa Y, Zhang X, Mo R, Crackower MA, Hui CC, 2006. Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia. Mol Cell Biol 26, 6609–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R, 2014. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep 6, 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikopensius T, Jagomagi T, Krjutskov K, Tammekivi V, Saag M, Prane I, Piekuse L, Akota I, Barkane B, Krumina A, Ambrozaityte L, Matuleviciene A, Kucinskiene ZA, Lace B, Kucinskas V, Metspalu A, 2010. Genetic variants in COL2A1, COL11A2, and IRF6 contribute risk to nonsyndromic cleft palate. Birth Defects Res A Clin Mol Teratol 88, 748–756. [DOI] [PubMed] [Google Scholar]
- Noden DM, Francis-West P, 2006. The differentiation and morphogenesis of craniofacial muscles. Dev Dyn 235, 1194–1218. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E, 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK, 2006. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444, 369–373. [DOI] [PubMed] [Google Scholar]
- Okubo T, Clark C, Hogan BL, 2009. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 27, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara S, Birjandi AA, Adel Al-Lami H, Sagai T, Amano T, Shiroishi T, Xavier GM, Liu KJ, Cobourne MT, Iseki S, 2019. Temporospatial sonic hedgehog signalling is essential for neural crest-dependent patterning of the intrinsic tongue musculature. Development 146. [DOI] [PubMed] [Google Scholar]
- Olsen CL, Hughes JP, Youngblood LG, Sharpe-Stimac M, 1997. Epidemiology of holoprosencephaly and phenotypic characteristics of affected children: New York State, 1984–1989. Am J Med Genet 73, 217–226. [DOI] [PubMed] [Google Scholar]
- Palm W, Swierczynska MM, Kumari V, Ehrhart-Bornstein M, Bornstein SR, Eaton S, 2013. Secretion and signaling activities of lipoprotein-associated hedgehog and non-sterolmodified hedgehog in flies and mammals. PLoS Biol 11, e1001505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Prochazka J, Martin A, Rothova M, Lambert A, Bernard L, Charles C, Viriot L, Peterkova R, Laudet V, 2008. Patterning of palatal rugae through sequential addition reveals an anterior/posterior boundary in palatal development. BMC Dev Biol 8, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C, Chai Y, 2015. Mandible and Tongue Development. Curr Top Dev Biol 115, 31–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C, Han D, Chai Y, 2012. Molecular and cellular regulatory mechanisms of tongue myogenesis. J Dent Res 91, 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski K, Taylor FR, Wang EA, Galdes A, 1998. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem 273, 14037–14045. [DOI] [PubMed] [Google Scholar]
- Pereira J, Johnson WE, O’Brien SJ, Jarvis ED, Zhang G, Gilbert MT, Vasconcelos V, Antunes A, 2014. Evolutionary genomics and adaptive evolution of the Hedgehog gene family (Shh, Ihh and Dhh) in vertebrates. PLoS One 9, e74132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Wolf A, Wagner M, Kuhlmann J, Waldmann H, 2004. The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc Natl Acad Sci U S A 101, 8531–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R, 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev 12, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov K, Wierbowski BM, Salic A, 2017. Sending and Receiving Hedgehog Signals. Annu Rev Cell Dev Biol 33, 145–168. [DOI] [PubMed] [Google Scholar]
- Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD, 2013. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat Chem Biol 9, 247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD, Herman GE, 2011. Malformation syndromes caused by disorders of cholesterol synthesis. J Lipid Res 52, 6–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA, 1996. Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259. [DOI] [PubMed] [Google Scholar]
- Qi X, Schmiege P, Coutavas E, Li X, 2018a. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Schmiege P, Coutavas E, Wang J, Li X, 2018b. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Cao P, Hu M, Gao S, Yan N, Gong X, 2019. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat Commun 10, 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT, 2011. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc Natl Acad Sci U S A 108, 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk J, van den Heuvel M, Henrique D, Marigo V, Jones TA, Tabin C, Ingham PW, 1997. The smoothened gene and hedgehog signal transduction in Drosophila and vertebrate development. Cold Spring Harb Symp Quant Biol 62, 217–226. [PubMed] [Google Scholar]
- Ray AT, Mazot P, Brewer JR, Catela C, Dinsmore CJ, Soriano P, 2020. FGF signaling regulates development by processes beyond canonical pathways. Genes Dev 34, 1735–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K, Kumari P, Sepulveda Rincon L, Gu R, Ji Y, Kumar S, Zhou CJ, 2019. Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis Model Mech 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K, Zhang S, Sun B, Garland MA, Ji Y, Zhou CJ, 2020. Genetics and signaling mechanisms of orofacial clefts. Birth Defects Res 112, 1588–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro LA, Quiezi RG, Nascimento A, Bertolacini CP, Richieri-Costa A, 2010. Holoprosencephaly and holoprosencephaly-like phenotype and GAS1 DNA sequence changes: Report of four Brazilian patients. Am J Med Genet A 152A, 1688–1694. [DOI] [PubMed] [Google Scholar]
- Rice R, Connor E, Rice DP, 2006. Expression patterns of Hedgehog signalling pathway members during mouse palate development. Gene Expr Patterns 6, 206–212. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP, 2004. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest 113, 1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach E, Demyer W, Conneally PM, Palmer C, Merritt AD, 1975. Holoprosencephaly: birth data, benetic and demographic analyses of 30 families. Birth Defects Orig Artic Ser 11, 294–313. [PubMed] [Google Scholar]
- Rodgers UR, Lanyon-Hogg T, Masumoto N, Ritzefeld M, Burke R, Blagg J, Magee AI, Tate EW, 2016. Characterization of Hedgehog Acyltransferase Inhibitors Identifies a Small Molecule Probe for Hedgehog Signaling by Cancer Cells. ACS Chem Biol 11, 3256–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M, 1996. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet 14, 357–360. [DOI] [PubMed] [Google Scholar]
- Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M, 2003. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci U S A 100, 13424–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, Velez JI, Solomon BD, Pineda-Alvarez DE, Lacbawan F, Zhou N, Ouspenskaia M, Paulussen A, Smeets HJ, Hehr U, Bendavid C, Bale S, Odent S, David V, Muenke M, 2009a. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat 30, E921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ma Y, Ouspenskaia MV, Lacbawan F, Bendavid C, Dubourg C, Beachy PA, Muenke M, 2009b. Truncating loss-of-function mutations of DISP1 contribute to holoprosencephaly-like microform features in humans. Hum Genet 125, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Muenke M, 1998. Holoprosencephaly: a paradigm for the complex genetics of brain development. J Inherit Metab Dis 21, 481–497. [DOI] [PubMed] [Google Scholar]
- Santagati F, Rijli FM, 2003. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci 4, 806–818. [DOI] [PubMed] [Google Scholar]
- Sarkar L, Cobourne M, Naylor S, Smalley M, Dale T, Sharpe PT, 2000. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc Natl Acad Sci U S A 97, 4520–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock EN, Struve JN, Chang CF, Williams TJ, Snedeker J, Attia AC, Stottmann RW, Brugmann SA, 2017. A tissue-specific role for intraflagellar transport genes during craniofacial development. PLoS One 12, e0174206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MM, DeBose-Boyd RA, 2021. Posttranslational Regulation of HMG CoA Reductase, the Rate-Limiting Enzyme in Synthesis of Cholesterol. Annu Rev Biochem 90, 659–679. [DOI] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, Fuchs E, Joyner A, Martin GR, de Sauvage FJ, Klein OD, 2010. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development 137, 3753–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala M, Depew MJ, Martinelli DC, Fan CM, Sharpe PT, Cobourne MT, 2007. Gas1 is a modifier for holoprosencephaly and genetically interacts with sonic hedgehog. J Clin Invest 117, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala M, Fraser GJ, Birjandi AA, Xavier GM, Cobourne MT, 2017. Sonic Hedgehog Signaling and Development of the Dentition. J Dev Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetani Y, Nobusada Y, Kuratani S, 2000. Ectodermally derived FGF8 defines the maxillomandibular region in the early chick embryo: epithelial-mesenchymal interactions in the specification of the craniofacial ectomesenchyme. Dev Biol 228, 73–85. [DOI] [PubMed] [Google Scholar]
- Signore IA, Jerez C, Figueroa D, Suazo J, Marcelain K, Cerda O, Colombo Flores A, 2016. Inhibition of the 3-hydroxy-3-methyl-glutaryl-CoA reductase induces orofacial defects in zebrafish. Birth Defects Res A Clin Mol Teratol 106, 814–830. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Bear KA, Wyllie A, Keaton AA, Dubourg C, David V, Mercier S, Odent S, Hehr U, Paulussen A, Clegg NJ, Delgado MR, Bale SJ, Lacbawan F, Ardinger HH, Aylsworth AS, Bhengu NL, Braddock S, Brookhyser K, Burton B, Gaspar H, Grix A, Horovitz D, Kanetzke E, Kayserili H, Lev D, Nikkel SM, Norton M, Roberts R, Saal H, Schaefer GB, Schneider A, Smith EK, Sowry E, Spence MA, Shalev SA, Steiner CE, Thompson EM, Winder TL, Balog JZ, Hadley DW, Zhou N, Pineda-Alvarez DE, Roessler E, Muenke M, 2012. Genotypic and phenotypic analysis of 396 individuals with mutations in Sonic Hedgehog. J Med Genet 49, 473–479. [DOI] [PubMed] [Google Scholar]
- Stanier P, Pauws E, 2012. Development of the lip and palate: FGF signalling. Front Oral Biol 16, 71–80. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim HJ, Cho MI, Gronostajski RM, 2003. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol 23, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B, Carmona-Fontaine C, Mayor R, 2005. Genetic network during neural crest induction: from cell specification to cell survival. Semin Cell Dev Biol 16, 647–654. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A, 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S, 2006. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 10, 187–197. [DOI] [PubMed] [Google Scholar]
- Tabler JM, Barrell WB, Szabo-Rogers HL, Healy C, Yeung Y, Perdiguero EG, Schulz C, Yannakoudakis BZ, Mesbahi A, Wlodarczyk B, Geissmann F, Finnell RH, Wallingford JB, Liu KJ, 2013. Fuz mutant mice reveal shared mechanisms between ciliopathies and FGF-related syndromes. Dev Cell 25, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler JM, Bolger TG, Wallingford J, Liu KJ, 2014. Hedgehog activity controls opening of the primary mouth. Dev Biol 396, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler JM, Rice CP, Liu KJ, Wallingford JB, 2016. A novel ciliopathic skull defect arising from excess neural crest. Dev Biol 417, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler JM, Rigney MM, Berman GJ, Gopalakrishnan S, Heude E, Al-Lami HA, Yannakoudakis BZ, Fitch RD, Carter C, Vokes S, Liu KJ, Tajbakhsh S, Egnor SR, Wallingford JB, 2017. Cilia-mediated Hedgehog signaling controls form and function in the mammalian larynx. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA, 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–897. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP, 2006. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell 10, 647–656. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P, 1997. Signalling networks regulating dental development. Mech Dev 67, 111–123. [DOI] [PubMed] [Google Scholar]
- Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF, Barlow LA, 2009. Fate mapping of mammalian embryonic taste bud progenitors. Development 136, 1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, Herron BJ, Chesebro AL, Qiu H, Scherz PJ, Shah JV, Yoder BK, Beier DR, 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet 40, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A, Sharpe P, 2004. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet 5, 499–508. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT, 1998. Transformation of tooth type induced by inhibition of BMP signaling. Science 282, 1136–1138. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT, 1999. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development 126, 51–61. [DOI] [PubMed] [Google Scholar]
- Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A, 2012. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep 2, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, Salic A, 2010. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol 191, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M, He M, Anderson KV, 2011. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development 138, 4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueharu H, Mishina Y, 2023. BMP signaling during craniofacial development: new insights into pathological mechanisms leading to craniofacial anomalies. Front Physiol 14, 1170511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urioste M, Valcarcel E, Gomez MA, Pinel I, Garcia de Leon R, Diaz de Bustamante A, Tebar R, Martinez-Frias ML, 1988. Holoprosencephaly and trisomy 21 in a child born to a nondiabetic mother. Am J Med Genet 30, 925–928. [DOI] [PubMed] [Google Scholar]
- Wassif CA, Maslen C, Kachilele-Linjewile S, Lin D, Linck LM, Connor WE, Steiner RD, Porter FD, 1998. Mutations in the human sterol delta7-reductase gene at 11q12–13 cause Smith-Lemli-Opitz syndrome. Am J Hum Genet 63, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, Romeijn GJ, Hennekam RC, Vreken P, Andersson HC, FitzPatrick DR, Kelley RI, Wanders RJ, 2001. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am J Hum Genet 69, 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh IC, O’Brien TP, 2009. Signaling integration in the rugae growth zone directs sequential SHH signaling center formation during the rostral outgrowth of the palate. Dev Biol 336, 53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierbowski BM, Petrov K, Aravena L, Gu G, Xu Y, Salic A, 2020. Hedgehog Pathway Activation Requires Coreceptor-Catalyzed, Lipid-Dependent Relay of the Sonic Hedgehog Ligand. Dev Cell 55, 450–467 e458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier GM, Seppala M, Papageorgiou SN, Fan CM, Cobourne MT, 2016. Genetic interactions between the hedgehog co-receptors Gas1 and Boc regulate cell proliferation during murine palatogenesis. Oncotarget 7, 79233–79246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Tang JJ, Peng C, Wang Y, Fu L, Qiu ZP, Xiong Y, Yang LF, Cui HW, He XL, Yin L, Qi W, Wong CC, Zhao Y, Li BL, Qiu WW, Song BL, 2017. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Mol Cell 66, 154–162 e110. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Adam M, Clouthier DE, Potter S, Jiang R, 2019. Hedgehog signaling patterns the oral-aboral axis of the mandibular arch. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang R, 2016. A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development. PLoS Genet 12, e1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Jiang R, 2022. The transcription factors Foxf1 and Foxf2 integrate the SHH, HGF and TGFbeta signaling pathways to drive tongue organogenesis. Development 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Zeng H, Ning G, Reiter JF, Liu A, 2014. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci U S A 111, 2164–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Chai Y, 2019. Regulatory mechanisms of jaw bone and tooth development. Curr Top Dev Biol 133, 91–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yu J, Fu G, Li J, Huang H, Liu J, Yu D, Qiu M, Li F, 2022. ISL1/SHH/CXCL12 signaling regulates myogenic cell migration during mouse tongue development. Development 149. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Beachy PA, 2023. Cellular and molecular mechanisms of Hedgehog signalling. Nat Rev Mol Cell Biol. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, Myers BR, Cho W, Cheng Y, Beachy PA, 2018. Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell 175, 1352–1364 e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y, 2002. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129, 4135–4146. [DOI] [PubMed] [Google Scholar]
- Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, Chai Y, 2014. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell 14, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gao Y, Lan Y, Jia S, Jiang R, 2013. Pax9 regulates a molecular network involving Bmp4, Fgf10, Shh signaling and the Osr2 transcription factor to control palate morphogenesis. Development 140, 4709–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


