SUMMARY
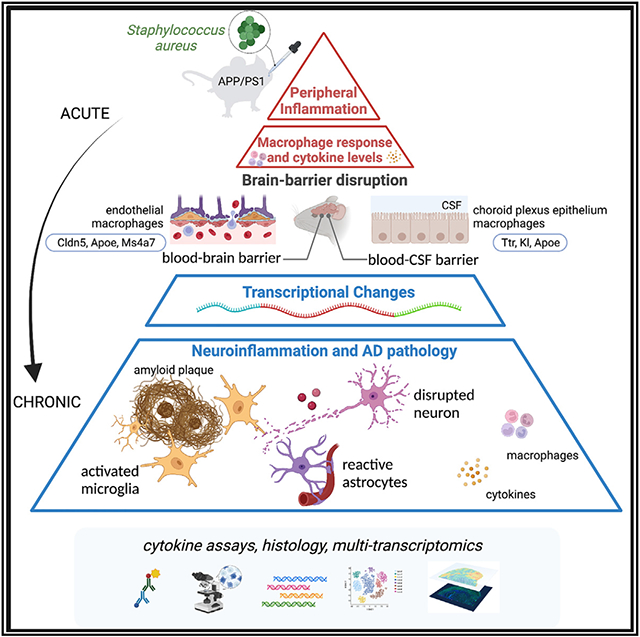
Peripheral inflammation has been linked to various neurodegenerative disorders, including Alzheimer’s disease (AD). Here we perform bulk, single-cell, and spatial transcriptomics in APP/PS1 mice intranasally exposed to Staphylococcus aureus to determine how low-grade peripheral infection affects brain transcriptomics and AD-like pathology. Chronic exposure led to increased amyloid plaque burden and plaque-associated microglia, significantly affecting the transcription of brain barrier-associated cells, which resulted in barrier leakage. We reveal cell-type- and spatial-specific transcriptional changes related to brain barrier function and neuroinflammation during the acute infection. Both acute and chronic exposure led to brain macrophage-associated responses and detrimental effects in neuronal transcriptomics. Finally, we identify unique transcriptional responses at the amyloid plaque niches following acute infection characterized by higher disease-associated microglia gene expression and a larger effect on astrocytic or macrophage-associated genes, which could facilitate amyloid and related pathologies. Our findings provide important insights into the mechanisms linking peripheral inflammation to AD pathology.
Graphical Abstract
In brief
Lu et al. studied how low-grade peripheral infection affects brain transcriptomics in a mouse model of AD using multi-transcriptomics. Chronic exposure affects brain-barrier-associated cells and causes barrier leakage. Unique transcriptional responses occur in the amyloid plaque microenvironment, affecting microglia, astrocyte, and macrophage-related genes.
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of dementia, characterized by extracellular amyloid plaques composed of aggregated amyloid beta (Ab) and intracellular neurofibrillary tangles.1 Over 95% of all AD cases are late onset with unknown etiology.
Studies have suggested that peripheral inflammation affects the central nervous system (CNS) and may be a factor in the pathophysiology of neurodegenerative diseases like AD.2 Clinical studies have demonstrated that the presence of chronic local or systemic inflammation in the elderly increases the likelihood of developing AD.3 A meta-analysis of 175 studies found that AD is accompanied by a peripheral inflammatory response, and IL-6 levels correlated with severity of cognitive impairment, making it a potential biomarker.4 Animal studies further explored potential mechanisms of periphery and CNS communication. In mice with increased peripheral inflammation induced by lipopolysaccharide (LPS), microglia underwent reprogramming associated with immune memory, which exacerbated cerebral β-amyloidosis.5 Systemic inflammation was shown to impair microglia clearance of Ab in APP/PS1 mice.6 Chronic infection with brain-colonizing parasites, Trypanosoma brucei, is associated with glial cell activation in the CNS, measured by the increased microglial and astrocytic markers.7
The CNS is mostly self-regulated and isolated from the periphery by the blood-brain barrier (BBB).8 The BBB is an interface between the brain and the systemic circulation, which protects the brain from toxins and pathogens by regulating the entry of molecules into brain. BBB integrity is mediated by its structural components, including tight junctions and the neurovascular unit, which is composed of brain microvascular endothelium, astrocytes, pericytes, neurons, and extracellular matrix.9 Dysregulation of the BBB is a significant contributor to the pathogenesis of many brain disorders.10 A variety of signaling factors originating in the periphery, such as inflammatory mediators, have been associated with BBB permeability.11 In AD, activated microglia and astrocytes also release proinflammatory factors, while amyloid deposits around microvessels directly affect brain endothelial cells.12-14
The blood-cerebrospinal fluid (CSF) barrier formed by the choroid plexus (CP) is critical in brain-periphery communication. In the brain, the ventricular surfaces are composed of a variety of cell types, including ependymal cells, microglia, astrocytes, blood vessel cells, immune cells, neural progenitor cells, and specialized CP epithelial cells (CPECs).15,16 The CP provides a crucial immune surveillance mechanism in the brain that regulates CSF secretion and composition influenced by peripheral blood and enables selective immune cell trafficking and regulation. CP disbalances are also associated with brain disorders triggered by peripheral inflammation.17 Systemic inflammation also activates the innate immune system and causes the release of proinflammatory cytokines, which can potentially affect the inflammatory response in the CNS, such as microglia activation and myelination.18 Other brain cell types, such as astrocytes and oligodendrocytes, also showed transcriptional changes in inflamed brains.19 However, little is known about how brain cells communicate with the periphery or barrier dysregulation in response to systemic inflammation.
Here, we hypothesized that peripheral inflammation dysregulates brain barrier integrity and induces neuroinflammation with consequences on amyloid pathology. To test the effect of peripheral inflammation on amyloid pathology and brain transcriptome, we used an AD mouse model nasally inoculated with Staphylococcus aureus. We show that chronic exposure for 16 weeks significantly increased amyloid pathology and blood cytokine levels. Single-cell RNA sequencing and spatial transcriptomics were conducted using mouse brain tissue after a short treatment to characterize transcriptional changes in major brain cell types and brain regions in response to acute peripheral inflammation. Our findings demonstrate cell-type- and spatial-specific molecular responses suggesting BBB and blood-CSF barrier dysregulation and molecular consequences in the amyloid plaque microenvironment.
RESULTS
Peripheral infection induced by nasal inoculation of S. aureus increases amyloid pathology in APP/PS1 mice
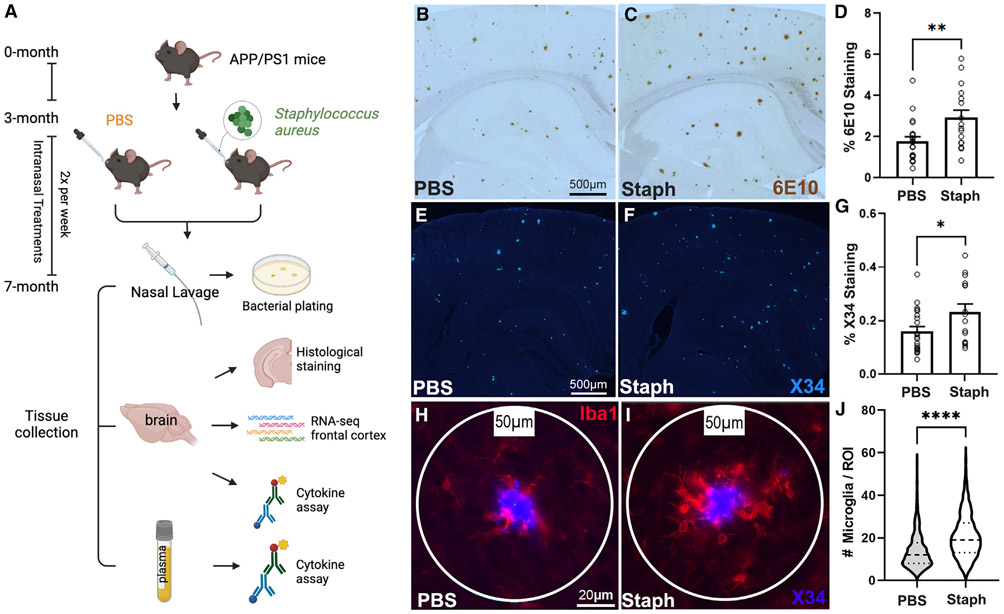
The initial goal of the study was to examine the effects of chronic peripheral inflammation on the progression of Aβ plaque pathology utilizing APP/PS1dE9 mice (referred to as APP/PS1). We used a low-dose S. aureus (referred to as Staph) infection by introducing the bacterial extract via nasal inoculation. The nares were inoculated two times per week with either Staph or PBS as a control starting at 3 until 7 month of age, as shown in Figure 1A. We chose 3 months of age as this is the age of onset of amyloid pathology in this model. Nasal lavage samples from both groups were plated and confirmed colonization of Staph in the nares of Staph mice but not PBS controls (Figures S1A and S1B). Brain sections of age- and sex-matched PBS-control and Staph-exposed mice were stained with both 6E10 antibody and X34 to visualize total Aβ (diffuse and fibrillar) and compact amyloid plaques, respectively. There was a significant increase in both total 6E10 (Figures 1B-1D) and X34 (Figures 1E-1G) staining in Staph-exposed animals compared with PBS controls, and the effect was similar in both sexes (Figures S1C and S1D). We also found a significant increase in the number of plaque-associated Iba1-positive microglia in the Staph-infected mice compared with PBS controls (Figures 1H-1J). Therefore, peripheral Staph infection aggravated amyloid pathology and inflammation around plaques in an AD mouse model.
Figure 1. Nasal inoculation of APP/PS1 mice with Staph shows increased amyloid pathology.
(A–C) Study outline (A). Representative images of 6E10 staining in the PBS (B) and the Staph group (C).
(D–F) Percentage coverage of 6E10 staining in Staph and PBS groups (D). Representative images of X34 staining show compact plaques in the PBS (E) and the Staph group (F). Scale bars in (B), (C), (E), and (F), 500 μm.
(G–I) Percentage coverage of X34 staining in Staph and PBS groups (G). For X34 and 6E10, Staph n = 16, 8 females and 8 males, and PBS n = 21, 11 females and 10 males, with 6 sections/mouse. Representative images of Iba1 and X34 staining in the PBS (H, n = 6, 6 sections/mouse, 352 plaques) and the Staph group (I, n = 8, 6 sections/mouse, 441 plaques). Scale bars, 20 μm.
(J) Number of microglia per region of interest (ROI). Statistical analysis was performed with unpaired t test. Bar plots are means ± SEM. ****p < 0.0001, **p < 0.01, *p < 0.05.
Chronic Staph infection significantly affects brain transcriptome of AD model mice
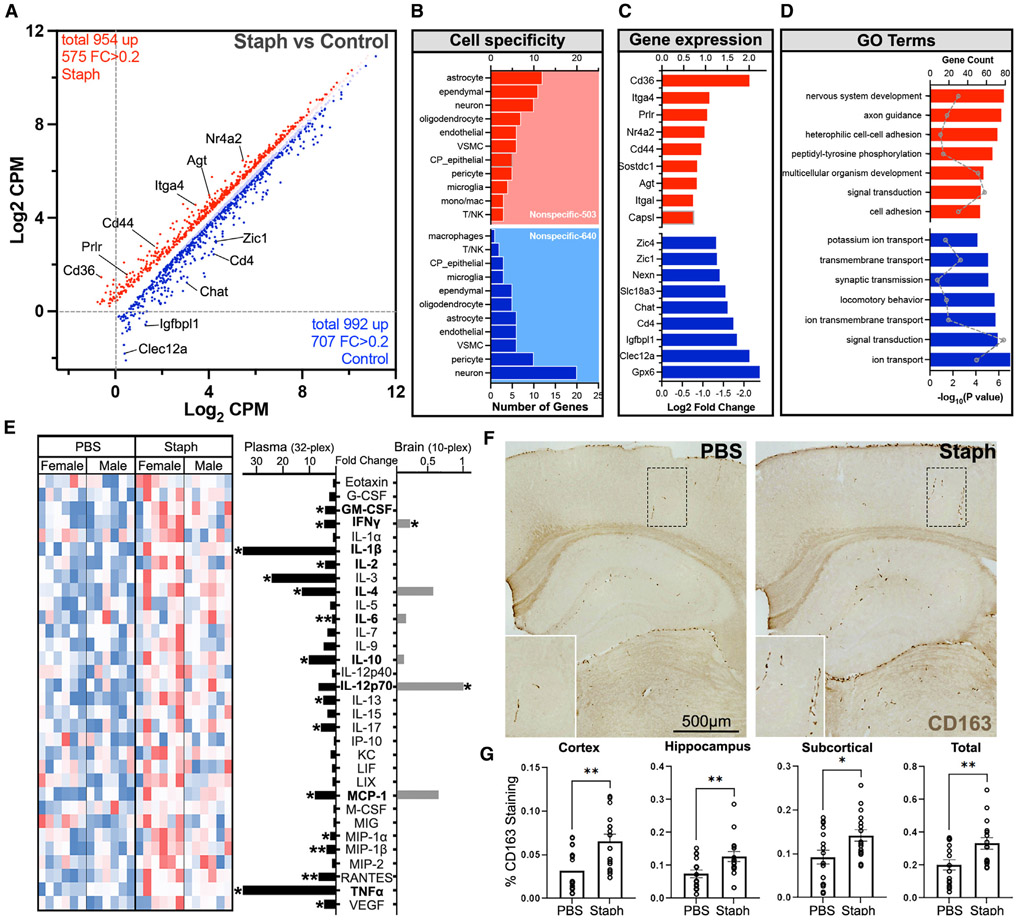
We assessed the effect of low-grade Staph-induced peripheral inflammation on the brain transcriptome by performing bulk RNA sequencing (RNA-seq) using the frontal cortex of mice at 7 months of age from Figure 1A. We identified a large number of differentially expressed genes (DEGs) at false discovery rate (FDR) < 0.05 and fold change (FC) > 0.2: 575 up- and 707 downregulated transcripts in the Staph group compared with controls (Figure 2A, Table S1). A significant number of DEGs represented cell-specific transcripts from cells composing brain barriers, namely BBB or blood-CSF barrier, such as astrocytes, endothelial cells, ependymal cells, CPECs, pericytes, and vascular smooth muscle cells (VSMCs) (Figure 2B). These vascular and perivascular cells are vital in maintaining brain barriers and establishing the neural milieu. Among the top upregulated genes were Cd36, Itga4, and Prlr, as well as genes associated with a reactive astrocytic state (Gfap, Agt, Gstm1, Synm, and Id2) (Figure 2C). Top upregulated categories were nervous system development, cell-cell adhesion, and signal transduction. Importantly, following Staph-induced peripheral inflammation there was a significant increase in the expression of genes encoding cell adhesion molecules, including Cldn1, which is important in sealing tight junctions at the brain barriers, and BBB integrins (Itga4 and Itgal). Associated with changes in brain barrier function, in the Staph group, we detected an increased expression of marker genes for infiltrating peripheral immune cells (Cd44 and Cd36). Consistent with defined pathways, multiple inflammation-related targets in the Staph group were upregulated (Prlr, Nr4a2, and Sostdc1), while others were downregulated (Clec12a and Cd4) (Figure 2D). Among the downregulated categories were ion transport, locomotory behavior, and synaptic transmission (Figure 2D). Top downregulated genes were astrocytic neuronal support genes (Id4, Foxg1, Pou3f2, Mapt, Nr2e1, and Wnt7a) and Cd163.
Figure 2. Chronic Staph infection significantly affects brain transcriptome of AD model mice.
Gene expression profiling was performed by RNA-seq on the frontal cortex of mice shown in Figure 1A.
(A) Scatterplot depicts differentially expressed genes between the Staph and the PBS group, with bright red/blue indicating FC (fold change) > 0.2.
(B) Bar plot showing number of different cell-type-specific genes: Staph (red) and PBS (blue).
(C) Bar plot showing the FC of the top differentially expressed genes in each treatment group: Staph (red) and PBS (blue).
(D) GO terms associated with the genes upregulated in Staph (red) or PBS (blue) group. Bars show −log10(p) and dots the count of genes. (A), (B), (C), and (D): Staph n = 16 and PBS n = 20.
(E) Heatmap (left) of plasma cytokine levels in PBS or Staph group, and bar plots (right) presenting the FC in terms of Staph versus PBS (n = 12 for both groups) in plasma (black) and brain tissue (gray). Cytokines in bold were assessed in both plasma and brain.
(F) Representative images of CD163-positive immunostaining. Scale bar, 500 μm.
(G) Bar plots showing the percentage of CD163 staining in different brain regions and total (n = 16 for both groups). Statistical analysis for cytokines and CD163 staining was performed with unpaired t test. *p < 0.05, **p < 0.01.
To confirm low-grade Staph-induced peripheral inflammation, we examined cytokine levels in the plasma using the mouse cytokine 32-plex assay. There was a significant increase in interferon-γ (IFN-γ), GM-CSF, TNF-α, and several interleukins in the plasma of Staph-treated mice compared with controls (Figure 2E). We also measured several cytokines in the brain of infected mice, but because most of the cytokines assessed in the plasma are secreted by peripheral immune cells, we used a smaller cytokine panel. We found that the level of most of the brain cytokines was very low and few were significantly increased (IFN-γ, IL-1270p, and MCP-1 [coded by the Ccl2 gene]). IL-4 was increased but not significantly (Figure 2E). Interestingly, this increase in cytokines and IFN-γ in brain and periphery by Staph treatment was accompanied by a significant decrease in Cd163 gene expression in the brain compared with controls. CD163 acts as a scavenger receptor for haptoglobin-hemoglobin and is expressed specifically in the monocyte/macrophage lineage. The large extracellular domain can undergo ADAM17-dependent cleavage, releasing soluble CD163 (sCD163) and downregulating surface expression. We performed immunohistochemistry (IHC) and observed a significant increase in the levels of CD163-positive immunostaining in the brains of the Staph compared with the PBS group (Figures 2F and 2G). CD163-staining pattern suggested an association with blood vessels and meninges, indicating an increased number of CD163+ macrophages or increased sCD163 in the perivascular space (Figure 2F). CD163 is suggested to serve an anti-inflammatory function,20 and CD163-positive macrophages are found in the resolution of inflammation.21 sCD163 is highly upregulated in plasma during infectious and inflammatory conditions22,23; however, the physiological role of sCD163 is unknown. sCD163 release suppresses the heme iron supply to hemolytic bacteria and trypanosomes and, important to this study, promotes phagocytosis of Staph.24 Several factors have been shown to downregulate Cd163 gene expression, including IFN-γ, GM-CSF, and TNF-α.23 We concluded that Cd163 gene expression and CD163 immunostaining in brain were differentiated during Staph-induced peripheral inflammation, with a positive correlation between CD163 immunostaining and increased levels of peripheral proinflammatory cytokines.
scRNA-seq distinguishes major brain cell types in APP/PS1 mice following acute peripheral inflammation
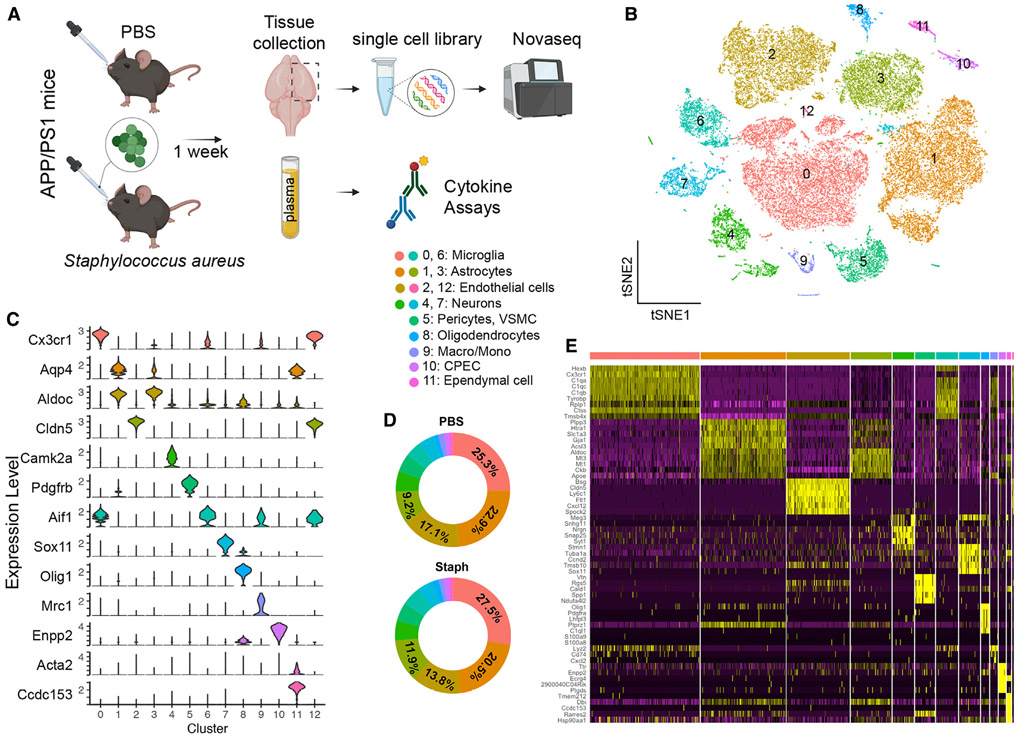
Chronic inflammation allows the organism to recover or adapt, thus obscuring the initial transcriptional responses that trigger the phenotypic changes we observed with chronic treatment. To investigate earlier transcriptome changes, we performed single-cell RNA-seq (scRNA-seq)25 (Figure 3A) and spatial transcriptome analysis (Figure 5A) on APP/PS1 mice following 1 week of Staph exposure. We also measured plasma cytokine levels for infected and control mice at an acute phase (Figure S1E). The number of significantly altered cytokines following acute exposure was less than in chronic infection (Figure S1E). Some interleukins, such as IL-3, IL-4, and MCP-1, are initial responders to acute infection and stay elevated in the periphery during chronic exposure (see the full list in Figure S1E). Other cytokines, like TNF-α, were increased only in chronic exposure, and G-CSF and IL-5 were increased only in the acute phase.
Figure 3. scRNA-seq distinguishes major brain cell types in APP/PS1 brains.
(A) Acute scRNA-seq study outline.
(B) t-SNE plot showing 13 distinguished clusters of 35,698 cells after filtering and pre-processing using Seurat.
(C) Violin plots of marker gene expression, identifying cell types for each cluster (one representative marker gene shown for each cell type).
(D) Donut plots showing proportion of cells in each cluster from PBS and Staph groups.
(E) Heatmap showing top 5 differentially expressed genes for each cluster. PBS n = 4 and Staph n = 3.
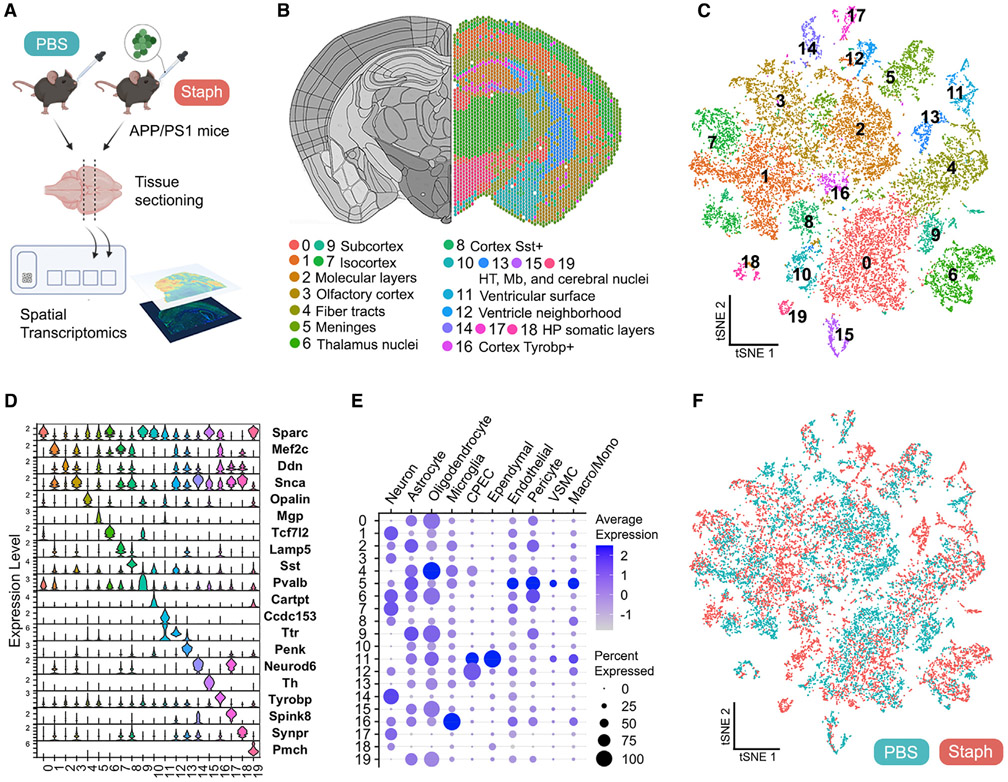
Figure 5. Spatial transcriptomics identifies discrete brain regions in APP/PS1 mice.
(A) Acute (1 week) spatial transcriptomics study outline. Two animals/group and two sections/mouse from a single hemisphere were used.
(B) Representative spatial plot of the 20 clusters identified (0–19) relative to the mouse brain atlas. HT, hypothalamus; Mb, midbrain; HP, hippocampus.
(C) t-SNE dimensional reduction of 20,800 spots labeled according to cluster identity.
(D) Expression levels (SCT normalized counts) of marker genes identified for each cluster.
(E) Presence of cell-type-specific gene set expression across clusters, showing gene set average expression (color intensity of dot) and percentage of spots where the gene set was detected (dot size).
(F) t-SNE plot showing 10,280 spots from the PBS (green) and 10,520 spots from the Staph (red) group.
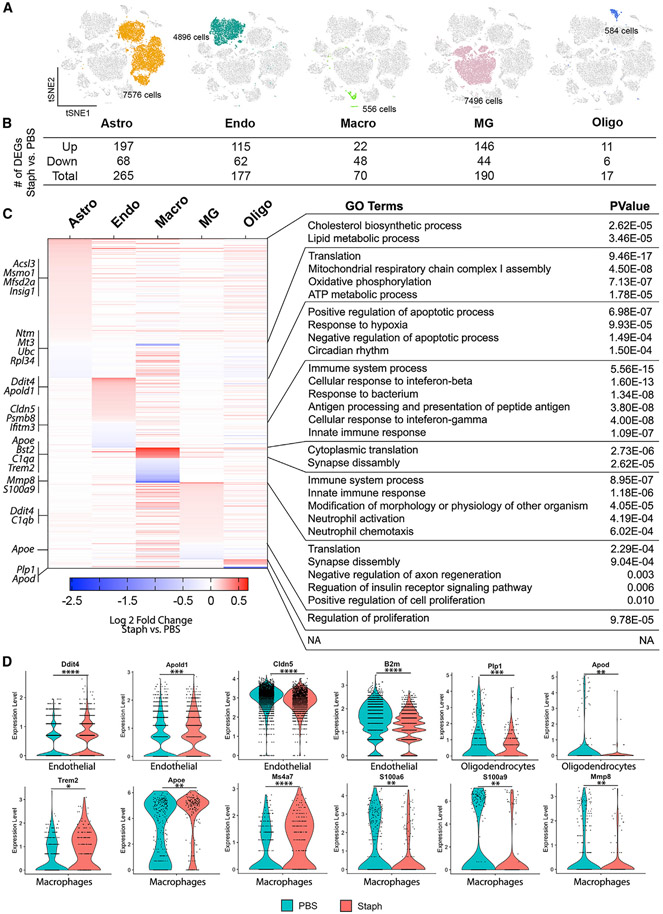
For scRNA-seq, cells were isolated from 7-month-old mice using the 10× Genomics platform (Figure 3A). After filtering, a total of 35,698 individual cells, with 1,667 genes and 3,893 UMI per cell on average, were arranged by t-SNE for visualization (Figure 3B). The infected and control groups had similar numbers of genes, reads, and percentages of mitochondrial genes (Figure S2A). Using unsupervised clustering, a total of 13 distinct clusters were identified across all samples. These clusters were manually identified by the expression of known cell-type-specific markers (Table S2) as microglia (clusters 0 and 6), astrocytes (clusters 1 and 3), endothelial cells (clusters 2 and 12), neurons (clusters 4 and 7), pericytes and VSMC (cluster 5), oligodendrocytes (cluster 8), macrophages and monocytes (cluster 9), CPECs (cluster 10), and ependymal cells (cluster 11) (Figures 3C and S2B-S2D). For most clusters, the Staph and PBS groups shared similar cell proportions with few exceptions. For example, the Staph-treated group had a higher proportion of cells in clusters 0 (microglia) and 3 (astrocytes) and fewer cells in clusters 1 (astrocytes) and 2 (endothelial cells) (Figure 3D). The expression heatmap (Figure 3E) displaying the top 5 markers of each cluster also confirmed the cell types assigned. Cluster 12 exhibited mainly endothelial and a few microglial markers, possibly due to unremoved doublets. For further analysis, cells expressing microglial markers were removed from cluster 12. Cluster 6 was excluded from further analysis because of cells with highly expressed ribosomal genes.
Staph infection triggers transcriptional changes in endothelial cells, microglia, macrophages, astrocytes, and oligodendrocytes
We next compared the Staph and PBS groups and examined transcriptional changes in different cell types. We examined in detail cell types in five clusters that showed the most robust response to infection, namely endothelial cells, microglia, macrophages, astrocytes, and oligodendrocytes (Figure 4A). scRNA-seq detected very limited transcriptional responses to Staph in neurons and other cell types (Figures S3A-S3D).
Figure 4. Staph treatment triggers transcriptional changes in endothelial cells, microglia, macrophages, astrocytes, and oligodendrocytes.
(A) t-SNE plots with each cell cluster and cell numbers labeled.
(B) Table showing the number of differentially up- or downregulated genes in each cell type.
(C) Heatmap (left) showing expression changes of DEGs in all five cell types in terms of Staph versus PBS and the GO terms (right) associated with them.
(D) Violin plots showing expression levels of example DEGs in the five cell types. The p value was determined by generalized linear model with Bonferroni correction. *adj. p < 0.05, **adj. p < 0.01, ***adj. p < 0.001, ****adj. p < 0.0001. PBS n = 4 and Staph n = 3.
First, in 4,896 high-quality endothelial cells, we identified 177 DEGs (Figures 4B and S3E) and gene ontology (GO) analysis demonstrated that functional categories upregulated by Staph were hypoxia response, apoptosis, and cell proliferation (Figure 4C). Top upregulated in the Staph group were endothelial-specific genes such as Tmem252, Ctla2a, Cavin2, Ddit4/Redd1, and Apold1. Ddit4/Redd1 (Figures 4D and S3E), a gene associated with hypoxia and apoptosis, was shown to promote macrophage infiltration and inflammation cytokine expression through NF-κB signaling.26 Apold1 (Figure 4D) is involved in endothelial functions, specifically regulating tight junctions,27 and its expression is affected by focal cerebral ischemia.28 Functional categories downregulated by Staph in the endothelial cell cluster were immune system process and response to IFN-β, with top downregulated genes B2m, Cldn5, Ly6a, and Ly6e (Figures 4D and S3E). Cldn5 is a key component of the tight junctions important for BBB permeability.29 Ly6a and Ly6e, also highly expressed in endothelial cells, were shown to mediate transport of adeno-associated viruses across the human BBB.30,31 A few IFN-signaling genes were downregulated in the Staph-infected group, however, by a small FC (Figure S3E).
Second, we investigated transcriptional changes in immune cell types: microglia and macrophages (Figures 4A-4C, S3F, and S3G). Overall, while there were more DEGs in microglia (190 DEGs) than in macrophages (57 DEGs), the effect measured by FC was higher in macrophages. For example, complement component 1 genes (C1qa, C1qb, and C1qc) and Ms4a6d were upregulated by Staph in both clusters, with much higher FC in macrophages than in microglia (Figures 4D, S3F, and S3G). In both clusters, synaptic assembly (encompassing the complement genes) was one of the top upregulated in the Staph group (Figure 4C). In addition to complement genes, top upregulated genes in macrophages (Figures 4C and 4D) were Ms4a genes, Apoe (log2FC = 0.67), and Bst2 (log2FC = 0.47). Interestingly, Apoe was among the downregulated DEGs in microglia, although with a small FC (Figure S3F). In contrast, downregulated categories and genes differed in macrophages and microglia. In macrophages, downregulated categories were immune response, with top genes such as S100as genes (Figures 4D and S3G). S100 proteins serve as damage-associated molecular pattern molecules (DAMPs) and are suggested to regulate chemotaxis and migration of macrophages,32 and S100a6, specifically, was found downregulated after peripheral inflammation. We conclude that the macrophage transcriptional response was stronger than in microglia probably because of direct contact with the infection.
Next, we examined the oligodendrocyte cluster (Figures 4A-4C and S3H), which showed 17 DEGs but with a significant FC of genes downregulated by Staph. Genes downregulated by Staph were Apod (Lipocalin, log2FC = −2.5) and Plp1 (Proteolipid protein [myelin] 1, log2FC = −1.9), both highly expressed in oligodendrocytes.33,34 A previous study demonstrated that the lack of Apod in mice results in altered compaction of the extracellular leaflet of myelin in both the CNS and the peripheral nervous system (PNS) and significant decrease in Plp1 expression.35 Among the upregulated genes were oligodendrocyte precursor genes, such as Pdgfra and Lhfpl3, in Staph versus PBS (Figure S3H).
Last, the astrocyte cluster was represented by 7,576 high-quality astrocytes and exhibited the highest number of DEGs (Figure 4B), although with a relatively small FC. GO analysis (Figure 4C) detected cholesterol sterol biosynthetic and lipid metabolic process as the top upregulated terms in response to Staph. Genes associated with these processes include Insig1, Lpcat, Msmo1, Crot, Acsl3, and Hmgcr (Figures 4C and S3I). Notably, Mfsd2a, a transporter involved in maintaining and regulating BBB,36 was also found upregulated with Staph treatment (Figure 4C), suggesting a possible interaction of astrocytes with endothelial cells. Consistent with endothelial cells and microglia, Ddit4 was upregulated in astrocytes (Figure S3I). In contrast, categories such as translation, mitochondrial respiratory chain complex, oxidative phosphorylation, and ATP metabolic process were downregulated by Staph in astrocytes.
Spatial transcriptomics reveals locally specific transcriptional signatures in APP/PS1 mouse brains
We next employed spatial transcriptomics to determine the brain region-resolved differential gene expression. We used brain coronal sections of APP/PS1 mice exposed to either PBS or Staph for 1 week (Figure 5A). The sections were processed for immunostaining followed by transcriptional analysis with spatial resolution through Visium spatial gene expression. After pre-processing and filtering, gene expression analysis was performed with 19,255 genes on 20,800 high-quality spots (Figure S4A). Graph-based clustering and t-SNE dimensional reduction identified 20 clusters (Figures 5B and 5C and Table S3). The spatial distribution of each cluster was associated with specific brain structures identified based on the Allen Mouse Brain Atlas37 (Figures 5B and S4B). We found that cortical regions were mostly represented by clusters 1 (inner layers of isocortex), 2 (molecular layers of hippocampus and cortex), 3 (olfactory/entorhinal areas and subcortical plate), and 7 (outer layers of isocortex). The somatic layers of the hippocampus (HP) were also resolved by clusters 14 (CA3), 17 (CA1), and 18 (DG). We combined the average expression of cell-type-specific markers on gene sets, which confirmed the enrichment of neuronal gene expression (Mef2c and Snca) in the cortical clusters (Figures 5D, 5E, and S4C). The analysis also resolved two clusters that were present in the cortical region but lacked spatial orientation: clusters 8 and 16. Cluster 8 was identified as Sst+ spots enriched in inhibitory neurons (Figures 5D and S4C). Cluster 16 expressed high levels of disease-associated microglia (DAM) genes,38 such as Tyrobp, Ctsb, and Ctss, and was enriched in microglial-specific genes (Figure 5E). We identified fiber tract spots (cluster 4) enriched in oligodendrocyte genes and surface meninges (cluster 5) (Figures 5D, 5E, and S4C). The ventricular regions, represented mainly by lateral ventricles (cluster 11), were enriched in CPECs and ependymal-specific genes (Ttr, Ccd153, and Enpp2); in addition, cluster 12, which was present in variable ventricle-neighborhood areas, was enriched in Ttr (Figures 5D and 5E).
Spots associated with subcortical regions relative to thalamus, hypothalamus, midbrain, and cerebral nuclei (clusters 0, 6, 9, 10, 15, and 19) generally expressed high levels of genes known for subcortical enrichment (Sparc, Resp18, and Nap1l5)39 40 (Figures 5D and S4C) and also presented with relatively high astrocyte- and oligodendrocyte-specific gene expression (Figure 5E). Overall, the t-SNE plots of PBS and Staph spots showed overlaying grouped projections that did not demonstrate major anatomical functional changes between groups (Figure 5F).
Gene expression at the ventricular surroundings is markedly responsive to Staph infection
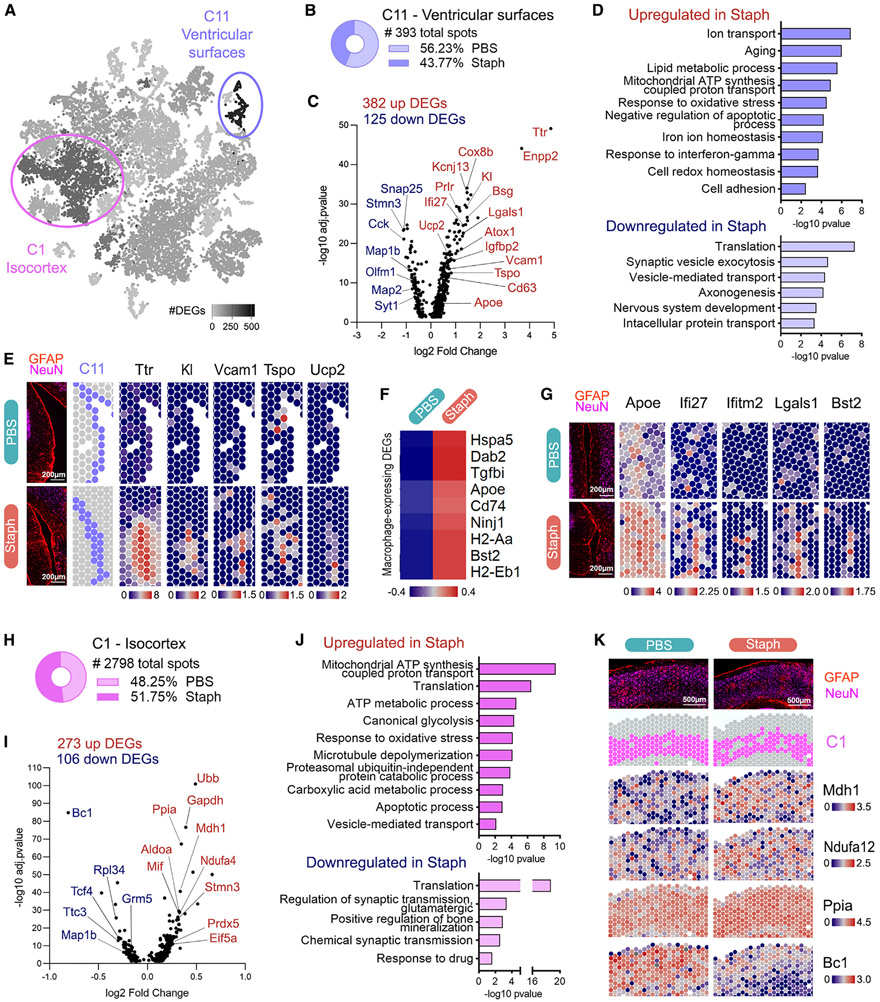
Next, we performed differential expression (DE) analysis at the cluster level comparing Staph and PBS groups. We found that the ventricular surface areas (cluster 11) had the highest numbers of DEGs between groups, followed by cortical areas (cluster 1) (Figure 6A and Table S3). Therefore, we first focused the analysis on cluster 11, which consisted of 393 spots similarly distributed across groups (Figure 6B). DE analysis showed 506 DEGs in the Staph versus PBS comparison (Figure 6C). GO analysis revealed that biological processes related to aging, ion transport, lipid metabolism, redox response, and inflammation were upregulated by Staph at the ventricular surfaces in brains of APP/PS1 mice (Figures 6D and S5A-S5E), and most of these genes were CPEC specific.
Figure 6. Molecular responses to Staph peripheral inflammation in ventricular surroundings and cortical areas of APP/PS1 mouse brains.
(A) t-SNE dimensional reduction of spatial dataset representing the number of DEGs per cluster in Staph versus PBS. The two most affected compartments are highlighted: cluster 11 (507 DEGs) and cluster 1 (379 DEGs).
(B) Number of spots in cluster 11 and distribution (%) of spots between groups.
(C) Staph versus PBS differential expression in ventricular surfaces (cluster 11).
(D) GO terms associated with DEGs in Staph versus PBS ventricular surfaces (cluster 11).
(E) Spatial expression plots of Ttr and aging-associated genes upregulated in ventricular surfaces. Tissue immunofluorescence (GFAP/NeuN) and cluster 11 (C11) spot location are shown for comparison. Scale bars, 200 μm.
(F) Heatmap showing expression levels of macrophage specific DEGs in cluster 11.
(G) Spatial expression plots of macrophage/monocyte-associated genes in ventricular surfaces compared with tissue image (GFAP/NeuN). Scale bars, 200 μm.
(H) Number of spots and distribution (%) between groups of cluster 1 relative to the inner layers of isocortex.
(I) Staph versus PBS local differential expression in isocortex (cluster 1).
(J) GO terms associated with DEGs in Staph versus PBS isocortex (cluster 1).
(K) Spatial expression plots of cluster 1 DEGs compared with tissue image (GFAP/NeuN) and with cluster 1 (C1) spot location. Scale bars, 500 μm. Volcano plots for cluster-specific DEGs adj. p < 0.05. GO terms are shown with −log10 p value for each term (bars). Spatial plots and heatmap depict SCTransform-corrected and gene-level-scaled expression values, with scales indicated.
The top genes upregulated by Staph were CPEC-specific genes such as Ttr, Kl, Igfbp2, Kcnj13, Prlr, and Cox8b, indicating a CP-mediated response to acute peripheral inflammation (Figures 6C and 6E). Top functional categories upregulated by Staph were ion transport, aging, lipid metabolic process, oxidative stress, and response to IFN-γ. Upregulation of ion transport categories in the Staph group (Figures 6C and S5A) included Slc4a2, Kcne2, and Kcnj13 solute carriers important in maintaining ion and pH during CSF production. The second most affected category was aging, including Kl, Vcam1,41 Tspo,42 Ucp2,43 and Apoe44 (Figures 6C and S5B). The Staph-induced upregulation of Apoe44 at the ventricles (Figures 6C and S5B) probably comes from its expression in perivascular macrophages, as suggested by our scRNA-seq data, where we detected increased Apoe expression only in macrophages and not in microglia and astrocytes. Kl encodes the anti-aging protein Klotho,45 reported to regulate communication between the periphery and the brain at the blood-CSF barrier.46 Tspo (Figure 6E) encodes a mitochondrial cholesterol translocator, and its expression in glial and endothelial cells has been widely used in neuroimaging to depict the neuroimmune endophenotype of AD.42 In the Staph group, we also observed upregulation of the aging-related mitochondrial uncoupling protein-coding gene Ucp243 (Figure 6E), expressed in microglia and macrophages/monocytes, which regulates reactive oxygen species production rates. Together with Ucp2, the gene categories response to oxidative stress, response to iron, and glutathione metabolic process were upregulated by Staph at the ventricles (Figure S5D). The cell adhesion category was upregulated in ventricles of Staph-exposed mice, with Vcam1 and Bsg as examples (Figures 6C and 6D). Vcam1 (Figure 6E) expression in endothelial cells is important for blood leukocyte-endothelial cell adhesion.
The lipid metabolic process category was upregulated by Staph infection and, concomitant with the increased expression of Apoe, it encompassed genes related to phospholipid biosynthesis, phospholipases, and phosphatidylinositol kinases (Figures 6D and S5C). Enpp2 is highly expressed in CPEC and is the second most upregulated gene in the Staph group (Figure 6C). Enpp2 encodes the CSF-soluble extracellular enzyme autotaxin that produces lipophilic signaling molecules regulating chronic neuroinflammation.47
Upregulated by Staph at the ventricular surfaces was response to IFN-γ and increased major histocompatibility complex (MHC) class II protein genes (Figures 6D and S5E). Antigen processing and presentation were similarly upregulated in the Staph group (Figure S5E). Many of those immunomodulatory players are expressed by blood-derived immune cells (shown on Figure 6F). Bst2 upregulation at the Staph ventricles also co-incides with the observed increased expression in macrophage cluster in single-cell data (see Figure 4). Together with the upregulation of macrophage markers like Dab2, Ninj1, and Tgfbi following Staph exposure (Figure 6F), our results suggest the initial stages of peripheral immune cell infiltration in the Staph group.41,48 We also observed upregulation of Lgals1 with Staph, expressed by CPECs and macrophages in scRNA data (Figure 6G and Table S1), which encodes an immunoregulatory lectin that reduces microglial activation through CD45 binding.49 Figure 6G highlights the Staph-induced upregulation of Apoe, Ifi27, Lgals1, and Bst2 genes relevant for immune regulation and macrophage responses at the ventricles.
Downregulated genes following Staph infection in cluster 11 were in categories related to synaptic vesicle exocytosis, axonogenesis, and nervous system development (Figures 6C and 6D).
Other highly vascularized brain areas can also be key players in the periphery-brain interplay. We identified 150 up- and 45 downregulated DEGs in the blood-vessel-enriched cluster 5 (related to meninges) (Figure S5F) when comparing Staph versus PBS. Hypoxia-associated genes (Cst3 and Itpr1) were upregulated and B2m was downregulated in Staph, similar to the observations for endothelial cells in the scRNA-seq analysis (see Figure 4).
To evaluate the dysregulation of brain barriers suggested by the transcriptomics data (bulk RNA-seq after chronic infection and single-cell and spatial transcriptomics data after acute infection), we next assessed the potential leakage of peripheral proteins, namely endogenous mouse IgG (150 kDa) and serum albumin (67 kDa), into the brain by IHC as previously described by Bien-Ly et al.50 We reasoned that these transcriptional changes initiated during the acute treatment will reflect on barrier function later in the disease course and thus selected chronically treated mice. We detected a significant increase in the level of IgG in Staph-infected mice compared with the PBS group (Figure S6A). The albumin level also showed a similar increasing trend following Staph exposure, although not significant (Figure S6B). These results confirmed disrupted functions of brain barriers, such as increased permeability.
Overall, results demonstrated that this brain area is strongly responsive to peripheral infection and inflammation, possibly due to disbalances in blood-CSF barrier, lipid, and redox homeostasis and neuroinflammation, with potential consequences to neuronal function.
Metabolic responses in brain cortex following Staph infection
Cluster 1, representing the cortical regions of the brain, had 379 DEGs in Staph versus PBS groups and consisted of a large population of 2,798 spots spatially distributed at the inner layers of the isocortical areas (Figures 6H-6K).51 The isocortex was also sampled by cluster 7, with 1,211 spots located in the outer layers (Figure S5K). The spatial cluster 1 had similar abundances for both groups and was mostly represented by neuronal transcripts (Table S3). Top DEGs upregulated in the Staph group included genes highly expressed in neurons, such as Gapdh, Cck, Snrpn, and Gng3 (Figure 6I). The top upregulated biological processes associated with Staph consisted of metabolic GO terms such as mitochondrial ATP synthesis, glycolysis, and carboxylic acid metabolic process (Figures 6J and S5G-S5J). The increased expression of seven members of the NDUF family of mitochondrial complex 1 and four members of the ATP synthase gene family demonstrates the consequences of Staph exposure to oxidative phosphorylation (Figures S5G and 6K). These changes triggered by Staph were accompanied by upregulation of the response to oxidative stress category and activation of Ppia, Cck, Ypel3, and Dynll1 genes linked to the energy-demanding apoptotic process (Figures 6J and S5H).
These cortical molecular responses were accompanied by downregulation of genes linked to neuronal function in the Staph group and categories such as regulation of synaptic transmission and translation (Figures 6J, S5I, and S5J). The brain cytoplasmic Bc1 non-coding RNA regulates protein synthesis and plays a role in plasticity and learning52 and was the top downregulated gene in cluster 1 following Staph infection (Figure 6K).
Spatial transcriptomics identifies Aβ plaque-associated changes in APP/PS1 brains after acute Staph infection
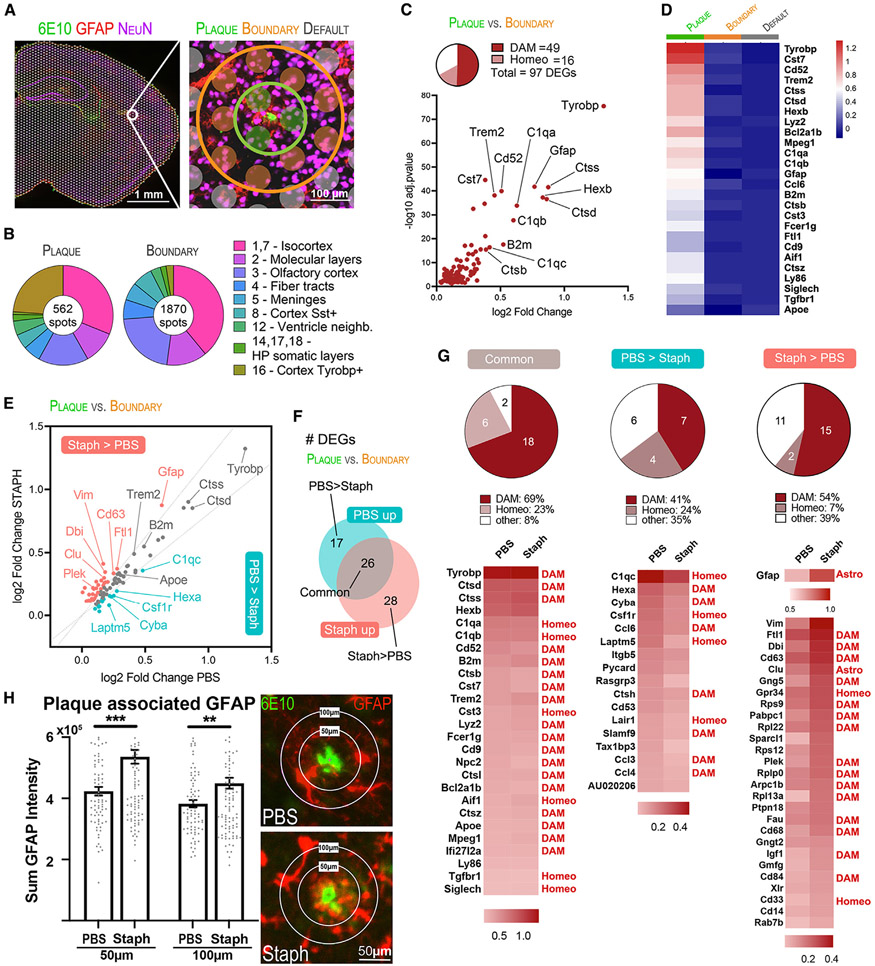
Since chronic inflammation triggered by long-term Staph infection resulted in increased amyloid pathology, we wondered if short-term infection could also indicate plaque alterations. We next investigated how the Aβ plaque niche transcriptome is affected by acute peripheral inflammation. To select plaque-associated spots and compare them with their neighboring areas, we stained Aβ deposits using 6E10 antibody and utilized the STUtility interactive interface to manually select spatially adjacent spots,53 thus identifying each spot according to its spatial distribution relative to Aβ plaques. Spots were labeled as plaque associated (plaque), plaque boundary associated (boundary), and independent of plaques (default) (Figure 7A). By comparing clusters and plaque-associated selections, we found that cortex Tyrobp+ (cluster 16) was spatially associated with Aβ plaques (Figures 7B and S7A).
Figure 7. Amyloid plaques promote a regional upregulation of disease-associated genes in Staph and PBS APP/PS1 mouse brains.
(A) On the left, immunostaining for Aβ (6E10, green), astrocytes (GFAP, red), and neurons (NeuN, purple). On the right, representative selection of plaque microenvironment (green), plaque boundaries (orange), and plaque-independent default spots (gray). Scale bar, 100 μm.
(B) Proportion of spots from each cluster for plaque and boundary selections.
(C) Scatterplot of plaque versus boundary independent of treatment DEGs.
(D) Expression levels of top plaque versus boundary DEGs in plaques, boundaries, and default spots. The heatmap depicts SCTransform-corrected and gene-level-scaled expression values, with scales indicated.
(E) Scatterplot showing plaque-associated DEGs in Staph and PBS groups. DEGs were categorized as higher in Staph than in PBS plaques (Staph/PBS log2FC ratio ≥ 1.20, shown in red), higher in PBS than in Staph plaques (Staph/PBS log2FC ratio ≤ 0.80, shown in green), or commonly affected in Staph and PBS (shown in gray).
(F) Venn diagram showing number of plaque-associated DEGs (plaque versus boundary) that are common in both groups, higher in PBS, or higher in Staph.
(G) Pie charts (top) and heatmaps (bottom) showing log2FC (plaque versus boundary) of DEGs categorized as common, higher in PBS, or higher in Staph.
(H) GFAP immunostaining in the brains of the PBS compared with the Staph group. Scale bars, 50 μm. Bar plots show the sum of GFAP intensity in 50 and 100 μm radii around plaques (n = 4 section/group). Statistical analysis was performed with unpaired t test. **p < 0.01, ***p < 0.001.
We then compared the gene expression of all plaque versus boundary spots to uncover Aβ plaque-associated molecular signature (Figures 7C and 7D and Table S3). In agreement with previous observations,51,54 we identified a clear DAM signature at the plaque microenvironment of APP/PS1 mice. Most of the 97 DEGs detected at plaques (all upregulated) were microglial associated: 16 homeostatic (homeo) and 49 DAM genes. The top DAM genes upregulated at plaques compared with boundaries were Tyropb, Ctss, Ctsd, B2m, Cd52, Trem2, Ctsb, and Cst7, with similar differences in expression level when comparing plaque to default spots. Plaque-associated upregulation of common markers for microglia (Hexb and Aif1), astrocytes (Gfap, Vim, and Clu), and macrophages (Lyz2 and Ccl6) was in line with the known recruitment of these cell types to Aβ plaque sites.38,54
Next, we performed plaque versus boundary DE analysis individually for each exposure group, which defined unique Aβ plaque-associated molecular signatures for both Staph and PBS. We then compared the FC values of all plaque-associated genes between the two groups to better understand if Staph induced differences in terms of effect size (Figures 7E-7G and Table S3). This demonstrated that there were 26 DEGs commonly upregulated at plaques in both conditions (Figure 7F). We also identified 17 and 28 DEGs that were relatively increased in PBS (PBS > Staph) and Staph (Staph > PBS) plaques, respectively (Figure 7F). Relative to microglial features, classical DAM genes38 such as Tyrobp, Trem2, B2m, Cst7, Mpeg1, and different cathepsins were very similarly upregulated at plaques in both groups (Figures 7E and 7G). The average log2FC values of Apoe were very similar, even though it was statistically significant only in Staph, and we consider it equally upregulated in both groups. C1qa, C1qb, Cst3, and Aif1 homeostatic genes were also similarly affected at plaque spots of both groups (Figure 7G). The list of DEGs in PBS > Staph included C1qc, Hexa, Cyba, Csf1r, and several cytokines (Figure 7G). We found that DEGs categorized as Staph > PBS included proportionally more DAM and fewer homeostatic genes relative to the group classified as PBS > Staph (Figure 7G). We also observed that a number of plaque-associated DAM genes had a higher FC near Staph than PBS plaques. These genes are involved in intracellular vesicular transport (Cd63 and Cd68) and immune signal transduction (Gng5, Cd63, Cd84, and Igf1). Interestingly, Gfap was one of the top plaque-associated genes that was upregulated at a higher FC in Staph compared with PBS and included in the Staph > PBS group together with other genes functionally associated with astrocytes, such as Sparcl1, Clu, and Vim (Figures 7E and 7G). Immunofluorescence analysis of GFAP staining confirmed its increased protein level at the Staph plaque niche relative to PBS, suggesting an increased astrogliosis following acute Staph exposure (Figure 7H). Cytoscape analysis (Figures S7C and S7D) of Staph-affected DEGs at the level of the plaque showed fewer connections centered around Rpl22, Rpl13a, Rplp0, Rps9, Ptpn18, Dbi, and Fau. There were also a larger number of non-microglial genes (astrocyte or macrophage) that were increased in Staph, suggesting recruitment of other cell types after infection. The cytoscape for the genes affected in the PBS group was mostly microglia specific and showed high levels of connectivity, suggesting a well-defined “classical” DAM response.38 This may explain why, even with these increased DAM responses in Staph, we still observe an increase in amyloid progression in the chronic model.
We conclude that the acute Staph peripheral infection leads to specific molecular changes at amyloid plaques, with increased DAM and astrocyte responses.
DISCUSSION
There is considerable evidence supporting the involvement of the central and peripheral immune systems in AD pathogenesis (reviewed in Bettcher et al.2 and Xie et al.3). Our results employing the APP/PS1 mouse model with intranasal infection reveal cell-type- and spatially specific transcriptional responses that indicate impacts on brain barriers of both the BBB and blood-CSF and may connect peripheral inflammation to AD pathology. Spatial transcriptomics allowed us to investigate molecular alterations in situ at selective brain niches, like the ventricular surfaces that play central roles in the brain-periphery interplay and Aβ plaque microenvironment. Together with scRNA-seq, spatial transcriptomics provides higher resolution to move the frontier of knowledge in AD research forward.
BBB
The brain barriers are fundamental players in the communication between brain parenchyma and periphery. The results of this study have shown that peripheral Staph inflammation triggers molecular responses in cell types with critical roles in BBB integrity. We found upregulation of BBB-related endothelial genes (Ddit4 and Apold1) and functions, such as apoptosis and hypoxia. Hypoxia response increases BBB permeability and is associated with blood flow disruption and rapid depletion of essential nutrients.8 Cldn5, encoding an important tight junction protein, was found downregulated with Staph treatment. Together with other claudins, claudin-5 forms different sizes of elliptic meshes to regulate macromolecules passing through the tissue barrier; thus, low levels of tight-junction proteins such as claudin-5 likely lead to increased BBB permeability. These endothelial-specific gene responses indicate dysregulated BBB transport and permeability with acute Staph treatment, likely to precede compensatory responses like the observed upregulation of claudins and integrins in the chronic Staph infection. Recently, dysfunctional claudin-5 and impaired BBB permeability have been implicated in the pathogenesis of schizophrenia, depression, and AD.55
Blood-CSF barrier
Spatial transcriptomics data demonstrated that the CP and its ventricular microenvironment also respond to acute Staph inflammation, showing upregulated CPEC-specific genes and ion transport genes. Upregulation of genes from the SLC and carbonic anhydrase families in CPECs has been reported in brain samples from patients with AD, while the expression levels of Slc12a2, Slc4a5, and Aqp1 in mouse CP are age dependent.16 CPEC-linked solute carrier transporters play fundamental roles in maintaining composition and ion and pH levels of the CSF in a variety of physiological and pathological conditions. CSF pH naturally decreases with age, and lower pH is associated with amyloid pathology in mice and humans.56 According to a recently published study, the infusion of young CSF in aged mice improved memory function.57 Among DEGs upregulated in response to acute Staph treatment, Ttr and Kl, both expressed in CP, deserve more attention. Ttr is upregulated in brain as an important stress response mechanism,58,59 and the protein inhibits Aβ fibril formation.60 The anti-aging gene Kl, whose levels are negatively correlated with age and neurodegenerative conditions like AD, participates in distinct signaling pathways (reviewed by Prud’homme et al.61). The expression levels of Kl in CP were also shown to influence multiple proinflammatory factors and macrophage infiltration at the blood-CSF barrier.46 Therefore, following Staph peripheral inflammation, the observed CPEC-associated gene responses in ventricles may indicate disbalances in blood-CSF barrier protein and CSF composition, suggesting the upregulation of protective genes such as Ttr and Kl, possibly as a compensatory mechanism. The ventricular areas also displayed upregulation of genes expressed by different cell types and associated with lipid metabolism (Apoe, Acaa2, and Acadm), oxidative stress response (Ppia, Ucp2, and Atox1), and immune regulation (Ifitm2, Ifi27, Lgals1, Tspo, Bst2, and Cd63), which occurred in parallel with the downregulation of neuronal-associated genes. Related to these transcriptional changes was our finding of an increased leakage of endogenous mouse IgG into the brain after the chronic treatment, confirming disrupted functions of brain barriers, such as increased permeability. Altogether, these changes indicate that the acute infection induces molecular disbalances in the blood-CSF barrier that lead to disturbance of brain barriers if the infection continues.
Inflammatory response
It has been suggested that peripheral immune cells can pass the blood-CSF barrier, a process facilitated by adhesion molecules such as VCAM1 secreted by CP cells.48 Spatial transcriptomic data demonstrated increased levels of Vcam1, similar to previous observations in conditions of infection,41,62 upregulation of macrophage markers and antigen-processing and presentation pathways, suggesting increased immune infiltration in the CSF-brain surfaces triggered by Staph. Recently, Yang et al. demonstrated that, upon COVID-19 infection, the human CP relayed peripheral inflammatory signals into the brain and facilitated the infiltration of peripheral immune cells, presumably through increased antiviral IFN and upregulation of antigen-processing pathways.17 In our study, we found significantly increased IFN-γ plasma and brain levels in response to chronic Staph exposure. Both spatial and single-cell data showed upregulation of the IFN-induced macrophage Bst2 gene, crucial for both innate and adaptative immunity. IFN-induced transmembrane proteins (IFITMs) are a family of small proteins that localize in the plasma- and endolysosomal membranes and inhibit viral entry into host cells and also reduce the production of infectious virions,63 and Ifitm3 was shown to modulate γ-secretase activity by increasing Aβ production.64 Here, we observed that Ifitm2 was upregulated by acute Staph exposure in ventricular areas and in bulk sequencing results following chronic infection. In contrast, Ifi47, Ifit1, and Ifitm3 genes were downregulated in endothelial cells, indicating that IFN signaling may not be homogeneous following peripheral inflammation and have a cell-specific effect. In line with the increased vascular or peripheral macrophage activation in scRNA-seq data is the increased expression of Apoe in macrophages, but not microglial clusters, suggesting that the infection has a direct effect on macrophages. Related to the inflammatory response is the increased CD163 immunostaining after chronic infection, which was localized mainly in close proximity to blood vessels, suggesting possible macrophage infiltration. Our scRNA-seq data placed Cd163 expression only in the macrophages, but recent single-nuclei RNA-seq studies utilizing human AD brain samples identified a distinct amyloid-responsive microglial subpopulation that expressed CD163 along with many of the DAM signature genes that could be unique to human AD brain.65,66
Effects on amyloid plaque
Chronic treatment with Staph aggravated amyloid pathology in APP/PS1 mouse brains, and spatial transcriptomics results showed that the acute treatment induced transcriptional responses in the Aβ plaque niche. Relative to Aβ plaque-associated microglia, spatial transcriptomics results demonstrated that Staph treatment diminished the contribution of homeostatic genes at the plaque niche and increased the DAM contributions. In a previous study, Chen et al. conducted spatial transcriptomics assays using AD mouse brains and identified 57 plaque-induced genes.54 In our spatial transcriptomics data, we found 97 plaque-associated DAM genes, and 37 of those were in the list reported by Chen et al.54 The list of common genes included Tyrobp, Apoe, Trem2, and Cst7 DAM genes. Our spatial data showed increased expression of astrocyte-associated genes Gfap, Sparcl1, Vim, and Clu at plaques in the Staph group. The increased expression of Gfap, which was at the top of the list Staph > PBS (Figure 7G), was validated using IHC, and collectively, the data indicate an increase in astrogliosis after short-term infection. Importantly, as demonstrated by the cytoscape analysis, even with the increased percentage of DAM responses in the Staph group, we still observed an increase in amyloid progression in the chronic model, suggesting the microglial response to Staph treatment is different compared with the classical response to neurodegeneration.38 Therefore, acute Staph peripheral infection in addition to classical DAM/homeo response is likely linked to plaque-associated reactive astrocyte responses, and the brain immune adaptations in our chronic infection model are associated with discontinued stimulus to astrocyte activation but increased microgliosis and amyloid pathology. We did not observe similar effects on astrocyte and microglial clusters of single-cell sequencing data, indicating that these gene responses are restricted to Aβ plaque niches. Thus, the results of our study demonstrated peripheral inflammation-induced changes in the plaque microenvironment that accompany the increase in amyloid pathology triggered by Staph.
Neuronal function
Chronic Staph treatment triggered downregulation of gene networks linked to neuronal function, which was similarly observed by spatial transcriptomics following acute treatment. This highlights the vulnerability of cortical neurons to peripheral inflammation in the AD context. The downregulation of neuron-specific genes was observed in cortical areas and ventricular surroundings. Such mechanisms were suggested also by astrocyte and oligodendrocyte signatures from scRNA-seq data and the upregulation of nervous system development and axon guidance terms observed in chronic Staph exposure. In addition, the cortical areas exhibited increased expression of ATP-, mitochondrial-, and oxidative phosphorylation-linked processes.
In conclusion, we demonstrate that low-grade peripheral infection triggers transcriptional responses linked to brain barriers and changes in brain-infiltrating/resident macrophages that indicate their fundamental importance in the mechanisms linking peripheral inflammation and AD pathogenesis. Finally, we showed that bacterial infection caused molecular changes restricted to specific cell types and/or brain spatial microenvironments, which highlights the relevance of studying brain molecular responses with high resolution to reveal new insights into AD etiology.
Limitations of the study
Our study has several limitations. First, we confined our investigation to an AD mouse model to assess the impact of peripheral inflammation on amyloid pathology and its correlation with brain transcriptome. As a result, we did not consider or investigate how peripheral infection would affect wild-type mice. Second, the short Staph exposure experiment helped distinguish acute effects from the potential compensation mechanisms following chronic exposure, but the dynamics of amyloid plaques after 1 week of treatment are technically challenging to assess. Instead, we found increased DAM signatures as well as transcriptional effects on astrocytic and macrophage-associated genes around the plaque niches following acute infection. Third, our bulk RNA-seq data showed a significant decrease in Cd163 gene expression following chronic Staph exposure, while the IHC staining displayed an increase in CD163. We were unable to conclude whether the CD163 signal in our IHC staining is a soluble form or membrane bound in macrophages. The biological significance of full-length membrane-bound versus soluble forms of CD163 remains unknown, and further research is needed before interpreting these findings.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Radosveta Koldamova (radak@pitt.edu).
Materials availability
The study did not generate new unique reagents.
Data and code availability
Raw data from bulk-, single-cell RNA-seq, and spatial transcriptomics were deposited at GEO and are publicly available as of the date of publication (GSE218346, GSE218352, and GSE218360). Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD163 antibody, mouse (IHC) | Abcam | Cat# ab201461 |
| Biotinylated anti-mouse antibody (IHC) | Vector | Cat# BA-2000; RRID:AB_2313581 |
| Anti-6E10 biotinylated antibody, mouse (IHC+Visium) | Biolegend | Cat# 803007; RRID:AB_2564657 |
| Mouse albumin polyclonal antibody (IHC) | Bethyl Laboratories | Cat# A90-134; RRID:AB_67120 |
| Goat anti-mouse IgG antibody conjugated with Alexa 488 (IHC) | Invitrogen | Cat# A-11001; RRID:AB_2534069 |
| Anti-IBA1 antibody, rabbit (IHC) | WAKO | Cat# 019-19741; RRID:AB_839504 |
| Donkey anti rabbit Alexa 594 (IHC) | Invitrogen | Cat#A-21207; RRID:AB_141637 |
| Donkey anti goat, Alexa 488 conjugated (IHC) | Invitrogen | Cat# A32814; RRID:AB_2762838 |
| Donkey anti mouse, Alexa488 conjugated (IHC+Visium) | Thermo Fisher Scientific | Cat # A-21202; RRID:AB_141607 |
| Alexa 594 Anti-GFAP antibody (Visium) | Abcam | Cat# ab302663 |
| Alexa 694 Anti-NEUN antibody (Visium) | Abcam | Cat# ab190565; RRID:AB_2732785 |
| Bacterial and virus strains | ||
| Staphylococcus aureus | Provided by Anatoliy Gashev, MD, PhD | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| BD BBL™ Trypticase™ Soy Agar | Fisher Scientific Edge | Cat# B11043 |
| Kanamycin | Thermo Fisher Scientific | Cat# J1792406 |
| Phosphate buffered saline (PBS) | Gibco | Cat# 10010-023 |
| Isoflurane | Covetrus | SKU# 029405 |
| Tribromoethanol | ACROS Organics | CAS# 75-80-9 |
| 2-Methyl-2-butanol | Sigma-Aldrich | CAS# 75-85-4 |
| EDTA | Fisher Chemical | Cat# S311500 |
| Phosphate-buffered paraformaldehyde | Electron Microscopy Sciences | Cat# 15714S |
| Ethanol | Decon laboratories | CAS# 64-17-5 |
| Sucrose | Fisher Scientific | Cat# S5-3 |
| OCT | Fisher Healthcare | Cat# 23-730-571 |
| Formic acid | MP Biomedicals | CAS# 64-18-6 |
| H2O2 | Fisher Scientific | CAS# 7722-84-1 |
| Normal Goat Serum | Jackson ImmunoResearch | Cat# 005-000-121 |
| Normal Horse Serum | Vector | Cat# S-2000 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat# 017-000-121 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat# BP1605-100 |
| TritonX-100 | Sigma-Aldrich | CAS# 9036-19-5 |
| Vector ABC kit | Vector | Cat# PK-6100 |
| DAB substrate kit | Vector | Cat# SK-4100 |
| Tween20 | Fisher Scientific | Cat# 337-500 |
| Hank’s Balanced Salt solution (HBSS) | Gibco | Cat# 41025-092 |
| X-34 | Provided by William E. Klunk, MD, PhD. | N/A |
| Sodium Citrate Dihydrate | Fisher Scientific | Cat# S279-500 |
| Critical commercial assays | ||
| Mouse Cytokine 32-Plex Discovery Assay (plasma) | Eve Technologies | SKU# MD32 |
| Mouse Cytokine 10-Plex Discovery Assay (brain) | Eve Technologies | SKU# MDF10 |
| RNeasy mini kit | Qiagen | Cat# 74104 |
| Neural Tissue Dissociation Kit (P) | Miltenyi | Cat# 130-092-628 |
| Myelin Removal Beads II | Miltenyi | Cat# 130-096-733 |
| Dead Cell Removal Kit | Miltenyi | Cat# 130-090-101 |
| Chromium Next GEM Single Cell 3’ Kit v3.1 | 10x Genomics | Cat# PN-1000269 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | Cat# PN-1000127 |
| Visium Spatial Gene Expression Slide & Reagents Kit | 10x Genomics | Cat# PN-1000187 |
| Bioanalyzer High Sensitivity DNA kit | Agilent Technologies | Cat# 5067-4626 |
| Bioanalyzer RNA 6000 Nano kit | Agilent Technologies | Cat# 5067-1511 |
| Deposited data | ||
| Raw bulk RNA-seq | This paper | [GEO]: [GSE218346] |
| Raw scRNA-seq | This paper | [GEO]: [GSE218352] |
| Raw Spatial Transcriptomics data | This paper | [GEO]: [GSE218360] |
| Experimental models: Organisms/strains | ||
| Transgenic mice: APP/PS1ΔE9 [B6.Cg-Tg(APPswe,PSEN1dE9)85Dbo/Mmjax] | The Jackson Laboratory | MMRRC Strain #034832-JAX |
| Software and algorithms | ||
| Prism v 8.2.0 and 9.4.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Nikon NIS-Elements Advanced Research software | Nikon Instruments Inc. | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research |
| Subread (v1.5.3) | Liao et al.67 | https://subread.sourceforge.net/ |
| Rsubread v1.34.2 | Liao et al.68 | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| DESeq2 v1.24.0 | Love et al.69 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| EdgeR v3.26.5 | Robinson et al.70 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8) | Huang da et al.71 | https://david.ncifcrf.gov |
| Cell Ranger v6.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-dna/software/downloads/latest |
| Seurat v4.1.0 | Hao et al.72 | https://satijalab.org/seurat/ |
| DoubletFinder v2.0 | McGinnis et al.73 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Space Ranger v1.3.1 | 10x Genomics | https://support.10xgenomics.com/spatial-gene-expression/software/downloads/latest |
| STutility | Bergenstråhle et al.53 | https://ludvigla.github.io/STUtility_web_site/ |
| GeneMANIA prediction server | Warde-Farley et al.74 | https://genemania.org/ |
| Other | ||
| Cryotome | Center for Biologic Imaging at University of Pittsburgh | N/A |
| Superfrost plus slides | Fisher Scientific | Cat# 12-550-15 |
| Nikon Eclipse 90i microscope | Nikon Instruments Inc. | Cat# 25248 |
| Keyence BZ-X810 All-in-One Fluorescence Microscope | Keyence Co. | Cat# BZ-X810/BZ-X800LE |
| 2100 Bioanalyzer | Agilent Technologies | Cat# G2939BA |
| EVOS M5000 Imaging System | Thermo Fisher Scientific | Cat# AMF5000 |
| Chromium Controller | 10X Genomics | Cat# 1000204 |
| Illumina NovaSeq | Novogen Co. Ltd. | N/A |
| Illumina NextSeq 550 | Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh | N/A |
| Illumina NovaSeq | MedGenome Inc. | N/A |
| Countess 2 FL Automated Cell Counter | Thermo Fisher Scientific | Cat# AMQAF1000 |
| 40μm Cell Strainer | Corning | Cat# 431750 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
APP/PS1ΔE9 [B6.Cg-Tg(APPswe,PSEN1dE9)85Dbo/Mmjax] transgenic mice (referred to as APP/PS1) were purchased from The Jackson Laboratory (USA) and animals for experimental use were bred in-house and used experimentally as heterozygous. These mice have the Mutations: APP K670_M671delinsNL (Swedish), PSEN1:deltaE9 mutations and display cortical amyloid plaques at 3 months, and increase in size and number with age. Experimental animals were housed with 5 females or 4 males per cage and breeders were house with 2 females and 1 male per cage. All animals were litter mates and housed with a 12-hour light/dark cycle with ad libitum food and water. All experimental animals entered the study as drug and test naïve and normal health status. Animals were housed in specific pathogen free rodent facility maintained by the University of Pittsburgh Animal Care and Use Program, through the DLAR. For all experiments, both male and female mice were used. All animal procedures were performed in accordance with the guidelines outlined in the Guide for the Care and Use of Laboratory Animals from the United States Department of Health and Human services and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
METHOD DETAILS
Bacterial growth
Staphylococcus aureus Xen29 isolated from 2-3 colonies grown on Trypticase Soy Agar (TSA) plates were grown overnight in Trypticase Soy Broth supplemented with 200 μg/mL kanamycin at 37°C with ambient aeration at 200 rpm. A 50 mL subculture was grown to log-phase (1.5-3 hours) until OD600 read 0.5 (1 x 108 CFU/mL). Bacteria was centrifuged for 10 minutes at 4000g and resuspended in a volume of sterile PBS equal to 10% of original sample (i.e. 50 mL culture resuspended in 5 mL). Bacteria was centrifuged again for 2 minutes at 12000g and resuspended in a volume of sterile PBS equal to 10% of original sample (i.e. 5 mL culture resuspended in 500 μL). Of the resulting bacteria suspension, 10 μL was used to inoculate each mouse at a 1 x 108 CFU. 50 μL was spot plated on TSA + kanamycin in duplicates to confirm dose administration.
Inoculations
For chronic inflammation, 3-month-old APP/PS1 mice were anesthetized with 5% isoflurane for 30 seconds and held in a supine position at a 45° angle to the floor. 5μL of S. aureus suspension or PBS control was introduced into each nostril 1 droplet at a time, for a total inoculation of 10μL per animal. Each droplet was inhaled by the animal before the next was added to insure inoculation of the proper area. Mice were placed back in their cage in a prone position and monitored until awake. Mice were treated twice a week for 16 weeks.
For acute inflammation, 6.8-month-old APP/PS1 mice were inoculated in the same manner as above but only given four inoculations, every other day, over an 8-day period.
Perfusions
Mice were anesthetized by IP injection of Avertin (1.25% tribromoethanol, 2.5% 2-methyl-2-butanol, 250mg/kg of body weight). Blood was collected via cardiac puncture of the right ventricle with EDTA treated syringes followed by transcardial perfusion with 20mL of 0.1M PBS, pH 7.4. The blood was centrifuged for separation of the plasma and a nasal lavage was performed with 500μL of sterile PBS and flash frozen on dry ice. For the nasal lavage, a longitudinal cut of the skin was made exposing the trachea followed by a transverse cut of the trachea half-way up. A cannulated syringe containing 500 μl of sterilized PBS was inserted into the trachea towards the nose and sample collection tube was placed beneath the nose of the mouse. To test correct placement of needle, 20 μl of PBS lavage fluid was administered and a drop of fluid visualized at the nares of the mouse. Then the sterile PBS was dispensed rapidly, allowing for displacement and maximal collection of bacteria, and contents were collected into the sample tube. The brain was removed and divided into hemispheres with one hemisphere dissected into the cerebellum, subcortical, hippocampus and cortex regions and flash frozen on dry ice. The other hemisphere was drop fixed in 4% phosphate-buffered paraformaldehyde at 4°C for 48 hours before being moved 30% sucrose until sectioning. Animals in the acute inflammation group had one hemisphere of their brain separated for spatial transcriptomics and the other for single cell RNA analysis.
Nasal lavage plating
Nasal lavage samples were frozen at −20°C until they were ready to be plated on TSA plates supplemented with 200μg/mL kanamycin. 500μL nasal lavage samples were centrifuged for 10 minutes at 5000g and resuspended in 50μL sterile PBS before plating. Each plated lavage sample was grown for 12 hours at 37°C before being imaged to confirm colonization of S. aureus in the nasal passage of the animals.
Cytokine profiling
Plasma collected from PBS controls and S. aureus exposed animals (both chronic and acute groups) were run with the Mouse Cytokine 32-Plex kit (Eve Technologies, Calgary, AB Canada) according to the manufacturer’s protocol. Brain tissues from the chronic exposed groups were run with the Mouse Cytokine 10-Plex Assay (Eve Technologies, Calgary, AB Canada) according to the manusfacturer’s protocol.
Histology
Fixed brains were mounted in OCT and cut on the coronal plane at 30μm using a frozen cryotome at the Center for Biologic Imaging at University of Pittsburgh. Series of 6 sections, 450μm apart, starting at approximately 150μm caudal to the first appearance of the dente gyrus were collected covering an area of the brain from bregma −1.25mm to bregma −3.95mm. Sections were stored in a glycol-based cryoprotectant at −20°C until used for histological staining.
One series of brain tissue sections were immunostained with Biotinylated 6E10 (Covance, USA) which identifies all Aβ species. Free floating sections were washed in PBS, then antigen retrieval was performed in 70% formic acid for 10 minutes followed by quenching in 0.3% H2O2. The tissues were then blocked in 3% Normal Goat Serum (Jackson ImmunoResearch, USA) in PBS with 0.2% TritonX-100 for 1 hour. Endogenous avidin and biotin were blocked and sections were incubated for two hours in 6E10 biotinylated antibody (1:1000). Tissue sections were then developed using the Vector ABC kit and DAB substrate kit (Vector, USA). Following immunostaining, sections were mounted onto superfrost plus slides (Fisher Scientific, USA) and coverslipped. Bright-field images were taken with a Nikon Eclipse 90i microscope at 4X magnification.
A second series of tissue was stained with X-34 to identify compacted Aβ plaques. Tissues were mounted onto superfrost plus sides (Fisher Scientific, USA) and washed in PBS before being stained with 100μM X-34. Slides were then destained for 3 minutes in 80% ethanol before being washed again and coverslipped. Fluorescent images were taken with a Nikon Eclipse 90i microscope at 10X magnification.
A third series of sections were stained with anti-CD163 antibody (Abcam, UK). The free-floating sections were washed, followed by antigen retrieval in Tris-EDTA buffer with Tween20 (10mM Tris base, 1mM EDTA solution, 0.05% Tween20) at 80°C for 30 min, then washed and quenched in 0.3% hydrogen peroxide for 10 min. The tissue was blocked in 5% Normal Horse Serum (Vector, USA) and incubated in CD163 antibody (1:100) overnight at 4°C. The sections were washed and transferred into secondary biotinylated antibody (1:250, Vector, USA) for 60min. Sections were washed again and developed with the Vector ABC kit and DAB substrate kit (Vector, USA) before mounting onto superfrost plus slides and cover slipped. Bright-field images were taken using a Keyence BZ-X810 All-in-One Fluorescence Microscope at 10x magnification.
A fourth and fifth series of sections were stained with either anti-IgG (Invitrogen, USA) or anti-albumin (Bethyl Laboratories, USA) antibodies, respectively. Free-floating brain sections were washed, then blocked in 5% donkey serum (Jackson ImmunoResearch Laboratories, USA) with 2% BSA (Fisher Scientifitic, USA) in PBS for 1 hour. For albumin staining, sections were incubated with goat anti-mouse albumin antibody (1:300) for 24 hours at 4°C, then washed and transferred into secondary donkey anti-goat Alexa 488 (Invitrogen, USA) for 1 hour. For IgG staining, sections were washed and incubated in goat anti-mouse IgG antibody conjugated with Alexa 488 (1:300) for 24 hours at 4°C. Finally, sections were washed and mounted onto superfrost plus slides (Fisher Scientific, USA) and coverslipped. Fluorescent images were taken using a Nikon Eclipse 90i microscope at 20X magnification.
All images were analyzed using Nikon NIS elements software (Nikon Instruments Inc., USA) with thresholds set to detect positive staining while not detecting background. Regions of interest (ROI) were drawn around the cortex, hippocampus, or subcortical for each section and ROI area, detected staining area and subsequent percent staining area for each ROI was determined.
A sixth series of sections were immunostained with anti-IBA1 antibody (Wako, USA). Free-floating sections were washed, then antigen retrieval performed in sodium citrate buffer at 80 °C for 60 min, blocked in normal donkey serum (Jackson Lab, USA) for 1 h, and finally incubated in IBA1 antibody (1:1000) overnight at 4 °C. Sections were washed and transferred into secondary donkey anti-rabbit Alexa 594 (Invitrogen, USA) for 1 h before being washed and mounted onto slides. Slides were stained with X34 as documented and cover slipped. Fluorescent images of individual plaques were taken using a Nikon Eclipse 90i microscope at 20X magnification. Plaques were chosen with an average area of 300 μm2. The number of IBA1 positive microglia was counted in circular radiating regions of interest with a diameter of 50 μm from the edge of the X34 positive plaque using Nikon NIS elements software.
Bulk mRNA-seq data
RNA was isolated from the frontal cortex using RNeasy mini kit (Qiagen, Germany) and RNA quality was assessed using 2100 Bioanalyzer (Agilent Technologies, USA). Samples (RIN > 8) were used for library generation and sequenced on Illumina NovaSeq PE 150 (Novogene Co. Inc., USA). Following initial processing and quality control, the sequencing data was aligned to the mouse genome mm10 using Subread (v1.5.3) with an average read depth of 50 million successfully aligned reads.67 Statistical analysis was carried out using Rsubread68 (v1.34.2), DEseq269 (1.24.0), and EdgeR70 (v3.26.5), in R environment (v3.6.0). Functional annotation clustering was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID).71 All DAVID terms are considered significant if p < 0.05 following multiplicity correction using the Benjamini-Hochberg method to control the FDR.
Sample preparation for single cell RNA-seq
Mice were perfused with cold PBS, and half of each brain tissue, excluding olfactory and cerebellum, were immediately dissected and dissociated using Neural Tissue Dissociation Kit (P) (Miltenyi, USA) following manufacturer’s manual with slight modification. Briefly, tissues were dissected into small pieces and washed in cold HBSS (Gibco, USA). Pellets were incubated at 37C with Enzyme P mix and Enzyme A mix stepwisely, followed by filtering through 40μm strainers (Corning, USA). Myelin was removed after tissue dissociation using Myelin Removal Beads II (Miltenyi, USA) according to manufacturer’s manual. Dead Cell Removal Kit (Miltenyi, USA) was applied to dead cells and debris. The single cell suspensions with viability >80% were adjusted to 1,000 cells/μl for library preparation.
For droplet-based scRNA-seq, libraries were prepared using the Chromium Next GEM Single Cell 3’ Kit v3.1 and Chip G Single Cell Kit (10x Genomics, USA) according to manufacturer’s protocol. Cell-RT mix were prepared, targeting 10,000 cells per sample, and applied to Chromium Controller (10x Genomics, USA) for GEM generation and barcoding. Samples were then purified from post GEM-RT cleanup and full-length cDNA were amplified for library construction. 11 PCR cycles were applied to amplify cDNA and 13 or 14 cycles for sample index PCR. Libraries were checked by Bioanalyzer High Sensitivity DNA kit (Agilent, USA) and sequenced on Illumina NovaSeq (MedGenome Inc., USA) following 10x Genomics recommendations.
Single cell RNA-seq data analysis
Reads were aligned to the mouse reference genome (GRCm38) using Cell Ranger pipeline v6.1(10x Genomics). Cells with over 200 unique molecular identifiers (UMIs) were selected for further analysis. Each of the scRNA-seq libraries were read into R (version 4.1.2) and processed using Seurat package (version 4.1.0).72 Cells were filtered by the following pre-processing criteria: (1) outliers with unique feature counts over 4000 or less than 200, and with total molecules over 50,000 were removed; (2) cells with > 10% mitochondrial counts were filtered; and (3) after SCTransform normalization and integration, multiplets were removed using DoubletFinder package (version 2.0) at 8% multiplet rate.73 After filtering, the dataset contained 35,698 high-quality, single cells with 1667 genes and 3893 UMI per cell in average. Genes were projected into principal component (PC) using the principal component analysis (RunPCA) in Seurat. The first 10 PCs were used as input for FindNeighbor, FindClusters (at 0.1 resolution) and RunTSNE functions. Differential expression was performed using “MAST” algorithm to identify cluster marker genes and help manually annotate cell types for each cluster. A list of marker genes for different cell types, generated based on our previous studies25 and other publications75-79 were used as a reference for cell type annotation (Table S1). A gene was considered differentially expressed if it had a Bonferroni-corrected p value <0.05. Cell types of interest were then subset as individual Seurat objects for further analysis. Functional annotation clustering was performed using DAVID v6.8.
Visium spatial gene expression
Visium spatial transcriptomic analysis was performed as before.51,54,80 One hemisphere of mouse brain used for Visium was embedded in optimal cutting temperature (OCT) in a cryomold and lowered into isopentane cooled with liquid nitrogen until the OCT was completely frozen. The OCT embedded tissue block were stored in a sealed container at −80°C until cryosectioning. Mouse brains were cryosectioned (Center for Biologic Imaging, University of Pittsburgh) at 10 μm and placed directly onto the Visium slides ensuring placement within the capture area. To ensure sampling of different brain regions, two sections were used from each mouse with the first section 500μm caudal to the first appearance of the dente gryus, and each 10-μm section was 500 μm apart. Visium tissue optimization and spatial gene expression protocols were followed exactly as described by 10x Genomics (https://10xgenomics.com/) using immunofluorescence as the counterstain. Briefly, tissues were methanol fixed at −20°C for 30 min and then blocked for 5 min with blocking buffer. Tissues were immunostained sequentially with anti-NEUN-694 antibody (1:500, Abcam, UK), anti-GFAP-594 (1:50, Agilent, USA) and 6E10- (1:100, Biolegend, USA) with Alexa488 labeled secondary incubations (1:500 Thermo Fisher, USA). All antibodies were incubated for 30 min followed by 4 washes. Optimal permeabilization time was obtained at 12-min incubation with permeabilization enzyme. Imaging was conducted on an EVOS M5000 Imaging System (Thermo Fisher Scientific, USA). Plaques were chosen based on 6E10 staining with an average area of 300 μm2. The sum GFAP intensity was counted in circular radiating regions of interest with a diameter of 50 and 100 μm from the edge of the plaque using Nikon NIS elements software. mRNA library preparation and sequencing using Illumina NextSeq 550 were done at the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh.
Spatial gene expression data analysis
Reads from eight capture areas (each one corresponding to a brain hemisphere) were aligned to the mouse reference genome (GRCm38) using Space Ranger v1.3.1 (10x Genomics). The immunofluorescence images and manual alignment files were also used as input to Space Ranger pipeline to generate matrices of spatial feature counts. An average of 2,688.5 spots under tissue per capture area (21508 total spots) were read and processed using Seurat v4.1.072 within R (version 4.1.2). High-quality spots were selected for further analysis: unique feature counts less than 5000, mitochondrial counts over 35%, and hemoglobin genes over 20% were filtered out. In addition, 403 spots from sample D_259 were filtered out based on abnormal counts and unique genes compared to respective tissue areas in the same chip. After spot quality control, the dataset contained 20,800 spots under tissue with 19,255 genes detected. SCtransform was used for data normalization, highly variable genes selection and gene expression scaling. Linear dimensional reduction analysis was performed with RunPCA and clusterization with FindNeighbors (1:30 PCA dimensions) and FindClusters (0.5 resolution). Spots’ gene expression and clusters were visualized within t-SNE projections and spatial plots. Plaque-associated spots were manually selected with the interactive tool STutility and the labels were added to Seurat object as metadata.53 Clusters’ markers and plaque-associated differential expression were identified using FindMarkers with default test options. Staph vs. PBS differential expression analysis within clusters was performed with FindMarkers using “MAST” as test. Differentially expressed genes (DEGs) were identified based on Bonferroni-corrected p value <0.05 criteria, and gene’s functional annotation was analyzed with DAVID v6.8. Gene network analysis was conducted using the GeneMANIA prediction server.74
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes (n) are indicated in the figure legends correspond to the number of biological replicates analyzed. Power analysis was performed (two groups, t test, G*Power 3.1) with individual estimated experimental effect size, alpha level 0.05, and to satisfy a 95% power. Mice were randomly assigned to experimental groups and all researchers were blinded to experimental group during each of the analysis. Mice were littermates chosen to balance sex and age and were in healthy condition for the experiments. Unless otherwise indicated, all results are reported and presented as means with error bars as standard error of the mean (SEM). Data were analyzed by unpaired t test. scRNA-seq and spatial transcriptomics data were normalized using SCTransform81 algorithm, which builds regularized negative binomial models of gene expression, and the differential expression were analyzed using the MAST82 algorithm of the Seurat package in R, which implements a two-part hurdle model. Unless otherwise indicated, all statistical analyses were performed in GraphPad Prism (v 8.2.0 and 9.4.1), or in R (v 3.6.0) and significance was determined as p < 0.05.
Supplementary Material
Highlights.
Peripheral bacterial infection affects brain transcriptomics and AD-like pathology
Transcriptional changes in brain-barrier-related cell types and leakage
Increased amyloid pathology and plaque-associated microglia
Unique transcriptional responses around amyloid plaques
ACKNOWLEDGMENTS
The authors thank William A. MacDonald and Amanda Poholek from the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh for their technical help with spatial transcriptomic experiments. We are grateful to our deceased friend and colleague Anatoliy A. Gashev, MD, for the idea to test the effect of the peripheral infection on amyloid pathology and for providing Staphylococcus aureus bacteria. This work was funded by the National Institutes of Health, USA (AG057565-IL, AG066198-RK, AG077636-RK, AG075992-IL, AG075069-NFF). C.S.-G. was supported by a scholarship from Coordination for the Improvement of Higher Education Personnel (CAPES), Brazil.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112785.
DECLARATION OF INTERESTS
C.S.-G. has an additional affiliation at Biotechnology Center, Federal University of Rio Grande do Sul, Porto Alegre, RS 91501-970, Brazil.
REFERENCES
- 1.Knopman DS, Amieva H, Petersen RC, Chételat G, Holtzman DM, Hyman BT, Nixon RA, and Jones DT (2021). Alzheimer disease. Nat. Rev. Dis. Prim 7, 33. 10.1038/s41572-021-00269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettcher BM, Tansey MG, Dorothée G, and Heneka MT (2021). Peripheral and central immune system crosstalk in Alzheimer disease - a research prospectus. Nat. Rev. Neurol 17, 689–701. 10.1038/s41582-021-00549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie J, Van Hoecke L, and Vandenbroucke RE (2021). The Impact of Systemic Inflammation on Alzheimer’s Disease Pathology. Front. Immunol 12, 796867. 10.3389/fimmu.2021.796867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai KSP, Liu CS, Rau A, Lanctôt KL, Köhler CA, Pakosh M, Carvalho AF, and Herrmann N (2017). Peripheral inflammatory markers in Alzheimer’s disease: a systematic review and meta-analysis of 175 studies. J. Neurol. Neurosurg. Psychiatry 88, 876–882. 10.1136/jnnp-2017-316201. [DOI] [PubMed] [Google Scholar]
- 5.Wendeln AC, Degenhardt K, Kaurani L, Gertig M, Ulas T, Jain G, Wagner J, Häsler LM, Wild K, Skodras A, et al. (2018). Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338. 10.1038/s41586-018-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tejera D, Mercan D, Sanchez-Caro JM, Hanan M, Greenberg D, Soreq H, Latz E, Golenbock D, and Heneka MT (2019). Systemic inflammation impairs microglial Abeta clearance through NLRP3 inflammasome. EMBO J. 38, e101064. 10.15252/embj.2018101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quintana JF, Chandrasegaran P, Sinton MC, Briggs EM, Otto TD, Heslop R, Bentley-Abbot C, Loney C, de Lecea L, Mabbott NA, and MacLeod A (2022). Single cell and spatial transcriptomic analyses reveal microglia-plasma cell crosstalk in the brain during Trypanosoma brucei infection. Nat. Commun 13, 5752. 10.1038/s41467-022-33542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persidsky Y, Ramirez SH, Haorah J, and Kanmogne GD (2006). Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J. Neuroimmune Pharmacol 1, 223–236. 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 9.Almutairi, Gong C, Xu YG, Chang Y, and Shi H (2016). Factors controlling permeability of the blood-brain barrier. Cell. Mol. Life Sci 73, 57–77. 10.1007/s00018-015-2050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbott NJ, Patabendige AAK, Dolman DEM, Yusof SR, and Begley DJ (2010). Structure and function of the blood-brain barrier. Neurobiol. Dis 37, 13–25. 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Obermeier B, Daneman R, and Ransohoff RM (2013). Development, maintenance and disruption of the blood-brain barrier. Nat. Med 19, 1584–1596. 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monje ML, Toda H, and Palmer TD (2003). Inflammatory blockade restores adult hippocampal neurogenesis. Science 302, 1760–1765. 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- 13.Nimmervoll B, White R, Yang JW, An S, Henn C, Sun JJ, and Luhmann HJ (2013). LPS-induced microglial secretion of TNFalpha increases activity-dependent neuronal apoptosis in the neonatal cerebral cortex. Cerebr. Cortex 23, 1742–1755. 10.1093/cercor/bhs156. [DOI] [PubMed] [Google Scholar]
- 14.Zlokovic BV (2002). Vascular disorder in Alzheimer’s disease: role in pathogenesis of dementia and therapeutic targets. Adv. Drug Deliv. Rev 54, 1553–1559. [DOI] [PubMed] [Google Scholar]
- 15.Bond AM, Ming GL, and Song H (2015). Adult Mammalian Neural Stem Cells and Neurogenesis: Five Decades Later. Cell Stem Cell 17, 385–395. 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dani N, Herbst RH, McCabe C, Green GS, Kaiser K, Head JP, Cui J, Shipley FB, Jang A, Dionne D, et al. (2021). A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell 184, 3056–3074.e21. 10.1016/j.cell.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, Fehlmann T, Stein JA, Schaum N, Lee DP, et al. (2021). Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595, 565–571. 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempuraj D, Thangavel R, Selvakumar GP, Zaheer S, Ahmed ME, Raikwar SP, Zahoor H, Saeed D, Natteru PA, Iyer S, and Zaheer A (2017). Brain and Peripheral Atypical Inflammatory Mediators Potentiate Neuroinflammation and Neurodegeneration. Front. Cell. Neurosci 11, 216. 10.3389/fncel.2017.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadick JS, O’Dea MR, Hasel P, Dykstra T, Faustin A, and Liddelow SA (2022). Astrocytes and oligodendrocytes undergo subtype-specific transcriptional changes in Alzheimer’s disease. Neuron 110, 1788–1805.e10. 10.1016/j.neuron.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austermann J, Roth J, and Barczyk-Kahlert K (2022). The Good and the Bad: Monocytes’ and Macrophages’ Diverse Functions in Inflammation. Cells 11. 10.3390/cells11121979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer-Riepe L, Daber N, Schulte-Schrepping J, Viéras De Carvalho BC, Russo A, Pohlen M, Fischer J, Chasan AI, Wolf M, Ulas T, et al. (2020). CD163 expression defines specific, IRF8-dependent, immune-modulatory macrophages in the bone marrow. J. Allergy Clin. Immunol 146, 1137–1151. 10.1016/j.jaci.2020.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Etzerodt A, Maniecki MB, Møller K, Møller HJ, and Moestrup SK (2010). Tumor necrosis factor alpha-converting enzyme (TACE/ADAM17) mediates ectodomain shedding of the scavenger receptor CD163. J. Leukoc. Biol 88, 1201–1205. 10.1189/jlb.0410235. [DOI] [PubMed] [Google Scholar]
- 23.Etzerodt A, and Moestrup SK (2013). CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxidants Redox Signal. 18, 2352–2363. 10.1089/ars.2012.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Etzerodt A, Rasmussen MR, Svendsen P, Chalaris A, Schwarz J, Galea I, Møller HJ, and Moestrup SK (2014). Structural basis for inflammation-driven shedding of CD163 ectodomain and tumor necrosis factor-alpha in macrophages. J. Biol. Chem 289, 778–788. 10.1074/jbc.M113.520213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitz NF, Nam KN, Wolfe CM, Letronne F, Playso BE, Iordanova BE, Kozai TDY, Biedrzycki RJ, Kagan VE, Tyurina YY, et al. (2021). Phospholipids of APOE lipoproteins activate microglia in an isoform-specific manner in preclinical models of Alzheimer’s disease. Nat. Commun 12, 3416. 10.1038/s41467-021-23762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunilkumar S, Toro AL, McCurry CM, VanCleave AM, Stevens SA, Miller WP, Kimball SR, and Dennis MD (2022). Stress response protein REDD1 promotes diabetes-induced retinal inflammation by sustaining canonical NF-kappaB signaling. J. Biol. Chem 298, 102638. 10.1016/j.jbc.2022.102638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stritt S, Nurden P, Nurden AT, Schved JF, Bordet JC, Roux M, Alessi MC, Trégouët DA, Mäkinen T, and Giansily-Blaizot M (2023). APOLD1 loss causes endothelial dysfunction involving cell junctions, cytoskeletal architecture, and Weibel-Palade bodies, while disrupting hemostasis. Haematologica 108, 772–784. 10.3324/haematol.2022.280816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regard JB, Scheek S, Borbiev T, Lanahan AA, Schneider A, Demetriades AM, Hiemisch H, Barnes CA, Verin AD, and Worley PF (2004). Verge: a novel vascular early response gene. J. Neurosci 24, 4092–4103. 10.1523/JNEUROSCI.4252-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia W, Lu R, Martin TA, and Jiang WG (2014). The role of claudin-5 in blood-brain barrier (BBB) and brain metastases (review). Mol. Med. Rep 9, 779–785. 10.3892/mmr.2013.1875. [DOI] [PubMed] [Google Scholar]
- 30.Ille AM, Kishel E, Bodea R, Ille A, Lamont H, and Amico-Ruvio S (2020). Protein LY6E as a candidate for mediating transport of adeno-associated virus across the human blood-brain barrier. J. Neurovirol 26, 769–778. 10.1007/s13365-020-00890-9. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Liang C, and Liu SL (2017). Interferon-inducible LY6E Protein Promotes HIV-1 Infection. J. Biol. Chem 292, 4674^685. 10.1074/jbc.M116.755819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia C, Braunstein Z, Toomey AC, Zhong J, and Rao X (2017). S100 Proteins As an Important Regulator of Macrophage Inflammation. Front. Immunol 8, 1908. 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linden JR, Flores C, Schmidt EF, Uzal FA, Michel AO, Valenzuela M, Dobrow S, and Vartanian T (2019). Clostridium perfringens epsilon toxin induces blood brain barrier permeability via caveolae-dependent transcytosis and requires expression of MAL. PLoS Pathog. 15, e1008014. 10.1371/journal.ppat.1008014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piatek P, Lewkowicz N, Michlewska S, Wieczorek M, Bonikowski R, Parchem K, Lewkowicz P, and Namiecinska M (2022). Natural fish oil improves the differentiation and maturation of oligodendrocyte precursor cells to oligodendrocytes in vitro after interaction with the blood-brain barrier. Front. Immunol 13, 932383. 10.3389/fimmu.2022.932383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Mateo N, Pascua-Maestro R, Pérez-Castellanos A, Lillo C, Sanchez D, and Ganfornina MD (2018). Myelin extracellular leaflet compaction requires apolipoprotein D membrane management to optimize lysosomal-dependent recycling and glycocalyx removal. Glia 66, 670–687. 10.1002/glia.23274. [DOI] [PubMed] [Google Scholar]
- 36.Huang B, and Li X (2021). The Role of Mfsd2a in Nervous System Diseases. Front. Neurosci 15, 730534. 10.3389/fnins.2021.730534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 38.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 39.Davies W, Smith RJ, Kelsey G, and Wilkinson LS (2004). Expression patterns of the novel imprinted genes Nap1l5 and Peg13 and their non-imprinted host genes in the adult mouse brain. Gene Expr. Patterns 4, 741–747. 10.1016/j.modgep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Vincent AJ, Lau PW, and Roskams AJ (2008). SPARC is expressed by macroglia and microglia in the developing and mature nervous system. Dev. Dynam 237, 1449–1462. 10.1002/dvdy.21495. [DOI] [PubMed] [Google Scholar]
- 41.Solár P, Zamani A, Kubíčková L, Dubový P, and Joukal M (2020). Choroid plexus and the blood-cerebrospinal fluid barrier in disease. Fluids Barriers CNS 17, 35. 10.1186/s12987-020-00196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nutma E, Ceyzériat K, Amor S, Tsartsalis S, Millet P, Owen DR, Papadopoulos V, and Tournier BB (2021). Cellular sources of TSPO expression in healthy and diseased brain. Eur. J. Nucl. Med. Mol. Imag 49, 146–163. 10.1007/s00259-020-05166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E, Nohara K, Wirianto M, Escobedo G Jr., Lim JY, Morales R, Yoo SH, and Chen Z (2021). Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model. Biomolecules 11, 1004. 10.3390/biom11071004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin C, Ackermann S, Ma Z, Mohanta SK, Zhang C, Li Y, Nietzsche S, Westermann M, Peng L, Hu D, et al. (2019). ApoE attenuates unresolvable inflammation by complex formation with activated C1q. Nat. Med 25, 496–506. 10.1038/s41591-018-0336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahu A, Clemens ZJ, Shinde SN, Sivakumar S, Pius A, Bhatia A, Picciolini S, Carlomagno C, Gualerzi A, Bedoni M, et al. (2021). Regulation of aged skeletal muscle regeneration by circulating extracellular vesicles. Nat. Aging 1, 1148–1161. 10.1038/s43587-021-00143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Stein LR, Kim D, Ho K, Yu GQ, Zhan L, Larsson TE, and Mucke L (2018). Klotho controls the brain-immune system interface in the choroid plexus. Proc. Natl. Acad. Sci. USA 115, E11388–E11396. 10.1073/pnas.1808609115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herr DR, Chew WS, Satish RL, and Ong WY (2020). Pleotropic Roles of Autotaxin in the Nervous System Present Opportunities for the Development of Novel Therapeutics for Neurological Diseases. Mol. Neurobiol 57, 372–392. 10.1007/s12035-019-01719-1. [DOI] [PubMed] [Google Scholar]
- 48.Demeestere D, Libert C, and Vandenbroucke RE (2015). Clinical implications of leukocyte infiltration at the choroid plexus in (neuro)inflammatory disorders. Drug Discov. Today 20, 928–941. 10.1016/j.drudis.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Starossom SC, Mascanfroni ID, Imitola J, Cao L, Raddassi K, Hernandez SF, Bassil R, Croci DO, Cerliani JP, Delacour D, et al. (2012). Galectin-1 deactivates classically activated microglia and protects from inflammation-induced neurodegeneration. Immunity 37, 249–263. 10.1016/j.immuni.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bien-Ly N, Boswell CA, Jeet S, Beach TG, Hoyte K, Luk W, Shihadeh V, Ulufatu S, Foreman O, Lu Y, et al. (2015). Lack of Widespread BBB Disruption in Alzheimer’s Disease Models: Focus on Therapeutic Antibodies. Neuron 88, 289–297. 10.1016/j.neuron.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 51.Castranio EL, Hasel P, Haure-Mirande JV, Ramirez Jimenez AV, Hamilton BW, Kim RD, Glabe CG, Wang M, Zhang B, Gandy S, et al. (2023). Microglial INPP5D limits plaque formation and glial reactivity in the PSAPP mouse model of Alzheimer’s disease. Alzheimers Dement. 19, 2239–2252. 10.1002/alz.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briz V, Restivo L, Pasciuto E, Juczewski K, Mercaldo V, Lo AC, Baatsen P, Gounko NV, Borreca A, Girardi T, et al. (2017). The non-coding RNA BC1 regulates experience-dependent structural plasticity and learning. Nat. Commun 3, 293. 10.1038/s41467-017-00311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergenstråhle J, Larsson L, and Lundeberg J (2020). Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genom. 21, 482. 10.1186/s12864-020-06832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen WT, Lu A, Craessaerts K, Pavie B, Sala Frigerio C, Corthout N, Qian X, Laláková J, Kühnemund M, Voytyuk I, et al. (2020). Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer’s Disease. Cell 132, 976–991.e19. 10.1016/j.cell.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 55.Greene C, Hanley N, and Campbell M (2019). Claudin-5: gatekeeper of neurological function. Fluids Barriers CNS 16, 3. 10.1186/s12987-019-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Decker Y, Németh E, Schomburg R, Chemla A, Fülöp L, Menger MD, Liu Y, and Fassbender K (2021). Decreased pH in the aging brain and Alzheimer’s disease. Neurobiol. Aging 101, 40–49. 10.1016/j.neurobiolaging.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Iram T, Kern F, Kaur A, Myneni S, Morningstar AR, Shin H, Garcia MA, Yerra L, Palovics R, Yang AC, et al. (2022). Young CSF restores oligodendrogenesis and memory in aged mice via Fgf17. Nature 605, 509–515. 10.1038/s41586-022-04722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinho A, Gonçalves I, Costa M, and Santos CR (2012). Stress and glucocorticoids increase transthyretin expression in rat choroid plexus via mineralocorticoid and glucocorticoid receptors. J. Mol. Neurosci 48, 1–13. 10.1007/s12031-012-9715-7. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Cattaneo F, Ryno L, Hulleman J, Reixach N, and Buxbaum JN (2014). The systemic amyloid precursor transthyretin (TTR) behaves as a neuronal stress protein regulated by HSF1 in SH-SY5Y human neuroblastoma cells and APP23 Alzheimer’s disease model mice. J. Neurosci 34, 7253–7265. 10.1523/JNEUROSCI.4936-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buxbaum JN, and Johansson J (2017). Transthyretin and BRICHOS: The Paradox of Amyloidogenic Proteins with Anti-Amyloidogenic Activity for Abeta in the Central Nervous System. Front. Neurosci 11, 119. 10.3389/fnins.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prud’homme GJ, Kurt M, and Wang Q (2022). Pathobiology of the Klotho Antiaging Protein and Therapeutic Considerations. Front. Aging 3, 931331. 10.3389/fragi.2022.931331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wewer C, Seibt A, Wolburg H, Greune L, Schmidt MA, Berger J, Galla HJ, Quitsch U, Schwerk C, Schroten H, and Tenenbaum T (2011). Transcellular migration of neutrophil granulocytes through the blood-cerebrospinal fluid barrier after infection with Streptococcus suis. J. Neuroinflammation 8, 51. 10.1186/1742-2094-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao X, Li J, Winkler CA, An P, and Guo JT (2018). IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front. Microbiol 9, 3228. 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hur JY, Frost GR, Wu X, Crump C, Pan SJ, Wong E, Barros M, Li T, Nie P, Zhai Y, et al. (2020). The innate immunity protein IFITM3 modulates gamma-secretase in Alzheimer’s disease. Nature 586, 735–740. 10.1038/s41586-020-2681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen AT, Wang K, Hu G, Wang X, Miao Z, Azevedo JA, Suh E, Van Deerlin VM, Choi D, Roeder K, et al. (2020). APOE and TREM2 regulate amyloid-responsive microglia in Alzheimer’s disease. Acta Neuropathol. 140, 477–93. 10.1007/s00401-020-02200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grubman A, Chew G, Ouyang JF, Sun G, Choo XY, McLean C, Simmons RK, Buckberry S, Vargas-Landin DB, Poppe D, et al. (2019). A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci 22, 2087–2097. 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- 67.Liao Y, Smyth GK, and Shi W (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108. 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, and Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, and Anders S (2014). Moderated estimation offold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26,139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 72.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 8, 329–337.e4. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, Hurd YL, Dracheva S, Casaccia P, Roussos P, and Zhang B (2018). Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci. Rep 8, 8868. 10.1038/s41598-018-27293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C, et al. (2019). Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci 22, 1696–1708. 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- 77.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini JL 2nd, Tran MN, Besich Z, Tippani M, et al. (2021). Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci 24, 425–436. 10.1038/s41593-020-00787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data from bulk-, single-cell RNA-seq, and spatial transcriptomics were deposited at GEO and are publicly available as of the date of publication (GSE218346, GSE218352, and GSE218360). Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD163 antibody, mouse (IHC) | Abcam | Cat# ab201461 |
| Biotinylated anti-mouse antibody (IHC) | Vector | Cat# BA-2000; RRID:AB_2313581 |
| Anti-6E10 biotinylated antibody, mouse (IHC+Visium) | Biolegend | Cat# 803007; RRID:AB_2564657 |
| Mouse albumin polyclonal antibody (IHC) | Bethyl Laboratories | Cat# A90-134; RRID:AB_67120 |
| Goat anti-mouse IgG antibody conjugated with Alexa 488 (IHC) | Invitrogen | Cat# A-11001; RRID:AB_2534069 |
| Anti-IBA1 antibody, rabbit (IHC) | WAKO | Cat# 019-19741; RRID:AB_839504 |
| Donkey anti rabbit Alexa 594 (IHC) | Invitrogen | Cat#A-21207; RRID:AB_141637 |
| Donkey anti goat, Alexa 488 conjugated (IHC) | Invitrogen | Cat# A32814; RRID:AB_2762838 |
| Donkey anti mouse, Alexa488 conjugated (IHC+Visium) | Thermo Fisher Scientific | Cat # A-21202; RRID:AB_141607 |
| Alexa 594 Anti-GFAP antibody (Visium) | Abcam | Cat# ab302663 |
| Alexa 694 Anti-NEUN antibody (Visium) | Abcam | Cat# ab190565; RRID:AB_2732785 |
| Bacterial and virus strains | ||
| Staphylococcus aureus | Provided by Anatoliy Gashev, MD, PhD | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| BD BBL™ Trypticase™ Soy Agar | Fisher Scientific Edge | Cat# B11043 |
| Kanamycin | Thermo Fisher Scientific | Cat# J1792406 |
| Phosphate buffered saline (PBS) | Gibco | Cat# 10010-023 |
| Isoflurane | Covetrus | SKU# 029405 |
| Tribromoethanol | ACROS Organics | CAS# 75-80-9 |
| 2-Methyl-2-butanol | Sigma-Aldrich | CAS# 75-85-4 |
| EDTA | Fisher Chemical | Cat# S311500 |
| Phosphate-buffered paraformaldehyde | Electron Microscopy Sciences | Cat# 15714S |
| Ethanol | Decon laboratories | CAS# 64-17-5 |
| Sucrose | Fisher Scientific | Cat# S5-3 |
| OCT | Fisher Healthcare | Cat# 23-730-571 |
| Formic acid | MP Biomedicals | CAS# 64-18-6 |
| H2O2 | Fisher Scientific | CAS# 7722-84-1 |
| Normal Goat Serum | Jackson ImmunoResearch | Cat# 005-000-121 |
| Normal Horse Serum | Vector | Cat# S-2000 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat# 017-000-121 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat# BP1605-100 |
| TritonX-100 | Sigma-Aldrich | CAS# 9036-19-5 |
| Vector ABC kit | Vector | Cat# PK-6100 |
| DAB substrate kit | Vector | Cat# SK-4100 |
| Tween20 | Fisher Scientific | Cat# 337-500 |
| Hank’s Balanced Salt solution (HBSS) | Gibco | Cat# 41025-092 |
| X-34 | Provided by William E. Klunk, MD, PhD. | N/A |
| Sodium Citrate Dihydrate | Fisher Scientific | Cat# S279-500 |
| Critical commercial assays | ||
| Mouse Cytokine 32-Plex Discovery Assay (plasma) | Eve Technologies | SKU# MD32 |
| Mouse Cytokine 10-Plex Discovery Assay (brain) | Eve Technologies | SKU# MDF10 |
| RNeasy mini kit | Qiagen | Cat# 74104 |
| Neural Tissue Dissociation Kit (P) | Miltenyi | Cat# 130-092-628 |
| Myelin Removal Beads II | Miltenyi | Cat# 130-096-733 |
| Dead Cell Removal Kit | Miltenyi | Cat# 130-090-101 |
| Chromium Next GEM Single Cell 3’ Kit v3.1 | 10x Genomics | Cat# PN-1000269 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | Cat# PN-1000127 |
| Visium Spatial Gene Expression Slide & Reagents Kit | 10x Genomics | Cat# PN-1000187 |
| Bioanalyzer High Sensitivity DNA kit | Agilent Technologies | Cat# 5067-4626 |
| Bioanalyzer RNA 6000 Nano kit | Agilent Technologies | Cat# 5067-1511 |
| Deposited data | ||
| Raw bulk RNA-seq | This paper | [GEO]: [GSE218346] |
| Raw scRNA-seq | This paper | [GEO]: [GSE218352] |
| Raw Spatial Transcriptomics data | This paper | [GEO]: [GSE218360] |
| Experimental models: Organisms/strains | ||
| Transgenic mice: APP/PS1ΔE9 [B6.Cg-Tg(APPswe,PSEN1dE9)85Dbo/Mmjax] | The Jackson Laboratory | MMRRC Strain #034832-JAX |
| Software and algorithms | ||
| Prism v 8.2.0 and 9.4.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Nikon NIS-Elements Advanced Research software | Nikon Instruments Inc. | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research |
| Subread (v1.5.3) | Liao et al.67 | https://subread.sourceforge.net/ |
| Rsubread v1.34.2 | Liao et al.68 | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| DESeq2 v1.24.0 | Love et al.69 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| EdgeR v3.26.5 | Robinson et al.70 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8) | Huang da et al.71 | https://david.ncifcrf.gov |
| Cell Ranger v6.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-dna/software/downloads/latest |
| Seurat v4.1.0 | Hao et al.72 | https://satijalab.org/seurat/ |
| DoubletFinder v2.0 | McGinnis et al.73 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Space Ranger v1.3.1 | 10x Genomics | https://support.10xgenomics.com/spatial-gene-expression/software/downloads/latest |
| STutility | Bergenstråhle et al.53 | https://ludvigla.github.io/STUtility_web_site/ |
| GeneMANIA prediction server | Warde-Farley et al.74 | https://genemania.org/ |
| Other | ||
| Cryotome | Center for Biologic Imaging at University of Pittsburgh | N/A |
| Superfrost plus slides | Fisher Scientific | Cat# 12-550-15 |
| Nikon Eclipse 90i microscope | Nikon Instruments Inc. | Cat# 25248 |
| Keyence BZ-X810 All-in-One Fluorescence Microscope | Keyence Co. | Cat# BZ-X810/BZ-X800LE |
| 2100 Bioanalyzer | Agilent Technologies | Cat# G2939BA |
| EVOS M5000 Imaging System | Thermo Fisher Scientific | Cat# AMF5000 |
| Chromium Controller | 10X Genomics | Cat# 1000204 |
| Illumina NovaSeq | Novogen Co. Ltd. | N/A |
| Illumina NextSeq 550 | Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh | N/A |
| Illumina NovaSeq | MedGenome Inc. | N/A |
| Countess 2 FL Automated Cell Counter | Thermo Fisher Scientific | Cat# AMQAF1000 |
| 40μm Cell Strainer | Corning | Cat# 431750 |