Abstract
For decades, plants have been the subject of genetic engineering to synthesize novel, value-added compounds. Polyhydroxyalkanoates (PHAs), a large class of biodegradable biopolymers naturally synthesized in eubacteria, are among the novel products that have been introduced to make use of plant acetyl-CoA metabolic pathways. It was hoped that renewable PHA production would help address environmental issues associated with the accumulation of nondegradable plastic wastes. However, after three decades of effort synthesizing PHAs, and in particular the simplest form polyhydroxybutyrate (PHB), and seeking to improve their production in plants, it has proven very difficult to reach a commercially profitable rate in a normally growing plant. This seems to be due to the growth defects associated with PHA production and accumulation in plant cells. Here, we review major breakthroughs that have been made in plant-based PHA synthesis using traditional genetic engineering approaches and discuss challenges that have been encountered. Then, from the point of view of plant synthetic biology, we provide perspectives on reprograming plant acetyl-CoA pathways for PHA production, with the goal of maximizing PHA yield while minimizing growth inhibition. Specifically, we suggest genetic elements that can be considered in genetic circuit design, approaches for nuclear genome and plastome modification, and the use of multiomics and mathematical modeling in understanding and restructuring plant metabolic pathways.
1. Introduction
As autotrophic organisms, plants have evolved sophisticated metabolic pathways to utilize sunlight and atmospheric carbon dioxide to produce a rich array of phytochemicals that are essential for plant growth and development. It has been estimated that there are 200,000 to 1 million distinct metabolites generated in plants [1]. The majority of the carbon fixed by plants, however, is lost due to respiration or is fixed in cell wall polymers [2]. To exploit plants for the production of customized compounds, considerable efforts have been made to engineer plant metabolic pathways [1–3]. In particular, biodegradable polyesters polyhydroxyalkanoates (PHAs), and especially its simplest form polyhydroxybutyrate (PHB), have been introduced as novel end products of acetyl-CoA anabolic metabolism. PHAs are being examined with the goals of mitigating the increasing dependence on plastic products in everyday life, the accumulation of a large body of petroleum-based nondegradable plastic wastes, and the consequent environmental and health issues [4–6]. To date, PHA yield accounting for up to 40% of dry weight (DW) has been demonstrated in Arabidopsis thaliana (Arabidopsis) [7]. Yet plant-based PHA production at a large scale remains challenging, largely due to the associated chlorosis and reduced growth observed in a number of cases [7–11].
In recent years, redirecting metabolic flux in microorganisms, including that for PHA synthesis, has been empowered by synthetic biology or “SynBio” [5, 12–14]. Inspired by integrated electric circuits that function in electronic devices, “SynBio” is aimed at building orthogonal biological parts into a genetic circuit that can predictably control the behavior of living organisms [15, 16]. “SynBio” relies on molecular technologies as much as traditional genetic engineering does. However, it distinguishes itself from classical genetic engineering in the emphasis on the ability to externally control gene expression and on the precision of gene expression sought in response to the external controls [16]. In addition, “SynBio” can be considered to be on a continuum with systems biology as it incorporates systems data and mathematical modeling to facilitate the understanding of the target organisms and to help ensure the resulting phenotype or behavior within set targets.
Compared with microorganisms, plants provide challenges in adapting some of the synthetic biology concepts. For example, quantitative prediction of behaviors of genetic parts (e.g., promoters, enhancers, and terminators), which are crucial for precise rewiring of metabolic pathways, is a daunting task, given the complex nature of land plants as multicellular organisms and the presence of multilevel regulation of gene expression [16]. Yet, proof-of-concept studies have proven the feasibility of identifying interchangeable genetic parts, delivering synthetic regulatory genetic circuits, and employing mathematical modeling in plant metabolic engineering [15, 17, 18]. With a focus on directing acetyl-CoA from endogenous metabolic pathways to PHA synthesis in plants, in this review, we provide our vision of how “SynBio” can be applied to the reconfiguration of plant metabolism for high levels of PHA production and minimal detrimental impacts on plant growth.
2. PHA Production in Plants: From Bud to Blossom
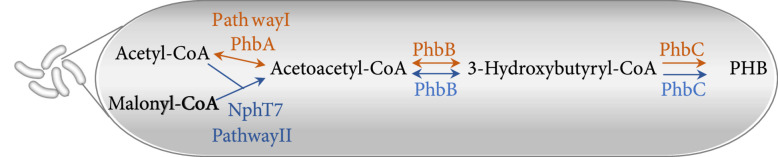
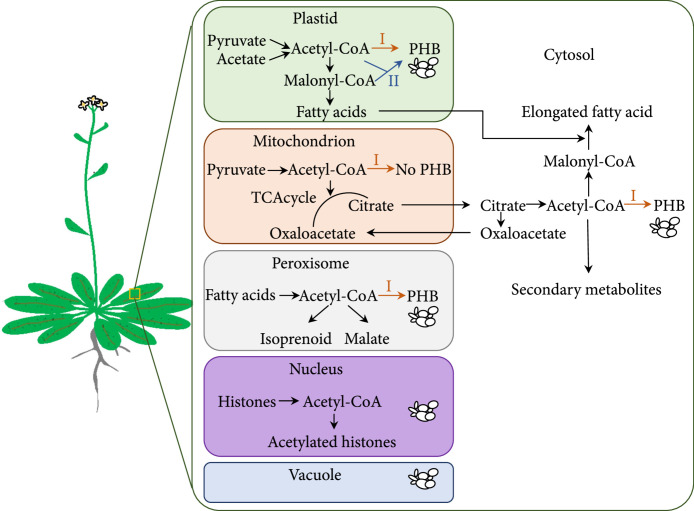
PHAs are a large group of polymers of 3-(R)-hydroxy fatty acids linked by an ester bond between the hydroxyl group and the carboxy group of an adjacent monomer. They are synthesized by most genera of eubacteria, typically under stress conditions to serve as carbon and energy storage compounds [5, 19]. As the simplest yet most representative form of PHAs, PHB has been actively studied since its initial discovery in the bacterium Bacillus megaterium in the early 1900s [20]. Biosynthesis of PHB requires acetyl-CoA as the substrate and three enzymes, β-ketothiolase (known as PhbA), acetoacetyl-CoA reductase (PhbB), and PHB synthase (PhbC), as catalysts (Figure 1(a), pathway I). Alternatively, with the presence of both acetyl-CoA and malonyl-CoA, acetoacetyl-CoA synthase (NphT7) from Streptomyces sp. can replace β-ketothiolase; it converts acetyl-CoA and malonyl-CoA to acetoacetyl-CoA for PHB synthesis (Figure 1(a), pathway II) [21]. The concept of synthesizing PHAs in plants at a cost that is comparable with petroleum-based plastics attracted the attention of the scientific community starting in about 1989, with the article “In search of the plastic potato” [22]. The required starting substrate for PHB synthesis—acetyl-CoA—is naturally produced in the plant cytosol and organelles, including plastids, mitochondria, peroxisomes, and the nucleus (Figure 1(b)). It serves as a key metabolite at the metabolic nexus—connecting catabolic and anabolic metabolism [23]. Compared with bacterial or yeast fermentation, crop plants, especially woody plants, are capable of producing large amounts of target compounds at large scale and low cost [19, 24]. It is estimated that a PHB production rate of 10-12.5% of DW in plants could be cost-competitive with petroleum-based plastics [17, 25–27].
Figure 1.
Biosynthesis of polyhydroxybutyrate (PHB) in bacteria and plants. (a) Bacterial pathways for PHB synthesis that have been engineered into plants. Pathway I (in orange) converts acetyl-CoA into PHB with three enzymes, PhbA (β-ketothiolase), PhbB (acetoacetyl-CoA reductase), and PhbC (PHB synthase). Pathway II (in blue) differs from pathway I in that it converts malonyl-CoA and acetyl-CoA to acetoacetyl-CoA with NphT7 (acetoacetyl-CoA synthase). Double arrows denote reversible reactions. (b) Visual summary of plant organelles that have been found to produce and/or accumulate PHB granules. Black arrows indicate endogenous acetyl-CoA metabolic pathways; orange and blue arrows indicate engineered PHB synthesis pathways using genes encoding enzymes in pathways I and II, respectively. Granules indicate the accumulation of PHB granules. PHB synthesis has been targeted in cytosol, plastids, and mitochondria, but no PHB has been produced in mitochondria. In addition, PHB has been observed in the nucleus and vacuoles in some studies, likely by transferring of the granules to these organelles. Figures and legend adapted from Oliver et al. [23] and Snell et al. [31].
(a).
(b).
The first successful plant-based PHA synthesis was reported in 1992, which demonstrated that constitutive expression of the bacterium Ralstonia eutropha (formerly known as Alcaligenes eutrophus) genes, PhbB and PhbC, can lead to PHB granule accumulation in cytosol, nucleus, and vacuoles, in Arabidopsis, at a yield of 0.1% of DW [28]. The omission of the PhbA in the construct took advantage of the presence of endogenous β-ketothiolase in the cytosol. The accumulation of PHB in nucleus and vacuoles was likely due to the transfer of the granules from the cytosol to these organelles. This proof-of-concept study initiated a series of efforts to promote PHA production in plants [29–32]. In an incremental manner, expression of plastid-targeted PhbA, PhbB, and PhbC in Arabidopsis (by adding DNA fragments encoding a pea chloroplast transit peptide to the three PHB synthesis genes from R. eutropha) increased PHB production to 14% of DW [33]. In these early research efforts, each of the PHB synthesis genes was transformed individually into Arabidopsis, then combined via sexual crosses. Later, single constructs containing all of the genes for plastidial PHB synthesis were created, which further promoted PHB production, for example, to 40% of DW in Arabidopsis [7]. This is the highest PHB in planta yield that has been achieved to date. Meanwhile, PHAs have been successfully synthesized in peroxisomes, where acetyl-CoA is produced as the end product of fatty acid β-oxidation (Figure 1(b)). In Arabidopsis, peroxisomal PHA production has been achieved by expressing modified PhaC1 from Pseudomonas aeruginosa [34, 35] or PhbA, PhbB, and PhbC from R. eutropha [36]. Besides the progresses made via nuclear genome modification, plastid transformation has been proven to be feasible for increasing PHB production, especially in tobacco [37–40]. By inserting native or modified forms of bacterial operons into the plastome (i.e., plastid genome), the PHB production rate has reached as high as 18.8% DW in tobacco [40], compared with a yield often below 0.3% of DW with nuclear transformation [41, 42].
In addition to model plant species Arabidopsis and tobacco, PHB and other forms of PHAs, including poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), short-chain-length (SCL) PHAs, medium-chain-length (MCL) PHAs, and SCL-MCL PHAs, have been synthesized in a number of crop plants which are more suitable for large-scale manufacture of these polymers [31, 32]. For example, industrial oilseed crops, including Brassica napus and Camelina sativa, have been examined as platforms for seed-based PHA production, with a yield up to 19.9% of DW [30, 43]. Sugarcane and switchgrass are two C4 biomass crops that have been most extensively tested for PHA production, particularly in the past decade [11, 17, 30, 31]. In sugarcane, the PHA synthesis pathways have been introduced to not only cytosol, plastids, and peroxisomes but also mitochondria [36, 44–48], where acetyl-CoA is used for energy generation (Figure 1(b)). However, no PHB accumulation was observed in mitochondria [44]. In C4 plants, it has been also found that PHA polymers preferentially accumulate in the plastids of bundle sheath cells with little to no polymers in mesophyll plastids [17, 31].
3. Factors Impeding Sufficient PHA Production
In the pioneering study reported in 1992, strong growth retardation and reduced seed production were found in PHB-producing Arabidopsis lines, despite low PHB accumulation at 0.1% of DW [28]. In this case, PHB was synthetized in the cytosol, where acetyl-CoA, the substrate for PHA production, is responsible for synthesis of other secondary metabolites (Figure 1(b)) that are essential for plant growth [23]. Because acetyl-CoA cannot be transported directly between the cytosol and other organelles, the diversion of cytosolic acetyl-CoA away from the endogenous metabolic pathways to PHA production has been proposed to be the main reason for disturbed plant growth and development [49]. In fact, the phenotypes of PHB-producing Arabidopsis plants have been found to resemble some of the phenotypes observed in plants with downregulated acetyl-CoA synthesis in the cytosol [50]. In agreement with these observations, overexpression of ATP citrate lyase (ACL), a cytosolic enzyme that generates acetyl-CoA and oxaloacetate from citrate and CoA, has been shown to be able to mitigate growth defects associated with cytosolic PHB synthesis in Arabidopsis [51].
Consequently, most of the subsequent efforts have been focused on synthesizing PHA in plastids, where higher abundance of acetyl-CoA is naturally available for fatty acid biosynthesis [33] (Figure 1(b)). Indeed, plastid-targeted synthesis has led to larger amount of PHA accumulation in general [30]. However, the presence of PHA granules in chloroplasts was also found to be problematic. For example, in the woody species poplar (Populus), negative effects on biomass growth and plant health were observed when PHB content exceeded 1% of DW [8, 19]. When produced in seed plastids of Camelina sativa, PHA accumulation can result in reduced seed oil content, germination rate, and seedling viability [43, 52, 53]. A series of C4 plant-based studies also found adverse phenotypes, such as stunting, chlorosis, and reduced biomass, especially with PHB yield above 1.5% of DW [9–11, 47]. It has been suggested that the accumulation of PHA granules in chloroplasts can shade thylakoids and disrupt the internal organization of chloroplasts, thereby leading to reduced photosynthetic efficiency and ATP starvation [11, 19]. To enhance photosynthesis, retransforming PHB-producing switchgrass with the bifunctional enzyme encoding gene FBPase/SBPase from Synechococcus was performed by Somleva et al. [30, 54]. The resulting PHA yield, although doubled what was observed in plants containing only the PHB genes, was only 7.7% DW, still lower than the commercially profitable yield (i.e., ≥10% of DW).
Another issue with C4 plant-based production is little accumulation of PHA in the plastids of mesophyll cells, which occupy 60%–70% of the total chlorenchyma area [31]. The uneven polymer accumulation was initially thought to be due to insufficient substrate availability or inefficient plastid targeting in mesophyll [44, 45, 55]. However, a nearly twofold increase in mesophyll production was accomplished in sugarcane and switchgrass by inhibiting the activity of acetyl-CoA carboxylase (ACCase), which catalyzes the first step of lipid biosynthesis and therefore competes with PhbA for acetyl-CoA [9, 48]. In addition, replacing PhbA with NphT7, which uses malonyl-CoA and acetyl-CoA for acetoacetyl-CoA production (Figure 1, pathway II), successfully promoted PHB synthesis in mesophyll and conferred up to 11.8% of DW PHB production in leaf samples of sugarcane [10]. Efficient plastid targeting in both bundle sheath and mesophyll cells with C3 dicot signals was proven to be successful by immunolocalization experiments [9]. These findings suggest that it is the availability and accessibility of acetyl-CoA in mesophyll cells, not the compatibility of the C3 dicot plastid targeting signal, that is the bottleneck to efficient PHB production in C4 plants.
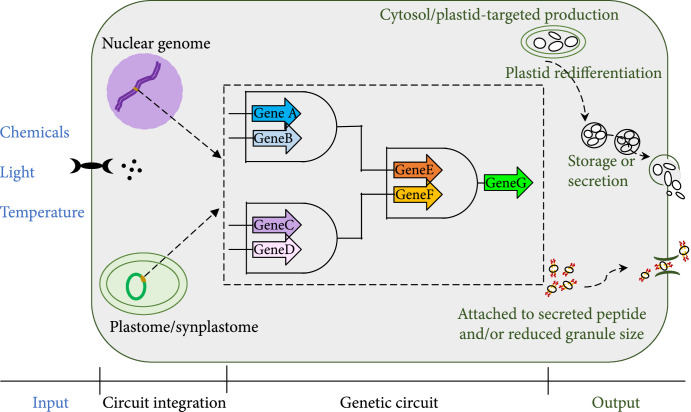
Guided by the groundbreaking studies mentioned above, in the sections below, we discuss several possible considerations for designing plant PHA synthesis constructs, delivering them into the plant genome (Figure 2), and utilizing systems data and metabolic modeling (Figure 3). We envision externally controllable PHA synthesis, by incorporating inducible or developmental stage-specific promoters into the genetic circuit with no or low impacts on plant growth and development. We also project targeted PHA synthesis, accumulation, relocation, and storage in dedicated organelles or attachments to secreted proteins.
Figure 2.
Elements to consider in genetic circuit design and genome modification for targeted synthesis, export, and storage.
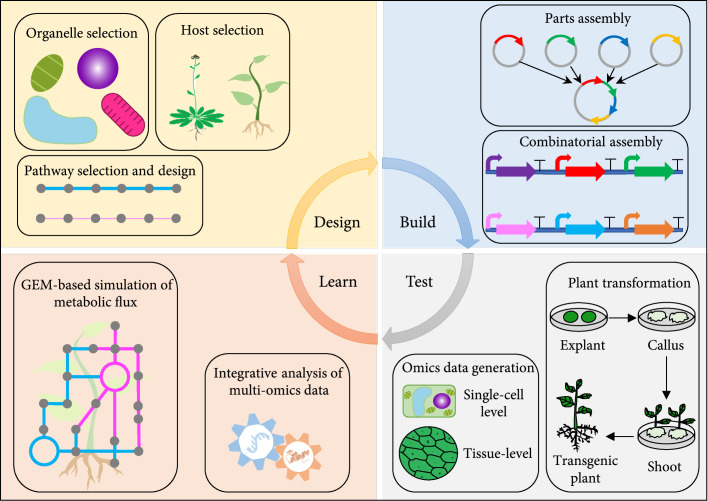
Figure 3.
Informing PHA engineering with systems biology. Systems biology approaches (e.g., omics technologies, integrative analysis tools, and genome-scale metabolic models (GEMs)) can be used to inform the design-build-test-learn cycle of PHA engineering in plants by identifying biological parts for genetic circuit design and assessing the metabolic performance of PHA-synthesizing plants at systems level.
4. Genetic Circuit for Controllable PHA Production and Storage
4.1. Precise Temporal Control of PHA Gene Expression
At present, growth defects associated with PHA synthesis seem to be inevitable. Future optimization efforts will require careful consideration of not only where in the plant the PHA is produced but also at what point in the plant’s lifecycle to avoid growth and development penalties. To date, several studies have suggested the superiority of inducible gene expression systems over constitutive gene expression systems in PHA biosynthesis. For example, replacing a constitutive promoter with a salicylic acid inducible promoter for plastid-targeted PhbA expression increased transformation efficiency in both tobacco and potato [42]. The use of an ethanol-inducible system for plastid-encoded PHB production solved previously observed growth deficiency and male sterility in tobacco plants engineered for constitutive PHB synthesis [38, 39]. In Arabidopsis, an ecdysone-inducible system led to improved health of PHB-producing plants in general [56]. In addition, a maize-derived light-inducible promoter was used for PHB synthesis in switchgrass [55]. However, these systems can be leaky [42, 56] and in some cases were not efficient in addressing the impaired plant health [42]. In addition, most of these studies reported a yield lower than the economically viable rate [39, 42, 55]. Despite that the ecdysone-inducible system was able to induce a yield up to 14% of DW in Arabidopsis [56], the system, when tested in poplar, only conferred a yield ranging 1-2% of DW [8].
Because there has been growing interest in synthesizing PHA in crop plants, and particularly in biomass crops, inducible systems suitable for use in field plantations are important. Among the inducible approaches discussed above, only the ecdysone-inducible system, which can be induced by certain types of commercially available pesticides [56], would attract broad interest. Clearly, additional systems need to be developed and tested. Developmental stage-specific promoters, such as senescence-associated promoters that have successfully altered biomass composition in Arabidopsis [57], hold promise for application in both greenhouse and field settings. However, if such approaches also reduce the development of cold and drought hardiness in perennial plants, this method might also have risks for plant health and productivity. Thus, it is essential to find the right promoters and to ensure their appropriate expression occurs (with regard to timing and intensity) even when inserted into a new genome position through transformation. High-throughput omics approaches now enable the identification of promoters at a large scale, providing many options for study. For example, in a poplar-based transcriptome study, a total of 68 sequence motifs were enriched among 1,712 genes found to be constantly upregulated during the onset and progression of seasonal leaf senescence [58]. These motifs hold considerable promise to be incorporated into inducible systems (i.e., genetic circuits) as genetic toggle switches to offer temporal control of PHA gene expression. Because leaf senescence is also regulated by multiple layers of epigenetic mechanisms [59, 60], one caveat in using senescence-specific promoters is that the effects of posttranscriptional modulation may give unexpected results. This includes mechanisms such as degradation by miRNAs and noncoding RNAs binding to cis-elements in mRNA [60]. Therefore, extensive fidelity testing in transgenic plants will be needed during promoter selection. It is possible to employ multiple tandem repeats in these promoters [61] to induce stronger senescence-specific PHA production. In addition, it may be possible to use multiple inducible or developmental stage-specific promoters to drive individual PHA synthesis genes in order to fine-tune the timing of gene expression. Finally, although plant insulator elements are not widely used, if effective they may be essential for providing reliable promoter expression, especially where the regulatory elements are nearby in constructs. A short transcriptional block from HIV [62] and/or the gypsy insulator with Hairy wing (Hw) binding protein from Drosophila melanogaster [63] should reduce promoter/enhancer interactions, though at the cost of increased construct size and complexity.
4.2. Increasing Production and Storage Capacity
When produced in plastids and other organelles, PHA accumulation is physically constrained by the size and the number of organelles within the cytoplasm and their storage capacity. Increasing the number and size of the organelles, therefore, is a logical path to promote PHA production. Plastids, like their free-living ancestors, cyanobacteria, proliferate through division of preexisting organelles. This process is orchestrated by ring-shaped contractile complexes with the coordination of nuclear gene encoding proteins, such as Plastid Division (PDV), Accumulation and Replication of Chloroplasts (ARC), Dynamin-Related Protein (DRP), and Min [64, 65]. In Arabidopsis, arc mutants have been found to have enlarged chloroplasts [66]. Suppression of the phosphatidylinositol 4-phosphate (PI4P) gene, which negatively regulates PDV1 and DRP5B expression, can accelerate chloroplast division and increase the number of chloroplasts per cell [67]. These functionally characterized chloroplast division genes serve as promising targets for customizing the size and number of chloroplasts for enhanced PHA production and accumulation. However, manipulation of chloroplast division can result in reduced photosynthetic efficiency [66] and therefore compromised plant fitness and productivity. Also, there is likely a trade-off between the number and size of chloroplasts, because the total chloroplast compartment size is often closely related to the size and type of the cell [68, 69]. These factors may limit our ability to simultaneously increase chloroplast number and size in individual cells.
Because plastids have the ability to differentiate or redifferentiate in response to developmental and environmental cues [70, 71], it might be possible to control the redifferentiation of plastids from a PHA production organelle into a PHA transport or storage organelle. Chloroplasts have been found to be able to form vesicles that transport flavonoids to the vacuole [72]. In tomato fruits, chloroplasts can convert into chromoplasts—the pigment storage organelles—as carotenoid accumulates [73]. These naturally occurring processes may serve as valuable sources for understanding how chloroplasts form transport vesicles and redifferentiate into storage organelles.
4.3. Extracellular Storage via Secretion
In both bacteria and plant cells, PHA polymers form spherical granules after their synthesis. In bacteria, these granules are naturally attached to phospholipids and proteins, with PhaP1, the phasin protein, being dominant [74]. Bacteria devoid of phasins have been found to produce less PHB and display a slightly reduced growth rate. It has been thought that phasin production is a protective mechanism against the highly hydrophobic PHB granule [75]. In Arabidopsis, although expressing PhaP from R. eutropha in parallel with the PHB synthesis genes was not able to repair plant growth defects, phasins were detected on the surface of the PHB granules with a similar abundance to that in bacteria [42]. Taking advantage of the presence of phasins in Escherichia coli (E. coli), Rahman et al. [76] created a secretion system for phasin-bound PHB granules by fusing the PhaP1 gene to the HlyA gene. Directed by the HlyA signal peptide, the resulting PhaP1 proteins and PHB granules were attached to these proteins and were able to be secreted via the type I secretion system. In addition, the presence of PhaP1 reduced the size of PHB granules, which facilitated the secretion of the granules together with the attached proteins. Although the secreted peptide-phasin-PHA fusion approach has not been tested in plants, similar fusion technology using oleosins, which are found in plants and have similar function to phasins, has been used in the production and isolation of hirudin—an anticoagulant for thrombosis treatment—in Brassica [77, 78]. Secreted peptides play essential roles in plant growth and development, and many of them, such as the Clavata3/Endosperm Surrounding Region- (ESR-) related root signal, are well characterized [79, 80]. By incorporating phasin protein and plant signal peptides into the PHA synthesis genetic circuit, it may be possible to relocate the PHA granules from within plant cells to extracellular spaces or even from production organs (e.g., leaves) to storage organs (e.g., roots).
5. Modification of the Nuclear and Plastid Genomes
5.1. Mitigating Chromosomal Context-Dependent Gene Expression
The integration of PHA synthesis constructs into the plant nuclear genome has relied on Agrobacterium tumefaciens-mediated transformation, which leads to random insertion of the PHA synthesis genes in the genome. Depending on the chromosomal context, the specificity of promoters, and consequently the expression of the transgenes, will vary widely. This context-dependent behavior presents a problem for PHA synthesis, which likely requires precise control, as discussed above. If endogenous promoters are used for driving PHA gene expression, inserting these genes into the plant genome precisely, as extension of native coding regions—for example, as protein fusions or inserting T2A or similar cleavable peptide linkers behind native protein coding regions [81]—may enable normal context-dependent expression. Site-directed insertion, though still challenging in plants, is feasible via genome editing approaches. For example, homologous recombination mediated by zinc-finger nuclease (ZFN), transcription activator-like effector nucleases (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) has been used to successfully deliver herbicide-resistant genes into tobacco [82, 83] and maize [84] by modifying the acetolactate synthase genes—endogenes already present in the plant genomes.
CRISPR/Cas9 has become the most universal and user-friendly tool for fine targeted genome editing and is applicable to almost all living species. Most commonly, CRISPR/Cas9 systems are used to create loss of function via knockout of protein coding genes (e.g., small insertion/deletion mutations in the nucleic acid sequence and consequently creation of early stop codons and frame shifts that severely disrupt the amino acids in the protein product). However, many uses of gene targeting demand the replacement of some alleles with others or the insertion of genes into particular genome regions. Therefore, technology that can create gain of function mutations is in high demand. To date, the CRISPR/Cas9-mediated gene knock-in allows for one-to-one substitution of DNA sequence or the insertion of a gene in a specific locus in the plant genome. For example, a single-stranded oligo with a KpnI+EcoRI site was introduced into the rice (Oryza sativa) phytoene desaturase gene OsPDS using protoplast transfection [85]. Also, gene replacement by homology-directed repair was accomplished with the presence of a DNA donor upon guide RNA (gRNA) combined with Cas9-mediated generation of a double-strand break in tobacco protoplasts [86]. However, in general, successful gene knock-ins in plants have been limited. One promising development, which generated in-frame gene knock-ins and amino acid substitutions, was accomplished by sequential transformation of Arabidopsis with a construct that induced germline-specific Cas9 expression together with gRNA and a donor sequence [87]. Other new precision gene editing tools include CRISPR-based prime editing which can perform targeted small insertions, deletions, and base swapping in a precise way without donor DNA [88]. Although prime editing was originally developed in yeast and human cells, the technique has been successfully applied to several plant species, such as rice and wheat [89–91]. Another new method is RNA-guided DNA insertion with CRISPR-associated transposases. Here, the reprogrammed transposases efficiently and specifically insert DNA both in vitro and into the Escherichia coli genome [92]. Similarly, another group reported a tool termed “insertion of transposable elements by gRNA-assisted targeting” that enabled highly specific, genome-wide DNA insertion across dozens of unique target sites in the E. coli genome [93]. Such CRISPR-mediated site-specific insertion, if applicable to plants and PHA biosynthesis regulons, may help mitigate context-variable behavior of promoters.
5.2. Plastome Modification
As an alternative to nuclear genome modification, plastome engineering provides a means to increase the expression level of PHA genes and consequently the yield of PHAs due to the polyploid nature of plastids. It has been estimated that there are approximately 500 to 5,000 copies of the plastid genome in a single plant cell [71]. Indeed, studies have confirmed that chloroplast transformation can lead to a very high level of transgene expression, which can exceed 70% of total soluble protein [94, 95]. Yet, reaching a profitable PHA yield via plastome modification is not as straightforward as expected. An early attempt, where the native R. eutropha phb operon was inserted into the tobacco plastome, showed only trace amounts (i.e., <0.05% of DW) of PHB accumulation [37], likely due to low genetic compatibility of the transgene cassette and poor transgene expression. Later efforts with optimized transgene expression cassette have been able to boost PHB yield. For example, the addition of a plastid-derived ribosome-binding site to each of the PHB synthesis genes in the native R. eutropha operon led to an increase in PHB yield to 0.016% of DW [96]. The fusion of a plastid promoter and plastid 5 UTR to the native bacterial operon was able to raise PHB yield to 1.7% of DW during early tissue culture stages [38]. Subsequent greenhouse studies, however, showed a much lower yield of approximately 0.1% of DW. In addition, the PHB-producing tobacco plants exhibited growth deficiency and male sterility. These growth defects could be circumvented with inducible synthesis at late tissue culture stages [39].
The highest level of PHB production achieved with plastome modification so far—18.8% of DW in leaves (and 8.8% of DW of the whole plant)—was reported by Bohmert-Tatarev et al. [40]. The authors created a synthetic operon with the PhbA and PhbC encoding genes from Acinetobacter sp., the PhbB encoding gene from Bacillus megaterium, an antibiotic selectable marker, and multiple regulatory elements known to yield high levels of plastidial recombinant protein production. The three PHB synthesis genes were chosen because they had a similar codon usage and GC content to the tobacco plastome. Meanwhile, regulatory elements with limited homology to the host plastome were used to reduce unwanted rearrangements between regions of the transgenic insert and the host plastome. Although the PHB-producing plants tended to be paler green and smaller than wild-type tobacco plants, they were fertile and produced viable seeds. These studies indicate that with proper optimizations, the plastome engineering-based system is capable of promoting PHB yield by nearly 60-fold compared with nucleus-encoded plastid targeting strategy in tobacco [42]. With a number of plant species, such as cotton, soybean, sugarcane, oilseed rape, and poplar that are now amenable to plastid transformation [71, 97], plastome-targeted transgene cassettes could have a wider application in enhancing plant-based PHB production.
One inherent problem with plastid transformation, however, is low transgene expression in nongreen plastids. This phenomenon was reported in plastome engineering-enabled PHB synthesis (e.g., when the phb operon was inserted into the tobacco plastome as an extension of the psbA operon [40]), creating a bottleneck to efficient PHA production especially when seed plastids are being targeted. To exploit nongreen plastids for PHA production, regulatory sequences that can improve transgene expression in chromoplasts, amyloplasts, and other nongreen plastid types need to be incorporated into the plastid transgene. One candidate could be the plastid gene clpP. It encodes the proteolytic subunit of the Clp ATP-dependent protease and can induce a high level of plastid transgene expression in amyloplasts in potato [98] and root plastids [99].
Plastome engineering could also be combined with nuclear genome transformation to further boost PHA biosynthesis efficiency. This approach has been adopted in the inducible PHB synthesis system created by Lössl et al. [39], which consisted of a nuclear-located, ethanol-inducible T7RNA polymerase and a plastid harboring phb operon under the control of T7 regulatory elements. Yet another direction to pursue would be to introduce genes that encode enzymes promoting the acetyl-CoA flux in plastids. Fuentes et al. [100] employed a similar design to increase the yield of artemisinic acid, the precursor for the antimalaria drug artemisinin, in tobacco. The authors first inserted the core pathway for artemisinic acid biosynthesis into the chloroplast genome. Then, they introduced gene cassettes encoding enzymes that affect the flux of the artemisinin pathway into the nuclear genome. The retransformation with nucleus-targeted gene cassettes led to a maximum 77-fold increase in artemisinic acid production in the transplastomic lines without noticeable negative impacts on plant growth and development.
In addition to modifying the endogenous plastome, building an exogenous, truly synthetic plastome, termed “synplastome,” and introducing it into plastids are now feasible in algae and land plant species [101, 102], which opens up more possibilities for exploiting the biosynthetic versatility of plastids for PHA production and storage. If it is smaller and engineered only for promotion of PHA production, perhaps it would have less adverse effects than modifying natural chloroplast functions or numbers, as discussed above.
6. The Promise of Systems Biology for Promoting Plant-Based PHA Biosynthesis
6.1. Omics Empowered Identification of Genetic Parts
Systems biology, aiming for a holistic description and understanding of a biological system, encompasses a diversity of technologies and methods. As a major area of systems biology, omics reflect the totality of a specific type of molecular constituents (for example, DNA, mRNA, proteins, metabolites, and ions) within a biological organism. Omics data can inform “SynBio,” including PHA engineering, in several ways. For example, the knowledge gained through omics-based studies can help to enlarge the number of genetic parts which synthetic biologists could build upon (Figure 3). In plants, several types of omics data covering transcriptomics, epigenomics, proteomics, and metabolomics have been generated while studying leaf senescence [103]. Likewise, transcriptomics and proteomics have been used to study plastid differentiation [104]. The understanding of how biological components, such as individual genes, interact with each other during these developmental processes could support efforts in building genetic circuits for senescence stage-specific PHA synthesis or improved PHA production and storage capacity in plastids, as discussed in sections above.
Most omics data generated in plants, however, are based on a tissue- or organ-scale, therefore unable to distinguish cells that are physically close together yet process different developmental properties. For example, different developmental regulatory programs have been suggested to exist between plastids in epidermal, mesophyll, guard, and pavement cells, which can result in variations in the total number and size of chloroplasts in these cell types. In addition, divergence in thylakoid membrane development has been found among different cell layers of shoot apical meristem [70]. Uncovering mechanisms governing the variations among these physically connected cells requires cell type-specific omics technologies. These technologies employ tissue digestion and cell sorting during sample collection and are capable of capturing omics data from a specific type of cell or a single cell [105]. Although mostly applied in animals, single-cell omics can be adapted to plants to elucidate mechanisms of plastid differentiation, as evidenced by studies on root development in Arabidopsis [106] and alkaloid localization in Catharanthus roseus [107]. In addition, single-cell omics, when used to examine plant cells that produce PHAs, might provide insights into toxicity of these polymers.
6.2. Evaluating the Performance of PHA-Producing Plants with Integrative Analysis of Multiomics Data
Omics data and associated analysis tools provide means to analyze the functioning of PHA-engineered plants as systems and therefore troubleshoot performance issues (Figure 3), such as the perturbation of endogenous acetyl-CoA pathways and the adverse effects on plant growth and development. In fact, to explore limitations to PHB accumulation in sugarcane chloroplasts, McQualter et al. [11] examined and compared changes in mRNA, proteins, and metabolites in PHB-producing lines and wild-type plants. The findings suggested that the presence of PHB granules scatters photosynthetically active radiation and physically disrupts thylakoid membranes, both of which could lead to ATP starvation in bundle sheath chloroplasts.
Integrative analysis of multiomics data serves as an effective way to assimilate multiple datasets into biologically meaningful interpretations. This has been highlighted in comparative analyses that also examined different omics datasets independently, for example, that adopted by McQualter et al. [11] in analyzing PHB-synthesizing sugarcane. Despite the analytical challenges, omics integration methods have been successfully applied to several studies of plants [108]. For example, in analyzing oxidative stress responses in cambium in poplar, Srivastava et al. [109] integrated three datasets, including transcriptomics, proteomics, and metabolomics, using a modified orthogonal partial least squares multivariate regression method. In another poplar-based study that was focused on lignin biosynthesis, Wang et al. [110] developed an integration method consisting of transcript/protein equations, mass balance kinetic equations, and multiple linear regression equations. The authors applied the integration analysis to four datasets—transcriptomic, proteomic, fluxomic, and phenomic data generated from 221 transgenic poplar lines and 18 wild-type plants. This integrative analysis enabled the prediction of how changing the expression of any pathway gene or gene combination can affect protein abundance, metabolic flux, and phenotypic traits including lignin content and composition, tree growth, wood density, and wood saccharification potential. Clearly, there are opportunities for integrating multiomics data derived from PHA-engineered and wild-type plants, estimating perturbations as a result of PHA biosynthesis, and identifying directions for future optimization efforts.
6.3. Simulating Metabolic Flux in PHA-Producing Plants Using Genome-Scale Metabolic Models
As one of the major systems-based approaches for metabolic studies, in silico genome-scale metabolic model (GEM) construction had been considered beyond reach for plant species. Yet in the past decade, several GEMs have been constructed for Arabidopsis, rice, tomato, maize, sorghum, and sugarcane [111, 112]. Although different GEMs were developed with unique domain considerations (for example, for organelles that represent plant metabolic network and interactions that occur between bundle sheath and mesophyll), generally speaking, these GEMs computationally describe a whole set of gene-protein-reaction associations for the entire metabolic network in a given organism [112]. A key advantage of GEMs is therefore that they can be used to predict metabolic fluxes for an entire set of metabolic reactions. For example, an Arabidopsis-based GEM (named “AraGEM”) was used in a six-tissue context to explore C/N partitioning and resource allocation across leaf, stem, and root systems within a diurnal cycle [113]. Recently, a multitissue GEM (named “MultiGEM”) was applied to four spatiotemporal compartments, namely, bundle sheath, mesophyll, day, and night, with the goal of elucidating causes of growth retardation and low PHB production in mesophyll plastids in C4 plants [17]. The results revealed that several factors, including photoassimilation capacity, carbon availability, ATP maintenance, relative photosynthetic activity of bundle sheath and mesophyll, and type of metabolites exchanged in the plasmodesmata, can affect PHB yield in leaf samples of C4 plants. With multiple GEMs available for the model species Arabidopsis [112], similar pipelines can be used to identify bottlenecks to efficient PHA production, at least in this model species. For a broader application of GEMs, however, the gap between the number of plant species with established GEMs and the type of crop plants suitable for industrial-scale manufacture of PHA polymers needs to be filled. Nonetheless, the progresses in GEM construction and application have made it possible to generate a wide range of hypotheses to test in future PHA engineering studies (Figure 3).
7. Concluding Remarks
Using traditional genetic engineering approaches, many laboratories—working in a number of plant species—have reliably overexpressed the bacterial PHA pathways. However, the goal of reaching economically viable yields (e.g., a minimum of 10% of DW) without substantially disturbing plant development does not appear to be feasible by simple ectopic expression. Plant-based PHA synthesis likely requires careful consideration of the timing and duration of biosynthesis for organelle-targeted PHA production, possible relocation, and storage. The use of inducible or native developmental stage-specific promoters in newly constructed genetic circuits could potentially help reach this goal. These genetic circuits may also include genes required for maximizing the production and storage capability of plastids and other targeted organelles for production or facilitating the relocation and secretion of the PHA polymers (Figure 2). Meanwhile, the advancement of CRISPR/Cas9-mediated site-specific insertion technologies makes it feasible to insert PHA genes into the plant genome to construct native promoter “operons,” which might help mitigate the context-variable behavior of promoters and promote precise external control of gene expression. A main risk of this approach, however, is that it may be difficult to get the desired high level of PHA synthesis. Compared with nuclear genome modification, plastome engineering, or combining it with nuclear genome modification, is likely to confer higher expression of PHA genes, due to the polyploid nature of plastids. Alternatively, synplastomes engineered specifically for PHA production might have less adverse effects than modifying natural chloroplast functions or numbers. The possibilities for identifying additional genes and genetic circuit structures for optimizing PHA production have been greatly expanded by the incorporation of omics analytical tools and mathematical modeling (Figure 3). As we are entering the era of “SynBio,” it is a good time to reconsider bioplastics engineering in plants using a biosystems design approach.
Acknowledgments
The writing of this manuscript was supported by U.S. Department of Energy (DOE), Office of Science, Biological and Environmental Research (BER), as part of the Plant-Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov), and the Center for Bioenergy Innovation (CBI), a U.S. DOE Bioenergy Research Center supported by the Office of Science. This manuscript has been authored by UT-Battelle, LLC, under Contract No. DE-AC05-00OR22725 with the U.S. DOE. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. DOE under Contract Number DE-AC05-00OR22725.
Contributor Information
Jin-Gui Chen, Email: chenj@ornl.gov.
Xiaohan Yang, Email: yangx@ornl.gov.
Disclosure
The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allows others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Authors’ Contributions
HL, SHS, XY, and JGC conceived the manuscript idea; HL and XY led the writing of the manuscript; HL and GY designed and draw the figures; all authors contributed to manuscript writing and revision. All authors read and approved the final version of the manuscript.
References
- 1.Wang S., Alseekh S., Fernie A. R., and Luo J., “The structure and function of major plant metabolite modifications,” Molecular Plant, vol. 12, no. 7, pp. 899–919, 2019 [DOI] [PubMed] [Google Scholar]
- 2.Pouvreau B., Vanhercke T., and Singh S., “From plant metabolic engineering to plant synthetic biology: the evolution of the design/build/test/learn cycle,” Plant Science, vol. 273, pp. 3–12, 2018 [DOI] [PubMed] [Google Scholar]
- 3.Capell T., and Christou P., “Progress in plant metabolic engineering,” Current Opinion in Biotechnology, vol. 15, no. 2, pp. 148–154, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Thompson R. C., Moore C. J., vom Saal F. S., and Swan S. H., “Plastics, the environment and human health: current consensus and future trends,” Philosophical Transactions of the Royal Society B: Biological Sciences, vol. 364, no. 1526, pp. 2153–2166, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi S. Y., Rhie M. N., Kim H. T., Joo J. C., Cho I. J., Son J., Jo S. Y., Sohn Y. J., Baritugo K. A., Pyo J., Lee Y., Lee S. Y., and Park S. J., “Metabolic engineering for the synthesis of polyesters: A 100-year journey from polyhydroxyalkanoates to non-natural microbial polyesters,” Metabolic Engineering, vol. 58, pp. 47–81, 2020 [DOI] [PubMed] [Google Scholar]
- 6.Critchell K., Bauer-Civiello A., Benham C., Berry K., Eagle L., Hamann M., Hussey K., and Ridgway T., “Plastic pollution in the coastal environment: current challenges and future solutions,” Coasts and Estuaries, Wolanski E., Day J. W., Elliott M., and Ramachandran R., Eds., Elsevier, pp. 595–609, 2019 [Google Scholar]
- 7.Bohmert K., Balbo I., Kopka J., Mittendorf V., Nawrath C., Poirier Y., Tischendorf G., Trethewey R. N., and Willmitzer L., “Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight,” Planta, vol. 211, no. 6, pp. 841–845, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Dalton D. A., Ma C., Shrestha S., Kitin P., and Strauss S. H., “Trade-offs between biomass growth and inducible biosynthesis of polyhydroxybutyrate in transgenic poplar,” Plant Biotechnology Journal, vol. 9, no. 7, pp. 759–767, 2011 [DOI] [PubMed] [Google Scholar]
- 9.McQualter R. B., Somleva M. N., Gebbie L. K., Li X., Petrasovits L. A., Snell K. D., Nielsen L. K., and Brumbley S. M., “Factors affecting polyhydroxybutyrate accumulation in mesophyll cells of sugarcane and switchgrass,” BMC Biotechnology, vol. 14, no. 1, p. 83, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McQualter R. B., Petrasovits L. A., Gebbie L. K., Schweitzer D., Blackman D. M., Chrysanthopoulos P., Hodson M. P., Plan M. R., Riches J. D., Snell K. D., Brumbley S. M., and Nielsen L. K., “The use of an acetoacetyl-CoA synthase in place of a β-ketothiolase enhances poly-3-hydroxybutyrate production in sugarcane mesophyll cells,” Plant Biotechnology Journal, vol. 13, no. 5, pp. 700–707, 2015 [DOI] [PubMed] [Google Scholar]
- 11.McQualter R. B., Bellasio C., Gebbie L. K., Petrasovits L. A., Palfreyman R. W., Hodson M. P., Plan M. R., Blackman D. M., Brumbley S. M., and Nielsen L. K., “Systems biology and metabolic modelling unveils limitations to polyhydroxybutyrate accumulation in sugarcane leaves; lessons for C4 engineering,” Plant Biotechnology Journal, vol. 14, no. 2, pp. 567–580, 2016 [DOI] [PubMed] [Google Scholar]
- 12.Choi K. R., Jang W. D., Yang D., Cho J. S., Park D., and Lee S. Y., “Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering,” Trends in Biotechnology, vol. 37, no. 8, pp. 817–837, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Meng D.-C., and Chen G.-Q., “Synthetic biology of polyhydroxyalkanoates (PHA),” Synthetic Biology – Metabolic Engineering, Zhao H., and Zeng A.-P., Eds., Springer International Publishing, pp. 147–174, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Lin Y., Wu Q., Wang Y., and Chen G.-Q., “Synthetic Biology and Genome-Editing Tools for Improving PHA Metabolic Engineering,” Trends in Biotechnology, vol. 38, no. 7, pp. 689–700, 2020 [DOI] [PubMed] [Google Scholar]
- 15.Medford J. I., and Prasad A., “Plant synthetic biology takes root,” Science, vol. 346, no. 6206, pp. 162–163, 2014 [DOI] [PubMed] [Google Scholar]
- 16.Kassaw T. K., Donayre-Torres A. J., Antunes M. S., Morey K. J., and Medford J. I., “Engineering synthetic regulatory circuits in plants,” Plant Science, vol. 273, pp. 13–22, 2018 [DOI] [PubMed] [Google Scholar]
- 17.de Oliveira Dal’Molin C. G., Quek L.-E., Saa P. A., Palfreyman R., and Nielsen L. K., “From reconstruction to C4 metabolic engineering: a case study for overproduction of polyhydroxybutyrate in bioenergy grasses,” Plant Science, vol. 273, pp. 50–60, 2018 [DOI] [PubMed] [Google Scholar]
- 18.Feike D., Korolev A. V., Soumpourou E., Murakami E., Reid D., Breakspear A., Rogers C., Radutoiu S., Stougaard J., Harwood W. A., Oldroyd G. E. D., and Miller J. B., “Characterizing standard genetic parts and establishing common principles for engineering legume and cereal roots,” Plant Biotechnology Journal, vol. 17, no. 12, pp. 2234–2245, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalton D. A., Murthy G., and Strauss S. H., “Production of traditional and novel biopolymers in transgenic woody plants,” Phytochemicals, Plant Growth, and the Environment, Gang D. R., Ed., Springer, New York, pp. 59–78, 2013 [Google Scholar]
- 20.Lemoigne M., “Produit de déshydratation et de polymérisation de l’acide b-oxybutyrique,” Bulletin de la Société de Chimie Biologique, vol. 8, pp. 770–782, 1926 [Google Scholar]
- 21.Okamura E., Tomita T., Sawa R., Nishiyama M., and Kuzuyama T., “Unprecedented acetoacetyl-coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway,” Proceedings of the National Academy of Sciences, vol. 107, no. 25, pp. 11265–11270, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pool R., “In search of the plastic potato: scientists in the emerging field of biopolymer engineering are aiming to produce bacteria and, eventually, food crops that are genetically tailored to yield a whole new breed of plastics,” Science, vol. 245, no. 4923, pp. 1187–1189, 1989 [DOI] [PubMed] [Google Scholar]
- 23.Oliver D. J., Nikolau B. J., and Wurtele E. S., “Acetyl-CoA—life at the metabolic nexus,” Plant Science, vol. 176, no. 5, pp. 597–601, 2009 [Google Scholar]
- 24.Poirier Y., Nawrath C., and Somerville C., “Production of Polyhydroxyalkanoates, a Family of Biodegradable Plastics and Elastomers, in Bacteria and Plants,” Bio/Technology, vol. 13, no. 2, pp. 142–150, 1995 [DOI] [PubMed] [Google Scholar]
- 25.Slater S., Mitsky T. A., Houmiel K. L., Hao M., Reiser S. E., Taylor N. B., Tran M., Valentin H. E., Rodriguez D. J., Stone D. A., Padgette S. R., Kishore G., and Gruys K. J., “Metabolic engineering of Arabidopsis and Brassica for poly(3-hydroxybutyrate- co -3-hydroxyvalerate) copolymer production,” Nature Biotechnology, vol. 17, no. 10, pp. 1011–1016, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Murthy G. S., Kumar D., Strauss S. H., Dalton D., and Vinocur J., “Feasibility analysis of poly-ß-hydroxybutyrate (PHB) extraction from hybrid poplar leaves,” 2010 Pittsburgh, Pennsylvania, June 20 - June 23, 2010, American Society of Agricultural and Biological Engineers, 2010 [Google Scholar]
- 27.Dalton D. A., Ma C., Murthy G. S., and Strauss S. H., “Bioplastic production by transgenic poplar,” Information Systems for Biotechnology News Report (Jan), 2012
- 28.Poirier Y., Dennis D. E., Klomparens K., and Somerville C., “Polyhydroxybutyrate, a Biodegradable Thermoplastic produced in transgenic plants,” Science, vol. 256, no. 5056, pp. 520–523, 1992 [DOI] [PubMed] [Google Scholar]
- 29.Poirier Y., and Brumbley S. M., “Metabolic engineering of plants for the synthesis of polyhydroxyalkanaotes,” Plastics from Bacteria: Natural Functions and Applications, Chen G. G.-Q., Ed., Springer, Berlin Heidelberg, pp. 187–211, 2010 [Google Scholar]
- 30.Somleva M. N., Peoples O. P., and Snell K. D., “PHA bioplastics, biochemicals, and energy from crops,” Plant Biotechnology Journal, vol. 11, no. 2, pp. 233–252, 2013 [DOI] [PubMed] [Google Scholar]
- 31.Snell K. D., Singh V., and Brumbley S. M., “Production of novel biopolymers in plants: recent technological advances and future prospects,” Current Opinion in Biotechnology, vol. 32, pp. 68–75, 2015 [DOI] [PubMed] [Google Scholar]
- 32.Dobrogojski J., Spychalski M., Luciński R., and Borek S., “Transgenic plants as a source of polyhydroxyalkanoates,” Acta Physiologiae Plantarum, vol. 40, no. 9, p. 162, 2018 [Google Scholar]
- 33.Nawrath C., Poirier Y., and Somerville C., “Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation,” Proceedings of the National Academy of Sciences, vol. 91, no. 26, pp. 12760–12764, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittendorf V., Robertson E. J., Leech R. M., Kruger N., Steinbuchel A., and Poirier Y., “Synthesis of medium-chain-length polyhydroxyalkanoates in arabidopsis thaliana using intermediates of peroxisomal fatty acid beta-oxidation,” Proceedings of the National Academy of Sciences of the United States of America, vol. 95, no. 23, pp. 13397–13402, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirier Y., Ventre G., and Caldelari D., “Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids,” Plant Physiology, vol. 121, no. 4, pp. 1359–1366, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tilbrook K., Gebbie L., Schenk P. M., Poirier Y., and Brumbley S. M., “Peroxisomal polyhydroxyalkanoate biosynthesis is a promising strategy for bioplastic production in high biomass crops,” Plant Biotechnology Journal, vol. 9, no. 9, pp. 958–969, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Nakashita H., Arai Y., Shikanai T., Doi Y., and Yamaguchi I., “Introduction of bacterial metabolism into higher plants by polycistronic transgene expression,” Bioscience, Biotechnology, and Biochemistry, vol. 65, pp. 1688–1691, 2014 [DOI] [PubMed] [Google Scholar]
- 38.Lössl A., Eibl C., Harloff H.-J., Jung C., and Koop H.-U., “Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction,” Plant Cell Reports, vol. 21, no. 9, pp. 891–899, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Lössl A., Bohmert K., Harloff H., Eibl C., Mühlbauer S., and Koop H. U., “Inducible trans-activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco,” Plant and Cell Physiology, vol. 46, no. 9, pp. 1462–1471, 2005 [DOI] [PubMed] [Google Scholar]
- 40.Bohmert-Tatarev K., McAvoy S., Daughtry S., Peoples O. P., and Snell K. D., “High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate,” Plant Physiology, vol. 155, no. 4, pp. 1690–1708, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arai Y., Nakashita H., Doi Y., and Yamaguchi I., “Plastid Targeting of Biosynthetic Pathway in Tobacco,” Plant Biotechnology, vol. 18, no. 4, pp. 289–293, 2001 [Google Scholar]
- 42.Bohmert K., Balbo I., Steinbüchel A., Tischendorf G., and Willmitzer L., “Constitutive Expression of the β-Gene in Transgenic Plants. A major obstacle for obtaining polyhydroxybutyrate-producing plants,” Plant Physiology, vol. 128, no. 4, pp. 1282–1290, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malik M. R., Yang W., Patterson N., Tang J., Wellinghoff R. L., Preuss M. L., Burkitt C., Sharma N., Ji Y., Jez J. M., Peoples O. P., Jaworski J. G., Cahoon E. B., and Snell K. D., “Production of high levels of poly-3-hydroxybutyrate in plastids of Camelina sativa seeds,” Plant Biotechnology Journal, vol. 13, no. 5, pp. 675–688, 2015 [DOI] [PubMed] [Google Scholar]
- 44.Petrasovits L. A., Purnell M. P., Nielsen L. K., and Brumbley S. M., “Production of polyhydroxybutyrate in sugarcane,” Plant Biotechnology Journal, vol. 5, no. 1, pp. 162–172, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Purnell M. P., Petrasovits L. A., Nielsen L. K., and Brumbley S. M., “Spatio-temporal characterization of polyhydroxybutyrate accumulation in sugarcane,” Plant Biotechnology Journal, vol. 5, no. 1, pp. 173–184, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Anderson D. J., Gnanasambandam A., Mills E., O’Shea M. G., Nielsen L. K., and Brumbley S. M., “Synthesis of short-chain-length/medium-chain length polyhydroxyalkanoate (PHA) copolymers in peroxisomes of transgenic sugarcane plants,” Tropical Plant Biology, vol. 4, no. 3-4, pp. 170–184, 2011 [Google Scholar]
- 47.Petrasovits L. A., Zhao L., McQualter R. B., Snell K. D., Somleva M. N., Patterson N. A., Nielsen L. K., and Brumbley S. M., “Enhanced polyhydroxybutyrate production in transgenic sugarcane,” Plant Biotechnology Journal, vol. 10, no. 5, pp. 569–578, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Petrasovits L. A., McQualter R. B., Gebbie L. K., Blackman D. M., Nielsen L. K., and Brumbley S. M., “Chemical inhibition of acetyl coenzyme A carboxylase as a strategy to increase polyhydroxybutyrate yields in transgenic sugarcane,” Plant Biotechnology Journal, vol. 11, no. 9, pp. 1146–1151, 2013 [DOI] [PubMed] [Google Scholar]
- 49.Beilen J. B. V., and Poirier Y., “Production of renewable polymers from crop plants,” The Plant Journal, vol. 54, no. 4, pp. 684–701, 2008 [DOI] [PubMed] [Google Scholar]
- 50.Fatland B. L., Nikolau B. J., and Wurtele E. S., “Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis,” Plant Cell, vol. 17, no. 1, pp. 182–203, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing S., van Deenen N., Magliano P., Frahm L., Forestier E., Nawrath C., Schaller H., Gronover C. S., Prüfer D., and Poirier Y., “ATP citrate lyase activity is post-translationally regulated by sink strength and impacts the wax, cutin and rubber biosynthetic pathways,” The Plant Journal, vol. 79, no. 2, pp. 270–284, 2014 [DOI] [PubMed] [Google Scholar]
- 52.Patterson N., Tang J., Cahoon E. B., Jaworski J. G., Yang W., Peoples O. P., and Snell K. D.. Generation of high polyhydroxybutrate producing oilseeds, 2012
- 53.Patterson N., Tang J., Han J., Tavva V., Hertig A., Zhang Z., Ramseier T. M., Bohmert-Tatarev K., Peoples O. P., and Snell K. D.. Generation of high polyhydroxybutyrate producing oilseeds, 2015
- 54.Somleva M., Chinnapen H., Ali A., Snell K. D., Peoples O. P., Patterson N., Tang J., and Bohmert-Tatarev K.. Increasing carbon flow for polyhydroxybutyrate production in biomass crops, 2012
- 55.Somleva M. N., Snell K. D., Beaulieu J. J., Peoples O. P., Garrison B. R., and Patterson N. A., “Production of polyhydroxybutyrate in switchgrass, a value-added co-product in an important lignocellulosic biomass crop,” Plant Biotechnology Journal, vol. 6, no. 7, pp. 663–678, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Kourtz L., Dillon K., Daughtry S., Peoples O. P., and Snell K. D., “Chemically inducible expression of the PHB biosynthetic pathway in Arabidopsis,” Transgenic Research, vol. 16, no. 6, pp. 759–769, 2007 [DOI] [PubMed] [Google Scholar]
- 57.Vega-Sánchez M. E., Loqué D., Lao J., Catena M., Verhertbruggen Y., Herter T., Yang F., Harholt J., Ebert B., Baidoo E. E. K., Keasling J. D., Scheller H. V., Heazlewood J. L., and Ronald P. C., “Engineering temporal accumulation of a low recalcitrance polysaccharide leads to increased C6 sugar content in plant cell walls,” Plant Biotechnology Journal, vol. 13, no. 7, pp. 903–914, 2015 [DOI] [PubMed] [Google Scholar]
- 58.Lu H., Gordon M. I., Amarasinghe V., and Strauss S. H., “Extensive transcriptome changes during seasonal leaf senescence in field-grown black cottonwood (Populus trichocarpa Nisqually-1),” Scientific Reports, vol. 10, no. 1, p. 6581, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yolcu S., Li X., Li S., and Kim Y. J., “Beyond the genetic code in leaf senescence,” Journal of Experimental Botany, vol. 69, no. 4, pp. 801–810, 2018 [DOI] [PubMed] [Google Scholar]
- 60.Kim J., Kim J. H., Lyu J. I., Woo H. R., and Lim P. O., “New insights into the regulation of leaf senescence in Arabidopsis,” Journal of Experimental Botany, vol. 69, no. 4, pp. 787–799, 2018 [DOI] [PubMed] [Google Scholar]
- 61.Tian S.-L., Li Z., Li L., Shah S. N. M., and Gong Z.-H., “Analysis of tandem repeat units of the promoter of capsanthin/capsorubin synthase (Ccs) gene in pepper fruit,” Physiology and Molecular Biology of Plants, vol. 23, no. 3, pp. 685–691, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaumberg K. A., Antunes M. S., Kassaw T. K., Xu W., Zalewski C. S., Medford J. I., and Prasad A., “Quantitative characterization of genetic parts and circuits for plant synthetic biology,” Nature Methods, vol. 13, no. 1, pp. 94–100, 2016 [DOI] [PubMed] [Google Scholar]
- 63.She W., Lin W., Zhu Y., Chen Y., Jin W., Yang Y., Han N., Bian H., Zhu M., and Wang J., “ThegypsyInsulator ofDrosophila melanogaster, Together With Its Binding Protein Suppressor of Hairy-Wing, facilitate high and precise expression of transgenes inArabidopsis thaliana,” Genetics, vol. 185, no. 4, pp. 1141–1150, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osteryoung K. W., and Pyke K. A., “Division and dynamic morphology of plastids,” Annual Review of Plant Biology, vol. 65, no. 1, pp. 443–472, 2014 [DOI] [PubMed] [Google Scholar]
- 65.Chen C., MacCready J. S., Ducat D. C., and Osteryoung K. W., “The molecular machinery of chloroplast division,” Plant Physiology, vol. 176, no. 1, pp. 138–151, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiong D., Huang J., Peng S., and Li Y., “A few enlarged chloroplasts are less efficient in photosynthesis than a large population of small chloroplasts in Arabidopsis thaliana,” Scientific Reports, vol. 7, pp. 1–12, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okazaki K., Miyagishima S., and Wada H., “Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis,” Plant Cell, vol. 27, no. 3, pp. 663–674, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pyke K. A., and Leech R. M., “A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana,” Plant Physiology, vol. 104, no. 1, pp. 201–207, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cole L. W., “The evolution of per-cell organelle number,” Frontiers in Cell and Development Biology, vol. 4, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liebers M., Grübler B., Chevalier F., Lerbs-Mache S., Merendino L., Blanvillain R., and Pfannschmidt T., “Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development,” Frontiers in Plant Science, vol. 8, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jensen P. E., and Scharff L. B., “Engineering of plastids to optimize the production of high-value metabolites and proteins,” Current Opinion in Biotechnology, vol. 59, pp. 8–15, 2019 [DOI] [PubMed] [Google Scholar]
- 72.Zhao J., “Flavonoid transport mechanisms: how to go, and with whom,” Trends in Plant Science, vol. 20, no. 9, pp. 576–585, 2015 [DOI] [PubMed] [Google Scholar]
- 73.Sun T., Yuan H., Cao H., Yazdani M., Tadmor Y., and Li L., “Carotenoid metabolism in plants: the role of plastids,” Molecular Plant, vol. 11, no. 1, pp. 58–74, 2018 [DOI] [PubMed] [Google Scholar]
- 74.Pötter M., Müller H., Reinecke F., Wieczorek R., Fricke F., Bowien B., Friedrich B., and Steinbüchel A., “The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha,” Microbiology, vol. 150, no. 7, pp. 2301–2311, 2004 [DOI] [PubMed] [Google Scholar]
- 75.Steinbüchel A., Aerts K., Liebergesell M., Wieczorek R., Babel W., Föllner C., Madkour M. H., Mayer F., Pieper-Fürst U., Pries A., and Valentin H. E., “Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions,” Canadian Journal of Microbiology, vol. 41, no. 13, pp. 94–105, 1995 [DOI] [PubMed] [Google Scholar]
- 76.Rahman A., Linton E., Hatch A. D., Sims R. C., and Miller C. D., “Secretion of polyhydroxybutyrate in Escherichia coli using a synthetic biological engineering approach,” Journal of Biological Engineering, vol. 7, no. 1, p. 24, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parmenter D. L., Boothe J. G., van Rooijen G. J. H., Yeung E. C., and Moloney M. M., “Production of biologically active hirudin in plant seeds using oleosin partitioning,” Plant Molecular Biology, vol. 29, no. 6, pp. 1167–1180, 1995 [DOI] [PubMed] [Google Scholar]
- 78.Chaudhary S., Parmenter D. L., and Moloney M. M., “Transgenic Brassica carinata as a vehicle for the production of recombinant proteins in seeds,” Plant Cell Reports, vol. 17, no. 3, pp. 195–200, 1998 [DOI] [PubMed] [Google Scholar]
- 79.Tabata R., and Sawa S., “Maturation processes and structures of small secreted peptides in plants,” Frontiers in Plant Science, vol. 5, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okamoto S., Tabata R., and Matsubayashi Y., “Long-distance peptide signaling essential for nutrient homeostasis in plants,” Current Opinion in Plant Biology, vol. 34, pp. 35–40, 2016 [DOI] [PubMed] [Google Scholar]
- 81.Liu Z., Chen O., Wall J. B. J., Zheng M., Zhou Y., Wang L., Vaseghi H. R., Qian L., and Liu J., “Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector,” Scientific Reports, vol. 7, pp. 1–9, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Townsend J. A., Wright D. A., Winfrey R. J., Fu F., Maeder M. L., Joung J. K., and Voytas D. F., “High-frequency modification of plant genes using engineered zinc-finger nucleases,” Nature, vol. 459, no. 7245, pp. 442–445, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Zhang F., Li X., Baller J. A., Qi Y., Starker C. G., Bogdanove A. J., and Voytas D. F., “Transcription Activator-Like Effector Nucleases Enable Efficient Plant Genome Engineering,” Plant Physiology, vol. 161, no. 1, pp. 20–27, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Svitashev S., Young J. K., Schwartz C., Gao H., Falco S. C., and Cigan A. M., “Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA,” Plant Physiology, vol. 169, no. 2, pp. 931–945, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., Zhang K., Liu J., Xi J. J., Qiu J. L., and Gao C., “Targeted genome modification of crop plants using a CRISPR-Cas system,” Nature Biotechnology, vol. 31, no. 8, pp. 686–688, 2013 [DOI] [PubMed] [Google Scholar]
- 86.Li J.-F., Norville J. E., Aach J., McCormack M., Zhang D., Bush J., Church G. M., and Sheen J., “Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9,” Nature Biotechnology, vol. 31, no. 8, pp. 688–691, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miki D., Zhang W., Zeng W., Feng Z., and Zhu J.-K., “CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation,” Nature Communications, vol. 9, pp. 1–9, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., Chen P. J., Wilson C., Newby G. A., Raguram A., and Liu D. R., “Search-and-replace genome editing without double-strand breaks or donor DNA,” Nature, vol. 576, no. 7785, pp. 149–157, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A. V., Raguram A., Doman J. L., Liu D. R., and Gao C., “Prime genome editing in rice and wheat,” Nature Biotechnology, vol. 38, no. 5, pp. 582–585, 2020 [DOI] [PubMed] [Google Scholar]
- 90.Xu R., Li J., Liu X., Shan T., Qin R., and Wei P., “Development of plant prime-editing systems for precise genome editing,” Plant Communications, vol. 1, no. 3, p. 100043, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H., Li J., Chen J., Yan L., and Xia L., “Precise modifications of both exogenous and endogenous genes in rice by prime editing,” Molecular Plant, vol. 13, no. 5, pp. 671–674, 2020 [DOI] [PubMed] [Google Scholar]
- 92.Strecker J., Ladha A., Gardner Z., Schmid-Burgk J. L., Makarova K. S., Koonin E. V., and Zhang F., “RNA-guided DNA insertion with CRISPR-associated transposases,” Science, vol. 365, no. 6448, pp. 48–53, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klompe S. E., Vo P. L. H., Halpin-Healy T. S., and Sternberg S. H., “Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration,” Nature, vol. 571, no. 7764, pp. 219–225, 2019 [DOI] [PubMed] [Google Scholar]
- 94.Oey M., Lohse M., Kreikemeyer B., and Bock R., “Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic,” The Plant Journal, vol. 57, no. 3, pp. 436–445, 2009 [DOI] [PubMed] [Google Scholar]
- 95.Ruhlman T., Verma D., Samson N., and Daniell H., “The role of heterologous chloroplast sequence elements in transgene integration and expression,” Plant Physiology, vol. 152, no. 4, pp. 2088–2104, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arai Y., Shikanai T., Doi Y., Yoshida S., Yamaguchi I., and Nakashita H., “Production of polyhydroxybutyrate by polycistronic expression of bacterial genes in tobacco plastid,” Plant & Cell Physiology, vol. 45, no. 9, pp. 1176–1184, 2004 [DOI] [PubMed] [Google Scholar]
- 97.Ahmad N., Michoux F., Lössl A. G., and Nixon P. J., “Challenges and perspectives in commercializing plastid transformation technology,” Journal of Experimental Botany, vol. 67, no. 21, pp. 5945–5960, 2016 [DOI] [PubMed] [Google Scholar]
- 98.Valkov V. T., Gargano D., Manna C., Formisano G., Dix P. J., Gray J. C., Scotti N., and Cardi T., “High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5’ and 3’ regulatory sequences,” Transgenic Research, vol. 20, no. 1, pp. 137–151, 2011 [DOI] [PubMed] [Google Scholar]
- 99.Zhang J., Ruf S., Hasse C., Childs L., Scharff L. B., and Bock R., “Identification ofcis-elements conferring high levels of gene expression in non-green plastids,” The Plant Journal, vol. 72, no. 1, pp. 115–128, 2012 [DOI] [PubMed] [Google Scholar]
- 100.Fuentes P., Zhou F., Erban A., Karcher D., Kopka J., and Bock R., “A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop,” eLife, vol. 5, article e13664, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O'Neill B. M., Mikkelson K. L., Gutierrez N. M., Cunningham J. L., Wolff K. L., Szyjka S. J., Yohn C. B., Redding K. E., and Mendez M. J., “An exogenous chloroplast genome for complex sequence manipulation in algae,” Nucleic Acids Research, vol. 40, no. 6, pp. 2782–2792, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Piatek A. A., Lenaghan S. C., and Neal Stewart C., “Advanced editing of the nuclear and plastid genomes in plants,” Plant Science, vol. 273, pp. 42–49, 2018 [DOI] [PubMed] [Google Scholar]
- 103.Kim J., Woo H. R., and Nam H. G., “Toward systems understanding of leaf senescence: an integrated multi-omics perspective on leaf senescence research,” Molecular Plant, vol. 9, no. 6, pp. 813–825, 2016 [DOI] [PubMed] [Google Scholar]
- 104.Sadali N. M., Sowden R. G., Ling Q., and Jarvis R. P., “Differentiation of chromoplasts and other plastids in plants,” Plant Cell Reports, vol. 38, no. 7, pp. 803–818, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chappell L., Russell A. J. C., and Voet T., “Single-cell (multi)omics technologies,” Annual Review of Genomics and Human Genetics, vol. 19, no. 1, pp. 15–41, 2018 [DOI] [PubMed] [Google Scholar]
- 106.Efroni I., Ip P.-L., Nawy T., Mello A., and Birnbaum K. D., “Quantification of cell identity from single-cell gene expression profiles,” Genome Biology, vol. 16, no. 1, p. 9, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yamamoto K., Takahashi K., Caputi L., Mizuno H., Rodriguez-Lopez C. E., Iwasaki T., Ishizaki K., Fukaki H., Ohnishi M., Yamazaki M., Masujima T., O'Connor S. E., and Mimura T., “The complexity of intercellular localisation of alkaloids revealed by single-cell metabolomics,” The New Phytologist, vol. 224, no. 2, pp. 848–859, 2019 [DOI] [PubMed] [Google Scholar]
- 108.Rajasundaram D., and Selbig J., “More effort — more results: recent advances in integrative ‘omics’ data analysis,” Current Opinion in Plant Biology, vol. 30, pp. 57–61, 2016 [DOI] [PubMed] [Google Scholar]
- 109.Srivastava V., Obudulu O., Bygdell J., Löfstedt T., Rydén P., Nilsson R., Ahnlund M., Johansson A., Jonsson P., Freyhult E., Qvarnström J., Karlsson J., Melzer M., Moritz T., Trygg J., Hvidsten T. R., and Wingsle G., “OnPLS integration of transcriptomic, proteomic and metabolomic data shows multi-level oxidative stress responses in the cambium of transgenic hipI- superoxide dismutase Populus plants,” BMC Genomics, vol. 14, no. 1, p. 893, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang J. P., Matthews M. L., Williams C. M., Shi R., Yang C., Tunlaya-Anukit S., Chen H. C., Li Q., Liu J., Lin C. Y., and Naik P., “Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis,” Nature Communications, vol. 9, pp. 1–16, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gomes de Oliveira Dal’Molin C., and Nielsen L. K., “Plant genome-scale reconstruction: from single cell to multi-tissue modelling and omics analyses,” Current opinion in biotechnology, vol. 49, pp. 42–48, 2018 [DOI] [PubMed] [Google Scholar]
- 112.Gu C., Kim G. B., Kim W. J., Kim H. U., and Lee S. Y., “Current status and applications of genome-scale metabolic models,” Genome Biology, vol. 20, no. 1, p. 121, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gomes de Oliveira Dal’Molin C., Quek L.-E., Saa P. A., and Nielsen L. K., “A multi-tissue genome-scale metabolic modeling framework for the analysis of whole plant systems,” Frontiers in plant science, vol. 6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]