Abstract
Carbapenems are atypical β-lactam antibiotics with a broade antibacterial spectrum and strong antibacterial activity; however, the emergence and spread of carbapenemases have led to a decline in their effectiveness. New Delhi metallo-β-lactamase (NDM) is an important carbapenemase that has attracted widespread attention and poses a major threat to public health. To investigate the epidemiological characteristics of blaNDM in swine and chicken farms in southwestern China, we isolated 102 blaNDM-positive Enterobacterales strains from 18 farms in Sichuan and Yunnan provinces in 2021, with Escherichia coli and Klebsiella spp. being the main reservoirs of blaNDM, variant blaNDM-5 being the most prevalent, and all strains being multi-drug resistant. Whole-genome sequencing analysis of 102 blaNDM-positive Enterobacterales strains revealed that blaNDM had spread primarily through its carriers on the same farm and among the 18 farms in this study. A high degree of genetic similarity between animal-derived blaNDM-positive Escherichia coli strains and human-derived strains was also identified, suggesting a potential mutual transmission between them. Nanopore sequencing results indicated that blaNDM is predominantly present on the IncX3 plasmid, that an insertion sequence might be important for recombination in the blaNDM genetic environment, and that most of the plasmids carrying blaNDM are transferable. Collectively, our results enrich the current epidemiological information regarding blaNDM in pig and chicken farms in Southwest China, revealing its transmission pattern, as well as the potential risk of transmission to humans, which could help to better understand and control the spread of blaNDM.
Keywords: antibiotic resistance, Enterobacterales, Carbapenemase, bla NDM , transmission
1. Introduction
The issue of antibiotic multi-resistance in bacteria is crucial because of the significant threat it poses to human health [1]. Carbapenems are considered important antibiotics for use against multi-drug-resistant (MDR) bacteria (resistant to at least three types of antibiotics). However, after carbapenems’s clinical use, associated resistant strains have started to become prevalent [2,3]. Three classes of carbapenemases play a major role in the prevalence of carbapenem-resistant bacteria, namely class A (KPC, SME, GES, etc.), class B (IMP-, VIM-, and NDM-like metallo-β-lactamases (MβLs)), and class D (OXA) [4]. New Delhi metallo-β-lactamase (NDM) is an MβL that hydrolyses most β-lactams, but not monobactams [5]. blaNDM-1-carrying strains were identified in hospital patients in India in 2009, and since then, strains carrying this gene or its variants have been widely identified in other countries [6,7]. Moreover, in recent studies, NDM appeared to be a significant cause of bacterial resistance to carbapenems [5,8,9,10,11].
Although the use of carbapenems has never been allowed in the animal breeding process, the current percentage of livestock farm animals carrying NDM-positive strains cannot be ignored [12]. However, because of the close relationship between farmed animals and humans, these blaNDM-positive strains can still be transmitted to humans, representing a potential threat to human health. Therefore, monitoring animal-derived blaNDM-positive strains is of public health importance.
In this study, to refine the investigations of animals carrying NDM-positive bacteria in pig and chicken farms in the Sichuan and Yunnan provinces of China and to assess the transmission pattern of the blaNDM gene among them and to assess the risk of transmission to humans via farmed animals, we collected samples from these locations. Here, 102 NDM-positive strains were isolated from 1512 samples, and we evaluated their prevalence and molecular epidemiological characteristics. We also assessed the transmission pattern of the blaNDM gene by combining the relevant information in the database.
2. Materials and Methods
The animal experiment operations involved in this study were maintained in compliance with the Approval from the Medical Ethics Committee of Sichuan University (License: K2022018).
2.1. Sample Collection, Strain Isolation, and Identification
The samples in this study (n = 1512) were obtained from pig (n = 942) and chicken farms (n = 570) in the Sichuan and Yunnan provinces, and the types included fecal and anal swabs (Supplementary Table S1). The samples were transported in a cryopreserved state, and anal swabs and 0.5 g stool samples were taken and placed in 10 mL of BHI broth medium (Luqiao, Beijing, China) at 37 °C, with shaking at 180 rpm, and incubated for 12 h. Then, 100 µL of medium was spread onto EMB (Eosin Methylene Blue, Luqiao, Beijing, China) agar medium containing 1 μg/mL meropenem and cultured at 37 °C for 16 h, and 1~2 colonies of different shapes were picked for purification and culture. The 16S rRNA and blaNDM genes of the purified strain were amplified using PCR and sequenced [13,14].
2.2. Antibiotic Susceptibility Testing
The broth microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) was used to determine the susceptibility of all strains to the following antibiotics: gentamicin (GEN), florfenicol (FFC), polymyxin B (PMB), ciprofloxacin (CIP), cefoxitin (FOX), fosfomycin (FOS), aztreonam (ATM), doxycycline (DOX), norfloxacin (NOR), tetracycline (TET), tigecycline (TGC), ceftazidime (CAZ), amoxicillin/clavulanate (2:1) (AMC), ceftriaxone (CRO), amikacin (AMK), trimethoprim-sulphamethoxazole 1:19 (SXT), and meropenem (MEM). Interpretation was conducted according to the relevant resistance criteria in the CLSI document M100-S22 [15,16]. Escherichia coli ATCC25922 was used as the quality control strain.
2.3. Whole Genome Sequencing and Assembly
Genomic DNA of 102 NDM-positive strains was extracted using the Tiangen Bacterial DNA Extraction Kit. The entire genomic DNA was fully sequenced using the Illumina HiSeq system (Novogene, Tianjin, China), and raw data were evaluated for quality and filtered by fastaQC v0.12.1 and Cutadapt v4.2 [17,18]. Clean reads were assembled into contigs using SAPdes v.3.15.2 [19]. Whole genome sequencing of strains was performed using the Oxford Nanopore GridION (Novogene, Tianjin, China) and Illumina platforms. Using unicycler v0.5.0 [20], Illumina platform sequencing data and Nanopore sequencing data were subjected to mixed stitching.
2.4. Bioinformatics Analysis
The nucleotide sequence files of strains classified according to different species were annotated using Prokka v1.14.5 [21], and the annotated files were used as the input for roary v3.13.0 [22] to obtain the core genome comparison sequences. The phylogenetic tree was further reconstructed randomly based on the maximum likelihood method using iqtree v2.1.2 [23] with the default parameters. MLST v2.23.0 (https://github.com/tseemann/mlst, 19 November 2022) was used to analyze the ST types of all of the strains [24]. The antibiotic resistance gene-related information of the isolates was predicted using ResFinder 4.0 [25]. The plasmid replicon species were determined via an analysis using PlasmidFinder 2.1 [26]. The minimum spanning tree of the multilocus sequence typing (MLST)-based isolates was constructed using GrapeTree [27]. GView Server (https://server.gview.ca, 9 December 2022) was used to generate pan-plasmid sequences of similar plasmids [28]. Visualization and embellishment of the phylogenetic tree were performed using ChiPlot [29]. Circle maps of the plasmids were generated using Proksee [30]. Gene cluster comparison maps were obtained using Clinker v0.0.27 [31].
2.5. Conjugation Experiment
The filter mating method was used to perform the conjugation experiment, and E. coli J53, which is resistant to sodium azide, was used as the recipient to assess the blaNDM gene transfer capacity from the blaNDM-positive strain to the recipient. The donor and recipient cells were cultivated in Luria–Bertani medium (Luqiao, Beijing, China) until they reached the logarithmic growth phase. They were then mixed at a 1:3 ratio and spread on a sterile 0.22-micron filter on a Luria–Bertani agar plate, where they were incubated for 24 h at 37 °C. Transformants were selected on LB agar plates with 4 mg/L meropenem and 100 mg/L sodium azide, and PCR-based confirmation was performed. The number of transformants divided by the total number of recipient bacteria was used to calculate the transfer frequency.
2.6. Data Availability
Information on the strains containing blaNDM-1 and blaNDM-5 variants (Dates to December 2022, Country: China, Species: E. coli, K. pneumoniae, K. quasipneumoniae, P. vermicola) obtained from the GenBank database is detailed in Supplementary Table S2. The nucleotide sequences of the bacterial genomes obtained from this study collection were deposited into the NCBI database and are publicly available under the accession number PRJNA909031. The NCBI database accession numbers of the plasmids carrying the blaNDM gene were OQ230782–OQ230789, OR233054-OR233062.
3. Results
3.1. Prevalence of blaNDM-Positive Bacteria in Farm Animals
Here, 102 blaNDM-positive strains (57 pigs and 45 chickens) were isolated from 1512 fecal or anal swab samples (942 pigs and 570 chickens) from 18 farms in the Sichuan and Yunnan provinces in 2021 (Detailed information in Supplementary Table S1). The total incidence of blaNDM-positive strains in animals was 6.75%, comprising 6.05% in pigs and 7.89% in chickens, which suggests that the blaNDM gene is carried at a high rate in farm animals. The 102 strains included 57 strains of E. coli, 31 strains of Klebsiella quasipneumoniae, 9 strains of Klebsiella pneumoniae, 4 strains of Providencia vermicola, and 1 strain of Proteus mirabilis. Two blaNDM subtypes were identified through further sequencing, namely blaNDM-1 (29, 28.5%) and blaNDM-5 (73, 71.6%). blaNDM-5 was determined to be the main common variant. Details of this are presented in Supplementary Table S1.
3.2. Antimicrobial Susceptibility, Antibiotic Resistance Genes, and Plasmids of NDM-Positive Strains
Phenotypic results of the 102 NDM-positive strains based on resistance to 17 antibiotics showed that these strains were MDR (Figure 1, Supplementary Table S3). These strains showed very high rates of resistance to GEN, AMK, CIP, TET, FFC, SXT, and NOR, reaching 91.2%, 97.1%, 86.3%, 85.3%, 86.3%, and 71.6%, respectively. All of the strains were resistant to FOX, CAZ, CRO, MEM, and AMC. Some isolates had MICs up to 64 µg/mL for meropenem. K. pneumoniae was completely resistant to GEN, AMK, TET, FFC, FOS, and SXT. K. quasipneumoniae was also resistant to AMK, NOR, CIP, and TET at a rate of approximately 100%. Of note, the rate of TGC resistance for K. quasipneumoniae strains in this study reached 58.1% (18/31), which is high. E. coli, K. pneumoniae, and K. quasipneumoniae were all found to be susceptible to PMB.
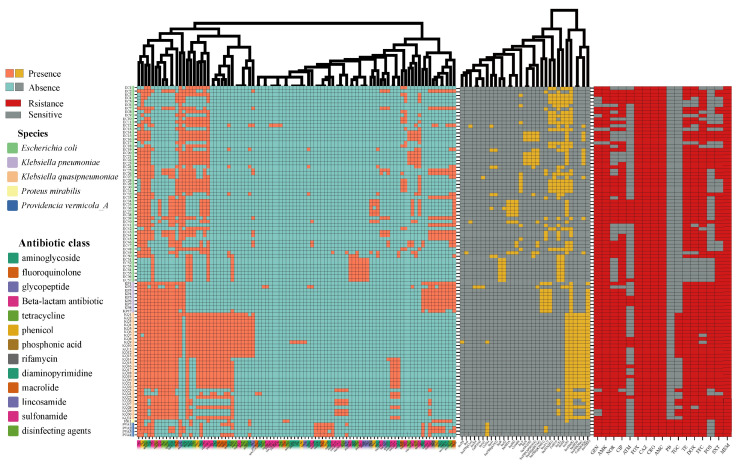
Figure 1.
Heat map of resistance genes, plasmid replicons, and antibiotic resistance phenotypes of 102 blaNDM-positive strains. Orange and yellow: present; blue and grey: absent; red: resistant; grey: sensitive. Clustering of resistance genes and plasmid replicons on the x axis.
According to the genome analysis results, 94 drug-resistant genes in 13 categories were found in the blaNDM-positive strains (Figure 1, Supplementary Table S4). We found that different bacterial species displayed different antibiotic-resistance genotypes. The antibiotic-resistance genes with a high prevalence in E. coli were aac(3)-Iid (63.2%), aadA2 (68.4%), aph(3’)-Ia (73.7%), tet(A) (77.2%), floR (78.9%), ble (84.2%), dfrA12 (68.4%), and sul3 (61.4%). Meanwhile, sul1, sul2, floR, tet(A), aph(3′)-Ia, fosA, blaLAP-2, oqxB, oqxA, aph(6)-Id, aph(3″)-Ib, qnrS1, ble, aac(3)-Iid, dfrA1, blaNDM-1, arr-3, catB3, dfrA27, aadA16, aac(6’)-Ib-cr5, qacEdelta1, and mph(A) were present in almost all K. pneumoniae genomes (>88.9%). Furthermore, sul1, sul2, floR, tet(A), fosA, blaLAP-2, oqxA, oqxB, aph(6)-Id, and aph(3″)-Ib resistance genes were present in almost all (>95%) K. quasipneumoniae isolates. Finally, the antibiotic resistance genes tet(X4) (16.1%, five strains from four farms) and tmexC1-tmexD1-toprJ1 (41.9%, 13 strains from 7 farms), related to tigecycline, an important clinical antibiotic, were highly prevalent in K. quasipneumoniae (Supplementary Table S4).
Furthermore, plasmid sequence analysis showed that the 102 blaNDM-positive strains carried 27 types of plasmids (Figure 1, Supplementary Table S4). Five plasmid types, IncFIB (66.7%), ColE10 (35.1%), IncFII (52.6%), IncX1 (61.4%), and IncX3 (66.7%) in E. coli were present at higher rates. Almost all K. pneumoniae isolates carried IncA/C2 (88.7%), IncFIA (HI1) (88.7%), IncFIB (100%), and IncFII (K) (88.7%) types. Moreover, IncFIB, IncFII(K), and IncX3 were detected in all K. quasipneumoniae isolates. Col (MG828) (87.1%) and IncHI1B (83.9%) also exhibited a high prevalence among the K. quasipneumoniae strain. Overall, IncFIB (76.5%), IncX3 (69.6%), Col (MG828) (38.2%), IncFII(K) (38.2%), and IncX1 (39.2%) were the most commonly carried plasmid types in blaNDM-positive strains in this study.
3.3. MLST and Phylogenetic Analysis
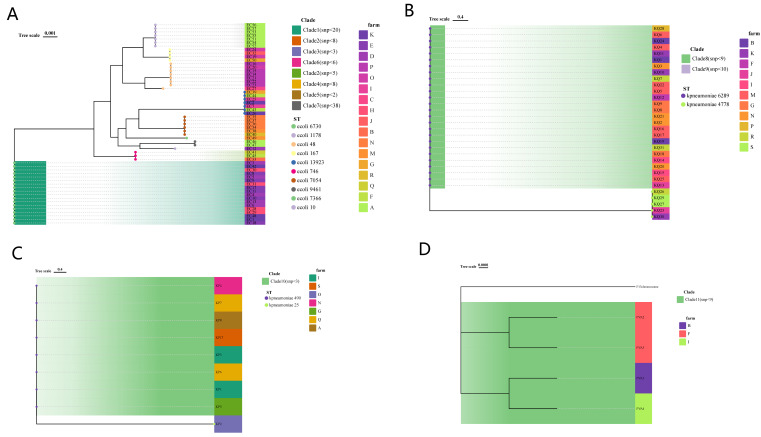
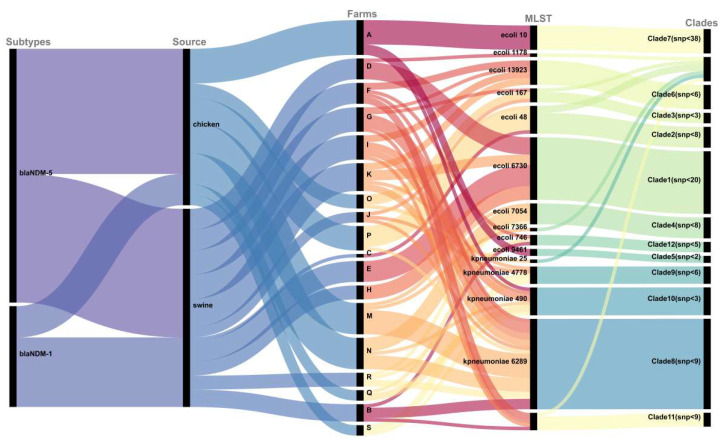
By performing MLST on 102 strains, it was found that 57 E. coli strains in this study could be classified into 10 different sequence types (STs), among which ST6730 (18/57), ST48 (8/57), ST10 (7/57), ST13923 (7/57), and ST7054 (6/57) were common, accounting for approximately 80.7% of the total. The remaining about one-fifth are ST167, ST7366, ST9461, ST1178, and ST746. K. pneumoniae and K. quasipneumoniae were assigned to four ST types, namely ST490 (8/9), ST5 (1/9), ST148 (26/31), and ST51 (5/31). E. coli ST13923 and K. pneumoniae ST51 and ST148 were considered new ST types. The constructed core genome phylogenetic tree was very similar to the MLST results, with the strains sharing the same ST type clustering together, and the number of SNPs was limited. Interestingly, we found that P. vermicola from three farms in Sichuan Province and Yunnan Province had a close evolutionary relationship (SNP < 35) with a strain (P13) isolated from the blood of Wenzhou Hospital patients in China (Figure 2 and Figure 3 and Supplementary Tables S1, S2 and S5) [32].
Figure 2.
SNP phylogenetic tree of E. coli (A), Klebsiella pneumoniae (B), Klebsiella quasipneumoniae (C), and Providencia vermicola (D) in this study. MLST, source farms were illustrated.
Figure 3.
Sankey diagram of blaNDM variants of 102 isolates, source farms, ST types, and phylogenetic tree branches. The diameter of the line is proportional to the number of isolates.
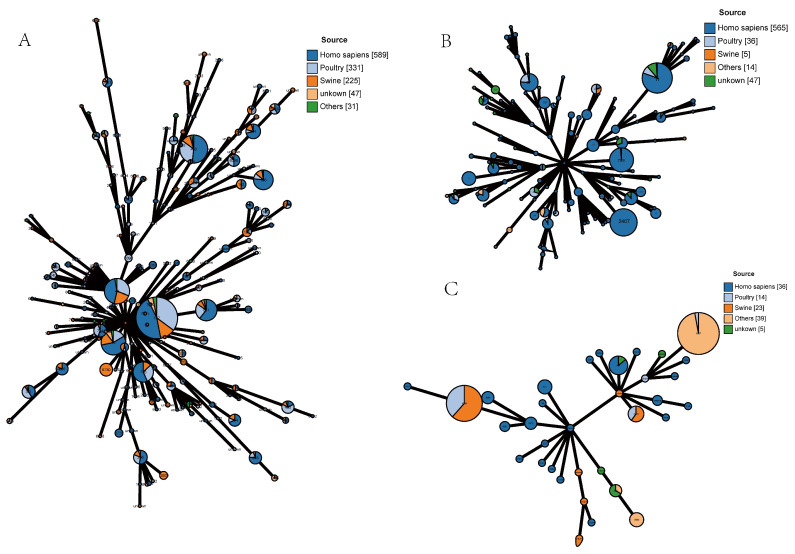
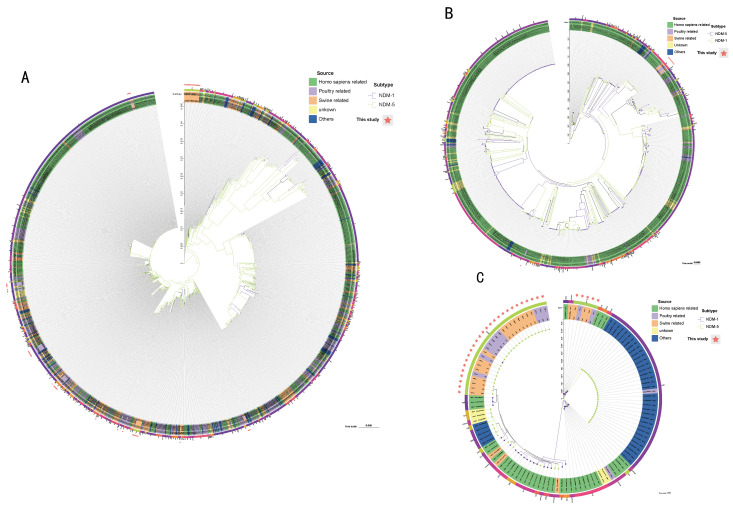
To further understand the epidemiological characteristics of blaNDM-positive strains, we analyzed the strains in this study together with blaNDM-positive strains from China in the NCBI database. Among E. coli isolates, ST167, ST10, ST48, ST746, ST617, and ST410 were the most prevalent STs, and they were all found to exist in humans, pigs, chickens, and other places. However, ST6730 was only isolated from pigs (Figure 4, Supplementary Table S2). The phylogenetic tree showed the phylogenetic relationships among strains from different sources, and some animal-derived isolates and human-derived isolates were found to have a very close evolutionary relationship (Figure 5, Supplementary Table S6). For example, there were only two SNP differences between the core genomes of strains EC51-EC57 in this study and strain Ec-03 (GCA_001893865.1) isolated from human urine in 2012. Strains EC19, EC24, EC32, and EC50 and strains E1-E6 (PRJNA298278, isolated in 2014 from the blood of a patient in a hospital in Zhejiang Province), strain SCEC020016 (GCA_002164985.1, isolated in 2016 from a human in a hospital in Sichuan Province), strain E4903 (GCA002265005.1, isolated from human blood of a hospital in Henan Province in 2015), and strain L743 (GCA_002870165.1, isolated from human feces of a hospital in Hangzhou City in 2017) share SNPs less than 24. For K. pneumoniae and K. quasipneumoniae, the prevailing STs were ST2407, ST789, ST11, ST37, ST6289, ST1697, ST414, and ST4778 (Figure 4). Combined with the corresponding phylogenetic tree (Figure 5), overall, unlike that with E. coli, both strains showed a strong host specificity, and the evolutionary relationship between strains from different sources was distant.
Figure 4.
MLST allele profiles of E. coli, Klebsiella pneumoniae, and Klebsiella quasipneumoniae in this study with corresponding blaNDM-positive strains from China in the NCBI gene assembly database. (A) 57 E. coli strains with 1166 blaNDM-positive E. coli isolates from the database; (B) 9 Klebsiella pneumoniae strains with 658 blaNDM-positive Klebsiella pneumoniae isolates from the database; (C) 31 Klebsiella quasipneumoniae strains and 86 blaNDM positive Klebsiella quasipneumoniae isolates from the database. Dark blue: human-associated source; light blue: avian-associated source; orange: pig-associated source; yellow: others, green: unknown.
Figure 5.
Maximum likelihood phylogenetic tree of E. coli, Klebsiella pneumoniae, and Klebsiella quasipneumoniae in this study with corresponding blaNDM-positive strains from China in the NCBI gene assembly database. (A) 57 E. coli strains with 1166 blaNDM-positive E. coli isolates in the database; (B) 31 Klebsiella pneumoniae strains with 658 blaNDM-positive Klebsiella pneumoniae isolates in the database; (C) 9 Klebsiella quasipneumoniae strains with 86 blaNDM-positive Klebsiella quasipneumoniae isolates in the database. The outermost circle is the MLST of the strain. Green: human-associated source; light pink: avian-associated source; orange: pig-associated source; yellow: others; blue: unknown.
3.4. Genetic Environments and Transferability of blaNDM
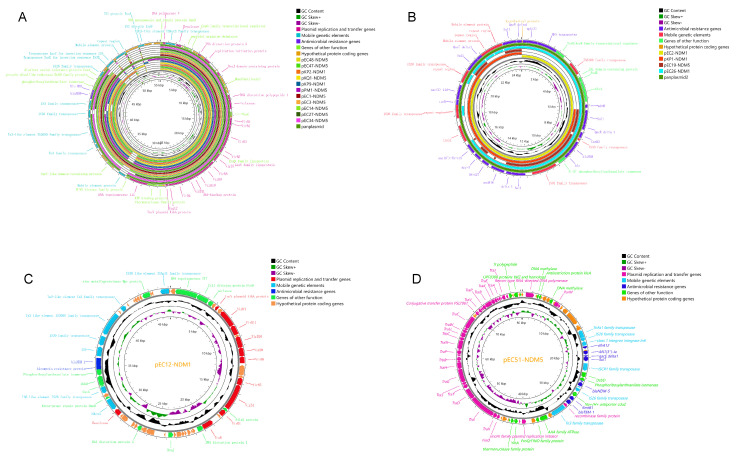
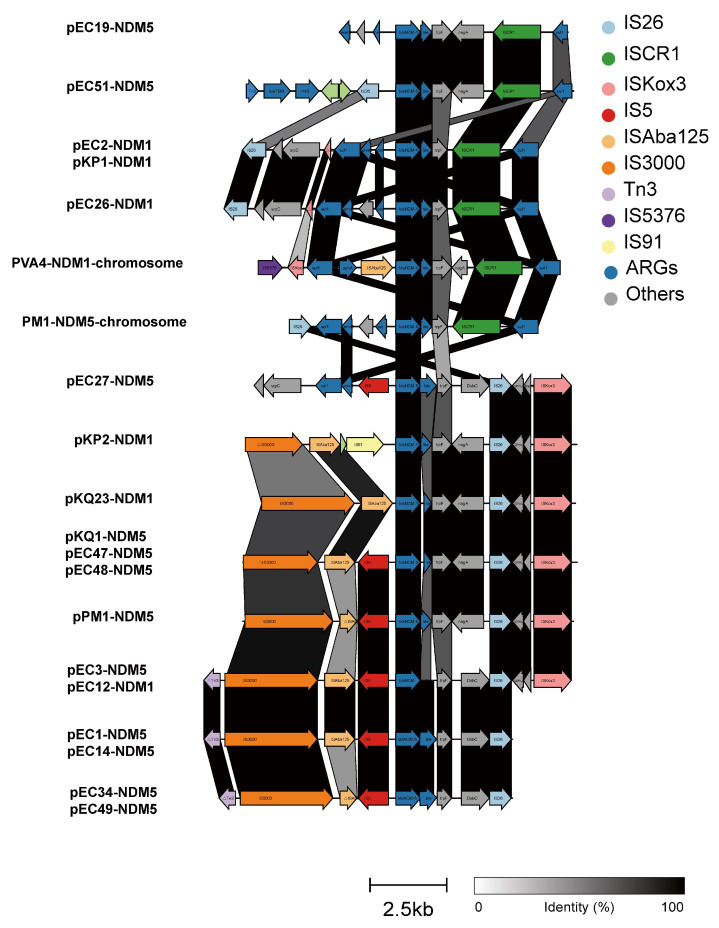
Based on the next-generation sequencing results of 102 strains, we selected the most similar plasmids by comparing the contigs containing the blaNDM gene with the NCBI database as a reference. The blaNDM genes in genetically distant strains share the same genetic environment. Complementary gaps were sequenced by PCR and nanopore long-read sequencing to obtain the complete genetic environment of the blaNDM genes. The blaNDM genes are located on the chromosome (Integrative Conjugative elements, ICEs) in P. vermicola and exists on both the chromosome (insertion sequence, IS) and the plasmid in P. mirabilis. The blaNDM genes in the other sequenced strains were found to all be located on the plasmids (Figure 6 and Figure 7). Of the 18 different plasmids, 13 belonged to the IncX3 type, pEC51-NDM-5 (80348 bp) belonged to the IncFII type, and the other four plasmids were not classified. The molecular weights of the complete plasmids range from 20817-80348bp (Supplementary Table S7). The genetic environment could be divided into 14 types, as follows: blaNDM-(Δ)ble-traF was the most conserved genetic structure, and (Δ)nagA often existed downstream of traF. In general, there were two types of mobile genetic elements downstream of blaNDM-(Δ)ble-traF, namely (I) ISCRⅠ and (II) IS26-umuD-(ISKox3). The upstream environment was found to be more complex, including IS26-ΔISKox3, IS5376-ΔISKox3-ISABa125, IS3000-ISABa125, ΔIS3000-ISABa125-IS91, IS26, and IS3000-(Δ) IS30-IS5 (Figure 7). We also determined the frequency of the conjugative transfer of blaNDM drug-resistance genes in the selected strains and found that the NDM gene from most strains could be transferred to the recipient strain EC600 (Supplementary Table S7), and the conjugative frequency ranged from 1.5 × 10−8 to 5.3 × 10−6. The non-conjugative plasmid pEC2-NDM-1(20817 bp) without conjugation-associated genes carried by E. coli could also be transferred to the recipient strain.
Figure 6.
Circle plots and comparative analysis of plasmids carrying blaNDM. (A) Comparative analysis of 11 IncX3 plasmids. (B) Comparative circle plots of pEC2-NDM1, pKP1-NDM1, pEC12-NDN5, and pEC26-NDM1 plasmids. (C) Circle plots of pEC12-NDM1 plasmid. (D) Circle plots of pEC51-NDM5 plasmids. GC content, GC backbone, plasmid replication and transfer-related genes, MGE-related genes, antibiotic resistance genes, other genes and pan-plasmids were illustrated.
Figure 7.
Comparative analysis and annotation of the gene environment of blaNDM in length-long sequenced strains. The degree of shading represents the identity of the nucleic acid sequences.
4. Discussion
Since its discovery in India in 2009, the blaNDM resistance gene has drawn much attention because it confers resistance to the therapeutic antibiotics carbapenems, which are crucial in clinical settings [6]. There have been numerous studies on the prevalence of the blaNDM gene in China, but reports on the prevalence of the blaNDM resistance gene in pig and chicken farms in the southwest are limited [9,32,33]. In 2021, we isolated and screened blaNDM-positive isolates from farms in the provinces of Sichuan and Yunnan and analyzed their epidemic characteristics.
The carriage rate of blaNDM-positive strains in pigs and chickens were 6.05% and 7.89%, respectively. These were higher than those reported in previous studies [12], indicating a high risk of blaNDM-positive strain transmission to humans via livestock and poultry. A possible reason for this high prevalence is that blaNDM can not only confer resistance to carbapenem antibiotics, but also to veterinary cephalosporin antibiotics. Therefore, the use of cephalosporins on farms may provide selection pressure. Moreover, their high occurrence could result from co-selection with other drug-resistance genes. Notably, blaNDM-positive strains are all multi-drug resistant bacteria that carry multiple antibiotic-resistance genes, and the antibiotic resistance situation is very serious. Given these circumstances, we need to strengthen the monitoring of blaNDM drug-resistance genes.
The transmission and spread of blaNDM genes are greatly facilitated by E. coli and Klebsiella spp., which are significant reservoirs of these drug resistance genes, in line with earlier research findings [9,12,34]. The results of antibiotic susceptibility experiments and the prediction of drug resistance genes showed that all bacterial strains were MDR and that different bacterial species had different patterns of antibiotic resistance. In previous studies, blaNDM-positive strains showed a low susceptibility to tigecycline; however, the high rate of tigecycline resistance in K. quasipneumoniae reported in this investigation suggests that this drug might become less effective [9,34]. Polymyxin B is also an effective antibiotic for the treatment of infections caused by blaNDM-positive strains.
There are many plasmid types in these bacteria, which is helpful for the potential transfer of blaNDM or other drug-resistance genes. For example, the blaNDM gene could be transferred between different plasmids through the IS. Furthermore, conjugative plasmids or ICEs can help non-conjugative plasmids containing the blaNDM gene move between bacteria; for example, the pEC2-NDM-1 plasmid (20817 bp) of strain EC2 in subsequent conjugative transfer experiments could be transferred in this way.
In this study, the same species of bacteria isolated from the same farm showed a limited genetic distance, indicating that vertical transmission is the main mode of transmission of the blaNDM gene within this farm. Moreover, the related strains were also common among different farms, indicating the transmission of blaNDM-positive strains in this region. The blaNDM-positive E. coli in this study had more genetic diversity than other bacteria, which indicates, to a certain extent, that the blaNDM gene might be transferred more frequently among this species, and this might also be caused by the larger base number of E. coli bacteria in the environment.
The close relationship between P. vermicola and human isolates in this study indicates that this strain might have spread widely throughout China [35]. The results of the joint analysis of strains in this study and blaNDM-positive strains in the database showed some interesting phenomena; for example, some strains of E. coli and Klebsiella spp. from human and animal sources were determined to have a high genetic similarity (SNP < 10), indicating that blaNDM-positive strains are transmitted between farm animals and humans. Moreover, unlike Klebsiella spp., more human- and animal-derived E. coli strains share the same ST and have a high genetic similarity, which might indicate that blaNDM-positive E. coli is more frequently transmitted between humans and farm animals. Considering these findings, the surveillance of E. coli carrying the blaNDM gene should be intensified.
Mobile genetic elements are important for the transmission of blaNDM. Plasmids and ICEs can carry and transfer blaNDM genes [33,36,37]. As in previous studies, IncX3 plasmids were found to be the main carriers of the blaNDM gene and drivers of its horizontal transmission, but other types of plasmids, ICEs, and ISs are also non-negligible drivers [15,18,19]. It appears from a more specific genetic context that the blaNDM gene, in addition to its most conserved structure (blaNDM-(Δ)ble-traF), is flanked by variable structures with multiple types of ISs on both sides. The results of this study suggest that ISs, such as IS26, ISCR1, and ISAba125, play important roles in the transfer and recombination of blaNDM between different genetic environments. Both ICEs and most plasmids in the experiments could be transferred between bacteria via conjugation, indicating that these blaNDM genes have a strong capacity for horizontal transmission. Although the horizontal transfer of blaNDM occurs readily under laboratory conditions, the reasons for not monitoring abundant and diverse blaNDM carriers within the same farm are worthy of further exploration.
5. Conclusions
Overall, this study provides an in-depth analysis of blaNDM-positive strains in pig and chicken farms in Southwest China. Carriage rates of blaNDM in farm animals on both pig and chicken farms are at a high level. E. coli and Klebsiella spp. were the main blaNDM carriers in the Enterobacterales, blaNDM-5 was the main subtype, and all strains were MDR. Moreover, the IncX3 plasmid was the main carrier of the blaNDM gene, and most of it was found to be transferred via conjugation. Phylogenetic analysis suggests that vertical transmission of host strains with limited genetic diversity is the main cause of the spread of blaNDM in Sichuan pig and chicken farm animals. The potential threat of reciprocal transmission between blaNDM-positive isolates of animal and human origins is worrisome. Therefore, further monitoring is urgently needed to explore this transmission and to disrupt the transmission network.
Acknowledgments
Thanks to the relevant farm staff for providing the sampling site and help.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11092304/s1. Table S1: Information for 102 Enterobacterales strains from blaNDM-positive strains in this study. Table S2: Details of 1911 Enterobacterales strains containing blaNDM-1 or blaNDM-1 resistance genes from China (database: NCBI, Dates to December 2022). Table S3: Resistance profiles of 102 blaNDM-positive Enterobacterales strains to 16 antibiotics in this study. Table S4: esistance genes and plasmid replicons profiles of 102 blaNDM-positive Enterobacterales strains in this study. Table S5: SNP matrix for 4 classes of strains in this study. Table S6: SNP matrix between strains in this study and homozygous strains from the database. Table S7: The frequency of conjugative transfer of blaNDM gene in the selected 19 strains.
Author Contributions
Conceptualization, R.W., H.W. (Hongcheng Wei), and H.W. (Hongning Wang); methodology, R.W. and H.W. (Hongcheng Wei); software, T.Z. and R.W.; validation, C.L. (Changwei Lei) and P.M.; formal analysis, H.W. (Hongcheng Wei) and C.L. (Chao Li); resources, H.W. (Hongning Wang); data curation, Q.W. and R.W.; writing—original draft preparation, R.W. and H.W. (Hongcheng Wei); writing—review and editing, R.W. and H.W. (Hongcheng Wei); visualization, R.W. and H.W. (Hongning Wang); supervision, Z.L., C.L. (Changwei Lei), and H.W. (Hongning Wang); project administration, T.Z. and P.M.; funding acquisition, C.L. (Changwei Lei) and H.W. (Hongning Wang). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant no. U21A20257), the National System for Layer Production Technology (CARS-40-K14) and Natural Science Foundation of Sichuan Province (2022NSFSC0076), the Central Government Guiding Local Science and Technology Development (2022ZYDF077).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Bassetti M., Peghin M., Vena A., Giacobbe D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019;6:74. doi: 10.3389/fmed.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karaiskos I., Giamarellou H. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: Current and emerging therapeutic approaches. Expert Opin. Pharmacother. 2014;15:1351–1370. doi: 10.1517/14656566.2014.914172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts L.W., Harris P.N.A., Forde B.M., Ben Zakour N.L., Catchpoole E., Stanton-Cook M., Phan M.D., Sidjabat H.E., Bergh H., Heney C., et al. Integrating multiple genomic technologies to investigate an outbreak of carbapenemase-producing Enterobacter Hormaechei. Nat. Commun. 2020;11:466. doi: 10.1038/s41467-019-14139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chudejova K., Kraftova L., Mattioni Marchetti V., Hrabak J., Papagiannitsis C.C., Bitar I. Genetic Plurality of OXA/NDM-Encoding Features Characterized from Enterobacterales Recovered from Czech Hospitals. Front. Microbiol. 2021;12:641415. doi: 10.3389/fmicb.2021.641415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P., Poirel L., Walsh T.R., Livermore D.M. The emerging NDM carbapenemases. Trends Microbiol. 2011;19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Kumarasamy K.K., Toleman M.A., Walsh T.R., Bagaria J., Butt F., Balakrishnan R., Chaudhary U., Doumith M., Giske C.G., Irfan S., et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugawara Y., Akeda Y., Hagiya H., Sakamoto N., Takeuchi D., Shanmugakani Rathina K., Motooka D., Nishi I., Zin Khwar N., Aye Mya M., et al. Spreading Patterns of NDM-Producing Enterobacteriaceae in Clinical and Environmental Settings in Yangon, Myanmar. Antimicrob. Agents Chemother. 2019;63:e01924-18. doi: 10.1128/AAC.01924-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R., Li J., Wang Y., Shen J., Shen Z., Wang S. Presence of NDM in non-E. coli Enterobacteriaceae in the poultry production environment. J. Antimicrob. Chemother. 2019;74:2209–2213. doi: 10.1093/jac/dkz193. [DOI] [PubMed] [Google Scholar]
- 9.Wang M.G., Zhang R.M., Wang L.L., Sun R.Y., Bai S.C., Han L., Fang L.X., Sun J., Liu Y.H., Liao X.P. Molecular epidemiology of carbapenemase-producing Escherichia coli from duck farms in south-east coastal China. J. Antimicrob. Chemother. 2021;76:322–329. doi: 10.1093/jac/dkaa433. [DOI] [PubMed] [Google Scholar]
- 10.Zhang R., Liu L., Zhou H., Chan E.W., Li J., Fang Y., Li Y., Liao K., Chen S. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Bi Z., Ma S., Chen B., Cai C., He J., Schwarz S., Sun C., Zhou Y., Yin J., et al. Inter-host Transmission of Carbapenemase-Producing Escherichia coli among Humans and Backyard Animals. Environ. Health Perspect. 2019;127:107009. doi: 10.1289/EHP5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z., et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 13.Clarridge J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordmann P., Poirel L., Carrër A., Toleman M.A., Walsh T.R. How to detect NDM-1 producers. J. Clin. Microbiol. 2011;49:718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2010. [Google Scholar]
- 16.Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2013. [Google Scholar]
- 17.Demond H., Hanna C.W., Castillo-Fernandez J., Santos F., Papachristou E.K., Segonds-Pichon A., Kishore K., Andrews S., D’Santos C.S., Kelsey G. Multi-omics analyses demonstrate a critical role for EHMT1 methyltransferase in transcriptional repression during oogenesis. Genome Res. 2023;33:18–31. doi: 10.1101/gr.277046.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kechin A., Boyarskikh U., Kel A., Filipenko M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- 19.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 20.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017;3:e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolley K.A., Maiden M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z., Alikhan N.F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carriço J.A., Achtman M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petkau A., Stuart-Edwards M., Stothard P., Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie J., Chen Y., Cai G., Cai R., Hu Z., Wang H. Tree Visualization by One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023;51:587–592. doi: 10.1093/nar/gkad359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant J.R., Enns E., Marinier E., Mandal A., Herman E.K., Chen C.Y., Graham M., Van Domselaar G., Stothard P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023;51:484–492. doi: 10.1093/nar/gkad326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilchrist C.L.M., Chooi Y.H. clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 32.Fu B., Yin D., Sun C., Shen Y., Liu D., Bai R., Zhang R., Shen J., Hu F., Wang Y. Clonal and Horizontal Transmission of blaNDM among Klebsiella pneumoniae in Children’s Intensive Care Units. Microbiol. Spectr. 2022;10:e0157421. doi: 10.1128/spectrum.01574-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong H., Li Y., Cheng J., Xia Z., Liu W., Yan T., Chen F., Wang Z., Li R., Shi J., et al. Genomic Epidemiology Insights on NDM-Producing Pathogens Revealed the Pivotal Role of Plasmids on blaNDM Transmission. Microbiol. Spectr. 2022;10:e0215621. doi: 10.1128/spectrum.02156-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei H.C., Kong L.H., Wang Y.L., Huang Z.R., Yang X., Zhou C.Y., Li C., Ma B.H., Li C., Lei C.W., et al. Characterization and Public Health Insights of the New Delhi Metallo-β-Lactamase-Producing Enterobacterales from Laying Hens in China. Microorganisms. 2022;10:800. doi: 10.3390/microorganisms10040800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng C., Gao M., Jiang W., Shi W., Li A., Liu S., Zhang L., Zhang X., Li Q., Lin H., et al. Identification of a novel aminoglycoside O-nucleotidyltransferase AadA33 in Providencia vermicola. Front. Microbiol. 2022;13:990739. doi: 10.3389/fmicb.2022.990739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong L.H., Xiang R., Wang Y.L., Wu S.K., Lei C.W., Kang Z.Z., Chen Y.P., Ye X.L., Lai Y., Wang H.N. Integration of the blaNDM-1 carbapenemase gene into a novel SXT/R391 integrative and conjugative element in Proteus vulgaris. J. Antimicrob. Chemother. 2020;75:1439–1442. doi: 10.1093/jac/dkaa068. [DOI] [PubMed] [Google Scholar]
- 37.He J., Sun L., Zhang L., Leptihn S., Yu Y., Hua X. A Novel SXT/R391 Integrative and Conjugative Element Carries Two Copies of the blaNDM-1 Gene in Proteus mirabilis. MSphere. 2021;6:e0058821. doi: 10.1128/mSphere.00588-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.