Abstract
The phenobarbital-inducible rat cytochrome P450 (CYP) 2B1 and 2B2 proteins are encoded by homologous genes whose promoters contain a mammalian-apparent long terminal repeat retrotransposon (MaLR). An NF-κB-like site within the MaLR forms multiple protein–DNA complexes with rat liver and HeLa cell nuclear extracts. Using antibody supershift assays, we have identified these complexes as NF-κB and RPB-Jκ/CBF1. Competition assays using a series of single site mutant oligonucleotides reveal that the recognition sites for these two factors overlap. We also show that the CYP2B1/2 NF-κB element, but not the Igκ NF-κB element, can repress transcription in vitro when positioned upstream of the heterologous adenovirus major late core promoter. In addition, RBP-Jκ overexpressed in COS-7 cells repressed expression in vivo from an SV40–luciferase reporter construct that contained the CYP2B1/2 NF-κB element. Finally, we observe similar levels of NF-κB and RBP-Jκ binding activities in nuclear extracts prepared from control and phenobarbital-induced rat livers. The results suggest that RBP-Jκ/CBF1 binds an atypical NF-κB site in the CYP2B1/2 promoters and may help to maintain a low level of expression in the absence of inducer.
INTRODUCTION
Cytochrome P450s (CYP) are heme-containing membrane proteins that metabolize both foreign (xenobiotic) chemicals and endogenous compounds. Individual members of the CYP450 gene superfamily are regulated in developmental-, tissue-, and gender-specific patterns and, in some cases, are activated by exposure to chemical inducers (1).
Transcription of the rat CYP4502B1 and 2B2 genes (CYP2B1/2) is driven by liver-specific promoters that are ~96% identical within 2.35 kb upstream of the transcription initiation site. These promoters are notable for their ability to be induced up to several hundred-fold by the barbiturate sedative phenobarbital (PB) or other ‘PB-like’ inducers. Several regions from the CYP2B1/2 promoters are proposed to contain PB-responsive elements (PBRE) (reviewed in 2–5) and the most compelling evidence points to a distal element between –2318 and –2155 identified by assaying the activity of CYP2B2 promoter constructs transfected into primary hepatocytes (6). Studies using transgenic mice confirm a role for this region (7,8) and in vivo footprinting shows PB-dependent alterations in chromatin structure (9). Genetic and biochemical characterization of the corresponding region from the homologous mouse Cyp2b10 promoter suggests a role for NF-1 and the orphan nuclear receptor CAR (10–12). These sequences are distinct from the ‘barbie box’ element responsible for PB-induction of the Bacillus megaterium CYP450BM-1 and CYP450BM-3 genes (13).
In the absence of inducer, the rat CYP2B1/2 and mouse CYP2b10 genes are expressed at very low basal levels and may be maintained in a constituitively repressed state via negative regulatory elements (7,10,14–16; reviewed in 3). For instance, a construct that contained rat CYP2B2 promoter proximal sequences (to –0.8 kb) was constituitively active in transgenic mice, whereas constructs that included additional upstream sequences (to –19 kb) suppressed basal activity (7). In addition, deletion analysis of the mouse CYP2b10 promoter revealed that sequences between –971 and –775 reduced thymidine kinase promoter–CAT activity (10). Similarly, a 3-fold reduction in luciferase activity was seen with CYP2B2 constructs containing sequences from –725 to –1400 that were transfected into HepG2 cells (16). Of additional interest is the fact that these reports focus on a region of the promoter that shares homology with a family of mammalian-apparent long terminal repeat retrotransposon (MaLR) elements (17).
In this report we identify and characterize an element within the CYP2B1/2 MaLR that is recognized by both NF-κB and the recombination signal sequence-binding protein RBP-Jκ (18), also known as CBF1 (19). The dual NF-κB/RBP-Jκ sequence element, when fused to a heterologous promoter, can inhibit transcription in vitro and in vivo. The results support a role for RBP-Jκ/CBF1 as a constitutively active nuclear repressor that can bind to a distinct class of NF-κB sites.
MATERIALS AND METHODS
Oligonucleotides
For band shift and competition experiments: NF-κB 2B1/2, 5′-ACTGTGGGAAATTCCACACC-3′; NF-κB Igκ, 5′-CAGAGGGGACTTTCCGAGAG-3′; AP-1 wt, 5′-GTGTCTGACTCATGCTT-3′; AP-1A, 5′-TCTCAAATGACTCTAGCTTG-3′; AP-1B, 5′-TCCATTTGACTCCTGAGCCT-3′; AP-1mut, 5′-CTCAAATTATTCTAGCTT-3′; AdML TATA, 5′-AAGGGGGGCTATAAAAGGGGGTGGG-3′; Sp1, 5′-TTCGATCGGGGCGGGGCGAG-3′; 2B1/2DR, 5′-CTGATTTCTTACAGAACCCAAGACTTTCTTACAGAAGTCC-3′; 2B1/2 core, 5′-GTGGAGGGGCGGATTCAGCATAAAAGATCCTGC-3′; C/EBP, 5′-TGCAGATTGCGCAATCTGCA-3′. For construction of transcription templates: NF-κB Igκ 5′-CAGAGGGGACTTTCCGAGAGTACTGCATGCAGAGGGGACTTTCCGAGAGTACTGCATG-3′; NF-κB 2B1/2, 5′-CATGCAGTACTGTGGGAAATTCCACACCGCATGCAGTACTGTGGGAAATTCCACACCG-3′. For construction of RBP-Jκ expression vectors: RBP-myc5′, 5′-CGCCGCGGATCCAGTAATGCCCTCCGGTTTTCCT-3′; RBP-myc3′, 5′-CCBCCBCTCGAGGGACACCACGGTTGCTGT-3′. For construction of pGL3-Pro-based reporter templates: Igκ promoter, 5′-AGCGAGCTCGATATCAGAGGGGACTTTCCGAGAGCTAGCCG-3′; CYP2B1/2 promoter, 5′-AGCGAGCTCGATATCACTGTGGGAAATTCCACACCGCTAGCTAG-3′. Oligos were obtained from Operon.
Preparation of nuclear extracts
Nuclear extracts were prepared essentially as described by Gorski et al. (20). Male Sprague–Dawley rats (~150–200 g; Zivic-Miller Laboratories) were given either no injection or i.p. injections of either saline (control) or 100 mg/kg PB (Sigma) 20 h prior to sacrifice. Livers were removed and homogenized in 10 ml/g ice-cold homogenization buffer (10 mM HEPES pH 7.6, 15 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, 2 M sucrose, 10% glycerol, 0.5 mM DTT, 0.5 mM PMSF and 1% Trasylol) through three passes in a glass–teflon homogenizer using a Glas-Col motor at a setting of 60. The resulting homogenate was layered onto 2 M sucrose cushions, centrifuged at 24 000 r.p.m. for 60 min and the cytoplasmic extract (supernatant) and pelleted nuclei were collected. Cytoplasmic extracts were dialyzed against dialysis buffer (25 mM HEPES pH 7.6, 40 mM KCl, 0.1 mM EDTA, 10% glycerol and 1 mM DTT) prior to use. Nuclei were resuspended in nuclear lysis buffer (10 mM HEPES, pH 7.6, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 0.1 mM PMSF and 1% Trasylol) and chromatin was pelleted by addition of 0.3 g/ml ammonium sulfate and centrifugation at 35 000 r.p.m. for 60 min. The upper aqueous nuclear extract was recovered and dialyzed. Extracts were aliquoted and snap frozen for storage at –70°C. Protein concentrations were typically 4–8 mg/ml and were determined using the Bradford method (Bio-Rad) with bovine serum albumin as the standard.
Electrophoretic mobility shift assays
Binding reactions (25 µl) were performed in 10 mM HEPES, pH 7.9, 60 mM KCl, 2% PEG-8000 (w/v), 0.2 mM EDTA, 8% glycerol and 0.1 mg/ml poly(dI-dC) as described (21). Reactions contained 6–10 µg of either nuclear or cytoplasmic proteins prepared from untreated animals, from the livers of saline-treated (control) or PB-induced animals, or from HeLa cells. Oligonucleotides were typically purified by Sephadex G-25 gel filtration and annealed by incubating at 75°C for 5 min and cooling slowly to room temperature. Purified oligonucleotides were radiolabeled with T4 polynucleotide kinase and [γ-32P]ATP and separated from unreacted nucleotides by purification with 1 ml Sephadex G-25 columns. Labeled oligonucleotides (~70 fmol) corresponding to 50 000–100 000 c.p.m. were added to reactions, which proceeded for 45 min at 4°C. The resulting protein–DNA complexes were loaded onto 0.5× TBE, 5% polyacrylamide gels and electrophoresed in 0.5× TBE running buffer. Unlabeled oligonucleotide competitors used in competition experiments were present in 100-fold excess over probe. In experiments where NF-κB was post-translationally activated (22), various combinations of Nonidet P-40 (NP-40) (2.4%), formamide (27%) and sodium deoxycholate (DOC) (0.4%) were added to the reaction mixtures as indicated. Results were visualized by drying the gels and exposing to X-OMAT AR film (Kodak). Supershift reactions contained 0.5–2 µl of antibodies specific for the p50, p52, or p65 subunit of NF-κB (Santa Cruz), or for mouse RBP-Jκ (K0043), and were added prior to the addition of probe.
In vitro transcription assays
Templates for in vitro transcription were based on pMLG4G, a G-free cassette reporter driven by the adenovirus major late (AdML) core promoter. Constructs were prepared by the insertion of synthetic double-stranded oligonucleotides containing either tandem NF-κB 2B1/2 sites (pMLG4G-2B1/2) or tandem NF-κB Igκ sites (pMLG4G-Igκ). In vitro transcription reactions (20,23) were performed in 25 mM HEPES, pH 7.6, 50 mM KCl, 6 mM MgCl2, 0.6 mM ATP, 0.6 mM UTP, 35 µM CTP, 1 µl [α-32P]CTP (800 Ci/mmol; ICN), 0.1 mM 3′-O-methyl-GTP (Pharmacia), 12% glycerol, 1 µl RNasin (∼30 U; Pharmacia). Reactions (50 µl) also contained 20 µg/ml HindIII-digested linear DNA template and ∼70 µg HeLa cell nuclear extract. After 45 min incubation at 30°C, the reactions were terminated by the addition 280 µl stop mix (300 mM sodium acetate, 50 mM EDTA and 1.2% SDS). Proteins were removed by proteinase K treatment (10 µl of 20 mg/ml proteinase K, 0.5 mg/ml tRNA) for 20 min at 55°C. After phenol/chloroform (1:1) extraction, labeled RNA was precipitated by addition of 350 µl of 100% ethanol. The RNA pellets were rinsed with 80% ethanol and dried for 5 min under vacuum. Pellets were resuspended in 5 µl of RNA loading dye (95% formamide, 1 mM EDTA and 0.05% bromophenol blue and xylene cyanol) and loaded onto 7 M urea–5% polyacrylamide sequencing gels. NF-κB 2B1/2 oligonucleotide competitor was added to 0.28 µM.
Transfections
The open reading frame of RBP-Jκ was PCR amplified from the RBP-2 cDNA using the RBP-myc5′ and RBP-myc3′ oligonucleotides. The product was digested with XbaI and BamHI and subcloned into pcDNA3.1/myc-His (Invitrogen). The insert produced by XhoI and BamHI digestion of this plasmid was then subcloned into pCMV-Tag 2 (Stratagene) to generate pCMV-RBP. Reporter constructs were generated by subcloning synthetic NF-κB binding sites from the CYP2B1/2 promoter or IgK enhancer into the pGL3 Pro vector (Promega) at the SacI–NheI site.
Transient transfections were performed using the calcium phosphate precipitate method (24). COS-7 cells were grown on 6-well plates containing Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone). When cells were 50% confluent they were co-transfected with 3 µg pGL3-Pro-based reporter constructs together with pCMV-RBP. An aliquot of 200 ng of pCMV-βGAL was added as a control for transfection efficiency. Cells were harvested 48 h after transfection, and luciferase assays using 20 µl of cellular extracts were performed as described by the manufacturer (Promega) using a Turner TD-20e luminometer. Experiments were performed in triplicate.
Measurement of RBP-Jκ in COS-7 cells was performed with the cellular lysates used for the luciferase assay. Immunoblotting was performed using the ECL system (Amersham) and an anti-myc monoclonal antibody (25) prepared from mouse cell line MYC1-9E10.2 (ATCC).
Other techniques
Other molecular biology techniques were performed as described (24). Quantitation of autoradiographs was performed using an LKB Ultroscan XL laser densitometer or a Molecular Dynamics phosphorimager. Sequences were analyzed using the Lasergene software package (DNASTAR), BLAST sequence homology searches at NCBI (http://www.ncbi.nlm.nih.gov/ ) and the TRANSFAC and TESS programs (http://dot.imgen.bcm.tmc.edu:9331/seq-search/gene-search.html ) to predict binding sites for transcription factors.
RESULTS
Structure of the PB-inducible rat cytochrome P450 2B1 and 2B2 gene promoters
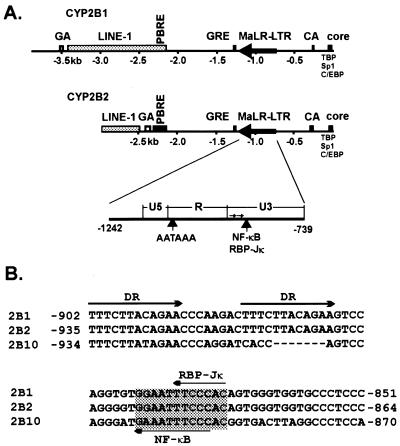
The direction and location of selected sequence elements in the CYP2B1/2 promoters are summarized in Figure 1A. These include a PBRE between –2318 and –2155 (6,10,11), an MaLR long terminal repeat (LTR) between –1242 and –739 (17), a glucocorticoid response element (26), a core promoter region that contains sites for C/EBP (16,27), TBP and Sp1 and a region of repetitive CA dinucleotides. Regions upstream of –2.35 kb are not homologous, but include distinct truncations of LINE-1 repetitive elements and GA dinucleotide repeats of different lengths. The available sequence of the PB-inducible mouse CYP2b10 (1.4 kb) promoter is ~83% identical to the CYP2B1/2 gene (10).
Figure 1.
(A) Schematic diagram of the rat CYP450 2B1 and 2B2 promoters. Core promoter binding factors, including TBP, C/EBP and Sp1, are indicated at the far right. CA refers to a repetitive CA sequence of five (2B1) or 19 (2B2) reiterations. The black arrow (–1242 to –739) pointing away from the core promoter indicates a solitary LTR from a family of mammalian apparant LTR retrotransposons (MaLR). The predicted boundaries between the U3, R and U5 regions of the LTR are shown in the expansion below, along with the position of putative NF-κB and polyadenylation (AATAAA) sites. GRE indicates the position of a glucocorticoid response element. The dark box centered around –2300 (PBRE) represents a distal enhancer that confers responsiveness to PB. Sequences further upstream consist of distinct regions of LINE-1 elements, as well as reiterated GA dinucleotide sequences. See text for references. (B) Comparison of selected sequences within the MaLR from the rat CYP2B1 and 2B2 and mouse CYP2b10 promoters. The rat CYP2B1 and 2B2 sequences are identical within the region shown and include a direct repeat (DR) of 12 nt. The putative NF-κB and RBP-Jκ sites are shaded and indicated with arrows. The MaLR element from the mouse CYP2b10 promoter lacks the DR, but contains NF-κB and RBP-Jκ sites.
Analysis of CYP2B1/2 sequences between –1242 and –739 showed ~68% identity to 400 nt of the mouse MTa repetitive element (28). This region is thus a member of a large family (40 000–100 000 estimated copies) of retrotransposon-like MaLR repetitive elements often found as solitary, inactive LTRs (17). We tentatively divided the CYP2B1/2 MaLR into U3, R and U5 regions oriented away from the direction of CYP2B1/2 transcription (Fig. 1A). Computer analysis of the U3 region revealed that it contains a short direct repeat of 12 nt, as well as two adjacent sequences that resemble the GGGRNNYYCC recognition site for the inducible transcription factor NF-κB (Fig. 1B). One of these sites (GGGAAATTCC) exactly matches a site found in the β-interferon enhancer (29). These sequences were of interest because little is known as to whether MaLR elements regulate expression of adjacent cellular genes and because of the possibility of a connection between CYP2B1/2 induction and the responsiveness of rel (NF-κB) family transcription factors to chemical stress (30).
The NF-κB site in the CYP2B1/2 LTR is recognized by multiple proteins in rat liver nuclear extracts
To determine if rat liver nuclei contained factors that would selectively recognize the NF-κB element within the CYP2B1/2 MaLR, we tested the ability of oligonucleotides that span this region (denoted NF-κB 2B1/2) to form complexes in electrophoretic mobility shift assays (Fig. 2A). For comparison we used a control oligonucleotide that contains the NF-κB site from the Igκ enhancer (denoted NF-κB Igκ) (31). The control probe detected up to three complexes (C2–C4) in this experiment (lane 1). The NF-κB 2B1/2 probe revealed complexes of approximately similar mobility, but also formed an abundant, faster migrating species (C1) (lane 4). Competition experiments using unlabeled oligonucleotides showed that the upper set of complexes were efficiently competed by both NF-κB 2B1/2 and NF-κB Igκ competitors (lanes 2, 3, 5 and 6), while formation of the major C1 complex was abolished only in the presence of the NF-κB 2B1/2 competitor (lane 6). To further establish that these results were not due to recognition of the NF-κB 2B1/2 probe by a non-specific factor, additional oligonucleotide competitors were tested for their ability to eliminate C1 complex formation (Fig. 2B). The results show that formation of the C1 complex was abolished only in the presence of NF-κB 2B1/2 competitor (lane 3).
Figure 2.
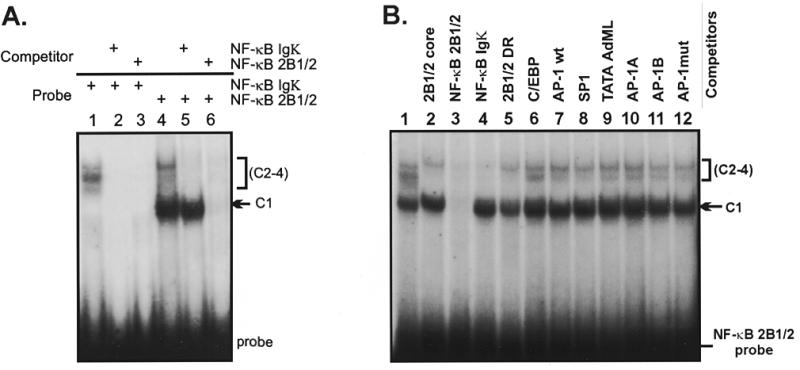
(A) The NF-κB element from the rat CYP2B1/2 MaLR LTR forms several shifted complexes in rat liver nuclear extracts. Band shift reactions contain probes for NF-κB Igκ (lanes 1–3) or NF-κB 2B1/2 (lanes 4–6), as shown. One major complex (C1) and up to three minor complexes (C2–C4) were observed. Addition of unlabeled oligonucleotide competitors NF-κB Igκ (lanes 2 and 5) and NF-κB 2B1/2 (lanes 3 and 6) indicate that the C1 complex is competed by the NF-κB 2B1/2 site, but not the NF-κB Igκ site. (B) C1 complex formation is specific for the NF-κB 2B1/2 element. Reactions were performed using the NF-κB 2B1/2 probe as described in (A). Oligonucleotide competitors include none (lane 1), 2B1/2 core –43 to –13 (lane 2), NF-κB 2B1/2 (lane 3), NF-κB Igκ (lane 4), 2B1/2 DR (lane 5), C/EBP (lane 6), AP-1 (lane 7), Sp1 (lane 8), AdML TATA (lane 9), AP-1-like site A (lane 10), AP-1-like site B (lane 11) and an AP-1 mutant (lane 12). The upper C2–C4 complexes are competed by both the NF-κB 2B1/2 and NF-κB Igκ oligonucleotides (lanes 3 and 4), while competition for the C1 complex is seen only with NF-κB 2B1/2 (lane 3).
Factors that recognize the NF-κB 2B1/2 element show differential localization and responsiveness to protein-dissociating reagents
Binding activities present in nuclear and cytoplasmic extracts prepared from rat livers were compared in the presence of dissociating agents such as NP-40, DOC and formamide that release active NF-κB from its cytoplasmic inhibitor I-κB (22). As shown in Figure 3, band shift experiments with the NF-κB 2B1/2 probe demonstrate that the upper complexes could be activated by various dissociating agents (for example lanes 5 and 12). In contrast, the C1 complex was restricted to the nuclear fraction and was unaffected by addition of protein-dissociating agents (Fig. 3B, lanes 1–7). These results suggest that the C1 complex contains a distinct nuclear factor that recognizes this particular NF-κB site, or an adjacent or overlapping sequence.
Figure 3.
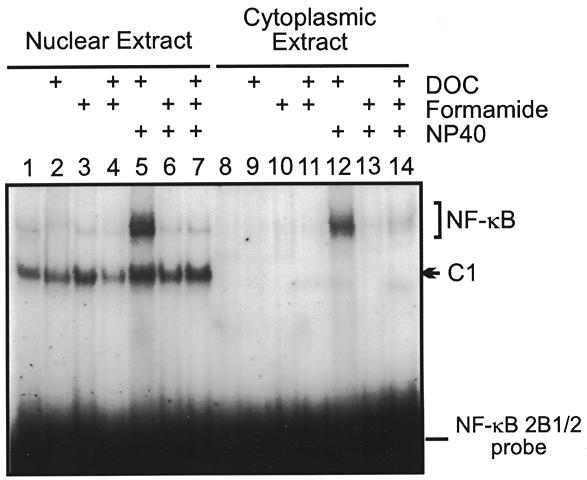
Localization and inducibility of protein–DNA complexes formed with the NF-κB 2B1/2 element. Extracts were prepared from rat liver nuclei (lanes 1–7) and cytoplasm (lanes 8–14) and tested for complex formation using the NF-κB 2B1/2 element as probe. Inducibility of complex formation was tested by the addition of DOC, formamide or NP-40, as indicated. The C1 complex (lanes 1–7) was restricted to nuclear extracts and was unaffected by treatment with dissociating reagents.
The CYP2B1/2 NF-κB element is recognized by NF-κB and RBP-Jκ
Interestingly, the NF-κB 2B1/2 sequence (but not the NF-κB Igκ control oligonucleotide) contains an overlapping site (GTGGGAA) that matches a concensus ag/cCGTGGGAActa/t RBP-Jκ recognition sequence (32; see Fig. 1B). To verify whether these proteins are present in the upper and lower complexes, we performed band shift experiments in the presence of factor-specific antibodies. As shown in Figure 4, addition of antibodies for the p50 (lanes 2 and 7) and p65 (lanes 3 and 8) subunits of NF-κB, but not for the p52 subunit (lanes 4 and 9), resulted in disappearance of the upper complex and formation of slower migrating complexes. Likewise, addition of a monoclonal antibody (K0043) specific for mouse RBP-Jκ resulted in disappearance of the lower C1 complex and formation of an upper complex (lanes 5 and 10) that migrated just above the position of the NF-κB complex (lane 1). The antibodies themselves do not shift the probe (lanes 11–14). This experiment demonstrates that the upper complexes contain NF-κB-like proteins, whereas the lower C1 complex is due to RBP-Jκ/CBF1. Interestingly, the corresponding sequence from the mouse Cyp2b10 gene (Fig. 1B) is also recognized by both NF-κB and RBP-Jκ (data not shown).
Figure 4.
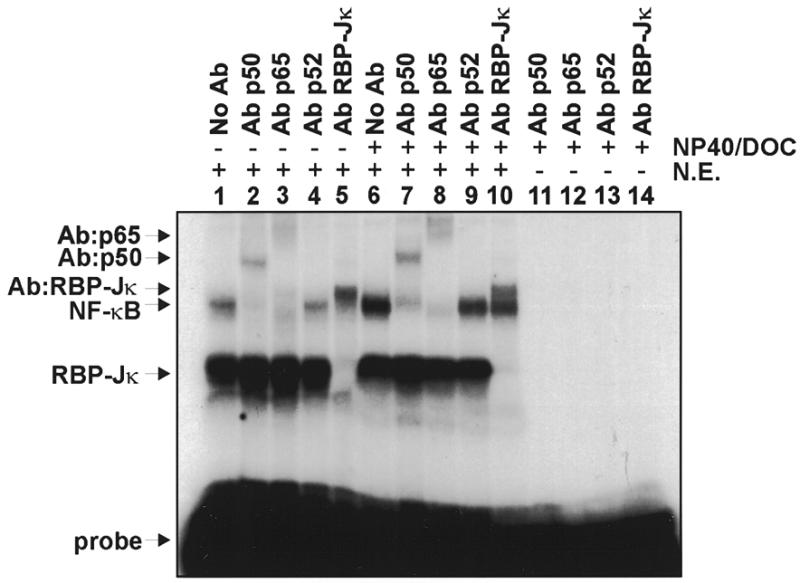
Supershift assays using antibodies against NF-κB subunits p50, p52 and p65 and against RBP-Jκ. Band shift reactions contain HeLa cell nuclear extract (N.E., lanes 1–10) and a kinase-labeled NF-κB 2B1/2 probe. Reactions were carried out in the absence (lanes 1–5) or presence (lanes 6–14) of DOC and NP-40. Lanes 11–14 contain antibody alone. The results indicate that Ab-p50 (lanes 2 and 7) and Ab-p65 (lanes 3 and 8) supershift the upper complex, while Ab-RBP-Jκ (lanes 5 and 10) supershifts the lower complex. The positions of RBP-Jκ, NF-κB and supershifted complexes are indicated to the left.
The CYP2B1/2 NF-κB element contains overlapping NF-κB and RBP-Jκ recognition sequences
To investigate the NF-κB 2B1/2 recognition sequence in more detail, we synthesized a series of oligonucleotides that contained mutations in the putative RBP-Jκ or NF-κB recognition sites (Fig. 5A). These oligonucleotides were used as competitors in band shift assays using the NF-κB 2B1/2 probe. The results (Fig. 5B) show that oligonucleotides with mutations in underlined positions of ACTGTGGGAAATTCCACACC competed poorly for NF-κB complex formation, consistent with the idea that these positions are important for NF-κB binding. Likewise, oligonucleotides with mutations in the underlined positions of ACTGTGGGAAATTCCACACC competed poorly for binding of RBP-Jκ, indicating that these positions are important for RBP-Jκ binding. The results are consistent with the idea that the recognition sequence for RBP-Jκ (GTGGGAA) overlaps that for NF-κB (GGGAATTCCC). This analysis, and the fact that slower migrating complexes that contain both NF-κB and RBP-Jκ proteins were not observed (data not shown), also suggests that binding of either factor is exclusive.
Figure 5.
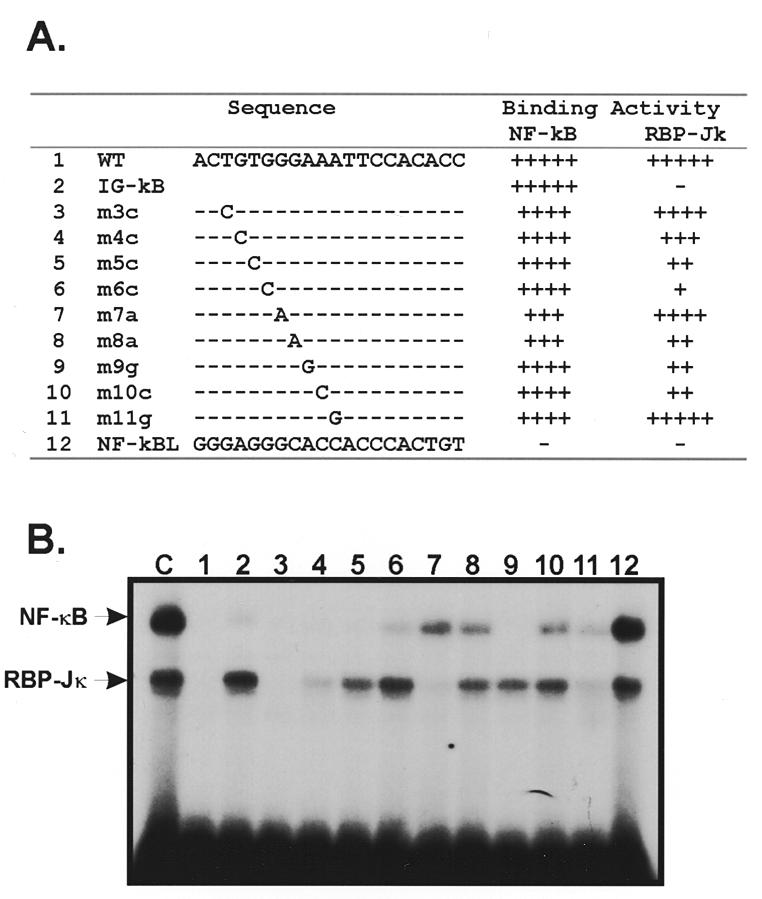
(A) Sequences of CYP2B1/2 NF-κB mutant oligonucleotides. Sequences (top strand) of NF-κB 2B1/2 (oligo 1) and NF-κB Igκ (oligo 2) oligonucleotides, and various point substitutions (oligos 3–12) are aligned. The relative ability of these oligos to compete for NF-κB and RBP-Jκ binding activity is indicated to the right (100%, +++++; 80–100%, ++++; 60–80%, +++; 40–60%, ++; 20–40%, +; <20%, –/+; no binding, –). (B) Competition for NF-κB and RBP-Jκ complex formation using mutant oligonucleotides. Oligonucleotides 1–12 were used as competitors in band shift reactions containing HeLa cell nuclear extracts and a labeled NF-κB 2B1/2 probe (lanes 1–12). Lane C shows a control experiment performed in the absence of competitor.
The CYP2B1/2 NF-κB element is a cis-acting repressor of transcription
To determine whether binding of RBP-Jκ to the NF-κB 2B1/2 element is responsible for a functionally distinct effect on gene expression compared to the NF-κB Igκ element, we performed in vitro transcription assays. Tandem NF-κB 2B1/2 or NF-κB Igκ sites were cloned into pMLG4G, a template that drives expression of a G-free cassette under the control of the AdML core promoter (–49 to +10). As shown in Figure 6A, transcription from pMLG4G-2B1/2 (lane 2) was ~6-fold lower compared to the pMLG4G-Igκ template (lane 1). This result suggests that the NF-κB site from the CYP2B1/2 gene, but not the NF-κB Igκ site, is a negative cis-acting regulatory element. Addition of an NF-κB 2B1/2 oligonucleotide competitor had no effect on pMLG4G-Igκ transcription (lane 3), but restored transcription of pMLG4G-2B1/2 to normal levels (lane 4). The selective reversal of pMLG4G-2B1/2 inhibition by this oligonucleotide and the protein–DNA interaction assays presented in Figures 2 and 4 suggest that RBP-Jκ is responsible for repression.
Figure 6.
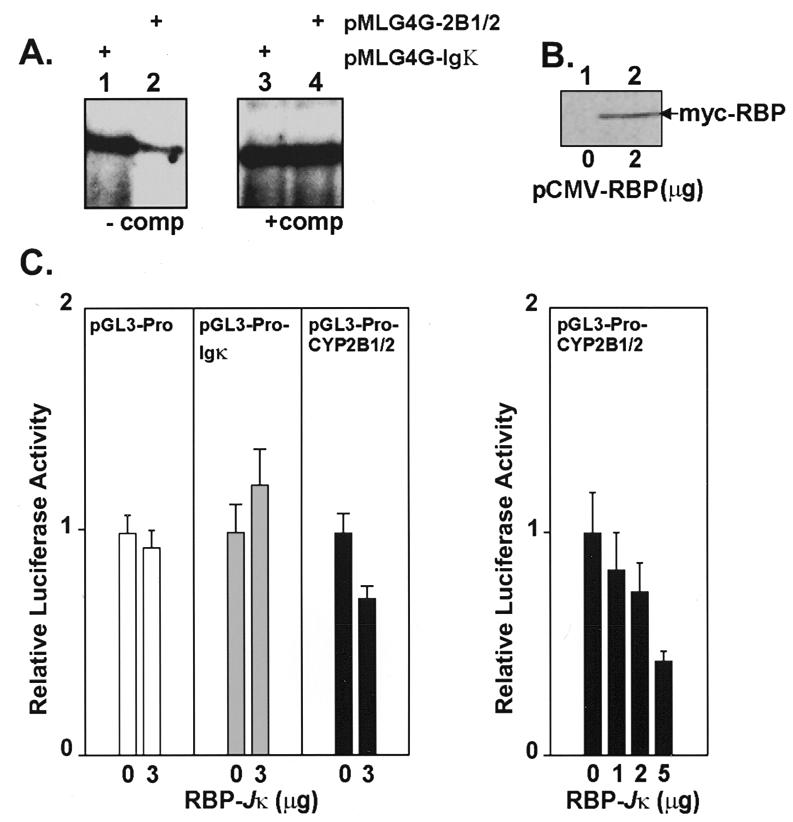
(A) The NF-κB element from the CYP2B1/2 promoter represses transcription from the adenovirus major late promoter in vitro. Adenovirus major late (AdML) promoter–G-free cassette in vitro transcription templates containing two NF-κB Igκ (pMLG4G-Igκ) (lanes 1 and 3) or NF-κB 2B1/2 (pMLG4G-2B1/2) (lanes 2 and 4) sites were tested for activity in in vitro transcription experiments using HeLa cell nuclear extracts. Reactions were performed in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of the NF-κB 2B1/2 oligonucleotide competitor. (B) A myc-tagged RBP-Jκ construct (pCMV-RBP-Jκ) overexpresses RBP-Jκ in COS-7 cells. RBP-Jκ protein (68 kDa) was detected in extracts prepared 48 h post-transfection using anti-myc antibodies. (C) Overexpressed RBP-Jκ protein reduces expression from an SV40 promoter construct that contains the CYP2B1/2 NF-κB site. The left panel shows that a control vector (pGL3-Pro) and an IgK NF-κB site vector (pGL3-Pro-Igκ) are not affected by co-transfection and overexpression of RBP-Jκ in vivo, whereas a construct with the CYP2B1/2 NF-κB site (pGL3-Pro-CYP2B1/2) is reduced. The right panel shows that repression of pGL3-Pro-CYP2B1/2 is dependent on the amount of co-transfected pCMV-RBP-Jκ.
To confirm that the CYP2B1/2 element, but not the IgK element, would down-regulate a heterologous promoter in vivo (as predicted by the experiment in Fig. 6A), CYP2B1/2 and Igκ NF-κB sites were positioned upstream of the SV40 promoter–luciferase reporter in the pGL3-Pro vector. The parental vector and each of the derived constructs were transfected into COS-7 cells with or without 3 µg of a pCMV-RBP expression vector that encoded a 68 kDa myc-tagged RBP protein (Fig. 6B, lane 2). The results in Figure 6C show that the pGL3-Pro-CYP2B1/2 promoter activity, as measured by luciferase activity, was repressed in the presence of pCMV-RBP. In contrast, the pGL3-Pro and pGL3-Pro-Igκ constructs, which do not contain RBP recognition sequences, were unaffected. The right hand panel in Figure 6C shows that repression increased (17, 28 and 57%) with increasing amounts of co-transfected pCMV-RBP (1, 2 and 5 µg).
Formation of NF-κB, RBP-Jκ and other protein–DNA complexes does not differ in control and PB-treated extracts
A unique characteristic of the CYP2B1/2 promoters is their transcriptional up-regulation in response to PB treatment. For this reason we wanted to test whether NF-κB or RBP-Jκ binding activities were altered in hepatic extracts prepared from saline- or PB-treated rats. To do so, we first normalized the amounts of each extract preparation on the basis of their ability to form complexes with oligonucleotide probes that spanned the CYP2B1/2 core promoter (–45 to –13) or with an AdML TATA element, as shown in Figure 7 (lanes 1–4). We then tested a panel of synthetic oligonucleotides that contained binding sites for several transcription factors present in adult rat liver. Extracts prepared from control (C) and induced (I) animals 20 h post-treatment showed ~2-fold or less change in the binding of RBP-Jκ (lanes 7 and 8), NF-κB (lanes 5–8), AP-1 (lanes 9 and 10), Sp1 (lanes 11 and 12) or C/EBP (lanes 13 and 14). Each of the shifted complexes observed in this assay was competed specifically (data not shown). It has been suggested elsewhere that NF-κB (33,34) and AP-1 (35,36) activities increase in hepatic nuclear extracts following PB treatment. Although the basis for these differences is not known, we conclude here that RBP-Jκ is a constituitive negative factor.
Figure 7.
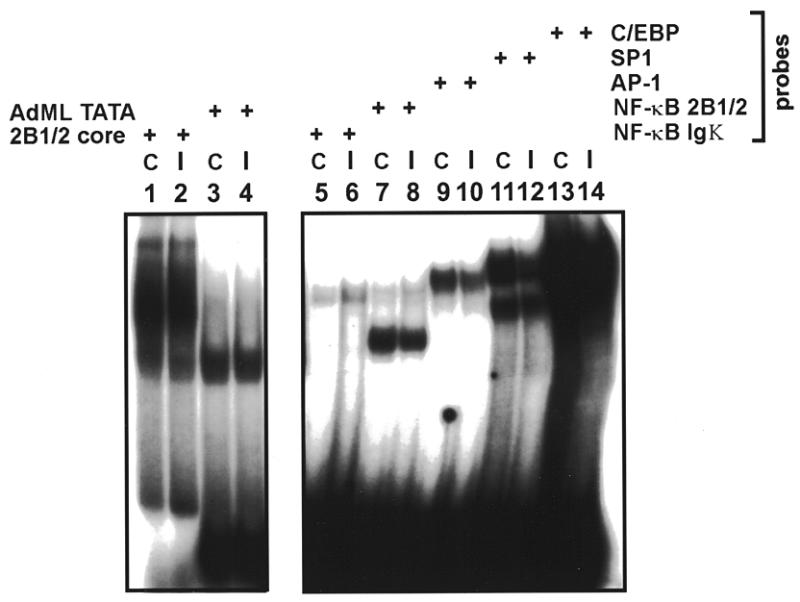
Formation of NF-κB, RBP-Jκ and several other protein–DNA complexes does not differ in rat hepatic extracts prepared from control (C) or PB-induced animals (I). Oligonucleotides spanning the CYP2B1/2 core (–45 to –13) (lanes 1 and 2) and the AdML TATA element (lanes 3 and 4) were used to normalize activity of control (C) and PB-induced (I) extracts with respect to TATA element binding activities. Oligos tested include NF-κB Igκ (lanes 5 and 6), NF-κB 2B1/2 (lanes 7 and 8), AP-1 (lanes 9 and 10), Sp1 (lanes 11 and 12) and C/EBP (lanes 13 and 14).
DISCUSSION
In this study we have identified an atypical NF-κB site within the rat CYP2B1/2 MaLR element that is recognized by the inducible NF-κB and constituitive RBP-Jκ/CBF1 transcription factors. Assuming a concensus RBP-Jκ recognition sequence of G/ATGGGAA (32), the occurrence of dual NF-κB/RBP-Jκ elements is predicted to occur at ~12.5% of NF-κB sites that contain the GGGAA NF-κB half site. This element, together with similar sites in the IL-6 (37,38) and NF-κB-2 genes (39), defines a functionally distinct subclass of NF-κB recognition sequences. Interestingly, the PB-inducible mouse CYP2b10 gene also contains a putative RBP-Jκ element that occurs in the context of a NF-κB site (Fig. 1B). However, a search for NF-κB and RBP-Jκ sites in the promoter regions of additional CYP450 genes did not reveal other examples of sites that overlap. It is interesting to note that another class of atypical NF-κB sites are G-C rich and are proposed to be the site of an interplay between NF-κB and Sp1 (40). The data also make the important point that a widely distributed family of repetitive elements, MaLRs (17), can contain sites that bind trans-acting transcriptional regulatory factors.
RBP-Jκ was originally isolated based on its recognition of an immunoglobulin κ gene recombination signal probe (18) and functions either as a cofactor-dependent positive regulator or, in the absence of additional proteins, as a negative regulator. For instance, RBP-Jκ recruits Epstein–Barr virus nuclear antigen-2 (EBNA-2) protein to sites in the viral latent membrane protein, BamHI C, terminal protein and cellular CD23 promoters (41–47), where it up-regulates transcription. The Drosophila counterpart of RBP-Jκ, Suppressor of Hairless, mediates transcriptional activation by Notch through a similar mechanism (reviewed in 48) and both EBNA-2 and mammalian Notch1 interact directly with RBP-Jκ to activate transcription (49). By itself, however, RBP-Jκ is able to repress gene expression. For instance, the adenovirus pIX (50), human IL-6 (37,38) and NF-κB-2 (39) promoters are repressed by RBP-Jκ. Moreover, Gal4–RBP-Jκ fusion proteins inhibit transcription from a herpes simplex virus thymidine kinase core promoter construct that contains upstream Gal4 sites (51). The mechanism of repression by RBP repression at sites near the TATA element may involve interference with the function of the general transcription factor TFIIA bound to TBP (52). RBP-Jκ is also associated with a histone deacetylase that could negatively modulate transcription factor accessibility to chromatin (53,54).
Induction of the CYP2B1/2 genes by chemical inducers involves a positively acting PBRE that binds a transcriptional activator(s) responsive to PB (reviewed in 2–5). The function of such factors may be to overcome, or derepress, the effects of negatively acting sequences that appear to maintain these genes in a relatively inactive state (reviewed in 3). Repression is strikingly illustrated by the ability of sequences between –971 and –775 of the mouse CYP2b10 promoter to reduce the activity of a heterologous thymidine kinase promoter (10). As the NF-κB/RBP-Jκ site in the rat CYP2B1/2 (–888 to –882) and mouse CYP2b10 (–894 to –888) promoters is within this negative regulatory region, our studies suggest a role for RBP-Jκ in down-regulating CYP2B1/2 gene expression. This idea is supported by several observations. First, RBP-Jκ is an abundant factor whose levels, as shown in this report, are not affected by PB treatment (Fig. 3). Moreover, subcellular fractionation (Fig. 3) and immunolocalization experiments (data not shown) show that RBP-Jκ is a nuclear factor. Second, the CYP2B1/2 site represses transcription from the heterologous AdML core promoter in a cell-free in vitro transcription system (Fig. 6). The degree of repression observed (~6-fold) is comparable to that observed with the adenovirus pIX promoter and a partially reconstituted in vitro transcription system to which purified RBP-Jκ was added (50). Third, the CYP2B1/2 NF-κB element also represses transcription in vivo from the heterologous SV40 promoter in COS-7 cells, an effect not seen with a similar NF-κB site from the IgK enhancer that does not bind RBP-Jκ (Fig. 6). Similarly, a related NF-κB/RBP-Jκ element from the IL-6 gene interleukin response element repressed expression in vivo when positioned ~1.2 kb away from the initiation site (37). Of course, it is possible that additional transcriptional regulatory proteins that bind the MaLR may also help to down-regulate CYP2B1/2 gene expression.
Several additional questions arise from our findings. For instance, are the CYP2B1/2 promoters activated by interactions between the mammalian Notch transcriptional activator or EBNA-2 proteins and promoter-bound RBP-Jκ? Also, are the CYP2B1/2 or other genes that contain dual NF-κB/RBP-Jκ elements controlled by an interplay of the cognate binding factors, or does competition from RBP-Jκ help mainly to re-establish repressed transcription levels more rapidly on dual NF-κB/RBP-Jκ sites? While the current study is limited to a demonstration that the CYP2B1/2 element is a negative regulatory element, in the case of the IL-6 gene RBP-Jκ is able to diminish the NF-κB-mediated response to TNFα and IL-1 (37). Displacement of RBP-Jκ by NF-κB would be facilitated by an increase in nuclear NF-κB concentrations following induction and by the higher affinity of NF-κB–DNA interactions (37,39). Indeed, Figure 3 also shows that the intensity of the C1 complex and the upper NF-κB complexes in nuclear extracts is ~11:1 in the absence of dissociating reagents (e.g. lane 1), but is nearly 1:1 following activation (lane 5). If these results reflect the abundance of NF-κB binding activities in the nucleus following the response to physiological inducers, they suggest the potential for an interplay between these factors on the CYP2B1/2 NF-κB site. However, future work will be required to investigate these issues in more detail.
In conclusion, the results here show that RBP-Jκ/CBF1 acts as a constituitively active nuclear repressor whose binding site can in some instances overlap sites for the NF-κB transcriptional activator. The activity of RBP-Jκ/CBF1 at such elements may be to constituitively diminish transcription from nearby core promoters, for instance the uninduced CYP2B1/2 genes, or to modulate the effects of signaling pathways that activate NF-κB.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Jing Hong Mu and Ashok Upadhyaya for reading the manuscript. We acknowledge the generous gift of anti-RBP-Jκ K0043 monoclonal antibody and RBP-2 cDNA from Dr Tasuku Honjo. Veterinary assistance was provided by Dr Tony Myers. This work was supported by the Welch Foundation.
REFERENCES
- 1.Okey A.B. (1990) Pharmacol. Ther., 45, 241–298. [DOI] [PubMed] [Google Scholar]
- 2.Waxman D.J. and Azaroff,L. (1992) Biochem. J., 281, 577–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honkakoski P. and Negishi,M. (1998) J. Biochem. Mol. Toxicol., 12, 3–9. [DOI] [PubMed] [Google Scholar]
- 4.Waxman D.J. (1999) Arch. Biochem. Biophys., 369, 11–23. [DOI] [PubMed] [Google Scholar]
- 5.Kemper B. (1999) Prog. Nucleic Acid Res. Mol. Biol., 61, 23–64. [PubMed] [Google Scholar]
- 6.Trottier E., Belzil,A., Stolz,C. and Anderson,A. (1995) Gene, 158, 263–268. [DOI] [PubMed] [Google Scholar]
- 7.Ramsden R., Sommer,K.M. and Omiecinski,C.J. (1993) J. Biol. Chem., 268, 21722–21726. [PubMed] [Google Scholar]
- 8.Ramsden R., Beck,N.B., Sommer,K.M. and Omiecinski,C.J. (1999) Gene, 228, 169–179. [DOI] [PubMed] [Google Scholar]
- 9.Kim J. and Kemper,B. (1997) J. Biol. Chem., 272, 29423–29425. [DOI] [PubMed] [Google Scholar]
- 10.Honkakoski P., Moore,R., Gynther,J. and Negishi,M. (1996) J. Biol. Chem., 271, 9746–9753. [DOI] [PubMed] [Google Scholar]
- 11.Honkakoski P. and Negishi,M. (1997) J. Biol. Chem., 272, 14943–14949. [DOI] [PubMed] [Google Scholar]
- 12.Honkakoski P., Zelko,I., Sueyoshi,T. and Negishi,M. (1998) Mol. Cell. Biol., 18, 5652–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Q., He,J.-S. and Fulco,A.J. (1995) J. Biol. Chem., 270, 4438–4450. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman M., Mager,W.H., Scholte,B.J., Civil,A. and Planta,R.J. (1992) Gene Expr., 2, 353–363. [PMC free article] [PubMed] [Google Scholar]
- 15.Ram N., Rao,M.V., Prabhu,L., Nirodi,C.S., Sultana,S., Vatsala,P.G. and Padmanaban,G. (1995) Arch. Biochem. Biophys., 317, 39–45. [DOI] [PubMed] [Google Scholar]
- 16.Park Y. and Kemper,B. (1996) DNA Cell Biol., 15, 693–701. [DOI] [PubMed] [Google Scholar]
- 17.Smit A.F.A. (1993) Nucleic Acids Res., 21, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunami N., Hamaguchi,Y., Yamamoto,Y., Kuze,K., Kangawa,K., Matsuo,H., Kawaichi,M. and Honjo,T. (1989) Nature, 342, 934–937. [DOI] [PubMed] [Google Scholar]
- 19.Ling P.D., Rawlins,D.R. and Hayward,S.D. (1993) Proc. Natl Acad. Sci. USA, 90, 9237–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorski K., Carneiro,M. and Schibler,U. (1986) Cell, 47, 767–776. [DOI] [PubMed] [Google Scholar]
- 21.DeJong J. and Roeder,R.G. (1993) Genes Dev., 7, 2220–2234. [DOI] [PubMed] [Google Scholar]
- 22.Baeuerle P.A., Lenardo,M., Pierce,J.W. and Baltimore,D. (1988) Cold Spring Harbor Symp. Quant. Biol., 13, 789–798. [DOI] [PubMed] [Google Scholar]
- 23.Sawadogo M. and Roeder,R.G. (1985) Proc. Natl Acad. Sci. USA, 82, 4394–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1996) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 25.Evan G.I., Lewis,G.K., Ramsay,G. and Bishop,J.M. (1985) Mol. Cell. Biol., 5, 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaiswal A.K., Haaparanta,T., Luc,P.-V., Schembri,J. and Adesnik,M. (1990) Nucleic Acids Res., 18, 4237–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luc P.-V.T., Adesnik,M., Ganguly,S. and Shaw,P.M. (1996) Biochem. Pharmacol., 51, 345–356. [DOI] [PubMed] [Google Scholar]
- 28.Bastien L. and Bourgaux,P. (1987) Gene, 57, 81–88. [DOI] [PubMed] [Google Scholar]
- 29.Goodbourn S. and Maniatis,T. (1988) Proc. Natl Acad. Sci. USA, 85, 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verma I.M., Stevenson,J.K., Schwarz,E.M., Van Antwerp,D. and Miyamoto,S. (1995) Genes Dev., 9, 2723–2735. [DOI] [PubMed] [Google Scholar]
- 31.Sen R. and Baltimore,D. (1986) Cell, 46, 705–716. [DOI] [PubMed] [Google Scholar]
- 32.Tun T., Hamaguchi,Y., Matsunami,N., Furukawa,T., Honjo,T. and Kawaichi,M. (1994) Nucleic Acids Res., 22, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Leung,L.K., Spear,B.T. and Glauert,H.P. (1996) Biochem. Biophys. Res. Commun., 229, 982–989. [DOI] [PubMed] [Google Scholar]
- 34.Mejdoubi N., Henriques,C., Bui,E. and Porquet,D. (1999) Biochem. Biophys. Res. Commun., 254, 93–99. [DOI] [PubMed] [Google Scholar]
- 35.Roe A.L., Blouin,R.A. and Howard,G. (1996) Biochem. Biophys. Res. Commun., 228, 110–114. [DOI] [PubMed] [Google Scholar]
- 36.Pinkus R., Bergelson,S. and Daniel,V. (1993) Biochem. J., 290, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plaisance S., Vanden Berghe,W., Boone,E., Fiers,W. and Haegeman,G. (1997) Mol. Cell. Biol., 17, 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannabiran C., Zeng,X. and Vales,L.D. (1997) Mol. Cell. Biol., 17, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oswald F., Liptay,S., Adler,G. and Schmid,R.M. (1998) Mol. Cell. Biol., 18, 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirano F., Tanaka,H., Hirano,Y., Hiramoto,M., Handa,H., Makino,I. and Scheidereit,C. (1998) Mol. Cell. Biol., 18, 1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahraeus R., Jansson,A., Ricksten,A., Sjoblom,A. and Rymo,L. (1990) Proc. Natl Acad. Sci. USA, 87, 7390–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grossman S.R., Johannsen,E., Tong,X., Yalamanchili,R. and Kieff,E. (1994) Proc. Natl Acad. Sci. USA, 91, 7568–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henkel T., Ling,P.D., Hayward,S.D. and Peterson,M.G. (1994) Science, 265, 92–95. [DOI] [PubMed] [Google Scholar]
- 44.Tsang S.-F., Wang,F., Izumi,K.M. and Kieff,E. (1991) J. Virol., 65, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin X.W. and Speck,S.H. (1992) J. Virol., 66, 2846–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimber-Strobl U., Kremmer,E., Grasser,F., Marschall,G., Laux,G. and Bornkamm,G.W. (1993) EMBO J., 12, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F., Kikutani,H., Tsang,S.-F., Kishimoto,T. and Kieff,E. (1991) J. Virol., 65, 4101–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Artavanis-Tsakonas S., Matsuno,K. and Fortini,M.E. (1995) Science, 268, 225–232. [DOI] [PubMed] [Google Scholar]
- 49.Hsieh J.J.-D., Henkel,T., Salmon,P., Robey,E., Peterson,M.G. and Hayward,S.D. (1996) Mol. Cell. Biol., 16, 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dou S., Zeng,X., Cortes,P., Erdjument-Bromage,H., Tempst,P., Honjo,T. and Vales,L.D. (1994) Mol. Cell. Biol., 14, 3310–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh J.J.-D. and Hayward,S.D. (1995) Science, 268, 560–563. [DOI] [PubMed] [Google Scholar]
- 52.Olave I., Reinberg,D. and Vales,L.D. (1998) Genes Dev., 12, 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kao H.-Y., Ordentlich,P., Koyano-Nakagawa,N., Tang,Z., Downes,M., Kintner,C.R., Evans,R.M. and Kadesch,T. (1998) Genes Dev., 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh J.J.-D., Zhou,S., Chen,L., Young,D.B. and Hayward,S.D. (1999) Proc. Natl Acad. Sci. USA, 96, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]