Abstract
In eukaryotic cells, mRNA synthesis is carried out by large, multifunctional complexes that are also involved in coordinating transcription with other nuclear processes. This survey focuses on the distribution and structural arrangement of these complexes within the nucleus, in relationship with the discrete positioning of particular chromosomal loci. To better understand the link between the spatial organization of the nucleus and the regulation of gene expression, it is necessary to combine information from biochemical studies with results from microscopic observations of preserved nuclear structures. Recent experimental approaches have made this possible. The subnuclear locations of specific chromosome loci, RNA transcripts, RNA polymerases, and transcription and pre-mRNA-processing factors can now be observed with computer-assisted microscopy and specific molecular probes. The results indicate that RNA polymerase II (RNAPII) transcription takes place at discrete sites scattered throughout the nucleoplasm, and that these sites are also the locations of pre-mRNA processing. Transcribing polymerases appear to be grouped into clusters at each transcription site. Cell cycle-dependent zones of transcription and processing factors have been identified, and certain subnuclear domains appear specialized for expression or silencing of particular genes. The arrangement of transcription in the nucleus is dynamic and depends on its transcriptional activity, with the RNAPII itself playing a central role in marshalling the large complexes involved in gene expression.
THE STRUCTURED NUCLEUS
The nucleus is a highly organized structure. Its most prominent feature is the nucleolus, a compartment specialized for transcription of RNA polymerase I (RNAPI) genes and initial ribosome assembly and now also known to be associated with maturation of several non-ribosomal RNAs (1,2). However, many other features have been identified in mammalian nuclei, by fluorescence in situ hybridization (FISH), immunolabeling and electron microscopy (reviewed in 3–6). Notably, the non-chromatin extranucleolar region of the nucleus known as the interchromatin granule cluster appears in the form of speckles when stained by immunofluorescence using antibodies against RNA-processing factors (6). Also of interest are the polymorphic interphase karyosomal association (PIKA) domains, which are nuclear bodies heterogeneous in number and size that vary in morphology with the cell cycle (7). Chromatin and non-chromatin structures such as the speckles do not diffuse freely in the nucleus, but instead undergo constrained motions suggesting they are tethered to a fixed structure (8). Microscopic imaging of live cells reveals that, in general, chromosome loci occupy defined, limited areas within the interphase nucleus (reviewed in 8).
ATTACHMENT AND POSITIONING OF CHROMOSOMES
The organization of chromatin, beyond the formation of nucleosomal arrays, is essential for both gene expression and cell function (9,10). Interphase chromatin is packaged with a stable, large-scale organization in which compaction of various regions of the genome is not uniform (10). The precise folding patterns and their correlation with chromosome biochemistry and function have not been fully elucidated. However, there is strong evidence for a role of condensed heterochromatic regions in transcriptional repression (8–10).
Current efforts are aimed at finding the proteins and DNA sequences responsible for the non-random arrangements of chromosomes in the interphase nucleus. Treatment of HeLa cell nuclei with nucleases under isotonic conditions permitted the identification of DNA sequences responsible for attaching the chromatin to the nucleus (11). These sequences were shown to be predominantly transcribed regions of the genome. Moreover, the DNA population isolated in this fashion was enriched to a lesser extent than predicted if particular sequences were permanent points of chromosome attachment in all cells (11). This observation would argue that the chromosomes are attached in the nucleus in a dynamic, functional manner requiring ongoing transcription (11).
Studies of silencing are revealing the importance of the subnuclear location of genes for their expression. It has been clear for some time that the position of genes with respect to heterochromatin and telomeres correlates with gene silencing (8,9,12,13). Since both telomeres and the Saccharomyces cerevisiae SIR proteins, which are critical for silencing, are concentrated at the nuclear periphery, subnuclear localization may play a role in silencing (8,12,13). This connection is well illustrated by an investigation of a HMR locus with a defective silencer in S.cerevisiae (13). When silencer elements at this locus were replaced by Gal4-binding sites, silencing of a nearby reporter gene could be restored by anchoring the defective locus to the nuclear periphery through the interaction of a protein fusion between the Gal4 DNA-binding domain and a transmembrane domain (13).
Other recent experiments indicate that certain transcription factors influence gene expression by controlling the relative positions of chromosomal loci within the nucleus. Ikaros, a lymphoid-specific transcription factor that binds to the promoters of several genes important for lymphoid development, was found localized with heterochromatin in the centromeric regions of chromosomes in lymphocyte nuclei (14). Transcriptionally inactive lymphoid genes colocalized with Ikaros at heterochromatic foci in a cell cycle-dependent fashion, consistent with a role for Ikaros as a recruiter of repressed genes targeted to centromeric domains (14). In a further study of the relationship between gene activity and nuclear position, the positioning of the Rag and TdT genes was followed in developing T cells (15). In immature thymocytes, the two genes are expressed and are not associated with centromeric regions. In contrast, downregulation and centromeric positioning of Rag and TdT occurred both in developing T cells upon induction of differentiation and in lymphocytes entering the cell cycle (15). Thus, centromeric positioning reflects a very different nuclear organization in cycling and non-cycling cells, with transcriptionally inactive genes localizing to centromeric domains as cells approach division. However, investigation of the time course of events indicates that the centromeric positioning does not appear to cause gene silencing in the first place. In immature T cells, locus repositioning occurred after the Rag and TdT expression shut down, rather than before. After induction of differentiation, centromeric locus positioning persists in the cycling progeny of the normal thymocytes. In contrast, in an immortalized thymic lymphoma cell line in which silencing was not heritable, locus repositioning fails to occur, suggesting that it is a feature of heritable gene silencing (15).
THE NUCLEAR MATRIX
A nuclear matrix was first identified as a nuclear fraction resistant to salt and DNase extraction (16, reviewed in 17). Subsequently, many experimental approaches to prepare and characterize the nuclear matrix have indicated the existence of a filamentous structure in the nucleus (18–22). However, because its biochemical isolation, and particularly chromatin elution, is so difficult, the existence of a nuclear matrix remains controversial. As pointed out by Pederson (17), procedures to isolate a structural framework from the highly condensed components of the nucleus provide opportunities for rearrangement and aggregation of proteins and nucleoproteins. Peter Cook’s group developed a gentle approach involving electroelution of nuclease-treated DNA from agarose-encapsulated cells; it reveals an insoluble protein network within the nucleus (23). In HeLa cells, this protein substructure contains most of the actively transcribing RNA polymerase II (RNAPII) as detected by nuclear run-on assay (11,24). This group also quantitated a number of proteins remaining in HeLa nuclei after detergent extraction of soluble components, and then again after nuclease digestion of the chromatin (24). Along with nascent RNA and almost all of the nuclear run-on activity, core RNAPII and general transcription factors were retained after chromatin elution. In contrast, the transcription factors TBP, C/EBP and Sp1 were released from the nucleus along with the DNA, consistent with these factors binding to active promoters in regions of nuclease-sensitive chromatin (24). These results suggest that RNAPII preinitiation complexes are bound to a nuclear matrix and that RNAPII remains bound to this nuclear framework after initiation (24).
The composition of the nuclear skeleton is not known. RNA and ribonucleoproteins have been identified in nuclear matrix preparations (21,22), suggesting that they are involved in its organization. Also, several candidate proteins have been identified as nuclear matrix constituents. Lamins, previously identified at the nuclear membrane, have been found in the interior of the nucleus as well (25). Another possible element for anchoring RNA polymerases is actin. First, actin is abundant in the nucleus, with nuclear extracts yielding both soluble and insoluble fractions (26). Second, there is biochemical evidence for an interaction between RNAPII and actin (27). Third, the existence of an actin scaffold associated with eukaryotic chromosomes has been demonstrated (28) and, in situ, removal of DNA from polytene chromosomes by micrococcal nuclease treatment does not displace RNAPII from the actin skeleton (28). Finally, a number of studies have pointed to a role for actin in transcription (26,29–31). In a recent analysis of one-cell mouse embryo nuclei microinjected with anti-actin antibodies, the actin distribution radically changed during the course of the first cell cycle from a few large patches in early one-cell embryos to hundreds of discrete foci (26). At this point, actin colocalized with transcription sites detected by immunofluorescence following BrUTP labeling (26). However, inhibition experiments did not point to a direct role for actin in the onset of transcription in these cells. Thus, the composition of the nuclear matrix and the role of nuclear actin in RNAPII function remain to be determined.
NUCLEAR DISTRIBUTIONS OF TRANSCRIPTION AND PRE-mRNA PROCESSING FACTORS
The development of confocal microscopy and the availability of specific probes for nuclear components now permits mapping of chromosomes, transcription and processing factors, and labeled nascent transcripts, thus allowing analysis of their relative locations in the nucleus (6,32,33, reviewed in 3). Many studies in mammalian nuclei have demonstrated that mRNA transcription occurs in hundreds to thousands of discrete foci (23,33–44). The RNAPII itself is also distributed widely throughout the nucleoplasm in many discrete sites that frequently coincide with the foci of labeled nascent transcripts (41,36–39,44) (Fig. 1). In general, factors associated with RNAPII transcription are also found distributed throughout the mammalian nucleus, often in discrete foci (37–39,41). For example, glucocorticoid receptors and mineralocorticoid receptors are each located in approximately 1000 nuclear clusters (37,41). GATA-1 immunofluorescence appears both as foci and also as diffuse staining in hematopoietic cells throughout the cell cycle (45). In a few cases, particular transcription factors have also been found localized in a limited number of subnuclear domains (41,46,47; see below).
Figure 1.
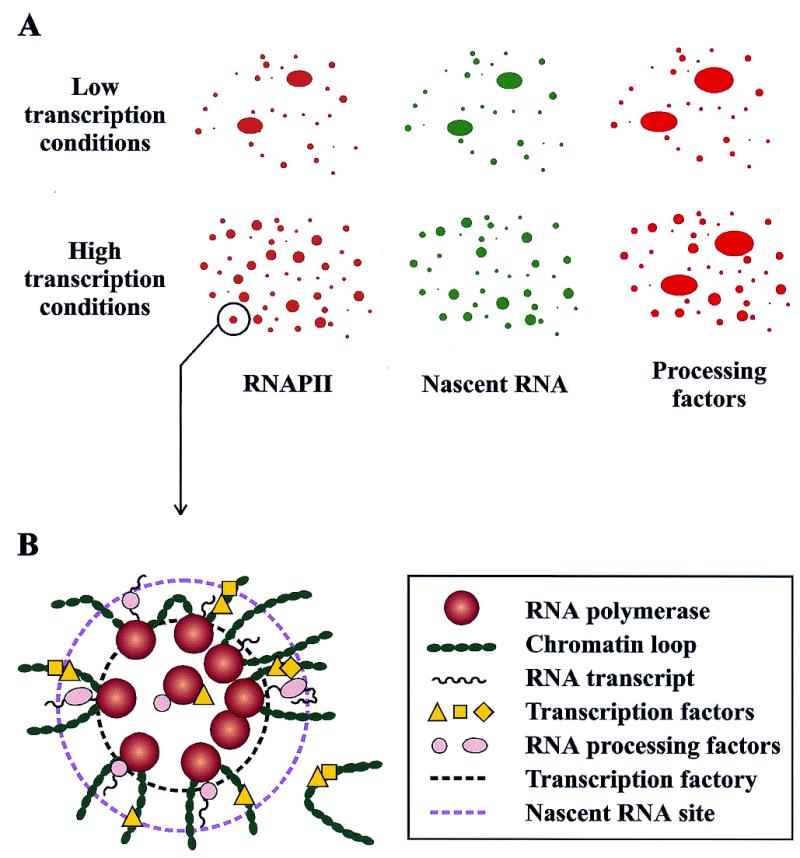
Schematic representation of the subnuclear localization of pre-mRNA transcription and processing components as observed by immunofluorescence labeling and confocal microscopy analyses. (A) Immunodetection with antibodies to the RNAPII phosphorylated CTD reveals RNAPII localized in the nucleus in thousands of tiny clusters. These foci, which occupy <1% of the nuclear volume, overlap with the sites of pre-mRNA synthesis detected by labeling the nascent transcripts with BrdU. In general, transcription and processing factors colocalize with the RNAPII foci, although particular factors can be observed at other locations that may represent storage pools. In cells that are actively transcribing, RNAPII transcription foci are diffusely distributed throughout the nucleus (bottom). When transcription is inhibited, sites contract to minimal domains (top). (B) Two-dimensional schematic representation of an RNAPII transcription factory (inner dashed circle) and its surrounding region of chromosome loops and transcripts with associated processing factors (outer dashed circle). In this model, genes are recruited to and transcribed by attached polymerases. Most genes are transcribed by a single RNAPII, although particular genes may have several transcribing polymerases.
Labeling of transcription and processing factors in the nucleus allows visualization of not only those factors that are in active complexes but also those that are in storage or at assembly sites, which can greatly complicate the interpretation of colocalization results (discussed in 42,48). In particular, specific probes recognize pools of inactive splicing factors and also splicing factors in association with RNAs that are in transit. Accordingly, the patterns observed by antibody labeling illustrate the association of these factors in various nuclear locations and not only at active sites. For example, the immunofluorescently labeled regions called speckles are thought to be storage locations for the splicing components and may be involved in the phosphorylation/dephosphorylation of processing factors (6,49). In general, transcriptionally active cells display, in addition to the speckles, a quite diffuse immunofluorescent staining of splicing components throughout the nucleoplasm (6,48), which is consistent with a wide distribution of coincident transcription and processing sites (Fig. 1A).
Another problem in determining the subnuclear distributions of individual components relative to the sites of active transcription is that multiple specific antibodies and labels must access the same small areas of active transcription in the nucleus (47). This technical difficulty, together with the requirement for exacting image analysis (discussed in 33,40,42), has so far restricted investigations of the colocalization of transcription factors with active transcription sites to only a few cases. In one study, the distribution of BRG1, an essential component of the human SWI–SNF chromatin remodeling complex, and TFIIH, a general RNAPII initiation factor, were shown to overlap significantly with transcription sites (41). The locations of several other factors did not coincide with RNAPII or nascent RNAs, however, and thus may be sites of storage or inhibitory complexes (41).
DYNAMIC SUBNUCLEAR ORGANIZATION OF TRANSCRIPTION AND PRE-mRNA PROCESSING
There is abundant evidence that the localization of transcription components in the nucleus is dynamic, reflecting both the cell-cycle stage and the overall transcriptional activity of the cell. A number of studies have revealed that RNAPII, splicing factors and transcription factors redistribute when the transcriptional activity of the nucleus is altered either by inhibitors or inducers of transcription or by the introduction of new transcription units (38,39,49–51) (Fig. 1). Introduction of new transcription sites by virus infection or induction, or by transient transfection, results in the relocalization of RNAPII and host splicing factors to the new sites of RNA synthesis (49–51). Misteli and colleagues transfected the splicing factor SF2/ASF into mammalian cells as a fusion with the green fluorescent protein and visualized the localization of this protein by time-lapse fluorescence microscopy (49). On transient expression of intron-containing genes, the labeled splicing factor accumulated at the new transcription sites, consistent with the idea that splicing occurs cotranscriptionally (see below). Furthermore, the time-lapse images showed trails of the tagged protein extending from pre-existing speckles to transcription sites, suggesting an active recruitment of splicing factors to transcription sites (49). Similarly, a dynamic transcription-dependent association of splicing factors at human cytomegalovirus immediate early genes was observed in rat 9G cells in which expression of the early viral genes had been induced (51).
There are other indications of a direct relationship between the transcriptional activity of the nucleus and its microscopic organization. For example, during periods of low transcription after inhibition of transcription by heat shock or drugs, the immunofluorescence from RNAPII and splicing factors contracted from a broadly distributed pattern to a pattern that corresponds to the nuclear speckles, forming minimal transcription domains (Fig. 1A) (38,39). The transcription factor HSF1, a key component for the inducible expression of the heat shock genes, also undergoes subnuclear redistribution, depending on environmental conditions. HSF1 staining distributes diffusely in the nucleoplasm before heat shock and then reversibly relocates to several nuclear punctate granules, termed stress granules, within 30 s after heat shock (46). In multiple cycles of heat shock and recovery, these stress granules rapidly form and disappear (46). The number of granules correlates with ploidy, suggesting specific chromosomal targets. However, the stress granules were not found to colocalize with the transcription sites of the hsp70 and hsp90 genes and appeared even in the absence of transcription in heat-shocked mitotic cells. Thus, the primary role of these granules may relate to the storage, release or regulation of HSF1 (46).
The effect of the RNAPII inhibitors 5,6-dichloro-β-d-ribofuranosyl-benzimidasole (DRB) and α-amanitin on localization of ribosomal RNA genes and heterochromatic DNA has been investigated by FISH (52). At levels that inhibit mRNA synthesis, these inhibitors reversibly dispersed heterochromatic DNA and euchromatic chromosomal structures, and also nucleolar structure, in the interphase nuclei of human fibroblasts and lymphocytes (52). This result indicates that ongoing transcription by RNAPII is essential for higher order organization of the interphase nucleus as a whole (52).
THE TRANSCRIPTION FACTORIES
Innovative experiments in Peter Cook’s laboratory have provided detailed information about the sites of transcription in mammalian nuclei (11,23–25,33,36,43,44,53,54). In HeLa cells, RNAPII transcription foci have diameters of ~40 nm and are scattered uniformly throughout the nucleoplasm, but occupy <1% of the nuclear volume (23,36,44). In determining numbers of RNAPII transcription sites, this laboratory has taken several experimental approaches, with different labeling, cell preparation and imaging methods, to ensure that only nascent transcripts are detected and to minimize the chances of missing relatively inactive sites. These experiments provided an estimate of several thousand RNAPII transcription foci in each HeLa cell nucleus (43,44,53). This initial measurement was combined with independent estimates of the numbers of active RNAPII molecules and nascent transcripts to determine that each focus contains 8–15 transcribing polymerases (44,53). Such a cluster of polymerases is termed a transcription factory (Fig. 1B) (23). Similar analyses indicated that transcription foci in the nucleoplasm are dedicated to either RNAPII or RNAPIII and that the two polymerases are not found together in any one factory (44).
By using modified Miller spreads of chromatin, the clustered polymerases in HeLa nuclei were separated and viewed by electron microscopy. The resulting micrographs showed that most non-ribosomal genes have only a single transcribing polymerase (43), consistent with the view that most protein-coding genes are not highly transcribed (55, reviewed in 53). A large number of these genes are expected to contain a promoter-paused RNAPII, a general feature of mammalian and Drosophila promoters (reviewed in 56). The promoter-paused polymerases are active in nuclear run-on assays, and will be counted in the total number of RNAPII in the nucleus, but will not be visible in Miller spreads as elongating polymerases with attached RNA. Even so, given the number of transcription foci in the nucleus, the predominance of lightly transcribed sequences and the number of transcribing polymerases within a cell at any given time, a cluster of polymerases must engage several different transcription units at once (33,40) (Fig. 1B).
Note that the factory model of RNAPII transcription is similar to the workings of the larger, better-characterized RNAPI transcription units in the nucleolus (3,53,57). Also in analogy to the events that are known to occur in the nucleolus, RNAPII transcription clusters would be predicted to rearrange to attach more genes during times of active transcription and perhaps fuse during inactive transcription periods (57) (Fig. 2). However, the mechanisms involved in such rearrangements of RNAPII groupings remain completely unknown.
Figure 2.
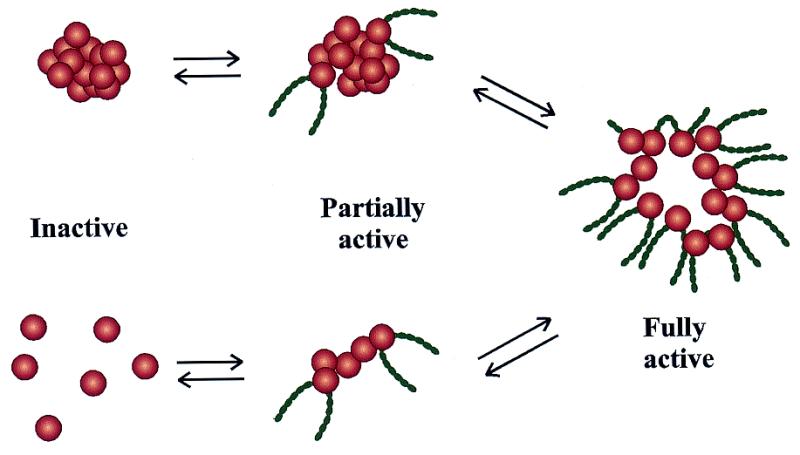
Possible models for the reorganization of RNAPII transcription factories as a function of the overall transcriptional activity of the cell. Nuclear components are known to undergo dynamic rearrangements as the transcriptional status of either endogenous or newly introduced genes changes. While transcriptionally active RNAPII molecules are clustered in transcription factories (right), the arrangement of inactive RNAPIIs is not known. Inactive RNAPIIs may be grouped (top left) or dissociated from any nuclear structure (bottom left).
The resistance of active RNAPII transcription units to electroelution, even after removal of most of the chromatin, supports a model in which the polymerases are attached in the nucleus (11,28,33). In this model, the loops of chromatin surround the transcription factories, and the DNA moves past the attached polymerases during transcription (33) (Fig. 1B). In agreement with this model, the RNAPII enzyme has been shown to be a molecular motor that is fully capable of supplying the force required for such chromosome motion (58).
GENE-SPECIFIC TRANSCRIPTION DOMAINS
The nucleolus, a specialized region dedicated to rRNA transcription and processing as well as nascent ribosomal subunit assembly, serves as a model of the spatial organization of gene expression (4,53,57,47). In the nucleolus, rRNA genes on different chromosomes are grouped to facilitate transcription. The RNA polymerases themselves are essential in organizing the nucleolar structure (59), and a major part of the rRNA processing occurs within the nucleoli in coordination with rRNA synthesis (1,2). Furthermore, maintenance of the nucleolar structure is dependent on ongoing RNAPI transcription (3).
Recent evidence suggests that organized regions analogous to nucleoli coordinate the expression of a subset of RNAPII-transcribed genes (47). A subnuclear domain enriched in transcription factors PTF and Oct1 (entitled the OPT domain for ‘Oct1, PTF and transcription’) was identified by immunostaining (41,47). The OPT domain colocalizes with a subset of the sites of active transcription in HeLa nuclei (47). Its presence is cell cycle-dependent, arising in G1 and disappearing in S phase (47). Most important, chromosomes were not found associated with the OPT domains as would be expected if they were randomly distributed throughout the nucleus (47). Instead, particular chromosomes preferentially colocalized with the OPT domain, indicating that this domain may constitute a specialized structure for transcription of certain PTF- and Oct1-dependent genes. The OPT domains always coincide with large PIKA domains (47), which are microscopically observable nuclear structures of previously unknown function (7). However, many smaller PIKAs did not coincide with OPT domains, and it remains to be determined whether these smaller PIKAs (or other known but uncharacterized nuclear substructures) are also gene-specific transcription domains.
RNA POLYMERASE II TRANSCRIPTION OCCURS WITHIN LARGE COMPLEXES
The discovery that RNAPII transcription sites are organized within the nucleus is consistent with biochemical and genetic data revealing the integration of RNAPII transcription with other nuclear processes. The approximately 12-subunit RNAPII enzyme is itself a subcomplex of various larger complexes, termed holoenzymes. Initiation-competent forms of the holoenzyme contain components of the general initiation machinery (60), chromatin remodeling proteins (61,62) and also transcriptional coactivators and repressors (62,63). Distinct forms of holoenzymes involved in elongation also exist (61,64). RNAPII holoenzymes isolated by a variety of approaches have also been shown to include pre-mRNA processing factors (65,66), repair proteins (67) and DNA replication proteins (68). Thus, these large complexes are likely to play a role in coordinating transcription with other nuclear events, including RNA processing, DNA repair and DNA replication. Many of the transcription factors involved in RNAPII transcription are themselves also large multiprotein complexes. Those include transcription factor TFIIH (69) and the promoter-recognizing factor TFIID (69). Following the pattern set for yeast, a number of mammalian chromatin-remodeling complexes have also been identified (70) and mammalian mediator complexes involved in transcriptional activation have been isolated (71,72).
In vitro studies indicate that RNAPII transcription initiation occurs through interaction between the promoter and a preassembled complex containing RNAPII and general initiation components (60). After initiation and upon the transition to the elongation mode of transcription, the set of proteins that comprises the holoenzyme changes (61,64,65). This switch correlates with increasing phosphorylation of the C-terminal domain (CTD) of the largest RNAPII subunit. The CTD, which consists of multiple heptapeptide repeats of the sequence YSPTSPS, can be phosphorylated by a variety of kinases, including the CDK7/cyclin H subcomplex of TFIIH and essential transcription factors such as P-TEFb (73). There is now strong evidence that CTD phosphorylation, prior to entry of RNAPII into the elongation mode and simultaneously with capping, is a key rate-limiting step in class II gene transcription (48,60,74,75). In addition, differences in the nature and extent of CTD phosphorylation occur during transcription elongation, indicating that these modifications serve multiple functions (see below; 76–79).
COORDINATION OF PRE-mRNA PROCESSING BY RNAPII
The need to consider transcription in a larger context is clearly illustrated by the tight association of pre-mRNA synthesis and its processing. The first unambiguous demonstration of cotranscriptional splicing was provided by Beyer and Osheim’s striking electron micrographs of Miller chromatin spreads of Drosophila embryo genes, where ribonucleoprotein particle formation and intron removal could be seen on nascent transcripts (80). There is now definitive biochemical, cytological and genetic evidence that much of the pre-mRNA processing is carried out concurrently with transcription (48,81,82), with the RNAPII elongation complex orchestrating the processing of its own transcripts (48,65,83).
As predicted by Greenleaf (84), the RNAPII CTD acts as a scaffold for the recruitment of processing factors. Many of these processing factors, including SR proteins and other splicing factors (65,76,85–87), cleavage-polyadenylation factors (66) and capping activities (74,88), bind to the phosphorylated form of the CTD present in the elongating RNAPII. Certain processing factors also interact with the RNAPII preinitiation complex (66,89). Both the polyadenylation factor (CstF) and the cleavage-polyadenylation specificity factor (CPSF) interact with the dephosphorylated CTD as well as with the phosphorylated CTD (66,90). CstF and CPSF copurify with an initiation-competent form of RNAPII holoenzyme (66), and CPSF has been shown to be a component of the RNAPII preinitiation complex at TATA box-containing promoters (89). After transcription initiation, CPSF dissociates from TFIID and then interacts with the CTD of the elongating polymerase (89). The association of these 3′-end-processing factors with both TFIID and the RNAPII dephosphorylated CTD in the preinitiation complex raises the possibility that these processing components may permanently reside with the RNAPII machinery at transcription sites.
The importance of RNAPII in the recruitment of pre-mRNA processing factors is clearly illustrated by the fact that truncation of the CTD prevents accumulation of SR proteins and snRNPs at transcription sites and also inhibits capping, splicing, 3′-end formation and termination (66,74,91). Furthermore, the RNAPII itself can be considered a polyadenylation factor, since in vitro experiments carried out in the absence of transcription reveal that RNAPII is required for polyadenylation of RNA (90). This active involvement of RNAPII in downstream events provides specificity for the processing of class II transcripts.
COREGULATION OF PRE-mRNA TRANSCRIPTION AND PROCESSING
The functional as well as physical interactions among factors that control transcription and processing permit coregulation of these two events. A number of instances of interdependent regulation of transcription and transcript processing have now been reported, with evidence for communication between the pre-mRNA processing complexes and the promoter of the corresponding gene (92–94). For example, the regulation of the thymidylate synthase gene during S phase at the level of pre-mRNA processing occurs in a promoter-specific fashion (92). Similarly, the choice of alternative exons in the fibronectin gene depends on its promoter structure, independently of promoter strength (93). Initiation from alternative promoters of the Fit-1 gene affects 3′-end processing, generating two different mRNA isoforms that code for either membrane or secreted Fit-1 proteins (94). Finally, expression of many genes is highly dependent on the presence of a generic intron, although the mechanisms involved are currently unknown (95).
SUBNUCLEAR DISTRIBUTION OF GENERAL AND GENE-SPECIFIC PROCESSING FACTORS
The link between processing and transcription has also been explored in experiments that preserve higher order structure in the nucleus (reviewed in 96). Consistent with the overwhelming biochemical and genetic evidence for cotranscriptional processing of pre-mRNA, microscopic imaging reveals that pre-mRNA processing occurs at transcription sites throughout the nucleus. Throughout various interphase mammalian nuclei, high concentrations of splicing factors (SR proteins) have been found at transcription sites labeled with antibodies to the large subunit of RNAPII or to BrUTP (39,42) (Fig. 1B). Images of HeLa nuclei showed that, for both an infected viral gene and an endogenous gene, splicing and transcription coincided spatially and temporally (82). Microscopy of the large, readily visualized insect polytene chromosomes demonstrated that splicing factors are localized at sites of active transcription but are not found at inactive genes (97). Evidence for the spatial association of polyadenylation components at the sites of transcription is provided by experiments in which subunits of the cleavage factors CstF and CPSF were shown to colocalize with nascent RNA (98). Polyadenylated transcripts were also detected at sites of cytomegalovirus immediate early gene transcription (51).
Different SR splicing proteins have distinct regulatory roles (48). Processing factors of this type that act cotranscriptionally and in a gene-specific fashion would therefore be expected to localize to only a subset of transcription sites. Recent results from Neugebauer and Roth are entirely consistent with this idea. In HeLa cells, considerable overlap is observed between transcription sites and SR proteins in general. In contrast, an antibody to a single SR protein, SRp20, reacted with only a subset of transcription sites (42). This restricted localization of SRp20 at particular transcription foci indicates that there is a degree of specificity in the association of specialized processing factors with a subset of genes and their transcripts. Although the mechanism of processing factor targeting is not known, it is intriguing to speculate that the specific interaction with certain RNAPII complexes may be mediated through CTD modification (76). A number of kinases are known to phosphorylate specific residues of the CTD, and heat-shock specific phosphorylation of the CTD has been observed in living cells (77,78). Thus, it is possible that the resulting varying phosphoisoforms of RNAPII may play a role in the subnuclear location of RNAPII and associated processing factors (76–78). In any case, the arrangement of gene-specific processing factors in the nucleus must depend not only on overall gene activity (39,97) but also on which particular genes are being expressed.
CONCLUSIONS
In light of the functional and genetic relationships among various nuclear events, and also the physical interactions of transcription components with proteins involved in other functions, transcriptional regulation must be considered in the context of the whole nucleus. Improved technology and innovative experimental approaches have revealed that the interphase nucleus is highly organized. Chromosomes are positioned in a non-random fashion, and the overall nuclear organization depends on gene expression. Transcription most likely occurs at polymerase foci that are attached to a nuclear scaffold, with subsets of these transcription foci organized around the expression of certain types of genes, forming microscopically visible structures.
It is well established that transcription factors contribute to RNAPII transcription by recruiting other factors to genes or by assisting one of the enzymatic steps of the reaction. The existence of transcription factories and subnuclear domains specialized in the transcription of particular classes of genes now suggests the possibility of additional roles for transcription factors. Some transcription factors may facilitate gene expression by seeding transcription factories, helping genes gain access to those factories, or directing the subnuclear localization of other transcription factors. Accordingly, certain activation or repression domains of gene-specific factors may well turn out to be targeting or localization domains. Similarly, the selective placement of repressed genes into specific environments suggests that certain transcription factors may control the grouping of chromosomes and transcription components in response to regulatory signals.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to William W. Mattox, Shona Murphy, Ana Pombo, Michael W. Van Dyke and Changqing Zeng for critical review of this manuscript and to the Robert A. Welch Foundation for financial support (grant G-1195 to M.S.)
REFERENCES
- 1.Pederson T. (1998) Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia S.N. and Pillus,L. (1999) Cell, 97, 825–828. [DOI] [PubMed] [Google Scholar]
- 3.Carmo-Fonseca M., Cunha,C., Custodio,N., Carvalho,C., Jordan,P., Ferreira,J. and Parreira,L. (1996) Exp. Cell Res., 229, 247–252. [DOI] [PubMed] [Google Scholar]
- 4.Strouboulis J. and Wolffe,A.P. (1996) J. Cell Sci., 109, 1991–2000. [DOI] [PubMed] [Google Scholar]
- 5.Lamond A.I. and Earnshaw,W.C. (1998) Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- 6.Misteli T. and Spector,D.L. (1998) Curr. Opin. Cell Biol., 10, 323–331. [DOI] [PubMed] [Google Scholar]
- 7.Saunders W.S., Cooke,C.A. and Earnshaw,W.C. (1991) J. Cell Biol., 115, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall W.F., Fung,J.C. and Sedat,J.W. (1997) Curr. Opin. Gen. Dev., 7, 259–263. [DOI] [PubMed] [Google Scholar]
- 9.Dernburg A.F., Broman,K.W., Fung,J.C., Marshall,W.F., Philips,J. Agard,D.A. and Sedat,J.W. (1996) Cell, 85,745–759. [DOI] [PubMed] [Google Scholar]
- 10.Belmont A.S., Dietzel,S., Nye,A.C., Strukov,Y.G. and Tumbar,T. (1999) Curr. Opin. Cell Biol., 11, 307–311. [DOI] [PubMed] [Google Scholar]
- 11.Jackson D.A., Bartlett,J. and Cook,P.R. (1996) Nucleic Acids Res., 24, 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamakaka R.T. (1997) Trends Biochem. Sci., 22, 124–128. [DOI] [PubMed] [Google Scholar]
- 13.Andrulis E.D., Neiman,A.M., Zappulia,D.C. and Sternglanz,R. (1998) Nature, 394, 592–595. [DOI] [PubMed] [Google Scholar]
- 14.Brown K.E., Guest,S.S., Smale,S.T., Hahm,K., Merkenschlager,M. and Fisher,A.G. (1997) Cell, 91, 845–854. [DOI] [PubMed] [Google Scholar]
- 15.Brown K.E., Baxter,J., Graf,D., Merkenschlager,M. and Fisher,A.G. (1999) Mol. Cell., 3, 207–217. [DOI] [PubMed] [Google Scholar]
- 16.Berezney R. and Coffey,D.S. (1977) J. Cell Biol., 73, 616–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pederson T. (1998) J. Mol. Biol., 277, 147–159. [DOI] [PubMed] [Google Scholar]
- 18.Hendzel M.J., Boisvert,F.-M. and Bazlett-Jones,D.P. (1999) Mol. Biol. Cell, 10, 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan K.M., Nickerson,J.A., Krockmalnic,G. and Penman,S. (1999) Proc. Natl Acad. Sci. USA, 96, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein G.S., van Wijnen,A.J., Stein,J.L., Lian,J.B., Pockwinse,S.M. and McNeil,S. (1998) J. Cell. Biochem., 70, 200–212. [PubMed] [Google Scholar]
- 21.He D.C., Nickerson,J.A. and Penman,S. (1990) J. Cell Biol., 110, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clemson C.M., McNeil,J.A., Willard,H.F. and Lawrence,J.B. (1996) J. Cell Biol., 146, 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson D.A., Hassan,A.B, Errington,R.J. and Cook,P.R. (1993) EMBO J., 12, 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura H., Tao,Y., Roeder,R.G. and Cook,P.R. (1999) Mol. Cell. Biol., 19, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hozak P., Sasseville,A.M., Raymond,Y. and Cook,P.R. (1995) J. Cell Sci., 108, 635–644. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen E., Besombes,D. and Debey,P. (1998) Mol. Rep. Dev., 50, 263–272. [DOI] [PubMed] [Google Scholar]
- 27.Smith S.S., Kelly,K.H. and Jockusch,B.M. (1979) Biochem. Biophys. Res. Commun., 86, 161–166. [DOI] [PubMed] [Google Scholar]
- 28.Sauman I. and Berry,S.J. (1994) Eur. J. Cell Biol., 64, 348–356. [PubMed] [Google Scholar]
- 29.Egly J.M., Miyamoto,N.G., Moncollin,V. and Chambon,P. (1984) EMBO J., 3, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y.W. and Stillman,D.J. (1996) Genes Dev., 10, 604–619. [DOI] [PubMed] [Google Scholar]
- 31.Scheer U., Hinssen,H., Franke,W.W. and Jockush,B.M. (1984) Cell, 39, 111–122. [DOI] [PubMed] [Google Scholar]
- 32.Schul W., de Jong,L. and van Driel,R. (1998) J. Cell. Biochem., 70, 159–171. [DOI] [PubMed] [Google Scholar]
- 33.Iborra F.J., Pombo,A., McManus,J., Jackson,D.A. and Cook,P.R. (1996) Exp. Cell Res., 229, 167–173. [DOI] [PubMed] [Google Scholar]
- 34.Wansink D.G., Schul,W., van der Kraan,I., van Steensel,B., van Driel,R. and de Jong,L. (1993) J. Cell Biol., 122, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wansink D.G., Sibon,O.C.M., Cremers,F.F.M., van Driel,R. and de Jong,L. (1996) J. Cell Biochem., 62, 10–18. [DOI] [PubMed] [Google Scholar]
- 36.Iborra F.J., Pombo,A., Jackson,D.A. and Cook,P.R. (1996) J. Cell Sci., 109, 1427–1436. [DOI] [PubMed] [Google Scholar]
- 37.Van Steensel B., Brink,M., van der Meulen,K., van Binnendijk,E.P., Wansink,D.G., de Jong, L, de Kloet,E.R. and Van Driel,R. (1995) J. Cell Sci., 108, 3003–3011. [DOI] [PubMed] [Google Scholar]
- 38.Bregman D.B., Du,L., van der Zee,S. and Warren,S.L. (1995) J. Cell Biol., 129, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng C., Kim,E., Warren,S.L. and Berget,S. (1997) EMBO J., 16, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fay F.S., Taneja,K.L., Shenoy,S., Lifshitz,L. and Singer,R.H. (1997) Exp. Cell Res. 231, 27–37. [DOI] [PubMed] [Google Scholar]
- 41.Grande M.A, van der Kraan,I., de Jong,L. and van Driel,R. (1997) J. Cell Sci., 110, 1781–1791. [DOI] [PubMed] [Google Scholar]
- 42.Neugebauer K.M. and Roth,M.B. (1997) Genes Dev. 11, 1148–1159. [DOI] [PubMed] [Google Scholar]
- 43.Jackson D.A., Iborra,F.J., Manders,E.M.M. and Cook,P.R. (1998) Mol. Biol. Cell, 9, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pombo A., Jackson,D.A., Hollinshead,M., Wang,Z., Roeder,R.G. and Cook,P.R. (1999) EMBO J., 18, 2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elefanty A.G., Antoniou,M., Custudio,N., Carmo-Fonseca,M. and Grosveld,F.G. (1996) EMBO J., 15, 319–333. [PMC free article] [PubMed] [Google Scholar]
- 46.Jolly C., Usson,Y. and Morimoto,R.I. (1999) Proc. Natl Acad. Sci. USA, 96, 6769–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pombo A., Cuello,P., Schul,W., Yoon,J.-B., Roeder,R.G., Cook,P.R. and Murphy,S. (1998) EMBO J., 17, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neugebauer K.M. and Roth,M.B. (1997) Genes Dev., 11, 3279–3285. [DOI] [PubMed] [Google Scholar]
- 49.Misteli T., Caceres,J.F. and Spector,D.L. (1997) Nature, 387, 523–527. [DOI] [PubMed] [Google Scholar]
- 50.Jiminez-Garcia L.F. and Spector,D.L. (1993) Cell, 73, 47–59. [DOI] [PubMed] [Google Scholar]
- 51.Dirks R.W., de Pauw,E.S.D. and Raap,A.K. (1997) J. Cell Sci., 110, 515–522. [DOI] [PubMed] [Google Scholar]
- 52.Haaf T. and Ward,D.C. (1996) Exp. Cell Res., 224, 163–173. [DOI] [PubMed] [Google Scholar]
- 53.Cook P.R. (1999) Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- 54.Cook P.R. (1995) J. Cell Sci., 108, 2927–2935. [DOI] [PubMed] [Google Scholar]
- 55.Foe V.E., Wilkinson,L.E. and Laird,C.D. (1976) Cell, 9, 131–146. [DOI] [PubMed] [Google Scholar]
- 56.Lis J.T. (1998) Cold Spring Harbor Symp. Quant. Biol., 63, 347–356. [DOI] [PubMed] [Google Scholar]
- 57.Jordan P., Mannervik,M., Tora,L. and Carmo-Fonseca,M. (1996) J. Cell Biol., 133, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin Y., Wang,M.D., Svoboda,K., Landick,R., Block,S.M. and Gelles,J. (1995) Science, 270, 1653–1657. [DOI] [PubMed] [Google Scholar]
- 59.Oakes M., Aris,J.P., Brockenbrough,J.S., Wai,H., Vu,L. and Nomura,M. (1998) J. Cell Biol., 143, 23–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenblatt J. (1997) Curr. Opin. Cell Biol., 9, 310–319. [DOI] [PubMed] [Google Scholar]
- 61.Cho H., Orphanides,G., Sun,X., Yang,X.J, Ogryzko,V., Lees,E., Nakatani,Y. and Reinberg,D. (1998) Mol. Cell. Biol., 18, 5355–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neish A.S., Anderson,S.F., Schlegel,B.P., Wei,W. and Parvin,J.D. (1998) Nucleic Acids Res., 26, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakajima T., Uchida,C., Anderson,S.F., Parvin,J.D. and Montminy,M. (1997) Genes Dev., 11, 738–747. [DOI] [PubMed] [Google Scholar]
- 64.Otero G., Fellows,J., Li,Y., de Bizemont,T., Dirac,A.M.G., Gustafsson,C.M., Erdjument-Bromage,H., Tempst,P. and Svejstrup,J.Q. (1999) Mol. Cell., 3, 109–118. [DOI] [PubMed] [Google Scholar]
- 65.Corden J.L. and Patturajan,M. (1997) Trends Biochem. Sci., 22, 413–416. [DOI] [PubMed] [Google Scholar]
- 66.McCracken S., Fong,N., Yankulov,K., Ballantyne,S., Pan,G., Greenblatt,J., Patterson,S., Wickens,M. and Bentley,D.L. (1997) Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- 67.Maldondo E., Shiekhattar,R., Sheldon,M., Cho,H., Drapkin,R., Rickert,P., Lees,E., Anderson,C.W., Linn,S. and Reinberg,D. (1996) Nature, 381, 86–89. [DOI] [PubMed] [Google Scholar]
- 68.Yankulov K., Todorov,I., Romanowski,P., Licatalosi,D., Cilli,K., McCracken,S., Laskey,R. and Bentley,D.L. (1999) Mol. Cell. Biol., 19, 6154–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orphanides G., Lagrange,T. and Reinberg,D. (1996) Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- 70.Ogryzko V.V., Kotani,T., Zhang,X., Schlitz,R.L., Howard,T., Yang,X.-J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- 71.Asturias F.J., Jiang,Y.W., Myers,L.C., Gustafsson,C.M. and Kornberg,R.D. (1999) Science, 283, 985–987. [DOI] [PubMed] [Google Scholar]
- 72.Ryu S., Zhou,S., Ladumer,A.G. and Tjian,R. (1999) Nature, 397, 446–450. [DOI] [PubMed] [Google Scholar]
- 73.Marshall N.F., Peng,J., Xie,Z. and Price,D.H. (1996) J. Biol. Chem., 271, 27176–27183. [DOI] [PubMed] [Google Scholar]
- 74.McCracken S., Fong,N., Rosonina,E., Yankulov,K., Brothers,G., Siderovski,D., Hessel,A., Foster,S. Amgen EST Program, Shuman,S. and Bentley,D.L. (1997) Genes Dev. 11, 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Brien T.S., Hardin,A., Greenleaf,A. and Lis,J.T. (1994) Nature, 370, 75–77. [DOI] [PubMed] [Google Scholar]
- 76.Patturajan M., Schulte,R.J., Sefton,B.M., Berezney,R., Vincent,M., Bensaude,O., Warren,S.L. and Corden,J. (1998) J. Biol. Chem., 273, 4689–4694. [DOI] [PubMed] [Google Scholar]
- 77.Rickert P., Corden,J.L. and Lees,E. (1999) Oncogene, 18, 1093–1102. [DOI] [PubMed] [Google Scholar]
- 78.Egyhazi E. Ossoinak,A., Lee,J.M., Greenleaf,A.L., Makela,T.P. and Pigon,A. (1998) Exp. Cell Res., 242, 211–221. [DOI] [PubMed] [Google Scholar]
- 79.Bensaude O., Bonnet,F., Casse,C., Dubois,M.-F., Nguyen,V.T. and Palancade,B. (1999) Biochem. Cell Biol., 77, 249–255. [PubMed] [Google Scholar]
- 80.Beyer A.L. and Osheim,Y.N. (1988) Genes Dev., 2, 754–765. [DOI] [PubMed] [Google Scholar]
- 81.Bauren G. and Wieslander,L. (1994) Cell, 76, 183–192. [DOI] [PubMed] [Google Scholar]
- 82.Zhang G., Taneja,K.L., Singer,R.H. and Green,M.R. (1994) Nature, 372, 809–812. [DOI] [PubMed] [Google Scholar]
- 83.Steinmetz E.J. (1997) Cell, 89, 491–494. [DOI] [PubMed] [Google Scholar]
- 84.Greenleaf A.L. (1993) Trends Biochem. Sci., 18, 117–119. [DOI] [PubMed] [Google Scholar]
- 85.Yuryev A., Patturajan,M., Litingtung,Y., Joshi,R.V., Gentile,C., Gebara,M. and Corden,J.L. (1996) Proc. Natl Acad. Sci. USA, 93, 6975–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim E., Du,L., Bregman,D.B. and Warren,S.L. (1997) J. Cell Biol., 136, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mortillaro M.J., Blencowe,B.J., Wei,X., Nakayasu,H., Du,L., Warren,S.L., Sharp,P.A. and Berezney,R. (1996) Proc. Natl Acad. Sci. USA, 93, 8253–8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho E.-J., Takagi,T., Moore,C.R. and Buratowski,S. (1997) Genes Dev., 11, 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dantonel J.-C., Murthy,K.G., Manley,J.L. and Tora,L. (1997) Nature, 389, 399–402. [DOI] [PubMed] [Google Scholar]
- 90.Hirose Y.R. and Manley,J.L. (1998) Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- 91.Misteli T. and Spector,D.L. (1999) Mol. Cell., 3, 697–705. [DOI] [PubMed] [Google Scholar]
- 92.Ke Y., Ash,J. and Johnson,L.F. (1996) Mol. Cell. Biol., 16, 376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cramer P., Pesce,C.G., Baralle,F.E. and Kornblihtt,A.R. (1997) Proc. Natl Acad. Sci. USA, 94, 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bergers G., Reikerstorfer,A., Braselmann,S., Graninger,P. and Busslinger,M. (1994) EMBO J., 13, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buchman A.R. and Berg,P. (1988) Mol. Cell. Biol., 8, 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sleeman J.E. and Lamond,A.I. (1999) Curr. Opin. Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- 97.Bauren G., Jiang,W.-Q., Bernholm,K., Gu,F. and Weislander,L. (1996) J. Cell Biol., 133, 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schul W., Groenhout,B., Koberna,K., Takagaki,Y., Jenny,A., Manders,E.M.M., Raska,I., Van Driel,R. and DeJong,L. (1996) EMBO J., 15, 2883–2892. [PMC free article] [PubMed] [Google Scholar]


