Abstract
Background.
While the clinical efficacy of antimalarial artemisinin-based combination therapies (ACTs) in Africa remains high, the recent emergence of Plasmodium falciparum artemisinin partial resistance (ART-R) on the continent is concerning, given the lack of alternative treatments.
Methods.
Using drug efficacy studies conducted in 2016–19 to evaluate artesunate-amodiaquine or artemether-lumefantrine treatment for uncomplicated malaria in Eritrea, we estimated the proportion of patients with persistent P. falciparum parasitemia at day3 (D3+) and assayed parasites for mutations in the Pfkelch13 gene as predictive markers of ART-R. We also screened for deletions in hrp2/hrp3 that result in variable performance of HRP2-based malaria rapid diagnostic tests (RDTs).
Results.
We noted increased proportions of D3+ patients from 2016 (0.4%, 1/273) to 2017 (1.9%, 4/209) and 2019 (4.2%, 15/359). A Pfkelch13 R622I variant, detected in 109/818 isolates prior to treatment, similarly showed a rise in prevalence from 2016 (8.6%, 24/278) to 2019 (21.0%, 69/329). The odds of D3+ increased by 6.2-fold (95% CI [2.5–15.5]) for patients carrying Pfkelch13 R622I variant. In vivo ART-R in Eritrea was observed with >5% of D3+ patients <15 years old harboring Pfkelch13 622I mutant parasites. In vitro, the 622I variant conferred low-level ART-R when edited into NF54 and Dd2 parasite lines. A proportion (16.9%) of Pfkelch13 622I parasites carried double hrp2/hrp3 deletions, possibly rendering these parasites undetectable by HRP2-based RDTs.
Conclusions.
The emergence and spread of P. falciparum lineages with both Pfkelch13-mediated ART-R and deletions in hrp2/hrp3 genes in Eritrea threaten to compromise regional malaria control and elimination campaigns. (ACTRN12618001223224, ACTRN12618000353291, ACTRN12619000859189.)
Artemisinin-based combination therapies (ACTs), which combine fast-acting and potent artemisinin derivatives with longer-acting partner drugs, are essential first-line treatments for uncomplicated Plasmodium falciparum malaria.1 Over the last fifteen years, P. falciparum parasites have developed artemisinin partial resistance (ART-R) in the Greater Mekong Subregion, manifested as delayed parasite clearance or persistence of parasites on day3 (D3+) following ACT treatment due to decreased susceptibility of intraerythrocytic ring-stage parasites.1–6 This Subregion has witnessed increasing rates of ACT treatment failure as parasites also acquire resistance to the partner drugs piperaquine or mefloquine.7
Although the clinical efficacy of ACTs in African settings is presently high, the recent emergence of ART-R in Rwanda and Uganda is of major concern.1,8–13 Molecular studies have confirmed the presence of nonsynonymous mutations in Pfkelch13 (PF3D7_1343700), the primary determinant of ART-R.14,15 These mutations include R561H in Rwanda, and C469Y and A675V in Uganda. All three variants, associated with delayed parasite clearance and/or D3+, have displayed increasing prevalence over time (7.8% in 2015 to 12.8% in 2018 in Rwanda, and 3.9% in 2015 to 19.8% in 2019 in Uganda).10,12 Ex vivo and in vitro assays measuring survival rates of Pfkelch13 R561H and C469Y parasites (either gene-edited lines or field isolates) support these mutations as markers of in vitro ART-R, in a parasite genetic background-dependent manner.10,11,16 Genomic analyses have demonstrated the independent emergence and local expansion of these Pfkelch13 mutants.10–12
In Eritrea, artesunate-amodiaquine (ASAQ), first introduced in 2007 as first-line treatment for uncomplicated falciparum malaria, is now available free of charge both at health facilities and the community level. In 2015, a single dose of primaquine was added to ASAQ as a transmission-blocking agent.13 Artemether-lumefantrine (AL), recommended as second-line treatment so far, was implemented in 2019 at health facilities as an alternative first-line treatment for uncomplicated malaria.
Here, we describe the results of therapeutic efficacy studies conducted between 2016–2019 at five sites in Eritrea evaluating ASAQ and AL treatment for uncomplicated falciparum malaria. We assessed the proportion of D3+ patients and assayed parasites for molecular signatures of ART-R. We also screened for deletions in hrp2 and hrp3 that result in variable performance of HRP2-based malaria rapid diagnostic tests (RDTs).
Methods
Study design, areas and population
Open-label, single-arm, multi-site clinical drug efficacy studies were designed to assess clinical ART-R in Eritrea, as determined by the proportion of D3+ patients after a 3-day ASAQ or AL treatments and conducted in 2016, 2017 and 2019 at health centers or hospitals at five sites in western Eritrea (all 3 parent protocols available at nejm.org). Studies were approved by the Eritrean Ethical Committee and the WHO Ethical Review Committee. Patients were at least six months old, and eligibility was determined according to WHO inclusion/exclusion criteria. Informed written consent was obtained from the adult patient or parent/caretaker.
Treatment, follow-up procedure and outcomes
Consenting patients were assigned a supervised standard 3-day course of ASAQ (2016 and 2019) or AL (2017) and monitored clinically throughout. Thick and thin blood smears were obtained by finger prick upon recruitment (day0), and during follow up visits on days 1, 2, 3, 7, 14, 21 and 28 to screen for P. falciparum and estimate parasite density. Additional follow up visits were scheduled if further symptoms occurred. Dried blood spot (DBS) filter papers were used for molecular studies. The primary outcome was the proportion of D3+ patients, as assessed by microscopic examination of thick blood smears after 3-day ACT treatment.1 We evaluated, as a secondary outcome, the PCR-adjusted clinical response to the designated treatment on day28.
Molecular analysis
Parasite DNA was extracted from pre- and post-treatment DBS (cases of recurrence) using the QIAamp DNA Blood Mini Kit (Qiagen). Genotyping of the polymorphic genetic markers msp1, msp2, and polyα was carried out by PCR, and post-treatment infections were either classified as recrudescence (same genotype as day0) or new infections (different genotype).17
Paired DNA samples (day0 and day of recurrence) were analyzed for mutations in the propeller domain of Pfkelch13 (codons 430–720) and in pfcrt, pfmdr-1, dhfr, and dhps, which are associated with decreased parasite susceptibility to artemisinin derivatives, 4-aminoquinolines (piperaquine and chloroquine), amino-alcohols (mefloquine and lumefantrine), pyrimethamine, and sulfadoxine, respectively.18 We also screened for hrp2 and hrp3 deletions that can cause false-negative results with HRP2-based rapid diagnostic tests (RDTs).19
Whole-genome sequencing was performed by Illumina paired-end sequencing after selective amplification of parasite DNA.20 Read alignments against the 3D7 genome (v45) were used to infer a phylogenetic tree. The Genome Analysis Toolkit was used to identify SNPs, genotype isolates, and assess the genetic identity of Pfkelch13 mutants from Eritrea. Principal Coordinate Analysis (PCoA), hierarchical clustering as well as AMOVA (Analysis of Molecular Variance) were performed based on pairwise Euclidean genetic distances between samples.
Generation of gene-edited lines and in vitro susceptibility assays.
The Pfkelch13 R622I mutation was introduced into African (NF54) and Asian (Dd2) parasite lines by CRISPR/Cas9-mediated gene editing. In vitro ART susceptibilities of edited parasites and wild-type controls were assessed using the Ring Stage Survival Assay (RSA0–3h)21 (Supplementary Information).
Statistical analysis
Data were analyzed with GraphPad Prism 9.3.1 (GraphPad Software). Because the analyses presented here were not originally specified in the protocols for the three component studies, all analyses are descriptive. 95% confidence intervals [CI] are provided for all estimates, but these have not been adjusted for multiple comparisons and were not used in place of hypothesis tests. The primary analysis used complete cases only and ignored data from participants with missing outcome data. The Kaplan-Meier analyses were conducted as an alternative to the complete-case analysis.
Results
Participants and study design
A total of 852 patients with uncomplicated falciparum malaria were enrolled (Table 1). Of these, 841 (98.7%) and 825 (96.8%) were assessed for D3+ rate and clinical efficacy outcome, respectively. Remaining patients either withdrew consent (n=10) or were lost to follow-up (n=17) (Fig. S1).
Table 1.
Characteristics of the participants with measured D3+ rate and detected Pfkelch13 genotypes from blood samples collected prior to artemisinin-based combination therapy according by study year.
| 2016 | 2017 | 2019 | Total | ||
|---|---|---|---|---|---|
| Baseline characteristics | |||||
| Study period | Jan-Dec | Sept-Dec | Aug-Nov | - | |
| No. of patients | 280 | 211 | 361 | 852 | |
| Antimalarial treatment | ASAQ | AL | ASAQ | - | |
| Site | Akordat | 58* | 19* | 88 | 165 |
| Ghindae | - | 15* | - | 15* | |
| Guluj | 73 | 25* | 97 | 195 | |
| Shambuko | 73 | 64 | 88 | 225 | |
| Tokombia | 76 | 88 | 88 | 252 | |
| Median age, years (IQR) | 13.0 (8.0–19.0) | 13.0 (9.0–22.7) | 17.5 (12.0–29.0) | 15.0 (10.0–25.0) | |
| Gender ratio (Female/Male) | 120/160 | 82/129 | 119/242 | 321/531 | |
| Median temperature, °C (IQR) | 38.0 (38.0–39.0) | 38.0 (38.0–38.3) | 38.0 (38.0–38.5) | 38.0 (38.0–39.0) | |
| Median parasite density, μL (IQR) | 7,530 (2,736–18,036) | 7,900 (2,677–22,335) | 9,312 (2,720–23,933) | 8,280 (2,715–21,902) | |
| Day3 positivity (D3+) rate, no. (%) | |||||
| Site | Akordat | 0/54 (0) | 0/19 (0) | 0/88 (0) | 0/161 (0) |
| Ghindae | - | 0/15 (0) | - | 0/15 (0) | |
| Guluj | 0/71 (0) | 0/23 (0) | 1/95 (1.1) | 1/189 (0.5) | |
| Shambuko | 1/73 (1.4) | 4/64 (6.2) | 5/88 (5.7) | 10/225 (4.4) | |
| Tokombia | 0/75 (0) | 0/88 (0) | 9/88 (10.2) | 9/251 (3.6) | |
| Total | 1/273 (0.4) | 4/209 (1.9) | 15/359 (4.2) | 20/841 (2.4) | |
| Pfkelch13 genotype, no. (%) | |||||
| Missing samples | 0/280 (0) | 0/211 (0) | 23/352 (6.5) | 23/852 (2.7) | |
| Missing data | 2/280 (0.7) | 0/211 (0) | 9/352 (2.5) | 11/852 (1.3) | |
| WT | 251/278 (90.3) | 193/211 (91.4) | 254/329 (77.2) | 698/818 (85.3) | |
| Mutant | 503 (K>W) | 1/211 (0.5) | 1/818 (0.1) | ||
| 515 (R>G) | 1/211 (0.5) | 1/818 (0.1) | |||
| 520 (V>A) | 1/278 (0.3) | 1/818 (0.1) | |||
| 532 (C>W) | 1/329 (0.3) | 1/818 (0.1) | |||
| 533 (G>N) | 1/329 (0.3) | 1/818 (0.1) | |||
| 543 (I>V) | 1/329 (0.3) | 1/818 (0.1) | |||
| 548 (G>C) | 1/329 (0.3) | 1/818 (0.1) | |||
| 556 (E>K) | 1/329 (0.3) | 1/818 (0.1) | |||
| 561 (R>H) | 1/329 (0.3) | 1/818 (0.1) | |||
| 591 (G>N) | 1/278 (0.3) | 1/818 (0.1) | |||
| 622 (R>I) | 24/278 (8.6) | 16/211 (7.6) | 69/329 (21.0) | 109/817 (13.3) | |
| 658 (K>E) | 1/278 (0.3) | 1/817 (0.1) | |||
The target number of patients to be enrolled at each study site was estimated, based on power calculations, to be 73. We note that lower than expected numbers of participants were enrolled in Akordat (2016 and 2017), Ghindae (2017 and 2019), and Guluj (2017), mainly due to the low number of malaria cases seen at health centers in this low malaria-transmission region during the study period.
Day3 positivity rate (D3+).
Twenty patients (2.4%, 20/841) from Guluj, Shambuko and Tokombia remained parasitemic on day3 post treatment (Table S1, Fig. 1A). D3+ rates increased from 2016 (1/273, 0.4%; 95% CI [0.01 to 2.0]) and 2017 (4/209, 1.9%; 95% CI [0.5 to 4.9]) to 2019 (15/359, 4.2%; 95% CI [2.3 to 6.9]). The highest D3+ rates were seen at Tokombia (10.2%) and Shambuko (5.7%) in 2019. In total, 825/852 (96.8%) participants were evaluated at day28 (Table S2). Of the 27 recurrent infections, 17 were classified as recrudescent. PCR-corrected complete-case and Kaplan-Meier estimates of ASAQ and AL treatment efficacies were >94%, above the threshold recommended by the WHO for treatment policy change (90%) (Supplementary Information, Tables S3–S5).
Fig. 1. Evidence of delayed parasite clearance associated with the expansion of the mutant Pfkelch13 622I in Eritrea.
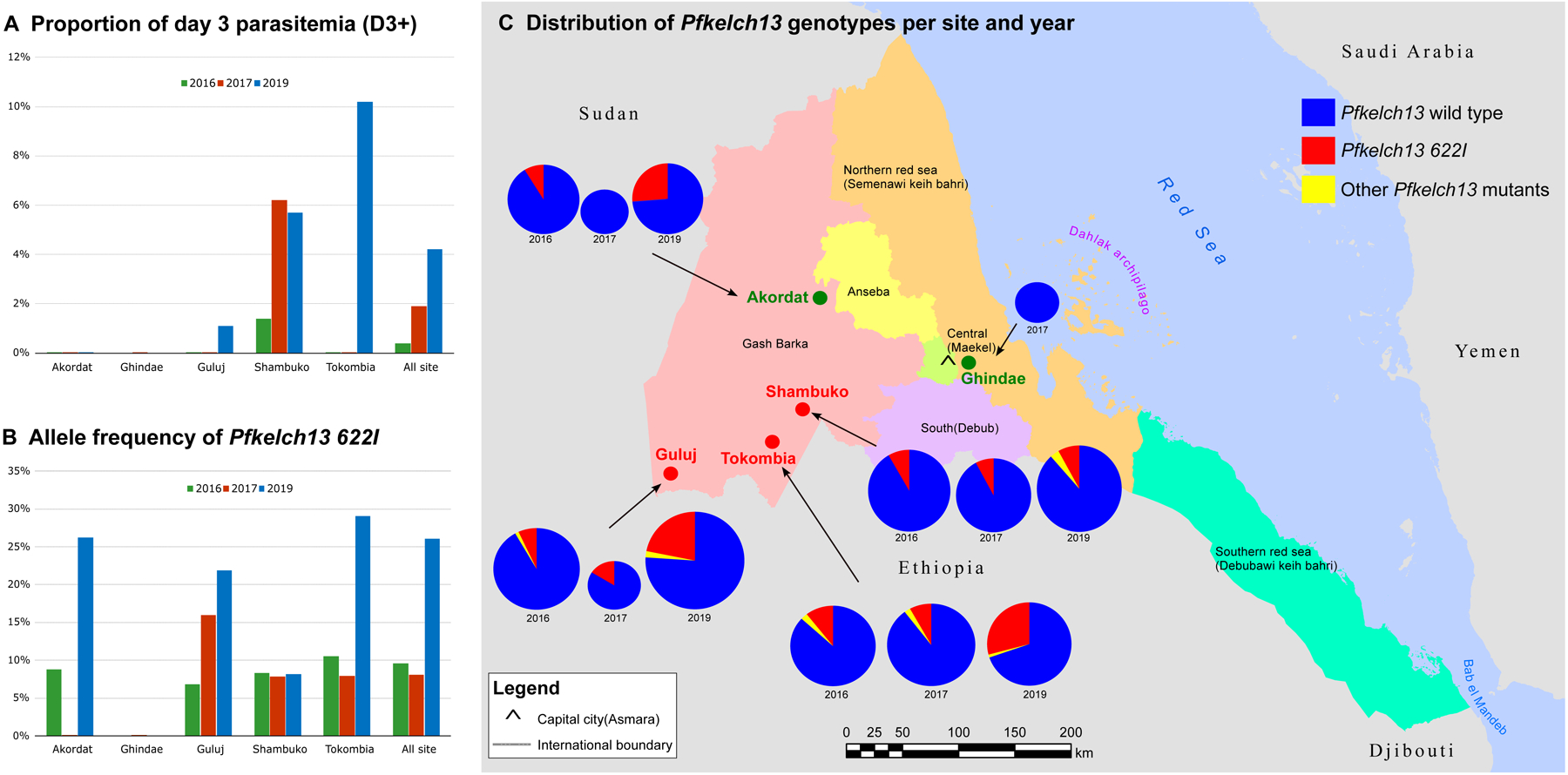
(A) Proportions of D3+ rate per site and year. D3+ cases were not observed in Akordat (2016, 2017 and 2019), Ghindae (2017, no data were available in 2016 and 2019), Guluj (2016 and 2017), and Tokombia ((2016 and 2017). (B) Allele frequency of P. falciparum kelch13 622I mutant parasites per site and year. Only Pfkelch13 wild-type parasites were observed in Akordat and Ghindae in 2017 (no data were available in 2016 and 2019). (C) Distribution of Pfkelch13 genotypes per site and year. The proportions of each Pfkelch13 allele are shown per year in pie charts (except for Ghindae where only 2017 data were available). The frequency for the Pfkelch13 wild type allele is shown in dark blue, the Pfkelch13 622I allele is in red and the other Pfkelch13 mutants are in yellow. The size of the pie chart is proportional to the sample size. The study sites colored in green correspond to areas where no D3+ case was observed. The study sites where D3+ cases where detected are colored in red (Guluj, Tokombia, and Shambuko). Additional information is presented in Tables S1, S6 and S7.
Pfkelch13 genotyping
Of the 828 available pre-treatment samples, 818 (98.8%) were successfully genotyped. Twelve Pfkelch13 non-synonymous mutations were detected in 120 samples (Table 1). The Pfkelch13 R561H mutation, a validated marker for ART-R, was observed in one isolate (Shambuko, 2019).11,12 A novel Pfkelch13 R622I mutation was detected in 13.3% (109/818). The prevalence of this variant increased from 2016 (24/278, 8.6%; 95% CI [5.5 to 12.8]) and 2017 (16/211, 7.6%; 95% CI [4.3 to 12.3]) to 2019 (69/329, 21.0%; 95% CI [16.3 to 26.5]) (Fig. 1B–1C, Table S6). In 2019, the prevalence was 8.1% in Shambuko, 21.9% in Guluj, 26.2% in Akordat, and 29.1% in Tokombia.
Validation of the Pfkelch13 R622I mutation as a novel marker of ART-R
Association with delayed parasite clearance (D3+).
The proportion of the Pfkelch13 622I variant prior to treatment was higher in D3+ patients (9/20, 45%; 95% CI [20.6–85.4]) compared to D3- patients (97/787, 12.3%; 95% CI [10.0–15.0] (risk ratio, 5.4; 95% CI [2.3 to 12.7]). In vivo ART-R was confirmed in Eritrea as the proportion of D3+ patients (aged <15 years) harboring Pfkelch13 622I mutant parasites at day0 was >5% in Tokombia (7.0% in 2019) and Shambuko (5.4% in 2017) (Table S7).22 The odds of D3+ were 6.2-fold higher (95% CI [2.5–15.5]) in patients carrying Pfkelch13 R622I variant in isolates prior to ACT treatment compared to patients carrying Pfkelch13 wild-type parasites (Table 2). While ASAQ treatment failure rates were similar between patients with Pfkelch13 622I mutant and wild-type parasites in day0 isolates (2/87, 2.3% vs. 15/495, 3.0%), Pfkelch13 genotyping of paired isolates from day0 and the day of recrudescence indicated selection of the 622I mutant following ASAQ administration (4.4-fold increase, from 12% at day0 to 53% at day of recrudescence). Using amplicon deep sequencing, we detected the presence of Pfkelch13 622I genotypes in minor proportions (1.4%−3.2%) in 7 of 9 day0 samples previously classified as wild-type, providing evidence of intra-host selection after administration of ASAQ (Table S8).
Table 2.
Multiple regression analysis for D3+ rate among 840 cases, Eritrea, 2016–2019.
Multiple regression was used to analyze the relationship between age (which is related to host immunity and the capacity of the immune system to clear parasites independent of treatment), sex, initial parasitemia (a high parasitemia on day0 can lead to parasite persistence on day3) and Pfkelch13 622 variant on the persistence parasitemia at day 3 (D3+).
| Covariate | Coefficient | Std. Error | Odds ratio (95% CI) |
|---|---|---|---|
| Age | −6.3E-03 | 0.016 | 0.99 (0.96–1.02) |
| Sex | −8.1E-03 | 0.487 | 0.98 (0.38–2.57) |
| Initial parasitemia | 8.7E-06 | 5.8E-06 | 1.00 (1.00–1.00) |
| Pfkelch13 622I | 1.824 | 0.466 | 6.2 (2.5–15.5) |
95% CI: the 95% confidence interval for the estimated odds ratio
We found that holding all other covariates constant, the odds of D3+ were 6.2-fold higher (95% CI [2.5–15.5]) in patients carrying the Pfkelch13 R622I variant in isolates prior to ACT treatment compared to patients carrying Pfkelch13 wild-type parasites.
The goodness of fit of our multiple regression model was evaluated both from the Hosmer-Lemeshow test (p-value >0.05) and the ROC curve analysis (Area under the ROC curve estimate of 0.726; SD 0.07, 95% CI [0.69–0.75]).
In vitro survival rate of the Pfkelch13 622I mutant.
The Pfkelch13 R622I mutation was edited into NF54 (African) and Dd2 (Asian) parasites. Recombinant clones were tested in the RSA0–3h that measures the survival of early ring-stage parasites exposed to 700 nM DHA for 6 hr. Survival >1% (relative to mock-treated parasites) indicates in vitro ART-R. Results showed that the Pfkelch13 R622I mutation conferred low-level ART-R in NF54R622I parasites compared to the isogenic wild-type (WT) control line (3.3% survival versus 0.6%). In vitro resistance was borderline in the Dd2R622I line (1.5% survival in the mutant versus 0.7% in the isogenic control). In NF54 parasites, the R622I mutation conferred somewhat lower levels of resistance than the C580Y mutation that predominates across Southeast Asia (RSA0–3h survival rate of 4.3%) (Fig. 2).
Fig. 2. Pfkelch13 R622I mediates low-level artemisinin resistance in P. falciparum parasites.
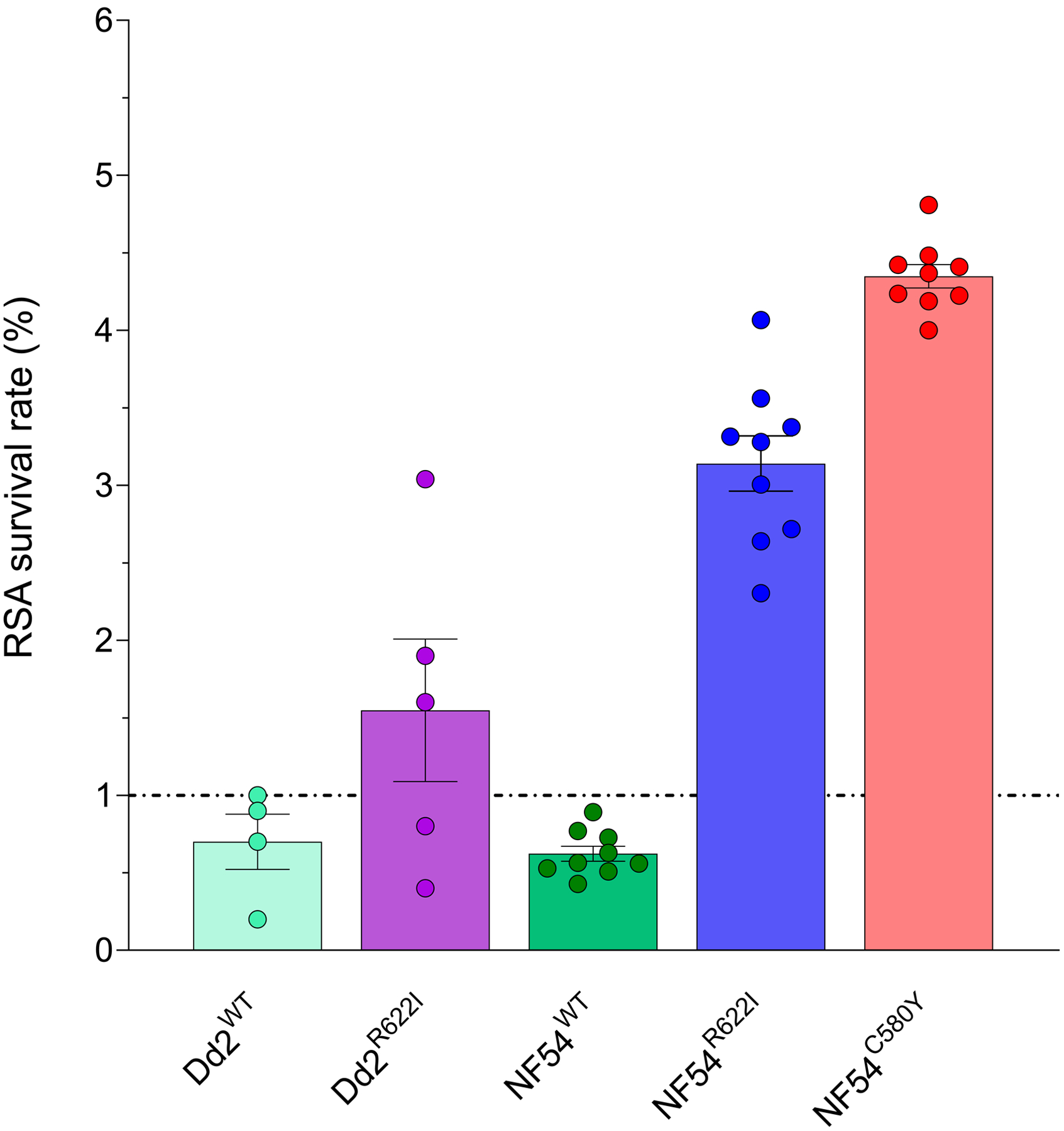
Results show the percentage of early ring-stage parasites (0–3 hours post-invasion) that survived a 6-hour pulse of 700 nM DHA, relative to DMSO-treated parasites assayed in parallel. Percent survival values are shown as means ± SEM. Results were obtained from two (Dd2WT) to three (NF54WT, Dd2R622I, NF54R622I and NF54C580Y) independent experiments, each performed in duplicate (Dd2WT and Dd2R622I) or triplicate (NF54WT, NF54R622I and NF54C580Y).
Origins of the Pfkelch13 622I genotype.
We compared whole-genome sequences of 291 samples, including 128 Eritrean P. falciparum sequences generated for this study, 162 publicly available sequences, and the 3D7 reference genome from Africa (Table S9). A maximum-likelihood phylogenetic tree showed that the Pfkelch13 622I mutants were scattered within East African wild-type isolates (Fig. 3).
Fig. 3. Genome-wide phylogenetic tree.
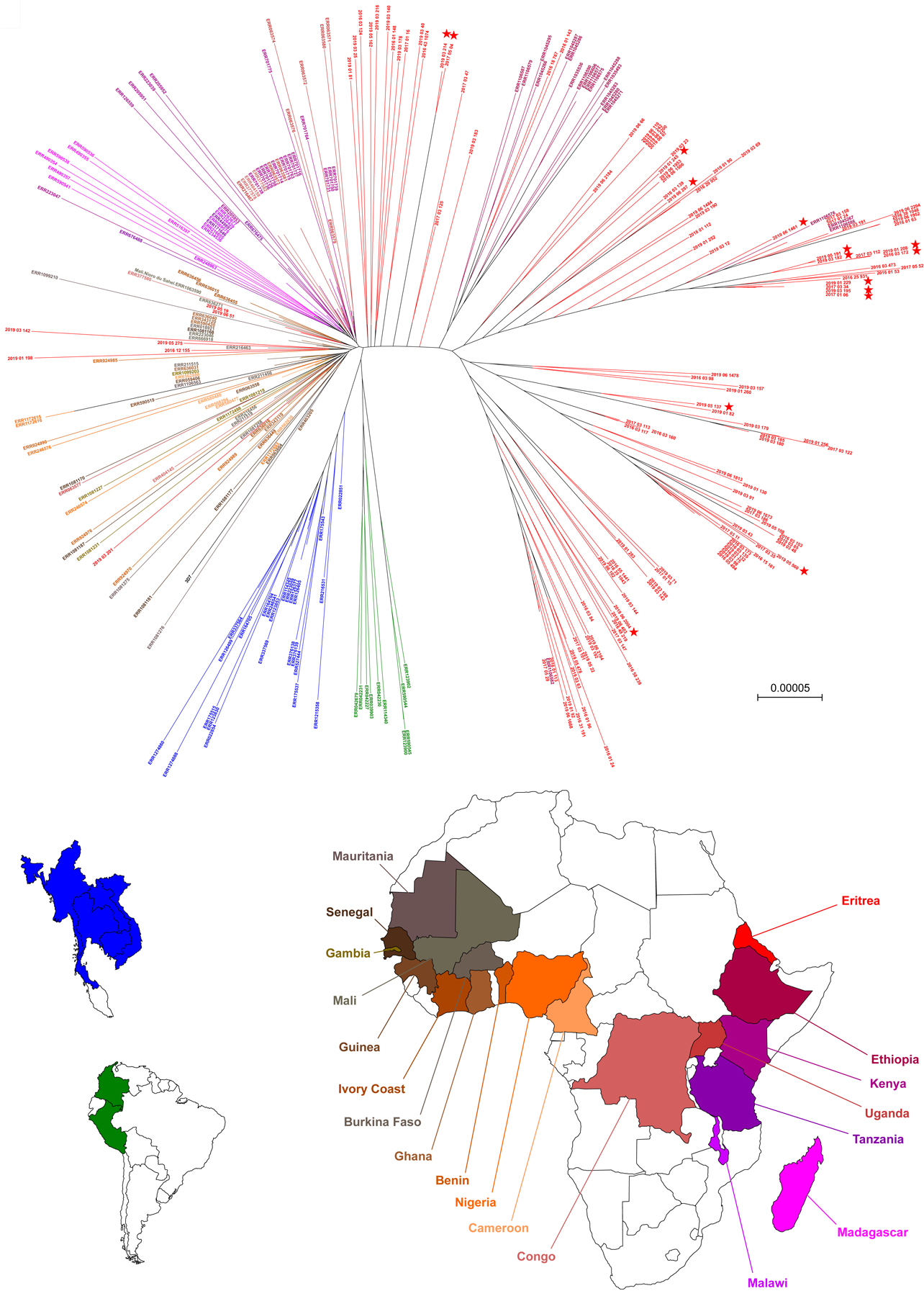
This maximum-likelihood tree is based on 128 P. falciparum Eritrean isolates (red), together with 162 isolates collected worldwide (Africa, Asia and South America) and the 3D7 reference genome from Africa. Labels of Eritrean isolates include the year of collection. The non-Eritrean isolates were sourced from the MalariaGEN P. falciparum Community Project (https://www.malariagen.net/apps/pf/4.0) and are labelled with their accession identifier. Each leaf represents one sample and is colored according to the country of collection (bottom). Eritrean parasites carrying the Pfkelch13 622I mutation are identified by filled red stars at the tip. Eritrean Pfkelch13 622I mutants are closely related to Pfkelch13 wild type parasites originating from East African countries. Scale bar, 0.00005 nucleotide substitutions per character.
We then explored haplotype diversity in the genomic regions flanking the R622I mutation. A PCoA based on a pairwise genetic distance matrix indicated a shared genetic background between Eritrean 622I mutants and wild-type isolates (Fig. S2–S3). Haplotype similarity in a ~300 kb region around the mutation pointed to a shared ancestry among mutants found across different sites (Fig. S4). In the absence of accurate estimates of recombination rates in populations of P. falciparum, the age of the R622I mutation could not be properly assessed. Nevertheless, the limited segment of haplotype homozygosity, as well as the lack of space/time structure in the distribution of haplotypes, do not suggest a recent clonal expansion of the Pfkelch13 R622I mutation (Fig. S5–S6, Table S10).
Genetic backgrounds of Eritrean Pfkelch13 622I variants.
We investigated the genetic background of Eritrean Pfkelch13 622I mutants by profiling both mutant and wild-type parasites at known antimalarial drug resistance loci, and by measuring the frequency of hrp2 and hrp3 gene deletions, a genomic feature previously observed in Eritrean P. falciparum (Table S11).23
We assessed 67 Pfkelch13 622I and 311 wild-type parasite samples for mutations in four genes. Differences in the proportions were observed in the pfcrt and dhfr genes, whose variants can confer resistance to chloroquine/piperaquine and pyrimethamine, respectively.24 Most of the Pfkelch13 622I mutants carried PfCRT M74I/N75E/K76T mutations (present in 92.5% of the 622I mutants vs. 66.9% of Pfkelch13 wild-type parasites), and N51I/S108N DHFR mutations (74.6% of mutants vs. 49.5% of wild-type parasites). We evaluated 29 Pfkelch13 622I and 139 wild-type parasites for amplification of plasmepsin2 or multidrug resistance-1, as these are considered markers of reduced susceptibility to piperaquine and lumefantrine/mefloquine, respectively.24 No parasites had pfmdr-1 amplification, and the proportion of isolates harboring ≥ 2 copies of plasmepsin-2 was similar in both mutant (31.0%) and wild-type (32.4%) parasites.
We also tested for deletions in the hrp2 and hrp3 genes amongst 65 Pfkelch13 622I mutant and 280 wild-type parasites. The majority (69.2%, 45/65) of Pfkelch13 622I parasites had an hrp3 deletion (vs. 22.5%, 63/280 in wild-type parasites). More worryingly, we detected a substantial proportion of 622I mutant parasites with both hrp2 and hrp3 deletions (16.9%, 9/65 vs. 21.8%, 61/280 for wild-type parasites), potentially threatening the efficacy of HRP2-based RDTs. No Pfkelch13 622I mutant parasites had only hrp-2 gene deletions (vs. 5.7%, 16/280 for wild-type parasites).
Discussion
P. falciparum ART-R is now firmly established in Africa. While to date ART-R has only been confirmed in Central (Rwanda) and East (Uganda) Africa,10–12 here we provide evidence of an additional hotspot of ART-R in the Horn of Africa. More worryingly, the emergence and spread of a novel Pfkelch13 622I mutant lineage was accompanied by deletions in the hrp2/hrp3 genes in a substantial proportion of parasites (16.9%), rendering these parasites likely undetectable by HRP2-based RDTs.
In vivo ART-R in Eritrea was evidenced by a substantial increase over time in the proportion of D3+ patients following ACT treatment (from 0.4% in 2016 to 4.2% in 2019). We also witnessed a substantial rise in the proportion of Pfkelch13 622I mutant parasites (from 8.6% in 2016 to 21.0% in 2019). We assessed that this mutation, which has not been observed previously in Southeast Asia, confers in vitro ART-R in the African parasite strain NF54, at slightly lower levels than the C580Y mutation that predominates in Southeast Asia (mean RSA0–3h survival rate of 3.3% vs. 4.3%, respectively). The R622I mutation conferred only low-level survival (1.5%) in Dd2 parasites (an Asian reference strain), consistent with prior evidence that Pfkelch13 mutations do not afford resistance across all strains, and that ART-R levels can be substantially modulated by the parasite genetic background.16 We also documented significant intra-host selection of the Pfkelch13 622I mutation in recrudescent infections following ASAQ treatment (from 12% at day0 to 53% at day28, a 4.4-fold increase). These findings suggest the Pfkelch13 622I mutation as a molecular marker of ART-R.
Eritrean Pfkelch13 622I mutants were phylogenetically closely related to other African parasites, clustering with both Eritrean and Ethiopian wild-type isolates. Hallmarks of the spread of a newly arisen mutation would be expected to include extended haplotype homozygosity in the genomic region flanking the Pfkelch13 622I mutation. By contrast, we observed a limited identity of core haplotypes among mutants. This might reflect the spread of a pre-existing Pfkelch13 resistance allele in P. falciparum populations from the Horn of Africa. This finding is supported by previous reports of low frequency detection of the Pfkelch13 622I variant in Eritrea and neighbouring countries (0.8% in Eritrea in 2013–14, 2.4% in Ethiopia in 2013–14, 0.7% in Somalia in 2016–17, and 0.3% in Zambia in 2012)25–28, and in Chinese travellers returning from Mozambique or Somalia (2016–18).29
Although recent data on treatment efficacy at sites with high prevalence of Pfkelch13 mutants are limited in Eritrea,30–32 we observed high cure rates with both ASAQ and AL for uncomplicated falciparum malaria (> 94%), presumably attributable to the continued efficacy of the partner drugs amodiaquine and lumefantrine. These data are concordant with recent reports in Rwanda for AL.12 Lower efficacy rates for AL (<90%), recently reported in Angola,33 the Democratic Republic of the Congo34 and East central Uganda35, remain highly questionable as the methodological deviation from WHO standard genotyping protocol might have potentially underestimated the efficacy of AL.
Our study also reports that a substantial proportion (16.9%) of Pfkelch13 622I mutant parasites had deletions in both the hrp2 and hrp3 genes (a similar proportion to Pfkelch13 wild-type parasites), potentially resulting in false-negative results in HRP2-based RDTs, which requires formal assessment.19 This genomic trait, frequently observed in Eritrea36 with prevalences ranging from 7% in Shambuko up to 81% in Ghindae23 resulted in a policy switch to pLDH-based RDTs in 2016. This emphasizes the need to conduct further research to evaluate the performance of pLDH-based RDTs for detecting Pfkelch13 622I mutant with dual hrp2/3 deletions.
Over the past two decades, Eritrea has achieved substantial reductions in malaria morbidity and mortality through active government engagement and effective implementation of Insecticide-Treated Nets (ITNs), Indoor Residual Spraying (IRS), larvicidal activities and malaria case management.37,38 However, decreased malaria prevalence may in turn have favored the emergence and spread of ART-R by reducing parasite genetic diversity and naturally acquired immunity, and increasing per-patient drug pressure as fewer infections are naturally cleared and therefore require treatment.39 These data suggest that in settings where strategies to reduce malaria transmission are efficiently implemented, surveillance of the emergence and spread of drug resistance should be prioritized.
Our findings find P. falciparum ART-R along with hrp2/hrp3 gene deletions in parasite populations from Eritrea. Strategies to contain the spread of these lineages across the Horn of Africa are needed, as the potential occurrence of partner drug resistance could lead to increased treatment failure rates and uncontrolled expansion of P. falciparum hrp2/hrp3-deleted parasites out of this region.
Supplementary Material
Acknowledgments
We thank all patients who contributed samples and their guardians in the communities of Akordat, Ghindae, Guluj, Shambuko and Tokombia, and all team members in the health centers. We are also grateful to the members of the national Malaria Control Programme in Eritrea for their support. We acknowledge the help HPC Core Facility of the Institut Pasteur for this work as well as the Biomics Platform, C2RT, Institut Pasteur, Paris, France (supported by France Génomique, ANR-10-INBS-09 and IBISA). We are grateful to ICAReB team of the CTS (Center for Translational Science) and CRBIP (BioResource Center of Institut Pasteur) for providing blood samples from healthy volunteers to Hélène Laude, Laurence Arowas, Ayla Zayoud and Marie Noelle Ungeheuer for managing the participants’ visits, to the healthy volunteers for their participation in the study, and to Emmanuel Roux, Alain Li, Dorian Cheval, Sophie Vacant, Sophie Chaouche, Elsa Lievin for preparing the blood samples from donors. We thank Aurélie Claës (Biology of Host-Parasite Interactions Unit, INSERM U1201, Institut Pasteur, Paris, France) for providing advice on P. falciparum genome editing.
Funding
Funding was obtained from the Bill and Melinda Gates Foundation through the World Health Organization (grant no. OPP1209843). The study was also supported by the Institut Pasteur, Paris, the French Government (Agence Nationale de la Recherche), Laboratoire d’Excellence (LabEx) “French Parasitology Alliance for Health Care” (ANR-11-15 LABX-0024-PARAFRAP), and the University of Strasbourg through the Programme IdEX 2022 to DM. DAF gratefully acknowledges funding from the US National Institutes of Health (R01 AI109023).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Contributor Information
Selam Mihreteab, National Malaria Control Program, Ministry of Health, Asmara, Eritrea.
Lucien Platon, Institut Pasteur, Université Paris Cité, Malaria Genetic and Resistance Unit, INSERM U1201, F-75015 Paris, France; Institut Pasteur, Université Paris Cité, Malaria Parasite Biology and Vaccines, F-75015 Paris; Sorbonne Université, Collège doctoral ED 515 Complexité du Vivant, F-75015 Paris, France.
Araia Berhane, Communicable Diseases Control Division, Ministry of Health, Asmara, Eritrea
Barbara H. Stokes, Columbia University Irving Medical Center, Department of Microbiology & Immunology, New York, NY 10032, USA
Marian Warsame, Gothenburg University, School of Public Health and Social Medicine, Gothenburg, Sweden
Pascal Campagne, Institut Pasteur, Université Paris Cité, Bioinformatics and Biostatistics Hub, F-75015 Paris, France
Alexis Criscuolo, Institut Pasteur, Université Paris Cité, Bioinformatics and Biostatistics Hub, F-75015 Paris, France
Laurence Ma, Institut Pasteur, Biomics Platform, C2RT, F-75015 Paris, France
Nathalie Petiot, Institut Pasteur, Université Paris Cité, Malaria Genetic and Resistance Unit, INSERM U1201, F-75015 Paris, France
Cécile Doderer-Lang, Université de Strasbourg, Institute of Parasitology and Tropical Diseases, UR7292 Dynamics of Host-Pathogen Interactions, F-67000 Strasbourg, France
Eric Legrand, Institut Pasteur, Université Paris Cité, Malaria Genetic and Resistance Unit, INSERM U1201, F-75015 Paris, France Institut Pasteur, Université Paris Cité, Malaria Parasite Biology and Vaccines, F-75015 Paris.
Kurt E. Ward, Columbia University Irving Medical Center, Department of Microbiology & Immunology, New York, NY 10032, USA
Assefash Zehaie Kassahun, World Health Organization office, Asmara, Eritrea
Pascal Ringwald, Global Malaria Programme, World Health Organization, Geneva, Switzerland
David A. Fidock, Columbia University Irving Medical Center, Department of Microbiology & Immunology, New York, NY 10032, USA Columbia University Irving Medical Center, Center for Malaria Therapeutics and Antimicrobial Resistance. Division of Infectious Diseases. Department of Medicine, New York, NY 10032, USA.
Didier Ménard, Institut Pasteur, Université Paris Cité, Malaria Genetic and Resistance Unit, INSERM U1201, F-75015 Paris, France Institut Pasteur, Université Paris Cité, Malaria Parasite Biology and Vaccines, F-75015 Paris; Université de Strasbourg, Institute of Parasitology and Tropical Diseases, UR7292 Dynamics of Host-Pathogen Interactions, F-67000 Strasbourg, France; CHU Strasbourg, Laboratory of Parasitology and Medical Mycology, F-67000 Strasbourg, France.
References
- 1.World Health Organization. Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019).2020.
- 2.Ashley EA, Dhorda M, Fairhurst RM, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2014;371:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2009;361:455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noedl H, Socheat D, Satimai W. Artemisinin-resistant malaria in Asia. N Engl J Med 2009;361:540–1. [DOI] [PubMed] [Google Scholar]
- 5.Imwong M, Suwannasin K, Srisutham S, et al. Evolution of Multidrug Resistance in Plasmodium falciparum: a Longitudinal Study of Genetic Resistance Markers in the Greater Mekong Subregion. Antimicrob Agents Chemother 2021;65:e0112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacob CG, Thuy-Nhien N, Mayxay M, et al. Genetic surveillance in the Greater Mekong subregion and South Asia to support malaria control and elimination. Elife 2021;10: e62997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Pluijm RW, Imwong M, Chau NH, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis 2019;19:952–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assefa DG, Yismaw G, Makonnen E. Efficacy of dihydroartemisinin-piperaquine versus artemether-lumefantrine for the treatment of uncomplicated Plasmodium falciparum malaria among children in Africa: a systematic review and meta-analysis of randomized control trials. Malar J 2021;20:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asua V, Conrad MD, Aydemir O, et al. Changing Prevalence of Potential Mediators of Aminoquinoline, Antifolate, and Artemisinin Resistance Across Uganda. J Infect Dis 2021;223:985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balikagala B, Fukuda N, Ikeda M, et al. Evidence of Artemisinin-Resistant Malaria in Africa. N Engl J Med 2021;385:1163–71. [DOI] [PubMed] [Google Scholar]
- 11.Uwimana A, Legrand E, Stokes BH, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 2020;26:1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uwimana A, Umulisa N, Venkatesan M, et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis 2021;21:1120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. World malaria report 2022. 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022
- 14.Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014;505:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straimer J, Gnadig NF, Witkowski B, et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 2015;347:428–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stokes BH, Dhingra SK, Rubiano K, et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. Elife 2021;10: e66277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization. Informal consultation on methodology to distinguish reinfection from recrudescence in high malaria transmission areas: report of a virtual meeting, 17–18 May 2021. 2021.
- 18.Menard D, Dondorp A. Antimalarial Drug Resistance: A Threat to Malaria Elimination. Cold Spring Harb Perspect Med 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. False-negative RDT results and implications of new reports of P. falciparum histidine-rich protein 2/3 gene deletions. 2017. [Google Scholar]
- 20.Oyola SO, Ariani CV, Hamilton WL, et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar J 2016;15:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witkowski B, Amaratunga C, Khim N, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 2013;13:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization. Artemisinin and artemisinin-based combination therapy resistance2016.
- 23.Mihreteab S, Anderson K, Pasay C, et al. Epidemiology of mutant Plasmodium falciparum parasites lacking histidine-rich protein 2/3 genes in Eritrea 2 years after switching from HRP2-based RDTs. Sci Rep 2021;11:21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wicht KJ, Mok S, Fidock DA. Molecular Mechanisms of Drug Resistance in Plasmodium falciparum Malaria. Annu Rev Microbiol 2020;74:431–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayih AG, Getnet G, Alemu A, Getie S, Mohon AN, Pillai DR. A unique Plasmodium falciparum K13 Gene Mutation in Northwest Ethiopia. Am J Trop Med Hyg 2016;94:132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.L’Episcopia M, Kelley J, Patel D, et al. Targeted deep amplicon sequencing of kelch 13 and cytochrome b in Plasmodium falciparum isolates from an endemic African country using the Malaria Resistance Surveillance (MaRS) protocol. Parasit Vectors 2020;13:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menard D, Khim N, Beghain J, et al. A Worldwide Map of Plasmodium falciparum K13-Propeller Polymorphisms. N Engl J Med 2016;374:2453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warsame M, Hassan AM, Hassan AH, et al. High therapeutic efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for the treatment of uncomplicated falciparum malaria in Somalia. Malar J 2019;18:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Ruan W, Zhou S, et al. Molecular surveillance of Pfcrt and k13 propeller polymorphisms of imported Plasmodium falciparum cases to Zhejiang Province, China between 2016 and 2018. Malar J 2020;19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohammed AO, Tewolde S, Estifanos D, Tekeste Y, Osman MH. Therapeutic efficacy of artesunate - amiodaquine for treating uncomplicated falciparum malaria at Ghindae Zonal Referral Hospital, Eritrea. Acta Trop 2018;177:94–6. [DOI] [PubMed] [Google Scholar]
- 31.Ghebremeskel T Therapeutic efficacy of sulfadoxine/pyrimethamine plus chloroquine and artesunate plus amodiaquine for the treatment of uncomplicated flaciparum malaria. J Eritrean Med Assoc 2007; 2, 14–16. [Google Scholar]
- 32.Mihreteab M, Berhane A, Berhane D, Zehaie A, Araia A, Banteyrga L. Therapeutic efficacy study on artesunate + Amodiaquine (AS + AQ) for the treatment of uncomplicated Plasmodium falciparum (Pf) malaria in Eritrea. J Eritrean Med Assoc 2014;1, 9–12. [Google Scholar]
- 33.Dimbu PR, Horth R, Candido ALM, et al. Continued Low Efficacy of Artemether-Lumefantrine in Angola in 2019. Antimicrob Agents Chemother 2021;65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriarty LF, Nkoli PM, Likwela JL, et al. Therapeutic Efficacy of Artemisinin-Based Combination Therapies in Democratic Republic of the Congo and Investigation of Molecular Markers of Antimalarial Resistance. Am J Trop Med Hyg 2021;105:1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebong C, Sserwanga A, Namuganga JF, et al. Efficacy and safety of artemether-lumefantrine and dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria and prevalence of molecular markers associated with artemisinin and partner drug resistance in Uganda. Malar J 2021;20:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berhane A, Anderson K, Mihreteab S, et al. Major Threat to Malaria Control Programs by Plasmodium falciparum Lacking Histidine-Rich Protein 2, Eritrea. Emerg Infect Dis 2018;24:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barat LM. Four malaria success stories: how malaria burden was successfully reduced in Brazil, Eritrea, India, and Vietnam. Am J Trop Med Hyg 2006;74:12–6. [PubMed] [Google Scholar]
- 38.Nyarango PM, Gebremeskel T, Mebrahtu G, et al. A steep decline of malaria morbidity and mortality trends in Eritrea between 2000 and 2004: the effect of combination of control methods. Malar J 2006;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott N, Ataide R, Wilson DP, et al. Implications of population-level immunity for the emergence of artemisinin-resistant malaria: a mathematical model. Malar J 2018;17:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


