Abstract
Background
Exosomes (Exos) can safely and effectively deliver therapeutic substances to glioma cells; however, their blood–brain barrier (BBB) crossing capacity remains limited. Focused ultrasound (FUS) can transiently, reversibly, and locally open the BBB, while the effects of FUS combined with Exos-miRNA on the treatment of glioma have not been explored to date.
Methods
Exos were extracted by differential centrifugation and the efficacy of miR-1208-loaded Exos combined with FUS in the treatment of glioma was detected by CCK-8, colony formation, flow cytometry, transwell and tumour xenografts assays. The METTL3-mediated regulation of IGF2BP2 on mRNA stability of NUP214 was determined by MeRIP-qPCR, half-life and RIP assays.
Results
We used Exos secreted by mesenchymal stem cells as carriers for the tumour suppressor gene miR-1208, and following FUS irradiation, more Exos carrying miR-1208 were allowed to pass through the BBB, and the uptake of miR-1208 in Exos by glioma cells was promoted, thereby achieving high-efficiency tumour-suppressive effects. Furthermore, the molecular mechanism underlying this effect was elucidated that miR-1208 downregulated the m6A methylation level of NUP214 mRNA by negatively regulating the expression of METTL3, thereby NUP214 expression and TGF-β pathway activity were suppressed.
Conclusions
MiR-1208-loaded Exos combined with FUS is expected to become an effective glioma treatment and deserves further clinical evaluation.
Subject terms: CNS cancer, Oncogenes
Background
Glioma is the most common central nervous system (CNS) malignancy with a high recurrence rate and poor prognosis [1]. Despite aggressive surgical treatment and the development of assisted adjuvant radiotherapy, chemotherapy, and immunotherapy, the prognosis of glioma patients remains poor, with a 5-year survival rate of only 3–5% and an average survival period of approximately 1 year [2, 3]. Therefore, the exploration of precise and effective treatment strategies for glioma has become a research hotspot in recent years.
The blood–brain barrier (BBB) effectively prevents harmful substances in the blood from entering the brain and maintains the stability of the internal environment of the CNS. However, the BBB also prevents the entry of drugs or tumour suppressor genes that have therapeutic effects on CNS diseases, which severely hinders treatment efficiency [4, 5]. Therefore, safe and effective transport of therapeutic substances across the BBB to tumour cells to exert an efficient therapeutic effect is the key to the treatment of glioma.
Exosomes (Exos) can encapsulate complex biomolecules, such as proteins, nucleic acids, lipids, and sugars, to form endogenous transport systems that allow cellular exchange [6]. Compared with traditional exogenous therapeutic carriers such as modified viruses and chemically synthesised nanoparticles, Exos, as endogenous nanocarriers, have the advantages of low toxicity, low immunogenicity, good biocompatibility, and the ability to pass through physiological and pathological barriers [7, 8]. Exos have attracted extensive attention in recent years. Mesenchymal stem cells (MSCs), characterised by immunosuppression and immune regulation functions, have a strong ability to secrete Exos and are an ideal cellular source for Exos [9]. Exos can be loaded with specific substances from the source cell and then transfer the contents to the recipient cell [10]. Recent studies have proved that MSC-derived Exos modified with tumour suppressor genes can exert suppressive effects on malignant biological behaviours of glioma cells [11, 12]. Although Exos have the intrinsic ability to cross biological barriers, their efficiency in delivering therapeutic substances across the BBB into tumours remains limited [13]. Focused ultrasound (FUS) is a non-invasive treatment that has been shown to transiently, reversibly, and locally access the BBB. Simultaneously, FUS can enhance the targeted delivery of therapeutic substances by focusing on the exposed area to promote their accumulation in the target area [14, 15]. For instance, Lin et al. [14] reported that FUS could induce BBB open of Parkinson’s disease mouse model and allow targeted delivery of therapeutic substances into the brain, then retard the progression of motor-related behavioural abnormalities. Shen et al. [15] proved that FUS in conjunction with microbubbles (MBs) increased the BBB permeability and drug concentration of glioblastoma in orthotopic tumour-bearing nude mice, then significantly prolonged the survival time of nude mice. Thereby FUS provides new opportunities for the treatment of CNS diseases and shows great clinical application prospects.
N6-methylated adenine (m6A) is the most common and abundant reversible epigenetic modification on eukaryotic mRNA; it has been reported to play an important regulatory role in the occurrence and malignant progression of glioma [16]. Methyltransferase-like 3 (METTL3) is an important “encoder” in the methyltransferase complex. Recent studies have reported that METTL3 is dysregulated in a variety of tumours and plays an important role in cancer by regulating the m6A methylation level of target gene mRNA [17]. In glioma, METTL3 was found to be highly expressed and exert oncogenic effects [18, 19]. MicroRNAs can specifically bind to the 3’ untranslated regions (3’UTR) of downstream mRNAs of target proteins, thereby inhibiting their expression at the post-transcriptional level [20]. MiR-1208 exerts a tumour-suppressive effect in malignant tumours such as non-small cell lung cancer, renal cancer, and liver cancer, but its expression and function in glioma remain unclear [21–23].
This study intends to use the Exos secreted by MSCs as the carrier carrying the tumour suppressor gene miR-1208, explore an effective strategy for the treatment of glioma by combined application of FUS, further clarify the regulatory effect and molecular mechanism of miR-1208/METTL3/NUP214/TGF-β pathway on the biological behaviours of glioma cells, and provide novel targets and ideas for the treatment of glioma.
Methods
Clinical specimens
A total of 8 human normal brain tissues (NBTs) and 56 glioma tissue specimens were used in this study. All specimens were collected at the Department of Neurosurgery, Shengjing Hospital of China Medical University. For more details, please see Supplementary Materials and methods.
Cell transfection
The miR-1208 overexpression and its negative control lentivirus were purchased from GenePharma (Shanghai, China). The METTL3 overexpression and its negative control plasmids were constructed by GenScript (Piscataway, NJ, USA). The short-hairpin RNA (shRNA) against METTL3, NUP214 and IGF2BP2 (every gene designed three target sequences) and their negative controls were purchased from GenePharma. The shRNA template sequence is provided in Supplementary Table S1. In pre-experiments, sh-METTL3-1, sh-NUP214-2, and sh-IGF2BP2-1 were the target sequences with the highest knockdown efficiency screened by western blot after 48 h of transient transfection (results showed in Supplementary Fig. S6), then the stable transfection was conducted with the above plasmids. For more details, please see Supplementary Materials and methods.
Separation, extraction, and identification of Exos
Exos were isolated from the supernatant of human bone marrow mesenchymal stem cells (hBMSCs) using density gradient centrifugation and performed as described previously [3]. The characterisation of Exos was proven through western blot assay, transmission electron microscopy and NTA analysis. Details of the experiment can be found in Supplementary Materials and methods.
Experiments on Exos uptake by cells
MiR-1208-Cy3 mimics and their negative controls were purchased from GenePharma, and hBMSCs with good growth status and a confluence of 70–80% were transfected using lipofectamine 3000 reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. After 6 h of transfection, the medium was replaced with fresh Exos-free serum medium, and the cells were continuously cultured for 48 h, following which the cell supernatant was collected to extract Exos. The extracted Exos were labelled using PKH67 (Sigma-Aldrich, USA) according to the manufacturer’s instructions and sterilised through a 0.22 µm filter. The labelled Exos were co-cultured for 24 h with U251 and U373 cells pre-plated on cell culture slides. The ultrasonic equipment used in the experiment was a low-power FUS experimental device developed by Ronghai Ultrasonic Medical Engineering Research Center. The ultrasonic parameters were set at a frequency of 650 KHz, an intensity of 0.5 W, and an irradiation time of 30 s based on pre-experiment results shown in Supplementary Fig. S1. After co-cultivation, cells were fixed, and their nuclei were stained using DAPI. Finally, cells were observed and photographed using a laser confocal microscope. The whole experiment should be conducted under dark conditions.
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted from clinically obtained tissue samples, normal human astrocytes (NHA), U251 and U373 cell lines using the Trizol reagent, and RNA in Exos was extracted using the Trizol LS reagent (Thermo Fisher Scientific, USA). Real-time quantitative PCR analysis was performed using the TB Green Fluorescence Quantitative PCR Kit of TaKaRa Company. The primers are exhibited in Supplementary Table S2. For more details, please see Supplementary Materials and methods.
Western blot analysis
Western blot assay was conducted as our previous research described [2, 3]. For more information and details of the experiment, please see Supplementary Materials and methods.
Cell counting kit-8 (CCK-8) assay
The CCK-8 kit (Beyotime, China) was used to detect the cell viability and performed as described previously [2]. For more details of the experiment, please see Supplementary Materials and methods.
Clone formation assay
Clone formation assay was conducted as our previous research described [3]. For more details of the experiment, please see Supplementary Materials and methods.
Cell migration and invasion experiments
In vitro migration and invasion abilities of cells were detected using Transwell chambers (Costar, Corning, USA) and conducted as our previous research described [2, 3]. For more details of the experiment, please see Supplementary Materials and methods.
Apoptosis assay
Cells in each experimental group were collected and double-stained with Annexin V-PE/7-AAD using the Annexin V-PE/7-AAD double-staining apoptosis kit (BD biosciences, USA) according to the manufacturer’s instructions. Sample detection was performed using a flow cytometer, and experimental data were analysed.
Immunohistochemical (IHC) experiments
IHC experiments were conducted as our previous research described [3]. For more details of the experiment, please see Supplementary Materials and methods.
Dual-luciferase reporter gene assay
Dual-luciferase reporter gene assay was carried out as previously described [2, 3]. Details of the experiment can be found in Supplementary Materials and methods.
RNA-binding protein immunoprecipitation assay (RIP)
The RIP experiment was performed using the EZ-Magna RIP kit (Millipore, USA) according to the manufacturer’s instructions. Details of the experiment can be found in Supplementary Materials and methods
RNA methylation co-immunoprecipitation (MeRIP)
The MeRIP Kit (BersinBio, China) was used to carry out the experiment according to the manufacturer’s instructions. For more details of the experiment, please see Supplementary Materials and methods.
RNA stability detection
RNA stability assays were performed using actinomycin D according to the manufacturer’s instructions. Details of the experiment can be found in Supplementary Materials and methods.
Nascent RNA capture assay
Nascent RNA capture assay was carried out as previously described [2, 3]. For more details of the experiment, please see Supplementary Materials and methods.
In vivo experiment on Exos combined with FUS on anti-tumour effect in nude mice
Female BALB/c nude mice (6 weeks old) were purchased from Vital River Laboratory Animal Technology (China) and housed in SPF-grade animal facility. All animal experiments were approved by the Laboratory Animal Management Committee of Shengjing Hospital of China Medical University. For more details of the experiment, please see Supplementary Materials and methods.
Xenograft experiment in nude mouse
The Xenograft subcutaneous and orthotopic tumour experiments were performed as previously described [2, 3]. For more information on the experiments, please see Supplementary Materials and methods.
Statistical methods
All experiments were repeated three times independently, and the data were expressed as mean ± standard deviation. The statistical software SPSS 22.0 was used to analyse and compare whether there were differences between groups. The t-test was used to compare the data between the two groups. One-way analysis of variance was used for statistical comparison of three or more groups, and statistical significance was set at P < 0.05.
Results
miR-1208 is low-expression in glioma, and it plays a role in cancer suppression
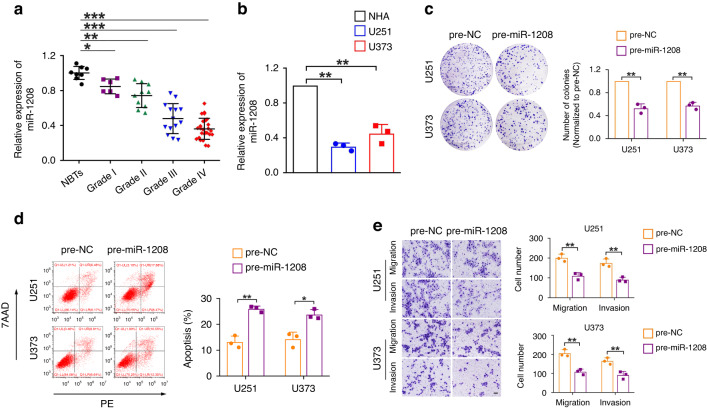
The expression levels of miR-1208 were detected using qRT-PCR in NBTs and glioma tissues collected clinically, NHA and U251 and U373 glioma cell lines. As shown in Fig. 1a, b, compared with that in NBTs, the expression level of miR-1208 in glioma tissues was significantly downregulated, and it decreased with the increase of glioma pathological grade; compared with those in NHA cells, the expression levels of miR-1208 in U251 and U373 glioma cell lines were also significantly downregulated. To further clarify the function of miR-1208 in glioma, we overexpressed miR-1208 in U251 and U373 glioma cell lines and investigated the biological behaviours. The results of the clone formation assay showed that overexpression of miR-1208 significantly inhibited the proliferation ability of glioma cells compared with that in the pre-NC group (Fig. 1c). The results of flow cytometry revealed that overexpression of miR-1208 significantly promoted the apoptosis of glioma cells (Fig. 1d). The results of Transwell experiments suggested that compared with those of the pre-NC group, the migration and invasion abilities of glioma cells were significantly decreased after the expression of miR-1208 was upregulated (Fig. 1e). The above experimental results indicate that miR-1208 is low-expression in glioma and acts as a tumour suppressor gene and that the overexpression of miR-1208 can significantly inhibit the malignant biological behaviours of glioma cells.
Fig. 1. miR-1208 exerts as a tumour suppressor gene in glioma.
a The expression levels of miR-1208 were downregulated in glioma tissues. Data are presented as the mean ± SD (n = 8, NBTs; n = 6, Grade I; n = 10, Grade II; n = 14, Grade III; n = 26, Grade IV). *P < 0.05, **P < 0.01, ***P < 0.001. b The expression levels of miR-1208 were detected in NHA, U251 and U373 cells. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01. c–e Colony formation assay, flow cytometry analysis and migration and invasion assays were used to detect the impacts of miR-1208 overexpression on biological behaviours of glioma cells. Data are presented as the mean ± SD (n = 3, each group). *P < 0.05, **P < 0.01. Scale bar of migration and invasion assays represents 40 μm.
Exos derived from hBMSCs can be used as a carrier carrying the tumour suppressor gene miR-1208
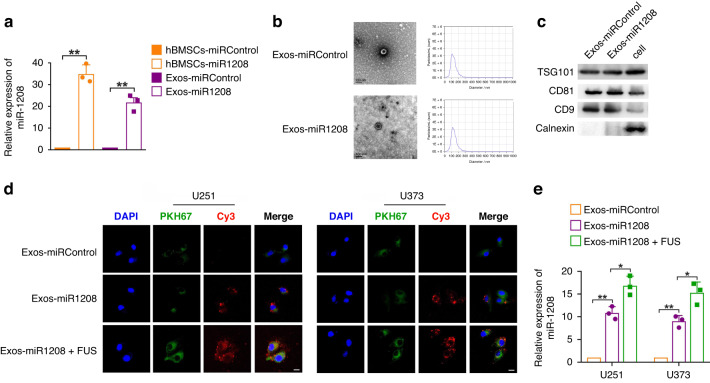
First, we established hBMSCs that stably overexpressed miR-1208. The results of qRT-PCR experiments showed that compared with the control group, the expression level of miR-1208 in the hBMSCs-miR-1208 group was significantly upregulated by approximately 35 times (Fig. 2a). Exos in the culture supernatant of hBMSCs overexpressing miR-1208 were isolated and extracted via differential centrifugation and identified. The results showed that Exos had the typical morphological structure of Exos, which were spherical vesicles with a double-layer membrane structure. The average particle size of Exos was about 130 nm, and Exos expressed Exo-specific marker proteins including TSG101, CD81 and CD9, lacking calnexin (Fig. 2b, c). The above results confirmed that the extracted vesicle structure was consistent with the definition of Exos. The expression level of miR-1208 in the extracted Exos was further detected by qRT-PCR, and the results showed that the expression level of miR-1208 in Exos extracted from the supernatant of the hBMSCs-miR-1208 group was upregulated by approximately 22 times compared with the control group (Fig. 2a). The above results indicate that overexpression of miR-1208 in hBMSCs can increase the expression of miR-1208 in Exos derived from hBMSCs, and Exos derived from hBMSCs can be used as a carrier carrying the tumour suppressor gene miR-1208.
Fig. 2. FUS promotes uptake of miR-1208 in Exos by glioma cells.
a qRT-PCR was used to detect the expression levels of miR-1208 in hBMSCs and hBMSCs-Exos after overexpression of miR-1208. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01. b Representative transmission electron micrograph of exosomes isolated from culture supernatants of hBMSCs (scale bar = 100 nm). Exosome concentration and size distribution were detected by ZetaView Particle analysis system. c Western blot analysis of the presence of TSG101, CD81, CD9 and the absence of calnexin in exosomes. d Representative images by confocal microscopy of the internalisation of PKH67-labelled exosomes and Cy3-labelled miR-1208 (green, PKH67; red, Cy3; blue, DAPI nuclear staining; scale bar = 10 μm). e qRT-PCR was used to detect the uptake of miR-1208 in Exos by glioma cells with or without FUS irradiation. Data are presented as the mean ± SD (n = 3, each group). *P < 0.05, **P < 0.01.
Cytotoxicity detection and ultrasound parameter selection for Exos in combination with FUS
The cell viability of U251 and U373 glioma cells co-cultured separately with different concentrations (5, 25, 50, and 100 μg/mL) of Exos-miRControl was detected using CCK-8 method. The results showed that cell viability was not affected, that is, Exos had no obvious toxic effect on cells (Supplementary Fig. S1a). Subsequently, the cell viability of U251 and U373 glioma cells separately irradiated with different ultrasonic parameters (0.5 W/15 s, 0.5 W/30 s, 0.5 W/45 s, 1.0 W/15 s, 1.0 W/30 s, and 1.0 W/45 s) was detected using CCK-8 method. The results demonstrated that with the increase in ultrasonic intensity and irradiation time, cell viability decreased (Supplementary Fig. S1b). Finally, we selected ultrasound parameters (0.5 W/15 s, 0.5 W/30 s, 1.0 W/15 s) with cell viability higher than 85%. The uptake of miR-1208 in Exos after co-culture of U251 and U373 glioma cells with Exos-miR-1208 under the above ultrasonic parameters was detected using qRT-PCR. As shown in Supplementary Fig. S1c, under the irradiation condition with ultrasound parameter of 0.5 W/30 s, glioma cells had the maximum uptake of miR-1208 in Exos. Therefore, this ultrasound parameter was chosen for subsequent experiments.
FUS promotes uptake of miR-1208 in Exos by glioma cells
We observed under confocal microscopy that after being co-cultured with Exos-miR-1208, U251 and U373 glioma cells had both green fluorescence signal PKH67 labelling Exos membrane structure and red fluorescence signal Cy3 labelling miR-1208, and the fluorescence signals of PKH67 and Cy3 were both obviously enhanced in the combined FUS irradiation group (Fig. 2d). Subsequently, qRT-PCR was used to detect the uptake of miR-1208 in Exos by U251 and U373 glioma cells with or without FUS irradiation. The results are shown in Fig. 2e. Compared with those in the non-FUS irradiation group, the uptake of miR-1208 by U251 and U373 glioma cells in the combined FUS group was increased. These results suggest that glioma cells can uptake miR-1208 in Exos, and FUS can promote the uptake of miR-1208 in Exos by glioma cells.
Exos-miR-1208 combined with FUS inhibits the malignant biological behaviours of glioma cells
Exos-miR-1208 and Exos-miRControl were respectively co-cultured with U251 and U373 glioma cells and irradiated under FUS, and the biological behaviours of U251 and U373 glioma cells were detected. The results of CCK-8 and Transwell experiments showed that Exos-miR-1208 inhibited the viability, migration and invasion of glioma cells, and the inhibitory effect was enhanced in combination with FUS. The results of flow cytometry suggested that Exos-miR-1208 significantly promoted the apoptosis of glioma cells and the apoptotic rate was further increased in combination with FUS (Supplementary Fig. S2a–c). The above experimental results indicate that Exos-miR-1208 inhibits the malignant biological behaviours of glioma cells, and the combined use of FUS further enhanced the inhibitory effect of Exos-miR-1208 on the malignant biological behaviours of glioma cells.
Exos-miR-1208 in combination with FUS inhibits the growth of orthotopic transplantation tumours in nude mice and prolongs the survival of tumour-bearing nude mice
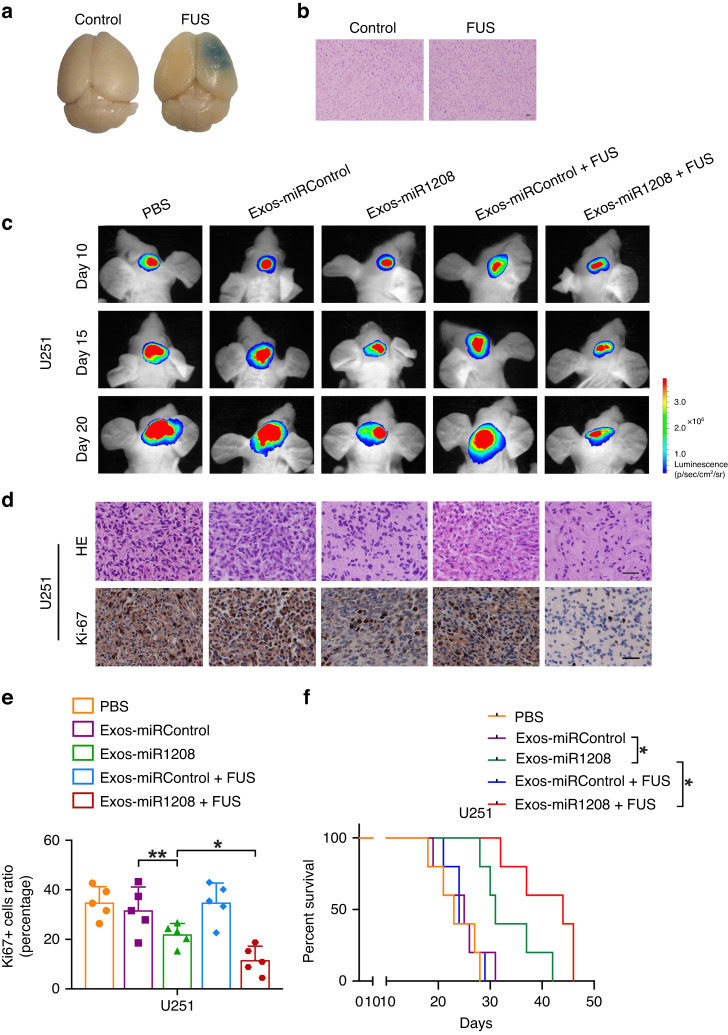
To clarify the effect and safety of FUS for the openness of the BBB, we used the EB extravasation experiment to detect the opening of the BBB in nude mice. As shown in Fig. 3a, obvious EB extravasation could be observed in the brain tissue after FUS irradiation in combination with MBs, confirming the opening of the BBB. Subsequently, results of HE staining suggested that no obvious erythrocyte extravasation or tissue damage was noticed in the brain tissue, indicating that opening the BBB was safe under FUS with chosen parameters (Fig. 3b).
Fig. 3. Exos-miR-1208 combined with FUS exerts tumour suppressor effects in vivo.
a EB extravasation experiment was used to examine the opening of the BBB in nude mice. b HE staining was used to evaluate the safety of BBB opening. Scale bar represents 50 μm. c In vivo bioluminescent imaging analysis of tumour growth in nude mice from respective groups was presented. Representative images on days 10, 15, and 20 post-implantation are shown. d HE staining of sections from glioma tissues of nude mice (scale bar = 50 μm). Representative images of IHC staining for Ki-67 in sections from the indicated nude mice (scale bar = 50 μm). e Quantification of IHC staining for Ki-67 in sections from indicated nude mice. Data are presented as the mean ± SD (n = 5, each group). *P < 0.05, **P < 0.01. f Survival curves from representative nude mice injected into the right striatum in orthotopic inoculations assay were exhibited. Log-rank test, *P < 0.05.
Next, we constructed luciferase-labelled U251 glioma cells and established an orthotopic tumour nude mice model for in vivo experiments. The growth of the tumour was dynamically observed by a small animal in vivo imaging system every five days. Finally, the survival analysis was performed, and the tumour tissue was subjected to HE staining and Ki-67 immunohistochemical experiments. As shown in Fig. 3c–f, injection of Exos-miR-1208 into nude mice significantly inhibited the size of intracranial tumours, the proliferation of cells, and prolonged the survival of nude mice. Moreover, the combination of Exos-miR-1208 and FUS further enhanced the anti-tumour effect, and the survival time of nude mice was longer. However, injection of PBS or Exos-miRControl, whether combined with FUS or not, had no inhibitory effect on the size of intracranial tumours and cell proliferation in nude mice; the glioma continued to grow, and the survival period of nude mice was short. Collectively, the experimental results showed that Exos-miR-1208 can cross the BBB to inhibit the tumour growth of orthotopic xenograft tumours in nude mice, prolong the survival time of tumour-bearing nude mice, and this anti-tumour effect was enhanced by combination with FUS.
Biosafety evaluation of Exos-miR-1208 combined with FUS
HE staining results showed that compared with those of the control group, the heart, liver, spleen, lung and kidney tissues of healthy nude mice treated with Exos-miR-1208 in combination with FUS demonstrated complete structure, neatly arranged cells, and no obvious abnormality (Supplementary Fig. S3a). Moreover, the general condition of the two groups of nude mice was good, and there was no significant difference in body weight change (Supplementary Fig. S3b). These results indicate that the combination of Exos-miR-1208 and FUS has no significant harmful effects on healthy nude mice and is relatively safe.
MiR-1208 targets the 3’UTR of METTL3 mRNA to inhibit the malignant biological behaviours of glioma cells
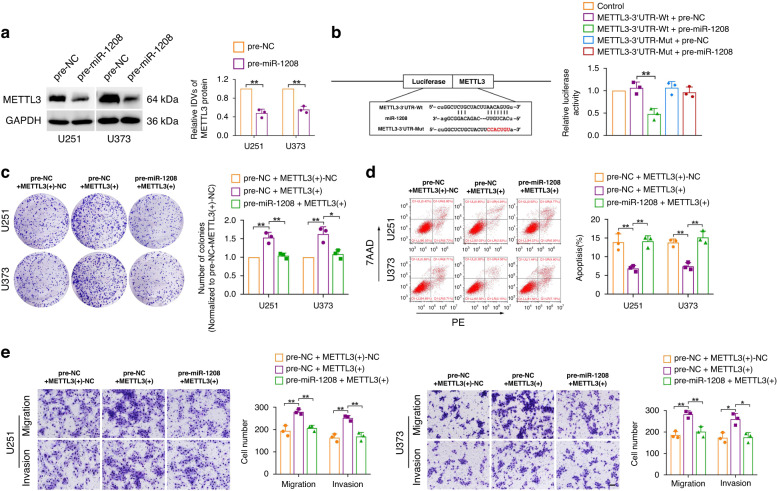
METTL3 was found to be a potential downstream target gene of miR-1208 by using bioinformatics software (TargetScan). The results of western blot showed that overexpression of miR-1208 significantly inhibited the expression of METTL3 in U251 and U373 glioma cells (Fig. 4a). The targeted binding between miR-1208 and METTL3 was further detected using the dual-luciferase reporter gene assay. The experimental results are shown in Fig. 4b. The luciferase activity was significantly decreased in the METTL3-3’-UTR-Wt+pre-miR-1208 group compared to that in the control group. However, the luciferase activity in the METTL3-3’-UTR-Mut+pre-miR-1208 group had no significant change. Finally, we respectively overexpressed miR-1208 and METTL3 in U251 and U373 glioma cells to examine their effects on the malignant biological behaviours of glioma cells. The results of clone formation assay, flow cytometry and Transwell assay suggested that overexpression of miR-1208 significantly reversed the promotion effect on glioma cell proliferation, migration, and invasion, as well as the inhibitory effect on apoptosis of glioma cell by overexpression of METTL3 alone (Fig. 4c–e). The above experimental results showed that miR-1208 can specifically bind to 3’UTR of METTL3 mRNA, and overexpression of miR-1208 can significantly inhibit METTL3 expression in glioma cells and reverse METTL3-mediated promotion of malignant biological behaviours in glioma cells.
Fig. 4. miR-1208 targets and negatively regulates METTL3 expression.
a The influence of overexpression of miR-1208 on METTL3 in glioma cells was detected by western blot. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01. b Dual-luciferase reporter assay verified the binding of METTL3 3’UTR with miR-1208. **P < 0.01. c–e Colony formation assay, flow cytometry analysis and migration and invasion assays were used to detect the impacts of co-transfection of METTL3 and miR-1208 on the biological behaviours of glioma cells. Data are presented as the mean ± SD (n = 3, each group). *P < 0.05, **P < 0.01. Scale bar of migration and invasion assays represents 40 μm.
NUP214 is a downstream target of METTL3 and plays a carcinogenic role in glioma
Recent studies have reported that silencing the expression of METTL3 in glioma stem cells reduces the level of NUP214 mRNA by as much as 5-fold [24]. The Cancer Genome Atlas (TCGA) data also showed a positive correlation between the expression of NUP214 and METTL3 in glioma dataset (Supplementary Fig. S4a). We found that silencing METTL3 significantly inhibited NUP214 expression in U251 and U373 glioma cells using western blot assay (Fig. 5a). These results confirmed that NUP214 was a downstream target of METTL3. NUP214 (Nucleoporin214) is an important member of the nuclear pore protein complex, which exerts essential biological functions by regulating the nucleus cytoplasm transport and distribution of specific proteins [25]. NUP214 has been found to promote cancer in a variety of cancers [26–28], but its expression and function in glioma have not been reported. TCGA data showed that the expression of NUP214 was upregulated in the glioma dataset (Supplementary Fig. S4b). Next, the results of western blot were shown in Supplementary Fig. S4c, d, NUP214 was highly expressed in glioma tissues and cells compared with that in NBTs and NHA cells, and its expression level was positively correlated with the pathological grade of glioma. The U251 and U373 glioma cells with silenced NUP214 expression were further established, and their biological behaviours were tested. The results showed that the downregulation of NUP214 expression significantly inhibited the proliferation, migration and invasion of glioma cells, and promoted cell apoptosis (Supplementary Fig. S4e–g). Finally, we simultaneously silenced the expression of NUP214 in METTL3 overexpressing U251 and U373 glioma cells and studied the effect on the malignant biological behaviours of glioma cells. The results revealed that silencing NUP214 reversed the promoting effects on the proliferation, migration, and invasion of glioma cells and the inhibitory effect on glioma cell apoptosis by METTL3 overexpression (Supplementary Fig. S5a‒c). The above results suggest that NUP214 is highly expressed in glioma tissues and cells, and silencing the expression of NUP214 significantly inhibits the malignant biological behaviours of glioma cells.
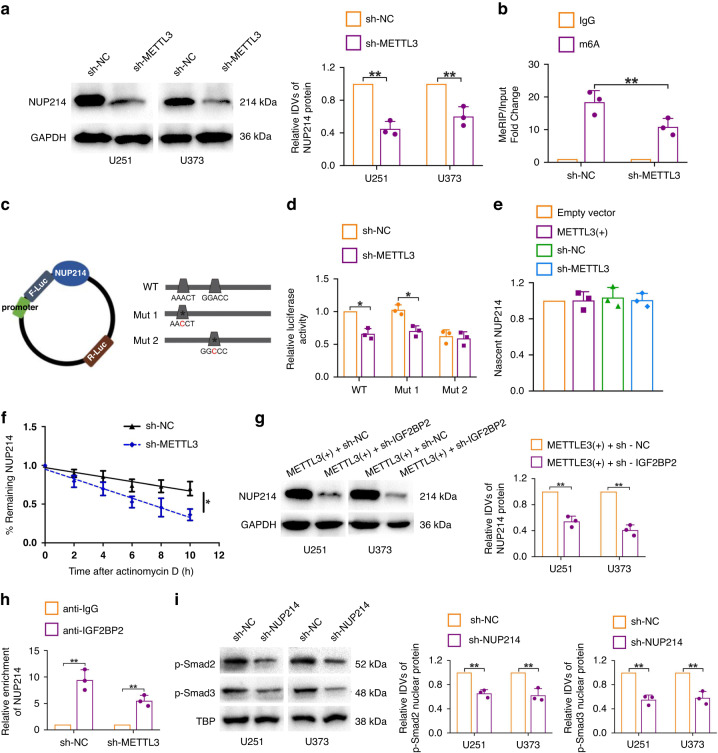
Fig. 5. METTL3 mediates NUP214 mRNA m6A and promotes NUP214 expression.
a Western blot results showed that METTL3 knockdown significantly inhibited NUP214 expression. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01. b MeRIP-qPCR results showed that METTL3 knockdown significantly reduced the m6A methylation level of NUP214 mRNA. **P < 0.01. c Potential m6A methylation sites of NUP214 3’UTR predicted by informatics software and designed mutant sequences. d Dual-luciferase reporter assay was applied to verify the METTL3-mediated m6A methylation site of NUP214 3’UTR. *P < 0.05. e Nascent RNA capture assay was used to evaluate the nascent NUP214 in METTL3 overexpression or METTL3 knockdown glioma cells. f The remaining levels of NUP214 at the different time points treated with actinomycin D were indicated. *P < 0.05. g Western blot results showed that IGF2BP2 knockdown significantly reversed the promotive effects of METTL3 overexpression on NUP214 expression. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01. h NUP214 mRNA levels in immunoprecipitates were determined by qRT-PCR. Expression levels of NUP214 were presented as fold enrichment relative to IgG immunoprecipitates. i Western blot results showed that NUP214 knockdown significantly reduced the expression of nuclear protein p-Smad2/3. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01.
METTL3 mediates NUP214 mRNA m6A and positively regulates NUP214 expression in glioma cells
To clarify the mechanism of how NUP214 is regulated by METTL3 in glioma cells, we first used the m6A bioinformatics analysis software SRAMP and RMVar for prediction and found that mRNA of NUP214 contains two potential m6A methylation sites (RRACH) located in the 3’UTR near the stop codon. Subsequently, the results of MeRIP-qPCR indicated that the enrichment extent of NUP214 mRNA in the anti-m6A group was significantly higher than that in the anti-IgG group, and silencing the expression of METTL3 in glioma cells significantly reduced the enrichment of NUP214 mRNA in the anti-m6A group (Fig. 5b). The METTL3-mediated m6A methylation site of NUP214 mRNA was further verified by dual-luciferase reporter gene experiment (Fig. 5c, d). The results showed that silencing METTL3 expression reduced luciferase activity of WT and Mut1 group in U251 glioma cells; however, there was no significant change in luciferase activity in the Mut2 group, suggesting that site 2 (GGACC) is the METTL3-mediated m6A methylation site of NUP214 mRNA (Fig. 5d).
IGF2BP2 binds and stabilises NUP214 mRNA
Similar to DNA methylation, RNA methylation needs to be recognised and regulated by specific binding proteins to perform biological functions. To further clarify the mechanism of how METTL3 regulates NUP214 mRNA expression, we first examined nascent NUP214 mRNA in U251 glioma cells with upregulated or silenced METTL3 expression, and found that upregulated or silenced METTL3 expression did not affect the nascent NUP214 mRNA, as shown in Fig. 5e. The results of RNA stability detection assays showed that silencing the expression of METTL3 shortened the half-life of NUP214 (Fig. 5f), suggesting that METTL3 enhanced the stability of NUP214 mRNA. It was found that IGF2BP2 may recognise and bind the m6A site of NUP214 to regulate the expression of NUP214 through the analysis of four sequencing chip sets (GSE90639, GSE90642, GSE90684 and GSE90686) from GEO dataset. Insulin-like growth factor 2 mRNA binding protein (IGF2BP2) has been proven to enhance the mRNA stability of target genes and upregulate their expression as m6A “reader” [29, 30]. In addition, the expression of IGF2BP2 is upregulated in glioma, playing the role of oncogene [31]. To confirm the above prediction results, we first conducted a western blot assay. The results showed that silencing IGF2BP2 significantly inhibited the expression of NUP214 in METTL3 overexpressing U251 and U373 glioma cells (Fig. 5g). Further RIP experiments showed that the enrichment extent of NUP214 mRNA in the anti-IGF2BP2 group was significantly higher than that in the anti-IgG group (Fig. 5h). The above results indicate that METTL3 positively regulates the expression of NUP214 by mediating the recognition of NUP214 mRNA m6A by IGF2BP2 and stabilising the mRNA of NUP214.
NUP214-mediated Smad2/3 nuclear translocation activates TGF-β signalling pathway activity
Recent studies have found that NUP214 can promote the malignant progression of cancer by regulating the activity of cell signalling pathways [32]. Transforming growth factor-beta (TGF-β) signalling pathway plays a role in promoting cancer in a variety of malignant tumours, whose abnormal activation in glioma substantially promotes the malignant biological behaviours and immune escape and stemness of glioma stem cells; besides, it is a research hotspot for development of molecular suppressor targets of glioma tumour [33, 34]. NUP214 is an important regulator of the TGF-β signalling pathway, mediating the translocation of p-Smad2/3 to the nucleus to activate the transcription of relevant target genes [35, 36]. We found that silencing the expression of NUP214 in U251 and U373 glioma cells significantly inhibited the expression of nuclear protein p-Smad2/3 from the results of the western blot assays (Fig. 5i), showing that NUP214 activates the activity of TGF-β signalling pathway by mediating the transport of p-Smad2/3 to the nucleus in glioma cells and plays a role in promoting cancer.
Double silencing of METTL3 and NUP214 combined with overexpression of miR-1208 significantly inhibited tumour growth in subcutaneous tumour-bearing nude mice and prolonged the survival of orthotopic tumour-bearing nude mice
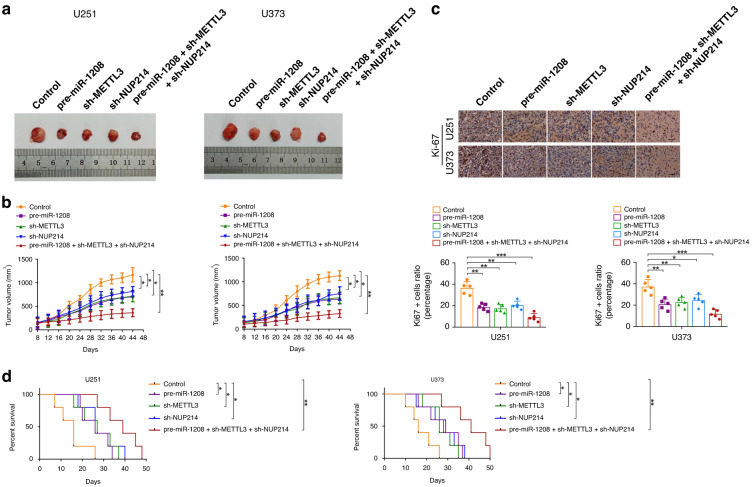
The effects of individually silencing METTL3 or NUP214, overexpressing miR-1208, or the combination of the three on tumour growth in subcutaneous tumour-bearing nude mice and survival in orthotopic tumour-bearing nude mice were analysed using in vivo experiments. The results are shown in Fig. 6a‒d, compared with that in the control group, the tumour volume of the subcutaneous tumour-bearing nude mice in the METTL3-silenced group or NUP214-silenced group or miR-1208-overexpressed group was reduced; the proliferation ability of glioma cells of orthotopic tumour-bearing nude mice in the METTL3-silenced group or NUP214-silenced group or miR-1208-overexpressed group was significantly inhibited, and the survival time was prolonged. In addition, the nude mice in the double-silenced METTL3 and NUP214 combined with miR-1208 overexpression group had the smallest tumour volume, worst cell proliferation ability and longest survival (Fig. 7).
Fig. 6. In vivo study.
a The sample tumours of nude mice from respective groups were presented. b Tumour xenograft growth curves in subcutaneous implantation assay were displayed. Data represent mean ± SD (n = 5, each group). *P < 0.05, **P < 0.01. c Representative images and quantification of IHC staining for Ki-67 in sections from the nude mice of orthotopic inoculations assay (scale bar = 50 μm). Data are presented as the mean ± SD (n = 5, each group). **P < 0.01, ***P < 0.001. d Survival curves from representative nude mice injected into the right striatum in orthotopic inoculations assay were exhibited. Log-rank test, *P < 0.05, **P < 0.01.
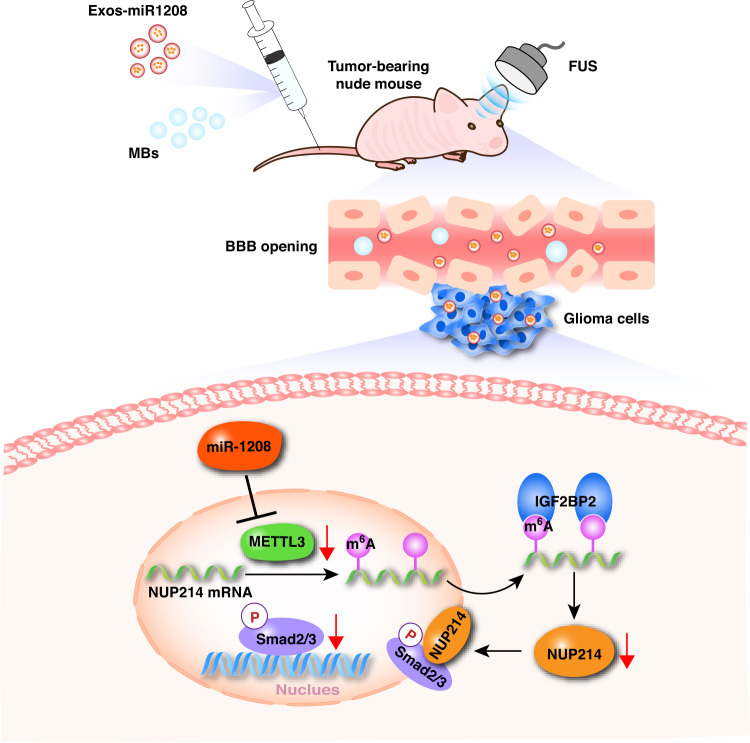
Fig. 7. Schematic diagram.
The schematic diagram underlying the mechanism of FUS combined with miR-1208-equipped Exos inhibit malignant biological behaviours of glioma cells.
Discussion
Our findings confirmed that the expression of miR-1208 was significantly downregulated in glioma tissues and cells and was negatively correlated with the pathological grade of glioma, thereby, further confirming that miR-1208 acts as a tumour suppressor gene. Overexpression of miR-1208 significantly inhibits the malignant biological behaviours of glioma cells. Furthermore, Exos secreted by hBMSCs can be used as a carrier for miR-1208 and can instantly penetrate the BBB and show significant tumour-suppressive effects when combined with FUS in the treatment of glioma. The subsequent molecular mechanism studies demonstrate that miR-1208 downregulated the m6A methylation level of NUP214 mRNA by negatively regulating METTL3, affecting the binding between IGF2BP2 and NUP214 and reducing the stability of NUP214 mRNA; thus, the expression of NUP214 was downregulated, which further inhibited the activity of TGF-β signalling pathway and the malignant biological behaviours of glioma cells. Moreover, simultaneous silencing of METTL3 and NUP214 combined with overexpressed miR-1208 significantly inhibited the growth of xenografted nude mice and prolonged the survival of tumour-bearing nude mice.
In recent years, although a variety of novel exogenous nanocarriers have been developed and applied in tumour therapy, several challenges, such as heterogeneity, immunogenicity and in vivo stability, remain to be overcome [37]. Compared with exogenous nanocarriers, Exos, as endogenous nanocarriers, have the advantages of abundant sources, low toxicity and immunogenicity, stable encapsulation of therapeutic drugs or genes, and the ability to pass through physiological and pathological barriers [38–40]. Recently, targeting tumour tissues with miRNA carried by Exos has become a novel therapeutic strategy; for instance, Exos loaded with miR-122 has significantly improved the chemotherapy sensitivity of hepatocellular carcinoma [41]. In the treatment of glioma, Exos loaded with miR-29a-3p and miR-124a also achieved good efficacy [11, 12]. In this study, we overexpressed miR-1208 in hBMSCs by lentiviral transfection and used hBMSCs as the source cells of Exos; therefore, the secreted Exos carried highly expressed miR-1208. The results showed that miR-1208-loaded Exos are not toxic to glioma cells and are naturally taken up by the cells. MiR-1208 was successfully transferred into glioma cells to exert its function, thereby confirming that Exos are ideal carriers for carrying therapeutic miRNAs. Although Exos loaded with miRNA have achieved great success in research, there are still many problems to be faced in clinical transformation. For example, more research is required to explore the homogeneity, quantitative standard, and mechanism of action of Exos in the future.
Researchers have been exploring therapeutic strategies to overcome the BBB because the presence of the BBB hinders the entry of therapeutic substances into the CNS. In recent years, low-power FUS combined with MBs has been demonstrated to transiently and non-invasively open the BBB to facilitate the delivery of therapeutic substances to glioma tissue, which greatly improved the therapeutic effect [42, 43]. Thus, for the first time, we investigated a novel glioma treatment modality of miR-1208-loaded Exos combined with FUS. The combination of FUS could open the BBB and promote the uptake of miR-1208 in Exos by glioma cells, thereby achieving a high-efficiency anti-tumour effect. The potential mechanism of action may be that FUS irradiation increases cerebral vascular permeability and enhances the enrichment of miR-1208-loaded Exos in the lesion site while simultaneously promoting the release of miR-1208 by Exos in glioma cells due to the generation of ultrasonic cavitation effects. Therefore, miR-1208-loaded Exos combined with FUS is a therapeutic strategy with great clinical application potential in the treatment of glioma.
METTL3 is one of the most important RNA methyltransferases in humans. Increasing research results have suggested that METTL3 can regulate the expression of target genes by mediating m6A modification of mRNA of different target genes. It plays an important role in regulating normal biological processes or the occurrence and development of diseases in different organs, tissues and various types of cells [44–46]. We confirmed that miR-1208 could negatively regulate the expression of METTL3 mRNA by targeting the 3’UTR of METTL3 mRNA and play a tumour-suppressive role in glioma cells through a series of experiments. In glioma, although METTL3 has been reported to possibly affect the malignant progression of glioma by regulating the stemness of glioma stem cells or the biological behaviours of glioma cells, the carcinogenesis mechanism of METTL3 in glioma remains not entirely clear [18, 19]. In this study, we found that there was an m6A methylation site in the 3’UTR region of NUP214 mRNA, and silencing the expression of METTL3 significantly downregulated the m6A level of NUP214 mRNA as well as the expression of NUP214, suggesting that NUP214 is the downstream target of METTL3 in glioma. METTL3-mediated m6A modification mainly regulates the expression of target genes by regulating RNA alternative splicing, RNA stability, and post-transcriptional translation [47]. Therefore, we further investigated the mechanism by which METTL3 regulates NUP214 expression. We found that in glioma cells, IGF2BP2 can recognise m6A of NUP214 mRNA and stabilise NUP214 mRNA, while silencing IGF2BP2 significantly inhibited the expression of NUP214 mRNA and NUP214 protein levels, indicating that IGF2BP2 positively regulates the expression of NUP214 in glioma, which is similar to results of previous studies. In oral squamous cell carcinoma, METTL3 mediated the recognition of SLC7A11 mRNA m6A by IGF2BP2, which enhances the stability of SLC7A11 mRNA and upregulates its expression, thereby promoting the progression of oral squamous cell carcinoma [48]. In colorectal cancer, IGF2BP2 promotes the malignant progression of colorectal cancer by recognising METTL3 and WTAP-mediated UCA1 mRNA m6A to stabilise UC mRNA and upregulate its expression [49]. Finally, we analysed the potential carcinogenic mechanism of NUP214 in glioma. Smad2/3 is an important molecule for intracellular transduction and regulation of TGF-β signal. Smad2/3 is phosphorylated by TGF-β signal in the cytoplasm to form p-Smad2/3, which is then transported into the nucleus to play a transcriptional regulatory role on downstream target genes [35, 36]. NUP214 is located in the nuclear membrane and has been demonstrated to be a key molecule for the transport of p-Smad2/3 into the nucleus [35, 36]. We detected p-Smad2/3 in the nuclear protein of glioma cells with silenced NUP214 expression and showed that the expression level of p-Smad2/3 was significantly downregulated. These results indicate that NUP214 can activate TGF-β signalling pathway activity by mediating p-Smad2/3 nuclear transport. To the best of our knowledge, this is the first study to demonstrate a novel mechanism by which miR-1208 regulates NUP214 expression in an m6A-dependent manner in glioma by negatively regulating METTL3, thereby inhibiting the malignant phenotype of glioma cells.
Conclusions
In conclusion, this study proposes the use of hBMSCs-derived Exos as carriers for the tumour suppressor gene miR-1208. Our results demonstrate that Exos carrying miR-1208 can effectively pass through the BBB when combined with FUS irradiation, which promotes the uptake of miR-1208 from Exos by glioma cells and results in a high-efficiency tumour-suppressive effect. In addition, the molecular mechanism of miR-1208-mediated inhibition of the malignant biological behaviours of glioma cells through miR-1208/METTL3/NUP214/TGF-β pathway was elucidated in this study. Our findings provide an experimental basis for the exploration of novel therapeutic strategies for the treatment of glioma and identify novel targets for the molecular therapy of glioma.
Supplementary information
Author contributions
WR contributed to the experiment design and implementation, manuscript draft, and data analysis. YZ contributed to the experiment implementation and data analysis. YS, WQ, BY, LJ, PS and LS performed the experiments. ZL, WZ, XW, and XY analysed the data. YZ, YS and ZL conceived or designed the experiments, performed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work is supported by grants from the Natural Science Foundation of China (81571686), Natural Science Foundation of Liaoning Province (2020-MS-170), Basic Scientific Research Project of Colleges and Universities of Liaoning Province (LJKMZ20221148), Science and Technology Plan of Shenyang City (22-321-33-18) and Outstanding Scientific Research Talent Plan of Shengjing hospital (No. 2020M0322).
Data availability
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
All clinical specimens used in this study were approved by the Ethics Committee of the Shengjing Hospital of China Medical University. All animal experiments were approved by the Laboratory Animal Management Committee of Shengjing Hospital of China Medical University.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ying Zhan, Yichen Song.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02393-w.
References
- 1.Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19:66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song Y, Shao L, Xue Y, Ruan X, Liu X, Yang C, et al. Inhibition of the aberrant A1CF-FAM224A-miR-590-3p-ZNF143 positive feedback loop attenuated malignant biological behaviors of glioma cells. J Exp Clin Cancer Res. 2019;38:248. doi: 10.1186/s13046-019-1200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhan Y, Qiao W, Yi B, Yang X, Li M, Sun L, et al. Dual role of pseudogene TMEM198B in promoting lipid metabolism and immune escape of glioma cells. Oncogene. 2022;41:4512–23. doi: 10.1038/s41388-022-02445-0. [DOI] [PubMed] [Google Scholar]
- 4.Grabrucker AM, Ruozi B, Belletti D, Pederzoli F, Forni F, Vandelli MA, et al. Nanoparticle transport across the blood brain barrier. Tissue Barriers. 2016;4:e1153568. doi: 10.1080/21688370.2016.1153568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gosselet F, Loiola RA, Roig A, Rosell A, Culot M. Central nervous system delivery of molecules across the blood-brain barrier. Neurochem Int. 2021;144:104952. doi: 10.1016/j.neuint.2020.104952. [DOI] [PubMed] [Google Scholar]
- 6.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841:108–20. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Yang S, Gao H. Nanoparticles for modulating tumor microenvironment to improve drug delivery and tumor therapy. Pharm Res. 2017;126:97–108. doi: 10.1016/j.phrs.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 8.You B, Xu W, Zhang B. Engineering exosomes: a new direction for anticancer treatment. Am J Cancer Res. 2018;8:1332–42. [PMC free article] [PubMed] [Google Scholar]
- 9.Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–41. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Tung SL, Boardman DA, Sen M, Letizia M, Peng Q, Cianci N, et al. Regulatory T cell-derived extracellular vesicles modify dendritic cell function. Sci Rep. 2018;8:6065. doi: 10.1038/s41598-018-24531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Guo X, Guo X, Yu R, Qian M, Wang S, et al. MicroRNA-29a-3p delivery via exosomes derived from engineered human mesenchymal stem cells exerts tumour suppressive effects by inhibiting migration and vasculogenic mimicry in glioma. Aging (Albany NY) 2021;13:5055–68. doi: 10.18632/aging.202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, et al. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro Oncol. 2018;20:380–90. doi: 10.1093/neuonc/nox152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall J, Prabhakar S, Balaj L, Lai CP, Cerione RA, Breakefield XO. Delivery of therapeutic proteins via extracellular vesicles: review and potential treatments for parkinson’s disease, glioma, and schwannoma. Cell Mol Neurobiol. 2016;36:417–27. doi: 10.1007/s10571-015-0309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CY, Hsieh HY, Chen CM, Wu SR, Tsai CH, Huang CY, et al. Non-invasive, neuron-specific gene therapy by focused ultrasound-induced blood-brain barrier opening in Parkinson’s disease mouse model. J Control Release. 2016;235:72–81. doi: 10.1016/j.jconrel.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Pi Z, Yan F, Yeh C, Zeng X, Diao X, et al. Enhanced delivery of daclitaxel liposomes using focused ultrasound with microbubbles for treating nude mice bearing intracranial glioblastoma xenografts. Int J Nanomed. 2017;12:5613–29. doi: 10.2147/IJN.S136401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Y, Feng R, Lu T, Chen X, Zhou X, Wang X. Novel insights into the m(6)A-RNA methyltransferase METTL3 in cancer. Biomark Res. 2021;9:27. doi: 10.1186/s40364-021-00278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–33. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 19.Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N6-methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 2019;79:5785–98. doi: 10.1158/0008-5472.CAN-18-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian Y, Nan Y, Han, Zhang A, Wang G, Jia Z, et al. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol. 2012;40:1105–12. doi: 10.3892/ijo.2011.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang M, Li T, Wang Q, Li C, Zhou H, Deng S, et al. Silencing circPVT1 enhances radiosensitivity in non-small cell lung cancer by sponging microRNA-1208. Cancer Biomark. 2021;31:263–79. doi: 10.3233/CBM-203252. [DOI] [PubMed] [Google Scholar]
- 22.Kim EA, Jang JH, Sung EG, Song IH, Kim JY, Lee TJ. MiR-1208 increases the sensitivity to cisplatin by targeting TBCK in renal cancer cells. Int J Mol Sci. 2019;20:3540. doi: 10.3390/ijms20143540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Wang D, Zhu T, Yu J, Wu X, Lin W, et al. CircPUM1 promotes hepatocellular carcinoma progression through the miR-1208/MAP3K2 axis. J Cell Mol Med. 2021;25:600–12. doi: 10.1111/jcmm.15998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visvanathan A, Patil V, Abdulla S, Hoheisel JD, Somasundaram K. N6-methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes (Basel) 2019;10:141. doi: 10.3390/genes10020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon DN, Rout MP. Cancer and the nuclear pore complex. Adv Exp Med Biol. 2014;773:285–307. doi: 10.1007/978-1-4899-8032-8_13. [DOI] [PubMed] [Google Scholar]
- 26.Mendes A, Fahrenkrog B. NUP214 in leukemia: it’s more than transport. Cells. 2019;8:76. doi: 10.3390/cells8010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharjya S, Roy KS, Ganguly A, Sarkar S, Panda CK, Bhattacharyya D, et al. Inhibition of nucleoporin member Nup214 expression by miR-133b perturbs mitotic timing and leads to cell death. Mol Cancer. 2015;14:42. doi: 10.1186/s12943-015-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv L, He L, Chen S, Yu Y, Che G, Tao X, et al. Long non-coding RNA LINC00114 facilitates colorectal cancer development through EZH2/DNMT1-induced miR-133b suppression. Front Oncol. 2019;9:1383. doi: 10.3389/fonc.2019.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu D, Pan M, Li Y, Lu T, Wang Z, Liu C, et al. RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner. J Exp Clin Cancer Res. 2022;41:6. doi: 10.1186/s13046-021-02212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, et al. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep. 2016;15:1634–47. doi: 10.1016/j.celrep.2016.04.086. [DOI] [PubMed] [Google Scholar]
- 32.Bindra D, Mishra RK. In pursuit of distinctiveness: transmembrane nucleoporins and their disease associations. Front Oncol. 2021;11:784319. doi: 10.3389/fonc.2021.784319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papachristodoulou A, Silginer M, Weller M, Schneider H, Hasenbach K, Janicot M, et al. Therapeutic targeting of TGFβ ligands in glioblastoma using novel antisense oligonucleotides reduces the growth of experimental gliomas. Clin Cancer Res. 2019;25:7189–201. doi: 10.1158/1078-0432.CCR-17-3024. [DOI] [PubMed] [Google Scholar]
- 34.Gong L, Ji L, Xu D, Wang J, Zou J. TGF-beta links glycolysis and immunosuppression in glioblastoma. Histol Histopathol. 2021;29:18366. doi: 10.14670/HH-18-366. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, Kang Y, Cöl S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol Cell. 2002;10:271–82. doi: 10.1016/S1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- 36.Xu L, Alarcón C, Cöl S, Massagué J. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem. 2003;278:42569–77. doi: 10.1074/jbc.M307601200. [DOI] [PubMed] [Google Scholar]
- 37.Zaki Ghali MG, Srinivasan VM, Kan P. Focused ultrasonography-mediated blood-brainbarrier disruption in the enhancement of delivery of brain tumor therapies. World Neurosurg. 2019;131:65–75. doi: 10.1016/j.wneu.2019.07.096. [DOI] [PubMed] [Google Scholar]
- 38.Hafiane A, Daskalopoulou SS. Extracellular vesicles characteristics and emerging roles in atherosclerotic cardiovascular disease. Metabolism. 2018;85:213–22. doi: 10.1016/j.metabol.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12:655–64. doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilligan KE, Dwyer RM. Engineering exosomes for cancer therapy. Int J Mol Sci. 2017;18:1122. doi: 10.3390/ijms18061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMahon D, Poon C, Hynynen K. Evaluating the safety profile of focused ultrasound and microbubble-mediated treatments to increase blood-brain barrier permeability. Expert Opin Drug Deliv. 2019;16:129–42. doi: 10.1080/17425247.2019.1567490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai L, Liu Y, Guo K, Zhang K, Liu Q, Wang P, et al. Ultrasound facilitates naturally equipped exosomes derived from macrophages and blood serum for orthotopic glioma treatment. ACS Appl Mater Interfaces. 2019;11:14576–87. doi: 10.1021/acsami.9b00893. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J, et al. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020;48:11083–96. doi: 10.1093/nar/gkaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L, Liu X, Hu X, Gao L, Zeng H, Wang X, et al. METTL3-mediated m6A modification of TIMP2 mRNA promotes podocyte injury in diabetic nephropathy. Mol Ther. 2022;30:1721–40. doi: 10.1016/j.ymthe.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 47.Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22. doi: 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- 48.Xu L, Li Q, Wang Y, Wang L, Guo Y, Yang R, et al. m6A methyltransferase METTL3 promotes oral squamous cell carcinoma progression through enhancement of IGF2BP2-mediated SLC7A11 mRNA stability. Am J Cancer Res. 2021;11:5282–98. [PMC free article] [PubMed] [Google Scholar]
- 49.He RZ, Jiang J, Hu X, Lei M, Li J, Luo W, et al. Stabilization of UCA1 by N6-methyladenosine RNA methylation modification promotes colorectal cancer progression. Cancer Cell Int. 2021;21:616. doi: 10.1186/s12935-021-02288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.