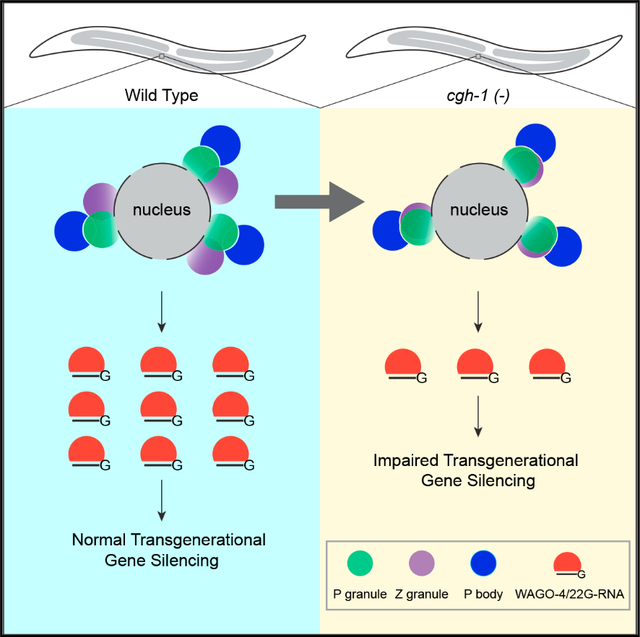
SUMMARY
Biomolecular condensates have been shown to interact in vivo, yet it is unclear whether these interactions are functionally meaningful. Here, we demonstrate that cooperativity between two distinct condensates—germ granules and P bodies—is required for transgenerational gene silencing in C. elegans. We find that P bodies form a coating around perinuclear germ granules and that P body components CGH-1/DDX6 and CAR-1/LSM14 are required for germ granules to organize into sub-compartments and concentrate small RNA silencing factors. Functionally, while the P body mutant cgh-1 is competent to initially trigger gene silencing, it is unable to propagate the silencing to subsequent generations. Mechanistically, we trace this loss of transgenerational silencing to defects in amplifying secondary small RNAs and the stability of WAGO-4 Argonaute, both known carriers of gene silencing memories. Together, these data reveal that cooperation between condensates results in an emergent capability of germ cells to establish heritable memory.
In brief
Du et al. characterize the function of P body-germ granule interactions in germ cells of C. elegans. They demonstrate that these interactions are critical for organizing small RNA factors within germ granules into distinct sub-compartments, which play a crucial role in maintaining small RNA-guided gene silencing across generations.
Graphical abstract
INTRODUCTION
Formation of biomolecular condensates has emerged as a critical mechanism for membraneless compartmentation in living cells.1 The formation of various condensates has been linked to diverse cellular functions, including cellular signaling, RNAi inheritance, RNA processing, and transcriptional regulation.2 Notably, many of these condensates are not homogeneous but instead organized to distinct sub-compartments.3 One remarkable example of an organized condensate is the perinuclear germ granule of C. elegans, where factors enriched in distinct layers of germ granules are associated with specific roles in small RNA-guided gene silencing, where P granule factors function in RNA surveillance,4–7 Z granule factors in the inheritance of RNAi,8,9 and Mutator foci factors in the production of secondary silencing small RNAs.10 However, the mechanisms contributing to the organization of germ granules or other cellular condensates remain largely unknown. It is also unclear whether the proper organization of the germ granule is critical for its function.
Distinct condensates are also found to interact in cells, including Cajal bodies with their attached B-snurposome, Cajal bodies with nucleoli, and processing bodies (P bodies) with stress granules.11–13 However, little is known about whether these interactions play a role in regulating the formation or function of their cognate interacting condensates. It has been reported that germ granules associate with P bodies in the early embryos of C. elegans and other animals.14–16 Germ granules and P bodies are two distinct condensates associated with different biological functions. In germ cells of diverse animals, many small RNA pathway factors are enriched in germ granules.17,18 P bodies in contrast are well known as condensates housing mRNA degradation factors and translationally inactive mRNAs.19,20 As some P bodies form perinuclear foci in the adult germline where perinuclear germ granules are found,21,22 we aimed to determine whether P bodies physically interact with perinuclear germ granules and, if so, to investigate the functional relevance of these interactions.
Here, we show that in the C. elegans germline, P bodies are situated specifically at the cytoplasmic side of perinuclear germ granules. We found that P body factors CGH-1 and CAR-1, the homologs of human DDX6 and LSM14, specifically interact with P granule and Z granule factors housed in germ granules. In addition, CGH-1’s interaction with germ granule factors relies on CAR-1. Notably, we found P body factor CGH-1 and its interaction with germ granules are critical for proper germ granule organization into discrete P/Z granules and for enrichment of small RNA factors in germ granules. We further showed that while cgh-1 mutants can initiate piRNA- and siRNA-mediated silencing, they exhibit defects in triggering transgenerational gene silencing. This defect in transgenerational gene silencing is associated with the inability of cgh-1 mutant to produce secondary small RNAs that associate with Argonaute WAGO-4. Together, our results suggest that highly specific P body-germ granule interactions are a key mode of regulation for germ granule formation and function.
RESULTS
P bodies are localized to the cytoplasmic side of perinuclear germ granules
As described above, previous studies have shown that some P body condensates are enriched at the nuclear periphery in the adult germline of C. elegans.21 Indeed, by monitoring the localization of different P body factors including CGH-1, LET-711, and IFET-1 using endogenously tagged fluorescent marker expressing strains, we confirmed that P body condensates present as perinuclear foci in nearly all stages of the adult germline (Figures S1A and S1B). When we simultaneously monitored two P body markers, we found that they are largely co-localized (Figure S1B), showing P body factors co-localize at the nuclear periphery. To examine the relationship between P bodies and germ granules, we examined the co-localization of P body markers with various germ granules markers, including factors enriched in distinct sub-compartments of germ granules, including P granules, Z granules, and Mutator foci. These sub-compartments of germ granules have shown to form a three-layer germ granule.8 When we simultaneously monitored the localization of P body and P granule markers, we found that P body markers, including CGH-1, LET-711, and IFET-1, are localized to the cytoplasmic side of perinuclear P granule markers, including PGL-1, PRG-1, and CSR-1 (Figures 1A and S1C). When we also monitored the localization of NPP-9 (a nuclear pore marker), we observed that P bodies are situated to the cytoplasmic side of P granules, which together are situated on top of nuclear pore clusters (Figure 1B).23 To further characterize the relationship between P bodies and adjacent germ granules, we identified and defined individual P bodies as the center and measured the vertical (outside/inside) and horizontal (left/right) distance between individual P body markers to their adjacent germ granule markers (Figure 1C). Using these analyses, we confirmed that the majority of CGH-1 condensates are located directly to the cytoplasmic side of PGL-1 or CSR-1 condensates (Figure 1C). In addition, we noticed that some of the P body and P granule markers exhibit overlapping signals (Figure 1A), indicating these two cellular bodies are partially co-localized. We then examined the relationship between P bodies and Z granules by simultaneously monitoring the localization of CGH-1 and ZNFX-1 or WAGO-4 (two Z granule markers). We found that while most CGH-1 condensates are located slightly toward the cytoplasmic side of Z granules, they are frequently found as adjacent condensates as indicated by the higher variance in horizontal distance between these two condensates (Figures 1C and 1D). We also observed some partial co-localization of CGH-1 condensates and WAGO-4 condensates (Figure 1D). We then monitored the relationship between P bodies and Mutator foci, and we found that CGH-1 condensates also tend to localize slightly toward the cytoplasmic side and are adjacent to Mutator marker MUT-16 (Figure 1C). Very little co-localizing signal between CGH-1 and MUT-16 is found (Figure S1D). Together with the previous report that PZM granules form tri-condensate assemblages,8 our image analyses suggest a spatial organization of P bodies, P granules, Z granules, and Mutator foci at the nuclear periphery (Figure 1E).
Figure 1. P bodies localize to the cytoplasmic side of perinuclear germ granules.
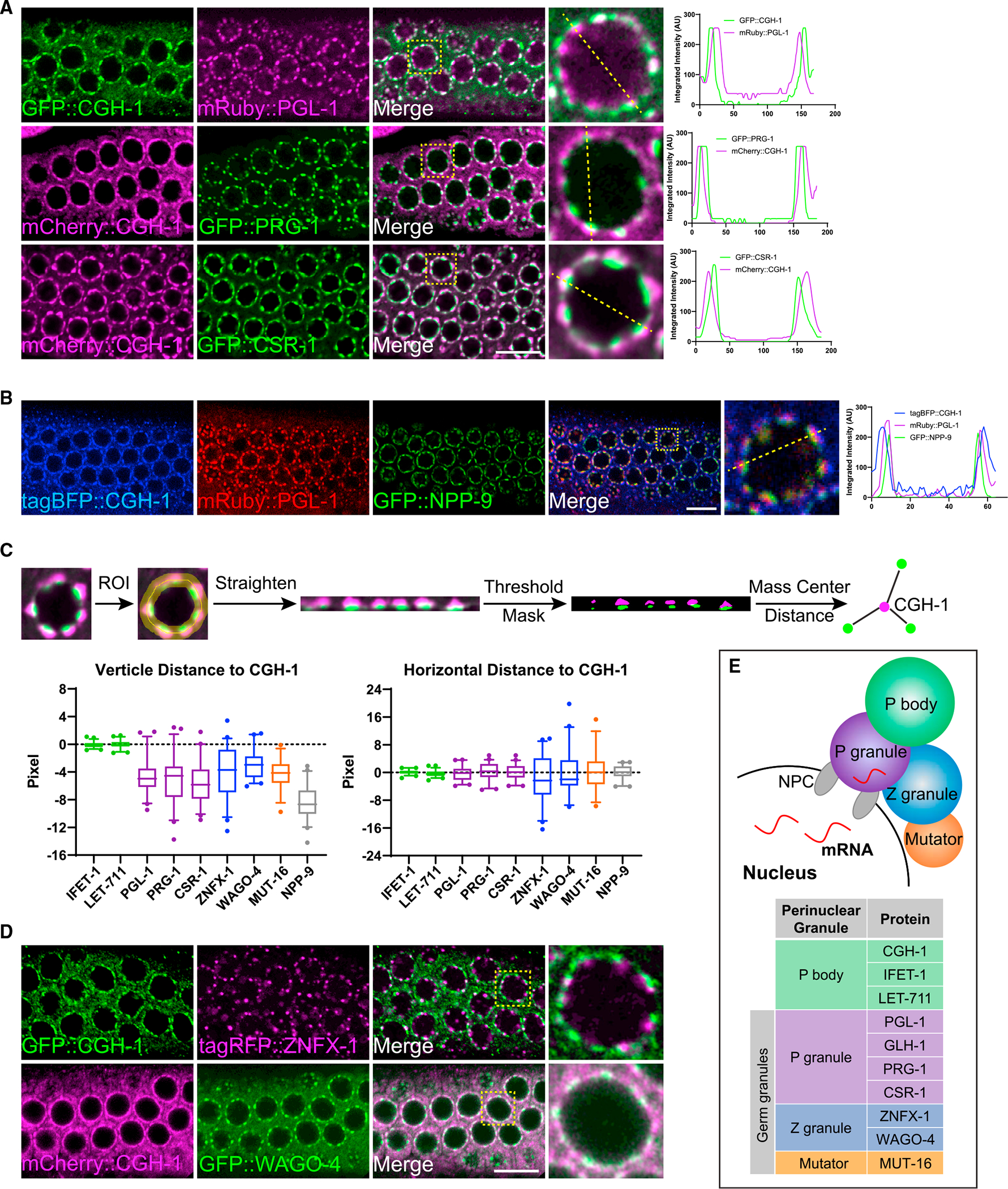
(A) Fluorescent micrographs show the localization of P body marker CGH-1 and the indicated P granule markers in the pachytene region of adult germlines. The line in the merged image indicates the position of the line scan for measuring fluorescent intensity across single germline nuclei (right). Note that white-colored foci in the merged image represent the co-localization of two makers. Bar: 10 μm.
(B) Fluorescent micrographs show the spatial arrangement of P body, P granule, and nuclear pore clusters at germline nuclei. The line in the merged image indicates the position of the line scan for measuring fluorescent intensity across single germline nuclei (right). Bar: 10 μm.
(C) A schematic (top) showing the measurements of vertical distance (bottom left) and horizontal (bottom right) distance between the indicated proteins to P body marker CGH-1. For boxplots, lines display median values, boxes display 1st and 3rd quartiles, and whiskers display 5th and 95th percentiles. Distributions represent data collected from 8 to 12 independent gonad images.
(D) Fluorescent micrographs show the localization of P body marker CGH-1 and the indicated Z granule proteins in the pachytene region of adult germlines. Note that white-colored foci in the merged image represent the co-localization of two markers. Bar: 10 μm.
(E) A model depicting spatial arrangements of the P body and distinct sub-compartments of the germ granule, including P granule, Z granule, and Mutator foci, at the nuclear periphery.
P body factors CGH-1 and CAR-1 interact with small RNA pathway factors enriched in germ granules
Our observations of partial co-localization between perinuclear P body markers and both P granule and Z granule markers raise the possibility that factors between these condensates might interact with each other. To test this hypothesis, we performed mass spectrometry (MS) analyses of CGH-1 complexes (Figure 2A). With CGH-1 immunoprecipitation (IP)-MS analyses, we identified several known P body factors but did not detect factors enriched in P granules or Z granules. To detect more transient interactions, we applied chemical crosslinking before performing IP-MS analyses, an approach that has successfully identified transient interactions between ribonucleoprotein complexes.24,25 Indeed, additional proteins were detected in the crosslinked CGH-1 complexes, including GLH-1 and Argonaute proteins PRG-1, CSR-1, and WAGO-1 (Figure 2A). The presence of these Argonaute proteins in crosslinked CGH-1 complexes was confirmed by IP-western analyses (Figure 2B). While we did not detect the presence of Z granule factors, such as ZNFX-1 or WAGO-4, in crosslinked CGH-1 complexes, we performed reciprocal IP-MS analyses of crosslinked ZFNX-1 and WAGO-4 complexes and were able to detect the presence of P body factors, specifically CGH-1 and CAR-1 (Figures 2C and 2D). No Mutator foci factors were identified in the IP complexes of CGH-1, ZNFX-1, or WAGO-4 (Table S1). Together, our MS analyses showed that P body factors CAR-1 and CGH-1 interact with the small RNA pathway factors enriched in P granules and Z granules.
Figure 2. Proteomic analyses of interactions between P body and germ granule factors.
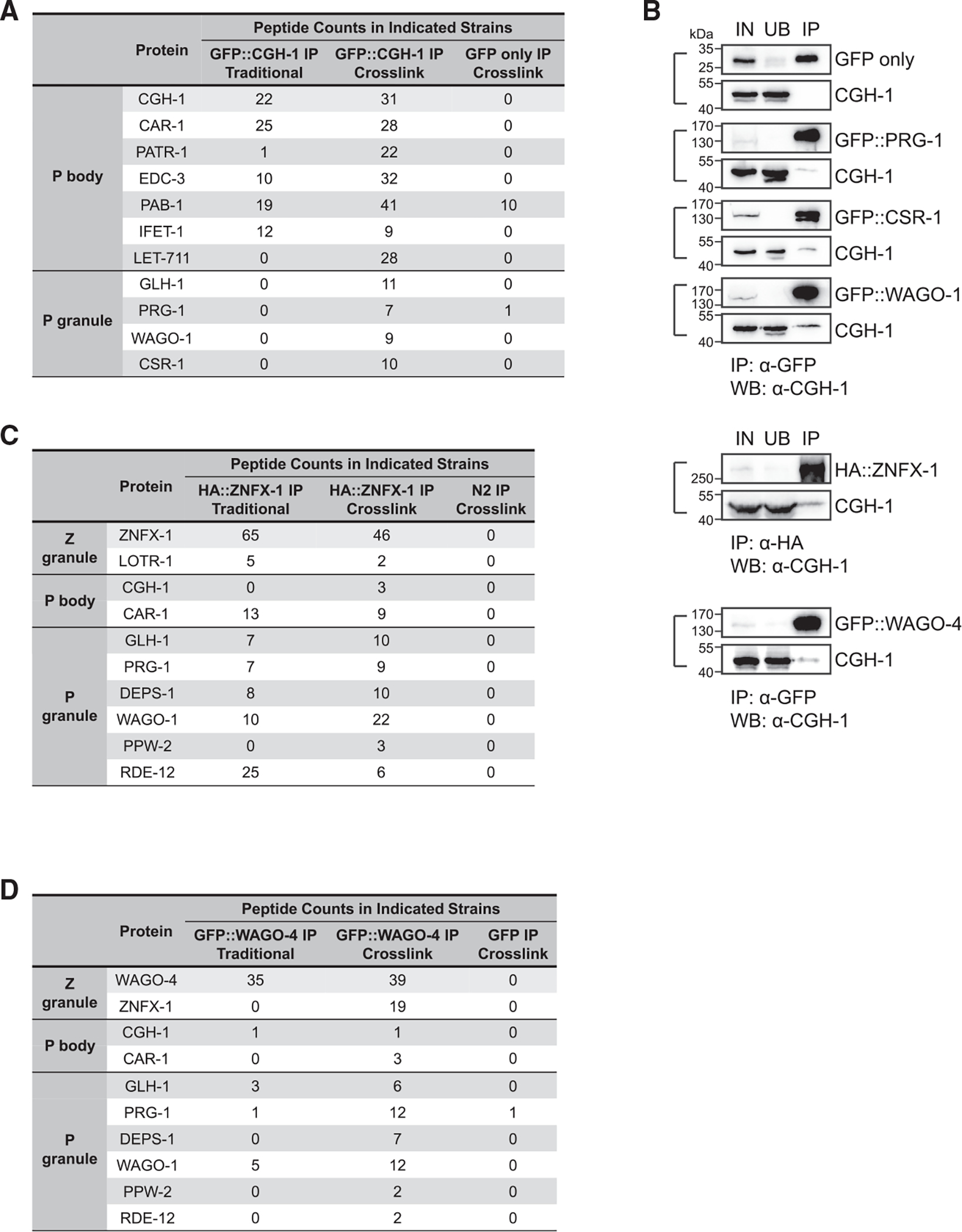
(A) Proteomic analyses of CGH-1 complex. The numbers of peptides identified in the crosslinked or non-crosslinked condition are shown. GFP-only IP experiment in the crosslink condition served as a control.
(B) Western blot analyses show the interactions between CGH-1 and the indicated germ granule factors. Immunoprecipitation of GFP- or HA-tagged germ granule factors was conducted. Subsequently, western blot analysis was performed using specific antibodies against GFP, HA, or CGH-1 to determine the presence of the indicated germ granule factors or CGH-1 in the indicated samples. IN, input; UB, unbound fraction; IP, immunoprecipitated fraction.
(C) Proteomic analyses of ZNFX-1 complex. The numbers of peptides identified in the crosslinked or non-crosslinked condition are shown. N2 control IP experiment in the crosslink condition served as a control.
(D) Proteomic analyses of WAGO-4 complex. The numbers of peptides identified in the crosslinked or non-crosslinked condition are shown. N2 control IP experiment in the crosslink condition served as a control.
CGH-1 and CAR-1 promote the localization of small RNA components in germ granules
Our analyses of P body and P granule interactions prompted us to examine whether P body factors, such as CGH-1 or CAR-1, regulate the localization of small RNA factors in germ granules. We first noticed that cgh-1 partial deletion mutant (ok492) animals exhibit a germline nuclei organization defect, where some germline nuclei detach from the surface and move to the center of the gonad (Figure 3A). Importantly, we found fewer perinuclear PRG-1 and CSR-1 perinuclear foci in cgh-1 mutants, including both in the partial deletion mutant (ok492) or a temperature-sensitive mutant (tn691) (Figures 3A, top, and S2A). Fluorescent recovery after photobleaching (FRAP) analyses further indicated that CGH-1 promotes the turnover of PRG-1 and CSR-1 in P granules (Figure S2B). We then examined the role of CGH-1 in regulating Z granules and Mutator foci. We found fewer perinuclear foci of Z granule factors, including ZNFX-1 and WAGO-4, as well as fewer MUT-16 foci, in cgh-1 mutants (Figures 3B, 3C, and S2C–S2D). Similar to cgh-1 mutants, the perinuclear localization of PRG-1 and WAGO-4 is also reduced in car-1 RNAi-treated animals (Figure S3A). Interestingly, the localization of P granule assembly factors, including PGL-1 and GLH-1, is not affected in cgh-1 (ok492), cgh-1 (tn691) mutants or in car-1 RNAi-treated animals (Figures 3A, bottom, S2A, and S3A). Except for WAGO-4 (see further characterizations of WAGO-4 later), the protein levels of PRG-1, PGL-1, CSR-1, ZNFX-1, GLH-1, and MUT-16 in cgh-1 mutants were found to be similar to those in the wild-type background (Figure S3B). Subsequently, we examined some of these factors in early embryos and observed that PRG-1 and WAGO-4, but not GLH-1, exhibited a reduced number of foci in the cgh-1 (tn691) mutant (Figure S4A). Together, our imaging analyses suggest that P body factors CGH-1 and CAR-1 specifically promote the localization of various small RNA pathway factors to germ granules.
Figure 3. P body factor CGH-1 promotes the localization of small RNA pathway factors at perinuclear germ granules.
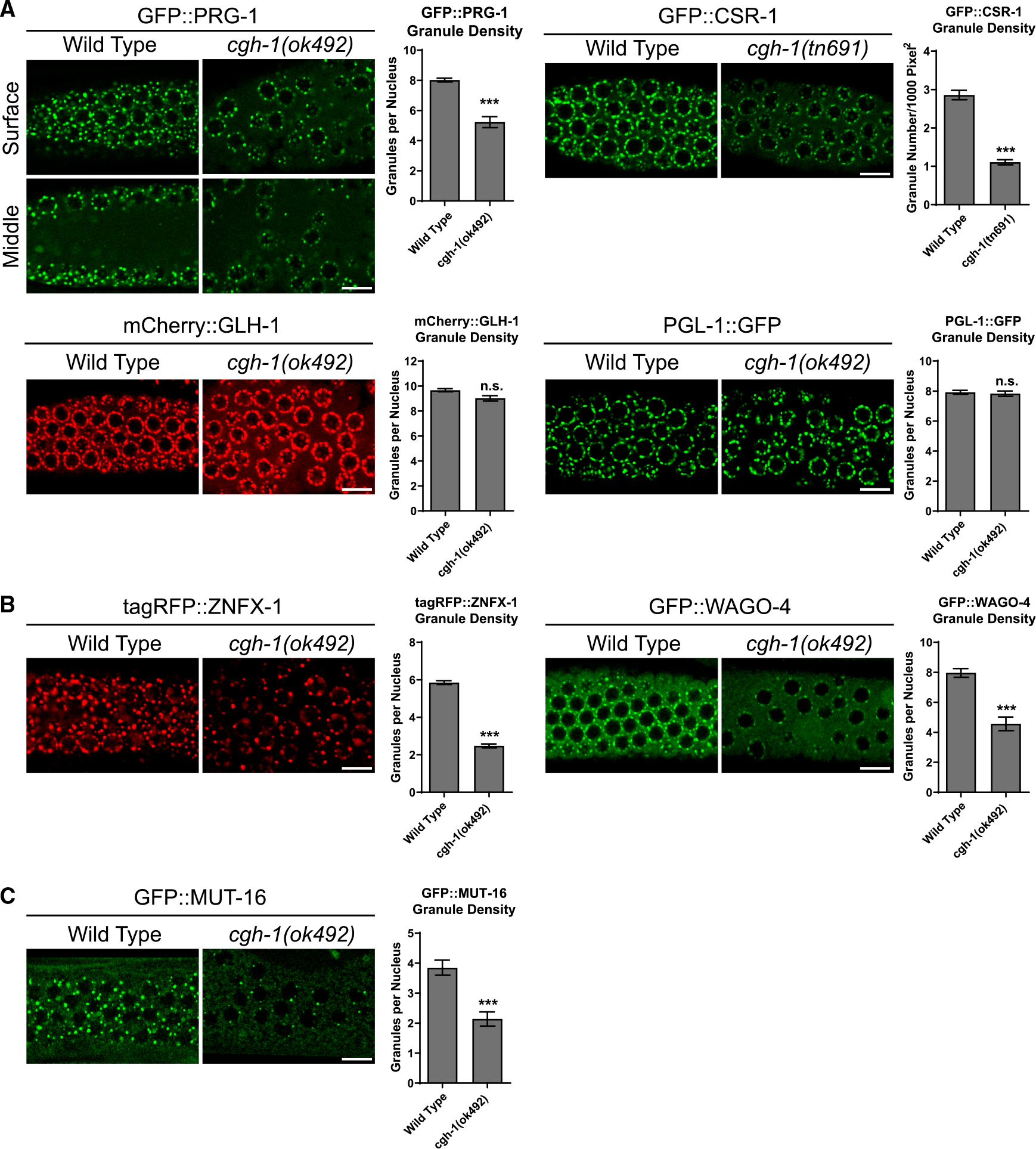
Representative fluorescent micrographs and corresponding quantification of granule density of indicated P granule markers (A), Z granule markers (B), and Mutator foci marker MUT-16 (C) in the pachytene region of adult germlines in wild-type or in cgh-1 (ok492) mutant animals. Bars: 10 μm. The images were captured from the surface section of the germline. Additionally, images of the middle section of the germline of GFP::PRG-1 were included to highlight the observed defects in germline nuclei organization in cgh-1 (ok492) mutant. Statistical analysis was performed using a one-tailed Student’s t test. Bars indicate the mean, and errors bars indicate the standard deviation. Distributions represent data collected from 8 to 12 independent gonad images.
We then examined whether the localization of P body factor CGH-1 to perinuclear foci requires P granule assembly. VASA-like helicases GLH-1 and GLH-4 have been reported to promote the perinuclear localization of various P granule factors.24,26 Surprisingly, while the localization of P granule factors, such as PRG-1, becomes diffuse upon glh-1 and glh-4 double RNAi treatment, the localization of CGH-1 is not affected (Figure S4B). In addition, we observed that CGH-1 perinuclear localization is not affected in mutants of the small RNA pathway, including prg-1, hrde-1, znfx-1, or mut-16 mutants (Figure S4C). This indicates that CGH-1’s perinuclear localization does not rely on P granule assembly factors GLH-1 and GLH-4 or piRNA pathway factors and implies that an alternative pathway contributes to the perinuclear localization of CGH-1.
CAR-1 promotes the interaction of CGH-1 with germ granules and supports CGH-1’s role in piRNA silencing
Our recent study has suggested that the P granule localization of small RNA pathway factors is critical for the proper recognition of mRNA targets by piRNAs.6 As the perinuclear localization of small RNA pathway factors is also compromised in cgh-1 mutants, we wondered whether piRNA-mediated gene silencing is defective in cgh-1 mutants. We used two sensitized piRNA reporters where the silencing of GFP transgenes by a synthetic GFP-targeting piRNA depends on both factors required for initiation and maintenance of piRNA silencing.27,28 We found that silencing of these piRNA reporters is compromised in cgh-1 (tn691) temperature-sensitive mutant animals, cgh-1 (dz407) null mutant animals, and in cgh-1 RNAi-treated animals (Figures 4A, 4B, S5A, and S5B). To examine the role of CGH-1’s ATPase activity in piRNA silencing, we generated gene-edited strains carrying CGH-1 mutation at its DEAD box motif (DEAD to DAAD or DQAD) known to impair its ATPase function.24,29,30 Nonetheless, these CGH-1 mutant proteins showed lower expression levels compared to wild-type CGH-1 (Figure S5C). We suspect these ATPase mutations may reduce the stability of CGH-1 protein. Consequently, further investigation is necessary to determine whether the ATPase activity of CGH-1 is essential for piRNA silencing or germ granule formation. We then examined the roles of other P body factors in piRNA silencing. While RNAi targeting P body factors appears to be highly effective, resulting in either sterile/embryonic lethal phenotypes or a reduction of over 90% in corresponding mRNA levels (Figure S5D), it is noteworthy that only RNAi of car-1 or cgh-1 resulted in the activation of our piRNA reporter (Figure 4C). Remarkably, only RNAi knockdown of car-1, but not of other P body factors examined, led to the dispersal of CGH-1 from perinuclear P bodies to the cytoplasm (Figure 4D). The levels of CGH-1 in car-1 RNAi treatments were not changed (Figure S5E). These observations show that CAR-1 promotes the formation of CGH-1 condensates and indicate that CGH-1’s perinuclear localization may be critical for CGH-1’s interaction with small RNA factors and for its function in piRNA silencing. We therefore examined whether the interactions between CGH-1 with P granule factors require CAR-1. Indeed, the interactions between CGH-1 with P granule factors WAGO-1 and PRG-1 were greatly reduced in car-1 RNAi-treated animals (Figure 4E). These observations show that the ability for P body factor CGH-1 to form perinuclear condensates correlates with its ability to interact with germ granules and to promote piRNA silencing.
Figure 4. P body factors CGH-1 and CAR-1 promote piRNA-dependent gene silencing.
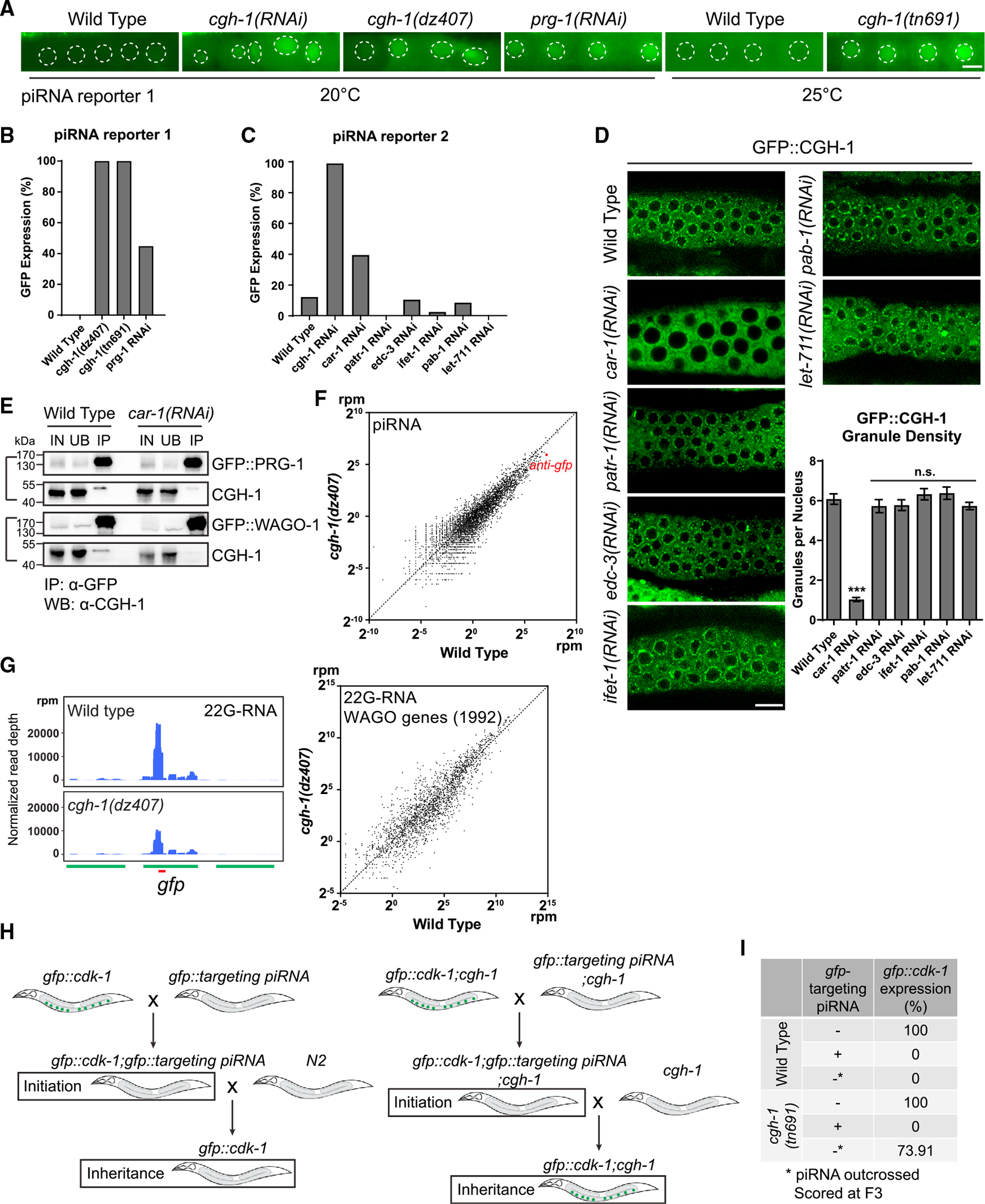
(A) GFP expression of the piRNA reporter #1 in the indicated strains. In this reporter, the expressions of both CDK-1:GFP (nuclear) and OMA-1:GFP (cytoplasm) are silenced by a synthetic GFP-targeting piRNA in wild-type animals. Worms were cultured at 20°C for imaging. For cgh-1 (tn691) mutant and its control wild-type animals, worms were grown till young adult stage at 20° and subsequently transferred to 25°C for a 12-h period before imaging. Dotted circles indicate the location of maturing oocyte nuclei. Bars: 10 μm.
(B) Percentage of screened animals showing GFP expression in the piRNA reporter #1 in the indicated strains. We define the expression of GFP (on/off) by observing whether the indicated mutant or RNAi-treated animals exhibit visible nuclear and cytoplasmic GFP signals in their germline.
(C) Percentage of screened animals showing GFP expression in the piRNA reporter #2 in the indicated strains. We define the expression of GFP (on/off) by observing whether the indicated RNAi-treated animals exhibit visible nuclear and cytoplasmic GFP signals in their germline.
(D) Fluorescent micrographs show the localization of P body marker CGH-1 in the indicated RNAi-treated animals. Bars: 10 μm. Granule density (number of granule/nuclei/section) of GFP::CGH-1 in wild-type or the indicated RNAi-treated animals was calculated. These quantifications correspond to micrographs in Figure 4D. Statistical analysis was performed using a one-tailed Student’s t test. Bars indicate the mean, and errors bars indicate the standard deviation. Distributions represent data collected from 8 to 12 independent gonad images.
(E) Western blot analyses showing that the interaction between CGH-1 and P granule factors PRG-1 or WAGO-1 is reduced in car-1 RNAi-treated animals. IN, input; UB, unbound fraction; IP, immunoprecipitated fraction.
(F) A scatterplot showing the abundance of piRNAs in wild-type worms compared to cgh-1 (dz407) mutant. The red dot indicates the levels of the synthetic GFP-targeting piRNA.
(G) 22G-RNAs distribution at GFP coding sequences in the indicated strains. The red bar indicates the location of the GFP sequence complementary to the GFP-targeting piRNA (left). A scatterplot shows the abundance of 22G-RNAs mapped to WAGO target genes in wild-type worms compared to cgh-1 (dz407) mutant (right).
(H) A schematic showing the GFP reporter assays to determine the role of CGH-1 in the initiation or maintenance of piRNA silencing.
(I) Percentage of GFP reporter expression from screened animals in the indicated strains. Note that cgh-1(tn691) mutant animals can initiate piRNA-directed silencing of the GFP reporter but exhibit defects in the inheritance of gene silencing.
CGH-1 promotes the inheritance of piRNA silencing
piRNAs trigger gene silencing against their targets through the production of secondary 22G-RNAs that are loaded onto WAGO Argonautes to mediate gene silencing.7,31 To examine how CGH-1 contributes to piRNA silencing, we compared the piRNA and downstream 22G-RNA levels in wild type versus cgh-1 mutants. In cgh-1 mutants, we observed a ~40% reduction of anti-GFP synthetic piRNA and secondary 22 RNAs at the synthetic piRNA targeting site (Figures 4F and 4G). However, analyses of small RNA sequencing data indicate that global piRNA and miRNA levels in the cgh-1 (dz407) mutant are predominantly unaffected (Figures 4F and S5F). Furthermore, unlike prg-1 mutant animals that display a global reduction in 22G-RNAs against genes regulated by WAGO Argonautes (WAGO targets),31 the levels of these 22G-RNAs remain largely unchanged in the cgh-1 (dz407) mutant (Figures 4G and, S6A). We observed that in the cgh-1 mutant, approximately 100 WAGO targets (5% of all WAGO targets) displayed a significant increase in 22G-RNA levels, while another 100 WAGO targets showed a significant decrease in 22G-RNA levels (Figure S6A). Although the underlying biological mechanism behind these changes in the cgh-1 mutant is currently unknown, they appear to be gene-specific alterations, as similar trends of 22G level changes are also observed in these genes in the biologically replicate data (Figure S6B). Collectively, these findings suggest that CGH-1 may play a role in regulating the levels of specific piRNAs or WAGO 22G-RNAs, but it is not essential for the biogenesis of either piRNA or WAGO 22G-RNAs.
As we observed that the sensitized piRNA reporter is activated following RNAi knockdown of znfx-1 (Figure S5B), which is required for transgenerational RNAi,8,9 we wondered if CGH-1 plays a regulatory role in piRNA silencing by promoting the inheritance of piRNA-induced gene silencing. Since CGH-1 is an essential gene, we cannot examine the role of CGH-1 in transgenerational gene silencing using a cgh-1-null allele. We reasoned that if the function of cgh-1 in the temperature-sensitive mutant (tn691) is partially compromised when these animals are grown at the permissive temperature, then this mutant would allow us to examine the role of CGH-1 in transgenerational gene silencing. In support of this hypothesis, we noticed that cgh-1 (tn691) mutant animals grown at the permissive temperature (20°C) already exhibit a more mild but similar phenotype to those grown at the non-permissive temperature (25°C), such as significantly reduced broods and the compromised formation of ZNFX-1 granules (Figures S7A and S7B). We further investigated the role of CGH-1 in the initiation of piRNA-induced silencing by crossing a strain containing a synthetic GFP-targeting piRNA with a strain carrying a GFP::CDK-1 transgene (Figure 4H). In the absence of the synthetic GFP-targeting piRNA, GFP::CDK-1 is expressed in both wild-type and cgh-1 (tn691) mutant animals grown at 20°C. However, when the GFP-targeting piRNA was introduced through the cross, we observed that 0% of either strain grown at 20°C exhibited detectable GFP::CDK-1 signals. This result suggests that cgh-1 (tn691) mutant animals are competent for triggering piRNA-directed gene silencing when raised at the permissive temperature. To examine the role of CGH-1 in the maintenance of gene silencing memory, we outcrossed the synthetic GFP-targeting piRNA from the wild-type or cgh-1 (tn691) mutant background and found that while all wild-type animals carrying GFP::CDK-1 transgene continue to exhibit no visible GFP::CDK-1 signals in the F3 generation, over 73% of cgh-1 (tn691) mutant animals carrying GFP::CDK-1 transgene exhibit expression (re-activation) of the GFP::CDK-1 (Figure 4I). These observations suggest that CGH-1 plays a critical role in promoting transgenerational gene silencing triggered by piRNAs.
CGH-1 promotes RNAi inheritance
Similar to piRNAs, double-stranded RNA (dsRNA) can also trigger transgenerational gene silencing lasting for several generations, a phenomenon known as RNAi inheritance.32 To determine whether CGH-1 plays a role in RNAi inheritance, we treated wild type and cgh-1 (tn691) mutants with GFP RNAi food (bacteria expressing dsRNA targeting GFP) and monitored the expression of the GFP reporter over generations (Figure 5A). We found that in the RNAi-treated generation (P0), both wild-type and cgh-1 (tn691) mutant animals exhibited robust GFP silencing, suggesting both strains are competent in triggering dsRNA-induced gene silencing (Figures 5B and 5C). Remarkably, while over 70% of the wild-type animals continued to keep the GFP reporter silenced for over three generations, less than 30% of the cgh-1 (tn691) mutants can maintain GFP silencing for a single generation in the absence of dsRNA (Figures 5B and 5C). In addition, in generations 2–6 (F2–F6) following the RNAi-treated generation (P0), we observed that a higher number of wild-type animals maintained GFP silencing compared to cgh-1 (tn691) mutant animals (Figures 5B and 5C). These observations suggest that CGH-1 promotes transgenerational gene silencing triggered by both piRNA and dsRNA.
Figure 5. CGH-1 promotes RNAi inheritance and 22G-RNA synthesis in inheriting generations.
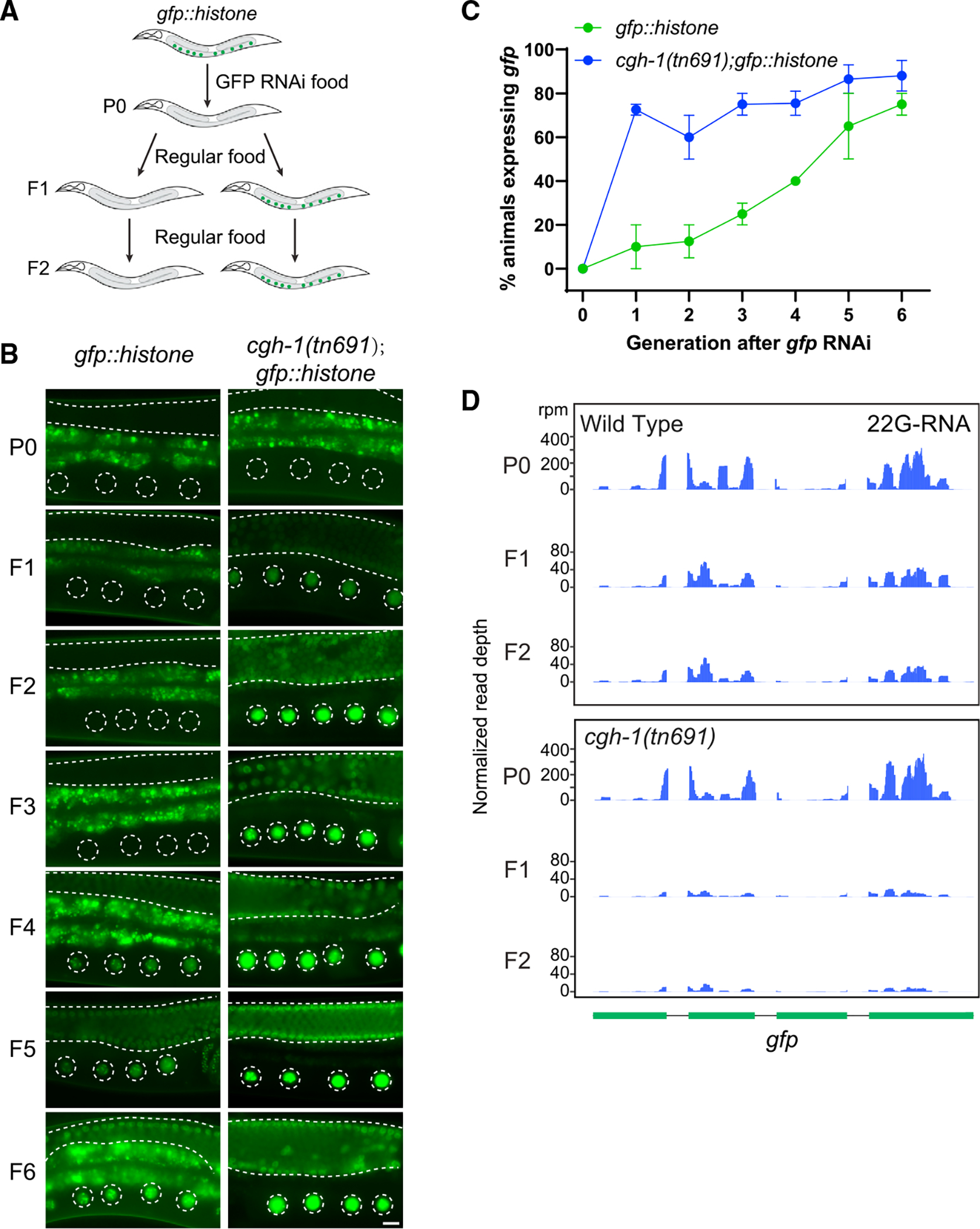
(A) A schematic showing the RNAi assay with a GFP::histone reporter to determine whether the strain can initiate RNAi and inherit gene silencing over generations. In these experiments, both wild-type and cgh-1 (tn691) mutant animals are grown at the permissive temperature (20°C).
(B) Fluorescent micrographs show representative images of the GFP::H2B reporter in the RNAi-treated generation (P0) or the following generations (F1 to F6) in wild-type and cgh-1 (tn691) mutant animals. Dashed circles indicate the location of maturing oocytes nuclei, and the area between two dashed lines indicates germline tissue. Note that the bright signals observed outside of these two highlighted areas are autofluorescence signals originating from C. elegans gut granules. Bars: 10 μm.
(C) Percentage of screened animals showing GFP reporter expression in wild-type and cgh-1 (tn691) mutant animals. Statistical analysis was performed using a one-tailed Student’s t test. Data points indicate the mean, and errors bars indicate the standard deviation. Distributions represent data collected from three independent experiments.
(D) Anti-sense 22G-RNAs distribution mapped to GFP coding sequences in the indicated strains from the RNAi-treated animals (P0) or the following generations (F1 or F2).
As previous studies have demonstrated the role of 22G-RNAs in RNAi inheritance,33,34 we performed small RNA sequencing to measure the levels of 22G-RNAs in the wild type and in the cgh-1 (tn691) mutants in the parental RNAi-treated animals as well as the untreated inheriting F1 and F2 progeny. In wild-type animals, we found that the production of anti-sense GFP-targeting 22G-RNAs are highest in the RNAi-treated generation (P0); then accumulation is reduced in the F1 and F2 generations in comparison to RNAi-treated animals (Figure 5D). Notably, the production of anti-sense GFP-targeting 22G-RNAs was largely normal in the RNAi-treated animals (P0) of cgh-1 (tn691) mutant animals but was much reduced in F1 and F2 generations compared to wild-type animals. Together, our data indicate that CGH-1 is uniquely required for the amplification of 22G-RNAs in the inherited generations and thus is critical for RNAi inheritance.
CGH-1 promotes the organization of germ granules into distinct sub-compartments
As described above, C. elegans germ granules are organized into distinct sub-compartments, including P granules, Z granules, and Mutator foci. As P granules, Z granules, and Mutator foci are enriched for factors involved in genome surveillance, RNAi inheritance, and 22G-RNA production, we wondered whether CGH-1 may regulate the organization of PZM granules to promote RNAi inheritance and 22G-RNA synthesis. We first confirmed that P granule marker PGL-1 and Z granule marker ZNFX-1 are partially separated in the adult germline, consistent with the previous report that these two granules are organized to distinct but partially overlapping sub-compartments (Figure 6A).8 Remarkably, we found that the separation of the Z and P granule markers is compromised in cgh-1 (tn691) mutants and in cgh-1 RNAi-treated animals, where they are nearly all co-localized (Figures 6A and 6B). The co-localization between PGL-1 and ZNFX-1 is also increased in car-1 RNAi-treated animals compared to control RNAi-treated animals (Figures 6A and 6B). It has been previously reported that P and Z granules are co-localized in P2 cells of early embryos (4-cell stage) before partially segregating from each other in Z2 and Z3 cells of late embryos (~100-cell stage).8 We therefore investigated whether the defects in P/Z granule separation observed in the adult germline of cgh-1 mutant animals could also be observed in embryos. We found that in P2 cells, both wild-type and cgh-1 (tn691) mutant animals exhibited a high degree of co-localization between P and Z granules. However, in Z2 and Z3 cells, we observed significantly more co-localization between P and Z granules in the cgh-1 (tn691) mutant compared to wild type (Figure S7C). These findings indicate that the separation of P and Z granules is compromised in cgh-1 (tn691) mutants, both in the late embryo and in the adult germline. Together, these observations suggest that CGH-1 and CAR-1 not only promote the formation of phase-separated condensates containing small RNA factors (Figures 3A–3C) but also promote the proper organization of germ granules.
Figure 6. CGH-1 promotes proper germ granule organization and WAGO-4 22G-RNA production.
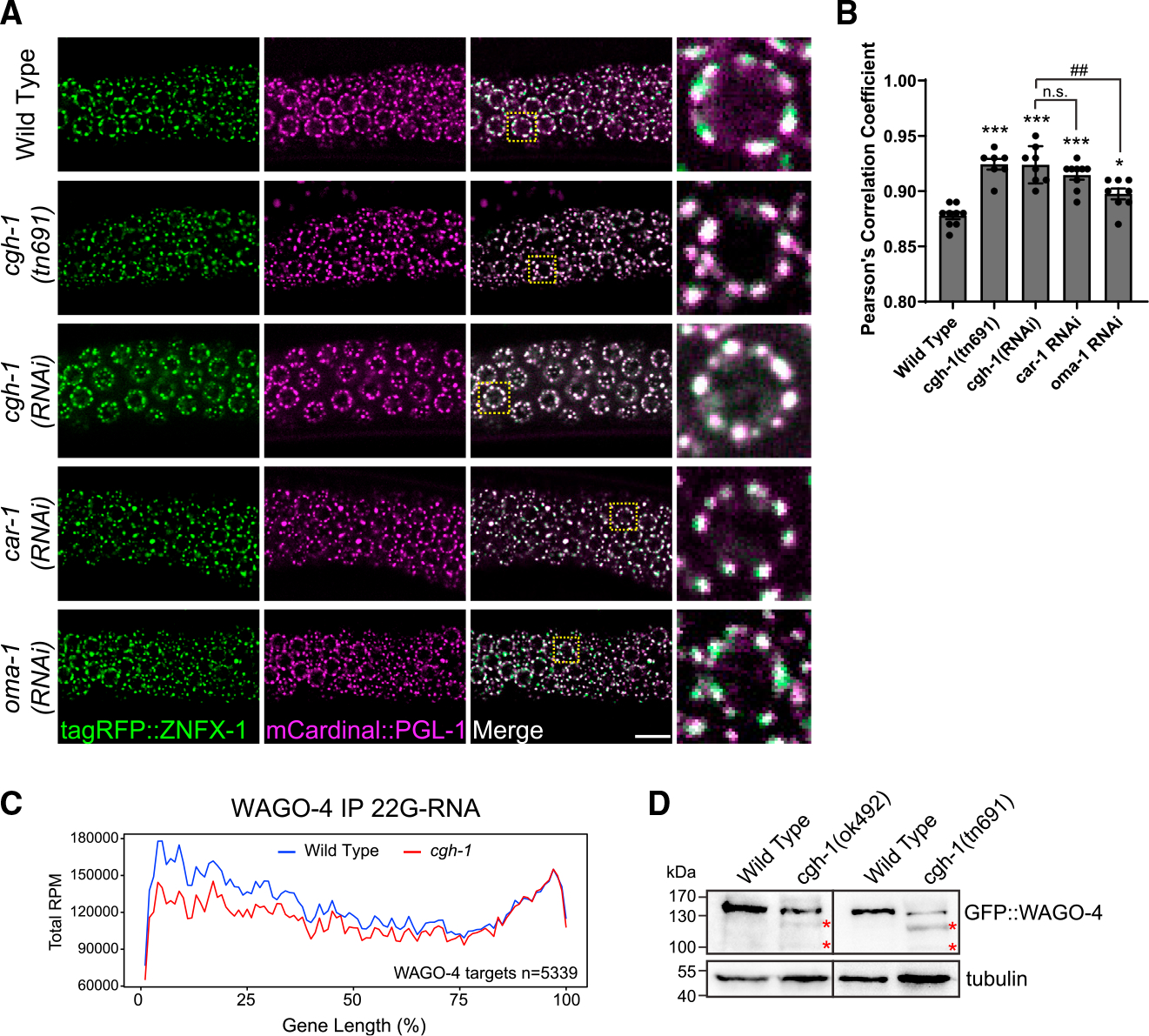
(A) Fluorescent micrographs show the localization of P granule marker PGL-1 and Z granule marker ZNFX-1 in the indicated mutant or RNAi-treated animals. Bars: 10 μm.
(B) Pearson’s correlation coefficient of PGL-1 and ZNFX-1 signals in the indicated mutant or RNAi-treated animals. Bars indicate the mean, and errors bars indicate the standard deviation. Distributions represent data collected from 8 to 12 independent gonad images.
(C) Metagene traces show the total accumulation of WAGO-4-associated 22G-RNAs by percentage of WAGO-4 target gene length in IP experiments. Reads from wild-type IP experiments are traced in blue, and reads from cgh-1 RNAi-treated IP experiments are traced in red.
(D) Western blots show the levels of WAGO-4 in wild type or in the indicated cgh-1 mutants. The red asterisks indicated partially degraded WAGO-4 proteins.
CGH-1 promotes the stability of WAGO-4 Argonaute and the accumulation of its associated small RNAs
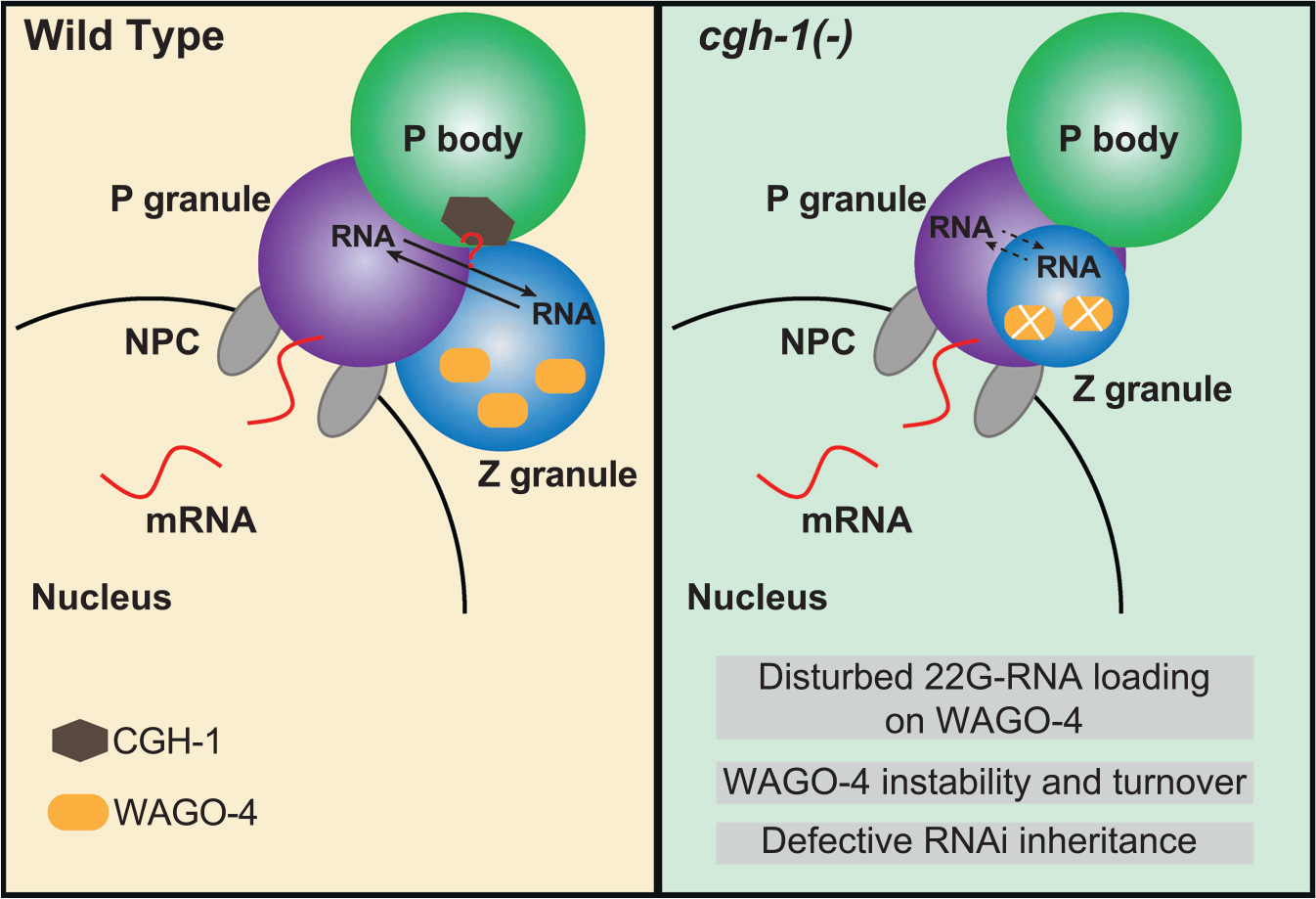
Previous studies have shown that WAGO-4 localizes to Z granules and plays a critical role in RNAi inheritance.8,34 As we found that CGH-1 is required for RNAi inheritance and promotes the organization of P and Z granules, we hypothesized that CGH-1 and CAR-1 regulate the trafficking of mRNA/protein complexes between P and Z granules so that 22G-RNAs can be made and associated with WAGO-4 to trigger transgenerational gene silencing. If so, we would expect the accumulation of WAGO-4 22G-RNAs to be compromised in CGH-1 mutants. To test this hypothesis, we performed WAGO-4 small RNA sequencing in wild-type and cgh-1 RNAi-treated animals. Since previous studies have shown that Z granule factor znfx-1 mutants exhibit a shift in 22G-RNA accumulation on mRNAs,9 we also monitored the distribution of 22G-RNAs. We noticed that the production of WAGO-4 22G RNAs is reduced in cgh-1 RNAi-treated animals, especially at the 5′ ends of mRNAs (Figure 6C). Notably, WAGO-4 protein levels were reduced and partially degraded in cgh-1 mutants (Figure 6D), while the levels of other P granule PRG-1, CSR-1, and GLH-1 protein remained normal (Figure S3B). As the absence of small RNAs can trigger the degradation of their associated Argonaute,35 the aberrant degradation of WAGO-4 proteins in cgh-1 mutants may be caused by the reduction in WAGO-4 small RNA levels. Together, our observations show the critical roles of CGH-1 in producing WAGO-4 small RNAs and further support a model where P body factor CGH-1 promotes proper germ granule organization that is critical for the production of small RNA-based gene silencing memories (Figure 7).
Figure 7. A model showing the roles of P body factor CGH-1 in germ granule organization and WAGO-4 22G-RNA production that promote transgenerational gene silencing.
DISCUSSION
While various interactions between distinct cellular condensates have been reported, the significance of such interactions is largely unexplored. In mice, flies, and worms, previous studies have demonstrated the association between P bodies and germ granules, which are distinct phase-separated condensates, in germ cells.12–14 In this study, we characterized the interactions between these two condensates in C. elegans with high-resolution imaging and proteomic approaches. We found that P bodies and germ granules interact with specific spatial arrangements in the germline in C. elegans, and this interaction promotes the proper formation and organization of germ granules. Specifically, our imaging analyses revealed that P bodies are situated at the cytoplasmic side of perinuclear germ granules (Figure 1B). Notably, previous studies have provided evidence that P body factors form complex structures at the outer surface of P granules in early embryos.14,36 These findings, along with our own observations, indicate that P bodies and germ granules exhibit a similar mode of interaction across different developmental stages in C. elegans. Using MS and IP-western analyses, we further identified the molecular interaction network between these two condensates, where P body factors, specifically CGH-1 and CAR-1, interact with small RNA factors enriched in P and Z granules, two sub-compartments of germ granules.8 Notably, our genetic analyses further revealed that P body factors, including CGH-1 and CAR-1, promote the formation and proper organization of germ granules. In particular, we found CGH-1 and CAR-1 promote the phase separation of small RNA pathway factors, such as PRG-1, CSR-1, and WAGO-4, but not of P granule assembly factors, such as PGL-1 or GLH-1 (Figures 3A, S2A, and S5A). In addition, in cgh-1- and car-1-depleted animals, the separation of P and Z granules within germ granules is compromised (Figure 6A). Together these findings suggest that P body factors CGH-1 and CAR-1 control the localization and organization of small RNA factors in germ granules. Interestingly, previous studies have demonstrated that CGH-1/DDX6 RNA helicase is involved in the formation of both P bodies and stress granules, two condensates known to interact in vivo.13,37 This suggests that CGH-1/DDX6, as a P body factor, may have a broader role in regulating the dynamics of various condensates that interact with P bodies. Together, our analyses demonstrate that the internal organization and formation of one condensate can be controlled by its interacting condensate, raising the possibility that various reported interactions between distinct condensates may also regulate condensate formation in space and time.
Importantly, our study suggests that P body-germ granule interaction is functionally important. Since distinct sub-compartments of germ granules, including P granule, Z granule, and Mutator foci, are enriched for factors involved in distinct steps of small RNA silencing,8,10 it was proposed that these special arrangements are critical for executing gene silencing processes occurring in sequential order: first in P granules and then in Z granules and Mutator foci.2 Our data provide experimental support for this model, as cgh-1 mutant animals exhibited defects where Z granules fail to separate from P granules. They also exhibit phenotypic defects associated with Z granule loss, where small RNA-guided gene silencing can be initiated but cannot be inherited.8,9 We further traced the transgenerational inheritance defects in cgh-1 mutants to defects in secondary 22G-RNAs synthesis and instability of Z granule factor WAGO-4, both known carriers of gene silencing signals over generations.8,34 It is currently unclear whether the mis-organization of P/Z granules observed in cgh-1 mutants is the cause or consequence of the defect in small RNA amplification. Nonetheless, our observations suggest that P body and germ granule interactions are critical for the function of germ granule factors to establish transgenerational gene silencing. One intriguing model is that CGH-1 is critical for the transfer of silenced mRNAs from P granules to Z granules, where these mRNAs can act as templates for small RNA. The absence of 22G-RNA amplification at the Z granule may consequently result in the destabilization of WAGO-4 and contribute to deficiencies in the transgenerational inheritance of gene silencing. Interestingly, it has been proposed that CGH-1 homolog DDX6 may regulate stress granule formation by promoting mRNA transfer from P bodies to stress granules.38 Future work is needed to examine whether condensate interactions are broadly involved in regulating RNA trafficking and processing between distinct condensates.
In summary, our study uncovers the critical role P body factors play in the formation and function of germ granules. piRNA silencing factors are enriched in germ granules across diverse animals, such as the nuage in Drosophila and the chromatoid body in mammals. Since it has been reported that Drosophila AGO3 (a PIWI Argonaute) associates with the CGH-1 homolog ME31B,39 it will be interesting to examine whether the conserved P body factors CGH-1 and CAR-1 also regulate germ granule formation/organization and contribute to piRNA silencing pathway in other animals in the future. As more cases of interactions between distinct cellular condensates are reported, our study raises the possibility that condensate interactions can provide an efficient way for cells to quickly evolve novel functions using factors that are enriched in distinct cellular compartments.
Limitations of the study
Although we have identified several defects in germ granule formation and function associated with car-1 RNAi-treated worms and cgh-1 mutants, the specific molecular mechanisms by which CGH-1 and CAR-1 contribute to these processes remain unknown. Unfortunately, due to the protein instability defects of CGH-1 DQAD and DAAD mutant proteins, our investigation into the roles of ATPase function of CGH-1 remains inconclusive. Future experiments are needed to investigate how CAR-1 promotes the localization of CGH-1 and to elucidate the molecular functions of CGH-1 and CAR-1 in gene silencing, including their potential role in regulating RNA dynamics in germ granules and/or P bodies. In addition, while our study has demonstrated that CGH-1 and CAR-1 are involved in regulating the localization of small RNA factors to germ granules, it is likely that interactions between CGH-1, CAR-1, and small RNA factors also occur outside of germ granules, and their interactions may vary in distinct stages during germ cell development. Therefore, further investigations are needed to determine the specific locations and developmental stages where the interactions between these two condensates contribute to transgenerational gene silencing.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Heng-Chi Lee (hengchilee@uchicago.edu).
Materials availability
C. elegans strains and plasmids generated in this study are available from the lead contact upon request.
Data and code availability
Small RNA sequencing data are deposited to Genome Sequence Archive (GSA) in National Genomics Data Center with the access number CRA008367. The proteomic data are deposited to Proteomics Identification database (PRIDE) with the access number of PXD037342. All custom scripts used for bioinformatic analysis are available at Zenodo: https://doi.org/10.5281/zenodo.8097062 or at Github: https://github.com/Uchicago-BSD-hlee-lab/Du_2023/. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained on standard nematode growth media (NGM) plates seeded with the Escherichia coli OP50 strain at 20°C or temperatures where indicated. Bristol N2 strain is used as the wild-type animal. Additional strains and alleles used in this study are listed in Table S2. The cgh-1 (dz407) allele used in this study is a gene-edited cgh-1 null allele that was generated in our research. It carries a deletion of the G nucleotide from the CGH-1 start codon (ATG), accompanied by the insertion of a short sequence (tccgatgtccgatgtcgacgctcc) at that site. As a consequence, this strain may produce an unrelated short peptide, but it does not produce any CGH-1 protein. In addition, strains carrying cgh-1 DQAD and cgh-1 DAAD mutations were immediately crossed into genetic balancer strains for maintenance. Some strains were obtained from the Caenorhabditis Genetics Center (CGC).
METHOD DETAILS
RNA interference
RNA interference (RNAi) was performed by feeding animals with E. coli HT115 (DE3) strains expressing the appropriate double-stranded RNA (dsRNA). RNAi bacterial strains were obtained from the Ahringer C. elegans RNAi Collection (Source BioScience) or the collection derived from C. elegans ORFeome Library (Horizon Discovery). Bacterial cμltures were grown in Luria broth supplemented with 100 μg/mL ampicillin overnight at 37°C. Cultures were seeded on NGM plates containing 100 μg/mL ampicillin and 1 mM IPTG and incubated at room temperature for 24 h. L1 or L4 hermaphrodites were picked onto the plates for feeding at 20°C prior to score for phenotypes. HT115 (DE3) expressing empty RNAi vector L4440 was used as the control.
Brood size
Single hermaphrodite L4s (P0s) were placed onto individual freshly seeded NGM plates and allowed to grow for 24 h at 16, 20 or 25°. P0 adults were transferred to new NGM plates every 24 h until they no longer laid eggs. All the F1 progeny on each plate were counted. The brood size of each P0 animal is the total sum of F1s for all plates where the P0 animal laid eggs. At least 10 P0 animals were used to calculate the brood size for each strain.
CRISPR CAS9-mediated gene editing
CRISPR-Cas9 guide RNAs were designed with the online IDT Alt-R Custom Cas9 crRNA Design Tool. DNA fragments were amplified by PCR and transcribed to sgRNAs in vitro with HiScribe Quick T7 High Yield RNA Synthesis Kit (New England Biolabs), and sgRNAs were purified with Monarch RNA Cleanup Kit (New England Biolabs). sgRNA sequences and PCR primers used in this study are listed in Table S3.
Single-stranded oligodeoxynucleotides (ssODNs) was used as donor templates for introducing cgh-1 DQAD and DAAD point mutations, and partially single-stranded DNAs was used as donor templates for in situ gene knock-in using a modified method from.40 GFP/mCherry/tagBFP coding sequences and the coding sequences with 40 bp homology arms were amplified by PCR. Purified PCR products were mixed as 1:1 and treated by a denaturation and renaturation process [95°C 2:00 min, 85°C 10 s, 75°C 10 s, 65°C 10 s, 55°C 1:00 min, 45° 30 s, 35° 10 s, 25° 10 s, 4° hold] to generate hybrid DNA donors.
CRISPR experiments were conducted with Cas9 RNP strategies. Final concentrations of injection components were as follows: 250 ng/μL of Alt-R Cas9 protein (IDT) and 200 ng/uL in vitro synthesized sgRNA. All components were mixed and incubated at 37°C for 10 min, then 200 ng/μL of ssODN donor or hybrid DNA donor and 40 ng/μL of pRF4 rol-6 (dm) were added. Injection mixtures were introduced into the germline of C. elegans young adults by microinjection. The F1 rollers were isolated and genotyped to identify the desired transgenes.
RNAi inheritance assay
Synchronized L1 animals (P0) were RNAi treated by feeding bacteria expressing gfp dsRNA or the empty vector L4440. F1 embryos were collected by bleaching P0 animals and grown on regular OP50 bacteria. Embryos from following generations were collected by the same method. The expression of GFP in ~80 animals in each generation was imaged and scored on a Zeiss Axio Scope A1 with a Plan-Neofluar 63x/1.25 Oil M27 objective and a Retiga R6 charge-coupled device camera. Total RNA from P0, F1 and F2 generations were extracted and small RNAs were cloned and sequenced as described above. All RNAi inheritance assays were performed at 20°C.
Immunoprecipitation
A total of ~100,000 synchronized young adult animals were used for immunoprecipitation. Synchronized animals were washed with M9 buffer three times before frozen in liquid nitrogen and stored at −80°C. Pellets were resuspended in equal volumes of immunoprecipitation buffer [20 mM Tris-HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 80 U ml−1 RNase inhibitor (Invitrogen), 1 mM dithiothreitol, and protease inhibitor cocktail without EDTA (Promega)], and grinded in a glass grinder for 8–10 times. The whole grinding process should not exceed 10 min. Lysates were clarified by spinning down at 15000 rpm, 4°C, for 15 min. Supernatants were incubated with GFP-Trap magnetic agarose beads (ChromoTek) or anti-HA-tag magnetic beads (MBL) at 4°C for 2 h. Beads were washed with IP wash buffer [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, and 1 mM dithiothreitol] six times for 10 min each time and PBS buffer twice for 5 min each time, and then resuspended in PBS buffer for mass spectrometry analysis, Western blot, or RNA extraction.
Chemical crosslink
Chemical cross-linking of proteins was performed with dithio-bismaleimidoethane (DTME) (Sigma-Aldrich) dissolved in dimethyl sulfoxide (DMSO). ~100,000 synchronized young adults were collected and washed three times with M9 buffer. M9 buffer was discarded to the same amount of the worm volume, then DTME was added to a final concentration of 2 mM. Samples were incubated for 30 min at room temperature with occasional shaking then washed three times with M9 buffer to remove excess cross-linker. Worm pellets were resuspended in equal volume of immunoprecipitation buffer [20 mM Tri-HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 0.5 mM dithiothreitol, 80 U/mL RNase inhibitor (Invitrogen), protease inhibitor cocktail without EDTA (Promega)]. Worm pellets were homogenized using glass homogenizer for 15–20 strokes on ice. Lysates were centrifuged at 14000 × g for 10 min to remove insoluble material. Supernatants were incubated with GFP-Trap magnetic agarose beads (ChromoTek) or anti-HA-tag magnetic beads (MBL) for 2 h at 4°C on an end-to-end rotator. Supernatant was removed and beads were washed with 1 mL of wash buffer [20 mM Tris-HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40] six times for 10 min each time and with a final wash of 0.05% NP-40. Beads were incubated at 37°C for 30 min in 50 μL de-crosslinking buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.2% Tween 20, 10 mM dithiothreitol]. The final samples were boiled in 2 x SDS loading buffer at 100°C for 5 min, then analyzed by mass spectrometry.
Mass spectrometry
The samples were subjected to SDS PAGE gel electrophoresis experiments, and gel slices were subjected to in-gel digestion with Trypsin (Promega) at 37°C overnight. Purified peptides were desalted using a C18 Stage Tip column (Pierce), concentrated and dried. Then the peptides are reconstituted with 0.1% formic acid aqueous solution for subsequent liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis using a Q Exactive HF-X mass spectrometer (Thermo Scientific) equipped with a nanoliter flow rate Easy-nLC 1200 chromatography system (Thermo Scientific). The buffers used in the liquid chromatography separation consisted of buffer A [0.1% formic acid] and buffer B [80% acetonitrile, 0.1% formic acid]. The liquid chromatography separation was achieved by linear gradient elution at a flow rate of 300 nL/min. The related liquid phase gradient was as follows: 0–3 min, linear gradient of liquid B from 2% to 8%; 3–43 min, linear gradient of liquid B from 8% to 28%; 43–51 min, linear gradient of liquid B from 28% to 40%; 51–52 min, linear gradient of liquid B from 40% to 100%; 52–60 min, linear gradient of liquid B maintained at 100%. The Q Exactive HF-X mass spectrometer was used in the data dependent acquisition (DDA) mode. The full scan survey mass spectrometry analysis was performed as detection mode of positive ion, analysis time of 60 min, scan range of 350–1800 m/z, resolution of 60,000 at m/z 20, AGC target of 3e6, and maximum injection time (IT) of 50 ms. The 20 highest intensity precursor ions were analyzed by the secondary mass spectrometry (MS2) with resolution of 15,000 at m/z 200, AGC target of 1e5, and maximum IT of 25 ms, activation type of HCD, isolation window of 1.6 m/z, and normalized collision energy of 28. The mass spectrometry data were analyzed using MaxQuant 1.6.1.0 and used the database from Uniprot Protein Database (species: Caenorhabditis elegans). The complete analyzed results described in this study can be found at Table S1.
Western blot
Immunoprecipitated proteins or lysates prepared from ~100 synchronized young adult worms by boiling worms in worm boiling buffer [100 Mm Tris-HCl pH 6.8, 8% SDS, 20 mM 2-mercaptoethanol] for 10 min and in 1× SDS sample buffer for 5 min were used for Western blot. Proteins were separated by standard SDS-PAGE and transferred to PVDF membranes (Roche) using a Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked in TBST (20 mM Tris-HCl pH 7.4, 150 mM NaCl, and 0.05% Tween 20) containing 5% skimmed milk for 2 h at room temperature, then probed with primary antibodies against CGH-1 (1:2000), GFP (1:500, Santa Cruz Biotechnology), mCherry (1:1000, Abcam), HA (1:1000, Cell Signaling Technology) or tubulin (1:2000, Sigma-Aldrich) overnight at 4°C. After five washes with TBST buffer, the membranes were incubated with secondary antibodies against the species of the primary antibodies at room temperature for 1 h and then washed five times with TBST. ECL substrates were used for detection of proteins by a Tanon 5200 Chemiluminescent Imaging System (Tanon).
Immunohistochemistry
CGH-1 antibody staining was performed on dissected adult C.elegans gonads. Staged young adult worms were washed with M9 and resuspended in Dissection Buffer (1xPBS, 1 mM EDTA) then deposited onto poly-L-lysine coated glass coverslips. Adults were dissected using hypodermic needles, and gonads were fixed in Fixation Buffer (1xPBS, 3.7% PFA) for 45 min at room temperature. Gonads were washed twice with Antibody Wash Buffer (1xPBS, 1 mM EDTA, 0.1% Tween 20) and stored in 75% ethanol for two nights at 4°C. Gonads were washed three times with Antibody Wash Buffer and incubated with custom-made anti-rabbit CGH-1 antibody (1:4000 diluted in 250 μL of Antibody Wash Buffer) overnight at 4°C.Gonads were washed three times with Antibody Wash Buffer and incubated with Alexa Fluor 546 donkey anti-rabbit (Invitrogen, 1:500 diluted in 500 μL of Antibody Wash Buffer) for 1 h at room temperature. Gonads were washed three times with Antibody Wash Buffer and stained with DAPI. Gonads were washed three times with 1xPBS, air dried for 20 min and mounted with ProLong Diamond Antifade Mountant (Invitrogen). Samples were imaged on a Zeiss LSM800 confocal microscope equipped with AiryScan detector and a Plan-Apochromat 63X/1.4 Oil DIC M27 objective.
Quantitative real-time PCR
1 μg of total RNA was reverse transcribed with HiScriptII Q Select RT SuperMix for qPCR (Vazyme). Each cDNA sample was amplified using ChamQ SYBR qPCR Master Mix (Vazyme) on a CFX Connect Thermal Cycler (Bio-Rad). The relative fold changes of related mRNAs were normalized to expression levels of actin. The experiments were repeated in three biological replicates. The PCR primers are listed in Table S3.
RNA extraction and small RNA libraries
Total RNA was extracted using the standard method with TRIzol reagent (Invitrogen) from immunoprecipitated samples or whole animals of ~100,000 synchronized young adults.
Small (<200nt) RNAs were enriched with mirVana miRNA Isolation Kit (Ambion). In brief, 80 μL (200–300 μg) of total RNA, 400 μL of mirVana lysis/binding buffer and 48 μL of mirVana homogenate buffer were mixed well and incubated at room temperature for 5 min. Then 176 μL of 100% ethanol was added and samples spun at 2500 × g for 4 min at room temperature to pellet large (>200nt) RNAs. The supernatant was transferred to a new tube and small (<200nt) RNAs were precipitated with pre-cooled isopropanol at −80°C. Small RNAs were pelleted at 20,000 × g at 4°C for 30 min, washed once with 70% pre-cooled ethanol, and dissolved with nuclease-free water. 10 μg of small RNA was fractionated on a 15% PAGE/7M urea gel, and RNA from 18 nt to 30 nt was excised from the gel. RNA was extracted by soaking the gel in 2 gel volumes of NaCl TE buffer [0.3 M NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 7.5] overnight. The supernatant was collected through a gel filtration column, RNA precipitated with isopropanol, washed once with 70% ethanol, and resuspended with 15 μL nuclease-free water. RNA samples were treated with RppH to convert 22G-RNA 5′ triphosphate to monophosphate in 1x reaction buffer, 10U RppH (New England Biolabs), and 20U RNase inhibitor (Invitrogen) for 3 h at 37°C, followed by 5 min at 65°C to inactivate RppH. RNA was then concentrated with the RNA Clean and Concentrator-5 Kit (Zymo Research). Small RNA libraries were prepared according to the manufacturer’s protocol of the NEBNext Multiplex Small RNA Sample Prep Set for Illumina-Library Preparation (New England Biolabs). The required number of PCR cycles to amplify library is determined by running PCR for 10–15 cycles and analyzing PCR products on a 6% PAGE gel. NEBNext Multiplex Oligos for Illumina Index Primers (New England Biolabs) were used for library preparation. Libraries were sequenced using Illumina NovaSeq 6000 system.
QUANTIFICATION AND STATISTICAL ANALYSIS
Bioinformatics
sRNA-seq
Fastq reads were trimmed using custom perl scripts. Trimmed reads were aligned to the C.elegans genome build WS230 with GFP transgenes added as new chromosomes using bowtie ver 1.2.1.141 with options -v 0 –best –strata. After alignment, reads that were between 17 and 40 nucleotides in length were overlapped with genomic features (rRNAs, tRNAs, snoRNAs, miRNAs, piRNAs, protein-coding genes, pseudogenes, transposons) using bedtools intersect. Annotations for miRNAs were according to miRBase 16, and piRNA annotations were as previously described using PRG-1 IP data.5 Sense and antisense reads mapping to individual miRNAs, piRNAs, protein-coding genes, pseudogenes, RNA/DNA transposons, simple repeats, and satellites were totaled and normalized to reads per million (RPM) by multiplying by 1e6 and dividing read counts by total mapped reads, minus reads mapping to structural RNAs (rRNAs, tRNAs, snoRNAs) because these sense reads likely represent degraded products. Reads mapping to multiple loci were penalized by dividing the read count by the number of loci they perfectly aligned to. 22G-RNAs were defined as 21 to 23 nucleotide long reads with a 5′G that aligned to protein-coding genes, pseudogenes, or transposons. 22G-RNAs against WAGO targets were considered if they met the above criteria and also aligned antisense to the coding region of WAGO targeted genes, defined as genes whose mapped 22G-RNAs exhibit over 2-fold enrichment from either WAGO-1 IP5 than that from input 22G-RNAs or WAGO-9 IP42 than that from input 22G-RNAs RPM values were then used in all downstream analyses using custom R scripts using R version 4.0.3,43 which rely on packages ggplot2,44 reshape2,45 ggpubr,46 dplyr.47 To determine the increased or decreased 22G-RNA gene list in cgh-1 mutant, a Bayesian approach as described previously was used: two models were compared to determine p values.48 The first model states that a given gene has the same probability of accumulating positive reads between two samples. The second model states that a given gene’s probability of accumulating positive reads is unique between two samples. The probability of each model for each gene was calculated and compared to yield the final probability (p value) for each gene’s read accumulation being the same between the two samples. Significance was determined with Bonferroni correction p < 0.05. Browser images were constructed by calculating normalized read depth using bedtools genomecov49 to obtain bedgraph files for each library, then a custom R script was used to build plots. All custom scripts can be found at Zenodo: https://doi.org/10.5281/zenodo.8097062 or at GitHub: https://github.com/Uchicago-BSD-hlee-lab/Du_2023/.
Metagene analysis
Metagene profiles across gene lengths were calculated by computing the depth at each genomic position using 21 to 23 nucleotide long small RNA reads with a 5′G using bedtools genomecov.49 A custom R script was then used to divide genes into 100 bins and sum the normalized depth within each bin. Groups of genes were then plotted using the sum of the normalized depth at each bin. Traces represent normalized depth for 22G-RNAs mapping to WAGO-4 targeted genes (n = 5339), defined as genes with at least 5 22G-RNA reads per million mapped reads in IP and input libraries, whose mapped 22G-RNAs exhibit over 2-fold enrichment from WAGO-4 IP than that from input 22G-RNAs. All custom scripts can be found at https://doi.org/10.5281/zenodo.8097062 or at GitHub through https://github.com/Uchicago-BSD-hlee-lab/Du_2023/.
Imaging
Fluorescent proteins were visualized in living nematodes by mounting young adult animals on 2% agarose pads with M9 buffer [22 mM KH2PO4, 42 mM Na2HPO4, and 86 mM NaCl] with 10–50 mM levamisole. Fluorescent images were captured using a Zeiss LSM800 confocal microscope equipped with AiryScan detector and a Plan-Apochromat 63X/1.4 Oil DIC M27 objective. Colocalization studies between CGH-1 and P granule proteins were performed by the 2D Superresolution mode for Zeiss Airyscan. Images were taken by ZEN 2.3 (blue edition) acquisition software.
Images were processed and germ granule fluorescence was quantified in ImageJ. For germ granule density analysis, 10 nuclei in each germline pachytene region were randomly selected and perinuclear puncta were counted manually. For quantitative co-localization analysis, all image manipulations were performed with ImageJ using the Coloc 2 plugin. Images of 8–12 gonads were collected and quantified.
Fluorescence recovery after photo bleaching
Photobleaching and fluorescence recovery of GFP::PRG-1 and GFP::CSR-1 were conducted and recorded using a Zeiss LSM 800 confocal microscope with a Plan-Apochromat 63×/1.4 Oil DIC M27 objective, controlled by the ZEN software. The ZEN built-in fluorescence recovery after photo bleaching (FRAP) module was used to perform FRAP experiments. Bleaching was performed using 100% laser power in the 488nm channel. Multiple granules were selected in each imaging field and fluorescence intensity of selected granules was bleached to less than 20% of the intensity before photobleaching. Images were acquired every 1 s during a recovery phase of 120 s after bleaching. Images were processed and quantified in ImageJ. The total fluorescence intensity was measured for areas containing bleached granules (I), unbleached granules (Inorm), or areas without granules (Ibkg) at each time point. Fluorescence recovery ratios were calculated as (In−Ibkgn)(Inormi/Inormn)/(Ii−Ibkgi), where n stands for time points after photobleaching and i stands for initial phase before photobleaching. Recovery ratios for >10 granules at different time points were calculated for each strain.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-CGH-1 | This paper | N/A |
| Mouse monoclonal anti-GFP (B-2) | Santa Cruz Biotechnology | Cat#sc-9996; AB_627695 |
| Mouse Monoclonal Anti-α-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T6199; RRID:AB_477583 |
| Rabbit monoclonal anti-HA-Tag | Cell Signaling Technology | Cat#3724S; RRID:AB_1549585 |
| Rabbit recombinant anti-mCherry | Abcam | Cat#ab213511; RRID:AB_2814891 |
| Peroxidase-conjugated Affinipure Goat Anti-Mouse | Proteintech | Cat#SA00001–1; RRID:AB_2722565 |
| HRP-Goat Anti Rabbit IgG (H + L) | ZSGB-Bio | Cat#ZB2301; RRID:AB_2747412 |
| Alexa Fluor 546 donkey anti-rabbit | Invitrogen | Cat#A10040; RRID:AB_2534016 |
| GFP-Trap Magnetic Agarose | Chromotek | Cat#gtma; RRID:AB_2631358 |
| Anti-HA-tag mAb-Magnetic Beads | Medical & Biological Laboratories | Cat#M180–11; RRID:AB_11124524 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Dithio-bis-maleimidoethane (DTME) | Sigma-Aldrich | Cat#803618; CAS:71865–37-7 |
| TRIzol Reagent | Invitrogen | Cat#AM9738 |
| SUPERase-in RNase Inhibitor | Invitrogen | Cat#AM2694 |
| Protease Inhibitor Cocktail (50X) | Promega | Cat#G6521 |
| RNA 5’Pyrophosphohydrolase (RppH) | New England Biolabs | Cat#M0356S |
| Isopropyl-beta-D-thiogalactopyranoside (IPTG) | Biotopped | Cat#I6070 CAS:367–93-1 |
| 1-Bromo-3-chloropropane (BCP) | Macklin | Cat#B802344 CAS:109–70-6 |
| SYBR Gold nucleic acid gel stain | Invitrogen | Cat#S11494 |
| Dithiothreitol (DTT) | Sangon Bioteh | Cat#A620058; CAS:3483–12-3 |
| Long AMP Taq 2X Master Mix | New England Biolabs | Cat#M0287S |
| 2X Taq Plus Master Mix II | Vazyme | Cat#P213–01 |
| Phanta Max Super-Fidelity DNA polymerase | Vazyme | Cat#P505-d1 |
|
| ||
| Critical commercial assays | ||
|
| ||
| NEBNext Multiplex Small RNA Library Prep Set for Illumina (Set 1) | New England Biolabs | Cat#E7300S |
| NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) | New England Biolabs | Cat#E7335S |
| mirVana miRNA Isolation Kit | Ambion | Cat#AM1561 |
| VAHTS Unversal V8 RNA-seq Library Prep kit for Illumina | Vazyme | Cat#NR605–00 |
| Library Preparation VAHTS RNA Adapters set 1 for Illumina | Vazyme | Cat#N803–01 |
| Library Preparation VAHTS mRNA Capture Beads | Vazyme | Cat#N401–01 |
| VAHTS DNA Clean Beads | Vazyme | Cat#N411–01 |
| HiScript II Q Select RT SuperMix for qPCR(+gDNA wiper) | Vazyme | Cat#R233 |
| 2X ChamQ SYBR qPCR Master Mix | Vazyme | Cat#Q311–02 |
| RNA Clean and Concentraor-5 | ZYMO RESEARCH | Cat#R1015 |
| Immobilon Western Chemiluminescent HRP Substrate | Millipore | Cat#WBKLS0100 |
|
| ||
| Deposited data | ||
|
| ||
| Small RNA-seq data (raw) | This paper | GSA: CRA008367 |
| Affinity mass spectrometry | This paper | PRIDE: PXD037342 |
| Experimental models: Organisms/strains | ||
| C. elegans strains | This paper | Table S2 |
| Oligonucleotides | ||
| Primers and oligos | This paper | Table S3 |
|
| ||
| Software and algorithms | ||
|
| ||
| ZEN 2.3 (blue edition) | https://www.zeiss.com.cn/microscopy/products/microscope-software/zen.htm | N/A |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
| Bowtie version 1.2.1.1 | https://bowtie-bio.sourceforge.net/index.shtml | N/A |
| Bedtools 2.30.0 | https://bedtools.readthedocs.io/ | N/A |
| R version 4.0.3 | https://www.r-project.org/ | N/A |
Highlights.
Many germ cell P bodies are located on top of perinuclear germ granules
P body factor CGH-1 promotes the segregation of germ granules into sub-compartments
cgh-1 mutants show impairments in small RNA-based transgenerational gene silencing
WAGO-4 instability is linked to transgenerational silencing defects in cgh-1 mutants
ACKNOWLEDGMENTS
We would like to thank David Pincus for critical comments on the manuscript. Some strains used in this study were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work is supported in part by NIH predoctoral training grant T32 GM07197 to J.B.; the Ministry of Science and Technology, Taiwan (MOST 108-2628-E-006-004-MY3 and MOST 110-2221-E-006-198-MY3 grants to W.-S.W.; the NIH grant R01-GM132457 to H.-C.L.; and the National Natural Science Foundation of China (grants 31771500 and 31922019) and the program for HUST Academic Frontier Youth Team (grant 2018QYTD11) to D.Z.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112859.
REFERENCES
- 1.Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyon AS, Peeples WB, and Rosen MK (2021). A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235. 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fare CM, Villani A, Drake LE, and Shorter J (2021). Higher-order organization of biomolecular condensates. Open Biol. 11, 210137. 10.1098/rsob.210137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. (2009). The Argonaute CSR-1 and Its 22G-RNA Cofactors Are Required for Holocentric Chromosome Segregation. Cell 139, 123–134. 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. (2009). Distinct Argonaute-Mediated 22G-RNA Pathways Direct Genome Surveillance in the C. elegans Germline. Mol. Cell. 36, 231–244. 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Brown JS, He T, Wu W-S, Tu S, Weng Z, Zhang D, and Lee H-C (2022). GLH/VASA helicases promote germ granule formation to ensure the fidelity of piRNA-mediated transcriptome surveillance. Nat. Commun. 13, 5306. 10.1038/s41467-022-32880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. (2008). PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Mol. Cell. 31, 67–78. 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan G, Fields BD, Spracklin G, Shukla A, Phillips CM, and Kennedy S (2018). Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683. 10.1038/s41586-018-0132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishidate T, Ozturk AR, Durning DJ, Sharma R, Shen E-Z, Chen H, Seth M, Shirayama M, and Mello CC (2018). ZNFX-1 Functions within Perinuclear Nuage to Balance Epigenetic Signals. Mol. Cell. 70, 639–649.e6. 10.1016/j.molcel.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips CM, Montgomery TA, Breen PC, and Ruvkun G (2012). MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444. 10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall JG, Bellini M, Wu Z, and Murphy C (1999). Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10, 4385–4402. 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pena E, Berciano MT, Fernandez R, Ojeda JL, and Lafarga M (2001). Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J. Comp. Neurol. . [DOI] [PubMed] [Google Scholar]
- 13.Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324.e28. 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin M-D, Fan S-J, Hsu W-S, and Chou T-B (2006). Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev. Cell 10, 601–613. 10.1016/j.devcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, and Sassone-Corsi P (2006). The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. USA 103, 2647–2652. 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo CM, Munro E, Rasoloson D, Merritt C, and Seydoux G (2008). Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 323, 76–87. 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Ouyang JPT, and Seydoux G (2022). Nuage condensates: accelerators or circuit breakers for sRNA silencing pathways? RNA 28, 58–66. 10.1261/rna.079003.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim AK, and Kai T (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104, 6714–6719. 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheth U, and Parker R (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science (New York, N.Y.) 300, 805–808. 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot J-B, Munier A, Fradet M, et al. (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell. 68, 144–157.e5. 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Boag PR, Atalay A, Robida S, Reinke V, and Blackwell TK (2008). Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182, 543–557. 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble SL, Allen BL, Goh LK, Nordick K, and Evans TC (2008). Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559–572. 10.1083/jcb.200802128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitt JN, Schisa JA, and Priess JR (2000). P Granules in the Germ Cells of Caenorhabditis elegans Adults Are Associated with Clusters of Nuclear Pores and Contain RNA. Dev. Biol. 219, 315–333. 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Hu Y, Lang CF, Brown JS, Schwabach S, Song X, Zhang Y, Munro E, Bennett K, Zhang D, and Lee HC (2020). The Dynamics of P Granule Liquid Droplets Are Regulated by the Caenorhabditis elegans Germline RNA Helicase GLH-1 via Its ATP Hydrolysis Cycle. Genetics 215, 421–434. 10.1534/genetics.120.303052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge DT, Wang W, Tipping C, Gainetdinov I, Weng Z, and Zamore PD (2019). The RNA-Binding ATPase, Armitage, Couples piRNA Amplification in Nuage to Phased piRNA Production on Mitochondria. Mol. Cell. 74, 982–995.e6. 10.1016/j.molcel.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, Kuznicki K, Yee C, Bennett K, and Strome S (2008). Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 178, 1973–1987. 10.1534/genetics.107.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown JS, Zhang D, Gaylord O, Chen W, and Lee H-C (2023). Sensitized piRNA reporter identifies multiple RNA processing factors involved in piRNA-mediated gene silencing. Geneticsiyad095. 10.1093/genetics/iyad095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordeiro Rodrigues RJ, de Jesus Domingues AM, Hellmann S, Dietz S, de Albuquerque BFM, Renz C, Ulrich HD, Sarkies P, Butter F, and Ketting RF (2019). PETISCO is a novel protein complex required for 21U RNA biogenesis and embryonic viability. Genes Dev. 33, 857–870. 10.1101/gad.322446.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putnam AA, and Jankowsky E (2013). DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta 1829, 884–893. 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, et al. (2014). RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157, 1698–1711. 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Lee HC, Gu W, Shirayama M, Youngman E, Conte D, and Mello CC (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcazar RM, Lin R, and Fire AZ (2008). Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 180, 1275–1288. 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang JPT, Zhang WL, and Seydoux G (2022). The conserved helicase ZNFX-1 memorializes silenced RNAs in perinuclear condensates. Nat. Cell Biol. 24, 1129–1140. 10.1038/s41556-022-00940-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu F, Feng X, Chen X, Weng C, Yan Q, Xu T, Hong M, and Guang S (2018). A Cytoplasmic Argonaute Protein Promotes the Inheritance of RNAi. Cell Rep. 23, 2482–2494. 10.1016/j.celrep.2018.04.072. [DOI] [PubMed] [Google Scholar]
- 35.Choudhary S, Lee H-C, Maiti M, He Q, Cheng P, Liu Q, and Liu Y (2007). A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell Biol. 27, 3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassani M, and Seydoux G (2022). Specialized Germline P-Bodies Are Required to Specify Germ Cell Fate in Caenorhabditis elegans Embryos. Development 149. 10.1242/dev.200920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majerciak V, Zhou T, and Zheng Z-M (2021). RNA helicase DDX6 in P-bodies is essential for the assembly of stress granules. Preprint at bioRxiv. 10.1101/2021.09.24.461736. [DOI] [Google Scholar]
- 38.Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, and Weis K (2019). DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148. 10.1038/s41586-019-1502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Qi H, Wang J, and Lin H (2011). PAPI, a novel TUDOR-domain protein, complexes with AGO3. ME31B and TRAL in the nuage to silence transposition 138, 1863–1873, Development. 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics 210, 781–787. 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, and Mello CC (2012). PiRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77. 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team (2020). R: A Language and Environment for Statistical Computing. [Google Scholar]
- 44.Wickham H (2016). ggplot2: Elegant Graphics for Data AnalysisTitle (Springer-Verlag). [Google Scholar]
- 45.Wickham H (2007). Reshaping Data with the {reshape} Package. J. Stat. Software 21, 1–20. [Google Scholar]
- 46.Kassambara A (2020). Ggpubr (“ggplot2” Based Publication Ready Plots). [Google Scholar]
- 47.Wickham H, François R, Henry L, and Müller K (2021). Dplyr: A Grammar of Data Manipulation. [Google Scholar]
- 48.Maniar JM, and Fire AZ (2011). EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short anti-sense RNAs. Curr. Biol. 21, 449–459. 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR, and Hall IM (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA sequencing data are deposited to Genome Sequence Archive (GSA) in National Genomics Data Center with the access number CRA008367. The proteomic data are deposited to Proteomics Identification database (PRIDE) with the access number of PXD037342. All custom scripts used for bioinformatic analysis are available at Zenodo: https://doi.org/10.5281/zenodo.8097062 or at Github: https://github.com/Uchicago-BSD-hlee-lab/Du_2023/. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


