Abstract
Cell science has made significant progress by focusing on understanding individual cellular processes through reductionist approaches. However, the sheer volume of knowledge collected presents challenges in integrating this information across different scales of space and time to comprehend cellular behaviors, as well as making the data and methods more accessible for the community to tackle complex biological questions. This perspective proposes the creation of next-generation virtual cells, which are dynamic 3D models that integrate information from diverse sources, including simulations, biophysical models, image-based models, and evidence-based knowledge graphs. These virtual cells would provide statistically accurate and holistic views of real cells, bridging the gap between theoretical concepts and experimental data, and facilitating productive new collaborations among researchers across related fields.
Keywords: Cell science, Virtual cells, 3D models, Spatial models, Biophysical models, Multiscale modeling, Simulations, Knowledge graphs, Integrating information, Community collaboration, Community modeling, Reproducibility
Significance
Cell science has made progress through reductionist approaches, but integrating vast knowledge and making it accessible is challenging. Next-generation virtual cells, which are dynamic 3D models that integrate information from diverse sources including simulations, biophysical models, image-based models, and evidence-based knowledge graphs, could bridge theory and data by integrating diverse information and facilitating collaboration among researchers.
Overview
Cell science has made enormous progress through the reductionist approach, which focuses on understanding individual cellular processes and their molecular machinery. However, the vast amount of knowledge collected is overwhelming and presents two challenges: understanding how these processes interact across vast scales of space and time to generate cellular behaviors (integration), and making the knowledge, data, and methods more accessible and useful (reproducible and reusable) to enable our field to take on increasingly complex biological questions as a community. This perspective suggests that we could work together to address these challenges by creating next-generation virtual cells—dynamic 3D models that integrate information from various sources with simulations and other methods to provide statistically accurate and holistic views of real cells. These virtual cells could be constructed by merging bottom-up biophysical models with top-down image-based models and dynamic evidence-based knowledge graphs (1) that connect decades worth of published biological concepts, principles, and theories directly to the evidence-based data, experiments, and analyses that support them (2,3).
Currently, there are many different types of virtual cells as well as platforms for storing and sharing models, but they lack consistency in format, accessibility methods, and are not designed to talk to each other, let alone interoperate their data or code. This makes it difficult to find, use, compare, integrate, reuse, or build upon different models of similar (or even the same) biological systems. They are also not typically connected directly to the data and methods used to create them, or to the higher-level knowledge derived from them as it is often dispersed in conclusions spread across journal web sites. However, by standardizing the inputs and outputs of different modeling systems and integrating the information, e.g., by breaking information into smaller modules and linking them, next-generation virtual cells can be created. These cells would enable diverse researchers to explore and discover, test hypotheses, communicate, and learn. They could ultimately become a reliable platform for experimentation, analysis, interpretation, and prediction.
Current state of virtual cells, simulation software, and platforms
Given predicted computer power and other limitations, current methods alone are unlikely to produce comprehensive virtual cells that can address multiscale biological questions ranging from the interactions and effects of molecular mechanics to daylong signaling cascade effects. At the same time, top-down approaches alone, such as machine learning analyses or other quantitative approaches, are unlikely to scale downward to bridge from whole-cell to molecular mechanical detail in any sort of intuitive manner in the foreseeable future. Our hope lies in integrating all of these methods. Creating full-blown virtual cells is a long-term goal, but progress has already been made in developing integrated models that cover multiple spatial scales and methods.
Phenomenological (or top-down) cell models mimic the behavior and architecture of cells
Top-down virtual cells, also known as “digital twins,” can characterize and imitate real cells by integrating multiple types of data gathered from experiments (4,5,6,7,8,9,10). Most digital twin projects aim to produce models that can move through space and time while remaining within the bounds of the input data. This type of model can be broadly categorized based on its phenomenological approach, which often involves using machine learning. While these models are useful for identifying and recreating patterns found in data, they do not directly explain the mechanisms behind the observed events.
Bottom-up cell models generated from biophysical components can be perturbed to probe how cells work
There are different methods to create more holistic and multiscale spatial models of complex systems that can include details down to the level of molecular structure. These methods use structural models with biophysical interactions and parameters. They allow for the simulation of perturbations that can be used to test hypotheses about how changes in components affect the overall system, outcomes, or other cause-and-effect relationships (11,12,13,14). There have also been significant efforts to create massively complicated time series models that incorporate contributions from all known gene products (without spatial constraints) to simulate complex long-term cell events, such as an entire bacterial cell life cycle, and to go the extra step to distill and present the models in a comprehensive and user-friendly manner (15).
Integrating biophysical details into phenomenological models to create dynamic spatial models
To begin to explain complex phenomena, the coming generation of virtual cells will likely need to incorporate various multiscale details, such as physical chemistry, structures, spatial interactions, and molecular mechanisms. Some published models have used various strategies to predict the outcomes of multiple interacting biophysical parameters that are spatially constrained. An example is adding multiscale spatial components and parameters to phenomenological models, which allows for dynamic simulations within the outputs of these models, such as cell segmentations. This helps to predict emergent outcomes based on the spatially constrained biophysical components. One recent study mapped the location and chemical characteristics of cellular components at an atomic scale in 3D space, enabling the model to track the movement of molecules within the cell, their chemical reactions, and the energy needed for each step (16). Other models have used various coarse graining or all-atom simulations to study crowding effects and other global influences on processes such as self-assembly, activation, or signal propagation (17,18).
Modeling software and model sharing platforms
Robust access to published models and their methods and data are crucial for reproducibility and reusability. Easy to use public platforms and community standards for documentation, testing, and minimal acceptable criteria can enhance this access. While some models may be initially generated using experimental code, if they gain more users in a public release, they should be required to meet reproducibility and usability standards. In many cases, experimental code has evolved into software that is intended for dissemination and use by the broader community. For example, SpringSaLaD is a software tool that explicitly models binding events and state changes while considering crowding effects (19). Cytosim is a cytoskeleton simulation suite designed to handle large systems of flexible filaments with associated proteins such as molecular motors (20), and MEDYAN software models cytoskeletons and their interactions with membranes in a multiscale/type manner, for example, by “iteratively switching between stochastic reaction-diffusion simulation and network mechanical equilibrations” (21,22). There are also software solutions for more general simulation needs; for example, NERDSS (23) and ReaDDY (24) provide relatively easy access to different reaction-diffusion systems. VCell is an example of a more comprehensive web-based platform that enables users to build and share models of cell biological systems using the VCell database. It supports multiple simulation types, including deterministic, stochastic (SSA), and spatial stochastic (reaction-diffusion), and includes features for membrane flux, lateral membrane diffusion, and electrophysiology. VCell has a user-friendly interface for modelers, and geometries can be generated from analytical expressions or phenomenological inputs such as microscope images (25,26). In a more generalized manner, the challenge of model integration is also being addressed by tools such as Vivarium, allowing users to connect different types of models and interpreting between their data formats (27). Vivarium is working on connecting to the Biosimulators database (28), which currently houses about 20 simulators that cover a broad range of simulations spanning multiple formats and algorithms. For example, these include BioNetGen, COBRA, COPASI, libRoadRunner, and Smoldyn. Simulation projects are stored on the Biosimulations (29) site, which has online simulation deployment options.
User interfaces reduce barriers to exploring and interrogating published models
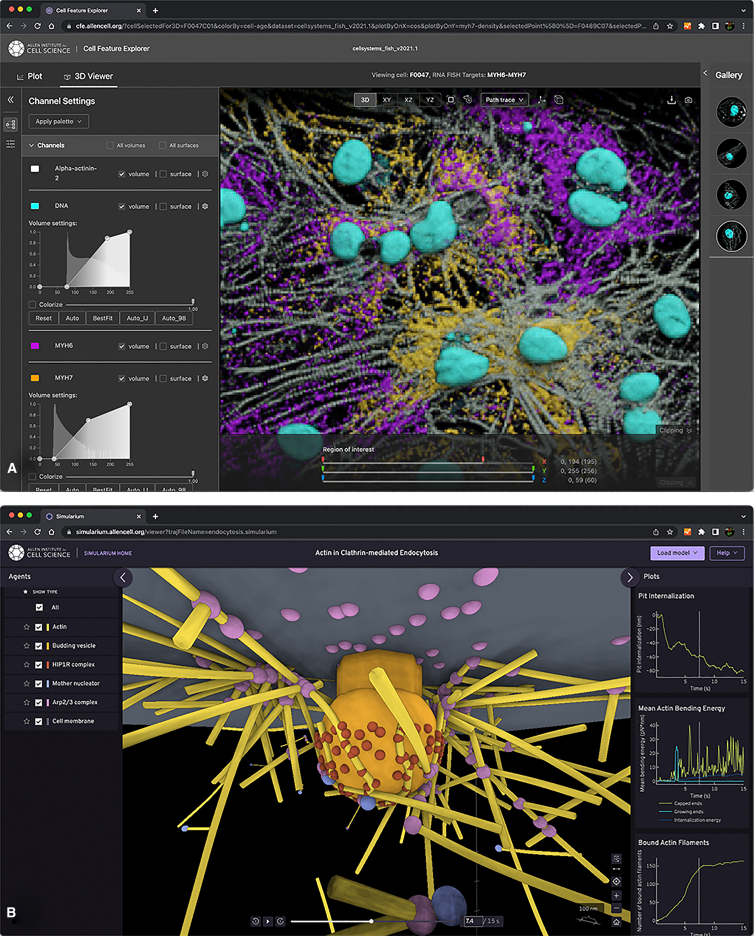
Tools have been developed to make it easier to explore and analyze published spatial models online. For example, screenshots from web browser windows in Fig. 1 show that the Cell Feature Explorer (30) allows the online plotting and 4D visual analysis of large numbers of 3D and time series microscopy images, published in standardized OME file formats (31). In a similar manner, the Simularium Viewer lets users share, visualize, and examine any spatial simulation results directly in a web browser once they have been converted to the Simularium format and hosted on the public internet (32). Rapidly evolving user interfaces for knowledge graphs, which provide access to underlying semantic networks that reveal relationships between historic literature and new information (e.g., (33)), can help reduce barriers to exploring and comparing published models (1), and web sites such as Bionumbers (34) have provided a great start to help the community gather and share bionumeric measurements that can often take a modeling expert days or weeks to scrape from the literature.
Figure 1.
Providing data, models, simulation software, and other connected knowledge in online interfaces can make it easy for researchers and students to find, access, use, and extend virtual cells or their contributing components. Example online interfaces already exist: (A) the Cell Feature Explorer enables the interactive exploration of hundreds of thousands of cells at a time on cfe.allencell.org; (B) the Simularium Viewer allows modelers to host their simulations online to provide interactive access to their models with a single URL that plays through the simularium.allencell.org user interface.
Existing models, methods, and platforms that were previously published and are foundational must live on to become essential building blocks for the next generation of virtual cells. The new virtual cells will also need to integrate and connect data, knowledge, and methods from these models and other published efforts with new and evolving approaches and outcomes. The goal is to enable the community, including various kinds of biologists, to ask different types of questions, regardless of their level of expertise in any particular method.
A vision for the next generation of virtual cells
To create next-generation virtual cells, one approach is to enhance the resolution, multiscale features, and accuracy of top-down digital twin cells and then to integrate them with different types of biophysical simulations that can ideally interoperate, or at least inform one another at adjacent scales. By accurately mimicking real cells, these twins can help us probe multiscale cell architecture and behavior more easily to better characterize what cells do. We could then use this information as a target or constraint for other types of biophysical models that explain how cells work (35), for example, to witness and characterize how complex behaviors captured in the digital twins might emerge from the interactions of many simpler building blocks and mechanisms that are now visible, adjustable, and easily measured in the hybrid system. Such integrative virtual cell modeling frameworks would expand and evolve, and the models they generate could be queried to address many different types of biological questions. For example, on a scale of microseconds, how might a particular modification to the actin monomer building blocks affect the speed or power with which actin filaments redirect forces from the cell membrane onto a budding vesicle in clathrin-mediated endocytosis (see Fig. 1 B); or, on a scale of hours or days, how might that modified endocytosis rate then affect the ability of the cells to interact with their environment (36)?
Improving phenomenological digital twins
Creating high-quality, multiscale digital twins is a long-term goal, but some progress has already been made, such as integrating models that provide holistic multiscale views of cell architecture. When assembled to integrate 100s–1000s of replicate images (views of the same system sampled across space and/or time), biologically meaningful variance and interrelations emerge among components (9,37). Advances in microscopy, artificial intelligence (AI), and image analysis promise to make it easier to integrate live-cell imaging (from micro- to millimeter resolution) with higher-resolution imaging such as x-ray tomography and electron microscopy (down to nanometer resolution) in the same instantiation of multiscale digital cells (38,39,40,41): picture the utility of a virtual 3D electron microscopy time series data set, captured at the temporal resolution and reduced toxicity of bright-field imaging with structures of functional regions further classified or segmented in an easy and useful manner. A major next step would be to include multiple types of localized single-cell measurements to further enhance these digital cells by connecting architectural relationships to highly detailed and spatially resolved physiological changes, such as: gene expression; protein activation, modification, and concentration; or signal-induced activation and propagation, all of which will improve the digital twin representations.
To explain how large-scale phenomena captured in the digital twins work in a more intuitive manner, it will be critical to integrate mechanistic spatial models. For example, reductionist cell science has successfully provided deep insight into the structure and function of many cellular components and processes. These include, for example, the motor protein kinesin’s structure, its function in organelle trafficking, its walking behavior along microtubule cytoskeletons, and its response to external forces (Fig. 2). While collective knowledge could be used to make discoveries and build models that advance our understanding, by and large this knowledge remains disconnected and spread overwhelmingly across publications, web sites, siloed databases, unusable models, and researchers’ memories. Therefore, despite the vast amount of existing data and theories, there is still much we do not understand about how these components and processes interact to create complex living systems. For example, we do not fully understand how kinesins and other components of the cytoskeleton and cytoplasm work together to regulate the spatial organizations and shapes of cellular membranes and subcellular compartments that we can observe and characterize using light microscopy techniques. Imagine the emergent phenotypes we could discover, predict, and explain if we combined new public cell image databases with the existing literature and multiple types of integrated modeling approaches made recently accessible.
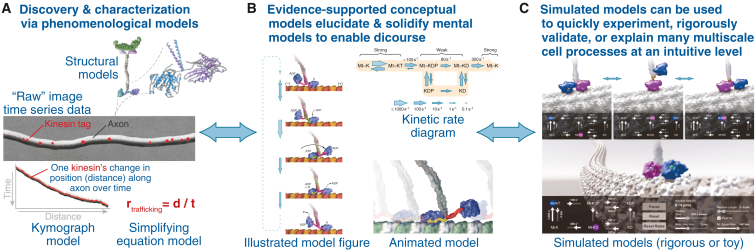
Figure 2.
Multiple types of useful models published over decades of research are exemplified with the well-known (although still evolving) function and mechanism of the motor protein kinesin. (A) Phenomenological models enable the discovery and characterization of cell structures and processes. Many are generated manually with observation and easy measurements made on raw data such as time series microscope movies, e.g., kymograph models (redrawn after (42)), which are graphical representations of position over time and predictive simplifying equation models. Structural models provide atomic-level detail. (B) Structural models facilitate conceptual and mechanistic hypotheses that combine phenomenological understandings with other types of data such as protein-protein interactomes or kinetic reaction rates. These often begin as mental models, but must be converted to evidence-based conceptual models such as illustrated or animated model figures to enable discourse (43). (C) Simulated models rigorously validate these hypotheses or test and explain detailed mechanisms and unexpected variations that are often too complex for a human to model in their mind (44,45).
Connecting information through knowledge graphs
Creating the next generation of virtual cells will likely require integrating databases and modeling technologies using, for example, dynamic knowledge graphs that provide user interfaces to connect biological concepts, principles, and theories to the data, analyses, tools, and models that support them in a reproducible and reusable manner. Even less comprehensive virtual cells in the form of knowledge graphs alone could serve as tools for exploration and discovery by establishing robust digital reference systems for biologically related spatial data and higher-level knowledge. This would allow researchers and students to study relationships between cellular organization, multiscale activities, and function at different spatial and temporal resolutions, focusing on specific questions by filtering and measuring different metadata parameter combinations. Whether generated from easy to ingest connected knowledge, or from the grander vision that further integrates more comprehensive spatial models, next-generation virtual cells hold promise for supporting applications from basic science to drug discovery, but some of their highest initial impact may be in engaging and training the next generation of scientists.
The challenges in constructing virtual cells
The construction of virtual cells faces several barriers, including the need to connect various types of data, evidence, and claims buried across the literature. Virtual cells would be like Google Maps in their spatial aspect and ability to provide or distill appropriate and useful levels of detail relative to the scale of interaction. But their intrinsic 3D and dynamic nature will make virtual cells more complicated to deliver, more akin to the challenges of 3D video games, which often go further by incorporating sophisticated interacting characters and objects. Furthermore, Google Map-type systems are based on relatively standardized instances of fixed and easily captured structural information from satellites, car cameras, and citizen annotations all pouring into a predetermined framework. In contrast, virtual cells will be based on statistical models that merge patterns and relationships from many imaging instances and modalities, with bottom-up modeling. In this regard, the utility of any unified virtual cell model will depend on the quality of the replicate data and the veracity of the simulation systems that generate it.
Constructing next generation virtual cells will require overcoming four major challenges: 1) interpreting and exploring across the vast spatial-temporal scales at which cell events occur, 2) finding useful frames of reference, 3) data quality, integration, and interpretation, and 4) uniting molecular structures and mechanisms with behaviors observed at the larger scales. Scaling from nanometer/nanosecond-level spatial-temporal events to the scales of whole cells or multiday events, as described in the clathrin-mediated endocytosis example above, is difficult. To combine spatial phenomena into a larger model, appropriate spatial reference coordinate systems must be identified. Models will also depend on the quality of data used to build them, how those data are integrated into a unified whole, and the quality of user experiences in visual analysis tools and reduced dimensionality representations that are used for interpretation. Overall, constructing virtual cells requires advanced technology and collaboration among researchers from different disciplines. To become comprehensive and to cover the knowledge landscape of cell biology (data, tools, claims/hypotheses, and conclusions), virtual cells must be community driven, likely enhanced by AI, and must embody contributions from diverse community members including both large and small labs.
Challenge of interpreting and exploring across many spatial-temporal scales
The challenge of studying complex cell structures and behaviors is that they emerge across a wide range of spatial and temporal scales, from tiny molecular interactions to large-scale cell transitions over hours, days, or years. It is impossible to use current computational resources and approaches to brute-force simulate whole-cell or whole-day events in nanometer/nanosecond-level detail. The appropriate scale for measuring phenomena depends on what is being studied (46). Experimentally, it is not necessary to collect data across all scales for each measurement; instead, data can be collected on the appropriate timescale, with the goal of developing methods to integrate the data later.
Challenge of finding useful frames of reference
Many breakthroughs in science came from uncovering or defining the most appropriate frame of reference, or coordinate system, for describing the phenomena of interest. Physicists, for example, routinely select coordinate systems to simplify calculation and promote understanding such as using angular instead of Cartesian coordinates to calculate the motion of a pendulum. Comparing or analyzing the relative locations of cells and their internal components will require similarly simplifying reference coordinate systems, which will likely vary depending on the tissue, cell type, and structure being studied. It is important to consider factors such as magnification and resolution when developing these systems. For well-stereotyped and polarized cells or tissues, simple overlay systems or orienting to an easy target such as the contractile apparatus have sufficed (47), while for mesenchymal and epithelia-like cells, the nucleus and more nuanced cell membrane has proven more effective (9,37). Reference systems must also be developed for the positions of organelles, substructures, and even individual cells within tissues. The robustness and predictive accuracy of these systems must be carefully assessed before implementation (8,37,48).
Challenges of data quality, integration, and interpretation
The creation of a digital twin cell involves merging measurements of a specific cell type into a unified image model that moves through virtual space and time while statistically accommodating all of the input image and measurement data that drives it. This model would need to capture variation by looking at both population means and specific individual digital twins along population distributions. Integrating multimodal and multiscale information is challenging due to the complexity and interoperability needed (46). Computational approaches are currently used to integrate different types of data into dimensionally reduced representations (4,5,6,7,8,9,10,37). Integrating data from multiple labs is also challenging due to differences in imaging platforms and settings. Establishing requirements that enable FAIR (findable, accessible, interoperable, reusable) data practices including community standards, quality assessments for image capture, useful metadata, and data accessibility will be not only be critical to enabling easy functionality and reproducibility in the development of virtual cells, but they should also become a mandate for public data sets (49). Algorithmic conversions and machine learning, such as label-free imaging, can help address these issues, but in the meantime we can make many other improvements as a community (50,51,52).
Uniting molecular structure and mechanisms with behaviors at the organelle, whole cell, signaling, and tissue scale
Virtual cells must integrate detailed spatially resolved functional, biophysical, structural, and proteomic data, along with computational models. This encompasses various biological factors such as metabolism, electrical activity, posttranslational modifications, force, calcium, pH fluxes, and other relevant data, where incorporating spatial information within the cell would enhance the accuracy and effectiveness of the model. Localized biophysical parameters, such as molecular transport rates, equilibrium constants, including on and off rates for binding, concentrations, and effective viscosities will help simulation and modeling efforts using virtual cells. The pioneering work of Ken Jacobson and others on FRAP and related microscopies (53) inspired the notion that physical chemical measurements could be done in living cells, themselves, thus providing localized biophysical data for integration into virtual cells.
A powerful approach to data integration is the use of deep learning methods applied to large data sets. These can be performant and generate highly accurate predictions. However, how they do this is typically specific to the input data set, somewhat mysterious to biologists, and produces little generalizable or mechanistic insight. This contrasts with other computational approaches, which may not be as highly predictive or accurate but can instead reveal deeper mechanistic insights by exposing the model rules and their more straightforward or understandable construction. In evaluating different approaches, this issue of balancing readily accessible and accurate predictions against mechanistic insight needs to be considered. Hopefully, in the future, AI will provide us with both predictive accuracy and insight into the driving multiscale mechanisms to aid our understanding.
A modular toolbox for exploration and discovery
In the future, a comprehensive, verified, and open access suite of computational tools must be integrated into the virtual cell platform to make it accessible to a wide range of users and use cases. These tools should evolve and improve over time, enabling analysis and discovery. Despite existing tools, challenges remain in coordinating diverse tools and bridging gaps in data translation and technical skills. Future tools should focus on user-friendly exploration of morphology-function relationships and framing mechanistic simulations in the context of whole cells for easy evaluation, use, and comparison of different models. This will democratize access and enhance the likelihood of discovery in virtual cell research.
Virtual cells will help capture, interpret, understand, communicate, and evolve knowledge of cells
Once a useful iteration of a virtual cell is built, what would we do with it? Like Google Maps, it is hard to fully anticipate its utility, except that it too, will likely greatly exceed expectation. Some immediate activities will likely include:
-
•
Studying interrelationships among molecular cellular machinery, localized cellular activities, and integrated cellular behaviors
-
•
Understanding emergent properties of cells
-
•
Providing a holistic and contextual view of cellular behaviors and processes
-
•
Allowing the public to observe cellular behaviors and processes as they would in a multiscale microscope
-
•
Providing a statistically integrated platform for enormous amounts of cellular data
-
•
Enabling the identification and detection of different cell types in developing or diseased tissues
-
•
Improving the prediction of emergent or longer-term outcomes from genomic data
-
•
Enabling rapid in situ diagnoses and prognoses
-
•
Developing new rules, principles, and theories for the integrated complex systems that comprise cells
If successful, these investigations and activities promise profound implications in many areas.
Transforming education
To truly revolutionize our understanding of how cells work, it will be critical to educate both current and future biologists on these methodologies and opportunities in both current and future biology. That future will likely require an intuitive grasp of emergent complexity based on reproducible evidence-based hypotheses and claims that support higher level conclusions, principles, rules, and theories as stated in the Vision and Change statement by the American Association for the Advancement of Science and the National Science Foundation (54). Virtual cells and accompanying analysis and modeling tools can provide a platform (a fearless playground) for students to gain intuition about cause-and-effect relationships and principles by changing input parameters in a simulation. Democratized access to this platform is critical, and hosting in the cloud or on open, public web sites could be a route to achieving this accessibility in an equitable manner, where dedicated training, low fees, and efficient mechanisms for efficiently microfunding these computational resources (regardless of an institution’s on-premises resources) will be crucial for enabling and encouraging general accessibility.
Engagement and motivation via the awe of discovery enhanced through visualization
The same awe that we sense when viewing the deep ocean, another planet, or a galaxy awaits us when we view next generation virtual cells. We have neither seen nor understood the complexity of a cell—its organization, dynamics, and interrelationships across scales—and this will be our window in.
The complexity and interrelationships of cells can be better understood through virtual cell models, which allow for real-time measurement and event plotting. The awe-inspiring experience of exploring virtual cells can engage and motivate biologists to gain intuition about cause-and-effect relationships and principles—perhaps even motivating them to dive deeper into cell systems that they do not normally focus on and thus expanding their specialized knowledge into the bigger picture of how different systems interact and affect one another across entire cells or tissues. By observing the effect of changes in one cell or subcellular structure on others, researchers can gain new insights and uncover underlying principles about cell behavior.
Improving human health
Next generation virtual cells will likely not only provide reliable initial predictions about what might be the best targets for mutation or drugs, but they can also be used for assessments of their downstream effects. They could predict how the phenotype will evolve over time and the impact of a mutant cell on its neighbors. While many mutations and drugs have known or predicted effects on a particular structure or activity, virtual cells will not only better predict how they might propagate throughout the cell as a whole; but they will also provide a deep understanding of the mechanisms and processes involved across all scales, leading to new potential avenues for intervention.
The principles of cellular morphogenesis
Virtual cells can provide insight into the principles of cellular morphogenesis, which has received little attention compared to the morphogenesis of tissues, organs, and embryos. Virtual cells can reveal interrelationships among various cellular structures, suggesting hypotheses for how cells organize and reorganize as they transition among states. The real power of virtual cells is to see how these interrelationships change over a wide range of cell types and contexts, leading to generalizations, rules, and an understanding of contextual differences. It is possible that the rules of organization may vary among cells and become a classification system, and we must be able to distinguish which changes in cell states are manifest in changes in variance rather than the mean, perhaps by testing various hypotheses across each of the scales accessible to these multiscale models.
Predicting cellular structure and emergent behaviors from 1D genomics
Virtual cells can incorporate spatially resolved, multiplexed single-cell gene expression data to explore the relationship between gene expression profiles and cellular organization. This can lead to the identification of cell types and states from microscopic observations, which can be used for rapid in situ diagnoses and prognoses. By combining biological activity, overall morphology, and gene expression information, virtual cells can lead to more nuanced and meaningful classifications of cells, as well as a better understanding of how cells change architecture and phenotype during complex processes. Much like weather forecasting, this can improve predicting emergent or longer-term outcomes from genomic data.
Conclusion
The construction of this next generation of virtual cells is an achievable goal that could one day provide a computational output that mimics real cells in every conceivable measure—enabling analysis, modeling, perturbation, and visualization of spatial and temporal data gathered together from both current and historical experimentation. In addition to exploring what an average cell looks like, the spectrum of variation could be accessed probabilistically. These outputs could integrate enormous amounts of cellular data and allow for the observation of cellular behaviors and processes in a virtual multiscale microscope, which could at once be useful for research and accessible to students, trainees, and even public audiences if properly presented. By studying interrelationships among cellular machinery, activities, and behaviors, emergent properties can be revealed. From these analyses, new rules, principles, and theory can emerge, moving cell science from a large collection of observations to viewing cells as the complex integrated systems that they are.
Author contributions
All authors contributed to discussions and draft documents that led to this evolution of the fully multiscale “next-generation virtual cell” concept described in this perspective. R.H., S.R., and G.T.J. assembled an initial draft of this perspective. G.T.J. illustrated the figures. All authors contributed edits, new text, citations, or clarifications in the final version.
Acknowledgments
G.T.J., B.L., M.R.-M., S.R. and R.H. thank the Allen Institute for Cell Science founder, Paul G. Allen, for his vision, encouragement, and support; E.A. is supported by P41 EB023912-01, W.O. is supported by SciLifeLab & Wallenberg Data Driven Life Science Program (grant no. KAW 2020.0239), and E.L. is supported by Knut and Alice Ellenberg Foundation 2021.0346 and Schmidt Futures.
R.H. wants to acknowledge Ken Jacobson’s inspiring work on membranes. Ken’s early work on FRAP gave me a first glimpse of what a virtual cell might look like. Our paths first crossed in 1966, when I was finishing my senior honors thesis in nuclear chemistry at UW Madison. Planning to attend graduate school in Biophysics in the fall, I got a summer job in the biophysics lab of W.W. Beeman and J.W. Anderegg in the UW physics department. I was told that I could have a desk in the lab that was presently occupied by someone who would be leaving shortly upon completion of his Masters. That someone was Ken Jacobson. Ten years later, Ken and some others published the FRAP methodology. It was inspirational, raising the prospect of doing localized biophysical measurements (transport, on-off rates, concentrations, etc.) in living cells, thereby inspiring the vision of a biophysical cellular map. Our common interest in membranes and cell migration intertwined our careers until Ken’s passing. In our recent conversations, Ken would tell me about the illnesses and passings of our mentors and colleagues. Unfortunately, he didn’t tell any of us about his illness, so I was unable to convey to him how much his friendship and research meant to me and so many others. Ken was a mensch with an impeccable character; his curiosity, insight, and collegiality will linger.
G.J. and the authors acknowledge Joel Chan, Peter Clark, Thao Do, Janani Gopalan, Brett Mensh, Irina Mueller, and Oyvind Tafjord with whom they have had numerous, stimulating, and informative scientific discussions about this topic.
Declaration of interests
The authors declare no competing interests.
Editor: Meyer Jackson.
References
- 1.Discourse graphs and the future of science; 2023. https://research.protocol.ai/blog/2023/discourse-graphs-and-the-future-of-scienceDiscourse.
- 2.Ji S., Pan S., et al. Yu P.S. A survey on knowledge graphs: representation, acquisition, and applications. IEEE Transact. Neural Networks Learn. Syst. 2022;33:494–514. doi: 10.1109/TNNLS.2021.3070843. [DOI] [PubMed] [Google Scholar]
- 3.Chan J., Chan J. Sustainable authorship models for a discourse-based scholarly communication Infrastructure. Common. Place. 2021;1:1. [Google Scholar]
- 4.Thul P.J., Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018;27:233–244. doi: 10.1002/pro.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gut G., Herrmann M.D., Pelkmans L. Multiplexed protein maps link subcellular organization to cellular states. Science. 2018;361 doi: 10.1126/science.aar7042. [DOI] [PubMed] [Google Scholar]
- 6.Qin Y., Huttlin E.L., et al. Ideker T. A multi-scale map of cell structure fusing protein images and interactions. Nature. 2021;600:536–542. doi: 10.1038/s41586-021-04115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray M.A., Singh S., et al. Carpenter A.E. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho N.H., Cheveralls K.C., et al. Leonetti M.D. OpenCell: endogenous tagging for the cartography of human cellular organization. Science. 2022;375 doi: 10.1126/science.abi6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viana M.P., Chen J., et al. Rafelski S.M. Integrated intracellular organization and its variations in human iPS cells. Nature. 2023;613:345–354. doi: 10.1038/s41586-022-05563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollingsworth P., Borkon L. Digital twins for cancer care: exploring a cross-disciplinary innovative approach. https://frederick.cancer.gov/news/digital-twins-cancer-care-exploring-cross-disciplinary-innovative-approach
- 11.Johnson G.T., Autin L., et al. Olson A.J. cellPACK: a virtual mesoscope to model and visualize structural systems biology. Nat. Methods. 2015;12:85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maritan M., Autin L., et al. Goodsell D.S. Building structural models of a whole Mycoplasma cell. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odell G.M., Foe V.E. An agent-based model contrasts opposite effects of dynamic and stable microtubules on cleavage furrow positioning. J. Cell Biol. 2008;183:471–483. doi: 10.1083/jcb.200807129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens J.A., Grünewald F., et al. Marrink S.J. Molecular dynamics simulation of an entire cell. Front. Chem. 2023;11 doi: 10.3389/fchem.2023.1106495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karr J.R., Sanghvi J.C., et al. Covert M.W. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornburg Z.R., Bianchi D.M., et al. Luthey-Schulten Z. Fundamental behaviors emerge from simulations of a living minimal cell. Cell. 2022;185:345–360.e28. doi: 10.1016/j.cell.2021.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casalino L., Seitz C., et al. Amaro R.E. Breathing and tilting: mesoscale simulations Illuminate Influenza glycoprotein vulnerabilities. ACS Cent. Sci. 2022;8:1646–1663. doi: 10.1021/acscentsci.2c00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao G., Perilla J.R., et al. Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalski P.J., Loew L.M. SpringSaLaD: a spatial, particle-based biochemical simulation platform with excluded volume. Biophys. J. 2016;110:523–529. doi: 10.1016/j.bpj.2015.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nedelec F., Foethke D. Collective Langevin dynamics of flexible cytoskeletal fibers. New J. Phys. 2007;9:427. [Google Scholar]
- 21.Ni H., Papoian G.A. Membrane-MEDYAN: simulating deformable vesicles containing complex cytoskeletal networks. J. Phys. Chem. B. 2021;125:10710–10719. doi: 10.1021/acs.jpcb.1c02336. [DOI] [PubMed] [Google Scholar]
- 22.Popov K., Komianos J., Papoian G.A. MEDYAN: mechanochemical simulations of contraction and polarity alignment in actomyosin networks. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varga M.J., Fu Y., et al. Johnson M.E. NERDSS A nonequilibrium simulator for multibody self-assembly at the cellular scale. Biophys. J. 2020;118:3026–3040. doi: 10.1016/j.bpj.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann M., Fröhner C., Noé F. ReaDDy 2: fast and flexible software framework for interacting-particle reaction dynamics. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The virtual cell. https://vcell.org
- 26.Blinov M.L., Schaff J.C., et al. Loew L.M. Compartmental and spatial rule-based modeling with virtual cell. Biophys. J. 2017;113:1365–1372. doi: 10.1016/j.bpj.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agmon E., Spangler R.K., et al. Covert M.W. Vivarium: an interface and engine for integrative multiscale modeling in computational biology. Bioinformatics. 2022;38:1972–1979. doi: 10.1093/bioinformatics/btac049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biosimulators: reproducing & reusing biomodels & simulations. https://biosimulators.org.
- 29.Biosimulations: reproducing & reusing biomodels & simulations. https://biosimulations.org.
- 30.Cell feature explorer. https://cfe.allencell.org.
- 31.Moore J., Allan C., et al. Swedlow J.R. OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies. Nat. Methods. 2021;18:1496–1498. doi: 10.1038/s41592-021-01326-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyons B., Isaac E., et al. Johnson G.T. The Simularium Viewer: an interactive online tool for sharing spatiotemporal biological models. Nat. Methods. 2022;19:513–515. doi: 10.1038/s41592-022-01442-1. [DOI] [PubMed] [Google Scholar]
- 33.Chan, C. (2021). Sustainable authorship models for a discourse-based scholarly communication Infrastructure. https://commonplace.knowledgefutures.org/pub/m76tk163/release/1.
- 34.B10NUMB3R5: the database of useful biological numbers. https://bionumbers.hms.harvard.edu/search.aspx.
- 35.Pollard T.D. No question about exciting questions in cell biology. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akamatsu M., Vasan R., et al. Drubin D.G. Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. Elife. 2020;9 doi: 10.7554/eLife.49840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majarian T.D., Cao-Berg I., et al. Murphy R.F. CellOrganizer: learning and using cell geometries for spatial cell simulations. Methods Mol. Biol. 2019;1945:251–264. doi: 10.1007/978-1-4939-9102-0_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu C.S., Pang S., et al. Hess H.F. An open-access volume electron microscopy atlas of whole cells and tissues. Nature. 2021;599:147–151. doi: 10.1038/s41586-021-03992-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman D.P., Shtengel G., et al. Hess H.F. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science. 2020;367 doi: 10.1126/science.aaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turk M., Baumeister W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 2020;594:3243–3261. doi: 10.1002/1873-3468.13948. [DOI] [PubMed] [Google Scholar]
- 41.Taraska J.W. Cell biology of the future: nanometer-scale cellular cartography. J. Cell Biol. 2015;211:211–214. doi: 10.1083/jcb.201508021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuster M., Lipowsky R., et al. Steinberg G. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. USA. 2011;108:3618–3623. doi: 10.1073/pnas.1015839108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard T., Earnshaw W., et al. Johnson G.J. Elsevier, NY; 2023. Cell Biology 4e. [Google Scholar]
- 44.Goldtzvik Y., Thirumalai D. Multiscale coarse-grained model for the stepping of molecular motors with application to kinesin. J. Chem. Theor. Comput. 2021;17:5358–5368. doi: 10.1021/acs.jctc.1c00317. [DOI] [PubMed] [Google Scholar]
- 45.https://allen-cell-animated.github.io/MolecularSimulationPrototypes/Kinesin/Unity/4/index.html
- 46.Iwasa J.H., Lyons B., Johnson G.T. The dawn of interoperating spatial models in cell biology. Curr. Opin. Biotechnol. 2022;78 doi: 10.1016/j.copbio.2022.102838. [DOI] [PubMed] [Google Scholar]
- 47.The Integrated Mitotic Stem Cell. https://imsc.allencell.org/
- 48.Donovan-Maiye R.M., Brown J.M., et al. Johnson G.R. A deep generative model of 3D single-cell organization. PLoS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1009155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bagheri N., Carpenter A.E., et al. Horwitz R. The new era of quantitative cell imaging-challenges and opportunities. Mol. Cell. 2022;82:241–247. doi: 10.1016/j.molcel.2021.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotte Y., Toy F., et al. Depeursinge C. Marker-free phase nanoscopy. Nat. Photonics. 2013;7:113–117. [Google Scholar]
- 51.Ounkomol C., Seshamani S., et al. Johnson G.R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods. 2018;15:917–920. doi: 10.1038/s41592-018-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christiansen E.M., Yang S.J., et al. Finkbeiner S. In silico labeling: predicting fluorescent labels in unlabeled images. Cell. 2018;173:792–803.e19. doi: 10.1016/j.cell.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lippincott-Schwartz J., Snapp E.L., Phair R.D. The development and enhancement of FRAP as a key tool for investigating protein dynamics. Biophys. J. 2018;115:1146–1155. doi: 10.1016/j.bpj.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The American Association for the Advancement of Science. Inclusive STEMM ecosystems for equity and diversity (ISEEED) https://visionandchange.org/wp-content/uploads/2013/11/aaas-VISchange-web1113.pdf