Abstract
Dectin-1A is a C-type lectin innate immunoreceptor that recognizes β-(1,3;1,6)-glucan, a structural component of Candida species cell walls. β-Glucans can adopt solution structures ranging from random coil to insoluble fiber due to tertiary (helical) and quaternary structure. Fungal β-glucans of medium and high molecular weight are highly structured, but low molecular weight glucan is much less structured. Despite similar affinity for Dectin-1, the ability of glucans to induce Dectin-1A-mediated signaling correlates with degree of structure. Glucan denaturation experiments showed that glucan structure determines agonistic potential, but not receptor binding affinity. We explored the impact of glucan structure on molecular aggregation of Dectin-1A. Stimulation with glucan signaling decreased Dectin-1A diffusion coefficient. Fluorescence measurements provided direct evidence of ligation-induced Dectin-1A aggregation, which positively correlated with increasing glucan structure content. In contrast, Dectin-1A is predominantly in a low aggregation state in resting cells. Molecular aggregates formed during interaction with highly structured, agonistic glucans did not exceed relatively small (<15 nm) clusters of a few engaged receptors. Finally, we observed increased molecular aggregation of Dectin-1A at fungal particle contact sites in a manner that positively correlated with the degree of exposed glucan on the particle surface. These results indicate that Dectin-1A senses the solution conformation of β-glucans through their varying ability to drive receptor dimer/oligomer formation and activation of membrane proximal signaling events.
Significance
“ITAM” signaling motifs are common in immunoreceptors. Dectin-1 is an immunoreceptor with a “hemITAM” motif (half-ITAM structure). HemITAMs are thought to require ligand-dependent aggregation for signaling. Dectin-1 recognizes β-glucan in fungal pathogens, activating inflammation. We find that high glucan molecular structure is critical for achieving Dectin-1 signal initiation. Only highly ordered glucan can induce Dectin-1 to shift to a higher state of aggregation, but poorly ordered glucan does not. During host-pathogen contact, Dectin-1 aggregation occurs at sites of glucan exposure on native cell walls of Candida albicans pathogenic yeast. This work directly validates the model that Dectin-1 requires ligand-dependent multimerization for signaling, and we extend that model by showing that ligand structure profoundly impacts multimerization and signaling via this important antifungal immunoreceptor.
Introduction
Overall, Candida infections have increased over the past 20 years in the United States (1,2,3,4,5). It is estimated that 46,000 cases of healthcare-associated invasive candidiasis occur in the United States annually. The fungal cell wall is composed of an inner layer of chitin, a middle layer of β(1,3;1,6)-D-glucan and an outer layer of N- and O-linked mannans (6). During infection, the cell wall of Candida is an important and relevant virulence factor, playing roles in adhesion, colonization, and immune recognition (7,8).
Due to the abundant amount of mannan in the outer cell wall, β-glucan exhibits a very limited, punctate pattern of nanoscale surface exposure. The extent of this glucan masking is influenced by environmental conditions such as intestinal pH or lactate levels (9,10). In addition, interactions with neutrophils have been shown to “unmask” the mannan layer through a neutrophil extracellular trap-mediated mechanism (11). Furthermore, our lab and others have determined that antifungal drugs unmask the fungal cell wall, which leads to increases in nanoscale regions of glucan exposure and correlates with enhanced host defense (12,13,14). Therefore, fungal species use masking as a way to evade immune recognition of β-glucan by the host’s immune system (15).
β-Glucans consist of a β-1,3-linked backbone with side chains of β-1,6-linked units that vary in length and degree of branching (16). β-Glucan forms triple-helical structures through intermolecular hydrogen bonds with two other strands (16,17,18,19,20). This triple helix conformation is shown to form with just the β-1,3-linked backbone; however, β-1,6-linked side chains play an important role in determination of the triple helix cavity formation through side chain/side-chain interactions (20).
β-Glucans are known for their biological activities such as enhancing antitumor, antibacterial, and antiviral immunity as well as wound healing (21,22,23,24). The biological activity of glucan is affected by its structure, size, structural modification, conformation, and solubility (25). Research has found that branching is not required to observe biological activity, but branching has been shown to enhance binding to the Dectin-1 receptor (26). In contrast, β-glucan size is thought to play a major role in biological activity with glucans that are shorter than 10,000 Da being generally inactive in vivo (27,28). However, despite having similar sizes, glucans can display differences in their biological activities (29,30,31). For example, studies have demonstrated that the immunoregulatory activity of variously sourced laminarin ranges from agonistic to antagonistic depending on its physicochemical properties, purity, and structure (32). Furthermore, β-glucans that have a triple-helical conformation are more potent agonists of host immune response than single helical glucan (26,33,34,35). We propose that the β-glucan triple helix conformation plays an important role in determining the biological activity of the β-glucan through modulating the degree of receptor oligomerization upon ligation.
During innate immune recognition of Candida, the organization of cell wall ligands and pattern recognition receptors is an important determinant of successful immune activation (7). The fungi-responsive C-type lectin (CTL) antifungal immunoreceptors play a central role in the detection of Candida (36). Human Dectin-1 is the main CTL that recognizes the β-glucan found in the fungal cell wall (37,38,39). Dectin-1A is the full-length isoform found in myeloid lineage cells and, once activated, it stimulates phagocytic activity, the production of reactive oxygen intermediates and inflammatory mediators. Dectin-1A contains a CTL-like domain, separated from the cell membrane by a glycosylated stalk region, a transmembrane domain and an intracellular cytosolic domain. Dectin-1 contains half an immunoreceptor tyrosine-based activation motif (a YXXL sequence with an upstream stretch of acidic amino acids) in its cytoplasmic tail, which is termed a (hem)ITAM domain (40,41). Monophosphorylated ITAM domains, which are anticipated to approximate the structure of phosphorylated (hem)ITAM domains, poorly recruit and activate Syk for downstream signaling because they cannot support bivalent engagement of both of Syk’s SH2 domains (42). Another (hem)ITAM bearing receptor, CLEC-2, is reported to require dimerization for its signaling (43). By analogy to this and other (hem)ITAM receptors, it is speculated that Dectin-1A must oligomerize to recapitulate a multivalent binding site for Syk to facilitate signal transduction (7,43,44). However, this prediction has not been directly explored for Dectin-1A in live cells at the molecular level with relation to signaling outcomes. Syk-dependent signaling of Dectin-1A involves activation of phospholipase C gamma. IP3 generation catalyzed by this enzyme triggers store-operated calcium entry, making observation of cytoplasmic calcium flux a good readout for Syk-dependent signaling downstream of Dectin-1.
In this study, we propose that factors that induce an aggregated membrane organization of Dectin-1A during activation are very important for determining signaling outcomes of Dectin-1A engagement by β-glucan (43,44). We hypothesize that ligand structure, at the levels of glucan tertiary and quaternary structure, as well as nanoscale glucan exposures on the pathogen surface, impacts signaling by determining the membrane organization and spatiotemporal clustering dynamics of Dectin-1A. To test this hypothesis, we stimulated HEK293 cells transfected with Dectin-1A with a variety of soluble β-glucans that have different structures and sizes. We chose to work in this model system because it provides a simplified platform necessary to investigate the physical biology of Dectin-1A activation by isolating Dectin-1A signaling from the complex milieu of other receptors and other Dectin-1 isoforms expressed in innate immunocytes. Also, this model facilitates the expression of multiple fluorescent protein-tagged Dectin-1A constructs necessary to the work. Using calcium imaging and western blotting assays, our results revealed that Dectin-1A activation is influenced by the β-glucan triple-helical structure. Furthermore, our subcellular Förster resonance energy transfer (FRET) measurements by fluorescence lifetime imaging microscopy (FLIM-FRET), as well as application of fluorescence correlation spectroscopy approaches, revealed shifts to higher states of aggregation of Dectin-1A when stimulated with highly structured β-glucans. In addition, these dimerization events occurred in fungal contact sites of fungal cells with high glucan exposure. Together, our findings indicate that β-glucan structure is required for Dectin-1A to form membrane aggregates.
Materials and methods
Cell culture
The HEK293 (ATCC, Manassas, Virginia, no. CRL-1573) cell line was maintained in Dulbecco’s minimum essential medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 2 mM L-glutamine, 11 mM sodium pyruvate, and 1% HEPES. Cells were grown in an incubator at 37°C at 5% CO2 and saturating humidity. Cells were maintained at 37°C, 5% CO2, and 75% relative humidity during imaging.
Plasmids and transfection of Dectin-1 constructs
Emerald-Dectin1A-N-10 (Addgene, Watertown, Massachusetts, plasmid no. 56291), Emerald-Dectin1A-C-10 (Addgene, plasmid no. 54057), mCherry-Dectin1A-C-10 (Addgene, plasmid no. 55025), and mCherry-Dectin1A-N-10 (Addgene, Watertown, Massachusetts, plasmid no. 55026) was a gift from Michael Davidson. pUNO1-hDectin-1A (InvivoGen, San Diego, California) was stably transfected into HEK293 cells for use in our calcium studies. Stable transfection of mEmerald-Dectin-1A was used for Syk immunoblotting experiments. To generate stable lines, HEK293 cells expressing either mEmerald-Dectin-1A or pUNO-hDectin-1A were selected using Geneticin (G418 Sulfate) (Thermo Fischer Scientific, Waltham, Massachusetts, no. 10131035) at 400 μg/mL or Blasticidin (Santa Cruz Biotechnology, Santa Cruz, California, no. SC-495389) at 20 μg/mL, respectively, for 2 weeks.
All other experiments involving exogenous protein expression used transient transfection. Transient transfection with plasmids was performed using standard manufacturer protocols with Fugene 6 (Promega, Madison, Wisconsin, no. E2691).
Fungal growth/preparation
Candida albicans SC5314 (ATCC, Manassas, Virginia, MYA-2876) or TRL035 yeast cells were grown from glycerol stock, and stored at −80°C. Samples were grown in YPD, for 16 h at 37°C in an orbital shaker at 250 rpm to mid log phase. Following a 3-min centrifugation at 6000 rpm, the supernatant was removed, and the cells were resuspended in 4% paraformaldehyde and sterile phosphate-buffered saline (PBS) for 15 min. The cells were centrifuged and washed with sterile PBS three times. The cell concentration was then determined using a disposable hemocytometer (C-Chip, Bulldog Bio, Portsmouth, New Hampshire, catalog no. DHC-N01). Cells (3.5 × 106) were resuspended in 1 mL of PBS. One hundred microliters of the solution was added to HEK293 cells in 35 mm dishes 15 min before imaging.
Glucan particles
Glucan microparticles were prepared from lyophilized C. albicans SC5314 yeast derived from stationary phase culture in YPD. Dry yeast were extracted thrice in boiling 0.75 N NaOH (15 min), then residue was extracted thrice in boiling 2 N H3PO4 (15 min), then residue was extracted thrice in boiling acidic ethanol (1%, v/v, H3PO4 in ethanol; 15 min), and residue slurry was adjusted to neutral pH and washed thrice with ultrapure water. Pyrogen-free reagent and glassware was used throughout preparation, and particles were stored at 4°C in sterile, pyrogen-free water.
Soluble glucan chromatographic analysis
Low molecular weight (LMW) (11 kDa), medium molecular weight (MMW) (145 kDa), and high molecular weight (HMW) (450 kDa β-(1,3;1-6)-glucan extracted from Saccharomyces cerevisiae cell wall was obtained from ImmunoResearch (Eagan, MN). The molecular weight was assessed by gel permeation chromatography and multiangle light scattering. Samples (100 μg) were injected and eluted with a mobile phase of 0.15 M sodium chloride containing 0.02% sodium azide at a flow rate of 0.5 mL/min using two Waters Ultrahydrogel 500 columns and one Waters Ultrahydrogel 250 column connected serially. The samples were run with the column temperature at 18°C. The Mw was calculated using Wyatt Astra software using data resulting from measurements of the angular variation of scattered light using the multiangle light scattering detector coupled with the concentration measured by the refractive index signal.
Soluble glucan linkage analysis
Desalted and lyophilized samples of the fractions were dissolved in dimethylsulfoxide (DMSO) and treated with NaOH and methyl iodide to methylate all free hydroxyl groups (45). The methylated material was purified by extraction with dichloromethane and washing with water. The purified material was then hydrolyzed with trifluoroacetic acid, the reducing ends of the resulting sugars were reduced with NaBD4, and then the resulting free hydroxyl groups were acetylated with acetic anhydride. The mixture of partially methylated alditol acetates was analyzed by gas chromatography. Each derivative corresponding to a particular linkage has been identified by a characteristic retention time and mass spectrum using a mass detector. The relative amount of each derivative was measured by gas chromatography with flame ionization detection. The areas obtained for each observed peak were used to calculate the relative amounts of each type of linkage found in the sample (Table 1). The 3,6-linked residues represent branch points.
Table 1.
Linkage analysis of glucans used in this study
| LMW | MMW | HMW | |
|---|---|---|---|
| Terminal glucose | 6.4 | 5.1 | 5.4 |
| 3-Glucose | 87.5 | 87.6 | 84.6 |
| 3,6-Glucose | 3.2 | 3.8 | 4.0 |
| 6-Glucose | 1.3 | 2.2 | 3.2 |
| 4-Glucose | 0.1 | 0.2 | 0.1 |
| Other | 1.5 | 1.1 | 2.7 |
1H NMR spectroscopy
The samples were dissolved in DMSO-d6/D2O (6:1 by volume) at 100°C for 1 h. 1H NMR spectra were recorded at the University of Minnesota Department of Chemistry NMR lab on a Varian UNITYplus-300 spectrometer at 80°C. The spectra were collected at 300 MHz with 32 scans, a relaxation delay of 1.5 s, a pulse of 45°, an acquisition time of 2.0 s, and a spectral width of 5999 Hz. Table 2 provides 1H NMR chemical shifts in all three glucans used in this study as well as literature values (46). These data, taken together with other characterization methods used, do confirm that the structure of the polysaccharides used in this study conforms to expected results from fungal cell wall glucans (Table 2; Fig. S3).
Table 2.
1H NMR chemical shifts for all glucans used in this study
| H-1 (1,3-Glucan) | H-2, 4, and 5 (1,3-) | H-3 and 6b (1,3-) | H-6a (1,3-) | |
|---|---|---|---|---|
| LMW | 4.52, d, J = 7.8 Hz, 1H | 3.20–3.33 m, 3H | ∼3.5, m, (hidden by H2O) | 3.72, d, J = 11 Hz, 1H |
| MMW | 4.52, d, J = 8.1 Hz, 1H | 3.21–3.33 m, 3H | ∼3.5, m, (hidden by H2O) | 3.72, d, J = 11 Hz, 1H |
| HMW | 4.52, d, J = 7.8 Hz, 1H | 3.20–3.32 m, 3H | ∼3.5, m, (hidden by H2O) | 3.72, d, J = 11 Hz, 1H |
| β-1,3/1,6-Glucan literature values | 4.52, d, J = 8 Hz, 1H | 3.25, m, 3H | 3.46, m, 2H | 3.7, d, J = 11 Hz, 1H |
Microscopy and image analysis (calcium imaging and RICS/N&B)
Confocal images were obtained on an Olympus FV1000 laser scanning confocal microscope (Olympus, Center Valley, PA) built around an IX81 inverted microscope. A 10× objective lens (0.40 NA) or a super corrected 60× oil objective lens (1.40 NA), Plan-Apochromat objective lens was used for imaging. Samples were excited with a 20 mW, 473 nm diode laser and a 20 mW, 635 nm diode laser. These lines were reflected to the specimen by a 405/473/559/635 multiedge main dichroic element followed by bandpass emission filters in front of two independent high-sensitivity GaAsP PMT detectors (HSD1/2). Specifically, the emission light passed by the main dichroic was directed to our first detector (HSD1) via reflection from an SDM560 filter cube and passage through a BA490-540 nm bandpass filter. Our second detector (HSD2) received light passed by the SDM560 filter cube and routed through a BA575-675 nm bandpass filter.
Calcium imaging
HEK293 cells expressing Dectin-1A were plated at 40,000 cells in 35 mm MatTek dishes 24 h before imaging. These cells were loaded with Fluo-4 and CellMask Deep Red (CMDR) at equimolar concentrations of 1 μM in 2 mL of medium for 1 h then washed before imaging. CellTracker Deep Red was used as a cell cytosolic volume control to account for cytosolic changes from cell contraction that occurs during stimulation. For Syk inhibition, plates were pretreated with 250 nM of Syk Inhibitor (Calbiochem, San Diego, California, no. 574711) for 30 min under normal growth environmental conditions. Images were taken at a resolution of 256 × 256 with a dwell time of 2 μs on a 10× objective lens (0.40 NA). A 20 mW, 473 nm diode laser operated at 4% power and CMDR was excited with a 20 mW, 635 nm diode laser operated at 4% power. Fluorescence of Fluo-4 was collected by a cooled GaAsP PMT set to 700 V, gain 1×, and offset of 0%. CMDR signal was collected by a cooled GaAsP PMT detector set to 700 V, gain 1×, and offset of 0%. Thirty frames before stimulation were used to set the basal fluorescence of the fluo-4 dye. After stimulation with 100 μL of 10 μg/mL of glucan, cells were imaged for 100 frames. To assess changes in intracellular calcium concentration, we measured the ratio of Fluo-4/CMDR intensity to correct for any variations in cytoplasmic volume within the confocal section across the field. This ratio was normalized to 1.0 based on mean prestimulation values (30 frames) and changes in calcium influx were measured as fold change of this normalized ratio (MFI fold change).
For MMW denaturation experiments, soluble β-glucans were weighed and resuspended in reverse osmosis purified H2O. To denature MMW glucan, we incubated MMW in DMSO or 1 M NaOH. To renature the glucan from DMSO, we placed denatured MMW into Slide-A-Lyzer Dialysis Cassettes (Thermo Fisher Scientific, no. 66203) of a molecular weight cut-off of 2000 Da and dialyzed against reverse osmosis purified H2O for 24 h. To renature glucan from 1 M NaOH, the solution was neutralized using 1 M HCl.
Protein isolation and immunoblotting
HEK293 cells stably expressing mEmerald-Dectin-1A were seeded at 5 × 105 in 6-well plates 24 h before the experiment. Cells were stimulated with LMW, MMW, and HMW β-glucan (1 mg/mL) for 5 min, then lysed. Cells were extracted in 1× lysis buffer (43.9 mM HEPES [pH 7.5], 131.7mM NaCl, 1.1% Triton X-100, 8.8% glycerol, 1× protease inhibitor cocktail, 1 mM PMSF, 1 mM EGTA). Samples were centrifuged at 12,000 × g for 20 min at 4°C and supernatants transferred to fresh tubes. Protein concentrations were determined by Bradford assay (Bio-Rad protein reagent, Hercules, California). NuPAGE LDS Sample Buffer (4×) with NuPAGE Sample Reducing Agent (10×) was added to samples (1× final concentration). Total proteins (typically 20–50 μg) were subjected to 4–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to Immobilon-FL PVDF transfer membrane (Millipore Sigma, Burlington, Massachusetts) using NuPAGE transfer buffer. Membranes were blocked with bovine serum albumin in Tris-buffered saline-Tween-20 (20 mM Tris, 137 mM NaCl, 0.1% Tween-20) and incubated with primary antibodies overnight at 4°C. Antibodies purchased from Cell Signaling Technology (Danvers, Massachusetts): rabbit mAb for p-SYK (Tyr525/526) and β-actin (13E5), and mouse mAb Syk (4D10) were used according to the manufacturer’s recommendations (1:1000). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies (Cell Signaling, Danvers, Massachusetts, or GE Healthcare, Chicago, Illinois) were used at a 1:10,000 dilution. Blots were visualized on a Li-Cor Odyssey FC imaging system and analyzed with Image Studio.
Biolayer interferometry
Advanced kinetics biolayer interferometry experiments were conducted using the Personal Assay BLItz System. Anti-human IgG Fc Capture biosensor tips were initially loaded with Dectin-1A:FC fusion protein (InvivoGen, no. fc-hdec1a) at 13 μg/mL. Binding kinetics were obtained for LMW, MMW, HMW, MMW (denatured), and MMW (renatured) at 0, 10, 50, 100, and 250 nM in triplicate. A global fitting was performed on the curves obtained using the BLItz software.
Congo red spectroscopic assay
A BioTek EonN Multiwell Spectrophotometer was used to analyze Congo red absorbances. A solution of 8.8 μM Congo red, 0–1 M NaOH solution (1, 0.75, 0.5, 0.25, 0.1, 0.075, 0.05, 0.025, 0.001, 0 M) and LMW, MMW, or HMW β-glucans at 1 mg/mL were analyzed for the denaturation experiments. For the renaturation experiments, 1 mg/mL of β-glucan was denatured at 1 M solution then renatured through neutralization with HCl 24 h before readings. DMSO denaturation conditions involved DMSO in water at 0, 5, and 10%. For the DMSO renaturation experiments, DMSO was removed by dialysis (see above) before spectrophotometer readings. Absorbance readings were taken at 400–700 nm with 1 nm steps. All experiments were conducted in technical triplication across three independent experimental replicates.
FLIM-FRET measurement
HEK293 cells were plated at 25,000 cells in 35 mm MatTek dishes 48 h before imaging. Cells were transfected with mEmerald-Dectin1A-N-10 and mCherry-Dectin1A-N-10 24 h before imaging. FLIM-FRET images were obtained using a Leica DMi8 inverted microscope. A Leica Harmonic Compound PL apochromatic CS2 63× water objective with a correction collar (1.2 NA) was used for imaging. A tunable and pulsed white light laser (470–670 nm) was operated at 80 MHz at 3% laser power using a 488 nm notch filter to excite our sample. A scan speed of 200 lines/s and a 256 × 256 pixel resolution (full field of view) was used for data acquisition. Two hybrid detectors collected photons at 512–540 and 650–700 nm, respectively, on the counting mode setting. Temperature was kept at 37°C using a Tokai Hit (Fujinomiya, Shizuoka, Japan) Stage Top Incubator for live cell imaging. Lifetime images were collected using a PicoHarp 300 Fluorescence lifetime microscopy time-correlated single-photon counting system. For our glucan-stimulated cells, 23 frames were collected before stimulation. A total of 230 frames were taken immediately after stimulated with β-glucans at a concentration of 10 μg/mL. Analysis was conducted on minute time points (1–5 min) by averaging 10 frames from 1 min intervals. For yeast contact site imaging studies, 3.5 × 106 fixed yeast cells were resuspended in 1 mL of PBS. One hundred microliters of the solution was added to HEK293 cells in 35 mm dishes 15 min before imaging. Twenty-three frames were collected per cell. Images were collected at a maximum of 45 min after the addition of yeast per plate.
Analysis was conducted on the plasma membrane by masking out internal cellular compartments on the images. For our fungal contact site studies, analysis was conducted on the plasma membrane that was in contact with the fungus and a separate masking for plasma membrane that was not in contact with any yeast. A biexponential fit was performed to the decay curve. For donor only as well as donor and acceptor on opposite sides of the plasma membrane (negative control), the decay curve indicated a negative amplitude for one of the components, thus indicating a monoexponential decay. Therefore, decay curves from these samples were analyzed using a monoexponential fit. For cells with donor-acceptor on the cytosolic tail, data were fit to a biexponential decay with the first lifetime component being locked at the donor only lifetime of 2.4 ns (τD). Lifetime values of the second component (τDA) of the decay curve were used to calculate FRET efficiency using the equation: . To determine the fraction of receptors undergoing an FRET process (donor-acceptor population), the amplitude ratio between the first component (AmpD) and the second component (AmpDA) from the biexponential decay curve fit was calculated according to the following formula: .
We characterized the expression of donor- and acceptor-tagged Dectin-1 by raster image correlation spectroscopy (RICS) analysis, we found that coexpression of donor- and acceptor-tagged Dectin-1 resulted in mean receptor density of 4612/μm2 (Fig. S4 A). We note that a small minority of outlier measurements were considerably higher than this average and may have been impacted by imaging artifacts, such as inadvertent inclusion of both dorsal and ventral plasma membrane. Within single cells, donor- and acceptor-tagged Dectin-1 was expressed at approximately similar levels (Fig. S4 B).
Raster image correlation analysis/number and brightness
Protocols on RICS and number and brightness (N&B) analysis have been previously described in more depth and analysis of diffusion coefficient and receptor density were performed using SimFCS software according to these previously published procedures (47,48). HEK293 cells expressing Emerald-Dectin1A-C-10 were plated at 40,000 cells in 35 mm glass-bottom MatTek dishes 24 h before imaging using equipment described in microscopy and image analysis above. Measurements were performed at the membrane facing the glass coverslip. Images were collected at 256 × 256 pixel resolution on a 60× 1.4 NA oil immersion objective lens with a scanning zoom of 16.4× (0.050 μm vertical and horizontal center to center distance between resulting image pixels). Data were collected using a GaAsP PMT detector operated in photon-counting mode. The 473 nm diode laser operated at 0.1% power was used in these images. The point spread function radial beam waist was estimated using 192 nM EFGP in solution and setting the diffusion coefficient to 90 μm2/s. Under these conditions the beam waist was determined to be 0.21 μm. Immobile features were removed using a four-frame moving average subtraction. Cells were stimulated with a final concentration of 1 μg/mL of glucan. After stimulation, 200 frames were collected at a pixel dwell time of 4 μs/pixel (line scan of 1.096 ms) with a pinhole size of 100 μm. All cell body pixels were used for analysis of N&B data from cells.
Images collected were also used for our numbers and brightness analysis; we used 192 nM EGFP in solution and purified mEmerald-Dectin-1 protein to set the average brightness of our monomeric protein (Fig. S4, C and D). Furthermore, the S-factor was calculated using the background image. We divided each brightness distribution by monomeric, dimeric, and oligomeric sections according to previous research (49). The cursors were scaled quadratically and centered at B values of 1.3, 1.6, and 2.35 for monomers, dimers, and oligomers, respectively.
dSTORM
HEK293 cells, were grown on cleaned and poly-L-lysine (0.1 mg/mL)-coated coverslips (∼5 × 104 cells/coverslip) within wells of a 6-well plate at 37°C 24 h before the experiment. The cells were then treated with MMW glucan at 1 μg/mL for 50 s. The cells were then fixed with paraformaldehyde (4%) for 5 min at 37°C followed by three washes of PBS.
Data acquisition was on an Olympus IX-71 microscope equipped with an objective-based TIRF illuminator using an oil-immersion objective (PlanApo N, 150×/1.45 NA, Olympus) in an oblique illumination configuration. Sample excitation was done using a 637 nm laser (Thorlabs, Newton, New Jersey, laser diode HL63133DG) with custom-built collimation optics (13). To minimize the drift that occurred during data acquisition, a self-registration algorithm was implemented (50,51).
The Dectin-1A nanodomain density by glucan exposure engagement was quantified by super-resolution imaging and analyzed using a hierarchical single-emitter hypothesis test (H-SET) as a clustering algorithm in MATLAB (The MathWorks, Natick, MA) (13). The data for 34 cells for each condition were run through the first pass of H-SET to collapse multiple observations of the blinking fluorophores into single estimates of the true fluorophore locations (13). The second H-SET pass determined clustering using the DBSCAN algorithm (52), which depends on the two parameters: (minPts), that is, the minimum number of objects composing a multicluster, and the maximum distance between the objects within a multicluster (epsilon). We optimized these parameters, defining them as 3 and 27 nm, respectively, according to optimization procedures described previously (53).
Software
For the RICS and N&B Data presentation and analysis we used the SimFCS Program (www.lfd.uci.edu). The calcium imaging was analyzed using ImageJ. The FLIM-FRET results were analyzed using Symphotime64 software. The biolayer interferometry analysis was done using BLItz Pro software. Statistical analysis was performed with GraphPad Prism version 8.2 (GraphPad Software). Direct stochastic optical reconstruction microscopy (dSTORM) analysis was performed using MATLAB using our own algorithms from previously cited works.
Statistical analysis and data summary
Sample sizes and number of data points are described in the figure legends. Biological replicates are defined as methodologically independent experimental results, such as experiments conducted on different days with independently prepared materials. Technical replicates are defined as experimental replicates conducted in parallel with one another, such as experiments performed with materials prepared at the same time and measured on the same day. No outlier exclusion was used for any of the datasets presented in this work. To best summarize the significant quantitative findings of this work, Table S1 reports all measured values as mean and standard deviation.
Results
Dectin-1A activation is dependent on the molecular weight of the soluble β-glucan
β-Glucans, existing as insoluble fibers in the cell wall, are likely to have a high degree of tertiary and quaternary structure. So, an encounter with highly structured glucan might be indicative of a pathogen cell wall structure. Furthermore, less structured glucans are encountered by Dectin-1A physiologically (40). Small soluble circulating glucan can derive from sloughed cell wall material or from dietary absorption (25,54,55,56,57,58,59). However, there is not much known about Dectin-1A’s ability to distinguish between highly structured β-glucan found on cell walls of fungal pathogens and less structured glucans found in circulation. Therefore, we examined how Dectin-1A activation is affected by glucans with different quaternary and tertiary structures. To accomplish this, we used HMW (450 kDa), MMW (145 kDa), and LMW (11 kDa) soluble glucans, in decreasing order of tertiary and quaternary structures, derived from S. cerevisiae cell walls. These S. cerevisiae glucans in soluble form or as particulate “zymosan” are common models for stimulation of innate immunocytes by fungal pathogen cell wall glucan. The above glucans have overall very similar composition and structures to C. albicans yeast glucan, although relatively minor differences in β-(1,6)-glucan side-chain length and branching frequency have been reported between these species (46,60).
Using these glucans, we performed intracellular calcium ([Ca2+]i) measurement experiments on HEK293 cells (lacking endogenous Dectin-1 expression) transfected with Dectin-1A. We stimulated the cells using LMW, MMW, or HMW glucan. All the glucans induced a significant increase in peak amplitude [Ca2+]i compared with unstimulated cells (Fig. 1 A and B). We found that large, highly ordered glucans (HMW and MMW) induced a significant, Dectin-1-dependent increase in peak amplitude of [Ca2+]i compared with LMW, with MMW having the highest peak amplitude (Fig. 1 C). Interestingly, the response to MMW glucan was uniform at the single-cell level. However, cells stimulated with HMW glucan exhibited a more heterogeneous response, with some cells achieving comparable maximum amplitudes as with MMW and others exhibiting little change from basal calcium levels (Fig. 1, A and B). We expect that HMW glucan is present as larger particles than MMW, so, at equal mass/volume concentrations, the HMW solution will have a lower concentration of particles. Nonresponder cells in HMW experiments may have stochastically encountered too few glucan particles to achieve a detectable signaling response. When we stimulated cells with MMW or HMW at equimolar concentrations, which should have similar glucan particulate concentrations, we observed a minor difference in peak amplitude, but we saw that the integrated Ca2+ flux over time was the same for MMW and HMW glucans (Fig. S1). Furthermore, single-cell data demonstrated a similarly uniform response of Dectin-1 to MMW and HMW glucans under these conditions. These results indicate that Dectin-1A drives differential Ca2+ flux to glucan ligands that vary in size and structure.
Figure 1.
Differential signaling response of Dectin-1A to soluble β-glucan. (A) HEK293 cells stably transfected with Dectin-1A were loaded with Fluo-4 and CellTracker Deep Red at equimolar concentrations. CellTracker Deep Red was simultaneously loaded to normalize for changes in cytosolic volume caused from cell contraction. The mean fluorescence intensity of 30 cells was averaged for Dectin-1A transfected (black) or untransfected (gray) HEK293 cells stimulated with LMW, MMW, or HMW glucan at 1 μg/mL (n = 30 per glucan from 3 independent experiments per glucan). Data shown as mean fold change in volume-normalized [Ca2+]intracellular. (B) Heatmaps of relative changes in intracellular calcium concentration of untransfected or Dectin-1A transfected individual cells upon addition of LMW, MMW, or HMW glucan. Each row represents the normalized ratio of Fluo-4 and CellTracker Deep Red for a single cell over time. (C) Maximum signal amplitude of single cells treated with LMW, MMW, or HMW glucan showing Dectin-1-dependent glucan responses. Statistical comparison of maximum amplitudes for Dectin-1-expressing cells treated with each of the three glucans is shown in the far right panel. Data shown as mean ± SD (n = 30 per glucan from 3 independent experiments per glucan). One-way ANOVA with multiple comparisons by Dunnett’s test, ∗∗∗p < 0.0001, ∗∗∗∗p < 0.000001. (D) Cell lysates were collected at 5 min after stimulation and analyzed by western blotting using antibodies against p-Syk and Syk. The intensity of p-Syk was normalized against total Syk. (E) Quantification of the western blot normalized p-Syk/Syk signal is shown as mean ± SD of n = 3 independent experimental replicates. (F) Untransfected (gray) and stably transfected Dectin-1A (black) HEK293 cells were loaded with Fluo-4 and CellTracker Deep Red at equimolar concentrations and treated with Syk inhibitor at 250 nM, then stimulated with MMW glucan. Average mean fluorescence intensity of 30 cells was observed for cells stimulated with MMW glucan at 1 μg/mL (n = 30 from 3 independent experiments).
To determine how these differently structured, soluble glucans impacted cellular patterns of Syk phosphorylation, we stimulated HEK293 cells expressing Dectin1A-mEmerald with H2O (vehicle), LMW, MMW, or HMW. Whole-cell lysate was collected and Syk phosphorylation was determined by western blot analysis. Likewise, our results show an increase in Syk phosphorylation in the larger, highly structured glucan MMW and HMW compared with unstimulated and LMW-stimulated cells (Fig. 1, D and E). In addition, Syk inhibitors abrogated calcium signaling of Dectin-1A when stimulated with MMW glucan (Fig. 1 F). These results indicate that glucans with higher-order structure are better able to activate Dectin-1A-mediated Ca2+ signaling and that this is a Syk-dependent process.
β-Glucan denaturation abrogates its potential for Dectin-1A activation
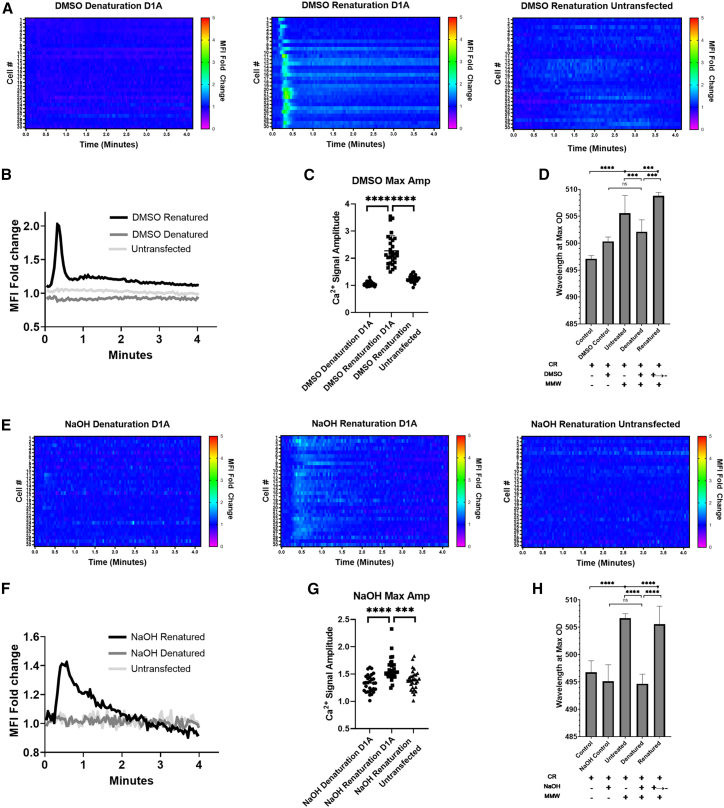
To determine if the glucan structure affects signaling outcomes, we denatured MMW (highly stimulatory glucan) using DMSO, a chaotrope that promotes a reduction in glucan tertiary structure, thus shifting MMW’s triple helix structure to a more single helical or random coil structure (16). The results showed that when we denatured MMW glucan, we did not observe calcium signaling in cells expressing Dectin-1A (Fig. 2, A and B). However, when we renatured the glucans by removing DMSO via dialysis we observe partial recovery of calcium signaling. We found that renatured glucans induce a significant increase in peak amplitude [Ca2+]i response compared with DMSO denatured MMW and renatured MMW stimulated untransfected HEK293 cells (Fig. 2 C). In addition, we confirmed the loss of helical structure via a Congo red assay. Congo red specifically binds to β-(1,3)-glucans with a triple helix conformation as their tertiary structure. This binding is detected by bathochromic shift in absorbance maximum from 488 to 516 nm (61). Our results indicated a loss in glucan structure after denaturation in a DMSO solution that was regained upon renaturation (Fig. 2 D). Moreover, we repeated these experiments by stimulating cells with glucan denatured with NaOH or neutralized renatured glucan (16). Similarly, our results show that cells lose the ability to activate Dectin-1A calcium signaling when stimulated with denatured MMW glucan but regain the ability to stimulate Dectin-1A activation when the glucan is renatured (Fig. 2, E and F). We found that renatured glucans induce a significant increase in peak amplitude [Ca2+]i response compared with NaOH denatured MMW and renatured MMW stimulated untransfected HEK293 cells (Fig. 2 G). In addition, we confirmed that glucan structure was lost when NaOH was added and regained when neutralized (Fig. 2 H). These results suggest that glucan structure is an important factor in activating a Dectin-1A response.
Figure 2.
Glucan higher-order structure is a critical determinant of its stimulatory potential. (A) HEK293 cells stably expressing Dectin-1A or untransfected were loaded with Fluo-4 and CellTracker Deep Red at equimolar concentrations. Heatmaps show relative changes in intracellular calcium concentration of untransfected or Dectin-1A transfected individual cells upon either addition of DMSO denatured MMW glucan or renatured MMW glucan. Each row represents the normalized ratio of Fluo-4 and CellTracker Deep Red for a single cell over time. (B) Average mean fluorescence intensity of 30 cells stimulated with DMSO denatured/renatured MMW glucan was observed (n = 30 from 3 independent experiments). (C) Maximum amplitude of single cells treated with DMSO denatured or renatured MMW glucan. Data shown as mean ± SD (n = 30 from 3 independent experiments). One-way ANOVA with multiple comparisons by Dunnett’s test, ∗∗∗∗p < 0.000001. (D) 1 mg/mL of MMW glucan was denatured using DMSO and incubated with Congo red. Control samples contained Congo red and DMSO. Renaturation was accomplished by dialyzing out DMSO 24 h before the experiment. Data shown as mean ± SD (n = 9 from 3 independent experiments). (E) HEK293 cells stably expressing Dectin-1A or untransfected were loaded with Fluo-4 and CellTracker Deep Red at equimolar concentrations. Heatmaps show relative changes in intracellular calcium concentration of untransfected or Dectin-1A transfected cells upon either addition of NaOH denatured or renatured MMW glucan. Each row represents the normalized ratio of Fluo-4 and CellTracker Deep Red for a single cell over time. (F) Average mean fluorescence intensity of 30 cells stimulated with NaOH denatured/renatured MMW glucan was observed (n = 30 from 3 independent experiments). (G) Maximum amplitude of single cells treated with NaOH denatured or renatured MMW glucan. Data shown as mean ± SD (n = 30 from 3 independent experiments). One-way ANOVA with multiple comparisons by Dunnett’s test, ∗∗∗∗p < 0.000001. (H) MMW glucan (1 mg/mL) was denatured using NaOH and incubated with Congo red. Control samples contained NaOH and Congo red. Renaturation was accomplished by neutralizing NaOH. Data shown as mean ± SD (n = 9 from 3 independent experiments). Welch’s t-test, ∗∗p < 0.001, ∗∗∗p < 0.0001, ∗∗∗∗p < 0.000001.
β-Glucan structure variation and manipulation does not alter affinity for Dectin-1
We considered the possibility that these soluble glucans might have different affinities for Dectin-1A, resulting in differential receptor activation. Thus, we conducted biolayer interferometry experiments to determine the binding affinity of these glucans to the carbohydrate recognition domain of Dectin-1A. This was accomplished using a chimeric fusion protein of the carbohydrate recognition domain of Dectin-1A and the human IgG Fc region. An anti-human IgG Fc Capture biosensor tip was used to load this fusion protein. Association and dissociation curves of the glucan and fusion protein where then collected. The results shown in Fig. 3 A indicate that all the glucans have approximately nanomolar dissociation constants for Dectin-1A carbohydrate recognition domain despite the previously described differences in signaling (26). Weight average molecular weights of purified S. cerevisiae β-(1,3)-glucan fractions vary over an approximate 40-fold range, but there is relatively little difference between these glucans’ apparent affinity for the Dectin-1 carbohydrate recognition domain.
Figure 3.
Characterization of Dectin-1 binding affinity and helical structure of fungal cell wall glucans used in this study. (A) Biolayer interferometry experiments were conducted on LMW, MMW, HMW, MMW denatured, and MMW renatured glucans using an anti-human IgG Fc capture biosensor tip and a Dectin-1A-Fc fusion protein. Data shown as mean ± SD (n = 3 from 3 independent experiments). Statistical comparison by one-way ANOVA. (B) LMW, MMW, and HMW β-glucans were denatured using 0–1 M NaOH in the presence of Congo red. Concentration of NaOH at which absorbance (516 nm) decreased to the half-maximal value was plotted. Data shown as mean ± SD (n = 9 from 3 independent experiments).
Furthermore, to determine differences in the structure of these glucans, we analyzed the conformational transition of triple helix to random coil of β-1,3-D-glucans through denaturation experiments. Experiments were conducted by denaturing glucans with NaOH at various concentrations in the presence of Congo red. Our results show that the amount of glucan tertiary structure scales with molecular weight as measured by the concentration of NaOH required to reduce Congo red binding to glucan (Fig. 3 B), suggesting that the size of the glucans is correlated with their higher-order structure. Together, these results indicate that downstream signaling of the receptor is determined by the structure of the glucan rather than affinity alone.
Dectin-1A decreases in diffusion coefficient when stimulated with highly structured β-glucans
We measured the diffusion coefficient of Dectin-1A pre/post glucan stimulation to determine whether the receptor diffusion coefficient decreased. We obtained average diffusion coefficients and spatial number density of our receptor using RICS. RICS allowed us to survey multiple areas of the cell for molecular parameters such as diffusion coefficient and receptor density. This section pertains to receptor diffusion measurement by RICS. Furthermore, because fluorescence was probed within a large cell area, RICS analyses suffered much less from photobleaching and location-specific artifacts than analogous single-point measurements. Previous research has described RICS in more detail (47,62). In brief, we generated a volume of excitation using focused laser illumination and calibrated a confocal observation volume using standard fluorophores of known diffusion coefficient. We observed fluorescent molecules (i.e., Dectin-1A-mEmerald) diffusing in and out of this excitation volume through fluctuation in the number of photons obtained. Experimentally observed fluorescence correlations at various spatiotemporal lags were then fit to a 2D autocorrelation function to obtain the receptor diffusion coefficient in the observed membrane (Fig. 4 A).
Figure 4.
Dectin-1A surface diffusion coefficient decreases when stimulated with highly structured β-glucans. (A) A representative example of raster image correlation spectroscopy (RICS) analysis. (Left) Representative RICS image of HEK293 cells expressing Dectin-1A-mEmerald. (Middle) Autocorrelation function (ACF) calculated from the time series. Red represents a high ACF value, blue represents a low ACF value. (Right) Fit of the ACF to a Gaussian diffusion model to calculate the diffusion coefficient. (B and C) RICS analysis of fluorescently tagged Dectin-1A expressed in HEK293 provided average diffusion coefficient (B) and receptor number (C) for cells that were unstimulated or stimulated with LMW, MMW, or HMW. Data shown as mean ± SD (n = 30). One-way ANOVA multiple comparison test, ∗∗∗∗p < 0.00001. To see this figure in color, go online.
Using HEK293 cells expressing Dectin-1A-mEmerald, we conducted RICS measurements before and after stimulation with soluble β-glucans. We determined that cells stimulated with MMW or HMW exhibited a significant decrease in mobility compared with LMW and unstimulated cells (Fig. 4 B). This finding may reflect some partial contribution of slower diffusion of larger ligand/receptor aggregates formed after glucan stimulation. However, changes in the membrane subjacent cytoskeleton or entry into nascent endocytic structures likely also influenced the reduced diffusion observed. It is possible that the decline in receptor density seen with MMW and HMW glucan stimulation (Fig. 4 C) could reflect the shift of diffusing Dectin-1 into more highly aggregated states (at lower density) and/or endocytic removal of receptor from the membrane. We proceeded to conduct additional membrane biophysical studies to further test for changes in the molecular aggregation state of Dectin-1 during stimulation with glucan.
Dectin-1A enters states of increased aggregation when stimulated with highly structured β-glucans
The results shown above indicate that the β-glucan structure is an important factor in signaling outcomes. Previous research has shown that other transmembrane CTLs that also contain a (hem)ITAM domain can form homodimers before or upon ligand recognition (63,64). Furthermore, crystallography studies of the carbohydrate recognition domain (CRD) have shown that Dectin-1A headgroups form dimers when laminaritriose is present (64). In addition, size-exclusion chromatography with multiangle light scattering analysis has described Dectin-1 ligand-induced tetramer (or dimer-of-dimers) formation in solution (64,65). In line with these ideas, we sought to examine how ligand structure impacts signaling by determining changes to the aggregation state of full-length Dectin-1A in living cell membranes.
To assess changing molecular proximity of Dectin-1 proteins, we employed FLIM-FRET. FRET-based imaging capitalizes on close proximity of two proteins to visualize protein-protein interactions, including receptor dimerization and receptor-ligand complex formation (66). FLIM characterizes the duration of a fluorophore’s excited state before returning to the ground state. The occurrence of FRET causes rapid quenching of donor fluorescence, so FRET can be determined by measuring the shortening of donor fluorescence lifetime when in proximity to acceptor. FLIM-FRET offers the opportunity of studying in vivo receptor interactions in a direct, spatially resolved manner. We examined Dectin-1A engagement using FLIM-FRET on HEK293 cells coexpressing two fluorescent Dectin-1 constructs—N-terminally tagged Dectin-1A-mEmerald (donor) and Dectin-1A-mCherry (acceptor)—both having their fluorophores on the cytoplasmic face of the plasma membrane, and then stimulated the cells with soluble β-glucan. Analysis was conducted on the plasma membrane itself by masking out internal cellular compartments (Fig. 5 A). FRET efficiency is a parameter that exhibits an inverse sixth power dependence upon donor-acceptor distance. Donor-acceptor percentage is simply the percentage of all donors that are involved in FRET interactions at a given time. These parameters were determined experimentally by performing biexponential curve fits to the observed frequency distribution of donor fluorophore lifetimes, wherein one exponential component represents the population of non-FRET (presumably monomeric) donors and the other exponential component represents the performance of the donors that are involved in FRET interactions (presumed receptor oligomers).
Figure 5.
Highly structured β-glucans induce dimerization/oligomerization of Dectin-1A. (A) Representative average lifetime image of HEK293 cell transfected with Dectin-1A-mEmerald or cotransfected with Dectin-1A-mEmerald and Dectin-1A-mCherry. The analysis was conducted on the plasma membrane by masking out internal cellular compartments on the images so only the masked plasma membrane signal used for analysis is shown in color. In these images, pixel hue indicates raw fluorescence lifetime over all populations observed and pixel intensity indicates total photon counts observed over all populations. (B) Lifetime and amplitudes of HEK293 cells expressing donor only on the cytoplasmic face, donor expressed on the cytoplasmic side and acceptor on the extracellular side, or donor and acceptor placed on the cytoplasmic side of the membrane. Fluorescence decay curves were mono- and biexponentially fit and individual fit components are shown. Data shown as mean ± SD (n = 15 cells). (C–F) FRET efficiency and donor-acceptor population of cells stimulated with (C) LMW, (D) MMW, (E) HMW, or (F) DMSO denatured MMW glucan. Data shown as mean ± SD (n = 15). One-way ANOVA with multiple comparisons by Dunnett’s test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. To see this figure in color, go online.
First, we characterized the average lifetimes of unstimulated cells expressing several configurations of fluorescent protein-tagged Dectin-1A: 1) receptor with donor tag only, 2) coexpression of separate donor- and acceptor-tagged receptors with tags placed on opposite sides of the plasma membrane (a negative control containing both fluorescent proteins but in a configuration that does not permit FRET), and 3) coexpression of separate donor- and acceptor-tagged receptors with both tags in the cytoplasmic tail of the receptors (configuration to be used for experimental determination of receptor aggregation by FRET) (Fig. 5 B). The observed decay curves were analyzed by performing a monoexponential and biexponential fit. For donor only and our negative control we observed a negative amplitude for the second component in the biexponential fit, indicating that a monoexponential fit was superior for these conditions. This was as anticipated since these conditions should have only a single, non-FRET donor signal. In addition, the lifetimes of the second component in these controls unrealistically exceed >150 ns, further justifying a monoexponential fit. Our results show that, when acceptor is not present, we see the expected lifetime of 2.4 ns using a monoexponential fit (Fig. 5 B). Negative controls with donor and acceptor on opposite sides of the membrane yield similar lifetime values. Data from coexpression of cytosolic donor- and acceptor-tagged receptor was fit biexponentially and the lifetime of both components is shown (Fig. 5 B). We observed a decrease in the lifetime of the donor to 0.41 ns (FRET-involved second fit component of a biexponential fit) indicating that some basal level of intermolecular Dectin-1 close proximity interactions were being observed in unstimulated cells. Of course, the first fit component lifetime (non-FRET involved donors) remained at ∼2.4 ns, as expected from controls above. The existence of this basal FRET signal is interesting, and the potential sources and interpretation of this observation are further considered below. However, we first focused on assessing ligation-dependent changes in Dectin-1A’s molecular aggregation state as influenced by various glucans and measured by FRET. For the remainder of the FLIM-FRET experiments, we fixed the lifetime of the first component to 2.4 ns, and we showed the second components’ lifetime and amplitude values as percentages, FRET efficiency and donor-acceptor population, respectively.
When we stimulate cells expressing Dectin-1A with donor and acceptor on the cytoplasmic face of the membrane using MMW (Fig. 5 D) or HMW (Fig. 5 E) glucan, we see a significant increase in the fraction of receptors undergoing FRET (donor-acceptor population) from 15% before stimulation to a maximum of 30% after 5 min of stimulation, with this trend starting at about 1 min poststimulation. However, there is no significant change in FRET efficiency before and after stimulation. On the other hand, when we stimulate with LMW (Fig. 5 C) or denatured MMW (Fig. 5 F), we see no significant change in FRET efficiency or donor-acceptor population. These data are consistent with a model wherein the population of receptors engaged in FRET interactions are in a close, ordered configuration (e.g., possibly a dimer or other oligomeric state) that does not permit a wide range of separations between donor and acceptor tags, leading to a constant FRET efficiency for the donor population that does attain this FRET-capable configuration. However, the fraction of the population of receptors engaged in these close molecular aggregates does change as a result of stimulation with glucan. These results suggest that the highly structured soluble glucans allow for an increase in Dectin-1A aggregation state, possibly including dimerization or oligomerization, to occur, which directly correlates with the amount of receptor activation and signaling observed.
In addition, to better characterize the change in aggregation state of Dectin-1A during activation by glucan, we conducted an N&B analysis on HEK293 cells expressing Dectin-1A-mEmerald. Previous research has described N&B more in depth (67,68,69). In brief, N&B analysis focuses on fluctuation of detected emission photons originating from fluorescent molecules that pass through a known observation volume. Statistics of fast fluctuations of the intensity at each pixel can be used to determine the number and intensity of the particles diffusing through the observation volume. For example, if the fluorescent proteins diffuse as a tetrameric protein, we expect to observe emission intensity fluctuation with four times more photons relative to a monomeric fluorescent protein diffusing through the excitation volume. Receptor aggregation was observed by stimulating these cells with soluble glucans. A brightness versus intensity 2D histogram of each pixel in a time series was developed and selection boxes were drawn to represent low (red box, presumed monomers), mid (green box), and high (blue box) (Fig. 6, A and B) aggregation states. Dectin-1 aggregation state maps of representative untreated (Fig. 6 C) and MMW-stimulated cells (Fig. 6 D) were generated using this color scheme. Our results indicated that unstimulated and LMW-stimulated cells contained a significantly higher amount of low aggregation state pixels compared with MMW and HMW glucan-treated cells (Fig. 6 E). Inversely, we observed a significant increase in pixels with a mid or high aggregation state in cells stimulated with MMW and HMW compared with cells that were unstimulated or LMW stimulated (Fig. 6, F and G). N&B analysis revealed an increase in Dectin-1 aggregation state during stimulation with larger, more ordered glucans.
Figure 6.
Number and brightness analysis shows shift to higher aggregation states of Dectin-1A when stimulated with highly structured β-glucans. (A) Brightness versus intensity 2D histogram for an unstimulated cell shows the selected pixels that contribute to the background (black), monomers (red), dimers (green), and oligomers (blue) in the image. (B) Similar to (A), but for a cell stimulated with MMW glucan. (C) Representative selection map of HEK293 cells expressing Dectin-1A-mEmerald shows receptor aggregation in unstimulated cells or those stimulated with (D) MMW β-glucan. Dectin-1 aggregation states are defined by colored boxes selected in the brightness versus intensity histogram. (E–G) Percentage of pixels with A-mEmerald Dectin-1 in (E) low-, (F) mid-, and (G) high-aggregation states as obtained from N&B analysis before or after stimulation. Data shown as mean ± SD (n = 30). One-way ANOVA with multiple comparisons by Dunnett’s test, ∗p < 0.05, ∗∗∗∗p < 0.00001.
β-Glucan induced Dectin-1A aggregates are below 15 nm in size
While FRET-based observations and N&B analysis clearly show the presence of ligand-induced, molecular-scale aggregation of Dectin-1A, these methods are not as well suited to discern the existence of larger-scale aggregation of the receptor (e.g., clusters of tens to hundreds of receptors). We used dSTORM coupled with H-SET analysis (13) to resolve aggregation of Dectin-1A before and after stimulation with MMW glucan. This localization super-resolution microscopy technique accurately resolves objects from the diffraction limit (∼300 nm, the resolution limit of conventional fluorescence microscopy methods) or above, down to ∼15 nm (a typical resolution limit of dSTORM using our configuration). H-SET analysis detected sites of Dectin-1 labeling as “singlet” objects or “multiple” clustered objects. Multiple clustered objects are those with three or more resolvable individual Dectin-1 molecules. Singlet objects are those that appear to contain only a single, resolvable Dectin-1-labeling event, although it is possible that multiple Dectin-1 molecules in very close proximity (<15 nm separation) would be unresolvable and appear as a singlet object. We detected no significant change in the density of singlet objects or multiple object clusters before versus after MMW glucan stimulation (Fig. 7, A and B and SFig. S2). Consistent with a Dectin-1 distribution of predominantly monomers or low-order oligomers (likely unresolvable by dSTORM), singlet exposures greatly outnumber multiple exposures on the cell wall surface (Fig. 7 C). Localization number per multiple cluster object did not change with stimulation (Fig. S2), suggesting no change in the number of receptors in this minority population of Dectin-1. In the context of the previous findings showing molecular aggregation at very small scales, potentially below the resolution limit of dSTORM, we concluded that dSTORM results indicated that the Dectin-1A aggregates formed upon glucan stimulation are quite small and remain below the resolution limit of dSTORM (<15 nm length scale). Because this places an upper bound on the size of ligand-induced Dectin-1 clusters and we can estimate that the CRD of Dectin-1 occupies an area of approximately 25 nm2 (PDB: 2BPD) (64,70), we conclude that Dectin-1 aggregation after MMW glucan stimulation most likely involves collections of not more than ∼7 receptors.
Figure 7.
Dectin-1A does not form large-scale aggregates when stimulated with highly structured β-glucan. (A) Representative immunofluorescence staining images of HEK293 cells expressing Dectin-1A and either unstimulated or stimulated with MMW β-glucan. Cells were stained with a conjugated anti-Dectin-1-Alexa 647 antibody. (B) Multiple cluster density of dSTORM analysis of HEK293 cells expressing Dectin-1A unstimulated or stimulated for 50 s with MMW glucan. (C) Singlet cluster density of dSTORM analysis of HEK293 cells expressing Dectin-1A unstimulated or stimulated for 50 s with MMW glucan. (D) Cluster density of singlet and multiple exposure of Dectin-1A expressing HEK293 cells treated for 50 s with MMW glucan. Data shown as mean ± SD (n = 34) with significance assessed by Student's t-test. To see this figure in color, go online.
Dectin-1A is predominantly in a low aggregation state in resting cell membranes
FRET-based measurements and N&B analysis reported that the large majority of Dectin-1 is distributed as low aggregation state receptors (i.e., likely monomers) in unstimulated cells. However, a minority population of apparent close-proximity receptor states was observed in resting cells by both techniques. This may represent density-dependent, close-proximity interactions between Dectin-1 molecules driven by random collisional interactions, without necessarily requiring receptor oligomer formation. Alternatively, it is possible that a small fraction of Dectin-1 does form low-order oligomers, even in the absence of glucan.
By RICS analysis, we found that coexpression of donor- and acceptor-tagged Dectin-1 resulted in mean receptor density of 4612/μm2 (Fig. S4 A). We note that a small minority of outlier measurements were considerably higher than this average and may have been impacted by imaging artifacts, such as inadvertent inclusion of both dorsal and ventral plasma membrane. Within single cells, donor- and acceptor-tagged Dectin-1 was expressed at approximately similar levels (Fig. S4 B).
Dectin-1A aggregation and contact site formation is more efficient with cells incubated with C. albicans containing high glucan exposure
Throughout the article, we have focused on experiments using soluble glucans. To show Dectin-1 aggregation occurs during fungal pathogen recognition, we used a high (TRL035) and low (SC5314) β-glucan exposing C. albicans yeast and examined Dectin-1 aggregation occurring at the contact sites. TRL035 has been shown previously to have high glucan exposure compared with SC5314 (53). In addition, our representative images show TRL035 inducing phagocytic cup formation more efficiently than SC5314 (Fig. 8 A). Our results show that HEK293 cells cotransfected with Dectin-1A-mEmerald and Dectin-1A-mCherry exhibit an increase in the proportion of aggregated-state receptors from approximately 15% in noncontact site membrane to about 25% in contact sites of high glucan exposing yeast TRL035 and C. albicans-derived glucan particles. Interestingly, we did not measure a significant increase in receptor aggregation between noncontact membranes and contact sites with low glucan exposing SC5314 (Fig. 8, B and C), which may indicate that the amount of aggregated Dectin-1 at SC5314 contacts was quite small and below the detection limit. Furthermore, we observed no significant difference in FRET efficiency between any conditions tested (Fig. 8 B), similarly to our observations with soluble glucan. The significance and interpretation of these findings is further discussed below. These results suggest that the higher magnitude of cell wall glucan exposure results in an increase of Dectin-1 in molecular aggregates with associated signaling, resulting in a more efficient recognition of yeast by Dectin-1A.
Figure 8.
Dectin-1A forms more oligomers at fungal contact sites with high β-glucan exposure. (A) Representative lifetime images of HEK293 cells cotransfected with Dectin-1A-mEmerald and Dectin-1A-mCherry incubated with SC5314 (left panel), TRL035 (middle panel), or particulate glucan (right panel). Transition from redder to greener pixels at contact sites is indicative of increased FRET interactions between Dectin-1 receptors. In these images, pixel hue indicates raw fluorescence lifetime over all populations observed and pixel intensity indicates total photon counts observed over all populations. (B) FRET efficiency and (C) donor-acceptor population of cell membranes with no fungal contact, SC5314 (low glucan exposure), TRL035 (high glucan exposure), or particulate glucan. Data shown as mean ± SD (n = 15). One-way ANOVA with multiple comparisons by Dunnett’s test, ∗∗∗∗p < 0.00001.
Discussion
Our results demonstrate that the structure of β-glucan impacts receptor signaling by determining the membrane organization and molecular aggregation state of Dectin-1A. We showed that glucans with higher-order structure are better able to activate Dectin-1A signaling. Upon activation by stimulatory soluble glucan, Dectin-1A enters aggregated states that may contain dimers and higher-order oligomers, but these appear to remain as very small (<15 nm) domains containing relatively small (∼7 or fewer) numbers of receptors. Our data suggest that Dectin-1A exists predominantly in a low aggregation state, likely monomeric, in resting cells. Furthermore, upon stimulation, these basal state receptors increase aggregation state in a fashion that is dependent upon glucan higher-order structure and correlated with the magnitude of membrane proximal signaling downstream of Dectin-1. Finally, we observed that a similar process of increasing Dectin-1 aggregation is seen at contact sites with yeast- and fungal-derived particles, and that the amount of aggregated state Dectin-1A correlates with the degree of glucan exposure on the surface of the particle.
The prevalence of ensemble-based studies of biological response to fungal glucans, using glucans from varying sources and with varying degrees of structural characterization, has complicated a thorough understanding of the impact of glucan physicochemical properties on their biological activity. Innate immunocytes are naturally exposed to β-glucan in both particulate and soluble forms. β-Glucan is widely distributed on various fungal species as an insoluble component of the cell wall. The soluble form is produced when macrophages recognize fungal surfaces and release enzymes that degrade cell wall glucans (25,54). These soluble glucans are commonly found in circulation in the serum of patients with fungal infections (58,59). LMW glucans typically possess a random coil structure while increasing single- or triple-helical structure is generally seen as molecular weight increases (71). In general, complexity of β-glucan correlates with immunostimulatory potency (16,26,28,71,72). Mueller et al. indicated a correlation between glucan triple-helical structure and binding affinity for receptors on human promonocytic cells, which is contrary to our finding of similar affinity across glucans tested herein (26). Discrepancies in the impact of glucan structure on affinity may be due to different cell backgrounds, preparative methods, or, more likely, to the fact that Mueller et al. used glucans from a wide range of sources, with complex differences in structure not merely limited to size or helical content. For instance, the size and frequency of β-(1-6)-glucan side chains could be an uncontrolled covariate in comparing our results. Our study used a carefully characterized series of fungal glucans to determine that Dectin-1A activation is specifically influenced by the degree of β-glucan triple-helical structure, not merely through its affinity or size. For glucan stimulation at equal mass/volume concentration, we saw consistent single-cell calcium response to MMW glucan, but some nonresponsive cells with HMW glucan. At equimolar concentrations, partial nonresponse to HMW glucan was eliminated, suggesting that ligand may have been a limiting factor for HMW glucan in the former case. Amplitude of calcium signal was similar in single cells, once successfully triggered.
Avidity of cell wall glucan/Dectin-1 interactions may also be important for signaling. Larger glucans may behave as particles with greater potential for multivalent binding to Dectin-1, but this topic requires further study. Our dSTORM work failed to detect resolvable (<15 nm) aggregations of Dectin-1 engaged with a stimulatory MMW glucan (see further discussion below). This argues that signaling does not require avidity of glucan/Dectin-1 engagement that would exceed the length scale likely associated with small oligomers.
Aggregate states of Dectin-1 relevant to signaling could exist in a preformed, ligation-independent state, be formed in a purely ligation-dependent manner, or a mixture of both models. We entertained the possibility of preformed aggregates because receptor oligomeric states exist in the basal state for some other CTL receptors, such as DCSIGN, DNGR-1, and NKp80 (73,74,75,76). However, Dectin-1A does not contain cysteine residues in its stalk region, which are important in the dimerization of some other CTL receptors (i.e., DNGR-1), so this specific mechanism of ligand-independent dimerization is not likely to be accessible to Dectin-1. Given our observation of basal FRET signal in unliganded Dectin-1, future research may productively address the above competing models.
We proceeded to test the hypothesis that Dectin-1 undergoes ligation-dependent aggregation. Specifically, we investigated whether glucan structure impacts signaling by shifting Dectin-1A to higher states of aggregation. Dectin-1 dimerization/oligomerization would create sites where Syk could be better recruited via interactions of its SH2 domains with the (hem)ITAM phosphorylated YXXL sequence in closely juxtaposed Dectin-1A cytosolic tails. In fact, this model is commonly cited in review literature in the field, but direct evidence in intact, live cells has been lacking (77,78). The plausibility of the Dectin-1 (hem)ITAM aggregation model is suggested by the fact that another CTL receptor, CLEC-2, forms a minimal signaling unit composed of a phosphorylated dimer, enabling recruitment of a single molecule of Syk (43). Our FLIM-FRET and N&B results reveal that Dectin-1A enters a state of greater molecular aggregation when stimulated with β-glucans, and that the degree of glucan helical structure correlates with its ability to induce Dectin-1 aggregation. FRET and N&B are very sensitive methods to identify receptor aggregation on the scale of small oligomers, but these methods are limited in their ability to distinguish such small aggregates from the formation of larger receptor nanodomains. dSTORM failed to detect ligand inducible Dectin-1 nanodomains on a length scale of ≥15 nm, suggesting that Dectin-1 aggregation events are limited to small collections of ≤7 receptors. Our core observation of ligation-inducible Dectin-1 aggregation is directly in line with previous crystallographic studies that show monomeric Dectin-1A CRD in the absence of glucan but able to form dimeric complexes in the presence of β-glucan (64). In addition, solution biophysical studies have shown a ligand-induced cooperative formation of Dectin-1 CRD tetramers (or dimers of dimers) (64,65). However, these studies were performed with truncated receptor ectodomain proteins outside the context of living cell membranes, so our findings better establish and define the relevance of ligation-dependent Dectin-1 aggregation in a more physiologically realistic context.
Nanoscale glucan exposures on Candida cell wall surfaces may be important determinants of the degree of Dectin-1 aggregation at host-pathogen contact sites (13,53). Dectin-1 is recruited to the “phagocytic synapse” between innate immunocytes and fungal particles. Here, Dectin-1A encounters fungal glucan and initiates signaling (79). In addition to a greater total magnitude of glucan exposure on TRL035, we have previously described that C. albicans TRL035 exhibits spatially larger sites of glucan nanoexposures than C. albicans SC5314, which primarily displays sites of glucan exposure that support only low valency interaction with Dectin-1 (53). Modulation of cell wall surface glucan exposure size is often attributed to the efficiency of glucan masking by the overlying mannan layer. Consistent with the presence of larger sites of glucan exposure, we observed greater Dectin-1 aggregation at contact sites with TLR035, relative to SC5314. Cell wall glucan nanoexposures are larger (∼20–200 nm) than the Dectin-1 aggregates generated by soluble glucans in the present work. It is most likely that the increase in Dectin-1 aggregation state we observed is substantially driven by the higher magnitude of total glucan exposure on the surfaces of TRL035 yeast. However, future studies could productively investigate the role of glucan nanoexposures in stabilizing larger aggregated collections of engaged Dectin-1, and the potential dependence of cellular activation on the scale of Dectin-1 aggregation at sites of cell wall glucan nanoexposures.
These and other studies improve our physical understanding of host-Candida interaction and highlight the exquisite sensitivity of the Dectin-1 system that drives innate immune fungal recognition. Our previous optical nanoscopy studies of Candida cell wall surfaces estimated multivalently engaging glucan exposure site density and area (per exposure site) as follows: SC5314—1 μm−2 density, 6.61 × 10−4 μm2 area; TRL035—4 μm−2 density, 9.62 × 10−4 μm2 area (53). The total area of contact sites between C. albicans and human immature dendritic cells is ∼10 μm2 (80). Finally, we estimate (see above) that one Dectin-1 CRD occupies a footprint of ∼25 nm2. From these figures, we calculate that a typical phagocytic synapse would contain a maximum of 264 multivalently engaged Dectin-1 proteins for C. albicans SC5314, and a maximum of 385 multivalently engaged Dectin-1 for TRL035 (at total ligand engagement). Based on our reported Dectin-1 density (Fig. 4), the contact sites we measured would contain ∼46,000 total Dectin-1 proteins. So, the Dectin-1 system is able to drive signaling responses when, at most, only a few hundred receptors, corresponding to less than 1% of the total contact site resident Dectin-1 proteins, are aggregated in the contact. These results and estimates suggest that fungal recognition requires the Dectin-1 system to engage in a search for rare sites of multivalent interaction with glucan. Signal initiation must be sensitive to activation of relatively small numbers of Dectin-1 proteins. In the future, it will be important to achieve a better understanding of Dectin-1’s collaboration with other antifungal receptors (e.g., DC-SIGN and CD206). Such receptors may be important for building and stabilizing a fungal contact that can effectively promote Dectin-1’s ability to search for and find its rare sites of glucan exposure.
Overall, these findings indicate that β-glucan structure is required for Dectin-1A to undergo Syk-dependent signaling. Here, we provide evidence in support of a model in which highly structured glucans induce stable dimerization and/or oligomerization of the receptor. This allows their (hem)ITAM domains to become close enough for a sufficient period of time to allow for the activation of Syk, leading to further signaling cascades. Greater understanding of receptor activation is required to better understand the role of Dectin-1A and its agonists as a potential way forward for adjuvant and immunotherapy development. Furthermore, given the worldwide burden of candidiasis, further experimentation is required to better understand the role of Dectin-1A in recognition of these pathogens.
Author contributions
E.U.A., A.E.A., and A.K.N. designed the research. E.U.A. and A.E.A. performed the research. E.U.A., A.E.A., M.E.D., and K.S.M. generated experimental/analytical tools. E.U.A., A.E.A., M.J.W., M.E.D., K.S.M., and A.K.N. wrote the manuscript.
Acknowledgments
We acknowledge the competent technical assistance of Ms. Zinia Pervin in relation to BLI determinations of Dectin-1A/glucan affinity. Glucans used in this study were a generous gift of ImmunoResearch Inc., which did not determine overall experimental design, data interpretation, or decision to publish. We acknowledge use of the University of New Mexico Comprehensive Cancer Center fluorescence microscopy shared facility and the assistance of Dr. Michael Paffett in conducting FLIM-FRET studies. We acknowledge the University of New Mexico Center for Spatiotemporal Modeling of Cell Signaling for training support and shared microscopy research resources used in this study. This research was supported by the National Institutes of Health (University of New Mexico Center for Spatiotemporal Modeling of Cell Signaling [STMC], P50GM085273, to A.K.N.; R01AI116894, to A.K.N.; UNM Infectious Disease and Inflammation Program, T32AI007538, to A.K.N.; New Mexico Comprehensive Cancer Center fluorescence microscopy shared facility, P30CA118100).
Declaration of interests
M.E.D. and K.S.M. are employees of HiberCell, Inc., which provided glucans used in this study.
Editor: Dylan Myers Owen.
Footnotes
Eduardo U. Anaya’s present address is Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland.
Akram Etemadi Amin’s present address is Intel Corp., Rio Rancho, New Mexico.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.07.021.
Supporting material
References
- 1.Kao A.S., Brandt M.E., et al. Hajjeh R.A. The Epidemiology of Candidemia in Two United States Cities: Results of a Population-Based Active Surveillance. Clin. Infect. Dis. 1999;29:1164–1170. doi: 10.1086/313450. [DOI] [PubMed] [Google Scholar]
- 2.Hajjeh R.A., Sofair A.N., et al. Warnock D.W. Incidence of Bloodstream Infections Due to Candida Species and In Vitro Susceptibilities of Isolates Collected from 1998 to 2000 in a Population-Based Active Surveillance Program. J. Clin. Microbiol. 2004;42:1519–1527. doi: 10.1128/JCM.42.4.1519-1527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaller M.A., Diekema D.J. Epidemiology of Invasive Candidiasis: a Persistent Public Health Problem. Clin. Microbiol. Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland A.A., Harrison L.H., et al. Park B.J. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008-2013: results from population-based surveillance. PLoS One. 2015;10:e0120452. doi: 10.1371/journal.pone.0120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland A.A., Farley M.M., et al. Chiller T.M. Changes in Incidence and Antifungal Drug Resistance in Candidemia: Results From Population-Based Laboratory Surveillance in Atlanta and Baltimore, 2008-2011. Clin. Infect. Dis. 2012;55:1352–1361. doi: 10.1093/cid/cis697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffin W.L. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008;72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-Herrera J., Elorza M.V., et al. Sentandreu R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006;6:14–29. doi: 10.1111/j.1567-1364.2005.00017.x. [DOI] [PubMed] [Google Scholar]
- 8.Gow N.A.R., Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012;15:406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Ballou E.R., Avelar G.M., et al. Brown A.J.P. Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2016;2:16238. doi: 10.1038/nmicrobiol.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherrington S.L., Sorsby E., et al. Hall R.A. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 2017;13:e1006403. doi: 10.1371/journal.ppat.1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopke A., Nicke N., et al. Wheeler R.T. Neutrophil Attack Triggers Extracellular Trap-Dependent Candida Cell Wall Remodeling and Altered Immune Recognition. PLoS Pathog. 2016;12:e1005644. doi: 10.1371/journal.ppat.1005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler R.T., Kombe D., et al. Fink G.R. Dynamic, Morphotype-Specific Candida albicans β-Glucan Exposure during Infection and Drug Treatment. PLoS Pathog. 2008;4:e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J., Wester M.J., et al. Neumann A.K. Nanoscopic cell-wall architecture of an immunogenic ligand in Candida albicans during antifungal drug treatment. Mol. Biol. Cell. 2016;27:1002–1014. doi: 10.1091/mbc.E15-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pappas H.C., Sylejmani R., et al. Neumann A.K. Antifungal Properties of Cationic Phenylene Ethynylenes and Their Impact on β-Glucan Exposure. Antimicrob. Agents Chemother. 2016;60:4519–4529. doi: 10.1128/AAC.00317-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler R.T., Fink G.R. A Drug-Sensitive Genetic Network Masks Fungi from the Immune System. PLoS Pathog. 2006;2:e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sletmoen M., Stokke B.T. Higher order structure of (1,3)-β-D-glucans and its influence on their biological activities and complexation abilities. Biopolymers. 2008;89:310–321. doi: 10.1002/bip.20920. [DOI] [PubMed] [Google Scholar]
- 17.Chuah C.T., Sarko A., et al. Marchessault R.H. Packing analysis of carbohydrates and polysaccharides. Part 14. Triple-helical crystalline structure of curdlan and paramylon hydrates. Macromolecules. 1983;16:1375–1382. doi: 10.1021/ma00242a020. [DOI] [Google Scholar]
- 18.Yoshioka Y., Uehara N., Saitô H. Conformation-dependent change in antitumor activity of linear and branched (1–--3)-beta-D-glucans on the basis of conformational elucidation by carbon-13 nuclear magnetic resonance spectroscopy. Chem. Pharm. Bull. 1992;40:1221–1226. doi: 10.1248/cpb.40.1221. [DOI] [PubMed] [Google Scholar]
- 19.Young S.-H., Dong W.-J., Jacobs R.R. Observation of a Partially Opened Triple-helix Conformation in 1→3-β-Glucan by Fluorescence Resonance Energy Transfer Spectroscopy. J. Biol. Chem. 2000;275:11874–11879. doi: 10.1074/jbc.275.16.11874. [DOI] [PubMed] [Google Scholar]
- 20.Okobira T., Miyoshi K., et al. Shinkai S. Molecular dynamics studies of side chain effect on the beta-1,3-D-glucan triple helix in aqueous solution. Biomacromolecules. 2008;9:783–788. doi: 10.1021/bm700511d. [DOI] [PubMed] [Google Scholar]
- 21.Chihara G., Hamuro J., et al. Fukuoka F. Fractionation and purification of the polysaccharides with marked antitumor activity, especially lentinan, from Lentinus edodes (Berk.) Sing. (an edible mushroom) Cancer Res. 1970;30:2776–2781. [PubMed] [Google Scholar]
- 22.Brown G.D., Gordon S. Fungal β-Glucans and Mammalian Immunity. Immunity. 2003;19:311–315. doi: 10.1016/S1074-7613(03)00233-4. [DOI] [PubMed] [Google Scholar]
- 23.Novak M., Vetvicka V. β -Glucans, History, and the Present: Immunomodulatory Aspects and Mechanisms of Action. J. Immunot. 2008;5:47–57. doi: 10.1080/15476910802019045. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L.d., Zhang Q.h., et al. Cao Y.m. The shiitake mushroom-derived immuno-stimulant lentinan protects against murine malaria blood-stage infection by evoking adaptive immune-responses. Int. Immunopharm. 2009;9:455–462. doi: 10.1016/j.intimp.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Kim H.S., Hong J.T., et al. Han S.-B. Stimulatory Effect of β-glucans on Immune Cells. Immune Netw. 2011;11:191–195. doi: 10.4110/in.2011.11.4.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller A., Raptis J., et al. Williams D.L. The influence of glucan polymer structure and solution conformation on binding to (1->3)- -D-glucan receptors in a human monocyte-like cell line. Glycobiology. 2000;10:339–346. doi: 10.1093/glycob/10.4.339. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L., Li X., et al. Zeng F. Correlation between antitumor activity, molecular weight, and conformation of lentinan. Carbohydr. Res. 2005;340:1515–1521. doi: 10.1016/j.carres.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 28.Elder M.J., Webster S.J., et al. Goodall J.C. β-Glucan Size Controls Dectin-1-Mediated Immune Responses in Human Dendritic Cells by Regulating IL-1β Production. Front. Immunol. 2017;8:791. doi: 10.3389/fimmu.2017.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Luzio N.R., Williams D.L., et al. Kitahama A. Comparative tumor-inhibitory and anti-bacterial activity of soluble and particulate glucan. Int. J. Cancer. 1979;24:773–779. doi: 10.1002/ijc.2910240613. [DOI] [PubMed] [Google Scholar]
- 30.Tzianabos A.O. Polysaccharide Immunomodulators as Therapeutic Agents: Structural Aspects and Biologic Function. Clin. Microbiol. Rev. 2000;13:523–533. doi: 10.1128/CMR.13.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Zhang L., et al. Zeng F. Correlation of structure to antitumor activities of five derivatives of a β-glucan from Poria cocos sclerotium. Carbohydr. Res. 2004;339:2567–2574. doi: 10.1016/j.carres.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Smith A.J., Graves B., et al. Williams D.L. Immunoregulatory Activity of the Natural Product Laminarin Varies Widely as a Result of Its Physical Properties. J. Immunol. 2018;200:788–799. doi: 10.4049/jimmunol.1701258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanaki T., Ito W., et al. Fujita H. Correlation between the antitumor activity of a polysaccharide schizophyllan and its triple-helical conformation in dilute aqueous solution. Biophys. Chem. 1983;17:337–342. doi: 10.1016/0301-4622(83)80018-0. [DOI] [PubMed] [Google Scholar]
- 34.Maeda Y.Y., Watanabe S.T., et al. Rokutanda M. Denaturation and renaturation of a beta-1,6;1,3-glucan, lentinan, associated with expression of T-cell-mediated responses. Cancer Res. 1988;48:671–675. [PubMed] [Google Scholar]
- 35.Suzuki T., Ohno N., et al. Yadomae T. Activation of the complement system by (1–--3)-beta-D-glucans having different degrees of branching and different ultrastructures. J. Pharmacobio-Dyn. 1992;15:277–285. doi: 10.1248/bpb1978.15.277. [DOI] [PubMed] [Google Scholar]
- 36.Duggan S., Leonhardt I., et al. Kurzai O. Host response to Candida albicans bloodstream infection and sepsis. Virulence. 2015;6:1–11. doi: 10.4161/21505594.2014.988096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown G.D., Taylor P.R., et al. Gordon S. Dectin-1 is a major beta-glucan receptor on macrophages. J. Exp. Med. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gow N.A.R., Netea M.G., et al. Kullberg B.J. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 2007;196:1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis S.E., Hopke A., et al. Reynolds T.B. Masking of β(1-3)-Glucan in the Cell Wall of Candida albicans from Detection by Innate Immune Cells Depends on Phosphatidylserine. Infect. Immun. 2014;82:4405–4413. doi: 10.1128/IAI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice P.J., Adams E.L., et al. Williams D.L. Oral delivery and gastrointestinal absorption of soluble glucans stimulate increased resistance to infectious challenge. J. Pharmacol. Exp. Therapeut. 2005;314:1079–1086. doi: 10.1124/jpet.105.085415. [DOI] [PubMed] [Google Scholar]
- 41.O’Neill S.K., Getahun A., et al. Cambier J.C. Monophosphorylation of CD79a and CD79b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy. Immunity. 2011;35:746–756. doi: 10.1016/j.immuni.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurosaki T., Johnson S.A., et al. Cambier J.C. Role of the Syk autophosphorylation site and SH2 domains in B cell antigen receptor signaling. J. Exp. Med. 1995;182:1815–1823. doi: 10.1084/jem.182.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes C.E., Pollitt A.Y., et al. Watson S.P. CLEC-2 activates Syk through dimerization. Blood. 2010;115:2947–2955. doi: 10.1182/blood-2009-08-237834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartel Y., Bauer B., Steinle A. Modulation of NK cell function by genetically coupled C-type lectin-like receptor/ligand pairs encoded in the human natural killer gene complex. Front. Immunol. 2013;4:362. doi: 10.3389/fimmu.2013.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Needs P.W., Selvendran R.R. Avoiding oxidative degradation during sodium hydroxide/methyl iodide-mediated carbohydrate methylation in dimethyl sulfoxide. Carbohydr. Res. 1993;245:1–10. doi: 10.1016/0008-6215(93)80055-J. [DOI] [Google Scholar]
- 46.Kim Y.-T., Kim E.-H., et al. Lim S.-T. Structural characterization of β-d-(1→3, 1→6)-linked glucans using NMR spectroscopy. Carbohydr. Res. 2000;328:331–341. doi: 10.1016/S0008-6215(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 47.Digman M.A., Stakic M., Gratton E. Raster Image Correlation Spectroscopy and Number and Brightness Analysis. Methods Enzymol. 2013;518:121–144. doi: 10.1016/B978-0-12-388422-0.00006-6. [DOI] [PubMed] [Google Scholar]
- 48.Youker R.T., Teng H. Measuring protein dynamics in live cells: protocols and practical considerations for fluorescence fluctuation microscopy. J. Biomed. Opt. 2014;19:90801. doi: 10.1117/1.JBO.19.9.090801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Planes N., Digman M.A., et al. Caballero-George C. Number and brightness analysis to study spatio-temporal distribution of the angiotensin II AT1 and the endothelin-1 ETA receptors: Influence of ligand binding. Biochim. Biophys. Acta Gen. Subj. 2019;1863:917–924. doi: 10.1016/j.bbagen.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Smith C.S., Joseph N., et al. Lidke K.A. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang F., Schwartz S.L., et al. Lidke K.A. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed. Opt Express. 2011;2:1377–1393. doi: 10.1364/BOE.2.001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ram A., Jalal S., et al. Kumar M. A Density Based Algorithm for Discovering Density Varied Clusters in Large Spatial Databases. Int. J. Comput. Appl. 2010;3:1–4. doi: 10.5120/739-1038. [DOI] [Google Scholar]
- 53.Graus M.S., Wester M.J., et al. Neumann A.K. Mannan Molecular Substructures Control Nanoscale Glucan Exposure in Candida. Cell Rep. 2018;24:2432–2442.e5. doi: 10.1016/j.celrep.2018.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hino S., Kito A., et al. Matsuda T. Discharge of solubilized and Dectin-1-reactive β-glucan from macrophage cells phagocytizing insoluble β-glucan particles: Involvement of reactive oxygen species (ROS)-driven degradation. Biochem. Biophys. Res. Commun. 2012;421:329–334. doi: 10.1016/j.bbrc.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Hong F., Yan J., et al. Ross G.D. Mechanism by which orally administered beta-1,3-glucans enhance the tumoricidal activity of antitumor monoclonal antibodies in murine tumor models. J. Immunol. 2004;173:797–806. doi: 10.4049/jimmunol.173.2.797. [DOI] [PubMed] [Google Scholar]
- 56.Gonzalez, J.A., J.D. Digby, … D.L. Williams. 2004. At low serum glucan concentrations there is an inverse correlation between serum glucan and serum cytokine levels in ICU patients with infections. Int. Immunopharm.. 4:1107–1115, doi: 10.1016/J.INTIMP.200405.010. [DOI] [PubMed]
- 57.Digby J., Kalbfleisch J., et al. Williams D. Serum glucan levels are not specific for presence of fungal infections in intensive care unit patients. Clin. Diagn. Lab. Immunol. 2003;10:882–885. doi: 10.1128/cdli.10.5.882-885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Digby J., Kalbfleisch J., et al. Williams D. Serum Glucan Levels Are Not Specific for Presence of Fungal Infections in Intensive Care Unit Patients. Clin. Diagn. Lab. Immunol. 2003;10:882–885. doi: 10.1128/CDLI.10.5.882-885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez J.A., Digby J.D., et al. Williams D.L. At low serum glucan concentrations there is an inverse correlation between serum glucan and serum cytokine levels in ICU patients with infections. Int. Immunopharm. 2004;4:1107–1115. doi: 10.1016/j.intimp.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Kogan G., Alföldi J., Masler L. 13 C-nmr spectroscopic investigation of two yeast cell wall. Biopolymers. 1988;27:1055–1063. doi: 10.1002/bip.360270702. [DOI] [Google Scholar]
- 61.Nitschke J., Modick H., et al. Mölleken H. A new colorimetric method to quantify β-1,3-1,6-glucans in comparison with total β-1,3-glucans in edible mushrooms. Food Chem. 2011;127:791–796. doi: 10.1016/j.foodchem.2010.12.149. [DOI] [PubMed] [Google Scholar]
- 62.Brown C.M., Dalal R.B., et al. Gratton E. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J. Microsc. 2008;229:78–91. doi: 10.1111/j.1365-2818.2007.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauer B., Steinle A. HemITAM: A single tyrosine motif that packs a punch. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aan3676. [DOI] [PubMed] [Google Scholar]
- 64.Brown J., O’Callaghan C.A., et al. Jones E.Y. Structure of the fungal β-glucan-binding immune receptor dectin-1: Implications for function. Protein Sci. 2007;16:1042–1052. doi: 10.1110/ps.072791207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dulal H.P., Adachi Y., et al. Yamaguchi Y. β-Glucan-induced cooperative oligomerization of Dectin-1 C-type lectin-like domain. Glycobiology. 2018;28:612–623. doi: 10.1093/glycob/cwy039. [DOI] [PubMed] [Google Scholar]
- 66.Singh D.R., Kanvinde P., et al. Hristova K. The EphA2 receptor is activated through induction of distinct, ligand-dependent oligomeric structures. Commun. Biol. 2018;1:15. doi: 10.1038/s42003-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Digman M.A., Dalal R., et al. Gratton E. Mapping the Number of Molecules and Brightness in the Laser Scanning Microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Unruh J.R., Gratton E. Analysis of Molecular Concentration and Brightness from Fluorescence Fluctuation Data with an Electron Multiplied CCD Camera. Biophys. J. 2008;95:5385–5398. doi: 10.1529/biophysj.108.130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trullo A., Corti V., et al. Zamai M. Application limits and data correction in number of molecules and brightness analysis. Microsc. Res. Tech. 2013;76:1135–1146. doi: 10.1002/jemt.22277. [DOI] [PubMed] [Google Scholar]
- 70.Wester M.J., Lin J., Neumann A.K. A computational model for regulation of nanoscale glucan exposure in Candida albicans. PLoS One. 2017;12:e0188599. doi: 10.1371/journal.pone.0188599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okazaki M., Adachi Y., et al. Yadomae T. Structure-activity relationship of (1-->3)-beta-D-glucans in the induction of cytokine production from macrophages, in vitro. Biol. Pharm. Bull. 1995;18:1320–1327. doi: 10.1248/bpb.18.1320. [DOI] [PubMed] [Google Scholar]
- 72.Cleary J.A., Kelly G.E., Husband A.J. The effect of molecular weight and beta-1,6-linkages on priming of macrophage function in mice by (1,3)-beta-D-glucan. Immunol. Cell Biol. 1999;77:395–403. doi: 10.1046/J.1440-1711.1999.00848.X. [DOI] [PubMed] [Google Scholar]
- 73.Vitale M., Falco M., et al. Moretta A. Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur. J. Immunol. 2001;31:233–242. doi: 10.1002/1521-4141(200101)31:1<233::AID-IMMU233>3.0.CO;2–4. [DOI] [PubMed] [Google Scholar]
- 74.Feinberg H., Guo Y., et al. Weis W.I. Extended Neck Regions Stabilize Tetramers of the Receptors DC-SIGN and DC-SIGNR. J. Biol. Chem. 2005;280:1327–1335. doi: 10.1074/jbc.M409925200. [DOI] [PubMed] [Google Scholar]
- 75.Itano M.S., Graus M.S., et al. Neumann A.K. Super-resolution imaging of C-type lectin spatial rearrangement within the dendritic cell plasma membrane at fungal microbe contact sites. Front. Physiol. 2014;2:46. doi: 10.3389/fphy.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanč P., Schulz O., et al. Reis e Sousa C. A pH- and ionic strength-dependent conformational change in the neck region regulates DNGR-1 function in dendritic cells. EMBO J. 2016;35:2484–2497. doi: 10.15252/embj.201694695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schorey J.S., Lawrence C. The Pattern Recognition Receptor Dectin-1: From Fungi to Mycobacteria. Curr. Drug Targets. 2008;9:123–129. doi: 10.2174/138945008783502430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marakalala M.J., Kerrigan A.M., Brown G.D. Dectin-1: a role in antifungal defense and consequences of genetic polymorphisms in humans. Mamm. Genome. 2011;22:55–65. doi: 10.1007/s00335-010-9277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodridge H.S., Reyes C.N., et al. Underhill D.M. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse.’. Nature. 2011;472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Graus M.S., Pehlke C., et al. Neumann A.K. A New Tool to Quantify Receptor Recruitment to Cell Contact Sites during Host-Pathogen Interaction. PLoS Comput. Biol. 2014;10:e1003639. doi: 10.1371/journal.pcbi.1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.