Abstract
Grapevine powdery mildew is caused by Erysiphe necator, which seriously harms grape production in the world. Stilbene synthase makes phytoalexins that contribute to the resistance of grapevine against powdery mildew. A novel VqNSTS3 was identified and cloned from Chinese wild Vitis quinquangularis accession Danfeng-2. The novel VqNSTS3 was transferred into susceptible ‘Thompson Seedless’ by Agrobacterium-mediated transformation. The transgenic plants showed resistance to the disease and activated other resistance-related genes. VqNSTS3 expression in grapevine is regulated by VqWRKY33, and which binds to TTGACC in the VqNSTS3 promoter. Furthermore, VqWRKY33 was phosphorylated by VqMAPK3/VqMAPK6 and thus led to enhanced signal transduction and increased VqNSTS3 expression. ProVqNSTS3::VqNSTS3-GFP of transgenic VqNSTS3 in Arabidopsis thaliana was observed to move to and wrap the pathogen’s haustoria and block invasion by Golovinomyces cichoracearum. These results demonstrate that stilbene accumulation of novel VqNSTS3 of the Chinese wild Vitis quinquangularis accession Danfeng-2 prevented pathogen invasion and enhanced resistance to powdery mildew. Therefore, VqNSTS3 can be used in generating powdery mildew-resistant grapevines.
Introduction
The grapevine is one of the ancient and most economically valuable fruits in the world [1]. Because of their high-quality fruit, European grape varieties (Vitis vinifera) are considered the world’s most prominent cultivars. However, this valuable species is highly susceptible to Erysiphe necator (previously Uncinula necator), a fungus that causes powdery mildew (PM) disease [2]. The obligate biotrophic fungus E. necator affects all parts of a plant, which leads to significant losses in fruit yield and quality in the viticulture industry [3–5]. The main measure for preventing and controlling grapevine PM in grape production is the spraying of chemical fungicides, which not only causes cost increases, fruit contamination, and environmental pollution, but also causes resistance in pathogenic bacteria and new variations in pathogenic bacteria, bringing new control difficulties to grape production [6–8]. Therefore, using the grapevine immune system to improve disease resistance is a fundamental way to solve the disease resistance problem in grape production. Obtaining PM-resistant grape varieties and elucidating the molecular mechanisms of disease resistance are vital steps in reducing the reliance on fungicides and breeding grapevine varieties for disease resistance.
An early study that first isolated resveratrol in grapevine leaves found that it had the effect of conferring disease resistance to grapevine [9]. Vitis vinifera was found to produce a stilbene phytoalexin, a derivative of resveratrol [10]. In later studies, resveratrol was isolated and obtained in grape berries [11] and wine [12]. There has been significant research on the role resveratrol plays in grapevine against Botrytis cinerea [13, 14], E. necator [15], Plasmopara viticola [16], and Neofusicoccum parvum [17]. Resveratrol also has benefit associated with human health [18, 19]. Stilbene synthase (STS) catalyzes the formation of resveratrol from three malonyl coenzyme A esters and one coenzyme A ester [20]. Heterologous expression of STS genes can improve the level of stilbene and enhance plant disease resistance. For example, transferring two STS genes from grapevine, where they are highly expressed, into tobacco plants, increased their resistance to B. cinerea [21]. Grapevine STS genes have been transferred to many plants, including rice [22], pea [23], lettuce [24], and kiwifruit [25], in each case resulting in significant improvements in accumulation of stilbene or pathogen resistance.
In recent years, several transcription factors were shown to regulate the expression of STS genes and stilbene accumulation, such as MYB, WRKY, ERF, AL, and bZIP [26–35]. Among them, WRKY transcription factors are important in regulating STS gene expression. VvWRKY24 can independently regulate the VvSTS29 promoter, while VvWRKY03 and VvMYB14 jointly upregulate the VvSTS29 promoter [35]. Recently, VvWRKY8 has been shown to repress VvSTS15/21 expression and resveratrol biosynthesis through interaction with VvMYB14 [31]. When grapes are under UV stress, they produce resveratrol through VvMYB14-VvWRKY8-VvMYB30, and prevent excessive accumulation of resveratrol [29]. VqWRKY53 positively regulates the expression of VqSTSs, and interacts with VqMYB14 and VqMYB15 to show stronger regulatory function [32]. VqWRKY31 can be induced after E. necator and directly regulates the promoters of STS9/48 [26]. However, it is currently unclear whether other WRKY transcription factors are involved in regulating STS gene expression.
In order to better study the traits and functions of grapevine genes, functional genome sequencing was conducted in different germplasms of grapes. In 2007, genome sequencing of PN40024 ‘Pinot Noir’ identified 48 VvSTS genes [1], of which 33 had potential functions [1, 36, 37]. Girollet et al. reported the de novo assembly of the Vitis riparia genome in 2019 [38], while an analysis of grapevine diversity and demographic history was performed using whole-genome resequencing of 472 Vitis accessions by Liang et al. [39]. The draft genome of V. riparia ‘Manitoba 37’, a native American cold-hardy grapevine, has been sequenced [40]. The genome of the grape interspecific hybrid ‘Shine Muscat’ (Vitis labruscana × V. vinifera) was sequenced and published in 2022 [41].
China is one of the main points of origin for grape varieties and has abundant germplasm resources that can be used for grape breeding [42]. Preliminary research in our laboratory found that Chinese wild Vitis pseudoreticulata accession Baihe-35-1 can provide a genetic resource to investigate the role of stilbene synthase genes in the PM interaction [43–45]. In total, 61 VpSTS genes have been isolated from Baihe-35-1 [46]. In particular, VpSTS29/STS2 contributes to basal resistance of grapevine and Arabidopsis thaliana to PM [47, 48]. Another important Chinese wild resource is Vitis quinquangularis accession Danfeng-2, containing 41 STS genes (GenBank accession numbers JQ868658–JQ868698) [49]. Many VqSTS genes from Danfeng-2 have been shown to significantly enhance resistance to PM [50–53]. Among them, overexpression of the fruit-specific and highly expressed gene VqSTS6 increases resveratrol content and pathogen resistance in V. vinifera ‘Thompson Seedless’ [53, 54]. Further analysis of Danfeng-2 novel transcriptome data (PRJNA306731) identified six novel STS transcripts: VqNSTS1–VqNSTS6 [55]. VqAL4 positively regulates VqNSTS4 expression, enhancing resistance to PM by activating salicylic acid (SA) signals in grapevine [34]. What is the mechanism of VqNSTS3 expression in disease resistance? This research elucidated the effect and regulation mechanism of VqNSTS3 in Chinese wild grape breeding for disease resistance.
Results
VqNSTS3 has conserved motifs of the stilbene synthase gene family and expresses resistance to E. necator
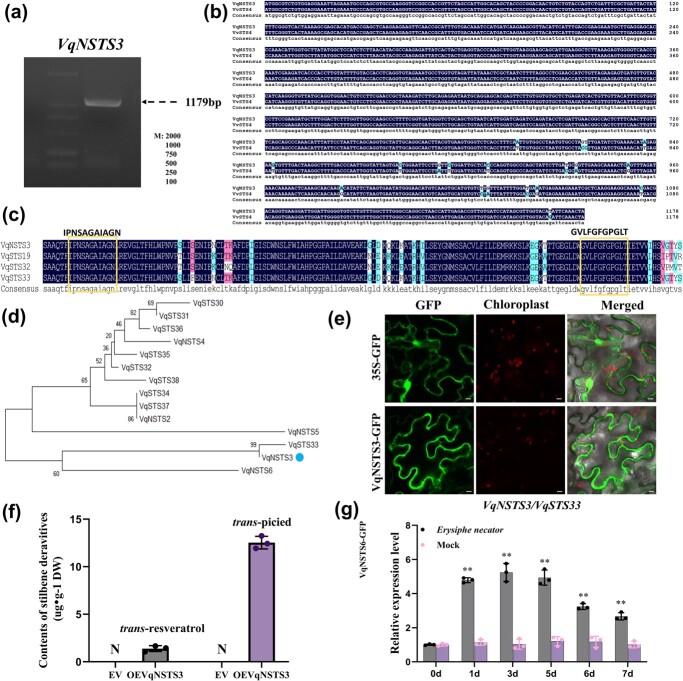
Six new STS transcripts were identified in our laboratory [55]. A homologous cloning method was used to identify VqNSTS3 (GenBank accession number OL589478) from Danfeng-2. The coding sequence of VqNSTS3 was 1179 bp (Fig. 1a) and showed 98.3% similarity to VvSTS4 from V. vinifera PN40024 (Fig. 1b). VqNSTS3 possessed the conserved domain of the STS family [56] (Fig. 1c) and showed 99% amino acid sequence identity with VqSTS33 [34] (Fig. 1d). VqNSTS3-GFP localized in the cytoplasm (Fig. 1e). Six new transcripts were transiently transformed into tobacco, in which we detected the highest content of stilbenes after overexpression of VqNSTS3 (Fig. 1f) (data not shown). To further explore whether the gene responds to the induction of E. necator in Danfeng-2, samples from Danfeng-2 plants were taken for qPCR analysis after artificial inoculation with E. necator. It was found that VqNSTS3 gene expression increased significantly on the first day after inoculation, and the trend continued to the third day after inoculation (Fig. 1g).
Figure 1.
Cloning and expression analysis of VqNSTS3 under E. necator inoculation in grapevine. a Amplification of VqNSTS3 from Chinese wild V. quinquangularis accession Danfeng-2. b DNA sequence alignment between VqNSTS3 and VvSTS4 (XM_003634021). c Multiple amino acid sequence alignments between VqNSTS3 and other VqSTS proteins. The yellow box indicates the STS conserved domain. d Phylogenetic analysis of VqNSTS3 and part of VqSTSs from Danfeng-2. VqNSTS3 is highlighted with a blue dot. e Subcellular localization of VqNSTS3 in N. benthamiana leaves. Scale bars, 10 μm. fVqNSTS3 was transformed into tobacco for 3 days. The content of stilbenes was determined by HPLC. Results are shown as mean ± standard error of the mean; n = 3. g qPCR analysis of VqNSTS3/VqSTS33 expression in Danfeng-2 leaves after infection with E. necator. Results are shown as mean ± standard error of the mean; n = 3. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (**P < .01).
Transgenic VqNSTS3 grapevine lines show enhanced resistance to E. necator and activation of resistance-related genes
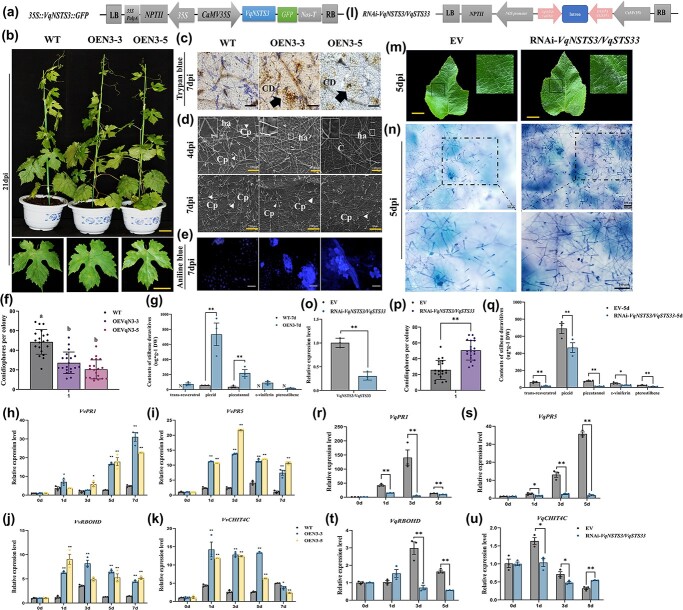
To determine whether VqNSTS3 is involved in grapevine resistance to E. necator, VqNSTS3 from Danfeng-2 was transferred into disease-susceptible European grape cultivar ‘Thompson Seedless’ using Agrobacterium tumefaciens-mediated transformation (Fig. 2a, Supplementary Data Fig. S3). Two independent VqNSTS3-transgenic overexpression lines were obtained (OEVqNSTS3-L3 and OEVqNSTS3-L5) (Supplementary Data Fig. S3i and j). Transgenic and wild-type (WT) plants were inoculated with E. necator to characterize the disease resistance function of VqNSTS3. WT plants were more susceptible, producing extensive fungal hyphae and conidiophores, whereas transgenic lines were not (Fig. 2b–d and f). Furthermore, transgenic lines showed enhanced callose deposition (Fig. 2e) and increased the expression of resistance-related genes after inoculation (Fig. 2h–k). HPLC assays indicated that after inoculation only piceid and piceatannol were detected in WT plants, while five stilbenes accumulated in transgenic plants. The contents of piceid and piceatannol in VqNSTS3-transgenic overexpression lines increased 13.0- and 6.3-fold, respectively, compared with WT plants (Fig. 2g, Supplementary Data Table S7). To study the role of VqNSTS3 in disease resistance further, we used RNA interference (RNAi) to study the resistance of VqNSTS3 to E. necator in Danfeng-2. Due to high sequence similarity between VqNSTS3 and VqSTS33, we were only able to interfere with both at the same time. RNAi-VqNSTS3/VqSTS33 plants showed contrasting results to the overexpressing (OE) plants. Following inoculation, trans-resveratrol, piceid, pterostilbene, ε-viniferin, and piceatannol levels in RNAi-VqNSTS3/VqSTS33 were 27, 42, 57, 59, and 14% lower than in empty vector (EV) controls (Fig. 2l–u; Supplementary Data Table S7). The above results show that VqNSTS3 plays an active role in the defense response after inoculation with E. necator.
Figure 2.
Transgenic VqNSTS3 grapevine plants show enhanced resistance to E. necator. a Diagram of the OEVqNSTS3 construct. b Photographs of VqNSTS3 overexpression and WT plants infected with E. necator at 21 days post-inoculation (dpi). Scale bars, 3 cm. c Trypan blue staining of OEVqNSTS3 and WT leaves at 7 dpi to observe the growth of hyphae. Scale bars, 100 μm. CD, cell death. d Scanning electron micrographs of WT and OEVqNSTS3 leaves at 4 and 7 dpi. C, conidium; ha, hyphal appressorium; Cp, conidiophore. e Aniline blue staining of WT and OEVqNSTS3 leaves at 7 dpi to detect callose deposition. Scale bars, 50 μm. f Number of conidiophores per colony at 7 dpi on WT and transgenic plants. Results are shown as mean ± standard error of the mean; n = 20; different letters represent significant differences (P < .05) as determined by one-way ANOVA followed by Tukey’s multiple comparisons test. g HPLC analysis of content of stilbenes in WT and OEVqNSTS3 leaves at 7 dpi. h–k Expression of defense-related genes determined by qPCR analysis in WT and transgenic plants after E. necator inoculation. Results are shown as mean ± standard error of the mean; n = 3. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (*P < .05; **P < .01). l Diagram of the RNAi-VqNSTS3/VqSTS33 construct. m Phenotypes of RNAi-VqNSTS3/VqSTS33 and EV leaves infected with E. necator for 5 days. Scale bars, 3 cm. n RNAi-VqNSTS3/VqSTS33 and EV leaves at 5 days stained with trypan blue. Scale bars, 50 μm. o qPCR analysis of VqNSTS3/VqSTS33 expression in RNAi-VqNSTS3/VqSTS33 and EV leaves. p Number of conidiophores per colony at 5 dpi on EV and RNAi leaves. Results are shown as mean ± standard error of the mean; n = 20. Significance was examined by Student’s t-test (**P < .01). q HPLC analysis of stilbenes in RNAi-VqNSTS3/VqSTS33 and EV leaves at 5 dpi. r–u Expression of defense-related genes determined by qPCR analysis in EV and RNAi leaves after E. necator inoculation. Results in (g, o, q-u) are shown as mean ± standard error of the mean; n = 3. Significance was examined by Student’s t-test (*P < .05; **P < .01).
VqNSTS3 expression enhances resistance to E. necator in grapevine due to regulation by VqWRKY33
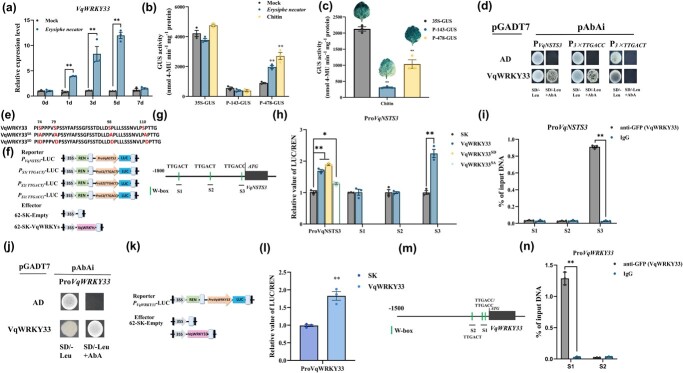
We analyzed the VqNSTS3 promoter cloned from Danfeng-2 to identify transcription factors that regulate VqNSTS3 expression. The VqNSTS3 promoter was found to contain three specific fungal elicitor-responsive elements: W-box elements (Supplementary Data Fig. S2, Supplementary Data Table S4), and the W-box element is the binding site of the WRKY transcription factor [57]. Previous studies in our laboratory found that 16 VqWRKY transcription factors responded to the induction of E. necator in Danfeng-2 [32]. To determine whether WRKY transcription factors could regulate the expression of VqNSTS3, we selected eight WRKY transcription factors that showed significant responses to the induction of E. necator using dual-luciferase assays to detect the promoter activity of VqNSTS3. The results show that VqWRKY2, VqWRKY18, VqWRKY33, and VqWRKY53 can positively activate the promoter activity of VqNSTS3 and that VqWRKY33 had the greatest regulatory activity on the VqNSTS3 promoter (Supplementary Data Fig. S5a and b). Therefore, we conducted further research on VqWRKY33. We artificially inoculated Danfeng-2 leaves with E. necator, and the expression of VqWRKY33 was significantly upregulated after inoculation (Fig. 3a). To further investigate the function of the transcription factor VqWRKY33, the characteristics of VqWRKY33 were analyzed. VqWRKY33 is a nuclear protein that encodes 561 amino acids and has two highly conserved WRKY domains (amino acid residues 230–285 and 392–449), predicted to be located on chromosome 8 (Supplementary Data Fig. S5c–f). The VqWRKY33 promoter also contained three W-box elements located 143–478 bp upstream of the start codon (Supplementary Data Fig. S4, SupplementaryData Table S5). Two GUS fragments, P-478-GUS (with the W-box) and P-143-GUS (without the W-box), were constructed to determine whether the W-box of the VqWRKY33 promoter could respond to chitin and E. necator (Supplementary Data Fig. S4a). Two GUS fragments were transiently transformed into the leaves of Danfeng-2 or tobacco, which were then artificially inoculated with E. necator or sprayed with chitin. The findings showed that the fragments containing W-box regions responded to E. necator and chitin (Fig. 3b and c). To test whether VqWRKY33 could bind to the VqNSTS3 promoter, yeast one-hybrid (Y1H) assays were developed. It was found that VqWRKY33 can regulate the promoter of VqNSTS3 by binding at TTGACC (Fig. 3d). Moreover, dual-luciferase assays were performed, which gave the same results (Fig. 3f–h). Phosphorylation sites in the SP cluster of VqWRKY33 are well known [58]. A mutation of the four Ser residues to Ala in VqWRKY33 blocked its binding to the VqNSTS3 promoter, while a mutation of Ser to Asp enhanced its binding to the VqNSTS3 promoter (Fig. 3e and h). Chromatin immunoprecipitation (ChIP)–qPCR was conducted to examine the binding of VqWRKY33 in the promoter of VqNSTS3 in vivo (Fig. 3g and i). Because of the presence of W-box elements in the VqWRKY33 promoter, it was speculated that VqWRKY33 could regulate its own expression. Through Y1H assays, VqWRKY33 was found to regulate its own expression by binding to its own promoter (Fig. 3j). Moreover, the dual-luciferase and ChIP–qPCR assays obtained the same results (Fig. 3k–n). The above results show that VqWRKY33 positively regulates VqNSTS3 expression by binding to TTGACC in the VqNSTS3 promoter and also regulates its own expression.
Figure 3.
VqWRKY33 responds to E. necator and regulates VqNSTS3 expression. a qPCR analysis of VqWRKY33 expression in Danfeng-2 leaves after inoculation with E. necator for 0, 1, 3, 5, and 7 days. b Measurement of GUS activity. Danfeng-2 leaves expressing P-478-GUS and P-143-GUS were inoculated with E. necator or treated with chitin. c Chitin-induced GUS activity in transient expression tobacco leaves. Tobacco leaves expressing P-478-GUS and P-143-GUS were treated with 1 mg/ml chitin for 30 minutes. d Y1H analysis using pGADT7-VqWRKY33 as the prey and PpVqNSTS3-AbAi, P3xTTGACT-AbAi, and P3xTTGACC-AbAi as baits to demonstrate VqWRKY33 can bind to ProVqNSTS3 and TTGACC. e Loss-of-phosphorylation VqWRKY33 mutant with all four Ser residues mutated to Ala (VqWRKY33SA), and the phospho-mimicking VqWRKY33 mutant with all four Ser residues mutated to Asp (VqWRKY33SD). f Structural diagrams of dual-luciferase assays. g Schematic diagram of the promoter region of VqNSTS3. h Ratio of luciferase activity of VqWRKY33, VqWRKY33SA, and VqWRKY33SD binding to the VqNSTS3 promoter. i VqWRKY33 binding to the promoter of VqNSTS3 in vivo after E. necator treatment verified by ChIP–qPCR assays. j Y1H analysis using pGADT7-VqWRKY33 as prey and PpVqWRKY33-AbAi as bait to demonstrate VqWRKY33 can bind to its own promoter. k Structural diagrams of dual-luciferase assays. l Ratio of luciferase activity of VqWRKY33 binding to its own promoter. m Schematic diagram of the promoter region of VqWRKY33. n VqWRKY33 binding to promoter of the VqWRKY33 in vivo after E. necator inoculation, shown by ChIP–qPCR assays. Results in (a-c, h) are shown as mean ± standard error of the mean; n = 3. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (*P < .05; **P < .01). Results in (i, l, n) are shown as values ± standard error of the mean; n = 3. Significance was examined by Student’s t-test (**P < .01).
Transfer of VqWRKY33 into ‘Thompson Seedless’ to promote resistance to E. necator through accumulation of stilbenes
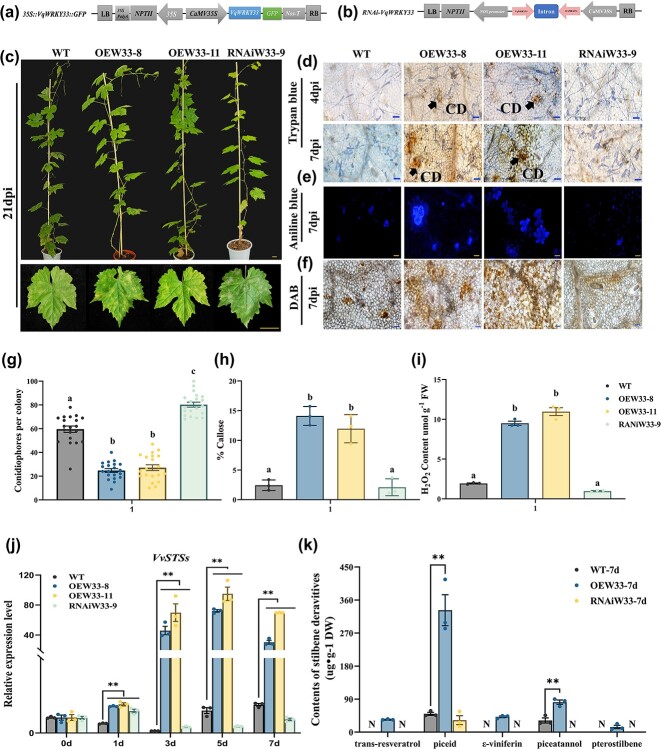
To determine the role of VqWRKY33 in the accumulation of stilbenes, two transgenic lines and one RNAi line were obtained by stable genetic transformation mediated by A. tumefaciens (Fig. 4a and b, Supplementary Data Fig. S6). Transgenic plants were identified by qPCR and western blot assays (Supplementary Data Fig. S6d and e), and the plants obtained were subjected to inoculation with E. necator to observe the phenotypes; WT plants were used as negative control. After artificial inoculation, WT and RNAi plants showed more colonies than those of OE lines. In transgenic lines a more obvious hypersensitive response (HR) cell death phenotype could be observed (Fig. 4c, d, and g). OE plants accumulated more callose and H2O2 than in WT and RNAi plants by histochemical staining (Fig. 4e–f and h–i). The expression of STSs after inoculation in transgenic plants showed a more significant response than in the WT and RNAi plants (Fig. 4j). VqWRKY33’s influence on the accumulation of stilbenes after E. necator inoculation was further investigated. The HPLC assay indicated that after inoculation, only piceid and piceatannol were detected in WT plants, while five stilbenes accumulated in transgenic plants. The contents of piceid and piceatannol in VqWRKY33 overexpression lines were 6.7 and 2.3 times higher, respectively, compared with WT. However, in RNAi-WRKY33 we only detected piceid after inoculation (Fig. 4k, Supplementary Data Table S8). Overall, these results showed that VqWRKY33 is an important transcription factor in regulating the stilbene synthesis pathway.
Figure 4.
Overexpression of VqWRKY33 in ‘Thompson Seedless’ promotes expression of STSs and resistance to E. necator. a, b Diagram of the (a) OEVqWRKY33 and (b) RNAiWRKY33 construct. c Photographs of OEW33-8, OEW33-11, RiW33-9, and WT plants at 21 dpi. Scale bars, 3 cm. d Trypan blue-stained OEW33-8, OEW33-11, RiW33-9, and WT leaves at 4 and 7 dpi. CD, cell death. Scale bars, 50 μm. e Aniline blue staining of OEW33-8, OEW33-11, RiW33-9, and WT leaves to detect callose deposition at 7 dpi. Scale bars, 50 μm. f DAB staining of OEW33-8, OEW33-11, RiW33-9, and WT leaves to detect H2O2 accumulation at 7 dpi. Scale bars, 50 μm. g Number of conidiophores per colony on OEW33-8, OEW33-11, RiW33-9, and WT leaves at 7 dpi. Results are shown as mean ± standard error of the mean; n = 20, and different letters represent significant differences (P < .05) as determined by one-way ANOVA followed by Tukey’s multiple comparisons test. h Quantification of callose deposition area on leaves at 7 dpi. i H2O2 content in OEW33-8, OEW33-11, RiW33-9, and WT leaves at 7 dpi. j qPCR analysis of VvSTS expression in OEW33-8, OEW33-11, RiW33-9, and WT plants after inoculation. Results are shown as mean ± standard error of the mean; n = 3. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (**P < .01). k HPLC analysis of stilbenes in (OE and RNAi) WRKY33 and WT after inoculation. Results are shown as mean ± standard error of the mean; n = 3. Significance was examined by Student’s t-test (**P < .01). Results in (h) and (i) are shown as mean ± standard error of the mean; n = 3, and different letters represent significant differences (P < .05) as determined by one-way ANOVA followed by Tukey’s multiple comparisons test.
VqWRKY33 induces enhanced expression of VqNSTS3 due to interaction with and phosphorylation by VqMAPK3/6
Our previous study found that MAPKKK38 responded significantly after E. necator induction [59]. We quantitatively analyzed MAPKKK38 and five MEKK genes after inoculation with E. necator in Danfeng-2. Here, MAPKKK38, MEKK3, and MEKK5 were significantly induced after E. necator inoculation (Supplementary Data Fig. S1). To further investigate VqWRKY33’s molecular role in regulating VqNSTS3 in response to E. necator, we focused on protein–protein interaction networks. The STRING database was used to predict the possible interacting proteins of VqWRKY33, including MAPK3 and MAPK6 (Supplementary Data Fig. S7a–c, Supplementary Data Table S6). VqMAPK3, VqMAPK4, and VqMAPK6 could be activated by E. necator and chitin [60] (Fig. 5a and b); meanwhile, VqWRKY33 could be phosphorylated after inoculation with E. necator (Fig. 5c). Next, to determine whether VqWRKY33 could interact with VqMAPK3 and VqMAPK6, a BiFC assay was performed, allowing direct interaction between VqMAPK3/6 and VqWRKY33 to be observed in nuclei (Fig. 5d). A split-luciferase complementation assay and a co-immunoprecipitation (CoIP) assay also confirmed the interaction between VqMAPK3/6 and VqWRKY33 (Fig. 5e–f). To determine whether VqMAPK3 and VqMAPK6 could phosphorylate VqWRKY33, we separately co-expressed VqMAPK3 and VqMAPK6 with VqWRKY33 in tobacco leaves and sprayed them with chitin. Phos-tag gel was used to separate the extracted protein. Under chitin treatment, VqWRKY33 can be phosphorylated by VqMAPK3 and VqMAPK6 (Fig. 5g). Constitutively active MAPK3 and MAPK6 (MAPK3CA and MAPK6CA), which result from two mutations (E198G/E202A and D220G/E224A) in each of the conserved domains, respectively, retain their substrate specificity and physiological functions [61] (Supplementary Data Fig. S7d and e). MAPK3/6CA could phosphorylate VqWRKY33 without chitin treatment (Fig. 5h), which confirms the previous report that MAPK3/6CA is a constitutively active form of MAPK3/6 [61]. To explore how VqMAPK3/6 affected VqWRKY33 function and regulated VqNSTS3 expression, transient transactivation was performed via assays using the VqNSTS3 promoter fused to GUS (ProVqNSTS3-GUS). Co-expression of VqMPK3/6 induced VqWRKY33-activated VqNSTS3 expression, and the VqMAPK3/6 constitutive activation form displayed enhanced VqNSTS3 expression activity (Fig. 5i and j).
Figure 5.
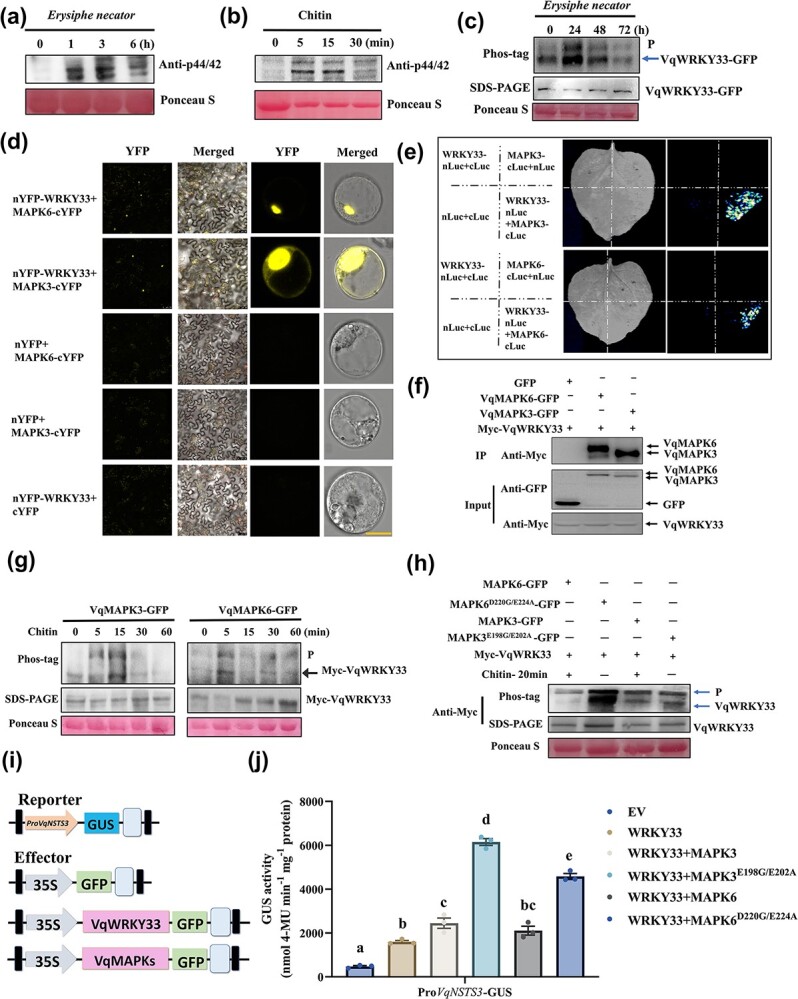
VqMAPK3/6 interact with and phosphorylate VqWRKY33, inducing the expression of VqNSTS3. a Activation of VqMAPKs in Danfeng-2 leaves treated with E. necator was verified by western blot assays. b Activation of VqMAPKs in Danfeng-2 leaves treated with 1 mg/ml chitin was verified by western blot assays. c Phosphorylation of VqWRKY33 was induced by E. necator. Proteins were separated by Phos-tag gel. d BiFC assays verified the interaction between VqWRKY33 and VqMAPK3/6 in tobacco leaves and grape protoplasts. Scale bars, 50/10 μm. e Split-luciferase complementation assays confirmed the interaction between VqWRKY33 and VqMAPK3/6. f CoIP assays validated that VqWRKY33 interacted with VqMAPK3 and VqMAPK6. g Phosphorylation of VqWRKY33 co-expressed with VqMAPK3 and VqMAPK6 after chitin treatment. Proteins were separated by Phos-tag gel, and then detected by immunoblotting with an anti-Myc antibody. h Phosphorylation of VqWRKY33 was induced by phospho-mimicking VqMAPK3/6 mutants. Proteins were separated by Phos-tag gel, and then detected by immunoblotting with an anti-Myc antibody. i Structural diagrams of GUS activity assays. j Measurement of GUS activity. ProVqNSTS3-GUS was co-transformed with 35S-GFP, 35S-VqWRKY33-GFP, and 35S-VqMAPKs-GFP in tobacco leaves. Results are shown as mean ± standard error of the mean; n = 3, and different letters represent significant differences (P < .05) as determined by one-way ANOVA followed by Tukey’s multiple comparisons test.
VqMAPK3/6 positively regulate the expression of VqSTSs and enhance resistance to E. necator in grapevine
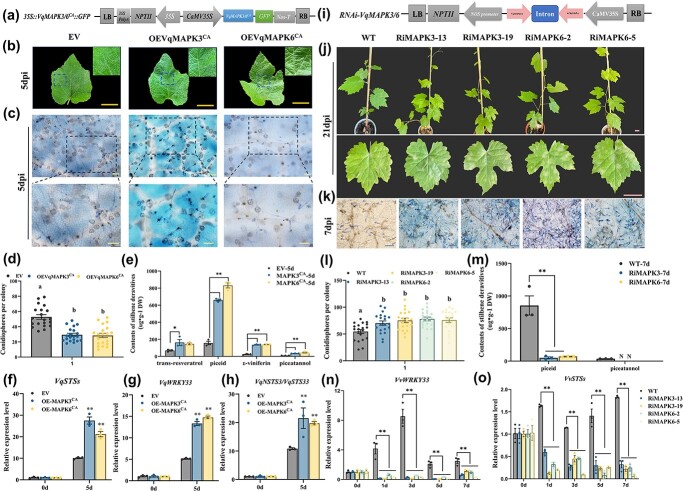
As VqMAPK3 and VqMAPK6 can be activated after E. necator inoculation (Fig. 5a), we speculated that VqMAPK3 and VqMAPK6 are involved in resistance to E. necator. 35S-VqMAPK3CA-GFP and 35S-VqMAPK6CA-GFP were transiently overexpressed in Danfeng-2 (EV as a negative control) (Fig. 6a, Supplementary Data S8c and d). We observed fewer spores on OE leaves than on the negative control after inoculation (Fig. 6b–d). The expression of VqWRKY33, VqNSTS3/VqSTS33, and VqSTSs in transient overexpression leaves was prominently higher than in the EV leaves (Fig. 6f–h). Contents of trans-resveratrol, piceid, ε-viniferin, and piceatannol in OEVqMAPK3CA leaves were 2.4-, 3.6-, 5.7-, and 1.3-fold compared with EV leaves after inoculation; in OEVqMAPK6CA leaves contents were 1.9-, 4.7-, 5.8-, and 1.6-fold compared with EV leaves after inoculation (Fig. 6e, Supplementary Data Table S9). These results indicated that phosphorylated VqMAPK3/6 could positively regulate the expression of VqWRKY33 and VqSTSs and the production of stilbenes. MAPK3 and MAPK6 were then silenced in grapevines (Fig. 6i, Supplementary Data Fig. S8e and f). Notably, spores of E. necator on RNAi-MAPK3 or RNAi-MAPK6 leaves were strikingly larger than those on WT after inoculation (Fig. 6j–l). RNAi-MAPK3 or RNAi-MAPK6 significantly reduced the expression levels of VvWRKY33 and VvSTSs (Fig. 6n and o). HPLC was performed to detect the accumulation of stilbenes in the RNAi lines, and only piceid was detected after inoculation. The contents of piceid in RNAi-MAPK3/6 increased 4.2- and 11.9-fold, respectively, after inoculation. The results showed that the non-toxic piceid in susceptible plants was the main stilbene accumulated (Fig. 6m, Supplementary Data Table S9). Collectively, these results indicate that VqMAPK3/6 positively regulate VqNSTS3 expression and increase the accumulation of stilbenes against E. necator.
Figure 6.
VqMAPK3/6 positively regulate the expression of STSs and enhance the disease resistance of grapevine. a Diagram of the OEVqMAPK3/6CA construct. b Photographs of OEVqMAPK3/6CA and EV leaves at 5 dpi. Scale bars, 3 cm. c Trypan blue-stained EV and OEVqMAPK3/6CA leaves at 5 dpi. Scale bars , 50 μm. d Number of conidiophores per colony on EV and OEVqMAPK3/6CA leaves at 5 dpi. e HPLC analysis of stilbenes in OEVqMAPK3/6CA and EV leaves. f–h qPCR analysis of VqWRKY33, VqNSTS3/VqNSTS33, and VqSTS in EV and OE leaves after E. necator inoculation. i Diagram of the RNAi-MAPK3/6 construct. j Photographs of RiMAPK3-13, RiMAPK3-19, RiMAPK6-2, RiMAPK6-5, and WT plants at 21 dpi. Scale bars, 3 cm. k Trypan blue-stained RNAi-MAPK3/6 and WT leaves at 7 dpi. Scale bars, 100 μm. l Number of conidiophores per colony on RNAi-MAPK3/6 and WT leaves at 7 dpi. m HPLC analysis of stilbenes in RNAi-MAPK3/6 and WT. n, o Expression of VvSTSs and VvWRKY33 analyzed by qPCR in WT and RNAi-MAPK3/6 plants after E. necator inoculation. In (d) and (l) results are shown as mean ± standard error of the mean; n = 20, and different letters represent significant differences (P < .05) as determined by one-way ANOVA followed by Tukey’s multiple comparisons test. In (e–h) and (m–o) results are shown as mean ± standard error of the mean; n = 3. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (*P < .05; **, P < .01).
Pro VqNSTS3::VqNSTS3-GFP moves to and wraps the pathogen haustoria, forming encasements to block the invasion of pathogens in A. thaliana
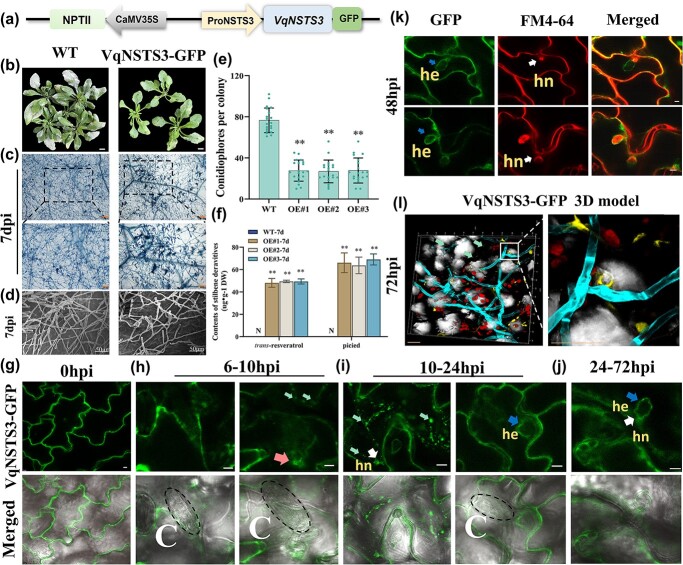
As the model plant A. thaliana does not contain STS genes, VqNSTS3 was linked to its own promoter to stably transform A. thaliana, thus revealing the expression and function of the VqNSTS3 gene in transgenic A. thaliana (Fig. 7a, Supplementary Data Fig. S9). Transgenic lines of the T3 generation were artificially inoculated with G. cichoracearum, and after trypan blue staining it was found that large areas of HR cell death appeared on transgenic A. thaliana after inoculation (Fig. 7b and c). Furthermore, the number of spores on the transgenic lines was less than on WT (Fig. 7d and e). Trans-resveratrol and piceid in transgenic lines accumulated after inoculation (Fig. 7f, Supplementary Data Table S10). Preliminary studies have shown that stilbenes can inhibit the growth of hyphae, and mainly accumulate at the place where G. cichoracearum invades [15]. However, there is no direct evidence of how the STS gene resists G. cichoracearum infection. Forty-eight hours after inoculation of the OE lines, green fluorescence of VqNSTS3 was observed on the plasma membrane and the intact haustorium encasement. The lipophilic dye FM4-64 mainly marked the cell phospholipid membrane. Green and red fluorescence had obvious fluorescence overlap on the haustorial neck and plasma membrane (Fig. 7k). The yellow fluorescence was observed gathered around the secondary haustorium developing from new appressoria along the hyphae 72 hours after inoculation with G. cichoracearum (Fig. 7l). To further determine when and how ProVqNSTS3::VqNSTS3-GFP accumulated at haustorium encasements, a time-process study was conducted. At the invasion site of G. cichoracearum, aggregation of GFP fluorescence could be seen 6–10 hours post-inoculation (hpi), accompanied by the germination of spores. With increasing invasion time, some small multivesicular body (MVB) structures aggregated in the infected sites and started accumulating at the haustorial neck (10–24 hpi). A cupular encasement then formed around the haustorium [62]. Finally, the haustorium was completely wrapped by ProVqNSTS3::VqNSTS3-GFP (24–72 hpi) (Fig. 7g–j). These results indicate that stilbene synthase directly interacts with spores and inhibits the germination and growth of spores.
Figure 7.
ProVqNSTS3::VqNSTS3-GFP moves to and wraps the pathogen haustoria to block the invasion of G. cichoracearum in A. thaliana. a Diagram of the ProVqNSTS3::VqNSTS3-GFP construct. b Photographs of ProVqNSTS3::VqNSTS3-GFP overexpression and Col-0 leaves infected with G. cichoracearum at 7 dpi. Scale bars, 1 cm. c Trypan blue staining of leaves from Col-0 and transgenic A. thaliana at 7 dpi. Scale bars, 100/50 μm. d Scanning electron micrographs of Col-0 and transgenic A. thaliana leaves inoculated with G. cichoracearum for 7 days. Scale bars, 50 μm. e Quantification of G. cichoracearum growth on A. thaliana leaves at 7 dpi. Results are shown as mean ± standard error of the mean; n = 20. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (**P < .01). f HPLC analysis of piceid and trans-resveratrol in ProVqNSTS3::VqNSTS3-GFP transgenic plants and Col-0. Results are shown as mean ± standard error of the mean; n = 3. Significance was examined by one-way ANOVA followed by Dunnett’s multiple comparisons test (**P < .01). g–j Confocal microscope images from single optical sections of A. thaliana leaf epidermal cells expressing ProVqNSTS3::VqNSTS3-GFP infected by G. cichoracearum. The top row shows ProVqNSTS3::VqNSTS3-GFP fluorescence and the bottom row shows the merged field images. The red arrow indicates the penetration site, green arrows indicate multivesicular bodies, white arrows indicate the haustorial necks, and blue arrows indicate haustorial encasements. hn, haustorial neck; he, haustorial encasement; C, conidium. Scale bars, 5 μm. kArabidopsis thaliana plants overexpressing ProVqNSTS3::VqNSTS3-GFP were inoculated with G. cichoracearum and haustorial encasements were analyzed by confocal microscopy at 48 hours post-inoculation (hpi) for PM. hn, haustorial neck; he, haustorial encasement. Scale bars, 5 μm. The left column shows ProVqNSTS3::VqNSTS3-GFP fluorescence, the middle column shows corresponding red fluorescence after staining with the membrane-specific tracer FM4-64, and the right column shows the corresponding merged images. l Z-projections through the epidermal cell layer were visualized by confocal microscopy at 72 hpi. They were 3D-reconstructed and displayed as maximum intensity projections. Image shows overlays of GFP fluorescence (yellow) and calcofluor white staining (cyan). Scale bars, 50 μm.
Discussion
On the stilbene synthase genes and novel VqNSTS3 resistance to disease in grapevine
STS is a key enzyme in the biosynthesis of resveratrol [63]. Resveratrol in grapevine plays a key role in plant disease resistance and is beneficial to human health [18, 19]. After infection or stress in grapevine, resveratrol, a major stilbene product, accumulates in large amounts in the stressed areas [64]. We recently found six novel transcripts of STSs by analyzing the transcriptome data of Danfeng-2 [55]. Based on sequence alignment, there are significant differences between VqNSTS2–6 and the reported VqSTSs, indicating that the VqNSTS genes are new members in Danfeng-2 [34]. Among them, VqNSTS3 produced the highest stilbene content after it was transiently transformed into tobacco (Fig. 1f). Therefore, we speculated that VqNSTS3 enhanced the resistance of Danfeng-2 to E. necator by accumulating stilbenes. In order to better study the function of VqNSTS3 from Chinese wild grapevine in European grapevine varieties, an A. tumefaciens-mediated genetic regeneration system for transgenic grapevines was used for the identification of gene function in transgenic lines [43, 65]. We genetically transformed VqNSTS3 into V. vinifera ‘Thompson Seedless’ and detected the accumulation of stilbenes after inoculation. The content of these chemicals in the VqNSTS3 transgenic lines exceeded that in WT plants (Fig. 2g). Similar results were observed in VpSTS29/STS2 [47] and VqNSTS4 overexpression lines [34]. Plants resist pathogens through two layers of innate immunity: PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) [66–68]. PTI is the basic defense of plants, characterized by activation of multiple immune responses [69–73]. Callose deposits, a sign of the plant PTI response [74], are accumulated at the sites of attack during early stages of pathogen invasion [75]. The HR may inhibit or delay further spread of the pathogen [76]. Compared with WT plants, VqNSTS3 overexpression plants exhibited more cell death and higher callose accumulation after E. necator infection (Fig. 2c–e). Consistent with this, VqNSTS3-transgenic A. thaliana plants showed HR cell death and limited spore growth and germination (Fig. 7b–e). These results were also found in previous studies [34, 47, 53]. SA signaling is another vital signal for plant immunity [77]. Overexpression of VqNSTS3 in transgenic grapevine lines activated SA-related signaling genes PR1 and PR5 (Fig. 2h and i) and disease resistance-related genes RBOHD and CHIT4C (Fig. 2j and k), which is similar to the findings in VqNSTS4 overexpression grapevines in that the plants showed enhanced disease resistance-related gene expression and enhanced resistance to E. necator [34]. In RNAi-VqNSTS3 plants, however, we observed the opposite results (Fig. 2l–u). Overall, these results suggest that overexpression of VqNSTS3 triggered several mechanisms after elicitor perception and regulated stilbene production in plant cells to enhance E. necator resistance [47, 78, 79]. Our results demonstrated that VqNSTS3 transgenic overexpression plants showed resistance-related gene expression, HR cell death, and callose deposition after inoculation by E. necator.
Transcription factor regulation and novel mechanism of VqWRKY33 in grapevine
Several transcription factors involved in the regulation of grape STS genes have been discovered, including MYB, WRKY, ERF, and bZIP [26–33, 80]. Höll et al. reported that the MYB transcription factors that regulate the STS genes in grapevine via a typical path, VvMYB14 and VvMYB15, can activate the promoters of VvSTS29/41 [30]. Jiang et al. revealed that VqMYB154 can promote polygene VqSTS9/32/42 expression by binding to their promoters [27]. Aside from MYB transcription factors, WRKY transcription factors are also vital regulators of STS genes [35, 81]. VvWRKY24 alone can activate the promoter of VvSTS29, but VvWRKY3 needs to form an integrated organization with VvMYB14 to regulate VvSTS29 [35]. VqSTS32/41 are positively regulated by VqWRKY53; meanwhile VqWRKY53 interacts with VqMYB14 and VqMYB15 to show a stronger regulatory function [32]. VqWRKY31 can directly bind to the promoters of STS9/48 [26]. The WRKY transcription factors are prominent signaling proteins participating in resistance to various fungal diseases in plants [82–85]. They are key regulatory components of plant disease resistance for A. thaliana [86], rice (Oryza sativa) [87], apple (Malus domestica) [85], Brassica napus [88], and rose (Rosa hybrida) [89]. Moreover, many WRKY transcription factors in grapevines have been demonstrated to be involved in plant resistance to disease. For example, heterologous expression of VpWRKY1, VpWRKY2, VpWRKY11, VqWRKY52, VqWRKY53, and VqWRKY56 enhances resistance to pathogens [32, 84, 90–92]. WRKY family members can be divided into three subfamilies [57]. WRKY33, belonging to the WRKY I family, is a pathogen-inducible transcription factor, the expression of which was shown to be essential for positively regulating resistance to B. cinerea [86, 93], Alternaria brassicicola [94], and the oomycete pathogen Plasmopara viticola [95]. In A. thaliana, AtWRKY33 induced camalexin biosynthesis after pathogen infection [86, 93, 94], and acted as an important node of the regulatory cascade [86]. In this study, a WRKY-type transcription factor, VqWRKY33, which can be induced by E. necator, was isolated from Danfeng-2 (Fig. 3a, Supplementary DataS5). WRKY transcription factors regulate target genes by binding to the W-box elements on target gene promoters [57]. The VqNSTS3 promoter contained both TTGACT and TTGACC (Supplementary Data Fig. S2), which indicates that VqNSTS3 may be directly regulated by VqWRKY33. Through Y1H, dual-luciferase, and ChIP–qPCR assays, the results showed that VqWRKY33 increased the activity of VqNSTS3 promoter by binding directly to TTGACC on the VqNSTS3 promoter (Fig. 3d, f–i). In this study, overexpression of VqWRKY33 in ‘Thompson Seedless’ plants enhanced E. necator tolerance and increased stilbene, callose, and H2O2 accumulation, and HR cell death after inoculation, whereas RNAi plants showed the opposite phenotype (Fig. 4a–i and k). Here, compared with WT and RNAi plants, the expression of the STS gene in plants overexpressing VqWRKY33 was significantly higher after inoculation with E. necator (Fig. 4j). Taken together, our data show that VqWRKY33 may be an important node for enhancing the expression of VqNSTS3 in grapevine, leading to stilbenes accumulation and consequently resistance to E. necator.
Phosphorylation signaling of the three-stage cascade of VqMAPK3/6-VqWRKY33 enhances VqNSTS3 stilbene accumulation and prevents infection by pathogens
In plants, MAPK cascades can regulate plant growth processes, hormonal signaling, and the response of the plant to various stresses [96–100]. This module typically consists of three protein kinases that activate each other through phosphorylation [101]. Two MAPK cascades are known to participate in plant immunity [61, 102–106]. The MAPK cascades communicate biological signals through phosphorylation of various transcription factors [107]. Among them, WRKYs are vital substrates of MAPK cascades. For example, AtMPK3/AtMPK6-AtWRKY33 functions against B. cinerea and MAPK-WRKY7/8/9/11 against Phytophthora [83, 108]. MdMMKK4-MdMPK3-MdWRKY17 increased susceptibility to Colletotrichum fructicola due to SA degradation in apple [85]. However, studies of grapevine MAPK signal transduction in response to E. necator have not been conducted. MAPK activation is one of the earliest signaling events in plants after perception of pathogen stress [109] and participates in signal transduction of multiple defense responses [96, 110]. In V. vinifera, there are 14 MAPKs, 5 MAPKKs, 62 MAPKKKs, and 7 MAPKKKKs [111]. Jiao et al. (2017) reported that stilbene accumulation can be positively regulated by VqMAPKKK38 by mediating the activation of VqMYB14 in grapevine [59]. In this study, expression of VqMAPKKK38, MEKK3, and MEKK5 was significantly induced after E. necator inoculation in Danfeng-2 (Supplementary Data Fig. S1). Then, VqMAPK3 and VqMAPK6 were activated following chitin and E. necator treatment (Fig. 5a and b). These results may suggest that VqMAPKKK38, VqMEKK3, and VqMEKK5 act upstream in Danfeng-2 in transducing signal downstream of MAPK3/6 after E. necator inoculation. AtMPK3/MPK6 was previously reported to phosphorylate AtWRKY33 and activate camalexin biosynthesis gene expression [58]. The OsMKK4-OsMPK6 cascade plays a vital role in biosynthesis of diterpenoid phytoalexins [112]. SIPK/NTF4/WIPK-phosphorylated, WRKY33-related NbWRKY8 induced a key gene for the production of isoprenoid phytoalexins [113]. In our study, we have shown that VqMAPK3/VqMAPK6 phosphorylation of VqWRKY33 forms an accessory pathway for the regulation of stilbene biosynthesis and resistance to E. necator in grapevine. WRKY I family members have SP clusters, which are thought to be phosphorylated by MAPKs at the N-terminal [58, 85, 108, 113]. Four sites in the SP clusters of VqWRKY33 are important phosphorylation sites that regulate VqWRKY33-mediated VqNSTS3 expression (Fig. 3e and h). In apple, the phosphorylation sites in the SP cluster are vital for regulating MdWRKY17-mediated MdDMR6 activation [85]. Phosphorylation of VqWRKY33 by VqMPK3/VqMPK6 enhances its activity in promoting the expression of downstream stilbene biosynthetic genes. However, other phosphorylation sites involved in phosphorylating VqWRKY33 by VqMAPK3/6 need to be further studied. In addition, RNAi-MAPK3/6 plants both showed increased susceptibility to E. necator and decreased the accumulation of stilbenes (Fig. 6). In Danfeng-2, both VqMAPK3 and VqMAPK6 showed vital functions in stilbene accumulation and E. necator resistance. Therefore, our study found that, under infection with E. necator, VqMAPK3/6 sense the stimulation of the pathogen and release phosphorylation signals, which cause downstream transcription factor VqWRKY33 to start the positive regulation of target gene VqNSTS3, which expresses and accumulates a large number of stilbenes, enhancing disease resistance.
ProVqNSTS3::VqNSTS3-GFP moves to and wraps the haustoria to prevent pathogen invasion in transgenic A. thaliana
Fungal conidia invade plants by forming haustoria, which secrete proteins that degrade the host cell wall, and then invade the host plant [114]. To prevent the growth of fungi, plant cells enclose the haustoria by forming an encasement [115]. Haustorial encasements may serve as a matrix in which to concentrate plant-derived antimicrobial compounds at the plant–fungal interface, thereby poisoning the haustorium [62, 116, 117]. Several proteins that are essential for encasement formation towards PM fungus have been found in previous studies. For example, the syntaxin PEN1 (SYP121) and its closest homolog, SYP122, are required for encasement formation [62, 118]. These syntaxins are required for mediating encasement formation at the site of a fungal attack [117]. Previous studies showed that STS proteins localized to the exocarp cell wall, secondary cell wall, chloroplast, endoplasmic reticulum, and the oil bodies using immune-histochemical, immunogold electron microscopy, and laser scanning confocal microscopy techniques [48, 119, 120]. In this study, ectopic expression of VqNSTS3 in A. thaliana under its own promoter showed that VqNSTS3 is crucial for postinvasive immunity against the PM pathogen by accumulating stilbenes in the transgenic lines after artificial inoculation with G. cichoracearum (Fig. 7f). By using laser scanning confocal microscopy, VqNSTS3 was found to localize on the plasma membrane in the absence of pathogen infection, but was actively translocated to the haustorial encasements and surrounded the haustoria when plants were challenged by G. cichoracearum (Fig. 7g–l). The expression enhancement of VqNSTS3 caused large amounts of the protein VqNSTS3 to be transported to the haustorium by vesicles (Fig. 7h and i). Therefore, our results imply that the VqNSTS3-containing MVBs move to the haustorial encasements and wrap around them, preventing the growth of fungal conidia and mycelium .
In summary, we have discovered and identified a novel VqNSTS3 from Danfeng-2 based on transcriptome sequencing. The novel VqNSTS3 has the conserved domain and sequence characteristics of the STS family. Transgenic VqNSTS3 plants not only rapidly produced phytoalexin but also showed HR cell death, callose accumulation, and resistance-related gene expression after E. necator inoculation. Grapevine VqMAPK3/6-VqWRKY33 positively regulates novel VqNSTS3 expression and resistance to E. necator in grapevine immunity. It has been found that in plants that have been attacked by pathogens VqNSTS3 is actively transported to the haustorium, then surrounds the haustorium and prevents it from invading the plant (Fig. 8). These results demonstrate that the Chinese wild grapevine integrates the three-stage cascade signal to positively regulate VqNSTS3 expression and stilbene accumulation, thereby enhancing the resistance to PM. Chinese wild grapes are valuable germplasm resources for grape disease resistance breeding.
Figure 8.
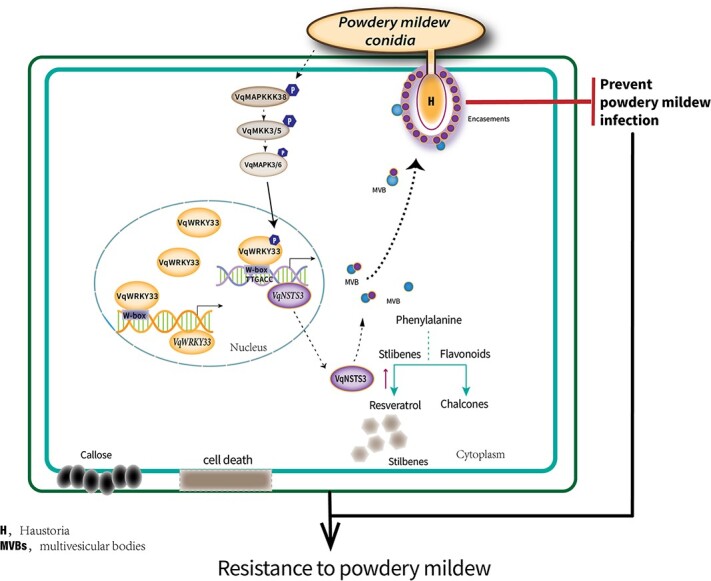
Model of the three-stage signaling cascade of VqMAPK3/6-VqWRKY33-VqNSTS3 in Danfeng-2, enhancing stilbene accumulation and preventing infection with pathogens. Erysiphe necator-triggered phosphorylation of VqWRKY33 by VqMAPK3/6 enhances the binding of VqWRKY33 to the VqNSTS3 promoter and activates VqNSTS3 expression to promote the accumulation of stilbenes. VqWRKY33 can also activate its own expression. VqNSTS3 can be carried by MVBs that positively accumulated at haustorial encasements to inhibit the growth of PM spores.
Materials and methods
Plant materials
All sample tissues of Chinese wild grapevine V. quinquangularis accession Danfeng-2 were gathered in 2020 from the grapevine resource nursery of Northwest A & F University, Yangling, Shaanxi, China (34°20′N, 108°24′E). Callus of V. vinifera cultivar ‘Thompson Seedless’ was used for genetic transformation. Arabidopsis thaliana Columbia WT (Col-0) was cultivated and used as a transgene receptor in a growth chamber. Tobacco plants (Nicotiana benthamiana) were cultivated in an incubator at 22 ± 2°C with light for 16 hours.
RNA extraction and reverse transcription–quantitative PCR
The Omega Plant RNA Kit (Omega, GA, USA) was used for RNA extraction. The FastKing RT Kit (TIANGEN, Beijing, China) was used for cDNA first-strand synthesis [92]. PerfectStart Green qPCR SuperMix (TransGen Biotech, Beijing, China) and the ABI QuantStudio 6 Flex (Applied Biosystems, Thermo Fisher, CA, USA) were used for qPCR. The 2−ΔΔc(t) method was used to calculate relative expression [121]. Primers are listed in Supplementary Data Tables S1–3.
Subcellular localization
The plasma membrane-localized marker PM-RK-mCherry [122] was co-expressed with 35S-VqMAPK3/6-GFP to validate the localization of these proteins and injected into 4-week-old tobacco leaves. And 35S-VqNSTS3-GFP construct was mobilized into the GV3101 strain of Agrobacterium tumefaciens and then transferred into tobacco leaves. [123]. In addition, 35S-VqWRKY33-GFP was co-expressed with 35S-AtHY5-mCherry (a marker of nucleus) into protoplasts of grapevine using the polyethylene glycol-mediated method [124]. GFP and mCherry signals were observed with confocal laser microscopy (Leica TCS SP8, Germany).
Grapevine transformation, E. necator infection, histochemical staining, and microscopy
Callus isolated from ‘Thompson Seedless’ was used for grapevine transformation by A. tumefaciens. The transgenic transformation method was similar to the method described previously [43, 65, 123]. The third to sixth healthy and newly developed leaves from the beginning of the shoot tip were selected for E. necator artificial inoculation [42].
After 7 days of inoculation, leaves were gathered for trypan blue staining and calculating the number of conidiophores per colony under a microscope as described [125]. Trypan blue staining was used to visualize hyphal growth and detect cell death [27]. Callose was stained by aniline blue and visualized by UV epifluorescence. Scanning electron microscopy observation was carried out following a previously described method [15]. For transient expression experiments in grapevine, A. tumefaciens containing 35S-VqMAPK3/6CA-GFP, RNAi-VqNSTS3, and empty vectors was cultured in LB liquid medium, and the OD600 was adjusted to 0.6–0.7. Leaves of Danfeng-2 were vacuumed for 30 minutes by immersion in bacterial solution [126]. Then, the leaves were placed in a growth chamber for moisturizing and cultivation for 3 days and gathered for subsequent research.
Stilbene content determination by high-performance liquid chromatography
Plant samples were freeze-dried for 48 hours, and dry weight was determined according to the volume-to-mass ratio of 1:10. Methanol was added to extract stilbene substances for 24 hours in dark. Then, the methanol extract was filtered through a 0.22-μm membrane film. High-performance liquid chromatography (HPLC) was conducted using a Nexera UHPLC LC-30A (Shimadzu, Japan). The gradient used was consistent with previous research methods [127]. Standard samples of trans-resveratrol (CAS: 501-36-0), piceid (CAS: 27208-80-6), piceatannol (CAS: 10083-24-6), pterostilbene (CAS: 537-42-8), and ε-viniferin (CAS: 62218-08-0) (Sigma–Aldrich, USA) were used to confirm retention times.
Acknowledgements
We are grateful to Cambridge Proofreading Company for editing the text language. The research was funded by the National Natural Science Foundation of China (grant no. 32272667).
Author contributions
Y.W. designed the research. R.L. analyzed Danfeng-2-specific novel transcripts and obtained six new stilbene synthase gene transcripts. C.Y. cloned six new STS genes, analyzed the gene structure and function, transiently transformed the new STS genes to tobacco and determined the expression of stilbenes. W.L. and C.Y. carried out the experiments, and W.L. analyzed the data. G.C. and X.W. helped with the experimental work. Y.W., C.Z., Y.X., and X.W. revised the manuscript. W.L. wrote and Y.W. reviewed and revised the manuscript.
Data availability
All relevant data can be found within the manuscript and its supporting information.
Conflict of interest statement
The authors declare that they have no competing interests.
Supplementary data
Supplementary data are available at Horticulture Research online.
Supplementary Material
Contributor Information
Wandi Liu, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Chaohui Yan, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Ruimin Li, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Guanyu Chen, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Xinqi Wang, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Yingqiang Wen, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Chaohong Zhang, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Xiping Wang, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Yan Xu, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
Yuejin Wang, College of Horticulture, Northwest A & F University, Yangling, Shaanxi, 712100, China; Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Yangling, Shaanxi, 712100, China; State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A & F University, Yangling, Shaanxi, 712100, China.
References
- 1. Jaillon O, Aury JM, Noel Bet al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–7. [DOI] [PubMed] [Google Scholar]
- 2. Gadoury DM, et al. Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol Plant Pathol. 2012;13:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brewer MT, Milgroom MG. Phylogeography and population structure of the grape powdery mildew fungus, Erysiphe necator, from diverse Vitis species. BMC Evol Biol. 2010;10:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cadle-Davidson L, Chicoine DR, Consolie NH. Variation within and among Vitis spp. for foliar resistance to the powdery mildew pathogen Erysiphe necator. Plant Dis. 2011;95:202–11. [DOI] [PubMed] [Google Scholar]
- 5. Qiu W, Feechan A, Dry I. Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic Res. 2015;2:15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kunova A, Pizzatti C, Saracchi Met al. Grapevine powdery mildew: fungicides for its management and advances in molecular detection of markers associated with resistance. Microorganisms. 2021;9:1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vielba-Fernández A, Polonio Á, Ruiz-Jiménez Let al. Fungicide resistance in powdery mildew fungi. Microorganisms. 2020;8:1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gadino AN, Walton VM, Dreves AJ. Impact of vineyard pesticides on a beneficial arthropod, Typhlodromus pyri (Acari: Phytoseiidae), in laboratory bioassays. J Econ Entomol. 2011;104:970–7. [DOI] [PubMed] [Google Scholar]
- 9. Langcake P, Pryce RJ. The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol Plant Pathol. 1976;9:77–86. [Google Scholar]
- 10. Langcake P, Pryce RJ. A new class of phytoalexins from grapevines. Experientia. 1977;33:151–2. [DOI] [PubMed] [Google Scholar]
- 11. Jeandet P, Bessis R, Gautheron B. The production of resveratrol (3,5,4′-trihydroxystilbene) by grape berries in different developmental stages. Am J Enol Vitic. 1991;42:41–6. [Google Scholar]
- 12. Siemann EH, Creasy LL. Concentration of the phytoalexin resveratrol in wine. Am J Enol Vitic. 1992;43:49–52. [Google Scholar]
- 13. Adrian M, Jeandet P, Veneau Jet al. Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J Chem Ecol. 1997;23:1689–702. [Google Scholar]
- 14. Pezet RGK, Gindro K, Viret Oet al. Effects of resveratrol, viniferins and pterostilbene on Plasmopara viticola zoospore mobility and disease development. Vitis. 2004;43:145–8. [Google Scholar]
- 15. Schnee S, Viret O, Gindro K. Role of stilbenes in the resistance of grapevine to powdery mildew. Physiol Mol Plant Pathol. 2008;72:128–33. [Google Scholar]
- 16. Alonso-Villaverde V, Voinesco F, Viret Oet al. The effectiveness of stilbenes in resistant Vitaceae: ultrastructural and biochemical events during Plasmopara viticola infection process. Plant Physiol Biochem. 2011;49:265–74. [DOI] [PubMed] [Google Scholar]
- 17. Khattab IM, Sahi VP, Baltenweck Ret al. Ancestral chemotypes of cultivated grapevine with resistance to Botryosphaeriaceae-related dieback allocate metabolism towards bioactive stilbenes. New Phytol. 2021;229:1133–46. [DOI] [PubMed] [Google Scholar]
- 18. Jang M, Cai L, Udeani GOet al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–20. [DOI] [PubMed] [Google Scholar]
- 19. Barger JL, Kayo T, Vann JMet al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chong J, Poutaraud A, Hugueney P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009;177:143–55. [Google Scholar]
- 21. Hain R, Reif HJ, Krause Eet al. Disease resistance results from foreign phytoalexin expression in a novel plant. Nature. 1993;361:153–6. [DOI] [PubMed] [Google Scholar]
- 22. Stark-Lorenzen P, Nelke B, Hänßler Get al. Transfer of a grapevine stilbene synthase gene to rice (Oryza sativa L.). Plant Cell Rep. 1997;16:668–73. [DOI] [PubMed] [Google Scholar]
- 23. Richter A, Jacobsen HJ, de Kathen Aet al. Transgenic peas (Pisum sativum) expressing polygalacturonase inhibiting protein from raspberry (Rubus idaeus) and stilbene synthase from grape (Vitis vinifera). Plant Cell Rep. 2006;25:1166–73. [DOI] [PubMed] [Google Scholar]
- 24. Liu S, Hu Y, Wang Xet al. High content of resveratrol in lettuce transformed with a stilbene synthase gene of Parthenocissus henryana. J Agric Food Chem. 2006;54:8082–5. [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi S, Ding CK, Nakamura Yet al. Kiwifruits (Actinidia deliciosa) transformed with a Vitis stilbene synthase gene produce piceid (resveratrol-glucoside). Plant Cell Rep. 2000;19:904–10. [DOI] [PubMed] [Google Scholar]
- 26. Yin W, Wang X, Liu Het al. Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic Res. 2022;9:uhab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang C, Wang D, Zhang Jet al. VqMYB154 promotes polygene expression and enhances resistance to pathogens in Chinese wild grapevine. Hortic Res. 2021;8:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fang L, Hou Y, Wang Let al. Myb14, a direct activator of STS, is associated with resveratrol content variation in berry skin in two grape cultivars. Plant Cell Rep. 2014;33:1629–40. [DOI] [PubMed] [Google Scholar]
- 29. Mu H, Li Y, Yuan Let al. MYB30 and MYB14 form a repressor-activator module with WRKY8 that controls stilbene biosynthesis in grapevine. Plant Cell. 2023;35:552–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Höll J, Vannozzi A, Czemmel Set al. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell. 2013;25:4135–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang J, Xi H, Dai Zet al. VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J Exp Bot. 2019;70:715–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang D, Jiang C, Liu Wet al. The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. J Exp Bot. 2020;71:3211–26. [DOI] [PubMed] [Google Scholar]
- 33. Wang D, Jiang C, Li Ret al. VqbZIP1 isolated from Chinese wild Vitis quinquangularis is involved in the ABA signaling pathway and regulates stilbene synthesis. Plant Sci. 2019;287:110202. [DOI] [PubMed] [Google Scholar]
- 34. Yan C, Yang N, Li Ret al. Alfin-like transcription factor VqAL4 regulates a stilbene synthase to enhance powdery mildew resistance in grapevine. Mol Plant Pathol. 2023;24:123–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vannozzi A, Wong DCJ, Höll Jet al. Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol. 2018;59:1043–59. [DOI] [PubMed] [Google Scholar]
- 36. Parage C, Tavares R, Réty Set al. Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol. 2012;160:1407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vannozzi A, Dry IB, Fasoli Met al. Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 2012;12:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Girollet N, Rubio B, Lopez-Roques Cet al. De novo phased assembly of the Vitis riparia grape genome. Sci Data. 2019;6:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang Z, Duan S, Sheng Jet al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat Commun. 2019;10:1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel S, Robben M, Fennell Aet al. Draft genome of the native American cold hardy grapevine Vitis riparia Michx. 'Manitoba 37′. Hortic Res. 2020;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shirasawa K, Hirakawa H, Azuma Aet al. De novo whole-genome assembly in an interspecific hybrid table grape, 'Shine Muscat'. DNA Res. 2022;29:dsac040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang, Liu, He Y, Y, P. et al. Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis spp. species. Vitis. 1995;34:159–64. [Google Scholar]
- 43. Dai L, Zhou Q, Li Ret al. Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. Chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant Cell Tissue Organ Cult. 2015;121:397–412. [Google Scholar]
- 44. Wang X. Cloning and analysing for the gene sequences of resistance to Uncinula necator in Chinese wild Vitis species. Ph.D. Dissertation,. China: Northwest A & F University; 2004. [Google Scholar]
- 45. Xu W. Cloning and functional analysis of stilbene synthase and its promoter from powdery mildew-resistant Chinese Vitis pseudoreticulata. Ph.D. Dissertation,. China: Northwest A & F University; 2010. [Google Scholar]
- 46. Cao J. Clone and characterization of stilbene synthase gene and genetic transformation in grape. Master’s Thesis,. China: Northwest A & F University; 2012. [Google Scholar]
- 47. Xu W, Ma F, Li Ret al. VpSTS29/STS2 enhances fungal tolerance in grapevine through a positive feedback loop. Plant Cell Environ. 2019;42:2979–98. [DOI] [PubMed] [Google Scholar]
- 48. Ma F, Wang L, Wang Y. Ectopic expression of VpSTS29, a stilbene synthase gene from Vitis pseudoreticulata, indicates STS presence in cytosolic oil bodies. Planta. 2018;248:89–103. [DOI] [PubMed] [Google Scholar]
- 49. Shi J, He M, Cao Jet al. The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera). Plant Physiol Biochem. 2014;74:24–32. [DOI] [PubMed] [Google Scholar]
- 50. Ding X. Study on the regulation of powdery mildew resistance by stilbene synthase genes VqSTS9 and VqSTS21 from Chinese wild Vitis quinquangularis. Master’s Thesis,. China: Northwest A & F University; 2020. [Google Scholar]
- 51. Wu F. The expression and function analysis of the stilbene synthase genes of the resistance to powdery mildew from Chinese wild Vitis quinquangularis. Master’s Thesis,. China: Northwest A & F University; 2019. [Google Scholar]
- 52. Zhao K. Study on stilbene synthase genes VqSTS11 and VqSTS23 regulating resistance to powdery mildew from Chinese wild Vitis quinquangularis. Master’s Thesis,. China: Northwest A & F University; 2020. [Google Scholar]
- 53. Liu M, Ma F, Wu Fet al. Expression of stilbene synthase VqSTS6 from wild Chinese Vitis quinquangularis in grapevine enhances resveratrol production and powdery mildew resistance. Planta. 2019;250:1997–2007. [DOI] [PubMed] [Google Scholar]
- 54. Cheng S, Xie X, Xu Yet al. Genetic transformation of a fruit-specific, highly expressed stilbene synthase gene from Chinese wild Vitis quinquangularis. Planta. 2016;243:1041–53. [DOI] [PubMed] [Google Scholar]
- 55. Li R. Regulation mechanism of stilbenes metabolism in Chinese wild Vitis quinquangularis using multi-omics analysis. Ph.D. Dissertation,. China: Northwest A & F University; 2019. [Google Scholar]
- 56. Xu W, Yu Y, Zhou Qet al. Expression pattern, genomic structure, and promoter analysis of the gene encoding stilbene synthase from Chinese wild Vitis pseudoreticulata. J Exp Bot. 2011;62:2745–61. [DOI] [PubMed] [Google Scholar]
- 57. Eulgem T, Rushton PJ, Robatzek Set al. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. [DOI] [PubMed] [Google Scholar]
- 58. Mao G, Meng X, Liu Yet al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23:1639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jiao Y, Wang D, Wang Let al. VqMAPKKK38 is essential for stilbene accumulation in grapevine. Hortic Res. 2017;4:17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brulé D, Villano C, Davies LJet al. The grapevine (Vitis vinifera) LysM receptor kinases VvLYK1-1 and VvLYK1-2 mediate chitooligosaccharide-triggered immunity. Plant Biotechnol J. 2019;17:812–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Genot B, Lang J, Berriri Set al. Constitutively active Arabidopsis MAP kinase 3 triggers defense responses involving salicylic acid and SUMM2 resistance protein. Plant Physiol. 2017;174:1238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meyer D, Pajonk S, Micali Cet al. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009;57:986–99. [DOI] [PubMed] [Google Scholar]
- 63. Rupprich N, Hildebrand H, Kindl H. Substrate specificity in vivo and in vitro in the formation of stilbenes. Biosynthesis of rhaponticin. Arch Biochem Biophys. 1980;200:72–8. [DOI] [PubMed] [Google Scholar]
- 64. Jeandet P, Clément C, Cordelier S. Regulation of resveratrol biosynthesis in grapevine: new approaches for disease resistance? J Exp Bot. 2019;70:375–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou Q, Dai L, Cheng Set al. A circulatory system useful both for long-term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant Cell Tissue Organ Cult. 2014;118:157–68. [Google Scholar]
- 66. Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–20. [DOI] [PubMed] [Google Scholar]
- 67. Yuan M, Ngou BPM, Ding Pet al. PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol. 2021;62:102030. [DOI] [PubMed] [Google Scholar]
- 68. Ngou BPM, Ding P, Jones JDG. Thirty years of resistance: zig-zag through the plant immune system. Plant Cell. 2022;34:1447–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nürnberger T, Brunner F, Kemmerling Bet al. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–66. [DOI] [PubMed] [Google Scholar]
- 70. Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–9. [DOI] [PubMed] [Google Scholar]
- 71. Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant. 2015;8:521–39. [DOI] [PubMed] [Google Scholar]
- 72. Zhang Y, Li X. Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol. 2019;50:29–36. [DOI] [PubMed] [Google Scholar]
- 73. Jiao Y, Xu W, Duan Det al. A stilbene synthase allele from a Chinese wild grapevine confers resistance to powdery mildew by recruiting salicylic acid signalling for efficient defence. J Exp Bot. 2016;67:5841–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Blümke A, Falter C, Herrfurth Cet al. Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol. 2014;165:346–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Luna E, Pastor V, Robert Jet al. Callose deposition: a multifaceted plant defense response. Mol Plant-Microbe Interact. 2011;24:183–93. [DOI] [PubMed] [Google Scholar]
- 76. Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004;23:3072–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li F, Chen X, Yang Ret al. Potato protein tyrosine phosphatase StPTP1a is activated by StMKK1 to negatively regulate plant immunity. Plant Biotechnol J. 2023;21:646–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dubrovina AS, Kiselev KV. Regulation of stilbene biosynthesis in plants. Planta. 2017;246:597–623. [DOI] [PubMed] [Google Scholar]
- 79. Chang X, Heene E, Qiao Fet al. The phytoalexin resveratrol regulates the initiation of hypersensitive cell death in Vitis cell. PLoS One. 2011;6:e26405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang L, Wang Y. Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Rep. 2019;38:1347–60. [DOI] [PubMed] [Google Scholar]
- 81. Wong DCJ, Matus JT. Constructing integrated networks for identifying new secondary metabolic pathway regulators in grapevine: recent applications and future opportunities. Front Plant Sci. 2017;8:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu J, Chen X, Liang Xet al. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016;171:1427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhou J, Wang X, He Yet al. Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell. 2020;32:2621–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang X, Guo R, Tu Met al. Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen Botrytis cinerea. Front Plant Sci. 2017;8:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shan D, Wang C, Zheng Xet al. MKK4-MPK3-WRKY17-mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 2021;186:1202–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zheng Z, Qamar SA, Chen Zet al. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. [DOI] [PubMed] [Google Scholar]
- 87. Qiu D, Xiao J, Ding Xet al. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant-Microbe Interact. 2007;20:492–9. [DOI] [PubMed] [Google Scholar]
- 88. Zhang K, Liu F, Wang Zet al. Transcription factor WRKY28 curbs WRKY33-mediated resistance to Sclerotinia sclerotiorum in Brassica napus. Plant Physiol. 2022;190:2757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu X, Zhou X, Li Det al. Rose WRKY13 promotes disease protection to Botrytis by enhancing cytokinin content and reducing abscisic acid signaling. Plant Physiol. 2023;191:679–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li H, Xu Y, Xiao Yet al. Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta. 2010;232:1325–37. [DOI] [PubMed] [Google Scholar]
- 91. Wang Y, Wang X, Fang Jet al. VqWRKY56 interacts with VqbZIPC22 in grapevine to promote proanthocyanidin biosynthesis and increase resistance to powdery mildew. New Phytol. 2023;237:1856–75. [DOI] [PubMed] [Google Scholar]
- 92. Yu Y, Xu W, Wang Jet al. The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 2013;200:834–46. [DOI] [PubMed] [Google Scholar]
- 93. Lai Z, Li Y, Wang Fet al. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell. 2011;23:3824–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tao H, Miao H, Chen Let al. WRKY33-mediated indolic glucosinolate metabolic pathway confers resistance against Alternaria brassicicola in Arabidopsis and Brassica crops. J Integr Plant Biol. 2022;64:1007–19. [DOI] [PubMed] [Google Scholar]
- 95. Merz PR, Moser T, Höll Jet al. The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol Plant. 2015;153:365–80. [DOI] [PubMed] [Google Scholar]
- 96. Galletti R, Ferrari S, De Lorenzo G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011;157:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shi H, Li Q, Luo Met al. BRASSINOSTEROID-SIGNALING KINASE1 modulates MAP KINASE15 phosphorylation to confer powdery mildew resistance in Arabidopsis. Plant Cell. 2022;34:1768–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhao C, Nie H, Shen Qet al. EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 2014;10:e1004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pitzschke A, Djamei A, Bitton Fet al. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol Plant. 2009;2:120–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bethke G, Unthan T, Uhrig JFet al. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA. 2009;106:8067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Berriri S, Garcia AV, Frey dit Frey Net al. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell. 2012;24:4281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Qiu JL, Zhou L, Yun BWet al. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008;148:212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gao M, Liu J, Bi Det al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–8. [DOI] [PubMed] [Google Scholar]
- 104. Asai T, Tena G, Plotnikova Jet al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–83. [DOI] [PubMed] [Google Scholar]
- 105. Lei L, Li Y, Wang Qet al. Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol. 2014;203:1146–60. [DOI] [PubMed] [Google Scholar]
- 106. Zou M, Guo M, Zhou Zet al. MPK3- and MPK6-mediated VLN3 phosphorylation regulates actin dynamics during stomatal immunity in Arabidopsis. Nat Commun. 2021;12:6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Xu J, Meng J, Meng Xet al. Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell. 2016;28:1144–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Adachi H, Nakano T, Miyagawa Net al. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell. 2015;27:2645–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–66. [DOI] [PubMed] [Google Scholar]
- 110. Bethke G, Pecher P, Eschen-Lippold Let al. Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol Plant-Microbe Interact. 2012;25:471–80. [DOI] [PubMed] [Google Scholar]
- 111. Çakır B, Kılıçkaya O. Mitogen-activated protein kinase cascades in Vitis vinifera. Front Plant Sci. 2015;6:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kishi-Kaboshi M, Okada K, Kurimoto Let al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010;63:599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ishihama N, Yamada R, Yoshioka Met al. Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell. 2011;23:1153–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Micali CO, Neumann U, Grunewald Det al. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol. 2011;13:210–26. [DOI] [PubMed] [Google Scholar]
- 115. Heath MC, Heath IB. Ultrastructure of an immune and a susceptible reaction of cowpea leaves to rust infection. Physiol Plant Pathol. 1971;1:277–87. [Google Scholar]
- 116. Ortmannová J, Sekereš J, Kulich Iet al. Arabidopsis EXO70B2 exocyst subunit contributes to papillae and encasement formation in antifungal defence. J Exp Bot. 2022;73:742–55. [DOI] [PubMed] [Google Scholar]
- 117. Liao W, Nielsen ME, Pedersen Cet al. Barley endosomal MONENSIN SENSITIVITY1 is a target of the powdery mildew effector CSEP0162 and plays a role in plant immunity. J Exp Bot. 2023;74:118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Rubiato HM, Liu M, O'Connell RJet al. Plant SYP12 syntaxins mediate an evolutionarily conserved general immunity to filamentous pathogens. eLife. 2022;11:e73487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Pan Q-H, Wang L, Li J-M. Amounts and subcellular localization of stilbene synthase in response of grape berries to UV irradiation. Plant Sci. 2009;176:360–6. [Google Scholar]
- 120. Wang W, Tang K, Yang HRet al. Distribution of resveratrol and stilbene synthase in young grape plants (Vitis vinifera L. cv. Cabernet Sauvignon) and the effect of UV-C on its accumulation. Plant Physiol Biochem. 2010;48:142–52. [DOI] [PubMed] [Google Scholar]
- 121. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 122. Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–36. [DOI] [PubMed] [Google Scholar]
- 123. Xie X, Agüero CB, Wang Yet al. Genetic transformation of grape varieties and rootstocks via organogenesis. Plant Cell Tissue Organ Cult. 2016;126:541–52. [Google Scholar]
- 124. Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72. [DOI] [PubMed] [Google Scholar]
- 125. Frye CA, Innes RW. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell. 1998;10:947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xu W, Yu Y, Ding Jet al. Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta. 2010;231:475–87. [DOI] [PubMed] [Google Scholar]
- 127. Zhou Q, Du Y, Cheng Set al. Resveratrol derivatives in four tissues of six wild Chinese grapevine species. N Z J Crop Hortic Sci. 2015;43:204–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data can be found within the manuscript and its supporting information.