Abstract
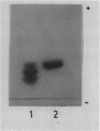
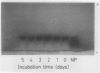
One cell strain with stable tolerance to allyl alcohol (AAr) was selected from 6 × 108 suspension cultured Nicotiana plumbaginifolia Viviani cells. The selected strain contained one-half the alcohol dehydrogenase (ADH) activity of the wild type (NP) due to the loss of two of three bands of ADH activity seen on starch gels following electrophoresis of wild-type cell extracts. Anaerobic conditions, simulated by not shaking the suspension cultures, increased the ADH specific activity to more than 3-fold the initial level in both strains but did not change the number of activity bands or the relative levels of activity. The cell strain with decreased ADH activity lost viability more rapidly than the wild type under the anaerobic conditions. The AAr cells were 10 times more tolerant to ethanol than the NP cells and were also somewhat more tolerant to acetaldehyde and antimycin A. The substrate specificities of the ADH enzymes from both strains were very similar. Further selection of AAr cells with allyl alcohol produced strains with even lower ADH activity and selection under anaerobic conditions produced strains with increased ADH activity. Genetic studies indicate that the N. plumbaginifolia ADH activity bands arise from subunits produced by two nonallelic genes. This is the first example of the use of allyl alcohol to select for decreased ADH using cultured plant cells.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Dolferus R., Jacobs M. Polymorphism of alcohol dehydrogenase in Arabidopsis thaliana (L.) Heynh.: genetical and biochemical characterization. Biochem Genet. 1984 Oct;22(9-10):817–838. doi: 10.1007/BF00499475. [DOI] [PubMed] [Google Scholar]
- Efron Y., Schwartz D. In vivo inactivation of maize alcohol dehydrogenase by a two-factor system. Proc Natl Acad Sci U S A. 1968 Oct;61(2):586–591. doi: 10.1073/pnas.61.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Bennett D. C. Maize Adh1. Annu Rev Genet. 1985;19:297–323. doi: 10.1146/annurev.ge.19.120185.001501. [DOI] [PubMed] [Google Scholar]
- Megnet R. Mutants partially deficient in alcohol dehydrogenase in Schizosaccharomyces pombe. Arch Biochem Biophys. 1967 Jul;121(1):194–201. doi: 10.1016/0003-9861(67)90024-0. [DOI] [PubMed] [Google Scholar]
- Roberts J. K., Callis J., Jardetzky O., Walbot V., Freeling M. Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6029–6033. doi: 10.1073/pnas.81.19.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M. M., Freeling M., Okimoto R. The anaerobic proteins of maize. Cell. 1980 Jul;20(3):761–767. doi: 10.1016/0092-8674(80)90322-0. [DOI] [PubMed] [Google Scholar]
- Schwartz D. Alcohol dehydrogenase in maize: genetic basis for multiple isozymes. Science. 1969 May 2;164(3879):585–586. doi: 10.1126/science.164.3879.585. [DOI] [PubMed] [Google Scholar]
- Schwartz D., Osterman J. A pollen selection system for alcohol-dehydrogenase-negative mutants in plants. Genetics. 1976 May;83(1):63–65. doi: 10.1093/genetics/83.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion J. P., Talbot B. Alcohol dehydrogenase mutants of Chinese hamster somatic cells resistant to allyl alcohol. Genetics. 1978 Feb;88(2):343–356. doi: 10.1093/genetics/88.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widholm J. M. The use of fluorescein diacetate and phenosafranine for determining viability of cultured plant cells. Stain Technol. 1972 Jul;47(4):189–194. doi: 10.3109/10520297209116483. [DOI] [PubMed] [Google Scholar]