Abstract
The clinical role of liquid biopsy in oncology is growing significantly. In gliomas and other brain tumours, targeted sequencing of cell-free DNA (cfDNA) from CSF may help differential diagnosis when surgery is not recommended and be more representative of tumour heterogeneity than surgical specimens, unveiling targetable genetic alterations. Given the invasive nature of lumbar puncture to obtain CSF, the quantitative analysis of cfDNA in plasma is a lively option for patient follow-up. Confounding factors may be represented by cfDNA variations due to concomitant pathologies (inflammatory diseases, seizures) or clonal haematopoiesis.
Pilot studies suggest that methylome analysis of cfDNA from plasma and temporary opening of the blood–brain barrier by ultrasound have the potential to overcome some of these limitations. Together with this, an increased understanding of mechanisms modulating the shedding of cfDNA by the tumour may help to decrypt the meaning of cfDNA kinetics in blood or CSF.
Keywords: liquid biopsy, brain tumours, cell-free DNA, methylome, targeted sequencing
Berzero et al. review the benefits and limitations of using cell-free DNA from plasma or cerebrospinal fluid in the differential diagnosis and follow-up of patients with brain tumours.
The challenges of liquid biopsy in neuro-oncology
In the late 1970s, evidence was found that cancer patients have higher levels of circulating cell-free DNA (cfDNA) in plasma than healthy subjects.1 It was later demonstrated that part of this cfDNA is tumour-derived2 and could be used for the non-invasive detection of cancer mutations,3,4 resulting in the term ‘liquid biopsy’ to refer to the non-invasive assessment of tumour genetic profiles using tumour-derived nucleic acids.
In the past decade, liquid biopsies have been increasingly used for response assessment, early detection of treatment resistance, prognostic prediction and treatment decisions, complementing or even replacing tissue biopsies in precision oncology.5 This is confirmed by the validation of commercial assays of liquid biopsy for clinical use.6,7 In colon cancer, tumour cfDNA-guided approaches reduced adjuvant chemotherapy use without compromising recurrence-free survival.8 In a prospective study of 1127 patients with non-small cell lung cancer, the detection of tumour cfDNA predicted shorter survival, therapeutic targets identified by sequencing of the cfDNA provided survival advantage, and mutations specific to cfDNA were discovered in a quarter of cases, representing subclonal resistance drivers.9
As a result of anatomical barriers (e.g. the blood–brain barrier) that are only partially disrupted by tumours, plasma-based liquid biopsy poses a greater challenge in neuro-oncology10 (Fig. 1). Among patients with gliomas, <10% had plasma-derived tumour-derived cfDNA, whereas 74–100% had CSF-derived tumour-derived cfDNA.11 The shedding of cfDNA into the CSF was especially informative in patients with high tumour burden, progressive tumours, or tumours adjacent to ventricles.12,13 CSF-cfDNA was more informative than plasma-cfDNA for targeted sequencing and mutation detection, identifying at least one tumour-derived genetic alteration in the majority of patients with gliomas or brain metastases.12–19 The lumbar puncture, however, is an invasive procedure and is not performed in the presence of intracranial hypertension. External ventricular drainage can be a source of CSF when ventriculostomy is required in brain tumour patients. Its use as a source of CSF, however, is hampered by the possibility of obstructions and infections that may increase the amount of cfDNA simulating tumour progression.20,21
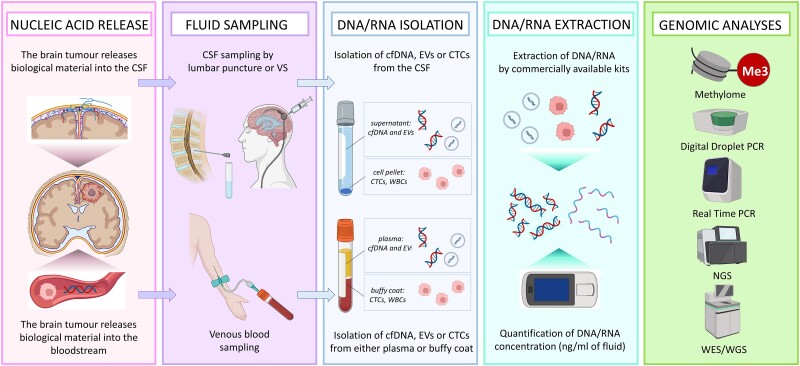
Figure 1.
The workflow of plasma-based and CSF-based liquid biopsy in neuro-oncology. Tumour-derived material (cfDNA, EVs, CTCs) is released in both CSF and plasma. Samples of both fluids can be obtained in the clinic by lumbar puncture or ventricular shunts (CSF) or venous blood sampling (plasma). Fresh biological fluids should be centrifuged at different speeds according to different protocols, depending on the material, in order to separate the supernatant from the pellet and isolate the material of interest. Nucleic acids can be extracted from either EVs or CTCs and then quantified by fluorometric assay or qPCR. Once good-quality material has been obtained, a plethora of genetic analyses can be performed, from whole-genome sequencing to methylomics. cfDNA = cell-free DNA; CTCs = circulating tumour cells; EVs = extracellular vesicles; NGS = next-generation sequencing; VS = ventricular shunt; WBCs = white blood cells; WES = whole exome sequencing; WGS = whole genome sequencing. Created with Biorender.com.
Thus, in the longitudinal sampling of patients in neuro-oncology, plasma is a preferable source of cfDNA (Fig. 2), as exemplified by patient follow-up with highly sensitive PCR techniques like digital PCR to detect TERT mutations, present in >60% of gliomas, with 90% specificity and 62.5% sensitivity.22 Plasma was also the source to set-up a sensitive, immunoprecipitation-based protocol to analyse the methylome of small amounts of circulating cfDNA, enabling detection of large-scale DNA methylation changes enriched for tumour-specific patterns.23 This approach was successfully expanded to tumours in the brain, as we will discuss later.24
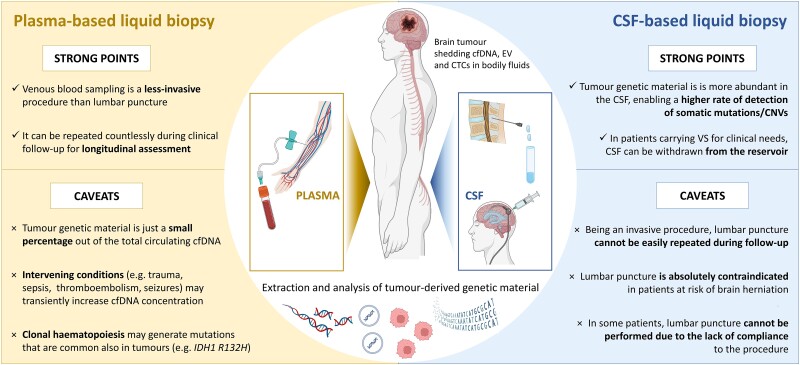
Figure 2.
Strong points and caveats of plasma-based (left) and CSF-based (right) liquid biopsy of cfDNA. Both plasma and CSF can be precious sources of genetic material shed by brain tumours in bodily fluids. On one hand, venous blood sampling is less invasive than CSF withdrawal, and can be easily repeated during clinical follow-up, and namely during routine outpatient visits. On the other hand, CSF generally contains larger amounts of tumour-derived genetic material and lesser amounts of genetic material released from normal cells, resulting in higher rates of mutation detection. Moreover, the total amount of cfDNA in plasma may also transiently increase due to intervening conditions (e.g. venous thromboembolism,90 inflammation,91 sepsis,91,92 focal epilepsy94), which should be accounted for during result interpretation. cfDNA = cell-free DNA; CNVs = copy number variations; CTCs = circulating tumour cells; EVs = extracellular vesicles; VS = ventricular shunt. Created with Biorender.com.
Detecting tumour cell-free DNA
Observations of cfDNA fragment size distributions identified peaks corresponding to DNA associated with nucleosomes (∼147 base pairs), suggesting that cfDNA is protected from nuclease digestion through associations with nucleosome core particles. As these associations are tissue-specific, comprehensive analysis of plasma-derived cfDNA allowed predicting nucleosome spacing and transcription factor footprints useful to track their tissue of origin, which in healthy individuals was lymphoid and myeloid cells.25
Cancer patients have higher levels of cfDNA in plasma compared to healthy subjects because of the high turnover of tumour cells.26 Necrotic and apoptotic cells shed DNA fragments in extracellular fluids including plasma and CSF, again reflecting the genetic and epigenetic features of the cell of origin.27 Increasing sequencing depth and using corrective algorithms allow to partially compensate for the presence of limited amounts of tumour cfDNA.28
Paediatric brain cancers
Liquid biopsy has made considerable progress in the follow-up of paediatric patients. In medulloblastomas, low-coverage whole-genome sequencing (WGS) of CSF-derived cfDNA used to detect minimal residual disease (MRD), unveiled tumour mutations at baseline in 83% of cases with metastatic spreading of the disease.29 During therapy, persistence of MRD in cfDNA was associated with increased risk of progression. A similar fraction of altered cfDNA in the CSF was found by targeted sequencing (MSK-IMPACT) in a cohort of paediatric patients with medulloblastoma and other brain tumours.30 However, in another cohort including 258 patients with different diagnosis (mostly high-grade gliomas) using ultra-low pass WGS copy number enabled the detection of molecular alterations in 9/46 CSF, 3/230 plasma and 0/153 urine samples.31
Diffuse midline gliomas, more frequent in childhood, are characterized by the H3K27M mutation of the H3 histone gene that has been analysed by digital PCR in cfDNA from CSF and plasma of patients enrolled in the NCT03416530 trial.32 Decrease of the mutant allelic frequencies (MAF) in the CSF was associated with prolonged progression-free survival (PFS) (P < 0.004), while 25% increase or more of MAF was predictive of progression in half of the patients, both in plasma and CSF.32
Extracellular vesicles
Machine-learning classification of plasma-derived extracellular vesicles (EVs) cargo, including immunoglobulins, revealed 95% sensitivity and 90% specificity in detecting different cancers not originating in the brain.33 Initial evidence supports a role for extracellular vesicles in the diagnosis and clinical follow-up of gliomas.34,35 Meningiomas release sizable amounts of extracellular vesicles and their amount decreases after surgery.36In vitro extracellular vesicle DNA from meningioma cultures reflect the genetic, epigenetic and mutational landscape of the tumour of origin.36
Extracellular vesicles from glioblastoma (GBM) patients can help predict prognosis,37–42 mirror modifications of tumour volume before and after surgery,43 and express specific GBM markers.44 They contained EGFRvIII45,46 and, intriguingly, amounts of the von Willebrand factor significantly higher than in healthy controls.47
Extracellular vesicles may play a role in conditioning the GBM immune milieu. Exosomes, one type of extracellular vesicles, can transport TGF-β, one major immune suppressive cytokine produced by GBM.48 In a cohort of patients receiving a dendritic cell vaccination, IL-8 (but also TGF-β) mRNA in plasma exosomes, positively correlated with immunological response to glioma antigens.49 On the other hand, exosomes in the CSF of GBM patients contain the LGALS9 ligand that inhibits antigen processing and presentation by dendritic cells by binding their TIM3 receptor.50 Recent data suggest extracellular vesicle-mediated interactions of GBM and natural killer cells showing that circulating immune vesicles ‘carry unique tumour-specific signals’.51
Clinical studies reporting on extracellular vesicles in liquid biopsy are summarized in Table 1. Overall evidence for a prominent role of EVs for diagnosis or clinical follow-up of brain tumours in the peripheral blood or the CSF requires more data. For instance, qalectin-3 binding protein in blood plasma and plasma-derived extracellular vesicles was at significantly higher levels in GBM patients than in healthy controls, but detection accuracy in predicting patient mortality was considerably higher in plasma than plasma extracellular vesicles (75% versus 45%, respectively).52
Table 1.
A selection of studies on liquid biopsy, listed based on order of publication, suggesting the potential use of extracellular vesicles for prognostic stratification and tumour monitoring in patients with primary and secondary CNS tumours
| Reference | n | Population | Tumour type | Biological fluid | Biomarker | Methods | Clinical application | Main findings of the study |
|---|---|---|---|---|---|---|---|---|
| Koch et al.37 | 11 | Adults | GBM | Plasma | MV count | Flow cytometry, Electron microscopy |
Response assessment | MV count helped to distinguish between tumour progression and pseudoprogression |
| Shi et al.38 | 70 | n.r. | Grade 2 astrocytoma, Grade 2 ependymoma, GBM |
Plasma CSF |
miR-21 | TEM, WB, RT-PCR |
Prognosis | miR-21 levels can predict PFS and OS |
| Evans et al.39 | 16 | Adults | GBM | Plasma | MV count | Flow cytometry, Cryoelectron microscopy |
Prognosis | MV count can predict PFS and OS |
| Manda et al.46 | 96 | Adults | HGG | Serum | EGFRvIII | BCA protein assay, TEM, flow cytometry, RT-PCR | Prognosis | EGFRvIII expression in EVs correlates with OS |
| Indira Chandran et al.44 | 82 | Adults | HGG and LGG | Plasma | SDC1 | NTA, TEM, ProSeek multiplex proximity extension assay, ELISA | Prognosis | SDC1 expression in EVs correlates with OS |
| Osti et al.43 | 68 | Adults | GBM and other CNS tumours | Plasma | EVs count and size, proteomic profile | NTA, TEM, mass spectrometry | Response assessment | EVs can assist in GBM diagnosis and monitoring |
| Zhong et al.40 | 147 | Adults | HGG | Serum | miR-29b | RT-PCR | Prognosis | miR-29b levels correlate with OS |
| Batool et al.35 | 40 | Adults | HGG | Plasma | EGFRvIII | ddPCR | Response assessment | EGFRvIII mutant copies assisted in response assessment |
| Khristov et al.41 | 82 | Adults | GBM | Plasma | IL13Ra2 | ELISA | Prognosis | IL13Ra2 levels in plasma correlate with OS |
| Dobra et al.42 | 222 | Adults | GBM, meningiomas, brain metastases |
Serum | MMP-9 | NTA, TEM, WB, LC-MS, ELISA | Prognosis | MMP-9 levels correlate with OS |
ddPCR = digital droplet PCR; ELISA = enzyme-linked immunosorbent assay; EVs = extracellular vesicles; GBM = glioblastoma; HGG = high-grade gliomas; LC-MS = liquid chromatography–mass spectrometry; LGG = low grade gliomas; MV = microvesicles; n = number of patients included; n.r. = information not reported; NTA = nanoparticle tracking analysis; OS = overall survival; PFS = progression-free survival; RT-PCR = reverse transcription PCR; TEM = transmission electron microscopy; WB = western blot.
Circulating tumour cells
The presence of circulating tumour cells (CTCs) in peripheral blood originating in primary brain tumours has been described years ago but their scarcity has limited their clinical use.53–56 On the contrary, selection of CTCs in CSF can play a relevant role in disease staging and follow-up in patients affected by medulloblastoma57 and leptomeningeal metastases (LM).58 In LM, CSF-CTCs were detected in 43/95 samples and one CSF-CTC/ml provided a diagnostic threshold with high sensitivity and specificity.58 A CTCs assay based on epithelial cell adhesion molecule (EpCAM) immunoflow cytometry in CSF of patients with suspected LM showed higher sensitivity than cytology (94% versus 76%).59 In LM patients treated by proton cranio-spinal irradiation, the presence of <53 CTCs in 3 ml of CSF was associated with longer PFS (12 versus 6 months; P < 0.01).60 The cfDNA, however, showed higher mutation rates (43.6% versus 19.8%) and MAF (41.1% versus 13.0%; P = 0.0001) than CTCs.61
Tracking glioma spatial heterogeneity and branched evolution
Gliomas and GBM are heterogeneous tumours composed of several clonal cell populations with different genetic and transcriptional profiles.62 Because of this, biopsy samples obtained from a single area may not represent the entire tumour while liquid biopsies might provide a more comprehensive representation of tumour molecular profiles. In 1 of 13 gliomas studied by shallow WGS, molecular alterations were detected in CSF cfDNA and not in one corresponding surgical specimen.63 In 17 gliomas, 34% of the mutations found by targeted sequencing were detected in the CSF only.64
Under the selective pressure of alkylating chemotherapy, GBM cells are subject to hypermutation and branched clonal evolution.65 Since only a minor proportion of GBM patients undergo second surgery at recurrence,66 liquid biopsies offer the chance to reassess tumour molecular profile and to monitor tumour evolution during treatment with MAF in plasma and CSF, as suggested by preliminary evidence in brainstem paediatric gliomas67–69 (Table 2). Along the same line, longitudinal liquid biopsies of plasma might support neuroimaging in response assessment (e.g. pseudoprogression versus true progression), a clinically relevant point raised by the liquid biopsy task-force of the RANO (Response Assessment in Neuro-Oncology) consortium.70
Table 2.
A selection of relevant studies, listed in order of publication, suggesting a potential application of cfDNA-based liquid biopsies for purposes of response assessment and prognostic stratification in patients with primary and secondary CNS tumours
| Reference | n | Population | Tumour type | Biological fluid | Biomarker | Sequencing method | Clinical application | Main findings of the study |
|---|---|---|---|---|---|---|---|---|
| De Mattos-Arruda et al.14 | 12 | Adults | Primary and secondary CNS tumours | CSF | Mutations and CNVs | NGS ddPCR |
Response assessment | MAF of CSF tcfDNA decreased after surgery and increased at progression |
| Panditharatna et al.12 | 48 | Children AYA |
Diffuse midline gliomas | Plasma | H3K27M mutations | ddPCR | Response assessment | MAF tracked tumour evolution and predicted relapse |
| Juratli et al.18 | 38 | Adults | Newly-diagnosed and recurrent GBM | CSF Plasma |
TERTp mutations | ddPCR | Prognosis | MAF are predictive of OS |
| Miller et al.15 | 85 | Adults | LGG and GBM | CSF | tcfDNA detection | NGS | Prognosis | The presence of tcfDNA was associated with shorter OS |
| Escudero et al.57 | 13 | Children Adolescents |
Medulloblastoma | CSF Plasma |
tcfDNA | WES ddPCR |
Response assessment | CSF tcfDNA allows the study of minimal residual disease |
| Muralidharan et al.22 | 157 | Adults | Lower grade gliomas and GBM | Plasma | TERTp mutations | ddPCR | Response assessment | MAF paralleled clinical and radiological evolution |
| Fontanilles et al.73 | 52 | Adults | Newly diagnosed GBM or gliosarcoma | Plasma | cfDNA concentration | – | Response assessment | Median cfDNA concentrations tracked tumour evolution |
| Bagley et al.72 | 62 | Adults | Newly diagnosed GBM | Plasma | cfDNA concentration | – | Prognosis | Preoperative cfDNA concentration correlated with PFS and OS |
| Liu et al.29 | 134 | Children | Newly diagnosed medulloblastoma | CSF | tcfDNA detection | NGS | Response assessment Prognosis | Patients with detectable tcfDNA during treatment had worse PFS |
| Li et al.78 | 92 | Adults | Newly diagnosed NSCLC with brain metastases | CSF Plasma |
tcfDNA detection | NGS | Response assessment Prognosis |
Patients with CSF tcfDNA at baseline had shorter OS |
| Cantor et al.32 | 28 | Children | H3K27M-mutant gliomas | CSF Plasma |
H3K27M mutations | ddPCR | Response assessment Prognosis | MAF variations correlate with PFS and predict targeted therapy response |
| Kojic et al.19 | 12 | Children Preadolecents |
Malignant primary brain tumours | CSF | tcfDNA | WES ddPCR |
Response assessment | ctDNA correlated with disease course and clinical outcomes |
AYA = adolescents and young adults; cfDNA = cell-free DNA; CNVs = copy number variations; ddPCR = digital droplet PCR; GBM = glioblastoma; LGG = lower grade gliomas; MAF = mutant allelic frequencies; n = number of patients included; NGS = next generation sequencing; NSCLC = non-small cell lung cancer; OS = overall survival; PFS = progression-free survival; tcfDNA = tumour-derived cell-free DNA; TERTp = TERT promoter; WES = whole exome sequencing.
Recently, evidence has emerged that liquid biopsies might also have a prognostic role, given that tumour cfDNA detection in the CSF is a strong independent predictor of reduced overall survival.15 In plasma, a first study in 42 GBM patients showed correlations between shorter PFS and cfDNA higher than 13.4 ng/ml.71 In a larger subsequent study on 62 GBM using a higher threshold (25. 2 ng/ml), higher values of cfDNA were also associated with shorter overall survival.72 This observation was challenged by data on another cohort of GBM patients, showing correlations between cfDNA levels and disease progression but not between cfDNA levels before surgery and survival73 (Table 2).
Identifying therapeutic targets
Although in glioma patients the number of actionable molecular alterations for targeted therapies is limited,74 the potential for their non-invasive identification is appealing. Studies of patient cohorts of primary brain tumours showed that actionable genetic alterations—including IDH1, BRAF, EGFR and NRAS mutations—could be identified from plasma cfDNA in 18–24% of patients.75,76
Liquid biopsies can also be considered for targeted therapy of brain metastases, which show divergence in terms of genetic profile77,78 and immune microenvironment79 with respect to the primary tumour. As an example, by identifying EGFR mutations as an intervening mechanism of resistance to EGFR inhibitors,80,81 liquid biopsies might provide a chance for a prompt switch in therapy.
The observation that MAF in the CSF decreases with response to treatment and increases at recurrence82,83 confirms further the potential of CSF-based liquid biopsies.
The ‘background noise’ in plasma-based liquid biopsy
The levels of cfDNA can change during the day and in the presence of stress.84,85
A confounding factor for interpreting the results of plasma liquid biopsies is represented by clonal haematopoiesis (CH), which may lead to the accumulation of non-tumoural somatic mutations in haematopoietic stem cells.86 If interpreted as tumour-associated genetic alterations in cfDNA analyses, they could steer toward inappropriate therapeutic decisions.86,87 CH mutations are quite common in the general population, associated with ageing (9.5–18.4% beyond 70 years old), prior radiotherapy or chemotherapy, and increased risk of developing haematological malignancies and cardiovascular diseases.87,88 They mainly include mutations in epigenetic modulators (DNMT3A, TET2 and ASXL), and in genes often altered in solid (KRAS, GNAS, NRAS and PIK3CA) and haematological tumours (JAK2, PPM1D, TP53, SF3B1, SRSF2, IDH1 and IDH2). Because of CH, the finding of IDH1 and IDH2 mutations in plasma-derived cfDNA of patients with gliomas may require investigation of their origin by sequencing the DNA from white blood cells (and tumour tissue, when available).86 The relevance of CH was demonstrated by the finding of mutations in peripheral blood but not in tumour tissue in 17 of 18 patients tested.89 Of note, CH is more frequent after chemoradiotherapy and is associated with shorter overall survival.
Other factors creating background noise in neuro-oncology include venous thromboembolism,90 inflammation,91 sepsis92 and trauma.92 In the plasma of septic patients, a positive correlation was present between the levels of myeloperoxidase, an enzyme released by activated neutrophils, and the amounts of cfDNA. NETosis, a specific form of cell death in which activated neutrophils release neutrophil extracellular traps (NETs) in response to inflammation, infection or hypoxia. Myeloperoxidase, released by activated neutrophils, was found significantly correlated to cfDNA levels in septic patients.92 NETosis can also be the underlying cause of the increased release of cfDNA in autoimmune diseases like lupus erythematosus and in rheumatoid arthritis.93
Of relevance in neuro-oncology is also the observation of increased cfDNA in the blood of patients with epileptic seizures. This was signalled in 2013: the difference in cfDNA concentrations between patients and controls was limited but reached statistical significance.94 The values, however, were considerably higher than expected (μg/ml rather than ng/ml), suggesting the presence of contaminating nuclear DNA. Confirmation in other patient cohorts would help to validate these observations.
Increasing the clinical translation of liquid biopsy in neuro-oncology
The large majority of tumour cfDNA does not contain tumour-defining mutations. Epigenetic profiles, on the other hand, are widely distributed in the genome95 and their evolution is modulated by the number of cell divisions as in ageing and cancer, where DNA methylation loss becomes more frequent.96,97 The potential of methylation analysis in brain tumour diagnostics is well illustrated by the development of a DNA methylation-based classification of CNS tumours founded on a machine-learning approach.98 The use of advanced methylation-based sequencing allowed the identification of specific methylation signatures and accurate classification of brain tumours from circulating cfDNA.24 A similar result was replicated by another group, who developed a machine learning algorithm to distinguish patients with or without glioma based on a cfDNA-derived methylation signature.99
In vitro, epigenetic profiling was also used to characterize the DNA cargo of glioma extracellular vesicles.100In vivo, extracellular vesicles may be released by neuronal-like subpopulation of glioma interconnecting with the surrounding normal astrocytes and neurons through microtubes, a key strategy favouring invasion.101 Extracellular vesicles are more abundant in GBM patients than in controls and their protein cargo may allude to GBM biology.43
Of note, blood platelets have the ability to take up secreted RNA-containing membrane vesicles derived from glioma cells.102 Recent data showed that tumour-educated platelet (TEP) RNA-based blood tests enable the detection of 18 cancer types. Specifically, the detection rate in GBM was 51%.103
As mentioned earlier, the blood–brain barrier limits the shedding of biomarkers, such as cfDNA, from brain tumours into the bloodstream, hampering their detection by conventional assays. Transcranial magnetic resonance-guided focused ultrasound (MRgFUS) can transiently open the blood–brain barrier, providing an opportunity for less invasive access to brain pathology. Meng et al.104 showed first-in-human proof-of-concept in nine GBM patients by finding that MRgFUS acutely enhances plasma cfDNA (2.6 ± 1.2-fold, P < 0.01, Wilcoxon signed-rank test).
Artificial intelligence
Artificial Intelligence (AI) and, specifically, machine learning shows considerable potential also for the elaboration of data gained by liquid biopsy.105 For instance, in patients with colorectal cancer (n = 72), a multimodal liquid biopsy resulting from a machine learning algorithm combining analysis of CTCs, exosomes and cfDNA outperformed each of the three biomarkers in predicting overall survival.106 Notably, the machine learning-generated classifier that uses whole genome methylation sequencing showed highest detection sensitivity out of 10 machine-learning classifiers trained on the same samples.107
An ‘upstream’ size selection (an example of cfDNA fragmentomics) can help enrich tumour cfDNA that are typically shorter than normal cfDNA fragments. By selecting in silico DNA fragments between 90 and 150 base pairs in length, Mouliere and colleagues108 achieved a more than 2-fold median enrichment of tumour cfDNA in ∼95% of cases, increasing the detection rate of clinically actionable mutations and copy number alterations in plasma cfDNA.
Furthermore, machine learning-guided extraction of MRI features of brain tumours, characterizing their texture,109 could complement quantitative data obtained by cfDNA improving the clinical follow-up and the rationale for therapeutic decisions.
Outstanding questions
In previous reports a negative association of baseline CSF cfDNA concentration with survival was independent of demographic or clinico-pathological data.15 A similar trend was reported with plasma cfDNA.72 This raises the intriguing possibility that cfDNA release is depending on intrinsic biological features of the tumour and not just on its size. Thus, to make progress in the interpretation of liquid biopsy data a deeper insight into mechanisms of cfDNA release into the CSF or the bloodstream is desirable.
The process of nucleosome eviction might be one relevant mechanism of cfDNA increase in the periphery worth further investigation. Different genes are responsible for nucleosome eviction and consequent chromatin reshaping that increases the availability of DNA stretches for interaction with transcription factors in promoter or enhancer regions of the genome. Bromodomain protein 4 (BRD4) is one such factor, facilitating nucleosome eviction by acetylating the critical residue for nucleosome stability H3 K122.110 Notably, BRD4, also implied in enhancing epistasis by maintaining 3D chromosomal interactions,111 is overexpressed in GBM and together with other BET bromodomain proteins is required for GBM proliferation.112
Another mechanism highly relevant to glioma biology is hypoxia.113,114 Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with nucleosome eviction.115 Nucleosome-free DNA regions (NFRs) can be established by nucleosome eviction in hypoxia-inducible promoters by hypoxia-inducible transcription factors and nucleosome re-assembly can take place hours after reoxygenation.116 NFRs are typical configurations of chromatin in active gene promoters, and each NFR in a promoter region encompasses 100–500 base pairs, corresponding to 1–2 nucleosomes.
Final remarks
The present status of liquid biopsy in neuro-oncology is summarized in Fig. 2. Analysis of CSF may provide information on differential diagnosis when surgery is not recommended or has not been informative. Plasma is more amenable to clinical follow-up, helping to decipher brain tumour modulation by tissue microenvironment and therapeutic challenges. Technical advances at three levels have the potential to improve the informative potential of plasma cfDNA: methylation profiling, temporary blood–brain barrier disruption, and increased knowledge of the biology of cfDNA shedding by the tumour.
Literature search
Data for this Update were identified by searches of PubMed, and references from relevant articles using the search terms ‘liquid biopsy’, ‘cell free DNA’, ‘glioma’, ‘brain metastases’, ‘liquid biopsy and CSF’, ‘liquid biopsy and plasma’. Abstracts and reports from meetings were not included. Only articles published in English between 1977 and March 2023 were included.
Acknowledgements
We thank Alleanza Contro il Cancro and the Fondazione Nadia Toffa for their support of the Liquid Biopsy Program at IRCCS Ospedale San Raffaele, Milano Italy.
Contributor Information
Giulia Berzero, Neurology Unit, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; Vita-Salute San Raffaele University, 20132 Milan, Italy.
Valentina Pieri, Neurology Unit, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy.
Pietro Mortini, Vita-Salute San Raffaele University, 20132 Milan, Italy; Department of Neurosurgery and Gamma Knife Radiosurgery, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy.
Massimo Filippi, Neurology Unit, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; Vita-Salute San Raffaele University, 20132 Milan, Italy; Neurorehabilitation Unit; Neurophysiology Unit; Neuroimaging Research Unit, Institute of Experimental Neurology, Division of Neuroscience, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy.
Gaetano Finocchiaro, Neurology Unit, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy.
Funding
No targeted funding was received towards this work.
Competing interests
The authors report no competing interests.
References
- 1. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 2. Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. [DOI] [PubMed] [Google Scholar]
- 3. Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86:774–779. [DOI] [PubMed] [Google Scholar]
- 4. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67–71. [PubMed] [Google Scholar]
- 5. Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020;1:276–290. [DOI] [PubMed] [Google Scholar]
- 6. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312. [DOI] [PubMed] [Google Scholar]
- 7. Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell. 2022;40:920–938. [DOI] [PubMed] [Google Scholar]
- 8. Tie J, Cohen JD, Lahouel K, et al. . Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med. 2022;386:2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jee J, Lebow ES, Yeh R, et al. . Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat Med. 2022;28:2353–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arvanitis CD, Ferraro GB, Jain RK. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat Rev Cancer. 2020;20:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224r.a24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Panditharatna E, Kilburn LB, Aboian MS, et al. . Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24:5850–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Springer S, Zhang M, et al. . Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Mattos-Arruda L, Mayor R, Ng CKY, et al. . Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller AM, Shah RH, Pentsova EI, et al. . Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019:565:654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan C, Diplas BH, Chen X, et al. . Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol. 2019;137:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pentsova EI, Shah RH, Tang J, et al. . Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34:2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Juratli TA, Stasik S, Zolal A, et al. . TERT Promoter mutation detection in cell-free tumor-derived DNA in patients with IDH wild-type glioblastomas: A pilot prospective study. Clin Cancer Res. 2018;24:5282–5291. [DOI] [PubMed] [Google Scholar]
- 19. Kojic M, Maybury MK, Waddell N, et al. . Efficient detection and monitoring of pediatric brain malignancies with liquid biopsy based on patient-specific somatic mutation screening. Neuro Oncol. Published online 9 February 2023. doi: 10.1093/neuonc/noad032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aten Q, Killeffer J, Seaver C, Reier L. Causes, complications, and costs associated with external ventricular drainage catheter obstruction. World Neurosurg. 2020;134:501–506. [DOI] [PubMed] [Google Scholar]
- 21. Li X, Yang L, Li D, et al. . Diagnosis of neurological infections in pediatric patients from cell-free DNA specimens by using metagenomic next-generation sequencing. Microbiol Spectr. 2023;11:e0253022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muralidharan K, Yekula A, Small JL, et al. . TERT Promoter mutation analysis for blood-based diagnosis and monitoring of gliomas. Clin Cancer Res. 2021;27:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen SY, Singhania R, Fehringer G, et al. . Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018:563:579–583. [DOI] [PubMed] [Google Scholar]
- 24. Nassiri F, Chakravarthy A, Feng S, et al. . Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat Med. 2020;26:1044–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1–2):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diaz LA, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wan JCM, Massie C, Garcia-Corbacho J, et al. . Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. [DOI] [PubMed] [Google Scholar]
- 28. Newman AM, Lovejoy AF, Klass DM, et al. . Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu APY, Smith KS, Kumar R, et al. . Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39:1519–1530.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller AM, Szalontay L, Bouvier N, et al. . Next-generation sequencing of cerebrospinal fluid for clinical molecular diagnostics in pediatric, adolescent and young adult brain tumor patients. Neuro Oncol. 2022;24:1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pagès M, Rotem D, Gydush G, et al. . Liquid biopsy detection of genomic alterations in pediatric brain tumors from cell-free DNA in peripheral blood, CSF, and urine. Neuro Oncol. 2022;24:1352–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cantor E, Wierzbicki K, Tarapore RS, et al. . Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro Oncol. 2022;24:1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoshino A, Kim HS, Bojmar L, et al. . Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Del Bene M, Osti D, Faletti S, Beznoussenko GV, DiMeco F, Pelicci G. Extracellular vesicles: The key for precision medicine in glioblastoma. Neuro Oncol. 2022;24:184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Batool SM, Hsia T, Khanna SK, et al. . Decoding vesicle-based precision oncology in gliomas. Neurooncol Adv. 2022;4(Suppl 2):ii53–ii60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ricklefs FL, Maire CL, Wollmann K, et al. . Diagnostic potential of extracellular vesicles in meningioma patients. Neuro Oncol. 2022;24:2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koch CJ, Lustig RA, Yang X-Y, et al. . Microvesicles as a biomarker for tumor progression versus treatment effect in radiation/temozolomide-treated glioblastoma patients. Transl Oncol. 2014;7:752–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi R, Wang P-Y, Li X-Y, et al. . Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6:26971–26981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans SM, Putt M, Yang X-Y, et al. . Initial evidence that blood-borne microvesicles are biomarkers for recurrence and survival in newly diagnosed glioblastoma patients. J Neurooncol. 2016;127:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhong F, Huang T, Leng J. Serum miR-29b as a novel biomarker for glioblastoma diagnosis and prognosis. Int J Clin Exp Pathol. 2019;12:4106–4112. [PMC free article] [PubMed] [Google Scholar]
- 41. Khristov V, Nesterova D, Trifoi M, et al. . Plasma IL13Rα2 as a novel liquid biopsy biomarker for glioblastoma. J Neurooncol. 2022;160(3):743–752. [DOI] [PubMed] [Google Scholar]
- 42. Dobra G, Gyukity-Sebestyén E, Bukva M, et al. . MMP-9 as prognostic marker for brain tumours: A comparative study on serum-derived small extracellular vesicles. Cancers (Basel). 2023;15:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osti D, Del Bene M, Rappa G, et al. . Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019;25:266–276. [DOI] [PubMed] [Google Scholar]
- 44. Indira Chandran V, Welinder C, Månsson AS, et al. . Ultrasensitive immunoprofiling of plasma extracellular vesicles identifies syndecan-1 as a potential tool for minimally invasive diagnosis of glioma. Clin Cancer Res. 2019;25:3115–3127. [DOI] [PubMed] [Google Scholar]
- 45. Figueroa JM, Skog J, Akers J, et al. . Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017;19:1494–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Manda SV, Kataria Y, Tatireddy BR, et al. . Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J Neurosurg. 2018;128:1091–1101. [DOI] [PubMed] [Google Scholar]
- 47. Sabbagh Q, André-Grégoire G, Alves-Nicolau C, et al. . The von Willebrand factor stamps plasmatic extracellular vesicles from glioblastoma patients. Sci Rep. 2021;11:22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Graner MW, Alzate O, Dechkovskaia AM, et al. . Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23:1541–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muller L, Muller-Haegele S, Mitsuhashi M, Gooding W, Okada H, Whiteside TL. Exosomes isolated from plasma of glioma patients enrolled in a vaccination trial reflect antitumor immune activity and might predict survival. Oncoimmunology. 2015;4:e1008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang M, Cai Y, Peng Y, Xu B, Hui W, Jiang Y. Exosomal LGALS9 in the cerebrospinal fluid of glioblastoma patients suppressed dendritic cell antigen presentation and cytotoxic T-cell immunity. Cell Death Dis. 2020;11:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ishwar D, Haldavnekar R, Das S, Tan B, Venkatakrishnan K. Glioblastoma associated natural killer cell EVs generating tumour-specific signatures: Noninvasive GBM liquid biopsy with self-functionalized quantum probes. ACS Nano. 2022;16:10859–10877. [DOI] [PubMed] [Google Scholar]
- 52. Rana R, Chauhan K, Gautam P, et al. . Plasma-derived extracellular vesicles reveal galectin-3 binding protein as potential biomarker for early detection of glioma. Front Oncol. 2021;11:778754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garzia L, Kijima N, Morrissy AS, et al. . A hematogenous route for medulloblastoma leptomeningeal metastases. Cell. 2018;172:1050–1062.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Müller C, Holtschmidt J, Auer M, et al. . Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med. 2014;6:247ra101. [DOI] [PubMed] [Google Scholar]
- 55. Sullivan JP, Nahed BV, Madden MW, et al. . Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macarthur KM, Kao GD, Chandrasekaran S, et al. . Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014;74:2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Escudero L, Llort A, Arias A, et al. . Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat Commun. 2020;11:5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin X, Fleisher M, Rosenblum M, et al. . Cerebrospinal fluid circulating tumor cells: A novel tool to diagnose leptomeningeal metastases from epithelial tumors. Neuro Oncol. 2017;19:1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Bussel MTJ, Pluim D, Milojkovic Kerklaan B, et al. . Circulating epithelial tumor cell analysis in CSF in patients with leptomeningeal metastases. Neurology. 2020;94:e521–e528. [DOI] [PubMed] [Google Scholar]
- 60. Wijetunga NA, Boire A, Young RJ, et al. . Quantitative cerebrospinal fluid circulating tumor cells are a potential biomarker of response for proton craniospinal irradiation for leptomeningeal metastasis. Neurooncol Adv. 2021;3:vdab181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bale TA, Yang S-R, Solomon JP, et al. . Clinical experience of cerebrospinal fluid-based liquid biopsy demonstrates superiority of cell-free DNA over cell pellet genomic DNA for molecular profiling. J Mol Diagn. 2021;23:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Patel AP, Tirosh I, Trombetta JJ, et al. . Single-cell RNA-Seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mouliere F, Mair R, Chandrananda D, et al. . Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med. 2018;10:e9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao Z, Zhang C, Li M, et al. . Applications of cerebrospinal fluid circulating tumor DNA in the diagnosis of gliomas. Jpn J Clin Oncol. 2020;50:325–332. [DOI] [PubMed] [Google Scholar]
- 65. Wang J, Cazzato E, Ladewig E, et al. . Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48:768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nava F, Tramacere I, Fittipaldo A, et al. . Survival effect of first- and second-line treatments for patients with primary glioblastoma: A cohort study from a prospective registry, 1997-2010. Neuro Oncol. 2014;16:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gojo J, Pavelka Z, Zapletalova D, et al. . Personalized treatment of H3K27M-mutant pediatric diffuse gliomas provides improved therapeutic opportunities. Front Oncol. 2019;9:1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mueller S, Jain P, Liang WS, et al. . A pilot precision medicine trial for children with diffuse intrinsic pontine glioma-PNOC003: A report from the Pacific pediatric neuro-oncology consortium. Int J Cancer. 2019;145:1889–1901. [DOI] [PubMed] [Google Scholar]
- 69. Izquierdo E, Proszek P, Pericoli G, et al. . Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neurooncol Adv. 2021;3:vdab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Soffietti R, Bettegowda C, Mellinghoff IK, et al. . Liquid biopsy in gliomas: A RANO review and proposals for clinical applications. Neuro Oncol. 2022;24:855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bagley SJ, Nabavizadeh SA, Mays JJ, et al. . Clinical utility of plasma cell-free DNA in adult patients with newly diagnosed glioblastoma: A pilot prospective study. Clin Cancer Res. 2020;26:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bagley SJ, Till J, Abdalla A, et al. . Association of plasma cell-free DNA with survival in patients with IDH wild-type glioblastoma. Neurooncol Adv. 2021;3:vdab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fontanilles M, Marguet F, Beaussire L, et al. . Cell-free DNA and circulating TERT promoter mutation for disease monitoring in newly-diagnosed glioblastoma. Acta Neuropathol Commun. 2020;8:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang K, Wu Z, Zhang H, et al. . Glioma targeted therapy: Insight into future of molecular approaches. Mol Cancer. 2022;21:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schwaederle M, Husain H, Fanta PT, et al. . Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. 2016;7:9707–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Piccioni DE, Achrol AS, Kiedrowski LA, et al. . Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol. 2019;8:CNS34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brastianos PK, Carter SL, Santagata S, et al. . Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li M, Chen J, Zhang B, et al. . Dynamic monitoring of cerebrospinal fluid circulating tumor DNA to identify unique genetic profiles of brain metastatic tumors and better predict intracranial tumor responses in non-small cell lung cancer patients with brain metastases: A prospective cohort study (GASTO 1028). BMC Med. 2022;20:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Song SG, Kim S, Koh J, et al. . Comparative analysis of the tumor immune-microenvironment of primary and brain metastases of non-small-cell lung cancer reveals organ-specific and EGFR mutation-dependent unique immune landscape. Cancer Immunol Immunother. 2021;70:2035–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ying S, Ke H, Ding Y, et al. . Unique genomic profiles obtained from cerebrospinal fluid cell-free DNA of non-small cell lung cancer patients with leptomeningeal metastases. Cancer Biol Ther. 2019;20:562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li YS, Jiang BY, Yang JJ, et al. . Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: A new medium of liquid biopsy. Ann Oncol. 2018;29:945–952. [DOI] [PubMed] [Google Scholar]
- 82. Li Y, Pan W, Connolly ID, et al. . Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol. 2016;128:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Song Y, Liu P, Huang Y, Guan Y, Han X, Shi Y. Osimertinib quantitative and gene variation analyses in cerebrospinal fluid and plasma of a non-small cell lung cancer patient with leptomeningeal metastases. Curr Cancer Drug Targets. 2019;19:666–673. [DOI] [PubMed] [Google Scholar]
- 84. Madsen AT, Hojbjerg JA, Sorensen BS, Winther-Larsen A. Day-to-day and within-day biological variation of cell-free DNA. EBioMedicine. 2019;49:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Herhaus B, Neuberger E, Juškevičiūtė E, Simon P, Petrowski K. Kinetics of plasma cell-free DNA under a highly standardized and controlled stress induction. Cells. 2023;12:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chan HT, Chin YM, Nakamura Y, Low SK. Clonal hematopoiesis in liquid biopsy: From biological noise to valuable clinical implications. Cancers (Basel). 2020;12:2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Coombs CC, Zehir A, Devlin SM, et al. . Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21:374–382.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366:eaan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Okamura R, Piccioni DE, Boichard A, et al. . High prevalence of clonal hematopoiesis-type genomic abnormalities in cell-free DNA in invasive gliomas after treatment. Int J Cancer. 2021;148:2839–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Martos L, Oto J, Fernández-Pardo Á, et al. . Increase of neutrophil activation markers in venous thrombosis-contribution of circulating activated protein C. Int J Mol Sci. 2020;21:5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zwirner K, Hilke FJ, Demidov G, et al. . Circulating cell-free DNA: A potential biomarker to differentiate inflammation and infection during radiochemotherapy. Radiother Oncol. 2018;129:575–581. [DOI] [PubMed] [Google Scholar]
- 92. Chornenki NL J, Coke R, Kwong AC, et al. . Comparison of the source and prognostic utility of cfDNA in trauma and sepsis. Intensive Care Med Exp. 2019;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Duvvuri B, Lood C. Cell-free DNA as a biomarker in autoimmune rheumatic diseases. Front Immunol. 2019;10:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liimatainen SP, Jylhävä J, Raitanen J, Peltola JT, Hurme MA. The concentration of cell-free DNA in focal epilepsy. Epilepsy Res. 2013;105:292–298. [DOI] [PubMed] [Google Scholar]
- 95. Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777–786. [DOI] [PubMed] [Google Scholar]
- 96. Klutstein M, Moss J, Kaplan T, Cedar H. Contribution of epigenetic mechanisms to variation in cancer risk among tissues. Proc Natl Acad Sci U S A. 2017;114:2230–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhou W, Dinh HQ, Ramjan Z, et al. . DNA Methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet. 2018;50:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Capper D, Jones DTW, Sill M, et al. . DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sabedot TS, Malta TM, Snyder J, et al. . A serum-based DNA methylation assay provides accurate detection of glioma. Neuro Oncol. 2021;23:1494–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Maire CL, Fuh MM, Kaulich K, et al. . Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol. 2021;23:1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Venkataramani V, Yang Y, Schubert MC, et al. . Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185:2899–2917.e31. [DOI] [PubMed] [Google Scholar]
- 102. Nilsson RJA, Balaj L, Hulleman E, et al. . Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. In ‘t Veld SGJG, Arkani M, Post E, et al. . Detection and localization of early- and late-stage cancers using platelet RNA. Cancer Cell. 2022;40:999–1009.e6. [DOI] [PubMed] [Google Scholar]
- 104. Meng Y, Pople CB, Suppiah S, et al. . MR-guided focused ultrasound liquid biopsy enriches circulating biomarkers in patients with brain tumors. Neuro Oncol. 2021;23:1789–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Swanson K, Wu E, Zhang A, Alizadeh AA, Zou J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell. 2023;186:1772–1791. [DOI] [PubMed] [Google Scholar]
- 106. Bu J, Lee TH, Poellmann MJ, et al. . Tri-modal liquid biopsy: Combinational analysis of circulating tumor cells, exosomes, and cell-free DNA using machine learning algorithm. Clin Transl Med. 2021;11:e499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jamshidi A, Liu MC, Klein EA, et al. . Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell. 2022;40:1537–1549.e12. [DOI] [PubMed] [Google Scholar]
- 108. Mouliere F, Chandrananda D, Piskorz AM, et al. . Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10:eaat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol. 2022;19:132–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Devaiah BN, Case-Borden C, Gegonne A, et al. . BRD4 Is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol. 2016;23:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lin X, Liu Y, Liu S, et al. . Nested epistasis enhancer networks for robust genome regulation. Science. 2022;377:1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pastori C, Daniel M, Penas C, et al. . BET Bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics. 2014;9:611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Colwell N, Larion M, Giles AJ, et al. . Hypoxia in the glioblastoma microenvironment: Shaping the phenotype of cancer stem-like cells. Neuro Oncol. 2017;19:887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bayat S, Mamivand A, Khoshnevisan A, et al. . Differential expression of hypoxia-related genes in primary brain tumors and correlation with clinicopathologic data. World Neurosurg. 2021;154:e465–e472. [DOI] [PubMed] [Google Scholar]
- 115. Fish JE, Yan MS, Matouk CC, et al. . Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J Biol Chem. 2010;285:810–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Suzuki N, Vojnovic N, Lee K-L, Yang H, Gradin K, Poellinger L. HIF-dependent and reversible nucleosome disassembly in hypoxia-inducible gene promoters. Exp Cell Res. 2018;366:181–191. [DOI] [PubMed] [Google Scholar]