Abstract
COQ7 encodes a hydroxylase responsible for the penultimate step of coenzyme Q10 (CoQ10) biosynthesis in mitochondria. CoQ10 is essential for multiple cellular functions, including mitochondrial oxidative phosphorylation, lipid metabolism, and reactive oxygen species homeostasis. Mutations in COQ7 have been previously associated with primary CoQ10 deficiency, a clinically heterogeneous multisystemic mitochondrial disorder. We identified COQ7 biallelic variants in nine families diagnosed with distal hereditary motor neuropathy with upper neuron involvement, expending the clinical phenotype associated with defects in this gene. A recurrent p.Met1? change was identified in five families from Brazil with evidence of a founder effect. Fibroblasts isolated from patients revealed a substantial depletion of COQ7 protein levels, indicating protein instability leading to loss of enzyme function. High-performance liquid chromatography assay showed that fibroblasts from patients had reduced levels of CoQ10, and abnormal accumulation of the biosynthetic precursor DMQ10. Accordingly, fibroblasts from patients displayed significantly decreased oxygen consumption rates in patients, suggesting mitochondrial respiration deficiency. Induced pluripotent stem cell-derived motor neurons from patient fibroblasts showed significantly increased levels of extracellular neurofilament light protein, indicating axonal degeneration. Our findings indicate a molecular pathway involving CoQ10 biosynthesis deficiency and mitochondrial dysfunction in patients with distal hereditary motor neuropathy. Further studies will be important to evaluate the potential benefits of CoQ10 supplementation in the clinical outcome of the disease.
Keywords: mitochondria, coQ10, charcot-Marie-Tooth disease, hereditary motor neuropathy, motor neuron
Rebelo et al. identify COQ7 biallelic variants in nine families diagnosed with distal hereditary motor neuropathy with upper motor neuron involvement, thereby expanding the clinical phenotype associated with defects in this gene.
See Desbats and Salviati (https://doi.org/10.1093/brain/awad302) for a scientific commentary on this article.
See Desbats and Salviati (https://doi.org/10.1093/brain/awad302) for a scientific commentary on this article.
Introduction
COQ7 encodes for a mitochondrial enzyme essential for the biosynthesis of coenzyme Q (CoQ, ubiquinone), a ubiquitous redox-lipid important for various cellular functions.1 In humans, CoQ10 is the predominant ubiquinone form and is present primarily in the mitochondria.2 CoQ10 is a main co-factor of the electron transport chain and essential for ATP production, where it shuttles electrons between mitochondrial respiratory complexes during respiration.1 It is also involved in lipid metabolism, and it functions as a powerful antioxidant.3-5 CoQ10 can be obtained from diet and food supplements; however, it is mainly produced endogenously in all cells of the body.2 The biosynthesis of CoQ10 requires over a dozen enzymes involving multiple steps. COQ7 is responsible for the penultimate step of the biosynthetic pathway, where it acts by hydroxylating the CoQ10 intermediate precursor, DMQ10.6 Mutations in genes encoding for several of these enzymes, including COQ7, cause primary CoQ10 deficiency, a mitochondrial disorder with a wide range of clinical phenotypes affecting multiple tissues.7 Mutations in COQ7 are very rare; and only seven isolated cases have been reported worldwide.8-13 The clinical manifestation of patients with COQ7 mutations is associated with a heterogeneous spectrum ranging from a multisystemic lethal phenotype9 to a less severe phenotype consisting of spasticity and hearing loss.8 CoQ10 supplementation has been suggested as a potential treatment for patients with CoQ10 deficiency; however, the clinical outcomes are variable. Treatment with CoQ10 supplements has shown promising results in stalling disease progression and in achieving pain reduction in a patient harbouring a COQ7 variant.10 In contrast, CoQ10 treatment has produced no apparent improvement in a patient harbouring a COQ7 variant, who was reported to have hereditary spastic paraplegia (HSP) and hearing loss.8 Likewise, no response to CoQ10 treatment was observed in a severe case of COQ7 mutation associated with progressive and fatal encephalo-myo-nephro-cardiopathy.9
Germline deletion of Mclk1 (mouse homologue of COQ7) in mice results in embryonic lethality14; therefore, an inducible adult knockout model was generated to bypass lethality. Similar to humans, the mice developed severe progressive mitochondrial disease phenotypes, growth retardation, and have a significantly shorter lifespan.14 Treatment with oral CoQ10 did not alleviate the phenotypes, likely because uptake was insufficient to elevate CoQ10 in most tissues except the liver. However, the mouse phenotype could be rescued with 2,4-DHB treatment, an unnatural precursor that can bypass COQ7 during biosynthesis of CoQ10, indicating that the phenotypes can be rescued when CoQ levels are restored.14
Here we report 11 patients from nine families diagnosed primarily with distal hereditary motor neuropathy (dHMN) with upper motor neuron involvement, expanding the clinical phenotype of patients with COQ7 variants. This is the largest cohort of patients with COQ7 mutations with detailed clinical descriptions and diverse ethnicity, including the first indigenous patient identified in the Amazon rainforest of Brazil. Although COQ7 mutations have been reported to affect multiple tissues, in this study, we describe the pathological effects of COQ7 predominantly targeting the motor neurons, both upper and lower. We, therefore, suggest COQ7 to be screened in patients diagnosed with dHMN.
Materials and methods
Distal hereditary motor neuropathy patients and families
Patients were studied by board certified neurologists, clinical data were obtained following clinical research standards in the field and family history was carefully assessed. All subjects consented to institutional review board-approved research protocols agreed by the institution at which the work was performed following standards of the Declaration of Helsinki for ethical principles for medical research involving human subjects.
Exome/genome and Sanger sequencing
The SureSelect Human All Exon Kit (Agilent) was used for in-solution enrichment, and a HiSeq 2500 instrument (Illumina) was used to produce 100 bp paired-end sequence reads. The Burrows–Wheeler aligner (version: 0.7.10) and Freebayes (version 1.1.0) were used for sequence alignment and variant calling. Exome and genome data were uploaded into the GENESIS software15,16 and analysed using strict filters for rare variants with minor allele frequency (MAFgnomAD < 0.001), effect on protein function prediction (Combined annotation dependent depletion, CADD) and high conservation across multiple species (genomic evolutionary rate profiling, GERP). The top variants were selected according to the highest machine learning algorithm scores (MAVERICK > 0.9).17COQ7 variants detected by next-generation sequencing were confirmed by Sanger sequencing (Eurofins) and analysed using Sequencher (Gene Codes).
Fibroblast culture
Fibroblast cells were obtained from affected probands from Families 1 and 2, and healthy controls, by skin biopsies. Fibroblasts were cultured in Dulbecco’s modified Eagle medium containing 10% (v/v) fetal bovine serum (FBS) and antibiotics (penicillin and streptomycin) under humidified air at 37°C in 5% CO2.
Western blot
Cells were lysed with RIPA buffer (ThermoFisher Scientific) containing protease inhibitors and sonicated with an ultrasonic generator (Hielscher). Thirty micrograms of protein were loaded into a 4–12% Bis-Tris acrylamide gel (ThermoFisher Scientific) and transferred to a PVDF membrane. Membranes were probed with the following antibodies: COQ7 (ThermoFisher Scientific, PA5-106348), TOM20 (Santa Cruz, sc-17764), GAPDH (Santa Cruz, sc-47724), β-tubulin (Cell Signaling, 2146) and neurofilament light (NFL, Cell signaling, 2837). Anti-rabbit and anti-mouse (Cell Signaling) secondary antibodies were used. Membranes were developed with SuperSignal chemiluminescence substrate (Pierce).
Generation of induced pluripotent stem cell-derived motor neurons
Induced pluripotent stem cell (iPSC)-derived lines from two patients harbouring the COQ7 variant 9 probands of Families 1 and 2 and two healthy controls were plated and grown to 95% confluency prior to starting the 24-day lower motor neuron differentiation protocol. Differentiation of the iPSCs into motor neurons was performed as previously described.18 Following the differentiation, motor neurons were sorted out of culture using a CD171-PE antibody (Invitrogen Cat No. 12-1719-42) and anti-PE MicroBeads (Multenye Biotec Cat. No. 130-048-801). Motor neurons were plated into dishes coated with poly-l-ornithine (10 μg/ml) and laminin (3.0 μg/ml). Motor neurons were allowed to mature for 10 days prior to cell pellet collection for western blot. Motor neurons used in CoQ10 treatment and NFL-enzyme linked immunosorbent assay (ELISA) experiments were plated onto 96-well plates after sorting and allowed to mature for 7 days prior to treatment. Motor neurons were treated with CoQ10 dissolved in ethanol at 1 μM and 10 μM concentrations at 7 days, and media were collected for NFL-ELISA measurements on Day 10 after sorting.
NFL-ELISA
A Uman Diagnostics NF-light ELISA kit (Quanterix/UmanDiagnostics, Cat. No. 10-7002) was used to quantitate NFL release from neurons, a marker of axonal damage, according to the manufacturer’s guidelines. Measurement was performed by collecting 100 μl of cell culture media hours after treatment with CoQ10. Two 50 μl technical replicates of five biological replicates were assayed. To normalize for variances in neuronal cell count between conditions, the total number of Homeobox B9 (HB9) positive-motor neurons was quantified via immunofluorescence. Cultures were fixed with 3.7% PFA (Sigma-Aldrich, 252549-100 ml) dissolved PBS (Corning, Cat. No. 21-030-CV) for 20 min, permeabilized using 0.2% Triton X-100 (Sigma-Aldrich, X100-100ML) in PBS for 2 min and blocked in 4% bovine serum albumin (BSA; Thermo Fisher, Cat. No. PI31873) for 1 h at room temperature. Cells were then incubated in 1:50 Rabbit anti-HB9 (Thermo Fisher, Cat. No. PA5-23407) diluted in 4% BSA in 0.2% Triton X-100 overnight at 4°C, and 1:400 Goat anti-Rabbit 555 (Invitrogen, Cat. No. A31572) diluted in 4% BSA for 1 h at room temperature. Nuclear counterstain was performed with 600 nM DAPI (Thermo Fisher, Cat. No. D1306) for 10 min. The entire area of each well was imaged at ×10 magnification and the number of highly positive motor neurons per well was quantitated using the CellInsight™ CX5 High Content Screening (HCS) Platform.
CoQ10 and DMQ10 quantification by high-performance liquid chromatography
Quinone detection and quantitation were carried out using high-performance liquid chromatography (HPLC). Briefly, cells were lysed in a radioimmunoprecipitation buffer (Tris-HCl, pH 7.5, 1% NP-40, 0.5% deoxycholate, 10 mM EDTA, 150 mM NaCl) and extracted with a mixture of 28.5% ethanol and 71.5% hexane (v/v) for 2 min by vigorously vortexing. After centrifugation, the upper organic layer was transferred to a new tube and hexane was evaporated by drying in a SpeedVac concentrator (Thermo Fisher Scientific). The remaining residue was finally redissolved in a mixture of methanol and ethanol (7:3, v/v) before injection into the HPLC system (Agilent 1260 Infinity). During HPLC, the samples were separated on a reverse-phase C18 column (2.1 × 50 mm, 1.8 µm, Agilent) and eluted with mixture of 70% methanol and 30% ethanol (v/v) at 1.8 ml/min. The detection wavelength was 275 nm. Peak identities were established by comparison with the profile obtained from a CoQ10 standard (Sigma-Aldrich). Protein content in the quinone extracts was determined by the BCA assay (Thermo Fisher Scientific) and used to normalize quinone levels.
Mitochondrial respiration
Endogenous cell respiration was measured polarographically at 37°C using a Clark-type electrode from Hansatech Instruments. Substrate-driven respiration was assayed in digitonin-permeabilized cultured cells, as reported previously.19 Briefly, trypsinized cells were washed with permeabilized-cell respiration buffer (PRB) containing 0.3 M mannitol, 10 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mg/ml BSA and 10 mM KH3PO4 (pH 7.4). The cells were resuspended at 1 × 106 cells/ml in 0.5 ml of the same buffer, air-equilibrated at 37°C and supplemented with 10 units of hexokinase. The cell suspension was immediately placed into the polarographic chamber to measure endogenous respiration. Subsequently, in some experiments, cells were permeabilized in-chamber with 0.02 mg/ml digitonin. The oxygen consumption rate under phosphorylating conditions was assessed using 5 mM glutamate plus 5 mM malate as the substrates in the presence of 2.5 mM ADP. The non-phosphorylating state was obtained after ATP synthesis inhibition using 0.75 μg/ml oligomycin. Mitochondrial respiration was uncoupled by successive addition of up to 0.4 μM CCCP to reach maximal oxygen consumption. Subsequently, complex IV activity was inhibited using 0.8 μM KCN to assess the mitochondrial specificity of the oxygen consumption measured. Values were normalized by total cell number.
Data availability
The data presented in this study are available from the corresponding author upon request.
Results
Identification of biallelic COQ7 variants in patients with distal hereditary motor neuropathy
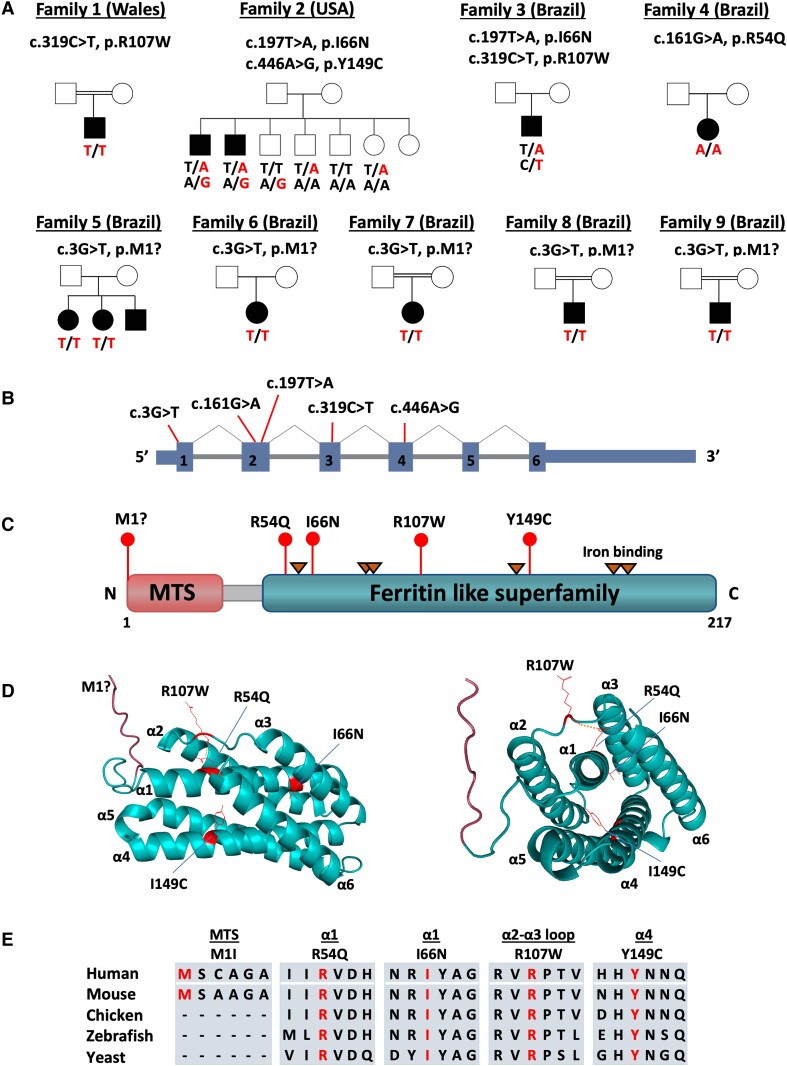
Exome/genome sequencing was performed in seven unrelated index individuals from recessive and sporadic families diagnosed with dHMN. Exome data were analysed by bioinformatic tools provided by the GENESIS platform, collaborative web-based software containing approximately 17 000 whole exomes/genomes from patients with different phenotypes, including about 2500 motor and motor-sensory neuropathy patients, such as those with Charcot-Marie-Tooth disease (CMT).15,16 Pathogenic variants in known CMT or dHMN genes were absent in all index individuals. Recessive variants were identified in GENESIS using a strict approach to filter for rare alleles predicted to impact protein function. The selected filters included MAFgnomAD < 0.001, high conservation score (GERP > 3), and predicted to be pathogenic according to multiple prediction tools, including our in-house developed machine learning algorithm (MAVERICK).17 We identified biallelic COQ7 (NM_016138) variants in nine families diagnosed with dHMN with upper neuron involvement (Fig. 1, Table 1 and Supplementary Table 1), expanding the clinical phenotype of COQ7 mutations. Three of these families (Families 1–3) were identified in GENESIS, while seven additional families (Families 4–9) were identified by a collaborative matchmaking approach. Family 1 originated from Wales, Family 2 from the US and Families 3–9 from Brazil, including an indigenous Amazonian patient (Family 3; Fig. 1A).
Figure 1.
Identification of biallelic COQ7 variants in families with distal hereditary motor neuropathy. (A) Pedigree of nine families with 11 affected individuals harbouring biallelic variants in COQ7. (B and C) The diagram illustrates the variants’ positions at the cDNA (B) and protein levels (C). (D) 3D structural model of human COQ7 protein predicted by AlphaFold (Q49A71_HUMAN) and illustrated by PyMOL. Residues mutated in patients are depicted in red. The mitochondrial targeting signal is labelled in red, and the ferritin-like superfamily in blue. (E) Protein alignment showing conservation across different species. Variants are highlighted in red. MTS = mitochondrial targeting signal.
Table 1.
Clinical phenotypes
| Family | ID/Sex/Age, y | Patient origin | Age at onset | First symptoms | Leading phenotype | Foot deformities |
|---|---|---|---|---|---|---|
| 1 | II.1, M, 54 | Wales | 8–10 y | Walking difficulties | CMT2 | ‘Wind-swept’ feet, contracted Achilles tendons |
| 2 | II.1, M, 60 | USA | Mid-teens | Walking and running difficulties | dHMN with prox. involvement | Hammer toes |
| 2 | II.2, M, MD | USA | Early childhood | Never able to run, walking difficulties | dHMN with prox. involvement | Club foot |
| 3 | II.1, M, 18 | Brazil | <10 y | Walking and running difficulties | dHMN | Pes cavus |
| 4 | II.1, F, 11 | Brazil | 3 y | Walking and running difficulties | dHMN with UMN involvement | Pes cavus, had tendon transfer surgery |
| 5 | II.1, F, 43 | Brazil | 5 y | Walking and running difficulties | dHMN with UMN involvement | Pes cavus |
| 5 | II.2, F, 41 | Brazil | 10 y | Walking and running difficulties | dHMN with UMN involvement | Pes cavus |
| 6 | II.1, F, 62 | Brazil | 1 y | Walking difficulties | dHMN | Pes cavus |
| 7 | II.1, F, 56 | Brazil | School age | Walking and running difficulties | dHMN | Pes cavus |
| 8 | II.1, M, 50 | Brazil | 4 y | Walking and running difficulties | dHMN | Normal |
| 9 | II.1, F, 52 | Brazil | 10 y | Walking difficulties | dHMN | Normal |
The given age refers to the age at the most recent examination. CMT2 = axonal Charcot-Marie-Tooth disease/hereditary motor and sensory neuropathy; dHMN = distal hereditary motor neuropathy; F = female; M = male; MD = missing data; UMN = upper motor neuron; y = years.
The index patient from Family 1 was homozygous for variant c.319C>T; Arg107Trp, which has previously been reported to be pathogenic in a compound heterozygous state with a loss-of-function variant in a fatal case of CoQ10 deficiency.9 The novel compound heterozygous variants, c.197T>A; Ile66Asn and c.446A>G; Tyr149Cys, segregated with the disease in two affected individuals from Family 2. Segregation analysis of the unaffected siblings confirmed the variants are in trans. The index patient from Family 3 is compound heterozygous for c.197T>A; Ile66Asn (the same variant as identified in Family 2) and c.319C>T; p.Arg107Trp (the same variant as identified in Family 1). The index patient from Family 4 was homozygous for c.161G>A; p.Arg54Gln; the same variant was recently reported in a case with a neuromuscular phenotype, however, electrophysiological studies were not performed to confirm neuropathy.12 Three patients from Families 5–9 shared the same novel homozygous loss-of-function variant c.3G>T; p.Met1?. The dHMN variants were located in different regions of the protein, including the mitochondrial targeting signal (MTS) and the ferritin-like superfamily domain. Most variants (p.Met1?, p.Arg107Trp and p.Ile66Asn) were shared between different families (Fig. 1B and C). Variant and segregation analyses were confirmed by Sanger sequencing.
Region of homozygosity and haplotype analysis
Because we identified five Brazilian families with the same homozygous variant, p.Met1?, which occurs at a very low allele frequency in the population (MAFgnomAD = 0.00000657), we decided to investigate the potential existence of a common ancestral haplotype. Regions of homozygosity (ROHs) spanning the entire COQ7 gene were identified in the affected individuals from Families 5 and 6. A large 5.6 Mb ROH was observed in the patients from Family 5 at chromosome (chr)16:15,135,071–20,791,093 (hg19), which was the largest ROH detected in that sample. The patient from Family 6 had 1.4 Mb and 1.6 Mb ROHs, chr16:16,832,774–20,493,224 (hg19), with a small gap in between at chr16:18,267,702–18,809,743. To investigate whether the variant p.Met1?, present in the three patients, was a result of a potential founder effect, we performed haplotype analysis. Using the exome/genome data from Families 5 and 6, we analysed a region expanding ∼10 Mb upstream and downstream of the p.Met1? variant. Our results revealed a shared haplotype in the two patients, strongly suggesting a founder effect (Supplementary Table 2).
COQ7 protein structure and dHMN variants’ locations
The 3D structure of COQ7 (PDB Q99807), obtained from the AlphaFold database,20 was modelled and visualized by PyMOL to analyse the dHMN variant’s locations. COQ7 is composed of an MTS (labelled in red) and six α-helices (α1–α6, labelled in green) (Fig. 1D). The COQ7 protein structure was recently solved by cryogenic electron microscopy, which revealed that the six helices are arranged to form a large hydrophobic channel leading to the active site. Three COQ7 variants identified in the dHMN patients affect highly conserved amino acids in the α-helices, important to channel structure. The variants Arg54Q and Ile66Asn are both located in α1, Y149C is located in α4 and Arg107Trp is located in the flexible α2-α3 loop (Fig. 1D). The residue Arg107 forms a polar interaction with R54Q, which is disrupted by the two dHMN variants targeting these residues. The Arg107 residue has also been shown to be part of the molecular interface between COQ7 and COQ9 during interaction, and point mutations in this interface, including, Arg107Glu, have been shown to disrupt COQ7–COQ9 interaction.21 All missense variants are located in highly conserved residues, from humans to yeast, except for the start-loss variant p.Met1?, which is predicted to cause loss of function (Fig. 1E).
Clinical description of patients with COQ7 variants
Our cohort consisted of 11 patients from nine families. The average age at the most recent clinical examination was 47 years, with a range of 17–60 years. All patients are still alive. Five individuals were male, six were female. In total, eight patients originated from Brazil, of which one was of indigenous descent. One patient was Welsh, and another family was of US American origin. A family history was positive in two cases, and all available individuals are reported in Table and Supplementary Table 1. Conversely, nine cases occurred sporadically. The most commonly reported diagnosis was dHMN (10 of 11 cases), partially with additional proximal (three cases) and upper motor neuron involvement (nine cases). One patient that presented with additional sensory deficits on nerve conduction studies was classified as having axonal CMT2 disease. As a sign of long-lasting, if not even congenital chronic motor neuropathy, 9 of 10 patients presented with foot deformities, typically high arched feet and hammer toes. Within the first two decades of life, all patients reported having developed difficulties walking and running, so that sports performance at school was impaired in all but one case. Additional difficulties with fine motor skills were reported in six of eight patients, mostly beginning in the third decade of life. Sensory plus or minus symptoms were not part of the frequently reported symptoms, and at the time of examination, none of the patients took any medication for neuropathic pain. The clinically leading phenotype consisted of distal pareses and atrophies, predominantly in the lower limbs. However, mild to moderate weakness of the intrinsic hand muscles was additionally observed in eight of nine examined patients, and the onset of proximal weakness became evident in three individuals. Deep tendon reflexes were absent at the ankles in seven of nine patients; however, patellar tendon and upper limb reflexes were found to be brisk in eight individuals. Increased muscle tone (spasticity) was reported in 4 of 10 patients, and pyramidal signs were positive in another four patients. In total, 9 of 11 individuals with biallelic COQ7 mutations showed some signs of upper motor neuron involvement. Four patients, all carriers of the homozygous c.3G>T; p.Met1? variant, presented with additional signs of cerebellar ataxia. Out of these, two patients showed MRI abnormalities consistent with cerebellar atrophy. Conversely, afferent ataxia was observed in one individual only. Other mild clinical signs of sensory involvement were hypopallaesthesia in four (of nine examined) patients, reduced distal position perception in three and reduced pinprick perception at distal lower limbs in another three individuals. As an additional characteristic, three unrelated patients developed chronic progressive hearing loss. In contrast to other mitochondrial spectrum diseases, none of the patients was reported with optic atrophy, cataracts or diabetes mellitus. Cognitive impairment and learning difficulties were described in two carriers of the homozygous c.3G>T; p.Met1? mutation. One patient had seizures of unspecified semiology in childhood. Nerve conduction studies, most recently performed at a mean age of 37 years (17–51 years), showed signs of a length-dependent, slowly progressive, axonal motor neuropathy with lower limb predominance. Sensory involvement was observed in one individual only, which was reproduced and shown to be progressive over time (observation interval: 13 years). Upon genetic diagnosis, one patient (Family 2, II.2) underwent an oral supplementation of low-dose COQ10, which she stopped at her own discretion after not observing any effect.
Evaluation of pathogenicity of COQ7 variants in fibroblasts from dHMN patients
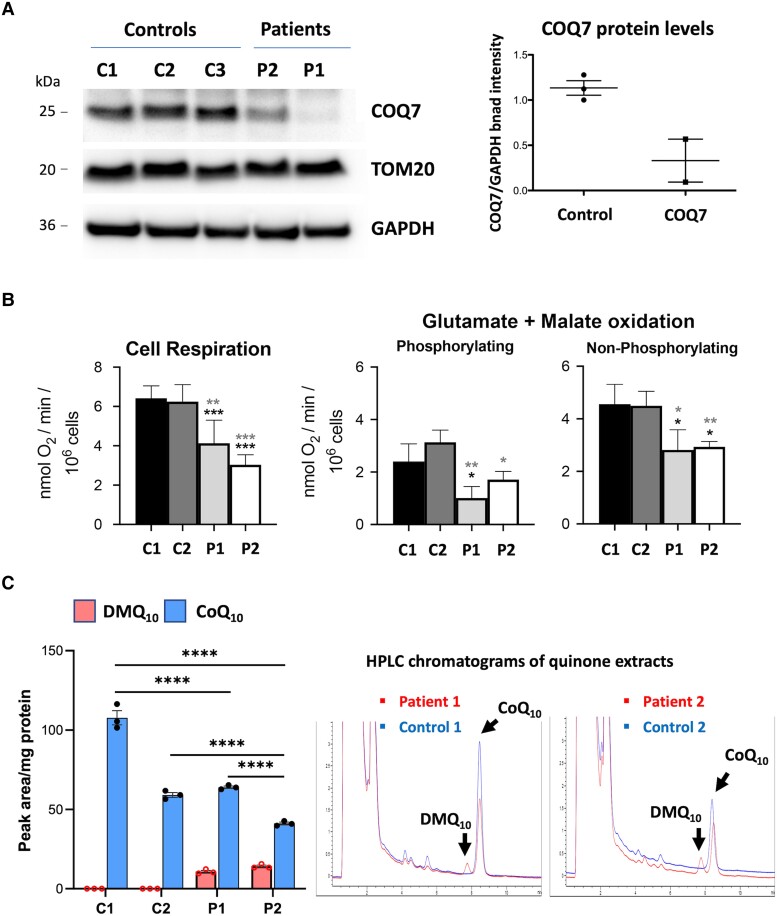
To evaluate the effects of the COQ7 variants, we obtained fibroblasts from two dHMN patients harbouring different COQ7 variants (index patients from Family 1, Patient 1, and Family 2, Patient 2). Western blot showed substantially reduced levels of COQ7 protein in fibroblasts from both patients, which indicates protein instability due to mutations leading to a loss-of-function mechanism (Fig. 2A). COQ7 protein appeared to be more severely depleted in Patient 1 compared with Patient 2. These results confirmed previously reported findings of pathogenic COQ7 variants leading to protein depletion, including the variant Arg54Gln also found in one of our patients.12 To assess mitochondrial function, we measured the oxygen consumption rate of fibroblasts. Cell respiration was measured in whole cells and substrate oxidation in digitonin-permeabilized cells in the presence of ADP (phosphorylating) or CCCP (non-phosphorylating). The basal mitochondrial respiration rate in whole cells was significantly decreased in both patients’ fibroblasts, indicating mitochondrial dysfunction (Fig. 2B). The maximal respiration obtained with mitochondrial uncoupler FCCP was also significantly reduced in the patients’ cells compared with controls (Fig. 2B).
Figure 2.
Investigation of pathogenicity of COQ7 variants. (A) Western blot using cultured fibroblasts from three healthy control (C1 and C2) and two COQ7 patients (Patient 1, proband of Family 1, and Patient 2, proband of Family 2). COQ7 protein is depleted in the patients’ fibroblasts. COQ7 band intensity was quantified and normalized by GAPDH. (B) Cell respiration in whole cells and substrate oxidation in digitonin-permeabilized cells in the presence of ADP (phosphorylating) or CCCP (non-phosphorylating); t-test (paired). (C) HPLC analysis of quinone extracts from control and patients’ fibroblasts. HPLC chromatograms detection of DMQ10 and CoQ10 peaks. Fibroblasts from patients show a slight reduction in the levels of CoQ10 and abnormal accumulation of the precursor DMQ10.
To analyse the CoQ10 biosynthetic pathway in patients, we used HPLC to quantify quinone extracts from fibroblasts. The results showed abnormal accumulation of the CoQ10 precursor DMQ10 in the patients’ fibroblasts, which was completely absent in controls (Fig. 2C). COQ7 is responsible for converting DMQ10 to the next COQ10 precursor; hence, accumulation of DMQ10 indicates COQ7 enzyme dysfunction. Therefore, DMQ10 is a known biomarker for CoQ10 biosynthesis deficiency, which confirms the pathogenicity of the dHMN variants. Consequently, CoQ10 levels appeared slightly decreased in the patients’ fibroblasts (Fig. 2C). It is not clear whether DMQ10 could play a pathological role in cells; however, there is no evidence that DMQ10 causes toxicity or influences phenotype severity. DMQ10 can function as an electron carrier in the mitochondrial respiratory chain, similar to CoQ10, but much less efficiently.22
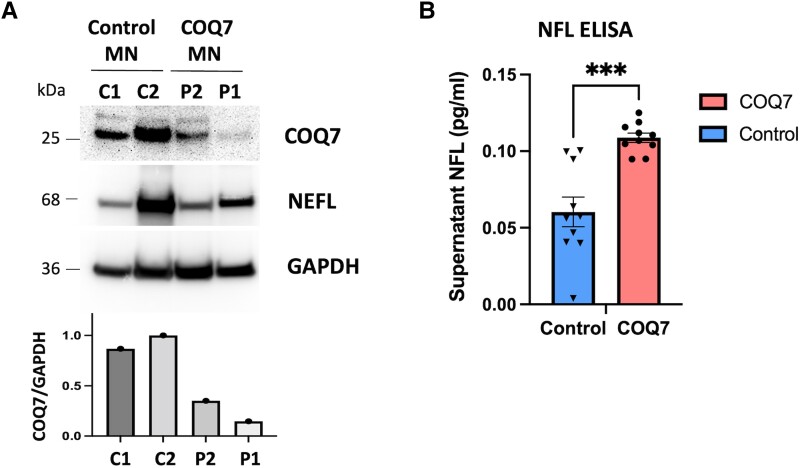
Effect of COQ7 variants in iPSC-derived motor neurons from patients
To develop a more suitable model to study the effect of COQ7 variants from the dHMN patients, we generated iPSC-derived motor neurons from the patients’ fibroblasts. Western blot showed decreased levels of COQ7 protein in the patients’ motor neurons (Fig. 3A). Motor neurons from the COQ7 patients showed significantly increased levels of supernatant (extracellular) NFL compared with the control, an indication of axonal degeneration, confirming the neuropathy phenotype in patients (Fig. 3B). NFL release from cells has been shown to be a potential biomarker for axonal degeneration in different neurogenerative diseases, including amyotrophic lateral sclerosis and CMT.23
Figure 3.
iPSC-derived motor neuron studies. (A) Western blot from motor neurons shows reduced levels of COQ7 in motor neurons derived from a COQ7 patient, proband from Family 2). (B) ELISA quantification of NFL protein release from iPSC-derived motor neurons. Motor neurons from COQ7 patients show significantly increased levels of cell supernatant NFL protein, indicating axonal degeneration (t-test, P < 0.001).
Discussion
In this study, we identified COQ7 as a novel candidate disease gene for dHMN with upper motor neuron involvement, expanding the clinical phenotype associated with COQ7 dysfunction. Our genetic findings from nine dHMN families are supported by functional studies with patients’ fibroblasts showing COQ7 protein depletion, reduced mitochondrial respiration, CoQ10 deficiency and abnormal accumulation of the COQ10 precursor DMQ10. Moreover, iPSC-derived motor neurons showed increased levels of supernatant NFL in a patient, confirming the axonal degeneration phenotype. Our findings indicate a molecular mechanism involving CoQ10 biosynthesis deficiency coupled with mitochondrial dysfunction leading to axonal degeneration. Several known CMT-related genes have previously been associated with mitochondrial dysfunction, including MFN2, GDAP1, SCO2 and POLG1, underlining the importance of optimal mitochondrial function in the peripheral nerve and its dependence on energy metabolism.24,25 However, COQ7 is the first identified dHMN gene to specifically be involved in the CoQ10 biosynthesis pathway. It is not clear why COQ7 variants lead to different clinical presentations, ranging from severe complex mitochondrial disorders to mild phenotypes involving spasticity with hearing loss, and now peripheral neuropathy. Although the phenotypes are distinct, the disease mechanism appears to be the same, which is caused by a loss-of-function mutation leading to CoQ10 deficiency, abnormal accumulation of the precursor DMQ10 and mitochondrial respiration deficiency. In general, peripheral neuropathy can be a common feature and, in rare cases, a predominant manifestation of mitochondrial disorders.25 In particular, mutations in MFN2 can cause CMT associated with more complex phenotypes, including optic atrophy, hearing loss and ataxia.26 POLG-associated disorders can also present with heterogeneous complex phenotypes.27 Therefore, the clinical variability observed in COQ7 patients might be attributed to the heterogeneous nature of mitochondrial diseases, since the mitochondrial composition highly varies in different tissues and individuals, due to the unorthodoxy of mitochondrial genetics. It would be important to determine whether COQ7-related disorders comprise a continuum of broad, overlapping phenotypes as part of the same disease spectrum, similar to other multisystem mitochondrial disorders.
Accordingly, our patients with the p.Met1? variant and motor neuropathy further showed symptoms of cerebellar ataxia (four of seven), cognitive impairment and learning difficulty (two of seven). This variant is predicted to cause loss of the start codon AUG, leading to protein loss of function. The 5′-UTR does not contain any alternative upstream AUG, and the next AUG in the transcript is out-of-frame and would completely disrupt the protein sequence. Because complete ablation of COQ7 might be embryonically lethal, and the phenotype of the patients was not severe, it is possible that alternative transcripts provide residual function in cells/tissues. This includes smaller transcripts, not affected by the mutation, such as transcript ENST00000569127.1, predicted to encode a protein with 194 amino acids, which exhibits low expression in the tibial nerve [GTEx, transcripts per million (TPM) = 1.68]. However, the canonical, transcript 1 (ENST00000321998.10), is the longest isoform with 217 amino acids, and it is also the predominant isoform in neuronal tissues, including the tibial nerve (TPM = 7.45).
Our studies with iPSC-derived motor neurons confirmed axonal degeneration. One of our patients (Patient 2, Family 2) received a short trial of low-dose oral coenzyme Q10 treatment, which was stopped by the patient after noting no benefit. Pharmacokinetic studies with different CoQ10 formulations would be necessary to investigate the effects of supplements in dHMN patients. The potential benefit of CoQ10 treatment in patients with CoQ10 deficiency has been explored extensively for several decades; however, this therapeutic approach remains paradoxical, due to conflicting reported outcomes.6 Oral CoQ10 treatment has been partially successful in some cases of primary CoQ deficiency and other human diseases28; however, it has not yet been approved by the US Food and Drug Administration (FDA). Some therapeutic challenges involve the delivery of CoQ10 to the target tissue, due to its hydrophobicity and high molecular weight, and in order to overcome these challenges, CoQ10 analogous have been developed, including idebenone and mitoquinone.29
Early diagnosis of COQ7 disorders and the development of therapeutic strategies for better delivery of CoQ10 or other analogs to the targeted disease tissue, including the peripheral nerve, should be explored further.
Supplementary Material
Contributor Information
Adriana P Rebelo, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Pedro J Tomaselli, Department of Neurology, University of São Paulo, Ribeirão Preto, 14048-900, Brazil.
Jessica Medina, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Ying Wang, Department of Biology, McGill University, Montreal, QC, H3A 1A1, Canada.
Maike F Dohrn, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA; Department of Neurology, Medical Faculty, RWTH Aachen University, Aachen 52074, Germany.
Eva Nyvltova, Department of Neurology, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Matt C Danzi, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Mark Garrett, Department of Neurology, Washington University, St. Louis, MO 63112, USA.
Sean E Smith, Department of Neurology, Washington University, St. Louis, MO 63112, USA.
Alan Pestronk, Department of Neurology, Washington University, St. Louis, MO 63112, USA.
Chengcheng Li, Department of Neurology, Washington University, St. Louis, MO 63112, USA.
Ariel Ruiz, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Elizabeth Jacobs, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Shawna M E Feely, Department of Neurology, Carver College of Medicine, University of Iowa, Iowa City, IA 52242, USA.
Marcondes C França, Jr, Department of Neurology, University of São Paulo, Ribeirão Preto, 14048-900, Brazil.
Marcus V Gomes, Department of Neurology, University of São Paulo, Ribeirão Preto, 14048-900, Brazil.
Diogo F Santos, Department of Neurology, Federal University of Uberlândia, Uberlândia, MG 38405-320, Brazil.
Surinder Kumar, Department of Pathology & Laboratory Medicine, Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
David B Lombard, Department of Pathology & Laboratory Medicine, Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Mario Saporta, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Siegfried Hekimi, Department of Biology, McGill University, Montreal, QC, H3A 1A1, Canada.
Antoni Barrientos, Department of Neurology, University of Miami Miller School of Medicine, Miami, FL 33136, USA.
Conrad Weihl, Department of Neurology, Washington University, St. Louis, MO 63112, USA.
Michael E Shy, Department of Neurology, Carver College of Medicine, University of Iowa, Iowa City, IA 52242, USA.
Wilson Marques, Department of Neurology, University of São Paulo, Ribeirão Preto, 14048-900, Brazil.
Stephan Zuchner, Dr. John T. Macdonald Foundation, Department of Human Genetics and John P. Hussman Institute for Human Genomics, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Funding
This study was supported by NINDS and NCATS grants (5U54NS065712 and 5R01NS105755 to M.S. and S.Z). We are also thankful for support from the Muscular Dystrophy Association, CMT Association and Hereditary Neuropathy Foundation. P.J.T. was supported by an MRC strategic award to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) MR/S005021/1. S.H. was supported by a Foundation grant from the Canadian Institutes of Health Research: FDN-159916. M.F.D. was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG, DO 2386/1-1).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Acosta MJ, Vazquez Fonseca L, Desbats MA, et al. . Coenzyme Q biosynthesis in health and disease. Biochim Biophys Acta. 2016;1857:1079–1085. [DOI] [PubMed] [Google Scholar]
- 2. Overvad K, Diamant B, Holm L, Holmer G, Mortensen SA, Stender S. Coenzyme Q10 in health and disease. Eur J Clin Nutr. 1999;53:764–770. [DOI] [PubMed] [Google Scholar]
- 3. Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta. 2004;1660(1-2):171–199. [DOI] [PubMed] [Google Scholar]
- 4. Littarru GP, Tiano L. Bioenergetic and antioxidant properties of coenzyme Q10: recent developments. Mol Biotechnol. 2007;37:31–37. [DOI] [PubMed] [Google Scholar]
- 5. Santoro MM. The antioxidant role of non-mitochondrial CoQ10: mystery solved! Cell Metab. 2020;31:13–15. [DOI] [PubMed] [Google Scholar]
- 6. Diaz-Casado ME, Quiles JL, Barriocanal-Casado E, et al. . The paradox of coenzyme Q10 in aging. Nutrients. 2019;11:2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desbats MA, Lunardi G, Doimo M, Trevisson E, Salviati L. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency. J Inherit Metab Dis. 2015;38:145–156. [DOI] [PubMed] [Google Scholar]
- 8. Wang Y, Smith C, Parboosingh JS, Khan A, Innes M, Hekimi S. Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J Cell Mol Med. 2017;21:2329–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwong AK, Chiu AT, Tsang MH, et al. . A fatal case of COQ7-associated primary coenzyme Q10 deficiency. JIMD Rep. 2019;47:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Freyer C, Stranneheim H, Naess K, et al. . Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J Med Genet. 2015;52:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashemi SS, Zare-Abdollahi D, Bakhshandeh MK, et al. . Clinical spectrum in multiple families with primary COQ10 deficiency. Am J Med Genet A. 2021;185:440–452. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y, Gumusb E, Hekimia S. A novel COQ7 mutation causing primarily neuromuscular pathology and its treatment options. Mol Genet Metab Rep. 2022;31:100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Hekimi S. The efficacy of coenzyme Q10 treatment in alleviating the symptoms of primary coenzyme Q10 deficiency: A systematic review. J Cell Mol Med. 2022;26:4635–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Oxer D, Hekimi S. Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat Commun. 2015;6:6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonzalez MA, Lebrigio RF, Van Booven D, et al. . Genomes management application (GEM.app): a new software tool for large-scale collaborative genome analysis. Hum Mutat. 2013;34:842–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez M, Falk MJ, Gai X, Postrel R, Schule R, Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat. 2015;36:950–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Danzi M, Dohrn M, Fazal S, et al. . Deep structured learning realizes variant prioritization for Mendelian diseases. Res Sq. [Preprint] doi: 10.21203/rs.3.rs-1602211/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saporta MA, Dang V, Volfson D, et al. . Axonal Charcot-Marie-Tooth disease patient-derived motor neurons demonstrate disease-specific phenotypes including abnormal electrophysiological properties. Exp Neurol. 2015;263:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barrientos A, Fontanesi F, Díaz F. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr Protoc Hum Genet. 2009;63:19.3.1–19.3.14. doi: 10.1002/0471142905.hg1903s63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jumper J, Evans R, Pritzel A, et al. . Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lohman DC, Aydin D, Von Bank HC, et al. . An isoprene lipid-binding protein promotes eukaryotic coenzyme Q biosynthesis. Mol Cell. 2019;73:763–774.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y, Hekimi S. Minimal mitochondrial respiration is required to prevent cell death by inhibition of mTOR signaling in CoQ-deficient cells. Cell Death Discov. 2021;7:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pareyson D, Shy ME. Neurofilament light, biomarkers, and Charcot-Marie-Tooth disease. Neurology. 2018;90:257–259. [DOI] [PubMed] [Google Scholar]
- 24. Rebelo AP, Saade D, Pereira CV, et al. . SCO2 mutations cause early-onset axonal Charcot-Marie-Tooth disease associated with cellular copper deficiency. Brain. 2018;141:662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pareyson D, Piscosquito G, Moroni I, Salsano E, Zeviani M. Peripheral neuropathy in mitochondrial disorders. Lancet Neurol. 2013;12:1011–1024. [DOI] [PubMed] [Google Scholar]
- 26. Rouzier C, Bannwarth S, Chaussenot A, et al. . The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain. 2012;135(Pt 1):23–34. [DOI] [PubMed] [Google Scholar]
- 27. Cohen BH, Chinnery PF, Copeland WC. POLG-related disorders. In: Adam MP, Mirzaa GM, Pagon RA, et al., eds. Genereviews®. University of Washington;1993-2003. [Google Scholar]
- 28. Stefely JA, Pagliarini DJ. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem Sci. 2017;42:824–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suárez-Rivero JM, Pastor-Maldonado CJ, Povea-Cabello S, et al. . Coenzyme Q10 analogues: benefits and challenges for therapeutics. Antioxidants (Basel). 2021;10:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available from the corresponding author upon request.