In this review, de Ruiter Swain et al. discuss the mechanisms of intercellular metabolite exchange underpinning glucose, fatty acid, and amino acid homeostasis in the healthy brain. They further illustrate how these metabolic processes are exploited by glioblastoma to provide essential nutrients for tumor growth and modulation of the tumor microenvironment, highlighting that targeting metabolic dependencies could be a potential therapeutic avenue against this aggressive brain cancer.
Keywords: brain metabolism, cancer metabolism, glioblastoma, glioma, glioma therapy, IDH mutation, immune suppression, tumor microenvironment
Abstract
The different cell types in the brain have highly specialized roles with unique metabolic requirements. Normal brain function requires the coordinated partitioning of metabolic pathways between these cells, such as in the neuron–astrocyte glutamate–glutamine cycle. An emerging theme in glioblastoma (GBM) biology is that malignant cells integrate into or “hijack” brain metabolism, co-opting neurons and glia for the supply of nutrients and recycling of waste products. Moreover, GBM cells communicate via signaling metabolites in the tumor microenvironment to promote tumor growth and induce immune suppression. Recent findings in this field point toward new therapeutic strategies to target the metabolic exchange processes that fuel tumorigenesis and suppress the anticancer immune response in GBM. Here, we provide an overview of the intercellular division of metabolic labor that occurs in both the normal brain and the GBM tumor microenvironment and then discuss the implications of these interactions for GBM therapy.
Glioblastoma multiforme (GBM; grade IV glioma) is the most common malignant primary brain tumor and among the most lethal of human cancers, with median survival after diagnosis of only ∼12–15 mo with intensive treatment and ∼4 mo without treatment (Tamimi and Juweid 2017; Baid et al. 2020). GBM is characterized by extensive invasion and infiltration of the brain parenchyma, a process facilitated by architectural features of the brain such as white matter tracts, making complete surgical resection nearly impossible (Seker-Polat et al. 2022). Uniquely, the vast majority of GBM cases are grade IV disease at initial diagnosis, with <20% of GBMs developing from a known lower-grade glioma (LGG) (Urbanska et al. 2014; Tamimi and Juweid 2017). Another conserved feature of GBM is a complex and highly immune-suppressive tumor microenvironment (TME) with heterogeneous cancer cell populations, vascular endothelial cells, central nervous system (CNS)-resident cells, and an abundance of dysfunctional immune cells (Antunes et al. 2020; Bikfalvi et al. 2023; Sharma et al. 2023). This heterogeneity contributes to the resistance of GBM to chemotherapies, targeted therapies, and immune therapies, such that the 5-yr survival rate of only ∼5% has not improved substantially for several decades (Tamimi and Juweid 2017). The failure of targeted and immune therapies to improve the prognosis for GBM patients has prompted investigation of nononcogene dependencies in GBM, including “metabolic addictions,” as opportunities for novel therapeutic interventions.
The cell of origin of GBM is still debated, with neural stem cells (NSCs) and oligodendrocyte precursor cells (OPCs) residing in the subventricular zone considered key candidates (Fan et al. 2019). Based on RNA sequencing (RNA-seq) analyses, GBM cells can exist in four distinct malignant states, with transcriptional programs reminiscent of either neural progenitor cells (NPC-like), OPCs (OPC-like), astrocytes (AC-like), or mesenchymal cells (MES-like) (Neftel et al. 2019; De Silva et al. 2023). A single GBM tumor typically contains cancer cells in more than one of these states, but the relative frequency of each state varies between tumors and correlates with genetic alterations in PDGFRA (primarily OPC-like), CDK4 (primarily NPC-like), EGFR (primarily AC-like), and NF1 (primarily MES-like) (Neftel et al. 2019; De Silva et al. 2023). There is extensive experimental evidence that GBM tumors contain a population of glioma stem cells (GSCs), which are proposed to underlie tumor initiation, evolution, and therapy resistance. However, GSCs have typically been functionally defined by their tumor-propagating and self-renewal potential, and a precise classification based on molecular profile is lacking (Suvà and Tirosh 2020). How the GSC model interfaces with the four malignant states of GBM is also poorly defined, as isolated cells from three of these states (NPC-like, OPC-like, and MES-like) are able to establish new GBM tumors that reacquire the heterogeneity observed in the primary tumor (Neftel et al. 2019). In contrast, the AC-like state is associated with decreased proliferation and low tumor-propagating potential and thus has been proposed as a goal for differentiation therapies in GBM (Suvà and Tirosh 2020).
A major distinguishing feature in gliomas of all grades is the presence or absence of oncogenic mutations in the gene encoding isocitrate dehydrogenase 1 (IDH1). These mutations lead to substitutions of arginine-132 (R132) in IDH1, most frequently (>90%) to histidine (R132H), resulting in a neomorphic enzyme that catalyzes the NADPH-dependent reduction of α-ketoglutarate (α-KG) into the oncometabolite D-2-hydroxyglutarate (2-HG) instead of the reversible interconversion of isocitrate and α-KG catalyzed by the native enzyme (Han et al. 2020). In IDH1 mutant glioma cells, 2-HG accumulates to concentrations as high as 30 mM and acts as a competitive inhibitor of α-KG-dependent dioxygenases, including histone demethylases and DNA demethylases, resulting in global epigenetic reprogramming that influences cellular differentiation (Han et al. 2020). Mutation of IDH1 is extremely common in LGG (>80% of cases) but relatively rare in high-grade glioma (∼10% of cases), where it is primarily found in secondary tumors that evolved from LGG. This has led to the recent reclassification of “IDH1 mutant GBM” as grade 4 astrocytoma to distinguish it from primary GBM, which is typically IDH1 wild type (Louis et al. 2021). For simplicity, in this review we use the term GBM to describe all grade 4 gliomas.
An important regulator of the normal brain tissue environment is the blood–brain barrier (BBB), which prevents the nonselective diffusion of molecules from the circulation into the brain interstitial fluid (ISF). The BBB is composed of vascular endothelial cells that form the capillary wall, pericytes embedded in the capillary basement membrane, and astrocytic end feet that completely ensheathe the blood vessel (Daneman and Prat 2015). GBM is typically highly vascularized and develops a “blood tumor barrier” (BTB), which shares many characteristics with the BBB including drug barrier functions but overall has more heterogeneous permeability (Arvanitis et al. 2019). The BBB also restricts the access of circulating immune cells into the brain parenchyma, such that the steady-state brain is relatively devoid of lymphocytes and peripheral monocytes (Louveau et al. 2015). This, along with other physiological features such as limited access to the lymphatic system, has led to the brain being considered an “immunologically privileged” site, although it is now recognized that the BBB is dynamically regulated during adaptive immunity, and infiltrating monocyte-derived macrophages, neutrophils, and T cells are all present in the GBM microenvironment (Brown et al. 2018). Another outcome of the regulated passage of molecules through the BBB is that the brain is partially isolated, metabolically, from the rest of the body and is strictly dependent on the endogenous synthesis of several metabolites that in other organs are supplied by the circulatory system. For example, the BBB prevents transfer of lipoprotein-bound cholesterol, and therefore almost all brain cholesterol must be synthesized de novo. Additionally, neuroactive metabolites including glutamate and aspartate must be maintained at extremely low levels in the brain ISF relative to blood plasma and similarly cannot efficiently cross the BBB (Dingledine and McBain 1999).
Intercellular cooperation across diverse physiological processes is an integral characteristic of healthy brain function and GBM pathology. The general principles of intercellular communication in the healthy brain, GBM tumor microenvironment, and GBM immune microenvironment have recently been reviewed elsewhere (Baumann and Pham-Dinh 2001; Benarroch 2005; Gieryng et al. 2017; Gillespie and Monje 2018; Szepesi et al. 2018; Grabowski et al. 2021; Crivii et al. 2022). In this review, we focus exclusively on metabolic interactions. The highly specialized roles of the different cell types in the brain impose markedly distinct metabolic requirements, and coordinated metabolic partitioning is consequently essential. An emerging theme is that GBM effectively integrates into and/or “hijacks” these metabolic interactions, co-opting nonmalignant brain cells to provide fuels for tumor growth and recycle waste products, and secreting metabolites that induce a more immune-suppressive TME. By defining these processes, opportunities for new therapeutic interventions that effectively starve GBM cells of critical nutrients or alleviate the immune-suppressive metabolic environment have begun to emerge. Here, we first summarize the metabolic division of labor between different cell types during normal brain function and then discuss how GBM integrates into or disrupts these interactions to drive tumorigenesis and immune suppression.
Intercellular metabolic compartmentation in the healthy brain
CNS function carries a massive energetic cost, with the adult human brain accounting for ∼20% of the body's resting energy expenditure despite comprising only ∼2% of total body weight. It is estimated that 75%–80% of the brain's energy budget is dedicated to neurons, primarily to power ATP-dependent ion pumps that re-establish and maintain the electrochemical gradients required for action potentials and synaptic potentials (Magistretti and Allaman 2015). The vast majority of neuronal ATP is generated in mitochondria by oxidative phosphorylation (OXPHOS), and defects in this pathway are associated with a spectrum of neurological diseases (Breuer et al. 2013). A unique characteristic of brain metabolism is that neurons do not consume energy at a consistent rate but instead can have long periods of low activity alternating with bursts of intense activity, imposing a need for a dynamic metabolic response to maintain homeostasis. Glucose is the obligatory metabolic fuel for the brain, but in specific physiological contexts monocarboxylates such as lactate, acetate, and ketone bodies also contribute, supplying up to 75% of the brain's energy needs during prolonged fasting (Dienel 2019). Despite the energetic demands of information processing, neurons have minimal energy storage capacity coupled with a lack of direct access to circulating nutrients due to the BBB (Barros et al. 2023). Consequently, neurons rely on coordinated intercellular metabolic cooperation to support their highly specialized roles (Hertz 2004). This metabolic support involves multiple cell types in the brain and includes the transfer of all of the major classes of metabolites. In the following sections we discuss key metabolic exchange processes that support brain function, including intercellular amino acid metabolism, redox support, partitioning of lipid metabolism, and the possible cycling of additional bioenergetic substrates such as lactate and ketone bodies.
The neuron–astrocyte glutamate–glutamine cycle
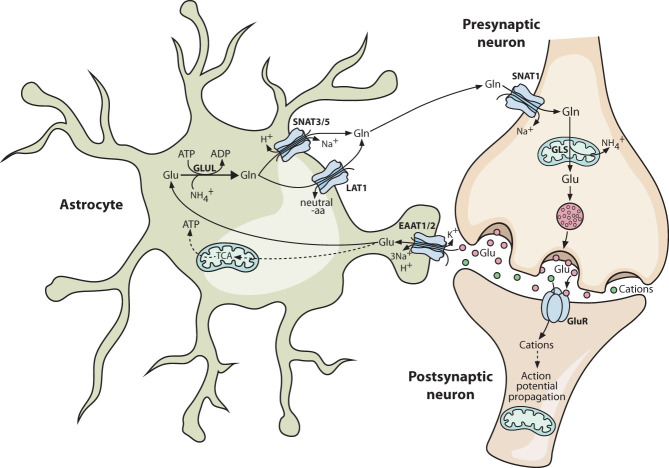
Glutamate is the most abundant excitatory neurotransmitter in the CNS and is generated from glutamine in glutamatergic neurons by mitochondrial glutaminase (GLS) prior to loading into synaptic vesicles. Synaptic transmission involves the Ca2+-dependent fusion of these vesicles with the plasma membrane of the presynaptic neuron to release glutamate into the synaptic cleft, where it binds and activates ionotropic (AMPA, NMDA, kainate, or δ) or metabotropic (mGluR1–8) glutamate receptors on the postsynaptic neuron (Meldrum 2000). Although some of this glutamate is subsequently transported back into presynaptic neurons by excitatory amino acid transporter 3 (EAAT3; encoded by SLC1A1), the majority is removed by astrocytic processes that ensheathe glutamatergic synapses to form the “tripartite synapse” (Fig. 1; Bergles and Jahr 1998; Kojima et al. 1999; Allen and Eroglu 2017; Mahmoud et al. 2019). These perisynaptic astrocytic processes contain high levels of EAAT1–2 transporters (encoded by SLC1A3 and SLC1A2, respectively) on their surface, and the tripartite synapse structure prevents spillage of glutamate out of the synaptic cleft, where glutamate concentrations transiently reach 100–1000 μM compared with only 1–3 μM in the brain ISF (de Groot and Sontheimer 2011; McKenna 2013). Remarkably, the concentrative EAAT family transporters are capable of maintaining a concentration gradient of up to 106-fold across biological membranes, and the glutamate concentration in bulk brain lysate (i.e., including intracellular glutamate) is 5–15 mM despite the exceptionally low level of this amino acid in the ISF (Featherstone 2010; Moussawi et al. 2011).
Figure 1.
Coupled neuron–astrocyte glutamate–glutamine metabolism supports glutamatergic neuron function. Glutamatergic neurons rely primarily on astrocyte-derived glutamine as a source of glutamate. Neuronal glutaminase deamidates glutamine to glutamate, which is loaded into presynaptic vesicles. In response to a presynaptic action potential, these vesicles fuse with the membrane and release the glutamate into the synaptic cleft. Here it activates glutamate receptors on the postsynaptic neuron, which propagates the action potential in the postsynaptic cell. Astrocytes form a tripartite synapse with the neurons and play an important role in clearing synaptic glutamate. Astrocytes take up glutamate via excitatory amino acid transporter (EAAT) family transporters in an energy-intensive process. Within astrocytes, a majority of the glutamate is reamidated to glutamine, while a minority is oxidized via the tricarboxylic acid (TCA) cycle to generate ATP. The astrocyte-synthesized glutamine is released into the extracellular space, where it is available for neuronal uptake once more. Solid lines indicate direct processes including movement of a molecule or single-step reactions, and dashed lines indicate indirect or multistep processes. (aa) Amino acid, (EAAT) excitatory amino acid transporter, (Gln) glutamine, (GLS) glutaminase, (Glu) glutamate, (GLUL) glutamine synthetase, (GluR) glutamate receptor, (SNAT) sodium-coupled neutral amino acid transporter, (TCA) tricarboxylic acid cycle.
Once taken up by astrocytes, ∼60%–85% of the glutamate is amidated to regenerate glutamine, a reaction catalyzed by the ATP-dependent enzyme glutamine synthetase (GLUL), which is abundant in astrocytes and not present in neurons (Norenberg and Martinez-Hernandez 1979). In parallel to glutamine synthesis, 15%–40% of the consumed glutamate is deaminated by glutamate dehydrogenase (GLUD1/2) or transaminases to generate α-KG, which is subsequently oxidized via the tricarboxylic acid (TCA) cycle (Fig. 1; Yu et al. 1982; Zielke et al. 1990; Farinelli and Nicklas 1992; Sonnewald et al. 1993). One function of this ATP-generating oxidative pathway is to offset the high energetic cost of the glutamate–glutamine cycle. However, glutamate oxidation along with additional metabolic fates such as glutathione biosynthesis constitutes a net loss of material from the glutamate–glutamine cycle. Glutamine derived from recycling of synaptic glutamate must therefore be supplemented by de novo glutamine synthesis, which is supplied by glucose-derived carbon (Schousboe et al. 2014). The ratio of glutamate oxidation to de novo synthesis in astrocytes is highly sensitive to exogenous glutamate levels, allowing homeostasis to be maintained despite continuous fluctuations in the level of glutamatergic neurotransmission (McKenna et al. 1996; Schousboe et al. 2014). To complete the glutamate–glutamine cycle, efflux of glutamine from astrocytes is coupled to glutamine uptake by neurons. The former is mediated by several astrocytic export systems including the Na+-dependent transporters SNAT3 and SNAT5 (encoded by SLC38A3 and SLC38A5) and Na+-independent antiporters such as LAT1 (an SLC7A5/SLC3A2 heterodimer) (Bak et al. 2006). Glutamine uptake by glutamatergic neurons occurs primarily through the concentrative Na+-dependent transporter SNAT1, encoded by SLC38A1 (Fig. 1; Bhutia and Ganapathy 2016).
The glutamate–glutamine cycle also imposes a need for intercellular ammonium (NH4+) cycling to maintain nitrogen balance, since NH4+ is generated in glutamatergic neurons by the glutaminase reaction and consumed in astrocytes during glutamine synthesis. Several questions remain over the transport mechanism for this nitrogen (Rothman et al. 2012). Briefly, any pathway besides direct NH4+ transfer or diffusion requires NH4+ capture in neurons, most likely by GLUD1/2 running in “reverse” to catalyze the reductive amination of α-KG. Transaminases would then use the nitrogen from glutamate to generate nonneuroactive amino acids, providing a safe route for transfer to astrocytes. There is evidence that metabolic cycles involving alanine and/or the branched chain amino acids (BCAAs) contribute to this process, but additional work is required to define the mechanism (Rothman et al. 2012).
An important outcome of the neuron–astrocyte glutamate–glutamine cycle is that the energetic burden of synaptic transmission is distributed between cell types. Glutamate uptake into astrocytes via EAATs occurs against an extreme concentration gradient and is powered by a Na+ electrochemical gradient (Beart and O'Shea 2007; Magi et al. 2019), which is maintained by the ATP-dependent Na+/K+ pump (Rose et al. 2009). Uptake of one glutamate is coupled to symport of three Na+ and one H+ along with antiport of one K+, with the result that every molecule of glutamate transported into astrocytes requires the hydrolysis of ∼1.3 ATPs, with an additional ATP required for glutamine synthesis (Attwell and Laughlin 2001; McKenna 2013). This division of labor preserves neuronal ATP for the energetically expensive maintenance of ion electrochemical gradients and for the concentrative loading of glutamate into synaptic vesicles (Attwell and Laughlin 2001).
Intercellular antioxidant support in the brain
The high demand for ATP in neurons necessitates rapid OXPHOS, an inevitable consequence of which is the production of damaging reactive oxygen species (ROS) (Halliwell 1992). The challenge of neutralizing ROS is exacerbated by a very low intrinsic antioxidant defense capacity in neurons (Murphy et al. 2001; Schmuck et al. 2002; Kraft et al. 2004). In contrast, astrocytes are proficient at mitigating oxidative stress and express high levels of all components of the de novo glutathione biosynthesis pathway along with ROS-scavenging enzymes such as NAD(P)H quinone dehydrogenase 1 (NQO1). Correspondingly, another function of astrocytes in brain metabolism is to support neuronal redox homeostasis via the indirect supply of glutathione (Dringen 2000). Glutathione, a modified tripeptide of glutamate, glycine, and cysteine, is the principal antioxidant and detoxifying metabolite in mammalian cells, serving as a cofactor for the glutathione peroxidase and the glutathione S-transferase families of enzymes (Dringen 2000). A bottleneck for glutathione biosynthesis in neurons is the acquisition of the rate-limiting metabolite cysteine, which exists primarily in the oxidized cystine form in the extracellular environment (Dringen et al. 1999; Dringen 2000). Neurons do not express the cystine/glutamate antiport system xCT, whereas this transporter is abundant on astrocytes, where it mediates glutamate-powered cystine uptake (Dringen 2000; Pow 2001; McGann and Mandel 2018; Combs and Denicola 2019). Transfer of reduced thiols from astrocytes to neurons involves astrocytic secretion of glutathione into the extracellular space, where γ-glutamyl transpeptidase (GGT1) on the astrocyte membrane cleaves glutathione to generate a cysteine–glycine dipeptide. This in turn is cleaved by aminopeptidase N (ANPEP) on the surface of neurons, yielding glycine and the reduced form of cysteine, which is then available for uptake by SLC1 family transporters (Dringen et al. 1999, 2001; Dringen 2000; Bélanger et al. 2011).
Intercellular partitioning of lipid metabolism in the brain
Polyunsaturated lipids readily react with ROS to form highly toxic lipid peroxides and aldehydes, which can initiate ferroptotic cell death (Gaschler and Stockwell 2017; Su et al. 2019). This is particularly relevant in the brain, as lipids account for ∼50% of the brain's dry mass, making it the most lipid-rich organ in the body after adipose tissue. Brain lipids are not only highly abundant but also extremely diverse, with ∼75% of the total number of lipid species in mammals found exclusively in the CNS (Fitzner et al. 2020). Each cell type in the brain has a unique lipid profile, and the expression of lipid metabolism genes varies accordingly. For example, neurons are enriched with cholesterol and ceramides, oligodendrocytes are enriched with the myelin lipids galactosylceramide and sulfatide, and astrocytes are enriched with phosphatidylinositol, phosphatidylserine, and diacylglycerol (Fitzner et al. 2020). These differences reflect intercellular partitioning of lipid metabolism in the brain, and high levels of lipoproteins, particularly apolipoprotein E (ApoE) and ApoJ, facilitate lipid transfer between cells. Lipid transport is highly dynamic and plays important roles in maintaining homeostasis in response to stress, including that caused by oxidative challenge.
In addition to their low cell-intrinsic antioxidant capacity, neurons have a limited ability to catabolize fatty acids or store them in lipid droplets as inert triacylglycerides, exacerbating their sensitivity to lipid peroxides (Ioannou et al. 2019). Astrocytes, in contrast, are highly proficient at fatty acid disposal via mitochondrial β-oxidation and can also detoxify peroxides and sequester excess fatty acids in lipid droplets (Fig. 2; Smolič et al. 2021; Mi et al. 2023). Evidence for neuron–astrocyte coupling of lipid metabolism includes the observation that neuronal oxidative stress triggers the apolipoprotein-dependent accumulation of astrocytic lipid droplets (Ioannou et al. 2019; Smolič et al. 2021). During periods of high neuronal activity with increased OXPHOS and ROS production, peroxidated fatty acids are generated in neurons but are rapidly expelled onto ApoE (Ioannou et al. 2019). The fatty acid-laden lipoprotein is subsequently endocytosed by astrocytes, where the cargo is initially processed into lipid droplets. Simultaneously, enhanced neuronal activity signals to activate β-oxidation in astrocytes, ultimately leading to turnover of lipid droplets (Ioannou et al. 2019). The benefits of minimizing β-oxidation in neurons include the avoidance of superoxide production associated with this pathway, as well as the intrinsic mismatch between the slow dynamics of fatty acid oxidation and the rapidly fluctuating bioenergetic needs of these cells (Schönfeld and Reiser 2013).
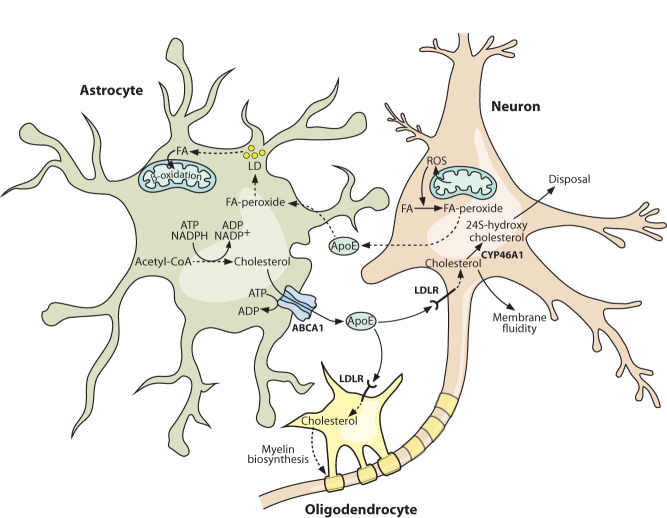
Figure 2.
Intercellular partitioning of lipid metabolism protects against activity-induced toxicity and supplies essential membrane components. Periods of intense neuronal activity lead to increased ROS generation in neurons, which in turn can generate highly damaging reactive lipid peroxide species. The lipid peroxides are efficiently exported from neurons via loading onto apolipoprotein E (ApoE) particles, which are subsequently taken up by astrocytes. Here, the lipid peroxides are detoxified and stored in lipid droplets before being oxidized to generate ATP. Astrocytes are also the major source of cholesterol in the brain, synthesizing it from acetyl-CoA building blocks in an ATP- and NADPH-intensive process. The cholesterol is exported from astrocytes and loaded onto ApoE particles before being taken up and used by neurons and oligodendrocytes. Cholesterol disposal in the brain occurs in neurons and involves the CYP46A1-catalyzed production of 24S-hydroxycholesterol, which can diffuse out across the blood–brain barrier. Solid lines indicate direct processes including movement of a molecule or single-step reactions, and dashed lines indicate indirect or multistep processes. (ABCA1) ATP-binding cassette transporter A1, (ApoE) apolipoprotein E, (FA) fatty acid, (LD) lipid droplet, (LDLR) low-density lipoprotein receptor, (ROS) reactive oxygen species.
In contrast to fatty acids, which are able to traverse the BBB regardless of their chain length and degree of saturation (Spector 1988; Guest et al. 2013), cholesterol is BBB-impermeable and must be synthesized in situ. Despite this isolation from circulating cholesterol supplies, the brain is the most cholesterol-rich organ in the body, containing ∼20% of the body's cholesterol pool, primarily in the unesterified form (Dietschy 2009). Approximately 70%–80% of brain cholesterol is present as a structural component of myelin sheaths (Zhang and Liu 2015), but cholesterol also has essential roles in cellular signal transduction and membrane fluidity (Hussain et al. 2019). Cholesterol metabolism in the brain is sharply partitioned between cell types, with ApoE again mediating intercellular cholesterol transfer (Fig. 2; Li et al. 2022). De novo cholesterol biosynthesis is metabolically costly, requiring ATP, reducing power in the form of NADPH, and a supply of acetyl-CoA, from which all 27 carbon atoms are derived. Fitting with their metabolic support role in the CNS, astrocytes are the major site of de novo cholesterol biosynthesis, supplying almost all of the cholesterol that is found in neurons (Camargo et al. 2017; Li et al. 2022). The ATP-powered transporter ABCA1 catalyzes cholesterol efflux from astrocytes onto extracellular ApoE, which is endocytosed by oligodendrocytes and neurons after binding the low-density lipoprotein receptor (LDLR) (Fig. 2; Li et al. 2022). Oligodendrocytes incorporate cholesterol into the myelin structures that ensheathe neurons, whereas neurons are the site of cholesterol turnover in the brain, carrying out the CYP46A1-dependent conversion of cholesterol into 24S-hydroxycholesterol, a more polar metabolite that can exit the CNS by diffusing across the BBB (Fig. 2; Li et al. 2022).
The N-acetylaspartate enigma
N-acetylaspartate is an enigmatic metabolite straddling lipid, glucose, and amino acid metabolism. After glutamate, it is the most abundant metabolite in bulk brain lysate at a concentration of ∼10 mM, but outside of the CNS it is present at high levels only in adipose tissue (Moffett et al. 2007; Amaral et al. 2013). In the brain, NAA is synthesized in neurons from aspartate and acetyl-CoA by the mitochondrial enzyme N-acetylaspartate synthetase (NAT8L) (Wiame et al. 2010). Conversely, hydrolysis of NAA back into aspartate and acetate occurs almost exclusively in oligodendrocytes and is catalyzed by the cytosolic enzyme aspartoacylase (ASPA) (Baslow et al. 1999; Madhavarao et al. 2004). NAA turnover in the brain is rapid, indicating a high flux of this metabolite between neuronal synthesis and oligodendrocytic disposal (Baslow 2002). The importance of maintaining NAA homeostasis has been highlighted by studies of Canavan disease, a lethal pediatric neurological disorder characterized by the degeneration of myelin structures throughout the CNS (Hoshino and Kubota 2014). Canavan disease is caused by homozygous loss-of-function mutations in ASPA, which abolish the ability of oligodendrocytes to catabolize NAA (Matalon and Matalon 2015). However, the function of NAA in the CNS has not been fully defined, and the exact pathological mechanism of Canavan disease remains elusive. Certain hallmarks of Canavan disease in Aspa-null mouse models, including demyelination and neuronal death, are reversed by concurrent Nat8l knockout to abolish NAA synthesis, suggesting that these phenotypes are caused by accumulation of NAA to toxic concentrations (Janson 2015; Maier et al. 2015; Gessler et al. 2017; Sohn et al. 2017). However, Nat8l deletion fails to rescue survival in these models, and the only known human patient with NAT8L deficiency has severe neurological symptoms, confirming that both neuronal NAA biosynthesis and neuron-to-oligodendrocyte NAA shuttling have critical physiological roles (Wiame et al. 2010). For now, NAA remains a poorly understood metabolite, and the various models for its role in the CNS have been discussed in detail elsewhere (Elliott et al. 2018).
Glucose and the astrocyte–neuron lactate shuttle hypothesis
In the resting brain, glycolysis and OXPHOS are tightly coordinated, allowing for almost complete oxidation of glucose to CO2. The oxygen–glucose index (OGI) describes the molar ratio of O2 used per glucose consumed and has an ideal value of 6 when glucose is fully oxidized in the absence of other metabolic substrates (i.e., six O2 are required for complete oxidation of one glucose, yielding six CO2 and six H2O, with ∼32 ATPs synthesized) (Flurkey 2010). The OGI of the resting brain is close to ideal stoichiometry at ∼5.5, but this value decreases to ∼5 upon neuronal stimulation, indicating a partial uncoupling of glycolysis and OXPHOS (Fox et al. 1988). Correspondingly, brain lactate levels transiently increase due to fermentative metabolism of a fraction of glucose-derived pyruvate to lactate instead of oxidation to CO2. The “astrocyte–neuron lactate shuttle” (ANLS) hypothesis proposes that glutamatergic signaling stimulates astrocytes to ferment glucose to lactate, which is then transferred to neurons for use as a respiratory fuel (Pellerin and Magistretti 1994). However, this model is controversial and has been contested on both theoretical and experimental grounds (Dienel 2017; Yellen 2018; Bonvento and Bolaños 2021). Briefly, it is clear that neurons have the capacity to use both lactate and glucose for ATP synthesis, and the debate centers on which nutrient constitutes the major neuronal fuel in vivo. Arguing against the ANLS, recent work has shown that neurons respond to stimulation by increasing glucose consumption and glycolysis without a requirement for lactate respiration (Díaz-García et al. 2017). Although astrocytic glycolysis is essential for neuronal health and overall brain function (Volkenhoff et al. 2015; Muraleedharan et al. 2020), this can be explained by many mechanisms that do not involve lactate supply to neurons. For example, glycolytic blockade in astrocytes would severely impair the glutamate–glutamine cycle. Despite the arguments against the ANLS hypothesis, lactate is relatively abundant in the brain ISF (∼1 mM), and its transfer between cell types might still be physiologically important in some contexts (Philips and Rothstein 2017; Yellen 2018; Wang et al. 2019a; Hollnagel et al. 2020; Karagiannis et al. 2021).
Ketone bodies as an alternative energy source
During fasting or on ketogenic diet, glucose is supplemented by the two principal ketone bodies, acetoacetate and β-hydroxybutyrate, as a fuel source for the brain (Morris 2005; Puchalska and Crawford 2017). The primary site of ketogenesis in the body is the liver, where the precursor metabolite acetyl-CoA is generated from fatty acids via β-oxidation (>95% of circulating ketone bodies) and from ketogenic amino acids (Evans et al. 2017). Matching their respective capacities for β-oxidation, neurons are incapable of ketogenesis, whereas astrocytes are proficient at this process (Takahashi et al. 2014; Takahashi 2020; Silva et al. 2022). An astrocyte–neuron ketone body shuttle has been proposed, but since peripheral ketone bodies can enter the brain through monocarboxylate transporters (MCTs) in the BBB, the physiological roles of astrocytic ketogenesis are not clear (Guzmán and Blázquez 2001; Takahashi et al. 2014). In Drosophila, glial-synthesized ketone bodies sustain memory formation under starvation conditions, and it will be of interest to determine whether these findings extend to the mammalian brain, particularly during aging when CNS glucose metabolism becomes impaired (Cunnane et al. 2020; Silva et al. 2022).
The nutrient environment influences microglia polarization
Microglia are the primary brain-resident immune cell population and perform a number of essential immune surveillance and homeostatic roles in the healthy brain (Tremblay et al. 2011). Microglia have a distinct yet plastic metabolic phenotype, which allows them to integrate into brain metabolic cycles and to function in a nutrient environment characterized by relatively low levels of key fuels such as glucose. One feature of resting microglial metabolism is a strong reliance on OXPHOS for ATP generation (Holland et al. 2018; Song et al. 2022), allowing for the efficient harnessing of energy from nutrients and simultaneously minimizing metabolic competition with other cell types. Microglia also display flexibility in their use of available nutrients. Glucose is the preferred fuel, and microglia express the glucose transporter GLUT1 (encoded by SLC2A1) in common with other glial cells, as well as the high-capacity, high-affinity, transporter GLUT3 (encoded by SLC2A3) in common with neurons (Kalsbeek et al. 2016; Aldana 2019; Wang et al. 2019b). However, in nonhomeostatic conditions when glucose levels are compromised, microglia can switch to glutamine as the primary bioenergetic substrate, which as with neurons is likely supplied by astrocytes (Bernier et al. 2020). This ability to switch between glucose and glutamine facilitates the continued surveillance function of microglia and is not associated with a change in cellular phenotype or polarization (Bernier et al. 2020).
Like other immune cells, microglia adopt different states in response to a variety of cues, ranging from anti-inflammatory wound healing phenotypes to proinflammatory phenotypes associated with many disease states. Microglia polarization has historically been viewed as involving a proinflammatory “M1-like” or anti-inflammatory “M2-like” state, but it should be noted that these categorizations were adopted from the study of peripheral macrophages and are often an oversimplification of microglial phenotypes in vivo (Cherry et al. 2014). Microglia polarization is regulated by proteins, cytokines, and metabolites, and metabolic processes also reinforce specific polarization states. Thus, there is cross-talk and feedback between metabolic activity in the brain and microglial phenotype, helping to ensure that these cells support normal brain function and the maintenance or restoration of homeostasis.
Collectively, the above examples illustrate how the division of metabolic processes across the multiple cell types in the brain allows for greater specialization of cellular function and allocates energy-intensive or toxic waste management tasks to the cells best equipped to deal with them. During tumorigenesis, these symbiotic relationships can be exploited or hijacked by GBM cells. An emerging theme is that, far from being metabolically isolated, GBM cells integrate into or disrupt diverse metabolic processes in the brain, obtaining metabolic fuels from noncancer cells and modulating the nutrient environment to advance tumor progression.
Metabolic interactions in the GBM microenvironment
The metabolic requirements of solid tumors are heterogeneous and distinct from those of their host tissue. Sustained activation of anabolic pathways in proliferative cancer cells requires a continuous supply of ATP and reducing equivalents within a TME characterized by fluctuating and limited levels of many nutrients, including O2 (De Berardinis and Chandel 2016). Although there is no single universal metabolic phenotype of cancer, certain characteristics are broadly conserved across a spectrum of malignancies (Pavlova and Thompson 2016; Pavlova et al. 2022). One such hallmark of cancer metabolism is the use of opportunistic modes of nutrient acquisition to obtain material from the TME (Pavlova et al. 2022). Cancer cells also co-opt stromal cells for metabolic support, which can involve stroma-to-cancer delivery of metabolic fuels and cancer-to-stroma transfer of waste products for disposal or recycling (Sousa et al. 2016; Wilde et al. 2017; Yan et al. 2018; Banh et al. 2020). In GBM, cancer cells interact with multiple brain-resident cell types, with astrocytes again having particularly important metabolic support roles. Indeed, many GBM–brain-resident cell metabolic interactions recapitulate or parasitize existing metabolic couplings in the healthy brain, as discussed further below. It should be noted that these stromal cells within the TME of GBM are frequently in altered nonhomeostatic states, such as reactive astrogliosis, which can be induced by changes in the metabolic environment and further contribute to disease progression. Relatedly, the metabolic secretome of GBM cells profoundly influences the immune microenvironment, promoting an immune-suppressive phenotype that might be pharmacologically targeted to potentiate immune therapies. In the following sections, we first cover examples of metabolic coupling that help to fuel GBM growth and then discuss metabolic suppression of the anticancer immune response and the therapeutic opportunities that arise from these interactions. While this review focuses on the metabolic interactions contributing to GBM progression, additional nonmetabolic aspects of the TME also impact GBM pathology and have recently been reviewed elsewhere (Gillespie and Monje 2018; Crivii et al. 2022).
Hijacking of the neuron–astrocyte glutamate–glutamine cycle by GBM
Many cancer cell lines are dependent on an exogenous supply of glutamine in ex vivo culture despite this being a nonessential amino acid that can be synthesized de novo by GLUL. In cultured GBM cell lines, glutamine is the second most rapidly consumed nutrient after glucose and serves as a major anaplerotic carbon source for the TCA cycle. This pathway is initiated by the GLS-catalyzed hydrolysis of glutamine to glutamate, underlying the sensitivity of cultured GBM cells to GLS inhibitors (Wise et al. 2008; Tardito et al. 2015; Yang et al. 2021). However, in GBM tumors in vivo, the pathways of glutamine metabolism differ from those in cultured cells, a phenomenon explained by differences in the nutrient environment of the brain ISF and additional features of the TME including tumor–stroma interactions (Natarajan and Venneti 2019; Oizel et al. 2020). There is evidence that GBM cells partially integrate into the neuron–astrocyte glutamate–glutamine cycle, exploiting astrocytes as an exogenous source of glutamine and even participating in glutamatergic neurotransmission (Fig. 3).
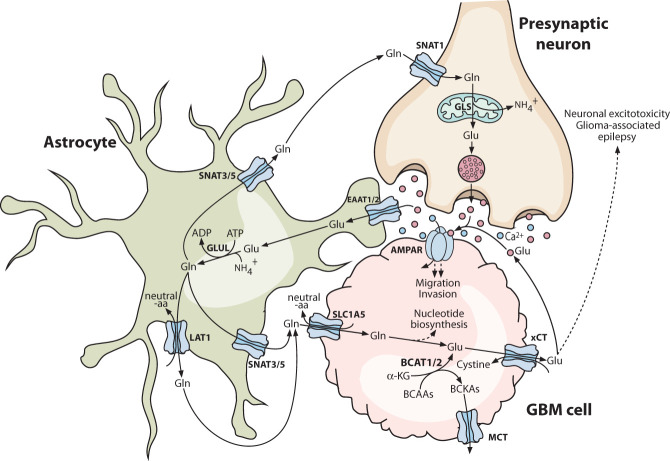
Figure 3.
Hijacking of the neuron–astrocyte glutamate–glutamine cycle by glioblastoma. Glioblastoma cells express glutamate receptors such as AMPAR and can form functional synapses with glutamatergic neurons, with calcium influx through glutamate receptors contributing to increased migration and invasion of glioblastoma cells. Simultaneously, glioblastoma cells can exploit astrocytes as a source of glutamine for anabolic reactions such as nucleotide biosynthesis. Many of these reactions convert glutamine into glutamate, which drives cystine uptake via the cystine/glutamate antiporter xCT, providing the glioblastoma cells with their primary source of cysteine. Glutamate export via xCT has other consequences, including autocrine glutamate receptor signaling and induction of neuronal excitotoxicity, contributing to the progression and symptomology of glioblastoma. Solid lines indicate direct processes including movement of a molecule or single-step reactions, and dashed lines indicate indirect or multistep processes. (aa) Amino acid, (α-KG) α-ketoglutarate, (AMPAR) α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor, (BCAA) branched chain amino acid, (BCKA) branched chain α-ketoacid, (EAAT) excitatory amino acid transporter, (GBM) glioblastoma multiforme, (Gln) glutamine, (GLS) glutaminase, (Glu) glutamate (GLUL) glutamine synthetase, (MCT) monocarboxylate transporter.
Stable isotope tracing studies have revealed a lack of net glutamine catabolism via the canonical GLS-initiated pathway in IDH1 wild-type GBM in vivo. Instead, an intratumoral glutamine pool accumulates, which is synthesized in situ and is important for sustaining de novo nucleotide biosynthesis, for which glutamine is the obligate nitrogen donor in five separate reactions (Marin-Valencia et al. 2012; Tardito et al. 2015). Some intratumoral glutamine production might occur in GSC cells, which have elevated GLUL expression relative to non-stem-like GBM cells. However, GLUL-negative GBM cells in tumors reside in close proximity to GLUL-positive astrocytes, implicating the latter as the primary source of glutamine for anabolic metabolism in GBM. Supporting this model, astrocyte-derived glutamine is sufficient to enable growth of GLUL-negative GBM cells in coculture experiments using glutamine-depleted medium (Tardito et al. 2015). Thus, GBM cells appear to exploit the normal physiological role of astrocytes as the glutamine supply cells of the CNS and potentially outcompete neurons for glutamine by expressing the high-affinity uptake transporter SLC1A5 (Fig. 3; Han et al. 2022).
In contrast to IDH1 wild-type GBM, the ∼12% of grade 4 gliomas that harbor oncogenic IDH1 mutations exhibit intratumoral glutamine depletion relative to the surrounding brain tissue (Andronesi et al. 2018). Here, glutamine serves as a carbon source for 2-HG production via its sequential conversion to glutamate and then α-KG, the substrate for neomorphic IDH1 variants. This is an extra burden in addition to the usual fates of glutamate, which include protein synthesis and sustaining glutathione biosynthesis both directly as a substrate and by driving cystine uptake via xCT (Fack et al. 2017). Consistent with these observations, oncogenic IDH1 mutations sensitize cells to GLS inhibitors (Seltzer et al. 2010). The feasibility of extending these findings into the clinic remains unclear, as the first-in-class GLS inhibitor CB-839 has poor BBB penetrance, and passage of a GLS inhibitor into the CNS would likely have undesirable impacts on glutamatergic neurotransmission due to the blockade of glutamate production in presynaptic neurons. While the canonical effect of 2-HG is to inhibit α-KG-dependent dioxygenases, it also blocks the active site of transaminase enzymes, particularly branched chain aminotransferase 1 and 2 (BCAT1/2) (McBrayer et al. 2018). The BCAT reaction reversibly converts BCAAs into their corresponding branched chain α-ketoacids (BCKAs), with the amine group transferred onto α-KG to generate glutamate (Fig. 3). Competitive inhibition of BCAT1/2 by 2-HG therefore prevents IDH1 mutant cells from using BCAAs for glutamate production, further increasing their dependence on exogenous glutamine and the GLS reaction (McBrayer et al. 2018). Correspondingly, in IDH1 mutant glioma, the expression of the BCAT1 gene is typically epigenetically silenced and GLS or GLS2 expression is up-regulated, whereas in IDH1 wild-type GBM, BCAT1 is almost universally highly expressed and its knockdown or inhibition suppresses tumorigenesis (Tönjes et al. 2013; McBrayer et al. 2018; Dekker et al. 2022). Both products of the BCAT1 reaction, glutamate and BCKAs, are secreted by IDH1 wild-type GBM cells via xCT and MCTs, respectively, and these metabolites modulate the TME through mechanisms including the activation of glutamatergic signaling (Fig. 3; Tönjes et al. 2013; Silva et al. 2017; Cai et al. 2022).
Glutamatergic signaling and synaptic integration of GBM
Since IDH1 wild-type GBM cells secrete copious amounts of glutamate, the extracellular glutamate concentration in the TME vastly exceeds that in normal brain tissue (Takano et al. 2001; Marcus et al. 2010). This triggers autocrine and paracrine glutamatergic signaling events in GBM, which promote tumorigenesis and contribute to additional pathologies that impact patient quality of life, such as tumor-associated epilepsy and excitotoxic neuronal cell death (Buckingham et al. 2011; Lange et al. 2021). GBM cells typically lack EAAT family glutamate uptake transporters and instead conduct net glutamate export coupled to cystine consumption via the xCT system (Fig. 3; Ye et al. 1999; Buckingham et al. 2011). High levels of xCT are a biomarker for epileptic seizures in GBM, supporting a direct connection between cancer cell glutamate secretion and neuronal hyperstimulation (Buckingham et al. 2011; Sørensen et al. 2018). Sustained stimulation of glutamatergic NMDA receptors also drives neuronal excitotoxicity in the vicinity of GBM tumors, and the resulting neuronal cell death increases inflammation in the TME and potentially drives more extensive necrosis (Takano et al. 2001; Noch and Khalili 2009). GBM cells frequently express AMPA family glutamate receptors lacking the GluA2 subunit that normally prevents Ca2+ permeability. This allows for glutamate-triggered autocrine Ca2+ signaling cascades, which stimulate GBM cell migration and invasion (Fig. 3; Lyons et al. 2007).
In addition to this autocrine signaling, some GBM cells are directly innervated by neurons and thereby also receive paracrine signals from glutamatergic neurotransmission (Fig. 3; Venkataramani et al. 2019, 2022; Venkatesh et al. 2019; Wirsching and Weller 2020). Specifically, callosal projection neurons (CPNs) projecting from the contralateral brain hemisphere innervate the tumor, and the activity of these neurons promotes overall GBM progression and invasion (Huang-Hobbs et al. 2023). Single-cell transcriptomics studies have shown that a distinct GBM cell subset expresses multiple synaptic genes, including selective up-regulation of AMPA- and kainate-type glutamate receptor subunits. Synaptic gene expression is particularly enriched in OPC-like cells (Venkataramani et al. 2019; Venkatesh et al. 2019), and it is notable that in normal brain development and function, glutamatergic synapses form between neurons and the OPC stem cell population (Bergles et al. 2000). Adding to this, spatial transcriptomics has shown that a population of GBM cells at the invasive edge of the tumor is activity dependent on and enriched in axon guidance gene expression, with semaphorin-4F (SEMA4F) a key regulator of activity-dependent GBM progression (Huang-Hobbs et al. 2023). As with the autocrine glutamatergic signaling described above, the functional chemical synapses that connect presynaptic neurons with postsynaptic GBM cells trigger Ca2+ currents that stimulate proliferation and invasion in GBM cells (Fig. 3).
Neuron-derived NAA as a potential metabolic fuel for GBM
In the healthy brain, NAA is synthesized and secreted almost exclusively by neurons and is then taken up by oligodendrocytes via the Na+-dependent dicarboxylate transporter SLC13A3 and catabolized by ASPA. GSCs also express SLC13A3 and ASPA and thus are capable of acquiring and hydrolyzing NAA into acetate and aspartate, which are potential fuels for proliferative metabolism (Long et al. 2013). Intriguingly, supplementation of culture medium with NAA not only boosts GSC proliferation but also suppresses differentiation, possibly due to the increased intracellular acetate level altering histone acetylation patterns (Long et al. 2013).
Lipid metabolism in the GBM microenvironment
Fatty acid metabolism in GBM is heterogenous and responsive to fluctuations in the nutrient environment, such that de novo fatty acid synthesis and the reverse process of fatty acid β-oxidation both contribute to GBM pathogenesis (Miska and Chandel 2023). Analysis of GBM tissue has revealed that lipid droplets are highly enriched in tumor cells relative to cells in the surrounding brain parenchyma (Taïb et al. 2019). Lipid droplets serve as inert energy storage organelles whose triacylglyceride contents can be hydrolyzed during nutrient starvation to yield free fatty acids, which are then catabolized via β-oxidation to provide substrates for OXPHOS. Consistent with this role, GBM cells with a high lipid droplet content are relatively resistant to glucose deprivation and adapt to this stress by increasing the lysosomal fusion of lipid droplets and subsequent autophagic processing of their contents to supply fatty acids (Hoang-Minh et al. 2018; Kant et al. 2020; Wu et al. 2020). As in astrocytes, this coupled use of lipid droplets and the β-oxidation pathway also protects GBM cells from the accumulation of toxic lipid peroxide species (Puca et al. 2021).
Several studies have identified a distinct lipid profile in GSCs relative to other GBM cells, resulting from expression of the genes encoding fatty acid synthase (FASN) and fatty acid desaturases, which in turn leads to increased production of polyunsaturated fatty acids (PUFAs) (Yasumoto et al. 2016; Shakya et al. 2021; Parik et al. 2022). Certain PUFAs, including arachidonic acid, are signaling metabolites that help to sustain self-renewal capacity in stem-like cells, and consequently pharmacological inhibition of FASN decreases the expression of stemness markers in GSCs (Yasumoto et al. 2016). A number of questions remain about the precise balance between fatty acid synthesis and β-oxidation in GBM. Since lipid droplets are enriched in hypoxic regions of the tumor (Shakya et al. 2021), we speculate that the availability of O2 for fatty acid catabolism is one factor influencing this balance. Another unanswered question, given the low concentration of free fatty acids in the brain ISF (Seyer et al. 2016), is how GBM cells that are engaged in β-oxidation and/or lipid droplet accumulation are supplied with these nutrients, including whether lipoprotein-mediated transport from other cell types such as neurons occurs within the TME (Fig. 4).
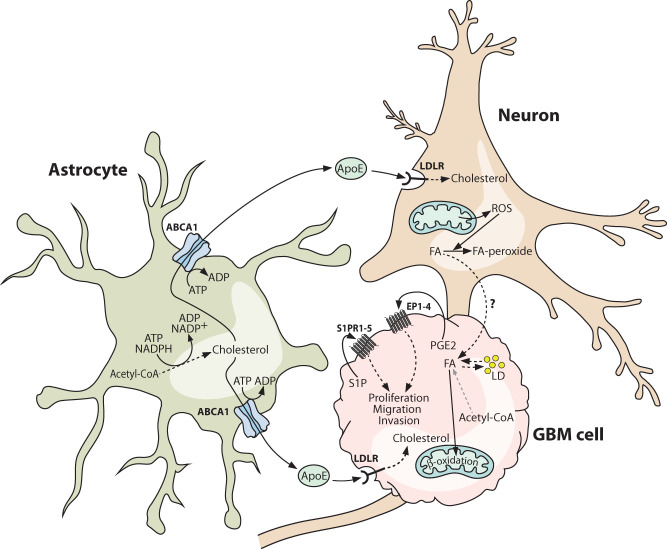
Figure 4.
Brain lipid metabolism is co-opted by glioblastoma cells. Glioblastoma cells exploit astrocytes as a source of cholesterol and may obtain fatty acids from neighboring neurons. Astrocyte-derived cholesterol is loaded onto apolipoprotein E particles and can be taken up by glioblastoma cells via the low-density lipoprotein receptor (LDLR). Glioblastoma cells typically suppress the endogenous cholesterol synthesis pathway, making this astrocyte-derived cholesterol essential for supporting proliferation and normal membrane functions. Glioblastoma cells may also scavenge fatty acids from neighboring neurons and oxidize them to generate ATP under nutrient-starved conditions. Under nutrient-replete conditions, glioblastoma cells are capable of synthesizing fatty acids, which are stored in lipid droplets or used for the synthesis of lipids for membrane biogenesis. A number of lipid species function as signaling metabolites, with prominent examples being sphingosine-1-phosphate and prostaglandin E2, both of which are enriched in glioblastoma. Both molecules engage in autocrine and paracrine signaling, which can remodel the tumor vasculature and promote the proliferation and invasion of glioblastoma cells. Solid lines indicate direct processes including movement of a molecule or single-step reactions, and dashed lines indicate indirect or multistep processes. (ABCA1) ATP-binding cassette transporter A1, (ApoE) apolipoprotein E, (EP1–4) prostaglandin E2 receptor 1–4, (FA) fatty acid, (GBM) glioblastoma multiforme, (LD) lipid droplet, (LDLR) low-density lipoprotein receptor, (PGE2) prostaglandin E2, (ROS) reactive oxygen species, (S1P) sphingosine-1-phosphate, (S1PR1–5) sphingosine-1-phosphate receptor 1–5.
Many tumors, including GBM, contain high levels of cholesterol, which is typically particularly enriched in mitochondria and protects cancer cells from apoptosis by lowering mitochondrial membrane fluidity to block Bax activation (Lucken-Ardjomande et al. 2008; Montero et al. 2008; Perelroizen et al. 2022). However, in contrast to their context-dependent capacity for fatty acid synthesis, GBM cells appear to be strictly auxotrophic for cholesterol. Expression of genes related to cholesterol biosynthesis are suppressed in GBM tissue relative to the normal brain, whereas levels of LDLR, which mediates the endocytosis of cholesterol-laden ApoE, are elevated (Villa et al. 2016). Consistent with a lack of de novo cholesterol biosynthesis, cultured GBM cells are unaffected by treatment with statins, which inhibit the rate-limiting step of this pathway, and instead are highly dependent on the presence of cholesterol-carrying lipoproteins in the culture medium. Reciprocally, normal human astrocytes are unaffected by removal of exogenous cholesterol but are killed by statin treatment, matching their known role as the site of cholesterol biosynthesis in the healthy brain (Villa et al. 2016; Perelroizen et al. 2022). Conditioned medium from GBM cells stimulates increased cholesterol biosynthesis and efflux by astrocytes, and astrocyte coculture completely rescues GBM cell proliferation in lipoprotein-depleted medium (Perelroizen et al. 2022). In vivo, the cholesterol efflux transporter ABCA1 is up-regulated in GBM-associated astrocytes, and targeted knockdown by RNAi depletes cholesterol in GBM tissue and triggers massive apoptotic cell death, leading to tumor regression and prolonged survival (Perelroizen et al. 2022). Collectively, these results implicate astrocyte-to-GBM cell cholesterol transfer as an essential metabolic coupling for GBM growth in vivo (Fig. 4).
Autocrine and paracrine lipid signaling in the GBM microenvironment
In addition to structural and metabolic roles, numerous lipid species are mediators of intracellular and intercellular signal transduction. Within the GBM microenvironment, autocrine and paracrine signals are transmitted by diverse bioactive lipid families including the sphingolipids and eicosanoids (Nathoo et al. 2004; Hawkins et al. 2020). The sphingolipid sphingosine-1-phosphate (S1P) is highly enriched in GBM relative to normal brain tissue, and up-regulation of the enzyme that catalyzes its production, sphingosine kinase 1 (SPHK1), correlates with poor prognosis (Van Brooklyn et al. 2005; Abuhusain et al. 2013). Autocrine S1P signaling via the G protein-coupled S1P receptors S1PR1–5 is mitogenic and also stimulates GBM cell migration and invasion (Fig. 4), and paracrine signaling to vascular endothelial cells constitutes a VEGF-independent angiogenic axis (Van Brooklyn et al. 2005; Abuhusain et al. 2013; Grassi et al. 2019).
Eicosanoids are oxidized derivatives of PUFAs and include the prostanoid and leukotriene subfamilies of lipid mediators. Arachidonic acid, which is generated from phospholipids by phospholipase A2 (PLA2), is a precursor for several eicosanoids, among which prostaglandin E2 (PGE2) is particularly important in GBM pathology. Up-regulation of cyclooxygenase-2 (COX-2), the enzyme that initiates the conversion of arachidonic acid into PGE2, correlates with poor prognosis in GBM patients (Dean and Hooks 2023). Analogous to S1P signaling, externalized PGE2 engages G protein-coupled PGE2 receptors (EP1–4), driving autocrine mitogenic and proinvasion signaling in GBM cells and stimulating angiogenesis (Fig. 4; Jiang et al. 2017; Dean and Hooks 2023). In addition to their roles in regulating proliferation, migration, and angiogenesis, both S1P and PGE2 contribute to immune suppression in the GBM microenvironment, as described in the following section.
Metabolic modulation of the GBM immune microenvironment
A hallmark of GBM is a profoundly immune-suppressive TME, with massive accumulation of immune cells in dysfunctional states. Tumor-associated myeloid cells (TAMCs), including microglia, macrophages, and myeloid-derived suppressor cells (MDSCs), are the most abundant immune cells in GBM, comprising up to 50% of the total number of cells in the tumor (Xuan et al. 2021). The extent of TAMC infiltration correlates positively with GBM progression, and this population of cells mediates immune suppression by impairing the activity of immune effector cells and promoting the induction and recruitment of immune-suppressive regulatory T cells (Tregs) (Gabrilovich and Nagaraj 2009; Thomas et al. 2015). As mentioned above, myeloid cells including microglia are highly plastic and can differentiate into distinct polarization states, which historically were categorized as proinflammatory “classically activated” (M1) and anti-inflammatory “alternatively activated” (M2). However, it is now appreciated that TAMCs exist in a broad spectrum of dynamic functional states, expressing transcripts linked to both M1 and M2 polarization. This results in the TAMC composition of GBM being highly heterogeneous but skewed toward a state enriched with M2-like characteristics, which is driven in part by immune-suppressive signaling metabolites such as S1P and PGE2 (Arseni et al. 2023; Dean and Hooks 2023). The tumor oncogenotype and its downstream impact on cytokine/chemokine signaling and metabolic phenotype are major determinants of immune suppression (Binnewies et al. 2018; Friedrich et al. 2021). In the context of grade IV glioma, the mutation status of IDH1 is a particularly important regulator of this aspect of the TME. In the following sections, we discuss the key metabolic mechanisms that modulate the anticancer immune response. Additional nonmetabolic factors contributing to immune suppression in GBM have recently been reviewed elsewhere (Gieryng et al. 2017; Grabowski et al. 2021).
IDH1 mutation status impacts glioma immune suppression
As described above, oncogenic mutation of IDH1 results in the production and accumulation of 2-HG to concentrations of 1–30 mM in the TME, with consequences including redox imbalance, metabolic reprogramming, and epigenetic dysregulation. The effects of 2-HG accumulation are not restricted to cancer cells but instead impact the broader TME, including resident immune cell populations. Caution is needed when comparing IDH1 wild-type with IDH1 mutant glioma, as the IDH1 mutation is far more common in LGG (∼80% of cases) than in grade IV disease (∼12% of cases), and differences in the immune microenvironment can be attributable to tumor grade as well as mutation status. Nevertheless, recent use of syngeneic mouse models supports a causative role for oncogenic IDH1 variants in mediating glioma immune suppression (Amankulor et al. 2017; Kohanbash et al. 2017; Friedrich et al. 2021).
Single-cell RNA-seq analysis of human gliomas shows that all cases contain an abundance of TAMCs in a mixture of polarization states, but that overall immune infiltration is more severely suppressed in IDH1 mutant tumors than in IDH1 wild-type GBM. In IDH1 mutant tumors, the vast majority of the myeloid compartment consists of microglia, whereas IDH1 wild-type tumors contain some microglia but are also infiltrated by peripheral monocyte-derived macrophages (MDMs), intermediate monocytes, and, to a lesser extent, neutrophils (Amankulor et al. 2017; Klemm et al. 2020; Yeo et al. 2022). Similarly, peripheral lymphocytes are largely excluded from IDH1 mutant gliomas, whereas IDH1 wild-type GBM has substantial infiltration, particularly by CD4+ T cells. Isogenic mouse glioma models differing only in IDH1 mutation status support the conclusion that these differences in the tumor immune landscape are directly attributable to the oncogenotype (Amankulor et al. 2017; Friedrich et al. 2021). Mechanistically, 2-HG is linked to activation of the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor that induces immune-suppressive cell polarization (Friedrich et al. 2021). Rather than binding AHR directly, 2-HG stimulates tryptophan catabolism to generate the AHR ligand kynurenine.
Tryptophan catabolism promotes immune suppression in GBM
Catabolism of the essential amino acid tryptophan via the kynurenine pathway is a central axis in the metabolic regulation of immunity (Opitz et al. 2011; Campesato et al. 2020). The kynurenine pathway is initiated by the rate-limiting oxidation of the indole ring of tryptophan to generate N-formylkynurenine, a reaction catalyzed by both indoleamine 2,3-dioxygenase 1/2 (IDO1/2) and tryptophan 2,3-dioxygenase (TDO2) (Fig. 5). N-formylkynurenine is then hydrolyzed by arylforamidase (AFMID) to generate kynurenine. Expression of IDO1 is induced by interferon and prostaglandin signaling, whereas TDO2 is a glucocorticoid-responsive gene normally expressed only in the liver (Nakamura et al. 1987; Bianchi et al. 1988; Braun et al. 2005). IDO1 is highly expressed in activated myeloid cells, but both IDO1 and TDO2 are also expressed by cancer cells in multiple tumor types including GBM (Liu et al. 2018; Pham et al. 2021). This tryptophan catabolism axis suppresses immune responses both by depleting tryptophan, which is required for T cell proliferation, and by the action of the kynurenine product on AHR. Binding of kynurenine triggers the release of AHR from an inactive cytosolic complex, allowing nuclear translocation and dimerization of AHR with the AHR nuclear translocator (ARNT) to form the active transcription factor complex (Fig. 5; Opitz et al. 2011).
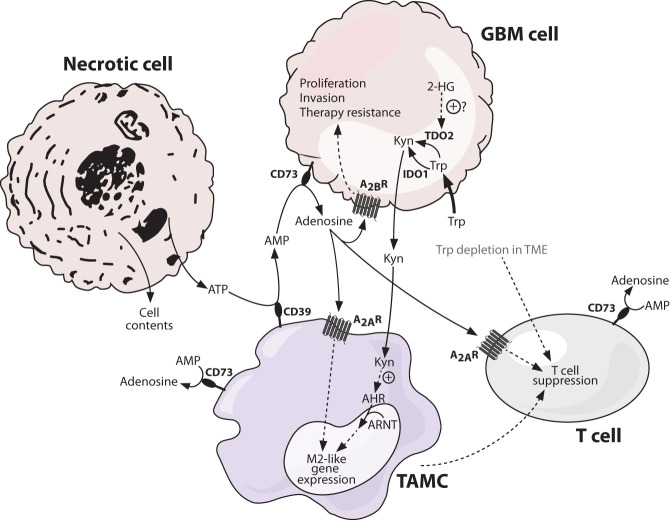
Figure 5.
Immune-suppressive metabolite signaling in the glioblastoma microenvironment. Metabolites play an important role in shaping the tumor immune microenvironment. Tryptophan is an essential amino acid, important for cancer cell proliferation as well as proper T cell functioning. Glioblastoma cells effectively compete with T cells for environmental tryptophan, depleting it and suppressing T cell function in turn. A key fate of tryptophan in glioblastoma cells is its breakdown to kynurenine, mediated by the enzymes IDO1 and TDO2, with the latter activated in isocitrate dehydrogenase 1 (IDH1) mutant glioma in a 2-hydroxyglutarate-dependent manner. Kynurenine thus accumulates in the tumor microenvironment and, as the endogenous ligand of the aryl hydrocarbon receptor (AHR), activates this receptor in neighboring tumor-associated myeloid cells (TAMCs). Here, AHR translocates to the nucleus and binds the aryl hydrocarbon receptor nuclear translocator (ARNT), forming an active complex that drives target gene transcription and contributes to an M2-like gene expression profile. One consequence of this is increased expression of CD39 on the surface of TAMCs. CD39 is a membrane-associated extracellular enzyme that converts ATP to AMP. ATP is not normally found in the extracellular milieu but is abundant in the tumor microenvironment due to the high level of necrotic cell death. AMP produced by CD39 can be converted by CD73 on the surface of GBM cells and various immune cells to adenosine, which has potent anti-inflammatory properties. Adenosine binds to adenosine receptors on the surface of TAMCs and T cells, promoting an M2-like gene expression profile and T cell suppression, respectively. Glioblastoma cells also express adenosine receptors, and signaling through these promotes proliferation, invasion, and therapy resistance. Solid lines indicate direct processes including movement of a molecule or single-step reactions, and dashed lines indicate indirect or multistep processes. (2-HG) 2-hydroxyglutarate, (A2AR) adenosine receptor 2A, (A2BR) adenosine receptor 2B, (AHR) aryl hydrocarbon receptor, (ARNT) aryl hydrocarbon receptor nuclear translocator, (CD39) cluster of differentiation 39 (ectonucleoside triphosphate diphosphohydrolase-1), (CD73) cluster of differentiation 73 (ecto-5′-nucleotidase), (GBM) glioblastoma multiforme, (IDO1) indoleamine 2,3-dioxygenase 1, (Kyn) kynurenine, (TAMC) tumor-associated myeloid cell, (TDO2) tryptophan 2,3-dioxygenase, (TME) tumor microenvironment, (Trp) tryptophan.
Both IDO1 and TDO2 are highly expressed in GBM, and cultured GBM cells avidly secrete kynurenine (Takenaka et al. 2019; Kudo et al. 2020; Zhai et al. 2021). GBM cells also secrete the cytokines IFNβ and interleukin-6 (IL-6), known activators of STAT1 and STAT3, respectively, which in turn drive increased AHR expression in TAMCs. Thus, GBM cells signal to TAMCs to up-regulate AHR gene expression and simultaneously provide kynurenine ligand to activate the AHR protein. Upon activation, AHR coordinates a transcriptional program that skews TAMCs toward an M2-like state, and this in turn blunts the antitumor T cell response (Fig. 5).
Immune-suppressive adenosine signaling in the TME
Yet another metabolic mechanism connects M2-like skewing of TAMCs with T cell suppression and tumor immune escape; namely, the expression of the ectonucleotidase CD39 (encoded by ENTPD1), which initiates production of the immune-suppressive metabolite adenosine. The major route for extracellular adenosine production is a two-step processing of ATP, which is normally present at very low levels in the healthy brain as a neurotransmitter and gliotransmitter but accumulates to micromolar concentrations in the TME due to release from necrotic cells (Fig. 5; Jennings et al. 2021). Extracellular ATP is first hydrolyzed to AMP by CD39, which is present on the surface of TAMCs and vascular endothelial cells. Then, AMP is hydrolyzed to adenosine by the membrane-anchored ecto-5′-nucleotidase CD73 (encoded by NT5E), which is expressed by both cancer cells and immune cells in the TME (Fig. 5; Goswami et al. 2019; Coy et al. 2022). Single-cell spatial transcriptomics analysis of GBM tissue shows that CD39 is expressed by both microglia and macrophage TAMCs and that these cells reside in close proximity to CD73-expressing cancer cells (Coy et al. 2022). Whereas extracellular ATP drives proinflammatory signaling, adenosine generated by the CD39/CD73 pathway potently suppresses inflammation via its activation of adenosine receptors.
There are four G protein-coupled adenosine receptors, A1R, A2AR, A2BR, and A3R, which have different affinities for adenosine (Sheth et al. 2014). All four receptors are expressed by GBM cells, but the low-affinity receptor A2BR, which is only activated by pathologically high adenosine concentrations as found in the TME, is selectively up-regulated relative to nontransformed cells (Yan et al. 2019). Cancer cell-intrinsic effects of A2BR activation include the increased expression of matrix metalloproteinases and multidrug resistance transporters, which promote cancer cell invasion and chemoresistance, respectively (Fig. 5). However, it is the role of the immune cell adenosine receptor A2AR that has received the most attention in GBM. Strikingly, CD39/CD73/A2AR is the most enriched immune-suppressive axis in human glioma samples, with TAMCs and infiltrating lymphocytes consistently expressing A2AR regardless of disease grade (Ott et al. 2020). Similar to the activation of AHR by kynurenine, adenosine-mediated activation of A2AR skews TAMCs toward an M2-like state, suppresses T cell effector activity, and induces Tregs (Fig. 5; Ott et al. 2020).
Targeting intercellular metabolic coupling for GBM therapy
Despite extensive exploration of therapies targeting oncogenic signaling pathways in GBM, long-term prognosis for patients has not improved substantially in decades, and treatment of this disease remains one of the most challenging problems in clinical oncology. This lack of progress has prompted investigation of nononcogene dependencies in GBM, including “metabolic addictions” that arise as a consequence of oncogenic signaling, epigenetic reprogramming, and/or features of the TME (Bi et al. 2019). As with any cancer therapy, a requirement of this approach is that it must achieve sufficient selectivity to eliminate malignant cells while sparing the patient from unacceptable side effects arising from off-target activities (Lee et al. 2018). A promising preclinical example is the targeting of intercellular cholesterol metabolism in GBM, which is discussed in more detail below. However, selective inhibition of oncogenic IDH1 variants represents the cleanest metabolism-targeted therapy for gliomas that harbor IDH1 mutations, as the neomorphic enzyme is present only in cancer cells. Since the initial observation of glioma IDH1 mutations in 2008, the development of such therapeutics has advanced rapidly, and clinical trials of neomorphic IDH1-targeted antibodies and inhibitors have recently yielded promising results in glioma patients (Parsons et al. 2008; Mellinghoff et al. 2021, 2023; Platten et al. 2021; Natsume et al. 2023). Given the connections between IDH1 mutation status and immune suppression in the TME, a logical extension of these studies will be to test whether neomorphic IDH1 inhibition potentiates immune therapies such as immune checkpoint blockade. Indeed, in preclinical models, combination of neomorphic IDH1 inhibition with anti-PDL1 therapy leads to complete tumor regression in 60% of IDH1 mutant glioma-bearing mice (Kadiyala et al. 2021).
Targeting intercellular metabolic dependencies in GBM—cholesterol as an example
Pharmacological blockade of anabolic metabolism is a widely used approach in cancer therapy, exemplified by antimetabolite inhibitors of nucleotide biosynthesis such as antifolates, antipyrimidines, and antipurines (Lukey et al. 2017). However, many of these agents nonselectively target all proliferative cell types, including activated immune cells, and therefore a challenge is to identify metabolic dependencies that are truly unique to cancer cells. As described above, GBM cells are strictly auxotrophic for cholesterol, requiring a supply of this nutrient from astrocytes. Inhibiting de novo cholesterol synthesis in the CNS would likely be detrimental for the host, as astrocytes are killed by blockade of this pathway, but a more selective approach for targeting cholesterol auxotrophy in GBM cells has recently been developed. Cholesterol homeostasis is regulated by the liver X receptors (LXRα/β), which upon activation by cholesterol catabolites heterodimerize with retinoid X receptors (RXRs). The LXR:RXR complex down-regulates LDLR and up-regulates ABCA1 in cholesterol-replete cells, thereby suppressing cholesterol uptake and simultaneously stimulating its efflux (Wang and Tontonoz 2018). Loss of this negative feedback loop due to low levels of endogenous LXR ligands is a unique feature of GBM cell metabolism and allows cholesterol to accumulate to levels that protect from apoptosis (Villa et al. 2016; Perelroizen et al. 2022). This renders GBM cells selectively sensitive to pharmacological LXR agonists, which spare normal astrocytes and neurons (Villa et al. 2016). The brain-penetrant agonist LXR-623 is able to drive cholesterol depletion-dependent death of GBM cells in vivo, leading to inhibition of tumor growth and extended survival in mice (Villa et al. 2016).
Pharmacological targeting of glutamatergic signaling in GBM
As described above, glutamatergic signaling contributes directly to GBM progression while also contributing to complications such as tumor-associated epilepsy via neuronal overactivation. Multiple steps of the GBM-specific glutamatergic signaling pathway can potentially be targeted, such as the cystine/glutamate antiporter xCT, as well as tumor-enriched glutamate receptors themselves. In this regard, several clinical trials have been conducted with talampanel and perampanel, which are noncompetitive inhibitors of the AMPA receptor. While talampanel monotherapy failed to show any clinical benefit for recurrent GBM (Iwamoto et al. 2010), a combination therapy of talampanel with radiation and temozolomide was well tolerated and showed encouraging improvements in survival time compared with radiation and temozolomide alone (Grossman et al. 2009). Additionally, perampanel has proven to be effective at controlling seizures in patients suffering GBM-associated epilepsy while also possibly restraining tumor progression (Izumoto et al. 2018).
Relieving the immune-suppressive metabolic environment in GBM
Immune-suppressive metabolic signaling axes in the GBM microenvironment include the kynurenine and adenosine pathways, both of which are highly amenable to pharmacological blockade. The first step of the kynurenine pathway of tryptophan catabolism can be targeted by inhibitors of IDO1 and/or TDO2. In immune-competent mouse models of GBM, monotherapy with a CNS-penetrant IDO1 inhibitor is ineffective at treating the disease, but trimodal therapy of IDO1 blockade, anti-PD1 therapy, and radiotherapy markedly increases survival, with complete tumor regression and durable survival in 40% of animals (Ladomersky et al. 2018). Given the redundancy between IDO1 and TDO2, dual IDO1/TDO2 inhibitors have also been developed and show improved efficacy relative to IDO1-selective inhibitors (Du et al. 2020). IDO1 and/or TDO2 inhibitors are currently being evaluated in numerous clinical trials, which to date have shown evidence of benefit in subsets of GBM patients (Platten et al. 2019; Opitz et al. 2020; Reardon et al. 2020; Modoux et al. 2021; Fujiwara et al. 2022; Peng et al. 2022). A potential caveat with competitive inhibitors of IDO1/TDO2 is that some of these compounds are structurally similar to kynurenine and can themselves be activating ligands for AHR, thus bypassing the intended disruption of this immune-suppressive axis (Moyer et al. 2017). An alternative approach that avoids this complication is to use direct antagonists of AHR. Such inhibitors show efficacy both as a monotherapy and in combination with anti-PD1 therapy in models of several solid tumors (Campesato et al. 2020; McGovern et al. 2022) but to our knowledge have not yet been extensively evaluated in animal models of GBM.
Preclinical studies also support the continued investigation of adenosine signaling blockade in GBM therapy (Jin et al. 2021; Bova et al. 2022). Inhibition of the adenosine receptor A2BR, which is highly up-regulated on GBM cells (Fig. 5), increases the chemosensitivity in ex vivo culture (Daniele et al. 2014; Yan et al. 2019; Erices et al. 2022). A brain-penetrant inhibitor of the immune cell adenosine receptor A2AR modestly extended survival in immune-competent mouse models of GBM but failed to relieve T cell exhaustion in the TME (Ott et al. 2020). A broader approach for adenosine blockade is to inhibit its production by CD39/CD73, a strategy that might simultaneously increase proinflammatory signaling by the precursor metabolite ATP (Fig. 5). Knockout of Nt5e (encloding CD73) inhibits tumor growth and invasion and modestly extends survival in mouse models (Goswami et al. 2019; Yan et al. 2019). However, supplying anti-CTLA4 and anti-PD1 combination therapy to Nt5e knockout animals leads to a remarkable increase in survival, with ∼40% of responses being durable (Goswami et al. 2019). These results point toward possible future clinical applications in GBM for adenosine pathway inhibitors, which are already being assessed in clinical trials for other cancer types (Leone and Emens 2018; Thompson and Powell 2021).
Concluding remarks
Coordinated intercellular partitioning of metabolism is a defining characteristic of the CNS and involves symbiotic relationships that allow brain-resident cells to direct their resources to highly specialized functions such as neuronal firing. In many of these processes, astrocytes play important metabolic support roles, powering energetically demanding neurotransmitter cycles, supplying biosynthetic precursors, and detoxifying and recycling metabolic waste products. An emerging theme in cancer biology is that tumors integrate into and/or hijack the metabolism of their host tissue, making use of nutrients in the ISF and co-opting stromal cells to provide additional metabolic support (Lyssiotis and Kimmelman 2017; Altea-Manzano et al. 2020). While these interactions contribute to cancer progression, in some cases they also lead to context-dependent metabolic dependencies that can be selectively targeted for cancer therapy (Muir and Vander Heiden 2018). In GBM, it is now clear that malignant cells exploit diverse aspects of brain metabolism and are likely dependent on astrocytes for both cholesterol and glutamine supply. GBM also hijacks the signaling roles of certain metabolites, even forming functional synapses with glutamatergic neurons that stimulate mitogenic and proinvasion signaling. Finally, the metabolic secretome of GBM cells induces immune suppression through complementary axes including the kynurenine and adenosine pathways. Despite the research to date, gaps in our understanding remain. In particular, there is a paucity of information on the role of oligodendrocytes in the GBM TME. On this topic and others, technological advances such as imaging mass spectrometry are likely to facilitate further gains in our understanding of the multidirectional metabolic interactions in the GBM microenvironment (Randall et al. 2020; Philipsen et al. 2023). The knowledge gained from this basic research has already been used to develop rational drug combinations, which in some cases lead to a high rate of durable remission in preclinical models of GBM. The challenge now is to translate these findings into new therapies that finally improve the long-term prognosis for patients diagnosed with this devastating disease.
Acknowledgments
We thank members of the Lukey and Van Aelst laboratories for their support and discussions. We further thank James Duffy for graphic artwork. Given the focus of this review on metabolic interactions, we apologize for not being able to cite all of our colleagues who have contributed to the field of GBM. J.d.R.S. is supported by the Leslie C. Quick Jr. fellowship from the Cold Spring Harbor Laboratory School of Biological Sciences. This work is supported by grants from the National Institutes of Health (NIH; R01MH119819 and 5P30CA045508) to L.V.A., and from the Department of Defense (BC200599) and the NIH (R01GM149957 and 5P30CA045508) to M.J.L.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350693.123.
Competing interest statement
The authors declare no competing interests.
References
- Abuhusain HJ, Matin A, Qiao Q, Shen H, Kain N, Day BW, Stringer BW, Daniels B, Laaksonen MA, Teo C, et al. 2013. A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis. J Biol Chem 288: 37355–37364. 10.1074/jbc.M113.494740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldana BI. 2019. Microglia-specific metabolic changes in neurodegeneration. J Mol Biol 431: 1830–1842. 10.1016/j.jmb.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Allen NJ, Eroglu C. 2017. Cell biology of astrocyte-synapse interactions. Neuron 96: 697–708. 10.1016/j.neuron.2017.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altea-Manzano P, Cuadros AM, Broadfield LA, Fendt S. 2020. Nutrient metabolism and cancer in the in vivo context: a metabolic game of give and take. EMBO Rep 21: e50635. 10.15252/embr.202050635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amankulor NM, Kim Y, Arora S, Kargl J, Szulzewsky F, Hanke M, Margineantu DH, Rao A, Bolouri H, Delrow J, et al. 2017. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev 31: 774–786. 10.1101/gad.294991.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AI, Meisingset TW, Kotter MR, Sonnewald U. 2013. Metabolic aspects of neuron-oligodendrocyte-astrocyte interactions. Front Endocrinol 4: 54. 10.3389/fendo.2013.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi OC, Arrillaga-Romany IC, Ly KI, Bogner W, Ratai EM, Reitz K, Iafrate AJ, Dietrich J, Gerstner ER, Chi AS, et al. 2018. Pharmacodynamics of mutant-IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2-hydroxyglutarate. Nat Commun 9: 1474. 10.1038/s41467-018-03905-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes ARP, Scheyltjens I, Duerinck J, Neyns B, Movahedi K, Van Ginderachter JA. 2020. Understanding the glioblastoma immune microenvironment as basis for the development of new immunotherapeutic strategies. Elife 9: e52176. 10.7554/eLife.52176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseni L, Sharma R, Mack N, Nagalla D, Ohl S, Hielscher T, Singhal M, Pilz R, Augustin H, Sandhoff R, et al. 2023. Sphingosine-1-phosphate recruits macrophages and microglia and induces a pro-tumorigenic phenotype that favors glioma progression. Cancers (Basel) 15: 479. 10.3390/cancers15020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis CD, Ferraro GB, Jain RK. 2019. The blood–brain barrier and blood–tumour barrier in brain tumours and metastases. Nat Rev Cancer 20: 26–41. 10.1038/s41568-019-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D, Laughlin SB. 2001. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab 21: 1133–1145. 10.1097/00004647-200110000-00001 [DOI] [PubMed] [Google Scholar]
- Baid U, Rane SU, Talbar S, Gupta S, Thakur MH, Moiyadi A, Mahajan A. 2020. Overall survival prediction in glioblastoma with radiomic features using machine learning. Front Comput Neurosci 14: 61. 10.3389/fncom.2020.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak LK, Schousboe A, Waagepetersen HS. 2006. The glutamate/GABA–glutamine cycle: aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem 98: 641–653. 10.1111/j.1471-4159.2006.03913.x [DOI] [PubMed] [Google Scholar]
- Banh RS, Biancur DE, Yamamoto K, Sohn ASW, Walters B, Kuljanin M, Gikandi A, Wang H, Mancias JD, Schneider RJ, et al. 2020. Neurons release serine to support mRNA translation in pancreatic cancer. Cell 183: 1202–1218.e25. 10.1016/j.cell.2020.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros LF, Ruminot I, Sotelo-Hitschfeld T, Lerchundi R, Fernández-Moncada I. 2023. Metabolic recruitment in brain tissue. Annu Rev Physiol 85: 115–135. 10.1146/annurev-physiol-021422-091035 [DOI] [PubMed] [Google Scholar]
- Baslow MH. 2002. Evidence supporting a role for N-acetyl-l-aspartate as a molecular water pump in myelinated neurons in the central nervous system: an analytical review. Neurochem Int 40: 295–300. 10.1016/S0197-0186(01)00095-X [DOI] [PubMed] [Google Scholar]
- Baslow MH, Suckow RF, Sapirstein V, Hungund BL. 1999. Expression of aspartoacylase activity in cultured rat macroglial cells is limited to oligodendrocytes. J Mol Neurosci 13: 47–53. 10.1385/JMN:13:1-2:47 [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. 2001. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev 81: 871–927. 10.1152/physrev.2001.81.2.871 [DOI] [PubMed] [Google Scholar]
- Beart PM, O'Shea RD. 2007. Transporters for L-glutamate: an update on their molecular pharmacology and pathological involvement. Br J Pharmacol 150: 5–17. 10.1038/sj.bjp.0706949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger M, Allaman I, Magistretti PJ. 2011. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14: 724–738. 10.1016/j.cmet.2011.08.016 [DOI] [PubMed] [Google Scholar]
- Benarroch EE. 2005. Neuron-astrocyte interactions: partnership for normal function and disease in the central nervous system. Mayo Clin Proc 80: 1326–1338. 10.4065/80.10.1326 [DOI] [PubMed] [Google Scholar]
- Bergles DE, Jahr CE. 1998. Glial contribution to glutamate uptake at Schaffer collateral-commissural synapses in the hippocampus. J Neurosci 18: 7709–7716. 10.1523/JNEUROSCI.18-19-07709.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Roberts JDB, Somogyl P, Jahr CE. 2000. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405: 187–191. 10.1038/35012083 [DOI] [PubMed] [Google Scholar]
- Bernier LP, York EM, Kamyabi A, Choi HB, Weilinger NL, MacVicar BA. 2020. Microglial metabolic flexibility supports immune surveillance of the brain parenchyma. Nat Commun 11: 1559. 10.1038/s41467-020-15267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutia YD, Ganapathy V. 2016. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim Biophys Acta - Mol Cell Res 1863: 2531–2539. 10.1016/j.bbamcr.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Chowdhry S, Wu S, Zhang W, Masui K, Mischel PS. 2019. Altered cellular metabolism in gliomas—an emerging landscape of actionable co-dependency targets. Nat Rev Cancer 20: 57–70. 10.1038/s41568-019-0226-5 [DOI] [PubMed] [Google Scholar]
- Bianchi M, Bertini R, Ghezzi P. 1988. Induction of indoleamine dioxygenase by interferon in mice: a study with different recombinant interferons and various cytokines. Biochem Biophys Res Commun 152: 237–242. 10.1016/S0006-291X(88)80705-8 [DOI] [PubMed] [Google Scholar]
- Bikfalvi A, da Costa CA, Avril T, Barnier JV, Bauchet L, Brisson L, Cartron PF, Castel H, Chevet E, Chneiweiss H, et al. 2023. Challenges in glioblastoma research: focus on the tumor microenvironment. Trends Cancer 9: 9–27. 10.1016/j.trecan.2022.09.005 [DOI] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. 2018. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24: 541–550. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvento G, Bolaños JP. 2021. Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab 33: 1546–1564. 10.1016/j.cmet.2021.07.006 [DOI] [PubMed] [Google Scholar]
- Bova V, Filippone A, Casili G, Lanza M, Campolo M, Capra AP, Repici A, Crupi L, Motta G, Colarossi C, et al. 2022. Adenosine targeting as a new strategy to decrease glioblastoma aggressiveness. Cancers 14: 4032. 10.3390/cancers14164032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D, Longman RS, Albert ML. 2005. A two-step induction of indoleamine 2,3 dioxygenase (IDO) activity during dendritic-cell maturation. Blood 106: 2375–2381. 10.1182/blood-2005-03-0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer ME, Koopman WJ, Koene S, Nooteboom M, Rodenburg RJ, Willems PH, Smeitink JAM. 2013. The role of mitochondrial OXPHOS dysfunction in the development of neurologic diseases. Neurobiol Dis 51: 27–34. 10.1016/j.nbd.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Brown NF, Carter TJ, Ottaviani D, Mulholland P. 2018. Harnessing the immune system in glioblastoma. Br J Cancer 119: 1171–1181. 10.1038/s41416-018-0258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham SC, Campbell SL, Haas BR, Montana V, Robel S, Ogunrinu T, Sontheimer H. 2011. Glutamate release by primary brain tumors induces epileptic activity. Nat Med 17: 1269–1274. 10.1038/nm.2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Li W, Brenner M, Bahiraii S, Heiss EH, Weckwerth W. 2022. Branched-chain ketoacids derived from cancer cells modulate macrophage polarization and metabolic reprogramming. Front Immunol 13: 966158. 10.3389/fimmu.2022.966158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo N, Goudriaan A, van Deijk ALF, Otte WM, Brouwers JF, Lodder H, Gutmann DH, Nave KA, Dijkhuizen RM, Mansvelder HD, et al. 2017. Oligodendroglial myelination requires astrocyte-derived lipids. PLOS Biol 15: e1002605. 10.1371/journal.pbio.1002605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campesato LF, Budhu S, Tchaicha J, Weng CH, Gigoux M, Cohen IJ, Redmond D, Mangarin L, Pourpe S, Liu C, et al. 2020. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat Commun 11: 4011. 10.1038/s41467-020-17750-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JD, Olschowka JA, O'Banion MK. 2014. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J Neuroinflammation 11: 98. 10.1186/1742-2094-11-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs JA, Denicola GM. 2019. The non-essential amino acid cysteine becomes essential for tumor proliferation and survival. Cancers 11: 678. 10.3390/cancers11050678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy S, Wang S, Stopka SA, Lin JR, Yapp C, Ritch CC, Salhi L, Baker GJ, Rashid R, Baquer G, et al. 2022. Single cell spatial analysis reveals the topology of immunomodulatory purinergic signaling in glioblastoma. Nat Commun 13: 4814. 10.1038/s41467-022-32430-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivii CB, Boșca AB, Melincovici CS, Constantin AM, Mărginean M, Dronca E, Suflețel R, Gonciar D, Bungărdean M, Șovrea A. 2022. Glioblastoma microenvironment and cellular interactions. Cancers (Basel) 14: 1092. 10.3390/cancers14041092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnane SC, Trushina E, Morland C, Prigione A, Casadesus G, Andrews ZB, Beal MF, Bergersen LH, Brinton RD, de la Monte S, et al. 2020. Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing. Nat Rev Drug Discov 19: 609–633. 10.1038/s41573-020-0072-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Prat A. 2015. The blood-brain barrier. Cold Spring Harb Perspect Biol 7: a020412. 10.1101/cshperspect.a020412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele S, Zappelli E, Natali L, Martini C, Trincavelli ML. 2014. Modulation of A1 and A2B adenosine receptor activity: a new strategy to sensitise glioblastoma stem cells to chemotherapy. Cell Death Dis 5: e1539. 10.1038/cddis.2014.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean PT, Hooks SB. 2023. Pleiotropic effects of the COX-2/PGE2 axis in the glioblastoma tumor microenvironment. Front Oncol 12: 1116014. 10.3389/fonc.2022.1116014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Berardinis RJ, Chandel NS. 2016. Fundamentals of cancer metabolism. Sci Adv 2: e1600200. 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot J, Sontheimer H. 2011. Glutamate and the biology of gliomas. Glia 59: 1181–1189. 10.1002/glia.21113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker LJM, Verheul C, Wensveen N, Leenders W, Lamfers MLM, Leenstra S, Luider TM. 2022. Effects of the IDH1 R132H mutation on the energy metabolism: a comparison between tissue and corresponding primary glioma cell cultures. ACS Omega 7: 3568–3578. 10.1021/acsomega.1c06121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva MI, Stringer BW, Bardy C. 2023. Neuronal and tumourigenic boundaries of glioblastoma plasticity. Trends Cancer 9: 223–236. 10.1016/j.trecan.2022.10.010 [DOI] [PubMed] [Google Scholar]
- Díaz-García CM, Mongeon R, Lahmann C, Koveal D, Zucker H, Yellen G. 2017. Neuronal stimulation triggers neuronal glycolysis and not lactate uptake. Cell Metab 26: 361–374. 10.1016/j.cmet.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienel GA. 2017. Lack of appropriate stoichiometry: strong evidence against an energetically important astrocyte-neuron lactate shuttle in brain. J Neurosci Res 95: 2103–2125. 10.1002/jnr.24015 [DOI] [PubMed] [Google Scholar]
- Dienel GA. 2019. Brain glucose metabolism: integration of energetics with function. Physiol Rev 99: 949–1045. 10.1152/physrev.00062.2017 [DOI] [PubMed] [Google Scholar]
- Dietschy JM. 2009. Central nervous system: cholesterol turnover, brain development and neurodegeneration. Biol Chem 390: 287–293. 10.1515/BC.2009.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, McBain CJ. 1999. Glutamate and aspartate are the major excitatory transmitters in the brain. In Basic neurochemistry: molecular, cellular and medical aspects, 6th ed. (ed. Siegel GJ, et al. ), pp. 315–334. Lippincott-Raven, Philadelphia. https://www.ncbi.nlm.nih.gov/books/NBK28252 [Google Scholar]
- Dringen R. 2000. Metabolism and functions of glutathione in brain. Prog Neurobiol 62: 649–671. 10.1016/S0301-0082(99)00060-X [DOI] [PubMed] [Google Scholar]
- Dringen R, Pfeiffer B, Hamprecht B. 1999. Synthesis of the antioxidant glutathione in neurons: supply by astrocytes of CysGly as precursor for neuronal glutathione. J Neurosci 19: 562–569. 10.1523/JNEUROSCI.19-02-00562.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dringen R, Gutterer JM, Gros C, Hirrlinger J. 2001. Aminopeptidase N mediates the utilization of the GSH precursor CysGly by cultured neurons. J Neurosci Res 66: 1003–1008. 10.1002/jnr.10042 [DOI] [PubMed] [Google Scholar]
- Du L, Xing Z, Tao B, Li T, Yang D, Li W, Zheng Y, Kuang C, Yang Q. 2020. Both IDO1 and TDO contribute to the malignancy of gliomas via the Kyn–AhR–AQP4 signaling pathway. Signal Transduct Target Ther 5: 10. 10.1038/s41392-019-0103-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C, Al-Humadi H, Gutierrez-Quintana R, Zarros A. 2018. Brain N-acetylaspartate accumulation in Canavan disease is not neurotoxic per se: the implications of the first gene replacement therapy study to demonstrate successful post-symptomatic treatment in mice. Ann Res Hosp 2: 4. 10.21037/arh.2018.04.01 [DOI] [Google Scholar]
- Erices JI, Niechi I, Uribe-Ojeda A, Toro MLÁ, García-Romero N, Carrión-Navarro J, Monago-Sánchez Á, Ayuso-Sacido Á, Martin RS, Quezada-Monrás C. 2022. The low affinity A2B adenosine receptor enhances migratory and invasive capacity in vitro and angiogenesis in vivo of glioblastoma stem-like cells. Front Oncol 12: 969993. 10.3389/fonc.2022.969993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M, Cogan KE, Egan B. 2017. Metabolism of ketone bodies during exercise and training: physiological basis for exogenous supplementation. J Physiol 595: 2857–2871. 10.1113/JP273185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fack F, Tardito S, Hochart G, Oudin A, Zheng L, Fritah S, Golebiewska A, Nazarov PV, Bernard A, Hau A-C, et al. 2017. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol Med 9: 1681–1695. 10.15252/emmm.201707729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Xiong Y, Wang Y. 2019. A reignited debate over the cell(s) of origin for glioblastoma and its clinical implications. Front Med 13: 531–539. 10.1007/s11684-019-0700-1 [DOI] [PubMed] [Google Scholar]
- Farinelli SE, Nicklas WJ. 1992. Glutamate metabolism in rat cortical astrocyte cultures. J Neurochem 58: 1905–1915. 10.1111/j.1471-4159.1992.tb10068.x [DOI] [PubMed] [Google Scholar]
- Featherstone DE. 2010. Intercellular glutamate signaling in the nervous system and beyond. ACS Chem Neurosci 1: 4–12. 10.1021/cn900006n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzner D, Bader JM, Penkert H, Bergner CG, Su M, Weil MT, Surma MA, Mann M, Klose C, Simons M. 2020. Cell-type- and brain-region-resolved mouse brain lipidome. Cell Rep 32: 108132. 10.1016/j.celrep.2020.108132 [DOI] [PubMed] [Google Scholar]
- Flurkey WH. 2010. Yield of ATP molecules per glucose molecule. J Chem Educ 87: 271. 10.1021/ed800102g [DOI] [Google Scholar]
- Fox PT, Raichle ME, Mintun MA, Dence C. 1988. Nonoxidative glucose consumption during focal physiologic neural activity. Science 241: 462–464. 10.1126/science.3260686 [DOI] [PubMed] [Google Scholar]
- Friedrich M, Sankowski R, Bunse L, Kilian M, Green E, Ramallo Guevara C, Pusch S, Poschet G, Sanghvi K, Hahn M, et al. 2021. Tryptophan metabolism drives dynamic immunosuppressive myeloid states in IDH-mutant gliomas. Nat Cancer 2: 723–740. 10.1038/s43018-021-00201-z [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Kato S, Nesline MK, Conroy JM, DePietro P, Pabla S, Kurzrock R. 2022. Indoleamine 2,3-dioxygenase (IDO) inhibitors and cancer immunotherapy. Cancer Treat Rev 110: 102461. 10.1016/j.ctrv.2022.102461 [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9: 162–174. 10.1038/nri2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschler MM, Stockwell BR. 2017. Lipid peroxidation in cell death. Biochem Biophys Res Commun 482: 419–425. 10.1016/j.bbrc.2016.10.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler DJ, Li D, Xu H, Su Q, Sanmiguel J, Tuncer S, Moore C, King J, Matalon R, Gao G. 2017. Redirecting N-acetylaspartate metabolism in the central nervous system normalizes myelination and rescues Canavan disease. JCI Insight 2: e90807. 10.1172/jci.insight.90807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. 2017. Immune microenvironment of gliomas. Lab Investig 97: 498–518. 10.1038/labinvest.2017.19 [DOI] [PubMed] [Google Scholar]
- Gillespie S, Monje M. 2018. An active role for neurons in glioma progression: making sense of Scherer's structures. Neuro Oncol 20: 1292–1299. 10.1093/neuonc/noy083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S, Walle T, Cornish AE, Basu S, Anandhan S, Fernandez I, Vence L, Blando J, Zhao H, Yadav SS, et al. 2019. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med 26: 39–46. 10.1038/s41591-019-0694-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski MM, Sankey EW, Ryan KJ, Chongsathidkiet P, Lorrey SJ, Wilkinson DS, Fecci PE. 2021. Immune suppression in gliomas. J Neurooncol 151: 3–12. 10.1007/s11060-020-03483-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi S, Mauri L, Prioni S, Cabitta L, Sonnino S, Prinetti A, Giussani P. 2019. Sphingosine 1-phosphate receptors and metabolic enzymes as druggable targets for brain diseases. Front Pharmacol 10: 807. 10.3389/fphar.2019.00807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SA, Ye X, Chamberlain M, Mikkelsen T, Batchelor T, Desideri S, Piantadosi S, Fisher J, Fine HA. 2009. Talampanel with standard radiation and temozolomide in patients with newly diagnosed glioblastoma: a multicenter phase II trial. J Clin Oncol 27: 4155–4161. 10.1200/JCO.2008.21.6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J, Garg M, Bilgin A, Grant R. 2013. Relationship between central and peripheral fatty acids in humans. Lipids Health Dis 12: 79. 10.1186/1476-511X-12-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán M, Blázquez C. 2001. Is there an astrocyte-neuron ketone body shuttle? Trends Endocrinol Metab 12: 169–173. 10.1016/S1043-2760(00)00370-2 [DOI] [PubMed] [Google Scholar]
- Halliwell B. 1992. Reactive oxygen species and the central nervous system. J Neurochem 59: 1609–1623. 10.1111/j.1471-4159.1992.tb10990.x [DOI] [PubMed] [Google Scholar]
- Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, Gilbert MR, Yang C. 2020. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer 122: 1580–1589. 10.1038/s41416-020-0814-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zhou J, Li L, Wu X, Shi Y, Cui W, Zhang S, Hu Q, Wang J, Bai H, et al. 2022. SLC1A5 enhances malignant phenotypes through modulating ferroptosis status and immune microenvironment in glioma. Cell Death Dis 13: 1071. 10.1038/s41419-022-05526-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CC, Ali T, Ramanadham S, Hjelmeland AB. 2020. Sphingolipid metabolism in glioblastoma and metastatic brain tumors: a review of sphingomyelinases and sphingosine-1-phosphate. Biomolecules 10: 1357. 10.3390/biom10101357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz L. 2004. Intercellular metabolic compartmentation in the brain: past, present and future. Neurochem Int 45: 285–296. 10.1016/j.neuint.2003.08.016 [DOI] [PubMed] [Google Scholar]
- Hoang-Minh LB, Siebzehnrubl FA, Yang C, Suzuki-Hatano S, Dajac K, Loche T, Andrews N, Schmoll Massari M, Patel J, Amin K, et al. 2018. Infiltrative and drug-resistant slow-cycling cells support metabolic heterogeneity in glioblastoma. EMBO J 37: e98772. 10.15252/embj.201798772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland R, McIntosh AL, Finucane OM, Mela V, Rubio-Araiz A, Timmons G, McCarthy SA, Gun'ko YK, Lynch MA. 2018. Inflammatory microglia are glycolytic and iron retentive and typify the microglia in APP/PS1 mice. Brain Behav Immun 68: 183–196. 10.1016/j.bbi.2017.10.017 [DOI] [PubMed] [Google Scholar]
- Hollnagel JO, Cesetti T, Schneider J, Vazetdinova A, Valiullina-Rakhmatullina F, Lewen A, Rozov A, Kann O. 2020. Lactate attenuates synaptic transmission and affects brain rhythms featuring high energy expenditure. iScience 23: 101316. 10.1016/j.isci.2020.101316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino H, Kubota M. 2014. Canavan disease: clinical features and recent advances in research. Pediatr Int 56: 477–483. 10.1111/ped.12422 [DOI] [PubMed] [Google Scholar]
- Huang-Hobbs E, Cheng Y-T, Ko Y, Luna-Figueroa E, Lozzi B, Taylor KR, McDonald M, He P, Chen H-C, Yang Y, et al. 2023. Remote neuronal activity drives glioma progression through SEMA4F. Nature 619: 844–850. 10.1038/s41586-023-06267-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain G, Wang J, Rasul A, Anwar H, Imran A, Qasim M, Zafar S, Kamran SKS, Razzaq A, Aziz N, et al. 2019. Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis 18: 1–12. 10.1186/s12944-019-0965-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou MS, Jackson J, Sheu SH, Chang CL, Weigel AV, Liu H, Pasolli HA, Xu CS, Pang S, Matthies D, et al. 2019. Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell 177: 1522–1535. 10.1016/j.cell.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Iwamoto FM, Kreisl TN, Kim L, Duic JP, Butman JA, Albert PS, Fine HA. 2010. Phase II trial of talampanel, a glutamate receptor inhibitor, for adults with recurrent malignant gliomas. Cancer 116: 1776–1782. 10.1002/cncr.24957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumoto S, Miyauchi M, Tasaki T, Okuda T, Nakagawa N, Nakano N, Kato A, Fujita M. 2018. Seizures and tumor progression in glioma patients with uncontrollable epilepsy treated with perampanel. Anticancer Res 38: 4361–4366. 10.21873/anticanres.12737 [DOI] [PubMed] [Google Scholar]
- Janson CG. 2015. Canavan disease: can two wrongs make a right? Sci Transl Med 7: 295ec115. 10.1126/scitranslmed.aac8103 [DOI] [Google Scholar]
- Jennings MR, Munn D, Blazeck J. 2021. Immunosuppressive metabolites in tumoral immune evasion: redundancies, clinical efforts, and pathways forward. J Immunother Cancer 9: e003013. 10.1136/jitc-2021-003013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Qiu J, Li Q, Shi Z. 2017. Prostaglandin E2 signaling: alternative target for glioblastoma? Trends Cancer 3: 75–78. 10.1016/j.trecan.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Mao C, Chen L, Wang L, Liu Y, Yuan J. 2021. Adenosinergic pathway: a hope in the immunotherapy of glioblastoma. Cancers 13: 229. 10.3390/cancers13020229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala P, Carney SV, Gauss JC, Garcia-Fabiani MB, Haase S, Alghamri MS, Núñez FJ, Liu Y, Yu M, Taher A, et al. 2021. Inhibition of 2-hydroxyglutarate elicits metabolic reprogramming and mutant IDH1 glioma immunity in mice. J Clin Invest 131: e139542. 10.1172/JCI139542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek MJT, Mulder L, Yi CX. 2016. Microglia energy metabolism in metabolic disorder. Mol Cell Endocrinol 438: 27–35. 10.1016/j.mce.2016.09.028 [DOI] [PubMed] [Google Scholar]
- Kant S, Kesarwani P, Prabhu A, Graham SF, Buelow KL, Nakano I, Chinnaiyan P. 2020. Enhanced fatty acid oxidation provides glioblastoma cells metabolic plasticity to accommodate to its dynamic nutrient microenvironment. Cell Death Dis 11: 253. 10.1038/s41419-020-2449-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis A, Gallopin T, Lacroix A, Plaisier F, Piquet J, Geoffroy H, Hepp R, Naudé J, Le Gac B, Egger R, et al. 2021. Lactate is an energy substrate for rodent cortical neurons and enhances their firing activity. Elife 10: e71424. 10.7554/eLife.71424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm F, Maas RR, Bowman RL, Kornete M, Soukup K, Nassiri S, Brouland JP, Iacobuzio-Donahue CA, Brennan C, Tabar V, et al. 2020. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181: 1643–1660.e17. 10.1016/j.cell.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanbash G, Carrera DA, Shrivastav S, Ahn BJ, Jahan N, Mazor T, Chheda ZS, Downey KM, Watchmaker PB, Beppler C, et al. 2017. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest 127: 1425–1437. 10.1172/JCI90644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Nakamura T, Nidaira T, Nakamura K, Ooashi N, Ito E, Watase K, Tanaka K, Wada K, Kudo Y, et al. 1999. Optical detection of synaptically induced glutamate transport in hippocampal slices. J Neurosci 19: 2580–2588. 10.1523/JNEUROSCI.19-07-02580.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft AD, Johnson DA, Johnson JA. 2004. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci 24: 1101–1112. 10.1523/JNEUROSCI.3817-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Prentzell MT, Mohapatra SR, Sahm F, Zhao Z, Grummt I, Wick W, Opitz CA, Platten M, Green EW. 2020. Constitutive expression of the immunosuppressive tryptophan dioxygenase TDO2 in glioblastoma is driven by the transcription factor C/EBPβ. Front Immunol 11: 657. 10.3389/fimmu.2020.00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomersky E, Zhai L, Lenzen A, Lauing KL, Qian J, Scholtens DM, Gritsina G, Sun X, Liu Y, Yu F, et al. 2018. IDO1 inhibition synergizes with radiation and PD-1 blockade to durably increase survival against advanced glioblastoma. Clin Cancer Res 24: 2559–2573. 10.1158/1078-0432.CCR-17-3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange F, Hörnschemeyer J, Kirschstein T. 2021. Glutamatergic mechanisms in glioblastoma and tumor-associated epilepsy. Cells 10: 1226. 10.3390/cells10051226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Tan YJ, Oon CE. 2018. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol 834: 188–196. 10.1016/j.ejphar.2018.07.034 [DOI] [PubMed] [Google Scholar]
- Leone RD, Emens LA. 2018. Targeting adenosine for cancer immunotherapy. J Immunother Cancer 6: 57. 10.1186/s40425-018-0360-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang J, Liu Q. 2022. Brain cell type-specific cholesterol metabolism and implications for learning and memory. Trends Neurosci 45: 401–414. 10.1016/j.tins.2022.01.002 [DOI] [PubMed] [Google Scholar]
- Liu M, Wang X, Wang L, Ma X, Gong Z, Zhang S, Li Y. 2018. Targeting the IDO1 pathway in cancer: from bench to bedside. J Hematol Oncol 11: 100. 10.1186/s13045-018-0644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long PM, Moffett JR, Namboodiri AMA, Viapiano MS, Lawler SE, Jaworski DM. 2013. N-acetylaspartate (NAA) and N-acetylaspartylglutamate (NAAG) promote growth and inhibit differentiation of glioma stem-like cells. J Biol Chem 288: 26188–26200. 10.1074/jbc.M113.487553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, et al. 2021. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23: 1231–1251. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau A, Harris TH, Kipnis J. 2015. Revisiting the mechanisms of CNS immune privilege. Trends Immunol 36: 569–577. 10.1016/j.it.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucken-Ardjomande S, Montessuit S, Martinou JC. 2008. Bax activation and stress-induced apoptosis delayed by the accumulation of cholesterol in mitochondrial membranes. Cell Death Differ 15: 484–493. 10.1038/sj.cdd.4402280 [DOI] [PubMed] [Google Scholar]
- Lukey MJ, Katt WP, Cerione RA. 2017. Targeting amino acid metabolism for cancer therapy. Drug Discov Today 22: 796–804. 10.1016/j.drudis.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SA, Chung WJ, Weaver AK, Ogunrinu T, Sontheimer H. 2007. Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res 67: 9463–9471. 10.1158/0008-5472.CAN-07-2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Kimmelman AC. 2017. Metabolic interactions in the tumor microenvironment. Trends Cell Biol 27: 863–875. 10.1016/j.tcb.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavarao CN, Moffett JR, Moore RA, Viola RE, Namboodiri MAA, Jacobowitz DM. 2004. Immunohistochemical localization of aspartoacylase in the rat central nervous system. J Comp Neurol 472: 318–329. 10.1002/cne.20080 [DOI] [PubMed] [Google Scholar]
- Magi S, Piccirillo S, Amoroso S, Lariccia V. 2019. Excitatory amino acid transporters (EAATs): glutamate transport and beyond. Int J Mol Sci 20: 5674. 10.3390/ijms20225674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti PJ, Allaman I. 2015. A cellular perspective on brain energy metabolism and functional imaging. Neuron 86: 883–901. 10.1016/j.neuron.2015.03.035 [DOI] [PubMed] [Google Scholar]
- Mahmoud S, Gharagozloo M, Simard C, Gris D. 2019. Astrocytes maintain glutamate homeostasis in the CNS by controlling the balance between glutamate uptake and release. Cells 8: 184. 10.3390/cells8020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Wang-Eckhardt L, Hartmann D, Gieselmann V, Eckhardt M. 2015. N-acetylaspartate synthase deficiency corrects the myelin phenotype in a Canavan disease mouse model but does not affect survival time. J Neurosci 35: 14501–14516. 10.1523/JNEUROSCI.1056-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus HJ, Carpenter KLH, Price SJ, Hutchinson PJ. 2010. In vivo assessment of high-grade glioma biochemistry using microdialysis: a study of energy-related molecules, growth factors and cytokines. J Neurooncol 97: 11–23. 10.1007/s11060-009-9990-5 [DOI] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang X-LL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. 2012. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab 15: 827–837. 10.1016/j.cmet.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalon R, Matalon KM. 2015. Canavan disease. In Rosenberg's molecular and genetic basis of neurological and psychiatric disease , 5th ed. (ed. Rosenberg RN, Pascual JM), pp. 695–701. Elsevier, London, UK. [Google Scholar]
- McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, Sarosiek KA, Briggs KJ, Robbins AK, Sewastianik T, et al. 2018. Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell 175: 101–116.e25 10.1016/j.cell.2018.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann JC, Mandel G. 2018. Neuronal activity induces glutathione metabolism gene expression in astrocytes. Glia 66: 2024–2039. 10.1002/glia.23455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern K, Castro AC, Cavanaugh J, Coma S, Walsh M, Tchaicha J, Syed S, Natarajan P, Manfredi M, Zhang XM, et al. 2022. Discovery and characterization of a novel aryl hydrocarbon receptor inhibitor, IK-175, and its inhibitory activity on tumor immune suppression. Mol Cancer Ther 21: 1261–1272. 10.1158/1535-7163.MCT-21-0984 [DOI] [PubMed] [Google Scholar]
- McKenna MC. 2013. Glutamate pays its own way in astrocytes. Front Endocrinol 4: 191. 10.3389/fendo.2013.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MC, Sonnewald U, Huang X, Stevenson J, Zielke HR. 1996. Exogenous glutamate concentration regulates the metabolic fate of glutamate in astrocytes. J Neurochem 66: 386–393. 10.1046/j.1471-4159.1996.66010386.x [DOI] [PubMed] [Google Scholar]
- Meldrum BS. 2000. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J Nutr 130: 1007S–1015S. 10.1093/jn/130.4.1007S [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Penas-Prado M, Peters KB, Burris HA, Maher EA, Janku F, Cote GM, de la Fuente MI, Clarke JL, Ellingson BM, et al. 2021. Vorasidenib, a dual inhibitor of mutant IDH1/2, in recurrent or progressive glioma; results of a first-in-human phase I trial. Clin Cancer Res 27: 4491–4499. 10.1158/1078-0432.CCR-21-0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Lu M, Wen PY, Taylor JW, Maher EA, Arrillaga-Romany I, Peters KB, Ellingson BM, Rosenblum MK, Chun S, et al. 2023. Vorasidenib and ivosidenib in IDH1-mutant low-grade glioma: a randomized, perioperative phase 1 trial. Nat Med 29: 615–622. 10.1038/s41591-022-02141-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y, Qi G, Vitali F, Shang Y, Raikes AC, Wang T, Jin Y, Brinton RD, Gu H, Yin F. 2023. Loss of fatty acid degradation by astrocytic mitochondria triggers neuroinflammation and neurodegeneration. Nat Metab 5: 445–465. 10.1038/s42255-023-00756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska J, Chandel NS. 2023. Targeting fatty acid metabolism in glioblastoma. J Clin Invest 133: e163448. 10.1172/JCI163448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modoux M, Rolhion N, Mani S, Sokol H. 2021. Tryptophan metabolism as a pharmacological target. Trends Pharmacol Sci 42: 60–73. 10.1016/j.tips.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AMA. 2007. N-acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol 81: 89–131. 10.1016/j.pneurobio.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero J, Morales A, Llacuna L, Lluis JM, Terrones O, Basañez G, Antonsson B, Prieto J, García-Ruiz C, Colell A, et al. 2008. Mitochondrial cholesterol contributes to chemotherapy resistance in hepatocellular carcinoma. Cancer Res 68: 5246–5256. 10.1158/0008-5472.CAN-07-6161 [DOI] [PubMed] [Google Scholar]
- Morris AAM. 2005. Cerebral ketone body metabolism. J Inherit Metab Dis 28: 109–121. 10.1007/s10545-005-5518-0 [DOI] [PubMed] [Google Scholar]
- Moussawi K, Riegel A, Nair S, Kalivas PW. 2011. Extracellular glutamate: functional compartments operate in different concentration ranges. Front Syst Neurosci 5: 94. 10.3389/fnsys.2011.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer BJ, Rojas IY, Murray IA, Lee S, Hazlett HF, Perdew GH, Tomlinson CR. 2017. Indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors activate the aryl hydrocarbon receptor. Toxicol Appl Pharmacol 323: 74–80. 10.1016/j.taap.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A, Vander Heiden MG. 2018. The nutrient environment affects therapy. Science 360: 962–963. 10.1126/science.aar5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraleedharan R, Gawali MV, Tiwari D, Sukumaran A, Oatman N, Anderson J, Nardini D, Bhuiyan MAN, Tkáč I, Ward AL, et al. 2020. AMPK-regulated astrocytic lactate shuttle plays a non-cell-autonomous role in neuronal survival. Cell Rep 32: 108092. 10.1016/j.celrep.2020.108092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TH, Yu J, Ng R, Johnson DA, Shen H, Honey CR, Johnson JA. 2001. Preferential expression of antioxidant response element mediated gene expression in astrocytes. J Neurochem 76: 1670–1678. 10.1046/j.1471-4159.2001.00157.x [DOI] [PubMed] [Google Scholar]
- Nakamura T, Niimi S, Nawa K, Noda C, Ichihara A, Takagi Y, Anai M, Sakaki Y. 1987. Multihormonal regulation of transcription of the tryptophan 2,3-dioxygenase gene in primary cultures of adult rat hepatocytes with special reference to the presence of a transcriptional protein mediating the action of glucocorticoids. J Biol Chem 262: 727–733. 10.1016/S0021-9258(19)75845-1 [DOI] [PubMed] [Google Scholar]
- Natarajan SK, Venneti S. 2019. Glutamine metabolism in brain tumors. Cancers (Basel) 11: 1628. 10.3390/cancers11111628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathoo N, Barnett GH, Golubic M. 2004. The eicosanoid cascade: possible role in gliomas and meningiomas. J Clin Pathol 57: 6–13. 10.1136/jcp.57.1.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume A, Arakawa Y, Narita Y, Sugiyama K, Hata N, Muragaki Y, Shinojima N, Kumabe T, Saito R, Motomura K, et al. 2023. The first-in-human phase I study of a brain-penetrant mutant IDH1 inhibitor DS-1001 in patients with recurrent or progressive IDH1-mutant gliomas. Neuro Oncol 25: 326–336. 10.1093/neuonc/noac155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. 2019. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178: 835–849.e21. 10.1016/j.cell.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noch E, Khalili K. 2009. Molecular mechanisms of necrosis in glioblastoma: the role of glutamate excitotoxicity. Cancer Biol Ther 8: 1791–1797. 10.4161/cbt.8.19.9762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norenberg MD, Martinez-Hernandez A. 1979. Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res 161: 303–310. 10.1016/0006-8993(79)90071-4 [DOI] [PubMed] [Google Scholar]
- Oizel K, Yang C, Renoult O, Gautier F, Do QN, Joalland N, Gao X, Ko B, Vallette F, Ge W-P, et al. 2020. Glutamine uptake and utilization of human mesenchymal glioblastoma in orthotopic mouse model. Cancer Metab 8: 9. 10.1186/s40170-020-00215-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. 2011. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478: 197–203. 10.1038/nature10491 [DOI] [PubMed] [Google Scholar]
- Opitz CA, Somarribas Patterson LF, Mohapatra SR, Dewi DL, Sadik A, Platten M, Trump S. 2020. The therapeutic potential of targeting tryptophan catabolism in cancer. Br J Cancer 122: 30–44. 10.1038/s41416-019-0664-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Tomaszowski KH, Marisetty A, Kong LY, Wei J, Duna M, Blumberg K, Ji X, Jacobs C, Fuller GN, et al. 2020. Profiling of patients with glioma reveals the dominant immunosuppressive axis is refractory to immune function restoration. JCI Insight 5: e134386. 10.1172/jci.insight.134386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parik S, Fernández-García J, Lodi F, De Vlaminck K, Derweduwe M, De Vleeschouwer S, Sciot R, Geens W, Weng L, Bosisio FM, et al. 2022. GBM tumors are heterogeneous in their fatty acid metabolism and modulating fatty acid metabolism sensitizes cancer cells derived from recurring GBM tumors to temozolomide. Front Oncol 12: 988872. 10.3389/fonc.2022.988872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. 2016. The emerging hallmarks of cancer metabolism. Cell Metab 23: 27–47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Zhu J, Thompson CB. 2022. The hallmarks of cancer metabolism: still emerging. Cell Metab 34: 355–377. 10.1016/j.cmet.2022.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. 1994. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci 91: 10625–10629. 10.1073/pnas.91.22.10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Zhao Z, Liu L, Bai L, Tong R, Yang H, Zhong L. 2022. Targeting indoleamine dioxygenase and tryptophan dioxygenase in cancer immunotherapy: clinical progress and challenges. Drug Des Devel Ther 16: 2639–2657. 10.2147/DDDT.S373780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelroizen R, Philosof B, Budick-Harmelin N, Chernobylsky T, Ron A, Katzir R, Shimon D, Tessler A, Adir O, Gaoni-Yogev A, et al. 2022. Astrocyte immunometabolic regulation of the tumour microenvironment drives glioblastoma pathogenicity. Brain 145: 3288–3307. 10.1093/brain/awac222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham QT, Taniyama D, Akabane S, Harada K, Babasaki T, Sekino Y, Hayashi T, Sakamoto N, Sentani K, Oue N, et al. 2021. TDO2 overexpression correlates with poor prognosis, cancer stemness, and resistance to cetuximab in bladder cancer. Cancer Rep 4: e1417. 10.1002/cnr2.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T, Rothstein JD. 2017. Oligodendroglia: metabolic supporters of neurons. J Clin Invest 127: 3271–3280. 10.1172/JCI90610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen MH, Hansson E, Manaprasertsak A, Lange S, Jennische E, Carén H, Gatzinsky K, Jakola A, Hammarlund EU, Malmberg P. 2023. Distinct cholesterol localization in glioblastoma multiforme revealed by mass spectrometry imaging. ACS Chem Neurosci 14: 1602–1609. 10.1021/acschemneuro.2c00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. 2019. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 18: 379–401. 10.1038/s41573-019-0016-5 [DOI] [PubMed] [Google Scholar]
- Platten M, Bunse L, Wick A, Bunse T, Le Cornet L, Harting I, Sahm F, Sanghvi K, Tan CL, Poschke I, et al. 2021. A vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature 592: 463–468. 10.1038/s41586-021-03363-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pow DV. 2001. Visualising the activity of the cystine-glutamate antiporter in glial cells using antibodies to aminoadipic acid, a selectively transported substrate. Glia 34: 27–38. 10.1002/glia.1037 [DOI] [PubMed] [Google Scholar]
- Puca F, Yu F, Bartolacci C, Pettazzoni P, Carugo A, Huang-Hobbs E, Liu J, Zanca C, Carbone F, Del Poggetto E, et al. 2021. Medium-chain acyl-CoA dehydrogenase protects mitochondria from lipid peroxidation in glioblastoma. Cancer Discov 11: 2904–2923. 10.1158/2159-8290.CD-20-1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalska P, Crawford PA. 2017. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab 25: 262–284. 10.1016/j.cmet.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall EC, Lopez BGC, Peng S, Regan MS, Abdelmoula WM, Basu SS, Santagata S, Yoon H, Haigis MC, Agar JN, et al. 2020. Localized metabolomic gradients in patient-derived xenograft models of glioblastoma. Cancer Res 80: 1258–1267. 10.1158/0008-5472.CAN-19-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon DA, Desjardins A, Rixe O, Cloughesy T, Alekar S, Williams JH, Li R, Taylor CT, Lassman AB. 2020. A phase 1 study of PF-06840003, an oral indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor in patients with recurrent malignant glioma. Invest New Drugs 38: 1784–1795. 10.1007/s10637-020-00950-1 [DOI] [PubMed] [Google Scholar]
- Rose EM, Koo JCP, Antflick JE, Ahmed SM, Angers S, Hampson DR. 2009. Glutamate transporter coupling to Na,K-ATPase. J Neurosci 29: 8143–8155. 10.1523/JNEUROSCI.1081-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman DL, De Feyter HM, Maciejewski PK, Behar KL. 2012. Is there in vivo evidence for amino acid shuttles carrying ammonia from neurons to astrocytes? Neurochem Res 37: 2597–2612. 10.1007/s11064-012-0898-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuck G, Röhrdanz E, Tran-Thi QH, Kahl R, Schlüter G. 2002. Oxidative stress in rat cortical neurons and astrocytes induced by paraquatin vitro. Neurotox Res 4: 1–13. 10.1080/10298420290007574 [DOI] [PubMed] [Google Scholar]
- Schönfeld P, Reiser G. 2013. Why does brain metabolism not favor burning of fatty acids to provide energy? Reflections on disadvantages of the use of free fatty acids as fuel for brain. J Cereb Blood Flow Metab 33: 1493–1499. 10.1038/jcbfm.2013.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe A, Scafidi S, Bak LK, Waagepetersen HS, McKenna MC. 2014. Glutamate metabolism in the brain focusing on astrocytes. Adv Neurobiol 11: 13–30. 10.1007/978-3-319-08894-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seker-Polat F, Degirmenci NP, Solaroglu I, Bagci-Onder T. 2022. Tumor cell infiltration into the brain in glioblastoma: from mechanisms to clinical perspectives. Cancers (Basel) 14: 443. 10.3390/cancers14020443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, et al. 2010. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 70: 8981–8987. 10.1158/0008-5472.CAN-10-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyer A, Boudah S, Broudin S, Junot C, Colsch B. 2016. Annotation of the human cerebrospinal fluid lipidome using high resolution mass spectrometry and a dedicated data processing workflow. Metabolomics 12: 91. 10.1007/s11306-016-1023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya S, Gromovsky AD, Hale JS, Knudsen AM, Prager B, Wallace LC, Penalva LOF, Brown HA, Kristensen BW, Rich JN, et al. 2021. Altered lipid metabolism marks glioblastoma stem and non-stem cells in separate tumor niches. Acta Neuropathol Commun 9: 101. 10.1186/s40478-021-01205-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Aaroe A, Liang J, Puduvalli VK. 2023. Tumor microenvironment in glioblastoma: current and emerging concepts. Neuro-Oncology Adv 5: vdad009. 10.1093/noajnl/vdad009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth S, Brito R, Mukherjea D, Rybak LP, Ramkumar V. 2014. Adenosine receptors: expression, function and regulation. Int J Mol Sci 15: 2024–2052. 10.3390/ijms15022024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LS, Poschet G, Nonnenmacher Y, Becker HM, Sapcariu S, Gaupel A-C, Schlotter M, Wu Y, Kneisel N, Seiffert M, et al. 2017. Branched-chain ketoacids secreted by glioblastoma cells via MCT1 modulate macrophage phenotype. EMBO Rep 18: 2172–2185. 10.15252/embr.201744154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B, Mantha OL, Schor J, Pascual A, Plaçais PY, Pavlowsky A, Preat T. 2022. Glia fuel neurons with locally synthesized ketone bodies to sustain memory under starvation. Nat Metab 4: 213–224. 10.1038/s42255-022-00528-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolič T, Tavčar P, Horvat A, Černe U, Halužan Vasle A, Tratnjek L, Kreft ME, Scholz N, Matis M, Petan T, et al. 2021. Astrocytes in stress accumulate lipid droplets. Glia 69: 1540–1562. 10.1002/glia.23978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Bannerman P, Guo F, Burns T, Miers L, Croteau C, Singhal NK, McDonough JA, Pleasure D. 2017. Suppressing N-acetyl-L-aspartate synthesis prevents loss of neurons in a murine model of Canavan leukodystrophy. J Neurosci 37: 413–421. 10.1523/JNEUROSCI.2013-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Yu L, Hasan MN, Paruchuri SS, Mullett SJ, Sullivan MLG, Fiesler VM, Young CB, Stolz DB, Wendell SG, et al. 2022. Elevated microglial oxidative phosphorylation and phagocytosis stimulate post-stroke brain remodeling and cognitive function recovery in mice. Commun Biol 5: 35. 10.1038/s42003-021-02984-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Westergaard N, Petersen SB, Unsgård G, Schousboe A. 1993. Metabolism of [U-13C]glutamate in astrocytes studied by 13C NMR spectroscopy: incorporation of more label into lactate than into glutamine demonstrates the importance of the tricarboxylic acid cycle. J Neurochem 61: 1179–1182. 10.1111/j.1471-4159.1993.tb03641.x [DOI] [PubMed] [Google Scholar]
- Sørensen MF, Heimisdóttir SB, Sørensen MD, Mellegaard CS, Wohlleben H, Kristensen BW, Beier CP. 2018. High expression of cystine-glutamate antiporter xCT (SLC7A11) is an independent biomarker for epileptic seizures at diagnosis in glioma. J Neurooncol 138: 49–53. 10.1007/s11060-018-2785-9 [DOI] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. 2016. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536: 479–483. 10.1038/nature19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector R. 1988. Fatty acid transport through the blood-brain barrier. J Neurochem 50: 639–643. 10.1111/j.1471-4159.1988.tb02958.x [DOI] [PubMed] [Google Scholar]
- Su LJ, Zhang JH, Gomez H, Murugan R, Hong X, Xu D, Jiang F, Peng ZY. 2019. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med Cell Longev 2019: 5080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Tirosh I. 2020. The glioma stem cell model in the era of single-cell genomics. Cancer Cell 37: 630–636. 10.1016/j.ccell.2020.04.001 [DOI] [PubMed] [Google Scholar]
- Szepesi Z, Manouchehrian O, Bachiller S, Deierborg T. 2018. Bidirectional microglia–neuron communication in health and disease. Front Cell Neurosci 12: 403531. 10.3389/fncel.2018.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taïb B, Aboussalah AM, Moniruzzaman M, Chen S, Haughey NJ, Kim SF, Ahima RS. 2019. Lipid accumulation and oxidation in glioblastoma multiforme. Sci Rep 9: 19593. 10.1038/s41598-019-55985-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S. 2020. Metabolic compartmentalization between astroglia and neurons in physiological and pathophysiological conditions of the neurovascular unit. Neuropathology 40: 121–137. 10.1111/neup.12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Iizumi T, Mashima K, Abe T, Suzuki N. 2014. Roles and regulation of ketogenesis in cultured astroglia and neurons under hypoxia and hypoglycemia. ASN Neuro 6: 1759091414550997. 10.1177/1759091414550997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Lin JHC, Arcuino G, Gao Q, Yang J, Nedergaard M. 2001. Glutamate release promotes growth of malignant gliomas. Nat Med 7: 1010–1015. 10.1038/nm0901-1010 [DOI] [PubMed] [Google Scholar]
- Takenaka MC, Gabriely G, Rothhammer V, Mascanfroni ID, Wheeler MA, Chao CC, Gutiérrez-Vázquez C, Kenison J, Tjon EC, Barroso A, et al. 2019. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 22: 729–740. 10.1038/s41593-019-0370-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamimi AF, Juweid M. 2017. Epidemiology and outcome of glioblastoma. In Glioblastoma (ed. De Vleeschouwer S), pp. 143–153. Codon Publications, Brisbane, Australia. 10.15586/codon.glioblastoma.2017.ch8 [DOI] [PubMed] [Google Scholar]
- Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PØ, Weinstock A, Wagner A, et al. 2015. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol 17: 1556–1568. 10.1038/ncb3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AA, Fisher JL, Rahme GJ, Hampton TH, Baron U, Olek S, Schwachula T, Rhodes CH, Gui J, Tafe LJ, et al. 2015. Regulatory T cells are not a strong predictor of survival for patients with glioblastoma. Neuro Oncol 17: 801–809. 10.1093/neuonc/nou363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EA, Powell JD. 2021. Inhibition of the adenosine pathway to potentiate cancer immunotherapy: potential for combinatorial approaches. Annu Rev Med 72: 331–348. 10.1146/annurev-med-060619-023155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tönjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AHCC, Karra D, Piro RM, et al. 2013. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med 19: 901–908. 10.1038/nm.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay MÈ, Stevens B, Sierra A, Wake H, Bessis A, Nimmerjahn A. 2011. The role of microglia in the healthy brain. J Neurosci 31: 16064–16069. 10.1523/JNEUROSCI.4158-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska K, Sokolowska J, Szmidt M, Sysa P. 2014. Glioblastoma multiforme – an overview. Contemp Oncol 18: 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Brooklyn JR, Jackson CA, Pearl DK, Kotur MS, Snyder PJ, Prior TW. 2005. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. J Neuropathol Exp Neurol 64: 695–705. 10.1097/01.jnen.0000175329.59092.2c [DOI] [PubMed] [Google Scholar]
- Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, Körber C, Kardorff M, Ratliff M, Xie R, et al. 2019. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573: 532–538. 10.1038/s41586-019-1564-x [DOI] [PubMed] [Google Scholar]
- Venkataramani V, Yang Y, Schubert MC, Reyhan E, Tetzlaff SK, Wißmann N, Botz M, Soyka SJ, Beretta CA, Pramatarov RL, et al. 2022. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell 185: 2899–2917.e31. 10.1016/j.cell.2022.06.054 [DOI] [PubMed] [Google Scholar]
- Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, Tam LT, Espenel C, Ponnuswami A, Ni L, et al. 2019. Electrical and synaptic integration of glioma into neural circuits. Nature 573: 539–545. 10.1038/s41586-019-1563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, Gu Y, Lum KM, Masui K, Yang H, et al. 2016. An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell 30: 683–693. 10.1016/j.ccell.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkenhoff A, Weiler A, Letzel M, Stehling M, Klämbt C, Schirmeier S. 2015. Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab 22: 437–447. 10.1016/j.cmet.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Wang B, Tontonoz P. 2018. Liver X receptors in lipid signalling and membrane homeostasis. Nat Rev Endocrinol 14: 452–463. 10.1038/s41574-018-0037-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cui Y, Yu Z, Wang W, Cheng X, Ji W, Guo S, Zhou Q, Wu N, Chen Y, et al. 2019a. Brain endothelial cells maintain lactate homeostasis and control adult hippocampal neurogenesis. Cell Stem Cell 25: 754–767. 10.1016/j.stem.2019.09.009 [DOI] [PubMed] [Google Scholar]
- Wang L, Pavlou S, Du X, Bhuckory M, Xu H, Chen M. 2019b. Glucose transporter 1 critically controls microglial activation through facilitating glycolysis. Mol Neurodegener 14: 2. 10.1186/s13024-019-0305-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiame E, Tyteca D, Pierrot N, Collard F, Amyere M, Noel G, Desmedt J, Nassogne MC, Vikkula M, Octave JN, et al. 2010. Molecular identification of aspartate N-acetyltransferase and its mutation in hypoacetylaspartia. Biochem J 425: 127–139. 10.1042/BJ20091024 [DOI] [PubMed] [Google Scholar]
- Wilde L, Roche M, Domingo-Vidal M, Tanson K, Philp N, Curry J, Martinez-Outschoorn U. 2017. Metabolic coupling and the reverse Warburg effect in cancer, implications for novel biomarker and anticancer agent development. Semin Oncol 44: 198–203. 10.1053/j.seminoncol.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching HG, Weller M. 2020. Does neuronal activity promote glioma progression? Trends Cancer 6: 1–3. 10.1016/j.trecan.2019.11.002 [DOI] [PubMed] [Google Scholar]
- Wise DR, Deberardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787. 10.1073/pnas.0810199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Geng F, Cheng X, Guo Q, Zhong Y, Cloughesy TF, Yong WH, Chakravarti A, Guo D. 2020. Lipid droplets maintain energy homeostasis and glioblastoma growth via autophagic release of stored fatty acids. iScience 23: 101569. 10.1016/j.isci.2020.101569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Lesniak MS, James CD, Heimberger AB, Chen P. 2021. Context-dependent glioblastoma–macrophage/microglia symbiosis and associated mechanisms. Trends Immunol 42: 280–292. 10.1016/j.it.2021.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, Liu X, Chen CH, Fadare O, Pizzo DP, et al. 2018. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol 20: 597–609. 10.1038/s41556-018-0083-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Joachims ML, Thompson LF, Miller AD, Canoll PD, Bynoe MS. 2019. CD73 promotes glioblastoma pathogenesis and enhances its chemoresistance via A2B adenosine receptor signaling. J Neurosci 39: 4387–4402. 10.1523/JNEUROSCI.1118-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Qiu Y, Stamatatos O, Janowitz T, Lukey MJ. 2021. Enhancing the efficacy of glutamine metabolism inhibitors in cancer therapy. Trends Cancer 7: 790–804. 10.1016/j.trecan.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto Y, Miyazaki H, Vaidyan LK, Kagawa Y, Ebrahimi M, Yamamoto Y, Ogata M, Katsuyama Y, Sadahiro H, Suzuki M, et al. 2016. Inhibition of fatty acid synthase decreases expression of stemness markers in glioma stem cells. PLoS One 11: e0147717. 10.1371/journal.pone.0147717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZC, Rothstein JD, Sontheimer H. 1999. Compromised glutamate transport in human glioma cells: reduction-mislocalization of sodium-dependent glutamate transporters and enhanced activity of cystine-glutamate exchange. J Neurosci 19: 10767–10777. 10.1523/JNEUROSCI.19-24-10767.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellen G. 2018. Fueling thought: management of glycolysis and oxidative phosphorylation in neuronal metabolism. J Cell Biol 217: 2235–2246. 10.1083/jcb.201803152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, Balaj L, Rogers VA, Uhlmann EJ, Varma H, et al. 2022. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol 23: 971–984. 10.1038/s41590-022-01215-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AC, Schousboec A, Hertz L. 1982. Metabolic fate of 14C-labeled glutamate in astrocytes in primary cultures. J Neurochem 39: 954–960. 10.1111/j.1471-4159.1982.tb11482.x [DOI] [PubMed] [Google Scholar]
- Zhai L, Bell A, Ladomersky E, Lauing KL, Bollu L, Nguyen B, Genet M, Kim M, Chen P, Mi X, et al. 2021. Tumor cell IDO enhances immune suppression and decreases survival independent of tryptophan metabolism in glioblastoma. Clin Cancer Res 27: 6514–6528. 10.1158/1078-0432.CCR-21-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu Q. 2015. Cholesterol metabolism and homeostasis in the brain. Protein Cell 6: 254–264. 10.1007/s13238-014-0131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke HR, Tildon JT, Landry ME, Max SR. 1990. Effect of 8-bromo-cAMP and dexamethasone on glutamate metabolism in rat astrocytes. Neurochem Res 15: 1115–1122. 10.1007/BF01101713 [DOI] [PubMed] [Google Scholar]