Abstract
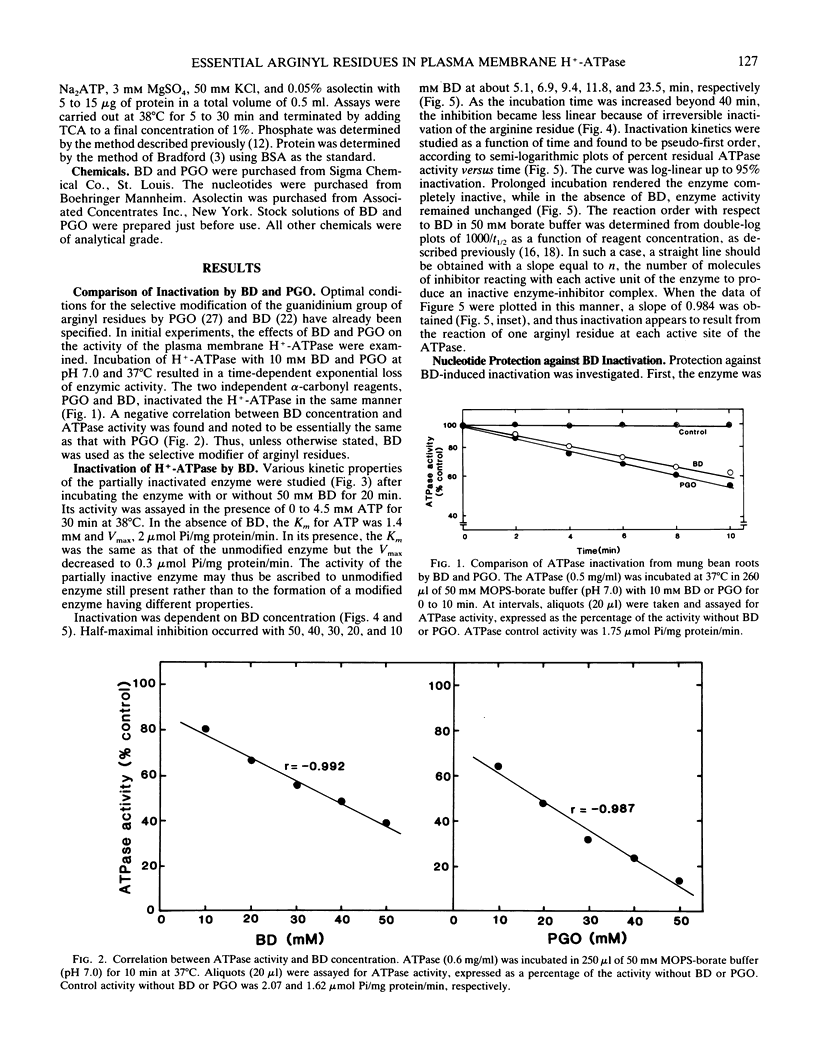
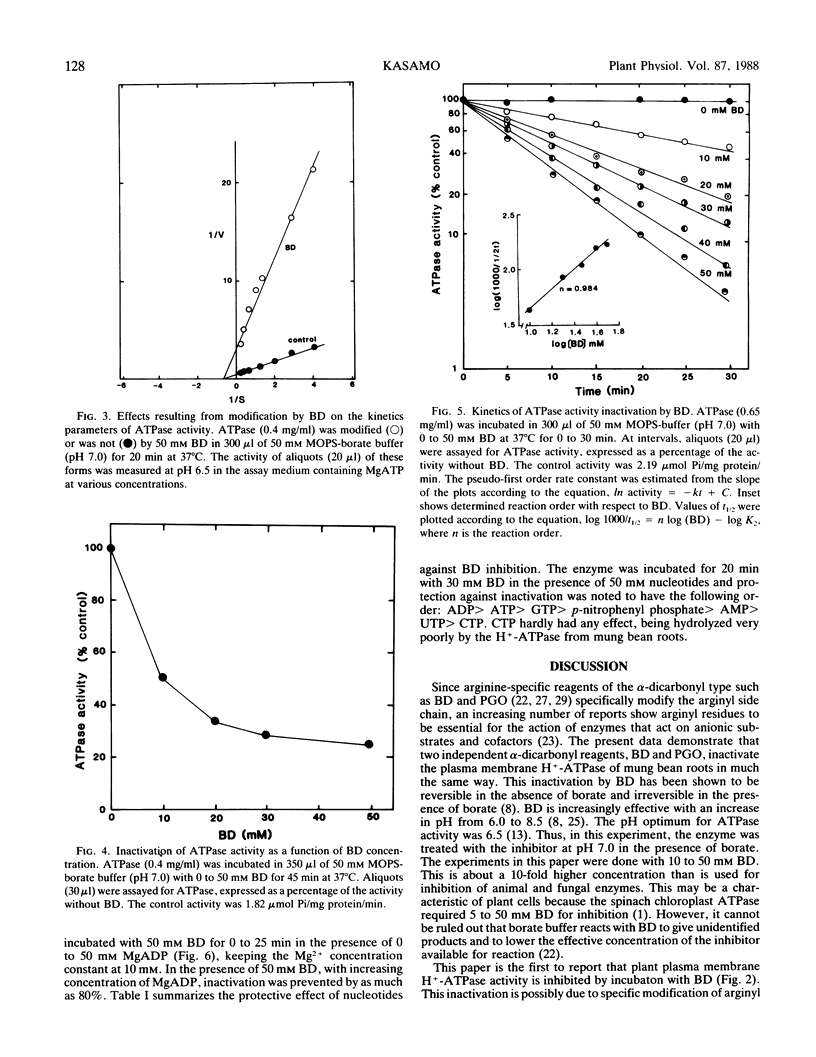
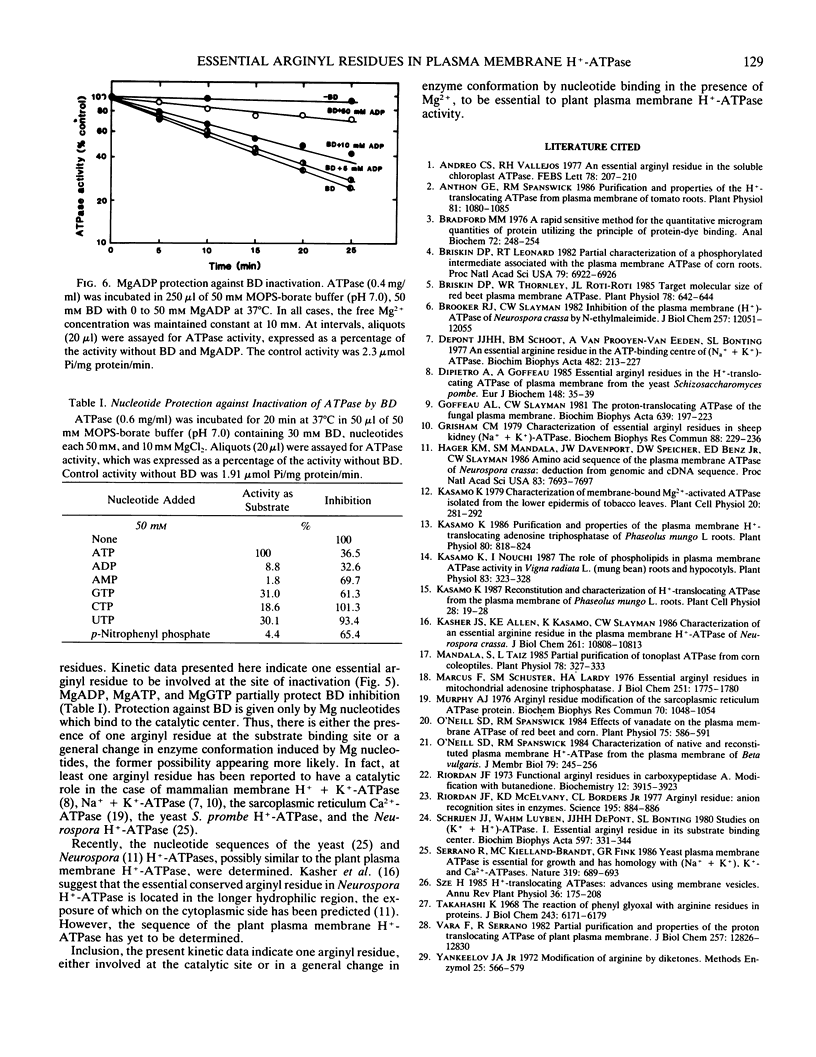
Proton-translocating ATPase (H+-ATPase) was purified from mung bean (Vigna radiata L.) roots. Treatment of this enzyme with the arginine-specific reagent 2,3-butanedione in the presence of borate at 37°C (pH 7.0), caused a marked decrease in its activity. Under this condition, half-maximal inhibition was brought about by 20 millimolar 2,3-butanedione at 12 minutes. MgATP and MgADP, the physiological substrate and competitive inhibitor of the ATPase, respectively, provided partial protection against inactivation. Loss of activity followed pseudo-first order kinetics with respect to 2,3-butanedione concentration, and double log plots of pseudo-first order rate constants versus reagent concentration gave a curve with a slope of 0.984. Thus, inactivation may possibly result from reaction of one arginine residue at each active site of the enzyme. The results obtained from the present study indicate that at least one arginyl residue performs an essential function in the plasma membrane H+-ATPase, probably at the catalytic site.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreo C. S., Vallejos R. H. An essential arginyl residue in the soluble chloroplast ATPase. FEBS Lett. 1977 Jun 15;78(2):207–210. doi: 10.1016/0014-5793(77)80307-4. [DOI] [PubMed] [Google Scholar]
- Anthon G. E., Spanswick R. M. Purification and properties of the h-translocating ATPase from the plasma membrane of tomato roots. Plant Physiol. 1986 Aug;81(4):1080–1085. doi: 10.1104/pp.81.4.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Briskin D. P., Leonard R. T. Partial characterization of a phosphorylated intermediate associated with the plasma membrane ATPase of corn roots. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6922–6926. doi: 10.1073/pnas.79.22.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briskin D. P., Thornley W. R., Roti-Roti J. L. Target molecular size of the red beet plasma membrane ATPase. Plant Physiol. 1985 Jul;78(3):642–644. doi: 10.1104/pp.78.3.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R. J., Slayman C. W. Inhibition of the plasma membrane [H+]-ATPase of Neurospora crassa by N-ethylmaleimide. Protection by nucleotides. J Biol Chem. 1982 Oct 25;257(20):12051–12055. [PubMed] [Google Scholar]
- Di Pietro A., Goffeau A. Essential arginyl residues in the H+-translocating ATPase of plasma membrane from the yeast Schizosaccharomyces pombe. Eur J Biochem. 1985 Apr 1;148(1):35–39. doi: 10.1111/j.1432-1033.1985.tb08803.x. [DOI] [PubMed] [Google Scholar]
- Goffeau A., Slayman C. W. The proton-translocating ATPase of the fungal plasma membrane. Biochim Biophys Acta. 1981 Dec 30;639(3-4):197–223. doi: 10.1016/0304-4173(81)90010-0. [DOI] [PubMed] [Google Scholar]
- Grisham C. M. Characterization of essential arginyl residues in sheep kidney (Na+ + K+) -ATPase. Biochem Biophys Res Commun. 1979 May 14;88(1):229–236. doi: 10.1016/0006-291x(79)91720-0. [DOI] [PubMed] [Google Scholar]
- Hager K. M., Mandala S. M., Davenport J. W., Speicher D. W., Benz E. J., Jr, Slayman C. W. Amino acid sequence of the plasma membrane ATPase of Neurospora crassa: deduction from genomic and cDNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7693–7697. doi: 10.1073/pnas.83.20.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamo K., Nouchi I. The Role of Phospholipids in Plasma Membrane ATPase Activity in Vigna radiata L. (Mung Bean) Roots and Hypocotyls. Plant Physiol. 1987 Feb;83(2):323–328. doi: 10.1104/pp.83.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamo K. Purification and Properties of the Plasma Membrane H-Translocating Adenosine Triphosphatase of Phaseolus mungo L. Roots. Plant Physiol. 1986 Apr;80(4):818–824. doi: 10.1104/pp.80.4.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasher J. S., Allen K. E., Kasamo K., Slayman C. W. Characterization of an essential arginine residue in the plasma membrane H+-ATPase of Neurospora crassa. J Biol Chem. 1986 Aug 15;261(23):10808–10813. [PubMed] [Google Scholar]
- Mandala S., Taiz L. Partial purification of a tonoplast ATPase from corn coleoptiles. Plant Physiol. 1985 Jun;78(2):327–333. doi: 10.1104/pp.78.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus F., Schuster S. M., Lardy H. A. Essential arginyl residues in mitochondrial adenosine triphosphatase. J Biol Chem. 1976 Mar 25;251(6):1775–1780. [PubMed] [Google Scholar]
- Murphy A. J. Arginyl residue modification of the sarcoplasmic reticulum ATPase protein. Biochem Biophys Res Commun. 1976 Jun 21;70(4):1048–1054. doi: 10.1016/0006-291x(76)91008-1. [DOI] [PubMed] [Google Scholar]
- O'neill S. D., Spanswick R. M. Effects of vanadate on the plasma membrane ATPase of red beet and corn. Plant Physiol. 1984 Jul;75(3):586–591. doi: 10.1104/pp.75.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J. F. Functional arginyl residues in carboxypeptidase A. Modification with butanedione. Biochemistry. 1973 Sep 25;12(20):3915–3923. doi: 10.1021/bi00744a020. [DOI] [PubMed] [Google Scholar]
- Riordan J. F., McElvany K. D., Borders C. L., Jr Arginyl residues: anion recognition sites in enzymes. Science. 1977 Mar 4;195(4281):884–886. doi: 10.1126/science.190679. [DOI] [PubMed] [Google Scholar]
- Schrijen J. J., Luyben W. A., De Pont J. J., Bonting S. L. Studies on (K+ + H+)-ATPase. I. Essential arginine residue in its substrate binding center. Biochim Biophys Acta. 1980 Apr 10;597(2):331–344. doi: 10.1016/0005-2736(80)90110-8. [DOI] [PubMed] [Google Scholar]
- Serrano R., Kielland-Brandt M. C., Fink G. R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986 Feb 20;319(6055):689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- Takahashi K. The reaction of phenylglyoxal with arginine residues in proteins. J Biol Chem. 1968 Dec 10;243(23):6171–6179. [PubMed] [Google Scholar]
- Vara F., Serrano R. Partial purification and properties of the proton-translocating ATPase of plant plasma membranes. J Biol Chem. 1982 Nov 10;257(21):12826–12830. [PubMed] [Google Scholar]
- de Pont J. J., Schoot B. M., van Prooijen-van Eeden A., Bonting S. L. An essential arginine residue in the ATP-binding centre of (Na+ + K+)-ATPase. Biochim Biophys Acta. 1977 May 12;482(1):213–227. doi: 10.1016/0005-2744(77)90367-9. [DOI] [PubMed] [Google Scholar]


